Origins and Mechanisms of miRNAs and siRNAs (original) (raw)
. Author manuscript; available in PMC: 2010 Feb 20.
Abstract
Over the last decade, ∼20−30 nucleotide RNA molecules have emerged as critical regulators in the expression and function of eukaryotic genomes. Two primary categories of these small RNAs— short interfering RNAs (siRNAs) and microRNAs (miRNAs)—act in both somatic and germline line-ages in a broad range of eukaryotic species to regulate endogenous genes and to defend the genome from invasive nucleic acids. Recent advances have revealed unexpected diversity in their biogenesis pathways and the regulatory mechanisms that they access. Our understanding of siRNA- and miRNA-based regulation has direct implications for fundamental biology as well as disease etiology and treatment.
In the last decade, few areas of biology have been transformed as thoroughly as RNA molecular biology. This transformation has occurred along many fronts, as detailed in this issue, but one of the most significant advances has been the discovery of small (∼20−30 nucleotide [nt]) noncoding RNAs that regulate genes and genomes. This regulation can occur at some of the most important levels of genome function, including chromatin structure, chromosome segregation, transcription, RNA processing, RNA stability, and translation. The effects of small RNAs on gene expression and control are generally inhibitory, and the corresponding regulatory mechanisms are therefore collectively subsumed under the heading of RNA silencing. The central theme that runs throughout is that the small RNAs serve as specificity factors that direct bound effector proteins to target nucleic acid molecules via base-pairing interactions. Invariably, the core component of the effector machinery is a member of the Argonaute protein superfamily. Because the small RNAs render the silencing machinery addressable in ways that can be predicted and in some cases controlled, the associated pathways have taken on great importance in practical and applied realms.
Although many classes of small RNAs have emerged, various aspects of their origins, structures, associated effector proteins, and biological roles have led to the general recognition of three main categories: short interfering RNAs (siRNAs), microRNAs (miRNAs), and piwi-interacting RNAs (piRNAs). These RNAs are only known to be present in eukaryotes, although the Argonaute proteins that function in eukaryotic silencing can also be found in scattered bacterial and archaeal species. The boundaries between the various small RNA classes are becoming increasingly difficult to discern as described in more detail below, but nonetheless some distinctions persist. siRNAs and miRNAs are the most broadly distributed in both phylogenetic and physiological terms and are characterized by the double-stranded nature of their precursors. In contrast, piRNAs are primarily found in animals, exert their functions most clearly in the germline, and derive from precursors that are poorly understood but appear to be single stranded (see Review by C.D. Malone and G. J. Hannon on page 656 of this issue). Most definitively, piRNAs and si/miRNAs associate with distinct subsets of effector proteins—siRNAs and miRNAs bind to members of the Ago clade of Argonaute proteins, whereas piRNAs bind to members of the Piwi clade.
This review will focus on siRNAs and miRNAs, with an emphasis on their biogenesis and silencing mechanisms. We will focus on developments over the last several years and will rely upon prior reviews to provide the reader with references to earlier discoveries in the field (also see Reviews in this issue by O. Voinnet on page 669, about the biological processes that are under siRNA and miRNA control in plants, and by C.D. Malone and G. J. Hannon on page 656, about piRNAs, Piwi proteins, and their roles in transposon control and genome defense). We will begin with the core aspects of the siRNA and miRNA pathways that are shared by both and then will discuss their unique features in turn.
siRNAs and miRNAs: Themes in Common
The first miRNA, lin-4 from Caenorhabditis elegans, was discovered by Ambros and coworkers in 1993 as an endogenous regulator of genes that control developmental timing (Bartel, 2004). Five years later, Fire, Mello, and colleagues reported that exogenous double-stranded RNA (dsRNA) specifically silences genes through a mechanism called RNA interference (RNAi) (Mello and Conte, 2004). In 1999, silencing in plants was shown to be accompanied by the appearance of ∼20−25 nt RNAs that match the sequence of the silencing trigger (Tomari and Zamore, 2005). Very shortly thereafter, the direct conversion of dsRNAs into ∼21−23 nt siRNAs was documented. In 2001, miRNAs were found to comprise a broad class of small RNA regulators, with at least dozens of representatives in each of several plant and animal species (Bartel, 2004). By this point, the two categories of small RNAs had become firmly embedded in our view of the gene regulatory landscape: miRNAs, as regulators of endogenous genes, and siRNAs, as defenders of genome integrity in response to foreign or invasive nucleic acids such as viruses, transposons, and transgenes. Single-stranded forms of both miRNAs and siRNAs were found to associate with effector assemblies (Meister and Tuschl, 2004) known as RNA-induced silencing complexes (RISCs) (Hammond et al., 2000). Both siRNA- and miRNA-programmed forms of RISC (siRISC and miRISC, respectively) were primarily associated with posttranscriptional modes of regulation, though this apparent limitation would soon be breached, at least in the case of siRNAs (Lippman and Martienssen, 2004).
In all cases, the identities of the genes to be silenced are spec-ified by the small RNA component, which recognizes each target by Watson-Crick base pairing. Accordingly, miRNA and siRNA silencing is readily reprogrammable. When changing circumstances require different expression patterns of endogenous genes, the silencing machinery can be redirected through the expression of new miRNAs and the dilution or removal of old ones. Similarly, when the genome faces new threats from novel invaders, it can exploit the foreign sequences themselves by co-opting them into the siRNA mechanism, thereby suppressing expression from invasive genes and responding adaptively to the threat.
Initially, miRNAs and siRNAs appeared to be distinguished in two primary ways. First, miRNAs were viewed as endogenous and purposefully expressed products of an organism's own genome, whereas siRNAs were thought to be primarily exogenous in origin, derived directly from the virus, transposon, or transgene trigger. Second, miRNAs appeared to be processed from stem-loop precursors with incomplete double-stranded character, whereas siRNAs were found to be excised from long, fully complementary double-stranded RNAs (dsRNAs) (Tomari and Zamore, 2005). Despite these differences, the size similarities and sequence-specific inhibitory functions of miRNAs and siRNAs immediately suggested relatedness in biogenesis and mechanism. Sure enough, both classes of small RNAs were quickly revealed to depend upon the same two families of proteins: Dicer enzymes to excise them from their precursors, and Ago proteins to support their silencing effector functions (Meister and Tuschl, 2004; Tomari and Zamore, 2005) (Figure 1A). Thus, these three sets of macromolecules—Dicers, Agos, and ∼21−23 nt duplex-derived RNAs—became recognized as the signature components of RNA silencing.
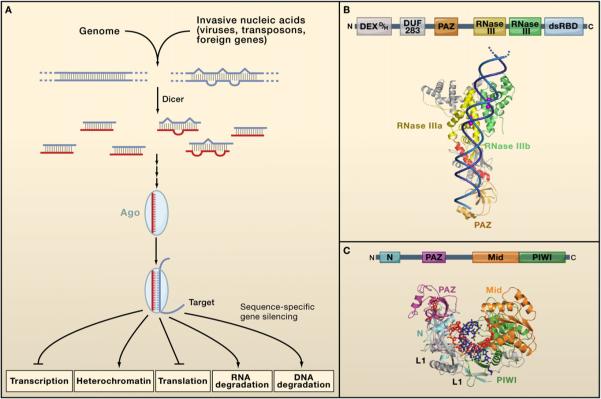
Figure 1. Core Features of miRNA and siRNA Silencing.
(A) Common aspects of all miRNA and siRNA pathways. Double-stranded RNA precursors of various kinds are processed by a Dicer protein into short (∼20−30 nt) fragments. One strand of the processed duplex is loaded into an Argonaute protein, enabling target RNA recognition through Watson-Crick base pairing. Once the target is recognized, its expression is modulated by one of several distinct mechanisms, depending on the biological context.
(B) Dicer proteins cleave dsRNA precursors into characteristic lengths through the action of two RNase III domains. The domain arrangement of most Dicer enzymes is shown at the top. Processing occurs most readily at dsRNA ends, which associate with the PAZ domain present in most Dicer enzymes. The substrate is then positioned within the active sites of the RNase III domains, which cleave the ∼20−30 nt miRNA/siRNA duplex from its precursor. This model is supported by the crystal structure of Giardia Dicer, shown with a dsRNA modeled into the structure (image kindly provided by J. Doudna). In addition to the canonical PAZ and RNase III domains, the structure shows active-site metal ions (purple) and a “ruler” helix (red) that helps to specify the length of the siRNA product.
(C) Argonaute proteins are RNA silencing effectors that are guided to their targets by short single-stranded nucleic acids. The canonical arrangement of Ago domains is given at the top. Below is a crystal structure of the Thermus thermophilus Ago protein, with a bound DNA guide strand base paired to an RNA target. The 5′ end of the guide strand associates with a binding pocket in the Mid domain, and the 3′ end binds the PAZ domain. The target cleavage site is juxtaposed with active-site residues in the PIWI domain, though in this case cleavage is suppressed by mismatches between the guide and the target. (Structure reprinted with permission from Macmillan Publishers Ltd.: Wang et al. [2008]. Nature 456, 921−926.)
