The Regulatory Role of MicroRNA in Hepatitis-B Virus-Associated Hepatocellular Carcinoma (HBV-HCC) Pathogenesis (original) (raw)
Abstract
The incidence and mortality of hepatitis B virus (HBV)-associated hepatocellular carcinoma (HBV-HCC) is an intractable public health problem in developing countries that is compounded by limited early detection and therapeutic options. Despite the early promise of utilizing the regulatory role of miRNA in liver cancer, this field remains largely in the work-in-progress phase. This exploratory review paper adopts a broad focus in order to collate evidence of the regulatory role of miRNA in each stage of the HBV-HCC continuum. This includes the regulatory role of miRNA in early HBV infection, chronic inflammation, fibrosis/cirrhosis, and the onset of HCC. The paper specifically investigates HBV dysregulated miRNA that influence the expression of the host/HBV genome in HBV-HCC pathogenesis and fully acknowledges that this does not cover the full spectrum of dysregulated miRNA. The sheer number of dysregulated miRNA in each phase support a hypothesis that future therapeutic interventions will need to consider incorporating multiple miRNA panels.
Keywords: regulatory, role, dysregulated, miRNA, hepatitis B infected hepatocellular carcinoma, HBV-HCC continuum
1. Hepatitis B Virus-Associated Hepatocellular Carcinoma (HBV-HCC)
An estimated 841,080 new liver cancers (LCs) were diagnosed worldwide in 2018, with hepatocellular carcinoma (HCC), the most common histological subtype, accounting for 75% to 90% of LCs [1]. A 12.4% increase of LC was recorded between 2008 (696,000) to 2012 (782,000) [2] and [3] 7.6% between 2012 and 2018 (841,080) [1]. HCC is listed globally as a leading cancer in men and women [4], with a recent report indicating that 83% of new cases diagnosed occurred in less well developed countries in East and south Eastern Asia and Western and Central Africa [5] [GLOBOCAN 2018 (http://gco.iarc.fr), accessed on 28 October 2019].
HBV and hepatitis C virus (HCV) infection is linked with more than 90% of all HCC incidence in developing countries compared to 40% in developed countries, with HBV being the major contributor in 66% of the cases [6]. According to the estimates of the global burden of disease study, deaths due to viral hepatitis increased by 63%, from 0.89 million to 1.45 million, between 1990 and 2013 [7,8]. HBV has been coined as “the second most dangerous carcinogen after tobacco” [8,9]. HBV, the prototype member of the family Hepadnaviridae, is hepatotropic and non-cytopathic. HBV enters the hepatocyte via the sodium-taurocholate cotransporting polypeptide (NTCP) receptor [10] and replicates by reverse transcription of a RNA intermediate, the pregenomic RNA (pgRNA). Although not required for replication of HBV, integration of HBV DNA can occur within the host genome and this has been implicated in hepatocarcinogenesis [11].
1.1. HBV-HCC Pathogenesis Continuum
HBV infection can result in a cascade of complex interactions between the host and the virus, which can lead to a spectrum of clinical manifestations including the asymptomatic carrier state, acute or fulminant hepatitis, and, ultimately, chronic hepatitis with progression to HCC with or without intermediary cirrhosis [12]. In 95% of infected adults and in only 10% of children, acute infection is self-limiting, with the virus persisting in 5% and 90% of the cases, respectively [12,13], with the risk of HCC increasing 100-fold if a patient is positive for both the hepatitis B surface antigen (HBsAg) and the hepatitis B e antigen (HBeAg). Even in the absence of HBsAg, there is a reduced and yet significant risk of HCC [14]. As viral load increases, the host immune response, triggered by HBV surface antigens (HBsAg), elicit T-cell responses accompanied by secondary inflammatory response [14], and an increase in free radicals, interferon, tumor necrosis factor (TNF), and hepatic injury. Moreover, if integration of HBV DNA into the host hepatocyte genome occurs, it results in the oncogenic disruption of cellular genes [15]. This disruption triggers apoptosis, regeneration, and early senescence [16], in addition to host cell deletions, cis/_trans_-activation, translocations, the production of fusion transcripts, aberrant epigenetic changes, and generalized genomic instability [17]. In parallel with these changes, the continuous destruction of well-differentiated hepatocytes and organized extracellular matrix results in their eventual replacement with undifferentiated liver stem cells and poorly organized fibrotic tissue [12].
HBV-HCC pathways typically include aberrant expression in the retinoblastoma-tumor protein 53 (RB1-TP53) suppressor networks, the Wingless-related integration site/beta-catenin (WNT/β-Catenin) pathway, and the phosphoinositide 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) and Janus kinase/signal transducer (JAK/STAT) pathways [18,19] (see Figure 1, Figure 2, Figure 3 and Figure 4).
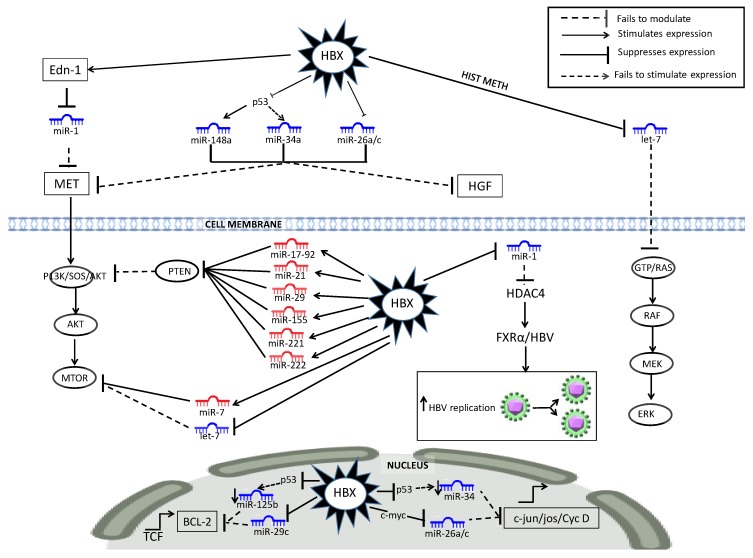
Figure 1.
HBx-dysregulated miRNA in the P13K/MAPK liver cancer pathway. HBV-specific abbreviations include endothelin-1 (Edn-1), HIST METH (histone methylation), Farnesoid X receptor (FXRα).
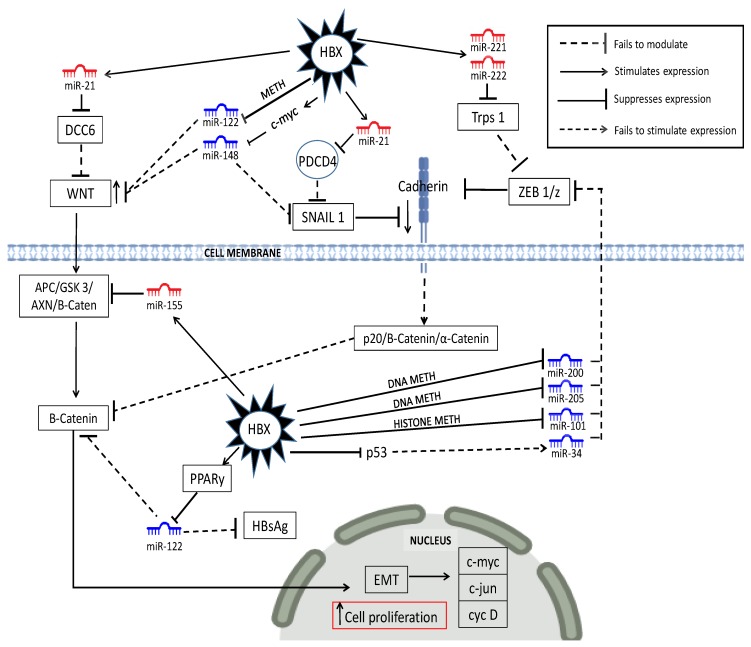
Figure 2.
Dysregulated miRNA in the WNT/β-catenin liver cancer pathway. HBV-specific abbreviations DCC6 (DCC-adaptor protein), DNA METH (DNA methylation), HISTONE METH ((histone methylation), PPARγ (peroxisome proliferator-activated receptor gamma), EMT (epithelial mesenchymal transition).
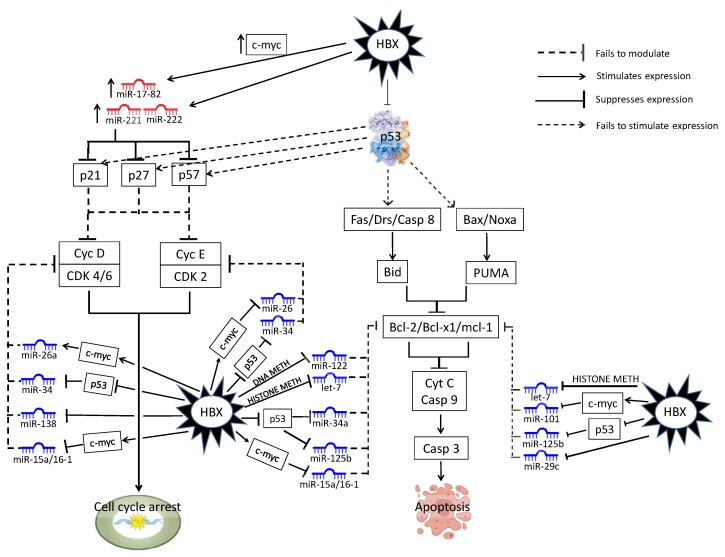
Figure 3.
Dysregulated miRNA in the TP53 liver cancer pathway. HBV-specific abbreviations include DNA METH (DNA methylation), HISTONE METH (histone methylation).
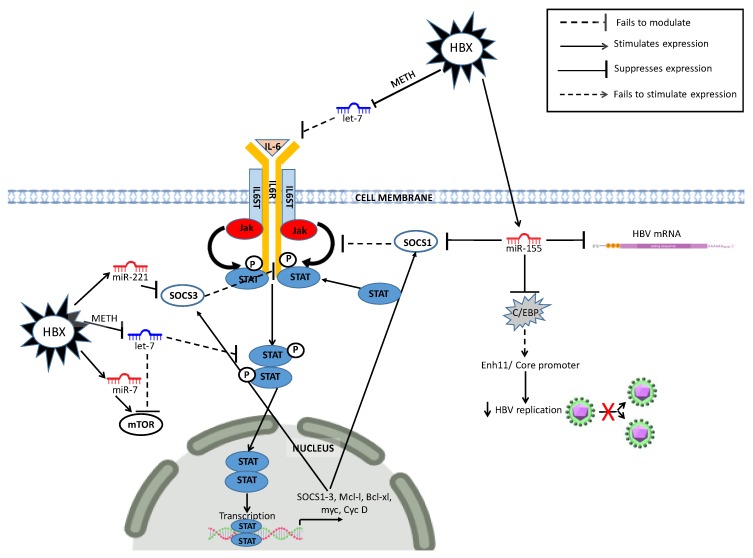
Figure 4.
Dysregulated miRNAs in the JAK/STAT liver cancer pathway. METH (Methylation) C/EBP (CCAAT/enhancer-binding protein) Enh11 (enhancer 11).
This paper specifically adopts a broad, exploratory review of the regulatory role of HBV dysregulated miRNA in the HBV-HCC continuum and miRNA dysregulation may provide a means of screening HBV carriers and developing HCC biomarkers and alternate therapeutic options.
1.2. The Deregulation of MiRNA in HBV-HCC Continuum
MicroRNA (miRNA) act as post-transcriptional gene silencers that collectively reduce or inhibit their target mRNA expression, thereby playing a homeostatic role that fine tunes the translation of proteins. The ancillary role of miRNA, as mild suppressors, has been explained by the inherently stochastic nature of gene transcription and environmental fluctuations [20]. In the case of transient reaction to environmental conditions, miRNA quantity become temporarily dysregulated until homeostasis is restored [21]. In the case of the continuum from asymptomatic HBV infection leading to HCC, multiple miRNA become increasingly permanently dysregulated (Table 1, Table 2, Table 3 and Table 4) as a result of HBV infection, epigenetic changes [22], inflammation [23], fibrosis [24], cirrhosis [16], and the onset of HCC. The increasing level of dysregulation in the HBV-HCC continuum is illustrated in a study that showed 37 miRNA deregulated in otherwise healthy controls (HC), 77 in asymptomatic HBV carriers (ASC), 101 in chronic hepatitis B infection (CHB) (Table 1), and 135 in acute liver failure (ALF) [25]. In general, dysregulated miRNA in the HBV-HCC continuum fall into two major categories, namely, those that target HBV transcripts and those that target the host cell genome expression.
Table 1.
Dysregulated miRNA in the HBV infection/inflammation stage (in miR alpha-numeric order).
| miRNA | HBV | Comment | Author |
|---|---|---|---|
| Let-7 fam | Down | Let 7a/b/c/d/e/f/g/I all downregulated by HBx protein which inhibits IL-6, IL-10, TLR4 expression | [40,51] |
| Let-7a-3p | Up | Upregulated in early stage HBV/targets DDX3X/expression can be downregulated in sera but upregulated in tissue | [57,58] |
| Let-7c | Up | Impairs immune response and HBV evasion/targets HDAC4/MET to enhance HBV replication via FXRA | [58,59,60] |
| miR-1 | Up | Targets HDAC4, MET to enhance (HBV replication)HBV RE by augmenting FXRA expression | [56] |
| miR-10a | Up | Upregulated > three-fold in HBV infection/upregulated in sera, Targets EphA4 which is an epithelial-mesynchymal-transition (EMT) suppressor | [58,59,61] |
| miR-106a | Down | Downregulated by HBx, targets IL-8 in CHB | [40,62] |
| miR-122 | Up | Upregulated in sera for HBeAg positive patients/correlates with HBV DNA/in sera and tissue | [32,60,63,64] |
| Up | Upregulated in CHB vs. Healthy Controls (HC)/upregulated in CHB vs. HC sera | [25,33,59,65] | |
| Up | Upregulated in HBV also HBeAg+ vs. HBeAg–/correlates with HBV DNA/HBsAg | [63,66] | |
| mir-122 | Down | Suppress HBV by targeting HBV mRNAs/targets Cyclin G1/HO-1 to promote HBV infection by inhibiting HO-1 | [28,35,36,67,68] |
| miR-122-5p | Up | Upregulated in CHB/biomarker for liver injury | [69] |
| Down | Decreased level in occult HBV infection (OBI) vs. HC | [70] | |
| miR-124 | Up | Serum miRNA correlates with necrosis/inflammation and upregulated in CHB vs. HC/targets AQP3 | [71,72] |
| miR-125 | Up | Impairs immune response and HBV evasion | [56] |
| miR-125a-5p | Up | Correlates with HBsAg/HBeAg and HBV-DNA/suppresses HBV expression by blocking HBsAg mRNA/targets KLF13 | [40,58,73,74] |
| miR-125a | Up | HBx upregulated in HBV infection | [40,75] |
| miR-125b | Up | Upregulated in CHB sera/correlated with HBV DNA, HBsAg, HBeAg | [59,63] |
| miR-125b-5p | Up | Upregulated 2.85 fold in CHB/targets STAT3 | [57,58,76] |
| miR-1275 | Up | Upregulated in HBV patients/correlated with serum ϒ-glutamyl transpeptidase | [63] |
| miR-130a | UpDown | Upregulated by NFκB/ Marker for OBI/upregulated vs. HC, ASC, CHBTargets PGC1α and PPARγ to regulate HBV replication/possible dual role in HBV | [70][77] |
| miR-15a | Down | Downregulated by HBx/Suppresses HBV infection by binding to HBx mRNA/targets TGF-β/Smad7 | [28,78,79] |
| miR-146 | Up | Targets/stimulates HBx/NFκB in HBV infection | [80] |
| miR-150 | Up | Upregulated > three-fold in HBV sera/downregulated in HBV asymptomatic vs. CHB | [59,60,81] |
| miR-151-3p | Up | Upregulated in CHB/biomarker for liver injury | [69] |
| miR-155 | Up | Blocks S_OCS1_ to upregulate JAK/STAT/targets C/EBP Suppresses HBV by augmenting INF signaling | [30,31] |
| miR-16-1 | Down | Downregulated by HBx RNA | [78] |
| miR-17 | Down | Downregulated in HBV-infected persons | [40] |
| miR-17-92 | Up | HBV transactivates c-Myc to upregulate miR-17-92 which suppresses HBV mRNA | [82] |
| miR-181a-d | Up | Correlated with HBV DNA and HBV disease progression/downregulates HLA-A, HBsAg by targeting HLA-A, E2F5 | [40,83,84,85,86] |
| miR-182 | Down | Downregulated in CHB leading to HBV-related cirrhosis | [87] |
| miR-191 | Up | Upregulated by HBx protein | [40] |
| miR-192 | Up | Upregulated in CHB vs. HC serum | [65,88] |
| miR-192-5p | Up | HBV activated/correlated with HBV Replication | [88,89] |
| miR-193a-5p | Down | Downregulated in A3 vs. A0 inflammation/targets SPOCK3 | [90,91] |
| miR-193b | Up | Upregulated by HBx protein/directly targets Mcl-1 | [40,92] |
| miR-196a | Down | Downregulated by HBx | [40,83,93] |
| miR-196c | Up | Upregulated in HBV-infected persons | [40] |
| miR-1974 | Up | Upregulated in CHB vs. normal liver (NL)/upregulated A3 vs. A0 inflammation | [90] |
| miR-199a-5p | Up | Upregulated in early HBV-immune tolerant/HBx upregulated/Target STAT2/low in blood sera but up in tissue | [40,57] |
| miR-20a | Down | Downregulated by HBx | [40] |
| miR-200b | Down | Downregulated in sera (ECVs) in CHB | [81] |
| miR-200c | Down | HBx reduced in HBV infection/targets nuclear factor IA to suppress HBV replication | [28,94] |
| miR-205 | Down | Targets HBx mRNA | [28,95] |
| miR-203a | Up | Upregulated in HBV infection/promotes inflammation targets IL-6, IL-8 | [96] |
| miR-21 | Up | Upregulated in A3 vs. A0 inflammation/impairs immune response and HBV evasion/up in CHB vs. HC serum | [56,65,90] |
| miR-210 | Up | Correlates with HBV DNA and HBsAg/marker for necroinflammation/suppresses HBV infection by targeting HBsAg pre-S1 region | [58,59,97,98] |
| miR-215-5p | Up | HBV activated miRNA/correlates with HBV replication | [89] |
| miR-221-3p | Up | Upregulated in early HBV/targets TBK1/upregulated in blood sera/tissue | [57] |
| miR-22 | Up | Upregulated in HBV-infected persons/correlates with HBeAg, HBeAg positive persons/correlated with miR-122 | [32,63] |
| Up | Upregulated in A0 vs. A3 inflammation/targets HDAC4, ERα to inhibit HBsAg/HBeAg | [90] | |
| miR-223-3p | Up | 5.55-fold upregulated in CHB in blood sera | [59,76] |
| miR-23a/b | Up | Upregulated > three-fold in HBV infection/upregulated in sera | [40,58,59,83] |
| miR-236 | Up | Upregulated in CHB vs. HC sera | [59] |
| miR-30c | Up | Upregulated by HBx protein | [40] |
| miR-3200 | Up | Upregulated in CHB leading to LC | [87] |
| HBV-mir-3 | Up | Mediates HBV RE by blocking HBc mRNA to downregulate HBV virions | [27] |
| miR-338-3p | Down | Downregulated in HBV | [40] |
| miR-339 | Up | Upregulated in CHB leading to LC | [87] |
| miR-34a | Up | Upregulated A3 vs. A0 inflammation/promotes Tregs that block effector T-cells | [28,29,90] |
| miR-342-3p | Up | Upregulated > three-fold in HBV infection in sera/HBx-induced deregulation | [40,58,59,87] |
| miR-3613-3p | Up | Upregulated in CHB/Target STAT3/down in sera | [57] |
| miR-3615 | Up | Upregulated in CHB leading to LC | [87] |
| miR-371 | Up | Upregulated by HBx protein | [40] |
| miR-372 | Up | Correlates with HBV DNA in CHB/deregulated by HBx/targets NFIB/ HNF4, RXR, and PPAR to regulate HBV transcription | [39,40,99] |
| miR-373 | Up | Correlates with HBV DNA in CHB/deregulated by HBx | [39,40] |
| miR-375 | Up | Upregulated > three-fold in HBV in sera | [58,59] |
| miR-378 | Down | Down-regulated in HBV | [40] |
| miR-423 | Up | Upregulated > three-fold in HBV infection/upregulated in sera | [58,59] |
| miR-4485 | Down | Downregulated in CHB re progression to HBV-LC | [87] |
| miR-451 | Up | Upregulated in CHB vs. NL/Upregulated in A3 vs. A0 inflammation | [90] |
| miR-4508 | Up | Upregulated in CHB leading to LC | [87] |
| miR-4717 | Down | Significant upregulation of PD-1 (programmed cell death) | [100] |
| miR-486-5p | Down | Downregulated in CHB vs. NL | [90] |
| MiR-501 | Up | Targets HBX1P to induce HBV RE | [101] |
| miR-548 | Down | Inhibits IFN-ϓ1, downregulates immune response/promotes HBV | [102] |
| miR-548d-5p | Up | Upregulated in A3 vs. A0 inflammation | [90] |
| miR-602 | Up | Upregulated in CHB vs. HC/targets RASSFIA/ascending upregulation on HBV-HCC continuum | [103] |
| miR-659 | Down | Downregulated in A3 vs. A0 inflammation | [90] |
| miR-711 | Down | Downregulated in A3 vs. A0 inflammation | [90] |
| miR-720 | Up | Upregulated in HBV patients also correlates with HBeAg+ HBeAg– | [63] |
| miR-760 | Up | Upregulated in A3 vs. A0 inflammation | [90] |
| miR-762 | Down | Downregulated in CHB vs. NL | [90] |
| miR-767-3p | Up | Upregulated in A3 vs. A0 inflammation | [90] |
| miR-885-5p | Up | Upregulated in CHB vs. HC | [104] |
| miR-92a | Up | Upregulated > three-fold in HBV in sera/ | [58,59,81] |
| miR-935 | Up | Upregulated in CHB leading to LC | [87] |
| miR-940 | Down | Downregulated in CHB/downregulated in sera and tissue | [57] |
| miR-9-5p | Down | Downregulated in CHB/even lower with higher fibrosis scores | [105] |
| miR-99a | Up | Upregulated in HBV+ patients also HBeAg+/upregulated in sera | [58,59,63] |
| mIr-99b | Up | Upregulated by HBx protein | [40] |
Table 2.
