The genome sequence of E. coli W (ATCC 9637): comparative genome analysis and an improved genome-scale reconstruction of E. coli (original) (raw)
…
42k Accesses
140 Citations
10 Altmetric
Abstract
Background
Escherichia coli is a model prokaryote, an important pathogen, and a key organism for industrial biotechnology. E. coli W (ATCC 9637), one of four strains designated as safe for laboratory purposes, has not been sequenced. E. coli W is a fast-growing strain and is the only safe strain that can utilize sucrose as a carbon source. Lifecycle analysis has demonstrated that sucrose from sugarcane is a preferred carbon source for industrial bioprocesses.
Results
We have sequenced and annotated the genome of E. coli W. The chromosome is 4,900,968 bp and encodes 4,764 ORFs. Two plasmids, pRK1 (102,536 bp) and pRK2 (5,360 bp), are also present. W has unique features relative to other sequenced laboratory strains (K-12, B and Crooks): it has a larger genome and belongs to phylogroup B1 rather than A. W also grows on a much broader range of carbon sources than does K-12. A genome-scale reconstruction was developed and validated in order to interrogate metabolic properties.
Conclusions
The genome of W is more similar to commensal and pathogenic B1 strains than phylogroup A strains, and therefore has greater utility for comparative analyses with these strains. W should therefore be the strain of choice, or 'type strain' for group B1 comparative analyses. The genome annotation and tools created here are expected to allow further utilization and development of E. coli W as an industrial organism for sucrose-based bioprocesses. Refinements in our E. coli metabolic reconstruction allow it to more accurately define E. coli metabolism relative to previous models.
Similar content being viewed by others
Background
Escherichia coli is a model prokaryotic organism, an important pathogen and commensal, and a popular host for biotechnological applications. Among thousands of isolates, only four strains (the common laboratory strains K-12, B, C, and W) and their derivatives are designated as Risk Group 1 organisms in biological safety guidelines [1, 2]. A fifth strain, E. coli Crooks (ATCC 8739), has also been used extensively in laboratories for over 70 years [3–5]; more recently, it has been used as a host for industrial biochemical production [6–8]. There have been no reported cases of the strain being pathogenic, suggesting that it is generally safe. When it was sequenced in 2007, ATCC 8739 was designated as a C strain [6], however, it is in fact a Crooks strain [4] and recent publications have reflected this correction [9, 10]. Of these five safe strains, K-12 [11], B [12] and Crooks [GenBank:CP000946] have been sequenced, but C and W have not.
E. coli W (ATCC 9637) was originally isolated from the soil of a cemetery near Rutgers University around 1943 by Selman A. Waksman, around the same time he and Alan Schatz discovered streptomycin (Eliora Ron, personal communication). Waksman coined the term 'antibiotic', and his discovery of streptomycin (and many other antibiotics) led to him being awarded the Nobel Prize in Physiology or Medicine in 1952. The strain was termed "Waksman's strain" or "W strain" because it showed the highest sensitivity to streptomycin compared to other isolated E. coli strains in Waksman's collection (Eliora Ron, personal communication).
The first reported use of W was as the standard E. coli strain in the assay for sensitivity to streptomycin and other antibiotics [13]. Bernard Davis, a prominent microbiologist from Harvard Medical School, developed a large auxotrophic mutant library from the strain [14] using his penicillin-based selection technique [15]. One of these mutants, vitamin B-12 auxotroph 113-3 (ATCC 11105), is well known as a production strain for penicillin G acyclase (PGA) [16] and for studies of aromatic compound degradation in bacteria [17]. It has also recently been discovered that the popular ethanol-producing strain KO11 [18] is a W strain rather than a B strain as previously thought [19]. Both W and KO11 have been engineered for the production of several chemicals, including ethanol [18, 20, 21], poly-3-hydroxybutyrate[22], lactic acid [23] and alanine [19]. The W strain has several properties that make it a preferred strain for industrial applications. It produces low amounts of acetate even without tight sugar control, and can be grown to high cell density during fed-batch culture with relative ease [22]. It also has good tolerance for environmental stresses such as high ethanol concentrations, acidic conditions, high temperatures and osmotic stress [24, 25]. It is a very fast growing strain; its superior growth rate on LB medium compared to classical K-12-derived strains has led to it being developed as a lab cloning strain [27]. These combined characteristics make W extremely attractive as a production strain. Significantly, W is the only safe E. coli strain which can utilize sucrose as a carbon source, and it grows as fast on sucrose as it does on glucose [22, 27, 28]. Sucrose is emerging as a preferred carbon source for industrial fermentation: life cycle analysis demonstrates that sucrose from sugarcane has a superior performance when compared to glucose from starch [29].
Modern development of good production strains entails application of metabolic engineering principles. Increasingly, metabolic engineering relies on a systems biology approach [30]; a key aspect of this approach is the integration of a metabolic model (genome-scale model, GEM). The first step in developing a GEM is to build an in silico genome-scale reconstruction (GSR) derived from the organism's genome sequence. In this paper, we present the complete genome sequence, detailed annotation of E. coli W. Comparative genome analyses were performed among safe E. coli strains and group B1 commensal/pathogenic E. coli strains. In addition, a comprehensive, W-specific GSR was developed to underpin construction of a GEM for engineering industrial production strains.
Results and Discussion
Annotation and comparative analysis with other safe laboratory strains
A combination of Roche/454 pyrosequencing, fosmid end sequencing and Sanger sequencing was used to obtain the complete genome sequence of E. coli W (ATCC 9637). The W genome consists of a circular chromosome [Genbank: CP002185] (Figure 1) and two plasmids, pRK1 [Genbank: CP002186] and pRK2 [Genbank: CP002187]. Detailed results of genome analysis can be found in Table 1. At 4,901 Kbp, the chromosome of E. coli W is the largest of all the sequenced safe laboratory strains. Comparison with available E. coli genome sequences in GenBank demonstrated that it is similar in size to the commensal E. coli strain SE11 (4,888 Kbp) [31], but smaller than most sequenced pathogenic strains. A total of 4,764 chromosomal genes (including 82 non-coding RNA genes) were predicted using Prodigal [32] and Glimmer[33]; these genes cover 89% of the chromosome.
Figure 1
Circular map of the E. coli W chromosome. The outer circle shows position in bp. The second, third and fourth circles (blue) show forward ORFs, reverse ORFs, and pseudogenes, respectively. The fifth circle (green) shows pseudoknots. The seventh circle shows large mobile elements (see Table 2 for details); pLEs are in green and prophages are in red. The inner circle shows a plot of G+C content, with purple being G+C and tan being A+T.
Table 1 Summary of genome features in safe strains.
A wide variety of algorithms were used to predict and annotate coding and non-coding genes (see Methods). Like the three other sequenced laboratory strains, W has 22 rRNA genes expressed from 7 rRNA operons; these operons are present at similar locations in all four genomes. The four strains share 85 tRNAs and there are four unshared tRNAs located in large mobile elements. W has thrX and tyrX, which occur within a variable region of the Rac*W prophage and are homologous to thrU and tyrU of E. coli K-12; due to separate IS-mediated deletions, W and B are both missing a tRNA which occurs upstream of ypjC in K-12; in K-12, ileY is present. In Crooks the sequence of a tRNA in the same location is identical to ileY of K-12 but has been mis-annotated as a tRNA-Met2 variant.
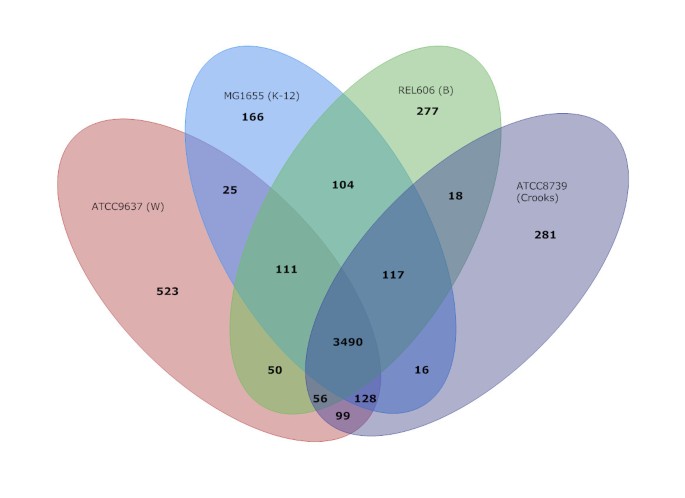
All-against-all BLASTP comparison of chromosomal protein-coding orthologs among the four safe laboratory strains (Figure 2, Additional File 1) showed that of 4,482 predicted CDSs in W, 3,490 are shared among these four strains. Another 413 are found in at least one other strain, leaving 523 CDSs that are unique to W. Consistent with the larger genome size, this is ~320-360 more CDSs than were found to be unique in any other safe strain. It should be noted that the number of shared orthologs between strains is not an indicator of overall relatedness, since increases in shared genes tends to arise from large insertion elements (for example, K-12 and B share a large genomic island encoding a restriction modification system while Crooks and W share two large gene clusters encoding excretion systems). Furthermore, differences in genome sizes bias this kind of relationship comparison.
Figure 2
Comparison of orthologous CDSs between W, K-12, B and Crooks strains. The number of shared genes, as well and the number of unique genes and genes shared between one, two, and three strains are shown. All-against-All BLASTP for amino acids (E-value ≤ 1E-5, identity ≥ 90%, coverage ≥ 80%) was used to assign orthologs. Total CDS counts for K-12, B & Crooks differ by 8, 14 & 5 respectively as some CDSs had more than one ortholog in another genome (Additional File 1).
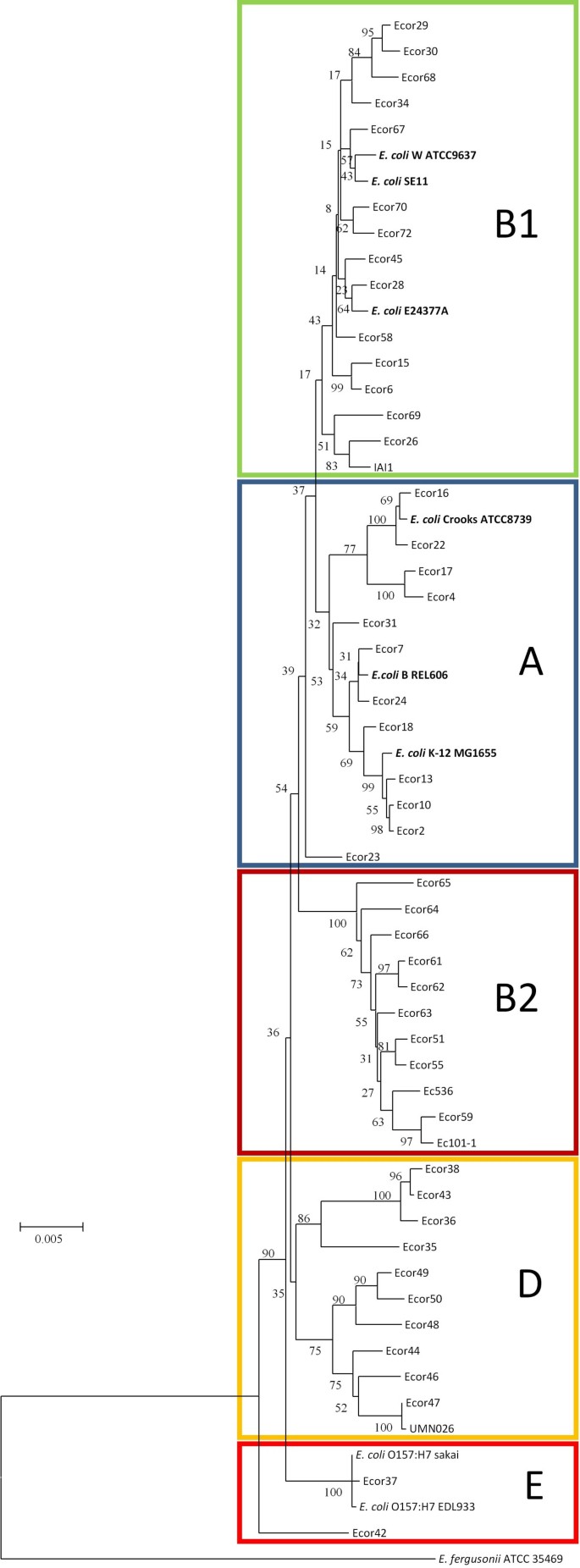
E. coli strains can be divided into five different ECOR phylogroups (A, B1, B2, D and E) based on the sequences of housekeeping genes [34]. Commensal strains are found primarily in group A or group B1, which are sister groups, while pathogenic strains are generally found in Group B2, D and E [31, 34, 35]. A phylogenetic tree was constructed by sequence concatenation of seven housekeeping genes [36] (Figure 3). Using this approach, W was assigned to group B1. Group B1 contains a large number of commensal strains [37]. The other three sequenced safe strains (K-12, B and Crooks), are all members of phylogroup A [31, 35]. Interestingly, these groupings are consistent with genome sizes of sequenced strains: group B1 strains have larger genomes than group A strains. W is arguably a more appropriate strain than K-12, B or Crooks for comparison with commensal and pathogenic strains of phylogroup B1.
Figure 3
Phylogenetic analysis of sequenced E. coli strains. Phylogenetic relationships based on seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA). Strains cluster into phylogroups; W can be found in group B1, whereas the other three laboratory strains are in group A. Escherichia fergusonii (ATCC 35469) was used as an out-group. The tree shows bootstrap values (percentage per 1000 replicates). The scale bar represents divergence time.
