Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism (original) (raw)
Abstract
The last few years have witnessed a rapid increase in our knowledge of the retinoid-related orphan receptors RORα, -β, and -γ (NR1F1-3), their mechanism of action, physiological functions, and their potential role in several pathologies. The characterization of ROR-deficient mice and gene expression profiling in particular have provided great insights into the critical functions of RORs in the regulation of a variety of physiological processes. These studies revealed that RORα plays a critical role in the development of the cerebellum, that both RORα and RORβ are required for the maturation of photoreceptors in the retina, and that RORγ is essential for the development of several secondary lymphoid tissues, including lymph nodes. RORs have been further implicated in the regulation of various metabolic pathways, energy homeostasis, and thymopoiesis. Recent studies identified a critical role for RORγ in lineage specification of uncommitted CD4+ T helper cells into Th17 cells. In addition, RORs regulate the expression of several components of the circadian clock and may play a role in integrating the circadian clock and the rhythmic pattern of expression of downstream (metabolic) genes. Study of ROR target genes has provided insights into the mechanisms by which RORs control these processes. Moreover, several reports have presented evidence for a potential role of RORs in several pathologies, including osteoporosis, several autoimmune diseases, asthma, cancer, and obesity, and raised the possibility that RORs may serve as potential targets for chemotherapeutic intervention. This prospect was strengthened by recent evidence showing that RORs can function as ligand-dependent transcription factors.
Introduction
The cloning of several steroid hormone receptors in the 1980s led to an intense search by many laboratories for additional, novel members of the steroid hormone superfamily [Aranda and Pascual, 2001; Desvergne and Wahli, 1999; Escriva et al., 2000; Evans, 1988; Giguere, 1999; Kumar and Thompson, 1999; Novac and Heinzel, 2004; Willy and Mangelsdorf, 1998]. This resulted in the identification of a number of orphan receptors, including members of the retinoid-related orphan receptor (ROR) subfamily, which consists of RORα (NR1F1, RORA or RZRα)[Becker-Andre et al., 1993; Giguere et al., 1995; Giguere et al., 1994], RORβ (NR1F2, RORB or RZRβ) [Andre et al., 1998b; Carlberg et al., 1994; Schaeren-Wiemers et al., 1997], and RORγ (NR1F3, RORC or TOR) [He et al., 1998; Hirose et al., 1994; Medvedev et al., 1996; Ortiz et al., 1995; Sun et al., 2000]. ROR genes have been cloned from several mammalian species [Becker-Andre et al., 1993; Carlberg et al., 1994; Giguere et al., 1994; He et al., 1998; Hirose et al., 1994; Jetten and Joo, 2006; Koibuchi and Chin, 1998; Medvedev et al., 1996; Ortiz et al., 1995] and zebrafish [Flores et al., 2007]. ROR orthologues have also been identified in several lower species, including Drosophila hormone receptor 3 (DHR3) in Drosophila melanogaster [Carney et al., 1997; Gates et al., 2004; Horner et al., 1995; Koelle et al., 1992; Sullivan and Thummel, 2003; Thummel, 1995], CHR3 in Caenorhabditis elegans [Kostrouch et al., 1995; Palli et al., 1997; Palli et al., 1996], and MHR3 in Manduca sexta [Hiruma and Riddiford, 2004; Lan et al., 1999; Lan et al., 1997; Langelan et al., 2000; Palli et al., 1992; Riddiford et al., 2003]. This article will review the current status of our knowledge of RORs, with emphasis on recent developments in their physiological functions, roles in pathophysiological processes, and their potential as pharmatherapeutical targets.
ROR structure and activity
ROR gene structure
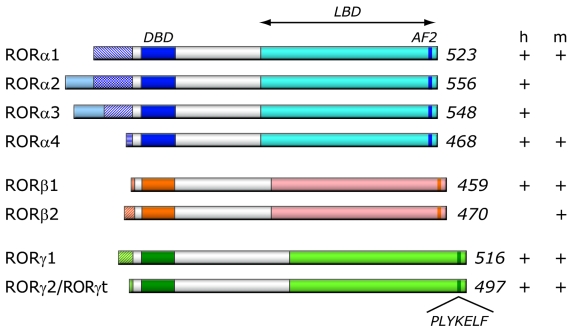
The RORα gene maps to human chromosome 15q22.2 and spans a relatively large 730 kb genomic region comprised of 15 exons. The RORβ and RORγ genes map to 9q21.13, and 1q21.3 and cover approximately 188 and 24 kb, respectively. As a result of alternative promoter usage and exon splicing, each ROR gene generates several isoforms that differ only in their amino-terminus [Andre et al., 1998b; Giguere et al., 1994; Hamilton et al., 1996; He et al., 1998; Hirose et al., 1994; Matysiak-Scholze and Nehls, 1997; Medvedev et al., 1996; Sun et al., 2000; Villey et al., 1999] (Figure 1). Four human RORα isoforms, referred to as RORα1-4, have been identified, while only two isoforms, α1 and α4, have been reported for mouse. The mouse RORβ gene generates two isoforms, β1 and β2, while humans appear to express only the RORβ1 isoform [Andre et al., 1998b]. Both the mouse and human RORγ gene generate two isoforms, γ1 and γ2 [He et al., 1998; Hirose et al., 1994; Medvedev et al., 1996; Villey et al., 1999]. Most isoforms exhibit a distinct pattern of tissue-specific expression and are involved in the regulation of different physiological processes and target genes. For example, human RORα3 is only found in human testis [Steinmayr et al., 1998]. RORα1 and RORα4 are both prominently expressed in mouse cerebellum, while other mouse tissues express predominantly RORα4 [Chauvet et al., 2002; Hamilton et al., 1996; Matysiak-Scholze and Nehls, 1997]. RORγ2, most commonly referred to as RORγt, is exclusively detected in a few distinct cell types of the immune system, while RORγ1 expression is restricted to several other tissues [Eberl et al., 2004; He et al., 1998; Hirose et al., 1994; Kang et al., 2007]. In the mouse, expression of RORβ2 is restricted to the pineal gland and the retina, while RORβ1 is the predominant isoform in cerebral cortex, thalamus, and hypothalamus [Andre et al., 1998b]. Although most ROR isoforms are under the control of different promoters, little is known about the transcriptional regulation of their tissue-specific expression.
Figure 1. Schematic representation of ROR family members.
Schematic structure of the various human (h) and mouse (m) ROR isoforms. The DNA binding domain (DBD) and ligand binding domain (LBD), and activation function 2 (AF2) are indicated. The variable regions at the N-terminus of each ROR generated by alternative promoter usage and/or alternative slicing are indicated by patterned boxes on the left. The numbers on the right refer to the total number of amino acids in RORs. The different ROR isoforms identified in human and mouse are shown on the right (+/-).
ROR protein structure
The ROR genes encode proteins of 459 to 556 amino acids (Figure 1). RORs exhibit a typical nuclear receptor domain structure consisting of four major functional domains: an N-terminal (A/B) domain followed by a highly conserved DNA-binding domain (DBD), a hinge domain, and a C-terminal ligand-binding domain (LBD) [Evans, 1988; Giguere, 1999; Jetten et al., 2001; Moras and Gronemeyer, 1998; Pike et al., 2000; Steinmetz et al., 2001; Willy and Mangelsdorf, 1998]. RORs regulate gene transcription by binding to specific DNA response elements (ROREs), consisting of the consensus RGGTCA core motif preceded by a 6-bp A/T-rich sequence, in the regulatory region of target genes [Andre et al., 1998a; Carlberg et al., 1994; Giguere et al., 1994; Greiner et al., 1996; Jetten et al., 2001; Medvedev et al., 1996; Moraitis and Giguere, 1999; Ortiz et al., 1995; Schrader et al., 1996]. RORs bind ROREs as a monomer and do not form heterodimers with retinoid-X receptors (RXRs) [Andre et al., 1998b; Carlberg et al., 1994; Giguere et al., 1994; Greiner et al., 1996; Medvedev et al., 1996; Moraitis and Giguere, 1999; Ortiz et al., 1995; Schrader et al., 1996] (Figure 2). The interaction of RORs with ROREs is mediated by the P-box, the loop between the last two cysteines within the first zinc finger, which recognizes the core motif in the major groove, and the C-terminal extension, a 30 residue region just downstream from the two zinc fingers, which interacts with the 5’-AT-rich segment of the RORE in the adjacent minor groove [Andre et al., 1998b; Giguere et al., 1995; Giguere et al., 1994; Jetten et al., 2001; McBroom et al., 1995; Sundvold and Lien, 2001; Vu-Dac et al., 1997]. Although RORα-γ and their different isoforms recognize closely-related ROREs, they exhibit distinct affinities for different ROREs. The amino-terminus (A/B domain) has been shown to play a critical role in conferring DNA binding specificity to the various ROR isoforms [Andre et al., 1998b; Giguere et al., 1995; Giguere et al., 1994; Sundvold and Lien, 2001; Vu-Dac et al., 1997]. In addition to the RORE sequence and the amino terminus, the promoter context may play an important factor in determining which ROR is recruited to a particular RORE.
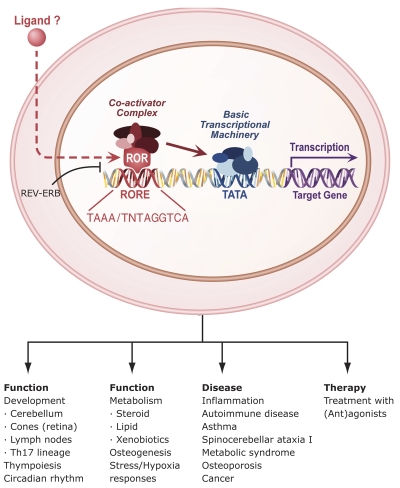
Figure 2. Mechanism of action of RORs, physiological functions and roles in disease.
RORs bind as a monomer to ROREs consisting of the GGTCA consensus core motif preceded by a 6A/T rich region. REV-ERBs can compete with RORs for binding to ROREs. RORs interact with coactivators or corepressors to positively or negatively regulate gene transcription. RORs are critical in the regulation of many physiological processes and may have a role in several pathologies. Although evidence has been provided indicating that certain ligands can modulate ROR transcriptional activity, whether ROR activity is modulated in vivo by endogenous ligands has yet to be determined. RORs might serve as potential novel targets for chemotherapeutic strategies to intervene in various disease processes.
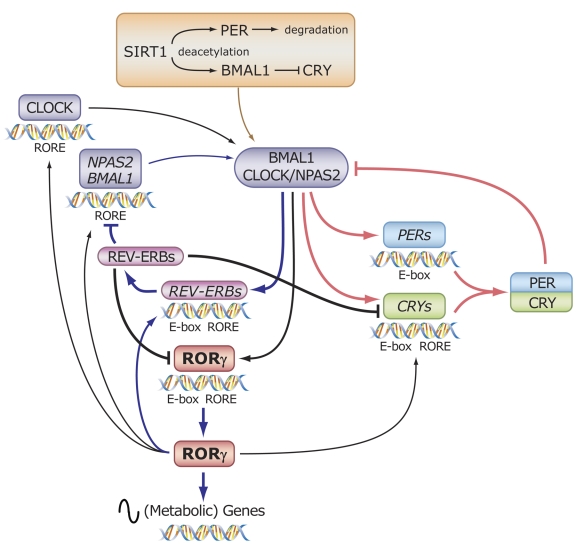
In several instances, crosstalk between nuclear receptor pathways involves competition between receptors for binding to the same response element. Such an interplay has been demonstrated between RORs and the nuclear receptors REV-ERBAα and REV-ERBβ (NR1D1 and D2, respectively), which recognize a subset of ROREs [Burris, 2008; Giguere et al., 1995]. Because REV-ERB receptors act as transcriptional repressors, they are able to inhibit ROR-mediated transcriptional activation by competing with RORs for RORE binding [Austin et al., 1998; Bois-Joyeux et al., 2000; Downes et al., 1996; Forman et al., 1994; Liu et al., 2007b; Medvedev et al., 1997; Retnakaran et al., 1994] (Figure 2). Positive and negative regulation of RORE-mediated gene transcription by RORs and REV-ERBs have been reported to play a role in the control of brain and muscle ARNT-like 1 (BMAL1 or ARNTL) expression and may be part of several other regulatory feedback loops [Akashi and Takumi, 2005; Albrecht, 2002; Gachon et al., 2004; Guillaumond et al., 2005; Nakajima et al., 2004; Preitner et al., 2002; Triqueneaux et al., 2004].
The LBDs of nuclear receptors are multifunctional and play a role in ligand binding, nuclear localization, receptor dimerization, and provide an interface for the interaction with coactivators and corepressors. X-ray structural analysis demonstrated that RORs have a secondary domain structure that is characteristic of that of nuclear receptors [Kallen et al., 2002; Stehlin-Gaon et al., 2003; Stehlin et al., 2001]. The LBDs of RORs contain, in addition to the typical 12 canonical α-helices (H1-12), two additional helices, H2’ and H11’. The activation function 2 (AF2) in H12 consists of PLYKELF, which is 100% conserved among RORs (Figure 1). Deletion of the H12 or point mutations within H12 causes loss of the ROR transactivation activity and results in a dominant-negative ROR [Kurebayashi et al., 2004; Lau et al., 1999]. It is believed that H10 plays a critical role in the homo- and heterodimerization of nuclear receptors. Structure analyses revealed the presence of a kink in H10 of the LBD of RORα and RORβ that would greatly affect the dimerization capability of RORs [Kallen et al., 2002; Stehlin-Gaon et al., 2003; Stehlin et al., 2001]. This is consistent with the conclusion that RORs do not form homodimers or heterodimers with other RORs or RXRs.
RORs: ligand-dependent transcription factors
Crystallography provided not only insights into the structure of the LBD, but also into size of the ligand binding pockets of RORs [Kallen et al., 2002; Stehlin et al., 2001]. The ligand-binding pocket of RORβ was calculated to be 766 Å3, similar to that of RORα (722 Å3). Homology modeling predicted that the ligand binding pocket of RORγ is similar in size (705 Å3), but different in shape. Cholesterol, 7-dehydrocholesterol, and cholesterol sulfate were identified as RORα agonists [Kallen et al., 2004; Kallen et al., 2002]. They were found to bind RORα in a reversible manner and to enhance RORE-dependent transcriptional activation by RORα in cells maintained in cholesterol-depleted medium. On this basis, it has been suggested that RORα might function as a lipid sensor and be implicated in the regulation of lipid metabolism. The latter would be consistent with reports indicating that RORα regulates the expression of several genes involved in lipid metabolism [Boukhtouche et al., 2004; Jakel et al., 2006; Kallen et al., 2002; Kang et al., 2007; Lau et al., 2008; Lau et al., 2004; Lind et al., 2005; Mamontova et al., 1998; Wada et al., 2008a]. However, whether cholesterol and cholesterol sulfate metabolites function as genuine physiological agonists of RORα, and whether other structurally-related lipids serve as endogenous ligands of RORα, needs to be established.
X-ray structure analysis of the RORβ(LBD) identified stearic acid as a fortuitously-captured ligand that appeared to act as a stabilizer by filling the ligand-binding pocket, rather than as a functional ligand [Stehlin et al., 2001]. Subsequently, several retinoids, including all-trans retinoic acid (ATRA) and the synthetic retinoid ALRT 1550 (ALRT), were identified as functional ligands for RORβ [Stehlin-Gaon et al., 2003]. ATRA and ALRT 1550 were able to bind RORβ(LBD) reversibly and with high affinity and reduced RORβ-mediated transcriptional activation, suggesting that they act as partial antagonists. These retinoids were also able to bind RORγ and inhibit RORγ-mediated transactivation, but did not bind RORα or affect RORα-induced transactivation [Stehlin-Gaon et al., 2003]. Interestingly, repression of RORβ-mediated transcription was observed only in neuronal cells and not in other cell types tested, indicating that this antagonism may be rather cell type-dependent and involve an interaction with (a) neuronal cell-specific RORβ repressor(s) [Stehlin-Gaon et al., 2003]. Future studies have to determine whether ATRA acts as a genuine physiological ligand of RORβ and RORγ and establish the physiological significance of such an interaction in neuronal and non-neuronal cells.
Although future research needs to determine whether in vivo ROR activity is regulated by endogenous ligands, these crystallographic and structural studies do support the concept that ROR activity can be modulated by specific endogenous and/or synthetic (ant)agonists [Kallen et al., 2004; Kallen et al., 2002; Stehlin-Gaon et al., 2003]. This conclusion is highly relevant to the emerging roles of RORs in several pathologies, including inflammation, various autoimmune diseases, obesity, and asthma, and the promise that these receptors might serve as potential targets for pharmacological intervention in these diseases (Figure 2).
Interaction with coregulatory proteins
For many receptors, binding of a ligand functions as a switch that induces a conformational change in the receptor that involves a repositioning of H12 (AF2) [Darimont et al., 1998; Glass and Rosenfeld, 2000; Harding et al., 1997; Heery et al., 2001; Heery et al., 1997; McInerney et al., 1998; Nagy et al., 1999; Nolte et al., 1998; Xu et al., 1999]. When RORs are in a transcriptionally-active conformation, H12 with H3 and H4 form a hydrophobic cleft and a charge clamp that involves the participation of a conserved Lys in H3 and a conserved Glu in H12 within RORs [Kallen et al., 2002; Stehlin-Gaon et al., 2003; Stehlin et al., 2001]. The clamp serves as an interaction interface for LXXLL motifs present in coactivators and related motifs in corepressors [Gold et al., 2003; Harris et al., 2002; Kurebayashi et al., 2004; Littman et al., 1999; Moraitis et al., 2002; Xie et al., 2005]. Receptor:coactivator complexes, through their histone acetylase activity, induce histone acetylation that results in decompaction of chromatin and increased transcription of target genes [Glass and Rosenfeld, 2000; McKenna and O'Malley, 2002; Wolf et al., 2008; Xu, 2005], while receptor association with corepressor complexes leads to histone deacetylation and subsequently compaction of chromatin and repression of gene expression [Horlein et al., 1995; Hu and Lazar, 2000; Nagy et al., 1999].
RORs interact with corepressors, as well as coactivators, suggesting that they can function both as repressors and activators of gene transcription. The coactivators NCOA1 (SRC1), NCOA2 (TIF2 or GRIP1), PGC-1α, p300, and CBP [Atkins et al., 1999; Gold et al., 2003; Harding et al., 1997; Harris et al., 2002; Jetten, 2004; Jetten and Joo, 2006; Kurebayashi et al., 2004; Lau et al., 1999; Lau et al., 2004; Littman et al., 1999; Liu et al., 2007b; Xie et al., 2005] and corepressors NCOR1, NCOR2, RIP140, and the neuronal interacting factor X (NIX1) [Greiner et al., 2000; Harding et al., 1997; Jetten, 2004; Jetten and Joo, 2006; Johnson et al., 2004; Littman et al., 1999] are among the coregulators found within ROR protein complexes. Recently, repression of ROR-mediated transcriptional activation by the forkhead box transcription factor p3 (FOXP3) was shown to depend on a direct interaction between RORs and FOXP3 [Du et al., 2008; Yang et al., 2008c; Zhou et al., 2008].
Posttranslational modifications
Posttranslational modifications, including phosphorylation, acetylation, ubiquitination and sumoylation, play an important role in regulating nuclear receptor activity and stability [Faus and Haendler, 2006; Popov et al., 2007]. The ubiquitin (Ub)-proteasome system is intimately involved in chromatin structure remodeling and transcriptional control by nuclear receptors [Dace et al., 2000; Dennis et al., 2001; Ismail and Nawaz, 2005; Kinyamu et al., 2005; Poukka et al., 1999; Wallace and Cidlowski, 2001]. Proteasome-mediated ubiquitination is also an integral part of the mechanism by which ROR receptor signaling is controlled. Inhibition of the 26S proteasome complex by the proteasome inhibitor MG-132 results in an accumulation of ubiquitinated RORα protein and reduces the transcriptional activity of this receptor [Moraitis and Giguere, 2003]. Similarly, the corepressor Hairless (Hr) [Cachon-Gonzalez et al., 1994; Thompson et al., 2006], which functions as an effective repressor of ROR-mediated transcriptional activation, enhances RORα protein stability by protecting it from proteasome-mediated degradation [Moraitis and Giguere, 2003; Moraitis et al., 2002]. Reports showing that various proteasome subunits, including proteasome subunit β type 6 (PSMB6) and the 26S ATPase subunit PSMC5, are part of ROR protein complexes, are consistent with a role of the proteasome system in the regulation of ROR transcriptional activity [Atkins et al., 1999; Jetten and Joo, 2006]. The ubiquitin-conjugating enzyme I (UBE2I or UBC9), which catalyzes sumoylation, was also found to interact with RORs. Whether UBE2I catalyzes ROR sumoylation and alters ROR activity needs to be established.
Recent studies demonstrated that activation of protein kinase C induces phosphorylation of RORα1 and inhibits RORα1-mediated transcriptional activation [Duplus et al., 2008], while calmodulin-dependent kinase IV (CaMKIV) has been shown to enhance the transcriptional activity of RORα by a mechanism that does not involve phosphorylation of RORα by CaMKIV [Kane and Means, 2000]. Extracellular signal-regulated kinase-2 (ERK2) has been reported to phosphorylate RORα4 in vitro at Thr128 [Lechtken et al., 2007]. Future studies are needed to determine the physiological significance of these different kinases on ROR activity and function.
RORs and development
Critical role of RORα in cerebellar development
Although RORα is expressed in a variety of tissues, including testis, kidney, adipose and liver, it is most highly expressed in the brain, particularly in the cerebellum and thalamus [Becker-Andre et al., 1993; Carlberg et al., 1994; Dussault et al., 1998; Hamilton et al., 1996; Matysiak-Scholze and Nehls, 1997; Nakagawa et al., 1997; Nakagawa et al., 1998; Nakagawa and O'Leary, 2003; Vogel et al., 2000]. The function of RORα in brain development has not been extensively studied beyond the cerebellum. A high level of RORα expression was observed in the cerebellar Purkinje cells, while RORα is not detected in the granule cell layer [Ino, 2004; Matsui et al., 1995; Nakagawa et al., 1997; Sashihara et al., 1996; Sotelo and Wassef, 1991]. During Purkinje cell development, RORα expression is first observed at E12.5, just after Purkinje cell precursors exit the mitotic cycle and leave the ventricular zone to migrate along radial glia to form a temporary cerebellar plate-like structure [Gold et al., 2003; Gold et al., 2007; Goldowitz and Hamre, 1998; Jetten and Joo, 2006].
RORα null mice, generated by targeted disruption of the RORα gene, display essentially an identical phenotype as homozygous staggerer mice (RORαsg/sg) [Dussault et al., 1998; Hamilton et al., 1996; Herrup and Mullen, 1981; Landis and Sidman, 1978; Sidman et al., 1962; Steinmayr et al., 1998]. This natural mutant strain carries a 6.4 kb intragenic insertion causing deletion of the fifth exon, encoding the beginning of the LBD, and a frameshift that results in a premature stop codon [Gold et al., 2007; Hamilton et al., 1996]. RORα-deficient mice exhibit several abnormalities, including thin long bones, suggesting a role for RORα in bone formation [Meyer et al., 2000]. In addition, these mice display ataxia and severe cerebellar atrophy, characterized by significantly fewer Purkinje cells and a loss of cerebellar granule cells [Doulazmi et al., 2001; Doulazmi et al., 1999; Dussault et al., 1998; Gold et al., 2007; Hamilton et al., 1996; Herrup and Mullen, 1979; Herrup and Mullen, 1981; Landis and Reese, 1977; Landis and Sidman, 1978; Sidman et al., 1962; Steinmayr et al., 1998]. Although the morphology of the cerebellar cortex in young heterozygous (RORαsg/+) mice appears normal, upon aging, an accelerated loss of Purkinje and granule cells was observed [Doulazmi et al., 1999; Hadj-Sahraoui et al., 2001; Hadj-Sahraoui et al., 1997; Zanjani et al., 1992].
At E17.5 of development, the number of Purkinje cells generated appears to be similar between WT and RORαsg/sg mice [Doulazmi et al., 2001; Vogel et al., 2000]. However, by PND5, the number of Purkinje cells is dramatically reduced and cells appear disorganized. Surviving RORαsg/sg Purkinje cells contain stunted dendritic arbors lacking distal spiny branchlets and are deficient in their assembly of mature synapses with granule cells, suggesting that dendritogenesis is impaired. This is accompanied by a lack of parallel fiber (PF) input and a persistence of multiple climbing fiber (CF) innervations [Boukhtouche et al., 2006a; Janmaat et al., 2009; Mariani, 1982; Mariani and Changeux, 1980]. Dysfunctional synapse formation between PF and Purkinje cells is associated with a delayed shift from the glutamate transporter Slc17a6 (VGluT2) into Slc17a7 (VGluT1) in PF terminals and to an altered distribution of the glutamate receptors, Grm1 and Grm2, in Purkinje cells [Janmaat et al., 2009]. Expression of RORα in RORαsg/sg Purkinje cells was shown to restore normal dendritogenesis in organotypic cultures [Boukhtouche et al., 2006a]. These observations indicated that RORα does not regulate the genesis of Purkinje cells, but rather their maturation, particularly dendritic differentiation (Figure 3). This concept was further supported by observations showing that several genes down-regulated in Purkinje cells of RORαsg/sg mice encode proteins involved in the maturation of Purkinje cells. These include genes involved in signal-dependent calcium release, such as the calmodulin inhibitor Pcp4, the IP3 receptor (Itpr1) and its interacting partner Cals1, the calcium-transporting ATPase Atp2a2, and major intracellular calcium buffer 1 (Calb1) [Gold et al., 2003; Gold et al., 2007; Hamilton et al., 1996; Messer et al., 1990]. In addition, several genes linked to the glutamatergic pathway were expressed at reduced levels in RORαsg/sg mice, including the glutamate receptor Grm1, the glutamate transporter Slc1a6, and its anchor Spnb3, encoding a brain-specific β-spectrin.
