Centromere Positioning and Dynamics in Living Arabidopsis Plants (original) (raw)
Abstract
The organization and dynamics of the genome have been shown to influence gene expression in many organisms. Data from mammalian tissue culture cells have provided conflicting conclusions with regard to the extent to which chromatin organization is inherited from mother to daughter nuclei. To gain insight into chromatin organization and dynamics, we developed transgenic Arabidopsis lines in which centromeres were tagged with a green fluorescent protein fusion of the centromere-specific histone H3. Using four-dimensional (4-D) live cell imaging, we show that Arabidopsis centromeres are constrained at the nuclear periphery during interphase and that the organization of endoreduplicated sister centromeres is cell type dependent with predominant clustering in root epidermal cells and dispersion in leaf epidermal cells. 4-D tracking of the entire set of centromeres through mitosis, in growing root meristematic cells, demonstrated that global centromere position is not precisely transmitted from the mother cell to daughter cells. These results provide important insight into our understanding of chromatin organization among different cells of a living organism.
INTRODUCTION
The centromere, the primary constriction of the chromosome, is a DNA-protein structure that directs the movement of chromosomes during mitosis and meiosis. All centromeric regions contain specialized nucleosomes in which histone 3 is replaced by the centromere-specific histone H3 (CENH3). CENH3 was known variously as CENP-A (human), Cse4 (Saccharomyces cerevisiae), Cnp1 (Schizosaccharomyces pombe), HCP-3 (Caenorhabditis elegans), and Cid (Drosophila) (Palmer et al., 1987; Stoler et al., 1995; Doe et al., 1998; Buchwitz et al., 1999; Henikoff et al., 2000). More recently, plant CENH3s were isolated from Arabidopsis (HTR12) (Talbert et al., 2002), Zea mays (maize CENH3) (Zhong et al., 2002), Oryza sativa (rice CENH3) (Nagaki et al., 2004), Saccharum officinarum (SoCENH3) (Nagaki and Murata, 2005), and from Luzula nivea (LnCENH3), which has holocentric chromosomes (Nagaki et al., 2005). Despite the divergence of centromeric DNA, domain organization (centromere core and pericentromere) seems to be conserved in plants, yeast, and mammals, and each domain has distinct functions for formation of the kinetochore structure or sister centromere cohesion (Kniola et al., 2001; Appelgren et al., 2003; Zhang et al., 2005). Arabidopsis centromeres consist of a core domain, which is characterized by HTR12- and Ser10-phosphohistone H3-containing nucleosomes as well as 180-bp satellite DNA repeats (Shibata and Murata, 2004). The centromere is flanked by pericentromeric heterochromatin domains that contain canonical histone H3 and middle repetitive elements, including retroelements and transposons (Copenhaver et al., 1999; reviewed in Heslop-Harrison et al., 2003; Jiang et al., 2003).
Similar to mammalian chromosomes, Arabidopsis chromosomes are organized as well-defined chromosome territories in interphase nuclei (Pecinka et al., 2004). Early observations with salamander cells suggested a preferentially polarized organization of chromosomes with centromeres clustered at one end of the nucleus (the apical side) and telomeres at the opposite end (the basal side), known as the Rabl configuration (Rabl, 1885). The Rabl configuration has been observed in trypanosomes (Chung et al., 1990), fission yeast (Funabiki et al., 1993), and Drosophila (Agard and Sedat, 1983; Hochstrasser et al., 1986; Marshall et al., 1996). Mammalian cells do not show such a simple polar organization of interphase chromosomes, as centromeres and telomeres can be found distributed throughout the nucleus (Luderus et al., 1996; Shelby et al., 1996). In plants, a Rabl configuration was observed in onion root tip cells (Stack and Clark, 1974) and other monocots with a large genome (Abranches et al., 1998; Dong and Jiang, 1998; Wegel and Shaw, 2005), but not in plant species with a small genome (Dong and Jiang, 1998; Fransz et al., 2002; Talbert et al., 2002). Arabidopsis centromere localization was observed in fixed cells in two dimensions (Talbert et al., 2002); however, the three-dimensional (3-D) spatial positioning of centromeres has not been characterized in living Arabidopsis plants.
Many studies have addressed chromatin dynamics in the interphase nucleus in mammalian cells as well as in Drosophila and yeast (reviewed in Spector, 2003). The general consensus has been that chromatin movement is constrained within the interphase nucleus. However, data regarding chromatin dynamics in living plants are limited (reviewed in Lam et al., 2004). Chromatin at three T-DNA insertions on the upper arm of chromosome 3 were observed to undergo diffusive movement in constrained areas, and the confinement area in endoreduplicated pavement cells was >6 times larger than in diploid guard cells (Kato and Lam, 2003). However, thus far, the dynamics of centromeres has not been examined in living Arabidopsis plants.
During mitosis, it is well-known that chromosomes with DNA are precisely transmitted from mother cell to daughter cells. Several studies have addressed the positioning of chromatin in mammalian daughter nuclei after mitosis, another important question regarding transmission of genetic/epigenetic information during cell division. By photobleaching green fluorescent protein (GFP)-tagged chromatin, Walter et al. (2003) found the location of chromosome territories to be stably maintained from mid-G1 to late G2, but major changes were observed from one cell cycle to the next. In addition, chromosome movement in early G1 was suggested to play a role in the final placement of chromosome territories in interphase nuclei. In contrast, Gerlich et al. (2003) concluded that global chromosome positions are transmitted through mitosis in mouse NRK cells. More recently, Thomson et al. (2004) found the radial positioning of chromatin is not inherited through mitosis in human HT1080 cells. Increased chromatin mobility was detected during the first 2 h of G1, and association with nuclear compartments was both gained and lost. However, it is not clear whether the observed mitotic transmission pattern of chromatin in mammalian cultured cells is representative of that found in a multicellular living organism where the mitotic cells are embedded in a living tissue and may be regulated by signals from adjacent cells or other cells in the same organism. To address this point, we studied in situ transmission of centromeres through mitosis in root meristematic cells, a diploid cell type in the flowering plant Arabidopsis thaliana.
Here, we have taken advantage of A. thaliana, with only 10 centromeres in diploid cells, to study the three-dimensional organization and dynamics of centromeres in interphase nuclei and through mitosis in living Arabidopsis plants. Centromeres were in vivo tagged by expressing a fusion of centromere-specific histone 3 (HTR12) with GFP variants. By in situ imaging of the transgenic plants, we addressed how the centromeres are organized in the interphase nucleus in three dimensions, whether this organization is different in endoreduplicated cells, and whether centromeres are dynamic in interphase nuclei. Furthermore, we followed the movement of each centromere through mitosis by four-dimensional (4-D) microscopy to address whether global centromere positions are transmitted from mother to daughter cells and whether the nuclei of the two daughter cells are symmetric.
