Epigenetic Mechanisms in Developmental Alcohol-Induced Neurobehavioral Deficits (original) (raw)
Abstract
Alcohol consumption during pregnancy and its damaging consequences on the developing infant brain are significant public health, social, and economic issues. The major distinctive features of prenatal alcohol exposure in humans are cognitive and behavioral dysfunction due to damage to the central nervous system (CNS), which results in a continuum of disarray that is collectively called fetal alcohol spectrum disorder (FASD). Many rodent models have been developed to understand the mechanisms of and to reproduce the human FASD phenotypes. These animal FASD studies have provided several molecular pathways that are likely responsible for the neurobehavioral abnormalities that are associated with prenatal alcohol exposure of the developing CNS. Recently, many laboratories have identified several immediate, as well as long-lasting, epigenetic modifications of DNA methylation, DNA-associated histone proteins and microRNA (miRNA) biogenesis by using a variety of epigenetic approaches in rodent FASD models. Because DNA methylation patterns, DNA-associated histone protein modifications and miRNA-regulated gene expression are crucial for synaptic plasticity and learning and memory, they can therefore offer an answer to many of the neurobehavioral abnormalities that are found in FASD. In this review, we briefly discuss the current literature of DNA methylation, DNA-associated histone proteins modification and miRNA and review recent developments concerning epigenetic changes in FASD.
Keywords: FASD, Synaptic plasticity, Learning and memory, FAS, DNA and histone modification
1. Introduction
A change of gene expression evoked by mechanisms other than alterations in DNA sequence contributes to the advancement of cell and tissue development and significantly aids in the maintenance of a regular cell-specific function. Thus, dynamically regulated, epigenetic mechanisms differing throughout life and between various cell populations have been shown to contribute to the development of neuronal circuits and brain function. The major epigenetic alterations that are known to regulate gene expression in the brain are as follows: (1) DNA methylation of cytosine residues in promoter-rich cytosine phosphate guanine (CpG) islands; (2) acetylation, methylation and other covalent posttranslational modification (PMT) of DNA-associated histone proteins; (3) chromatin remodeling proteins that indirectly affect gene transcription; (4) pre-mRNA editing and splicing by non-coding, small nucleolar RNA (snoRNAs); (5) mRNA processing, translation, and stability of binding proteins, long non-coding RNAs (lncRNAs) and microRNAs (miRNAs); and (6) mRNA translation that is controlled by cellular signaling molecules. These epigenetic mechanisms can selectively respond to environmental factors and alter transcriptional machinery leading to altered physiological and pathological processes in the brain. An increasing number of environmental factors, such as maternal care, stress, chemical agents, drugs of abuse, alcohol and nutritional disparity [1,2,3,4,5,6,7,8], are undoubtedly shown to remodel epigenetics. However, the mechanism by which epigenetic remodeling events are linked to environmentally induced diseases is poorly understood. The review will provide a current understanding of epigenetic mechanisms and how alcohol exposure during development, as a type of environmental teratogen, can influence epigenetic programs to alter gene transcription and induce persistent early alcohol-associated abnormalities.
2. Epigenetic Mechanisms
2.1. DNA Methylation
Our modern understanding of epigenetic marks on DNA has provided insight into an additional regulatory system in the cell that operates to regulate when and where a particular gene should be expressed during development and normal cellular homeostasis. Historically, the presence of methylated DNA was identified in mammals as soon as DNA was recognized as the primary genetic material [9,10]. It was in 1948 when the presence of modified cytosine in the calf thymus and the existence of this modified cytosine naturally in DNA was established [11]. Later, studies have confirmed that methylation of cytosine (DNA methylation) regulate gene activity during development [12,13]. Currently, DNA methylation along with other remodeling factors is considered to be the major epigenetic remodeling event known to control gene activities (for references see [14,15]).
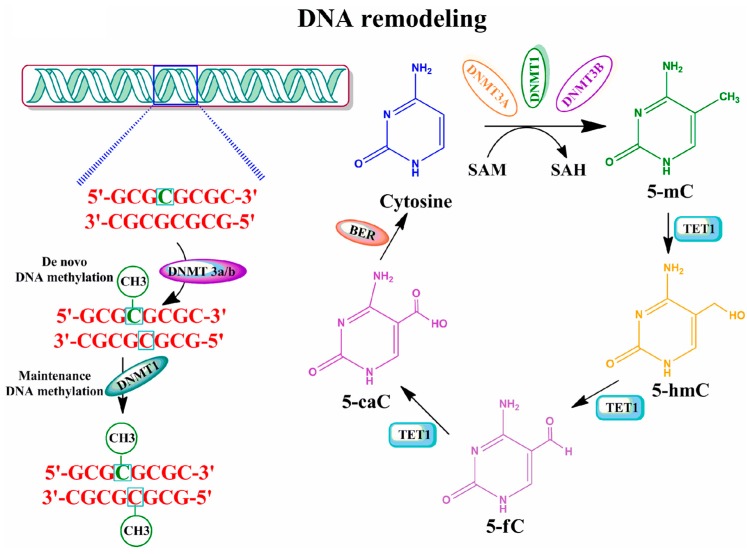
The methylation of cytosine (mC) is a covalent modification of DNA (Figure 1). A family of DNA methyltransferases (DNMTs) transfer a methyl group from S-adenyl methionine (SAM) to the cytosine residue to form 5-mC [16,17]. DNMT3a and -3b are characterized as de novo DNMTs partly because they can establish a new methylation pattern for unmodified DNA. Conversely, DNMT1 copies the DNA methylation pattern from the parental DNA strand onto the newly produced daughter DNA strand during DNA replication [18]. These unique functions of DNMTs supposedly ensure that the DNA methylation pattern is maintained and preserved in a tissue-specific manner across different individuals [19,20]. All three DNMTs are expressed extensively during embryo development, and a significant level is found in postmitotic neurons in the mature mammalian brain [21,22,23,24,25]. These observations suggest that DNMTs and DNA methylation patterns have an important novel role not only in early development but also in mature brain function [26,27]. Additionally, several recent studies have described at least two mechanisms that actively remove mC. One mechanism is through the activity of deaminases that catalyze the conversion of mC to thymidine [28]. A second mechanism is through the action of ten-eleven translocation (TET) proteins **(**α-ketoglutarate-dependent dioxygenases). TET proteins oxidize 5-mC to 5-hydroxymethylcytosine (5-hmC) using an oxygen- and α-ketoglutarate-dependent mechanism. This is then oxidized to 5-formylcytosine and finally to 5-carboxylcytosine (5-caC), which is then followed by the removal of the modified base through base excision repair and glycosylase activity [15,28]. DNA demethylation processes via 5-hmC were shown to operate in both developmental, as well as in an age-dependent manner in the mammalian brain [29], thereby providing the basis for an important epigenetic regulator of gene expression [30]. These discoveries suggest that external factors, such as environmental exposure/experiences, have the ability to modify the DNA methylation pattern. Indeed, research over the last decade suggests that DNA methylation responds to environmental exposure/experience, thereby resulting in stable phenotypes [31,32,33]. It has been suggested that these additional changes in DNA methylation that are due to environmental influences may then be inherited in a transgenerational manner [34,35]. The vast majority of recent data suggest that rapid and dynamic methylation and demethylation of specific genes in the brain have a direct role in synaptic plasticity, learning and memory formation [14,15,26,36,37,38,39,40].
Figure 1.
Graphic representation of DNA methylation and its regulation by enzymatic mechanisms. Methylation of DNA begins with the covalent addition of a methyl group from s-adenyl methionine (SAM) [41] to the fifth carbon of the cytosine pyrimidine ring to form 5-methylcytosine (5-mC), a process that is catalyzed by a family of DNA methyltransferases (DNMTs). The majority of DNA methylation usually occurs at genes on cytosines that precede a guanine nucleotide or CpG islands. De novo methyltransferases (e.g., DNMT3a/b) transfer methyl groups to naked DNA CpG pairs (e.g., CpG/GpC to mCpG/GpC) [42,43]. DNMT1 is the maintenance methyltransferase that transfers methyl groups to hemimethylated DNA strands (e.g., mCpG/GpC to mCpG/GpCm) and maintains the parental DNA methylation pattern during replication [44]. 5-mC undergoes sequential oxidation to 5-caC by TET1 activities. 5-caC, through base-excision-repair (BER) mechanisms, results in the regeneration of cytosine [39,45,46]. 5-methylcytosine (5-mC); 5-hydroxymethylcytosine (5-hmC); 5-formylcytosine (5-fC); 5-carboxylcytosine (5-caC).
Another group of proteins that work closely with methylated DNA to control gene transcription in the brain is the family of methyl-binding proteins. Methyl-binding proteins are persistently expressed in the adult brain and often act as gene repressors through binding to methylated cytosines [47,48]. The methyl-binding protein 2 (MeCP2) is expressed at high levels in the brain, specifically in neurons, but not in glia, and correlates with neuronal maturation [49,50]. The MeCP2 recognizes and binds to DNA that contains single 5-mC sites. Additionally, the binding of MeCP2 to DNA further recruits transcriptional corepressor complexes, such as Sin3a and histone deacetylases (HDACs) 1 and 2 [47]. During neuronal activity, the activity of MeCP2 often results in the release of promoters due to the removal of the methylation mark on the DNA [51]. Posttranslational modifications of MeCP2, such as its phosphorylation, affect its ability to bind to DNA and alter gene expression [52,53]. Inhibition of MeCP2 phosphorylation is associated with improved synapse formation, synaptic plasticity and learning and memory behavior [54,55]. This is partly because activity-dependent phosphorylation may release MeCP2 from promoters, thereby making the gene sequence available for demethylation process. Furthermore, approximately 95% of cases of neurodevelopmental disorders, such as Rett Syndrome, are due to MeCP2 mutations [56,57]. In an animal model, a 50% increase in MeCP2 in the brain induces impairments in motor coordination, anxiety, long-term potentiation (LTP), learning and memory [58]. Therefore, DNA methylation followed by the binding of MeCP2 appears to have a critical role in synaptic plasticity and cognitive function.
2.2. Post-Translational Modification of DNA-Associated Histone Proteins
Nuclear DNA does not appear in free straight strands. It is highly condensed and wrapped around nuclear proteins in the form of chromatin to accommodate chromosomes in the nuclei [59]. The chromatin consists of the nucleosomes. Each nucleosome core consists of two copies of each H2A, H2B, H3 and H4 proteins as well as 147 base pairs of superhelical DNA wrapped around this octamer [60,61]. These four histone proteins form a central structured globular domain that closely contact with the DNA and a less well-structured amino-terminal tail domain [62,63]. Specific amino acid residues of the N-terminal tail domain of these histone proteins undergo chemical modification through PTMs, including acetylation, methylation, phosphorylation, and sumoylation [64]. This distinctive chemical posttranslational modification influences the overall chromatin structure and recruitment of DNA binding proteins and allows for the loosening or tightening of the chromatin around particular genetic loci, thereby resulting in the activation or repression of specific gene expression [64]. As these PTM can alter gene expression without changing the DNA sequence, they have frequently been termed “epigenetic” modifications, although many changes are not maintained across the cell cycle [65].
2.2.1. Histone Acetylation and Deacetylation
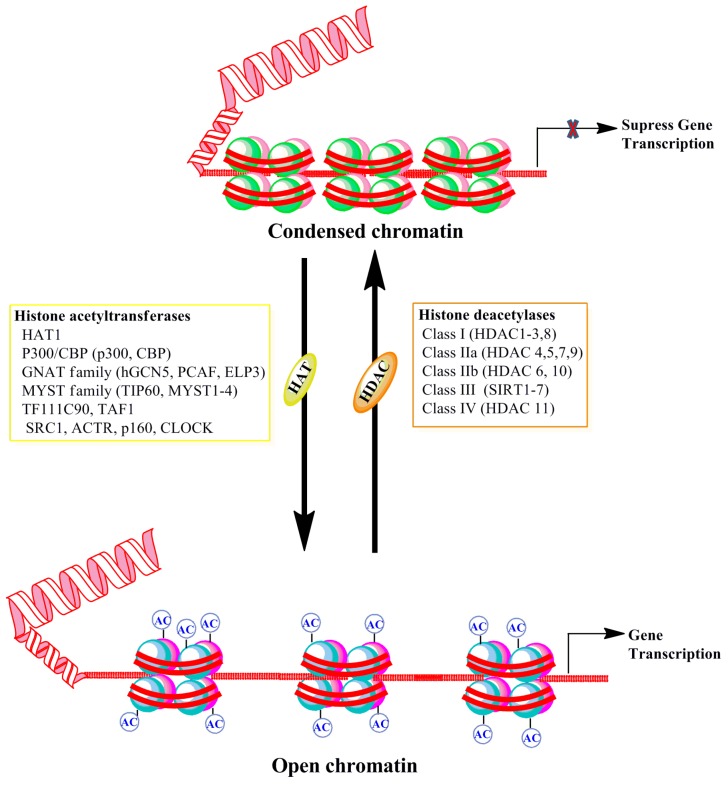
Histone acetyltransferases (HATs) transfer the acetyl group from acetyl-coA to the ε-amino group of lysine residues of histone proteins (Figure 2). Three main families of HATs, general control nonderepressible-5 (Gcn5), MYST, and p300/cAMP response element-binding protein (CREB) binding protein (CBP), share a conserved acetyl-coA binding site and often work in large multi-protein complexes [66,67]. The acetylation of histone proteins affects histone-histone protein interactions between neighboring nucleosomes and also interactions between histones and transcription factors (for references see [67]). These alterations can influence the individual nucleosome structure as well as the higher-order chromatin structure, thereby causing a more open and permissive chromatin environment for transcription. While HATs are often not specific to individual lysine amino acid residues, they fulfill specific functions. The broad catalytic role of HATs means that its localization to the proper genomic loci is essential for its particular function. The non-catalytic domains of HATs direct the protein to the appropriate location [68]. Acetylated histones provide a more relaxed chromatin conformation due to hydrophobicity and reduced electrostatic interactions. Additionally, acetylated histones further recruit bromodomain-containing proteins, which are primarily transcription factors and cofactors, and facilitate gene expression [69]. Among the HATs, CBP/p300 is critically involved in development. Genetic mutations and deletions of either enzyme cause Rubinstein-Taybi syndrome, which is characterized by intellectual disabilities, among other impairments [70]. The elevated expression of p300 mRNA was found in neural tissue and the genetic deletion of CBP or p300 resulted in an open neural tube through the impairment of paired box 3 (PAX3) and the activation of enhancer binding protein 2 (AP2) expression, thereby delaying embryo development [71,72]. Both Gcn5/Myc HATs co-regulate transcriptional programs in neural stem cells (NSCs) that are required for stem cell proliferation and brain development [73]. A number of memory tasks have been shown to enhance specific histone H3 and H4 acetylation immediately after memory performance through the enhanced expression and activity of several HATs (Taf1/Kat4, Gcn5/Kat2a, PCAF, Tip60, p300 and CBP) in rodents (for references see [74,75,76,77]). Recently, specific activators of HATs (SPV106, CSP-TTK21, etc.) [78,79] have been developed and used in certain neurological disorders for their beneficial function in augmenting neurodegeneration and cognitive function (for references see [80,81,82]). Thus, well-characterized HAT activators may be useful in treating cognitive dysfunction found in FASD children.
Figure 2.
Schematic representation of DNA-associated histone protein acetylation and deacetylation by histone acetyltransferase (HAT) and histone deacetylase (HDAC) enzyme families. The net acetylation at lysine residues of histone proteins of nucleosomes is decided by the interplay between acetylation and deacetylation processes through HAT and HDAC enzyme activities, respectively. The box provides different families and classes of HAT and HDAC enzymes. CBP, cyclic adenomonophosphate response element-binding (CREB) binding protein; GNAT, Gcn5-related _N_-acetyltransferases; hGCN5, human general control of amino acid synthesis protein 5-like 2; PCAF, p300/CBP-associated factor; ELP3, elongation protein 3; TIP60, TAT interacting proteins 60; TFIIIC90, transcription factor IIIC 90kDa; TAF1, TATA Box Binding Protein-Associated Factor; SRC1, steroid receptor coactivator 1; ACTR, activator of thyroid receptor; p160, receptor coactivators proteins 160; CLOCK, Clock Circadian Regulator.
