Biochemical and genetic analysis of the yeast proteome with a movable ORF collection (original) (raw)
Abstract
Functional analysis of the proteome is an essential part of genomic research. To facilitate different proteomic approaches, a MORF (moveable ORF) library of 5854 yeast expression plasmids was constructed, each expressing a sequence-verified ORF as a C-terminal ORF fusion protein, under regulated control. Analysis of 5573 MORFs demonstrates that nearly all verified ORFs are expressed, suggests the authenticity of 48 ORFs characterized as dubious, and implicates specific processes including cytoskeletal organization and transcriptional control in growth inhibition caused by overexpression. Global analysis of glycosylated proteins identifies 109 new confirmed N-linked and 345 candidate glycoproteins, nearly doubling the known yeast glycome.
Keywords: Biochemical genomics, protein microarray, proteome, high-throughput expression, galactose lethality, glycosylation
The wealth of sequenced genomes has spawned a variety of powerful genomic-scale approaches to identify the genes, proteins, and RNAs in an organism, to define individual functions of genes, and to map the interactions between them that underlie cell and organismal biology (Bader et al. 2003; Phizicky et al. 2003; Hughes et al. 2004). High-throughput genomic analyses have provided unprecedented views of gene function and of networks of interactions connecting genes (Uetz et al. 2000; Gavin et al. 2002; Giaever et al. 2002; Ho et al. 2002; Tong et al. 2004). The study of entire genomes has also highlighted some of the complexities involved in gene and protein characterization, such as the difficulties in identification of bona fide genes from genomic sequence (Basrai et al. 1997; Blandin et al. 2000; Kumar et al. 2002; Oshiro et al. 2002; Cliften et al. 2003; Kellis et al. 2003; Kessler et al. 2003) and the need to examine proteins for processing and multiple post-translational modifications (Zhu et al. 2000; Huang et al. 2004; Kus et al. 2005).
The parallel assay of whole proteomes, using genomic collections of purified proteins derived from cloned genes (Martzen et al. 1999; Zhu et al. 2000), has provided a powerful approach for searching for proteins (and their cognate genes) with particular biochemical activities (Alexandrov et al. 2002; Gu et al. 2003; Jackman et al. 2003; Ma et al. 2003; Bieganowski and Brenner 2004), for characterizing the global sets of proteins that bind particular ligands (Zhu et al. 2001; Hazbun and Fields 2002) and for identifying substrates of enzymes that mediate post-translational modifications of proteins (Zhu et al. 2000; Kafadar et al. 2003; Ubersax et al. 2003; Huang et al. 2004). However, previous genomic collections of this type have been deficient with respect to both gene and protein coverage due to the introduction of mutations during cloning, incomplete/incorrect annotation of genes in the collections, and fusion of affinity tags to the N termini of cloned genes, which is likely to interfere with targeting of proteins destined for the secretory pathway. This latter problem is particularly acute since as many as 20%–30% of eukaryotic proteins have been estimated to be membrane or secreted proteins (Krogh et al. 2001).
We describe here a new library of yeast ORFs for use in high-throughput biochemical and genetic analyses that overcomes the most significant limitations of previous libraries and extends the uses of these types of libraries. This collection consists of two parts: a library of extensively sequence-analyzed plasmids, which provides a versatile collection of yeast ORFs that can be rapidly transferred to new vectors for different applications (hence the name MORF, for moveable ORF), and a parallel library of yeast strains, each expressing the corresponding ORF as a C-terminal ORF fusion protein under tightly regulated transcriptional control. The new collection is based on recent annotation of the yeast genome and is made with high-efficiency and high-fidelity cloning procedures, providing the most complete collection of ORFs available for any organism. The use of a C-terminal tandem affinity tag allows efficient purification of ORF products, including transmembrane and secreted proteins. The utility of the MORF collection was demonstrated by analysis of expression of the genomic collection of ORFs, by examination of growth inhibition caused by overexpression, and by global analysis of glycosylation of the expressed proteins, a test of the usefulness of the collection for proteins that transit the secretory pathway.
Results
The MORF collection contains 5854 sequence-analyzed ORFs
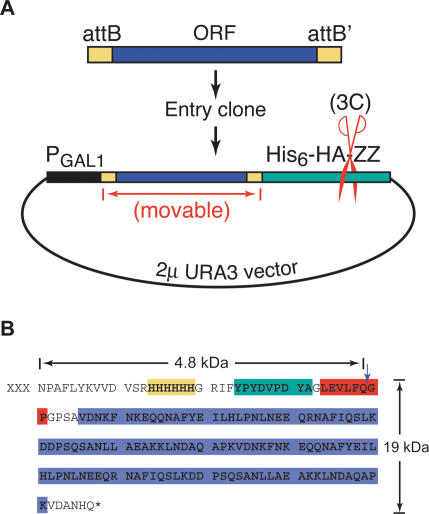
The MORF collection was designed to maximize gene and protein representation in a high-quality expression library featuring movable ORFs. A collection of 6426 yeast ORFs including all ORFs annotated in SGD (Balakrishnan et al., August 2002) and 46 tORFs identified by transposon insertions (Kumar et al. 2002) was amplified by PCR and cloned into a yeast expression vector with directional att sites via Gateway recombination (Fig. 1); ORFs from this vector can be transferred by site-specific recombination to other att_-containing vectors (hence, the name MORF, moveable ORF). Yeast proteins are expressed under P_GAL promoter control, starting at the natural N-terminal methionine and ending with a fusion of the C-terminal amino acid to a tag consisting of His6, an HA epitope, a protease 3C cleavage site, and the IgG-binding domain from protein A (Fig. 1).
Figure 1.
MORF plasmid structure. (A) Diagram of MORF expression vector. PCR amplification of ORFs results in addition of directional attB sequences directly abutting the initiating ATG and the final sense codon. After two rounds of recombination, the ORF, again flanked by directional attB sequences, is cloned into vector pBG1805 (described in Supplemental Material) in frame with a triple affinity tag comprised of His6-HAepitope-3Cprotease site-ZZproteinA. (B) C-terminal affinity tag. Purified proteins have a 4.8-kDa tag including His6 (gold) and a single HA epitope (green) after cleavage with 3C protease. Before cleavage, the entire tag is 19 kDa, including an IgG-binding ZZ domain (blue) and a 3C protease cleavage site (red). MORF clones are available from Open Biosystems (http://www.openbiosystems.com).
The MORF collection currently contains 5854 ORFs, consisting of 91.1% of the target ORFs and 93.2% of the currently verified ORFs in SGD (Balakrishnan et al., May 2005). The identity of the cloned ORFs, as well as information on their sequencing, size, and expression in yeast and the primers used to amplify them, are shown in Supplementary Table S1. Each insert was sequenced from both ends, yielding an average of 1078 base pairs (bp) per ORF, and resulting in the complete sequence verification of 3217 ORFs (55%). Although all clones with mutations were remade, 45 ORFs had identical mutations isolated repeatedly from independent PCR amplifications. Twenty-nine of these were reproducible missense mutations and were included in the collection, while 16 ORFs had reproducible insertions or deletions and were not included in the collection (Supplementary Table S2). These differences presumably represent either errors in the yeast sequence or polymorphisms between our template strain (BY4700) and the sequenced strain. Based on our sequence analysis, the proofreading polymerase introduces ∼0.7 errors per 10,000 bp; thus, <200 (188) of the incompletely sequenced clones should contain a mutation.