Dysregulated miRNA in HBV-associated liver fibrosis/cirrhosis (in miR numeric order).
| MiRNA | Deregulation | Comment | Author |
|---|---|---|---|
| miR-1 | Up | Upregulated in late-stage fibrosis (f3–4)/targets ATF2/E2F3/CREB3L2/up in tissue and sera | [57] |
| miR-10b-5p | Up | Upregulated advanced fibrosis vs. early fibrosis/targets ATF2/E2F3/CREB3L2/up in tissue and sera | [57] |
| miR-101 | Down | Downregulated in HBV fibrosis/HBx downregulated via _EZH2_-induced methylation/suppresses TGF-B signaling to block collagen | [58,119] |
| Up | Suppresses TGF-β signaling to block collagen/upregulated in cirrhosis versus CHB | [120] | |
| miR-103 | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-122 | Up | Upregulated in early fibrosis/significant decrease as f0–2 progresses to f3–4 | [33,58,81,121] |
| Down | Downregulated in advanced f4 vs. f0–1/suppresses TGF-β pathway to block collagen | [33] | |
| miR-122-5p | Up | Correlates with degree of fibrogenic damage | [69,122,123] |
| miR-1224-3p | Up | Upregulated in early fibrosis (f1–2)/target ELK1/AKT2/upregulated in sera/tissue | [57] |
| miR-1227-3p | Up | Upregulated early fibrosis (f1–2)/target HSPG2/PTEN/downregulated in sera | [57] |
| miR-125b-5p | Up | Upregulated in cirrhosis | [76] |
| miR-125a-5p | Up | Targets F1H1 in HBV-related fibrosis vs. H. Controls | [124,125] |
| miR-126 | Up | Upregulated in cirrhosis vs. CHB stage | [118] |
| miR-1275 | Down | Downregulated in cirrhosis vs. CHB stage | [118] |
| miR-128 | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-130a | Up | Upregulated in fibrosis/f3 > f0/upregulated in cirrhosis vs. CHB/IFNs trigger JAK/STAT | [118] |
| miR-133a | Down | Downregulated in cirrhosis | [126] |
| miR-133b | Up | Upregulated in advanced vs. early fibrosis/targets ATF2/E2F3/CREB3L2/upregulated in sera and tissue | [57] |
| miR-140-5p | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-141-3p | Down | Downregulated in cirrhosis vs. HCC | [123] |
| miR-142-3p | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-143 | Down | Downregulated in advanced fibrosis vs. early fibrosis | [127] |
| miR-146a | Up | Upregulated in CHB fibrosis/upregulated in cirrhosis vs. HC/HBx upregulated > CFH fiber proteins | [104,118,121,128] |
| miR-148a/b | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-149-5p | Down | Downregulated in HBV-induced cirrhosis | [129] |
| miR-150 | Down | Inhibits collagen-1 expression and HSC activation/targets c-myb | [130] |
| miR-151a-3p | Down | Downregulated as injury increases | [69] |
| miR-151-5p | Up | Upregulated in cirrhosis vs. CHB/predictor of activated HSCs | [118] |
| miR-17 | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-17-3p | Down | Downregulated as injury increases/downregulated in cirrhosis vs. fibrosis | [129] |
| miR-17-92 | Down | MiR-17-92 family (miR-19a/b/92) downregulated in activated HSCs | [131] |
| miR-181b | Up | Promote fibrosis via TGF-β or NF- κ B pathways/correlates with HBV DNA/downregulated in f4 vs. f1/upregulated in cirrhosis | [58,126] |
| miR-185 | Down | TGF-1 > miR-185 > RICTOR/RHEB > HSCs > ECM > fibrosis/downregulated in HBV-induced fibrosis | [111] |
| miR-1915 | Down | Downregulated in cirrhosis vs. CHB | [118] |
| miR-19b | Down | Downregulated in activated HSCs/CHB led fibrosis/f3 > f0 | [131] |
| miR-192 | Down | Downregulated in sera of CHB patients re ECVs | [81] |
| miR-193-5p | Down | Downregulated in cirrhosis vs. HBV-HCC | [123] |
| miR-194 | Down | Inhibits collagen-1 expression and HSC/targets rac1 | [130] |
| Up | Upregulated as injury increases/targets ACVR28 | [132] | |
| miR-199 | Down | Downregulated as injury/cirrhosis develops | [126] |
| miR-199-5p | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-199a-3p | Down | Upregulated in cirrhosis vs. CHB | [118] |
| miR-199a | Up | Upregulated in HBV-led fibrosis | [40] |
| miR-200a/b | Up | Upregulated in early HBV-fibrosis | [40,81] |
| miR-20a | Up | Upregulated in CHB fibrosis/f3–4 < f0–2 | [121] |
| miR-20b-5p | Down | Downregulated in advanced fibrosis versus early fibrosis/Targets p21/upregulated in sera | [57] |
| miR-206 | Down | Downregulated in cirrhosis vs. HBV-HCC | [123] |
| miR-21 | Down | Downregulated in advanced fibrosis vs. early fibrosis/acts via TGF-B/NF-kB pathways/downregulated f0–2 > f3–4 in CHB fibrosis | [58,118,127] |
| Down | Downregulated in cirrhosis progression | [118,133] | |
| miR-214-3p | Down | AUC 0.87 predictor of fibrosis/suppresses TGF-B pathway blocks collagen | [58,134] |
| miR-214-5p | Up | Upregulated in cirrhosis | [126] |
| miR-215 | Up | Upregulated in cirrhosis vs. HC | [104,128] |
| miR-221-3p | Up | Promotes fibrosis via TGF-B/NF-kB paths/f4 down vs. F1 | [57,118] |
| miR-221 | Down | Downregulated in CHB fibrosis but/f3–4 < f0–2 | [58,121] |
| Up | Upregulated in cirrhosis | [118,126] | |
| miR-222 | Up | CHB fibrosis via TGF-β/NF-κB pathways/increasing significant upregulation from f0–4Upregulation in cirrhosis | [58,121,126] |
| miR-222-3p | Up | Upregulated 13.88-fold in cirrhosis | [76,118] |
| miR-223 | Down | Downregulated in advanced fibrosis vs. early fibrosis/see article for path | [127] |
| miR-224 | Down | Downregulated in CHB led fibrosis/f3–4 significantly less downregulated than f0–2 | [121] |
| Up | Upregulated in cirrhosis vs. HC | [104,128] | |
| miR-23a/b | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-26a | Down | Increasingly downregulated in cirrhosis continuum | [133] |
| miR-26a-5p | Up | Upregulated in cirrhosis vs. HBV-HCC | [123] |
| miR-27-3p | Up/Down | Biomarker for cirrhosis | [129] |
| miR-27a | Up | TGF-1 > miR-27a > PPARγ, FOXO1, APC, P53 and RXRα α-SMA and COL1A2 (activated HSCs) | [118] |
| miR-27b | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-2861 | Up | Upregulated stage f4 vs. f0 fibrosis | [135] |
| miR-29a | Up | Upregulated in CHB fibrosis/f3–4 < f0–2 | [121] |
| Down | Increasingly downregulated as cirrhosis develops via TGF-B, NF κ B | [40] | |
| miR-29b | Down | Blocks collagen/TGF-B/targets COL1A1, COL4A5, COL5A3 | [58,117] |
| miR-30b/c | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-30e | Down | HBx-led downregulation of miR-30e >> P4HA2 fibrosis/cirrhosis | [118,123,136] |
| Down | Targets IL-6R in fibrogenic pathway | [134] | |
| miR-301a | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-324-5p | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-33a | Up | Stimulates TGF-B, HSCs and promotes fibrosis/targets SMAD7 to stimulate TGF-B HSCs/upregulated in cirrhosis vs. CHB | [116,118] |
| miR-331-3p | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-338-3p | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-34a/b/c | Up | Upregulated in HBV-led fibrosis | [40] |
| miR-34b-3p | Up | Upregulated in early fibrosis/targets GRB10/PIK3CA/up in sera/tissue | [57] |
| miR-340 | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-345-3p | Up | Upregulated in stage f4 vs. f0 fibrosis | [135] |
| miR-346-3p | up | Upregulated in early CHB induced fibrosis stages f1–2 | [57] |
| miR-3620-3p | Up | Upregulated in stage f4 vs. f0 fibrosis/cirrhosis | [135] |
| miR-3656 | Up | upregulated stage f4 vs. f0 fibrosis/cirrhosis | [135] |
| miR-371a-5p | Up | Upregulated in stage f4 vs. f0 fibrosis/cirrhosis | [135] |
| miR-374 | Down | Downregulated in advanced fibrosis vs. early fibrosis | [127] |
| miR-374b | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-377-3p | Up | Biomarker for cirrhosis | [129] |
| miR-410-3p | Up | Biomarker for cirrhosis | [129] |
| miR-424 | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-455-3p | Down | Downregulated in advanced fibrosis vs. early fibrosis/Targets p21/upregulated in sera | [57] |
| miR-4646-5p | Up | Upregulated f4 vs. f0 fibrosis/cirrhosis | [135] |
| miR-4651 | Up | Upregulated f4 vs. f0 fibrosis/cirrhosis | [135] |
| miR-4695-5p | Up | Upregulated f4 vs. f0 fibrosis/cirrhosis | [135] |
| miR-4800-5p | Up | Upregulation f4 vs. f0 fibrosis/cirrhosis | [135] |
| miR-483-5p | Down | Downregulated in f3 vs. f0 fibrosis/cirrhosis | [90] |
| miR-486-3p | Down | Downregulation f4 vs. f0 fibrosis/cirrhosis | [135] |
| miR-486-5p | Down | Downregulated in cirrhosis vs. HBV-HCC | [123] |
| miR-497-5p | Down | Downregulation f4 vs. f0 fibrosis/cirrhosis | [135] |
| miR-499a-5p | Down | Downregulated in early HBV-induced fibrosis/target CDKNIA/IKBKB/upregulated in sera | [57] |
| miR-513-3p | Up | Upregulated in early cirrhosis | [58,118] |
| miR-571 | Up | Upregulated in early cirrhosis | [58,118] |
| miR-574-3p | Up | Upregulated in cirrhosis vs. HC | [104,128] |
| Upregulated early cirrhosis/differentiates between cirrhosis and HBV-HCC | [118] | ||
| miR-602 | Up | Upregulated in cirrhosis vs. HC/increasing in HBV-HCC | [103,128] |
| miR-615-3p | Up | Promotes hypersplenism/cirrhosis | [137] |
| miR-638 | Up | Upregulated f4 vs. f0 fibrosis/cirrhosis | [135] |
| miR-652 | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-671-5p | Down | Downregulated in advanced versus early fibrosis/targets ATF2/E2F3/CREB3L2/up in sera and tissue | [57] |
| miR-744 | Up | Upregulated in cirrhosis vs. CHB | [118] |
| miR-885-5p | Up | Upregulated in cirrhosis versus HC | [104,128] |
| miR-92 | Down | Downregulated in CHB fibrosis/f3–f4 > f0–f2 | [121] |
| miR-939 | Down | Downregulated in cirrhosis vs. CHB | [118] |
| miR-940 | Down | Downregulated in cirrhosis vs. CHB | [129] |
| miR-942 | Up | Upregulated in activated HSCs/TGF and LPS induced miR-942 | [138] |
| miR-9-5p | Down | Fibrosis/activates HSCs via TGFBR1/TGFBR2 | [105] |
| miR-96-5p | Up | Upregulated in advanced fibrosis vs. early fibrosis/targets ATF2/E2F3/CREB3L2/up in sera and tissue | [57] |
Table 3.
HBV–HBx-downregulated miRNA in HBV-HCC (in alpha-numeric order).
| miRNA | Expression | Target | Gene Function | Reference |
|---|---|---|---|---|
| let-7 fam | Down | STAT3/RAS/HMGA2C-MYC/IL-6/IL-10/TLR-4/COL1A2/NGF, BCL-XL | Angiogenesis/growth/migration/inflammation/HBx repression | [51,52,145,146] |
| miR-1 | Down | EDN1/PI3K/AKT/HDAC4/MET | Angiogenesis, migration, invasion/modulates HBV replication | [147,148,149] |
| miR-101 | Down | DNMT3A/RASSF1/PRDM2/GSTP1/FOS/MCL-1/EZH2 | Proliferation/migration/invasion, increases HBV replication/induce DNA methylation | [120,150,151,152,153] |
| miR-101-3p | Down | ND, RAP1B/MCL-1,SOX9 | Inhibits HBV, proliferation/migration/promotes apoptosis | [154,155] |
| miR-122 | Down | β-CATENIN | EMT, cell migration, invasion and metastasis. Downregulated by HBx DNA methylation | [28,156] |
| Down | CCNG1 modulated p53/GLD2 | Promotes cell cycle progression/increases HBV replication | [35,157,158,159] | |
| Down | NDRG3/GALNT10 Cyclin G1/PTTG1 | Promotes apoptosis/blocks cell proliferation/invasion | [68,143,160,161] | |
| Down | PBF/ADAM10/Cyclin G1/Igf1R/ADAM 17/BCL-W/NDRG3 | Modulates proliferation, invasion, apoptosis | [28,35,68,162,163] | |
| miR -124 | Down | STAT3 and PIK3CA | Suppresses cell proliferation | [164] |
| miR-125b | Down | SMAD2/4/SIRTUIN7/SUV39H1/LIN28B/PIGF | Modulates EMT, growth, migration, and invasion | [28,165] |
| miR-132 | Down | AKT | Enhances cell proliferation/HBx DNA methylation downregulates miR-132 | [166] |
| miR-136 | Down | AEG-1 | Cell migration, invasion, and metastasis | [167] |
| miR-138 | Down | CCND3 | Promotes cell cycle progression/reduces HBV replication | [168] |
| miR-139-5p | Down | ZEB1/2 | Modulates EMT/metastasis | [28,169,170] |
| miR-145 | Down | MAP3K/CUL5/HDAC2/ADAM17 | Enhances proliferation, cell cycle progression, anti-apoptosis | [54,171,172] |
| miR-148a | Down | HPIP/AKT/ERK/FOXO4/ATF5/mTOR/MET/ACVR1 | Down in HCC tissue/Cell proliferation, EMT, and cell migration | [28,173,174,175] |
| miR-152 | Down | DNMT1/GSTP/CDH1/KIT | Modulates DNA methylation/cell proliferation/migration/invasion, blocks HBV | [176,177,178,179] |
| miR-15a/16-1 | Down | CCND1/BCL-2 | Promotes cell cycle progression/proliferation | [28,78,180,181] |
| miR-15b | Down | FUT2/GloboH/HNFα | Enhances cell proliferation | [182,183] |
| mIR-16 | Down | Cyclin D1, NCOR2 | Promotes apoptosis, decreases proliferation | [28,180] |
| miR-18a | Down | ERα/CTGF | Enhances proliferation, regulates connective tissue growth | [184,185] |
| miR-192 | Down | G1-G2 arrest_/SLC39A6/SNAIL_ | Cell cycle control/regulates metastasis | [28,186,187] |
| miR-193b | Down | ING5/CCND1/ETS1 | Regulate CDK2, proliferation, invasion | [188,189] |
| miR-200 fam | Down | ZEB1/2 | Modulates ZEB1/2 regulated E-cadherin | [28,190] |
| miR-205 | Down | ACSL4/E2F1/ZEB1/2 | Lipogenesis and cell proliferation/promotes EMT | [28,95,191] |
| miR-21 | Down | PTEN/PIP3/AKT | Down in serum/inhibits cell proliferation | [54,192] |
| miR-216b | Down | IGF2BP2/IGF2/AKT/mTOR/MAPK/ERK | Cell proliferation and cell migration | [54,193] |
| miR-222 | Down | p27 | Inhibits cell cycle progression | [54] |
| miR-23a | Down | Myc/E-CADHERIN/SPROUTY2 | Regulates EMT/metastasis | [28,194,195,196] |
| miR-26a/c | Down | IL-6/IFNα/ERα/IL-6/Cyclin D2/Cyclin E2 | Inhibits cell proliferation and metastasis | [197,198,199] |
| miR-29c | Down | BCL-2/MCL-1/TNFA1P3 | Inhibits cell proliferation and promotes apoptosis | [200,201] |
| miR-338-3p | Down | CCND1 | Promotes cell cycle progression | [202,203] |
| miR-34a | Down | CCL22/MAP4K4/SIRT1/CCND1/CDK4/6/MET | Modulates metastasis/growth/apoptosis | [29,204,205,206,207] |
| miR-363-3p | Down | SP1 | Modulates tumor growth | [28,208] |
| miR-373 | Down | E-CADHERIN | EMT, cell migration, invasion, and metastasis | [49] |
| miR-375 | Down | AEG-1 | Cell migration, invasion, and metastasis | [167] |
| miR-429 | Down | RAB18, NOTCH1 | Lipogenesis/proliferation, apoptosis | [209,210] |
| miR-520b | Down | HBXIP | Enhancing cell proliferation | [211] |
| miR-548p | Down | HBXIP, IFN-λ1 | Increases growth, blocks apoptosis/immune response | [102,212] |
| miR-661 | Down | MTA1/NK-κB/iNOS/NO | Angiogenesis, cell proliferation, and migration | [213] |
Table 4.
