Rapid and transient recruitment of DNMT1 to DNA double-strand breaks is mediated by its interaction with multiple components of the DNA damage response machinery (original) (raw)
Abstract
DNA methylation is an epigenetic mark critical for regulating transcription, chromatin structure and genome stability. Although many studies have shed light on how methylation impacts transcription and interfaces with the histone code, far less is known about how it regulates genome stability. We and others have shown that DNA methyltransferase 1 (DNMT1), the maintenance methyltransferase, contributes to the cellular response to DNA damage, yet DNMT1's exact role in this process remains unclear. DNA damage, particularly in the form of double-strand breaks (DSBs), poses a major threat to genome integrity. Cells therefore possess a potent system to respond to and repair DSBs, or to initiate cell death. In the current study, we used a near-infrared laser microirradiation system to directly study the link between DNMT1 and DSBs. Our results demonstrate that DNMT1 is rapidly but transiently recruited to DSBs. DNMT1 recruitment is dependent on its ability to interact with both PCNA and the ATR effector kinase CHK1, but is independent of its catalytic activity. In addition, we show for the first time that DNMT1 interacts with the 9-1-1 PCNA-like sliding clamp and that this interaction also contributes to DNMT1 localization to DNA DSBs. Finally, we demonstrate that DNMT1 modulates the rate of DSB repair and is essential for suppressing abnormal activation of the DNA damage response in the absence of exogenous damage. Taken together, our studies provide compelling additional evidence for DNMT1 acting as a regulator of genome integrity and as an early responder to DNA DSBs.
INTRODUCTION
Methylation of cytosines within CpG dinucleotides in mammalian DNA is an epigenetic modification essential for proper control of transcription, embryonic development, genomic imprinting, chromatin structure and genomic stability. DNA methylation patterns are established and maintained by a family of three enzymatically active DNA methyltransferases, DNMT1, DNMT3A and DNMT3B and an inactive DNMT3-like protein known as DNMT3L. DNMT1 is traditionally viewed as the maintenance DNA methyltransferase, copying methylation patterns after replication, and the DNMT3s as de novo enzymes that establish new methylation patterns during development (1). This division, however, has become increasingly blurred as both overlapping and separable functions within the DNMT family have been identified (2,3). It remains unclear exactly how DNA methylation patterns are established but an increasing number of interactions between DNA methyltransferases and chromatin-associated proteins suggests that chromatin structure and the histone code are important determinants (4). What has become clear is that DNA methylation patterns are globally disrupted in cancer cells and this aberration contributes directly to tumorigenesis and to the genomic instability that is hallmark of transformed cells (5).
The cellular response to DNA damage is a highly orchestrated process leading to the activation of cell-cycle checkpoints, followed by a decision to resume cycling if damage can be repaired, or commit to apoptosis if it is irreparable. The high-molecular-weight PI(3)-like kinases ATM and ATR sense damaged or abnormal DNA structures and initiate a cascade of phosphorylation events that transmit and amplify the damage response and recruit repair machinery to damaged regions (6). ATR, complexed with ATRIP, is activated via RPA-coated single-stranded DNA (ssDNA) formed by replication fork stalling or collapse (7). A PCNA-like sliding clamp known as the 9-1-1 complex (RAD9, RAD1, HUS1) is also loaded onto DNA ends adjacent to the ssDNA independently of ATR. The 9-1-1 complex, and its clamp loader RAD17, appears to enable ATR to recognize its substrates (8). ATM is recruited primarily to double-strand breaks (DSBs) induced by DNA damage via the MRN complex (MRE11, RAD50, NBS1) (9). Two key downstream effectors of ATR and ATM are the checkpoint kinases CHK1 and CHK2, respectively (10,11). CHK1 is of particular interest because it, like its upstream activator ATR, is essential for cell viability under conditions of unperturbed growth and because it mediates both the intra-S and G2/M DNA damage checkpoints (12). Under normal growth conditions, CHK1 regulates replication fork stability and sustains rapid fork progression (13). Following DNA damage, CHK1 phosphorylates CDC25A, triggering its degradation, leading to the inactivation of CDK1/cyclin B1 and G2/M arrest (10,14).
DNA damage recognition and repair are intimately tied to chromatin structure and modulation of epigenetic marks since repair factors must content with DNA packaged into nucleosomes and higher order structures. Damage needs to be recognized, the repair machinery must gain access to DNA (both within the context of chromatin) and finally proper chromatin structure and epigenetic marks must be restored once repair is complete. This linkage is perhaps most dramatically exemplified by the histone H2A variant H2AX. H2AX, which substitutes for H2A within the nucleosome core, is very rapidly phosphorylated on serine 139 by ATM, ATR and DNA-PKcs upon DNA damage (15). Phosphorylated H2AX, termed γH2AX, accumulates in mega-base size regions flanking DSBs that are readily visible by immunofluorescence microscopy (also termed ionizing radiation-induced foci or IRIF). These foci of γH2AX are critical for the recruitment and/or maintenance of certain components of the DNA repair machinery at break sites (16). For example, MDC1 (mediator of the DNA damage checkpoint 1) contains a BRCT domain that specifically recognizes and binds phosphorylated H2AX and subsequently amplifies recruitment of other important repair-related factors such as ATM, CHK2 and 53BP1 to damage foci (17).
Interestingly, a large fraction of CHK1 is associated with chromatin in unperturbed cells, where its kinase activity is constitutively active. Therefore, not only is redistribution of CHK1 from a chromatin-bound to a soluble form within the nucleus upon DNA damage critical for its checkpoint function (18), this finding also suggested that CHK1 had substrates within its chromatin environment. Interestingly, chromatin-bound CHK1 was recently shown to phosphorylate histone H3 at the threonine 11 (H3T11) position. This mark appears to help recruit the GCN5 histone acetyltransferase complex leading to H3K9 acetylation and transcription of critical cell-cycle regulatory genes like cyclin B1. Upon DNA damage, CHK1 rapidly dissociates from chromatin, H3T11 phosphorylation and H3K9 acetylation levels are reduced and expression of GCN5 target genes is downregulated. Thus, CHK1 may be viewed as a transcriptional regulator in both the unperturbed and DNA damaged states in addition to its roles in checkpoint control (14,19).
Although one established connection between DNA methylation and genomic stability lies in the ability of DNA methylation to suppress homologous recombination between repetitive sequences and silence transcription of transposable elements (20), other less well-characterized links specific to DNMT1 have also emerged. For example, _Dnmt1-_deficient ES cells display an ∼10-fold increase in mutation frequency, primarily in the form of deletions and mutations, demonstrating that DNMT1 deficiency results in a mutator phenotype (21). A genetic screen for genes involved in mismatch repair (MMR) uncovered DNMT1. _Dnmt1_-deficient murine ES cells exhibited a 4-fold increase in the rate of microsatellite instability (compared with an 8-fold increase in Msh6 mutant cells) (22). More direct evidence for a role of DNMT1 in regulating genome integrity came from a study where DNA damage was induced with an ultraviolet laser microirradiation system, revealing that DNMT1 was recruited to irradiated regions (23). In addition, our prior studies placed DNMT1 at γH2AX-positive foci in 5-aza-2′-deoxycytidine (5-azadC)-treated cells by immunofluorescence microscopy (24). These studies suggest that DNMT1 plays a role in sensing and/or repairing DNA damage or that it is involved in one or more checkpoint responses.
We have shown previously that HCT116 human colorectal cancer cells expressing a hypomorphic mutant form of DNMT1 (1KO) lacking its N-terminal PCNA-binding domain (PBD) (25) exhibited an altered DNA damage response when treated with the DNA methylation inhibitor 5-azadC, including constitutive activation (phosphorylation) of CHK1 (S345) and ATM (S1981) and reduced phosphorylation of p53 (S15) and CHK1 (S317). HCT116 1KO cells displayed increased sensitivity to DNA-damaging agents and elevated levels of activated DNA damage markers under unperturbed growth conditions. We also showed that DNMT1 interacts with CHK1 (24). Although our studies with 5-azadC were consistent with those of others suggesting a possible function for DNMT1 in checkpoint control and DNA repair (23,26,27), it left unanswered questions about whether DNMT1 plays a more general role in DNA damage sensing and/or repair especially since the mechanism of action of 5-azadC is becoming increasingly complex (28). In the current study, we sought to directly address the role of DNMT1 in DNA DSB repair, using a near-infrared (NIR) laser microirradiation system. NIR laser induction of DNA damage resulted in rapid but transient recruitment of DNMT1 to DSB regions in a PBD-dependent, catalytic activity-independent manner. DNMT1 accumulated at DSBs by 1 min post-laser treatment and was largely lost from DSBs by 3 h. DNMT1 recruitment was dependent primarily on ATR but not on DNA-PKcs signaling. Short hairpin RNA (shRNA) knockdown experiments revealed that CHK1 and PCNA, both DNMT1-interacting proteins, were essential for efficient recruitment of DNMT1 to DSBs. We demonstrate for the first time that DNMT1 interacts with the RAD9 subunit of the 9-1-1 complex and that this interaction is also important for the localization of DNMT1 to DSBs. Finally, we showed that cells deficient in DNMT1 displayed an altered time course of recruitment and/or decay of damage-signaling proteins such as ATM, ATR and γH2AX, reduced levels of CHK1-mediated H3T11 phosphorylation and a slower rate of DSB repair. Taken together, our results demonstrate that DNMT1 is recruited early and transiently to DSBs by multiple DNA damage-related interactions and that it plays a role in modulating the cellular response to DNA damage.
