Discovery of adapalene and dihydrotachysterol as antiviral agents for the Omicron variant of SARS-CoV-2 through computational drug repurposing (original) (raw)
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been significantly paralyzing the societies, economies and health care systems around the globe. The mutations on the genome of SARS-CoV-2 led to the emergence of new variants, some of which are classified as “variant of concern” due to their increased transmissibility and better viral fitness. The Omicron variant, as the latest variant of concern, dominated the current COVID-19 cases all around the world. Unlike the previous variants of concern, the Omicron variant has 15 mutations on the receptor-binding domain of spike protein and the changes in the key amino acid residues of S protein can enhance the binding ability of the virus to hACE2, resulting in a significant increase in the infectivity of the Omicron variant. Therefore, there is still an urgent need for treatment and prevention of variants of concern, particularly for the Omicron variant. In this study, an in silico drug repurposing was conducted through the molecular docking of 2890 FDA-approved drugs against the mutant S protein of SARS-CoV-2 for Omicron variant. We discovered promising drug candidates for the inhibition of alarming Omicron variant such as quinestrol, adapalene, tamibarotene, and dihydrotachysterol. The stability of ligands complexed with the mutant S protein was confirmed using MD simulations. The lead compounds were further evaluated for their potential use and side effects based on the current literature. Particularly, adapalene, dihydrotachysterol, levocabastine and bexarotene came into prominence due to their non-interference with the normal physiological processes. Therefore, this study suggests that these approved drugs can be considered as drug candidates for further in vitro and in vivo studies to develop new treatment options for the Omicron variant of SARS-CoV-2.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s11030-022-10440-6.
Keywords: SARS-CoV-2, Omicron variant, Adapalene, Vitamin D, Drug repurposing
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to a severe global outbreak and impacted negatively the societies, economies and public health care systems all over the globe. Based on the World Health Organization (WHO) current data, as of February 6, 2022, this pandemic has infected almost 400 million people, resulting in around 6 million deaths. SARS-CoV-2 belongs to the Coronaviridae family as an enveloped, positive polarity, single-stranded RNA betacoronavirus. Similar to SARS-CoV, its genome includes the genes for non-structural proteins (Nsps), structural proteins, and several accessory proteins [1, 2].
The availability of millions of genomes for SARS-CoV-2 on public databases led to the characterization of mutation profile in SARS-CoV-2 genome. The mutations resulted in the emergence of new variants, some of which are classified as “variant of concern” due to their increased transmissibility and better viral fitness [3]. Table 1 summarizes the number of mutations in the variants of concerns, including the very recent Omicron variant. The mutations in spike protein (S) and receptor-binding domain (RBD) of S protein are particularly important because the cell entry of SARS-CoV-2 occurs with the binding of S protein to its receptor human ACE2 (hACE2) through RBD and is activated by human proteases [4]. The Alpha and Beta variants of SARS-CoV-2 had a mutation (N501Y) in the RBD of S protein. This mutation resulted in a high transmission (40–70% increased transmissibility) in the context of high population immunity, which promoted the emergence and spread of the variants [5, 6]. Two additional mutations (E484K and K417N) were found in S protein of the Beta variant and provided a potential immune escape from the neutralization of antibodies [7]. The Gamma variant had three mutations (N501Y, E484K and K417T) in RBD of S protein as well as five mutations (L18F, T20N, P26S, D138Y and R190S) in the N-terminal domain (NTD) of S protein [8]. In particular, L18F mutation was reported to be associated with escape from multiple NTD-binding antibodies [9]. The Delta variant had two mutations (L452R, T478K) in the RBD and four mutations (T19R, G142D, Δ156–157 and R158G) in the NTD of S protein. The Delta variant was believed to be 60% more transmissible than the Alpha variant [10]. Due to high transmission of this variant, it spread to 54 countries and rapidly replaced the Alpha variant in the UK and the USA [11]. The rapid spread of the Delta variant of SARS-CoV-2 was reconciled with its ability to escape from antibodies targeting non-RBD and RBD epitopes of S protein [10].
Table 1.
Number of mutations in the five variants of concern of SARS-CoV-2
| Variant | Lineage | Country of first identification | Number of amino acid mutations | ||
|---|---|---|---|---|---|
| S protein | RBD | Total | |||
| Alpha | B.1.1.7 | UK | 9 | 1 | 21 |
| Beta | B.1.351 | South Africa | 8 | 3 | 16 |
| Gamma | P1 | Brazil | 12 | 3 | 22 |
| Delta | B.1.617.2 | India | 8 | 2 | 20 |
| Omicron | B.1.1.529 | South Africa | 33 | 15 | 51 |
WHO designated another strain of SARS-CoV-2 (B.1.1.529) as a variant of concern on November 26, 2021. This strain was named as the Omicron variant and was extensively mutated as compared to previous variants of concern. It totally includes 51 mutations, 33 of which are located on the S protein. Particularly, RDB of S protein has 15 mutations in the Omicron variant, which are listed as G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y and Y505H [12]. Some of the mutations (K417N, Q493R, N501Y, and Y505H) are among the nine key residues of S protein, which are quite important for the binding of S protein to hACE2 [13]. Therefore, it is considered that the changes in the key amino acid residues of S protein can enhance the binding ability of the virus to hACE2, resulting in a significant increase in the infectivity of the Omicron variant [14]. A model to predict the spread of the Omicron variant revealed that rate of infection of the Omicron variant will be 100 times higher than that of the Delta variant [15]. Recently, the higher infectivity of the new variant was confirmed to a degree, in which the binding affinity of RBD of the Omicron variant to hACE2 was investigated. It revealed the stronger binding affinity of RBD to hACE2 for the Omicron variant as compared other variants of concern [16, 17]. It has thus been predicted that the Omicron variant of SARS-CoV-2 is more contagious and infectious as opposed to other variants. What is more concerning is that the efficacies of monoclonal antibodies from Eli Lilly, Celltrion and Rockefeller University may be seriously diminished by the Omicron variant, while the Regeneron monoclonal antibody cocktail can be mildly impacted by Omicron variant [16]. The Omicron variant led to a widespread escape from neutralizing antibody responses due to the mutations in the Omicron variant, which diminished or substantially reduced neutralization by potent monoclonal antibodies and antibodies under commercial development [18]. The escape of Omicron variant from the antibody neutralization elicited by the Pfizer BNT162b2 mRNA vaccine was also reported [19].
