Next-generation Sequencing of Advanced Prostate Cancer Treated with Androgen-deprivation Therapy (original) (raw)
Abstract
Background
Androgen-deprivation therapy (ADT) is standard treatment for locally advanced or metastatic prostate cancer (PCa). Many patients develop castration resistance (castration-resistant PCa [CRPC]) after approximately 2–3 yr, with a poor prognosis. The molecular mechanisms underlying CRPC progression are unclear.
Objective
To undertake quantitative tumour transcriptome profiling prior to and following ADT to identify functionally important androgen-regulated pathways or genes that may be reactivated in CRPC.
Design, setting, and participants
RNA sequencing (RNA-seq) was performed on tumour-rich, targeted prostatic biopsies from seven patients with locally advanced or metastatic PCa before and approximately 22 wk after ADT initiation. Differentially regulated genes were identified in treatment pairs and further investigated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) on cell lines and immunohistochemistry on a separate CRPC patient cohort. Functional assays were used to determine the effect of pathway modulation on cell phenotypes.
Outcome measurements and statistical analysis
We searched for gene expression changes affecting key cell signalling pathways that may be targeted as proof of principle in a CRPC in vitro cell line model.
Results and limitations
We identified ADT-regulated signalling pathways, including the Wnt/β-catenin signalling pathway, and observed overexpression of β-catenin in a subset of CRPC by immunohistochemistry. We validated 6 of 12 (50%) pathway members by qRT-PCR on LNCaP/LNCaP-AI cell RNAs, of which 4 (67%) demonstrated expression changes consistent with RNA-seq data. We show that the tankyrase inhibitor XAV939 (which promotes β-catenin degradation) reduced androgen-independent LNCaP-AI cell line growth compared with androgen-responsive LNCaP cells via an accumulation of cell proportions in the G0/G1 phase and reduction in the S and G2/M phases. Our biopsy protocol did not account for tumour heterogeneity, and pathway inhibition was limited to pharmacologic approaches.
Conclusions
RNA-seq of paired PCa samples revealed ADT-regulated signalling pathways. Proof-of-principle inhibition of the Wnt/β-catenin signalling pathway specifically delays androgen-independent PCa cell cycle progression and proliferation and warrants further investigation as a potential target for therapy for CRPC.
Keywords: Prostate cancer, Androgen-deprivation therapy, Castration resistant, Wnt, β-catenin
Take Home Message
A comprehensive RNA sequencing analysis of the prostate cancer (PCa) transcriptome during androgen-deprivation therapy identifies gene expression changes within several cell-signalling pathways, including the Wnt/β-catenin signalling pathway, which has a potential role in androgen-independent PCa cell growth.
1. Introduction
At diagnosis, many men have “incurable” locally advanced or metastatic prostate cancer (PCa), the most common cancer in Europe [1]. PCa progression is initially driven by androgens acting via the cognate androgen receptor (AR) transcription factor. The initial treatment standard for patients with locally advanced or metastatic PCa is androgen-deprivation therapy (ADT), which inactivates the AR for a period of time. After approximately 2–3 yr, these patients can develop castration-resistant PCa (CRPC), for which the prognosis is poor despite newer second-line cytotoxic chemotherapy and endocrine therapies [2], [3], [4]. There is an urgent, unmet need for novel therapies for CRPC led by a better understanding of the biology underlying treatment resistance.
The mechanisms underlying CRPC are unclear and may be the result of cellular adaptation to or clonal selection by ADT [5]. AR signalling pathways and transcriptional activity may be reactivated [6], or cell growth may be supported by AR-independent outlaw cell signalling pathways [7]. Hence, a greater understanding of ADT-driven molecular changes may yield information on the mechanisms underlying progression to CRPC. Although previously published transcriptome-wide studies have successfully identified ADT-driven transcriptional events [8], [9], these analyses have been limited by the inherent bias associated with microarrays [10].
In this study, we undertake quantitative transcriptome profiling of prostate tumours from patients prior to and following ADT using next-generation sequencing (RNA-seq) to identify functionally important novel androgen-regulated pathways and specific gene products that may be reactivated in CRPC as potential targets for therapy.
2. Materials and methods
2.1. Patient samples for RNA sequencing
Clinical samples for RNA-seq were prospectively collected as part of the GenTax study [11]. Illumina RNA-seq was performed with complementary DNA sample library normalisation using the Illumina duplex-specific nuclease protocol prior to cluster generation and library sequencing on the HiSeq 2000 sequencer (Illumina, San Diego, CA, USA) with a paired-end sequencing strategy. Further details are given in the Supplement.
