Conserved and divergent features of kinetochores and spindle microtubule ends from five species (original) (raw)
A comprehensive, cross-species electron tomography analysis of kinetochore–microtubule interfaces has provided insight into shared structural features and their likely functional consequences.
Abstract
Interfaces between spindle microtubules and kinetochores were examined in diverse species by electron tomography and image analysis. Overall structures were conserved in a mammal, an alga, a nematode, and two kinds of yeasts; all lacked dense outer plates, and most kinetochore microtubule ends flared into curved protofilaments that were connected to chromatin by slender fibrils. Analyses of curvature on >8,500 protofilaments showed that all classes of spindle microtubules displayed some flaring protofilaments, including those growing in the anaphase interzone. Curved protofilaments on anaphase kinetochore microtubules were no more flared than their metaphase counterparts, but they were longer. Flaring protofilaments in budding yeasts were linked by fibrils to densities that resembled nucleosomes; these are probably the yeast kinetochores. Analogous densities in fission yeast were larger and less well-defined, but both yeasts showed ring- or partial ring-shaped structures girding their kinetochore microtubules. Flaring protofilaments linked to chromatin are well placed to exert force on chromosomes, assuring stable attachment and reliable anaphase segregation.
Introduction
Interactions between mitotic kinetochores and spindle microtubules (MTs) are important because they contribute to accurate chromosome segregation. A fruitful union of genetics, cell biology, and biochemistry has recently identified many of the proteins of kinetochores in several species (Welburn and Cheeseman, 2008; DeLuca and Musacchio, 2012). Most of these are organized into functionally significant complexes: some bind MTs, some bind centromeric chromatin, and some connect these two significant sites or play regulatory roles. A few of these protein complexes, like Ndc80, have been found in all species so far described (Cheeseman et al., 2004). High-resolution light microscopy has ordered these and other components along the centromere-to-pole axis and found similar relative positions in budding yeast, fruit flies, and humans (Schittenhelm et al., 2007; Joglekar et al., 2008; Wan et al., 2009), revealing considerable conservation in the molecular organization of kinetochore composition.
There are, however, some significant differences in the molecules that connect kinetochore-associated MTs (KMTs) with centromeric chromatin. Dynein is prevalent in vertebrate kinetochores (Howell et al., 2001) but absent from kinetochores in yeasts, although this motor contributes to other aspects of yeast mitosis (Yeh et al., 2000; Grishchuk et al., 2007). Kinesin 13s are important for mitosis in metazoans (Rogers et al., 2004) but are lacking from yeasts. Meanwhile, MT-encircling rings formed by the Dam1 complex (also known as DASH) are an essential component of budding yeast kinetochores (Westermann et al., 2005), but they are absent from organisms other than fungi. In mammals, the Ska1 complex, which is not found in yeast, may play a mitotic role analogous to Dam1 (Schmidt et al., 2012), but structural studies of kinetochores suggest additional possibilities: fibrils appear by electron tomography to connect chromatin with the bending tubulin protofilaments (PFs) seen at the plus ends of mammalian KMTs (McIntosh et al., 2008). These structures have been proposed to serve as couplers between the spindle and its “payload.” Such differences motivate a critical comparison of kinetochore–MT attachments in various species to identify conserved and divergent structural features of this important interface.
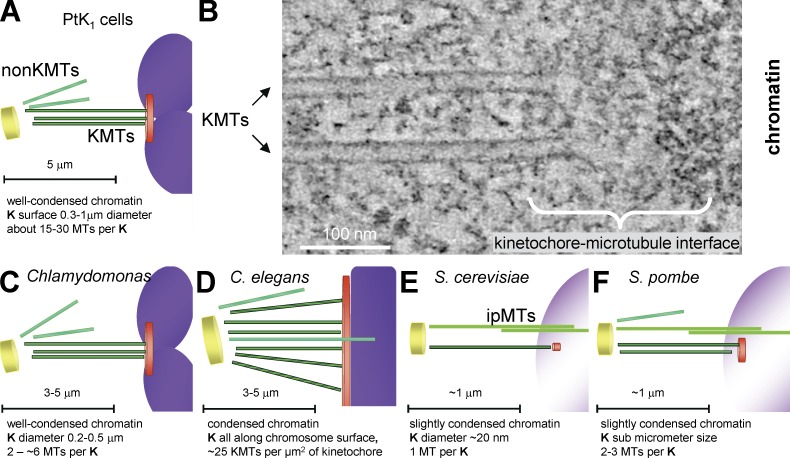
Structural comparison of kinetochores is further warranted because published works show inconsistencies, depending on the species examined or fixation method used. Classical electron microscope studies of cells from diverse organisms have described the kinetochore as a trilaminar structure and identified its outermost layer as the site of kinetochore–MT interaction (for review see Rieder, 1982). However, studies on both higher plants (Bajer and Mole-Bajer, 1972; Jackson and Doyle, 1982) and numerous microorganisms (Kubai, 1973, 1975) have revealed considerable diversity in kinetochore structure, opening a question about the generality of this layered organization. Because the relevant studies have been conducted over many years using diverse methods, the observed structural differences are hard to interpret. Methods are now available to compare kinetochore fine structure reliably. Rapid freezing preserves biological structure so well that many cells frozen in this way are viable upon thawing. Freeze-substitution fixation preserves much of this structure, making it available for study; electron tomography can then provide 3D views of these samples at 4–5-nm resolution (McIntosh, 2007). Indeed, electron tomography has already been used to characterize kinetochores in some mammalian cells (Fig. 1, A and B; McEwen et al., 1998b). With these methods, the kinetochore of mammalian cells, strain PtK1, looks less like a set of plates than a fibrous web that connects KMTs with each other and the underlying chromatin (Dong et al., 2007).
Figure 1.
Structural variety among the kinetochore–MT interfaces studied here. (A) Diagram of a mammalian chromosome (purple) and its kinetochore (K; orange disk) interacting with KMTs (dark green), which connect it with a spindle pole (yellow disk). KMT numbers are from McEwen et al. (1998a). Spindles also contain MTs that do not encounter kinetochores (non-KMTs; light green). (B) 4-nm slice from a tomogram of a kinetochore from a metaphase NRK cell. (C–F) Diagrams of the kinetochores analyzed in this paper. Images are colored as in A; the structures and numbers indicated are based on this paper, Figs. S1–S4.
To obtain a broader perspective on the fine structure of the kinetochore–MT interface, we have used electron tomography to analyze four species that cover a range of phylogeny. The green alga Chlamydomonas reinhardtii represents the lower plants (Fig. 1 C). The nematode Caenorhabditis elegans represents invertebrates and introduces a “holocentric” kinetochore (Fig. 1 D), i.e., one in which chromosome–MT interactions occur along most of the pole-facing chromosomal surface (Albertson and Thomson, 1982). Saccharomyces cerevisiae (Fig. 1 E) and Schizosaccharomyces pombe (Fig. 1 F) are distantly related fungi. The former has a small, well-studied kinetochore in which only 110 base pairs of centromeric DNA bind a special nucleosome, which associates with many protein complexes that connect it to a single MT (Westermann et al., 2007). In S. pombe, tens of kilobases of heterochromatic DNA define a protein-binding platform that ultimately associates with two to three KMTs (Pidoux and Allshire, 2000).
Here we report that trilaminar kinetochore plates are faint or absent in all these organisms, but that we found considerable similarity in the structures of KMT plus ends. These are flared in all mitotic stages, a fact of significant interest, as bending PFs can drive the motion of objects to which they are attached (McIntosh et al., 2010). As in PtK1 cells, the bending PFs of KMTs are connected by slender fibrils with nearby chromatin in all these species. These observations are assembled into a synthetic view of kinetochore–MT interaction that highlights its conserved and divergent features.
Results
Kinetochores in these diverse organisms look similar; a trilamellar structure is either missing or subtle
Tomographic slices of metaphase spindles from the four species examined here testify to the high quality of our sample preservation and provide an overview of each spindle (Fig. 2, A–D). Graphic models of MTs traced through many tomographic slices from these and other spindles have allowed an unambiguous identification of each cell’s mitotic stage (Figs. S1–S4). These models also identify different MT classes: KMTs, spindle MTs not associated with kinetochores or antiparallel bundles (non-KMTs), and interpolar MTs (ipMTs), which form an interdigitating framework between the spindles poles (for details see Figs. S1–S4).
Figure 2.