Dicer: A Portal into RNA Silencing
Because the double-stranded nature of miRNA and siRNA precursors was evident, and because RNase III enzymes had long been characterized as dsRNA-specific nucleases, enzymes with RNase III domains were promptly recognized as primary candidates in the search for miRNA/siRNA biogenesis factors, and confirmation of this role came quickly (Meister and Tuschl, 2004; Tomari and Zamore, 2005). One class of large RNase III enzymes is characterized by several domains in a specific order from the amino-to-carboxy terminus: a DEXD/H ATPase domain, a DUF283 domain, a PAZ domain, two tandem RNase III domains, and a dsRNA-binding domain (dsRBD) (Figure 1B). Some members of this family differ slightly from this arrangement, for instance in the apparent lack of a functional ATPase domain or PAZ domain or in the presence of anywhere from zero to two C-terminal dsRBDs. C. elegans and D. melanogaster orthologs of these proteins were shown to be required for RNAi and for miRNA biogenesis and function (Meister and Tuschl, 2004; Tomari and Zamore, 2005), and the members of this protein family came to be known as Dicer enzymes. Some organisms including mammals and nematodes have only a single Dicer that does double duty in the biogenesis of both miRNAs and siRNAs, whereas other organisms divide the labor among multiple Dicer proteins. For instance, Drosophila melanogaster expresses two distinct Dicers, and Arabidopsis thaliana produces four. As a general rule, organisms with multiple Dicers exhibit functional specialization between them, as exemplified by the fruit fly: Drosophila Dicer-1 is required for miRNA biogenesis, whereas Dicer-2 is devoted mostly to the siRNA pathway (Tomari and Zamore, 2005).
How do Dicer proteins work in dsRNA processing? Biochemical, genetic, and structural studies have converged on a model in which the PAZ and RNase III domains play central roles in excising siRNAs preferentially from ends of dsRNA molecules (Zhang et al., 2004; Macrae et al., 2006). PAZ domains are shared with Argonaute proteins (see below) and are specialized to bind RNA ends, especially duplex ends with short (∼2 nt) 3′ overhangs. An end engages the Dicer PAZ domain, and the substrate dsRNA then extends approximately two helical turns along the surface of the protein before it reaches a single processing center (Figure 1B). The center resides in a cleft of an intramolecular dimer involving the RNase III domains. Each of the two RNase III active sites cleaves one of the two strands, leading to staggered duplex scission to generate new ends with ∼2 nt 3′ overhangs. The reaction leaves a 5′ monophosphate on the product ends, consistent with a requirement for this group during later stages of silencing (Tomari and Zamore, 2005). This general model pertains equally to pre-miRNA stem-loop substrates and to long, perfectly base-paired dsRNAs. In some species, different functional categories of small RNAs exhibit slightly different lengths, and, not surprisingly, this appears to be dictated by the distance between the PAZ domain and the processing center in the relevant Dicer enzyme (Macrae et al., 2007).
The roles of the ATPase domain have proven enigmatic and probably vary among different forms of Dicer. ATP promotes dsRNA processing by Drosophila Dicer-2 and C. elegans Dcr-1, and mutations predicted to cripple ATPase activity in Drosophila Dicer-2 specifically abolish dsRNA processing (Tomari and Zamore, 2005). In contrast, ATP is dispensable for dsRNA processing by human Dcr (hDcr), and an ATPase-defective mutant exhibits no processing defect (Tomari and Zamore, 2005). Recently, the ATPase domain of hDcr was shown to have an autoinhibitory effect on dsRNA processing by diminishing the enzyme's catalytic efficiency (Ma et al., 2008). The roles of dsRNA processing autoinhibition, the conditions under which its regulatory potential might be used, and the basis for the apparent differences in ATPase domain function among distinct Dicers remain unknown.
Dicers isolated from their natural sources are generally found in a heterodimeric complex with a protein that contains two or three dsRBDs (Tomari and Zamore, 2005). Both hDcr and Drosophila Dcr-2 process dsRNAs effectively in the absence of the heterodimeric partner (TRBP and R2D2, respectively). In at least some cases, the role of Dicer in silencing extends beyond dsRNA processing and into the pathway of RISC assembly, as discussed in more detail below, and this activity is much more dependent on the dsRBD partner protein.
Argonaute: At the Core of RNA Silencing
The Argonaute superfamily can be divided into three separate subgroups: the Piwi clade that binds piRNAs, the Ago clade that associates with miRNAs and siRNAs, and a third clade that has only been described thus far in nematodes (Yigit et al., 2006). All gene-regulatory phenomena involving ∼20−30 nt RNAs are thought to require one or more Argonaute proteins, and these proteins are the central, defining components of the various forms of RISC. The double-stranded products of Dicer enter into a RISC assembly pathway that involves duplex unwinding, culminating in the stable association of only one of the two strands with the Ago effector protein (Meister and Tuschl, 2004; Tomari and Zamore, 2005). This guide strand directs target recognition by Watson-Crick base pairing, whereas the other strand of the original small RNA duplex (the passenger strand) is discarded.
Argonaute proteins are defined by the presence of four domains: the PAZ domain (shared with Dicer enzymes), the PIWI domain that is unique to the Argonaute superfamily, and the N and Mid domains (Figure 1C). Crystallographic studies of bacterial and archaeal Argonaute proteins have greatly illuminated many aspects of Argonaute function (Parker et al., 2004, 2005; Song et al., 2004; Ma et al., 2005; Yuan et al., 2005). These structural studies have recently been extended to Thermus thermophilus Argonaute loaded with a guide nucleic acid (in this case, a short DNA molecule), with and without a base-paired target RNA (Figure 1C) (Wang et al., 2008b, 2008c). The overall protein structure is bilobed, with one lobe consisting of the PAZ domain and the other lobe consisting of the PIWI domain flanked by N-terminal (N) and middle (Mid) domains. The Argonaute PAZ domain has RNA 3′ terminus binding activity, and the co-crystal structures reveal that this function is used in guide strand binding. The other end of the guide strand engages a 5′-phosphate binding pocket in the Mid domain, and the remainder of the guide tracks along a positively charged surface to which each of the domains contributes. The protein-DNA contacts are dominated by sugar-phosphate backbone interactions, as expected for a protein that can accommodate a wide range of guide sequences. Guide strand nucleotides 2−6, which are especially important for target recognition, are stacked with their Watson-Crick faces exposed and available for base pairing.
A critical breakthrough was the demonstration that the PIWI domain adopts an RNase H-like fold that in some cases can catalyze guide strand-dependent endonucleolytic cleavage of a base-paired target (Parker et al., 2004; Song et al., 2004). This initial cut represents the critical first step in a subset of small RNA silencing events that proceed through RNA destabilization. Not all Argonaute proteins have endonucleolytic activity, and those that lack it usually also lack critical active-site residues that coordinate a presumptive catalytic metal ion. The protein structures have been less informative thus far in explaining nonendonucleolytic modes of silencing, which is not surprising given the substantial roster of other factors that are necessary in those cases.
Although some species such as Schizosaccharomyces pombe express only a single Argonaute protein, most contain multiple Argonaute genes. For example, five, eight, and 27 paralogs exist in Drosophila, humans, and C. elegans, respectively. Functional specialization (for instance, between siRNA and miRNA silencing) is very clear in plants, flies, and worms, even among members of the same clade (e.g., Yigit et al., 2006). In humans, four of the eight proteins are from the Ago clade and associate with both siRNAs and miRNAs (Meister and Tuschl, 2004; Tomari and Zamore, 2005), but little difference has been reported thus far in the populations of small RNAs that they bind, so the degree of functional specialization in mammals remains unclear.
siRNAs
Sources of siRNA Precursors
The canonical inducer of RNAi is long, linear, perfectly base-paired dsRNA, introduced directly into the cytoplasm or taken up from the environment (Mello and Conte, 2004). These dsRNAs are processed by Dicer into the siRNAs that direct silencing (Meister and Tuschl, 2004; Tomari and Zamore, 2005). siRNAs were originally observed during transgene- and virus-induced silencing in plants (Mello and Conte, 2004), consistent with a natural role in genome defense (Figure 2). In 2002 and 2003, centromeres, transposons, and other repetitive sequences were uncovered as another wellspring of siRNAs (Lippman and Martienssen, 2004). Shortly thereafter, functional studies in plants led to the discovery of _trans_-acting siRNAs (ta-siRNAs) that are diced from specific genomic transcripts and regulate discrete sets of target genes (Vazquez et al., 2004; Allen et al., 2005). More recently, other sources of endogenous siRNAs (endo-siRNAs) have been identified (Golden et al., 2008, and references therein). These include convergent mRNA transcripts and other natural sense-antisense pairs, duplexes involving pseudogene-derived antisense transcripts and the sense mRNAs from their cognate genes, and hairpin RNAs (hpRNAs). Thus, it has become clear that siRNAs are not solely the products of foreign nucleic acid but arise from endogenous genomic loci as well (Figure 2). As such, they differ from many exogenous siRNAs (exo-siRNAs) in that they or their precursors have an obligate nuclear phase.
