B lymphocytes: how they develop and function (original) (raw)
Abstract
The discovery that lymphocyte subpopulations participate in distinct components of the immune response focused attention onto the origins and function of lymphocytes more than 40 years ago. Studies in the 1960s and 1970s demonstrated that B and T lymphocytes were responsible primarily for the basic functions of antibody production and cell-mediated immune responses, respectively. The decades that followed have witnessed a continuum of unfolding complexities in B-cell development, subsets, and function that could not have been predicted. Some of the landmark discoveries that led to our current understanding of B lymphocytes as the source of protective innate and adaptive antibodies are highlighted in this essay. The phenotypic and functional diversity of B lymphocytes, their regulatory roles independent of antibody production, and the molecular events that make this lineage unique are also considered. Finally, perturbations in B-cell development that give rise to certain types of congenital immunodeficiency, leukemia/lymphoma, and autoimmune disease are discussed in the context of normal B-cell development and selection. Despite the significant advances that have been made at the cellular and molecular levels, there is much more to learn, and cross-disciplinary studies in hematology and immunology will continue to pave the way for new discoveries.
Introduction
Despite a monotonous microscopic appearance that belies their remarkable developmental and functional heterogeneity, lymphocytes have stimulated the intellectual curiosity and challenged the experimental skills of investigators in many disciplines. We know them as T (_t_hymus-derived) and B (_b_ursal or _b_one marrow–derived) lymphocytes, and the latter is the focus of this historical essay. A simple definition of B lymphocytes is a population of cells that express clonally diverse cell surface immunoglobulin (Ig) receptors recognizing specific antigenic epitopes. Their origin can be traced to the evolution of adaptive immunity in jawed vertebrates beginning more than 500 million years ago.1 As shown in Figure 1, mammalian B-cell development encompasses a continuum of stages that begin in primary lymphoid tissue (eg, human fetal liver and fetal/adult marrow), with subsequent functional maturation in secondary lymphoid tissue (eg, human lymph nodes and spleen). The functional/protective end point is antibody production by terminally differentiated plasma cells. However, these B-cell development stages and their functional identities were essentially discovered in reverse chronologic order.
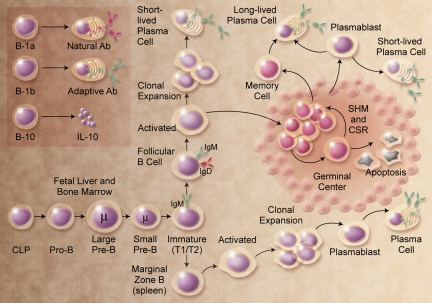
Figure 1.
B-cell development. The figure shows the broad outline of B-cell developmental stages in mice and humans. B-1a, B-1b, and B10 populations are less well characterized in humans. CLP indicates common lymphoid progenitor; SHM, somatic hypermutation; and CSR, class switch recombination. Illustration by A. Y. Chen.
The discovery of B cells did not originate in the identification of a cell, but rather the identification of a protein (ie, Ig or antibody). Identification of serum gammaglobulin as the source of antibodies2 was a launching point for the eventual discovery of antibody-producing cells. Plasma cells were suggested as a source of antibody production as early as 1948.3 The 2 competing views of antibody formation, the “natural selection” theory and the “clonal selection” theory, were proposed by 2 giants of 20th century immunology: Niels Jerne4 and Sir Macfarlane Burnet.5 Both men subsequently won the Nobel Prize in Physiology or Medicine—Burnet in 1960 and Jerne in 1984. Shortly thereafter, the first experimental evidence to support the clonal selection theory was revealed by studying single cells.6 A historical vignette describing those times was recently published by Sir Gustav Nossal.7 The chemical structure of the antibody molecule was elucidated by Gerald Edelman8 and Rodney Porter,9 who went on to share the Nobel Prize in Physiology or Medicine in 1972.
The 1960s and 1970s yield the identity of B cells
The discovery and characterization of B cells occurred in the mid-1960s and early 1970s using experimental animal models, clinical evaluation of patients with immune deficiency diseases,10 and the nascent technology of cell surface molecule characterization. B and T cells were essentially discovered simultaneously. Max Cooper and Robert Good suggested a functional division of labor between cells in the chicken bursa of Fabricius responsible for antibody production and cells that required an intact thymus for manifestation of delayed-type hypersensitivity (Cooper et al11,12). Murine transplant models were used to demonstrate that cells derived from the marrow were the subset that mediated antibody responses.13,14 A developmental link between B cells and antibody production was then revealed by studies showing that surface Ig expression could be used as a marker to characterize normal and leukemic B cells.15,16 The first major review of B and T lymphocytes was published in Nature17 followed by a textbook on the subject.18
Blood published one of the earliest papers in this nascent field, reporting that surface Ig was expressed on peripheral blood lymphocytes from patients with chronic lymphocytic leukemia.19 Characterization of cell surface marker expression by normal and malignant B (and T) cells spawned a new method of immunophenotyping for the classification of leukemias and lymphomas, a critical diagnostic adjunct that to this day represents a gold standard for the diagnosis and treatment of these malignancies. These seminal studies also facilitated the identification and characterization of normal B cells, since purification of peripheral blood or tonsillar B cells was technically difficult at the time.
Following in the footsteps of their initial independent contributions, Martin Raff and Cooper collaborated to identify precursor (pre)-B cells in murine fetal liver and marrow (Raff et al20). The existence of a rare population of lymphoid cells expressing cytoplasmic μ H chains suggested that pre-B cells were the precursors of newly formed B cells expressing cell surface IgM. Shortly thereafter, pre-B cells with similar characteristics were described in human fetal liver and marrow.21 Application of this discovery to studies of hematopoietic malignancies revealed that cells expressing cytoplasmic μ H chains were the dominant subclone in some cases of childhood acute lymphoblastic leukemia (ALL)22 and chronic myeloid leukemia in lymphoid blast crisis.23
What makes a lymphocyte a B cell?
B-cell development in mice24 and humans25 has been extensively studied, and the functional rearrangement of the Ig loci is a sine qua non. This occurs via an error-prone process involving the combinatorial rearrangement of the V, D, and J gene segments in the H chain locus and the V and J gene segments in the L chain loci.26 Susumu Tonegawa was awarded the Nobel Prize in Physiology or Medicine in 1987 for this discovery. In mice and humans, this occurs primarily in fetal liver and adult marrow, culminating in the development of a diverse repertoire of functional VDJH and VJL rearrangements encoding the B-cell receptor (BCR). However, in other species (eg, chickens and rabbits) the development of the preimmune Ig repertoire occurs primarily in GALT, and diversification of the repertoire uses the mechanism of gene conversion.27,28 The discovery of the recombination activating genes 1/2 (RAG-1/2) in the late 1980s by David Baltimore and colleagues provided the mechanistic explanation for the initial steps of DNA strand breakage in both Ig and T-cell receptor rearrangement (Schatz et al29 and Oettinger et al30).
Early B-cell development is characterized by the ordered rearrangement of Ig H and L chain loci, and Ig proteins themselves play an active role in regulating B-cell development.31 Pivotal to understanding how early B-cell development is regulated was the discovery of surrogate L chains (SLCs). Originally identified in murine B-lineage cells,32 the SLC is a heterodimer consisting of 2 distinct proteins originally designated λ5 and VpreB. These 2 proteins pair with the μ H chain to form the so-called pre-BCR in murine and human pre-B cells.33 Pre-B cells arise from progenitor (pro-B) cells that express neither the pre-BCR or surface Ig (Figure 1). Whether pre-BCR interactions with ligand(s) can serve as a proliferative stimulus and thereby expand pre-B cells with functional μ H chain rearrangements remains unknown. Although potential pre-BCR ligands have been described,34,35 the recent crystal structure solution of a soluble form of the human pre-BCR suggests that ligand-independent oligomerization is a likely mechanism of pre-BCR–mediated signaling.36 Subsequently, BCR expression is requisite for B-cell development and survival in the periphery.37
Antigen-induced B-cell activation and differentiation in secondary lymphoid tissues are mediated by dynamic changes in gene expression that give rise to the germinal center (GC) reaction (see section on B-cell maturation). The GC reaction is characterized by clonal expansion, class switch recombination (CSR) at the IgH locus, somatic hypermutation (SHM) of VH genes, and selection for increased affinity of a BCR for its unique antigenic epitope through affinity maturation. CSR, also known as isotype-switching, was first demonstrated in the chicken.38 A decade after the discovery of RAG1/2, Honjo and his colleagues (Muramatsu et al39) demonstrated that CSR and SHM are mediated by an enzyme designated activation-induced cytidine deaminase (AID). Expectedly, B-cell AID expression is induced in GCs where CSR and SHM occur. There are 2 theories on how AID functions to promote antibody diversification. One suggests that AID carries out an “RNA editing” function—not being the source of hypermutator activity, per se, but cooperating with another protein to mediate SHM.40 A more prevailing view posits that AID participates more directly to effect mutation of IgH genes at the DNA level.41 Unfortunately, generating a diverse protective Ig repertoire can be deleterious because AID can collaborate with other enzymes to generate chromosomal translocations involving c-myc and the IgH locus in some B-cell lymphomas.42
Lymphocyte development requires the concerted action of a network of cytokines and transcription factors that positively and negatively regulate gene expression. Marrow stromal cell–derived interleukin-7 (IL-7) is a nonredundant cytokine for murine B-cell development that promotes V to DJ rearrangement and transmits survival/proliferation signals.43 FLT3-ligand and TSLP play important roles in fetal B-cell development.24 The cytokine(s) that regulate human B-cell development are not as well understood.25 However, the presence of normal numbers of circulating B cells in primary immune deficiency patients with mutations in genes encoding the IL-7R argues that B-cell development at this stage of life does not require IL-7R signaling.44 An informative experiment of nature would be a patient with a null mutation in the IL-7 gene, but no such patient has yet been described. The cytokine (or cytokines) that promote marrow B-cell development at all stages of human life remains unknown.