High expression of proteins in the MORF collection, including membrane proteins
The presence, amount, size, and quality of 5573 ORF fusion proteins were examined after transformation into yeast and galactose induction. Fusion proteins were detected from 5188 yeast ORFs (93%) in whole yeast cell lysates by immunoblot analysis using antibody to the HA epitope. Most fusion proteins appear to be intact in vivo, based on detection of a single prominent protein of the expected size (Fig. 2A,B). Only 130 proteins migrate significantly smaller than expected, and 95 of these are larger than 80 kDa, a size class that is not accurately measured on SDS-PAGE. Others in this class may be proteolyzed or may have transferred poorly during immunoblotting, resulting in a failure to detect the full-size protein. Many proteins that migrate significantly larger than expected in SDS-PAGE are likely to be glycoproteins, based on enrichment for proteins localized to the cell wall (9.3% of this set compared with 1.2% in the MORF collection), known glycoproteins (8.6% compared with 3%), and secreted proteins (15% compared with 5.3%).
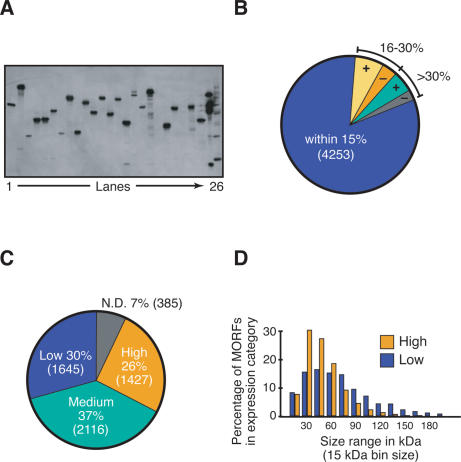
Figure 2.
MORF expression. (A) Detection of MORF fusion protein expression. Yeast cells containing different MORFs were induced for expression of fusion proteins, and whole-cell lysates were subjected to SDS-PAGE and analyzed by immunoblot with anti-HA antibody (Materials and Methods). (Lanes 1–24) Different MORF clones. (Lane 25) MORF fusion proteins purified by immobilized metal ion affinity chromatography (Supplemental Material): Ura1-His6-HA-ZZp (54 kDA), Tkl1-His6-HA-ZZp (93 kDa), and Lys1-His6-HA-ZZp (60 kDa). (Lane 26) Invitrogen MagicMark XP Western Protein Standards. (B) Comparison of predicted MORF protein size to observed SDS-PAGE migration. A total of 349 MORF proteins run 16%–30% slower than predicted, while 177 run 16%–30% faster; 278 MORF proteins run >30% slower than predicted, while 130 run >30% faster. (C) Classification of MORFs based on expression levels. (N.D.) Not detected. (D) Molecular weight distribution of MORF proteins in high and low expression categories. Proteins in high (yellow) and low (blue) expression categories were binned according to predicted native molecular weight, without the tag. The upper limit of the size range is indicated on the _X_-axis. To remove ORFs whose expression might be compromised by an unstable polypeptide, only ORFs that are classified as verified and uncharacterized by SGD were included in the analysis.
We find substantial variations in the levels of protein expression of different ORFs (Fig. 2A,C). The expression levels of the ORFs were classified into three categories: high (∼1 + mg/L), medium (∼0.1 mg/L), and low (∼0.01 mg/L). Fully 3543 ORF fusion proteins (63%) are expressed at medium or high levels (Fig. 2C), facilitating biochemical analysis. In general, larger ORFs are expressed at lower levels than smaller ORFs (Fig. 2D), perhaps due to lack of translational processivity (Arava et al. 2003). In addition, proteins with a lower pI, as well as those with increased codon bias and codon adaptation index, exhibit better expression on average (Balakrishnan et al., May 2005; Supplementary Table S3; Supplementary Fig. S1).
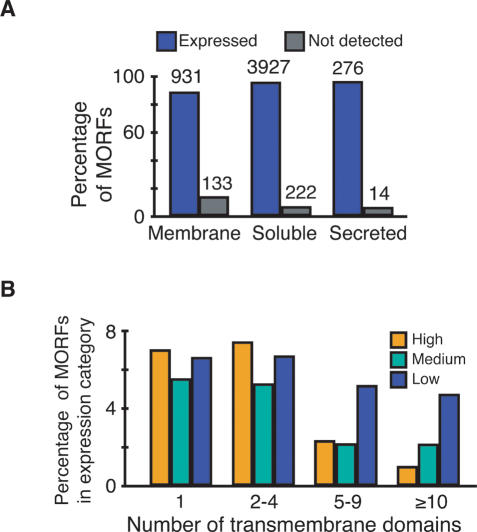
Because of the preservation of N-terminal signal peptides, the MORF collection is well suited for the expression of membrane-associated and secreted proteins, which have been generally understudied due to their lack of solubility and difficulties in their purification. Based on the program TMHMM, nearly 20% of the MORF proteins (1064 out of 5503 examined) are potential integral membrane proteins (Krogh et al. 2001; Kall and Sonnhammer 2002); of these, 931 (87%) are expressed in the MORF collection (Fig. 3A). Contrary to the prevailing view that it is difficult to express membrane proteins at high levels, almost the same fraction of membrane proteins (23%, 247) are in the high-expression category, as observed for the entire collection; for 164 of these, there is independent localization data consistent with membrane character (Huh et al. 2003; Balakrishnan et al., May 2005). However, proteins with large numbers of transmembrane domains exhibit reduced expression (Fig. 3B). Similarly, 95% of the 290 secreted proteins, predicted by the SignalP program (Nielsen et al. 1997; Bendtsen et al. 2004), are expressed (Fig. 3A). Thus, the vast majority of yeast membrane and secreted proteins can be readily expressed using the MORF collection.
Figure 3.
Membrane protein expression. (A) Expression of MORF fusion proteins predicted to encode soluble, secreted, or membrane proteins. (Blue) Expressed proteins; (gray) not detected. (B) Effect of transmembrane domains on expression levels. TMHMM was used to predict the number of transmembrane domains, and ORFs in each expression category were sorted into the bins indicated. (Yellow) High expression; (green) medium expression; (blue) low expression.
Intrinsic ORF/protein properties affect native cellular abundance
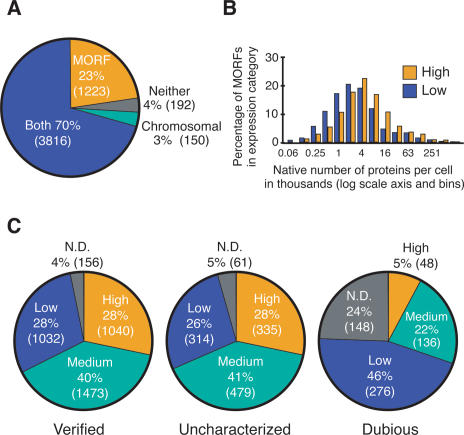
Because transcription of the MORF collection is driven by a strong regulated promoter, we expected to find more comprehensive expression of MORF proteins than was found with two chromosomally tagged collections (Ghaemmaghami et al. 2003; Huh et al. 2003) that also employ C-terminal ORF fusions. Almost all (3816, 96%) of the 3966 ORFs that were detectable with either a GFP or a TAP chromosomal C-terminal tag and were assayed for expression in the MORF collection (Ghaemmaghami et al. 2003; Huh et al. 2003) are also expressed in the MORF collection (Fig. 4A). An additional 1223 ORFs express detectable amounts of protein only in the MORF collection, but not in either chromosomal collection, indicating that the use of a strong inducible promoter can enhance expression of normally rare cellular proteins. Moreover, over half of the proteins that were not detectable in the MORF collection were also not detected with either chromosomal C-terminal tag (192 of 342 common targets). It is possible that the C-terminal tag itself may destabilize this set of proteins, consistent with the observation that 7%–13% of essential proteins could not be tagged at their C terminus in the chromosome (Ghaemmaghami et al. 2003; Huh et al. 2003). Of the 150 proteins that could not be detected by immunoblotting from the MORF expressing strains, but were detected in the chromosomal collections, 42 were only visualized with the GFP fluorescent tag and were not detectable with the chromosomal TAP tag.
Figure 4.
