Self-assembling chimeric polypeptide-doxorubicin conjugate nanoparticles that abolish tumors after a single injection (original) (raw)
. Author manuscript; available in PMC: 2010 Jun 1.
Published in final edited form as: Nat Mater. 2009 Nov 8;8(12):993–999. doi: 10.1038/nmat2569
Abstract
New strategies to self-assemble biocompatible materials into nanoscale, drug-loaded packages with improved therapeutic efficacy are needed for nanomedicine. To address this need, we developed artificial recombinant chimeric polypeptides (CPs) that spontaneously self-assemble into sub-100 nm size, near monodisperse nanoparticles upon conjugation of diverse hydrophobic molecules, including chemotherapeutics. These CPs consist of a biodegradable polypeptide that is attached to a short Cys-rich segment. Covalent modification of the Cys residues with a structurally diverse set of hydrophobic small molecules, including chemotherapeutics leads to spontaneous formation of nanoparticles over a range of CP compositions and molecular weights. When used to deliver chemotherapeutics to a murine cancer model, CP nanoparticles have a four-fold higher maximum tolerated dose than free drug, and induce nearly complete tumor regression after a single dose. This simple strategy can promote co-assembly of drugs, imaging agents, and targeting moieties into multifunctional nanomedicines.
Keywords: biomolecular engineering, elastin-like polypeptide, nanoparticle, drug delivery, ELP, nanomedicine, chimeric polypeptide
Text
Packaging clinically approved drugs into nanoscale delivery vehicles (10-100 nm diameter)1-3 is of particular interest for cancer therapy, as numerous studies have shown that objects within this size range accumulate within solid tumors due to the enhanced permeability and retention (EPR) effect, which results from abnormalities of tumor blood and lymphatic vasculature4. In our view, drug-loaded nanoparticles for cancer drug delivery should: (1) be easy to synthesize in a few steps with high yield and purity; (2) self-assemble into monodisperse drug-loaded nanoparticles with a size below 100 nm; (3) allow encapsulation of diverse drugs; (5) exhibit favorable pharmacokinetics and tumor accumulation; (6) release the drug with controlled and tunable kinetics; (7) lead to a therapeutic response; and (8) degrade into non-toxic components to enable clearance from the body without adverse toxicity. Although a number of different nanoscale delivery systems have been proposed for cancer therapy5, most do not satisfy these criteria, which are critical to move these systems into clinical practice.
Motivated by this rationale, we report herein the first example of chimeric polypeptides (CPs) that self-assemble into near-monodisperse, sub-100 nm size nanoparticles upon drug attachment, and which are biodegradable and display good pharmacokinetics and tumor accumulation, low toxicity, and excellent in vivo efficacy in a murine tumor model. The CPs consist of two segments, a hydrophilic, biodegradable elastin-like polypeptide (ELP) and a short segment for the attachment of drugs including a cancer chemotherapeutic –Doxorubicin (Dox)– through a pH-labile linker (Fig. 1a). ELPs are a class of artificial peptide polymers composed of a Val-Pro-Gly-Xaa-Gly repeat derived from human tropoelastin, where the “guest residue,” Xaa, can be any mixture of amino acids except proline6. We chose ELPs as one of the segments of the CP for multiple reasons. First, ELPs undergo an inverse phase transition in aqueous solutions at a characteristic transition temperature (Tt), above which they desolvate and phase separate from bulk water7. For recombinant ELP block copolymers, this phase transition behavior promotes self-assembly into nanostructures, driven by selective desolvation of one block8-10. These observations led us to hypothesize that the attachment of multiple copies of a hydrophobic agent, such as Dox or other hydrophobic moieties, would impart sufficient amphiphilicity to the polypeptide to drive its self-assembly into nanoparticles9, 10. Second, ELPs are useful biopolymers, being non-toxic11, 12, biodegradable, and displaying good pharmacokinetics13. Third, because ELPs can be produced via genetic engineering, their composition, MW, and polydispersity can be precisely controlled. Fourth, ELPs can be produced with high yield (~100-200 mg/L) in E. coli, and can be easily and rapidly purified by exploiting their phase transition behavior14, so that high-purity, clinical grade material is easily and cheaply obtained. As one component of the CP system, these attributes of ELPs satisfy many of the proposed requirements for a nanoscale drug carrier.
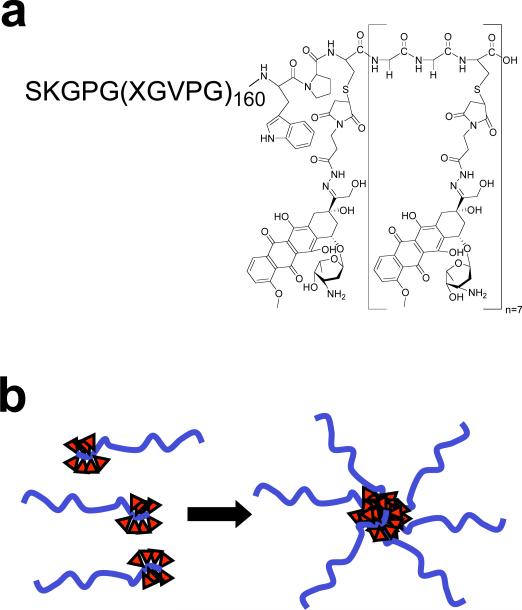
Figure 1. Structure of CP-Dox conjugate.
a) High molecular weight ELPs were synthesized by genetically encoded biosynthesis and conjugated to Dox at Cys residues via a heterobifunctional linker. b) The hydrophobic-drug block triggers self-assembly of chimeric polypeptide (CP) nanoparticles with a drug ( ) rich core surrounded by a hydrophilic polypeptide corona.
) rich core surrounded by a hydrophilic polypeptide corona.
