Imaging proteins inside cells with fluorescent tags (original) (raw)
. Author manuscript; available in PMC: 2013 Jan 1.
Abstract
Watching biological molecules provides clues to their function and regulation. Some of the most powerful methods of labeling proteins for imaging use genetically encoded fluorescent fusion tags. There are four standard genetic methods of covalently tagging a protein with a fluorescent probe for cellular imaging. These use I) auto-fluorescent proteins, II) self-labeling enzymes, III) enzymes that catalyze the attachment of a probe to a target sequence, and IV) biarsenical dyes that target tetracysteine motifs. Each of these techniques has advantages and disadvantages. In this review, we cover new developments in these methods and discuss practical considerations for their use in imaging proteins inside living cells.
Protein labeling methods
What do biological molecules do? Where are they located? How do they behave and how are they regulated? These are central questions for modern cellular and molecular biology. The parallel development of new imaging probes and advanced imaging technologies useful for tracking and measuring the properties of biological molecules has opened a new era of biological discovery. By watching the behavior of molecules at nanometer resolution in living cells in real time, the underlying complexities and regulatory mechanisms of biological systems are being unraveled. The standard tools to label molecules in biological systems are based almost exclusively on the genetic fusion of fluorescent tags. These tags fused in frame to proteins of interest have ushered in the modern era of intracellular molecular imaging.
There are four standard ways to covalently label a protein inside a cell for fluorescent imaging. The most common and conventional method is the use of intrinsically fluorescent proteins (FPs) related in structure or sequence to green fluorescent protein (GFP) [1]. These 25 kD proteins were first isolated from the small 10 cm jellyfish Aequorea Victoria and have been engineered to spontaneously form dozens of relatively bright, stable, chromatically diverse fluorophores that can be imaged with conventional fluorescence microscopy [1]. Second, a series of self-labeling enzymes have been developed that can covalently attach a fluorescent ligand to one of its own amino acid residues. These enzymes are similar in size to FPs and are commonly called Halo tags and SNAP/CLIP tags [2–4]. If these enzyme domains are fused in frame to a protein the pair can be labeled by introducing a cell-permeable fluorescent ligand, which covalently reacts with the fusion tag. Third, an enzyme can be used to covalently attach a fluorophore-ligand to another protein or peptide [5]. Fourth, small cell-permeable biarsenical dyes FLAsH and ReAsH have been developed that can covalently react with a short peptide sequence [6, 7]. The amino acid sequence that specifically recognizes the bisarsenic is small, only 4 amino acids in length. However, optimized sequences of 12 amino acids have been shown to increase the specificity of labeling [8].
This review evaluates recent developments related to these four covalent labeling methods. We focus on fluorescent tags that can be used to image proteins inside cells and review practical considerations for their application in biological systems. We discuss the advantages and disadvantages of each method, their photo-physical characteristics and chemical structures, and practical considerations for determining which probe will provide the optimal labeling scheme. For example, we discuss issues such as probe size, toxicity, cellular and molecular behavior, linkers, background fluorescence, and brightness.
Fluorescent proteins
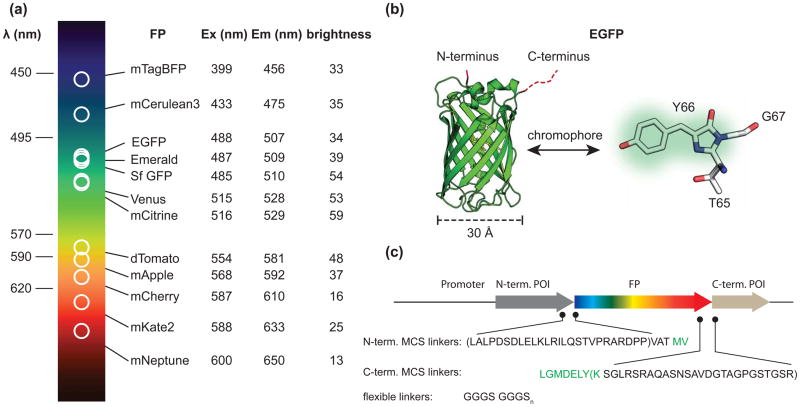
Tracking cellular proteins in vivo with fluorescent tags was made routine by the development of GFP and its family members (Figure 1). Creating a genetic in-frame fusion of an FP to a protein of interest allows that protein to be localized in time and space to specific tissues, cells, or sub-cellular compartments. The development and refinement of these tags for biological applications has intensified over the last decade, providing researchers with numerous choices in FP excitation and emission spectra, brightness, pH sensitivity, and photo physical properties [1]. Indeed, a palette of FP proteins is now available to genetically “paint’ a protein in vivo for light microscopy. These tags can—in large part—be minimally disruptive to most proteins when attached to the N- or C-terminus, can fold well within biological temperature ranges, can mature quickly, and can remain fluorescent in many sub-cellular compartments.
Figure 1.
Auto-fluorescent proteins as fusion tags. a) Table of excitation and emission wavelengths for twelve optimized fluorescent proteins (FP). The comparative brightness of each FP is listed as the product of the quantum yield and the extinction coefficient at the peak absorbance wavelength divided by 1000. Values were generating from the literature [1, 23, 26, 28]. b) Structure of EGFP [39]. A magnified view of the cyclized chromophore (TYG) is shown to the right. c) Cartoon of N- and C- terminal linkers introduced during genetic fusion of a protein of interest (POI) to an FP in standard Clontech vectors. The minimum and maximum linkers are indicated by parenthesis. The unstructured amino acids from EGFP are colored green. A common flexible peptide linker is shown below.