Management of COVID-19 is still a huge challenge due to the emergence of variants of concern including the recent Omicron variant. Even though there are various available vaccines and treatment options, people are concerned due to the substantial reduction in the effectiveness of treatments or vaccines for Omicron variant [15]. Therefore, there is an urgent need to come up with alternative solutions for treatment and prevention of variants of concern. In this regard, the inhibition of virus entry into human cells is important target to develop alternative treatment solutions for COVID-19 [20, 21]. In this study, in silico analysis of FDA-approved drugs against the RBD of S protein of SARS-CoV-2 was conducted to discover promising drug candidates for the treatment of alarming Omicron variant. Our docking results indicated that several drugs, such as quinestrol, tamibarotene, adapalene and dihydrotachysterol, exhibited a strong binding affinity to the mutant S protein of Omicron variant. Our results were further confirmed through Molecular Dynamics (MD) simulations, in which the stability of the ligand in the binding pocket of RBD was validated. Our study suggests that these drugs, particularly adapalene, can be considered as a potent alternative anti-SARS-CoV-2 drug, especially for the Omicron variant.
Material and methods
Macromolecular antiviral target
The access of SARS-CoV-2 within the human cell is regulated by the viral S protein, which intermingles with hACE2 receptor. The involvement of S protein, hACE2, and transmembrane protease serine-2 (TMPRSS2) receptor in pathogenicity was confirmed by the investigation of viral entry mechanism. Competitive clampdown of the S protein can restrict its complexation with the hACE2 and TMPRSS2 and hence dismiss the viral entry within the human cell, thereby preventing the infection [4]. The X-ray diffraction model of the S protein of SARS-CoV-2 was obtained from the Protein Databank (PDB ID: 7BNN). The acquired model is a complex structure having the S glycoprotein complexed with N-acetyl glucosamine (NAG).
Mutations
Currently circulating SARS-CoV-2 variants have mutant S glycoproteins with a common D614G mutation together with some other mutations. Sequential investigation of the mutant forms indicated that the essential amino acids of the viral S protein's S1 subunit, which are critical in binding process with the hACE2 and TMPRSS2, were altered by mutations. The SARS-CoV-2 was highly dynamic in nature with continuous mutations for the development of newer strains. The recently reported Omicron variant was found to be highly mutated. Some of the important mutations reported in the Omicron variant were G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, Q954H, N969K, and L981F [12, 22]. These amino acid residues were among the key residues interacting with the hACE2 and TMPRSS2 receptors as well as the reported inhibitor molecules; therefore, these mutations were found to have a significant involvement in the development of drug/antibody resistance by the viral pathogen. To obtain the mutant version of S protein, the reported-mutations on the S protein of the omicron variant were introduced into the original SARS-CoV-2 S protein model by Swiss PDB Viewer (SPDBV) and the energy minimization of the mutated macromolecular model were conducted through applying AMBER force field. Energy-minimized structural model of the mutant S protein of Omicron variant was further utilized for virtual screening process [22].
Molecular docking simulation
The preparation of the mutant viral S protein structure model for docking studies was performed with the addition of the polar hydrogens, and applying Gasteiger charge for individual atoms followed by assigning an Autodock4 (AD4) atom type to every macromolecular atom. AutoDock-based docking of the viral target protein with the ligand library was performed by simulating the temperature and pressure conditions of the human body at molecular level to predict best binding conformation to reveal the strength of association between them [23, 24].
Virtual screening
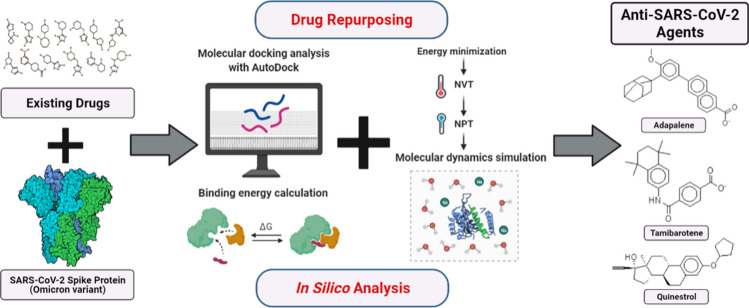
For the discovery of possible lead compounds as viral entry inhibitors, the mutant S protein of the Omicron variant was computationally screened against a ligand library containing 2890 approved drug molecules downloaded from the Zinc is Not Commercial (ZINC) database [25]. The prospective leads were determined based on the binding energy and their structure–activity relationship. The clinical utility of the shortlisted molecules was determined based on their conventional therapeutic action, and it was assured that their established therapeutic effect does not influence the normal human physiology and lead to any undesirable effects on humans while administering as an adjunctive therapy for the cure of COVID-19 [22, 24]. Figure 1 depicts the full architecture used in this study for in silico repurposing of existing drugs against the viral S protein of the Omicron variant.
Fig. 1.
Framework for in silico repurposing of existing drugs against viral S protein of the Omicron variant to temper the pathogenicity of viral infection in humans
Molecular dynamics simulation
The lead drug from the virtual screening was further marked off to shortlist tamibarotene, adapalene, dihydrotachysterol, caprofen, levocabastine, and sulindac, based on their safety profile and decent docking score to advance further into MD simulations. To validate the complex stability with respect to time, all six shortlisted leads were initially subjected to a simulation of a shorter period of 10 ns. According to the findings, the viral S protein complexed with adapalene is found to be most stable complex, which was further validated by magnified simulations for longer periods of 100 ns. The Desmond module of Maestro software was used to perform dynamics simulation of the ligand bound within a macromolecular protein at a constant temperature of 300 K. OPLS3e force field was used for the MD simulation of the macromolecular complex of ligand bound with the target receptor by solvating it in an explicit water box of size 10. The single point charge (SPC) model was used to infer the water molecules [26]. The usage of the OPLS3e force field and the SPC water model for macromolecular complexes with small ligands were previously described to yield the optimum reproducible results [27]. The complex was neutralized by adding the oppositively charged ions followed by their energy minimization [28]. The long-range electrostatic connections between the macromolecule and the complexed ligand were computed by applying the particle-mesh Ewald (PME) method with 0.8 grid spacing, and a cutoff radius of 9.0 for Coulomb interactions after the NPT ensemble MD simulation was run for 100 ns [29]. The ligand's precise binding interactions with the viral S protein were identified by using simulation interaction diagram module of the Desmond 2019–4 package.