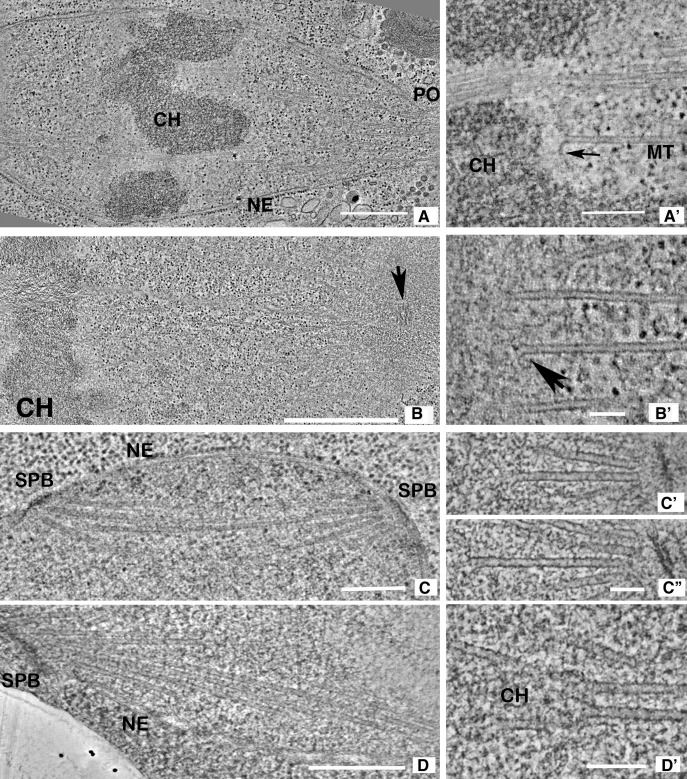
Tomographic slices of the spindles studied here. (A) 10-nm slice from a serial tomogram of a C. reinhardtii metaphase (CrM1, see Fig. S1 for model). Chromosomes (CH), the nuclear envelope (NE), and an opening in the envelope (PO) through which MTs project toward the spindle pole are evident. (A′) 4-nm slice through a different metaphase spindle (CrM2 in Fig. S1). One MT ends near a chromosome (a KMT) but others continue. The arrow indicates a flared PF. (B) 10-nm slice from a serial tomogram of a one-cell embryo of C. elegans. Chromosomes (CH) are evident, and a centriole (arrow) marks the spindle pole. (B′) 4-nm slice through the chromosome–MT interface of a different C. elegans metaphase (CeM in Fig. S2). Chromatin, at left, is separated by a fibrous zone from the KMTs with flaring plus ends. No outer plate is seen. (C) 10-nm slice from a tomogram of a metaphase budding yeast (ScM1, Fig. S3); SPB, spindle pole body; NE, nuclear envelope. (C′ and C″) Two 4-nm slices from a prometaphase budding yeast (ScPM1); slender filaments connect the capped MT minus ends with the spindle pole. (D) 10-nm slice from a prometaphase fission yeast (SpPM in Fig. S4 C); SPB and the NE are evident, but chromosomes are not. (D′) A 4-nm slice from the same cell. Some MTs end in pairs where the texture of nucleoplasm is different (CH). Such MTs have been interpreted as KMTs. Bars: (A) 0.5 µm; (A′) 200 nm; (B) 1 µm; (B′) 100 nm; (C) 0.5 µm; (C′ and C″) 100 nm; (D) 0.5 µm; (D′) 100 nm.
Higher magnification images of the places where spindle MTs end on or near chromatin show KMTs and the structures of chromosome–MT interfaces (Fig. 2, A′–D′). All these MTs end with a conspicuous flaring of PFs. This gives the kinetochores a similar appearance overall, but the structures differ from many published descriptions, based on conventional electron microscopy. None of these species displays either a dense “outer plate” or the more darkly staining chromatin subregion of an inner plate. Kinetochores in C. reinhardtii, as in PtK1 cells, sometimes show a hint of a plate, even in rapidly frozen, freeze-substitution fixed material, especially when examined with a thicker section (Fig. S5). Kinetochores in S. pombe have previously been seen to have a slight extra density around the chromosomal ends of KMTs in conventional thin sections of spindles (Ding et al., 1993), but in tomographic reconstructions we found no indication of plates, regardless of sample thickness. Neither C. elegans nor S. cerevisiae showed the slightest hint of kinetochore plates (Fig. S5).
Several factors may explain why our images of kinetochores are different from the classical trilaminar kinetochore. Methods of sample preparation account for part of the difference, because some cells, like PtK1, show beautiful plates after conventional chemical fixation but only subtle plates after rapid freezing and freeze substitution (McEwen et al., 1998b). Other cells, like blastomeres of C. elegans and S. cerevisiae, show no kinetochore plates, regardless of sample preparation. We have noticed a loose correlation between the visibility of a plate and the close proximity of neighboring KMTs (unpublished data), which suggests that flaring PFs and the connections between KMTs may contribute to plate appearance. However, these MT-dependent structures cannot be the whole story, as some organisms show kinetochore plates after treatment with colchicine to remove their MTs (Maiato et al., 2006). We conclude that plates are not a conserved feature of kinetochore structure, but the reasons for their presence in some cells but absence in others remain elusive.
The plus ends of all types of spindle MTs display flared PFs
Fig. 3 shows typical plus ends of metaphase KMTs (left) and non-KMTs (right). All these MT ends appear flared, meaning that one or more of their PFs curves outward, extending a considerable distance from the MT axis before terminating. This appearance closely resembles our previous observations on the MTs of PtK1 cells. The observations are striking because spindle MTs are dynamic polymers that elongate as well as shorten. In vitro studies with purified tubulin have found that whereas shortening MTs displayed flared PFs, elongating MTs did not (Mandelkow et al., 1991). We have examined 691 KMTs and 338 non-KMTs in these species (details shown in Table 1), and almost all ends displayed flaring PFs, which suggests that MTs growing in vivo display flaring PFs. In this context, the two rightmost images of non-KMTs from budding and fission yeast in Fig. 3 (C′ and D′) are of particular interest. They depict the plus ends of non-KMTs from anaphase interzones, i.e., ipMTs. These polymers were elongating by tubulin addition at their plus ends at the time they were frozen, yet they too show flaring PFs. Although correlations with in vitro MT structures have been seen in a plant mitotic interzone (Austin et al., 2005), both the ipMTs described here and elongating MTs in the cytoplasm of interphase fission yeast (Höög et al., 2011) showed flared ends. It follows that PF flare is not a simple reporter for the dynamic state of MTs in cells.
Figure 3.
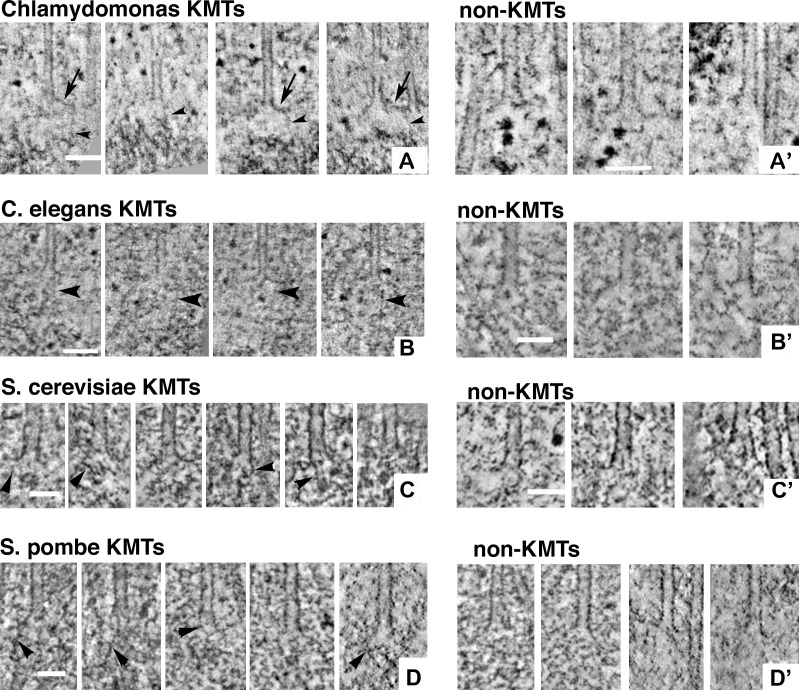
Galleries of spindle MT plus ends. (A) 4-nm slices that contain the ends of four algal metaphase KMTs. In all panels of this figure, arrows indicate flaring PFs and arrowheads demark fibrils that run from the PFs to nearby chromatin. (A′) Analogous images of non-KMTs. (B) 4-nm slices of four metaphase KMTs from C. elegans. (B′) Analogous images of non-MTs. (C) 4-nm slices from the plus ends of KMTs from S. cerevisiae. Here, no chromatin is evident, but there are small circular densities not far from each MT end. (C′) Budding yeast non-KMTs. Leftmost is the plus end of an “astral” MT that projected from the spindle pole body into the cytoplasm. The center and right images are from ipMTs in the anaphase interzone. (D and D′) 4-nm slices of KMTs and non-KMTs from fission yeast. The electron densities near the tips of the KMTs in D are suggestive of chromatin, but a well-formed chromosome is not seen. The non-KMTs of D′ are pole-distal ends of MTs that diverged widely from the spindle axis (two leftmost images) and from MTs that ended in the anaphase interzone (two rightmost images). Bars: (A) 100 nm; (A′) 75 nm; (B) 100 nm; (B′) 50 nm; (C and C′) 50 nm; (D and D′) 50 nm.
Table 1.