Histone lysine deacetylases (HDACs) are a family of chromatin remodeling enzymes that remove acetyl groups from proteins and restrict the access of transcription factors to the DNA, thereby inhibiting gene expression (Figure 2). There are 11 well-described HDACs that are grouped into the following four classes, based upon their sequence, subcellular distribution and function: Class I (HDAC1, 2, 3, and 8), Class IIa (HDAC4, 5, 7, 9), Class IIb (HDAC6, 10), and Class IV HDACs (HDAC11) [83]. These HDACs are zinc-dependent enzymes. Class I HDACs (1 to 3 and 8) are largely localized to the nucleus, but they can also be found in the cytoplasm. Class II HDACs (4 to 7, 9, and 10) and IV HDAC (11) are either found as nucleocytoplasmic shuttling enzymes or primarily cytoplasmic. Class IIa HDACs (4, 5, 7, and 9) have been shown to exhibit low intrinsic deacetylase activity [84]. A separate family of HDACs, which are referred to as sirtuins, are classified as Class III HDACs. These are nicotinamide adenine dinucleotide (NAD)-dependent enzymes, which have mechanisms of action that are distinct from those of other HDACs [85,86]. Not much is presently known about HDAC11, as more studies have yet to emerge about its role in the adult brain. Class I HDACs were expressed at more elevated levels throughout the adult brain in comparison with many other HDACs [87,88,89]. The precise localization and cell-specific expression of individual HDACs in the brain suggests that each HDAC may have a broader, yet specific, significant role in neuronal development and function. Indeed, studies of a conditional knockout of both HDAC1 and HDAC2 suggest that neuronal progenitors fail to differentiate into mature neurons and lead to cell death, thereby resulting in animal death by Postnatal Day 7 (P7) [90]. Additionally, HDAC2 was also critically required to suppress progenitor transcripts during neuronal differentiation of adult-generated neurons [91]. Furthermore, HDAC 1/2 support the stability of synaptic networks during early brain development by suppressing the maturation of excitatory synapses [92]. The genetic deletion of HDAC3 suppresses the translocation of thyroid hormone receptors (SMRT) that regulate the neurogenic pathway to the nucleus [93]. The class II HDAC5 has also been suggested to have an important role in neuronal differentiation [94]. HDAC10 has also been shown to interact with and maintain a deacetylated state of Pax3, a key transcription factor in stem cell maintenance and neurogenesis [95]. Together, these findings suggest that HDAC family members control gene expression and cell fate during brain development into adulthood. Some HDAC families function independently of their catalytic activity function in neurite formation, dendrite and axonal growth (for references see [96]). Methods that involve pharmacological (HDAC inhibitors) and genetic manipulations of HDACs have shown a role of an individual family of HDACs in learning and memory process (for references see [92]). The conditional, brain-specific genetic deletion of HDAC2 in the mouse brain display enhanced acquisition of associative fear learning, whereas the overexpression of HDAC2, but not HDAC1, in the brain led to deficits in fear learning and aversively motivated spatial memory performance in the Morris water maze task [97]. Administration of an HDAC inhibitor, in addition to improving learning and memory, also enhanced the induction and maintenance of LTP at hippocampal and amygdala synapses, two brain regions that are essential for associative learning [76,98,99,100]. It should be noted that many learning and memory studies have utilized trichostatin (TSA) or suberoylanilide hydroxamic acid (SAHA) as an HDAC inhibitor. In doing so, researchers have observed that they appear to exhibit specificity, as these drugs act on class I, but not class II, HDACs [101]. The preclinical use of HDAC inhibitors as therapeutic agents was effective in many animal models of neurological disorders and the study of the benefits of HDAC inhibition for treating several cognitive disorders is underway [102]. Therefore, well-characterized HDAC inhibitors may be valuable for the improvement of the cognitive function of FASD children.
2.2.2. Histone Methylation and Demethylation
The methylation of histones usually occurs at the arginine or lysine N-terminal region of histones, which causes multiple effects, including the activation or inhibition of gene transcription. Each arginine residue in histone proteins can be subjected to monomethylation and symmetric/asymmetric dimethylation. Similarly, each lysine residue can be monomethylated, dimethylated and trimethylated and also adjacent residues can form different methylation combinations [103,104,105,106]. Arginine residue methylation is catalyzed by the family of arginine methyltransferases (PRMT). PRMT1 catalyzes the monomethylation and asymmetric dimethylation of arginine residues, thereby resulting in gene activation. PRMT2 catalyzes monomethylation and symmetric dimethylation of arginine residues, which results in gene suppression. Furthermore, the cofactor-associated arginine methyltransferase (CARM1) catalyzes the formation of monomethylation and asymmetric dimethylation of arginine residues, which promotes gene expression.
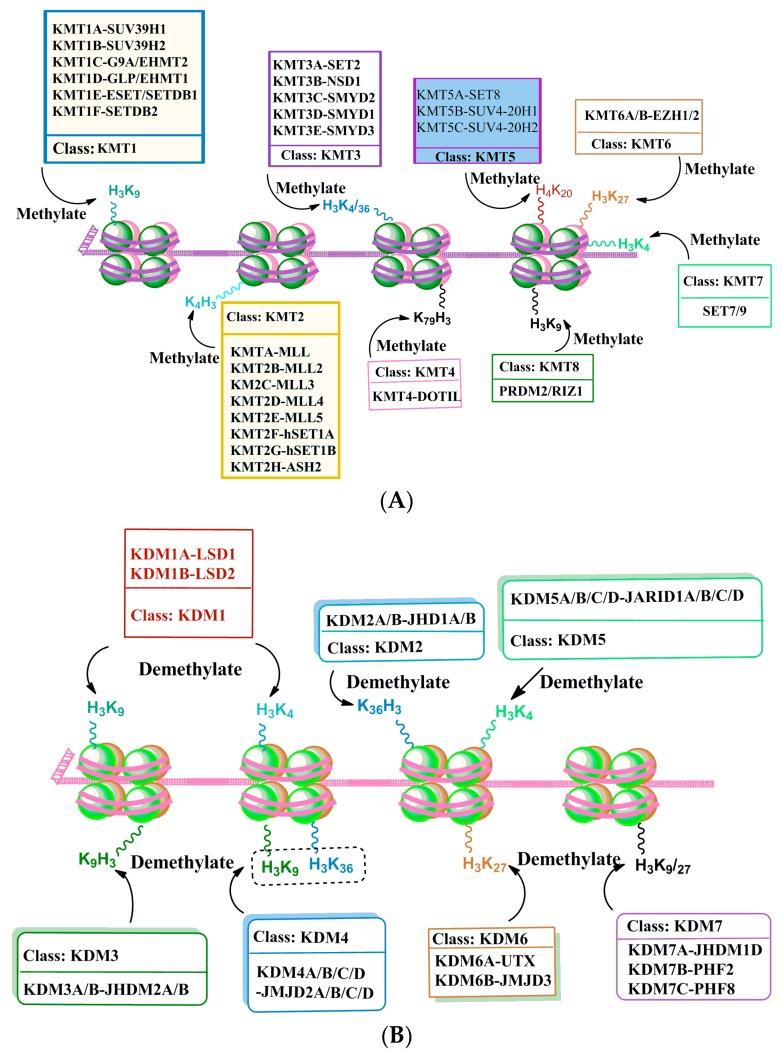
Histone methyltransferases (HMTs) that methylate not only lysine amino acids of histone proteins but also other proteins, are called lysine methyltransferases (KMTs) [107] for their broad enzymatic activity and substrate specificity (Figure 3A). All KMTs, except for KMT4, contain a conserved Su (var)3-9, Enhancer of Zeste, Trithorax (SET) domain that exhibits catalytic activity (for references see [108]). The presence of a characteristic homologous sequence [107,109] has helped to classify KMTs into distinct subcategories. Lysine methylation of histone H3K4 is catalyzed by a family (KMT2) of mixed lineage leukemia (MLL) proteins, such as SET1A, SET1B, MLL1, MLL2, MLL3, MLL4 and ASH1 [110,111,112]. H3K4 methylation closely correlates with the activation of gene expression. H3K9 methylation is catalyzed by a group of methyltransferases (KMT1: SUV39H1, SUV39H2, Eu-HMTase/GLP, G9A and ESET/SETDB1; KMT8:RIZ1), whose catalyzing substrates and resulting products are different. The trimethylation of H3K9 is catalyzed by SUV39 and results in the formation of heterochromatin and transcriptional inhibition [113]. The dimethylation of H3K9 is catalyzed by G9a and results in a euchromatin structure and the suppression of gene expression [6,114]. G9a-like protein (GLP) can form a hetero polypeptide complex with G9a, and together they catalyze the formation of H3K9 dimethylation [115]. ESET/SETDB1 catalyze the trimethylation of H3K9 enzymes [116], which results in gene inhibition. H3K27 methylation is mediated by the EZH2 enzyme (KMT6) and results in the inhibition of gene transcription and has been shown to have a significant role in fetal development and cell differentiation [117,118]. H3K36 methylation activity is performed by (KMT3) SET2, SYMD2 and NSD1 enzymes and is associated with active transcription [119,120,121]. H3K4 methylation by SYMD1-3 enzymes is related to gene activation [122]. H3K79 di- and tri-methylation are performed by DOT1L enzymes (KMT4) and are universally associated with active transcription [123]. H4K20 mono- or tri-methylation is catalyzed by SET8 and SUV420H1/2 enzymes (KMT5), respectively, and have been linked with transcriptional repression [124].
Figure 3.
DNA-associated histone protein lysine methylation (A) and demethylation (B) by histone methyltransferase (KMTs) and histone demethylase (KDMs) enzyme families. Histone H3 and H4 tails with known lysine methyltransferases (KMT1-8) and demethylases (KDM1-7) are shown under each specific lysine residue.
Experimental data concerning enzymatic histone demethylation were not known until the discovery of the lysine-specific demethylase 1 (LSD1) (KDM1 family). LSD1 (KDM1A) is characterized as a component of the v-raf murine sarcoma viral oncogene homolog B1 (BRAF)-histone deacetylase (HDAC) (or BHC) complex that contains the restrictive silencer factor (REST) co-repressor, CoREST. CoREST has been shown to function as a repressor of neuronal genes in non-neuronal cells [125,126,127]. LSD1, through its amine oxidase domain, has been shown to act as a transcriptional repressor and to demethylate K3K4, thereby providing the first evidence that histone methylation is a dynamic and reversible event [128]. LSD2 (KDM1B) was recently described as an additional H3K4 demethylase [129]. Similar to KDM1A, KDM1B was shown to demethylate the mono- and di-methylated histone H3K9 [130]. Soon after discovery of the KDM1 family, a protein containing a Fe2+ dioxygenase Jumonji-C (JmjC) domain (KDM2 family proteins) was characterized as a specific H3K36me1/2 demethylase (KDM2A and B), which does not demethylate H3K36me3 [131]. Now, accumulating evidence suggests the presence of specific demethylases for all of the major methylated histones that have so far been studied (Figure 3B), with the exception of methylated histone H3K79, despite its characterization as a reversible process [123]. The KDM3 family of proteins was characterized as the second family of JmjC histone demethylases (KDM3A/JHDM2A and KDM3B/JHDM2B) and was shown to demethylate mono- and di-methylated H3K9, thereby resulting in active gene expression [132]. The KDM4 family proteins consist four histone demethylases (KDM4A–4D or JMJD2A–2D, respectively). All KDM4 demethylases remove di- and tri-methylation from histone H3K9 and H3K36 proteins [133,134,135]. In addition to the JmjC domain, they all contain a conserved JmjN domain. All, except KDM4D, have tandem plant homeodomain (PHD) fingers and Tudor domains that help to recognize specific histone methylation marks [136,137]. These families of proteins were activated in response to hormonal activity. For example, KDM4A and KDM4D act as androgen receptor (AR) coactivators: estrogen receptor (ER) activation results in KDM4B-mediated demethylation of H3K9me3 proteins [138,139]. KDM5 (KDM5A–5D/JARID1A–1D) specifically catalyzed the demethylation of histone H3K4me2/3 proteins [140,141,142,143,144]. These proteins are characterized as multi domain-containing proteins and include a combination of JmjC/JmjN catalytic domains, with an AT-rich interaction domain (ARID) DNA-binding domain, a C5HC2 zinc finger and two to three PHD fingers. The PHD finger domains of KDM5A and KDM5C were shown to facilitate binding to methylated H3K4 or H3K9, respectively [141,145]. KDM6 family proteins contain KDM6A/UTX and KDM6B/JMJD3 demethylases. These enzymes catalyze the removal of di- and tri-methyl groups form histone H3K27me2/3 proteins [146,147,148,149,150] and result in transcriptional activation. KDM6A functions in development via its association with the KMT2/MLL proteins to regulate H3K4 methylation followed by HOX gene expression [149]. The KDM7 (PHF2) family enzymes contain three members (KDM7A/JHDM1D, KDM7B/PHF8, and KDM7C/PHF2). This enzyme family removes mono and di methyl group from histone H3K91/2 and H3K27me1/2 proteins [139,151,152,153,154,155,156,157]. Additionally, they regulate the expression of genes involved in X-linked mental retardation and expression of ribosomal RNA [154,155,157]. Additionally, PHF8 demethylates histone H4K20me1 proteins [155,157,158]. KDM7 family proteins have a PHD finger domain that helps to recognize and bind to histone the H3K4me3 mark. This ensures substrate specificity, genomic occupancy, and the regulation of target gene expression by KDM7 demethylases [152,156,159,160,161]. Few studies have explored the participation of histone demethylase in developmental alcohol studies, except in one study in which acute prenatal alcohol exposure at GD7 was shown to disrupt the transcripts that encoded KDM1A and KDM4A proteins [162] (see Section 3 for details).
2.3. MicroRNAs
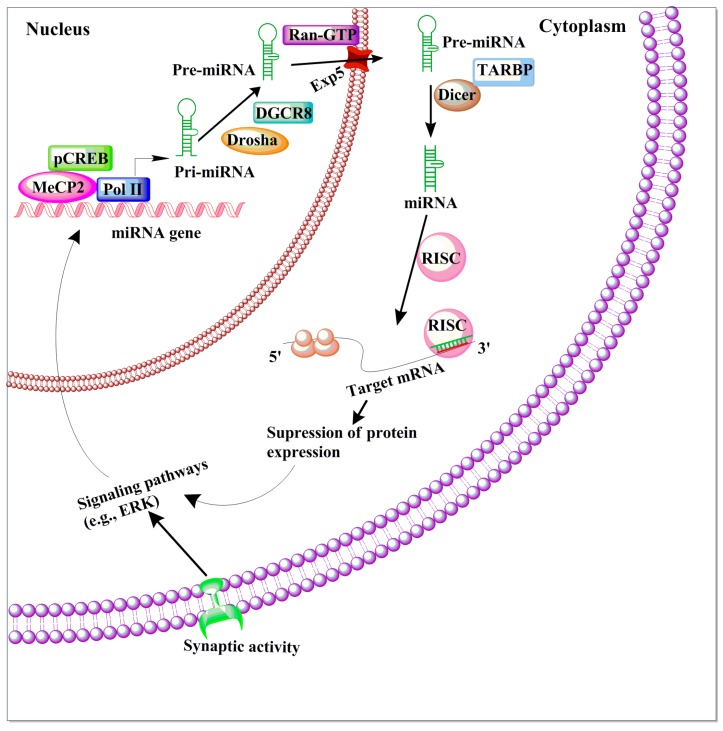
MicroRNAs (miRNAs) are a family of small, non-coding RNA molecules that facilitate the degradation of untranslated regions of target mRNAs. Thus, miRNAs play a critical epigenetic function by inhibiting the translation and transcription of a network of genes encoding for proteins. MicroRNAs are generated from longer precursor miRNA (pre-miRNAs) in sequential biogenesis pathways (Figure 4). MicroRNA genes primarily bind to the imperfect complementary sequence that forms a secondary stem-loop structure within the pre-miRNAs. This structure, along with its guide strand [163,164], act as a substrate for two double-stranded RNases, Drosha (nucleus) and Dicer (cytoplasmic) [165]. This specific targeting occurs through a partial complementary association between the mRNA’s 3′UTR and 5′ end of the microRNA (a 6–8 nucleotides sequence long) regions. This partial association facilitates the targeting of a single miRNA to many mRNAs at the same time and vice versa. This provides a mechanism wherein a single mRNA can be influenced by diverse miRNAs [166,167]. To date, a few miRNAs have been shown to be expressed in mammalian brains and recent estimates suggest the presence of 1100 or more human miRNAs, which regulate the expression of nearly two-thirds of all genes [168]. Thus, they can control the expression of networks of genes and whole pathways and therefore are considered as master regulators of gene expression [169]. Therefore, miRNAs are critically essential for the proper development and functioning of the adult brain. Indeed, many recent studies suggest the involvement of several miRNAs in an array of cellular homeostatic processes, including developmental timing, cellular differentiation, neural patterning, apoptosis and synaptic plasticity [170,171,172,173].
Figure 4.
The schematic diagram of miRNA biogenesis and function. In the nucleus, RNA polymerase II (Pol II)-dependent transcription of a miRNA-encoding gene, which may include both intron- and exon-coding regions, results in the formation of a long primary miRNA transcript (pri-miRNA) that is 50-capped and 30-polyadenylated. This pri-miRNA transcript is subject to nuclear processing by the microprocessor complex, which includes DCGR8 and Drosha, into the precursor miRNA (pre-miRNA) transcript. Pre-miRNAs are transported out of the nucleus to the cytoplasm by Exp5 and Ran-GTP. Pre-miRNA can then be further cleaved by Dicer/TARBP to generate a mature miRNA. The mature miRNA is incorporated into the RNA-induced silencing complex (RISC), which will select mRNA transcripts for the down-regulation of protein expression. In the synapses, miRNA can regulate the local down-regulation of protein expression. Certain conditions, such as neuronal activity, could affect the signaling events as well as miRNA formation. Additionally, genes regulated by miRNA can act on synaptic activity-dependent signaling pathways that promote the activation of epigenetic factors (e.g., CREB and MeCP2), which in turn can control miRNA transcription in the nucleus.