Comparison of MORF expression with native protein expression and ORF status. (A) Comparison of expression of proteins in the MORF collection with two chromosomally tagged collections. All ORFs tagged and analyzed in both collections were compared. An additional 551 ORFs from which expression was detected using chromosomal tags are not included in the analysis since they were not tested in the yeast MORF collection; 208 of these have been cloned in the MORF collection, but not examined in yeast. (B) Comparison of expression levels of chromosomally tagged collection and MORF collection. Proteins in high (yellow) and low (blue) expression categories were binned based on the estimates of the native number of molecules per cell, as measured with chromosomal TAP tags (Ghaemmaghami et al. 2003). The upper limits of these estimates are shown on the _X_-axis, which is a log scale. Dubious ORFs, as well as MORF clones, severely compromised for growth on raffinose or raffinose + galactose were discarded prior to analysis. (C) Analysis of expression of verified, uncharacterized, and dubious ORFs.
Analysis of the expression data from the MORF and chromosomal-tagged collections reveals a strong correlation between the native abundance of the protein (Ghaemmaghami et al. 2003) and its expression in the MORF collection (Fig. 4B). The median number of endogenous molecules per cell measured with the chromosomal TAP tag (Ghaemmaghami et al. 2003) is almost twofold higher for ORFs of the MORF library that are in the high-expression category compared with those in the low-expression category (3030 vs. 1590 molecules per cell) (Supplementary Table S3). We conclude that most ORFs and/or proteins contain intrinsic information in their DNA, RNA, and/or protein sequence that influences their overall level of expression.
Forty-eight dubious ORFs are efficiently expressed and likely authentic
Despite more than 40 years of genetic analysis and almost 10 years since the yeast genome was sequenced (Goffeau et al. 1996), for many ORFs, both the identity and existence of their encoded proteins is still in doubt. SGD classifies yeast ORFs into three categories: verified and uncharacterized ORFs, which are supported by strong evidence including the existence of orthologs in other species (Cliften et al. 2003; Kellis et al. 2003; Balakrishnan et al., May 2005); and dubious ORFs, which lack orthologs in other Saccharomyces species and for which there is only limited evidence for their existence. We find that most dubious ORFS are expressed very poorly, if at all. As shown in Figure 4C, 70% of dubious ORFs are either not detected or are expressed at low levels (24% and 46%, respectively), whereas only 31%–32% of verified or uncharacterized ORFs are poorly expressed. Since most dubious ORFs probably do not encode cellular proteins, we infer, as have others, that these nonnative polypeptides are rapidly degraded (Friedlander et al. 2000). Strikingly, 48 dubious ORFs are expressed at high levels (Supplementary Table S4), which demonstrates that they encode stable polypeptides and provides some evidence that these ORFs encode functional proteins. For 12 of these highly expressed dubious ORFs, there is some independent evidence of their existence: (1) Two ORFs (YGR151C and YOR105W) were observed with a chromosomal tag (Ghaemmaghami et al. 2003; Huh et al. 2003); (2) two ORFs (YBL077W and YDR396W) are essential (Giaever et al. 2002; Hazbun et al. 2003); (3) two ORFs (YBR126W-A and YDL114W-A) were identified in other studies as having recognizable homologs (Kumar et al. 2002; Blandin et al. 2000); (4) two ORFs (YER087C-A and YOR300W) have been associated with phenotypes and given names (Toikkanen et al. 1996; Kang and Jiang 2005); and (5) four ORFs (YDL240C-A, YGR011W, YNR005C, and YOL099C) cause slow growth on galactose, glycerol, and ethanol.
Inhibition of cell growth due to overexpression of proteins affecting the cytoskeleton, transport, and transcription
Although numerous previous screens have identified genes whose overproduction (or misexpression) is deleterious to cell growth (Liu et al. 1992; Ramer et al. 1992; Espinet et al. 1995; Akada et al. 1997; Stevenson et al. 2001; Boyer et al. 2004), a systematic analysis of a genomic collection of ORFs that are represented equally has not been carried out previously. We therefore screened the MORF collection for genes that severely inhibit growth in a dosage-dependent manner by growing the strains on carbon sources expected to induce protein expression to different degrees: glucose, in which repression is nearly complete; raffinose, in which there is very low expression due to partial loss of glucose repression; raffinose + galactose, the standard condition causing moderate induction of the MORF proteins; and galactose + glycerol + ethanol, resulting in maximal induction of the MORF proteins.
Almost all strains grow well under repressing conditions, and only a single MORF encoding TOM22, an essential component of the mitochondrial outer membrane translocase (Wiedemann et al. 2003), causes severe growth defects on raffinose. However, incubation under moderately inducing conditions (raffinose plus galactose) resulted in lack of growth for 88 (1.6%) MORF strains (Fig. 5A; Supplementary Table S5) and an even larger number, 371 strains, fail to grow (or are very sick) in galactose + glycerol + ethanol medium. The fact that relatively few strains fail to grow under protein-induction conditions (88) demonstrates that lethality caused by protein induction is not a significant problem for production of proteins. Because 67 of these strains still produce fusion protein, ORF-induced lethality is not significant during the relatively short induction time used for protein production (6 h).
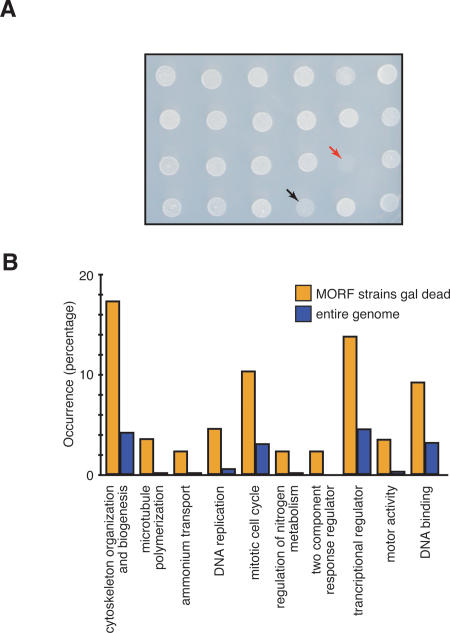
Figure 5.
Examination of the effects of MORF expression on growth. (A) Growth of yeast strains on medium containing raffinose + galactose. Arrows indicate strains with no growth (red) or slow growth (black) 48 h after transfer of strains from minimal medium containing glucose to medium containing raffinose and galactose. (B) Functions defined by MORF strains that fail to grow in galactose + raffinose. The 88 genes of MORF strains that fail to grow in galactose + raffinose are grouped according to biological or molecular function as defined using the SGD Gene Ontology Term Finder at http://db.yeastgenome.org/cgi-bin/GO/goTermFinder. The percentage of ORFs in a particular GO category is shown for the 88 lethal genes (yellow) and the genome (blue). The data are plotted in order of decreasing _P_-values in the Biological GO groups through regulation of nitrogen metabolism; Molecular Function GO groups are shown next. The _P_-values range from 1.99E-6 to 6.5E-3, and individual values are reported in Supplementary Table S6 together with categories enriched by growth on galactose, glycerol, and ethanol. The _P_-values for the transcription regulators category are 4.8E-04 among the 88 galactose + raffinose set and 1.97E-09 among the galactose, glycerol, and ethanol set, although they are enriched to nearly the same representation in both sets of genes.
Surprisingly, 36 of the 74 characterized genes that inhibit growth on galactose + raffinose are enriched in a subset of biological processes, molecular functions, or locations, as determined by gene ontology (GO) analysis (Fig. 5B; Balakrishnan et al., May 2005). For example, 16.6% of these 74 genes are classified with the GO term “cytoskeleton organization and biogenesis” compared with 4% overall in the genome (_P_-value = 2.8 E-06). As pointed out by Liu et al. (1992), who noted a similar enrichment for cytoskeletal components in a previous screen, lethality can be explained if stoichiometric ratios of these proteins are important for growth. The lethality associated with specific functions is also indicated by the inclusion in this group of both components of the two-component response regulator, two of three proteins involved in ammonia transport, and three of four involved in microtubule depolymerization. These examples clearly demonstrate the specificity of overproduction lethality as a phenotype.