At least 10 distinct transcription factors regulate the early stages of B-cell development, with E2A, EBF, and Pax5 being particularly important in promoting B-lineage commitment and differentiation.45 Pax5, originally characterized by its capacity to bind to promoter sequences in Ig loci, may be the most multifunctional transcription factor for B cells.46 Pax5-deficient mice have an arrest in B-cell development at the transition from DJ to VDJ rearrangement. An important revelation came from the discovery that Pax5 can activate genes necessary for B-cell development and repress genes that play critical roles in development of non–B-lineage cells. Thus, Pax5-deficient pro-B cells harbor the capacity to adapt non–B-lineage fates and develop into other hematopoietic lineages.47 Pax5 also regulates expression of at least 170 genes, a significant number of them important for B-cell signaling, adhesion, and migration of mature B cells.46 Conditional Pax5 deletion in mature murine B cells can result in dedifferentiation to an uncommitted hematopoietic progenitor and subsequent differentiation into T-lineage cells under certain conditions.48 It is remarkable that eliminating the function of this single transcription factor can lead to such a profound change in developmental fate. One obvious question is the whether such dedifferentiation occurs in normal mice (or healthy humans)? Alterations in the Pax5 locus may also have dire consequences; mice lacking Pax5 frequently develop high-grade lymphomas.48 Moreover, up to 30% of newly diagnosed cases of childhood B-lineage ALL harbor somatic PAX5 mutations, representing primarily monoallelic deletions.49 This provides a fresh perspective on the well-known maturation arrest in early B-cell development that characterizes essentially all childhood ALL cases.
Charting the B-cell surface
Up until approximately 1980, the molecular architecture of the B-cell surface was known to consist of membrane-bound Ig, complement component receptors, and Fc receptors; beyond that, the molecular constitution of the cell surface was completely uncharacterized. That all changed with the advent of monoclonal antibody (mAb) technology, a classic example of how the field of B-cell immunology has contributed important experimental tools that influence diverse areas of biology.50 The authors' involvement in characterizing B-lineage cell surface molecules with mAbs began as a postdoctoral fellow (T.W.L.) with John Kersey, and a graduate student (T.F.T.) with Max Cooper. In parallel, with Lee Nadler and the team of Ed Clark and Jeff Ledbetter, most of these B cell–restricted cell surface molecules were identified. Fittingly, the first B cell–specific molecule described was termed B1 by Nadler and colleagues (Stashenko et al51) and is now known as CD20.
Over the past 25 years, approximately 10 B cell–specific cell surface molecules have been identified by mAbs, with non–B-cell expression identified for some following their original characterization (Table 1). To bring a common nomenclature to the field, these mAbs were given WHO-sanctioned designations as “clusters of differentiation” (CD) by a series of international workshops on human leukocyte differentiation antigens. The first workshop held in Paris in 1982 was organized by Alain Bernard, Laurence Boumsell, Jean Dausset, César Milstein, and Stuart Schlossman. The CD nomenclature facilitated the classification of mAbs generated by different laboratories around the world against leukocyte cell surface epitopes. The mAbs were assigned a CD number once 2 independent mAbs were shown to bind to the same molecule. Since then, CD numbers have been assigned to more than 320 unique clusters and subclusters of mAbs. The CD designation is now universally embraced as a label for the target molecule rather than just a grouping of mAbs with common reactivity.
Table 1.
Cell surface CD molecules that are preferentially expressed by B cells
| Name | Original names | Cellular reactivity | Structure |
|---|---|---|---|
| CD19 | B4 | Pan-B cell, FDCs? | Ig superfamily |
| CD20 | B1 | Mature B cells | MS4A family |
| CD21 | B2, HB-5 | Mature B cells, FDCs | Complement receptor family |
| CD22 | BL-CAM, Lyb-8 | Mature B cells | Ig superfamily |
| CD23 | FcϵRII | Activated B cells, FDCs, others | C-type lectin |
| CD24 | BA-1, HB6 | Pan-B cell, granulocytes, epithelial cells | GPI anchored |
| CD40 | Bp50 | B cells, epithelial cells, FDCs, others | TNF receptor |
| CD72 | Lyb-2 | Pan-B cell | C-type lectin |
| CD79a,b | Igα,β | Surface Ig+ B cells | Ig superfamily |
Following their identification using mAbs, the structures of B cell–restricted target molecules have been determined and gene-targeted mice lacking expression of these CDs have been generated. Most of these target molecules regulate B-cell development and function, facilitate communication with the extracellular environment, or provide a cellular context in which to interpret BCR signals. Critical to the latter function are CD79a (Igα) and CD79b (Igβ), which are noncovalently associated components of the multiprotein cell surface BCR complex first described by John Cambier (Campbell et al52). CD79a and CD79b cytoplasmic domains contain highly conserved motifs for tyrosine phosphorylation and Src family kinase docking that are essential for initiating BCR signaling and B-cell activation.53
Studies from many laboratories evaluating murine and human B-lineage cells allow us to draw general conclusions regarding the function of specific cell surface molecules. CD19 is expressed by essentially all B-lineage cells and regulates intracellular signal transduction by amplifying Src-family kinase activity. CD20 is a mature B cell–specific molecule that functions as a membrane-embedded Ca2+ channel. Importantly, ritixumab is a chimeric CD20 mAb, which was the first mAb approved by the Food and Drug Administration (FDA) for clinical use in cancer therapy (eg, follicular lymphoma). CD21 is the C3d and Epstein-Barr virus receptor that interacts with CD19 to generate transmembrane signals and inform the B cell of inflammatory responses within microenvironments. CD22 functions as a mammalian lectin for α2,6-linked sialic acid that regulates follicular B-cell survival and negatively regulates signaling. CD23 is a low-affinity receptor for IgE expressed on activated B cells that influences IgE production. CD24 was among the first pan-B-cell molecules to be identified, but this unique GPI-anchored glycoprotein's function remains unknown. CD40 serves as a critical survival factor for GC B cells and is the ligand for CD154 expressed by T cells. CD72 functions as a negative regulator of signal transduction and as the B-cell ligand for Semaphorin 4D (CD100). There may be other unidentified molecules preferentially expressed by B cells, but the cell surface landscape is likely dominated by molecules shared with multiple leukocyte lineages. For example, David Pisetsky demonstrated in 1991 that B cells proliferate in response to bacterial DNA stimulation (Messina et al54), indicating the existence of B-cell surface receptors for DNA. These are now known to represent Toll-like receptors that are expressed by multiple leukocyte lineages.
B-cell maturation and subset development
B cells outside the marrow are morphologically homogenous, but their cell surface phenotypes, anatomic localization, and functional properties reveal still-unfolding complexities. Immature B cells exiting the marrow acquire cell surface IgD as well as CD21 and CD22, with functionally important density changes in other receptors. Immature B cells are also referred to as “transitional” (T1 and T2) based on their phenotypes and ontogeny (Figure 1), and have been characterized primarily in the mouse.55 Immature B cells respond to T cell–independent type 1 antigens such as lipopolysaccharides, which elicit rapid antibody responses in the absence of MHC class II–restricted T-cell help.56 The majority of mature B cells outside of the GALT reside within lymphoid follicles of the spleen and lymph nodes, where they encounter and respond to T cell–dependent foreign antigens bound to follicular dendritic cells (DCs), proliferate, and either differentiate into plasma cells or enter GC reactions.
