Intermediate Neuronal Progenitors (Basal Progenitors) Produce Pyramidal–Projection Neurons for All Layers of Cerebral Cortex (original) (raw)
Abstract
The developing cerebral cortex contains apical and basal types of neurogenic progenitor cells. Here, we investigated the cellular properties and neurogenic output of basal progenitors, also called intermediate neuronal progenitors (INPs). We found that basal mitoses expressing transcription factor Tbr2 (an INP marker) were present throughout corticogenesis, from embryonic day 10.5 through birth. Postnatally, Tbr2+ progenitors were present in the dentate gyrus, subventricular zone (SVZ), and posterior periventricle (pPV). Two morphological subtypes of INPs were distinguished in the embryonic cortex, “short radial” in the ventricular zone (VZ) and multipolar in the SVZ, probably corresponding to molecularly defined INP subtypes. Unexpectedly, many short radial INPs appeared to contact the apical (ventricular) surface and some divided there. Time-lapse video microscopy suggested that apical INP divisions produced daughter INPs. Analysis of neurogenic divisions (_Tis21_-green fluorescent protein [GFP]+) indicated that INPs may produce the majority of projection neurons for preplate, deep, and superficial layers. Conversely, proliferative INP divisions (_Tis21_-GFP−) increased from early to middle corticogenesis, concomitant with SVZ growth. Our findings support the hypothesis that regulated amplification of INPs may be an important factor controlling the balance of neurogenesis among different cortical layers.
Keywords: Eomes, intermediate progenitor cells, Pax6, radial glia, Tbr2
Introduction
Projection neurons of the cerebral neocortex are generated from progenitor cells located in the ventricular zone (VZ) and subventricular zone (SVZ) of the cortical neuroepithelium (Boulder Committee 1970). Various progenitor types have been distinguished on the basis of morphology, molecular expression, lineage relations, symmetric or asymmetric modes of cell division, and apical or basal location during M-phase (reviewed by Götz and Huttner 2005; Guillemot et al. 2006; Kriegstein et al. 2006; Pontious et al. 2008). In rodents, at least 3 or 4 types of neurogenic progenitors have been identified, whose relative abundance changes markedly between stages of neurogenesis (Götz and Huttner 2005; Gal et al. 2006; Martínez-Cerdeño et al. 2006). Even more progenitor types may be present in the primate SVZ, especially in humans (Letinic et al. 2002; Smart et al. 2002; Zecevic et al. 2005; Bayatti et al. 2008; Bystron et al. 2008).
The first neurogenic progenitors in the cortex are neuroepithelial cells (NECs), which produce mainly preplate neurons (reviewed by Götz and Huttner 2005). NECs then give rise to radial glial progenitors (RGPs), which not only act as guides to neuron migration but also give rise to the majority of cortical neurons, directly or indirectly (Heins et al. 2002; Malatesta et al. 2003; Anthony et al. 2004; Götz and Barde 2005). NECs and RGPs divide at the apical surface and give rise to neurons as well as intermediate neuronal progenitors (INPs), also known as basal progenitors (Haubensak et al. 2004; Miyata et al. 2004; Noctor et al. 2004; Attardo et al. 2008). INPs have limited proliferative potential (1–3 mitotic cycles), divide symmetrically, and generate neurons exclusively, suggesting that one of their functions is to transiently amplify the production of projection neurons from NECs and RGPs (Noctor et al. 2004; Wu et al. 2005). A fourth type of neurogenic progenitors, designated short neural precursors (SNPs), has been described in rodents (Gal et al. 2006). SNPs have short radial morphology within the VZ, divide at the apical surface, and express markers of neurogenic fate (Gal et al. 2006). Recently, 3 types of progenitor cells were defined by single-cell gene profiling in the developing mouse cortex (Kawaguchi et al. 2008). They were identified as RGPs and 2 subtypes of INPs, the latter classified as VZ and SVZ subtypes.
The existence of multiple progenitor types and their dynamic contributions to neurogenesis suggest the possibility that each might produce different classes of neurons. INPs have attracted particular interest because of their unique properties and association with the SVZ. Only INPs divide basally; all others are thought to be strictly apical progenitors. Unlike NECs and RGPs (but similar to SNPs), INPs appear to produce only neurons and not glia (Noctor et al. 2004; Wu et al. 2005; Gal et al. 2006). Indeed, INPs may be committed to neurogenic fate (Attardo et al. 2008; Noctor et al. 2008). INPs are further distinguished by solely symmetric modes of division (proliferative or neurogenic).
Regarding specialized neuron production, evidence that upper layer neurons (late born) are produced mainly in the SVZ has prompted the hypothesis that INPs are fated to produce upper layer neurons (Tarabykin et al. 2001; Zimmer et al. 2004). However, the reported prevalence of INPs or basal mitoses during early stages, before the SVZ develops (Smart 1973; Haubensak et al. 2004; Attardo et al. 2008), suggests that INPs may also produce neurons for lower layers (early born). Besides producing specialized types of neurons, INPs have also been proposed to play central roles in the development and evolution of cortical surface area and gyration (Kriegstein et al. 2006), as well as area-specific laminar patterns (Pontious et al. 2008).
Until now, hypotheses concerning INP roles in cortical development have been difficult to evaluate because of limited data on INP numbers and neuronal output at different stages of neurogenesis. In particular, no previous studies have surveyed changes in INP abundance or compared the contributions with neurogenesis from INPs and other progenitor types, using consistent methods across developmental stages. Our goal in the present study was to characterize INPs and their contributions to neuron production comprehensively, throughout mouse corticogenesis. Our results indicate that INPs are present throughout cortical neurogenesis, display distinct morphological types in the VZ and SVZ, and contribute a substantial fraction, probably the majority of projection neurons to preplate, as well as deep and superficial cortical plate layers.
Materials and Methods
Animals
Wild-type C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and maintained as a breeding colony. Tis21(Mouse Genome Informatics [MGI]:Btg2)-green fluorescent protein (GFP) knock-in mice (Haubensak et al. 2004) were maintained as a breeding colony on the C57BL/6J background and were studied as heterozygotes. Tbr2(MGI:Eomes)-GFP bacterial artificial chromosome (BAC) transgenic mice were obtained from GENSAT (Gong et al. 2003; www.gensat.org), maintained on a CD1 background, and studied as heterozygotes. Animals were housed under standard conditions (12 h light/dark cycle). All experimental procedures were approved by Institutional Animal Care and Use Committees at the University of Washington and Seattle Children's Hospital Research Institute. Timed-pregnant embryos were identified by vaginal plugs, with noon on the day of plug appearance designated embryonic day (E) 0.5. Noon on the day of birth was designated postnatal day (P) 0.5.
Bromodeoxyuridine Labeling
Dividing cells (S-phase) were labeled by intraperitoneal injection of bromodeoxyuridine (BrdU) (40 mg/kg) 30 min before death as described previously (Englund et al. 2005).
Tissue Preparation
Animals were killed by cervical dislocation under Avertin anesthesia (pregnant females) or cryoanesthesia (neonatal mice), as described previously (Hevner et al. 2004; Englund et al. 2005). Embryos were rapidly removed and evaluated for GFP fluorescence under an epifluorescence dissecting microscope (Nikon SMZ1500 stereomicroscope). Embryonic brains were fixed by immersion in cold (4 °C) fixative consisting of freshly prepared 4% paraformaldehyde in phosphate-buffered saline (PBS) for 4–6 h (for immunofluorescence) or overnight (for 1,1′-dioctadecyl-3,3,3′3′-tetramethylindocarbocyanine perchlorate [DiI] labeling). Postnatal pups were perfused with cold fixative, and then the brains were removed and postfixed by immersion in cold fixative for an additional 4–6 h. For all immunofluorescence antigens except O4, brains were rinsed and cryoprotected in increasing concentrations of cold-buffered sucrose (4%, 10%, 20%, and 30% in 0.1 M sodium phosphate, pH 7.0), frozen in optimum cutting temperature compound (Sakura Finetek, Torrance, CA), cryosectioned at 10–12 μm, and mounted on Superfrost Plus slides (Thermo Fisher Scientific, Waltham, MA). Slides were stored at −80 °C until needed. For O4 detection, fixed postnatal brains were embedded in 4% low–melting point agarose and sliced coronally (100 μm) on a vibratory microtome (Leica VT1000).
DiI Labeling
_Tbr2_-GFP+ embryonic brains were removed from fixative into a plastic Petri dish and kept wet with drops of fixative as necessary. Under the dissecting microscope, 2–5 small crystals (<100 μm diameter) of DiI (Invitrogen, Carlsbad, CA) were placed firmly on the pial surface of the cortex bilaterally. The brain was then placed in PBS with 4% sucrose and 0.05% sodium azide and stored at room temperature for 1–2 days for DiI diffusion. Brains were embedded in 4% low–melting point agarose, cooled on ice, and sectioned at 80–100 μm into cold PBS on the vibratory microtome. Sections were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (0.01%, Invitrogen) and mounted in PBS on Superfrost Plus slides.
