Structure of the histone chaperone CIA/ASF1–double bromodomain complex linking histone modifications and site-specific histone eviction (original) (raw)
Abstract
Nucleosomes around the promoter region are disassembled for transcription in response to various signals, such as acetylation and methylation of histones. Although the interactions between histone-acetylation-recognizing bromodomains and factors involved in nucleosome disassembly have been reported, no structural basis connecting histone modifications and nucleosome disassembly has been obtained. Here, we determined at 3.3 Å resolution the crystal structure of histone chaperone cell cycle gene 1 (CCG1) interacting factor A/antisilencing function 1 (CIA/ASF1) in complex with the double bromodomain in the CCG1/TAF1/TAF(II)250 subunit of transcription factor IID. Structural, biochemical, and biological studies suggested that interaction between double bromodomain and CIA/ASF1 is required for their colocalization, histone eviction, and pol II entry at active promoter regions. Furthermore, the present crystal structure has characteristics that can connect histone acetylation and CIA/ASF1-mediated histone eviction. These findings suggest that the molecular complex between CIA/ASF1 and the double bromodomain plays a key role in site-specific histone eviction at active promoter regions. The model we propose here is the initial structure-based model of the biological signaling from histone modifications to structural change of the nucleosome (hi-MOST model).
Keywords: chromatin, transcription, transcription factor IID, x-ray crystallography
The nucleosome, a fundamental repeating unit of chromatin, consists of approximately 200 base pairs of DNA and a histone octamer, which is composed of two copies each of four histone proteins (H2A, H2B, H3, and H4) (1–4). Since the associations of the DNA with the histones within the nucleosome make it difficult to access the DNA by protein molecules, the nucleosome structure should be disassembled before nuclear events such as transcription, DNA replication, and DNA repair (5–7). Histone eviction from the sites of the reactions is therefore an essential and critical process to these nuclear events (8–11) and is mainly mediated by histone chaperones in the eukaryotic nucleus (8–10).
Histone chaperone CIA/ASF1 [cell cycle gene 1 (CCG1)-interacting factor A or antisilencing function 1] is the most conserved histone chaperone in eukaryotes (12–14). Biochemical and biological studies have revealed a wide variety of CIA/ASF1 functions (8–10) in transcription (15–20), DNA replication (12, 13, 15, 21, 22), and DNA repair (12, 13, 22–24). The variety of biological functions of CIA/ASF1 has been attributed to diverse macromolecular-histone-chaperone regulatory complexes containing CIA/ASF1 (25). For example, the CIA/ASF1 activities of the DNA-replication-dependent and -independent nucleosome structural change require complex formations with chromatin assembly factor-1 (CAF-1) and histone regulatory homolog A (HIRA) proteins, respectively (8–10, 21). Interaction with other factor(s) is likely to cause additional biological activity of CIA/ASF1.
One of the critical biological functions of CIA/ASF1 involves activity in the transcription initiation step. Biological analyses have revealed that CIA/ASF1 is required for site-specific histone eviction and/or the entry of RNA polymerase II (pol II) to the transcription starting point at various promoter regions (17–19, 26, 27). The molecular mechanism underlying histone eviction from the nucleosome by CIA/ASF1 was proposed on the basis of its histone (H3–H4)2 tetramer-disrupting activity in vitro (28) and its crystal structure in complex with the histone-H3–H4 dimer (28, 29). The site-specific histone eviction at the transcription initiation process generally seems to be activated by the histone acetylations around the promoter region (30–32). Despite the importance and generality of this process, the molecule that recruits CIA/ASF1 to the promoter region(s) and the molecular mechanism connecting histone acetylations and the CIA/ASF1-mediated histone eviction remain elusive.
We previously revealed that CIA/ASF1 physically and genetically interacts with the double bromodomain (DBD) in the CCG1/TAF1/TAF(II)250 subunit [DBD(CCG1)] of the general transcription initiation factor TFIID (16). Since DBD(CCG1) recognizes the acetylated histone tail (33), our previous results led us to hypothesize that DBD(CCG1) recruits CIA/ASF1 to active promoter regions (16). However, no direct evidence supporting this hypothesis has been obtained. To prove this hypothesis at the atomic level, we determined the crystal structure of histone chaperone CIA/ASF1 in complex with DBD(CCG1) and performed structure-based biochemical and biological analyses.
Results
Localization of CIA/ASF1 and DBD(CCG1) on the Promoters.
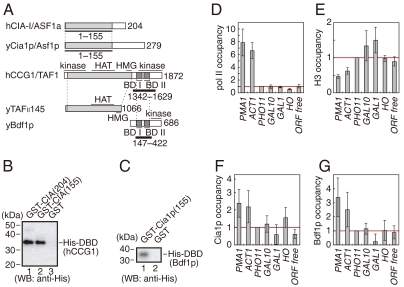
Of the two members of the CIA family—CIA-I/ASF1a and CIA-II/ASF1b—human CIA-I/ASF1a was utilized in the present study (14, 34). Although the two CIA/ASF1s are similar in primary sequences, their mRNA distributions are different. CIA-I/ASF1a mRNA is widely distributed throughout the body, while CIA-II/ASF1b mRNA is highly cell-specific (34). We used CIA-I/ASF1a for our analyses in order to elucidate the basic and general roles of CIA/ASF1 activity. The physical interaction between the core domain of human CIA-I/ASF1a [residues 1 to 155; CIA/ASF1(155)] and the human CCG1 double bromodomain [residues 1342 to 1629; DBD(hCCG1)] was shown by the GST pull-down assay (Fig. 1A and B). The physical interaction of their functional counterparts in _Saccharomyces cerevisiae_—Cia1p/Asf1p and Bdf1p, respectively—was also confirmed (Fig. 1C). In S. cerevisiae, Bdf2p has also been thought to functionally correspond to DBD(CCG1) (35–37). Since Bdf1p is mainly involved in the recognition of acetylated histones in yeast (36), Bdf1p was utilized in this analysis.
