Integrative Structure and Functional Anatomy of a Nuclear Pore Complex (original) (raw)
. Author manuscript; available in PMC: 2018 Sep 14.
Published in final edited form as: Nature. 2018 Mar 14;555(7697):475–482. doi: 10.1038/nature26003
Summary
Despite the central role of Nuclear Pore Complexes (NPCs) as gatekeepers of RNA and protein transport between the cytoplasm and nucleoplasm, their large size and dynamic nature have impeded a full structural and functional elucidation. Here, we have determined a subnanometer precision structure for the entire 552-protein yeast NPC by satisfying diverse data including stoichiometry, a cryo-electron tomography map, and chemical cross-links. The structure reveals the NPC’s functional elements in unprecedented detail. The NPC is built of sturdy diagonal columns to which are attached connector cables, imbuing both strength and flexibility, while tying together all other elements of the NPC, including membrane-interacting regions and RNA processing platforms. Inwardly-directed anchors create a high density of transport factor-docking Phe-Gly repeats in the central channel, organized in distinct functional units. Taken together, this integrative structure allows us to rationalize the architecture, transport mechanism, and evolutionary origins of the NPC.
Introduction
Nuclear Pore Complexes (NPCs) are large proteinaceous assemblies studded through the nuclear envelope (NE), the double-membraned barrier surrounding the nucleus; they are the sole mediators of macromolecular transport between the nucleus and the cytoplasm, and carry key regulatory platforms for numerous nuclear processes1. NPCs are also major targets for viral manipulation, and defects in this transport machine are directly linked to human diseases, including cancers2. Each NPC is an 8-fold symmetric, cylindrical assembly consisting of ~500 copies of ~30 different proteins (nucleoporins or Nups). These Nups assemble into subcomplexes that form higher order structures called spokes. Eight spokes assemble into even larger modules: coaxial outer and inner rings form a symmetric core scaffold, which is connected to a membrane ring, a nuclear basket, and cytoplasmic RNA export complexes3. The scaffold surrounds a central channel, which is formed in part by multiple intrinsically disordered Phe-Gly (FG) repeat motifs extending from nucleoporins termed FG Nups. These FG motifs mediate selective nucleocytoplasmic transport through specific interactions with nuclear transport factors (NTRs) carrying their cognate macromolecular cargoes4. It has also been suggested that the central channel contains a feature called the central transporter5. Although partial structures have been described3,6,7, a full, high-resolution structure for the entire NPC in any organism has been lacking, leaving open key questions as to how the NPC is organized and functions, and how it evolved. To address these questions, we have determined an integrative structure of the yeast NPC at sub-nanometer precision.
Results and Discussion
Solving the structure of the S. cerevisiae NPC
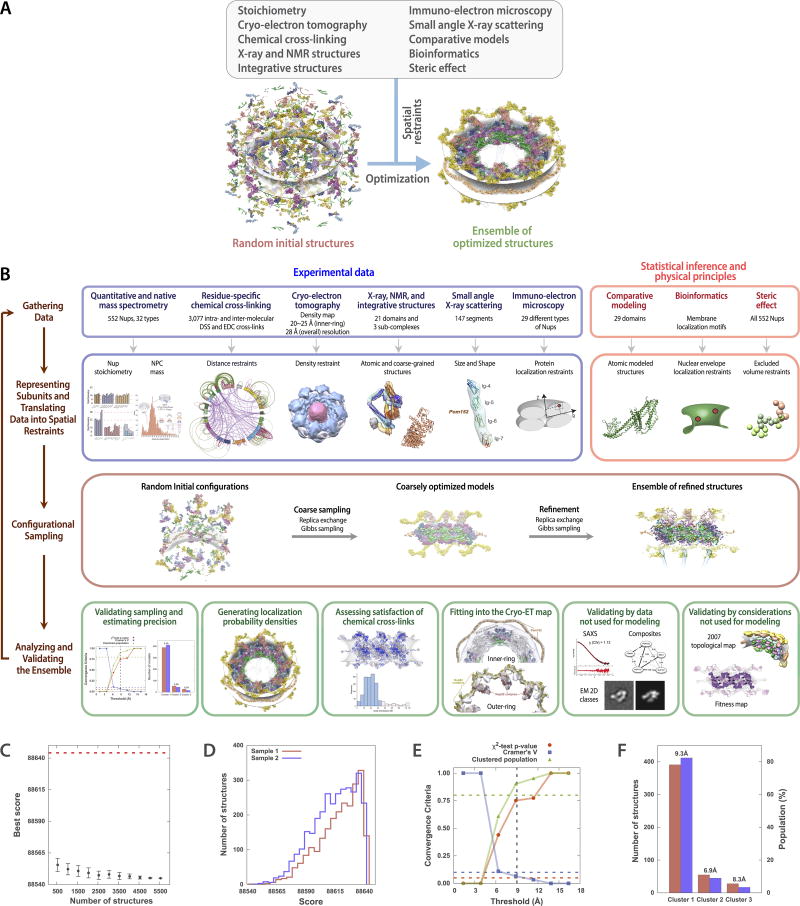
We developed a method to rapidly and gently isolate native yeast NPCs, allowing us to determine the type and amount of each Nup in the NPC, the proximities between Nups resolved to the amino acid residue level, as well as the mass and detailed morphology of the entire NPC. These data were then used to solve the structure using an integrative modeling approach8,9 (Extended Data Fig. 1; Methods; Supplementary Results and Discussion).
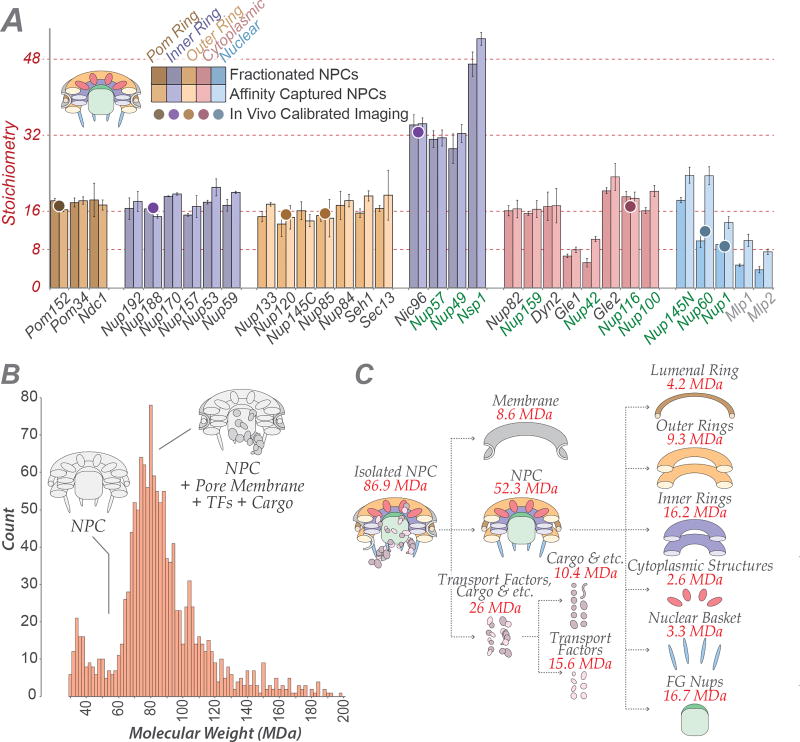
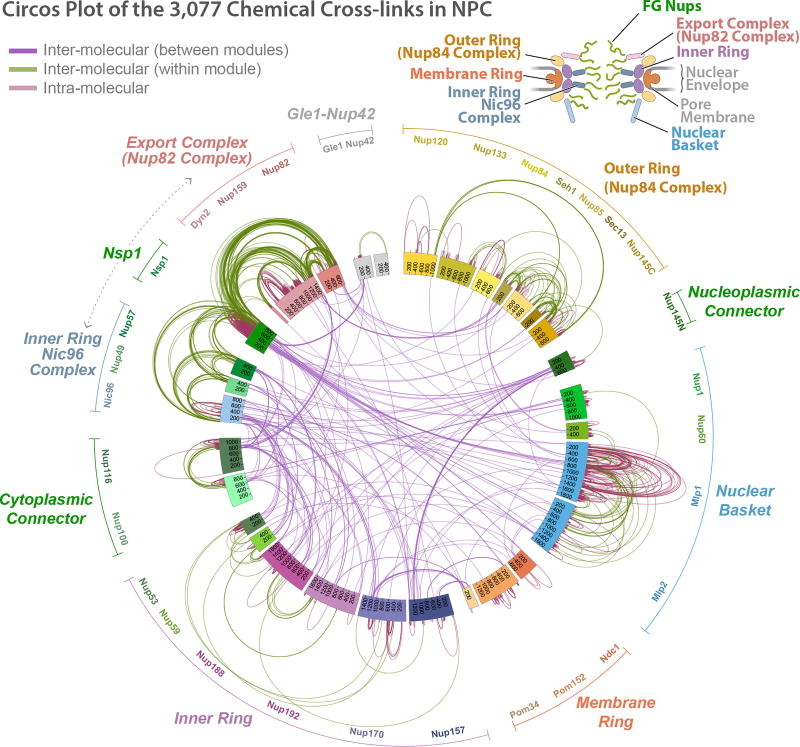
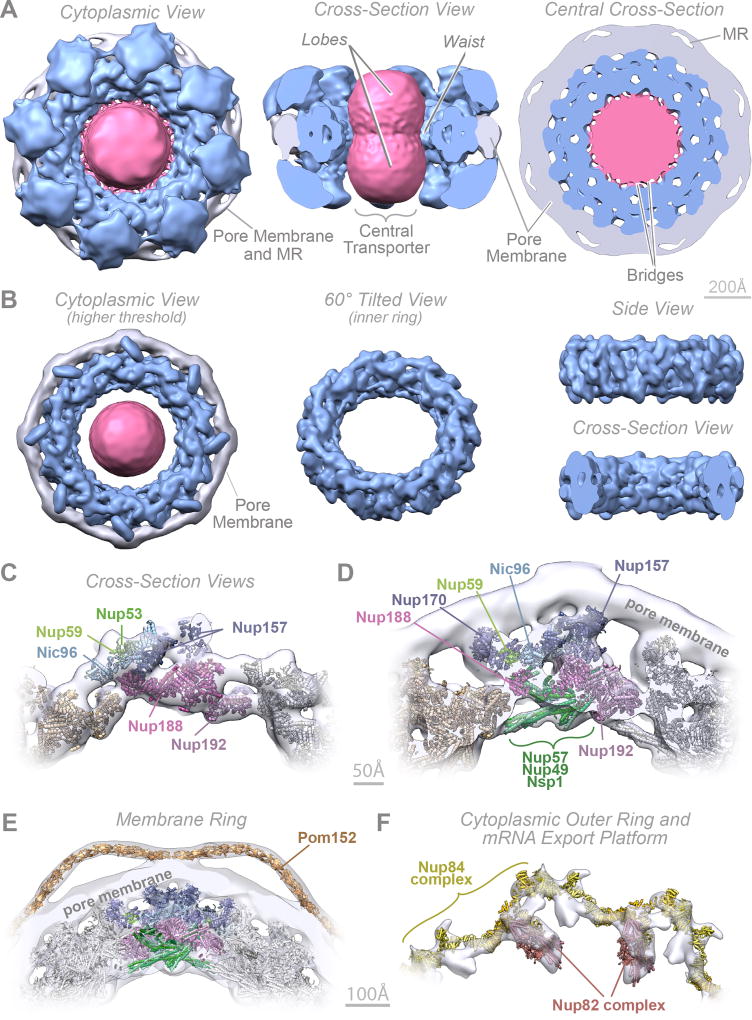
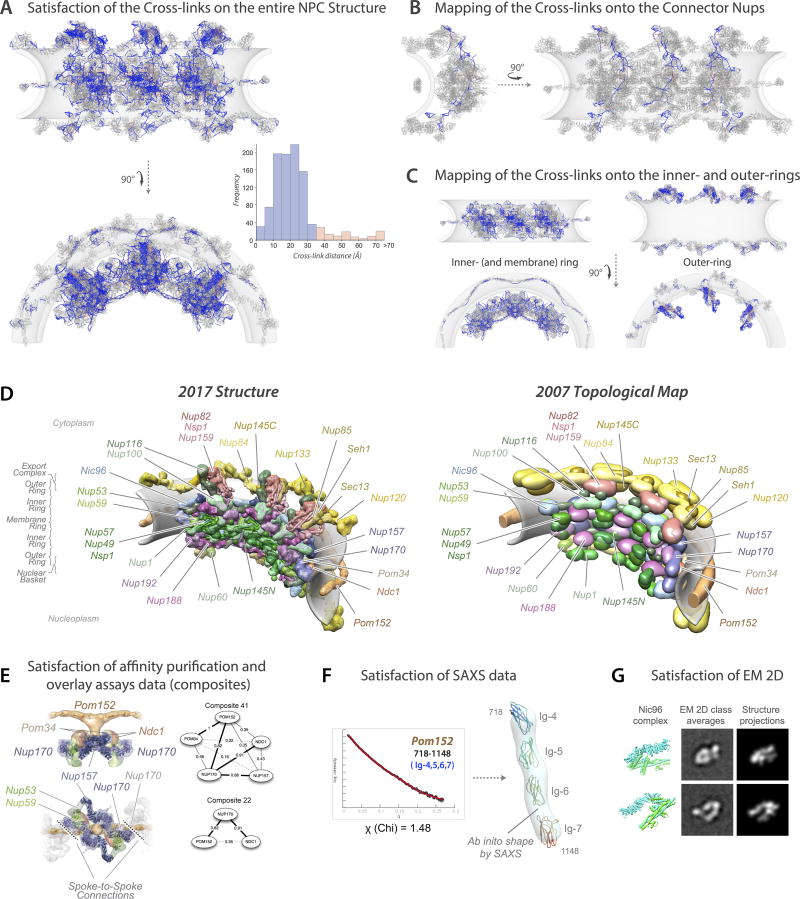
We determined the native mass of the entire NPC and a definitive stoichiometry for every Nup and associated molecules using mass spectrometric (MS) and in vivo imaging methods. The native NPC has a mass of 52 MDa, or ~87 MDa when considering membrane, cargo, and NTRs (Fig. 1; Extended Data Figs. 2–3C). To inform the proximities, orientations, and conformations of the Nups, isolated NPCs were subjected to cross-linking with MS readout9,10. This approach identified 3,077 unique cross-linked pairs of residues, providing distance restraints between them, both within and between Nups (Fig. 2; Supplementary Table 1; Methods). The morphology of the NPC was determined using cryo-electron tomography (Cryo-ET) and sub-tomogram averaging11 (Methods). This approach provided a final 3D map at ~28Å resolution, with a local resolution of 20–25Å for the inner ring, which has ~C2-symmetry (Fig. 3; Extended data Figs. 4–6). The NPCs retained a significant amount of NE membrane that forms a continuous belt around the midline of the structure (Fig. 3A–B,D–E). Though largely absent from recent EM maps6, a membrane protein ring interconnects adjacent spokes within the NE lumen (Fig. 3A,E). A cylindrically averaged, bi-lobed density fills the central channel (“central transporter” in Figs. 3A–B, 6A). Individual Nups, their domains, and subcomplexes were represented based on published crystallographic structures, integrative structures, and comparative models9,10,12 (Supplementary Table 2; Methods), validated by SAXS profiles for 18 Nups (147 constructs; Supplementary Table 6). An ensemble of structural solutions for the NPC that sufficiently satisfied all experimental data was calculated by extensive configurational sampling8,9 (Supplementary Table 3; Methods). Variability among these solutions defines the structure’s precision, as quantified by the average root-mean-square deviation between solutions in the final ensemble9. Our final structure defines the positions of 552 Nups (Fig. 4; Supplementary Videos 1–3), with an overall precision of ~9 Å (Extended Data Fig. 1E–F). The centroid solution is used as the representative structure. The structure was validated by numerous independent tests (Extended Data Figs. 1, 7–8; Supplementary Tables 3–4; Methods).
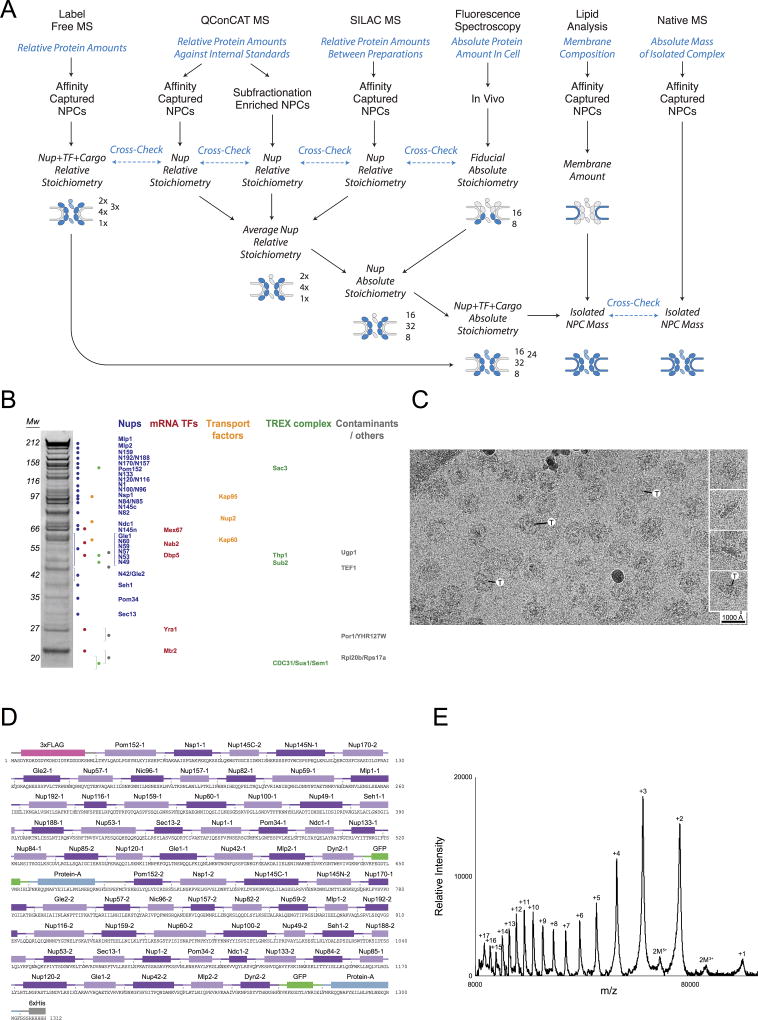
Figure 1. Defining the mass, composition and stoichiometry of the native NPC.
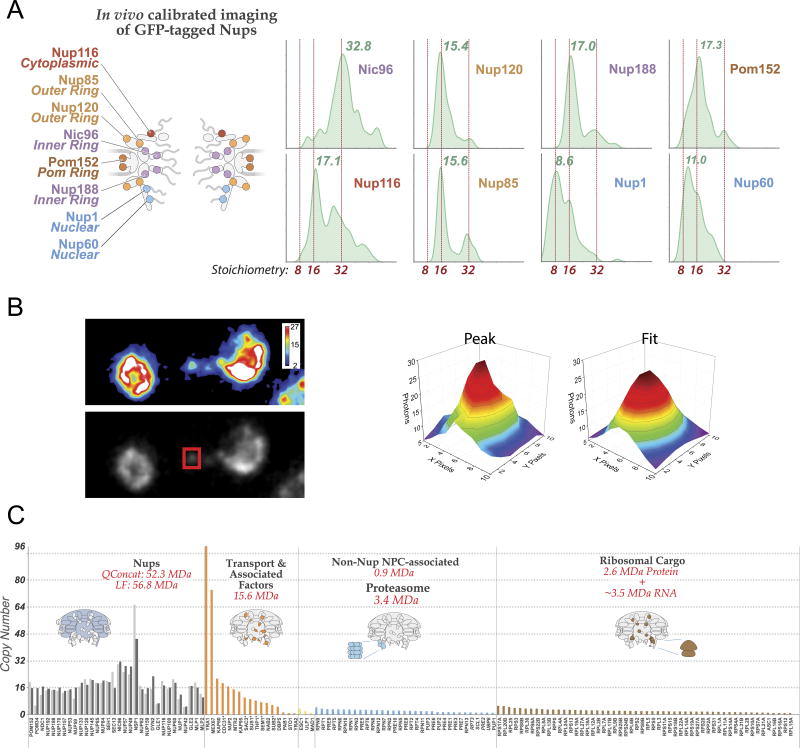
(A) Stoichiometry of the entire complement of NPC components determined by QConCat mass spectrometry (bar plot) and by in vivo calibrated imaging of Nup-GFP reporters (dots) (Extended Data Fig. 3). Darker and lighter colour bars (average ± SD) represent measurements from a diploid, non-tagged, S. uvarum strain (n=2–3 technical and 2 biological replicas) and haploid, tagged, S. cerevisiae strains (n=1–3 technical and 4 biological replicas), respectively. Each Nup is coloured based on their localization, as depicted in the cartoon. FG repeat-containing Nups are labeled in green.
(B) Affinity captured whole NPCs were analyzed intact by charge detection MS, and a representative MS spectrum is shown. 2 biological replicas, >3 runs and >1500 individual NPCs per run.
(C) Dissection of the mass and composition of an NPC.
Figure 2. Chemical cross-linking and mass spectrometry reveals nucleoporin connectivity in the NPC.
Circular plot showing the distribution of chemical cross-links (Supplementary Table 1), mapped to each nucleoporin represented as a coloured segment, with the amino acid residues indicated. The identity of each module and Nup is shown in the periphery of the plot. Types of cross-links are indicated in the top left diagram. Top right, diagram illustrating the relative positions of modules in the NPC.