MATERIALS AND METHODS
Constructs
The endogenous promoter and terminator of the centromere-specific histone H3 variant gene HTR12 (At1g01370) and a histone H2B gene HTB1 (At1g07790) (A. thaliana, ecotype Columbia) were used to regulate the expression of the fusion proteins. A 2.5-kb genomic DNA fragment of HTR12 including upstream regulatory sequences, 5′ untranslated region, coding sequences, and introns was PCR amplified by using primers 5′-NNNAAGCTTAGGGCATTGAAGTGAAGTATC-3′ and 5′-NNNACCGGTCGCCATGGTCTGCCTTTTCCTCC-3′; the fragment contains a 5′ _Hin_dIII and a 3′ _Age_I restriction site. A 1.8-kb genomic DNA fragment of HTR12 downstream sequence was PCR amplified using primers 5′-NNNGCGGCCGCAAAACTCACTCACTATTCACA-3′ and 5′-NNNGAATTCCTCAAAAGACACCTTAGTACA-3′; the fragment contains 5′ _Not_I and 3′ _Eco_RI restriction sites. A 1.9-kb genomic DNA fragment of HTB1 including upstream regulatory sequences, 5′ untranslated region, and coding sequence was PCR amplified using primers 5′-NNNGGATCCTGGTGCCGGTCTCATCTCAAC-3′ and 5′-NNNACCGGTCGAGAGCTAGTAAACTTAGTAAC-3′; the fragment contains 5′ _Bam_HI and 3′ _Age_I restriction sites. A 1.3-kb genomic DNA fragment of HTB1 downstream sequence was amplified using primers 5′-NNNGCGGCCGCGTTTAGCCGTTTAGGTTTCGA-3′ and 5′-TATGAATTCTTCACCATTTCT-3′; the fragment contains 5′ _Not_I and 3′ _Eco_RI restriction sites.
The coding sequence of GFP, yellow fluorescent protein (YFP) variant (Venus), and cyan fluorescent protein (CFP) was amplified from pGFP-N1(BD Biosciences Clontech, Palo Alto, CA), Venus/pCS2 (Nagai et al., 2002), and pCFP-N1 (BD Biosciences Clontech), respectively, using primers 5′-NNNACCGGTCGGAGGTGGAGGTGGAGCTGTGAGCAAGGGCGAGGAGCTG-3′ and 5′-NNNGCGGCCGCGTTACTTGTACAGCTCGTCCAT-3′. A flexible linker, Gly5-Ala (Fang et al., 2004), was added at the N terminus of GFP, Venus, and CFP; these fragments contain a 5′-_Age_I and a 3′-_Not_I restriction site.
These PCR fragments were cloned into pCR2.1 vector (Invitrogen, Carlsbad, CA), confirmed by sequencing and then released by treatment with the respective restriction enzymes. HTR12 was directionally subcloned into the _Hin_dIII/_Eco_RI-digested vector pFGC5941 (http://www.chromdb.org) with the direction: _Hin_dIII-HTR12 upstream regulatory sequence-HTR12 coding DNA with introns-_Age_I-Gly5-Ala linker-Venus (or GFP)-_Not_I-HTR12 terminator-_Eco_RI to obtain binary vectors 1) PHTR12-HTR12-Venus-THTR12 and 2) PHTR12-HTR12-GFP-THTR12. HTB1 was directionally subcloned into the _Bam_HI/_Eco_RI-digested vector pCambia2300 (CAMBIA, Canberra, Australia) with the direction _Bam_HI-HTB1 upstream regulatory sequence-HTB1 coding DNA-_Age_I-Gly5-Ala linker-CFP-_Not_I-HTB1 terminator-_Eco_RI to obtain binary vector PHTB1-HTB1-CFP-THTB1.
Plant Transformation and Cotransformation
The vectors PHTR12-HTR12-Venus-THTR12, PHTR12-HTR12-GFP-THTR12, and PHTB1-HTB1-CFP-THTB1 were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation. A. thaliana (ecotype Columbia) plants were transformed by the floral dip method (Clough and Bent, 1998). Cotransformation was performed by mixing equal amounts of GV3101(PHTR12-HTR12-Venus-THTR12) and GV3101(PHTB1-HTB1-CFP-THTB1) suspension before dipping. Transgenic plant selection and growth conditions were described in Fang et al. (2004). Selective agents used were 12 mg/l glufosinate-ammonium (Aldrich Chemical, Milwaukee, WI) for HTR12-GFP, and 50 mg/l kanamycin and 12 mg/l glufosinate-ammonium for cotransformants of HTR12-Venus and HTB1-CFP. Transgenic lines carrying one copy of each transgene were used for imaging.
Fluorescence Deconvolution Microscopy and Time-Lapse Three-dimensional (4-D) Live Cell Imaging
Fluorescence deconvolution microscopy and settings were performed as described in Fang et al. (2004). Filters used for Venus were excitation 500/20 and emission 535/30 nm; for CFP 436/10 and 465/30 nm; and for GFP 490/20 and 528/38 nm, respectively, and 86006bs beamsplitter (Chroma Technology, Brattleboro, VT).
T2 transgenic plants of HTR12-Venus and HTB1-CFP cotransformants were used for imaging. Rosette leaves (∼0.5 × 0.5 cm2), sepal, and petal were cut and mounted in MS medium between two 50 × 24-mm no. 1.5 coverslips and immediately used for imaging (Invitrogen). The image stacks of nuclei were collected with a Z step size of 0.20 μm.
HTR12-GFP transgenic T2 seeds were used for 4-D imaging. For imaging the root meristematic cells, the transgenic seeds were sown onto chambered coverglass system (Nalge Nunc International, Naperville, IL), the growing roots in MS medium was used for imaging (Fang et al., 2004). Time-lapse 4-D images up to 2 h with a time point interval of 45 s were collected in seven sections with a image size of 512 × 512 pixels, an exposure time of 0.06 s for each section, and a Z step size of 0.60 μm. Deconvolution, measurement of distances, and volume rendering of the image stacks were performed using SoftWoRx software (Applied Precision, Issaquah, WA). The DeltaVision files were saved as TIF images and processed using Adobe Photoshop 7.0 (Adobe Systems, Mountain View, CA).
Quantitative Analyses of Centromere Positions
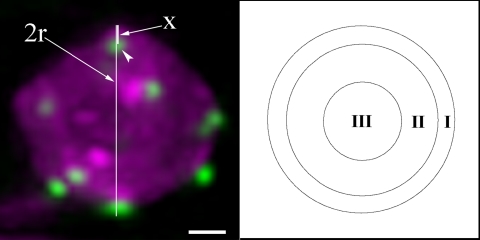
Deconvolved 3-D stacks were rotated to an angle where the centromere of interest is closest to the nuclear periphery as observed in projection (nuclei are defined by HTB1-CFP or diffuse HTR12-GFP signals). Centromere positions were then quantified similar to analyses of telomere position described in Hediger et al. (2002). To determine the position of the centromere of interest, the centromere spot-to-periphery distance (x) was divided by the nucleus radius (r). Each centromere fell into one of three concentric spherical zones of equal surface (zone I, x < 0.184 r; zone II, x = 0.184 r to 0.422 r; and zone III, x > 0.422 r) (Figure 2).