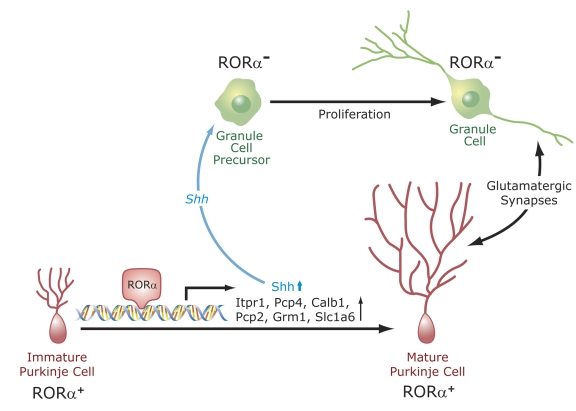
Figure 3. Regulation of Purkinje cell maturation and cerebellar development by RORα.
RORα regulates the expression of several genes in Purkinje cells. RORα becomes highly expressed in postmitotic Purkinje cells. It regulates their maturation, particularly dendritic differentiation. Dendritogenesis and the expression of several genes, including Shh, Itpr1, Pcp4, Calb1, Pcp2, and Slc1a6, normally expressed in mature Purkinje cells, are inhibited in RORα-deficient mice. The transcription of several of these genes is under direct control of RORα. Shh released by Purkinje cells interacts with Patched (Ptch) receptors on granule cell precursors, leading to activation of GLI transcription factors and the subsequent induction of proliferation-promoting genes. Reduced Shh expression in Purkinje cells from RORα-deficient mice is a major factor in the cerebellar atrophy observed in these mice. Cerebellar granule cells and Purkinje cells mutually interact (e.g., glutamatergic synapses).
The promoter regulatory regions of several genes down-regulated in RORαsg/sg Purkinje cells, including those of Pcp2, Pcp4, Itpr1, Shh, and Slc1a6, contain ROREs that bind RORα [Gold et al., 2003; Gold et al., 2007; Serinagaoglu et al., 2007]. Chromatin immunoprecipitation (ChIP) analysis demonstrated that RORα is associated with these ROREs in vivo, suggesting that these genes are directly regulated by RORα. Moreover, these studies revealed the identity of several transcriptional mediators that are part of RORα transcription activation complexes at these ROREs. Interestingly, the composition of these RORα:coactivator complexes associated with different promoters exhibited a certain promoter specificity. For example, the RORα transactivation complex associated with the _Pcp2_-RORE contained TIP60 (Tat-interactive protein, 60 kD), and SRC-1; the _Slc1a6_-RORE bound complex contained CBP, TIP60, SRC-1, and GRIP-1; while the _Pcp4_-RORE associated complex contained β-catenin, SRC-1, p300, and TIP60 [Gold et al., 2003]. These results suggest that the sequence and the promoter context of the RORE play a critical role in determining the composition of the RORα:coactivator complex recruited to each promoter.
The development of Purkinje cells and granule cells is mutually dependent. Positioning and morphological differentiation of Purkinje cells depends on granule cells, while proliferation of granule precursors requires factors released by Purkinje cells [Goldowitz and Hamre, 1998; Vogel et al., 2000]. At birth, the number of granule cells is somewhat reduced, and this becomes more prominent during the first weeks after birth. In adult RORα-deficient mice, the granular layer is very thin and depleted of granule cells [Doulazmi et al., 2001; Doulazmi et al., 1999; Dussault et al., 1998; Gold et al., 2007; Hamilton et al., 1996; Herrup and Mullen, 1979; Herrup and Mullen, 1981; Landis and Reese, 1977; Landis and Sidman, 1978; Sidman et al., 1962; Steinmayr et al., 1998; Yoon, 1972]. Studies using WT:staggerer chimeras indicated that the depletion of cerebellar granule cells in RORαsg/sg mice is related to abnormalities inherent to RORαsg/sg Purkinje cells [Herrup and Mullen, 1979; Herrup and Mullen, 1981]. This is consistent with the high level of expression of RORα in Purkinje cells and the lack of RORα in cerebellar granule cells. Sonic hedgehog (Shh) released by Purkinje cells plays a critical role in the regulation of the proliferation of cerebellar granule cells [Dahmane and Ruiz i Altaba, 1999; Gold et al., 2003; Kenney et al., 2003; Wallace, 1999] (Figure 3). Interaction of Shh with Patched (Ptch) receptors on cerebellar granule precursors results in the activation of GLI Krüppel-like zinc finger proteins and the subsequent transcriptional activation of N-MYC and other cell cycle regulatory genes. The expression of Shh in RORαsg/sg Purkinje cells was found to be reduced several fold, suggesting that the repression of Shh may be a critical factor in the diminished proliferation of granule precursors [Gold et al., 2003; Gold et al., 2007]. This is consistent with the observed repression of several Shh target genes, including N-MYC, in the cerebellum of RORαsg/sg mice. This hypothesis is further supported by experiments showing that Shh was able to enhance proliferation of granule precursor cells in sections of cerebellum from PND4 RORαsg/sg mice and partially prevented the reduction in granule cells [Boukhtouche et al., 2006a]. These observations indicate that the down-regulation of Shh in RORαsg/sg mice is an important contributing factor in the development of cerebellar hypoplasia in these mice and that RORα functions as a positive regulator of Shh expression. ChIP analysis indicated that RORα regulates Shh expression directly through ROREs in the promoter regulatory region of the Shh gene. Moreover, these studies identified β-catenin and p300 as constituents of the RORα transactivation complex associated with the RORE in Shh. The association of β-catenin with RORα transactivation complexes suggests a link between Wnt and ROR signaling, and is consistent with a reported role for Wnt signaling in cerebellar development [Salinas et al., 1994; Wang et al., 2001].
Link between RORα and spinocerebellar ataxia type 1 (SCA1)
Comparison of gene expression profiles indicated a possible link between RORα and spinocerebellar ataxia type 1 (SCA1), an autosomal dominant inherited neurodegenerative disorder that is characterized by progressive loss of motor coordination, speech impairment, and problems swallowing [Serra et al., 2006; Zoghbi and Orr, 2000]. SCA1 is caused by aberrant effects triggered by an expanded polyglutamine (polyQ) tract within the protein ataxin 1 (ATXN1), resulting in variable degrees of neurodegeneration, predominantly in the cerebellum (due to loss of Purkinje cells), brainstem, and spinal cord. Sca1[82Q] mutant mice, which express an Atxn1 protein containing an 82 glutamine repeat, develop severe ataxia and degeneration of Purkinje cells [Burright et al., 1995]. A considerable overlap was observed between genes repressed in the cerebellum of Sca1[82Q] mice and genes found to be regulated by RORα in Purkinje cells, suggesting a possible connection between the two [Gold et al., 2003; Serra et al., 2004]. Immunohistochemical analysis indicated that the level of RORα was significantly reduced in Purkinje cells from Sca1[82Q] mice [Serra et al., 2006]. This reduction was found to be, at least in part, due to a decrease in RORα protein stability. In addition, it was demonstrated that RORα and ATXN1 are part of the same protein complex. This association was indirect and mediated by a direct interaction of ATXN1 with the coactivator TIP60, which has been show to be part of several RORα-coactivator complexes in Purkinje cells [Gold et al., 2003; Serra et al., 2004]. It was concluded that decreased levels of RORα protein in SCA1 Purkinje cells causes reduced expression of RORα-target genes that are critical for Purkinje cell development and function, and consequently is an important contributory factor in the neurodegeneration observed in SCA1 [Serra et al., 2006].
Physiological Function of RORβ in brain and retina
RORβ exhibits a restricted pattern of expression that is limited to certain regions of the brain and retina [Andre et al., 1998a; Andre et al., 1998b; Azadi et al., 2002; Nakagawa and O'Leary, 2003; Schaeren-Wiemers et al., 1997]. During the period between E12.5 and PND5 of mouse development, RORβ is expressed in the neocortex and dorsal thalamus in a highly dynamic, spatiotemporal manner [Jetten and Joo, 2006; Nakagawa and O'Leary, 2003]. In adult brain, RORβ mRNA is most highly expressed in the cerebral cortex, thalamus, hypothalamus, suprachiasmatic nucleus (SCN), and the pineal gland, while little expression was detected in the cerebellum or hippocampus [Andre et al., 1998a; Andre et al., 1998b; Schaeren-Wiemers et al., 1997; Sumi et al., 2002]. RORβ is also expressed in the anterior olfactory nucleus posterior and sensory cortices. These observations suggested that RORβ is highly restricted to several regions involved in the processing of sensory information.
Study of RORβ null mice provided several clues about the physiological function of RORβ. RORβ null mice exhibit several motor defects, including a ‘duck-like’ gait and an impairment in several neurological reflexes, e.g., hind paw clasping reflex [Andre et al., 1998a; Masana et al., 2007]. RORβ null mice display several behavioral changes, including increased exploratory activity and reduced anxiety behavior, and olfactory deficits [Masana et al., 2007]. In addition, RORβ-/- mice show a delayed onset of male fertility [Andre et al., 1998a].
The relatively high expression in the retina and the development of retinal degeneration in RORβ-/- mice suggested an important regulatory function for RORβ in this tissue [Andre et al., 1998a]. At E17.5 of mouse and rat development, RORβ expression is detected in the formative ganglion cell layer and the neuroblastic layer [Andre et al., 1998a; Schaeren-Wiemers et al., 1997; Srinivas et al., 2006]. By PND6, the neuroblastic layer has split into the inner nuclear layer of interneurons and the outer nuclear layer, where the photoreceptor cells, rods and cones, reside. At PND16 and older, RORβ is still expressed in all retinal layers, but most highly in the outermost part of the outer nuclear layer. RORβ-/- mice exhibit several retinal developmental defects. At PND6, cell loss was observed in the ganglion layer, while the interneurons in the inner layer appeared in disarray. Although an outer nuclear layer was formed, it was significantly thinner than in WT retina, but maintained for several weeks. Rods and cones photoreceptors in the outer layer appeared disorganized and lacked outer and inner segments. These observations suggested that RORβ plays a key role in the maturation of photoreceptors, particularly the formation of the outer segments, the light sensing structures of cones and rods that contain opsin and rhodopsin, respectively [Srinivas et al., 2006].
In most mammals, color vision depends on the photopigments, S and M opsin, which are synthesized by distinct cone populations [Ebrey and Koutalos, 2001]. In WT mice, S opsin expression is observed at PND6, and by PND13 is concentrated to the outer segments of maturing cones; however, in RORβ-/- mice, induction of S opsin (Opn1sw) is significantly delayed. After PND23, S opsin is detected at greatly reduced levels. Little difference in the induction of M opsin (Opn1mw) and thyroid hormone receptor TRβ2, which is essential for Opn1mw expression, was observed between WT and RORβ-/- mice. The transcription factor CRX (cone-rod homeobox factor), which is critical for both S and M opsin gene expression, is normally expressed in rods and cones [Srinivas et al., 2006]. These data and histological studies suggested that RORβ is not a requirement for the initiation of differentiation of progenitors into photoreceptors, but specifically regulates their maturation, as illustrated by the reduced level of S opsin.
Study of the Opn1sw promoter provided further evidence that RORβ is an important regulator of Opn1sw expression. The proximal promoter region of Opn1sw contains two putative ROREs that are able to bind RORβ. Optimal activation of this promoter region was found to require a synergy between RORβ and CRX [Srinivas et al., 2006]. The latter is consistent with the co-expression of both transcription factors in the photoreceptor layer during the period when Opn1sw expression is induced. The activation of the Opn1sw promoter by RORβ was mediated principally through interaction with the most proximal RORE and, as expected, required both the DBD and AF2 domains of RORβ. Analysis of a transgene consisting of LacZ controlled by the Opn1sw proximal promoter region in mice showed that its expression was restricted to the photoreceptors with features of cones in the outer nuclear layer, while mutations in the ROREs totally abolished this expression. These observations indicate that RORβ is required for the induction of S opsin and support the conclusion that RORβ regulates Opn1sw transcription in a direct manner through ROREs within its proximal promoter region. In addition, they explain the greatly diminished expression of Opn1sw observed in the retina of RORβ-/- mice.
Role for RORα in cone development
Recent studies have indicated that RORα also plays a role in mouse retinal development [Fujieda et al., 2009; Ino, 2004]. In the retina, RORα expression is highly dynamic. It is first observed at E17 in the developing ganglion layer, while it is not detectable in the neuroblastic layer. At PND3, RORα was also found in the outer and inner border of the neuroblastic layer. In retina of adult mice, RORα is intensely expressed in the ganglion cell layer, the inner nuclear layer, and in cone photoreceptors in the outer layer, as indicated by its co-localization with both S and M opsin. Analysis of Opn1sw and Opn1mw expression showed that their level of expression was significantly reduced in RORαsg/sg retina. Expression of cone arrestin (Arr3) was also diminished, whereas the expression of several other cone genes, including phosphodiesterases 6c and 6h (Pde6c and Pde6h) and cyclic nucleotide gated channel b3 (Gngb3), and the expression of the rod-specific gene rhodopsin (Rho) were unchanged. Expression of RORβ was also unaffected in RORαsg/sg retina. As demonstrated for RORβ, RORα was shown to act in synergy with CRX to induce Opn1sw expression [Fujieda et al., 2009]. In vivo ChIP analysis supported the conclusion that Opn1sw, Opn1mw, and Arr3 are direct targets of RORα regulation. Thus, both RORα and RORβ function as regulators of cone development and Opn1sw; however, in contrast to RORα, RORβ does not affect Opn1mw expression. These data suggest that although RORα and RORβ exhibit some functional redundancy, their functions in cone development appear largely distinct from each other.
Role of RORγ in the development of secondary lymphoid tissues
RORγ1 and RORγt (RORγ2) exhibit distinct patterns of tissue-specific expression. RORγ1 is expressed in many tissues, including liver, adipose, skeletal muscle, and kidney, while the expression of RORγt is exclusively expressed in a few distinct cell types of the immune system [Eberl and Littman, 2003; Eberl and Littman, 2004; Jetten, 2004; Jetten and Joo, 2006; Lipp and Muller, 2004]. Mice deficient in RORγ expression lack lymph nodes and Peyer's patches (PPs), suggesting that RORγ is indispensable for lymph node organogenesis and development of PPs [Eberl and Littman, 2003; Eberl et al., 2004; He, 2002; Jetten and Joo, 2006; Jetten et al., 2001; Jetten and Ueda, 2002; Kurebayashi et al., 2000; Lipp and Muller, 2004; Littman et al., 1999; Sun et al., 2000]. Lymphoid tissue inducer (LTi) cells play a critical role in the development of lymph nodes and PPs. LTi cells originate from hematopoietic precursor cells in the fetal liver and have been recently characterized as CD45intCD4+CD3-CD127(IL-7Rα)+Lin- cells in mice and in humans as lineage-negative CD45intCD4-CD3-CD127hiLin- cells [Cupedo et al., 2009; Eberl and Littman, 2003; Mebius et al., 2001; Mebius et al., 1997]. LTi cells are recruited to the lymph node anlagen through their interaction with mesenchymal organizer cells, specialized cells derived from differentiating mesenchymal cells [Cupedo et al., 2002; Cupedo and Mebius, 2005; Eberl, 2005; Eberl and Littman, 2003; Lipp and Muller, 2004] (Figure 4). This cell:cell interaction involves several positive feedback loops and amplifying mechanisms. Lymph node organogenesis is initiated by interaction of lymphotoxin α1β2 at the surface of LTi cells with their respective receptors on mesenchymal organizer cells. This leads to the induction of several adhesion molecules, including vascular cell adhesion molecule 1 (VCAM-1) and mucosal addressin cellular adhesion molecule 1 (MadCAM-1) and the subsequent production of homeostatic chemokines by mesenchymal organizer cells [Cupedo et al., 2002; Cupedo and Mebius, 2005; Eberl, 2005; Lipp and Muller, 2004; Muller et al., 2003; Yoshida et al., 2002] (Figure 4). In addition to reinforcing the interaction between LTi and mesenchymal organizer cells, these interactions amplify the recruitment of additional LTi cells and promote the recruitment of other cells, such as monocytes and T and B lymphocytes, from the circulation [Cupedo and Mebius, 2005; Muller et al., 2003]. Subsequent studies showed that LTi cells are absent from spleen, mesentery, and intestine of RORγ-/- E18.5 embryos, indicating that RORγt plays a critical role in the generation and/or survival of LTi cells [Eberl, 2005; Eberl and Littman, 2003]. As a consequence, the lack of lymph nodes and PPs in RORγt- or RORγ-deficient mice is due to the absence of LTi cells.
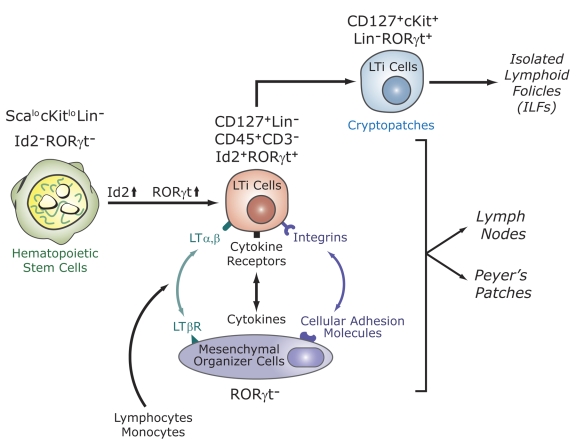
Figure 4. RORγt is essential for the development of secondary lymphoid tissues.
Lymphoid tissue inducer (LTi) cells are derived from fetal liver hematopoietic stem cells. This differentiation is accompanied by induction of the transcription factors Id2 and RORγt. LTi cells are recruited from the circulation by mesenchymal organizer cells. This is mediated by multiple receptor-ligand interactions. LTi cells are required for the development of lymph nodes, Peyer’s patches, cryptopatches, and isolated lymphoid follicles (ILFs), which are thought to be derived from cryptopatches after the colonization of the intestine by bacteria. Recently, a novel lymphocyte population (NKp46+NK1.1intCD127+RORγthi) was identified in the gut that may be derived from LTi-like cells in cryptopatches. RORγt is required for the generation and/or survival of LTi cells. The absence of LTi cells in RORγ-deficient mice is responsible for the lack of lymph nodes, Peyer’s patches, cryptopatches, ILFs, and NKp46+NK1.1intCD127+RORγthi cells.
Besides PPs and mesenteric lymph nodes, the intestinal immune system contains several other lymphoid cell compartments, including cryptopatches and isolated lymphoid follicles (ILFs) [Eberl, 2005; Taylor and Williams, 2005] (Figure 4). ILFs develop from cryptopatches in response to inflammatory innate immune signals generated by the colonization of the intestine by bacteria. Lin-cKit+CD127+CD44+ cells in cryptopatches and in ILFs express high levels of RORγt and appear to constitute the adult counterpart of LTi cells [Eberl, 2005; Eberl and Littman, 2004]. The deficiency in cryptopatches and ILFs observed in RORγ-/- mice indicate that RORγt is essential for the development of these cell compartments and is due to the absence of these LTi-like cells in these mice.
Recent studies identified a novel lymphocyte population in the intestinal lamina propria that produces IL-22 and co-expresses RORγt and CD127, and natural killer (NK) cell markers NKp46 and NK1.1 (CD3-RORγthiCD127+NKp46+NK1.1int) [Cupedo et al., 2009; Luci et al., 2009; Sanos et al., 2009] (Figure 4). These cells are absent in RORγt-deficient mice. Because of its dependence on RORγt and its CD127+c-Kit+ phenotype, these NK cells resemble LTi cells, suggesting a possible link between the two cell types. LTi cells have been reported to be able to differentiate into NK and antigen presenting cells [Mebius et al., 2001; Mebius et al., 1997]. The basic helix-loop-helix transcription factor Id2, which as RORγt is expressed in LTi cells, is essential for both LTi and NK cell development [Yokota et al., 1999]. In contrast, peripheral NK cells do not express RORγt and their number is unaffected in RORγ-deficient mice [Sun et al., 2000]. It is has been suggested that RORγthiNKp46+NK1.1int cells do not belong to the NK cell lineage, but may be derived from the Lin-Kit+CD127+RORγthi LTi-like cells in cryptopatches [Cupedo et al., 2009; Luci et al., 2009; Sanos et al., 2009]. The precise role of these cells in normal intestinal mucosa immunity and pathology, including colitis, needs further study.
In contrast to lymph nodes and Peyer’s patches, the development of several other lymphoid tissues, including nasal- and bronchial-associated lymphoid tissue (BALT and NALT), was undisturbed in RORγ-/- mice, suggesting that the organogenesis of these lymphoid tissues involves a different mechanism [Harmsen et al., 2002].
Critical functions of RORγ in thymopoiesis
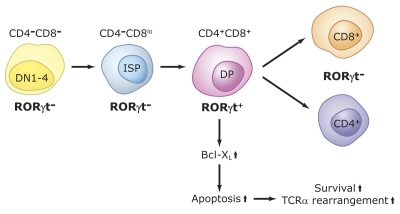
Several studies have demonstrated that RORγt plays a critical role in the regulation of thymopoiesis [Guo et al., 2002; He, 2000; He et al., 1998; Hirose et al., 1994; Jetten, 2004; Jetten and Joo, 2006; Kurebayashi et al., 2000; Medvedev et al., 1996; Ortiz et al., 1995; Sun et al., 2000]. During thymopoiesis, T cell precursor CD25-CD44+CD4-CD8- cells (DN1) differentiate successively via two intermediate stages, CD25+CD44+ (DN2) and CD25+CD44- (DN3), into CD44-CD25- (DN4) thymocytes (Figure 5). These cells then differentiate via immature single positive (ISP) cells (CD3-CD4-CD8low) into double positive CD4+CD8+ (DP) thymocytes [Miyazaki, 1997]. After successful T cell receptor α (TCRα) gene rearrangement, DP cells expressing TCRαβ receptor undergo a careful selection process to eliminate thymocytes expressing nonfunctional or autoreactive TCR [Starr et al., 2003; Zhang et al., 2005]. The positive selected DP thymocytes mature into single positive (SP) CD4+CD8- helper and CD4-CD8+ cytotoxic T cells that then colonize the secondary lymphoid organs, including the spleen, lymph nodes, and PPs.
Figure 5. Role of RORγt in thymopoiesis.
CD4-CD8-CD25-CD44+ (DN1) cells differentiate via DN2, DN3, DN4 into immature single positive (ISP) cells (CD3-CD4-CD8low). These cells subsequently differentiate into CD3+CD4+CD8+, DP thymocytes. RORγt, as well as Bcl-XL, are induced during the ISP-DP transition and again down-regulated during the differentiation of DP into CD4+ and CD8+ single positive cells. RORγt promotes the differentiation of ISP into DP cells and positively regulates the expression of the anti-apoptotic gene Bcl-XL. The latter enhances cell survival that subsequently promotes TCR rearrangements. Lack of RORγt expression inhibits the ISP-DP transition and reduces expression of Bcl-XL in DP thymocytes. The latter results in accelerated apoptosis, reduced lifespan of DP thymocytes and, consequently, impaired TCRα rearrangements
RORγt is transiently expressed during thymopoiesis [Guo et al., 2002; He, 2002; He et al., 2000; He et al., 1998; Jetten, 2004; Jetten and Joo, 2006; Jetten et al., 2001; Sun et al., 2000] (Figure 5). It is undetectable in DN thymocytes and highly induced when ISP cells differentiate into DP thymocytes and again down-regulated when DP thymocytes differentiate into mature T lymphocytes. At birth and during early stages of life, RORγ null mice have a significantly smaller thymus compared to wild type mice as a result of a drastic reduction in the number of double positive DP and SP thymocytes, while the percentage of ISP thymocytes is greatly increased [Eberl and Littman, 2003; Jetten et al., 2001; Jetten and Ueda, 2002; Kurebayashi et al., 2000; Sun et al., 2000]. The accumulation of ISP cells in RORγ-/- mice appears to be due to a delay in the differentiation of ISP into DP cells, suggesting a role for RORγt in the regulation of the ISP-DP transition [Guo et al., 2002]. In addition, DP thymocytes undergo massive apoptosis in vivo and in vitro [Jetten, 2004; Jetten and Joo, 2006; Jetten et al., 2001; Kurebayashi et al., 2000; Sun et al., 2000]. The accelerated apoptosis of RORγ-/- DP thymocytes is related to reduced expression of the anti-apoptotic gene Bcl-XL. This repression is an early and key event in accelerated apoptosis in RORγ-/- thymocytes [Sun et al., 2000; Xie et al., 2005]. Thus, RORγ functions as a positive regulator of Bcl-XL expression and, as a result, promotes the survival of DP thymocytes, thereby enabling gene rearrangement [Eberl et al., 2004; He et al., 1998; Jetten, 2004; Jetten and Joo, 2006; Jetten et al., 2001; Jetten and Ueda, 2002; Kurebayashi et al., 2000; Ma et al., 1995].
Little is still understood about the mechanism by which Bcl-XL and RORγt expression are regulated during thymopoiesis. The reduced expression of Bcl-XL in RORγ-/- mice and the observation that the induction and down-regulation of Bcl-XL and RORγt expression occur in parallel, are consistent with the concept that RORγt regulates Bcl-XL expression. Whether this regulation involves a direct or indirect mechanism has yet to be established. A recent study provided evidence for a link between early growth response gene 3 (EGR3) expression and the regulation of RORγt and Bcl-XL [Xi and Kersh, 2004; Xi et al., 2006]. Pre-TCR signals transiently induce EGR3, which promotes proliferation, whereas the subsequent down-regulation of EGR3 leads to growth cessation and induction of RORγt and Bcl-XL needed for cell survival and proper gene rearrangement.
