A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells (original) (raw)
Abstract
microRNA (miRNA) expression profiles are often characteristic of specific cell types. The mouse mammary epithelial cell line, Comma-Dβ, contains a population of self-renewing progenitor cells that can reconstitute the mammary gland. We purified this population and determined its miRNA signature. Several microRNAs, including miR-205 and miR-22, are highly expressed in mammary progenitor cells, while others, including let-7 and miR-93, are depleted. Let-7 sensors can be used to prospectively enrich self-renewing populations, and enforced let-7 expression induces loss of self-renewing cells from mixed cultures.
[Keywords: RNAi, microRNA, stem cell]
MicroRNAs (miRNAs) hone patterns of gene expression by programming the RNA interference (RNAi) machinery to regulate complementary targets. While miRNAs have been integrated into numerous biological circuits, seminal studies in Caenorhabditis elegans implicated these small RNAs in regulation of differentiation and development (Reinhart et al. 2000). In this regard, some miRNAs are characteristic of particular differentiated cell types (Lagos-Quintana et al. 2002), while others are specifically expressed during early development and in stem and progenitor cells (Houbaviy et al. 2003; Giraldez et al. 2006; Neilson et al. 2007).
Evidence for a role of miRNAs in stem cell maintenance and differentiation is accumulating from analysis of mutations in key RNAi components. For example, Dicer-mutant mice die early in development with a loss of Oct4-positive multipotent stem cells (Bernstein et al. 2003). Even in the presence of a strong differentiation inducer, DGCR8/pasha knockout embryonic stem (ES) cells fail to inactivate self-renewal programs (Wang et al. 2007). In Drosophila ovaries, dcr-1 mutant germline stem cells are depleted within 3 wk of Dicer loss (Jin and Xie 2007), and homozygous mutation of loqs, an obligate Dicer partner, causes defects in egg chamber development (Förstemann et al. 2005; Jiang et al. 2005).
In many tissues, stem and progenitor cell populations are becoming increasingly well defined. In the mammary gland, this was elegantly demonstrated by the reconstitution of a functional gland from a single stem cell, which was isolated using cell surface markers CD49f, CD29, and CD24 (Shackleton et al. 2006 Stingl et al. 2006). Recently, ALDH (aldehyde dehydrogenase) has been used to isolate and characterize normal human mammary stem cells (Ginestier et al. 2007). Hematopoietic stem cells and neuronal progenitor cells have also been isolated on the basis of ALDH activity (Corti et al. 2006; Hess et al. 2006). Interestingly, ALDH-positive cells derived from AML patients have increased NOD/SCID engraftment potential relative to ALDH-negative cells, suggesting that these cells represent primitive leukemic stem cells (Cheung et al. 2007). ALDH-positive cells from human breast tumors were also shown to be the tumorigenic population, with as few as 500 cells being capable of initiating disease (Ginestier et al. 2007).
Comma-Dβ cells harbor a permanent population of undifferentiated basal cells that are able to reconstitute the mammary tree (Deugnier et al. 2006), thus providing an excellent system in which to study the role of miRNAs in stem cell maintenance, self-renewal, and differentiation. By combining ALDH and Sca-1 (stem cell antigen) expression criteria, we performed an unbiased characterization of miRNAs in mammary progenitor populations using deep sequencing. These studies identified miRNAs that are highly expressed in the progenitor fraction as well as miRNAs that are relatively depleted in this population. By manipulating expression of at least one of these miRNAs, we link miRNAs to progenitor self-renewal.
Results and Discussion
ALDH is a marker of mammary progenitor cells
Sca-1high Comma-Dβ cells have retained the ability to reconstitute a functional mammary gland upon transplantation of as few as 1000 cells into the fat pad of a syngeneic virgin female (Deugnier et al. 2006). Two-dimensional (2D) and three-dimensional (3D) cultures, including mammosphere assays, have provided evidence of the self-renewal and differentiation capacity of these cells as they can generate both myoepithelial and luminal cells in vitro (Deugnier et al. 2006; Chen et al. 2007). Since Sca-1 expression was not enriched in the recently defined murine mammary stem/progenitor cells (Shackleton et al. 2006), we asked whether ALDH expression could be used to isolate progenitor populations from Comma-Dβ. We also tested whether a combination of ALDH and Sca-1 markers provided increased specificity for progenitor cells, at least in cultured populations.