To ensure favorable pharmacokinetics, the predominant CP described here was engineered to have 160 pentameric repeats where the guest residues Xaa = Val:Ala:Gly [1:8:7]. This CP is a hydrophilic polymer (MW=62.6 kD) with a Tt >> 37 °C (Supplementary Fig. 1) so that it exhibits high solubility at body temperature, has long plasma circulation as seen by its area under the concentration-time curve (AUC)13, and degrades in serum at the rate of 2.5 weight % day-1 13. A second, shorter (Gly-Gly-Cys)8 segment was appended at the C-terminal end of the CP to provide drug attachment sites and impart sufficient amphiphilicity to the polymer. This segment provides eight drug attachment points –unique Cys residues– that are clustered at the end of the CP, with embedded diglycine spacers between the attachment sites to minimize steric hindrance during drug conjugation.
The CP was over-expressed from a plasmid-borne synthetic gene in E. coli using shaker flask cultures and purified with a yield of > 100 mg L-1 from bacterial lysate by inverse transition cycling (ITC), a simple, non-chromatographic method15. 5 rounds of ITC provided a monodisperse product with > 95% purity, as verified by SDS-PAGE (Supplementary Fig. 1) and MALDI-MS (Supplementary Table 1). To activate Dox for conjugation, it was reacted with n-ß-maleimidopropionic acid hydrazide tri-fluoroacetic acid to incorporate an internal, acid labile hydrazone moiety with a terminal maleimide16, 17, and this activated drug was covalently attached to the Cys residues of the CP (Fig. 1a). Purified CP-Dox had 4.8 ± 1.3 (SD, n=3) drug molecules per polypeptide (Supplementary Table 1).
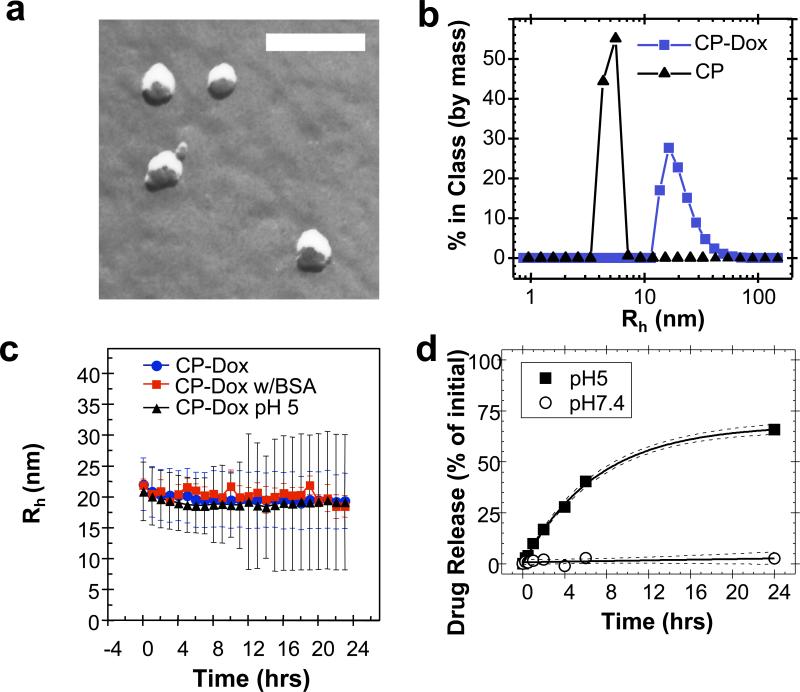
Results of both transmission electron microscopy (TEM) and dynamic light scattering (DLS) confirmed the spontaneous assembly of CP-Dox into nanoparticles (Fig. 2). Spherical structures of the CP-Dox nanoparticles were observed by freeze-fracture TEM (Fig. 2a) to have a mean particle radius of 19.3 ± 0.9 nm (SD, n=67; 25th, 50th, 75th percentile= 14, 18, 22 nm). DLS measurements further confirmed the narrow size distribution of CP-Dox nanoparticles with a mean hydrodynamic radius, Rh, of 21.1 ± 1.5 nm (SD, n=11; 25th, 50th, 75th percentile = 14.3, 16.9, 21.1 nm) (Fig. 2b). In contrast, unmodified CPs have a Rh of 5.5 ± 0.9 nm (SD, n=6; 25th, 50th, 75th % = 3.9, 4.5, 5.0 nm). DLS results also showed that CP-Dox nanoparticles have a critical aggregation concentration (CAC) below 3 μM CP (14.4 μM Dox Equivalents (Equiv), Supplementary Fig. 1).
Figure 2. Characterization of CP-Dox nanoparticles.
a) Freeze fracture transmission electron microscopy image of CP-Dox nanoparticles (bar = 200 nm). b) Distribution of hydrodynamic radii for CP and CP-Dox nanoparticles at 25 μM in PBS at 37 °C by dynamic light scattering (DLS). c) CP-Dox nanoparticles have a stable hydrodynamic radius over time in PBS at pH 7.4, PBS with 0.1 mM BSA, and in buffer at pH 5.0; however, the distribution of hydrodynamic radii (error bars) increases over time at low pH. (Mean ± SD). d) The kinetics of particle diameter broadening are in agreement with the pH dependent release of Dox from CP-Dox nanoparticles as determined using size exclusion chromatography at pH 7.4 and 5.0. The fit line (solid) and 95% confidence interval lines (dash) are provided.
The low CAC of the CP-Dox conjugate suggests that it will circulate as self-assembled nanoparticles in vivo for an extended period of time. This is because the maximum tolerated dose (MTD) of the CP-Dox nanoparticles in mice is 20 mg Dox Equiv kg-1 body weight (BW) (Supplementary information), which corresponds to a plasma drug concentration of ~600 μM Dox upon injection. The conjugate is hence well above the CAC after i.v. injection. We also observed that the nanoparticles were stable in size and polydispersity at pH 7.4 over a 24 h period, both in the presence and absence of bovine serum albumin (Fig. 2c) indicating that they should exist in systemic circulation as nanoparticles for a significant period of time. Furthermore, the ~40 nm diameter of these nanoparticles is below the pore size of the permeable vasculature found in many solid tumors, suggesting that CP-Dox nanoparticles should be able to selectively accumulate in a solid tumor via the EPR effect1, 3, 4. In contrast, at pH 5.0, the polydispersity of CP-Dox nanoparticles increased with time, which is suggestive of the release of Dox from the CP-Dox conjugate via acid-catalyzed cleavage of the hydrazone linkages, and subsequent dis-assembly of the nanoparticles (Fig. 2c).