Figure 2.
Diagram of the approach used for quantitative analyses of centromere positions. A projection image of a small leaf epidermal cell nucleus is shown in the left panel. Centromeres were labeled by HTR12-Venus in green, and nucleus was labeled by HTB1-CFP in magenta. The distance of the centromere of interest (arrowhead) to the nuclear periphery (x) is divided by the nuclear radius (r). Centromere position can be mapped to three concentric zones of equal surface (I, II, and III) as described in Materials and Methods. Bar, 1 μm.
Measurement of Centromere Movements
To exclude error in measurement caused by z_-axis resolution and nuclear rotation, we selected nuclei in which the two centromeres remained in the same z-section, an approach used previously in studies on budding yeast, Drosophila, and human cells (Heun et al., 2001; Vazquez et al., 2001; Chubb et al., 2002). The signal intensities of each pixel in deconvolved nuclear images were obtained in the Data Inspector Module, and the distances between centromeres were measured by Standard Two Point method in SoftWoRx software. The position of a centromere was defined as the brightest pixel in the local fluorescent spot. The changes in distance between centromeres relative to t = 0 (<Δ_d_>) were calculated at each time point. The squared Δd values (<Δ_d_2>) were plotted against the elapsed time (<_Δt_>). Standard normal deviates were calculated to assess the significance of the apparent differences between <Δ_d_2> values. The diffusion coefficient (D) was calculated from the linear slopes of <Δ_d_2> plots with the formula D = Δd2/4_Δt (Berg, 1993). As a control, roots of transgenic seedlings were fixed in phosphate-buffered saline containing 4% formaldehyde prepared fresh from paraformaldehyde followed by washing and 4-D microscopic visualization.
Cell Tracking and Centromere Tracking through Mitosis
Cell and centromere tracking were performed using SoftWoRx software. Selected cells were tracked by centering the cell in the image stacks to eliminate the effect of root growth on data analysis. Image stacks containing the cell of interest were then cut time point by time point from the raw data and assembled into 4-D images. After deconvolution, centromeres were segmented and tracked in individual sections, partial projection images (projection of only a part of sections), and total maximum projection images (projection of all sections). The same centromere in a section and the total maximum projection image at a given time point were defined by comparing the signal intensity and its x and y positions in pixel numbers. At least three identical results were obtained from independent centromere tracking processes for each mitotic event.
RESULTS
Three-dimensional Localization of Centromeres in Nuclei of Living Arabidopsis Plants
To visualize centromeres in living Arabidopsis plants, amplified HTR12 genomic DNA including the promoter region and 5′ untranslated region was fused in frame with Venus, a brighter variant of yellow fluorescent protein (YFP) (Nagai et al., 2002). In addition, an endogenous terminator was used to control the stability of the fusion mRNA. For double labeling of bulk chromatin, a histone H2B gene (HTB1; http://www.chromdb.org) was fused in frame with CFP, which was also under control of its endogenous promoter and terminator. Transgenic plants and progeny coexpressing HTR12-Venus and HTB1-CFP grow normally without any obvious phenotypic changes, indicating the cells expressing these fusions have functional centromeres and normal cell cycles.
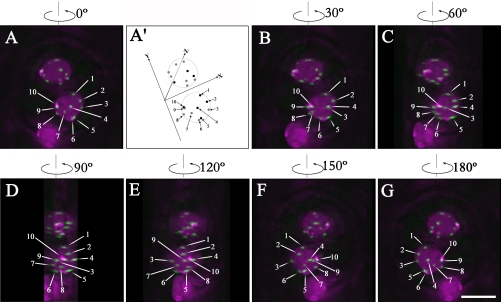
Arabidopsis centromeres were observed as small, bright HTR12-Venus foci in nuclei labeled with HTB1-CFP (Figure 1, projection images of z-stacks). Ten spots (a range of 8–10) were usually detected in diploid cell types, corresponding to the 10 centromeres in these cells. Signal intensity variation was also observed, especially in leaves, where the strongest signals were detected in guard cells. It is not clear whether the observed differences in signal intensity in different cell types are correlated to cell type-specific expression or to differences in HTR12 deposition at centromeres. To investigate the localization of centromeres in 3-D, optical sections were collected from different cell types in living Arabidopsis plants by 3-D optical sectioning microscopy followed by image restoration using iterative deconvolution on a DeltaVision microscope (Applied Precision). Centromere positions in the nuclei were examined by analyzing individual image sections, maximum signal intensity projections from different angles, and volume-view rotation videos. Figure 1 shows projection images of guard cell nuclei at different angles (also see Supplemental Video 1). Because HTR12-Venus labels all centromeres we could not define centromeres of specific chromosomes; therefore, in Figure 1A (0°) the centromeres are arbitrarily numbered 1–10. This figure represents a projection image of a 20 image stack through the _z_-axis of the cells shown at different rotation angles. The results reveal that all of the 10 centromeres in a given guard cell nucleus localize at the nuclear periphery and are distributed all around the nucleus. Although centromere 4 seems to be in the center of the nucleus in the 0° rotation, the 90° rotation shows that it is at the periphery (top) of the nucleus. Importantly, no obvious symmetric pattern can be observed in the two guard cells (Figures 1A′ and S1), which are descendents of the same mother cell.
Figure 1.
Maximum intensity projections of guard cells from different angles with centromere labeled by HTR12-Venus in green and chromatin labeled by Histone HTB1-CFP in magenta (see also Supplemental Video 1). (A) Projection image of guard cells from 20 image sections with z-interval 0.2 μm. Centromeres in one guard cell were arbitrarily numbered from 1 to 10. (A′) Diagram of positions of centromeres in A. The _x_-axis represents cell wall between two guard cells, the _y_-axis is vertical to the _x_-axis, and the _z_-axis is directed to the bottom of the guard cell nuclei. Centromeres were defined as brightest spot in the local fluorescent locus. Centromeres from sections 1–10 are represented by dark gray-scaled dots and highlighted by numbered arrows (1, 2, 4, 5, and 10). Centromeres from sections 11–20 are represented by light gray-scaled dots and highlighted by numbered arrows (3, 6, 7, 8, and 9). Also see Supplemental FigureS1. (B) Projection image of guard cells after 30° counterclockwise rotation along vertical axis. Centromeres (3, 6, 7, and 9) at the bottom side of the nuclei move left. Centromere 9 can be observed at the periphery of the projected nuclei. Centromeres (4 and 10) at the topside of the nuclei move right. (C) Projection image of guard cells after 60° counterclockwise rotation along the vertical axis. (D) Projection image of guard cells after 90° counterclockwise rotation along the vertical axis. Centromere 4 and centromere 7, which were at the center of the projected image at 0° (A), can be observed at the nuclear periphery of the projected nucleus. (E) Projection image of guard cells after 120° counterclockwise rotation along the vertical axis. (F) Projection image of guard cells after 150° counterclockwise rotation along the vertical axis. (G) Projection image of guard cells after 180° counterclockwise rotation along vertical axis. Bar, 5 μm.