2.2. Functional assays
All cells were grown at 37 °C in 5% carbon dioxide. LNCaP (CRL-1740, ATCC) cells were maintained in RPMI-1640 medium (Life Technologies, Carlsbad, CA, USA; 31870-025) with 20 mM L-glutamine (Life Technologies, 25030-024) supplemented with 10% foetal bovine serum (PAA Laboratories, Yeovil Somerset, UK; A15-101). LNCaP-AI cells were derived from LNCaP parental cells and maintained as previously described [12]. Proliferation assays were carried out using the WST-1 reagent (Roche Diagnostics, Indianapolis, IN, USA; 05015944001) as per the manufacturer's instructions in medium containing either 10 μm XAV939 (Novartis Pharmaceuticals, Plantation, FL, USA) in 0.1% dimethyl sulfoxide (DMSO) [13] or vehicle. Cell cycle analysis was performed following treatment with 10 μm XAV939 in 0.1% DMSO or vehicle, as previously described [14]. Further details are given in the Supplement.
3. Results
3.1. The transcriptional landscape of androgen-deprivation therapy in clinical prostate cancer
RNA-seq was performed on 16 paired pre- and post-ADT samples from eight patients with locally advanced or metastatic PCa (Gleason score >7 [15]; Table 1). The post-ADT sample from patient 8 performed markedly worse on multiple quality control measures, and so both samples from this patient were excluded from further analysis (Supplemental Table 1). Recently, genomic rearrangements rendering ETS-family transcription factors under the control of androgen-responsive or other promoters have been hypothesised as a mechanism driving prostate carcinogenesis [16]. The TMPRSS2/ERG translocation yields the most common PCa-associated gene fusion product, reported in >50% cases [16]. Consistent with this, three of the seven (43%) patients expressed transcripts with sequences corresponding to this fusion event in the pre-ADT samples alone. We observed, on average, a sixfold downregulation of ERG expression following ADT, but expression of TMPRSS2/ERG did not correlate with time to biochemical relapse (data not shown). We also identified 12 additional candidate fusion products, 9 of which were only detectable in pre-ADT samples (Supplemental Table 2).
Table 1.
Patient demographics
| Patient | Age, yr | KPS | GSS | TNM stage | iPSA, ng/ml | nPSA, ng/ml (% iPSA) | PFS, d | ||
|---|---|---|---|---|---|---|---|---|---|
| T | N | M | |||||||
| 1 | 64.7 | 100 | 8 | 3b | 0 | 0 | 370 | 3.1 (0.8) | 942 |
| 2 | 65.4 | 90 | 9 | 3b | 0 | 0 | 7.6 | 0.7 (0.1) | 155 |
| 3 | 69.6 | 100 | 8 | 3b | 1 | 0 | 5.9 | 0.7 (0.1) | 223 |
| 4 | 64.6 | 90 | 8 | 3a | 0 | 1 | 47.7 | 0.5 (1.6) | N/P |
| 5 | 51.8 | 90 | 7 | 3a | 0 | 0 | 158 | 0.4 (0.3) | N/P |
| 6 | 58.6 | 100 | 7 | 3b | 1 | 0 | 69 | 0.13 (0.2) | 489 |
| 7 | 62.9 | 90 | 7 | 3b | 1 | 0 | 32.7 | <0.02 (0.06) | N/P |
| 8* | 54.9 | 90 | 7 | 3a | 2 | 1 | 7.4 | 0.06 (0.8) | 154 |
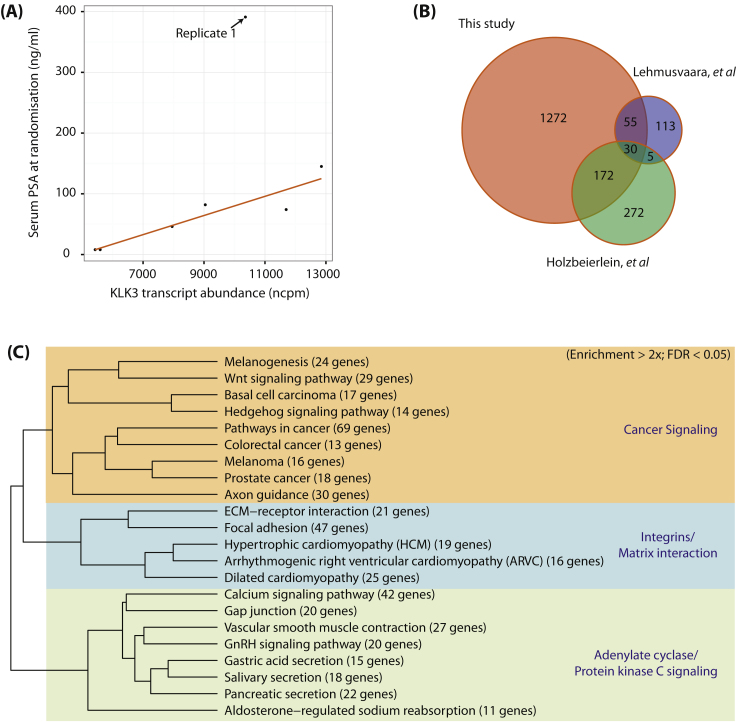
We identified a total of 774 genes upregulated at least twofold (false discovery rate [FDR] <0.05) in response to ADT and 755 genes similarly downregulated (Table 2 and Supplemental Table 3). Examples of genes differentially regulated and unchanged by ADT are shown in Supplemental Figure 1A. Levels of expression of KLK3, which encodes prostate-specific antigen (PSA), detected by RNA-seq correlated well with serum PSA levels for all but one patient (Fig. 1A). Comparing against data from two previously published microarray-based gene expression analyses prior to and following ADT [8], [9], we confirmed approximately 42% of the genes each of these previous studies had found and additionally identified a large number of previously unknown ADT-regulated genes (Fig. 1B).