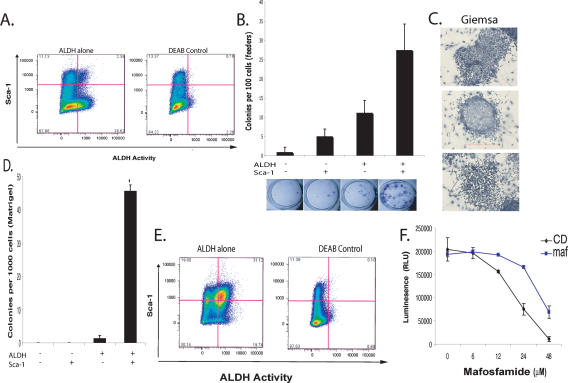
ALDH activity can be measured in living cells by using a fluorogenic substrate, ALDEFLUOR (Corti et al. 2006; Hess et al. 2006; Ginestier et al. 2007). ALDH induces retention of this substrate, resulting in increased florescence. Truly positive cells can be identified by comparison with cells cultured in ALDEFLUOR in the presence of DEAB, an ALDH inhibitor. The Comma-Dβ cell line contains ALDHbrightSca-1high cells that comprise ∼2% of the total population (Fig. 1A). This number is consistent with the number of side population (SP) cells we observed in this cell line (Supplementary Fig. 1).
Figure 1.
Characterization of ALDH as a marker for progenitor cells in Comma-Dβ. (A) A FACS pseudo-color dot plot showing ALDH activity and Sca-1 expression in Comma-Dβ cells. (Left) Cells incubated with ALDEFLOUR substrate and stained with Sca-1. (Right) Cells stained with ALDEFLOUR and costained with Sca-1 and incubated with DEAB to establish background fluorescence. Shown are 100,000 events. (B) Histogram showing the colony-forming capacity of four sorted populations based on ALDH activity and Sca-1 expression seeded at clonal density on irradiated NIH3T3 feeders. Data represent the mean of four independent experiments. (C) Giemsa staining of ALDHbrScahi colonies grown on irradiated feeders for 6 d. Based on morphology, myoepithelial (top), luminal (middle), and mixture (bottom) colonies were observed. (D) Histogram showing the colony-forming capacity of four sorted populations embedded at clonal density in Matrigel (n = 4). (E) FACS profile of Comma-Dβ cells treated with a 6 μM dose of MAF for 4 d. (Left) Cells incubated with ALDEFLOUR and stained with Sca-1. (Right) DEAB control. (F) Cell viability assay after a 24-h treatment with various doses of MAF of Comma-Dβ cells (black) and MAF-resistant cells (blue). Data represent the mean ± SD (error bar) of two independent experiments done in triplicate.
Colony formation on irradiated feeders or Matrigel is commonly used to assess the proliferative capacity of purified epithelial stem and progenitor cells. In several studies, this capacity has been shown to correlate with in vivo morphogenic potential (Deugnier et al. 2006; Shackleton et al. 2006). We therefore examined the colony-forming capacity of four sorted populations (ALDHbrightSca-1high, ALDHbrightSca-1neg, ALDHnegSca-1high, and ALDHnegSca-1neg). Only the two ALDHbright populations yielded significant numbers of colonies, with the ALDHbrightSca-1high subset exhibiting a threefold greater colony-forming frequency and substantially larger colonies (Fig. 1B). ALDHbright cells gave rise to both luminal and myoepithelial colonies, based on morphology (Fig. 1C). A third colony morphology was also observed that fit neither the dispersed teardrop shape characteristic of myoepithelial cells nor the tightly arranged cells with distinct cell borders that indicate luminal cells (Stingl et al. 1998).
ALDHbrightSca-1high cells plated at clonogenic density in Matrigel expanded and formed spheroids (average 46 per well, n = 4; P < 0.001), whereas the ALDHbrightSca-1neg cells grew poorly under these conditions (Fig. 1D). ALDHneg cells were unable to form colonies. These results are consistent with previous studies showing the inability of Sca-1neg cells to grow in Matrigel (Deugnier et al. 2006).
Resistance to a group of anti-cancer drugs called oxazaphosphorines has been linked to ALDH activity (Bunting and Townsend 1996). We therefore reasoned that mafosfamide (MAF) treatment might enrich the population of ALDHbright progenitor cells. We treated cells for 4 d and analyzed the surviving population by FACS (fluorescence-activated cell sorting). This resulted in a 15-fold enrichment in ALDHbrightSca-1high cells. Thus, the progenitor population resident within Comma-Dβ can be selected by this method, and these progenitors are intrinsically resistant to at least some anti-cancer drugs (Fig. 1E,F). Following selection, we also noted a twofold expansion in the ALDHnegSca-1hi compartment. We could not distinguish between resistance of this population to MAF and its arising by differentiation of selected ALDHbrightSca-1high cells.
