Regulation of mTOR and Cell Growth in Response to Energy Stress by REDD1 (original) (raw)
Abstract
The tuberous sclerosis tumor suppressors TSC1 and TSC2 regulate the mTOR pathway to control translation and cell growth in response to nutrient and growth factor stimuli. We have recently identified the stress response REDD1 gene as a mediator of tuberous sclerosis complex (TSC)-dependent mTOR regulation by hypoxia. Here, we demonstrate that REDD1 inhibits mTOR function to control cell growth in response to energy stress. Endogenous REDD1 is induced following energy stress, and _REDD1_−/− cells are highly defective in dephosphorylation of the key mTOR substrates S6K and 4E-BP1 following either ATP depletion or direct activation of the AMP-activated protein kinase (AMPK). REDD1 likely acts on the TSC1/2 complex, as regulation of mTOR substrate phosphorylation by REDD1 requires TSC2 and is blocked by overexpression of the TSC1/2 downstream target Rheb but is not blocked by inhibition of AMPK. Tetracycline-inducible expression of REDD1 triggers rapid dephosphorylation of S6K and 4E-BP1 and significantly decreases cellular size. Conversely, inhibition of endogenous REDD1 by short interfering RNA increases cell size in a rapamycin-sensitive manner, and _REDD1_−/− cells are defective in cell growth regulation following ATP depletion. These results define REDD1 as a critical transducer of the cellular response to energy depletion through the TSC-mTOR pathway.
Regulation of cell growth and proliferation in response to cellular stress is critical for survival of normal cells, while dysregulation of appropriate stress responses can promote tumorigenesis (29). The tuberous sclerosis complex (TSC) is composed of two proteins, TSC1 (also known as hamartin) and TSC2 (known as tuberin), which function to integrate growth factor and cell stress responses (35, 39). Loss or mutation of either TSC1 or TSC2 in humans causes tuberous sclerosis syndrome, characterized by benign tumors (hamartomas) of multiple tissues, including the kidney, brain, heart, lung, and skin, and a small but significant increased risk of malignant clear-cell tumors of the kidney (7). Recent work both in Drosophila melanogaster and in mammalian cells has demonstrated that the major function of the TSC1/2 complex is to inhibit the checkpoint protein kinase target of rapamycin (TOR), a major regulator of cell growth and proliferation (15, 27, 39).
In mammalian cells, the TSC1/2 complex integrates upstream signals from growth factor receptors via the phosphatidylinositol 3-kinase (PI3K)/AKT pathway (18, 40, 54) and from the cellular energy-sensing apparatus via the AMP-activated kinase (AMPK) (32). In response to growth factor stimuli, the serine/threonine kinase AKT is activated by PI3K and phosphorylates TSC2, inhibiting its function by a mechanism that is currently controversial (10, 31, 37, 41). AMPK functions as the key energy-sensing kinase by virtue of its exquisite sensitivity to increases in the cellular AMP/ATP ratio (6, 24). Increases in this ratio promote AMPK phosphorylation and activation by the upstream kinase LKB1, a human tumor suppressor mutated in Peutz-Jeghers syndrome (46). Activated AMPK in turn phosphorylates TSC2 (on residues distinct from those phosphorylated by AKT), apparently promoting its activation (8, 45). TSC2 acts as a GTPase-activating protein (GAP) toward the small GTPase Rheb, a positively acting upstream regulator of TOR (30, 36, 55). Therefore, activation of TSC2 converts Rheb to its inactive GDP-bound configuration, leading to decreased mTOR activity (19, 43, 50). The precise mechanism by which phosphorylation by AMPK activates TSC2 is unknown, and no additional regulators of TSC activity in response to energy stress have yet been identified. Nevertheless, _TSC2_−/− cells are highly defective in the regulation of mTOR-dependent phosphorylation and in the suppression of cell growth following energy depletion, demonstrating the pivotal role of TSC1/2 complex in responding to energy stress (32).
Downregulation of mTOR activity following energy stress results in dephosphorylation of its two best-studied substrates, ribosomal S6 kinase 1 (S6K1) and eIF4E binding protein 1 (4E-BP1) (15, 27). Although S6K1 displays a complex pattern of phosphorylation consistent with its activation by a wide variety of extracellular signals, the primary rapamycin-sensitive (i.e., mTOR-dependent) site is Thr389 (4). Phosphorylated S6K1 in turn phosphorylates ribosomal protein S6, which is thought to promote preferential translation of essential components of the translational machinery (49), although this model has recently been called into question (51, 53). Phosphorylation of S6 by S6K occurs primarily at S235 and S236, the major rapamycin-sensitive sites (11). 4E-BP1 is a negative regulator of eIF4E, the rate-limiting factor that binds to the Cap structure at the 5′ end of RNA messages to initiate Cap-dependent translation (22). Phosphorylation of 4E-BP1 induces its dissociation from eIF4E. The 4E-BP1 protein exhibits a stepwise pattern of phosphorylation, giving rise to at least three differentially migrating protein species (α, β, and γ) (20, 21). Through their effects on translation regulation and perhaps other mechanisms, regulation of both S6K and 4E-BP1 phosphorylation by mTOR contributes to mammalian cell size control (17).
We initially identified mammalian REDD1 (also known as RTP801/Dig2/DDIT4) as a gene induced following DNA damage by both p53-dependent and -independent mechanisms (14). Additional studies reported regulation of REDD1 in response to other cellular stresses including hypoxia and glucocorticoid treatment (48, 56). We recently demonstrated that REDD1 functions as a negative regulator of mTOR substrate phosphorylation in response to hypoxia (3). Specifically, _REDD1_−/− cells did not exhibit normal dephosphorylation of S6K and S6 in response to hypoxia. In addition, short interfering RNA (siRNA) directed against TSC2 abrogated the ability of REDD1 expression to downregulate S6K phosphorylation, implying that REDD1 functions via the TSC/mTOR pathway (3).
This study was complemented by work in Drosophila that identified the _REDD1_-like genes scylla and charybdis as suppressors of phenotypes induced following overexpression of the PI3K proteins PKB and PDK-1 (42). In this study, overexpression of the two putative REDD1 orthologues reversed the increase in cell growth resulting from PKB/PDK-1 overexpression. This effect correlated with decreased S6K activity, consistent with negative regulation of mTOR function. While overexpression of scylla and charybdis produced smaller flies and smaller cell size, loss of both genes was associated with increased cell size and larger flies. Both genes are induced by hypoxia, and genetic data placed their effects downstream of PKB but upstream of TSC. Interestingly, both _REDD1_-like genes were also induced in response to starvation. In addition, flies mutant for charybdis were abnormally sensitive to starvation, while overexpression of both genes strongly extended life span under starvation conditions (42). These data imply a role for Drosophila REDD1 orthologues in response both to hypoxia and to energy depletion.
Here we demonstrate that REDD1 is essential for the TSC-dependent cellular energy stress response that controls cell growth. REDD1 is induced in response to energy stress, and _REDD1_−/− cells are markedly defective in downregulation of S6K, S6, and 4E-BP1 phosphorylation following energy depletion. REDD1 expression induces rapid dephosphorylation of these three proteins and decreases cell size. These effects of REDD1 depend on the presence of the TSC1/2 complex, even though AMPK activation and AMPK-dependent phosphorylation of TSC2 are unaffected in the absence of REDD1. Most significantly, siRNA-mediated inhibition of REDD1 increases cellular size in a rapamycin-sensitive fashion, and _REDD1_−/− cells are defective in cell size regulation following energy stress. Thus, REDD1 is an essential regulator of mTOR and cell growth in response to energy stress.
MATERIALS AND METHODS
Materials.
Anti-4E-BP1 (R-113), anti-β-tubulin (D-10), anti-TSC2 (C-20), and antiactin (C-11) were purchased from Santa Cruz Biotechnology, and antihemagglutinin (anti-HA) (16B12) was purchased from Covance. Affinity-purified REDD1 polyclonal antiserum was used as described previously (14). All other antibodies were purchased from Cell Signaling Technologies. The REDD1 cDNA was subcloned, and REDD1-dC was generated by PCR mutagenesis as described previously (3). HA-S6K1 and Rheb cDNAs were generously provided by Diane Fingar and David Kwiatkowski, respectively. AICAR (5-aminoimidazole-4-carboxamide riboside) was obtained from Covance; compound C was obtained from Merck; 2-deoxyglucose (2DG), rapamycin, insulin, and tetracycline were all obtained from Sigma.
