Quorum-Sensing Regulation of the Production of Blp Bacteriocins in Streptococcus thermophilus (original) (raw)
Abstract
The blp gene cluster identified in the genome sequences of Streptococcus thermophilus (_blp_St) LMG18311, CNRZ1066, and LMD-9 displays all the characteristics of a class II bacteriocin locus. In the present study, we showed that the _blp_St locus is only fully functional in strain LMD-9 and regulates the production of antimicrobial peptides that inhibit strains LMG18311 and CNRZ1066. The _blp_St cluster of LMD-9 contains 23 genes that are transcriptionally organized in six operons: _blpABC_St (peptide transporter genes and pheromone gene); _blpRH_St (two-component regulatory system genes); _blpD_St-_orf1, blpU_St-orf3, and _blpE-F_St (bacteriocin precursors and immunity genes); and _blpG-X_St (unknown function). All the operons, except the regulatory unit _blpRH_St, were shown to be coregulated at the transcriptional level by a quorum-sensing mechanism involving the mature S. thermophilus pheromone BlpC* (BlpC*St), which was extracellularly detected as two active forms (30 and 19 amino acids). These operons are differentially transcribed depending on growth phase and pheromone concentration. They all contain a motif with two imperfect direct repeats in their mapped promoter regions that could serve as binding sites of the response regulator BlpRSt. Through the construction of deletion mutants, the _blp_St locus of strain LMD-9 was shown to encode all the essential functions associated with bacteriocin production, quorum-sensing regulation, and immunity.
Many lactic acid bacteria (LAB) secrete antimicrobial peptides called bacteriocins. In general, these peptides are small, are cationic, and have hydrophobic/amphiphilic properties. They kill susceptible strains by the formation of poration complexes through the membrane (24, 40). Most bacteriocins identified in LAB belong to the class II bacteriocins that include non-posttranslationally modified peptides (41). This class is further subdivided into two main subcategories: IIa, the pediocin-like bacteriocins with strong antilisteria effects, which contain a conserved N-terminal YGNGVXC sequence (17); and IIb, bacteriocins whose activity depends on the complementary activity of two peptides (21, 24). All other nonmodified bacteriocins are classified as class IIc (41). Production of class II bacteriocins is usually under the control of a dedicated three-component regulatory system (induction factor [IF], histidine kinase [HK], and response regulator) that acts as a quorum-sensing (QS) device, coupling bacteriocin production to cell density (30).
Numerous reports on the regulation of LAB bacteriocins are available, but relatively little is known about bacteriocins from Streptococcus thermophilus, a species extensively used in the manufacture of yogurt and hard, “cooked” cheese. To our knowledge, eight thermophilins produced by industrial strains have been purified and characterized (1, 2, 22, 29, 37, 38, 47, 48), but no specific genetic locus has been associated with their production, except for thermophilin 13, for which structural genes have been identified (37). Recently, we identified a common locus displaying characteristics of a class II bacteriocin gene cluster in the genome of the sequenced S. thermophilus strains LMG18311, CNRZ1066, and LMD-9, which are regarded as non-bacteriocin producers (27). This locus strongly resembles the blp (for bacteriocin-like peptide) locus of Streptococcus pneumoniae (_blp_Sp) (12) and was therefore designated the S. thermophilus blp locus (_blp_St). A similar locus was also found in Streptococcus salivarius (P. Renault, personal communication), Streptococcus mutans (bsm locus) (45), Streptococcus pyogenes (sil locus) (25), and Streptococcus equi (31). Recently, functional Blp bacteriocin systems were reported in S. mutans (mutacin IV and mutacin V) (23, 45) and S. pneumoniae (BlpM and BlpN) (10). Among S. thermophilus strains, LMD-9 harbors the most complex locus (Fig. 1A). Genes encoding products specific to a three-component QS system are common in the three strains: _blpH_St and _blpR_St that encode proteins similar to HKs and response regulators, respectively, and _blpC_St encoding the corresponding putative IF precursor that contains the typical double-glycine (2-Gly) cleavage site. The _blp_St gene cluster also encodes a potential bacteriocin/IF ABC-transporter (_blpA_St) and an accessory transporter protein (_blpB_St) that is truncated in strains LMG18311 and CNRZ1066; the cluster also includes a variable number of bacteriocin-like peptides containing a 2-Gly leader (_bac_St genes): _blpD_St, _blpU_St, _blpE_St, and _blpF_St in LMD-9; _blpK_St in CNRZ1066; and _blpU_St and _blpK_′St (pseudogene) in LMG18311 (27) (Fig. 1A). We also identified a range of genes (_blpQ_St, _blpX_St, and orf genes) that encode proteins that show structural similarities to immunity proteins and _blpG_St, encoding a protein containing a CXXC motif, which could act as a thioredoxin isomerase in the formation of disulfide bonds (Fig. 1A).
FIG. 1.
The _blp_St locus of S. thermophilus and study of the functionality of bacteriocin production in S. thermophilus. (A) Schematic representation of the _blp_St locus in strains CNRZ1066 (genes str1691 to str1673), LMG18311 (stu1691 to stu1673), and LMD-9 (STER_1634 to STER_1653). Genes encoding peptides with predicted functions are represented by patterned arrows as follows: ABC transporter, small squares; transport accessory protein, large squares; response regulator, vertical lines; HK, horizontal lines; bacteriocin-like peptide, light gray; inducing factor, dark gray; hydrophobic peptide of unknown function, black; hydrophilic peptide of unknown function, white; peptide similar to the immunity protein SakIX of the Class IIa bacteriocin sakacin X, chess squares) (46); and modification protein, points. Genes encoding peptides with a 2-Gly leader are represented by gray arrows (light and dark). Letters and numbers in italics refer to the corresponding blp genes and orf genes, respectively. (B) Detection of bacteriocin production using the spot-on-lawn method from strains LMD-9 (pGILF002), CNRZ1066 (pGILF001), and LMG18311 (pGILF001) overexpressing their cognate _blpC_St gene. Strains carrying the empty expression vector (pMG36e) are used as control or indicator strains. Five microliters of culture (OD600 of 1) of each producer strain (names of strains and their corresponding numbers are indicated on the right) was spotted directly on a soft agar layer containing 108 CFU of the indicator strain (names of strains are indicated above each picture).