Spatial positioning of Arabidopsis centromeres was quantified in several diploid cell types (see Materials and Methods and Figure 2), including guard cells (I, 278; II, 3; and III, 2) (nuclear number n = 30), small leaf epidermal pavement cells (I, 212; II, 3; and III, 4) (n = 25), root meristematic cells (I, 234; II, 3; and III, 0) (n = 25), and sepal and petal epidermal pavement cells (I, 167; II, 2; and III, 1) (n = 18). The results demonstrated that centromeres in all of these cell types localize predominantly at the nuclear periphery.
Organization of Centromeres in Endoreduplicated Cell Types
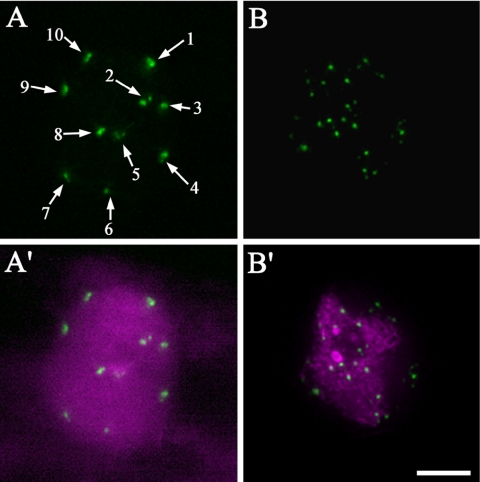
Because endoreduplicated cells contain a significantly larger number of chromosomes, it is of interest to investigate how sister centromeres are organized in such cells. We studied the organization of centromeres in larger leaf epidermal pavement cells and root epidermal cells in the differentiated region. These cells have larger and elongated nuclei with ploidy levels more than 2C (Melaragno et al., 1993). In the nuclei of endoreduplicated root epidermal cells, typically 10 fluorescent foci could be detected (Figure 3A, arrows). Compared with the size of the fluorescent spots in 2C cell types (Figure 1), the tagged centromere foci in root epidermal cells are normally bigger, and some foci with irregular shapes are composed of two to three clustered fluorescent spots (Figure 3A, arrows 1, 4, and 10), indicating that the sister centromeres originating from endomitosis are clustered in root epidermal cells. Interestingly, in larger leaf epidermal pavement cells, 20–80 smaller foci could be detected (Figure 3, B and B′), indicating that endoreduplicated sister centromeres in these cells are more disassociated than those in the root epidermal cells (Figure 3, A and A′).
Figure 3.
Centromere organization in endoreduplicated cell types. (A) Centromeres in a larger root epidermal cell. (A′) Overlay of centromeres in green and chromatin in magenta. (B) Centromeres in a larger leaf epidermal pavement cell. (B′) Overlay of centromeres in green and chromatin in magenta. Bar, 5 μm.
Dynamics of Arabidopsis Centromeres in Interphase
Time-lapse 3-D image stacks were collected from root tips of transgenic seedlings growing in slide chambers to study the dynamics of centromeres during interphase. In initial experiments, we found that long-term excitation in the CFP channel caused bleaching of centromeric signals in the Venus channel. Therefore, we labeled the centromeres with HTR12-GFP, which is more stable for imaging. For GFP single-channel imaging, the low degree of nucleoplasmic signal of HTR12-GFP served as a reference for nuclear shape.
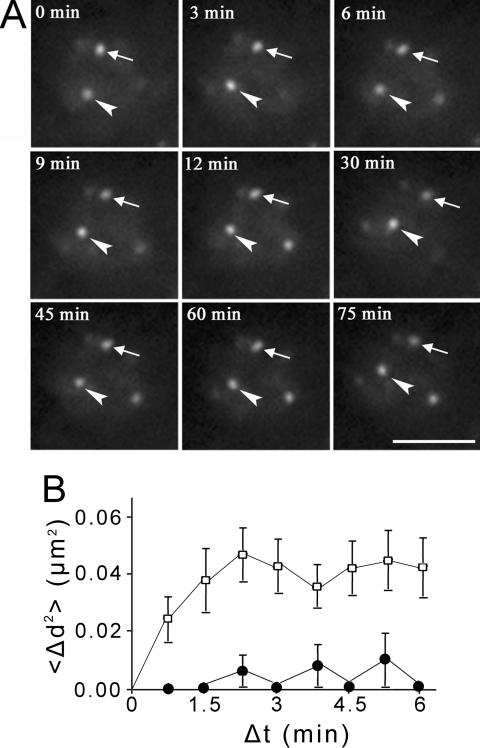
Because roots were growing during the imaging time, cell tracking was applied to center the cells of interest. The small nuclear size and peripheral positioning of centromeres in Arabidopsis resulted in larger relative movements in projection images due to nuclear rotation. To characterize centromere dynamics in living plants with severe nuclear rotations, we measured the distance between two centromeres in a single image section, in which the centromeres were sustained in the focal plane (Figure 4A and Supplemental Video 2). Centromere motion was represented by plotting the overall mean square change in distance <Δ_d_2> between two centromeres against elapsed time interval Δt (Vazquez et al., 2001). If the two centromeres move freely, a plot of <_Δd2_> against Δt should increase continuously. If the movement is constrained to a certain area, the plot will plateau in a limited time. We measured 24 pairs of centromeres in a total of 15 nuclei. The <Δ_d_2> values reach an average plateau of ∼0.042 μm2 in ∼2 min 15 s (Figure 4B), reflecting that the centromeres undergo diffusive movement in constrained areas within the nucleus with a mean change in distance between centromeres of ∼0.21 μm and a diffusion coefficient of 7.78 × 10-5 μm2/s. A plot of <Δ_d_2> for fixed seedlings indicates that the experimental noise in our microscope system is minimal. We then observed centromere movements in other cell types including leaf guard cells and sepal and petal pavement cells. Unlike cells in root tips growing on chambered coverglass, these cell types are not compatible with long-term imaging when mounted in water or MS medium. However, during an imaging period of up to 10 min, we found that centromeres in these cell types are also essentially stationary (our unpublished data), consistent with the results from root meristematic cells.
Figure 4.
Dynamics of centromeres in interphase. (A) Time-lapse single z-section images of a root meristematic cell in interphase. The two centromeres remaining in the focal plane during the imaging period are highlighted with an arrow and arrowhead. The elapsed time is indicated at the top left corner of each image (also see Supplemental Video 2). Bar, 5 μm. (B) Overall mean squared displacement in distance between two centromeres <Δ_d_2> plotted against elapsed time <_Δt_>. Unfilled squares (□) represent data from living cells and filled circles (•)represent data from fixed samples. Vertical bars at each time point show the SEs for the particular set of data collected in five independent experiments.