HBV–HBx-upregulated miRNA in HBV-HCC (in miR numeric order).
| miRNA | Expression | Target | Gene Function | Reference |
|---|---|---|---|---|
| miR-1 | Up | MASPIN | HBx-induced HBV-HCC progression | [214] |
| miR-107 | Up | AXIN2/MASPIN | Cell proliferation/HBV-HCC progression | [214,215] |
| miR-125a | Up | ERBB2, HBsAg | Suppresses HBsAg | [74,178,216] |
| miR-143 | Up | FNDC3B | Cell migration, invasion and metastasis | [28,217] |
| miR-146a | Up | CFH/STAT1 | Regulates inflammation/IFNα mediated anti- HBV efficiency | [104,218,219] |
| miR-155 | Up | PTEN/SOX6/ZHX2/SOCS1 | Promotes cell growth | [220,221,222] |
| miR-17-92 | Up | E2F1 (C-MYC-repressor), Cyclin G1 | Promotes HCC, blocks HBV replication, enhances proliferation and anchorage-independent growth | [82,223,224] |
| miR-181a | Up | FAS, E2F5 | Inhibits apoptosis, promotes cell growth | [85,86] |
| miR-199a-5p | Up | CHC | Promoting cell proliferation | [168] |
| miR-203a | Up | RAP1A | Increases inflammation/alters MAPK signaling | [96] |
| miR-21 | Up | PDCD4, PTEN | Up in HCC tissue/Targets tumor suppressors/Stops apoptosis/immune response/HBV evasion/promotes cell growth/up in HBV-HCC vs. CHB-L Cirrhosis serum | [54,55,56,65,214,225,226] |
| miR-215 | Up | PTPRP | Proliferation of hepatoma cells | [104,186,227] |
| miR-221 | Up | ERα, DDIT4/BMF/p27/p57 | Cell cycle progression (G1/S) and cell proliferation | [228,229,230] |
| miR-222 | Up | P27(kip1)/PTEN/PPP2R2A/p27/p57 | Promotes cell growth, migration | [54,230,231,232] |
| miR-224 | Up | PAK4/MMP9 inhibitor_-5/SMAD4_ | Enhance HBV replication, cell growth and invasion | [233,234,235] |
| miR-27a | Up | PPARγ, FOXO1, APC, P53 and RXRα | Enhances proliferation, migration, invasion | [118,236] |
| miR-29a | Up | PTEN/PI3K/AKT/MMP-2 | Enhancing cell migration | [54,237] |
| miR-30c | Up | HMBOX1 | Induces NK activity | [146,238] |
| miR-331-3p | Up | ING5 | Cell proliferation and apoptosis | [239] |
| miR-545/374a | Up | ESRRG | Cell proliferation and cell migration | [240,241] |
| miR-602 | Up | RASSF1a/STAT3/MYC | Cell apoptosis and proliferation, increases HBV | [13,103] |
| miR-7 | Up | EGFR/RAF/EKERK/PI3K-AKT/MASPIN | Inhibits cell growth | [214,241,242,243] |
2. MiRNA Dysregulation in the HBV-HCC Continuum
This section illustrates the regulatory role of HBV dysregulated miRNA in the different stages of the HBV-HCC continuum (Table 1, Table 2, Table 3 and Table 4). A brief description of each stage is provided before describing the regulatory role of miRNA.
2.1. Early/Chronic HBV Infection
In this stage the pivotal role of HBV, in cases where viral clearance does not occur, appears mainly to be due to the continuous destruction of hepatocytes by T-cells, which attempt to eliminate the infection in conditions of chronic inflammation and increasing oxidative stress [18]. If viral clearance does not occur, patients that progress to CHB will elicit a weak peripheral cytotoxic T-lymphocyte (CTL) response. Typically, an activated humoral response in CHB involves the production of interleukin-4 (IL-4), IL-5, and IL-10 secreted by type-2 helper T lymphocytes that promote antibody production rather than viral clearance. It is also hypothesized that low levels of intrahepatic HBV-specific CTLs are the cause of hepatic inflammation flares that occur in CHB patients without viral clearance [26]. Host cell miRNA modulate HBV expression by either targeting own cell transcription factors required for HBV genome transcription or by directly binding to HBV transcripts [13]. Many host cell miRNAs are also modulated by HBV proteins in order to ensure the persistence and replication of the virus.
MiRNA Regulating HBV Genome Expression
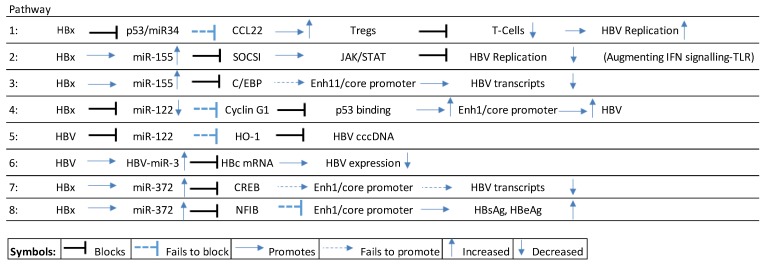
HBV genome replication and survival is self-managed as a result of modulating (up/down) both host cell miRNA expression, as well as coding for its own miRNA [27]. The hepatitis B virus x protein (HBx), for example, blocks p53 stimulated miR-34 expression in hepatocytes leading to the upregulation of macrophage-derived chemokine (CCL22) that stimulates regulatory T-cells (Tregs) that, in turn, block effector T-cells allowing HBV expression to increase [28,29] HBV genome transcription or by directly binding to HBV transcripts [13] (see Scheme 1, pathway 1).
Scheme 1.
HBx-mediated miRNA pathways in early/chronic HBV infection. CCL22: C-C motif chemokine 22; SOCS1: Suppressor of cytokine signaling 1; C/EBP: CCAAT/enhancer-binding protein; HO1: heme oxygenase 1; CREB: c-AMP-response element binding protein.
Upregulation of miR-155 by HBx results in the downregulation of suppressor of cytokine signaling-1 (SOCSI) expression contributing to increased JAK/STAT signaling, leading to the suppression of HBV infection by augmenting interferon (IFN) signaling [30] (see Scheme 1, pathway 2). Alternatively, HBx upregulated miR-155 also subdues HBV replication by blocking the CCAAT/enhancer-binding protein (C/EBP) that binds and activates the HBV Enhancer (Enh) 11/core promoter [31] (see Scheme 1, pathway 3).
The liver miR-122 is widely reported as upregulated in serum following HBV infection (references in Table 1) and is regarded as a key suppressor of HBV expression [32,33,34]. However, it has been reported that HBx downregulates miR-122 and facilitates increases in HBV transcripts by failing to block Cyclin G1, which then blocks p53 from binding to HBV Enh1/core promoter thus facilitating an increase in HBV expression [35] (see Scheme 1, pathway 4). Other studies have demonstrated that miR-122 blocks HBV pgRNA that encodes the hepatitis B core antigen (HBcAg) and viral polymerase, and that HBV downregulated miR-122 blocks HBV replication by failing to modulate heme-oxygenase (HO-1), which blocks HBV covalently closed circular DNA (cccDNA) [36] (see Scheme 1, pathway 5).
Multiple miRNA directly target HBV transcripts and examples of these include miR-184/-185/-196a/-199a-3p/-210/-217 that are all significantly upregulated [28]. Recently it was also discovered that the HBV virion generates its own HBV-miR-2/-3 [37]. HBV-miR-3 suppresses the hepatitis B core protein (HBc) mRNA to self-regulate HBV expression downwards [27], possibly in order to promote its survival by way of avoiding the host immune system [28,38] (see Scheme 1, pathway 6).
Some miRNA, like miR-372, can influence HBV expression positively and negatively depending on their target pathways. HBx upregulated miR-372 [39,40] targets the c-AMP-response element binding protein (CREB) from binding to HBV enhancer 1/core promoter (Enh 1/ENI-Cp) to reduce HBV transcripts. It also targets the nuclear factor 1 B-type protein (NF1B) that fails to modulate HBV Enh 1 (ENI-Cp), thus promoting HBV expression [28,39] (see Scheme 1, pathway 7–8).
2.2. HBV-Induced Inflammation Pathways
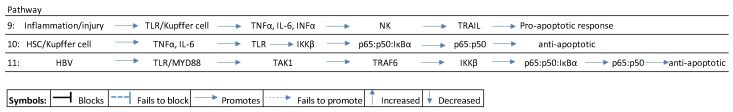
In this stage, chronic inflammation, accompanied by T-cell response, causes tissue damage where the continuous replacement of hepatocytes is initially achieved by differentiated hepatocytes, which eventually become depleted and are replaced by the recruitment of non-epithelial cells in the liver [18]. While their activation and proliferation initially serves to support the immune response and the regeneration of the tissue, during chronic liver damage and with increasing inflammation, their expansion predominates and they gradually replace epithelial structures in the organ. Inflammation/injury induces pro-inflammatory cytokines like tumor-necrosis factor-alpha (TNFα) and interleukin 6 (IL-6) by activating Kupffer cells and liver-derived macrophages. The activation of pro-inflammatory cytokines like TNFα and Il-6 can induce both pro-apoptotic and anti-apoptotic effects in injured liver tissue [41,42]. A pro-apoptotic response would, typically, be triggered by inflammation/necrosis that triggers Toll-like receptor (TLR) signaling to induce Kupffer cells to synthesize pro-inflammatory cytokines like TNFα, IL-6, and interferon alpha (IFNα), that recruit natural killer cells (NKs) to promote TNF-related apoptosis-inducing ligand (TRAIL) [43,44,45] (see Scheme 2, pathway 9).
Scheme 2.
Inflammation pathways in HBV-induced pathways. HSC: hepatocyte stellate cell; TLR: toll-like receptor; TNF: tumor necrosis factor; NK: natural killer cells; TRAIL: TNF-related apoptosis-inducing ligand; TAK1: transforming growth factor beta-activated kinase 1; IKKβ: inhibitor of NFκB kinase.
Alternatively, an anti-apoptotic response results from the activation of the canonical nuclear factor-κB (NFκB) inflammation pathway [46]. Injury triggers HSC/Kupffer cell stimulation of chemokines like TNFα and IL-6 that induce TLR signaling in hepatocytes. TLR signaling then activates the inhibitor of kappa kinase (IKK) complex to phosphorylate the IαKα/p65:p50 bound complex in the cytoplasm that leads to the accumulation of transcription factors p65:p50 in the nucleus. This accumulation, in turn, promotes an anti-apoptotic response [41,47,48] (see Scheme 2 pathway 10).
In CHB, a deficiency in interferon can block the immune response by subduing TLR expression, and the HBx protein can stimulate TGF-β1 to induce T-cell regulators to subdue the immune response [49,50]. Persistent activation of the NFκB pathway in HBV infection is orchestrated by way of TLRs that bind to HBV proteins (e.g., HBx) and activate the NFκB pathway. Typically HBV proteins bind to TLR-4/7/9 and recruit the myeloid differentiation primary response adaptor protein (MYD88) to activate tumor necrosis factor receptor (TNF-R)-associated factor 6 (TRAF6) via transforming growth factor beta-activated kinase 1 (TAK1), to activate the inhibitor of nuclear factor kappa-B kinase (IKK complex), to induce nuclear translocation of p65:p50 accumulation in the nucleus, and this results in an anti-apoptotic effect [41,49,51] (see Scheme 2 pathway 11).
MiRNA Regulating HBV-Induced Inflammation Pathways
At the commencement of the inflammation–fibrosis continuum in persistent HBV infection, a wide range of host miRNA are deregulated (see Table 1). Many of these miRNA subtly manipulate the host immune response to ensure both the survival and proliferation of the virus [28]. Complex interactions between HBV, the immune system, and miRNA trigger TLR pathways that interact with cytokines, interferons, and TNFs to modulate innate immune responses [51].
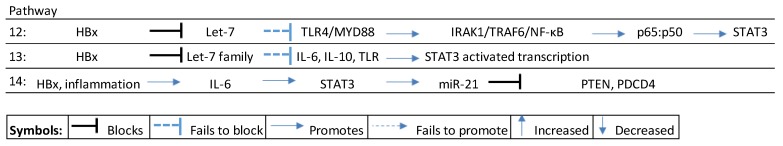
Examples of miRNA-influenced inflammation include miR-145/-148a/-200b/-200c/-455 and the Let-7 family [51]. Typically the Let-7 family, for instance, is downregulated in CHB by the HBx protein (or alternatively “mopped” up by HBV mRNA), thus preventing it from suppressing TLR4 signaling along the interleukin-1 receptor associated kinase (IRAK1)/TRAF6/NFKB pathway, to induce nuclear accumulation of p65:p50 [51] that promotes STAT3, resulting in cell proliferation and survival [48,52] (see Scheme 3, pathway 12). Alternatively, HBx downregulates Let-7 that can also activate STAT3 signaling by failing to modulate the expression of inflammatory cytokines, to influence miRNA-mediated suppression of key tumor suppressors [53] (see Scheme 3, pathway 13). HBx-induced upregulation of miR-21 by Il-6 mediated STAT3 signaling typically blocks tumor suppressors like PTEN and PDCD4 at an early stage in the inflammation–fibrosis axis and continues in the HBV-HCC stage [54,55,56] (see Scheme 3, pathway 14).
Scheme 3.
HBx-induced miRNA in inflammation pathways. TLR: toll-like receptor; IRAK1: interleukin-1 receptor associated kinase 1; TRAF6: tumor necrosis factor receptor associated factor 6; IL: interleukein; STAT3: signal transducer and activator of transcription 3; PTEN: phosphatase and tensin gene; PDCD4: programmed cell death protein 4 gene.
2.3. MiRNA Regulating HBV-Induced Fibrosis/Cirrhosis
In this stage, chronic tissue damage and inflammation are accompanied by the activation of non-epithelial cells in the liver, which proliferate. Initially this activation supports the immune response to regenerate damaged tissue, as these non-epithelial cells gradually replace epithelial structures in the liver. This process; however, eventually manifests as fibrosis/cirrhosis where well-organized parenchymal tissue is increasingly replaced by disorganized and dysfunctional fibrotic tissue [18]. In the inflammation–fibrosis axis, fibrogenesis is orchestrated by a complex network of common cytokine-mediated signaling pathways that regulate the activation of hepatic stellate cells (HSCs) and downstream extracellular matrix (ECM) proteins. These cytokines include TGF-β, platelet-derived growth factor (PDGF), TNF-α, interferons (IFNα/β), and interleukins (IL-1/6/17) [41,42]. Of all the different cytokine mediated pathways, upregulated TGF-B signaling is thought to be the principal fibrogenic pathway that activates HSCs to synthesize fibrogenic materials like collagen and alpha smooth muscle actin (α-SMA) [106,107,108,109]. In HBV-infected patients, the HBx protein has been identified as an activator of cytokine signaling [110], notwithstanding the synergistic effect induced by chronic inflammation, oxidative stress, and hepatocyte loss that triggers the activation of quiescent HSCs into myofibroblasts, which are the main source of ECM production (e.g., collagen 1/111, α-SMA) in the liver [106,107].
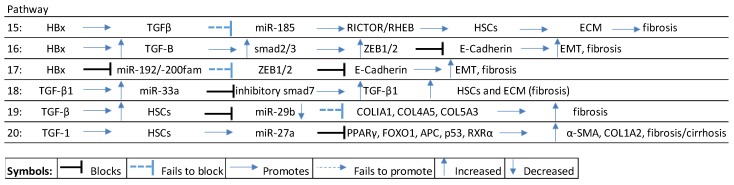
A hypothesized HBx-directed fibrogenic miRNA pathway involves miR-185 as follows: HBx promotes TGFβ signaling, that blocks miR-185 that, in turn, fails to modulate Rapamycin-insensitive companion of mammalian target of rapamycin (RICTOR/RHEB), resulting in the activation of HSCs, ECM proteins, and the development of fibrotic tissue [111,112] (see Scheme 4, pathway 15).
Scheme 4.
HBx-mediated fibrosis/cirrhosis pathways. TGFβ: transforming growth factor beta; RICTOR: rapamycin-insensitive companion of mammalian target of rapamycin; RHEB: Ras homolog enriched in brain; HSC: hepatocyte stellate cells; ECM: extracellular matrix; ZEB1/2: zinc finger E-box-binding homeobox1/2.
A wide range of miRNA that modulate fibrogenesis are deregulated in HBV-induced liver injury from early stage reversible fibrosis to irreversible fibrosis, loss of liver function and cirrhosis (See Table 2). miRNA directly targeting the TGF-β pathway, for example, include miR-122/-181b/-21/-214-3p/-221-3p/-222/-29/-33a/-942. Typically, HBx-induced TGF-β signaling induces fibrogenesis by sequestering the transcriptional ability of coSmad4 to form a complex to activate smad2/3, thus promoting the transcription of zinc finger E-box-binding homeobox proteins (ZEB1/2), which blocks E-cadherin formation to reduce cell adhesion and promote epithelial mesenchymal transition (EMT) [28,113,114] (see Scheme 4, pathway 16).