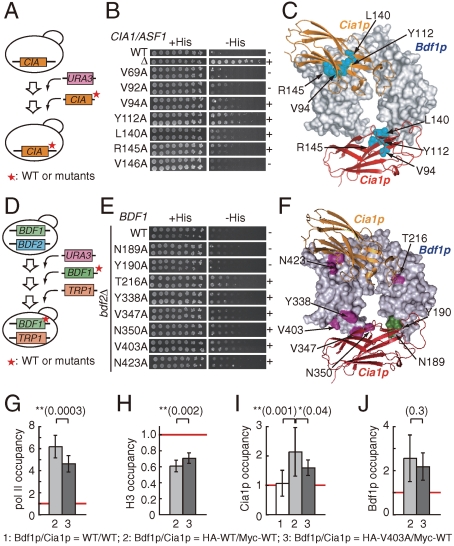
Fig. 1.
Physical and functional interactions between CIA/ASF1 and DBD(CCG1). (A) Schematic drawing of human CIA/ASF1, the CCG1 subunit containing DBD(hCCG1), and their yeast homologs, yCia1p/Asf1p, yTAFII145, and yBdf1p. The evolutionarily conserved N-terminal region (colored in gray) of CIA/ASF1 can bind the histone H3–H4 dimer (28, 29). BD stands for bromodomain. (B) Physical interaction between human GST-CIA/ASF1(155) and His-DBD(hCCG1). CIA/ASF1(155) and the full-length CIA/ASF1 [CIA/ASF1(204)] have nearly the same binding activities for DBD(hCCG1). (C) Physical interaction between yeast GST-Cia1p/Asf1p(155) and His-DBD(Bdf1p). (D_–_G) Relative occupancies of pol II (D), histone H3 (E), Cia1p/Asf1p (F), and Bdf1p (G) on the promoter regions of the PMA1, ACT1, PHO11, GAL10, GAL1, and HO genes as well as one ORF-free region. The occupancy of each protein on the PHO11 promoter region (indicated by a red line in each graph) was utilized as a reference.
To investigate the functional relationship between Cia1p/Asf1p and Bdf1p in vivo, localization of Cia1p/Asf1p and Bdf1p was examined by chromatin immunoprecipitation (ChIP) analysis using the promoter regions of the PMA1, ACT1, PHO11, GAL10, GAL1, and HO genes, as well as one ORF-free region. The occupancies of pol II, histone H3, Bdf1p, and Cia1p/Asf1p in the promoter regions of PHO11, GAL10, GAL1, and HO were comparable to those of the ORF-free region, suggesting that these genes were inactive in the present conditions (Fig. 1D_–_G). On the other hand, higher occupancy of pol II and lower occupancy of histone H3 were observed in the promoter regions of PMA1 and ACT1, suggesting that the genes are in a transcriptionally active state (Fig. 1D and E). In these promoter regions, significantly higher occupancies of Cia1p/Asf1p and Bdf1p were also observed (Fig. 1F and G). Both Cia1p/Asf1p and Bdf1p therefore seem to be localized in the promoter regions of transcriptionally active genes. Considering the fact that CIA/ASF1 physically interacts with DBD(CCG1) in both yeast and human (16), the CIA/ASF1–DBD(CCG1) complex could exist on active promoter regions.
Crystal Structure of the CIA/ASF1(155)–DBD(hCCG1) Complex.
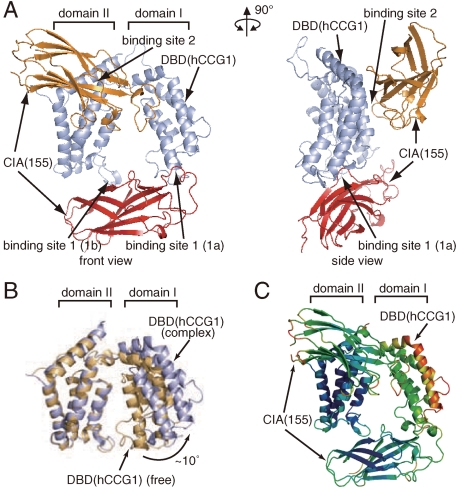
To analyze the functional implications of the CIA/ASF1–DBD(CCG1) complex at the atomic level, the crystal structure of the CIA/ASF1(155)–DBD(hCCG1) complex was determined at 3.3 Å resolution (Fig. 2A, Fig. S1, and Table S1). The crystal of the CIA/ASF1(155)–DBD(hCCG1) complex showed strong anisotropy. The crystal diffracted to more than 3.0 Å resolution along the c axis; the averaged I/σ(I) value for the reflections located around the _c_∗ axis (reflections with h + k < 10) was more than 25 between 3.1 and 3.0 Å resolution. However, the crystal diffracted to significantly lower resolution in the direction perpendicular to the c axis. When considering the reflections with l < 40, the averaged I/σ(I) value became less than 3 at more than 3.3 Å resolution. From these observations, we decided to refine the crystal structure at 3.3 Å resolution. Before the crystallographic refinement, the anisotropic scaling was applied to the diffraction data using the diffraction anisotropy server (38).