Plasmids
An early report suggested that E. coli W contains three plasmids [38]. However, it was later suggested that W contains only two plasmids [26]. Our sequence data confirmed the latter report: W contains two plasmids, pRK1 and pRK2. pRK1 is a circular plasmid of 102,536 bp. It encodes 118 genes: 114 protein coding genes, one pseudogene and three ncRNAs (Table 1). BLAST analysis demonstrated that it belongs to Incompatibility Group I1 (IncI1) and has high structural similarity with the IncI plasmids pR64 (a reference IncI1 plasmid), pSE11-1 (a plasmid of roughly 100 Kbp isolated from E. coli SE11), and pColIb-P9. Analysis of inc, a marker for IncI designations [39], showed that inc in pRK1 differed by only one base pair from the reference inc of Inc I1 subgroup Iγ [40]. IncI1 plasmids are characterized by the presence of genes encoding a thick pilus, a thin type IVB pilus, the pilus-associated protein gene pilV, and the DNA primase gene _sog_[41].
Genes for antibiotic resistance are found on most sequenced IncI plasmids, including IncI1 plasmids [42] and IncIγ-type R621a [43]; however, pRK1 does not encode any antibiotic resistance genes. This is desirable in industrial strains as genetic manipulation for strain improvement often involves the use of antibiotic selection. In addition, an IS_91_ insertion has interrupted two genes involved in colicin production (cib and imm). This insertion also resulted in the introduction of genes involved in κ-type fimbriae (see further comments below).
The _trbA_-exc region in IncI1 plasmids is a diverse region and includes genes that are involved in plasmid maintenance and transfer. pRK1 contains a complete trb regulon, which is required for plasmid transfer. Two other genes are of interest: excAB, which controls surface exclusion and thus determines which plasmid types can conjugate into the host cell, and pndCA, which controls plasmid stability [44]. In pRK1, pndCA has been lost, suggesting that plasmid stability might be affected even though there is no direct evidence that pRK1 is unstable in W. In addition, the 3' region of exc differs greatly from other exc genes on IncI1 plasmids, suggesting that this gene encodes a protein which determines different mating specificity than other IncI plasmids.
Plasmid pRK2 has been sequenced previously [45] and our analysis is in agreement with the reported information. Briefly, pRK2 is a cryptic ColE1-type plasmid; it is 5,360 bp and encodes 16 predicted genes including 15 protein-coding genes and one non-coding RNA. It is stably inherited and contains four putative mobilisation genes and a gene encoding a Rom protein. It shares 99% identity with pSE11-4, a plasmid isolated from the group B1 commensal E. coli SE11 [31].
Finally, there is some evidence that E. coli W once harbored a third plasmid. An IS_91_ insertion in pRK1 (see below for further details) is homologous to a region in pSE11-3, an IncF plasmid from E. coli SE11 [31]. The insertion has deleted a region of pRK1 which is normally found in IncI plasmids. Additionally, the partial fimbrial gene cluster which was transferred with the insertion is known to be plasmid-encoded [46]. W and SE11 belong to the same phylogroup and therefore might share a common ancestry; furthermore, two of the SE11 plasmids are highly similar to pRK1 and pRK2 (pSE11-1 and pSE11-4, respectively). Thus, it seems likely that an ancestral W strain might have harbored a plasmid similar to pSE11-3.
Mobility elements and defence systems
E. coli genomes consist of a conserved core interspersed with variable regions encoding accessory functions [47]. The conserved core is shared with closely related genera such as _Citrobacter_[48], _Shigella_[49] and _Salmonella_[50]. The accessory genome encodes lifestyle-specific functions which are often found in large clusters or related genes (so called 'genomic islands') [51–53]. These clusters contain a different G+C content compared to the rest of the genome (see Figure 1) and are acquired through horizontal gene transfer (HGT) via natural transformation, bacteriophage-mediated transduction or conjugation.
Mobility elements
Large genomic islands which are flanked by mobility elements are known as large mobile elements (LMEs), and include prophages or phage-like elements (pLEs) [54]. Differentiation between prophages and pLEs can be difficult; in general, a prophage will contain specific metabolic and structural genes associated with a prophage, while a pLE will contain an integrase and very few regions which are homologous to known prophages. LMEs carry large complements of genes which might confer a variety of metabolic attributes. E. coli W has six prophages and three pLEs, the latter of which we have designated 'E. coli W phage Like Elements' (WpLEs). A detailed list of LMEs in E. coli W and other safe strains can be found in Table 2.
Table 2 Large mobile elements found in safe strains.
A total of twenty-eight LMEs are annotated amongst the safe E. coli strains. They are spread out over nineteen different sites in the chromosome and all but one can be classified as either a pLE or one of three different prophages (P2-like, P4-like or λ-like). The exception is the Mu prophage, a transpositional phage that inserts into almost random chromosomal locations [55]; among the four strains, Mu prophage is only found in W. None of the LMEs in W encode any genes of particular note. In the other strains, a few genes of interest are encoded on prophages. Rybb*B carries retron Ec86 [6], which encodes a reverse transcriptase that is missing from Rybb*C and Rybb*W. The P4 prophage CP4-44 is absent in W and Crooks but present in K-12; the flu gene is encoded on this prophage in K-12 and is encoded on Phev*B in B. The λ prophage is the most promiscuous prophage element among the four strains.
Ten pLEs are found among safe strains. Only KpLE2 is shared (being found in both K-12 and B). E. coli Crooks might have harboured KpLE2: it contains a 259 bp pseudogene, the first 137 bp of which shares 72% identity with the P4-integrases of KpLE2 in K-12 and B. KpLE2 contains the fec regulon (discussed below) and the sgc operon, which is involved in pentose and pentitol sugar breakdown [56]. K-12 contains KpLE1, which includes the gtrAB regulon encoding a bactoprenol glucosyl-transferase involved in O-antigen modification. The Crooks strain harbours CpLE1, which contains an endonuclease, and CpLE3 which also contains a fec regulon. The WpLE3 of W appears to comprise two separate pLEs, as a second P4-integrase is found with distinct regions of DNA following each integrase. The first region contains a toxin-antitoxin system while the second region contains a putative 5-methylcytosine restriction system.
Insertion sequences (ISs) play an important role in the cell's ability to evolve and adapt to new environments [57]. A complete description of the IS elements in safe strains can be found in Table 3. Only two ISs are conserved among all four strains; as previously reported [58], no copies of IS_1_ were found within the W genome. The W genome contains 24 IS elements, which is significantly fewer than K-12, B or Crooks; as a consequence, W has no IS-related gene inactivation occurring in the chromosome, whereas K-12 and B both have a number of genes inactivated. These include genes involved in lipopolysaccharide (LPS) and capsular polysaccharide (CPS) synthesis, as well as large deletions such as the 41 Kbp region between uvrY and hchA in B which removes the Flag-1 flagella-encoding gene cluster (see below for further details).
Table 3 Insertion sequences found in safe strains.
Restriction modification and CRISPR systems
Restriction modification and clustered regularly interspaced short palindromic repeat (CRISPR) systems play an important role in antiviral defence against invasive foreign genetic material (e.g., bacteriophages and integrative elements) and hence control the extent of HGT [59]. Restriction capabilities are conferred by the immigration control region [60]. Both W and Crooks are restriction minus as they lack hsdMRS, mcrBC and mrr, which encode the restriction modification complexes. In W, this cluster has been replaced by the pac gene encoding a penicillin G acyclase (PGA), which catalyses the breakdown of penicillin G into phenylacetic acid and 6-aminopenicillanic acid [17]. This capability has been exploited for the industrial production of PGA using E. coli W [16]. In Crooks, the immigration control region has undergone multiple changes due to IS element insertions. The lack of restriction modification systems in W and Crooks suggests that these strains are less able to inactivate foreign DNA.
CRISPR systems inhibit horizontal gene transfer. The detailed mechanisms have just begun to be exposed [61]. Recently, two CRISPR systems have been described in E. coli: CRISPR2 and CRISPR4 [62]. These systems differ by the presence or absence of CRISPR associated sequence (CAS) proteins (the function of which is unknown), and by the location, number and sequence of repeats. E. coli W contains three CRISPR2 arrays, CRISPR2.1, 2.2, and 2.3 (Table 4). Genes encoding E. coli Cas proteins are present next to CRISPR2.1. W also contains the CRISPR4.1-2 array but not the associated Yersinia pestis Cas proteins, which are found in many E. coli strains [62]. Each safe strain has the same number of arrays, but the sequences and number of repeat regions varies (Table 4). There are two cas gene clusters found in E. coli which vary in the _cas3_-cse3 region; it is unclear if they have the same function [63]. One is found in K-12 and Crooks and the other is found in W and O157. Multiple insertions and deletions have destroyed the cas gene cluster in E. coli B.
Table 4 CRISPR arrays found in safe strains.
Virulence/Fitness Factors
Virulence factors are classically considered to be associated with host interactions and pathogenicity. However, it should be noted that many of these so-called virulence factors can also be considered fitness factors in a non-virulence context [64]. For example, adhesins are important for colonizing all manner of niches; colonisation does not necessarily lead to infection and disease.
Serotypic antigens
E. coli serotypes are defined according to the polysaccharide component of LPS molecules [65–67]. These include CPSs, which can be either K-antigen or colonic acid (M-antigen) and O-polysaccharides (O-antigen). The H-antigen is used for serotyping, and its type is usually determined by FliC, a flagellar structural protein [68]. HGT of the gene regions responsible for production of O-antigen, K-antigen, H-antigen, and the LPS core has lead to a high degree of variability [69]. There are 167 different O-antigen types and 80 K-antigen types currently recorded amongst E. coli. Whereas other safe E. coli strains have accumulated IS-mediated deletions in antigenic clusters (Table 1), W has intact clusters. It has an R1 type LPS core and an O6 type O-antigen. Type O6 is widely distributed and found both in uropathogenic E. coli (UPEC) strains and in commensal strains [70]. W does not produce a K-antigen, but it has the gene cluster involved in colonic acid synthesis; colonic acid resembles K-antigen group IA capsular polysaccharides [66]. It also has the phosphorelay regulon (encoded by rcsA and rcsDBC) which activates production of colonic acid. FliC homology suggests that E. coli W produces an H49 type H-antigen [71]. W can thus be antigenically characterised as E. coli W (O6:K-:H49) CA+.
Adhesins
Fimbriae and other adhesins determine whether E. coli can bind to and colonise specific environments, including different types of cells. They are associated with virulence in pathogenic strains of E. coli such as enteroaggregative E. coli 55989 (EAEC) [72] but are also key to the fitness of probiotic E. coli strains such as strain Nissle 1917, as they allow it to colonize the human intestine [73]. In W, there are thirteen chromosomal gene clusters involved in fimbrial biosynthesis, and most of these are conserved among the safe strains of E. coli (Table 5). Differences arise in genes encoding the fimbrial usher protein and the tip adhesins. Tip adhesins are important determinants of host cell specificity during pathogenesis; the usher protein is a membrane protein which is involved in the assembly of a fimbria and determines which group the fimbria belongs to [74].
Table 5 Fimbrial gene clusters found in safe strains and in representative Group B1 strains.
There are 2 α-type fimbrial gene clusters in W: ecpABC-yagW-ecpE, and a novel fimbrial gene cluster found between exuT and exuR. We have designated this novel cluster E. coli α-type fimbria, eafABCD. However, neither of the clusters in W contains a gene encoding for the tip adhesin protein, which is found in other α-type fimbrial clusters and is responsible for cell binding [75]. Thus, it is unlikely that the W α-type fimbriae can function in pathogenesis or colonisation of cells in general.
W contains five γ1-type fimbrial gene clusters. One of these is E. coli YcbQ laminin-binding fimbria (ELF, formerly ycbQRST) [76] which is shared between group B1 strains. In W, the major subunit protein ElfA is relatively different (84% identity) from that found in K-12 and O157:H7 EDL933. Deletion of this gene in O157:H7 EDL933 has been shown to lead to a significant reduction in ability to adhere to HEK293 cells [76]. A γ1-type cluster found in E. coli O157:H7 and annotated as ECs2113-ECs2107, is also present in W. This cluster is also present in E. coli K-12 (annotated as ydeQRST), but a deletion removes ECs2113-ECs2112 and truncates ECs2111 (which normally encodes the usher protein). We have designated this gene cluster E. coli γ-type 1, with the operon consequently designated egoABCDEF. Information on the other three γ1-type fimbrial gene clusters is limited but all are found in K-12 and are cryptic or poorly expressed under classic laboratory conditions [77].
Two groups of fimbriae closely related to γ1-type fimbriae and known as long polar fimbriae [78] are also found in E. coli W. They are commonly found in both pathogenic and commensal strains of E. coli and consist of 3-6 genes. The first cluster, lpfA1-E1, is found in other E. coli group B1 strains (Table 5) and shows 44-77% amino acid identity to the lpf gene cluster of Salmonella enterica. The adherence of lpfA1-E1 homologs in other E. coli strains is known to vary depending on both the sequence of the gene cluster and on its regulation [78–80]. The second cluster, lpfA2-D2, is identical to the lpf operon found in E. coli 789. This lpf operon has been shown to produce the fimbria responsible for adherence to human HEK293 cells [81].
