Chromosome Territories (original) (raw)
Abstract
Chromosome territories (CTs) constitute a major feature of nuclear architecture. In a brief statement, the possible contribution of nuclear architecture studies to the field of epigenomics is considered, followed by a historical account of the CT concept and the final compelling experimental evidence of a territorial organization of chromosomes in all eukaryotes studied to date. Present knowledge of nonrandom CT arrangements, of the internal CT architecture, and of structural interactions with other CTs is provided as well as the dynamics of CT arrangements during cell cycle and postmitotic terminal differentiation. The article concludes with a discussion of open questions and new experimental strategies to answer them.
Chromosomes occupy distinct territories in the cell nucleus. The major components of chromosome territories are chromatin domains separated by an interchromatin compartment.
Impressive progress has been achieved during the last decade with regard to the functional implications of DNA methylation, histone modifications, and chromatin remodeling events for gene regulation (Fuks 2005; Kouzarides 2007; Maier et al. 2008; Jiang and Pugh 2009). It has, however, also become obvious that decoding the chromatin language does not suffice to fully understand the ways in which the diploid genome contributes to the formation of the different epigenomes present in the various cell types of a multicellular organism.
Different epigenomes and their functional implications also depend on differences in higher‐order chromatin organization and nuclear architecture at large. Epigenomic research aims for an integrated understanding of the structural and functional aspects of epigenetics with nuclear architecture during the differentiation of toti- or pluripotent cells to functionally distinct cell types.
The territorial organization of chromosomes in interphase (chromosome territories, CTs) constitutes a basic feature of nuclear architecture. This article starts with a brief historical account of the CT concept and the compelling experimental evidence in favor of a territorial organization of chromosomes in all eukaryotes studied to date. A survey of what is presently known about nonrandom arrangements of CTs, about changes of such arrangements in cycling cells as a result of internal or external influences and about the internal architecture of CTs and their structural interactions with each other is provided. The article concludes with a discussion of open questions on CT organization and new experimental strategies to answer them.
ORIGIN OF THE CHROMOSOME TERRITORY CONCEPT
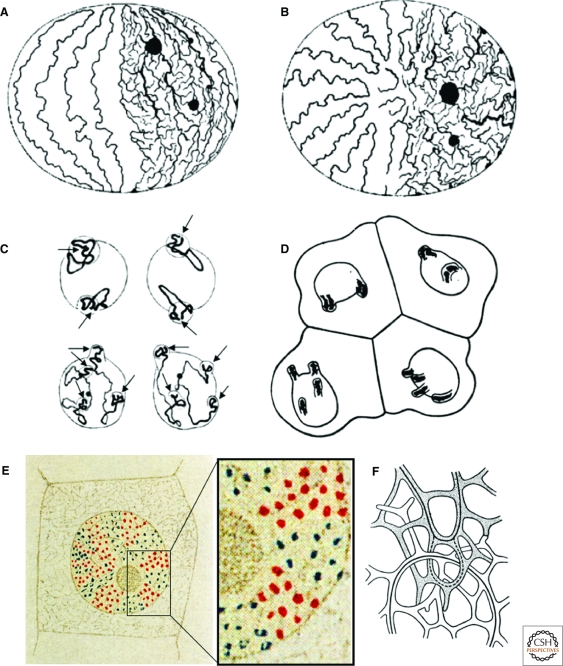
Since the late 19th century, an uncounted number of microscopic studies has appeared on numerous aspects of nuclear structure and on the observation of mitotic chromosomes. A territorial organization of interphase chromosomes was first suggested for animal cell nuclei by Carl Rabl (Rabl 1885) (Fig. 1A,B), but it was Theodor Boveri who introduced the term chromosome territory (CT) in his seminal studies of blastomere stages of the horse roundworm Parascaris equorum or Ascaris megalocephala, as the species was called at his time (Boveri 1909). The worm exists in two varieties, one with two pairs (Ascaris bivalens), the other with one pair of germ line chromosomes (Ascaris univalens). Boveri argued that each chromosome visible in mitosis retains its individuality during interphase and occupies a distinct part of the nuclear space. As can be seen in his drawings, Boveri was able to distinguish chromosome ends sticking out in protrusions of prophase nuclei (Fig. 1C) and used these protrusions as markers for the nuclear position of the asserted CTs in interphase nuclei (Fig. 1D). In fixed two-cell embryos, Boveri noted that their nuclear topography was strikingly similar during interphase and prophase, when chromosomes became visible as distinct entities. In four cell embryos, however, he typically observed two pairs of nuclei each carrying distinctly different protrusion patterns (Fig. 1D). His ingenious speculative talent led him to the following predictions on chromosome arrangements in Ascaris nuclei during the first steps of postzygotic development (for a comprehensive review, see Cremer and Cremer 2006a): (I) CT order is stably maintained during interphase; (II) Chromosome neighborhood patterns change from prophase to metaphase; and (III) New chromosome neighborhood arrangements established in the metaphase plate are conserved to a considerable extent throughout anaphase and telophase, resulting in rather symmetrical arrangements of CTs in the two daughter nuclei. Different opinions on the structural organization of CTs were put forward in the early days of the 20th century. Eduard Strasburger (1905) published a colored cartoon claiming that CTs are built up from little chromatin clumps (Fig. 1E), whereas Theodor Boveri discussed a sponge-like CT structure built up from networks of anastomizing chromatin bundles (Fig. 1F).
Figure 1.
Early concepts in favor of a territorial organization of chromosomes in interphase nuclei. (A–B) Carl Rabĺs hypothetical view (Rabl 1885) of a territorial chromosome arrangement in the interphase nucleus was based on studies of Proteus and Salamandra, in particular on epithelial cells from Salamandra maculata larvae. (A) Side view; supposed CTs are built up from primary chromatin threads (left side), from which secondary and tertiary threads branch out and form a chromatin network (right side). Spindle attachment sites, now known as centromeres, congress at one site of the nucleus (top, Rabl’s Polfeld), whereas the telomeres cluster at the opposite site (bottom, Rabl’s Gegenpolseite). (B) View from above on the Polfeld. (C) Drawings made by Theodor Boveri (1909) from two Ascaris megalocephala univalens embryos fixed during prophase of the two-cell stage. Arrows point to nuclear protrusions containing the distal parts of the two germ line chromosomes. (D) Boveri’s drawing of a fixed four-cell embryo shows two pairs of cells with a distinctly different arrangement of these protrusions in interphase nuclei. Boveri argued that the two upper and two lower cells, respectively, represent daughter cells and explained the strikingly different nuclear protrusion patterns observed in the two cell pairs as a result of chromosome movements during prometaphase. (E) Eduard Strasburger's colored model view of a tissue cell nucleus from the plant Galtonia candicans (Strasburger 1905). Chromosome territories supposedly are built up from higher‐order chromatin foci delineated in red and blue. (F) The white and gray shaded bundles in Boveri’s sketch from 1909 reflect two neighboring CTs with sponge-like structures built up from networks of anastomizing chromatin bundles. The continuous lines reflect Rabl’s primary threads, and the dotted line depicts the possibility of a rare pathological situation, in which secondary chromatin threads from one CT encompass the primary thread of the other, a situation possibly leading to an exchange of chromatin material between the chromosomes or a segregation failure during the next mitosis.
Despite light microscopic evidence in favor of chromosome territories at least in some species and cell types (for review see Stack et al. 1977) the weight of electron microscopic evidence established since the 1950s apparently argued for an unraveling of chromosomes in interphase nuclei into intermingling chromatin fibers of 10–30 nm in diameter with no sign of individual chromosomes. As a consequence, the concept of chromosome territories fell into oblivion or was even considered to be experimentally disproved (Wischnitzer 1973). During the 1970s and 1980s, most researchers seemed content with the assumption that the nucleus is filled with intermingling chromatin fibers and loops like a dish of spaghetti, an assumption widely reflected by textbooks of cell biology.
EXPERIMENTAL EVIDENCE FOR CHROMOSOME TERRITORIES
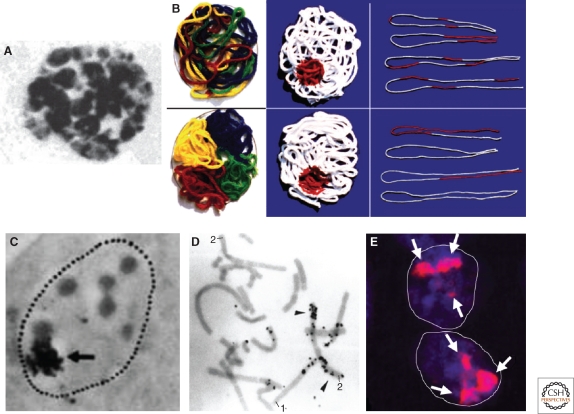
Stephen M. Stack, David B. Brown, and William C. Dewey were among the few researchers in the 1970s that still adhered to Boveri’s seemingly outdated concept (Stack et al. 1977; for review, see Cremer and Cremer 2006b). They squashed fixed cells from Allium cepa root tips, as well as Chinese hamster cells in acetic acid, and subjected the air-dried cells to salt solutions containing NaOH, and finally performed a Giemsa staining. This treatment yielded clumps of condensed chromatin arguably reflecting interphase chromosomes (Fig. 2A). The authors concluded that “chromosomes … remain in distinct domains throughout interphase.” Our group obtained early evidence for CTs in nuclei of diploid Chinese hamster cells with the help of laser-UV-microirradiation experiments (Zorn et al. 1976; Zorn et al. 1979; Cremer et al. 1982a; Cremer et al. 1982b; for review, see Cremer and Cremer 2006b; Meaburn and Misteli 2007). A laser microbeam (λ = 257 nm) was used to induce UV-damaged DNA within a small part of the nucleus. It was predicted that DNA damage inflicted within a small volume of the nucleus would yield different results depending on how chromosomes were arranged. Figure 2B exemplifies the experimental rationale with the example of woolen threads assembled within a “nuclear” space (Cremer et al. 1982a). Each thread reflects a chromatin fiber constituting an individual chromosome. In case of threads distributed throughout the whole nuclear space (upper panel), the “damage” label would become scattered over many threads. If individual threads occupy distinct territories (lower panel), localized label would mark only a small subset of threads. This experimental rationale was realized in the following way (Fig. 2C–E): Nuclei of living cells were microirradiated in G1. Locally damaged DNA was pulse-labeled with 3H-thymidine reflecting unscheduled DNA synthesis during excision repair of DNA photolesions. 3H-thymidine incorporation was detected by autoradiography in nuclei fixed immediately after the pulse (Fig. 2C) or in metaphase spreads prepared from cells that were allowed to proceed to the next mitosis (Fig. 2D). Alternatively, microirradiated DNA was visualized by immunostaining with antibodies raised against UV-damaged DNA (Cremer et al. 1984b) (Fig. 2E). As predicted by the CT concept, both approaches clearly showed that microirradiation of a small part of the nucleus damaged only a small subset of the mitotic chromosome complement (Fig. 2D). The labeled parts of mitotic chromosomes revealed the UV damaged segments of neighboring chromosome territories hit by the microbeam during the preceding interphase. Consistent with this result, microirradiation of a small part of the metaphase plate of a living cell yielded a mirror-like pattern of distinctly labeled domains in the resulting daughter nuclei (Fig. 2F). These experiments provided the first compelling, although still indirect, evidence for the existence of chromosome territories.
