Promoter CpG methylation contributes to ES cell gene regulation in parallel with Oct4/Nanog, Polycomb binding and histone H3 lys4/lys27 trimethylation (original) (raw)
. Author manuscript; available in PMC: 2011 Apr 4.
Published in final edited form as: Cell Stem Cell. 2008 Feb 7;2(2):160–169. doi: 10.1016/j.stem.2007.12.011
Abstract
We report here genome-wide mapping of DNA methylation patterns at proximal promoter regions in mouse embryonic stem cells (mESCs). Most methylated genes are differentiation-associated and repressed in mESCs. By contrast, the unmethylated gene set includes many housekeeping and pluripotency genes. By cross- referencing methylation patterns to genome-wide mapping of histone H3 lysine (K) 4/27 trimethylation and binding of Oct4, Nanog and Polycomb proteins on gene promoters, we found that promoter DNA methylation is the only marker of this group present on approximately 30% of genes, many of which are silenced in mESCs. In demethylated mutant mESCs, we saw upregulation of a subset of X-linked genes and developmental genes that are methylated in wild-type mESCs, but lack either H3 K4 and K27 trimethylation or association with Polycomb, Oct4 or Nanog. Our data suggest that in mESCs promoter methylation represents a unique epigenetic program that complements other regulatory mechanisms to ensure appropriate gene expression.
Introduction
Embryonic stem cells (ESCs), which are derived from the inner cell mass of blastocyst embryos, have the potential to differentiate into all cell types including germ cells in vitro and in vivo, thus representing an ideal system for studying regenerative medicine (Keller, 2005). Factors influencing ESC self renewal and differentiation include extracellular matrix, growth factors and cytokines, intracellular signaling molecules, transcription factors, and epigenetic regulators such as histone modification and DNA methylation (Keller, 2005). For example, key transcription factors such as Oct4 (encoded by Pou5f1), Nanog and Sox2 form a transcription regulatory network in ESCs that activates genes essential for ESC survival and proliferation while concurrently represses those target genes that will be only activated during cell differentiation (Boyer et al., 2005; Loh et al., 2006), thus playing an essential role in maintaining the pluripotency and self-renewal of ESCs (Avilion et al., 2003; Chambers et al., 2003; Mitsui et al., 2003; Nichols et al., 1998).
More recently, the role of chromatin structure and epigenetic modifications in controlling gene expression during ESC self-renewal and differentiation has been under intensive investigation (Bernstein et al., 2007, Guenther et al., 2007). For example, gene repression mediated by the Polycomb group (PcG) protein complex and the associated histone H3 lysine (K) 27 trimethylation is required for ESC self-renewal and pluripotency (Boyer et al., 2006; Lee et al., 2006). In addition, the genome of ESCs contains domains with “bivalent” histone modifications of both H3K4 and K27 trimethylation (H3K4me3 and H3K27me3) that mark a number of differentiation genes, including many transcription factors, which are repressed in ESCs but “poised” to be activated upon differentiation (Bernstein et al., 2006; Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007). However, the remaining approximately one third of genes are not marked by histone modifications of either H3K4me or H3K27me3, and yet are mostly repressed in ESCs (Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007). Our knowledge is also very limited as to how multiple regulatory mechanisms including transcriptional factors and epigenetic factors, such as histone modification and DNA methylation, are coordinated to control the “on” and “off” of pluripotent versus developmental genes in ESCs and during in vitro differentiation of ESCs.
DNA methylation in mammalian cells is postulated to play multiple roles in cell physiology including genome stability, repression of endogenous retroviral and transposable elements, genomic imprinting and developmental gene regulation (Bird, 2002; Jaenisch and Bird, 2003; Li, 2002 ; Robertson, 2005; Feng et al., 2007). During embryogenesis, levels of DNA methylation are dynamically regulated by the de novo DNA methyltransferase (Dnmt) 3a, Dnmt3b and maintenance enzyme Dnmt1 (Chen and Li, 2004; Goll and Bestor, 2005). Failure to place or maintain the patterns of DNA methylation leads to early embryonic lethality in mice (Li et al., 1992; Okano et al., 1999) and also many human diseases including cancer, Fragile-X, ICF, and ATRX syndromes (reviewed by Robertson, 2005). Interestingly, mESCs deficient for DNA methylation can survive and proliferate in an undifferentiated state (Meissner et al., 2005; Tsmura et al., 2006), but undergo rapid apoptotic cell death upon in vitro differentiation (Panning and Jaenisch, 1996). Therefore, it is still unclear as to whether DNA methylation plays any role in gene expression and the maintenance of pluripotency in mESCs.