Another important miRNA family that modulates fibrogenesis is the miR-200 cluster. In HBV-related fibrogenesis, the HBx protein suppresses p53-led transcription of miR-192/-200, which then fails to modulate ZEB1/2, blocking the transcription of E-cadherin in the WNT/β-catenin pathway. A loss of E-cadherin is a key feature of fibrosis/EMT and occurs early in the HBV-HCC continuum [18,28,115] (see Scheme 4, pathway 17).
HBx-induced TGF signaling in the early phase of liver disease also promotes the increase in ECM, as a result of TGF-β1 upregulation of miR-33a to block inhibitory SMAD7, thus promoting Rsmad-induced TGF-β1, HSCs, ECM, and fibrosis [116] (see Scheme 4, pathway 18).
Collagen is a key downstream fibrogenic product of TGF signaling and cytokines like TFG-β, IL4/13, for instance, trigger TGF-β1-activated smad3, Stat6 to transcribe collagen type 1 alpha chain 2 (COL1A2) [112]. TGF signaling typically employs miRNA to amplify the production of collagen. For example, TGF-β activation of HSCs can downregulate miR-29b that then fails to modulate the expression of collagen [117] (see Scheme 4, pathway 19). TGF-β1-activated HSCs also enhance the expression of miR-27a, which suppresses antagonists of α-SMA and collagen like PPARϒ, FOXOI, APC, p53, and RXRα to promote fibrogenesis [118] (see Scheme 4, pathway 20).
2.4. HBV Deregulated MiRNA in HBV-HCC
In this stage, persistent HBV infection, chronic inflammation, oxidative stress, and cirrhosis can dysregulate a wide range of host gene expression by initiating deletions, amplifications, mutations, epigenetic changes, or by targeting miRNA loci or their transcription factors [139]. In this disrupted tissue, HBV-HCC may develop from cells that are able to survive in cirrhotic livers and that are more resistant to adverse conditions, viral infection, and apoptosis [18]. In this regard, the HBx protein is thought to play a key role in the development of HCC because it can inhibit the TP53 function in early carcinogenesis, and may contribute to the accumulation of aberrant replacement cells by downregulating apoptosis [18,140]. The HBx transactivating protein promotes cell cycle progression, inactivates negative growth regulators, tumor suppressors, and senescence-related factors [141,142].
2.4.1. HBV–HBx-Downregulated MiRNA in HBV-HCC
In general, downregulated miRNAs lose their ability to modulate oncoprotein expression. In HBV-infected persons, the HBx protein downregulates a wide range of miRNA (see Table 3) that then fail to modulate host oncogenic target genes and/or epigenetic mechanisms [143,144]. In Table 3, for example, key HBx-downregulated liver miRNA, like miR-122, typically fail to modulate oncogenic cell cycle proteins (cyclins/ β-catenin/ADAM 10 and 17/BCL-W), while Let-7 fails to suppress oncogenic proteins involved in angiogenesis, growth, and migration (STAT3/RAS/C-MYC/BCL-Xl). Another well cited miRNA that is downregulated by the HBx protein is miR-148a, which also fails to suppress oncogenic expression in the P13/MAPK pathway (see Figure 1).
2.4.2. HBx-Upregulated MiRNA in HBV-HCC
In general, many HBx-upregulated miRNAs typically block tumor suppressor networks (see Table 4). For example, the phosphatase and tensin homologue (PTEN) tumor suppressor is downregulated by HBx upregulated miR-21, miR-29a, miR-221, and miR-222. Upregulated miR-21 is a key liver miRNA that has been consistently cited as a silencer of tumor suppressors like PTEN and PDCD4 (see references in Table 4).
3. HBV–HBx-Dysregulated MiRNA in the Principal HBV-HCC Cancer Pathways
Understanding of the molecular etiology of HCC remains incomplete [244]. Evidence to date shows that HCC generally involves a range of disruptions of the PI3K/MAPK pathways and the p53 network that includes cell cycle controls (RB1), as well as increased WNT signaling and the inactivation of key tumor suppressors (SOCSI) in the JAK/STAT network [18,19]. Alterations of RB1, p53, and WNT pathways in HCC are frequently associated with HCV, HBV, and alcoholic liver cirrhosis [245]. This section focuses on examples of HBx-induced dysregulation of miRNA in HBV-HCC pathways.
3.1. Dysregulated MiRNA in the p13K/MAPK Pathway in HBV-HCC
A wide range of HBx-dysregulated miRNA (see Figure 1) play a complex regulatory role in the activation of the PI3K/AKT/mTOR and MAPK (RAF/MEK/ERK) pathways, which are a key feature of HCC [246,247]. Typically, a range of HBx-upregulated miRNA suppress tumor suppressor regulation and HBx-downregulated miRNA fail to modulate oncogenic proteins. Examples of HBx dysregulation include the upregulation of miR-17-92, miR-21 (via IL-6 activation) [55,226,248], miR-29, miR-155-5p [222], and miR-221/-222 [54,231] to suppress PTEN regulation of AKT/mTOR expression. Simultaneously, HBx downregulates miR1 (via EDN-1), miR-148a/-34a/26a/c and Let-7, which then fail to modulate MET/HGF [29,147,149,173,197] and RAS/RAF/mTOR expression respectively [145,249]. The subtle HBx counter modulation of mTOR is illustrated by HBx-upregulated miR-7 that increases inhibition of mTOR signaling [241,250], while HBx-downregulated Let-7 fails to modulate mTOR signaling. Simultaneously, HBx-induced downregulation of miR-1 can promote HBV replication via HDAC4-mediated stimulation of FXRα. HBx also downregulates miR-29c/-125b, resulting in reduced controls for the transcription of BCL-2 [28,200,201] and miR-26a/-34a that modulate C-JUN/FOS/cyclinD expression [28,197].
3.2. Dysregulated MiRNA in the WNT/β-Catenin Pathway in HBV-HCC
The upregulation of the WNT/β-catenin pathway is a frequent event in early HCC [251]. It yields an aggressive phenotype that is implicated in the proliferation, migration, invasion, and survival of cancer cells [244]. Figure 2 illustrates some examples of HBx-dysregulated miRNAs and their target genes in this pathway. The HBx protein typically plays a role in enhancing WNT and β-catenin expression while suppressing the expression of E-Cadherin. WNT signaling, for instance, is not modulated as a result of HBx led downregulation of miR-122/148a/b [157,173,174] and Cadherin expression is suppressed because these miRNA fail to suppress cadherin suppressors like SNAIL 1 [174]. HBx-induced upregulation of mIR-21 also enhances WNT signaling because it reduces DCC6 suppression of WNT signaling [252] and contributes to the suppression of E-cadherin by suppressing PDCD4 expression [55]. HBx-upregulated miR-221/-222 also contributes to the suppression of cadherin by enhancing ZEB 1/2 via blocking one of its suppressors like TRPS-1 [54,157]. Cadherin expression is also suppressed as a result of HBx-induced downregulation of miR-200/-205/-101/34 that fails to regulate ZEB 1/2 [95,150,190]. HBx-induced downregulation of miR-122 reduces β-catenin-led transcription in the cytoplasm [156], while HBx upregulation of miR-155 blocks the APC tumor suppressor in the WNT pathway, as well as suppresses HBV replication by blocking enhancer 11 to potentially hide the virus from the host immune system [28,31].
3.3. Dysregulated MiRNA in the TP 53 Pathway in HBV-HCC
The deregulation of multiple p53 pathways is a central event in the progression of HBV-HCC [253]. The INK4alpha/ARF locus, that encodes p14(ARF) and p16(INK4alpha) to arrest the cell cycle in the p53 and RB pathways, is frequently disrupted in HCC [254]. Figure 3 illustrates how HBx directly targets p53 expression [28,241] to influence the expression of miRNAs in this pathway. Typically, the HBx protein downregulates miR-26a/-34a/-138/15a/16-1, that fail to modulate cyclinD/CDK 4/6 and cyclinE/CDK2 expression [78,168,197,207], as well as miR-122/-Let-7/-34a/-125b/15a/16-1 [78,145,255,256,257,258], that then fail to modulate BCL-2-mediated suppression of CASP9/3-induced apoptosis. Similarly, HBx downregulation of miR-101/-125b/-29a/-Let-7 exerts an anti-apoptotic effect by failing to modulate MCL-1-mediated suppression of CASP9/3-induced apoptosis [200,255,258,259]. HBx-induced C-MYC upregulation of miR-17-92 and miR-221/222 also contribute to the E2F-mediated downregulation of p21/p27/p57 expression as a regulator of cyclins and cyclin-dependent kinases (CDKs) in the network [82,223,230].
3.4. Dysregulated MiRNAs in the JAK/STAT Pathway in HBV-HCC
The aberrant methylation in the CpG island of the SOCS-1 gene is a common feature in HBV-HCC and its silencing demonstrates its important tumor suppressor role in the JAK/STAT pathway [260]. Suppressor of cytokine signaling (SOCS-1) switches this signaling ‘off’ by means of its direct interaction with (JAK). The loss of function of SOCS-1 is a common feature in HCC and the HBx-mediated upregulation of miR-155 is a contributing factor in HBV-HCC (see Figure 4) [261,262]. HBx upregulated miR-221/-203 contribute to a reduction in SOCS3 regulation of JAK/STAT signaling [263]. HBx also downregulates Let-7, which reduces its modulation of IL-6 induced activation of JAK/STAT signaling and mTOR mediated transcription of oncogenic proteins like C-MYC/MCL-1 [145,250,264]. However, HBx upregulation of miR-7 [241], has been demonstrated as a control by way of suppressing mTOR signaling in JAK/STAT pathway [250].
4. Discussion and Conclusions
This exploratory review deliberately adopts a broad focus to demonstrate the complex regulatory role of miRNAs in the HBV-HCC continuum. Multiple knowledge gaps, exposed in this paper, prompt further research, to clarify the complex regulatory roles of the same miRNA across the HBV-HCC continuum. In addition, numerous miRNA target the same genes and cascades of miRNA respond to injury and disease with differing levels of expression in tissue, serum and cell-lines. Circulating miRNA, for instance, may originate from different cells (e.g., blood-immune) rather than an HBV or cancer specific origin by way of secretion or cell death [265,266,267]. The broad focus of the paper is, therefore, specifically adopted to highlight this complexity rather than to explain it. The key focus of this exploratory review was, therefore, to illustrate the multiplicity of dysregulated miRNA in clearly defined stages of pathogenesis rather than to attempt to try and explain their role in mediating multiple targets or summarize their role across every stage of HCC pathogenesis (see miR-122/-21). A key limitation of the study, therefore, is that it conveys an overly simplistic role of specific miRNA in HBV-HCC pathogenesis.
The potential roles of HBx downregulated miR-124, illustrates this limitation (see Table 3). Its various roles could include interaction with lncRNA-MALAT1 to regulate HBx-induced cancer stem cell properties in HepG2 through PI3K/AKT signaling [268]. miR-124 also suppresses cell proliferation and tumor growth in HCC in vitro and in vivo models by direct targeting STAT3 and PIK3CA thereby repressing both JAK/STAT and PI3K/AKT pathways [164]. Other studies demonstrate that miR-124 can suppress cell proliferation in HCC by targeting PIK3CA [269], as well as extracellular matrix protein laminin gamma 1 (LAMC1), which is a key feature in HCC progression [270]. However, LAMC1 mRNA promote malignancy by competing with miR-124 by binding with CD151 [270] and the overexpression of LAMC1 promotes HCC progression pathway by interaction with integrin receptors on a cell surface to promote proliferation and metastasis in HCC [271]. Elevated LAMC1 mRNA also acts as a sponge for miR-124 thus preventing its binding to another oncogenic membrane protein (CD151) that contributes to its elevated expression in HCC [270].
Similarly, the figures illustrating isolated roles of HBx-dysregulated miRNA in the main HBV-HCC pathways are clearly simplistic and only serve to demonstrate the interaction of the HBV dysregulated miRNA and their respective targets, thus ignoring the fact that they only represent a fraction of miRNA targeting these cancer pathways. The present review illustrates the complex range of miRNA regulatory roles in HBV-HCC pathogenesis. It also demonstrates how miRNA manipulation of HBV expression can be used as a tool to dissect HCC molecular pathways, and be harnessed to improve diagnosis, prognosis, anti-viral, and anti-tumor therapeutic modalities.
Author Contributions
K.S.—conceptualization of article, preparation of first draft, writing and review; J.M.—writing and review; B.S.—writing, review, development of figures; P.A.—writing and review; C.W.—writing and review; A.C.—writing and review; A.K.—writing and review.
Funding
This project has been funded in part with Federal funds from the Frederick National Laboratory for Cancer Research, National Institutes of Health, under contract HHSN261200800001E and Russian Science Foundation (project 17- 14-01338). This research was supported in part by the Intramural Research Program of NIH, Frederick National Lab, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 4.Cancer I.A.f.R.O. World Health Organization. Globocan 2012: Estimated cancer incidence, mortality and prevalence worldwide in 2012. [(accessed on 1 September 2014)]; Available online: http://globocan.iarc.fr/Pages/fact_sheets_cancer.Aspx.
