The SPRY domain of Pyrin, mutated in familial Mediterranean fever patients, interacts with inflammasome components and inhibits proIL-1β processing (original) (raw)
Main
Hereditary recurrent fevers are disorders characterized by recurrent episodes of fever and serosal inflammation, associated with more specific symptoms.1, 2, 3 This group of diseases comprises three syndromes associated with mutations in NALP3, a key component of the inflammasome4: Muckle–Wells syndrome, familial cold urticaria5and chronic infantile neurological cutaneous and articular syndrome/OMID,6 as well as other syndromes like hyper-IgD,7, 8 PAPA syndrome,9 TRAPS10, 11 and familial Mediterranean fever (FMF).12, 13 The latter is a recessively inherited disorder, in which mutations occur in the pyrin gene.12, 13 Pyrin is expressed in neutrophils and monocytes, and its expression is tightly regulated by cytokines.14 Several spliced isoforms of Pyrin have been described so far.15, 16 The subcellular localization of Pyrin has been well studied in overexpression studies as well as at the endogenous level. It appears that according to the cell type or to the spliced isoform studied, Pyrin can be localized diffusely or as specks, in the cytoplasm, but also in the nucleus.15, 16, 17, 18, 19, 20
Pyrin contains at the N-terminus a Pyrin domain (PYD), a member of the death effector-fold domains,21, 22, 23 two B-boxes and a coiled-coil domain (BBCC), as well as a SPRY domain (also called B30.2 domain). The PYD found in Pyrin is also present in NALP proteins. The NALP proteins, and more precisely NALP1, NALP2 and NALP3, are involved in the formation of molecular platforms that trigger activation of caspase-1 followed by processing and release of active interleukin (IL)-1_β_, a major inflammatory cytokine.4, 24 These platforms have been named inflammasomes, and are formed by the recruitment of several proteins to the NALPs, like ASC, Cardinal, caspase-1 and caspase-5. In addition to Pyrin and NALPs, the PYD is also found in the adaptor ASC that makes the link between NALPs and caspase-1 in the inflammasome formation, through PYD-PYD interactions between ASC and NALPs and CARD-CARD interactions between ASC and caspase-1.4, 24 Several groups have reported that the PYD of Pyrin can interact with the PYD of ASC.19, 25, 26 However the relevance and function of this interaction is not clear and controversial. Some reports suggest that this interaction negatively regulates caspase-1,27, 28 and another indicates that it can promote the formation of an active caspase-1 activating complex.26 These contradictory data are probably due to the use of different experimental systems.
The other domains of Pyrin, the BBCC and the SPRY domains, are found in many proteins of the TRIM (tripartite motif) family,29 which often contain, instead of the amino-terminal PYD, an E3 ubiquitin-ligase-like RING domain. BBCC and SPRY domains are thought to be protein–protein interaction domains, but for the great majority of the TRIM proteins, no binding partners have been described and their functions are not yet established. Interestingly, TRIM5 has been implicated in HIV-1 restriction in primates, and this antiviral effect is mediated through the SPRY domain.30, 31 Antiviral effects have also been reported for other TRIM members, suggesting that the SPRY domain has evolved for anti-viral immunity. Interestingly, the mutations found in FMF patients are also mainly localized in the SPRY domain of Pyrin.
The presence of a common domain in Pyrin and NALPs, as well as the similarities of the syndromes associated with the mutations in Pyrin and NALP3, suggest that these proteins could be involved in related pathways. In accord with this hypothesis, recent evidence suggests that the SPRY domain of Pyrin directly binds to caspase-1.32 Therefore we decided to investigate the role of Pyrin in inflammasome regulation in more detail.
Results
Pyrin interacts with NALPs and caspases
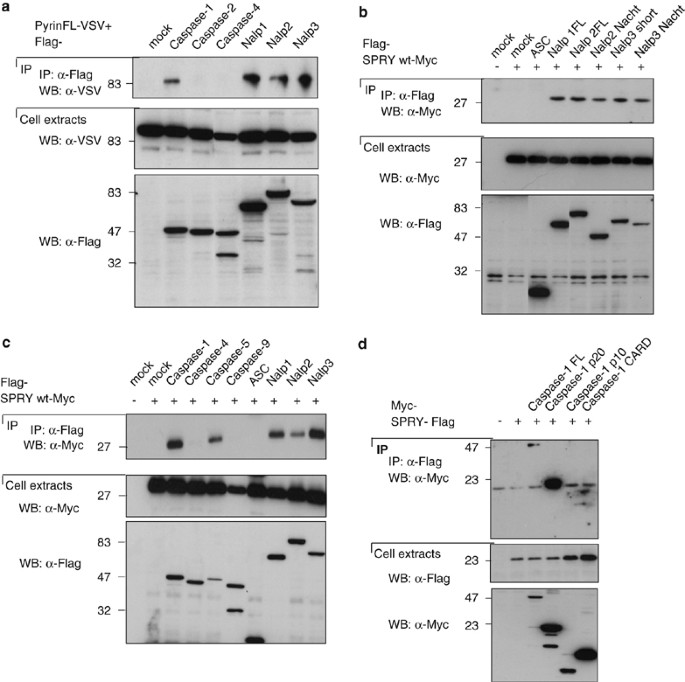
To determine whether Pyrin can interact with inflammasome components, co-immunoprecipitation experiments were performed in HEK293T cells transfected with plasmids coding for tagged proteins. Upon overexpression of VSV-tagged Pyrin together with Flag-tagged caspases and NALPs, we detected a specific interaction with caspase-1 (in accordance with recently published data32) and caspase-5, but not with the other pro-inflammatory caspase-4 (Figure 1a and c). Interactions between Pyrin and the inflammasome components NALP1, NALP2 and NALP3 were equally strong (Figure 1a).
Figure 1
Pyrin interacts with NALPs, caspase-1 and caspase-5. (a–d) 293T cells were transfected with 5 _μ_g of each cDNA encoding Flag-, Myc- or VSV-tagged proteins. After 24 h, cells were collected, and the lysates were subjected to immunoprecipitation using anti-Flag agarose beads. Cell extracts and immunoprecipitates were then analyzed by Western blot
To identify the domains involved in these interactions, immunoprecipitation experiments using only the SPRY domain of Pyrin and different domains of NALP proteins were performed. They revealed that the SPRY domain of Pyrin is sufficient for binding to the NALPs. Furthermore, using NALP constructs containing only the NACHT domain, it became evident that the SPRY domain of Pyrin interacts with NALPs through their NACHT domain (Figure 1b).