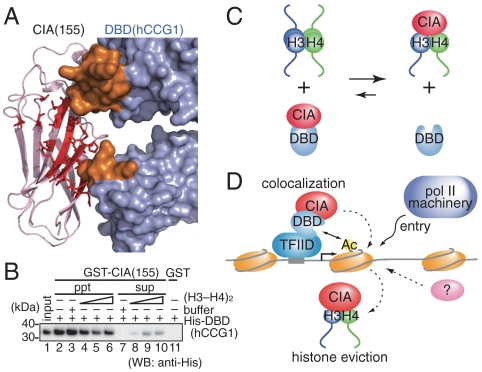
Fig. 2.
Crystal structure of the CIA/ASF1(155)–DBD(hCCG1) complex. (A) Crystal structure of the CIA/ASF1(155)–DBD(hCCG1) complex (front and side views). CIA/ASF1(155) at binding sites 1 and 2, and DBD(hCCG1) are shown in red, orange, and light blue, respectively. All molecular graphics were prepared by PyMOL (http://www.pymol.org). (B) Conformational change of DBD(hCCG1) induced by the CIA/ASF1(155) binding. The Cα atoms in domain II of DBD(hCCG1) were superposed by the least-square fittings. The DBD(hCCG1) of CIA/ASF1-free form (PDB ID code 1EQF) (33) and that in the present complex are shown in brown and light blue, respectively. (C) The result of the TLS-tensor analysis of the CIA/ASF1(155)–DBD(hCCG1) complex. Temperature factors calculated from the TLS tensors are displayed in the crystal structure of the CIA/ASF1(155)–DBD(hCCG1) complex (blue, low; red, high).
The crystal structure of the CIA/ASF1(155)–DBD(hCCG1) complex showed that one DBD(hCCG1) molecule binds two CIA/ASF1(155) molecules using two spatially separated regions, binding sites 1 and 2 (Fig. 2A). CIA/ASF1(155) showed no significant structural changes between the DBD(hCCG1) free and complex forms. However, DBD(hCCG1) underwent a global conformational change upon CIA/ASF1(155) binding; the CIA/ASF1 binding induced an increase of the angle between the principal axes of domains I and II by approximately 10° (Fig. 2B) (33), although no significant conformational changes were observed in either domain.
Physical Interaction Between CIA/ASF1(155) and DBD(hCCG1) in the Crystal.
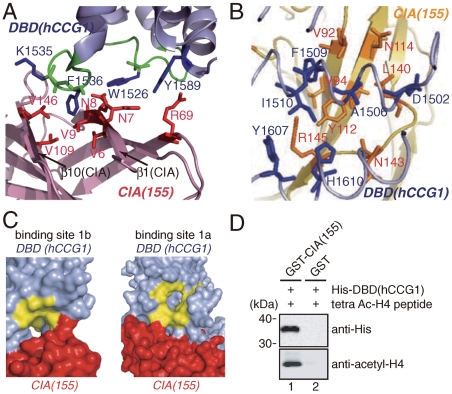
Binding site 1 is composed of two subsites, binding sites 1a and 1b, which are located at domains I and II of DBD(hCCG1), respectively (Fig. 2A and Table S2). Since the analysis of the TLS parameters used in the crystallographic refinement revealed that domain I of DBD(hCCG1) is more mobile than domain II (Fig. 2C), the interaction between CIA/ASF1(155) and binding site 1b of DBD(hCCG1) seems to be the main contributor to the interaction at binding site 1. At binding site 1b, Trp1526(DBD), Phe1536(DBD), and Tyr1589(DBD) are involved in the interaction with CIA/ASF1(155) (Fig. 3A). Trp1526(DBD) and Phe1536(DBD) are located at the ZA loop (1521–1538), and Tyr1589(DBD) is located at the αC helix (1588–1606). The region around the ZA and BC loops (BC loop: 1584–1587) forms a part of an acetylated-lysine-binding site (33). Binding site 2 is also composed of domains I and II of DBD(hCCG1). Residues in domain II of DBD(hCCG1) mainly interact with CIA/ASF1(155) (Table S3).
Fig. 3.
Interacting surface among CIA/ASF1(155), DBD(hCCG1), and acetylated-histone N tail. (A) Interaction between CIA/ASF1(155) (purple) and DBD(hCCG1) (light blue) at binding site 1b. The residues of CIA/ASF1(155) and DBD(hCCG1) are shown in red and blue, respectively. The main chain of the ZA loop is indicated by green. (B) Interaction between CIA/ASF1(155) (yellow) and DBD(hCCG1) (light blue) at binding site 2. The residues of CIA/ASF1(155) and DBD(hCCG1) are shown in orange and blue, respectively. (C) Surface representation of the binding sites 1b (Left) and 1a (Right) of DBD(hCCG1) (light blue) with CIA/ASF1(155) (red). The predicted acetylated lysine-binding residues on DBD(CCG1) (40) shown in yellow are mostly exposed to the solvent. (D) In vitro binding assay between DBD(hCCG1) and CIA/ASF1(155) in the presence of tetraacetylated histone H4 N-tail peptide (Ac-Lys5/8/12/16) (lane 1). The upper and lower images show the bound His-DBD(hCCG1) and tetraacetylated histone H4 N-tail peptide as detected by Western blotting analysis, respectively.