Figure 2. A Diversity of siRNA Sources.
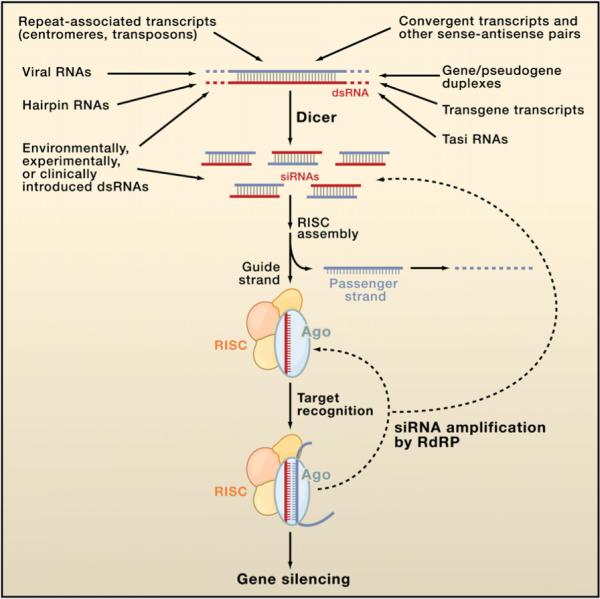
Several different categories of transcripts can adopt dsRNA structures that can be processed by Dicer into siRNAs. These duplexes can be intraor intermolecular, and although most are perfectly base paired, some (e.g., hairpin RNAs and gene/pseudogene duplexes) are not. An siRNA consists of a guide strand (red), which assembles into functional siRISC, and a passenger strand (blue), which is ejected and degraded. All forms of siRISC contain the siRNA bound to an Ago protein, and many if not most forms of siRISC contain additional factors. Target RNAs are then recognized by base pairing, and silencing ensues through one of several mechanisms. In many species, the siRNA populations that engage a target can be amplified by the action of RNA-dependent RNA polymerase (RdRP) enzymes, strengthening and perpetuating the silencing response.
RISC Assembly and siRNA Strand Selection
Although single-stranded siRNAs can load directly into purified Argonaute proteins (Rivas et al., 2005), the double-stranded siRNAs that are generated by Dicer cannot and rely instead upon siRISC assembly pathways (Figure 2). These pathways have been best characterized in Drosophila and in humans. siRISC assembly in Drosophila is nucleated by the R2D2/Dicer-2 heterodimer, which binds an siRNA duplex (Tomari and Zamore, 2005) and then progresses by the addition of unknown factors to form the RISC-loading complex (RLC). The RLC then assembles into pre-RISC, with the siRNA still in duplex form (Kim et al., 2007). pre-RISC formation is the first step that requires Ago2, the Drosophila Ago protein that is primarily dedicated to the siRNA pathway. Ago2 then cleaves the passenger strand (Matranga et al., 2005; Miyoshi et al., 2005; Rand et al., 2005), leading to its ejection and the conversion of the entire assembly into the 80S holo-RISC (Tomari and Zamore, 2005). Smaller forms of cleavage-competent Drosophila siRISC have also been reported (Tomari and Zamore, 2005). RISC assembly in humans has also been characterized biochemically and appears to be a simpler process. Three proteins—Dicer, TRBP, and Ago-2—associate with each other even in the absence of the dsRNA trigger (Gregory et al., 2005; Maniataki and Mourelatos, 2005). This trimer, also referred to as the RISC-loading complex, is capable of binding dsRNA, dicing it into an siRNA, loading the siRNA into Ago-2, and discarding the passenger strand to generate functional RISC (Macrae et al., 2008). Additional proteins associate with Ago complexes from human cells (Meister et al., 2005; Hock et al., 2007; Landthaler et al., 2008), but they do not appear to be essential for RISC loading or target cleavage. Surprisingly, mouse cells carrying a null allele of Dicer can still assemble siRNAs into functional RISCs (Kanellopoulou et al., 2005; Murchison et al., 2005), indicating that Dicer is not required for RISC loading in mammals.
The double-stranded nature of Dicer products contrasts with the single guide strand that eventually finds its way into functional RISC (Figure 2). Strand selection does not require or involve the presence of a cognate mRNA target, as preformed RISC programmed to cleave a heterologous target can do so when the target is added later. In vitro and in vivo experiments revealed that strand selection is dictated by the relative thermodynamic stabilities of the two duplex ends: whichever strand has its 5′ terminus at the less stably base-paired end will be favored as the guide strand (Tomari and Zamore, 2005). This thermodynamic asymmetry is graded rather than all-or-none, and siRNAs with equal base-pairing stabilities at their ends will incorporate either strand into RISC with approximately equal frequency. In Drosophila, the R2D2/Dicer-2 heterodimer appears to sense siRNA asymmetry during the earliest phase of RISC assembly (Tomari and Zamore, 2005). In contrast, Dicer null mouse cells exhibit no obvious deficit in siRNA loading (Kanellopoulou et al., 2005; Murchison et al., 2005), so the mechanism of strand selection is unclear in mammals.
Different categories of siRNAs can depend upon different proteins for their function, indicating that they rely on different biogenesis and RISC assembly pathways. This is particularly true in plants, where viral siRNAs, transgene siRNAs, and tasiRNAs have highly distinct cofactor requirements. This theme has recently been extended to animals with the observation that Drosophila Dicer-2 relies upon different dsRBD proteins for endo- and exo-siRNAs (Golden et al., 2008). Although Dicer-2's partnership with R2D2 has been well established for exosiRNAs (Tomari and Zamore, 2005), R2D2 depletion has little effect on the accumulation and function of most endo-siRNAs. Instead, endo-siRNAs depend upon Loquacious (Loqs), which had previously been characterized as a Dicer-1 partner that functions primarily in the miRNA pathway (Forstemann et al., 2005; Jiang et al., 2005; Saito et al., 2005). The mechanistic basis for the differential Loqs or R2D2 requirement with endo-siRNAs, exo-siRNAs, and miRNAs is not known, but it is one of a growing number of examples of blurred functional boundaries between different categories of small silencing RNAs.
Posttranscriptional Silencing by siRNAs
During the canonical RNAi pathway, the siRNA guide strand directs RISC to perfectly complementary RNA targets, which are then degraded. RNA degradation is induced by the PIWI domain of the Ago protein (Figure 3, lower right). This “slicer” activity is very precise: the phosphodiester linkage between the target nucleotides that are base paired to siRNA residues 10 and 11 (counting from the 5′ end) is cleaved to generate products with 5′-monophosphate and 3′-hydroxyl termini (Tomari and Zamore, 2005). Once this initial cut is made, cellular exonucleases attack the fragments to complete the degradative process (Orban and Izaurralde, 2005). The newly generated 3′ end of RISC cleavage products is also a substrate for oligouridylation, which can promote exonucleolytic targeting (Shen and Goodman, 2004). The target dissociates from the siRNA after cleavage, freeing RISC to cleave additional targets. In some cases, highly purified forms of RISC fail to cleave their targets with multiple turnover (Rivas et al., 2005; Förstemann et al., 2007), suggesting that extrinsic factors promote product release, which is likely to be driven by ATP hydrolysis (Tomari and Zamore, 2005).
Figure 3. Mechanisms of siRNA Silencing.