The SPRY domain of Pyrin was also sufficient for the interaction between Pyrin and caspases. Indeed, strong interactions of the Pyrin SPRY domain with caspase-1 and caspase-5 were observed. We did not detect interaction of the Pyrin SPRY domain with ASC, caspase-4 and caspase-9 (Figure 1c).
To map the interaction between Pyrin and caspase-1 more precisely, we overexpressed the SPRY domain of Pyrin together with different constructs of caspase-1: the full-length protein, the p20 subunit, the p10 subunit or the CARD domain. We detected a weak interaction with the full-length caspase-1, and a very strong interaction with the p20 subunit. These data suggest that Pyrin can interact weakly with the proform of caspase-1, but that the interaction with the cleaved active form of caspase-1 is much stronger (Figure 1d).
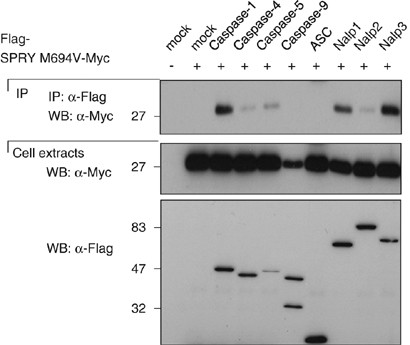
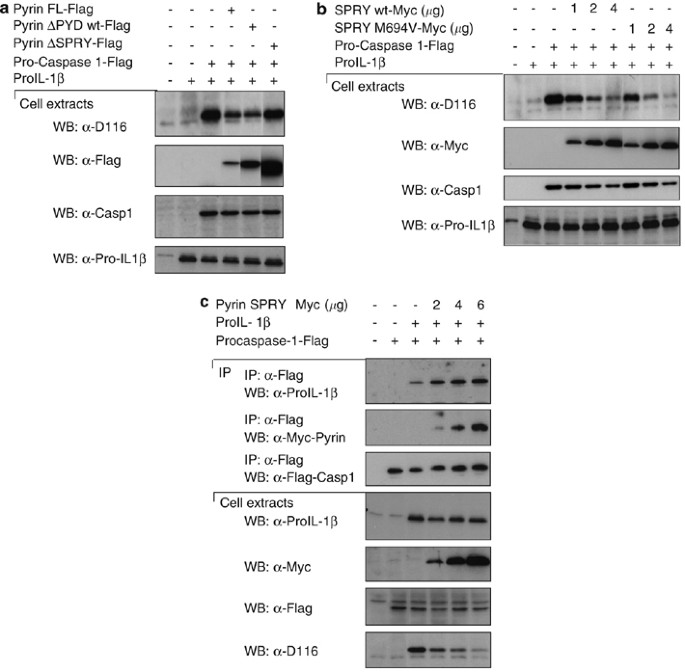
The mutations in Pyrin found in FMF patients are mainly localized in the SPRY domain of the protein. They have been recently proposed to affect the interaction between caspase-1 and Pyrin interaction.32 We therefore tested whether the mutations would also affect the binding of Pyrin to the inflammasome components and co-expressed the SPRY domain of Pyrin containing the most frequent mutation found in FMF patients (M694V) together with caspases and NALPs. As shown in Figure 2, the interactions of caspase-1, caspase-5 and NALPs with the mutated SPRY domain of Pyrin was equally strong as the one observed with wild-type SPRY (Figure 1c), suggesting that this mutation does not affect the binding of Pyrin to its partners, at least if assayed in our overexpression system. The reduced binding of caspase-5 and NALP2 seen in this experiment was not consistently seen.
Figure 2
The most frequent mutation of Pyrin found in patients with FMF does not affect Pyrin binding to the NALPs and caspases. 293T cells were transfected with 5 _μ_g of each cDNA encoding Flag- or Myc-tagged proteins. After 24 h, cells were collected, lysed, and the lysates were subjected to immunoprecipitation using anti-Flag agarose beads. Cell extracts and immunoprecipitates were then analyzed by Western blot
Pyrin interacts with the inflammasome substrate proIL-1_β_
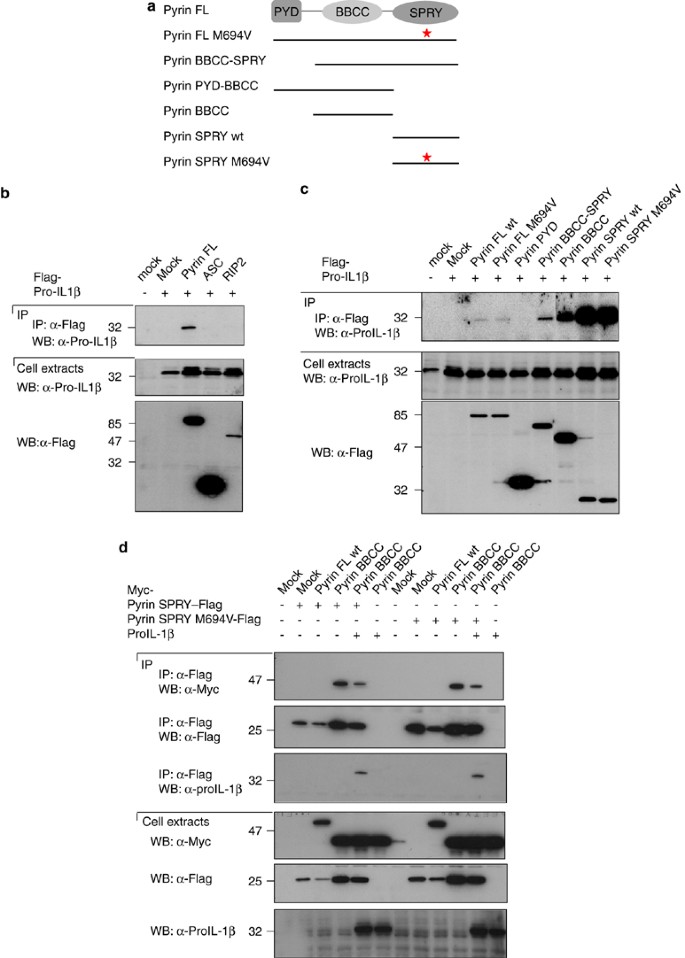
We further investigated if Pyrin could interact with the substrate of the active inflammasome: proIL-1_β_. By co-immunoprecipitation in HEK293T cells an interaction of the full-length Pyrin with proIL-1_β_ was detected, but no interaction of proIL-1_β_ with ASC, RIP2 (Figure 3a and b). When mapping this interaction we found that both the BBCC and the SPRY domain of Pyrin interact with proIL-1_β_ (Figure 3a and c), although the interaction with the SPRY domain was clearly stronger. In this experiment we did not detect the interaction with the full-length Pyrin as shown in Figure 3b, indicating that proIL-1_β_ interacts much more strongly with the individual domains than with the full-length protein. Furthermore, when expressing constructs containing the two domains (BBCC-SPRY), we no longer observed the interaction with proIL-1_β_, suggesting that these two domains interact with each other thereby masking the proIL-1_β_ interaction site.
