ExpressYourself: a modular platform for processing and visualizing microarray data (original) (raw)
Abstract
DNA microarrays are widely used in biological research; by analyzing differential hybridization on a single microarray slide, one can detect changes in mRNA expression levels, increases in DNA copy numbers and the location of transcription factor binding sites on a genomic scale. Having performed the experiments, the major challenge is to process large, noisy datasets in order to identify the specific array elements that are significantly differentially hybridized. This normally requires aggregating different, often incompatible programs into a multi-step pipeline. Here we present ExpressYourself, a fully integrated platform for processing microarray data. In completely automated fashion, it will correct the background array signal, normalize the Cy5 and Cy3 signals, score levels of differential hybridization, combine the results of replicate experiments, filter problematic regions of the array and assess the quality of individual and replicate experiments. ExpressYourself is designed with a highly modular architecture so various types of microarray analysis algorithms can readily be incorporated as they are developed; for example, the system currently implements several normalization methods, including those that simultaneously consider signal intensity and slide location. The processed data are presented using a web-based graphical interface to facilitate comparison with the original images of the array slides. In particular, Express Yourself is able to regenerate images of the original microarray after applying various steps of processing, which greatly facilities identification of position-specific artifacts. The program is freely available for use at http://bioinfo.mbb.yale.edu/expressyourself.
INTRODUCTION
Microarrays are widely employed, among other uses, to compare mRNA expression levels (1–5), DNA copy number (6–9) and transcription factor binding in biological samples (10–13). The concept underlying these experiments is straightforward; fluorescence-labeled nucleic acids in ‘test’ and ‘reference’ samples are probed simultaneously on a microarray slide, and their relative abundance is derived from the comparative fluorescence of the probe molecules hybridized to individual array elements. Though the technology is relatively new, several aspects of data analysis beyond the experimental stage are now well established; these include scanning the arrays to measure fluorescence intensity, quantifying the array images via densitometry algorithms (14,15), clustering similarly expressed genes (16–20) and integrating microarray data with genomic information (21–28). However, a topic still under much discussion is how to treat the raw numerical data immediately after scanning and quantifying the array images (27,29).
Data processing aims to fill this gap. In particular it serves three purposes: (i) to detect and minimize the level of noise associated with the experiments; (ii) to assess the quality of the data once the noise has been reduced; and (iii) to identify the array elements that are actually differentially hybridized.
Here we present ExpressYourself, an automated platform for processing microarray data that is freely available over the web (http://bioinfo.mbb.yale.edu/expressyourself). The software performs correction of the background array signal, normalization, scoring, combination of replicate experiments, filtering problematic regions of the array and quality assessment of hybridizations. We incorporate novel and published algorithms that are reasonable, understandable and make minimal assumptions about the data. The program can handle gene expression, chromatin immunoprecipitated DNA probings (ChIp-chip) and most comparative genomic hybridization (CGH) data. The results are clear and easy to understand, and the graphical interface allows users to compare each processing step with the original slide images.
DATA PROCESSING IN ExpressYourself
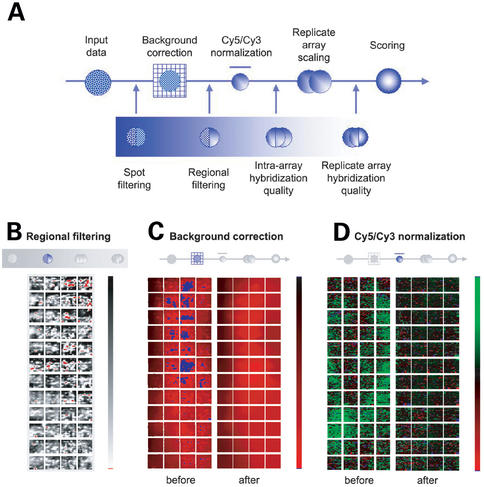
ExpressYourself processes the data in a sequential manner using the major steps shown in Figure 1A. The stages can be broadly grouped into: (i) noise reduction; (ii) quality control; and (iii) differential hybridization scoring. We demonstrate the use of ExpressYourself using the data from a ChIp-chip experiment of the HCM1 transcription factor (30).