MAF is a cyclophosphamide derivative that is active in cultured cells. Cyclophosphamide is commonly used as part of a first-line therapy for breast cancer (Smith et al. 2003). Thus, the fact that treatment with MAF can enrich ALDH-positive cells has profound implications, since recent data show that tumor-initiating cells can be isolated from human breast tumors on the basis of ALDH activity (Ginestier et al. 2007).
A miRNA fingerprint of mouse mammary epithelial progenitor cells
To probe potential roles for miRNAs in the maintenance and differentiation of mammary epithelial progenitor cells, we constructed small RNA libraries from Sca-1high, Sca-1neg, ALDHbrightSca-1high, and MAF-treated Comma-Dβ cells. These were deeply sequenced on the Illumina 1G platform and mapped to the mouse genome using a customized bioinformatics pipeline. Reads were annotated by BLAT (Kent 2002) to a unified database comprised of mouse entries from miRbase (Griffiths-Jones et al. 2006), NONCODE (Liu et al. 2005), tRNAs in “The RNA Modification Database” (Limbach et al. 1994), and rRNA entries in the Entrez Nucleotide Database.
Approximately 50% of all sequences that mapped to the genome corresponded to known miRNAs (Supplementary Table 1) for Sca-1high, ALDHbrightSca-1high, and Sca-1neg libraries. In the MAF library, 80% of reads mapped to miRNAs. Breakdown products of noncoding RNAs such as rRNAs, tRNAs, snRNAs, snoRNAs, and others represented <0.5% of total sequences for all four libraries. An estimated 25% of sequences mapped neither to known miRNAs nor other annotated small RNAs in the sorted libraries, whereas only 5% remained unidentified for the MAF library. The top 50 miRNAs sorted based on abundance in the ALDHbrightSca-1high library are shown in Supplementary Table 2.
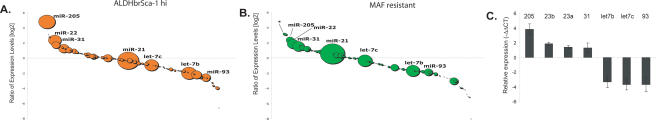
Expression signatures are often presented as heat maps, illustrating the relative signal for an individual species in two samples. Although there are undoubtedly biases in the cloning of specific RNAs, the available sequence data permitted us to examine both differential expression and approximate abundance. We reasoned that focusing on highly expressed miRNAs would maximize the possibility of identifying those that are biologically relevant. A “bubble plot” can be used to depict both the abundance of a particular miRNA (given as the sum of the reads in the two libraries) and its relative expression (plotted as a log2 of the ratio of reads in each library).
The ALDHbrightSca-1high (Fig. 2A) and the MAF libraries (Fig. 2B) were compared with the Sca-1neg library to identify differentially expressed miRNAs. Two abundant miRNAs, miR-205 and miR-22, were consistently enriched in the progenitor population. Both were also abundant in the Sca-1hi library, suggesting that they may be important for the basic physiology or identity of basal cells. MiRNA expression profiling of various tissues showed that miR-205 was preferentially expressed in breast and thymus (Baskerville and Bartel 2005). In human ES cells, Nanog- and Sox2-binding sites are located near the miR-205 and miR-22 promoters (Boyer et al. 2005). However, in comparing our data set to ES cell-specific miRNAs, no consistent overlap in patterns was found.
Figure 2.
MiRNAs are differentially expressed in self-renewing compartments. (A,B) Bubble plots depicting the relative abundance and log2 ratio of miRNAs in ALDHbrScahi (A) and MAF-resistant (B) cells relative to Sca-1negative cells. (C) qRT–PCR for the mature forms of selected differentially expressed miRNAs. Shown are relative expression levels ΔΔCT of each miRNA from sorted Sca-1hi and Sca-1neg Comma-Dβ cells.
Other miRNAs showed substantially lower expression in the progenitor compartment. Let-7b, let-7c, and miR-93 were the most abundant miRNAs that showed preferential expression in Sca-1neg cells. Collectively, let-7b and let-7c represented only 8.8% of the total miRNA sequences in the ALDHbrightSca-1high library compared with 24% of miRNA sequences in Sca-1neg cells. Interestingly, miR-20a is part of a polycistronic cluster containing 17-5p, miR-18a, and miR-19b. These are also underrepresented in the progenitor compartment. miR-21 was the most abundant miRNA found in relatively equal amounts in all four libraries constituting a consistent average of 30% of mapped miRNAs sequences.
Overall, the trends in miRNA representation seen upon comparison of ALDHbrightSca-1high to Sca-1neg cells were reproduced upon examination of the MAF-treated library. However, miR-200a and miR-429, both of which are part of the miR-8 family, were found at substantial levels in the MAF library only.