The formation of nanoparticles is not restricted to the conjugation of multiple copies of Dox to the CP. It is notable that the attachment of five structurally diverse small molecules (Supplementary Fig. 2) to CPs also triggered their spontaneous self-assembly into near monodisperse nanoparticles with a size range (Rh = 11.2 to 41.7 nm) (Supplementary Table 2). Furthermore, this behavior is not restricted to a single CP, and five different CPs with a range of MWs and compositions also self-assemble into nanoparticles upon conjugation to Dox (Supplementary Table 2). The self-assembly of nanoparticles upon conjugation of multiple copies of diverse hydrophobic small molecules suggests that the attachment-triggered self-assembly of CPs is potentially a robust and general principle for the formation of nanoparticles with a core that sequesters hydrophobic drugs and a corona comprised of hydrophilic polypeptide chains (Fig. 1b).
The liberation of free drug from the CP-Dox nanoparticles requires the pH-dependent cleavage of the hydrazone linkage (Fig. 1a). To verify the kinetics of drug release, CP-Dox nanoparticles were incubated either at pH 7.4 or pH 5.0 at 37 °C for 24 h (Fig. 2d). The release of free drug was monitored by size exclusion high performance liquid chromatography. At pH 7.4, the hydrazone linkage was stable, and no significant release of free drug was observed over 24 h. In contrast, at pH 5.0, free drug was generated with a first order half-life of 4.9 ± 0.5 h (CI 95%) and reached a maximum release of 68 ± 3 % (CI 95%) of the initial amount of Dox (Supplementary Table 1), similar to the levels observed with other polymeric hydrazones18. These data confirm that the covalent linkage to Dox is exceptionally stable at the pH of blood, but that the linkage cleaves at an appreciable rate at a pH that is relevant to endo-lysosomal trafficking. Because CP-Dox conjugates have previously been demonstrated to localize within low pH compartments in cellular uptake assays19, the pH dependent release of Dox from the CP-Dox conjugate suggests endo-lysosomal release of the drug following cellular uptake of the CP-Dox nanoparticles.
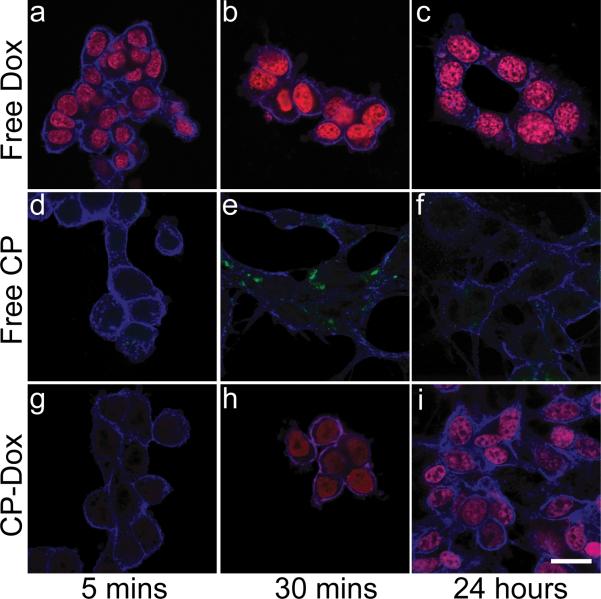
To explore the mechanism of CP-Dox internalization and intracellular drug delivery, cells were observed by laser scanning confocal fluorescence microscopy at various time points after incubation with free Dox (Fig. 3a-c), free CP (Fig. 3d-f), and CP-Dox (Fig. 3g-i). Free Dox rapidly accumulated in the nucleus after incubation for 5 min (Fig. 3a). To observe the cellular fate of free CP, the polypeptide was labeled with Oregon Green via a pH-insensitive thio-ether linkage (Supplementary Table 2). CP-Oregon Green led to intense punctate fluorescence in the cytoplasm after 30 min (Fig. 3e), suggesting the localization of the CP to endosomes and lysosomes. In contrast, Dox delivered via CP nanoparticles showed intermediate behavior between free Dox and free CP. There was very low accumulation of Dox in the nucleus at 5 minutes following incubation with CP-Dox, indicative of the slower uptake of CP-Dox compared to free Dox; however, there was significant accumulation of Dox within the nucleus 30 min after incubation with CP-Dox (Fig. 3h). These results suggest that Dox accumulation in the nucleus after incubation of cells with CP-Dox is a result of cellular uptake of CP-Dox nanoparticles, intracellular drug release, and subsequent trafficking of drug to the nucleus.
Figure. 3. Internalization of CP-Dox and delivery of drug to the nucleus.
Confocal laser scanning microscopy images through C26 cells (blue) that show cellular uptake for free Dox, free CP-Oregon Green, and CP-Dox at 5 mins, 30 mins, and 24 hrs. a-c) Free Dox (red) rapidly localizes to the nucleus at all time points. d-f) CP-Oregon Green (green) internalizes into the cytoplasm with intense punctate staining after 30 mins. g-i) CP-Dox (red) nanoparticles produce intense nuclear staining with drug after 30 mins. (bar = 20 μm).
