Activated GTPase movement on an RNA scaffold drives cotranslational protein targeting (original) (raw)
. Author manuscript; available in PMC: 2013 Jun 13.
Published in final edited form as: Nature. 2012 Dec 13;492(7428):271–275. doi: 10.1038/nature11726
Abstract
Roughly one third of the proteome is initially destined for the eukaryotic endoplasmic reticulum or the bacterial plasma membrane1. The proper localization of these proteins is mediated by a universally conserved protein targeting machinery, the signal recognition particle (SRP), which recognizes ribosomes carrying signal sequences2–4 and, via interactions with the SRP receptor5,6, delivers them to the protein translocation machinery on the target membrane7. The SRP is an ancient ribonucleoprotein particle containing an essential, elongated SRP RNA whose precise functions have remained elusive. Here, we used single molecule fluorescence microscopy to demonstrate that the SRP-receptor GTPase complex, after initial assembly at the tetraloop end of SRP RNA, travels over 100 Å to the distal end of this RNA where rapid GTP hydrolysis occurs. This movement is negatively regulated by the translating ribosome and, at a later stage, positively regulated by the SecYEG translocon, providing an attractive mechanism to ensure the productive exchange of the targeting and translocation machineries at the ribosome exit site with exquisite spatial and temporal accuracy. Our results show that large RNAs can act as molecular scaffolds that enable the facile exchange of distinct factors and precise timing of molecular events in a complex cellular process; this concept may be extended to similar phenomena in other ribonucleoprotein complexes.
Cotranslational protein targeting face fundamental challenges in both spatial and temporal coordination. Spatially, both the SRP2–4 and SecYEG (or Sec61p) translocon7 contact the L23 ribosomal protein and the signal sequence, raising puzzling questions about how the translating ribosome is transferred from the targeting to translocation machinery. Temporally, guanosine-5′-triphosphate (GTP) hydrolysis by the SRP-SRP receptor complex, which drives its irreversible disassembly8, must be accurately timed during cargo delivery and unloading to avoid abortive reactions9. Such accurate spatial and temporal coordination is required in all protein targeting pathways, but its underlying molecular mechanism is not understood. Here, single molecule experiments reveal large-scale rearrangements in the SRP, providing a unifying molecular mechanism to explain how such coordination is achieved during co-translational protein targeting.
The bacterial SRP is comprised of an SRP54 protein subunit, Ffh, and a 114-nucleotide SRP RNA1. Ffh contains two domains connected by a flexible linker: a methionine-rich M-domain, which recognizes the signal sequence10 and binds the SRP RNA11, and a GTPase, NG-domain that interacts with a homologous NG-domain in the SRP receptor, FtsY5,6 (Fig. 1a). The SRP RNA is a universally conserved and essential SRP component, but its precise roles are not completely understood. Most previous work12–15 focused on the GGAA tetraloop that caps one end of this RNA, which accelerates the initial SRP-FtsY assembly by electrostatically interacting with FtsY13. These findings, however, do not explain why the SRP RNA has a conserved elongated structure16. Valuable clues come from a recent crystal structure that found the Ffh-FtsY GTPase complex at another docking site near the 5′,3′-distal end of this RNA, where mutations disrupt GTPase activation17 (Fig. 1a, distal state). This posits an attractive hypothesis in which the Ffh-FtsY GTPase complex, after initial assembly near the tetraloop12–15, can relocalize to the distal site of the SRP RNA ~100Å away17. Nevertheless, no functional evidence for the relocalization is available, nor are the importance, timing, mechanism, and regulation of such a large-scale movement understood.
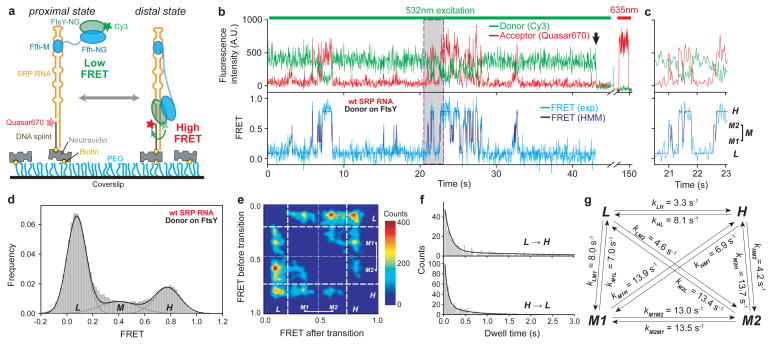
Figure 1. smFRET-TIRF microscopy reveals dynamic movements of the SRP-FtsY complex on the SRP RNA.
a, smFRET setup for the SRP-FtsY complex. FtsY C345 is labeled with Cy3. The 5′-end of the DNA splint (2 nt from the 3′-end of SRP RNA) is labeled with Quasar670. b, Fluorescent signals (upper) and FRET trajectory (lower) of the SRP-FtsY complex in GMPPNP. Hidden Markov Modeling (HMM) of the FRET trajectory is in navy. The arrow denotes the bleaching of Cy3, after which Quasar670 was excited using a 635nm laser to confirm the presence of the complex. c, Magnification of the grey box in b to depict the four FRET states resolved by HMM. d, smFRET histogram depicting the distribution of molecules in different states. In M state, the M1 and M2 states are binned together. e, Transition density plot (TDP) for the GTPase movements. f, Analysis of the transition kinetics between L and H states. Exponential fits of the data gave the transition rate constants in g.