To interact with DBD(hCCG1), CIA/ASF1(155) mainly utilizes the two hydrophobic regions, the side-edge pocket and the concave surface of a β-sheet. The side-edge pocket of CIA/ASF1(155) accommodates Phe1536(DBD) in binding site 1 (Fig. 3A and Table S2), and Tyr112(CIA) in the concave surface of CIA/ASF1(155) interacts with a hydrophobic patch formed by Ala1506(DBD), Phe1509(DBD), Ile1510(DBD), and His1610(DBD) in binding site 2 (Fig. 3B and Table S3). These hydrophobic regions of CIA/ASF1(155) are also utilized in forming a complex with the histone H3–H4 dimer (28, 29); the side-edge pocket accommodates Phe100 of histone H4, and the concave surface of the β-sheet accommodates the α3 helix of histone H3. The interaction surfaces of CIA/ASF1(155) for the histone H3–H4 dimer and DBD(hCCG1) are overlapped, but are not exactly the same.
The present crystal structure showed that the acetylated-lysine-binding sites in both domains (33, 39, 40) are exposed to the solvent, suggesting that the CIA/ASF1(155)–DBD(hCCG1) complex is able to interact with an acetylated lysine (Fig. 3C). The biochemical analysis showed that a tetraacetylated histone H4 N-tail peptide coprecipitated with CIA/ASF1(155) and DBD(hCCG1) (Fig. 3D). Furthermore, the existence of an acetylated histone H4 N tail does not inhibit the interaction between CIA/ASF1(155) and DBD(hCCG1) (Fig. S2_A_ and B).
Physical Interaction Between CIA/ASF1(155) and DBD(hCCG1) in Solution.
The relatively small interacting surface areas of both binding sites on DBD(hCCG1) (818 _Å_2 and 719 _Å_2, Tables S2 and S3) suggested marginally weak interactions between CIA/ASF1(155) and DBD(hCCG1). Since we could not exclude the possibility of crystallographic artifacts of these interactions, further analyses of the interactions were performed in solution.
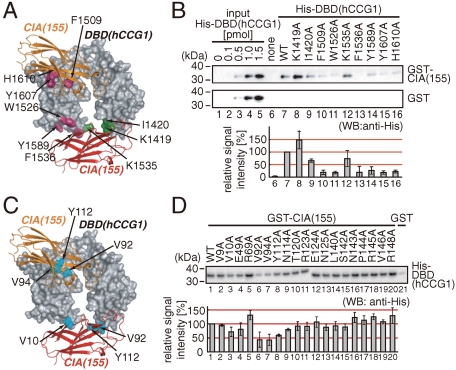
We analyzed the interaction between CIA/ASF1(155) and DBD(hCCG1) by the GST pull-down assay using 19 and 9 mutants for CIA/ASF1(155) and DBD(hCCG1), respectively. Residues for the mutations were selected from a list of residue pairs at distances of less than 4 Å (Tables S2 and S3). Among them, the residues whose side chains are involved in the interaction between the two molecules were substituted with alanine. The GST pull-down assay with the DBD(hCCG1) mutants showed that the mutations of residues at binding sites 1 (Trp1526Ala, Phe1536Ala, and Tyr1589Ala) and 2 (Phe1509Ala, Tyr1607Ala, and His1610Ala) of DBD(hCCG1) affected the interaction with CIA/ASF1(155) (Fig. 4A and B). The GST pull-down assay with the CIA/ASF1(155) mutants showed that the mutants Val10Ala, Val92Ala, Val94Ala, and Tyr112Ala of CIA/ASF1(155) had decreased interaction with DBD(hCCG1) (Fig. 4C and D). These amino acids of CIA/ASF1(155) are involved in the interaction with binding site 1 or 2, or both, of DBD(hCCG1) in the crystal structure (Fig. 4C). The present GST pull-down assays suggested that DBD(hCCG1) has two binding sites for CIA/ASF1(155) in solution.
Fig. 4.
In vitro physical interaction between CIA/ASF1(155) and DBD(hCCG1). (A) Positions of the mutations of DBD(hCCG1) used in the in vitro binding assay. Residues with and without significant effects from mutation are shown in violet and green, respectively. (B) Result of the GST pull-down assay using GST-CIA/ASF1(155) (WT: wild type) and His-DBD(hCCG1) (WT and mutants). Result of densitometric analysis showing means with error bars (SD) of triplicate experiments. (C) CIA/ASF1(155) residues showing significant effects from mutation are shown in cyan. (D) Result of the GST pull-down assay using GST-CIA/ASF1(155) (WT and mutants) and His-DBD(hCCG1) (WT). The result of densitometric analysis showing means with error bars (SD) of triplicate experiments.