Average properties of MT PFs
| Organism/cell type | Stages studied | No. cells | No. KMTs | No. PFs | Length | Curve | No. non- KMTs | No. PFs | Length | Curve |
|---|---|---|---|---|---|---|---|---|---|---|
| nm | nm | |||||||||
| C. reinhardtii | Metaphases | 2 | 77 | 702 | 42 ± 15 | 19 ± 9 | 64 | 407 | 39 ± 12 | 18 ± 10 |
| Anaphases | 3 | 40 | 362 | 54 ± 14 | 17 ± 7 | 50 | 407 | 38 ± 16 | 21 ± 9 | |
| C. elegans | Metaphases | 1 | 49 | 321 | 44 ± 17 | 18 ± 8 | 171 | 579 | 38 ± 14 | 21 ± 11 |
| Blastomeres | Anaphases | 1 | 66 | 467 | 49 ± 18 | 18 ± 8 | 18 | 123 | 34 ± 12 | 20 ± 10 |
| S. pombe | Prometa/metaphases | 3 | 49 | 332 | 33 ± 12 | 24 ± 9 | 40 | 346 | 30 ± 12 | 20 ± 11 |
| Anaphases | 3 | 8 | 60 | 40 ± 21 | 18 ± 8 | 30 | 214 | 32 ± 13 | 12 ± 8 | |
| S. cerevisiae | Prometaphases | 3 | 113 | 823 | 29 ± 12 | 24 ± 14 | NA | NA | NA | NA |
| Early metaphases | 5 | 141 | 939 | 37 ± 14 | 20 ± 11 | NA | NA | NA | NA | |
| Metaphases | 3 | 63 | 456 | 27 ± 11 | 23 ± 13 | NA | NA | NA | NA | |
| Anaphases | 3 | 85 | 546 | 31 ± 12 | 26 ± 16 | 15 | 88 | 30 ± 12 | 21 ± 13 | |
| Totals | 27 | 691 | 5,008 | 338 | 2,164 |
When metaphase KMTs shorten in anaphase, their PFs elongate but curvature is unchanged
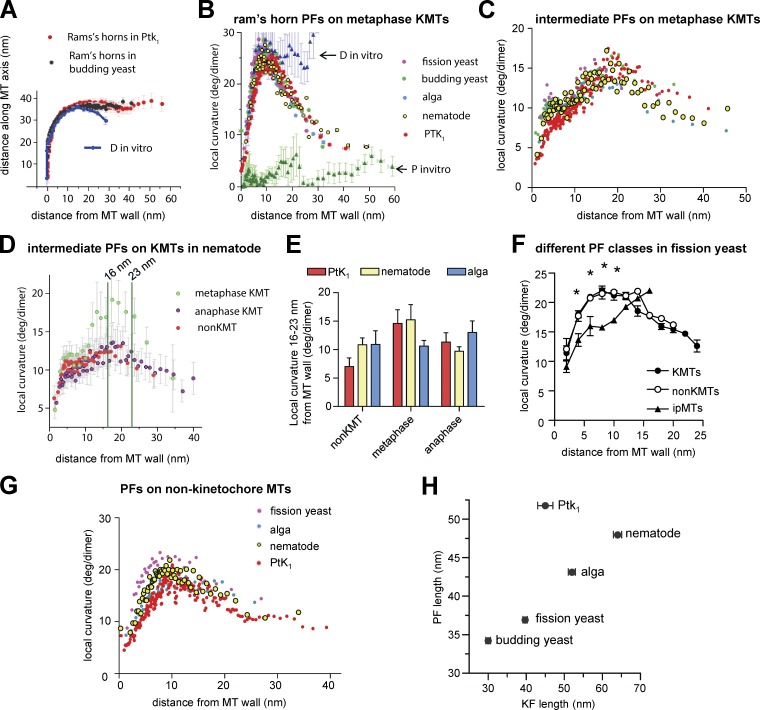
To study with more depth the relationship between PF shape and MT dynamics in vivo, we have digitized the paths of curving PFs from many MTs in cells at known stages of mitosis. All visible PFs from the KMTs in one C. reinhardtii metaphase cell are displayed in Fig. 4 A; clearly these shapes vary. To quantify and compare PF shapes, we have determined the mean positions for all the PFs of all MTs of a given kind from each cell as a function of distance from the MT wall (Fig. 4 A, dark line). Fig. 4 B illustrates that the shape of KMT PFs from metaphase cells of the species described here are strikingly consistent, although they differ from those in PtK1 cells and are markedly different from PFs on either growing or shrinking MTs in vitro.
Figure 4.
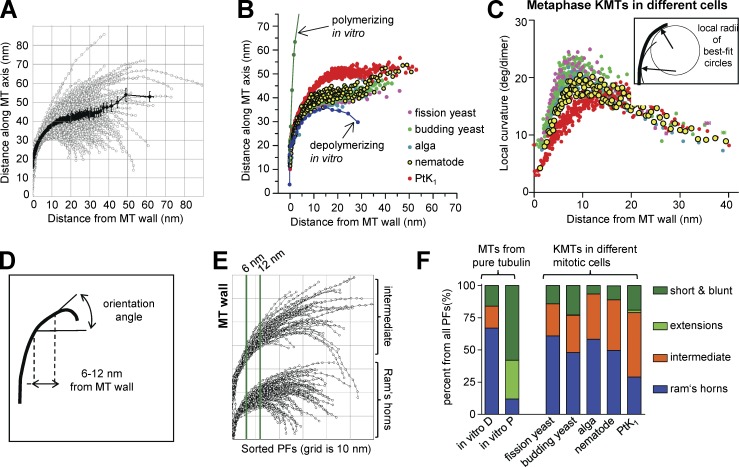
Analysis of PF curvatures. (A) Tracings of all visible PFs from all the KMTs imaged in one algal metaphase spindle. The dark line displays the mean position of these PFs as a function of distance from the MT wall. Error bars indicate SEMs. (B) Average shapes of KMT PFs from PtK1 cells (data are from McIntosh et al. [2008] whenever PtK1 numbers are given) and the species described here, all in metaphase. (C) Mean local curvatures, measured as described in Materials and methods, based on circles fit to 10 adjacent points at each position along every PF (inset). KMT PFs from all organisms are compared, colors are as in B. (D) Diagram showing how PF slopes were determined for sorting. (E) Metaphase KMT PFs from a C. reinhardtii metaphase sorted by their slope in the region between the two green vertical lines (see Materials and methods). The lower cluster displays the “ram’s horns,” the upper cluster displays the “intermediate” PFs. There were no “short/blunt” or “extension” PFs in this cell. (F) Fractions of KMT PFs from each organism that fall into the four categories defined in Materials and methods. Comparisons include MTs polymerizing (P) or depolymerizing (D) in vitro, as characterized in McIntosh et al. (2008).
In all species studied, the mean curvatures of KMT PFs are not measurably different in metaphase and anaphase, but in C. reinhardtii and C. elegans the flared portions of KMT PFs become longer as the chromosomes segregate (Table 1). The Mann-Whitney test identifies low probabilities for the null hypothesis: P < 0.0001 for the alga and P < 0.0008 for the nematode, so these differences are highly significant, results that mirror data from PtK1 cells (McIntosh et al., 2008). We did not find significant length differences between metaphase and anaphase KMT PFs in budding and fission yeast (Table 1), but anaphase A in these organisms is so brief in duration and extent that documenting such a difference by electron tomography would be difficult.
Perhaps the greater length of anaphase PFs, rather than an increase in PF curvature, accounts for the fact that others have found more KMTs with flaring PFs in anaphase than in metaphase (VandenBeldt et al., 2006). Another factor in this discrepancy may be our method of viewing MT ends from multiple orientations; with our approach, ∼100% of MT ends displayed at least some flaring PFs in both metaphase and anaphase, whereas the other study found fewer. Certainly, all relevant results from our group are consistent: there is no anaphase-dependent change in the mean curvature of KMT PFs, only in their length.
PFs at the ends of the MTs studied here commonly had the “ram’s horn” shape, but the elongated PF “extensions” found on growing MTs in vitro were almost never seen
To look for subtle differences between PFs on MTs from different species, we determined the local curvature of every PF at each point along its length (Fig. 4 C, inset). The means of these local curvatures, plotted as a function of distance from the MT wall, showed a sharper increase in curvature near the MT wall in the newly studied organisms relative to what was previously seen in PtK1 (Fig. 4 C). To learn whether all KMT PFs in the new cell types were more bent than in PtK1, we sorted the PFs on the bases of their slope in the region between 6 and 12 nm from the MT wall (Fig. 4 D). We could then put each PF from a given cell type into one of four classes, as done previously (McIntosh et al., 2008): those that resembled PFs on depolymerizing MTs in vitro (ram’s horns), those that were more like polymerizing MTs in vitro (short/blunt end or with gently curving extensions; Chrétien et al., 1995), and those with intermediate curvature that did not fit either category (Fig. 4 E). All species contained ram’s horn and short blunt PFs, as well as many of intermediate shapes, but only PtK1 cells contained MTs with the extensions found on MTs growing rapidly in vitro, and even here the number of extensions was small (Fig. 4 F). Moreover, ipMTs from anaphase cells, which are known to be growing, displayed some curved PFs and lacked extensions. Although issues of fixation artifact can never be totally excluded, we interpret this to mean that extensions are not part of the pathway for normal MT growth in cells.
Some aspects of PF shape are conserved, but there is a higher fraction of ram’s horn PFs in the newly studied cells than in PtK1
Mean local curvatures of PFs from the same class were highly similar on mitotic MTs from different species. For example, the ram’s horn PFs on KMTs from budding yeast were almost identical to those in PtK1, though they differed from ram’s horns on MTs depolymerizing in vitro (Fig. 5 A); starting ∼15 nm from the MT wall they became straight, whereas those from MTs in vitro continued to curve. We asked whether this aspect of PF shape was conserved and found that local curvatures for ram’s horn PFs are virtually identical for all species studied (Fig. 5 B); all show the marked drop in curvature that is not seen in vitro. These data suggest that the tendency of shortening polymers of pure tubulin to form PFs of constant curvature is altered in vivo by some cellular factors that straighten PFs at a distance from the MT wall. The same tendency is also seen in the ram’s horn PFs of non-KMTs at both metaphase and anaphase in all species studied (unpublished data), although the effect is less obvious because they are even shorter than the PFs of metaphase KMTs.
Figure 5.
Conserved and divergent features of PF curvatures. (A) Average shapes of KMT ram’s horn PFs from PtK1 and budding yeast compared with analogous PFs from MTs depolymerizing in vitro (D). (B) Mean local curvatures as a function of distance from the MT wall for all ram’s horn PFs from the KMTs of each species studied versus those from polymerizing (P-MTs) and depolymerizing MTs (D-MTs) in vitro. Colors are as in Fig. 4 B, both here and in C and G. (C) Analogous graph of the “intermediate” PFs from all species. (D) Comparison of mean local curvatures for intermediate PFs from nematode KMTs in metaphase and anaphase versus from non-KMTs. (E) Histograms of mean curvatures of intermediate PFs from KMTs 16–23 nm from the MT wall. (F) Local PF curvatures for three classes of MTs from S. pombe. Curvatures of KMTs and non-KMT PFs are similar, but ipMTs are different, both visually and in a test for significance (asterisks) using the unpaired Student’s t test with 95% confidence intervals. (G) Average local curvatures of all PF classes for non-KMTs compared with C. (H) Lengths of PFs versus fibrils in all species studied. For each new species, 100 fibrils were measured on each of two metaphase cells; the numbers of PFs studied are as in Table 1. Error bars indicate SEMs.
Intermediate PFs from spindle MTs are also similar to one another, with one intriguing exception: ∼5 nm from the MT wall, the KMT PFs from the species described here all show a local peak in curvature, which corresponds to a shoulder seen on the analogous plot for intermediate PFs from PtK1 KMTs (Fig. 5 C). Elsewhere, these curves are essentially superimposable. The only major difference in PF shape across species was that KMTs in PtK1 cells included fewer ram’s horn PFs than the species described here. This distinction is likely to account for the difference in local curvatures of the unsorted PFs shown in Fig. 4 (B and C).