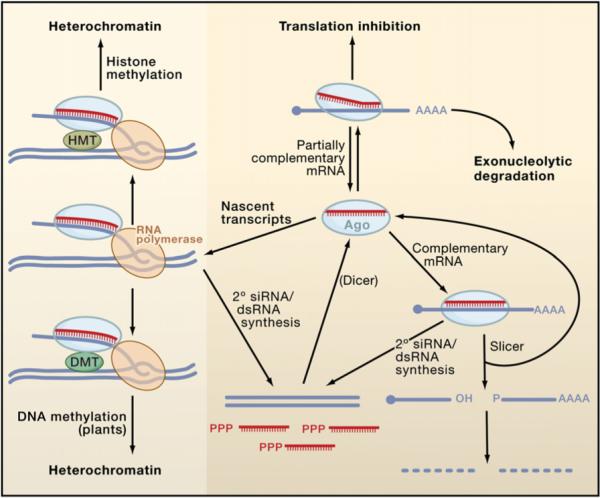
During canonical RNAi (lower right), siRISC recognizes a perfectly complementary mRNA, leading to Ago-catalyzed mRNA cleavage at a single site within the duplex. After cleavage, functional siRISC is regenerated, whereas the mRNA fragments are further degraded. siRNAs are also capable of recognizing targets with imperfect complementarity (upper right). In some cases, they can silence targets by miRNA-like mechanisms involving translational repression and exonucleolytic degradation, though the frequency with which natural siRNAs use these pathways is not clear. Finally, siRISC can direct heterochromatin formation (left) by associating with nascent transcripts and RNA polymerases (RNA Pol II in S. pombe and RNA Pol IV/V in A. thaliana). In plants, target engagement leads to the association or activation of a DNA methyltransferase (DMT) that methylates the DNA (lower left), leading to heterochromatin formation. In S. pombe and probably in animals (upper left), the pathway involves a histone methyltransferase (HMT) that methylates Lys9 of histone H3 (data not shown), thereby inducing heterochromatinization. In most eukaryotes other than insects and mammals, target recognition by siRISC induces the synthesis of secondary dsRNAs and siRNAs by RdRP enzymes (lower middle). The secondary dsRNAs are processed by Dicer into siRNAs, which add to the pool of siRISC. In nematodes, many of the secondary siRNAs arise as single-stranded, unprimed transcripts with 5′-triphosphates and do not require Dicer processing.
Mismatches at or near the center of the siRNA/target duplex suppress endonucleolytic cleavage; furthermore, some siRNA-programmed Ago proteins lack endonuclease activity even with perfectly paired targets (Tomari and Zamore, 2005). Nonetheless, targets that are partially mismatched or are recognized by endonuclease-inactive siRISCs can still be silenced at a post-transcriptional level. In such cases, silencing can involve translational repression or exonucleolytic degradation in a manner similar to miRNA silencing (Figure 3, upper right), as discussed in detail below. In mammalian cells, siRNAs specifically designed to engage targets with imperfect complementarity are virtually indistinguishable from miRNAs in their silencing effects. Silencing of imperfectly matched mRNAs in a miRNA-like fashion appears to account for most “off-target” effects of siRNAs and is therefore of considerable practical importance. The extent to which natural endo-siRNAs silence imperfectly matched targets is not known, but it would be surprising if this regulatory potential were not tapped with some frequency.
Effector phases of posttranscriptional siRNA silencing are thought to occur primarily in the cytoplasm. siRNA binding induces the localization of Ago proteins into subcellular foci called P bodies (Liu et al., 2005b) that are enriched in mRNA degradation factors. However, P body localization does not appear to be strictly required for RNAi (Chu and Rana, 2006), and the nuclear environment is also amenable to RNAi (Robb et al., 2005). The recent application of fluorescence correlation and crosscorrelation spectroscopic approaches has led to the direct observation of a nuclear RISC that is much smaller than its cytoplasmic counterpart (Ohrt et al., 2008). A genetic screen for factors required for nuclear RNAi in C. elegans identified the Ago protein Nrde-3, which was found to reside in the cytoplasm until the induction of nuclear translocation by siRNA binding (Guang et al., 2008). These observations indicate that the localization of RNAi factors is dynamic and that the silencing machinery can be broadly distributed within the confines of the cell.
Priming the Pump: siRNA Amplification
One of the most striking features of RNAi is its potency: only a few molecules of dsRNA per cell can induce a robust response (Mello and Conte, 2004). In some organisms, such as C. elegans, the primary dsRNA trigger induces synthesis of secondary siRNAs (if the target mRNA is present) (Figure 3, bottom) through the action of RNA-dependent RNA polymerase (RdRP) enzymes (Meister and Tuschl, 2004). This secondary pool of siRNAs can greatly amplify and sustain the response, and in some organisms, such as plants and nematodes, they can lead to systemic silencing that spreads throughout the organism. Recognizable RdRP-encoding genes are present in the genomes of many RNAi-competent eukaryotes, with the notable exceptions of insect and vertebrate species. One functional consequence of siRNA amplification by RdRPs is known as transitive RNAi and involves the appearance of siRNAs corresponding to regions of the mRNA that were not targeted by the initial dsRNA trigger. This can in turn lead to the silencing of multiple transcripts, especially if they share a highly conserved sequence or a common exon. The apparent lack of transitive RNAi in vertebrates and insects has a positive effect on its specificity and allows the targeting of individual alternatively spliced mRNA isoforms from a common locus.
Secondary siRNAs have been cataloged most extensively in worms (Pak and Fire, 2007; Sijen et al., 2007), and their characterization led to some surprises. First, nearly all of them correspond to the antisense strand of the mRNA targeted by the primary dsRNA trigger. This would be difficult to rationalize if the secondary siRNAs were generated through a dsRNA intermediate that is processed by Dicer, since thermodynamic siRNA asymmetry would then lead to a mixture of sense and antisense guide strands. Second, they carry di- or triphosphate groups at their 5′ termini, indicating that they are likely to be primary, unprimed RdRP products, again consistent with a lack of processing by Dicer. Thus, it appears that not all siRNAs are directly generated from double-stranded precursors. It is possible that secondary siRNAs bypass the RISC assembly pathway that is followed by primary siRNAs and instead load directly into Ago proteins.
siRNAs Can Induce Heterochromatin Formation
siRNAs are not restricted to posttranscriptional modes of repression. In 2002, siRNAs were shown to induce heterochromatin formation in S. pombe, consistent with earlier reports of transcriptional gene silencing (TGS) in plants (Lippman and Martienssen, 2004). Direct links between RNA silencing and heterochromatin were quickly established in animals, plants, and ciliates as well. The process has been best characterized in S. pombe. The Ago1-containing effector in fission yeast is referred to as the RNA-induced transcriptional silencing (RITS) complex and is guided to specific chromosomal loci such as centromeric repeats by its bound siRNAs (Figure 3, left). Current models involve siRNA recognition of nascent transcripts (Bühler et al., 2006), facilitated by direct interaction between RITS and RNA polymerase II (Djupedal et al., 2005; Kato et al., 2005). RITS association promotes histone H3 methylation on lysine 9 (H3K9) by the histone methyltransferases (HMTs), leading to the recruitment of the chromodomain-containing protein Swi6 and subsequent chromatin compaction (Lippman and Martienssen, 2004)(Figure 3, upper left). Engagement of nascent transcripts by RITS also activates the RNA-dependent RNA polymerase complex (RDRC) that uses its RdRP subunit (Rdp1) to generate secondary siRNAs (Sugiyama et al., 2005) that reinforce and spread silencing (Figure 3, left and bottom). TGS has also been extensively investigated in plants and exhibits many similarities with the process in fission yeast. Key differences also exist, however, prominent among them the direct methylation of DNA by DNA methyltransferases (DMTs) in addition to histone methylation (Lippman and Martienssen, 2004)(Figure 3, left) and the involvement of dedicated DNA-dependent RNA polymerases, RNA Pol IV and Pol V, in the synthesis of siRNA precursors (Herr et al., 2005; Onodera et al., 2005; Wierzbicki et al., 2008). siRNA-directed transcriptional silencing is a fascinating and fast-moving field that has recently been reviewed in depth elsewhere (Grewal and Elgin, 2007; Henderson and Jacobsen, 2007; Moazed, 2009).
MicroRNAs
Although we had learned a great deal about miRNAs by mid-decade (Bartel, 2004; Kim, 2005), the past few years have seen some surprising new discoveries. Far from following a few simple rules of production and action, miRNAs show diverse features that are defying simple classification. Diversification is a key feature of life processes, and miRNAs are no exception.
MicroRNA Biogenesis
MicroRNAs are found in the plant and animal branches of Eukaryota and are encoded by a bewildering array of genes. Transcription of miRNAs is typically performed by RNA polymerase II, and transcripts are capped and polyadenylated (Kim, 2005). Although some animal miRNAs are individually produced from separate transcription units, many more miRNAs are produced from transcription units that make more than one product (Bartel, 2004). A transcript may encode clusters of distinct miRNAs, or it may encode a miRNA and protein. The latter type of transcript is organized such that the miRNA sequence is located within an intron. Many new animal miRNAs are thought to arise from accumulation of nucleotide sequence changes and not from gene duplication (Lu et al., 2008). If the new miRNA sequence should appear within an existing transcription unit, it immediately expresses its new product without invention or duplication of enhancers and promoters. This enables new miRNA genes to more easily appear without gene duplication and may account for the abundance of miRNA genes with multiple products.