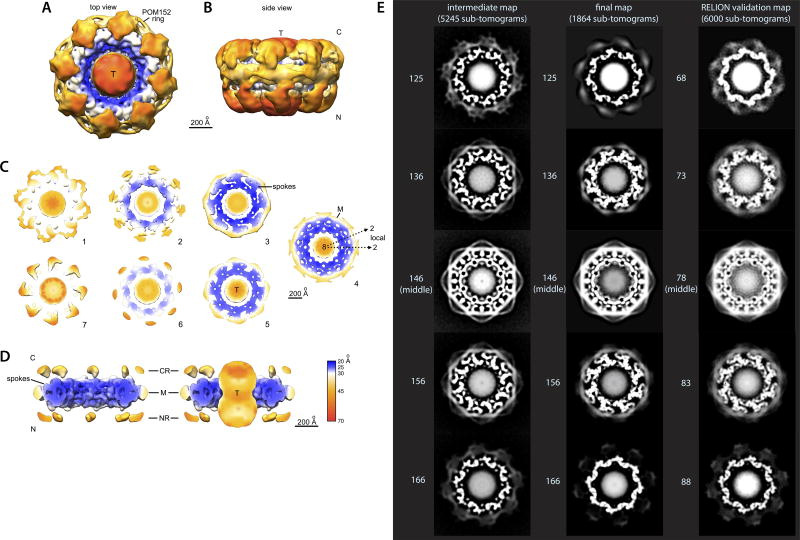
Figure 3. Morphology of the NPC.
(A–B) Cryo-ET map of the NPC: core scaffold, blue; membrane region, grey; central transporter, pink. MR: membrane ring. (A) (Left) top, cytoplasmic view; (middle) cross-section side view; (right) central cross section top view. (B) Cryo-ET map presented at a higher threshold. (Left) top view; (middle) inner ring 60° tilted view; (right) inner ring side (top) and cross-section (bottom) views. Scale bar 200 Å.
(C–F) Cross-section views show a representative structure embedded within the Cryo-ET density (grey) presented with different filtering and thresholding, to show the good fit to the Cryo-ET map in the inner (C, D) and membrane ring (E) and the cytoplasmic outer ring and mRNA export platform (F). Nups indicated as in Fig. 4. Scale bar 50 Å (C, D); 100 Å (E, F).
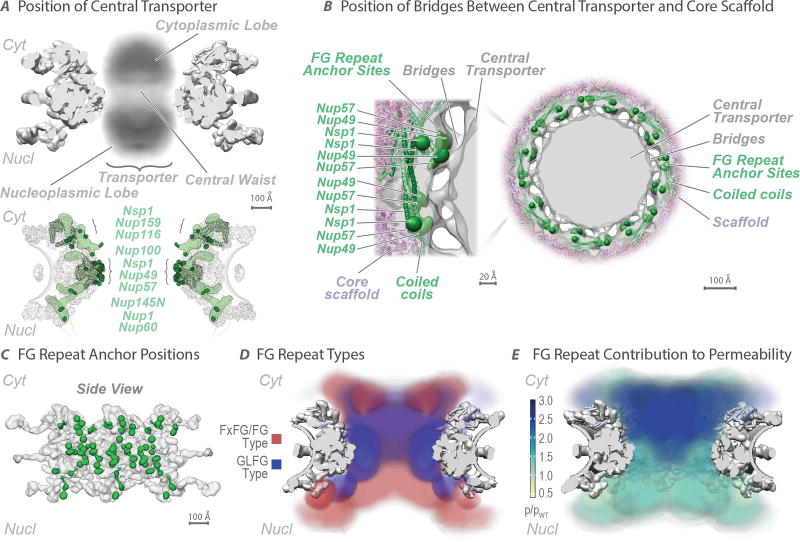
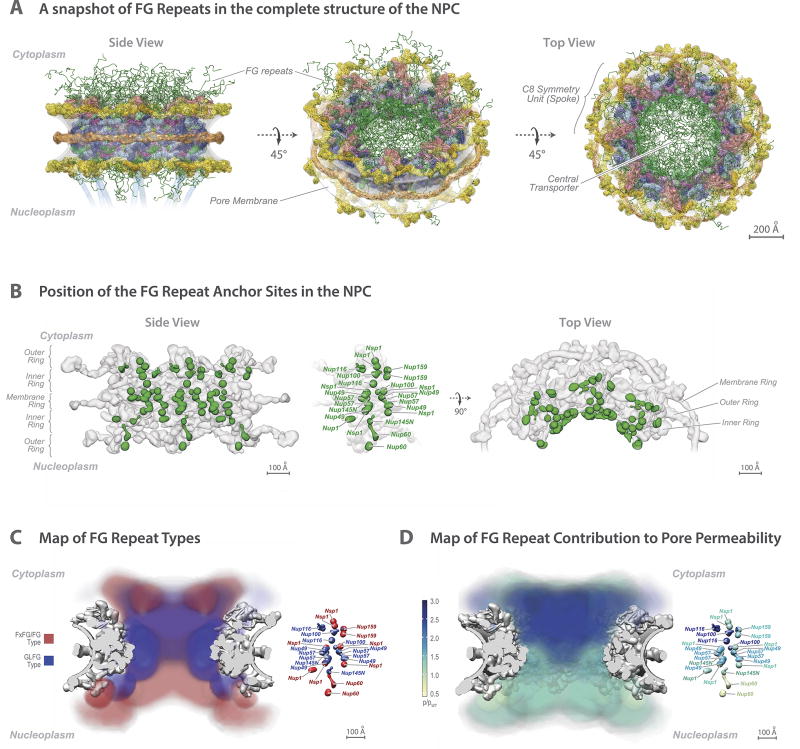
Figure 6. The distribution of the FG repeats informs the NPC transport gating mechanism.
(A) (Top) Central transporter density from the Cryo-ET map (Fig. 3) is shown within the structure of the NPC scaffold (grey). Features of the central transporter are indicated. (Bottom) Anchors (light green) in FG Nups largely direct the FG repeat emanating points (dark green) towards the central channel. Scale bar 100 Å.
(B) Central cross-section of the Cryo-ET map (grey) with embedded representative NPC structure (Fig. 4), showing the central transporter and the bridges connecting it to the core scaffold in top view (scale bar 100 Å), with a magnified view of one spoke on the left (scale bar 20 Å). The anchor points for the FG repeats of Nup49, Nup57, and Nsp1 are depicted as green densities.
(C) Position of FG repeat anchor points (green) within a side view of three spokes of the scaffold (grey). Scale bar 100 Å.
(D) Heat mapping of repeats of FxFG/FG type (red) and GLFG type (blue), from Brownian dynamics simulations (Methods), showing partitioning to different regions of the central channel. Scale bar 100 Å.
(E) Heat mapping of the effect of FG repeat region truncations on NPC permeability; the severity of the permeability defect (p/pWT)32 is indicated in increasing shades from minor (light green) to severe (dark blue). Scale bar 100 Å.
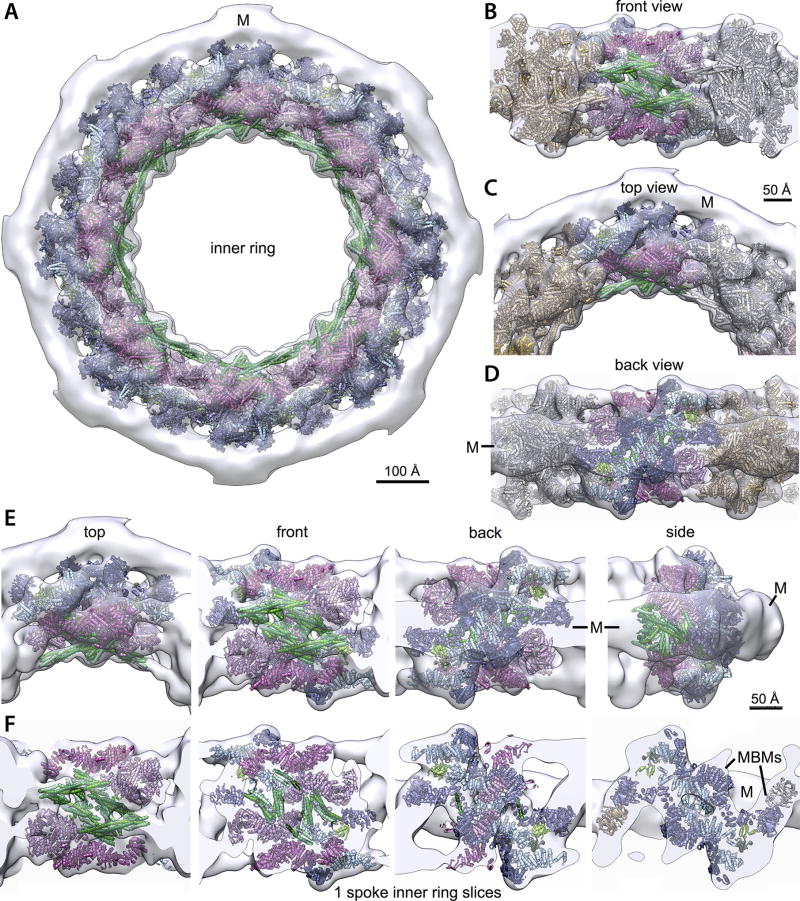
Figure 4. Structural dissection of the NPC.
The complete structure of the NPC and its components. For each Nup, the localization probability density of the ensemble of structures is shown with a representative structure from the ensemble embedded within it (Supplementary Table 2). The structure is shown in different orientations, with a model of the pore membrane region shown in grey (Supplementary Videos 1–3).
(A) Two views of three consecutive NPC spokes (C8-symmetry units), showing how the coaxial outer, inner, and membrane rings run continuously between spokes.
(B) Top cytoplasmic view of the complete NPC structure with modeled FG repeat regions (green). Scale bar 200 Å.
(C) Front view of a single NPC spoke. Scale bar 100 Å.
(D) Relative position of major NPC components and connections both within and between spokes. Top (left column) and side (right column) views are shown. The membrane ring (beige) is included for reference. Flexible connectors between outer and inner rings are shown on the top and bottom panels, with the inner and membrane rings shown as faded grey densities.
(E) Exploded view of three consecutive spokes, spanning from the cytoplasmic face (top) to the nuclear face (bottom), with blue dashed lines connecting neighboring rings.
(F) The cytoplasmic mRNA export complex (top), the Nup84 complex (center) and the inner ring complex, including the Nic96 complex (bottom), from a single spoke. The complexes are shown as an exploded diagram, with blue dashed lines connecting neighboring components.
The NPC’s multiple functionalities and enormous size present unique and significant structural challenges: it must form a stable passageway with a fixed inner diameter; it must be anchored to the NE and stabilize the pore membrane within which it resides, with a height appropriate for the thickness of the NE; it must correctly position the transport machinery; and, it must resist stresses that might lead to disassembly or malfunction. Our structure suggests how each of these challenges are met and, by comparison with the vertebrate scaffold6, how different organisms may meet these challenges (below).
Forming a stable and defined passageway
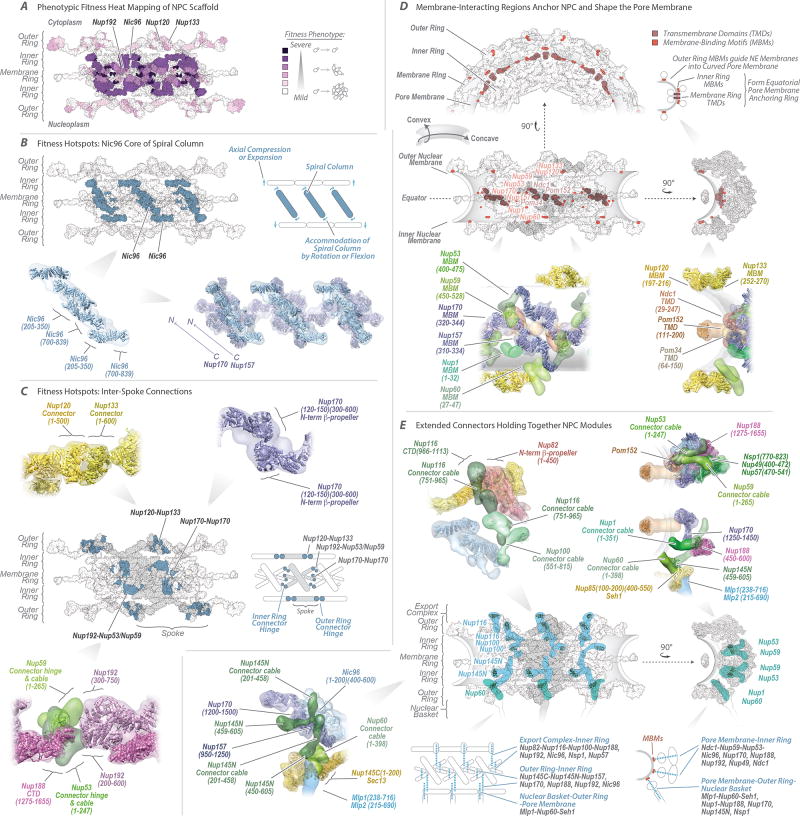
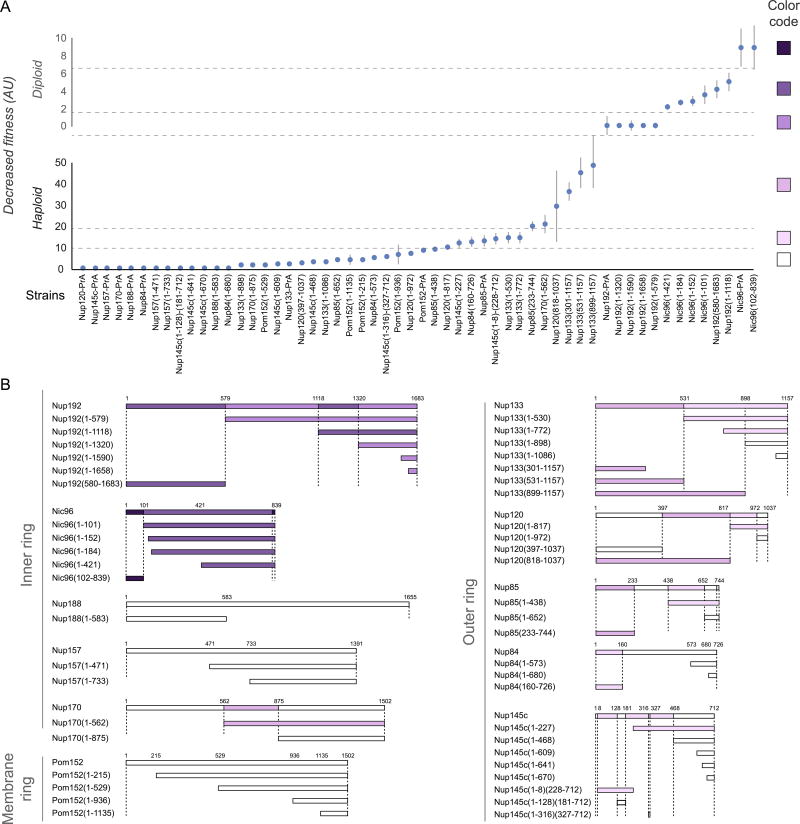
The fitness defects of strains containing Nup truncations provide an estimate of the structural importance of the truncated regions9,13. Thus, we quantified the fitness defect of strains containing systematic truncations of every major symmetric Nup using ODELAY14 (an automated phenotypic analysis platform; Extended Data Fig. 12). Results were heat-mapped onto the NPC structure to reveal critical elements of NPC stability (Fig. 5A). Crucial stabilizing elements are found in the inner ring, including Nic96, which forms the heart of a diagonally oriented column within each spoke (Fig. 5B) and interacts with every other protein in the inner ring (Fig. 4D–F). This high connectivity explains why Nic96 is an essential keystone holding much of the NPC’s scaffold in place. The remainder of each diagonal column is made of Nup157 and Nup170, which flank Nic96 (Fig. 5B); Nup157 and Nup170 are functionally redundant, but are synthetically lethal15 and together form another essential element of the diagonal column. Inter-spoke connections represent a second crucial stabilizing element. Nup192 likely serves as a cross-brace between adjacent spokes (Fig. 5A,C), while the N-termini of Nup170 and disordered regions of Nup53 and Nup59 also form key connections between adjacent spokes16 (Figs. 4D, 5C). The inter-spoke connections are established largely through small, hinge-like contacts that may confer flexibility to the interface between adjacent spokes. Similarly, the diagonal arrangement of the central columns may also allow rotation or local flexing (Fig. 5B), accommodating compression and expansion forces from NE distortions and from the central transporter and the transit of cargoes. Nup188 and Nup192 act as radial separators between the Nic96 column and the triple coiled-coil domains of Nsp1, Nup57, and Nup49, which form a discontinuous ring that defines the narrowest part of the passageway, and may allow some dilation of the NPC (Fig. 4D–F). This architecture sets a soft upper limit for the size of cargoes (~40nm) that can transit the NPC4.
Figure 5. Key NPC architectural features and principles.
(A) Severity of fitness defects, indicated in increasing shades of purple for specific truncations of nucleoporins (Extended Data Fig. 12), mapped onto three spokes of the NPC.
(B–C) Structures corresponding to the position of the most severe defects (dark blue). (B) Diagonally oriented columns reinforcing the core scaffold may accommodate NPC compression and expansion (diagram to right). Molecular details of Nup arrangement are shown at bottom (relevant residue numbers indicated). (C) The position of hotspots also coincides with spoke-to-spoke connections (central spoke in grey, flanking spokes white; diagram to right). Top and bottom, molecular details of spoke-to-spoke connector hinges.
(D) Top and center left, three spokes shown as top and front views; center right, one spoke, side view. A diagram indicates convex and concave pore membrane curvatures. Positions of transmembrane domains (TMDs) and membrane-binding motifs (MBMs) are depicted and their proteins labeled in brown and orange, respectively. Top right, diagrammatic side view showing how the MBMs and TMDs curve the pore membrane. Bottom, molecular details of the Nups containing the TMDs and MBMs.
(E) Second row left, three spokes in front view, showing how vertical connector Nups (cyan) spanning from the cytoplasmic to nuclear sides of the NPC connect the rings. Second row right, one spoke in side view, showing how horizontal connector Nups (aquamarine) connect modules spanning from the pore membrane to the central channel. First row and third row left show the molecular details of the connectors within the NPC. Bottom row center-right, diagrammatic views of the connectors depicted as blue dotted lines and the modules they connect labeled in blue; major Nups being contacted by connectors listed in grey.
How the NPC shapes the NE
The pore membrane, where the inner and outer membranes of the NE join, defines the inner surface of a torus, and so has both concave and convex curvatures (Fig. 5D). The inner ring is anchored to the pore membrane through membrane-binding motifs (MBMs) on the β-propellers at the N-termini of Nup157 and Nup170, and on the C-termini of Nup53 and Nup5917–19. These proteins also interact with the scaffold-facing domains of Pom152, Ndc1, and Pom34, each of which carry transmembrane domains (TMDs) (Fig. 4D). Together, these MBMs and TMDs form an NPC-anchoring girdle of membrane-associated motifs around the scaffold equator, defining the concave curve of the pore membrane. The convex curvature is defined by both outer and inner rings (Figs. 4A,E, 5D). Each outer ring is formed by eight Y-shaped Nup84 complexes arranged head-to-tail and joined by an interaction between the N-termini of Nup120 and Nup13320, creating another hinged spoke-to-spoke interface and a minor fitness hotspot (Fig. 5A,C). The outer rings also help define the overall height of the NPC, such that it is appropriate for the width of the NE. Each Nup84 complex is anchored to the pore membrane by MBMs situated within the N-terminal β-propellers of Nup133 and Nup12010,12,21 (Fig. 5D). The convex curvature of the pore membrane is thus defined and stabilized by both a ring of MBMs underneath the outer rings, and the thick girdle of MBMs and TMDs around the NPC equator. At the nuclear side, MBMs from Nup1 and Nup60 help anchor the basket to the NE (Fig. 5D–E).
In the membrane ring, the luminal domain of Pom152 is composed of 9 Ig-like fold repeats22 that oligomerize in an anti-parallel fashion to form 8 circumferential arches within the NE lumen, forming additional connections between adjacent spokes. Pom152 appears to be pre-stressed by assembly into these arches (Fig. 4E); the resulting tension may minimize elliptical distortion of the NPC22. Each arch also delimits a channel (300 × 120 Å wide) between itself and the underlying pore membrane (Figs. 3E and 4E). The outer rings form a series of circumferential arches that align with the Pom152 luminal arches (Fig. 4B,E). These arches align with hinges in the inner ring (Fig. 5C) that could flex to form lateral openings between spokes. This juxtaposition of arches and transient openings may delineate conduits for nucleocytoplasmic transport of transmembrane proteins23, potentially resolving the issue of how membrane proteins transit the NPC24.
Positioning the RNA processing platforms
Whereas the core scaffold is symmetric about the plane of the NE, two machineries associated with RNA processing and transport, termed the basket and export platform, are located at the nuclear and cytoplasmic faces of the NPC, respectively (Fig. 4D–F). At the core of the export platform is the Nup82 complex, whose coiled-coil bundle is attached to the Nup85/Seh1 arm and hub region of the Nup84 complex in the cytoplasmic outer ring (Fig. 4F). Together, they form a lateral gantry facing the central channel. Gle1 extends from the Nup82 complex by an α-helical rod that holds itself, the RNA helicase Dbp5, and the FG repeat-carrying Nup42 over the middle of the central channel9,24,25. As a result, numerous transport factor docking sites and the ATP-dependent RNA remodeling proteins are aligned above the cytoplasmic exit of the NPC to efficiently receive exporting RNAs to remodel and then release them into the cytoplasm. Likewise, Mlp1 and Mlp2 in the nuclear basket are anchored to the core scaffold mainly via the Nup85/Seh1 arm, much like the Nup82 complex (Figs. 4D–E, 5E). The nuclear basket serves as a platform for the first stages of RNA processing and export26, while the export platform organizes the last stages of export25. Similarities between the export platform and basket suggest that these structures are ancient homologs (Extended Data Fig. 11), whose asymmetric localization directs unidirectional export of transcripts out of the nucleus.
Flexible connectors tie the NPC together
Recently, certain disordered connectors have been shown to be important for holding parts of the scaffold together6,7,16. However, the full extent to which such connectors are critical to NPC integrity can only be appreciated in light of the present structure. Remarkably, flexible connectors run the entire length of each spoke, tying together every major element in the NPC (Fig. 5C,E). They link the periphery and outer rings to the inner rings, both inner rings to the pore membrane, and adjacent spokes to each other. We identified two types of connectors (Supplementary Results and Discussion). First, there are vertical connections, aligned parallel to the cylindrical axis of the NPC, constituting the main anchor points between the export platform and the inner ring. On the nuclear side, similar connections are present between the nuclear basket and the inner ring, with an additional connection between the basket and outer ring (Figs. 4D, 5E). Secondly, there are horizontal flexible connectors that link the central channel to the pore membrane between adjacent spokes (Fig. 5E). Collectively, these flexible connectors may serve to permit limited movement of the more rigid modules with respect to each other, thereby providing the NPC with another degree of flexibility in response to deformation27.