GCs containing rapidly proliferating cells (ie, centroblasts) were first described in 1884, but were identified as the main site for high-affinity antibody-secreting plasma cell and memory B-cell generation a century later.57 It is within GCs where SHM and purifying selection produce the higher affinity B-cell clones that form the memory compartments of humoral immunity.57,58 Significantly, affinity maturation in GCs does not represent an intrinsic requirement for BCR signal strength but rather a local, Darwinian competition. The dynamics of lymphocyte entry into follicles and their selection for migration into and within GCs represents a complex ballet of molecular interactions orchestrated by chemotactic gradients and BCR engagement that is only now being elucidated.59
B-cell subsets with individualized functions such as B-1 and marginal zone (MZ) B cells have also been identified. First described in 1983 by Lee Herzenberg, murine B-1 cells are a unique CD5+ B-cell subpopulation (Hayakawa et al60) distinguished from conventional B (B-2) cells by their phenotype, anatomic localization, self-renewing capacity, and production of natural antibodies.61 Peritoneal B-1 cells are further subdivided into the B-1a (CD5+) and B-1b (CD5−) subsets. Their origins, and whether they derive from the same or distinct progenitors compared with B-2 cells, have been controversial.62 However, a B-1 progenitor that appears distinct from a B-lineage progenitor that develops primarily into the B-2 population has been identified in murine fetal marrow, and to a lesser degree in adult marrow.63 B-1a cells and their natural antibody products provide innate protection against bacterial infections in naive hosts, while B-1b cells function independently as the primary source of long-term adaptive antibody responses to polysaccharides and other T cell–independent type 2 antigens during infection.64 The function and potential subpopulation status of human B-1 cells is less understood.62 MZ B cells are a unique population of murine splenic B cells with attributes of naive and memory B cells,65 and constitute a first line of defense against blood-borne encapsulated bacteria. Uncertainty regarding the identity of human MZ B cells partially reflects the fact that the microscopic anatomy of the human splenic MZ differs from rodents.66 Likewise, the microscopic anatomy of human follicular mantle zones is not recapitulated in mouse spleen and lymph nodes.
The B1, MZ, and GC B-cell subsets all contribute to the circulating natural antibody pool, thymic-independent IgM antibody responses, and adaptive immunity by terminal differentiation into plasma cells, the effector cells of humoral immunity.67 Antigen activation of mature B cells leads initially to GC development, the transient generation of plasmablasts that secrete antibody while still dividing, and short-lived extrafollicular plasma cells that secrete antigen-specific germ line–encoded antibodies (Figure 1). GC-derived memory B cells generated during the second week of primary antibody responses express mutated BCRs with enhanced affinities, the product of SHM. Memory B cells persist after antigen challenge, rapidly expand during secondary responses, and can terminally differentiate into antibody-secreting plasma cells.68 In a manner similar to the early stages of B-cell development in fetal liver and adult marrow, plasma cell development is tightly regulated by a panoply of transcription factors, most notably Bcl-6 and BLIMP-1.69
Persistent antigen-specific antibody titers derive primarily from long-lived plasma cells.67 Primary and secondary immune responses generate separate pools of long-lived plasma cells in the spleen, which migrate to the marrow where they occupy essential survival niches and can persist for the life of the animal without the need for self-replenishment or turnover.67,68 The marrow plasma cell pool does not require ongoing contributions from the memory B-cell pool for its maintenance, but when depleted, plasma cells are replenished from the pool of memory B cells.70 Thereby, persisting antigen, cytokines, or Toll-like receptor signals may drive the memory B-cell pool to chronically differentiate into long-lived plasma cells for long-lived antibody production.
B-cell functions in addition to antibody production
In addition to their essential role in humoral immunity, B cells also mediate/regulate many other functions essential for immune homeostasis (Figure 2). Of major importance, B cells are required for the initiation of T-cell immune responses, as first demonstrated in mice depleted of B cells at birth using anti-IgM antiserum.71 However, this has not been without controversy as an absence of B cells impairs CD4 T-cell priming in some studies, but not others. Nonetheless, antigen-specific interactions between B and T cells may require the antigen to be first internalized by the BCR, processed, and then presented in an MHC-restricted manner to T cells.72–74 Recent studies depleting B cells from normal adult mice demonstrated that B cells are essential for optimal CD4 T-cell activation during immune responses to low-dose foreign antigens and autoantigens, balancing this responsibility with DCs.75 The differential activation and expansion of CD4 T cells by B cells and/or DCs may impart characteristic features on immune responses, making this a critical area for future studies of host defense and autoimmunity.
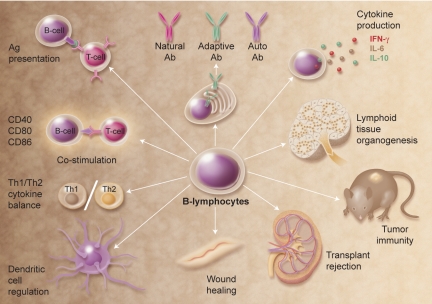
Figure 2.
Multifunctional attributes of B cells. Selected examples of how B cells regulate immune homeostasis are shown; many of these functions are independent of Ig production. Illustration by A. Y. Chen.
The congenital absence of B cells during mouse development also leads to abnormalities within the immune system, including a profound decrease in thymocyte numbers and diversity, significant defects within spleen DC and T-cell compartments, an absence of Peyer patch organogenesis and follicular DC networks, and an absence of MZ and metallophilic macrophages with decreased chemokine expression. While critical for normal immune system development, B cells are also important for its maintenance. For example, B cells can release immunomodulatory cytokines that can influence a variety of T-cell, DC, and antigen-presenting cell functions, regulate lymphoid tissue organization and neogenesis, regulate wound healing and transplanted tissue rejection, and influence tumor development and tumor immunity. B cells can also function as polarized cytokine-producing effector cells that influence T-cell differentiation.76 In addition, regulatory B cells have been described.77 One phenotypically distinct subset, designated B10 cells, has been shown to uniquely regulate T cell–mediated inflammatory responses through the production of IL-10 (Figure 1).78 The further characterization of B-cell subsets that carry out each of these functions and the molecular mediators for these diverse activities are exciting areas for future study.
Abnormalities in B-cell development: immunodeficiencies
The importance of genes encoding the pre-BCR and downstream signaling molecules has been demonstrated in gene-targeted mice and patients with primary immunodeficiencies. Perhaps the best studied is the Bruton tyrosine kinase (BTK) gene mutated in X-linked agammaglobulinemia (XLA).79,80 XLA was originally observed in a single male patient with recurrent bacterial sepsis and no detectable serum gammaglobulin.81 All XLA patients have a block at the pro-B to large pre-B-cell transition in marrow, and a substantial reduction in the percentage of normal peripheral blood B cells.82 Btk plays a central role in signaling downstream of pre-BCR and BCR activation, primarily by promoting calcium flux.83 The BTKbase has cataloged more than 600 distinct mutations in 1100 patients,84 with many mutations leading to changes in Btk protein folding or stability. Although the perturbation in marrow B-cell development is a universal characteristic of XLA patients, susceptibility to specific pathogens, age at diagnosis, number of circulating B cells, and level of serum Ig are more variable.82 Interestingly, the Btk mutation in the xid mouse and mice with a targeted disruption of Btk82 have a milder form of B-cell immunodeficiency compared with most XLA patients. Future studies may reveal the contribution of individual genetic variation in either compensating for or exacerbating the phenotypic effect of a BTK mutation.85
Two heterogeneous groups of immunodeficiencies impact primarily later stages of B-cell development. Individuals with common variable immune deficiency (CVID) exhibit low serum Ig and an increased susceptibility to infection, accompanied by variable reductions in memory B cells, CSR, and B-cell activation. Mutations in several genes have been identified in CVID patients, including the activated T-cell stimulatory molecule ICOS, the B-cell surface receptor CD19, and the TNF receptor superfamily member TACI.86 However, it is estimated that mutations in these 3 genes occur in only 10% to 15% of CVID patients. The age at diagnosis ranges from 3 to 78 years and is probably influenced by a host of genetic and environmental factors. A second group includes individuals with so-called hyper-IgM syndrome. These patients have recurrent infections, elevated serum IgM, a relative paucity of other serum Ig isotypes, and a general failure of B cells to undergo CSR and SHM.87 Mutations in several genes have been identified, the most common being a mutation in CD154 (CD40 ligand) expressed on activated T cells, which is inherited as an X-linked mutation. Rare patients with mutations in CD40, IKK-gamma/NEMO, and uracil-DNA glycosylase have also been reported. The critical function of AID in development of a functional Ig repertoire is underscored by the fact that patients with an autosomal recessive form of the hyper-IgM syndrome harbor a deficiency of this enzyme.88
Abnormalities in B-cell regulation: autoimmunity
A series of checkpoints normally controls B-cell selection, both centrally in the marrow and in the peripheral lymphoid tissues; in the latter a second screening process for reactivity with peripheral self-antigens results in apoptosis, receptor editing, or anergy.89 The outcome is the production of a diverse population of B cells capable of producing high-affinity effector antibodies that have been purged of pathological autoreactivity. However, disruption of the delicate balance between activating and inhibitory signals that regulate normal B-cell activation and longevity can predispose to pathogenic autoantibody production and autoimmunity.