The resulting primary or pri-miRNA transcript extends both 5′ and 3′ from the miRNA sequence, and two sequential processing reactions trim the transcript into the mature miRNA (Figure 4). Processing depends on the miRNA sequence folding into a stem-loop structure. A typical animal pri-miRNA consists of an imperfectly paired stem of ∼33 bp, with a terminal loop and flanking segments (Bartel, 2004). The first processing step, which occurs in the nucleus, excises the stem-loop from the remainder of the transcript to create a pre-miRNA product. For most pri-miRNAs, a nuclear member of the RNase III family (Dcl1 in plants and Drosha in animals) carries out this cleavage reaction (Kim, 2005). Although Drosha catalyzes pri-miRNA processing (Lee et al., 2003), it depends upon a protein cofactor for efficient and precise processing. This cofactor contains two dsRBD domains and stably associates with the ribonuclease to form the Microprocessor complex (Denli et al., 2004). However, Microprocessor-mediated cleavage is not the only way to produce pre-miRNAs in animals. An alternative pathway uses splicing of pri-miRNA transcripts to liberate introns that precisely mimic the structural features of pre-miRNAs (Okamura et al., 2007; Ruby et al., 2007). These mirtrons then enter the miRNA processing pathway without the aid of the Microprocessor. Mirtrons are not common, but they are found throughout the animal kingdom.
Figure 4. Biogenesis of miRNAs and Assembly into miRISC in Plants and Animals.
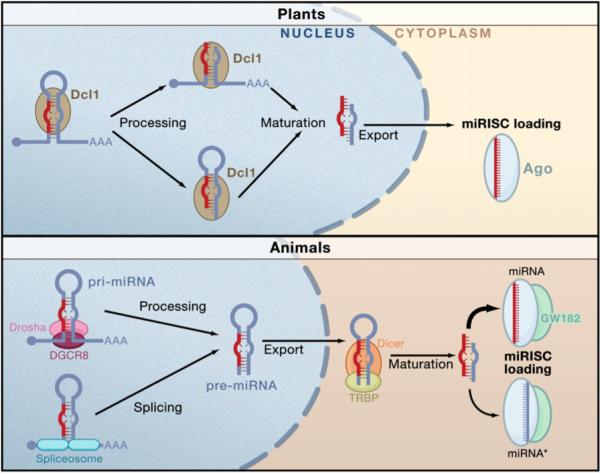
Nuclear transcription leads to capped and polyadenylated pri-miRNAs. In plants, Dcl1 processes the RNA in succession. The order of processing is not certain. The terminal loop may first be excised or it might be the flanking segments that are cleaved first. The second processing step by Dcl1 yields a mature miRNA/miRNA* duplex that becomes methylated and exported from the nucleus. In animals, the pri-miRNA is processed by Drosha with the aid of DGCR8 to generate a pre-miRNA species. This is exported from the nucleus and processed by Dicer to form the mature miRNA/miRNA* duplex. After processing, miRNAs are assembled into miRISC. Only one strand of the duplex is stably associated with an miRISC complex—the miRNA strand is usually more strongly favored than the miRISC* strand, although there are exceptions.
The second processing step excises the terminal loop from the pre-miRNA stem to create a mature miRNA duplex of approximately 22 bp length (Bartel, 2004). In plants, Dcl1 carries out this reaction in the nucleus. In animals, the pre-miRNA is first exported from the nucleus, and the canonical Dicer enzyme carries out the cleavage reaction in the cytoplasm (Kim, 2005).
One of the key differences between miRNAs and most siRNAs is in the precision of their ends. MicroRNAs behave like traditional polymeric products of gene activity, such that most species of a miRNA have highly exact ends, although there is a little variation. In contrast, siRNAs tend to be much more heterogeneous in end composition. It is this feature of miRNAs that has probably allowed them to interact with greater specificity on substrate mRNAs without a need for stringent complementarity or large overlap. Consequently, the processing machinery is constructed to produce miRNA duplexes with highly exact ends. The first cut is most critical. Drosha carries this out with the aid of its dsRBD domain binding partner protein, called DGCR8 in mammals. DGCR8 directly interacts with the pri-miRNA stem and flanking single-stranded segments (Han et al., 2006). Indeed, the flanking segments are critical for processing since the cleavage site is determined by the distance from the stem-flank junction. This distance is precisely one turn of a dsRNA helix (11 bp) and is the minimal processing length for an RNase III enzyme. Although Drosha carries out the cleavage reaction, it is not sufficient to directly bind the pri-miRNA. Instead, Drosha relies upon DGCR8 to serve as a molecular anchor that properly positions Drosha's catalytic site the correct distance from the stem-flank junction. Thus, the endpoint of the stem is a critical determinant for one end of the mature miRNA.
The second cut performed by Dicer defines the other end of the mature miRNA. Dicer will cleave anywhere along a dsRNA molecule, but it has strong preference for the terminus (Kim, 2005; Vermeulen et al., 2005). The PAZ domain of Dicer interacts with the 3′ overhang at the terminus and determines the cleavage site in a ruler-like fashion. The RNase III catalytic sites are positioned two helical turns or 22 bp away from the terminus/PAZ portion of the Dicer-RNA complex.
Interestingly, the Drosha-Dicer double measure is not the only mechanism to produce RNAs with miRNA-like end fidelity. The ACA45 small nucleolar RNA (snoRNA) is a double-hairpin RNA that can be processed by Dicer to generate a 20−22 nt product (Ender et al., 2008). This becomes associated with Ago and exerts miRNA-like repression on an endogenous target gene. It will be interesting to see how many other noncoding RNAs can perform such dual functions.
Regulation of miRNA biogenesis is clearly an important issue but has not been extensively studied. However, an interesting trend has emerged. A surprising number of miRNA genes are formed under the control of the very targets that they regulate. For example, transcription of the Drosophila miR-7 gene is repressed by an ETS domain transcription factor called Yan (Li and Carthew, 2005). However, translation of Yan is repressed by miR-7, thus forming a double-negative feedback loop. Another example of a double-negative feedback loop involves posttranscriptional regulation of miRNA biogenesis. In C. elegans, the let-7 miRNA inhibits translation of Lin28, and let-7 in turn is inhibited by Lin28 protein (Seggerson et al., 2002). Recently, a mechanism for Lin28 action has been described. The Lin28 protein specifically associates with both pri-let-7 and pre-let-7 RNAs (Heo et al., 2008; Newman et al., 2008; Rybak et al., 2008). Sequences in the terminal loop that are unique to let-7 appear to be required for association (Newman et al., 2008). Lin28 is weakly localized in the nucleus and strongly localized in the cytoplasm, and indeed it affects both nuclear and cytoplasmic processing of let-7. Drosha-mediated cleavage is inhibited by Lin28 (Heo et al., 2008; Newman et al., 2008; Viswanathan et al., 2008), as is Dicer-mediated cleavage (Heo et al., 2008; Rybak et al., 2008). In addition, Lin28 promotes polyuridylation of the 3′ terminus of pre-let-7 (Heo et al., 2008). Polyuridylated pre-let-7 is highly resistant to cleavage by Dicer, suggesting a possible mechanism by which Lin28 inhibits processing. Alternatively, polyuridylation might occur on lingering pre-let-7 RNAs that are unable to be cleaved by Dicer because of Lin28.
A rationale behind these double-negative regulatory relationships is that tight regulation of miRNA biogenesis is crucial. Misexpression of miRNAs frequently mimics loss-of-function phenotypes for their targets. This would be prevented if biogenesis of an miRNA is strictly controlled by its targets. The restriction would also explain how off-targeting effects by wayward miRNAs are carefully limited.
MicroRNA Associations
The mature miRNA duplex is a short-lived entity; it is rapidly unwound when it associates with an Ago protein. As with siRNAs, miRNA unwinding is accompanied by differential strand retention; one strand is retained while the other strand is lost. As with siRNAs, strand retention is based on the relative thermodynamic stability of the duplex's ends. The 5′ terminus of the retained strand is at the less stably base-paired end of the duplex (Kim, 2005). However, this rule is not absolute. The other strand is appreciably detected in Ago complexes, lending ambiguity to the notion of strand asymmetry (Okamura et al., 2008). Although either strand can become stably associated with Ago proteins, the more commonly associated strand is called the miRNA strand; the other strand is called the miRNA* strand. miRNA unwinding is still a mysterious process that is not accompanied by cleavage of the ejected strand by the associated Ago, in contrast to the siRNA unwinding mechanism. This may be because of the multiple bulges and mismatches typical of miRNA duplexes, which presumably block cleavage by Ago (Matranga et al., 2005).