Analysis of gene expression profiles of thymocytes from WT and RORγ-/- mice identified additional changes in gene expression, including repression of the transcription factor EGR1, Ngfi-A binding protein 2 (NAB2), and the glucocorticoid-induced transcript 1 (GLCCI1) [Kang et al., 2006]. One of the genes most dramatically repressed in thymocytes from RORγ-/- mice is oligonucleotide/oligosaccharide-binding fold-containing 2a (Obfc2a, also named NABP1). The pattern of Obfc2a expression during thymopoiesis correlates with that of RORγt. Therefore, Obfc2a might be a potential RORγ target gene; alternatively, its down-regulation is just a consequence of the reduction in DP cells observed in RORγ-/- mice. In summary, these studies indicate that RORγ plays a critical role in thymocyte homeostasis by regulating differentiation, proliferation, and apoptosis of thymocytes [He, 2002; Jetten, 2004; Jetten and Joo, 2006; Jetten et al., 2001; Winoto and Littman, 2002].
Role of RORα in lymphocyte development
Both the spleen and the thymus are significantly smaller in RORα-deficient mice, suggesting that RORα may have a role in the regulation of thymopoiesis and lymphocyte development [Dzhagalov et al., 2004; Trenkner and Hoffmann, 1986]. RORα mRNA is expressed at low levels in DP thymocytes and at high levels in SP thymocytes, while it is decreased in mature CD8+ and increased in mature CD4+ T lymphocytes. B220+ B lymphocytes express low levels of RORα. The total number of thymocytes and splenocytes is greatly diminished in RORα-/- mice and is associated with a loss of DP thymocytes and a corresponding increase in the percentage of DN and SP thymocytes [Dzhagalov et al., 2004]. In RORα-/- spleen, both mature T and B lymphocytes were significantly reduced, suggesting that RORα plays a critical role in lymphocyte development.
No differences were observed between RORα-/- and WT mice in the induction of CD4+ and CD8+ T lymphocyte proliferation in response to anti-CD3 or lipopolysaccharide (LPS), nor in the induction of cytokines by TNFα [Dzhagalov et al., 2004]. In contrast, the induction of IL-6 and tumor necrosis factor α was greatly increased in RORα-/- mast cells and macrophages after LPS treatment [Dzhagalov et al., 2004; Kopmels et al., 1992]. These results indicate that at least in certain immune cells, RORα functions as a negative regulator of cytokine expression. Although the mechanism of this repression is not fully understood, RORα has been reported to positively regulate the expression of IKBα, a negative regulator of the NF-κB signaling pathway [Delerive et al., 2001]. On this basis, it was concluded that RORα might function as a negative regulator of inflammation.
RORs and T cell lineage specification
Role of RORs in Th17 cell differentiation
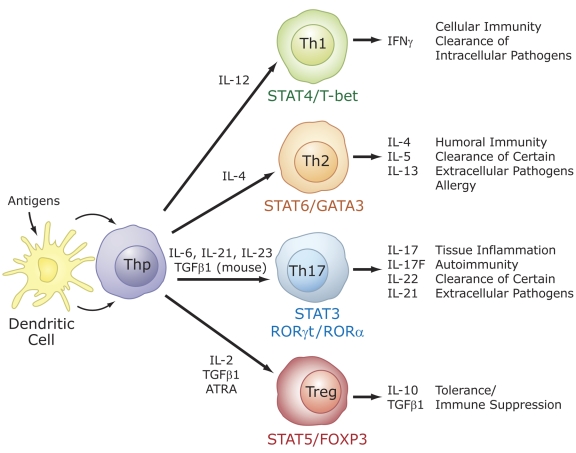
Recent studies identified a critical role for RORs in the regulation of lineage specification in helper T cells [Chen et al., 2007; Chen and O'Shea, 2008; Dong, 2008; Furuzawa-Carballeda et al., 2007; Huang et al., 2007; Ivanov et al., 2006; Ivanov et al., 2007; Laurence and O'Shea, 2007; Weaver et al., 2007; Yang et al., 2008d]. Differentiation into different effector CD4+ T cell lineages, T helper (Th) 1, Th2, Th17, and T regulatory (Treg or Th3) cells is initiated through an interaction of dendritic cells with uncommitted (naïve) CD4+ T helper cells (Thp). Th1, Th2, Treg, and Th17 are characterized by their synthesis of specific cytokines and their immuno-regulatory functions (Figure 6). Interferon γ is the signature cytokine produced by Th1 cells, while IL-4, IL-5 and IL-13 are major cytokines produced by Th2 cells. The recently discovered Th17 cells produce IL-17A (IL-17), IL-17F, IL-21, and IL-22 as major cytokines, while Treg cells synthesize IL-10 and TGFβ1 [Bettelli et al., 2008; Chen and O'Shea, 2008; Dong, 2008; Harrington et al., 2006; Park et al., 2005]. T helper and Treg cells play a critical role in several inflammatory responses, including adaptive immune responses to various pathogens. Host defense is coordinated by the proinflammatory Th1, Th2, and Th17 cells, while Treg cells are involved in the down regulation and contraction of an immune inflammatory response. Th17 cells are believed to be the major proinflammatory cells involved in autoimmunity, while Treg cells protect against autoimmunity [Dong, 2008; Furuzawa-Carballeda et al., 2007; Ivanov et al., 2007; Laurence and O'Shea, 2007; Weaver et al., 2007]. Differentiation along different lineages is initiated by dendritic cells, which process and present (microbial) antigens to TCRs on Thp cells. Interaction of dendritic cells with different classes of pathogens results in the activation of different pathways and the release of different cytokines that subsequently interact with receptors on Thp cells and either guide or repress their differentiation along different lineages (Figure 6). The latter involves the activation of distinct signaling cascades and transcription factors and the synthesis of other cyto/chemokines and cyto/chemokine receptors that are part of additional positive and negative feedback loops. For example, induction of Th1 cells by IL-12 involves the transcription factors STAT4, STAT1, and T-bet, while differentiation along the Th2 lineage in response to IL-4 requires STAT6 and GATA3 [Harrington et al., 2006; Moulton and Farber, 2006]. Differentiation into Treg cells and Th17 cells is often reciprocal, involving several positive and negative regulatory networks that favor one or the other lineage [Bettelli et al., 2006; Bettelli et al., 2008; Dong, 2008; Furuzawa-Carballeda et al., 2007; Harrington et al., 2006; Ivanov et al., 2006; Ivanov et al., 2007; Laurence and O'Shea, 2007; Park et al., 2005; Veldhoen et al., 2006; Weaver et al., 2007]. This includes a yin-yang relationship between RORγt and FOXP3 expression and their regulation by several cytokines, transforming growth factor β (TGFβ), and ATRA.
Figure 6. Specific role for RORs in T cell lineage specification.
Differentiation into different effector CD4+ T cell lineages, T helper (Th) 1, Th2, Th17, and T regulatory (Treg) cells is initiated through an interaction of dendritic cells with uncommitted (naïve) CD4+ T helper cells (Thp). The effector cell types are characterized by their synthesis of specific cytokines and their immuno-regulatory functions, as indicated on the right. The differentiation along different lineages involves different cytokines and the activation of distinct signaling cascades and transcription factors that result in the induction of additional cyto/chemokines and cyto/chemokine receptors, which may be part of positive and negative feedback loops. These cytokines and transcription factors may favor one cell lineage, while inhibiting another (not indicated). RORα and RORγt are induced during differentiation of Thp into the Th17 lineage and are critical for this lineage specification.
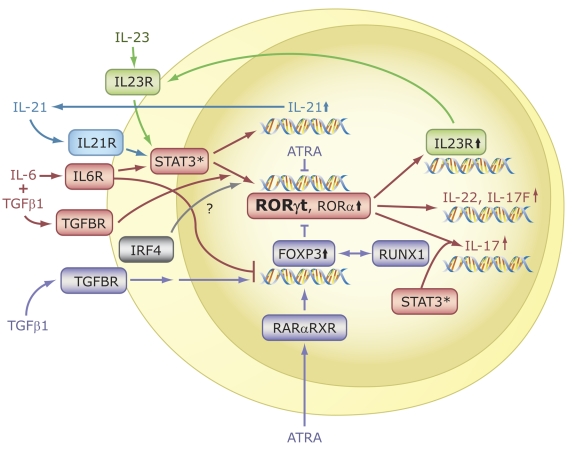
Littman, Cua, and their colleagues were first to report that RORγt is required for the differentiation of naïve CD4+ T cells into Th17 cells [Ivanov et al., 2006; Ivanov et al., 2007; Manel et al., 2008]. This was supported by findings from several other laboratories [Dong, 2008; Laurence et al., 2007; Yang et al., 2007b; Yang et al., 2008d]. RORγt is induced during differentiation of antigen-stimulated Thp cells along the Th17 lineage in response to IL-6 or IL-21 and TGFβ (Figure 7). Cells deficient in IL-6 do neither express RORγt nor IL-17F and IL23R [Korn et al., 2007a; Laurence et al., 2007; Yang et al., 2007b]. IL-6 mediates its action through activation of STAT3. Deficiency in STAT3 greatly impaired the activation of RORγt expression and Th17 differentiation, suggesting that this induction is STAT3-dependent [Harris et al., 2007; Nurieva et al., 2007; Yang et al., 2007b]. Inversely, deficiency in suppressor of cytokine signaling 3 (SOCS3), a negative regulator of STAT3 activity, enhances Th17 differentiation [Chen et al., 2006]. Whether STAT3 regulates RORγt expression directly by binding to the RORγt promoter needs to be established. Additional evidence for a role of RORγt in Th17 differentiation came from studies showing that Thp cells isolated from RORγ null mice exhibited a marked reduction in their ability to undergo differentiation along the Th17 lineage [Ivanov et al., 2006; Nurieva et al., 2007; Yang et al., 2007b]. Conversely, exogenous expression of RORγt in Thp cells greatly increased the expression of IL-17 cytokines and IL23R in the absence of cytokines. Subsequent studies showed that, like RORγt, RORα is highly induced during Th17 cell differentiation in a STAT3-dependent manner [Yang et al., 2008d] (Figure 7). Deficiency in RORα reduced IL-17 and IL23R, but not IL-17F or IL-22 expression, while exogenous expression of RORα in Thp cells or Jurkat cells enhanced IL-17, IL-17F, IL-22, and IL23R expression [Du et al., 2008; Yang et al., 2008d]. These observations indicate that RORα also functions as a positive regulator of Th17 differentiation, suggesting a degree of functional redundancy between RORγt and RORα. This was consistent with findings showing that Th17 differentiation was completely impaired in Thp cells deficient in both RORα and RORγ [Yang et al., 2008c; Yang et al., 2008d]. Thus, both RORα and RORγt are required for optimal differentiation of Thp cells into Th17 cells; however, RORγt appears to be the major player in this process because RORγt deficiency has a more pronounced effect on the expression of Th17 cytokines than loss of RORα. Whether RORα and RORγ have any unique roles in the regulation of gene expression during or after Th17 differentiation needs further study.
Figure 7. RORs are essential for Th17 cell differentiation and IL-17 expression.
In the presence of TCR engagement, treatment of Thp cells with IL-6 plus TGFβ1 induces differentiation along the Th17 lineage and the activation of several genes, including IL-21, RORα, RORγt, IL-17, IL-17F, IL-22, and IL23R. Interaction of IL-23 and IL-21 with, respectively, IL23R and IL21R, reinforce Th17 differentiation and ROR expression. RORα and RORγt are required for the induction of IL-17, IL-17F, and IL23R, but not IL-21. A balance between FOXP3, which is induced by treatment with TGFβ alone, determines whether Thp cells differentiate into Th17 or Treg cells. ATRA, through activation of RARα-RXR complexes, leads to increased FOXP3 and reduced ROR expression. FOXP3 inhibits RORα and RORγt transcriptional activity by interacting directly with RORs, thereby promoting differentiation along the Treg lineage and inhibiting Th17 differentiation. Transcriptional regulators IRF4 and RUNX1 have also been implicated in the regulation of Treg and Th17 differentiation.
In addition to the induction of RORγt, IL-17, and IL-17F, differentiation along the Th17 lineage is associated with increased expression of IL-21 and IL23R [Acosta-Rodriguez et al., 2007; Korn et al., 2007a; Laurence and O'Shea, 2007; Nurieva et al., 2007; Wilson et al., 2007; Zhou et al., 2007]. The induction of IL-21 in response to IL-6 involves direct binding of STAT3 to the IL-21 promoter. IL-21, through its activation of STAT3, synergizes with IL-6 and TGFβ to induce RORγt and IL-17 expression [Nurieva et al., 2007; Palmer and Weaver, 2007; Zhou et al., 2007] (Figure 7). IL-21 expression is not affected in RORγ-deficient mice, suggesting that this regulation occurs upstream of RORγ. IL-21 is also expressed in T follicular helper T (Tfh) cells, another novel T cell subset that develops independent of RORs [Nurieva et al., 2008]. In contrast to IL-21, the induction of IL23R is greatly reduced in RORγ-deficient mice, suggesting that its expression is regulated by RORγt. IL23R itself is part of a positive feedback loop; it allows IL-23 to bind, thereby reinforcing Th17 differentiation through activation of STAT3 [Ivanov et al., 2007; McGeachy et al., 2007; Yang et al., 2007b; Zhou et al., 2007].
Suppression of ROR transcriptional activity by FOXP3
Treg and Th17 cells are reciprocally regulated, involving several positive and negative regulatory networks [Bettelli et al., 2008; Dong, 2008; Furuzawa-Carballeda et al., 2007; Ivanov et al., 2007; Laurence and O'Shea, 2007; Nurieva et al., 2007; Weaver et al., 2007; Zheng and Rudensky, 2007; Zhou et al., 2008]. The balance between the expression of RORγt and the forkhead box transcription factor p3 (FOXP3) plays a critical role in determining whether uncommitted CD4+ T helper cells differentiate into Treg or Th17 cells (Figure 6). FOXP3 contains a repressor domain in the N-terminal half and a C2H2 zinc finger, a leucine zipper, and a forkhead domain at its C-terminal half, allowing the protein to function as an activator or repressor of gene transcription [Campbell and Ziegler, 2007; Carson et al., 2006; Li et al., 2007; Lopes et al., 2006]. FOXP3 is predominantly expressed in CD4+CD25+ Treg cells, where it plays an important role in directing differentiation [Campbell and Ziegler, 2007; Lochner et al., 2008; Zheng and Rudensky, 2007]. In addition, it regulates several functional properties and contributes to the survival of Treg cells. FOXP3-deficient mice lack Treg cells and are highly susceptible to inflammatory disease [Brunkow et al., 2001; Fontenot et al., 2005]. Conversely, induction or exogenous expression of FOXP3 inhibits Th17 differentiation.
Regulation of FOXP3 expression by TGFβ and IL-6 plays a critical role in Treg and Th17 lineage determination [Bettelli et al., 2008; Chen et al., 2007; Dong, 2008; Ivanov et al., 2007; Weaver et al., 2007]. In the presence of TCR engagement, TGFβ induces FOXP3 expression and Treg differentiation in murine Thp cells. Although TGFβ can moderately enhance the expression of RORα and RORγt, FOXP3 is induced at much higher levels, thereby shifting the ROR/FOXP3 balance towards FOXP3 and Treg differentiation [Bettelli et al., 2006; Chen et al., 2003; Fantini et al., 2004; Korn et al., 2007a; Nurieva et al., 2007; Veldhoen et al., 2006; Wan and Flavell, 2007; Yang et al., 2008a; Zhou et al., 2008] (Figure 7). Addition of IL-6 or IL-21 inhibits the induction of FOXP3 by TGFβ and activates RORα and RORγt expression, thereby shifting the ROR/FOXP3 balance in favor of Th17 differentiation. These observations indicate that FOXP3 represses Th17 differentiation by antagonizing RORγt function rather than inhibiting its expression [Du et al., 2008; Ichiyama et al., 2008; Ivanov et al., 2007; Yang et al., 2008c; Zhou et al., 2008]. This antagonism is mediated through a direct interaction of FOXP3 with RORα and RORγt and involves the AF2 domain of RORs and an LXXLL motif within a region encoded by exon 2 of FOXP3. Mutation of this motif greatly inhibits the interaction of FOXP3 with RORs, but did not completely prevent FOXP3-mediated suppression of ROR activity. Transcriptional repression by FOXP3 involves an interaction with the histone acetyltransferase TIP60 and histone deacetylase HDAC7 through the 106-190 amino acid region of FOXP3 [Li et al., 2007]. Deletion of this region diminishes the inhibitory effect of FOXP3 on ROR-mediated transactivation, while lack of both the LXXLL motif and the TIP60-HDAC7 interaction domain totally abolished the FOXP3-mediated repression [Yang et al., 2008c]. Thus, the interaction of FOXP3, possibly in complex with TIP60 and HDAC, with RORs represses ROR transcriptional activity and consequently inhibits the activation of IL-17 expression and Th17 differentiation by RORα and RORγt.
A number of other cytokines and transcription factors influence the balance between Treg and Th17 differentiation. In contrast to IL-6, IL-21, and IL23, IL-2 inhibits RORγt expression and Th17 differentiation through a STAT5-dependent mechanism and promotes the generation of Treg cells [Ivanov et al., 2007; Laurence et al., 2007]. IL-10, in addition to inhibiting Th1 differentiation, also suppresses Th17 differentiation and the expression of RORγt and IL-17 [Gu et al., 2008]. Conversely, RORγt expression and Th17 differentiation is enhanced in IL-10-/- spleen cells. Moreover, treatment of IL-10-/- macrophages with LPS greatly induces RORγt and IL-17 expression. The increased expression of RORγ and IL-17 in IL-10-/- spleen cells and LPS-treated macrophages was repressed by the addition of IL-10. In addition to the STAT transcription factors, interferon regulatory factor 4 (IRF4) has been reported to regulate T lineage commitment and to be required for Th17 differentiation [Brustle et al., 2007; Huber et al., 2008]. IRF4 is essential for IL21-induced Th17 differentiation. The expression of RORα, RORγt, and IL-17 are significantly reduced in IRF4-deficient T cells, whereas the expression of FOXP3 is increased, indicating that IRF4 also plays a role in regulating the balance between Th17 and Treg cells. Thus, the balance between Th17 and Treg differentiation is under a complex control, involving multiple transcription factors.
Suppression of Th17 differentiation and RORγt expression by ATRA
Retinoids play an important role in the control of several immune functions, including the commitment of Thp cells to different lineages [Iwata et al., 2003; Iwata et al., 2004]. ATRA produced from retinol by dendritic cells has been demonstrated to negatively regulate Th1 and Th2 cell differentiation [Iwata et al., 2003]. More recent studies showed that retinoic acid is a critical factor in controlling the balance between Th17 and Treg cells [Coombes et al., 2007; Mucida et al., 2007; Sun et al., 2007]. Upon TCR activation, the differentiation of Thp cells into Treg cells was shown to depend on both TGFβ and ATRA [Benson et al., 2007; Coombes et al., 2007; Elias et al., 2008; Sun et al., 2007]. ATRA inhibited the induction of Th17 differentiation and down-regulated RORγt expression in response to TGFβ and IL-6, and instead promoted FOXP3 expression and the subsequent generation of Treg cells (Figure 7). In addition to IL-17, ATRA also represses the expression of IL-23R and IL-6Rα. These findings are consistent with the concept that differentiation of TCR-activated Thp cells into Th17 or Treg is decided by the balance between FOXP3 and RORγt.
The precise mechanism of the inhibition of RORγt and induction of FOXP3 expression by ATRA still needs to be elucidated. Because IL-2 induces FOXP3 expression through the activation of STAT5, ATRA could enhance FOXP3 through increased IL-2 expression and STAT5 activation. However, FOXP3 induction by ATRA was found to be independent of IL-2 and STAT5 [Elias et al., 2008]. Binding to and activation of retinoic acid receptors α, β, and γ (RARs), in a heterodimeric complex with RXRs, is the major mechanism by which ATRA regulates gene transcription [Minucci and Ozato, 1996]. The retinoid (4-[E-2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl] benzoic acid (TTNPB), a synthetic RAR pan-agonist, effectively induced FOXP3 expression and Treg differentiation, while an RAR-antagonist inhibited this induction by ATRA. These observations suggested that these effects of ATRA are mediated by RARs. Study of RAR-selective retinoids showed that RARα rather than RARγ was found to be the key player in the induction of FOXP3 and suppression of Th17 differentiation [Elias et al., 2008]. This was supported by data demonstrating that ATRA was able to promote the conversion of RARβ- and RARγ-deficient Thp cells into FOXP3+ cells, but not in Thp cells isolated from RARα-deficient mice [Hill et al., 2008]. It has been suggested that stimulation of the TGFβ signaling pathway might partly explain the increased Treg differentiation by ATRA in the presence of TGFβ. This is supported by findings showing that ATRA increases the expression and phosphorylation of SMAD3 [Xiao et al., 2008]. However, recent studies indicated that the induction of Treg cells by ATRA is mediated by an indirect mechanism involving alleviation of the inhibition of FOXP3 expression by CD4+CD44hi memory cells [Hill et al., 2008].
The inhibition of Th17 differentiation by ATRA might explain the protective effects of retinoids in animal models of autoimmune disease [Wang et al., 2000; Xiao et al., 2008]. Supplementation with ATRA suppresses the development of experimental autoimmune encephalomyelitis (EAE) in mice by inhibiting proinflammatory responses. This suppression appears to be related to the inhibition of Th17 cell generation rather than to an increase in Treg cells.
Regulation of IL-17 by RORs
Although the expression of IL-17, IL-17F, IL-22, and IL23R during Th17 differentiation is dependent on RORs, there is little understanding of whether these genes are regulated directly by RORs. Several studies have identified the presence of functional ROREs in a 6 kb promoter flanking region of the IL-17 gene [Ichiyama et al., 2008; Yang et al., 2008d; Zhang et al., 2008b]. However, there is no consensus on which ROREs are most important in driving IL-17 expression by RORγt. Recent studies indicated that the CNS2 region at the il17-il17f locus (between -4 and -6 kb) undergoes chromatin remodeling in response to IL-6/TGFβ [Akimzhanov et al., 2007]. Subsequent studies showed that the regulation of IL-17 by RORs involves this CNS2 regulatory region [Yang et al., 2008d]. This putative enhancer sequence contains two ROREs that are able to bind ROR directly and mediate transcriptional activation by RORα and RORγt, while the 1kb proximal promoter region was nonresponsive to ROR. ChIP analysis showed that RORs are associated in vivo with a region containing the CNS2 site [Yang et al., 2008d]. These observations indicate that the induction of IL-17 by RORs is mediated through this region and suggest that IL-17 is a direct target of ROR transcriptional regulation.
In addition to RORs, several other transcription factors, including STAT3, have been reported to positively regulate IL17 transcription [Yang et al., 2008d; Zhou et al., 2007]. The induction by STAT3 is mediated through binding to the promoter region of the IL17 gene. Expression of constitutively active STAT3 induces IL17 when co-expressed with RORγt, but has little effect in RORγ-deficient T cells, indicating that the induction of IL17 expression by STAT3 is dependent on the presence of RORγt.
Runt-related transcription factor 1 (RUNX1) is another transcription regulator that promotes Th17 differentiation by enhancing RORγt and IL17 expression [Zhang et al., 2008b]. The activation of IL17 expression by RUNX1 is mediated through two Runx-binding sites that are near the ROREs in the IL17 promoter region. In addition, it was shown that RUNX1 interacts directly with RORγt and acts synergistically with RORγt to optimally activate IL17 expression. Expression of FOXP3 inhibits this activation by interacting directly with RORγt and RUNX1, thereby repressing their transcriptional activity.
RORs and autoimmune disease
Th17 cells have been implicated in autoimmune disease and other inflammatory conditions in humans, including lupus, arthritis, multiple sclerosis, atopic dermatitis, psoriasis, asthma, and inflammatory bowel disease [Bi et al., 2007; Huang et al., 2007; Ivanov et al., 2007]. IL-17 induces the secretion of a variety of chemokines, cytokines, metalloproteinases, and other proinflammatory mediators and promotes the recruitment of neutrophils. Mice lacking IL-17 are resistant to collagen-induced arthritis (CIA) and EAE, a model for multiple sclerosis. Neutralizing IL-17 with antibodies suppresses CNS autoimmune inflammation [Dong, 2008; Furuzawa-Carballeda et al., 2007; Huang et al., 2007; Korn et al., 2007b; Stockinger et al., 2007]. Inversely, transfer of Th17 cells aggravates the disease. Rag1-/- mice reconstituted with CD4+ splenocytes or bone marrow from RORγ- or RORα-deficient mice are less susceptible to EAE than mice reconstituted with WT CD4+ splenocytes or bone marrow [Ivanov et al., 2006; Yang et al., 2008d]. Similar experiments with Rag1-/- mice reconstituted with cells from mice deficient in both RORα and RORγ showed a complete resistance to EAE. Recent studies using the adoptive transfer model of colitis showed that transfer of RORγ-null T cells failed to increase mucosal IL-17 cytokine levels and did not induce colitis, while treatment with IL-17A restored colitis after transfer of RORγ-null T cells [Leppkes et al., 2009]. These studies indicate that loss of RORα/γt function greatly reduces the susceptibility to the development of autoimmune disease in mice. These observations suggest that by controlling Th17 differentiation and IL-17 expression, RORγt and RORα play a critical role in the regulation of inflammatory responses (Figure 2). Therefore, RORs might function as potential drug targets for therapeutic intervention into autoimmune disease, including chronic colitis and multiple sclerosis.