Figure 2.
Experimental evidence for a territorial organization of interphase chromosomes. (A) Giemsa stained interphase nucleus of a fixed Chinese hamster cell (CHO line) reveals chromatin clumps likely representing individual CTs (reprinted with permission from Stack et al. 1977). (B) Experimental rationale of laser-UV-microbeam experiments to distinguish between a nonterritorial (upper row) and a territorial (bottom row) chromosome arrangement in cell nuclei (compare Cremer et al. 1982a). (C) Autoradiograph of a diploid Chinese Hamster cell. The nucleus of the living cell was microirradiated in G1, pulse-labeled with 3H thymidine and fixed immediately thereafter. The arrow points to a cluster of silver grains detected over the site of microirradiation. (D) Metaphase spread from the same experiment obtained about 40 hours after microirradiation. One chromosome 1 and one chromosome 2 are intensely marked with silver grains, indicating that the microbeam hit the respective territories during interphase, whereas their homologs are unlabeled, arguing against the spatial association of the homolog territories. (E) Immunocytochemical identification of microirradiated DNA (arrows) in a pair of Chinese hamster daughter nuclei fixed around 4 hours after microirradiation of a small part of a metaphase plate. The mirror-like distribution of microirradiated chromatin in the two daughter nuclei argues for a similar arrangement of chromosome territories (reprinted from Cremer et al. [1984b] with permission).
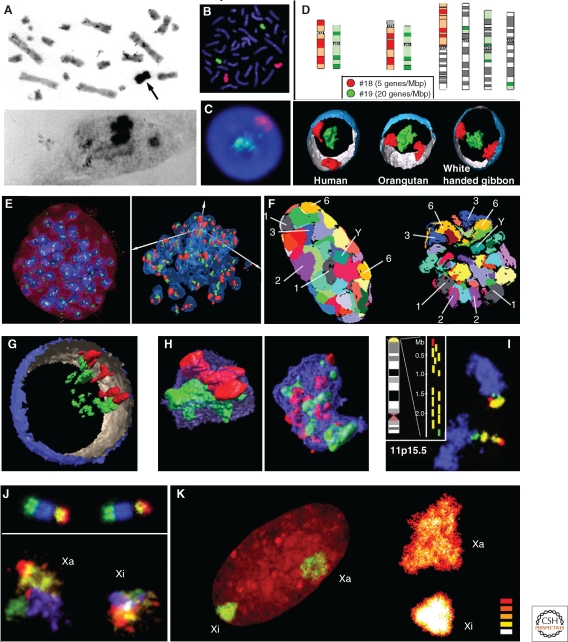
The direct visualization of individual CTs was made possible by in situ hybridization techniques developed during the mid 1980s. Initial experiments were performed with total human genomic DNA on cell hybrids that contained only one or few human chromosomes in a mouse or hamster genome complement (Manuelidis 1985; Schardin et al. 1985) (Fig. 3A). The achievement of chromosome sorting by flow cytometry of fluorescently labeled mitotic chromosomes (Cremer et al. 1984a; Gray et al. 1987; Fawcett et al. 1994) enabled the generation of chromosome specific painting probes for a large number of species. Subsequent amplification of DNA by cloning in bacterial vectors or by universal PCR (Telenius et al. 1992) and novel techniques for the suppression of ubiquitous repetitive sequences by COT-1 DNA (Cremer et al. 1988; Pinkel et al. 1988; Lichter et al. 1988a; Lichter et al. 1988b) or by depletion of these sequences from the respective probes (Bolzer et al. 1999) made it possible to delineate individual chromosomes in metaphase plates (Fig. 3B) and their territories in the interphase nucleus (Fig. 3C–F,K). To study the spatial arrangement of CTs, elaborate three-dimensional (3D) FISH protocols were developed (Cremer et al. 2008). 3D FISH in combination with light optical serial sectioning of nuclei by laser confocal microscopy and 3D image reconstruction has become the method of choice for studies of higher‐order arrangements of CTs. The increasing availability of DNA probes for specific subchromosomal regions from chromosome arms down to bands and single gene loci enabled studies of CT substructures (Fig. 3G–J).
Figure 3.
Direct evidence for chromosome territories (CTs) by in situ hybridization experiments. (A) In situ hybridization with biotinylated human genomic DNA of a Chinese hamster × human hybrid cell line carrying a single human X chromosome reveals the specifically labeled human X chromosome (arrow) in metaphase spreads (top; other chromosomes are Giemsa stained) and its respective human X territory in interphase nuclei (bottom); for details, see Schardin et al. (1985). (B) Visualization of individual chromosomes in a human (HSA) metaphase plate (chr. HSA18 in red, chr. HSA 19 in green) after fluorescence in situ hybridization (FISH) using labeled chromosome painting probes. (C) A single light optical mid-section through the nucleus of a human lymphoblastoid cell after 3D FISH with the same painting probes shows a HSA 19 CT (green) in the nuclear center and a HSA 18 CT (red) at the periphery; for details, see Tanabe et al. (2002). (D) Top: Idiogramatic illustration of primate chromosomes or subchromosomal regions from the Orangutan (pongus pygmaeus, PPY, middle) and white handed gibbon (Hylobates lar, HLA, right) orthologous to human chromosomes 18 (red) and 19 (green). Note the pronounced chromosomal rearrangements between the human and the HLA karyotype: The arm of a large HLA chromosome is orthologous to HSA 18, whereas four HSA 19 orthologous chromosome segments are distributed on three other large HLA chromosomes. Bottom: 3D reconstructions of representative HSA (left), PPY (middle), and HLA (right) lymphoblastoid cell nuclei reveal the same nonrandom radial nuclear positions of orthologous gene dense HSA 19 (green) and gene poor chromatin HSA 18 (red); for details, including quantitative evaluation, see Tanabe et al. (2002). (E) Painted CTs of the gene dense chromosome BTA 19 (green) and gene poor BTA 20 chromosome (red) in a domestic cattle (Bos taurus, BTA) embryo during blastocyst stage. Left: Maximum intensity projections of painted CTs in DAPI stained nuclei (blue). Right: 3D reconstructions of nuclei of the same embryo from a different perspective. Gene dense BTA 19 chromatin is preferentially distributed in the nuclear interior and gene poor BTA 20 chromatin at the nuclear periphery: for details, including quantitative evaluation, see Koehler et al. (2009). (F) Simultaneous delineation of all chromosomes in a human fibroblast nucleus (left) and a prometaphase rosette (right) by multi-color FISH. Light optical mid-sections with false color representation of all CTs and prometaphase chromosomes, respectively, are shown. Examples of individual CTs and mitotic chromosomes are denoted with their respective karyotypic number; for details, see Bolzer et al. (2005). (G) Partial 3D reconstruction of a human lymphocyte nucleus after 3D-FISH of two differently labeled sets of BAC clones from HSA 12 carrying sequences from several gene-dense (green) and several gene-poor chromosome segments (red), respectively. This nucleus illustrates two neighboring HSA 12 CTs with distinct gene density correlated radial nuclear arrangement. (H) Left: 3D reconstruction of a single painted HSA 12 territory (blue) showing the distinctly different, polarized arrangements of these two sets of BAC-clones. Right: 3D reconstruction of a HSA 12 CT recorded after 3D FISH with two differentially labeled sets of BAC clones containing sequences from highly expressed (green) and repressed genes (red), respectively, shows that active and silent genes are distributed throughout the CT; for details, see Kupper et al. (2007). (I) Multicolor 3D FISH to a human fibroblast reveals the two HSA 11 CTs (blue) together with the particular gene dense region 11p15.5 (green, yellow, and red). For the delineation of this region, 15 BACs were used and differentially labeled as shown in the inset. Z-projections of light optical serial sections illustrate different shapes of this region: a finger-like chromatin protrusion in the lower CT and a much more compact shape in the upper CT; for details, see Albiez et al. (2006). (J) Top: Two human X chromosomes in a human fibroblast metaphase plate are shown after multicolor FISH with four differentially labeled BAC pools representing four segments from qter to pter (q-arm: green, blue; p-arm: yellow, red). Bottom: Projections of light optical sections through the Xa- and Xi-territory of a human fibroblast nucleus following 3D FISH with the same BAC pools show four separate domains of these segments within the Xa- and Xi-territory (images courtesy of Kathrin Teller, Univ of Munich). (K) Left: Painting of the Xa- and Xi-territory in a female human fibroblast nucleus (46,XX) exemplifies the different shape and painting intensity of the two X-territories. The Xi-territory was independently identified by Barr body staining (not shown). Right: intensity profiles of enlarged images of the Xi- and Xa-territory. The color code white, yellow, and red reflects high, medium, and low intensities (images courtesy of Irina Solovei, Univ of Munich).
NONRANDOM ARRANGEMENTS OF CHROMOSOME TERRITORIES AND CHROMOSOMAL SUBREGIONS
Higher-order chromatin arrangements may primarily reflect geometrical constraints, which obviously affect the 3D distribution of larger and smaller CTs crowded together in the nuclear space (Neusser et al. 2007). Occasionally, however, proximity patterns assembled by chance, may have provided functional advantageous and consequently were favored by natural selection. The search for nonrandom chromatin assemblies, the mechanisms responsible for their formation and their functional implications is one of the major goals of nuclear architecture research. This search is still in its beginning.