In mESCs, each of the highly expressed Dnmts plays a specific role in the establishment and/or the maintenance of DNA methylation (Chen et al., 2003). The total level of methylcytosine in mESCs is similar to that in differentiated tissues such as kidney and liver cells in vivo (Biniszkiewicz et al., 2002). Several recent studies have attempted to identify changes in methylation patterns during long-term cultures or upon cell differentiation of ESCs. In both mouse and human ESCs, a subset of CpG islands are subject to de novo methylation during in vitro differentiation (Hattori et al., 2004; Kremenskoy et al., 2003; Shen et al., 2006). A more recent study demonstrated that an increase in DNA methylation occurs in selected CpG islands in a few lines of hESCs during long-term passages (Allegrucci et al., 2007). Overall, these studies suggest that methylation patterns in ESCs may be distinctly different from differentiated somatic cells and the methylation status of ESCs or ESC derivatives should be monitored carefully when they are used in regenerative medicine (Allegrucci et al., 2007; Shen et al., 2006).
We report here the first comprehensive genome-wide mapping of promoter methylation patterns in undifferentiated mouse ESCs. We further examine the relationship between DNA methylation, histone modifications, and the promoter occupancy of pluripotent regulators such as Polycomb group proteins (PcG) and Oct4/Nanog in regulating gene expression in mouse ES cells. Our results reveal that CpG methylation patterns complement other regulatory mechanisms in maintaining the unique transcriptional program of undifferentiated mESCs.
Results
Genome-wide profiling of promoter DNA methylation in mESCs
With the knowledge of genome-wide patterns of histone modifications, PcG binding, and Sox2/Nanog/Oct4 binding in mouse and human ESCs (Boyer et al., 2005; Boyer et al., 2006; Bernstein et al., 2006; Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al, 2007), we wanted to map DNA methylation patterns in gene promoters to address the role of DNA methylation in ESCs in the context of other regulatory mechanisms. One high-throughput method involves the enrichment of the methylated DNA through the use of immunoprecipitation with methylcytosine antibodies or methyl-binding domain from MeCP2 in a column (Cross et al., 1994; Weber et al., 2005). By coupling methylated DNA immunoprecipitation with DNA microarray chip technology (mDIP-Chip), genome-wide methylation profiles have been examined for human normal and cancer cell lines, as well as Arabidopsis thaliana (Keshet et al., 2006; Weber et al., 2005; Weber et al., 2007; Zhang et al., 2006; Zilberman et al., 2007). These studies have provided valuable insights into the function and evolution of DNA methylation in different types of cells and organisms.
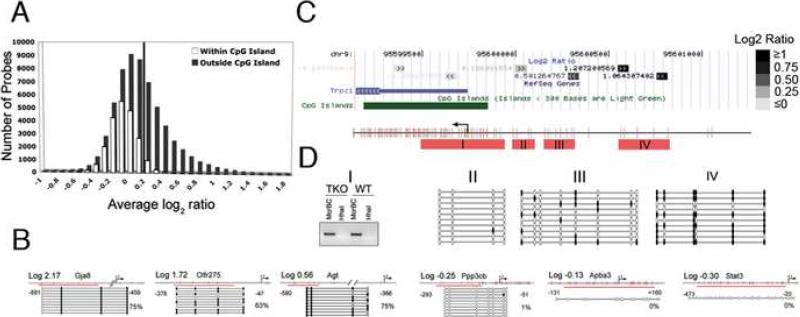
Using a previously described mDIP protocol (see Experimental Procedures) (Keshet et al., 2006, Zhang et al., 2006), we immunoprecipitated methylated DNA from a line of male wild-type (WT) mESCs (J1 cells) as well as J1-derived mutant ESCs [Dnmt3a-/-, Dnmt3b -/-; Dnmt1 KD (TKO)] that are virtually demethylated across the entire genome (Meissner et al., 2005). We performed cross-hybridization of pulldown DNA from WT and TKO cells with Agilent microarrays, which contain 15,561 annotated gene promoters with a resolution of one 60mer probe for every 200bp region (Table S1). To annotate the methylation status of each probe on each promoter, we first classified promoter probes into two subgroups that are either within a CpG island (>200bp, GC>50%, observed/expected CpG ratio>0.6) or in a region outside of CpG island. By this classification, approximately 25% of total probes are within CpG islands and 75% of probes are in non-CpG island promoter regions (Figure 1A). After plotting the averaged log2 ratios for each probe (WT over TKO) from seven replicates of independent mDIP/hybridization experiments, we found a clear bimodal distribution of probe sets within or outside CpG islands (Figure 1A). A majority of probes within CpG islands showed log2 ratios