Identification and Characterization of alcR, a Gene Encoding an AraC-Like Regulator of Alcaligin Siderophore Biosynthesis and Transport in Bordetella pertussis and Bordetella bronchiseptica (original) (raw)
Abstract
A Bordetella bronchiseptica iron transport mutant was isolated following an enrichment procedure based on streptonigrin resistance. The mutant displayed a growth defect on iron-restricted medium containing ferric alcaligin as the sole iron source. In addition to the apparent inability to acquire iron from the siderophore, the mutant failed to produce alcaligin as well as two known iron-regulated proteins, one of which is the AlcC alcaligin biosynthesis protein. A 1.6-kb _Kpn_I-_Pst_I Bordetella pertussis DNA fragment mapping downstream of the alcaligin biosynthesis genes alcABC restored both siderophore biosynthesis and expression of the iron-regulated proteins to the mutant. Nucleotide sequencing of this complementing 1.6-kb region identified an open reading frame predicted to encode a protein with strong similarity to members of the AraC family of transcriptional regulators, for which we propose the gene designation alcR. Primer extension analysis localized an iron-regulated transcription initiation site upstream of the alcR open reading frame and adjacent to sequences homologous to the consensus Fur repressor binding site. The AlcR protein was produced by using an Escherichia coli expression system and visualized in electrophoretic gels. In-frame alcR deletion mutants of B. pertussis and B. bronchiseptica were constructed, and the defined mutants exhibited the alcR mutant phenotype, characterized by the inability to produce and transport alcaligin and express the two iron-repressed proteins. The cloned alcR gene provided in trans restored these siderophore system activities to the mutants. Together, these results indicate that AlcR is involved in the regulation of Bordetella alcaligin biosynthesis and transport genes and is required for their full expression.
The ability of microorganisms to multiply depends on their ability to acquire essential nutrients, among which iron is almost universally limiting in availability. The level of freely available iron in the environment and the extracellular fluids of mammalian hosts is many orders of magnitude below the iron concentration of 4 × 10−7 to 4 × 10−6 M required for the growth of most microorganisms (13, 56). Microbial strategies for overcoming iron restriction can be grouped into two classes: those mediated by siderophores (36, 42) and siderophore-independent processes involving direct iron removal from host molecules such as transferrin, lactoferrin, and heme-containing compounds (38).
Upon iron starvation, bacteria may produce siderophores and the cognate ferric siderophore transport machinery, as well as express transport systems for heterologous microbial siderophores and for siderophore-independent iron retrieval from host compounds (36, 42). Strict regulation of iron assimilation is necessary due to the formation of damaging oxygen radical species catalyzed by excess intracellular iron. For most bacteria studied to date, regulation of iron transport systems is mediated primarily by the ferric uptake regulator protein, Fur, which acts as a corepressor with ferrous iron under conditions of iron abundance (4, 27). However, in addition to the negative transcriptional regulator Fur, other transcriptional regulators which positively regulate the expression of siderophore biosynthesis and transport genes and genes involved in the uptake of other iron compounds have been identified. The genes encoding these positive regulators are themselves members of the Fur regulon. The positively regulated iron transport systems respond to the presence of the cognate siderophore or iron compound and represent three mechanistic classes: (i) alternative sigma factors, such as the FecI regulator of Escherichia coli ferric citrate transport genes (35); (ii) classical two-component sensory transduction systems, as described for the Pseudomonas aeruginosa PfeR-PfeS regulators of the ferric enterobactin receptor PfeA (16); and (iii) AraC-like transcriptional regulators, including the P. aeruginosa PchR pyochelin biosynthesis and transport regulator protein (29, 30) and the Yersinia pestis YbtA yersiniabactin receptor gene regulator (21).
Both Bordetella pertussis and Bordetella bronchiseptica, the causative agents of respiratory diseases in mammals (6, 44), produce the macrocyclic dihydroxamate siderophore alcaligin under iron-depleted growth conditions (12, 40). Alcaligin biosynthesis requires the action of an ornithine decarboxylase encoded by the odc gene in a reaction yielding putrescine from ornithine (10), as well as the alcABC gene products, which have strong primary amino acid sequence similarity to the E. coli IucD, IucB, and IucC aerobactin biosynthesis enzymes, respectively (25, 33). On the basis of the similarities with the Iuc enzymes, we have proposed that AlcA is an oxygenase catalyzing the hydroxylation of putrescine, and AlcB is postulated to function in an acylation step involving succinate. AlcC is similar to the IucC aerobactin synthetase and may be involved in one of the final steps in alcaligin biosynthesis. The Bordetella alcABC genes are cotranscribed and comprise part of an iron-regulated operon (32) with predicted Fur repressor binding sites upstream of alcA (11, 25, 33). B. bronchiseptica fur mutants selected on the basis of manganese resistance were deregulated for the production of alcaligin; the B. pertussis fur gene restored iron repressibility of siderophore expression to the mutants, confirming a role for the Fur repressor in regulation of alcaligin biosynthesis (9).
In this paper, we report the identification of the Bordetella alcR gene, encoding a protein predicted to be a member of the AraC family of transcriptional regulators. Mutations in alcR resulted in defects in both alcaligin biosynthesis and transport, a pleiotropic phenotype which was restored to wild type by genetic complementation using the alcR gene alone. The accompanying article by Pradel and colleagues (44a) describes an independent study in which the Bordetella alcR gene was identified by a different experimental approach.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
E. coli DH5α (Bethesda Research Laboratories, Gaithersburg, Md.), DH10B (Bethesda Research Laboratories), and HB101 (7) were used as hosts for general DNA cloning procedures and as donor strains in triparental matings. E. coli BL21(DE3) was used as the host for bacteriophage T7 RNA polymerase-promoter protein expression studies (46, 53). Wild-type B. bronchiseptica B013N and alcaligin siderophore biosynthesis mutant derivative BRM3 (odc) have been described previously (3, 10). The spontaneous streptomycin-resistant derivative of wild-type B. pertussis UT25 (22), UT25Sm1, has been described elsewhere (33); isogenic B. pertussis mutant PM-4, in which the 105-bp alcA promoter-operator chromosomal DNA region is deleted (32), was used in primer extension experiments.