The aim of the present study was to establish the functional role of the _blp_St gene cluster of S. thermophilus with respect to bacteriocin production, to reveal its transcriptional organization, and to elucidate the subjacent regulation mechanisms.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in the present study are listed in Table 1. Plasmids derived from pMG36e (44) and pGhost9 (35) were constructed in strains TG1 (42) and EC1000 (33), respectively, of Escherichia coli. E. coli was grown in LB medium with shaking at 37°C (42). S. thermophilus was grown anaerobically (BBL GasPak systems; Becton Dickinson, Franklin Lakes, NJ) in M17 broth (Difco Laboratories Inc., Detroit, MI) with 1% (wt/vol) glucose (M17G) at 42°C. When required, erythromycin (250 μg/ml for E. coli and 2.5 μg/ml for S. thermophilus) was added to the medium. Solid agar plates were prepared by adding 2% (wt/vol) agar to the medium.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| S. thermophilus | ||
| LMD-9 | Wild type | ATCCb |
| LF101 | LMD-9 Δ_blpR_St | This study |
| LF102 | LMD-9 Δ_blpH_St | This study |
| LF103 | LMD-9 Δ(_blpR_St-_blpH_St) | This study |
| LF104 | LMD-9 Δ(_blpD_St-_blpX_St) | This study |
| LF105 | LMD-9 Δ(_blpD_St-_blpF_St) | This study |
| LF106 | LMD-9 Δ(_blpG_St-_blpX_St) | This study |
| LF107 | LMD-9 Δ_blpB_St | This study |
| LF108 | LMD-9 Δ(_blpA_St-_blpB_St) | This study |
| LMG18311 | Wild type | 8 |
| CNRZ1066 | Wild type | 8 |
| E. coli | ||
| TG1 | supE hsd_Δ_5 thi Δ(lac-proAB) F′[_traD36 proAB+ lacI_q_lacZ_Δ_M15_] | 42 |
| EC1000 | Kmr RepA+; MC1000 containing a copy of the repA gene of pWV01 in its chromosome | 33 |
| Plasmids | ||
| pMG36e | Emr; E. coli-S. thermophilus shuttle vector; contains the P32 promoter | 44 |
| pGIBG001 | Emr; pMG36e where a NcoI restriction site has been introduced in order to allow the cloning of genes in translational fusion with the P32 promoter | This study |
| pGILF001 | Emr; pGIBG001 with a 0.16-kb insert containing the St_blpC_ ORF of S. thermophilus LMG18311 in translational fusion the P32 promoter | This study |
| pGILF002 | Emr; pGIBG001 with a 0.16-kb insert containing the St_blpC_ ORF of S. thermophilus LMD-9 in translational fusion the P32 promoter | This study |
| pGhost9 | Emr Ts | 35 |
Analysis of antimicrobial activity and immunity of S. thermophilus.
Synthetic S. thermophilus BlpC (BlpCSt) mature forms of D9C-30, D9C-19, and D9C-11 peptides (purity of >95%) were supplied by Sigma-Genosys Ltd. (Haverhill, United Kingdom). Activity was assayed by two methods.
(i) Spot-on-lawn method.
An overnight culture of the producer strain was diluted 100-fold in fresh medium and incubated anaerobically at 42°C. When necessary, synthetic IF was added to the culture at an optical density at 600 nm (OD600) of 0.1 (unless otherwise stated), and at the desired growth phase (OD600 of 1 unless otherwise stated), 5 μl of the growing culture was spotted directly on a 6-ml soft M17G layer (0.8% agar) containing 108 CFU of the indicator strain (100 μl of a culture at OD600 of 1). Cell-free culture supernatants were obtained by centrifugation and subsequent filter sterilization. Plates were incubated anaerobically at 42°C overnight for detection of inhibition zones surrounding the producer cells.
(ii) Overlay (multilayer) method.
An overnight culture of the producer strain was diluted 100-fold in fresh medium and incubated anaerobically at 42°C. At an OD600 of 1, 100 μl of the culture was diluted 106-fold in 6-ml of prewarmed soft M17G medium (0.8% agar) and poured on a plate containing a supporting layer of 25 ml of solid M17G medium (agar 2%). A second 6-ml soft M17G layer, without IF or with the appropriate concentration of IF, was poured on the layer of producer cells. Plates were incubated for 10 h anaerobically at 42°C, and a third 6-ml layer of soft M17G medium containing 108 CFU of the indicator strain (100 μl of a culture at OD600 of 1) was poured on the top. Plates were incubated for 10 h anaerobically at 42°C for detection of inhibition zones surrounding the producer colonies.
DNA techniques and transformation.
General molecular biology techniques were performed according to the instructions given by Sambrook et al. (42). Electrotransformation of E. coli was performed as described by Dower et al. (15). Electrocompetent S. thermophilus cells were prepared as previously described (7). After transformation with 1 μg of plasmid DNA, cells were immediately resuspended in 1 ml of M17G medium and incubated anaerobically for 6 h at 37°C (pMG36e derivatives) or 29°C (pGhost9 derivatives). S. thermophilus chromosomal DNA was prepared as described by Ferain et al. (19). PCRs were performed with Taq DNA polymerase (Promega, Madison, Wis.) in a GeneAmp PCR system 2400 (Applied Biosystems, Lennik, Belgium). The primers used in this study are listed in Table S1 in the supplemental material.
Construction of overexpression vectors for the strain-specific _blpC_St genes.
The pGIBG001 overexpression vector contains a P32-ribosome binding site-ATG expression cassette amplified by PCR with primers P32U and P32D, which is translationally fused to the ospA open reading frame amplified by PCR with primers POsp2 and POsp3. The fusion construct was cloned as an MfeI-SacI restriction fragment into pMG36e digested with EcoRI and SacI. The entire open reading frames of the _blpC_St gene of strain LMG1831 (_blpC_St LMG1831) and of _blpC_St LMD-9 were amplified by PCR with primers BlpC1/LMGBlpC2 and BlpC1/LMDBlpC2, respectively. These 0.16-kb fragments were then digested with NcoI and StyI and cloned into pGIBG001, digested with NcoI and XbaI. The resulting plasmids were designated pGILF001 (_blpC_St LMG18311) and pGILF002 (_blpC_St LMD-9).
Construction of deletion mutants in the _blp_St locus.
The deletion plasmids were constructed by cloning in the thermosensitive pGhost9 vector (35) two fragments of approximately 1 kb, containing the upstream region and the downstream region of the gene or genes of interest, respectively. Deletions in the _blp_St locus were performed by double homologous recombination after two steps of temperature shift, as previously described (36). Both recombination steps (plasmid integration and excision) were confirmed by PCR with primers located upstream and downstream of the recombination regions. For details on the strategy used for the construction of the different deletion vectors and the corresponding S. thermophilus mutant strains, see Text S1 and Table S1 in the supplemental material.
RNA extraction, Northern blotting, and primer extension.
For the time course experiment, the LMD-9 culture at an OD600 of 0.1 received 400 ng/ml of D9C-30. Aliquots (50 ml) were collected before IF addition (time zero sample) and every 30 min (during 180 min) after peptide addition. For the dose-response experiment with D9C-30, an LMD-9 culture (OD600 of 0.1) was separated in different subcultures, and increasing concentrations of D9C-30 were added. After a 2-h induction, 50-ml aliquots were collected. Cells were harvested by centrifugation (6,000 × g for 4 min) and mechanically broken with 0.18-mm-diameter glass beads in a Braun Homogenizer (three 1-min periods of homogenization with 1-min intervals on ice). Total RNA was extracted using a High Pure RNA isolation kit (Roche, Basel, Switzerland).
Northern blotting experiments were carried out as described by Lorquet et al. (34). Hybridization was performed as previously reported (34), using [α-32P]dCTP-radiolabeled PCR fragments (400 to 800 bp) as specific probes with a Rediprime II labeling kit (Amersham Biosciences, Little Chalfont, Buckinghamshire, United Kingdom). Probes 1 to 7 were amplified by PCR with primer pairs ND1/rt8 (P1, orf1 and orf2), rt15/NU2 (P2, orf3 and orf4), Norf41/Norf42 (P3, orf4 and orf5), NE1/NE2 (P4, orf7), rt12/rt17 (P5, part of _blpG_St and orf8), NA1/NA2 (P6, blpA St) and NRH1/NRH2 (P7, part of _blpR_St and _blpH_St). Radioactive bands were visualized by autoradiography and quantified with an Instant Imager (Packard Instruments, Meriden, CT). The relative amounts of transcripts were standardized by hybridization with an S. thermophilus 16S rRNA-specific probe (primers 16S1/16S2).