There are also three π-type fimbrial gene clusters in W and the other safe strains. One of these, located between sixA-yfcN and consisting of seven genes, shows >95% sequence identity with a fimbrial gene cluster located in the same chromosomal position in O157:H7. In O157:H7, this cluster is annotated as _ECs3222_-ECs3216; we have designated it E. coli π-type one, with the operon consequently designated epoA-H.
Due to an insertion event on pRK1, W has five of the eight genes from the κ-type csh fimbrial gene cluster. However, the lack of the terminal three genes most likely renders this cluster non-functional.
Antigen-43 is a protein which works synergistically with fimbriae to promote adhesion [82]. It is encoded by the flu gene on the prophage CP4-44 [77], which is present in E. coli K-12 and B, but is absent in W; consequently, antigen-43 is also absent in W.
Pili are involved in gene transfer and thus in obtaining pathogenicity factors and other elements. They also affect biofilm formation, which is an important consideration for industrial fermentation. Plasmid pRK1 contains the 14-gene pil cluster which encodes a type IVB thin pilus involved in liquid mating [83]. In contrast to R64 and ColIb-P9, pRK1 does not contain the recombinase gene rci or repeat-flanked shufflon regions that increase the host adhesion variability of the thin pilus [84]. In addition, there are mutations in pilS and pilU, which encode essential functions for pilus activity. The resulting PilS protein has three amino acid mutations at positions where mutations have been shown to limit or inactivate pilus function [85]. PilU has three amino acid mutations at positions which severely affect transfer frequency [86]. Furthermore, the PilS and PilU proteins have an additional 33 and 12 amino acid changes, respectively, at positions which have not been previously characterised. Additionally, E. coli C producing the PilVA-type thin pilus forms cell aggregates in liquid culture due to the pilus activity [87], whereas E. coli W does not (data not shown). All of these considerations suggest that E. coli W does not form thin pili.
Plasmid pRK1 also contains a set of transfer genes, comprising 29 genes over 3 operons, which encode a thick pilus involved in both surface and liquid mating [88]. The pRK1 complement includes all but one of the tra genes: the traABCD operon is incomplete as it is missing traD, a non-essential thick pilus protein of unknown function [89].
Secretion Systems
Secretion systems are required for the transport of proteins across the cell membrane and play a role in virulence [90] and fitness [91]. The conservation of core genes between flagellar systems and Type III secretion systems has led some authors to recognise the flagellar export mechanism as a type of secretion system [92]. Consequently, there are seven secretion systems in _E. coli_[90].
Flagella are required for cellular propulsion. There are two flagella systems in _E. coli_[93]. In addition to the well known Flag-1 flagellar cluster common in E. coli, W has a Flag-2 gene cluster. The Flag-2 locus has been found in many genera of gammaproteobacteria, including _Vibrio parahaemolyticus_[94], _Escherichia coli_[93], _Yersinia enterolitica_[95], _Citrobacter rodentium_[48] and _Aeromonas hydrophila_[96]. The V. parahaemolyticus and A. hydrophilia Flag-2 systems have been shown to be active experimentally [94, 96]. In E. coli, it is found in some strains but not others; it was originally assigned in E. coli 042 by homology [93] but has never been shown experimentally to be functional. In E. coli 042, lfgC (flgC in other genera), which encodes a rod protein required for protein export through the outer membrane, has a frameshift mutation, suggesting that the Flag-2 system is not functional. In support of this, a swarming motility assay was negative [97]. E. coli W and Crooks both contain a Flag-2 locus. The lfgC genes are not mutated, but a two-gene toxin/anti-toxin system found in 042 between lafW and lafZ is absent. Both strains are missing motY, which encodes a motor protein essential for swarming in V. parahaemolyticus; in addition, they do not contain maf-5, a modification accessory factor essential for a functional lateral flagellar in _A. hydrophilia_[96]. W (but not Crooks) contains a Mu prophage located in a non-coding region of the Flag-2 locus (between EcolC_3376 and EcolC_3377). Together, these observations suggest that the Flag-2 locus is not functional in E. coli W or in Crooks. In K-12 and B, all that is left of the Flag-2 system are the two terminal remnants, fhiA (lfhA pseudogene) and mbhA (lafU pseudogene) [93].
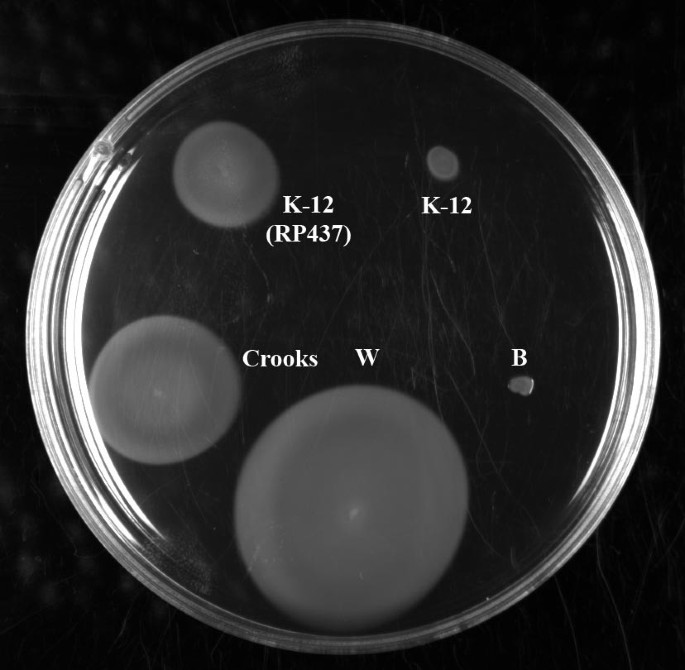
A swarming motility assay was performed to examine functionality of the Flag-2 locus (Figure 4). Consistent with loss of the Flag-2 locus, E. coli B does not swarm. However, despite the loss of what appear to be essential Flag-2 genes, W and Cooks strains both swarm. Although the swarming assay has been used to assess Flag-2 activity [93, 96], it should be stressed that the test is not specific to Flag-2. E. coli K-12, which has clearly lost the Flag-2 locus, shows very limited swarming; however a K-12 mutant (RP437) exhibits a swarming phenotype even though it does not contain a Flag-2 locus [98]. Further analysis by specific deletion will be required to determine whether or not the Flag-2 locus is active in W.
Figure 4
Swarming motility assay. A swarming motility assay was performed using E. coli strains W, Crooks, K-12 (MG1655), K-12 (RP437), and B. B was negative; K-12 (MG1655) showed very minimal swarming, while K-12 (RP437), Crooks and W were positive. Assays were performed in triplicate at 25°C and at 37°C; results were similar at both temperatures (figure shows representative results from 25°C incubation).
There are two Type II secretion systems (T2SSs) in E. coli. T2SSs are required for toxin export from cells [99] as well as a variety of other proteins which affect fitness for specific environments [64]. E. coli K-12, B, and Crooks all carry a repressed 14-gene T2SS gene cluster (gspA-O, located between rpsJ and bfr) [100]. This T2SS has been lost in W due to a gspO-rpsJ deletion. Both W and B (but not K-12 or Crooks) carry the second T2SS gene cluster (yghJ-pppA-yghG-gspC-M). Unlike E. coli B, in which gspL is truncated, all genes in W appear functional. However, it should be noted that unlike K-12, which can export chitinase through an expressed T2SS [100], the W genome does not contain any known genes encoding enzymes or toxins that can be exported through T2SSs.
Type III secretion systems (T3SSs) inject effector proteins into recipient cells leading to pathogenic or pro-survival responses [101]. There are two T3SSs in E. coli: the E. coli Type III secretion systems 1 and 2 (ETT1 and 2) [102]. ETT1 is absent in all four sequenced laboratory strains. Remnants of the ETT2 locus can be found in all of them, but they do not have a functional ETT2. Mutational attrition of ETT2 is common in E. coli strains [103].
Type VI secretion system (T6SS) gene clusters consist of 15 to 25 genes and have been identified in numerous Gram-negative Proteobacteria [104]. In some T6SSs, the genes encoding the secreted proteins, Vgr and Hcp, are found in different locations of the genome [105], but commonly next to rhs genes [106]. This is the case in W, which contains two T6SSs. The structure of the first gene cluster is homologous to the system previously described in E. coli O157:H7 Sakai [107]. It consists of 17 genes and is termed the 'enterohaemorrhagic E. coli type six secretion system cluster' (EHS) [48]. However, this system is found in numerous other non-pathogenic strains, including SE11 and HS (data not shown). A second T6SS is located downstream of metV and is homologous to the T6SS found in E. coli CFT073 [108], also located downstream of metV. We have designated this cluster Escherichia coli type six secretion system cluster 2 (ETSS2) as the EHS is cluster 1. In W, it is most likely deactivated due to an IS621-mediated insertion. W is the only safe strain which contains a T6SS, although none of the effector molecules which are transported into host cells [104] are present. Therefore, this system is unlikely to function in pathogenicity.
Rearrangement hot spot (Rhs) elements
Rhs elements are large highly repetitive regions; they constitute roughly 1% of the E. coli genome [109]. They are composed of four elements: a clade-specific N-terminal domain, a core domain, a hyperconserved domain, and a variable C-terminal domain [106]. Often, partial core domain and variable C-termini regions (called C-terminal tips) are observed downstream of intact rhs genes. These are proposed to play a role in intra-rhs variability [106]. C-terminal tips have occasionally been annotated as insertion sequences in the ISFinder database due to the presence of an H-repeat (H-rpt), although transposition activity has not been observed [110]. E. coli W contains seven rhs genes (rhs1-rhs7; Table 6), two of which are deactivated due to frame-shift mutations. Of the remaining five, four have downstream C-terminal tips of varying number. Both Crooks and W also possess type IV Rhs elements; these are missing in K-12 and B.
Table 6 Rearrangement hot spot (Rhs) elements found in safe strains.
Comparison with other group B1 strains
We performed a comparison between W and other sequenced group B1 strains, including the commensal strains SE11 and IAI1, and a variety of pathogenic strains: EAEC strain 55989, ETEC strain E24377A, and EHEC strains O26, O103, and O111 (Table 7). The chromosome size is relatively variable, ranging from 4.7 Mbp (IAI1) to 5.7 Mbp (O26). A backbone genome can be defined for each strain by subtracting the LMEs (including plasmids and integrative elements) from the total genome size (Table 7). Interestingly, the size of this backbone genome is very similar (ca. 4.5 Mbp +/- 83 Kbp) for all strains. The backbone sequences are not identical; differences are found primarily in the presence or absence of large structural elements encoding secretion systems (including flagella) and adhesins. For example, the Flag-2 is found W and the two EHEC strains O26 and O111 (but not in the EHEC strain O103 or in other pathogenic strains, or in the commensal strains) (Table 8). W has the largest backbone genome (4.588 Mbp) as it has the largest number of large structural elements (T2SS, T3SS, T6SS and flagella). No group B1 strain contained the T2SS gspA-gspO which is present in group A. E. coli. W contains the smallest number of insertion sequences of all B1 strains; these sequences also play a role in attrition, since recombination between them may result in loss of large regions of DNA [111]. Additionally, each of the group B1 strains examined contains the csc regulon for permease-mediated sucrose utilisation.
Table 7 Comparison between sequenced Group B1 strain genome features.
Table 8 Large structural components found in Group B1 strains.
A key observation arising from the Group B1 comparison is that most virulence factors are found in LMEs outside the backbone genome (Additional File 2, Additional File 3, Additional File 4). For example, in the EHEC strains, the LEE is encoded on an LME, while shiga toxins are encoded on lambdoid phages; and in E24377A, the enterotoxin and CS3 fimbriae are encoded on plasmid pE24377A_79; and in 55989, the aggregative adhesion fimbrial operon is also plasmid-borne. While each strain had a number of lambdoid prophages present in its genome, only EHEC strains contained lambdoid prophages which encode the T3SS effectors which enhance virulence in these strains (Additional File 4). The presence of essential virulence factors on LMEs is consistent with previous findings, which have shown that non-pathogenic strains can be made pathogenic by introduction of elements found on LMEs [72, 112]. Fitness factors related to colonisation of ecological niches not related to pathogenicity can also be found encoded on LMEs.
Genome-scale reconstruction and metabolic profiling
GSMs are in silico metabolic models built using the collection of reactions that can be predicted from the annotated genome of an organism together with experimental data. They are used for many applications, including production strain design, examining evolutionary relationships, and linking phenotype and genotype information [113, 114]. GSMs can be used to examine theoretical flux phenotypes, ATP maintenance, and redox balance requirements of cells under various genotypic and environmental conditions. These considerations allow prediction of growth rates and other characteristics such as organic acid production under specific conditions of interest. GSMs allow one to examine the effect of network alterations by performing in silico gene knock-out and gain-of-function experiments prior to labour-intensive and expensive wet-lab experiments. The first step in building a GSM is to reconstruct the metabolic network using the annotated genome (genome-scale reconstruction, GSR).
