DNA interrogation by the CRISPR RNA-guided endonuclease Cas9 (original) (raw)
. Author manuscript; available in PMC: 2014 Sep 6.
Published in final edited form as: Nature. 2014 Jan 29;507(7490):62–67. doi: 10.1038/nature13011
Abstract
The CRISPR-associated enzyme Cas9 is an RNA-guided endonuclease that uses RNA:DNA base-pairing to target foreign DNA in bacteria. Cas9:guide RNA complexes are also effective genome engineering agents in animals and plants. Here we use single-molecule and bulk biochemical experiments to determine how Cas9:RNA interrogates DNA to find specific cleavage sites. We show that both binding and cleavage of DNA by Cas9:RNA require recognition of a short trinucleotide protospacer adjacent motif (PAM). Non-target DNA binding affinity scales with PAM density, and sequences fully complementary to the guide RNA but lacking a nearby PAM are ignored by Cas9:RNA. DNA strand separation and RNA:DNA heteroduplex formation initiate at the PAM and proceed directionally towards the distal end of the target sequence. Furthermore, PAM interactions trigger Cas9 catalytic activity. These results reveal how Cas9 employs PAM recognition to quickly identify potential target sites while scanning large DNA molecules, and to regulate dsDNA scission.
RNA-mediated adaptive immune systems in bacteria and archaea rely on Clustered Regularly Interspaced Short Palindromic Repeats (CRISPRs) and CRISPR-associated (Cas) proteins to provide protection from invading viruses and plasmids1. Bacteria harbouring CRISPR-Cas loci respond to viral and plasmid challenge by integrating short fragments of the foreign nucleic acid (protospacers) into the host chromosome at one end of the CRISPR array2. Transcription of the CRISPR array followed by enzymatic processing yields short CRISPR RNAs (crRNAs) that direct Cas protein-mediated cleavage of complementary target sequences within invading viral or plasmid DNA3-5. In Type II CRISPR-Cas systems, Cas9 functions as an RNA-guided endonuclease that uses a dual-guide RNA consisting of crRNA and _trans_-activating crRNA (tracrRNA) for target recognition and cleavage by a mechanism involving two nuclease active sites that together generate dsDNA breaks (DSBs)6,7.
RNA-programmed Cas9 can be a versatile tool for genome engineering in multiple cell types and organisms8-19. Guided by either a natural dual-RNA complex or a chimeric single-guide RNA6, Cas9 generates site-specific DSBs that are repaired either by non-homologous end joining or homologous recombination. In addition, catalytically inactive Cas9 alone or fused to transcriptional activator or repressor domains can be used to alter transcription levels at sites targeted by guide RNAs20-25. Despite the ease in applying this technology, the fundamental mechanism that enables Cas9:RNA to locate specific 20 base-pair (bp) DNA targets within the vast sequence space of genomes remains unknown.
Single-molecule visualization of Cas9
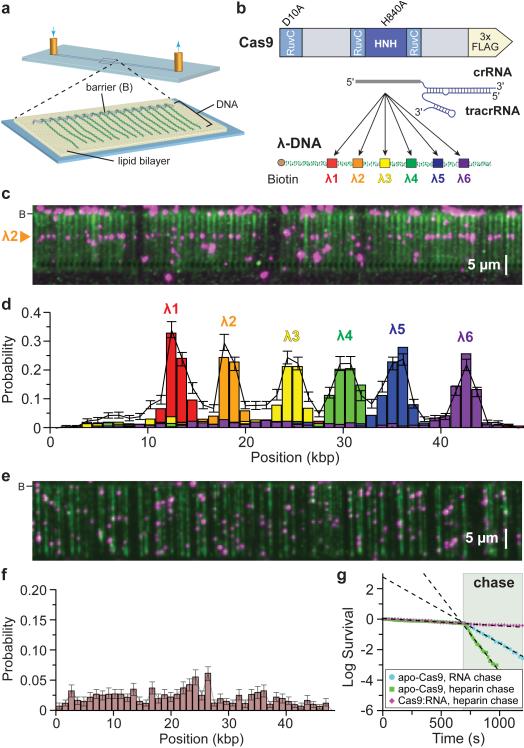
To determine how Cas9:RNA complexes locate targets, we used a single-tethered DNA curtain assay and total internal reflection fluorescence microscopy (TIRFM) to visualize the binding site distribution of single Cas9:RNA molecules on λ-DNA substrates (48,502 bp) (Fig. 1a)26,27. We purified S. pyogenes Cas9 containing a C-terminal 3x-FLAG tag that enabled fluorescent labeling using anti-FLAG antibody-coated quantum dots (QDs)27,28, and generated guide RNAs (dual crRNA:tracrRNA) bearing complementarity to six different 20-bp sites within the λ-DNA (Fig. 1b and Extended Data Table 1). Neither the 3x-FLAG tag nor QDs inhibited DNA cleavage by Cas9:RNA, and all guide RNAs were functional (Extended Data Fig. 1). Initial experiments used a nuclease-inactive Cas9 containing D10A/H840A mutations (dCas9) that binds but does not cleave DNA6. QD-tagged dCas9:RNA localized almost exclusively to the expected target site (Fig. 1c and Supplementary Video 1). Furthermore, Cas9 could be directed to any region of the DNA by redesigning the RNA guide sequence (Fig. 1d and Extended Data Fig. 2)6,8,9. Thus, DNA targeting by Cas9:RNA is faithfully recapitulated in the DNA curtain assays.