Immunofluorescence
Slide-mounted cryostat sections were thawed into cold PBS. Single- and double-label immunofluorescence were done routinely as described (Hevner et al. 2004; Englund et al. 2005; Hodge et al. 2008). For triple-label immunofluorescence detection of GFP or Neurogenin2 (Ngn2) in combination with other antigens, nonspecific binding was first reduced by preincubation with cold (4 °C) blocking solution, consisting of PBS with 0.1% Triton X-100 (PBS-T), 10% normal goat serum, and 2% bovine serum albumin for 30 min. Slides were then incubated overnight at 4 °C with mouse monoclonal anti-GFP or anti-Ngn2 (antibodies listed below), rinsed 3 times in PBS for 5 min, and then incubated for 2 h at room temperature with goat anti-mouse immunoglobulin G conjugated to Alexa Fluor 687 (1:200 in PBS-T). Sections were rinsed 3 times in PBS for 10 min, postfixed in cold fixative (4% paraformaldehyde in PBS) for 30 min, and again rinsed 3 times in PBS for 5 min. Sections were then treated for antigen enhancement by microwave heating for 90 s in 10 mM sodium citrate (pH 6.0), taking care to avoid excessive evaporation and tissue drying. This was followed by cooling for 5 min in PBS on ice and then rinsing 3 times for 5 min in cold PBS. In experiments involving BrdU immunodetection, sections were then incubated in 2N HCl for 30 min at 37 °C and rinsed 5 times in PBS for 5 min. Sections were again treated with blocking solution for 30 min at room temperature and further processed as described for routine single- or double-label immunofluorescence (Hevner et al. 2004; Englund et al. 2005; Hodge et al. 2008). For immunofluorescence of O4, vibratome sections were processed without detergent, as described previously (Hevner et al. 2001).
Primary and Secondary Antibodies
The following primary antibodies were used: rat monoclonal anti-BrdU (1:200, OBT0030, Accurate, Westbury, NY); goat polyclonal anti-doublecortin (1:250, sc-8066, Santa Cruz Biotechnology, Santa Cruz, CA); rabbit polyclonal anti-pan-Dlx (1:75, courtesy Dr J. Kohtz, Northwestern University, Chicago, IL); mouse monoclonal anti-GFP (1:1000, MAB3580, Millipore, Billerica, MA); rabbit polyclonal anti-GFP (1:500, AB3080, Millipore); mouse monoclonal anti-glial fibrillary acidic protein (GFAP) (1:1000, MAB360, Millipore/Chemicon); mouse monoclonal anti-Ngn2 (1:2, courtesy Dr D. Anderson, California Institute of Technology, Pasadena, CA); mouse monoclonal anti-O4 (1:50, MAB345, Millipore); mouse monoclonal anti-phospho[Ser10]-histone H3 (pH3) (1:800, ab7031, Abcam, Cambridge, MA); rabbit polyclonal anti-pH3 (1:400, 06-570, Millipore); and rabbit polyclonal anti-Tbr2 (1:2000, R.F.H. laboratory). Secondary antibodies were conjugates of Alexa Fluors 488, 594, and 647 (1:200, Invitrogen). DAPI (0.01%) and TO-PRO-3 (0.1 μM, Invitrogen) were used as nuclear counterstains.
In Situ Hybridization
Slide-mounted cryostat sections were processed to detect GFP mRNA by in situ hybridization using digoxigenin-labeled probes as described previously (Fink et al. 2006). Double labeling by in situ hybridization and immunofluorescence in the same section was also done as described previously (Fink et al. 2006). For figures, the in situ hybridization color reaction was photographed in bright field gray scale, inverted using Adobe Photoshop CS2, and pasted in the red color channel for dark field pseudocolor in combination with Pax6, Tbr2, or Tbr1 immunofluorescence.
Fixed Tissue Microscopy and Image Processing
Digital immunofluorescence images were obtained on a Zeiss Axio Imager Z1 epifluorescence microscope with Apotome or on a Bio-Rad LS2000 confocal microscope. Images were adjusted for brightness and contrast using Adobe Photoshop CS2. For images in Supplementary Figure 1, single-color channels were converted to gray scale and digitally inverted to appear as bright field.
Cell Counting and Bin Analysis
To quantitatively analyze dividing progenitor cells, epifluorescence images were collected from coronal sections through the developing parietal cortex (presumed prospective primary somatosensory area) using the 20× objective on the Zeiss Axio Imager Z1. For each experiment, 3 or 4 brains were studied at every age, and 3–4 images were collected from each brain. From each image, one columnar region of cortex measuring 100 μm wide was selected for analysis (as illustrated in Supplementary Fig. 1). Mitotic figures were identified by pH3 labeling or by chromatin condensation as revealed by nuclear counterstains. Each mitotic figure was scored for expression of transcription factors (Tbr2 or Pax6) and, in some experiments, _Tis21_-GFP. Expression of Tbr2 or pH3 was considered positive if clearly above background levels; Pax6 was considered high level if detectable over ≥50% of the nucleus. Low-level Pax6 is detectable in many INPs, but high-level Pax6 is relatively specific for apical progenitors (Englund et al. 2005).
Mitotic divisions were analyzed for position by bin analysis, using the same principles as described previously (Hevner et al. 2004). Briefly, the cortical wall was divided into 20-μm bins according to distance from the ventricular surface, with bin 1 nearest the ventricle. Mitoses were scored in the bin containing the center of the nucleus. Thus, bin 1 contained divisions centered within 20 μm of the ventricular surface. Divisions in bin 1 were considered to be apical or subapical (Haubensak et al. 2004), and divisions in other bins were considered to be basal. Divisions in bin 1 (apical or subapical) showed no differences of outcome by time-lapse imaging; thus 20-μm bins (no smaller) were considered suitable for this analysis.
Two-Photon Time-Lapse Video Microscopy and Analysis
Procedures were similar to previous studies using _Tis21_-GFP embryos (Attardo et al. 2008). Heterozygous _Tbr2_-GFP embryos (E10.5–E12.5) were dissected at room temperature in Dulbecco's PBS (Invitrogen) supplemented with 10% heat-inactivated fetal calf serum and 100 U/mL penicillin/streptomycin (Invitrogen). The head without skin (E10.5) or the dissected brain (E12.5) was immersed in 3% low–melting point agarose in PBS (Sigma) at 37 °C, the sample was cooled to room temperature, and 250- to 500-μm slices of telencephalon were cut using a manual tissue chopper (Vibratome 600 McIlvain, Ted Pella, Inc., Redding, CA), with the cutting plane oriented perpendicular to the anteroposterior axis of the neural tube. Imaging of the dorsal telencephalon was carried out with the X_–_Y plane oriented perpendicular to the ventricular surface. Only those slices in which, upon proper orientation in the microscope, most of the radial migration of cells occurred in the focal plane were selected for further study.
Slices (on average 7) were immersed at room temperature in ∼2.5 mL of type Ia collagen (Cellmatrix, Nitta Gelatin, Osaka, Japan) (diluted to 1.5 mg/mL with Dulbecco’s modified Eagle’s medium and neutralizing buffer according to the manufacturer's protocol and kept on ice until use) and then transferred onto a circular 42-mm glass cover slip, which constituted the bottom of the incubation chamber of a POC-Chamber-System (Saur, Reutlingen, Germany). After 40 min at 37 °C (the time necessary for the collagen to solidify), the slices were cultured at 37 °C in 5–7 mL of Neurobasal medium (Invitrogen) supplemented with 10% immediately centrifuged mouse serum (Harlan, Indianapolis, IN), 1× N2 supplement (Invitrogen), 1× B27 supplement (Invitrogen), and 100 U/mL penicillin/streptomycin. The POC-Chamber-System was gassed with 40% O2, 5% CO2, and 55% N2, with the incubation chamber sealed with a semipermeable membrane that allowed gas exchange while preventing evaporation. Slices were superfused with medium (0.1 mL/min) recirculated using a peristaltic pump (Gilson, Middleton, WI).
Time-lapse analysis was usually started ∼1 h after the beginning of the culture and after visual verification of the integrity of the slice. Images were recorded on an inverted Radiance 2100 2-photon microscope (Bio-Rad, Hercules, CA) equipped with Nikon optics (ECLIPSE TE300) and a Mira 900 Titanium:Sapphire laser (Coherent, Santa Clara, CA) tuned to 880–900 nm, which was pumped by a Verdi 5 or 10 W solid-state laser (Coherent). A 60× water immersion lens, which was heated to 37 °C using an objective heater (Bioptechs, Butler, PA), was used to record the GFP fluorescence. Stacks of optical sections, spaced at 2.7-μm intervals, were collected every 12 min, starting at 20 μm from the cut surface, yielding optical cuboids with a z axis of 60–130 μm. Recording was performed using a single scanning run at 512 × 512 pixel resolution with a scanning speed of typically 166 lines per second. Images were acquired using the Laser 2000 software (Bio-Rad/Zeiss, Thornwood, NY); 3D reconstructions were computed using Imaris software (Bitplane AG, Saint Paul, MN) in the Maximum Intensity Projection rendering mode.
Seven 2-photon time-lapse imaging experiments were carried out on slice cultures of dorsal telencephalon from E10.5 (n = 5) and E12.5 (n = 2) heterozygous _Tbr2_-GFP embryos. For each experiment, data were obtained from a single slice, so that each experiment is independent from the others and reflects a separate embryo. Only experiments in which _Tbr2_-GFP+ cells were observed to undergo migration and mitosis and in which there was no significant apoptosis (as judged by the appearance of pyknotic nuclei) throughout the entire time of acquisition were considered for analysis.