A large part of the success in applying FPs to biological imaging applications results from their unique chemical structure (Figure 1)[9]. Members of the GFP family are relatively small, compact, chemically inert proteins. These proteins are ~240 amino acids long (~27 kD) and fold into a 11 β-sheet barrel surrounding an internal distorted helix (Figure 1) [10]. FPs are autofluorescent because the central portion of this internal helix contains three amino acids (TYG for EGFP) that undergo a spontaneous chemical reaction to generate the stable chromophore. The βbarrel shields the internal helix from the solvent and provides a chemically complex environment for the flurophore. Indeed, the chemical properties of the residues near the chromophore greatly influence the photochemistry of each FP variant [11, 12]. The diversity of these residues generates the wide range of photo-physical characteristics of the different FP isoforms. Unfolding GFP, which maintains the covalent chemical structure of the central chromophore, but radically repositions the amino acids surrounding the chromophore, completely abolishes fluorescence [13]. The sensitivity of GFP to unfolding illustrates the idea that the fluorophore of an FP can be thought of as a distributed fluorescent system that contains both the cyclized amino acid chromophore and the neighboring amino acids. In this review we cover the most commonly used and best behaved variants along with four practical considerations for their application to cellular imaging. For a more complete survey, a number of excellent reviews have been written that exhaustively cover the entire palette of fluorescent proteins currently available [1, 10, 14–16].
When making the choice of what FP to use in an imaging experiment, one of the first considerations is the color variant to use. The most common fluorescent protein used in cellular imaging applications is the optimized S65T mutant of the original GFP. The preferred variant of this tag is the enhanced version (EGFP) that contains additional folding mutations along with the monomeric A206K substitution (commonly called mGFP) [17, 18]. The A206K mutation has been shown to reduce the intrinsic dimerization of FPs at high protein concentrations [17]. Two recent high performance mutants of GFP are superfolder GFP (sfGFP) and Emerald, which contain additional mutations to enhance folding, brightness, and reduce environmental sensitivity [19, 20]. These green fluorophores are particularly bright; however, cellular autofluorescence is close to the emission color of blue/green FPs and this background can complicate the interpretation of fluorescence images [21, 22]. Furthermore, Emerald has a large and rapid initial bleaching component which can further complicate quantitative measurements. Several other high performance FPs are TagBFP (blue), mCerulean3 (cyan), mCitrine/mVenus (green-yellow), tdTomato (orange), mCherry and mApple (red), and mKate2 and mNeptune (far-red). These FPs span the visible spectrum and in some cases provide enough spectral separation for multiplexed labeling of several targets (Figure 1) [23–27]. Orange/red fluorophores in particular have great potential because cellular autofluorescence is lower in this part of the visible spectrum and red light can penetrate more deeply into thick biological samples than shorter wavelength light [21, 22, 28]. These proteins are, however, not derived from the original GFP and greater incidences of mis-folding and mis-targeting have been reported with variants of red FPs [29–32]. Aside from color, it is also important to match the pKa of an FP to the particular application. Indeed, some intracellular organelles have a low pH (<5.5) and certain FP isoforms will be quenched at these pH values [11, 33]. There are dozens of other FPs, but the variants mentioned above have been shown to perform relatively well in standard biological imaging applications, tend to be well behaved when attached to partner proteins, are bright and photo-stable, and have in general survived the rigors of testing in multiple labs across multiple experimental platforms.
The second consideration when tagging a protein of interest with an FP is to which side to attach the FP sequence. The choice can be guided by what is known about the protein; however, it is recommended that both sides be tested for functionality and sub-cellular localization. Some proteins are sensitive to the sidedness of GFP attachment. For example, placing an FP before a signal sequence in a transmembrane protein or secreted protein will likely disrupt the sub-cellular targeting or expression of the protein. In some instances, FPs have been added within the sequence of a protein. However, in this case the size of an FP and the resulting perturbations to the overall fold and function of the underlying protein can be significant. For internal fusions, careful considerations and planning must be done to ensure that the GFP is added within loops or flexible domains. In one example, GFP was inserted randomly into the protein and a functional screen was used to test for effects of the inserted tag [34].
A third consideration when making an FP fusion is the linkers used to join the two sequences. Generally, it is prudent to add a flexible linker between the two partners. The standard flexible linkers consist of runs of glycines interspersed with serines or threonines (GGGSGGGS)[35]. Glycines provide conformational flexibility while the hydrophilic residues allow hydrogen bonding with the solvent and prevent the linker from interacting with hydrophobic protein-binding interfaces. These linkers have been used to create flexible biosensors and tandem dimers of FPs [36, 37]. While adding a linker is good practice, most cloning vectors have multiple cloning sites (MCS) that introduce a random sequence between the fusion partner and the FP sequence. These nucleotides can add upwards of 30 amino acids between the protein of interest and the FP (Figure 1). These amino acids are generally not solely serines, threonines, or glycines, and have the potential to affect the behavior of an attached protein [38]. Furthermore, the first two and last eight amino acids of an FP are unstructured [9, 39, 40]. Thus, any proteins fused to an FP incorporate these unstructured residues between the FP and the sequence of interest. The combination of these unstructured regions and amino acids added from cloning procedures can add close to 40 amino acids of sequence space between the two proteins. Due to the potential effects of such long chemically complex linkers, it is prudent to limit the unnecessary sequences space between the FP and fusion partner.
Quite often the attachment of an FP to a protein of interest has no observable effects on a protein’s structure, function, and localization [41]. However, this is not always the case, particularly when proteins are over-expressed [32, 42, 43]. Thus, the fourth and final consideration when using an FP as a fusion tag is the nature of the expression system. Most plasmid vectors used to transiently transfect and express FP fusion proteins contain strong promoters such as the CMV or EF-1α promoter. These promoters can drive a hundreds-fold increase in the amount of a protein present in a cell [44, 45]. Furthermore, promoters can act differently in different cells types. This potentially large increase in a protein’s expression level can have a dramatic influence on the sub-cellular localization of a fusion protein and its cellular function. The selection of cells expressing the smallest amount of a fluorescent protein that can still be imaged clearly with the desired imaging modality is preferred. Along these lines, the use of stable cell lines or promoters with disrupted sequences, tunable activity, or the direct integration of fluorescent proteins into the genome, can mitigate the effects of extreme over-expression [43, 46, 47]. For example, a severely truncated CMV promoter (sometimes called the speckle promoter) has been used to express very low amounts of protein necessary for single molecule imaging [46, 48].