less than 0, suggestive of an unmethylated state. We confirmed the unmethylated status of nine promoter regions with probe log2 ratios<0 using established methylation assays such as bisulfite sequencing and McrBC/HpaII genomic PCRs (Figure 1B and Figure S1). In contrast, a large fraction of non-CpG island probes exhibit log2 ratios > 0, suggesting a methylated state. As shown in Figure 1B and summarized in Table S2, we confirmed that all 32 promoter regions we analyzed with probe log2 ratios > 0 were methylated. We further examined the methylation status of the entire promoter region by using the transient receptor potential channel 1 (Trpc1) gene locus on chromosome 9 as an example. The Trpc1 promoter contains probes both inside and outside of CpG islands with probe log2 ratios range from -0.20 to +1.06. We found that significant hybridization signals (log2 >0.2) are over the non-CpG island promoter regions (-250bp to -1200bp), but not near or over the CpG island (<0.2) (Figure 1C). Bisulfite genomic sequencing confirmed that the region containing the probes with signals less than 0.2 was either unmethylated or sparsely methylated. Moreover, a gradient of increasing methylation overlays the non-CpG island region in the promoter with hybridization signals >0.2, which is confirmed to be heavily methylated (Figure 1D). The distribution of averaged probe log2 ratios for each of 15,561 gene promoters can be visualized through an online genome browser as seen in Figure 1C.
Figure 1. Using mDIP-CHIP Assay to Profile the Promoter Methylation Pattern in Wild Type mESCs.
(A) All probes for the array are classified as either inside or within 50bp of a CpG island (white) or outside a CpG island (grey) and then grouped by their average log2 ratio. The solid line is placed at log2 ratio of +0.2 value to annotate the proportion of methylated gene promoters. (B) Using the average Log2 ratio, we selected 3 genes thought to be methylated as well as 3 that were thought to be unmethylated. For the bisulfite confirmation, each line represents an individual clone and each circle represents a CpG dinucleotide. The red line indicates the region we analyzed. Filled circles are methylated CpGs and open circles are unmethylated. (C) The average signals (Log2 ratio) for each probe (block with double arrowheads) in the promoter region are plotted in the UCSC genome browser – here we take the Trpc1 gene as an example. Note the dark probes annotate probes with high Log2 ratios whereas light color probes represent probes with low or negative Log2 ratios. (D) The confirmation of methylation status for the probes in the Trpc1 gene either inside or outside of a CpG island. Red bars (I-IV) indicate regions analyzed. Region I shows a gel picture of a McrBC-HhaI genomic PCR, indicating the unmethylated state of the CpG island of the Trpc1 transcription start site. II-IV show bisulfite genomic sequencing results of methylated probes upstream of the CpG islands, indicating a gradient of methylation towards the probes with high log2 ratios (filled dots are methylated CpG sites).
With the above data, we made a large-scale annotation of proximal promoters as either methylated or unmethylated by setting up the following stringent criterion. To be annotated as a methylated region, the average probe log2 ratios of WT over TKO in that promoter region should be ≥0.2 and the statistical significance of the difference between hybridization intensities of WT and TKO is P<0.01 (t-test). Using this criterion, we have annotated 6,127 genes (39.4%) as methylated, which contain at least one methylated domain surrounding the hybridized probe(s) in their proximal promoter (e.g. TrpC1, see Table S1). To be annotated as an unmethylated promoter, all probes on a promoter region should have average probe log2 ratios<0 with P<0.01 (t-test). Using this conservative criterion, we annotated 5,074 unique genes (32.6%) that are unmethylated in the entire proximal promoter region (Table S1). The remaining genes (28% out of total 15,561 promoters) that do not meet our designated criteria are excluded from further analysis.