To address these questions, we used single molecule fluorescence resonance energy transfer (smFRET) and total internal reflection fluorescence (TIRF) microscopy to directly detect conformational dynamics of individual SRPs18,19. Migration of the SRP-FtsY GTPase complex on the SRP RNA was tracked using FRET between a donor (Cy3) attached to the FtsY NG-domain and an acceptor (Quasar670) labeled near the RNA distal end (Fig. 1a). Stable SRP-FtsY complexes, formed with the non-hydrolyzable GTP analogue 5′-guanylyl-imidodiphosphate (GMPPNP), displayed rapid transitions among multiple FRET states (Fig. 1b,c). A low FRET (~0.1) state, L, was assigned to the proximal state in which the GTPase complex resides near the SRP RNA tetraloop13. A high FRET (~0.8) state, H, was attained ~20% of the time and assigned to the distal state in which the GTPase complex stably docks at the distal site, as verified below. Cy3 attached to the Ffh NG-domain showed similar transitions but with a lower FRET value in the **H**-state (Supplementary Fig. 2a,b), consistent with Ffh being further from the distal site than FtsY17. These results directly demonstrate dynamic movements of the SRP-FtsY GTPase complex on the SRP RNA that span over 100Å.
We used Hidden Markov Modeling (HMM)-based statistical analyses to determine the most likely sequence of FRET transitions20. This revealed an ensemble of additional states with intermediate FRET values (0.3–0.6; M1 and M2) and extremely short lifetimes (Fig. 1b–d and Supplementary Fig. 2b–g, 3a–c), representing alternative binding modes of the GTPase complex on the SRP RNA. The transition information was pooled into a transition density map (TDP) that describes the number of distinct FRET states, their FRET values, and their transition frequencies (Fig. 1e and Supplementary Fig. 2h). Additionally, the kinetics of FRET transitions was obtained from dwell time analyses (Fig. 1f–g and Supplementary Fig. 2i–o,3d–h). While molecules leaving L rapidly transitioned to all the other states, the H state had a longer lifetime than M1 and M2 and was hence more populated (Fig. 1d), indicating more stable docking of the GTPase complex in this state. 58% of transitions to H occurred directly from L, whereas molecules in the intermediate FRET states transitioned primarily back to L (Fig. 1e and Supplementary Fig. 2h). Thus, correct docking at the RNA distal site requires extensive searching that involves frequent trial-and-error.
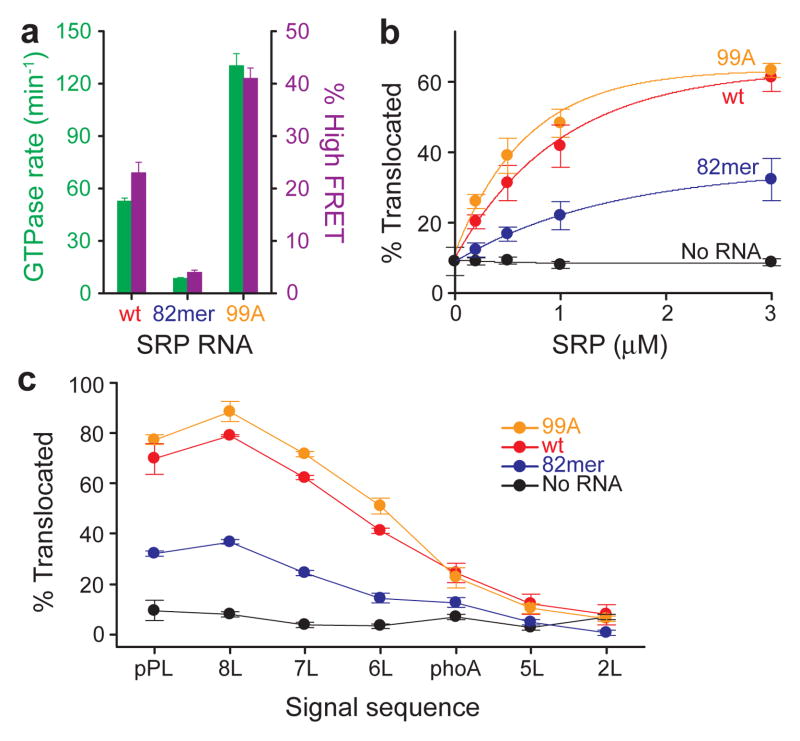
To test whether the H state is responsible for GTPase activation, we isolated mutant RNAs that specifically perturb the distal docking site. The 82mer RNA, which lacks this site17, reduced GTPase activation 6-fold, whereas a ‘superactive’ mutant, 99A, enhanced GTP hydrolysis 2.5-fold (Fig. 2a, green bars and Supplementary Figs. 1b,4a). The GTPase activity of these mutants quantitatively correlated with their efficiency of reaching the H state (Fig. 2a and Supplementary Figs. 5,6), strongly suggesting that activated GTP hydrolysis occurs at the RNA distal site.
