A functional study of miR-124 in the developing neural tube (original) (raw)
Abstract
Neural development is a highly orchestrated process that entails precise control of gene expression. Although microRNAs (miRNAs) have been implicated in fine-tuning gene networks, the roles of individual miRNAs in vertebrate neural development have not been studied in vivo. We investigated the function of the most abundant neuronal miRNA, miR-124, during spinal cord development. Neither inhibition nor overexpression of miR-124 significantly altered the acquisition of neuronal fate, suggesting that miR-124 is unlikely to act as a primary determinant of neuronal differentiation. Two endogenous targets of miR-124, laminin γ1 and integrin β1, were identified, both of which are highly expressed by neural progenitors but repressed upon neuronal differentiation. Thus miR-124 appears to ensure that progenitor genes are post-transcriptionally inhibited in neurons.
Keywords: microRNA, chick neural tube, neurogenesis, ventricular zone, basal lamina
Whole-genome sequence annotations indicate that there is no simple correlation between the morphological and behavioral complexity of an organism and the number of genes the organism bears. It has been suggested that the key driving force for the increase of organism complexity during evolution is the more elaborate regulation of gene expression, manifested by the expansion of _cis_-regulatory sequences, core transcription complexes, cofactor complexes, and chromatin modification complexes (Levine and Tjian 2003). MicroRNAs (miRNAs), along with several classes of regulatory noncoding RNAs, add a new dimension to the repertoire of gene regulation machineries (Zamore and Haley 2005). Mature miRNAs are ∼22-nucleotide (nt) single-stranded RNAs that negatively regulate gene expression post-transcriptionally. By binding to complementary sequences located at the 3′-untranslated region (3′-UTR) of target mRNAs, miRNAs inhibit translation and/or lead to the degradation of target mRNAs. Target recognition by a miRNA in many cases only requires a continuous 6-base-pair (bp) “seed match” between the 5′ end of the miRNA and its target. Therefore, one miRNA can theoretically coordinate the regulation of hundreds of genes. Comparative genomics analyses have predicted that one third of human genes could be miRNA targets (Lewis et al. 2005).
Both the prevalence of miRNAs in higher eukaryotes and their striking evolutionary conservation point to their functional importance. Several hypotheses have been formulated regarding the general role of miRNAs as a distinct class of gene regulators. Based on the phenotypes of a limited number of miRNAs identified in forward genetic screens, it was speculated that miRNAs acted as developmental switches by regulating a few target genes. The finding that a single miRNA can potentially regulate a large set of genes suggests that miRNAs are involved in more complex gene networks (Bartel and Chen 2004). Recent data further demonstrate that miRNAs and target genes are preferentially expressed in neighboring tissues, whereas genes coexpressed with miRNAs selectively lack miRNA-binding sites (Farh et al. 2005; Stark et al. 2005), leading to the proposal that miRNAs often function to prevent stochastic variations in gene expression and confer robustness to organism development and physiology (Hornstein and Shomron 2006).
Such analyses are insightful but await more experimental testing, especially in vivo testing. We set out to study the in vivo function of miR-124, the most abundant miRNA in the adult brain (Lagos-Quintana et al. 2002). The sequence of mature miR-124 is completely conserved from worm to human. Conserved seed match sequences to miR-124 are abundant in mammalian genomes; one miRNA target prediction algorithm identified >1000 potential targets for miR-124 (Lewis et al. 2005). During neurogenesis, miR-124 is present at undetectable or very low levels in neural progenitors but is highly expressed in differentiating and mature neurons (Deo et al. 2006). Overexpression of miR-124 in HeLa cells decreased the transcripts of 174 genes, whose expression levels are generally lower in the brain than in other tissues, suggesting that miR-124 may help maintain the gene expression profile of the brain by repressing unwanted transcripts (Lim et al. 2005). However, whether the down-regulated transcripts represent the physiological targets of miR-124 and whether the findings reflect its in vivo function remain to be tested.
We used the chick neural tube as a model system to study the role of miR-124 in vivo. In the developing neural tube, the cell bodies of neural progenitors reside medially within the ventricular zone and extend a long radial process to the basal, or lateral, side of the neural tube. Neural progenitors give rise to differentiating cells, which migrate laterally toward the mantle zone and start to acquire generic as well as subtype-specific neuronal features (Gotz and Huttner 2005). Since miR-124 is repressed in progenitor cells but becomes robustly expressed when neuronal differentiation begins, we hypothesized that miR-124 is involved in suppressing certain progenitor functions and/or promoting neuronal properties as cells differentiate. We found that neither inhibition nor ectopic expression of miR-124 had a significant effect on neuronal differentiation. However, by repressing laminin γ1 and integrin β1, two endogenous targets of miR-124, it may ensure the transition from neural progenitors to neurons.