Methodological Problems in Distinguishing Random and Nonrandom Higher-Order Chromatin Arrangements
An overview on methods and problems for the spatial quantitative analysis of fluorescently labeled CTs and other nuclear structures is provided in Ronneberger et al. (2008). To decide about the random or nonrandom distribution of a given target, such as a CT, chromosomal subregion, or gene, it is important to define a single or sometimes multiple 3D reference points, which represent the target in question in the nuclear space. For a painted CT, its intensity gravity center can be chosen as a single reference point or the CT surface may be used to define multiple reference points. Next, proper reference structures must be defined to decide whether the chosen target is distributed randomly or nonrandomly with respect to them. For reference structures, one can choose other chromatin targets or distinct nuclear structures, e.g., the nuclear lamina, nucleoli, or splicing speckles. Two cases of nonrandom arrangements of specific chromatin targets are considered here: nonrandom radial arrangements and nonrandom neighborhood arrangements, also referred to as proximity patterns. For studies of the radial arrangement, the nuclear space can be divided into a number of concentric shells with equal volume. 3D distances measured in a random sample of nuclei between the fluorescence intensity gravity center of a given labeled target and either the nuclear center or the closest point of the nuclear lamina provide information about its preferred radial nuclear position. Alternatively, all voxels contributing to a given target and reference structure, respectively, may be used to measure 3D distances. An equal frequency of voxels for a given chromatin target in all nuclear shells strongly argues for its random radial nuclear interior or peripheral arrangement of the target. Nonrandom proximity patterns between sets of targets are indicated by 3D distance measurements, which are significantly smaller than expected in case of their random distribution in the nuclear space. It is important to note that nonrandom proximity patterns between genes located at distant sites of the same chromosome or on different chromosomes provide hints for but do not prove functional implications. For example, an apparently nonrandom proximity pattern of certain targets may simply reflect their nonrandom radial distribution in an interior nuclear shell, resulting in significantly smaller mean distances and a significantly higher frequency of contacts compared with targets randomly arranged in a peripheral nuclear shell. The importance of clearly defining targets and reference structures for which possible random or nonrandom arrangements are analyzed, can hardly be overemphasized. Certain targets may be arranged highly nonrandomly in the nuclear space, but still entirely at random in an internal nuclear shell or with respect to other more limited reference volumes or other reference structures. Before a detected nonrandom proximity pattern between certain targets gives rise to speculations about functional interactions, it should always be shown that this pattern cannot be simply explained as a consequence of a nonrandom radial distribution. As another caveat, it should be noted that the size of samples of nuclei subjected to a detailed 3D analysis is typically small (in the order of 20–50). It is of utmost importance to avoid a biased selection of such nuclei.
Radial Nuclear Arrangements of Chromosome Territories and Chromosome Subregions
Early evidence for a nonrandom radial distribution of entire CTs was based on (3D) FISH experiments in human lymphocyte nuclei using chromosome painting probes for human (HSA) chromosome 19, the chromosome with the highest gene density and for HSA 18, a gene poor chromosome. HSA 19 CTs were consistently found in the interior of human lymphocyte nuclei and of numerous other cell types, whereas the territories of the gene poor HSA 18 were located at the nuclear periphery (Fig. 3C) (Croft et al. 1999; Cremer et al. 2001; Cremer et al. 2003). These observations of a gene density correlated radial arrangement in the nucleus were completed and confirmed by analyses comprising all human chromosomes (Boyle et al. 2001). An evolutionary comparison of lymphoblastoid cells from various primate species showed that this nonrandom radial nuclear distribution has been evolutionary conserved despite major evolutionary chromosome rearrangements. Orthologous segments of HSA 19 are positioned in the nuclear interior, whereas segments corresponding to human chromosome 18 locate at the nuclear periphery (Fig. 3D) (Tanabe et al. 2002). Nonrandom radial nuclear arrangements of CTs, depending on their gene density, were also observed in rodents (Mayer et al. 2005; Neusser et al. 2007), cattle (Koehler et al. 2009), and birds (Habermann et al. 2001). Recently, it was shown in bovine preimplantion embryos that this difference was not yet present in nuclei of early blastomere stages. Its first appearance correlated with major genome activation and was fully established in blastocysts (Koehler et al. 2009) (Fig. 3E).
A multicolor 3D FISH approach was established for diploid human fibroblasts (46,XY) and allowed the colorful discrimination of the 22 pairs of autosmal CTs and the CTs of the two sex chromosomes (Bolzer et al. 2005) (Fig. 3F). For the flat-ellipsoid fibroblast nuclei, a radial arrangement of CTs, mainly according to chromosome size or DNA content, was shown in contrast to the clearly gene density correlated radial pattern observed in spherically shaped nuclei, such as nuclei in lymphocytes. Still, gene density correlated patterns on the subchromosomal level were also present in fibroblast nuclei, as shown by the preference of gene dense, Alu sequence‐rich chromatin in the nuclear interior. 3D FISH experiments using sets of BAC clones with inserts from gene dense and gene poor chromosome segments confirmed such a distinct gene density correlated radial nuclear arrangement (Fig. 3G,H). Present data provide strong evidence that the local gene density within windows of 2–10 Mb is apparently a strong and likely pivotal key player for the radial position of chromatin in the nucleus (Kozubek et al. 2002; Cremer et al. 2003; Murmann et al. 2005; Kupper et al. 2007). Other parameters, in particular transcriptional activity, replication timing, and GC content have also been correlated with nonrandom radial nuclear arrangements of CTs and chromosomal subregions (Mayer et al. 2005; Federico et al. 2006; Goetze et al. 2007; Grasser et al. 2008; Hepperger et al. 2008). However, the interdependence of these parameters within a given DNA segment makes it difficult to dissect their impact on radial chromatin distribution in the nucleus (Kupper et al. 2007). The observation that radial positions of specific gene regions can differ significantly between cell types argues for additional cell-type-specific factors (Hepperger et al. 2008). The picture gets even more complex when we consider movements toward the nuclear interior shown for individual genes on transcriptional activation (see later). These movements emphasize that radial nuclear positions of genes do not solely depend on the local DNA sequence environment, where a given gene is embedded.
Proximity Patterns of Chromosome Territories
Whereas little evidence for nonrandom neighborhood/proximity patterns was found between specific homologous and heterologous chromosomes in nuclei of human fibroblasts (Bolzer et al. 2005), nonrandom proximity patterns for smaller subsets of nonhomologous CTs were described by Misteli and coworkers in cell nuclei from various mouse tissues (Parada et al. 2002; Roix et al. 2003; Parada et al. 2004) and in human lymphocytes (Brianna Caddle et al. 2007; Khalil et al. 2007). These proximity patterns, however, were of a rather probabilistic nature, i.e., their presence was shown by an excess of certain heterologous (and occasionally also homologous) CTs in a population of nuclei, not as an event consistently observed in each nucleus. Differences of statistically preferred proximity patterns were recently also described for CT subsets in different fibroblast-derived cell lines (Zeitz et al. 2009).
It should be emphasized that a pronounced cell-to-cell variation of CT neighborhood arrangements was found in all cell types studied so far. Neither functional implications of cell type or cell-line-specific probabilistic neighborhood arrangements nor mechanisms responsible for their establishment are presently understood. Such studies resemble snap-shots of individuals present in a room. A single snap-shot does not allow conclusions about the dynamic behavior of these individuals. Accordingly, a decision whether certain CTs are permanently close to each other, have met before or will meet after the snap-shot was taken, would require many snap-shots of well defined cell types in fixed samples at the same stage of interphase or postmitotic terminal differentiation, which are presently not available.
CT STRUCTURE, SHAPES, AND PLASTICITY
Apparently, ∼1 Mb-chromatin domains, first detected in S-phase nuclei as replication foci (Ma et al. 1998) and later shown to be persistent higher-order chromatin structures, are basic structural units, which build up CTs (Jackson and Pombo 1998; Visser and Aten 1999; Berezney et al. 2005; Albiez et al. 2006). To date, neither their ultrastructural organization nor the packaging of chromatin connecting these domains has been fully clarified. In addition, chromonema fibers may play an important role in the higher-order organization of CTs (Belmont and Bruce 1994) and their structural interconnection to global chromatin networks (Albiez et al. 2006). Single ∼1-Mb domains are likely built up from smaller loop domains, whereas larger chromatin clumps may be composed of clusters of ∼1-Mb domains. CTs visualized by 3D FISH appear as structures with manifold shapes composed of higher-order chromatin domains (Dietzel et al. 1998; Khalil et al. 2007; Kupper et al. 2007) (Fig. 3H–K). Separate arm domains were disclosed by painting of chromosome arms or parts of them in human cell nuclei (Dietzel et al. 1998) (Fig. 3K).
The outer surface of an individual CT apparently does not generally provide a particular compartment for gene dense and/or transcriptionally active chromatin as originally suggested (Zirbel et al. 1993). A few chromatin regions with particular high gene density and/or transcriptional activity, such as the 11p15.5 segment (Fig. 3I), the MHC and EDC loci, or the HOX gene cluster have been consistently found looping out as protrusions from the core territory (Volpi et al. 2000; Williams et al. 2002; Chambeyron et al. 2005; Kupper et al. 2007). On a more global level, however, gene-dense and/or highly expressed sequences were found equally distributed throughout their respective territories (Mahy et al. 2002; Kupper et al. 2007) (Fig. 3H). The dissociation of gene localization relative to their CTs and gene regulation was shown for the murine _Hox_d locus during differentiation and development: Although both decondensation and movement outside of the CT occur during gene activation in ES cells and the tail bud of the embryo, in the limb bud, gene activation and chromatin condensation occur without any looping out from the CT (Morey et al. 2007). This topic is described in detail in Heard and Bickmore (2007) and Morey et al. (2009).
A particular staining intensity of the inactive X chromosome (Xi) with DNA-specific dyes was observed decades ago (so called Barr body [Lyon 1962]), indicating a higher compactness in Xi compared with the active X chromosome (Xa) or other autosomes (Fig. 3K, L). Previous studies showed distinct differences between Xi and Xa in shape and surface structure but surprisingly little differences in volume (Eils et al. 1996). In Xi, Clemson et al. (2006) observed a preferential positioning of genes —irrespective of their transcriptional activity— at the nuclear periphery, whereas repetitive sequences were found rather in the nuclear interior. Such a distinct radial arrangement was not reported for Xa, possibly because of larger invaginations in Xa. At large, however, structural differences between Xi and Xa on the subchromosomal level are poorly defined to date, although the CTs X provide an excellent model system for structure/function relationships in homologous chromosomes with different functional allocation.