Three-dimensional Positioning of Centromeres through Mitosis
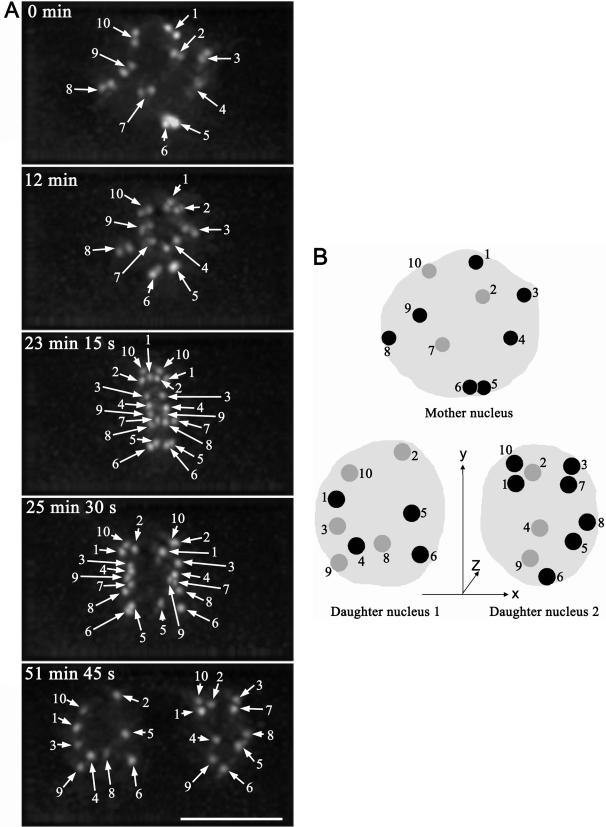
The three-dimensional positions of centromeres in the mother cell and the two daughter cells were investigated to ascertain whether global centromeric position is transmitted to daughter cells. To track centromere positions through mitosis, we collected time-lapse 3-D image stacks of Arabidopsis root meristematic cells. To minimize photo-toxicity due to the excitation light, we collect data using a short exposure time (0.06 s) for each section (see Materials and Methods). The cells were followed as they progressed through mitosis and the root growth rate at the beginning of imaging was similar to that at the end of imaging (our unpublished data), indicating that our imaging conditions were appropriate. During mitosis, possibly due to anchoring of microtubules, we observed much less rotation of centromeres than that in interphase nuclei, making centromere tracking less complicated. We could unambiguously follow the movements of all the visible centromeres through mitosis in about half of the mitotic events observed.
We analyzed nine complete in vivo mitotic events. Figure 5A shows a typical distribution of centromeres through mitosis in Arabidopsis root meristemic cells (also see Supplemental Video 3). From prophase to metaphase, upon breaking down of nuclear envelope, each pair of sister centromeres rotated at a different angle gradually to become oriented perpendicular to the metaphase plate (compare the angles of sister centromeres in Figure 5A from 0 to 23 min 15 s and corresponding Supplemental Video 3). Centromere order perpendicular to the spindle axis was partially preserved along the metaphase plate (Figure 5A). For example, in the G2/prophase mother cell the sequence of centromeres from top to bottom is 1–10-2–3-9–8-4(7)-5(6) (Figure 5A, 0 min), whereas in metaphase (Figure 5A, 23 min 15 s), the sequence is 10–1-2–3-4–9-7–8-5–6, indicating some repositioning of chromosomes upon formation of the metaphase plate. In contrast, the spatial information of centromeres along the spindle axis was totally lost upon superimposition of these sister centromeres to the metaphase plate (Figure 5A, 23 min 15 s, compare with Figure 5A, 0 min).
Figure 5.
Tracking centromeres through mitosis in 3-D (also see Supplemental Video 3). (A) Projection images of nuclei of a mitotic cell at elapse time of 0 min, 12 min, 23 min 15 s, 25 min 30 s, and 51 min 45 s. Centromeres are highlighted with numbered arrows at each time point. Bar, 5 μm. (B) Diagram of positions of centromeres in nuclei of mother cell and two daughter cells. The _z_-axis is directed to the bottom of the nuclei. Centromeres from the top four sections of the nuclei are represented by numbered dark gray-scaled dots, and centromeres from the bottom three sections of the nuclei are represented by numbered light gray-scaled dots (also see Supplemental Figure S2).
The movements of the same set of centromeres were followed until early G1 to locate where the paired sister centromeres resided after they were pulled apart (Figure 5A, from 23 min 15 s to 51 min 45 s; also see Supplemental Video 3). In early anaphase, sister centromeres start to move toward opposite poles; however, movement is not synchronous because some move early (for example, Figure 5A, 25 min 30 s, centromere 1 in left and centromeres 3 and 8 in right daughter nuclei), and others move later (for example, Figure 5A, 25 min 30 s, centromeres 2 and 5 in left and centromeres 1 and 5 in right daughter nuclei). Importantly, the behavior of sister centromeres is not the same, as shown in Figure 5A (25 min 30 s): centromere 1 moves first toward the left daughter nucleus, whereas it moves later toward the right daughter nucleus; centromere 2 moves first toward the right daughter nucleus, whereas it moves later toward the left daughter nucleus. Although the lagging centromeres in early anaphase normally move to the proximal areas of the daughter nuclei (41/51) and advancing centromeres move to distal parts of daughter nuclei (44/53), lagging centromeres can move to the distal part of the daughter nuclei (see centromere 5 in the right daughter nuclei in Figure 5A, 25 min 30 s and 51 min 45 s), and the advancing centromeres can move to proximal parts of the daughter nuclei. In later telophase and early G1, centromeres expand isometrically upon the enlargement of the nuclear envelope of daughter nuclei without changing their neighborhood (Supplemental Video 3).
Figure 5B shows a cartoon of centromere localization in a mother cell nucleus (Figure 5A, 0 min) and its two daughter nuclei (Figure 5A, 51 min 45 s) in 3-D (see Supplemental Figure S2, which shows image sections of mother and daughter nuclei). Centromeres do not exhibit similar positioning between mother and daughter cells. For example, in the mother nucleus centromeres 3 and 4 are on one side of the nucleus and centromeres 8 and 9 are at the opposite side along the spindle axis, whereas in the nucleus of daughter cell 1, these four centromeres are on the same side of the nucleus, and in the nucleus of daughter cell 2, centromeres 3 and 8 are on one side and centromeres 4 and 9 are on the opposite side. Significant differences in 3-D centromere distribution between mother and daughter cell nuclei and clear asymmetry between the two daughter cell nuclei are observed (Figure 5B) (also see Supplemental Figure S3, A and B, and Video 4, which shows another example of the tracking of centromeres through mitosis). We conclude that the organization of centromeres in Arabidopsis nuclei is not passed down precisely from one cell to its descendents, but instead it is more plastic.