Numerous metabolic differences were observed between E. coli W and the other safe E. coli strains. In order to capture these differences, a GSR was constructed for E. coli W. Protein-coding genes from W were compared with those annotated in the E. coli K-12 MG1655 model, iAF1260 [115] using AUTOGRAPH [116]. Additional reactions were added or removed based on analyses of growth phenotypes, in silico simulations, and bibliomics (in-depth literature search). The resulting W model, iCA1273, includes 1,273 genes represented by 1,111 metabolites and 2,477 reactions (Additional File 5, Additional File 6). Relative to the K-12 model, iCA1273 is missing 41 genes that were not present in the W genome (Additional File 7). Conversely, iCA1273 contains 61 new genes, including 28 found in K-12 which had not previously been annotated (Additional File 8). Forty-eight genes found in the K-12 model, representing 155 reactions, were not included in iCA1273 as no functional orthologs were present in the W genome. In terms of modelling biomass formation, the most important difference between the two models was found in the production of membrane components. Fourteen genes involved in LPS synthesis in K-12 were not found in W and twelve LPS genes found in W were not found in K-12. Several genes common to both strains but not previously represented in the K-12 model were found. These included seven genes involved in the modification of LPS, specifically the inner core consisting of Kdo2-lipid A; two genes involved in the transport of peptidoglycan from the cytoplasm into the periplasmic space; and twelve genes involved the phenylacetic acid degradation pathway. Seven genes in the K-12 model were located on phage regions, whereas no genes encoding metabolic reactions relevant to the model were found in phage regions in the W genome. The localisation of gene-protein-reaction information was also refined relative to the K-12 model. Carbon and nitrogen source utilization were investigated using Biolog™ phenotype arrays (Additional File 9) in order to characterise the metabolism of the strain and further refine the GSR. All of these refinements allow improved resolution of pathways involved in metabolism in our model. Comparative analyses between K-12 and W were made both at genome and phenome levels [115, 117] (Additional File 10). In addition, comparative studies were done between all four safe strains where appropriate. Key differences are detailed below.
Carbon and nitrogen source utilization
Sugars are ubiquitous throughout the environment and their breakdown supplies a key source of carbon and energy for bacteria. Sucrose is the main carbohydrate transport molecule in plants, and is therefore the most abundant disaccharide encountered in most environments. A key metabolic difference between E. coli W and the other three safe strains is the ability of E. coli W to grow on sucrose. This is due to the presence of the csc regulon, which was originally described in E. coli EC3132 and encodes a regulator (cscR), a sucrose transporter (cscB), an invertase (cscA) and a fructokinase (cscK) [118]. The csc regulon has been inserted between the highly variable argW gene region and the dsdX gene of the D-serine regulon [119, 120]. Due to the insertion in dsdX, a D-serine transporter, E. coli W has lost the ability to utilize D-serine.
Several operons have been identified in E. coli strains for uptake and metabolism of cellobiose, a glucose disaccharide formed by hydrolysis of cellulose. The four safe strains contain only the six gene bgl regulon for cellobiose metabolism. This operon has been reported to be silenced in wild-type E. coli strains [121] and K-12 is unable to grow on cellobiose [122]. In contrast, W displays weak growth on cellobiose, indicating that the bgl genes are not silenced. Uptake of the β-glycosides salicin and arbutin is generally seen in conjunction with cellobiose uptake [122], though E. coli W exhibited growth only on salicin. The absence of the arbutin transporter gene _arbT_[122] is the most likely explanation for lack of growth on arbutin.
The pentose monosaccharide D-ribose is a key component of DNA and RNA; D-allose is a ribose analog. Ribose can be transported into the cell [123] and enter amino acid and pentose phosphate pathways after it is phosphorylated; allose can be converted to fructose-6-phosphate [124] for entry into central carbon metabolism. The D-allose transporter can also transport D-ribose [125]. In contrast to the other safe strains, W is unable to catabolise ribose or allose; this is explained by the absence of the _rbsDACBKR_[123, 124] and _alsBACEK_[125] regulons in W.
Many environmental applications require industrial strains to break down aromatic compounds, which are typically found in soil and water. This capability varies between safe strains. W is able to break down the widest range of aromatic compounds among four strains [17]. Unlike the other strains, K-12 is unable to break down 3- and 4-hydroxyphenylacetic acids as it does not contain the eleven-gene hpa gene cluster [17].
Both K-12 and W can break down phenylacetic acid due to the presence of paa gene cluster. E. coli B has lost this cluster due to an IS_3_-mediated insertion while Crooks has an intact paa gene cluster and can presumably also break down phenylacetic acid. E. coli W was isolated from soil, which may help explain its capability to break down diverse aromatic compounds. In addition, loss of extraneous carbon source genes can be observed in strains maintained for long periods on laboratory carbon sources [127]. Since W was archived shortly after isolation, it is less likely to have undergone this selective pressure.
D-Galactosamine is a constituent of animal glycoprotein hormones while _N_-acetyl-D-galactosamine (NAG) is a core component of peptidoglycan. Both are important nitrogen sources. W shares with B and Crooks the agaV-I gene cluster, which is involved in D-galactosamine and NAG catabolism [128, 129]. This cluster has been partially lost in K-12 due to deletion of agaEF.
In K-12, two separate base pair insertions in ilvG result in valine sensitivity [130]. When K-12 is grown with valine as a nitrogen source, valine accumulation results in positive inhibition of the branched chain amino acid synthesis pathway and a subsequent deficit of isoleucine and leucine. IlvG is intact in W, B and Crooks; consequently, these strains are likely to have high L-valine tolerance.
There are a number of discrepancies between model predictions and phenotype array data (Additional File 10). In some cases, C and N sources which can be used by W and K-12 according to the phenotype array data are not supported by model predictions. This can be explained by insufficient annotation of metabolic pathways for many of these C and N sources. In other cases, the models predict utilization of C and N sources which do not support growth (or support only poor growth) in phenotype arrays; in these cases, it is likely that specific conditions (e.g. anaerobic growth, requirement for cofactors) are not met in the phenotype assay.
Other metabolic considerations
Inorganic ions such as iron and cobalt play important roles in many biological processes, and there are many uptake systems available for different ionic forms. W differs from other safe strains in two ion transport systems. Firstly, it does not contain the seven-gene _tonB_-dependant diferric dicitrate uptake system, fecIRABCDE. In K-12 and B, this gene cluster is located within the phage-like element KpLE2. Secondly, it has a cobalt transport system, cbiQ-O2, located in the region epd-yggC; this transport system is not present in the other three strains.
Conclusions
E. coli W has been used in research laboratories and for industrial applications for almost seventy years. Because of this long history, the strain is considered a 'safe' laboratory strain. The safety of a strain is an important consideration both for laboratory research and for industrial applications. Containment and handling in both environments is less complex for safe strains, and safety requirements can significantly impact on the economics of production. Like other safe strains, W harbors genes which encode pathogenicity determinants. W has more such genes than other safe strains; however, many have been mutationally inactivated or are missing key components required for pathogenicity. These observations confirm the historical attribution of W as a safe strain.
Amongst the four safe laboratory strains, W has several unique features: it belongs to phylogroup B1 rather than A; it has a larger genome size; and the period of time between isolation and strain archiving was relatively short. The two latter features are probably related: strains that are maintained under laboratory conditions for extended time periods are subject to specific selection pressures, and tend to lose genes which are not required for survival under laboratory conditions [127]. In line with this, and consistent with its larger genome size, the W genome encodes more genes than other safe strains. Additionally, it has fewer ISs, which tend to multiply in genomes of organisms maintained under laboratory conditions [131]. Overall, W is more similar to other pathogenic and commensal strains than it is to the other safe laboratory strains. Furthermore, it has the largest backbone sequence of the Group B1 strains, suggesting that it has the most complete complement of ancestral genes. These considerations place W as the preferred laboratory strain for use in genomic comparisons aimed at investigating genes involved in pathogenicity and commensalism.
Like other wild-type isolates [132], W encodes a large number of carbon source utilization genes, and it grows on a much broader range of carbon substrates than K-12 strains (Additional File 9). Of particular interest is the ability of W to utilize sucrose as a carbon source. For industrial production applications, in particular for large-scale production of commodity biochemicals (e.g., biofuels, industrial polymers, and other industrial feedstocks), sucrose from sugarcane is the preferred carbon source [29]. It is abundant, it is cheaper than glucose [133] and it is also 'greener' than glucose; for example, greenhouse gas emissions for ethanol production are reduced by 85% relative to petrochemicals when using sugarcane sucrose as a carbon source, whereas use of glucose from corn reduces emissions by only 30% [133]. The growth of W on sucrose, in combination with its many other desirable industrial traits (fast growth rate, growth to high cell densities, lack of adhesins which result in clumping, lack of antibiotic markers, and relative resistance to environmental stresses) also place E. coli W as a preferred strain for industrial biotechnology applications. Some of these characteristics (e.g. sucrose utilisation and lack of adhesins/antibiotic markers) are easily explained by genome analysis. However, the raw sequence data does not shed any light on why W exhibits the other characteristics. Further experimental analysis using a systems biology approach might shed light on this.
An annotated genome sequence is an important step in characterisation of an organism, and allows construction of genome scale models which can be used to (a) interrogate the metabolic attributes of organisms and (b) facilitate strain development for industrial applications. Our W GSR includes a number of genes which were not annotated in the original K-12 GEM; this includes both genes that are unique to W and genes that were omitted from the K-12 model. Our improved model more accurately reflects the metabolism of an E. coli cell. There is good agreement between genome data, phenome data, and model data; the combination of these allows us to define the metabolic capabilities of E. coli W both in vitro and in silico. The W strain exhibits many industrially desirable traits, including fast growth, stress tolerance, growth to high cell densities, and the ability to utilise sucrose efficiently [22, 24–28]. With the availability of an annotated genome and GSR, the W strain can now be used as a platform organism for developing sucrose-based bioprocesses to replace current unsustainably-produced industrial chemicals.
Methods
Sequencing and assembly
E. coli W (ATCC 9637) was obtained from NCIMB Ltd (Aberdeen, Scotland; Accession Number 8666. The NCIMB stock was supplied by ATCC). Roche/454 pyrosequencing and fosmid end sequencing followed by manual gap-filling were used to construct the E. coli W genome. The shotgun reads in SFF files that were produced from GS 20 (707,210 reads, 81.8 Mb; MWG Biotech, Germany) and GS FLX (236,190 reads, 56.5 Mb; National Instrument Center for Environmental Management, Korea), totalling ca. 27.7× genome coverage, were assembled into 209 contigs by Roche's gsAssembler. CONSED [[134](/article/10.1186/1471-2164-12-9#ref-CR134 "CONSED. [ http://www.phrap.org/
]")\] was used for sequence manipulation that included read/contig editing, primer design, and finish read processing. Specifically, 127 large contigs with accompanying quality scores produced by the gsAssembler were imported into CONSED as single-read contigs. 2,479 paired-end reads of pCC1FOS (EPICENTRE Biotechnologies, United States) off from ABI 3700 (1.98 Mb, ca. 9.9× clone coverage; GenoTech Co., Korea) were then aligned on the contigs and the resulting scaffolds were validated using the mate information derived from the fosmid end reads.The remaining sequence gaps were filled by Sanger sequencing of the gap-spanning PCR products or fosmid clones. Repeat-induced over-collapsed short contigs were resolved by reproducing contigs according to the copy number deduced from the read depth of contigs and by ordering them using 'from/to' information given by the gsAssembler. The most difficult assembly was with two highly similar copies of P2-like prophages (31,005 bp and 32,732 bp); each was reconstructed into the relevant sequences after disentangling the over-collapsed contigs. Ambiguous sequences resulting from the differences of the two prophages were refined by primer walks on fosmid clones containing each prophage segment. The overall error rate of the assembled genome sequence was calculated as 0.09 bp/10 kb, and verification of the assembly came from the consistency of fosmid end reads on the final contig.
The sequence was validated by comparison against independent sequence data generated using a GAII platform. The 65-bp reads were assembled by scaffolding against the original sequence using Burrows-Wheeler Aligner (BWA) [135]. SNPs and INDELS relative to original sequence were identified using SAMtools [136]. Corrections were made based on confidence (related to depth of local sequencing) for each reported discrepancy.