Figure 3
Pyrin interacts with proIL-1_β_ through its BBCC and SPRY domains. (a) Schematic representation of the various Pyrin constructs used in this experiment. (b and c) 293T cells were transfected with 5 μ_g of each cDNA encoding Flag-tagged proteins or proIL-1_β. After 24 h, cells were collected, lysed, and the lysates were subjected to immunoprecipitation using anti-Flag agarose beads. Cell extracts and immunoprecipitates were then analyzed by Western blot. (d) 293T cells were transfected with 4 μ_g of each cDNA encoding Flag-, Myc-tagged proteins and proIL-1_β. After 24 h, cells were collected, lysed, and the lysates were subjected to immunoprecipitation using anti-Flag agarose beads. Cell extracts and immunoprecipitates were then analyzed by Western blot
To test this hypothesis, the SPRY domain of Pyrin, either wild type or mutated, together with different domains of Pyrin coupled to another tag were expressed in HEK293T cells. The SPRY domain indeed interacted with the BBCC domain of Pyrin (Figure 3d). The mutated SPRY domain also interacted with the BBCC, suggesting that the mutation does also not affect the auto-inhibition of Pyrin. Based on the proposed model, we predicted that proIL-1_β_ could compete for binding of the BBCC domain to the SPRY domain, which was indeed the case (Figure 3d).
We did not detect any interaction of Pyrin with mature IL-1_β_ (data not shown), indicating that the interaction of Pyrin with proIL-1_β_ depends on the pro-domain of proIL-1_β_.
Endogenous Pyrin can be recruited to the inflammasome
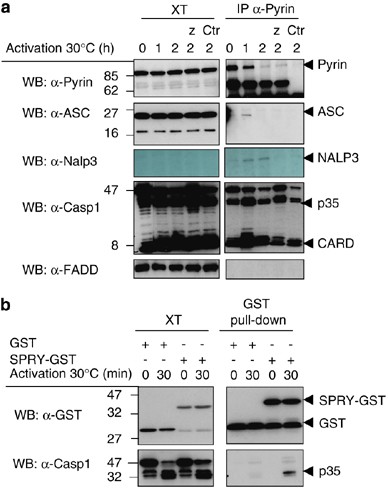
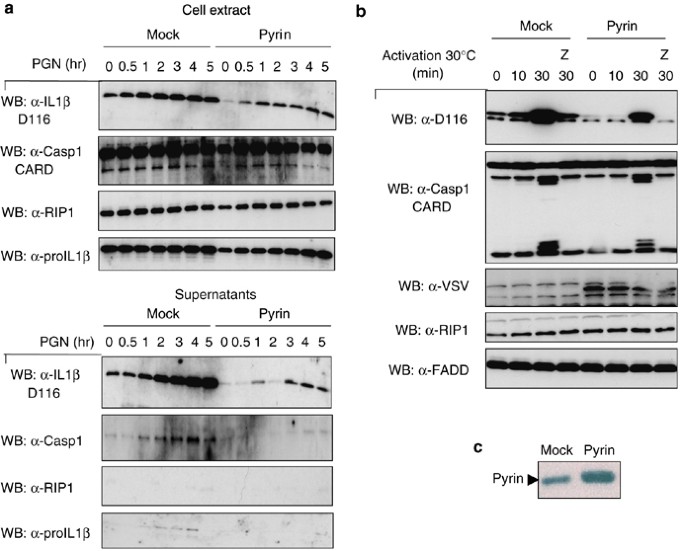
To determine a possible interaction of Pyrin and the inflammasome components at the endogenous level, we used the in vitro assembly system described previously for the identification of the inflammasome.33 Immunoprecipitations using an anti-Pyrin antibody were performed and as shown in Figure 4a, full-length Pyrin at the expected size (85 kDa) was readily detected in the cell extracts and in the immunoprecipitates. Interestingly, in immunoprecipitates but not in cell extracts, the full-length Pyrin disappeared during the activation at 30°C, suggesting that it is degraded. This degradation is a caspase-independent process, as it was not inhibited by the caspase inhibitor z-VAD-fmk.
Figure 4
Interaction of Pyrin with endogenous inflammasome components in THP1 cells. (a) THP1 cells were lysed in the Wang hypotonic buffer (buffer W, see methods), as described. The cell extracts were filtered and activated at 30°C for different times, allowing the activation of the inflammasome, and then subjected to immunoprecipitation using an anti-Pyrin antibody, or control antibody (Ctr). Z-VAD-fmk (Z) was added at some time points. The immunoprecipitates and the cell extracts were then analyzed by Western blot. (b) The recombinant GST-SPRY domain was added to THP1 cell extract generated as described under (a) and the interaction with caspase-1 in the assembling inflammasome investigated
In this experiment we detected a transient recruitment of ASC in Pyrin immunoprecipitates upon 1 h of inflammasome activation. Binding of NALP3 to Pyrin was also detected after 1 and 2 h of activation. Finally, an interaction with processed caspase-1 was also detected at 1 and 2 h. No recruitment of FADD was seen, demonstrating the specificity of complex formation.
To further demonstrate the interaction of the SPRY domain of Pyrin with caspase-1 in this more physiological system, we produced the recombinant GST-SPRY domain and incubated it with THP1 extracts, activated or not at 30°C. As shown in Figure 4b, the SPRY domain associated with the active caspase-1, present only in preactivated THP1 cell extracts. It is possible that the observed Pyrin–caspase-1 interaction is not direct but mediated by (a) linker protein(s).