Table 2.
Differentially expressed genes following androgen-deprivation therapy
| Gene set | Up | Down |
|---|---|---|
| Protein coding | 774 | 755 |
| Noncoding | 35 | 116 |
Fig. 1.
Differential expression of genes after androgen-deprivation therapy. (A) Correlation between KLK3 messenger RNA transcript expression levels (_x_-axis) normalised by trimmed means of M value in normalised counts per million and serum prostate-specific antigen levels (nanograms per millilitre; _y_-axis). (B) Overlap between genes identified as twofold differentially expressed and previously reported microarray experiments [8], [9]. (C) Hierarchical clustering of Kyoto Encyclopaedia of Genes and Genomics (KEGG) pathways enriched in upregulated gene sets. Dendrograms show clustering of KEGG terms based on the number of differentially expressed genes two pathways have in common as a proportion of the total number of differentially regulated genes in the two pathways combined. This shows, for example, that the set of cardiac-related pathways cluster with pathways connected to adhesion and matrix interaction. These categories are driven by differential expression of the integrin genes, which are known to be involved in cell–cell and cell–matrix adhesion [37].
FDR = false discovery rate; PSA = prostate-specific antigen.
To understand how many of these expression changes might directly result from changes in AR activity, we used a list of genes whose regulatory regions were bound by the AR from published chromatin immunoprecipitation sequencing data on patient tissue [17]. Seven (1.7%) of the 331 genes with AR binding peaks [17] within 2 kb of the transcription start site were significantly upregulated in expression twofold, but 35 (8.8%) were significantly downregulated twofold, which is greater than twice as many as would be expected by chance (p < 1 × 10−5). In line with previous reports, we conclude that direct AR binding is a significant but minor determinant of overall expression changes. Nevertheless, we do note that most AR binding events occur outside promoters, presumably within enhancers.
To identify biologic pathways perturbed following ADT, an enrichment analysis was performed on our lists of up- and downregulated genes (FDR <0.05) using the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database [18]. We identified 24 pathways significantly enriched for the upregulated genes (enrichment more than twofold; FDR <0.05) and 19 enriched for the downregulated genes (Supplemental Table 4 and Fig. 1). To understand themes associated with enriched pathways, we clustered pathways by the fraction of differentially regulated genes shared by two pathways (Fig. 1C and Supplemental Fig. 1B). For pathways enriched for upregulated genes, three clusters became apparent, including a cluster containing cancer-related pathways, such as Prostate cancer, Pathways in cancer, and Wnt signalling pathway (Fig. 1C). Pathways enriched for downregulated genes consisted mostly of metabolic pathways (Supplemental Fig. 1B).