Results and Discussion
Inhibiting miR-124 does not affect neuronal differentiation
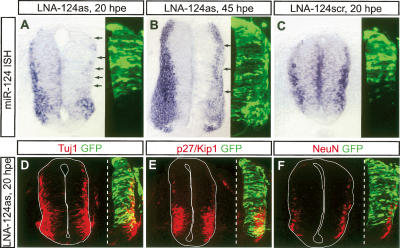
To inhibit miR-124 in vivo, we introduced “locked” nucleic acid (LNA)-modified miR-124 antisense oligonucleotides (LNA-124as) into Hamburger-Hamilton (HH) stage 12–13 (Hamburger and Hamilton 1992) chick spinal cords via in ovo electroporation. A GFP plasmid was coelectroporated to mark transfected cells. LNA-modified antisense oligonucleotides form stable hybrids with endogenous miRNAs, stoichiometrically sequestering miRNAs from their targets and thereby inhibiting their functions (Fazi et al. 2005). To monitor the extent of inhibition, we performed in situ hybridizations. At 20 h post-electroporation (hpe), miR-124 in situ hybridization signals were markedly reduced on the transfected side of the spinal cord. A detectable but weaker reduction was also observed at 45 hpe. Such an effect was not seen with scrambled oligonucleotides (Fig. 1A–C; Supplementary Fig. S1A), indicating sequence-specific inhibition of miR-124. 2′-_O_-methyl antisense oligonucleotides are also commonly used as miRNA inhibitors (Krutzfeldt et al. 2006). We, however, found that they were much less effective than LNA-oligonucleotides (Supplementary Fig. S1B–D). For both types of oligonucleotides, reductions in in situ hybridization signals were no longer detectable at 70 hpe (data not shown), which may be due to the oligonucleotides being cleared from the embryo and/or being titrated out by high levels of endogenous miR-124.
Figure 1.
Inhibiting miR-124 does not affect neuronal differentiation. (A–C) miR-124 in situ hybridization (ISH) on chick embryos transfected with LNA-modified miR-124 antisense oligonucleotides (LNA-124as, 100 μM) showed decreased hybridization signals on the transfected side of the spinal cord at 20 and 45 hpe (arrows). Scrambled oligonucleotides (LNA-124scr) did not cause such a decrease. The image next to each ISH photograph shows the expression of the cotransfected GFP plasmid on an adjacent section. (D–F) Transfection of LNA-124as did not affect the expression of neuronal markers Tuj1, p27/Kip1, and NeuN at 20 hpe. Images on the left of the dashed lines show the whole neural tube sections, those on the right show only the transfected side.
The above in situ hybridizations also confirmed that miR-124 was highly expressed by differentiating and mature neurons located in the mantle zone. We thus examined whether inhibiting miR-124 affected neuronal differentiation. Immunocytochemistry analyses with various generic and subtype-specific neuronal markers, including Tuj1, p27/Kip1, NeuN, Neurofilament, Lim1/2, Lim3, MNR2, and Isl1/2 (Farah et al. 2000; for review, see Shirasaki and Pfaff 2002), on spinal cords transfected with either LNA- or 2′-_O_-methyl antisense oligonucleotides did not detect any noticeable effect at 20 and 45 hpe (Fig. 1D–F; data not shown). The expression of the neural progenitor marker Sox2 also appeared normal (Bylund et al. 2003; data not shown). These results suggest that at least during early stages of neural tube development, miR-124 is not essential for generic neuronal differentiation or neuronal subtype specification.
Overexpression of miR-124 in the chick neural tube
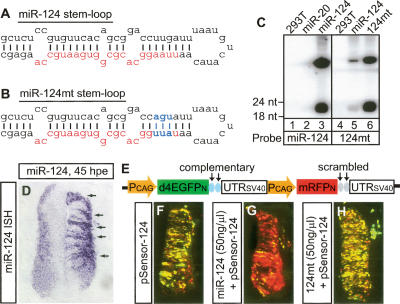
Since miR-124 expression is up-regulated when cells begin neuronal differentiation, it is conceivable that miR-124 may repress transcripts left over from progenitor stages. We therefore sought to test the effect of overexpressing miR-124 in neural progenitors. We cloned a 300-bp fragment of mir-124a genomic DNA, with the stem-loop sequence located in the middle of the fragment, downstream from a mouse U6 promoter (Chen et al. 2004). As the negative control, we introduced point mutations corresponding to nucleotides 2–4 of mature miR-124 (miR-124mt), which should disrupt seed matches between miR-124 and its targets. The nucleotides at the opposite arm were also mutated to preserve the stem-loop structure (Fig. 2A,B). Northern blots showed that both plasmids could overexpress the cloned miRNA at high levels when transfected into 293T cells (Fig. 2C). Overexpression of miR-124 in the chick spinal cord via electroporation was confirmed by in situ hybridization (Fig. 2D).