Root-mean-square deviation (RMSD) was used to identify the atomic dislodgement of the macromolecular receptor as well as the ligand to their initial position with respect to a specific time frame during their complexation. Root-mean-square fluctuation (RMSF) for the proteineous residues was calculated in terms of their initial state as present in their crystalline model. The spreading of the secondary structure elements (SSE) within the macromolecular model like alpha-helices and beta-strands was represented as the residue index, and these structural features were supposed to be unchanged throughout the simulation. The interactions between the macromolecule and the complexed ligand during the simulation were calculated by considering four main categories, i.e., water bridges, hydrogen bonds, ionic interactions, and hydrophobic interactions. The RMSD value of the ligand molecule was computed for the whole simulation duration in relation to its beginning frame. The ligand's extended length, which corresponds to its principal moment of inertia, was measured in terms of rGyr MolSA, which corresponds to the van der Waals surface area. rGyr MolSA was calculated with a 1.4 probe radius, whereas PSA was calculated by factoring in the contribution of oxygen and nitrogen atoms [22].
Results
Macromolecular antiviral target
The X-ray diffraction model of the S protein was revealed at a resolution of 3.50 Å. The macromolecular model of the viral S protein is a trimeric form consisting of an amino acids sequence of 1287 residues in each chain complexed with a N-acetyl glucosamine (NAG). The three monomers in the structural model were interconnected via a glucopyranose moiety via the N-glycosylation process [30]. S protein is tangled in the chemical interaction with hACE2, while entering the host and the inhibition of the S protein is supposed to hinder the pathogenic entry into the host cell. Thus, the viral S protein is used for the development of viral entry inhibitors to stop the pathogenic entry within the human cells.
Virtual screening with molecular docking
Chain A of the viral S protein was kept intact and remaining chains were detached by UCSF Chimera tool to retain the monomeric subunit. Non-standard ligands were deleted to get the nascent receptor required for performing docking studies. The monomeric subunit of S protein was incorporated with a charge value of 3.0083 as per Gasteiger computation and saved in the *.pdbqt format after assigning the Autodock4 (AD4) atom type. The used grid box coordinates were x = 201.546, y = 210.946 and z = 268.993 with sizes of 60, 60 and 60 Å.
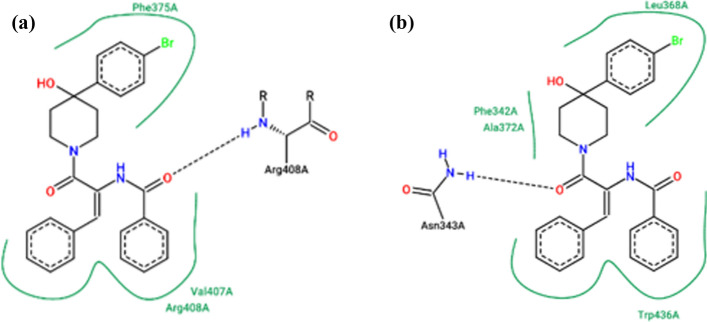
The comparative docking studies of the mutant viral S protein with respect to its non-mutant wild-type variant with an existing S protein inhibitor were performed to confirm the engagement of the mutated residues of the viral S protein with the inhibitor molecule. The engagement of the residues out of the twenty-nine mutated residues confirmed the involvement of the mutational changes in the viral S protein in the generation of the resistance against the existing viral S protein inhibitor molecules. Low binding energy of the mutated S protein against the non-mutant form of S protein strongly indicated the engagement of the mutated residues of the viral S protein in the binding of the inhibitor molecule. Molecular docking results for the ligand K22 against the mutated as well as non-mutated viral S proteins are tabulated in Table 2. Obviously, the binding affinity of K22 molecule to the S protein was significantly impaired with the mutations. The binding interactions of the mutant and wild-type monomeric subunit of the S protein are shown in Fig. 2.
Table 2.
Molecular docking results of the ligand K22 against the S protein of SARS-CoV-2
| Proteins | Interacting residues | Binding energy (kcal/mol) |
|---|---|---|
| 7BNN (Non-mutated) | Asn343, Phe342, Ala372, Leu368, Trp436 | − 8.87 |
| 7BNN (Mutated) | Phe375, Val407, Arg408 | − 4.07 |
Fig. 2.
Binding interactions of mutated monomeric subunit of the viral S protein (a) and original non-mutant variant (b) with the reported inhibitor K22
After confirming the key impact of the mutations in the S protein of the Omicron variant on the efficacy of known viral entry inhibitor, the molecular docking experiment for 2890 FDA-approved drug molecules was conducted against the mutated S protein. The top ten promising lead ligands with low binding affinities toward the mutated viral S protein are tabulated in Table 3. Also, the receptor and ligand interactions are illustrated in Figs. S1–S20 and the interacting amino acid residues are listed Table S1. Some of the interacting amino acid residues are among the specific mutations on RBD of S protein of the Omicron variant, such as Asp339, Leu371, Pro373, Lys440 and Phe375. Particularly, Lys440 was the common interacting amino acid in the mutated S protein for all shortlisted molecules, while it was not observed in the non-mutated S protein. Additional few other amino acids were interacting with the receptor, yet shortlisted molecules do not have significant differences among each other. Thus, short MD simulations were performed to find the most stable ligand-receptor complex, which was further analyzed in the longer MD simulation.
Table 3.