Figure 1.
(A) Flow chart of data processing. Schematic images of a microarray slide depicting different stages of data processing: (B) filtering flawed array regions, (C) the effects of background correction, and (D) outcome of normalization.
Input data
The data input to ExpressYourself comprise text files generated by image analysis software. Currently, the program recognizes files from Axon GenePix versions 2.0–4.0 (http://www.axon.com/GN_GenePixSoftware.html), Scanalyze version 2.0 (http://rana.lbl.gov/EisenSoftware.htm) and UCSF SPOT version 2.0 (15) and produces the best results if input files are left intact (i.e. no data is deleted). Multiple files are accepted and may represent information for replicate experiments. The processing steps to be applied to the data may be changed by altering the parameters at this stage.
We interpret the Cy5 and Cy3 signals of an array element as the median foreground minus background intensities for each dye (_S_=_I_foreground−_I_background). The foreground intensity is the fluorescence of a spot within a defined area, usually described by a circle enclosing the spot, and the background is that of the immediate area surrounding the spot, usually described by a bounding box. The level of differential hybridization at each array element is determined as the relative signal between the two dyes.
Noise reduction
Individual spot and regional filtering.
Technological limitations in array production and experimental techniques mean that microarray slides are often imperfect. Nearly every experiment contains individual array elements of poor quality, comprising spots that are small compared to the rest of the array, have unusual morphology (i.e. non-round), exhibit uneven hybridization (i.e. doughnut or crescent-moon patterns) or have saturated signal intensity. Most image analysis software permits users to flag such array elements manually. But with up to 40 000 spots per slide, this is very time-consuming and difficult to perform in a consistent manner. ExpressYourself automatically flags and, if necessary, removes poor quality spots. Manual flagging is therefore unnecessary, although the program will consider such flags if instructed by the user. Imperfections on the array can also extend beyond individual spots. Large dust particles, printing inconsistencies and scratches sometimes render entire regions of the array unusable. ExpressYourself detects and removes these flaws automatically (Fig. 1B).
Background correction.
Although we remove obvious defects before further processing, it is important, if possible, to correct minor imperfections confined to small areas so that we preserve the maximum amount of usable data (14,15,31,32). As mentioned above, the background signal is commonly defined as the average intensity of the immediate area surrounding each array element. Minor imperfections (very small specks, dust and scratches limited to the vicinity of a spot) often distort background signals, making them extremely variable even between adjacent spots. Therefore the aim of background correction is to reduce the local background distortions that are restricted to single array elements, while maintaining the overall variability represented by gradual changes between bright and dark regions across the slide. We overcome this problem by calculating the average background signal from a wider area, typically spanning 3×3 to 5×5 spots (31). In doing so, we minimize the contribution of minor flaws to the background signal, and we remove most of the local distortions. Figure 1C displays the effects of correcting the Cy5 background intensity. Many regions of local variability are removed, but overall variation in array intensity remains.
Cy5/Cy3 normalization.
Once the signal intensities have been calculated using the corrected background, we can compare the relative contributions of the Cy5 and Cy3 signals. Ideally, the signals of the two dyes should be equal for nucleic acid probes that have equal concentration in the test and reference samples (i.e. the ratio, _R_=_S_1/_S_2, of the two signals should approach 1 for probes hybridizing to an equal degree in both fluorescence channels). In practice, the signals can be quite different. Dyes have different molecular characteristics, hybridization to the arrays can be non-specific or incomplete, and there is spatial heterogeneity in the probing conditions across the slide. Normalization aims to compensate for these effects by applying a scale factor such that signals of probes with unchanged concentration are equal (29,31,33–39). The signals of the remaining array elements are scaled relative to the baseline set for the constant probes. Figure 1D shows a schematic of the example array before and after it has been normalized.
A major issue in microarray normalization lies in defining the set of constant probes and this is reflected in the many approaches that have been published, including the use of house-keeping genes, spiked controls and total nucleic acid concentrations (see 31 for an overview). We prefer to use the ‘constant majority’ method, which assumes that the majority of probes do not change in concentration. The method is generally applicable to many experiments as it is valid even in cases where up to 50% of probes have altered concentrations, does not require prior knowledge of which probes remain constant and allows for intensity and spatial considerations (see below).