Figure 2.
Overexpressing miR-124 in the chick neural tube. (A,B) The stem-loop structures of miR-124 and miR-124mt with red letters designating the sequences of mature miRNAs. Blue letters are the mutations introduced that should disrupt seed matches between miR-124 and its targets. (C) Northern blots probed for miR-124 or miR-124mt showed similar levels of expression from the overexpression plasmids in 293T cells. Lanes 1 and 4 are RNAs from untransfected 293T cells. Lanes 2, 3, 5, and 6 are RNAs from 293T cells transfected with the plasmid expressing miR-20, miR-124, miR-124, and miR-124mt, respectively. (D) miR-124 ISH showed high levels of expression (arrows) from the miR-124 plasmid in the transfected neural tube at 45 hpe. (E) The schematic drawing of a miRNA sensor plasmid. The two blue ovals represent two copies of complementary sequences to the miRNA that the sensor is designed to detect. The gray ovals represent scrambled sequences. (Pcag) Chick β-actin promoter; (d4EGFPn) nuclear-localized destabilized EGFP with a half-life of 4 h; (mRFPn) nuclear-localized monomeric red fluorescent protein. (F–H) At 45 hpe, most cells transfected with the sensor for miR-124 (pSensor-124) were yellow, expressing both GFP and RFP (F). Cotransfection of the miR-124 plasmid strongly repressed GFP expression (G), which was not observed with the miR-124mt plasmid (H).
To test if the overexpressed miR-124 was functional, we developed a miRNA functional readout. We modified a previously published “miRNA responsive sensor” (Mansfield et al. 2004) into a two-colored “sensor” that consists of a reporter and a control (Fig. 2E). The reporter is composed of a destabilized GFP gene driven by a β-actin promoter and followed by two copies of miRNA complementary sequences at the 3′-UTR. In addition, the sensor encodes an RFP driven by the same promoter but followed by scrambled sequences, thus serving as an internal control. Both GFP and RFP are targeted to the nucleus to facilitate the identification of transfected cells. At 45 h after transfection of the sensor for miR-124 into the chick neural tube, most transfected cells appeared yellow, expressing both GFP and RFP. Even at very low concentrations, cotransfection of the miR-124 plasmid strongly repressed GFP expression and cells appeared red, which was not observed with miR-124mt, suggesting that the overexpression plasmids are highly effective and specific (Fig. 2F–H).
Overexpressing miR-124 does not promote overt neuronal differentiation
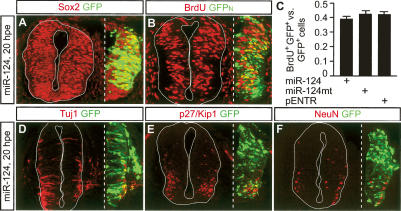
We next transfected miR-124 or miR-124mt plasmids into HH12–13 neural tubes. At this stage, the neural tube is composed mostly of neural progenitors. To evaluate if overexpressing miR-124 reduced the number of progenitor cells, we examined the expression of Sox2 but failed to detect any obvious effect at either 20 or 45 hpe (Fig. 3A; Supplementary Fig. S2A). Because neuronal differentiation is accompanied by cell cycle exit, we assessed cell proliferation by labeling dividing cells with the nucleotide analog BrdU 2 h before harvesting the embryos. Comparing embryos transfected with miR-124, miR-124mt, or the cloning vector pENTR, there was no significant difference in the fraction of transfected cells (GFP+ cells) that remained mitotically active (BrdU+GFP+ cells), suggesting that overexpressing miR-124 in neural progenitors did not enhance or suppress cell proliferation (Fig. 3B,C). Expressions of neuronal markers Tuj1, p27/Kip1, and NeuN were not markedly affected by ectopic miR-124 at 20 or 45 hpe (Fig. 3D–F; Supplementary Fig. S2B–D). In comparison, overexpression of the proneural transcription factor Ngn2 (Ma et al. 1996) led to a dramatic up-regulation of Tuj1 (Supplementary Fig. S2E,F). Together, these data suggest that ectopic expression of miR-124 in neural progenitors is unable to promote overt neuronal differentiation.
Figure 3.