Pyrin inhibits caspase-1-dependent IL-1_β_ processing
Since the SPRY domain of Pyrin can interact with inflammasome components and with proIL-1_β_, we investigated its effect on proIL-1_β_ processing. To this end, HEK293T cells were reconstituted with proIL-1_β_ and pro-caspase-1, because these proteins are not present endogenously, and we measured caspase-1-dependent IL-1_β_ processing. In this system, overexpression of pro-caspase-1 leads to its auto-activation and subsequent processing of proIL-1_β_ to mature IL-1_β_. By co-overexpressing different Pyrin constructs, lower levels of IL-1_β_ were detected in the presence of full-length Pyrin, and Pyrin that lacks the PYD domain was observed (Figure 5a). However, Pyrin lacking the SPRY domain did not inhibit IL-1_β_ processing, indicating that the inhibitory effect is mediated through the SPRY domain in accordance with the binding studies shown above. To corroborate this finding, we expressed the SPRY domain of Pyrin, either wild type or mutated. A dose-dependent inhibition of caspase-1-dependent IL-1_β_ processing, by both the wild-type or the mutated SPRY domain of Pyrin (Figure 5b) was observed, confirming that the inhibitory effect of Pyrin on proIL-1_β_ processing mostly depended on the SPRY domain, and is not affected by the mutations found in FMF patients. Here the inhibitory effect of Pyrin is observed on caspase-1 self-activation, but we could also observe an inhibitory effect of the SPRY on ASC-dependent activation of caspase-1 and subsequent IL-1_β_ processing (data not shown).
Figure 5
Pyrin inhibits caspase-1-dependent IL-1_β_ processing through its SPRY domain in reconstituted 293T cells. (a) 293T cells were transfected with 2 μ_g of each cDNA encoding proIL-1_β, pro-caspase-1 and Flag-tagged proteins. After 24 h, cells were collected, lysed, and the lysates were then analyzed by Western blot. (b) 293T cells were transfected with 2 μ_g of each cDNA encoding proIL-1_β, pro-caspase-1 and the indicated amount of Myc- tagged proteins. After 24 h, cells were harvested, lysed and analyzed by Western blot. The D116 antibody detects the cleaved, active IL-1_β_. (c) As (b), but the interaction between the transfected components were assayed by immunoprecipitation
We expected that the SPRY domain would interfere with the proIL-1_β_ caspase-1 interaction. To demonstrate this mechanism, we co-expressed caspase-1, proIL-1_β_ and different amounts of the SPRY domain. Surprisingly, higher concentrations of the SPRY domain resulted in slightly increased recruitment of proIL-1_β_ in caspase-1 immunoprecipitates (Figure 5c), while inhibiting the processing of the cytokine. We have to conclude that the SPRY domain does not compete with proIL-1_β_ for the binding to caspase-1, but that it freezes the enzyme/substrate in an inactive state, through the formation of a trimolecular complex.
To investigate the effect of Pyrin on proIL-1_β_ processing in a more physiological setting, a THP1 cell line was generated by retroviral infection that stably expresses VSV-tagged full-length Pyrin. THP1 cells producing relatively low levels of exogenous Pyrin were only obtained, as shown in Figure 6c, by increasing endogenous Pyrin levels approximately three times. In this context, it is noteworthy that cells infected with viruses expressing the SPRY domain of Pyrin were not viable, suggesting that the SPRY domain may be toxic. Cells were differentiated using PMA to induce the synthesis of proIL-1_β_, followed by stimulation with peptidoglycans (PGN) to activate the processing and secretion of mature IL-1_β_ (Figure 6a). Although the level of proteins in the cell extracts was comparable between Pyrin and mock-stable cell lines, there was a lower amount of IL-1_β_ and caspase-1 secreted by the Pyrin overexpressing cell line (Figure 6a, lower panel).
Figure 6
THP1 stably expressing Pyrin show reduced IL-1_β_ processing. (a) Mock- and Pyrin-infected THP1 cells were differentiated 3 h with PMA. After 24 h, cells were stimulated with peptidoglycan (PGN) or uric acid crystals (MSU) for 1–5 h. Cell extracts and the precipitated supernatants were then analyzed by Western blot. (b) Differentiated mock- and Pyrin-infected THP1 cells were lysed in the Wang hypotonic buffer, and the cell extracts were activated at 30°C for different times, with or without z-VAD-fmk (Z). Cell extracts were then subjected to Western-blot analysis. (c) Mock-infected THP1 cells and Pyrin-infected THP1 cells were lysed and cell extracts were analyzed by Western blot to determine the level of Pyrin expression
We also used the THP1 Pyrin cell line for in vitro inflammasome activation experiments. As in intact cells, activation of caspase-1 and processing of proIL-1_β_ was reduced (Figure 6b) in cell extracts originating from cells expressing exogenous Pyrin.
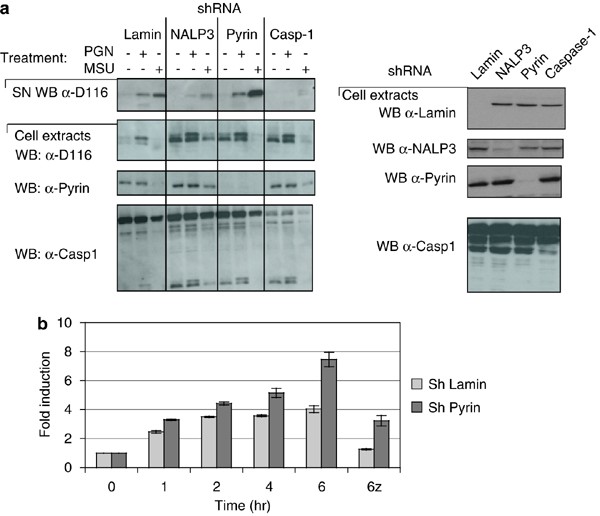
In view of the above-described inhibitory effect of the SPRY domain on proIL-1_β_ processing, we anticipated increased processing and secretion in cells in which Pyrin was absent. To this end, we generated THP1 cells in which expression of Pyrin and various inflammasome components were knocked down by shRNA (Figure 7a). The deficient cell lines were treated with the two potent NALP3 inflammasome activators, uric acid crystals (MSU) and PGN. Whereas cells lacking NALP3 or caspase-1 showed highly reduced secretion of mature IL-1_β_ as expected, increased levels of the cytokine were observed in cells that had the Pyrin protein knocked down (Figure 7a and b).