Autoantibodies represent important hallmarks for adaptive immune responses that are inappropriately directed against self-tissues.90 Autoantibody and rheumatoid factor identification during the 1950s to 1970s triggered a paradigm shift that some “connective tissue diseases” such as rheumatoid arthritis were actually autoimmune diseases. The identification of autoantibodies in glomerulonephritis patients in 1949 provided the first clinical and experimental evidence that disease is due to a continuous organ-specific antigen-antibody reaction.91 Equally remarkable is that rheumatoid factor was identified and biochemically characterized in 1957, prior to the identification of IgM and IgG.92 Although a close correlation between autoantibody production and systemic autoimmune disease is universally acknowledged, the precise contributions of autoantibodies and their autoantigen targets to disease pathogenesis remain to be elucidated in many human syndromes. Furthermore, B cells may contribute to autoimmune pathogenesis by presentation of autoantigen to T cells, or through production of proinflammatory cytokines.93
A striking example of how dysregulated apoptotic regulatory genes can lead to autoimmunity is in Bcl-2 transgenic mice. Enforced Bcl-2 expression allows the inappropriate survival of autoreactive B-cell clones that normally are lost through negative selection and apoptosis.94 Bcl-2 transgenic mice develop antinuclear antibodies, immune complex deposition, and glomerulonephritis. MRL mice homozygous for mutations in Fas or FasL (frequently referred to as the lpr and gld phenotypes, respectively) spontaneously develop an autoimmune disease resembling human systemic lupus erythematosus (SLE).95 Subtle alterations in signaling pathways that influence BCR responses to autoantigen are also sufficient to predispose mice and possibly humans to autoantibody production and autoimmunity. For example, mice with lymphoproliferation phenotypes share similarities with Canale-Smith syndrome, a childhood disorder characterized by lymphadenopathy and autoimmunity due to novel FAS mutations.96 In most cases of human autoimmune disease, however, the combination of environmental factors and polygenic effects collectively dysregulate normal B-cell function and thereby confer disease susceptibility.
As it has become more obvious that B cells contribute substantially to multiple human autoimmune diseases, highly targeted less toxic therapies focused on restoring normal B-cell function and eliminating pathogenic autoantibodies are beginning to reduce the reliance on immunosuppressive drugs. Rituximab (CD20 mAb) effectively reduces B-cell numbers without significant toxicity, and has become a standard therapy for rheumatoid arthritis and increasingly for other autoimmune diseases as well.97 Studies in the mouse have shown that rituximab leads to B-cell elimination by monocyte-mediated Ab-dependent cellular cytotoxicity,98 but likely inhibits autoimmune disease initiation by regulating CD4+ T-cell expansion, thereby delaying the subsequent inflammatory phase of disease that leads to tissue pathology.75
The BAFF/BlyS cytokine produced by DCs and monocyte/macrophages is particularly important in B-cell survival and differentiation.99 BAFF binds 3 receptors—BCMA, TACI, and BAFF-R100—and induces immature B-cell survival and mature B-cell proliferation within peripheral lymphoid tissues when coupled with BCR ligation. BAFF transgenic mice have SLE-like disease, and elevated BAFF levels are found in a significant number of SLE patients. Such findings suggest that antagonizing BAFF function may disrupt B-cell survival. Disrupting CD22 ligand binding also disrupts B-cell survival and may thereby provide an additional therapeutic strategy for patients with autoimmune disease.101
B-cell abnormalities: leukemia and lymphomas
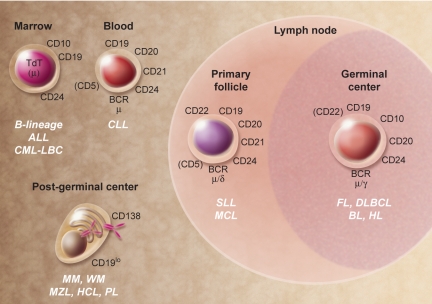
Studies at the interface of hematology and immunology are exemplified by interchangeable lessons learned from parallel evaluation of B-cell ontogeny and hematologic malignancies. Broadly speaking, the normal B-cell developmental stages shown in Figure 1 have malignant counterparts that reflect the expansion of a dominant subclone leading to development of leukemia and lymphoma. Figures 3 and 4 show some specific subtypes of human leukemia/lymphoma and their approximate stage of B-cell development, and selected morphologic images. The application of mAbs in multiparameter flow cytometry,102 detection of Ig gene rearrangements, detection of cytogenetic/molecular genetic abnormalities, and gene expression profiling103 have revealed an incredibly detailed portrait of gene expression and gene product function in normal and malignant B-lineage cells. For example, the marriage of Ig gene rearrangement detection and mAb phenotyping resulted in the so-called (at the time) “non-T, non-B” ALL being reclassified as a malignancy of B-cell precursors.104 The antiapoptotic Bcl-2 gene was discovered as the translocation partner with the IgH locus in the t(14;18)(q32;q21); frequently occurring in follicular lymphoma.105 Not only are the biologic and clinical consequences of Bcl-2 overexpression in follicular lymphoma profound, but the concept that increased expression of a single protein could promote cell survival has had an impact in areas of biology that go far beyond this lymphoma. The discovery that a substantial number of cases of diffuse large B-cell lymphoma exhibit dysregulated expression of the transcriptional repressor Bcl-6106 has led to new insight into the normal developmental program of antigen-activated B cells and the transformation events that underlie this lymphoma. The developmental origin of the Hodgkin/Reed-Sternberg cell in Hodgkin lymphoma, a controversy that was unsettled for many years, was resolved by single-cell analyses indicating a B-lymphocyte origin based on the demonstration of clonal Ig gene rearrangements.107
Figure 3.
Human B-cell malignancies. Selected cell surface, cytoplasmic, and nuclear markers expressed during normal B-cell development that are generally expressed in malignancies. Molecules within parentheses are variable in their expression at the indicated stage of development. ALL indicates acute lymphoblastic leukemia; BL, Burkitt lymphoma; CLL, chronic lymphocytic leukemia; CML-LBC, chronic myelocytic leukemia–lymphoid blast crisis; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma; HCL, hairy cell leukemia; HL, Hodgkin lymphoma; MCL, mantle cell lymphoma; MM, multiple myeloma; MZL, marginal zone lymphoma; PL, plasmablastic lymphoma; SLL, small lymphocytic lymphoma; and WM, Waldenström macroglobulinemia. Illustration by A. Y. Chen.
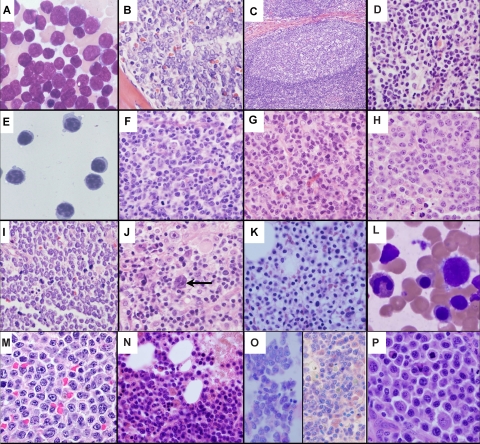
Figure 4.