Generation of the _REDD1_-null allele.
Synthesis of the REDD1 targeting construct, which replaces the entire REDD1 coding sequence and 3′ untranslated region (UTR) with the β-galactosidase/neo fusion cDNA driven by the endogenous REDD1 promoter, has been described previously (14). The 12-kb linearized construct was electroporated into J1 embryonic stem (ES) cells, followed by selection in G418-containing medium as described previously (34). Successful replacement of the targeted allele was determined by Southern blot analysis. ES cells were injected into C57BL/6 blastocysts, followed by transfer to pseudopregnant hosts to generate chimeras according to standard procedures (28). Germ line transmission of the _REDD1_-null allele was verified in offspring of chimeric animals by Southern analysis.
Cell culture, stimulation, and transfection.
All cell lines were grown in Dulbecco modified Eagle medium (Cambrex) with 10% fetal bovine serum (Sigma). Primary mouse embryo fibroblasts (MEFs) were isolated from 14.5 day embryos using standard procedures (26) and were used at passage 2 to 5. Tetracycline-inducible cell lines were derived from the T-REx-U2OS line (Invitrogen). For glucose starvation, the cells were cultured with d-glucose-free Dulbecco modified Eagle medium, 25 mM HEPES, and 10% dialyzed fetal bovine serum. All transfections were performed using FuGene (Roche) according to the manufacturer's instructions. Where indicated, primary MEFs were immortalized by expression of simian virus 40 large T antigen.
Retroviral and adenoviral infections.
For retroviral infection of primary MEFs, the REDD1 cDNA was subcloned into pLPC-Puro. Retroviral supernatants were generated by cotransfection with the packaging and pseudotyping plasmids as described previously (12). Cells underwent two rounds of infection and were split once prior to serum starvation or 2DG treatment. Replication-defective adenoviral stocks expressing dominant-negative rat α2-AMPK (K45R) or a control vector were generously provided by Kenneth Walsh. Viruses were amplified in HEK293 cells and plaque titered. Cells were infected at a multiplicity of infection of 10 to 20 PFU.
Northern and Southern analyses.
Northern analysis was performed as previously described (13) using 15 μg of total RNA per lane. Northern blots were probed with 32P-labeled REDD1 or glyceraldehyde-3-phosphate dehydrogenase cDNA. Southern analysis was performed using 10 μg of restriction-digested DNA from primary MEFs. Southern blots were probed with a 32P-labeled probe adjacent to the REDD1 genomic locus.
Immunoprecipitation and Western blot analyses.
Cells were lysed in PS6 buffer (50 mM Tris [pH 7.5], 150 mM NaCl, and 0.5% NP-40) with protease inhibitor cocktail (Roche) and phosphatase inhibitor cocktails I and II (Sigma). For phospho-AMPK and phospho-acetyl coenzyme A-carboxylase (phospho-ACC) immunoblots, cells were lysed in phosphate-buffered saline, 1% NP-40, and 1 mM dithiothreitol with protease and phosphatase inhibitors. Fifteen to twenty-five micrograms of protein per lane was run on a sodium dodecyl sulfate-polyacrylamide gel (Cambrex). Phosphoprotein blots were stripped with the Restore Western blot stripping buffer (Pierce), absence of signal was verified, and then blots were reprobed with the appropriate antibody to quantitate total protein loading. Where indicated, lysates in PS6 buffer were immunoprecipitated with anti-HA antibody and protein G Sepharose beads (Amersham BioSciences), washed 3× with PS6 buffer, and eluted with 2× sodium dodecyl sulfate loading buffer. Five percent of the lysates was run to quantitate REDD1 expression levels.
RNA interference (RNAi).
REDD1-targeted small hairpin RNAs were designed against the following sequences and cloned into pBS/U6 as described previously (52): siR1-1 (5′-GGGCAAAGAACTACTGCGCCTG-3′) and siR1-2 (5′-GGGTGTCGTTGCCCGACTTCGA-3′).
FACS analysis for cell size and cell cycle; statistical analysis.
Cells were fixed in 70% ethanol, treated with RNase (Sigma), and stained with propidium iodide (PI) as described previously (12). Mean forward scatter height (FSC-H) was determined on G1-gated cells identified by PI fluorescence (FL2-A). Cell size was assayed in transfected cells by cotransfection of the CD20 cell surface marker and then staining with fluorescein isothiocyanate-conjugated anti-CD20 (Becton Dickinson) or isotype control prior to fixing and PI staining. In this case cell size was determined by FSC-H of G1-gated, fluorescein isothiocyanate-positive cells. All fluorescence-activated cell sorting (FACS) analyses were carried out using the FACSCalibur flow cytometer and Cell Quest Pro software (Becton Dickinson). All statistical analyses were done using the two-tailed Student t test.
RESULTS
REDD1 is required for dephosphorylation of mTOR substrates in response to energy stress.
Genetic data from Drosophila and our own work have linked hypoxia-dependent induction of REDD1 to regulation of mTOR substrate phosphorylation through the TSC1/2 complex (3, 42). Given that the TSC1/2 complex is also known to regulate translation and cell size control following energy deprivation, we wished to determine whether REDD1 contributes to the energy stress response. First, we asked whether REDD1 is induced in response to energy stress. Figure 1A shows that the REDD1 mRNA is significantly upregulated in fibroblasts following either glucose withdrawal or depletion of cellular ATP by the glucose analog 2DG, which blocks cellular glucose utilization. Our previous work demonstrated that REDD1 is induced in a p53-dependent manner following ionizing radiation (14). However, induction of REDD1 by energy stress is p53 independent, as it occurs both in primary cells and in cells immortalized by inactivation of p53 (Fig. 1A and data not shown).
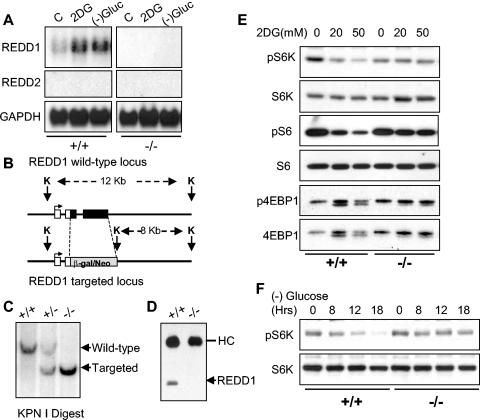
FIG. 1.
REDD1 is induced following energy stress and is required for mTOR substrate dephosphorylation. (A) Induction of the REDD1 message. Immortal MEFs of the indicated genotypes were washed, followed by addition of control medium with 10% fetal calf serum (C), the same medium/fetal calf serum with 2DG (25 mM), or glucose-free medium with 10% dialyzed fetal calf serum. Cells were harvested for Northern analysis at 4 h and probed with a REDD1 or REDD2 cDNA fragment. REDD2 expression is not detectable in MEFs, and no REDD1 expression is observed in _REDD1_−/− MEFs. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (B) REDD1 wild-type and targeted genomic alleles. White boxes, REDD1 3′ UTR; black boxes, REDD1 coding region and 5′ UTR; grey box, β-galactosidase/neo fusion cDNA. The sizes of KpnI (K) fragments detected in panel C are shown. (C) Southern blot analysis of KpnI-digested MEF DNA of the indicated genotype, probed with a genomic fragment 3′ of the REDD1 targeting construct (not shown). (D) Absence of REDD1 protein in _REDD1_−/− MEFs. Extracts from cells treated with 2DG to induce REDD1 protein were analyzed by immunoprecipitation-Western analysis using affinity-purified polyclonal REDD1 antisera. The immunoglobulin heavy chain (HC) is shown as a control for immunoprecipitation. (E) Dephosphorylation of S6K (T389), S6 (S235/236), and 4EBP1 (T70) is defective in _REDD1_−/− MEFs following ATP depletion. Western blot analysis of lysates from primary MEFs treated with 2DG for 4 h, probed with phosphospecific antibodies. The same blots were stripped and reprobed to determine the respective total protein levels. (F) S6K dephosphorylation (T389) in response to glucose starvation requires REDD1. Glucose withdrawal in MEFs was performed as in panel A for the indicated times prior to harvest for Western blot analysis as above.
