LIS1 controls mitosis and mitotic spindle organization via the LIS1–NDEL1–dynein complex (original) (raw)
Abstract
Heterozygous LIS1 mutations are responsible for the human neuronal migration disorder lissencephaly. Mitotic functions of LIS1 have been suggested from many organisms throughout evolution. However, the cellular functions of LIS1 at distinct intracellular compartments such as the centrosome and the cell cortex have not been well defined especially during mitotic cell division. Here, we used detailed cellular approaches and time-lapse live cell imaging of mitosis from Lis1 mutant mouse embryonic fibroblasts to reveal critical roles of LIS1 in mitotic spindle regulation. We found that LIS1 is required for the tight control of chromosome congression and segregation to dictate kinetochore–microtubule (MT) interactions and anaphase progression. In addition, LIS1 is essential for the establishment of mitotic spindle pole integrity by maintaining normal centrosome number. Moreover, LIS1 plays crucial roles in mitotic spindle orientation by increasing the density of astral MT plus-end movements toward the cell cortex, which enhances cortical targeting of LIS1–dynein complex. Overexpression of NDEL1–dynein and MT stabilization rescues spindle orientation defects in Lis1 mutants, demonstrating that mouse LIS1 acts via the LIS1–NDEL1–dynein complex to regulate astral MT plus-ends dynamics and establish proper contacts of MTs with the cell cortex to ensure precise cell division.
INTRODUCTION
Mitotic cell divisions are essential for the accurate partitioning of genetic material into two daughter cells. Inappropriate segregation of chromosomes during mitosis leads to aneuploidy and genomic instability (1). During the mitotic phase of the cell cycle (M phase), microtubules (MTs) undergo dynamic reorganization to coordinate chromosome separation. Mitotic spindles are assembled by dramatic MT remodeling and emanate from the centrosome, an MT-organizing center, also called the spindle pole (2). The centrosome participates in MT nucleation and anchoring MT minus-ends. The core component of the centrosome is a centriole pair composed of a mother centriole and a daughter centriole, which recruits pericentriolar material components (3,4). The centrosome duplication cycle is precisely controlled to preserve centrosome number and proper centriole assembly (5). Importantly, mammalian cell division planes are mainly determined by the positioning of bipolar mitotic spindles (6). In addition, the spatiotemporal interactions between the cell cortex and astral MT plus-ends have critical roles in mitotic spindle regulation (7–9). Several MT plus-end binding proteins mediate dynamic contacts of astral MT plus-ends to the cell cortex by interacting with cortical force generators on the membrane (10).
Many of the proteins important for mitosis have been discovered, although much of the detailed mechanisms employed by each protein involved in cell division remains to be understood. Among those mitotically important proteins, LIS1 is part of a complex that interacts with diverse cortical factors and centrosomal proteins at kinetochores on the chromosomes, the mitotic spindles and the cell cortex, and it has been implicated in the regulation of the mitotic spindles and chromosome segregation during mitosis (11–13). Human LIS1 was first identified as a causative gene of human lissencephaly (‘smooth brain’), a severe neuro-developmental disease (14,15). Heterozygous mutation or deletion of human LIS1 leads to this brain malformation due to defects in neuronal migration. LIS1 is also part of a highly conserved protein complex first discovered in Aspergillus nidulans that is responsible for nuclear distribution (NUD) and functions in cytoplasmic dynein regulation (16,17). LIS1 homologues from Aspergillus to mammals form a complex with cytoplasmic dynein and NUD proteins (18–20). Cytoplasmic dynein is a MT minus-end-directed motor involved in mitotic spindle assembly by regulating MT dynamics especially at astral MTs and mediating poleward transport of spindle assembly checkpoint proteins (21–25). Through its motor activity, cytoplasmic dynein exerts pulling forces on the chromosomes. Dynactin, an accessory linker protein complexed with dynein subunits, also contributes to these cellular functions by assisting cargo loading and increasing processivity (26). Cortically anchored cytoplasmic dynein/dynactin complexes are important cortical force generators along MTs (24,27) that are essential for mitotic spindle formation and positioning in M phase (21,28), and LIS1 has been implicated in dynein targeting at MT plus-ends along astral MTs during cell division of various cell types (11,20,29). In addition, several NUD family proteins associate with both LIS1 and cytoplasmic dynein. Two mammalian NudE homologues, NDE1 and NDEL1, interact with LIS1/cytoplasmic dynein complex (19,30–34). NDE1 and NDEL1 display prominent centrosomal localization, as does LIS1 (19,32,35). NDE1 is required for targeting of LIS1 to the cytoplasmic dynein complex to generate persistent motor forces (36,37), while NDEL1 has been implicated in the process of LIS1/dynein recruitment, serving as a scaffold (11,38,39). In addition, a subset of NDE1 and NDEL1 proteins is observed in close proximity to the cell cortex where LIS1 accumulates (13,40,41). These previous studies support the notion that the LIS1–NDE1/NDEL1–dynein/dynactin complex is likely part of the protein machinery needed to coordinate various signals from the cell cortex to the mitotic spindles by generating pulling forces on spindle MTs. Despite these studies, the precise functions of LIS1 and its complex during mitosis remain elusive. Furthermore, it is unclear whether other components of LIS1 protein complex are involved in LIS1-dependent mitotic spindle regulation during mammalian cell division.
We took advantage of genetic null (knock-out, KO) and hypomorphic-conditional (HC) alleles of Lis1 (42,43) to uncover the critical dose-dependent roles of LIS1 in mouse embryonic fibroblasts (MEFs) and mouse neural progenitors (NPs). Previously, we found that Lis1 deficiency in mouse brains resulted in apoptosis and mitotic spindle orientation defects in NPs, while in MEFs loss of Lis1 led to severe defects in cell growth and MT capture at the cell cortex in interphase cells (13). In the current study, we used Lis1 mutant MEFs and performed time-lapse live cell imaging of mitotic progression compared with WT MEFs, to examine the functions of LIS1 during mitosis. We also analyzed mitotic spindle organization in detail by examining centrosome integrity and number. To address the mechanism of LIS1-dependent spindle regulation, we overexpressed several candidate protein complexed with LIS1 in Lis1 mutant MEFs and tested whether any of them can rescue spindle misorientation defects caused by _Lis1_-deficiency. Importantly, we performed time-lapse live cell imaging of MT plus-ends in Lis1 mutant MEFs to explore changes in the dynamics of astral MTs reaching to the cell cortex. Thus, by using both genetic and conditional Lis1 mutant MEFs with reduced LIS1 protein amount, we investigated essential functions of LIS1 complex to form proper mitotic spindle during mammalian cell division.
RESULTS
Perturbed mitotic progression in mitosis of Lis1 mutant MEFs
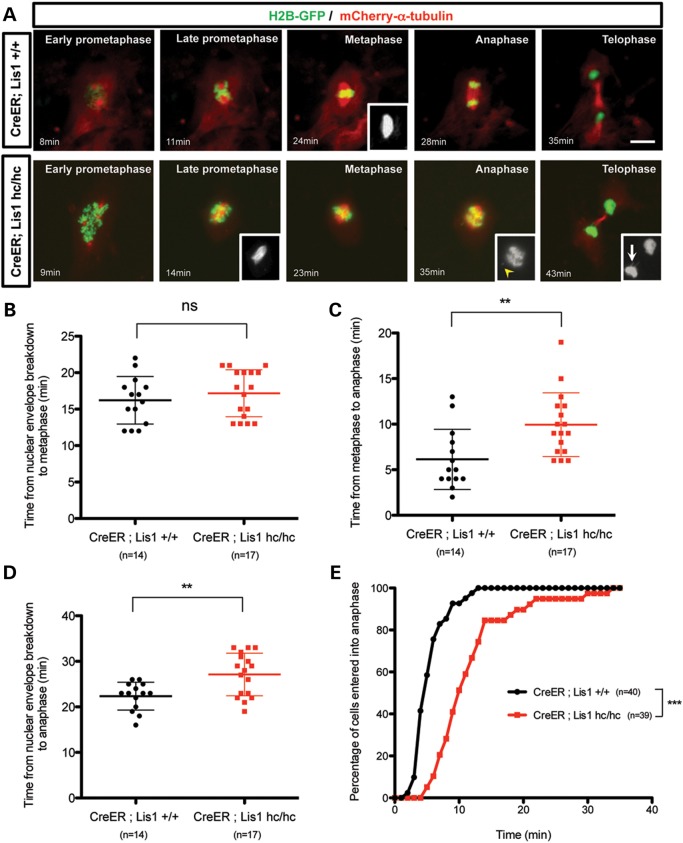
To investigate cellular functions of mouse LIS1 during mitotic cell division, we performed time-lapse live cell imaging of mitosis of Lis1 mutant MEFs isolated from Lis1 mutant conditional knock-out (CKO) mice that harbor HC alleles of mouse Lis1. Tamoxifen (TM)-inducible Cre (44) mice were mated to produce homozygous Lis1 CKO mice (CreERTM;Lis1hc/hc, hereafter termed _Lis1_-CKO-Cre) to acutely delete the Lis1 gene upon TM treatment, which reduced the LIS1 protein level to <10% of wild-type (WT) levels after 72 h incubation, as described in our previous study (13). Control mice contained two normal WT Lis1 alleles with the Cre transgene (CreERTM;Lis1+/+, hereafter termed WT-Cre). To visualize dynamic movements of chromosomes and MTs simultaneously during mitotic progression, we infected MEFs with retroviruses encoding histone 2B (H2B)-GFP (45) and mCherry–α-tubulin, followed by treatment with 4-hydroxy-TM for 12 h. Live cell imaging revealed that _Lis1_-CKO-Cre MEFs treated with TM (CreERTM;Lis1hc/hc + TM) exhibited a high frequency of abnormalities during mitosis that were not observed in WT-Cre MEFs (Supplementary Material, Video S1). In prometaphase, _Lis1_-CKO-Cre MEFs often formed extra centrosomes (e.g. four centrosomes) (Fig. 1A and Supplementary Material, Video S2) (See below). Loss of LIS1 frequently resulted in a kinked and curved morphology of the mitotic spindle. As the mitotic cell cycle eventually proceeded into metaphase, _Lis1_-CKO-Cre MEFs displayed chromosomes roughly aligned near the metaphase plate along with pseudo-bipolar spindles. In anaphase, _Lis1_-CKO-Cre MEFs retained several misaligned and unattached chromosomes, which ultimately resulted in chromosome missegregation and lagging chromosomes in telophase. The average time from nuclear envelope breakdown to metaphase plate formation was not significantly extended in _Lis1_-CKO-Cre MEFs (17.2 ± 0.8 min) compared with those of WT-Cre MEFs (16.2 ± 0.9 min), indicating that acute loss of LIS1 did not severely impair the timing of metaphase plate formation (Fig. 1B). However, the average time from metaphase to anaphase onset was significantly delayed from _Lis1_-CKO-Cre MEFs (9.9 ± 0.9 min) compared with those of WT-Cre MEFs (6.1 ± 0.9 min) (Fig. 1C). We obtained similar results of mitotic delay from a different analysis of anaphase onset timing that determines the accumulative percentage of cells that enter anaphase. The time when 50% cells entered anaphase was longer in _Lis1_-CKO-Cre MEFs (_t_50% = ∼10 min) than WT-Cre MEFs (_t_50% = ∼5 min) by 2-fold (Fig. 1E). The total duration of mitosis from nuclear envelope breakdown to anaphase onset was extended in _Lis1_-CKO-Cre MEFs (27.1 ± 1.1 min) compared with WT-Cre (22.4 ± 0.8 min) by ∼1.2-fold (Fig. 1D).
Figure 1.
Loss of LIS1 results in a prolonged mitotic cell cycle to induce anaphase onset delay. (A) Frame series of time-lapse live imaging movies from _Lis1-_CKO-Cre MEFs (CreER ; Lis1hc/hc) and control WT-Cre MEFs (CreER; Lis1+/+). MEFs were co-infected with H2B-GFP (green) and mCherry–α-tubulin (red) retroviruses. Cre activation was induced by treatment of 4-hydroxy-TM for 12 h. Inset from WT-Cre; Metaphase—normal spindle morphology (mCherry–α-tubulin). Insets from _Lis1_-CKO-Cre; Late prometaphase—kinked and curved spindle morphology (mCherry–α-tubulin), Anaphase—arrowhead: unattached chromosomes (H2B-GFP), Telophase—arrow: lagging chromosomes (H2B-GFP). Minutes indicate the time from nuclear envelope breakdown. Scale bar: 10 μm. (B) Time from nuclear envelope breakdown to metaphase plate formation. (C) Time from metaphase plate formation to anaphase onset. (D) Time from nuclear envelope breakdown to anaphase onset. (E) Accumulated percentage of cells entered into anaphase from metaphase. Lines in (B–D): mean ± SD, asterisks: **P < 0.01, ***P < 0.001 by Student's _t_-test. ns, not significant.