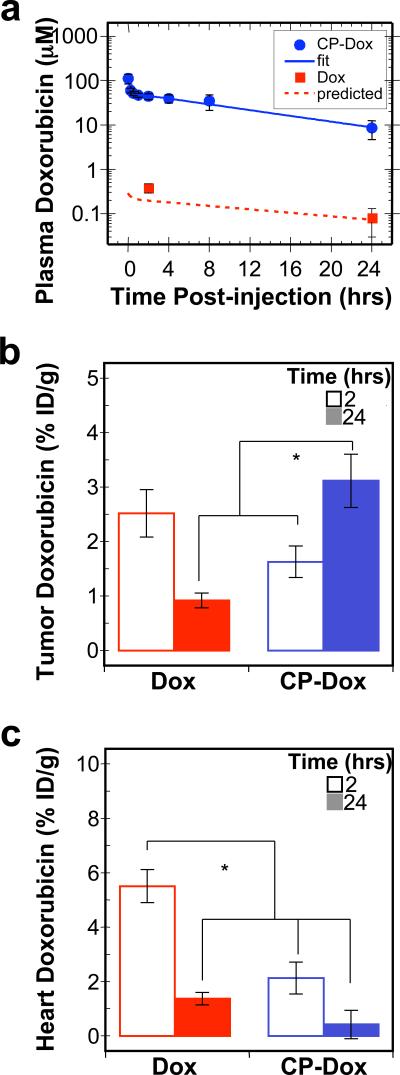
To evaluate the behavior of CP-Dox nanoparticles in mice, the conjugates were administered systemically and the plasma drug concentration was measured as a function of time post-injection. The data were fit to a two compartment pharmacokinetic model (Supplementary information), yielding a terminal half-life of 9.3 ± 2.1 hr (95% CI) and a plasma AUC of 716 ± 139 μM hr (95% CI) (Fig. 4a, Supplementary Table 3). The AUC for a mouse treated with the same dose of free Dox is only 4.7 μM hr20. Based on this significant increase in plasma AUC, these results suggested that CP-Dox nanoparticles are likely to preferentially accumulate in solid tumors as compared to free drug.
Figure 4. Plasma pharmacokinetics and tissue biodistribution.
a) Plasma Dox concentrations as a function of time post-injection. A two-compartment model was fit to the plasma Dox concentration, which yielded a terminal half-life of 9.3 ± 2.1 hrs (CI 95%) for CP-Dox. The concentration of free Dox has been confirmed at two time points experimentally; furthermore, the observed concentrations correlate with the prediction of a pharmacokinetic model of free Dox in mice (Supplementary Table 3)20. (Mean ± 95% CI; n=5-8). b-c) The Dox concentration in b) tumor tissue and c) heart tissue at 2 and 24 hours post-administration was determined. *indicates p<0.0005 (ANOVA, Tukey HSD). (Mean ± SD; n=4 to 6).
Motivated by these findings, we next evaluated tissue exposure to Dox. Mice were administered free drug or CP-Dox nanoparticles, and tissues samples were obtained after 2 or 24 h (Fig. 4b, c). Notably, 24 h after administration of CP-Dox, the tumor had a significant 3.5 fold increase in drug concentration as compared to free drug at the same dose (Fig. 4b) (Tukey HSD; p=2×10-6). Equally important, the CP-Dox nanoparticles significantly reduced drug concentration at several non-tumor sites in the body, including the muscle, the lung (Supplementary Fig. 4), and the heart (Fig. 4c). In particular, the incorporation of Dox into CP nanoparticles decreased the peak concentration in the heart by 2.6 fold compared to free Dox (Tukey HSD; p=1×10-6). The decreased accumulation in the heart is notable, as cardiomyopathy is the dose-limiting side effect of free Dox21. The increase in tumor exposure and decrease in heart exposure is one mechanism which explains why the MTD of CP-Dox nanoparticles is higher than that of free Dox (Supplementary Fig. 4, Table 4), and suggested that CP-Dox encapsulation may improve the therapeutic index of Dox.
To compare the therapeutic effect of free Dox and CP-Dox nanoparticles, both formulations were administered in a dose escalation study to determine their MTD. The MTD of Dox was 5 mg kg-1 BW and the MTD for CP-Dox was 20 mg Dox Equiv kg-1 BW (Supplementary Fig. 4). Thus, by administration of the MTD of CP-Dox nanoparticles, which is four-fold greater than the MTD of free drug, it is potentially possible to increase the absolute concentration of drug in the tumor at 24 hours by an estimated 4 × 3.5 = 14 fold over free drug.
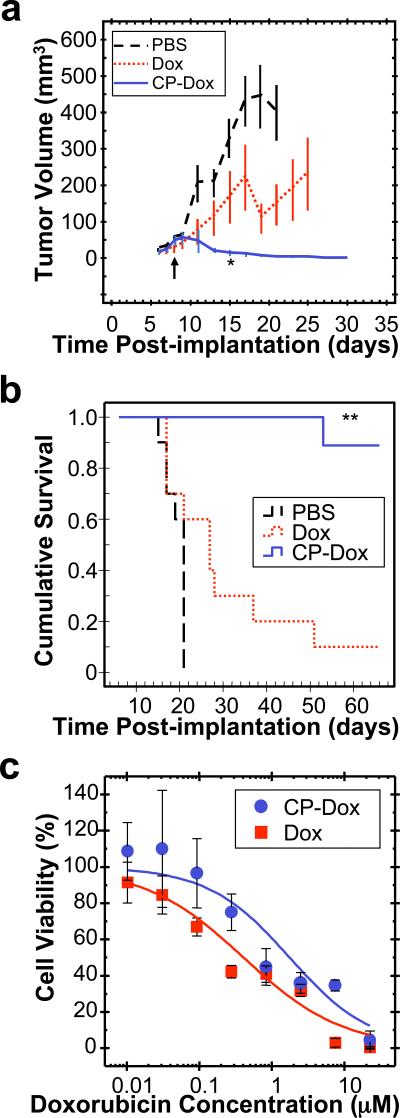
CP-Dox nanoparticles were next evaluated for their anti-tumor activity at their MTD. Mice with 8-day old C26 tumors (25th, 50th, 75th percentile = 17, 21, 66 mm3 tumor volume) were treated with PBS, Dox, or CP-Dox nanoparticles (Fig. 5a). 15 days after tumor implantation, CP-Dox treated mice had a mean tumor volume of 13 mm3 (n=9) vs. 329 mm3 (n=10) for PBS (Mann-Whitney; p=0.00002) and vs. 166 mm3 (n=10) for free drug (Mann-Whitney; p=0.03) treated controls. Clearly, the CP-Dox formulation at MTD outperforms free drug in reducing tumor volume, which correlated with a substantial increase in animal survival (Fig. 5b). The median survival time for mice treated with PBS (n=10) was 21 days, and treatment with Dox (n=10) slightly increased this survival to 27 days (Kaplan-Meier, p=0.03). In contrast, the CP-Dox nanoparticles cured 8 of 9 mice for up to 66 days after tumor implantation, a significant improvement over free drug (Kaplan-Meier, p=0.0002). Thus, with only a single dose injection, CP-Dox nanoparticles provide a substantial curative effect.