Genetic Interaction Between CIA1/ASF1 and Bromodomain Factor (BDF).
To reveal the functional significance of the CIA/ASF1–DBD(CCG1) interaction observed in the crystal structure, the genetic interaction between CIA/ASF1 and DBD(CCG1) was analyzed at the single-residue level using S. cerevisiae. Although the functional significance of each protein can be examined in vivo by deleting a particular protein using a technique such as RNAi (41), this type of analysis cannot yield information about the functional significance of the interaction at the single-residue level.
The residues corresponding to those mutated in the in vitro binding analysis of CIA/ASF1(155) or DBD(hCCG1) were mutated in the yeast genetic analysis (Tables S4 and S5). Four mutations on Cia1p/Asf1p (Val94Ala, Tyr112Ala, Leu140Ala, and Arg145Ala) showed the Spt(-) phenotype (Fig. 5A_–_C). In the case of Bdf1p, four and two mutations of binding sites 1 (Tyr338Ala, Val347Ala, Asn350Ala, and Val403Ala) and 2 (Thr216Ala and Asn423Ala), respectively, showed the Spt(-) phenotype in _bdf2_-deletion mutant strains (Fig. 5D_–_F). These results suggested that Cia1p/Asf1p and Bdf1p have a genetic interaction through the residues located in the molecular interfaces of the CIA/ASF1–DBD(CCG1) complex (Fig. 5C and F).
Fig. 5.
In vivo functional interaction between CIA1/ASF1 and BDF1. (A) Construction of the cia1/asf1 point mutant strains for the Spt phenotype analysis. (B) The Spt phenotype analysis of cia1/asf1 point mutant strains. The mutants showing and not showing the Spt(-) phenotype are indicated by “+” and “-,” respectively. (C) Summary of the yeast genetic analyses of cia1/asf1 depicted on the corresponding residues of the CIA/ASF1—DBD(CCG1) complex (Table S5). Cia1p/Asf1p residues showing the Spt(-) phenotype by mutation are shown in cyan. (D) Construction of the bdf1 point mutant strains lacking BDF2 for the Spt phenotype analysis. (E) The Spt phenotype analysis of yeast bdf1 point mutant strains lacking BDF2 (_bdf2_Δ). (F) Summary of the yeast genetic analyses on Bdf1p depicted on the corresponding residues of the CIA/ASF1–DBD(CCG1) complex (Table S4). Bdf1p residues showing the Spt(-) phenotype and not showing the Spt(-) phenotype by mutation in the _BDF2_-deletion strain are shown in violet and green, respectively. (G_–_J) ChIP analysis of pol II, histone H3, Cia1p/Asf1p, and Bdf1p on the ACT1 promoter. Relative occupancies of pol II (G), histone H3 (H), Cia1p/Asf1p (I), and Bdf1p (J) at the promoter region of the ACT1 gene in the wild-type and mutant (Val403Ala) strains for Bdf1p. The results of the two-sided t test are shown above the graph.
We also examined the genetic interaction between CIA1/ASF1 and BDF2. Since Bdf1p and Bdf2p seem in part to function redundantly (35, 37), it is reasonable to expect that CIA1/ASF1 and BDF2 have a genetic interaction. In fact, the Spt(-) phenotype caused by the Tyr112Ala mutation of CIA1/ASF1 can be suppressed by the overexpression of Bdf2p. Furthermore, mutations of the Bdf2p residues presumably located in the Cia1p/Asf1p-interacting surface of binding site 1 (Leu354Ala and Val408Ala) cannot suppress the Spt(-) phenotype, even when the mutants were overexpressed (Fig. S3). These results suggested that the interaction between Cia1p/Asf1p and binding site 1 of Bdf2p is functional in vivo, probably in the transcription process.
Functional Interaction Between Cia1p/Asf1p and Bdf1p on the ACT1 Promoter.
Next, the in vivo functional significance of a residue located in binding site 1 was analyzed at the promoter region of ACT1 using ChIP analysis (Fig. 5G_–_J and Fig. S4). Both Cia1p/Asf1p and Bdf1p are localized on the promoter region of the ACT1 gene (Fig. 1F and G). The occupancies of pol II, histone H3, Bdf1p, and Cia1p/Asf1p were compared between the wild-type and Val403Ala(Bdf1p) mutant strains. Val403 of Bdf1p, which is located at binding site 1, was involved in the interaction between GST-Cia1p/Asf1p(155) and His-DBD(Bdf1p) (Fig. S4_A_–C). The functional significance of Val403 in Bdf1p and its corresponding residue Val408 in Bdf2p were also suggested from genetic analyses (Fig. 5D_–_F and Fig. S3_A_–C).
The results of the ChIP analysis showed that the Val403Ala mutation of Bdf1p induces statistically significant differences in the pol II, histone H3, and Cia1p/Asf1p occupancies (Fig. 5G_–_I). Compared to the wild-type strain, the Val403Ala mutant strain had lower occupancies of pol II (Fig. 5G) and Cia1p/Asf1p (Fig. 5I) and a higher occupancy of histone H3 (Fig. 5H). In contrast, a marginal difference was observed for Bdf1p occupancy between the wild type and mutant (p = 0.3, Fig. 5J). These results suggest that the mutation of Val403(Bdf1p), which induces the inhibition of the Cia1p/Asf1p–Bdf1p interaction (Fig. S4_A_–C), hampers the localization of pol II and Cia1p/Asf1p and the delocalization of histone H3.