B. bronchiseptica and E. coli strains were cultured on Luria-Bertani (LB) agar or in LB broth (39), and B. pertussis was cultured on Bordet-Gengou agar (6). Bacteriophage T7 RNA polymerase-promoter protein expression experiments with E. coli used M9 medium (39) supplemented with 0.2% glucose and 0.01% l-amino acids (minus cysteine and methionine) or 0.01% Casamino Acids (Difco, Detroit, Mich.). The defined culture medium for Bordetella strains was Stainer-Scholte (SS) medium (51) modified as described previously (49); iron-replete SS medium contained 36 μM FeSO4, and iron-depleted culture conditions were achieved by deionization of the medium with Chelex100 (Bio-Rad, Hercules, Calif.) as described elsewhere (3). Growth was monitored as optical density with a spectrophotometer or a Klett-Summerson colorimeter equipped with a no. 54 filter (Klett Manufacturing Co., Long Island City, N.Y.). The following antibiotics were used at the indicated concentrations (in micrograms per milliliter): kanamycin, 50; gentamicin, 10; tetracycline, 15; ampicillin, 100; and streptomycin, 50.
Plasmids and genetic methods.
Cosmid pCP1.11 was isolated from a B. pertussis UT25 chromosomal DNA library and has been described previously (33). Plasmid pBRM6 carries an approximately 20-kb chromosomal alc DNA insert fragment encompassing the mini-Tn_5 lacZ1_ marker from B. bronchiseptica alcC mutant BRM6 (3). Plasmid pRK2013 (23) supplied transfer functions in matings. Plasmid cloning vectors pGEM3Z (Promega, Madison, Wis.), pRK415 (34), pBluescript SK+ and pBluescript KS+ (Stratagene, La Jolla, Calif.), and pET-3 (46) were used in the construction of recombinant plasmids. Suicide plasmid vectors pSS1129 (52) and pEG7 and pEG18.3 (both kindly provided by Peggy Cotter and Jeffrey F. Miller) were used in allelic-exchange procedures in the construction of Bordetella mutants. Conjugal transfer of plasmids to Bordetella strains was accomplished by methods described previously (10).
General genetic techniques were performed essentially as described previously (47). DNA probes used in nucleic acid hybridizations were radiolabelled by the random priming method (20) using the Random Primers DNA Labeling System (Gibco BRL) and [α-32P]dCTP (ICN Radiochemicals, Irvine, Calif.). The nucleotide sequence of alcR was determined on both strands by the dideoxy chain termination method (48) using a Sequenase version 2.0 kit (United States Biochemical Corp., Cleveland, Ohio) and double-stranded plasmid DNA templates as described previously (19). Nucleotide sequencing was accomplished by using a set of nested deletion derivatives of alc DNA generated with the Erase-a-base system (Promega) and synthetic DNA oligonucleotide primers. Nucleotide sequence data management and analysis employed the EditSeq and Seqman modules of a demonstration version of the Lasergene sequence analysis software system for the Macintosh PowerPC computer (DNASTAR, Inc., Madison, Wis.). Database searches and data retrievals employed the BLAST (2) server provided by the National Center for Biotechnology Information at the National Library of Medicine. For protein database BLASTP searches, Bordetella DNA sequences were translated in all possible reading frames and the amino acid sequences were submitted to the National Center for Biotechnology Information for analysis using the nonredundant database mandatory DATALIB search parameter. Multiple amino acid sequence alignments were performed by the Clustal method (31) with the MegAlign module of the Lasergene sequence analysis software system (DNASTAR, Inc.). Putative Bordetella Fur-binding sequences were identified by using the MegAlign software to locate DNA regions of at least 50% identity over a 30-nucleotide search window with the dyad sequence 5′-GATAATGATAATCATTATC-3′, representing the proposed consensus E. coli Fur binding site (14, 18).
Primer extension was performed as described previously (47), with 50 μg of total RNA and 100 fmol of 32P-end-labelled oligonucleotide primer used per reaction mixture. RNA was purified from B. pertussis strains grown in iron-replete and iron-depleted SS media as described previously (33). RNA was denatured at 85°C for 10 min and hybridized with radiolabelled antisense primer overnight at 42°C. Extension reactions used 400 U of Moloney murine leukemia virus reverse transcriptase (Bethesda Research Laboratories) at 37°C. The 35-mer DNA oligonucleotide primer used to map the transcription initiation site immediately upstream of alcR (5′-GGTGGGGGGAGCGTCGGTTGTGTCATTGGCGTTGC-3′) was antisense to nucleotides 148 to 182 of the sequence of the 1.6-kb _Kpn_I-_Pst_I DNA fragment (see Fig. 3). The DNA primer was also used to generate the accompanying DNA sequencing ladder from an alc plasmid template.
FIG. 3.
Nucleotide sequence of alcR. The nucleotide sequence (upper strand) of the 1,200-bp region of the B. pertussis 1.6-kb _Kpn_I-_Pst_I DNA fragment and the predicted amino acid sequence of AlcR in the one-letter code are shown. The transcription initiation sites (+1) determined by primer extension analysis using the designated antisense primer (arrow) are indicated. Sequences similar to the consensus Fur binding site (Fur Box) and the positions of the _Ngo_AIV restriction endonuclease sites used to construct deletion mutants PM10 and BRM11 are shown.
Mutational analysis. (i) Isolation of B. bronchiseptica iron transport mutant BRM10.
A pool of B. bronchiseptica B013N cells randomly mutagenized with mini-Tn_5 lacZ1_ (17) was cultured in iron-replete SS medium containing 100 μg of the nonutilizable iron chelator ethylenediaminedi-[(_o_-hydroxyphenyl)acetic acid] (EDDA) per ml at 37°C for 4 h to effect iron starvation status by restriction of iron availability. The cultures were then provided with purified alcaligin siderophore to a 20 μM concentration and streptonigrin (Sigma Chemical Co., St. Louis, Mo.) at concentrations ranging from 0 to 6 μg/ml in replicate cultures. The cells were cultured for an additional 3 h and then spread onto LB agar containing 50 μg of kanamycin per ml for growth of the survivor population enriched for iron transport mutants. The resulting kanamycin-resistant colonies were replica plated onto LB agar (iron replete), LB agar containing 30 μg of EDDA per ml (LB-EDDA agar) (iron restricted), and LB-EDDA agar containing 20 μM alcaligin. One mutant, BRM10, which displayed a growth defect on the two iron-restricted media compared with growth on iron-replete LB agar, a potential iron transport mutant, was selected for further analysis.
(ii) Construction of defined Bordetella alcR deletion mutants.
The 264-bp _Ngo_AIV fragment internal to the B. pertussis alcR gene was deleted by restriction endonuclease cleavage followed by religation. The _Kpn_I-_Pst_I DNA subfragment encompassing the mutated alcR gene was subcloned to the allelic-exchange plasmid vector pSS1129, the resulting plasmid was mated to B. pertussis UT25Sm1, and the mutation was transferred to the chromosome by homologous recombination as described previously (52). Allelic exchange in B. pertussis mutant PM10 was verified by Southern hybridization analysis.