A histopathological montage of normal and malignant human B-lineage cells. (A) Bone marrow aspirate of ALL cells exhibiting a high nuclear to cytoplasmic ratio, finely dispersed chromatin, indistinct nucleoli, and blue-gray cytoplasm. (B) Bone marrow biopsy showing effacement of the marrow architecture by a monotonous population of ALL lymphoblasts. (C) Representative normal lymph node follicle showing a reactive GC surrounded by mantle zone. (D) MCL cells display a heterogeneous morphology ranging from small round (true mantle zone/CLL-like) cells to those showing irregular cleaved nuclei (FL-like). (E) A peripheral blood sample of CLL cells exhibiting closely condensed chromatin, indistinct nucleoli, and scant basophilic cytoplasm with a regular outline. (F) Normal GC centrocytes with vesicular chromatin and irregular nuclear outlines, along with larger round centroblasts showing a similar open chromatin. (G) FL with a predominance of small cleaved centrocyte-like cells demonstrating significant nuclear irregularity. (H) DLBCL with classic post-GC immunoblastic morphology exhibiting a large nucleus, eosinophilic nucleoli, vesicular chromatin, and relatively abundant cytoplasm. (I) Biopsy from an abdominal mass showing BL cells with a monomorphic infiltrate of small to intermediate-sized cells with round nuclei, indistinct nucleoli, clumped chromatin, a basophilic cytoplasm, and a high apoptotic cell index. (J) A characteristic Reed-Sternberg cell (arrow) in HL. The cells are large and binucleated, with mirror image nuclei, demonstrating prominent eosinophilic nucleoli. Many of the other cells in the background are reactive lymphocytes. (K) Bone marrow biopsy showing hairy cell leukemia. The cells are evenly spaced from each other with round nuclei and clear abundant cytoplasm, imparting a “fried egg” appearance to the cells. (L) Peripheral blood smear of hairy cell leukemia (HCL) demonstrating classic concentric hairlike cytoplasmic projections. (M) MZL cells with a heterogeneous appearance ranging from small round cells to those manifesting a cleaved morphology. (N) WM/lymphoplasmacytic lymphoma showing considerable morphologic overlap with MZL. The cells demonstrate significant plasmacytoid features with pseudointranuclear inclusions (ie, Dutcher bodies). Many of the plasmacytoid cells contain globular collections of paraprotein in the cytoplasm, imparting a mulberry-like appearance to the cell. (O) Two cases of MM within the marrow. The cells show typical plasma cell features including the eccentrically disposed nucleus and abundant Ig-containing cytoplasm. (P) PL comprising enlarged plasmacytoid cells with enhanced atypia; mitoses are easily identifiable. All photographs were taken with an Olympus BX41 microscope and DP71 camera (Olympus America, Center Valley, PA). The images were acquired using the following UPlanFL N objective lenses: panels A,E,J,L,M,O, P: 100×/1.30, oil immersion; panels B,F,G,H,I,K,N: 40×/0.75; panels C,D: 20×/0.50. Panels A, L, and O are Giemsa stains; all others are H and E stains. Images were assembled using Adobe Photoshop version 9.0.2 (Adobe Systems, San Jose, CA). Panels A and E were provided by Dr Amy Chadburn, and all other images were provided by Dr Cynthia Magro, both from the Weill Medical College of Cornell University, New York, NY.
Concluding remarks
B cells are blood cells. Studies of normal and abnormal B-cell development/function have crossed the 2 prominent disciplines of hematology and immunology since the inception of Blood 50 years ago. A prominent accomplishment of this cross-disciplinary effort is the identification of somatic mutations in B-lineage cells that can result in immunodeficiency and leukemia/lymphoma. However, the underlying genetic abnormalities that predispose to many autoimmune diseases are much less well characterized, and likely are more complex. Continuing characterization of how B cells communicate with and respond to their microenvironment will provide additional insight into the pathophysiology of autoimmune disease. High-throughput SNP analysis and modifier gene characterization will likely lead to revised etiologies for these disorders. Progress in basic and clinical B-cell research has been a gradual continuum, with notable spikes of discovery frequently driven by technical advances. This will continue. For example, continuing technical advancements in intravital and subcellular imaging, genomics, and proteomics will be drivers of discovery. Imaging tools at the tissue/cellular and molecular levels will characterize critical interactions and protein topologies not feasible with conventional biochemical/biophysical methods. We expect that the proteome of B-lineage cells at various stages of development will be quantified in the not-too-distant future. This will require assistance from our bioinformatics colleagues to make sense of it all. The recently discovered noncoding micro RNAs (miR) have already emerged as a new centerpiece of understanding. One species, miR-155, regulates GC formation108 and plasma cell development.109 Alterations in miR expression in leukemia and lymphoma are being rapidly characterized. The target genes regulated by miR will reveal another layer of developmental complexity that may also provide new insight into B-cell development and the pathogenesis of leukemia/lymphoma, immunodeficiency, and autoimmunity.
Acknowledgments
This paper is dedicated to Dr Max Cooper who we believe, as much as anyone, deserves the title “The Father of B-cell Immunology.” Max is a true gentleman who can claim 4 decades of monumental contributions to our understanding of the normal and abnormal development of B lymphocytes. The authors are inspired by his science and privileged to be his friends. We are also grateful to our many students who provided daily inspiration over the years, and to current colleagues who made comments on this review. We wish to particularly acknowledge Drs Cynthia Magro and Amy Chadburn of Weill Cornell Medical College, who contributed Figure 4.
Biography
Thomas F. Tedder
T.F.T.: My interest in immune system development was sparked when I applied to graduate school with the specific intention of working with Max Cooper at the University of Alabama in Birmingham (UAB). Joining Max's laboratory in 1980 turned a spark into a passion as I learned of the unique developmental program of B cells that was unfolding in the early 1980s. With the goal of developing reagents to identify human B-cell subsets and their differentiation stages, I began making mAbs that were preferentially reactive with B-cell surface molecules that might also regulate B-cell function. The naive concept was that we would identify molecular switches to turn on and off for the treatment of B-cell malignancies, immunodeficiency, and autoimmunity. These early studies focused on molecules now known as CD21, CD24, CD45RB, and others. Remarkably, I am still using many of the mAbs that we developed during that time in my laboratory today.
My career path was cemented when I joined the laboratory of Stuart Schlossman in the Division of Tumor Immunology at the Dana-Farber Cancer Institute and Harvard Medical School in 1984. Lee Nadler and other members of the Division were also developing mAbs to other B-cell surface molecules, with the shared goal of deciphering the function of each target molecule. This was an exciting time in human B-cell biology where these collaborative efforts not only identified new molecules such as CD19, CD20, CD21, and CD22, but also characterized their biochemical features and associations, and their functions in vitro. When Haruo Sato joined the division, he taught me molecular biology and how to clone cDNAs encoding CD20 and the other surface molecules that we were studying. This molecular approach also allowed us to identify new molecules with interesting functions, such as L-selectin. This was the perfect environment for starting an independent career, with a wealth of close collaborators and supportive colleagues, and the generous support of Stuart Schlossman.
When I left the division in 1993 to become the first chairman of the new Department of Immunology at Duke University Medical Center, we made a complete shift from studying human B-cell development into mouse models. We began knocking out the genes encoding the proteins we had been studying, and obtained the first clear views of how these molecules were working in vivo. It was amazing to see how much faster experiments could be done when you were not purifying B cells from human blood each day for experiments. Over the past 15 years, we have remained steadfast in our focus on understanding how B-lymphocyte cell surface molecules regulate humoral immunity, and more recently innate and adaptive immunity in a broader context. Finally, we now know enough about the functions of CD19, CD20, and CD22 that we can take our mAbs back into humans with the potential for new treatments for oncology and autoimmune disease. It has been an amazing journey from growing up with dirt roads, to UAB, Harvard, and now Duke. Along the way, I have always remembered the sobering reality that I am still working on the subject of my PhD thesis, and calling Max Cooper for advice!
The authors' friendship goes back more than 20 years when T.W.L. took a sabbatical to work in Cox Terhorst's laboratory at the Dana-Farber Cancer Institute in 1986. We were aware of each other's work long before that, and decided we might learn from each other. The 2 of us would get together to discuss mutual interests in B lymphocytes, publishing papers, submitting National Institutes of Health (NIH) applications—all the good stuff. The current essay is our first collaboration, but hopefully also the first of more to follow.