Binding energies of screened drug molecules attained after computational screening of a library containing 2890 approved drugs against mutated S protein of the Omicron variant
MD simulations
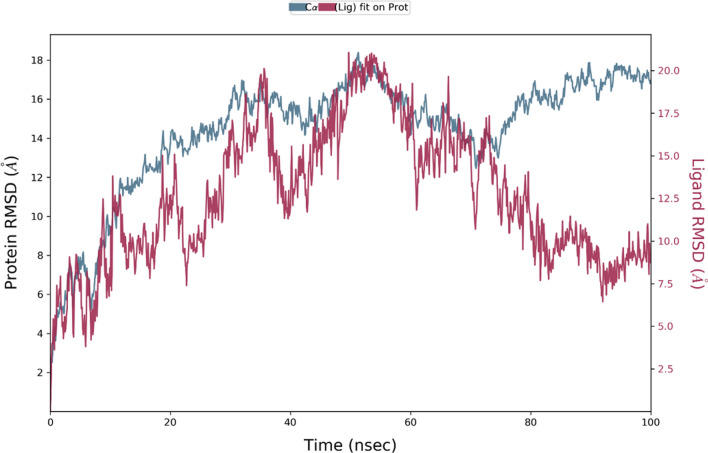
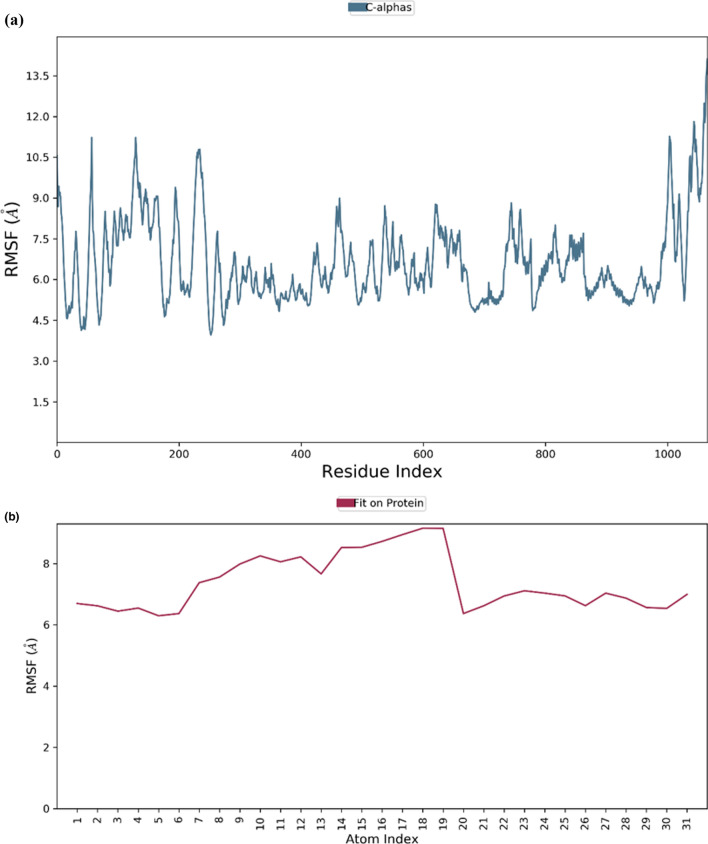
To validate the complex stability, the shortlisted molecules from molecular docking were initially subjected to MD simulations for a shorter period of 10 ns. According to the findings, the viral S protein complexed with adapalene was the most stable complex, which was further validated by magnified simulations for longer periods of 100 ns. From the MD simulations, RMSD for macromolecular residues was determined based upon the atomic selection by realigning all macromolecular frames over their stationary frame of the backbone. Based on structural validation throughout the process, RMSD analysis validates the smooth execution of the equilibrated simulation process. By aligning the heavy metals of the macromolecular binding residues, the ligand RMSD demonstrates the ligand's stability with respect to the macromolecular residues throughout the simulation. The macromolecular RMSD in this study was found to be fluctuating up to the range of 16 Å and confirmed that most of the macromolecular residues fluctuated well within the allowed limits from their initial state throughout the complexation with the ligand. Despite some early oscillations up to 10 ns through the initial adjustment of ligand within the macromolecular cavity, both ligand and the macromolecule preserved their RMSD in between 10–20 Å, signifying the effective binding of the ligand in the macromolecular cavity during the simulation. Adapalene had a number of vibrating moves after reaching the active site of the mutant S protein to attain the most stabilized confirmation within the active site. Thus, the early fluctuations in the ligand’s RMSD value till 20 ns were due to the continuous vibrations while attaining the most stabile confirmation within the active site of the macromolecule, but after 75 ns, the macromolecular backbone is supposed to achieve more stabilized conformation which has been characterized by the presence of extended RMSD values within the range of 14–18 Å. RMSDs of the protein and ligand detected during 100 ns simulation are represented in Fig. 3. RMSF value for the macromolecular residues was also recorded well within the acceptable range of 4.5–10.5 Å with an average value of 7.5 Å. Few residues were found to fluctuate somewhat outside from the given RMSF value, otherwise, the majority of the ligand residues were found to have fewer fluctuations within 6–8 Å. RMSF of the monomeric subunit of the S protein complexed with adapalene detected during MD simulation of 100 ns is represented in Fig. 4.
Fig. 3.
RMSD of the mutated S protein and complexed adapalene recorded during the MD simulation of 100 ns
Fig. 4.
RMSF of the monomeric subunit of the mutated S protein of the Omicron variant (a) and complexed adapalene (b) recorded during the MD simulation for 100 ns
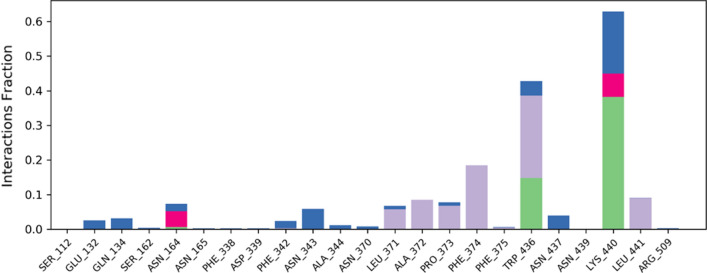
SSE analysis during whole simulation process revealed that the macromolecular structure had a total of 38.19% of SSE, out of which 16.09% of alpha-helices as well as 22.10% of beta-sheets, which may remain conserved during the simulation. The interaction analysis of macromolecular complex during the whole simulation showed that LYS440 and TRP436 interacted with the ligand during the most of the simulation time by hydrogen bond, hydrophobic interaction or via a water-bridge, while residues PHE374, LEU441, PRO373, ALA372 and LEU371 were found to only interact with the ligand via hydrophobic interactions. During the simulation, more than 3 amino acids were continually interacting with the complexed adapalene. The detailed macromolecular contacts with the complexed ligand are shown in Fig. 5. RMSD value of the complexed adapalene was well within the acceptable range of 1–3 Å, indicating the least oscillation of the adapalene during the simulation. rGyr value of the adapalene was within the range of 4.0–4.8 Å. The MolSA of the ligand was found to be within a range of 375–400 Å2 with an average value of 390 Å2 during the whole simulation process. The SASA of the ligand was in between 240 and 320 Å2 after some fluctuations. The PSA for the complexed adapalene was within a range of 70–90 Å2 throughout the simulation.