At its simplest the method calculates the scale factor from the robust mean of all _S_1/_S_2 ratios, i.e. the distribution of all ratios is transformed so that it centers about 1. However, two particular issues must be addressed: signal intensity and array position. First, because the two dyes differ in fluorescent properties, the bias in ratios often depends on the signal intensity (29,37–40). Therefore, different scale factors must be used for array elements at different intensities. Second, the positional issue is due to differences in hybridization conditions across the slide (31,35,40), and it is common to observe array images in which hybridization of entire regions is dominated by one dye. Thus different scale factors must be used for different regions of the physical slide. To determine scale factors in each situation we employ local regression to determine a ‘best fit’ for the data, using the LOWESS and LOESS packages (41–43). In the former case, we calculate the local mean intensity ratio as it varies over a range of signals in two dimensions (Cy5 versus Cy3). In the latter case, we determine the mean ratio as it varies across the surface of the microarray slide by fitting a three-dimensional curve to the data points.
Replicate array scaling.
Many array experiments are conducted in replicates; however, differences in sample concentrations, probing conditions and scanner settings mean that the range of signal intensities can be quite variable. Prior to combining replicate experiments, we calculate the robust standard deviations of signals in each experiment and scale each so the widths of signal distributions are equal.
Quality control
It is useful to have an objective measure of data quality (Fig. 1A) (31,33,39,44,45). Firstly, it allows the user to see how well the experiment has performed as this is not usually obvious from visual inspection of the slide images. Secondly, it assesses the degree to which the noise reduction steps have improved the data. Finally, by identifying the most serious problems, the user can modify future experiments to improve results. Here we introduce some of the data quality measures that we have incorporated into ExpressYourself to date.
Percentage of good quality array elements.
The simplest quality metric is a basic calculation of the proportion of array elements and regions the filtering process has removed; the larger the proportion, the poorer the quality of experiment. By breaking down the numbers according to error type (e.g. spot diameter, homogeneity, saturation), we can determine the defective properties that are most problematic for a given array.
Intra-array hybridization quality.
Many microarrays are designed with spots printed in duplicate, side-by-side. We gauge the consistency of hybridizations within the array by measuring the difference in signals between these duplicates [e.g. _D_=(_R_dup1−_R_dup2)/(_R_dup1+_R_dup2)]. The mean of _D_2, 〈_D_2〉, then summarizes the consistency of hybridization within the array. Since we expect 〈_D_〉=0 then Var(D)=∑(D _i_−0)2/_N_=〈_D_2〉 so the consistency of hybridization can also be visualized as the width of the distribution of D.
Replicate array hybridization quality.
We extend this measure to determine the consistency of replicate experiments, by calculating the difference in signals between equivalent spots across multiple slides. We construct an analogous quality score D _i_′=(_R_α,_i_−_R_β,i)/(_R_α,i+_R_β,i) for spot i on slides α and β. Again, the width of the distribution of _D_′ measures the quality of an experiment with respect to others and allows users to decide whether the entire experiment should be removed from the dataset. Values for _D_′ can also be used to identify regions of a slide that are of particularly poor quality.
Scoring differentially hybridized array elements
The final step is to identify array elements that exhibit differential hybridization (Fig. 1A). These ultimately correspond to those genes that have altered expression levels, chromosomal regions that have changed copy number or the locations of transcription factor binding sites, depending on the nature of the experiment. The major issue is to single out spots whose relative Cy5-Cy3 signals stand out from the experimental noise at sufficient statistical significance.
ExpressYourself currently incorporates three scoring methods. The most simplistic and widely used approach is to define a ratio cut-off and identify the probes that exhibit fold changes greater than this threshold (3,46–48). Another popular approach is to use variations of Student's paired _t_-test to compare all signals from the test and reference samples (49–51). Differentially hybridized spots are identified as those exhibiting a _p_-value less than a user-specified cut-off. We also include a novel method for scoring differential hybridization (Fig. 2B; manuscript in preparation). We standardize each spot's ratio by dividing it with a local standard deviation; this deviation is determined as a function of the spot's total intensity (_S_1+_S_2). The standardized ratios are fit to a distribution and outliers at a user-defined _p_-value are identified as being differentially hybridized. The outliers are removed from the dataset and the entire process is repeated with the new, smaller set of spots. The iteration continues until no new outliers are detected.