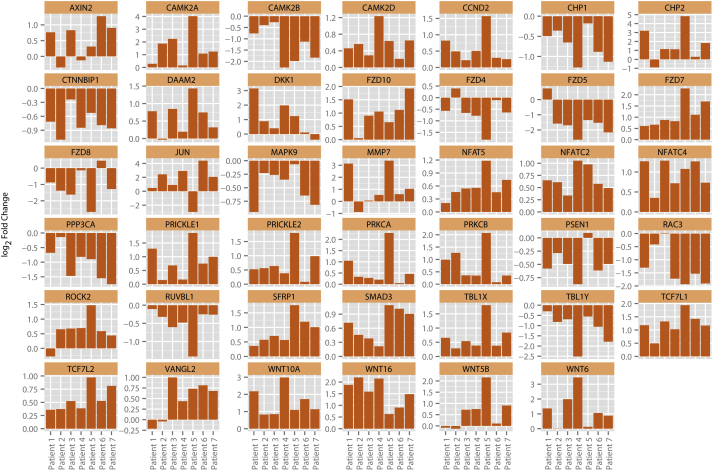
In light of evidence implicating cross-talk of AR signalling with other pathways (eg, PI3K/Akt/mTOR) in PCa [19], we focused on KEGG terms relating to cell signalling pathways. With the exception of the general Pathways in cancer, the KEGG term Wnt signalling pathway contained the largest number of significantly upregulated genes (Supplemental Table 4 and Fig. 2; 29 of 150; enrichment was twofold; FDR <0.05) among the cluster of upregulated cancer pathways. The Wnt/β-catenin signalling pathway is a proproliferative pathway in PCa [20], and enrichment for this pathway was an unexpected finding in the upregulated gene set. Fourteen of the upregulated genes were also found to be upregulated in one or other of the previously published studies on ADT-driven gene expression changes (34 patients) [8], [9].
Fig. 2.
Differential expression of genes encoding components of the Wnt/β-catenin–signalling pathway in individual patients. Fold change (logarithmic base 2) after androgen-deprivation therapy of genes in both up- and downregulated gene sets, annotated as being part of the Kyoto Encyclopaedia of Genes and Genomes term Wnt signalling pathway.
Taken together, these data suggest that the Wnt/β-catenin signalling pathway is regulated, either directly or indirectly, by androgens in vivo. Specific components of the Wnt/β-catenin signalling pathway have been implicated in AR signalling in vitro [21] and with ADT in clinical PCa [22] as well as in murine prostate development and PCa progression [23]. Recent exome-sequencing studies have identified mutations in genes encoding components of the pathway in PCa [24], [25], but its role in clinical CRPC remains unclear. To determine whether the Wnt/β-catenin signalling pathway contributes to CRPC, we used immunohistochemistry to investigate β-catenin protein expression in a panel of 36 matched pairs of hormone-naïve PCa (HNPC) and CRPC. Among the 29 informative tumour pairs, 16 pairs demonstrated β-catenin overexpression in CRPC samples (Supplemental Fig. 2). In addition, a statistically significant correlation exists between β-catenin and nuclear AR protein expression in CRPC (r = 0.440; p = 0.017) but not HNPC (r = 0.15; p = 0.375). These data are consistent with the notion that β-catenin is involved in PCa progression in a cohort of tumours and may be associated with AR reactivation in CRPC.
3.2. Inhibition of the Wnt/β-catenin signalling pathway limits androgen-independent prostate cancer cell growth via a delay in cell cycle progression
We next investigated whether perturbations within the Wnt/β-catenin signalling pathway may be functionally important in CRPC. To verify the RNA-seq data, we took advantage of two in vitro models: the LNCaP PCa cell line as a model for HNPC and the LNCaP-AI subline, derived from continued culture of LNCaP cells in steroid-deplete medium (DCC), as a model for CRPC. To check the validity of these models, we searched for significantly downregulated genes within the KEGG term Wnt signalling pathway in a previously published exon-level analysis of the androgen-responsive LNCaP transcriptome [26]. We observed androgen-dependent downregulation of expression of 35 pathway genes (FDR <0.05), of which 7 were significantly upregulated in the RNA-seq data set, suggesting that LNCaP cells may be suitable for modelling our in vivo findings.
Twelve candidate canonical Wnt/β-catenin signalling pathway components differentially expressed between pre- and post-ADT samples in the initial RNA-seq analysis were further investigated by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis on RNAs extracted from LNCaP and LNCaP-AI cells (Supplemental Table 5). Of the 12 candidates, expression of 6 (50%) genes could be detected by qRT-PCR in these cell lines, which may be the result of cell line–specific gene expression. Of the six validated candidates, four (67%) demonstrated expression changes consistent with RNA-seq data. These included the signal-transducing Frizzled family serpentine receptors (FZD4/FZD5/FZD7) and the pathway downstream effector JUN.