DISCUSSION
Chromosome Organization in Arabidopsis Interphase Nuclei
By in situ visualization, 3-D restoration, and quantitative analysis, we revealed that all centromeres localize predominantly at the nuclear periphery in different cell types in living Arabidopsis plants. Interestingly, Fransz et al. (2002) demonstrated most telomeres localize in the vicinity of the nucleolus. Together, the arrangement of Arabidopsis chromosomes in interphase nuclei seems predominantly like a radial pattern. In this distribution, centromeres and their associated pericentromeric heterochromatin localize at the nuclear periphery, and chromosome arms are directed toward the vicinity of the nucleolus where telomeres localize, resulting in the organization of chromosome territories. Chromosomal loops might be a feature of the euchromatin in chromosome territories, as observed for chromosome 4 (Fransz et al., 2002). The nucleolar-organizing regions of chromosomes 2 and 4 (and the knob in ecotype Columbia in our experiments) might result in more complicated arrangements of chromosomes 2 and 4 than that of chromosomes 1, 3, and 5. Tagging telomeres by telomere repeat-binding protein-GFP fusions has been a good approach to study the localization and dynamics of telomeres in human cells (Mattern et al., 2004); however, in vivo tagging of plant homologues of telomere repeat-binding proteins (AtTRP1 or AtTBP1) (Chen et al., 2001; Hwang et al., 2001) shows no detectable signal at telomeres (our unpublished data). Labeling individual chromatin loci by LacI-GFP/LacO tagging and quantitatively analyzing their localization at the genomic level will further elucidate genome organization in living plant cells (reviewed in Lam et al., 2004).
Although centromeres are localized at the nuclear periphery in all cell types examined, clear differences were observed in the organization of endoreduplicated sister centromeres between larger leaf and root epidermal cells, implying a tissue-specific centromere/chromatin organization because plant leaves and roots have distinct functions. Cell-type dependent differences in the organization of RNA processing factors in Arabidopsis further support this concept (Fang et al., 2004). In addition, a tissue-specific spatial organization of the genome was previously observed in animal tissues (Parada et al., 2005).
Stability of Centromere Position in Arabidopsis Interphase Nuclei
In mammalian cells, individual loci or centromeric regions undergo slow diffusional movement that is confined to a radius of <1 μm (Shelby et al., 1996; Abney et al., 1997; Chubb et al., 2002) and only rarely were single domains found to move 1–3 μm (Shelby et al., 1996; Tumbar and Belmont, 2001). Here, we have demonstrated that plant centromeres are confined to the nuclear periphery, and the movement of individual tagged chromatin loci also is constrained in diploid and endoreduplicated _Arabidopsis_ cells (Kato and Lam, 2003). Thus, a rather “structured” chromatin organization can be proposed for _Arabidopsis_ interphase nuclei. We observed a lower diffusion coefficient of centromeres at the nuclear envelope (7.78 × 10-5 μm2/s) than what was observed for chromatin loci at internal nuclear regions (>1.25 × 10-4μm2/s) (Kato and Lam, 2003), implying a possible interaction between centromeres and the nuclear envelope. Reduced chromatin mobility near the nucleoli and nuclear periphery has also been reported in mammalian cells and yeast (Heun et al., 2001; Chubb et al., 2002). The nuclear lamina in mammalian cells and Drosophila has been suggested to anchor chromatin domains (Paddy et al., 1990). However, no bona fide lamina seems to exist in yeast and plants, and Arabidopsis does not encode identifiable homologues of genes encoding for metazoan lamins or lamina-associated integral inner nuclear membrane proteins (Rose et al., 2004). Future studies should identify the interaction partners between centromeres and the Arabidopsis nuclear envelope and clarify whether disruption to normal centromere positioning affects gene expression, chromosome segregation, or other biological processes.
Flexibility of Centromere Positions through Mitosis
Individual Arabidopsis chromosomes occupy discrete nuclear regions termed chromosomal territories with little intermixing among territories (Pecinka et al., 2004), similar to what has been observed in mammalian cells (Manuelidis, 1985; Cremer et al., 1988; Lichter et al., 1988; Pinkel et al., 1988) and other plant species (Schwarzacher et al., 1989; Leitch et al., 1990). To assess chromosomal position, we tracked the set of Arabidopsis centromeres in 3-D through mitosis in root meristematic cells of living plants. Our results demonstrated that global centromere positions are not transmitted through mitosis, because significant changes in the organization of centromeres were observed during transmission of genetic information to daughter cells. This is supported by a recent report showing that chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis cells are predominantly random, except for NOR-bearing chromosomes (Pecinka et al., 2004). Moreover, as these plant cells are fully functional, both mother cell and daughter cells have the potential to develop into whole plants, suggesting that the epigenetic information is not lost upon changes in chromosome position during mitosis. Therefore, the relative centromere/chromosome territory positions at the nuclear periphery might not serve as important epigenetic markers, but instead they may function in anchoring the genome.
Although somewhat controversial in mammalian cells (Gerlich et al., 2003; Walter et al., 2003), centromere position in Arabidopsis does not seem to be absolutely transmitted to daughter cells during cell division. However, the global position of Arabidopsis centromeres at the nuclear periphery and their constrained movement is more highly conserved. In human cells, it was found that gene-dense chromosome 19 is localized at a more internal nuclear position compared with gene-poor chromosome 18 (Croft et al., 1999; Cremer et al., 2001; Tanabe et al., 2002). In a more detailed study of all human chromosomes, Bolzer et al. (2005) found that gene-poor chromatin domains form a layer beneath the nuclear envelope, whereas gene-dense chromatin is enriched in the nuclear interior. Therefore, the radial arrangement of chromosomes may play a role in gene regulation. It will be interesting to investigate the radial position of plant chromosomes between mother and daughter cells, and the possible role of nuclear positioning on gene expression. Our data also raise an intriguing possibility that flexibility in chromosome positioning may provide an opportunity for different chromosomes to be closely positioned allowing for recombination to occur among a broader range of the genome. This in turn would accommodate acceleration in the evolution of the genome and the biogenesis of new species.
In summary, our findings have demonstrated a specific positioning of Arabidopsis centromeres at the nuclear periphery in all cell types examined. Centromere movement is constrained suggesting that centromeres anchor the chromosomes within the nucleus. In addition, centromere position between mother and daughter nuclei was found to be variable, indicating that precise positioning of centromeres/chromosomes is not essential for gene expression. However, positioning of subchromosomal domains and nuclear bodies may exist, resulting in distinct local environments, which may affect the activities of individual gene or gene clusters.