The use of FPs for biological studies is now ubiquitous. They are used for imaging proteins inside cells, tissues, and animals and to monitor binding and association between proteins with techniques such as fluorescence resonance energy transfer, fluorescence correlation spectroscopy, and single molecule imaging [49]. They are used to watch proteins move on the molecular scale [50]. They are used to build biosensors that monitor cellular signaling pathways [51]. All of these methods continue to be developed and improved. Of particular note are the recent developments in photo-switchable FPs such as mEOS2, Dronpa, PS-CFP2, PA-mCherry, and PA-GFP [52, 53]. These probes can be used to highlight and then track fusion proteins over time. They are also powerful probes for super-resolution optical methods [54, 55]. These new probes are helping to drive the rapid developments in super-resolution techniques, and as these probes improve and develop with brighter, more stable, and chromatically diverse variants, multicolor and live cell methods for super-resolution imaging will likewise improve [53]. Furthermore, alternative autofluorescent proteins are being developed for conventional imaging. These include the flavin-based fluorescent proteins (iLOV-FMN) and bacterial phytochrome-biliverdin proteins [56, 57]. These new probes have great potential as they have either far-red excitation wavelengths (phytochrome) or are ~20 kD smaller than standard FPs (iLOV).
Self-labeling proteins and enzyme targeting peptide tags
Fluorescent proteins have limitations. For example, their photo-physical properties are not as good as organic dyes. They can blink, they are not very bright, some are not photo-stable, and their colors and chemistry are limited. To overcome these issues, another class of protein-based fusion tags has been developed that can catalyze the covalent auto-attachment of an organic fluorophore inside living cells [58]. These tags are near to the size of an FP (Figure 2). When expressed, the proteins are not innately fluorescent and only become fluorescent when the cells are exposed to the fluorescent ligand. This scenario lends several advantages to these systems over standard FP labeling. For example, labeling can be restricted in both time and space, and sequential labeling schemes can be employed. For example, pulse chase experiments, where one fluorophore is added and then at some time later a second (different) fluorophore is added, can be performed. Furthermore, the development of these tags is rapidly progressing and future variants and their ligands will certainly complement the current palate of FPs with additional chemical or spectral characteristics difficult or impossible to obtain from fluorescent proteins [59]. For example, these probes could be brighter and more chromatically diverse, and will likely have specific chemical or photonic properties such as photoswitching, environmental sensitivity, or free radical generation.
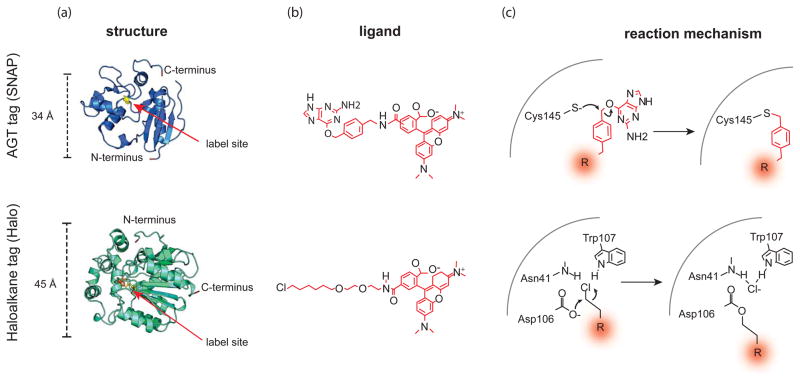
Figure 2.
Self-labeling enzymes as fluorescent fusion tags. a) The structure of AGT/SNAP tag (top) and the Haloalkane dehydrogenase tag (bottom). The residues that are labeled by the fluorescent ligand are shown in yellow. In Halo, the mutated catalytic histidine is shown in orange. b) (top) Structure of a benzyl guanine-linked TMR ligand for SNAP lag labeling and a (bottom) TMR-linked haloalkane for Halotag labeling along with c) the corresponding reaction mechanisms for covalent attachment.
The first protein used to catalyze the covalent auto-attachment of a fluorophore within a cell was the 20 kD DNA repair protein human O6-alkylguanine-DNA alkytransferase (AGT) (marketed as SNAP tag) (Figure 2)[4, 60]. This protein catalyzes the attachment of O6-alkylguanine or O6-benzylguanine to a cysteine on the protein. The modified AGT is then degraded to protect the cell from the mutagenic effects of O6-alkylguanine. To achieve labeling of AGT, fluorescent membrane permeable O6-alkylguanine substrates were synthesized and added to cells expressing proteins with genetic in-frame fusion of AGT. These proteins were shown to be specifically labeled by these cell permeable fluorescent O6-benzylguanine substrates. Recently, a new variant of this enzyme has been developed that reacts specifically with O6-benzylcytosine substrates (CLIP tag) [3]. Thus, SNAP and CLIP tags can be used together in one cell to label two proteins with two different fluorescent ligands.
Since the development of AGT, this system has been used for localizing proteins, tracking their expression and transport, and sensing and measuring structures. Furthermore, these enzymes have opened the door for the improvement of super-resolution methods including stimulated emission depletion (STED) and pointillism microscopy for locating proteins to sub-diffraction limited positions within cells [61–63]. For example, dye labeled SNAP tags have been shown to provide improved resolution to pointillism imaging of clathrin coated pits [61].
Aside from AGT, the bacterial enzyme haloalkane dehalogenase (marketed as Halo tag) has been developed as a self-labeling fusion tag (Figure 2) [2]. Like AGT, this enzyme covalently attaches a modified flurophore-ligand to an active site residue. Specifically, haloalkane dehalogenase removes halides from hydrocarbon chains by nucleophilic displacement. During the reaction a covalent ester is formed in the active site between the hydrocarbon chain and an active site aspartate. In the native enzyme a conserved histidine reacts with the covalent protein-chemical intermediate, releasing the ligand and regenerating the aspartate nucleophile [64, 65]. When this histidine is mutated, the enzyme proceeds to the covalent intermediate but cannot release the substrate. Thus, substrates containing a haloalkane chain are covalently attached to the mutant enzyme in a one-to-one ratio. Similar to AGT (SNAP) tags, Halo tags have been used in studies to investigate the localization and trafficking of proteins in living cells. Being organic dyes, Halo tag fluorophores are bright and relatively photo-stable. This has allowed the targeted labeling of proteins for single molecule studies [66]. Additionally, the ability to pulse label proteins allows protein populations to be tracked over time.