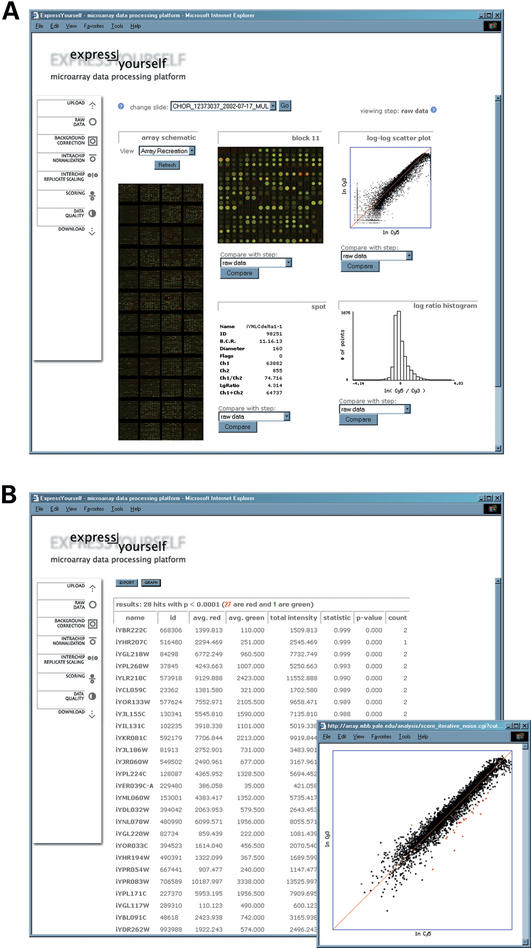
Figure 2.
Screenshot of: (A) the main, and (B) scoring pages of ExpressYourself.
THE USER INTERFACE FOR ExpressYourself
ExpressYourself is accessed using a web browser and Figure 2 displays elements of the user interface. The toolbar allows users to view the data at different stages of processing and the corresponding output is presented in the main area of the web page (Fig. 2A). In the centre of the display, we recreate the slide image using values from the input file, and it is updated through each processing step. Specific regions of the slide can be viewed in detail by clicking on the area of interest. Selecting individual spots can access data associated with each array element (e.g. name of array element, diameter, signal intensities and data quality flagging). Distributions of the Cy5 and Cy3 signals are displayed at the right side of the page. The scoring page lists the differentially hybridized spots that are considered statistically significant and also displays them as graphical plots (Fig. 2B). The user can download the results in a text file for further analysis. The aim of the graphical interface is to enable users to visualize the data in the context of a microarray slide and statistical distributions. It facilitates comparisons of the processed data with the original slide images and allows them to track changes to spots of interest. In additional data quality pages, the schematics are particularly useful for uncovering position specific artifacts on the microarray slide.
DATA DOWNLOAD
Processed data can be downloaded by the user as text files; these include array signal intensities after each processing step, a list of array elements that are differentially hybridized along with significance scores, and the results of data quality analyses including flagging (−50 for poor quality array elements and 0 for good quality elements).
CONCLUSIONS
Summary
Here we presented ExpressYourself, a web-based program for processing microarray data. We have incorporated novel and published algorithms to reduce the experimental noise, assess the quality of the data and identify differentially hybridized array elements. The program can process data from most gene expression, ChIp-chip and CGH experiments. The results are clear and the graphical interface allows immediate identification of the most important features of the experiment.
Future improvements
ExpressYourself is continually updated as better processing methods are developed both within and outside our laboratory. Immediate plans include addition of alternative normalization methods, clustering and a visual tool linking array images to genomic features, given a corresponding microarray designed to map chromosomal loci. We also have future plans for improved scoring schemes and more advanced methods for combining the data from replicate experiments.
AVAILABILITY
ExpressYourself is freely accessible for use at http://bioinfo.mbb.yale.edu/expressyourself. The program is written in C and Perl and may be installed on any web server for local use. Enquiries can be made to nicholas.luscombe@yale.edu.