Each primer extension analysis was performed on 1 μg of RNA extracted from an induced LMD-9 culture (2-h induction with 400 ng/ml D9C-30) as previously described (11). The radiolabeled primers (with T4 polynucleotide kinase) used to map the 5′ termini of _blp_St mRNAs were EXT67D and EXT71D for _blpD_St, EXT60U and EXT73U for _blpU_St, EXT72E and EXT77E for _blpE_St, EXT66A for _blpA_St, and EXT73R for _blpR_St. cDNAs were generated using Superscript III reverse transcriptase (Invitrogen), and the extension products were analyzed on 6% (wt/vol) polyacrylamide-urea sequencing gels, next to DNA sequencing reactions (AmpliCycle sequencing kit; Applied Biosystem, Foster City, CA) performed with the same primers.
MALDI-TOF MS analysis.
Bacteria were collected by sweeping sterile loops across colonies and were transferred to a target plate (26). Each sample was overlaid with 0.5 μl of a matrix solution containing 10 mg/ml of sinapinic acid (3,5-dimethoxy-4-hydroxy-cinnamic acid) in 50% acetonitrile-0.15% trifluoroacetic acid-water and allowed to dry. When hydrolysis of surface polypeptides was required, each bacterial sample of the target plate was overlaid with 0.5 μl of a solution containing 10 μg/ml of trypsin (Promega) in ammonium carbonate buffer (50 mM; pH 8.0) and was allowed to dry for 4 or 8 min at room temperature before addition of the matrix. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) spectra were collected with a Voyager DE STR instrument (Applied Biosystems, Framingham, CA). The mass spectra were acquired in the reflector mode with the following parameters: 25 kV accelerating voltage, 62% grid voltage, and 120-ns delay on different mass ranges.
RESULTS
Functionality of the blp loci for bacteriocin production in three S. thermophilus strains.
The involvement of IF in QS-regulated mechanisms is well documented (9, 13, 16), and _blpC_Sp was shown to encode a communication molecule that regulates the expression of bacteriocin-like blp genes in S. pneumoniae (12). We thus hypothesized that BlpCSt could act as a pheromone governing bacteriocin production in S. thermophilus. The sequences of the three BlpCSt peptides (30 amino acids [aa]) differ only in the C-terminal amino acid: Ala in the case of strain LMD-9 and Val in strains LMG18311 and CNRZ1066 (Fig. 2A). In order to test the functional role of BlpCSt as an inducer of bacteriocin production, the strain-specific _blpC_St genes were constitutively expressed on a multicopy plasmid in strains LMG18311 and CNRZ1066 (pGILF001 [_blpC_St LMG18311/CNRZ1066]) and in strain LMD-9 (pGILF002 [_blpC_St LMD-9]) (Table 1). The antimicrobial activity of the _blpC_St-overexpressing strains was assayed by the spot-on-lawn method using LMD-9, LMG18311, or CNRZ1066 carrying the empty expression vector (pMG36e) as an indicator strain. Expression of _blpC_St LMD-9 induced bacteriocin production in LMD-9 (Bac+ phenotype) that could inhibit growth of LMG18311 and CNRZ1066 (Imm− phenotype) (Fig. 1B). LMD-9 itself was resistant to its own antimicrobial compound(s) (Imm+ phenotype). The same phenotypes were observed when _blpC_St CNRZ1066/LMG18311 was expressed in LMD-9 (data not shown). In contrast, strains LMG18311 and CNRZ1066 expressing _blpC_St CNRZ1066/LMG18311 did not display antimicrobial activity against any of the indicator strains (Fig. 1B).
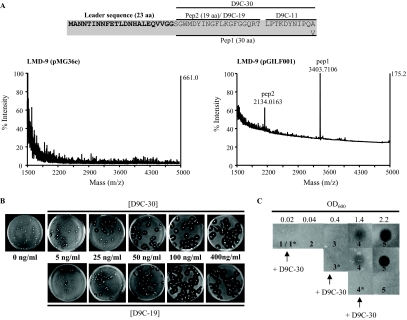
FIG. 2.
Identification of mature BlpCSt forms and their role in bacteriocin production of strain LMD-9. (A) Detection of secreted forms of BlpCSt (Pep1 and Pep2) by MALDI-TOF MS experiments on whole LMD-9(pMG36e) (left panel) and LMD-9(pGILF001) (right panel) colonies. The two peptides were identified in four independent MALDI-TOF experiments. The sequences of BlpCSt LMG18311 and BlpCSt LMD-9 are given above the MALDI-TOF spectra. The 2-Gly leader is in bold characters. The sequence of the mature forms Pep1 and Pep2 of BlpCSt LMG18311 and of the synthetic peptides D9C-30, D9C-19, and D9C-11 of BlpCSt LMD-9 are indicated with black lines. (B) Induction of bacteriocin production by S. thermophilus LMD-9 in soft growth medium (overlay method). A soft M17G overlay containing increasing concentrations of D9C-30 (upper panel) or D9C-19 (lower panel) was poured on a soft M17G overlay containing 20 to 30 growing cells of strain LMD-9. After a 10-h incubation at 42°C, a third overlay containing 108 CFU of the indicator strain CNRZ1066 was poured on the top. (C) Induction of bacteriocin production by S. thermophilus LMD-9 in liquid growth medium (spot-on-lawn method). An overnight culture of strain LMD-9 was diluted 100-fold in fresh medium and separated into three cultures. The asterisk superscript indicates the time at which 200 ng/ml D9C-30 peptide was added in the LMD-9 culture: 1*, beginning of growth (OD600 of 0.02); 3*, mid-log phase (OD600 of 0.4); and 4*, late-log phase (OD600 of 1.4). After the addition of the peptides, cell-free supernatant samples (5 μl) were collected at different times during growth: 1, beginning of growth; 2, early log phase (OD600 of 0.04); 3, mid-log phase (OD600 of 0.4); 4, late log phase (OD600 of 1.4); and 5, stationary phase (OD600 of 2.2); samples were then spotted on a soft agar layer containing 108 CFU of strain CNRZ1066.
These results clearly show that S. thermophilus is able to produce antimicrobial compounds and that this phenotype is related to the expression of _blpC_St, which most likely encodes IF-regulating bacteriocin production. The rest of our study was focused on the _blp_St locus of LMD-9.
Two secreted mature forms of BlpCSt induce bacteriocin production.
To fulfill its signaling function, the pheromone must be matured and secreted in order to interact with its cognate HK (30). The secretion and maturation of BlpCSt was investigated by performing MALDI-TOF MS at the surface of whole LMD-9 cells grown on solid medium as reported previously by Hindré et al. (26). We compared the peptide content at the surface of LMD-9 cells either overexpressing or not overexpressing the _blpC_St LMG18311 gene in order to study whether both the endogenous and heterologous BlpCSt peptides could be secreted.