Detailed knowledge of the folding structure (conformation) of small contiguous chromatin segments within a given CT is limited so far to a few regions in autosomes and further studies are definitely required before a comprehensive picture can be drawn on the internal structure and local conformation of CTs. Shopland et al. (2006) analyzed the 3D structure of a highly conserved 4.3-Mb region on mouse chromosome 14 containing four clusters of genes separated by gene “deserts.” In Drosophila melanogaster, gene dense and gene poor segments within a 7-Mb region of chromosome 2 were described to form spatially segregated clusters of different stability and folding structure (Boutanaev et al. 2005), and Goetze et al. (2007) showed for several cell lines that ridges (contiguous genomic regions harboring adjacent genes with high ubiquitous transcriptional activity) are in general less condensed and more irregularly shaped than “antiridges”, arguing that the structure of these two types of genomic domains is largely independent of tissue-specific variations in gene expression. Mateos-Langerak et al. (2009) provide data on the folding structure of subregions of CT 1 and 11 that are in accordance with a suggested random loop model with 10–30 loops/Mb.
The relevance of 3D FISH studies for conclusions regarding the fine structure of CTs in vivo has to consider some technical issues. Differences in the fixation procedure of cells, amplification, and labeling efficiency of chromosome painting probes may have an impact on CT delineation. The necessary suppression or depletion of interspersed repetitive sequences prevents a complete visualization of CTs even under optimal hybridization conditions. The most crucial step for CT fine structure occurs during the denaturation of the nuclear DNA, a step necessary for successful probe hybridization (Solovei et al. 2002). These limitations emphasize the need for the visualization of CTs in living cells under conditions which do not interfere with chromatin or other nuclear functions (Zink et al. 1998; Walter et al. 2003). Although urgently required, in vivo approaches have their own technical problems, such as an interference of in vivo labeling strategies with the function of labeled chromatin and phototoxicity effects during prolonged observation periods.
Different approaches have been established for the quantitative assessment of CT conformation. These include density, volume, roundness, and smoothness factors, as well as principal component analyses, which are also potentially useful to assess CT orientation within the nucleus (Eils et al. 1996; Roix et al. 2003; Ronneberger et al. 2008). The subjective choice of threshold setting for confocal image stacks influences the determination of CT borders and accordingly of measured CT volumes. Low threshold settings can result in the inclusion of background, leading to overestimation of CT volumes and apparent chromatin intermingling between neighboring CTs, whereas high thresholds can lead to the visual loss of fine DNA structures looping out from the chromatin bulk of a given territory.
STABILITY AND CHANGES OF CT ARRANGEMENTS IN CYCLING CELLS
The question of to what extent a given proximity pattern established between CTs during a given interphase may be transmitted through mitosis to the next interphase has remained controversial (Gerlich et al. 2003; Walter et al. 2003; Thomson et al. 2004; Kalmarova et al. 2008). These studies were based on live-cell approaches using transgenic cell lines with either FP-tagged or photoconvertible tagged histones. This allowed following up fluorescently labeled chromatin through mitosis into the next interphase. Present evidence argues for the stability of a given CT neighborhood arrangement once established at the onset of interphase until the next prophase, whereas chromosome movements during prometaphase necessary to establish the metaphase plate can lead to major changes of side-by-side chromosome arrangements in the metaphase plate compared with the side-by-side arrangements of the respective CTs during the preceding interphase (Walter et al. 2003; Cvackova et al. 2009).
DYNAMICS OF CT ARRANGEMENTS DURING POSTMITOTIC CELL DIFFERENTIATION AND IN TERMINALLY DIFFERENTIATED CELLS
In a seminal investigation, Barr and Bertram described that on electric stimulation of cat motor neurons, a “nucleolar satellite” (now known as the Barr body) moved from its usual position adjacent to the nucleolus toward the nuclear membrane within a time course of several days (Barr and Bertram 1949). Borden and Manuelidis (1988) showed a pronounced repositioning of the human X-territories in neurons of both males and females in electrophysiologically defined seizure foci. Repositioning of CTs during cellular differentiation of murine cerebellar Purkinje neurons was described by Martou and De Boni (2000) in terms of changes of centromere positions that adopted their final position around day 5 post partum.
A striking example of CT reorganization during terminal differentiation was recently shown by Solovei et al. (2009) in a study of the mammalian retina. In mammals adapted to nocturnal life, all heterochromatin becomes located in the nuclear interior during postmitotic terminal differentiation of rod cells, whereas all euchromatin is shifted toward the nuclear periphery. This transformation starts around day 6 post partum and takes several weeks for completion. Rod cells of mammals with diurnal life styles do not show such a chromatin reorganization. Their nuclei reveal the conventional pattern with heterochromatin enriched at the nuclear periphery and around the nucleoli, whereas euchromatin is mostly distributed in the nuclear interior. This global nuclear reorganization in rod cells of nocturnal species necessitates a profound reorganization of radial chromatin arrangements rather than a change of CT proximity patterns. Unexpectedly, the inverted pattern of rod cell nuclei in nocturnal mammals reflects an adaptation to vision in low light conditions. Because of the somewhat higher refractive index of heterochromatin compared with less condensed euchromatin, inverted nuclei act as micro-lenses, which help to channel photons to the photoreceptors. This may be the first example in which a specific adaptive advantage of a cell-type-specific nuclear architecture could be shown. Interestingly, this functional specialty shows to what extent the nuclear architecture can be modified under an overriding selective pressure.
DYNAMICS AND INTERACTIONS OF SPECIFIC GENE LOCI LOCATED ON THE SAME OR ON DIFFERENT CTs
This question is particularly important with respect to long-range chromatin movements involved in the congression of coregulated genes. Live-cell experiments performed with cultured mammalian and Drosophila cells showed locally constrained movements of subchromosomal domains (Abney et al. 1997; Marshall et al. 1997; Bornfleth et al. 1999; Edelmann et al. 2001).
The typical prevalence of heterochromatin localized at the lamina and the observation of silenced genes in this peripheral nuclear subcompartment has supported the concept that the nuclear periphery is a largely repressive environment for transcription, and vice versa, the nuclear interior a compartment for transcriptional activity (Schneider and Grosschedl 2007). This would require corresponding movements of gene loci in correlation to their (cell type) correlated transcriptional activity. Recent studies, however, support a more complex picture (Taddei et al. 2006; Akhtar and Gasser 2007; Deniaud and Bickmore 2009). Several groups succeeded to tether specific chromatin segments to the nuclear envelope in living cells. They found that some genes were suppressed when closely associated with the envelope, but that others were not (Finlan et al. 2008; Kumaran and Spector 2008; Reddy et al. 2008).
Of particular interest are hints that a long-range spatial nuclear convergence of genes, which are located many megabases apart in cis, i.e., on the same chromosome, or trans, i.e., on different chromosomes, might be involved in mechanisms of gene activation or silencing (Zuckerkandl and Cavalli 2007; Bartkuhn and Renkawitz 2008). This phenomenon has been referred to as “gene kissing” (Kioussis 2005) or “chromosome kissing” (Cavalli 2007). As an early example, LaSalle and Lalande (1996) presented 3D FISH evidence for the transient spatial association of the AS/PWS loci during late S phase. These loci comprise the genes involved in two imprinting disorders, the Angelman syndrome and the Prader–Willi syndrome. The authors argued that transient “kissing” between the two loci is required for maintaining opposite imprints in cycling cells. This specific case of “kissing,” however, could not be confirmed in a later study (Teller et al. 2007).
A “nonmicroscopic” biochemical approach with a great potential for the disclosure of spatial interactions of specific genomic loci is the technique of “chromosome conformation capture” (3C) introduced by Dekker et al. (2002) and its recent extension to 4C (chromosome conformation capture-on-chip [Simonis et al. 2006] and “circular chromosome conformation capture” [Zhao et al. 2006]), respectively. With this “high throughput” approach, it has become possible to determine genome wide nonrandom spatial interactions between specific DNA segments in cis and trans to a reference locus on a given CT (for review, see Dekker 2008). This method is based on in situ formaldehyde cross-linking of proximal DNA–protein interactions with a distance of several Ångström, subsequent fragmentation of cross linked DNA, recircularization (ligation) and amplification, and finally identification of these products by microarray techniques. To date, numerous spatial interactions of CTs were reported in cis and trans (Ling et al. 2006; Lomvardas et al. 2006; Babu et al. 2008). These assays demand strict controls to escape the danger of false positive or negative findings (Simonis et al. 2007). By combining this proximity based ligation assay with massive parallel sequencing (Hi-C), the construction of spatial proximity maps of an entire genome down to a resolution level of 1 Mb was recently achieved as shown for a human lymphoblastoid cell line (Lieberman-Aiden et al. 2009). These maps confirmed the presence of chromosome territories, the spatial proximity of small, gene dense chromosomes, and the spatial segregation of open and closed chromatin, a parameter that strongly correlates with gene density (Gilbert et al. 2004). At the megabase scale, a chromatin conformation consistent with a fractal globule was suggested.
High throughput assays with a superior resolution and microscopic approaches are complementary to further elucidate CT conformation and spatial interactions in trans (Simonis and de Laat 2008). Ligation techniques to date require millions of cells and their principle excludes an application at the single cell level. More important, microscopic approaches are the only way to reveal the entire structure of nuclear components and to determine their topography with respect to each other.
MODELS OF NUCLEAR ARCHITECTURE: OPEN QUESTIONS AND EXPERIMENTAL STRATEGIES TO ANSWER THEM
The territorial organization of interphase chromosomes is now generally accepted as a basic principle of nuclear organization in both animals (Cremer and Cremer 2001) and plants (Shaw et al. 2002; Pecinka et al. 2004; Berr et al. 2006), and may even hold for single-cell eukaryotes, such as budding and fission yeast (Bystricky et al. 2005; Molnar and Kleckner 2008). However, severe limitations in our present knowledge of the functional architecture of CTs and the nucleus at large become obvious when we consider currently proposed models (Fig. 4).
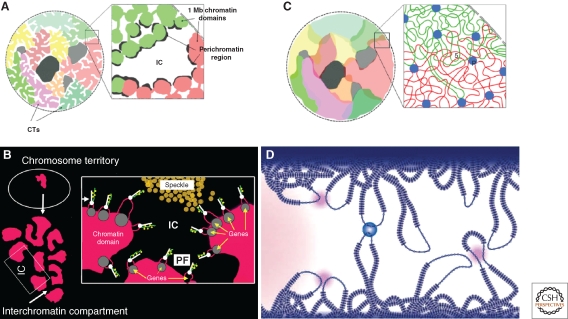
Figure 4.