- 5.Wong M.C., Jiang J.Y., Goggins W.B., Liang M., Fang Y., Fung F.D., Leung C., Wang H.H., Wong G.L., Wong V.W. International incidence and mortality trends of liver cancer: A global profile. Sci. Rep. 2017;7:45846. doi: 10.1038/srep45846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. Ca: A Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 7.Stanaway J.D., Flaxman A.D., Naghavi M., Fitzmaurice C., Vos T., Abubakar I., Abu-Raddad L.J., Assadi R., Bhala N., Cowie B. The global burden of viral hepatitis from 1990 to 2013: Findings from the Global Burden of Disease Study 2013. Lancet. 2016;388:1081–1088. doi: 10.1016/S0140-6736(16)30579-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Serag H.B. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–1273.e1261. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomaa A.I., Khan S.A., Toledano M.B., Waked I., Taylor-Robinson S.D. Hepatocellular carcinoma: Epidemiology, risk factors and pathogenesis. World J. Gastroenterol. WJG. 2008;14:4300. doi: 10.3748/wjg.14.4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan H., Zhong G., Xu G., He W., Jing Z., Gao Z., Huang Y., Qi Y., Peng B., Wang H. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. elife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tu T., Budzinska M.A., Shackel N.A., Jilbert A.R. Conceptual models for the initiation of hepatitis B virus-associated hepatocellular carcinoma. Liver Int. 2015;35:1786–1800. doi: 10.1111/liv.12773. [DOI] [PubMed] [Google Scholar]
- 12.Ganem D., Prince A.M. Hepatitis B virus infection—Natural history and clinical consequences. New Engl. J. Med. 2004;350:1118–1129. doi: 10.1056/NEJMra031087. [DOI] [PubMed] [Google Scholar]
- 13.Liu W.-H., Yeh S.-H., Chen P.-J. Role of microRNAs in hepatitis B virus replication and pathogenesis. Biochim. Et Biophys. Acta. 2011;1809:678–685. doi: 10.1016/j.bbagrm.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 14.Mak D., Babb de Villiers C., Chasela C., Urban M.I., Kramvis A. Analysis of risk factors associated with hepatocellular carcinoma in black South Africans: 2000–2012. PLoS ONE. 2018;13:e0196057. doi: 10.1371/journal.pone.0196057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bréchot C. Pathogenesis of hepatitis B virus—Related hepatocellular carcinoma: Old and new paradigms. Gastroenterology. 2004;127:S56–S61. doi: 10.1053/j.gastro.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 16.Jiang J., Gusev Y., Aderca I., Mettler T.A., Nagorney D.M., Brackett D.J., Roberts L.R., Schmittgen T.D. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin. Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ringelhan M., O’connor T., Protzer U., Heikenwalder M. The direct and indirect roles of HBV in liver cancer: Prospective markers for HCC screening and potential therapeutic targets. J. Pathol. 2015;235:355–367. doi: 10.1002/path.4434. [DOI] [PubMed] [Google Scholar]
- 18.Schulz W.A. Molecular Biology of Human Cancers: An Advanced Student’s Textbook. Springer; New York, NY, USA: 2005. [Google Scholar]
- 19.Garzon R., Marcucci G., Croce C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vidigal J.A., Ventura A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015;25:137–147. doi: 10.1016/j.tcb.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bushati N., Cohen S.M. microRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 22.Furuta M., Kozaki K.-i., Tanaka S., Arii S., Imoto I., Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2009;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 23.Hatziapostolou M., Polytarchou C., Aggelidou E., Drakaki A., Poultsides G.A., Jaeger S.A., Ogata H., Karin M., Struhl K., Hadzopoulou-Cladaras M. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami Y., Yasuda T., Saigo K., Urashima T., Toyoda H., Okanoue T., Shimotohno K. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 25.Ji F., Yang B., Peng X., Ding H., You H., Tien P. Circulating microRNAs in hepatitis B virus–infected patients. J. Viral Hepat. 2011;18 doi: 10.1111/j.1365-2893.2011.01443.x. [DOI] [PubMed] [Google Scholar]
- 26.Locarnini S. Molecular virology of hepatitis B virus. Semin.Liver Dis. 2004;24(Suppl. 1):3–10. doi: 10.1055/s-2004-828672. [DOI] [PubMed] [Google Scholar]
- 27.Yang X., Li H., Sun H., Fan H., Hu Y., Liu M., Li X., Tang H. Hepatitis B virus-encoded microRNA controls viral replication. J. Virol. 2017;91:e01919-16. doi: 10.1128/JVI.01919-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie K.-L., Zhang Y.-G., Liu J., Zeng Y., Wu H. MicroRNAs associated with HBV infection and HBV-related HCC. Theranostics. 2014;4:1176. doi: 10.7150/thno.8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang P., Li Q.-J., Feng Y., Zhang Y., Markowitz G.J., Ning S., Deng Y., Zhao J., Jiang S., Yuan Y. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Su C., Hou Z., Zhang C., Tian Z., Zhang J. Ectopic expression of microRNA-155 enhances innate antiviral immunity against HBV infection in human hepatoma cells. Virol. J. 2011;8:354. doi: 10.1186/1743-422X-8-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang B., Majumder S., Nuovo G., Kutay H., Volinia S., Patel T., Schmittgen T.D., Croce C., Ghoshal K., Jacob S.T. Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid–defined diet in C57BL/6 mice. Hepatology. 2009;50:1152–1161. doi: 10.1002/hep.23100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arataki K., Hayes C.N., Akamatsu S., Akiyama R., Abe H., Tsuge M., Miki D., Ochi H., Hiraga N., Imamura M. Circulating microRNA-22 correlates with microRNA-122 and represents viral replication and liver injury in patients with chronic hepatitis B. J. Med Virol. 2013;85:789–798. doi: 10.1002/jmv.23540. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura M., Kanda T., Jiang X., Haga Y., Takahashi K., Wu S., Yasui S., Nakamoto S., Yokosuka O. Serum microRNA-122 and Wisteria floribunda agglutinin-positive Mac-2 binding protein are useful tools for liquid biopsy of the patients with hepatitis B virus and advanced liver fibrosis. PLoS ONE. 2017;12:e0177302. doi: 10.1371/journal.pone.0177302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bandiera S., Pfeffer S., Baumert T.F., Zeisel M.B. miR-122—A key factor and therapeutic target in liver disease. J. Hepatol. 2015;62:448–457. doi: 10.1016/j.jhep.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Wang S., Qiu L., Yan X., Jin W., Wang Y., Chen L., Wu E., Ye X., Gao G.F., Wang F. Loss of microRNA 122 expression in patients with hepatitis B enhances hepatitis B virus replication through cyclin G1-modulated P53 activity. Hepatology. 2012;55:730–741. doi: 10.1002/hep.24809. [DOI] [PubMed] [Google Scholar]
- 36.Qiu L., Fan H., Jin W., Zhao B., Wang Y., Ju Y., Chen L., Chen Y., Duan Z., Meng S. miR-122-induced down-regulation of HO-1 negatively affects miR-122-mediated suppression of HBV. Biochem. Biophys. Res. Commun. 2010;398:771–777. doi: 10.1016/j.bbrc.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 37.Yao L., Sui Z.-H., Liu Y.-K., Xie H., Gao H.-J., Fan H.-X., Zhang Y., Liu M., Li S., Tang H. HBV-Encoded miR-2 Functions as an Oncogene by Downregulating TRIM35 But Upregulating RAN in Liver Cancer Cells. [(accessed on 1 October 2019)]; doi: 10.1016/j.ebiom.2019.09.012. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6838411/ [DOI] [PMC free article] [PubMed] [Retracted]
- 38.Mahajan V.S., Drake A., Chen J. Virus-specific host miRNAs: Antiviral defenses or promoters of persistent infection? Trends Immunol. 2009;30:1–7. doi: 10.1016/j.it.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo H., Liu H., Mitchelson K., Rao H., Luo M., Xie L., Sun Y., Zhang L., Lu Y., Liu R. MicroRNAs-372/373 promote the expression of hepatitis B virus through the targeting of nuclear factor I/B. Hepatology. 2011;54:808–819. doi: 10.1002/hep.24441. [DOI] [PubMed] [Google Scholar]
- 40.Li G., Cai G., Li D., Yin W. MicroRNAs and liver disease: Viral hepatitis, liver fibrosis and hepatocellular carcinoma. Postgrad. Med J. 2014;90:106–112. doi: 10.1136/postgradmedj-2013-131883. [DOI] [PubMed] [Google Scholar]
- 41.Elsharkawy A.M., Mann D.A. Nuclear factor-κB and the hepatic inflammation-fibrosis-cancer axis. Hepatology. 2007;46:590–597. doi: 10.1002/hep.21802. [DOI] [PubMed] [Google Scholar]
- 42.Zhou W.-C., Zhang Q.-B., Qiao L. Pathogenesis of liver cirrhosis. World J. Gastroenterol. WJG. 2014;20:7312. doi: 10.3748/wjg.v20.i23.7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tu Z., Bozorgzadeh A., Pierce R.H., Kurtis J., Crispe I.N., Orloff M.S. TLR-dependent cross talk between human Kupffer cells and NK cells. J. Exp. Med. 2008;205:233–244. doi: 10.1084/jem.20072195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunn C., Brunetto M., Reynolds G., Christophides T., Kennedy P.T., Lampertico P., Das A., Lopes A.R., Borrow P., Williams K. Cytokines induced during chronic hepatitis B virus infection promote a pathway for NK cell–mediated liver damage. J. Exp. Med. 2007;204:667–680. doi: 10.1084/jem.20061287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Protzer U., Maini M.K., Knolle P.A. Living in the liver: Hepatic infections. Nat. Rev. Immunol. 2012;12:201. doi: 10.1038/nri3169. [DOI] [PubMed] [Google Scholar]
- 46.Baker R.G., Hayden M.S., Ghosh S. NF-κB, inflammation, and metabolic disease. Cell Metab. 2011;13:11–22. doi: 10.1016/j.cmet.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tacke F., Luedde T., Trautwein C. Inflammatory pathways in liver homeostasis and liver injury. Clin. Rev. Allergy Immunol. 2009;36:4–12. doi: 10.1007/s12016-008-8091-0. [DOI] [PubMed] [Google Scholar]
- 48.He G., Karin M. NF-κB and STAT3–key players in liver inflammation and cancer. Cell Res. 2011;21:159. doi: 10.1038/cr.2010.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arzumanyan A., Friedman T., Kotei E., Ng I.O., Lian Z., Feitelson M.A. Epigenetic repression of E-cadherin expression by hepatitis B virus x antigen in liver cancer. Oncogene. 2012;31:563. doi: 10.1038/onc.2011.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stoop J.N., van der Molen R.G., Baan C.C., van der Laan L.J., Kuipers E.J., Kusters J.G., Janssen H.L. Regulatory T cells contribute to the impaired immune response in patients with chronic hepatitis B virus infection. Hepatology. 2005;41:771–778. doi: 10.1002/hep.20649. [DOI] [PubMed] [Google Scholar]
- 51.Jiang X., Kanda T., Wu S., Nakamura M., Miyamura T., Nakamoto S., Banerjee A., Yokosuka O. Regulation of microRNA by hepatitis B virus infection and their possible association with control of innate immunity. World J. Gastroenterol. WJG. 2014;20:7197. doi: 10.3748/wjg.v20.i23.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takata A., Otsuka M., Ohno M., Kishikawa T., Yoshikawa T., Koike K. Mutual antagonism between hepatitis B viral mRNA and host microRNA let-7. Sci. Rep. 2016;6:23237. doi: 10.1038/srep23237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li C.H., Xu F., Chow S., Feng L., Yin D., Ng T.B., Chen Y. Hepatitis B virus X protein promotes hepatocellular carcinoma transformation through interleukin-6 activation of microRNA-21 expression. Eur. J. Cancer. 2014;50:2560–2569. doi: 10.1016/j.ejca.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 54.Bandopadhyay M., Banerjee A., Sarkar N., Panigrahi R., Datta S., Pal A., Singh S.P., Biswas A., Chakrabarti S., Chakravarty R. Tumor suppressor micro RNA miR-145 and onco micro RNAs miR-21 and miR-222 expressions are differentially modulated by hepatitis B virus X protein in malignant hepatocytes. BMC Cancer. 2014;14:721. doi: 10.1186/1471-2407-14-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu X., Dong S., Qiao F., Lu S., Song Y., Lao Y., Li Y., Zeng T., Hu J., Zhang L. HBx-mediated miR-21 upregulation represses tumor-suppressor function of PDCD4 in hepatocellular carcinoma. Oncogene. 2013;32:3296. doi: 10.1038/onc.2013.150. [DOI] [PubMed] [Google Scholar]
- 56.Momeni M., Hassanshahi G., Arababadi M.K., Kennedy D. Ectopic expression of micro-RNA-1, 21 and 125a in peripheral blood immune cells is associated with chronic HBV infection. Mol. Biol. Rep. 2014;41:4833–4837. doi: 10.1007/s11033-014-3355-7. [DOI] [PubMed] [Google Scholar]
- 57.Singh A.K., Rooge S.B., Varshney A., Vasudevan M., Bhardwaj A., Venugopal S.K., Trehanpati N., Kumar M., Geffers R., Kumar V. Global microRNA expression profiling in the liver biopsies of hepatitis B virus–infected patients suggests specific microRNA signatures for viral persistence and hepatocellular injury. Hepatology. 2018;67:1695–1709. doi: 10.1002/hep.29690. [DOI] [PubMed] [Google Scholar]
- 58.Hayes C., Chayama K. MicroRNAs as biomarkers for liver disease and hepatocellular carcinoma. Int. J. Mol. Sci. 2016;17:280. doi: 10.3390/ijms17030280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li L.-M., Hu Z.-B., Zhou Z.-X., Chen X., Liu F.-Y., Zhang J.-F., Shen H.-B., Zhang C.-Y., Zen K. Serum microRNA profiles serve as novel biomarkers for HBV infection and diagnosis of HBV-positive hepatocarcinoma. Cancer Res. 2010 doi: 10.1158/0008-5472.CAN-10-1001. [DOI] [PubMed] [Google Scholar]
- 60.Chen Y., Li L., Zhou Z., Wang N., Zhang C.-Y., Zen K. A pilot study of serum microRNA signatures as a novel biomarker for occult hepatitis B virus infection. Med. Microbiol. Immunol. 2012;201:389–395. doi: 10.1007/s00430-011-0223-0. [DOI] [PubMed] [Google Scholar]
- 61.Wang Y., Liu Z., Yao B., Dou C., Xu M., Xue Y., Ding L., Jia Y., Zhang H., Li Q. Long non-coding RNA TUSC7 acts a molecular sponge for miR-10a and suppresses EMT in hepatocellular carcinoma. Tumor Biol. 2016;37:11429–11441. doi: 10.1007/s13277-016-4892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hong Z., Hong H., Liu J., Zheng X., Huang M., Li C., Xia J. miR-106a is downregulated in peripheral blood mononuclear cells of chronic hepatitis B and associated with enhanced levels of interleukin-8. Mediat. Inflamm. 2015;2015:629862. doi: 10.1155/2015/629862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Akamatsu S., Hayes C.N., Tsuge M., Miki D., Akiyama R., Abe H., Ochi H., Hiraga N., Imamura M., Takahashi S. Differences in serum microRNA profiles in hepatitis B and C virus infection. J. Infect. 2015;70:273–287. doi: 10.1016/j.jinf.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 64.Xing T., Jiang D., Huang J., Xu Z. Expression and clinical significance of miR-122 and miR-29 in hepatitis B virus-related liver. Genet. Mol. Res. 2014;13:7912–7918. doi: 10.4238/2014.September.29.4. [DOI] [PubMed] [Google Scholar]
- 65.Trung N.T., Duong D.C., Van Tong H., Hien T.T.T., Hoan P.Q., Bang M.H., Binh M.T., Ky T.D., Tung N.L., Thinh N.T. Optimisation of quantitative miRNA panels to consolidate the diagnostic surveillance of HBV-related hepatocellular carcinoma. PLoS ONE. 2018;13:e0196081. doi: 10.1371/journal.pone.0196081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waidmann O., Bihrer V., Pleli T., Farnik H., Berger A., Zeuzem S., Kronenberger B., Piiper A. Serum microRNA-122 levels in different groups of patients with chronic hepatitis B virus infection. J. Viral Hepat. 2012;19:e58–e65. doi: 10.1111/j.1365-2893.2011.01536.x. [DOI] [PubMed] [Google Scholar]
- 67.Chen Y., Shen A., Rider P.J., Yu Y., Wu K., Mu Y., Hao Q., Liu Y., Gong H., Zhu Y. A liver-specific microRNA binds to a highly conserved RNA sequence of hepatitis B virus and negatively regulates viral gene expression and replication. Faseb J. 2011;25:4511–4521. doi: 10.1096/fj.11-187781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li C., Wang Y., Wang S., Wu B., Hao J., Fan H., Ju Y., Ding Y., Chen L., Chu X. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J. Virol. 2013;87:2193–2205. doi: 10.1128/JVI.02831-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng J.-L., Zhao H., Yang S.-G., Chen E.-M., Chen W.-Q., Li L.-J. Plasma miRNA-122-5p and miRNA-151a-3p identified as potential biomarkers for liver injury among CHB patients with PNALT. Hepatol. Int. 2018:1–11. doi: 10.1007/s12072-018-9871-0. [DOI] [PubMed] [Google Scholar]
- 70.Wang Y., Zhu P., Qiu J., Wang J., Zhu H., Zhu Y., Zhang L., Zhu J., Liu X., Dong C. Identification and characterization of interferon signaling-related microRNAs in occult hepatitis B virus infection. Clin. Epigenetics. 2017;9:101. doi: 10.1186/s13148-017-0404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang J.Y., Mao R.C., Zhang Y.M., Zhang Y.J., Liu H.Y., Qin Y.L., Lu M.J., Zhang J.M. Serum micro RNA-124 is a novel biomarker for liver necroinflammation in patients with chronic hepatitis B virus infection. J. Viral Hepat. 2015;22:128–136. doi: 10.1111/jvh.12284. [DOI] [PubMed] [Google Scholar]
- 72.Chen G., Shi Y., Liu M., Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Coppola N., Potenza N., Pisaturo M., Mosca N., Tonziello G., Signoriello G., Messina V., Sagnelli C., Russo A., Sagnelli E. Liver microRNA hsa-miR-125a-5p in HBV chronic infection: Correlation with HBV replication and disease progression. PLoS ONE. 2013;8:e65336. doi: 10.1371/journal.pone.0065336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Potenza N., Papa U., Mosca N., Zerbini F., Nobile V., Russo A. Human microRNA hsa-miR-125a-5p interferes with expression of hepatitis B virus surface antigen. Nucleic Acids Res. 2011;39:5157–5163. doi: 10.1093/nar/gkr067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H., Huang C., Wang Y., Lu Z., Zhuang N., Zhao D., He J., Shi L. Hepatitis B virus X protein sensitizes TRAIL-induced hepatocyte apoptosis by inhibiting the E3 ubiquitin ligase A20. PLoS ONE. 2015;10:e0127329. doi: 10.1371/journal.pone.0127329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Giray B.G., Emekdas G., Tezcan S., Ulger M., Serin M.S., Sezgin O., Altintas E., Tiftik E.N. Profiles of serum microRNAs; miR-125b-5p and miR223-3p serve as novel biomarkers for HBV-positive hepatocellular carcinoma. Mol. Biol. Rep. 2014;41:4513–4519. doi: 10.1007/s11033-014-3322-3. [DOI] [PubMed] [Google Scholar]