Studies involving individual genetic deletion of miRNA have suggested that miR-124, miR-125b, miR-132, miR-134, miR-137, and miR-138 in neurons regulate synaptic development and dendritic branching (for references see [174]). The disruption of miRNA biogenesis pathways, such as Dicer, which affects the expression of all miRNAs, can induce impaired cell differentiation, reduction in neuronal size, loss of dendritic branching, disturbed axonal guidance [175], impairments in inhibitory synaptic transmission and cognitive dysfunction [176]. It should be noted that the activity-dependent regulation of gene expression is essential for synaptic plasticity and memory formation and similar neuronal activity was shown to regulate miR-132 expression in a CREB signaling pathway-dependent manner [177,178]. In a recent study, reduced miR-125a levels were found in synaptoneurosomes after activation of mGluR1/ 5 receptors, thereby indicating that neuronal activity may influence the miRNA turnover [179]. Additionally, miRNAs in neurons decay much faster than in non-neuronal cells, and their turnover rate is activity-dependent [180]. Therefore, similar to many learning and memory genes, miRNAs can be regulated in a neuronal activity-dependent manner. It has been suggested that neuronal activity is one mechanism for the high turnover rate of miRNAs in neurons. Consistent with their neuronal function, increasing evidence suggests that the activity of miRNAs is directly implicated in the pathogenesis of complex neuropsychiatric disorders (for references see [181]) that are linked to abnormalities in synaptic plasticity as well as in neurodegenerative diseases [182,183]. Limited studies have explored miRNA dysregulation in FASD animal models, as described later in this section. Therefore, an understanding of the signaling mechanisms that regulate the patterns and activity of miRNA expression have the potential to serve as potential targets for FASD treatment.
3. Alcohol’s Influence on Epigenetic Mechanisms in the Developing Brain
The presence of fetal alcohol syndrome was recognized after the initial study [184] suggesting series of deformities in the offspring of alcoholic mothers. Among the several possible consequences of developmental alcohol exposure, defects in brain maturation and the considerable lifelong behavioral impairments have been the most catastrophic. The most serious consequences of early developmental alcohol exposure are fetal alcohol syndrome (FAS) [184], which is characterized by pre- and post-natal growth defects, craniofacial anomalies and persistent impairments of CNS function. FAS occur in 1–2 of every 1000 newborns. It has been acknowledged as an important cause of non-genetic intellectual disabilities and behavioral impairments in the world [185,186,187,188,189]. Nevertheless, now, it has become clear that FAS is not the only enduring defect that results from developmental alcohol exposure. Unquestionably, the term fetal alcohol spectrum disorders (FASD) [190] (estimated the prevalence of as high as 2%–5%) has been used to represent the persistent nature of developmental alcohol effects, with FAS on the stronger side of the spectrum. Though the behavioral profile of FASD is somewhat limited [191], the available literature suggests that FASD is associated with many neuropsychological impairments, such as verbal learning/recall abilities, learning and memory [187,192,193,194,195,196]. These neuropsychological deficits create many daily challenges for individuals with FASD and are also responsible for causing a majority of intellectual disability in the Western world [194,196,197,198]. The neurobehavioral outcome of FASD is also dependent on the amount [199,200], pattern (continuous vs. binge drinking) and developmental timing [201] of alcohol exposure. Additionally, the pattern of alcohol exposure can often undermine alcohol dose effects. For example, binge-like exposures result in more severe behavioral outcomes than does chronic exposure [202,203]. Furthermore, several other factors, such as maternal metabolism, genetic susceptibility, and variation in the vulnerability of different brain regions, may contribute to behavioral outcomes. Nonetheless, factors that alter the peak blood alcohol levels (BALs) experienced by the embryo or the fetus (e.g., maternal metabolism) are expected to affect the occurrence and severity of FASD. Unfortunately, this information is often hard to obtain, mainly in retrospectively recruited samples, and individual studies provide varying degrees of detail concerning the amount and patterns of exposure. These issues highlight the modifying variables that contribute to the vast range of phenotypes presented in FASD children.
Persistent data from all areas of biological science, such as epidemiology, genetics, epigenetics, neuroscience and neuroimaging studies, strongly suggest that many adult neuropsychiatric illnesses have a neurodevelopmental origin [204]. Recent technological advances and experimental evidence have provided more precise mechanistic insights on the developmental origin of adult diseases. Disturbance of brain development at sensitive and critical periods by adverse environmental exposure, along with genetic programming, results in the early or late manifestation of heterogeneous clinical conditions [205,206,207,208,209]. The range of developmental deficits varies from learning or cognitive functions to global impairments of social skills or intellect. To date, the etiological basis of the majority of developmentally related adult diseases, such as autism spectral disorder (ASD), attention deficit hyperactivity disorder (ADHD) and FASD conditions, are still unknown. Data from several studies support their polygenic and multifactorial, including epigenetic etiology [4,6,210,211,212,213].
Using animal models of FASD, several studies have reported epigenetics changes, including DNA methylation [214,215,216,217,218,219], DNA-associated histone modification [4,5,6,213,220,221,222,223,224] and miRNAs [225,226,227,228,229,230,231]. Acute alcohol exposure at Gestational Days (GD) 9 through 11 was found to induce hypomethylation of DNA in the fetus [215], and this study was the first one to indicate that DNA methylation may contribute to abnormalities found in FAS. In the prenatal alcohol model, alcohol was shown to significantly decrease astroglial-specific marker, glial fibrillary acidic protein (GFAP) immunoreactivity and its mRNA levels in both astrocytes in primary culture and brains of pups from alcohol-fed mothers. The reduced GFAP transcript found in fetal brains was due to the hypermethylated state of the GFAP DNA [232]. These findings suggest that alcohol-induced DNA hypermethylation, followed by impaired GFAP expression and astroglial development, may underlie the deficits in brain functions that are observed after prenatal alcohol exposure. In some instances, male alcohol exposure before breeding resulted in reduced cytosine methyltransferase mRNA levels in paternal sperm, thereby suggesting that distorted genomic imprinting caused by DNA hypomethylation leads to the expression of normally silent paternal alleles [233]. This phenomenon was also demonstrated between alcohol use in men and hypomethylation of paternally imprinted loci in sperm DNA in genomic regions that are essential for embryonic development, therefore providing a mechanism for paternal effects in the etiology of FASD [218].
The imprinted genes are expressed mainly from either the paternal or maternal allele and are important regulators of fetal growth and development [234]. Additionally, imprinted genes have been found to cause some birth defect syndromes in humans [235,236] and in animals [237,238,239]. In a whole-embryo culture, alcohol enhanced DNA methylation, particularly in genes on chromosome 7, 10 and X, and a neural tube defect phenotype. Enhanced DNA methylation was found in imprinted genes, genes that are essential for cell cycle, growth, apoptosis, cancer, and in a significant number of genes important for olfaction, and with associated changes in gene expression [217]. In monolayer cultures of NSCs, alcohol was shown to increase DNA methylation of multiple cell cycle genes and increased the expression and activity of DNA methyltransferases [240]. In another study, prenatal alcohol-exposed embryos exhibited decreased DNA methylation at insulin-like growth factor 2 (IGF2) transcripts, which was reversed by methyl supplementation [214]. In NSC culture, alcohol exposure retarded the migration, neuronal formation, and growth processes of NSCs. Additionally, alcohol exposure prevented DNA methylation of specific genes related to neural development, eye development and developmental disorders, thereby suggesting that developmental alcohol alters DNA methylation of genes that are critical for regulating neural development [219]. Furthermore, in studies using a whole-embryo culture, alcohol delayed the DNA methylation program, which paralleled the developmental impediment of the neural tube in a spatial and temporal manner [241]. A delay in hippocampus development was also associated with altered 5-mC and 5-hmC in DG and CA region of the hippocampus in prenatally alcohol-exposed embryos or P7 mice [242]. Although these studies have demonstrated the presence of altered methylation program that correlated well with a developmental delay in the formation of the neuronal tube and the hippocampus, whether the abnormal DNA methylation is responsible for the developmental delay and behavioral deficits remains to be established in further studies. In another animal model of FASD (third-trimester equivalent pregnancy), studies have suggested a direct link of this phenomenon. Alcohol administration in Postnatal Days 2–10 (P2–10) mice significantly caused hypermethylation of DNA in the hippocampus (HP) and prefrontal cortex (PFC) at P21. Administration of choline (P2–20), a nutrient supplement that has been shown in a rat model to rescue some of the alcohol’s behavioral effects [243,244], prevented DNA methylation in alcohol-treated P21 mice HP and PFC tissues [245]. In a gestational alcohol model, alcohol increased the methylation of the proopiomelanocortin (Pomc) DNA in a gene- and cell-specific manner in the adult rat hypothalamus despite the exposure to alcohol having ended during the prenatal period [221]. Interestingly, the hypermethylated Pomc gene was found in the sperm of prenatal alcohol-exposed F1 offspring that was transmitted through the F3 generation through the male germline [221]. Global DNA methylation was found to be reduced by alcohol in an in vitro murine embryonic fibroblast (MEF) model. Reduction in DNA methylation was associated with alcohol-induced proteasomal-mediated degradation of DNA methyltransferases (DNMT-1, DNMT-3a, and DNMT-3b) [246]. In a neonatal animal model of FASD (third-trimester equivalent pregnancy), the administration of alcohol to P7 mice resulted in reduced global DNA methylation in neonatal HP and neocortical (NC) tissues [23] and impaired learning and memory in adult animals [4,213,247]. This reduction in DNA methylation was linked with alcohol-induced caspase-3 mediated degradation of DNMT-1, DNMT-3a proteins [23] and impaired learning and memory [4,213,247]. Combined pre-(GD1-21) and postnatal (P2–10) alcohol exposure resulted in a significant increase in DNA methyltransferase activity (DNMT), as well as Dnmt1 and Dnmt3a gene expression in the HP of P21 rats [248]. These studies warrant future investigation to understand how these events affect global or gene-specific DNA methylation and learning and memory. Prenatal alcohol exposure decreased DNA methylation in the solute carrier family 17 member 6 (SLC17A6) promoter region, which encodes vesicular glutamate transporter 2 (VGLUT2). In contrast to Slc17a6 mRNA levels, HP VGLUT2 protein levels were significantly reduced in adult alcohol-exposed offspring and suggest that DNA methylation-regulated glutamate neurotransmission in the hippocampus could contribute to the cognitive and behavioral phenotypes that are observed in FASD [249]. Another important observation was made using gestational alcohol exposure in epigenetically sensitive alleles (metastable epialleles) in the Agouti viable yellow (Avy) mice model [250]. The activity of Avy is changeable among genetically identical mice, thereby resulting in mice with a range of coat colors [from yellow to mottled to agouti (termed pseudoagouti)] [251]. In this model, exposure to alcohol increases the probability of transcriptional silencing of the Ayy locus through DNA methylation, thereby resulting in more mice with an agouti-colored coat [35]. A genome-wide DNA methylome analysis revealed widespread alcohol-induced alterations with significant hypermethylation of many differentiation-related genes in human embryonic stem cells (hESCs) [252]. In a Japanese rice fish embryogenesis model, alcohol was shown to have the capacity to specifically alter the developmental pattern of DNA methylation machinery [253]. A genome-wide analysis of DNA methylation in adult mice exposed to prenatal alcohol using voluntary alcohol exposure paradigm indicated that a large number of gene promoter regions are differentially affected. Furthermore, network analysis of these promoter regions has suggested that behavior-, neurological disease-, and psychological disorder-associated networks are the most significantly affected ones [254]. Enrichment analysis using the genes that were up-methylated in FASD children from 5 to 18 years of age showed a significant enrichment in genes associated with neurodevelopment and diseases [255]. It is clear from the above-discussed studies that the alteration of global DNA methylation or methylation (Table 1) of specific genes/promoter regions is dependent upon several factors, including the specific gene or tissue, amount and pattern of alcohol exposure, as well as the developmental timing of exposure. DNA methylation or demethylation (5-hCm) can influence gene expression through enhancing or preventing the recruitment and binding of transcriptional regulatory proteins with decreased [256] or increased [257,258] transcription activity. Findings also suggest that increased DNA methylation in the gene body can lead to an increased transcription, thus implicating a bidirectional regulation of transcription, depending upon the site of the DNA methylation mark [259]. The mechanisms of the interplay between DNA methylation and transcription activity are therefore crucial. Furthermore, the signaling events that are responsible for DNA methylation-related chromatin remodeling are largely unknown and warrant future research in this line to develop potential therapeutic approaches to treat FASD.
Table 1.
Summary of the developmental alcohol-induced DNA methylation modifications.
| Alcohol Exposure | Tissue Examined | Effects |
|---|---|---|
| 1. GD 9–11 | GD 12 fetus | Reduced DNA methylation [215] |
| 2. GD | Cultured astrocytes | BDNF gene hypermethylation [260] |
| 3. GD 1–18 | PD 10 | Hypermethylation of the GFAP gene [232] |
| 4. Oryzias latipes embryos | 2–6 dpf | No change in Aldh1A2 gene promoter methylation [261] |
| 5. Embryo (E8.25) cultures | Embryo cultures | Both hyper/hypomethylation of gene promoter [217] |
| 6. Neural stem cells in culture | NSCs culture | Hypermethylation of cell cycle genes [240] |
| 7. Neural stem cells in culture | NSCs culture | Hypomethylation of NSC genes [219] |
| 8. GD 9 | E-15 and 17 | Decreased Igf2 gene DNA methylation [214] |
| 9. PD 2–10 | PD 21 | Increased DNA methylation [245] |
| 10. GD 1–22 | PD 21 | Enhanced DNMTs activity [248] |
| 11. hESC | Cell lines | No change in gene methylation [262] |
| 12. hESC | Cell lines | Hypermethylation of many regions of chromosomes [252] |
| 13. GD 1–17 | Cultured neural progenitor cells | Disrupt DNA methylation machinery and delays the maturation of dentate gyrus [242] |
| 14. Murine embryonic fibroblasts | Fibroblasts | Impaired DNA methylation and DNMT1, DNMT3a and DNMT3b proteins [246] |
| 15. GD 7–21 | PD 60–65 | Increased DNMT1protein and Pomc gene methylation [220] |
| 16. PD 7 | PD 7 | Reduced DNA methylation, DNMT1and DNMT3a proteins [23] |
| 17. GD 0.5–8.5 | PD 28 | Decreased mRNA levels of Dnmt1 and Dnmt3a genes [263] |
| 18. GD 0.5–8.5 | PD 28 | Both up/down regulation of DNA methylation [264] |
| 19. GD 0.5–8.5 | PD 87 | Decreased Slc17a6 gene promoter [249] |
| 20. Oryzias latipes embryos | 2–6 dpf | Altered DNMT1 mRNA [265] |
Limited laboratories using many animal models of FASD have demonstrated the participation of DNA-associated histone proteins modifications in responses to alcohol exposure during the different sensitive period of brain development. The earliest report suggests that binge-like alcohol exposure reduces the level of histone H2A family proteins in endothelial caveolae cells [266]. Another study demonstrated the reduced expression of the CREB binding protein (CBP), a histone acetyltransferase, in the rat cerebellum exposed to alcohol during the third trimester-equivalent of human pregnancy. These studies also demonstrated the global reduction of both acetylated histone H3 and H4 proteins in the cerebellum [222]. In a prenatal alcohol model, alcohol was shown to affect the hypothalamus function, such as stress axis responsiveness, which may be due to reduced expression of Pomc gene and production of β-endorphin protein in the arcuate (ARC) nucleus of the hypothalamus. In the same arcuate nucleus, reduced staining of H3K4me2,3, acetylated H3K9 and increased H3K9me2 was also observed in alcohol-exposed male offspring. Consistent with acetylated or methylated histone H3 and H4 staining data, the transcriptional activity of enzymes responsible for methylation or acetylation of histone H3 and H4 proteins (G9a, Set7/9, Setdb1 and HDAC2) were also altered in alcohol-exposed offspring [221]. In another study, during the third-trimester equivalent human pregnancy animal model, alcohol was shown to enhance the expression of G9a mRNA and protein in NC and HP tissues. This resulted in enhanced dimethylation of H3K9 and H3K27 followed by caspase-3 mediated degradation of these proteins. Further studies using G9a (Bix 01294), as well as caspase-3 (Q-VD-Oph) inhibitors, have suggested that the activation of G9a results in robust neurodegeneration in neonatal mice and deficits in synaptic plasticity and learning and memory in adult mice [6,213] as observed in FASD children [193,195,198]. These neurobehavioral impairments were rescued by preventing the alcohol activated G9a [213]. Similarly, alcohol also enhanced the transcriptional activity of cannabinoid receptor 1 (CB1R) through promoter (exon1) specific enrichment of H4K8ace protein in the mouse NC and HP. Transcriptional activation of CB1R results in impaired CB1R-mediated signal transduction (pERK1/2/pCREB/activity-regulated cytoskeletal protein (Arc)), resulting in neurodegeneration in neonatal mice and learning and memory deficits in adult mice [4,247]. These neurobehavioral deficits were prevented by CB1R antagonist or CB1R knockout mice [4,247]. Furthermore, a promoter specific analysis of histone H3 proteins using low dose alcohol (elicit mild neurodegeneration) showed an enhanced specific acetylation of H3K14 and no change in H3K9me2 or H4K8ace levels, as observed in the G9a exon1 region [5]. Transcriptional activation of G9a results in increased expression of the G9a gene and protein, thereby resulting in enhanced global dimethylation of H3K9 and H3K27 proteins without any significant degradation of H3 proteins due to the low activity of caspase-3, as found in a low-dose alcohol model [5]. In another prenatal alcohol model, alcohol was shown to enhance the expression of genes that have a role in the fetal heart through promoter-specific H3K14 acetylation in mice [267]. In a similar fetal heart study, alcohol increased global HAT activity without affecting global HDAC activity. Additionally, alcohol increased the binding of P300, CBP, PCAF, SRC1, but not GCN5, at the Gata4 promoter region in comparison to the saline-treated group. The increases in acetylation of H3K9, as well as in Gata4, α-myosin heavy chain (α-MHC) and cardiac troponin T (cTnT) mRNA levels, were also evident in these fetal hearts [268]. Studies in a prenatal alcohol model suggest that alcohol enhanced the enrichment of histone H3K4me3 at the Slc17a6 promoter, which encodes VGLUT2 [249]. Acute exposure to alcohol at GD 7 in mice exhibit significantly increased H3K9me2 and H3K9ace levels and reduced H3K27me3 levels at GD 17 and that these histone modifications strongly correlate with the development of craniofacial and CNS defects in this animal model. These observations indicate that an epigenetic signature persists long after the window of alcohol exposure and is mostly linked to alterations in a repressive chromatin structure, such as H3K9me2 [162].