RESULTS
DNMT1 is rapidly but transiently recruited to regions of DNA DSBs
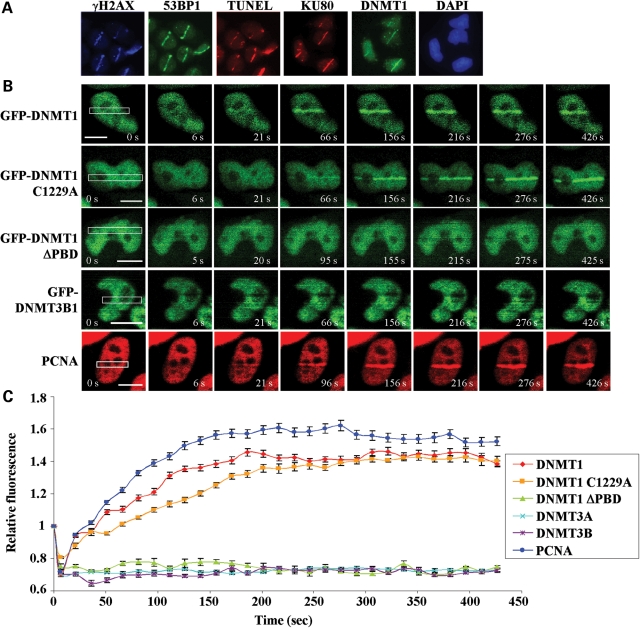
Laser microirradiation has become an invaluable technique for studying the DNA damage response. It allows for analysis of the temporal sequence of recruitment and decay of DNA damage-related factors to sites of damage in live or fixed cells and identification of novel factors. Several wavelengths may be utilized to induce DNA damage and their advantages and disadvantages have been compared (29). For example, a UVA laser was used to study DNMT1 localization (23), however this method requires pre-sensitization of cells by growth in BrdU and, depending on the conditions used, may not result in efficient recruitment of some DSB repair-associated factors (29). Many forms of DNA damage (e.g. ssDNA breaks) will, if unrepaired prior to the passage of a replication fork, lead to DSBs, the most deleterious form of DNA damage (6). In the current study, we employed an NIR (800 nm) laser, which does not require pre-sensitization, to create DSBs in a line pattern across the nucleus. NIR lasers have been shown to induce DNA damage that efficiently recruits DSB repair-associated factors with minimal activation of other pathways, such as the nucleotide excision repair response (29,30). We validated this system by showing that several DNA damage response proteins known to be recruited to DSBs (IRIF-associated factors), including KU80, γH2AX and 53BP1, were localized to laser-microirradiated (laser-IR) regions (line pattern across the nucleus, 5 min post-laser, Fig. 1A). We also validated the presence of free DNA ends at irradiated regions, using TUNEL staining. Interestingly, using these same conditions, we readily detected green fluorescent protein (GFP)-tagged DNMT1 at DSB regions at 5 min post-irradiation (Fig. 1A).
Figure 1.
DNMT1 is rapidly recruited to sites of DNA DSBs independent of its catalytic activity. (A) DNA damage response proteins known to be recruited to IRIF (KU80, γH2AX and 53BP1) and the presence of broken DNA ends (shown by TUNEL staining) demonstrate the validity of the NIR laser microirradiation system used in subsequent experiments. DNMT1 is also recruited to NIR laser-irradiated regions. Images are taken 5 min post-NIR laser-induced damage (in a line pattern) using HCT116 cells. (B) Time course of factor recruitment to laser-irradiated regions in live cells. HCT116 cells were transfected with the indicated GFP-tagged DNMT1 or DNMT3 constructs and subjected to NIR laser-induced DNA damage 48 h later. DNMT1 is visible at DSBs within 30 s of laser microirradiation. ΔPBD, deletion of the PCNA-binding domain; C1229A, catalytic center mutation of DNMT1. PCNA, HCT116 cells transfected with RFP-PCNA. Representative images are shown with the irradiated region denoted by the white box and time (in seconds) after irradiation indicated in white text. Bar: 10 µm. (C) Graphical summary of factor recruitment over ∼7.5 min based on the live-cell imaging in (B). At least 10 cells were quantified for each time point (values are the average, and error bars represent the standard deviation). Accumulation curves of fluorescent proteins at sites of damage were obtained by measuring average fluorescence within the laser target area using Image J. Data are presented as the fluorescence level relative to the pre-NIR laser damage level for each construct/transfection.
To more precisely examine DNMT1 recruitment to regions of DNA DSBs, we laser-irradiated cells transfected with GFP-DNMT1 and monitored its localization in live cells. Wild-type DNMT1 was rapidly recruited to the DSB region, showing clear accumulation by ∼1 min, faint accumulation as early as 21 s and peaking by 3–4 min post-laser-IR (Fig. 1B top row, C). GFP-DNMT1 containing a mutation of the catalytic site cysteine residue (DNMT1 C1229A) was recruited with similar but slightly slower kinetics at the earliest time points. In contrast, DNMT1 lacking the PBD (ΔPBD) in exon 5 did not accumulate at DSBs. We also monitored PCNA recruitment because it is a known DNMT1-associated factor and it is involved in the repair of many types of DNA damage (31,32). RFP-tagged PCNA accumulated slightly faster than DNMT1 at DSBs. Neither DNMT3A nor DNMT3B accumulated at laser-induced DSB regions (Fig. 1B, C and data not shown).
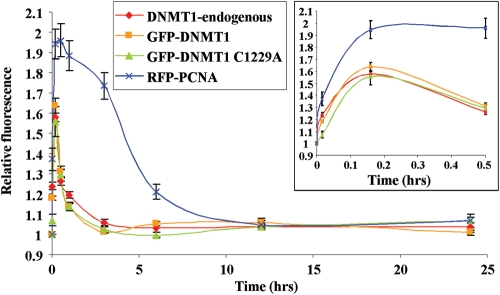
To determine the residence time of DNMT1 at DNA damage, we performed a longer time course where cells were fixed at specific points after laser-IR out to 24 h and localization patterns determined by immunofluorescence staining. This experiment showed that DNMT1 was completely lost from DSB regions by ∼3 h after laser-IR (Fig. 2). The residence time of catalytically inactive DNMT1 was identical to that of WT-DNMT1. Since fixed cells were used, we also compared the kinetics of recruitment of GFP-DNMT1 with that of endogenous DNMT1 (compare red and orange lines in Fig. 2). No difference in recruitment or decay was detectable between exogenous GFP-DNMT1 and endogenous DNMT1. Finally, we examined PCNA localization over a 24 h period. Interestingly, PCNA remained at regions of DNA damage much longer than DNMT1, being almost completely lost from DNA damage sites by ∼6 h (Fig. 2). Taken together, these data demonstrate that DNMT1, but not DNMT3A/3B, is rapidly recruited to DNA DSBs in a catalytic activity-independent but PBD-dependent manner. In addition, although DNMT1 and PCNA have similar recruitment kinetics, their rate of loss from DSB regions is quite distinct, demonstrating that DNMT1-PCNA recruitment to regions of DNA damage is not strictly linked.
Figure 2.
DNMT1 and PCNA are recruited to DSBs with similar kinetics but are lost at different rates. Graphical summary of long-term recruitment/decay kinetics of the indicated DNMT1 constructs compared with PCNA at regions of DSBs. Following transfection of HCT116 cells and NIR laser irradiation 48 h later, cells were fixed at the indicated time points and the degree of fluorescence at regions of DNA damage quantified and set relative to the pre-damage level. Damaged regions were identified by co-staining all slides with γH2AX antibody (not shown). Also shown is a summary of immunostaining for endogenous DNMT1 to demonstrate that GFP-DNMT1 behaves similarly to native DNMT1. DNMT1 is essentially undetectable at DSBs by 3 h post-laser IR, whereas PCNA persists until ∼6–8 h. A blow-up of the early time points is shown in the inset and is similar to the rapid live-cell kinetics in Figure 1C.
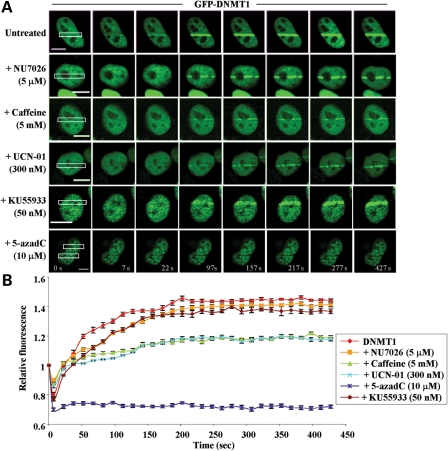
Pharmacologic inhibition of the DNA damage response reveals a role for ATR signaling in DNMT1 recruitment to DSBs
In order to better understand the mechanisms by which DNMT1 is recruited to DNA DSBs, we employed several pharmacologic inhibitors of the DNA damage response. Caffeine is a potent inhibitor of ATM and ATR but has relatively little effect on DNA-PKcs (33). NU7026 preferentially inhibits DNA-PKcs and is essentially inactive against ATM and ATR (34). KU-55933 was developed specifically to inhibit ATM (35). UCN-01 is a potent inhibitor of CHK1 (36). 5-azadC, the DNA methyltransferase inhibitor that covalently traps DNMT1 on DNA and immobilizes it (37), was used as a control. HCT116 cells were transfected with WT GFP-DNMT1 and then treated with each inhibitor as described in Materials and Methods. Following drug treatment, cells were subjected to laser-IR and DNMT1 localization monitored in live cells. The effect of each agent on DNMT1 is shown in representative live-cell images and summarized graphically in Figure 3. NU7026, the DNA-PK inhibitor, had little effect on DNMT1 recruitment to regions of DSBs, however caffeine dramatically reduced DNMT1 recruitment. Interestingly, UCN-01, the CHK1 inhibitor, reduced DNMT1 recruitment to DSBs as effectively as caffeine. In contrast, inhibition of ATM by KU-55933 had little effect on DNMT1 recruitment. As expected, treatment of cells with 5-azadC completely immobilized DNMT1 and no recruitment to DNA damage sites was observed (Fig. 3A and B). These results therefore suggest that ATR signaling in particular is important for the recruitment of DNMT1 to regions of DNA DSBs. The equivalency of recruitment inhibition by caffeine and UCN-01 further implicates CHK1 as an important player in DNMT1's ability to localize to regions of DNA damage.