Overexpressing miR-124 does not promote overt neuronal differentiation. (A) Ectopic miR-124 did not affect the expression of the neural progenitor marker Sox2. (B) Cell cycle analysis by BrdU labeling. miR-124 was cotransfected with a plasmid expressing a nuclear localized GFP (GFPn) to facilitate cell counting. (C) Quantifications of the fraction of transfected cells (GFP+) that were mitotically active (BrdU+GFP+) showed no difference in embryos transfected with miR-124, miR-124mt, or the empty vector pENTR. (D–F) Overexpression of miR-124 did not increase the expression of neuronal markers Tuj1, p27/Kip1, or NeuN at 20 hpe.
Despite the lack of noticeable effect on neuronal differentiation, ectopic miR-124 did cause increased cell death (Supplementary Fig. S3). At 20 hpe, the apoptotic effect appeared to be a combination of both sequence-specific and nonspecific effects, as miR-124mt also caused increased apoptosis, albeit to a lesser extent. At 45 hpe, however, the apoptotic effect was no longer associated with miR-124mt and became sequence-specific to miR-124 (Supplementary Fig. S3B). At present, we do not know which target(s) of miR-124 may be responsible for the increased cell death. We also found that at 45 hpe, miR-124-transfected neural tubes were frequently disorganized: The adherens junctions that form the inner barrier of the neural tube were disrupted and some cells were mispositioned inside the ventricle (Supplementary Figs. S2A–D, S4, arrowheads). We cannot yet rule out that this defect may, at least partially, be secondary to the increased apoptosis caused by ectopic miR-124.
Ectopic miR-124 causes basal lamina defects by repressing LAMC1 and ITGB1
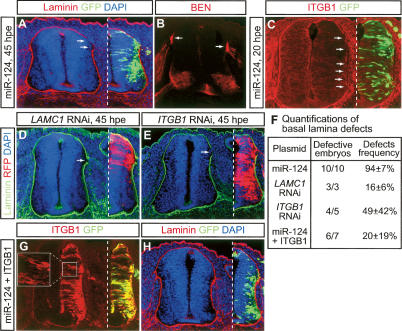
During our analyses, we noticed that the boundary between miR-124-transfected neural tubes and adjacent tissues was often ambiguous (data not shown). As the neural tube is ensheathed by basal laminae to separate it from surrounding tissues, we examined the integrity of basal laminae by immunostaining for laminin-1, a major component of basal laminae. Obvious disruptions in basal laminae were observed at 45 hpe (cf. Fig. 4A, arrows, and a miR-124mt-transfected section in Supplementary Fig. S5B). While disruptions sometimes occurred across the electroporated side (Supplementary Fig. S5A), most often they were located near the dorsal root entry sites where sensory afferents invade the spinal cord (Fig. 4A,B, arrows). It has been reported that basal laminae are partially degraded into a “loose meshwork” at dorsal root entry sites, presumably allowing sensory afferents to enter the spinal cord (Golding and Cohen 1997), which may explain why the basal lamina defect was more prominent in this region.
Figure 4.
Ectopic miR-124 causes basal lamina defects. (A,B) Immunostaining for laminin-1 showed disintegration of basal laminae upon the overexpression of miR-124 (A, arrows). (B) The defect was located near the dorsal root entry site, shown on an adjacent section by immunostaining with BEN, an antibody that labels sensory axons (arrows). (C) The line of integrin β1 (ITGB1) that surrounds the outer surface of the neural tube was disrupted by ectopic miR-124 (arrows). (D,E) LAMC1 and ITGB1 RNAi caused similar, albeit milder, basal lamina defects (arrows). Transfected cells were marked by RFP encoded in RNAi vectors. (F) Quantifications of basal lamina defects. “Defects frequency” refers to the percentage of defective sections among all the sections examined in each embryo. (G,H) Cotransfection of an ITGB1-expressing plasmid with miR-124 partially rescued the basal lamina defect at 45 hpe (H). (G) High levels of ITGB1 expression were achieved from the transfected plasmid, with the overexpressed protein properly localized to the plasma membrane (inset).
Laminin-1 is a heterotrimeric molecule formed by laminin α1, β1, and γ1 subunits. Interestingly, both laminin γ1 (LAMC1) and integrin β1 (ITGB1)—which encodes an essential subunit of laminin receptors—have been found to be potential targets of miR-124 based on computational predications and cell culture assays (Lewis et al. 2005; Lim et al. 2005). We therefore investigated the expression of integrin β1. At HH20–21, integrin β1 was highly enriched in the outer surface of the neural tube, with moderate levels of expression throughout the neural tube (Fig. 4C; Supplementary Fig. S5C). However, in miR-124-transfected spinal cords, the continuous outer “line” of integrin β1 was disrupted and the immunoreactivity within the neural tube decreased ∼20% (Fig. 4C, arrows; Supplementary Fig. S5D,E).