Two additional products were detected at the surface of LMD-9 (pGILF001) cells with average m/z values (z = 1) of 3,403.71 and 2,134.01 (Fig. 2A), compared to the control LMD-9 (pMG36e). Trypsic cleavage performed on the cell surface, followed by time course MALDI-TOF experiments, confirmed that these two products were derived from the 53-aa BlpCSt peptide (data not shown). The 30-aa peptide 1 (Pep1) corresponds to the predicted mature sequence of BlpCSt LMG18311 (downstream of the first 2-Gly motif). The mass of peptide 2 (Pep2) corresponds to the first 19 N-terminal residues located downstream of the 2-Gly residues (Fig. 2A). The predicted mature form of the endogenous BlpCSt LMD-9 peptide (30-aa peptide ending with Ala) could not be detected at the surface of LMD-9 (pGILF001) colonies.
The observed secreted forms of BlpCSt LMG18311 in strain LMD-9 suggest that BlpCSt LMD-9 can be processed into three peptides: the 30-aa peptide (D9C-30), the 19-aa peptide (D9C-19), and the C-terminal 11-aa peptide (D9C-11) (Fig. 2A). These peptides were thus synthesized in order to investigate their functionality as inducers of the antimicrobial activity of LMD-9. To assess dose dependence of the induction, increasing concentrations of each peptide were added in a soft agar layer containing isolated LMD-9 cells (Fig. 2B). No bacteriocin production was observed upon addition of D9C-11 (up to 400 ng/ml) (data not shown). In contrast, the other two forms of D9C were found to induce antimicrobial activity at similar levels (Fig. 2B). Induction of bacteriocin production was also investigated in liquid cultures. Induction in S. thermophilus LMD-9 was observed only when IF was added either at the start of growth or during the exponential phase of growth (Fig. 2C; data shown only for D9C-30). In both cases, the highest antimicrobial activity was detected in cell-free supernatants taken from the stationary phase of growth (Fig. 2C), indicating that bacteriocin production is more efficient at a high cell density.
From these data, it can be concluded that BlpCSt is the precursor of two pheromones that regulate the antimicrobial activity of S. thermophilus LMD-9 in a dose-dependent manner, which strongly suggests a QS mechanism of regulation. The D9C-19 peptide is not more efficient as a bacteriocin inducer than the D9C-30 peptide. Successive processing steps without any apparent biological role have already been reported for the maturation of IF involved in the regulation of bacteriocin loci, such as plantaricin A (PlnA 26-, 23-, and 22-mer peptides) of Lactobacillus plantarum (13). However, we cannot rule out here that the induction effect observed with the full-length mature peptide of BlpCSt is indirectly caused by its conversion into its shorter form, D9C-19.
All the genetic determinants of bacteriocin production, immunity, and regulation are restricted to the _blp_St locus.
In order to investigate the dedicated functions of the _blp_St gene products, mutants bearing single or multiple deletions in the _blp_St locus were constructed (Table 1), and their phenotypes were analyzed upon D9C-30 induction using the spot-on-lawn method (Table 2) or the overlay method (Fig. 3 and data not shown). Similar results were obtained upon D9C-19 induction with all mutant strains (data not shown).
TABLE 2.
Phenotype of S. thermophilus LMD-9 derivativesa
| LMD-9 strain | Bacteriocin productionb | Immunityc |
|---|---|---|
| Wild type | + | + |
| Δ_blpR_St strain | − | − |
| Δ_blpH_St strain | − | − |
| Δ(_blpR_St-_blpH_St) strain | − | − |
| Δ(_blpD_St-_blpX_St) strain | − | − |
| Δ(_blpD_St-_blpF_St) strain | − | − |
| Δ(_blpG_St-_blpX_St) strain | + | + |
| Δ_blpB_St strain | +d | + |
| Δ(_blpA_St-_blpB_St) strain | − | + |
FIG. 3.
Content of the _blp_St locus in 2-Gly-containing peptides and analysis of the Bac phenotype of deletion mutants in _blpAB_St transporter genes of strain LMD-9 (overlay method). (A) Alignment of the 2-Gly-containing peptides of strain LMD-9. Light gray residues are identical amino acids (100% conservation), and blocks of conserved identical residues are shown in dark gray. Residues occurring in at least three peptides are included in the consensus sequence. (B) Antimicrobial activity of deletion mutants in the transporter-encoding genes. Producer strains containing the pGILF002 expression plasmid were grown in the absence of D9C-30, while plasmid-free producer strains were induced with 400 ng/ml D9C-30. The names of the producer strains and indicator strains are indicated above and below the images, respectively.
The presence of genes encoding a two-component system (TCS) in the _blp_St locus strongly suggests that BlpHSt and BlpRSt constitute the receptor for mature BlpCSt and the effector of bacteriocin production, respectively. The Bac− Imm− phenotype of the LMD-9 Δ_blpH_St, LMD-9 Δ_blpR_St, and LMD-9 Δ(_blpR_St-_blpH_St) strains (Table 2) showed that the BlpRHSt TCS is absolutely required for the BlpCSt-dependent induction of antimicrobial activity and immunity. Additionally, these results indicate that BlpCSt-derived peptides act solely as communication molecules and have no intrinsic antimicrobial activity.
Deletion of regions comprised between _blpD_St and _blpF_St [LMD-9 Δ(_blpD_St-_blpF_St)] and between _blpD_St and _blpX_St [LMD-9 Δ(_blpD_St-_blpX_St)] resulted in the loss of the Bac+ Imm+ phenotype of the mutant strains, while the Δ(_blpG_St-_blpX_St) strain retained its Bac+ Imm+ phenotype (Table 2). This indicates that the genetic determinants of bacteriocin production and immunity are located in the region between _blpD_St and _blpF_St. The genes located downstream of _blpF_St do not seem to be involved in either bacteriocin production or immunity against S. thermophilus.
BlpCSt and BacSt peptides display similar leader sequences (Fig. 3A), suggesting that they are secreted through a common transport system. The Bac− Imm+ phenotype of the LMD-9 Δ(_blpA_St-_blpB_St) strain indicates that the antimicrobial compounds produced by S. thermophilus LMD-9 are secreted through the BlpABSt transport system (Table 2 and Fig. 3B). In gram-positive bacteria, accessory transporter proteins are believed to facilitate the externalization of 2-Gly peptides, but their necessity for the secretion of class II bacteriocins was shown to vary among the bacteriocin systems (5, 23, 43, 46). The contribution of the accessory protein BlpBSt to transport was thus investigated. The single _blpB_St knockout strain did not suppress the Bac+ phenotype, but the size of the inhibition zone produced by this strain was reduced compared to strain LMD-9 (Fig. 3B). In contrast, when BlpCSt LMD-9 was overproduced intracellularly using plasmid pGILF002, the Δ_blpB_St mutant was unable to inhibit growth of the indicator strain, suggesting that an active BlpBSt accessory transport protein is essential for the secretion of the induction factor BlpCSt (Fig. 3B). The specific involvement of accessory transporters in IF secretion has also been shown for ComB, which, together with ComA, is required for the transport of the competence-stimulating peptide in S. pneumoniae (28). These data thus show that the BlpABSt proteins constitute the transport machinery involved in the secretion of BlpCSt and BacSt peptides.