To investigate the effects of stably reduced LIS1 protein levels on its intracellular distribution during mitotic cell cycle progression, we compared endogenous LIS1 localization in WT MEFs and genetically mutated Lis1hc/ko MEFs with 35% of WT LIS1 protein levels (Supplementary Material, Fig. S1A and B) (13). In WT MEFs, LIS1 was enriched at the centrosomes and the kinetochores from prometaphase to metaphase. A majority of LIS1 was cytosolic, but a small fraction of LIS1 was also observed near the cell cortex as puncta. From metaphase to anaphase, LIS1 immunostaining overlapped with MTs along the mitotic spindles. Prior to telophase, a majority of LIS1 co-localized with the centrosomes in WT MEFs. In contrast, Lis1hc/ko MEFs displayed significant reduction in LIS1 distribution in the cytoplasm, although centrosome-specific localization of LIS1 appeared to be maintained. In metaphase, LIS1 puncta accumulated at several kinetochores near the metaphase plate in Lis1hc/ko MEFs. Importantly, mitotic spindle-associated LIS1 mostly disappeared in these Lis1hc/ko MEFs (Supplementary Material, Fig. S1A). Together, these observations support critical mitotic functions of mouse LIS1 at the centrosome, the mitotic spindle and the cell cortex in M phase (12), suggesting that the LIS1 protein complex may mediate mitotic functions in these specific intracellular compartments.
Formation of extra centrosomes caused by loss of Lis1
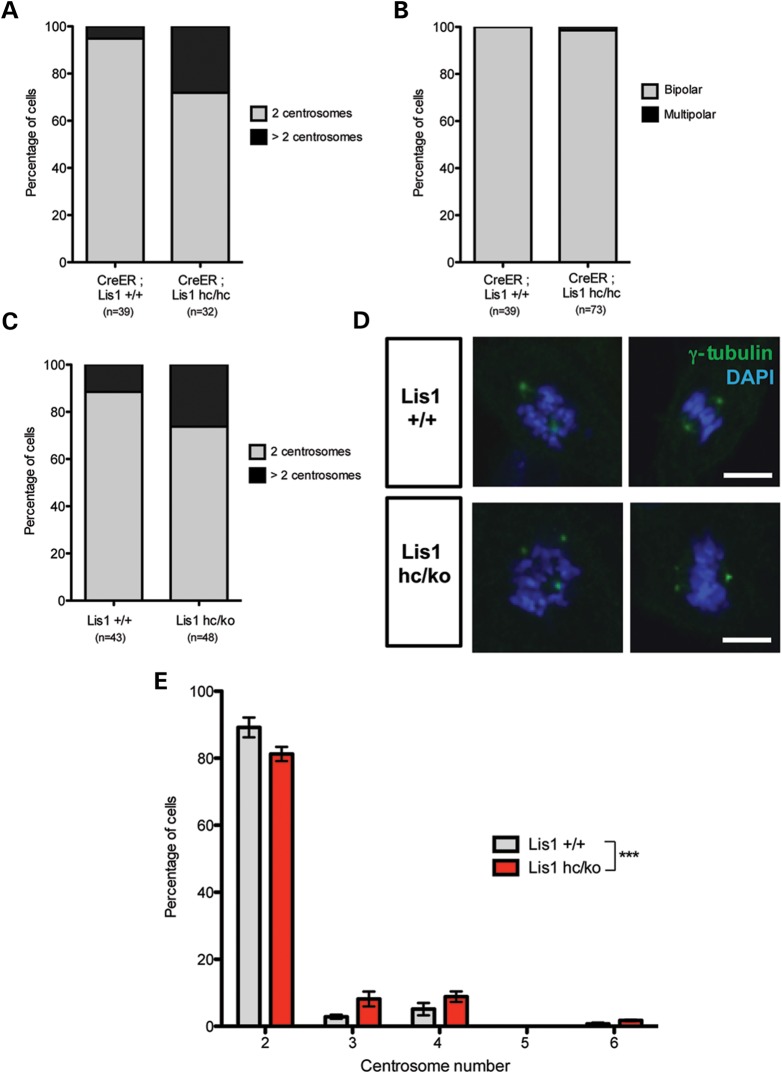
Time-lapse live cell imaging revealed that the incidence of multiple centrosomes (more than two centrosomes) was increased in Lis1 mutant MEFs (Fig. 2A). _Lis1_-CKO-Cre MEFs had a greater than 5-fold increase in the occurrence of centrosome amplification (_Lis1_-CKO-Cre: 28.1% versus WT-Cre: 5.1%). However, this rarely resulted in multipolar spindle formation during mitosis, since only 1.4% of _Lis1-_CKO-Cre MEFs displayed multipolar division (1/73) compared with none in WT-Cre MEFs (0/39) (Fig. 2B).
Figure 2.
Loss of LIS1 leads to extra centrosomes formation with centrosome number abnormality. (A) Time-lapse live cell imaging analysis reveals the centrosome number abnormality in _Lis1_-CKO-Cre MEFs compared with WT-Cre MEFs. (B) Multipolar division was found in time-lapse live cell imaging of _Lis1_-CKO-Cre MEFs (1/73). (C) Fixed sample analysis of centrosome number from WT and Lis1hc/ko MEFs. (D) WT MEFs with normal two centrosomes and Lis1hc/ko MEFs with extra centrosomes. MEFs were stained with γ-tubulin (green) and DAPI (blue). Scale bars: 10 μm. (E) Centrosome number distributions in WT and Lis1hc/ko MEFs. More than three slides were analyzed, n > 250 cells from each genotype. Bars: mean ± SEM, asterisk: ***P < 0.001 by the two-way ANOVA test.
To determine whether centrosome amplification is a common feature resulting from the reduction in LIS1, we performed γ-tubulin staining in WT and Lis1hc/ko MEFs. Similar to _Lis1_-CKO-Cre, Lis1hc/ko MEFs frequently displayed centrosome amplification. Lis1hc/ko had a nearly 2-fold increase in the occurrence of multiple centrosomes (Lis1hc/ko: 18.7% versus WT: 10.8%) (Fig. 2C and D). Notably, the occurrence of cells with three and four centrosomes was increased about 3-fold and 2-fold in Lis1hc/ko MEFs, respectively. The frequency of six centrosomes was also increased in Lis1hc/ko MEFs (Fig. 2E). Thus, LIS1 reduction results in centrosome amplification.
Abnormal centrosome integrity caused by clustering of amplified centrosomes in Lis1 mutant MEFs
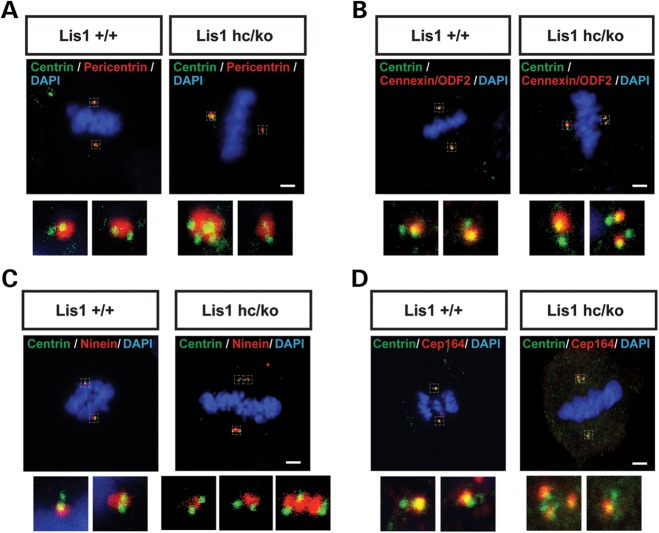
To test whether _Lis1_-deficiency impairs centrosome structure in Lis1 mutants, we performed pericentrin immunostaining and found that the size and shape of percentriolar material in Lis1hc/ko MEFs were abnormal compared with WT MEFs. Lis1hc/ko MEFs frequently had apparently enlarged spindle poles with larger amounts of percentriolar material than WT, even in cells undergoing bipolar division with two clearly distinguished centrosomes. This generated asymmetry of the two spindle poles in Lis1hc/ko MEFs, while symmetric bipolar spindles were maintained in WT MEFs (Fig. 3A). In addition to asymmetric spindles, Lis1hc/ko MEFs displayed an elongated, cylindrical shape of the pericentriolar material rather than a normal spherical shape (Fig. 3C), suggesting that LIS1 is required for maintaining proper centrosome integrity and assembly.
Figure 3.
Lis1 mutant MEFs exhibit centrosome clustering phenotype during mitosis. (A–D) Centrosome clustering phenotype with normal centrosome maturation in Lis1hc/ko MEFs. (A) Co-staining with pericentrin (pericentriolar material marker) and centrin (each centriole). Co-staining with centrin and several mature centriole markers: (B) cennexin/ODF2, (C) ninein, (D) Cep164, respectively. Insets in (A–D): high magnification images of centrosomes. Scale bars: 5 μm.
We assessed whether LIS1 is necessary for the regulation centrosome organization or centrosome maturation by co-immunostaining with centrosome or centriole markers and several centrosome maturation markers that are associated with centriole distal/subdistal appendages. Mature centriole appendage proteins such as cennexin/ODF2 (46), ninein (47) and Cep164, another mother centriole marker (48), displayed intact localization at the centrosomes in Lis1hc/ko MEFs (Fig. 3B–D). Only one centrin-positive centriole among a centriole pair had cennexin/ODF2 or ninein-immunoreactivity, suggesting that the centrosomes in Lis1hc/ko MEFs undergo normal centrosome maturation. However, a significant fraction of Lis1hc/ko MEFs with bipolar spindles displayed a ‘centrosome clustering’ phenotype (with more than two pairs of centrioles with normal configuration in one pole), a phenotype reported in the cells with extra centrosomes (49,50). Clustered spindle poles displayed a large amount of pericentrin-positive pericentriolar materials (Fig. 3C).
The cennexin/ODF2-positive mother centriole functions as a basal body, an anchor of non-motile primary cilium originating in interphase (51). We hypothesized that the frequent appearance of extra centrosomes in Lis1hc/ko MEFs may result in the formation of multiple primary cilia in these cells. Lis1hc/ko MEFs produced multiple cilia from amplified centrosomes under serum starvation, as identified by immunostaining of acetylated-α-tubulin and pericentrin, a primary cilia marker and a basal body marker, respectively (Supplementary Material, Fig. S2A). There was a 3-fold increase in the percentage of cells with multiple cilia from Lis1hc/ko MEFs (5.1 ± 1.4%) compared with WT MEFs (1.7 ± 0.5%) (Supplementary Material, Fig. S2B). Given the 2-fold increase in incidence of extra centrosomes in Lis1hc/ko versus WT MEFs, we concluded that the formation of multiple centrosomes in M phase of Lis1hc/ko MEFs ultimately leads to the generation of ectopic primary cilia in interphase, suggesting that amplified extra centrosomes in Lis1 mutant MEFs are fully functional.