Tucker W. LeBien
T.W.L.: I started my postdoctoral training with John Kersey at the University of Minnesota in 1977. One of my projects was to generate rabbit heteroantisera against human leukemic cells, with the hope that such reagents could be used to discriminate childhood leukemia from normal marrow and peripheral blood cells. I well remember immunizing rabbits with a pre-B acute lymphoblastic leukemia cell line (NALM-6) or crude membrane fractions, and “adsorbing” the hyperimmune sera with an autologous B-lymphoblastoid cell line to eliminate components of the polyclonal sera that would cross-react with membrane molecules on normal cells. Although this was a logical approach at the time, there was endless variability in antibody titer and specificity from rabbit to rabbit. However, a change in direction was prompted by a chance conversation I had when the late Robert Good was visiting in the summer of 1978. I remember Good describing the new technology of monoclonal antibody (mAb) production, and we became enthralled with the possibility of applying this technology to our studies of leukemia. Carlo Croce (who was then at the Wistar Institute) was invited to Minnesota in early 1979 (as I recall) to present a seminar and discuss the technical nuances of mAb production. With these events as a catalyst, I set out to immunize mice with NALM-6 and produce novel reagents. It took me several months to resolve the technical problems, one of which was the variability in lots of polyethylene glycol used in the delicate procedure of fusing immunized spleen cells with murine myeloma cell lines. But in the end it worked and we were off and running. Although we failed in our naive goal of producing “leukemia-specific” mAbs, we did produce reagents that were instrumental in identifying and defining CD9, CD10, CD11a, and CD24 in the First International Workshop on Leukocyte Differentiation Antigens in 1982. So it was really the development of mAbs and my interests in applying these reagents to studying human leukemia that catalyzed my initial interests in hematology.
My continuing education in hematology was greatly influenced by numerous distinguished investigators in the Departments of Pediatrics and Medicine at the University of Minnesota. With a tradition of excellence in areas such as childhood leukemia and marrow transplantation, I came to appreciate the importance of translational research long before the term was popularized. In fact, working with my colleagues John Kersey and Norma Ramsay we developed an ex vivo protocol to purge autologous bone marrow of residual leukemic cells using the mAbs BA-1 (anti-CD24), BA-2 (anti-CD9), and BA-3 (anti-CD10). Although only a small number of patients underwent transplantation following this procedure, it really opened my eyes to the possibility of applying basic science to attack complex clinical problems. My involvement with ASH ramped up considerably during my term as an Associate Editor of Blood from 1990 to 1997. This was a time-consuming but greatly rewarding experience. I learned an extraordinary amount about hematology from the approximately 3000 reviewer critiques I handled over that 7-year period! Blood continues to be one of my favorite reads, and I am amazed at how every issue is chock full of terrific science. Currently, I juggle my research interests in human B-cell development with other responsibilities as Associate Director of Basic Sciences in the University of Minnesota Cancer Center, and Director of the University of Minnesota Medical Scientist Training Program.
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tucker W. LeBien, MMC 806, University of Minnesota Cancer Center, 420 Delaware St SE, Minneapolis, MN 55455; e-mail: lebie001@umn.edu; or Thomas F. Tedder, Box 3010, Department of Immunology, Duke University Medical Center, Durham, NC 27710; e-mail: thomas.tedder@duke.edu.
References
- 1.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Tiselius A, Kabat EA. Electrophoresis of immune serum. Science. 1938;87:416–417. doi: 10.1126/science.87.2262.416-a. [DOI] [PubMed] [Google Scholar]
- 3.Fagraeus A. The plasma cellular reaction and its relation to the formation of antibodies in vitro. J Immunol. 1948;58:1–13. [PubMed] [Google Scholar]
- 4.Jerne NK. The natural-selection theory of antibody formation. Proc Natl Acad Sci U S A. 1955;41:849–857. doi: 10.1073/pnas.41.11.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnet FM. A modification of Jerne's theory of antibody production using the concept of clonal selection. Aust J Sci. 1957;20:67–69. doi: 10.3322/canjclin.26.2.119. [DOI] [PubMed] [Google Scholar]
- 6.Nossal GJ, Lederberg J. Antibody production by single cells. Nature. 1958;181:1419–1420. doi: 10.1038/1811419a0. [DOI] [PubMed] [Google Scholar]
- 7.Nossal GJ. One cell-one antibody: prelude and aftermath. Nat Immunol. 2007;8:1015–1017. doi: 10.1038/ni1007-1015. [DOI] [PubMed] [Google Scholar]
- 8.Edelman GM, Gally JA. A model for the 7S antibody molecule. Proc Natl Acad Sci U S A. 1964;51:846–853. doi: 10.1073/pnas.51.5.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Porter RR. Structure of gamma-globulin and antibodies. Br Med Bull. 1963;19:197–201. doi: 10.1093/oxfordjournals.bmb.a070056. [DOI] [PubMed] [Google Scholar]
- 10.Good RA, Zak SJ. Disturbances in gamma globulin synthesis as experiments of nature. Pediatrics. 1956;18:109–149. [PubMed] [Google Scholar]
- 11.Cooper MD, Peterson RD, Good RA. Delineation of the thymic and bursal lymphoid systems in the chicken. Nature. 1965;205:143–146. doi: 10.1038/205143a0. [DOI] [PubMed] [Google Scholar]
- 12.Cooper MD, Raymond DA, Peterson RD, South MA, Good RA. The functions of the thymus system and the bursa system in the chicken. J Exp Med. 1966;123:75–102. doi: 10.1084/jem.123.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller JF, Mitchell GF. Cell to cell interaction in the immune response, I: hemolysin-forming cells in neonatally thymectomized mice reconstituted with thymus or thoracic duct lymphocytes. J Exp Med. 1968;128:801–820. doi: 10.1084/jem.128.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell GF, Miller JF. Cell to cell interaction in the immune response, II: the source of hemolysin-forming cells in irradiated mice given bone marrow and thymus or thoracic duct lymphocytes. J Exp Med. 1968;128:821–837. doi: 10.1084/jem.128.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coombs RR, Feinstein A, Wilson AB. Immunoglobulin determinants on the surface of human lymphocytes. Lancet. 1969;2:1157–1160. doi: 10.1016/s0140-6736(69)92484-2. [DOI] [PubMed] [Google Scholar]
- 16.Froland S, Natvig JB, Berdal P. Surface-bound immunoglobulin as a marker of B lymphocytes in man. Nat New Biol. 1971;234:251–252. doi: 10.1038/newbio234251a0. [DOI] [PubMed] [Google Scholar]
- 17.Raff MC. T and B lymphocytes and immune responses. Nature. 1973;242:19–23. doi: 10.1038/242019a0. [DOI] [PubMed] [Google Scholar]
- 18.Greaves MF, Owen JJT, Raff MC. Amsterdam, The Netherlands: Excerpta Medica; 1974. T and B lymphocytes: Origins. Properties, and Roles in Immune Responses. [Google Scholar]
- 19.Preud'homme JL, Seligmann M. Surface bound immunoglobulins as a cell marker in human lymphoproliferative diseases. Blood. 1972;40:777–794. [PubMed] [Google Scholar]
- 20.Raff MC, Megson M, Owen JJ, Cooper MD. Early production of intracellular IgM by B-lymphocyte precursors in mouse. Nature. 1976;259:224–226. doi: 10.1038/259224a0. [DOI] [PubMed] [Google Scholar]
- 21.Gathings WE, Lawton AR, Cooper MD. Immunofluorescent studies of the development of pre-B cells, B lymphocytes and immunoglobulin isotype diversity in humans. Eur J Immunol. 1977;7:804–810. doi: 10.1002/eji.1830071112. [DOI] [PubMed] [Google Scholar]
- 22.Vogler LB, Crist WM, Bockman DE, Pearl ER, Lawton AR, Cooper MD. Pre-B-cell leukemia: a new phenotype of childhood lymphoblastic leukemia. N Engl J Med. 1978;298:872–878. doi: 10.1056/NEJM197804202981603. [DOI] [PubMed] [Google Scholar]