The first evidence of histone acetylation modification by developmental alcohol was found in cardiac progenitor cells where alcohol enhanced H3K9ace and augmented the mRNA expression of heart development-related genes [224]. Exposure to alcohol during the third trimester-equivalent of human pregnancy reduces CBP levels, followed by reduced histones H3 and H4 acetylation in the neonatal cerebellum [222]. Prenatal alcohol exposure enhanced long-lasting H3K9ace marks and CBP mRNA levels in β-endorphin (BEP)-positive cells in the ARC of the hypothalamus of male rat offspring [221]. The transient exposure of immature P7 mice to alcohol, which is comparable with a time point within the third trimester of human pregnancy, enhances specific acetylation of H3K14 in the G9a exon1 gene, which results in enhanced G9a transcription [5]. In a similar mouse model, it was shown that alcohol enhances acetylation of H4K8 in the CB1R exon1 gene, thereby leading to enhanced CB1R expression and impaired synaptic plasticity, learning and memory in adult mice [4].
Prenatal alcohol exposure reduced both mRNA levels of KMT7/9 (Set7/9) and the number of H3K4me2, 3-positively stained BEP cells. Additionally, enhanced KMT1C (G9a) and KMT1F (Setdb1) mRNA, as well as H3K9me2-positively stained BEP cells, was observed in the ARC of the hypothalamus of rat offspring exposed to prenatal alcohol [221]. In another study, binge-like alcohol exposure in P7 mice, which is comparable with the third trimester of human pregnancy, enhanced KMT1C (G9a) expression, as well as in H3K9me2 and H3K27me2, in both neocortical and hippocampal tissues of neonatal mice [5]. This particular dimethylation of both histones marks them for proteolytic degradation by activated caspase-3 in this animal model [6]. The inhibition of KMT1C (G9a) activity before alcohol treatment not only rescued neurodegeneration in neonatal mice but also synaptic plasticity and learning memory in adult mice [213]. Gestational choline treatment, which has been shown to prevent pre/postnatal alcohol-induced learning and memory [244,269], also normalized the alcohol-suppressed hypothalamic levels of H3K4me3, as well as the NA levels of KMT7/9, and increased the H3K9me2 and mRNA levels of KMT1C and KMT1F in the hypothalamus of adult male offspring rats [220]. In another study, prenatal alcohol exposure enhanced SLC17A6 gene expression, which codes for VGLUT2 through the enrichment of H3K4me3 at the SLC17A6 promoter region in adult mice [249]. In another study, acute exposure of GD7 mice to alcohol significantly disrupts transcripts that encode for KMT1C (G9a), KMT6 (EzH2) and KMT7 (SETdB1) proteins [162]. Together, these findings suggest that these epigenetic mechanisms (Table 2) may have a direct role in cognitive dysfunction observed in FASD.
Table 2.
Influence of developmental alcohol on histone modifications.
| Alcohol Exposure | Tissue Examined | Effects |
|---|---|---|
| 1. Gestational Day (GD) 7 | GD 17 | Increased H3K9me2, H3K9ace and decreased H3K27me3 [162] Altered G9a, Setdb1, Kdm1a, Kdm4c, Uhrf1, Ezh2 and Dnmt1 mRNA levels [162] |
| 2. Postnatal Days (PD) 2–12 | PD 2–12 | Decreased AcH3, AcH4, H3K23ace and increased HAT (CBP) [222] |
| 3. GD 7–21 | PD 60–80 from F1–F3 generation | Decreased H3K4me2, H3K4me3, H3K9ace, pH3S10 and mRNA Set7/9 [220]; Increased mRNA G9a, setdb1 H3K9me2 [220] Decreased H3K4me2,H3K4me3, H3K9ace and Decreased mRNA levels of Set7/9; Increased G9a mRNA H3K9me2 and HDAC2 [221] |
| 4. GD 1–14.5 | Embryonic days (ED) 7.0–14.5 | Increased H3K14ace [267] |
| 5. PD 7 | PD 7 | Increased H4K8ace [4], H3K14ace [5] Decreased H3K9me2, H3K27me2 [4] Decreased H3K9me2, H3K27me2 [6], H3K9me2 [213] Increased G9a mRNA and Protein [6] |
| 6. ED 8.5–E16.5 | ED14.5–PD 7 | Increased p300 and SRC1 protein. No Change in HDAC. Increased HAT (CBP and PACF) [268] |
| 7. ED 8.5–16.5 | ED 14.5–16.5 | Increased H3K14ace [267] |
| 8. Days of post cotium (dpc) 0.5–8.5 | PD 87 | Increased H3K4me3 and Slcl7a6 gene expression [249] |
The first evidence to show that early alcohol alters miRNA (Table 3) was shown in NSCs using ex vivo fetal cerebral cortical neuroepithelium model [228]. In this study, alcohol, which was shown to promote cell proliferation and aberrant differentiation, did not cause the death of NSCs, but rather suppressed miR-9, -21, -153, and -335 levels [228]. Furthermore, it has been shown that the lack of cell death by alcohol in this study was due to the combined loss of miR-335 and miR-21 and the loss of miR-335 alone was responsible for the increased NSC proliferation in response to alcohol [228]. In another study using a prenatal mouse model, it has been shown that alcohol increased certain miRNAs (miR-10a, -10b, -9, -145, -30a-3p and miRNA-152) and decreased other miRNAs (miR-200a, -496, -296, -30e-5p, -362, -339, -29c and -154) in the fetal brain. This resulted in impaired task acquisition in the Morris water maze in P35 mice [231].
Table 3.
Summary of the developmental alcohol elicited changes in miRNAs.
| Alcohol Exposure | Tissue Examined | Effects |
|---|---|---|
| 1. GD 12.5 | Neurosphere cultures | Reduced the expression of miR-9, 21, 335 and -135 [228] |
| 2. GD 6–15 | GD 17 embryo culture | Increased miR-10a, -9, -145, -30a-3p and -152. Also decreased miR-200a, -496, -296, -30e-5p, -362, -339, -29c and -154 [231] |
| 3. Zebrafish | Embryos (4–96 hpf) | Enhanced miR-153a, -30d, -736, -183 and reduced -23a [229] |
| 4. GD 12.5 | Neurosphere cultures | Reduced the expression of miR-140-3p [225] |
| 5. PD 7 | PD 60 | Increased expression of miR-26b [230] |
| 6. Zebrafish | Embryos (24–72 hpf) | Suppressed miR-9a and increased the accumulation of pre-miR-9-3 [270] |
| 7. PD 7 | PD 60 | Enhanced miR-302c [271] |
| 8. GD 1–22 | PD 42 | Decreased miR-874-5p, 1843a-3p, -221-5p, -29c-3p, -384-5p, -412-3p, 129-1, -138-2, -322-2, -496, -9a-2. Increased miR-155, -34c, -let-7c-1, -let-7c-2-3p, -542-1 [272] |
| 9. GD 0.5–8.5 | PD 87 | Enhanced miR-135a, -135b, -467b-5b and -487b [249] |
| 10. GD 4–132 (Ewes) | Plasma (GD 147) | Decreased miR-572, -720, -9, -15b, -17-92 and increased miR-34b [273] |
In another study, Zebrafish embryos were exposed to alcohol, which was then followed by the evaluation of miRNA levels and deformation. This study showed increases in miR-153a, -725, -30d, let-7k, -100, -738, and -732 and decreases in miR-128 and -193b and induced cell death along the dorsal axis of the embryo [229]. In another study, using P7 mouse model of FASD, alcohol was shown to enhance miR-26b levels in adult whole brain tissues [230]. MicroRNA-26b, which is implicated in the differentiation of NSCs [227], has also been shown to control the expression of genes that encode for brain-derived neurotrophic factor (BDNF), which is important for neurodevelopment [226,227]. Exposure of GD12.5 mouse fetal murine cerebral cortical-derived neurosphere cultures to alcohol suppressed the expression of several miRNAs, including miR-9, -21, -153, and -335, as well as miR-140-3p [225]. In Zebrafish embryos, alcohol suppresses miR-9 biogenesis. This early embryonic loss of miR-9 function recapitulates the severe range of teratology that is associated with early alcohol exposure [270]. In another study that uses an acute prenatal model (second-trimester equivalent), alcohol was shown to enhance miR-302c in the adult whole brain [271]. MicroRNA-302 is suggested to have a critical function in neural tube closure and embryonic survival [274]. Furthermore, rats treated with acute alcohol at GD 12 exhibit both upward and downward changes in several miRNAs in the amygdala and ventral striatum in association with decreased social motivation behavior at P42 [272]. Social enrichment of GD12 alcohol-exposed rats rescued both miRNA and social motivation behavior in P42 mice [272]. Furthermore, it has been shown that miR-125b can rescue alcohol-induced apoptosis in NCCs by decreasing the expression of Bcl-2 antagonist killer 1 (BAK1) and p53-upregulated modulator of apoptosis (PUMA) genes (pro-apoptotic genes). Additionally, the microinjection of miR-125b mimic into cultured mouse embryos prevented alcohol-induced embryotoxicity [275]. Prenatal exposure of mice on GD1-8 resulted in enhanced miR-135a, -135b, -467b-5p and -487b in the P87 murine hippocampus. Additionally, miR-467b-5p and the 3′UTR of Slc17a6 exhibited specific interactions in vitro and suggested that a similar interaction in vivo may contribute to reduced HP VGLUT2 protein levels in alcohol-exposed adult males [249]. In this model, alcohol not only enhanced HP levels of miRNAs but also increased miRNAs, miR-135a, miR-135b and miR-467b-5p levels in the serum [249]. These findings suggest that miRNA may have an important role in HP dysfunction found in FASD and circulating miRNAs could be used as biomarkers of early developmental alcohol exposure [249]. A similar change in circulating miRNAs (miR-9, -15b, -19b, and -20a) was also found in pregnant ewes and in newborn lambs (GD 147) that were exposed to alcohol between GD 4 and GD 132 [273]. Alcohol exposure in P7 mice was shown to alter several miRNA transcripts and was found to be associated with genes that are important for neural connectivity [276] and may contribute to cognitive deficits, as found in FASD children.
4. Conclusions
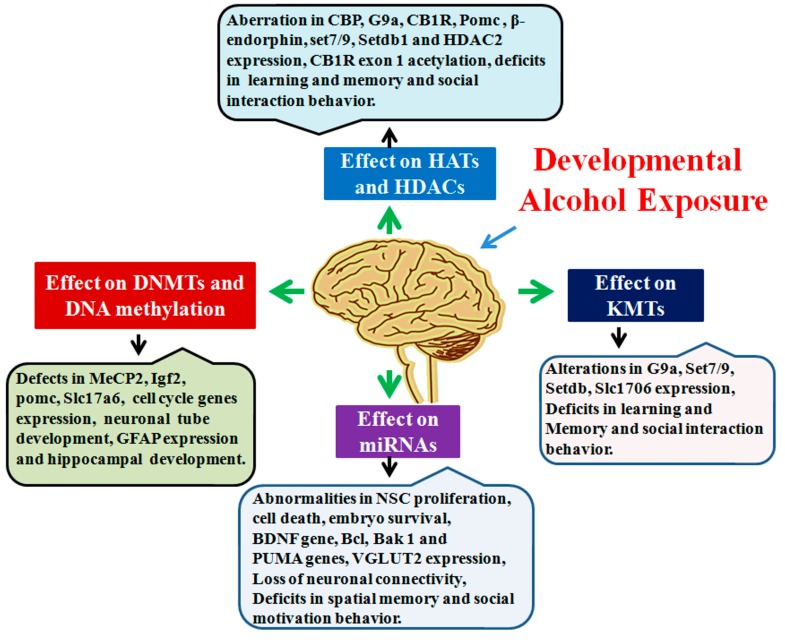
In summary, several epigenetic mechanisms are suggested to contribute to the harmful consequences of alcohol abuse during pregnancy in the developing fetus (Figure 5). Epigenetic modifications, such as altered DNA methylation, specific histone protein modification and dysregulation of miRNA, in response to developmental alcohol exposure, can contribute to impaired neurogenesis, neuronal communication and neural circuit assembly. These alcohol-induced neuronal deficits can be long-lasting and could result in abnormalities in synaptic plasticity and cognitive function and can provide bases for many of the neurobehavioral abnormalities found in FASD. The broad role of DNA methylation and chromatin modification in learning and memory has not been fully understood. This is because learning and memory is regulated by a dynamic interplay between the various signaling molecules, enzymes and proteins that are capable of modifying the chromatin structure. Additionally, there is a lack of understanding of the mechanisms that both predicate and result from epigenetic modifications that regulate learning and memory process. Modulation of epigenetic marks using pharmacological and gene knock-in or knockout approaches support a role for the epigenetic process in learning and memory, therefore making it a promising target for the future development of potential therapeutic agents to treat FASD.
Figure 5.
Graphical summary of developmental alcohol-induced epigenetic defects. Developmental alcohol exposure has been shown to affect DNA methylation, HATs/HDACs, KMTs and miRNAs, followed by several changes in genes and protein expression that are important for cognitive and other brain function.
Acknowledgements
This work was supported by National Institute of Alcohol and Alcoholism grant (RO1-AA019443) to Balapal S. Basavarajappa.
Abbreviations
The following abbreviations are used in this manuscript:
FAS, fetal alcohol syndrome; FASD, fetal alcohol spectrum disorder; miRNA, microRNA; CpG, cytosine phosphate guanine islands; PMT, posttranslational modification; snoRNA, small nucleolar RNA; lncRNA, long non-coding RNA; DNMTs, DNA methyltransferases; SAM, S-adenyl methionine; TET, ten-eleven translocation; mC, methylcytosine; 5-hmC, 5-hydroxymethylcytosine; 5-caC, 5-carboxylcytosine; MeCP2, methyl-binding protein 2; HDACs, histone deacetylases; LTP, long-term potentiation; HAT, Histone acetyltransferases; Gcn5, general control nonderepressible-5; CREB, cAMP response element-binding protein; CBP, CREB binding protein; PAX3, paired box 3; AP2, activation of enhancer binding protein 2; TSA, trichostatin; SAHA, suberoylanilide hydroxamic acid; PRMT, arginine methyltransferases; CARM1, cofactor-associated arginine methyltransferase; SET, Su (var)3-9, Enhancer of Zeste, Trithorax; MLL, mixed lineage leukemia; GLP, G9A-like protein; KMTs, lysine methyltransferases; HMTs, Histone methyltransferases; KDM, lysine demethylase; BRAF, v-raf murine sarcoma viral oncogene homolog B1; REST, restrictive silencer factor; PHD, plant homeodomain; AR, androgen receptor; ER, estrogen receptor; ARID, AT-rich interaction domain; BAL, blood alcohol level; ASD, autism spectral disorder; ADHD, attention deficit hyperactivity disorder; GD, gestational day; GFAP, glial fibrillary acidic protein; NSC, neural stem cell; IGF2, insulin-like growth factor 2; HP, hioppocampus; NC, neocortex; PFC, prefrontal cortex; SLC17A6, solute carrier family 17 member 6; VGLUT2, vesicular glutamate transporter 2; hESCs, human embryonic stem cells; ARC, activity-regulated cytoskeletal protein; CB1R, cannabinoid receptor type 1; α-MHC, α-myosin heavy chain; cTnT, cardiac troponin T; BEP, β-endorphin; BAK1, Bcl-2 antagonist killer 1; PUMA, p53-upregulated modulator of apoptosis; hGCN5, human general control of amino acid synthesis protein 5-like 2; PCAF, p300/CBP-associated factor; ELP3, elongation protein 3; TIP60, TAT interacting proteins 60; TFIIIC90, transcription factor IIIC 90kDa; TAF1, TATA Box Binding Protein-Associated Factor; SRC1, steroid receptor coactivator 1; ACTR, activator of thyroid receptor; p160, receptor coactivators proteins 160; CLOCK, Clock Circadian Regulator.