A similar analysis of the 371 ORFs that inhibited growth on galactose + glycerol + ethanol both confirmed specific categories seen with the galactose + raffinose genes and added new categories. Lethal genes are significantly enriched for transcriptional regulators (13.7% in the raffinose set and 12.3% in the glycerol + ethanol set, compared with 4.4% in the genome) indicating the importance of maintaining proper expression of such proteins in the cell (Supplementary Table S6), presumably because they regulate batteries of other genes. In the larger set, we also observe significant enrichment of proteins involved in transport (74 genes) and in protein import (17 genes) as well as those localized to the mitochondria (105 genes, P = 8 E-14), which might be suspected since growth on glycerol and ethanol relies on aerobic respiration and the mitochondrion.
Global analysis of protein glycosylation
We have used the MORF collection to globally analyze protein glycosylation, a post-translational modification integral to the function and regulation of many proteins (Helenius and Aebi 2004). Although 20%–50% of proteins in eukaryotes are predicted to be glycosylated (Apweiler et al. 1999), the number of known glycosylated proteins is quite small (171 in the Saccharomyces cerevisiae genome [Csank et al. 2002]). The difficulties with studying glycoproteins are significant and include heterogeneous glycan populations created by stepwise and nontemplated processing, difficulty in preparation of the proteins, and the lack of strong predictions of glycosylation (particularly for O-linked). The C-terminally tagged MORF collection is particularly well suited to the large-scale analysis of glycosylation because it preserves the native N terminus of the proteins, allowing native processing to occur.
Using the MORF collection, we prepared a protein chip of 5573 proteins and probed it using a polyclonal antibody that recognizes yeast glycans (Fig. 6A; see Materials and Methods). Five-hundred-nine putative glycosylated proteins that reproducibly reacted with the antibody were identified (Supplementary Table S7). We evaluated effectiveness of the detection of glycoproteins with two metrics: coverage (defined as percentage of known glycoproteins identified) and fold-enrichment (defined as the percentage of known glycoproteins in the candidate list divided by the percentage of known glycoproteins in the entire collection). Significant coverage was achieved despite using a stringent cutoff, identifying 40% (55 of 136) of known glycoproteins present on the chip and 67% (20 of 30) of known GPI-anchored proteins (the GPI anchor is a glycolipid). Furthermore, known glycoproteins constitute 10.8% of the antibody-reacting set of 509 proteins, but only 2.4% of the entire collection. This dramatic enrichment of known glycoproteins and GPI-anchored proteins in the antibody-reactive set (4.5-fold and 7.8-fold, respectively) (Fig. 6B) suggests that the proteins identified by the antibody are likely to be glycosylated. Additionally, the antibody-reactive proteins are 2.7-fold enriched for cell-wall proteins (Balakrishnan et al., May 2005), 2.9-fold enriched for proteins containing predicted signal peptides (Nielsen et al. 1997; Bendtsen et al. 2004), and 1.9-fold enriched for proteins that migrate ≥30% slower than expected by SDS-PAGE.
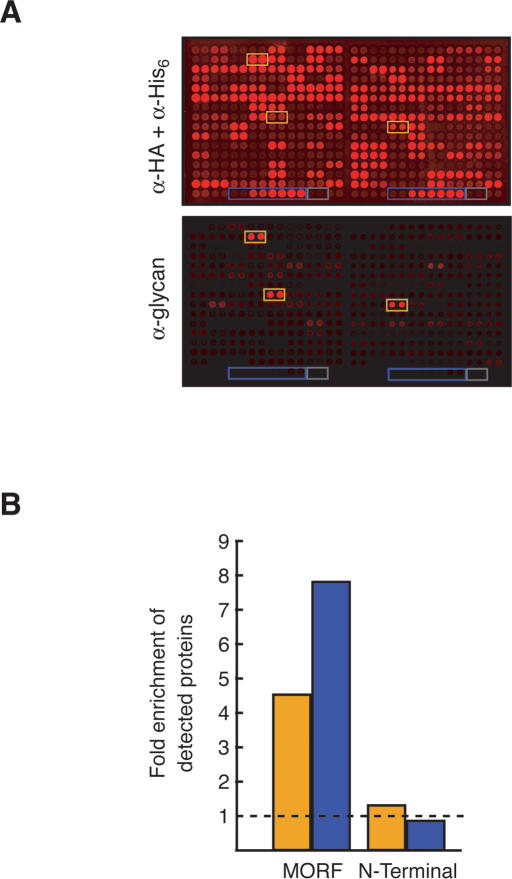
Figure 6.
Identification of yeast glycoproteins. (A) Detection of glycoproteins on a protein chip. Two blocks (out of 48) of a protein chip of 5573 C-terminally tagged proteins printed in duplicate are shown, probed with anti-HA and anti-His6 antibodies (top) or anti-yeast glycan antibody (bottom). Each block contains a dilution series of Leu2p (boxed in blue) and elution buffer alone (gray). Representative reactive candidate glycoproteins are boxed in orange. (B) Known glycoprotein enrichment in candidate list. There is a 4.5-fold enrichment of known glycoproteins (orange) in the MORF candidate list compared with the proteins on the chip. Such proteins comprise 10.8% of the candidates, but only 2.4% of the proteins on the chip. Similar calculations were done for GPI-linked proteins (blue, 7.8-fold). In contrast, no enrichment was seen when probing an N-terminally tagged collection (1.3-fold for glycoproteins, 0.8-fold for GPI-linked proteins).
To compare glycosylation of the C-terminally tagged MORF collection with an N-terminally tagged collection, we probed a yeast protein chip containing 4300 unique proteins tagged at their N termini with GST-His6, using conditions identical to those used with the MORF protein chip. In contrast to the MORF collection, the N-terminally tagged collection had inefficient coverage of known glycoproteins; of 269 antibody-reactive proteins, we observed only 8% coverage (seven of 87) of known glycoproteins, and 5% coverage of known GPI-anchored proteins (one of 19). Furthermore, antibody-reactive proteins from the N-terminal library were not significantly enriched for known glycoproteins (1.3-fold) or GPI-anchored proteins (0.8-fold) (Fig. 6B).
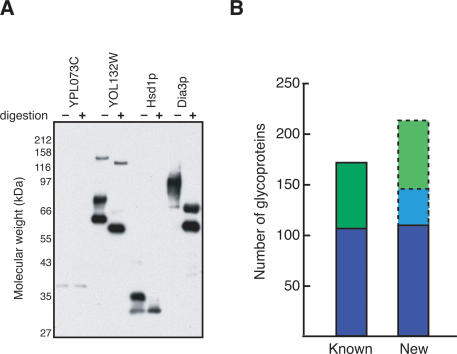
To independently assess whether or not the antibody-reacting proteins are authentic glycoproteins, we digested individual proteins in solution with two enzymes that remove N-linked glycans (Endo H and PNGase F), and screened for a mobility shift of the protein on SDS-PAGE (Fig. 7A; Supplementary Table S7). We initially set up the test using 49 individually purified, known N- and O-linked glycoproteins that reacted with the antibody on the protein array as positive controls and 19 nonreactive, soluble MORF proteins as negative controls. Thirty-three of 49 known glycoproteins exhibit a mobility shift after digestion, including 21 of 25 (84%) known N-linked glycoproteins. None of the 19 negative control proteins exhibited a mobility shift after digestion, as expected. Thus, overall we find that ∼67% of known glycoproteins exhibit altered mobility after enzyme treatment.
Figure 7.