We performed an independent verification of differential miRNA expression using quantitative stem–loop PCR (qRT–PCR) as described previously (Fig. 2C; Chen et al. 2005). We examined expression of let-7b, let-7c, miR-93, miR-23b, miR-23a, miR-205, and miR-31 in the Sca-1hi fraction versus Sca-1neg libraries. In all seven cases, differential expression was confirmed, although the absolute magnitudes of expression ratios did not precisely agree with those determined from sequencing data.
let-7 depletes the self-renewing ALDH compartment in Comma-Dβ
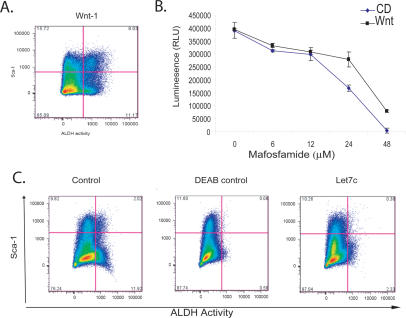
We sought to investigate a role for reduced let-7 expression in mammary progenitor cells. However, it was first necessary to investigate whether the ALDHbrSca-1hi compartment was receptive to signals known to expand stem/progenitor populations and whether miRNA expression patterns responded similarly. Enforced expression of β-catenin in Comma-Dβ cells was shown to expand the Sca-1hi compartment and increase mammosphere-forming capacity (Chen et al. 2007). Similarly, we observed a 3.5-fold increase in the ALDHbrightSca-1high population upon the ectopic expression of Wnt-1 (Fig. 3A). Wnt-1-expressing cells survived higher doses of MAF than empty vector control cells (Fig. 3B), consistent with the ALDHbrightSca-1high compartment having intrinsically higher drug resistance.
Figure 3.
let-7c depletes the self-renewing compartment in Comma-Dβ. (A) Ectopic expression of Wnt expands the ALDHbr Sca-1hi compartment. FACS plot of Comma-Dβ overexpressing Wnt-1 costained with ALDEFLOUR and Sca-1. (B) Cell viability assay after a 48-h treatment with various doses of MAF of Comma-Dβ cells (blue) and Wnt-1-expressing cells (black). Data represent the mean ± SD (error bar) of two independent experiments done in triplicate. (C) FACS profile of empty vector control Comma-Dβ cells costained with ALDEFLOUR and Sca-1 (left), DEAB control for empty vector cells (middle), and Comma-Dβ cells ectopically expressing Let7c (right) and also stained with ALDEFLOUR and Sca-1. A depletion of the ALDH compartment is observed upon introduction of Let7c.
In concert with changes in the progenitor compartment, we observed that Wnt-1-expressing cells expressed sixfold higher levels of miR-205 when compared with empty vector control cells with no observed reduction in let-7b, let-7c, or miR-93 expression, as might be expected since the differentiated compartments were still prominent in this mixed population (data not shown).
To probe the functional relevance of differential miRNA expression patterns, we examined the consequences of enforced expression of let-7c. Comma-Dβ let-7c cells showed a substantial, sixfold reduction in the ALDHbright compartment (n = 4). In concert, we observed the emergence of distinctly Sca-1neg and Sca-1lo populations (Fig. 3C). These results suggest that differences in miRNA expression between differentiated and self-renewing populations within Comma-Dβ cells have substantial impacts on cell identity and physiology.
A let-7 sensor marks the progenitor compartment
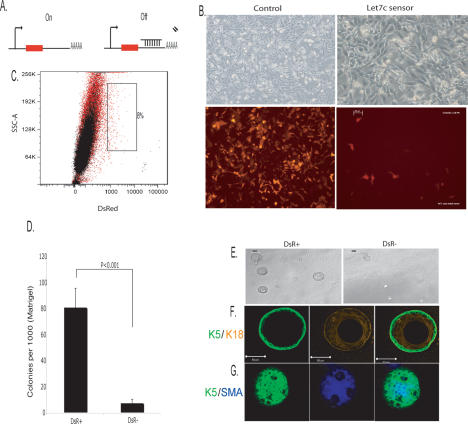
Convenient markers for rare cell populations have proven difficult to identify. miRNA sensors have been used in plants and animals to visualize the expression patterns of individual small RNA species. We wished to investigate the general possibility that miRNA sensors, as directed by our observed expression patterns, could be used to mark rare cell populations and permit their isolation. We constructed a let-7c sensor by introducing its perfect complement into the 3′ untranslated region of DsRed, thus specifying silencing by RNAi in the presence of the miRNA (Fig. 4A).
Figure 4.