ExpressYourself currently accepts input files in GenePix Pro versions 2.0–4.0, Scanalyze version 2.0, or UCSF SPOT version 2.0 format. The processing steps to be applied to the data may be changed by altering the parameters at the input stage. The program and its outputs are accessible using any modern web browser (Explorer 6.0, Netscape 7.0 or Mozilla 1.3) and text-based results can be downloaded for further analysis.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to the Snyder and Stern laboratories for sample datasets and their assistance in testing the software, Paul Lizardi and Michelle Lacey for useful discussions. N.M.L. is sponsored by the Anna Fuller Fund and M.G. acknowledges support from the Keck Foundation. P.B., M.S. and M.G. are supported in part by NIH grants P50 HG02357 and R01 CA77808.
REFERENCES
- 1.Schena M., Shalon,D., Davis,R.W. and Brown,P.O. (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science, 270, 467–470. [DOI] [PubMed] [Google Scholar]
- 2.DeRisi J., Penland,L., Brown,P.O., Bittner,M.L., Meltzer,P.S., Ray,M., Chen,Y., Su,Y.A. and Trent,J.M. (1996) Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nature Genet., 14, 457–460. [DOI] [PubMed] [Google Scholar]
- 3.DeRisi J.L., Iyer,V.R. and Brown,P.O. (1997) Exploring the metabolic and genetic control of gene expression on a genomic scale. Science, 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 4.Watson A., Mazumder,A., Stewart,M. and Balasubramanian,S. (1998) Technology for microarray analysis of gene expression. Curr. Opin. Biotechnol., 9, 609–614. [DOI] [PubMed] [Google Scholar]
- 5.Brown P.O. and Botstein,D. (1999) Exploring the new world of the genome with DNA microarrays. Nature Genet., 21, 33–37. [DOI] [PubMed] [Google Scholar]
- 6.Forozan F., Karhu,R., Kononen,J., Kallioniemi,A. and Kallioniemi,O.P. (1997) Genome screening by comparative genomic hybridization. Trends Genet., 13, 405–409. [DOI] [PubMed] [Google Scholar]
- 7.Pollack J.R., Perou,C.M., Alizadeh,A.A., Eisen,M.B., Pergamenschikov,A., Williams,C.F., Jeffrey,S.S., Botstein,D. and Brown,P.O. (1999) Genome-wide analysis of DA copy-number changes using cDNA microarrays. Nature Genet., 23, 41–46. [DOI] [PubMed] [Google Scholar]
- 8.Forozan F., Mahlamaki,E.H., Monni,O., Chen,Y., Veldman,R., Jiang,Y., Gooden,G.C., Ethier,S.P., Kallioniemi,A. and Kallioniemi,O.P. (2000) Comparative genomic hybridization analysis of 38 breast cancer cell lines: a basis for interpreting complementary DNA microarray data. Cancer Res., 60, 4519–4525. [PubMed] [Google Scholar]
- 9.Kashiwagi H. and Uchida,K. (2000) Genome-wide profiling of gene amplification and deletion in cancer. Hum. Cell, 13, 135–141. [PubMed] [Google Scholar]
- 10.Ren B., Robert,F., Wyrick,J.J., Aparicio,O., Jennings,E.G., Simon,I., Zeitlinger,J., Schreiber,J., Hannett,N., Kanin,E. et al. (2000) Genome-wide location and function of DNA binding proteins. Science, 290, 2306–2309. [DOI] [PubMed] [Google Scholar]
- 11.Iyer V.R., Horak,C.E., Scafe,C.S., Botstein,D., Snyder,M. and Brown,P.O. (2001) Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature, 409, 533–538. [DOI] [PubMed] [Google Scholar]
- 12.Horak C.E. and Snyder,M. (2002) ChIP-chip: a genomic approach for identifying transcription factor binding sites. Methods Enzymol., 350, 469–483. [DOI] [PubMed] [Google Scholar]
- 13.Lee T.I., Rinaldi,N.J., Robert,F., Odom,D.T., Bar-Joseph,Z., Gerber,G.K., Hannett,N.M., Harbison,C.T., Thompson,C.M., Simon,I. et al. (2002) Transcriptional regulatory networks in Saccharomyces cerevisiae. Science, 298, 799–804. [DOI] [PubMed] [Google Scholar]
- 14.Yang Y.H., Buckley,M.J. and Speed,T.P. (2001) Analysis of cDNA microarray images. Brief Bioinform., 2, 341–349. [DOI] [PubMed] [Google Scholar]