RORs and lung inflammation
RORαsg/sg and RORγ-/- mice are less susceptible to ovalbumin (OVA)-induced lung inflammation, which serves as an experimental model for allergic airway disease [Jaradat et al., 2006; Tilley et al., 2007]. The infiltration of inflammatory cells, including eosinophils, neutrophils, and lymphocytes into peribronchiolar and perivascular regions in lungs and in bronchial alveolar lavage fluid, was significantly less in OVA-challenged RORαsg/sg mice compared to WT mice, and the induction of Th2 cytokines IL-4, IL-5, and IL-13 was greatly compromised. Analysis of gene expression profiles demonstrated that the expression of many genes implicated in inflammatory responses, including the chemokines TARC, eotaxin-2 (Ccl24), MIP-1a (Ccl3), MCP-2 (Ccl8), and RANTES (Ccl5), acute response proteins, mucins MUC5b and MUC5ac, are induced to a significantly smaller degree in lungs from OVA-challenged RORαsg/sg mice than those from WT mice. Together, these observations show that RORαsg/sg mice develop a less severe allergic inflammatory response. Interestingly, RORαsg/sg mice are more susceptible to LPS-induced inflammation [Stapleton et al., 2005]. The mechanism of this increased sensitivity is not quite yet understood.
The allergic response in RORγ-/- mice is also severely impaired, as evidenced by attenuated eosinophilic pulmonary inflammation and reduction in CD4+ lymphocytes and level of Th2 cytokines/chemokines [Tilley et al., 2007]. OVA sensitized and challenged RORγ-/- animals showed unexpected increases in TNF-α, IL-2, IL-10, and IFN-γ and reciprocal reductions in IL-4, IL-5, and IL-13. This shift in cytokine profiles, coupled with reduced IgG1 and elevated IgG2c levels in the sera of OVA-exposed RORγ-/- mice, reveals a critical role for RORγ in the regulation of immunoglobulin production and balance between different T helper cell populations. The decreased inflammatory response in RORγ-/- mice might be in part related to the role RORγ plays in Th17 lineage determination [Ivanov et al., 2006; Ivanov et al., 2007; Yang et al., 2008d]. Interestingly, mice lacking IL-17 have been reported to exhibit an attenuated eosinophilic lung inflammation and Th2 cytokine production, while IL-10, which is produced by Treg cells, has been reported to inhibit allergic inflammation [Umemura et al., 2007; Urry et al., 2006]. Moreover, mice deficient in the IL17 gene, but not IL17F, also exhibited reduced Th2 responses in a similar asthma model [Yang et al., 2008b]. Thus, the reduction in Th17 cells in RORγ-/- mice and the observed increase in IFN-γ (Th1) and IL-10 (Treg) in OVA-challenged RORγ-/- mice is consistent with the hypothesis that the decreased susceptibility to allergic lung inflammation in these mice might be related to shifts in the commitment to different Th lineages. This hypothesis is consistent with the concept that crosstalk between lineage-specific transcription factors plays an important role in lineage specification.
RORs and circadian rhythm
Circadian behavior of ROR expression
Circadian rhythms are daily cycles of behavioral and physiological changes that are regulated by endogenous circadian clocks [Hastings et al., 2007; Isojima et al., 2003; Ko and Takahashi, 2006; Liu et al., 2007a; Schibler and Naef, 2005; Schibler and Sassone-Corsi, 2002]. The circadian clock impacts virtually all physiological functions and behavior. It is therefore not surprising that abnormalities in circadian rhythm are implicated in an increasing number of diseases, including sleep and mood disorders, diabetes, obesity, and cancer [Barnard and Nolan, 2008; Gery and Koeffler, 2007; Ishida, 2007; Liu et al., 2007a; Maywood et al., 2006; Turek et al., 2005].
A number of studies have established links between nuclear receptor function and expression, the circadian regulatory circuitry, and the regulation of various physiological processes [Akashi and Takumi, 2005; Duez and Staels, 2008; Guillaumond et al., 2005; Horard et al., 2004; Jetten and Joo, 2006; Liu et al., 2008; Preitner et al., 2003; Ueda et al., 2002b; Yang et al., 2006; Yang et al., 2007a]. Many nuclear receptors, including RORs, display, at least in some tissues, rhythmic patterns of expression during the circadian cycle, indicating that their expression is controlled by the circadian clock. RORα expression shows a weak circadian oscillation in liver, kidney, retina, and lung [Akashi and Takumi, 2005; Kamphuis et al., 2005; Tosini et al., 2007; Ueda et al., 2002b; Yang et al., 2006]. In the SCN, RORα exhibits a rhythmic pattern of expression with a peak at CT6-8. A similar circadian profile was observed for RORβ, while RORγ is not expressed in the SCN [Schaeren-Wiemers et al., 1997; Sumi et al., 2002; Ueda et al., 2002b]. RORβ2 displays a rhythmic expression pattern in mouse pineal gland and retina, with a maximum at CT18 [Andre et al., 1998a; Ueda et al., 2002b]. RORγ exhibits an oscillatory expression pattern in liver, brown adipose tissue, and kidney, but not in skeletal muscle and thymus [Guillaumond et al., 2005; Jetten and Joo, 2006; Kang et al., 2007; Ueda et al., 2002b; Ueda et al., 2005; Yang et al., 2006]. In these tissues, RORγ is expressed at low levels during the day and at optimum levels at night.
Mice deficient in RORα or RORβ exhibit an aberrant circadian behavior [Akashi and Takumi, 2005; Masana et al., 2007; Sato et al., 2004; Schaeren-Wiemers et al., 1997], while no abnormalities in circadian behavior have been noticed in RORγ-/- mice [Liu et al., 2008]. RORαsg/sg mice display an anomalous, slightly decreased, free-running period of locomotor activity and an altered feeding pattern during the circadian cycle [Akashi and Takumi, 2005; Guastavino et al., 1991; Sato et al., 2004], while RORβ-/- mice display an extended period of free-running rhythmicity under conditions of constant darkness [Andre et al., 1998a; Masana et al., 2007].
Regulation of clock genes by ROREs
Nearly every mammalian cell type contains its own circadian oscillator. The oscillators in peripheral tissues operate in a self-sustained, cell-autonomous manner [Isojima et al., 2003; Ko and Takahashi, 2006; Liu et al., 2007a; Reppert and Weaver, 2001; Reppert and Weaver, 2002; Ripperger and Schibler, 2001; Schibler and Sassone-Corsi, 2002]. Their synchronization is coordinated by signals originating from the master circadian clock residing in the SCN of the anterior hypothalamus. The phase of the master oscillator is set by the light-dark cycle input received by both visual and non-visual photoreceptor systems in the retina.
At the molecular level, the clockwork consists of an integral network of several interlocking negative feedback and positive feedforward loops. The basic molecular circuitry of circadian clocks is analogous between tissues. A major positive loop consists of the basic helix-loop-helix/PAS-type transcriptional activators BMAL1 and CLOCK or its paralog NPAS2, while two cryptochrome (CRY) and three period proteins (PER) are involved in the negative control of the oscillator [Albrecht, 2002; DeBruyne et al., 2007a; DeBruyne et al., 2007b; Isojima et al., 2003; Reppert and Weaver, 2002; Schibler and Naef, 2005; Schibler and Sassone-Corsi, 2002] (Figure 8). BMAL1-CLOCK heterodimers positively regulate the circadian expression of many genes, including PER and CRY, through interaction with E-box enhancers (CACGTG) in the promoter regulatory region of target genes. When PER and CRY proteins accumulate to a critical level, they in turn repress BMAL1:CLOCK-mediated transcription, including their own transcription, by interacting directly with BMAL1-CLOCK complexes. This subsequently leads to reduced levels of PER and CRY protein and to a new cycle of activation and repression.
Figure 8. RORs function as integrators of circadian oscillators and the rhythmic expression of downstream (metabolic) genes.
The rhythmic expression of RORs is greatly dependent on the ROR isoform and tissue type. The core loop of the oscillator consists of the heterodimeric activators BMAL1 and CLOCK that activate the expression of PERs and CRYs. PER and CRY heterodimers interact with the BMAL1-CLOCK complex and negatively regulate their transcriptional activity. BMAL1 and CLOCK also activate the expression of REV-ERBs, which through their interaction with ROREs repress the transcription of several genes, including BMAL1, CRY1, and RORγ. REV-ERBs are critical in the rhythmic expression of BMAL1, CRY1 and RORγ in liver and several other tissues. RORγ does not affect the rhythmic expression, but increases the peak level of expression of BMAL1, CRY1, NPAS2, CLOCK, and REV-ERBα. The major function of RORγ appears to be to regulate the oscillatory pattern of expression of downstream target genes and, therefore, the rhythmic nature of several physiological processes. RORα exhibits a very weak rhythmic pattern of expression in liver and its major function also appears to be in regulating the (rhythmic) expression of downstream target genes. SIRT, through its deacetylase activity, promotes the degradation of PER and inhibits the interaction of CRY with BMAL1/CLOCK complex, thereby enhancing BMAL1/CLOCK transcriptional activity.
The core loop is further regulated by several accessory pathways, including one in which the D box-binding proteins, the activator D site-binding protein (DBP) and the repressor E4 promoter binding-protein 4 (E4BP4 or NFIL3), play a role, and another that involves regulation by RORs and REV-ERBs through ROREs [Burris, 2008; Ko and Takahashi, 2006; Liu et al., 2008; Preitner et al., 2003; Preitner et al., 2002; Ueda et al., 2005]. ROREs have been identified in several clock genes, including BMAL1, CLOCK, and CRY1 [Guillaumond et al., 2005; Kumaki et al., 2008; Nakajima et al., 2004; Preitner et al., 2002; Sato et al., 2004; Ueda et al., 2002b]. The BMAL1 gene, which contains two ROREs in its promoter region, is positively regulated by RORs, whereas REV-ERBs repress BMAL1 transcription (Figure 8). Mutation of these ROREs abolishes the circadian rhythmicity of the BMAL1 promoter activation, indicating that these response elements are essential for rhythmic transcriptional regulation of BMAL1. Further study showed that BMAL1 expression is elevated and its rhythmicity impaired in fibroblasts deficient in both REV-ERBα and β, but fibroblasts deficient in only one of the REV-ERB receptors still retain BMAL1 rhythmicity [Liu et al., 2008]. These observations suggested that REV-ERBα and β are functionally redundant and essential for the rhythmic expression of BMAL1. However, REV-ERBα and β are not required for the rhythmicity of several other clock genes, including PER and CRY.
In contrast to REV-ERBs, RORs are not required for the rhythmicity of BMAL1 expression [Liu et al., 2008]. RORαsg/sg fibroblasts, which lack RORα and do not express either RORβ or γ, display rhythmic oscillations of BMAL1 or PER2 promoter-regulated luciferase reporter expression that are similar to that observed in WT fibroblasts. These findings are in agreement with studies showing that the rhythmic expression of various clock proteins, including PER2, CRY1, BMAL1, and CLOCK, was not altered in RORαsg/sg or RORγ null mice [Liu et al., 2008; Sato et al., 2004]. However, the peak expression levels of BMAL1, as well as that of several other RORE-containing clock genes, including CLOCK, CRY1, and NPAS2, were reduced in both RORαsg/sg and RORγ null mice, while those of PER2 and DBP were not changed. Regulation of BMAL1 by RORs is supported by findings showing that RORs interact with ROREs in the BMAL1 promoter and induce transcriptional activation through this promoter region [Akashi and Takumi, 2005; Guillaumond et al., 2005; Sato et al., 2004; Ueda et al., 2005]. This activation of BMAL1 expression by RORs appears to be mediated through the coactivator PGC-1α [Liu et al., 2007b]. Thus, RORs are not essential for the rhythmic expression of BMAL1 or other clock genes, but do regulate their level of expression. Moreover, in contrast to common regulators of the circadian clock, which appear to be ubiquitously expressed, RORs exhibit a much more restricted pattern of expression, consistent with the view that they are not common constituents of clock regulatory circuitry mechanisms. In addition to modulating the expression of several components of the circadian clock, the major function of RORs might be regulating the rhythmic expression of downstream target genes. Therefore, RORs may function as intermediates that couple the circadian oscillators with the cyclic control of various physiological processes, including energy homeostasis, lipid, and xenobiotic metabolism [Kang et al., 2007; Liu et al., 2008; Sato et al., 2004] (Figure 8).
Regulation of circadian expression of RORs
As mentioned above, in several tissues RORs display a rhythmic expression during the circadian cycle [Liu et al., 2008; Panda et al., 2002; Sato et al., 2004; Ueda et al., 2002b; Ueda et al., 2005; Ukai-Tadenuma et al., 2008]. Several functional elements relevant to circadian rhythm have been identified in the promoter of RORs. Functional D-boxes were identified in RORα and RORβ, suggesting regulation by members of the bZIP-family. The proximal promoter of RORγ1 and RORα1 contain one functional E-box, while that of RORα4 contains two E-boxes, suggesting regulation by BMAL1/CLOCK. Based on the presence of functional ROREs in the promoter regions of RORs, it was proposed that their expression might be controlled by REV-ERBs proteins. The observation that in liver and several other tissues the expression of RORγ1 and REV-ERBs is inversely related, is consistent with the view that REV-ERBs function as repressors of RORγ1 transcription and regulate the rhythmicity of RORγ1 expression (‘repressor-antiphasic-to-activator’ mechanism) [Liu et al., 2008; Ueda et al., 2005; Ukai-Tadenuma et al., 2008]. RORγ1 appears to have a minor role in controlling the transcription of REV-ERBs because their expression is only moderately affected in liver from RORγ null mice. Analysis of RORγ1 mRNA expression in liver of BMAL1-/- mice showed that the rhythmicity of RORγ1 expression was almost totally abolished and that RORγ1 expression at all CTs was greatly elevated [Grechez-Cassiau et al., 2008; Liu et al., 2008]. It was concluded that, at least in liver, the loss of REV-ERB repressor expression observed in BMAL1-/- mice is likely responsible for the lack of rhythmicity and relieved or elevated RORγ1 expression in these mice [Liu et al., 2008]. Expression was particularly increased at CT6-8, a time at which REV-ERB is normally highly expressed. How then is RORγ1 transcription activated in liver of BMAL1-/- mice? It was suggested that the expression of RORs might be driven by non-circadian, tonic signals, such as the cAMP:cAMP response element-binding (CREB) pathway [Liu et al., 2007a; Liu et al., 2008; O'Neill et al., 2008].
SIRT1 and RORγ expression
Posttranslational modifications, including phosphorylation by casein kinases and other kinases and acetylation, play a critical role in regulating protein-protein interactions, the nuclear localization, transcriptional activity, and turnover of clock proteins [Eide et al., 2005; Gallego and Virshup, 2007; Ko and Takahashi, 2006; Ripperger and Schibler, 2001]. CLOCK, which functions as a histone acetyltransferase, has been implicated in chromatin remodeling during circadian rhythm [Doi et al., 2006]. In addition to histones, CLOCK also acetylates other proteins, including BMAL1 [Hirayama et al., 2007]. Recently, sirtuin 1 (SIRT1), a NAD+-dependent deacetylase, was identified as an important regulator of circadian gene expression. It was suggested that it provides a functional link between metabolic activity and circadian control [Asher et al., 2008; Belden and Dunlap, 2008; Nakahata et al., 2008]. SIRT1 deacetylates histones and a number of transcriptional regulators, and was shown to bind BMAL1-CLOCK-PER2 complexes in a circadian-dependent manner. SIRT1 catalyzes deacetylation of PER2, thereby facilitating its degradation by the proteasome system. Consequently, absence of SIRT1 expression leads to high levels of PER2, resulting in diminished BMAL1-CLOCK transcriptional activity and repression of clock-controlled target genes, including reduced expression of PER2 and CRY1 (Figure 8). In addition, SIRT1 induces deacetylation of BMAL1 at Lys537, which reduces the efficacy of CRY to interact with and silence CLOCK-BMAL1 [Nakahata et al., 2008]. Although RORγ1 is expressed at low levels in mouse embryo fibroblasts (MEFs), its expression is greatly diminished in SIRT1 KO MEFs. Because RORγ1 expression is positively regulated by BMAL1-CLOCK, its repression in SIRT1 KO MEFs may be related to the silencing of BMAL1-CLOCK activity. Inversely, the down-regulation of RORγ1 in SIRT1 KO MEFs may, at least in part, account for the reduced expression of BMAL1 observed in these cells.
It has been become increasingly clear that SIRT1 functions as a link between the circadian rhythm circuitry, cellular metabolism, and nuclear receptors, including RORs [Asher et al., 2008; Nakahata et al., 2008; Schwer and Verdin, 2008]. One such link involves PGC-1α. SIRT1 has been reported to induce deacetylation and activation of PGC-1α, which functions as a coactivator of several nuclear receptors [Rodgers et al., 2008]. This, together with the circadian regulation of nuclear receptor expression, is part of the complex mechanism by which nuclear receptors regulate cellular metabolism. It is interesting to note that SIRT1 activity peaks around the same time as RORγ1 mRNA expression (at CT16). Further studies are needed to elucidate the precise molecular and physiological connections between SIRT1 and ROR activity.
RORs and cellular metabolism
Changes in lipid metabolism and ROR-deficient mice
Accumulating evidence indicates that RORs play an important role in the regulation of several metabolic pathways, particularly lipid and steroid metabolism [Boukhtouche et al., 2004; Duez and Staels, 2008; Genoux et al., 2005; Jetten, 2004; Jetten and Joo, 2006; Kang et al., 2007; Lau et al., 2008; Lau et al., 2004; Lind et al., 2005; Mamontova et al., 1998; Raichur et al., 2007; Raspe et al., 2001; Vu-Dac et al., 1997; Wada et al., 2008a; Wada et al., 2008b]. RORαsg/sg mice on a normal diet display a pronounced hypo-α-lipoproteinemia and have lower levels of total plasma cholesterol, high density lipoprotein (HDL), ApoA1, the major constituent of HDL, ApoC3, ApoA2, and triglycerides compared to WT mice. The reduced expression of the reverse cholesterol transporters ABCA1 and ABCA8/G1 in the liver and of ApoA1 in the intestine may in part be responsible for the reduced HDL levels. Functional ROREs have been identified in the promoter regulatory region of APOC2 and APOA1 [Raspe et al., 2001; Vu-Dac et al., 1997]. The RORE within the APOAI promoter was shown to interact selectively with RORα1, but not with RORα2 or α3. Although human ApoA5 was identified as a RORα target gene, its expression was not altered in liver of RORαsg/sg mice [Genoux et al., 2005; Lind et al., 2005]. Upon aging, RORαsg/sg mice are less susceptible to the development of hepatic steatosis than WT mice [Lau et al., 2008]. In addition, despite their higher food consumption, aging RORαsg/sg mice exhibit a reduced body fat index. Fat cells in brown and white adipose tissue are smaller and the level of triglycerides is lower in liver of RORαsg/sg mice than WT mice. The increased food intake correlates with an observed reduction in leptin expression. As observed for aging RORαsg/sg mice, 2-3 months-old RORαsg/sg mice are less susceptible to high fat diet-induced obesity and steatosis [Lau et al., 2008].
In contrast to RORαsg/sg mice, cholesterol and triglyceride levels were unaltered in RORγ-/- mice; however, these mice have a somewhat reduced blood glucose level [Kang et al., 2007]. Mice deficient in both RORα and RORγ (DKO) exhibit similar changes in cholesterol, triglycerides, and glucose as observed for single KO mice. Comparison of gene expression profiles from livers of WT, RORα-deficient, RORγ-deficient, and DKO mice revealed that RORα and RORγ can function as positive and negative regulators of gene expression. In addition to regulating the expression of a distinct set of genes, suggesting unique regulatory functions, some genes are only up- or down-regulated in DKO mice, suggesting a degree of functional redundancy between RORα and RORγ [Kang et al., 2007]. The latter is likely related to their similarities in RORE binding affinities. However, the main functions of RORα and RORγ appear to be distinct from each other.
RORs in the regulation of (lipid) metabolism
The effect of RORα on triglyceride homeostasis can be due to the changes in the regulation of a number of genes, including those involved in the control of lipogenesis and fatty acid oxidation. The expression of sterol regulatory element-binding protein 1, isoform c (SREBP1c), which is a critical regulator of lipogenesis and several lipogenic genes, was significantly reduced in liver and skeletal muscle of RORαsg/sg mice [Lau et al., 2008; Wada et al., 2008a]. This was associated with a suppression of several SREBP1c target genes, including fatty acid synthase (FAS). LXRs and carbohydrate response element-binding protein (ChREBP) have been reported to be critical factors in the regulation of SREBP1c [Cha and Repa, 2007; Pegorier et al., 2004; Wagner et al., 2003]. However, the expression of LXRs and ChREBP was not significantly altered in liver from RORαsg/sg mice. ChIP and promoter analysis indicated that SREBP1c transcription is under the selective and direct control of RORα4, the predominant isoform of RORα in liver [Lau et al., 2008]. It was concluded that RORα positively regulates lipogenesis through increased SREBP1c expression. The latter might, at least in part, explain the reduced adiposity in RORαsg/sg mice.
The expression of estrogen-related receptor α (ERRα) and peroxisome proliferators-activated receptors (PPARs) α, β, and γ, which are implicated in the control of lipid homeostasis as well, was not changed in RORαsg/sg mice [Desvergne et al., 2006; Tremblay and Giguere, 2007]. However, the expression of the coactivators PGC-1α and β, which are involved in the regulation of oxidative metabolism and gluconeogenesis, was increased in liver and brown adipose tissue, respectively [Liu et al., 2008]. Expression of lipin 1, which has multiple functions in lipid metabolism and glucose homeostasis in several tissues, was found to be enhanced in liver and adipose tissues of RORαsg/sg mice [Lau et al., 2008]. Loss of lipin 1 function leads to hepatic steatosis and hypertriglyceridemia, but reduced adipose tissue [Peterfy et al., 2001]. Lipin 1 has been reported to interact directly with the nuclear receptor PPARα and coactivator PGC-1α in a complex that modulates fatty acid oxidation gene expression in hepatocytes [Finck et al., 2006]. Thus, increased levels of PGC-1 and lipin 1 in RORαsg/sg liver might enhance oxidative metabolism and account for the increased expression of acyl-coenzyme A dehydrogenase ACADM. However, expression of carnitine palmitoyltransferases 1A and 1B (CPT1A/B), which are also involved in fatty acid oxidation, was not significantly affected in liver of RORαsg/sg mice. Clearly, further studies are needed to understand the complex relationships between RORα, its regulation of lipid metabolism, and its effects on obesity and triglyceride levels.
Regulation of CYP7b1 and sulfotransferases
Several of the genes regulated by RORs encode proteins involved in Phase I and II enzymes that are important in the metabolism of xenobiotics, drugs, environmental chemicals, and various endogeneous compounds, including steroids and bile acids [Kang et al., 2007; Wada et al., 2008a]. These enzymes play an important role in elimination and detoxification of endogenous and exogenous compounds. One of the genes regulated by RORs is oxysterol 7alpha-hydroxylase (Cyp7b1), which plays a role in the alternative pathway of cholesterol metabolism, whereas the expression of Cyp7a1 and Cyp27, which play a role in the major cholesterol metabolic pathway, was not affected in ROR-deficient mice. Cyp7b1, which is preferentially expressed in liver of male mice, was down-regulated in RORαsg/sg mice and was affected only to a small degree in RORγ-/- mice [Kang et al., 2007; Wada et al., 2008a]. RORα regulates Cyp7b1 expression directly by binding to an RORE at nt -951 in the promoter regulatory region of the Cyp7b1 gene. This RORE preferentially binds RORα rather than RORγ. These observations suggest that RORα functions as a positive modulator of Cyp7b1 expression. Recent studies identified CYP2C8, which play an important role in drug metabolism, as a putative ROR target gene in human hepatocytes [Chen et al., 2009]. Both RORα and RORγ were able to enhance the activation through a RORE within the CYP2C8 promoter and to enhance CYP2C8 expression in human primary hepatocytes.
Sulfotransferase 1e1 (Sult1e1) is among the genes most dramatically induced in liver of RORαsg/sg mice, while the expression is not significantly changed in RORγ-/- mice, suggesting that it is under the selective control of the RORα signaling pathway [Kang et al., 2007]. Sult1e1 expression in liver is restricted to female mice. It catalyzes the sulfonation of estrogen and estrone, thereby increasing their solubility and facilitating their transport and excretion [Duanmu et al., 2006]. Sult2a1 expression is induced in liver of both RORαsg/sg and RORγ-/- mice, suggesting that both RORα and RORγ function as negative modulators of Sult2a1 expression. It catalyzes the sulfonation of procarcinogens, xenobiotics, hydroxysteroids, and bile acids, including dehydroepiandrosterone and lithocholic acid [Kitada et al., 2003]. In addition to these sulfotransferases, the expression of several members of the hydroxysteroid dehydrogenase family, many of which are involved in the biosynthesis or inactivation of steroid hormones, are regulated by RORs [Kang et al., 2007]. These observations suggest that RORs, and particularly RORα, play an important role in the regulation of specific steps in bile acid and steroid metabolism. The control of several sulfotransferases by RORα is interesting with regard to reports showing that cholesterol sulfate functions as a ligand of RORα [Kallen et al., 2004; Kallen et al., 2002]. Possibly other sulfated steroid or cholesterol metabolites might function as high affinity RORα ligands and be part of regulatory feedback loops that control specific steps in steroid/cholesterol metabolism.
Several metabolic genes regulated by RORs have been reported to be target genes of other nuclear receptors [Akiyama and Gonzalez, 2003; Kalaany and Mangelsdorf, 2006; Kliewer, 2003; Qatanani and Moore, 2005; Runge-Morris and Kocarek, 2005]. For example, Sult2a1 expression is regulated by several receptors, including constitutive androstane receptor (CAR), hepatocyte nuclear factor 4α (HNF4α), and pregnane X receptor (PXR) [Fang et al., 2007; Maglich et al., 2004]. An overlap between genes regulated by RORα and liver X receptors (LXRs) was particularly evident [Gong et al., 2007; Wada et al., 2008b]. CD36, Sult1e1, and Sult2a1 are induced while Cyp7b1 expression is inhibited by activated LXR, a pattern that is the inverse of that observed for RORα [Kang et al., 2007; Wada et al., 2008a; Wada et al., 2008b]. This raised the possibility of potential crosstalk between the RORα and LXR pathways. However, this crosstalk does not occur at the level of the regulation of LXR or ROR expression because LXRα and β expression were not altered in liver of RORαsg/sg mice, while RORα expression was unchanged in liver of LXR DKO mice [Lau et al., 2008; Wada et al., 2008a]. Although in cultured cells, competition between LXR and RORα for binding common coactivators has been observed, it is not clear whether such a competition is physiologically significant. In vivo, the inverse regulation of common targets by RORα and LXR may involve several mechanisms, including crosstalk between ROR and LXR transcriptional complexes bound to their respective response elements within the promoter of the same target gene.