Author Contributions
Balapal S. Basavarajappa conceived, designed and wrote the sections on epigenetics and FASD. Shivakumar Subbanna contributed literature on alcohol’s influence on epigenetic mechanisms and provided tables for the review.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Heijmans B.T., Tobi E.W., Stein A.D., Putter H., Blauw G.J., Susser E.S., Slagboom P.E., Lumey L.H. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl. Acad. Sci. USA. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar A., Choi K.H., Renthal W., Tsankova N.M., Theobald D.E., Truong H.T., Russo S.J., Laplant Q., Sasaki T.S., Whistler K.N., et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 3.Madrigano J., Baccarelli A., Mittleman M.A., Wright R.O., Sparrow D., Vokonas P.S., Tarantini L., Schwartz J. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ. Health Perspect. 2011;119:977–982. doi: 10.1289/ehp.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Subbanna S., Nagaraja N.N., Umapathy N.S., Pace B.S., Basavarajappa B.S. Ethanol exposure induces neonatal neurodegeneration by enhancing CB1R Exon1 histone H4K8 acetylation and up-regulating CB1R function causing neurobehavioral abnormalities in adult mice. Int. J. Neuropsychopharmacol. 2015 doi: 10.1093/ijnp/pyu028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subbanna S., Nagre N.N., Shivakumar M., Umapathy N.S., Psychoyos D., Basavarajappa B.S. Ethanol induced acetylation of histone at G9a Exon1 and G9a-mediated histone H3 dimethylation leads to neurodegeneration in neonatal mice. Neuroscience. 2014;258:422–432. doi: 10.1016/j.neuroscience.2013.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Subbanna S., Shivakumar M., Umapathy N.S., Saito M., Mohan P.S., Kumar A., Nixonc R.A., Verin A.D., Psychoyos D., Basavarajappa B.S. G9a-mediated histone methylation regulates ethanol-induced neurodegeneration in the neonatal mouse brain. Neurobiol. Dis. 2013;54:475–485. doi: 10.1016/j.nbd.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suderman M., McGowan P.O., Sasaki A., Huang T.C., Hallett M.T., Meaney M.J., Turecki G., Szyf M. Conserved epigenetic sensitivity to early life experience in the rat and human hippocampus. Proc. Natl. Acad. Sci. USA. 2012;109:17266–17272. doi: 10.1073/pnas.1121260109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang T.Y., Hellstrom I.C., Bagot R.C., Wen X., Diorio J., Meaney M.J. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J. Neurosci. 2010;30:13130–13137. doi: 10.1523/JNEUROSCI.1039-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avery O.T., Macleod C.M., McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarty M., Avery O.T. Studies on the chemical nature of the substance inducing transformation on pneumococcal types; an improved method for the isolation of the transforming substance and its application to Pneumococcus Types II, III, and VI. J. Exp. Med. 1946;83:97–104. doi: 10.1084/jem.83.2.97. [DOI] [PubMed] [Google Scholar]
- 11.Hotchkiss R.D. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J. Biol. Chem. 1948;175:315–332. [PubMed] [Google Scholar]
- 12.Compere S.J., Palmiter R.D. DNA methylation controls the inducibility of the mouse metallothionein-I gene lymphoid cells. Cell. 1981;25:233–240. doi: 10.1016/0092-8674(81)90248-8. [DOI] [PubMed] [Google Scholar]
- 13.Holliday R., Pugh J.E. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. doi: 10.1126/science.1111098. [DOI] [PubMed] [Google Scholar]
- 14.Guo J.U., Ma D.K., Mo H., Ball M.P., Jang M.H., Bonaguidi M.A., Balazer J.A., Eaves H.L., Xie B., Ford E., et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011;14:1345–1351. doi: 10.1038/nn.2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J.U., Su Y., Zhong C., Ming G.L., Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams R.L., Burdon R.H., Gibb S., McKay E.L. DNA methylase during Xenopus laevis development. Biochim. Biophys. Acta. 1981;655:329–334. doi: 10.1016/0005-2787(81)90042-3. [DOI] [PubMed] [Google Scholar]
- 17.Adams R.L., McKay E.L., Craig L.M., Burdon R.H. Mouse DNA methylase: Methylation of native DNA. Biochim. Biophys. Acta. 1979;561:345–357. doi: 10.1016/0005-2787(79)90143-6. [DOI] [PubMed] [Google Scholar]
- 18.Gruenbaum Y., Cedar H., Razin A. Substrate and sequence specificity of a eukaryotic DNA methylase. Nature. 1982;295:620–622. doi: 10.1038/295620a0. [DOI] [PubMed] [Google Scholar]
- 19.Razin A., Riggs A.D. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- 20.Razin A., Szyf M. DNA methylation patterns. Formation and function. Biochim. Biophys. Acta. 1984;782:331–342. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- 21.Brown S.E., Weaver I.C., Meaney M.J., Szyf M. Regional-specific global cytosine methylation and DNA methyltransferase expression in the adult rat hippocampus. Neurosci. Lett. 2008;440:49–53. doi: 10.1016/j.neulet.2008.05.028. [DOI] [PubMed] [Google Scholar]
- 22.Feng J., Chang H., Li E., Fan G. Dynamic expression of de novo DNA methyltransferases DNMT3a and DNMT3b in the central nervous system. J. Neurosci. Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 23.Nagre N.N., Subbanna S., Shivakumar M., Psychoyos D., Basavarajappa B.S. CB1-receptor knockout neonatal mice are protected against ethanol-induced impairments of DNMT1, DNMT3a and DNA methylation. J. Neurochem. 2015;132:429–442. doi: 10.1111/jnc.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson K.D., Uzvolgyi E., Liang G., Talmadge C., Sumegi J., Gonzales F.A., Jones P.A. The human DNA methyltransferases (DNMTs) 1, 3a and 3b: Coordinate mRNA expression in normal tissues and overexpression in tumors. Nucleic Acids Res. 1999;27:2291–2298. doi: 10.1093/nar/27.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simmons R.K., Stringfellow S.A., Glover M.E., Wagle A.A., Clinton S.M. DNA methylation markers in the postnatal developing rat brain. Brain Res. 2013;1533:26–36. doi: 10.1016/j.brainres.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng J., Zhou Y., Campbell S.L., Le T., Li E., Sweatt J.D., Silva A.J., Fan G. DNMT1 and DNMT3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goto K., Numata M., Komura J.I., Ono T., Bestor T.H., Kondo H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- 28.Nabel C.S., Kohli R.M. Molecular biology. Demystifying DNA demethylation. Science. 2011;333:1229–1230. doi: 10.1126/science.1211917. [DOI] [PubMed] [Google Scholar]
- 29.Szulwach K.E., Li X., Li Y., Song C.X., Wu H., Dai Q., Irier H., Upadhyay A.K., Gearing M., Levey A.I., et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat. Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kubiura M., Okano M., Kimura H., Kawamura F., Tada M. Chromosome-wide regulation of euchromatin-specific 5mC to 5hmC conversion in mouse ES cells and female human somatic cells. Chromosome Res. 2012;20:837–848. doi: 10.1007/s10577-012-9317-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Szyf M. The early-life social environment and DNA methylation. Clin. Genet. 2012;81:341–349. doi: 10.1111/j.1399-0004.2012.01843.x. [DOI] [PubMed] [Google Scholar]
- 32.Szyf M. How do environments talk to genes? Nat. Neurosci. 2013;16:2–4. doi: 10.1038/nn.3286. [DOI] [PubMed] [Google Scholar]
- 33.Szyf M. DNA methylation, behavior and early life adversity. J. Genet. Genom. 2013;40:331–338. doi: 10.1016/j.jgg.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 34.Daxinger L., Whitelaw E. Transgenerational epigenetic inheritance: More questions than answers. Genome Res. 2010;20:1623–1628. doi: 10.1101/gr.106138.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaminen-Ahola N., Ahola A., Maga M., Mallitt K.A., Fahey P., Cox T.C., Whitelaw E., Chong S. Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS Genet. 2010;6:12. doi: 10.1371/journal.pgen.1000811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morris M.J., Monteggia L.M. Role of DNA methylation and the DNA methyltransferases in learning and memory. Dialogues Clin. Neurosci. 2014;16:359–371. doi: 10.31887/DCNS.2014.16.3/mmorris. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rudenko A., Dawlaty M.M., Seo J., Cheng A.W., Meng J., Le T., Faull K.F., Jaenisch R., Tsai L.H. TET1 is critical for neuronal activity-regulated gene expression and memory extinction. Neuron. 2013;79:1109–1122. doi: 10.1016/j.neuron.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergman Y., Cedar H. DNA methylation dynamics in health and disease. Nat. Struct. Mol. Biol. 2013;20:274–281. doi: 10.1038/nsmb.2518. [DOI] [PubMed] [Google Scholar]
- 39.He Y.F., Li B.Z., Li Z., Liu P., Wang Y., Tang Q., Ding J., Jia Y., Chen Z., Li L., et al. TET-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333:1303–1307. doi: 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller C.A., Gavin C.F., White J.A., Parrish R.R., Honasoge A., Yancey C.R., Rivera I.M., Rubio M.D., Rumbaugh G., Sweatt J.D. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010;13:664–666. doi: 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miranda T.B., Jones P.A. DNA methylation: The nuts and bolts of repression. J. Cell. Physiol. 2007;213:384–390. doi: 10.1002/jcp.21224. [DOI] [PubMed] [Google Scholar]
- 42.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases DNMT3a and DNMT3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 43.Okano M., Xie S., Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998;19:219–220. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- 44.Hermann A., Goyal R., Jeltsch A. The DNMT1 DNA-(cytosine-C5)-methyltransferase methylates DNA processively with high preference for hemimethylated target sites. J. Biol. Chem. 2004;279:48350–48359. doi: 10.1074/jbc.M403427200. [DOI] [PubMed] [Google Scholar]
- 45.Ito S., D’Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by mll partner tet1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 48.Sarraf S.A., Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol. Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 49.Akbarian S., Chen R.Z., Gribnau J., Rasmussen T.P., Fong H., Jaenisch R., Jones E.G. Expression pattern of the Rett syndrome gene MeCP2 in primate prefrontal cortex. Neurobiol. Dis. 2001;8:784–791. doi: 10.1006/nbdi.2001.0420. [DOI] [PubMed] [Google Scholar]
- 50.Shahbazian M.D., Antalffy B., Armstrong D.L., Zoghbi H.Y. Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum. Mol. Genet. 2002;11:115–124. doi: 10.1093/hmg/11.2.115. [DOI] [PubMed] [Google Scholar]
- 51.Martinowich K., Hattori D., Wu H., Fouse S., He F., Hu Y., Fan G., Sun Y.E. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 52.Tao J., Hu K., Chang Q., Wu H., Sherman N.E., Martinowich K., Klose R.J., Schanen C., Jaenisch R., Wang W., et al. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc. Natl. Acad. Sci. USA. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou Z., Hong E.J., Cohen S., Zhao W.N., Ho H.Y., Schmidt L., Chen W.G., Lin Y., Savner E., Griffith E.C., et al. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen S., Gabel H.W., Hemberg M., Hutchinson A.N., Sadacca L.A., Ebert D.H., Harmin D.A., Greenberg R.S., Verdine V.K., Zhou Z., et al. Genome-wide activity-dependent MeCP2 phosphorylation regulates nervous system development and function. Neuron. 2011;72:72–85. doi: 10.1016/j.neuron.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Zhong X., Chau K.F., Williams E.C., Chang Q. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat. Neurosci. 2011;14:1001–1008. doi: 10.1038/nn.2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amir R.E., van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MeCP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 57.Na E.S., Nelson E.D., Kavalali E.T., Monteggia L.M. The impact of MeCP2 loss- or gain-of-function on synaptic plasticity. Neuropsychopharmacology. 2013;38:212–219. doi: 10.1038/npp.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Na E.S., Nelson E.D., Adachi M., Autry A.E., Mahgoub M.A., Kavalali E.T., Monteggia L.M. A mouse model for MeCP2 duplication syndrome: MeCP2 overexpression impairs learning and memory and synaptic transmission. J. Neurosci. 2012;32:3109–3117. doi: 10.1523/JNEUROSCI.6000-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luger K., Dechassa M.L., Tremethick D.J. New insights into nucleosome and chromatin structure: An ordered state or a disordered affair? Nature Rev. Mol. Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kornberg R.D., Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98:285–294. doi: 10.1016/S0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 61.Lorch Y., Zhang M., Kornberg R.D. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/S0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 62.Goll M.G., Bestor T.H. Histone modification and replacement in chromatin activation. Genes Dev. 2002;16:1739–1742. doi: 10.1101/gad.1013902. [DOI] [PubMed] [Google Scholar]
- 63.Grant P.A. A tale of histone modifications. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-4-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smolle M., Workman J.L. Transcription-associated histone modifications and cryptic transcription. Biochim. Biophys. Acta. 2013;1829:84–97. doi: 10.1016/j.bbagrm.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petruk S., Sedkov Y., Johnston D.M., Hodgson J.W., Black K.L., Kovermann S.K., Beck S., Canaani E., Brock H.W., Mazo A. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 2012;150:922–933. doi: 10.1016/j.cell.2012.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marmorstein R., Roth S.Y. Histone acetyltransferases: Function, structure, and catalysis. Curr. Opin. Genet. Dev. 2001;11:155–161. doi: 10.1016/S0959-437X(00)00173-8. [DOI] [PubMed] [Google Scholar]
- 67.Roth S.Y., Denu J.M., Allis C.D. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 68.Yun M., Wu J., Workman J.L., Li B. Readers of histone modifications. Cell Res. 2011;21:564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zeng L., Zhou M.M. Bromodomain: An acetyl-lysine binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/S0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 70.Kalkhoven E., Roelfsema J.H., Teunissen H., den Boer A., Ariyurek Y., Zantema A., Breuning M.H., Hennekam R.C., Peters D.J. Loss of CBP acetyltransferase activity by PHD finger mutations in Rubinstein-Taybi syndrome. Hum. Mol. Genet. 2003;12:441–450. doi: 10.1093/hmg/ddg039. [DOI] [PubMed] [Google Scholar]
- 71.Tanaka Y., Naruse I., Hongo T., Xu M., Nakahata T., Maekawa T., Ishii S. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech. Dev. 2000;95:133–145. doi: 10.1016/S0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 72.Yao T.P., Oh S.P., Fuchs M., Zhou N.D., Ch’ng L.E., Newsome D., Bronson R.T., Li E., Livingston D.M., Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/S0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 73.Martinez-Cerdeno V., Lemen J.M., Chan V., Wey A., Lin W., Dent S.R., Knoepfler P.S. N-Myc and GCN5 regulate significantly overlapping transcriptional programs in neural stem cells. PLoS ONE. 2012;7:12. doi: 10.1371/journal.pone.0039456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bousiges O., Vasconcelos A.P., Neidl R., Cosquer B., Herbeaux K., Panteleeva I., Loeffler J.P., Cassel J.C., Boutillier A.L. Spatial memory consolidation is associated with induction of several lysine-acetyltransferase (histone acetyltransferase) expression levels and H2B/H4 acetylation-dependent transcriptional events in the rat hippocampus. Neuropsychopharmacology. 2010;35:2521–2537. doi: 10.1038/npp.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gehrking K.M., Andresen J.M., Duvick L., Lough J., Zoghbi H.Y., Orr H.T. Partial loss of Tip60 slows mid-stage neurodegeneration in a spinocerebellar ataxia type 1 (SCA1) mouse model. Hum. Mol. Genet. 2011;20:2204–2212. doi: 10.1093/hmg/ddr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Levenson J.M., O’Riordan K.J., Brown K.D., Trinh M.A., Molfese D.L., Sweatt J.D. Regulation of histone acetylation during memory formation in the hippocampus. J. Biol. Chem. 2004;279:40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- 77.Peleg S., Sananbenesi F., Zovoilis A., Burkhardt S., Bahari-Javan S., Agis-Balboa R.C., Cota P., Wittnam J.L., Gogol-Doering A., Opitz L., et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science. 2010;328:753–756. doi: 10.1126/science.1186088. [DOI] [PubMed] [Google Scholar]