Some cells show subtle changes in the shape of intermediate PFs between metaphase and anaphase
Given that anaphase KMTs in many cells shorten by loss of tubulin at the kinetochore, the curvatures of these PFs are of particular interest in seeking a relationship between PF shape and MT dynamics in vivo. Looking at PFs by class we have found that neither the number nor the curvature of KMT ram’s horn PFs change in the transition from metaphase to anaphase, which is consistent with our finding about mean curvature of KMT PFs.
Intermediate PFs are also highly similar between metaphase and anaphase, but there is one potentially important difference. In C. elegans, as in PtK1 cells (McIntosh et al., 2008), metaphase intermediate PFs showed a distinctive rise in curvature in the region 16–23 nm from the MT wall (Fig. 5 D). This rise is not seen in the intermediate PFs of non-KMTs or of anaphase KMTs, nor is it obvious in an alga (Fig. 5 E). This rise in higher eukaryotes may be related to the differences in KMT dynamics at different times of mitosis.
The ipMTs of anaphase also deserve attention because they grow throughout anaphase B. The distribution of local curvature for ipMT PFs in S. pombe is distinct from those of the KMTs and of other non-KMTs in this organism (Fig. 5 F). Their PFs also contain a smaller fraction of ram’s horns (unpublished data), so in vivo there are quantitative differences between the end morphologies of growing MTs versus others, although these differences are much less marked than those seen in vitro.
Curvatures of non-KMTs are similar among species
Quantifications of curvatures for PFs from non-KMTs show that they have the same general shape as PFs from KMTs (Fig. 5 G compared with Fig. 4 C) and that there is considerable conservation among species; only fission yeast non-KMTs have PFs with significantly different curvatures (same tests as in Fig. 5 F; unpublished data). The only feature of non-KMT PFs that distinguishes them from KMT PFs is their slightly lesser lengths (Table 1).
Slender fibrils connect KMT PFs to nearby chromatin in all species studied
Previous work in PtK1 detected fibrils connecting the flaring PFs of KMTs with chromatin. Here we report similar structures on KMTs from the four newly studied species (Fig. 3, left, arrowheads; and Video 1). Importantly, such fibrils are not seen at the plus ends of flared non-KMTs (Fig. 3, right). Furthermore, we found a linear relation between the mean length of the curved portions of metaphase KMT PFs and the length of their associated fibrils (Fig. 5 H, see Materials and methods). The data from PtK1 cells fall off this line, but the significance of these findings is not known.
These fibrils are at the edge of what can reliably be described in individual tomographic slices. Limitations are imposed by issues of sample preservation, the large amount of background material in a well-fixed cell, and the reliability of stain for describing biological structure. Previously, we improved the image signal-to-noise ratio by aligning and averaging images of individual PFs. Intermediate class PFs clearly showed attached fibrils, but this alignment was done manually (McIntosh et al., 2008). To get a more objective view of the KMT-associated material, we averaged volumes containing the plus end of each MT of a given class from a given cell (see Materials and methods). Representative 2D slices cut from these 3D averages are shown in Figs. 6 and 7, and full 3D averages are shown in Videos 2–4). One or more reasonably distinct fibrils can be seen running from the end of each averaged KMT into the region of the chromatin (Figs. 6 and 7, arrowheads). Such fibrils are visible in averages from KMTs of all these species, but similar averages of non-KMTs produced no such enhancement (Figs. 6 and 7; and Video 5), which suggests that the fibrils are a conserved aspect of the KMT–chromatin interface.
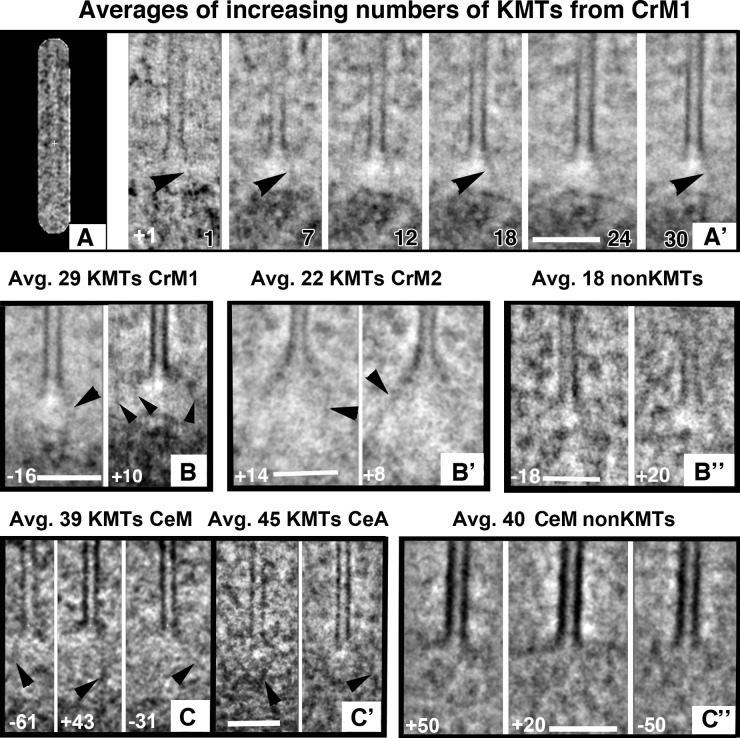
Figure 6.
Structures of averaged plus ends. The program PEET was used to align and average multiple KMT plus ends from each cell (number and kind of MTs plus cell name are given in black for each picture). Averaged structures were sampled with Slicer by cutting a lamina that contained the MT axis at an angle specified in degrees (white in each image). Orientations were selected to show fibrous connections between the averaged MT end and nearby chromatin (arrowheads). (A) Algal KMT shown with the mask used to occlude non-MT features during alignment. (A′) Leftmost is the slice of the reference volume used for MT alignment. Others panels show stages in averaging as more MTs are added. All images are slices at +1° (number in white). Arrowheads indicate a fibril that becomes more distinct with averaging. Bars, 100 nm. (B) Slices from the same averaged algal KMT at different angles. Arrowheads indicate apparent fibrils on all panels. (B′) Analogous images cut at angles shown in white from averaged KMTs from a different metaphase algal cell. (B″) Similar averages of MTs ending far from the chromatin in CrM1. No fibers are seen. (C) Averages from KMTs of one C. elegans metaphase (C) and one anaphase (C′). (C″) Averages of non-KMTs from blastomere spindles.
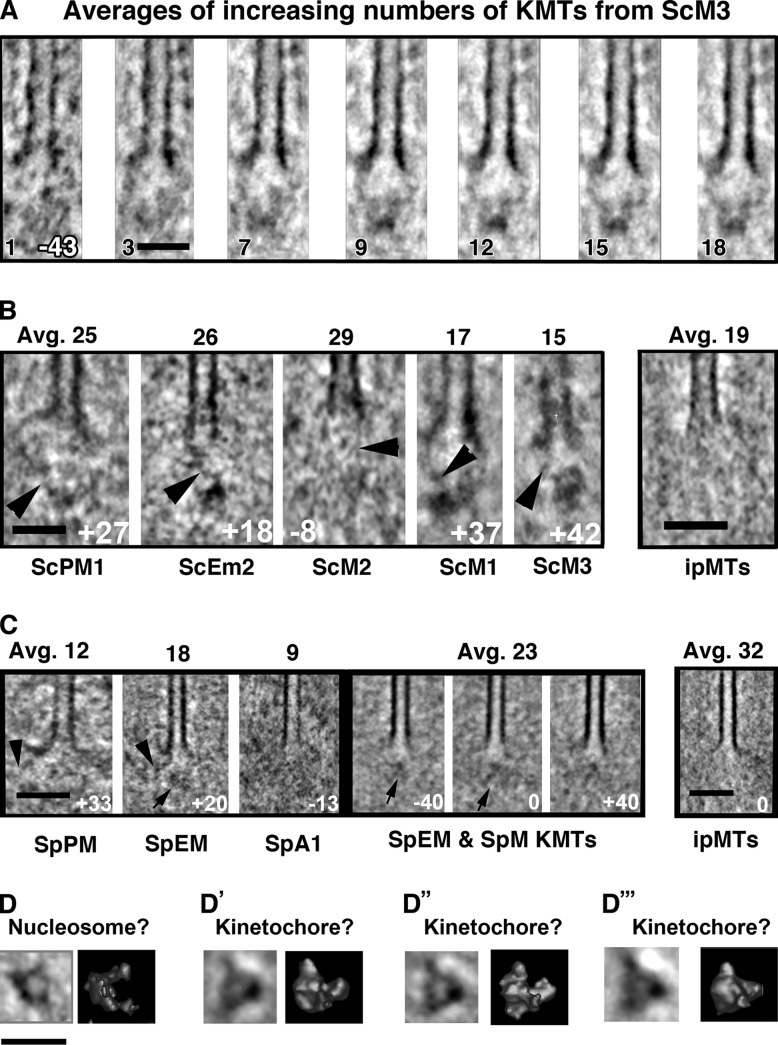
Figure 7.
Structures of averaged plus ends from yeast spindle MTs. (A) A slice from the single KMT (left) used as an alignment reference. No mask was used. Cell name and number of MTs averaged are shown in black. Slices are all at −43°. Strong development of image features is seen, including both a puck-shaped structure ∼55 nm from the MT tip and fibrils connecting this with flaring PFs. (B) Averages of KMTs from the S. cerevisiae cell named under each panel. Numbers of MTs are averaged at the top, the angle of the sampling slice is shown in white, and fibrils are indicated with arrowheads. Fibers associated with bending PFs are again visible, but not on an ipMT (B′). (C) Averages of KMTs from S. pombe. Fibers are again evident in averages (arrowheads), but they are oriented at diverse angles, perhaps because of the size of the fission yeast kinetochore. A mass of ill-defined shape ∼40 nm from the MT tip accompanies all KMTs (arrows) not ipMTs (C′). (D) PEET averages of particles from different regions of budding yeast nuclei. In each part of the figure, the left image is a slice cut through the average, where the right is a surface rendering. (D) Average of 100 particles far from the spindle, which we interpret as nucleosomes. (D′) Average of 64 particles near the plus ends of spindle MTs in a diploid prometaphase. These we interpret as kinetochores. (D″) Similar average of 58 particles from two haploid prometaphases. (D‴) Similar average from 68 particles from three haploid metaphases. Bars: (A–C) 50 nm; (D) 22 nm.