Figure 1. DNA curtains assay for target binding by Cas9:RNA.
a, Schematic of a single-tethered DNA curtain26,27. b, Wild-type Cas9 or dCas9 was programmed with crRNA:tracrRNA targeting one of six sites. c, YOYO1-stained DNA (green) bound by QD-tagged dCas9 (magenta) programmed with λ2 guide RNA. d, dCas9:RNA binding distributions; error bars represent 95% confidence intervals obtained through bootstrap analysis28. e, Image of apo-Cas9 bound to DNA curtains bound to apo-Cas9. f, Binding distribution of apo-Cas9; error bars represent 95% confidence intervals. g, Lifetimes of DNA-bound apo-Cas9 and Cas9:RNA after injection of λ2 crRNA:tracrRNA (100 nM) or heparin (10 μg mL−1).
We next used apo-Cas9 protein to confirm that the binding observed in DNA curtain assays was due to Cas9:RNA and not apo-Cas9 lacking guide RNA. Interestingly, apo-Cas9 also bound DNA but exhibited no apparent sequence specificity (Fig. 1e,f). Attempts to measure the dissociation rate of DNA-bound apo-Cas9 were hampered by their exceedingly long lifetimes (a lower limit of at least 45 min was calculated; Fig. 1g). Biochemical experiments revealed an upper limit of ~25 nM for the equilibrium dissociation constant (_K_d) of the apo-Cas9:DNA complex, compared to ~0.5 nM for the Cas9:RNA complex bound to a bona fide target (Extended Data Fig. 3).
We test whether DNA-bound apo-Cas9 could be distinguished from Cas9:RNA based on a differential response to chases with free guide RNAs, we measured the lifetime of apo-Cas9 on DNA curtains before and after injection of crRNA:tracrRNA or heparin. Apo-Cas9 rapidly dissociated from non-specific sites in the presence of either competitor, and this result was verified with bulk biochemical assays (Fig. 1g and Extended Data Fig. 3). In contrast, target-bound Cas9:RNA was unaffected by either heparin or excess crRNA:tracrRNA (Fig. 1g and Extended Data Fig. 3). These findings show that non-specifically bound apo-Cas9 has properties distinct from those of Cas9:RNA complexes bound to cognate targets.
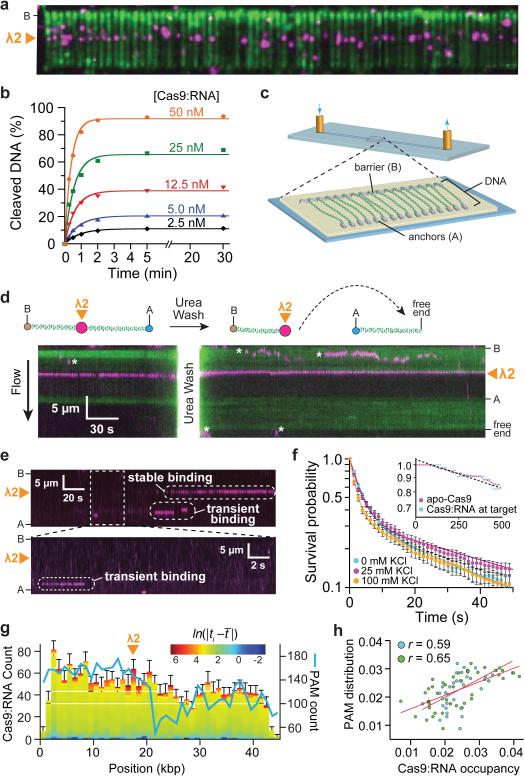
Initially we used catalytically inactive dCas9 to avoid DNA cleavage. Surprisingly, experiments with wild-type Cas9 also failed to reveal DNA cleavage; rather, Cas9:RNA molecules remained bound to their target sites, yielding identical results to those obtained using dCas9:RNA (Fig. 2a). We confirmed that the imaging conditions did not inhibit Cas9:RNA cleavage activity (Extended Data Fig. 1). These results suggested that Cas9:RNA might cleave DNA but remain tightly bound to both cleavage products, which was confirmed with biochemical gel shift assays using 72-bp duplex DNA substrates (Extended Data Fig. 4). To determine whether stable product binding would prevent Cas9:RNA from performing multiple turnover cleavage, we conducted plasmid DNA cleavage assays at varying molar ratios of Cas9:RNA and target DNA and measured the rate and yield of product formation. Surprisingly, the amount of product rapidly plateaued at a level proportional to the molar ratio of Cas9:RNA to DNA, indicating that Cas9:RNA does not follow Michaelis-Menten kinetics (Fig. 2b). Turnover also does not occur with short duplex DNA substrates and is not stimulated by either elevated temperature or an excess of free crRNA:tracrRNA (Extended Data Fig. 5).
Figure 2. Cas9:RNA remains bound to cleaved products and localizes to PAM-rich regions during the target search.
a, Wild-type Cas9:RNA bound to DNA curtains. b, Cleavage yield of 25 nM plasmid DNA is proportional to [Cas9:RNA]. c, Schematic of a double-tethered DNA curtain29-31. d, Liberation of the cleaved DNA with 7 M urea; asterisks denote QDs that are attached to the lipid bilayer but not bound to the DNA. e, Kymographs illustrating distinct binding events. f, Survival probabilities for non-target binding events; solid lines represent double-exponential fits. Inset: survival probabilities of DNA-bound apo-Cas9 and target DNA-bound Cas9:RNA. g, Distribution of Cas9:RNA binding events (N = 2,330) and PAM density. Colour-coding reflects the binding dwell time (t i) relative to the mean dwell time (t̄). h, Correlation of PAM distribution and non-target Cas9:RNA binding for λ2 (blue) and spacer 2 (green) guide RNAs.