Results
Tbr2+ Basal Mitoses Are Present throughout Cortical Neurogenesis
To survey the abundance and distribution of INPs and compare them with RGPs, we identified progenitor types according to both transcription factor expression and the location of mitotic division. INPs were identified by Tbr2 expression and basal mitosis, whereas apical progenitors were identified by high-level Pax6 expression and apical mitosis (Haubensak et al. 2004; Miyata et al. 2004; Noctor et al. 2004; Englund et al. 2005). To first confirm that Pax6 and Tbr2 expression identify most progenitor cells, we studied sections of acute BrdU-labeled cerebral cortex by triple immunofluorescence. In E12.5 cortex, 99.7% of BrdU+ cells expressed Pax6 and/or Tbr2 (n = 645 BrdU+ cells); in E14.5 cortex, 99.0% expressed Pax6 and/or Tbr2 (n = 384 BrdU+ cells). Dividing cells were recognized by expression of pH3, a marker of late G2/M-phase (Scott et al. 2003), or by chromatin condensation. Immunofluorescence across all ages revealed that pH3 and Tbr2 were detectable throughout neurogenesis from E10.5 to P0.5, but Pax6 was not consistently detectable until E12.5 (Supplementary Fig. 1). Thus, for histological studies of E10.5 and E11.5 cortex, Tbr2 and pH3 were studied but not Pax6.
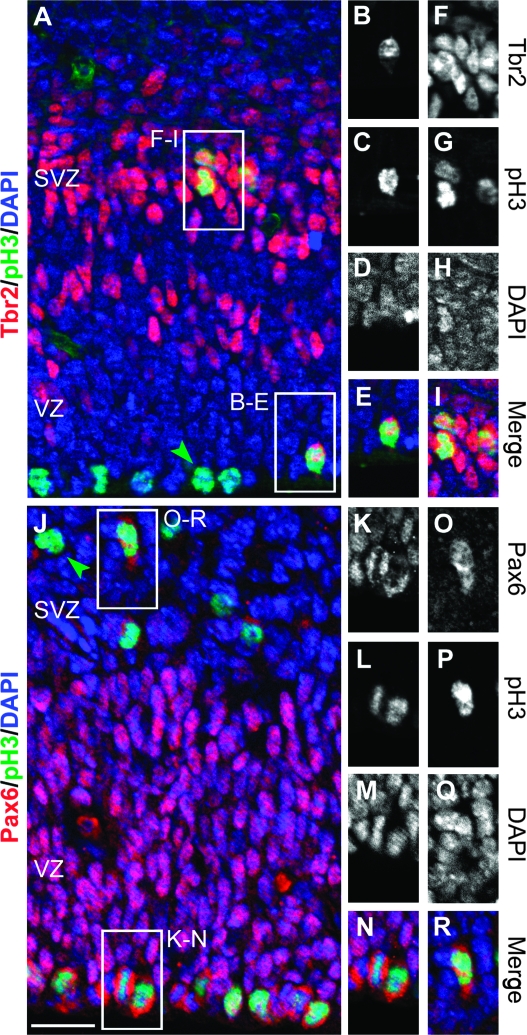
Transcription factor expression and mitotic division were assessed together in the same cells by double- or triple-label immunofluorescence detection of pH3, Tbr2, and Pax6. Figure 1 shows representative double-labeled sections from E14.5 cortex. As expected, Tbr2 expression was highly correlated with basal divisions and high-level Pax6 expression with apical divisions. Occasionally, Tbr2 was detected in divisions at or near the ventricular surface (apical or subapical, respectively) (Fig. 1_A_–E). Conversely, Pax6 was detectable at low to moderate levels in many basal divisions (Fig. 1_J_,O_–_R). Overall, across all ages, 7.0% of Tbr2+ mitoses were located in apical or subapical locations; this ratio fluctuated in the range of 5–20% at different ages (Fig. 2_A_). The expression of Tbr2 in mitoses very close to the ventricular surface suggested that some INPs might undergo apical divisions. For this reason, we refer to Tbr2+ dividing cells as INPs rather than basal progenitors. Another possibility, suggested by the asymmetric distribution of Tbr2 protein in some mitotic figures, was that a subset of apical progenitors underwent asymmetric division to produce Tbr2+ and Tbr2− daughter cells, with differential distribution of Tbr2 preceding the completion of mitosis (Fig. 1_A_–E).
Figure 1.
M-phase location and transcription factor expression distinguish progenitor types in E14.5 mouse cortex. (A_–_I) Tbr2+ (red) INPs divided in both the SVZ and the VZ, including rare divisions at the ventricular (apical) surface. Some divisions showed uneven distribution of Tbr2 protein, possibly suggestive of asymmetric division (B_–_E). Most apical mitoses (green arrowhead, A) showed no Tbr2 protein. (J_–_R) Highly Pax6+ (red) RGPs divided at the apical surface (K_–_N). Pax6 was present at low levels in some SVZ divisions (green arrowhead, J) and rarely reached levels comparable to apical divisions (O_–_R). Scale bar: 20 μm.
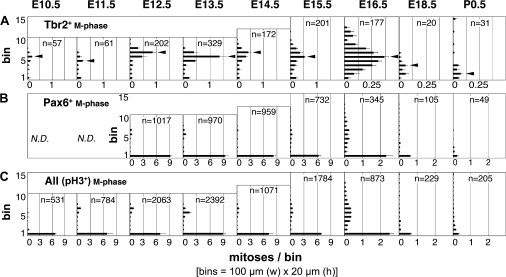
Figure 2.
M-phase divisions (pH3+) binned by distance from the ventricular surface (20-μm bins) in parietal cortex from E10.5 through P0.5. (A) Tbr2+ mitoses (INPs) were most abundant along the basal edge of the VZ (arrowheads), equivalent to the SVZ visible histologically on E13.5 and later. At the end of neurogenesis (E16.5–P0.5), the SVZ broadened and shifted closer to the ventricular surface, as the VZ involuted. (B) Highly Pax6+ mitoses were most numerous at the apical surface (bin 1) representing RGPs; few basal divisions (INPs) contained high Pax6. (C) Mitoses were most abundant at apical/subapical positions (bin 1) throughout neurogenesis.
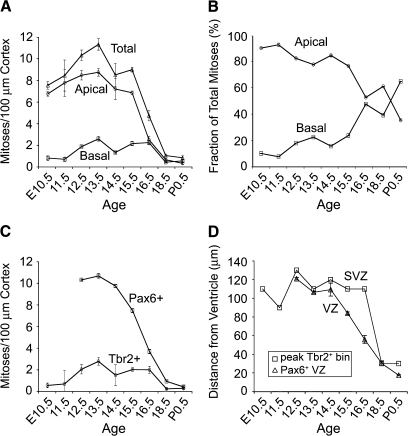
To map the abundance and distribution of basal and apical progenitor divisions, we counted Tbr2+ and Pax6+ M-phase cells, plotted their distance from the ventricular surface by bin analysis (Fig. 2), and graphed changes in their abundance throughout corticogenesis (Fig. 3). The results revealed several findings. First, INPs (mainly basal Tbr2+ divisions) were present at all ages (Figs 2_A_ and 3_A_–C), including the onset of cortical neurogenesis on E10.5 (Figs 3 and 4_A_). Second, M-phase Tbr2+ divisions were most abundant along the basal edge of the VZ, corresponding to the SVZ (defined histologically as the distinct proliferative zone at the junction of the VZ and intermediate zone; Boulder Committee 1970). Although the SVZ was not visible as a distinct histological layer until E13.5, bin analysis revealed accumulation of Tbr2+ INPs along the basal VZ edge as early as E10.5 (Figs 2_A_ and 3_D_). However, Tbr2+ INP divisions were also scattered in the VZ, including at or near the apical surface (bin 1), at all ages (Fig. 2_A_). Third, apical mitoses were more abundant than basal mitoses until the end of neurogenesis (E16.5–P0.5), when apical mitoses decreased to approximately the same numbers as basal mitoses (Figs 2_C_ and 3_A_,B). Fourth, the abundance profiles of Tbr2+ mitoses and Pax6+ (high level) mitoses were virtually identical to basal and apical mitoses, respectively, reinforcing the correlation between transcription factor expression and mitotic location (Fig. 3_A_,C). Finally, the abundance of INP divisions (defined by either basal position or Tbr2 expression) fluctuated, with peaks on E12.5–E13.5 and E15.5–E16.5, and an intervening decline on E14.5 (Fig. 3_A_,C). The significance of decreased INP divisions on E14.5 was not immediately clear but could relate to rapid expansion of the cortical surface area and/or cell cycle slowing (Takahashi et al. 1995a, 1995b).
Figure 3.
Dynamics of INP and RGP abundance. (A) Apical/subapical mitoses (bin 1 of Fig. 2) were more numerous than basal mitoses until late in neurogenesis. Basal progenitors fluctuated, with a significant dip on E14.5. (B) As neurogenesis progressed, the relative proportion of apical progenitors declined and basal progenitors rose. (C) Divisions identified by Pax6 and Tbr2 expression generally paralleled apical and basal mitoses, respectively. (D) Tbr2+ mitoses were most abundant (squares, representing peak bins identified by arrowheads in Fig. 2_A_) at the basal edge of the VZ (triangles), identified as the basal edge of the zone of high Pax6 expression. Note SVZ broadening and VZ involution beginning on E15.5.