Organization of the nucleocytoplasmic transport machinery
Despite its critical function, the central gating machinery has been largely excluded from recent NPC maps, and its properties have remained controversial. Here, we confirm the existence of a large central transporter, with two high-density ‘lobes’ connected by a narrower ‘waist’ of lower density5 (Figs. 3, 6; Extended Data Figs. 4–6). This central transporter is comprised of multiple FG repeats that account for ~9 MDa, together with on average ~26 MDa of NTRs and their cargoes caught in transit (though likely somewhat averaged out in our map) (Fig. 1B–C; Extended Data Fig. 3C). Indeed, even after isolation each NPC carried 10–80 copies of each of the major NTRs28, reflecting the huge and varied transport flux through NPCs.
The localization of FG repeat anchor points reveals three patterns. First, a vertical path is formed along each spoke by a continuous array of FG repeats (Fig. 6A,C; Extended Data Fig. 9B–D). By binding to these repeats, NTRs may follow these paths across the entire NPC. Second, the FG anchor points of Nsp1/Nup57/Nup49 form a central ring on the equator of the NPC (Fig. 6B). Thin bridges in our Cryo-ET map coincide with the location of these FG anchor points, indicating that these bridges are comprised of the FG repeats themselves emanating from their anchor points (Fig. 6B). Third, instead of projecting from the NPC towards the cytoplasm and nucleoplasm as often represented, the NPC’s structured regions largely direct the FG repeat regions inwards, towards the axis of the central channel (Fig. 6A). This geometry generates a highly concentrated (25–150 mM) and dynamic FG repeat phase through which cargo-carrying NTRs readily pass, facilitated by their specific FG interactions, while nonspecific macromolecular diffusion is hindered by this same dense phase29.
It has been suggested that the two main FG repeat types (“FxFG/FG” and “GLFG”) are segregated in the NPC to define functionally distinct zones of the gating machinery30. In agreement, we find the FxFG/FG-type repeats to be enriched in the nuclear and cytoplasmic peripheries of the NPC, where the RNA-associating export platform and basket reside (Fig. 6D; Extended Data Fig. 9C), consistent with the known role of FxFG/FG-type repeats in docking exporting RNAs31. In contrast, the GLFG-type repeats are enriched in regions adjacent to the inner ring and near the cytoplasmic entrance to the central channel. This cytoplasmic localization coincides with the position of those FG repeats that are most important for limiting the passage of nonspecific macromolecules (Fig. 6E; Extended Data Fig. 9D), and is consistent with the known role for the GLFG-type repeats in maintaining the passive permeability barrier32,33.
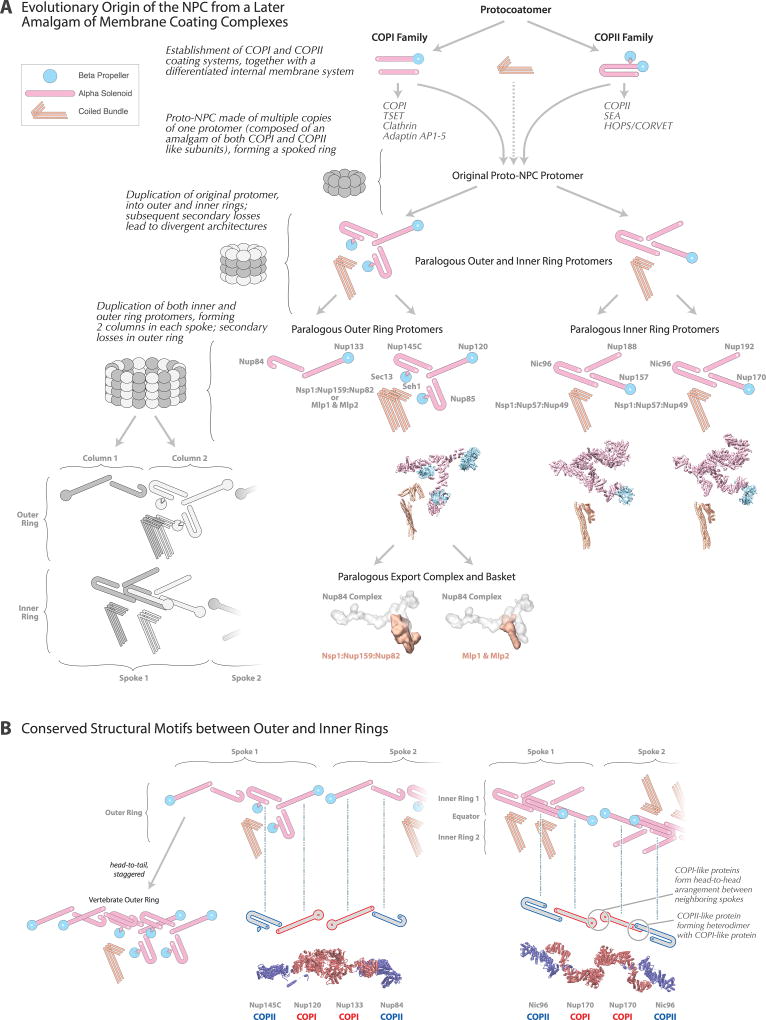
Evolutionary origin and diversity of the NPC
NPCs share architectural features with vesicle coating complexes (Extended Data Fig. 11), which led us to hypothesize that they share a common evolutionary ancestor termed the “protocoatomer”34. Two major families of coating complexes exist: COPI/clathrin and COPII, with each having discrete vesicle recognition and trafficking roles35,36. We find both COPI/clathrin-like and COPII-like features in the NPC, suggesting that ancestral COPI and COPII coating families evolved first, followed by the NPC, which may have evolved through a partnership of COPI and COPII coats. This hypothesis implies that the nucleus was a later addition on the evolutionary path of the first eukaryotes (Supplementary Results and Discussion).
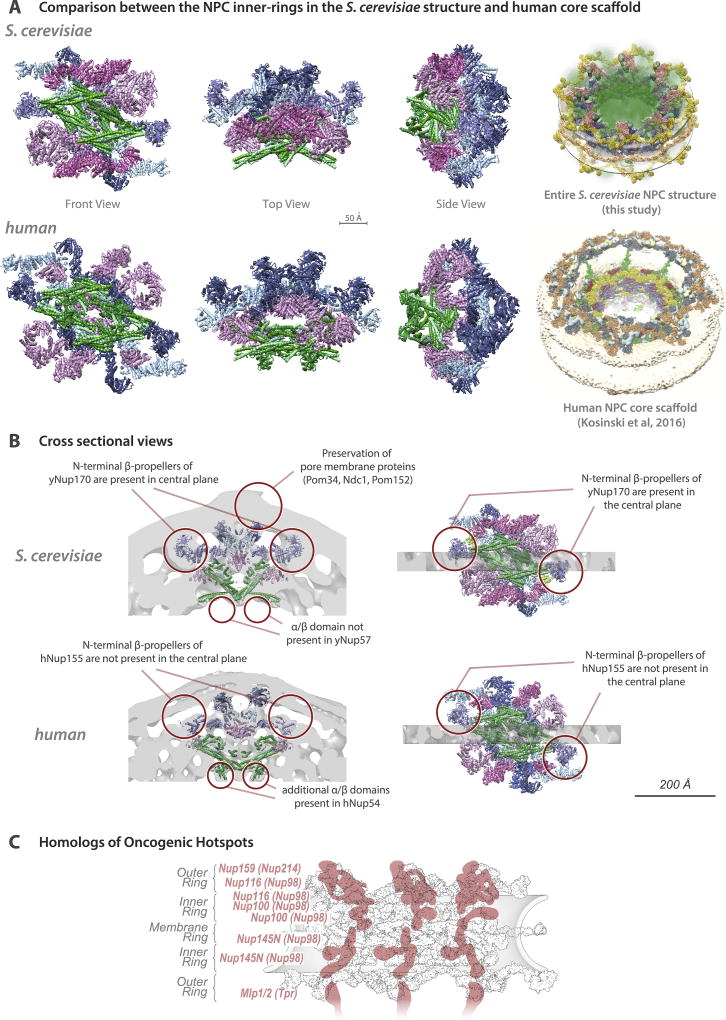
Despite significant conservation of some elements of NPC architecture, other elements can vary widely between species. Generally, the inner ring appears most conserved37,38, as seen in a comparison of our yeast structure with the human scaffold6, although the latter is more expanded (Extended Data Fig. 10). In contrast, peripheral elements exhibit significant lineage-specific losses and duplications38,39. In yeast, each outer ring is formed by 8 copies of the Nup84 complex (Figs. 3F, 4), whereas in vertebrates each outer ring contains 16 copies of the equivalent Y-shaped complex arranged in two interlocked rings17,40. Moreover, we see neither an additional copy of Nup157/Nup170 connecting the outer and inner rings nor Nup188/Nup192 in the outer rings, as indicated in humans6 (Fig. 4D–F). Another model assumed that fungal and human core scaffolds have essentially identical structures7. Our data invalidate this assumption (Fig. 1; Supplementary Table 2A; Supplementary Results and Discussion) as well as an earlier “fencepost” model41. In summary, there is no single universal NPC structure; instead, similar structural elements are used in somewhat different arrangements to generate many lineage-specific adaptations.
Conclusions
We described the structure of the entire yeast NPC at sub-nanometer precision. At the heart of the inner ring, rigid diagonal columns reinforce the NPC’s structural integrity. Membrane-binding and transmembrane Nups are strategically placed throughout the core scaffold to stabilize pore membrane curvature and clamp the NPC to the NE. Connectors run the length of each spoke, flexibly tying together all the major modules in the NPC. The NPC’s architecture is reminiscent of a suspension bridge, in which rigid supporting columns are firmly anchored to a substrate while flexible suspension cables connect the columns and roadway, to provide a strong and resilient structure. We show that most FG Nup anchor points face inward, towards the NPC central channel, to generate a highly concentrated milieu of FG repeats: FxFG/FG repeats form mRNA docking “traps” at the entrance and exit of the channel, whilst GLFG repeats help form a cytoplasmically biased permeability barrier.
Despite differences, yeast and human NPCs retain a striking degree of structural conservation (Extended Data Fig. 10). As a result, many of the conclusions drawn here should be applicable to the human NPC. To illustrate this point, we mapped the positions of yeast homologs of the oncogenic hotspot human Nup214, Nup98, and Tpr2 (Extended Data Fig. 10C). Rather than being randomly scattered, these positions coincide with RNA-binding platforms on the cytoplasmic and nucleoplasmic faces of the NPC as well as with several critical connectors and associated FG regions. This conservation suggests that alterations in RNA export, and changes in NPC architecture induced by defective connectors, may underlie the altered behavior of NPCs in cancer cells. Thus, our yeast structure provides a roadmap with the potential to advance our understanding of NPC physiology and nuclear transport in general.
METHODS
1. Yeast strains and materials
All S. cerevisiae strains used in this study are listed in Supplementary Table 5, with the exception of the Nup84 complex truncation mutants42 and the Pom152 truncation mutants43. Unless otherwise stated, strains were grown at 30°C in YPD media (1% yeast extract, 2% bactopeptone, and 2% glucose). The diploid Saccharomyces uvarum strain (ATCC 9080) was grown and processed for NE purification as previously described44.
The following materials were used in this study: Dynabeads M-270 Epoxy (143.02D; Invitrogen); rabbit IgG (55944; MP Biomedicals); protease inhibitor cocktail (P-8340; Sigma-Aldrich); and Solution P (2 mg Pepstatin A, 90 mg PMSF, and 5 mL of absolute ethanol).
2. Immuno-purification of the endogenous S. cerevisiae NPC
A novel immuno-purification protocol for the isolation of endogenous whole NPCs from S. cerevisiae was developed using previously published methodology45–50. S. cerevisiae Mlp1, Nup84, or Nup82 encoding genes were genomically tagged with PrA preceded by the human rhinovirus 3C protease (ppx) target sequence (GLEVLFQGPS). Cells were grown in YPD media at 30°C until early log phase (~2×107 cells/ml), harvested, frozen in liquid nitrogen and cryogenically lysed in a planetary ball mill PM 100 (Retsch) (http://lab.rockefeller.edu/rout/protocols). Frozen cell powder was resuspended in 9 volumes of resuspension buffer (20 mM Hepes/KOH pH 7.4, 50 mM potassium acetate, 20 mM NaCl, 2 mM MgCl2, 0.5% (w/v) Triton X-100, 0.1% (w/v) Tween-20, 1 mM DTT, 10% (v/v) glycerol, 1/500 (v/v) Protease Inhibitor Cocktail (Sigma)). Cell lysate was clarified by centrifugation at 2,500 RCF for 5 minutes followed by filtration through 1.6 µm filters (Whatman glass microfiber syringe filters). Magnetic beads (Invitrogen) conjugated to rabbit IgG antibodies (http://lab.rockefeller.edu/rout/protocols) were added to the clarified cell lysate at a concentration of 50 µL slurry per 1 gram of frozen cell powder and incubated for 30 minutes at 4°C. Beads were washed once with 1 mL of elution buffer without protease inhibitors (20 mM Hepes/KOH pH 7.4, 50 mM potassium acetate, 20 mM NaCl, 2 mM MgCl2, 0.1% (w/v) Tween-20, 1 mM DTT, 10% (v/v) glycerol). For native elution of the complex, the desired volume of elution buffer with PreScission protease (GE Healthcare) (1/15 (v/v)) was added to the beads and incubated for 45 minutes at 4°C. A magnet was used to remove the beads and collect the supernatant. Beads were subsequently washed with the desired volume of elution buffer containing 1/500 (v/v) Protease Inhibitor Cocktail (Sigma). The total elution volume was centrifuged at 20,000 g for 5 minutes to remove the residual magnetic beads. Typical yield of the immuno-purification is ~4 µg of isolated NPCs per 1 gram frozen cell powder (see Extended Data Fig. 2B for SDS-PAGE analysis; for gel source data, see Supplementary Fig. 1).
3. Mass and Stoichiometry of the native S. cerevisiae NPC
Quantification of the absolute stoichiometry of each nucleoporin in the native NPCs was performed using a strategy that combined several orthologous methods (Extended Data Fig. 2A): (1) use of synthetic concatemers of tryptic peptides or QconCATs51 to define the relative stoichiometry of each component by quantitative MS in affinity-captured NPCs; (2) In vivo calibrated imaging analysis of GFP-tagged Nups52, to quantify the absolute copy number per NPC of Nups selected to represent each major module of the NPC; and (3) Charge Detection MS to measure the total mass of affinity-captured NPCs53. For the calculation of the integrative NPC structure, the final copy numbers were rounded to fit the known NPC C8-symmetry and these values are indicated in Supplementary Table 2A.
3.1 NPC QconCat design and purification
Mass spectrometry quantification of the relative amounts of each nucleoprotein in the purified NPC complex was performed using two specifically designed, heavy-labeled synthetic internal standards or QconCATs51,54 (Extended Data Fig. 2D–E) formed by concatenated quantotypic nucleoporin peptides. To minimize the potential effect of having different residues flanking the trypsin cleavage site on the cleavage efficiency, we included the native, three-residue flanking sequences framing the trypsin cleavage site for each peptide55. For QconCAT-A (Extended Data Fig. 2D), two peptides for each of the nucleoporins and one peptide for S. aureus Protein A and A. victoria GFP proteins, were selected (Supplementary Table 7) based on their favorable signal responses in LC-MS analyses of NPC samples and by fitting to the following criteria (when possible): i) the native three-residue flanking sequences at both sides of the trypsin cleavage sequence do not contain Lys or Arg; ii) avoid the presence of Cys or Met residues within the peptide; iii) avoid the presence of potential internal trypsin cleavage sites (Lys or Arg residues); iv) peptides should be less than 3,000 Da (small size); and v) avoid peptides showing obvious interferences from co-eluting peptides during the liquid chromatography separation for MS analyses. QconCAT-B included two quantotypic peptides for Nup159, Mlp2, Nup192, Nup84, Nup85, Nup120, Nup49, Nup57, Pom152, Nic96 and the same GFP peptide as in QconCAT-A (Supplementary Table 7). As an internal control, both QconCAT A and B included the same peptides for Nic96, Pom152 and GFP. Each synthetic gene was designed by concatenation of the sequences encoding the selected peptides and addition of a 6×His C-terminal tag (Extended Data Fig. 2D). A sacrificial 3×FLAG peptide was also included at the N-terminus of QconCAT-A, resulting in a protein of 148.2 kDa. The E. coli codon optimized sequences were cloned into: i) plasmid pET15-b (as a NcoI-XhoI fragment) in the case of QconCAT-A; and ii) pGEX6p-1 (as a BamHI-XhoI fragment) in the case of QconCAT-B, resulting in the expression of a 68.1 kDa protein with a n-terminal GST tag that was mainly used as a sacrificial peptide56. The QconCAT proteins were expressed by growing 300 mL of BL21 E. coli cells at 37°C to OD600 = 0.6 in minimal M9 media without ammonium chloride51,54 supplemented with light amino acids and 0.5 mg/mL of heavy arginine and lysine (L-arginine:HCl 13C6; L-lysine:2HCl 13C6, Cambridge Isotope Laboratories Inc.). IPTG (1 mM) was used to induce expression of the constructs for 3 hours at 37°C. Harvested cells were processed using BugBuster Extraction Reagent (Novagen) as indicated by the manufacturer to isolate the inclusion bodies were the QconCAT protein is accumulated. The full-length QconCAT-A was then purified by resuspending the inclusion bodies pellet in binding buffer (20 mM sodium phosphate pH 7.4, 45 mM imidazole, 500 mM NaCl, 6 M guanidinium chloride, 10 mM TCEP (0.5 M Bond-Breaker TCEP solution, Thermo-Scientific), 1/500 protease inhibitor cocktail (PIC, Roche)) and passed through an equilibrated His-Trap HP (GE Healthcare) at room temperature. The retained NPC QconCAT-A was then eluted in 20 mM sodium phosphate pH 7.4, 500 mM imidazole, 500 mM NaCl, 6 M guanidinium hydrochloride, 1 mM TCEP, 1/500 PIC. 100 µL aliquots of the resulting elution were precipitated to eliminate the guanidinium hydrochloride by adding ice-cold ethanol to a final concentration of 90% an incubating the samples at −20°C for 2 hours. Samples were then centrifuged for 10 minutes at 14,000 rpm and 4°C to pellet the precipitated protein. The resulting pellet was washed with ice-cold 90% ethanol and let air dry until most of the liquid was evaporated, leaving a wet pellet. These pellets were solubilized with 5% SDS, 500 mM Tris-HCl pH 8.0, 5 mM TCEP buffer, by incubating for 5 minutes at RT and 5 minutes at 72°C and centrifuged for 10 minutes at 14,000 rpm and RT. The supernatants were recovered and two of them combined and injected into a TSKgel G4000SWxl size exclusion column (TOSOH Bioscience) coupled to a TSKgel SWxl guard column (TOSOH Bioscience), pre-equilibrated in running buffer (40 mM Hepes-KOH pH 7.0, 150 mM NaCl, 0.1% SDS, 5 mM TCEP, 1 mM EDTA). 200 µL fractions were collected and analyzed by SDS-PAGE to detect the presence of the QconCAT-A peak. Fractions containing the full-length, pure protein were supplemented with a final 20% glycerol (v/v), aliquoted, flash-frozen in liquid nitrogen and stored at −80°C for further use. In the case of QconCAT-B, the protein was purified using His-Trap HP and the elution precipitated and prepared as described for QconCAT-A. The resulting sample was injected into a TSKgel Super SW3000 size exclusion column (TOSOH Bioscience) pre-equilibrated in running buffer (40 mM Hepes-KOH pH 7.0, 150 mM NaCl, 0.1% SDS, 5 mM TCEP, 1 mM EDTA). 100 µL fractions were collected and analyzed by SDS-PAGE to detect the presence of the QconCAT-B peak. Fractions containing the full-length, pure protein were stored as indicated for QconCAT-A.
For the quantitative MS analysis, the native NPCs from Saccharomyces cerevisiae PPX-PrA-tagged haploid strains were affinity captured as described above, or purified as enriched NPCs from a diploid Saccharomyces uvarum strain using a subfractionation method previously described in detail44,57–59 (http://lab.rockefeller.edu/rout/protocols), using 0.035 mg heparin per mg of fraction protein. For affinity captured NPCs, the natively eluted NPCs (5 µg) were concentrated by pelleting at 40,000 rpm for 20 minutes at 4°C in a TLA 55 rotor (Beckman). In the case of subfractionation enriched NPCs, a volume of the 1.45 M / 1.85 M sucrose gradient fraction that contained an estimated 5 µg of NPCs was diluted 1/5 (v/v) in bt-DMSO buffer (10 mM bis-Tris-HCl pH 6.50, 0.1 mM MgCl2, 20% DMSO) and pelleted at 15,000 rpm for 450 minutes at 4°C in a TLA 55 rotor (Beckman). For in-solution MS analysis of subfractionation enriched NPCs, 0.1 µg of QconCAT-A were immobilized on Dynabeads His-Tag Isolation and Pulldown resin (Thermo Fisher Scientific) pre-equilibrated in binding buffer (20 mM Hepes, 150 mM NaCl, 8 M urea, 5 mM TCEP). The purified protein sample was incubated with the resin for 20 minutes at RT and washed with binding buffer 5 × 200 µL to eliminate residual SDS (note: in-solution and in-gel analyses showed consistent results (not shown), so most of the further analyses were performed in-gel to improve consistency, speed and throughput). For the solid state, in-gel MS analyses, pelleted NPCs were solubilized in 10 µL of 0.5 M Tris-HCl pH 8.0, 5% SDS by incubating at 72°C for 5 minutes and then diluted 1:1 with 20% glycerol, 50 mM TCEP, 0.5 mM EDTA, 0.05% (w/v) bromophenol blue. To each 5 µg NPC sample, approximately equimolar amounts of 0.1 µg of purified QconCAT-A or 0.045 µg of purified QconCAT-B were added. Samples were then incubated at 72°C for 10 minutes, cooled down to RT and treated with a final 30 mM of iodoacetamide (Sigma), at RT in the dark for 30 minutes. Samples was then loaded into a 4% (37.5:1) in-house prepared stacking acrylamide SDS-PAGE gel. The resulting bands, containing a mixture of whole NPCs and stable-isotopically labeled QconCAT protein, were excised and processed for quantitative MS analyses.