Using a double-tethered DNA curtain (Fig. 2c)29-31, we confirmed that although Cas9:RNA catalyzed DNA cleavage when bound to target sites on λ-DNA, it did not dissociate even in the presence of 10 μg ml−1 heparin (Fig. 1g) or up to 0.5 M NaCl (not shown). However, injection of 7 M urea caused Cas9:RNA to release the downstream end containing the PAM, confirming that the DNA was cleaved at the expected target site (Fig. 2d). The finding that Cas9:RNA remains tightly bound to both ends of the cleaved DNA, suggests it acts as a single-turnover enzyme.
Cas9:RNA locates targets by 3D diffusion
To determine how Cas9:RNA locates DNA targets, we visualized the target search process using double-tethered DNA curtains. Site-specific DNA-binding proteins can locate target sites by three-dimensional (3D) collisions or through facilitated diffusion processes including one-dimensional (1D) sliding, hopping, and/or intersegmental transfer32; these mechanisms can be distinguished by single-molecule imaging30,31,33. For these assays, Cas9 programmed with λ2 guide RNA was injected into the sample chamber, buffer flow was terminated, and reactions were visualized in real-time. These experiments revealed long-lived binding events at the target site and transient binding events at other sites on the DNA (Fig. 2e,f). We saw no evidence of Cas9:RNA associating with target sites by facilitated diffusion (either 1D sliding and/or hopping); instead, all target association appeared to occur directly through 3D collisions (Fig. 2e and Supplementary Video 2).
The shorter-lived, non-specific binding events exhibited complex dissociation kinetics, and the simplest model that describes the data is double-exponential decay with lifetimes of ~3.3 and ~58 seconds (at 25 mM KCl) (Fig. 2f). These lifetimes were readily distinguished from the long lifetimes of apo-Cas9 (Fig. 2f, inset). Furthermore, the experiments were conducted in the presence of a saturating (10-fold) molar excess of crRNA:tracrRNA to exclude contamination from apo-Cas9. This result indicates that at least two and possibly more binding intermediates exist on the pathway towards cognate target recognition. Although non-specific DNA binding typically involves electrostatic interactions with the phosphate backbone such that non-specific lifetimes tend to decrease with increasing ionic strength34, the lifetimes of Cas9:RNA bound at non-specific DNA sites were not appreciably affected by salt concentration (Fig. 2f). It is remarkable that these Cas9:RNA non-target binding events have characteristics more commonly attributed to site-specific association34,35.
To gain further insight into the nature of the target search mechanism, we measured the locations and corresponding lifetimes of all binding events (Fig. 2g). The off-target binding lifetime distributions did not vary substantially at different regions of the DNA. However, the number of observed binding events was not uniformly distributed along the substrate, suggesting that some underlying feature of the λ-DNA might influence the target search. The λ-phage genome contains 5,677 PAM sites (~1 PAM per 8.5 bp), but it also has an unusual polar distribution of A/T- and G/C-rich sequences27, which leads to an asymmetric distribution of PAMs (5'-NGG-3' for S. pyogenes Cas9) (Fig. 2g). Pearson correlation analysis revealed that the Cas9:RNA binding site distribution was positively correlated with the PAM distribution (r = 0.59, P <0.05) (Fig. 2h). When we repeated this experiment using a guide RNA having no complementary target sites within λ-DNA (spacer 2 crRNA)6, we saw no change in the binding lifetimes and an even stronger correlation with the PAM distribution (Fig. 2h and Extended Data Fig. 2b,c). These results, together with the insensitivity of short-lived binding events to ionic strength, suggested that while searching for potential targets Cas9:RNA might bind specifically to PAMs and minimize interactions with non-PAM DNA.
A PAM is required for DNA interrogation
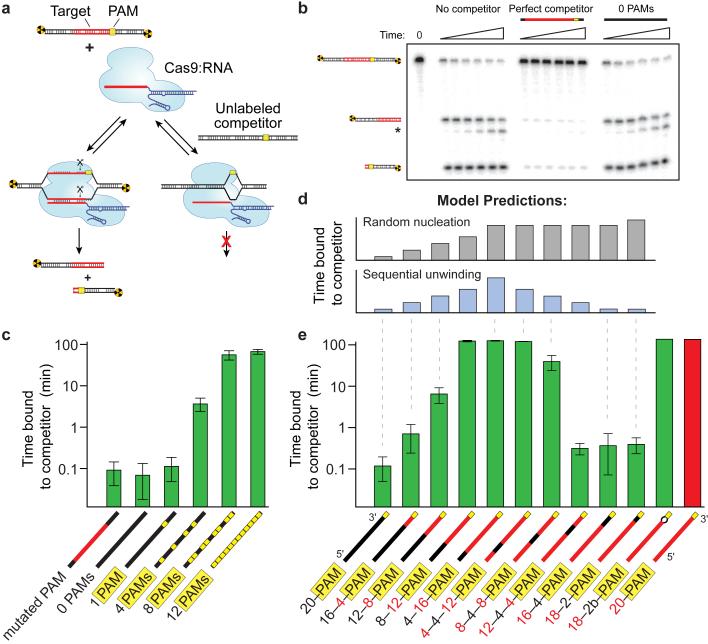
To test whether Cas9:RNA uses PAM recognition as an obligate precursor to interrogation of flanking DNA for potential guide-RNA complementarity, we used competition assays to monitor the rate of Cas9:RNA-mediated DNA cleavage (Fig. 3a,b). From these data we extracted the average time Cas9:RNA spends sampling each competitor DNA prior to locating and cleaving a radiolabeled substrate (Extended Data Fig. 6). In control experiments, reaction kinetics were not perturbed by the presence of an unlabeled competitor DNA lacking PAMs and bearing no sequence relationship to the crRNA, whereas a competitor containing a PAM and fully complementary target sequence substantially reduced the cleavage rate of the radiolabeled substrate (Fig. 3b).