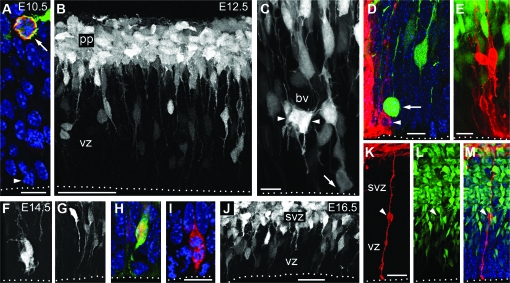
Figure 4.
INP divisions and morphological subtypes. (A) In E10.5 cortex, Tbr2 (red) and _Tbr2_-GFP (green) were expressed by virtually all basal progenitors (arrow). A small proportion of apical progenitors expressed Tbr2, but most did not (arrowhead). Basal and apical divisions of _Tbr2_-GFP+ cells are also documented in Supplementary Movies 1 and 2. (B_–_C) On E12.5, _Tbr2_-GFP (white) was detected in a subset of VZ cells and in most preplate (pp) neurons. In the VZ, _Tbr2_-GFP+ cells exhibited mainly short radial, but also some multipolar morphologies (arrowheads, C). The latter were often clustered around blood vessels (bv). Some _Tbr2_-GFP+ cells appeared to divide at the apical surface (arrow, C). (D_–_E) DiI (red) labeling (E12.5) from the pial surface–labeled RGPs (including a mitotic RGP, arrowhead) but not _Tbr2_-GFP+ (green) basal progenitors in the VZ (arrow indicates a mitotic INP). (F_–_I) In E14.5 VZ, _Tbr2_-GFP (green or white), and Tbr2 (red) expression revealed multipolar (F) and short radial (G_–_I) INP morphologies. The latter sometimes contacted the ventricular surface (H_–_I). A Tbr2+ mitotic figure is shown in (I). (J_–_M) E16.5 cortex, showing radial and multipolar morphologies of _Tbr2_-GFP+ cells (white in J, green in K_–_M), and absence of _Tbr2_-GFP expression in DiI-labeled (red) RGPs (arrowhead, K_–_M). Dotted lines indicate the ventricular surface. Scale bars: (A) 10 μm; (B) 50 μm; (C) 10 μm; (D) 10 μm; (E) 15 μm; (F_–_G) 20 μm (in I); (H) 15 μm (in I); (I) 10 μm; (J) 20 μm; (K_–_M) 25 μm.
Together, these results (Figs 1–3) indicated that INPs are present throughout corticogenesis, divide predominantly in the SVZ but also the VZ, and are overall less abundant than apical progenitors (Tbr2−) except at the end of neurogenesis.
INPs Display Radial and Multipolar Morphologies
To investigate the morphological characteristics of INPs, we studied _Tbr2_-GFP BAC transgenic mice (Gong et al. 2003), in which GFP fills the cytoplasm of Tbr2+ cells, including neurogenic progenitors in the developing and adult brain (Kwon and Hadjantonakis 2007; Hodge et al. 2008). First, we verified that GFP expression accurately reports Tbr2 expression in the VZ and SVZ. Whereas the enhanced green fluorescent protein mRNA was detected only in VZ and SVZ (Supplementary Fig. 2), GFP protein was additionally detected in postmitotic cells that lacked Tbr2 protein (Fig. 4_A_ and Supplementary Fig. 3). This was interpreted as GFP perdurance in projection neurons derived from _Tbr2_-GFP+ lineages. Consistent with this interpretation, GFP was not detectable in Dlx+ interneurons (Supplementary Fig. 4).
From the earliest stages, _Tbr2_-GFP+ INPs exhibited 2 principal morphologies. In the VZ, most _Tbr2_-GFP+ INPs exhibited radial processes (apical and basal), which sometimes appeared to contact the ventricular surface (Fig. 4). Among the latter, some appeared to divide at or near the apical surface (arrow, Fig. 4_C_), thus resembling the previously described “short radial” SNPs (Gal et al. 2006). Fewer _Tbr2_-GFP+ cells in the VZ exhibited multipolar morphologies (Fig. 4_C_,F). In the SVZ, multipolar morphologies were predominant (Fig. 4_B_,J). Interestingly, multipolar _Tbr2_-GFP+ INPs were often seen in close apposition to blood vessels (Fig. 4_C_), suggesting that the developing vasculature might be a specialized niche for INPs. In contrast to radial glia, no _Tbr2_-GFP+ INPs contacted the basal (pial) surface, as determined by DiI labeling on E12.5 and E16.5 (Fig. 4_D_,E,K_–_M; more than 100 DiI-labeled cells examined on each age). In sum, INP morphology in the VZ was mainly short radial with or without ventricular surface contact, but only multipolar in the SVZ.
Neurogenic Tbr2+ Progenitors in the Early Postnatal Cortex
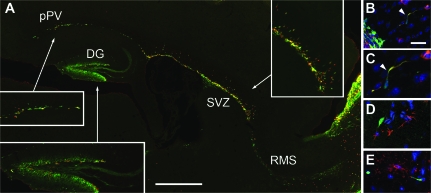
To determine if Tbr2+ progenitors remain neurogenic or shift to gliogenic fates during late embryonic and early postnatal stages, _Tbr2_-GFP+ lineages were studied in P7 cortex (Fig. 5). Sagittal sections revealed 3 distinct regions containing numerous Tbr2+/_Tbr2_-GFP+ cells: the dentate gyrus (DG), the SVZ and rostral migratory stream (RMS), and the posterior periventricle zone (pPV) near occipital cortex (Fig. 5_A_). Tbr2+ mitotic figures were observed in each of these regions, confirming their progenitor identity (not shown). The DG and postnatal SVZ/RMS are well-documented neurogenic zones in adult brain (reviewed by Zhao et al. 2008), and Tbr2+ cells in the adult DG have been confirmed as neurogenic (Hodge et al. 2008). The pPV, although not so well known, has also been identified as a reservoir of neurogenic progenitors in adult rodents (Nakatomi et al. 2002). Thus, Tbr2+ cells in the P7 cortex occupied locations consistent with neurogenic fate. To test for neuronal or glial differentiation of _Tbr2_-GFP+ cells, we studied specific markers (Fig. 5_B_–E). Of more than 100 _Tbr2_-GFP+ cells examined in neocortex adjacent to the SVZ and pPV, many showed coexpression of doublecortin, a neuronal marker (Fig. 5_B_–C), but none showed coexpression of GFAP (Fig. 5_D_), an astrocytic marker, or O4, an oligodendroglial marker (Fig. 5_E_).
Figure 5.
Tbr2 and _Tbr2_-GFP expression in P7 cortex. (A) Double labeling for Tbr2 (red) and _Tbr2_-GFP (green) showed double-labeled cells in the DG, the pPV adjacent to hippocampus (Nakatomi et al. 2002), the SVZ, and the RMS. Insets show selected zones at higher magnification. Parasagittal. (B_–_C) Double labeling for doublecortin (red) and _Tbr2_-GFP (green) in neocortex adjacent to SVZ confirmed neuronal differentiation. Panel (C) shows higher magnification of a cell with migratory morphology from (B). (D) Double labeling for GFAP (red) and _Tbr2_-GFP (green) in the cerebral cortex showed no astrocytic differentiation. (E) Double labeling for O4 (red) and _Tbr2_-GFP (green) showed no oligodendroglial differentiation. Scale bars: (A) 500 μm (250 μm for insets); (B, D_–_E) 20 μm (in B); (C) 10 μm (in B).
Time-Lapse 2-Photon Microscopy of Tbr2-GFP+ Progenitors
To further investigate INP morphologies and mitotic divisions in live tissue slices, we studied _Tbr2_-GFP transgenic cortex using time-lapse 2-photon microscopy. Slices of E10.5 (n = 5) and E12.5 (n = 2) cortex were imaged for periods of 420–864 min, at 12-min intervals. Similar to results in fixed tissue sections (Fig. 4), _Tbr2_-GFP+ cells in slice cultures exhibited short radial and multipolar morphologies (Supplementary Movies 1 and 2). Short radial _Tbr2_-GFP+ cells were located in the VZ and sometimes appeared to contact the ventricular surface (Supplementary Movie 2). Multipolar _Tbr2_-GFP+ cells were abundant at or near the interface between VZ and preplate. In addition, preplate neurons were brightly fluorescent in _Tbr2_-GFP cortex slices, due (as noted above) to GFP perdurance as well as transient expression of Tbr2 in preplate neurons on E10.5–E12.5 (Englund et al. 2005).
Mitotic divisions (n = 111) of _Tbr2_-GFP+ cells were observed in basal (n = 57), apical (n = 31), and subapical (n = 23) locations. Basal mitoses were underrepresented by this sampling technique because they were often difficult to distinguish and track among the numerous brightly fluorescent cells in the preplate and incipient SVZ. In contrast, the ventricular surface region contained few _Tbr2_-GFP+ cells, and thus, apical mitoses (although generally exhibiting weaker fluorescence than basal divisions) were more easily observed. Basal divisions gave rise to daughter cells that remained in the vicinity of the preplate, did not contact the apical surface (despite sometimes extending a process toward the ventricle), and presumably differentiated as neurons or reentered the cell cycle as multipolar INPs (Supplementary Movie 1). Apical divisions of _Tbr2_-GFP+ cells produced daughter cells with dynamic morphology, initially short radial (usually with apical surface contact; in 5 of 7 apical divisions where daughter cell tracking was feasible), followed by detachment from the apical surface and migration toward the preplate (Supplementary Movie 2). The daughter cells of apical _Tbr2_-GFP+ divisions did not adopt neuronal morphologies within the imaging periods, suggesting that they could have become multipolar INPs. Daughter cells from subapical divisions behaved similarly to those from apical divisions.