While both Halo and SNAP tags appear to fold well and are efficiently labeled by cell-permeant fluorescent ligands, an additional step of washing the free dye from the cells is required to ensure a low fluorescent background. Furthermore, the labeling of Halo tag is inhibited by both detergents and fixation. Thus, the labeling must be done before the cells are fixed or permeabilized with detergents. For AGT tags, native AGT in mammalian cells is able to react with the fluorescent ligand [60]. For this reason, the use of cell lines with depleted or no endogenous AGT is recommended to keep off-target background labeling low. Additionally, ligand-bound AGT has been shown to be targeted for degradation. In this regard, it is possible that labeled AGT fusion proteins will have a reduced half-life compared to the turnover of native proteins. However, newer AGT variants with faster reaction kinetics have been shown to be better behaved in living systems [3]. Furthermore, these tags are relatively large proteins. Thus, issues similar to the ones mentioned above for FPs including attachment site, linker composition, internal fusions, expression system choice, and disruptions of the overall fold, function, or localization of the protein should be considered.
An alternative method for labeling proteins is the use of an enzyme that catalyzes the attachment of a probe to a specific peptide sequence. For example, the small 9 kD acyl carrier protein (ACP) tag can be covalently labeled with CoA derivatives by the enzyme AcpS [67]. However, substrates for this system are not membrane permeable and restrict its use to extracellular proteins. Recently, a new enzyme-peptide system was developed that allows the covalent modification of a peptide tag in living cells [5]. This system uses an engineered fluorophore ligase derived from the bacterial enzyme lipoic acid ligase (LplA). This mutant enzyme can catalyze the attachment of a cell permeant fluorophore (7-hydroxycoumarin) to the 13-amino acid LplA acceptor peptide tag (LAP). To label an intracellular target, the LplA enzyme and LAP target must be co-expressed in the same cell. An advantage of this system is that the location of the enzyme can be restricted to subcellular compartments, spatially restricting the addition of the fluorophore to specific populations of the acceptor.
Covalent organic fluorescent ligands
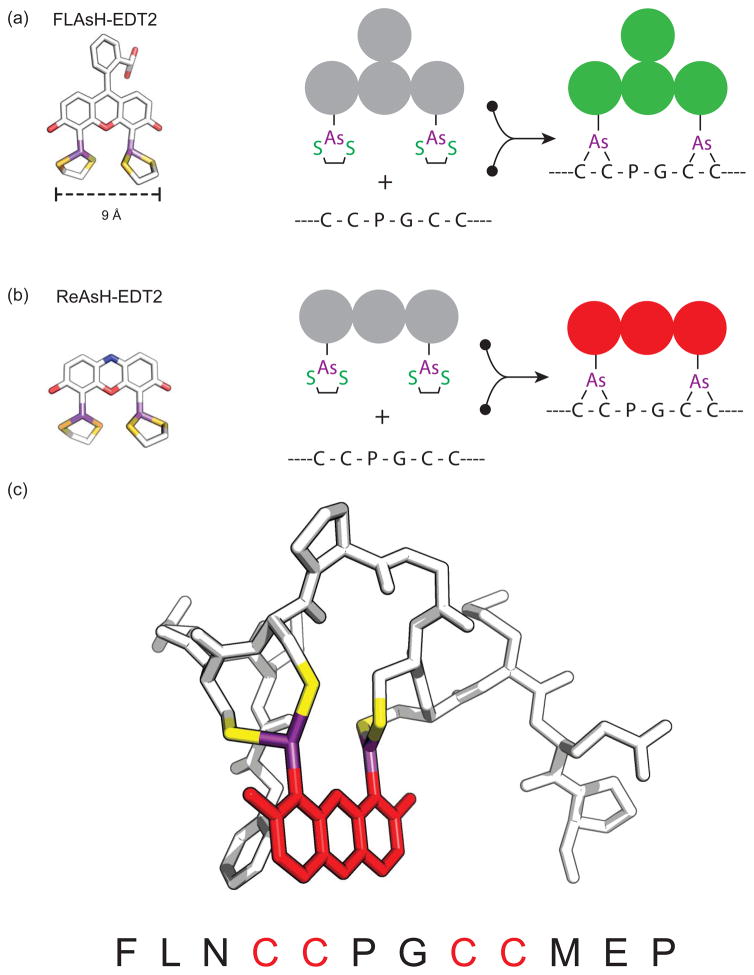
GFP and self-labeling proteins are quite large on the molecular scale. Currently, the smallest and most successful genetically encoded tag for covalent small fluorophore labeling is the tetracysteine-biarsenical system (Table 1 & Figure 3). With this system, a short peptide sequence (Cys-Cys-Xaa-Xaa-Cys-Cys, where X is any amino acid other than cysteine) is genetically introduced into the sequence of a target protein [68]. This sequence, while rare in native proteins, can specifically react with a membrane-permeable biarsenical dye. There are two standard biarsenical dyes, green and red (Table 1). The green emitting variant is FlAsH-EDT2 (fluorescein arsenical hairpin binder). This dye has a peak absorbance around 508 nm and a peak emission at 528 nm [6, 69]. The red variant, ReAsH (resorufin arsenical hairpin binder), is a derivative of the red fluorophore resorufin; it absorbs at 593nm and emits at 608nm. In both dyes, the reaction of the biarsenical ligands with the cysteine thiols results in covalent bonds between the organoarsenicals and the tetracysteine motif. This binding is thought to involve consecutive cysteine residues in the target sequence (i and i+1 and i+4 and i+5). Other attachment schemes including i and i+4 and i+1 and i+5, however, are possible [6].
Table 1.