- 15.Jain A.N., Tokuyasu,T.A., Snijders,A.M., Segraves,R., Albertson,D.G. and Pinkel,D. (2002) Fully automatic quantification of microarray image data. Genome Res., 12, 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eisen M.B., Spellman,P.T., Brown,P.O. and Botstein,D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA, 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alon U., Barkai,N., Notterman,D.A., Gish,K., Ybarra,S., Mack,D. and Levine,A.J. (1999) Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissues probed by oligonucleotide arrays. Proc. Natl Acad. Sci. USA, 96, 6745–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamayo P., Slonim,D., Mesirov,J., Zhu,Q., Kitareewan,S., Dmitrovsky,E., Lander,E.S. and Golub,T.R. (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl Acad. Sci. USA, 96, 2907–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tavazoie S., Hughes,J.D., Campbell,M.J., Cho,R.J. and Church,G.M. (1999) Systematic determination of genetic network architecture. Nature Genet., 22, 281–285. [DOI] [PubMed] [Google Scholar]
- 20.Toronen P., Kolehmainen,M., Wong,G. and Castren,E. (1999) Analysis of gene expression data using self-organizing maps. FEBS Lett., 451, 142–146. [DOI] [PubMed] [Google Scholar]
- 21.Drawid A., Jansen,R. and Gerstein,M. (2000) Genome-wide analysis relating expression level with protein subcellular localization. Trends Genet., 16, 426–430. [DOI] [PubMed] [Google Scholar]
- 22.Gerstein M. and Jansen,R. (2000) The current excitement in bioinformatics-analysis of whole-genome expression data: how does it relate to protein structure and function? Curr. Opin. Struct. Biol., 10, 574–584. [DOI] [PubMed] [Google Scholar]
- 23.Jansen R. and Gerstein,M. (2000) Analysis of the yeast transcriptome with structural and functional categories: characterizing highly expressed proteins. Nucleic Acids Res., 28, 1481–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenbaum D., Luscombe,N.M., Jansen,R., Qian,J. and Gerstein,M. (2001) Interrelating different types of genomic data, from proteome to secretome: ‘oming in on function. Genome Res., 11, 1463–1468. [DOI] [PubMed] [Google Scholar]
- 25.Greenbaum D., Jansen,R. and Gerstein,M. (2002) Analysis of mRNA expression and protein abundance data: an approach for the comparison of the enrichment of features in the cellular population of proteins and transcripts. Bioinformatics, 18, 585–596. [DOI] [PubMed] [Google Scholar]
- 26.Jansen R., Greenbaum,D. and Gerstein,M. (2002) Relating whole-genome expression data with protein-protein interactions. Genome Res., 12, 37–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Slonim D.K. (2002) From patterns to pathways: gene expression data analysis comes of age. Nature Genet., 32(Suppl. 2), 502–508. [DOI] [PubMed] [Google Scholar]
- 28.Reinke V. (2002) Functional exploration of the C. elegans genome using DNA microarrays. Nature Genet., 32(Suppl. 2), 541–546. [DOI] [PubMed] [Google Scholar]
- 29.Quackenbush J. (2002) Microarray data normalization and transformation. Nature Genet., 32(Suppl. 2), 496–501. [DOI] [PubMed] [Google Scholar]
- 30.Horak C.E. and Snyder,M. (2002) Global analysis of gene expression in yeast. Funct. Integr. Genomics, 2, 171–180. [DOI] [PubMed] [Google Scholar]
- 31.Goryachev A.B., Macgregor,P.F. and Edwards,A.M. (2001) Unfolding of microarray data. J. Comput. Biol., 8, 443–461. [DOI] [PubMed] [Google Scholar]
- 32.Kim J.H., Shin,D.M. and Lee,Y.S. (2002) Effect of local background intensities in the normalization of cDNA microarray data with a skewed expression profiles. Exp. Mol. Med., 34, 224–232. [DOI] [PubMed] [Google Scholar]