Figure 7
THP1 cells stably expressing Pyrin shRNA show reduced IL-1_β_ processing. (a) THP1 cells were infected with lentivirus carrying various shRNA. The efficacy of knockdown was determined by Western blots. PMA differentiated mock- and THP1 cells with various shRNA were activated with PGN or MSU for 4 h and the amount of active IL-1_β_ in the supernatant detected with the D116 antibody. (b) The levels of IL-1_β_ were determined by ELISA in supernatants of MSU-activated THP1 cells
Discussion
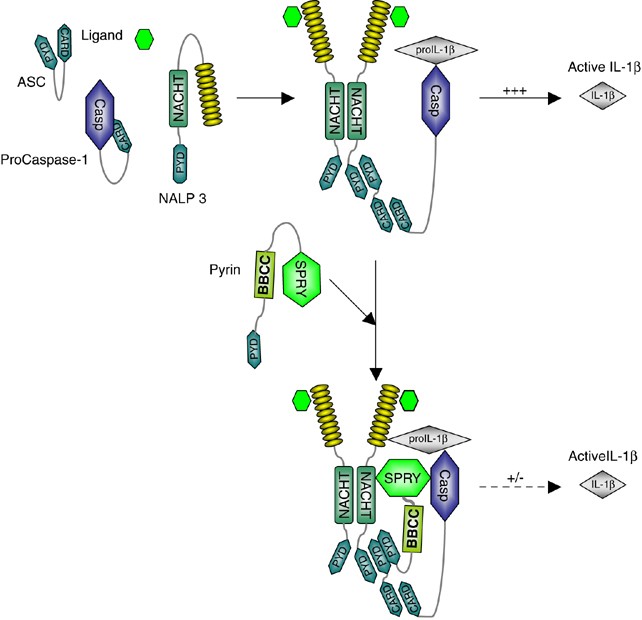
The findings in this article support the hypothesis for the function of Pyrin as an inhibitor of caspase-1 and subsequent processing of IL-1_β_.34 Pyrin, through its SPRY domain, can interact with the inflammasome components caspase-1, NALP1, NALP2 and NALP3. In addition to its interaction with different inflammasome components, the same domain also interacts with the substrate of the inflammasomes: proIL-1_β_. It is important to note that no interaction could be detected between Pyrin and mature IL-1_β_, indicating that this interaction requires the pro-domain of IL-1_β_. Considering that Pyrin also interacts with ASC via the PYD domain,19, 26 we speculate that Pyrin serves as a inhibitory protein for the inflammasome/proIL-1_β_ complex (Figure 8). In this model the proIL-1_β_ processing activity of the assembling inflammasome, through interaction of its NACHT and its PYD domain with the SPRY and PYD domain of Pyrin, respectively, is inhibited. The blockade of inflammasome activity is further strengthened by the fact that Pyrin also binds to the inflammasome substrate, that is, proIL-1_β_.
Figure 8
Schematic model of NALP3-induced caspase-1 activation and proIL-1_β_ processing by the SPRY domain of Pyrin. Pyrin is initially autoinhibited through the interaction between the Pyrin and BBCC domain. Following the activation of the inflammasome, Pyrin is recruited into the assembling complex, where it interacts with processed caspase-1 and its substrate proIL-1_β_
This model is supported by experiments in which we show that the caspase-1/proIL-1_β_ processing inhibitory activity of Pyrin is mostly confined to the SPRY domain. We detected this inhibitory effect of Pyrin in overexpression experiments in 293T cells, by transfection of the full-length Pyrin or only the SPRY domain of Pyrin. But we also see this inhibitory effect of Pyrin in a more physiological system, in THP1 cells that were genetically modified to express exogenous Pyrin and in which a dampening of IL-1_β_ processing and secretion is observed. Most convincingly, dowregulation of endogenous Pyrin in THP1 cells by shRNA led to a substantial increase in inflammasome activity. As we could detect interaction of Pyrin with NALP1, NALP2 and NALP3, we can speculate that Pyrin not only modulates the NALP3-associated inflammasome activity, but also of others NALPs-associated inflammasomes. Interestingly, another SPRY containing protein, the estrogen-responsive B box protein, was recently also shown to modulate inflammasome activity.35
As this caspase-1 inhibiting activity is associated with the SPRY domain of Pyrin, it was of interest to test whether mutations found in FMF patients in the SPRY domain of Pyrin36, 37 may affect SPRY binding to NALPs, caspases or proIL-1_β_. However, our experiments failed to demonstrate that the most frequent mutation M694V affects one of these interactions. We cannot exclude that the strength of the interactions is only mildly affected and cannot be measured with the experiments shown here that mostly rely on overexpression. Alternatively, the FMF mutations may affect binding of an as yet unidentified inhibitory interaction partner or influence the stability or localization of Pyrin, thereby leading to a higher level of secreted IL-1_β_ and exacerbated inflammatory responses, as observed in FMF patients.
Our results are in direct contradiction with results recently published by Chae et al.32 While these authors describe a Pyrin–SPRY domain interaction with caspase-1, in full agreement with the results presented here, they also report that this interaction is reduced in the SPRY domain carrying the M694V mutation. Although we could find dampened interaction in some experiments, this was not consistently seen. Thus, unlike Chae et al.,32 we cannot propose that the FMF mutations affect Pyrin–caspase-1 interaction, considering also that this conclusion is based on overexpression studies. We are presently co-crystallizing the SPRY-domain of Pyrin with active caspase-1 to further investigate this important issue.
These data generate a mechanistic link between two autoinflammatory diseases, FMF and Muckle–Wells syndrome as both diseases appear to be associated with increased inflammasome activity. A connection between FMF and yet another inflammatory disorders has already been described. The protein mutated in PAPA syndrome, named PSTPIP1, interacts with Pyrin through the BBCC domain of Pyrin.38 This interaction is enhanced in mutated PSTPIP1, suggesting that PAPA syndrome is due to abnormal interaction with Pyrin with PSTPIP1, thus indirectly modulating inflammasome activity.