RORs and muscle
RORα and RORγ mRNA are highly expressed in skeletal muscle, suggesting a regulatory role for these receptors in myogenesis or muscle function [Becker-Andre et al., 1993; Hirose et al., 1994; Lau et al., 1999]. This is supported by observations showing that RORαsg/sg mice develop muscular atrophy; however, the mechanism underlying this phenotype has not been clearly identified. C2C12 skeletal muscle cells have been used to examine the function of RORα and RORγ in myogenesis and lipid homeostasis. Expression of a dominant-negative RORα, lacking the LBD, delays the morphological differentiation of myotubes and induction of muscle-specific genes, such as the helix-loop-helix transcription factors MyoD and myogenin, transcription factors that are critical for myotubule differentiation [Lau et al., 1999]. In addition, the expression of fatty acid translocase (FAT/CD36) and fatty acid binding protein 3 (FABP3), which are involved in lipid and fatty acid uptake, and of lipoprotein lipase (LPL), CPT-1, and acyl-CoA-synthetase-4 (ACS4) involved in triglyceride hydrolysis and hydroxylation, respectively, were reduced in these cells [Lau et al., 2004]. Expression of RORγ in C2C12 cells was shown to increase the expression of several genes, including CD36, FABP4, LPL, UCP3, fructose transporter GLUT5, adiponectin receptor 2 (ADPR2), and IL-15, suggesting a role for RORγ in the regulation of lipid metabolism in muscle [Raichur et al., 2007].
RORs and cancer
RORs and increased cancer susceptibility
A number of studies have provided evidence suggesting a role for RORs in cancer. Mice deficient in the expression of RORγ exhibit a high incidence of thymic lymphomas that metastasize frequently to liver and spleen [Jetten and Ueda, 2002; Ueda et al., 2002a]. As a consequence, the lifespan of RORγ null mice is greatly reduced. The enhanced lymphoma formation may be related to changes in thymic homeostasis observed in RORγ KO mice [Kurebayashi et al., 2000; Sun et al., 2000]. Although the molecular mechanism underlying enhanced lymphoma formation has not yet been established, it may be related to the dysregulation of differentiation and proliferation in RORγ-/- thymocytes. Interestingly, type B leukemogenic virus (TBLV), which causes T-cell lymphomas in mice, was found to be frequently integrated at the RORγ locus [Broussard et al., 2004]. However, in contrast to RORγ null mice, RORγ expression correlated positively with lymphomas. Whether RORγ is implicated in human lymphomas requires further study.
Another potential link between RORγ and cancer is emerging from studies showing increased expression of Th17-associated genes, including RORγ, IL-17, and IL-23, in gastric tumors, an increase in the population of circulating Th17 cells in gastric cancer patients [Zhang et al., 2008a], and a high incidence of Th17 cells at sites of ovarian cancer [Miyahara et al., 2008]. These elevated levels of proinflammatory cytokines may contribute to cancer pathogenesis. Alterations in RORγt expression or activity may cause changes in the Th17 cell population and production of proinflammatory cytokines, thereby affecting cancer progression positively or negatively.
Although no explicit links between RORα and cancer have been established, a number of studies have indicated a possible a role for RORα in cancer development. The RORα gene spans a 730 kb genomic region that is located in the middle of the common fragile site FRA15A within chromosomal band 15q22.2 [Smith et al., 2006; Zhu et al., 2006]. Common fragile sites are highly unstable genomic regions found in all individuals and are hotspots for deletions and other genetic alterations that may lead to altered expression and function of genes encoded within these regions. Common fragile sites have been implicated in several human diseases and are associated with a number of different cancer types [Smith et al., 2006]. Genomic instability within FRA15A might lead to changes in the expression of RORα and play a role in the development of certain cancers. This hypothesis is consistent with observations showing that RORα mRNA expression is often down-regulated in tumor cell lines and primary cancer samples [Zhu et al., 2006]. Moreover, studies examining gene expression profiles in various cancers identified RORα as a gene commonly down-regulated in several tumor types, particularly breast and lung cancer [Lu et al., 2007]. Analysis of the methylation status of a series of genes identified RORα as one of methylation-silenced genes in gastric cancer cell lines [Yamashita et al., 2006]. The latter is in agreement with the concept that reduced expression of RORα expression positively correlates with tumor formation.
RORs, hypoxia and stress
Several studies have revealed a connection between RORα expression and hypoxia. Hypoxia is important in normal physiology and in several pathologies, including ischemia, stroke, heart attack, chronic kidney disease, and cancer progression [Pouyssegur et al., 2006; Semenza, 2000]. Oxygen sensing is a key control mechanism of vasculogenesis and tumor growth. Hypoxia triggers a multifaceted cellular response in which the transcription factor hypoxia-inducible factor HIF-1α plays a central role. Several studies have reported that RORα4, but not RORα1, expression is highly induced under hypoxic conditions in a variety of cell types [Besnard et al., 2002; Chauvet et al., 2004; Chauvet et al., 2002; Miki et al., 2004]. Conditions that mimic effects of hypoxia, including exposure to cobalt chloride and the iron chelator, desferrioxamine, also enhance RORα4 expression [Chauvet et al., 2002]. In addition to hypoxia, several other stress conditions induce RORα4 expression, including H2O2 and several inducers of endoplasmic reticulum (ER) stress, including farnesol, thapsigargin, tunicamycin, and brefeldin [Joo et al., 2007; Zhu et al., 2006]. These observations indicate a regulatory role for RORα4 in hypoxia and stress responses. Studies in vivo demonstrated that ischemia-induced angiogenesis was enhanced in RORα-deficient mice [Besnard et al., 2001]. These observations suggest that RORα functions as an important negative modulator of angiogenesis. Angiogenesis is controlled by pro- and anti-angiogenic factors. Inflammatory cells and cytokines have also been implicated in the regulation of angiogenesis. Thus, changes in the expression of pro- and/or anti-angiogenic or inflammatory genes in RORαsg/sg mice might account for the observed increased angiogenesis. The expression of the pro-angiogenic vascular endothelial growth factor (VEGF) was not changed in ischemia-induced angiogenesis in RORαsg/sg mice; however, the level of interleukin 12 (IL-12), an anti-angiogenic cytokine, was found to be reduced. Clearly, additional studies are needed to understand the role of RORα in angiogenesis.
The induction of RORα4 expression by hypoxia is mediated by HIF-1α through an interaction with a hypoxia response element (HRE) in the proximal promoter region of RORα4 [Chauvet et al., 2004]. Subsequent studies showed that activation of RORα4 by hypoxia was dependent on an interaction between HIF-1α and the transcription factors Sp1/Sp3 bound to GC-rich sequences adjacent to the HRE [Miki et al., 2004]. Recently, exogenous expression of RORα was reported to increase the level of HIF1α and VEGF protein, while knockdown of RORα expression by siRNAs reduced HIF1α and VEGF expression [Kim et al., 2008]. Moreover, RORα was found to enhance capillary tube formation by human umbilical vein endothelial cells. These observations suggest that RORα may function as an amplifier of HIF1α-mediated responses by hypoxia. RORα was reported to stimulate hypoxia response element (HRE)-dependent transcriptional activation of a reporter. The enhancement in HIF1α-mediated transactivation by RORα was found to be related to increased stability of HIF-1α protein and to involve a direct interaction between HIF-1α and the DBD of RORα [Kim et al., 2008]. Several of the experiments in this study were based on activation of RORα by melatonin, a rather controversial putative ligand of RORα. Thus, additional studies are needed to understand the role of RORα and melatonin in hypoxia.
RORα1 and RORγ1 have also been implicated in other stress-related regulatory functions. RORα1 has been reported to provide protection to oxidative stress-induced apoptosis in cortical neurons by increasing the expression of antioxidant proteins glutathione peroxidase 1 (GPX1) and peroxiredoxin 6 (Prx6), thereby reducing the accumulation of stress-related reactive oxygen species (ROS) [Boukhtouche et al., 2006b]. Ectopic expression of RORγ in C2C12 cells has been reported to enhance UCP3 expression and decrease ROS production [Raichur et al., 2007]. Identification of genes regulated by RORα might shed light on its precise role in hypoxia- and stress-induced responses.
Summary
Recent studies of RORs have greatly widened our understanding of the physiological roles of this nuclear receptor subfamily and provided exciting clues about their critical functions in embryonic development, cellular differentiation and proliferation, immunity, cellular metabolism, and circadian rhythm. The rhythmic pattern of expression of RORs in certain tissues and the regulation of several components of the circadian clock and metabolic pathways by RORs are consistent with the emerging view that the controls of these processes are coupled. RORs function as a subcomponent of the circadian oscillator and integrate the control of the circadian clocks and the rhythmic pattern of expression of (metabolic) genes and, as such, regulate the cyclic nature of several physiological processes, including energy homeostasis, lipid and xenobiotic metabolism. The reduced susceptibility of RORα-deficient mice to hepatic steatosis and obesity suggests a role for RORα in energy homeostasis. The critical regulatory roles that RORs have in thymopoiesis, development of several secondary lymphoid tissues, and Th17 lineage specification are highly relevant to a variety of immune responses and inflammatory disorders, including autoimmune diseases and asthma. The greatly decreased susceptibility to allergic airway inflammation, experimental autoimmune encephalomyelitis, and colitis in ROR-deficient mice raises the possibility that RORs might serve as potential novel targets for chemotherapeutic strategies to intervene in these disease processes. The discovery that ROR activity can be modulated by ligands strongly supports this prospect. Hopefully, the next decade will see the discovery of ROR-specific, clinically-useful (ant)agonists.
Acknowledgments
The author would like to thank Drs. Chen Dong (M.D. Anderson Cancer Center), Andrew Liu (University of Memphis), Christina Teng, Donald Cook, Gary Zeruth, and Yukimasa Takeda (NIEHS) for their comments on the manuscript. This research was supported by the Intramural Research Program of the NIEHS, NIH (Z01-ES-101586).
Glossary
AF2
activation function 2
ApoA
apolipoprotein
ATRA
all-trans retinoic acid
ATXN1
ataxin 1
BMAL1
brain and muscle ARNT-like 1
CaMKIV
calmodulin-dependent kinase IV
ChIP
Chromatin immunoprecipitation
CRX
cone-rod homeobox factor
CRY
cryptochrome
CT
circadian time
Cyp7b1
oxysterol 7alpha-hydroxylase
DBD
DNA-binding domain
DBP
D site-binding protein
DHR3
Drosophila hormone receptor 3
DKO
double knockout
DP
double positive
EAE
experimental autoimmune encephalomyelitis
EGR
early growth response gene
FOXP3
forkhead box transcription factor p3
HDAC
histone deacetylase
HDL
high density lipoprotein
HIF1α
hypoxia-inducible factor α
IL
interleukin
IRF4
interferon regulatory factor 4
ISP
immature single positive
LBD
ligand binding domain
LPS
lipopolysaccharide
LTi
lymphoid tissue inducer cells
LXR
liver X receptor
NCOA
nuclear receptor coactivator
NCOR
nuclear receptor corepressor
NK
natural killer
Obfc2a
oligonucleotide/oligosaccharide-binding fold-containing 2a
Opn
opsin
OVA
ovalbumin
PER
period protein
PGC-1
peroxisome proliferator-activated receptors coactivator
PND
postnatal day
PPAR
peroxisome proliferators-activated receptor
PPs
Peyer’s patches
RAR
retinoic acid receptor
RIP140
receptor-interacting protein 140
ROR
RAR- or retinoid-related orphan receptor
RORE
ROR response element
RUNX1
runt-related transcription factor 1
RXR
retinoid X receptor
sg
staggerer
SCA1
spinocerebellar ataxia type 1
SCN
suprachiasmatic nucleus
Shh
Sonic hedgehog
SIRT1
sirtuin 1
SP
single positive
SRC
steroid receptor coactivator
SREBP1c
sterol regulatory element-binding protein 1, isoform c
STAT
signal transducer and activator of transcription
SULT
sulfotranferase
TGFβ
transforming growth factor β
Th
T helper
Thp
uncommitted (naïve) CD4+ T helper cells
TIP60
Tat-interactive protein, 60 kD
Treg
T regulatory
UBE2I
ubiquitin-conjugating enzyme I
WT
wild type
References
- Acosta-Rodriguez E. V., Napolitani G., Lanzavecchia A., Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-β are essential for the differentiation of interleukin 17-producing human T helper cells. Nat Immunol. 2007;8:942–9. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- Akashi M., Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–8. doi: 10.1038/nsmb925. [DOI] [PubMed] [Google Scholar]
- Akimzhanov A. M., Yang X. O., Dong C. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J Biol Chem. 2007;282:5969–72. doi: 10.1074/jbc.C600322200. [DOI] [PubMed] [Google Scholar]
- Akiyama T. E., Gonzalez F. J. Regulation of P450 genes by liver-enriched transcription factors and nuclear receptors. Biochim Biophys Acta. 2003;1619:223–34. doi: 10.1016/s0304-4165(02)00480-4. [DOI] [PubMed] [Google Scholar]
- Albrecht U. Invited review: regulation of mammalian circadian clock genes. J Appl Physiol. 2002;92:1348–55. doi: 10.1152/japplphysiol.00759.2001. [DOI] [PubMed] [Google Scholar]
- Andre E., Gawlas K., Becker-Andre M. A novel isoform of the orphan nuclear receptor RORbeta is specifically expressed in pineal gland and retina. Gene. 1998a;216:277–83. doi: 10.1016/s0378-1119(98)00348-5. [DOI] [PubMed] [Google Scholar]
- Andre E., Conquet F., Steinmayr M., Stratton S. C., Porciatti V., Becker-Andre M. Disruption of retinoid-related orphan receptor β changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. Embo J. 1998b;17:3867–77. doi: 10.1093/emboj/17.14.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda A., Pascual A. Nuclear hormone receptors and gene expression. Physiol Rev. 2001;81:1269–304. doi: 10.1152/physrev.2001.81.3.1269. [DOI] [PubMed] [Google Scholar]
- Asher G., Gatfield D., Stratmann M., Reinke H., Dibner C., Kreppel F., Mostoslavsky R., Alt F. W., Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–28. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- Atkins G. B., Hu X., Guenther M. G., Rachez C., Freedman L. P., Lazar M. A. Coactivators for the orphan nuclear receptor RORalpha. Mol Endocrinol. 1999;13:1550–7. doi: 10.1210/mend.13.9.0343. [DOI] [PubMed] [Google Scholar]
- Austin S., Medvedev A., Yan Z. H., Adachi H., Hirose T., Jetten A. M. Induction of the nuclear orphan receptor RORgamma during adipocyte differentiation of D1 and 3T3-L1 cells. Cell Growth Differ. 1998;9:267–76. [PubMed] [Google Scholar]
- Azadi S., Zhang Y., Caffe A. R., Holmqvist B., van Veen T. Thyroid-beta2 and the retinoid RAR-α, RXR-γ and ROR-beta2 receptor mRNAs; expression profiles in mouse retina, retinal explants and neocortex. Neuroreport. 2002;13:745–50. doi: 10.1097/00001756-200205070-00003. [DOI] [PubMed] [Google Scholar]
- Barnard A. R., Nolan P. M. When clocks go bad: neurobehavioural consequences of disrupted circadian timing. PLoS Genet. 2008;4:e1000040. doi: 10.1371/journal.pgen.1000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker-Andre M., Andre E., DeLamarter J. F. Identification of nuclear receptor mRNAs by RT-PCR amplification of conserved zinc-finger motif sequences. Biochem Biophys Res Commun. 1993;194:1371–9. doi: 10.1006/bbrc.1993.1976. [DOI] [PubMed] [Google Scholar]
- Belden W. J., Dunlap J. C. SIRT1 is a circadian deacetylase for core clock components. Cell. 2008;134:212–4. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson M. J., Pino-Lagos K., Rosemblatt M., Noelle R. J. All-trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co-stimulation. J Exp Med. 2007;204:1765–74. doi: 10.1084/jem.20070719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard S., Heymes C., Merval R., Rodriguez M., Galizzi J. P., Boutin J. A., Mariani J., Tedgui A. Expression and regulation of the nuclear receptor RORalpha in human vascular cells. FEBS Lett. 2002;511:36–40. doi: 10.1016/s0014-5793(01)03275-6. [DOI] [PubMed] [Google Scholar]
- Besnard S., Silvestre J. S., Duriez M., Bakouche J., Lemaigre-Dubreuil Y., Mariani J., Levy B. I., Tedgui A. Increased ischemia-induced angiogenesis in the staggerer mouse, a mutant of the nuclear receptor Roralpha. Circ Res. 2001;89:1209–15. doi: 10.1161/hh2401.101755. [DOI] [PubMed] [Google Scholar]
- Bettelli E., Korn T., Oukka M., Kuchroo V. K. Induction and effector functions of T(H)17 cells. Nature. 2008;453:1051–7. doi: 10.1038/nature07036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettelli E., Carrier Y., Gao W., Korn T., Strom T. B., Oukka M., Weiner H. L., Kuchroo V. K. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–8. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Bi Y., Liu G., Yang R. Th17 cell induction and immune regulatory effects. J Cell Physiol. 2007;211:273–8. doi: 10.1002/jcp.20973. [DOI] [PubMed] [Google Scholar]
- Bois-Joyeux B., Chauvet C., Nacer-Cherif H., Bergeret W., Mazure N., Giguere V., Laudet V., Danan J. L. Modulation of the far-upstream enhancer of the rat α-fetoprotein gene by members of the ROR α, Rev-erb α, and Rev-erb β groups of monomeric orphan nuclear receptors. DNA Cell Biol. 2000;19:589–99. doi: 10.1089/104454900750019344. [DOI] [PubMed] [Google Scholar]
- Boukhtouche F., Vodjdani G., Jarvis C. I., Bakouche J., Staels B., Mallet J., Mariani J., Lemaigre-Dubreuil Y., Brugg B. Human retinoic acid receptor-related orphan receptor alpha1 overexpression protects neurones against oxidative stress-induced apoptosis. J Neurochem. 2006b;96:1778–89. doi: 10.1111/j.1471-4159.2006.03708.x. [DOI] [PubMed] [Google Scholar]
- Boukhtouche F., Janmaat S., Vodjdani G., Gautheron V., Mallet J., Dusart I., Mariani J. Retinoid-related orphan receptor α controls the early steps of Purkinje cell dendritic differentiation. J Neurosci. 2006a;26:1531–8. doi: 10.1523/JNEUROSCI.4636-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhtouche F., Mariani J., Tedgui A. The "CholesteROR" protective pathway in the vascular system. Arterioscler Thromb Vasc Biol. 2004;24:637–43. doi: 10.1161/01.ATV.0000119355.56036.de. [DOI] [PubMed] [Google Scholar]
- Broussard D. R., Lozano M. M., Dudley J. P. Rorgamma (Rorc) is a common integration site in type B leukemogenic virus-induced T-cell lymphomas. J Virol. 2004;78:4943–6. doi: 10.1128/JVI.78.9.4943-4946.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow M. E., Jeffery E. W., Hjerrild K. A., Paeper B., Clark L. B., Yasayko S. A., Wilkinson J. E., Galas D., Ziegler S. F., Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- Brustle A., Heink S., Huber M., Rosenplanter C., Stadelmann C., Yu P., Arpaia E., Mak T. W., Kamradt T., Lohoff M. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat Immunol. 2007;8:958–66. doi: 10.1038/ni1500. [DOI] [PubMed] [Google Scholar]
- Burright E. N., Clark H. B., Servadio A., Matilla T., Feddersen R. M., Yunis W. S., Duvick L. A., Zoghbi H. Y., Orr H. T. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–48. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- Burris T. P. Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22:1509–20. doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cachon-Gonzalez M. B., Fenner S., Coffin J. M., Moran C., Best S., Stoye J. P. Structure and expression of the hairless gene of mice. Proc Natl Acad Sci U S A. 1994;91:7717–21. doi: 10.1073/pnas.91.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D. J., Ziegler S. F. FOXP3 modifies the phenotypic and functional properties of regulatory T cells. Nat Rev Immunol. 2007;7:305–10. doi: 10.1038/nri2061. [DOI] [PubMed] [Google Scholar]
- Carlberg C., Hooft van Huijsduijnen R., Staple J. K., DeLamarter J. F., Becker-Andre M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol Endocrinol. 1994;8:757–70. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- Carney G. E., Wade A. A., Sapra R., Goldstein E. S., Bender M. DHR3, an ecdysone-inducible early-late gene encoding a Drosophila nuclear receptor, is required for embryogenesis. Proc Natl Acad Sci U S A. 1997;94:12024–9. doi: 10.1073/pnas.94.22.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson B. D., Lopes J. E., Soper D. M., Ziegler S. F. Insights into transcriptional regulation by FOXP3. Front Biosci. 2006;11:1607–19. doi: 10.2741/1908. [DOI] [PubMed] [Google Scholar]
- Cha J. Y., Repa J. J. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J Biol Chem. 2007;282:743–51. doi: 10.1074/jbc.M605023200. [DOI] [PubMed] [Google Scholar]
- Chauvet C., Bois-Joyeux B., Danan J. L. Retinoic acid receptor-related orphan receptor (ROR) alpha4 is the predominant isoform of the nuclear receptor RORalpha in the liver and is up-regulated by hypoxia in HepG2 human hepatoma cells. Biochem J. 2002;364:449–56. doi: 10.1042/BJ20011558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvet C., Bois-Joyeux B., Berra E., Pouyssegur J., Danan J. L. The gene encoding human retinoic acid-receptor-related orphan receptor α is a target for hypoxia-inducible factor 1. Biochem J. 2004;384:79–85. doi: 10.1042/BJ20040709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Jin W., Hardegen N., Lei K. J., Li L., Marinos N., McGrady G., Wahl S. M. Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–86. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Coulter S., Jetten A. M., Goldstein J. A. Identification of Human CYP2C8 as a Retinoic Acid Receptor-Related Orphan Receptor (ROR) Target Gene. J Pharmacol Exp Ther. 2009:In Press. doi: 10.1124/jpet.108.148916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Laurence A., Kanno Y., Pacher-Zavisin M., Zhu B. M., Tato C., Yoshimura A., Hennighausen L., O'Shea J. J. Selective regulatory function of Socs3 in the formation of IL-17-secreting T cells. Proc Natl Acad Sci U S A. 2006;103:8137–42. doi: 10.1073/pnas.0600666103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Laurence A., O'Shea J. J. Signal transduction pathways and transcriptional regulation in the control of Th17 differentiation. Semin Immunol. 2007;19:400–8. doi: 10.1016/j.smim.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., O'Shea J. J. Th17 cells: a new fate for differentiating helper T cells. Immunol Res. 2008;41:87–102. doi: 10.1007/s12026-007-8014-9. [DOI] [PubMed] [Google Scholar]
- Coombes J. L., Siddiqui K. R., Arancibia-Carcamo C. V., Hall J., Sun C. M., Belkaid Y., Powrie F. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–64. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cupedo T., Mebius R. E. Cellular interactions in lymph node development. J Immunol. 2005;174:21–5. doi: 10.4049/jimmunol.174.1.21. [DOI] [PubMed] [Google Scholar]
- Cupedo T., Crellin N. K., Papazian N., Rombouts E. J., Weijer K., Grogan J. L., Fibbe W. E., Cornelissen J. J., Spits H. Human fetal lymphoid tissue-inducer cells are interleukin 17-producing precursors to RORC+ CD127+ natural killer-like cells. Nat Immunol. 2009;10:66–74. doi: 10.1038/ni.1668. [DOI] [PubMed] [Google Scholar]
- Cupedo T., Kraal G., Mebius R. E. The role of CD45+CD4+CD3- cells in lymphoid organ development. Immunol Rev. 2002;189:41–50. doi: 10.1034/j.1600-065x.2002.18905.x. [DOI] [PubMed] [Google Scholar]
- Dace A., Zhao L., Park K. S., Furuno T., Takamura N., Nakanishi M., West B. L., Hanover J. A., Cheng S. Hormone binding induces rapid proteasome-mediated degradation of thyroid hormone receptors. Proc Natl Acad Sci U S A. 2000;97:8985–90. doi: 10.1073/pnas.160257997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmane N., Ruiz i Altaba A. Sonic hedgehog regulates the growth and patterning of the cerebellum. Development. 1999;126:3089–100. doi: 10.1242/dev.126.14.3089. [DOI] [PubMed] [Google Scholar]