- 78.Chatterjee S., Mizar P., Cassel R., Neidl R., Selvi B.R., Mohankrishna D.V., Vedamurthy B.M., Schneider A., Bousiges O., Mathis C., et al. A novel activator of CBP/p300 acetyltransferases promotes neurogenesis and extends memory duration in adult mice. J. Neurosci. 2013;33:10698–10712. doi: 10.1523/JNEUROSCI.5772-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei W., Coelho C.M., Li X., Marek R., Yan S., Anderson S., Meyers D., Mukherjee C., Sbardella G., Castellano S., et al. P300/CBP-associated factor selectively regulates the extinction of conditioned fear. J. Neurosci. 2012;32:11930–11941. doi: 10.1523/JNEUROSCI.0178-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Johnson A.A., Sarthi J., Pirooznia S.K., Reube W., Elefant F. Increasing Tip60 HAT levels rescues axonal transport defects and associated behavioral phenotypes in a Drosophila Alzheimer’s disease model. J. Neurosci. 2013;33:7535–7547. doi: 10.1523/JNEUROSCI.3739-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pirooznia S.K., Elefant F. Targeting specific HATs for neurodegenerative disease treatment: Translating basic biology to therapeutic possibilities. Front. Cell. Neurosci. 2013;7 doi: 10.3389/fncel.2013.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schneider A., Chatterjee S., Bousiges O., Selvi B.R., Swaminathan A., Cassel R., Blanc F., Kundu T.K., Boutillier A.L. Acetyltransferases (HATs) as targets for neurological therapeutics. Neurotherapeutics. 2013;10:568–588. doi: 10.1007/s13311-013-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haberland M., Montgomery R.L., Olson E.N. The many roles of histone deacetylases in development and physiology: Implications for disease and therapy. Nature reviews. Genetics. 2009;10:32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lahm A., Paolini C., Pallaoro M., Nardi M.C., Jones P., Neddermann P., Sambucini S., Bottomley M.J., Lo Surdo P., Carfi A., et al. Unraveling the hidden catalytic activity of vertebrate class IIa histone deacetylases. Proc. Natl. Acad. Sci. USA. 2007;104:17335–17340. doi: 10.1073/pnas.0706487104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Finkel T., Deng C.X., Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Michan S., Sinclair D. Sirtuins in mammals: Insights into their biological function. Biochem. J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Morris M.J., Monteggia L.M. Unique functional roles for class I and class II histone deacetylases in central nervous system development and function. Int. J. Dev. Neurosci. 2013;31:370–381. doi: 10.1016/j.ijdevneu.2013.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yeh H.H., Young D., Gelovani J.G., Robinson A., Davidson Y., Herholz K., Mann D.M. Histone deacetylase class II and acetylated core histone immunohistochemistry in human brains with Huntington’s disease. Brain Res. 2013;1504:16–24. doi: 10.1016/j.brainres.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 89.Yoo J.Y., Larouche M., Goldowitz D. The expression of HDAC1 and HDAC2 during cerebellar cortical development. Cerebellum. 2013;12:534–546. doi: 10.1007/s12311-013-0459-x. [DOI] [PubMed] [Google Scholar]
- 90.Montgomery R.L., Hsieh J., Barbosa A.C., Richardson J.A., Olson E.N. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc. Natl. Acad. Sci. USA. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jawerka M., Colak D., Dimou L., Spiller C., Lagger S., Montgomery R.L., Olson E.N., Wurst W., Gottlicher M., Gotz M. The specific role of histone deacetylase 2 in adult neurogenesis. Neuron Glia Biol. 2010;6:93–107. doi: 10.1017/S1740925X10000049. [DOI] [PubMed] [Google Scholar]
- 92.Morris M.J., Karra A.S., Monteggia L.M. Histone deacetylases govern cellular mechanisms underlying behavioral and synaptic plasticity in the developing and adult brain. Behav. Pharmacol. 2010;21:409–419. doi: 10.1097/FBP.0b013e32833c20c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Soriano F.X., Hardingham G.E. In cortical neurons HDAC3 activity suppresses RD4-dependent SMRT export. PLoS ONE. 2011;6:12. doi: 10.1371/journal.pone.0021056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schneider J.W., Gao Z., Li S., Farooqi M., Tang T.S., Bezprozvanny I., Frantz D.E., Hsieh J. Small-molecule activation of neuronal cell fate. Nat Chem Biol. 2008;4:408–410. doi: 10.1038/nchembio.95. [DOI] [PubMed] [Google Scholar]
- 95.Lai I.L., Lin T.P., Yao Y.L., Lin C.Y., Hsieh M.J., Yang W.M. Histone deacetylase 10 relieves repression on the melanogenic program by maintaining the deacetylation status of repressors. J. Biol. Chem. 2010;285:7187–7196. doi: 10.1074/jbc.M109.061861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Cho Y., Cavalli V. HDAC signaling in neuronal development and axon regeneration. Curr. Opin. Neurobiol. 2014;27:118–126. doi: 10.1016/j.conb.2014.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guan J.S., Haggarty S.J., Giacometti E., Dannenberg J.H., Joseph N., Gao J., Nieland T.J., Zhou Y., Wang X., Mazitschek R., et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Barrett R.M., Wood M.A. Beyond transcription factors: The role of chromatin modifying enzymes in regulating transcription required for memory. Learn. Mem. 2008;15:460–467. doi: 10.1101/lm.917508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vecsey C.G., Hawk J.D., Lattal K.M., Stein J.M., Fabian S.A., Attner M.A., Cabrera S.M., McDonough C.B., Brindle P.K., Abel T., et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. J. Neurosci. 2007;27:6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yeh S.H., Lin C.H., Gean P.W. Acetylation of nuclear factor-kappaB in rat amygdala improves long-term but not short-term retention of fear memory. Mol. Pharmacol. 2004;65:1286–1292. doi: 10.1124/mol.65.5.1286. [DOI] [PubMed] [Google Scholar]
- 101.Kilgore M., Miller C.A., Fass D.M., Hennig K.M., Haggarty S.J., Sweatt J.D., Rumbaugh G. Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer’s disease. Neuropsychopharmacology. 2010;35:870–880. doi: 10.1038/npp.2009.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Didonna A., Opal P. The promise and perils of HDAC inhibitors in neurodegeneration. Ann. Clin. Transl. Neurol. 2015;2:79–101. doi: 10.1002/acn3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi X., Hong T., Walter K.L., Ewalt M., Michishita E., Hung T., Carney D., Pena P., Lan F., Kaadige M.R., et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shi X., Kachirskaia I., Walter K.L., Kuo J.H., Lake A., Davrazou F., Chan S.M., Martin D.G., Fingerman I.M., Briggs S.D., et al. Proteome-wide analysis in Saccharomyces cerevisiae identifies several PHD fingers as novel direct and selective binding modules of histone H3 methylated at either lysine 4 or lysine 36. J. Biol. Chem. 2007;282:2450–2455. doi: 10.1074/jbc.C600286200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Wysocka J., Swigut T., Xiao H., Milne T.A., Kwon S.Y., Landry J., Kauer M., Tackett A.J., Chait B.T., Badenhorst P., et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 106.Xiao B., Jing C., Wilson J.R., Walker P.A., Vasisht N., Kelly G., Howell S., Taylor I.A., Blackburn G.M., Gamblin S.J. Structure and catalytic mechanism of the human histone methyltransferase SET7/9. Nature. 2003;421:652–656. doi: 10.1038/nature01378. [DOI] [PubMed] [Google Scholar]
- 107.Allis C.D., Berger S.L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R., et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 108.Qian C., Zhou M.M. SET domain protein lysine methyltransferases: Structure, specificity and catalysis. Cell. Mol. Life Sci. 2006;63:2755–2763. doi: 10.1007/s00018-006-6274-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Aravind L., Abhiman S., Iyer L.M. Natural history of the eukaryotic chromatin protein methylation system. Prog. Mol. Biol. Transl. Sci. 2011;101:105–176. doi: 10.1016/B978-0-12-387685-0.00004-4. [DOI] [PubMed] [Google Scholar]
- 110.Glaser S., Schaft J., Lubitz S., Vintersten K., van der Hoeven F., Tufteland K.R., Aasland R., Anastassiadis K., Ang S.L., Stewart A.F. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- 111.Goo Y.H., Sohn Y.C., Kim D.H., Kim S.W., Kang M.J., Jung D.J., Kwak E., Barlev N.A., Berger S.L., Chow V.T., et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Milne T.A., Briggs S.D., Brock H.W., Martin M.E., Gibbs D., Allis C.D., Hess J.L. Mll targets SET domain methyltransferase activity to hox gene promoters. Mol. Cell. 2002;10:1107–1117. doi: 10.1016/S1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- 113.Peters A.H., O’Carroll D., Scherthan H., Mechtler K., Sauer S., Schofer C., Weipoltshammer K., Pagani M., Lachner M., Kohlmaier A., et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 114.Tachibana M., Sugimoto K., Nozaki M., Ueda J., Ohta T., Ohki M., Fukuda M., Takeda N., Niida H., Kato H., et al. G9a histone methyltransferase plays a dominant role in euchromatic histone H3 lysine 9 methylation and is essential for early embryogenesis. Genes Dev. 2002;16:1779–1791. doi: 10.1101/gad.989402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tachibana M., Ueda J., Fukuda M., Takeda N., Ohta T., Iwanari H., Sakihama T., Kodama T., Hamakubo T., Shinkai Y. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schultz D.C., Ayyanathan K., Negorev D., Maul G.G., Rauscher F.J., 3rd SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev. 2002;16:919–932. doi: 10.1101/gad.973302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lund A.H., van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr. Opin. Cell Biol. 2004;16:239–246. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- 118.Otte A.P., Kwaks T.H. Gene repression by Polycomb group protein complexes: A distinct complex for every occasion? Curr. Opin. Genet. Dev. 2003;13:448–454. doi: 10.1016/S0959-437X(03)00108-4. [DOI] [PubMed] [Google Scholar]
- 119.Huang J., Perez-Burgos L., Placek B.J., Sengupta R., Richter M., Dorsey J.A., Kubicek S., Opravil S., Jenuwein T., Berger S.L. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 120.Lu T., Jackson M.W., Wang B., Yang M., Chance M.R., Miyagi M., Gudkov A.V., Stark G.R. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl. Acad. Sci. USA. 2010;107:46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Saddic L.A., West L.E., Aslanian A., Yates J.R., 3rd, Rubin S.M., Gozani O., Sage J. Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. 2010;285:37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kunizaki M., Hamamoto R., Silva F.P., Yamaguchi K., Nagayasu T., Shibuya M., Nakamura Y., Furukawa Y. The lysine 831 of vascular endothelial growth factor receptor 1 is a novel target of methylation by SMYD3. Cancer Res. 2007;67:10759–10765. doi: 10.1158/0008-5472.CAN-07-1132. [DOI] [PubMed] [Google Scholar]
- 123.Nguyen A.T., Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Karachentsev D., Sarma K., Reinberg D., Steward R. PR-SET7-dependent methylation of histone H4 lys 20 functions in repression of gene expression and is essential for mitosis. Genes Dev. 2005;19:431–435. doi: 10.1101/gad.1263005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hakimi M.A., Bochar D.A., Chenoweth J., Lane W.S., Mandel G., Shiekhattar R. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. USA. 2002;99:7420–7425. doi: 10.1073/pnas.112008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hakimi M.A., Dong Y., Lane W.S., Speicher D.W., Shiekhattar R. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 2003;278:7234–7239. doi: 10.1074/jbc.M208992200. [DOI] [PubMed] [Google Scholar]
- 127.You A., Tong J.K., Grozinger C.M., Schreiber S.L. CoREST is an integral component of the CoREST-human histone deacetylase complex. Proc. Natl. Acad. Sci. USA. 2001;98:1454–1458. doi: 10.1073/pnas.98.4.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 129.Ciccone D.N., Su H., Hevi S., Gay F., Lei H., Bajko J., Xu G., Li E., Chen T. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature. 2009;461:415–418. doi: 10.1038/nature08315. [DOI] [PubMed] [Google Scholar]
- 130.Van Essen D., Zhu Y., Saccani S. A feed-forward circuit controlling inducible NF-kappaB target gene activation by promoter histone demethylation. Mol. Cell. 2010;39:750–760. doi: 10.1016/j.molcel.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 131.Tsukada Y., Fang J., Erdjument-Bromage H., Warren M.E., Borchers C.H., Tempst P., Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 132.Yamane K., Toumazou C., Tsukada Y., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–495. doi: 10.1016/j.cell.2006.03.027. [DOI] [PubMed] [Google Scholar]
- 133.Fodor B.D., Kubicek S., Yonezawa M., O’Sullivan R.J., Sengupta R., Perez-Burgos L., Opravil S., Mechtler K., Schotta G., Jenuwein T. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Klose R.J., Yamane K., Bae Y., Zhang D., Erdjument-Bromage H., Tempst P., Wong J., Zhang Y. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 135.Whetstine J.R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 136.Adams-Cioaba M.A., Min J. Structure and function of histone methylation binding proteins. Biochem. Cell Biol. 2009;87:93–105. doi: 10.1139/O08-129. [DOI] [PubMed] [Google Scholar]
- 137.Huang Y., Fang J., Bedford M.T., Zhang Y., Xu R.M. Recognition of histone H3 lysine-4 methylation by the double tudor domain of JMJD2A. Science. 2006;312:748–751. doi: 10.1126/science.1125162. [DOI] [PubMed] [Google Scholar]
- 138.Kawazu M., Saso K., Tong K.I., McQuire T., Goto K., Son D.O., Wakeham A., Miyagishi M., Mak T.W., Okada H. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS ONE. 2011;6:12. doi: 10.1371/journal.pone.0017830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shi L., Sun L., Li Q., Liang J., Yu W., Yi X., Yang X., Li Y., Han X., Zhang Y., et al. Histone demethylase JMJD2B coordinates H3K4/H3K9 methylation and promotes hormonally responsive breast carcinogenesis. Proc. Natl. Acad. Sci. USA. 2011;108:7541–7546. doi: 10.1073/pnas.1017374108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Christensen J., Agger K., Cloos P.A., Pasini D., Rose S., Sennels L., Rappsilber J., Hansen K.H., Salcini A.E., Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 141.Iwase S., Lan F., Bayliss P., de la Torre-Ubieta L., Huarte M., Qi H.H., Whetstine J.R., Bonni A., Roberts T.M., Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- 142.Lee M.G., Norman J., Shilatifard A., Shiekhattar R. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell. 2007;128:877–887. doi: 10.1016/j.cell.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 143.Tahiliani M., Mei P., Fang R., Leonor T., Rutenberg M., Shimizu F., Li J., Rao A., Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 144.Yamane K., Tateishi K., Klose R.J., Fang J., Fabrizio L.A., Erdjument-Bromage H., Taylor-Papadimitriou J., Tempst P., Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol. Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 145.Wang G.G., Song J., Wang Z., Dormann H.L., Casadio F., Li H., Luo J.L., Patel D.J., Allis C.D. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Agger K., Cloos P.A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A.E., Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 147.De Santa F., Totaro M.G., Prosperini E., Notarbartolo S., Testa G., Natoli G. The histone H3 lysine-27 demethylase JMJD3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 148.Hong S., Cho Y.W., Yu L.R., Yu H., Veenstra T.D., Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc. Natl. Acad. Sci. USA. 2007;104:18439–18444. doi: 10.1073/pnas.0707292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lee M.G., Villa R., Trojer P., Norman J., Yan K.P., Reinberg D., di Croce L., Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 150.Xiang Y., Zhu Z., Han G., Lin H., Xu L., Chen C.D. Jmjd3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- 151.Baba A., Ohtake F., Okuno Y., Yokota K., Okada M., Imai Y., Ni M., Meyer C.A., Igarashi K., Kanno J., et al. PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat. Cell. Biol. 2011;13:668–675. doi: 10.1038/ncb2228. [DOI] [PubMed] [Google Scholar]