Supplementary Material
[Supplementary Material]
Acknowledgments
We acknowledge Atsushi Miyawaki for providing the Venus/pCS2 clone, and Natalie Doetsch and Richard Jorgensen for providing vector pFGC5941. We thank Robert Martienssen, Tatsuya Hirano, and Jason Swedlow for critical reading the manuscript; and Christian Bacher, Constantin Kappel, Roland Eils, and members of the Spector laboratory for suggestions and discussions. This work was supported by Plant Genome Research Program Grant 0077617 from the National Science Foundation.
References
- Abney, J. R., Cutler, B., Fillbach, M. L., Axelrod, D., and Scalettar, B. A. (1997). Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J. Cell Biol. 137, 1459-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abranches, R., Beven, A. F., Aragon-Alcaide, L., and Shaw, P. J. (1998). Transcription sites are not correlated with chromosome territories in wheat nuclei. J. Cell Biol. 143, 5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agard, D. A., and Sedat, J. W. (1983). Three-dimensional architecture of a polytene nucleus. Nature 302, 676-681. [DOI] [PubMed] [Google Scholar]
- Appelgren, H., Kniola, B., and Ekwall, K. (2003). Distinct centromere domain structures with separate functions demonstrated in live fission yeast cells. J. Cell Sci. 116, 4035-4042. [DOI] [PubMed] [Google Scholar]
- Berg, H. C. (1993). Random Walks in Biology, Princeton, NJ: Princeton University Press.
- Bolzer, A., et al. (2005). Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol. 3, e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwitz, B. J., Ahmad, K., Moore, L. L., Roth, M. B., and Henikoff, S. (1999). A histone-H3-like protein in C. elegans. Nature 401, 547-548. [DOI] [PubMed] [Google Scholar]
- Chen, C. M., Wang, C. T., and Ho, C. H. (2001). A plant gene encoding a Myb-like protein that binds telomeric GGTTTAG repeats in vitro. J. Biol. Chem. 276, 16511-16519. [DOI] [PubMed] [Google Scholar]
- Chubb, J. R., Boyle, S., Perry, P., and Bickmore, W. A. (2002). Chromatin motion is constrained by association with nuclear compartments in human cells. Curr. Biol. 12, 439-445. [DOI] [PubMed] [Google Scholar]
- Chung, H. M., Shea, C., Fields, S., Taub, R. N., Van der Ploeg, L. H., and Tse, D. B. (1990). Architectural organization in the interphase nucleus of the protozoan Trypanosoma brucei: location of telomeres and mini-chromosomes. EMBO J. 9, 2611-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for _Agrobacterium_-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G. P., et al. (1999). Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286, 2468-2474. [DOI] [PubMed] [Google Scholar]
- Cremer, M., von Hase, J., Volm, T., Brero, A., Kreth, G., Walter, J., Fischer, C., Solovei, I., Cremer, C., and Cremer, T. (2001). Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res. 9, 541-567. [DOI] [PubMed] [Google Scholar]
- Cremer, T., Lichter, P., Borden, J., Ward, D. C., and Manuelidis, L. (1988). Detection of chromosome aberrations in metaphase and interphase tumor cells by in situ hybridization using chromosome specific library probes. Hum. Genet. 80, 235-246. [DOI] [PubMed] [Google Scholar]
- Croft, J. A., Bridger, J. M., Boyle, S., Perry, P., Teague, P., and Bickmore, W. A. (1999). Differences in the localization and morphology of chromosomes in the human nucleus. J. Cell Biol. 145, 1119-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doe, C. L., Wang, G., Chow, C., Fricker, M. D., Singh, P. B., and Mellor, E. J. (1998). The fission yeast chromo domain encoding gene chp1(+) is required for chromosome segregation and shows a genetic interaction with α-tubulin. Nucleic Acids Res. 26, 4222-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, F., and Jiang, J. (1998). Non-Rabl patterns of centromere and telomere distribution in the interphase nuclei of plant cells. Chromosome Res. 6, 551-558. [DOI] [PubMed] [Google Scholar]
- Fang, Y., Hearn, S., and Spector, D. L. (2004). Tissue-specific expression and dynamic organization of SR splicing factors in Arabidopsis. Mol. Biol. Cell 15, 2664-2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransz, P., De Jong, J. H., Lysak, M., Castiglione, M. R., and Schubert, I. (2002). Interphase chromosomes in Arabidopsis are organized as well defined chromocenters from which euchromatin loops emanate. Proc. Natl. Acad. Sci. USA 99, 14584-14589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki, H., Hagan, I., Uzawa, S., and Yanagida, M. (1993). Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J. Cell Biol. 121, 961-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlich, D., Beaudouin, J., Kalbfuss, B., Daigle, N., Eils, R., and Ellenberg, J. (2003). Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112, 751-764. [DOI] [PubMed] [Google Scholar]
- Hediger, F., Neumann, F. R., Van Houwe, G., Dubrana, K., and Gasser, S. M. (2002). Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr. Biol. 12, 2076-2089. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., Ahmad, K., Platero, J. S., and van Steensel, B. (2000). Heterochromatic deposition of centromeric histone H3-like proteins. Proc. Natl. Acad. Sci. USA 97, 716-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heslop-Harrison, J. S., Brandes, A., and Schwarzacher, T. (2003). Tandemly repeated DNA sequences and centromeric chromosomal regions of Arabidopsis species. Chromosome Res. 11, 241-253. [DOI] [PubMed] [Google Scholar]
- Heun, P., Laroche, T., Shimada, K., Furrer, P., and Gasser, S. M. (2001). Chromosome dynamics in the yeast interphase nucleus. Science 294, 2181-2186. [DOI] [PubMed] [Google Scholar]
- Hochstrasser, M., Mathog, D., Gruenbaum, Y., Saumweber, H., and Sedat, J. W. (1986). Spatial organization of chromosomes in the salivary gland nuclei of Drosophila melanogaster. J. Cell Biol. 102, 112-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, M. G., Chung, I. K., Kang, B. G., and Cho, M. H. (2001). Sequence-specific binding property of Arabidopsis thaliana telomeric DNA binding protein 1 (AtTBP1). FEBS Lett. 503, 35-40. [DOI] [PubMed] [Google Scholar]
- Jiang, J., Birchler, J. A., Parrott, W. A., and Dawe, R. K. (2003). A molecular view of plant centromeres. Trends Plant Sci. 8, 570-575. [DOI] [PubMed] [Google Scholar]
- Kato, N., and Lam, E. (2003). Chromatin of endoreduplicated pavement cells has greater range of movement than that of diploid guard cells in Arabidopsis thaliana. J. Cell Sci. 116, 2195-2201. [DOI] [PubMed] [Google Scholar]
- Kniola, B., O'Toole, E., McIntosh, J. R., Mellone, B., Allshire, R., Mengarelli, S., Hultenby, K., and Ekwall, K. (2001). The domain structure of centromeres is conserved from fission yeast to humans. Mol. Biol. Cell 12, 2767-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, E., Kato, N., and Watanabe, K. (2004). Visualizing chromosome structure/organization. Annu. Rev. Plant Biol. 55, 537-554. [DOI] [PubMed] [Google Scholar]