To determine the physiological role of REDD1 in the cellular energy response, we generated _REDD1_−/− cells. The REDD1 gene is comprised of three exons and two introns, and the entire transcription unit is contained within 2 kb of genomic DNA. We generated a targeting construct that replaces the entire REDD1 coding and 3′ untranslated regions with the β-galactosidase/neomycin resistance fusion protein (Fig. 1B) (14). MEFs derived from crosses of heterozygous mice harboring the mutant allele exhibit the predicted restriction enzyme pattern on Southern analysis of genomic DNA (Fig. 1C). We confirmed that _REDD1_−/− MEFs express no REDD1 coding sequence or protein, as assessed by Northern analysis (Fig. 1A) and Western blot analysis (Fig. 1D), respectively. Of note, expression of the _REDD1_-like gene REDD2 is undetectable by Northern analysis in either wild-type or _REDD1_−/− MEFs (Fig. 1A).
Under nutrient-replete conditions, _REDD1_−/− MEFs exhibit a doubling time and saturation density essentially equivalent to those of wild-type MEFs (data not shown). To examine the potential role of REDD1 in the energy stress response, we first examined phosphorylation of mTOR substrates 4E-BP1 and S6K following ATP depletion in wild-type or _REDD1_−/− MEFs (Fig. 1E). Cells were treated with 2DG, and we assayed phosphorylation of S6K Thr389, the major rapamycin-sensitive site, and 4E-BP1 Thr70, one of several rapamycin-sensitive sites (4, 22). We also examined phosphorylation of the S6K substrate ribosomal protein S6 at S235/S236, the major sites phosphorylated in a rapamycin-sensitive manner by activated S6K (11). As a loading control, in each case blots were stripped and reprobed for total S6K, S6, or 4E-BP1 protein. Wild-type cells exhibit the predicted downregulation in phosphorylation of S6K and S6 proteins following 2DG treatment, as indicated by a decrease in the signal observed using the respective phosphospecific antibodies. Of note, in these primary MEFs 4E-BP1 is almost exclusively in the hyperphosphorylated state (gamma form), such that dephosphorylation is demonstrated most clearly by the appearance of one or more faster-migrating bands (20). Remarkably, _REDD1_−/− cells are resistant to dephosphorylation of S6K, S6, and 4E-BP1 following ATP depletion (Fig. 1E). This difference is apparent even at relatively high doses of 2DG (50 mM). We next examined S6K phosphorylation following glucose withdrawal. Similar to results following 2DG treatment, dephosphorylation of S6K is essentially absent in _REDD1_−/− MEFs compared to wild-type cells (Fig. 1F). Importantly, these and subsequent results presented regarding _REDD1_−/− cells were reproduced using three independently derived batches of litter-matched MEFs (data not shown). Thus, REDD1 is induced and is essential for appropriate mTOR substrate dephosphorylation following energy stress.
REDD1 regulates mTOR-dependent phosphorylation following AMPK activation.
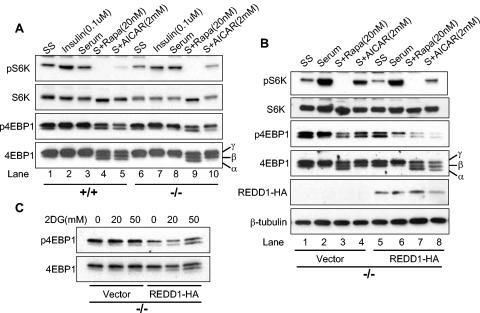
To begin to understand how REDD1 functions within the energy stress response pathway, we examined phosphorylation of mTOR substrates in primary wild-type and _REDD1_−/− MEFs under various physiological conditions that modulate mTOR activity. Primary MEFs were serum starved and then treated with serum, insulin, rapamycin, or the AMP mimetic AICAR, a direct activator of AMPK. Wild-type and _REDD1_−/− cells do not exhibit consistent differences in basal (serum-starved) S6K or 4E-BP1 phosphorylation (Fig. 2A, lanes 1 and 6). In addition, increased phosphorylation of these proteins is similar in the two genotypes following insulin treatment or serum addition, both of which are predicted to activate AKT signaling (lanes 2 and 7 and lanes 3 and 8). As anticipated, rapamycin potently inhibits phosphorylation of both mTOR substrates irrespective of genotype. This is evidenced by the complete disappearance of phosphorylated S6K (lanes 4 and 9), and the appearance of a third, faster-migrating 4E-BP1 band following probing for total protein, which corresponds to the α 4E-BP1 phosphorylation form (bottom panel, lanes 4 and 9) (20). Following AMPK activation, wild-type cells exhibit a pattern of dephosphorylation similar to that observed following rapamycin treatment. In contrast, _REDD1_−/− cells are significantly defective in dephosphorylation of both S6K and 4E-BP1 following AMPK activation by AICAR, compared to wild-type cells. Thus, the ratio of phospho-S6K to total S6K is dramatically different in wild-type versus _REDD1_−/− cells (lane 5 versus 10). The defect is also apparent with regard to 4E-BP1 phosphorylation, as the rapamycin-induced α 4E-BP1 form, visible on probing for total 4E-BP1 protein, is readily detectable in wild-type cells but is entirely absent in _REDD1_−/− cells (bottom panel, lane 5 versus 10). These differences are highly reproducible, and REDD1 heterozygous MEFs appear to have an intermediate phenotype with respect to mTOR substrate regulation (data not shown).
FIG. 2.
Signaling to mTOR substrates is dysregulated in _REDD1_−/− cells following AMPK, but not AKT, activation. (A) Downregulation of S6K (T389) and 4E-BP1 (T70) phosphorylation is defective in _REDD1_−/− cells following AMPK activation. Primary MEFs were serum starved (SS) for 24 h and then treated as indicated (S, serum) for 90 min prior to Western blot analysis. The phosphorylation forms of 4E-BP1 are indicated by convention as α, β, and γ (bottom panel) (19). Note in particular the absence of the α 4E-BP1 form in _REDD1_−/− cells (lane 5 versus 10). (B) Reconstitution of REDD1 expression restores S6K (T389) and 4E-BP1 (T70) dephosphorylation following AMPK activation (lanes 4 and 8). Primary _REDD1_−/− MEFs were infected with control or REDD1-expressing retrovirus prior to treatment as in panel A for 90 min. (C) REDD1 reconstitution restores 4E-BP1 (T70) dephosphorylation following ATP depletion. Primary _REDD1_−/− MEFs were retrovirally infected as in panel B and then treated with 2DG as indicated for 4 h.
Next, we wished to establish whether abnormal mTOR substrate regulation following energy stress in _REDD1_−/− cells is a direct consequence of REDD1 loss. Therefore we sought to “rescue” the defect by ectopic REDD1 expression. Primary _REDD1_−/− MEFs were infected with a control retroviral vector or HA-tagged REDD1-expressing virus, and cells were propagated in the absence of selection. REDD1 expression was documented by a Western blot probed for HA (Fig. 2B). These cells were then serum starved followed by treatment as in Fig. 2A. REDD1 expression at physiological levels does not significantly affect regulation of S6K and 4E-BP1 phosphorylation following either serum starvation, subsequent serum stimulation, or rapamycin treatment (Fig. 2B). However, REDD1 restores regulation following AMPK activation: S6K phosphorylation is downregulated compared to control _REDD1_−/− cells (compare the ratio of phospho-S6K to total S6K in lane 4 versus 8), and dephosphorylation of 4E-BP1 is restored, as evidenced by the appearance of the rapamycin-sensitive α 4E-BP1 form (total 4E-BP1 panel, lane 4 versus 8). To confirm that these results could be extended to include the cellular response to ATP depletion, we treated vector or REDD1-expressing MEFs with 2DG. As shown in Fig. 2C, expression of REDD1 restores 4E-BP1 dephosphorylation following 2DG treatment. Together these data imply that REDD1 plays a direct role in the energy stress response that regulates mTOR-dependent phosphorylation.
REDD1 is not required for AMPK activation or AMPK-dependent TSC2 phosphorylation.