Self-renewal and differentiation of let-7c-negative cells in vitro. (A) Cartoon depicting the let-7c sensor construct. (B) Phase-contrast images of Comma-Dβ cells expressing a construct with no let-7c-binding sites (control) and Comma-Dβ cells expressing a sensor construct containing let-7c complementary sites. (C) Overlay FACS dot plot of let-7c sensor cells (red) and uninfected Comma-Dβ cells (black) as an unstained control. DsR+ cells constitute 0.8% of the total population. (D) Histogram showing the colony-forming ability of DsR+ and DsR− cells embedded at clonal density in Matrigel (n = 4). (E) Phase-contrast images of resultant DsR+ and DsR− spheroids grown on Matrigel. DsR+ cells gave rise to substantially larger colonies (>50 μm), whereas DsR− cells never exceeded this size. (F,G) Confocal images of spheroids derived from DsR+ cells. (F) Representative cross-section through the middle of a sphere costained with basal K5 and luminal K18 antibodies. (G) Representative image through the top of a spheroid costained with basal K5 and α-Sma antibodies.
Since let-7c expression is low in ALDHbrightSca-1high cells, we predicted that the sensor would not be silenced, thus marking the progenitor compartment by DsRed expression. Where let-7c expression is high in the more differentiated cell types, we predicted that the sensor would be silenced (Fig. 4B). Indeed, we found that overall, DsRed-positive cells (DsR+) constituted ∼0.8% of the population (Fig. 4C). DsRed-positive cells are enriched for Sca-1high- and ALDH-expressing cells, as expected (data not shown).
We tested DsRed+ cells for their ability to self-renew and differentiate in vitro. DsR+ cells formed spheroids with 10-fold greater efficacy than DsR− cells (Fig. 4D), with only DsR+ cells forming spheroids >50 μm in size (Fig. 4E). Confocal images of spheroids costained with Keratin 5 (K5) and Keratin 18 (K18) revealed that a single DsR+ cell was able to give rise to a K5-positive, basal, outer layer and an inner layer of luminal, K18-positive cells (Fig. 4F), although, consistent with previous observations, not all spheres had such an apparent luminal structure (Deugnier et al. 2006). To probe the ability of DsR+ cells to differentiate into myoepithelial cells, we costained spheroids with K5 and smooth muscle actin (SMA) and indeed observed spheroids with an outer layer of K5- and SMA-positive cells (Fig. 4G).
These studies demonstrate that a lack of let-7c expression can be used to prospectively isolate mammary progenitor cells. Perhaps in combination with additional sensors, this allows the experimentally determined miRNA expression signature to be converted into a functional tool that can augment existing markers of murine progenitors and likely also tumor-initiating cells.
Overall, our results support the notion that miRNA expression patterns form both a characteristic signature of a given cell type and help to reinforce cell fate specification. Even within a single cell line, distinct compartments containing progenitor cells and more differentiated cells have unique miRNA patterns, suggesting that such signatures can be used not only to define and track rare cell populations in vitro and in vivo, but that manipulation of these signatures might be used to expand or deplete stem cell and tumor-initiating cell populations for therapeutic benefit.
Materials and methods
Cell culture
Comma-Dβ cells were grown in DMEM:F12 (HyClone) supplemented with 2% FCS, 5 ng/mL murine EGF (Sigma), 10 μg/mL human insulin (Sigma), and 50 μg/mL gentamicin (GIBCO). Cells were only used within passages 17–35. Phoenix cells were maintained in DMEM supplemented with 10% FBS (Hyclone) and penicillin-streptomycin (GIBCO).
Constructs and infections
For construction of Let7c stable expression vectors, the following primers were used: forward, 5′-GGCCAGATCTGTGTGGTCAAGGAGAT GTTAG-3′, and reverse, 5′-GATCCTCGAGTAACAGCCCGTGAGAAA TAG-3′, containing Bgl-II/Xho1 restriction sites. A 500-base-pair fragment was PCR-amplified from mouse genomic DNA and cloned into an MSCV vector carrying a hygromycin cassette (Clontech). Phoenix cell transfections were performed using LT-1 transfection reagent (Mirus) according to the manufacturer’s instructions. To construct Wnt-1MSCV, the 1.9-kb fragment of wingless cDNA (nucleotides 284–2181) in pMV7 (kind gift of Anthony Brown) was subcloned into an MSCV-hygro vector. For construction of the Let7c sensor, miRNA-complementary oligonucleotides were annealed and cloned into a Marx vector that directs dsRED expression (a kind gift of Scott Hammond).