Overall, these time-lapse imaging studies confirmed that Tbr2+ INPs are present at the earliest stages of cortical neurogenesis and most often divide at basal locations, less frequently at apical/subapical locations. The data furthermore suggested that apical divisions of _Tbr2_-GFP+ cells may be proliferative in most cases, although further studies will be necessary to confirm the daughter cell fates over longer time periods or with molecular markers of specific cell types.
INPs Express Variable Levels of Pax6 and Ngn2
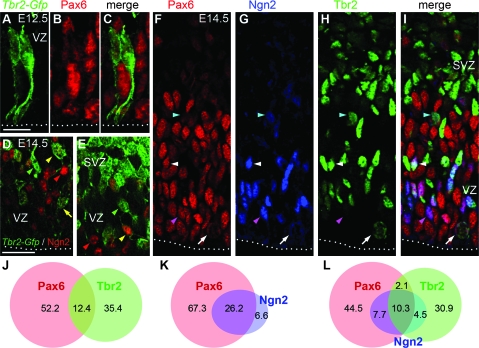
To investigate the expression of additional key transcription factors in INPs, we quantified the number of cells that coexpressed Tbr2 or _Tbr2_-GFP with transcription factors Pax6 and Ngn2 in cortex on E12.5 and E14.5. Pax6 (high level) is a marker of RGPs (Götz et al. 1998), whereas Ngn2 identifies a subset of VZ cells with prospective INP fates (Britz et al. 2006). We found that INPs with short radial morphology, like INPs generally, expressed only low to absent levels of Pax6 (Fig. 6_A_–C). Because Ngn2 promotes INP production from apical progenitors (Miyata et al. 2004), we expected to find Ngn2 in a subset of Pax6+/Tbr2− apical progenitors, presumably committed to INP fate, and in a subset of Tbr2+ INPs, presumably newly generated from apical progenitors. Results were consistent this prediction (Fig. 6_D_–I). Cell counting of triple-labeled sections in E14.5 cortex (Fig. 6_J_–L) indicated that Ngn2 was present in 15% of RGPs (Pax6+/Tbr2−), 83% of new INPs (Pax6+/Tbr2+), and 13% of differentiated INPs (Pax6−/Tbr2+). Among Ngn2+ cells, 34% were characterized as RGPs, 46% as new INPs, and 20% as differentiated INPs (Fig. 6_J_–L). Ngn2 was rarely detected in M-phase mitotic figures (arrow, Fig. 6_F_–I), consistent with the reported bias of Ngn2 expression to G1 and G0 (Miyata et al. 2004; Britz et al. 2006). Interestingly, Ngn2 expression also appears to oscillate throughout the cell cycle (Shimojo et al. 2008).
Figure 6.
Ngn2 is expressed by subsets of RGPs and INPs. (A_–_C) Double labeling of _Tbr2_-GFP (green) and Pax6 (red) in E12.5 cortex (merged image in C). SNP-like INPs expressed low to absent levels of Pax6. (D_–_E) Double labeling of _Tbr2_-GFP (green) and Ngn2 (red) in E14.5 cortex. Ngn2 was expressed by some _Tbr2_-GFP+ INPs, including multipolar (yellow arrowheads) and SNP-like (yellow arrow) types. Some _Tbr2_-GFP+ INPs lacked Ngn2 (green arrowheads). Conversely, some Ngn2+ cells (likely RGPs) did not express _Tbr2_-GFP (red arrowhead). (F_–_I) Triple labeling to detect Pax6 (red), Ngn2 (blue), and Tbr2 (green) in E14.5 cortex (merged image in I). Various combinations of transcription factor coexpression were observed. Some Tbr2+ INPs had no detectable Pax6 or Ngn2, especially in the SVZ; some expressed Ngn2 but little or no Pax6 (blue arrowhead); others expressed all 3 transcription factors, including interphase (white arrowhead) and, rarely, M-phase basally dividing cells (white arrow). Cells expressing Pax6 and Ngn2, but not Tbr2, were presumed RGPs (purple arrowhead). (J_–_L) Overlapping expression of Pax6, Ngn2, and Tbr2 in VZ and SVZ cells, illustrated by Venn diagrams (n = 863 cells counted by triple-label immunofluorescence). Numbers indicate the percentage of cells in each area of the graph. Scale bars: (A_–_C) 10 μm; (D_–_E) 30 μm; (F_–_I) 20 μm (in D).
Neurogenic Divisions Identified by Tis21-GFP Expression
As shown above (Fig. 3_A_–C), apical progenitor divisions outnumbered INP divisions throughout corticogenesis. However, the number of progenitor divisions is not necessarily indicative of neuronal output, which depends on additional factors, namely, the “neurogenic fraction” and the “mode of division.” The neurogenic fraction is the proportion of mitoses that are neurogenic rather than proliferative and varies from 0 to 100%. The mode of division can be symmetric or asymmetric: symmetric neurogenic divisions produce 2 neurons, whereas asymmetric neurogenic divisions produce only one neuron and one progenitor cell. Arithmetically, neuronal output (N) equals the number of progenitors (p) multiplied by the neurogenic fraction (f) and the daughter neurons per division (d): N = (p) × (f) × (d). Previous studies have shown that apical progenitor neurogenic divisions are asymmetric, whereas INP neurogenic divisions are symmetric (Haubensak et al. 2004; Miyata et al. 2004; Noctor et al. 2004; Attardo et al. 2008).
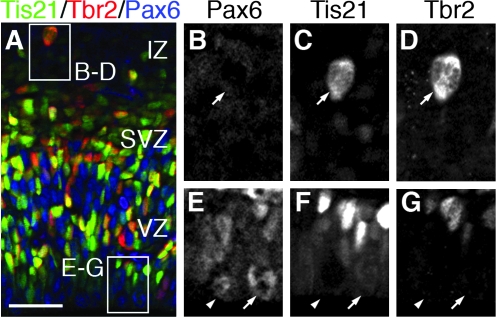
To identify neurogenic divisions, we studied _Tis21_-GFP mice, in which GFP is expressed only in neurogenic, and not proliferative divisions (Haubensak et al. 2004). To determine the neurogenic fate of RGPs and INP divisions, we studied _Tis21_-GFP in combination with Pax6 and Tbr2 by triple-label immunofluorescence and used nuclear counterstains to identify M-phase cells. Typical results are shown in Figure 7. As the figure shows, neurogenic apical divisions were recognized by modest increases of _Tis21_-GFP expression (Fig. 7_E_–G), whereas INP neurogenic divisions showed more robust increases (Fig. 7_B_–D). Similar distinctions were reported previously, in studies of early corticogenesis (Haubensak et al. 2004; Attardo et al. 2008).
Figure 7.
Tis21-GFP expression distinguishes proliferative (_Tis21_-GFP−) and neurogenic (_Tis21_-GFP+) INP and apical progenitor divisions. (A_–_G) Triple labeling for Tbr2 (red), _Tis21_-GFP (green), and Pax6 (blue) in E14.5 cortex. (A) Merged image. (B_–_D) Higher magnification of basal mitosis with neurogenic INP expression profile (_Tis21_-GFP+/Pax6−/Tbr2+), located in the intermediate zone (IZ). (E_–_G) Higher magnification of 2 apical progenitor mitoses: one proliferative (_Tis21_-GFP−/Pax6+/Tbr2−) and one neurogenic (_Tis21_-GFP+/Pax6+/Tbr2−). Scale bar: (A) 50 μm; (B_–_G) 20 μm.
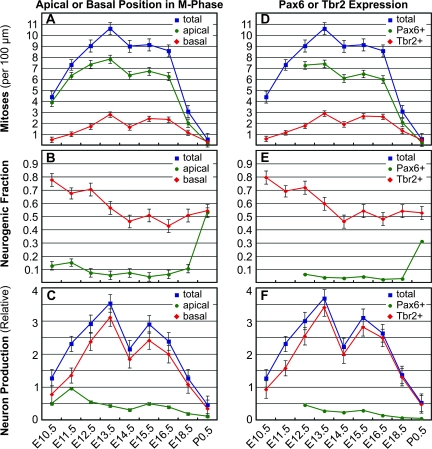
To quantify changes in the neurogenic fractions of INP and apical progenitor mitoses, we counted total as well as neurogenic mitoses of each type (apical, basal, Pax6+, and Tbr2+) and graphed the results by age and progenitor type (Fig. 8). Importantly, the profiles of mitotic activity in _Tis21_-GFP mice (Fig. 8) were highly consistent with the data set obtained in wild-type C57BL/6 mice (Fig. 3). For example, results from _Tis21_-GFP mice replicated the finding that apical progenitor mitoses were more numerous than INP mitoses, whether counted on the basis of mitotic location (Fig. 8_A_–C) or transcription factor expression (Fig. 8_D_–F). Likewise, both data sets showed a transient decrease in the abundance of INPs on E14.5 (Figs 3_A_,C and 8_A_,D).
Figure 8.