The present ChIP data showed relatively large deviations (Fig. 5I and J). Other groups also pointed out the same problem (19, 42, 43). There are two main reasons for the relatively large deviations of data, which arise from the low signal intensities of Cia1p/Asf1p and Bdf1p. First, since Cia1p/Asf1p and Bdf1p cannot directly interact with DNA, the immunoprecipitation efficiencies were less than those of DNA-interacting factors such as pol II and histones (Fig. 5G and H) (42). Second, transient interactions of Cia1p/Asf1p and Bdf1p on promoter regions would be another reason for the low immunoprecipitation efficiencies of these factors (42). These problems are inevitable in the ChIP analysis with Bdf1p and Cia1p/Asf1p.
CIA/ASF1(155) Changes Its Interacting Partner.
In order to analyze the relationship between CIA/ASF1(155)–DBD(hCCG1) and CIA/ASF1–histone-H3–H4 complexes, we considered the structural effect of the histone H3–H4 dimer on the CIA/ASF1(155)–DBD(hCCG1) complex. In both binding sites (binding sites 1 and 2), some histone-interacting residues of CIA/ASF1 are exposed to the solvent (Fig. 6A and Tables S6 and S7). However, some histone-interacting residues are buried in the complex. Thus, the binding of the histone H3–H4 dimer to CIA/ASF1 in the CIA/ASF1(155)–DBD(hCCG1) complex causes close contacts between the histone H3–H4 dimer and DBD(hCCG1) (Fig. 6A and Fig. S5). The CIA/ASF1–DBD(CCG1) and CIA/ASF1–histone-H3–H4 complexes are mutually exclusive.
Fig. 6.
The effect of the histone H3–H4 dimer on the CIA/ASF1(155)–DBD(hCCG1) interaction. (A) CIA/ASF1(155) residues that interact with the histone H3–H4 dimer are shown by a stick model (red). CIA/ASF1(155) (purple) and DBD(hCCG1) (light blue) are shown by ribbon and surface models, respectively. CIA/ASF1(155) in the CIA/ASF1—histone-H3–H4 complex was superposed onto CIA/ASF1(155) in the CIA/ASF1(155)–DBD(hCCG1) complex, and close contacts were analyzed. The DBD(hCCG1) residues (in binding site 1) that make close contacts with the histone H3–H4 dimer are shown in orange. (B) In vitro competition assay between the histone H3–H4 dimer and DBD(hCCG1) for CIA/ASF1(155). (C) Summary of the competition assay in (B). (D) The colocalization and histone eviction model of CIA/ASF1 and DBD(CCG1).
Next, the interacting surface areas of these complexes were compared. The structural data showed that the interacting surface area of CIA/ASF1 for the histone H3–H4 dimer (1358 _Å_2; ref. 28) is substantially larger than those for DBD(hCCG1) (818 _Å_2 and 719 _Å_2 for binding sites 1 and 2, respectively). The structural analyses suggested that the change from the CIA/ASF1(155)–DBD(hCCG1) complex to the CIA/ASF1–histone-H3–H4 complex is a favorable process. Indeed, the biochemical analysis showed that the DBD(hCCG1) in the CIA/ASF1(155)–DBD(hCCG1) complex was dissociated from CIA/ASF1(155) by adding the histone (H3–H4)2 tetramer to the complex (Fig. 6B and C). The histone H3–H4 dimer dissociation from CIA/ASF1(155) by adding DBD(hCCG1) to the CIA/ASF1(155)–histone-H3–H4 complex was, however, not observed under our experimental conditions (Fig. 6C and Fig. S5_C_).
Discussion
In the present study, we determined the crystal structure of the CIA/ASF1(155)–DBD(hCCG1) complex (Fig. 2). This is the initial crystal structure of a histone chaperone in complex with a histone-modification-recognizing domain. Taking into account the function of each molecule, the CIA/ASF1–DBD(CCG1) complex can be considered molecular machinery connecting the histone acetylations to nucleosome structural change. Indeed, the crystal structure and the biochemical analysis showed that the CIA/ASF1(155)–DBD(hCCG1) complex can interact with an acetylated histone peptide (Fig. 3D). Furthermore, our structure-based biochemical (Fig. 4) and biological (Fig. 5) studies suggested that CIA/ASF1 colocalized with DBD(CCG1) at the promoter region through the interaction with DBD(CCG1) and is transferred to the histone (H3–H4)2 tetramer complex to disassemble the nucleosome structure in a site-specific manner (Fig. 6D). This is the initial structure-based model of the biological signaling from histone modifications to structural change of the nucleosome (hi-MOST model).