Chromosome missegregation and less recruitment of kinetochore proteins in Lis1 mutant MEFs
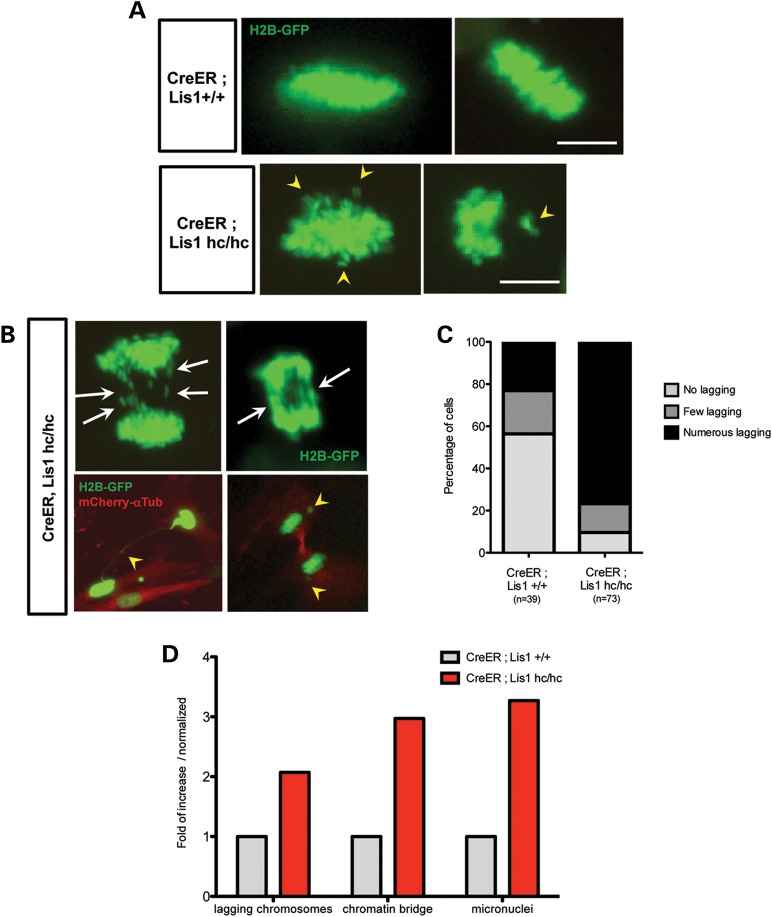
Time-lapse live cell imaging revealed congression of chromosomes at the metaphase plate in WT and _Lis1_-CKO-Cre MEFs, but metaphase plates were less compact and displayed distorted morphology in _Lis1_-CKO-Cre MEFs compared with those of control WT-Cre MEFs (Fig. 4A). We also observed an increase in the number of misaligned and unattached chromosomes scattered away from the metaphase plate in _Lis1_-CKO-Cre MEFs. _Lis1_-CKO-Cre MEFs displayed a 2-fold increase of lagging chromosomes in anaphase (Fig. 4B) compared with WT-Cre MEFs (Fig. 4D). We categorized the occurrence of lagging chromosomes into three groups: none, few and numerous lagging chromosomes. Numerous lagging chromosomes were frequently observed (76.7%) in _Lis1_-CKO-Cre MEFs compared with WT-Cre MEFs (Fig. 4C). In addition, other types of severe chromosome segregation defects were also found in _Lis1_-CKO-Cre MEFs. There was a 3-fold increase in intracellular chromatin bridges and a 3-fold increase in micronuclei found in daughter cells derived from mitosis of _Lis1_-CKO-Cre MEFs compared with WT-Cre MEFs (Fig. 4D).
Figure 4.
Loss of LIS1 induces chromosome misalignment in metaphase and chromosome missegregation in anaphase during mitosis. (A) Time-lapse live cell imaging analysis of chromosomal behavior (labeled with H2B-GFP, green) from _Lis1_-CKO-Cre MEFs and WT-Cre MEFs. Arrowhead: misaligned chromosomes in _Lis1_-CKO-Cre MEFs. Scale bars: 5 μm. (B) Chromosome missegregation phenotypes in mitosis of _Lis1_-CKO-Cre MEFs. Arrow: numerous lagging chromosomes; arrowhead: (left) the chromosomes stuck in intercellular chromatin bridge, (right) micronuclei formation in telophase. MT networks were visualized by mCherry–α-tubulin (red). (C) Quantification of lagging chromosome appearance in time-lapse live cell imaging of _Lis1-_CKO-Cre MEFs and WT-Cre MEFs. Numbers of lagging chromosomes—no lagging: 0, few lagging: 1–3, numerous lagging: ≥4. (D) Increased incidence of various chromosome segregation defects _Lis1-_CKO-Cre MEFs compared with WT-Cre MEFs.
We reasoned that these chromosome congression and separation defects in _Lis1_-deficiency were caused by misregulation of protein targeting to kinetochores, since LIS1 is a known kinetochore-binding protein in prophase and prometaphase (12). Targeting of LIS1 to kinetochores (identified with CREST staining) was reduced to 50% in Lis1hc/ko MEFs, as expected due to an overall 35% reduction of LIS1 protein in these cells compared with WT (Supplementary Material, Fig. S3A). In addition, there was an overall reduction in kinetochore recruitment of dynein subunits, including the dynein intermediate chain (DIC70.1) and the dynactin subunit p 150Glued, to 40 and 50%, respectively, compared with WT (Supplementary Material, Fig. S3A and B). Targeting of CLIP170 to kinetochores was also significantly reduced in Lis1hc/ko MEFs to 30% of WT levels (Supplementary Material, Fig. S3C). Next, we determined interkinetochore distances that reflect the strength of tension from kinetochore-bound MTs to the spindle poles. Lis1hc/ko MEFs displayed a decrease in interkinetochore distance (Lis1hc/ko: 0.48 ± 0.01 μm versus WT: 0.65 ± 0.17 μm) (Supplementary Material, Fig. S3D). These data suggest that aberrant and/or unstable MT attachments to kinetochores in Lis1hc/ko MEFs may result from a depletion of kinetochore-targeted proteins such as LIS1, the dynein/dynactin complex and CLIP170.
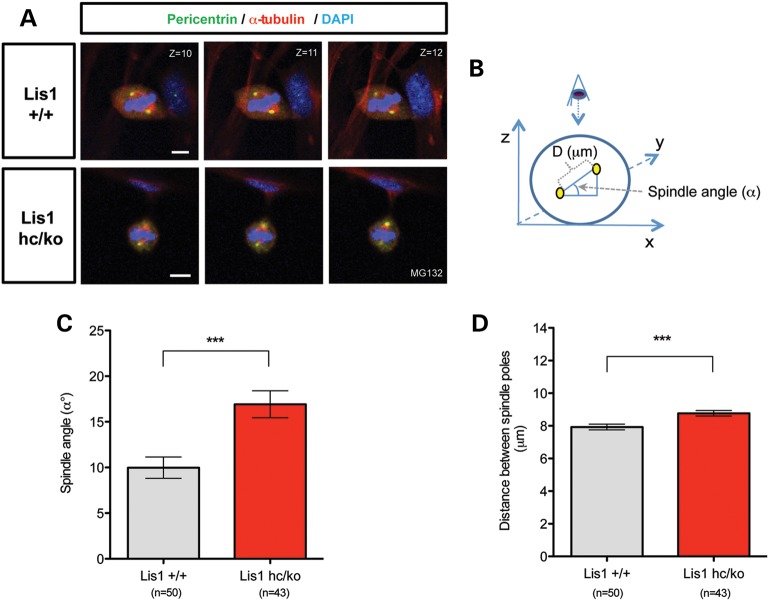
Impairment of mitotic spindle formation and spindle misorientation in Lis1 mutant MEFs
Previously, it was shown that LIS1 overexpression results in spindle misorientation in MDCK cells (12), and that Drosophila Dlis1 mutants as well as mouse Lis1 mutants display mitotic spindle defects in NP division (13,52). Consistent with these findings, we uncovered defects in spindle formation and positioning from time-lapse live cell imaging of _Lis1_-CKO-Cre MEFs (Fig. 1A). To determine mitotic spindle orientation parallel to the cell-substrate adhesion plane, which occurs in adherent cells (53), we arrested MEFs in metaphase by treatment with MG132. Pericentrin, a pericentriolar marker of centrosomes (54), was used to identify spindle poles. Confocal images from WT MEFs displayed a relatively narrow centrosomal distribution in the same or adjacent confocal planes along the cell axis. In contrast, Lis1hc/ko MEFs exhibited a high degree of spindle tilting compared with the substrate plane (Fig. 5A). Spindle angle (α°, measured by an amplitude of angle between centrosomes and the plane of cell substrate, shown in Fig. 5B) was significantly increased in Lis1hc/ko MEFs. The average spindle angle in Lis1hc/ko MEFs (α° = 16.9 ± 1.5) was increased about 2-fold compared with WT MEFs (α° = 10.0 ± 1.2) (Fig. 5C). Since Drosophila Dlis1 mutants displayed defects in centrosome separation during mitosis of NPs (52), we expected that the distance between the two spindle poles may be impaired in Lis1hc/ko MEFs. Lis1hc/ko MEFs displayed a moderate change in pole distance (8.8 ± 0.2 μm) compared with WT MEFs (7.9 ± 0.2 μm), reflecting alteration in the pulling forces on two spindle poles (Fig. 5D).
Figure 5.
Loss of LIS1 impairs spindle orientation during mitosis of MEFs. (A) Confocal Z-stack image series of mitotic spindles stained with pericentrin (centrosomal marker, green), α-tubulin (mitotic spindle, red) and DAPI (blue). MEFs were arrested in metaphase with MG132 treatment for 2 h. Scale bars: 5 μm. (B) Schematic representation of spindle orientation in mitotic cells. Spindle angle (α°) and distance between spindle poles (D, μm) were measured by taking Z-stack confocal images from 0.5 μm thick sections. Centrosomes were identified with co-localization of pericentrin and α-tubulin (marked in yellow). (C) Average spindle angles of Lis1hc/ko MEFs and WT MEFs. (D) Average distance between spindle poles. Bars in (C) and (D): mean ± SEM, Asterisks in (C) and (D): ***P < 0.001 by Student's _t_-test.
To determine whether spindle misorientation in Lis1hc/ko MEFs results from cell shape changes, we analyzed the cell height and the longest cell axis of the metaphase cells from Lis1hc/ko MEFs and WT MEFs. Neither was significantly altered in Lis1hc/ko MEFs compared with WT MEFs (cell height Lis1hc/ko: 12.7 ± 0.6 μm versus WT: 13.7 ± 0.4 μm, and long axis Lis1hc/ko: 17.9 ± 0.9 μm versus WT: 17.5 ± 0.7 μm) (Supplementary Material, Fig. S4A and B).
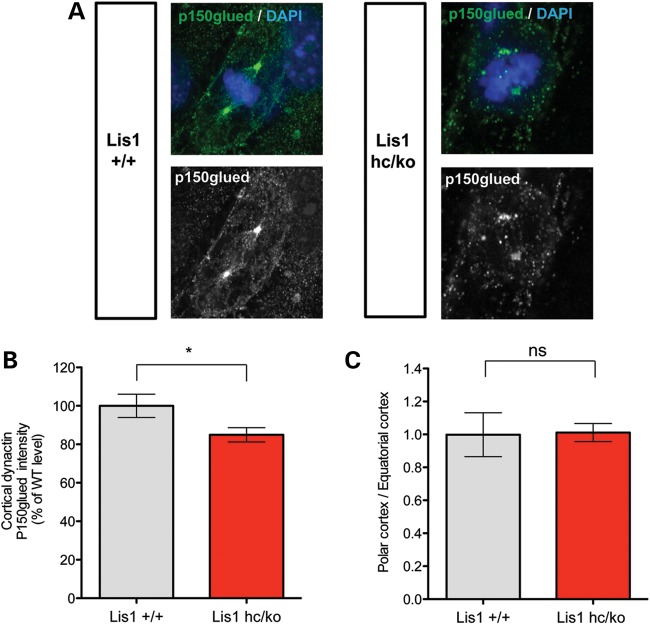
Reduced cortical dynein/dynactin complex in Lis1 mutant MEFs during mitosis
To examine cortically located dynein/dynactin complexes in Lis1hc/ko MEFs, we performed immunostaining with the p150Glued subunit of dynactin. Cortical staining of p150Glued was significantly reduced in Lis1hc/ko MEFs to 85% compared with WT MEFs 100% (Lis1hc/ko: 85 ± 3.7% versus WT: 100 ± 6.1%) (Fig. 6A and B). We also examined the distribution ratio of dynactin subunit pools between equatorial cortex-associated and polar cortex-associated p150Glued, but this ratio was not significantly altered in Lis1hc/ko MEFs (Lis1hc/ko: 1.0 ± 0.1% versus WT: 1.0 ± 0.1%) (Fig. 6C). In addition, astral or central MT-associated p150Glued signals were prominent in WT MEFs, but they were severely reduced in Lis1hc/ko MEFs. Reduced localization of cortical dynein/dynactin complex in M phase may ultimately result in spindle misorientation in Lis1hc/ko MEFs. These findings suggest that LIS1 mediates dynein/dynactin targeting to the cell cortex in this cell cycle, contributing to spindle orientation and positioning to ensure proper cell division.
Figure 6.