Different models of nuclear architecture. (A) Chromosome territory‐interchromatin compartment (CT-IC) model (for description, see text). (B) Hypothetical view of the functional nuclear architecture according to the CT-IC model. Chromatin domains are considered the major constituents of a CT. The IC expands between these domains as a rather DNA free nuclear compartment carrying splicing speckles and nuclear bodies. The width of the IC space is highly variable depending on Brownian movements of chromatin domains and allowing transient contacts of domain surfaces in cis and trans. During ongoing transcription, genes are at least partially decondensed at any given time into the perichromatin region (PR) located at the domain periphery. Perichromatin fibrils (PF) are generated there. Each PF carries a nascent transcript (green) from a different gene. White dots with a line symbolize RNA Pol II molecules with their CTD domain, which may play a role in the structural organization of splicing events. Splicing speckles located in the IC provide the splicing factors to PFs, which also represent the structures in which cotranscriptional splicing occurs. (C) According to the interchromatin network (ICN) model (Branco and Pombo 2006), intermingling chromatin fibers/loops from the same CT, as well as from neighboring CTs, can make contact in cis and trans. Blue dots represent sites of intrachromosomal and interchromosomal contacts with unknown composition. Although there is extensive space between chromatin fibers/loops, this space should not be confused with the functional relationship of the IC and PR predicted by the CT-IC model. (D) Model suggested by Fraser and Bickmore (Fraser and Bickmore 2007, figure reprinted with permission from Macmillan Publishers Ltd). These authors review evidence arguing for the colocalization of genes in the nucleus for expression or coregulation. Transcription factories (dark pink) can recruit genes in cis and trans located on decondensed chromatin loops that extend outside chromosome territories. The pale pink area on the left represents a splicing-factor enriched speckle. The blue circle exemplifies an interaction for coregulation in trans, which can occur between regulatory elements and/or gene loci.
The chromosome territory-interchromatin compartment (CT-IC) model (Fig. 4A,B) argues that nuclei are built up from two principal components, chromosome territories (CTs) and the interchromatin compartment (IC). Individual CTs form an interconnected higher-order chromatin network (Visser et al. 2000; Albiez et al. 2006). According to the CT-IC model, this chromatin network is spatially associated with a second contiguous 3D spatial network, the interchromatin compartment (IC), which was observed both at the light and electron microscopic level (Albiez et al. 2006; Rouquette et al. 2009). The IC concept asserts a DNA free or at least largely free, contiguous space of channels, which start at the nuclear pores and expand as larger channels and lacunes between the higher-order chromatin network described earlier. The IC harbors splicing speckles and a variety of nonchromatin nuclear bodies (Verschure et al. 1999; Visser et al. 2000; Albiez et al. 2006). The IC concept evolved from the interchromosomal domain (ICD) concept originally proposed by Peter Lichter and co-workers (Zirbel et al. 1993), who defined the ICD as a network-like space expanding mainly around CTs with little penetration into the CT interior (Cremer et al. 1993). Supposedly, genes were preferentially transcribed in a region of decondensed chromatin at the CT periphery and RNA transcripts would be directly released into the ICD compartment. This concept was supported by a series of studies from this group (Bridger et al. 1998; Reichenzeller et al. 2000; Bridger et al. 2005; Richter et al. 2005). Accumulating evidence for genes transcribed both outside and in the interior of CTs (Cmarko et al. 1999; Verschure et al. 1999; Mahy et al. 2002; Kupper et al. 2007) is consistent with electron microscopic evidence for a (nearly) network-like DNA free space both outside and inside CTs (Visser et al. 2000; Rouquette et al. 2009). Based on this evidence, the hypothetical CT structure suggested by the CT-IC model can be compared with a sponge of chromatin permeated by intraterritorial IC channels (Fig. 4B). The entire IC is separated from the more condensed interior of chromatin domains and/or higher-order chromatin fibers by a thin (<200 nm) layer of rather decondensed chromatin, termed the perichromatin region (PR) (Fakan and van Driel 2007). EM evidence has supported the view that the PR topographically represents the utmost periphery of a given chromatin domain bordering the IC and functionally represents the major nuclear subcompartment for transcription, cotranscriptional RNA splicing (Fakan and Bernhard 1971; Cmarko et al. 1999; Trentani et al. 2003), as well as DNA replication (Jaunin and Fakan 2002) and possibly also DNA repair (Solimando et al. 2009). Transcription yields perichromatin fibrils, which are generated in the PR as nascent pre-mRNA transcripts of single genes complexed with hnRNPs and are served by nearby speckles with factors for cotranscriptional splicing.
The PR concept is a decisive part of the CT-IC model but has been disregarded by proponents of other models (Dehghani et al. 2005; Branco and Pombo 2006; Fraser and Bickmore 2007; Alberts et al. 2008; Fedorova and Zink 2008; Fedorova and Zink 2009). The “lattice” model of interphase chromatin proposed by Dehghani et al. (2005) suggests a lattice-like network of 10- and 30-nm fibers. This structure yields a porous organization of chromatin with fibers intermingling at the borders of neighboring CTs. The interchromatin network (ICN) model (Fig. 4C) (Branco and Pombo 2006) predicts that chromatin fibers and loops intermingle in a rather uniform way both in the interior of individual CTs and between differentially labeled neighboring CTs, making any distinction between the interior or periphery of distinct chromatin domains functionally meaningless. In this interchromatin network, loops may expand from one CT to meet loops from another CT. Arguably, active genes on decondensed chromatin loops that extend outside chromosome territories can colocalize both in cis in so called expression hubs (Kosak and Groudine 2004) or be transcribed at preassembled transcription factories (Fig. 4D) (Fraser and Bickmore 2007).
Some models emphasize the functional importance of giant chromatin loops, which emanate from chromosome territories (Chubb and Bickmore 2003; Fraser and Bickmore 2007). Supposedly, giant loops may even expand across the nuclear space (Alberts et al. 2008). According to these models, giant loops may carry genes to even very remote sites in the nuclear space for coregulation in an expression hub (Kosak and Groudine 2004). Alternatively, giant loops may transport genes to a remote repressive nuclear compartment (Alberts et al. 2008).
The lack of quantitative rigor with respect to the predicted numbers, length, and compaction levels of such fibers and their 3D distribution has limited the usefulness of the models described earlier. To overcome this limitation, chromatin polymer models have been developed, which make experimentally testable, quantitative predictions about functionally important features of the nuclear architecture, such as the expected size distribution of chromatin loops and chromatin compaction levels (van den Engh et al. 1992; Munkel and Langowski 1998; Munkel et al. 1999). A recent chromatin polymer model has assumed a broad range of loop sizes (Mateos-Langerak et al. 2009).
None of the present models for functional nuclear architecture is fully supported by compelling experimental evidence. In particular, two problems need urgent clarification: 1) the possible speed and extent of chromatin movements driven by Brownian motion in case of random walks and by unknown mechanisms in case of directed chromatin movements; 2) the topography of the major nuclear functions and the validation or experimental falsifications of the functional marriage between the IC and the PR as predicted by the CT-IC model (Fig. 4B) but not by other models (Fig. 4C, D). Although estimates of the number of transcription factories vary widely, the number of genes transcribed at any given time seems to be much larger. Accordingly, it has been suggested that a single transcription factory is able to transcribe several genes simultaneously in cis and trans and that an extraordinarily large part of the genome passes through the limited number of transcription factories in a cell nucleus (Chakalova et al. 2005; Sutherland and Bickmore 2009). As a consequence, one would also expect an extraordinary mobility of chromatin in the nucleus. In contrast, the concept of perichromatin fibrils argues that most transcription takes place in the PR and that simultaneous transcription in a single factory is rather the exception than the rule, if this concept can finally be proven (Sutherland and Bickmore 2009).
On the basis of FISH experiments with differentially labeled chromosome painting probes on cryosections (140–180 nm) from cell nuclei, Branco and Pombo (2006) detected zones of color overlap between pained CTs (Fig. 5A–C), which they interpreted as the result of intermingling chromatin fibers. Evidence for intermingling was substantiated by transmission electron microscopy with colloidal gold particles of different size for “intermingling” CTs. In control experiments, Branco and Pombo showed that the positions of gold grains reflecting the location of histone H2B molecules did not significantly change when cryosections were studied before and after mock FISH, excluding the possibility that chromatin intermingling was an artifact of chromatin denaturation. Examination of published electron micrographs does not, however, allow a morphological orientation in the section with regard to chromatin domains and other nuclear structures.
Figure 5.
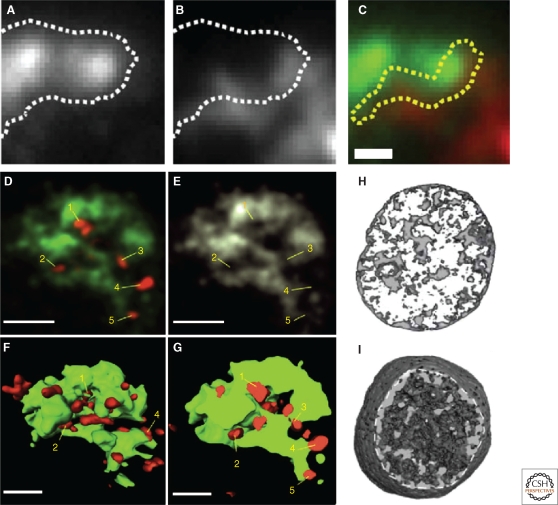
Experimental evidence against and in favor of an interchromatin space. Fluorescence microscopic images of a painted HSA 5 CT (A) and HSA 7 CT (B) in a cryosection of approximately 150 nm thickness from a human lymphocyte nucleus (Branco and Pombo 2006). (C) Overlap of images A and B shows the two CTs in different colors. The area of assumed intermingling between the two CTs is delineated by the yellow line. (D–G) Topographical relationships between double minute (DM) chromosomes carrying active MYCN genes (red) and the painted 3q-arm domain (green) observed in a nucleus of the human neuroblastoma cell HDN-16 (for details, see Solovei et al. 2000). (D) Laser confocal section. DMs denoted by 1–5 are located both at the periphery of the 3q-arm domain and in channel-like invaginations of the interchromatin compartment (IC) penetrating into the interior of the arm domain. (E) IC invaginations are emphasized in this gray image of the same 3q-arm domain section. Numbers indicate the location of DMs 1–5 in D. (F–G) 3D reconstruction of the entire 3q-domain (green) with all associated DMs (red). (F) _top_-view and (G) section through the reconstructed domain. (H,I) 3D reconstruction of a rat liver cell nucleus specifically stained for DNA and imaged by Serial Block-Face—Scanning Electron Microscopy (Rouquette et al. 2009). (H) A reconstructed mid part of the nucleus with 250-nm thickness shows a strongly inhomogeneous distribution of DNA in higher-order chromatin clusters (gray) and the wide mostly DNA free interchromatin compartment (white) expanding between these clusters. (I) Reconstruction of the major part of the nucleus. The dotted line indicates the removal of the nuclear periphery exposed to the viewer. Here, the nuclear volume seems to be entirely filled with chromatin, most likely—as we suggest—as a consequence of the sponge-like organization of CTs built up from interconnected chromatin domains/bundles permeated by the IC.