Figure 3.
DNMT1 recruitment to regions of DNA damage is reduced by pharmacologic inhibition of the ATR/CHK1 pathway but not by DNA-PKcs or ATM inhibition. (A) HCT116 cells were transfected with wild-type GFP-DNMT1 for 24 h, followed by treatment with the indicated inhibitors for an additional 24 h. Cells were then subjected to NIR laser irradiation and the effect on DNMT1 recruitment to DSBs monitored by live-cell imaging. Representative images are shown. NU7026 preferentially inhibits DNA-PKcs, caffeine inhibits ATM and ATR, KU-55933 inhibits ATM and UCN-01 inhibits CHK1. (B) Graphical summary of DNMT1 recruitment to sites of DNA DSBs and the impact of DNA damage-signaling inhibitors. 5-aza-2′-deoxycytidine (5-azadC) was used as a control and is known to completely block DNMT1 mobility. Bar:10 µm.
DNMT1 interacts with multiple components of the DNA damage machinery
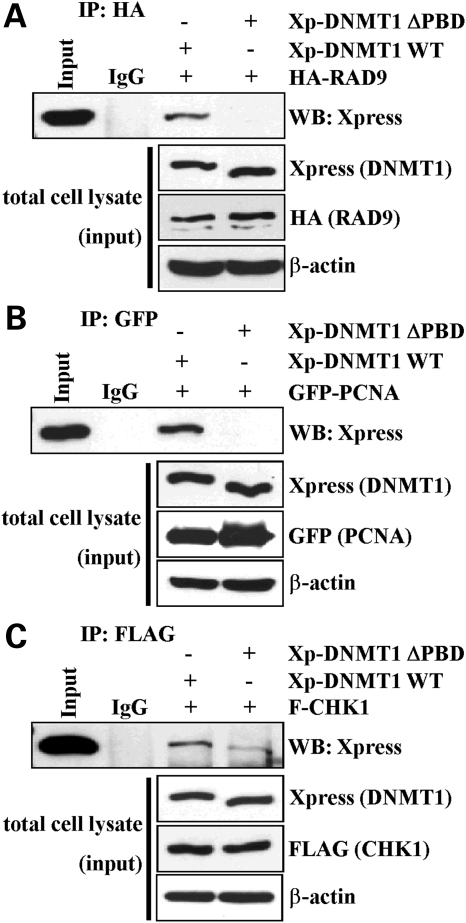
We have shown previously that DNMT1 interacts with CHK1, but not with CHK2, in both unperturbed and DNA-damaged cells (24). In addition, several studies have demonstrated that DNMT1 interacts with PCNA (31,38). These studies, however, left several unanswered questions. Given that DNMT1 interacts with CHK1, and the 9-1-1 DNA damage clamp plays an important role in CHK1-ATR/ATRIP-mediated DNA damage signaling (11), we asked whether DNMT1 was capable of interacting, directly or indirectly, with the 9-1-1 complex. Although relatively divergent at the primary sequence level, PCNA and 9-1-1 are quite similar at the three-dimensional structural level, both forming clamps that encircle DNA and act as assembly platforms for factors that operate on DNA (11,39). We therefore co-transfected plasmids expressing WT DNMT1 fused to an Xpress antibody epitope tag and RAD9 (a 9-1-1 subunit) fused to an HA tag. Upon immunoprecipitation (IP) of RAD9, we readily detected DNMT1 in the specifically bound material. PCNA served as a control and also interacted with DNMT1 (Fig. 4A and B). Owing to the poor quality of several commercially available RAD9 antibodies, we were unable to test for co-IP of endogenous DNMT1 and RAD9 (data not shown). Given the importance of the PDB in the recruitment of DNMT1 to sites of DNA damage as shown in our results (Fig. 1) and those of others (23), we asked next whether deletion of this domain affected the DNMT1–9-1-1 and DNMT1–CHK1 interactions. Interestingly, although deletion of the PBD in exon 5 eliminated the ability of DNMT1 to interact with RAD9 and PCNA, it had a more minor effect on the DNMT1–CHK1 interaction (we estimate an ∼5-fold reduced DNMT1–CHK1 interaction by loss of the PBD, Fig. 4C). These data therefore demonstrate for the first time that DNMT1 interacts with the 9-1-1 complex (RAD9) and that, although the PBD plays some role in the DNMT1–CHK1 interaction, CHK1 also interacts with regions of DNMT1 outside of the PBD independently of the PCNA–RAD9 interaction.
Figure 4.
DNMT1 interacts with multiple components of the DNA damage machinery. Wild-type epitope-tagged DNMT1 or DNMT1 lacking the PCNA-binding domain (DNMT1 ΔPBD) was used in co-immunoprecipitation (IP) reactions to examine their ability to interact with (A) RAD9, (B) PCNA and (C) CHK1. The ability of DNMT1 to interact with PCNA and the 9-1-1 (RAD9-RAD1-HUS1) PCNA-like clamp complex subunit RAD9 is dependent on the PBD region; however, the interaction between DNMT1 and CHK1 is not mediated solely by the PBD. Inputs and a loading control (β-actin) are shown below each IP panel. Xp, Xpress epitope tag; IgG, negative control species-matched IP.
Interactions with multiple components of the DNA damage machinery mediate DNMT1 recruitment to DNA DSBs
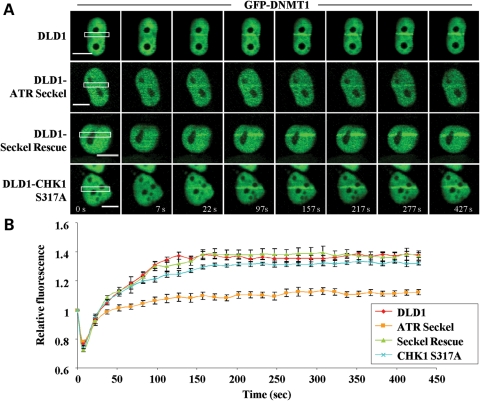
Our results using pharmacologic inhibitors of the DNA damage-signaling machinery implicated the ATR/CHK1 pathway as being important for DNMT1 recruitment to regions of DSBs, however these inhibitors lack complete specificity. For example, UCN-01 also inhibits CHK2, the downstream effector of ATM (40). In addition, there is significant crosstalk between ATM and ATR (41,42). We therefore more directly examined the role of factors that may be involved in DNMT1 recruitment to DSBs in two ways. In the first set of experiments, we made use of the DLD1 colorectal cancer cell line and three isogenic derivatives of this line generated by homologous recombination: (i) DLD1 ATR-Seckel, which expresses a hypomorphic mutant form of ATR derived from a Seckel syndrome patient, (ii) DLD1 CHK1 S317A, which expresses a mutant form of CHK1 in which serine 317, a critical residue phosphorylated by ATR in response to DNA damage, is mutated to alanine and (iii) DLD1 ATR-Seckel cells in which WT ATR has been stably re-expressed (DLD1-Seckel rescue) (43,44). Phosphorylation on S317 of CHK1 is essential for its DNA damage functions, whereas phosphorylation on S345 is important for CHK1's regulation of DNA replication under normal growth conditions. Cells are viable with CHK1 S317 mutations but not with CHK1 S345 mutations (44,45), allowing us to examine only the role of S317 phosphorylation in DNMT1 recruitment using this system. DLD1 cell lines were transfected with GFP-DNMT1, subjected to laser-IR DSB induction and DNMT1 localization monitored in live cells. Although DNMT1 mobilization to regions of DSBs in the parental DLD1 line was comparable with what we observed in HCT116 cells, DLD1 ATR-Seckel cells displayed a marked reduction in DNMT1 recruitment to laser-IR-induced DSBs (Fig. 5). In contrast, mutation of serine 317 of CHK1 had only a minimal effect on DNMT1 recruitment. DLD1 ATR-Seckel cells engineered to express WT ATR showed normal DNMT1 recruitment. These results therefore demonstrate that ATR signaling, but not CHK1 S317 phosphorylation, is essential for the recruitment of DNMT1 to regions of DNA DSBs. Interestingly, since DLD1 cells lack CHK2 activity (46), our results also show that CHK2 is not essential for DNMT1 recruitment.
Figure 5.
ATR status influences DNMT1 recruitment to regions of DNA DSBs. (A) DLD1 parental or isogenic DLD1 lines expressing a hypomorphic mutant form of ATR (ATR Seckel) or a mutant form of CHK1 that cannot be phosphorylated on the serine 317 position (CHK1 S317A) generated by homologous recombination were used for laser microirradiation studies. DLD1 ATR Seckel cells engineered to re-express WT ATR (Seckel Rescue) were used as a control. Each cell line was transfected with GFP-DNMT1 and laser-irradiated; representative live-cell images are shown. (B) Graphical summary of the effect of each mutant on DNMT1 localization to DSBs. CHK1 S345 was not examined because mutation of this site is incompatible with cell viability.