- Darimont B. D., Wagner R. L., Apriletti J. W., Stallcup M. R., Kushner P. J., Baxter J. D., Fletterick R. J., Yamamoto K. R. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–56. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne J. P., Weaver D. R., Reppert S. M. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007a;10:543–5. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBruyne J. P., Weaver D. R., Reppert S. M. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007b;17:R538–9. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- Delerive P., Monte D., Dubois G., Trottein F., Fruchart-Najib J., Mariani J., Fruchart J. C., Staels B. The orphan nuclear receptor ROR α is a negative regulator of the inflammatory response. EMBO Rep. 2001;2:42–8. doi: 10.1093/embo-reports/kve007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis A. P., Haq R. U., Nawaz Z. Importance of the regulation of nuclear receptor degradation. Front Biosci. 2001;6:D954–9. doi: 10.2741/dennis. [DOI] [PubMed] [Google Scholar]
- Desvergne B., Wahli W. Peroxisome proliferator-activated receptors: nuclear control of metabolism. Endocr Rev. 1999;20:649–88. doi: 10.1210/edrv.20.5.0380. [DOI] [PubMed] [Google Scholar]
- Desvergne B., Michalik L., Wahli W. Transcriptional regulation of metabolism. Physiol Rev. 2006;86:465–514. doi: 10.1152/physrev.00025.2005. [DOI] [PubMed] [Google Scholar]
- Doi M., Hirayama J., Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Dong C. TH17 cells in development: an updated view of their molecular identity and genetic programming. Nat Rev Immunol. 2008;8:337–48. doi: 10.1038/nri2295. [DOI] [PubMed] [Google Scholar]
- Doulazmi M., Frederic F., Capone F., Becker-Andre M., Delhaye-Bouchaud N., Mariani J. A comparative study of Purkinje cells in two RORalpha gene mutant mice: staggerer and RORalpha(-/-) Brain Res Dev Brain Res. 2001;127:165–74. doi: 10.1016/s0165-3806(01)00131-6. [DOI] [PubMed] [Google Scholar]
- Doulazmi M., Frederic F., Lemaigre-Dubreuil Y., Hadj-Sahraoui N., Delhaye-Bouchaud N., Mariani J. Cerebellar Purkinje cell loss during life span of the heterozygous staggerer mouse (Rora(+)/Rora(sg)) is gender-related. J Comp Neurol. 1999;411:267–73. [PubMed] [Google Scholar]
- Downes M., Burke L. J., Muscat G. E. Transcriptional repression by Rev-erbA α is dependent on the signature motif and helix 5 in the ligand binding domain: silencing does not involve an interaction with N-CoR. Nucleic Acids Res. 1996;24:3490–8. doi: 10.1093/nar/24.18.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duanmu Z., Weckle A., Koukouritaki S. B., Hines R. N., Falany J. L., Falany C. N., Kocarek T. A., Runge-Morris M. Developmental expression of aryl, estrogen, and hydroxysteroid sulfotransferases in pre- and postnatal human liver. J Pharmacol Exp Ther. 2006;316:1310–7. doi: 10.1124/jpet.105.093633. [DOI] [PubMed] [Google Scholar]
- Duez H., Staels B. The nuclear receptors Rev-erbs and RORs integrate circadian rhythms and metabolism. Diab Vasc Dis Res. 2008;5:82–8. doi: 10.3132/dvdr.2008.0014. [DOI] [PubMed] [Google Scholar]
- Du J., Huang C., Zhou B., Ziegler S. F. Isoform-specific inhibition of ROR α-mediated transcriptional activation by human FOXP3. J Immunol. 2008;180:4785–92. doi: 10.4049/jimmunol.180.7.4785. [DOI] [PubMed] [Google Scholar]
- Duplus E., Gras C., Soubeyre V., Vodjdani G., Lemaigre-Dubreuil Y., Brugg B. Phosphorylation and transcriptional activity regulation of retinoid-related orphan receptor α 1 by protein kinases C. J Neurochem. 2008;104:1321–32. doi: 10.1111/j.1471-4159.2007.05074.x. [DOI] [PubMed] [Google Scholar]
- Dussault I., Fawcett D., Matthyssen A., Bader J. A., Giguere V. Orphan nuclear receptor ROR α-deficient mice display the cerebellar defects of staggerer. Mech Dev. 1998;70:147–53. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- Dzhagalov I., Giguere V., He Y. W. Lymphocyte development and function in the absence of retinoic acid-related orphan receptor α. J Immunol. 2004;173:2952–9. doi: 10.4049/jimmunol.173.5.2952. [DOI] [PubMed] [Google Scholar]
- Eberl G., Marmon S., Sunshine M. J., Rennert P. D., Choi Y., Littman D. R. An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol. 2004a;5:64–73. doi: 10.1038/ni1022. [DOI] [PubMed] [Google Scholar]
- Eberl G. Inducible lymphoid tissues in the adult gut: recapitulation of a fetal developmental pathway? Nat Rev Immunol. 2005;5:413–20. doi: 10.1038/nri1600. [DOI] [PubMed] [Google Scholar]
- Eberl G., Littman D. R. The role of the nuclear hormone receptor RORgammat in the development of lymph nodes and Peyer's patches. Immunol Rev. 2003;195:81–90. doi: 10.1034/j.1600-065x.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- Eberl G., Littman D. R. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004b;305:248–51. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- Ebrey T., Koutalos Y. Vertebrate photoreceptors. Prog Retin Eye Res. 2001;20:49–94. doi: 10.1016/s1350-9462(00)00014-8. [DOI] [PubMed] [Google Scholar]
- Eide E. J., Woolf M. F., Kang H., Woolf P., Hurst W., Camacho F., Vielhaber E. L., Giovanni A., Virshup D. M. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias K. M., Laurence A., Davidson T. S., Stephens G., Kanno Y., Shevach E. M., O'Shea J. J. Retinoic acid inhibits Th17 polarization and enhances FoxP3 expression through a Stat-3/Stat-5 independent signaling pathway. Blood. 2008;111:1013–20. doi: 10.1182/blood-2007-06-096438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escriva H., Delaunay F., Laudet V. Ligand binding and nuclear receptor evolution. Bioessays. 2000;22:717–27. doi: 10.1002/1521-1878(200008)22:8<717::AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Evans R. M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–95. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang H. L., Strom S. C., Ellis E., Duanmu Z., Fu J., Duniec-Dmuchowski Z., Falany C. N., Falany J. L., Kocarek T. A., Runge-Morris M. Positive and negative regulation of human hepatic hydroxysteroid sulfotransferase (SULT2A1) gene transcription by rifampicin: roles of hepatocyte nuclear factor 4alpha and pregnane X receptor. J Pharmacol Exp Ther. 2007;323:586–98. doi: 10.1124/jpet.107.124610. [DOI] [PubMed] [Google Scholar]
- Fantini M. C., Becker C., Monteleone G., Pallone F., Galle P. R., Neurath M. F. Cutting edge: TGF-β induces a regulatory phenotype in CD4+CD25- T cells through Foxp3 induction and down-regulation of Smad7. J Immunol. 2004;172:5149–53. doi: 10.4049/jimmunol.172.9.5149. [DOI] [PubMed] [Google Scholar]
- Faus H., Haendler B. Post-translational modifications of steroid receptors. Biomed Pharmacother. 2006;60:520–8. doi: 10.1016/j.biopha.2006.07.082. [DOI] [PubMed] [Google Scholar]
- Finck B. N., Gropler M. C., Chen Z., Leone T. C., Croce M. A., Harris T. E., Lawrence J. C., Jr., Kelly D. P. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Flores M. V., Hall C., Jury A., Crosier K., Crosier P. The zebrafish retinoid-related orphan receptor (ror) gene family. Gene Expr Patterns. 2007;7:535–43. doi: 10.1016/j.modgep.2007.02.001. [DOI] [PubMed] [Google Scholar]
- Fontenot J. D., Rasmussen J. P., Williams L. M., Dooley J. L., Farr A. G., Rudensky A. Y. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–41. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Forman B. M., Chen J., Blumberg B., Kliewer S. A., Henshaw R., Ong E. S., Evans R. M. Cross-talk among ROR α 1 and the Rev-erb family of orphan nuclear receptors. Mol Endocrinol. 1994;8:1253–61. doi: 10.1210/mend.8.9.7838158. [DOI] [PubMed] [Google Scholar]
- Fujieda H., Bremner R., Mears A. J., Sasaki H. Retinoic acid receptor-related orphan receptor α regulates a subset of cone genes during mouse retinal development. J Neurochem. 2009;108:91–101. doi: 10.1111/j.1471-4159.2008.05739.x. [DOI] [PubMed] [Google Scholar]
- Furuzawa-Carballeda J., Vargas-Rojas M. I., Cabral A. R. Autoimmune inflammation from the Th17 perspective. Autoimmun Rev. 2007;6:169–75. doi: 10.1016/j.autrev.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Gachon F., Nagoshi E., Brown S. A., Ripperger J., Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113:103–12. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- Gallego M., Virshup D. M. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–48. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Gates J., Lam G., Ortiz J. A., Losson R., Thummel C. S. rigor mortis encodes a novel nuclear receptor interacting protein required for ecdysone signaling during Drosophila larval development. Development. 2004;131:25–36. doi: 10.1242/dev.00920. [DOI] [PubMed] [Google Scholar]
- Genoux A., Dehondt H., Helleboid-Chapman A., Duhem C., Hum D. W., Martin G., Pennacchio L. A., Staels B., Fruchart-Najib J., Fruchart J. C. Transcriptional regulation of apolipoprotein A5 gene expression by the nuclear receptor RORalpha. Arterioscler Thromb Vasc Biol. 2005;25:1186–92. doi: 10.1161/01.ATV.0000163841.85333.83. [DOI] [PubMed] [Google Scholar]
- Gery S., Koeffler H. P. The role of circadian regulation in cancer. Cold Spring Harb Symp Quant Biol. 2007;72:459–64. doi: 10.1101/sqb.2007.72.004. [DOI] [PubMed] [Google Scholar]
- Giguere V., Tini M., Flock G., Ong E., Evans R. M., Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR α, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–53. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- Giguere V. Orphan nuclear receptors: from gene to function. Endocr Rev. 1999;20:689–725. doi: 10.1210/edrv.20.5.0378. [DOI] [PubMed] [Google Scholar]
- Giguere V., Beatty B., Squire J., Copeland N. G., Jenkins N. A. The orphan nuclear receptor ROR α (RORA) maps to a conserved region of homology on human chromosome 15q21-q22 and mouse chromosome 9. Genomics. 1995;28:596–8. doi: 10.1006/geno.1995.1197. [DOI] [PubMed] [Google Scholar]
- Glass C. K., Rosenfeld M. G. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–41. [PubMed] [Google Scholar]
- Goldowitz D., Hamre K. The cells and molecules that make a cerebellum. Trends Neurosci. 1998;21:375–82. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- Gold D. A., Gent P. M., Hamilton B. A. ROR α in genetic control of cerebellum development: 50 staggering years. Brain Res. 2007;1140:19–25. doi: 10.1016/j.brainres.2005.11.080. [DOI] [PubMed] [Google Scholar]
- Gold D. A., Baek S. H., Schork N. J., Rose D. W., Larsen D. D., Sachs B. D., Rosenfeld M. G., Hamilton B. A. RORalpha coordinates reciprocal signaling in cerebellar development through sonic hedgehog and calcium-dependent pathways. Neuron. 2003;40:1119–31. doi: 10.1016/s0896-6273(03)00769-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H., Guo P., Zhai Y., Zhou J., Uppal H., Jarzynka M. J., Song W. C., Cheng S. Y., Xie W. Estrogen deprivation and inhibition of breast cancer growth in vivo through activation of the orphan nuclear receptor liver X receptor. Mol Endocrinol. 2007;21:1781–90. doi: 10.1210/me.2007-0187. [DOI] [PubMed] [Google Scholar]
- Grechez-Cassiau A., Rayet B., Guillaumond F., Teboul M., Delaunay F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J Biol Chem. 2008;283:4535–42. doi: 10.1074/jbc.M705576200. [DOI] [PubMed] [Google Scholar]
- Greiner E. F., Kirfel J., Greschik H., Huang D., Becker P., Kapfhammer J. P., Schule R. Differential ligand-dependent protein-protein interactions between nuclear receptors and a neuronal-specific cofactor. Proc Natl Acad Sci U S A. 2000;97:7160–5. doi: 10.1073/pnas.97.13.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner E. F., Kirfel J., Greschik H., Dorflinger U., Becker P., Mercep A., Schule R. Functional analysis of retinoid Z receptor β, a brain-specific nuclear orphan receptor. Proc Natl Acad Sci U S A. 1996;93:10105–10. doi: 10.1073/pnas.93.19.10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastavino J. M., Bertin R., Portet R. Effects of the rearing temperature on the temporal feeding pattern of the staggerer mutant mouse. Physiol Behav. 1991;49:405–9. doi: 10.1016/0031-9384(91)90064-u. [DOI] [PubMed] [Google Scholar]
- Guillaumond F., Dardente H., Giguere V., Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- Gu Y., Yang J., Ouyang X., Liu W., Li H., Yang J., Bromberg J., Chen S. H., Mayer L., Unkeless J. C., Xiong H. Interleukin 10 suppresses Th17 cytokines secreted by macrophages and T cells. Eur J Immunol. 2008;38:1807–13. doi: 10.1002/eji.200838331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Hawwari A., Li H., Sun Z., Mahanta S. K., Littman D. R., Krangel M. S., He Y. W. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol. 2002;3:469–76. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- Hadj-Sahraoui N., Frederic F., Zanjani H., Delhaye-Bouchaud N., Herrup K., Mariani J. Progressive atrophy of cerebellar Purkinje cell dendrites during aging of the heterozygous staggerer mouse (Rora(+/sg)) Brain Res Dev Brain Res. 2001;126:201–9. doi: 10.1016/s0165-3806(01)00095-5. [DOI] [PubMed] [Google Scholar]
- Hadj-Sahraoui N., Frederic F., Zanjani H., Herrup K., Delhaye-Bouchaud N., Mariani J. Purkinje cell loss in heterozygous staggerer mutant mice during aging. Brain Res Dev Brain Res. 1997;98:1–8. doi: 10.1016/s0165-3806(96)00153-8. [DOI] [PubMed] [Google Scholar]
- Hamilton B. A., Frankel W. N., Kerrebrock A. W., Hawkins T. L., FitzHugh W., Kusumi K., Russell L. B., Mueller K. L., van Berkel V., Birren B. W., Kruglyak L., Lander E. S. Disruption of the nuclear hormone receptor RORalpha in staggerer mice. Nature. 1996;379:736–9. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- Harding H. P., Atkins G. B., Jaffe A. B., Seo W. J., Lazar M. A. Transcriptional activation and repression by RORalpha, an orphan nuclear receptor required for cerebellar development. Mol Endocrinol. 1997;11:1737–46. doi: 10.1210/mend.11.11.0002. [DOI] [PubMed] [Google Scholar]
- Harmsen A., Kusser K., Hartson L., Tighe M., Sunshine M. J., Sedgwick J. D., Choi Y., Littman D. R., Randall T. D. Cutting edge: organogenesis of nasal-associated lymphoid tissue (NALT) occurs independently of lymphotoxin-α (LT α) and retinoic acid receptor-related orphan receptor-γ, but the organization of NALT is LT α dependent. J Immunol. 2002;168:986–90. doi: 10.4049/jimmunol.168.3.986. [DOI] [PubMed] [Google Scholar]
- Harrington L. E., Mangan P. R., Weaver C. T. Expanding the effector CD4 T-cell repertoire: the Th17 lineage. Curr Opin Immunol. 2006;18:349–56. doi: 10.1016/j.coi.2006.03.017. [DOI] [PubMed] [Google Scholar]
- Harris J. M., Lau P., Chen S. L., Muscat G. E. Characterization of the retinoid orphan-related receptor-α coactivator binding interface: a structural basis for ligand-independent transcription. Mol Endocrinol. 2002;16:998–1012. doi: 10.1210/mend.16.5.0837. [DOI] [PubMed] [Google Scholar]
- Harris T. J., Grosso J. F., Yen H. R., Xin H., Kortylewski M., Albesiano E., Hipkiss E. L., Getnet D., Goldberg M. V., Maris C. H., Housseau F., Yu H., Pardoll D. M., Drake C. G. Cutting edge: An in vivo requirement for STAT3 signaling in TH17 development and TH17-dependent autoimmunity. J Immunol. 2007;179:4313–7. doi: 10.4049/jimmunol.179.7.4313. [DOI] [PubMed] [Google Scholar]
- Hastings M., O'Neill J. S., Maywood E. S. Circadian clocks: regulators of endocrine and metabolic rhythms. J Endocrinol. 2007;195:187–98. doi: 10.1677/JOE-07-0378. [DOI] [PubMed] [Google Scholar]
- He Y. W., Beers C., Deftos M. L., Ojala E. W., Forbush K. A., Bevan M. J. Down-regulation of the orphan nuclear receptor ROR γ t is essential for T lymphocyte maturation. J Immunol. 2000b;164:5668–74. doi: 10.4049/jimmunol.164.11.5668. [DOI] [PubMed] [Google Scholar]
- Heery D. M., Kalkhoven E., Hoare S., Parker M. G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–6. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Heery D. M., Hoare S., Hussain S., Parker M. G., Sheppard H. Core LXXLL motif sequences in CREB-binding protein, SRC1, and RIP140 define affinity and selectivity for steroid and retinoid receptors. J Biol Chem. 2001;276:6695–702. doi: 10.1074/jbc.M009404200. [DOI] [PubMed] [Google Scholar]
- He Y. W. Orphan nuclear receptors in T lymphocyte development. J Leukoc Biol. 2002;72:440–6. [PubMed] [Google Scholar]
- He Y. W., Deftos M. L., Ojala E. W., Bevan M. J. RORgamma t, a novel isoform of an orphan receptor, negatively regulates Fas ligand expression and IL-2 production in T cells. Immunity. 1998;9:797–806. doi: 10.1016/s1074-7613(00)80645-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K., Mullen R. J. Role of the Staggerer gene in determining Purkinje cell number in the cerebellar cortex of mouse chimeras. Brain Res. 1981;227:475–85. doi: 10.1016/0165-3806(81)90002-x. [DOI] [PubMed] [Google Scholar]
- Herrup K., Mullen R. J. Staggerer chimeras: intrinsic nature of Purkinje cell defects and implications for normal cerebellar development. Brain Res. 1979;178:443–57. doi: 10.1016/0006-8993(79)90705-4. [DOI] [PubMed] [Google Scholar]
- He Y. W. The role of orphan nuclear receptor in thymocyte differentiation and lymphoid organ development. Immunol Res. 2000a;22:71–82. doi: 10.1385/IR:22:2-3:71. [DOI] [PubMed] [Google Scholar]
- Hill J. A., Hall J. A., Sun C. M., Cai Q., Ghyselinck N., Chambon P., Belkaid Y., Mathis D., Benoist C. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity. 2008;29:758–70. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama J., Sahar S., Grimaldi B., Tamaru T., Takamatsu K., Nakahata Y., Sassone-Corsi P. CLOCK-mediated acetylation of BMAL1 controls circadian function. Nature. 2007;450:1086–90. doi: 10.1038/nature06394. [DOI] [PubMed] [Google Scholar]
- Hirose T., Smith R. J., Jetten A. M. ROR γ: the third member of ROR/RZR orphan receptor subfamily that is highly expressed in skeletal muscle. Biochem Biophys Res Commun. 1994;205:1976–83. doi: 10.1006/bbrc.1994.2902. [DOI] [PubMed] [Google Scholar]
- Hiruma K., Riddiford L. M. Differential control of MHR3 promoter activity by isoforms of the ecdysone receptor and inhibitory effects of E75A and MHR3. Dev Biol. 2004;272:510–21. doi: 10.1016/j.ydbio.2004.04.028. [DOI] [PubMed] [Google Scholar]
- Horard B., Rayet B., Triqueneaux G., Laudet V., Delaunay F., Vanacker J. M. Expression of the orphan nuclear receptor ERRalpha is under circadian regulation in estrogen-responsive tissues. J Mol Endocrinol. 2004;33:87–97. doi: 10.1677/jme.0.0330087. [DOI] [PubMed] [Google Scholar]
- Horlein A. J., Naar A. M., Heinzel T., Torchia J., Gloss B., Kurokawa R., Ryan A., Kamei Y., Soderstrom M., Glass C. K. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Horner M. A., Chen T., Thummel C. S. Ecdysteroid regulation and DNA binding properties of Drosophila nuclear hormone receptor superfamily members. Dev Biol. 1995;168:490–502. doi: 10.1006/dbio.1995.1097. [DOI] [PubMed] [Google Scholar]
- Huang Z., Xie H., Wang R., Sun Z. Retinoid-related orphan receptor γ t is a potential therapeutic target for controlling inflammatory autoimmunity. Expert Opin Ther Targets. 2007;11:737–43. doi: 10.1517/14728222.11.6.737. [DOI] [PubMed] [Google Scholar]
- Huber M., Brustle A., Reinhard K., Guralnik A., Walter G., Mahiny A., von Low E., Lohoff M. IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proc Natl Acad Sci U S A. 2008;105:20846–51. doi: 10.1073/pnas.0809077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Lazar M. A. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol Metab. 2000;11:6–10. doi: 10.1016/s1043-2760(99)00215-5. [DOI] [PubMed] [Google Scholar]
- Ichiyama K., Yoshida H., Wakabayashi Y., Chinen T., Saeki K., Nakaya M., Takaesu G., Hori S., Yoshimura A., Kobayashi T. Foxp3 inhibits RORgammat-mediated IL-17A mRNA transcription through direct interaction with RORgammat. J Biol Chem. 2008;283:17003–8. doi: 10.1074/jbc.M801286200. [DOI] [PubMed] [Google Scholar]
- Ino H. Immunohistochemical characterization of the orphan nuclear receptor ROR α in the mouse nervous system. J Histochem Cytochem. 2004;52:311–23. doi: 10.1177/002215540405200302. [DOI] [PubMed] [Google Scholar]
- Ishida N. Circadian clock, cancer and lipid metabolism. Neurosci Res. 2007;57:483–90. doi: 10.1016/j.neures.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Ismail A., Nawaz Z. Nuclear hormone receptor degradation and gene transcription: an update. IUBMB Life. 2005;57:483–90. doi: 10.1080/15216540500147163. [DOI] [PubMed] [Google Scholar]
- Isojima Y., Okumura N., Nagai K. Molecular mechanism of mammalian circadian clock. J Biochem. 2003;134:777–84. doi: 10.1093/jb/mvg219. [DOI] [PubMed] [Google Scholar]
- Ivanov, II, McKenzie B. S., Zhou L., Tadokoro C. E., Lepelley A., Lafaille J. J., Cua D. J., Littman D. R. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–33. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- Ivanov, II, Zhou L., Littman D. R. Transcriptional regulation of Th17 cell differentiation. Semin Immunol. 2007;19:409–17. doi: 10.1016/j.smim.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M., Hirakiyama A., Eshima Y., Kagechika H., Kato C., Song S. Y. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 2004;21:527–38. doi: 10.1016/j.immuni.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Iwata M., Eshima Y., Kagechika H. Retinoic acids exert direct effects on T cells to suppress Th1 development and enhance Th2 development via retinoic acid receptors. Int Immunol. 2003;15:1017–25. doi: 10.1093/intimm/dxg101. [DOI] [PubMed] [Google Scholar]
- Jakel H., Nowak M., Helleboid-Chapman A., Fruchart-Najib J., Fruchart J. C. Is apolipoprotein A5 a novel regulator of triglyceride-rich lipoproteins? Ann Med. 2006;38:2–10. doi: 10.1080/07853890500407488. [DOI] [PubMed] [Google Scholar]
- Janmaat S., Frederic F., Sjollema K., Luiten P., Mariani J., van der Want J. Formation and maturation of parallel fiber-Purkinje cell synapses in the Staggerer cerebellum ex vivo. J Comp Neurol. 2009;512:467–77. doi: 10.1002/cne.21910. [DOI] [PubMed] [Google Scholar]
- Jaradat M., Stapleton C., Tilley S. L., Dixon D., Erikson C. J., McCaskill J. G., Kang H. S., Angers M., Liao G., Collins J., Grissom S., Jetten A. M. Modulatory role for retinoid-related orphan receptor α in allergen-induced lung inflammation. Am J Respir Crit Care Med. 2006;174:1299–309. doi: 10.1164/rccm.200510-1672OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten A. M. Recent advances in the mechanisms of action and physiological functions of the retinoid-related orphan receptors (RORs) Curr Drug Targets Inflamm Allergy. 2004;3:395–412. doi: 10.2174/1568010042634497. [DOI] [PubMed] [Google Scholar]
- Jetten A. M., Ueda E. Retinoid-related orphan receptors (RORs): roles in cell survival, differentiation and disease. Cell Death Differ. 2002;9:1167–71. doi: 10.1038/sj.cdd.4401085. [DOI] [PubMed] [Google Scholar]
- Jetten A. M., Joo J. H. Retinoid-related Orphan Receptors (RORs): Roles in Cellular Differentiation and Development. Adv Dev Biol. 2006;16:313–355. doi: 10.1016/S1574-3349(06)16010-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jetten A. M., Kurebayashi S., Ueda E. The ROR nuclear orphan receptor subfamily: critical regulators of multiple biological processes. Prog Nucleic Acid Res Mol Biol. 2001;69:205–47. doi: 10.1016/s0079-6603(01)69048-2. [DOI] [PubMed] [Google Scholar]
- Johnson D. R., Lovett J. M., Hirsch M., Xia F., Chen J. D. NuRD complex component Mi-2beta binds to and represses RORgamma-mediated transcriptional activation. Biochem Biophys Res Commun. 2004;318:714–8. doi: 10.1016/j.bbrc.2004.04.087. [DOI] [PubMed] [Google Scholar]
- Joo J. H., Liao G., Collins J. B., Grissom S. F., Jetten A. M. Farnesol-induced apoptosis in human lung carcinoma cells is coupled to the endoplasmic reticulum stress response. Cancer Res. 2007;67:7929–36. doi: 10.1158/0008-5472.CAN-07-0931. [DOI] [PubMed] [Google Scholar]