Validation of candidate glycoproteins. (A) Western blot of candidate glycoproteins in gel-shift assay. Purified proteins were mock-treated (-) or digested with Endo H and PNGase F (+) to remove N-linked glycans before Western blot analysis with anti-HA antibody to detect mobility shifts. (B) New glycoproteins identified. Known N-linked glycoproteins (blue) and total known glycoproteins (green) in the genome are shown. Newly identified N-linked glycoproteins confirmed by Endo H and PNGase gel-shift (109 of 344 tested) are shown in blue. Based on the rate of gel-shift for known glycoproteins and known N-linked glycoproteins tested (see text), the projected number of additional new N-linked glycoproteins (35, light blue) and projected total new glycoproteins (217, light green) after testing of the 110 untested candidates are shown.
Endo H/PNGase treatment of 344 individual candidate glycoproteins resulted in direct confirmation (by mobility shift) of 109 new glycoproteins, more than doubling the number of known N-linked glycoproteins in yeast. Based on the observation that 31.7% of 344 candidates were confirmed by mobility shifts after Endo H/PNGase treatment, we extrapolate that ∼35 of the 110 remaining candidates would be confirmed by this assay as N-linked glycoproteins (Fig. 7B). Moreover, because only 67% of known glycoproteins exhibit an altered mobility shift, it is likely that nearly half (47%) of the 454 previously unknown candidates identified in our screen are bona fide glycoproteins. In summary, we demonstrate a robust method for identifying post-translational modifications and greatly increase the number of known glycoproteins present in yeast.
Biochemical activities are efficiently detected using MORF protein pools
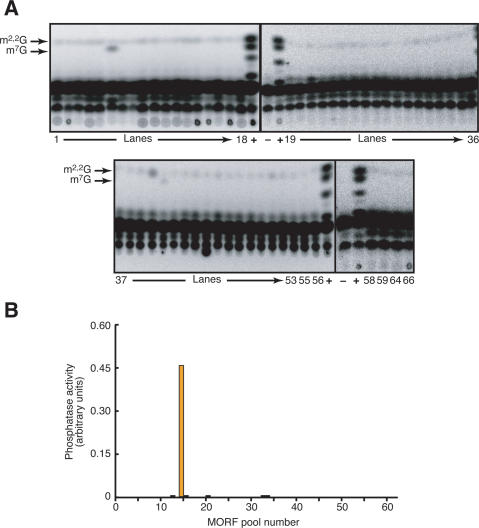
The MORF library is also highly useful as a resource for parallel enzymatic analysis of the proteome by the biochemical genomics approach (Martzen et al. 1999; Phizicky et al. 2003). Using pools containing 96 different MORF proteins purified on IgG Sepharose, we could easily detect the activity of three proteins known to catalyze tRNA modification reactions (Fig. 8A). Both members of a two-component tRNA methyltransferase complex were detected in different pools, demonstrating that there was sufficient sensitivity to detect copurification of an active complex when only one component is overproduced, as also reported previously for a GST-ORF fusion library (Alexandrov et al. 2002). In addition, we detected another methyltransferase responsible for m2,2G formation that could not be detected earlier (Fig. 8A). Detection of generic phosphatase activity using paranitrophenylphosphate was so sensitive that we could observe hydrolysis from the Pho13p-expressing yeast strain with protein derived from as little as 80 nL of culture (Fig. 8B). This high sensitivity is almost entirely due to the near absence of background using the highly selective IgG purification procedure.
Figure 8.
Biochemical activities are detected in the correct, individual protein pools with high sensitivity. MORF strains from each 96-well plate were pooled as described previously (Martzen et al. 1999; Phizicky et al. 2002) and grown, and proteins were purified on IgG sepharose followed by cleavage with 3C protease as described in the Supplemental Material. (A) Detection of two activities that modify single specific nucleotides in tRNAPhe: m2,2G formation catalyzed by Trm1p and m7G formation catalyzed by Trm8p/Trm82p. Plates 54, 57, 60–63, and 65 were eliminated from the yeast strain collection in the process of resorting good clones. (B) Assay for phosphatase activity using paranitrophenylphosphate and colorimetric detection.
Discussion
Genomic collections of ORFs have been central to the development of technologies in functional genomics (Hudson Jr. et al. 1997; Uetz et al. 2000). Findings from these studies have led not only to the identification of individual gene functions, but also to an appreciation of the overall organization of cellular networks. The MORF collection is the most complete set of cloned ORFs described for any eukaryote to date (Lamesch et al. 2004), containing 93% of verified yeast ORFs. The collection is also of very high quality, since over half of the ORFs in the collection (3217) are completely sequenced with an average of 1078 bp of sequence for each ORF in the collection. In addition, we examined protein expression from a yeast transformant for each ORF, and in 187 cases in which expression was not detected in the first transformant, or the size of the expressed protein was sufficiently different than expected, a second transformant that behaved better was tested and saved. We suspect that for many of these cases, rearrangements of the clones might have occurred as reported previously (Clancy et al. 1984). The use of both DNA sequencing and protein expression has helped produce a high-quality collection of yeast clones and expression strains. The yeast-expression collection is expected to be superior to previous collections that fused GST to the N terminus of each yeast ORF in that it uses a new genome annotation and has higher quality-control checks. Nonetheless, the use of both collections is expected to be particularly useful for yeast research, since for any protein, a particular tag or the location of that tag may affect its function.
In this work, we have demonstrated the value of the MORF protein collection both as a source of the yeast proteome, allowing the direct analysis of the glycome, and as a source of biochemically active proteins, facilitating linkage of activities to genes. The benefits of homologous expression in yeast are underscored by comparison to collections from Caenorhabditis elegans, in which proteins are expressed in Escherichia coli, resulting in expression of ∼50% of ORFs and solubility of ∼20% of ORFs (Luan et al. 2004). In contrast, 93% of the proteins in the MORF collection are expressed in yeast, and the large majority of these are soluble (Supplementary Fig. S2; data not shown). Many other factors that impact protein function are also preserved by expression in yeast, including normal post-translational modifications such as phosphorylation or glycosylation, correct subcellular localization, and association with interacting proteins that may be essential for enzymatic function (see Alexandrov et al. 2002). Thus, the MORF library can be used for large-scale preparation of highly purified, correctly processed, and modified proteins at quantities suitable for functional studies or structural biology (Supplementary Fig. S2).
The MORF collection creates a permanent source of the yeast ORFs and is itself useful for a number of different purposes. Since the yeast ORFs are cloned into a movable vector, they can be used for multiple different purposes by shuttling them into appropriate vectors using the Gateway system, which has also been exploited for the C. elegans collection (Reboul et al. 2003). This is of particular importance for genomic studies, since new approaches and vectors are constantly being generated. Moreover, due to the clonal nature of the yeast and E. coli collections, this library will be useful as a collection of verified cloned ORFs for systematic genetic analyses such as multicopy suppressor screens. Both the yeast and E. coli MORF collections are currently available from Open Biosystems http://www.openbiosystems.com.
This is the first study in which protein and ORF differences that may affect expression in the host organism were examined. Factors correlated with higher expression include an increase in measured native abundance, codon bias and codon adaptation index, and a decrease in median pI and median size (Supplementary Table S3). Since many of these properties change to a relatively minor degree relative to the range of these variables, we also examined the distribution of ORF/protein properties of just the verified and uncharacterized ORFs in each expression category. This analysis revealed that decreased protein size (Fig. 2D) and increased native abundance (Fig. 4B) were the major factors affecting expression level. Our results suggest that intrinsic parameters affect protein abundance in yeast. Such factors might include transcription elongation, mRNA decay, protein synthesis, or degradation; these factors can be investigated in future studies.