Neurogenic output of INP and apical progenitor divisions as indicated by _Tis21_-GFP expression. (A_–_C) Comparison of apical and basal mitoses. Apical divisions were more numerous than basal throughout neurogenesis (A), but basal divisions were more often neurogenic (B). (D_–_F) Comparison of Tbr2+ and highly Pax6+ mitoses. Pax6+ divisions were more numerous (D), but Tbr2+ divisions were more often neurogenic (E). Neurogenic output from INPs was found to be higher than from RGPs, regardless of whether progenitor types were defined by apical or basal division (C) or by transcription factor expression (F).
_Tis21_-GFP analysis revealed a profound difference of neurogenic activity between INPs and apical progenitors. Throughout the period from E10.5 to E18.5, the neurogenic fraction of INP (basal or Tbr2+) divisions remained much higher than the neurogenic fraction of RGP (apical or highly Pax6+) divisions (Fig. 8_B_,E). During these ages, the INP neurogenic fraction varied between 40% and 80%, whereas the apical progenitor neurogenic fraction remained below 10–20% (Fig. 8_B_,E). Only at the end of neurogenesis, on P0.5, did the neurogenic fraction of RGPs approach that of INPs (converging to 30–55%), consistent with terminal neurogenesis (Fig. 8_B_,E).
The overall higher neurogenic fraction of INP divisions, combined with their symmetric production of neuron pairs (vs. asymmetric production of single neurons from RGPs), indicated that despite the numerical predominance of RGP divisions, INPs actually produced the majority of cortically derived projection neurons at all ages (Fig. 8_C_,F). Even during preplate neurogenesis on E10.5–E11.5, a majority of neurons were produced by INPs. Throughout subsequent stages encompassing neurogenesis of deep, middle, and superficial layers, INPs produced the vast majority (>80%) of cortical projection neurons (Fig. 8_C_,F). These results suggested that, contrary to the hypothesis that INPs are fated to produce upper layer (late born) neurons, INPs actually generate the majority of pyramidal–projection neurons for all layers.
Increased Proliferation of INPs during SVZ Histogenesis
Interestingly, our analysis in _Tis21_-GFP mice revealed a progressive decline in the neurogenic fraction of INP divisions from ∼80% on E10.5 to ∼50% on E14.5 and later (Fig. 8_B_,E). The declining neurogenic fraction implied a reciprocal increase in the INP proliferative fraction. Previous studies have found that proliferative INP divisions occur by symmetric progenitor division, producing 2 daughter INPs (Miyata et al. 2004; Noctor et al. 2004). This suggests that from E10.5 to E14.5, increasing numbers of INPs reentered the cell cycle to expand the pool of INPs. This shift toward increasing proliferation may account in part for the rapid increase of INP numbers from E11.5 to E13.5 (Fig. 3_A_,C), as well as histological expansion of the SVZ, which becomes visible around E13.5 in mice (Viti et al. 2003; Götz and Barde 2005). However, because INPs exhibit limited proliferative capacity (Wu et al. 2005), production of new INPs from apical progenitors must also be an important factor regulating INP numbers. Because the INP neurogenic fraction in M-phase appeared to remain steady near ∼50% in later stages of neurogenesis (Fig. 8_B_,E), it is likely that decreasing production of new INPs accounted for the precipitous decline of INP numbers observed from E16.5 to E18.5 (Fig. 3_A_,C).
Discussion
The present study confirmed that INPs are present in the developing cortex at all stages and divide in not only the SVZ but also the VZ (Figs 1–3). In addition, several new conclusions were suggested: 1) VZ and SVZ INP subtypes exhibit short radial and multipolar morphologies, respectively; 2) the SVZ subtype of INPs are present early in corticogenesis, before the SVZ becomes visible as a distinct histological compartment; 3) INPs may produce the majority of pyramidal–projection neurons; and 4) the proliferative fraction of INP divisions increases from early to middle stages while the SVZ expands.
VZ and SVZ Subtypes of INPs
Confirming previous reports (Englund et al. 2005; Noctor et al. 2008), we observed that significant numbers of INPs were located in the VZ, and some divided at or near the apical surface (Figs 1–3 and 4_C_,I). Moreover, INPs in the VZ and SVZ exhibited distinct morphologies: mainly short radial in the VZ (with fewer multipolar forms) and multipolar in the SVZ (Figs 4 and 5). Possibly, these morphological variations could be imposed by extrinsic differences in the VZ and SVZ environments or could reflect intrinsic differences between INP subtypes. The latter interpretation is suggested by a recent single-cell gene-profiling study (Kawaguchi et al. 2008), in which INPs were clustered into 2 molecular subtypes, located in the VZ and SVZ. In that study, both subtypes were found to express Tbr2, but other molecules were expressed differentially. For example, VZ INPs highly expressed Hes6 and Gadd45g, whereas SVZ INPs selectively expressed Elavl2 and Lrp8 (Kawaguchi et al. 2008). Notably, SVZ INPs also expressed Svet1 (Kawaguchi et al. 2008), recently identified as an intronic sequence spliced from Unc5d mRNA (Sasaki et al. 2008). The Svet1 RNA, Unc5d mRNA, and Und5d protein were all localized to SVZ cells with multipolar morphology (Sasaki et al. 2008). Thus, our results confirm that SVZ INPs have multipolar morphology and furthermore reveal that VZ INPs have short radial morphology. Notably, the molecular profiles of VZ and SVZ INPs were both more closely related to postmitotic neurons than to RGPs, reinforcing the conclusion that both INP subtypes are committed to neurogenic fate (Kawaguchi et al. 2008).
These new findings suggest that SNPs (Gal et al. 2006) may be identical to VZ INPs. If the embryonic rodent cortex during midcorticogenesis contains only RGPs and 2 subtypes of INPs (Kawaguchi et al. 2008), then SNPs should correspond to one of these 3 types. (NECs, a fourth type of neurogenic progenitor, are present only during very early corticogenesis.) Among the 3 types, SNPs most resemble VZ INPs. Like VZ INPs, SNPs exhibit short radial morphology and express markers of neurogenic fate. Whereas some SNPs were observed to divide at the apical surface (Gal et al. 2006), the possibility that some SNPs divide more basally within the VZ was not excluded. Tbr2+ INPs can also divide at or near the apical surface (Fig. 4_C_,I; also see Noctor et al. 2008). The hypothesis that SNPs are VZ INPs could be tested by determining whether SNPs express Tbr2 and other INP markers.
INPs and Transcriptional Programs in Cortical Neurogenesis
Previous studies suggested that Pax6, Ngn2, and Tbr2 are part of a transcription factor cascade (Pax6 → Ngn2 → Tbr2 → Tbr1) that is central to the gene expression program for glutamatergic neurogenesis (reviewed in Hevner 2006; Hevner et al. 2006). In developing cortex, Pax6 is highly expressed in RGPs (Götz et al. 1998) and activates transcription of the Neurogenin2 (MGI:Neurog2) gene (Scardigli et al. 2003). Expression of Ngn2 is thought to then drive the transition from apical progenitors to INPs (Miyata et al. 2004; Britz et al. 2006), which in turn specifically express Tbr2 (Englund et al. 2005). This schema of sequential transcription factor expression in INP genesis was supported by patterns of Pax6, Ngn2, and Tbr2 colocalization observed in the present study (Fig. 6). A recent bioinformatic analysis was also consistent with this view (Gohlke et al. 2008). In future studies, it will be interesting to characterize the transcriptional programs implemented by these factors. Along this line, a recent study found that Ngn2 regulates radial cell migration by directly activating transcription of Rnd2, a small guanosine-5′-triphosphate-binding protein (Heng et al. 2008). Tbr2 also activated Rnd2 transcription but less efficiently than Ngn2, evidently maintaining the radial migration program initiated by Ngn2.
INPs Contribution to Cortical Neurogenesis
Our analysis of neuron production utilized _Tis21_-GFP as a neurogenic marker. The validity of _Tis21_-GFP as a neurogenic marker is supported by several observations: 1) Tis21 is not expressed until neurogenesis begins (Iacopetti et al. 1999); 2) virtually all early-born neurons are derived from _Tis21_-GFP+ progenitors (Haubensak et al. 2004); and 3) the vast majority of _Tis21_-GFP+ basal progenitors in early neurogenesis produce daughter cells with neuronal features (Attardo et al. 2008). However, recent studies suggest that _Tis21_-GFP+ apical progenitors infrequently generate neurons directly and more often generate basal progenitors (Attardo et al. 2008). This fate was also suggested by time-lapse imaging in the present study.
Additional support for our findings regarding the contribution of INPs to all cortical layers is provided by several recent studies. Insm1 is a zinc finger transcription factor gene recently identified as a panneurogenic marker in the developing and adult central nervous system (Duggan et al. 2008; Farkas et al. 2008). In the developing neocortex, Insm1 is expressed mainly by basal progenitors, although some apical divisions also express Insm1 (Farkas et al. 2008). Cell counting in E10.5 mouse cortex determined that Insm1 was expressed in ∼90% of basal mitoses but only in 10–15% of apical mitoses (Farkas et al. 2008). These values closely match our estimates of apical and basal progenitor neurogenic fractions based on _Tis21_-GFP expression (Fig. 8_B_). Inactivation of the Insm1 gene caused a marked reduction of INPs and cortical neurons, evident by E13.5 (Farkas et al. 2008). Likewise, Tbr2 gene inactivation was found to cause deficiencies of both early and late cortical neurogenesis (Arnold et al. 2008; Sessa et al. 2008). These studies utilized conditional inactivation because constitutive Tbr2 mutant embryos die of early lethality preceding brain development (Russ et al. 2000). Both studies found a marked reduction of deep layer neurons in the cortical plate by E13–E15, as well as defects of upper layer neurogenesis. The neuron loss was associated with depletion of INPs but no evidence of increased cell death. Interestingly, axon guidance defects were also prominent.