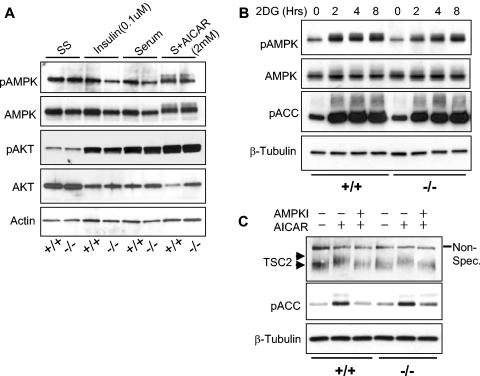
Two general possibilities could account for the observation that _REDD1_−/− cells are defective in mTOR substrate regulation following either energy depletion or AMPK activation by AICAR. REDD1 may be required for AMPK activation, or alternatively it may function downstream or in a parallel pathway. To address this issue, we first examined AMPK activation in wild-type and _REDD1_−/− cells that were serum starved and then treated with insulin, serum, or AICAR as in Fig. 2. AMPK is a heterotrimeric complex composed of catalytic α and regulatory β and γ subunits (6). The primary mechanism of AMPK activation involves phosphorylation of the α subunit at Thr172 by the upstream kinase LKB1 (46). Increases in cellular AMP activate AMPK by multiple mechanisms, most importantly by promoting its phosphorylation by LKB1 (24). Therefore, we assayed AMPK activation by Thr172 phosphorylation. We also examined AKT activation by assessing phosphorylation at S473, one of two major sites for PI3K-dependent AKT phosphorylation (5).
We observed no activation of AMPK by insulin or serum treatment, as expected (Fig. 3A). In contrast, AICAR treatment increased the signal and induced a significant shift in the migration of α-AMPK, indicative of activation. However, little difference was observed between wild-type and _REDD1_−/− MEFs. In addition, AKT was predictably activated by insulin and serum; however, there was again little difference between wild-type and _REDD1_−/− cells (Fig. 3A).
FIG. 3.
REDD1 is not required for AKT or AMPK activation. (A) AMPK (T172) is phosphorylated following AICAR treatment in both wild-type and _REDD1_−/− MEFs (last two lanes). Primary MEFs of the indicated genotype were serum starved for 24 h (SS) and then treated as indicated (S, serum) for 90 min prior to Western blot analysis. (B) AMPK is functionally activated by ATP depletion in both wild-type and _REDD1_−/− MEFs. Primary MEFs were treated with 2DG (50 mM) for the indicated times, prior to Western blot analysis for phospho-AMPK (T172) or its substrate phospho-ACC (S79). (C) AMPK-dependent phosphorylation of TSC2 does not require REDD1. MEFs were serum starved as in panel A, pretreated with the AMPK inhibitor (AMPKI) compound C (10 μM) for 30 min where indicated, and then treated with AICAR (2.0 mM) for 30 min prior to Western blotting for endogenous TSC2. The phosphorylation-induced shift of TSC2 (upper versus lower arrowhead) is evident compared to the uniform migration of the nonspecific band (Non-Spec.). TSC2 phosphorylation correlates with AMPK activation, as evidenced by ACC (S79) phosphorylation.
We next examined AMPK activation in primary MEFs following ATP depletion by 2DG treatment (Fig. 3B). Similar to our observations following AMPK activation by AICAR, we found that AMPK was activated in both wild-type and _REDD1_−/− cells, as assayed by Thr172 phosphorylation. To confirm functional activation of AMPK in these cells, we examined phosphorylation of the AMPK substrate ACC at the AMPK-dependent site S79 (23). We found that phosphorylation of ACC paralleled that of AMPK in both wild-type and _REDD1_−/− cells following 2DG treatment. Together, these experiments demonstrated that AMPK activation and function are unaffected in _REDD1_−/− cells under conditions in which mTOR substrate phosphorylation is dysregulated. Thus, REDD1 is not required for AMPK activation following energy stress.
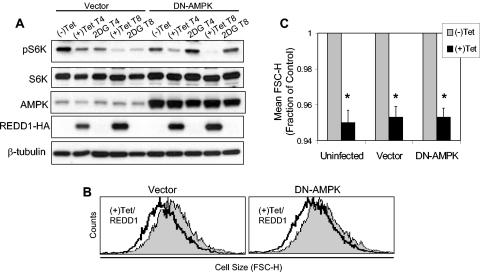
Phosphorylation of TSC2 by activated AMPK is required for TSC-dependent regulation of mTOR substrate phosphorylation (32). In _REDD1_−/− cells, TSC2 might be phosphorylated by AMPK and yet be unable to regulate mTOR, or alternatively REDD1 may be required for AMPK-dependent phosphorylation of TSC2. To distinguish between these possibilities, we examined phosphorylation of TSC2 following AMPK activation in wild-type and _REDD1_−/− fibroblasts. Previous studies have demonstrated that phosphorylation of TSC2 by AMPK is evidenced by a mobility shift in the TSC2 protein and that this shift can be blocked by inhibition of AMPK (8, 32, 45). We found that wild-type and _REDD1_−/− cells expressed equivalent levels of endogenous TSC2 and that AMPK activation by AICAR consistently induced an identical mobility shift in cells of the two genotypes (Fig. 3C). To demonstrate that this shift corresponded to AMPK-dependent phosphorylation, we pretreated cells with the specific AMPK inhibitor compound C prior to AICAR treatment. As expected, AMPK inhibition completely abolished the mobility shift in the TSC2 protein in both wild-type and _REDD1_−/− cells (Fig. 3C). Finally, to confirm that these findings correlated with endogenous AMPK activity, we examined phosphorylation of the AMPK substrate ACC in the same lysates. As predicted, AICAR induced ACC phosphorylation that was blocked following AMPK inhibition (Fig. 3C). Together, these findings demonstrated that REDD1 does not affect AMPK activation or AMPK-dependent phosphorylation of TSC2. Thus, REDD1 likely functions in a pathway parallel to AMPK in regulating mTOR function. This conclusion is supported by data presented in Fig. 6, demonstrating that expression of a dominant-negative AMPK does not block REDD1-mediated effects on mTOR substrate phosphorylation or cell growth.
FIG. 6.
AMPK activation is not required for REDD1 function. (A) DN-AMPK blocks phospho-S6K (T389) regulation by 2DG but not by REDD1. U2-REDD1 cells were infected with DN-AMPK or vector control adenovirus, and 24 h later cells were treated with tetracycline (Tet) to induce REDD1 or with 2DG (25 mM) for the indicated times (T4 and T8, 4 and 8 h of treatment, respectively). Note that DN-AMPK comigrates with endogenous AMPK. (B) DN-AMPK does not inhibit cell size regulation by REDD1. Histogram showing cell size (FSC-H) distribution of G1-gated U2-REDD1 cells infected as in panel A and then induced to express REDD1 (black line) or untreated (grey shading) for 48 h. (C) Quantitation of cell size decrease following induction of REDD1, which is unchanged in the presence or absence of DN-AMPK. The average of mean FSC-H values is shown for three independent experiments performed as in panel B. *, P < 0.005 versus (−)Tet for all comparisons. Error bars show 1 standard deviation.
REDD1 functions in a TSC-dependent pathway upstream of Rheb.
We then turned to address the downstream pathway mediating REDD1 function. Genetic data from Drosophila place the function of the REDD1 orthologues scylla and charybdis in a _TSC-_dependent pathway upstream of Rheb, the target of TSC2 GAP activity (42). Therefore, we wished to determine whether this model could be validated in mammalian cells. Our previous work demonstrated that siRNA directed against TSC2 abrogated the effect of REDD1 on S6K phosphorylation (3). To confirm these data in a genetic model, we transfected REDD1 into wild-type or _TSC2_−/− fibroblasts. (These cells are derived on a _p53_−/− background in order to overcome premature senescence that results from loss of TSC2 [58].) As a control, we transfected a REDD1 deletion mutant (REDD1-dC) that lacks amino acids 96 to 156 within a central phylogenetically conserved region common to known REDD1 orthologues (14). Our previous studies had shown this mutant to be inactive with respect to regulation of S6K phosphorylation (3). To evaluate S6K phosphorylation in transfected cells, we cotransfected HA-tagged S6K1 along with REDD1 or REDD1-dC and performed immunoprecipitation for the tagged protein prior to Western blot analysis. As a control for the immunoprecipitation, the membrane was stripped and reprobed for total S6K protein following analysis of phospho-S6K.