Loss of LIS1 results in less cortical dynein/dynactin complex recruitment to the cell cortex during mitosis. (A) Distribution of a dynactin subunit, p150glued (green) in mitosis of WT and Lis1hc/ko MEFs. Chromosomes were stained with DAPI (blue). Scale bars: 5 μm. (B) Relative fluorescence intensity of cortically located dynactin p150glued in WT and Lis1hc/ko MEFs. (C) Ratio of polar cortex-associated dynactin normalized by equatorial cortex-associated dynactin p150glued (more than eight cells were analyzed from each genotype). Asterisk in (A): *P < 0.05, (B) ns: not significant by Student's _t_-test.
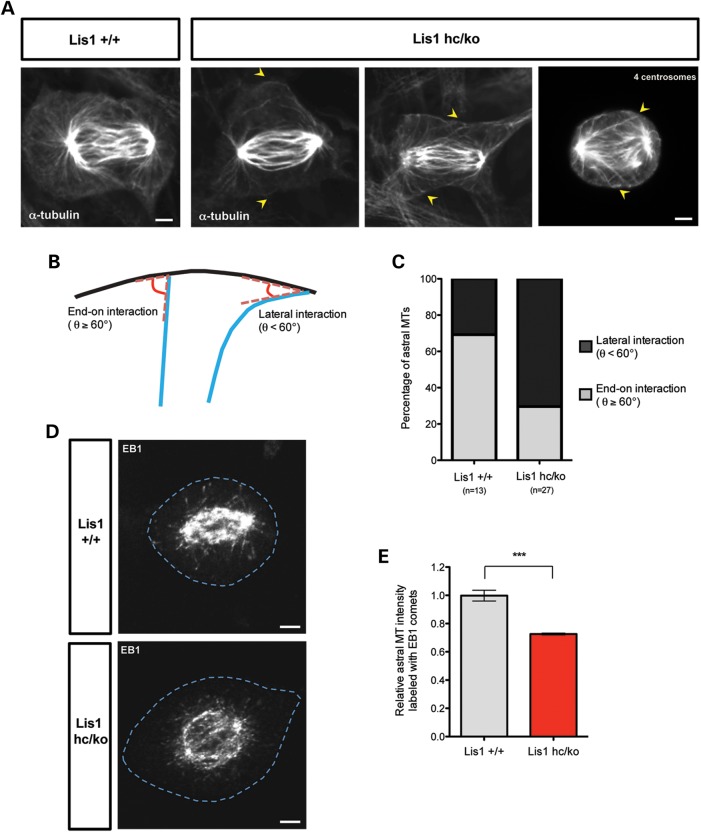
Abnormal interaction between plus-ends of astral MTs and the cell cortex in Lis1 mutant MEFs
Previous studies (12,13) and the dynein/dynactin studies described above (Fig. 6) suggested that astral MTs may be reduced in Lis1hc/ko MEFs, so we examined astral MTs in great detail. We found that WT MEFs in early anaphase displayed clear astral MTs–cortex interactions that maintained a wide angle compared with lateral surface of the cell cortex [end-on interaction, Fig. 7A and B, see (55)]. Each lobe of the polar cortex was filled with straightened astral MT tips, suggesting there were pulling forces between spindle poles and the cell cortex. In contrast, the radial arrays of astral MTs were reduced in Lis1hc/ko MEFs. These astral MTs were often curled and contacted the cell cortex laterally with a shallow angle [lateral or side-on interaction, MT sliding, Fig. 7A and B, see (55)], suggesting that these astral MTs were not under strong tension. The angles between astral MT tips and the cortical membrane were measured (θ°). The lateral interaction (θ° < 60) of astral MTs to the polar cortex was significantly increased in Lis1hc/ko MEFs (Lis1hc/ko: 29.6% versus WT: 69.2%) (Fig. 7B and C). In addition, astral MTs in WT MEFs never crossed the equatorial region where the cleavage furrow forms, but in the case of Lis1hc/ko MEFs with extra centrosomes, a fraction of astral MTs connected to the opposite polar cortex far from the closer pole (Fig. 7A, right panel).
Figure 7.
Loss of LIS1 causes aberrant interactions between astral MT plus-ends and the cell cortex. (A) The astral MTs in early anaphase from Lis1hc/ko MEFs and WT MEFs stained with α-tubulin after glutaraldehyde fixation. Arrow: aberrant astral MT tips in Lis1hc/ko MEFs. (Right panel) Misattachment of astral MTs to the opposite polar cortex in Lis1hc/ko MEFs harboring extra centrosomes (four centrosomes). (B) Schematic representation of interaction between astral MTs and the cell cortex–end-on interaction (θ° ≥ 60) versus lateral interaction (θ° < 60). (C) Quantification of types of astral MT interaction with the cell cortex in MEFs. (D) Localization of MT plus-ends stained with EB1 (MT plus-end binding protein) in metaphase of Lis1hc/ko MEFs and WT MEFs. Blue dashed lines indicate the cell membranes. Scale bars: 5 μm. (E) Relative intensity of astral MTs labeled with EB1 in metaphase-arrested MEFs (more than eight cells were analyzed from each genotype). Asterisk in (E): ***P < 0.001 by Student's _t_-test.
The dynein/dynactin complex is targeted to MT plus-end tips by EB1, a MT plus-end binding protein (56,57), and is necessary for establishment of astral MTs and appropriate spindle orientation in M phase (53). To investigate the interaction between MT plus-end tips and the cell cortex, we examined EB1 localization and distribution in WT and Lis1hc/ko MEFs. In metaphase, WT MEFs displayed many discrete EB1-labeled MT plus-end tips nearly touching the cell cortex (Fig. 7D), whereas fewer MT plus-end tips were found in the vicinity of the cell cortex and the length of each astral MT strand was reduced in Lis1hc/ko MEFs (Fig. 7D). We analyzed the relative astral MTs signal intensity with EB1 comets in metaphase-arrested cells from each genotype. Lis1hc/ko MEFs had less astral MT comets compared with WT MEFs by 0.73-fold (Lis1hc/ko: 1.00 ± 0.04 versus WT: 0.73 ± 0.01) (Fig. 7E). Together, these data support the notion of reduced spindle pulling forces in Lis1hc/ko MEFs by aberrant and weakened interactions between astral MTs and the cell cortex.
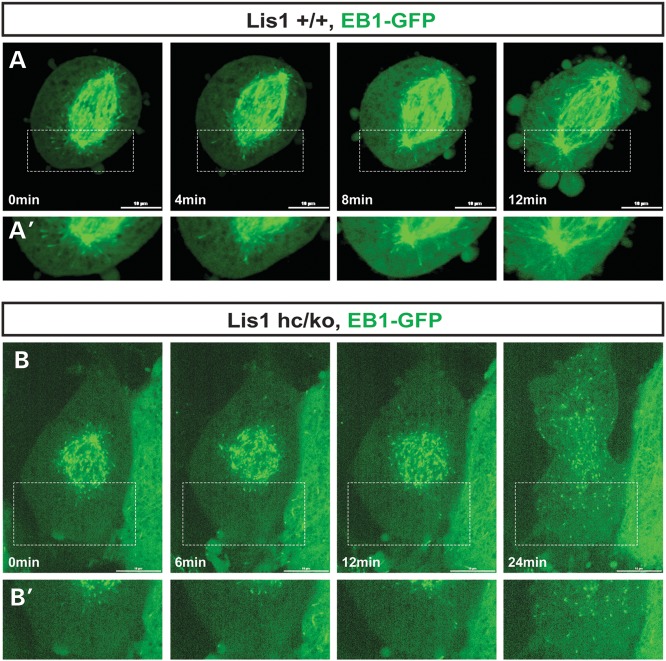
Reduced density of astral MT plus-ends near the cell cortex in Lis1 mutant MEFs
To further investigate how LIS1 affects the dynamics of MT plus-ends, we performed time-lapse live cell imaging experiments in EB1-GFP expressing MEFs undergoing M phase and traced MT plus-end comet movements by confocal microscopy. In WT MEFs, the growing MT plus-end comets (EB1-GFP) were generated mainly from the centrosomes and EB1 comets established transient contacts to the cell cortex in metaphase (Fig. 8A and Supplementary Material, Video S3). The frequency of EB1 comet contacts with the cortex was further increased at the onset of anaphase with spindle pole elongation, suggesting that pulling forces were generated from this MT–cortex interaction. However, Lis1hc/ko MEFs displayed significantly decreased numbers of MT asters with EB1 comets, and the frequency of transient interactions of MT plus-end tips with the cell cortex was also reduced (Fig. 8B and Supplementary Material, Video S4). These findings demonstrate that cortical interactions of MT asters were significantly impaired in _Lis1_-deficient mitosis.
Figure 8.
Reduced frequency of movements of EB1-labeled astral MT plus-ends near the cell cortex. (A) Frame series of time-lapse live cell imaging movies from WT MEF infected with EB1-GFP adenovirus. (B) Frame series of time-lapse live cell imaging movies from Lis1hc/ko MEF. (A′, B′) High magnification images from insets in (A) and (B). Scale bars: 10 μm. These montages are generated from the representative movies of dividing MEFs from each genotype (more than ten cells were imaged for live cell imaging).
Spindle misorientation phenotype in Lis1 mutant MEFs is rescued by MT stabilization, as well as LIS, NDEL1 and dynein overexpression
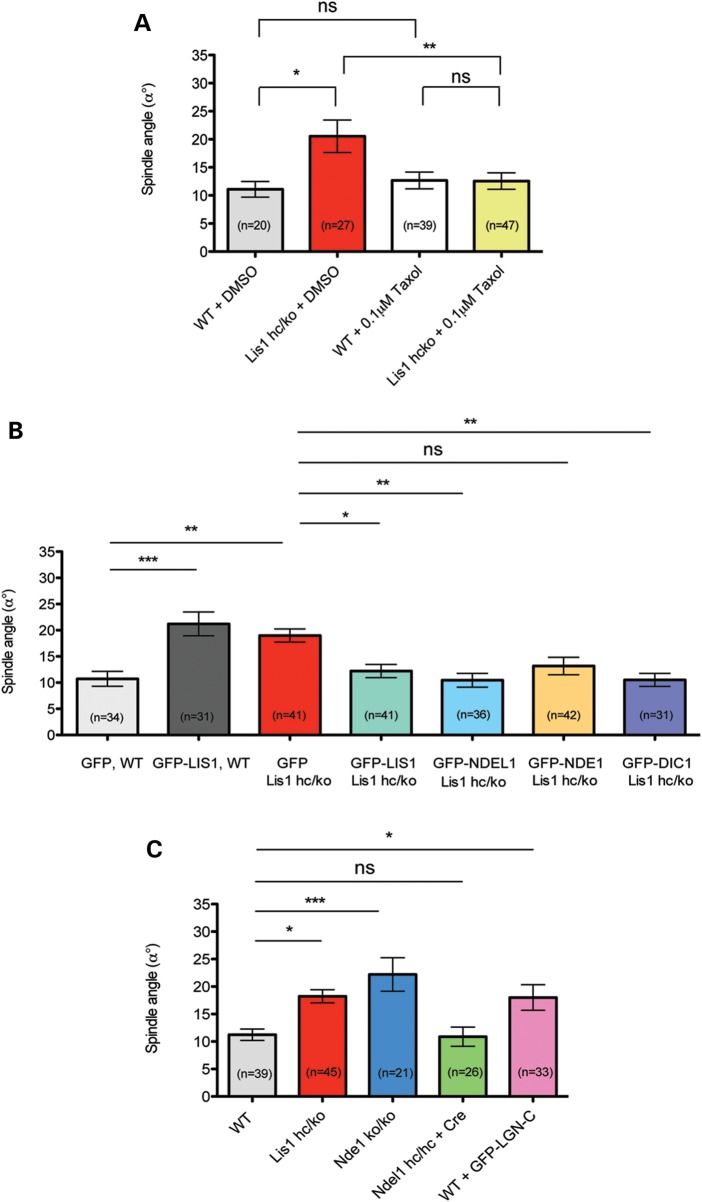
We first hypothesized that spindle misorientation may result from misregulation of MTs in Lis1hc/ko MEFs. To test this possibility, we treated WT MEFs and Lis1hc/ko MEFs with taxol, a MT stabilizing reagent, and then analyzed spindle orientation in metaphase cells. DMSO-treated Lis1hc/ko MEFs displayed significant spindle misorientation compared with DMSO-treated WT control MEFs (Lis1hc/ko: α° = 20.5 ± 2.9 versus. WT: α° = 11.1 ± 1.4) (Fig. 9A). In a previous study, a low dose of taxol treatment had been used to restore astral MTs by MT stabilization (58). We noted that a low dose of taxol treatment in Lis1hc/ko MEFs led to astral MT enrichment (based on EB1 staining, data not shown). Importantly, taxol rescued spindle misorientation. The average angle of spindle orientation from taxol-treated Lis1hc/ko MEFs was similar to those of taxol-treated WT MEFs (taxol-treated Lis1hc/ko: α° = 12.6 ± 1.5 versus taxol-treated WT: α° = 12.7 ± 1.5) (Fig. 9A). These results suggest that taxol treatment can restore MT plus-end movements near the cell cortex by stabilizing astral MTs in Lis1hc/ko MEFs.