Annotation
ORF prediction was performed using Prodigal [32] and Glimmer [33]. AutoFACT [137], an automatic annotation pipeline, was employed to score predicted ORFs against existing databases, including non-redundant protein sequences (nr) in GenBank [[138](/article/10.1186/1471-2164-12-9#ref-CR138 "GenBank. [ http://www.ncbi.nlm.nih.gov/genbank/
]")\], KEGG \[[139](/article/10.1186/1471-2164-12-9#ref-CR139 "Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al: KEGG for linking genomes to life and the environment. Nucl Acids Res. 2008, 36 (suppl_1): D480-484.")\] and COG \[[140](/article/10.1186/1471-2164-12-9#ref-CR140 "Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al: The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003, 4: 41-10.1186/1471-2105-4-41.")\], using homology search. Where the AutoFACT annotation differed from the K-12 annotation for shared orthologs, the difference was resolved through manual curation. In particular, if AutoFACT proposed a less ambiguous annotation, experimental evidence for the AutoFACT annotation was sought in the literature. tRNA genes were predicted using tRNAscan-SE \[[141](/article/10.1186/1471-2164-12-9#ref-CR141 "Lowe T, Eddy S: tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 1997, 25 (5): 955-964. 10.1093/nar/25.5.955.")\], rRNA genes were predicted using rnammer \[[142](/article/10.1186/1471-2164-12-9#ref-CR142 "Lagesen K, Hallin P, Andreas Rodland E, Staerfeldt H-H, Rognes T, Ussery DW: RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl Acids Res. 2007, 35 (9): 3100-3108. 10.1093/nar/gkm160.")\], and ncRNA genes were predicted using INFERNAL \[[143](/article/10.1186/1471-2164-12-9#ref-CR143 "Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A: Rfam: annotating non-coding RNAs in complete genomes. Nucl Acids Res. 2005, 33 (suppl_1): D121-124.")\]. These predictions were integrated into the annotation using Artemis \[[144](/article/10.1186/1471-2164-12-9#ref-CR144 "Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, Barrell B: Artemis: sequence visualization and annotation. Bioinformatics. 2000, 16 (10): 944-945. 10.1093/bioinformatics/16.10.944.")\]. ORFs which resided within rRNA genes and ncRNAs covering rRNA or tRNA genes were removed. Transcriptional start sites were further curated using Artemis and modified based on matches to homologous genes from _E. coli_ K-12, B and Crooks. CRISPR regions were predicted using a combination of CRT \[[145](/article/10.1186/1471-2164-12-9#ref-CR145 "Bland C, Ramsey T, Sabree F, Lowe M, Brown K, Kyrpides N, Hugenholtz P: CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007, 8 (1): 209-10.1186/1471-2105-8-209.")\] and PILER \[[146](/article/10.1186/1471-2164-12-9#ref-CR146 "Edgar R, Myers E: PILER: identification and classification of genomic repeats. Bioinformatics. 2005, 21 (Suppl 1): i152-158. 10.1093/bioinformatics/bti1003.")\].Comparative Genome Analysis
Comparative genome analysis was based on protein-coding sequences predicted from the E. coli W (ATCC 9637) annotation and three other safe E. coli strains: K-12 MG1655 [GenBank:U00096], B REL606 [GenBank:CP000819], and Crooks ATCC 8739 [GenBank: CP000946]. Comparative analysis of the E. coli W plasmids pRK1 and pRK2 was based on protein-coding sequences and was performed against five representative plasmids: pSE11-1 [GenBank: AP009241], pSE11-3 [GenBank: AP009243], ColIb-P9 [GenBank:AB021078], R64 [GenBank:AP005147, and pSE11-5 [GenBank: AP009245]. All-against-All BLASTP for amino acids was used to assign orthologs; these were further curated using gene context data, analysis of orthologs provided by the E. coli B REL606 genome annotation, and literature data.
Protein-coding genes and pseudogenes were mapped to orthologs in each of the three other sequenced laboratory strains by BLAST to attain the bi-directional best hit (BBH) relationships. Genes with high sequence similarities to a gene in another strain but differing significantly in length were inspected manually to establish the cause of variation.
Insertion Sequences (ISs) for E. coli W, Crooks and SE11 were annotated using BLASTN against the ISFinder database [147, [148](/article/10.1186/1471-2164-12-9#ref-CR148 "ISFinder. [ http://www-is.biotoul.fr/
]")\]. Large mobile elements and rearrangement hot spot (Rhs) elements were identified during the annotation using BLASTP against the nr database in GenBank. Labels for _rhs_ genes were assigned using nomenclature described by Jackson et. al. (2009).Phylogenetic analysis was performed using the gene concatenation method [36]. Concatenated sequences of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, recA) and sequence types (STs) of E. coli reference (ECOR) collection strains and related organisms were downloaded from the E. coli MLST Database [[149](/article/10.1186/1471-2164-12-9#ref-CR149 "E. coli MLST Database. [ http://mlst.ucc.ie/mlst/dbs/Ecoli
]")\]. W gene sequences were aligned using ClustalW \[[150](/article/10.1186/1471-2164-12-9#ref-CR150 "Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al: Clustal W and Clustal X version 2.0. Bioinformatics. 2007, 23 (21): 2947-2948. 10.1093/bioinformatics/btm404.")\] then concatenated. A phylogenetic tree was generated by the neighbour joining method with 1000 bootstrap iterations using MEGA4 \[[151](/article/10.1186/1471-2164-12-9#ref-CR151 "Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007, 24 (8): 1596-1599. 10.1093/molbev/msm092.")\].Motility Assay
Motility assays was performed as described previously [95] with the following alterations: assays were performed at 25°C and 37°C only, and antibiotics were not included in the medium.
GSR Construction
The GSR was created using AUTOGRAPH [116] to generate a database of predicted ORFs against the E. coli K-12 GSR, iAF1260 [115]. Additional reactions were added or removed based on an in-depth literature search, high-throughput carbon/nitrogen/phosphorous/sulphur source growth assays (PM Kit, Biolog, Hayward, CA) and in silico validation using the COBRA toolbox [152] to ensure all biomass components could be synthesized. In silico simulations used the biomass composition of iAF1260 [115].
Gene-protein-reaction associations were curated and assigned a confidence score based on experimental data and information from the E. coli K-12 iAF1260 GEM. Boolean logic was employed to denote the relationships between proteins and whether they formed complexes; isozymes were described as an 'OR' relationship and protein complexes were represented as 'AND' relationships linked to other peptides required for a functional protein. In cases where different combinations of proteins can form a complex which catalyses the same reaction, each complex was represented by an 'AND' relationship and 'OR' relationships were made between complexes. Gaps in the metabolic network, resulting from missing genes which are essential for the synthesis of biomass components and production of waste products, were filled by incorporating reactions from the iAF1260 and KEGG database.
Abbreviations
BBH:
bi-directional best hit
CAS:
CRISPR associated sequence
COG:
clusters of orthologous groups of proteins
CPS:
capsular polysaccharide
CRISPR.:
clustered regularly interspaced short palindromic repeat
ECOR:
Escherichia coli Reference Collection
EHS:
enterohaemorrhagic E. coli type six secretion system cluster
ELF:
E. coli YcbQ laminin-binding fimbria
ETEC:
enterotoxigenic E. coli
ETT1:
E. coli Type III secretion system 1
ETT2:
E. coli Type III secretion system 2
GEM:
genome-scale model
GSR:
genome-scale reconstruction
HGT:
horizontal gene transfer
H-rpt:
H-repeat
IncI1:
Incompatability group I1
IS:
insertion sequence
KEGG:
Kyoto Encyclopaedia of Genes and Genomes
LME:
large mobile element
LPS:
lipopolysaccharide
NAG:
_N_-acetyl-D-galactosamine
ORF:
open reading frame
PGA:
penicillin G acyclase
pLE:
phage-like element
Rhs:
rearrangement hot spot
T2SS:
type II sectrtion system
T3SS:
type III secretion system
T6SS:
type VI secretion system
UPEC:
uropathogenic E. coli
WpLE:
E. coli W phage Like Elements
References
- Bauer A, Dieckmann S, et al: Rapid identification of Escherichia coli safety and laboratory strain lineages based on Multiplex-PCR. FEMS Microbiology Letters . 2007, 269 (1): 36-40. 10.1111/j.1574-6968.2006.00594.x.
Article CAS PubMed Google Scholar - Bauer A, Ludwig W, et al: A novel DNA microarray design for accurate and straightforward identification of Escherichia coli safety and laboratory strains. Systematic and Applied Microbiology. 2008, 31 (1): 50-61. 10.1016/j.syapm.2008.01.001.
Article CAS PubMed Google Scholar - Esselen WB, Fuller JE: The oxidation of ascorbic acid as influenced by intestinal bacteria. J Bacteriol. 1939, 37 (5): 501-521.
CAS PubMed PubMed Central Google Scholar - Gunsalus IC, Hand DB: The use of bacteria in the chemical determination of total vitamin C. J Biol Chem. 1941, 141 (3): 853-858.
CAS Google Scholar - Gunsalus CF, Tonzetich J: Transaminases for pyridoxamine and purines. Nature. 1952, 170 (4317): 162-10.1038/170162a0.
Article CAS PubMed Google Scholar - Jantama K, Haupt MJ, Svoronos SA, Zhang X, Moore JC, Shanmugam KT, Ingram LO: Combining metabolic engineering and metabolic evolution to develop nonrecombinant strains of Escherichia coli C that produce succinate and malate. Biotechnol Bioeng. 2008, 99 (5): 1140-1153. 10.1002/bit.21694.
Article CAS PubMed Google Scholar - Jantama K, Zhang X, Moore JC, Shanmugam KT, Svoronos SA, Ingram LO: Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol Bioeng. 2008, 101 (5): 881-893. 10.1002/bit.22005.
Article CAS PubMed Google Scholar - Alterthum F, Ingram LO: Efficient ethanol production from glucose, lactose, and xylose by recombinant Escherichia coli. Appl Environ Microbiol. 1989, 55 (8): 1943-1948.
CAS PubMed PubMed Central Google Scholar - Zhang X, Jantama K, Moore JC, Jarboe LR, Shanmugam KT, Ingram LO: Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proceedings of the National Academy of Sciences. 2009, 106 (48): 20180-20185. 10.1073/pnas.0905396106.
Article CAS Google Scholar - Zhang X, Jantama K, Shanmugam KT, Ingram LO: Reengineering Escherichia coli for Succinate Production in Mineral Salts Medium. Appl Environ Microbiol. 2009, 75 (24): 7807-7813. 10.1128/AEM.01758-09.
Article CAS PubMed PubMed Central Google Scholar - Blattner FR: The Complete Genome Sequence of Escherichia coli K-12. Science. 1997, 277 (5331): 1453-1462. 10.1126/science.277.5331.1453.
Article CAS PubMed Google Scholar - Jeong B, Barbe V: Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). Journal of Molecular Biology . 2007, 394 (4): 644-652. 10.1021/i560145a007.
Article CAS Google Scholar - Waksman SA, Reilly HC: Agar-streak method for assaying antibiotic substances. Ind Eng Chem. 1945, 17 (9): 556-558. 10.1021/i560145a007.
CAS Google Scholar - Davis BD: The isolation of biochemically deficient mutants of bacteria by means of penicillin. Proc Natl Acad Sci USA. 1949, 35 (1): 1-10. 10.1073/pnas.35.1.1.
Article CAS PubMed PubMed Central Google Scholar - Davis BD: Isolation of biochemically deficient mutants of bacteria by penicillin. J Am Chem Soc. 1948, 70 (12): 4267-4267. 10.1021/ja01192a520.
Article CAS PubMed Google Scholar - Sobotková L, Stepánek V, Plhácková K, Kyslík P: Development of a high-expression system for penicillin G acylase based on the recombinant Escherichia coli strain RE3(pKA18). Enzyme Microb Technol. 1996, 19 (5): 389-397.
Article Google Scholar - Diaz E, Ferrandez A, Prieto MA, Garcia JL: Biodegradation of aromatic compounds by Escherichia coli. Microbiol Mol Biol Rev. 2001, 65 (4): 523-569. 10.1128/MMBR.65.4.523-569.2001.
Article CAS PubMed PubMed Central Google Scholar - Ohta K, Beall DS, Mejia JP, Shanmugam KT, Ingram LO: Genetic improvement of Escherichia coli for ethanol production: chromosomal integration of Zymomonas mobilis genes encoding pyruvate decarboxylase and alcohol dehydrogenase II. Appl Environ Microbiol. 1991, 57 (4): 893-900.
CAS PubMed PubMed Central Google Scholar - Zhang X, Jantama K, Moore J, Shanmugam K, Ingram L: Production of l-alanine by metabolically engineered Escherichia coli. Appl Microbiol Biotechnol. 2007, 77 (2): 355-366. 10.1007/s00253-007-1170-y.
Article CAS PubMed Google Scholar - Zhou S, Iverson AG, Grayburn WS: Engineering a native homoethanol pathway in Escherichia coli B for ethanol production. Biotechnol Lett. 2008, 30 (2): 335-342. 10.1007/s10529-007-9544-x.
Article CAS PubMed Google Scholar - Yomano L, York S, Zhou S, Shanmugam K, Ingram L: Re-engineering Escherichia coli for ethanol production. Biotechnol Lett. 2008, 30 (12): 2097-2103. 10.1007/s10529-008-9821-3.
Article CAS PubMed Google Scholar - Lee SY, Chang HN: High cell density cultivation of Escherichia coli W using sucrose as a carbon source. Biotechnol Lett. 1993, 15 (9): 971-974. 10.1007/BF00131766.
Article CAS Google Scholar - Shukla VB, Zhou S, Yomano LP, Shanmugam KT, Preston JF, Ingram LO: Production of D(-)-lactate from sucrose and molasses. Biotechnol Lett. 2004, 26 (9): 689-693. 10.1023/B:BILE.0000024088.36803.4e.
Article CAS PubMed Google Scholar - Alterthum F, Ingram LO: Efficient ethanol production from glucose, lactose, and xylose by recombinant Escherichia coli. Appl Environ Microbiol. 1989, 55 (8): 1943-1948.
CAS PubMed PubMed Central Google Scholar - Nagata S: Growth of Escherichia coli ATCC 9637 through the uptake of compatible solutes at high osmolarity. J Biosci Bioeng. 2001, 92 (4): 324-329. 10.1263/jbb.92.324.
Article CAS PubMed Google Scholar - Bloom FR, Pfau J, Yim H: Rapidly growing microorganisms for biotechnology applications. patent U. United States. 2004
Google Scholar - Shiloach J, Bauer S: High-yield growth of E. coli at different temperatures in a bench scale fermentor. Biotechnol Bioeng. 1975, 17 (2): 227-239. 10.1002/bit.260170208.
Article CAS Google Scholar - Gleiser IE, Bauer S: Growth of E. coli W to high cell concentration by oxygen level linked control of carbon source concentration. Biotechnol Bioeng. 1981, 23 (5): 1015-1021. 10.1002/bit.260230509.