Figure 3. Cas9:RNA searches for PAMs and unwinds dsDNA in a directional manner.
a, Schematic of the competition cleavage assay. b, Cleavage assay with and without competitor DNAs. c, Quantitation of competition data (mean ± s.d.). Competitor cartoon representations show PAMs (yellow) and regions complementary to the crRNA (red). d, Predicted data trends for the random nucleation or sequential unwinding models aligned with the corresponding data in (e). e, Competition assays using substrates with variable degrees of crRNA complementarity, shown as in (c). Numeric descriptions of the competitor DNAs indicate the regions of complementarity (red) or mismatches (black) to the crRNA sequence.
A series of competitors that bore no crRNA guide sequence complementarity (Extended Data Table 1) but contained increasing numbers of PAMs (Fig. 3c) were tested. There was a direct correspondence between the number of PAMs and the ability of a DNA competitor to interfere with target cleavage, indicating that the lifetime of Cas9:RNA on competitor DNA increased with PAM density (Fig. 3c). This result persisted over a range of competitor DNA concentrations (Extended Data Fig. 6), and the same pattern of competition was observed for DNA binding by dCas9:RNA (Extended Data Fig. 7). These results demonstrate that the residence time of Cas9:RNA on non-target DNA lacking PAMs is negligible, and support the hypothesis that transient, non-target DNA binding events observed on the DNA curtains likely occurred at PAM sequences. While Cas9:RNA complexes undoubtedly sample DNA lacking PAMs, these rapid binding events are detectable in neither single-molecule assays nor bulk binding experiments (Extended Data Fig. 7), and they do not appreciably influence overall reaction kinetics in bulk biochemical assays.
We repeated the competition assay with a competitor bearing perfect complementarity to the crRNA, but a single point mutation in the adjacent PAM (5'-TCG-3'); like similarly mutated substrates6, this competitor cannot be cleaved by Cas9:RNA (Extended Data Fig. 7). This competitor failed to inhibit DNA cleavage by Cas9:RNA and behaved comparably to the non-target competitor DNA lacking PAMs, despite the fact that it contained perfect complementarity to the crRNA (Fig. 3c). Together, these results demonstrate that PAM recognition is an obligate first step during target recognition by Cas9:RNA, as previously hypothesized6.
Mechanism of RNA:DNA heteroduplex formation
After PAM recognition, Cas9:RNA must destabilize the adjacent duplex and initiate strand separation to enable base-pairing between the target DNA strand and the crRNA guide sequence. Because Cas9 has no energy-dependent helicase activity, the mechanism of local DNA unwinding has been enigmatic, but must rely upon thermally available energy. One possibility is that PAM binding could induce a general destabilization of the duplex along the length of the entire target sequence, leading to random nucleation of the RNA:DNA heteroduplex (Fig. 3d, top). Alternatively, PAM binding may cause only local melting of the duplex, with the RNA:DNA heteroduplex nucleating at the 3' end of the target sequence next to the PAM and proceeding sequentially towards the distal 5' end of the target sequence (Fig. 3d, bottom).
To distinguish between these models, we conducted cleavage assays with a panel of DNA competitors in which the length and position of complementarity to the guide RNA was systematically varied (Extended Data Table 1). These competitors were designed to distinguish between the random nucleation and sequential unwinding models for heteroduplex formation based upon the predicted patterns of cleavage inhibition for each model (Fig. 3d). The ability of a competitor DNA to inhibit substrate cleavage by Cas9:RNA increased as the extent of complementarity originating at the 3' end of the target sequence adjacent to the PAM increased (Fig. 3e). Inhibition increased dramatically when 12 or more base-pairs were complementary to the crRNA guide sequence, which agrees with the requirement for an 8-12 nt seed sequence for the Cas9:RNA DNA cleavage reaction6,36. Strikingly, although competitors containing mismatches to the crRNA at the 5' end of the target sequence competed effectively for Cas9:RNA binding, competitors containing mismatches to the crRNA at the extreme 3' end immediately adjacent to the PAM were completely inert to binding (Fig. 3e). This was true even with a 2-bp mismatch followed by 18 bp of contiguous sequence complementarity to the crRNA. Therefore, when mismatches to the crRNA are encountered within the first two nucleotides of the target sequence, Cas9:RNA loses the ability to interrogate and recognize the remainder of the DNA. The pattern of inhibition observed with the different competitor DNAs indicates that sequence homology adjacent to the PAM is necessary to initiate target duplex unwinding until the reaction has proceeded sufficiently far (~12 bp, approximately one turn of an A-form RNA:DNA helix), such that the energy necessary for further propagation of the RNA:DNA heteroduplex falls below the energy needed for the reverse reaction. These findings suggest that formation of the RNA:DNA heteroduplex initiates at the PAM and proceeds through the target sequence by a sequential, step-wise unwinding mechanism consistent with a Brownian ratchet37.