The SPRY domain of Pyrin has undergone remarkable changes during evolution. In zebrafish and fugu, a SPRY domain has been fused directly to the NALP homolog, suggesting an important function of the SPRY domain in inflammasome regulation. During evolution, the SPRY domain separated from the NALPs and acquired other regulatory domains such as the BBCC and PYD. Interestingly, in rodents the SPRY domain is completely lost, indicating a strong variation in the function of Pyrin.39 Furthermore, the mutations found in FMF patients, except the M694V mutation, correspond to the wild-type amino-acids of Pyrin of lower primates.40 This suggests that Pyrin in patients with FMF corresponds to an ancestral state of Pyrin structure and function. If Pyrin is necessary in humans to reduce unnecessary inflammatory response against certain pathogen X, whereas in lower primates the response against pathogen X is still required, the mutations in Pyrin could allow a loss of inhibition of this response. In rodents, where the SPRY is lost, the inhibitory role of Pyrin is probably completely abrogated allowing a complete inflammatory response against the pathogen X. FMF is very frequent only in the Mediterranean area, so one can speculate that the reappearance or disappearance of a Mediterranean endemic pathogen X has stimulated selective pressure in favor of Pyrin with the mutated allele leading to a response against this pathogen. As such heterozygotes could be quite protected against this pathogen and bear a selective advantage, whereas homozygotes show an excessive response to this agent X, leading to the disease. Indeed it is expected, given the very high frequency of heterozygotes in the Mediterranean area, that heterozygotes have a yet unknown selective advantage.
Experimental Procedures
Antibodies
Polyclonal antibodies against human Pyrin (AL196) and human proIL-1_β_ were generated in rabbits (Eurogentec, Belgium). Monoclonal antibodies against NALP3 were generated from GST-NALP3_-immunized mice. Four monoclonal antibodies were purified (Nalpy3a, Nalpy3b, Nalpy3c and Nalpy3d). Other antibodies were purchased from the following companies: cleaved IL-1_β, D116 (Cell Signaling), Flag antibody (M2, Sigma), MYC antibody (9E10, Sigma), VSV antibody (P5D4, Sigma), mAbs to ASC and FADD (MBL), anti-RIP1 (Transduction), anti-caspase-1 (Santa-Cruz Biotech) and polyclonal anti-ASC (AL177, Apotech). Peroxidase-conjugated secondary anti-mouse and anti-rabbit antibodies were from Jackson IR and isotype-specific anti-IgG1 from Southern Biotech.
Cell lines
THP1 and 293T were cultured in RPMI 1640 and DMEM, respectively, supplemented with 10% fetal calf serum, 50 mM _β_-mercaptoethanol for THP1 and penicillin/streptomycin (100 _μ_g/ml of each). THP1 cells stably expressing VSV-Pyrin and shRNA were cultured with medium supplemented with 5 _μ_g/ml puromycin.
Transient transfection and immunoprecipitation
Transient transfection of 293T cells was performed as described previously.41 For co-immunoprecipitation experiments, transfected 293T cells were suspended in 500 _μ_l of lysis buffer (50 mM Tris (pH 7.8), 200 mM NaCl, 1% Nonidet P-40, 5 mM EDTA and protease inhibitors) and incubated at 4°C 15 min. The lysates were then cleared by centrifugation at 13 000 r.p.m. for 5 min and incubated with 3 _μ_g of anti-Flag agarose (Sigma) bound to protein G at 4°C overnight. The agarose beads were washed four times with lysis buffer, and the precipitated proteins were then fractionated on 8 and 15% SDS-PAGE and analyzed by Western blotting.
Immunoprecipitation of the inflammasome
Immunoprecipitations of endogenous Pyrin were performed using 108 THP-1 cells/time points, as described.24. Cell extracts prepared in buffer W24 were incubated with 3 _μ_g of the indicated antibodies (anti-Pyrin) conjugated to protein G at 4°C for overnight incubation. The immunoprecipitates were recovered by centrifugation and washed five times with buffer W before SDS-PAGE and Western analysis.
Generation of cell lines stably expressing Pyrin
The Pyrin stable cell line was generated by retroviral infection of THP1 cells. The Pyrin cDNA was cloned into the pMSCV retroviral vector that contains the puromycin resistance gene. Briefly, 293T cells were transfected with a mock retroviral vector, or a retroviral vector encoding Pyrin, as well as with cDNA encoding the proteins allowing the synthesis of the viral particles (Hit60 and VSV-G). At 8 h after transfection, cells were incubated overnight with sodium butyrate. After 48 h, retroviral supernatants were collected, filtered and used to perform four infection cycles (1 h incubation at 37°C and 1 h centrifugation) of THP1 cells. THP1-infected cells were then selected by culturing them in the presence of puromycin.
shRNA
nt GCTGGAGCAGGTGTACTACTTC of the human Pyrin coding sequence, nt CAGGTTTGACTATCTGTTCT of the human NALP3 coding sequence and nt GTGAAGAGATCCTTCTGTA of the 3′ UTR of the human caspase-1 were inserted into pSUPER. The Pol III promoter shRNA cassettes from these vectors and from a lamin A/C-specific pSUPER control construct were inserted into the lentiviral vector pAB286.1, a derivative of pHR that contains a SV40-puromycin acetyl transferase cassette for antibiotic selection (lamin A/C-pSUPER and pAB286.1 were gifts of Richard Iggo, Lausanne, Switzerland). Second generation packaging plasmids pMD2-VSVG and pCMV-R8.91 (Naldini et al., 1996) were used for lentivirus production and infection as described elsewhere (http://www.tronolab.unige.ch).
Processing and secretion of IL-1_β_
To study IL-1_β_ processing and secretion in THP1 cells, the following method was used. Mock- or Pyrin-infected THP1 cells were differentiated with PMA for 3 h, allowing the synthesis of the precursor of IL-1_β_. After an overnight incubation, THP1 cells become adherent, and they were stimulated with bacterial PGN(10 μ_g/ml, Alexis) or uric acid crystals (200 μ_g/ml, Alexis), with or without z-VAD-fmk, for 1–6 h. Then the supernatants were collected and precipitated, and the cell extracts were prepared, and both were subjected to Western-blot analysis to detect the presence of cleaved IL-1_β. On the other hand, the in vitro system previously described for the identification of the inflammasome33 was used to study the processing of IL-1_β in mock and Pyrin-infecetd cells. Briefly, THP1 cells were differentiated by an overnight PMA incubation, and then lysed in the Wang hypotonic buffer. The cell extracts were then activated at 30°C, a temperature allowing the activation of the inflammasome, and subjected to Western-blot analysis.