Figure 2. The distal site of SRP RNA is crucial for GTPase activation and protein targeting.
a, Correlation between GTPase rate constants in the SRP-FtsY complex (green bars) and the frequency of reaching the high FRET state (purple bars) for wildtype (red), 82mer (blue), and 99A SRP RNA (orange). Data represent mean±s.d. (n=5). b–c, Co-translational targeting and translocation of pPL (b) and its signal sequence variants (c) mediated by the wildtype and mutant SRPs. Color codings are the same as in a. Reactions in the absence of SRP RNA are in black. Data represent mean±s.d. (n=3).
To test the importance of the RNA distal site in protein targeting, we measured the ability of SRP and FtsY to deliver a model substrate, preprolactin (pPL), to ER microsomes21. Translocation of pPL results in cleavage of its signal sequence, allowing the targeting and translocation efficiency to be quantified (Supplementary Fig. 4b). Further, the specificity of targeting was tested using pPL variants in which the signal sequence is systematically varied9 (Supplementary Fig. 4d). Mutant 82mer RNA significantly reduced the targeting of correct substrates (wildtype-, 8L- and 7L-pPL; Fig. 2b,c and Supplementary Figs. 4c,e). In contrast, the superactive 99A RNA targeted these substrates more efficiently than the wildtype SRP, without compromising the discrimination against incorrect substrates (Figs. 2b,c and Supplementary Figs. 4c,e). Thus the SRP RNA distal site, though not essential for cell survival11, does enhance efficient and accurate co-translational protein targeting.
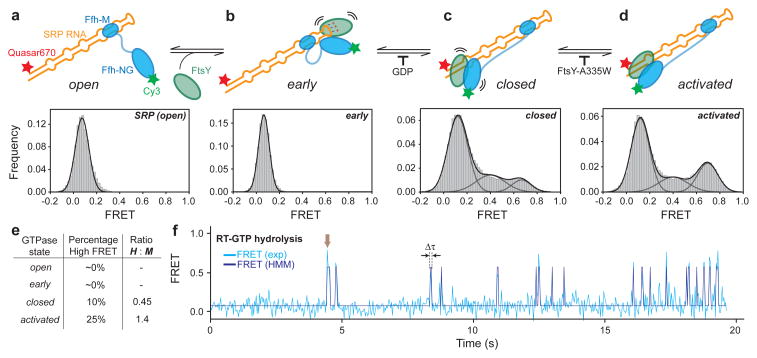
SRP and FtsY undergo an unusual GTPase cycle, driven by multiple conformational rearrangements in their heterodimer that culminate in GTPase activation (Figs. 3a–d, cartoon)22–24. We asked how these rearrangements within the GTPase complex drive its global movements on the SRP RNA, using conditions that block the GTPase cycle at distinct stages22,23. SRP by itself exhibited no movements on the RNA (Fig. 3a and Supplementary Fig. 7a). Recruitment of FtsY begins with a transient early intermediate, which lacks close contacts between the G-domains and hence can be isolated by leaving out GTP analogues12,24. No GTPase movement was observed at this stage either (Fig. 3b and Supplementary Fig. 7b). Subsequently, GTP-dependent rearrangements give a stable closed complex, which lacks optimal positioning of the catalytic loops and can be isolated by a mutation, FtsY(A335W), in the catalytic loop (Fig. 3c,d)22,23. Although GTPase movements were observed in the closed complex, most of them only reached M1 and M2 but did not significantly populate the H state (Figs. 3c–e and Supplementary Figs. 7c,d). Thus, GTP-induced rearrangements within the NG-domain complex drive its global movements on the SRP RNA. Moreover, stable GTPase docking at the RNA distal site requires optimal positioning of the catalytic loops, explaining why mutants that block GTPase activation, such as FtsY(A335W), severely impair protein targeting21.
Figure 3. Conformational rearrangements within the SRP-FtsY GTPase complex drive its movement to the RNA distal site.
a–d, smFRET histograms of free SRP in the open state (a) and of the SRP-FtsY complex in the early (b), closed (c), and activated (d) states. Conditions for isolating each conformational state are described in the text and Methods. e, Summary of the FRET distributions. f, A representative smFRET trajectory of the complex incubated in GTP. The arrow denotes a burst of high FRET that results from GTPase docking at the distal site terminated by rapid GTP hydrolysis driving complex disassembly. Δτ denotes the duration of the high FRET burst.