For construction of a B. bronchiseptica alcR mutant, the 264-bp _Ngo_AIV fragment internal to the B. bronchiseptica alcR gene was deleted from the subcloned 2.3-kb _B. bronchiseptica Eco_RI-_Pst_I fragment of pBRM6 (3). From this plasmid, a 2.0-kb _Eco_RI-_Hin_dIII fragment was subcloned to the allelic-exchange plasmid vector pEG7, and the deletion mutation was introduced to the chromosome of B. bronchiseptica B013N by allelic exchange employing positive selection on sucrose-containing medium as described previously (1). Presumptive mutants were initially identified by in situ DNA hybridization analysis using the 264-bp _Ngo_AIV DNA fragment as the probe. Southern hybridization analysis confirmed that the deletion had been transferred to the chromosome, resulting in B. bronchiseptica mutant BRM11.
Measurement of siderophore activity.
The chrome azurol S (CAS) universal siderophore detection assay (50) was used to monitor siderophore production by Bordetella cells grown in SS medium by measurement of the decrease in absorbance of the CAS dye reaction at 630-nm wavelength relative to that of uninoculated culture medium. All siderophore assays were performed in triplicate. CAS agar (50), modified as described previously (3), was also used for qualitative assessment of B. bronchiseptica siderophore activity.
Alcaligin bioassays.
Utilization of alcaligin for iron acquisition was determined for B. bronchiseptica mutants in quantitative bioassays performed as described previously (12), with the transport-proficient alcaligin biosynthesis mutant B. bronchiseptica BRM3 used as a control indicator strain. Briefly, cells were cultured on LB agar for 24 to 30 h, harvested, and suspended in LB to an _A_600 of 1.0, and a 100-μl volume of the suspension was added to 25 ml of molten iron-restricted LB-EDDA agar (50°C) prior to being poured into a petri dish. Wells were punched into the solidified agar, and 100-μl volumes of purified alcaligin diluted serially in distilled water were added. The plates were incubated at 37°C, and the diameters of bacterial growth zones surrounding the wells were measured after 21 h.
Cell fractionation and SDS-PAGE.
Bordetella cells harvested from iron-replete and iron-depleted SS medium cultures were disrupted with a French pressure cell (American Instrument Company, Silver Spring, Md.), and the soluble and insoluble cell fractions were prepared as described previously (33). Protein samples were treated in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) solubilization buffer at 100°C for 7 min, and the proteins were resolved by SDS-PAGE on 10 or 12% polyacrylamide gels containing 0.5 M urea as described previously (49). For analysis of total cellular proteins, Bordetella cells were harvested from SS medium cultures by centrifugation and resuspended directly in SDS-PAGE solubilization buffer. After electrophoresis, proteins were visualized by Coomassie blue staining.
Expression of plasmid-encoded proteins.
Plasmid-encoded proteins were conditionally expressed in E. coli BL21(DE3) by using a bacteriophage T7 polymerase-promoter system (46, 53). Induction was achieved by addition of the lac inducer isopropyl-β-d-thiogalactopyranoside (IPTG) to bacterial cultures. Cells were harvested by centrifugation and analyzed by SDS-PAGE on 12% polyacrylamide gels. Proteins were visualized by staining with Coomassie blue or intrinsic radiolabelling using Tran35S-label (ICN Biochemicals, Inc.) and autoradiography subsequent to SDS-PAGE.
Nucleotide sequence accession number.
The GenBank accession number assigned to the B. pertussis UT25 alcR gene is AF018255.
RESULTS
Characterization of B. bronchiseptica mutant BRM10.
A random mutant pool of B. bronchiseptica cells carrying mini-Tn_5 lacZ1_ chromosomal insertions was exposed to streptonigrin to enrich the population for cells defective in alcaligin-mediated iron uptake. Mutant BRM10 displayed poor growth on low-iron media, despite provision of exogenous alcaligin as a normally utilizable source of iron, suggesting that the mutant was defective in alcaligin transport or utilization. Interestingly, further characterization revealed that BRM10 was also defective in alcaligin production.
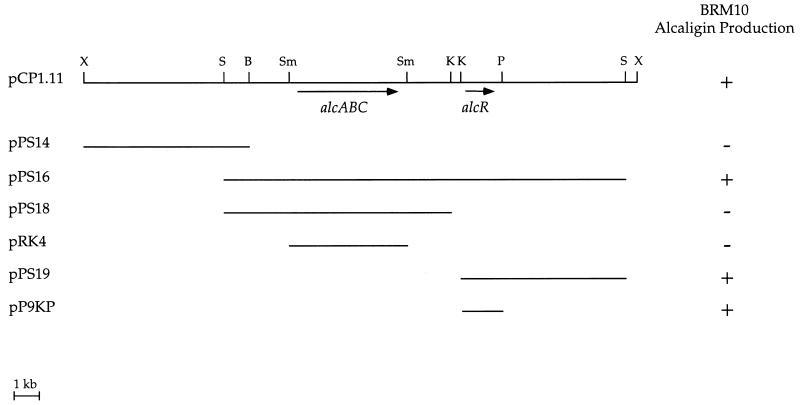
B. pertussis recombinant cosmid pCP1.11, which carries a ca. 21-kb genetic region including the known alcABC alcaligin biosynthesis operon, functionally restored siderophore biosynthesis to BRM10 (Fig. 1). Plasmid pRK4, a subclone of pCP1.11 which carries the alcaligin biosynthesis genes alcABC (33), did not complement BRM10, indicating that the defective BRM10 gene was not one of the known alcaligin biosynthesis genes. Further deletion analysis of pCP1.11 localized the smallest complementing genetic region to a 1.6-kb _Kpn_I-_Pst_I DNA fragment approximately 2 kb downstream of alcABC (plasmid pP9KP).
FIG. 1.
Identification of the B. pertussis DNA region restoring alcaligin production to B. bronchiseptica mutant BRM10. Phenotypic complementation by B. pertussis recombinant cosmid pCP1.11 and plasmid subclones was evaluated on CAS siderophore indicator agar. BRM10 carrying the designated plasmids was scored as follows: +, siderophore activity was produced; −, no siderophore activity was detected. The alcABC alcaligin biosynthesis genes and the newly identified alcR gene are indicated. Abbreviations: B, _Bam_HI; K, _Kpn_I; P, _Pst_I; S, _Sal_I; Sm, _Sma_I; X, _Xho_I.