Physical parameters of biarsenical dyes and optimized tetracysteine binding sequences
| Pro-fluorophore | Abs./Em. (nm) | Peptide ligand | Kapp (μM) | QY | Extinction coefficient | pKa | Dithiol resistance | Ref. |
|---|---|---|---|---|---|---|---|---|
| FLAsH-EDT2 | 508/528 | FLNCCPGCCMEP | 0.48±0.16 | 0.85 | 70,000 M-1cm-1 | 5.5 | 5.6±0.3mM BAL | [8, 71, 86] |
| HRWCCPGCCKTF | 0.65 | 2.3±0.1mM BAL | [8, 86] | |||||
| CCPGCC | 0.7±0.1mM BAL | [8, 86] | ||||||
| FLNCCPSQPTYPGDDAPVED LIRFYDNLQQYLNVCCMEP | 5.8±0.7 | [71, 72] | ||||||
| FLNCCEWTWDDATKTWTWT CCMEP | 0.62±0.12 | [71, 72] | ||||||
| ReAsH-EDT2 | 593/608 | FLNCCPGCCMEP | 0.37±0.20 | 0.48 | 69,000 M-1cm-1 | 4.7 | 2.0±0.3mM BAL | [8, 71, 86] |
| HRWCCPGCCKTF | 0.40 | 0.8±0.1mM BAL | [8, 86] | |||||
| CCPGCC | 0.2±0.1mM BAL | [8, 86] | ||||||
| FLNCCPSQPTYPGDDAPVED LIRFYDNLQQYLNVCCMEP | 3.4±1.2 | [71, 72] | ||||||
| FLNCCEWTWDDATKTWTWT CCMEP | 0.38±0.17 | [71, 72] |
Figure 3.
Structure of the biarsenical dyes FLAsH-EDT2 (a) and ReAsH-EDT2 (b). (c) Model of the optimized tetracysteine peptide bound to ReAsH based on the NMR structure of the complex [70].
In model peptides, rate constants for FlAsH labeling are between 105 M−1s−1 and 10−6s−1 and the dissociation constant is 10−11M [68]. While the minimal peptide sequence for flash labeling is CCXXCC, an optimized sequence CCPGCC has been shown to form a more stable complex with faster association kinetics. This sequence has been further optimized resulting in two high affinity 12 amino acid biarsenical binding motifs (FLNCCPGCCMEP and HRWCCPGCCKTF) (Table 1) [8]. In peptides where the cysteines were separated by one amino acid the binding to dye was reduced and in peptides with four consecutive cysteines (CCCC) the affinity for the fluorophore was extremely low and the complex had a low quantum yield. These data along with NMR studies suggest that the peptide-dye complex does not adopt a straight alpha helix and instead adopts a hairpin conformation (Figure 3) [69, 70]. Alternative bipartite sequences have been developed that space the two di-cysteine motifs between a longer sequence that can fold back onto itself, thus positioning the four cysteines in the correct orientation to bind the biarsenical dye [71]. Furthermore, intermolecular sequences have been shown to bind a biarsenaical dye between two independent di-cystein motifs [71]. These alternative methods could be used to develop new biosensors or as structural probes for protein conformational changes[72].
Both FLAsH-EDT2 and ReAsH-EDT2 are essentially non-fluorescent before they react. In the excited state, the free molecules are quenched by either vibrational deactivation or photo-induced electron transfer mechanisms [6]. This is thought to occur because in the unbound dye the EDT groups allow free rotation of the aryl-arsenic bonds. When the dye interacts with a tetracysteine motif, each EDT is displaced and the rigidity of the complex impedes conjugation of the arsenic and chromophore orbitals. Thus, the rigid molecule can evade quenching due to photo-induced electron transfer or rotational decay pathways present in the freely rotating dye [6]
The advantages of biarsenical dyes are two-fold. First, the targeting motifs are small. Thus, the introduced sequences have less opportunity to disrupt the overall fold and function of the labeled protein. The second advantage of these probes is the ease of pulse-labeling procedures. One of the first applications of biarsenical dyes was in vivo pulse labeling of the gap junction protein connexin. In this work, gap junction assembly was monitored by sequentially labeling connexin proteins with FlAsH-EDT2and ReAsH-EDT2 [68]. More recently, pulse labeling of a viral vesicular stomatitis protein with FlAsH and ReAsH was used to map the rate of plasma membrane trafficking of newly synthesized virus proteins [32]. Interestingly, the same protein tagged with mRFP was not functional, illustrating that FP fusions are sometimes not well tolerated, whereas smaller tetracysteine tags can support the underlying biological function of the protein [32]. Aside from pulse labeling experiments, the ability to add a fluorophore to an expressed protein in vivo has made biarsenical dyes useful tools for FRET. In these studies, the fluorescence of a donor fluorophore attached to a protein is measured. Next, the acceptor biarsenical dye is reacted to a tetracysteine motif and the quantity of donor quenching is measured [49]. This method provides a direct measurement of FRET efficiency [49]. This technique has been used to map structural transitions in membrane receptors during ligand activation [73, 74].
The three major concerns when using biarsenical dyes are the level of background labeling, the overall cellular toxicity of the ligands, and the effect of the tag on the proteins function or localization. One of the causes of background labeling is the interaction of these dyes with the membrane or off-target cellular proteins. These are generally proteins that are rich in cysteines, including zinc finger proteins, RING finger proteins, protein kinases, and cytoskeletal proteins [75, 76]. Other non-thiol binding sites such as hydrophobic pockets, membranes, or other cellular structures could contribute to nonspecific background fluorescence [75]. To reduce this background, washing the cells with British anti-Lewisite (BAL), a chemical that binds to arsenic, free EDT2, or the dye disperse blue 3, have been found to reduce off-target labeling and suppress background fluorescence [7, 69, 77, 78]. Further methods to increase the specific signal over the noise of the fluorescent background have included using robust promoters to increase the expression level of target proteins or engineering tandem repeats of the tetracysteine motifs to increase the total number of dyes per fusion partner [79].