- 23.LeBien TW, Hozier J, Minowada J, Kersey JH. Origin of chronic myelocytic leukemia in a precursor of pre-B lymphocytes. N Engl J Med. 1979;301:144–147. doi: 10.1056/NEJM197907193010307. [DOI] [PubMed] [Google Scholar]
- 24.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 25.LeBien TW. Fates of human B-cell precursors. Blood. 2000;96:9–23. [PubMed] [Google Scholar]
- 26.Brack C, Hirama M, Lenhard-Schuller R, Tonegawa S. A complete immunoglobulin gene is created by somatic recombination. Cell. 1978;15:1–14. doi: 10.1016/0092-8674(78)90078-8. [DOI] [PubMed] [Google Scholar]
- 27.Reynaud CA, Bertocci B, Dahan A, Weill JC. Formation of the chicken B-cell repertoire: ontogenesis, regulation of Ig gene rearrangement, and diversification by gene conversion. Adv Immunol. 1994;57:353–378. doi: 10.1016/s0065-2776(08)60676-8. [DOI] [PubMed] [Google Scholar]
- 28.Becker RS, Knight KL. Somatic diversification of immunoglobulin heavy chain VDJ genes: evidence for somatic gene conversion in rabbits. Cell. 1990;63:987–997. doi: 10.1016/0092-8674(90)90502-6. [DOI] [PubMed] [Google Scholar]
- 29.Schatz DG, Oettinger MA, Baltimore D. The V(D)J recombination activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 30.Oettinger MA, Schatz DG, Gorka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 31.Alt FW, Blackwell TK, DePinho RA, Reth MG, Yancopoulos GD. Regulation of genome rearrangement events during lymphocyte differentiation. Immunol Rev. 1986;89:5–30. doi: 10.1111/j.1600-065x.1986.tb01470.x. [DOI] [PubMed] [Google Scholar]
- 32.Sakaguchi N, Melchers F. Lambda 5, a new light-chain-related locus selectively expressed in pre-B lymphocytes. Nature. 1986;324:579–582. doi: 10.1038/324579a0. [DOI] [PubMed] [Google Scholar]
- 33.Melchers F. The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol. 2005;5:578–584. doi: 10.1038/nri1649. [DOI] [PubMed] [Google Scholar]
- 34.Bradl H, Wittmann J, Milius D, Vettermann C, Jack HM. Interaction of murine precursor B cell receptor with stroma cells is controlled by the unique tail of lambda 5 and stroma cell-associated heparan sulfate. J Immunol. 2003;171:2338–2348. doi: 10.4049/jimmunol.171.5.2338. [DOI] [PubMed] [Google Scholar]
- 35.Gauthier L, Rossi B, Roux F, Termine E, Schiff C. Galectin-1 is a stromal cell ligand of the pre-B cell receptor (BCR) implicated in synapse formation between pre-B and stromal cells and in pre-BCR triggering. Proc Natl Acad Sci U S A. 2002;99:13014–13019. doi: 10.1073/pnas.202323999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bankovich AJ, Raunser S, Juo ZS, Walz T, Davis MM, Garcia KC. Structural insight into pre-B cell receptor function. Science. 2007;316:291–294. doi: 10.1126/science.1139412. [DOI] [PubMed] [Google Scholar]
- 37.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–1083. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 38.Kincade PW, Lawton AR, Bockman DE, Cooper MD. Suppression of immunoglobulin G synthesis as a result of antibody-mediated suppression of immunoglobulin M synthesis in chickens. Proc Natl Acad Sci U S A. 1970;67:1918–1925. doi: 10.1073/pnas.67.4.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 40.Honjo T, Nagaoka H, Shinkura R, Muramatsu M. AID to overcome the limitations of genomic information. Nat Immunol. 2005;6:655–661. doi: 10.1038/ni1218. [DOI] [PubMed] [Google Scholar]
- 41.Reynaud CA, Aoufouchi S, Faili A, Weill JC. What role for AID: mutator, or assembler of the immunoglobulin mutasome? Nat Immunol. 2003;4:631–638. doi: 10.1038/ni0703-631. [DOI] [PubMed] [Google Scholar]
- 42.Okazaki IM, Kotani A, Honjo T. Role of AID in tumorigenesis. Adv Immunol. 2007;94:245–273. doi: 10.1016/S0065-2776(06)94008-5. [DOI] [PubMed] [Google Scholar]
- 43.Milne CD, Paige CJ. IL-7: a key regulator of B lymphopoiesis. Semin Immunol. 2006;18:20–30. doi: 10.1016/j.smim.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 44.Buckley RH. Molecular defects in human severe combined immunodeficiency and approaches to immune reconstitution. Annu Rev Immunol. 2004;22:625–655. doi: 10.1146/annurev.immunol.22.012703.104614. [DOI] [PubMed] [Google Scholar]
- 45.Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26:715–725. doi: 10.1016/j.immuni.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 46.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 47.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- 48.Cobaleda C, Jochum W, Busslinger M. Conversion of mature B cells into T cells by dedifferentiation to uncommitted progenitors. Nature. 2007;449:473–477. doi: 10.1038/nature06159. [DOI] [PubMed] [Google Scholar]
- 49.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 50.Kohler G, Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 51.Stashenko P, Nadler LM, Hardy R, Schlossman SF. Characterization of a human B lymphocyte-specific antigen. J Immunol. 1980;125:1678–1685. [PubMed] [Google Scholar]
- 52.Campbell KS, Hager EJ, Friedrich RJ, Cambier JC. IgM antigen receptor complex contains phosphoprotein products of B29 and mb-1 genes. Proc Natl Acad Sci U S A. 1991;88:3982–3986. doi: 10.1073/pnas.88.9.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reth M. Antigen receptor tail clue. Nature. 1989;338:383–384. [PubMed] [Google Scholar]
- 54.Messina JP, Gilkeson GS, Pisetsky DS. Stimulation of in vitro murine lymphocyte proliferation by bacterial DNA. J Immunol. 1991;147:1759–1764. [PubMed] [Google Scholar]
- 55.Chung JB, Silverman M, Monroe JG. Transitional B cells: step by step towards immune competence. Trends Immunol. 2003;24:343–349. doi: 10.1016/s1471-4906(03)00119-4. [DOI] [PubMed] [Google Scholar]
- 56.Coutinho A, Moller G. Thymus-independent B-cell induction and paralysis. Adv Immunol. 1975;21:113–236. doi: 10.1016/s0065-2776(08)60220-5. [DOI] [PubMed] [Google Scholar]
- 57.Jacob J, Kelsoe G, Rajewsky K, Weiss U. Intraclonal generation of antibody mutants in germinal centres. Nature. 1991;354:389–392. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- 58.Kelsoe G. Life and death in germinal centers (redux). Immunity. 1996;4:107–111. doi: 10.1016/s1074-7613(00)80675-5. [DOI] [PubMed] [Google Scholar]
- 59.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hayakawa K, Hardy RR, Parks DR, Herzenberg LA. The “Ly-1 B” cell subpopulation in normal immunodefective, and autoimmune mice. J Exp Med. 1983;157:202–218. doi: 10.1084/jem.157.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Immunol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 62.Dorshkind K, Montecino-Rodriguez E. Fetal B-cell lymphopoiesis and the emergence of B-1-cell potential. Nat Rev Immunol. 2007;7:213–219. doi: 10.1038/nri2019. [DOI] [PubMed] [Google Scholar]
- 63.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 64.Haas KM, Poe JC, Steeber DA, Tedder TF. B-1a and B-1b cells exhibit distinct developmental requirements and have unique functional roles in innate and adaptive immunity to S. pneumoniae. Immunity. 2005;23:7–18. doi: 10.1016/j.immuni.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 65.Pillai S, Cariappa A, Moran ST. Marginal zone B cells. Annu Rev Immunol. 2005;23:161–196. doi: 10.1146/annurev.immunol.23.021704.115728. [DOI] [PubMed] [Google Scholar]
- 66.Steiniger B, Timphus EM, Barth PJ. The splenic marginal zone in humans and rodents: an enigmatic compartment and its inhabitants. Histochem Cell Biol. 2006;126:641–648. doi: 10.1007/s00418-006-0210-5. [DOI] [PubMed] [Google Scholar]
- 67.Radbruch A, Muehlinghaus G, Luger EO, et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Rev Immunol. 2006;6:741–750. doi: 10.1038/nri1886. [DOI] [PubMed] [Google Scholar]
- 68.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 69.Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- 70.Dilillo DJ, Hamaguchi Y, Ueda Y, et al. Maintenance of long-lived plasma cells and serological memory despite mature and memory B cell depletion during CD20 immunotherapy in mice. J Immunol. 2008;180:361–371. doi: 10.4049/jimmunol.180.1.361. [DOI] [PubMed] [Google Scholar]