Figure 5. Anti-tumor activity of CP-Dox nanoparticles.
a-b)Tumors cells (C26) were implanted subcutaneously on day zero. Mice were treated on day 8 (↑) at MTD with PBS (n=10), free Dox (5 mg kg-1 BW; n=10), and CP-Dox (20 mg Dox Equiv kg-1 BW; n=9). a) Tumor volume up to day 30 (Mean ± SD; n=6 to 10). *indicates p= 0.03, 0.00002 for CP-Dox vs. Dox and PBS (day 15) respectively (Mann Whitney). b) Cumulative survival of mice **indicates p=0.0001, 0.00004 for CP-Dox vs. Dox and PBS respectively (Kaplan Meier). c) Cell viability for CP-Dox (n=4; IC50 =1.8 μM) and free Dox (n=14; IC50 =0.41 μM) in C26 cells. (Mean ± 95% CI).
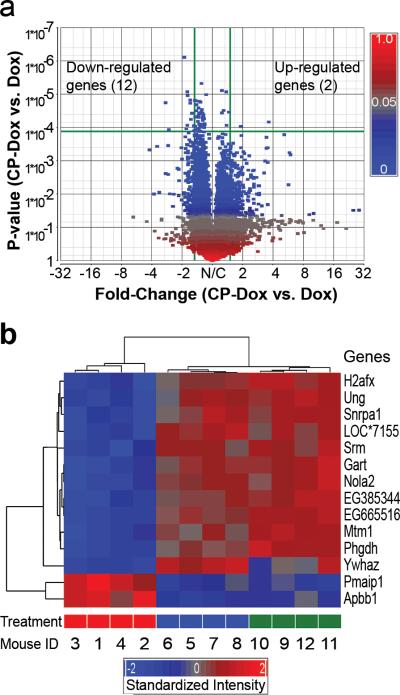
To explore the mechanism by which CP-Dox nanoparticles outperform the free drug, we next compared the genomic profiles for tumors obtained from mice administered with PBS, free Dox or CP-Dox, two days after treatment. The comparison between CP-Dox and PBS produced many differences, and the detailed analysis of these results exceeds the scope of this paper and will be reported elsewhere. Instead, we focused on comparison of the gene array data between CP-Dox and free Dox to elucidate any potential differences in the molecular mechanism(s) of tumor cell death elicited by the CP-Dox nanoparticles. We identified 14 genes that differed significantly (p<0.00012) between CP-Dox and free Dox given at the MTD (Fig. 6a). A hierarchical clustering analysis (Fig. 6b) was performed whereby genes were linked together according to their expression patterns (dendrogram on left) and individual mice were clustered (dendrogram on top). For all mice, the clustering results matched their treatment, suggesting that these genes may be useful biomarkers to indicate tumor responses to CP nanoparticles (Fig. 6b).
Figure 6. Genomic profiles of CP-Dox and free Dox treated tumor tissues.
a) Volcano plot of gene expression contrasting CP-Dox treated mice vs. Dox treated mice. Expression level is denoted by a square colored to reflect the P-value. The horizontal green line indicates the cutoff of P-value (P<0.00012), which corrects for multiple comparisons. The vertical green lines indicate the cutoff for significant fold change (>1.5 fold). In CP-Dox treated mice, 12 genes were significantly down-regulated, and 2 genes were up-regulated in contrast to Dox treated mice. b) A hierarchical clustering analysis was performed based on the expression pattern. The genes were linked together according to their expression patterns (dendrogram on left). Individual mice were also clustered (dendrogram on top). For all mice, the clustering results match their treatment: CP-Dox (red bar); Dox (blue bar); and PBS (green bar). The gene expression intensities were standardized between -2.0 (blue) and 2.0 (red). LOC*7155 is an abbreviation of an untitled gene LOC100047155.
Unlike either free Dox or PBS, the CP-Dox formulation up-regulated programmed cell death and negative regulators of the cell cycle (Supplementary Table 5). A primary mechanism of Dox cytotoxicity is mediated by topoisomerase-II, which cleaves genomic DNA, allows adjacent DNA strands to crossover, relieving rotational strain, and permits the successful repair of breakages. Dox intercalates with DNA and freezes topoisomerase-II to the ends of freshly cut DNA, which leads to permanent DNA damage and exit from the cell cycle22. While numerous genes are involved in DNA repair23, this analysis identified one particular gene, uracil-DNA glycosylase (Ung), that is down-regulated only by CP-Dox treatment (Fig. 6b); furthermore, Ung is associated with resistance to chemotherapeutics, including Dox24, 25. By down-regulating DNA repair, CP-Dox may directly enhance the antitumor drug effect. These data provide mechanistic insights into CP nanoparticle delivery and support the contention that CP nanoparticles may overcome drug resistance, as has been suggested for other drug delivery systems26-28.
The closest analog to the present system are monoblock PEG-Dox conjugates that appear to form micelles, but their self-assembly was only explored with a single hydrophobic drug (Dox), so it remains unclear whether this approach will apply to other hydrophobic drugs and polymers29. Furthermore, in comparison with CP-Dox, the PEG-Dox conjugate displayed limited efficacy in extending survival time and did not produce long-term cures in an animal model. Other methods to create drug loaded polymer nanoparticles have relied upon: (1) the chemical synthesis of amphiphilic PEG-based biohybrid block copolymers to encapsulate the drug18, 30-32; (2) the synthesis of complex terpolymers in which the co-polymerization or attachment of fatty acids or cholesterol to the polymer chain drives its self-assembly33, 34; or (3) the design of sophisticated and complex dendrimer architectures3. While all of these approaches have their merits, they are far more complicated than the genetically encoded delivery system described herein, as they rely upon complex, multi-step synthesis of polymers, which can be difficult to scale-up. Many synthetic polymeric vehicles are not degradable, nor have they been shown to self-assemble across a broad range of hydrophobic molecules. A high degree of pharmaceutical complexity typifies many methods of nano-encapsulation. For example, methods that rely on physical encapsulation and size fractionation of nanoparticles, such as polymer micelles3, liposomes1, and emulsion polymerization35, are highly multicomponent systems (i.e. drugs, buffers, mixtures of amphiphiles, monomers, solvents, and excipients), which can hinder pharmaceutical viability.