Conclusions
Brightness, photo-stability, background, toxicity, and spectral overlap, ion sensitivity, along with structural and cellular effects on the targeted partner, are all important issues to weigh when choosing a fluorescent tag for in vivo imaging of cells. The four covalent labeling modalities mentioned in this review all provide specific advantages and disadvantages in this context. Indeed, each can provide robust fluorescent signals within cells (Figure 4). FPs are autofluorescent and thus have essentially no background. However, their photophysical characteristics are not optimal and they are large. Self-labeling enzyme and enzyme tags provide more robust organic fluorophores, but there are issues with background fluorescence and probe stability, and the fusion tags are of similar size to FPs. Enzyme attachment to a small protein domain is possible but requires the expression of a second protein in the cell or only works on extracellular proteins. Biarsenical dyes and tetracysteine motifs are small and their targeting sequence is short. There are, however, significant issues with background fluorescence. Additionally, while this review focused on covalent methods to label proteins inside cells several developing technologies for labeling proteins rely on non-covalent methods. These methods include the use the protein dihydrofolate reductase as a fusion tag which can bind to trimethoprim-linked fluorophores with nanomolar affinites, and fluorogen-actived proteins which bind to and activate the fluorescence of chemicals such as malachite green [80, 81]. Furthermore, several methods have been developed which cannot easily be used inside cells but have be used predominantly to label extracellular proteins. These methods include histidine tags to bind nickel-NTA-linked fluorophores or quantum dots or the use biotin ligase to covalently attach a fluorescent biotin to an acceptor peptide [82–84]. As these additional methods improve they will further add functionality to experimental imaging systems.
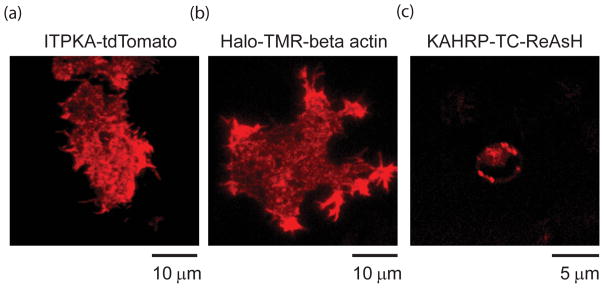
Figure 4.
Examples of fluorescent labeling methods in cells. a) Total Internal Reflection Fluorescence image (TIRF) of a living PC12 cells expressing the F-actin binding protein ITPKA tagged with tdTomato [85]. b) TIRF image of a PC12 cell expressing Halotag-Beta actin labeled with the red fluorophore TMR-halotag ligand. c) Confocal image of an erythrocyte infected with transgenic Plasmodium falciparum expressing the tetracysteine tag (TC)-containing protein KAHRP (+His)-TC labeled with ReAsH.
Clearly, there is not one answer to which probe or method will provide the best signal for all cellular imaging applications. The development and refinement of tags for live cell imaging is progressing rapidly and the molecular toolkit continues to grow. Future developments might include I) smaller genetic tags, II) probes with interesting chemistries such as multi-color photo-switchable or photo-activating dyes, III) probes which respond to voltage, temperature, or ligands including ions or small molecules, IV) additional cell permeable ligands for self-labeling enzymes, tetracysteine tags, or enzyme targets and, V) multi-functional probes that can be used together for fluorescence, electron, magnetic, or x-ray imaging. These developments, along with similar advances in image processing and acquisition methods will provide new views of biological structure and function.
Acknowledgments
We would like to thank M.W. Davidson (FSU) for helpful discussions and information presented at the 2011 analytical and quantitative light microscopy course (Marine Biological Lab, Woods Hole, MA). The authors would also like to thank J. Hwang (NIST), T. Wellems, and F. Tokumasu (NIAID/NIH) for helpful discussions and for providing the image of ReAsH labeled transgenic parasites and M.J. Schell (Uniformed Services University) for the ITPKA-tdTomato construct. The authors would also like to thank J. Silver (NHLBI/NIH), G. Shtengel, H. Hess, and L.D. Lavis (Janelia Farm/HHMI) for helpful discussions. We also thank F. Madani and A.O. Graslund (Stockholm University) for providing the NMR structure of the ReAsH bound peptide. J.W. Taraska is supported by the intramural program at the National Heart Lung and Blood Institute, National Institutes of Health. G. Crivat was supported by the National Research Council Research Award (NIST) and the National Institute of Allergy and Infectious Diseases, National Institutes of Health. Funding for the award was provided by NIST and the Intramural Program of the National Institute for Biomedical Imaging and Bioengineering of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Day RN, Davidson MW. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Los GV, Wood K. The HaloTag: a novel technology for cell imaging and protein analysis. Methods Mol Biol. 2007;356:195–208. doi: 10.1385/1-59745-217-3:195. [DOI] [PubMed] [Google Scholar]
- 3.Gautier A, et al. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Juillerat A, et al. Directed evolution of O6-alkylguanine-DNA alkyltransferase for efficient labeling of fusion proteins with small molecules in vivo. Chem Biol. 2003;10:313–317. doi: 10.1016/s1074-5521(03)00068-1. [DOI] [PubMed] [Google Scholar]
- 5.Uttamapinant C, et al. A fluorophore ligase for site-specific protein labeling inside living cells. Proc Natl Acad Sci U S A. 2010;107:10914–10919. doi: 10.1073/pnas.0914067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griffin BA, et al. Specific covalent labeling of recombinant protein molecules inside live cells. Science. 1998;281:269–272. doi: 10.1126/science.281.5374.269. [DOI] [PubMed] [Google Scholar]
- 7.Griffin BA, et al. Fluorescent labeling of recombinant proteins in living cells with FlAsH. Methods Enzymol. 2000;327:565–578. doi: 10.1016/s0076-6879(00)27302-3. [DOI] [PubMed] [Google Scholar]
- 8.Martin BR, et al. Mammalian cell-based optimization of the biarsenical-binding tetracysteine motif for improved fluorescence and affinity. Nat Biotechnol. 2005;23:1308–1314. doi: 10.1038/nbt1136. [DOI] [PubMed] [Google Scholar]
- 9.Ormo M, et al. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 10.Chudakov DM, et al. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol Rev. 2010;90:1103–1163. doi: 10.1152/physrev.00038.2009. [DOI] [PubMed] [Google Scholar]