As a further test of this model, we used a DNA competitor that contained mismatches to the crRNA at positions 1-2 but was itself mismatched at the same two positions, forming a small bubble in the duplex. Despite the absence of sequence complementarity to the crRNA within the DNA bubble, this substrate was a robust competitor and bound Cas9:RNA with an affinity nearly indistinguishable from that of an ideal substrate (Fig. 3e and Extended Data Fig. 8). Remarkably, this DNA could also be cleaved with near wild-type rates (Extended Data Fig. 8). We speculate that the presence of the DNA bubble allowed Cas9:RNA to bypass the mismatches and reinitiate nucleation of the RNA:DNA heteroduplex downstream of the bubble, thereby propagating strand separation through the remainder of the target.
The PAM triggers Cas9 nuclease activity
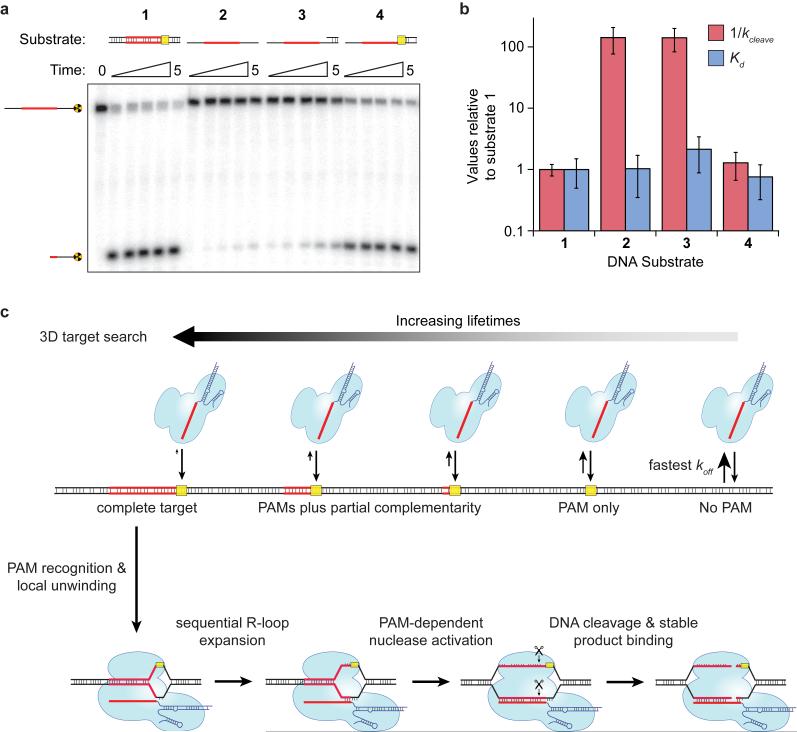
The results presented above indicate that PAM recognition plays a central role in target recognition, and that introduction of a small bubble in the DNA target eliminates the need for RNA:DNA heteroduplex formation immediately adjacent to the PAM. One might expect PAM recognition to be dispensable for Cas9:RNA-mediated recognition and cleavage of a single-stranded DNA (ssDNA) target. Surprisingly, however, a ssDNA substrate was cleaved more than two orders of magnitude slower than a dsDNA substrate (Fig. 4a,b), despite the fact that dCas9:RNA bound both dsDNA and ssDNA substrates with similar affinities (Fig. 4b and Extended Data Fig. 9).
Figure 4. PAM recognition regulates Cas9 nuclease activity.
a, Cleavage assay with single-stranded, double-stranded, and partially double-stranded substrates. b, Relative affinities and cleavage rates; (mean ± s.d.). c, Model for target search, recognition and cleavage by Cas9:RNA. The search initiates through random 3D collisions. Cas9:RNA rapidly dissociates from non-PAM DNA, but binds PAMs for longer times and samples adjacent DNA for guide RNA complementarity, giving rise to a heterogeneous population of intermediates. At correct targets, Cas9:RNA initiates formation of an RNA:DNA heteroduplex, and R-loop expansion propagates via sequential unwinding. The DNA is cleaved and Cas9:RNA remains bound to the cleaved products.
Importantly, Cas9:RNA recognizes the 5'-NGG-3' PAM on the non-target DNA strand6, so the ssDNA substrates did not contain a PAM but rather the complement to the PAM sequence. We hypothesized that the absence of the PAM on the ssDNA might explain why an otherwise fully complementary target is resistant to cleavage. To test this possibility, we prepared hybrid substrates with varying lengths of dsDNA at the 3' flanking sequence (Fig. 4a). Cleavage assays revealed that the ssDNA target strand could be activated for cleavage in the presence of flanking dsDNA that extended across the PAM sequence, but that this activating effect was lost when the dsDNA was truncated immediately before the PAM (Fig. 4a,b and Extended Data Fig. 9). Binding experiments confirmed these results were not a consequence of discrimination at the level of binding (Fig. 4b). Rather, the presence of the 5'-NGG-3' PAM on the non-target strand was critical for some step of the reaction that occurred after binding. These data suggest that the PAM acts as an allosteric regulator of Cas9:RNA nuclease activity.