One reason unwinding occurs so rapidly after duplex formation is that the two processes are physically coupled due to Ago2's presence in a complex with Dicer and TRBP. Such a trimeric complex is capable of generating mature duplexes from pre-miRNAs and transferring one strand to Ago2 (Gregory et al., 2005; Maniataki and Mourelatos, 2005; Macrae et al., 2008). In Drosophila, the situation is more complicated, owing to the partitioning of siRNAs and miRNAs into different Ago complexes. As in mammals, Ago loading is highly dependent upon Dicer and its partner. However, partitioning is not absolute since small RNA processing is not physically coupled to an Ago. Processing of pre-miRNAs is performed by Dicer-1 and its partner Loquacious, whereas Dicer-2 and its partner R2D2 generate siRNAs. Although Dicer-2/R2D2 load siRNAs with their cognate Ago protein (Ago2), they do so without stably associating with Ago2 (Tomari and Zamore, 2005). Dicer-2/R2D2 tend not to bind miRNA duplexes and load them into Ago2 because of the central mismatches often found in miRNAs (Tomari et al., 2007). Instead, an independent mechanism loads miRNAs with their cognate Ago protein, Ago1.
The mammalian Dicer/Ago/miRNA complex is associated with other proteins. Gemin3, Gemin4, Mov10, and Imp8 associate with Ago2 (Bartel, 2004; Meister et al., 2005; Weinmann et al., 2009). The mammalian protein GW182 also associates with Ago2 (Liu et al., 2005a; Meister et al., 2005; Till et al., 2007). In Drosophila, the ortholog of GW182 associates with the miRNA effector Ago1 (Behm-Ansmant et al., 2006). Functional analysis in humans, C. elegans, and Drosophila indicates that GW182 is both necessary and sufficient for miRNA-bound Ago to silence gene expression (Jakymiw et al., 2005; Liu et al., 2005a; Eulalio et al., 2008; Ding and Grosshans, 2009). Thus, miRNA-bound Ago in association with GW182 can be thought of as the miRISC complex.
Posttranscriptional Repression by miRNAs
The miRNA acts as an adaptor for miRISC to specifically recognize and regulate particular mRNAs. If miRISC is tethered to a heterologous RNA recognition factor, the factor enables miRISC to recognize and repress mRNAs that lack miRNA-binding sites (Pillai et al., 2004). With few exceptions, miRNA-binding sites in animal mRNAs lie in the 3′ UTR and are usually present in multiple copies. Most animal miRNAs bind with mismatches and bulges, although a key feature of recognition involves Watson-Crick base pairing of miRNA nucleotides 2−8, representing the seed region. In contrast, most plant miRNAs bind with near-perfect complementarity to sites within the coding sequence of their targets.
The degree of miRNA-mRNA complementarity has been considered a key determinant of the regulatory mechanism. Perfect complementarity allows Ago-catalyzed cleavage of the mRNA strand, whereas central mismatches exclude cleavage and promote repression of mRNA translation. It has been thought that perfect complementarity excludes translational repression because it enables cleavage, and it has contributed to the notion that plant and animal miRNAs act in fundamentally different ways. However, genetic analysis has recently upset the applecart. In Arabidopsis, two mutants have normal miRNA-directed cleavage but exhibit a defect in miRNA-directed translation repression (Brodersen et al., 2008). The defective translational repression is widespread and irrespective of the degree of complementarity or location of target sites within mRNAs. This suggests that translational repression is the default mechanism by which miRNAs repress gene expression, both in animals and plants. Perfectly complementary miRNAs may additionally engage in mRNA cleavage such that their effects are a result of both mechanisms.
The mechanisms by which miRISC regulates translation have been subject to ongoing debate. The fundamental issue of whether repression occurs at translation initiation or postinitiation has not yet been resolved. Initiation begins with the recognition of the mRNA 5′ terminal cap by eIF4E, a subunit of the eIF4F complex. This complex also contains eIF4A and eIF4G. Interaction of eIF4G with another initiation factor, eIF3, recruits the 40S ribosomal subunit to the 5′ end. The 40S preinitiation complex joins with the 60S ribosomal subunit at the AUG codon to begin elongation. eIF4G also interacts with the polyA-binding protein PABP1 that decorates the 3′ end of the message. The ability of eIF4G to interact simultaneously with eIF4E and PABP1 effectively circularizes the mRNA molecule. This circularization greatly enhances translation efficiency, especially under circumstances where mRNAs are abundant. Some viral mRNAs initiate translation without any initiation factors or they require only a subset of initiation factors due to the presence of an internal ribosome entry site (IRES).
The issue of initiation versus postinitiation responsiveness to miRISC has been judged in two ways. One criterion has been whether density gradient centrifugation shows repressed mRNAs located in the free mRNP pool (repressed initiation) or in large polysomes (repressed elongation or termination). A second criterion has been whether repressed mRNAs that contain an IRES are resistant to repression. On the basis of these criteria, some studies have found evidence for miRISC-repressed initiation (Humphreys et al., 2005; Pillai et al., 2005; Kiriakidou et al., 2007; Mathonnet et al., 2007; Wakiyama et al., 2007; Ding and Grosshans, 2009). Other studies have found evidence for repression of postinitiation processes (Seggerson et al., 2002; Maroney et al., 2006; Nottrott et al., 2006; Petersen et al., 2006). There are no simple technical or experimental explanations to account for the discrepancy. Rather, the inconsistencies suggest that repression may be exerted either at the initiation step or at some subsequent stage. Divergent results might be explained by different steps limiting overall translation in the various experimental systems.
Petersen et al. (2006) suggest a possible means by which miRISC represses elongation. A repressed mRNA can be associated with polysomes, but when translation initiation is rapidly blocked with hippuristanol, then ribosomes rapidly dissociate in a miRNA-dependent manner. Their results suggest that miRISC promotes premature ribosome dissociation from mRNAs (Figure 5).
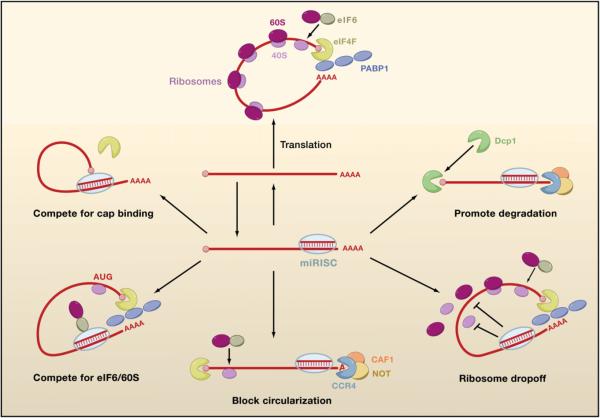
Figure 5. Possible Mechanisms of miRISC-Mediated Repression.
Nonrepressed mRNAs recruit initiation factors and ribosomal subunits and form circularized structures that enhance translation (top). When miRISCs bind to mRNAs, they can repress initiation at the cap recognition stage (upper left) or the 60S recruitment stage (lower left). Alternatively, they can induce deadenylation of the mRNA and thereby inhibit circularization of the mRNA (bottom). They can also repress a postinitiation stage of translation by inducing ribosomes to drop off prematurely (lower right). Finally, they can promote mRNA degradation by inducing deadenylation followed by decapping.
Cell-free systems have been crucial in providing insights into how miRISC represses initiation, but even these studies are controversial. Currently, there are three competing models for how miRISC represses initiation, and each is fundamentally different from the others (Figure 5). One model proposes that there is competition between miRISC and eIF4E for binding to the mRNA 5′ cap structure. eIF4E binds to the cap in part by stacking the methylated base of the cap between two tryptophan residues. If miRISC competes with eIF4E, then one prediction is that providing excess eIF4F complex (containing eIF4E) would alleviate repression. This indeed is the case when purified eIF4F is added to an ascites cell-free system (Mathonnet et al., 2007). Additional support comes from Drosophila embryo lysates, where miRISC inhibits loading of the 40S preinitiation complex onto mRNA (Thermann and Hentze, 2007). One possible means by which miRISC competes with eIF4E has been proposed. The Mid domain of human Ago2 has been proposed to resemble eIF4E, with two phenylalanine residues in the Mid sequence adopting equivalent positions to the eIF4E tryptophans (Kiriakidou et al., 2007). Mutation of the phenylalanines impairs the ability of Ago2 to repress translation and bind to m7GTP-coupled beads in vitro. Hence, it was speculated that Ago2 itself competes with eIF4E for cap binding.
This proposal has been challenged by other studies in Drosophila. Human Ago2 is not the only Ago protein to contain these important residues; indeed, they are found in Ago family members throughout the animal kingdom. The Drosophila Ago1 Mid domain contains the phenylalanines, and mutagenesis abolishes the ability of Ago1 to repress translation (Eulalio et al., 2008). However, mutagenesis does not affect Ago1 binding to m7GTP-coupled beads in vitro, and instead the mutant Ago1 is impaired for binding to GW182. Moreover, GW182 bound to mRNAs is sufficient to repress their translation without Ago1, arguing against an obligatory Ago1-cap binding mechanism. This data suggests that GW182 or a downstream factor could be the eIF4E competitor.