Mice with mutations in either LAMC1 or ITGB1 gene showed similar disruptions in their cortical pial basal laminae as did miR-124-transfected spinal cords (Graus-Porta et al. 2001; Halfter et al. 2002), suggesting that repressions of LAMC1 and ITGB1 by the overexpressed miR-124 may be responsible for the basal lamina phenotype we observed. To confirm this, we tried to recapitulate the phenotype by silencing LAMC1 and ITGB1 through RNA interference (RNAi) (Das et al. 2006). Similar basal lamina defect was observed when each gene was knocked down (Fig. 4D–F; Supplementary Table S1). Meanwhile, neither RNAi plasmid was as potent as miR-124-expressing plasmid in terms of causing basal lamina defects, which may be because the RNAi plasmids were not very effective in silencing the target genes or, more likely, each RNAi plasmid repressed only one gene, whereas the miR-124 plasmid repressed both.
We next attempted to rescue the basal lamina defect by overexpressing LAMC1 and ITGB1. We cloned the coding sequence of ITGB1 upstream of an SV40 3′-UTR, making it insensitive to miR-124. Cotransfection of this ITGB1 plasmid with miR-124 significantly reduced the occurrence of the defect; <50% of the sections examined showed the defect in double-transfected embryos, compared with >80% in miR-124-transfected ones (Fig. 4F–H; Supplementary Table S1). Due to the size of LAMC1 coding sequence, we were unable to clone it. Despite this, the RNAi and ITGB1 rescue experiments strongly indicate that the basal lamina phenotype had resulted from repressions of LAMC1 and ITGB1 by ectopic miR-124.
LAMC1 and ITGB1 are endogenous targets of miR-124
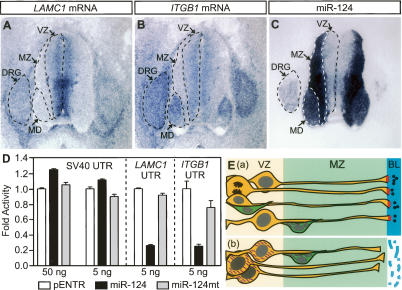
To find out whether LAMC1 and ITGB1 are endogenous targets of miR-124, we compared their expression patterns in HH27–28 spinal cords. Both mRNAs showed largely complementary patterns of expression to that of miR-124; whereas miR-124 was much more abundant in the mantle zone than in the ventricular zone, both mRNAs were present at higher levels in the ventricular zone (Fig. 5A–C). While LAMC1 mRNA and miR-124 were almost mutually exclusive, ITGB1 mRNA and miR-124 did exhibit overlapping expressions in motor domains and dorsal root ganglia. Interestingly, these two areas also showed lower levels of miR-124 than did other regions in the mantle zone. We next analyzed miR-124-binding sites in human, mouse, and chick LAMC1 and ITGB1 3′-UTRs (Supplementary Fig. S6). Three conserved binding sites were found in LAMC1 3′-UTRs. For each site, base-pairings to miR-124 include not only seed matches but also the 3′ end of miR-124. Two binding sites were found in ITGB1 3′-UTRs and the base-pairings were largely limited to seed matches. From this sequence analysis, it is conceivable that LAMC1 might be a stronger target of miR-124 than ITGB1, which correlates remarkably with their expression patterns.
Figure 5.
LAMC1 and ITGB1 mRNAs are endogenous targets of miR-124. (A–C) LAMC1, ITGB1, and miR-124 ISH on HH27–28 brachial-level transverse sections. (VZ) Ventricular zone; (MZ) mantle zone; (MD) motor domain; (DRG) dorsal root ganglion. (D) Luciferase assays with the luciferase gene fused to SV40, chick LAMC1, or chick ITGB1 3′-UTR. pENTR, miR-124, or miR-124mt plasmids (amounts shown in the _X_-axis) were cotransfected with luciferase–UTR constructs. Luciferase activities obtained from the cotransfection of pENTR and each luciferase–UTR construct were used as denominators. Cotransfection of miR-124, but not miR-124mt, specifically repressed LAMC1 and ITGB1 UTRs but not SV40 UTR. (E, panel a) During neural tube development, neural progenitors (orange) contribute to the formation and maintenance of basal laminae (BL) by producing laminin γ1 (black balls) and integrin β1 (red bars). (Panel b) Upon ectopic expression of miR-124 (short purple lines) in neural progenitors, both genes were repressed, leading to the disintegration of basal laminae. Green cells represent differentiating neurons.