If the GTPase complex only transiently reaches the SRP RNA distal site where GTPase activation occurs, previous ensemble measurements8 would have significantly underestimated the hydrolysis rate. We therefore performed real-time GTPase assays using the smFRET setup. If GTP hydrolysis at the distal site, which drives irreversible SRP-FtsY dissociation, occurred faster than their return to the proximal state, we would observe high FRET ‘bursts’ with GTP instead of the reversible transitions with GMPPNP. This was indeed observed (Fig. 3f). The duration between these bursts has a rate constant (0.59s−1; Supplementary Fig. 8b) expected for rearrangement to the activated complex (~1s−1)12 and is similar to the ensemble GTPase rate (0.7s−1)8, strongly suggesting that the latter is rate-limited by GTPase movement to the RNA distal site. The duration of the high FRET bursts includes GTP hydrolysis and subsequent SRP-FtsY disassembly and exhibits a rate constant of 7.1s−1 (Supplementary Fig. 8a), providing a lower limit for the actual hydrolysis rate and is at least 10-fold faster than ensemble measurements8.
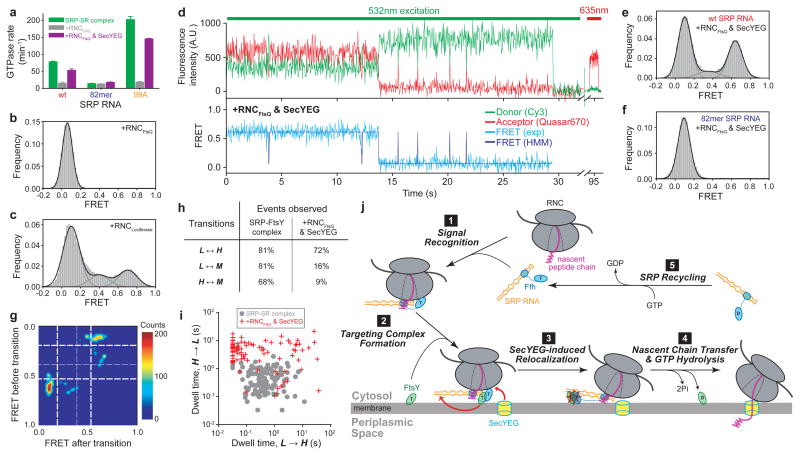
These results also show that GTP drives almost irreversible movement of the GTPases to the RNA distal site, necessitating accurate control of the timing of this movement. Indeed, ribosome-nascent chain complexes (RNC or cargo) delay GTPase activation in the SRP-FtsY complex9,23 (Fig. 4a, wt). This effect, termed ‘pausing’, prevents premature GTP hydrolysis and is essential for ensuring the efficiency and specificity of the SRP pathway9. We asked whether the RNC negatively regulates the GTPase movement to the SRP RNA distal site. RNCFtsQ, which carries an obligate SRP substrate FtsQ, completely abolished the GTPase movements on the RNA (Fig. 4b and Supplementary Figs. 9a–e). This is specific to the correct cargo, as RNCLuciferase, which contains no signal sequence, exerted no effects (Fig. 4c and Supplementary Fig. 9f). Further, GTP hydrolysis in the presence of RNC is no longer affected by mutations in the RNA distal end (Fig. 4a and Supplementary Figs. 10a,b), but is still reduced by a mutation in FtsY active site22 (Supplementary Figs. 10d,e). These results demonstrate that correct cargos stabilize the GTPase complex in the proximal state and prevent its relocalization to the RNA distal site, thus exerting the ‘pausing’ effect.
Figure 4. RNC and SecYEG regulate GTPase movements on the SRP RNA.
a, Effect of RNCFtsQ (grey bars) and SecYEG (purple bars) on the GTPase activity of the SRP-FtsY complex assembled with the wildtype, 82mer, and 99A SRP RNA. Data represent mean±s.d. (n=3). b–c, smFRET histograms of the SRP-FtsY complex bound to RNCFtsQ (b) or RNCLuciferase (c). d, Fluorescent signals (upper) and FRET trajectory (lower) of the RNCFtsQ-SRP-FtsY complex in the presence of SecYEG. Color coding is the same as in Fig. 1b. e–f, smFRET histograms of the RNCFtsQ-SRP-FtsY complex in the presence of SecYEG with the wildtype (e) or 82mer (f) SRP RNA. g, TDP of the GTPase movements in the presence of RNCFtsQ and SecYEG. h, Summary of the percentage of molecules that exhibit the specified transitions. In the presence of RNCFtsQ and SecYEG, transitions to intermediate FRET states are significantly reduced. i, Scatter plot of the transition dwell times of individual molecules in the absence (grey circles) and presence of RNCFtsQ and SecYEG (red crosses). j, Model for the role of the SRP RNA-mediated GTPase relocalization in co-translational protein targeting.
On the target membrane, RNC must be transferred from the targeting to the translocation machinery. The mechanism of this transfer and its timing have remained long-standing challenges. To test whether the translocon helps regulate these events, we added the SecYEG complex to the RNCFtsQ-SRP-FtsY complex (Supplementary Fig. 11a). SecYEG restored the high FRET state (Fig. 4d,e). It also reversed the cargo-induced ‘pausing’ and restored efficient GTP hydrolysis (Fig. 4a and D.A. & S.S., manuscript under revision). Neither effect was observed with DDM alone (Supplementary Fig. 11b) nor with mutant 82mer RNA (Fig. 4f and Supplementary Fig. 10c) or FtsY(A335W) (Supplementary Fig. 11c). Thus, SecYEG drives productive docking of the GTPase complex at the RNA distal site and thus re-activates GTP hydrolysis.