Altogether, these results show that the _blp_St locus of strain LMD-9 encodes all the essential functions associated with bacteriocin production, regulation, and immunity (Fig. 4A).
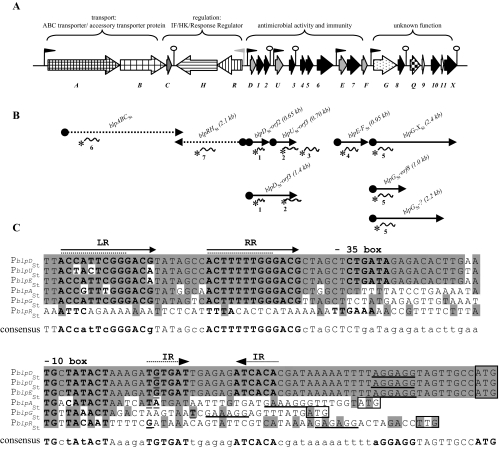
FIG. 4.
The _blp_St locus of S. thermophilus LMD-9, its transcriptional organization, and analysis of the upstream sequences of the main operons. (A) Schematic representation of the _blp_St locus of strain LMD-9. Genes encoding peptides with predicted functions are represented as described in the legend of Fig. 1. The functions of the genes deduced from the phenotypes of deletion mutants (Table 2) in the _blp_St gene cluster are indicated above the locus. Black flags represent promoter sequences containing DR, and the gray flag represents a vegetative promoter sequence. Hairpin structures indicate the presence of IR that could serve as transcription terminators. (B) Transcriptional organization of the _blp_St locus of strain LMD-9 determined by Northern blotting experiments. The plain arrows represent the mRNAs detected in the Northern blotting experiments of Fig. 5A and B. The names and sizes are indicated above the arrows. The dashed arrows indicate that the corresponding mRNAs were degraded. The specific radiolabeled probes used (probes 1 to 7) are represented by curved lines with an asterisk. (C) Promoter (P) mapping and sequence alignment of the upstream regions of _blpD_St, _blpU_St, _blpE_St, _blpF_St, _blpA_St, _blpG_St, and _blpR_St. The consensus sequence is shown below the alignment: nucleotides present in each of these sequences are in uppercase, and nucleotides occurring in three or four of these sequences are in lowercase. The plain and dashed arrows represent DR (left repeat, LR; right repeat, RR) and IR, respectively. The −35 and −10 boxes are indicated by bold characters. The transcription start site and the Shine-Dalgarno sequence are underlined, and the start codons are boxed. The +1 nucleotides were localized by primer extension analysis with two independent specific primers for each _blp_St operon, except for the _blpABC_St and _blpRH_St operons, for which only one primer gave results. Dashed lines indicate the DR proposed by Blomqvist et al. (7).
Transcriptional organization and analysis of _blp_St promoters.
The transcriptional arrangement of the LMD-9 _blp_St locus was analyzed by Northern blotting experiments (Fig. 4B and 5). The LMD-9 _blp_St locus is organized into six independent transcription units coding specific functions related to bacteriocin production: _blpABC_St, encoding the induction factor and the secretion apparatus; _blpRH_St, encoding the TCS; three operons involved in bacteriocin synthesis and immunity (_blpD_St-_orf2, blpU_St-orf3, and _blpE-F_St); and _blpG-X_St (three transcripts: _blpG-X_St, _blpG_St-?, and _blpG_St-orf8; the question mark indicates an unknown locus), whose function remains unclear. Transcripts containing orf4, orf5, and/or orf6 were not detected (data not shown). Intriguingly, hybridization of _blpRH_St and _blpABC_St mRNAs gave poor results: a smeared signal was detected, indicating that the corresponding mRNAs were degraded (Fig. 5A and B). This instability may have biological relevance since it was observed in four independent experiments.
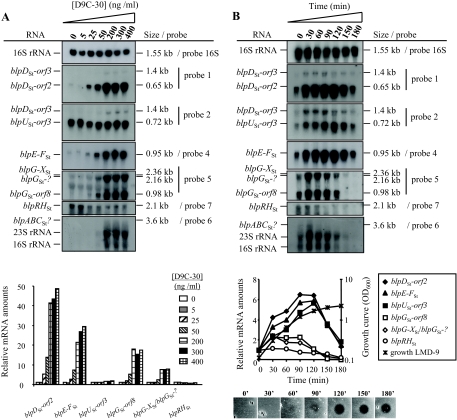
FIG. 5.
Transcriptional regulation of the _blp_St locus. (A) Analysis of the BlpCSt dose-response on transcription of _blp_St genes. The amount of D9C-30 added to LMD-9 cultures (OD600 of 0.1) ranged from 0 to 400 ng/ml. After a 2-h induction, total RNA was extracted, and Northern blotting experiments were performed. Total RNAs were extracted from each sample, and equal amounts were separated on formaldehyde gels and hybridized with radiolabeled probes (Fig. 4B). The same membranes were rehybridized with the different probes. The relative mRNA amounts of the various _blp_St transcripts are shown below the blots and were calculated at each D9C-30 concentration with respect to the RNA amount in noninduced cultures (0 ng/ml D9C-30). The radioactivity levels corresponding to _blpG-X_St and _blpG_St-? transcripts were added together. (B) Time course expression of the various _blp_St transcripts upon D9C-30 induction and correlation with bacteriocin production during growth. Culture samples were collected before the addition of 400 ng/ml D9C-30 in the mid-log growing culture (OD600 of 0.1; time zero), and every 30 min for 180 min after the addition. Northern blotting experiments were performed as for panel A. The relative mRNA amounts (shown below the blots) were calculated from the radioactivity measured in the transcript bands at each time point with respect to that found before the addition of the inducer peptide (time zero). The radioactivity levels from _blpG-X_St and _blpG_St-? transcripts were added together. The induction of bacteriocin production is shown at the bottom of panel B. Five microliters of cell-free supernatants from the same samples used for the RNA extraction was spotted on a lawn of the indicator strain CNRZ1066. For experiments presented in panels A and B, one representative result of two individual experiments performed with different RNAs is shown.
The transcription start site of the main transcripts (mRNAs containing _blpABC_St, _blpRH_St, _blpD_St-_orf2, blpU_St-_orf3, blpE-F_St, and _blpG_St) was mapped by primer extension (Fig. 4C). The promoter regions of the three _bac_St operons (_PblpD_St, _PblpU_St, and _PblpE_St) share 97% identity. The corresponding transcripts display the same G as the transcription start site, which is located 30 bp upstream of the putative ribosome binding site (Fig. 4C). An extended −10 box, TGNTATACT, found in vegetative promoters is conserved upstream of the transcriptional start, and a suboptimal −35 box (CTGATA) could be identified at the noncanonical distance of 16 bp from the putative −10 box. Compared to the other _blp_St operons, the _bac_St transcripts also display a relatively long (44 bp) 5′ untranslated leader region, containing perfect 6-bp inverted repeats (IR) that overlap the transcription start nucleotide. The promoter regions of _blpA_St and _blpG_St share a low degree of identity (50 to 60%) with the corresponding regions of the _bac_St operons. An extended −10 box is also present, but the promoter regions lack a −35 sequence. Also, they do not contain the IR motif present in the _bac_St sequences. The most striking feature common to the _bac_St, _blpA_St, and _blpG_St promoter sequences is the presence of a highly conserved motif consisting of two imperfect direct repeats (DR): ACCATTCGGGACG-7-ACTTTTTGGGACG) (Fig. 4C). This consensus sequence overlaps the DR motif (underlined) of _blpU_St LMG18311, previously identified by Blomqvist et al. (Fig. 4C) (7). The conserved sequence is absent from the _blpR_St upstream region, which consists of a typical vegetative promoter. The DR motif found in all _blp_St promoters except that of _blpRH_St could serve as a putative binding site for BlpRSt, mediating the QS-regulated expression of the _blp_St operons.