3.2 Characterization of stable isotopically labeled Qconcat by MS
The mass of purified intact stable isotopically labeled Qconcat A protein was analyzed by MALDI (Extended Data Fig. 2E) on a JEOL JMS-S3000 SpiralTOF mass spectrometer using the ultra-thin-layer sample preparation method60,61 in which α-cyano-4-hydroxycinnamic acid (Sigma) was used as the matrix. The mass of Qconcat A was internally calibrated with horse myoglobin ([m+H]+ = 16,952.5 Da). Mass calibration and background subtraction were carried out with the JEOL msTornado control software, while additional analyses were carried out with the MoverZ software62. The Qconcat A protein was also characterized by peptide mapping, wherein tryptic peptides from in-gel digestion were loaded onto a PicoFrit® column (New Objective, Woburn, MA) with an integrated emitter tip (360 mm O.D., 50 mm I.D., 10 mm tip) self-packed with 6 cm of reverse-phase C18 material (ReproSil-Pur C18-AQ, 3 mm beads from Dr. Maisch GmbH), and analyzed with a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific), with a Agilent 1200 series HPLC system (Agilent) and a homebuilt micro electrospray source. The purified Qconcat B was characterized by peptide mapping on a Thermo Orbitrap Fusion mass spectrometer, with a Thermo Easy-nLC 1000 HPLC and a Thermo Easy-Spray electrospray source.
3.3 Stoichiometry quantification of NPC using Qconcat and by MS
Mixtures of yeast nuclear pore complex (NPC) proteins and stable isotopically labeled Qconcat were either enzymatically digested in solution in the presence of Urea or inside a SDS-PAGE gel matrix. (a) In solution digestion: A mixture of the NPCs and immobilized Qconcats on His-dynabeads were sequentially digested at room temperature by Endoproteinase LysC in 8 M Urea for 66 hours and by trypsin in 2 M Urea for 3 hours. (b) In-gel digestion: Proteins in the gel matrix were digested in 100 mM Tris-HCl at room temperature either sequentially by 0.25–2 µg endoproteinase LysC for 66 hours and by 3–25 µg trypsin for 3 hours, or, in later experiments, by 25 µg trypsin alone for 3 hours. The resulting peptides were analyzed in duplicate by LCMS using a Thermo Fusion or a Thermo Q Exactive Plus mass spectrometer, with a Thermo Easy-nLC 1000 HPLC and a Thermo Easy-Spray electrospray source. L/H ratios for standard peptides were obtained using MaxQuant63, complemented with manual determination.
We incorporated two standard peptides from each nucleoporin into the Qconcat standard to allow us to check for internal consistency of the measured L/H ratios for each nucleoporin. Our check required that the relative standard deviations of L/H ratios for two standard peptides from two duplicate LCMS runs - i.e., for a total of four measurements per nucleoporin - be ≤ 25%. When deriving relative stoichiometry for any given preparation of NPCs analyzed in different replication experiments, we corrected for variations in the mixing ratio of light nucleoporins and heavy Qconcat proteins by scaling the measured L/H ratios to minimize the sum, over all nucleoporins, of the relative standard deviations of the L/H ratios. The resulting scaled L/H ratios from different experiments were used to derive the average L/H ratios and standard deviations. To assay for potential nucleoporin stoichiometry bias arising from capture through particular affinity handles, we used SILAC-MS analysis of these preparations versus the nuclear envelope preparation. We performed n=2–3 technical and 2 biological replicas for NPCs purified by subfractionation procedures from a diploid, non-tagged, S. uvarum strain, and n=1–3 technical and 4 biological replicas for the NE-corrected, affinity captured NPCs from haploid, tagged, S. cerevisiae strains (Fig. 1A).
The absolute stoichiometry (Fig. 1; Supplementary Table 2A) was then determined by normalizing the summed copies of Nup188, Nup120 and Nic96 per NPC to 64 copies (i.e., 16 for Nup188 and Nup120, and 32 for Nic96).
3.4 SILAC-MS analyses of the NPC stoichiometry
A preparation of yeast nuclear envelopes (NE) obtained by a well-established subfractionation method44 does not involve disruption of the NE membrane by detergents and generates sheets of NE studded with intact NPCs. To assess the degree to which the affinity captured NPCs were intact, we used SILAC-MS to compare the levels of each Nup in the affinity captured preparation relative to those in the NE preparation. To do this, the light isotopically labelled NE sample was mixed with a heavy isotopically labelled (L-Lysine:2HCL 13C6) Mlp1-ppxPrA affinity captured NPC sample in a SILAC experiment. Mixtures of nuclear envelope proteins and stable isotopically labelled NPCs purified via the Mlp1-PrA handle were digested sequentially in gel matrix by endoproteinase LysC and by trypsin. Resulting peptides were analyzed by LCMS on a Thermo Q Exactive Plus mass spectrometer, with a Thermo Easy-nLC 1000 HPLC and a Thermo Easy-Spray electrospray source. H/L ratios for all peptides were obtained using MaxQuant63, complemented with manual examinations. Peptides’ H/L ratios were used to derive nucleoporins’ H/L ratios and standard deviations (Data not shown). The result showed that the affinity capture process does not significantly affect the overall ratios of the major Nups and NPC modules relative to the NE samples (data not shown), indicating that the affinity capture procedure generates intact, complete NPCs. We also used this comparison relative to NEs to correct for the slight increases observed in the ratios of Nups closely associated with the Mlp1 handle in the affinity captured NPCs (Fig. 1A).
3.5 In vivo calibrated imaging analysis of GFP-tagged Nups
Calibrated imaging data was acquired as described in a previous publication52. Using the avalanche photodiode (APD) imaging module of a Zeiss confocor 3, confocal z-stacks of live yeast were acquired with a 40× 1.2 NA Plan-Apochromat water objective. The 488 nm laser line was used to excite GFP, with a 405/488/561 dichroic. Emission was reflected with a LP580 emission dichroic and collected through a BP 505–540 nm emission filter. The pinhole was set to 1 airy unit. Zoom was set to maintain a pixel size of 55 nm, and a z-step size of 400 nm was used. After acquisition, images were binned in XY by 2, resulting in an effective pixel size of 110 nm, and anaphase cells were analyzed for diffraction limited Nup spots along the anaphase bridge. These spots, when present, were fit to a 2 dimensional Gaussian to obtain the amplitude of the signal. The z-slice with the maximum signal intensity of the spot was analyzed. FCS was used to convert the amplitude of the Gaussian fit of the Nup spot number of molecules of GFP. Briefly, using a strain expressing only cytosolic GFP, fluorescence correlation spectroscopy determined the average number of molecules in the focal volume, as described previously52. Then, the amplitude of the signal of the Nup spot was compared to the intensity of cytosolic GFP, taken with the same imaging setup. For all measurements, number 1.5 coverslips were measured for uniformity, and the correction collar of the water objective was optimized for this thickness using signal intensity of alexa fluor 488 in solution. For each day data was acquired, the calibration using cytosolic monomer GFP was obtained.
3.6 Phospholipid analysis
These analyses were kindly performed by Avanti Polar Lipids, Inc, using their standard protocols.
3.7 Label-free MS quantitation of the NPC and associated proteins
Raw MS files from QConCat Mlp1 immunoisolation experiments were analyzed by the MaxQuant iBAQ method64. Only non-isotopically labeled (not QConCat) peptides were considered. Proteins were filtered to require more than three unique peptides per protein, and stoichiometries normalized to the absolute minimum value of the difference between label-free and the QconCAT stoichiometry for all the Nups (Extended Data Fig. 3C; Supplementary Table 8). Stoichiometries were multiplied by molecular weight to obtain mass per NPC complex and the results summed to obtain total mass of the NPC (Fig. 1C; Extended Data Fig. 3C).
3.8 Living Mass of the NPC with Charge Detection Mass Spectrometry (CDMS)
The CDMS instrument is described in detail elsewhere53,65. Briefly, the measurements are made by trapping single ions in a linear electrostatic ion trap. As the ions oscillate back and forth in the trap they pass through a cylindrical electrode. The charge induced on the electrode is detected by a charge sensitive preamplifier. The resulting signal is amplified and digitized, and then analyzed using fast Fourier transforms. The fundamental frequency provides the m/z and the magnitude is proportional to the charge. The mass of each ion is then obtained by multiplying the charge and m/z. Each NPC sample was characterized by measuring the masses of several thousand ions individually and then binning the masses to yield a true mass spectrum (Fig. 1B–C).
4. Chemical cross-linking and MS (CX-MS) analysis of the cross-linked NPC
NPCs were immuno-purified from Mlp1, Nup82, and Nup84 tagged S. cerevisiae strains. After native elution, 0.5 or 1.0 mM disuccinimidyl suberate (DSS) was added and sample incubated at room temperature for 30 min with gentle shaking (~1,000 rpm). The reaction was quenched with 50 mM ammonium bicarbonate or SDS PAGE buffer containing 100 mM Tris-HCl. The sample was then precipitated using 90% methanol at −80°C or concentrated in a speed vacuum before separation by SDS electrophoresis.
The sample was reduced by 10 mM tris-(2-carboxyethyl)-phosphine (Invitrogen) at 80°C for 15–20 mins, cooled to room temperature, and alkylated by 50 mM iodoacetamide for 20 min in the dark to block the formation of disulfide bonds. After reduction and alkylation, the cross-linked complexes were separated by 3–8% SDS-PAGE (NuPAGE Tris-Acetate Fisher) to reduce the complexity of the sample. For in-gel digestion, the high molecular weight region gel bands (> 460 kDa, estimated by the high MW protein markers, Invitrogen) corresponding to the cross-linked NPC proteins were sliced and proteolysed by trypsin as previously described66,67. Briefly, gel plugs were crushed into small pieces, ~5–10 µg of sequencing grade trypsin (Promega) per ~100 µg protein was added with subsequent 6 – 8 hour incubation. This proteolysis was repeated once more to ensure optimal results. Peptides were extracted by formic acid and acetonitrile, desalted on C18 cartridges (Waters), and snap-freezed prior to fractionation.
To reduce the complexity of the sample, proteolyzed mixtures were separated by an orthogonal, two-step fractionation strategy. First, peptide size chromatography68 was used for size-based separation into 2 – 4 fractions (~2–10 kDa). Then, a secondary fractionation, using a self-packed basic (pH10) C18 resins (Dr. Masch GmbH), resulted in 10 – 12 peptide fractions which were subsequently analyzed by LC/MS.
Each peptide fraction was dissolved in the sample loading buffer (5% MeOH, 0.2% FA) and analyzed either by an Orbitrap Q Exactive (QE) Plus mass spectrometer or a LTQ Velos Orbitrap Pro mass spectrometer(Thermo Fisher). The QE instrument was directly coupled to an easy-nLC system (Thermo Fisher) for electrospray. The cross-linked peptides were loaded onto the Easy-Spray columns (15 cm prepacked columns that are filled with C18 reverse phase material of 2 or 3 µm particle size, 200 Å pore size and 50 µm inner diameter, Thermo fisher) that were heated to 35 °C. Mobile phase A consisted of 0.1% formic acid and mobile phase B of 100% ACN with 0.1% formic acid. Peptides were eluted in LC gradients of 120 minutes (e.g., a LC gradient of 3–7% B, 0–6 minutes, 7–28% B, 6–101 minutes, 28–100%B, 101–113 minutes, followed by equilibration with 100% A until 120 minutes). Flow rates were set at ~250–275 nl/min. Other instrumental parameters for CX-MS analyses include: capillary temperature: 250–275 °C; target mass resolutions (at 200 Th): 70,000 for MS and 17,500 for MS/MS; AGC targets: 1–3 × 106 (full mass) and 2 × 105 (MS/MS); MS range of 300–1,700 Th; isolation window: 1.3–1.7 Th; HCD normalized energy: 24–29; dynamic exclusion allowed once per 75–90 s. The top 8 most abundant ions (with charge stage of 3–7 and intensity thresholds of 3,000–7,500 ions) were selected for fragmentation by HCD. The max injection times were set at 200 ms (for MS) and 500–800 ms (for MS/MS). For samples that were analyzed by Orbitrap Velos, the cross-linked peptide mixtures were pressure-loaded onto a self-packed PicoFrit® column with integrated electrospray ionization emitter tip (360 O.D, 75 I.D with 15 µm tip, New Objective). The column was packed with 10–15 cm reverse-phase C18 material (3 µm porous silica, 200 Å pore size, Dr. Maisch GmbH). Mobile phase A consisted of 0.5% acetic acid and mobile phase B of 70% ACN with 0.5% acetic acid. The peptides were eluted in a 120 or a 140-minute LC gradient (8% B to 50% B, 0–93 minutes, followed by 50% B to 100% B, 93–110 minutes and equilibrated with 100% A until 120 or 150 minutes) using a HPLC system (Agilent), and analyzed with a LTQ Velos Orbitrap Pro mass spectrometer using similar parameters to the QE instrument.
The raw data were searched by pLink69 using a FASTA database containing 34 NPC protein sequences. An initial MS1 search window of 5 Da was allowed to cover all isotopic peaks of the cross-linked peptides. The data were automatically filtered using a mass accuracy of MS1 ≤ 10 ppm (parts per million) and MS2 ≤ 20 ppm of the theoretical monoisotopic (A0) and other isotopic masses (A+1, A+2, A+3, and A+4) as specified in the software. Other search parameters include cysteine carbamidomethyl as a fixed modification, and methionine oxidation as a variable modification. A maximum of two trypsin missed-cleavage sites was allowed. The initial search results were obtained using a default 5% false discovery rate (FDR) expected by target-decoy search strategy. All spectra were manually verified as previously described66,67,70–72. The cross-linking data was analyzed and plotted by an on-line software tool of CX-Circos (http://cx-circos.net; manuscript in preparation) (Fig. 2).
5. Cryo-Electron Tomography (Cryo-ET) of whole NPCs
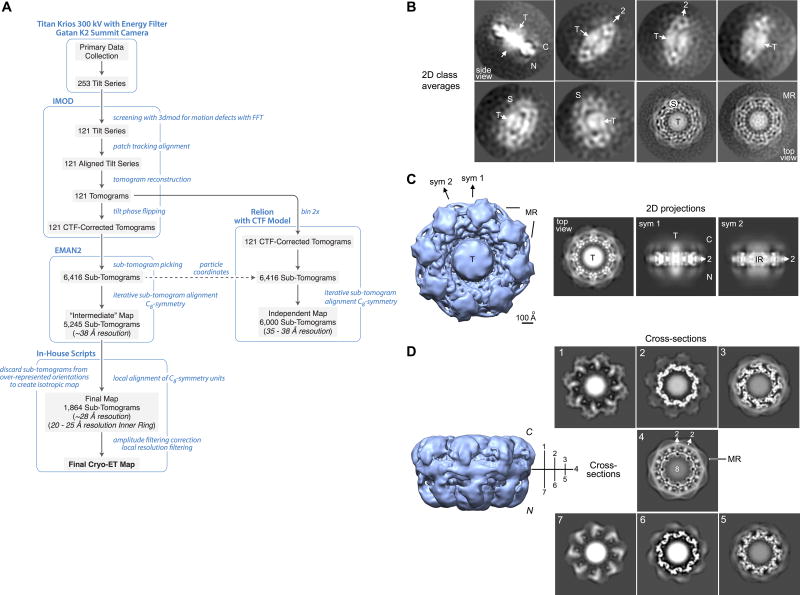
We used Cryo-electron tomography (Cryo-ET) and sub-tomogram averaging (Extended Data Fig. 4A) to obtain a final map with a global resolution of 28 Å, while the inner ring was solved at 20–25 Å (Extended Data Fig. 5; Supplementary Table 9). To create this map, NPCs were immuno-purified from Mlp1 tagged S. cerevisiae strain, in a final buffer of 20 mM Hepes (pH 7.5), 50 mM Potassium Acetate, 20 mM NaCl, 2 mM MgCl2, 0.1% Tween 20 and 1 mM DTT (see Methods section 2 for details). The concentration was estimated by SDS-PAGE to be ~0.3 – 0.4 mg/ml. Freshly cleaned Quantafoil 300 mesh copper grids with 2 µm holes in the support film were prepared with a continuous carbon support film that spanned the holes. Before use, the grids were glow discharged in air, floated on 5 µL sample drops for 45 minutes and then washed by serial transfer on 4 × 20 µL drops of elution buffer without glycerol. Each grid was mounted on forceps in a Mark III Vitrobot (FEI) at room temperature and 100% relative humidity. Buffer on the grid was removed by blotting from the bottom with a tool that held a filter paper wedge, using access through the left-hand port. Then 2 µL of freezing buffer was added to the grid from the right-hand port and the grid was plunge frozen in liquid ethane after blotting.
Cryo-ET data collection was done on a Titan-Krios electron microscope operating at 300 kV, equipped with an X-FEG, a post-column energy filter set to 20 eV, and a spherical aberration (Cs) corrector (Supplementary Table 9). Images were recorded with a Gatan K2 Summit direct electron detector in integration mode, with single frames taken at each tilt with UCSF Tomo73, at a nominal spacing of 5.6 Å per pixel. A total of 253 tilt series were collected in steps between −60°, 0° and 60° in increments of 2.5–4° for different tilt series. While the full tilt range was used for tomogram reconstruction, in the final sub-tomogram averaging step only data up to ± 45° tilt from each sub-tomogram were included in the final average. The dose target for each tilt series was 90–100 electrons/Å2 and followed a cosine α dose curve with a flux of 20 electrons/pixel/second, and a dose of 3.5 electrons/Å2 for the zero tilt image. Extended Data Fig. 4A presents the strategy we employed to reconstruct the 3D map of the whole yeast NPC. The 3dmod viewer in IMOD74 was used to screen tilts visually for defects with the Fourier transform to gauge image motion. In total 120 tilt series with a defocus range of −4.6 to −7.5 µm were kept for further processing (Raw data tilt series deposited in EMPIAR with accession number EMPIAR-10155; Supplementary Power Point Presentation, slides 1–2). After interactive test runs with etomo74, we processed the tilt series in an automated fashion with batchtomo using a 7 × 7 patch tracking to create aligned tilt series and calculated back-projection and SIRT tomograms which were CTF-corrected by phase-flipping each image in the tilt series (Supplementary Power Point Presentation, slides 3–6). The final SIRT tomograms were binned 3× and used for interactive sub-tomogram "particle" picking with e2spt_boxer.py in the EMAN2 single particle tomography package75,76 with a low pass filter of 100 Å. In total 6,416 fully sampled unfiltered sub-tomograms were extracted from the back-projection tomograms in 300 × 300 × 300 voxel volumes.
In the alignment and averaging process, new algorithms for high-speed 3D alignment with automatic missing wedge compensation and averaging76, were employed throughout, and were critical for processing such large sub-tomograms. An initial reference was prepared by averaging a small subset of sub-tomograms, to produce a low-resolution reference using the C8-symmetry of the complex77. Due to the large size and distinct shape of the particles, alignments were unambiguous. The alignment and averaging strategy for the final map was adapted from described procedures75, and applied iteratively. The observed flexibility of the NPC ring initially limited the overall resolution to ~38 Å with 5,245 sub-tomograms (data not shown); the sub-tomograms discarded at this stage were those with the worst quality as compared to the average, generally due to higher noise levels, but in some cases due to particle damage or a false positives during sub-tomogram picking. We realized that observed flexibility of the NPCs limited our resolution to ~38 Å and therefore employed a tactic to locally align all individual spokes (C8-symmetry units) to the reference rather than aligning whole rings which could contain long-range deviations from a perfect toroid. It is important to note that these deviations are not large, across the entire NPC they are on the order of the 38 Å resolution achieved without local alignment. This approach has been used before in the NPC Cryo-ET field and essentially all NPC Cryo-ET maps to date were done using this approach of dividing the NPC into subunits78–81. Briefly, two reference volumes were prepared, one consisting of the entire NPC and a second, masked volume, in which one-fourth of the ring had been retained, centered roughly on the mass of a single subunit. Each NPC was rotationally and translationally aligned to the reference ring. Using this initial alignment, each NPC was replicated into its 8 pseudo-symmetric orientations, then a translation-only alignment against the masked reference was performed. This has the effect of bringing one asymmetric unit per replicated ring into register with the reference at a consistent radius. While small per-subunit rotations might occur, this possibility was not included in the local alignment. The average from the 8 subunits was then used to construct a symmetric ring by applying an azimuthal linear ramp mask centered on the mask used for alignment, which fell to zero at an angle of 45° in both directions, and then imposing the C8-symmetry. This interpolates smoothly from one side of the subunit to the other symmetry-related side, to produce a complete symmetrized ring. This processing dramatically reduces the blurring caused by local fluctuations in subunit position and the resulting 3D volume was used as a reference in the next cycle of iterative refinement, which was repeated until no further improvement was observed.