- 71.Ron Y, De Baetselier P, Gordon J, Feldman M, Segal S. Defective induction of antigen-reactive proliferating T cells in B cell-deprived mice. Eur J Immunol. 1981;11:964–968. doi: 10.1002/eji.1830111203. [DOI] [PubMed] [Google Scholar]
- 72.Ron Y, Sprent J. T cell priming in vivo: a major role for B cells in presenting antigen to T cells in lymph nodes. J Immunol. 1987;138:2848–2856. [PubMed] [Google Scholar]
- 73.Janeway CA, Ron J, Jr, Katz ME. The B cell is the initiating antigen-presenting cell in peripheral lymph nodes. J Immunol. 1987;138:1051–1055. [PubMed] [Google Scholar]
- 74.Lanzavecchia A. Antigen-specific interaction between T and B cells. Nature. 1985;314:537–539. doi: 10.1038/314537a0. [DOI] [PubMed] [Google Scholar]
- 75.Bouaziz JD, Yanaba K, Venturi GM, et al. Therapeutic B cell depletion impairs adaptive and autoreactive CD4+ cell activation in mice. Proc Natl Acad Sci U S A. 2007;104:20882–20887. doi: 10.1073/pnas.0709205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Harris DP, Haynes L, Sayles PC, et al. Reciprocal regulation of polarized cytokine production by effector B and T cells. Nat Immunol. 2000;1:475–482. doi: 10.1038/82717. [DOI] [PubMed] [Google Scholar]
- 77.Mizoguchi A, Bhan AK. A case for regulatory B cells. J Immunol. 2006;176:705–710. doi: 10.4049/jimmunol.176.2.705. [DOI] [PubMed] [Google Scholar]
- 78.Yanaba K, Bouaziz J-D, Haas KM, Poe JC, Fujimoto M, Tedder TF. A regulatory B cell subset with a unique phenotype controls T cell-dependent inflammatory responses. Immunity. doi: 10.1016/j.immuni.2008.03.017. In press. [DOI] [PubMed] [Google Scholar]
- 79.Vetrie D, Vorechovsky I, Sideras P, et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature. 1993;361:226–233. doi: 10.1038/361226a0. [DOI] [PubMed] [Google Scholar]
- 80.Tsukada S, Saffran DC, Rawlings DJ, et al. Deficient expression of a B cell cytoplasmic tyrosine kinase in human X-linked agammaglobulinemia. Cell. 1993;72:279–290. doi: 10.1016/0092-8674(93)90667-f. [DOI] [PubMed] [Google Scholar]
- 81.Bruton OC. Agammaglobulinemia. Pediatrics. 1952;9:722–728. [Google Scholar]
- 82.Conley ME, Broides A, Hernandez-Trujillo V, et al. Genetic analysis of patients with defects in early B-cell development. Immunol Rev. 2005;203:216–234. doi: 10.1111/j.0105-2896.2005.00233.x. [DOI] [PubMed] [Google Scholar]
- 83.Rawlings DJ. Bruton's tyrosine kinase controls a sustained calcium signal essential for B lineage development and function. Clin Immunol. 1999;91:243–253. doi: 10.1006/clim.1999.4732. [DOI] [PubMed] [Google Scholar]
- 84.Valiaho J, Smith CI, Vihinen M. BTKbase: the mutation database for X-linked agammaglobulinemia. Hum Mutat. 2006;27:1209–1217. doi: 10.1002/humu.20410. [DOI] [PubMed] [Google Scholar]
- 85.Casanova JL, Abel L. Primary immunodeficiencies: a field in its infancy. Science. 2007;317:617–619. doi: 10.1126/science.1142963. [DOI] [PubMed] [Google Scholar]
- 86.Bacchelli C, Buckridge S, Thrasher AJ, Gaspar HB. Translational mini-review series on immunodeficiency: molecular defects in common variable immunodeficiency. Clin Exp Immunol. 2007;149:401–409. doi: 10.1111/j.1365-2249.2007.03461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Durandy A, Taubenheim N, Peron S, Fischer A. Pathophysiology of B-cell intrinsic immunoglobulin class switch recombination deficiencies. Adv Immunol. 2007;94:275–306. doi: 10.1016/S0065-2776(06)94009-7. [DOI] [PubMed] [Google Scholar]
- 88.Revy P, Muto T, Levy Y, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 89.Goodnow CC, Adelstein S, Basten A. The need for central and peripheral tolerance in the B cell repertoire. Science. 1990;248:1373–1379. doi: 10.1126/science.2356469. [DOI] [PubMed] [Google Scholar]
- 90.Lipsky PE. Systemic lupus erythematosus: an autoimmune disease of B cell hyperactivity. Nat Immunol. 2001;2:764–766. doi: 10.1038/ni0901-764. [DOI] [PubMed] [Google Scholar]
- 91.Lange K, Gold MM, Weiner D, Simon V. Autoantibodies in human glomerulonephritis. J Clin Invest. 1949;28:50–55. doi: 10.1172/JCI102052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Franklin EC, Holman HR, Muller-Eberhard HJ, Kunkel HG. An unusual protein component of high molecular weight in the serum of certain patients with rheumatoid arthritis. J Exp Med. 1957;105:425–438. doi: 10.1084/jem.105.5.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shlomchik MJ. Sites and stages of autoreactive B cell activation and regulation. Immunity. 2008;28:18–28. doi: 10.1016/j.immuni.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 94.McDonnell TJ, Deane N, Platt FM, et al. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell. 1989;57:79–88. doi: 10.1016/0092-8674(89)90174-8. [DOI] [PubMed] [Google Scholar]
- 95.Cohen PL, Eisenberg RA. Lpr and gld: single gene models of systemic autoimmunity and lymphoproliferative disease. Ann Rev Immunol. 1991;9:243–269. doi: 10.1146/annurev.iy.09.040191.001331. [DOI] [PubMed] [Google Scholar]
- 96.Drappa J, Vaishnaw AK, Sullivan KE, Chu JL, Elkon KB. Fas gene mutations in the Canale-Smith syndrome, an inherited lymphoproliferative disorder associated with autoimmunity. N Engl J Med. 1996;335:1643–1649. doi: 10.1056/NEJM199611283352204. [DOI] [PubMed] [Google Scholar]
- 97.Edwards JC, Cambridge G. B-cell targeting in rheumatoid arthritis and other autoimmune diseases. Nat Rev Immunol. 2006;6:394–403. doi: 10.1038/nri1838. [DOI] [PubMed] [Google Scholar]
- 98.Uchida J, Hamaguchi Y, Oliver JA, et al. The innate mononuclear phagocyte network depletes B lymphocytes through Fc receptor-dependent mechanisms during anti-CD20 antibody immunotherapy. J Exp Med. 2004;199:1659–1669. doi: 10.1084/jem.20040119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Moore PA, Belvedere O, Orr A, et al. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science. 1999;285:260–263. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 100.Mackay F, Browning JL. BAFF: a fundamental survival factor for B cells. Nat Rev Immunol. 2002;2:465–475. doi: 10.1038/nri844. [DOI] [PubMed] [Google Scholar]
- 101.Poe JC, Fujimoto Y, Hasegawa M, et al. CD22 regulates B lymphocyte function in vivo through both ligand-dependent and ligand-independent mechanisms. Nat Immunol. 2004;5:1078–1087. doi: 10.1038/ni1121. [DOI] [PubMed] [Google Scholar]
- 102.Craig FE, Foon KA. Flow cytometric immunophenotyping for hematologic neoplasms. Blood. 2008;111:3941–3967. doi: 10.1182/blood-2007-11-120535. [DOI] [PubMed] [Google Scholar]
- 103.Staudt LM, Dave S. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol. 2005;87:163–208. doi: 10.1016/S0065-2776(05)87005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Korsmeyer SJ, Arnold A, Bakhshi A, et al. Immunoglobulin gene rearrangement and cell surface antigen expression in acute lymphocytic leukemias of T cell and B cell precursor origins. J Clin Invest. 1983;71:301–313. doi: 10.1172/JCI110770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tsujimoto Y, Finger LR, Yunis J, Nowell PC, Croce CM. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science. 1984;226:1097–1099. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 106.Ye BH, Lista F, Lo Coco F, et al. Alterations of a zinc finger-encoding gene, Bcl-6, in diffuse large-cell lymphoma. Science. 1993;262:747–750. doi: 10.1126/science.8235596. [DOI] [PubMed] [Google Scholar]
- 107.Kuppers R, Rajewsky K, Zhao M, et al. Hodgkin disease: Hodgkin and Reed-Sternberg cells picked from histological sections show clonal immunoglobulin gene rearrangements and appear to be derived from B cells at various stages of development. Proc Natl Acad Sci U S A. 1994;91:10962–10966. doi: 10.1073/pnas.91.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thai TH, Calado DP, Casola S, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 109.Vigorito E, Perks KL, Abreu-Goodger C, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]