A second model has proposed that miRISC stimulates dead-enylation of the mRNA tail. In this model, translation is repressed because the cap and PABP1-free tail of the deadenylated mRNA are unable to circularize. In support of the model, many repressed mRNAs are deadenylated by miRNAs in vivo (Behm-Ansmant et al., 2006; Giraldez et al., 2006; Wu et al., 2006) and in vitro (Wakiyama et al., 2007). Deadenylation is not simply a consequence of impaired initiation because IRES-containing mRNAs are also deadenylated even though they are resistant to translational repression by miRISC (Wakiyama et al., 2007). Instead, deadenylation is promoted by GW182, which triggers deadenylation and translation repression in Drosophila cells (Behm-Ansmant et al., 2006).
Again, this model suffers from contradictory evidence. Nonpolyadenylated mRNAs can be translationally silenced by miRNAs (Pillai et al., 2005; Wu et al., 2006; Eulalio et al., 2008). This effect requires Ago and GW182 since depletion of both factors or expression of a dominant-negative GW182 abrogates the effect (Eulalio et al., 2008). Moreover, depletion of a component of the deadenylase enzyme has little effect on translation repression by GW182.
A third model has proposed that miRISC blocks association of the 60S ribosomal subunit with the 40S preinitiation complex. Human Ago2 physically associates with eIF6 and 60S ribosomal subunits in vitro (Chendrimada et al., 2007). eIF6 is involved in the biogenesis and maturation of 60S ribosomal subunits and prevents their premature association with 40S subunits. Depletion of eIF6 in either human cells or C. elegans rescues mRNAs from miRNA inhibition. In contrast, depletion of eIF6 in Drosophila cells has little or no effect on silencing (Eulalio et al., 2008). A reticulocyte cell-free system provides additional evidence for the model (Wang et al., 2008a). Targeted mRNAs become enriched for 40S but not 60S ribosomal subunits after addition to miRNA-programmed lysate. A toeprint of these mRNAs shows relative protection over the initiating codon, consistent with 40S ribosomal subunits paused at the start codon. Thus, the recruitment of eIF6 by miRISC may repress translation by preventing the assembly of translationally competent ribosomes at the start codon.
Degradation of mRNAs by miRNAs
Early studies of animal miRNAs indicated that translational repression is not accompanied by mRNA destabilization. However, for some miRNA-target interactions, there is a signifi-cant reduction in mRNA abundance due to an increase in mRNA degradation (Bagga et al., 2005; Lim et al., 2005; Behm-Ansmant et al., 2006; Giraldez et al., 2006; Wu et al., 2006). This increased degradation is not because of Ago-catalyzed mRNA cleavage but rather because of deadenylation, decapping, and exonucleolytic digestion of the mRNA (Behm-Ansmant et al., 2006; Giraldez et al., 2006; Wu et al., 2006). It requires Ago, GW182, and the cellular decapping and deadenylation machinery (Behm-Ansmant et al., 2006). A critical question is whether degradation is a consequence of a primary effect on translation. Some evidence suggests that degradation can be uncoupled from translation. Messages whose translation is prevented are nevertheless deadenylated in an miRNA-dependent manner (Wu et al., 2006; Wakiyama et al., 2007). Furthermore, miRNA-mediated mRNA degradation can occur in vitro without active translation (Wakiyama et al., 2007). This suggests that degradation might be an independent mechanism of repression for some targets. At present it is unclear why some targets are degraded and others are not. It has been suggested that the number, type, and position of mismatches in the miRNA/mRNA duplex play an important role in triggering degradation or translation arrest (Aleman et al., 2007).
Blind Men and the Elephant?
It is difficult to reconcile these diverse accounts of the miRNA mechanism with each other. Although it is formally possible that different experimental approaches are responsible, other explanations seem more likely. One explanation is that silencing proceeds through a single unifying mechanism and that the accounted differences reflect secondary events downstream. Another explanation is that multiple mechanisms exist, and particular mechanisms take pre-eminence according to conditions that we do not yet understand. In this way, we might be in a present situation analogous to the fable of the blind men describing the elephant. We do not yet see the trick that makes the phenomenon whole and unified.
In this regard, it is important to consider this phenomenon in the context of the whole cell and the myriad activities that affect posttranscriptional gene regulation. One of these is the cell cycle. The miRNA let-7 and an artificial miRNA (CXCR4) repress translation in proliferating human cells but change into translational activators when the cell cycle is arrested at the G1 checkpoint by serum starvation (Vasudevan et al., 2007). The mode of regulation (repression versus activation) is dependent on the stage of the cell cycle and not whether the cells are arrested or free cycling. Aphidicolin-induced arrest at G1 also generates translational activation, whereas nocodazole-induced arrest at G2/M generates translational repression. This mode of regulation is not universal since G1-arrested cells in the Drosophila eye use miRNA-mediated repression (Li and Carthew, 2005). Nevertheless, it appears to have a physiological role in regulating expression of the cytokine TNFα. Lymphocyte growth arrest induces TNFα expression that is required for macrophage maturation; miR-369−3p switches from a repressor to an activator of TNFα translation when cells in culture are growth arrested (Vasudevan et al., 2007).
Another example of miRNA-mediated activation highlights the possible importance of binding site position. Interaction of miR-10a with the 5′ UTR of certain ribosomal subunit mRNAs leads to their activated translation, whereas interaction with the 3′ UTR leads to repression (Orom et al., 2008). Although this might suggest a UTR rule, other studies show that miRNA interaction with the 5′ UTR can repress a postinitiation step of translation (Lytle et al., 2007).
Other Cell Contexts
How small RNA regulation is organized and modulated within the cell is an important issue. Ago proteins are frequently associated with membrane trafficking compartments such as the Golgi and ER (Cikaluk et al., 1999). Cytosolic extraction experiments support this notion (Pillai et al., 2005). In this regard, it is worth considering that miRISC factors might become anchored in certain subcellular compartments. Among these compartments are P bodies or GW bodies, discrete cytoplasmic foci in which translationally inactive mRNAs often accumulate. Subunits of miRISC (miRNAs, Ago, and GW182) and their repressed targets are also enriched in GW bodies (Jakymiw et al., 2005; Liu et al., 2005b; Pillai et al., 2005). GW bodies per se are not essential for miRNA repression; depletion of certain GW body proteins results in dispersal of the GW bodies with no effect on miRNA activities (Eulalio et al., 2007). Rather, GW body formation requires an intact miRNA pathway. These observations imply that silencing is initiated elsewhere in the cytoplasm, and the localization into GW bodies is a consequence of silencing.
Photobleaching experiments indicate that Ago proteins in GW bodies exchange very slowly with other Ago molecules (Leung et al., 2006). Are these bodies or other locales graveyards for miRISC machinery bound to their targets? There might be ways in which miRISC is recycled to allow engagement with more targets. Interestingly, the importin protein Imp8 stimulates miRISC-mediated silencing by enhancing association of Ago2 complexes to target mRNAs (Weinmann et al., 2009).
Conclusions
It is an exciting time in RNAi research. We are witnessing a description of the chemical mechanism that just a few years ago was unimagined. We are rethinking our views as to what constitutes an siRNA or miRNA. The rules about biogenesis and action are much more fluid than we thought. Perhaps the most unwavering distinction between miRNAs and siRNAs has been whether they silence their own expression. Almost all siRNAs, whether endo-siRNAs or virus siRNAs, silence the same locus from which they were derived. They sometimes have the ability to silence other loci as well. But their capacity to strike at their encoding DNA (in the case of some plant endosiRNAs) and RNA (in the case of virus or animal endo-siRNAs) makes their production a continual struggle against themselves. This of course is perfectly suited to their roles in defense. It also means that they are highly adaptable, allowing for little or no selective pressure to maintain their sequence conservation. In contrast, most miRNAs do not silence their own loci but silence other genes. This is attributable to the nuclear processing of miRNA precursor RNAs coupled with nuclear exclusion of mature effector miRNAs. Even this rule, however, is not absolute; human miR-29b is a nuclear miRNA (Hwang et al., 2007), and Imp8 stimulates nuclear localization of Ago2 in cells (Weinmann et al., 2009). Nevertheless, the overwhelming number of miRNAs that exert strictly heterotypic silencing are more severely constrained in the precision of their sequence structure because of the necessary coupling of their sequence to heterologous targets. This feature makes miRNAs less adaptable for functions such as biological defense.
The leap of small RNAs from laboratory into the marketplace is ongoing, and much of what has been learned in the past few years is acting as a springboard (see Analysis by L. Bonetta in this issue of Cell). Biotechnology will have to contend with issues such as specificity that directly relate to mechanistic studies pursued in the lab. Other hurdles such as delivery remain, and one hopes that in the next few years as we learn more about the cell biology of RNAi, these and other issues will be addressed as rapidly as we have witnessed to date.