Article CAS Google Scholar - Renouf MA, Wegener MK, Nielsen LK: An environmental life cycle assessment comparing Australian sugarcane with US corn and UK sugar beet as producers of sugars for fermentation. Biomass Bioenerg. 2008, 32 (12): 1144-1155. 10.1016/j.biombioe.2008.02.012.
Article CAS Google Scholar - Lee SY, Lee D-Y, Kim TY: Systems biotechnology for strain improvement. Trends Biotechnol. 2005, 23 (7): 349-358. 10.1016/j.tibtech.2005.05.003.
Article CAS PubMed Google Scholar - Oshima K, Toh H, Ogura Y, Sasamoto H, Morita H, Park S-H, Ooka T, Iyoda S, Taylor TD, Hayashi T, et al: Complete genome sequence and comparative analysis of the wild-type commensal Escherichia coli strain SE11 isolated from a healthy adult. DNA Res. 2008, 15 (6): 375-386. 10.1093/dnares/dsn026.
Article CAS PubMed PubMed Central Google Scholar - Hyatt D, Chen G-L, LoCascio P, Land M, Larimer F, Hauser L: Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010, 11 (1): 119-10.1186/1471-2105-11-119.
Article PubMed PubMed Central CAS Google Scholar - Delcher AL, Bratke KA, Powers EC, Salzberg SL: Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics. 2007, 23 (6): 673-679. 10.1093/bioinformatics/btm009.
Article CAS PubMed PubMed Central Google Scholar - Gordon DM, Clermont O, Tolley H, Denamur E: Assigning Escherichia coli strains to phylogenetic groups: multi-locus sequence typing versus the PCR triplex method. Environmental Microbiology. 2008, 10 (10): 2484-2496. 10.1111/j.1462-2920.2008.01669.x.
Article CAS PubMed Google Scholar - Dobrindt U, Agerer F, Michaelis K, Janka A, Buchrieser C, Samuelson M, Svanborg C, Gottschalk G, Karch H, Hacker J: Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J Bacteriol. 2003, 185 (6): 1831-1840. 10.1128/JB.185.6.1831-1840.2003.
Article CAS PubMed PubMed Central Google Scholar - Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MCJ, Ochman H, et al: Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol. 2006, 60 (5): 1136-1151. 10.1111/j.1365-2958.2006.05172.x.
Article CAS PubMed PubMed Central Google Scholar - Duriez P, Clermont O, Bonacorsi S, Bingen E, Chaventre A, Elion J, Picard B, Denamur E: Commensal Escherichia coli isolates are phylogenetically distributed among geographically distinct human populations. Microbiology. 2001, 147 (6): 1671-1676.
Article CAS PubMed Google Scholar - Sobotkova L, Grafkova J, Stepanek V, Vacik T, Maresova H, Kyslik P: Indigenous plasmids in a production line of strains for penicillin G acylase derived from Escherichia coli W. Folia Microbiol (Praha). 1999, 44 (3): 263-266. 10.1007/BF02818544.
Article CAS Google Scholar - Couturier M, Bex F, Bergquist PL, Maas WK: Identification and classification of bacterial plasmids. Microbiol Mol Biol Rev. 1988, 52 (3): 375-395.
CAS Google Scholar - Nikoletti S, Bird P, Praszkier J, Pittard J: Analysis of the incompatibility determinants of I-complex plasmids. J Bacteriol. 1988, 170 (3): 1311-1318.
CAS PubMed PubMed Central Google Scholar - Komano T, Funayama N, Kim SR, Nisioka T: Transfer region of IncI1 plasmid R64 and role of shufflon in R64 transfer. J Bacteriol. 1990, 172 (5): 2230-2235.
CAS PubMed PubMed Central Google Scholar - Garcia-Fernandez A, Chiaretto G, Bertini A, Villa L, Fortini D, Ricci A, Carattoli A: Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum β-lactamases in Escherichia coli and Salmonella of human and animal origin. J Antimicrob Chemother. 2008, 61 (6): 1229-1233. 10.1093/jac/dkn131.
Article CAS PubMed Google Scholar - Bird PI, Pittard J: An unexpected incompatibility interaction between two plasmids belonging to the I compatibility complex. Plasmid. 1982, 8 (2): 211-214. 10.1016/0147-619X(82)90059-2.
Article CAS PubMed Google Scholar - Furuya N, Komano T: Nucleotide sequence and characterization of the trbABC region of the IncI1 Plasmid R64: existence of the pnd gene for plasmid maintenance within the transfer region. J Bacteriol. 1996, 178 (6): 1491-1497.
CAS PubMed PubMed Central Google Scholar - Stepánek V, Valesová R, Kyslík P: Cryptic plasmid pRK2 from Escherichia coli W: sequence analysis and segregational stability. Plasmid. 2005, 54 (1): 86-91.
Article PubMed CAS Google Scholar - Klemm P: Fimbrial adhesins of Escherichia coli. Rev Infect Dis. 1985, 7 (3): 321-340.
Article CAS PubMed Google Scholar - Kolisnychenko V, Plunkett G, Herring CD, Fehér T, Pósfai J, Blattner FR, Pósfai G: Engineering a reduced Escherichia coli genome. Genome Res. 2002, 12 (4): 640-647. 10.1101/gr.217202.
Article CAS PubMed PubMed Central Google Scholar - Petty NK, Bulgin R, Crepin VF, Cerdeno-Tarraga AM, Schroeder GN, Quail MA, Lennard N, Corton C, Barron A, Clark L, et al: The Citrobacter rodentium Genome Sequence Reveals Convergent Evolution with Human Pathogenic Escherichia coli. J Bacteriol. 2010, 192 (2): 525-538. 10.1128/JB.01144-09.
Article CAS PubMed Google Scholar - Wei J, Goldberg MB, Burland V, Venkatesan MM, Deng W, Fournier G, Mayhew GF, Plunkett G, Rose DJ, Darling A, et al: Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect Immun. 2003, 71 (5): 2775-2786. 10.1128/IAI.71.5.2775-2786.2003.
Article CAS PubMed PubMed Central Google Scholar - Anjum MF, Marooney C, Fookes M, Baker S, Dougan G, Ivens A, Woodward MJ: Identification of Core and Variable Components of the Salmonella enterica Subspecies I Genome by Microarray. Infect Immun. 2005, 73 (12): 7894-7905. 10.1128/IAI.73.12.7894-7905.2005.
Article CAS PubMed PubMed Central Google Scholar - Lawrence JG, Ochman H: Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci USA. 1998, 95 (16): 9413-9417. 10.1073/pnas.95.16.9413.
Article CAS PubMed PubMed Central Google Scholar - Ochman H, Lawrence JG, Groisman EA: Lateral gene transfer and the nature of bacterial innovation. Nature. 2000, 405 (6784): 299-304. 10.1038/35012500.
Article CAS PubMed Google Scholar - Langille MGI, Brinkman FSL: IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009, 25 (5): 664-665. 10.1093/bioinformatics/btp030.
Article CAS PubMed PubMed Central Google Scholar - Feil EJ: Small change: keeping pace with microevolution. Nat Rev Micro. 2004, 2 (6): 483-495. 10.1038/nrmicro904.
Article CAS Google Scholar - Morgan GJ, Hatfull GF, Casjens S, Hendrix RW: Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J Mol Biol. 2002, 317 (3): 337-359. 10.1006/jmbi.2002.5437.
Article CAS PubMed Google Scholar - Reizer J, Ramseier TM, Reizer A, Charbit A, Saier MH: Novel phosphotransferase genes revealed by bacterial genome sequencing: a gene cluster encoding a putative N-acetylgalactosamine metabolic pathway in Escherichia coli. Microbiology. 1996, 142 (2): 231-250. 10.1099/13500872-142-2-231.
Article CAS PubMed Google Scholar - Schneider D, Lenski RE: Dynamics of insertion sequence elements during experimental evolution of bacteria. Res Microbiol. 2004, 155 (5): 319-327. 10.1016/j.resmic.2003.12.008.
Article CAS PubMed Google Scholar - Nyman K, Nakamura K, Ohtsubo H, Ohtsubo E: Distribution of the insertion sequence IS1 in Gram-negative bacteria. Nature. 1981, 289 (5798): 609-612. 10.1038/289609a0.
Article CAS PubMed Google Scholar - Labrie SJ, Samson JE, Moineau S: Bacteriophage resistance mechanisms. Nat Rev Micro. 2010, 8 (5): 317-327. 10.1038/nrmicro2315.
Article CAS Google Scholar - Sibley MH, Raleigh EA: Cassette-like variation of restriction enzyme genes in Escherichia coli C and relatives. Nucl Acids Res. 2004, 32 (2): 522-534. 10.1093/nar/gkh194.
Article CAS PubMed PubMed Central Google Scholar - Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J: Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science. 2008, 321 (5891): 960-964. 10.1126/science.1159689.
Article CAS PubMed Google Scholar - Diez-Villasenor C, Almendros C, Garcia-Martinez J, Mojica FJM: Diversity of CRISPR loci in Escherichia coli. Microbiology. 2010, 156 (5): 1351-1361. 10.1099/mic.0.036046-0.
Article CAS PubMed Google Scholar - Chakraborty S, Waise TMZ, Hassan F, Kabir Y, Smith MA, Arif M: Assessment of the Evolutionary Origin and Possibility of CRISPR-Cas (CASS) Interference Pathway in Vibrio cholerae O395. In Silico Biol. 2009, 9 (4): 245-254.
PubMed Google Scholar - Cianciotto NP: Type II secretion: a protein secretion system for all seasons. Trends Microbiol. 2005, 13 (12): 581-588. 10.1016/j.tim.2005.09.005.
Article CAS PubMed Google Scholar - Orskov I, Orskov F, Jann B, Jann K: Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol Rev. 1977, 41 (3): 667-710.
CAS PubMed PubMed Central Google Scholar - Stevenson G, Andrianopoulos K, Hobbs M, Reeves P: Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996, 178 (16): 4885-4893.
CAS PubMed PubMed Central Google Scholar - Whitfield C, Roberts IS: Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999, 31 (5): 1307-1319. 10.1046/j.1365-2958.1999.01276.x.
Article CAS PubMed Google Scholar - Reid SD, Selander RK, Whittam TS: Sequence Diversity of Flagellin (fliC) Alleles in Pathogenic Escherichia coli. J Bacteriol. 1999, 181 (1): 153-160.
CAS PubMed PubMed Central Google Scholar - Milkman R, Jaeger E, McBride RD: Molecular Evolution of the Escherichia coli Chromosome VI. Two Regions of High Effective Recombination. Genetics. 2003, 163 (2): 475-483.
CAS PubMed PubMed Central Google Scholar - Brzuszkiewicz E, Brüggemann H, Liesegang H, Emmerth M, Ölschläger T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emödy L, et al: How to become a uropathogen: Comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proceedings of the National Academy of Sciences. 2006, 103 (34): 12879-12884. 10.1073/pnas.0603038103.
Article CAS Google Scholar - Wang L, Rothemund D, Curd H, Reeves PR: Species-Wide Variation in the Escherichia coli Flagellin (H-Antigen) Gene. J Bacteriol. 2003, 185 (9): 2936-2943. 10.1128/JB.185.9.2936-2943.2003.
Article CAS PubMed PubMed Central Google Scholar - Bernier C, Gounon P, Le Bouguenec C: Identification of an aggregative adhesion fimbria (AAF) type III-encoding operon in enteroaggregative Escherichia coli as a sensitive probe for detecting the AAF-encoding operon family. Infect Immun. 2002, 70 (8): 4302-4311. 10.1128/IAI.70.8.4302-4311.2002.
Article CAS PubMed PubMed Central Google Scholar - Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U: Analysis of the Genome Structure of the Nonpathogenic Probiotic Escherichia coli Strain Nissle 1917. J Bacteriol. 2004, 186 (16): 5432-5441. 10.1128/JB.186.16.5432-5441.2004.
Article CAS PubMed PubMed Central Google Scholar - Nuccio S-P, Baumler AJ: Evolution of the Chaperone/Usher Assembly Pathway: Fimbrial Classification Goes Greek. Microbiol Mol Biol Rev. 2007, 71 (4): 551-575. 10.1128/MMBR.00014-07.
Article CAS PubMed PubMed Central Google Scholar - Gaastra W, Svennerholm A-M: Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol. 1996, 4 (11): 444-452. 10.1016/0966-842X(96)10068-8.
Article CAS PubMed Google Scholar - Samadder P, Xicohtencatl-Cortes J, Saldaña Z, Jordan D, Tarr PI, Kaper JB, Girón JA: The Escherichia coli ycbQRST operon encodes fimbriae with laminin-binding and epithelial cell adherence properties in Shiga-toxigenic E. coli O157:H7. Environmental Microbiology. 2009, 11 (7): 1815-1826. 10.1111/j.1462-2920.2009.01906.x.
Article CAS PubMed PubMed Central Google Scholar - Korea C-G, Badouraly R, Prevost M-C, Ghigo J-M, Beloin C: Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environmental Microbiology. 2010, 12 (7): 1957-1977. 10.1111/j.1462-2920.2010.02202.x.
Article CAS PubMed Google Scholar - Torres AG, Lopez-Sanchez GN, Milflores-Flores L, Patel SD, Rojas-Lopez M, Martinez de la Pena CF, Arenas-Hernandez MMP, Martinez-Laguna Y: Ler and H-NS, Regulators Controlling Expression of the Long Polar Fimbriae of Escherichia coli O157:H7. J Bacteriol. 2007, 189 (16): 5916-5928. 10.1128/JB.00245-07.