At this stage we realized that the preferred orientation of the particles within our tomograms was leading to anisotropic resolution in the final structure, with 2/3 of the NPC rings oriented within 30° of the C8-symmetry axis (as clearly observed in our raw data). Producing an isotropic average required balancing the various ring orientations by discarding the lowest contrast rings in the over-populated orientations to more evenly balance the orientation distribution82–84. This normalizing of orientations, and not 3D classification, was what led to discarding a fraction of our data (Extended Data Fig. 4A). Naturally when doing this, we elected to discard noisier sub-tomograms. This was achieved by comparing the agreement of each sub-tomogram with the overall average. In each angular range, we then retained roughly the same number of sub-tomograms, keeping those with the best quality. The discarded sub-tomograms were nearly as good, meaning we could have equally well used the next best subset of the sub-tomograms with virtually no impact on the final structure. Indeed, in the less common orientations, we were forced to use virtually all of the sub-tomograms irrespective of quality. Thus, the reason for discarding a significant fraction of the sub-tomograms in our work was not due to poor quality or conformational variability, but due to preferred orientation of the particles within the tomogram. The final reconstruction used 1,864 (of the 6,416 initial) sub-tomograms. These sub-tomograms were further divided randomly into two groups for resolution assessment. “Gold standard” refinement was used for resolution testing and to ensure self-consistency (Supplementary Power Point Presentation, slides 7–8). Both global and local resolution assessments were done using a set of tiled Gaussian masks to estimate the local resolution and reproducibility of the structure, which is one of the standard methods for local resolution assessment. Briefly, a 3-D Gaussian is generated with a FWHM of at least 2× the anticipated resolution (generally even larger). This Gaussian is then applied as a mask to both maps and an FSC curve is computed. This process is repeated in a tiled pattern throughout the volume. This provides a resolution for each sampled location in the volume. This procedure involves a trade-off. The smaller the Gaussian, the less precision we have in the FSC curves, the larger the Gaussian, the less localized the resolution estimate is. The Gaussians overlap to provide better sampling of the volume. The resulting resolution map is similar to what is produced by ResMap85, but measures FSC (which is filter-independent) unlike ResMap85, which requires unfiltered volumes and wouldn’t work well on subtomogram averages. The global resolution of our Cryo-ET map at the standard FSC0.143 cutoff is ~28 Å and the local resolution distribution ranges from 20 to 38 Å, with the inner ring being in the 20–25 Å range (Extended Data Fig. 5). This local resolution estimate was used to locally filter the density map, to produce a map with the appropriate level of detail in each area (Extended Data Fig. 5). The size of the Gaussian window was 140 Å, indicating the smallest region over which the resolution is considered to vary. While this may seem large, it is quite small compared to the size of the overall NPC (Extended Data Fig. 5). CTF phase-flipping was applied during tomographic reconstruction and a final approximate amplitude correction was applied to the averaged NPC ring. Hence, theoretical CTF curves for the mean defocus values present in the tomograms were averaged assuming 10% amplitude contrast. The reciprocal of this curve was then applied as a filter to the final uncorrected map. The Cryo-ET density map was refined at 5.3 Å/pixel based on a recalibration of the map with known structures.
In parallel, and as an additional validation of our final map, we also carried out a tomographic analysis of the yeast NPC data set (same 6,416 sub-tomograms) using Relion 1.4 and incorporated a CTF model86,87. In brief, we calculated back-projection tomograms without phase-flipping corrections for the individual tilted images, and binned the output sub-tomograms 2-fold to 10.6 Å/pixel. The data sets underwent sequential rounds of 2D classification using Z-projections of the sub-tomograms to eliminate poor particles. A subsequent 2D classification identified near top, tilted and side views; the latter provided an independent estimate of NPC thickness perpendicular to the NE (620–640 Å). A 3D reconstruction with the best sub-tomograms produced a map at ~35 Å resolution (data not shown) with similar features to those obtained with the e2spt package75,76, including distinct connections between each spoke and the transporter, further validating the features in our Cryo-ET map (Fig. 3). Finally, Z-projections of original sub-tomograms that were roughly aligned along the C8-symmetry axis were used for an additional unsupervised 2D classification, which produced classes with central transporters without using the C8-symmetry restraint (Extended Data Fig. 6A–B). Differences in the apparent resolution of the class averages in Extended Data Fig. 6A–B reflect different particle numbers in the classes. As mentioned above, the data set of particles have a strong orientational bias in which the NPCs tend to bind to the carbon support film with a range of 0–30 degrees of tilt. The class averages are based on 2-dimensional projections along the z-axis of the original sub-tomograms, to avoid issues with the missing wedge. Hence, there is a disparity in particle numbers in the classes. Tilting in the tomographic data collection helped to fill in the missing data, but as mentioned previously we took great care to ensure an equal coverage of Fourier space in the calculation of our final map to avoid distortions, and took a number of other steps to ensure that radiation damage and loss of data quality in later tilts was minimized by utilizing only information in Fourier space at ±45° from each particle sub-tomogram when they were combined to form the final map. The Relion map serves as a strong validation of our final map, since if it was flawed, a reconstruction with Relion would have resulted in a different map (Extended Data Fig. 5E). Additionally, the fact that a Relion reconstruction resulted in a ~35 Å resolution map, virtually the same resolution of our ‘intermediate’ map described above (~38 Å) validates our methodology and the quality of our final map (Extended Data Fig. 5E). An additional point providing prima facie evidence that our Cryo-ET map was calculated correctly lies in the fact that the local 2-fold symmetry (C2-symmetry), which was expected to show up in the inner ring of the NPC, does emerge without any enforcement, while the overall map shows a clear asymmetry, with large and distinctly different features on the nuclear and cytoplasmic face of the yeast NPC (which were also observed in the Relion map) and a slightly tapered appearance, as is shown in Fig. 3A and Extended Data Figs 4D and 5B.
6. Small angle X-ray scattering (SAXS)
SAXS measurements for 147 constructs of 18 Nups43,72,88–92 (Supplementary Table 6; manuscript in preparation; source data are provided with this article) were carried out both at the Stanford Synchrotron Radiation Lightsource Beamline 4-2 in the SLAC National Accelerator Laboratory (Menlo Park, CA) and at the SIBYLS Beamline 12.3.1 of the Advanced Light Source in the Lawrence Berkeley National Laboratory (Berkeley, CA). SAXS data were collected at concentrations ranging from 0.5 to 5.0 (or higher, depending on the sample) mg/mL, using the previously defined standard protocol43,88,90; ~20 one-second exposures were used for each sample and buffers maintained at 15°C. Further details of the SAXS experiments are provided in our previous publications43,88,90.
7. Phenotypic analysis by One-Cell Doubling Evaluation by Living Arrays of Yeast (ODELAY)
Yeast growth phenotypes were quantified using the ODELAY assay as described previously93. Briefly, yeast was cultured in YPD media in 96 well plates overnight. Cultures were diluted to an OD600 of 0.09 and allowed to grow for 6 hours at 30°C. The cultures were then diluted again to an OD600 of 0.02 and spotted onto YPD agarose media. The resulting cultures were then observed using time-lapse microscopy for 48 hours with 30 minute intervals between images. All images were collected on Leica DMI6000 microscopes with a 10× 0.3NA lens using bright field microscopy. MATLAB scripts utilizing the Micro-Manager interface controlled the image collection process94. 6 independent experiments were performed. The population growth rates were scored against each other using the following equation:
, where di is the ith decile of query population doubling time, μi is the mean of the ith decile of the parent strains doubling time, and σi is the standard deviation of the ith decile of the parent strain’s doubling time. The mean and standard deviation deciles (μi and σi) were calculated from at least 4 separate populations containing at least 200–300 individuals. All calculations were performed using MATLAB scripts. Following Z-scoring of the populations, an additional weight was added to the scoring for truncation strains that occurred in haploid versus diploid strains of yeast.
8. Negative-stain electron microscopy of the native Nic96 complex
An affinity captured and natively eluted sample of the endogenous Nic96 complex (composed of Nic96, Nsp1, Nup49, and Nup57) was applied to a glow-discharged grid and stained with 1% uranyl formate. Images were collected on a Philips CM200 transmission electron microscope (FEI) operating at 200 kV at 50,000× magnification and a defocus of ~1.5 µm (2.03 pixels/Å). Images were recorded on a Gatan UltraScan 1000 2×2 CCD camera (Gatan Inc., Pleasanton, CA). Particles were selected using Boxer from EMAN95, normalized and then phase-flipped using ctfit from EMAN. In total, 34 class averages (selected classes shown in Extended Data Fig. 7G) were generated through ISAC96 that classified ~86% of the original set of 5,458 particles.
9. Integrative structure determination of the S. cerevisiae NPC
The structure of the S. cerevisiae NPC, including the scaffold, membrane rings, cytoplasmic export platform, and nuclear baskets in the context of the pore membrane but excluding the flexible FG regions, was solved by integrative structure determination (9.1 Integrative structure determination of the S. cerevisiae NPC scaffold, membrane rings, cytoplasmic export platform, and nuclear basket). Moreover, the distributions of the FG regions and the cargo-bound Nuclear Transport Receptors (NTRs), comprising the central transporter, were computed by Brownian dynamics simulation (9.2 Brownian dynamics simulation of FG repeats and NTRs).
9.1 Integrative structure determination of the S. cerevisiae NPC scaffold, membrane rings, cytoplasmic export platform, and nuclear basket
Integrative structure determination of the S. cerevisiae NPC proceeded through four stages97–100 (Extended Data Fig. 1; Supplementary Table 3; Supplementary Videos 1–3): (1) gathering data, (2) representing subunits and translating data into spatial restraints, (3) configurational sampling to produce an ensemble of structures that satisfies the restraints, and (4) analyzing and validating the ensemble structures and data (Supplementary Tables 2 to 4; Extended Data Figs. 1, 7, and 8). The integrative structure modeling protocol (i.e., stages 2, 3, and 4) was scripted using the Python Modeling Interface (PMI) package, version 4d97507, a library for modeling macromolecular complexes based on our open-source Integrative Modeling Platform (IMP) package98, version 2.6 (https://integrativemodeling.org). The current procedure is an updated version of previously described protocols66,72,90,101–104. Files containing the input data, scripts, and output results are available at https://salilab.org/npc2018.
9.1.1 Stage 1: Gathering data
The stoichiometry of Nups in the NPC was determined via native mass spectrometry and biochemical quantitation of the purified NPC complex (Fig. 1; Extended Data Figs. 2 and 3). In total, 3,077 intra- and inter-molecular DSS and EDC unique cross-links were identified via mass spectrometry (Fig. 2; Supplementary Table 1), informing the spatial proximities among the 32 Nups and their conformations. The density map of the entire NPC was determined by cryo-electron tomography (Cryo-ET) at an average resolution of 28 Å, with the local resolution as high as ~20 Å for the inner-ring, informing the shape of the NPC (Fig. 3; Extended Data Figs. 4 to 6). Re-interpreted immuno-EM data48,97 informed the positions of 29 Nups. Predictions of the transmembrane domains obtained from SGD105 (Saccharomyces Genome Database; http://yeastgenome.org) and predictions of membrane binding motifs by the HeliQuest webserver90,106 informed about their proximity to the pore membrane. Previous immuno-EM measurements107 informed the end-to-end distance for Mlp1 and Mlp2. Low-resolution EM images of the NPC44 informed the diameter of the distal basket ring (formed by Mlp1 and Mlp2).
Representations of individual Nups and some of their subcomplexes (Supplementary Table 2 and references therein) relied on (1) atomic structures of 21 yeast Nup domains and 3 sub-complexes determined by X-ray crystallography or NMR spectroscopy; (2) our structures of Nup116, Nup133, Nup145N, Nup192, and Pom152, as well as the Nup82 and Nup84 sub-complexes solved by integrative structure determination43,66,72,88–92; (3) 29 comparative models built with MODELLER 9.13108 based on the known structure(s) detected by HHPred109,110 and the literature; (4) SAXS profiles for 147 constructs of 18 Nups43,72,88–92 (Supplementary Table 6; manuscript in preparation); (5) secondary structure, disordered regions, and domain boundaries predicted by PSIPRED111,112, DISOPRED113, and DomPred114, respectively; (6) coiled-coil regions of Nup82, Nup159, Nsp1, Nup49, Nup57, Mlp1, and Mlp2 predicted by COILS/PCOILS115 and Multicoil2116; (7) an atomic structure of the Nup53229–365 RRM domain from S. cerevisiae determined by X-ray crystallography (manuscript in preparation); and (8) the negative-stain EM density maps of full-length Nup192 (EMD-555692) and Pom152 (EMD-854343). See Supplementary Table 2 and references therein.
Our previous topological map of the NPC48 published in 2007 and the 82 composites determined by affinity purification and overlay assay97 were not used for computing the current NPC structure, but only for its validation.
9.1.2 Stage 2: Representing subunits and translating data into spatial restraints
Information about the modeled system (above) can in general be used for defining its representation, defining the scoring function that guides sampling of alternative structural models, limiting sampling, filtering of good-scoring structures obtained by sampling, and final validation of the structures. Here, the NPC representation relies primarily on stoichiometry as well as atomic structures, integrative structures, comparative models, and SAXS profiles of Nups and their subcomplexes (Supplementary Tables 2 and 6; references therein); the scoring function relies on chemical cross-links, Cryo-ET density map, immuno-EM localizations, excluded volume, sequence connectivity, the shape of the pore membrane, and 4 types of sequence-based localization relative to the membrane (below); the sampling benefits from symmetry constraints (below); and the validation of the final structure relies in part on the SAXS profiles (Supplementary Table 6) and composites determined by affinity purification and overlay assays97 (below).
To improve computational efficiency and avoid too coarse a representation, we represented the NPC in a multi-scale fashion. A rigid-body consisting of multiple beads was defined for each X-ray structure, NMR structure, comparative model, and integrative structure of the NPC components (Supplementary Table 2). The remainders of the Nup sequences not in rigid bodies (36.8% of residues, excluding FG repeats) were represented as flexible strings of beads. In a rigid-body, the beads have their relative distances constrained during configurational sampling, whereas in a flexible string, the beads are restrained by the sequence connectivity, excluded volume, and potentially additional restraints, such as chemical cross-links, as exemplified in our previous studies43,66,72,101,117.
Rigid bodies (63.2% of residues, excluding FG repeats) were coarse-grained using two resolutions, where beads represented either individual residues or segments of up to 10 residues. The coordinates of a 1-residue bead were those of the corresponding Cα atom. The coordinates of a 10-residue bead were the center of mass of the 10 constituent 1-residue beads. Finally, the remaining regions without an atomic representation (i.e., the predicted transmembrane and disordered regions) were represented by a flexible string of beads encompassing 25 to 100 residues each; the low-resolution representation of these regions is justified because their conformations are likely “decoupled” from the structure of the rest of the NPC48,118.
We used the SAXS data to confirm the rigid-body representations of 8 Nups with X-ray structures, comparative models, and previously published atomic integrative structures43,72,88–92 (Supplementary Tables 2 and 6; Extended Data Fig. 7F). The rigid-body representation of a Nup construct was validated by a chi-value that quantifies the difference between the computed (from an atomic rigid-body representation using FoXS119) and experimental SAXS profiles, except for several constructs of Nup133 and Nup192 that were flexible during integrative modeling and were thus evaluated as described in detail in our original publications90,92,120. The chi-value validation assumes that each Nup construct, corresponding in most cases to a single domain (not the whole protein), has the same conformation in solution and in complex; this assumption is consistent with other data (e.g. the chemical cross-links and Cryo-ET map). The SAXS validation is necessarily limited to Saccharomyces cerevisiae constructs of Nups that exist as a rigid monomer in solution and do not contain FG repeats; rigid-body representations of the constructs from other species, constructs that oligomerize in solution, and constructs that include FG repeats can not be easily used for validation, because of the sensitivity of a computed SAXS profile to the differences in the sequence and stoichiometry, as well as to potential errors in comparative modeling (especially of insertion and deletion).
With this validated representation in hand, we next encoded the spatial restraints based on the information gathered in Stage 1, as follows (Supplementary Table 4; the scoring function consisting of these restraints is defined in 9.1.3.2 Scoring function).
(1) Cross-link restraints
1,643 of the 3,077 unique cross-links (Fig. 2; Supplementary Table 1A) were used to restrain the distances spanned by the cross-linked residues, relying on a Bayesian scoring function66. The evaluation takes into account the ambiguity due to multiple copies of identical subunits and, for cross-links involving the same protein type, due to the lack of knowledge of whether they are intra- or inter-molecular72,101,117; the ambiguous cross-link restraint considers all intra- and inter-molecular assignments in multiple copies of identical subunits, with only the least violated distance contributing to the score. The remaining 1,434 DSS and EDC cross-links (Fig. 2; Supplementary Table 1B–F) were already used as restraints to build the integrative structures of the Nup8466 and Nup82 sub-complexes72, represented here as rigid bodies. The two homo-dimer DSS cross-links between two copies of residue 62 of Pom15243 and two copies of residue 151 in Nup60121 were transformed into harmonic upper distance bounds, enforcing the homo-dimer configuration.
(2) Cryo-ET density restraint
The Cryo-ET density restraint corresponded to the cross-correlation between the Gaussian Mixture Model (GMM) representation of most Nups and the GMM representation of the Cryo-ET density map103,122–124 (Fig. 3; Extended Data Figs. 4 to 6); we used a GMM representation for the sake of computational efficiency, necessitated by the large size of the NPC. An assessment of a given structure against a density map is much faster when both are represented with a mixture model (because the number of components in a mixture model is much smaller than the number of grid points covering the maps). However, these two scores are very strongly correlated. Thus, the structures obtained with a grid representation, if we had sufficient computational power, would certainly be indistinguishable from the current NPC structures124.
A 90° arc of the Cryo-ET density map was approximated by the GMM containing 1,750 components computed using the expectation-maximization algorithm as implemented in scikit-learn (http://scikit-learn.org); the Cryo-ET GMM appeared to be sufficient to reproduce the significant features of the density map (excluding the central transporter region). To use a comparable number of GMM components for Nups, a Nup was approximated by a GMM component for each of its 100 to 500 residues. The cross-correlation quantified the degree of overlap between the Nup GMM components and the Cryo-ET GMM components.
(3) Immuno-EM localization restraints
The immuno-EM localization restraint was used to localize the C-terminal beads of 29 of the 32 Nups based on prior immuno-EM data48,97,125. This goal was achieved by imposing upper and lower harmonic bounds on the axial and radial coordinates of the restrained bead, reflecting the uncertainty in the immuno-EM data97. The 3 remaining Nups (Nsp1, Sec13, and Seh1) were not restrained by the immuno-EM data because of its high uncertainty, presumably due to the positional heterogeneity of the tagged Nup in the multiple superposed EM images of the NPC (this heterogeneity is more likely to occur for Nups with multiple copies per C2-symmetry unit, which are unlikely to share the same radial and axial coordinates).
(4) Excluded volume restraints
The protein excluded volume restraints were applied to each 10-residue bead, using the statistical relationship between the volume and the number of residues that it covered66,72,97,126.
(5) Sequence connectivity restraints
We applied sequence connectivity restraints, using a harmonic upper bound on the distance between consecutive beads in a subunit, with a threshold distance equal to three times the sum of the radii of the two connected beads. The bead radius was calculated from the excluded volume of the corresponding bead, assuming standard protein density66,72,97,126.
(6) Membrane exclusion restraints
The membrane exclusion restraints were applied to beads in the non-membrane spanning Nups or their segments to prevent these beads from penetrating the pore membrane. A lower harmonic bound at 0 Å was applied to the distance between a bead and the closest point on the pore-side membrane surface48,97 (modeled as a half-torus with the large and small radii of 390 and 150 Å, respectively), for all coarse beads (10 residues or more per bead) in all Nups but Pom152, Ndc1, and Pom34; the restraint was also applied to all non-membrane coarse beads of Pom1521–110, Ndc11–28, Ndc1248–655, Pom341–63, and Pom34151–299.
(7) Transmembrane domain restraints
The transmembrane domain restraint was used to localize the coarse beads in the predicted transmembrane domains (Pom152111–200, Ndc129–247, and Pom3464–150; Supplementary Table 2 and references therein) within the pore membrane, which is 45 Å thick48,97. This aim was achieved by imposing an upper harmonic bound at 45 Å and a lower harmonic bound at 0 Å on the distance between the bead and the closest point on the pore-side membrane surface.
(8) Membrane surface binding restraints
The membrane surface binding restraint was used to localize the coarse beads in the predicted membrane binding motifs (Nup11–32, Nup6027–47, Nup120135–152, Nup120197–216, Nup133252–270, Nup157310–334, Nup170320–344, Nup53475–475, and Nup59528–528; Supplementary Table 2 and references therein), within the pore membrane up to 12 Å from the pore-side membrane surface127. This aim was achieved by imposing an upper harmonic bound at 12 Å within the pore membrane and a lower harmonic bound at 0 Å on the distance between the bead and the closest point on the pore-side membrane surface. For Nup120, only the best satisfied of the Nup120135–152 and Nup120197–216 restraints was used66,90 (conditional restraint).
(9) Pom152 perinuclear volume restraint
Only the C-terminal region of Pom152 (residues 201–1337) was restrained to the perinuclear lumen of the pore membrane43. This aim was achieved by imposing a lower harmonic bound at 0 Å on the distance between the Pom152 beads and the closest point on the perinuclear-side of the membrane surface.
(10) Distal basket ring restraints
The conformations of Mlp1 and Mlp2 each were restrained by an upper harmonic bound at 350 Å and a lower harmonic bound at 230 Å on the distance between the N-terminal and C-terminal beads, based on immuno-EM measurements107. In addition, the radius of the distal basket ring was restrained by an upper harmonic bound at 170 Å and a lower harmonic bound at 130 Å on the radial coordinates of the C-terminal beads of Mlp1 and Mlp2, based on low-resolution EM images of the NPC44. As an aside, the nuclear basket is also informed by cross-linking restraints and the C8-symmetry constraint (9.1.3.1 Sampling space with symmetry constraints).
9.1.3 Stage 3: Configurational sampling to produce an ensemble of structures that satisfies the restraints
9.1.3.1 Sampling space with symmetry constraints
We aimed to maximize the efficiency of the configurational sampling. More precisely, we aimed to maximize the precision at which the sampling of good-scoring solutions was exhaustive (9.1.4. Stage 4: Analyzing and validating the ensemble structures and data). Therefore, we reduced the number of independently moving parts in the NPC structure by explicitly considering the C8- and C2-symmetries of the NPC, as follows. The entire NPC consists of 8 clones of the C8-symmetry unit, related by multiples of a 45° rotation around the Z-axis (Fig. 3; Extended Data Figs. 4 to 6). The C8-symmetry unit was further broken into two C2-symmetry units and non-C2-symmetric Nups (Supplementary Table 2A); the C2-symmetry unit contains Nups that occur equally on both the cytoplasmic and nucleoplasmic sides48,97,125. For computational efficiency, we defined the coordinate system such that the C2-symmetry is imposed simply by cloning a bead in the C2-symmetry unit at (x,y,z) to (x,-y,-z) (equivalent to a rotation of 180° around the X-axis). This aim was achieved by fitting both copies of Pom15243 into the Cryo-ET density map, followed by moving the center of the map to the origin of the coordinate system and orienting the map such that the (x,-y,-z) transformation applies to Pom152.