- 152.Feng W., Yonezawa M., Ye J., Jenuwein T., Grummt I. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat. Struct. Mol. Biol. 2010;17:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 153.Fortschegger K., de Graaf P., Outchkourov N.S., van Schaik F.M., Timmers H.T., Shiekhattar R. PHF8 targets histone methylation and RNA polymerase II to activate transcription. Mol. Cell. Biol. 2010;30:3286–3298. doi: 10.1128/MCB.01520-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kleine-Kohlbrecher D., Christensen J., Vandamme J., Abarrategui I., Bak M., Tommerup N., Shi X., Gozani O., Rappsilber J., Salcini A.E., et al. A functional link between the histone demethylase PHF8 and the transcription factor ZNF711 in X-linked mental retardation. Mol. Cell. 2010;38:165–178. doi: 10.1016/j.molcel.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Qiu J., Shi G., Jia Y., Li J., Wu M., Dong S., Wong J. The X-linked mental retardation gene PHF8 is a histone demethylase involved in neuronal differentiation. Cell Res. 2010;20:908–918. doi: 10.1038/cr.2010.81. [DOI] [PubMed] [Google Scholar]
- 156.Wen H., Li J., Song T., Lu M., Kan P.Y., Lee M.G., Sha B., Shi X. Recognition of histone H3K4 trimethylation by the plant homeodomain of PHF2 modulates histone demethylation. J. Biol. Chem. 2010;285:9322–9326. doi: 10.1074/jbc.C109.097667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhu Z., Wang Y., Li X., Xu L., Wang X., Sun T., Dong X., Chen L., Mao H., Yu Y., et al. PHF8 is a histone H3K9me2 demethylase regulating rrna synthesis. Cell Res. 2010;20:794–801. doi: 10.1038/cr.2010.75. [DOI] [PubMed] [Google Scholar]
- 158.Liu W., Tanasa B., Tyurina O.V., Zhou T.Y., Gassmann R., Liu W.T., Ohgi K.A., Benner C., Garcia-Bassets I., Aggarwal A.K., et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Horton J.R., Upadhyay A.K., Qi H.H., Zhang X., Shi Y., Cheng X. Enzymatic and structural insights for substrate specificity of a family of jumonji histone lysine demethylases. Nat. Struct. Mol. Biol. 2010;17:38–43. doi: 10.1038/nsmb.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Qi H.H., Sarkissian M., Hu G.Q., Wang Z., Bhattacharjee A., Gordon D.B., Gonzales M., Lan F., Ongusaha P.P., Huarte M., et al. Histone H4K20/H3K9 demethylase PHF8 regulates zebrafish brain and craniofacial development. Nature. 2010;466:503–507. doi: 10.1038/nature09261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yu L., Wang Y., Huang S., Wang J., Deng Z., Zhang Q., Wu W., Zhang X., Liu Z., Gong W., et al. Structural insights into a novel histone demethylase PHF8. Cell Res. 2010;20:166–173. doi: 10.1038/cr.2010.8. [DOI] [PubMed] [Google Scholar]
- 162.Veazey K.J., Parnell S.E., Miranda R.C., Golding M.C. Dose-dependent alcohol-induced alterations in chromatin structure persist beyond the window of exposure and correlate with fetal alcohol syndrome birth defects. Epigenet. Chromatin. 2015;8 doi: 10.1186/s13072-015-0031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Denli A.M., Tops B.B., Plasterk R.H., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 164.Han J., Lee Y., Yeom K.H., Nam J.W., Heo I., Rhee J.K., Sohn S.Y., Cho Y., Zhang B.T., Kim V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 165.Carmell M.A., Hannon G.J. RNase III enzymes and the initiation of gene silencing. Nat. Struct. Mol. Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- 166.Klein M.E., Impey S., Goodman R.H. Role reversal: The regulation of neuronal gene expression by microRNAs. Curr. Opin. Neurobiol. 2005;15:507–513. doi: 10.1016/j.conb.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 167.Kosik K.S. The neuronal microRNA system. Nat. Rev. Neurosci. 2006;7:911–920. doi: 10.1038/nrn2037. [DOI] [PubMed] [Google Scholar]
- 168.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nature Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 169.Martinez N.J., Gregory R.I. MicroRNA gene regulatory pathways in the establishment and maintenance of esc identity. Cell Stem Cell. 2010;7:31–35. doi: 10.1016/j.stem.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kuwabara T., Hsieh J., Nakashima K., Taira K., Gage F.H. A small modulatory dsRNA specifies the fate of adult neural stem cells. Cell. 2004;116:779–793. doi: 10.1016/S0092-8674(04)00248-X. [DOI] [PubMed] [Google Scholar]
- 171.Poole R.J., Hobert O. Early embryonic programming of neuronal left/right asymmetry in C. elegans. Curr. Biol. 2006;16:2279–2292. doi: 10.1016/j.cub.2006.09.041. [DOI] [PubMed] [Google Scholar]
- 172.Ronshaugen M., Biemar F., Piel J., Levine M., Lai E.C. The Drosophila microRNA iab-4 causes a dominant homeotic transformation of halteres to wings. Genes Dev. 2005;19:2947–2952. doi: 10.1101/gad.1372505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schratt G.M., Tuebing F., Nigh E.A., Kane C.G., Sabatini M.E., Kiebler M., Greenberg M.E. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 174.Olde Loohuis N.F., Kos A., Martens G.J., van Bokhoven H., Nadif Kasri N., Aschrafi A. MicroRNA networks direct neuronal development and plasticity. Cell. Mol. Life Sci. 2012;69:89–102. doi: 10.1007/s00018-011-0788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Davis T.H., Cuellar T.L., Koch S.M., Barker A.J., Harfe B.D., McManus M.T., Ullian E.M. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Stark K.L., Xu B., Bagchi A., Lai W.S., Liu H., Hsu R., Wan X., Pavlidis P., Mills A.A., Karayiorgou M., et al. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat. Genet. 2008;40:751–760. doi: 10.1038/ng.138. [DOI] [PubMed] [Google Scholar]
- 177.Nudelman A.S., DiRocco D.P., Lambert T.J., Garelick M.G., Le J., Nathanson N.M., Storm D.R. Neuronal activity rapidly induces transcription of the CREB-regulated microRNA-132, in vivo. Hippocampus. 2010;20:492–498. doi: 10.1002/hipo.20646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Wayman G.A., Davare M., Ando H., Fortin D., Varlamova O., Cheng H.Y., Marks D., Obrietan K., Soderling T.R., Goodman R.H., et al. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl. Acad. Sci. USA. 2008;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Muddashetty R.S., Nalavadi V.C., Gross C., Yao X., Xing L., Laur O., Warren S.T., Bassell G.J. Reversible inhibition of PSD-95 mRNA translation by miR-125a, FMRP phosphorylation, and mGluR signaling. Mol. Cell. 2011;42:673–688. doi: 10.1016/j.molcel.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Krol J., Busskamp V., Markiewicz I., Stadler M.B., Ribi S., Richter J., Duebel J., Bicker S., Fehling H.J., Schubeler D., et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 181.Wang W., Kwon E.J., Tsai L.H. MicroRNAs in learning, memory, and neurological diseases. Learn. Mem. 2012;19:359–368. doi: 10.1101/lm.026492.112. [DOI] [PubMed] [Google Scholar]
- 182.Shafi G., Aliya N., Munshi A. MicroRNA signatures in neurological disorders. Can. J. Neurol. Sci. 2010;37:177–185. doi: 10.1017/S0317167100009902. [DOI] [PubMed] [Google Scholar]
- 183.Xu B., Karayiorgou M., Gogos J.A. MicroRNAs in psychiatric and neurodevelopmental disorders. Brain Res. 2010;1338:78–88. doi: 10.1016/j.brainres.2010.03.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Jones K.L., Smith D.W. Recognition of the fetal alcohol syndrome in early infancy. Lancet. 1973;2:999–1001. doi: 10.1016/S0140-6736(73)91092-1. [DOI] [PubMed] [Google Scholar]
- 185.Alati R., Al Mamun A., Williams G.M., O’Callaghan M., Najman J.M., Bor W. In utero alcohol exposure and prediction of alcohol disorders in early adulthood: A birth cohort study. Arch. Gen. Psychiatry. 2006;63:1009–1016. doi: 10.1001/archpsyc.63.9.1009. [DOI] [PubMed] [Google Scholar]
- 186.Autti-Ramo I., Fagerlund A., Ervalahti N., Loimu L., Korkman M., Hoyme H.E. Fetal alcohol spectrum disorders in Finland: Clinical delineation of 77 older children and adolescents. Am. J. med. Genet. A. 2006;140:137–143. doi: 10.1002/ajmg.a.31037. [DOI] [PubMed] [Google Scholar]
- 187.Ceccanti M., Alessandra Spagnolo P., Tarani L., Luisa Attilia M., Chessa L., Mancinelli R., Stegagno M., Francesco Sasso G., Romeo M., Jones K.L., et al. Clinical delineation of fetal alcohol spectrum disorders (FASD) in Italian children: Comparison and contrast with other racial/ethnic groups and implications for diagnosis and prevention. Neurosci. Biobehav. Rev. 2007;31:270–277. doi: 10.1016/j.neubiorev.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 188.Spohr H.L., Willms J., Steinhausen H.C. Fetal alcohol spectrum disorders in young adulthood. J. Pediatr. 2007;150:175–179. doi: 10.1016/j.jpeds.2006.11.044. [DOI] [PubMed] [Google Scholar]
- 189.Streissguth A.P., Aase J.M., Clarren S.K., Randels S.P., LaDue R.A., Smith D.F. Fetal alcohol syndrome in adolescents and adults. JAMA. 1991;265:1961–1967. doi: 10.1001/jama.1991.03460150065025. [DOI] [PubMed] [Google Scholar]
- 190.Sokol R.J., Delaney-Black V., Nordstrom B. Fetal alcohol spectrum disorder. JAMA. 2003;290:2996–2999. doi: 10.1001/jama.290.22.2996. [DOI] [PubMed] [Google Scholar]
- 191.Burd L., Klug M.G., Martsolf J.T., Kerbeshian J. Fetal alcohol syndrome: Neuropsychiatric phenomics. Neurotoxicol. Teratol. 2003;25:697–705. doi: 10.1016/j.ntt.2003.07.014. [DOI] [PubMed] [Google Scholar]
- 192.Aragon A.S., Kalberg W.O., Buckley D., Barela-Scott L.M., Tabachnick B.G., May P.A. Neuropsychological study of FASD in a sample of American Indian children: Processing simple versus complex information. Alcohol. Clin. Exp. Res. 2008;32:2136–2148. doi: 10.1111/j.1530-0277.2008.00802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Green C.R., Mihic A.M., Nikkel S.M., Stade B.C., Rasmussen C., Munoz D.P., Reynolds J.N. Executive function deficits in children with fetal alcohol spectrum disorders (FASD) measured using the Cambridge Neuropsychological Tests Automated Battery (CANTAB) J. Child Psychol. Psychiatry. 2009;50:688–697. doi: 10.1111/j.1469-7610.2008.01990.x. [DOI] [PubMed] [Google Scholar]
- 194.Mattson S.N., Crocker N., Nguyen T.T. Fetal alcohol spectrum disorders: Neuropsychological and behavioral features. Neuropsychol. Rev. 2011;21:81–101. doi: 10.1007/s11065-011-9167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mattson S.N., Roesch S.C., Fagerlund A., Autti-Ramo I., Jones K.L., May P.A., Adnams C.M., Konovalova V., Riley E.P. Toward a neurobehavioral profile of fetal alcohol spectrum disorders. Alcohol. Clin. Exp. Res. 2010;34:1640–1650. doi: 10.1111/j.1530-0277.2010.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.May P.A., Blankenship J., Marais A.S., Gossage J.P., Kalberg W.O., Joubert B., Cloete M., Barnard R., de Vries M., Hasken J., et al. Maternal alcohol consumption producing fetal alcohol spectrum disorders (FASD): Quantity, frequency, and timing of drinking. Drug Alcohol Depend. 2013;133:502–512. doi: 10.1016/j.drugalcdep.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lebel C., Mattson S.N., Riley E.P., Jones K.L., Adnams C.M., May P.A., Bookheimer S.Y., O’Connor M.J., Narr K.L., Kan E., et al. A longitudinal study of the long-term consequences of drinking during pregnancy: Heavy in utero alcohol exposure disrupts the normal processes of brain development. J. Neurosci. 2012;32:15243–15251. doi: 10.1523/JNEUROSCI.1161-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Norman A.L., O’Brien J.W., Spadoni A.D., Tapert S.F., Jones K.L., Riley E.P., Mattson S.N. A functional magnetic resonance imaging study of spatial working memory in children with prenatal alcohol exposure: Contribution of familial history of alcohol use disorders. Alcohol. Clin. Exp. Res. 2013;37:132–140. doi: 10.1111/j.1530-0277.2012.01880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Sood B., Delaney-Black V., Covington C., Nordstrom-Klee B., Ager J., Templin T., Janisse J., Martier S., Sokol R.J. Prenatal alcohol exposure and childhood behavior at age 6 to 7 years: I. dose-response effect. Pediatrics. 2001;108:E34. doi: 10.1542/peds.108.2.e34. [DOI] [PubMed] [Google Scholar]
- 200.Streissguth A.P., Bookstein F.L., Sampson P.D., Barr H.M. Neurobehavioral effects of prenatal alcohol: Part III. PLS analyses of neuropsychologic tests. Neurotoxicol. Teratol. 1989;11:493–507. doi: 10.1016/0892-0362(89)90026-3. [DOI] [PubMed] [Google Scholar]
- 201.Guerri C., Bazinet A., Riley E.P. Foetal alcohol spectrum disorders and alterations in brain and behaviour. Alcohol Alcohol. 2009;44:108–114. doi: 10.1093/alcalc/agn105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bailey B.N., Delaney-Black V., Covington C.Y., Ager J., Janisse J., Hannigan J.H., Sokol R.J. Prenatal exposure to binge drinking and cognitive and behavioral outcomes at age 7 years. Am. J. Obstet. Gynecol. 2004;191:1037–1043. doi: 10.1016/j.ajog.2004.05.048. [DOI] [PubMed] [Google Scholar]
- 203.Bonthius D.J., Goodlett C.R., West J.R. Blood alcohol concentration and severity of microencephaly in neonatal rats depend on the pattern of alcohol administration. Alcohol. 1988;5:209–214. doi: 10.1016/0741-8329(88)90054-7. [DOI] [PubMed] [Google Scholar]
- 204.Levitt P., Veenstra-VanderWeele J. Neurodevelopment and the origins of brain disorders. Neuropsychopharmacology. 2015;40:1–3. doi: 10.1038/npp.2014.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Giedd J.N., Raznahan A., Alexander-Bloch A., Schmitt E., Gogtay N., Rapoport J.L. Child psychiatry branch of the National Institute of Mental Health longitudinal structural magnetic resonance imaging study of human brain development. Neuropsychopharmacology. 2015;40:43–49. doi: 10.1038/npp.2014.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hammock E.A. Developmental perspectives on oxytocin and vasopressin. Neuropsychopharmacology. 2015;40:24–42. doi: 10.1038/npp.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Hartley C.A., Lee F.S. Sensitive periods in affective development: Nonlinear maturation of fear learning. Neuropsychopharmacology. 2015;40:50–60. doi: 10.1038/npp.2014.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ross E.J., Graham D.L., Money K.M., Stanwood G.D. Developmental consequences of fetal exposure to drugs: What we know and what we still must learn. Neuropsychopharmacology. 2015;40:61–87. doi: 10.1038/npp.2014.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Washbourne P. Synapse assembly and neurodevelopmental disorders. Neuropsychopharmacology. 2015;40:4–15. doi: 10.1038/npp.2014.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Archer T., Oscar-Berman M., Blum K. Epigenetics in developmental disorder: ADHD and endophenotypes. J. Genet. Syndr. Gene Ther. 2011;2 doi: 10.4172/2157-7412.1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sakurai T., Dorr N.P., Takahashi N., McInnes L.A., Elder G.A., Buxbaum J.D. Haploinsufficiency of Gtf2i, a gene deleted in Williams Syndrome, leads to increases in social interactions. Autism Res. 2011;4:28–39. doi: 10.1002/aur.169. [DOI] [PubMed] [Google Scholar]
- 212.Schanen N.C. Epigenetics of autism spectrum disorders. Hum. Mol. Genet. 2006;15:R138–R150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- 213.Subbanna S., Basavarajappa B.S. Pre-administration of G9a/GLP inhibitor during synaptogenesis prevents postnatal ethanol-induced LTP deficits and neurobehavioral abnormalities in adult mice. Exp. Neurol. 2014;261:34–43. doi: 10.1016/j.expneurol.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Downing C., Johnson T.E., Larson C., Leakey T.I., Siegfried R.N., Rafferty T.M., Cooney C.A. Subtle decreases in DNA methylation and gene expression at the mouse Igf2 locus following prenatal alcohol exposure: Effects of a methyl-supplemented diet. Alcohol. 2011;45:65–71. doi: 10.1016/j.alcohol.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Garro A.J., McBeth D.L., Lima V., Lieber C.S. Ethanol consumption inhibits fetal DNA methylation in mice: Implications for the fetal alcohol syndrome. Alcohol Clin. Exp. Res. 1991;15:395–398. doi: 10.1111/j.1530-0277.1991.tb00536.x. [DOI] [PubMed] [Google Scholar]
- 216.Haycock P.C., Ramsay M. Exposure of mouse embryos to ethanol during preimplantation development: Effect on DNA methylation in the H19 imprinting control region. Biol. Reprod. 2009;81:618–627. doi: 10.1095/biolreprod.108.074682. [DOI] [PubMed] [Google Scholar]
- 217.Liu Y., Balaraman Y., Wang G., Nephew K.P., Zhou F.C. Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics. 2009;4:500–511. doi: 10.4161/epi.4.7.9925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ouko L.A., Shantikumar K., Knezovich J., Haycock P., Schnugh D.J., Ramsay M. Effect of alcohol consumption on CpG methylation in the differentially methylated regions of H19 and IG-DMR in male gametes: Implications for fetal alcohol spectrum disorders. Alcohol Clin. Exp. Res. 2009;33:1615–1627. doi: 10.1111/j.1530-0277.2009.00993.x. [DOI] [PubMed] [Google Scholar]