Fig. 5.
The detailed contacts observed between macromolecular complex during 100 ns MD simulation. Green-colored bars: hydrogen bonds, blue-colored bars: water bridges, purple-colored bars: hydrophobic interactions, and pink-colored bars: ionic interactions
Discussion
Pathogenic viruses are highly dynamic and transformative in nature, enabling them to achieve malleability for their enhanced survival. SARS-CoV-2 has already caused the global pandemic condition referred to as COVID-19. Since the identification of the newer viral strain accountable for the COVID-19 pandemic in 2019, the pathogenic virus has been found to be transforming itself by continuous mutational changes to increase its adaptability and pathogenicity [22]. The genome of the Omicron variant has over 50 mutations, along with more than 30 mutations in the S protein alone. Some of the identified mutational alterations in the Omicron variation had previously been detected in other variants, but never all together in one virus, and it also contains unique mutations. The mutations in the viral S protein are of great concern since it interacts with host cells for cellular entrance, which is the principal target of existing vaccinations [31]. Speculations based on observed mutations and initial observations, which should be inferred with caution, suggest that Omicron may spread faster and escape antibodies more easily than previous variants, resulting in an increase in cases of reinfection and mild breakthrough infections in vaccinated people [14]. Multiple variants of the SARS-CoV-2 have been reported till date through mutational amendments with the intent to increase their virulence in the human host [32].
Even after 2 years since the emergence of this deadly virus, numerous mutational variations were discovered in the different variants of the pathogenic genome, making it a more diverse and complex variant with respect to the initially reported original variant. Point mutations within the viral genome lead to alterations in the virus's functional and structural proteins. Changes in the protein structure (including the S protein) resulted in an enhanced virulence of the virus mainly due to more favorable penetrance into the human host cells through a stronger contact with the hACE2 [33]. Furthermore, modifications of many more viral proteins such as major protease (Mpro), RNA dependent RNA polymerase (RdRp), and RNA replicase were also found to be responsible for reduced susceptibility to medications [34]. This is attributed to the mutations that constantly arise and affect the sites of the enzymes responsible for the binding of designed pharmacological inhibitors [22].
The Omicron variant of SARS-CoV-2 is the most recent form of the virus that accumulated numerous genetic change compared to the previous ones [35]. Most of the mutations are located on the RBD of S protein, which compromises the efficacy of the current vaccines. It was also reported that the Omicron variant can diminish the efficacy of drugs and monoclonal antibodies [16, 36]. The Omicron variant was estimated to be more contagious than the previous variants of concern and in fact the current pandemic wave due to the Omicron variant proves that it is indeed more contagious than any other variants [16]. This situation indicates the urgent need for alternative drug candidates and for the new variant-adapted vaccines to treat and prevent COVID-19 due to this variant of concern.
In this study, we performed molecular modelling of the mutant S protein for the Omicron variant to compare the binding of existing spike inhibitor (K22) with respect to the original S protein. The comparative molecular docking for K22 clearly indicated the impairment of K22 against the mutated S protein of the Omicron variant. Then, the molecular docking experiments of mutant S viral protein with 2890 FDA-approved drugs as ligand library were carried out. This virtual screening was accompanied with the validation using MD simulation in order to find possible inhibitors of the Omicron S protein. Based upon the binding potential against the target macromolecule, quinestrol, tamibarotene, adapalene, dihydrotachysterol, carprofen, levocabastine, bexarotene, troglitazone, sulindac, and lutein were shortlisted as potential inhibitors for viral spike protein of Omicron variant. Out of these shortlisted leads tamibarotene, adapalene, dihydrotachysterol, caprofen, levocabastine, and sulindac were found to be safe for the human administration despite of their primary pharmacological role. Based upon the initial stability studies of these shortlisted leads, adapalene was found to be most stable compound. For instance, adapalene is a retinoic acid analog used in the treatment of acne vulgaris. It is supposed to deliver its biological action via interacting with beta and gamma subunits of retinoic acid receptor (RAR) receptors without causing any adverse effects [37].
Quinestrol is an estrogen analog used for menopausal hormonal therapy, hormonal birth control, for the treatment of breast and prostate cancers. However, its use is associated with dysregulation of hormonal homeostasis that may lead to undesirable side effects. Tamibarotene is a synthetic derivative of retinol that is used for the management of acute promyelocytic leukemia (APL). Due to its anticancer activity, it may be highly toxic and cause undesirable side effects in the normal healthy individuals. Dihydrotachysterol (DHT2) is a synthetic prodrug of vitamin D, which gets activated in the liver and does not require renal hydroxylation as that in the case of vitamin D2 and vitamin D3. Being vitamin D analog, it is not supposed to cause any undesirable side effects in the healthy individuals. Carprofen is a nonsteroidal anti-inflammatory agent (NSAID) that is used to relieve pain and reduce inflammation in patients with osteoarthritis. Carprofen may have associated side effects related to other NSAIDs in the healthy individuals. Levocabastine is a potential antiallergic agent, which selectively inhibits histaminic H1-receptor. Its use is not associated with undesirable side effects in the normal healthy individuals. Bexarotene is a retinoid analog used for the T-cell lymphoma. Troglitazone is an oral hypoglycemic agent used in treatment of diabetes and also used as an anti-inflammatory agent. Sulindac is an anticancer agent that is derived from the arylalkanoic acid class of drugs. It works via inhibition of cyclic guanosine monophosphate-phosphodiesterase (cGMP-PDE), an enzyme that is engaged in the regulation of the physiological apoptotic process. It is widely utilized for the treatment of several types of cancer. The administration of sulindac sulfoxide may block the enzyme linked with the normal apoptotic process, resulting in its disruption and the development of some adverse effects.