BlpCSt-mediated QS regulation of _blp_St genes and bacteriocin production.
To gain insight into the transcriptional response of the _blp_St operons to different inducer concentrations, Northern blotting experiments were performed on total RNA isolated from LMD-9 cultures grown for 2 h in the presence of increasing amounts of D9C-30 (0 to 400 ng/ml). In this range of concentrations, D9C-30 had no effect on S. thermophilus LMD-9 growth (data not shown). The various operons responded differently to increasing inducer concentrations (Fig. 5A). Expression of operons containing _blpD_St-_orf2, blpE-F_St, and _blpG_St displayed a dose response to the amount of D9C-30 but with different strengths (induction factors with 200 ng/ml of D9C-30 were 41 for _blpD_St-orf2, 21 for _blpE-F_St, 18 for _blpG_St-orf8, and 8 for _blpG-X_St-_blpG_St-?) (Fig. 5A). Concentrations higher than 200 ng/ml did not significantly further increase the expression of the above-mentioned transcripts, indicating the saturation of D9C-30 induction. The _blpABC_St transcript was degraded, but the amount of hybridized RNA smears also increased with the D9C-30 concentration (Fig. 5A). In contrast to the other transcripts, _blpU_St-orf3 and _blpRH_St mRNAs showed a very poor response to D9C-30. The amount of _blpRH_St mRNA even decreased slightly in the presence of high D9C-30 concentrations (1.6-fold decrease with 200 ng/ml).
Since bacteriocin production observed in an IF-induced culture of S. thermophilus LMD-9 depends on the growth phase and is more efficient at high cell densities, the temporal expression of the _blp_St transcripts upon addition of D9C-30 (400 ng/ml) was investigated. As shown in Fig. 5B, the _blp_St mRNAs can be divided into three main groups that are differentially regulated by IF during growth: (i) the _bac_St transcripts (_blpD_St-_orf2, blpU_St-orf3, and _blpE-F_St), which are strongly induced (about sixfold) and show a peak of induction in the early stationary phase of growth (90 to 120 min after induction) before a sharp decrease; (ii) the _blpG_St-containing transcripts, which are less induced (twofold) and display a maximum of induction during the log phase (30 min after addition of D9C-30) before a slow decrease starting in early stationary phase; and (iii) the noninduced _blpRH_St mRNA, whose abundance also decreases when cells enter in the stationary phase (90 min after induction). The _blpABC_St transcript was degraded but the time course profile of the amount of hybridized RNA smears was similar to that of the _blpG_St-containing transcripts.
The antimicrobial activity of cell-free supernatants from the same cultures was tested concomitantly during growth (Fig. 5B). It became detectable 1 h after induction and increased progressively during the log phase, with a maximum at the entry of stationary phase (120 min of induction). This induction profile was very similar to that of the _bac_St mRNAs (Fig. 5B), providing further evidence for the involvement of the _bac_St operons in antimicrobial activity. However, the antimicrobial activity remained constant during the stationary phase even after the decrease in the level of the _bac_St mRNAs (data not shown).
Altogether these results show that bacteriocin production in S. thermophilus LMD-9 is regulated at the transcriptional level by the concentration of the induction factor BlpCSt and by the growth phase. The groups of transcripts defined on the basis of the temporal expression profiles correlate remarkably with the presence and the conservation of the putative BlpRSt-binding site (DR motif) in their corresponding promoters. The presence of an auto-induction loop via the induction of the _blpABC_St transcript, as well as the regulation of _blp_St operons and antimicrobial activity by the amount of BlpCSt and by cell density, are typical features of QS-regulated loci.
DISCUSSION
The _blp_St gene cluster of S. thermophilus LMD-9 was characterized in detail. This locus contains all the genetic information required for the production of bacteriocin and is regulated at the transcriptional level by a QS mechanism in which the mature form(s) of the induction factor BlpCSt trigger(s) the expression of the bacteriocin and immunity genes through the BlpHSt-BlpRSt TCS.
The mechanism of regulation by cell density implies that there is a basal level of secretion of IF and that a critical concentration of IF triggers its auto-induction, resulting in the amplification of the response (30). S. thermophilus LMD-9 does not produce bacteriocins in the absence of added IF, probably because the level of secreted BlpCSt is too low under our culture conditions. Interestingly, Northern blotting results suggest an intrinsic instability of the _blpABC_St transcript, which could act as a control to limit the secretion of pheromones, an energy-costly process. The instability of operons encoding the transport machinery has been previously reported for the production of plantaricin E/F (14) and sakacin P (9), which are regulated by a similar pheromone-based signaling pathway. The mechanism responsible for the instability of the _blpABC_St mRNA could be partially explained by the readthrough of these transcripts through the _blpRH_St operon. Indeed, reverse transcription-PCR experiments showed that the _blpABC_St transcript includes the 3′ terminal part of _blpH_St, suggesting that the transcription terminator found between _blpC_St and _blpH_St is leaky (data not shown). Since the two operons are transcribed in opposite directions, this would result in a two-stranded RNA, a structure known to induce RNase III-mediated degradation (6, 32). The slight degradation of _blpRH_St transcripts observed in Northern blotting experiments supports this hypothesis.
All BlpCSt-induced operons were found to contain a conserved imperfect DR motif in their upstream region, suggesting that this sequence could act as a binding site for BlpRSt. DNA motif searches performed in the three available S. thermophilus genomes indicate that this putative regulatory motif is exclusively found in the _blp_St loci. In agreement with this observation, transcriptome analyses suggest that BlpCSt regulates the transcription of only genes located within the _blp_St locus of S. thermophilus LMD-9 (data not shown). In the course of the present study, Blomqvist et al. (7) showed that a reporter fusion between the _blpU_St LMG18311 promoter and gusA was induced 10-fold by the predicted mature form of BlpCSt LMG18311 in S. thermophilus LMG18311. These authors reported that the complete deletion of the DR motif from _blpU_St LMG18311 abolished the BlpCSt-dependent induction of glucuronidase activity, further supporting the hypothesis that the DR motif is the binding site of BlpRSt. Additionally, various mutations in the left repeat, the right repeat, and the spacer of the DR motif affected induction of the promoter by BlpCSt (7). In this context, the observed differences in the strength of the response to BlpCSt among the DR-containing _blp_St operons _blpD_St-_orf2, blpE-F_St, _blpU_St-orf3, and _blpABC_St might result from different affinities of the regulatory protein BlpRSt for the corresponding promoters as a consequence of mutations in the DR motif (Fig. 4C). In contrast to the other _blp_St operons, the TCS-encoding genes seem to be negatively regulated by BlpCSt since the amount of the _blpRH_St mRNAs decreases in the presence of high D9C-30 concentrations (Fig. 5A). A similar observation was recently reported for the IF SilCR concentration and expression of the TCS-encoding operon silAB of the _blp_-like locus (sil locus) of S. pyogenes (18). This peculiar transcriptional response apparently constitutes an atypical regulation mechanism for class II bacteriocin systems since, in most cases, the TCS is induced by its dedicated pheromone, which is encoded on the same transcript (5, 9, 14).