Budding yeast kinetochores appear different from kinetochores in other species
While examining the aligned and averaged images of budding yeast KMTs, we found a small, puck-shaped density ∼55 nm from the MT tip (Fig. 7, A and B; and Video 3). We surmised that these densities might correspond to yeast kinetochores, which are thought to be one, or a part of one, nucleosome that associates with specific centromere-binding proteins. Fig. 7 D compares averages of particles picked by eye from near the tips of KMTs of cells from three mitotic stages (putative kinetochores, see Materials and methods) with particles from regions of the nucleus that contained chromatin but no MTs (putative nucleosomes). The similarity of the different “kinetochore” images is marked, and the differences between these and the putative nucleosomes may reflect the material that binds the budding yeast centromere to make it a kinetochore. These “kinetochores” may be analogous to the isolated budding yeast kinetochores seen by negative staining (Gonen et al., 2012). Their morphology is different from the kinetochore regions in other organisms studied here, but this may reflect a unitary structure whose presence in other cells is obscured by associated material. Indeed, a close examination of the chromatin near KMT plus ends in fission yeast (Fig. 3 D and Video 6) shows some similar disks.
Partial rings surround the KMTs of some mitotic cells
We have used our 3D reconstructions of KMTs in budding yeast to look for structures that might correspond to ring-shaped oligomers formed by the Dam1 complex (also known as DASH; Miranda et al., 2005; Fig. 8, A and B). We rotated individual KMTs to align their axes perpendicularly to the plane of view and examined slices ∼5 nm thick at multiple places along their axes. As expected, the MT cross sections in these images were not symmetrical (Fig. 8 C, arrows) because of the wedge of information that is missing in tomographic reconstructions. Nonetheless, as the image plane was moved close to a KMT plus end, the PFs flared out from the MT axis (Videos 7–9). With this approach we did find some complete rings, but such images were rarer than partial rings (Fig. 8 C, bottom, arrowheads). Many but not all of these structures lay near KMT plus ends (Fig. 8 C, top, plus signs), implying that they were kinetochore associated. When similar viewing was done with averaged KMTs (labeled “Avg.”), the results were mixed; Fig. 8 C includes the clearest out of 12 tries. In these images one can see rings, which is remarkable because the rings we did see were not always perpendicular to the MT axis, and there was no obvious need for the rings on multiple KMTs to lie at exactly the same spot along the MT axis. The structures seen are similar to what one might expect for oligomers of Dam1 heterodecamers (Fig. 8, A and B), but our methods did not allow us to determine whether the observed structures were partial rings or simply imperfect images of full rings.
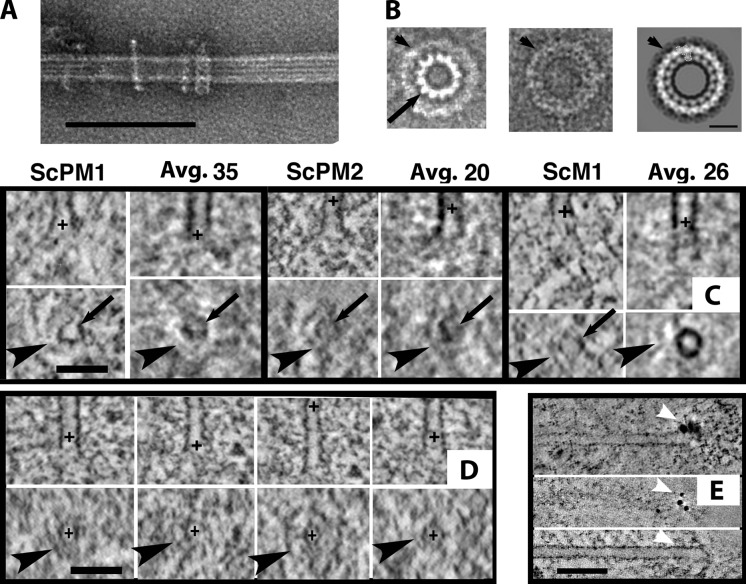
Figure 8.
Ring-shaped structures on KMTs in vivo. (A) MT polymerized in vitro from bovine tubulin, then mixed with recombinant Dam1 and negatively stained. (B) Negatively stained Dam1 rings. (B1) Unpublished image of the Dam1 ring (arrowhead) surrounding an MT (arrow), courtesy of E. Nogales (personal communication). (B2 and B3) Single (B2) and average of multiple (B3) negatively stained Dam1 rings in isolation (Wang et al. [2007], with permission from Nature Structural & Molecular Biology). (C) 4-nm slices cut from single KMTs and from averaged KMTs (labeled “Avg.,” then the number of MTs was averaged) from three budding yeast cells. Bottom panels are cross sections at the positions marked by black plusses in the top panels. The arrow indicates the MT wall in cross section; arrowheads indicate the ring and partial rings surrounding the MTs. (D) Comparable images for KMTs from fission yeast. (E) Localization of the S. pombe Dam1 complex, triply tagged with GFP, was accomplished with anti-GFP and a secondary anti-rabbit Fab coupled to 5 nm colloidal gold (arrowheads). The top image is a tomographic slice showing the stain and MT in the same slice. The bottom two images show a different tomogram; the top image slices the section’s surface where the antibody is bound (arrowheads), and the bottom image shows the MT in perfect alignment with the top image, so the position of the gold can be identified. Bars: (A) 100 nm; (B) 25 nm; (C and D) 50 nm; (E) 75 nm.
Components of the Dam1 complex are also found in fission yeast. Here the protein contributes to the fidelity of chromosome segregation but is not essential for chromosome segregation, as long as kinesin 8s are present (Sanchez-Perez et al., 2005). We found partial rings in mitotic S. pombe cells (Fig. 8 D and Video 9). To ask whether these might be assemblies of the Dam1 complex, we performed immuno-EM on S. pombe expressing GFP fusions with three separate subunits of the Dam1 complex (Liu et al., 2005). Anti-GFP, labeled indirectly with colloidal gold, gave clear signals near the pole-distal ends of spindle MTs (Fig. 8 E), confirming the presence of the Dam1 complex at this location. However, antibody labeling occurred only at the surfaces of our sections, making it impossible to establish the 3D arrangements of these antigens. C. reinhardtii, C. elegans, and PtK1 provided no evidence to support the generality of rings.
Discussion
The observations reported here, compared with analogous images from the literature, have identified several conserved and divergent structural features of the kinetochore–MT interface (summarized in Table S1). This mixture at the fine structural level resembles the molecular conservation of kinetochores. Indeed, the differences in PF structure in different species is probably a result of the different molecules involved in kinetochore–MT attachment.
Yeast kinetochore–MT interfaces differ from those of other organisms but may give clues about the fundamentals of centromere organization
Budding yeast kinetochores appear to be small disks connected to PFs by fibrils. There is no direct evidence that these disks are actually kinetochores, though the idea is supported by their position, structure, and similarity to kinetochores isolated from budding yeasts (Akiyoshi et al., 2010). Although our results are right at the limit of what electron tomography of well-preserved cells can tell us, the average structures described here represent a promising starting point for further investigation. For example, the chromatin near ends of fission yeast KMTs in Fig. 3 D reveals some darkly stained toroids that resemble the putative budding yeast kinetochores, and there are hints of similar structures in the alga (Fig. 3 A), which suggests that centromeric chromatin in both another yeast and an alga may be built up from units analogous to the budding yeast centromere. Further work with different methods may refine this possibility.
PFs from KMTs in the yeast cells examined here are also most different from mammalian PFs: they are shorter, slightly more curved, and begin curving closer to the MT wall. Unlike in other cells, these KMTs appear partially surrounded by ringlike structures, probably made from oligomers of the Dam1 protein complex. Sequence information suggests that this kinetochore component is limited to the fungi; it may be particularly well suited to organisms with only one or a very few MTs per kinetochore. The curvatures of yeast PFs may be manifestations of this molecular strategy for forming firm kinetochore–MT connections.
Bending PFs at the plus ends of metaphase MTs are heterogeneous in shape but overall similar across species
The shapes of KMT PFs vary in detail, but on average they are similar, both across species and over time in mitosis. The only clear difference seen is between the local curvatures of KMT PFs in PtK1 cells and in all cells of the current study (Figs. 4, B and C); this we can attribute to the relative frequency of ram’s horn PFs in the different populations (Fig. 4 F). We hypothesize that this difference may be a result of KMT flux: in PtK1 cells the MTs are moving toward the spindle pole throughout metaphase (Mitchison, 1989); the associated polymerization may reduce the number of ram’s horn PFs. The MTs of yeast spindles do not flux (Maddox et al., 2000). No equivalent data are available for C. reinhardtii or the blastomeres of C. elegans, but it will be interesting to see in the future whether the differences in spindle MT PFs seen here will correlate with MT flux.
The lengths of bending PFs on anaphase KMTs suggest that PF peeling is the rate-limiting step for chromosome-to-pole motion
In a mammal, an alga, and a nematode, KMT PFs in anaphase cells are longer than those in metaphase. Because these MTs were shortening at the time they were frozen, the data suggest that PFs on depolymerizing mitotic MTs peel away from their neighbors faster than they lose subunits from their distal ends. Apparently, the rate of subunit loss is not rate-limiting for KMT shortening in anaphase. We propose that PF peeling is the rate-limiting step for anaphase A. MT-surrounding structures, like those formed by the Dam1 complex, could easily regulate the rate of PF peeling. Indeed, in vitro work on the rates of MT shortening in the presence or absence of Dam1 shows that this complex is good at slowing tubulin depolymerization (Grishchuk et al., 2008). Analogous molecular functions may be found in organisms that lack Dam1; such components will probably be important parts of the system that controls the speed of anaphase chromosome motion.