Adapalene, dihydrotachysterol, levocabastine and bexarotene use have been proved to be safe and well-tolerated in individuals without other comorbidities due to their selective activity toward pathogens and cancer-associated molecular targets. This limits the occurrence of undesirable adverse effect associated with their use. Therefore, their repurposing for the treatment of COVID-19 is legitimate. As a result, adapalene, dihydrotachysterol, levocabastine, and bexarotene were supposed to not interfere with human normal well-being because their pharmacological role is linked with pathogenic microbes or cancer causing polypeptides, and it was convinced that their established therapeutic role does not impede with the normal biological process and does not lead to any unwanted effect on human health during the administration as an antiviral agent to inhibit SARS-CoV-2.
We previously performed repurposing of tamibarotene against triple mutant variant of SARS-CoV-2 and found that this drug presents low binding energy and stable mode of binding for triple mutated viral S1 of S protein. The safety profile of tamibarotene with the best docking score among the screened compounds and its stability against the macromolecular target indicated that tamibarotene is a safe and effective therapeutic candidate in the potential treatment of the triple mutated SARS-CoV-2 [22]. With respect to the present findings, quinestrol was identified as one of the top molecules with the lowest binding energies for RdRp [38]. Furthermore, non-steroidal anti-inflammatory drugs (NSAIDs) were estimated as a significant source of potential Mpro inhibitors [39, 40]. Among the compounds chosen for in vitro investigation by the COVID Moonshot effort, carprofen and celecoxib were identified as potential SARS-CoV-2 Mpro inhibitors, which showed 3.97 and 11.90% inhibition at 50 µM, respectively [41]. In other study, levocabastine showed free binding energy values lesser than − 10.5 kcal/mol for the nonstructural protein (Nsp15) of SARS-CoV-2 [42]. Bexarotene has also been previously identified as a potential SARS-CoV-2 drug with the highest Cmax:EC50 ratio (1.69), which exceeded the estimated values for chloroquine, hydroxychloroquine, and ivermectin that were known for their anti-SARS-CoV-2 activity in in vitro studies or human studies involving small patient groups [43]. Moreover, bexarotene inhibited Mpro and ACE2 receptors more profoundly than hydroxychloroquine and represented superior binding mode for Mpro than remdesivir, which was considered as a potential remedy for COVID-19 and was approved by FDA for COVID-19 treatment [44]. In silico studies involving FDA approved drugs indicated troglitazone, alvesco, dihydroergotoxine and avodart as potential inhibitors of Nsp9 SARS-CoV-2 replicase [45]. Similar results were obtained using network analysis based on shared gene signatures between males and females. Identified hub proteins were used for drug repositioning studies and revealed roscovitine, curcumin, simvastatin, fulvestrant, troglitazone, alvocidib, l-alanine, tamoxifen, serine, and 2-butanone as potential drug candidates [46]. Troglitazone was also been identified as a potential drug candidate for SARS-CoV-2 in computational studies based on protein–protein topological similarities. Troglitazone, niclosamide, and chloroquine were the best negatively correlated medications that can potentially reverse the consequences of SARS-CoV-2 infection on the cell transcriptome [47]. Furthermore, Wu et al. indicated troglitazone, losartan, ergotamine, cefmenoxime, and silybin as potential ACE2 inhibitors [48]. Using omics-based methods, sulindac was found to be one of the 30 top drugs candidates for COVID-19 [49]. Sulindac, which was previously utilized as an anti-inflammatory medicine, was discovered to be phosphodiesterases (PDE5 and PDE10) inhibitor. This compound, which was shown to be effective in inhibiting cell proliferation, may also be effective in the management of COVID-19-related inflammation [50].
Conclusion
Molecular docking for the 2890 FDA-approved drug molecules against mutated S protein for the recent variant of concern of SARS-CoV-2 revealed that a few drugs such as quinestrol, adapalene, tamibarotene, dihydrotachysterol, and carprofen had strong binding affinities toward the mutant S protein. The stability of ligands in the binding site of mutant S protein was confirmed using MD simulations for further validation of in silico results. The lead compounds from this in silico drug repurposing study were further evaluated for their potential use and side effects based on the current literature. Particularly, adapalene, dihydrotachysterol, levocabastine and bexarotene came into prominence due to their non-interference with the normal physiological processes. It is therefore suggested that these approved drugs can be considered as alternative treatment options for the Omicron variant of SARS-CoV-2 after conducting basic in vitro and in vivo studies followed by pre-clinical and clinical trials.
Supplementary Information
Below is the link to the electronic supplementary material.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harvey WT, Carabelli AM, Jackson B, Gupta RK, Thomson EC, Harrison EM, Ludden C, Reeve R, Rambaut A, Consortium C-GU, Peacock SJ, Robertson DL. SARS-CoV-2 variants, spike mutations and immune escape. Nat Rev Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shang J, Wan Y, Luo C, Ye G, Geng Q, Auerbach A, Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O'Toole Á. Transmission of SARS-CoV-2 Lineage B. 1.1. 7 in England: Insights from linking epidemiological and genetic data. MedRxiv. 2021 doi: 10.1101/2020.12.30.20249034. [DOI] [Google Scholar]