The production of bacteriocins by S. thermophilus LMD-9 is dependent on the growth phase and the concentration of IF, in agreement with the transcriptional regulation of the _bac_St genes. However, while the bacteriocin activity remained constant during the stationary phase, a rapid decrease in the amount of _bac_St mRNAs was observed. This could result from a rapid turnover of mRNAs commonly observed at this stage of growth (4) and/or from a specific regulatory mechanism that prevents an overshooting of bacteriocin production. In the latter case, different levels of control are possible. First, the available pool of BlpRSt could limit the expression of _bac_St transcripts in the late stages of growth. Indeed, the amount of TCS-encoding mRNA does not increase during growth but, instead, decreases constantly during the stationary phase until it becomes undetectable. Alternatively, the IR overlapping the transcription start site of _blpD_St, _blpU_St, and _blpE_St may serve as a binding site for a repressor produced in the early stationary growth phase.
By constructing deletion mutants in the _blp_St locus, we determined that the bacteriocin structural genes and immunity genes are located within the region from _blpD_St to _blpF_St. This region encodes four putative bacteriocin precursors (BlpDSt, BlpUSt, BlpESt, and BlpFSt) with a 2-Gly leader sequence, and each _bac_St gene is cotranscribed with one or two orf gene(s). The predicted mature BacSt peptides and Orf peptides share several characteristics with the class IIb two-component bacteriocins (e.g., ABP-118 [20], thermophilin 13 [37], brochocin C [39], and lactacin F [3]) and their immunity peptides, respectively. A genetic dissection by deletion of the region from _blpD_St to _blpF_St was recently achieved. The analysis of the resulting mutant strains showed that each _bac_St operon contributes to the antimicrobial activity of S. thermophilus LMD-9 (L. Fontaine and P. Hols, unpublished data).
Future work will be dedicated to the study of the activity of BacSt peptides and their corresponding immunity/modification proteins in order to provide further insights into their implication in the intra- and interspecies antimicrobial activity of S. thermophilus.
Supplementary Material
[Supplemental material]
Acknowledgments
This research was carried out with financial support from the Wallon Region (Bioval no. 981/3866 and -3845 and First Europe no. EPH3310300R0082) and FNRS. L.F. and C.B. hold doctoral fellowships from FRIA. P.H. is research associate at FNRS.
We are grateful to E. Maguin and J. Kok for providing the pGhost9 and pMG36e vectors, respectively. We acknowledge P. Renault for the communication of the blp locus sequence in advance of the publication of the genome of Streptococcus salivarius. We thank P. Goffin for critically reading the manuscript. We warmly thank J. Delcour for fruitful discussions and scientific advice.
Footnotes
▿
Published ahead of print on 10 August 2007.
REFERENCES
- 1.Aktypis, A., and G. Kalantzopoulos. 2003. Purification and characterization of thermophilin ST-1, a novel bacteriocin produced by Streptococcus thermophilus ACA-DC 0001. Lait 83**:**365-378. [DOI] [PubMed] [Google Scholar]
- 2.Aktypis, A., G. Kalantzopoulos, J. H. J. Huis in't Veld, and B. ten Brink. 1998. Purification and characterization of thermophilin T, a novel bacteriocin produced by Streptococcus thermophilus ACA-DC 0040. J. Appl. Microbiol. 84**:**568-576. [DOI] [PubMed] [Google Scholar]
- 3.Allison, G. E., C. Fremaux, and T. R. Klaenhammer. 1994. Expansion of bacteriocin activity and host range upon complementation of two peptides encoded within the lactacin F operon. J. Bacteriol. 176**:**2235-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrade, J. M., F. Cairrao, and C. M. Arraiano. 2006. RNase R affects gene expression in stationary phase: regulation of ompA. Mol. Microbiol. 60**:**219-228. [DOI] [PubMed] [Google Scholar]
- 5.Axelsson, L., and A. Holck. 1995. The genes involved in production of and immunity to sakacin A, a bacteriocin from Lactobacillus sake Lb706. J. Bacteriol. 177**:**2125-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomberg, P., E. G. Wagner, and K. Nordstrom. 1990. Control of replication of plasmid R1: the duplex between the antisense RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 9**:**2331-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blomqvist, T., H. Steinmoen, and L. S. Havarstein. 2006. Pheromone-induced expression of recombinant proteins in Streptococcus thermophilus. Arch. Microbiol. 186**:**465-473. [DOI] [PubMed] [Google Scholar]
- 8.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22**:**1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brurberg, M. B., I. F. Nes, and V. G. Eijsink. 1997. Pheromone-induced production of antimicrobial peptides in Lactobacillus. Mol. Microbiol. 26**:**347-360. [DOI] [PubMed] [Google Scholar]
- 10.Dawid, S., A. M. Roche, and J. N. Weiser. 2007. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect. Immun. 75**:**443-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derzelle, S., B. Hallet, K. P. Francis, T. Ferain, J. Delcour, and P. Hols. 2000. Changes in cspL, cspP, and cspC mRNA abundance as a function of cold shock and growth phase in Lactobacillus plantarum. J. Bacteriol. 182**:**5105-5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Saizieu, A., C. Gardes, N. Flint, C. Wagner, M. Kamber, T. J. Mitchell, W. Keck, K. E. Amrein, and R. Lange. 2000. Microarray-based identification of a novel Streptococcus pneumoniae regulon controlled by an autoinduced peptide. J. Bacteriol. 182**:**4696-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diep, D. B., L. S. Havarstein, and I. F. Nes. 1995. A bacteriocin-like peptide induces bacteriocin synthesis in Lactobacillus plantarum C11. Mol. Microbiol. 18**:**631-639. [DOI] [PubMed] [Google Scholar]
- 14.Diep, D. B., L. S. Havarstein, and I. F. Nes. 1996. Characterization of the locus responsible for the bacteriocin production in Lactobacillus plantarum C11. J. Bacteriol. 178**:**4472-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dower, W. J., J. F. Miller, and C. W. Ragsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16**:**6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eijsink, V. G., M. B. Brurberg, P. H. Middelhoven, and I. F. Nes. 1996. Induction of bacteriocin production in Lactobacillus sake by a secreted peptide. J. Bacteriol. 178**:**2232-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ennahar, S., T. Sashihara, K. Sonomoto, and A. Ishizaki. 2000. Class IIa bacteriocins: biosynthesis, structure and activity. FEMS Microbiol. Rev. 24**:**85-106. [DOI] [PubMed] [Google Scholar]