Discussion
Our results suggest a model for target binding and cleavage by Cas9:RNA involving an unanticipated importance for PAM sequences at each stage of the reaction (Fig. 4c). Although details may differ, we hypothesize that PAM interactions may function similarly for other CRISPR RNA-guided surveillance complexes38-44. The Cas9:RNA target search begins with random collisions with DNA. However, rather than sampling all DNA equivalently, Cas9:RNA accelerates the search by rapidly dissociating from non-PAM sites, thereby reducing the amount of time spent at off-targets. Only upon binding to a PAM site does Cas9:RNA interrogate the flanking DNA for guide RNA complementarity, as previously hypothesized for Cas96 and a distinct CRISPR RNA-guided complex (Cascade)42. A requirement for initial PAM recognition also eliminates the potential for suicidal self-targeting, since perfectly matching targets within the bacterial CRISPR locus are not flanked by PAMs. Our results suggest that PAM recognition coincides with initial destabilization of the adjacent sequence, as evidenced from experiments using a bubble-containing DNA substrate, followed by sequential extension of the RNA:DNA heteroduplex. This mechanism explains the emergence of seed sequences, because mismatches encountered early in a directional melting-in process would prematurely abort target interrogation. Moreover, the complex dissociation kinetics observed on non-target λ-DNA would arise from heterogeneity in the potential target sites as Cas9:RNA probes sequences adjacent to PAMs for guide RNA complementarity. Binding to a correct target then leads to activation of both nuclease domains. This step also requires PAM recognition, providing an unanticipated level of PAM-dependent regulation that may ensure further protection against self-cleavage of the CRISPR locus. Interestingly, Cas9:RNA does not dissociate from the cleaved DNA except under extremely harsh conditions, suggesting that Cas9:RNA may remain bound to the cleaved site in vivo4 and require other cellular factors to promote recycling. Finally, our data indicate that efforts to minimize off-target effects during genome engineering using Cas9:RNA complexes need only consider off-targets adjacent to a PAM, because potential targets lacking a PAM are unlikely to be interrogated36,45-47.
METHODS
Cas9 and RNA preparation
Wild-type Cas9 and D10A/H840A dCas9 from S. pyogenes were purified as described6, and a 3x-FLAG tag was cloned onto the C-terminus of Cas9 for single-molecule experiments. crRNAs (42 nucleotides in length) were either ordered synthetically (Integrated DNA Technologies) or transcribed in vitro with T7 polymerase using single-stranded DNA templates (Extended Data Table 1), as described48. tracrRNA was also transcribed in vitro and contained nucleotides 15–87 following the numbering scheme used previously6. crRNA:tracrRNA duplexes were prepared by mixing equimolar concentrations of each RNA in Hybridization Buffer (20 mM Tris-HCl pH 7.5, 100 mM KCl, 5 mM MgCl2), heating to 95 °C for 30 seconds, and slow-cooling.
DNA curtains post-steady state binding measurements
Post steady-state binding assays in Fig. 1 were performed with single-tethered DNA curtains26,27. First, 100 nM 3x-FLAG-tagged dCas9 was reconstituted with 1 μM crRNA:tracrRNA targeting the desired region of λ-DNA by incubating for ~10 min at 37 °C in Reaction Buffer (20 mM Tris-HCl pH 7.5, 100 mM KCl, 5 mM MgCl2, 5% glycerol, 1 mM DTT). 10 nM dCas9:RNA was then incubated with λ-DNA (100 pM) for ~15 min at 37 °C in 40 mM Tris-HCl pH 7.5, 25 mM KCl, 1 mg mL−1 BSA, 1 mM MgCl2, and 1 mM DTT, before being diluted to 1 nM and injected into the flow cell. The flow cell was then washed with 3–5 mL of Imaging Buffer containing 40 mM Tris-HCl, 25 mM KCl, 1mg mL−1 BSA, 1 mM MgCl2, 1 mM DTT, 0.75 nM YOYO1 (Life Technologies), 0.8% glucose, and 0.2X glucose oxidase/catalase. Finally, 0.5 nM anti-FLAG antibody-coated QDs were incubated in the flow cell for 5 min, followed by a wash of 1–2 mL of Imaging Buffer. Curtains were imaged and dCas9:RNA positions determined by fitting a 2D Gaussian to individual molecules31,33. The data from all six dCas9:RNA complexes (λ1–λ6) were combined, and error bars for the combined set were generated by bootstrap methods31,33.
DNA curtains equilibrium binding measurements
Binding position and lifetime measurements in Fig. 2 were performed using the λ2 crRNA:tracrRNA and double-tethered DNA curtains31,33. Cas9 was reconstituted with a 10X excess of crRNA:tracrRNA and incubated with anti-FLAG antibody-coated QDs for ~10 min. Cas9:RNA was then diluted to 2 nM in Imaging Buffer containing 0–100 mM KCl as indicated, and injected into the flow cell. The approximate ionic strength for Imaging Buffer containing 0, 25, and 100 mM KCl is 32, 57, and 132 mM, respectively, given the expected ionization of Tris-HCl at pH 7.5 and presence of 1 mM MgCl2. Videos were recorded at 50, 25, or 10 Hz, and the position of each binding event was determined from the y-coordinate within kymographs generated for each DNA molecule. The lifetime of each binding event was defined as the difference between the first frame and last frame in which the QD-tagged Cas9:RNA was observed. To analyze lifetimes, all binding events were synchronized, and the probability that a binding event survived up to a particular time was determined as the number of Cas9:RNA complexes bound at time t divided by the number initially bound. Position data were pooled to generate the binding distribution histogram, which was binned at 1,078 bp per bin27. Error bars for the binding distributions and survival probabilities were determined by bootstrap methods31,33.