- 219.Zhou F.C., Balaraman Y., Teng M., Liu Y., Singh R.P., Nephew K.P. Alcohol alters DNA methylation patterns and inhibits neural stem cell differentiation. Alcohol Clin. Exp. Res. 2011;35:735–746. doi: 10.1111/j.1530-0277.2010.01391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Bekdash R.A., Zhang C., Sarkar D.K. Gestational choline supplementation normalized fetal alcohol-induced alterations in histone modifications, DNA methylation, and proopiomelanocortin (POMC) gene expression in beta-endorphin-producing pomc neurons of the hypothalamus. Alcohol. Clin. Exp. Res. 2013;37:1133–1142. doi: 10.1111/acer.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Govorko D., Bekdash R.A., Zhang C., Sarkar D.K. Male germline transmits fetal alcohol adverse effect on hypothalamic proopiomelanocortin gene across generations. Biol. Psychiatry. 2012;72:378–388. doi: 10.1016/j.biopsych.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Guo W., Crossey E.L., Zhang L., Zucca S., George O.L., Valenzuela C.F., Zhao X. Alcohol exposure decreases CREB binding protein expression and histone acetylation in the developing cerebellum. PLoS ONE. 2011;6:12. doi: 10.1371/journal.pone.0019351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Subbanna S., Psychoyos D., Shan X., Basavarajappa B.S. Postnatal ethanol exposure alters levels of 2-arachidonylglycerol-metabolizing enzymes and pharmacological inhibition of monoacylglycerol (MAGL) does not cause neurodegeneration in neonatal mice. J. Neurochem. 2015;134:276–287. doi: 10.1111/jnc.13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Zhong L., Zhu J., Lv T., Chen G., Sun H., Yang X., Huang X., Tian J. Ethanol and its metabolites induce histone lysine 9 acetylation and an alteration of the expression of heart development-related genes in cardiac progenitor cells. Cardiovasc. Toxicol. 2010;10:268–274. doi: 10.1007/s12012-010-9081-z. [DOI] [PubMed] [Google Scholar]
- 225.Balaraman S., Winzer-Serhan U.H., Miranda R.C. Opposing actions of ethanol and nicotine on microRNAs are mediated by nicotinic acetylcholine receptors in fetal cerebral cortical-derived neural progenitor cells. Alcohol Clin. Exp. Res. 2012;36:1669–1677. doi: 10.1111/j.1530-0277.2012.01793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Caputo V., Sinibaldi L., Fiorentino A., Parisi C., Catalanotto C., Pasini A., Cogoni C., Pizzuti A. Brain derived neurotrophic factor (BDNF) expression is regulated by microRNAs miR-26a and miR-26b allele-specific binding. PLoS ONE. 2011;6:12. doi: 10.1371/journal.pone.0028656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Dill H., Linder B., Fehr A., Fischer U. Intronic miR-26b controls neuronal differentiation by repressing its host transcript, ctdsp2. Genes Dev. 2012;26:25–30. doi: 10.1101/gad.177774.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sathyan P., Golden H.B., Miranda R.C. Competing interactions between micro-RNAs determine neural progenitor survival and proliferation after ethanol exposure: Evidence from an ex vivo model of the fetal cerebral cortical neuroepithelium. J. Neurosci. 2007;27:8546–8557. doi: 10.1523/JNEUROSCI.1269-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Soares A.R., Pereira P.M., Ferreira V., Reverendo M., Simoes J., Bezerra A.R., Moura G.R., Santos M.A. Ethanol exposure induces upregulation of specific microRNAs in zebrafish embryos. Toxicol. Sci. 2012;127:18–28. doi: 10.1093/toxsci/kfs068. [DOI] [PubMed] [Google Scholar]
- 230.Stringer R.L., Laufer B.I., Kleiber M.L., Singh S.M. Reduced expression of brain cannabinoid receptor 1 (Cnr1) is coupled with an increased complementary micro-RNA (miR-26b) in a mouse model of fetal alcohol spectrum disorders. Clin. Epigenet. 2013;5 doi: 10.1186/1868-7083-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wang L.L., Zhang Z., Li Q., Yang R., Pei X., Xu Y., Wang J., Zhou S.F., Li Y. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum. Reprod. 2009;24:562–579. doi: 10.1093/humrep/den439. [DOI] [PubMed] [Google Scholar]
- 232.Valles S., Pitarch J., Renau-Piqueras J., Guerri C. Ethanol exposure affects glial fibrillary acidic protein gene expression and transcription during rat brain development. J. Neurochem. 1997;69:2484–2493. doi: 10.1046/j.1471-4159.1997.69062484.x. [DOI] [PubMed] [Google Scholar]
- 233.Bielawski D.M., Zaher F.M., Svinarich D.M., Abel E.L. Paternal alcohol exposure affects sperm cytosine methyltransferase messenger RNA levels. Alcohol Clin. Exp. Res. 2002;26:347–351. doi: 10.1111/j.1530-0277.2002.tb02544.x. [DOI] [PubMed] [Google Scholar]
- 234.Tycko B., Morison I.M. Physiological functions of imprinted genes. J. Cell. Physiol. 2002;192:245–258. doi: 10.1002/jcp.10129. [DOI] [PubMed] [Google Scholar]
- 235.Bliek J., Terhal P., van den Bogaard M.J., Maas S., Hamel B., Salieb-Beugelaar G., Simon M., Letteboer T., van der Smagt J., Kroes H., et al. Hypomethylation of the H19 gene causes not only Silver-Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am. J. Hum. Genet. 2006;78:604–614. doi: 10.1086/502981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Reik W., Walter J. Genomic imprinting: Parental influence on the genome. Nature Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 237.Dean W., Bowden L., Aitchison A., Klose J., Moore T., Meneses J.J., Reik W., Feil R. Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: Association with aberrant phenotypes. Development. 1998;125:2273–2282. doi: 10.1242/dev.125.12.2273. [DOI] [PubMed] [Google Scholar]
- 238.Khosla S., Dean W., Brown D., Reik W., Feil R. Culture of preimplantation mouse embryos affects fetal development and the expression of imprinted genes. Biol. Reprod. 2001;64:918–926. doi: 10.1095/biolreprod64.3.918. [DOI] [PubMed] [Google Scholar]
- 239.Sinclair K.D., Young L.E., Wilmut I., McEvoy T.G. In-utero overgrowth in ruminants following embryo culture: Lessons from mice and a warning to men. Hum. Reprod. 2000;15(Suppl. 5):68–86. doi: 10.1093/humrep/15.suppl_5.68. [DOI] [PubMed] [Google Scholar]
- 240.Hicks S.D., Middleton F.A., Miller M.W. Ethanol-induced methylation of cell cycle genes in neural stem cells. J. Neurochem. 2010;114:1767–1780. doi: 10.1111/j.1471-4159.2010.06886.x. [DOI] [PubMed] [Google Scholar]
- 241.Zhou F.C., Chen Y., Love A. Cellular DNA methylation program during neurulation and its alteration by alcohol exposure. Birth Defects Res. A Clin. Mol. Teratol. 2011;91:703–715. doi: 10.1002/bdra.20820. [DOI] [PubMed] [Google Scholar]
- 242.Chen Y., Ozturk N.C., Zhou F.C. DNA methylation program in developing hippocampus and its alteration by alcohol. PLoS ONE. 2013;8:12. doi: 10.1371/journal.pone.0060503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Ryan S.H., Williams J.K., Thomas J.D. Choline supplementation attenuates learning deficits associated with neonatal alcohol exposure in the rat: Effects of varying the timing of choline administration. Brain Res. 2008;1237:91–100. doi: 10.1016/j.brainres.2008.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Thomas J.D., Biane J.S., O’Bryan K.A., O’Neill T.M., Dominguez H.D. Choline supplementation following third-trimester-equivalent alcohol exposure attenuates behavioral alterations in rats. Behav. Neurosci. 2007;121:120–130. doi: 10.1037/0735-7044.121.1.120. [DOI] [PubMed] [Google Scholar]
- 245.Otero N.K., Thomas J.D., Saski C.A., Xia X., Kelly S.J. Choline supplementation and DNA methylation in the hippocampus and prefrontal cortex of rats exposed to alcohol during development. Alcohol. Clin. Exp. Res. 2012;36:1701–1709. doi: 10.1111/j.1530-0277.2012.01784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Mukhopadhyay P., Rezzoug F., Kaikaus J., Greene R.M., Pisano M.M. Alcohol modulates expression of DNA methyltranferases and methyl CpG-/CpG domain-binding proteins in murine embryonic fibroblasts. Reprod. Toxicol. 2013;37:40–48. doi: 10.1016/j.reprotox.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Subbanna S., Shivakumar M., Psychoyos D., Xie S., Basavarajappa B.S. Anandamide-CB1 receptor signaling contributes to postnatal ethanol-induced neonatal neurodegeneration, adult synaptic and memory deficits. J. Neuosci. 2013;33:6350–6366. doi: 10.1523/JNEUROSCI.3786-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Perkins A., Lehmann C., Lawrence R.C., Kelly S.J. Alcohol exposure during development: Impact on the epigenome. Int. J. Dev. Neurosci. 2013;31:391–397. doi: 10.1016/j.ijdevneu.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zhang C.R., Ho M.F., Vega M.C., Burne T.H., Chong S. Prenatal ethanol exposure alters adult hippocampal VGLUT2 expression with concomitant changes in promoter DNA methylation, H3K4 trimethylation and miR-467b-5p levels. Epigenet. Chromatin. 2015;8 doi: 10.1186/s13072-015-0032-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Rakyan V.K., Blewitt M.E., Druker R., Preis J.I., Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/S0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 251.Wolff G.L. Influence of maternal phenotype on metabolic differentiation of agouti locus mutants in the mouse. Genetics. 1978;88:529–539. doi: 10.1093/genetics/88.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Khalid O., Kim J.J., Kim H.S., Hoang M., Tu T.G., Elie O., Lee C., Vu C., Horvath S., Spigelman I., et al. Gene expression signatures affected by alcohol-induced DNA methylomic deregulation in human embryonic stem cells. Stem Cell Res. 2014;12:791–806. doi: 10.1016/j.scr.2014.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Dasmahapatra A.K., Khan I.A. Modulation of DNA methylation machineries in Japanese rice fish (Oryzias latipes) embryogenesis by ethanol and 5-azacytidine. Comp. Biochemistry Physiol. Toxicol. Pharmacol. 2016;179:174–183. doi: 10.1016/j.cbpc.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 254.Laufer B.I., Mantha K., Kleiber M.L., Diehl E.J., Addison S.M., Singh S.M. Long-lasting alterations to DNA methylation and ncRNAs could underlie the effects of fetal alcohol exposure in mice. Dis. Models Mech. 2013;6:977–992. doi: 10.1242/dmm.010975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Portales-Casamar E., Mah S.M., MacIsaac J.L., Barhdadi A., Provost S., Dube M.P., Reynolds J.N., Pavlidis P., Kobor M.S. DNA methylation changes in fetal alcohol spectrum disorder. Int. J. Dev. Neurosci. 2015;47 doi: 10.1016/j.ijdevneu.2015.04.336. [DOI] [Google Scholar]
- 256.Wood M.A., Attner M.A., Oliveira A.M., Brindle P.K., Abel T. A transcription factor-binding domain of the coactivator CBP is essential for long-term memory and the expression of specific target genes. Learn. Mem. 2006;13:609–617. doi: 10.1101/lm.213906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Weber M., Hellmann I., Stadler M.B., Ramos L., Paabo S., Rebhan M., Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 259.Rishi V., Bhattacharya P., Chatterjee R., Rozenberg J., Zhao J., Glass K., Fitzgerald P., Vinson C. CpG methylation of half-CRE sequences creates C/EBPalpha binding sites that activate some tissue-specific genes. Proc. Natl. Acad. Sci. USA. 2010;107:20311–20316. doi: 10.1073/pnas.1008688107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Maier S.E., Cramer J.A., West J.R., Sohrabji F. Alcohol exposure during the first two trimesters equivalent alters granule cell number and neurotrophin expression in the developing rat olfactory bulb. J. Neurobiol. 1999;41:414–423. doi: 10.1002/(SICI)1097-4695(19991115)41:3<414::AID-NEU9>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 261.Hu Y., Willett K.L., Khan I.A., Scheffler B.E., Dasmahapatra A.K. Ethanol disrupts chondrification of the neurocranial cartilages in medaka embryos without affecting aldehyde dehydrogenase 1A2 (Aldh1A2) promoter methylation. Comp. Biochem. Physiol. Toxicol. Pharmacol. 2009;150:495–502. doi: 10.1016/j.cbpc.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Krishnamoorthy M., Gerwe B.A., Scharer C.D., Sahasranaman V., Eilertson C.D., Nash R.J., Usta S.N., Kelly S., Rose M., Peraza R., et al. Ethanol alters proliferation and differentiation of normal and chromosomally abnormal human embryonic stem cell-derived neurospheres. Birth Defects Res. B Dev. Reprod. Toxicol. 2013;98:283–295. doi: 10.1002/bdrb.21063. [DOI] [PubMed] [Google Scholar]
- 263.Tyler C.R., Allan A.M. Prenatal alcohol exposure alters expression of neurogenesis-related genes in an ex vivo cell culture model. Alcohol. 2014;48:483–492. doi: 10.1016/j.alcohol.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Marjonen H., Sierra A., Nyman A., Rogojin V., Grohn O., Linden A.M., Hautaniemi S., Kaminen-Ahola N. Early maternal alcohol consumption alters hippocampal DNA methylation, gene expression and volume in a mouse model. PLoS ONE. 2015;10:12. doi: 10.1371/journal.pone.0124931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Dasmahapatra A.K., Khan I.A. DNA methyltransferase expressions in Japanese rice fish (Oryzias latipes) embryogenesis is developmentally regulated and modulated by ethanol and 5-azacytidine. Comp. Biochem. Physiol. Toxicol. Pharmacol. 2015;176–177:1–9. doi: 10.1016/j.cbpc.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 266.Ramadoss J., Liao W.X., Chen D.B., Magness R.R. High-throughput caveolar proteomic signature profile for maternal binge alcohol consumption. Alcohol. 2010;44:691–697. doi: 10.1016/j.alcohol.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Zhang W., Peng C., Zheng M., Gao W., Zhu J., Lv T., Liu L., Liu Z., Li H., Xv Y., et al. Prenatal alcohol exposure causes the over-expression of dhand and ehand by increasing histone H3K14 acetylation in C57 BL/6 mice. Toxicol. Lett. 2014;228:140–146. doi: 10.1016/j.toxlet.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 268.Peng C., Zhu J., Sun H.C., Huang X.P., Zhao W.A., Zheng M., Liu L.J., Tian J. Inhibition of histone H3K9 acetylation by anacardic acid can correct the over-expression of Gata4 in the hearts of fetal mice exposed to alcohol during pregnancy. PLoS ONE. 2014;9:12. doi: 10.1371/journal.pone.0104135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Thomas J.D., Idrus N.M., Monk B.R., Dominguez H.D. Prenatal choline supplementation mitigates behavioral alterations associated with prenatal alcohol exposure in rats. Birth Defects Res. A Clin. Mol. Teratol. 2010;88:827–837. doi: 10.1002/bdra.20713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Pappalardo-Carter D.L., Balaraman S., Sathyan P., Carter E.S., Chen W.J., Miranda R.C. Suppression and epigenetic regulation of miR-9 contributes to ethanol teratology: Evidence from zebrafish and murine fetal neural stem cell models. Alcohol. Clin. Exp. Res. 2013;37:1657–1667. doi: 10.1111/acer.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Mantha K., Laufer B.I., Singh S.M. Molecular changes during neurodevelopment following second-trimester binge ethanol exposure in a mouse model of fetal alcohol spectrum disorder: From immediate effects to long-term adaptation. Dev. Neurosci. 2014;36:29–43. doi: 10.1159/000357496. [DOI] [PubMed] [Google Scholar]
- 272.Ignacio C., Mooney S.M., Middleton F.A. Effects of acute prenatal exposure to ethanol on microRNA expression are ameliorated by social enrichment. Front. Pediatrics. 2014;2 doi: 10.3389/fped.2014.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Balaraman S., Lunde E.R., Sawant O., Cudd T.A., Washburn S.E., Miranda R.C. Maternal and neonatal plasma microRNA biomarkers for fetal alcohol exposure in an ovine model. Alcohol. Clin. Exp. Res. 2014;38:1390–1400. doi: 10.1111/acer.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Parchem R.J., Moore N., Fish J.L., Parchem J.G., Braga T.T., Shenoy A., Oldham M.C., Rubenstein J.L., Schneider R.A., Blelloch R. MiR-302 is required for timing of neural differentiation, neural tube closure, and embryonic viability. Cell Rep. 2015;12:760–773. doi: 10.1016/j.celrep.2015.06.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Chen X., Liu J., Feng W.K., Wu X., Chen S.Y. MiR-125b protects against ethanol-induced apoptosis in neural crest cells and mouse embryos by targeting Bak 1 and PUMA. Exp. Neurol. 2015;271:104–111. doi: 10.1016/j.expneurol.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kleiber M.L., Laufer B.I., Stringer R.L., Singh S.M. Third trimester-equivalent ethanol exposure is characterized by an acute cellular stress response and an ontogenetic disruption of genes critical for synaptic establishment and function in mice. Dev. Neurosci. 2014;36:499–519. doi: 10.1159/000365549. [DOI] [PubMed] [Google Scholar]