- 6.Gu H, Chen Q, Yang G, He L, Fan H, Deng YQ, Wang Y, Teng Y, Zhao Z, Cui Y, Li Y, Li XF, Li J, Zhang NN, Yang X, Chen S, Guo Y, Zhao G, Wang X, Luo DY, Wang H, Yang X, Li Y, Han G, He Y, Zhou X, Geng S, Sheng X, Jiang S, Sun S, Qin CF, Zhou Y. Adaptation of SARS-CoV-2 in BALB/c mice for testing vaccine efficacy. Science. 2020;369:1603–1607. doi: 10.1126/science.abc4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wibmer CK, Ayres F, Hermanus T, Madzivhandila M, Kgagudi P, Oosthuysen B, Lambson BE, De Oliveira T, Vermeulen M, Van der Berg K. SARS-CoV-2 501Y. V2 escapes neutralization by South African COVID-19 donor plasma. Nat Med. 2021;27:622–625. doi: 10.1038/s41591-021-01285-x. [DOI] [PubMed] [Google Scholar]
- 8.Faria NR, Mellan TA, Whittaker C, Claro IM, Candido DdS, Mishra S, Crispim MA, Sales FC, Hawryluk I, McCrone JT. Genomics and epidemiology of the P. 1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCallum M, De Marco A, Lempp FA, Tortorici MA, Pinto D, Walls AC, Beltramello M, Chen A, Liu Z, Zatta F. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184(2332–2347):e2316. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Planas D, Veyer D, Baidaliuk A, Staropoli I, Guivel-Benhassine F, Rajah MM, Planchais C, Porrot F, Robillard N, Puech J, Prot M, Gallais F, Gantner P, Velay A, Le Guen J, Kassis-Chikhani N, Edriss D, Belec L, Seve A, Courtellemont L, Pere H, Hocqueloux L, Fafi-Kremer S, Prazuck T, Mouquet H, Bruel T, Simon-Loriere E, Rey FA, Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596:276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 11.Tao K, Tzou PL, Nouhin J, Gupta RK, de Oliveira T, Kosakovsky Pond SL, Fera D, Shafer RW. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat Rev Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cameroni E, Bowen JE, Rosen LE, Saliba C, Zepeda SK, Culap K, Pinto D, VanBlargan LA, De Marco A, di Iulio J, Zatta F, Kaiser H, Noack J, Farhat N, Czudnochowski N, Havenar-Daughton C, Sprouse KR, Dillen JR, Powell AE, Chen A, Maher C, Yin L, Sun D, Soriaga L, Bassi J, Silacci-Fregni C, Gustafsson C, Franko NM, Logue J, Iqbal NT, Mazzitelli I, Geffner J, Grifantini R, Chu H, Gori A, Riva A, Giannini O, Ceschi A, Ferrari P, Cippà PE, Franzetti-Pellanda A, Garzoni C, Halfmann PJ, Kawaoka Y, Hebner C, Purcell LA, Piccoli L, Pizzuto MS, Walls AC, Diamond MS, Telenti A, Virgin HW, Lanzavecchia A, Snell G, Veesler D, Corti D. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2021;602:664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karim SSA, Karim QA. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet. 2021;398:2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao S, Singh M (2021) The Newly Detected B 11 529 (Omicron) Variant of SARS-CoV-2 With Multiple Mutations: Implications for transmission, diagnostics, therapeutics, and immune evasion. J DHR 1:7–10. 10.47488/dhrp.v1iS5.35
- 16.Chen J, Wang R, Gilby NB, Wei GW (2021) Omicron (B.1.1.529): infectivity, vaccine breakthrough, and antibody resistance. https://arxiv.org/pdf/2112.01318v1.pdf [DOI] [PMC free article] [PubMed]
- 17.Ortega JT, Jastrzebska B, Rangel HR. Omicron SARS-CoV-2 variant spike protein shows an increased affinity to the human ACE2 receptor: an in silico analysis. Pathogens. 2022;11:45. doi: 10.3390/pathogens11010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dejnirattisai W, Huo J, Zhou D, Zahradnik J, Supasa P, Liu C, Duyvesteyn HME, Ginn HM, Mentzer AJ, Tuekprakhon A, Nutalai R, Wang B, Dijokaite A, Khan S, Avinoam O, Bahar M, Skelly D, Adele S, Johnson SA, Amini A, Ritter TG, Mason C, Dold C, Pan D, Assadi S, Bellass A, Omo-Dare N, Koeckerling D, Flaxman A, Jenkin D, Aley PK, Voysey M, Costa-Clemens SA, Naveca FG, Nascimento V, Nascimento F, Fernandes da Costa C, Resende PC, Pauvolid-Correa A, Siqueira MM, Baillie V, Serafin N, Kwatra G, Da Silva K, Madhi SA, Nunes MC, Malik T, Openshaw PJM, Baillie JK, Semple MG, Townsend AR, Huang KA, Tan TK, Carroll MW, Klenerman P, Barnes E, Dunachie SJ, Constantinides B, Webster H, Crook D, Pollard AJ, Lambe T, Consortium O, Consortium IC, Paterson NG, Williams MA, Hall DR, Fry EE, Mongkolsapaya J, Ren J, Schreiber G, Stuart DI, Screaton GR. SARS-CoV-2 Omicron-B11529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467–484. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cele S, Jackson L, Khan K, Khoury D, Moyo-Gwete T, Tegally H, Scheepers C, Amoako D, Karim F, Bernstein M. SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. MedRxiv. 2021 doi: 10.1101/2021.12.08.21267417. [DOI] [Google Scholar]
- 20.Gil C, Ginex T, Maestro I, Nozal V, Barrado-Gil L, Cuesta-Geijo MA, Urquiza J, Ramirez D, Alonso C, Campillo NE, Martinez A. COVID-19: drug targets and potential treatments. J Med Chem. 2020;63:12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 21.Saxena A. Drug targets for COVID-19 therapeutics: ongoing global efforts. J Biosci. 2020;45:1–24. doi: 10.1007/s12038-020-00067-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mujwar S. Computational repurposing of tamibarotene against triple mutant variant of SARS-CoV-2. Comput Biol Med. 2021;136:104748. doi: 10.1016/j.compbiomed.2021.104748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mujwar S, Deshmukh R, Harwansh RK, Gupta JK, Gour A. Drug repurposing approach for developing novel therapy against mupirocin-resistant Staphylococcus aureus. Assay Drug Dev Technol. 2019;17:298–309. doi: 10.1089/adt.2019.944. [DOI] [PubMed] [Google Scholar]
- 25.Irwin JJ, Shoichet BK. ZINC–a free database of commercially available compounds for virtual screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Toukan K, Rahman A. Molecular-dynamics study of atomic motions in water. Phys Rev B Condens Matter. 1985;31:2643–2648. doi: 10.1103/physrevb.31.2643. [DOI] [PubMed] [Google Scholar]
- 27.Gahtori J, Pant S, Srivastava HK. Modeling antimalarial and antihuman African trypanosomiasis compounds: a ligand- and structure-based approaches. Mol Divers. 2020;24:1107–1124. doi: 10.1007/s11030-019-10015-y. [DOI] [PubMed] [Google Scholar]
- 28.Posch HA, Hoover WG, Vesely FJ. Canonical dynamics of the nose oscillator: stability, order, and chaos. Phys Rev A Gen Phys. 1986;33:4253–4265. doi: 10.1103/physreva.33.4253. [DOI] [PubMed] [Google Scholar]