How does SecYEG restore the GTPase movements? Although SecYEG could simply remove the RNC from the SRP-FtsY complex, the following strongly suggests that this is not the case. Compared to the SRP-FtsY complex alone, GTPase movements in the presence of RNCFtsQ and SecYEG displayed a distinct pattern, characterized by fewer transitions to intermediate FRET states, more frequent docking (Figs. 4g,h) and longer dwell times in the H state (Figs. 4d,i). These SecYEG-induced changes were not observed without RNC (Supplementary Figs. 11d,e). To directly test if RNC remains on the targeting complex, we labeled the RNC with Alexa647, which was found to colocalize with labeled SRP (Supplementary Fig. 12). These colocalized spots remain after incubation with SecYEG (Supplementary Figs. 12c,d), indicating that RNC was not displaced by SecYEG. These data imply that SecYEG forms a quaternary complex with RNC, SRP, and FtsY; which could represent a transient intermediate in the targeting and translocation reaction. These results also suggest that SecYEG drives the GTPase movement via two mechanisms: (i) displacing the GTPase complex from the proximal site, as indicated by the reappearance of H state even with RNC present (cf. Fig. 4d vs. Supplementary Fig. 9b); and (ii) prolonging productive docking at the RNA distal site (Fig. 4i). Finally, nonproductive movements to intermediate FRET states are minimized with RNC and SecYEG present (Fig. 4g,h). Considering the size of SRP RNA relative to the ribosome, the RNC possibly masks nonproductive GTPase docking sites on the SRP RNA, which could also explain the conserved length of this RNA.
In summary, we demonstrate that the SRP RNA provides a molecular scaffold that mediates large-scale movements of the SRP-FtsY complex, which are tightly regulated by the GTPase cycle of SRP and FtsY, the translating ribosome, and the SecYEG translocon. Together with previous studies, we propose a molecular model for co-translational protein targeting (Fig. 4j). Upon cargo recognition (step 1), the SRP RNA tetraloop is optimally positioned adjacent to the Ffh NG-domain, allowing efficient recruitment of FtsY near the ribosome exit site12–15 (step 2). GTP-induced rearrangements primes23,25 but is insufficient to release the SRP-FtsY GTPase complex from the vicinity of ribosome due to the RNC’s ‘pausing’ effect. SecYEG is required to drive GTPase relocalization to the SRP RNA distal site (step 3). This vacates the ribosome exit site and allows SecYEG to initiate contacts with L23, thus enabling the coordinated transfer of RNC from the targeting to translocation machinery (step 4). Concomitantly, GTPase docking at the RNA distal site triggers rapid GTP hydrolysis, driving the disassembly and recycling of SRP and FtsY (step 4–5). This provides an attractive mechanism to allow the concerted exchange of SRP and SecYEG at the ribosome and the precise timing of GTP hydrolysis, thus minimizing abortive reactions due to premature SRP-FtsY disassembly or nonproductive loss of cargo.
Nucleic acid-mediated protein movement is a widespread phenomenon and has been observed with the spliceosome26, helicases27,28, and type I restriction endonucleases29,30. Our results here enrich these findings and further suggest that large RNA molecules can provide useful molecular scaffolds to coordinate multiple protein interactions and large-scale protein rearrangements, thus enabling productive exchange of different factors and precise timing of molecular events in a cellular pathway. This may provide general principles for understanding similar phenomena in other ribonucleoprotein particles.
Methods
Plasmids
Plasmids for in vivo expression of Ffh, full-length FtsY, and SRP RNA and for in vitro transcription of FtsQ, luciferase, and pPL and its signal sequence variants have been described8,9,31. The pEK20 construct for SecYEG expression was a kind gift of Arnold Driessen32. Plasmids for mutant SRP RNAs and mutant proteins were constructed using the QuikChange mutagenesis protocol (Stratagene) following manufacturer’s instructions. The plasmid for in vitro transcription of Hammerhead-SRP RNA-HDV was a generous gift from Adrian Ferre-D’Amare33. The hammerhead coding sequence was removed and the 5′-end of SRP RNA was extended using the QuikChange mutagenesis protocol (Stratagene) to make in vitro transcription constructs for smRNA.
Protein preparations
Wildtype and single cysteine mutants of Ffh and FtsY were expressed and purified as described previously8. Briefly, Ffh expression was induced in logarithmically growing BL21(DE3)pLysE cells with 1 mM IPTG. The soluble fraction from lysed cells were purified by cation-exchange chromatography on the SP Sepharose Fast Flow resin (GE Healthcare) using a gradient of 0.25–1 M NaCl, and was further purified by gel filtration chromatography on the Superose12 column (Amersham Biosciences). His6-tagged full-length FtsY was expressed in BL21(DE3)pLysS cells by induction with 0.5 mM IPTG in logarithmically growing cells. The soluble fraction from lysed cells was purified by anion exchange chromatography using Q Sepharose Fast Flow resin (GE Healthcare) with a gradient of 150–500 mM NaCl, followed by affinity purification using Ni-NTA resin (Qiagen). For GTPase assays, FtsY was further purified by anion exchange chromatography on the MonoQ column (Amersham Biosciences) using a gradient of 150 –350 mM NaCl. All proteins were exchanged into SRP buffer (50 mM KHEPES, pH 7.5, 150 mM KOAc, 2 mM Mg(OAc)2, 2 mM DTT, 0.01% Nikkol) before use.