Figure 9.
Spindle misorientation phenotype in Lis1 mutant MEFs is rescued by MT stabilization and overexpression of several components of the LIS1–NDEL1–dynein complex. (A) Average spindle angles in WT and Lis1hc/ko MEFs treated with DMSO and taxol. (B) MEFs infected with retroviruses encoding GFP, GFP-LIS1, GFP-NDEL1, GFP-NDE1 and GFP-DIC1. (C) Average spindle angles in NDE1 KO MEFs (Nde1ko/ko), NDEL1 CKO MEFs (Ndel1hc/hc + Cre) and GFP-LGN-C overexpressing WT MEFs. Bars in (A)–(C): mean ± SEM, Asterisks in (A)–(C): *P < 0.05, **P < 0.01, ***P < 0.001 by ANOVA with Bonferroni's post hoc test. ns, not significant.
Since LIS1 and its binding partner proteins, NDE1, NDEL1 and cytoplasmic dynein, are all localized close to the cell cortex in WT MEFs, we hypothesized that ectopic overexpression of these proteins may rescue spindle misorientation of Lis1hc/ko MEFs during mitosis by sequestering the remaining LIS1 to help transport it to the critical sites such as the cell cortex. We generated a series of retroviruses encoding GFP, GFP-LIS1, GFP-NDEL1, GFP-NDE1 and GFP-DIC1. MEFs were transduced with each virus and spindle orientation was measured from metaphase-arrested GFP-positive cells. As expected, spindles were severely misoriented in GFP-infected Lis1hc/ko MEFs (Lis1hc/ko + GFP, control) (Fig. 9B). Overexpression of ectopic LIS1 in WT MEFs resulted in severe spindle misorientation (Fig. 9B), suggesting that precise levels of LIS1 are critical for normal spindle orientation. As a control, normal spindle orientation in Lis1hc/ko MEFs was restored by overexpression of GFP-LIS1. Interestingly, Lis1hc/ko MEFs infected with GFP-NDEL1 and GFP-DIC1 displayed significantly reduced and rescued spindle angles compared with GFP-infected control Lis1hc/ko MEFs to the spindle angles similar to normal WT MEFs (Fig. 9B). However, GFP-NDE1 overexpression did not rescue spindle misorientation in Lis1hc/ko MEFs. These results support the interpretation that LIS1 acts cooperatively with NDEL1 and dynein, but not NDE1, to regulate precise spindle orientation via a LIS1–NDEL1–dynein complex to regulate MTs. In contrast, overexpression of GFP-LIS1 did not result in corrections of the abnormal numbers of centrosomes in the Lis1hc/ko MEFs. This phenotype may be the result of chronic reduction of LIS1 protein levels, and may not be corrected by expression of LIS1 over the short time frame of this experiment.
To test whether the loss of function of each protein component of LIS1 protein complex has similar effects on spindle orientation, we measured spindle angles in MEFs derived from NDE1 KO (Nde1ko/ko) and NDEL1 CKO (Ndel1hc/hc) mice. To obtain NDEL1 complete KO MEFs, Ndel1hc/hc MEFs were infected with Cre–mCherry retrovirus and only mCherry-positive MEFs were selected for spindle measurement. Loss of NDE1 in MEFs severely disrupted spindle orientation, consistent with a previous report that NDE1 siRNA-mediated knockdown in HeLa cells resulted in severe spindle misorientation with reduced cortical dynein/dynactin (59). However, loss of NDEL1 did not affect spindle orientation (Fig. 9C).
To address whether the cortical recruitment of dynein/dynactin is a critical common mechanism to regulate mitotic spindle, we performed experiments that modulate the activity of LGN (Leu-Gly-Asn-enriched protein). LGN mediates cortical targeting of dynein/dynactin complexes (60,61). Similar to LIS1, LGN is enriched at the cell cortex during mitosis of various mammalian cells (60) and LGN acts in upstream pathway of cortical dynein/dynactin recruitment to regulate proper spindle positioning (61). Overexpression of a GFP-LGN-C terminal fusion protein (hereafter, termed GFP-LGN-C) led to exclusive targeting of LGN specifically at the cell cortex (61,62) to disrupt cortical recruitment of dynein/dynactin. We introduced a GFP-LGN-C retrovirus in WT MEFs. Similar to Lis1 mutant MEFs (Lis1hc/ko), GFP-LGN-C overexpression in WT MEFs displayed severe defects in spindle orientation (Fig. 9C).
DISCUSSION
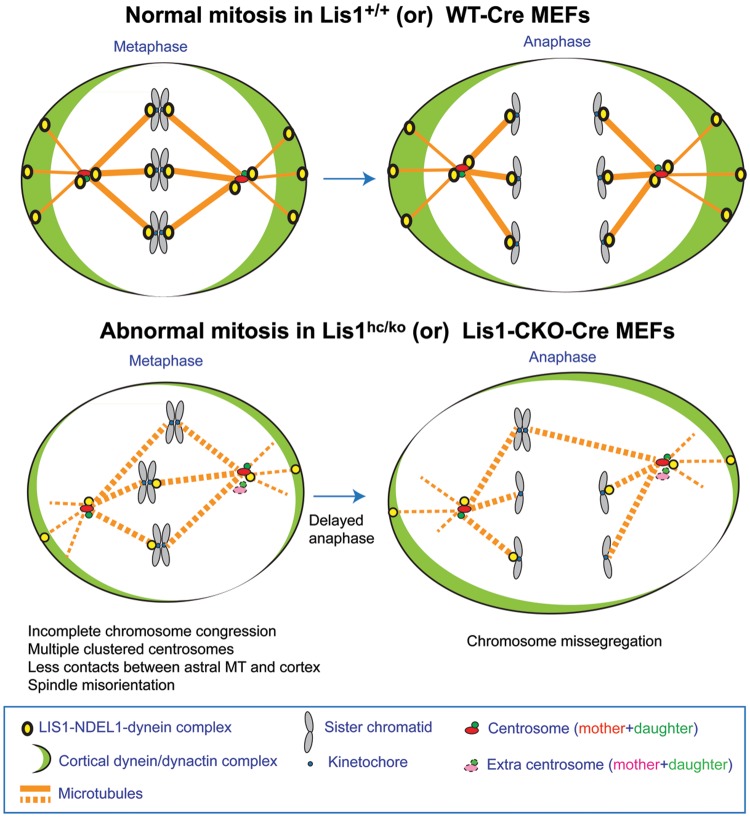
Heterozygous loss of function of human LIS1 results in the severe neurodevelopmental brain malformation disorder lissencephaly, a neuronal migration disorder. We and others demonstrated that LIS1 plays important roles during mitosis in mammalian cells (12,13,20,63). Using Lis1 mutant mice, we previously showed that _Lis1_-deficiency causes growth defects of MEFs and cell death of self-renewing NPs (13). However, the exact cellular mechanisms regulated by LIS1 at intracellular compartments during mitosis are not well understood. Due to a severe prometaphase arrest in M phase resulting from complete loss of function by siRNA-mediated knockdown or antibody injection against LIS1 (12,20,63), the dose-dependent mitotic functions of mammalian LIS1 protein have not been determined in previous studies. Here, we studied the detailed cellular processes regulated by LIS1 at different intracellular compartments of MEFs. We took advantage of our Lis1 genetic allelic series to control LIS1 protein levels by using Lis1 mutant MEFs, employing both TM-inducible acute conditional KOs (CreER, Lis1hc/hc + TM) and conventional KOs (Lis1hc/ko) with 35% LIS1 protein compared with WT. We found that mouse LIS1 is essential for maintaining normal centrosome numbers and preserving centrosomal integrity in mitotic cycle (Fig. 10). LIS1 has kinetochore-specific functions to complete chromosome congression by recruiting several kinetochore proteins to ensure anaphase progression with chromosome segregation. Time-lapse live cell imaging of MEFs undergoing M phases revealed that LIS1 plays crucial roles in mitotic spindle organization and structure. We also found that LIS1 is an important regulator of LIS1–NDEL1–dynein complex to enhance astral MT plus-end dynamics and to mediate offloading of dynein/dynactin on the cell cortex to establish proper spindle orientation during mitosis (Fig. 10). Interestingly, detailed functions of mouse LIS1 in mitotic MEFs in the present study are consistent with the roles of Pac1, the yeast Lis1 homolog. Dynein offloading to the stationary cell cortex from dynamic MT plus-end asters is mediated by an increase in the frequencies of cortical targeting of dynein/dynactin complex (64–66), suggesting evolutionarily conserved functions of LIS1 in the regulation of cortical dynein complex.
Figure 10.
Proposed working model of LIS1 function in mitotic cell divisions. See text in discussion.
Centrosome amplification at MT minus-ends in _Lis1_-deficient mitosis
Lis1 CKO MEFs frequently displayed aberrant centrosome number, suggesting that LIS1 is indispensible for centrosome maintenance. The extra centrosomes in Lis1hc/ko MEFs transiently clustered into two distinct opposite poles to form a pseudo-bipolar spindle. Since the presence of extra centrosomes delays mitotic progression (67), LIS1 may be involved in mitotic checkpoint control by maintaining a consistent centrosome number. Three possible mechanisms can cause abnormal centrosome numbers: (i) misregulation of centrosome assembly, (ii) failure of cytokinesis, and (iii) dysfunction of the centrosome duplication cycle (68,69). Centrosome morphology was abnormal in a majority of Lis1hc/ko MEFs because of centrosome clustering, suggesting that there are minor defects in centrosome assembly in Lis1hc/ko MEFs. However, the configuration of amplified centrosomes as mother–daughter centriole pairs and centrosome maturation were normal in Lis1hc/ko MEFs. Due to centrosome amplification, the over-production of multiple mother centrioles in M phase led to the formation of multiple primary cilia in interphase. We recently found defects in cytokinesis in Lis1hc/ko MEFs (unpublished data), which may partially contribute to centrosome number abnormality in _Lis1_-deficiency. Finally, dysregulation of the centrosome duplication cycle may be evoked by loss of LIS1 by misregulating a signaling pathway. For example, aberrant activation of Rho GTPases enhances ROCK2 kinase activity and promotes centrosome amplification (70–72). Lis1 heterozygous MEFs (Lis1ko/+) displayed elevated RhoA GTPase activity (73), suggesting that a high level of LIS1 protein expression is required for inhibiting RhoA GTPase. Therefore, misregulated RhoA signaling in Lis1hc/ko MEFs may cause ROCK activation to induce centrosome over-duplication.
Missegregation of chromosomes and fate determinants in _Lis1_-deficient mitosis
Appropriate levels of LIS1 protein at kinetochores are required for spindle checkpoint control to inhibit improper kinetochore-MT attachments and to execute chromosome segregation and inheritance by recruiting other kinetochore proteins (dynein and CLIP170). Extra centrosomes in Lis1hc/ko MEFs may lead to merotelic kinetochore-chromosome attachments by interfering with kinetochore-MT capture (74). Improper kinteochore-MT attachments and extra centrosomes promote genomic instability by generating aneuploid daughter cells. The frequent formation of chromatin bridges and micronuclei in _Lis1_-deficient mitosis reflect severe chromosome segregation defects. It has been shown that a human hepatocellular carcinoma cell type displays downregulation of LIS1 mRNA and protein, and the severity of tumor formation is strongly correlated with LIS1 downregulation (75). This suggests that LIS1 may act as a tumor suppressor under certain conditions. Together with our data, this study supports the notion that LIS1 is required for maintaining genomic stability to prevent the loss of chromosomes during mitosis.