To test for cleavage in our single molecule assays, Cas9:RNA bound to double-tethered DNA was exposed to a 7 M urea wash in the presence of flow (Fig 2d). Similar experiments were conducted on single-tethered curtains in the absence of YOYO1, and 42% (N = 150) of target-bound molecules remained bound to the upstream DNA product following the urea wash. The remaining 58% (N = 206) either remained bound to the downstream product containing the PAM, dissociated from the DNA altogether, or remained bound to the upstream fragment but lost their florescent tag.
Bulk binding and cleavage experiments
The plasmid DNA substrate used in Fig. 2b contained a λ2 target sequence cloned into the EcoRI and BamHI sites on pUC19. Oligoduplex DNA substrates (Extended Data Table 1) were 55-bp in length and were prepared by mixing together complementary synthetic oligonucleotides (Integrated DNA Technologies) in Hybridization Buffer, heating to 95 °C for 1–2 min followed by slow-cooling, and purifying on a 5% native polyacrylamide gel (0.5X TBE buffer with 5 mM MgCl2) run at 4 °C. When assayed directly, DNA substrates were 5'-radiolabeled using [γ-32P]-ATP (Promega) and T4 polynucleotide kinase (New England Biolabs) or 3'-radiolabeled using [α-32P]-dATP (Promega) and terminal transferase (New England Biolabs) (Extended Data Fig. 4). The substrates presented in Fig. 4b were prepared by 5'-radiolabeling only the target strand, hybridizing it to a 10X excess of the indicated unlabeled complementary strand, and gel purifying the partial/full duplex by 10% native gel electrophoresis.
Cas9:RNA complexes were reconstituted prior to cleavage and binding experiments by incubating Cas9 and the crRNA:tracrRNA duplex for 10 min at 37 °C in Reaction Buffer. Binding experiments used dCas9 (except as indicated in Extended Data Fig. 4) and either equimolar crRNA:tracrRNA or a 10X molar excess of crRNA:tracrRNA over dCas9 (Extended Data Fig. 3 & 8). Binding reactions contained 0.1–1 nM DNA and increasing apo-dCas9 or dCas9:RNA concentrations, and were incubated at 37 °C for one hour before being resolved by 5% native polyacrylamide gel electrophoresis (0.5X TBE buffer with 5 mM MgCl2) run at 4 °C. DNA was visualized by phosphorimaging, quantified with ImageQuant (GE Healthcare), and analyzed with Kaleidagraph (Synergy Software).
Cleavage assays were conducted in Reaction Buffer at room temperature and analyzed by 1% agarose gel electrophoresis and ethidium bromide staining (Fig. 2b) or 10% denaturing polyacrylamide gel electrophoresis and phosphorimaging. Aliquots were removed at each time point and quenched by the addition of gel loading buffer supplemented with 25 mM EDTA (at 1X). Reactions contained ~1 nM radiolabeled DNA substrate and 10 nM Cas9:RNA (competition experiments, Fig. 3b,c,e, Extended Data Fig. 6) or 100 nM Cas9:RNA (Fig. 4a,b, Extended Data Fig. 1, 7, 8, 9). Competition experiments used λ1 target DNA and were supplemented with 500 nM unlabeled competitor DNAs (Fig. 3c,e) or an extended concentration range of competitor DNAs (Extended Data Fig. 6). All oligoduplex DNA cleavage experiments were visualized by phosphorimaging and quantified with ImageQuant (GE Healthcare)
Analysis of cleavage competition assays
The competition experiments were analyzed to determine the survival probability of the radiolabeled target DNA, S(t). In principle, the survival probability should begin at 1 and go to 0, but in practice the reaction rarely proceeds to completion. Therefore, we conditioned the survival against the probability of a particular amount of target DNA being cleaved. This conditional survival probability, S*(t), relates to the survival probability as follows:
All reactions in the presence of competitor DNA that reached ~90% completion were conditioned against their final values, whereas reactions uncompleted after 2 hrs were conditioned to the reaction in the absence of competitor DNA. For each reaction, we then obtained the change in the survival probability of the target DNA, Δ_Ps_(t), in the presence of competitor DNA. Finally, Δ_Ps_(t) was integrated over the 2 hr reaction time. For reactions that reached completion in the presence of competitor within 2 hrs, this analysis yields the change in the mean relaxation time of the reaction (or the inverse of the average reaction rate). In cases where the reaction did not reach completion by the 2 hr time point, this analysis instead yields a mean time spent on competitor DNA during the 2 hr reaction. Notably, this analysis makes no assumptions about the nature of the reaction or the dynamic changes in the reactive species.