Controversial issues both with respect to CT structure and the interchromatin space are in part because of the limited resolution of present light microscopy techniques and the difficulties to obtain 3D data sets by electron microscopy. Recent developments of laser-based light microscopic techniques with ultra-high resolution has now opened possibilities for light optical nanoscopy, which will help to overcome these limitations (Hell 2007; Schermelleh et al 2008; Gunkel et al. 2009). The combination of a focused ion beam with high resolution scanning electron microscopy is an additional, very promising tool for 3D analysis of chromosome architecture (Schroeder-Reiter et al. 2009).
ACKNOWLEDGMENTS
This work was supported by DFG (Cr 59/28-1) and CIPSM. The authors are grateful to the many colleagues of the group of the Cremer laboratory who contributed with their experiments, ideas, and enthusiasm to unravel the nature of CTs.
Footnotes
Editors: David Spector and Tom Misteli
REFERENCES
- Abney JR, Cutler B, Fillbach ML, Axelrod D, Scalettar BA 1997. Chromatin dynamics in interphase nuclei and its implications for nuclear structure. J Cell Biol 137:1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar A, Gasser SM 2007. The nuclear envelope and transcriptional control. Nat Rev Genet 8:507–517 [DOI] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P 2008. Molecular biology of the cell Garland Science, New York [Google Scholar]
- Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Kupper K, Joffe B, Thormeyer T, von Hase J, et al. 2006. Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res 14:707–733 [DOI] [PubMed] [Google Scholar]
- Babu DA, Chakrabarti SK, Garmey JC, Mirmira RG 2008. Pdx1 and β2/NeuroD1 participate in a transcriptional complex that mediates short-range DNA looping at the insulin gene. J Biol Chem 283:8164–8172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr ML, Bertram EG 1949. A morphological distinction between neurons of the male and female, and the behavior of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163:676–677 [DOI] [PubMed] [Google Scholar]
- Bartkuhn M, Renkawitz R 2008. Long range chromatin interactions involved in gene regulation. Biochim Biophys Acta 1783:2161–2166 [DOI] [PubMed] [Google Scholar]
- Belmont AS, Bruce K 1994. Visualization of G1 chromosomes: A folded, twisted, supercoiled chromonema model of interphase chromatid structure. J Cell Biol 127:287–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Malyavantham KS, Pliss A, Bhattacharya S, Acharya R 2005. Spatio-temporal dynamics of genomic organization and function in the mammalian cell nucleus. Adv Enzyme Regul 45:17–26 [DOI] [PubMed] [Google Scholar]
- Berr A, Pecinka A, Meister A, Kreth G, Fuchs J, Blattner FR, Lysak MA, Schubert I 2006. Chromosome arrangement and nuclear architecture but not centromeric sequences are conserved between Arabidopsis thaliana and Arabidopsis lyrata. Plant J 48:771–783 [DOI] [PubMed] [Google Scholar]
- Bolzer A, Craig JM, Cremer T, Speicher MR 1999. A complete set of repeat-depleted, PCR-amplifiable, human chromosome-specific painting probes. Cytogenet Cell Genet 84:233–240 [DOI] [PubMed] [Google Scholar]
- Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauth C, Muller S, Eils R, Cremer C, Speicher MR, et al. 2005. Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes. PLoS Biol 3:e157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden J, Manuelidis L 1988. Movement of the X chromosome in epilepsy. Science 242:1687–1691 [DOI] [PubMed] [Google Scholar]
- Bornfleth H, Edelmann P, Zink D, Cremer T, Cremer C 1999. Quantitative motion analysis of subchromosomal foci in living cells using four-dimensional microscopy. Biophys J 77:2871–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev AM, Mikhaylova LM, Nurminsky DI 2005. The pattern of chromosome folding in interphase is outlined by the linear gene density profile. Mol Cell Biol 25:8379–8386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boveri T 1909. Die Blastomerenkerne von Ascaris megalocephala und die Theorie der Chromosomenindividualität. Arch Zellforsch 3:181–268 [Google Scholar]
- Boyle S, Gilchrist S, Bridger JM, Mahy NL, Ellis JA, Bickmore WA 2001. The spatial organization of human chromosomes within the nuclei of normal and emerin-mutant cells. Hum Mol Genet 10:211–219 [DOI] [PubMed] [Google Scholar]
- Branco MR, Pombo A 2006. Intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol 4:e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brianna Caddle L, Grant JL, Szatkiewicz J, van Hase J, Shirley BJ, Bewersdorf J, Cremer C, Arneodo A, Khalil A, Mills KD 2007. Chromosome neighborhood composition determines translocation outcomes after exposure to high-dose radiation in primary cells. Chromosome Res 15:1061–1073 [DOI] [PubMed] [Google Scholar]
- Bridger JM, Herrmann H, Munkel C, Lichter P 1998. Identification of an interchromosomal compartment by polymerization of nuclear-targeted vimentin. J Cell Sci 111:1241–1253 [DOI] [PubMed] [Google Scholar]
- Bridger JM, Kalla C, Wodrich H, Weitz S, King JA, Khazaie K, Krausslich HG, Lichter P 2005. Nuclear RNAs confined to a reticular compartment between chromosome territories. Exp Cell Res 302:180–193 [DOI] [PubMed] [Google Scholar]
- Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM 2005. Chromosome looping in yeast: telomere pairing and coordinated movement reflect anchoring efficiency and territorial organization. J Cell Biol 168:375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli G 2007. Chromosome kissing. Curr Opin Genet Dev 17:443–450 [DOI] [PubMed] [Google Scholar]
- Chakalova L, Debrand E, Mitchell JA, Osborne CS, Fraser P 2005. Replication and transcription: shaping the landscape of the genome. Nat Rev Genet 6:669–677 [DOI] [PubMed] [Google Scholar]
- Chambeyron S, Da Silva NR, Lawson KA, Bickmore WA 2005. Nuclear re-organisation of the Hoxb complex during mouse embryonic development. Development 132:2215–2223 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Bickmore WA 2003. Considering nuclear compartmentalization in the light of nuclear dynamics. Cell 112:403–406 [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hall LL, Byron M, McNeil J, Lawrence JB 2006. The X chromosome is organized into a gene-rich outer rim and an internal core containing silenced nongenic sequences. Proc Natl Acad Sci 103:7688–7693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu XD, van Driel R, Fakan S 1999. Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell 10:211–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer C, Rappold G, Gray JW, Muller CR, Ropers HH 1984a. Preparative dual-beam sorting of the human Y chromosome and in situ hybridization of cloned DNA probes. Cytometry 5:572–579 [DOI] [PubMed] [Google Scholar]
- Cremer M, Grasser F, Lanctot C, Muller S, Neusser M, Zinner R, Solovei I, Cremer T 2008. Multicolor 3D fluorescence in situ hybridization for imaging interphase chromosomes. Methods Mol Biol 463:205–239 [DOI] [PubMed] [Google Scholar]
- Cremer M, Kupper K, Wagler B, Wizelman L, von Hase J, Weiland Y, Kreja L, Diebold J, Speicher MR, Cremer T 2003. Inheritance of gene density-related higher order chromatin arrangements in normal and tumor cell nuclei. J Cell Biol 162:809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremer M, von Hase J, Volm T, Brero A, Kreth G, Walter J, Fischer C, Solovei I, Cremer C, Cremer T 2001. Non-random radial higher-order chromatin arrangements in nuclei of diploid human cells. Chromosome Res 9:541–567 [DOI] [PubMed] [Google Scholar]
- Cremer T, Baumann H, Nakanishi K, Cremer C 1984b. Correlation between interphase and metaphase chromosome arrangements as studied by laser-uv-microbeam experiments. Chromosomes Today 8:203–212 [Google Scholar]
- Cremer T, Cremer C 2001. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2:292–301 [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C 2006a. Rise, fall and resurrection of chromosome territories: a historical perspective. Part I. The rise of chromosome territories. Eur J Histochem 50:161–176 [PubMed] [Google Scholar]
- Cremer T, Cremer C 2006b. Rise, fall and resurrection of chromosome territories: a historical perspective. Part II. Fall and resurrection of chromosome territories during the 1950s to 1980s. Part III. Chromosome territories and the functional nuclear architecture: experiments and models from the 1990s to the present. Eur J Histochem 50:223–272 [PubMed] [Google Scholar]
- Cremer T, Cremer C, Baumann H, Luedtke EK, Sperling K, Teuber V, Zorn C 1982a. Rabl’s model of the interphase chromosome arrangement tested in Chinese hamster cells by premature chromosome condensation and laser-UV-microbeam experiments. Hum Genet 60:46–56 [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C, Schneider T, Baumann H, Hens L, Kirsch-Volders M 1982b. Analysis of chromosome positions in the interphase nucleus of Chinese hamster cells by laser-UV-microirradiation experiments. Hum Genet 62:201–209 [DOI] [PubMed] [Google Scholar]
- Cremer T, Kurz A, Zirbel R, Dietzel S, Rinke B, Schrock E, Speicher MR, Mathieu U, Jauch A, Emmerich P, et al. 1993. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harb Symp Quant Biol 58:777–792 [DOI] [PubMed] [Google Scholar]
- Cremer T, Lichter P, Borden J, Ward DC, Manuelidis L 1988. Detection of chromosome aberrations in metaphase and interphase tumor cells by in situ hybridization using chromosome-specific library probes. Hum Genet 80:235–246 [DOI] [PubMed] [Google Scholar]
- Croft JA, Bridger JM, Boyle S, Perry P, Teague P, Bickmore WA 1999. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol 145:1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvackova Z, Masata M, Stanek D, Fidlerova H, Raska I 2009. Chromatin position in human HepG2 cells: although being non-random, significantly changed in daughter cells. J Struct Biol 165:107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehghani H, Dellaire G, Bazett-Jones DP 2005. Organization of chromatin in the interphase mammalian cell. Micron 36:95–108 [DOI] [PubMed] [Google Scholar]
- Dekker J 2008. Gene regulation in the third dimension. Science 319:1793–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N 2002. Capturing chromosome conformation. Science 295:1306–1311 [DOI] [PubMed] [Google Scholar]