Abbreviations
BBCC:
B-boxes and a coiled-coil
FMF:
familial Mediterranean fever
IL:
interleukin
PYD:
Pyrin domain
References
- McDermott MF, Aksentijevich I . The autoinflammatory syndromes. Curr Opin Allergy Clin Immunol 2002; 2: 511–516.
Article PubMed Google Scholar - Grateau G, Drenth JP, Delpech M . Hereditary fevers. Curr Opin Rheumatol 1999; 11: 75–78.
Article CAS PubMed Google Scholar - Stojanov S, Kastner DL . Familial autoinflammatory diseases: genetics, pathogenesis and treatment. Curr Opin Rheumatol 2005; 17: 586–599.
Article CAS PubMed Google Scholar - Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J . NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle–Wells autoinflammatory disorder. Immunity 2004; 20: 319–325.
Article CAS PubMed Google Scholar - Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD . Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle–Wells syndrome. Nat Genet 2001; 29: 301–305.
Article CAS PubMed PubMed Central Google Scholar - Feldmann J, Prieur AM, Quartier P, Berquin P, Certain S, Cortis E et al. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet 2002; 71: 198–203.
Article CAS PubMed PubMed Central Google Scholar - Drenth JP, Cuisset L, Grateau G, Vasseur C, van de Velde-Visser SD, de Jong JG et al. Mutations in the gene encoding mevalonate kinase cause hyper-IgD and periodic fever syndrome. International Hyper-IgD Study Group. Nat Genet 1999; 22: 178–181.
Article CAS PubMed Google Scholar - Houten SM, Kuis W, Duran M, de Koning TJ, van Royen-Kerkhof A, Romeijn GJ et al. Mutations in MVK, encoding mevalonate kinase, cause hyperimmunoglobulinaemia D and periodic fever syndrome. Nat Genet 1999; 22: 175–177.
Article CAS PubMed Google Scholar - Wise CA, Gillum JD, Seidman CE, Lindor NM, Veile R, Bashiardes S et al. Mutations in CD2BP1 disrupt binding to PTP PEST and are responsible for PAPA syndrome, an autoinflammatory disorder. Hum Mol Genet 2002; 11: 961–969.
Article CAS PubMed Google Scholar - Hull KM, Drewe E, Aksentijevich I, Singh HK, Wong K, McDermott EM et al. The TNF receptor-associated periodic syndrome (TRAPS): emerging concepts of an autoinflammatory disorder. Medicine (Baltimore) 2002; 81: 349–368.
Article CAS Google Scholar - McDermott M, Aksentijevich I, Galon J, McDermott E, Ogunkolade B, Centola M et al. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell 1999; 97: 133–144.
Article CAS PubMed Google Scholar - The French FMF Consortium. A candidate gene for familial Mediterranean fever. Nat Genet 1997; 17: 25–31.
Article Google Scholar - The International FMF Consortium. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell 1997; 90: 797–807.
Article Google Scholar - Centola M, Wood G, Frucht DM, Galon J, Aringer M, Farrell C et al. The gene for familial Mediterranean fever, MEFV, is expressed in early leukocyte development and is regulated in response to inflammatory mediators. Blood 2000; 95: 3223–3231.
CAS PubMed Google Scholar - Diaz A, Hu C, Kastner DL, Schaner P, Reginato AM, Richards N et al. Lipopolysaccharide-induced expression of multiple alternatively spliced MEFV transcripts in human synovial fibroblasts: a prominent splice isoform lacks the C-terminal domain that is highly mutated in familial Mediterranean fever. Arthritis Rheum 2004; 50: 3679–3689.
Article CAS PubMed Google Scholar - Papin S, Duquesnoy P, Cazeneuve C, Pantel J, Coppey-Moisan M, Dargemont C et al. Alternative splicing at the MEFV locus involved in familial Mediterranean fever regulates translocation of the marenostrin/pyrin protein to the nucleus. Hum Mol Genet 2000; 9: 3001–3009.
Article CAS PubMed Google Scholar - Jeru I, Papin S, L'Hoste S, Duquesnoy P, Cazeneuve C, Camonis J et al. Interaction of pyrin with 14.3.3 in an isoform-specific and phosphorylation-dependent manner regulates its translocation to the nucleus. Arthritis Rheum 2005; 52: 1848–1857.
Article CAS PubMed Google Scholar - Mansfield E, Chae JJ, Komarow HD, Brotz TM, Frucht DM, Aksentijevich I et al. The familial Mediterranean fever protein, pyrin, associates with microtubules and colocalizes with actin filaments. Blood 2001; 98: 851–859.
Article CAS PubMed Google Scholar - Richards N, Schaner P, Diaz A, Stuckey J, Shelden E, Wadhwa A et al. Interaction between pyrin and the apoptotic speck protein (ASC) modulates ASC-induced apoptosis. J Biol Chem 2001; 276: 39320–39329.
Article CAS PubMed Google Scholar - Tidow N, Chen X, Muller C, Kawano S, Gombart AF, Fischel-Ghodsian N et al. Hematopoietic-specific expression of MEFV, the gene mutated in familial Mediterranean fever, and subcellular localization of its corresponding protein, pyrin. Blood 2000; 95: 1451–1455.
CAS PubMed Google Scholar - Fairbrother WJ, Gordon NC, Humke EW, O'Rourke KM, Starovasnik MA, Yin JP et al. The Pyrin domain: a member of the death domain-fold superfamily. Protein Sci. 2001; 10: 1911–1918.
Article CAS PubMed PubMed Central Google Scholar - Kohl A, Grutter MG . Fire and death: the pyrin domain joins the death-domain superfamily. C R Biol 2004; 327: 1077–1086.
Article CAS PubMed Google Scholar - Martinon F, Hofmann K, Tschopp J . The pyrin domain: a possible member of the death domain-fold family implicated in apoptosis and inflammation. Curr Biol 2001; 11: R118–R120.
Article CAS PubMed Google Scholar - Martinon F, Burns K, Tschopp J . The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 2002; 10: 417–426.