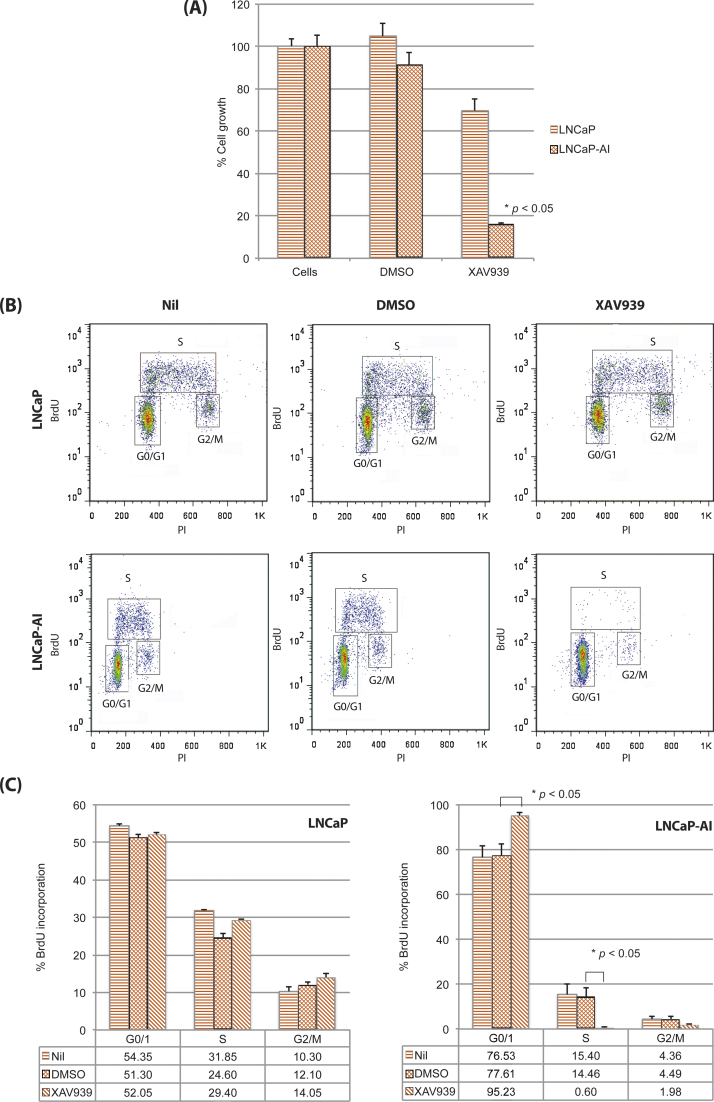
Extracellular Wnt signals are transduced by two receptors, a Frizzled family member and a transmembrane protein LRP5 or LRP6, which together form a complex following binding by cysteine-rich Wnt proteins. The Frz-LRP5/6 complex then inactivates the β-catenin degradation complex (which includes axin and GSK3β), thus allowing β-catenin nuclear translocation [20]. As a proof-of-principle experiment, we tested the functional effects of the Wnt/β-catenin signalling in CRPC using LNCaP-AI cells [27] by WST-1 proliferation assays in the presence of a well-characterised Wnt/β-catenin signalling inhibitor [13] (Fig. 3A). We treated LNCaP or LNCaP-AI cells (grown in full medium or DCC, respectively) with 10 μm XAV939, an inhibitor of tankyrase 1 and 2 (which stabilise axin and promote β-catenin degradation) [13], or vehicle. Compared with controls, treatment with XAV939 reduced growth of LNCaP cells in full medium by approximately 30% and had even stronger effects on LNCaP-AI cells, potently diminishing cell growth by approximately 75% (p < 0.05; Fig. 3A).
Fig. 3.
Inhibition of the Wnt/β-catenin signalling pathway limits growth and delays cell cycle progression of LNCaP-AI cells. (A) WST-1 proliferation assays were performed using either LNCaP cells grown in full medium or LNCaP-AI cells grown in DCC. Cells were treated with 10 μm XAV939 in 0.1% dimethyl sulfoxide (DMSO) or vehicle. Values were normalised to the growth of LNCaP cells in full medium without any treatment. Data from at least three independent experiments were used to obtain the mean relative growth plus or minus standard deviation (SD) for each cell type and treatment condition. (B) Cell cycle analysis was performed using either LNCaP cells grown in full medium or LNCaP-AI cells grown in DCC. Cells were treated with 10 μm XAV939 in 0.1% DMSO or vehicle. Plots shown are representative of at least three independent experiments, from which (C) percentages of cells in each phase of the cell cycle were calculated for each cell type and treatment condition to obtain mean percentage plus or minus SD.
BrdU = BrdU fluorescence intensity.