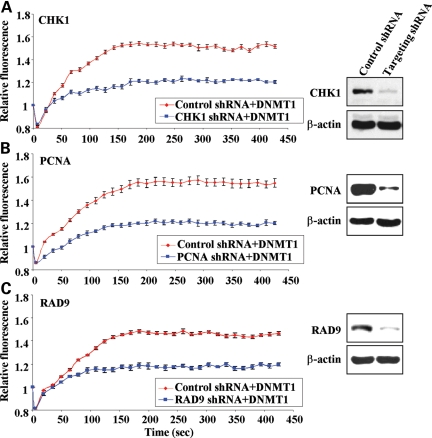
CHK1 S317A mutation in DLD1 cells had little effect on DNMT1 recruitment to DSBs, yet ATR mutation and UCN-01 treatment had marked effects (Figs 3 and 5). These data, coupled with the fact that CHK1 S345 mutation could not be examined in the DLD1 system because it is incompatible with cell growth (44), motivated us to use alternative techniques. In the second set of experiments to directly address the role of DNA damage-related DNMT1-interacting factors and ATR pathway components in DNMT1 recruitment to DSBs, we made use of shRNA to specifically knockdown expression of CHK1, 9-1-1 (RAD9) and PCNA. HCT116 cells were transfected with a CHK1 shRNA plasmid, which also constitutively expresses RFP, enabling microscopic identification of transfected shRNA-expressing cells (Supplementary Material, Fig. S1). Therefore, only transfected cells with both green (DNMT1) and red (CHK1 shRNA) fluorescence were subjected to laser-IR to induce DSBs. Compared with an irrelevant shRNA plasmid, knockdown of CHK1 alone markedly reduced DNMT1 localization to regions of DNA damage, using live-cell imaging (Figs 6A; Supplementary Material, Fig. S2). On the basis of western blot data, we estimate an ∼80% reduction in CHK1 protein levels under our shRNA knockdown conditions (Fig. 6A).
Figure 6.
CHK1, the 9-1-1 complex and PCNA influence recruitment of DNMT1 to DSBs. (A) GFP-DNMT1 and an shRNA plasmid targeting CHK1 were co-transfected into HCT116 cells. The shRNA plasmid also expressed RFP, enabling microscopic identification of transfected cells (Supplementary Material, Fig. S1). Red (shRNA-positive) and green (DNMT1-positive) cells were laser-irradiated and the localization of DNMT monitored by live-cell imaging (Supplementary Material, Fig. S2). Results are summarized in graphical form as recruitment versus time (left panel). Western blotting of whole-cell extract from transfected HCT116 cells demonstrates the efficiency of shRNA knockdown (right panel). β-Actin serves as a loading control. Identical studies were performed to monitor GFP-DNMT1 recruitment in cells transfected with (B) PCNA shRNA plasmid and (C) RAD9 shRNA plasmid.
Our previous results showed that PCNA and RAD9 (9-1-1) interact with DNMT1 via a domain at least partially distinct from that responsible for the CHK1 interaction and that deletion of the PBD eliminated DNMT1 recruitment to DSBs. Since both PCNA and 9-1-1 have important non-overlapping roles in the DNA damage response, we also separately analyzed the effect of RAD9 and PCNA shRNA knockdown on DNMT1 recruitment to sites of damage. After identifying shRNAs that effectively targeted PCNA or RAD9 (Fig. 6B and C), we co-transfected each shRNA with GFP-DNMT1 in HCT116 cells and then subjected doubly transfected cells to DSB induction by laser-IR treatment (Supplementary Material, Figs S1 and S2). Interestingly, knockdown of either PCNA or RAD9 resulted in a marked reduction in DNMT1 recruitment to regions of DNA damage, to a level equal to or greater than what we observed upon CHK1 knockdown (Figs 6B and C; Supplementary Material,Fig. S2). Therefore, DNMT1 recruitment to DSBs is not mediated by a simple bimolecular constitutive interaction between PCNA (or RAD9) and DNMT1 via the PBD. Rather, these results demonstrate that multiple factors are involved in enabling rapid and efficient recruitment of DNMT1 to sites of DNA DSBs and that at least two of these factors (CHK1 and RAD9) act via the ATR pathway.
Cells expressing a hypomorphic PBD-deleted form of DNMT1 display altered DNA damage signaling in both unperturbed and DNA-damaged states and delayed DSB repair
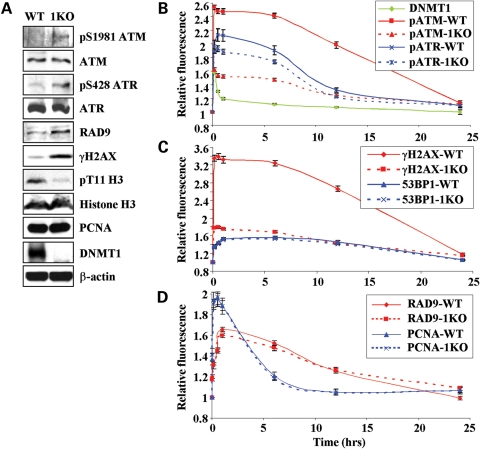
HCT116 DNMT1 KO (hypomorph) cells, which express very low levels of a DNMT1 isoform lacking exons 2–5 [including the PBD (47,48)], are more sensitive to a number of DNA-damaging agents, although their growth under unperturbed conditions is only slightly slower than that of parental HCT116 cells (24). We first assayed levels of several DNA damage markers, including phosphorylated ATM and H2AX (among the first to be activated by and recruited to DNA DSBs), 53BP1 (recruited slightly later) and ATR (recruited later still) (49). We also analyzed levels of RAD9, PCNA, total ATR and ATM and H3T11 phosphorylation [a CHK1 kinase target in unperturbed chromatin (19)]. Western blotting of HCT116 parental and 1KO cells revealed a marked elevation in the level of activated ATM (phosphorylated on serine 1981) and γH2AX, and a similarly marked reduction in H3T11 phosphorylation in 1KO cells (Fig. 7A). The change in γH2AX levels in 1KO cells is consistent with our previous microscopic quantitation of γH2AX-foci in 5-azadC-treated cells (24). ATR phosphorylated at serine 428 was also markedly elevated in 1KO cells, although the significance of this mark in terms of ATR function remains obscure (11). Total ATM and ATR levels were essentially unchanged by DNMT1 mutation. RAD9 levels were slightly elevated while PCNA and 53BP1 levels were unchanged in 1KO cells. Equal loading of cell extracts and chromatin is indicated by the comparable western blot signals for β-actin and histone H3, respectively, in parental and 1KO HCT116 cells (Fig. 7A). These data demonstrate a marked activation of DNA damage signaling, particularly the early-phase components, in cells lacking WT DNMT1 under conditions of unperturbed growth. The interaction between DNMT1 status and H3T11 phosphorylation further underscores the functional interconnections between DNMT1 and CHK1.
Figure 7.
DNMT1 modulates the DNA damage response in unperturbed and DNA-damaged cells. (A) Western blotting for levels of DNA damage response factors or their activated (phosphorylated) forms in unperturbed HCT116 and HCT116 hypomorphic mutant DNMT1 (1KO) cells. The recently discovered CHK1 kinase target in chromatin, histone H3 threonine 11 (H3T11) (19), was also examined. Total histone H3, DNMT1 and β-actin are shown as controls for loading and identifying each cell line. Chromatin extracts were used for histone protein westerns. HCT116 and 1KO cells were transfected with GFP-DNMT1, subject to DSB generation by laser irradiation, and the recruitment and decay of (B) ATM, ATR (phosphorylated forms), DNMT1, (C) γH2AX and 53BP1 and (D) the replicative and damage clamps PCNA and 9-1-1 (RAD9) were monitored over 24 h. Irradiated cells were fixed at the indicated time points and immunostained. Regions of laser-induced damage were identified by co-staining with γH2AX antibody (not shown). Fluorescence values are set relative to those in the same cell line before DSB induction.
We next examined the recruitment and decay kinetics of ATM, ATR, γH2AX, 53BP1, PCNA and RAD9 from regions of DSBs generated by laser-IR in parental compared with 1KO HCT116 cells. One observation of initial interest in parental HCT116 cells is the markedly different time course of DNMT1 compared with all the other factors examined. Although all were rapidly recruited to DSBs, DNMT1 was lost much more quickly (Fig. 7B–D). ATM and γH2AX display very similar decay kinetics, as would be expected (Fig. 7B and C). When comparing the two cell lines, damage-induced levels of activated ATM and γH2AX were markedly reduced in 1KO cells. This was due, in large part, to the highly elevated basal level of these marks pre-laser-IR treatment (which damage-induced levels are set relative to, Fig. 7B and C). In addition, phosphorylated ATM and ATR appeared to return to their baseline levels more rapidly in 1KO compared with parental HCT116 cells (the _t_1/2 for the decay of phosphorylated ATM was 15.2 h in HCT116 compared with 10.5 h in 1KO cells), suggesting a role for DNMT1 in maintaining the activated state. In contrast, however, 53BP1, PCNA and RAD9 behaved identically in parental and 1KO cells, showing that mutation of DNMT1 does not globally disrupt DNA damage signaling (Fig. 7C and D). Although CHK1 is a critical player in the early DNA damage response, it does not appear to accumulate at DSB foci (50) and our results using a YFP-tagged CHK1 construct are consistent with this observation (data not shown). This property precluded us from performing a similar time course experiment for CHK1.