With these symmetries in hand, we sampled only the positions of rigid bodies and beads corresponding to the Nups in the C2-symmetry unit and non-C2-symmetric Nups. There are no Nups that occur on both the cytoplasmic and nuclear sides and are not related by the C2-symmetry; there are no Nups that occur with a different stoichiometry on both sides. In addition, the luminal domain of Pom152 was considered already well-positioned given its fit into the Cryo-ET density map (above) and peripheral location in the NPC43, and was not sampled further.
9.1.3.2 Scoring function
The scoring function included restraints on the sampled Nups and the Pom152 luminal domain as well as restraints across the interfaces with neighboring symmetry units: (1) the Cryo-ET density restraint and distal basket ring restraint applied to the Nups in the sampled C8-symmetry unit; (2) sequence connectivity, immuno-EM localization, and the 4 types of sequence-based localizations relative to the membrane applied to the Nups in the sampled C2-symmetry unit and non-C2-symmetric Nups; and (3) cross-link and excluded volume restraints applied to the pairs of beads for Nups within the sampled C8-symmetry unit and across the interfaces with neighboring symmetry units.
9.1.3.3 Sampling algorithm
The search for good-scoring structures relied on Replica Exchange Gibbs sampling, based on the Metropolis Monte Carlo algorithm66,72 (Supplementary Table 3). The Monte Carlo moves included random translation and rotation of rigid bodies (up to 4 Å and 0.04 radians, respectively) and random translation of individual beads in the flexible segments (up to 4 Å). As indicated above, these operations were only applied to the sampled rigid bodies and beads. The remaining, symmetry-constrained rigid bodies and units were moved in lockstep to maintain the exact C8- and C2-symmetries at each sampling step, as described above. Up to 64 replicas were used, with temperatures ranging between 1.0 and 5.0. 42 independent sampling calculations were performed, each one starting with a random initial configuration. The coordinates were saved every 10 Gibbs sampling steps, each consisting of a cycle of Monte Carlo steps that moved every rigid-body and flexible bead once.
To further increase the efficiency of sampling, we first applied the above Monte Carlo algorithm separately to the following four subsets of Nups, which are likely co-localized based on prior characterizations43,48,72,97 and the current Cryo-ET density map: (1) Nup82 and Nup84 subcomplexes, (2) Pom152, (3) inner ring Nups (Nup157, Nup170, Nup188, Nup192, Pom34, Ndc1, Nup53, Nup59, and Nic96 CTD), and (4) Mlp1 and Mlp2. Next, the best-scoring solution from sampling each of the first three subsets were combined; they already were in the same reference frame, because they were all obtained by fitting the same Cryo-ET density map and immuno-EM data. The rest of the Nups and the Mlp1-Mlp2 heterodimer were then added in random positions and orientations, followed by another application of the above Monte Carlo algorithm to all sampled Nups. This sampling produced a total of 100,453 modeled structures in 42 independent runs (the score ranges from 88,545.0 to 103,589.5, with the mean and standard deviation of 88,831.5 and 187.4, respectively), requiring ~10 weeks on a cluster of ~2,500 cores. For the most detailed specification of the sampling procedure, see the IMP modeling script (https://salilab.org/npc2018).
We only considered for further analysis the 5,529 modeled structures with the score better than 88,644.1 (1 standard deviation below the mean value); this threshold implies satisfaction of the input datasets within their uncertainties (Supplementary Table 4; 9.1.4.3 Fit to input information). These structures are already superposed because they were fit into the same Cryo-ET map and sampling did not move the luminal domain of Pom152 (above).
9.1.4 Stage 4: Analyzing and validating the ensemble structures and data
Input information and output structures need to be analyzed to estimate structure precision and accuracy, detect inconsistent and missing information, and to suggest more informative future experiments. We used the analysis and validation protocol published earlier72,97: Assessment began with a test of the thoroughness of structural sampling, followed by structural clustering of the modeled structures and estimating their precision based on the variability in the ensemble of good-scoring structures, quantification of the structure fit to the input information, and structure assessment by data not used to compute it; structure assessment by cross-validation was not performed in this case, because it takes ~10 weeks on approximately 2,500 cores to compute an ensemble of structures for a single set of input datasets. These validations are based on the nascent wwPDB effort on archival, validation, and dissemination of integrative structures100. We now discuss each one of these validations in turn.
9.1.4.1 Thoroughness of the configurational sampling
We must first estimate the precision at which sampling found most good-scoring solutions (sampling precision); the sampling precision must be at least as high as the precision of the structure ensemble consistent with the input data (structure precision). As a proxy for testing the thoroughness of sampling, we performed four tests of sampling convergence128, as follows.
The first convergence test confirmed that the scores of refined structures do not continue to improve as more structures are computed essentially independently (Extended Data Fig. 1C).
The second convergence test confirmed that the good-scoring structures in independent sampling runs 1–21 (structure sample 1; nsample1=2,359 structures) and 22–42 (structure sample 2; nsample2=3,170 structures) satisfied the data equally well. The non-parametric Kolmogorov-Smirnov two-sample test129,130 (two-sided) indicates that the difference between the two score distributions is insignificant (p-value (1.0) > 0.05). In addition, the magnitude of the difference is small, as demonstrated by the Kolmogorov-Smirnov two-sample test statistic, D, of 0.045 (Extended Data Fig. 1D). Thus, the two score distributions are effectively equal.
Next, we considered the 5,529 good-scoring structures themselves, not their scores as in the two tests described above. For stochastic sampling methods, thoroughness of sampling can be assessed by showing that multiple independent runs (e.g., using random starting configurations and different random number generator seeds, as is the case for structure samples 1 and 2) do not result in significantly different structures42,66,72,97. We tested the similarity between structure samples 1 and 2 in the following two ways.
The third convergence test128 relied on the χ2-test (one-sided) for homogeneity of proportions131 between structure samples 1 and 2 (Extended Data Fig. 1E–F). The test involves clustering structures from both samples, followed by comparing the proportions of structures from each sample in each cluster. No adjustment was made for multiple comparisons. A comparison of two NPC structures considered only the beads representing Nups with a single copy per C2-symmetry unit and the Nic96 complex (including all Nups in the inner, outer, and membrane rings, but excluding Nup100, Nup116, Nup145N, Nup1, Nup60, Gle1, Nup42, Mlp1, and Mlp2), to avoid the combinatorially explosive identification of topologically equivalent Nup copies. The sampling precision is defined as the largest RMSD value between a pair of NPC structures within any cluster, in the finest clustering for which each sample contributes structures proportionally to its size (considering both significance and magnitude of the difference) and for which a sufficient proportion of all structures occur in sufficiently large clusters. The sampling precision for our NPC structure is 9 Å (Extended Data Fig. 1E).
Threshold-based clustering132 results in a single dominant cluster containing 80.3% of the good-scoring structures (Extended Data Fig. 1E–F) whose root-mean-square fluctuation (RMSF) is 9 Å (cluster precision). The remaining 19.7% of the structures are similar to those in the dominant cluster; the largest RMSD value from a structure in the dominant cluster is 17 Å (the mean and standard deviation of the RMSD values are 13.3 and 1.3 Å, respectively). Therefore, there is effectively a single good-scoring solution, at the structure precision of 9 Å (equal to the cluster precision). The sampling precision of 9 Å (RMSD) is sufficiently high for computing a structure at 9 Å precision (RMSF; RMSD is approximately square root of 2 times RMSF133). For the remainder of our analysis, we only use the structures in the dominant cluster.
The fourth convergence test relied on a comparison of two localization probability density maps for each Nup, obtained for dominant cluster structures in samples 1 and 2. A localization probability density map defines the probability of any voxel (here, 6×6×6 Å3) being occupied by a specific protein in a set of structure densities, which in turn are obtained by convolving superposed structures with a Gaussian kernel (here, with a standard deviation of 5.4 Å). The average cross-correlation coefficient between the two maps for each Nup is 0.90, indicating that the positions of most Nups in the two samples are nearly identical at the structure precision of 9 Å.
In conclusion, all four sampling tests indicate that the sampling was exhaustive at 9 Å precision (Supplementary Table 3). The caveat is that passing these tests is only necessary but not sufficient as evidence of thorough sampling; a positive outcome of the tests may be misleading if, for example, the landscape contains only a narrow, and thus difficult to find, pathway to the pronounced minimum corresponding to the correct structure. Moreover, our sampling was not completely stochastic because it proceeded in two steps, the first of which prepared the starting configuration for the second step. As a result, the actual structure precision might be worse134–137 than the estimated 9 Å.
9.1.4.2 Clustering and structure precision
An ensemble of good-scoring structures needs to be analyzed in terms of the precision of its structural features48,72,97. The precision of a component position can be quantified by its variation in an ensemble of superposed good-scoring structures. It can also be visualized by the localization probability density for each of the components of the NPC structure.
As described above, integrative structure determination of the NPC resulted in effectively a single good-scoring solution, at the precision of ~9 Å. This precision is sufficiently high to pinpoint the locations and orientations of the constituent Nups, demonstrating the quality of the input data, including the chemical cross-links (Fig. 2; Extended Data Fig. 7A–C) and the Cryo-ET density map (Fig. 3; Extended Data Fig. 8).
9.1.4.3 Fit to input information
An accurate structure needs to satisfy the input information used to compute it. Because the sampling was exhaustive (at 9 Å precision), overfitting is not a problem (at this precision); all structures (at this precision) that are consistent with the data are provided in the ensemble.
The dominant cluster satisfies 90% of the DSS cross-links (Extended Data Fig. 7A–C; Supplementary Tables 1 and 4); a cross-link restraint is satisfied by a cluster of structures if the corresponding Cα–Cα distance in any of the structures in the cluster (considering restraint ambiguity) is < 35 Å (Extended Data Fig. 7A–C; shown in blue). Therefore, the dominant cluster essentially satisfies the cross-linking data within its uncertainty (the false detection rate is approximately 5% to 10%138,139). Most of the cross-link violations are small, and can be rationalized by local structural fluctuations, coarse-grained representations of some Nup domains, and/or finite structural sampling, as shown in Extended Data Fig. 7A (a histogram presenting the distribution of the cross-linked Cα–Cα distances).
The localization densities for the dominant cluster overlap well with the Cryo-ET density map, with the cross-correlation coefficient of 0.92 (Fig. 3; Extended Data Fig. 8; Supplementary Table 4). Additional density is present in the Cryo-ET map for the Nup82 complex (cytoplasm) and basket attachment sites (nucleoplasm). This density may arise from local flexibility of these modules or may be due to the presence of cargo/TF associated with the NPC (Fig. 1B–C; Extended Data Fig. 3C). For visualization, the localization probability densities are typically contoured at the threshold that results in approximately twice the protein volume estimated from its sequence (Fig. 4).
The remainder of the restraints are harmonic, with a specified standard deviation. The dominant cluster generally satisfied at least 95% of restraints of each type (Supplementary Table 4); a restraint is satisfied by a cluster of structures if the restrained distance in any structure in the cluster (considering restraint ambiguity) is violated by less than 3 standard deviations, specified for the restraint. Most of the violations are small, and can be rationalized by local structural fluctuations, coarse-grained representations of some Nup domains, and/or finite structural sampling.
9.1.4.4 Satisfaction of data and considerations that were not used to compute structures
The most direct test of a modeled structure is by comparing it to the data that were not used to compute it (a generalization of cross-validation).
First, our current NPC structure is consistent with our previously published data and structure48,97 (Extended Data Fig. 7D–E). Our current structure satisfies all 82 composites determined by affinity purification and overlay assays48,97, even though they were not used in this calculation. For example, Pom152, Pom34, Ndc1, Nup157, and Nup170 are connected with each other (left panel in Extended Data Fig. 7E), consistent with the composites determined by the affinity purification data48,97 published in 2007 (right panel in Extended Data Fig. 7E). Moreover, the position of each Nup in the current structure is generally similar to that in the 2007 topological map48,97, albeit the current structure is determined at an order of magnitude higher precision (Extended Data Fig. 7D).
Second, the atomic structures of 8 Nups are consistent with the corresponding SAXS profiles for their constructs (Supplementary Tables 2 and 6; Extended Data Fig. 7F), as discussed in 9.1.2 Stage 2: Representing subunits and translating data into spatial restraints. For example, the SAXS profile calculated from the atomic structure of Pom152718–1148 (red curve in Extended Data Fig. 7F) using FoXS119 is well matched (χ=1.48) to the corresponding experimental SAXS profile43 (black dots in Extended Data Fig. 7F; n=20 exposures). For visualization purposes, the Pom152718–1148 structure (represented as a ribbon) is shown along with the best fit of the ab initio shape (represented as a transparent envelope) computed from the experimental SAXS profile, in Extended Data Fig. 7F.
Third, the localization density of the Nic96 complex (composed of Nic96, Nsp1, Nup49, and Nup57) in the dominant cluster can be projected well on most of the 2D class averages obtained for the natively isolated complex (Extended Data Fig. 7G; 8. Negative-stain electron microscopy of the native Nic96 complex in Methods above). More specifically, the EM 2D validation fits the structure of the Nic96 complex in the whole NPC context to the EM class averages of the Nic96 complex and computes a score that quantifies the match. The computation proceeds in three stages: (i) generation of alternative model projections, (ii) alignment of the class average and each model projection, and (iii) calculation of the fitting score for each projection, as follows. First, 1,000 uniformly distributed projections of the low-pass filtered structure of the NPC on the sphere (stage i) were generated. Second, each projection was optimally aligned to each of the class averages in Fourier space (stage ii). Finally, a score, corresponding to the cross-correlation coefficient, was computed (stage iii). For example, the experimental class averages were satisfied by the structure with cross-correlation coefficients of 0.81 and 0.83, respectively (Extended Data Fig. 7G).
Fourth, the structure was also validated by its comparison to the core scaffold maps of the Homo sapiens NPC, based primarily on EM density maps79,80,140,141 (Extended Data Fig. 10). Overall, the inner ring architecture is similar in both yeast and vertebrates, in agreement with it being the most conserved part of the NPC142.
Finally, the structure allows us to rationalize the functional fitness (Fig. 5), the transport through the NPC (Fig. 6; Extended Data Fig. 9), and the evolution (Extended Data Fig. 11), therefore increasing our confidence in the structure compared to not being able to rationalize these aspects of the NPC97,143,144.
9.2 Brownian dynamics simulation of FG repeats and nuclear transport receptors (NTRs)
Distributions of the FG repeats and NTRs were computed by Brownian dynamics simulation145, using our protocol146 implemented in IMP98, version 2.6. The simulated system included the static NPC ring determined in this study, the pore membrane, disordered and flexible FG repeat domains, as well as freely diffusing NTRs and inert macromolecules, all enclosed within a bounding box of 2,000 × 2,000 × 2,000 Å3.
The pore membrane was represented as a 250 Å slab with a cylindrical pore of radius 375 Å that contains the static NPC ring (this pore membrane representation is simplified compared to the toroidal pore used for solving the structure of the static NPC ring, for reasons of computational efficiency). Each of the FG repeat domains was represented as a flexible string of beads; a bead had a radius of 6 Å and encompassed 20 residues to achieve a compromise between computational efficiency and accuracy118,146–149. Consecutive beads were restrained by a bond with an equilibrium length of 18 Å and a constant force of 1.0 kcal/mol/Å, approximating the spring-like nature of flexible polymers150 in general and FG repeat domains118,147–149,151–153 in particular. The freely diffusing molecules included 1,600 NTRs and 1,600 inert macromolecules (0.33 mM each), each consisting of eight subgroups of 200 macromolecules, ranging in radius from 4 to 28 Å in 2 Å increments (10 to 75 kDa, assuming constant protein density of 1.38 g/cm2). Excluded volume interactions were applied to pairs of overlapping beads and to beads penetrating the pore membrane or the bounding box, using a constant repulsive force of 10.0 kcal/mol/Å. The potential binding energy between a binding site on an FG motif and a binding site on an NTR was modeled by an anisotropic harmonic potential dependent on the distance and orientation between the two sites that reproduces measured apparent dissociation constants in our simulations (scripts available at https://salilab.org/npc2018; manuscript in preparation).
The Brownian dynamics of the entire system were simulated at 297.15 K for 40 microseconds with a time step of 1,047 femtoseconds, independently 400 times; the first 10 microseconds of each trajectory were considered equilibration time and ignored in subsequent analysis. The distributions of the FG Nup and NTR positions were then computed from the total of 12 milliseconds of simulations over a cubic grid with a voxel size of 10×10×10 Å3, at time intervals of 9.5 picoseconds, from all 400 trajectories; these distributions were averaged by relying on the C8-symmetry of the NPC.
10. Code Availability
Files containing integrative structure modeling scripts, as well as the input data and output results are available at http://salilab.org/npc2018.
11. Data Availability
Source data for Fig. 1A is provided as an excel file.
Raw data for the chemical cross-links (source data for Fig. 2; Supplementary Table 1) is available via Zenodo data repository with a DOI (10.5281/zenodo.1149746).
The Cryo-ET density map (source data for Fig. 3) is deposited into the EMDB, accession code EMD-7321.
The Cryo-ET raw data (120 tilt series) are deposited into the EMPIAR, accession code EMPIAR-10155.
The integrative NPC structure (source data for Fig. 4) is deposited into the nascent public PDB repository, PDB-dev (https://pdb-dev.rcsb.rutgers.edu), under the accession codes of PDBDEV_00000010, PDBDEV_00000011, and PDBDEV_00000012.
Source data for Extended Data Fig. 2B is provided in the Supplementary Information (Supplementary Fig. 1).
SAXS data (source data for Extended Data Fig. 7F) are deposited into SASBDB, under the accession codes of SASDBV9, SASDBW9, SASDBX9, SASDBY9, and SASDBZ9. In addition, all SAXS data (Supplementary Table 6) are provided as source data with the article.
Raw data are available from the corresponding author upon request.
Extended Data
Extended Data Figure 1. Integrative structure determination of the S. cerevisiae NPC at 9 Å precision.
(A) A simplified schematic diagram of integrative structure determination of the S. cerevisiae NPC. Random initial structures of the Nups and their sub-complexes were optimized by satisfying spatial restraints implied by the input information.
(B) The full description of Integrative structure determination of the S. cerevisiae NPC, proceeded through four stages154–157 (Supplementary Table 3): (1) gathering data, (2) representing subunits and translating data into spatial restraints, (3) configurational sampling to produce an ensemble of structures that satisfies the restraints, and (4) analyzing and validating the ensemble structures and data (Supplementary Tables 2 to 4; Extended Data Figs. 7 and 8; Methods). The integrative structure modeling protocol (i.e., stages 2, 3, and 4) was scripted using the Python Modeling Interface (PMI) package, version 4d97507, a library for modeling macromolecular complexes based on our open-source Integrative Modeling Platform (IMP) package155, version 2.6 (https://integrativemodeling.org).
(C) Convergence of the structure score, for the 5,529 good-scoring NPC structures; the scores do not continue to improve as more structures are computed essentially independently. The error bar represents the standard deviations of the best scores, estimated by repeating sampling of NPC structures ten times (n=10, mean score values plotted). The red dotted line indicates the total score threshold (88,644.1) that defines the good-scoring NPC structures (Methods).
(D) Distribution of scores for structure samples 1 (red) and 2 (blue), comprising the 5,529 good-scoring NPC structures (nsample1=2,359 and nsample2=3,170 structures). The non-parametric Kolmogorov-Smirnov two-sample test158,159 (two-sided) indicates that the difference between the two score distributions is insignificant (p-value (1.0) > 0.05). In addition, the magnitude of the difference is small, as demonstrated by the Kolmogorov-Smirnov two-sample test statistic, D, of 0.045. Thus, the two score distributions are effectively equal.
(E) Three criteria for determining the sampling precision (Y-axis), evaluated as a function of the RMSD clustering threshold160 (X-axis) (n=5,529 structures). First, the p-value is computed using the χ2-test (one-sided) for homogeneity of proportions161 (red dots). Second, an effect size for the χ2-test is quantified by the Cramer’s V value (blue squares). Third, the population of structures in sufficiently large clusters (containing at least 10 structures from each sample) is shown as green triangles. The vertical dotted grey line indicates the RMSD clustering threshold at which three conditions are satisfied (χ2-test p-value (0.75) > 0.05 (red, horizontal dotted line), Cramer’s V (0.065) < 0.10 (blue, horizontal dotted line), and the population of clustered structures (0.90) > 0.80 (green, horizontal dotted line)), thus defining the sampling precision of 9 Å. The three solid curves (in red, blue, and green) were drawn through the points, to help visualize the results.
(F) Population of sample 1 and 2 structures in the three clusters obtained by threshold-based clustering160 using a RMSD threshold of 12 Å. The dominant cluster (cluster 1) contains 80.3% of the structures. Cluster precision is shown for each cluster. The precision of the dominant cluster defines the structure precision.
Extended Data Figure 2. Quantitative analysis of the mass and stoichiometry of the endogenous NPC (I).
(A) A multipronged approach to accurately define the mass, stoichiometry, and composition of native macromolecular assemblies. Schematic representation is shown of the multiple orthologous methods that are integrated in our strategy for the analysis of native assemblies. The main experimental methods are listed on top, followed by the characteristic that they help to quantify (in blue) and the type of sample to which they were applied. The final outcome of each method is indicated (black arrows); the steps of each method that are compared to serve as a control cross-check are indicated (blue dashed lines). At bottom, the integration of the different data points into a final comprehensive description of the endogenous assembly is depicted. Small cartoon insets of the NPC are used to illustrate the analysis.
(B) SDS-PAGE analysis of the affinity captured S. cerevisiae NPCs isolated from an MLP1-ppxPrA tagged strain (n>20 independent experiments). Molecular weight marker values are indicated to the left of the gel lane. Dots signify the main protein components of the isolated NPCs as identified by LC-MS (Extended Data Fig. 3C). Proteins are grouped and coloured by functional categories or membership of discrete macromolecular assemblies (blue - Nups; red - mRNA transport factors (TF); orange - Transport factors; green - TREX complex; grey - contaminants/others). For gel source data, see Supplementary Figure 1.