Bending PFs of KMTs are well positioned to participate in mitotic force generation
Given their homogeneous composition, oligomers of GDP-tubulin should be uniformly curved. However, the PFs we have seen in vivo are straight in their portions distal to the MT (Fig. 5, A and B). This implies the presence of factors that modify PF shape, making even the ram’s horn class straighter than expected for pure tubulin. A plausible explanation for this morphology is a class of MT-associated proteins, not yet discovered, that associates with the tips of shortening MTs. These must differ from EB1, which binds only to growing MT ends (Jiang and Akhmanova, 2011). We therefore posit a new class of tip-interacting proteins that prefer association with GDP tubulin oligomers.
Another factor that may shape KMT PFs is the fibrils that connect chromosomes directly to the bending PFs of KMTs. If these structures provide a mechanical as well as structural link, they would allow bending PFs to pull on a chromosome and vice versa, much as we proposed previously (McIntosh et al., 2008). It is noteworthy that a recent study of the sites at which pole-directed forces act on mitotic chromosome in PtK1 cells identified a place nearer to the chromatin than proteins of the outer kinetochore, e.g., Hec1 and Cdc20 (Dumont et al., 2012). This is exactly the site of bending PFs; it is closer to the chromosomes than the site of any known mitotic motor enzyme. That work and our own combine to support the hypothesis that bending PFs are the place where the majority of pole-directed force is generated in metaphase and anaphase. The conservation of this structural feature suggests that this mechanism may be widespread. The fact that minus end–directed motor enzymes are dispensable for chromosome-to-pole motion, at least in yeast (Grishchuk and McIntosh, 2006; Tanaka et al., 2007), is consistent with this hypothesis.
There are many possibilities for the protein composition of these chromosome–KMT connections. The Ska1 complex might bind to bending PFs and couple them to a fibrous protein, like the NDC80 complex (Schmidt et al., 2012). CENP-E, CENP-F, and homologues of XMAP215 all have the right shape. Dynein can help to hold on to MTs that are trying to shorten (Laan et al., 2012). The CENP-T-W-S-X complex (Nishino et al., 2012) or complexes of EB1 with adenomatous polyposis coli, or centromeric Formin, which bind to and stabilize MTs (Cheng et al., 2011), are all interesting possibilities. Identification of protein components in these fibrils remains an important area for further work.
We have seen little evidence for fibrils running from chromatin to the outer MT walls, where the N terminus of the conserved NDC80 complex is known to bind (Alushin et al., 2010) and where all motor enzymes and other known MT-associated proteins interact with MTs. Why not? Our methods might not be sufficiently sensitive, but the observations of both fibrils and ringlike structures around yeast KMTs argue against this possibility, and Video 1 does show a nice example. Another possibility is that during metaphase and anaphase, the stages studied here, connections to the MT wall are not a major part of chromosome–spindle coupling. Motors and NDC80 complexes are certainly important for the formation of kinetochore–MT attachments, but perhaps there is a significant difference between initial kinetochore–MT interactions and the arrangements that form the stable, bipolar attachments at metaphase. For initial attachments, the MT walls are ideal, because the number of tubulin molecules in this condition is high; wall-binding kinetochore proteins are therefore ideal couplers during the initial stages of chromosome–spindle interaction. Stable attachments that assure accurate chromosome segregation may be different, relying on interactions that are specific to KMT plus ends. The structures we see may be the underpinnings of such interactions. Proteins important at earlier mitotic stages, such as the NDC80 complex, may still be part of these later connections, but they may show modified behavior because of chemical changes or different binding partners.
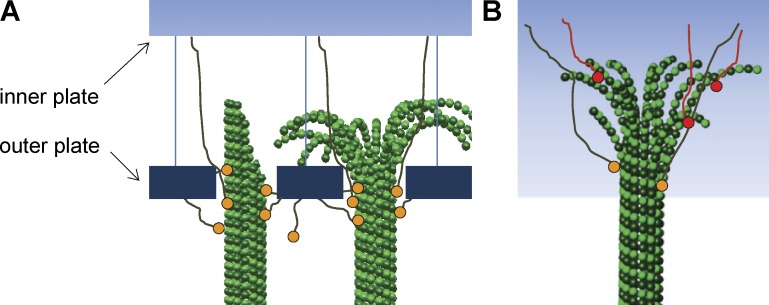
Our work suggests a modification of current views about the kinetochore–KMT interface. Most published models for this interaction posit linkages between either kinetochore plates or centromeric chromatin and the MT wall (Fig. 9 A). They also propose different structures for MTs that are growing or shrinking. Our work suggests that all states of the KMTs include flaring PFs and that the majority of chromatin–KMTs links are through PFs (Fig. 9 B). This model recognizes the potential differences between wall-binding and end-binding MT attachments: the latter may prefer bending PFs and/or the inward-facing surface of the tubulin molecule, both structural features that are unique to MT ends. In this way, the stable connections of metaphase and anaphase could be biochemically distinct from earlier connections, providing the stability necessary for accurate chromosome segregation.
Figure 9.
Models for chromosome–KMT attachment. (A) Diagram of customary views about kinetochore–MT interaction: kinetochore plates are linked to KMTs via connections with the MT wall. Some models see the outer plate as a sleeve that can diffuse on the MT wall. (B) Diagram based on our results showing no plate-like organization and a majority of connections running from the chromosome to the PFs at the KMT end.
Materials and methods
Culture and preparation of mitotic cells for microscopy
C. reinhardtii (strain 137c) was grown in liquid cultured at 25°C on a 12-h light/dark cycle to enrich for mitotic cells (Umen and Goodenough, 2001). The cells were collected by centrifugation after shifting to the dark, then high-pressure frozen essentially as described previously (Preble et al., 2001). This material was freeze-substituted in 1% OsO4 plus 0.1% uranyl acetate in acetone and embedded in epon/Araldite as described previously (O’Toole et al., 2003). Serial, 250-nm-thick sections were collected onto Formvar-coated copper slot grids and poststained in 2% uranyl acetate and Reynolds lead citrate. C. elegans whole worms or isolated embryos were prepared for electron microscopy as described previously (Müller-Reichert et al., 2008). In brief, hermaphrodites were high pressure frozen in M9 worm buffer containing 20% BSA, freeze substituted in 1% OsO4 and 0.1% uranyl acetate in acetone, then flat embedded in epon/Araldite. Alternatively, isolated embryos were released from hermaphrodites into M9 buffer containing 20% BSA and collected in cellulose capillary tubes for high-pressure freezing. The frozen embryos were then freeze substituted and flat embedded. Haploid or diploid strains of S. cerevisiae were prepared for electron microscopy by high-pressure freezing as described previously (Winey et al., 2005). The frozen cells were freeze-substituted in acetone containing either 2% glutaraldehyde and 0.1% uranyl acetate or 1% OsO4 and 0.1% uranyl acetate, then embedded in either Lowicryl HM20 or epon/Araldite. S. pombe cells were grown in liquid culture until early-to-mid log phase, quickly collected by filtration, high-pressure frozen, and processed as described previously (Ding et al., 1997). After freeze substitution in 2% glutaraldehyde and 0.1% uranyl acetate in acetone, they were embedded in Lowicryl HM20 and sectioned at 250 nm thickness. For immuno-EM, we cut sections 75–100 nm thick, blocked them with fish gelatin, and applied antibodies as described previously (Morphew et al., 2008). S. pombe expressing the Dam1 complex conjugated to three GFPs was supplied by X. He (Baylor College of Medicine, Houston, TX). Sections of all species were prepared for electron tomography by affixing 10- or 15-nm colloidal gold particles to both surfaces (tilt alignment fiducials).
Electron imaging
Mitotic cells were identified in serial sections at low magnification, then one or more sections were imaged as a tilt series about two orthogonal axes. Data were collected with 1–1.5° increments over a ±60° range using the automatic tilt-acquisition software, SerialEM (Mastronarde, 2005). A Tecnai microscope (TF30; FEI Inc.) was used for most applications, but some nematode and fission yeast samples were imaged on a JEM1000 electron microscope (Japanese Electron Optical Laboratories). Images were recorded on charged-couple device cameras at a magnification that sampled the specimen with 1.0–1.4 nm pixels. The tilt series data were aligned and tomograms computed using the IMOD software package as described in Kremer et al. (1996) and O’Toole et al. (2002). Often the area needed to cover a significant fraction of a given spindle required a montage of adjacent areas; these were blended into single images by routines in IMOD. When multiple serial tomograms were used, they were aligned into a single volume by tracing MTs across the section interfaces, using the points where they began and ended on adjacent sections as points of alignment to define general linear transformations that would morph these point clusters into as close an alignment as possible. These transformations were the applied to the sections, and the MTs traced across the section boundary. Chromosomes, spindle poles, and MTs were identified by inspection and then traced in 3D using the Slicer module from IMOD; the resulting models were used to identify stages in mitosis (Figs. S1–S4). In the alga and the two yeasts these models also helped to identify KMTs. For electron images of MTs surrounded by rings of the Dam1 complex, tubulin and Dam1 were prepared as described previously (Grishchuk et al., 2008), allowed to settle on Formvar-carbon-coated EM grids, and negatively stained with uranyl acetate.
Making models and classifying spindle MT
Models of spindle organization (Figs. S1–S4) were made by tracing each MT through the tomographic volumes reconstructed. This defined the path of each MT, and the position of its pole-distal end could then be used to assign it to a class. If several MTs ended in a bunch on a region of chromatin, they were called kinetochore associated (KMTs); if their ends were free in the volume of the spindle, they were non-KMTs. Many MTs left the volume of a single tomogram, but with serial tomograms the MTs in one could be linked with MTs in reconstructions of the adjacent sections, permitting their assignment to different categories. MTs whose pole-distal ends lay outside the volume reconstructed were not considered further.