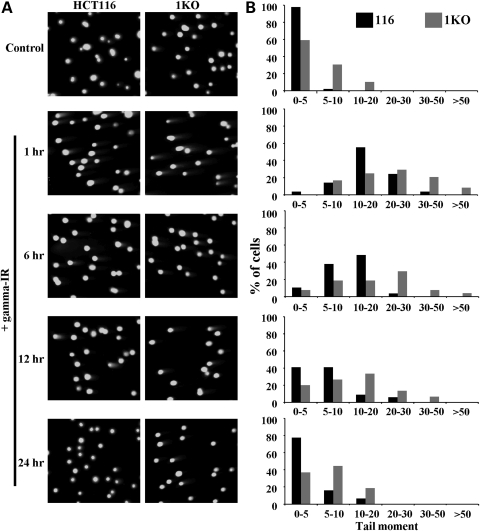
Since DNMT1 localized to sites of laser-induced DNA damage and because the hypomorphic mutant HCT116 DNMT1 KO cells displayed altered localization kinetics of some DSB-related factors, we hypothesized that the repair rate of DNA damage may be altered in 1KO cells. We therefore measured DNA repair efficiency in HCT116 parental and 1KO cells for 24 h after γ-irradiation (10 Gy) using the neutral comet assay. Representative comet images are shown in Figure 8A and quantitation of comet data as tail moments is shown in Figure 8B. Under normal growth condition, we observed greater levels of DNA damage in 1KO cells than in HCT116 parental cells (Fig. 8B, top graph). This result correlated with our observation of increased activation of DNA damage markers in 1KO cells (Fig. 7). Following γ-irradiation, a much higher level of DNA damage was induced in 1KO compared with parental HCT116 cells (see tail moments in Fig. 8B). Furthermore, 1KO cells exhibited lower repair efficiency (or a reduced rate of repair) when the tail moments were compared with those of parental HCT116 cells over the entire 24 h period of the experiment. Levels of DNA strand breaks in HCT116 cells were nearing their pre-irradiated levels after 24 h but higher levels of DNA damage persisted in 1KO cells (Fig. 8A and B). Taken together, these results demonstrate that DNMT1 plays a role in suppressing DNA damage signaling, promoting genome stability under conditions of unperturbed cell growth and regulating the rate of DSB repair. These data also show that DNMT1 is important for the maximal retention of some of the most rapidly recruited damage-signaling components at DSBs.
Figure 8.
DNMT1 status influences the rate of DSB repair. (A) Parental and hypomorphic DNMT1 mutant 1KO HCT116 cells before (control) and 1, 6, 12 and 24 h after induction of DNA DSBs with 10 Gy of γ-irradiation (+IR) were used for neutral comet assays. Representative micrographs of cells stained with SYBR green from each treatment are shown. Images are taken at 20× magnification. (B) Quantitation of comet assay data as groups of tail moment scores (an indicator of the level of DNA damage) versus the percentage of cells with each tail moment for each treatment. Fifty randomly selected cells from each treatment were photographed using an Axioplan 3 fluorescent microscope. Tail moment (tail length × percentage of DNA in the tail) was scored by using TriTek Comet Score software.
DISCUSSION
In the present study, we used NIR laser microirradiation to examine the role of DNMT1 in the DNA DSB response. DNMT1 was unique among DNA methyltransferases in being rapidly recruited to DSBs in a PBD-dependent but catalytic activity-independent manner. DNMT1 was also distinct from several established players in the DNA repair response because of its short residence time at sites of damage. We confirmed our previous findings that DNMT1 interacts with CHK1 and extended these results to show that CHK1 does not interact with the same region of DNMT1 as PCNA, suggesting that PCNA is not simply serving as a bridge between DNMT1 and CHK1. In addition, we show for the first time that DNMT1 interacts with the PCNA-like DNA damage sliding clamp component RAD9 (of the 9-1-1 complex) via the PBD region, further linking DNMT1 to the ATR/CHK1 arm of the DNA damage response. Use of pharmacologic inhibitors of the damage response, stable knockin mutant cell lines, and specific shRNA knockdowns revealed that ATR, CHK1, 9-1-1 and PCNA are important for the localization of DNMT1 to DNA DSBs. Using HCT116 cells expressing a DNMT1 isoform lacking the PBD, we showed that wild-type DNMT1 is essential for suppressing basal activation of the DNA damage response under normal growth conditions and for maintaining CHK1-mediated phosphorylation of H3T11. DNMT1 status also influenced the rate of DSB repair following γ-irradiation. In addition, in cells with NIR laser-induced DSBs, DNMT1 was important for maintaining activated ATM and ATR at sites of damage. Taken together, our results firmly place DNMT1 in the DNA DSB response pathway and suggest that DNMT1 is important for genome stability under normal growth conditions and for cells to mount a full response to DSBs by virtue of its interaction with CHK1, PCNA and 9-1-1.
Although the recruitment kinetics of DNMT1 and PCNA were both very rapid, PCNA was recruited more quickly to DSBs than DNMT1. In contrast, the decay kinetics of these two proteins were markedly different, with PCNA resident on DSBs much longer than DNMT1. Given that the PBD of DNMT1 is essential for its recruitment to DSBs, the role of PCNA in directly recruiting DNMT1 to sites of DNA damage appears more complicated than an isolated bimolecular DNMT1–PCNA interaction. There are clearly PCNA-independent mechanisms of targeting DNMT1 to chromatin in unperturbed cells (51). DNMT1's interaction with CHK1, which is also essential for efficient localization of DNMT1 to DSBs, represents at least one additional mediator of DNMT1 recruitment to damaged DNA. CHK1 interacts with full-length and, to a lesser extent, ΔPBD forms of DNMT1 and shRNA knockdown of CHK1 markedly reduced DNMT1 localization to DSBs, comparable with 9-1-1 and PCNA knockdown. It is therefore possible that CHK1 is required to act upon DNMT1 to ‘license’ its recruitment to DSBs, perhaps by making it competent to bind PCNA or 9-1-1 (Supplementary Material, Fig. S3). An obvious candidate for this licensing activity is phosphorylation of DNMT1 by CHK1's kinase activity. Indeed, an in silico search for CHK1 consensus phosphorylation sites [M/I/L/V-X-R/K-X-X-S/T, with phosphorylation occurring at S or T (52)] within the DNMT1 protein sequence using the PHOSIDA database (53) reveals 11 good matches that are conserved in the chimp, dog and murine Dnmt1 homologues. Intriguingly, a tight cluster of four of these sites lies in the region immediately flanking the PBD (two serines and two threonines between amino acids 85–162 of DNMT1). Although speculative at this time, these findings suggest that phosphorylation of DNMT1 by CHK1, or CHK1 serving as a bridge between DNMT1 and another repair associated factor, is required for efficient localization of DNMT1 to DSBs. In further support of this notion, regulation of the DNMT1–PCNA interaction by AKT and PKC phosphorylation of the N-terminal region of DNMT1, unrelated to DNA damage, was recently reported (54).
Recent findings from several laboratories clearly show that DNMT1's interaction with PCNA is not required for maintenance methylation (47,48,51,55). These data then raise the question as to the true function of the DNMT1–PCNA interaction in mammalian cells. Genomic DNA methylation levels are reduced by only ∼20% in cells expressing low levels of a PBD-deleted form of DNMT1 (HCT116 hypomorphic 1KO cells) (56) but 1KO cells do exhibit elevated genomic instability (57). In contrast, disruption of the recently described interaction between UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) and DNMT1 results in massive genomic hypomethylation (58,59). UHFR1 is a chromatin-associated factor that not only interacts with DNMT1, but also has an intrinsic binding preference for hemimethylated CpG sites, like DNMT1. DNMT1 and UHRF1 also co-localize extensively during S-phase. On the basis of these results, UHRF1 appears to play a much more important role in targeting DNMT1 to replication foci to maintain DNA methylation patterns during replication than does PCNA. Therefore, the principal role for the DNMT1–PCNA interaction, and likely also the DNMT1–RAD9 (9-1-1) interaction we identified here, is for DNA repair and/or replication fork stability-related functions rather than maintenance methylation. As all cells in culture undergo some replication stress under normal growth conditions, the PCNA–DNMT1 (and DNMT1–9-1-1) interaction would be predicted to be readily detected as part of any standard biochemical purification or IP experiment from tissue culture cells [Fig. 4 and (31,38)].
Our data showing that DNMT1 is rapidly and actively recruited to sites of DNA damage, that DNMT1 modulates CHK1 function [Fig. 7A and (24)] and that DNMT1 status influences the susceptibility to, and repair rate of, DSBs (Figs 7B and 8) firmly implicates DNMT1 as a modulator of the cellular response to DSBs and as a regulator of genomic stability under unperturbed growth conditions. Our results, however, raise new questions, such as the function of DNMT1 at sites of DNA damage. An obvious possibility is for DNMT1 to re-methylate newly synthesized DNA as part of the repair process to restore normal chromatin structure at damaged regions. Although this remains possible, our observations that DNMT1 is very rapidly recruited, likely before re-synthesis is completed, and that it is retained at sites of damage only transiently, with many other repair-associated factors (including PCNA) remaining at DSBs much longer, suggest that genomic methylation is not the main function of DNMT1, at least in the early part of the DNA damage response. In addition, a catalytically inactive but otherwise full-length form of DNMT1 displayed almost identical recruitment kinetics as WT DNMT1. Further support for a non-methylation role for DNMT1 in the damage response comes from a study showing that CHK1/CHK2 activation and γH2AX foci formation by DNMT1 siRNA depletion were rescued by expression of a catalytically inactive form of DNMT1 (26). Induction of p53-dependent apoptosis and a G2 damage checkpoint by DNMT1 depletion in Xenopus laevis embryos was also rescued by expression of catalytically inactive DNMT1 (27). An alternative methylation-independent function worthy of consideration is that mobilization of DNMT1, like the transcriptional co-repressor TIF1β (KAP-1), is involved in the global chromatin decondensation required for efficient DNA repair (60). Phosphorylated TIF1β is rapidly recruited to DSBs, like DNMT1, and disassociates from damaged regions more rapidly than most DSB repair-related proteins (TIF1β dissociates even more rapidly than DNMT1—by ∼15 min). DNMT1, like TIF1β, may have an essential function in the earliest stages of the DNA damage response that needs to be relayed throughout the nucleus. ATM-dependent phosphorylation and mobilization of TIF1β appears to mediate global chromatin relaxation (60). Similarly, it may be necessary for DNMT1 to depart the damaged region to allow other factors access or to create a more open chromatin configuration by removal of DNMT1-associated co-repressors [e.g. HDACs (61)]. Previous studies have in fact shown that HCT116 hypomorphic 1KO cells possess a significantly more open chromatin structure than parental HCT116 cells with intact DNMT1 (56), suggesting that DNMT1 mobilization following damage could contribute to chromatin relaxation. It remains to be determined, however, whether damage-induced chromatin relaxation is altered in HCT116 1KO cells and whether DNMT1 is phosphorylated in a DNA damage-specific manner. Alternatively, dissociation of DNMT1 from sites of damage prior to the repair/re-synthesis step might be advantageous to prevent methylation errors. DNMT1's propensity for methylating abnormal DNA structures (62) could prove a liability to the cell and permit the establishment of aberrant DNA methylation patterns if allowed access to damaged DNA or DNA actively undergoing resection and repair. Such aberrant de novo methylation events at sites of DNA damage in experimental DSB model systems have been recently documented (63,64). One of these studies showed that DNMT1 mediated aberrant de novo methlylation of substrates undergoing repair specifically by homologous recombination (63), consistent with our pharmacologic and knockin cell-line data showing that ATR, but not DNA-PKcs (critical for non-homologous end-joining type repair), mediates DNMT1 recruitment to DSBs (65). Hemimethylated DNA remaining after repair, or fully methylated DNA adjacent to the repaired/resected region, may be sufficient to target DNMT1 for remethylation during a subsequent normal S-phase by virtue of its innate substrate preference, its interaction with UHRF1 or its ability to bind methylated DNA via an allosteric site in the N-terminal regulatory domain (66).