- Leitch, A. R., Mosgrller, W., Sehwarzacher, T., Bennett, M. D., and Heslop-Harrison, J. S. (1990). Genomic in situ hybridization to sectioned nuclei shows chromosome domains in grass hybrids. J. Cell Sci. 95, 335-341. [DOI] [PubMed] [Google Scholar]
- Lichter, P., Cremer, T., Borden, J., Manuelidis, L., and Ward, D. C. (1988). Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum. Genet. 80, 224-234. [DOI] [PubMed] [Google Scholar]
- Luderus, M. E., van Steensel, B., Chong, L., Sibon, O. C., Cremers, F. F., and de Lange, T. (1996). Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J. Cell Biol. 135, 867-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuelidis, L. (1985). Individual interphase chromosome domains revealed by in situ hybridization. Hum. Genet. 71, 288-293. [DOI] [PubMed] [Google Scholar]
- Marshall, W. F., Dernburg, A. F., Harmon, B., Agard, D. A., and Sedat, J. W. (1996). Specific interactions of chromatin with the nuclear envelope: positional determination within the nucleus in Drosophila melanogaster. Mol. Biol. Cell 7, 825-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattern, K. A., Swiggers, S. J., Nigg, A. L., Lowenberg, B., Houtsmuller, A. B., and Zijlmans, J. M. (2004). Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function. Mol. Cell. Biol. 24, 5587-5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melaragno, J. E., Mehrotra, B., and Coleman, A. W. (1993). Relationship between endopolyploidy and cell size in epidermal tissue of Arabidopsis. Plant Cell 5, 1661-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, T., Ibata, K., Park, E. S., Kubota, M., Mikoshiba, K., and Miyawaki, A. (2002). A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87-90. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., and Murata, M. (2005). Characterization of CENH3 and centromere-associated DNA sequences in sugarcane. Chromosome Res. 13, 195-203. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Cheng, Z., Ouyang, S., Talbert, P. B., Kim, M., Jones, K. M., Henikoff, S., Buell, C. R., and Jiang, J. (2004). Sequencing of a rice centromere uncovers active genes. Nat. Genet. 36, 138-145. [DOI] [PubMed] [Google Scholar]
- Nagaki, K., Kashihara, K., and Murata, M. (2005). Visualization of diffuse centromeres with centromere-specific histone H3 in the holocentric plant Luzula nivea. Plant Cell 17, 1886-1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddy, M. R., Belmont, A. S., Saumweber, H., Agard, D. A., and Sedat, J. W. (1990). Interphase nuclear envelope lamins form a discontinuous network that interacts with only a fraction of the chromatin in the nuclear periphery. Cell 62, 89-106. [DOI] [PubMed] [Google Scholar]
- Palmer, D. K., O'Day, K., Wener, M. H., Andrews, B. S., and Margolis, R. L. (1987). A 17-kD centromere protein (CENP-A) copurifies with nucleosome core particles and with histones. J. Cell Biol. 104, 805-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada, L. A., McQueen, P. G., and Misteli, T. (2005). Tissue-specific spatial organization of genomes. Genome Biol. 5, R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecinka, A., Schubert, V., Meister, A., Kreth, G., Klatte, M., Lysak, M. A., Fuchs, J., and Schubert, I. (2004). Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113, 258-269. [DOI] [PubMed] [Google Scholar]
- Pinkel, D., Landegent, J., Collins, C., Fuscoe, J., Segraves, R., Lucas, J., and Gray, J. W. (1988). Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc. Natl. Acad. Sci. USA 85, 9138-9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl, C. (1885). Ueber Zelltheilung. Morphol. Jahrbuch 10, 214-330. [Google Scholar]
- Rose, A., Patel, S., and Meier, I. (2004). The plant nuclear envelope. Planta 218, 327-336. [DOI] [PubMed] [Google Scholar]
- Schwarzacher, T., Leitch, A. R., Bennett, M. D., and Heslop-Harrison, J. S. (1989). In situ localization of parental genomes in a wide hybrid. Ann. Bot. 64, 315-324. [Google Scholar]
- Shelby, R. D., Hahn, K. M., and Sullivan, K. F. (1996). Dynamic elastic behavior of alpha-satellite DNA domains visualized in situ in living human cells. J. Cell Biol. 135, 545-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata, F., and Murata, M. (2004). Differential localization of the centromere-specific proteins in the major centromeric satellite of Arabidopsis thaliana. J. Cell Sci. 117, 2963-2970. [DOI] [PubMed] [Google Scholar]
- Spector, D. L. (2003). The dynamics of chromosome organization and gene regulation. Annu. Rev. Biochem. 72, 573-608. [DOI] [PubMed] [Google Scholar]
- Stack, S. M., and Clark, C. R. (1974). Chromosome polarization and nuclear rotation in Allium cepa roots. Cytologia 39, 553-560. [Google Scholar]
- Stoler, S., Keith, K. C., Curnick, K. E., and Fitzgerald-Hayes, M. (1995). A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9, 573-586. [DOI] [PubMed] [Google Scholar]
- Talbert, P. B., Masuelli, R., Tyagi, A. P., Comai, L., and Henikoff, S. (2002). Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell 14, 1053-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe, H., Muller, S., Neusser, M., von Hase, J., Calcagno, E., Cremer, M., Solovei, I., Cremer, C., and Cremer, T. (2002). Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc. Natl. Acad. Sci. USA 99, 4424-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson, I., Gilchrist, S., Bickmore, W. A., and Chubb, J. R. (2004). The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr. Biol. 14, 166-172. [DOI] [PubMed] [Google Scholar]
- Tumbar, T., and Belmont, A. S. (2001). Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat. Cell Biol. 3, 134-139. [DOI] [PubMed] [Google Scholar]
- Vazquez, J., Belmont, A. S., and Sedat, J. W. (2001). Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 11, 1227-1239. [DOI] [PubMed] [Google Scholar]
- Walter, J., Schermelleh, L., Cremer, M., Tashiro, S., and Cremer, T. (2003). Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J. Cell Biol. 160, 685-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegel, E., and Shaw, P. J. (2005). Chromosome organization in wheat endosperm and embryo. Cytogenet. Genome Res. 109, 175-180. [DOI] [PubMed] [Google Scholar]
- Zhang, X., Li, X., Marshall, J. B., Zhong, C. X., and Dawe, R. K. (2005). Phosphoserines on maize CENH3 and histone H3 define the centromere and pericentromere during chromosome segregation. Plant Cell 17, 572-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, C. X., Marshall, J. B., Topp, C., Mroczek, R., Kato, A., Nagaki, K., Birchler, J. A., Jiang, J., and Dawe, R. K. (2002). Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell 14, 2825-2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Material]