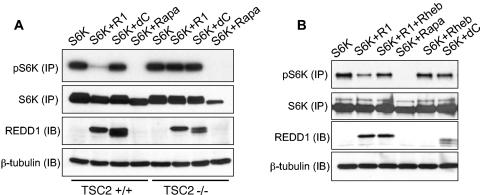
REDD1 dramatically downregulates S6K phosphorylation in cells expressing TSC2; however, it has essentially no effect in matched _TSC2_−/− cells (Fig. 4A). As predicted, REDD1-dC has no effect on S6K1 phosphorylation in either cell population. Thus, REDD1 function requires TSC2 expression.
FIG. 4.
REDD1 function requires TSC2 and is suppressed by Rheb expression. (A) REDD1 regulation of S6K (T389) phosphorylation requires TSC2. Fibroblasts of the indicated genotype were transfected with HA-S6K, with or without REDD1 (R1) or REDD1-dC (dC) (1:1 molar ratio), treated as indicated with rapamycin (Rapa, 2 nM, 90 min), and then immunoprecipitated with anti-HA, followed by Western blot analysis (IP). Following phospho-S6K analysis, the same membrane was stripped and reprobed for total S6K. Four percent of the lysate was probed for REDD1 or β-tubulin as a loading control (IB). (B) Rheb suppresses regulation of S6K (T389) phosphorylation by REDD1. Fibroblasts were transfected with S6K; S6K plus REDD1, REDD1-dC, or Rheb (2:1 molar ratio); or S6K plus REDD1 and Rheb (2:1:1 ratio) and then treated with rapamycin and immunoprecipitated as in panel A. Four percent of the lysate was probed for REDD1 or loading control as in panel A.
In Drosophila, ectopic expression of REDD1 orthologues is unable to rescue Rheb-dependent increased cell growth (42). To determine whether Rheb expression abrogates the effect of REDD1 in mammalian cells, we cotransfected Rheb along with S6K and either REDD1 or REDD1-dC into murine fibroblasts. We find that coexpression of Rheb substantially blocks the effect of REDD1 expression on S6K1 phosphorylation (Fig. 4B). Together these data are consistent with a model in which REDD1 regulates mTOR-dependent phosphorylation by promoting activation of the TSC1/2 complex (see Fig. 8).
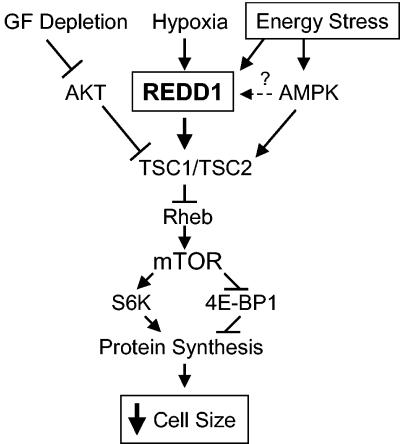
FIG. 8.
Proposed model of REDD1 function in response to energy stress. Energy stress induces expression of REDD1, which functions in a TSC-dependent manner upstream of Rheb to inhibit mTOR activity. REDD1 induction therefore promotes dephosphorylation of S6K and 4E-BP1, which inhibits protein synthesis and decreases cellular growth/cell size. GF, growth factor.
Inducible REDD1 expression regulates mTOR substrate phosphorylation and cell size.
To study REDD1-mediated functions directly, we established cell lines with tetracycline-inducible expression of REDD1 (Fig. 5A). As a control, we generated cells with inducible expression of the REDD1 deletion mutant (REDD1-dC). These lines exhibit undetectable levels of REDD1 or REDD1-dC in the absence of tetracycline, rapid induction of protein expression following tetracycline addition, and equivalent induced levels of the two proteins (Fig. 5A). We first examined phosphorylation of 4E-BP1, S6K, and S6 at various time points following addition of tetracycline. REDD1 protein expression is detectable within 1 hour following induction, at which time we observe essentially maximal downregulation of both S6K1 and 4E-BP1 phosphorylation (Fig. 5A; note the appearance of the hypophosphorylated α 4E-BP1 form). We also observe a slight lag between dephosphorylation of S6K and S6, consistent with the requirement for downregulation of S6K activity prior to S6 dephosphorylation. In contrast, we observe no significant change in phosphorylation of any of these proteins following induction of REDD1-dC. Of note, these findings are reproducible in two independently derived cell lines expressing either inducible REDD1 or REDD1-dC (data not shown). While these experiments cannot precisely define the time required for functional effects of REDD1, the rapid effect that we observe is consistent with our model proposing direct involvement of REDD1 within this pathway. Significantly, we do not observe changes in AKT or AMPK phosphorylation following inducible expression of REDD1 (data not shown). This finding is in agreement with the normal AKT and AMPK activation that we observe in _REDD1_−/− cells (Fig. 3) and with Drosophila data placing the function of the REDD1 orthologues scylla and charybdis downstream of PI3K signaling (42). Together these findings support our model that REDD1 is a potent inhibitor of mTOR activity.
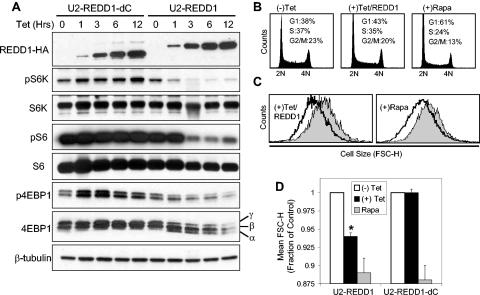
FIG. 5.
REDD1 regulation of mTOR substrate phosphorylation decreases cell size. (A) REDD1, but not REDD1-dC, downregulates phosphorylation of S6K (T389), S6 (S235/236), and 4E-BP1 (T70). Expression of REDD1-dC or REDD1 was induced by addition of tetracycline (1 μg/ml) for the indicated time, and lysates were analyzed by Western blot analysis. The α, β, and γ phosphorylation forms of 4E-BP1 are indicated. (B) Cell cycle profiles of U2-REDD1 cells treated with tetracycline as in panel A or with rapamycin (Rapa, 20 nM) for 48 h and then fixed and stained with PI and analyzed by flow cytometry. (C) REDD1 induction decreases cell size. Representative histogram showing cell size distribution (FSC-H) of G1-gated U2-REDD1 cells treated as in panel B for 48 h (black line) or untreated (grey shading) and then stained with PI and analyzed by flow cytometry. (D) Quantitation of cell size decrease following REDD1 induction. The average of mean FSC-H values from five independent experiments is shown for U2-REDD1 or U2-REDD1-dC cells treated and analyzed as in panel C. *, P = 0.001 versus (−) Tet. Error bars show 1 standard deviation.
The critical role of the TOR pathway in cell size regulation and the involvement of REDD1 orthologues in Drosophila cell size control prompted us to evaluate effects on cell size following inducible expression of REDD1 or REDD1-dC. For cell size evaluation, we used standard techniques involving FACS analysis of mean FSC-H. To exclude potential confounding by cell cycle effects, we analyzed FSC-H on G1-gated cells following PI staining (17, 32). Inducible expression of REDD1 leads to a modest but reproducible increase in the G1 fraction of cells (Fig. 5B) and a decrease in cell proliferation as measured by serial cell counts (data not shown). These data are consistent with the ability of mTOR to regulate both cell growth and cell cycle progression (16). Most significantly, REDD1 induction leads to a highly reproducible mean 6% decrease in cellular size, while inducible expression of REDD1-dC has no effect on cell size (Fig. 5C and D). The magnitude of this effect is comparable to that of rapamycin treatment at a concentration (20 nM) that induces maximal inhibition of mTOR function (17). At this dose, rapamycin decreases cell size by approximately 10% in both inducible cell lines (Fig. 5C and D). Using a similar approach, Fingar et al. (17) observed the same effect (a 10% decrease in cell size) following rapamycin treatment at this dose in U2OS cells, from which the inducible lines were derived. Thus, REDD1-mediated inhibition of mTOR substrate phosphorylation is reflected in its effects on cell growth.
REDD1-mediated regulation of mTOR substrates and cell growth does not require AMPK activation.