LIS1-dependent centrosome number maintenance may play critical roles in the maintenance of genomic stability essential for asymmetric NP division during development. In Drosophila, PLK4 homologue mutants induce extra centrosome formation and misalignment of apical cell fate determinants during NP divisions (76). Multiple centrosomes-harboring NPs misorient the mitotic spindle and the relationship between mitotic spindles and apical markers was disrupted. We also frequently observed three centrosomes in apical NPs from Lis1 mutant embryonic mouse brains (unpublished data), suggesting that mammalian NP cells may employ similar mechanisms.
Spindle misorientation and reduced interaction between astral MT plus-ends and the cell cortex during mitosis in Lis1 mutant MEFs
Mitotic spindle orientation is critical for the proper segregation of chromosomes to the daughter cells. Here, we demonstrate that Lis1 mutant MEFs showed significant spindle misorientation with severe tilting compared with adhesion plane. Similarly, we previously found spindle orientation defects in NPs from Lis1 mutants (13). NDEL1- (77) and NDE1-depleted NPs (78) also displayed severe spindle misorientation. In the present study, we showed that _Lis1_-deficiency results in reduced cortical dynactin p150Glued in M phase of MEFs. The number of astral MTs and the frequency of movements of EB1-labeled MT plus-ends reaching to the cortex were significantly reduced in Lis1hc/ko MEFs. Due to the loss of LIS1, a key component of the LIS1–NDEL1–dynein/dynactin complex, MT asters may become destabilized at the cell cortex. By overexpression experiments, we confirmed that NDEL1 and dynein are the core mediators acting in LIS1-dependent pathways of mitotic spindle regulation. Most recently, it was shown that stationary barrier- or bead-attached dynein controls the dynamics of MT plus-ends in vitro (79,80). In addition, it has been shown that LGN functions as an upstream regulator to target cortical dynein/dynactin in HeLa cells (61). Taken together, these data indicate that LIS1- (or LGN-) mediated cortical targeting of dynein/dynactin complex is a key cellular mechanism to regulate contacts between astral MT plus-ends and the cell cortex during mitosis.
In addition to the function of the LIS1–NDEL1–dynein complex near the cell cortex that controls MT dynamics in mitotic spindles, other cytoskeletal components may contribute to the mitotic function of LIS1. Subcortical actin cytoskeletal components as well as actin retraction fibers at the adhesion sites have been implicated in mitotic spindle formation (81,82). Interestingly, Lis1 heterozygous mutant neurons (Lis1ko/+) displayed misregulated actin fibers at the leading edges during cell migration (83). This finding raises the possibility that dysfunction of the actin cytoskeleton affected by reduced LIS1 has at least an indirect impact on actin-dependent mitotic spindle regulation. The cell cortex is a critically important site for vigorous crosstalk and interactions between MTs and the actin cytoskeleton to integrate signal cues into mitotic spindle regulatory pathway. In the current study, we conclude that LIS1, as a central component of LIS1–NDEL1–dynein complex, participates in the regulation of MT plus-ends dynamics on astral MTs near the cell cortex to ensure mitotic spindle positioning and orientation. In future studies, it will be interesting to determine whether LIS1 directly regulates the actin cytoskeleton to promote actin fiber assembly at the cortical adhesion-binding sites during mitosis.
MATERIALS AND METHODS
Mice
Lis1 null (Lis1ko, formerly referred to as Pafah1b1tm1Awb) and HC (Lis1hc, formerly referred to as Pafah1b1tm2Awb(loxP)) alleles were used, which were described previously (13,42). Previously reported Nde1 null (Nde1ko, formerly referred to as Nde1tm1Caw) allele (78) and Ndel1 HC KO (Ndel1hc, formerly referred to as Ndel1tm1Shr) allele (84) were used for spindle orientation analysis. To induce Cre recombinase activation to delete Lis1 conditional allele, the TM-inducible CreERTM line (44) was used to mate with Lis1 mutant mice.
Cell culture
Primary MEFs were derived from E14.5 embryos and were cultured in DMEM (Mediatech) supplemented with 12.5% FBS (Gibco), penicillin/streptomycin and l-glutamine at 37°C in a 5% CO2 incubator. All MEFs used in this study were less than passage 4 (13). CreERTM recombinase activity was induced by administrating 4-hydroxy TM (Sigma, 100 nm) dissolved in culture media. H293T cells for viral packaging were maintained in 10% FBS (Gibco) in DMEM medium (Mediatech).
Retrovirus production and MEF infection
To produce retroviruses, pMV-GP (gag, pol), pCMV-G (VSV-G env) and pCX retroviral vectors were purified by Endofree Plasmid Maxiprep kit (Qiagen) and transfected in H293T cells with TransIT-LT1 transfection reagent (Mirus). Packaging vectors (pMV-GP, pCMV-G) were gifts from Atsushi Miyanohara (UCSD). pCLNR-H2BG (H2B-GFP) was a gift from Geoffrey Wahl (Salk institute, Addgene plasmid #17735). To construct pCX–mCherry–α-tubulin retroviral vector, cDNA of mCherry was amplified by PCR from pcDNA3.1-mCherry derived from an mCherry expression vector (a gift from Roger Tsien, UCSD). The mCherry cDNA PCR product was subcloned into pEGFP-α-tubulin (Clontech) digested with _Sal_I/_Not_I to generate mCherry–α-tubulin. Then, the PCR product of cDNA encoding mCherry–α-tubulin was ligated into the linearized pCX backbone vector derived from pCX-Centrin2-DsRed (gift from Joseph Gleeson, UCSD) with _Bam_HI/_Pac_I. Other GFP–fusion proteins used for rescue experiments (pCX-GFP-LIS1, pCX-GFP-NDEL1, pCX-GFP-NDE1 and pCX-GFP-DIC1) were generated by cDNA amplification from mammalian expression vectors pCMV-GFP-NDEL1 [described in (85)], pCMV-GFP-LIS1, pCMV-GFP-NDE1 [gifts from Shinji Hirotsune, Osaka City University, Japan, described in (84)] and pCMV-GFP-DIC1 [gifts from Shinji Hirotsune, described in (86)], followed by _Bam_HI/_Pac_I digestion for cloning into the pCX retroviral vector. To construct the pCX–Cre–mCherry retroviral vector, Cre cDNA PCR product was amplified from pBS500-CreGFP (a gift from Brian Sauer, NIH, Addgene plasmid #11920) and inserted into pcDNA3.1-mCherry with _Nhe_I/_Kpn_I. Then Cre–mCherry fusion cDNA was derived from this construct and subcloned into pCX vector with _Eco_RI/_Pac_I to generate pCX–Cre–mCherry. Protein expression of both Cre and mCherry was confirmed by Cre antibody staining and mCherry fluorescence. To construct pCX-GFP-LGN-C, GFP-LGN-C fragment cDNA was amplified from pIC389 [pBabe-GFP-LGN-C, a gift from Iain Cheeseman, MIT, described in Kiyomitsu and Cheeseman (61)]. We generated pCX-GFP-LGN-C construct by serial introductions of PCR products digested with _Bam_HI/_Pac_I and _Bam_HI, respectively. Viral supernatants (in DMEM without antibiotics and FBS) were collected at 48 h post-transfection and filtered through 0.45 μm filter (Sartorius). MEFs were infected with retrovirus by co-incubating the mixture of viral supernatant and fresh media for 24 h. During infection, 12.5% FBS was supplemented in the viral supernatant media along with 4 μg/ml polybrene (Sigma).
Western blotting
MEFs were lysed in Tris-Triton buffer (10 mm Tris pH 7.4, 100 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 10% glycerol, 0.1% SDS, 0.5% deoxycholate) with protease/phosphatase inhibitors. Lysates were collected on ice and centrifuged. Supernatants with protein extracts were transferred into new tubes, boiled at 95°C for 5 min and then stored at −20°C before use. Protein concentrations were determined by the BCA protein assay kit (Pierce) and equal amounts were loaded on a 10% SDS–PAGE resolving gel (Bio-Rad). After electrophoresis, proteins were transferred to a nitrocellulose membrane (Bio-Rad). The membrane was blocked in TBST (TBS with 0.1% Tween 20) with 2.5% skim milk for 1 h at RT and incubated with the following primary antibodies at 4°C overnight: rabbit anti-LIS1 (1:1000, a gift from Shinji Hirotsune), mouse anti-α-tubulin (Sigma, 1:8000) and mouse anti-β-actin (Sigma, 1:5000). Secondary antibodies used were HRP-conjugated goat anti-rabbit and goat anti-mouse (Jackson Lab, 1:10 000) which were incubated for 1 h. Antibody binding was detected via the ECL kit (Pierce). Relative protein amount was measured using the ImageJ software after background subtraction.
Immunocytochemistry
MEFs were grown on acid-washed and 0.2% gelatin (Millipore)-coated glass cover slips and fixed with 4% PFA in PBS for 20 min. For centrosomal protein immunostaining and EB1 staining, MEFs were fixed with −20°C cold methanol for 2 min. 2.5% normal goat serum (or FBS) and 0.1% Triton X-100 in PBS were used for blocking for 1 h at RT. The primary antibodies were diluted in blocking buffer and incubated overnight at 4°C. The primary antibodies used were: rabbit anti-LIS1 (Abcam, 1:250); mouse anti-α-tubulin (Sigma, 1:500); rat anti-α-tubulin (AbD Serotec, 1:1000); mouse anti-γ-tubulin (Sigma, 1:500); rabbit anti-pericentrin (Covance, 1:1000); mouse anti-p150Glued (BD bioscience, 1:200); mouse anti-EB1 (BD bioscience, 1:200); mouse anti-GFP (Invitrogen, 1:400); rabbit anti-GFP (Invitrogen, 1:400); mouse anti-centrin (Millipore, 1:200); rabbit anti-cennexin/ODF2 (Abcam, 1:200); rabbit anti-ninein (Abcam, 1:200); and rabbit anti-Cep164 (1:1000, a gift from Erich Nigg, University of Basel, Switzerland). The following goat secondary antibodies were incubated for 1 h at RT: anti-mouse AlexaFluor-488; anti-mouse AlexaFluor-594; anti-rabbit AlexaFluor-488; anti-rabbit Alexafluor-568; anti-rat AlexaFluor-488; and anti-rat AlexaFluor-647 (Invitrogen). ProLong gold antifade reagent with DAPI (Invitrogen) was used to counterstain the nucleus and as mounting medium. All the fixed sample images were captured using a Nikon C1si laser-scanning confocal microscope with a ×60 1.4 NA PlanApo oil objective lense (Nikon).
For primary cilia staining, MEFs were serum-starved for 24 h with 0.5% FBS in DMEM and fixed in 4% PFA in PBS. Primary mouse anti-acetylated α-tubulin (Sigma, 1:1000) was used to identify primary cilia. For visualization of astral MTs, MEFs were fixed in 0.25% glutaraldehyde in BRB80 buffer (80 mm PIPES pH 6.8, 1 mm MgCl2, 1 mm EGTA) for 10 min, and then treated with 0.2% sodium borohydride in PBS for 20 min, changing the borohydride solution two times. Primary rat anti-α-tubulin antibody was diluted in blocking buffer (2% FBS, 0.1% Triton X-100 in PBS). To visualize cortical dynactin p150Glued, MEFs were pre-extracted with 0.5% Triton X-100 in PHEM buffer (120 mm PIPES, 50 mm HEPES, 20 mm EGTA, 8 mm MgSO4) with 5 μm taxol (Sigma) for 1 min and then fixed with −20°C cold methanol for 2 min. To compare cortical dynactin p150Glued, we quantified the amount of staining in metaphase cells from each genotype by measuring fluorescence intensity at two equatorial cortex locations and four polar cortex locations per cell.
Mitotic spindle analysis and cell shape analysis
For analysis of the mitotic spindle, MEFs were arrested in metaphase by treatment with proteosome inhibitor, 10 μm MG132 (EMD biosciences) for 2 h. From the cover slips of MEFs, a series of Z stack images (0.5 μm apart) of metaphase mitotic spindles stained with anti-pericentrin, anti-α-tubulin antibodies were obtained using the Nikon C1si laser-scanning confocal microscope (Biological Imaging Development Center, UCSF). The linear (x–y plane) and vertical (z-axis) distance between spindle poles was measured from Nikon EZ-C1 imaging software. Mitotic spindle length (D) and spindle angles (α°) were calculated by 3D trigometric functions. The drug treatment in MEFs was performed in following conditions: 1 μm taxol (Sigma) were added to the medium and incubated for 30 min after the treatment of 10 μm MG132 for 2 h. Cell shape was analyzed from the MEFs incubated with lipophilic dye, DHCC (3,3′-dihexyoxacarbo cyanine inodide, Sigma) for 10 min and then treated with 10 μm MG132 for 2 h.