- 77.Huang J.-Y., Chou S.-F., Lee J.-W., Chen H.-L., Chen C.-M., Tao M.-H., Shih C. MicroRNA-130a can inhibit hepatitis B virus replication via targeting PGC1α and PPARγ. Rna. 2015;21:385–400. doi: 10.1261/rna.048744.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y., Jiang L., Ji X., Yang B., Zhang Y., Fu X.-D. Hepatitis B viral RNA directly mediates down-regulation of the tumor suppressor microRNA miR-15a/miR-16-1 in hepatocytes. J. Biol. Chem. 2013;288:18484–18493. doi: 10.1074/jbc.M113.458158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu N., Jiao T., Huang Y., Liu W., Li Z., Ye X. Hepatitis B virus regulates apoptosis and tumorigenesis through the microRNA-15a-Smad7-transforming growth factor beta pathway. J. Virol. 2015;89:2739–2749. doi: 10.1128/JVI.02784-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lee H.-M., Kim T.S., Jo E.-K. MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep. 2016;49:311. doi: 10.5483/BMBRep.2016.49.6.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lambrecht J., Jan Poortmans P., Verhulst S., Reynaert H., Mannaerts I., van Grunsven L.A. Circulating ECV-associated miRNAs as potential clinical biomarkers in early stage HBV and HCV induced liver fibrosis. Front. Pharmacol. 2017;8:56. doi: 10.3389/fphar.2017.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jung Y.J., Kim J.W., Park S.J., Min B.Y., Jang E.S., Kim N.Y., Jeong S.H., Shin C.M., Lee S.H., Park Y.S. c-Myc-mediated overexpression of miR-17-92 suppresses replication of hepatitis B virus in human hepatoma cells. J. Med Virol. 2013;85:969–978. doi: 10.1002/jmv.23534. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Z.-Z., Liu X., Wang D.-Q., Teng M.-K., Niu L.-W., Huang A.-L., Liang Z. Hepatitis B virus and hepatocellular carcinoma at the miRNA level. World J. Gastroenterol. WJG. 2011;17:3353. doi: 10.3748/wjg.v17.i28.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu F., Zhou G., Li G., Chen B., Dong P., Zheng J. Serum miR-181b is correlated with hepatitis B virus replication and disease progression in chronic hepatitis B patients. Dig. Dis. Sci. 2015;60:2346–2352. doi: 10.1007/s10620-015-3649-1. [DOI] [PubMed] [Google Scholar]
- 85.Zou C., Li Y., Cao Y., Zhang J., Jiang J., Sheng Y., Wang S., Huang A., Tang H. Up-regulated MicroRNA-181a induces carcinogenesis in Hepatitis B virus-related hepatocellular carcinoma by targeting E2F5. BMC Cancer. 2014;14:97. doi: 10.1186/1471-2407-14-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zou C., Chen J., Chen K., Wang S., Cao Y., Zhang J., Sheng Y., Huang A., Tang H. Functional analysis of miR-181a and Fas involved in hepatitis B virus-related hepatocellular carcinoma pathogenesis. Exp. Cell Res. 2015;331:352–361. doi: 10.1016/j.yexcr.2014.11.007. [DOI] [PubMed] [Google Scholar]
- 87.Riazalhosseini B., Mohamed R., Apalasamy Y.D., Langmia I.M., Mohamed Z. Circulating microRNA as a marker for predicting liver disease progression in patients with chronic hepatitis B. Rev. Da Soc. Bras. De Med. Trop. 2017;50:161–166. doi: 10.1590/0037-8682-0416-2016. [DOI] [PubMed] [Google Scholar]
- 88.Brunetto M.R., Cavallone D., Oliveri F., Moriconi F., Colombatto P., Coco B., Ciccorossi P., Rastelli C., Romagnoli V., Cherubini B. A serum microRNA signature is associated with the immune control of chronic hepatitis B virus infection. PLoS ONE. 2014;9:e110782. doi: 10.1371/journal.pone.0110782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nielsen K.O., Jacobsen K.S., Mirza A.H., Winther T.N., Størling J., Glebe D., Pociot F., Hogh B. Hepatitis B virus upregulates host microRNAs that target apoptosis-regulatory genes in an in vitro cell model. Exp. Cell Res. 2018;371:92–103. doi: 10.1016/j.yexcr.2018.07.044. [DOI] [PubMed] [Google Scholar]
- 90.Murakami Y., Toyoda H., Tanahashi T., Tanaka J., Kumada T., Yoshioka Y., Kosaka N., Ochiya T., Taguchi Y. Comprehensive miRNA expression analysis in peripheral blood can diagnose liver disease. PLoS ONE. 2012;7:e48366. doi: 10.1371/journal.pone.0048366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li P., Xiao Z., Luo J., Zhang Y., Lin L. MiR-139-5p, miR-940 and miR-193a-5p inhibit the growth of hepatocellular carcinoma by targeting SPOCK1. J. Cell. Mol. Med. 2019 doi: 10.1111/jcmm.14121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ninomiya M., Kondo Y., Kimura O., Funayama R., Nagashima T., Kogure T., Morosawa T., Tanaka Y., Nakayama K., Shimosegawa T. The expression of miR-125b-5p is increased in the serum of patients with chronic hepatitis B infection and inhibits the detection of hepatitis B virus surface antigen. J. Viral Hepat. 2016;23:330–339. doi: 10.1111/jvh.12522. [DOI] [PubMed] [Google Scholar]
- 93.Liu Y., Zhao J.-J., Wang C.-M., Li M.-Y., Han P., Wang L., Cheng Y.-Q., Zoulim F., Ma X., Xu D.-P. Altered expression profiles of microRNAs in a stable hepatitis B virus-expressing cell line. Chin. Med J. 2009;122:10–14. doi: 10.3901/jme.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 94.Tian H., He Z. miR-200c targets nuclear factor IA to suppress HBV replication and gene expression via repressing HBV Enhancer I activity. Biomed. Pharmacother. 2018;99:774–780. doi: 10.1016/j.biopha.2018.01.141. [DOI] [PubMed] [Google Scholar]
- 95.Zhang T., Zhang J., Cui M., Liu F., You X., Du Y., Gao Y., Zhang S., Lu Z., Ye L. Hepatitis B virus X protein inhibits tumor suppressor miR-205 through inducing hypermethylation of miR-205 promoter to enhance carcinogenesis. Neoplasia. 2013;15:IN24–IN26. doi: 10.1593/neo.131362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wu A., Chen H., Xu C., Zhou J., Chen S., Shi Y., Xu J., Gan J., Zhang J. miR-203a is involved in HBx-induced inflammation by targeting Rap1a. Exp. Cell Res. 2016;349:191–197. doi: 10.1016/j.yexcr.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 97.Yu F., Yang J., Ouyang J., Zheng Y., Chen B., Li G., Lu Z., Dong P., Zheng J. Serum microRNA-210 levels in different groups of chronic hepatitis B patients. Clin. Chim. Acta. 2015;450:203–209. doi: 10.1016/j.cca.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 98.Zhang G.-L., Li Y.-X., Zheng S.-Q., Liu M., Li X., Tang H. Suppression of hepatitis B virus replication by microRNA-199a-3p and microRNA-210. Antivir. Res. 2010;88:169–175. doi: 10.1016/j.antiviral.2010.08.008. [DOI] [PubMed] [Google Scholar]
- 99.Raney A.K., Johnson J.L., Palmer C., McLachlan A. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J. Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang G., Li N., Li Z., Zhu Q., Li F., Yang C., Han Q., Lv Y., Zhou Z., Liu Z. microRNA-4717 differentially interacts with its polymorphic target in the PD1 3′ untranslated region: A mechanism for regulating PD-1 expression and function in HBV-associated liver diseases. Oncotarget. 2015;6:18933. doi: 10.18632/oncotarget.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jin J., Tang S., Xia L., Du R., Xie H., Song J., Fan R., Bi Q., Chen Z., Yang G. MicroRNA-501 promotes HBV replication by targeting HBXIP. Biochem. Biophys. Res. Commun. 2013;430:1228–1233. doi: 10.1016/j.bbrc.2012.12.071. [DOI] [PubMed] [Google Scholar]
- 102.Li Y., Xie J., Xu X., Wang J., Ao F., Wan Y., Zhu Y. MicroRNA-548 down-regulates host antiviral response via direct targeting of IFN-λ1. Protein Cell. 2013;4:130–141. doi: 10.1007/s13238-012-2081-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang L., Ma Z., Wang D., Zhao W., Chen L., Wang G. MicroRNA-602 regulating tumor suppressive gene RASSF1A is over-expressed in hepatitis B virus-infected liver and hepatocellular carcinoma. Cancer Biol. Ther. 2010;9:803–808. doi: 10.4161/cbt.9.10.11440. [DOI] [PubMed] [Google Scholar]
- 104.Gui J., Tian Y., Wen X., Zhang W., Zhang P., Gao J., Run W., Tian L., Jia X., Gao Y. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin. Sci. 2011;120:183–193. doi: 10.1042/CS20100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yu F., Chen B., Fan X., Li G., Dong P., Zheng J. Epigenetically-Regulated MicroRNA-9-5p Suppresses the Activation of Hepatic Stellate Cells via TGFBR1 and TGFBR2. Cell. Physiol. Biochem. 2017;43:2242–2252. doi: 10.1159/000484303. [DOI] [PubMed] [Google Scholar]
- 106.Puche J.E., Lee Y.A., Jiao J., Aloman C., Fiel M.I., Muñoz U., Kraus T., Lee T., Yee H.F., Jr., Friedman S.L. A novel murine model to deplete hepatic stellate cells uncovers their role in amplifying liver damage in mice. Hepatology. 2013;57:339–350. doi: 10.1002/hep.26053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kan F., Ye L., Yan T., Cao J., Zheng J., Li W. Proteomic and transcriptomic studies of HBV-associated liver fibrosis of an AAV-HBV-infected mouse model. BMC Genom. 2017;18:641. doi: 10.1186/s12864-017-3984-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brenner D.A. Molecular pathogenesis of liver fibrosis. Transactions of the American Clinical and Climatological Association. 2009;120:361. [PMC free article] [PubMed] [Google Scholar]
- 109.Moreira R.K. Hepatic stellate cells and liver fibrosis. Arch. Pathol. Lab. Med. 2007;131:1728–1734. doi: 10.5858/2007-131-1728-HSCALF. [DOI] [PubMed] [Google Scholar]
- 110.Suhail M., Abdel-Hafiz H., Ali A., Fatima K., Damanhouri G.A., Azhar E., Chaudhary A.G., Qadri I. Potential mechanisms of hepatitis B virus induced liver injury. World J. Gastroenterol. WJG. 2014;20:12462. doi: 10.3748/wjg.v20.i35.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou L., Liu S., Han M., Ma Y., Feng S., Zhao J., Lu H., Yuan X., Cheng J. miR-185 Inhibits Fibrogenic Activation of Hepatic Stellate Cells and Prevents Liver Fibrosis. Mol. Ther. -Nucleic Acids. 2018;10:91–102. doi: 10.1016/j.omtn.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bhogal R.K., Stoica C.M., McGaha T.L., Bona C.A. Molecular aspects of regulation of collagen gene expression in fibrosis. J. Clin. Immunol. 2005;25:592. doi: 10.1007/s10875-005-7827-3. [DOI] [PubMed] [Google Scholar]
- 113.Mizuguchi Y., Takizawa T., Yoshida H., Uchida E. Dysregulated miRNA in progression of hepatocellular carcinoma: A systematic review. Hepatol. Res. 2015 doi: 10.1111/hepr.12606. [DOI] [PubMed] [Google Scholar]
- 114.Lee D.K., Park S.H., Yi Y., Choi S.-G., Lee C., Parks W.T., Cho H., De Caestecker M.P., Shaul Y., Roberts A.B. The hepatitis B virus encoded oncoprotein pX amplifies TGF-β family signaling through direct interaction with Smad4: Potential mechanism of hepatitis B virus-induced liver fibrosis. Genes Dev. 2001;15:455–466. doi: 10.1101/gad.856201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Murakami Y., Toyoda H., Tanaka M., Kuroda M., Harada Y., Matsuda F., Tajima A., Kosaka N., Ochiya T., Shimotohno K. The progression of liver fibrosis is related with overexpression of the miR-199 and 200 families. PLoS ONE. 2011;6:e16081. doi: 10.1371/journal.pone.0016081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Huang C.-F., Sun C.-C., Zhao F., Zhang Y.-D., Li D.-J. miR-33a levels in hepatic and serum after chronic HBV-induced fibrosis. J. Gastroenterol. 2015;50:480–490. doi: 10.1007/s00535-014-0986-3. [DOI] [PubMed] [Google Scholar]
- 117.Roderburg C., Urban G.W., Bettermann K., Vucur M., Zimmermann H., Schmidt S., Janssen J., Koppe C., Knolle P., Castoldi M. Micro-RNA profiling reveals a role for miR-29 in human and murine liver fibrosis. Hepatology. 2011;53:209–218. doi: 10.1002/hep.23922. [DOI] [PubMed] [Google Scholar]
- 118.Zhang H., Yan X.-L., Guo X.-X., Shi M.-J., Lu Y.-Y., Zhou Q.-M., Chen Q.-L., Hu Y.-Y., Xu L.-M., Huang S. MiR-27a as a predictor for the activation of hepatic stellate cells and hepatitis B virus-induced liver cirrhosis. Oncotarget. 2018;9:1075. doi: 10.18632/oncotarget.23262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tu X., Zhang H., Zhang J., Zhao S., Zheng X., Zhang Z., Zhu J., Chen J., Dong L., Zang Y. MicroRNA-101 suppresses liver fibrosis by targeting the TGF β signalling pathway. J. Pathol. 2014;234:46–59. doi: 10.1002/path.4373. [DOI] [PubMed] [Google Scholar]
- 120.Xie Y., Yao Q., Butt A.M., Guo J., Tian Z., Bao X., Li H., Meng Q., Lu J. Expression profiling of serum microRNA-101 in HBV-associated chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Cancer Biol. Ther. 2014;15:1248–1255. doi: 10.4161/cbt.29688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Appourchaux K., Dokmak S., Resche-Rigon M., Treton X., Lapalus M., Gattolliat C.-H., Porchet E., Martinot-Peignoux M., Boyer N., Vidaud M. MicroRNA-based diagnostic tools for advanced fibrosis and cirrhosis in patients with chronic hepatitis B and C. Sci. Rep. 2016;6:34935. doi: 10.1038/srep34935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang W.J., Lai R.T., Lu J., Xiang X.G., Zhao G.D., Tang W.L., Cai W., Wang H., Zhou H.J., Xie Q. Correlation between circulating miR-122 and prognosis of chronic HBV-related liver failure. J. Dig. Dis. 2016;17:334–339. doi: 10.1111/1751-2980.12348. [DOI] [PubMed] [Google Scholar]
- 123.Tan Y., Lin B., Ye Y., Wen D., Chen L., Zhou X. Differential expression of serum microRNAs in cirrhosis that evolve into hepatocellular carcinoma related to hepatitis B virus. Oncol. Rep. 2015;33:2863–2870. doi: 10.3892/or.2015.3924. [DOI] [PubMed] [Google Scholar]
- 124.Li G., Li J., Li C., Qi H., Dong P., Zheng J., Yu F. MicroRNA-125a-5p contributes to hepatic stellate cell activation through targeting FIH1. Cell. Physiol. Biochem. 2016;38:1544–1552. doi: 10.1159/000443095. [DOI] [PubMed] [Google Scholar]
- 125.Zheng J., Zhou Z., Xu Z., Li G., Dong P., Chen Z., Lin D., Chen B., Yu F. Serum microRNA-125a-5p, a useful biomarker in liver diseases, correlates with disease progression. Mol. Med. Rep. 2015;12:1584–1590. doi: 10.3892/mmr.2015.3546. [DOI] [PubMed] [Google Scholar]
- 126.Sarkar N., Chakravarty R. Hepatitis B virus infection, microRNAs and liver disease. Int. J. Mol. Sci. 2015;16:17746–17762. doi: 10.3390/ijms160817746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bao S., Zheng J., Li N., Huang C., Chen M., Cheng Q., Yu K., Chen S., Zhu M., Shi G. Serum MicroRNA Levels as a Noninvasive Diagnostic Biomarker for the Early Diagnosis of Hepatitis B Virus-Related Liver Fibrosis. Gut Liver. 2017;11:860. doi: 10.5009/gnl16560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xin X., Zhang Y., Liu X., Xin H., Cao Y., Geng M. MicroRNA in hepatic fibrosis and cirrhosis. Front. Biosci. (Landmark Ed.) 2014;19:1418–1424. doi: 10.2741/4292. [DOI] [PubMed] [Google Scholar]
- 129.Liu Y., Wang M., Chen C., Shi Y., Tang C., Zhou Q., Zhang H., Hu Y., Su S., Chen Q. MiRNA-target network analysis identifies potential biomarkers for Traditional Chinese Medicine (TCM) syndrome development evaluation in hepatitis B caused liver cirrhosis. Sci. Rep. 2017;7:11054. doi: 10.1038/s41598-017-11351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Venugopal S.K., Jiang J., Kim T.-H., Li Y., Wang S.-S., Torok N.J., Wu J., Zern M.A. Liver fibrosis causes downregulation of miRNA-150 and miRNA-194 in hepatic stellate cells, and their overexpression causes decreased stellate cell activation. Am. J. Physiol. -Gastrointest. Liver Physiol. 2009;298:G101–G106. doi: 10.1152/ajpgi.00220.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lakner A.M., Steuerwald N.M., Walling T.L., Ghosh S., Li T., McKillop I.H., Russo M.W., Bonkovsky H.L., Schrum L.W. Inhibitory effects of microRNA 19b in hepatic stellate cell-mediated fibrogenesis. Hepatology. 2012;56:300–310. doi: 10.1002/hep.25613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao X., Zhao P., Hu J., Zhu H., Zhang J., Zhou Z., Zhao J., Tang F. Micro RNA-194 protects against chronic hepatitis B-related liver damage by promoting hepatocyte growth via ACVR 2B. J. Cell. Mol. Med. 2018;22:4534–4544. doi: 10.1111/jcmm.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jiang S., Jiang W., Xu Y., Wang X., Mu Y., Liu P. Serum miR-21 and miR-26a Levels Negatively Correlate with Severity of Cirrhosis in Patients with Chronic Hepatitis B. Microrna (Shariqahunited Arab Emir.) 2018 doi: 10.2174/2211536607666180821162850. [DOI] [PubMed] [Google Scholar]
- 134.Chen R., Wu J.C., Liu T., Qu Y., Lu L.G., Xu M.Y. Micro RNA profile analysis in the liver fibrotic tissues of chronic hepatitis B patients. J. Dig. Dis. 2017;18:115–124. doi: 10.1111/1751-2980.12452. [DOI] [PubMed] [Google Scholar]
- 135.Zhang Q., Xu M., Qu Y., Li Z., Zhang Q., Cai X., Lu L. Analysis of the differential expression of circulating microRNAs during the progression of hepatic fibrosis in patients with chronic hepatitis B virus infection. Mol. Med. Rep. 2015;12:5647–5654. doi: 10.3892/mmr.2015.4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Feng G., Li J., Yang Z., Zhang S., Liu Y., Zhang W., Ye L., Zhang X. Hepatitis B virus X protein promotes the development of liver fibrosis and hepatoma through downregulation of miR-30e targeting P4HA2 mRNA. Oncogene. 2017;36:6895. doi: 10.1038/onc.2017.291. [DOI] [PubMed] [Google Scholar]
- 137.Li Z., Zhang S., Huang C., Zhang W., Hu Y., Wei B. MicroRNAome of splenic macrophages in hypersplenism due to portal hypertension in hepatitis B virus-related cirrhosis. Exp. Biol. Med. 2008;233:1454–1461. doi: 10.3181/0711-RM-321. [DOI] [PubMed] [Google Scholar]