Detergent-solubilized SecYEG was expressed in BL21(DE3) cells with 0.5 mM IPTG and was purified following published procedures32,34,35. Cells were lysed by sonication and the membranes were collected by ultracentrifugation. SecYEG was extracted and purified by cation exchange chromatography on the SP-Sepharose Fast Flow resin (GE Healthcare) followed by affinity purification with Ni-NTA (Qiagen). DDM (Affimetrix) was used for purification of solubilized SecYEG, which has been shown to be fully functional in binding RNC (D.A. & S.S. manuscript)36, in mediating nascent peptide translocation36, and stimulating SecA ATPase activity37.
Fluorescence labeling
Single cysteine mutants of Ffh and FtsY were labeled with Cy3-maleimide (GE Healthcare) as described12. Protein concentration during labeling was 50 – 100 μM, and the dye was in 10-fold molar excess. Labeling reaction was carried out in buffer A (50 mM KHEPES, pH 7.0, 300 mM NaCl, 2 mM EDTA, 10% glycerol) with gentle shaking at room temperature for 2 hours. Unconjugated dyes were removed by gel filtration chromatography using Sephadex G-25 resin (Sigma). Mass-spectrometry confirmed >95% labeling efficiencies. Fluorescence labeling and modifications of the SRP RNA for surface immobilization (Fig. 1a and Supplementary Fig. 1b) did not affect the activity of SRP and FtsY (Supplementary Fig. 1d).
Fluorescent DNA probes for hybridization with the mRNA on RNC were prepared by incubating NH2-modified DNA oligo (IDT) with a 10-fold excess of Alexa Fluor 647 carboxylic acid succinimidyl ester (Invitrogen) for an hour at 37 °C. Excess dyes were removed by HPLC.
RNA preparation
Wildtype SRP RNA was expressed in vivo and purified as described8. SRP RNAs for smFRET experiments (smRNA; Supplementary Fig. 1b) were prepared by in vitro transcription using T7 polymerase according to the Megascript protocol (Ambion). The 3′-end of SRP RNA coding sequence was fused to that of an HDV ribozyme (sequence: GGG CGG CAT GGT CCC AGC CTC CTC GCT GGC GCC GCC TGG GCA ACA TTC CGA GGG GAC CGT CCC CTC GGT AAT GGC GAA TGG GAC C). Self-cleavage of the HDV ribozyme occurred during in vitro transcription to generate a homogeneous 3′-end of the SRP RNA. Purified smRNA was annealed to a complementary DNA splint by the following procedures: (1) heat the TE buffer (10 mM Tris-HCl, pH 7.0, 2 mM EDTA) containing 10 μM DNA and 20 μM smRNA for 5 minutes at 75 °C. (2) Gradually cool to 50 °C over a period of 30 minutes. (3) Add 12 mM MgCl2 to the mixture. (4) Gradually cool to room temperature over a period of 30 minutes. The annealed DNA-smRNA hybrids were stored at −80 °C.
Messenger RNAs for in vitro translation were generated by in vitro transcription using T7 (for RNC prep) or SP6 (for targeting assays) polymerase following the Megascript protocol (Ambion).
Preparation of RNCs
Synchronized RNCs with defined nascent chain length and sequence were prepared as described previously31. In short, mRNAs encoding a Strep3 tag at the N-terminus, the first 74 amino acids of FtsQ or luciferase, and a SecM translation stall sequence at the C-terminus were translated by S100 extract as described31. RNC from the translation mixture was purified by affinity chromatography using the streptactin resin (IBA), collected by ultracentrifugation, and re-dissolved in SRP buffer and stored at −80 °C. RNCs used for GTPase assay were further purified by ultracentrifugation and fractionation on a 10 – 50% sucrose gradient as described31.
RNCFtsQ was fluorescently labeled by incubation with fluorescent DNA probes complementary to the mRNA for 3 hours at room temperature. Labeled RNC was isolated by ultracentrifugation and re-dissolved in SRP buffer.
Single molecule instrument
Objective-type TIRF microscope was home-built based on an Olympus IX-81 model as described19. Green (532nm) and red (635nm) lasers were focused in a 100x oil immersed objective. Scattering light was removed by 560 nm and 660 nm long pass filters (Chroma) for the green and red lasers, respectively. Cy3 and Quasar670 signals were split by a dichroic mirror and simultaneously focused onto the Ixon 897 camera (Andor) through DV2 Dualview (Photometrics). Data were recorded at 30 ms time resolution.