In contrast, the CP nanoparticle system is, we believe, the first example of a simple and rationally designed recombinant polypeptide that is extraordinarily efficient to synthesize and purify from E. coli and self-assembles into nearly monodisperse sub-100 nm nanoparticles in water, thereby obviating the need for organic solvents during processing. This attachment-triggered self-assembly of CPs is conserved across a range of small hydrophobic molecules, which further distinguishes it from previous studies with synthetic polymers that are restricted to one or a few hydrophobic molecules (Supplementary Table 2). In addition, the degree of drug loading and particle diameter is conveniently controlled at the design level by the number of unique reactive sites that are appended to chain termini of a CP and its MW, orthogonal variables that are precisely and trivially encoded into the gene-level design of the polymer. Also, attachment of drug solely at the chain end ensures that the drug is sequestered in the nanoparticles’ core– unlike other nanoparticle drug carriers, such as dendrimers3, metal nanoparticles36, or carbon nanotubes37. The ability to deterministically place the drug molecules at the end of the polymer chain also differentiates this system from many protein and polymer conjugates where the reactive sites are distributed along the polymer chain or protein surface. Such arrangements leave the drug exposed to the solution environment and do not necessarily impart amphiphilic self-assembly of a nanostructures2, 3, 38. In common with other peptide-drug conjugates2, our approach permits control over release (rate, mechanism, in vivo location) via the design of linker chemistry. For example, the linker described here contains an internal, acid-labile hydrazone bond3, 16, 17 that triggers intracellular drug release in endosomes and lysosomes. Similarly, peptide linkers that are substrates for tumor specific proteases (cathepsins and matrix metalloproteases) may enable preferential release in the tumor2, 39. Both approaches can be easily incorporated into a CP via recombinant DNA methodology.
In conclusion, the recombinant CP nanoparticles described herein, hence present four important points of novelty: (1) Generality: the assembly of these polypeptides into near monodisperse nanoparticles is conserved across a range of small molecules and polypeptides (Supplementary Table 2); (2) Genetically encoded synthesis: the use of recombinant DNA methodology provides a trivial mechanism for controlling particle diameter, degree of drug loading, and incorporation of other biologically active peptides; (3) Simplicity: these self-assembling subunits result from a simple two-step synthesis consisting of polypeptide biosynthesis followed by covalent drug conjugation; and (4) Biodegradability: these polypeptides may be used at molecular weights above the renal filtration cutoff, beyond which non-biodegradable polymers undergo chronic accumulation. We believe that this combination of features make the CP formulation uniquely attractive for the development of advanced nanoparticle drug carriers.
Methods
CPs were synthesized by heterologous expression of a plasmid-borne synthetic gene in E. coli as described previously14. Dox (MW= 580 g mol-1) was covalently linked to cysteine residues on CPs (Supplementary information). The molar extinction coefficient for Dox was determined in PBS at 495 nm (ε = 1.00×104 M-1 cm-1) and used to determine drug concentration.
Dynamic light scattering was used to measure particle radius at 37 °C after filtration through an Anotop™ syringe filter with 0.1 μm size pores (Whatman; Florham Park, NJ) using a DynaPro™ Plate Reader (Wyatt Technology; Santa Barbara, CA). Freeze fracture electron microscopy (TEM) was also used to estimate particle radius on a JEOL 100 CX electron microscope (Nano Analytical Laboratory; San Francisco, CA). The radii for particles were analyzed using ImageJ 1.34i (NIH; Bethesda, MD) using the measurement tool.
High performance liquid chromatography (HPLC) was used to determine the fraction of drug remaining bound using a LC10 HPLC (Shimadzu Scientific Instruments; Columbia, MD) and a Shodex OHPak KB-804 column (New York, NY). To assay for the release of drug, samples of CP-Dox in PBS (10 mM NaH2 PO4, 140 mM NaCl, pH 7.4) were diluted into either pH 5.0 (0.1 M Na acetate) or pH 7.4 (0.1 M NaH2 PO4). Samples were incubated at 37 °C, quenched at pH 7.4, and measured for free Dox at 14.7 min (CP-Dox eluted at 8.8 min). The percent of drug released, F %,released was fit to a first order model:
| F%,released=a[1−e(−ln(2)tt1∕2)] | (1) |
|---|
where t is the time after incubation, t1/2 is the half-life of release, and a is the maximum cleavage.
A murine colon carcinoma cell line, C26, was used to evaluate the potency of Dox both in vitro and in vivo. I_n vitro_ cytotoxicity was determined in a 96-well format, where cells were incubated for 1 day before addition of drug. Blank wells and PBS-treated wells were defined as 0 and 100% viability. Dilutions of drug were incubated with cells for 3 days. 20 μL of CellTiter 96 AQueous™ (Promega; Madison, WI) reagent was added to each well (120 μL total) and incubated before measurement. The 50% inhibitory concentration, IC50, was determined by fitting to the following equation:
| V%=100%/[1+(CDoxIC50)p] | (2) |
|---|
where V% is the viability, CDox is the Dox concentration, and p defines the slope of the curve.
To study cellular uptake, cells were seeded in 8-well Lab-Tek II chamber Slides (Thermo Fisher Scientific; Rochester, NY) at the density of 8×104 cells per chamber and allowed to attach overnight. Cells were then treated with free Dox, CP-Oregon Green, or CP-Dox at the concentration of 40 μM Equiv of Dox or Oregon green at 37 °C for different time periods. After treatment, the cells were washed with Dulbecco's Phosphate Buffered Saline (DPBS), fixed in/ 2% paraformaldehyde, stained with Alexa Fluor 594 Wheat germ agglutinin (W11262; Invitrogen; Carlsbad, CA), washed with DPBS 5 times, and imaged using an LSM5 upright laser-scanning confocal microscope (Zeiss; Oberkochen, Germany) with an 100X oil-immersion objective. The following wavelengths were used: excitation at 545 nm and detection through a 595 nm high-pass filter for W11262, excitation at 488 nm and detection through a 505 - 550 nm band-pass filter for Oregon Green, and excitation at 488 nm and detection through a 560 nm low-pass filter for Dox.