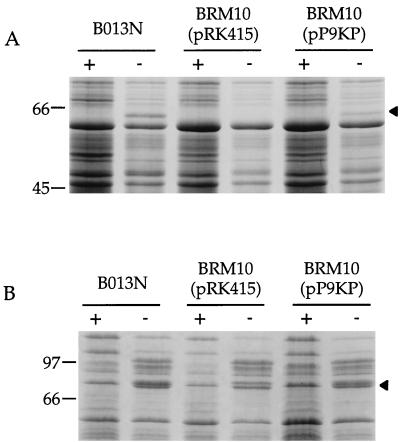
BRM10 total cellular proteins and fractions from cells cultured in high- and low-iron media were analyzed by SDS-PAGE. Wild-type iron-starved B. bronchiseptica B013N cells expressed the soluble AlcC protein with an apparent molecular mass of 59 kDa (33), but mutant BRM10 carrying the plasmid vector control pRK415 failed to produce detectable levels of the protein (Fig. 2A). When pP9KP was provided to BRM10, production of AlcC was restored. Plasmid pRK4 carrying alcABC failed to restore expression of AlcC to BRM10, although previous studies have shown that it restored both siderophore biosynthesis and AlcC production to an alcC mutant (33). An iron-repressed membrane protein with an approximate molecular mass of 79 kDa was observed in the insoluble fraction prepared from the wild-type parent strain B013N, but this protein was absent in mutant BRM10 samples (Fig. 2B). Expression of this 79-kDa protein was also restored when BRM10 was supplied with pP9KP, containing the B. pertussis 1.6-kb _Kpn_I-_Pst_I DNA fragment.
FIG. 2.
SDS-PAGE analysis of wild-type B. bronchiseptica B013N and mutant derivative BRM10. Strain B013N, streptonigrin-resistant mutant BRM10 harboring the plasmid vector pRK415, and the complementing 1.6-kb _B. pertussis Kpn_I-_Pst_I DNA fragment as pP9KP were grown in parallel under iron-replete (+) and iron-depleted (−) conditions, and cell fractions were prepared as described in Materials and Methods. (A) Soluble cell fractions showing the iron-repressed ca. 59-kDa AlcC protein (arrowhead). (B) Total membrane fractions; the 79-kDa iron-repressed protein, which is absent in mutant BRM10(pRK415), migrates as the middle species of a protein triplet (arrowhead). The migration positions of molecular mass protein standards are shown on the left in kilodaltons.
To map the location of the mini-Tn_5 lacZ1_ transposon mutation in BRM10, relative to the B. pertussis 1.6-kb _Kpn_I-_Pst_I DNA fragment which complemented the mutant phenotype, the BRM10 transposon marker and flanking chromosomal DNA sequences were cloned. Restriction endonuclease mapping of the cloned BRM10 chromosomal DNA fragment did not reveal any similarity to the complementing B. pertussis pCP1.11 cosmid or pP9KP DNA. Southern hybridizations demonstrated no genetic homology between the BRM10 chromosomal DNA region flanking the transposon and B. pertussis pCP1.11 or pP9KP (data not shown). These results indicated that the transposon was not located within the chromosomal region of the mutant represented by either pCP1.11 or pP9KP, although each plasmid was able to complement the BRM10 mutant phenotype. Nucleotide sequencing from the transposon ends of the cloned BRM10 DNA did not reveal any sequence similarity with the 1.6-kb _Kpn_I-Pst_I DNA fragment from pCP1.11 or any known alc gene (data not shown). These data suggested that the observed BRM10 siderophore-defective phenotype may not be related to the mini-Tn_5 lacZ1 transposon insertion and that the BRM10 phenotype may be attributable to a spontaneous mutation selected during streptonigrin enrichment affecting the activity encoded within the 1.6-kb _Kpn_I-_Pst_I genetic region. To address this hypothesis, the homologous 1.6-kb _Kpn_I-_Pst_I DNA fragment was cloned from the BRM10 chromosome, and its identity was verified by nucleotide sequencing analysis. In contrast to the corresponding wild-type region, when provided in trans and in multicopy to BRM10, this BRM10-derived DNA fragment failed to complement the alcaligin-defective phenotype of the mutant (data not shown), providing suggestive evidence for a spontaneous mutation in the putative BRM10 alc gene, supporting the hypothesis that the transposon mutation is irrelevant to the iron transport phenotype of BRM10.
Nucleotide sequence analysis of the 1.6-kb _Kpn_I-_Pst_I B. pertussis DNA fragment.
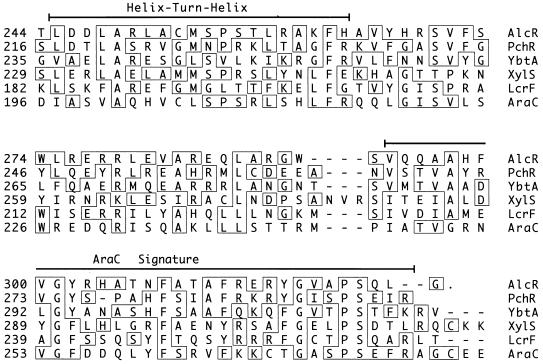
Nucleotide sequence analysis of the 1.6-kb B. pertussis wild-type DNA fragment which complemented the BRM10 mutant phenotype identified an open reading frame, alcR, predicted to encode a protein with a molecular mass of 36.4 kDa (Fig. 3). Two overlapping potential Fur-binding DNA sequences which precede alcR, sharing 13 and 12 of the 19 bases of the consensus E. coli Fur-binding sequence (14, 18), were identified. BLAST database searches showed that the deduced AlcR protein exhibited remarkable similarity to members of the AraC family of transcriptional regulators (24) (Fig. 4). Similarity among the AraC family members, including AlcR, is most pronounced at the carboxy terminus (24). Most significantly, over the carboxy-terminal 82-amino-acid sequence, the deduced AlcR protein was 41% identical to the carboxy-terminal region of PchR, the P. aeruginosa AraC-like protein involved in regulation of pyochelin biosynthesis and transport functions (29, 30). AlcR also exhibited 32% identity over the same sequence interval with YbtA, an AraC-like transcriptional regulator of pesticin/yersiniabactin siderophore receptor expression in Y. pestis (21).
FIG. 4.
Primary amino acid sequence alignments of translated B. pertussis DNA alcR sequences with selected high-scoring AraC family members identified in BLAST database searches. The partial sequences shown represent the highest-scoring carboxy-terminal segments of similarity. The helix-turn-helix DNA binding and AraC signature motifs (lines), amino acid positions in the protein sequences as reported in GenBank (numbers), and residues which match the consensus (boxes) are indicated. The GenBank accession numbers are as follows: B. pertussis AlcR, AF018255; P. aeruginosa PchR, L11657; Y. pestis YbtA, U50452; P. putida XylS, M10143, M15819, and M20635; Y. pestis LcrF, M86690; and E. coli K-12 AraC, V00259.