- 33.Tseng G.C., Oh,M.K., Rohlin,L., Liao,J.C. and Wong,W.H. (2001) Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acids Res., 29, 2549–2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bilban M., Buehler,L.K., Head,S., Desoye,G. and Quaranta,V. (2002) Normalizing DNA microarray data. Curr. Issues Mol. Biol., 4, 57–64. [PubMed] [Google Scholar]
- 35.Colantuoni C., Henry,G., Zeger,S. and Pevsner,J. (2002) Local mean normalization of microarray element signal intensities across an array surface: quality control and correction of spatially systematic artifacts. Biotechniques, 32, 1316–1320. [DOI] [PubMed] [Google Scholar]
- 36.Hoffmann R., Seidl,T. and Dugas,M. (2002) Profound effect of normalization on detection of differentially expressed genes in oligonucleotide microarray data analysis. Genome Biol., 3, RESEARCH0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kepler T.B., Crosby,L. and Morgan,K.T. (2002) Normalization and analysis of DNA microarray data by self-consistency and local regression. Genome Biol., 3, RESEARCH0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Workman C., Jensen,L.J., Jarmer,H., Berka,R., Gautier,L., Nielser,H.B., Saxild,H.H., Nielsen,C., Brunak,S. and Knudsen,S. (2002) A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol., 3, research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang I.V., Chen,E., Hasseman,J.P., Liang,W., Frank,B.C., Wang,S., Sharov,V., Saeed,A.I., White,J., Li,J. et al. (2002) Within the fold: assessing differential expression measures and reproducibility in microarray assays. Genome Biol., 3, research0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y.H., Dudoit,S., Luu,P., Lin,D.M., Peng,V., Ngai,J. and Speed,T.P. (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res., 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cleveland W.S. (1981) LOWESS: A program for smoothing scatterplots by robust locally weighted regression. Amer. Stat., 35, 54. [Google Scholar]
- 42.Cleveland W.S. and Devlin,S.J. (1988) Locally-weighted regression: an approach to regression analysis by local fitting. J. Amer. Stat. Assoc., 83, 596–610. [Google Scholar]
- 43.Cleveland W.S. and Grosse,E. (1991) Computational methods for local regression. Stat. Comp., 1, 47–62. [Google Scholar]
- 44.Wang X., Ghosh,S. and Guo,S.W. (2001) Quantitative quality control in microarray image processing and data acquisition. Nucleic Acids Res., 29, E75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jenssen T.K., Langaas,M., Kuo,W.P., Smith-Sorensen,B., Myklebost,O. and Hovig,E. (2002) Analysis of repeatability in spotted cDNA microarrays. Nucleic Acids Res., 30, 3235–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schena M., Shalon,D., Heller,R., Chai,A., Brown,P.O. and Davis,R.W. (1996) Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc. Natl Acad. Sci. USA, 93, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wodicka L., Dong,H., Mittmann,M., Ho,M.H. and Lockhart,D.J. (1997) Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol., 15, 1359–1367. [DOI] [PubMed] [Google Scholar]
- 48.Chu S., DeRisi,J., Eisen,M., Mulholland,J., Botstein,D., Brown,P.O. and Herskowitz,I. (1998) The transcriptional program of sporulation in budding yeast. Science, 282, 699–705. [DOI] [PubMed] [Google Scholar]
- 49.Golub T.R., Slonim,D.K., Tamayo,P., Huard,C., Gaasenbeek,M., Mesirov,J.P., Coller,H., Loh,M.L., Downing,J.R., Caligiuri,M.A. et al. (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science, 286, 531–537. [DOI] [PubMed] [Google Scholar]
- 50.Model F., Adorjan,P., Olek,A. and Piepenbrock,C. (2001) Feature selection for DNA methylation based cancer classification. Bioinformatics, 17(Suppl. 1), S157–164. [DOI] [PubMed] [Google Scholar]
- 51.Tusher V.G., Tibshirani,R. and Chu,G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA, 98, 5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]