The data presented above suggest that REDD1 may function independently of AMPK to control cell growth in response to energy stress. If so, then activation of endogenous AMPK may not be required for REDD1 function. To test this model directly, we examined the ability of REDD1 to regulate mTOR substrate phosphorylation and cell size following expression of a dominant-negative AMPK (DN-AMPK) construct. We used adenoviral transduction to express a mutant α-AMPK subunit (K45R) that has been well validated as blocking endogenous AMPK activation (38, 47). We expressed this mutant or the control vector in cells with inducible REDD1 expression and then treated the cells either with tetracycline or with 2DG as a control for the efficacy of the DN-AMPK. As predicted, DN-AMPK completely abolishes downregulation of S6K phosphorylation following 2DG treatment (Fig. 6A). However, blocking AMPK activity has no effect on the ability of REDD1 to inhibit S6K phosphorylation in a parallel culture. To determine the physiologic significance of this observation, we examined the effect of REDD1 induction on cell size in the presence or absence of DN-AMPK expression. Consistent with its lack of effect on S6K phosphorylation, expression of DN-AMPK has no effect on the ability of REDD1 to decrease cell size compared to vector or uninfected cells (Fig. 6B and C). Together with our observation that AMPK is appropriately activated to phosphorylate TSC2 in _REDD1_−/− cells (Fig. 3), these data strongly argue that REDD1 functions in a parallel pathway to AMPK in the energy stress response.
Endogenous REDD1 regulates cell growth in response to energy stress.
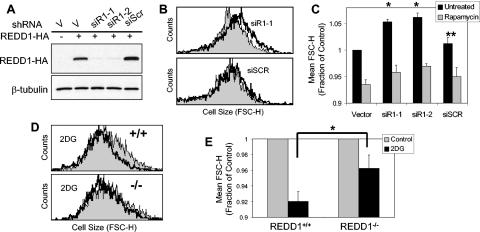
Finally, we tested whether endogenous REDD1 is involved in regulation of cell growth. First, we first examined effects of REDD1 RNAi in U2OS cells. We designed multiple siRNA species that target the REDD1 mRNA, expressed them as plasmid-based small hairpin RNAs, and demonstrated that they potently inhibit REDD1 compared to a control nonspecific siRNA (Fig. 7A) (52). To examine cell size effects of REDD1 RNAi in transfected cells, we cotransfected these siRNA plasmids along with the cell surface marker CD20 and then performed FACS analysis for cell size on G1-gated, CD20+ cells (see Materials and Methods) (17). Within 48 h following transfection, cell size is consistently increased by a mean of 5.3% and 6.2% in cells expressing the two REDD1 siRNA constructs, while expression of a nonspecific siRNA species has an insignificant effect on cell size compared to the vector backbone (Fig. 7B and C). To address whether these changes in cell size can be attributed to regulation of mTOR function, we asked whether rapamycin treatment would suppress the effect of REDD1 RNAi. As predicted, rapamycin treatment decreases cell size in all populations. However, rapamycin has a larger effect in cells treated with REDD1 RNAi, as it decreases the cell size to approximately that observed in vector-transfected, rapamycin-treated cells (Fig. 7C). This finding supports the presence of increased mTOR activity following REDD1 RNAi. Of note, these data are consistent with those recently reported by Coradetti et al. (9), who demonstrated that REDD1 RNAi increases endogenous S6K and S6 phosphorylation in HEK293 cells (9).
FIG. 7.
Endogenous REDD1 regulates cell size in response to energy stress. (A) Efficient inhibition of REDD1 by RNAi. Western blot showing REDD1-HA expression in U2OS cells transfected with REDD1-HA along with the indicated siRNA-expressing plasmid, the backbone vector (V), or a nonspecific siRNA (siScr). (B) REDD1 RNAi increases cell size. Histogram showing cell size distribution (FSC-H) of G1-gated U2OS cells expressing the indicated siRNA (black lines) or control vector plasmid (grey shading), 48 h following transfection (see Materials and Methods). (C) Increased cell size following REDD1 RNAi is reversed following rapamycin treatment. Quantitation of cell size (average of mean FSC-H values) for U2OS cells 48 h following transfection with the indicated siRNA construct, analyzed as in panel B. Where indicated, rapamycin (20 nM) was added 12 h following transfection. Mean values for four independent experiments are shown. *, P < 0.002 versus vector; **, P = NS versus vector. (D) REDD1 is required for cell size regulation following ATP depletion. Histogram showing cell size distribution (FSC-H) of G1-gated MEFs, treated with 2DG (2 mM) for 48 h. (E) Quantitation of altered cell size regulation in _REDD1_−/− MEFs following ATP depletion. The average of mean FSC-H values is shown for four independent experiments. *, P = 0.02 for wild-type versus _REDD1_−/− cells. Error bars show 1 standard deviation.
We next addressed whether endogenous REDD1 plays a role in cell growth control following energy stress. Unlike acute downregulation of REDD1 in transformed cells, germ line inactivation of REDD1 does not lead to consistent differences in the size of primary or immortal _REDD1_−/− MEFs compared to wild-type cells (data not shown). This observation is consistent with our finding that wild-type and _REDD1_−/− MEFs show similar basal levels of mTOR substrate phosphorylation in nutrient-replete conditions (Fig. 1 and 2). To study cell size control following ATP depletion, we treated MEFs with 2DG for 48 h prior to FACS analysis for cell size as described above. While we observe similar, dose-dependent decreases in proliferation and S-phase fraction between wild-type and _REDD1_−/− cells following 2DG treatment (data not shown), _REDD1_−/− cells consistently show a less pronounced decrease in cell size following energy stress than do wild-type cells (Fig. 7D and E). These differences are in agreement with the defective regulation of mTOR substrate phosphorylation that we observe following treatment of these cells with 2DG (Fig. 1E). Together, these data demonstrate that REDD1 is an essential mediator of cell growth regulation in response to energy stress (Fig. 8).
DISCUSSION
The data presented here demonstrate a critical role for REDD1 in the cellular response to energy stress through the TSC/mTOR pathway. Previously, we determined that REDD1 functions in TSC-dependent mTOR regulation in response to hypoxia (3). These findings were further supported by Corradetti et al. (9), who also demonstrated REDD1-mediated regulation of mTOR substrate phosphorylation. Together these observations begin to elucidate how the cellular responses to hypoxia and energy depletion converge to control metabolism. ATP depletion ultimately results from both prolonged hypoxia, through loss of oxidative metabolism, and decreased glucose availability. Interestingly, however, Arsham et al. (1) demonstrated that mTOR substrate regulation by hypoxia does not require a decrease in the ATP/ADP ratio. This finding is consistent with our recent work (3) and implies that REDD1 itself may be regulated by distinct mechanisms following hypoxia and energy stress. Nevertheless, it seems apparent that organisms have evolved a common REDD1-dependent response to these two forms of stress in order to protect themselves from impending energetic crisis (1).
Our findings are strongly supported by genetic data derived from Drosophila, as both _REDD1_-like genes scylla and charybdis are induced in response to starvation, and Charybdis mutants exhibit increased sensitivity to starvation conditions. Drosophila studies also support our placement of REDD1 function upstream of TSC2, rather than in a downstream or parallel pathway. We find that, in the absence of TSC2, REDD1 is unable to regulate mTOR substrate phosphorylation. In addition, expression of the downstream biochemical target of the TSC1/2 complex, Rheb, efficiently blocks the effect of REDD1 expression. In the fly, overexpression of scylla and charybdis is unable to rescue TSC1/2 or TSC2 mutant phenotypes, while overexpression of Rheb is dominant to scylla and charybdis expression, and scylla/charybdis/TSC2 triple mutants exhibit the same cell size increase as that of TSC2 single mutants. Although the precise mechanism of REDD1 function awaits further studies, the rapid downregulation of mTOR substrate phosphorylation that we observe following inducible expression of REDD1 is consistent with a direct effect of the REDD1 protein in promoting activation of the TSC1/2 complex.
AMPK is reported to phosphorylate TSC2 directly, and this phosphorylation is important for TSC activation and mTOR inhibition following energy stress (32). It is unknown how AMPK phosphorylation regulates TSC function. Interestingly, we find that AMPK-dependent phosphorylation of TSC2 is unaffected in the absence of REDD1, suggesting that this event is not sufficient to fully activate the TSC1/2 complex in response to energy stress. While these observations suggest parallel pathways for mTOR regulation by REDD1 and AMPK, they do not rule out the possibility that AMPK might contribute to regulation of REDD1 itself. However, in a series of pilot experiments we found that activation of AMPK by 2DG does not promote stabilization or other obvious modification of the REDD1 protein, and expression of DN-AMPK does not block induction of the REDD1 message following energy stress (data not shown). Thus, REDD1 induction and AMPK activation may be regulated by distinct pathways, and they converge to promote activation of the TSC1/2 complex (Fig. 8).