The 452-bp region downstream of alcR on the 1.6-kb _Kpn_I-_Pst_I fragment contains a partial open reading frame predicted to encode the amino terminus of a protein with high similarity to members of the multidrug efflux system family (data not shown) (43). The best similarity scores were with Pur8 (puromycin resistance) of Streptomyces alboniger (now Streptomyces anulatus) (GenBank accession no. X76855) (54) and E. coli proteins Bcr (bicyclomycin resistance) (GenBank accession no. X63703) (5) and EmrD (resistance to phenylmercury acetate and carbonyl cyanide _m_-chlorophenylhydrazone) (Swiss-Prot accession no. P31442) (41).
Determination of the transcription initiation site of alcR.
Other studies in this laboratory used a B. pertussis chromosomal mutant, PM-4, in which the alcA promoter-operator region was deleted, to demonstrate cotranscription of alcABC and to determine the 3′ limit of the alc operon (32). Quantitative RNA hybridization analyses using an _alcR_-specific probe and RNA isolated from the wild type versus alcA promoter-operator mutant PM-4 showed that alcR was transcribed primarily from the upstream iron-regulated alcA promoter but was also independently transcribed from a weaker secondary iron-regulated promoter which retained its activity when the alcA promoter-operator region was deleted. This secondary promoter was analyzed in the present study by primer extension experiments, and the transcriptional initiation site was determined.
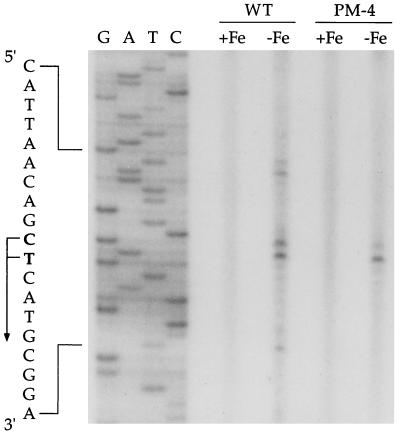
Reverse transcription of mRNA from B. pertussis wild-type strain UT25Sm1 and isogenic alcA promoter-operator deletion mutant PM-4 using a synthetic primer corresponding to the antisense of nucleotide positions 148 to 182 of the B. pertussis DNA sequence (Fig. 3) revealed two extension products corresponding to transcription initiation sites at the C residue and the adjacent T residue at positions 124 and 125, respectively (Fig. 5). These iron-regulated transcripts from the wild-type and mutant strains were mapped to the same initiation positions and were present in similar abundances, indicating that an iron-regulated promoter, P_alcR_, resides immediately upstream of alcR and functions independently of the alcA promoter. The alcR transcription initiation sites were located adjacent to the putative Fur repressor binding site in a spatial organization remarkably similar to that of the alcA promoter-operator region (33). The RNA samples from wild-type cells also yielded several longer iron-regulated extension products presumably derived from reverse transcription of mRNA species originating at the alcA promoter which were absent in samples from mutant PM-4.
FIG. 5.
Localization of the iron-regulated P_alcR_ promoter of alcR. RNA was isolated from B. pertussis wild-type strain UT25Sm1 (WT) and B. pertussis mutant PM-4, in which the alcA promoter-operator region is deleted, grown in iron-replete (+Fe) and iron-depleted (−Fe) conditions. Transcriptional initiation sites (+1) corresponding to two nucleotides, C and T (at positions 124 and 125, respectively), were mapped by primer extension analysis using the antisense primer shown in Fig. 3.
Inspection of the region upstream of the alcR transcription start site revealed sequences with similarity to E. coli ς70 promoter determinants (28). The transcription initiation site is optimally spaced 6 nucleotides from the hexameric sequence 5′-TATCAT-3′ (positions −12 to −7 with respect to the transcription start site +1), which shares five of six of the most highly conserved nucleotides with the E. coli ς70 −10 consensus sequence TATAAT, including the invariant T (28) at the last position.
Characterization of defined B. pertussis and B. bronchiseptica alcR mutants.
Chromosomal deletions in alcR were constructed in wild-type Bordetella parental backgrounds to yield mutations exerting little or no polarity on downstream genes. Removal of a 264-bp _Ngo_AIV DNA fragment internal to alcR (Fig. 3) was predicted to result in an in-frame deletion altering the amino-terminal region of the AlcR protein, thus avoiding premature translation termination that may result in polar effects on unknown downstream genes. Allelic exchange of mutated DNA subfragments lacking 264-bp _Ngo_AIV fragments with wild-type alleles of B. pertussis UT25Sm1 and B. bronchiseptica B013N resulted in mutants PM10 and BRM11, respectively.
As was observed for B. bronchiseptica mutant BRM10, defined B. pertussis mutant PM10 failed to produce alcaligin and did not express AlcC (data not shown), and introduction of the 1.6-kb _Kpn_I-_Pst_I B. pertussis alcR DNA fragment in trans restored both siderophore production (Table 1) and AlcC expression to wild-type levels. Likewise, defined B. bronchiseptica alcR mutant BRM11 was defective in siderophore production but was fully complemented by B. pertussis alcR (pP9KP) as well as by the cloned wild-type B. bronchiseptica DNA region containing alcR (data not shown). When BRM11 was supplied in trans with a derivative of the B. bronchiseptica alcR gene in which the internal 264-bp _Ngo_AIV fragment was deleted, no complementation of the siderophore production defect was observed on CAS agar.
TABLE 1.
Alcaligin siderophore activity of B. pertussis
| Strain | Siderophore activitya | |
|---|---|---|
| With iron | Without iron | |
| UT25Sm1 (wild type) | 0.018 ± 0.014 | 0.592 ± 0.014 |
| PM10/pP9KP (alcR+) | 0.000 ± 0.028 | 0.747 ± 0.006 |
| PM10/pRK415 (vector) | 0.000 ± 0.026 | 0.047 ± 0.005 |
T7 promoter-directed expression of AlcR.