ALDEFLUOR and SP cell staining and flow cytometry
Cells were stained at 106 cells per milliliter in assay buffer containing 1 μmol of BAAA for 1 h at 37°C. The ALDEFLUOR kit was purchased from StemCell Technologies. An aliquot of stained cells was treated with 50 mmol/L DEAB as a negative control. After ALDEFLUOR staining, cells were costained with anti-Sca-1-PE (BD Pharmigen) for 20 min on ice. For small RNA cloning, cells were FACS-sorted directly into Trizol LS reagent (Invitrogen). ALDEFLUOR was excited at 488 nm, and fluorescence emission was detected using a standard fluorescein isothiocyanate (FITC) 530/30-nm band-pass filter. For SP analysis, cells were stained with Hoescht33342 Dye as described previously (Goodell et al. 1996).
In vitro assays
Colony formation assays on feeders were essentially performed as described by Shackleton et al. (2006). 3D cultures were performed as described in Debnath et al. (2003).
Antibodies and immunofluorescence
The following primary antibodies were used: anti-Sca-1 PE (BD Pharmigen), mouse anti-cytokertain peptide 18 (Sigma), mouse anti-α-Sma (Sigma), and rabbit polyclonal anti-cytokeratin 5 (Covance). Flurochrome-conjugated secondary antibodies included anti-rabbit IgG-Alexa488 and anti-mouse IgG-Alexa647 (Molecular Probes).
Small RNA cloning
One to four micrograms of total RNA from sorted cells were used for small RNA cloning performed as described in Pfeffer et al. (2005), and a detailed protocol is available on request. Illumina 1G sequencing and analysis was performed as described (Stark et al. 2007).
miRNA expression analyses
Mature miRNAs were quantified using the TaqMan MicroRNA Assays described previously by Chen et al. (2005) (Applied Biosystems). Data were normalized to Actin using the SuperScript III SYBR Green One-Step qRT–PCR system (Invitrogen). The experiments were repeated twice, and all reactions were run in triplicate.
Cell viability assay
To assess the cytotoxic effects of MAF, cells were seeded at 5000 cells per well in a 96-well format. Twenty-four hours or 48 h later, cells were treated with various doses of MAF L-lysine salt (D-17930) (NIOMECH in der IIT GmbH) freshly dissolved in water. Cell viability was measured using the CellTiter-Glo Luminescent Cell Viability Assay (Promega).
Acknowledgments
We are grateful to Pamela Moody, Frances Kittrell, and Daniel Medina for help with flow cytometry and culturing Comma-Dβ cells. We thank Avi Rosenberg (CSHL), Max Wicha, Christopher Ginestier (University of Michigan), Susan Ludeman, and Mike Colvin for advice and for sharing protocols and reagents. We also thank Monica Dus for help in confocal imaging. This work was supported in part by grants from the NIH (I.I., S.K.M., and G.J.H.) and a kind gift from Kathryn W. Davis (G.J.H.). Y.E. is a Goldberg-Lindsay Fellow of the Watson School of Biological Sciences.
Footnotes
References
- Baskerville S., Bartel D.P., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J., Elledge S.J., Anderson K.V., Hannon G.J., Anderson K.V., Hannon G.J., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., Kumar R.M., Murray H.L., Jenner R.G., Murray H.L., Jenner R.G., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunting K.D., Townsend A.J., Townsend A.J. De novo expression of transfected human class 1 aldehyde dehydrogenase (ALDH) causes resistance to oxazaphosphorine anti-cancer alkylating agents in hamster V79 cell lines. Elevated class 1 ALDH activity is closely correlated with reduction in DNA interstrand cross-linking and lethality. J. Biol. Chem. 1996;27:11884–11890. doi: 10.1074/jbc.271.20.11884. [DOI] [PubMed] [Google Scholar]
- Chen C., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Ridzon D.A., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Broomer A.J., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Zhou Z., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Lee D.H., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Nguyen J.T., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Barbisin M., Xu N.L., Mahuvakar V.R., Andersen M.R., Xu N.L., Mahuvakar V.R., Andersen M.R., Mahuvakar V.R., Andersen M.R., Andersen M.R., et al. Real-time quantification of microRNAs by stem–loop RT–PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.S., Woodward W.A., Behbod F., Peddibhotla S., Alfaro M.P., Buchholz T.A., Rosen J.M., Woodward W.A., Behbod F., Peddibhotla S., Alfaro M.P., Buchholz T.A., Rosen J.M., Behbod F., Peddibhotla S., Alfaro M.P., Buchholz T.A., Rosen J.M., Peddibhotla S., Alfaro M.P., Buchholz T.A., Rosen J.M., Alfaro M.P., Buchholz T.A., Rosen J.M., Buchholz T.A., Rosen J.M., Rosen J.M. Wnt/β-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J. Cell Sci. 2007;120:468–477. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]