Kinetochore staining and quantification
MEFs were treated with 10 μm nocodazole (Sigma) for 1 h under normal incubation conditions to accumulate proteins at the kinetochores. Then MEFs were fixed with −20°C cold methanol for 2 min. Confocal images were obtained from an Olympus FV1000 laser scanning confocal microscope. From each genotype, 10 cells were analyzed and compared in softWoRx Explorer (Applied Precision) using 10 × 10 window in Data Inspector. Interkinetochore distance was determined using the line segment tool across Z-stacks. The total immunofluorescence of 10 kinetochores was averaged for each cell. Primary antibodies were: goat anti-LIS1 (Abcam, 1:200); mouse anti-p150glued (BD bioscience, 1:200); rabbit anti-CLIP170 (Holly Goodson, University of Norte Dame, 1:200); mouse anti-DIC70.1 (Sigma, 1:25); human SH-CREST autoimmune serum (a gift from William Brinkley, Baylor College of Medicine, 1:10 000); and Mad2 (a gift from Don Cleveland, UCSD, 1:200). Secondary antibodies were: donkey anti-rabbit FITC; donkey anti-goat Cy3; donkey anti-human Cy2, and donkey anti-human Cy3 (Jackson Laboratory).
Time-lapse live cell imaging
Glass-bottom six-well tissue culture plates (MatTek) were used for live cell imaging of mitosis. The plates were pre-incubated with 0.2% gelatin (Millipore) solution before plating MEFs on glass-bottom dishes 24 h prior to live cell imaging. Retroviruses were transduced into primary MEFs by co-incubating with viral supernatants for 24 h. 4-hydroxy TM (100 nm, Sigma) was used for 12 h for CreERTM for Cre transgene activation. MEFs expressing both H2B-GFP and mCherry–α-tubulin retroviruses were recorded with a time-lapse Nikon Ti microscope equipped with 5% CO2 and a 37°C temperature-controlled chamber (Nikon imaging center, UCSF). To monitor long-term mitosis of MEFs, multiple points were selected for acquiring each fluorescence image (GFP/mCherry) with a motorized stage and perfect focus function. Fluorescence was captured with a Coolsnap camera (Roper Scientific) with exposure time of 100 ms for GFP and 200 ms for mCherry. Time-lapse images of cell division were taken every 1 min (up to 12 h). Filters and multipoint scanning were controlled by NIS-Elements imaging software (Nikon).
Tracking EB1-GFP
EB1-GFP was amplified using oligos (fwd: caccatggcagtgaacgtatactca and rev: ttacttgtacagctcgtccat) and inserted into pTOPO vector and subsequently cloned into pAd/CMV/V5-DEST with Gateway cloning (Invitrogen). Adenovirus particles were purified as described previously (87). One microliter of EB1-GFP adenovirus was added to MEF growth media 24 h prior to imaging. Cells were treated with 9 μm RO-3306 (EMD bioscience), a CDK1 inhibitor, for 18 h to arrest cells at the G2/M transition of the cell cycle. Mitosis of MEFs was imaged after drug release by changing to fresh media containing 20 mm HEPES (Sigma).
EB1-GFP protein dynamics were imaged at 37°C with a ×100 NA 1.49 oil-immersed objective lens (CFI Apo TIRF, Nikon) equipped with a spinning-disk confocal scanning unit (Borealis-modified CSU-X1, Spectral Applied Research) using a Nikon Ti microscope (Nikon) with a 488 nm laser, electronic shutters, a cool charged-coupled device camera (CoolSnap HQ, Photometrics) and controlled by NIS-Elements software (Nikon). MT plus-ends were tracked in time-lapse images from EB1-GFP expressing MEFs acquired every 30 s using perfect focus function. Three sections of Z stacks (2 μm apart) were acquired at every time point.
Online supplementary materials
Supplementary Material, Figure S1 shows LIS1 localization during mitosis from WT and Lis1 mutant MEFs and the amount of protein expression in each genotype. Supplementary Material, Figure S2 shows primary cilia formation from WT and Lis1 mutant MEFs and quantification of multiple cilia. Supplementary Material, Figure S3 shows kinetochore protein staining in WT and Lis1 mutant MEFs and quantification of fluorescence intensity at kinetochores. Supplementary Material, Figure S4 shows quantification of metaphase cell morphology in WT and Lis1 mutant MEFs. Supplementary Material, Video S1 shows the time-lapse live cell imaging of mitotic cell division from CreERTM;Lis1+/+ MEFs treated with 4-hydroxy TM. H2B-GFP and mCherry–α-tubulin labeled fluorescence signals were acquired with a 1 min interval (as shown in Fig. 1A, upper panel). Supplementary Material, Video S2 shows the time-lapse live cell imaging of mitotic cell division from CreERTM;Lis1hc/hc MEFs treated with 4-hydroxy TM. H2B-GFP and mCherry-α-tubuin labeled fluorescence signals were acquired with a 1 min interval (as shown in Fig. 1A, lower panel). Supplementary Material, Video S3 shows the dynamic movements of MT plus-ends comets labeled with EB1-GFP in WT MEFs. Confocal images were acquired with 30 s interval (as shown in Fig. 8A). Supplementary Material, Video S4 shows the dynamic movements of MT plus-ends comets labeled with EB1-GFP in Lis1hc/ko mutant MEFs. Confocal images were acquired with 30 s interval (as shown in Fig. 8B).
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by National Institutes of Health, R01 NS41030andNational Institutes of Health,R01 HD047380. H.M.M. was supported by a UCSF Graduate Student Research Award.
Supplementary Material
Supplementary Data
REFERENCES
- 1.Holland A.J., Cleveland D.W. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev.. Mol. Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 3.Zimmerman W., Doxsey S.J. Construction of centrosomes and spindle poles by molecular motor-driven assembly of protein particles. Traffic. 2000;1:927–934. [PubMed] [Google Scholar]
- 4.Doxsey S., McCollum D., Theurkauf W. Centrosomes in cellular regulation. Ann. Rev. Cell Dev. Biol. 2005;21:411–434. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- 5.Nigg E.A., Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat. Cell Biol. 2011;13: 1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glotzer M. Mitosis: don't get mad, get even. Curr. Biol. 1996;6:1592–1594. doi: 10.1016/s0960-9822(02)70781-1. [DOI] [PubMed] [Google Scholar]
- 7.Kirschner M.W., Mitchison T. Microtubule dynamics. Nature. 1986;324:621. doi: 10.1038/324621a0. [DOI] [PubMed] [Google Scholar]
- 8.Kline-Smith S.L., Walczak C.E. Mitotic spindle assembly and chromosome segregation: refocusing on microtubule dynamics. Mol. Cell. 2004;15:317–327. doi: 10.1016/j.molcel.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 9.Glotzer M. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 2009;10:9–20. doi: 10.1038/nrm2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moore J.K., Cooper J.A. Coordinating mitosis with cell polarity: molecular motors at the cell cortex. Semin. Cell Dev. Biol. 2010;21:283–289. doi: 10.1016/j.semcdb.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coquelle F.M., Caspi M., Cordelieres F.P., Dompierre J.P., Dujardin D.L., Koifman C., Martin P., Hoogenraad C.C., Akhmanova A., Galjart N., et al. LIS1, CLIP-170's key to the dynein/dynactin pathway. Mol. Cell Biol. 2002;22:3089–3102. doi: 10.1128/MCB.22.9.3089-3102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faulkner N.E., Dujardin D.L., Tai C.Y., Vaughan K.T., O'Connell C.B., Wang Y., Vallee R.B. A role for the lissencephaly gene LIS1 in mitosis and cytoplasmic dynein function. Nat. Cell Biol. 2000;2:784–791. doi: 10.1038/35041020. [DOI] [PubMed] [Google Scholar]
- 13.Yingling J., Youn Y.H., Darling D., Toyo-Oka K., Pramparo T., Hirotsune S., Wynshaw-Boris A. Neuroepithelial stem cell proliferation requires LIS1 for precise spindle orientation and symmetric division. Cell. 2008;132:474–486. doi: 10.1016/j.cell.2008.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reiner O., Carrozzo R., Shen Y., Wehnert M., Faustinella F., Dobyns W.B., Caskey C.T., Ledbetter D.H. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature. 1993;364:717–721. doi: 10.1038/364717a0. [DOI] [PubMed] [Google Scholar]
- 15.Hattori M., Adachi H., Tsujimoto M., Arai H., Inoue K. Miller-Dieker lissencephaly gene encodes a subunit of brain platelet-activating factor acetylhydrolase [corrected] Nature. 1994;370: 216–218. doi: 10.1038/370216a0. [DOI] [PubMed] [Google Scholar]
- 16.Xiang X., Osmani A.H., Osmani S.A., Xin M., Morris N.R. Nudf, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol. Biol. Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris N.R., Efimov V.P., Xiang X. Nuclear migration, nucleokinesis and lissencephaly. Trends Cell Biol. 1998;8:467–470. doi: 10.1016/s0962-8924(98)01389-0. [DOI] [PubMed] [Google Scholar]
- 18.Smith D.S., Niethammer M., Ayala R., Zhou Y., Gambello M.J., Wynshaw-Boris A., Tsai L.H. Regulation of cytoplasmic dynein behaviour and microtubule organization by mammalian Lis1. Nat. Cell Biol. 2000;2:767–775. doi: 10.1038/35041000. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki S., Shionoya A., Ishida M., Gambello M.J., Yingling J., Wynshaw-Boris A., Hirotsune S. A LIS1/NUDEL/cytoplasmic dynein heavy chain complex in the developing and adult nervous system. Neuron. 2000;28:681–696. doi: 10.1016/s0896-6273(00)00146-x. [DOI] [PubMed] [Google Scholar]
- 20.Tai C.Y., Dujardin D.L., Faulkner N.E., Vallee R.B. Role of dynein, dynactin, and CLIP-170 interactions in LIS1 kinetochore function. J. Cell Biol. 2002;156:959–968. doi: 10.1083/jcb.200109046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Busson S., Dujardin D., Moreau A., Dompierre J., De Mey J.R. Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr. Biol. 1998;8:541–544. doi: 10.1016/s0960-9822(98)70208-8. [DOI] [PubMed] [Google Scholar]
- 22.Howell B.J., McEwen B.F., Canman J.C., Hoffman D.B., Farrar E.M., Rieder C.L., Salmon E.D. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J. Cell Biol. 2001;155:1159–1172. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merdes A., Ramyar K., Vechio J.D., Cleveland D.W. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 1996;87:447–458. doi: 10.1016/s0092-8674(00)81365-3. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen-Ngoc T., Afshar K., Gonczy P. Coupling of cortical dynein and G alpha proteins mediates spindle positioning in Caenorhabditis elegans. Nat. Cell Biol. 2007;9:1294–1302. doi: 10.1038/ncb1649. [DOI] [PubMed] [Google Scholar]
- 25.O'Connell C.B., Wang Y.L. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol. Biol. Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schroer T.A. Dynactin. Ann. Rev. Cell Dev. Biol. 2004;20:759–779. doi: 10.1146/annurev.cellbio.20.012103.094623. [DOI] [PubMed] [Google Scholar]
- 27.Gonczy P. Mechanisms of spindle positioning: focus on flies and worms. Trends Cell Biol. 2002;12:332–339. doi: 10.1016/s0962-8924(02)02306-1. [DOI] [PubMed] [Google Scholar]
- 28.Dujardin D.L., Vallee R.B. Dynein at the cortex. Curr. Opin. Cell Biol. 2002;14:44–49. doi: 10.1016/s0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- 29.Sheeman B., Carvalho P., Sagot I., Geiser J., Kho D., Hoyt M.A., Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr. Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 30.Derewenda U., Tarricone C., Choi W.C., Cooper D.R., Lukasik S., Perrina F., Tripathy A., Kim M.H., Cafiso D.S., Musacchio A., et al. The structure of the coiled-coil domain of Ndel1 and the basis of its interaction with Lis1, the causal protein of Miller-Dieker lissencephaly. Structure. 2007;15:1467–1481. doi: 10.1016/j.str.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 31.Efimov V.P., Morris N.R. The LIS1-related NUDF protein of Aspergillus nidulans interacts with the coiled-coil domain of the NUDE/RO11 protein. J. Cell Biol. 2000;150:681–688. doi: 10.1083/jcb.150.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niethammer M., Smith D.S., Ayala R., Peng J., Ko J., Lee M.S., Morabito M., Tsai L.H. NUDEL Is a novel Cdk5 substrate that associates with LIS1 and cytoplasmic dynein. Neuron. 2000;28:697–711. doi: 10.1016/s0896-6273(00)00147-1. [DOI] [PubMed] [Google Scholar]