Whereas it seems clear that INPs are necessary to produce neurons for all layers of cortex, our data suggest a larger fractional contribution from INPs versus RGPs than reported in some previous studies. Regarding early neurogenesis (E10.5–E12.5), our data are actually in excellent agreement with previous work (Haubensak et al. 2004; Farkas et al. 2008). In contrast, previous estimates of INP neurogenesis during middle and late neurogenesis were sometimes lower than in the present study. Studying middle neurogenesis, Miyata et al. (2004) estimated that “almost 100%” of neurons were derived from nonsurface mitoses on E13, but only 32–44% on E14. In their study of late neurogenesis (rat E17–E19), Noctor et al. (2004) did not compare the contributions from INPs and RGPs but did report that 65.8% of VZ divisions were neurogenic. This is markedly higher than the ∼10% neurogenic fraction of apical divisions observed in the present study (Fig. 8_B_). However, the previous studies on middle and late neurogenesis were published before VZ INPs (or SNPs) were identified; most or all VZ progenitors were thought to be RGPs (Noctor et al. 2004). Also, the previous studies of middle and late neurogenesis were based on progenitor labeling with DiI or retroviruses, and fewer cells were counted than in the present study. Finally, species differences between rats and mice could be significant.
INP Proliferation and the SVZ
Previous studies have proposed that the SVZ may contain a self-renewing population of progenitors devoted to either gliogenic (Takahashi et al. 1995b) or upper layer neurogenic (Zimmer et al. 2004) fate. However, progenitor lineage tracing suggests that INPs divide only 1–3 times before neuronal differentiation (Noctor et al. 2004; Wu et al. 2005). In the present study, the proliferative fraction of M-phase INPs reached ∼50% during E14.5–P0.5 (Fig. 8_B_,E), which might appear to support self-renewal. However, whereas M-phase divisions accurately report new cell production, they may not accurately indicate progenitor numbers because M-phase accounts for different fractions of the cell cycle in different division types. The length of M-phase remains essentially constant, but other phases (especially G1 and G2) can vary substantially, depending on the type of division. In E14.5 mouse cortex, neurogenic VZ divisions have ∼29% longer cell cycle than do nonneurogenic VZ divisions (Calegari et al. 2005). The cell cycle lengths of SVZ divisions have not been determined but could be even longer than VZ divisions. In the present study, M-phase counts likely underestimated neurogenic INP numbers by 30% or more relative to nonneurogenic INPs, due to the cell cycle lengthening associated with neurogenic divisions (Calegari et al. 2005). Thus, our results suggest that the majority of INPs are neurogenic and thus insufficient for self-renewal.
Tbr2 and Postnatal Neurogenesis
Our studies in P7 mice (Fig. 5) indicated that Tbr2+ progenitors play an ongoing role in postnatal neurogenesis in the DG (Hodge et al. 2008), SVZ/RMS, and pPV. The adult SVZ produces γ-aminobutyric acid (GABA)ergic olfactory bulb neurons (reviewed by Zhao et al. 2008). Previously, Tbr2 has only been associated with glutamatergic neurogenesis (reviewed by Hevner et al. 2006); thus, our results suggest that either 1) Tbr2 may sometimes be expressed in GABAergic neurogenesis or 2) the SVZ/RMS system may produce some glutamatergic neurons, at least in the postnatal period. We are currently working to resolve this issue. The pPV was previously identified as a source of new pyramidal neurons in adult rats with ischemic damage to hippocampus (Nakatomi et al. 2002).
INP Functions in Cortical Development
Several hypotheses have been proposed concerning the function of INPs. Smart (1973) suggested that basal divisions provide a mechanism to relieve crowding in the VZ. Kriegstein et al. (2006) suggested that INPs provide neurons important to evolutionary expansion of the cerebral cortex surface area. Molnár et al. (2006) proposed that INPs are a mammalian specialization to produce upper cortical layers. We proposed the “radial amplification hypothesis” (Pontious et al. 2008) stating that INPs 1) amplify neuron production from radial unit progenitors and 2) provide a control point to modulate neuron amplification across temporal (laminar) and spatial (areal) coordinates, thus shaping the laminar structure of different cortical areas.
In showing that INP proliferation changes during cortical development (Fig. 8_B_,E), the present study supports the proposition that INP proliferation is regulated dynamically. Factors that may selectively regulate INP proliferation include transcription factors, such as Id4 (Yun et al. 2004); delta-notch signaling (Mizutani et al. 2007); secreted morphogens, such as Wnts (Zhou et al. 2006); glutamate and GABA (Haydar et al. 2000); and thalamic innervation (Dehay et al. 2001). Because INPs account for a large fraction of cortical neurogenesis, the factors that regulate INP proliferation should be an important subject for future investigation.
Funding
National Institutes of Health (K02 NS045018, R01 NS050248 to RFH); Deutsche Forschungsgemeinschaft (SPP 1109 Hu 275/7-3, SPP 1111 Hu 275/8-3, SFB/TR 13 B1, SFB 655 A2 to WBH); Deutsche Forschungsgemeinschaft–funded Center for Regenerative Therapies Dresden; the Fonds der Chemischen Industrie; Federal Ministry of Education and Research in the framework of the National Genome Research Network (NGFN-2, SMP RNAi, 01GR0402, PRI-S08T05).
Supplementary Material
Supplementary material can be found at http://www.cercor.oxfordjournals.org/.
[Supplementary Data]
Acknowledgments
We thank Dr Jhumku Kohtz (Northwestern University, Chicago) for anti-pan-Dlx and Dr David Anderson (Caltech, Pasadena) for anti-Ngn2. Conflict of Interest: None declared.
References
- Anthony TE, Klein C, Fishell G, Heintz N. Radial glia serve as neuronal progenitors in all regions of the central nervous system. Neuron. 2004;41:881–890. doi: 10.1016/s0896-6273(04)00140-0. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, Molnár Z, Robertson EJ, Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attardo A, Calegari F, Haubensak W, Wilsch-Bräuninger M, Huttner WB. Live imaging at the onset of cortical neurogenesis reveals differential appearance of the neuronal phenotype in apical versus basal progenitor progeny. PLoS ONE. 2008;3:e2388. doi: 10.1371/journal.pone.0002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayatti N, Moss JA, Sun L, Ambrose P, Ward JF, Lindsay S, Clowry GJ. A molecular neuroanatomical study of the developing human neocortex from 8 to 17 postconceptional weeks revealing the early differentiation of the subplate and subventricular zone. Cereb Cortex. 2008;18:1536–1548. doi: 10.1093/cercor/bhm184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulder Committee. Embryonic vertebrate central nervous system: revised terminology. Anat Rec. 1970;166:257–262. doi: 10.1002/ar.1091660214. [DOI] [PubMed] [Google Scholar]
- Britz O, Mattar P, Nguyen L, Langevin L-M, Zimmer C, Alam S, Guillemot F, Schuurmans C. A role for proneural genes in the maturation of cortical progenitor cells. Cereb Cortex. 2006;16:i138–i151. doi: 10.1093/cercor/bhj168. [DOI] [PubMed] [Google Scholar]
- Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- Calegari F, Haubensak W, Haffner C, Huttner WB. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci. 2005;25:6533–6538. doi: 10.1523/JNEUROSCI.0778-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehay C, Savatier P, Cortay V, Kennedy H. Cell-cycle kinetics of neocortical precursors are influenced by embryonic thalamic axons. J Neurosci. 2001;21:201–214. doi: 10.1523/JNEUROSCI.21-01-00201.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan A, Madathany T, de Castro SC, Gerrelli D, Guddati K, García-Añoveros J. Transient expression of the conserved zinc finger gene INSM1 in progenitors and nascent neurons throughout embryonic and adult neurogenesis. J Comp Neurol. 2008;507:1497–1520. doi: 10.1002/cne.21629. [DOI] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RAM, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas LM, Haffner C, Giger T, Khaitovich P, Nowick K, Birchmeier C, Pääbo S, Huttner WB. Insulinoma-associated 1 has a panneurogenic role and promotes the generation and expansion of basal progenitors in the developing mouse neocortex. Neuron. 2008;60:40–55. doi: 10.1016/j.neuron.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Fink AJ, Englund C, Daza RAM, Pham D, Lau C, Nivison M, Kowalczyk T, Hevner RF. Development of the deep cerebellar nuclei: transcription factors and cell migration from the rhombic lip. J Neurosci. 2006;26:3066–3076. doi: 10.1523/JNEUROSCI.5203-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gal JS, Morozov YM, Ayoub AE, Chatterjee M, Rakic P, Haydar TF. Molecular and morphological heterogeneity of neural precursors in the mouse neocortical proliferative zones. J Neurosci. 2006;26:1045–1056. doi: 10.1523/JNEUROSCI.4499-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohlke JM, Armant O, Parham FM, Smith MV, Zimmer C, Castro DS, Nguyen L, Parker JS, Gradwohl G, Portier CJ, et al. Characterization of the proneural gene regulatory network during mouse telencephalon development. BMC Biol. 2008;6:15. doi: 10.1186/1741-7007-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Götz M, Barde Y-A. Radial glial cells: defined and major intermediates between embryonic stem cells and CNS neurons. Neuron. 2005;46:369–372. doi: 10.1016/j.neuron.2005.04.012. [DOI] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat Rev Mol Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Götz M, Stoykova A, Gruss P. Pax6 controls radial glia differentiation in the cerebral cortex. Neuron. 1998;21:1031–1044. doi: 10.1016/s0896-6273(00)80621-2. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Molnár Z, Tarabykin V, Stoykova A. Molecular mechanisms of cortical differentiation. Eur J Neurosci. 2006;23:857–868. doi: 10.1111/j.1460-9568.2006.04626.x. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydar TF, Wang F, Schwartz ML, Rakic P. Differential modulation of proliferation in the neocortical ventricular and subventricular zones. J Neurosci. 2000;20:5764–5774. doi: 10.1523/JNEUROSCI.20-15-05764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins N, Malatesta P, Cecconi F, Nakafuku M, Tucker KL, Hack MA, Chapouton P, Barde Y-A, Götz M. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci. 2002;5:308–315. doi: 10.1038/nn828. [DOI] [PubMed] [Google Scholar]