PEGylated slides and coverslips
PEGylated slides and coverslips were prepared based on an existing protocol19. Briefly, quartz slides and coverslips were treated sequentially with 10% alconox, acetone, and 10 M KOH. The surfaces were then burnt with a propane torch to remove autofluorescence. Aminosilation reactions were carried out in methanol with 5% (v/v) HOAc and 1% (v/v) aminopropylsilane. PEGylation reactions were carried out in 100 mM NaHCO3 buffer containing 20% (w/v) PEG and 0.6% (w/v) biotin-PEG. PEGylated slides and coverslips were stored in vacuum at −20 °C and assembled into flow chambers before use.
Single molecule assay
To remove aggregates, all protein samples were ultracentrifuged at 100,000 rpm (Optima TLX, Beckman Coulter) for an hour before use. PEGylated slides and coverslips were assembled to form a flow chamber. 0.2 mg/ml neutravidin was applied to the chamber and incubated for 10 minutes before flowing in molecules of interest.
SRP complexes were assembled in SRP buffer under the following conditions. SRP-FtsY complex with labeled Ffh: 1 μM DNA-smRNA hybrid, 2 μM Ffh-Cy3, 5 μM FtsY, 100 μM GMPPNP. SRP-FtsY complex with labeled FtsY: 1 μM DNA-smRNA hybrid, 2 μM Ffh, 3 μM FtsY-Cy3, 100 μM GMPPNP. RNC-SRP-FtsY complexes: 200 nM DNA-smRNA hybrid, 400 nM Ffh-Cy3, 500 nM RNCFtsQ or 1 μM RNCLuciferase, 1 μM FtsY, 100 μM GMPPNP. SecYEG solubilized in 0.02% DDM was added to RNCFtsQ-SRP-FtsY complex at 10 μM. The samples were then diluted to 50 pM in imaging buffer (SRP buffer supplemented with 0.4% glucose and 1% Gloxy in Trolox), flowed onto the sample chamber and incubated for 5 minutes before imaging. Movies were recorded at 30 ms intervals for up to 3 minutes until most fluorescent molecules were photobleached. A red laser was applied at the end of the movie to confirm the presence of immobilized SRP.
Data analysis
Single molecule data were processed by scripts written in IDL and Matlab. Briefly, fluorescent peaks in the images were identified and traced throughout the trajectory. Traces that showed a single donor bleaching event were used for data analysis. Hidden Markov Modeling was calculated using the HaMMy program20. Transition density map was generated by TDP program20 using the output from HaMMy. FRET histograms were generated using home-written script in Matlab19. Transition kinetics between different states was obtained by exponential fits to dwell time histograms. Two-dimensional scatter plots of the average dwell time of individual molecules during transitions were generated using the home-written script in Matlab.
GTPase assay
GTPase rate constants were determined using a well-established GTPase assay8. In general, reactions contained 100 nM Ffh, 200 nM SRP RNA, 100 μM GTP (doped with γ-32P-GTP), varying concentrations of FtsY, and 250 nM RNCFtsQ and 10 μM SecYEG where applicable. Reactions were quenched with 0.75 M KH2PO4 (pH 3.3) at different time points, separated by thin layer chromatography (TLC), and quantified by autoradiography.
Translocation assay
Assays for co-translational protein targeting and translocation were carried out as described21,38. Reactions contained 10 μL in vitro translation mixtures synthesizing 35S-methionine labeled pPL or pPL signal sequence variants, to which 200 nM Ffh, 333 nM wildtype or mutant SRP RNA, 300 nM FtsY and 0.5 eq/μL of salt washed, trypsin digested microsomal membrane was added to a total volume of 15 μL. Reactions were analyzed by SDS-PAGE followed by autoradiography.
Supplementary Material
1
Acknowledgments
We thank N. Ban and members of the Shan group for helpful comments on the manuscript, C. Richards, L. Cai, T. Zhiyentayev, K. Lee, and R. Zhou for help with RNC labeling and the instrument and software setup, and C.L. Guo, S. Kou, and H. Lester for helpful discussions. This work is supported by NIH grant GM078024 to S.-o.S., an NIH instrument supplement to grant GM45162 to D.C. Rees, and Caltech matching fund 350270 for the single molecule instruments. S.-o.S. was supported by the Beckman Young Investigator award, the David and Lucile Packard Fellowship in science and engineering, and the Henry Dreyfus teacher-scholar award. T.H. was supported by NSF Physics Frontiers Center program (08222613) and NIH grant GM065367.
Footnotes
Author contributions
K.S., S.A., T.H., and S.-o.S. conceived the experiments. K.S. purified and labeled Ffh, FtsY, DNA, RNA, and RNC. D.A. purified SecYEG and performed the GTPase assay in Fig. 4a. K.S. and S.A. carried out smFRET measurements under the direction of T.H. K.S. and S.A. analyzed the data. K.S. and S.-o.S. wrote the paper with inputs from all the authors.