To evaluate the in vivo activity of CP-Dox, animals were treated in accordance with National Institute of Health Guide for the Care and Use of Laboratory Animals as approved by the Duke University Institutional Animal Care and Use Committee. A Balb/C mouse tumor model was developed by subcutaneous injection of 2.5-5×105 cells giving > 95% tumor-take. For the pharmacokinetic study, CP-Dox was administered systemically (5 mg Dox Equiv kg-1 BW) and sampled via the tail vein, and the plasma concentration of CP-Dox was fit to a two-compartment pharmacokinetic model using SAAMII™ (University of Washington, Seattle, WA). For the biodistribution study, drug was administered systemically 8 days after implantation. At 2 and 24 hrs post-administration, tissue samples were assayed for drug concentration using calibrated fluorometry (Supplementary information). Biodistribution data were compared using ANOVA followed by Tukey Honestly Significant Differences. In studies of therapeutic efficacy, mice (6-8 weeks old) bearing dorsal tumors were treated 8 days after implantation. Tumor dimensions and body weight (BW) were determined 3-4 times a week, and the tumor volume was calculated:
| Volume=length×width2×π∕6 | (3) |
|---|
Mice exhibiting more than 15% BW loss or tumors greater than 1,000 mm3 were euthanized Tumor volumes were compared using a Kruskal-Wallis test followed by Mann-Whitney test. Cumulative survival curves were compared using Kaplan-Meier analysis followed by the Log Rank Test. Statistics were calculated using SPSS™ 15.0 (Chicago, IL).
Supplementary Material
1
Acknowledgements
This work was supported with NIH grant 5F32-CA-123889 to JA MacKay and NIH grant R01-EB-00188 to A Chilkoti. We thank M Dewhirst, M Zalutsky, and M Dreher for advice regarding experimental design and analysis. We thank Prof. FC Szoka Jr, for the use of C26 cells and B Papahadjopoulos-Sternberg for preparation of freeze-fracture electron microscopy images. We thank S Morales, M Schneiderman, K Fitzgerald, and K Liang for expression and purification of CPs used in this study. A Chilkoti is a cofounder of a start-up company, Phase Bioscience, in Durham, NC, USA that is commercializing the ELP phase transition technology for application in biotechnology and medicine.
Abbreviations
AUC
Area under the curve
BW
Body weight
CAC
Critical Aggregation Concentration
CP
Chimeric Polypeptide
ELP
Elastin-like polypeptide
Equiv
Equivalents
Dox
Doxorubicin
References
- 1.Drummond DC, Meyer O, Hong K, Kirpotin DB, Papahadjopoulos D. Optimizing liposomes for delivery of chemotherapeutic agents to solid tumors. Pharmacol Rev. 1999;51:691–743. [PubMed] [Google Scholar]
- 2.Duncan R. Polymer conjugates as anticancer nanomedicines. Nature Reviews Cancer. 2006;6:688–701. doi: 10.1038/nrc1958. [DOI] [PubMed] [Google Scholar]
- 3.Lee CC, MacKay JA, Frechet JM, Szoka FC. Designing dendrimers for biological applications. Nat Biotechnol. 2005;23:1517–1526. doi: 10.1038/nbt1171. [DOI] [PubMed] [Google Scholar]
- 4.Matsumura Y, Maeda H. A new concept for macromolecular therapeutics in cancer chemotherapy: mechanism of tumoritropic accumulation of proteins and the antitumor agent smancs. Cancer Res. 1986;46:6387–6392. [PubMed] [Google Scholar]
- 5.Brannon-Peppas L, Blanchette JO. Nanoparticle and targeted systems for cancer therapy. Adv Drug Deliv Rev. 2004;56:1649–1659. doi: 10.1016/j.addr.2004.02.014. [DOI] [PubMed] [Google Scholar]
- 6.Urry DW. Physical chemistry of biological free energy transduction as demonstrated by elastic protein-based polymers. Journal of Physical Chemistry B. 1997;101:11007–11028. [Google Scholar]
- 7.Yamaoka T, et al. Mechanism for the phase transition of a genetically engineered elastin model peptide (VPGIG)(40) in aqueous solution. Biomacromolecules. 2003;4:1680–1685. doi: 10.1021/bm034120l. [DOI] [PubMed] [Google Scholar]
- 8.Cappello J, et al. In-situ self-assembling protein polymer gel systems for administration, delivery, and release of drugs. Journal of Controlled Release. 1998;53:105–117. doi: 10.1016/s0168-3659(97)00243-5. [DOI] [PubMed] [Google Scholar]
- 9.Dreher MR, et al. Temperature triggered self-assembly of polypeptides into multivalent spherical micelles. J Am Chem Soc. 2008;130:687–694. doi: 10.1021/ja0764862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wright ER, Conticello VP. Self-assembly of block copolymers derived from elastinmimetic polypeptide sequences. Advanced Drug Delivery Reviews. 2002;54:1057–1073. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 11.Megeed Z, Cappello J, Ghandehari H. Genetically engineered silk-elastinlike protein polymers for controlled drug delivery. Advanced Drug Delivery Reviews. 2002;54:1075–1091. doi: 10.1016/s0169-409x(02)00063-7. [DOI] [PubMed] [Google Scholar]
- 12.Urry DW, Parker TM, Reid MC, Gowda DC. Biocompatibility of the Bioelastic Materials, Poly(Gvgvp) and Its Gamma-Irradiation Cross-Linked Matrix - Summary of Generic Biological Test-Results. Journal of Bioactive and Compatible Polymers. 1991;6:263–282. [Google Scholar]
- 13.Liu W, et al. Tumor accumulation, degradation and pharmacokinetics of elastin-like polypeptides in nude mice. J Control Release. 2006;116:170–178. doi: 10.1016/j.jconrel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Chilkoti A, Dreher MR, Meyer DE. Design of thermally responsive, recombinant polypeptide carriers for targeted drug delivery. Adv Drug Deliv Rev. 2002;54:1093–1111. doi: 10.1016/s0169-409x(02)00060-1. [DOI] [PubMed] [Google Scholar]