The reduction in cleavage rate in the presence of competitor DNA is directly proportional to the time that Cas9:RNA spends bound to each competitor. In each reaction, Cas9:RNA encounters competitor DNA on average more frequently than the target DNA, and the time Cas9:RNA spends interrogating a competitor has the cumulative effect of slowing the overall reaction. The presented models merely state that the amount of time spent on competitor DNA will be proportional to the “observed” complementarity between crRNA and bound DNA, i.e. the number of canonical Watson-Crick base-pairs that can be formed. It then directly follows that in the case where the R-loop is randomly nucleated (regardless of nucleation size), the time bound to competitor DNA will simply scale with the total amount of complementarity between competitor DNA and crRNA (Fig. 3d, top). However, in the case where the R-loop is nucleated from a particular site, i.e. the 3' end of the target sequence directly adjacent to the PAM, the time bound to competitor DNA will scale proportionally to the length of contiguous complementarity between the crRNA and DNA beginning from the nucleation site (Fig. 3d, bottom).
Supplementary Material
1
6
7
8
9
01
10
11
12
13
2
3
4
5
ACKNOWLEDGEMENTS
We thank Prashant Bhat, Alison Marie Smith, and Kaihong Zhou for technical assistance, and members of the Doudna and Greene laboratories and Jamie Cate for helpful discussions and critical reading of the manuscript. S.H.S. acknowledges support from the National Science Foundation and National Defense Science & Engineering Graduate Research Fellowship programs. Funding was provided by the National Institutes of Health (GM074739 to E.C.G.) and the National Science Foundation (MCB-1154511 to E.C.G. and MCB-1244557 to J.A.D.). M.J. was a Research Specialist, E.C.G. is an Early Career Scientist, and J.A.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
EXTENDED DATA and SUPPLEMENTARY VIDEOS are available with this paper.
AUTHOR CONTRIBUTIONS
S.H.S. generated RNAs, conducted biochemical and single-molecule experiments, and assisted with single-molecule data analysis. S.R. conducted single-molecule experiments and data analysis, and assisted with the design and analysis of biochemical assays. M.J. cloned and purified 3x-FLAG-Cas9, and assisted with the design and interpretation of initial single-molecule experiments. S.H.S., S.R., M.J., E.C.G., and J.A.D. discussed the data and wrote the manuscript.
REFERENCES
- 1.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou R, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 3.Brouns SJJ, et al. Small CRISPR RNAs Guide Antiviral Defense in Prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 5.Deltcheva E, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinek M, et al. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–86. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jinek M, et al. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang WY, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang H, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bassett AR, Tibbit C, Ponting CP, Liu J-L. Highly Efficient Targeted Mutagenesis of Drosophila with the CRISPR/Cas9 System. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gratz SJ, et al. Genome Engineering of Drosophila with the CRISPR RNA-Guided Cas9 Nuclease. Genetics. 2013;194:1029–1035. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedland AE, et al. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Meth. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shan Q, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 17.Li J-F, et al. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- 19.Xie K, Yang Y. RNA-guided Genome Editing in Plants Using A CRISPR-Cas System. Mol Plant. 2013 doi: 10.1093/mp/sst119. doi:10.1093/mp/sst119. [DOI] [PubMed] [Google Scholar]
- 20.Qi LS, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bikard D, et al. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert LA, et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nat Meth. 2013 doi: 10.1038/nmeth.2598. doi:10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Meth. 2013 doi: 10.1038/nmeth.2600. doi:10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mali P, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fazio T, Visnapuu M-L, Wind S, Greene EC. DNA curtains and nanoscale curtain rods: high-throughput tools for single molecule imaging. Langmuir. 2008;24:10524–10531. doi: 10.1021/la801762h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visnapuu M-L, Greene EC. Single-molecule imaging of DNA curtains reveals intrinsic energy landscapes for nucleosome deposition. Nat Struct Mol Biol. 2009;16:1056–1062. doi: 10.1038/nsmb.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ilya J Finkelstein M-LVECG. Single-molecule imaging reveals mechanisms of protein disruption by a DNA translocase. Nature. 2010;468:983–987. doi: 10.1038/nature09561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorman J, Fazio T, Wang F, Wind S, Greene EC. Nanofabricated racks of aligned and anchored DNA substrates for single-molecule imaging. Langmuir. 2010;26:1372–1379. doi: 10.1021/la902443e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gorman J, Plys AJ, Visnapuu M-L, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17:932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, et al. The promoter-search mechanism of Escherichia coli RNA polymerase is dominated by three-dimensional diffusion. Nat Struct Mol Biol. 2013;20:174–181. doi: 10.1038/nsmb.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hippel, von PH, Berg OG. Facilitated target location in biological systems. J Biol Chem. 1989;264:675–678. [PubMed] [Google Scholar]
- 33.Gorman J, et al. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proc Natl Acad Sci USA. 2012;109:E3074–83. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hippel, von PH, Berg OG. On the specificity of DNA-protein interactions. Proc Natl Acad Sci USA. 1986;83:1608–1612. doi: 10.1073/pnas.83.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohs R, et al. Origins of specificity in protein-DNA recognition. Annu Rev Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abbondanzieri EA, Greenleaf WJ, Shaevitz JW, Landick R, Block SM. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438:460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (Reading, Engl) 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 39.Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Semenova E, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wiedenheft B, et al. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci USA. 2011;108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hou Z, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Meth. 2013 doi: 10.1038/nmeth.2681. doi:10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pattanayak V, et al. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu Y, et al. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA. 2012;18:661–672. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
6
7
8
9
01
10
11
12
13
2
3
4
5