- 138.Tao L., Xue D., Shen D., Ma W., Zhang J., Wang X., Zhang W., Wu L., Pan K., Yang Y. MicroRNA-942 mediates hepatic stellate cell activation by regulating BAMBI expression in human liver fibrosis. Arch. Toxicol. 2018;92:2935–2946. doi: 10.1007/s00204-018-2278-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Croce C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Reviews. Genet. 2009;10:704. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lamontagne R.J., Bagga S., Bouchard M.J. Hepatitis B virus molecular biology and pathogenesis. Hepatoma Res. 2016;2:163. doi: 10.20517/2394-5079.2016.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kew M.C. Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. J. Gastroenterol. Hepatol. 2011;26:144–152. doi: 10.1111/j.1440-1746.2010.06546.x. [DOI] [PubMed] [Google Scholar]
- 142.Park I.Y., Sohn B.H., Yu E., Suh D.J., Chung Y.H., Lee J.H., Surzycki S.J., Lee Y.I. Aberrant epigenetic modifications in hepatocarcinogenesis induced by hepatitis B virus X protein. Gastroenterology. 2007;132:1476–1494. doi: 10.1053/j.gastro.2007.01.034. [DOI] [PubMed] [Google Scholar]
- 143.Wu Q., Liu H.-O., Liu Y.-D., Liu W.-S., Pan D., Zhang W.-J., Yang L., Fu Q., Xu J.-J., Gu J.-X. Decreased expression of hepatocyte nuclear factor 4α (Hnf4α)/microRNA-122 (miR-122) axis in hepatitis B virus-associated hepatocellular carcinoma enhances potential oncogenic GALNT10 protein activity. J. Biol. Chem. 2015;290:1170–1185. doi: 10.1074/jbc.M114.601203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Neuveut C., Wei Y., Buendia M.A. Mechanisms of HBV-related hepatocarcinogenesis. J. Hepatol. 2010;52:594–604. doi: 10.1016/j.jhep.2009.10.033. [DOI] [PubMed] [Google Scholar]
- 145.Wu G., Huang P., Ju X., Li Z., Wang Y. Lin28B over-expression mediates the repression of let-7 by hepatitis B virus X protein in hepatoma cells. Int. J. Clin. Exp. Med. 2015;8:15108. [PMC free article] [PubMed] [Google Scholar]
- 146.Wang Y., Lu Y., Toh S.T., Sung W.-K., Tan P., Chow P., Chung A.Y., Jooi L.L., Lee C.G. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J. Hepatol. 2010;53:57–66. doi: 10.1016/j.jhep.2009.12.043. [DOI] [PubMed] [Google Scholar]
- 147.Lu J.-W., Liao C.-Y., Yang W.-Y., Lin Y.-M., Jin S.-L.C., Wang H.-D., Yuh C.-H. Overexpression of endothelin 1 triggers hepatocarcinogenesis in zebrafish and promotes cell proliferation and migration through the AKT pathway. PLoS ONE. 2014;9:e85318. doi: 10.1371/journal.pone.0085318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang X., Zhang E., Ma Z., Pei R., Jiang M., Schlaak J.F., Roggendorf M., Lu M. Modulation of hepatitis B virus replication and hepatocyte differentiation by MicroRNA-1. Hepatology. 2011;53:1476–1485. doi: 10.1002/hep.24195. [DOI] [PubMed] [Google Scholar]
- 149.Datta J., Kutay H., Nasser M.W., Nuovo G.J., Wang B., Majumder S., Liu C.-G., Volinia S., Croce C.M., Schmittgen T.D. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 150.Wei X., Xiang T., Ren G., Tan C., Liu R., Xu X., Wu Z. miR-101 is down-regulated by the hepatitis B virus x protein and induces aberrant DNA methylation by targeting DNA methyltransferase 3A. Cell. Signal. 2013;25:439–446. doi: 10.1016/j.cellsig.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 151.Au S.L.K., Wong C.C.L., Lee J.M.F., Fan D.N.Y., Tsang F.H., Ng I.O.L., Wong C.M. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56:622–631. doi: 10.1002/hep.25679. [DOI] [PubMed] [Google Scholar]
- 152.Li S., Fu H., Wang Y., Tie Y., Xing R., Zhu J., Sun Z., Wei L., Zheng X. MicroRNA-101 regulates expression of the v-fos FBJ murine osteosarcoma viral oncogene homolog (FOS) oncogene in human hepatocellular carcinoma. Hepatology. 2009;49:1194–1202. doi: 10.1002/hep.22757. [DOI] [PubMed] [Google Scholar]
- 153.Fu Y., Wei X., Tang C., Li J., Liu R., Shen A., Wu Z. Circulating microRNA-101 as a potential biomarker for hepatitis B virus-related hepatocellular carcinoma. Oncol. Lett. 2013;6:1811–1815. doi: 10.3892/ol.2013.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sheng Y., Ding S., Chen K., Chen J., Wang S., Zou C., Zhang J., Cao Y., Huang A., Tang H. Functional analysis of miR-101-3p and Rap1b involved in hepatitis B virus-related hepatocellular carcinoma pathogenesis. Biochem. Cell Biol. 2014;92:152–162. doi: 10.1139/bcb-2013-0128. [DOI] [PubMed] [Google Scholar]
- 155.Shen S., Lin Y., Yuan X., Shen L., Chen J., Chen L., Qin L., Shen B. Biomarker MicroRNAs for diagnosis, prognosis and treatment of hepatocellular carcinoma: A functional survey and comparison. Sci. Rep. 2016;6:38311. doi: 10.1038/srep38311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Liang H.-W., Wang N., Wang Y., Wang F., Fu Z., Yan X., Zhu H., Diao W., Ding Y., Chen X. Hepatitis B virus-human chimeric transcript HBx-LINE1 promotes hepatic injury via sequestering cellular microRNA-122. J. Hepatol. 2016;64:278–291. doi: 10.1016/j.jhep.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 157.Song K., Han C., Zhang J., Lu D., Dash S., Feitelson M., Lim K., Wu T. Epigenetic regulation of MicroRNA-122 by peroxisome proliferator activated receptor-gamma and hepatitis b virus X protein in hepatocellular carcinoma cells. Hepatology. 2013;58:1681–1692. doi: 10.1002/hep.26514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yu G., Chen X., Chen S., Ye W., Hou K., Liang M. MiR-19a, miR-122 and miR-223 are differentially regulated by hepatitis B virus X protein and involve in cell proliferation in hepatoma cells. J. Transl. Med. 2016;14:122. doi: 10.1186/s12967-016-0888-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Peng F., Xiao X., Jiang Y., Luo K., Tian Y., Peng M., Zhang M., Xu Y., Gong G. HBx down-regulated Gld2 plays a critical role in HBV-related dysregulation of miR-122. PLoS ONE. 2014;9:e92998. doi: 10.1371/journal.pone.0092998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fan C.-G., Wang C.-M., Tian C., Wang Y., Li L., Sun W.-S., Li R.-F., Liu Y.-G. miR-122 inhibits viral replication and cell proliferation in hepatitis B virus-related hepatocellular carcinoma and targets NDRG3. Oncol. Rep. 2011;26:1281–1286. doi: 10.3892/or.2011.1375. [DOI] [PubMed] [Google Scholar]
- 161.Fornari F., Gramantieri L., Giovannini C., Veronese A., Ferracin M., Sabbioni S., Calin G.A., Grazi G.L., Croce C.M., Tavolari S. MiR-122/cyclin G1 interaction modulates p53 activity and affects doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2009;69:5761–5767. doi: 10.1158/0008-5472.CAN-08-4797. [DOI] [PubMed] [Google Scholar]
- 162.Gramantieri L., Ferracin M., Fornari F., Veronese A., Sabbioni S., Liu C.-G., Calin G.A., Giovannini C., Ferrazzi E., Grazi G.L. Cyclin G1 is a target of miR-122a, a microRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–6099. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 163.Coulouarn C., Factor V.M., Andersen J.B., Durkin M.E., Thorgeirsson S.S. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28:3526–3536. doi: 10.1038/onc.2009.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Lu Y., Yue X., Cui Y., Zhang J., Wang K. MicroRNA-124 suppresses growth of human hepatocellular carcinoma by targeting STAT3. Biochem. Biophys. Res. Commun. 2013;441:873–879. doi: 10.1016/j.bbrc.2013.10.157. [DOI] [PubMed] [Google Scholar]
- 165.Liu W., Hu J., Zhou K., Chen F., Wang Z., Liao B., Dai Z., Cao Y., Fan J., Zhou J. Serum exosomal miR-125b is a novel prognostic marker for hepatocellular carcinoma. Oncotargets Ther. 2017;10:3843. doi: 10.2147/OTT.S140062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wei X., Tan C., Tang C., Ren G., Xiang T., Qiu Z., Liu R., Wu Z. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell. Signal. 2013;25:1037–1043. doi: 10.1016/j.cellsig.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 167.Zhao J., Wang W., Huang Y., Wu J., Chen M., Cui P., Zhang W., Zhang Y. HBx elevates oncoprotein AEG-1 expression to promote cell migration by downregulating miR-375 and miR-136 in malignant hepatocytes. DNA Cell Biol. 2014;33:715–722. doi: 10.1089/dna.2014.2376. [DOI] [PubMed] [Google Scholar]
- 168.Wang W., Zhao L.J., Tan Y.-X., Ren H., Qi Z.-T. Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J. Gastroenterol. WJG. 2012;18:5442. doi: 10.3748/wjg.v18.i38.5442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wong C.C.L., Wong C.M., Tung E.K.K., Au S.L.K., Lee J.M.F., Poon R.T.P., Man K., Ng I.O.L. The microRNA miR-139 suppresses metastasis and progression of hepatocellular carcinoma by down-regulating Rho-kinase 2. Gastroenterology. 2011;140:322–331. doi: 10.1053/j.gastro.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 170.Qiu G., Lin Y., Zhang H., Wu D. miR-139-5p inhibits epithelial–mesenchymal transition, migration and invasion of hepatocellular carcinoma cells by targeting ZEB1 and ZEB2. Biochem. Biophys. Res. Commun. 2015;463:315–321. doi: 10.1016/j.bbrc.2015.05.062. [DOI] [PubMed] [Google Scholar]
- 171.Gao F., Sun X., Wang L., Tang S., Yan C. Downregulation of microRNA-145 caused by hepatitis B virus X protein promotes expression of CUL5 and contributes to pathogenesis of hepatitis B virus-associated hepatocellular carcinoma. Cell. Physiol. Biochem. 2015;37:1547–1559. doi: 10.1159/000438522. [DOI] [PubMed] [Google Scholar]
- 172.Yang X.W., Zhang L.J., Huang X.H., Chen L.Z., Su Q., Zeng W.T., Li W., Wang Q. mi R-145 suppresses cell invasion in hepatocellular carcinoma cells: Mi R-145 targets ADAM 17. Hepatol. Res. 2014;44:551–559. doi: 10.1111/hepr.12152. [DOI] [PubMed] [Google Scholar]
- 173.Xu X., Fan Z., Kang L., Han J., Jiang C., Zheng X., Zhu Z., Jiao H., Lin J., Jiang K. Hepatitis B virus X protein represses miRNA-148a to enhance tumorigenesis. J. Clin. Investig. 2013;123 doi: 10.1172/JCI64265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhang J., Zeng C., Xu L., Gong J., Fang J., Zhuang S. MicroRNA-148a suppresses the epithelial-mesenchymal transition and metastasis of hepatoma cells by targeting Met/Snail signaling. Oncogene. 2014;33:4069. doi: 10.1038/onc.2013.369. [DOI] [PubMed] [Google Scholar]
- 175.Jung K.H., Zhang J., Zhou C., Shen H., Gagea M., Rodriguez-Aguayo C., Lopez-Berestein G., Sood A.K., Beretta L. Differentiation therapy for hepatocellular carcinoma: Multifaceted effects of miR-148a on tumor growth and phenotype and liver fibrosis. Hepatology. 2016;63:864–879. doi: 10.1002/hep.28367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Huang J., Wang Y., Guo Y., Sun S. Down-regulated microRNA-152 induces aberrant DNA methylation in hepatitis B virus–related hepatocellular carcinoma by targeting DNA methyltransferase 1. Hepatology. 2010;52:60–70. doi: 10.1002/hep.23660. [DOI] [PubMed] [Google Scholar]
- 177.Huang H., Hu M., Li P., Lu C., Li M. Mir-152 inhibits cell proliferation and colony formation of CD133+ liver cancer stem cells by targeting KIT. Tumor Biol. 2015;36:921–928. doi: 10.1007/s13277-014-2719-x. [DOI] [PubMed] [Google Scholar]
- 178.Chen G., Liu M., Jiang Z., Yu M., Wei S. MicroRNAs Play Significant Roles in Pathogenesis of HBV-Related Diseases. J. Biomed. Sci. Eng. 2016;9:78. doi: 10.4236/jbise.2016.910B011. [DOI] [Google Scholar]
- 179.Zhao Z., Hu Y., Shen X., Lao Y., Zhang L., Qiu X., Hu J., Gong P., Cui H., Lu S. HBx represses RIZ1 expression by DNA methyltransferase 1 involvement in decreased miR-152 in hepatocellular carcinoma. Oncol. Rep. 2017;37:2811–2818. doi: 10.3892/or.2017.5518. [DOI] [PubMed] [Google Scholar]
- 180.Wu G., Yu F., Xiao Z., Xu K., Xu J., Tang W., Wang J., Song E. Hepatitis B virus X protein downregulates expression of the miR-16 family in malignant hepatocytes in vitro. Br. J. Cancer. 2011;105:146. doi: 10.1038/bjc.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu N., Zhang J., Jiao T., Li Z., Peng J., Cui Z., Ye X. Hepatitis B virus inhibits apoptosis of hepatoma cells by sponging the MicroRNA 15a/16 cluster. J. Virol. 2013;87:13370–13378. doi: 10.1128/JVI.02130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Wu C.S., Yen C.J., Chou R.H., Chen J.N., Huang W.C., Wu C.Y., Yu Y.L. Downregulation of microRNA-15b by hepatitis B virus X enhances hepatocellular carcinoma proliferation via fucosyltransferase 2-induced Globo H expression. Int. J. Cancer. 2014;134:1638–1647. doi: 10.1002/ijc.28501. [DOI] [PubMed] [Google Scholar]
- 183.Dai X., Zhang W., Zhang H., Sun S., Yu H., Guo Y., Kou Z., Zhao G., Du L., Jiang S. Modulation of HBV replication by microRNA-15b through targeting hepatocyte nuclear factor 1α. Nucleic Acids Res. 2014;42:6578–6590. doi: 10.1093/nar/gku260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Liu W.H., Yeh S.H., Lu C.C., Yu S.L., Chen H.Y., Lin C.Y., Chen D.S., Chen P.J. MicroRNA-18a prevents estrogen receptor-α expression, promoting proliferation of hepatocellular carcinoma cells. Gastroenterology. 2009;136:683–693. doi: 10.1053/j.gastro.2008.10.029. [DOI] [PubMed] [Google Scholar]
- 185.Liu X., Zhang Y., Wang P., Wang H., Su H., Zhou X., Zhang L. HBX protein-induced downregulation of microRNA-18a is responsible for upregulation of connective tissue growth factor in HBV infection-associated hepatocarcinoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2016;22:2492. doi: 10.12659/MSM.895943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Georges S.A., Biery M.C., Kim S.-y., Schelter J.M., Guo J., Chang A.N., Jackson A.L., Carleton M.O., Linsley P.S., Cleary M.A. Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res. 2008;68:10105–10112. doi: 10.1158/0008-5472.CAN-08-1846. [DOI] [PubMed] [Google Scholar]
- 187.Lian J., Jing Y., Dong Q., Huan L., Chen D., Bao C., Wang Q., Zhao F., Li J., Yao M. miR-192, a prognostic indicator, targets the SLC39A6/SNAIL pathway to reduce tumor metastasis in human hepatocellular carcinoma. Oncotarget. 2016;7:2672. doi: 10.18632/oncotarget.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Yip W.K. Ph.D. Thesis. Chinese University of Hong Kong; Hong Kong, China: 2009. MicroRNA Profiling of Human Hepatocytes Induced by HBx in Hepatocarcinogenesis. [Google Scholar]
- 189.Xu C., Liu S., Fu H., Li S., Tie Y., Zhu J., Xing R., Jin Y., Sun Z., Zheng X. MicroRNA-193b regulates proliferation, migration and invasion in human hepatocellular carcinoma cells. Eur. J. Cancer. 2010;46:2828–2836. doi: 10.1016/j.ejca.2010.06.127. [DOI] [PubMed] [Google Scholar]
- 190.Hill L., Browne G., Tulchinsky E. ZEB/miR-200 feedback loop: At the crossroads of signal transduction in cancer. Int. J. Cancer. 2013;132:745–754. doi: 10.1002/ijc.27708. [DOI] [PubMed] [Google Scholar]
- 191.Cui M., Wang Y., Sun B., Xiao Z., Ye L., Zhang X. MiR-205 modulates abnormal lipid metabolism of hepatoma cells via targeting acyl-CoA synthetase long-chain family member 1 (ACSL1) mRNA. Biochem. Biophys. Res. Commun. 2014;444:270–275. doi: 10.1016/j.bbrc.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 192.Ge W., Yu D.-C., Li Q.-G., Chen X., Zhang C.-Y., Ding Y.-T. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clin. Lab. 2014;60:427–434. doi: 10.7754/Clin.Lab.2013.130133. [DOI] [PubMed] [Google Scholar]
- 193.Liu F., Zhou S., Deng Y., Zhang Z., Zhang E., Wu Z., Huang Z., Chen X. MiR-216b is involved in pathogenesis and progression of hepatocellular carcinoma through HBx-miR-216b-IGF2BP2 signaling pathway. Cell Death Dis. 2016;6:e1670. doi: 10.1038/cddis.2015.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Cao M., Seike M., Soeno C., Mizutani H., Kitamura K., Minegishi Y., Noro R., Yoshimura A., Cai L., Gemma A. MiR-23a regulates TGF-β-induced epithelial-mesenchymal transition by targeting E-cadherin in lung cancer cells. Int. J. Oncol. 2012;41:869–875. doi: 10.3892/ijo.2012.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Li X., Liu X., Xu W., Zhou P., Gao P., Jiang S., Lobie P.E., Zhu T. c-MYC-regulated miR-23a/24-2/27a cluster promotes mammary carcinoma cell invasion and hepatic metastasis by targeting Sprouty2. J. Biol. Chem. 2013;288:18121–18133. doi: 10.1074/jbc.M113.478560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Huang S., He X., Ding J., Liang L., Zhao Y., Zhang Z., Yao X., Pan Z., Zhang P., Li J. Upregulation of miR-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int. J. Cancer. 2008;123:972–978. doi: 10.1002/ijc.23580. [DOI] [PubMed] [Google Scholar]
- 197.Chen L., Zheng J., Zhang Y., Yang L., Wang J., Ni J., Cui D., Yu C., Cai Z. Tumor-specific expression of microRNA-26a suppresses human hepatocellular carcinoma growth via cyclin-dependent and-independent pathways. Mol. Ther. 2011;19:1521–1528. doi: 10.1038/mt.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yang X., Liang L., Zhang X.F., Jia H.L., Qin Y., Zhu X.C., Gao X.M., Qiao P., Zheng Y., Sheng Y.Y. MicroRNA-26a suppresses tumor growth and metastasis of human hepatocellular carcinoma by targeting interleukin-6-Stat3 pathway. Hepatology. 2013;58:158–170. doi: 10.1002/hep.26305. [DOI] [PubMed] [Google Scholar]
- 199.Ji J., Shi J., Budhu A., Yu Z., Forgues M., Roessler S., Ambs S., Chen Y., Meltzer P.S., Croce C.M. MicroRNA expression, survival, and response to interferon in liver cancer. New Engl. J. Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Xiong Y., Fang J.H., Yun J.P., Yang J., Zhang Y., Jia W.H., Zhuang S.M. Effects of MicroRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology. 2010;51:836–845. doi: 10.1002/hep.23380. [DOI] [PubMed] [Google Scholar]
- 201.Wang C.-M., Wang Y., Fan C.-G., Xu F.-F., Sun W.-S., Liu Y.-G., Jia J.-H. miR-29c targets TNFAIP3, inhibits cell proliferation and induces apoptosis in hepatitis B virus-related hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2011;411:586–592. doi: 10.1016/j.bbrc.2011.06.191. [DOI] [PubMed] [Google Scholar]
- 202.Fu X., Tan D., Hou Z., Hu Z., Liu G. miR-338-3p is down-regulated by hepatitis B virus X and inhibits cell proliferation by targeting the 3′-UTR region of cyclinD1. Int. J. Mol. Sci. 2012;13:8514–8539. doi: 10.3390/ijms13078514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Huang X.H., Wang Q., Chen J.S., Fu X.H., Chen X.L., Chen L.Z., Li W., Bi J., Zhang L.J., Fu Q. Bead-based microarray analysis of microRNA expression in hepatocellular carcinoma: miR-338 is downregulated. Hepatol. Res. 2009;39:786–794. doi: 10.1111/j.1872-034X.2009.00502.x. [DOI] [PubMed] [Google Scholar]