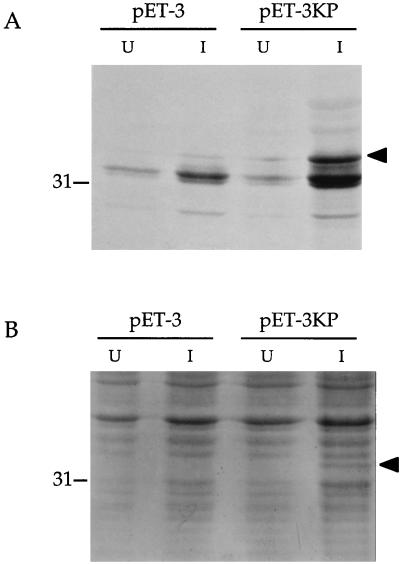
AlcR production programmed by the 1.6-kb _Kpn_I-_Pst_I B. pertussis DNA fragment was accomplished by using a bacteriophage T7 polymerase-promoter protein expression system in E. coli. The alcR open reading frame was directionally cloned positioned in the sense orientation with respect to the bacteriophage T7 promoter in plasmid pET-3. Under inducing conditions, cells carrying the alcR plasmid expressed a protein with a molecular mass of approximately 34 kDa, similar to that predicted for AlcR, which was absent in uninduced cells and host cells carrying the plasmid vector alone (Fig. 6). The AlcR protein was readily visualized in electrophoretic gels as a 35S-radiolabelled polypeptide or visualized by staining with Coomassie blue.
FIG. 6.
Expression of the alcR protein product in E. coli. A T7 polymerase-promoter system was used to express alcR from the 1.6-kb _Kpn_I-_Pst_I B. pertussis DNA fragment as described in Materials and Methods. (A) Autoradiogram of 35S-labelled translational products. (B) Proteins visualized by Coomassie blue staining. Lanes: pET-3, BL21(DE3) cells carrying the plasmid vector control; pET-3KP, cells carrying the alcR gene. U, protein samples from uninduced cells; I, protein samples after induction of cells by addition of IPTG. The migration position of the 31-kDa molecular mass standard is indicated on the left. The AlcR polypeptide is indicated on the right (arrowheads). The labelled protein in panel A migrating at approximately 32 kDa is presumed to be the vector-encoded β-lactamase.
Involvement of AlcR in alcaligin transport and/or utilization.
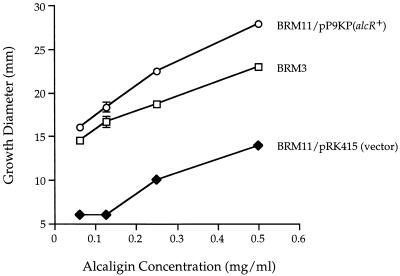
The ability of B. bronchiseptica alcR mutant BRM11 to transport and/or utilize alcaligin for growth on LB medium made iron restricted by addition of EDDA was determined in quantitative alcaligin growth stimulation assays using exogenously supplied purified alcaligin and B. bronchiseptica alcaligin biosynthesis mutant BRM3 as a transport-proficient positive-control indicator strain (3, 12) (Fig. 7). Growth stimulation of alcR mutant BRM11 carrying the plasmid vector control was significantly impaired at all alcaligin concentrations tested, with only 30% of the growth stimulation level of the positive-control strain achieved. B. bronchiseptica streptonigrin-resistant mutant BRM10 also displayed a similar alcaligin utilization defect in quantitative siderophore bioassays (data not shown). When the B. pertussis alcR gene was supplied in trans, BRM11 alcaligin-mediated growth stimulation levels exceeded those of the transport-proficient positive-control indicator strain, perhaps attributable to multicopy effects. Together, the data indicate that alcR is involved in ferric alcaligin transport and/or utilization as well as alcaligin biosynthesis.
FIG. 7.
Alcaligin utilization bioassays. Promotion of growth of B. bronchiseptica by exogenously supplied alcaligin in iron-restricted medium was performed as detailed in Materials and Methods. Alcaligin biosynthesis mutant BRM3 is wild type with respect to ferric alcaligin transport. The diameters of the zones of growth include the 6 mm contributed by the sample well. Bacteria supplied with diluent alone showed no growth stimulation. Standard deviations are indicated as vertical bars; no standard deviation bars are shown if the values were less than 0.4.
DISCUSSION
Treatment of a pool of B. bronchiseptica transposon mutants with the redox-activated quinone antibiotic streptonigrin in iron-restricted medium containing alcaligin was expected to enrich the population for cells defective in ferric alcaligin transport or utilization functions (8, 57). B. bronchiseptica mutant BRM10 exhibited a pleiotropic phenotype: it failed to produce alcaligin, was defective in alcaligin utilization, and lacked two iron-repressed proteins, AlcC and an iron-repressible membrane protein of approximately 79 kDa. In phenotypic complementation analyses, the 1.6-kb _Kpn_I-_Pst_I DNA subfragment of B. pertussis cosmid pCP1.11 restored alcaligin production, as well as expression of AlcC and the 79-kDa protein, to BRM10. However, the 1.6-kb _Kpn_I-_Pst_I DNA fragment does not encode the AlcC protein and has insufficient coding capacity for a 79-kDa protein. The BRM10 pleiotropic phenotype, along with the fact that alcC expression was prevented by a mutation located outside and downstream of alcC (a phenotype that cannot be attributed to polarity on alcC), suggested the involvement of a positive regulatory mechanism controlling alcaligin gene expression.
While all of the Bordetella alcR mutants lacked detectable AlcC protein, only in B. bronchiseptica (mutants BRM10 and BRM11) could production of the 79-kDa iron-repressed membrane protein be determined conclusively. The 79-kDa protein was not readily observed in either wild-type B. pertussis cells or B. pertussis alcR mutant PM10 due to interference from numerous comigrating protein species in SDS-PAGE. Because the alcR mutants were defective in ferric alcaligin transport and/or utilization, it is possible that the iron-repressible 79-kDa membrane protein may function in this capacity.
Analysis of the nucleotide sequence of the 1.6-kb _Kpn_I-_Pst_I DNA fragment revealed an open reading frame, alcR, preceded by a DNA sequence with similarity to known Fur repressor binding sites and which binds purified B. pertussis Fur protein with high affinity in gel mobility shift assays (11). Two potential AlcR translation initiation codons which would result in an AlcR polypeptide with a molecular mass of either 36.4 or 33.9 kDa (sizes consistent with that observed for AlcR expressed in the T7 polymerase-promoter experiments) were identified. AlcR is a new member of the AraC family of transcriptional regulators, each of which exhibits the greatest similarity at the carboxy terminus containing characteristic family signature motifs and the helix-turn-helix region implicated in the binding of DNA (24). AlcR showed the highest similarities to the AraC family members PchR of P. aeruginosa (29, 30) and YbtA of Y. pestis (21), both of which are involved in siderophore system gene regulation. AlcR, PchR, and YbtA were clustered on a hypothetical phylogenetic tree generated with the complete amino acid sequences of the AraC family members noted in Fig. 4 and those of 16 other family members retrieved in our database searches (data not shown).