- Kalaany N. Y., Mangelsdorf D. J. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–91. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- Kallen J., Schlaeppi J. M., Bitsch F., Delhon I., Fournier B. Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem. 2004;279:14033–8. doi: 10.1074/jbc.M400302200. [DOI] [PubMed] [Google Scholar]
- Kallen J. A., Schlaeppi J. M., Bitsch F., Geisse S., Geiser M., Delhon I., Fournier B. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure. 2002;10:1697–707. doi: 10.1016/s0969-2126(02)00912-7. [DOI] [PubMed] [Google Scholar]
- Kamphuis W., Cailotto C., Dijk F., Bergen A., Buijs R. M. Circadian expression of clock genes and clock-controlled genes in the rat retina. Biochem Biophys Res Commun. 2005;330:18–26. doi: 10.1016/j.bbrc.2005.02.118. [DOI] [PubMed] [Google Scholar]
- Kane C. D., Means A. R. Activation of orphan receptor-mediated transcription by Ca(2+)/calmodulin-dependent protein kinase IV. Embo J. 2000;19:691–701. doi: 10.1093/emboj/19.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H. S., Angers M., Beak J. Y., Wu X., Gimble J. M., Wada T., Xie W., Collins J. B., Grissom S. F., Jetten A. M. Gene expression profiling reveals a regulatory role for ROR α and ROR γ in phase I and phase II metabolism. Physiol Genomics. 2007;31:281–94. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- Kang H. S., Beak J. Y., Kim Y. S., Petrovich R. M., Collins J. B., Grissom S. F., Jetten A. M. NABP1, a novel RORgamma-regulated gene encoding a single-stranded nucleic-acid-binding protein. Biochem J. 2006;397:89–99. doi: 10.1042/BJ20051781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney A. M., Cole M. D., Rowitch D. H. Nmyc upregulation by sonic hedgehog signaling promotes proliferation in developing cerebellar granule neuron precursors. Development. 2003;130:15–28. doi: 10.1242/dev.00182. [DOI] [PubMed] [Google Scholar]
- Kim E. J., Yoo Y. G., Yang W. K., Lim Y. S., Na T. Y., Lee I. K., Lee M. O. Transcriptional activation of HIF-1 by RORalpha and its role in hypoxia signaling. Arterioscler Thromb Vasc Biol. 2008;28:1796–802. doi: 10.1161/ATVBAHA.108.171546. [DOI] [PubMed] [Google Scholar]
- Kinyamu H. K., Chen J., Archer T. K. Linking the ubiquitin-proteasome pathway to chromatin remodeling/modification by nuclear receptors. J Mol Endocrinol. 2005;34:281–97. doi: 10.1677/jme.1.01680. [DOI] [PubMed] [Google Scholar]
- Kitada H., Miyata M., Nakamura T., Tozawa A., Honma W., Shimada M., Nagata K., Sinal C. J., Guo G. L., Gonzalez F. J., Yamazoe Y. Protective role of hydroxysteroid sulfotransferase in lithocholic acid-induced liver toxicity. J Biol Chem. 2003;278:17838–44. doi: 10.1074/jbc.M210634200. [DOI] [PubMed] [Google Scholar]
- Kliewer S. A. The nuclear pregnane X receptor regulates xenobiotic detoxification. J Nutr. 2003;133:2444S–2447S. doi: 10.1093/jn/133.7.2444S. [DOI] [PubMed] [Google Scholar]
- Koelle M. R., Segraves W. A., Hogness D. S. DHR3: a Drosophila steroid receptor homolog. Proc Natl Acad Sci U S A. 1992;89:6167–71. doi: 10.1073/pnas.89.13.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koibuchi N., Chin W. W. ROR α gene expression in the perinatal rat cerebellum: ontogeny and thyroid hormone regulation. Endocrinology. 1998;139:2335–41. doi: 10.1210/endo.139.5.6013. [DOI] [PubMed] [Google Scholar]
- Ko C. H., Takahashi J. S. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15 Spec No 2:R271–7. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Kopmels B., Mariani J., Delhaye-Bouchaud N., Audibert F., Fradelizi D., Wollman E. E. Evidence for a hyperexcitability state of staggerer mutant mice macrophages. J Neurochem. 1992;58:192–9. doi: 10.1111/j.1471-4159.1992.tb09295.x. [DOI] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Gao W., Awasthi A., Jager A., Strom T. B., Oukka M., Kuchroo V. K. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 2007a;448:484–7. doi: 10.1038/nature05970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Oukka M., Kuchroo V., Bettelli E. Th17 cells: effector T cells with inflammatory properties. Semin Immunol. 2007b;19:362–71. doi: 10.1016/j.smim.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostrouch Z., Kostrouchova M., Rall J. E. Steroid/thyroid hormone receptor genes in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:156–9. doi: 10.1073/pnas.92.1.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumaki Y., Ukai-Tadenuma M., Uno K. D., Nishio J., Masumoto K. H., Nagano M., Komori T., Shigeyoshi Y., Hogenesch J. B., Ueda H. R. Analysis and synthesis of high-amplitude Cis-elements in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2008;105:14946–51. doi: 10.1073/pnas.0802636105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Thompson E. B. The structure of the nuclear hormone receptors. Steroids. 1999;64:310–9. doi: 10.1016/s0039-128x(99)00014-8. [DOI] [PubMed] [Google Scholar]
- Kurebayashi S., Ueda E., Sakaue M., Patel D. D., Medvedev A., Zhang F., Jetten A. M. Retinoid-related orphan receptor γ (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci U S A. 2000;97:10132–7. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurebayashi S., Nakajima T., Kim S. C., Chang C. Y., McDonnell D. P., Renaud J. P., Jetten A. M. Selective LXXLL peptides antagonize transcriptional activation by the retinoid-related orphan receptor RORgamma. Biochem Biophys Res Commun. 2004;315:919–27. doi: 10.1016/j.bbrc.2004.01.131. [DOI] [PubMed] [Google Scholar]
- Lan Q., Hiruma K., Hu X., Jindra M., Riddiford L. M. Activation of a delayed-early gene encoding MHR3 by the ecdysone receptor heterodimer EcR-B1-USP-1 but not by EcR-B1-USP-2. Mol Cell Biol. 1999;19:4897–906. doi: 10.1128/mcb.19.7.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landis D. M., Sidman R. L. Electron microscopic analysis of postnatal histogenesis in the cerebellar cortex of staggerer mutant mice. J Comp Neurol. 1978;179:831–63. doi: 10.1002/cne.901790408. [DOI] [PubMed] [Google Scholar]
- Landis D. M., Reese T. S. Structure of the Purkinje cell membrane in staggerer and weaver mutant mice. J Comp Neurol. 1977;171:247–60. doi: 10.1002/cne.901710208. [DOI] [PubMed] [Google Scholar]
- Langelan R. E., Fisher J. E., Hiruma K., Palli S. R., Riddiford L. M. Patterns of MHR3 expression in the epidermis during a larval molt of the tobacco hornworm Manduca sexta. Dev Biol. 2000;227:481–94. doi: 10.1006/dbio.2000.9895. [DOI] [PubMed] [Google Scholar]
- Lan Q., Wu Z., Riddiford L. M. Regulation of the ecdysone receptor, USP, E75 and MHR3 mRNAs by 20-hydroxyecdysone in the GV1 cell line of the tobacco hornworm, Manduca sexta. Insect Mol Biol. 1997;6:3–10. doi: 10.1046/j.1365-2583.1997.00151.x. [DOI] [PubMed] [Google Scholar]
- Lau P., Bailey P., Dowhan D. H., Muscat G. E. Exogenous expression of a dominant negative RORalpha1 vector in muscle cells impairs differentiation: RORalpha1 directly interacts with p300 and myoD. Nucleic Acids Res. 1999;27:411–20. doi: 10.1093/nar/27.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurence A., Tato C. M., Davidson T. S., Kanno Y., Chen Z., Yao Z., Blank R. B., Meylan F., Siegel R., Hennighausen L., Shevach E. M., O'Shea J J. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007b;26:371–81. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Laurence A., O'Shea J. J. T(H)-17 differentiation: of mice and men. Nat Immunol. 2007a;8:903–5. doi: 10.1038/ni0907-903. [DOI] [PubMed] [Google Scholar]
- Lau P., Nixon S. J., Parton R. G., Muscat G. E. RORalpha regulates the expression of genes involved in lipid homeostasis in skeletal muscle cells: caveolin-3 and CPT-1 are direct targets of ROR. J Biol Chem. 2004;279:36828–40. doi: 10.1074/jbc.M404927200. [DOI] [PubMed] [Google Scholar]
- Lau P., Fitzsimmons R. L., Raichur S., Wang S. C., Lechtken A., Muscat G. E. The orphan nuclear receptor, RORalpha, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem. 2008;283:18411–21. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- Lechtken A., Hornig M., Werz O., Corvey N., Zundorf I., Dingermann T., Brandes R., Steinhilber D. Extracellular signal-regulated kinase-2 phosphorylates RORalpha4 in vitro. Biochem Biophys Res Commun. 2007;358:890–6. doi: 10.1016/j.bbrc.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Leppkes M., Becker C., Ivanov II, Hirth S., Wirtz S., Neufert C., Pouly S., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Becher B., Littman D. R., Neurath M. F. RORgamma-expressing Th17 cells induce murine chronic intestinal inflammation via redundant effects of IL-17A and IL-17F. Gastroenterology. 2009;136:257–67. doi: 10.1053/j.gastro.2008.10.018. [DOI] [PubMed] [Google Scholar]
- Li B., Samanta A., Song X., Iacono K. T., Bembas K., Tao R., Basu S., Riley J. L., Hancock W. W., Shen Y., Saouaf S. J., Greene M. I. FOXP3 interactions with histone acetyltransferase and class II histone deacetylases are required for repression. Proc Natl Acad Sci U S A. 2007;104:4571–6. doi: 10.1073/pnas.0700298104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind U., Nilsson T., McPheat J., Stromstedt P. E., Bamberg K., Balendran C., Kang D. Identification of the human ApoAV gene as a novel RORalpha target gene. Biochem Biophys Res Commun. 2005;330:233–41. doi: 10.1016/j.bbrc.2005.02.151. [DOI] [PubMed] [Google Scholar]
- Lipp M., Muller G. Lymphoid organogenesis: getting the green light from RORgamma(t) Nat Immunol. 2004;5:12–4. doi: 10.1038/ni0104-12. [DOI] [PubMed] [Google Scholar]
- Littman D. R., Sun Z., Unutmaz D., Sunshine M. J., Petrie H. T., Zou Y. R. Role of the nuclear hormone receptor ROR γ in transcriptional regulation, thymocyte survival, and lymphoid organogenesis. Cold Spring Harb Symp Quant Biol. 1999;64:373–81. doi: 10.1101/sqb.1999.64.373. [DOI] [PubMed] [Google Scholar]
- Liu A. C., Lewis W. G., Kay S. A. Mammalian circadian signaling networks and therapeutic targets. Nat Chem Biol. 2007b;3:630–9. doi: 10.1038/nchembio.2007.37. [DOI] [PubMed] [Google Scholar]
- Liu A. C., Tran H. G., Zhang E. E., Priest A. A., Welsh D. K., Kay S. A. Redundant function of REV-ERBalpha and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Li S., Liu T., Borjigin J., Lin J. D. Transcriptional coactivator PGC-1alpha integrates the mammalian clock and energy metabolism. Nature. 2007a;447:477–81. doi: 10.1038/nature05767. [DOI] [PubMed] [Google Scholar]
- Lochner M., Peduto L., Cherrier M., Sawa S., Langa F., Varona R., Riethmacher D., Si-Tahar M., Di Santo J. P., Eberl G. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J Exp Med. 2008;205:1381–93. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes J. E., Torgerson T. R., Schubert L. A., Anover S. D., Ocheltree E. L., Ochs H. D., Ziegler S. F. Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J Immunol. 2006;177:3133–42. doi: 10.4049/jimmunol.177.5.3133. [DOI] [PubMed] [Google Scholar]
- Luci C., Reynders A., Ivanov II, Cognet C., Chiche L., Chasson L., Hardwigsen J., Anguiano E., Banchereau J., Chaussabel D., Dalod M., Littman D. R., Vivier E., Tomasello E. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009;10:75–82. doi: 10.1038/ni.1681. [DOI] [PubMed] [Google Scholar]
- Lu Y., Yi Y., Liu P., Wen W., James M., Wang D., You M. Common human cancer genes discovered by integrated gene-expression analysis. PLoS ONE. 2007;2:e1149. doi: 10.1371/journal.pone.0001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A., Pena J. C., Chang B., Margosian E., Davidson L., Alt F. W., Thompson C. B. Bclx regulates the survival of double-positive thymocytes. Proc Natl Acad Sci U S A. 1995;92:4763–7. doi: 10.1073/pnas.92.11.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglich J. M., Watson J., McMillen P. J., Goodwin B., Willson T. M., Moore J. T. The nuclear receptor CAR is a regulator of thyroid hormone metabolism during caloric restriction. J Biol Chem. 2004;279:19832–8. doi: 10.1074/jbc.M313601200. [DOI] [PubMed] [Google Scholar]
- Mamontova A., Seguret-Mace S., Esposito B., Chaniale C., Bouly M., Delhaye-Bouchaud N., Luc G., Staels B., Duverger N., Mariani J., Tedgui A. Severe atherosclerosis and hypoalphalipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORalpha. Circulation. 1998;98:2738–43. doi: 10.1161/01.cir.98.24.2738. [DOI] [PubMed] [Google Scholar]
- Manel N., Unutmaz D., Littman D. R. The differentiation of human T(H)-17 cells requires transforming growth factor-β and induction of the nuclear receptor RORgammat. Nat Immunol. 2008;9:641–9. doi: 10.1038/ni.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani J. Extent of multiple innervation of Purkinje cells by climbing fibers in the olivocerebellar system of weaver, reeler, and staggerer mutant mice. J Neurobiol. 1982;13:119–26. doi: 10.1002/neu.480130204. [DOI] [PubMed] [Google Scholar]
- Mariani J., Changeux J. P. Multiple innervation of Purkinje cells by climbing fibers in the cerebellum of the adult staggerer mutant mouse. J Neurobiol. 1980;11:41–50. doi: 10.1002/neu.480110106. [DOI] [PubMed] [Google Scholar]
- Masana M. I., Sumaya I. C., Becker-Andre M., Dubocovich M. L. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORbeta knockout. Am J Physiol Regul Integr Comp Physiol. 2007;292:R2357–67. doi: 10.1152/ajpregu.00687.2006. [DOI] [PubMed] [Google Scholar]
- Matsui T., Sashihara S., Oh Y., Waxman S. G. An orphan nuclear receptor, mROR α, and its spatial expression in adult mouse brain. Brain Res Mol Brain Res. 1995;33:217–26. doi: 10.1016/0169-328x(95)00126-d. [DOI] [PubMed] [Google Scholar]
- Matysiak-Scholze U., Nehls M. The structural integrity of ROR α isoforms is mutated in staggerer mice: cerebellar coexpression of ROR alpha1 and ROR alpha4. Genomics. 1997;43:78–84. doi: 10.1006/geno.1997.4757. [DOI] [PubMed] [Google Scholar]
- Maywood E. S., O'Neill J., Wong G. K., Reddy A. B., Hastings M. H. Circadian timing in health and disease. Prog Brain Res. 2006;153:253–69. doi: 10.1016/S0079-6123(06)53015-8. [DOI] [PubMed] [Google Scholar]
- McBroom L. D., Flock G., Giguere V. The nonconserved hinge region and distinct amino-terminal domains of the ROR α orphan nuclear receptor isoforms are required for proper DNA bending and ROR α-DNA interactions. Mol Cell Biol. 1995;15:796–808. doi: 10.1128/mcb.15.2.796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeachy M. J., Bak-Jensen K. S., Chen Y., Tato C. M., Blumenschein W., McClanahan T., Cua D. J. TGF-β and IL-6 drive the production of IL-17 and IL-10 by T cells and restrain T(H)-17 cell-mediated pathology. Nat Immunol. 2007;8:1390–7. doi: 10.1038/ni1539. [DOI] [PubMed] [Google Scholar]
- McInerney E. M., Rose D. W., Flynn S. E., Westin S., Mullen T. M., Krones A., Inostroza J., Torchia J., Nolte R. T., Assa-Munt N., Milburn M. V., Glass C. K., Rosenfeld M. G. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes Dev. 1998;12:3357–68. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N. J., O'Malley B. W. Minireview: nuclear receptor coactivators--an update. Endocrinology. 2002;143:2461–5. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- Mebius R. E., Rennert P., Weissman I. L. Developing lymph nodes collect CD4+CD3- LTbeta+ cells that can differentiate to APC, NK cells, and follicular cells but not T or B cells. Immunity. 1997;7:493–504. doi: 10.1016/s1074-7613(00)80371-4. [DOI] [PubMed] [Google Scholar]
- Mebius R. E., Miyamoto T., Christensen J., Domen J., Cupedo T., Weissman I. L., Akashi K. The fetal liver counterpart of adult common lymphoid progenitors gives rise to all lymphoid lineages, CD45+CD4+CD3- cells, as well as macrophages. J Immunol. 2001;166:6593–601. doi: 10.4049/jimmunol.166.11.6593. [DOI] [PubMed] [Google Scholar]
- Medvedev A., Yan Z. H., Hirose T., Giguere V., Jetten A. M. Cloning of a cDNA encoding the murine orphan receptor RZR/ROR γ and characterization of its response element. Gene. 1996;181:199–206. doi: 10.1016/s0378-1119(96)00504-5. [DOI] [PubMed] [Google Scholar]
- Medvedev A., Chistokhina A., Hirose T., Jetten A. M. Genomic structure and chromosomal mapping of the nuclear orphan receptor ROR γ (RORC) gene. Genomics. 1997;46:93–102. doi: 10.1006/geno.1997.4980. [DOI] [PubMed] [Google Scholar]
- Messer A., Plummer-Siegard J., Eisenberg B. Staggerer mutant mouse Purkinje cells do not contain detectable calmodulin mRNA. J Neurochem. 1990;55:293–302. doi: 10.1111/j.1471-4159.1990.tb08851.x. [DOI] [PubMed] [Google Scholar]
- Meyer T., Kneissel M., Mariani J., Fournier B. In vitro and in vivo evidence for orphan nuclear receptor RORalpha function in bone metabolism. Proc Natl Acad Sci U S A. 2000;97:9197–202. doi: 10.1073/pnas.150246097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki N., Ikuta M., Matsui T. Hypoxia-induced activation of the retinoic acid receptor-related orphan receptor alpha4 gene by an interaction between hypoxia-inducible factor-1 and Sp1. J Biol Chem. 2004;279:15025–31. doi: 10.1074/jbc.M313186200. [DOI] [PubMed] [Google Scholar]
- Minucci S., Ozato K. Retinoid receptors in transcriptional regulation. Curr Opin Genet Dev. 1996;6:567–74. doi: 10.1016/s0959-437x(96)80085-2. [DOI] [PubMed] [Google Scholar]
- Miyahara Y., Odunsi K., Chen W., Peng G., Matsuzaki J., Wang R. F. Generation and regulation of human CD4+ IL-17-producing T cells in ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:15505–10. doi: 10.1073/pnas.0710686105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki T. Two distinct steps during thymocyte maturation from CD4-CD8- to CD4+CD8+ distinguished in the early growth response (Egr)-1 transgenic mice with a recombinase-activating gene-deficient background. J Exp Med. 1997;186:877–85. doi: 10.1084/jem.186.6.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraitis A. N., Giguere V., Thompson C. C. Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants. Mol Cell Biol. 2002;22:6831–41. doi: 10.1128/MCB.22.19.6831-6841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moraitis A. N., Giguere V. The co-repressor hairless protects RORalpha orphan nuclear receptor from proteasome-mediated degradation. J Biol Chem. 2003;278:52511–8. doi: 10.1074/jbc.M308152200. [DOI] [PubMed] [Google Scholar]
- Moraitis A. N., Giguere V. Transition from monomeric to homodimeric DNA binding by nuclear receptors: identification of RevErbAalpha determinants required for RORalpha homodimer complex formation. Mol Endocrinol. 1999;13:431–9. doi: 10.1210/mend.13.3.0250. [DOI] [PubMed] [Google Scholar]
- Moras D., Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–91. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- Moulton V. R., Farber D. L. Committed to memory: lineage choices for activated T cells. Trends Immunol. 2006;27:261–7. doi: 10.1016/j.it.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Mucida D., Park Y., Kim G., Turovskaya O., Scott I., Kronenberg M., Cheroutre H. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 2007;317:256–60. doi: 10.1126/science.1145697. [DOI] [PubMed] [Google Scholar]
- Muller G., Hopken U. E., Lipp M. The impact of CCR7 and CXCR5 on lymphoid organ development and systemic immunity. Immunol Rev. 2003;195:117–35. doi: 10.1034/j.1600-065x.2003.00073.x. [DOI] [PubMed] [Google Scholar]
- Nagy L., Kao H. Y., Love J. D., Li C., Banayo E., Gooch J. T., Krishna V., Chatterjee K., Evans R. M., Schwabe J. W. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 1999;13:3209–16. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., Watanabe M., Isobe T., Kondo H., Inoue Y. Cytological compartmentalization in the staggerer cerebellum, as revealed by calbindin immunohistochemistry for Purkinje cells. J Comp Neurol. 1998;395:112–20. doi: 10.1002/(sici)1096-9861(19980525)395:1<112::aid-cne8>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Nakagawa Y., O'Leary D. D. Dynamic patterned expression of orphan nuclear receptor genes RORalpha and RORbeta in developing mouse forebrain. Dev Neurosci. 2003;25:234–44. doi: 10.1159/000072271. [DOI] [PubMed] [Google Scholar]
- Nakagawa S., Watanabe M., Inoue Y. Prominent expression of nuclear hormone receptor ROR α in Purkinje cells from early development. Neurosci Res. 1997;28:177–84. doi: 10.1016/s0168-0102(97)00042-4. [DOI] [PubMed] [Google Scholar]
- Nakahata Y., Kaluzova M., Grimaldi B., Sahar S., Hirayama J., Chen D., Guarente L. P., Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–40. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y., Ikeda M., Kimura T., Honma S., Ohmiya Y., Honma K. Bidirectional role of orphan nuclear receptor RORalpha in clock gene transcriptions demonstrated by a novel reporter assay system. FEBS Lett. 2004;565:122–6. doi: 10.1016/j.febslet.2004.03.083. [DOI] [PubMed] [Google Scholar]
- Nolte R. T., Wisely G. B., Westin S., Cobb J. E., Lambert M. H., Kurokawa R., Rosenfeld M. G., Willson T. M., Glass C. K., Milburn M. V. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature. 1998;395:137–43. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- Novac N., Heinzel T. Nuclear receptors: overview and classification. Curr Drug Targets Inflamm Allergy. 2004;3:335–46. doi: 10.2174/1568010042634541. [DOI] [PubMed] [Google Scholar]
- Nurieva R., Yang X. O., Martinez G., Zhang Y., Panopoulos A. D., Ma L., Schluns K., Tian Q., Watowich S. S., Jetten A. M., Dong C. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 2007;448:480–3. doi: 10.1038/nature05969. [DOI] [PubMed] [Google Scholar]
- Nurieva R. I., Chung Y., Hwang D., Yang X. O., Kang H. S., Ma L., Wang Y. H., Watowich S. S., Jetten A. M., Tian Q., Dong C. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 2008;29:138–49. doi: 10.1016/j.immuni.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill J. S., Maywood E. S., Chesham J. E., Takahashi J. S., Hastings M. H. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–53. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz M. A., Piedrafita F. J., Pfahl M., Maki R. TOR: a new orphan receptor expressed in the thymus that can modulate retinoid and thyroid hormone signals. Mol Endocrinol. 1995;9:1679–91. doi: 10.1210/mend.9.12.8614404. [DOI] [PubMed] [Google Scholar]
- Palli S. R., Hiruma K., Riddiford L. M. An ecdysteroid-inducible Manduca gene similar to the Drosophila DHR3 gene, a member of the steroid hormone receptor superfamily. Dev Biol. 1992;150:306–18. doi: 10.1016/0012-1606(92)90244-b. [DOI] [PubMed] [Google Scholar]
- Palli S. R., Ladd T. R., Retnakaran A. Cloning and characterization of a new isoform of Choristoneura hormone receptor 3 from the spruce budworm. Arch Insect Biochem Physiol. 1997;35:33–44. doi: 10.1002/(SICI)1520-6327(1997)35:1/2<33::AID-ARCH4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Palli S. R., Ladd T. R., Sohi S. S., Cook B. J., Retnakaran A. Cloning and developmental expression of Choristoneura hormone receptor 3, an ecdysone-inducible gene and a member of the steroid hormone receptor superfamily. Insect Biochem Mol Biol. 1996;26:485–99. doi: 10.1016/0965-1748(96)00004-5. [DOI] [PubMed] [Google Scholar]
- Palmer M. T., Weaver C. T. Immunology: narcissistic helpers. Nature. 2007;448:416–8. doi: 10.1038/448416a. [DOI] [PubMed] [Google Scholar]
- Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., Hogenesch J. B. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–20. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Park H., Li Z., Yang X. O., Chang S. H., Nurieva R., Wang Y. H., Wang Y., Hood L., Zhu Z., Tian Q., Dong C. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat Immunol. 2005;6:1133–41. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegorier J. P., Le May C., Girard J. Control of gene expression by fatty acids. J Nutr. 2004;134:2444S–2449S. doi: 10.1093/jn/134.9.2444S. [DOI] [PubMed] [Google Scholar]
- Peterfy M., Phan J., Xu P., Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–4. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- Pike A. C., Brzozowski A. M., Hubbard R. E. A structural biologist's view of the oestrogen receptor. J Steroid Biochem Mol Biol. 2000;74:261–8. doi: 10.1016/s0960-0760(00)00102-3. [DOI] [PubMed] [Google Scholar]
- Popov V. M., Wang C., Shirley L. A., Rosenberg A., Li S., Nevalainen M., Fu M., Pestell R. G. The functional significance of nuclear receptor acetylation. Steroids. 2007;72:221–30. doi: 10.1016/j.steroids.2006.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukka H., Aarnisalo P., Karvonen U., Palvimo J. J., Janne O. A. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J Biol Chem. 1999;274:19441–6. doi: 10.1074/jbc.274.27.19441. [DOI] [PubMed] [Google Scholar]