It was intriguing that DBD(hCCG1) has two binding sites for CIA/ASF1(155) (Fig. 2). The structure-based biological analysis suggested that the interaction between Cia1p/Asf1p and binding site 1 of Bdf1p is required for their colocalization at active promoter sites (Fig. 5I). The binding site on DBD(hCCG1) for the acetylated histone-N-tail peptide could not be determined in this study. However, the acetylated histone-N-tail peptide may bind to the acetylated-lysine-binding sites of DBD(hCCG1) in the CIA/ASF1(155)–DBD(hCCG1) complex, because both acetylated-lysine-binding sites are exposed to the solvent (Fig. 3C). When the acetylated histone tail binds to these binding sites, the bound peptide might affect the interaction between CIA/ASF1(155) and DBD(hCCG1) at binding site 1, because the bound peptide could be located close to CIA/ASF1 in the complex (Fig. 3C). It is of note that binding site 2 of BDF1 seems to have a genetic interaction with CIA1/ASF1 in yeast (Fig. 5 E and F). Further analysis is needed to clarify the functional implication of this binding site.
We showed that CIA/ASF1 localizes at the ACT1 promoter through the interaction with DBD(CCG1). This is consistent with the fact that the ACT1 gene is dominantly regulated by TFIID; TFIID predominates in the expression of about 90% of measurable genes in yeast (44). Our proposed mechanism, however, can be applied to some, but not all, genes since the remaining 10% of genes, most of which seem to be inducible genes in yeast, are dominated by the Spt-Ada-Gcn5-acetyltransferase (SAGA) complex (44). Other combinations of one or more histone chaperones and their recruiting factors could be involved in the histone eviction, particularly in the SAGA-dependent genes. In fact, it has been suggested that Bdf1p is not involved in Cia1p/Asf1p localization in the PHO5 gene (17), which is induced by low phosphate conditions.
After the colocalization of CIA/ASF1 and DBD(CCG1) to the promoter region, site-specific histone eviction should occur through the interaction between CIA/ASF1 and histone proteins. In this process, CIA/ASF1 seems to change its interacting partner from DBD(CCG1) to the core domains of the histone H3–H4 dimer. This molecular mechanism can be explained on the basis of the present crystallographic and biochemical results. The crystal structure of the CIA/ASF1(155)–DBD(hCCG1) complex showed that some of the histone-H3–H4-interacting residues on CIA/ASF1 (at binding site 1) are exposed to the solvent (Fig. 6A). Interaction between these exposed residues and the histone H3–H4 complex in the acetylated nucleosome or adjacent ones could be facilitated by the local compartmentalization of CIA/ASF1 through the interaction with DBD(CCG1) and is likely to be an initial step of the interacting partner change. Since the interacting surfaces of CIA/ASF1 for the histone H3–H4 dimer and DBD(CCG1) partially overlap (Fig. 6A and Fig. S5_A_), DBD(CCG1) and the histone H3–H4 dimer compete for CIA/ASF1. Since the interacting surfaces of CIA/ASF1 for the histone H3–H4 dimer are significantly larger than those for DBD(CCG1), the CIA/ASF1–histone-H3–H4 complex would be formed after the competition, resulting in the site-specific histone eviction. Since CIA/ASF1 changes its functional role by changing its interacting partner (8–10, 21), the resultant CIA/ASF1–histone-H3–H4 complex seems to induce another CIA/ASF1-relevant process. The exposed HIRA-interacting surface in the CIA/ASF1–histone-H3–H4 complex (28, 29, 45) might be a key to initiation of the subsequent process.
In the present study, we determined the crystal structure of the CIA/ASF1(155)–DBD(hCCG1) complex. The interactions between CIA/ASF1 and DBD(CCG1) found in the complex structure are functional in vivo (Fig. 5), and the complex structure possesses critical characteristics that can connect histone acetylation and histone eviction (Figs. 3C and 6A). Taken together, the findings indicate that CIA/ASF1–DBD(CCG1) complex can be considered a functional complex occurring in site-specific histone eviction (Fig. 6D). In this study, however, histones H2A and H2B and DNA were not involved in the analysis. Indeed, facilitate chromatin transcription (FACT) and Bdf1p seem to be involved in nucleosome structural change through interaction with histone H2A (27, 37). A structure-based study including all these factors and an analysis of DBD(CCG1) specificity for the modification patterns of histone proteins are required in order to fully understand the molecular mechanisms connecting acetylation-signal recognition and site-specific histone eviction.
Materials and Methods
The protein solution (15–20 mg ml-1) containing CIA/ASF1(155) and DBD(hCCG1) was prepared by mixing equimolar amounts of each component. The complex was crystallized by the hanging-drop vapor diffusion method at 293 K. The crystallization solution was 0.1 M Mes (pH 5.65), 1.1 M ammonium sulfate, and 0.25 M lithium sulfate. The crystal structure was determined by the molecular replacement method with the coordinates of CIA/ASF1(155) (PDB ID code 1ROC) and DBD(hCCG1) (PDB ID code 1EQF) as search models.
The details of expression, purification, crystallography, GST pull-down assay, Spt phenotype analysis, and ChIP analysis are described in the SI Text published online.
Supplementary Material
Supporting Information
Acknowledgments.
We thank Drs. F. Arisaka, T. Makino, S. Kidokoro, and T. Ishida for discussion. We also thank Dr. K. Struhl for the gift of plasmids and primer information and Dr. A. G. Ladurner for the anti-Bdf1p antibody and primer information. Our thanks also go to K. Hasegawa for critical reading of the manuscript. This work was supported in part by a Grant-in-Aid for JSPS Fellows, the New Energy and Industrial Technology Development Organization (NEDO) of Japan, the Ministry of Education, Culture, Sports, Science, and Technology of Japan, the Mitsubishi Foundation, the Uehara Memorial Foundation, and the Exploratory Research for Advanced Technology (ERATO) program of the Japan Science and Technology Agency (JST).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3AAD).