- 29.Petersen HG. Accuracy and efficiency of the particle mesh Ewald method. J Chem Phys. 1995;103:3668–3679. doi: 10.1063/1.470043. [DOI] [Google Scholar]
- 30.Benton DJ, Wrobel AG, Roustan C, Borg A, Xu P, Martin SR, Rosenthal PB, Skehel JJ, Gamblin SJ. The effect of the D614G substitution on the structure of the spike glycoprotein of SARS-CoV-2. Proc Natl Acad Sci U S A. 2021;118:e2022586118. doi: 10.1073/pnas.2022586118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torjesen I. Covid-19: omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ. 2021;375:n2943. doi: 10.1136/bmj.n2943. [DOI] [PubMed] [Google Scholar]
- 32.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodcroft EB, Zuber M, Nadeau S, Crawford KHD, Bloom JD, Veesler D, Vaughan TG, Comas I, Candelas FG, Stadler T, Neher RA. Emergence and spread of a SARS-CoV-2 variant through Europe in the summer of 2020. medRxiv. 2020 doi: 10.1101/2020.10.25.20219063. [DOI] [Google Scholar]
- 34.Gil C, Ginex T, Maestro I, Nozal V, Barrado-Gil L, Cuesta-Geijo MÁ, Urquiza J, Ramírez D, Alonso C, Campillo NE. COVID-19: drug targets and potential treatments. J Med Chem. 2020;63:12359–12386. doi: 10.1021/acs.jmedchem.0c00606. [DOI] [PubMed] [Google Scholar]
- 35.Callaway E, Ledford H. How bad is omicron? What scientists know so far. Nature. 2021;600:197–199. doi: 10.1038/d41586-021-03614-z. [DOI] [PubMed] [Google Scholar]
- 36.Li P, Wang Y, Lavrijsen M, Lamers MM, de Vries AC, Rottier RJ, Bruno MJ, Peppelenbosch MP, Haagmans BL, Pan Q. SARS-CoV-2 Omicron variant is highly sensitive to molnupiravir, nirmatrelvir, and the combination. Cell Res. 2022;32:322–324. doi: 10.1038/s41422-022-00618-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wishart DS, Knox C, Guo AC, Shrivastava S, Hassanali M, Stothard P, Chang Z, Woolsey J. DrugBank: a comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006;34:D668–672. doi: 10.1093/nar/gkj067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akinlalu AO, Chamundi A, Yakumbur DT, Afolayan FID, Duru IA, Arowosegbe MA, Enejoh OA. Repurposing FDA-approved drugs against multiple proteins of SARS-CoV-2: an in silico study. Sci Afr. 2021;13:e00845. doi: 10.1016/j.sciaf.2021.e00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sisakht M, Solhjoo A, Mahmoodzadeh A, Fathalipour M, Kabiri M, Sakhteman A. Potential inhibitors of the main protease of SARS-CoV-2 and modulators of arachidonic acid pathway: non-steroidal anti-inflammatory drugs against COVID-19. Comput Biol Med. 2021;136:104686. doi: 10.1016/j.compbiomed.2021.104686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abo Elmaaty A, Hamed MIA, Ismail MI, Khattab M, Al-Karmalawy AA. Computational insights on the potential of some NSAIDs for treating COVID-19: Priority set and lead optimization. Molecules. 2021;26:3772. doi: 10.3390/molecules26123772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gimeno A, Mestres-Truyol J, Ojeda-Montes MJ, Macip G, Saldivar-Espinoza B, Cereto-Massague A, Pujadas G, Garcia-Vallve S. Prediction of novel inhibitors of the main protease (M-pro) of SARS-CoV-2 through consensus docking and drug reposition. Int J Mol Sci. 2020;21:3793. doi: 10.3390/ijms21113793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sixto-Lopez Y, Martinez-Archundia M. Drug repositioning to target NSP15 protein on SARS-CoV-2 as possible COVID-19 treatment. J Comput Chem. 2021;42:897–907. doi: 10.1002/jcc.26512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan S, Chan JFW, Chik KKH, Chan CCY, Tsang JOL, Liang R, Cao J, Tang K, Chen LL, Wen K, Cai JP, Ye ZW, Lu G, Chu H, Jin DY, Yuen KY. Discovery of the FDA-approved drugs bexarotene, cetilistat, diiodohydroxyquinoline, and abiraterone as potential COVID-19 treatments with a robust two-tier screening system. Pharmacol Res. 2020;159:104960. doi: 10.1016/j.phrs.2020.104960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shahabadi N, Zendehcheshm S, Mahdavi M, Khademi F. Inhibitory activity of FDA-approved drugs cetilistat, abiraterone, diiodohydroxyquinoline, bexarotene, remdesivir, and hydroxychloroquine on COVID-19 main protease and human ACE2 receptor: a comparative in silico approach. Inform Med Unlock. 2021;26:100745. doi: 10.1016/j.imu.2021.100745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandra A, Gurjar V, Ahmed MZ, Alqahtani AS, Qamar I, Singh N. Exploring potential inhibitor of SARS-CoV2 replicase from FDA approved drugs using insilico drug discovery methods. J Biomol Struct Dyn. 2021 doi: 10.1080/07391102.2020.1871416. [DOI] [PubMed] [Google Scholar]
- 46.Shahjaman M, Rezanur Rahman M, Rabiul Auwul M. A network-based systems biology approach for identification of shared gene signatures between male and female in COVID-19 datasets. Inform Med Unlocked. 2021;25:100702. doi: 10.1016/j.imu.2021.100702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pérez-Moraga R, Forés-Martos J, Suay-García B, Duval J-L, Falcó A, Climent J. A COVID-19 drug repurposing strategy through quantitative homological similarities using a topological data analysis-based framework. Pharmaceutics. 2021;13:488. doi: 10.3390/pharmaceutics13040488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, Wang Q, Xu Y, Li M, Li X, Zheng M, Chen L, Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barh D, Tiwari S, Weener ME, Azevedo V, Goes-Neto A, Gromiha MM, Ghosh P. Multi-omics-based identification of SARS-CoV-2 infection biology and candidate drugs against COVID-19. Comput Biol Med. 2020;126:104051. doi: 10.1016/j.compbiomed.2020.104051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giorgi M, Cardarelli S, Ragusa F, Saliola M, Biagioni S, Poiana G, Naro F, Massimi M. Phosphodiesterase inhibitors: could they be beneficial for the treatment of COVID-19? Int J Mol Sci. 2020;21:5338. doi: 10.3390/ijms21155338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.