- Cheung A.M., Wan T.S., Leung J.C., Chan L.Y., Huang H., Kwong Y.L., Liang R., Leung A.Y., Wan T.S., Leung J.C., Chan L.Y., Huang H., Kwong Y.L., Liang R., Leung A.Y., Leung J.C., Chan L.Y., Huang H., Kwong Y.L., Liang R., Leung A.Y., Chan L.Y., Huang H., Kwong Y.L., Liang R., Leung A.Y., Huang H., Kwong Y.L., Liang R., Leung A.Y., Kwong Y.L., Liang R., Leung A.Y., Liang R., Leung A.Y., Leung A.Y. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia. 2007;21:1423–1430. doi: 10.1038/sj.leu.2404721. [DOI] [PubMed] [Google Scholar]
- Corti S., Locatelli F., Papadimitriou D., Donadoni C., Salani S., Del Bo R., Strazzer S., Bresolin N., Comi G.P., Locatelli F., Papadimitriou D., Donadoni C., Salani S., Del Bo R., Strazzer S., Bresolin N., Comi G.P., Papadimitriou D., Donadoni C., Salani S., Del Bo R., Strazzer S., Bresolin N., Comi G.P., Donadoni C., Salani S., Del Bo R., Strazzer S., Bresolin N., Comi G.P., Salani S., Del Bo R., Strazzer S., Bresolin N., Comi G.P., Del Bo R., Strazzer S., Bresolin N., Comi G.P., Strazzer S., Bresolin N., Comi G.P., Bresolin N., Comi G.P., Comi G.P. Identification of a primitive brain-derived neural stem cell population based on aldehyde dehydrogenase activity. Stem Cells. 2006;24:975–985. doi: 10.1634/stemcells.2005-0217. [DOI] [PubMed] [Google Scholar]
- Debnath J., Muthuswamy S.K., Brugge J.S., Muthuswamy S.K., Brugge J.S., Brugge J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Deugnier M.A., Faraldo M.M., Teuliere J., Thiery J.P., Medina D., Glukhova M.A., Faraldo M.M., Teuliere J., Thiery J.P., Medina D., Glukhova M.A., Teuliere J., Thiery J.P., Medina D., Glukhova M.A., Thiery J.P., Medina D., Glukhova M.A., Medina D., Glukhova M.A., Glukhova M.A. Isolation of mouse mammary epithelial progenitor cells with basal characteristics from the Comma-Dβ cell line. Dev. Biol. 2006;293:414–425. doi: 10.1016/j.ydbio.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Förstemann K., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Tomari Y., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Du T., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Vagin V.V., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Denli A.M., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Bratu D.P., Klattenhoff C., Theurkauf W.E., Zamore P.D., Klattenhoff C., Theurkauf W.E., Zamore P.D., Theurkauf W.E., Zamore P.D., Zamore P.D. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestier C., Hur M.H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C., Liu S., Hur M.H., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C., Liu S., Charafe-Jauffret E., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C., Liu S., Monville F., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C., Liu S., Dutcher J., Brown M., Jacquemier J., Viens P., Kleer C., Liu S., Brown M., Jacquemier J., Viens P., Kleer C., Liu S., Jacquemier J., Viens P., Kleer C., Liu S., Viens P., Kleer C., Liu S., Kleer C., Liu S., Liu S., et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007 doi: 10.1016/j.stem.2007.08.014. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Van Dongen S., Inoue K., Enright A.J., Schier A.F., Inoue K., Enright A.J., Schier A.F., Enright A.J., Schier A.F., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Goodell M.A., Brose K., Paradis G., Conner A.S., Mulligan R.C., Brose K., Paradis G., Conner A.S., Mulligan R.C., Paradis G., Conner A.S., Mulligan R.C., Conner A.S., Mulligan R.C., Mulligan R.C. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J. Exp. Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J., Grocock R.J., van Dongen S., Bateman A., Enright A.J., van Dongen S., Bateman A., Enright A.J., Bateman A., Enright A.J., Enright A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34 (Database issue):D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D.A., Wirthlin L., Craft T.P., Herrbrich P.E., Hohm S.A., Lahey R., Eades W.C., Creer M.H., Nolta J.A., Wirthlin L., Craft T.P., Herrbrich P.E., Hohm S.A., Lahey R., Eades W.C., Creer M.H., Nolta J.A., Craft T.P., Herrbrich P.E., Hohm S.A., Lahey R., Eades W.C., Creer M.H., Nolta J.A., Herrbrich P.E., Hohm S.A., Lahey R., Eades W.C., Creer M.H., Nolta J.A., Hohm S.A., Lahey R., Eades W.C., Creer M.H., Nolta J.A., Lahey R., Eades W.C., Creer M.H., Nolta J.A., Eades W.C., Creer M.H., Nolta J.A., Creer M.H., Nolta J.A., Nolta J.A. Selection based on CD133 and high aldehyde dehydrogenase activity isolates long-term reconstituting human hematopoietic stem cells. Blood. 2006;107:2162–2169. doi: 10.1182/blood-2005-06-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houbaviy H.B., Murray M.F., Sharp P.A., Murray M.F., Sharp P.A., Sharp P.A. Embryonic stem cell-specific microRNAs. Dev. Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Jiang F., Ye X., Liu X., Fincher L., McKearin D., Liu Q., Ye X., Liu X., Fincher L., McKearin D., Liu Q., Liu X., Fincher L., McKearin D., Liu Q., Fincher L., McKearin D., Liu Q., McKearin D., Liu Q., Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes & Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Xie T., Xie T. Dcr-1 maintains Drosophila ovarian stem cells. Curr. Biol. 2007;17:539–544. doi: 10.1016/j.cub.2007.01.050. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-Like Alignment Tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Meyer J., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Limbach P.A., Crain P.F., McCloskey J.A., Crain P.F., McCloskey J.A., McCloskey J.A. Summary: The modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Bai B., Skogerbø G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Bai B., Skogerbø G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Skogerbø G., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Cai L., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Deng W., Zhang Y., Bu D., Zhao Y., Chen R., Zhang Y., Bu D., Zhao Y., Chen R., Bu D., Zhao Y., Chen R., Zhao Y., Chen R., Chen R. NONCODE: An integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33 (Database issue):D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neilson J.R., Zheng G.X., Burge C.B., Sharp P.A., Zheng G.X., Burge C.B., Sharp P.A., Burge C.B., Sharp P.A., Sharp P.A. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes & Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., Sewer A., Lagos-Quintana M., Sheridan R., Sander C., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., Lagos-Quintana M., Sheridan R., Sander C., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., Sheridan R., Sander C., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., Sander C., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., Grasser F.A., van Dyk L.F., Ho C.K., Shuman S., Chien M., van Dyk L.F., Ho C.K., Shuman S., Chien M., Ho C.K., Shuman S., Chien M., Shuman S., Chien M., Chien M. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Rougvie A.E., Horvitz H.R., Ruvkun G., Horvitz H.R., Ruvkun G., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Shackleton M., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E., Vaillant F., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E., Simpson K.J., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E., Stingl J., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E., Smyth G.K., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E., Asselin-Labat M.L., Wu L., Lindeman G.J., Visvader J.E., Wu L., Lindeman G.J., Visvader J.E., Lindeman G.J., Visvader J.E., Visvader J.E. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- Smith R.E., Bryant J., DeCillis A., and Anderson S., Bryant J., DeCillis A., and Anderson S., DeCillis A., and Anderson S., and Anderson S. Acute myeloid leukemia and myelodysplastic syndrome after doxorubicin-cyclophosphamide adjuvant therapy for operable breast cancer: The National Surgical Adjuvant Breast and Bowel Project Experience. J. Clin. Oncol. 2003;21:1195–1204. doi: 10.1200/JCO.2003.03.114. [DOI] [PubMed] [Google Scholar]
- Stark A., Kheradpour P., Parts L., Brennecke J., Hodges E., Hannon G.J., Kellis M., Kheradpour P., Parts L., Brennecke J., Hodges E., Hannon G.J., Kellis M., Parts L., Brennecke J., Hodges E., Hannon G.J., Kellis M., Brennecke J., Hodges E., Hannon G.J., Kellis M., Hodges E., Hannon G.J., Kellis M., Hannon G.J., Kellis M., Kellis M. Systematic discovery and characterization of fly microRNA using 12 Drosophila genomes. Genome Res. 2007 doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl J., Eaves C.J., Kuusk U., Emerman J.T., Eaves C.J., Kuusk U., Emerman J.T., Kuusk U., Emerman J.T., Emerman J.T. Phenotypic and functional characterization in vitro of a multipotent epithelial cell present in the normal adult human breast. Differentiation. 1998;63:201–213. doi: 10.1111/j.1432-0436.1998.00201.x. [DOI] [PubMed] [Google Scholar]
- Stingl J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H.I., Eaves C.J., Eirew P., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H.I., Eaves C.J., Ricketson I., Shackleton M., Vaillant F., Choi D., Li H.I., Eaves C.J., Shackleton M., Vaillant F., Choi D., Li H.I., Eaves C.J., Vaillant F., Choi D., Li H.I., Eaves C.J., Choi D., Li H.I., Eaves C.J., Li H.I., Eaves C.J., Eaves C.J. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R., Medvid R., Melton C., Jaenisch R., Blelloch R., Melton C., Jaenisch R., Blelloch R., Jaenisch R., Blelloch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]