To determine whether the above reduction in cell proliferation reflected a delay in cell cycle progression, LNCaP or LNCaP AI cells treated with 10 μm XAV939 or vehicle were subjected to cell cycle analysis (Fig. 3B and 3C). In the absence of treatment, synchronised LNCaP cells were predominantly in the G0/G1 phase, with a smaller proportion of cells in the S or G2/M phase. Compared with LNCaP cells, a larger proportion of synchronised LNCaP-AI cells were in the G0/G1 phase, with a smaller proportion in the S or G2/M-phase, demonstrating that the cell cycle of LNCaP-AI cells progresses more slowly than the parental cell line, as previously described [28]. Following treatment with XAV939, there was no statistically significant difference in the proportion of LNCaP cells in the G0/G1 phase compared with control (p > 0.05) and only small (approximately 3–5%) differences in the proportion of cells in the S and G2/M phases. However, there was a 17% increase in the proportion of LNCaP-AI cells in the G0/G1 phase (p < 0.05) and a 10% fall in the proportion of cells in the S phase (p < 0.05). These data suggest that, in LNCaP-AI cells, XAV939 treatment specifically causes an accumulation of cells in the G0/1 phase and a reduction of cells in the S and G2/M phases, thereby delaying cell cycle progression. Taken together, these data suggest that the LNCaP-AI subline is particularly dependent on the Wnt/β-catenin signalling pathway for cell growth.
4. Discussion
We report the first quantitative transcriptome profiling of clinical PCa from patients prior to and following ADT by using RNA-seq and have substantially enlarged the ADT-regulated gene set, as is expected from a more sensitive sequencing study [29], [30], [31]. The substantially better overlap between our and the two previously published microarray gene sets [8], [9] contrasts with the poor individual overlap of the gene sets between these two studies, which may be the result of a lack of power of these studies to identify differentially regulated genes. Specifically, we identified several novel pathways perturbed by ADT, including the Wnt/β-catenin signalling pathway. Although activation of Wnt/β-catenin signalling in carcinogenesis is well known in breast and colorectal cancers [32], evidence for this pathway in clinical PCa has been conflicting. Recent exome-sequencing studies have identified mutations with genes encoding components of the Wnt/β-catenin signalling pathway [24], [25], but there is no clear consensus on the significance of increased nuclear β-catenin expression in primary PCa based on immunohistochemistry [20].
The current focus of novel therapies for CRPC is the targeting of renewed AR signalling [2], [3], [19]. In CRPC, an inverse correlation has been observed between β-catenin nuclear localisation and AR expression [33], yet others report no statistically significant difference in expression following progression from hormone-naïve disease to CRPC [34]. We demonstrate overexpression of β-catenin in a subset of CRPC and correlation with expression of AR consistent with AR reactivation. Functional or physical interactions between AR and Wnt/β-catenin signalling have been reported in vitro, with β-catenin functioning as an AR coactivator [20], [32] to drive ligand-independent cell growth.
Despite conflicting results from immunohistochemistry experiments in clinical PCa, which may be caused by experimental variability [34], the above reports together with our own observations suggest that Wnt/β-catenin signalling may be active in CRPC. In keeping with this hypothesis, expression of putative inhibitors of Wnt/β-catenin signalling and downstream transcription factors were downregulated following progression to CRPC in an LNCaP xenograft model [35]. It is thought that these expression changes may lead to an increase cytoplasmic pool of β-catenin, allowing potential interaction with unliganded AR and other transcription factors.
We observed a potent inhibitory effect of XAV939 treatment on LNCaP-AI cell growth, which appeared to be caused by an accumulation of cells in the G0/1 phase of the cell cycle (Fig. 3), suggesting a functional role for Wnt/β-catenin signalling in CRPC. The molecular mechanisms underlying our observations remain unclear, and an exhaustive investigation thereof was outside the scope of this proof-of-principle study. Despite using a well-validated inhibitor of Wnt/β-catenin signalling, a potential limitation of this approach is off-target effects.
We identified ADT-driven changes in components of cell signalling pathways, including the Wnt/β-catenin signalling pathway, approximately 22 wk following ADT initiation. Although our patients had not yet developed CRPC at the time of the second biopsy, some did develop early biochemical relapse. A caveat of our approach is the targeted biopsy regime, which, although it yields tumour-rich tissue, could have included a heterogeneous population of epithelial and stromal cells. However, a surprising observation was that our KEGG analysis identified upregulation of expression of cancer gene–related pathways, which would have been expected to be downregulated if stromal expression signatures were overtly expressed.
5. Conclusions
Our observations suggest that CRPC may represent repopulation of a tumour with androgen-independent clones reliant on Wnt/β-catenin signalling activity after sustained ADT. In light of recent data suggesting that Wnt ligand expression by the tumour microenvironment may attenuate cytotoxic chemotherapy and confer treatment resistance [36], Wnt/β-catenin signalling may represent a potential target for therapy in PCa. Further mechanistic insights using genetic approaches such as in vitro RNA interference and genetically engineered autochthonous murine cancer models will determine whether this pathway is a potential therapeutic target for CRPC.
Author contributions: Hing Y. Leung and Prabhakar Rajan had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Leung, Pedley, Rajan.