References
- 1.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 2.Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B. Distinct modes of signal recognition particle interaction with the ribosome. Science. 2002;297:1345–1348. doi: 10.1126/science.1072366. [DOI] [PubMed] [Google Scholar]
- 3.Halic M, et al. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 4.Schaffitzel C, et al. Structure of the E-coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 5.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Egea PF, et al. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 7.Becker T, et al. Structure of monomeric yeast and mammalian Sec61 complexes interacting with the translating ribosome. Science. 2009;326:1369–1373. doi: 10.1126/science.1178535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peluso P, Shan SO, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of ffh and FtsY. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 9.Zhang X, Rashid R, Wang K, Shan SO. Sequential checkpoints govern substrate selection during cotranslational protein targeting. Science. 2010;328:757–760. doi: 10.1126/science.1186743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janda CY, et al. Recognition of a signal peptide by the signal recognition particle. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X, Kung S, Shan SO. Demonstration of a multistep mechanism for assembly of the SRP.SRP receptor complex: Implications for the catalytic role of SRP RNA. J Mol Biol. 2008;381:581–593. doi: 10.1016/j.jmb.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen K, Shan SO. Transient tether between the SRP RNA and SRP receptor ensures efficient cargo delivery during cotranslational protein targeting. Proc Natl Acad Sci U S A. 2010;107:7698–7703. doi: 10.1073/pnas.1002968107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen K, Zhang X, Shan SO. Synergistic actions between the SRP RNA and translating ribosome allow efficient delivery of the correct cargos during cotranslational protein targeting. RNA. 2011;17:892–902. doi: 10.1261/rna.2610411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Estrozi LF, Boehringer D, Shan S, Ban N, Schaffitzel C. Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nat Struct Mol Biol. 2011;18:88–90. doi: 10.1038/nsmb.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Althoff S, Selinger D, Wise JA. Molecular Evolution of Srp Cycle Components -Functional Implications. Nucleic Acids Research. 1994;22:1933–1947. doi: 10.1093/nar/22.11.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ataide SF, et al. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;331:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha T, et al. Probing the interaction between two single molecules: Fluorescence resonance energy transfer between a single donor and a single acceptor. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:6264–6268. doi: 10.1073/pnas.93.13.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan SO, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of the signal reception particle and its initiation of protein translocation. Journal of Cell Biology. 2007;178:611–620. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan SO, Stroud RM, Walter P. Mechanism of association and reciprocal activation of two GTPases. Plos Biology. 2004;2:1572–1581. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Schaffitzel C, Ban N, Shan SO. Multiple conformational switches in a GTPase complex control co-translational protein targeting. Proc Natl Acad Sci U S A. 2009;106:1754–1759. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, et al. Direct visualization reveals dynamics of a transient intermediate during protein assembly. Proc Natl Acad Sci U S A. 2011;108:6450–6455. doi: 10.1073/pnas.1019051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halic M, Gartmann M, Schlenker O, Mielke T, Pool MR, Sinning I, Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- 26.Hoskins AA, Gelles J, Moore MJ. New insights into the spliceosome by single molecule fluorescence microscopy. Curr Opin Chem Biol. 2011;15:864–870. doi: 10.1016/j.cbpa.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lohman TM, Bjornson KP. Mechanisms of helicase-catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 28.Yodh JG, Schlierf M, Ha T. Insight into helicase mechanism and function revealed through single-molecule approaches. Q Rev Biophys. 2010;43:185–217. doi: 10.1017/S0033583510000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan R. Structure and mechanism of multifunctional restriction endonucleases. Annu Rev Biochem. 1981;50:285–319. doi: 10.1146/annurev.bi.50.070181.001441. [DOI] [PubMed] [Google Scholar]
- 30.Murray NE. Type I restriction systems: sophisticated molecular machines (a legacy of Bertani and Weigle) Microbiol Mol Biol Rev. 2000;64:412–434. doi: 10.1128/mmbr.64.2.412-434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schaffitzel C, Ban N. Generation of ribosome nascent chain complexes for structural and functional studies. Journal of Structural Biology. 2007;158:463–471. doi: 10.1016/j.jsb.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 32.van der Sluis EO, Nouwen N, Driessen AJ. SecY-SecY and SecY-SecG contacts revealed by site-specific crosslinking. FEBS Lett. 2002;527:159–165. doi: 10.1016/s0014-5793(02)03202-7. [DOI] [PubMed] [Google Scholar]
- 33.Ferre-D’Amare AR, Doudna JA. Use of cis- and trans-ribozymes to remove 5′ and 3′ heterogeneities from milligrams of in vitro transcribed RNA. Nucleic Acids Res. 1996;24:977–978. doi: 10.1093/nar/24.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dalal K, Duong F. Reconstitution of the SecY translocon in nanodiscs. Methods Mol Biol. 2010;619:145–156. doi: 10.1007/978-1-60327-412-8_9. [DOI] [PubMed] [Google Scholar]
- 35.Van den Berg B, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 36.Mothes W, Jungnickel B, Brunner J, Rapoport TA. Signal sequence recognition in cotranslational translocation by protein components of the endoplasmic reticulum membrane. Journal of Cell Biology. 1998;142:355–364. doi: 10.1083/jcb.142.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. Embo Journal. 2003;22:4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Powers T, Walter P. Co-translational protein targeting catalyzed by the Escherichia coli signal recognition particle and its receptor. EMBO J. 1997;16:4880–4886. doi: 10.1093/emboj/16.16.4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1