Article CAS PubMed PubMed Central Google Scholar - Torres AG, Kanack KJ, Tutt CB, Popov V, Kaper JB: Characterization of the second long polar (LP) fimbriae of Escherichia coli O157:H7 and distribution of LP fimbriae in other pathogenic E. coli strains. FEMS Microbiol Lett. 2004, 238 (2): 333-344.
CAS PubMed Google Scholar - Tatsuno I, Mundy R, Frankel G, Chong Y, Phillips AD, Torres AG, Kaper JB: The lpf Gene Cluster for Long Polar Fimbriae Is Not Involved in Adherence of Enteropathogenic Escherichia coli or Virulence of Citrobacter rodentium. Infect Immun. 2006, 74 (1): 265-272. 10.1128/IAI.74.1.265-272.2006.
Article CAS PubMed PubMed Central Google Scholar - Ideses D, Biran D, Gophna U, Levy-Nissenbaum O, Ron EZ: The lpf operon of invasive Escherichia coli. International Journal of Medical Microbiology. 2005, 295 (4): 227-236. 10.1016/j.ijmm.2005.04.009.
Article CAS PubMed Google Scholar - Henderson IR, Nataro JP: Virulence Functions of Autotransporter Proteins. Infect Immun. 2001, 69 (3): 1231-1243. 10.1128/IAI.69.3.1231-1243.2001.
Article CAS PubMed PubMed Central Google Scholar - Kim S, Komano T: The plasmid R64 thin pilus identified as a type IV pilus. J Bacteriol. 1997, 179 (11): 3594-3603.
CAS PubMed PubMed Central Google Scholar - Gyohda A, Komano T: Purification and Characterization of the R64 Shufflon-Specific Recombinase. J Bacteriol. 2000, 182 (10): 2787-2792. 10.1128/JB.182.10.2787-2792.2000.
Article CAS PubMed PubMed Central Google Scholar - Horiuchi T, Komano T: Mutational Analysis of Plasmid R64 Thin Pilus Prepilin: the Entire Prepilin Sequence Is Required for Processing by Type IV Prepilin Peptidase. J Bacteriol. 1998, 180 (17): 4613-4620.
CAS PubMed PubMed Central Google Scholar - Akahane K, Sakai D, Furuya N, Komano T: Analysis of the pilU gene for the prepilin peptidase involved in the biogenesis of type IV pili encoded by plasmid R64. Molecular Genetics and Genomics. 2005, 273 (4): 350-359. 10.1007/s00438-005-1143-8.
Article CAS PubMed Google Scholar - Yoshida T, Furuya N, Ishikura M, Isobe T, Haino-Fukushima K, Ogawa T, Komano T: Purification and Characterization of Thin Pili of IncI1 Plasmids ColIb-P9 and R64: Formation of PilV-Specific Cell Aggregates by Type IV Pili. J Bacteriol. 1998, 180 (11): 2842-2848.
CAS PubMed PubMed Central Google Scholar - Komano T, Yoshida T, Narahara K, Furuya N: The transfer region of IncI1 plasmid R64: similarities between R64 tra and Legionella icm/dot genes. Mol Microbiol. 2000, 35 (6): 1348-1359. 10.1046/j.1365-2958.2000.01769.x.
Article CAS PubMed Google Scholar - Kim SR, Funayama N, Komano T: Nucleotide sequence and characterization of the traABCD region of IncI1 plasmid R64. J Bacteriol. 1993, 175 (16): 5035-5042.
CAS PubMed PubMed Central Google Scholar - Tseng T-T, Tyler B, Setubal J: Protein secretion systems in bacterial-host associations, and their description in the Gene Ontology. BMC Microbiol. 2009, 9 (Suppl 1): S2-10.1186/1471-2180-9-S1-S2.
Article PubMed PubMed Central CAS Google Scholar - Preston GM, Haubold B, Rainey PB: Bacterial genomics and adaptation to life on plants: implications for the evolution of pathogenicity and symbiosis. Curr Opin Microbiol. 1998, 1 (5): 589-597. 10.1016/S1369-5274(98)80094-5.
Article CAS PubMed Google Scholar - Pallen MJ, Gophna U: Bacterial flagella and Type III secretion: case studies in the evolution of complexity. Genome Dyn. 2007, 3: 30-47. full_text.
Article CAS PubMed Google Scholar - Ren C-P, Beatson SA, Parkhill J, Pallen MJ: The Flag-2 Locus, an Ancestral Gene Cluster, Is Potentially Associated with a Novel Flagellar System from Escherichia coli. J Bacteriol. 2005, 187 (4): 1430-1440. 10.1128/JB.187.4.1430-1440.2005.
Article CAS PubMed PubMed Central Google Scholar - Stewart BJ, McCarter LL: Lateral Flagellar Gene System of Vibrio parahaemolyticus. J Bacteriol. 2003, 185 (15): 4508-4518. 10.1128/JB.185.15.4508-4518.2003.
Article CAS PubMed PubMed Central Google Scholar - Bresolin G, Trcek J, Scherer S, Fuchs TM: Presence of a functional flagellar cluster Flag-2 and low-temperature expression of flagellar genes in Yersinia enterocolitica W22703. Microbiology. 2008, 154 (1): 196-206. 10.1099/mic.0.2007/008458-0.
Article CAS PubMed Google Scholar - Canals R, Altarriba M, Vilches S, Horsburgh G, Shaw JG, Tomas JM, Merino S: Analysis of the Lateral Flagellar Gene System of Aeromonas hydrophila AH-3. J Bacteriol. 2006, 188 (3): 852-862. 10.1128/JB.188.3.852-862.2006.
Article CAS PubMed PubMed Central Google Scholar - Ren C-P, Beatson SA, Parkhill J, Pallen MJ: The Flag-2 Locus, an Ancestral Gene Cluster, Is Potentially Associated with a Novel Flagellar System from Escherichia coli. Journal of Bacteriology. 2005, 187 (4): 1430-1440. 10.1128/JB.187.4.1430-1440.2005.
Article CAS PubMed PubMed Central Google Scholar - Niu C, Graves JD, Mokuolu FO, Gilbert SE, Gilbert ES: Enhanced swarming of bacteria on agar plates containing the surfactant Tween 80. J Microbiol Methods. 2005, 62 (1): 129-132. 10.1016/j.mimet.2005.01.013.
Article CAS PubMed Google Scholar - Sandkvist M: Type II Secretion and Pathogenesis. Infect Immun. 2001, 69 (6): 3523-3535. 10.1128/IAI.69.6.3523-3535.2001.
Article CAS PubMed PubMed Central Google Scholar - Francetic O, Belin D, Badaut C, Pugsley AP: Expression of the endogenous type II secretion pathway in Escherichia coli leads to chitinase secretion. EMBO J. 2000, 19 (24): 6697-6703. 10.1093/emboj/19.24.6697.
Article CAS PubMed PubMed Central Google Scholar - Shames SR, Deng W, Guttman JA, De Hoog CL, Li Y, Hardwidge PR, Sham HP, Vallance BA, Foster LJ, Finlay BB: The pathogenic E. coli type III effector EspZ interacts with host CD98 and facilitates host cell prosurvival signalling. Cell Microbiol. 2010, 12 (9): 1322-1339. 10.1111/j.1462-5822.2010.01470.x.
Article CAS PubMed Google Scholar - Perna NT, Plunkett G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, et al: Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001, 409 (6819): 529-533. 10.1038/35054089.
Article CAS PubMed Google Scholar - Ren C-P, Chaudhuri RR, Fivian A, Bailey CM, Antonio M, Barnes WM, Pallen MJ: The ETT2 Gene Cluster, Encoding a Second Type III Secretion System from Escherichia coli, Is Present in the Majority of Strains but Has Undergone Widespread Mutational Attrition. J Bacteriol. 2004, 186 (11): 3547-3560. 10.1128/JB.186.11.3547-3560.2004.
Article CAS PubMed PubMed Central Google Scholar - Pukatzki S, McAuley SB, Miyata ST: The type VI secretion system: translocation of effectors and effector-domains. Curr Opin Microbiol. 2009, 12 (1): 11-17. 10.1016/j.mib.2008.11.010.
Article CAS PubMed Google Scholar - Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ: Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci USA. 2006, 103 (5): 1528-1533. 10.1073/pnas.0510322103.
Article CAS PubMed PubMed Central Google Scholar - Jackson A, Thomas G, Parkhill J, Thomson N: Evolutionary diversification of an ancient gene family (rhs) through C-terminal displacement. BMC Genomics. 2009, 10 (1): 584-10.1186/1471-2164-10-584.
Article PubMed PubMed Central CAS Google Scholar - Shrivastava S, Mande SS: Identification and functional characterization of gene components of Type VI Secretion system in bacterial genomes. PLoS ONE. 2008, 3 (8): e2955-10.1371/journal.pone.0002955.
Article PubMed PubMed Central CAS Google Scholar - Lloyd AL, Rasko DA, Mobley HLT: Defining Genomic Islands and Uropathogen-Specific Genes in Uropathogenic Escherichia coli. J Bacteriol. 2007, 189 (9): 3532-3546. 10.1128/JB.01744-06.
Article CAS PubMed PubMed Central Google Scholar - Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al: The Complete Genome Sequence of Escherichia coli K-12. Science. 1997, 277 (5331): 1453-1462. 10.1126/science.277.5331.1453.
Article CAS PubMed Google Scholar - Zhao S, Sandt CH, Feulner G, Vlazny DA, Gray JA, Hill CW: Rhs elements of Escherichia coli K-12: complex composites of shared and unique components that have different evolutionary histories. J Bacteriol. 1993, 175 (10): 2799-2808.
CAS PubMed PubMed Central Google Scholar - Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi S-H, Couloux A, Lee S-W, Yoon SH, Cattolico L, et al: Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol. 2009, 394 (4): 644-652. 10.1016/j.jmb.2009.09.052.
Article CAS PubMed Google Scholar - McDaniel TK, Kaper JB: A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol Microbiol. 1997, 23 (2): 399-407. 10.1046/j.1365-2958.1997.2311591.x.
Article CAS PubMed Google Scholar - Feist AM, Palsson BO: The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotech. 2008, 26 (6): 659-667. 10.1038/nbt1401.
Article CAS Google Scholar - Oberhardt MA, Palsson BO, Papin JA: Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009, 5: 10.1038/msb.2009.77.
Google Scholar - Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, Karp PD, Broadbelt LJ, Hatzimanikatis V, Palsson BO: A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007, 3: 10.1038/msb4100155.
Google Scholar - Notebaart RA, van Enckevort FH, Francke C, Siezen RJ, Teusink B: Accelerating the reconstruction of genome-scale metabolic networks. BMC Bioinformatics. 2006, 7: 296-10.1186/1471-2105-7-296.
Article PubMed PubMed Central CAS Google Scholar - AbuOun M, Suthers PF, Jones GI, Carter BR, Saunders MP, Maranas CD, Woodward MJ, Anjun MF: Genome scale reconstruction of a Salmonella metabolic model: comparison of similarity and differences with a commensal Escherichia coli strain. J Biol Chem. 2009, M109.005868
Google Scholar - Bockmann J, Heuel H, Lengeler JW: Characterization of a chromosomally encoded, non-PTS metabolic pathway for sucrose utilization in Escherichia coli EC3132. Molecular and General Genetics MGG. 1992, 235 (1): 22-32. 10.1007/BF00286177.
Article CAS PubMed Google Scholar - Moritz RL, Welch RA: The Escherichia coli argW-dsdCXA Genetic Island Is Highly Variable, and E. coli K1 Strains Commonly Possess Two Copies of dsdCXA. J Clin Microbiol. 2006, 44 (11): 4038-4048. 10.1128/JCM.01172-06.
Article CAS PubMed PubMed Central Google Scholar - Alaeddinoglu NG, Charles HP: Transfer of a Gene for Sucrose Utilization into Escherichia coli K-12, and Consequent Failure of Expression of Genes for D-Serine Utilization. J Gen Microbiol. 1979, 110 (1): 47-59.
Article CAS PubMed Google Scholar - Neelakanta G, Sankar TS, Schnetz K: Characterization of a β-Glucoside Operon (bgc) Prevalent in Septicemic and Uropathogenic Escherichia coli Strains. Appl Environ Microbiol. 2009, 75 (8): 2284-2293. 10.1128/AEM.02621-08.
Article CAS PubMed PubMed Central Google Scholar - Hall BG, Betts PW: Cryptic Genes for Cellobiose Utilization in Natural Isolates of Escherichia coli. Genetics. 1987, 115 (3): 431-439.
CAS PubMed PubMed Central Google Scholar - Bell AW, Buckel SD, Groarke JM, Hope JN, Kingsley DH, Hermodson MA: The nucleotide sequences of the rbsD, rbsA, and rbsC genes of Escherichia coli K-12. J Biol Chem. 1986, 261 (17): 7652-7658.
CAS PubMed Google Scholar - Gibbins LN, Simpson FJ: The Incorporation of D-Allose into the Glycolytic Pathway by Aerobacter Aerogenes. Can J Microbiol. 1964, 10: 829-836. 10.1139/m64-108.
Article CAS PubMed Google Scholar - Kim C, Song S, Park C: The D-allose operon of Escherichia coli K-12. J Bacteriol. 1997, 179 (24): 7631-7637.