(C) Cryo-EM analysis of the affinity captured NPCs (n>20 independent experiments). The particles have a clear preferred orientation (Methods). Some side views are presented in the inset. The central transporter is present in every NPC (indicated by “T”). Scale bar 1000 Å.
(D) Schematic showing the primary amino acid sequence of the 148.2 kDa synthetic QconCAT-A. It includes two peptides for each Nup (thick bars), arranged in the indicated order. The native three amino acid residues flanking regions (thin bars) of each peptide were included to preserve the native trypsin target sequence. A sacrificial N-terminal 3×FLAG tag was included, as well as a C-terminal 6×His tag for purification under denaturing conditions. The stringent criteria used for the selection of the QconCAT peptides are described in detail in the Methods section.
(E) MALDI-MS spectrum of intact, purified full-length stable isotopically labeled QconCAT-A, showing that a single species was detected. n+ (n=1,2,…,17) denotes the Qconcat-A protein species with n positive charges and 2M Qconcat-A protein dimer. The measured molecular weight of the stable isotopically labeled QconCAT-A was 149,049 ± 38 Da, agreeing with its calculated molecular weight of 148,200 Da (Methods).
Extended Data Figure 3. Quantitative analysis of the mass and stoichiometry of the endogenous NPC (II).
(A) Left, schematic localization of the Nup-GFP reporters selected for the in vivo calibrated imaging stoichiometry analyses. Nups were selected to represent every major NPC module and to provide comprehensive coverage of the assembly. Right, from the calibrated imaging data, Kernel density estimation of distributions of GFP proteins per Nup were calculated. n = 48 – 178.
(B) Heat map of a yeast cell expressing Nup85-GFP. Image (right) was acquired as described in Methods. In addition, for illustration purposes, a max projection over z was done, and the image was smoothed with a Gaussian blur of radius 1. A heat map was used to illustrate intensity units, in raw photon counts. For the spot outlined in red, a 2 dimensional distribution of photon counts, and the corresponding Gaussian fit, are shown (left).
(C) Stoichiometries of main components associated with the affinity captured NPCs, as determined by label-free MS quantitation (at least 3 peptides per protein). Proteins are grouped and coloured by functional categories or membership of discrete macromolecular assemblies. TREX complex components are included in the “Transport Factors” category and labeled with an asterisk. QConCAT-derived stoichiometries for all the Nups (dark grey bars) are shown for comparison.
Extended Data Figure 4. Cryo-ET strategy and the resulting 3D Cryo-ET map of the NPC exhibiting non-enforced local C2-symmetry axes in the inner ring.
Labels throughout the figure are: C for cytoplasm, N for nucleoplasm, T for central transporter, S for core scaffold, MR for membrane ring, IR for inner ring. Scale bar 100 Å.
(A) A diagram describing the methodology used to obtain the whole S. cerevisiae NPC Cryo-ET map (Methods).
(B) 2D class averages are shown (protein white) which were calculated using the original unaligned sub-tomograms projected along the Z-axis. The overall thickness of the yeast NPC is apparent in a side view class, and the local C2-symmetry axes in the inner ring are also apparent (indicated with “2”). The transporter density is present in every class.
(C) Left, top view of the Cryo-ET map with the two local C2-symmetry axes indicated by arrows and labels (sym 1 and sym 2). They are 22.5° apart, due to the C8-symmetry axis. Right, 2D projections of the top view, and two side views along the two local C2-symmetry axes (side 1 and side 2 projected along axes sym 1 and sym 2, respectively).
(D) Seven cross-sections of the Cryo-ET map are shown on the right (1–7) with their positions in the 3D map indicated in the side view on the left. The local C2-symmetry of the inner-ring is apparent in cross-sections 2–6 mirrored about the central section in panel 4.
Extended Data Figure 5. Resolution estimates for the Cryo-ET map of the NPC and comparison of cross-sections between the intermediate, final and Relion Cryo-ET maps.
(A) Top and (B) side views of the Cryo-ET map are colour-coded according to local resolution estimates (colour bar) and are shown at a low threshold to reveal weaker density features at the periphery of the NPC, which are more flexible.
(C) Cross-sections are shown at a reduced scale, colour coded according to local resolution estimates (colour bar). A remnant of the pore membrane is present (M), encircling the entire mid-line of the NPC. Sections 3–5 are shown at a higher threshold. In section 3, the inner ring region is indicated by “spokes”. In section 4, local C2-symmetry axes are indicated by dashed arrows.
(D) Thick sections of the inner ring are shown at a higher threshold, as viewed along the membrane plane. Note that the inner ring (indicated) is almost entirely in the 20–25 Å resolution range. C, cytoplasmic side; N, nuclear side; CR, cytoplasmic ring; NR, nuclear ring; M, pore membrane; T, central transporter.
(E) Comparison of 5 cross-sections (cross-section number on left) in the inner ring region of the NPC between Cryo-ET maps in different stages of the reconstruction process (Extended Data Fig. 4A): intermediate map (left column), final map (middle column) and an independent validation map, reconstructed with Relion at a twice reduced Å/pixel size (right column). Details on how each map was reconstructed are provided in the Methods section.
Extended Data Figure 6. 2D classification of projections from 1,864 original NPC sub-tomograms aligned with their C8-symmetry axis nearly along the Z-axis.
(A) In total, 18 good class averages are shown after Maximum Likelihood classification (using RELION 1.4162,163) without symmetry imposed. Each class average (on the left) is paired with a C8-symmetry enforced image of itself (on the right). Central transporter densities are present in each of the class averages (both with and without imposed C8-symmetry), indicating that the central transporter is generally present in these particles.
(B) An expanded view of a large class from panel A (marked by white dots) shows bridges (indicated) between the core scaffold and the central transporter, both before and after averaging using the C8-symmetry of the NPC. S, core scaffold; MR, membrane ring; T, central transporter. Matching panels in A and B are marked with white dots.
(C,D) The Cryo-ET 3D map is presented as in Fig. 3 and is zoomed in to show the meshwork of bridges between the scaffold and the central transporter, as viewed from the cytoplasm and the nucleoplasm, respectively.
Extended Data Figure 7. Validation of the NPC structure (I).
(A–C) Satisfaction of the chemical cross-links
(A) Identified cross-links were mapped onto the integrative structure of the entire NPC, as shown in front (upper) and top (lower) views. Satisfied cross-links, whose Cα–Cα distances fall within the distance threshold of 35 Å in at least one good-scoring solution, are shown in blue. Violated cross-links, whose Cα–Cα distances are larger than 35 Å, are shown in orange. The histogram on the right shows the distribution of the cross-linked Cα–Cα distances, validating the NPC structure.
(B) Mapping of the cross-links onto the cytoplasmic and nucleoplasmic connector Nups (Nup116, Nup100, Nup145N, Nup1, and Nup60). Front (right) and side (left) views show how the NPC outer rings are connected to the inner ring through a network of connector Nups across the length of the spoke.
(C) Mapping of the cross-links onto the inner- (and membrane) and outer-rings, in front (upper) and top (lower) views.
(D–G) Satisfaction of data and considerations that were not used to compute the structure
(D) The integrative structure of the NPC (this study, left panel) was compared to the 2007 topological map154,164 (right panel). The two structures are consistent with each other, albeit the current structure is defined at an order of magnitude higher precision.
(E) Satisfaction of affinity purification and overlay assays data (composites); our current structure satisfies all 82 composites determined by affinity purification and overlay assays154,164, even though they were not used in its determination. For example, Pom152, Pom34, Ndc1, Nup157, and Nup170 are connected with each other (left panel), consistent with the composites determined by the affinity purification data154,164 published in 2007 (right panel).
(F) Satisfaction of SAXS data; the atomic structures of 8 Nups are consistent with the corresponding SAXS profiles for their constructs165–171 (Supplementary Tables 2 and 6; Methods). For example, the SAXS profile calculated from the atomic structure of Pom152718–1148 (red curve) using FoXS172 is well matched (χ=1.48) to the corresponding experimental SAXS profile170 (black dots; n=20 exposures). For visualization purposes, the Pom152718–1148 structure (represented as a ribbon) is shown along with the best fit of the ab initio shape (represented as a transparent envelope) computed from the experimental SAXS profile.
(G) Satisfaction of the negative-stain EM 2D class averages for the Nic96 complex; the localization density of the Nic96 complex (composed of Nic96, Nsp1, Nup49, and Nup57) in the dominant cluster can be projected well on 2D class averages obtained for the natively isolated complex (n=5,458 particles; Methods). The experimental class averages were satisfied by the structure with cross-correlation coefficients of 0.81 and 0.83, respectively (Methods).
Extended Data Figure 8. Validation of the NPC structure (II) - consistency between the NPC structure and the Cryo-ET density map.
The Cryo-ET density map is shown at a high density threshold (grey), to reveal details of the inner ring. A representative structure of the inner ring is shown docked into the density, showing the excellent fit. All Nups are coloured as in Fig. 4. The pore membrane is indicated by M. (A) Full 8-spoke inner ring (scale bar 100 Å); front (B), top (C), and back (D) views of 3 spokes with neighbors coloured brown and grey (scale bar 50 Å). (E) Different views of a single spoke (scale bar 50 Å) are shown within the density map. (F) Thick cross-sections are shown through a single spoke in the inner ring, as viewed from the central C8-symmetry axis (scale bar 50 Å); MBM’s (membrane-binding motifs; Fig. 5) are indicated.
Extended Data Figure 9. Position of the FG repeat anchor points and heat mapping of the FG repeats.
(A) Three views of the complete structure of the NPC are shown with major structural features (coloured as in Fig. 4 and Supplementary Table 2) and a snapshot of modeled FG repeat regions (indicated in green). For each Nup, the localization probability density of the ensemble of structures is shown with a representative structure from the ensemble embedded within it. See also Supplementary Videos 1–3. Scale bar 200 Å.
(B) Position of FG repeat anchor points within the ensemble of solutions are depicted as green surfaces; the Nups to which they belong are labeled accordingly in the center image. Three spokes side view (left), one spoke side view (center), three spokes top view (right). Scale bar 100 Å.
(C) Heat mapping of the type of FG repeat region of each FG Nup (FxFG/FG type, red; GLFG type, blue), showing partitioning of the FG types to different regions of the central transporter. Identity of mapped Nups is shown in the diagram on the right. Scale bar 100 Å.
(D) Heat mapping of the effect on NPC permeability of the truncation of an FG repeat in each FG Nup, relative to the WT strain (p/pWT); the severity of the permeability defect is indicated in increasing shades of blue from minor defect (light green) to severe defect (dark blue), thereby defining the most important FG repeats needed to maintain the passive permeability barrier. Identity of mapped Nups is shown in the diagram on the right. Scale bar 100 Å.
Extended Data Figure 10. Comparison of the S. cerevisiae NPC structure and human NPC core scaffold.
(A) Comparison between the inner-rings in the structure of the S. cerevisiae NPC (first and third rows) and the core scaffold of the human NPC (second and fourth rows) (PDB code 5IJN173). Yeast Nups are coloured as in Fig. 4; human Nup homologs are coloured as their yeast counterparts. All copies of human Nup155 are coloured as yeast Nup157, and all copies of human Nup205/Nup188 are coloured as yeast Nup192. Only homolog Nups present in both yeast and human are shown. The human NPC core scaffold includes two additional copies of Nup155 that are absent in yeast (due to the different stoichiometry between organisms). Yeast Nup53 and Nup59 are not shown because their counterparts are not present in the human NPC core scaffold.
(B) Major differences in the inner-ring between the S. cerevisiae and human NPCs are highlighted, in the cross-sectional view near the equator.
(C) Positions of yeast Nups homologous to oncogenic human Nups (in parentheses) are shown in red, mapped onto three spokes of the NPC.
Extended Data Figure 11. Proposed evolutionary origin of the NPC from a later amalgam of membrane coating complexes.
(A) Diagram depicting how the NPC may have originated from an ancestral coatomer module through a series of duplications, divergence, and secondary loss events.
(Top) The origin of an ancestral proto-NPC coatomer module from an amalgamation of COPI-like and COPII-like complexes. Middle, the initial duplication leading to the origin of the inner and outer rings and their associated coiled bundles. Presumed secondary losses removed the inner ring protomer’s additional COPII-like subunit; loss of the outer ring’s adaptin-like subunit may have occurred here, or later in only certain lineages.
(Bottom) Another duplication and divergence within each spoke may then have generated two parallel and laterally-offset paralogous columns; in the outer ring, a COPII-like subunit was then lost from one of the duplicates. The coiled bundles of the outer rings gave rise to the cytoplasmic export complex and nuclear basket by subsequent duplication; the export complex itself is a duplicate with a dimer of trimeric coiled bundles in its core. Outer ring duplications are not shown. Relevant nucleoporin domains are depicted as follows: β-propellers (cyan circles), α-solenoids (pink bars), and coiled-coil domains (orange sticks). On the left, a series of diagrams (grey) exemplify the path of duplications within the whole NPC. Examples of ribbon representations for each module are presented. The anchoring points of the coiled-coil cytoplasmic Nup82 complex and the nuclear basket (orange densities) into an equivalent region of the outer ring Nup84 complex (grey density) are shown.
(B) Conserved structural motifs connecting spokes in the outer and inner rings. Diagram showing how the spoke-to-spoke connection is established through similar head-to-head connections of COPI-like/COPII like heterodimer in both the outer (left side) and the inner (right side) NPC rings. Top, nucleoporin domains coloured as in (A); bottom, COPI-like nups highlighted in red, COPII-like nups highlighted in blue.
Extended Data Figure 12. Functional analysis of nucleoporin mutants’ fitness using ODELAY.
(A) The fitness defect phenotype was quantified and plotted (mean Z-score; n=6 experiments, containing at least 200–300 individuals per point; see Methods for details) for each nucleoporin truncation or C-terminal Protein-A tagged mutant in order of decreasing fitness (increasing number of units) as observed by ODELAY assay174 (Methods). Strains for which truncations in a haploid background were found to lead to lethality after tetrad dissection (Nic96 and Nup192) were assigned the maximum level of defect and plotted on top of the rest (diploid), based on the fitness phenotype observed for the indicated Nic96 and Nup192 mutants in a diploid background (where a wild type copy of the nucleoporin is also present and expressed). Six divisions were assigned based on decreasing level of fitness171,175 (white = wild type, to dark purple = severe defect). AU, arbitrary unit; Error = standard deviation.
(B) Mapping of the colour code described in (A) into the NPC components. Horizontal lines represent the amino acid residue length of each protein and truncated version; amino acid residue positions are shown on top of the lines.
Supplementary Material
1
Supp Table 6
Supp Table 7
Supp Table 8
Supp Table 9
Supp Video 1
Supp Video 2
Supp Video 3
2
3
4
Supp Table 1
Supp Table 2
Supp Table 3
Supp Table 4
Supp Table 5
Acknowledgments
We thank B. Webb (UCSF) for help with IMP, the Rockefeller University Outreach Program for support for A.S.C., the NYULMC OCS Microscopy Core, K. Uryu, and the EMRC Resource Center (Rockefeller University) for assistance with negative-stain EM, and F. Alber, M.C. Field, N. Ketaren, S. Obado, R. Hayama, and D. Simon for feedback and critical reading of the manuscript. The work was supported by a NSF GRF 1650113 (I.E.C.), a NSF grant CHE-1531823 (M.F.J.), the SIMR (J.L.G.), NIH grants of R01 GM080477 (J.L.G.), U54 GM103511 (B.T.C., A.S., J.D.A., and M.P.R.), R01 GM112108 (M.P.R., J.D.A.), P41 GM109824 (M.P.R., A.S., J.D.A., and B.T.C.), P50 GM076547 (J.D.A.), R01 GM063834 (C.W.A.), R01 GM080139 (S.J.L.), P41 GM103314 (B.T.C.), R01 GM083960 (A.S.), and U54 DK107981 (M.P.R, J.D.A.), and a donation from Louis Herlands.
Footnotes
Supplementary information is available in the online version of the paper.