References
- 1.Kornberg RD. Chromatin structure: A repeating unit of histones and DNA. Science. 1974;184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Arents G, Burlingame RW, Wang BC, Love WE, Moudrianakis EN. The nucleosomal core histone octamer at 3.1 Å resolution: A tripartite protein assembly and a left-handed superhelix. Proc Natl Acad Sci USA. 1991;88:10148–10152. doi: 10.1073/pnas.88.22.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arents G, Moudrianakis EN. Topography of the histone octamer surface: Repeating structural motifs utilized in the docking of nucleosomal DNA. Proc Natl Acad Sci USA. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 5.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–733. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 6.Kornberg RD, Lorch Y. Chromatin rules. Nat Struct Mol Biol. 2007;14:986–988. [Google Scholar]
- 7.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 8.De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: An escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. doi: 10.1038/nsmb1318. [DOI] [PubMed] [Google Scholar]
- 9.Eitoku M, Sato L, Senda T, Horikoshi M. Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci. 2008;65:414–444. doi: 10.1007/s00018-007-7305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Park YJ, Luger K. Histone chaperones in nucleosome eviction and histone exchange. Curr Opin Struct Biol. 2008;18:282–289. doi: 10.1016/j.sbi.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns BR. Chromatin remodeling: Insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le S, Davis C, Konopka JB, Sternglanz R. Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast. 1997;13:1029–1042. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1029::AID-YEA160>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 13.Tyler JK, et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 14.Munakata T, Adachi N, Yokoyama N, Kuzuhara T, Horikoshi M. A human homologue of yeast anti-silencing factor has histone chaperone activity. Genes Cells. 2000;5:221–233. doi: 10.1046/j.1365-2443.2000.00319.x. [DOI] [PubMed] [Google Scholar]
- 15.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 16.Chimura T, Kuzuhara T, Horikoshi M. Identification and characterization of CIA/ASF1 as an interactor of bromodomains associated with TFIID. Proc Natl Acad Sci USA. 2002;99:9334–9339. doi: 10.1073/pnas.142627899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adkins MW, Howar SR, Tyler JK. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol Cell. 2004;14:657–666. doi: 10.1016/j.molcel.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Korber P, Hörz W. In vitro assembly of the characteristic chromatin organization at the yeast PHO5 promoter by a replication-independent extract system. J Biol Chem. 2004;279:35113–35120. doi: 10.1074/jbc.M405446200. [DOI] [PubMed] [Google Scholar]
- 19.Schwabish MA, Struhl K. Asf1 mediates histone eviction and deposition during elongation by RNA polymerase II. Mol Cell. 2006;22:415–422. doi: 10.1016/j.molcel.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 20.Williams SK, Truong D, Tyler JK. Acetylation in the globular core of histone H3 on lysine-56 promotes chromatin disassembly during transcriptional activation. Proc Natl Acad Sci USA. 2008;105:9000–9005. doi: 10.1073/pnas.0800057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- 22.Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emili A, Schieltz DM, Yates JR, 3rd, Hartwell LH. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 24.Hu F, Alcasabas AA, Elledge SJ. Asf1 links Rad53 to control of chromatin assembly. Genes Dev. 2001;15:1061–1066. doi: 10.1101/gad.873201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Y, et al. Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat Struct Mol Biol. 2008;15:738–745. doi: 10.1038/nsmb.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 27.Takahata S, Yu Y, Stillman DJ. FACT and Asf1 regulate nucleosome dynamics and coactivator binding at the HO promoter. Mol Cell. 2009;34:405–415. doi: 10.1016/j.molcel.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Natsume R, et al. Structure and function of the histone chaperone CIA/ASF1 complexed with histones H3 and H4. Nature. 2007;446:338–341. doi: 10.1038/nature05613. [DOI] [PubMed] [Google Scholar]
- 29.English CM, Adkins MW, Carson JJ, Churchill ME, Tyler JK. Structural basis for the histone chaperone activity of Asf1. Cell. 2006;127:495–508. doi: 10.1016/j.cell.2006.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 31.Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 32.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 34.Umehara T, Horikoshi M. Transcription initiation factor IID-interactive histone chaperone CIA-II implicated in mammalian spermatogenesis. J Biol Chem. 2003;278:35660–35667. doi: 10.1074/jbc.M303549200. [DOI] [PubMed] [Google Scholar]
- 35.Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 36.Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol Cell. 2003;11:353–363. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 37.Durant M, Pugh BF. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol Cell Biol. 2007;27:5327–5335. doi: 10.1128/MCB.00468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Strong M, et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 40.Owen DJ, et al. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright KJ, Marr MT, Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci USA. 2006;103:12347–12352. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aparicio O, et al. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol. 2005;21:21.3.1–21.3.33. doi: 10.1002/0471142727.mb2103s69. [DOI] [PubMed] [Google Scholar]
- 43.Ladurner AG, Inouye C, Jain R, Tjian R. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol Cell. 2003;11:365–376. doi: 10.1016/s1097-2765(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 44.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 45.Tang Y, et al. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat Struct Mol Biol. 2006;13:921–929. doi: 10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information