We show here for the first time that DNMT1 interacts with another component of the DSB sensing and repair machinery, RAD9. RAD9 is a subunit of the heterotrimeric PCNA-like DNA damage sliding clamp known as 9-1-1. 9-1-1 appears to act as a sensor of DNA damage in concert with ATR/ATRIP and RPA (7). The exact function of 9-1-1, relative to PCNA, remains unclear. It has been speculated that 9-1-1 substitutes for PCNA during times of cellular stress or in the repair of certain types of DNA damage (67). Phosphorylation of the 9-1-1 clamp loader, RAD17-RFC, by ATM and ATR results in enhanced 9-1-1 loading onto DNA. ssDNA also enhances loading by RAD17 via its interaction with RPA. The RAD17-RFC–9-1-1 complex and ATM/ATR independently accumulate at sites of DNA damage (7,8). RAD17 interacts with Claspin and RAD9 interacts with and recruits TopBP1. Both Claspin and TopBP1 promote phosphorylation and activation of CHK1 and other ATR substrates (68,69). Although the exact function of the DNMT1–RAD9 interaction is currently unknown, it may play a role in regulating CHK1 activity. Our previous work demonstrated that DNMT1 modulated the release of CHK1 from chromatin following 5-azadC treatment of cells (24) and our current results show that DNMT1 influences CHK1 phosphorylation of H3T11 under normal growth conditions. Intriguingly, PCNA also interacts with CHK1, and the PCNA-interacting region of CHK1 is essential for its checkpoint function and release from chromatin following DNA damage (70). Therefore, DNMT1 may modulate CHK1 function by virtue of its ability to interact not only with CHK1 itself, but also with at least two proteins involved in controlling CHK1 activity, the 9-1-1 complex and PCNA. Finally, a role for the long known PCNA–DNMT1 interaction in DNA repair, rather than maintenance methylation, should not be understated. PCNA is an extremely important player in a growing number of DNA repair-related pathways. For example, PCNA is critical for base excision repair, nucleotide excision repair, MMR and the response to damaged or stalled replication forks. In the latter, PCNA ubiquitination regulates the decision between error-prone or error-free translesion bypass (32). The relative roles of PCNA and 9-1-1 in DNA repair, and the mechanism by which DNMT1 affects their functions, will be an interesting area of future research.
In summary, we have shown that DNMT1 localization is dynamically regulated in response to DNA DSBs and that DNMT1 interacts with multiple components of the repair machinery via its PBD and other domains. DNMT1 appears essential for CHK1 function under normal and genotoxic conditions, and CHK1, along with PCNA and 9-1-1, is essential for DNMT1's localization to sites of DNA damage. Our results, and the results of others (23,26,27), therefore strongly suggest that DNMT1 functions in sensing and/or mobilizing the response to certain forms of DNA damage. Complete inactivation of DNMT1, unlike the hypomorphic mutation we studied here in HCT116 1KO cells, results in rapid induction of a G2/M checkpoint and typical hallmarks of a DNA damage response with minimal DNA demethylation, indicating that this role for DNMT1 does not depend on its DNA methylation activity (71). As we have hypothesized previously, DNMT1 may participate in sensing DNA damage, in one of the cell-cycle checkpoints, or in both (72). This novel function of DNMT1 may be observable only under extreme depletion conditions not normally achieved in typical knockdown experiments. These data have important implications for therapeutic strategies aimed at targeting DNA methylation and DNMT1 in tumor cells. Understanding the role of DNMT1 in the DNA damage response may allow for better use of DNA methylation inhibitors and, more excitingly, the development of novel combination therapies that will exploit DNMT1's role not only as a maintenance methyltransferase, but also as a modulator of the DNA damage response, to more efficiently target tumor cells for destruction.
MATERIALS AND METHODS
Cell culture and transient transfection
Parental HCT116 and isogenic HCT116 cells expressing a hypomorphic mutant form of DNMT1 (referred to as 1KO, provided by Dr Bert Vogelstein, The Johns Hopkins University) (25,47,48) were cultured in McCoy's-5a medium (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin–streptomycin solution (Hyclone) at 37°C in a 5% CO2 incubator. DLD1 and the isogenic knockin lines DLD1-ATR Seckel, DLD1-Seckel Rescue and DLD1-CHK1 S317A (43,44) were provided by Dr Fred Bunz (The Johns Hopkins University) and cultured under the same conditions as HCT116 cells. For laser microirradiation and IP experiments, cells were transfected using Lipofectamine 2000 according to the manufacturer's protocol (Invitrogen).
Chemicals, plasmids and antibodies
NU7026, caffeine, UCN-01, KU-55933 and 5-aza-2′-deoxycytidine were purchased from Sigma. Plasmids expressing GFP-tagged DNMT1, DNMT3A and DNMT3B1 have been described previously (24,73,74). GFP-DNMT1 with a deletion of the PBD (GFP-DNMT1 ΔPBD or WpKR452) was created by PCR using full-length DNMT1 as a template to create an in-frame deletion of exon 5 (bp 235–546 of human DNMT1, which contains the PBD). The active site point mutant DNMT1 C1229A was created using the QuikChange Site-directed Mutagenesis Kit (Stratagene). Both mutants were confirmed by DNA sequencing. Full-length DNMT1 and DNMT1 ΔPBD were subcloned into pcDNA4HisMax, which contains the Xpress antibody epitope tag (Invitrogen) for IP and western blotting. The HA-tagged human RAD9 expression vector was provided by Dr Laura Lindsey-Boltz (University of North Carolina, Chapel Hill, NC, USA), and the RFP-PCNA expression construct was provided by Dr M Cristina Cardoso (Max Delbruck Center for Molecular Medicine, Berlin, Germany). FLAG-tagged human CHK1 was provided by Dr Yolanda Sanchez (Dartmouth Medical School). For drug treatment-microirradiation experiments, cells were first transfected with GFP-DNMT1, then subsequently treated with 5 μm NU7026, 5 mm caffeine, 50 nm KU-55933 or 300 nm UCN-01 for 24 h, or 10 μm 5-azadC for 48 h. Plasmids expressing shRNA targeting human CHK1 (5′-GTG CCT ATG GAG AAG TTC AAC TTG CTG TG-3′), PCNA (5′-GAT ATT AGC TCC AGC GGT GTA AAC CTG CA-3′), RAD9 (5′-TGG TCA GGA CCT GCT GCG CTG TAA GAT CC-3′), or negative control non-effective shRNA was purchased from Origene. These plasmids also constitutively express RFP as a marker of transfected cells. Cells were collected 2 days after transfection to test shRNA knockdown efficiency at the protein level. Expression of CHK1, PCNA and RAD9 was determined by western blotting. Antibodies used for western blot analysis and immunofluorescence staining are listed in Supplementary Material, Table S1.
Laser microirradiation and cell imaging
DNA damage was introduced into defined subnuclear volumes by microirradiation with a pulsed Ti:Sapphire laser (800 nm, Mira 900, Coherent, Inc.). Real-time images were collected using an LSM510 META confocal microscope equipped with 543 nm HeNe and multiline (458, 488 and 514 nm) argon lasers and a 40× C-Apochromat 1.2 N.A. water immersion lens (Carl Zeiss MicroImaging, Inc.). A heated stage (Carl Zeiss MicroImaging, Inc.) and an objective lens heater (Physitemp) were used to maintain cells at 37°C. Relative fluorescence levels at sites of DNA damage were measured using Image J software (NIH).