ACKNOWLEDGMENTS
We are grateful to members of our laboratories for stimulating discussions. This work was supported by National Institutes of Health grants GM077581 and GM068743 to R.W.C. and GM072830 to E.J.S.
REFERENCES
- Aleman LM, Doench J, Sharp PA. Comparison of siRNA-induced off-target RNA and protein effects. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen P, Sakvarelidze-Achard L, Bruun-Rasmussen M, Dunoyer P, Yamamoto YY, Sieburth L, Voinnet O. Widespread translational inhibition by plant miRNAs and siRNAs. Science. 2008;320:1185–1190. doi: 10.1126/science.1159151. [DOI] [PubMed] [Google Scholar]
- Bühler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Finn KJ, Ji X, Baillat D, Gregory RI, Liebhaber SA, Pasquinelli AE, Shiekhattar R. MicroRNA silencing through RISC recruitment of eIF6. Nature. 2007;447:823–828. doi: 10.1038/nature05841. [DOI] [PubMed] [Google Scholar]
- Chu CY, Rana TmM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cikaluk DE, Tahbaz N, Hendricks LC, DiMattia GE, Hansen D, Pilgrim D, Hobman TC. GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol. Biol. Cell. 1999;10:3357–3372. doi: 10.1091/mbc.10.10.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- Ding XC, Grosshans H. Repression of C. elegans microRNA targets at the initiation level of translation requires GW182 proteins. EMBO J. 2009 doi: 10.1038/emboj.2008.275. Published online January 8, 2009. 10.1038/emboj.2008.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djupedal I, Portoso M, Spåhr H, Bonilla C, Gustafsson CM, Allshire R, Ekwall K. RNA Pol II subunit Rpb7 promotes centromeric transcription and RNAi-directed chromatin silencing. Genes Dev. 2005;19:2301–2306. doi: 10.1101/gad.344205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with micro-RNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat. Struct. Mol. Biol. 2008;15:346–353. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- Förstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct Argonaute protein complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Tomari Y, Du T, Vagin VV, Denli AM, Bratu DP, Klattenhoff C, Theurkauf WE, Zamore PD. Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol. 2005;3:e236. doi: 10.1371/journal.pbio.0030236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Golden DE, Gerbasi VR, Sontheimer EJ. An inside job for siRNAs. Mol. Cell. 2008;31:309–312. doi: 10.1016/j.molcel.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123:631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guang S, Bochner AF, Pavelec DM, Burkhart KB, Harding S, Lachowiec J, Kennedy S. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- Hock J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys DT, Westman BJ, Martin DI, Preiss T. Micro-RNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. USA. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- Jakymiw A, Lian S, Eystathioy T, Li S, Satoh M, Hamel JC, Fritzler MJ, Chan EK. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005;7:1267–1274. doi: 10.1038/ncb1334. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Goto DB, Martienssen RA, Urano T, Furukawa K, Murakami Y. RNA polymerase II is required for RNAi-dependent heterochromatin assembly. Science. 2005;309:467–469. doi: 10.1126/science.1114955. [DOI] [PubMed] [Google Scholar]
- Kim K, Lee YS, Carthew RW. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA. 2007;13:22–29. doi: 10.1261/rna.283207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M, Tan GS, Lamprinaki S, De Planell-Saguer M, Nelson PT, Mourelatos Z. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–1151. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Landthaler M, Gaidatzis D, Rothballer A, Chen P, Soll SJ, Dinic L, Ojo T, Hafner M, Zavolan M, Tuschl T. Molecular characterization of human Argonaute-containing ribonucleoprotein complexes and their bound target mRNAs. RNA. 2008;14:2580–2596. doi: 10.1261/rna.1351608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl. Acad. Sci. USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Carthew RW. A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell. 2005;123:1267–1277. doi: 10.1016/j.cell.2005.10.040. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- Liu J, Rivas FV, Wohlschlegel J, Yates JR, 3rd, Parker R, Hannon GJ. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005a;7:1261–1266. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. Micro-RNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005b;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Shen Y, Wu Q, Kumar S, He B, Shi S, Carthew RW, Wang SM, Wu CI. The birth and death of microRNA genes in Drosophila. Nat. Genet. 2008;40:351–355. doi: 10.1038/ng.73. [DOI] [PubMed] [Google Scholar]
- Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc. Natl. Acad. Sci. USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E, Macrae I, Kirsch JF, Doudna J. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- Macrae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc. Natl. Acad. Sci. USA. 2008;105:512–517. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that micro-RNAs are associated with translating messenger RNAs in human cells. Nat. Struct. Mol. Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Mathonnet G, Fabian MR, Svitkin YV, Parsyan A, Huck L, Murata T, Biffo S, Merrick WC, Darzynkiewicz E, Pillai RS, et al. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science. 2007;317:1764–1767. doi: 10.1126/science.1146067. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel D, Zamore P. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Mello CC, Conte D., Jr. Revealing the world of RNA interference. Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon G. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat. Struct. Mol. Biol. 2006;13:1108–1114. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- Ohrt T, Mütze J, Staroske W, Weinmann L, Höck J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat. Struct. Mol. Biol. 2008;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onodera Y, Haag JR, Ream T, Nunes PC, Pontes O, Pikaard CS. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005;120:613–622. doi: 10.1016/j.cell.2005.02.007. [DOI] [PubMed] [Google Scholar]
- Orban TI, Izaurralde E. Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome. RNA. 2005;11:459–469. doi: 10.1261/rna.7231505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and enhances their translation. Mol. Cell. 2008;30:460–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315:241–244. doi: 10.1126/science.1132839. [DOI] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. EMBO J. 2004;23:4727–4737. doi: 10.1038/sj.emboj.7600488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pillai RS, Artus CG, Filipowicz W. Tethering of human Ago proteins to mRNA mimics the miRNA-mediated repression of protein synthesis. RNA. 2004;10:1518–1525. doi: 10.1261/rna.7131604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rivas FV, Tolia NH, Song JJ, Aragon JP, Liu J, Hannon GJ, Joshua-Tor L. Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 2005;12:340–349. doi: 10.1038/nsmb918. [DOI] [PubMed] [Google Scholar]
- Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak A, Fuchs H, Smirnova L, Brandt C, Pohl EE, Nitsch R, Wulczyn FG. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- Saito K, Ishizuka A, Siomi H, Siomi M. Processing of premicroRNAs by the Dicer-1-Loquacious complex in Drosophila cells. PLoS Biol. 2005;3:e235. doi: 10.1371/journal.pbio.0030235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seggerson K, Tang L, Moss EG. Two genetic circuits repress the Caenorhabditis elegans heterochronic gene lin-28 after translation initiation. Dev. Biol. 2002;243:215–225. doi: 10.1006/dbio.2001.0563. [DOI] [PubMed] [Google Scholar]
- Shen B, Goodman HM. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Sijen T, Steiner FA, Thijssen KL, Plasterk RH. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007;315:244–247. doi: 10.1126/science.1136699. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon G, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Cam H, Verdel A, Moazed D, Grewal S. RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production. Proc. Natl. Acad. Sci. USA. 2005;102:152–157. doi: 10.1073/pnas.0407641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–878. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- Till S, Lejeune E, Thermann R, Bortfeld M, Hothorn M, Enderle D, Heinrich C, Hentze MW, Ladurner AG. A conserved motif in Argonaute-interacting proteins mediates functional interactions through the Argonaute PIWI domain. Nat. Struct. Mol. Biol. 2007;14:897–903. doi: 10.1038/nsmb1302. [DOI] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Zamore PD. Perspective: machines for RNAi. Genes Dev. 2005;19:517–529. doi: 10.1101/gad.1284105. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert J-L, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005;11:674–682. doi: 10.1261/rna.7272305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–1862. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yanez A, Novina CD. MicroRNA-repressed mRNAs contain 40S but not 60S components. Proc. Natl. Acad. Sci. USA. 2008a;105:5343–5348. doi: 10.1073/pnas.0801102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Tuschl T, Patel DJ. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008b;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand-containing argonaute silencing complex. Nature. 2008c;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinmann L, Höck J, Ivacevic T, Ohrt T, Mütze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets Argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AT, Haag JR, Pikaard C. Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell. 2008;135:635–648. doi: 10.1016/j.cell.2008.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yigit E, Batista PJ, Bei Y, Pang KM, Chen CC, Tolia NH, Joshua-Tor L, Mitani S, Simard MJ, Mello CC. Analysis of the C. elegans Argonaute family reveals that distinct Argonautes act sequentially during RNAi. Cell. 2006;127:747–757. doi: 10.1016/j.cell.2006.09.033. [DOI] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma J, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, Patel D. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kolb F, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]