- Deniaud E, Bickmore WA 2009. Transcription and the nuclear periphery: Edge of darkness? Curr Opin Genet Dev 19:187–191 [DOI] [PubMed] [Google Scholar]
- Dietzel S, Jauch A, Kienle D, Qu G, Holtgreve-Grez H, Eils R, Munkel C, Bittner M, Meltzer PS, Trent JM, et al. 1998. Separate and variably shaped chromosome arm domains are disclosed by chromosome arm painting in human cell nuclei. Chromosome Res 6:25–33 [DOI] [PubMed] [Google Scholar]
- Edelmann P, Bornfleth H, Zink D, Cremer T, Cremer C 2001. Morphology and dynamics of chromosome territories in living cells. Biochim Biophys Acta 1551:M29–39 [DOI] [PubMed] [Google Scholar]
- Eils R, Dietzel S, Bertin E, Schrock E, Speicher MR, Ried T, Robert-Nicoud M, Cremer C, Cremer T 1996. Three-dimensional reconstruction of painted human interphase chromosomes: Active and inactive X chromosome territories have similar volumes but differ in shape and surface structure. J Cell Biol 135:1427–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakan S, Bernhard W 1971. Localisation of rapidly and slowly labelled nuclear RNA as visualized by high resolution autoradiography. Exp Cell Res 67:129–141 [DOI] [PubMed] [Google Scholar]
- Fakan S, van Driel R 2007. The perichromatin region: A functional compartment in the nucleus that determines large-scale chromatin folding. Semin Cell Dev Biol 18:676–681 [DOI] [PubMed] [Google Scholar]
- Fawcett JJ, Longmire JL, Martin JC, Deaven LL, Cram LS 1994. Large-scale chromosome sorting. Methods Cell Biol 42:319–330 [DOI] [PubMed] [Google Scholar]
- Federico C, Scavo C, Cantarella CD, Motta S, Saccone S, Bernardi G 2006. Gene-rich and gene-poor chromosomal regions have different locations in the interphase nuclei of cold-blooded vertebrates. Chromosoma 115:123–128 [DOI] [PubMed] [Google Scholar]
- Fedorova E, Zink D 2008. Nuclear architecture and gene regulation. Biochim Biophys Acta 1783:2174–2184 [DOI] [PubMed] [Google Scholar]
- Fedorova E, Zink D 2009. Nuclear genome organization: common themes and individual patterns. Curr Opin Genet Dev 19:166–171 [DOI] [PubMed] [Google Scholar]
- Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, Ylstra B, Chubb JR, Bickmore WA 2008. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet 4:e1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser P, Bickmore W 2007. Nuclear organization of the genome and the potential for gene regulation. Nature 447:413–417 [DOI] [PubMed] [Google Scholar]
- Fuks F 2005. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev 15:490–495 [DOI] [PubMed] [Google Scholar]
- Gerlich D, Beaudouin J, Kalbfuss B, Daigle N, Eils R, Ellenberg J 2003. Global chromosome positions are transmitted through mitosis in mammalian cells. Cell 112:751–764 [DOI] [PubMed] [Google Scholar]
- Gilbert N, Boyle S, Fiegler H, Woodfine K, Carter NP, Bickmore WA 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118:555–566 [DOI] [PubMed] [Google Scholar]
- Goetze S, Mateos-Langerak J, Gierman HJ, de Leeuw W, Giromus O, Indemans MH, Koster J, Ondrej V, Versteeg R, van Driel R 2007. The three-dimensional structure of human interphase chromosomes is related to the transcriptome map. Mol Cell Biol 27:4475–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasser F, Neusser M, Fiegler H, Thormeyer T, Cremer M, Carter NP, Cremer T, Muller S 2008. Replication-timing-correlated spatial chromatin arrangements in cancer and in primate interphase nuclei. J Cell Sci 121:1876–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JW, Dean PN, Fuscoe JC, Peters DC, Trask BJ, van den Engh GJ, Van Dilla MA 1987. High-speed chromosome sorting. Science 238:323–329 [DOI] [PubMed] [Google Scholar]
- Gunkel M, Erdel F, Rippe K, Lemmer P, Kaufmann R, Hormann C, Amberger R, Cremer C 2009. Dual color localization microscopy of cellular nanostructures. Biotechnol J 4:927–938 [DOI] [PubMed] [Google Scholar]
- Habermann FA, Cremer M, Walter J, Kreth G, von Hase J, Bauer K, Wienberg J, Cremer C, Cremer T, Solovei I 2001. Arrangements of macro- and microchromosomes in chicken cells. Chromosome Res 9:569–584 [DOI] [PubMed] [Google Scholar]
- Heard E, Bickmore W 2007. The ins and outs of gene regulation and chromosome territory organisation. Curr Opin Cell Biol 19:311–316 [DOI] [PubMed] [Google Scholar]
- Hell SW 2007. Far-field optical nanoscopy. Science 316:1153–1158 [DOI] [PubMed] [Google Scholar]
- Hepperger C, Mannes A, Merz J, Peters J, Dietzel S 2008. Three-dimensional positioning of genes in mouse cell nuclei. Chromosoma 117:535–551 [DOI] [PubMed] [Google Scholar]
- Jackson DA, Pombo A 1998. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140:1285–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaunin F, Fakan S 2002. DNA replication and nuclear architecture. J Cell Biochem 85:1–9 [PubMed] [Google Scholar]
- Jiang C, Pugh BF 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10:161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmarova M, Smirnov E, Kovacik L, Popov A, Raska I 2008. Positioning of the NOR-bearing chromosomes in relation to nucleoli in daughter cells after mitosis. Physiol Res 57:421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A, Grant JL, Caddle LB, Atzema E, Mills KD, Arneodo A 2007. Chromosome territories have a highly nonspherical morphology and nonrandom positioning. Chromosome Res 15:899–916 [DOI] [PubMed] [Google Scholar]
- Kioussis D 2005. Gene regulation: Kissing chromosomes. Nature 435:579–580 [DOI] [PubMed] [Google Scholar]
- Koehler D, Zakhartchenko V, Froenicke L, Stone G, Stanyon R, Wolf E, Cremer T, Brero A 2009. Changes of higher order chromatin arrangements during major genome activation in bovine preimplantation embryos. Exp Cell Res 315:2053–2063 [DOI] [PubMed] [Google Scholar]
- Kosak ST, Groudine M 2004. Form follows function: The genomic organization of cellular differentiation. Genes Dev 18:1371–1384 [DOI] [PubMed] [Google Scholar]
- Kouzarides T 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- Kozubek S, Lukasova E, Jirsova P, Koutna I, Kozubek M, Ganova A, Bartova E, Falk M, Pasekova R 2002. 3D Structure of the human genome: order in randomness. Chromosoma 111:321–331 [DOI] [PubMed] [Google Scholar]
- Kumaran RI, Spector DL 2008. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol 180:51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupper K, Kolbl A, Biener D, Dittrich S, von Hase J, Thormeyer T, Fiegler H, Carter NP, Speicher MR, Cremer T, et al. 2007. Radial chromatin positioning is shaped by local gene density, not by gene expression. Chromosoma 116:285–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaSalle JM, Lalande M 1996. Homologous association of oppositely imprinted chromosomal domains. Science 272:725–728 [DOI] [PubMed] [Google Scholar]
- Lichter P, Cremer T, Borden J, Manuelidis L, Ward DC 1988a. Delineation of individual human chromosomes in metaphase and interphase cells by in situ suppression hybridization using recombinant DNA libraries. Hum Genet 80:224–234 [DOI] [PubMed] [Google Scholar]
- Lichter P, Cremer T, Tang CJ, Watkins PC, Manuelidis L, Ward DC 1988b. Rapid detection of human chromosome 21 aberrations by in situ hybridization. Proc Natl Acad Sci 85:9664–9668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326:289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312:269–272 [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R 2006. Interchromosomal interactions and olfactory receptor choice. Cell 126:403–413 [DOI] [PubMed] [Google Scholar]
- Lyon MF 1962. Sex chromatin and gene action in the mammalian X-chromosome. Am J Hum Genet 14:135–148 [PMC free article] [PubMed] [Google Scholar]
- Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R 1998. Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol 143:1415–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA 2002. Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J Cell Biol 157:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier VK, Chioda M, Becker PB 2008. ATP-dependent chromatosome remodeling. Biol Chem 389:345–352 [DOI] [PubMed] [Google Scholar]
- Manuelidis L 1985. Individual interphase chromosome domains revealed by in situ hybridization. Hum Genet 71:288–293 [DOI] [PubMed] [Google Scholar]
- Marshall WF, Straight A, Marko JF, Swedlow J, Dernburg A, Belmont A, Murray AW, Agard DA, Sedat JW 1997. Interphase chromosomes undergo constrained diffusional motion in living cells. Curr Biol 7:930–939 [DOI] [PubMed] [Google Scholar]
- Martou G, De Boni U 2000. Nuclear topology of murine, cerebellar Purkinje neurons: changes as a function of development. Exp Cell Res 256:131–139 [DOI] [PubMed] [Google Scholar]
- Mateos-Langerak J, Bohn M, de Leeuw W, Giromus O, Manders EM, Verschure PJ, Indemans MH, Gierman HJ, Heermann DW, van Driel R, et al. 2009. Spatially confined folding of chromatin in the interphase nucleus. Proc Natl Acad Sci 106:3812–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer R, Brero A, von Hase J, Schroeder T, Cremer T, Dietzel S 2005. Common themes and cell type specific variations of higher order chromatin arrangements in the mouse. BMC Cell Biol 6:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaburn KJ, Misteli T 2007. Cell biology: chromosome territories. Nature 445:379–781 [DOI] [PubMed] [Google Scholar]
- Molnar M, Kleckner N 2008. Examination of interchromosomal interactions in vegetatively growing diploid Schizosaccharomyces pombe cells by Cre/loxP site-specific recombination. Genetics 178:99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morey C, Da Silva NR, Perry P, Bickmore WA 2007. Nuclear reorganisation and chromatin decondensation are conserved, but distinct, mechanisms linked to Hox gene activation. Development 134:909–919 [DOI] [PubMed] [Google Scholar]
- Morey C, Kress C, Bickmore WA 2009. Lack of bystander activation shows that localization exterior to chromosome territories is not sufficient to up-regulate gene expression. Genome Res 19:1184–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munkel C, Eils R, Dietzel S, Zink D, Mehring C, Wedemann G, Cremer T, Langowski J 1999. Compartmentalization of interphase chromosomes observed in simulation and experiment. J Mol Biol 285:1053–1065 [DOI] [PubMed] [Google Scholar]