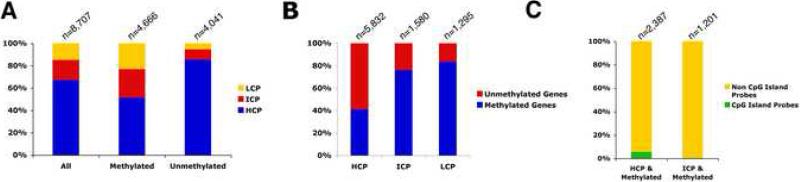
Based on their entire CpG contents across the genomic region, gene promoters have been recently classified into one of three categories: HCP (high CpG promoter that contains a 500 bp region with a GC content ≥0.55 and a CpG observed to expected ratio ≥0.6), LCP (low CpG promoter containing no 500 bp interval and with a CpG observed to expected ratio of ratio≥0.4, and ICP (intermediate CpG content promoter with CpG density between HCP and ICP) (Mikkelsen et al., 2007; Weber et al., 2007). Using the dataset generated by Mikkelsen et al. (2007), we further examined the CpG content in the pool of methylated versus unmethylated promoters in mESCs. We found that in the pool of methylated genes, 51% of them belong to the HCP cluster, which is much lower than the genome average (67 %) (Figure 2A). In contrast, in the pool of unmethylated genes, over 85% of genes are considered HCP promoters (Figure 2A). Detailed analysis of the distribution of methylated probes over the HCP promoters, which should contain at least a CpG island by the definition, indicated that only 3% of HCP genes have a methylated probe that overlaps with the CpG island itself (Figure 2C and Table S1). In contrast, in the pool of ICP and LCP genes, approximately 80% are annotated as methylated genes (Figure 2B). We conclude that DNA methylation in mESCs primarily takes place on ICP and LCP promoters or on non-CpG island regions of HCP promoters.
Figure 2. CpG Density of Methylated and Unmethylated Promoters.
(A) Classification of all promoters, methylated promoters or unmethylated promoters with High (HCP), Intermediate (ICP) and Low (LCP) CpG content. It should be noted that these classifications were assigned for V6.5 mouse ES cells which are similar but no the same as the J1 mouse ES cells used in our study. (B) Breakdown of methylation status for HCP, ICP and LCP promoters. (C) Percentage of genes with methylated CpG islands in HCP and ICP containing genes. By definition, there are no CpG islands in LCP containing genes.
Gene ontology analysis of unmethylated versus methylated promoters in mESCs
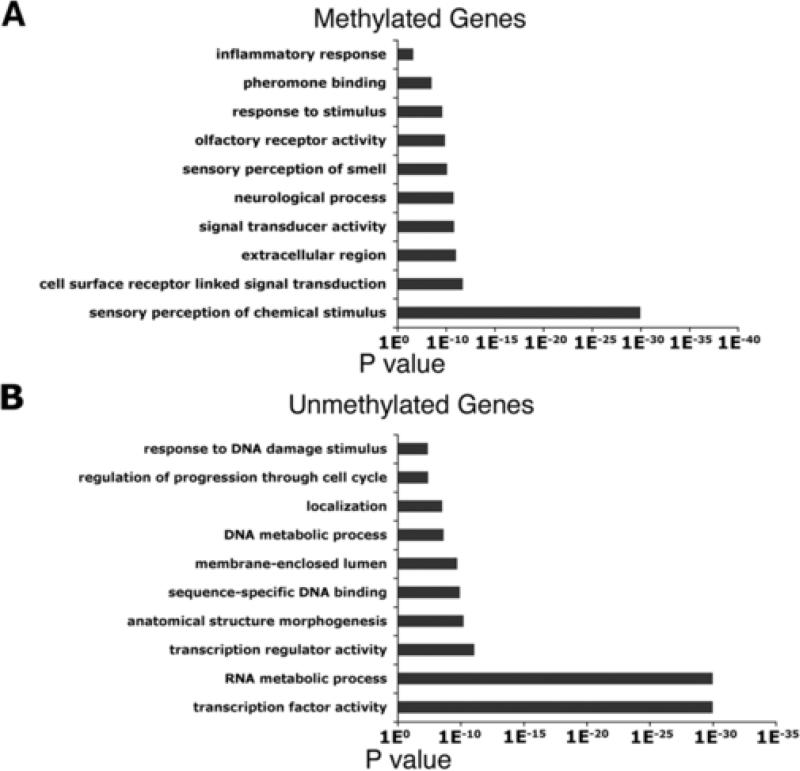
To further understand the role of DNA methylation in mESCs, we performed gene ontology analysis of the 6,127 methylated genes in mESCs. We found that methylated genes can be classified into response to stimuli and cell signaling molecules (Figure 3A and Table S3). This suggests that genes that exhibit DNA methylation are late differentiation-associated and signal transduction genes. Comparing these genes with gene expression datasets for mESCs in the GEO database also indicated that a majority of these genes are not expressed in mESCs (data not shown).
Figure 3. Comparision of Methylated and Unmethylated Genes with H3K4me3 and H3K27me3.
Gene ontology classifications for Methylated (A) or Unmethylated (B) genes. The GO term is on the y-axis and the p value indicating significance of enrichment is on the x-axis.