To further confirm that the 3′-UTRs of LAMC1 and ITGB1 are miR-124 targets, we cloned the chick UTRs downstream from a luciferase gene and transfected the constructs into 293T cells. Cotransfection of miR-124 specifically inhibited luciferase expression when it was fused to LAMC1 and ITGB1 UTRs, suggesting that miR-124 could directly repress chick LAMC1 and ITGB1 3′-UTRs (Fig. 5D).
In summary, our data showed that neural progenitors residing in the ventricular zone produce and deposit laminin, thereby contributing to the formation of basal laminae (see also Hatakeyama et al. 2004). Progenitor cells also synthesize integrin β1 proteins, most of which are transported to the endfeet to anchor radial processes onto basal laminae and to maintain the structural integrity of basal laminae (Graus-Porta et al. 2001). When miR-124 was ectopically expressed in neural progenitors, the syntheses of laminin γ1 and integrin β1 were decreased, leading to the disintegration of basal laminae (Fig. 5E).
Concluding remarks
We employed the chick spinal cord as an in vivo system to investigate the role of miR-124 during neural development. We found that neither inhibition nor ectopic expression of miR-124 markedly altered general neuronal differentiation, suggesting that miR-124 does not act as a determinant of neuronal differentiation. These results were, at first thought, unexpected, given the abundance of miR-124 in neurons and its ability to shift the gene expression profile of HeLa cells toward that of the brain. Our finding, however, is consistent with two reports where Dicer, the enzyme responsible for generating all mature miRNAs, was deleted: Dicer mutant zebrafish underwent fairly normal early stages of embryonic development with many cell types properly specified (Giraldez et al. 2005), and tissue-specific deletion of Dicer in mouse limb mesoderm did not affect basic patterning or differentiation of limb buds (Harfe et al. 2005). Both studies indicate that miRNAs do not have widespread crucial roles in embryonic patterning or tissue differentiation. On the other hand, our data do not exclude that miR-124 may be involved in more subtle or restricted aspects of neural development. Furthermore, miR-124 may have more prominent functions in mature neurons—where it is most abundantly expressed—such as maintaining neuron-specific gene expression profiles (Conaco et al. 2006). Our loss-of-function approach using antisense oligonucleotides could only generate temporary knockdown effect. Genetic deletion of mir-124 genes in mice and continuous administration of antisense inhibitors are feasible approaches to address these possibilities.
By ectopically expressing miR-124 in neural progenitors, we identified LAMC1 and ITGB1 as two endogenous targets of miR-124. Both genes are highly expressed in neural progenitors but are repressed upon neuronal differentiation. The temporal relationship between miR-124 and these two targets resembles that between muscle-specific miRNAs miR-1 and miR-133 and their targets during myotube differentiation (Farh et al. 2005). In all three cases, miRNA targets are preferentially expressed at high levels before miRNA expression and are down-regulated as the miRNA starts to accumulate. Our data thus support the hypothesis that miRNAs induced during differentiation function to ensure proper developmental transitions by repressing leftover transcripts from the previous developmental stage (Farh et al. 2005).
Recent evidence suggests that miRNAs often act as fine-tuning devices rather than as primary gene regulators (Hornstein and Shomron 2006); genes targeted by a miRNA are usually transcribed at lower levels in miRNA-expressing tissues than in other tissues (Farh et al. 2005). Thus, inhibiting a miRNA may not generate any strong or even detectable phenotype, as expressions of its targets are already repressed at the transcriptional level. Conversely, misexpressing a miRNA in cells where its conserved targets are highly expressed—thus presumably functioning—may render the action of the miRNA more detectable, and thus may be an informative way to decipher miRNA functions.
Fly miR-124 is also neuron-specific and has been shown to preferentially target genes expressed in the epidermis, whereas the nervous system is enriched for genes evolved to avoid miR-124 sites, the so-called “antitargets.” Given that both the neural and epidermal tissues are derived from the neuroectoderm, by repressing epidermal genes in neurons, fly miR-124 may ensure the separation of sibling tissues (Stark et al. 2005). Our conclusion is consistent with this hypothesis, implicating the evolutionary conservation of miR-124 function. At the same time, similar “targets and antitargets” analysis for vertebrate miRNAs will likely aid in uncovering their functions.