- 11.Shaner NC, et al. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 12.Shu X, et al. Novel chromophores and buried charges control color in mFruits. Biochemistry. 2006;45:9639–9647. doi: 10.1021/bi060773l. [DOI] [PubMed] [Google Scholar]
- 13.Baldini G, et al. Pre-unfolding resonant oscillations of single green fluorescent protein molecules. Science. 2005;309:1096–1100. doi: 10.1126/science.1115001. [DOI] [PubMed] [Google Scholar]
- 14.Kremers GJ, et al. Fluorescent proteins at a glance. J Cell Sci. 2011;124:157–160. doi: 10.1242/jcs.072744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davidson MW, Campbell RE. Engineered fluorescent proteins: innovations and applications. Nat Methods. 2009;6:713–717. doi: 10.1038/nmeth1009-713. [DOI] [PubMed] [Google Scholar]
- 16.Shaner NC, et al. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 17.Zacharias DA, et al. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
- 18.Cubitt AB, et al. Understanding structure-function relationships in the Aequorea victoria green fluorescent protein. Methods Cell Biol. 1999;58:19–30. doi: 10.1016/s0091-679x(08)61946-9. [DOI] [PubMed] [Google Scholar]
- 19.Tsien RY. Building and breeding molecules to spy on cells and tumors. FEBS Lett. 2005;579:927–932. doi: 10.1016/j.febslet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 20.Pedelacq JD, et al. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 21.Benson RC, et al. Cellular autofluorescence--is it due to flavins? J Histochem Cytochem. 1979;27:44–48. doi: 10.1177/27.1.438504. [DOI] [PubMed] [Google Scholar]
- 22.Aubin JE. Autofluorescence of viable cultured mammalian cells. J Histochem Cytochem. 1979;27:36–43. doi: 10.1177/27.1.220325. [DOI] [PubMed] [Google Scholar]
- 23.Subach OM, et al. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 25.Griesbeck O, et al. Reducing the environmental sensitivity of yellow fluorescent protein. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- 26.Markwardt ML, et al. An improved cerulean fluorescent protein with enhanced brightness and reduced reversible photoswitching. PLoS One. 2011;6:e17896. doi: 10.1371/journal.pone.0017896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 28.Lin MZ, et al. Autofluorescent proteins with excitation in the optical window for intravital imaging in mammals. Chem Biol. 2009;16:1169–1179. doi: 10.1016/j.chembiol.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizuno H, et al. Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer. Biochemistry. 2001;40:2502–2510. doi: 10.1021/bi002263b. [DOI] [PubMed] [Google Scholar]
- 30.Jakobs S, et al. EFGP and DsRed expressing cultures of Escherichia coli imaged by confocal, two-photon and fluorescence lifetime microscopy. FEBS Lett. 2000;479:131–135. doi: 10.1016/s0014-5793(00)01896-2. [DOI] [PubMed] [Google Scholar]
- 31.Yanushevich YG, et al. A strategy for the generation of non-aggregating mutants of Anthozoa fluorescent proteins. FEBS Lett. 2002;511:11–14. doi: 10.1016/s0014-5793(01)03263-x. [DOI] [PubMed] [Google Scholar]
- 32.Das SC, et al. Biarsenical labeling of vesicular stomatitis virus encoding tetracysteine-tagged m protein allows dynamic imaging of m protein and virus uncoating in infected cells. J Virol. 2009;83:2611–2622. doi: 10.1128/JVI.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kneen M, et al. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys J. 1998;74:1591–1599. doi: 10.1016/S0006-3495(98)77870-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giraldez T, et al. Generation of functional fluorescent BK channels by random insertion of GFP variants. J Gen Physiol. 2005;126:429–438. doi: 10.1085/jgp.200509368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huston JS, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci U S A. 1988;85:5879–5883. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campbell RE, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alfthan K, et al. Properties of a single-chain antibody containing different linker peptides. Protein Eng. 1995;8:725–731. doi: 10.1093/protein/8.7.725. [DOI] [PubMed] [Google Scholar]
- 38.Michael DJ, et al. Fluorescent cargo proteins in pancreatic beta-cells: design determines secretion kinetics at exocytosis. Biophys J. 2004;87:L03–05. doi: 10.1529/biophysj.104.052175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Royant A, Noirclerc-Savoye M. Stabilizing role of glutamic acid 222 in the structure of Enhanced Green Fluorescent Protein. J Struct Biol. 2011;174:385–390. doi: 10.1016/j.jsb.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 40.Dopf J, Horiagon TM. Deletion mapping of the Aequorea victoria green fluorescent protein. Gene. 1996;173:39–44. doi: 10.1016/0378-1119(95)00692-3. [DOI] [PubMed] [Google Scholar]
- 41.Clyne PJ, et al. Green fluorescent protein tagging Drosophila proteins at their native genomic loci with small P elements. Genetics. 2003;165:1433–1441. doi: 10.1093/genetics/165.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Agbulut O, et al. GFP expression in muscle cells impairs actin-myosin interactions: implications for cell therapy. Nat Methods. 2006;3:331. doi: 10.1038/nmeth0506-331. [DOI] [PubMed] [Google Scholar]
- 43.Doyon JB, et al. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat Cell Biol. 2011;13:331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin JY, et al. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Running Deer J, Allison DS. High-level expression of proteins in mammalian cells using transcription regulatory sequences from the Chinese hamster EF-1alpha gene. Biotechnol Prog. 2004;20:880–889. doi: 10.1021/bp034383r. [DOI] [PubMed] [Google Scholar]
- 46.Watanabe N, Mitchison TJ. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 2002;295:1083–1086. doi: 10.1126/science.1067470. [DOI] [PubMed] [Google Scholar]
- 47.Taylor MJ, et al. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barg S, et al. Syntaxin clusters assemble reversibly at sites of secretory granules in live cells. Proc Natl Acad Sci U S A. 2010;107:20804–20809. doi: 10.1073/pnas.1014823107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taraska JW, Zagotta WN. Fluorescence applications in molecular neurobiology. Neuron. 2010;66:170–189. doi: 10.1016/j.neuron.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taraska JW, Zagotta WN. Structural dynamics in the gating ring of cyclic nucleotide-gated ion channels. Nat Struct Mol Biol. 2007;14:854–860. doi: 10.1038/nsmb1281. [DOI] [PubMed] [Google Scholar]