Article CAS PubMed Google Scholar - Masumoto J, Taniguchi S, Nakayama J, Shiohara M, Hidaka E, Katsuyama T et al. Expression of apoptosis-associated speck-like protein containing a caspase recruitment domain, a pyrin N-terminal homology domain-containing protein, in normal human tissues. J Histochem Cytochem 2001; 49: 1269–1275.
Article CAS PubMed Google Scholar - Yu JW, Wu J, Zhang Z, Datta P, Ibrahimi I, Taniguchi S et al. Cryopyrin and pyrin activate caspase-1, but not NF-kappaB, via ASC oligomerization. Cell Death Differ 2005; 13: 236–249.
Article Google Scholar - Dowds TA, Masumoto J, Chen FF, Ogura Y, Inohara N, Nunez G . Regulation of cryopyrin/Pypaf1 signaling by pyrin, the familial Mediterranean fever gene product. Biochem Biophys Res Commun 2003; 302: 575–580.
Article CAS PubMed Google Scholar - Masumoto J, Dowds TA, Schaner P, Chen FF, Ogura Y, Li M et al. ASC is an activating adaptor for NF-kappa B and caspase-8-dependent apoptosis. Biochem Biophys Res Commun 2003; 303: 69–73.
Article CAS PubMed Google Scholar - Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L et al. The tripartite motif family identifies cell compartments. EMBO J 2001; 20: 2140–2151.
Article CAS PubMed PubMed Central Google Scholar - Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J . The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 2004; 427: 848–853.
Article CAS PubMed Google Scholar - Yap MW, Nisole S, Lynch C, Stoye JP . Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci USA 2004; 101: 10786–10791.
Article CAS PubMed PubMed Central Google Scholar - Chae JJ, Wood G, Masters SL, Richard K, Park G, Smith BJ et al. The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1beta production. Proc Natl Acad Sci USA 2006; 103: 9982–9987.
Article CAS PubMed PubMed Central Google Scholar - Martinon F, Burns K, Tschopp J . The inflammasome. A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol Cell 2002; 10: 417.
Article CAS PubMed Google Scholar - Chae JJ, Komarow HD, Cheng J, Wood G, Raben N, Liu PP et al. Targeted disruption of Pyrin, the FMF protein, causes heightened sensitivity to endotoxin and a defect in macrophage apoptosis. Mol Cell 2003; 11: 591–604.
Article CAS PubMed Google Scholar - Munding C, Keller M, Niklaus G, Papin S, Tschopp J, Werner S et al. The estrogen-responsive B box protein: a novel enhancer of interleukin-1beta secretion. Cell Death Differ 2006; 13: 1938–1949.
Article CAS PubMed Google Scholar - Cazeneuve C, Ajrapetyan H, Papin S, Roudot-Thoraval F, Genevieve D, Mndjoyan E et al. Identification of MEFV-independent modifying genetic factors for familial Mediterranean fever. Am J Hum Genet 2000; 67: 1136–1143.
CAS PubMed PubMed Central Google Scholar - Samuels J, Aksentijevich I, Torosyan Y, Centola M, Deng Z, Sood R et al. Familial Mediterranean fever at the millennium. Clinical spectrum, ancient mutations, and a survey of 100 American referrals to the National Institutes of Health. Medicine (Baltimore) 1998; 77: 268–297.
Article CAS Google Scholar - Shoham NG, Centola M, Mansfield E, Hull KM, Wood G, Wise CA et al. Pyrin binds the PSTPIP1/CD2BP1 protein, defining familial Mediterranean fever and PAPA syndrome as disorders in the same pathway. Proc Natl Acad Sci USA 2003; 100: 13501–13506.
Article CAS PubMed PubMed Central Google Scholar - Chae JJ, Centola M, Aksentijevich I, Dutra A, Tran M, Wood G et al. Isolation, genomic organization, and expression analysis of the mouse and rat homologs of MEFV, the gene for familial Mediterranean fever. Mamm Genome 2000; 11: 428–435.
Article CAS PubMed Google Scholar - Schaner P, Richards N, Wadhwa A, Aksentijevich I, Kastner D, Tucker P et al. Episodic evolution of pyrin in primates: human mutations recapitulate ancestral amino acid states. Nat Genet 2001; 27: 318–321.
Article CAS PubMed Google Scholar - Thome M, Hofmann K, Burns K, Martinon F, Bodmer JL, Mattmann C et al. Identification of CARDIAK, a RIP-like kinase that associates with caspase-1. Curr Biol 1998; 8: 885–888.
Article CAS PubMed Google Scholar
Acknowledgements
We thank Helen Everett for critical reading of the article. This work was supported by grants from the Swiss National Science foundation and CTI (to JT and HDB). SP is a recipient of a Marie-Curie fellowship.
Author information
Authors and Affiliations
- Department of Biochemistry, University of Lausanne, Chemin des Boveresses 155, CH-1066, Epalinges, Switzerland
S Papin, S Cuenin, L Agostini, F Martinon & J Tschopp - Department of Biology, Institute of Cell Biology, ETH Zürich, CH-8093, Zürich, Switzerland
S Werner & H-D Beer - Department of Biochemistry, University of Zürich, Winterthurerstr 160, CH-8057, Zürich, Switzerland
C Grütter & M Grütter
Authors
- S Papin
You can also search for this author inPubMed Google Scholar - S Cuenin
You can also search for this author inPubMed Google Scholar - L Agostini
You can also search for this author inPubMed Google Scholar - F Martinon
You can also search for this author inPubMed Google Scholar - S Werner
You can also search for this author inPubMed Google Scholar - H-D Beer
You can also search for this author inPubMed Google Scholar - C Grütter
You can also search for this author inPubMed Google Scholar - M Grütter
You can also search for this author inPubMed Google Scholar - J Tschopp
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toJ Tschopp.
Additional information
Edited by P Mehlen
Rights and permissions
About this article
Cite this article
Papin, S., Cuenin, S., Agostini, L. et al. The SPRY domain of Pyrin, mutated in familial Mediterranean fever patients, interacts with inflammasome components and inhibits proIL-1_β_ processing.Cell Death Differ 14, 1457–1466 (2007). https://doi.org/10.1038/sj.cdd.4402142
- Received: 06 October 2006
- Revised: 07 March 2007
- Accepted: 07 March 2007
- Published: 13 April 2007
- Issue Date: August 2007
- DOI: https://doi.org/10.1038/sj.cdd.4402142