Author Contributions
I.N., J.F-M., A.S.C., R.W., M.P.R. performed the affinity purifications; W.Z., J.F-M., R.W., R.M., E.Y.J., M.P.R., B.T.C. performed the quantitative MS; M.S., B.D.S., J.R.U., J.L.G. performed the calibrated imaging; J.H., B.T.C., M.F.J. performed the charge detection MS; Y.S., J.F-M., R.W., I.N., J.W., B.T.C. performed the CX-MS; C.W.A., S.J.L., I.N. Z.Y., M.J.C. performed the Cryo-ET; S.J.K. performed the SAXS; T.H., J.F-M., J.D.A. performed the phenotypic profiling; P.U., D.L.S. performed the negative-stain EM; S.J.K., B.R., I.E.C., R.P., I.E., C.H.G., A.S. performed the integrative structure computations; S.J.L., C.W.A., B.T.C., A.S., M.P.R. supervised the project; S.J.K., J.F-M., I.N., Y.S., W.Z., B.R., S.J.L., C.W.A., B.T.C., A.S., M.P.R. wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Ptak C, Aitchison JD, Wozniak RW. The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr Opin Cell Biol. 2014;28:46–53. doi: 10.1016/j.ceb.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nofrini V, Di Giacomo D, Mecucci C. Nucleoporin genes in human diseases. European journal of human genetics : EJHG. 2016;24:1388–1395. doi: 10.1038/ejhg.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 4.Stanley GJ, Fassati A, Hoogenboom BW. Biomechanics of the transport barrier in the nuclear pore complex. Seminars in cell & developmental biology. 2017 doi: 10.1016/j.semcdb.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Akey CW. Interactions and structure of the nuclear pore complex revealed by cryo-electron microscopy. The Journal of cell biology. 1989;109:955–970. doi: 10.1083/jcb.109.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosinski J, et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science. 2016;352:363–365. doi: 10.1126/science.aaf0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin DH, et al. Architecture of the symmetric core of the nuclear pore. Science. 2016;352:aaf1015. doi: 10.1126/science.aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alber F, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 9.Fernandez-Martinez J, et al. Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell. 2016;167:1215–1228. e1225. doi: 10.1016/j.cell.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y, et al. Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Molecular & cellular proteomics : MCP. 2014;13:2927–2943. doi: 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Briggs JA. Structural biology in situ--the potential of subtomogram averaging. Current opinion in structural biology. 2013;23:261–267. doi: 10.1016/j.sbi.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Kim SJ, et al. Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Molecular & cellular proteomics : MCP. 2014;13:2911–2926. doi: 10.1074/mcp.M114.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Martinez J, et al. Structure-function mapping of a heptameric module in the nuclear pore complex. The Journal of cell biology. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herricks T, et al. One-Cell Doubling Evaluation by Living Arrays of Yeast, ODELAY! G3 (Bethesda) 2017;7:279–288. doi: 10.1534/g3.116.037044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. The Journal of cell biology. 1995;131:1133–1148. doi: 10.1083/jcb.131.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer J, Teimer R, Amlacher S, Kunze R, Hurt E. Linker Nups connect the nuclear pore complex inner ring with the outer ring and transport channel. Nature structural & molecular biology. 2015;22:774–781. doi: 10.1038/nsmb.3084. [DOI] [PubMed] [Google Scholar]
- 17.von Appen A, et al. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marelli M, Lusk CP, Chan H, Aitchison JD, Wozniak RW. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. Journal of Cell Biology. 2001;153:709–723. doi: 10.1083/Jcb.153.4.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vollmer B, et al. Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J. 2012;31:4072–4084. doi: 10.1038/emboj.2012.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seo HS, et al. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14281–14286. doi: 10.1073/pnas.0907453106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drin G, et al. A general amphipathic alpha-helical motif for sensing membrane curvature. Nature structural & molecular biology. 2007;14:138–146. doi: 10.1038/nsmb1194. [DOI] [PubMed] [Google Scholar]
- 22.Upla P, et al. Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex. Structure. 2017;25:434–445. doi: 10.1016/j.str.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meinema AC, et al. Long unfolded linkers facilitate membrane protein import through the nuclear pore complex. Science. 2011;333:90–93. doi: 10.1126/science.1205741. [DOI] [PubMed] [Google Scholar]
- 24.Knockenhauer KE, Schwartz TU. The Nuclear Pore Complex as a Flexible and Dynamic Gate. Cell. 2016;164:1162–1171. doi: 10.1016/j.cell.2016.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Folkmann AW, Noble KN, Cole CN, Wente SR. Dbp5, Gle1-IP6 and Nup159: a working model for mRNP export. Nucleus. 2011;2:540–548. doi: 10.4161/nucl.2.6.17881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saroufim MA, et al. The nuclear basket mediates perinuclear mRNA scanning in budding yeast. The Journal of cell biology. 2015;211:1131–1140. doi: 10.1083/jcb.201503070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meseroll RA, Cohen-Fix O. The Malleable Nature of the Budding Yeast Nuclear Envelope: Flares, Fusion, and Fenestrations. J Cell Physiol. 2016;231:2353–2360. doi: 10.1002/jcp.25355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paradise A, Levin MK, Korza G, Carson JH. Significant proportions of nuclear transport proteins with reduced intracellular mobilities resolved by fluorescence correlation spectroscopy. J Mol Biol. 2007;365:50–65. doi: 10.1016/j.jmb.2006.09.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adams RL, Wente SR. Uncovering nuclear pore complexity with innovation. Cell. 2013;152:1218–1221. doi: 10.1016/j.cell.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel SS, Belmont BJ, Sante JM, Rexach MF. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell. 2007;129:83–96. doi: 10.1016/j.cell.2007.01.044. [DOI] [PubMed] [Google Scholar]
- 31.Adams RL, Terry LJ, Wente SR. Nucleoporin FG domains facilitate mRNP remodeling at the cytoplasmic face of the nuclear pore complex. Genetics. 2014;197:1213–1224. doi: 10.1534/genetics.114.164012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timney BL, et al. Simple rules for passive diffusion through the nuclear pore complex. The Journal of cell biology. 2016;215:57–76. doi: 10.1083/jcb.201601004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada J, et al. A bimodal distribution of two distinct categories of intrinsically disordered structures with separate functions in FG nucleoporins. Molecular & cellular proteomics : MCP. 2010;9:2205–2224. doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Devos D, et al. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faini M, Beck R, Wieland FT, Briggs JA. Vesicle coats: structure, function, and general principles of assembly. Trends Cell Biol. 2013;23:279–288. doi: 10.1016/j.tcb.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 36.Rout MP, Field MC. The Evolution of Organellar Coat Complexes and Organization of the Eukaryotic Cell. Annual review of biochemistry. 2017;86:637–657. doi: 10.1146/annurev-biochem-061516-044643. [DOI] [PubMed] [Google Scholar]
- 37.Obado SO, et al. Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex. PLoS Biol. 2016;14:e1002365. doi: 10.1371/journal.pbio.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Iwamoto M, et al. Compositionally distinct nuclear pore complexes of functionally distinct dimorphic nuclei in the ciliate Tetrahymena. J Cell Sci. 2017;130:1822–1834. doi: 10.1242/jcs.199398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obado SO, Field MC, Rout MP. Comparative interactomics provides evidence for functional specialization of the nuclear pore complex. Nucleus. 2017;8:340–352. doi: 10.1080/19491034.2017.1313936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bui KH, et al. Integrated structural analysis of the human nuclear pore complex scaffold. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- 41.Debler EW, et al. A fence-like coat for the nuclear pore membrane. Mol Cell. 2008;32:815–826. doi: 10.1016/j.molcel.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez-Martinez J, et al. Structure-function mapping of a heptameric module in the nuclear pore complex. The Journal of cell biology. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Upla P, et al. Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex. Structure. 2017;25:434–445. doi: 10.1016/j.str.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rout MP, Blobel G. Isolation of the yeast nuclear pore complex. The Journal of cell biology. 1993;123:771–783. doi: 10.1083/jcb.123.4.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.LaCava J, Fernandez-Martinez J, Hakhverdyan Z, Rout MP. Protein Complex Purification by Affinity Capture. Cold Spring Harb Protoc. 2016;2016 doi: 10.1101/pdb.top077545. pdb top077545. [DOI] [PubMed] [Google Scholar]
- 46.LaCava J, Fernandez-Martinez J, Hakhverdyan Z, Rout MP. Optimized Affinity Capture of Yeast Protein Complexes. Cold Spring Harb Protoc. 2016;2016 doi: 10.1101/pdb.prot087932. pdb prot087932. [DOI] [PubMed] [Google Scholar]
- 47.LaCava J, Fernandez-Martinez J, Rout MP. Native Elution of Yeast Protein Complexes Obtained by Affinity Capture. Cold Spring Harb Protoc. 2016;2016 doi: 10.1101/pdb.prot087940. pdb prot087940. [DOI] [PubMed] [Google Scholar]
- 48.Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 49.Oeffinger M, et al. Comprehensive analysis of diverse ribonucleoprotein complexes. Nature methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- 50.Hakhverdyan Z, et al. Rapid, optimized interactomic screening. Nature methods. 2015;12:553–560. doi: 10.1038/nmeth.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beynon RJ, Doherty MK, Pratt JM, Gaskell SJ. Multiplexed absolute quantification in proteomics using artificial QCAT proteins of concatenated signature peptides. Nature methods. 2005;2:587–589. doi: 10.1038/nmeth774. [DOI] [PubMed] [Google Scholar]
- 52.Shivaraju M, et al. Cell-cycle-coupled structural oscillation of centromeric nucleosomes in yeast. Cell. 2012;150:304–316. doi: 10.1016/j.cell.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Keifer DZ, Motwani T, Teschke CM, Jarrold MF. Measurement of the accurate mass of a 50 MDa infectious virus. Rapid Commun Mass Spectrom. 2016;30:1957–1962. doi: 10.1002/rcm.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pratt JM, et al. Multiplexed absolute quantification for proteomics using concatenated signature peptides encoded by QconCAT genes. Nat Protoc. 2006;1:1029–1043. doi: 10.1038/nprot.2006.129. [DOI] [PubMed] [Google Scholar]
- 55.Kito K, Ota K, Fujita T, Ito T. A synthetic protein approach toward accurate mass spectrometric quantification of component stoichiometry of multiprotein complexes. Journal of proteome research. 2007;6:792–800. doi: 10.1021/pr060447s. [DOI] [PubMed] [Google Scholar]
- 56.Ding C, et al. Quantitative analysis of cohesin complex stoichiometry and SMC3 modification-dependent protein interactions. Journal of proteome research. 2011;10:3652–3659. doi: 10.1021/pr2002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. The Journal of cell biology. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strambio-de-Castillia C, Blobel G, Rout MP. Isolation and characterization of nuclear envelopes from the yeast Saccharomyces. The Journal of cell biology. 1995;131:19–31. doi: 10.1083/jcb.131.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rout MP, Strambio-De-Castillia C. In: Cell Biology: A Laboratory Handbook 2. Celis JE, editor. Academic Press; 1998. pp. 143–151. [Google Scholar]
- 60.Cadene M, Chait BT. A robust, detergent-friendly method for mass spectrometric analysis of integral membrane proteins. Anal Chem. 2000;72:5655–5658. doi: 10.1021/ac000811l. [DOI] [PubMed] [Google Scholar]
- 61.Fenyo D, et al. MALDI sample preparation: the ultra thin layer method. Journal of visualized experiments : JoVE. 2007;192 doi: 10.3791/192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Field HI, Fenyo D, Beavis RC. RADARS, a bioinformatics solution that automates proteome mass spectral analysis, optimises protein identification, and archives data in a relational database. Proteomics. 2002;2:36–47. [PubMed] [Google Scholar]
- 63.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 64.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 65.Contino NC, Pierson EE, Keifer DZ, Jarrold MF. Charge detection mass spectrometry with resolved charge states. Journal of the American Society for Mass Spectrometry. 2013;24:101–108. doi: 10.1007/s13361-012-0525-5. [DOI] [PubMed] [Google Scholar]
- 66.Shi Y, et al. Structural characterization by cross-linking reveals the detailed architecture of a coatomer-related heptameric module from the nuclear pore complex. Molecular & cellular proteomics : MCP. 2014;13:2927–2943. doi: 10.1074/mcp.M114.041673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi Y, et al. A strategy for dissecting the architectures of native macromolecular assemblies. Nature methods. 2015;12:1135–1138. doi: 10.1038/nmeth.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Leitner A, et al. Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Molecular & cellular proteomics : MCP. 2012;11:M111 014126. doi: 10.1074/mcp.M111.014126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang B, et al. Identification of cross-linked peptides from complex samples. Nature methods. 2012;9:904–906. doi: 10.1038/nmeth.2099. [DOI] [PubMed] [Google Scholar]
- 70.Cevher MA, et al. Reconstitution of active human core Mediator complex reveals a critical role of the MED14 subunit. Nature structural & molecular biology. 2014;21:1028–1034. doi: 10.1038/nsmb.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun J, et al. The architecture of a eukaryotic replisome. Nature structural & molecular biology. 2015;22:976–982. doi: 10.1038/nsmb.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fernandez-Martinez J, et al. Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell. 2016;167:1215–1228 e1225. doi: 10.1016/j.cell.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zheng SQ, et al. UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. Journal of structural biology. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 74.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. Journal of structural biology. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 75.Galaz-Montoya JG, Flanagan J, Schmid MF, Ludtke SJ. Single particle tomography in EMAN2. Journal of structural biology. 2015;190:279–290. doi: 10.1016/j.jsb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Galaz-Montoya JG, et al. Alignment algorithms and per-particle CTF correction for single particle cryo-electron tomography. Journal of structural biology. 2016;194:383–394. doi: 10.1016/j.jsb.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–234. doi: 10.1016/s1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 78.Beck M, et al. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- 79.Eibauer M, et al. Structure and gating of the nuclear pore complex. Nat Commun. 2015;6:7532. doi: 10.1038/ncomms8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kosinski J, et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science. 2016;352:363–365. doi: 10.1126/science.aaf0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.von Appen A, Beck M. Structure Determination of the Nuclear Pore Complex with Three-Dimensional Cryo electron Microscopy. J Mol Biol. 2016;428:2001–2010. doi: 10.1016/j.jmb.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ludtke SJ. Single-Particle Refinement and Variability Analysis in EMAN2.1. Methods in enzymology. 2016;579:159–189. doi: 10.1016/bs.mie.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Iwanczyk J, et al. Structure of the Blm10–20 S proteasome complex by cryo-electron microscopy. Insights into the mechanism of activation of mature yeast proteasomes. J Mol Biol. 2006;363:648–659. doi: 10.1016/j.jmb.2006.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Elad N, et al. The dynamic conformational landscape of gamma-secretase. J Cell Sci. 2015;128:589–598. doi: 10.1242/jcs.164384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nature methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bharat TA, Scheres SH. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat Protoc. 2016;11:2054–2065. doi: 10.1038/nprot.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bharat TA, Russo CJ, Lowe J, Passmore LA, Scheres SH. Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging. Structure. 2015;23:1743–1753. doi: 10.1016/j.str.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sampathkumar P, et al. Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata. Proteins. 2012;80:2110–2116. doi: 10.1002/prot.24102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sampathkumar P, et al. Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex. Proteins. 2011;79:1672–1677. doi: 10.1002/prot.22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim SJ, et al. Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Molecular & cellular proteomics : MCP. 2014;13:2911–2926. doi: 10.1074/mcp.M114.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sampathkumar P, et al. Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145. Proteins. 2010;78:1992–1998. doi: 10.1002/prot.22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sampathkumar P, et al. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure. 2013;21:560–571. doi: 10.1016/j.str.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Herricks T, et al. One-Cell Doubling Evaluation by Living Arrays of Yeast, ODELAY! G3 (Bethesda) 2017;7:279–288. doi: 10.1534/g3.116.037044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Edelstein AD, et al. Advanced methods of microscope control using μManager software. J Biol Methods. 2014;1 doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-particle reconstructions. Journal of structural biology. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 96.Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Alber F, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 98.Russel D, et al. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Schneidman-Duhovny D, Pellarin R, Sali A. Uncertainty in integrative structural modeling. Current opinion in structural biology. 2014;28:96–104. doi: 10.1016/j.sbi.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sali A, et al. Outcome of the First wwPDB Hybrid/Integrative Methods Task Force Workshop. Structure. 2015;23:1156–1167. doi: 10.1016/j.str.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.LoPiccolo J, et al. Assembly and Molecular Architecture of the Phosphoinositide 3-Kinase p85alpha Homodimer. J Biol Chem. 2015;290:30390–30405. doi: 10.1074/jbc.M115.689604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Luo J, et al. Architecture of the Human and Yeast General Transcription and DNA Repair Factor TFIIH. Mol Cell. 2015;59:794–806. doi: 10.1016/j.molcel.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Robinson PJ, et al. Molecular architecture of the yeast Mediator complex. eLife. 2015:4. doi: 10.7554/eLife.08719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Webb B, et al. Modeling of proteins and their assemblies with the Integrative Modeling Platform. Methods in molecular biology. 2014;1091:277–295. doi: 10.1007/978-1-62703-691-7_20. [DOI] [PubMed] [Google Scholar]
- 105.Cherry JM, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic acids research. 2012;40:D700–705. doi: 10.1093/nar/gkr1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gautier R, Douguet D, Antonny B, Drin G. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics (Oxford, England) 2008;24:2101–2102. doi: 10.1093/bioinformatics/btn392. [DOI] [PubMed] [Google Scholar]
- 107.Niepel M, et al. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol Biol Cell. 2013;24:3920–3938. doi: 10.1091/mbc.E13-07-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 109.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic acids research. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics (Oxford, England) 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 111.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic acids research. 2013;41:W349–357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 113.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics (Oxford, England) 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 114.Marsden RL, McGuffin LJ, Jones DT. Rapid protein domain assignment from amino acid sequence using predicted secondary structure. Protein science : a publication of the Protein Society. 2002;11:2814–2824. doi: 10.1110/ps.0209902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 116.Trigg J, Gutwin K, Keating AE, Berger B. Multicoil2: predicting coiled coils and their oligomerization states from sequence in the twilight zone. PLoS One. 2011;6:e23519. doi: 10.1371/journal.pone.0023519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Algret R, et al. Molecular architecture and function of the SEA complex, a modulator of the TORC1 pathway. Molecular & cellular proteomics : MCP. 2014;13:2855–2870. doi: 10.1074/mcp.M114.039388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Raveh B, et al. Slide-and-exchange mechanism for rapid and selective transport through the nuclear pore complex. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E2489–2497. doi: 10.1073/pnas.1522663113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic acids research. 2010;38:W540–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schneidman-Duhovny D, Kim SJ, Sali A. Integrative structural modeling with small angle X-ray scattering profiles. BMC structural biology. 2012;12:17. doi: 10.1186/1472-6807-12-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Meszaros N, et al. Nuclear pore basket proteins are tethered to the nuclear envelope and can regulate membrane curvature. Dev Cell. 2015;33:285–298. doi: 10.1016/j.devcel.2015.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kawabata T. Multiple subunit fitting into a low-resolution density map of a macromolecular complex using a gaussian mixture model. Biophys J. 2008;95:4643–4658. doi: 10.1529/biophysj.108.137125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jonic S, et al. Denoising of high-resolution single-particle electron-microscopy density maps by their approximation using three-dimensional Gaussian functions. Journal of structural biology. 2016;194:423–433. doi: 10.1016/j.jsb.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 124.Hanot S, et al. Multi-scale Bayesian modeling of cryo-electron microscopy density maps. 2017 https://doi.org/10.1101/113951Preprint.
- 125.Rout MP, et al. The yeast nuclear pore complex: composition, architecture, and transport mechanism. The Journal of cell biology. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen MY, Sali A. Statistical potential for assessment and prediction of protein structures. Protein science : a publication of the Protein Society. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Campelo F, Kozlov MM. Sensing membrane stresses by protein insertions. PLoS Comput Biol. 2014;10:e1003556. doi: 10.1371/journal.pcbi.1003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Viswanath S, Chemmama IE, Cimermancic P, Sali A. Assessing Exhaustiveness of Stochastic Sampling for Integrative Modeling of Macromolecular Structures. Biophys J. 2017;113:2344–2353. doi: 10.1016/j.bpj.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Siegel S. Nonparametric statistics for the behavioral sciences. McGraw-Hill; 1956. [Google Scholar]
- 130.McCarroll D. Simple statistical tests for geography [Google Scholar]
- 131.McDonald JH. Handbook of Biological Statistics. 3. Baltimore: Sparky House Publishing; 2014. [Google Scholar]
- 132.Daura X, et al. Peptide folding: When simulation meets experiment. Angew Chem Int Edit. 1999;38:236–240. doi: 10.1002/(Sici)1521-3773(19990115)38:1/2<236::Aid-Anie236>3.3.Co;2-D. [DOI] [Google Scholar]
- 133.Kuzmanic A, Zagrovic B. Determination of ensemble-average pairwise root mean-square deviation from experimental B-factors. Biophys J. 2010;98:861–871. doi: 10.1016/j.bpj.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Read RJ, et al. A new generation of crystallographic validation tools for the protein data bank. Structure. 2011;19:1395–1412. doi: 10.1016/j.str.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Montelione GT, et al. Recommendations of the wwPDB NMR Validation Task Force. Structure. 2013;21:1563–1570. doi: 10.1016/j.str.2013.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Henderson R, et al. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Trewhella J, et al. Report of the wwPDB Small-Angle Scattering Task Force: data requirements for biomolecular modeling and the PDB. Structure. 2013;21:875–881. doi: 10.1016/j.str.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 138.Leitner A, et al. Chemical cross-linking/mass spectrometry targeting acidic residues in proteins and protein complexes. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:9455–9460. doi: 10.1073/pnas.1320298111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Erzberger JP, et al. Molecular architecture of the 40S-eIF1-eIF3 translation initiation complex. Cell. 2014;158:1123–1135. doi: 10.1016/j.cell.2014.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.von Appen A, et al. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Lin DH, et al. Architecture of the symmetric core of the nuclear pore. Science. 2016;352:aaf1015. doi: 10.1126/science.aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Obado SO, et al. Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex. PLoS Biol. 2016;14:e1002365. doi: 10.1371/journal.pbio.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Alber F, Forster F, Korkin D, Topf M, Sali A. Integrating diverse data for structure determination of macromolecular assemblies. Annual review of biochemistry. 2008;77:443–477. doi: 10.1146/annurev.biochem.77.060407.135530. [DOI] [PubMed] [Google Scholar]
- 144.Alber F, Chait BT, Rout MP, Sali A. In: Protein-protein interactions and networks: identification, characterization and prediction. Panchenko A, Przytycka T, editors. Springer-Verlag; 2008. pp. 99–114. [Google Scholar]
- 145.Ermak DL, Mccammon JA. Brownian Dynamics with Hydrodynamic Interactions. J Chem Phys. 1978;69:1352–1360. doi: 10.1063/1.436761. [DOI] [Google Scholar]
- 146.Timney BL, et al. Simple rules for passive diffusion through the nuclear pore complex. The Journal of cell biology. 2016;215:57–76. doi: 10.1083/jcb.201601004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hough LE, et al. The molecular mechanism of nuclear transport revealed by atomic-scale measurements. eLife. 2015;4 doi: 10.7554/eLife.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Milles S, et al. Plasticity of an ultrafast interaction between nucleoporins and nuclear transport receptors. Cell. 2015;163:734–745. doi: 10.1016/j.cell.2015.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sakiyama Y, Mazur A, Kapinos LE, Lim RY. Spatiotemporal dynamics of the nuclear pore complex transport barrier resolved by high-speed atomic force microscopy. Nat Nanotechnol. 2016;11:719–723. doi: 10.1038/nnano.2016.62. [DOI] [PubMed] [Google Scholar]
- 150.Maarel vd. Introduction to Biopolymer Physics. World Scientic, J.R.C; 2008. p. 247. [Google Scholar]
- 151.Denning DP, Patel SS, Uversky V, Fink AL, Rexach M. Disorder in the nuclear pore complex: the FG repeat regions of nucleoporins are natively unfolded. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2450–2455. doi: 10.1073/pnas.0437902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lemke EA. The Multiple Faces of Disordered Nucleoporins. J Mol Biol. 2016;428:2011–2024. doi: 10.1016/j.jmb.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lim RY, et al. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:9512–9517. doi: 10.1073/pnas.0603521103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Alber F, et al. Determining the architectures of macromolecular assemblies. Nature. 2007;450:683–694. doi: 10.1038/nature06404. [DOI] [PubMed] [Google Scholar]
- 155.Russel D, et al. Putting the pieces together: integrative modeling platform software for structure determination of macromolecular assemblies. PLoS Biol. 2012;10:e1001244. doi: 10.1371/journal.pbio.1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Schneidman-Duhovny D, Pellarin R, Sali A. Uncertainty in integrative structural modeling. Current opinion in structural biology. 2014;28:96–104. doi: 10.1016/j.sbi.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Sali A, et al. Outcome of the First wwPDB Hybrid/Integrative Methods Task Force Workshop. Structure. 2015;23:1156–1167. doi: 10.1016/j.str.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Siegel S. Nonparametric statistics for the behavioral sciences. McGraw-Hill; 1956. [Google Scholar]
- 159.McCarroll D. Simple statistical tests for geography [Google Scholar]
- 160.Daura X, et al. Peptide folding: When simulation meets experiment. Angew Chem Int Edit. 1999;38:236–240. doi: 10.1002/(Sici)1521-3773(19990115)38:1/2<236::Aid-Anie236>3.3.Co;2-D. [DOI] [Google Scholar]
- 161.McDonald JH. Handbook of Biological Statistics 3rd ed. Baltimore: Sparky House Publishing; 2014. [Google Scholar]
- 162.Bharat TA, Scheres SH. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat Protoc. 2016;11:2054–2065. doi: 10.1038/nprot.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bharat TA, Russo CJ, Lowe J, Passmore LA, Scheres SH. Advances in Single-Particle Electron Cryomicroscopy Structure Determination applied to Sub-tomogram Averaging. Structure. 2015;23:1743–1753. doi: 10.1016/j.str.2015.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Alber F, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 165.Sampathkumar P, et al. Atomic structure of the nuclear pore complex targeting domain of a Nup116 homologue from the yeast, Candida glabrata. Proteins. 2012;80:2110–2116. doi: 10.1002/prot.24102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sampathkumar P, et al. Structure of the C-terminal domain of Saccharomyces cerevisiae Nup133, a component of the nuclear pore complex. Proteins. 2011;79:1672–1677. doi: 10.1002/prot.22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kim SJ, et al. Integrative structure-function mapping of the nucleoporin Nup133 suggests a conserved mechanism for membrane anchoring of the nuclear pore complex. Molecular & cellular proteomics : MCP. 2014;13:2911–2926. doi: 10.1074/mcp.M114.040915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sampathkumar P, et al. Structures of the autoproteolytic domain from the Saccharomyces cerevisiae nuclear pore complex component, Nup145. Proteins. 2010;78:1992–1998. doi: 10.1002/prot.22707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sampathkumar P, et al. Structure, dynamics, evolution, and function of a major scaffold component in the nuclear pore complex. Structure. 2013;21:560–571. doi: 10.1016/j.str.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Upla P, et al. Molecular Architecture of the Major Membrane Ring Component of the Nuclear Pore Complex. Structure. 2017;25:434–445. doi: 10.1016/j.str.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fernandez-Martinez J, et al. Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell. 2016;167:1215–1228. e1225. doi: 10.1016/j.cell.2016.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schneidman-Duhovny D, Hammel M, Sali A. FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic acids research. 2010;38:W540–544. doi: 10.1093/nar/gkq461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kosinski J, et al. Molecular architecture of the inner ring scaffold of the human nuclear pore complex. Science. 2016;352:363–365. doi: 10.1126/science.aaf0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Herricks T, et al. One-Cell Doubling Evaluation by Living Arrays of Yeast, ODELAY! G3 (Bethesda) 2017;7:279–288. doi: 10.1534/g3.116.037044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Fernandez-Martinez J, et al. Structure-function mapping of a heptameric module in the nuclear pore complex. The Journal of cell biology. 2012;196:419–434. doi: 10.1083/jcb.201109008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
Supp Table 6
Supp Table 7
Supp Table 8
Supp Table 9
Supp Video 1
Supp Video 2
Supp Video 3
2
3
4
Supp Table 1
Supp Table 2
Supp Table 3
Supp Table 4
Supp Table 5