- 204.Ou Q., Wang G., Li B., Li W.-F. Decreased miR-34a promotes growth by regulating MAP4K4 in hepatitis B virus related hepatocellular carcinoma. Int. J. Clin. Exp. Med. 2017;10:2523–2531. [Google Scholar]
- 205.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 206.Corney D.C., Flesken-Nikitin A., Godwin A.K., Wang W., Nikitin A.Y. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8438. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 207.Sun F., Fu H., Liu Q., Tie Y., Zhu J., Xing R., Sun Z., Zheng X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582:1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 208.Ying J., Yu X., Ma C., Zhang Y., Dong J. MicroRNA-363-3p is downregulated in hepatocellular carcinoma and inhibits tumorigenesis by directly targeting specificity protein 1. Mol. Med. Rep. 2017;16:1603–1611. doi: 10.3892/mmr.2017.6759. [DOI] [PubMed] [Google Scholar]
- 209.Gao H., Liu C. miR-429 represses cell proliferation and induces apoptosis in HBV-related HCC. Biomed. Pharmacother. 2014;68:943–949. doi: 10.1016/j.biopha.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 210.You X., Liu F., Zhang T., Li Y., Ye L., Zhang X. Hepatitis B virus X protein upregulates oncogene Rab18 to result in the dysregulation of lipogenesis and proliferation of hepatoma cells. Carcinogenesis. 2013;34:1644–1652. doi: 10.1093/carcin/bgt089. [DOI] [PubMed] [Google Scholar]
- 211.Zhang W., Lu Z., Kong G., Gao Y., Wang T., Wang Q., Cai N., Wang H., Liu F., Ye L. Hepatitis B virus X protein accelerates hepatocarcinogenesis with partner survivin through modulating miR-520b and HBXIP. Mol. Cancer. 2014;13:128. doi: 10.1186/1476-4598-13-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Hu X.M., Yan X.H., Hu Y.W., Huang J.L., Cao S.W., Ren T.Y., Tang Y.T., Lin L., Zheng L., Wang Q. miRNA-548p suppresses hepatitis B virus X protein associated hepatocellular carcinoma by downregulating oncoprotein hepatitis B x-interacting protein. Hepatol. Res. 2016;46:804–815. doi: 10.1111/hepr.12618. [DOI] [PubMed] [Google Scholar]
- 213.Bui-Nguyen T.M., Pakala S.B., Sirigiri D.R., Martin E., Murad F., Kumar R. Stimulation of inducible nitric oxide by hepatitis B virus transactivator protein HBx requires MTA1 coregulator. J. Biol. Chem. 2016;291:1198. doi: 10.1074/jbc.A109.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Chen W.-S., Yen C.-J., Chen Y.-J., Chen J.-Y., Wang L.-Y., Chiu S.-J., Shih W.-L., Ho C.-Y., Wei T.-T., Pan H.-L. miRNA-7/21/107 contribute to HBx-induced hepatocellular carcinoma progression through suppression of maspin. Oncotarget. 2015;6:25962. doi: 10.18632/oncotarget.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhang J.-J., Wang C.-Y., Hua L., Yao K.-H., Chen J.-T., Hu J.-H. miR-107 promotes hepatocellular carcinoma cell proliferation by targeting Axin2. Int. J. Clin. Exp. Pathol. 2015;8:5168. [PMC free article] [PubMed] [Google Scholar]
- 216.Mosca N., Castiello F., Coppola N., Trotta M.C., Sagnelli C., Pisaturo M., Sagnelli E., Russo A., Potenza N. Functional interplay between hepatitis B virus X protein and human miR-125a in HBV infection. Biochem. Biophys. Res. Commun. 2014;449:141–145. doi: 10.1016/j.bbrc.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 217.Zhang X., Liu S., Hu T., Liu S., He Y., Sun S. Up-regulated microRNA-143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology. 2009;50:490–499. doi: 10.1002/hep.23008. [DOI] [PubMed] [Google Scholar]
- 218.Li J.-F., Dai X.-P., Zhang W., Sun S.-H., Zeng Y., Zhao G.-Y., Kou Z.-H., Guo Y., Yu H., Du L.-Y. Upregulation of microRNA-146a by hepatitis B virus X protein contributes to hepatitis development by downregulating complement factor H. MBio. 2015;6:e02459-14. doi: 10.1128/mBio.02459-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hou Z.H., Han Q.J., Zhang C., Tian Z.G., Zhang J. miR146a impairs the IFN-induced anti-HBV immune response by downregulating STAT1 in hepatocytes. Liver Int. 2014;34:58–68. doi: 10.1111/liv.12244. [DOI] [PubMed] [Google Scholar]
- 220.Xie Q., Chen X., Lu F., Zhang T., Hao M., Wang Y., Zhao J., McCrae M.A., Zhuang H. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer. 2012;118:2431–2442. doi: 10.1002/cncr.26566. [DOI] [PubMed] [Google Scholar]
- 221.Song X., Tan S., Wu Z., Xu L., Wang Z., Xu Y., Wang T., Gao C., Gong Y., Liang X. HBV suppresses ZHX2 expression to promote proliferation of HCC through miR-155 activation. Int. J. Cancer. 2018;143:3120–3130. doi: 10.1002/ijc.31595. [DOI] [PubMed] [Google Scholar]
- 222.Fu X., Wen H., Jing L., Yang Y., Wang W., Liang X., Nan K., Yao Y., Tian T. Micro RNA-155-5p promotes hepatocellular carcinoma progression by suppressing PTEN through the PI 3K/Akt pathway. Cancer Sci. 2017;108:620–631. doi: 10.1111/cas.13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Aguda B.D., Kim Y., Piper-Hunter M.G., Friedman A., Marsh C.B. MicroRNA regulation of a cancer network: Consequences of the feedback loops involving miR-17-92, E2F, and Myc. Proc. Natl. Acad. Sci. USA. 2008;105:19678–19683. doi: 10.1073/pnas.0811166106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Connolly E., Melegari M., Landgraf P., Tchaikovskaya T., Tennant B.C., Slagle B.L., Rogler L.E., Zavolan M., Tuschl T., Rogler C.E. Elevated expression of the miR-17–92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am. J. Pathol. 2008;173:856–864. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Damania P., Sen B., Dar S.B., Kumar S., Kumari A., Gupta E., Sarin S.K., Venugopal S.K. Hepatitis B virus induces cell proliferation via HBx-induced microRNA-21 in hepatocellular carcinoma by targeting programmed cell death protein4 (PDCD4) and phosphatase and tensin homologue (PTEN) PLoS ONE. 2014;9:e91745. doi: 10.1371/journal.pone.0091745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Liu C., Yu J., Yu S., Lavker R.M., Cai L., Liu W., Yang K., He X., Chen S. MicroRNA-21 acts as an oncomir through multiple targets in human hepatocellular carcinoma. J. Hepatol. 2010;53:98–107. doi: 10.1016/j.jhep.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 227.Liu F., You X., Chi X., Wang T., Ye L., Niu J., Zhang X. Hepatitis B virus X protein mutant HBxΔ127 promotes proliferation of hepatoma cells through up-regulating miR-215 targeting PTPRT. Biochem. Biophys. Res. Commun. 2014;444:128–134. doi: 10.1016/j.bbrc.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 228.Chen J.-J., Tang Y.-S., Huang S.-F., Ai J.-G., Wang H.-X., Zhang L.-P. HBx protein-induced upregulation of microRNA-221 promotes aberrant proliferation in HBV-related hepatocellular carcinoma by targeting estrogen receptor-α. Oncol. Rep. 2015;33:792–798. doi: 10.3892/or.2014.3647. [DOI] [PubMed] [Google Scholar]
- 229.Rong M., Chen G., Dang Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer. 2013;13:21. doi: 10.1186/1471-2407-13-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yang Y.-F., Wang F., Xiao J.-J., Song Y., Zhao Y.-Y., Cao Y., Bei Y.-H., Yang C.-Q. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HepG2 cells by downregulating p27. Int. J. Clin. Exp. Med. 2014;7:893. [PMC free article] [PubMed] [Google Scholar]
- 231.Wong Q.W., Ching A.K., Chan A.W., Choy K.-W., To K.-F., Lai P.B., Wong N. MiR-222 overexpression confers cell migratory advantages in hepatocellular carcinoma through enhancing AKT signaling. Clin. Cancer Res. 2010;16:867–875. doi: 10.1158/1078-0432.CCR-09-1840. [DOI] [PubMed] [Google Scholar]
- 232.Gramantieri L., Fornari F., Callegari E., Sabbioni S., Lanza G., Croce C.M., Bolondi L., Negrini M. MicroRNA involvement in hepatocellular carcinoma. J. Cell. Mol. Med. 2008;12:2189–2204. doi: 10.1111/j.1582-4934.2008.00533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Wang Y., Lee A.T., Ma J.Z., Wang J., Ren J., Yang Y., Tantoso E., Li K.-B., Ooi L.L.J., Tan P. Profiling microRNA expression in hepatocellular carcinoma reveals microRNA-224 up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific target. J. Biol. Chem. 2008;283:13205–13215. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 234.Lan S.H., Wu S.Y., Zuchini R., Lin X.Z., Su I.J., Tsai T.F., Lin Y.J., Wu C.T., Liu H.S. Autophagy suppresses tumorigenesis of hepatitis B virus-associated hepatocellular carcinoma through degradation of microRNA-224. Hepatology. 2014;59:505–517. doi: 10.1002/hep.26659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Scisciani C., Vossio S., Guerrieri F., Schinzari V., De Iaco R., de Meo P.D.O., Cervello M., Montalto G., Pollicino T., Raimondo G. Transcriptional regulation of miR-224 upregulated in human HCCs by NFκB inflammatory pathways. J. Hepatol. 2012;56:855–861. doi: 10.1016/j.jhep.2011.11.017. [DOI] [PubMed] [Google Scholar]
- 236.Wu X.-J., Li Y., Liu D., Zhao L.-D., Bai B., Xue M.-H. miR-27a as an oncogenic microRNA of hepatitis B virus-related hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2013;14:885–889. doi: 10.7314/APJCP.2013.14.2.885. [DOI] [PubMed] [Google Scholar]
- 237.Kong G., Zhang J., Zhang S., Shan C., Ye L., Zhang X. Upregulated microRNA-29a by hepatitis B virus X protein enhances hepatoma cell migration by targeting PTEN in cell culture model. PLoS ONE. 2011;6:e19518. doi: 10.1371/journal.pone.0019518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Gong J., Liu R., Zhuang R., Zhang Y., Fang L., Xu Z., Jin L., Wang T., Song C., Yang K. miR-30c-1* promotes natural killer cell cytotoxicity against human hepatoma cells by targeting the transcription factor HMBOX 1. Cancer Sci. 2012;103:645–652. doi: 10.1111/j.1349-7006.2012.02207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Cao Y., Chen J., Wang D., Peng H., Tan X., Xiong D., Huang A., Tang H. Upregulated in Hepatitis B virus-associated hepatocellular carcinoma cells, miR-331-3p promotes proliferation of hepatocellular carcinoma cells by targeting ING5. Oncotarget. 2015;6:38093. doi: 10.18632/oncotarget.5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhao Q., Li T., Qi J., Liu J., Qin C. The miR-545/374a cluster encoded in the Ftx lncRNA is overexpressed in HBV-related hepatocellular carcinoma and promotes tumorigenesis and tumor progression. PLoS ONE. 2014;9:e109782. doi: 10.1371/journal.pone.0109782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zhang B., Han S., Feng B., Chu X., Chen L., Wang R. Hepatitis B virus X protein-mediated non-coding RNA aberrations in the development of human hepatocellular carcinoma. Exp. Mol. Med. 2017;49:e293. doi: 10.1038/emm.2016.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Chen Y.-J., Chien P.-H., Chen W.-S., Chien Y.-F., Hsu Y.-Y., Wang L.-Y., Chen J.-Y., Lin C.-W., Huang T.-C., Yu Y.-L. Hepatitis B virus-encoded X protein downregulates EGFR expression via inducing microRNA-7 in hepatocellular carcinoma cells. Evid. -Based Complement. Altern. Med. 2013;2013 doi: 10.1155/2013/682380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Zhang X., Hu S., Zhang X., Wang L., Zhang X., Yan B., Zhao J., Yang A., Zhang R. MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2014;443:1078–1084. doi: 10.1016/j.bbrc.2013.12.095. [DOI] [PubMed] [Google Scholar]
- 244.Waisberg J., Saba G.T. Wnt-/-β-catenin pathway signaling in human hepatocellular carcinoma. World J. Hepatol. 2015;7:2631. doi: 10.4254/wjh.v7.i26.2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Edamoto Y., Hara A., Biernat W., Terracciano L., Cathomas G., Riehle H.M., Matsuda M., Fujii H., Scoazec J.Y., Ohgaki H. Alterations of RB1, p53 and Wnt pathways in hepatocellular carcinomas associated with hepatitis C, hepatitis B and alcoholic liver cirrhosis. Int. J. Cancer. 2003;106:334–341. doi: 10.1002/ijc.11254. [DOI] [PubMed] [Google Scholar]
- 246.Whittaker S., Marais R., Zhu A. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene. 2010;29:4989. doi: 10.1038/onc.2010.236. [DOI] [PubMed] [Google Scholar]
- 247.El–Serag H.B., Rudolph K.L. Hepatocellular carcinoma: Epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 248.Murakami Y., Kawada N. MicroRNAs in hepatic pathophysiology. Hepatol. Res. 2017;47:60–69. doi: 10.1111/hepr.12730. [DOI] [PubMed] [Google Scholar]
- 249.Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F.J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 250.Wang Y., Wang Q., Song J. Inhibition of autophagy potentiates the proliferation inhibition activity of microRNA-7 in human hepatocellular carcinoma cells. Oncol. Lett. 2017;14:3566–3572. doi: 10.3892/ol.2017.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Vilchez V., Turcios L., Marti F., Gedaly R. Targeting Wnt/β-catenin pathway in hepatocellular carcinoma treatment. World J. Gastroenterol. 2016;22:823. doi: 10.3748/wjg.v22.i2.823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Callegari E., Gramantieri L., Domenicali M., D’abundo L., Sabbioni S., Negrini M. MicroRNAs in liver cancer: A model for investigating pathogenesis and novel therapeutic approaches. Cell Death Differ. 2015;22:46. doi: 10.1038/cdd.2014.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Meng X., Franklin D.A., Dong J., Zhang Y. MDM2–p53 pathway in hepatocellular carcinoma. Cancer Res. 2014;74:7161–7167. doi: 10.1158/0008-5472.CAN-14-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Peng C.-Y., Chen T.-C., Hung S.-P., Chen M.-F., Yeh C.-T., Tsai S.-L., Chu C.-M., Liaw Y.-F. Genetic alterations of INK4alpha/ARF locus and p53 in human hepatocellular carcinoma. Anticancer Res. 2002;22:1265–1271. [PubMed] [Google Scholar]
- 255.Shimizu S., Takehara T., Hikita H., Kodama T., Miyagi T., Hosui A., Tatsumi T., Ishida H., Noda T., Nagano H. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J. Hepatol. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 256.Lin C.J.-F., Gong H.-Y., Tseng H.-C., Wang W.-L., Wu J.-L. miR-122 targets an anti-apoptotic gene, Bcl-w, in human hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 2008;375:315–320. doi: 10.1016/j.bbrc.2008.07.154. [DOI] [PubMed] [Google Scholar]
- 257.Yang F., Li Q.-J., Gong Z.-B., Zhou L., You N., Wang S., Li X.-L., Li J.-J., An J.-Z., Wang D.-S. MicroRNA-34a targets Bcl-2 and sensitizes human hepatocellular carcinoma cells to sorafenib treatment. Technol. Cancer Res. Treat. 2014;13:77–86. doi: 10.7785/tcrt.2012.500364. [DOI] [PubMed] [Google Scholar]
- 258.Zhao A., Zeng Q., Xie X., Zhou J., Yue W., Li Y., Pei X. MicroRNA-125b induces cancer cell apoptosis through suppression of Bcl-2 expression. J. Genet. Genom. 2012;39:29–35. doi: 10.1016/j.jgg.2011.12.003. [DOI] [PubMed] [Google Scholar]
- 259.He H., Tian W., Chen H., Deng Y. MicroRNA-101 sensitizes hepatocellular carcinoma cells to doxorubicin-induced apoptosis via targeting Mcl-1. Mol. Med. Rep. 2016;13:1923–1929. doi: 10.3892/mmr.2015.4727. [DOI] [PubMed] [Google Scholar]
- 260.Yoshikawa H., Matsubara K., Geng-Sun Q., Jackson P., Groopman J.D., Manning J.E., Harris C.C., Herman J.G. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat. Genet. 2001;28:29. doi: 10.1038/ng0501-29. [DOI] [PubMed] [Google Scholar]
- 261.Chen Z., Ma T., Huang C., Hu T., Li J. The Pivotal Role of microRNA-155 in the Control of Cancer. J. Cell. Physiol. 2014;229:545–550. doi: 10.1002/jcp.24492. [DOI] [PubMed] [Google Scholar]
- 262.Gui Y., Yeganeh M., Donates Y., Tobelaim W., Chababi W., Mayhue M., Yoshimura A., Ramanathan S., Saucier C., Ilangumaran S. Regulation of MET receptor tyrosine kinase signaling by suppressor of cytokine signaling 1 in hepatocellular carcinoma. Oncogene. 2015;34:5718. doi: 10.1038/onc.2015.20. [DOI] [PubMed] [Google Scholar]
- 263.Huang S., Zhou D., Li Y.X., Ming Z.Y., Li K.Z., Wu G.B., Chen C., Zhao Y.N. In vivo and in vitro effects of microRNA-221 on hepatocellular carcinoma development and progression through the JAK–STAT3 signaling pathway by targeting SOCS3. J. Cell. Physiol. 2019;234:3500–3514. doi: 10.1002/jcp.26863. [DOI] [PubMed] [Google Scholar]
- 264.Yu L.-X., Ling Y., Wang H.-Y. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. NPJ Precis. Oncol. 2018;2:6. doi: 10.1038/s41698-018-0048-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Pritchard C.C., Kroh E., Wood B., Arroyo J.D., Dougherty K.J., Miyaji M.M., Tait J.F., Tewari M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. 2012;5:492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Nagy Z.B., Barták B.K., Kalmár A., Galamb O., Wichmann B., Dank M., Igaz P., Tulassay Z., Molnár B. Comparison of circulating miRNAs expression alterations in matched tissue and plasma samples during colorectal cancer progression. Pathol. Oncol. Res. 2019;25:97–105. doi: 10.1007/s12253-017-0308-1. [DOI] [PubMed] [Google Scholar]
- 267.Ma R., Jiang T., Kang X. Circulating microRNAs in cancer: Origin, function and application. J. Exp. Clin. Cancer Res. 2012;31:38. doi: 10.1186/1756-9966-31-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.He B., Peng F., Li W., Jiang Y. Interaction of lncRNA-MALAT1 and miR-124 regulates HBx-induced cancer stem cell properties in HepG2 through PI3K/Akt signaling. J. Cell. Biochem. 2019;120:2908–2918. doi: 10.1002/jcb.26823. [DOI] [PubMed] [Google Scholar]
- 269.Lang Q., Ling C. MiR-124 suppresses cell proliferation in hepatocellular carcinoma by targeting PIK3CA. Biochem. Biophys. Res. Commun. 2012;426:247–252. doi: 10.1016/j.bbrc.2012.08.075. [DOI] [PubMed] [Google Scholar]
- 270.Yang Z.P., Ma H.S., Wang S.S., Wang L., Liu T. LAMC1 mRNA promotes malignancy of hepatocellular carcinoma cells by competing for MicroRNA-124 binding with CD151. Iubmb Life. 2017;69:595–605. doi: 10.1002/iub.1642. [DOI] [PubMed] [Google Scholar]
- 271.Maltseva D.V., Rodin S.A. Laminins in metastatic cancer. Mol. Biol. 2018;52:350–371. doi: 10.1134/S0026893318030093. [DOI] [PubMed] [Google Scholar]