Recent RNA hybridization experiments indicated that alcR is transcribed as part of the Fur-controlled alcABC_-containing operon, as well as from its own iron-regulated secondary promoter (32). In the present study, primer extension analysis identified this iron-regulated promoter and the alcR transcription initiation site. By using RNA from wild-type cells and the alcA promoter-operator deletion mutant PM-4, further evidence which is consistent with the hybridization data indicating that alcR is also transcribed as part of a larger RNA species (i.e., from the alcA promoter) was obtained. Although alcR upstream sequences resembling a consensus ς70 −10 promoter region were identified, there were no obvious −35 promoter determinants. The absence of strong −35 determinants is consistent with the observation that genes encoding positive regulators are usually expressed at low levels in the cell (45). In addition, it is possible that alcR requires a positively acting regulatory factor (perhaps AlcR itself) for optimal transcription from P_alcR .
It was determined that the transposon in the BRM10 chromosome was not located in alcR and that the BRM10 alcR gene most likely suffered a spontaneous mutation that conferred streptonigrin resistance. Because of the undefined nature of the BRM10 mutation, defined nonpolar chromosomal alcR mutations in B. pertussis (PM10) and B. bronchiseptica (BRM11) were constructed. Each mutation resulted in an in-frame deletion of an 88-amino-acid sequence within the amino-terminal region of AlcR. The amino termini of the AraC-like regulators are presumed to be involved in substrate recognition (24); for AraC, the amino terminus functions in both dimerization and binding of the inducer arabinose (37). Therefore, these Bordetella mutants may produce AlcR proteins lacking the domain required for dimerization or inducer binding. The two alcR deletion mutants exhibited phenotypes similar to that of BRM10, consistent with defects in alcaligin synthesis and ferric alcaligin transport. Function was restored by supplying either the B. pertussis or the B. bronchiseptica wild-type alcR alleles in trans.
The Fur repressor protein and its role in transcriptional regulation of iron acquisition genes have been well documented (4, 14, 18, 26). The activities of both Fur and the more recently described positive regulators ensure that the genes encoding siderophore biosynthesis and transport functions are expressed maximally only under appropriate environmental conditions when the cognate siderophore is an effective iron scavenger. This general type of priority regulation is an established function of positive regulators (45). In the E. coli ferric citrate system, the presence of ferric citrate bound to its receptor, FecA, activates transcription of the transport genes fecABCD (35). A cytoplasmic membrane protein and the TonB system appear to transduce the FecA receptor occupancy signal to the regulator FecI, a cytoplasmic protein bearing significant similarity to alternative sigma factors. Other siderophore systems positively regulated through the function of alternative sigma factor-like proteins include the Pseudomonas putida WCS358 native and heterologous pseudobactin systems (55) as well as the native pyoverdin system of P. aeruginosa (15). A second type of positive regulation of siderophore utilization genes has been described only for the PfeR-PfeS system of P. aeruginosa, which allows the organism to respond to the heterologous siderophore enterobactin, resulting in up-regulated expression of the ferric enterobactin receptor PfeA (16). PfeR and PfeS are similar to proteins of classical two-component sensory transduction systems. The third type of positive regulation of siderophore gene expression involves proteins which are members of the AraC family of transcriptional regulators, some of which can act positively or negatively, depending on the presence or absence of inducer and the position of the regulator binding site on the DNA (24, 45). These AraC-like regulators include the P. aeruginosa PchR and Y. pestis YbtA proteins and now Bordetella AlcR. Pyochelin biosynthesis and transport are regulated in P. aeruginosa both positively and negatively by PchR; the system responds to pyochelin and requires the pyochelin receptor (29, 30). In Y. pestis, YbtA acts both positively and negatively in the regulation of the yersiniabactin receptor (21). The specific mechanisms of action of these AraC-like regulators of siderophore genes are undefined at present but may involve direct activation by the cognate siderophore (or a degradation product thereof) functioning as an inducer. The alcR mutant phenotype, together with the similarities between AlcR, PchR, YbtA, and other members of the AraC protein family, indicates that alcaligin biosynthesis and transport genes in Bordetella spp. can not only be repressed by Fur but also can be controlled by the AlcR regulator protein.
ACKNOWLEDGMENTS
We thank Peggy Cotter, Jeff F. Miller, Scott Stibitz, and F. William Studier for providing strains and plasmids. We acknowledge Chantel Sabus and Genell Pridgen for technical assistance.