Immunofluorescence, TUNEL staining and neutral comet assays
Cells were grown on glass coverslips overnight. After induction of DNA damage, cells were washed with PBS and fixed in 4% paraformaldehyde, 0.5% Triton X-100 for 10 min. Coverslips were washed three times with 1X PBS and incubated in block-permeabilization solution [15% goat serum (Sigma), 0.2% fish skin gelatin (Sigma), 0.03% NaN3 and 0.5% Triton X-100 in 1X PBS] for 10 min. Cells were rinsed and incubated with blocking solution (15% goat serum, 0.2% fish skin gelatin and 0.03% NaN3 in 1X PBS) overnight. Following this, cells were incubated with diluted primary antibodies in blocking solution for 2 h at 37°C, washed three times with PBS and incubated with appropriate fluorophore-conjugated secondary antibodies in blocking solution. Cells were washed three times with 1X PBS and counterstained for DNA using DAPI (4′-6-diamidino-2-phenylindol, Invitrogen). Images were collected using an Axioplan 3 fluorescent microscope (Zeiss). The TUNEL reaction was performed using TAMRA-dUTP and terminal deoxynucleotidyl transferase according to the manufacturer's protocol (Roche Diagnostics). DNA repair efficiency was measured by neutral comet assay following the manufacturer's instructions (Trevigen). Briefly, cells were γ-irradiated (10 Gy), harvested, mixed with low-melting agarose and layered onto glass slides. Slides were immersed in lysis solution for 30 min, washed with TBE buffer for three times and incubated for 30 min in electrophoresis buffer. After electrophoresis (1 V/cm and 20 mA for 30 min), slides were immersed in 70% ethanol for 5 min and stained with SYBR green dye for 10 min in the dark. Fifty randomly selected cells per sample were captured using an Axioplan 3 fluorescent microscope. Tail moment (tail length × percentage of DNA in the tail) indicating DNA damage levels was scored by using TriTek Comet Score software (TriTek Corp.).
Western blot analysis and IP
Cells were washed with 1X PBS and lysed in RIPA buffer (Pierce) containing Complete EDTA-free protease inhibitor cocktail (Roche Diagnostics) on ice for 30 min. Preparation of chromatin extracts for western blotting of histone modifications was performed as described previously (75). Protein concentrations were determined using the BCA Protein Assay Kit (Thermo Scientific). Total cellular lysates were separated on SDS–PAGE gels and transferred to nitrocellulose membrane. Membranes were incubated with primary antibody (Supplementary Material, Table S1) overnight at 4°C, washed with TBS–T three times and incubated with HRP-conjugated secondary antibody for 2 h at room temperature. After washing with TBS-T three times, blots were visualized by enhanced chemiluminescence (GE Healthcare). For IPs, cell lysates (prepared as for western blot analyses) were mixed with lysis buffer [25 mm Tris-phosphate (pH 7.8), 2 mm DTT, 2 mm EDTA, 10% glycerol and 0.2% Triton X-100] and the indicated antibodies (Supplementary Material, Table S1) and incubated overnight at 4°C with rotation. Immune complexes were washed with lysis buffer three times, incubated with sample buffer for 10 min at 95°C and bound proteins resolved by SDS–PAGE.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
FUNDING
This work was supported by NIH grant R01CA116028 and the Georgia Cancer Coalition (K.D.R.). K.D.R. is a Georgia Cancer Coalition Distinguished Cancer Scholar.
Supplementary Material
Supplementary Data
ACKNOWLEDGEMENTS
We thank Laura Lindsey-Boltz, Yolanda Sanchez and Cristina Cardoso for providing plasmids and Bert Vogelstein and Fred Bunz for providing cell lines.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Goll M.G., Bestor T.H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- 2.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 3.Liang G., Chan M.F., Tomigahara Y., Tsai Y.C., Gonzales F.A., Li E., Laird P.W., Jones P.A. Cooperativity between DNA methyltransferases in the maintenance methylation of repetitive elements. Mol. Cell. Biol. 2002;22:480–491. doi: 10.1128/MCB.22.2.480-491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gopalakrishnan S., Van Emburgh B.O., Robertson K.D. DNA methylation in development and human disease. Mutat. Res. 2008;647:30–38. doi: 10.1016/j.mrfmmm.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma S., Kelly T.K., Jones P.A. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jackson S.P., Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA–ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 8.Zou L., Cortez D., Elledge S.J. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes. Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carson C.T., Schwartz R.A., Stracker T.H., Lilley C.E., Lee D.V., Weitzman M.D. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003;22:6610–6620. doi: 10.1093/emboj/cdg630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shiloh Y. ATM and related protein kinases: safeguarding genome integrity. Nat. Rev. Cancer. 2003;3:155–168. doi: 10.1038/nrc1011. [DOI] [PubMed] [Google Scholar]
- 11.Cimprich K.A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Q., Guntuku S., Cui X.S., Matsuoka S., Cortez D., Tamai K., Luo G., Carattini-Rivera S., De Mayo F., Bradley A., et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes. Dev. 2000;14:1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 13.Petermann E., Maya-Mendoza A., Zachos G., Gillespie D.A., Jackson D.A., Caldecott K.W. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol. Cell. Biol. 2006;26:3319–3326. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimada M., Nakanishi M. Checkpoints meet the transcription at a novel histone milestone (H3-T11) Cell Cycle. 2008;7:1555–1559. doi: 10.4161/cc.7.11.6062. [DOI] [PubMed] [Google Scholar]
- 15.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 16.Paull T.T., Rogakou E.P., Yamazaki V., Kirchgessner C.U., Gellert M., Bonner W.M. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 2000;10:886–895. doi: 10.1016/s0960-9822(00)00610-2. [DOI] [PubMed] [Google Scholar]
- 17.Jungmichel S., Stucki M. MDC1: the art of keeping things in focus. Chromosoma. 2010;119:337–349. doi: 10.1007/s00412-010-0266-9. [DOI] [PubMed] [Google Scholar]
- 18.Smits V.A.J., Reaper P.M., Jackson S.P. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr. Biol. 2006;16:150–159. doi: 10.1016/j.cub.2005.11.066. [DOI] [PubMed] [Google Scholar]
- 19.Shimada M., Ninida H., Zineldeen D.H., Tagami H., Tanaka M., Saito H., Nakanishi M. Chk1 is a histone H3 threonine 11 kinase that regulates DNA damage-induced transcriptional repression. Cell. 2008;132:221–232. doi: 10.1016/j.cell.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 20.Yoder J.A., Walsh C.P., Bestor T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 21.Chen R.Z., Pettersson U., Beard C., Jackson-Grusby L., Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 22.Guo G., Wang W., Bradley A. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature. 2004;429:891–895. doi: 10.1038/nature02653. [DOI] [PubMed] [Google Scholar]
- 23.Mortusewicz O., Schermelleh L., Walter J., Cardoso M.C., Leonhardt H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl Acad. Sci. USA. 2005;102:8905–8909. doi: 10.1073/pnas.0501034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palii S.S., Van Emburgh B.O., Sankpal U.T., Brown K.D., Robertson K.D. DNA methylation inhibitor 5-aza-2′-deoxycytidine (5-azadC) induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B. Mol. Cell. Biol. 2008;28:752–771. doi: 10.1128/MCB.01799-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhee I., Bachman K.E., Park B.H., Jair K.-W., Yen R.-W.C., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W., et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 26.Unterberger A., Andrews S.D., Weaver I.C.G., Szyf M. DNA methyltransferase 1 knockdown activates a replication stress checkpoint. Mol. Cell. Biol. 2006;26:7575–7586. doi: 10.1128/MCB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ruzov A., Shorning B., Mortusewicz O., Dunican D.S., Leonhardt H., Meehan R.R. MBD4 and MLH1 are required for apoptotic induction in xDNMT1-depleted embryos. Development. 2009;136:2277–2286. doi: 10.1242/dev.032227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel K., Dickson J., Din S., Macleod K., Jodrell D., Ramsahoye B. Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated DNMT1 induces proteosomal degradation of the free enzyme. Nucleic Acids Res. 2010;38:4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong X., Mohanty S.K., Stephens J., Heale J.T., Gomez-Godinez V., Shi L.Z., Kim J.-S., Yokomori K., Berns M.W. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009;37:e68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mari P.-O., Florea B.I., Persengiev S.P., Verkaik N.S., Bruggenwirth H.T., Modesti M., Giglia-Mari G., Bezstarosti K., Demmers J.A.A., Luider T.M., et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl Acad. Sci. USA. 2006;103:18597–18602. doi: 10.1073/pnas.0609061103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chuang L.S.-H., Ian H.-I., Koh T.-W., Ng H.-H., Xu G., Li B.F.L. Human DNA-(cytosine-5) methyltransferase-PCNA complex is a target for p21Waf1. Science. 1997;277:1996–2000. doi: 10.1126/science.277.5334.1996. [DOI] [PubMed] [Google Scholar]
- 32.Moldovan G.-L., Pfander B., Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Sarkaria J.N., Busby E.C., Tibbetts R.S., Roos P., Taya Y., Karnitz L.M., Abraham R.T. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 34.Veuger S.J., Curtain N.J., Richardson C.J., Smith G.C.M., Durkacz B.W. Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 2003;63:6008–6015. [PubMed] [Google Scholar]