KMTs were easy to identify in C. reinhardtii and C. elegans, but chromatin was more difficult to identify in fission yeast. There we used the ending of two or more MTs in close proximity to a characteristic staining of nucleoplasm (Fig. 2 D′) as our identifier of KMTs. In budding yeast, the number of MTs per pole was usually only a few more than the number of chromosomes (16 in haploid cells), so we assumed that all prometaphase and metaphase MTs were KMTs. In anaphase, a core bundle became clear, especially as the KMTs shortened, so all MTs that did not interdigitate with a counterpart from the opposite pole (ipMTs) were called KMTs.
Extraction and analysis of PF curvatures at MT plus ends
MT plus ends were characterized in rectangular volumes cut from the tomograms by routines in IMOD. These programs used the axis of the terminal segment of each modeled MT to define the y axis of a volume that contained the MT end and the material around it. Commonly, the x, y, and z dimensions were 120, 240, and 90 nm. These volumes were examined with Slicer to place the MT axis precisely along y, then a sampling plane 4 nm thick was rotated about the y axis until a curving PF was clearly visible on one side of the MT. The MT wall leading to this PF was represented by two points, one on a straight part of the wall, the second at the point where the PF began to bend outwards. From this point to the end of the PF, a cursor was used to deposit points at ∼2 nm intervals, providing sufficient data to describe the trajectory of each PF in 3D. The plane of sampling was then rotated about the MT axis in increments of ∼27°, and additional PFs were sought on the same side of the MT. The process was then repeated for the other MT side. The resulting tracings were used to determine structural properties of each PF from its point of divergence from the MT wall to its end, e.g., its length and mean curvature (initial angle of PF relative to the MT axis minus final PF angle divided by PF length, as in Table 1).
A special-purpose program (McIntosh et al., 2008) was used to assemble data from all PFs from a given class of MTs, e.g., KMTs from an S. pombe cell in metaphase. Local curvatures were measured at points along a flaring PF by fitting a circle to 10 adjacent points and expressing its reciprocal in degrees of bending per 8 nm of distance, i.e., degrees/dimer. The program then produced graphs of PF curvature as a function of distance from the MT wall. We also used the orientation of each PF at a distance interval from 6 to 12 nm from the MT wall to classify them into groups that resembled PFs on polymerizing and depolymerizing MTs in vitro: “extensions” that continued beyond the MT end with orientations 72°–90° relative to the MT normal; “short-blunt,” meaning those that did not extend beyond the MT end; and “ram’s horns” that had orientations <40°. PFs in vivo included many “intermediates” that were oriented between these extreme groups (McIntosh et al., 2008). The curvatures of PFs in each group were heterogeneous, so we pooled the curvatures of many PFs over an interval of distances from the MT wall to obtain useful averages. The mean values of these binned curvatures, together with the SEM, allowed statistically valid distinctions between PFs from analogous groups in different stages of mitosis in other cells at a similar stage. The D’Agostino and Pearson omnibus normality test in the Prism GraphPad package was used to ask whether the distributions of lengths and curvatures in these populations were Gaussian; the answer was most commonly no. The significance of differences has therefore been assessed by statistical tests that make no assumption of normality.
Determining the length of MT-associated fibrils
The lengths of fibrils were assessed visually by measuring the 3D distance between the point where a fibril associated with a PF and where it appeared to end on chromatin. Given the subtlety of these structural features, the measurements cannot be taken as precise, although observations by any one investigator were nicely reproducible.
Averaging images to improve their signal-to-noise ratio
The Slicer routine in IMOD provided useful images of PF at MT ends (e.g., Fig. 2 C), but to improve the signal-to-noise ratio of structures associated with MT ends, we averaged the 3D volumes of all MTs of a given type from a given cell. Initially, we aligned the MT axes visually and placed the position on the MT axis where the PFs began to flare at the volume’s center. An azimuthal orientation for each volume was chosen by putting a nicely curving PF in the xy plane on the left side of a plane that contained the y axis. We then usually masked the image data with a cylinder coaxial with the MT and as long as the MT segment studied; its diameter was ∼7/5 the MT outer diameter, and it was capped by hemispherical ends (Fig. 6 A). A reference MT was chosen for the clarity of its walls (e.g., Fig. 7 A, 1), then the MTs were aligned with the masked reference computationally using the program Particle Estimation by Electron Tomography (PEET) from the IMOD package (Heumann et al., 2011). PEET aligned each volume relative to the reference by small changes in 3D position and orientation, seeking the position that maximized the cross-correlation of pixel values between the reference and each volume. Given the careful initial alignments, we limited the range of possible translations to 6 pixels (a distance of 6–9 nm). Rotations about the initial x and z axes were limited to ±2° and those around y to ±10°. After the program had optimized alignments of all KMTs in the set, the mask was removed and the volumes were averaged. These averages were then sampled manually, using Slicer to cut them along the y axis at a wide range of azimuthal orientations. The paper’s figures display many examples of these images, and QuickTime movies of such averages are shown as Videos 2–6.
The averages of “nucleosomes” and “kinetochores” (Fig. 7 D) were made by picking many particles of each kind and excising them as cubes 30 nm on a side. All such images were aligned with one reference nucleosome then averaged to produce the volumes shown here.
Online supplemental material
Figs. S1–S4 show graphic models of the spindles used in this study. Fig. S5 shows the change in kinetochore appearance that results from projecting laminae of different thicknesses into a single image. Table S1 summarizes conserved and divergent structural features of the kinetochore–MT interface. Videos 1–9 were made either from individual tomograms or from averages of multiple KMTs that were generated with PEET. Each video was made by opening the relevant 3D reconstruction in the Slicer window of IMOD, then saving successive JPEG images as the Slicer window was moved through the volumes of interest. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201209154/DC1. Additional data are available in the JCB DataViewer at http://dx.doi.org/10.1083/jcb.201209154.dv, including several tomograms used for this analysis: a PtK1 prometaphase spindle (complementary to the data in Fig. 1 B); a fission yeast prometaphase spindle (shown in Figs. 2 D and S4, labeled “SpPM”); a fission yeast metaphase spindle (shown in Fig. S4, labeled “M”); a fission yeast early anaphase spindle (shown in Fig. S4, labeled “A1”); and 51 examples of C. reinhardtii metaphase KMT plus ends (taken from the cell shown in Figs. 2 A and S1, labeled “CrM1”); these represent the source of data shown in Fig. 3 A).
Supplementary Material
Supplemental Material
Acknowledgments
We thank all the members of our laboratories for innumerable helpful discussions, A. Zaytsev and P. Zakharov for help with figures, Mark Ladinsky and Erin White for Fig.1 B and Fig.8 E, respectively, and Eva Nogales for an unpublished micrograph, Fig.8 B1. Fig.8 (B2 and B3) are included with permission.
The work was supported in part by National Institutes of Health (NIH)grant GM033787 to J.R. McIntosh, grant 07 from the MCB program of the Russian Academy of Sciences to F. Ataullakhanov, NIH grant GM098389 to E.L. Grishchuk, who is a Kimmel scholar, and and NIH grant P41GM103431 to A. Hoenger.
Footnotes
Abbreviations used in this paper:
ipMT
interpolar MT
KMT
kinetochore-associated MT
MT
microtubule
non-KMT
spindle MT not associated with kinetochores or antiparallel bundles
PEET
Particle Estimation by Electron Tomography
PF
protofilament
References
- Akiyoshi B., Sarangapani K.K., Powers A.F., Nelson C.R., Reichow S.L., Arellano-Santoyo H., Gonen T., Ranish J.A., Asbury C.L., Biggins S. 2010. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 468:576–579 10.1038/nature09594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson D.G., Thomson J.N. 1982. The kinetochores of Caenorhabditis elegans. Chromosoma. 86:409–428 10.1007/BF00292267 [DOI] [PubMed] [Google Scholar]
- Alushin G.M., Ramey V.H., Pasqualato S., Ball D.A., Grigorieff N., Musacchio A., Nogales E. 2010. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 467:805–810 10.1038/nature09423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin J.R., II, Seguí-Simarro J.M., Staehelin L.A. 2005. Quantitative analysis of changes in spatial distribution and plus-end geometry of microtubules involved in plant-cell cytokinesis. J. Cell Sci. 118:3895–3903 10.1242/jcs.02512 [DOI] [PubMed] [Google Scholar]
- Bajer A.S., Mole-Bajer J. 1972. Spindle dynamics and chromosome movements. International Review of Cytology, Supplement 3. Academic Press, New York: 271 pp [Google Scholar]