- Pouyssegur J., Dayan F., Mazure N. M. Hypoxia signalling in cancer and approaches to enforce tumour regression. Nature. 2006;441:437–43. doi: 10.1038/nature04871. [DOI] [PubMed] [Google Scholar]
- Preitner N., Brown S., Ripperger J., Le-Minh N., Damiola F., Schibler U. Orphan nuclear receptors, molecular clockwork, and the entrainment of peripheral oscillators. Novartis Found Symp. 2003;253:89–99; discussion 99-109. [PubMed] [Google Scholar]
- Preitner N., Damiola F., Lopez-Molina L., Zakany J., Duboule D., Albrecht U., Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–60. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Qatanani M., Moore D. D. CAR, the continuously advancing receptor, in drug metabolism and disease. Curr Drug Metab. 2005;6:329–39. doi: 10.2174/1389200054633899. [DOI] [PubMed] [Google Scholar]
- Raichur S., Lau P., Staels B., Muscat G. E. Retinoid-related orphan receptor γ regulates several genes that control metabolism in skeletal muscle cells: links to modulation of reactive oxygen species production. J Mol Endocrinol. 2007;39:29–44. doi: 10.1677/jme.1.00010. [DOI] [PubMed] [Google Scholar]
- Raspe E., Duez H., Gervois P., Fievet C., Fruchart J. C., Besnard S., Mariani J., Tedgui A., Staels B. Transcriptional regulation of apolipoprotein C-III gene expression by the orphan nuclear receptor RORalpha. J Biol Chem. 2001;276:2865–71. doi: 10.1074/jbc.M004982200. [DOI] [PubMed] [Google Scholar]
- Reppert S. M., Weaver D. R. Coordination of circadian timing in mammals. Nature. 2002;418:935–41. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Reppert S. M., Weaver D. R. Molecular analysis of mammalian circadian rhythms. Annu Rev Physiol. 2001;63:647–76. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- Retnakaran R., Flock G., Giguere V. Identification of RVR, a novel orphan nuclear receptor that acts as a negative transcriptional regulator. Mol Endocrinol. 1994;8:1234–44. doi: 10.1210/mend.8.9.7838156. [DOI] [PubMed] [Google Scholar]
- Riddiford L. M., Hiruma K., Zhou X., Nelson C. A. Insights into the molecular basis of the hormonal control of molting and metamorphosis from Manduca sexta and Drosophila melanogaster. Insect Biochem Mol Biol. 2003;33:1327–38. doi: 10.1016/j.ibmb.2003.06.001. [DOI] [PubMed] [Google Scholar]
- Ripperger J. A., Schibler U. Circadian regulation of gene expression in animals. Curr Opin Cell Biol. 2001;13:357–62. doi: 10.1016/s0955-0674(00)00220-9. [DOI] [PubMed] [Google Scholar]
- Rodgers J. T., Lerin C., Gerhart-Hines Z., Puigserver P. Metabolic adaptations through the PGC-1 α and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge-Morris M., Kocarek T. A. Regulation of sulfotransferases by xenobiotic receptors. Curr Drug Metab. 2005;6:299–307. doi: 10.2174/1389200054633871. [DOI] [PubMed] [Google Scholar]
- Salinas P. C., Fletcher C., Copeland N. G., Jenkins N. A., Nusse R. Maintenance of Wnt-3 expression in Purkinje cells of the mouse cerebellum depends on interactions with granule cells. Development. 1994;120:1277–86. doi: 10.1242/dev.120.5.1277. [DOI] [PubMed] [Google Scholar]
- Sanos S. L., Bui V. L., Mortha A., Oberle K., Heners C., Johner C., Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashihara S., Felts P. A., Waxman S. G., Matsui T. Orphan nuclear receptor ROR α gene: isoform-specific spatiotemporal expression during postnatal development of brain. Brain Res Mol Brain Res. 1996;42:109–17. doi: 10.1016/s0169-328x(96)00118-0. [DOI] [PubMed] [Google Scholar]
- Sato T. K., Panda S., Miraglia L. J., Reyes T. M., Rudic R. D., McNamara P., Naik K. A., FitzGerald G. A., Kay S. A., Hogenesch J. B. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–37. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Schaeren-Wiemers N., Andre E., Kapfhammer J. P., Becker-Andre M. The expression pattern of the orphan nuclear receptor RORbeta in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur J Neurosci. 1997;9:2687–701. doi: 10.1111/j.1460-9568.1997.tb01698.x. [DOI] [PubMed] [Google Scholar]
- Schibler U., Sassone-Corsi P. A web of circadian pacemakers. Cell. 2002;111:919–22. doi: 10.1016/s0092-8674(02)01225-4. [DOI] [PubMed] [Google Scholar]
- Schibler U., Naef F. Cellular oscillators: rhythmic gene expression and metabolism. Curr Opin Cell Biol. 2005;17:223–9. doi: 10.1016/j.ceb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Schrader M., Danielsson C., Wiesenberg I., Carlberg C. Identification of natural monomeric response elements of the nuclear receptor RZR/ROR. They also bind COUP-TF homodimers. J Biol Chem. 1996;271:19732–6. doi: 10.1074/jbc.271.33.19732. [DOI] [PubMed] [Google Scholar]
- Schwer B., Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–12. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Semenza G. L. HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol. 2000;88:1474–80. doi: 10.1152/jappl.2000.88.4.1474. [DOI] [PubMed] [Google Scholar]
- Serinagaoglu Y., Zhang R., Zhang Y., Zhang L., Hartt G., Young A. P., Oberdick J. A promoter element with enhancer properties, and the orphan nuclear receptor RORalpha, are required for Purkinje cell-specific expression of a Gi/o modulator. Mol Cell Neurosci. 2007;34:324–42. doi: 10.1016/j.mcn.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Serra H. G., Byam C. E., Lande J. D., Tousey S. K., Zoghbi H. Y., Orr H. T. Gene profiling links SCA1 pathophysiology to glutamate signaling in Purkinje cells of transgenic mice. Hum Mol Genet. 2004;13:2535–43. doi: 10.1093/hmg/ddh268. [DOI] [PubMed] [Google Scholar]
- Serra H. G., Duvick L., Zu T., Carlson K., Stevens S., Jorgensen N., Lysholm A., Burright E., Zoghbi H. Y., Clark H. B., Andresen J. M., Orr H. T. RORalpha-mediated Purkinje cell development determines disease severity in adult SCA1 mice. Cell. 2006;127:697–708. doi: 10.1016/j.cell.2006.09.036. [DOI] [PubMed] [Google Scholar]
- Sidman R. L., Lane P. W., Dickie M. M. Staggerer, a new mutation in the mouse affecting the cerebellum. Science. 1962;137:610–2. doi: 10.1126/science.137.3530.610. [DOI] [PubMed] [Google Scholar]
- Smith D. I., Zhu Y., McAvoy S., Kuhn R. Common fragile sites, extremely large genes, neural development and cancer. Cancer Lett. 2006;232:48–57. doi: 10.1016/j.canlet.2005.06.049. [DOI] [PubMed] [Google Scholar]
- Sotelo C., Wassef M. Cerebellar development: afferent organization and Purkinje cell heterogeneity. Philos Trans R Soc Lond B Biol Sci. 1991;331:307–13. doi: 10.1098/rstb.1991.0022. [DOI] [PubMed] [Google Scholar]
- Srinivas M., Ng L., Liu H., Jia L., Forrest D. Activation of the blue opsin gene in cone photoreceptor development by retinoid-related orphan receptor β. Mol Endocrinol. 2006;20:1728–41. doi: 10.1210/me.2005-0505. [DOI] [PubMed] [Google Scholar]
- Stapleton C. M., Jaradat M., Dixon D., Kang H. S., Kim S. C., Liao G., Carey M. A., Cristiano J., Moorman M. P., Jetten A. M. Enhanced susceptibility of staggerer (RORalphasg/sg) mice to lipopolysaccharide-induced lung inflammation. Am J Physiol Lung Cell Mol Physiol. 2005;289:L144–52. doi: 10.1152/ajplung.00348.2004. [DOI] [PubMed] [Google Scholar]
- Starr T. K., Jameson S. C., Hogquist K. A. Positive and negative selection of T cells. Annu Rev Immunol. 2003;21:139–76. doi: 10.1146/annurev.immunol.21.120601.141107. [DOI] [PubMed] [Google Scholar]
- Stehlin-Gaon C., Willmann D., Zeyer D., Sanglier S., Van Dorsselaer A., Renaud J. P., Moras D., Schule R. All-trans retinoic acid is a ligand for the orphan nuclear receptor ROR β. Nat Struct Biol. 2003;10:820–5. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- Stehlin C., Wurtz J. M., Steinmetz A., Greiner E., Schule R., Moras D., Renaud J. P. X-ray structure of the orphan nuclear receptor RORbeta ligand-binding domain in the active conformation. Embo J. 2001;20:5822–31. doi: 10.1093/emboj/20.21.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmayr M., Andre E., Conquet F., Rondi-Reig L., Delhaye-Bouchaud N., Auclair N., Daniel H., Crepel F., Mariani J., Sotelo C., Becker-Andre M. staggerer phenotype in retinoid-related orphan receptor α-deficient mice. Proc Natl Acad Sci U S A. 1998;95:3960–5. doi: 10.1073/pnas.95.7.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz A. C., Renaud J. P., Moras D. Binding of ligands and activation of transcription by nuclear receptors. Annu Rev Biophys Biomol Struct. 2001;30:329–59. doi: 10.1146/annurev.biophys.30.1.329. [DOI] [PubMed] [Google Scholar]
- Stockinger B., Veldhoen M., Martin B. Th17 T cells: linking innate and adaptive immunity. Semin Immunol. 2007;19:353–61. doi: 10.1016/j.smim.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Sullivan A. A., Thummel C. S. Temporal profiles of nuclear receptor gene expression reveal coordinate transcriptional responses during Drosophila development. Mol Endocrinol. 2003;17:2125–37. doi: 10.1210/me.2002-0430. [DOI] [PubMed] [Google Scholar]
- Sumi Y., Yagita K., Yamaguchi S., Ishida Y., Kuroda Y., Okamura H. Rhythmic expression of ROR β mRNA in the mice suprachiasmatic nucleus. Neurosci Lett. 2002;320:13–6. doi: 10.1016/s0304-3940(02)00011-3. [DOI] [PubMed] [Google Scholar]
- Sundvold H., Lien S. Identification of a novel peroxisome proliferator-activated receptor (PPAR) γ promoter in man and transactivation by the nuclear receptor RORalpha1. Biochem Biophys Res Commun. 2001;287:383–90. doi: 10.1006/bbrc.2001.5602. [DOI] [PubMed] [Google Scholar]
- Sun Z., Unutmaz D., Zou Y. R., Sunshine M. J., Pierani A., Brenner-Morton S., Mebius R. E., Littman D. R. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–73. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- Sun C. M., Hall J. A., Blank R. B., Bouladoux N., Oukka M., Mora J. R., Belkaid Y. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–85. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. T., Williams I. R. Lymphoid organogenesis in the intestine. Immunol Res. 2005;33:167–81. doi: 10.1385/IR:33:2:167. [DOI] [PubMed] [Google Scholar]
- Thompson C. C., Sisk J. M., Beaudoin G. M., 3rd. Hairless and Wnt signaling: allies in epithelial stem cell differentiation. Cell Cycle. 2006;5:1913–7. doi: 10.4161/cc.5.17.3189. [DOI] [PubMed] [Google Scholar]
- Thummel C. S. From embryogenesis to metamorphosis: the regulation and function of Drosophila nuclear receptor superfamily members. Cell. 1995;83:871–7. doi: 10.1016/0092-8674(95)90203-1. [DOI] [PubMed] [Google Scholar]
- Tilley S. L., Jaradat M., Stapleton C., Dixon D., Hua X., Erikson C. J., McCaskill J. G., Chason K. D., Liao G., Jania L., Koller B. H., Jetten A. M. Retinoid-related orphan receptor γ controls immunoglobulin production and Th1/Th2 cytokine balance in the adaptive immune response to allergen. J Immunol. 2007;178:3208–18. doi: 10.4049/jimmunol.178.5.3208. [DOI] [PubMed] [Google Scholar]
- Tosini G., Davidson A. J., Fukuhara C., Kasamatsu M., Castanon-Cervantes O. Localization of a circadian clock in mammalian photoreceptors. Faseb J. 2007;21:3866–71. doi: 10.1096/fj.07-8371com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay A. M., Giguere V. The NR3B subgroup: an ovERRview. Nucl Recept Signal. 2007;5:e009. doi: 10.1621/nrs.05009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trenkner E., Hoffmann M. K. Defective development of the thymus and immunological abnormalities in the neurological mouse mutation "staggerer". J Neurosci. 1986;6:1733–7. doi: 10.1523/JNEUROSCI.06-06-01733.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triqueneaux G., Thenot S., Kakizawa T., Antoch M. P., Safi R., Takahashi J. S., Delaunay F., Laudet V. The orphan receptor Rev-erbalpha gene is a target of the circadian clock pacemaker. J Mol Endocrinol. 2004;33:585–608. doi: 10.1677/jme.1.01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., Bass J. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda H. R., Chen W., Adachi A., Wakamatsu H., Hayashi S., Takasugi T., Nagano M., Nakahama K., Suzuki Y., Sugano S., Iino M., Shigeyoshi Y., Hashimoto S. A transcription factor response element for gene expression during circadian night. Nature. 2002a;418:534–9. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- Ueda E., Kurebayashi S., Sakaue M., Backlund M., Koller B., Jetten A. M. High incidence of T-cell lymphomas in mice deficient in the retinoid-related orphan receptor RORgamma. Cancer Res. 2002b;62:901–9. [PubMed] [Google Scholar]
- Ueda H. R., Hayashi S., Chen W., Sano M., Machida M., Shigeyoshi Y., Iino M., Hashimoto S. System-level identification of transcriptional circuits underlying mammalian circadian clocks. Nat Genet. 2005;37:187–92. doi: 10.1038/ng1504. [DOI] [PubMed] [Google Scholar]
- Ukai-Tadenuma M., Kasukawa T., Ueda H. R. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat Cell Biol. 2008;10:1154–63. doi: 10.1038/ncb1775. [DOI] [PubMed] [Google Scholar]
- Umemura M., Yahagi A., Hamada S., Begum M. D., Watanabe H., Kawakami K., Suda T., Sudo K., Nakae S., Iwakura Y., Matsuzaki G. IL-17-mediated regulation of innate and acquired immune response against pulmonary Mycobacterium bovis bacille Calmette-Guerin infection. J Immunol. 2007;178:3786–96. doi: 10.4049/jimmunol.178.6.3786. [DOI] [PubMed] [Google Scholar]
- Urry Z., Xystrakis E., Hawrylowicz C. M. Interleukin-10-secreting regulatory T cells in allergy and asthma. Curr Allergy Asthma Rep. 2006;6:363–71. doi: 10.1007/s11882-996-0005-8. [DOI] [PubMed] [Google Scholar]
- Veldhoen M., Hocking R. J., Atkins C. J., Locksley R. M., Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–89. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Villey I., de Chasseval R., de Villartay J. P. RORgammaT, a thymus-specific isoform of the orphan nuclear receptor RORgamma / TOR, is up-regulated by signaling through the pre-T cell receptor and binds to the TEA promoter. Eur J Immunol. 1999;29:4072–80. doi: 10.1002/(SICI)1521-4141(199912)29:12<4072::AID-IMMU4072>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- Vogel M. W., Sinclair M., Qiu D., Fan H. Purkinje cell fate in staggerer mutants: agenesis versus cell death. J Neurobiol. 2000;42:323–37. doi: 10.1002/(sici)1097-4695(20000215)42:3<323::aid-neu4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Vu-Dac N., Gervois P., Grotzinger T., De Vos P., Schoonjans K., Fruchart J. C., Auwerx J., Mariani J., Tedgui A., Staels B. Transcriptional regulation of apolipoprotein A-I gene expression by the nuclear receptor RORalpha. J Biol Chem. 1997;272:22401–4. doi: 10.1074/jbc.272.36.22401. [DOI] [PubMed] [Google Scholar]
- Wada T., Kang H. S., Angers M., Gong H., Bhatia S., Khadem S., Ren S., Ellis E., Strom S. C., Jetten A. M., Xie W. Identification of oxysterol 7alpha-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor α (RORalpha) (NR1F1) target gene and a functional cross-talk between RORalpha and liver X receptor (NR1H3) Mol Pharmacol. 2008a;73:891–9. doi: 10.1124/mol.107.040741. [DOI] [PubMed] [Google Scholar]
- Wada T., Kang H. S., Jetten A. M., Xie W. The emerging role of nuclear receptor RORalpha and its crosstalk with LXR in xeno- and endobiotic gene regulation. Exp Biol Med (Maywood) 2008b;233:1191–201. doi: 10.3181/0802-MR-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner B. L., Valledor A. F., Shao G., Daige C. L., Bischoff E. D., Petrowski M., Jepsen K., Baek S. H., Heyman R. A., Rosenfeld M. G., Schulman I. G., Glass C. K. Promoter-specific roles for liver X receptor/corepressor complexes in the regulation of ABCA1 and SREBP1 gene expression. Mol Cell Biol. 2003;23:5780–9. doi: 10.1128/MCB.23.16.5780-5789.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A. D., Cidlowski J. A. Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J Biol Chem. 2001;276:42714–21. doi: 10.1074/jbc.M106033200. [DOI] [PubMed] [Google Scholar]
- Wallace V. A. Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr Biol. 1999;9:445–8. doi: 10.1016/s0960-9822(99)80195-x. [DOI] [PubMed] [Google Scholar]
- Wang Y., Huso D., Cahill H., Ryugo D., Nathans J. Progressive cerebellar, auditory, and esophageal dysfunction caused by targeted disruption of the frizzled-4 gene. J Neurosci. 2001;21:4761–71. doi: 10.1523/JNEUROSCI.21-13-04761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Niwa S., Bouda K., Matsuura S., Homma T., Shudo K., Nagai H. The effect of Am-80, one of retinoids derivatives on experimental allergic encephalomyelitis in rats. Life Sci. 2000;67:1869–79. doi: 10.1016/s0024-3205(00)00776-1. [DOI] [PubMed] [Google Scholar]
- Wan Y. Y., Flavell R. A. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–70. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- Weaver C. T., Hatton R. D., Mangan P. R., Harrington L. E. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–52. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- Willy P. J., Mangelsdorf D. J. Hormones and signaling. San Diego: Academic Press; 1998. Nuclear orphan receptors: the search for novel ligands and signaling pathways; pp. 308–358. [Google Scholar]
- Wilson N. J., Boniface K., Chan J. R., McKenzie B. S., Blumenschein W. M., Mattson J. D., Basham B., Smith K., Chen T., Morel F., Lecron J. C., Kastelein R. A., Cua D. J., McClanahan T. K., Bowman E. P., de Waal Malefyt R. Development, cytokine profile and function of human interleukin 17-producing helper T cells. Nat Immunol. 2007;8:950–7. doi: 10.1038/ni1497. [DOI] [PubMed] [Google Scholar]
- Winoto A., Littman D. R. Nuclear hormone receptors in T lymphocytes. Cell. 2002;109 Suppl:S57–66. doi: 10.1016/s0092-8674(02)00710-9. [DOI] [PubMed] [Google Scholar]
- Wolf I. M., Heitzer M. D., Grubisha M., DeFranco D. B. Coactivators and nuclear receptor transactivation. J Cell Biochem. 2008;104:1580–6. doi: 10.1002/jcb.21755. [DOI] [PubMed] [Google Scholar]
- Xiao S., Jin H., Korn T., Liu S. M., Oukka M., Lim B., Kuchroo V. K. Retinoic acid increases Foxp3+ regulatory T cells and inhibits development of Th17 cells by enhancing TGF-β-driven Smad3 signaling and inhibiting IL-6 and IL-23 receptor expression. J Immunol. 2008;181:2277–84. doi: 10.4049/jimmunol.181.4.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Sadim M. S., Sun Z. RORgammat recruits steroid receptor coactivators to ensure thymocyte survival. J Immunol. 2005;175:3800–9. doi: 10.4049/jimmunol.175.6.3800. [DOI] [PubMed] [Google Scholar]
- Xi H., Schwartz R., Engel I., Murre C., Kersh G. J. Interplay between RORgammat, Egr3, and E proteins controls proliferation in response to pre-TCR signals. Immunity. 2006;24:813–26. doi: 10.1016/j.immuni.2006.03.023. [DOI] [PubMed] [Google Scholar]
- Xi H., Kersh G. J. Sustained early growth response gene 3 expression inhibits the survival of CD4/CD8 double-positive thymocytes. J Immunol. 2004;173:340–8. doi: 10.4049/jimmunol.173.1.340. [DOI] [PubMed] [Google Scholar]
- Xu L., Glass C. K., Rosenfeld M. G. Coactivator and corepressor complexes in nuclear receptor function. Curr Opin Genet Dev. 1999;9:140–7. doi: 10.1016/S0959-437X(99)80021-5. [DOI] [PubMed] [Google Scholar]
- Xu W. Nuclear receptor coactivators: the key to unlock chromatin. Biochem Cell Biol. 2005;83:418–28. doi: 10.1139/o05-057. [DOI] [PubMed] [Google Scholar]
- Yamashita S., Tsujino Y., Moriguchi K., Tatematsu M., Ushijima T. Chemical genomic screening for methylation-silenced genes in gastric cancer cell lines using 5-aza-2'-deoxycytidine treatment and oligonucleotide microarray. Cancer Sci. 2006;97:64–71. doi: 10.1111/j.1349-7006.2006.00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Anderson D. E., Baecher-Allan C., Hastings W. D., Bettelli E., Oukka M., Kuchroo V. K., Hafler D. A. IL-21 and TGF-β are required for differentiation of human T(H)17 cells. Nature. 2008c;454:350–2. doi: 10.1038/nature07021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. O., Nurieva R., Martinez G. J., Kang H. S., Chung Y., Pappu B. P., Shah B., Chang S. H., Schluns K. S., Watowich S. S., Feng X. H., Jetten A. M., Dong C. Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity. 2008a;29:44–56. doi: 10.1016/j.immuni.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Downes M., Yu R. T., Bookout A. L., He W., Straume M., Mangelsdorf D. J., Evans R. M. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–10. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- Yang X., Lamia K. A., Evans R. M. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2007b;72:387–94. doi: 10.1101/sqb.2007.72.058. [DOI] [PubMed] [Google Scholar]
- Yang X. O., Chang S. H., Park H., Nurieva R., Shah B., Acero L., Wang Y. H., Schluns K. S., Broaddus R. R., Zhu Z., Dong C. Regulation of inflammatory responses by IL-17F. J Exp Med. 2008d;205:1063–75. doi: 10.1084/jem.20071978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. O., Panopoulos A. D., Nurieva R., Chang S. H., Wang D., Watowich S. S., Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem. 2007a;282:9358–63. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- Yang X. O., Pappu B. P., Nurieva R., Akimzhanov A., Kang H. S., Chung Y., Ma L., Shah B., Panopoulos A. D., Schluns K. S., Watowich S. S., Tian Q., Jetten A. M., Dong C. T helper 17 lineage differentiation is programmed by orphan nuclear receptors ROR α and ROR γ. Immunity. 2008b;28:29–39. doi: 10.1016/j.immuni.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota Y., Mansouri A., Mori S., Sugawara S., Adachi S., Nishikawa S., Gruss P. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 1999;397:702–6. doi: 10.1038/17812. [DOI] [PubMed] [Google Scholar]
- Yoon C. H. Developmental mechanism for changes in cerebellum of "staggerer" mouse, a neurological mutant of genetic origin. Neurology. 1972;22:743–54. doi: 10.1212/wnl.22.7.743. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Naito A., Inoue J., Satoh M., Santee-Cooper S. M., Ware C. F., Togawa A., Nishikawa S., Nishikawa S. Different cytokines induce surface lymphotoxin-alphabeta on IL-7 receptor-α cells that differentially engender lymph nodes and Peyer's patches. Immunity. 2002;17:823–33. doi: 10.1016/s1074-7613(02)00479-x. [DOI] [PubMed] [Google Scholar]
- Zanjani H. S., Mariani J., Delhaye-Bouchaud N., Herrup K. Neuronal cell loss in heterozygous staggerer mutant mice: a model for genetic contributions to the aging process. Brain Res Dev Brain Res. 1992;67:153–60. doi: 10.1016/0165-3806(92)90216-j. [DOI] [PubMed] [Google Scholar]
- Zhang F., Meng G., Strober W. Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat Immunol. 2008a;9:1297–306. doi: 10.1038/ni.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Rong G., Wei H., Zhang M., Bi J., Ma L., Xue X., Wei G., Liu X., Fang G. The prevalence of Th17 cells in patients with gastric cancer. Biochem Biophys Res Commun. 2008b;374:533–7. doi: 10.1016/j.bbrc.2008.07.060. [DOI] [PubMed] [Google Scholar]
- Zhang N., Hartig H., Dzhagalov I., Draper D., He Y. W. The role of apoptosis in the development and function of T lymphocytes. Cell Res. 2005;15:749–69. doi: 10.1038/sj.cr.7290345. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Rudensky A. Y. Foxp3 in control of the regulatory T cell lineage. Nat Immunol. 2007;8:457–62. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- Zhou L., Ivanov II, Spolski R., Min R., Shenderov K., Egawa T., Levy D. E., Leonard W. J., Littman D. R. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat Immunol. 2007;8:967–74. doi: 10.1038/ni1488. [DOI] [PubMed] [Google Scholar]
- Zhou L., Lopes J. E., Chong M. M., Ivanov II, Min R., Victora G. D., Shen Y., Du J., Rubtsov Y. P., Rudensky A. Y., Ziegler S. F., Littman D. R. TGF-β-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–40. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., McAvoy S., Kuhn R., Smith D. I. RORA, a large common fragile site gene, is involved in cellular stress response. Oncogene. 2006;25:2901–8. doi: 10.1038/sj.onc.1209314. [DOI] [PubMed] [Google Scholar]
- Zoghbi H. Y., Orr H. T. Glutamine repeats and neurodegeneration. Annu Rev Neurosci. 2000;23:217–47. doi: 10.1146/annurev.neuro.23.1.217. [DOI] [PubMed] [Google Scholar]