Acquisition of data: Rajan, Sudbery, Villasevil, Mui, Ahmad, Edwards.
Analysis and interpretation of data: Rajan, Sudbery, Edwards, Leung.
Drafting of the manuscript: Rajan, Sudbery, Leung.
Critical revision of the manuscript for important intellectual content: Sansom, Sims, Ponting, Heger, McMenemin, Pedley.
Statistical analysis: Rajan, Sudbery, Edwards, Sims, Heger.
Obtaining funding: Rajan, Ponting, Pedley, Leung.
Administrative, technical, or material support: Villasevil, Mui, Fleming, Davis, Ahmad, Edwards, Sansom, Sims, Ponting, Heger, McMenemin, Pedley.
Supervision: Sims, Ponting, Heger, Leung.
Financial disclosures: Hing Y. Leung and Prabhakar Rajan certify that all conflicts of interest, including specific financial interests and relationships and affiliations relevant to the subject matter or materials discussed in the manuscript (eg, employment/affiliation, grants or funding, consultancies, honoraria, stock ownership or options, expert testimony, royalties, or patents filed, received, or pending), are the following: None.
Funding/Support and role of the sponsor: The study was supported by grants from the Sanofi-Aventis, NHS Greater Glasgow and Clyde Endowments, Medical Research Council, Cancer Research UK, and Royal College of Surgeons of England, but these bodies did not have any involvement in the analysis, preparation of the manuscript, or decision regarding publication.
Acknowledgement statement: We are grateful to the patients recruited to GenTax, without whom this work would not have been possible, and staff at the Department of Urology and the Northern Centre for Cancer Care, Newcastle upon Tyne Hospitals NHS Foundation Trust for help with patient recruitment and clinical care. We thank Colin Nixon and David Huels (CR-UK Beatson Institute) for technical assistance and advice, respectively, and Rachana Patel (CR-UK Beatson Institute) and Jacqueline Stockley (University of Glasgow) for critical appraisal of the manuscript.
Footnotes
Contributor Information
Prabhakar Rajan, Email: p.rajan@beatson.gla.ac.uk.
Hing Y. Leung, Email: h.leung@beatson.gla.ac.uk.
Appendix A. Supplementary data
References
- 1.Center M.M., Jemal A., Lortet-Tieulent J. International variation in prostate cancer incidence and mortality rates. Eur Urol. 2012;61:1079–1092. doi: 10.1016/j.eururo.2012.02.054. [DOI] [PubMed] [Google Scholar]
- 2.de Bono J.S., Logothetis C.J., Molina A., COU-AA-301 Investigators Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scher H.I., Fizazi K., Saad F., AFFIRM Investigators Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–1197. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 4.Tannock I.F., de Wit R., Berry W.R., TAX 327 Investigators Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004;351:1502–1512. doi: 10.1056/NEJMoa040720. [DOI] [PubMed] [Google Scholar]
- 5.Zong Y., Goldstein A.S. Adaptation or selection—mechanisms of castration-resistant prostate cancer. Nat Rev Urol. 2013;10:90–98. doi: 10.1038/nrurol.2012.237. [DOI] [PubMed] [Google Scholar]
- 6.Sharma N.L., Massie C.E., Ramos-Montoya A. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23:35–47. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 7.Feldman B.J., Feldman D. The development of androgen-independent prostate cancer. Nat Rev Cancer. 2001;1:34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 8.Holzbeierlein J., Lal P., LaTulippe E. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol. 2004;164:217–227. doi: 10.1016/S0002-9440(10)63112-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehmusvaara S., Erkkila T., Urbanucci A. Chemical castration and anti-androgens induce differential gene expression in prostate cancer. J Pathol. 2012;227:336–345. doi: 10.1002/path.4027. [DOI] [PubMed] [Google Scholar]
- 10.Rajan P., Elliott D.J., Robson C.N., Leung H.Y. Alternative splicing and biological heterogeneity in prostate cancer. Nat Rev Urol. 2009;6:454–460. doi: 10.1038/nrurol.2009.125. [DOI] [PubMed] [Google Scholar]
- 11.Pedley I.D., Frew J.A., Wilson J.M. Tolerability and efficacy of anti-androgen manipulation versus taxotere and anti-androgen manipulation in patients with hormone-naive, high-risk/metastatic prostate cancer: a phase II, open-labeled, randomized study [abstract 147] J Clin Oncol. 2009;29(Suppl 7) [Google Scholar]
- 12.Halkidou K., Gnanapragasam V.J., Mehta P.B. Expression of Tip60, an androgen receptor coactivator, and its role in prostate cancer development. Oncogene. 2003;22:2466–2477. doi: 10.1038/sj.onc.1206342. [DOI] [PubMed] [Google Scholar]
- 13.Huang S.M., Mishina Y.M., Liu S. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]; Natl Acad Sci U S A 2010;107:15473–8.