We took advantage of the native N termini of proteins expressed from the MORF collection to perform the first systematic survey of glycosylated proteins in yeast. We identified 454 new candidate glycoproteins, 109 of which were confirmed to be modified with N-linked glycans, nearly doubling in one experiment the known yeast glycome. As expected, N-linked glycoproteins are strongly enriched for components of the cell wall and membranes, as determined by GO analysis (Balakrishnan et al.), consistent with the known roles of glycosylation in the secretory system (Helenius and Aebi 2004). Surprisingly, the confirmed N-linked glycoproteins also include three transcription factors (YLR266C, Adr1p, and Sok2p) and a cytosolic kinase (YNR047W), suggesting that glycosylation has other important functional roles, as also described earlier (Guinez et al. 2005). We speculate that the identification of this large set of glycosylated proteins will stimulate rapid definition of their functions, and that there will be a varied spectrum of unanticipated roles of this modification, as has proven true of the phosphorylome.
Materials and methods
Construction, analysis, and transformation of the MORF collection
Each ORF was amplified using primers listed in Supplementary Table S1 (Illumina) (0.5 μM) with 2.5 U either Pfx (Invitrogen) or Pfu Ultra (Stratagene) polymerase, 0.2 mM dNTPs, and 100 ng of S. cerevisiae genomic DNA (strain BY4700, _MAT_a ura3_Δ_0, [Brachmann et al. 1998]) in 50 μL reactions. Two cycling conditions were used: (1) 95°C for 2 min, 5 cycles of 95°C for 30 sec, 54°C for 30 sec, 72°C for 6.5 min, 28 cycles of 95°C for 30 sec, 62°C for 30 sec, and 72°C for 6.5 min, followed by 10 min at 72°C; or (2) 95°C for 2 min, 5 cycles of 95°C for 30 sec, 62°C (-4°C/cycle) for 1 min, 72°C for 8 min, 5 cycles of 95°C for 30 sec, 47°C (+4°C/cycle), 72°C for 8 min, 27 cycles of 95°C for 30 sec, 57°C for 1 min, and 72°C for 8 min, followed by 10 min at 72°C. PCR products of the correct size, visualized with ethidium bromide and long-wave UV, were excised from 1% agarose-TAE gels and gel-purified (Qiaquick 96, QIAGEN).
BP recombination reactions (5 μL) into the Gateway entry vector pDONR221 (150 ng) were done according to the manufacturer's instructions (Invitrogen) and incubated overnight at 25°C. Half of these were incubated directly in LR reactions (6 μL) containing BP recombination reactions (3 μL), 150 ng BG1805 DNA, 0.6 μL LR clonase, 0.6 μL 10× LR buffer (Invitrogen) at room temperature overnight, treated with Proteinase K according to manufacturer's directions (Invitrogen) and used to transform competent DH5α E. coli (Invitrogen). The other half were directly transformed into competent DH5α E. coli, followed by selection on Terrific Broth (Invitrogen) with 50 μg/mL kanamycin at 30°C. Candidate clones, identified from mini-prep DNA, either by restriction digestion with BsrGI or analytical PCR with Taq polymerase and flanking primers (see Supplemental Material) were sequenced. ORFs from sequence-verified entry plasmids were recombined into the expression vector BG1805 by LR reaction (Invitrogen), followed by transformation, DNA preparation, and BsrGI restriction analysis to verify inserts.
Candidate clones were sequenced (Genaissance Pharmaceuticals) with BG1805 primers, F5 (5′-CATTTTCGGTTTGTATTACTTCTTATTC-3′) and R3 (5′-GGACCTTGAAAAAGAACTTC-3′), or pDONR221 primers BPF (5′-GTAAAACGACGGCCAG-3′) and BPR (5′-CAGGAAACAGCTATGA-3′). Clones with correct sequence of both vector and ORF (≥100 bp) in both directions were acceptable; silent mutations as well as sequence coverage were recorded. In most cases, missing clones failed at the sequence verification step despite multiple attempts, including synthesis of new primers. The ORFs missing from the collection are enriched for longer ORFs (median length 1413 bp vs. 1089 bp for all targets) and for ORFs sharing a high degree of sequence identity with other ORFs (13.7% of uncloned vs. 4.6% of all targets). Expression plasmids containing sequence-verified ORFs were transformed into yeast (Y258: _Mat_a, pep4-3, his4-580, ura3-52, leu2-3, 112) (see Zhu et al. 2001); two individual transformants were saved and either one or both were analyzed for protein expression and size by immunoblotting.
Detection of fusion proteins by immunoblot analysis
Yeast MORF strains, grown in 0.8 mL of SD-ura medium overnight, were washed with SC-ura/Raffinose, and 5 μL was diluted into 0.8 mL of SC-ura/Raffinose in a 96-well box (2 mL/well) with a 3.5-mm glass ball (PGC Scientific) to facilitate mixing. After growth at 30°C for 15 h, fusion protein expression was induced by addition of 0.4 mL of 3× YEP-Gal (3% yeast extract, 6% peptone, 6% galactose) for 6 h, followed by centrifugation of cells, washing with ice-cold water, and storage at -80°C. Crude lysates were obtained by lysing cells in 200 μL of Lysis Buffer 150 (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1 mM EGTA, 10% glycerol, 0.1% Triton X-100, 0.5 mM DTT, 1 mM PMSF, 1× Complete protease inhibitors [Roche]) by shaking for 6 min in a paint-shaker (5G-HD, Harbil) at 4°C with 250 μL of acid-washed glass beads (0.5 mm, Sigma), followed by centrifugation for 5 min at 2500 × g. Crude lysates (50 μL) and 5× SDS-loading buffer (12.5 μL) were heated for 5 min at 95°C, centrifuged at 2500 × g (5 min.), and resolved (12 μL) on SDS-PAGE gels (PANTERA-W, B-Bridge International or Criterion, Bio-Rad). Transfer of proteins to PVDF membranes (Immobilon-P, Millipore) with a semi-dry transfer apparatus (Fisher) was followed by staining with amido black, blocking of the membrane for ≥30 min in TBS-Tween containing 5% nonfat dry milk, overnight incubation in TBS-Tween with 1% milk and anti-HA antibodies (16B12, 1:1000 dilution, Covance), washing five times for 5 min in TBS-Tween, incubation with HRP-conjugated sheep anti-mouse IgG (Amersham) (1 h), washing five times for 5 min in TBS-Tween, and development with Supersignal West Chemiluminescent substrate (2:1 ratio of pico:femto reagent, Pierce).
Analysis of MORF strain growth on different carbon sources
Growth of yeast MORFs on SC-ura medium containing four different carbon sources (2% dextrose; 2% raffinose; 2% raffinose + 2% galactose: 2% galactose + 3% glycerol + 2% ethanol) was compared by plating 3 μL of yeast strains from 96-well frozen stocks that had been grown on SD-ura plates and transferred to 50 μL sterile H20. Growth was scored following incubation at 30°C for 2–3 d.
Printing and probing proteins on slides
Purification of MORF proteins, both individually and in 96-well format, is described in the Supplemental Material. Purified proteins were printed onto FAST slides (catalog no. 10 486 111, Schleicher and Schuell) with a 48-pin contact printer (Bio-Rad ChipWriter Pro) as described in Zhu et al. (2001). Control spots were included on each block of the array, consisting of elution buffer alone, GST-3C protease alone, and a threefold dilution series of purified Leu2p-His6-HA. In addition, fluorescently labeled proteins were placed in the corners of each block to aid in aligning the blocks. Slides were blocked with Superblock (Pierce) for 1 h at 4°C before incubating with primary antibody (16B12 anti-HA, Covance) at 1:4,000, or rabbit anti-yeast glycan at 1:10,000 in 1:4 Superblock:TBS-Tween for 1 h at 4°C. The anti-yeast glycan antiserum was a kind gift of Susan Ferro-Novick (Yale University, New Haven, CT) and was raised against intact _mnn2_- mutant cells. Since _mnn2_- mutant strains produce core glycans lacking α-1,2 mannose and α-1,3 mannose linkages, glycoproteins from this strain have exposed α-1,6 mannose linkages (see Ballou 1990, and references therein). Slides were washed five times with TBS-Tween for 5 min before addition of secondary antibody (Alexa 647-coupled goat anti-mouse IgG; Molecular Probes) diluted 1:3,000 in 1:4 Superblock:TBS-Tween. After 1 h of incubation at 4°C, slides were washed again five times for 5 min in TBS-Tween, spun dry, and scanned in a Genepix 4200A slide scanner (Axon Instruments). Spot intensities were first background normalized by subtracting the median background of a local 22 × 22-spot sliding window from the foreground intensity of each spot. The corrected intensity of each spot was then compared with the distribution of intensities in a surrounding 8 × 8-spot window and assigned a standard deviation. The standard deviation scores for the duplicate spots of each protein were averaged and ranked; the cut-off score of 1.5 standard deviations in three of four slides was empirically chosen based on coverage and fold enrichment of known glycoproteins. Proteins that react with secondary antibody alone were discarded.