In contrast, gene ontology analysis on the list of 5,074 unmethylated genes further indicated that over 50% of unmethylated genes are associated with transcription machinery, protein and RNA metabolic process, and other cellular machinery essential for cell survival and proliferation (Figure 3B and Table S3). This result suggests that the unmethylated status of the proximal gene promoter is a good indicator for those genes that would be expressed in mESCs. In addition, gene ontology analysis showed that approximately 10-15% of unmethylated genes are classified as genes involved in cell differentiation and developmental process, which are not expressed in mESCs. Potentially, the repression of this small subset of development genes in mESCs could be through other mechanisms such as PcG and Oct4/Nanog complexes-mediated gene inhibition (see below) (Boyer et al. 2005, 2006; Lee et al. 2005; Loh et al. 2006).
Relationships between DNA methylation and histone modifications in mESCs
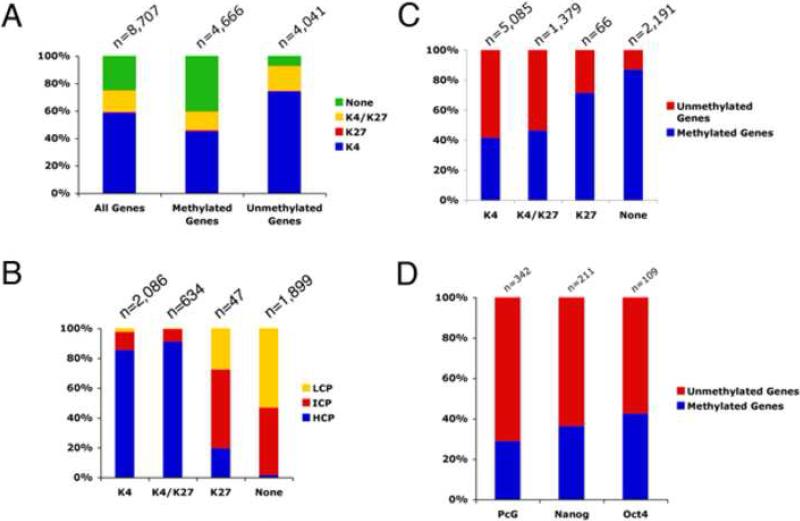
To further understand the role of epigenetic regulation in mouse ES cells, we compared our methylation data to recent whole genome histone mapping (Mikkelsen et al., 2007). Mapping of histone modifications in the promoter region of both mouse and human ES cells (Guenther et al., 2007; Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007) has shown four distinct populations of genes that are associated with either H3K4me3 or H3K27me3 alone, bivalent H3K4me3 and H3K27me3, or neither of these marks. H3 K4 and K27 trimethylation can be explained by the actions of Trithorax (trxG) and PcG complexes and possibly the occupancy of key transcription factors, but the absence of both H3K4me3 and H3K27me3 in a significant portion of genes (27-33% of all annotated genes) suggests the presence of other unique epigenetic marker(s) (Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007). By comparing the distribution across the genome (Mikkelsen et al. 2007), we found the subset of genes lacking both H3K4 and K27me3 marks are significantly enriched in methylated genes, but under-represented in unmethylated genes (Figure 4A). We noticed that the majority of genes without either H3 K4 or K27me3 are found in ICP and LCP genes (Mikkelsen et al. 2007), which are enriched in the pool of methylated genes in our study (Figures 4B and 2B). Moreover, examination of the overlap between methylated genes and the subset of genes lacking both H3K4 and K27me3 showed that approximately 87% of genes with neither of these marks are methylated in the proximal gene promoter regions (Figure 4C). In fact, the gene ontology terms for those genes without H3K4 and K27me3 are similar to the terms in our Gene Ontology analysis of methylated promoters in mESCs (data not shown; compare Figure 3A here with Fig. 5 in Pan et al. 2007). Thus, for the roughly one third of genes without H3K4 and K27me3, our analysis suggests that DNA methylation could be considered a distinguishing epigenetic mark.
Figure 4. Comparison of Methylated and Unmethylated Genes With H3K4me3 and H3K27me3 as Well as Nanog, Oct4, and PcG Bound Genes.
(A) Classification of all promoters, methylated promoters or unmethylated promoters with H3K4me3, H3k27me3, H3K4me3 & H3K27me3 (bivalent) or neither mark. Methylated genes are enriched for genes without either histone mark. (B) Percentage of K4, K4/K27, K27 or neither bound genes that are HCP, ICP or LCP. (C) Breakdown of methylation status for genes with H3K4me3 only, H3k27me3 only, H3K4me3 & H3K27me3 (bivalent) or neither mark. (D) Bar graph showing the breakdown of methylation status for PcG, Nanog or Oct4 bound genes in mESCs. The Nanog, Oct4 and PcG bound genes that are either methylated or unmethyalted are listed in Table S1.