- 18.Eran, Y., Y. Getter, M. Baruch, I. Belotserkovsky, G. Padalon, I. Mishalian, A. Podbielski, B. Kreikemeyer, and E. Hanski. 2007. Transcriptional regulation of the sil locus by the SilCR signalling peptide and its implications on group A streptococcus virulence. Mol. Microbiol. 63**:**1209-1222. [DOI] [PubMed] [Google Scholar]
- 19.Ferain, T., J. N. Hobbs, Jr., J. Richardson, N. Bernard, D. Garmyn, P. Hols, N. E. Allen, and J. Delcour. 1996. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J. Bacteriol. 178**:**5431-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flynn, S., D. van Sinderen, G. M. Thornton, H. Holo, I. F. Nes, and J. K. Collins. 2002. Characterization of the genetic locus responsible for the production of ABP-118, a novel bacteriocin produced by the probiotic bacterium Lactobacillus salivarius subsp. salivarius UCC118. Microbiology 148**:**973-984. [DOI] [PubMed] [Google Scholar]
- 21.Garneau, S., N. I. Martin, and J. C. Vederas. 2002. Two-peptide bacteriocins produced by lactic acid bacteria. Biochimie 84**:**577-592. [DOI] [PubMed] [Google Scholar]
- 22.Gilbreth, S. E., and G. A. Somkuti. 2005. Thermophilin 110: a bacteriocin of Streptococcus thermophilus ST110. Curr. Microbiol. 51**:**175-182. [DOI] [PubMed] [Google Scholar]
- 23.Hale, J. D., Y. T. Ting, R. W. Jack, J. R. Tagg, and N. C. Heng. 2005. Bacteriocin (mutacin) production by Streptococcus mutans genome sequence reference strain UA159: elucidation of the antimicrobial repertoire by genetic dissection. Appl. Environ. Microbiol. 71**:**7613-7617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Héchard, Y., and H. G. Sahl. 2002. Mode of action of modified and unmodified bacteriocins from gram-positive bacteria. Biochimie 84**:**545-557. [DOI] [PubMed] [Google Scholar]
- 25.Hidalgo-Grass, C., M. Ravins, M. Dan-Goor, J. Jaffe, A. E. Moses, and E. Hanski. 2002. A locus of group A Streptococcus involved in invasive disease and DNA transfer. Mol. Microbiol. 46**:**87-99. [DOI] [PubMed] [Google Scholar]
- 26.Hindré, T., S. Didelot, J. P. Le Pennec, D. Haras, A. Dufour, and K. Vallée-Réhel. 2003. Bacteriocin detection from whole bacteria by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 69**:**1051-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hols, P., F. Hancy, L. Fontaine, B. Grossiord, D. Prozzi, N. Leblond-Bourget, B. Decaris, A. Bolotin, C. Delorme, E. S. Dusko, E. Guédon, V. Monnet, P. Renault, and M. Kleerebezem. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29**:**435-463. [DOI] [PubMed] [Google Scholar]
- 28.Hui, F. M., L. Zhou, and D. A. Morrison. 1995. Competence for genetic transformation in Streptococcus pneumoniae: organization of a regulatory locus with homology to two lactococcin A secretion genes. Gene 153**:**25-31. [DOI] [PubMed] [Google Scholar]
- 29.Ivanova, I., V. Miteva, T. Stefanova, A. Pantev, I. Budakov, S. Danova, P. Moncheva, I. Nikolova, X. Dousset, and P. Boyaval. 1998. Characterization of a bacteriocin produced by Streptococcus thermophilus 81. Int. J. Food Microbiol. 42**:**147-158. [DOI] [PubMed] [Google Scholar]
- 30.Kleerebezem, M., and L. E. Quadri. 2001. Peptide pheromone-dependent regulation of antimicrobial peptide production in gram-positive bacteria: a case of multicellular behavior. Peptides 22**:**1579-1596. [DOI] [PubMed] [Google Scholar]
- 31.Kotel'nikova, E. A., and M. S. Gel'fand. 2002. Regulation of transcription in the system of genes responsible for bacteriocins production in Streptococcus equi. Genetika 38**:**911-915. [PubMed] [Google Scholar]
- 32.Lamontagne, B., and S. A. Elela. 2004. Evaluation of the RNA determinants for bacterial and yeast RNase III binding and cleavage. J. Biol. Chem. 279**:**2231-2241. [DOI] [PubMed] [Google Scholar]
- 33.Law, J., G. Buist, A. Haandrikman, J. Kok, G. Venema, and K. Leenhouts. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177**:**7011-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lorquet, F., P. Goffin, L. Muscariello, J. B. Baudry, V. Ladero, M. Sacco, M. Kleerebezem, and P. Hols. 2004. Characterization and functional analysis of the poxB gene, which encodes pyruvate oxidase in Lactobacillus plantarum. J. Bacteriol. 186**:**3749-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maguin, E., P. Duwat, T. Hege, D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174**:**5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178**:**931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marciset, O., M. C. Jeronimus-Stratingh, B. Mollet, and B. Poolman. 1997. Thermophilin 13, a nontypical antilisterial poration complex bacteriocin, that functions without a receptor. J. Biol. Chem. 272**:**14277-14284. [DOI] [PubMed] [Google Scholar]
- 38.Mathot, A. G., E. Beliard, and D. Thuault. 2003. Streptococcus thermophilus 580 produces a bacteriocin potentially suitable for inhibition of Clostridium tyrobutyricum in hard cheese. J. Dairy Sci. 86**:**3068-3074. [DOI] [PubMed] [Google Scholar]
- 39.McCormick, J. K., A. Poon, M. Sailer, Y. Gao, K. L. Roy, L. M. McMullen, J. C. Vederas, M. E. Stiles, and M. J. van Belkum. 1998. Genetic characterization and heterologous expression of brochocin-C, an antibotulinal, two-peptide bacteriocin produced by Brochothrix campestris ATCC 43754. Appl. Environ. Microbiol. 64**:**4757-4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moll, G. N., W. N. Konings, and A. J. Driessen. 1999. Bacteriocins: mechanism of membrane insertion and pore formation. Antonie Leeuwenhoek 76**:**185-198. [PubMed] [Google Scholar]
- 41.Nes, I. F., and H. Holo. 2000. Class II antimicrobial peptides from lactic acid bacteria. Biopolymers 55**:**50-61. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 43.Stoddard, G. W., J. P. Petzel, M. J. van Belkum, J. Kok, and L. L. McKay. 1992. Molecular analyses of the lactococcin A gene cluster from Lactococcus lactis subsp. lactis biovar diacetylactis WM4. Appl. Environ. Microbiol. 58**:**1952-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van de Guchte, M., J. M. van der Vossen, J. Kok, and G. Venema. 1989. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 55**:**224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Ploeg, J. R. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187**:**3980-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughan, A., V. G. Eijsink, and D. van Sinderen. 2003. Functional characterization of a composite bacteriocin locus from malt isolate Lactobacillus sakei 5. Appl. Environ. Microbiol. 69**:**7194-7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villani, F., O. Pepe, G. Mauriello, G. Salzano, G. Moschetti, and S. Coppola. 1995. Antilisterial activity of thermophilin 347, a bacteriocin produced by Streptococcus thermophilus. Int. J. Food Microbiol. 25**:**179-190. [DOI] [PubMed] [Google Scholar]
- 48.Ward, D. J., and G. A. Somkuti. 1995. Characterization of a bacteriocin produced by Streptococcus thermophilus ST134. Appl. Microbiol. Biotechnol. 43**:**330-335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]