In contrast to cells with loss of TSC1 or TSC2, _REDD1_−/− MEFs do not exhibit markedly elevated mTOR activity in nutrient-replete conditions (33, 58). This finding is consistent with our model that REDD1 functions as a stress-induced regulator of mTOR and is therefore not essential for basal control of mTOR activity under nonstressed conditions. Similarly, we observe normal serum and insulin-induced AKT activation in these cells, unlike _TSC1_−/− or _TSC2_−/− cells, which are resistant to AKT activation as a result of an mTOR-mediated negative feedback mechanism (25, 44). The lack of basal cell size differences that we observed in wild-type and _REDD1_−/− MEFs is also in agreement with this model. Normal basal mTOR activity in _REDD1_−/− cells is also likely to explain our observation, and that of others, that _REDD1_−/− mice are born viable (2). Nevertheless, normal development and environmental adaptation present multiple stresses that could trigger REDD1-dependent phenotypes in these mice as they age.
Our previous study showed that both REDD1 and its paralog REDD2 are able to downregulate mTOR substrate phosphorylation when expressed individually and that there may be an additive effect of overexpressing both genes (3). However, our data imply that REDD1 and REDD2 act independently. We detect little or no REDD2 expression in fibroblasts, yet REDD1 clearly functions in the energy stress response within these cells. In addition, we observe a largely nonoverlapping expression pattern of REDD1 and REDD2 in adult tissues (K. Lei and L. W. Ellisen, unpublished data). It remains to be determined whether potentially divergent functions of these genes result from differences in tissue-specific expression or from distinct biochemical properties. Notably, the two _Drosophila REDD1_-like genes also exhibit nonoverlapping expression patterns, and genetic data provide additional evidence that they function independently (42).
Recent evidence has provided a clear link between dysregulation of mTOR-dependent cell growth pathways and tumorigenesis. Multiple human tumor suppressors, including the LKB1 gene responsible for Peutz-Jeghers syndrome, the tuberous sclerosis genes TSC1 and TSC2, and the PTEN gene responsible for multiple tumor syndromes, function in converging pathways to regulate cellular metabolism through the mTOR kinase (29). In turn, mTOR controls downstream effectors such as 4E-BP1, which has recently been linked to tumor formation through its downstream target eIF4E (57). Like REDD1, these proteins respond to changes in nutrient, growth factor, and oxygen status, functioning in part to dictate the activity level of the cellular translational machinery. Additional regulatory genes in these pathways are yet to be identified, and a subset of these will likely be implicated in human cancer. Our previous finding that REDD1 functions as an important TSC-dependent regulator of mTOR in response to hypoxia and this report demonstrating the role of REDD1 in cell growth regulation in response to energy stress suggest that dysregulation of REDD1 function may contribute to human tumorigenesis.
Acknowledgments
We thank David Kwiatkowski for use of _TSC2_−/− cells, Jeffrey Settleman and Diane Fingar for critical review of the manuscript, and James Brugarolas for helpful advice.
This work was supported by grants from the American Cancer Society and the National Institutes of Health to L.W.E.
REFERENCES
- 1.Arsham, A. M., J. J. Howell, and M. C. Simon. 2003. A novel hypoxia-inducible factor-independent hypoxic response regulating mammalian target of rapamycin and its targets. J. Biol. Chem. 278**:**29655-29660. [DOI] [PubMed] [Google Scholar]
- 2.Brafman, A., I. Mett, M. Shafir, H. Gottlieb, G. Damari, S. Gozlan-Kelner, V. Vishnevskia-Dai, R. Skaliter, P. Einat, A. Faerman, E. Feinstein, and T. Shoshani. 2004. Inhibition of oxygen-induced retinopathy in RTP801-deficient mice. Investig. Ophthalmol. Vis. Sci. 45**:**3796-3805. [DOI] [PubMed] [Google Scholar]
- 3.Brugarolas, J., K. Lei, R. L. Hurley, B. D. Manning, J. H. Reiling, E. Hafen, L. A. Witters, L. W. Ellisen, and W. G. Kaelin, Jr. 2004. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18**:**2893-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnett, P. E., R. K. Barrow, N. A. Cohen, S. H. Snyder, and D. M. Sabatini. 1998. RAFT1 phosphorylation of the translational regulators p70 S6 kinase and 4E-BP1. Proc. Natl. Acad. Sci. USA 95**:**1432-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantley, L. C. 2002. The phosphoinositide 3-kinase pathway. Science 296**:**1655-1657. [DOI] [PubMed] [Google Scholar]
- 6.Carling, D. 2004. The AMP-activated protein kinase cascade—a unifying system for energy control. Trends Biochem. Sci. 29**:**18-24. [DOI] [PubMed] [Google Scholar]
- 7.Cheadle, J. P., M. P. Reeve, J. R. Sampson, and D. J. Kwiatkowski. 2000. Molecular genetic advances in tuberous sclerosis. Hum. Genet. 107**:**97-114. [DOI] [PubMed] [Google Scholar]
- 8.Corradetti, M. N., K. Inoki, N. Bardeesy, R. A. DePinho, and K. L. Guan. 2004. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 18**:**1533-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corradetti, M. N., K. Inoki, and K. L. Guan. 2005. The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J. Biol. Chem. 280**:**9769-9772. [DOI] [PubMed] [Google Scholar]
- 10.Dan, H. C., M. Sun, L. Yang, R. I. Feldman, X. M. Sui, C. C. Ou, M. Nellist, R. S. Yeung, D. J. Halley, S. V. Nicosia, W. J. Pledger, and J. Q. Cheng. 2002. Phosphatidylinositol 3-kinase/Akt pathway regulates tuberous sclerosis tumor suppressor complex by phosphorylation of tuberin. J. Biol. Chem. 277**:**35364-35370. [DOI] [PubMed] [Google Scholar]
- 11.Dufner, A., and G. Thomas. 1999. Ribosomal S6 kinase signaling and the control of translation. Exp. Cell Res. 253**:**100-109. [DOI] [PubMed] [Google Scholar]
- 12.Ellisen, L. W., N. Carlesso, T. Cheng, D. T. Scadden, and D. A. Haber. 2001. The Wilms tumor suppressor WT1 directs stage-specific quiescence and differentiation of human hematopoietic progenitor cells. EMBO J. 20**:**1897-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellisen, L. W., R. E. Palmer, R. G. Maki, V. B. Truong, P. Tamayo, J. D. Oliner, and D. A. Haber. 2001. Cascades of transcriptional induction during human lymphocyte activation. Eur. J. Cell Biol. 80**:**321-328. [DOI] [PubMed] [Google Scholar]
- 14.Ellisen, L. W., K. D. Ramsayer, C. M. Johannessen, A. Yang, H. Beppu, K. Minda, J. D. Oliner, F. McKeon, and D. A. Haber. 2002. REDD1, a developmentally regulated transcriptional target of p63 and p53, links p63 to regulation of reactive oxygen species. Mol. Cell 10**:**995-1005. [DOI] [PubMed] [Google Scholar]
- 15.Fingar, D. C., and J. Blenis. 2004. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23**:**3151-3171. [DOI] [PubMed] [Google Scholar]
- 16.Fingar, D. C., C. J. Richardson, A. R. Tee, L. Cheatham, C. Tsou, and J. Blenis. 2004. mTOR controls cell cycle progression through its cell growth effectors S6K1 and 4E-BP1/eukaryotic translation initiation factor 4E. Mol. Cell Biol. 24**:**200-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16**:**1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao, X., and D. Pan. 2001. TSC1 and TSC2 tumor suppressors antagonize insulin signaling in cell growth. Genes Dev. 15**:**1383-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garami, A., F. J. Zwartkruis, T. Nobukuni, M. Joaquin, M. Roccio, H. Stocker, S. C. Kozma, E. Hafen, J. L. Bos, and G. Thomas. 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol. Cell 11**:**1457-1466. [DOI] [PubMed] [Google Scholar]
- 20.Gingras, A. C., S. G. Kennedy, M. A. O'Leary, N. Sonenberg, and N. Hay. 1998. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev. 12**:**502-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gingras, A. C., B. Raught, S. P. Gygi, A. Niedzwiecka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieszynska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 15**:**2852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gingras, A. C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15**:**807-826. [DOI] [PubMed] [Google Scholar]