- 15.Meyer DE, Chilkoti A. Purification of recombinant proteins by fusion with thermally-responsive polypeptides. Nature Biotechnology. 1999;17:1112–1115. doi: 10.1038/15100. [DOI] [PubMed] [Google Scholar]
- 16.Furgeson DY, Dreher MR, Chilkoti A. Structural optimization of a “smart” doxorubicin-polypeptide conjugate for thermally targeted delivery to solid tumors. J Control Release. 2006;110:362–369. doi: 10.1016/j.jconrel.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Rodrigues PC, et al. Acid-sensitive polyethylene glycol conjugates of doxorubicin: preparation, in vitro efficacy and intracellular distribution. Bioorg Med Chem. 1999;7:2517–2524. doi: 10.1016/s0968-0896(99)00209-6. [DOI] [PubMed] [Google Scholar]
- 18.Bae Y, et al. Preparation and biological characterization of polymeric micelle drug carriers with intracellular pH-triggered drug release property: tumor permeability, controlled subcellular drug distribution, and enhanced in vivo antitumor efficacy. Bioconjug Chem. 2005;16:122–130. doi: 10.1021/bc0498166. [DOI] [PubMed] [Google Scholar]
- 19.Dreher MR, et al. Evaluation of an elastin-like polypeptide-doxorubicin conjugate for cancer therapy. J Control Release. 2003;91:31–43. doi: 10.1016/s0168-3659(03)00216-5. [DOI] [PubMed] [Google Scholar]
- 20.Lopes de Menezes DE, Mayer LD. Pharmacokinetics of Bcl-2 antisense oligonucleotide (G3139) combined with doxorubicin in SCID mice bearing human breast cancer solid tumor xenografts. Cancer Chemother Pharmacol. 2002;49:57–68. doi: 10.1007/s00280-001-0385-3. [DOI] [PubMed] [Google Scholar]
- 21.Singal PK, Iliskovic N. Doxorubicin-induced cardiomyopathy. N Engl J Med. 1998;339:900–905. doi: 10.1056/NEJM199809243391307. [DOI] [PubMed] [Google Scholar]
- 22.Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. 1984;226:466–468. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 23.Nitiss JL. Targeting DNA topoisomerase II in cancer chemotherapy. Nat Rev Cancer. 2009;9:338–350. doi: 10.1038/nrc2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drummond JT, Anthoney A, Brown R, Modrich P. Cisplatin and adriamycin resistance are associated with MutLalpha and mismatch repair deficiency in an ovarian tumor cell line. J Biol Chem. 1996;271:19645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- 25.Hao XY, Bergh J, Brodin O, Hellman U, Mannervik B. Acquired resistance to cisplatin and doxorubicin in a small cell lung cancer cell line is correlated to elevated expression of glutathione-linked detoxification enzymes. Carcinogenesis. 1994;15:1167–1173. doi: 10.1093/carcin/15.6.1167. [DOI] [PubMed] [Google Scholar]
- 26.Kabanov AV, et al. Polymer genomics: shifting the gene and drug delivery paradigms. Journal of Controlled Release. 2005;101:259–271. doi: 10.1016/j.jconrel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Kabanov AV. Polymer genomics: An insight into pharmacology and toxicology of nanomedicines. Advanced Drug Delivery Reviews. 2006;58:1597–1621. doi: 10.1016/j.addr.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batrakova EV, et al. Alteration of genomic responses to doxorubicin and prevention of MDR in breast cancer cells by a polymer excipient: Pluronic P85. Mol Pharmaceut. 2006;3:113–123. doi: 10.1021/mp050050g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Veronese FM, et al. PEG-doxorubicin conjugates: influence of polymer structure on drug release, in vitro cytotoxicity, biodistribution, and antitumor activity. Bioconjug Chem. 2005;16:775–784. doi: 10.1021/bc040241m. [DOI] [PubMed] [Google Scholar]
- 30.Kwon G, et al. Block copolymer micelles for drug delivery: loading and release of doxorubicin. Journal of Controlled Release. 1997;48:195–201. [Google Scholar]
- 31.Kataoka K, Kwon GS, Yokoyama M, Okano T, Sakurai Y. Block-Copolymer Micelles as Vehicles for Drug Delivery. Journal of Controlled Release. 1993;24:119–132. [Google Scholar]
- 32.Kataoka K, Harada A, Nagasaki Y. Block copolymer micelles for drug delivery: design, characterization and biological significance. Adv Drug Deliv Rev. 2001;47:113–131. doi: 10.1016/s0169-409x(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 33.Ulbrich K, et al. Polymeric anticancer drugs with pH-controlled activation. International Journal of Pharmaceutics. 2004;277:63–72. doi: 10.1016/j.ijpharm.2003.02.001. [DOI] [PubMed] [Google Scholar]
- 34.Chytil P, et al. New HPMA copolymer-based drug carriers with covalently bound hydrophobic substituents for solid tumour targeting. J Control Release. 2008;127:121–130. doi: 10.1016/j.jconrel.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Anton N, Benoit JP, Saulnier P. Design and production of nanoparticles formulated from nano-emulsion templates-a review. J Control Release. 2008;128:185–199. doi: 10.1016/j.jconrel.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Huang X, Jain PK, El-Sayed IH, El-Sayed MA. Gold nanoparticles: interesting optical properties and recent applications in cancer diagnostics and therapy. Nanomed. 2007;2:681–693. doi: 10.2217/17435889.2.5.681. [DOI] [PubMed] [Google Scholar]
- 37.Martin CR, Kohli P. The emerging field of nanotube biotechnology. Nat Rev Drug Discov. 2003;2:29–37. doi: 10.1038/nrd988. [DOI] [PubMed] [Google Scholar]
- 38.Nori A, Kopecek J. Intracellular targeting of polymer-bound drugs for cancer chemotherapy. Adv Drug Deliv Rev. 2005;57:609–636. doi: 10.1016/j.addr.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Kratz F, et al. Development and in vitro efficacy of novel MMP2 and MMP9 specific doxorubicin albumin conjugates. Bioorg Med Chem Lett. 2001;11:2001–2006. doi: 10.1016/s0960-894x(01)00354-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1