- 14.Wiltshire C., Singh B.L., Stockley J. Docetaxel-resistant prostate cancer cells remain sensitive to S-trityl-L-cysteine-mediated Eg5 inhibition. Mol Cancer Ther. 2010;9:1730–1739. doi: 10.1158/1535-7163.MCT-09-1103. [DOI] [PubMed] [Google Scholar]
- 15.Gleason D.F., Mellinger G.T. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J Urol. 1974;111:58–64. doi: 10.1016/s0022-5347(17)59889-4. [DOI] [PubMed] [Google Scholar]
- 16.Kumar-Sinha C., Tomlins S.A., Chinnaiyan A.M. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu J., Yu J., Mani R.S. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carver B.S., Chapinski C., Wongvipat J. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–586. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kypta R.M., Waxman J. Wnt/beta-catenin signalling in prostate cancer. Nat Rev Urol. 2012;9:418–428. doi: 10.1038/nrurol.2012.116. [DOI] [PubMed] [Google Scholar]
- 21.Zhuo M., Zhu C., Sun J., Weis W.I., Sun Z. The beta-catenin binding protein ICAT modulates androgen receptor activity. Mol Endocrinol. 2011;25:1677–1688. doi: 10.1210/me.2011-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson V.C., Hurtado-Coll A., Turbin D. Relaxin drives Wnt signaling through upregulation of PCDHY in prostate cancer. Prostate. 2010;70:1134–1145. doi: 10.1002/pros.21148. [DOI] [PubMed] [Google Scholar]
- 23.Francis J.C., Thomsen M.K., Taketo M.M., Swain A. Beta-catenin is required for prostate development and cooperates with Pten loss to drive invasive carcinoma. PLoS Genet. 2013;9:e1003180. doi: 10.1371/journal.pgen.1003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grasso C.S., Wu Y.M., Robinson D.R. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barbieri C.E., Baca S.C., Lawrence M.S. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet. 2012;44:685–689. doi: 10.1038/ng.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rajan P., Dalgliesh C., Carling P.J. Identification of novel androgen-regulated pathways and mRNA isoforms through genome-wide exon-specific profiling of the LNCaP transcriptome. PLoS One. 2011;6:e29088. doi: 10.1371/journal.pone.0029088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark E.L., Hadjimichael C., Temperley R., Barnard A., Fuller-Pace F.V., Robson C.N. P68/DdX5 supports beta-catenin & RNAP II during androgen receptor mediated transcription in prostate cancer. PLoS One. 2013;8:e54150. doi: 10.1371/journal.pone.0054150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu G., Wu J., Zhou L. Characterization of the small RNA transcriptomes of androgen dependent and independent prostate cancer cell line by deep sequencing. PLoS One. 2010;5:e15519. doi: 10.1371/journal.pone.0015519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marioni J.C., Mason C.E., Mane S.M., Stephens M., Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradford J.R., Hey Y., Yates T., Li Y., Pepper S.D., Miller C.J. A comparison of massively parallel nucleotide sequencing with oligonucleotide microarrays for global transcription profiling. BMC Genomics. 2010;11:282. doi: 10.1186/1471-2164-11-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.‘t Hoen P.A., Ariyurek Y., Thygesen H.H. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms. Nucleic Acids Res. 2008;36:e141. doi: 10.1093/nar/gkn705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anastas J.N., Moon R.T. WNT signalling pathways as therapeutic targets in cancer. Nat Rev Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
- 33.Wan X., Liu J., Lu J.F. Activation of beta-catenin signaling in androgen receptor-negative prostate cancer cells. Clin Cancer Res. 2012;18:726–736. doi: 10.1158/1078-0432.CCR-11-2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitaker H.C., Girling J., Warren A.Y., Leung H., Mills I.G., Neal D.E. Alterations in beta-catenin expression and localization in prostate cancer. Prostate. 2008;68:1196–1205. doi: 10.1002/pros.20780. [DOI] [PubMed] [Google Scholar]
- 35.Wang G., Wang J., Sadar M.D. Crosstalk between the androgen receptor and beta-catenin in castrate-resistant prostate cancer. Cancer Res. 2008;68:9918–9927. doi: 10.1158/0008-5472.CAN-08-1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Y., Campisi J., Higano C. Treatment-induced damage to the tumor microenvironment promotes prostate cancer therapy resistance through WNT16B. Nat Med. 2012;18:1359–1368. doi: 10.1038/nm.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desgrosellier J.S., Cheresh D.A. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.