Materials and methods
DNA constructs
The miR-124 expression construct was generated by cloning a 300-bp fragment of the mouse mir-124a-2 genomic sequence into pENTR/U6 (Invitrogen). The mature chick and mouse miR-124 sequences are identical. Point mutations were introduced using the QuikChange mutagenesis kit (Stratagene). LAMC1 and ITGB1 RNAi constructs were generated using pRFPRNAi (Das et al. 2006). ITGB1 coding sequence was amplified from chick cDNAs and cloned into a modified pMIW vector (Muramatsu et al. 1997). pTK-Luc was generated by subcloning the thymidine kinase promoter from pRL-TK into pGL3-Basic (Promega). LAMC1 and ITGB1 3′-UTRs were amplified from chick genomic DNA and cloned into pTK-Luc. See Supplemental Material for details.
Northern blot and in situ hybridization
Northern blot was performed as described (Lau et al. 2001). In situ hybridization with LNA-modified probes (Proligo) was done as described (Kloosterman et al. 2006). See Supplemental Material for details.
In ovo electroporation, immunocytochemistry, TUNEL assay, and BrdU labeling
In ovo electroporation and immunocytochemistry were performed as described (Lee et al. 2005). See Supplemental Material for antibody sources. TUNEL assay was performed using the ApopTag kit (Chemicon). For BrdU labeling, 50 μL of BrdU (5 mg/mL) were added on top of embryos at 18 hpe, which were further incubated for 2 h and harvested.
Cell culture, transfection, and luciferase assay
293T cells were cultured in DMEM/F12 medium (Invitrogen) supplemented with 10% fetal bovine serum. Transfection was performed using Fugene 6 (Roche). Luciferase assay was performed with the Dual-luciferase assay system (Promega).
Acknowledgments
We thank Gordon Gill, Ying Liu, Hoonkyo Suh, Ahmet Denli, Till Marquardt, and members of the Gage and Pfaff laboratories for advice and discussions, Roger Tsien for mRFP plasmid, and Mary Lynn Gage for editorial assistance. This work is supported by NIH/NINDS (NS052842). X.C. is supported by the Pioneer Fund and Lookout Fund.
Footnotes
References
- Bartel D.P., Chen C.Z., Chen C.Z. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- Bylund M., Andersson E., Novitch B.G., Muhr J., Andersson E., Novitch B.G., Muhr J., Novitch B.G., Muhr J., Muhr J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- Chen C.Z., Li L., Lodish H.F., Bartel D.P., Li L., Lodish H.F., Bartel D.P., Lodish H.F., Bartel D.P., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Conaco C., Otto S., Han J.J., Mandel G., Otto S., Han J.J., Mandel G., Han J.J., Mandel G., Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R.M., Van Hateren N.J., Howell G.R., Farrell E.R., Bangs F.K., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Van Hateren N.J., Howell G.R., Farrell E.R., Bangs F.K., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Howell G.R., Farrell E.R., Bangs F.K., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Farrell E.R., Bangs F.K., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Bangs F.K., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Porteous V.C., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., Manning E.M., McGrew M.J., Ohyama K., Sacco M.A., McGrew M.J., Ohyama K., Sacco M.A., Ohyama K., Sacco M.A., Sacco M.A. A robust system for RNA interference in the chicken using a modified microRNA operon. Dev. Biol. 2006;294:554–563. doi: 10.1016/j.ydbio.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Deo M., Yu J.Y., Chung K.H., Tippens M., Turner D.L., Yu J.Y., Chung K.H., Tippens M., Turner D.L., Chung K.H., Tippens M., Turner D.L., Tippens M., Turner D.L., Turner D.L. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev. Dyn. 2006;235:2538–2548. doi: 10.1002/dvdy.20847. [DOI] [PubMed] [Google Scholar]
- Farah M.H., Olson J.M., Sucic H.B., Hume R.I., Tapscott S.J., Turner D.L., Olson J.M., Sucic H.B., Hume R.I., Tapscott S.J., Turner D.L., Sucic H.B., Hume R.I., Tapscott S.J., Turner D.L., Hume R.I., Tapscott S.J., Turner D.L., Tapscott S.J., Turner D.L., Turner D.L. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lim L.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., De Marchis M.L., Nervi C., Bozzoni I., Nervi C., Bozzoni I., Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Hammond S.M., Bartel D.P., Schier A.F., Bartel D.P., Schier A.F., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Golding J.P., Cohen J., Cohen J. Border controls at the mammalian spinal cord: Late-surviving neural crest boundary cap cells at dorsal root entry sites may regulate sensory afferent ingrowth and entry zone morphogenesis. Mol. Cell. Neurosci. 1997;9:381–396. doi: 10.1006/mcne.1997.0647. [DOI] [PubMed] [Google Scholar]