This work was supported by Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Akerley B J, Cotter P A, Miller J F. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 3.Armstrong S K, Clements M O. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J Bacteriol. 1993;175:1144–1152. doi: 10.1128/jb.175.4.1144-1152.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagg A, Neilands J B. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 5.Bentley J, Hyatt L S, Ainley K, Parish J H, Herbert R B, White G R. Cloning and sequence analysis of an Escherichia coli gene conferring bicyclomycin resistance. Gene. 1993;127:117–120. doi: 10.1016/0378-1119(93)90625-d. [DOI] [PubMed] [Google Scholar]
- 6.Bordet J, Gengou O. Le microbe de la coqueluche. Ann Inst Pasteur (Paris) 1906;20:731–741. [Google Scholar]
- 7.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 8.Braun V, Gross R, Koster W, Zimmermann L. Plasmid and chromosomal mutants in the iron(III)-aerobactin system of Escherichia coli. Use of streptonigrin for selection. Mol Gen Genet. 1983;192:131–139. doi: 10.1007/BF00327658. [DOI] [PubMed] [Google Scholar]
- 9.Brickman T J, Armstrong S K. Bordetella pertussis fur gene restores iron repressibility of siderophore and protein expression to deregulated Bordetella bronchiseptica mutants. J Bacteriol. 1995;177:268–270. doi: 10.1128/jb.177.1.268-270.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brickman T J, Armstrong S K. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J Bacteriol. 1996;178:54–60. doi: 10.1128/jb.178.1.54-60.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brickman T J, Armstrong S K. Abstracts of the 97th General Meeting of the American Society for Microbiology 1997. Washington, D.C: American Society for Microbiology; 1997. Fur repressor-operator interactions at the iron-responsive alcaligin siderophore biosynthesis operon control region of Bordetella pertussis, abstr. B-241; p. 70. [Google Scholar]
- 12.Brickman T J, Hansel J-G, Miller M J, Armstrong S K. Purification, spectroscopic analysis, and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. BioMetals. 1996;9:191–203. doi: 10.1007/BF00144625. [DOI] [PubMed] [Google Scholar]
- 13.Bullen J J. The significance of iron in infection. Rev Infect Dis. 1981;3:1127–1138. doi: 10.1093/clinids/3.6.1127. [DOI] [PubMed] [Google Scholar]
- 14.Calderwood S B, Mekalanos J J. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J Bacteriol. 1987;169:4759–4764. doi: 10.1128/jb.169.10.4759-4764.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunliffe H E, Merriman T R, Lamont I L. Cloning and characterization of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J Bacteriol. 1995;177:2744–2750. doi: 10.1128/jb.177.10.2744-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean C R, Poole K. Expression of the ferric enterobactin receptor PfeA of Pseudomonas aeruginosa: involvement of a two-component regulatory system. Mol Microbiol. 1993;8:1095–1103. doi: 10.1111/j.1365-2958.1993.tb01654.x. [DOI] [PubMed] [Google Scholar]
- 17.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeShazer D, Wood G E, Friedman R L. Boiling eliminates artifact banding when sequencing double-stranded templates. BioTechniques. 1994;17:288–290. [PubMed] [Google Scholar]
- 20.Feinberg A P, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 21.Fetherston J D, Bearden S W, Perry R D. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 22.Field L H, Parker C D. Differences observed between fresh isolates of Bordetella pertussis and their laboratory-passaged derivatives. In: Manclark C R, Hill J C, editors. International Symposium on Pertussis. U.S. Washington, D.C: Department of Health, Education, and Welfare; 1979. pp. 124–132. [Google Scholar]
- 23.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallegos M-T, Michan C, Ramos J L. The XylS/AraC family of regulators. Nucleic Acids Res. 1993;4:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giardina P C, Foster L-A, Toth S I, Roe B A, Dyer D W. Identification of alcA, a Bordetella bronchiseptica gene necessary for alcaligin production. Gene. 1995;167:133–136. doi: 10.1016/0378-1119(95)00659-1. [DOI] [PubMed] [Google Scholar]
- 26.Guerinot M L. Microbial iron transport. Annu Rev Microbiol. 1994;48:743–772. doi: 10.1146/annurev.mi.48.100194.003523. [DOI] [PubMed] [Google Scholar]
- 27.Hantke K. Regulation of ferric iron transport in Escherichia coli K-12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 28.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinrichs D E, Poole K. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175:5882–5889. doi: 10.1128/jb.175.18.5882-5889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heinrichs D E, Poole K. PchR, a regulator of ferripyochelin receptor gene (fptA) expression in Pseudomonas aeruginosa, functions both as an activator and as a repressor. J Bacteriol. 1996;178:2586–2592. doi: 10.1128/jb.178.9.2586-2592.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. CABIOS. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 32.Kang H Y, Armstrong S K. Transcriptional analysis of the Bordetella alcaligin siderophore biosynthesis operon. J Bacteriol. 1998;180:855–861. doi: 10.1128/jb.180.4.855-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang H Y, Brickman T J, Beaumont F C, Armstrong S K. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J Bacteriol. 1996;178:4877–4884. doi: 10.1128/jb.178.16.4877-4884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 35.Kim I, Stiefel A, Plantör S, Angerer A, Braun V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol Microbiol. 1997;23:333–344. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- 36.Lankford C E. Bacterial assimilation of iron. Crit Rev Microbiol. 1973;2:273–331. [Google Scholar]
- 37.Lobell R, Schleif R. DNA looping and unlooping by AraC protein. Science. 1990;250:528–532. doi: 10.1126/science.2237403. [DOI] [PubMed] [Google Scholar]
- 38.Mietzner T A, Morse S A. The role of iron-binding proteins in the survival of pathogenic bacteria. Annu Rev Nutr. 1994;14:471–493. doi: 10.1146/annurev.nu.14.070194.002351. [DOI] [PubMed] [Google Scholar]
- 39.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- 40.Moore C H, Foster L-A, Gerbig J G, Dyer D W, Gibson B W. Identification of alcaligin as the siderophore produced by Bordetella pertussis and Bordetella bronchiseptica. J Bacteriol. 1995;177:1116–1118. doi: 10.1128/jb.177.4.1116-1118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naroditshaya V, Schlosser M J, Fang N Y, Lewis K. An E. coli gene emrD is involved in adaptation to low energy shock. Biochem Biophys Res Commun. 1994;196:803–809. doi: 10.1006/bbrc.1993.2320. [DOI] [PubMed] [Google Scholar]
- 42.Neilands J B. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 43.Nikaido H. Prevention of drug access to bacterial targets: permeability barriers and active efflux. Science. 1994;264:382–388. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 44.Pittman M. Genus Bordetella. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: Williams & Wilkins; 1984. pp. 388–393. [Google Scholar]
- 44a.Pradel E, Guiso N, Locht C. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J Bacteriol. 1998;180:871–880. doi: 10.1128/jb.180.4.871-880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raibaud O, Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;18:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- 46.Rosenberg A H, Lade B L, Chui D, Lin S-W, Dunn J J, Studier F W. Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene. 1987;56:125–135. doi: 10.1016/0378-1119(87)90165-x. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 48.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneider D R, Parker C D. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect Immun. 1982;38:548–553. doi: 10.1128/iai.38.2.548-553.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 51.Stainer D W, Scholte M J. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol. 1970;63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- 52.Stibitz S. Use of conditionally counterselectable suicide vectors for allelic exchange. Methods Enzymol. 1994;235:458–465. doi: 10.1016/0076-6879(94)35161-9. [DOI] [PubMed] [Google Scholar]
- 53.Studier F W, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 54.Tercero J A, Lacalle R A, Jimenez A. The pur8 gene from the pur cluster of Streptomyces alboniger encodes a highly hydrophobic polypeptide which confers resistance to puromycin. Eur J Biochem. 1993;218:963–971. doi: 10.1111/j.1432-1033.1993.tb18454.x. [DOI] [PubMed] [Google Scholar]
- 55.Venturi V, Weisbeek P, Koster M. Gene regulation of siderophore-mediated iron acquisition in Pseudomonas: not only the Fur repressor. Mol Microbiol. 1995;17:603–610. doi: 10.1111/j.1365-2958.1995.mmi_17040603.x. [DOI] [PubMed] [Google Scholar]
- 56.Weinberg E D. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeowell H N, White J R. Iron requirement in the bactericidal mechanism of streptonigrin. Antimicrob Agents Chemother. 1982;22:961–968. doi: 10.1128/aac.22.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]