- 33.Liang Y., Yu W., Li Y., Yang Z., Yan X., Huang Q., Zhu X. Nudel functions in membrane traffic mainly through association with Lis1 and cytoplasmic dynein. J. Cell Biol. 2004;164:557–566. doi: 10.1083/jcb.200308058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stehman S.A., Chen Y., McKenney R.J., Vallee R.B. Nude and NudEL are required for mitotic progression and are involved in dynein recruitment to kinetochores. J. Cell Biol. 2007;178:583–594. doi: 10.1083/jcb.200610112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng Y., Olson E.C., Stukenberg P.T., Flanagan L.A., Kirschner M.W., Walsh C.A. LIS1 regulates CNS lamination by interacting with mNudE, a central component of the centrosome. Neuron. 2000;28:665–679. doi: 10.1016/s0896-6273(00)00145-8. [DOI] [PubMed] [Google Scholar]
- 36.McKenney R.J., Vershinin M., Kunwar A., Vallee R.B., Gross S.P. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenney R.J., Weil S.J., Scherer J., Vallee R.B. Mutually exclusive cytoplasmic dynein regulation by NudE-Lis1 and dynactin. J. Biol. Chem. 2011;286:39615–39622. doi: 10.1074/jbc.M111.289017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J., Lee W.L., Cooper J.A. NudEL targets dynein to microtubule ends through LIS1. Nat. Cell Biol. 2005;7:686–690. doi: 10.1038/ncb1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zylkiewicz E., Kijanska M., Choi W.C., Derewenda U., Derewenda Z.S., Stukenberg P.T. The N-terminal coiled-coil of Ndel1 is a regulated scaffold that recruits LIS1 to dynein. J. Cell Biol. 2011;192:433–445. doi: 10.1083/jcb.201011142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam C., Vergnolle M.A., Thorpe L., Woodman P.G., Allan V.J. Functional interplay between LIS1, NDE1 and NDEL1 in dynein-dependent organelle positioning. J. Cell Sci. 2010;123:202–212. doi: 10.1242/jcs.059337. [DOI] [PubMed] [Google Scholar]
- 41.Sumigray K.D., Chen H., Lechler T. Lis1 is essential for cortical microtubule organization and desmosome stability in the epidermis. J. Cell Biol. 2011;194:631–642. doi: 10.1083/jcb.201104009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hirotsune S., Fleck M.W., Gambello M.J., Bix G.J., Chen A., Clark G.D., Ledbetter D.H., McBain C.J., Wynshaw-Boris A. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat. Genet. 1998;19:333–339. doi: 10.1038/1221. [DOI] [PubMed] [Google Scholar]
- 43.Gambello M.J., Darling D.L., Yingling J., Tanaka T., Gleeson J.G., Wynshaw-Boris A. Multiple dose-dependent effects of Lis1 on cerebral cortical development. J. Neurosci. 2003;23:1719–1729. doi: 10.1523/JNEUROSCI.23-05-01719.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayashi S., McMahon A.P. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 45.Kanda T., Sullivan K.F., Wahl G.M. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 1998;8:377–385. doi: 10.1016/s0960-9822(98)70156-3. [DOI] [PubMed] [Google Scholar]
- 46.Nakagawa Y., Yamane Y., Okanoue T., Tsukita S., Tsukita S. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol. Biol. Cell. 2001;12:1687–1697. doi: 10.1091/mbc.12.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mogensen M.M., Malik A., Piel M., Bouckson-Castaing V., Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 2000;113:3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- 48.Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quintyne N.J., Reing J.E., Hoffelder D.R., Gollin S.M., Saunders W.S. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- 50.Kwon M., Godinho S.A., Chandhok N.S., Ganem N.J., Azioune A., Thery M., Pellman D. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22: 2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishikawa H., Kubo A., Tsukita S., Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat. Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 52.Siller K.H., Serr M., Steward R., Hays T.S., Doe C.Q. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol. Biol. Cell. 2005;16: 5127–5140. doi: 10.1091/mbc.E05-04-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toyoshima F., Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmerman W.C., Sillibourne J., Rosa J., Doxsey S.J. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell. 2004;15:3642–3657. doi: 10.1091/mbc.E03-11-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gusnowski E.M., Srayko M. Visualization of dynein-dependent microtubule gliding at the cell cortex: implications for spindle positioning. J. Cell Biol. 2011;194:377–386. doi: 10.1083/jcb.201103128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuyler S.C., Pellman D. Microtubule "plus-end-tracking proteins": the end is just the beginning. Cell. 2001;105:421–424. doi: 10.1016/s0092-8674(01)00364-6. [DOI] [PubMed] [Google Scholar]
- 57.Mimori-Kiyosue Y., Shiina N., Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr. Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- 58.Thoma C.R., Toso A., Gutbrodt K.L., Reggi S.P., Frew I.J., Schraml P., Hergovich A., Moch H., Meraldi P., Krek W. VHL loss causes spindle misorientation and chromosome instability. Nat. Cell Biol. 2009;11: 994–1001. doi: 10.1038/ncb1912. [DOI] [PubMed] [Google Scholar]
- 59.Chan Y.W., Fava L.L., Uldschmid A., Schmitz M.H., Gerlich D.W., Nigg E.A., Santamaria A. Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 2009;185:859–874. doi: 10.1083/jcb.200812167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kaushik R., Yu F., Chia W., Yang X., Bahri S. Subcellular localization of LGN during mitosis: evidence for its cortical localization in mitotic cell culture systems and its requirement for normal cell cycle progression. Mol. Biol. Cell. 2003;14:3144–3155. doi: 10.1091/mbc.E03-04-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kiyomitsu T., Cheeseman I.M. Chromosome- and spindle-pole-derived signals generate an intrinsic code for spindle position and orientation. Nat. Cell Biol. 2012;14:311–317. doi: 10.1038/ncb2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du Q., Macara I.G. Mammalian Pins is a conformational switch that links NuMA to heterotrimeric G proteins. Cell. 2004;119:503–516. doi: 10.1016/j.cell.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 63.Tsai J.W., Chen Y., Kriegstein A.R., Vallee R.B. LIS1 RNA interference blocks neural stem cell division, morphogenesis, and motility at multiple stages. J. Cell Biol. 2005;170:935–945. doi: 10.1083/jcb.200505166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee W.L., Kaiser M.A., Cooper J.A. The offloading model for dynein function: differential function of motor subunits. J. Cell Biol. 2005;168:201–207. doi: 10.1083/jcb.200407036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Markus S.M., Punch J.J., Lee W.L. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr. Biol. 2009;19:196–205. doi: 10.1016/j.cub.2008.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Markus S.M., Plevock K.M., St Germain B.J., Punch J.J., Meaden C.W., Lee W.L. Quantitative analysis of Pac1/LIS1-mediated dynein targeting: implications for regulation of dynein activity in budding yeast. Cytoskeleton (Hoboken) 2011;68:157–174. doi: 10.1002/cm.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang Z., Loncarek J., Khodjakov A., Rieder C.L. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat. Cell Biol. 2008;10:748–751. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nigg E.A. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev.. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 69.Kramer A., Maier B., Bartek J. Centrosome clustering and chromosomal (in)stability: a matter of life and death. Mol. Oncol. 2011;5: 324–335. doi: 10.1016/j.molonc.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma Z., Kanai M., Kawamura K., Kaibuchi K., Ye K., Fukasawa K. Interaction between ROCK II and nucleophosmin/B23 in the regulation of centrosome duplication. Mol. Cell. Biol. 2006;26:9016–9034. doi: 10.1128/MCB.01383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kanai M., Crowe M.S., Zheng Y., Vande Woude G.F., Fukasawa K. Rhoa and RhoC are both required for the ROCK II-dependent promotion of centrosome duplication. Oncogene. 2010;29:6040–6050. doi: 10.1038/onc.2010.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fukasawa K. Aberrant activation of cell cycle regulators, centrosome amplification, and mitotic defects. Horm. Cancer. 2011;2:104–112. doi: 10.1007/s12672-010-0060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kholmanskikh S.S., Koeller H.B., Wynshaw-Boris A., Gomez T., Letourneau P.C., Ross M.E. Calcium-dependent interaction of Lis1 with IQGAP1 and Cdc42 promotes neuronal motility. Nat. Neurosci. 2006;9:50–57. doi: 10.1038/nn1619. [DOI] [PubMed] [Google Scholar]
- 74.Ganem N.J., Godinho S.A., Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Xing Z., Tang X., Gao Y., Da L., Song H., Wang S., Tiollais P., Li T., Zhao M. The human LIS1 is downregulated in hepatocellular carcinoma and plays a tumor suppressor function. Biochem. Biophys. Res. Commun. 2011;409:193–199. doi: 10.1016/j.bbrc.2011.04.117. [DOI] [PubMed] [Google Scholar]
- 76.Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J.W. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pramparo T., Youn Y.H., Yingling J., Hirotsune S., Wynshaw-Boris A. Novel embryonic neuronal migration and proliferation defects in Dcx mutant mice are exacerbated by Lis1 reduction. J. Neurosci. 2010;30:3002–3012. doi: 10.1523/JNEUROSCI.4851-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feng Y., Walsh C.A. Mitotic spindle regulation by Nde1 controls cerebral cortical size. Neuron. 2004;44:279–293. doi: 10.1016/j.neuron.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 79.Laan L., Pavin N., Husson J., Romet-Lemonne G., van Duijn M., Lopez M.P., Vale R.D., Julicher F., Reck-Peterson S.L., Dogterom M. Cortical dynein controls microtubule dynamics to generate pulling forces that position microtubule asters. Cell. 2012;148:502–514. doi: 10.1016/j.cell.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hendricks A.G., Lazarus J.E., Perlson E., Gardner M.K., Odde D.J., Goldman Y.E., Holzbaur E.L. Dynein tethers and stabilizes dynamic microtubule plus ends. Curr. Biol. 2012;22:632–637. doi: 10.1016/j.cub.2012.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Thery M., Jimenez-Dalmaroni A., Racine V., Bornens M., Julicher F. Experimental and theoretical study of mitotic spindle orientation. Nature. 2007;447:493–496. doi: 10.1038/nature05786. [DOI] [PubMed] [Google Scholar]
- 82.Fink J., Carpi N., Betz T., Betard A., Chebah M., Azioune A., Bornens M., Sykes C., Fetler L., Cuvelier D., et al. External forces control mitotic spindle positioning. Nat. Cell Biol. 2011;13:771–778. doi: 10.1038/ncb2269. [DOI] [PubMed] [Google Scholar]
- 83.Kholmanskikh S.S., Dobrin J.S., Wynshaw-Boris A., Letourneau P.C., Ross M.E. Disregulated RhoGTPases and actin cytoskeleton contribute to the migration defect in Lis1-deficient neurons. J. Neurosci. 2003;23:8673–8681. doi: 10.1523/JNEUROSCI.23-25-08673.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sasaki S., Mori D., Toyo-oka K., Chen A., Garrett-Beal L., Muramatsu M., Miyagawa S., Hiraiwa N., Yoshiki A., Wynshaw-Boris A., et al. Complete loss of Ndel1 results in neuronal migration defects and early embryonic lethality. Mol. Cell. Biol. 2005;25:7812–7827. doi: 10.1128/MCB.25.17.7812-7827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Toyo-oka K., Shionoya A., Gambello M.J., Cardoso C., Leventer R., Ward H.L., Ayala R., Tsai L.H., Dobyns W., Ledbetter D., et al. 14–3–3epsilon Is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat. Genet. 2003;34: 274–285. doi: 10.1038/ng1169. [DOI] [PubMed] [Google Scholar]
- 86.Yamada M., Toba S., Yoshida Y., Haratani K., Mori D., Yano Y., Mimori-Kiyosue Y., Nakamura T., Itoh K., Fushiki S., et al. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 2008;27:2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar P., Lyle K.S., Gierke S., Matov A., Danuser G., Wittmann T. GSK3beta phosphorylation modulates CLASP-microtubule association and lamella microtubule attachment. J. Cell. Biol. 2009;184: 895–908. doi: 10.1083/jcb.200901042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data