Figure 5. Gene Expression Profiling of DNA Methylation Deficient mESCs.
(A) Gene ontology analysis for up-regulated genes in TKO cells shows enrichment of transcription factor activity, protein binding, extracellular region, and developmental genes. (B) Tissue specificity of over expression genes in TKO cells. Up-regulated genes were analyzed for the tissues they are normally expressed in using the GNF database. Note the over-representation of genes that are expressed in reproductive tissues including ovary/testis (16.2%) and placenta/umbilical cord (11.3%). (C) The chromosomal location of the 390 genes that are upregulated in TKO mESC compared to wild type mESC (clockwise from Chromosome 1 to X and Y sex chromosomes). The percentage of genes upregulated on the X chromosome is 14.5%. (D) Confirmation of Rhox2 and Magea3 that are upregulated in DNA methylation deficient mESCs by Q-PCR analysis. * P <0.05
To further analyze the significance of the presence of DNA methylation on the subpopulation of genes without H3K4 or K27me3, we used previously published expression array data (Mikkelsen et al., 2007) to assay whether the presence of DNA methylation is correlated with gene repression in mESCs. We found that approximately 80% of methylated genes lacking both histone H3K4 and K27me3 marks are not expressed in mouse ES cells (data not shown). We conclude that DNA methylation in proximal gene promoters is highly correlated with gene silencing for genes that lack both H3 K4 and K27 trimethylation.
For the pool of genes carrying H3K4me3 only, we found that unmethylated promoters make up approximately 60% (Figure 4C), consistent with the possibility that these genes are the most actively transcribed in mESCs (Mikkelsen et al., 2007). For the pool of genes carrying H3K4/K27me3 bivalent marks, we found that 53% are unmethylated (Figure 4C). This result suggests that repressive mechanisms associated with bivalent H3K4/K27 such as Polycomb-mediated gene silencing may be a predominant factor in controlling gene activities for this subset of genes in mESCs (see next section). However, DNA methylation could serve as a secondary repressive mechanism to further modulate the activities of these bivalent genes in mESCs or upon cell differentiation. Finally, for genes with only the H3K27me3 mark, although the pool of genes may be too small (n=66) to draw definitive conclusions, approximately 70% are methylated (Figure 4C).
Relationships between DNA methylation and PcG- or Oct4/Nanog- complex mediated gene regulation in mESCs
In mouse and human ESCs, Polycomb proteins were found to bind and repress a subset of developmental genes, rendering them “poised” for expression upon differentiation (Boyer et al., 2006; Lee et al., 2006). To determine directly whether polycomb targeted genes in mESCs also show promoter methylation, we compared the PcG-targeted genes in mESCs with our list of methylated (n=6,127) and unmethylated (n=5,074) promoters and found that only 28.7% (98 out of 342) exhibit promoter DNA methylation (Figure 4D and Tables S1,S4). This result suggests that the pool of genes targeted by DNA methylation and PcG are distinctively different. That a majority of PcG complex-repressed genes are unmethylated (71.3%) in mESCs is more compatible with the possibility that the PcG-targeted genes are poised be activated upon cell differentiation.
It is also known that transcription factors Oct4, Nanog, Sox2, and Stat3 are required for the pluripotency and self-renewal of mESCs. The gene promoters of Oct4, Nanog, Sox2, and Stat3 are all unmethylated, allowing for high levels of expression in mESCs (Table S1, Figure 1B) (Imamura et al., 2006). Oct4, Nanog, and Sox2 form a regulatory circuit that maintains their own expression and that of many other genes essential for ESC self-renewal, and at the same time represses differentiation genes (Boyer et al., 2005; Loh et al., 2006; Pan and Thomson, 2007, Walker et al., 2007). We therefore examined whether promoter methylation is correlated with either the activation or repression of genes by the Oct4/Nanog/Sox2 complex in mESCs. Because we were focusing on the relationship between proximal promoter methylation and occupancy of Oct4/Nanog, we confined our search to genes that are bound by Oct4/Nanog within 10 kb of the transcription initiation site and either methylated or unmethylated on our promoter array (109 genes bound by Oct4 and 211 target genes bound by Nanog). We found that approximately 42% and 36% of Oct4 and Nanog targeted genes, respectively, contain methylation domain(s) in the proximal promoter regions in mESCs (Figure 4D and Tables S1,S4). Conversly, 64% of Nanog targeted genes and 58% of Oct4-targeted genes are totally unmethylated in mESCs (Figure 4D and Tables S1, S4). To further confirm these results, we analyzed all bound loci for Nanog and Oct4, some of which are up to 500kb away from a known transcript. Again, we found similar results for all Nanog and Oct4 bound loci (46% of genes for both Nanog and Oct4 are methylated). When we looked to see if there was any association with gene expression, we found that 91-93% of unmethylated Nanog/Oct4 proximal bound genes were expressed (data not shown). When looking at genes that are methylated and bound by either Nanog or Oct4, we found that ~75% are expressed (data not shown). Thus, DNA methylation might play a small role in the dampening the expression of Nanog/Oct4 bound genes, but overall does not appear to have a strong effect. Taken together, our data favor the hypothesis that methylation-mediated repression is independent of Oct/Nanog-mediated gene expression. This suggests that DNA methylation could be a separate epigenetic regulator that works in parallel, but generally in a non-overlapping fashion with the action of the Polycomb-repression complexes and transcription factors such as Oct4 and Nanog in regulation of the pluripotency and differentiation program of mESCs.