- Cheeseman I.M., Niessen S., Anderson S., Hyndman F., Yates J.R., III, Oegema K., Desai A. 2004. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 18:2255–2268 10.1101/gad.1234104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Zhang J., Ahmad S., Rozier L., Yu H., Deng H., Mao Y. 2011. Aurora B regulates formin mDia3 in achieving metaphase chromosome alignment. Dev. Cell. 20:342–352 10.1016/j.devcel.2011.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrétien D., Fuller S.D., Karsenti E. 1995. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J. Cell Biol. 129:1311–1328 10.1083/jcb.129.5.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J.G., Musacchio A. 2012. Structural organization of the kinetochore-microtubule interface. Curr. Opin. Cell Biol. 24:48–56 10.1016/j.ceb.2011.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., McDonald K.L., McIntosh J.R. 1993. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J. Cell Biol. 120:141–151 10.1083/jcb.120.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., West R.R., Morphew D.M., Oakley B.R., McIntosh J.R. 1997. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol. Biol. Cell. 8:1461–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Vanden Beldt K.J., Meng X., Khodjakov A., McEwen B.F. 2007. The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat. Cell Biol. 9:516–522 10.1038/ncb1576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S., Salmon E.D., Mitchison T.J. 2012. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 337:355–358 10.1126/science.1221886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen S., Akiyoshi B., Iadanza M.G., Shi D., Duggan N., Biggins S., Gonen T. 2012. The structure of purified kinetochores reveals multiple microtubule-attachment sites. Nat. Struct. Mol. Biol. 19:925–929 10.1038/nsmb.2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., McIntosh J.R. 2006. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 25:4888–4896 10.1038/sj.emboj.7601353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Spiridonov I.S., McIntosh J.R. 2007. Mitotic chromosome biorientation in fission yeast is enhanced by dynein and a minus-end-directed, kinesin-like protein. Mol. Biol. Cell. 18:2216–2225 10.1091/mbc.E06-11-0987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk E.L., Efremov A.K., Volkov V.A., Spiridonov I.S., Gudimchuk N., Westermann S., Drubin D., Barnes G., McIntosh J.R., Ataullakhanov F.I. 2008. The Dam1 ring binds microtubules strongly enough to be a processive as well as energy-efficient coupler for chromosome motion. Proc. Natl. Acad. Sci. USA. 105:15423–15428 10.1073/pnas.0807859105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heumann J.M., Hoenger A., Mastronarde D.N. 2011. Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J. Struct. Biol. 175:288–299 10.1016/j.jsb.2011.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höög J.L., Huisman S.M., Sebö-Lemke Z., Sandblad L., McIntosh J.R., Antony C., Brunner D. 2011. Electron tomography reveals a flared morphology on growing microtubule ends. J. Cell Sci. 124:693–698 10.1242/jcs.072967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., Salmon E.D. 2001. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 155:1159–1172 10.1083/jcb.200105093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson W.T., Doyle B.G. 1982. Membrane distribution in dividing endosperm cells of Haemanthus. J. Cell Biol. 94:637–643 10.1083/jcb.94.3.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Akhmanova A. 2011. Microtubule tip-interacting proteins: a view from both ends. Curr. Opin. Cell Biol. 23:94–101 10.1016/j.ceb.2010.08.008 [DOI] [PubMed] [Google Scholar]
- Joglekar A.P., Bouck D., Finley K., Liu X., Wan Y., Berman J., He X., Salmon E.D., Bloom K.S. 2008. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J. Cell Biol. 181:587–594 10.1083/jcb.200803027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., McIntosh J.R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kubai D.F. 1973. Unorthodox mitosis in Trichonympha agilis: kinetochore differentiation and chromosome movement. J. Cell Sci. 13:511–552 [DOI] [PubMed] [Google Scholar]
- Kubai D.F. 1975. The evolution of the mitotic spindle. Int. Rev. Cytol. 43:167–227 10.1016/S0074-7696(08)60069-8 [DOI] [PubMed] [Google Scholar]
- Laan L., Pavin N., Husson J., Romet-Lemonne G., van Duijn M., López M.P., Vale R.D., Jülicher F., Reck-Peterson S.L., Dogterom M. 2012. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 148:502–514 10.1016/j.cell.2012.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., McLeod I., Anderson S., Yates J.R., III, He X. 2005. Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24:2919–2930 10.1038/sj.emboj.7600762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox P.S., Bloom K.S., Salmon E.D. 2000. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat. Cell Biol. 2:36–41 10.1038/71357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Hergert P.J., Moutinho-Pereira S., Dong Y., Vandenbeldt K.J., Rieder C.L., McEwen B.F. 2006. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells. Chromosoma. 115:469–480 10.1007/s00412-006-0076-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E.M., Mandelkow E., Milligan R.A. 1991. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J. Cell Biol. 114:977–991 10.1083/jcb.114.5.977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- McEwen B.F., Ding Y., Heagle A.B. 1998a. Relevance of kinetochore size and microtubule-binding capacity for stable chromosome attachment during mitosis in PtK1 cells. Chromosome Res. 6:123–132 10.1023/A:1009239013215 [DOI] [PubMed] [Google Scholar]
- McEwen B.F., Hsieh C.E., Mattheyses A.L., Rieder C.L. 1998b. A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma. 107:366–375 10.1007/s004120050320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R. 2007. Cellular Electron Microscopy. Methods in Cell Biology. Vol. 79 Wilson L., Matsudaira P., Academic Press, San Diego: xxi–xxv [Google Scholar]
- McIntosh J.R., Grishchuk E.L., Morphew M.K., Efremov A.K., Zhudenkov K., Volkov V.A., Cheeseman I.M., Desai A., Mastronarde D.N., Ataullakhanov F.I. 2008. Fibrils connect microtubule tips with kinetochores: a mechanism to couple tubulin dynamics to chromosome motion. Cell. 135:322–333 10.1016/j.cell.2008.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh J.R., Volkov V., Ataullakhanov F.I., Grishchuk E.L. 2010. Tubulin depolymerization may be an ancient biological motor. J. Cell Sci. 123:3425–3434 10.1242/jcs.067611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda J.J., De Wulf P., Sorger P.K., Harrison S.C. 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12:138–143 10.1038/nsmb896 [DOI] [PubMed] [Google Scholar]
- Mitchison T.J. 1989. Polewards microtubule flux in the mitotic spindle: evidence from photoactivation of fluorescence. J. Cell Biol. 109:637–652 10.1083/jcb.109.2.637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morphew M., He W., Bjorkman P.J., McIntosh J.R. 2008. Silver enhancement of Nanogold particles during freeze substitution for electron microscopy. J. Microsc. 230:263–267 10.1111/j.1365-2818.2008.01983.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller-Reichert T., Mäntler J., Srayko M., O’Toole E. 2008. Electron microscopy of the early Caenorhabditis elegans embryo. J. Microsc. 230:297–307 10.1111/j.1365-2818.2008.01985.x [DOI] [PubMed] [Google Scholar]
- Nishino T., Takeuchi K., Gascoigne K.E., Suzuki A., Hori T., Oyama T., Morikawa K., Cheeseman I.M., Fukagawa T. 2012. CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell. 148:487–501 10.1016/j.cell.2011.11.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E.T., Winey M., McIntosh J.R., Mastronarde D.N. 2002. Electron tomography of yeast cells. Methods Enzymol. 351:81–95 10.1016/S0076-6879(02)51842-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E.T., Giddings T.H., McIntosh J.R., Dutcher S.K. 2003. Three-dimensional organization of basal bodies from wild-type and delta-tubulin deletion strains of Chlamydomonas reinhardtii. Mol. Biol. Cell. 14:2999–3012 10.1091/mbc.E02-11-0755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux A.L., Allshire R.C. 2000. Centromeres: getting a grip of chromosomes. Curr. Opin. Cell Biol. 12:308–319 10.1016/S0955-0674(00)00094-6 [DOI] [PubMed] [Google Scholar]
- Preble A.M., Giddings T.H., Jr, Dutcher S.K. 2001. Extragenic bypass suppressors of mutations in the essential gene BLD2 promote assembly of basal bodies with abnormal microtubules in Chlamydomonas reinhardtii. Genetics. 157:163–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L. 1982. The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int. Rev. Cytol. 79:1–58 10.1016/S0074-7696(08)61672-1 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rogers S.L., Schwimmer T.A., Ems-McClung S.C., Walczak C.E., Vale R.D., Scholey J.M., Sharp D.J. 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 427:364–370 10.1038/nature02256 [DOI] [PubMed] [Google Scholar]
- Sanchez-Perez I., Renwick S.J., Crawley K., Karig I., Buck V., Meadows J.C., Franco-Sanchez A., Fleig U., Toda T., Millar J.B. 2005. The DASH complex and Klp5/Klp6 kinesin coordinate bipolar chromosome attachment in fission yeast. EMBO J. 24:2931–2943 10.1038/sj.emboj.7600761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittenhelm R.B., Heeger S., Althoff F., Walter A., Heidmann S., Mechtler K., Lehner C.F. 2007. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 116:385–402 10.1007/s00412-007-0103-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt J.C., Arthanari H., Boeszoermenyi A., Dashkevich N.M., Wilson-Kubalek E.M., Monnier N., Markus M., Oberer M., Milligan R.A., Bathe M., et al. 2012. The kinetochore-bound Ska1 complex tracks depolymerizing microtubules and binds to curved protofilaments. Dev. Cell. 23:968–980 10.1016/j.devcel.2012.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Kitamura E., Kitamura Y., Tanaka T.U. 2007. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 178:269–281 10.1083/jcb.200702141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen J.G., Goodenough U.W. 2001. Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev. 15:1652–1661 10.1101/gad.892101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBeldt K.J., Barnard R.M., Hergert P.J., Meng X., Maiato H., McEwen B.F. 2006. Kinetochores use a novel mechanism for coordinating the dynamics of individual microtubules. Curr. Biol. 16:1217–1223 10.1016/j.cub.2006.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan X., O’Quinn R.P., Pierce H.L., Joglekar A.P., Gall W.E., DeLuca J.G., Carroll C.W., Liu S.T., Yen T.J., McEwen B.F., et al. 2009. Protein architecture of the human kinetochore microtubule attachment site. Cell. 137:672–684 10.1016/j.cell.2009.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.-W., Ramey V.H., Westermann S., Leschziner A.E., Welburn J.P.I., Nakajima Y., Drubin D.G., Barnes G., Nogales E. 2007. Architecture of the Dam1 kinetochore ring complex and implications for microtubule-driven assembly and force-coupling mechanisms. Nat. Struct. Mol. Biol. 14:721–726 10.1038/nsmb1274 [DOI] [PubMed] [Google Scholar]
- Welburn J.P., Cheeseman I.M. 2008. Toward a molecular structure of the eukaryotic kinetochore. Dev. Cell. 15:645–655 10.1016/j.devcel.2008.10.011 [DOI] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H.W., Niederstrasser H., Wong J., Drubin D.G., Nogales E., Barnes G. 2005. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol. Cell. 17:277–290 10.1016/j.molcel.2004.12.019 [DOI] [PubMed] [Google Scholar]
- Westermann S., Drubin D.G., Barnes G. 2007. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 76:563–591 10.1146/annurev.biochem.76.052705.160607 [DOI] [PubMed] [Google Scholar]
- Winey M., Morgan G.P., Straight P.D., Giddings T.H., Jr, Mastronarde D.N. 2005. Three-dimensional ultrastructure of Saccharomyces cerevisiae meiotic spindles. Mol. Biol. Cell. 16:1178–1188 10.1091/mbc.E04-09-0765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Yang C., Chin E., Maddox P., Salmon E.D., Lew D.J., Bloom K. 2000. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol. Biol. Cell. 11:3949–3961 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material