Assays for methyltransferase activity
Reaction mixtures of 10 μL, containing 50 mM Tris-Cl (pH 8), 2.5 mM MgCl2, 1 mM DTT, 50 mM NH4Ac, 0.05 mM EDTA, 1 mM spermidine, 0.5 mM S-adenosyl methionine, 50,000 cpm [α-32P]GTP tRNAPhe, were incubated at 30°C for 2 h with 1 μL MORF proteins purified on IgG sepharose in pools from 96 strains (or buffer equivalents). P1 digestion and analysis of modified nucleotides were as described (Alexandrov et al. 2002).
Acknowledgments
We thank Erin K. O'Shea and Jonathan S. Weissman for many discussions during the initial construction of the library, Asya Sklyar (Invitrogen) for sequence analysis, Susan Ferro-Novick for the anti-yeast glycan antibody, Erin O'Shea for plasmid pRSAB1234 from which pBG1805 was constructed, Richard Insel for initial support of robotics, Lukas Käll and Erik Sonnhammer for identification of yeast proteins containing a signal sequence, Ken Nelson for help with robotics, Xiaowei Zhu for protein chip analysis, and Mike Hudson for help purifying proteins. E.M.P. is supported by NIH grant HG02311; M.S. is supported by NIH grants GM62480-04, GM36494-17, and CA77808-09.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1362105.
Corresponding authors.
References
- Akada, R., Yamamoto, J., and Yamashita, I. 1997. Screening and identification of yeast sequences that cause growth inhibition when overexpressed. Mol. Gen. Genet. 254**:** 267-274. [DOI] [PubMed] [Google Scholar]
- Alexandrov, A., Martzen, M.R., and Phizicky, E.M. 2002. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8**:** 1253-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apweiler, R., Hermjakob, H., and Sharon, N. 1999. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim. Biophys. Acta 1473**:** 4-8. [DOI] [PubMed] [Google Scholar]
- Arava, Y., Wang, Y., Storey, J.D., Liu, C.L., Brown, P.O., and Herschlag, D. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 100**:** 3889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader, G.D., Heilbut, A., Andrews, B., Tyers, M., Hughes, T., and Boone, C. 2003. Functional genomics and proteomics: Charting a multidimensional map of the yeast cell. Trends Cell. Biol. 13**:** 344-356. [DOI] [PubMed] [Google Scholar]
- Balakrishnan, R., Christie, K.R., Costanzo, M.C., Dolinski, K., Dwight, S.S., Engel, S.R., Fisk, D.G., Hirschman, J.E., Hong, E.L., Nash, R., et al. Saccharomyces Genome Database. ftp://ftp.yeastgenome.org/yeast. (See citations for dates of access.)
- Ballou, C.E. 1990. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 185**:** 440-470. [DOI] [PubMed] [Google Scholar]
- Basrai, M.A., Hieter, P., and Boeke, J.D. 1997. Small open reading frames: Beautiful needles in the haystack. Genome Res. 7**:** 768-771. [DOI] [PubMed] [Google Scholar]
- Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340**:** 783-795. [DOI] [PubMed] [Google Scholar]
- Bieganowski, P. and Brenner, C. 2004. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117**:** 495-502. [DOI] [PubMed] [Google Scholar]
- Blandin, G., Durrens, P., Tekaia, F., Aigle, M., Bolotin-Fukuhara, M., Bon, E., Casaregola, S., de Montigny, J., Gaillardin, C., Lepingle, A., et al. 2000. Genomic exploration of the hemiascomycetous yeasts: 4. The genome of Saccharomyces cerevisiae revisited. FEBS Lett. 487**:** 31-36. [DOI] [PubMed] [Google Scholar]
- Boyer, J., Badis, G., Fairhead, C., Talla, E., Hantraye, F., Fabre, E., Fischer, G., Hennequin, C., Koszul, R., Lafontaine, I., et al. 2004. Large-scale exploration of growth inhibition caused by overexpression of genomic fragments in Saccharomyces cerevisiae. Genome Biol. 5**:** R72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14**:** 115-132. [DOI] [PubMed] [Google Scholar]
- Clancy, S., Mann, C., Davis, R.W., and Calos, M.P. 1984. Deletion of plasmid sequences during Saccharomyces cerevisiae transformation. J. Bacteriol. 159**:** 1065-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften, P., Sudarsanam, P., Desikan, A., Fulton, L., Fulton, B., Majors, J., Waterston, R., Cohen, B.A., and Johnston, M. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science 29**:** 29. [DOI] [PubMed] [Google Scholar]
- Csank, C., Costanzo, M.C., Hirschman, J., Hodges, P., Kranz, J.E., Mangan, M., O'Neill, K., Robertson, L.S., Skrzypek, M.S., Brooks, J., et al. 2002. Three yeast proteome databases: YPD, PombePD, and CalPD (MycoPathPD). Methods Enzymol. 350**:** 347-373. [DOI] [PubMed] [Google Scholar]
- Espinet, C., de la Torre, M.A., Aldea, M., and Herrero, E. 1995. An efficient method to isolate yeast genes causing overexpression-mediated growth arrest. Yeast 11**:** 25-32. [DOI] [PubMed] [Google Scholar]
- Friedlander, R., Jarosch, E., Urban, J., Volkwein, C., and Sommer, T. 2000. A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell. Biol. 2**:** 379-384. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415**:** 141-147. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami, S., Huh, W.K., Bower, K., Howson, R.W., Belle, A., Dephoure, N., O'Shea, E.K., and Weissman, J.S. 2003. Global analysis of protein expression in yeast. Nature 425**:** 737-741. [DOI] [PubMed] [Google Scholar]
- Giaever, G., Chu, A.M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418**:** 387-391. [DOI] [PubMed] [Google Scholar]
- Goffeau, A., Barrell, B.G., Bussey, H., Davis, R.W., Dujon, B., Feldmann, H., Galibert, F., Hoheisel, J.D., Jacq, C., Johnston, M., et al. 1996. Life with 6000 genes. Science 274**:** 546, 563-567. [DOI] [PubMed] [Google Scholar]
- Gu, W., Jackman, J.E., Lohan, A.J., Gray, M.W., and Phizicky, E.M. 2003. tRNAHis maturation: An essential yeast protein catalyzes addition of a guanine nucleotide to the 5′ end of tRNAHis. Genes & Dev. 17**:** 2889-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinez, C., Morelle, W., Michalski, J.C., and Lefebvre, T. 2005. O-GlcNAc glycosylation: A signal for the nuclear transport of cytosolic proteins? Int. J. Biochem. Cell. Biol. 37**:** 765-774. [DOI] [PubMed] [Google Scholar]
- Hazbun, T.R. and Fields, S. 2002. A genome-wide screen for site-specific DNA-binding proteins. Mol. Cell. Proteomics 1**:** 538-543. [DOI] [PubMed] [Google Scholar]
- Hazbun, T.R., Malmstrom, L., Anderson, S., Graczyk, B.J., Fox, B., Riffle, M., Sundin, B.A., Aranda, J.D., McDonald, W.H., Chiu, C.H., et al. 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12**:** 1353-1365. [DOI] [PubMed] [Google Scholar]
- Helenius, A. and Aebi, M. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73**:** 1019-1049. [DOI] [PubMed] [Google Scholar]
- Ho, Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415**:** 180-183. [DOI] [PubMed] [Google Scholar]