- 35.Hickson I., Zhao Y., Richardson C.J., Green S.J., Martin N.M.B., Orr A.I., Reaper P.M., Jackson S.P., Curtin N.J., Smith G.C.M. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- 36.Graves P.R., Yu L., Schwarz J.K., Gales J., Sausville E.A., O'Connor P.M., Piwnica-Worms H. The Chk1 protein kinase and the Cdc25c regulatory pathways are targets of the anticancer agent UCN-01. J. Biol. Chem. 2000;275:5600–5605. doi: 10.1074/jbc.275.8.5600. [DOI] [PubMed] [Google Scholar]
- 37.Schermelleh L., Spada F., Easwaran H.P., Zolghadr K., Margot J.B., Cardoso M.C., Leonhardt H. Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat. Meth. 2005;10:751–756. doi: 10.1038/nmeth794. [DOI] [PubMed] [Google Scholar]
- 38.Iida T., Suetake I., Tajima S., Morioka H., Ohta S., Obuse C., Tsurimoto T. PCNA clamp facilitates action of DNA cytosine methyltransferase 1 on hemimethylated DNA. Genes Cells. 2002;7:997–1007. doi: 10.1046/j.1365-2443.2002.00584.x. [DOI] [PubMed] [Google Scholar]
- 39.Parrilla-Castellar E.R., Arlander S.J.H., Karnitz L. Dial 9-1-1 for DNA damage: the Rad9-Hus1-Rad1 (9-1-1) clamp complex. DNA Repair. 2004;3:1009–1014. doi: 10.1016/j.dnarep.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 40.Yu Q., La Rose J., Zhang H., Takemura H., Kohn K.W., Pommier Y. UCN-01 inhibits p53 upregulation and abrogates γ-radiation-induced G2-M checkpoint independently of p53 by targeting both the checkpoint kinases, Chk2 and Chk1. Cancer Res. 2002;62:5743–5748. [PubMed] [Google Scholar]
- 41.Adams K.E., Medhurst A.L., Dart D.A., Lakin N.D. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN complex. Oncogene. 2006;25:3894–3904. doi: 10.1038/sj.onc.1209426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jazayeri A., Falck J., Lukas C., Bartek J., Smith G.C.M., Lukas J., Jackson S.P. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 43.Hurley P.J., Wilsker D., Bunz F. Human cells require ATR for cell cycle progression following exposure to ionizing radiation. Oncogene. 2007;26:2535–2542. doi: 10.1038/sj.onc.1210049. [DOI] [PubMed] [Google Scholar]
- 44.Wilsker D., Petermann E., Helleday T., Bunz F. Essential function of Chk1 can be uncoupled from DNA damage checkpoint replication control. Proc. Natl Acad. Sci. USA. 2008;105:20752–20757. doi: 10.1073/pnas.0806917106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Niida H., Katsuno Y., Banerjee B., Hande M.P., Nakanishi M. Specific role of Chk1 phosphorylations in cell survival and checkpoint activation. Mol. Cell. Biol. 2007;27:2572–2581. doi: 10.1128/MCB.01611-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Falck J., Lukas C., Protopova M., Lukas J., Selivanova G., Bartek J. Functional impact of concomitant versus alternative defects in the Chk2-p53 tumour suppressor pathway. Oncogene. 2001;20:5503–5510. doi: 10.1038/sj.onc.1204811. [DOI] [PubMed] [Google Scholar]
- 47.Egger G., Jeong S., Escobar S.G., Cortez C.C., Li T.W.H., Saito Y., Yoo C.B., Jones P.A., Liang G. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc. Natl Acad. Sci. USA. 2006;103:14080–14085. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spada F., Hemmer A., Kuch D., Rothbauer U., Schermelleh L., Kremmer E., Carell T., Langst G., Leonhardt H. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J. Cell. Biol. 2007;176:656–571. doi: 10.1083/jcb.200610062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 50.Bekker-Jensen S., Lukas C., Kitagawa R., Melander F., Kastan M.B., Bartek J., Lucas K. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell. Biol. 2006;173:195–206. doi: 10.1083/jcb.200510130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Easwaran H.P., Schermelleh L., Leonhardt H., Cardoso M.C. Replication-independent chromatin loading of Dnmt1 during G2 and M phases. EMBO Rep. 2004;5:1–6. doi: 10.1038/sj.embor.7400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hutchins J.R., Hughes M., Clarke P.R. Substrate specificity determinants of the checkpoint protein kinase Chk1. FEBS Lett. 2000;466:91–95. doi: 10.1016/s0014-5793(99)01763-9. [DOI] [PubMed] [Google Scholar]
- 53.Gnad F., Ren S., Cox J., Olsen J.V., Macek B., Oroshi M., Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hervouet E., Lalier L., Debien E., Cheray M., Geairon A., Rogniaux H., Loussouarn D., Martin S.A., Vallette F.M., Cartron P.-F. Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells. PLos ONE. 2010;5:e11333. doi: 10.1371/journal.pone.0011333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schermelleh L., Haemmer A., Spada F., Rosing N., Meilinger D., Rothbauer U., Cardoso M.C., Leonhardt H. Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation. Nucleic Acids Res. 2007;35:4301–4312. doi: 10.1093/nar/gkm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Espada J., Ballestar E., Fraga M.F., Villar-Garea A., Juarranz A., Stocker J.C., Robertson K.D., Fuks F., Esteller M. Human DNA methyltransferase 1 is required for maintenance of the histone H3 modification pattern. J. Biol. Chem. 2004;279:37175–37184. doi: 10.1074/jbc.M404842200. [DOI] [PubMed] [Google Scholar]
- 57.Karpf A.R., Matsui S.-I. Genetic disruption of cytosine DNA methyltransferase enzymes induces chromosomal instability in human cancer cells. Cancer Res. 2005;65:8635–8639. doi: 10.1158/0008-5472.CAN-05-1961. [DOI] [PubMed] [Google Scholar]
- 58.Sharif J., Muto M., Takebayashi S.-I., Suetake I., Iwamatsu A., Endo T.A., Shinga J., Mizutani-Koseki Y., Toyoda T., Okamura K., et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 59.Bostick M., Kim J.K., Esteve P.-O., Clark A., Pradhan S., Jacobsen S.E. UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science. 2007;317:1760–1764. doi: 10.1126/science.1147939. [DOI] [PubMed] [Google Scholar]
- 60.Ziv Y., Bielopolski D., Galanty Y., Lukas C., Taya Y., Schultz D.C., Lukas J., Bekker-Jensen S., Bartek J., Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM-and KAP-1 dependent pathway. Nat. Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
- 61.Robertson K.D., Ait-Si-Ali S., Yokochi T., Wade P.A., Jones P.L., Wolffe A.P. DNMT1 forms a complex with Rb, E2F1, and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- 62.Smith S.S. Stalling of DNA methyltransferase in chromosome stability and chromosome remodelling. Int. J. Mol. Med. 1998;1:147–156. [PubMed] [Google Scholar]
- 63.Cuozzo C., Porcellini A., Angrisano T., Morano A., Lee B., DiPardo A., Messina S., Iuliano R., Fusco A., Santillo M.R., et al. DNA damage, homology-directed repair, and DNA methylation. PLoS Genet. 2007;3:e110. doi: 10.1371/journal.pgen.0030110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.O'Hagan H.M., Mohammad H.P., Baylin S.B. Double strand breaks can initiate gene silencing and SIRT1-dependent onset of DNA methylation in an exogenous promoter CpG island. PLoS Genet. 2008;4:e1000155. doi: 10.1371/journal.pgen.1000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shrivastav M., DeHaro L.P., Nickoloff J.A. Regulation of the DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 66.Fatemi M., Hermann A., Pradhan S., Jeltsch A. The activity of the murine DNA methyltransferase Dnmt1 is controlled by interaction of the catalytic domain with the N-terminal part of the enzyme leading to an allosteric activation of the enzyme after binding to methylated DNA. J. Mol. Biol. 2001;309:1189–1199. doi: 10.1006/jmbi.2001.4709. [DOI] [PubMed] [Google Scholar]
- 67.Helt C.E., Wang W., Keng P.C., Bambara R.A. Evidence that DNA damage detection machinery participates in DNA repair. Cell Cycle. 2005;4:529–532. doi: 10.4161/cc.4.4.1598. [DOI] [PubMed] [Google Scholar]
- 68.Kumagai A., Dunphy W.G. Repeated phophopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat. Cell Biol. 2003;5:161–165. doi: 10.1038/ncb921. [DOI] [PubMed] [Google Scholar]
- 69.Bermudez V.B., Lindsey-Boltz L.A., Cesare A.J., Maniwa Y., Griffith J.D., Hurwitz J., Sancar A. Loading of the human 9-1-1 checkpoint complex onto DNA by the checkpoint clamp loader hRAD17-replication factor C complex in vitro. Proc. Natl Acad. Sci. USA. 2003;100:1633–1638. doi: 10.1073/pnas.0437927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scorah J., Dong M.-Q., Yates J.R., III, Scott M., Gillespie D., McGowan C.H. A conserved proliferating cell nuclear antigen-interacting protein sequence in Chk1 is required for checkpoint function. J. Biol. Chem. 2008;283:17250–17259. doi: 10.1074/jbc.M800369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen T., Hevi S., Gay F., Tsujimoto N., He T., Zhang B., Ueda Y., Li E. Complete inactivation of DNMT1 leads to mitotic catastrophe in human cancer cells. Nat. Genet. 2007;39:391–396. doi: 10.1038/ng1982. [DOI] [PubMed] [Google Scholar]
- 72.Brown K.D., Robertson K.D. DNMT1 knockout delivers a strong blow to genome stability and cell viability. Nat. Genet. 2007;39:289–290. doi: 10.1038/ng0307-289. [DOI] [PubMed] [Google Scholar]
- 73.Geiman T.M., Sankpal U.T., Robertson A.K., Chen Y., Mazumdar M., Heale J.T., Schmiesing J.A., Kim W., Yokomori K., Zhao Y., et al. Isolation and characterization of a novel DNA methyltransferase complex linking DNMT3B with components of the mitotic chromosome condensation machinery. Nucleic Acids Res. 2004;32:2716–2729. doi: 10.1093/nar/gkh589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ling Y., Sankpal U.T., Robertson A.K., McNally J.G., Karpova T., Robertson K.D. Modification of de novo DNA methyltransferase 3a (Dnmt3a) by SUMO-1 modulates its interaction with histone deacetylases (HDACs) and its capacity to repress transcription. Nucleic Acids Res. 2004;32:598–610. doi: 10.1093/nar/gkh195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jin B., Yao B., Li J.-L., Fields C.R., Delmas A.L., Liu C., Robertson K.D. DNMT1 and DNMT3B modulate distinct polycomb-mediated histone modifications in colon cancer. Cancer Res. 2009;69:7412–7421. doi: 10.1158/0008-5472.CAN-09-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data