- Munkel C, Langowski J 1998. Chromosome structure predicted by a polymer model. Phys Rev E57:5888–5896 [Google Scholar]
- Murmann AE, Gao J, Encinosa M, Gautier M, Peter ME, Eils R, Lichter P, Rowley JD 2005. Local gene density predicts the spatial position of genetic loci in the interphase nucleus. Exp Cell Res 311:14–26 [DOI] [PubMed] [Google Scholar]
- Neusser M, Schubel V, Koch A, Cremer T, Muller S 2007. Evolutionarily conserved, cell type and species-specific higher order chromatin arrangements in interphase nuclei of primates. Chromosoma 116:307–320 [DOI] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Misteli T 2004. Tissue-specific spatial organization of genomes. Genome Biol 5:R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parada LA, McQueen PG, Munson PJ, Misteli T 2002. Conservation of relative chromosome positioning in normal and cancer cells. Curr Biol 12:1692–1697 [DOI] [PubMed] [Google Scholar]
- Pecinka A, Schubert V, Meister A, Kreth G, Klatte M, Lysak MA, Fuchs J, Schubert I 2004. Chromosome territory arrangement and homologous pairing in nuclei of Arabidopsis thaliana are predominantly random except for NOR-bearing chromosomes. Chromosoma 113:258–269 [DOI] [PubMed] [Google Scholar]
- Pinkel D, Landegent J, Collins C, Fuscoe J, Segraves R, Lucas J, Gray J 1988. Fluorescence in situ hybridization with human chromosome-specific libraries: detection of trisomy 21 and translocations of chromosome 4. Proc Natl Acad Sci 85:9138–9142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C 1885. Über Zelltheilung. Morph Jb 10:214–330 [Google Scholar]
- Reddy KL, Zullo JM, Bertolino E, Singh H 2008. Transcriptional repression mediated by repositioning of genes to the nuclear lamina. Nature 452:243–247 [DOI] [PubMed] [Google Scholar]
- Reichenzeller M, Burzlaff A, Lichter P, Herrmann H 2000. In vivo observation of a nuclear channel-like system: evidence for a distinct interchromosomal domain compartment in interphase cells. J Struct Biol 129:175–185 [DOI] [PubMed] [Google Scholar]
- Richter K, Reichenzeller M, Gorisch SM, Schmidt U, Scheuermann MO, Herrmann H, Lichter P 2005. Characterization of a nuclear compartment shared by nuclear bodies applying ectopic protein expression and correlative light and electron microscopy. Exp Cell Res 303:128–137 [DOI] [PubMed] [Google Scholar]
- Roix JJ, McQueen PG, Munson PJ, Parada LA, Misteli T 2003. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet 34:287–291 [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Baddeley D, Scheipl F, Verveer PJ, Burkhardt H, Cremer C, Fahrmeir L, Cremer T, Joffe B 2008. Spatial quantitative analysis of fluorescently labeled nuclear structures: Problems, methods, pitfalls. Chromosome Res 16:523–562 [DOI] [PubMed] [Google Scholar]
- Rouquette J, Genoud C, Vazquez-Nin GH, Kraus B, Cremer T, Fakan S 2009. Revealing the high-resolution three-dimensional network of chromatin and interchromatin space: A novel electron-microscopic approach to reconstructing nuclear architecture. Chromosome Res 17:801–810 [DOI] [PubMed] [Google Scholar]
- Schardin M, Cremer T, Hager HD, Lang M 1985. Specific staining of human chromosomes in Chinese hamster x man hybrid cell lines demonstrates interphase chromosome territories. Hum Genet 71:281–287 [DOI] [PubMed] [Google Scholar]
- Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, et al. 2008. Subdiffraction multicolor imaging of the nuclear periphery with 3D structured illumination microscopy. Science 320:1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R 2007. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 21:3027–3043 [DOI] [PubMed] [Google Scholar]
- Schroeder-Reiter E, Perez-Willard F, Zeile U, Wanner G 2009. Focused ion beam (FIB) combined with high resolution scanning electron microscopy: A promising tool for 3D analysis of chromosome architecture. J Struct Biol 165:97–106 [DOI] [PubMed] [Google Scholar]
- Shaw PJ, Abranches R, Paula Santos A, Beven AF, Stoger E, Wegel E, González-Melendi P 2002. The architecture of interphase chromosomes and nucleolar transcription sites in plants. J Struct Biol 140:31–38 [DOI] [PubMed] [Google Scholar]
- Shopland LS, Lynch CR, Peterson KA, Thornton K, Kepper N, Hase J, Stein S, Vincent S, Molloy KR, Kreth G, et al. 2006. Folding and organization of a contiguous chromosome region according to the gene distribution pattern in primary genomic sequence. J Cell Biol 174:27–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis M, de Laat W 2008. FISH-eyed and genome-wide views on the spatial organisation of gene expression. Biochim Biophys Acta 1783:2052–2060 [DOI] [PubMed] [Google Scholar]
- Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet 38:1348–1354 [DOI] [PubMed] [Google Scholar]
- Simonis M, Kooren J, de Laat W 2007. An evaluation of 3C-based methods to capture DNA interactions. Nat Methods 4:895–901 [DOI] [PubMed] [Google Scholar]
- Solimando L, Luijsterburg MS, Vecchio L, Vermeulen W, van Driel R, Fakan S 2009. Spatial organization of nucleotide excision repair proteins after UV-induced DNA damage in the human cell nucleus. J Cell Sci 122:83–91 [DOI] [PubMed] [Google Scholar]
- Solovei I, Cavallo A, Schermelleh L, Jaunin F, Scasselati C, Cmarko D, Cremer C, Fakan S, Cremer T 2002. Spatial preservation of nuclear chromatin architecture during three-dimensional fluorescence in situ hybridization (3D-FISH). Exp Cell Res 276:10–23 [DOI] [PubMed] [Google Scholar]
- Stack SM, Brown DB, Dewey WC 1977. Visualization of interphase chromosomes. J Cell Sci 26:281–299 [DOI] [PubMed] [Google Scholar]
- Solovei I, Kreysing M, Lanctot C, Kosem S, Peichl L, Cremer T, Guck J, Joffe B 2009. Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137:356–368 [DOI] [PubMed] [Google Scholar]
- Strasburger E 1905. Die stofflichen Grundlagen der Vererbung im organischen Reich. Gustav Fischer, Jena, Germany [Google Scholar]
- Sutherland H, Bickmore WA 2009. Transcription factories: gene expression in unions? Nat Rev Genet 10:457–466 [DOI] [PubMed] [Google Scholar]
- Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, Gasser SM 2006. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 441:774–778 [DOI] [PubMed] [Google Scholar]
- Tanabe H, Muller S, Neusser M, von Hase J, Calcagno E, Cremer M, Solovei I, Cremer C, Cremer T 2002. Evolutionary conservation of chromosome territory arrangements in cell nuclei from higher primates. Proc Natl Acad Sci 99:4424–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telenius H, Pelmear AH, Tunnacliffe A, Carter NP, Behmel A, Ferguson-Smith MA, Nordenskjold M, Pfragner R, Ponder BA 1992. Cytogenetic analysis by chromosome painting using DOP-PCR amplified flow-sorted chromosomes. Genes Chromosomes Cancer 4:257–263 [DOI] [PubMed] [Google Scholar]
- Teller K, Solovei I, Buiting K, Horsthemke B, Cremer T 2007. Maintenance of imprinting and nuclear architecture in cycling cells. Proc Natl Acad Sci 104:14970–14975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson I, Gilchrist S, Bickmore WA, Chubb JR 2004. The radial positioning of chromatin is not inherited through mitosis but is established de novo in early G1. Curr Biol 14:166–172 [DOI] [PubMed] [Google Scholar]
- Trentani A, Testillano PS, Risueno MC, Biggiogera M 2003. Visualization of transcription sites at the electron microscope. Eur J Histochem 47:195–200 [DOI] [PubMed] [Google Scholar]
- Van den Engh G, Sachs R, Trask BJ 1992. Estimating genomic distance from DNA sequence location in cell nuclei by a random walk model. Science 257:1410–1412 [DOI] [PubMed] [Google Scholar]
- Verschure PJ, van Der Kraan I, Manders EM, van Driel R 1999. Spatial relationship between transcription sites and chromosome territories. J Cell Biol 147:13–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser AE, Aten JA 1999. Chromosomes as well as chromosomal subdomains constitute distinct units in interphase nuclei. J Cell Sci 112:3353–3360 [DOI] [PubMed] [Google Scholar]
- Visser AE, Jaunin F, Fakan S, Aten JA 2000. High resolution analysis of interphase chromosome domains. J Cell Sci 113:2585–2593 [DOI] [PubMed] [Google Scholar]
- Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, et al. 2000. Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci 113:1565–1576 [DOI] [PubMed] [Google Scholar]
- Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T 2003. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol 160:685–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RR, Broad S, Sheer D, Ragoussis J 2002. Subchromosomal positioning of the epidermal differentiation complex (EDC) in keratinocyte and lymphoblast interphase nuclei. Exp Cell Res 272:163–175 [DOI] [PubMed] [Google Scholar]
- Wischnitzer S 1973. The submicroscopic morphology of the interphase nucleus. Int Rev Cytol 34:1–48 [DOI] [PubMed] [Google Scholar]
- Zeitz MJ, Mukherjee L, Bhattacharya S, Xu J, Berezney R 2009. A probabilistic model for the arrangement of a subset of human chromosome territories in WI38 Human fibroblasts. J Cell Physiol 221:120–129 [DOI] [PubMed] [Google Scholar]
- Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, et al. 2006. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet 38:1341–1347 [DOI] [PubMed] [Google Scholar]
- Zink D, Cremer T, Saffrich R, Fischer R, Trendelenburg MF, Ansorge W, Stelzer EH 1998. Structure and dynamics of human interphase chromosome territories in vivo. Hum Genet 102:241–251 [DOI] [PubMed] [Google Scholar]
- Zirbel RM, Mathieu UR, Kurz A, Cremer T, Lichter P 1993. Evidence for a nuclear compartment of transcription and splicing located at chromosome domain boundaries. Chromosome Res 1:93–106 [DOI] [PubMed] [Google Scholar]
- Zorn C, Cremer C, Cremer T, Zimmer J 1979. Unscheduled DNA synthesis after partial UV irradiation of the cell nucleus. Distribution in interphase and metaphase. Exp Cell Res 124:111–119 [DOI] [PubMed] [Google Scholar]
- Zorn C, Cremer T, Cremer C, Zimmer J 1976. Laser UV microirradiation of interphase nuclei and post-treatment with caffeine. A new approach to establish the arrangement of interphase chromosomes. Hum Genet 35:83–89 [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E, Cavalli G 2007. Combinatorial epigenetics, “junk DNA”, and the evolution of complex organisms. Gene 390:232–242 [DOI] [PubMed] [Google Scholar]