- Huang, J., Zhu, H., Haggarty, S.J., Spring, D.R., Hwang, H., Jin, F., Snyder, M., and Schreiber, S.L. 2004. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. 101**:** 16594-16599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson Jr., J.R., Dawson, E.P., Rushing, K.L., Jackson, C.H., Lockshon, D., Conover, D., Lanciault, C., Harris, J.R., Simmons, S.J., Rothstein, R., et al. 1997. The complete set of predicted genes from Saccharomyces cerevisiae in a readily usable form. Genome Res. 7**:** 1169-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T.R., Robinson, M.D., Mitsakakis, N., and Johnston, M. 2004. The promise of functional genomics: Completing the encyclopedia of a cell. Curr. Opin. Microbiol. 7**:** 546-554. [DOI] [PubMed] [Google Scholar]
- Huh, W.K., Falvo, J.V., Gerke, L.C., Carroll, A.S., Howson, R.W., Weissman, J.S., and O'Shea, E.K. 2003. Global analysis of protein localization in budding yeast. Nature 425**:** 686-691. [DOI] [PubMed] [Google Scholar]
- Jackman, J.E., Montange, R.K., Malik, H.S., and Phizicky, E.M. 2003. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9**:** 574-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafadar, K.A., Zhu, H., Snyder, M., and Cyert, M.S. 2003. Negative regulation of calcineurin signaling by Hrr25p, a yeast homolog of casein kinase I. Genes & Dev. 17**:** 2698-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kall, L. and Sonnhammer, E.L. 2002. Reliability of transmembrane predictions in whole-genome data. FEBS Lett. 532**:** 415-418. [DOI] [PubMed] [Google Scholar]
- Kang, C.M. and Jiang, Y.W. 2005. Genome-wide survey of non-essential genes required for slowed DNA synthesis-induced filamentous growth in yeast. Yeast 22**:** 79-90. [DOI] [PubMed] [Google Scholar]
- Kellis, M., Patterson, N., Endrizzi, M., Birren, B., and Lander, E.S. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423**:** 241-254. [DOI] [PubMed] [Google Scholar]
- Kessler, M.M., Zeng, Q., Hogan, S., Cook, R., Morales, A.J., and Cottarel, G. 2003. Systematic discovery of new genes in the Saccharomyces cerevisiae genome. Genome Res. 13**:** 264-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh, A., Larsson, B., von Heijne, G., and Sonnhammer, E.L. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305**:** 567-580. [DOI] [PubMed] [Google Scholar]
- Kumar, A., Harrison, P.M., Cheung, K.H., Lan, N., Echols, N., Bertone, P., Miller, P., Gerstein, M.B., and Snyder, M. 2002. An integrated approach for finding overlooked genes in yeast. Nat. Biotechnol. 20**:** 58-63.11753363 [Google Scholar]
- Kus, B.M., Gajadhar, A., Stanger, K., Cho, R., Sun, W., Rouleau, N., Lee, T., Chan, D., Wolting, C., Edwards, A.M., et al. 2005. A high throughput screen to identify substrates for the ubiquitin ligase Rsp5. J. Biol. Chem. 280**:** 29470-29478. [DOI] [PubMed] [Google Scholar]
- Lamesch, P., Milstein, S., Hao, T., Rosenberg, J., Li, N., Sequerra, R., Bosak, S., Doucette-Stamm, L., Vandenhaute, J., Hill, D.E., et al. 2004. C. elegans ORFeome version 3.1: Increasing the coverage of ORFeome resources with improved gene predictions. Genome Res. 14**:** 2064-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., Krizek, J., and Bretscher, A. 1992. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics 132**:** 665-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, C.H., Qiu, S., Finley, J.B., Carson, M., Gray, R.J., Huang, W., Johnson, D., Tsao, J., Reboul, J., Vaglio, P., et al. 2004. High-throughput expression of C. elegans proteins. Genome Res. 14**:** 2102-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, X., Zhao, X., and Yu, Y.T. 2003. Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J. 22**:** 1889-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martzen, M.R., McCraith, S.M., Spinelli, S.L., Torres, F.M., Fields, S., Grayhack, E.J., and Phizicky, E.M. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286**:** 1153-1155. [DOI] [PubMed] [Google Scholar]
- Nielsen, H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10**:** 1-6. [DOI] [PubMed] [Google Scholar]
- Oshiro, G., Wodicka, L.M., Washburn, J.R., Yates III, M.P., Lockhart, D.J., and Winzeler, E.A. 2002. Parallel identification of new genes in Saccharomyces cerevisiae. Genome Res. 12**:** 1210-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky, E.M., Martzen, M.R., McCraith, S.M., Spinelli, S.L., Xing, F., Shull, N.P., Van Slyke, C., Montagne, R.K., Torres, F.M., Fields, S., et al. 2002. Biochemical genomics approach to map activities to genes. Methods Enzymol. 350**:** 546-559. [DOI] [PubMed] [Google Scholar]
- Phizicky, E., Bastiaens, P.I., Zhu, H., Snyder, M., and Fields, S. 2003. Protein analysis on a proteomic scale. Nature 422**:** 208-215. [DOI] [PubMed] [Google Scholar]
- Ramer, S.W., Elledge, S.J., and Davis, R.W. 1992. Dominant genetics using a yeast genomic library under the control of a strong inducible promoter. Proc. Natl. Acad. Sci. 89**:** 11589-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34**:** 35-41. [DOI] [PubMed] [Google Scholar]
- Stevenson, L.F., Kennedy, B.K., and Harlow, E. 2001. A large-scale overexpression screen in Saccharomyces cerevisiae identifies previously uncharacterized cell cycle genes. Proc. Natl. Acad. Sci. 98**:** 3946-3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toikkanen, J., Gatti, E., Takei, K., Saloheimo, M., Olkkonen, V.M., Soderlund, H., De Camilli, P., and Keranen, S. 1996. Yeast protein translocation complex: Isolation of two genes SEB1 and SEB2 encoding proteins homologous to the Sec61 β subunit. Yeast 12**:** 425-438. [DOI] [PubMed] [Google Scholar]
- Tong, A.H., Lesage, G., Bader, G.D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G.F., Brost, R.L., Chang, M., et al. 2004. Global mapping of the yeast genetic interaction network. Science 303**:** 808-813. [DOI] [PubMed] [Google Scholar]
- Ubersax, J.A., Woodbury, E.L., Quang, P.N., Paraz, M., Blethrow, J.D., Shah, K., Shokat, K.M., and Morgan, D.O. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature 425**:** 859-864. [DOI] [PubMed] [Google Scholar]
- Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., et al. 2000. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403**:** 623-627. [DOI] [PubMed] [Google Scholar]
- Wiedemann, N., Kozjak, V., Chacinska, A., Schonfisch, B., Rospert, S., Ryan, M.T., Pfanner, N., and Meisinger, C. 2003. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 424**:** 565-571. [DOI] [PubMed] [Google Scholar]
- Zhu, H., Klemic, J.F., Chang, S., Bertone, P., Casamayor, A., Klemic, K.G., Smith, D., Gerstein, M., Reed, M.A., and Snyder, M. 2000. Analysis of yeast protein kinases using protein chips. Nat. Genet. 26**:** 283-289. [DOI] [PubMed] [Google Scholar]
- Zhu, H., Bilgin, M., Bangham, R., Hall, D., Casamayor, A., Bertone, P., Lan, N., Jansen, R., Bidlingmaier, S., Houfek, T., et al. 2001. Global analysis of protein activities using proteome chips. Science 293**:** 2101-2105. [DOI] [PubMed] [Google Scholar]