Comparison of gene expression in methylation proficient- and deficient- mESCs
To examine whether DNA methylation directly regulates the expression of genes in mESCs, we carried out genome-wide gene expression analysis (see material and methods) in wild-type and methylation-deficient mESCs that lack all three Dnmts (TKO cells) (Meissner et al., 2005). The TKO ESCs are virtually demethylated across the entire genome, but exhibit similar cell proliferation properties and embryonic stem cell markers to the parental WT J1 mESCs. In the first round of bioinformatics analysis with a fold change cutoff of 2 (see Experimental Procedures), our genome-wide expression profiling yielded a list of 337 genes that are up-regulated and 113 genes that are down regulated in the TKO cells compared to wild type (Table S5). Using a less strict filter that accounts for genes that may be turned completely off in the wild type condition but slightly expressed in the TKO cells (Ohm et al., 2007) we found an additional 53 genes that were up-regulated upon loss of DNA methylation. The inability of methylation-deficient ESCs to differentiate can result in potential inaccuracies when analyzing genes that are down regulated. For example, the existence of a small percentage of partially differentiated cells found in wild-type ESC cultures but not in the TKO cultures (due to cell death) could contribute to the list of down-regulated genes in TKO cells. Therefore, the analysis of up-regulation of genes in TKO cells versus wild-type cells is more robust for ascertaining the effect of DNA demethylation on gene expression.
Gene ontology analysis of these up-regulated genes shows an over representation of tissue specific genes, such as transcription factors and signaling molecules (Figure 5A). We then examined the tissue specificity of each up-regulated gene and found that testis- and oocyte-specific genes were highly enriched in the TKO cell line (Figure 5B, Table S6). We were interested to see whether there are any genomic loci that were enriched in deregulated genes. When we mapped the list of genes that were up-regulated in TKO mESCs to their loci, we found that a high percentage are located on the X chromosome (14.5% of observed values versus 4% of expected) (Figure 5C). We confirmed the microarray data by real time PCR for a few genes, including the X-linked genes Rhox2 and Magea3 (Figure 5D and unpublished data). The up-regulation of many X-linked genes was also found in a separate line of methylation-deficent mESCs due to Dnmt1 gene deletion (Lei, et al., 1996) (Figure S2), consistent with the notion that DNA demethylation is linked to the up-regulation of this subset of X-linked genes.
We next looked to see what histone marks the TKO upregulated genes had and whether they were also marked by DNA methylation. When we look at the breakdown of histone marks amongst the TKO upregulated genes, we find that 40% are lacking both H3K4 and K27me3, 35% are bivalent K4/K27me3, 23% are K4me3, and 2% are K27me3 only. Of the TKO up regulated that are not marked by either histone mark, we find that 85% are methylated in wildtype mESCs (Table S7). In fact, the X-linked Rhox family genes that are up-regulated in the absence of DNA methylation fall in this category (Figure 5D). This result suggests that DNA methylation is one of the major repressive mechanisms for a subset of genes that lack both H3K4 and K27me3 in mESCs. In contrast, when we examine the methylation status for the subset of genes with bivalent H3K4/K27me3 or H3K4me3 only in wildtype mESCs that would be upregulated in TKO cells, we found they are not as frequently methylated as genes without H3K4 or K27me3 (37% and 50%, respectively).