- 51.Frommer WB, et al. Genetically encoded biosensors based on engineered fluorescent proteins. Chem Soc Rev. 2009;38:2833–2841. doi: 10.1039/b907749a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lippincott-Schwartz J, Patterson GH. Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging. Trends Cell Biol. 2009;19:555–565. doi: 10.1016/j.tcb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patterson G, et al. Superresolution imaging using single-molecule localization. Annu Rev Phys Chem. 2010;61:345–367. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shtengel G, et al. Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proc Natl Acad Sci U S A. 2009;106:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shu X, et al. Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science. 2009;324:804–807. doi: 10.1126/science.1168683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chapman S, et al. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc Natl Acad Sci U S A. 2008;105:20038–20043. doi: 10.1073/pnas.0807551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin MZ, Wang L. Selective labeling of proteins with chemical probes in living cells. Physiology (Bethesda) 2008;23:131–141. doi: 10.1152/physiol.00007.2008. [DOI] [PubMed] [Google Scholar]
- 59.Lavis LD, Raines RT. Bright ideas for chemical biology. ACS Chem Biol. 2008;3:142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 61.Jones SA, et al. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011 doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klein T, et al. Live-cell dSTORM with SNAP-tag fusion proteins. Nat Methods. 2011;8:7–9. doi: 10.1038/nmeth0111-7b. [DOI] [PubMed] [Google Scholar]
- 63.Toomre D, Bewersdorf J. A new wave of cellular imaging. Annu Rev Cell Dev Biol. 2010;26:285–314. doi: 10.1146/annurev-cellbio-100109-104048. [DOI] [PubMed] [Google Scholar]
- 64.Verschueren KH, et al. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature. 1993;363:693–698. doi: 10.1038/363693a0. [DOI] [PubMed] [Google Scholar]
- 65.Newman J, et al. Haloalkane dehalogenases: structure of a Rhodococcus enzyme. Biochemistry. 1999;38:16105–16114. doi: 10.1021/bi9913855. [DOI] [PubMed] [Google Scholar]
- 66.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.George N, et al. Specific labeling of cell surface proteins with chemically diverse compounds. J Am Chem Soc. 2004;126:8896–8897. doi: 10.1021/ja048396s. [DOI] [PubMed] [Google Scholar]
- 68.Gaietta G, et al. Multicolor and electron microscopic imaging of connexin trafficking. Science. 2002;296:503–507. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 69.Adams SR, et al. New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. J Am Chem Soc. 2002;124:6063–6076. doi: 10.1021/ja017687n. [DOI] [PubMed] [Google Scholar]
- 70.Madani F, et al. Hairpin structure of a biarsenical-tetracysteine motif determined by NMR spectroscopy. J Am Chem Soc. 2009;131:4613–4615. doi: 10.1021/ja809315x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luedtke NW, et al. Surveying polypeptide and protein domain conformation and association with FlAsH and ReAsH. Nat Chem Biol. 2007;3:779–784. doi: 10.1038/nchembio.2007.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Scheck RA, Schepartz A. Surveying Protein Structure and Function Using Bis-Arsenical Small Molecules. Acc Chem Res. 2011 doi: 10.1021/ar2001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Granier S, et al. FRET-based measurement of GPCR conformational changes. Methods Mol Biol. 2009;552:253–268. doi: 10.1007/978-1-60327-317-6_18. [DOI] [PubMed] [Google Scholar]
- 74.Hoffmann C, et al. A FlAsH-based FRET approach to determine G protein-coupled receptor activation in living cells. Nat Methods. 2005;2:171–176. doi: 10.1038/nmeth742. [DOI] [PubMed] [Google Scholar]
- 75.Stroffekova K, et al. The protein-labeling reagent FLASH-EDT2 binds not only to CCXXCC motifs but also non-specifically to endogenous cysteine-rich proteins. Pflugers Arch. 2001;442:859–866. doi: 10.1007/s004240100619. [DOI] [PubMed] [Google Scholar]
- 76.Donoghue N, et al. Presence of closely spaced protein thiols on the surface of mammalian cells. Protein Sci. 2000;9:2436–2445. doi: 10.1110/ps.9.12.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Adams SR, Tsien RY. Preparation of the membrane-permeant biarsenicals FlAsH-EDT2 and ReAsH-EDT2 for fluorescent labeling of tetracysteine-tagged proteins. Nat Protoc. 2008;3:1527–1534. doi: 10.1038/nprot.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Machleidt T, et al. Protein labeling with FlAsH and ReAsH. Methods Mol Biol. 2007;356:209–220. doi: 10.1385/1-59745-217-3:209. [DOI] [PubMed] [Google Scholar]
- 79.Van Engelenburg SB, et al. FACS-based selection of tandem tetracysteine peptides with improved ReAsH brightness in live cells. Chembiochem. 2010;11:489–493. doi: 10.1002/cbic.200900689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fitzpatrick JA, et al. STED nanoscopy in living cells using Fluorogen Activating Proteins. Bioconjug Chem. 2009;20:1843–1847. doi: 10.1021/bc900249e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miller LW, et al. In vivo protein labeling with trimethoprim conjugates: a flexible chemical tag. Nat Methods. 2005;2:255–257. doi: 10.1038/nmeth749. [DOI] [PubMed] [Google Scholar]
- 82.Zhao C, et al. Hexahistidine-tag-specific optical probes for analyses of proteins and their interactions. Anal Biochem. 2010;399:237–245. doi: 10.1016/j.ab.2009.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Howarth M, Ting AY. Imaging proteins in live mammalian cells with biotin ligase and monovalent streptavidin. Nat Protoc. 2008;3:534–545. doi: 10.1038/nprot.2008.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boeneman K, et al. Intracellular bioconjugation of targeted proteins with semiconductor quantum dots. J Am Chem Soc. 2010;132:5975–5977. doi: 10.1021/ja100201w. [DOI] [PubMed] [Google Scholar]
- 85.Johnson HW, Schell MJ. Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol Biol Cell. 2009;20:5166–5180. doi: 10.1091/mbc.E09-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zurn A, et al. Site-Specific, Orthogonal Labeling of Proteins in Intact Cells with Two Small Biarsenical Fluorophores. Bioconjugate Chemistry. 2010;21:853–859. doi: 10.1021/bc900394j. [DOI] [PubMed] [Google Scholar]