- 23.Ha, J., S. Daniel, S. S. Broyles, and K. H. Kim. 1994. Critical phosphorylation sites for acetyl-CoA carboxylase activity. J. Biol. Chem. 269**:**22162-22168. [PubMed] [Google Scholar]
- 24.Hardie, D. G. 2004. The AMP-activated protein kinase pathway—new players upstream and downstream. J. Cell Sci. 117**:**5479-5487. [DOI] [PubMed] [Google Scholar]
- 25.Harrington, L. S., G. M. Findlay, A. Gray, T. Tolkacheva, S. Wigfield, H. Rebholz, J. Barnett, N. R. Leslie, S. Cheng, P. R. Shepherd, I. Gout, C. P. Downes, and R. F. Lamb. 2004. The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J. Cell Biol. 166**:**213-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harvey, M., A. T. Sands, R. S. Weiss, M. E. Hegi, R. W. Wiseman, P. Pantazis, B. C. Giovanella, M. A. Tainsky, A. Bradley, and L. A. Donehower. 1993. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene 8**:**2457-2467. [PubMed] [Google Scholar]
- 27.Hay, N., and N. Sonenberg. 2004. Upstream and downstream of mTOR. Genes Dev. 18**:**1926-1945. [DOI] [PubMed] [Google Scholar]
- 28.Hogan, G., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo, a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 29.Inoki, K., M. N. Corradetti, and K. L. Guan. 2005. Dysregulation of the TSC-mTOR pathway in human disease. Nat. Genet. 37**:**19-24. [DOI] [PubMed] [Google Scholar]
- 30.Inoki, K., Y. Li, T. Xu, and K. L. Guan. 2003. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 17**:**1829-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Inoki, K., Y. Li, T. Zhu, J. Wu, and K. L. Guan. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat. Cell Biol. 4**:**648-657. [DOI] [PubMed] [Google Scholar]
- 32.Inoki, K., T. Zhu, and K. L. Guan. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115**:**577-590. [DOI] [PubMed] [Google Scholar]
- 33.Kwiatkowski, D. J., H. Zhang, J. L. Bandura, K. M. Heiberger, M. Glogauer, N. el-Hashemite, and H. Onda. 2002. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum. Mol. Genet. 11**:**525-534. [DOI] [PubMed] [Google Scholar]
- 34.Li, E., T. H. Bestor, and R. Jaenisch. 1992. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69**:**915-926. [DOI] [PubMed] [Google Scholar]
- 35.Li, Y., M. N. Corradetti, K. Inoki, and K. L. Guan. 2004. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem. Sci. 29**:**32-38. [DOI] [PubMed] [Google Scholar]
- 36.Li, Y., K. Inoki, and K. L. Guan. 2004. Biochemical and functional characterizations of small GTPase Rheb and TSC2 GAP activity. Mol. Cell. Biol. 24**:**7965-7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manning, B. D., A. R. Tee, M. N. Logsdon, J. Blenis, and L. C. Cantley. 2002. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol. Cell 10**:**151-162. [DOI] [PubMed] [Google Scholar]
- 38.Minokoshi, Y., T. Alquier, N. Furukawa, Y. B. Kim, A. Lee, B. Xue, J. Mu, F. Foufelle, P. Ferre, M. J. Birnbaum, B. J. Stuck, and B. B. Kahn. 2004. AMP-kinase regulates food intake by responding to hormonal and nutrient signals in the hypothalamus. Nature 428**:**569-574. [DOI] [PubMed] [Google Scholar]
- 39.Pan, D., J. Dong, Y. Zhang, and X. Gao. 2004. Tuberous sclerosis complex: from Drosophila to human disease. Trends Cell Biol. 14**:**78-85. [DOI] [PubMed] [Google Scholar]
- 40.Potter, C. J., H. Huang, and T. Xu. 2001. Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105**:**357-368. [DOI] [PubMed] [Google Scholar]
- 41.Potter, C. J., L. G. Pedraza, and T. Xu. 2002. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 4**:**658-665. [DOI] [PubMed] [Google Scholar]
- 42.Reiling, J. H., and E. Hafen. 2004. The hypoxia-induced paralogs Scylla and Charybdis inhibit growth by down-regulating S6K activity upstream of TSC in Drosophila. Genes Dev. 18**:**2879-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saucedo, L. J., X. Gao, D. A. Chiarelli, L. Li, D. Pan, and B. A. Edgar. 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5**:**566-571. [DOI] [PubMed] [Google Scholar]
- 44.Shah, O. J., Z. Wang, and T. Hunter. 2004. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr. Biol. 14**:**1650-1656. [DOI] [PubMed] [Google Scholar]
- 45.Shaw, R. J., N. Bardeesy, B. D. Manning, L. Lopez, M. Kosmatka, R. A. DePinho, and L. C. Cantley. 2004. The LKB1 tumor suppressor negatively regulates mTOR signaling. Cancer Cell 6**:**91-99. [DOI] [PubMed] [Google Scholar]
- 46.Shaw, R. J., M. Kosmatka, N. Bardeesy, R. L. Hurley, L. A. Witters, R. A. DePinho, and L. C. Cantley. 2004. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA 101**:**3329-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shibata, R., N. Ouchi, M. Ito, S. Kihara, I. Shiojima, D. R. Pimentel, M. Kumada, K. Sato, S. Schiekofer, K. Ohashi, T. Funahashi, W. S. Colucci, and K. Walsh. 2004. Adiponectin-mediated modulation of hypertrophic signals in the heart. Nat. Med. 10**:**1384-1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shoshani, T., A. Faerman, I. Mett, E. Zelin, T. Tenne, S. Gorodin, Y. Moshel, S. Elbaz, A. Budanov, A. Chajut, H. Kalinski, I. Kamer, A. Rozen, O. Mor, E. Keshet, D. Leshkowitz, P. Einat, R. Skaliter, and E. Feinstein. 2002. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol. Cell. Biol. 22**:**2283-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sonenberg, N., and A. C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10**:**268-275. [DOI] [PubMed] [Google Scholar]
- 50.Stocker, H., T. Radimerski, B. Schindelholz, F. Wittwer, P. Belawat, P. Daram, S. Breuer, G. Thomas, and E. Hafen. 2003. Rheb is an essential regulator of S6K in controlling cell growth in Drosophila. Nat. Cell Biol. 5**:**559-565. [DOI] [PubMed] [Google Scholar]
- 51.Stolovich, M., H. Tang, E. Hornstein, G. Levy, R. Cohen, S. S. Bae, M. J. Birnbaum, and O. Meyuhas. 2002. Transduction of growth or mitogenic signals into translational activation of TOP mRNAs is fully reliant on the phosphatidylinositol 3-kinase-mediated pathway but requires neither S6K1 nor rpS6 phosphorylation. Mol. Cell. Biol. 22**:**8101-8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sui, G., C. Soohoo, E. B. Affar, F. Gay, Y. Shi, W. C. Forrester, and Y. Shi. 2002. A DNA-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl. Acad. Sci. USA 99**:**5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang, H., E. Hornstein, M. Stolovich, G. Levy, M. Livingstone, D. Templeton, J. Avruch, and O. Meyuhas. 2001. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol. 21**:**8671-8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tapon, N., N. Ito, B. J. Dickson, J. E. Treisman, and I. K. Hariharan. 2001. The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105**:**345-355. [DOI] [PubMed] [Google Scholar]
- 55.Tee, A. R., B. D. Manning, P. P. Roux, L. C. Cantley, and J. Blenis. 2003. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Curr. Biol. 13**:**1259-1268. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Z., M. H. Malone, M. J. Thomenius, F. Zhong, F. Xu, and C. W. Distelhorst. 2003. Dexamethasone-induced gene 2 (dig2) is a novel pro-survival stress gene induced rapidly by diverse apoptotic signals. J. Biol. Chem. 278**:**27053-27058. [DOI] [PubMed] [Google Scholar]
- 57.Wendel, H. G., E. De Stanchina, J. S. Fridman, A. Malina, S. Ray, S. Kogan, C. Cordon-Cardo, J. Pelletier, and S. W. Lowe. 2004. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature 428**:**332-337. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, H., G. Cicchetti, H. Onda, H. B. Koon, K. Asrican, N. Bajraszewski, F. Vazquez, C. L. Carpenter, and D. J. Kwiatkowski. 2003. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. J. Clin. Investig. 112**:**1223-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]