CAS PubMed PubMed Central Google Scholar - Burland V, Plunkett G, Daniels DL, Blattner FR: DNA Sequence and Analysis of 136 Kilobases of the Escherichia coli Genome: Organizational Symmetry around the Origin of Replication. Genomics. 1993, 16 (3): 551-561. 10.1006/geno.1993.1230.
Article CAS PubMed Google Scholar - Funchain P, Yeung A, Stewart JL, Lin R, Slupska MM, Miller JH: The Consequences of Growth of a Mutator Strain of Escherichia coli as Measured by Loss of Function Among Multiple Gene Targets and Loss of Fitness. Genetics. 2000, 154 (3): 959-970.
CAS PubMed PubMed Central Google Scholar - Brinkkötter A, Klöß H, Alpert C-A, Lengeler JW: Pathways for the utilization of N-acetyl-galactosamine and galactosamine in Escherichia coli. Mol Microbiol. 2000, 37 (1): 125-135.
Article PubMed Google Scholar - Mukherjee A, Mammel MK, LeClerc JE, Cebula TA: Altered Utilization of N-Acetyl-D-Galactosamine by Escherichia coli O157:H7 from the 2006 Spinach Outbreak. J Bacteriol. 2008, 190 (5): 1710-1717. 10.1128/JB.01737-07.
Article CAS PubMed Google Scholar - Park JH, Lee KH, Kim TY, Lee SY: Metabolic engineering of Escherichia coli for the production of L-valine based on transcriptome analysis and in silico gene knockout simulation. Proceedings of the National Academy of Sciences. 2007, 104 (19): 7797-7802. 10.1073/pnas.0702609104.
Article CAS Google Scholar - Naas T, Blot M, Fitch WM, Arber W: Insertion Sequence-Related Genetic Variation in Resting Escherichia coli K-12. Genetics. 1994, 136 (3): 721-730.
CAS PubMed PubMed Central Google Scholar - Chaudhuri RR, Sebaihia M, Hobman JL, Webber MA, Leyton DL, Goldberg MD, Cunningham AF, Scott-Tucker A, Ferguson PR, Thomas CM, et al: Complete Genome Sequence and Comparative Metabolic Profiling of the Prototypical Enteroaggregative Escherichia coli Strain 042. PLoS ONE. 2010, 5 (1): e8801-10.1371/journal.pone.0008801.
Article PubMed PubMed Central CAS Google Scholar - IEA: Biofuels for Transport: An International Perspective. 2004, OECD Publications, Paris: International Energy Agency
Google Scholar - CONSED. [http://www.phrap.org/]
- Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009, 25 (14): 1754-1760. 10.1093/bioinformatics/btp324.
Article CAS PubMed PubMed Central Google Scholar - Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing Subgroup: The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009, 25 (16): 2078-2079. 10.1093/bioinformatics/btp352.
Article PubMed PubMed Central CAS Google Scholar - Koski L, Gray M, Lang BF, Burger G: AutoFACT: An Automatic Functional Annotation and Classification Tool. BMC Bioinformatics. 2005, 6 (1): 151-10.1186/1471-2105-6-151.
Article PubMed PubMed Central CAS Google Scholar - GenBank. [http://www.ncbi.nlm.nih.gov/genbank/]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al: KEGG for linking genomes to life and the environment. Nucl Acids Res. 2008, 36 (suppl_1): D480-484.
CAS PubMed Google Scholar - Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al: The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003, 4: 41-10.1186/1471-2105-4-41.
Article PubMed PubMed Central Google Scholar - Lowe T, Eddy S: tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 1997, 25 (5): 955-964. 10.1093/nar/25.5.955.
Article CAS PubMed PubMed Central Google Scholar - Lagesen K, Hallin P, Andreas Rodland E, Staerfeldt H-H, Rognes T, Ussery DW: RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucl Acids Res. 2007, 35 (9): 3100-3108. 10.1093/nar/gkm160.
Article CAS PubMed PubMed Central Google Scholar - Griffiths-Jones S, Moxon S, Marshall M, Khanna A, Eddy SR, Bateman A: Rfam: annotating non-coding RNAs in complete genomes. Nucl Acids Res. 2005, 33 (suppl_1): D121-124.
CAS PubMed Google Scholar - Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream M-A, Barrell B: Artemis: sequence visualization and annotation. Bioinformatics. 2000, 16 (10): 944-945. 10.1093/bioinformatics/16.10.944.
Article CAS PubMed Google Scholar - Bland C, Ramsey T, Sabree F, Lowe M, Brown K, Kyrpides N, Hugenholtz P: CRISPR Recognition Tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics. 2007, 8 (1): 209-10.1186/1471-2105-8-209.
Article PubMed PubMed Central CAS Google Scholar - Edgar R, Myers E: PILER: identification and classification of genomic repeats. Bioinformatics. 2005, 21 (Suppl 1): i152-158. 10.1093/bioinformatics/bti1003.
Article CAS PubMed Google Scholar - Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M: ISfinder: the reference centre for bacterial insertion sequences. Nucl Acids Res. 2006, 34 (suppl_1): D32-36. 10.1093/nar/gkj014.
Article CAS PubMed Google Scholar - ISFinder. [http://www-is.biotoul.fr/]
- E. coli MLST Database. [http://mlst.ucc.ie/mlst/dbs/Ecoli]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al: Clustal W and Clustal X version 2.0. Bioinformatics. 2007, 23 (21): 2947-2948. 10.1093/bioinformatics/btm404.
Article CAS PubMed Google Scholar - Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007, 24 (8): 1596-1599. 10.1093/molbev/msm092.
Article CAS PubMed Google Scholar - Becker SA, Feist AM, Mo ML, Hannum G, Palsson BO, Herrgard MJ: Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox. Nat Protocols. 2007, 2 (3): 727-738. 10.1038/nprot.2007.99.
Article CAS PubMed Google Scholar - Genome Encyclopedia of Microbes. [http://www.gem.re.kr]
Acknowledgements
We would like to thank Simon Boyes, Haryadi Sugiarto, Sarah Bydder, Jennifer Steen, Alex Waidmann and Rainier Wolfcastle for assistance with curation of the genome annotation, and members of the Genome Encyclopedia of Microbes [[153](/article/10.1186/1471-2164-12-9#ref-CR153 "Genome Encyclopedia of Microbes. [ http://www.gem.re.kr
]")\] at KRIBB for technical assistance. We thank Robin Palfreyman for useful discussions and assistance with bioinformatics analyses, and Eliora Ron for discussions about the history of the W strain. We also thank Guy Plunkett III for useful correspondence regarding _E. coli_ C and Crooks. This research was supported by a Queensland State Government grant under the National and International Research Alliances Program (LKN, CEV), the Cooperative Research Centre for Sugar Industry Innovation through Biotechnology (CTN), Korea-Australia Collaborative Research Project on Sucrose-based Biorefinery Platform Development from the Ministry of Knowledge Economy (J.H.P. and S.Y.L.), the KRIBB Research Initiative Program (J.F.K. and H.J.), and the 21C Frontier Microbial Genomics and Applications Centre Program of the Korean Ministry of Education, Science and Technology (J.F.K.)Author information
Authors and Affiliations
- Australian Institute for Bioengineering and Nanotechnology, Cnr Cooper and College Rds, The University of Queensland, St Lucia, Queensland, 4072, Australia
Colin T Archer, Claudia E Vickers & Lars K Nielsen - Industrial Biotechnology and Bioenergy Research Center, Korea Research Institute of Bioscience and Biotechnology, 111 Gwahangno, Yuseong-gu, Daejeon, Korea
Jihyun F Kim & Haeyoung Jeong - Department of Chemical and Biomolecular Engineering (BK21 program) and Center for Systems and Synthetic Biotechnology, Institute for the BioCentury, KAIST, 335 Gwahangno, Yuseong-gu, Daejeon, 305-701, Republic of Korea
Jin Hwan Park & Sang Yup Lee
Authors
- Colin T Archer
You can also search for this author inPubMed Google Scholar - Jihyun F Kim
You can also search for this author inPubMed Google Scholar - Haeyoung Jeong
You can also search for this author inPubMed Google Scholar - Jin Hwan Park
You can also search for this author inPubMed Google Scholar - Claudia E Vickers
You can also search for this author inPubMed Google Scholar - Sang Yup Lee
You can also search for this author inPubMed Google Scholar - Lars K Nielsen
You can also search for this author inPubMed Google Scholar
Corresponding author
Correspondence toClaudia E Vickers.
Additional information
Authors' contributions
LKN and SYL conceived the idea for the project. LKN and CEV were responsible for project management and supervision. Genome sequencing and automated annotation was performed by JFK and HJ. CTA did the manual curation of the annotation, comparative anlayses, and genome scale reconstruction. CEV, CTA and LKN wrote the manuscript. All authors contributed to revision of the manuscript. All authors have read and approved the final manuscript.
Electronic supplementary material
12864_2010_3148_MOESM1_ESM.XLS
Additional file 1:List of CDSs which occur once in the genome of one safe strain but more than once in genomes of other safe strains. A list of CDSs which have only one copy in one safe strain, but have more than one ortholog in one or more other safe strains. For example, hokE occurs once in the K-12 genome but multiple times in the W genome. The CDS count of each strain does not reconcile unless these one-to-many and many-to-many relationships are considered. Detailed CDS counts are provided within the file. The counts explain the CDS skew which occurs when counting the number of CDSs in Figure 2 for K-12, B, or ATCC 8739. For example, in ATCC 8739 one copy of EcolC_3064 is present, while two are present in W as ECW_m0635 and ECW_m0636. When shared orthologs are counted the number in the ATCC 8739-W region can be one or two, depending on whether the number of orthologs is taken from W or ATCC 8739s context. We have thus detailed all orthologous CDSs which are found in different copy numbers in the other safe strains genomes. (XLS 24 KB)
12864_2010_3148_MOESM2_ESM.DOC
Additional file 2:Description of supplementary files and instructions for use thereof. Detailed description of the contents of each additional file. (DOC 30 KB)
12864_2010_3148_MOESM3_ESM.XLS
Additional file 3:Plasmids found in Group B1 strains. Overview and analysis of the integrative elements which are present in each sequenced group B1 strain. Sheet "Group B1 IEs" presents the attachment sites and significant fitness or virulence factors which are present in each integrative element. Sheet "IE sizes" shows the assumed start and finish sites of each integrative element and the elements size. These sizes were used to calculate each group B1 strains genome backbone size. (XLS 20 KB)
12864_2010_3148_MOESM4_ESM.XLS
Additional file 4:Integrative elements found in Group B1 strains. Analysis of the plasmids which are found in sequenced group B1 strains including plasmid size and fitness/virulence factors which are present on each plasmids genome. (XLS 71 KB)
12864_2010_3148_MOESM5_ESM.XLS
Additional file 5:iCA1273 GSR. A list of the reactions, including GPR associations and constraints (lower bound, upper bound, objective functions) which are present in iCA1273. (XLS 739 KB)
Additional file 6:iCA1273 GSR. iCA1273 in xml format for use with the COBRA Toolbox. (XML 3 MB)
12864_2010_3148_MOESM7_ESM.XLS
Additional file 7:List of unique iAF1260 features compared to iCA1273. A list of reactions which are present in iAF1260 but either do not occur in iCA1273 or do occur but have different gene-protein-reaction associations. Data columns are as follows: 1. Reaction abbreviation 2. Function of the reaction 3. Reaction catalysed 4. The genes necessary for the reaction to be catalysed in Boolan format 5. Notes about the reaction including reference to literature which details experimental evidence for the reaction and the PubMed ID of the paper. (XLS 42 KB)
12864_2010_3148_MOESM8_ESM.XLS
Additional file 8:List of unique iCA1273 reactions and metabolites compared to iAF1260. A list of new reactions and metabolites in iCA1273 which are not found in iAF1260. This file contains the following: 1. "Missing iAF1260 reactions" details reactions which occur in iAF1260 that are not present in W 2. "iCA1273 rxns miss K12 ortho" details reactions from iAF1260 which still occur in iCA1273 but are missing genes which are not present in the W genome. e.g. reaction "RPE" from iAF1260 can be catalyzed by the enzyme encoded by b3386 or b4301. However, in W, an ortholog for b4301 is not present while an ortholog for b3386 is present so the reaction still occurs within the cell. (XLS 246 KB)
12864_2010_3148_MOESM9_ESM.XLS
Additional file 9:Growth phenotype data for E. coli W (ATCC 9637). Results of the Biolog™ growth phenotype assays for E. coli W and E. coli K-12 on a wide range of carbon and nitrogen sources. (XLS 54 KB)
12864_2010_3148_MOESM10_ESM.XLS
Additional file 10:Comparison between predictions and experimental growth data for K-12 GEM and W GSR. A comparison between K-12 GEM (iAF1260) predicted growth phenotypes and Biolog™ data growth, and between W GEM (iCA1273) predicted growth phenotypes and Biolog™ data growth. Overlap between predicted and actual growth phenotypes is higher in W than in K-12. (XLS 36 KB)
Authors’ original submitted files for images
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License (https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Archer, C.T., Kim, J.F., Jeong, H. et al. The genome sequence of E. coli W (ATCC 9637): comparative genome analysis and an improved genome-scale reconstruction of E. coli.BMC Genomics 12, 9 (2011). https://doi.org/10.1186/1471-2164-12-9
- Received: 26 May 2010
- Accepted: 06 January 2011
- Published: 06 January 2011
- DOI: https://doi.org/10.1186/1471-2164-12-9