- Gotz M., Huttner W.B., Huttner W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Graus-Porta D., Blaess S., Senften M., Littlewood-Evans A., Damsky C., Huang Z., Orban P., Klein R., Schittny J.C., Muller U., Blaess S., Senften M., Littlewood-Evans A., Damsky C., Huang Z., Orban P., Klein R., Schittny J.C., Muller U., Senften M., Littlewood-Evans A., Damsky C., Huang Z., Orban P., Klein R., Schittny J.C., Muller U., Littlewood-Evans A., Damsky C., Huang Z., Orban P., Klein R., Schittny J.C., Muller U., Damsky C., Huang Z., Orban P., Klein R., Schittny J.C., Muller U., Huang Z., Orban P., Klein R., Schittny J.C., Muller U., Orban P., Klein R., Schittny J.C., Muller U., Klein R., Schittny J.C., Muller U., Schittny J.C., Muller U., Muller U. β1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- Halfter W., Dong S., Yip Y.P., Willem M., Mayer U., Dong S., Yip Y.P., Willem M., Mayer U., Yip Y.P., Willem M., Mayer U., Willem M., Mayer U., Mayer U. A critical function of the pial basement membrane in cortical histogenesis. J. Neurosci. 2002;22:6029–6040. doi: 10.1523/JNEUROSCI.22-14-06029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V., Hamilton H.L., Hamilton H.L. A series of normal stages in the development of the chick embryo. 1951. Dev. Dyn. 1992;195:231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]
- Harfe B.D., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J., Mansfield J.H., Hornstein E., Tabin C.J., Hornstein E., Tabin C.J., Tabin C.J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama J., Bessho Y., Katoh K., Ookawara S., Fujioka M., Guillemot F., Kageyama R., Bessho Y., Katoh K., Ookawara S., Fujioka M., Guillemot F., Kageyama R., Katoh K., Ookawara S., Fujioka M., Guillemot F., Kageyama R., Ookawara S., Fujioka M., Guillemot F., Kageyama R., Fujioka M., Guillemot F., Kageyama R., Guillemot F., Kageyama R., Kageyama R. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development. 2004;131:5539–5550. doi: 10.1242/dev.01436. [DOI] [PubMed] [Google Scholar]
- Hornstein E., Shomron N., Shomron N. Canalization of development by microRNAs. Nat. Genet. 2006;38:S20–S24. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- Kloosterman W.P., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H., Wienholds E., de Bruijn E., Kauppinen S., Plasterk R.H., de Bruijn E., Kauppinen S., Plasterk R.H., Kauppinen S., Plasterk R.H., Plasterk R.H. In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods. 2006;3:27–29. doi: 10.1038/nmeth843. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J., Poy M.N., Stoffel M., Poy M.N., Stoffel M., Stoffel M. Strategies to determine the biological function of microRNAs. Nat. Genet. 2006;38:S14–S19. doi: 10.1038/ng1799. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Meyer J., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P., Lim L.P., Weinstein E.G., Bartel D.P., Weinstein E.G., Bartel D.P., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Lee B., Ruiz E.C., Pfaff S.L., Lee B., Ruiz E.C., Pfaff S.L., Ruiz E.C., Pfaff S.L., Pfaff S.L. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes & Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M., Tjian R., Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Bartel D.P., Linsley P.S., Johnson J.M., Linsley P.S., Johnson J.M., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Ma Q., Kintner C., Anderson D.J., Kintner C., Anderson D.J., Anderson D.J. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Mansfield J.H., Harfe B.D., Nissen R., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Harfe B.D., Nissen R., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Nissen R., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Obenauer J., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Srineel J., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Chaudhuri A., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Farzan-Kashani R., Zuker M., Pasquinelli A.E., Ruvkun G., Zuker M., Pasquinelli A.E., Ruvkun G., Pasquinelli A.E., Ruvkun G., Ruvkun G., et al. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat. Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- Muramatsu T., Mizutani Y., Ohmori Y., Okumura J.-i., Mizutani Y., Ohmori Y., Okumura J.-i., Ohmori Y., Okumura J.-i., Okumura J.-i. Comparison of three nonviral transfection methods for foreign gene expression in early chicken embryos in ovo. Biochem. Biophys. Res. Commun. 1997;230:376–380. doi: 10.1006/bbrc.1996.5882. [DOI] [PubMed] [Google Scholar]
- Shirasaki R., Pfaff S.L., Pfaff S.L. Transcriptional codes and the control of neuronal identity. Annu. Rev. Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Bushati N., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Zamore P.D., Haley B., Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]