- Heng JI, Nguyen L, Castro DS, Zimmer C, Wildner H, Armant O, Skowronska-Krawczyk D, Bedogni F, Matter JM, Hevner R, et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–118. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- Hevner RF. From radial glia to pyramidal-projection neuron: transcription factor cascades in cerebral cortex development. Mol Neurobiol. 2006;33:33–50. doi: 10.1385/MN:33:1:033. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Daza RAM, Englund C, Kohtz J, Fink A. Postnatal shifts of interneuron position in the neocortex of normal and reeler mice: evidence for inward radial migration. Neuroscience. 2004;124:605–618. doi: 10.1016/j.neuroscience.2003.11.033. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Hodge RD, Daza RAM, Englund C. Transcription factors in glutamatergic neurogenesis: conserved programs in neocortex, cerebellum, and adult hippocampus. Neurosci Res. 2006;55:223–233. doi: 10.1016/j.neures.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Hodge RD, Kowalczyk TD, Wolf SA, Encinas JM, Rippey C, Enikolopov G, Kempermann G, Hevner RF. Intermediate progenitors in adult hippocampal neurogenesis: Tbr2 expression and coordinate regulation of neuronal output. J Neurosci. 2008;28:3707–3717. doi: 10.1523/JNEUROSCI.4280-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacopetti P, Michelini M, Stuckmann I, Oback B, Aaku-Saraste E, Huttner WB. Expression of the antiproliferative gene TIS21 at the onset of neurogenesis identifies single neuroepithelial cells that switch from proliferative to neuron-generating division. Proc Natl Acad Sci USA. 1999;96:4639–4644. doi: 10.1073/pnas.96.8.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi A, Ikawa T, Kasukawa T, Ueda HR, Kurimoto K, Saitou M, Matsuzaki F. Single-cell gene profiling defines differential progenitor subclasses in mammalian neurogenesis. Development. 2008;135:3113–3124. doi: 10.1242/dev.022616. [DOI] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Hadjantonakis AK. Eomes::GFP—a tool for live imaging cells of the trophoblast, primitive streak, and telencephalon in the mouse embryo. Genesis. 2007;45:208–217. doi: 10.1002/dvg.20293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letinic K, Zoncu R, Rakic P. Origin of GABAergic neurons in the human neocortex. Nature. 2002;417:645–649. doi: 10.1038/nature00779. [DOI] [PubMed] [Google Scholar]
- Malatesta P, Hack MA, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, Götz M. Neuronal or glial progeny: regional differences in radial glia fate. Neuron. 2003;37:751–764. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Martínez-Cerdeño V, Noctor SC, Kriegstein AR. The role of intermediate progenitor cells in the evolutionary expansion of the cerebral cortex. Cereb Cortex. 2006;16:i152–i161. doi: 10.1093/cercor/bhk017. [DOI] [PubMed] [Google Scholar]
- Miyata T, Kawaguchi A, Saito K, Kawano M, Muto T, Ogawa M. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–3145. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- Mizutani K, Yoon K, Dang L, Tokunaga A, Gaiano N. Differential notch signalling distinguishes neural stem cells from intermediate progenitors. Nature. 2007;449:351–355. doi: 10.1038/nature06090. [DOI] [PubMed] [Google Scholar]
- Molnár Z, Métin C, Stoykova A, Tarabykin V, Price DJ, Francis F, Meyer G, Dehay C, Kennedy H. Comparative aspects of cerebral cortical development. Eur J Neurosci. 2006;23:921–934. doi: 10.1111/j.1460-9568.2006.04611.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatomi H, Kuriu T, Okabe S, Yamamoto S, Hatano O, Kawahara N, Tamura A, Kirino T, Nakafuku M. Regeneration of hippocampal pyramidal neurons after ischemic brain injury by recruitment of endogenous neural progenitors. Cell. 2002;110:429–441. doi: 10.1016/s0092-8674(02)00862-0. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–144. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martínez-Cerdeño V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30:24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, Barton SC, Surani MA, Ryan K, Nehls MC, et al. Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature. 2000;404:95–99. doi: 10.1038/35003601. [DOI] [PubMed] [Google Scholar]
- Sasaki S, Tabata H, Tachikawa K, Nakajima K. The cortical subventricular zone-specific molecule Svet1 is part of the nuclear RNA coded by the putative Netrin receptor gene Unc5d and is expressed in multipolar migrating cells. Mol Cell Neurosci. 2008;38:474–483. doi: 10.1016/j.mcn.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Scardigli R, Bäumer N, Gruss P, Guillemot F, Le Roux I. Direct and concentration-dependent regulation of the proneural gene Neurogenin2 by Pax6. Development. 2003;130:3269–3281. doi: 10.1242/dev.00539. [DOI] [PubMed] [Google Scholar]
- Scott IS, Morris LS, Bird K, Davies RJ, Vowler SL, Rushbrook SM, Marshall AE, Laskey RA, Miller R, Arends MJ, et al. A novel immunohistochemical method to estimate cell-cycle phase distribution in archival tissue: implications for the prediction of outcome in colorectal cancer. J Pathol. 2003;201:187–197. doi: 10.1002/path.1444. [DOI] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimojo H, Ohtsuka T, Kageyama R. Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron. 2008;58:52–64. doi: 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Smart IHM. Proliferative characteristics of the ependymal layer during the early development of the mouse neocortex: a pilot study based on recording the number, location and plane of cleavage of mitotic figures. J Anat. 1973;116:67–91. [PMC free article] [PubMed] [Google Scholar]
- Smart IHM, Dehay C, Giroud P, Berland M, Kennedy H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex. 2002;12:37–53. doi: 10.1093/cercor/12.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS., Jr The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 1995a;15:6046–6057. doi: 10.1523/JNEUROSCI.15-09-06046.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Nowakowski RS, Caviness VS., Jr Early ontogeny of the secondary proliferative population of the embryonic murine cerebral wall. J Neurosci. 1995b;15:6058–6068. doi: 10.1523/JNEUROSCI.15-09-06058.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarabykin V, Stoykova A, Usman N, Gruss P. Cortical upper layer neurons derive from the subventricular zone as indicated by Svet1 gene expression. Development. 2001;128:1983–1993. doi: 10.1242/dev.128.11.1983. [DOI] [PubMed] [Google Scholar]
- Viti J, Gulacsi A, Lillien L. Wnt regulation of progenitor maturation in the cortex depends on Shh or fibroblast growth factor 2. J Neurosci. 2003;23:5919–5927. doi: 10.1523/JNEUROSCI.23-13-05919.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-X, Goebbels S, Nakamura K, Nakamura K, Kometani K, Minato N, Kaneko T, Nave K-A, Tamamaki N. Pyramidal neurons of upper cortical layers generated by NEX-positive progenitor cells in the subventricular zone. Proc Natl Acad Sci USA. 2005;102:17172–17177. doi: 10.1073/pnas.0508560102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun K, Mantani A, Garel S, Rubenstein J, Israel MA. Id4 regulates neural progenitor proliferation and differentiation in vivo. Development. 2004;131:5441–5448. doi: 10.1242/dev.01430. [DOI] [PubMed] [Google Scholar]
- Zecevic N, Chen Y, Filipovic R. Contributions of cortical subventricular zone to the development of the human cerebral cortex. J Comp Neurol. 2005;491:109–122. doi: 10.1002/cne.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Zhou CJ, Borello U, Rubenstein JL, Pleasure SJ. Neuronal production and precursor proliferation defects in the neocortex of mice with loss of function in the canonical Wnt signaling pathway. Neuroscience. 2006;142:1119–1131. doi: 10.1016/j.neuroscience.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Zimmer C, Tiveron M-C, Bodmer R, Cremer H. Dynamics of Cux2 expression suggest that an early pool of SVZ precursors is fated to become upper cortical layer neurons. Cereb Cortex. 2004;14:1408–1420. doi: 10.1093/cercor/bhh102. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplementary Data]