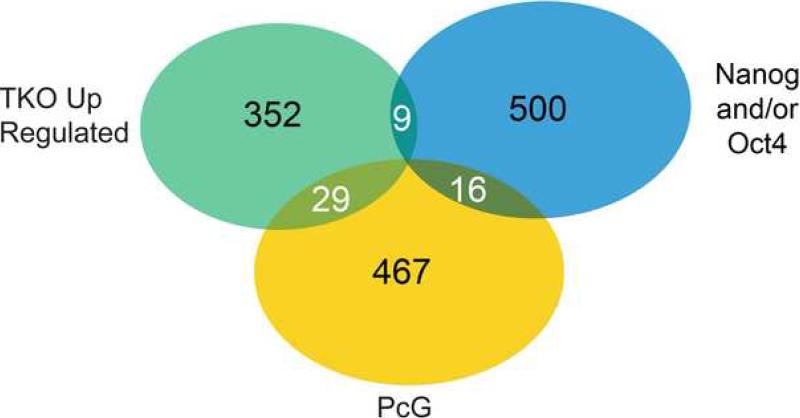
To determine whether DNA demethylation also affects the expression of any genes that are repressed by PcG or Oct4/Nanog/Sox2, we compared the genes that are up-regulated in the DNA methylation deficient mouse ES cells to the list of genes that are bound by the PcG complex and Oct4/Nanog (Boyer et al., 2006; Loh et al., 2006). Interestingly, only 5.7% (29/512) of up-regulated genes in TKO mESCs overlap with the PcG bound genes (Figure 6 and Table S7). This result provides further evidence that the genes repressed by PcG are distinct from the DNA methylated gene set. Similarly, we found that only 1.7% (9/525) of Nanog and/or Oct4 proximal promoter (within 10kb) bound genes [or 1.2% (37/3,080) of all Nanog and/or Oct4 bound genes (up to 500kb from the transcript)] (Loh et al., 2006) overlap with the up-regulated genes in demethylated ESCs (Figure 6 and Table S7), although it should be noted that the majority of genes that are bound by Oct4/Nanog and methylated are expressed even in WT cells. Nevertheless, these results also support the conclusion that DNA methylation and the transcriptional factor complex containing Oct4/Nanog play distinct roles in gene regulation in pluripotent mESCs.
Figure 6. Comparison of Upregulated Genes in Demethylated mESCs With Oct4, Nanog and PcG Bound Genes.
Venn diagram showing the minimal overlap between the TKO upregulated genes (green), Nanog and/or Oct4 bound genes (within 10kb proximal promoter region) (blue), and PcG bound genes (yellow). It should be noted that there is one gene, Podxl, which overlaps with all three categories. This analysis includes all genes, not just those present on the promoter methylation array. The Nanog, Oct4 (Loh et al., 2006) and PcG (Boyer et al., 2006) genes that overlap with TKO upregulated genes are listed in Table S7.
Discussion
ESCs represent a unique type of stem cell that can undergo indefinite cycles of self-renewal while maintaining pluripotency. Previous studies have identified many crucial gene transcription factors and regulatory networks that are required for maintaining the “stemness” of ESCs (Avilion et al., 2003; Chambers et al., 2003; Ivanova et al.,2006; Mitsui et al., 2003; Nichols et al., 1998; Walker et al, 2007). One of the insightful conclusions is that Oct4, Nanog, and Sox2 form transcriptional circuitry to activate their own expression in a forward feedback manner; furthermore, the Oct4/Nanog/Sox2 complex promotes expression of those genes required for the self-renewal of ESCs, but represses the developmental genes that will only be activated upon cell differentiation (Boyer et al., 2006; Loh et al., 2006). In addition, along with Oct4/Nanog/Sox2, Trithorax- and Polycomb-complex mediated histone modifications are also involved in the activation or repression genes, as well as in maintaining the “poised” nature of some genes (Mikkelsen et al., 2007; Walker et al., 2007; Pan et al., 2007; Zhao et al., 2007). In this report, we have comprehensively mapped CpG methylation in the proximal gene promoter regions and identified 6,127 methylated and 5,074 unmethylated proximal gene promoters. Our data help refine the emerging epigenetic landscape of mESCs. By comparing promoter DNA methylation with histone modifications, we can gain insights into how overlapping or independent epigenetic regulators regulate particular sets of genes (Figure 7). One of the novel findings in this study is that DNA methylation occurs in approximately 87% of the genes in ESCs that lack either H3K4me3 and H3K27me3 (Figure 4B). This population of methylated genes could potentially constitute close to one-third of all annotated genes in mouse and human ESCs (Mikkelsen et al., 2007; Pan et al., 2007; Zhao et al., 2007). Therefore, we conclude that DNA methylation in proximal gene promoter regions represents another major epigenetic marker that can distinguish different classes of genes in undifferentiated ESCs (Figure 7).
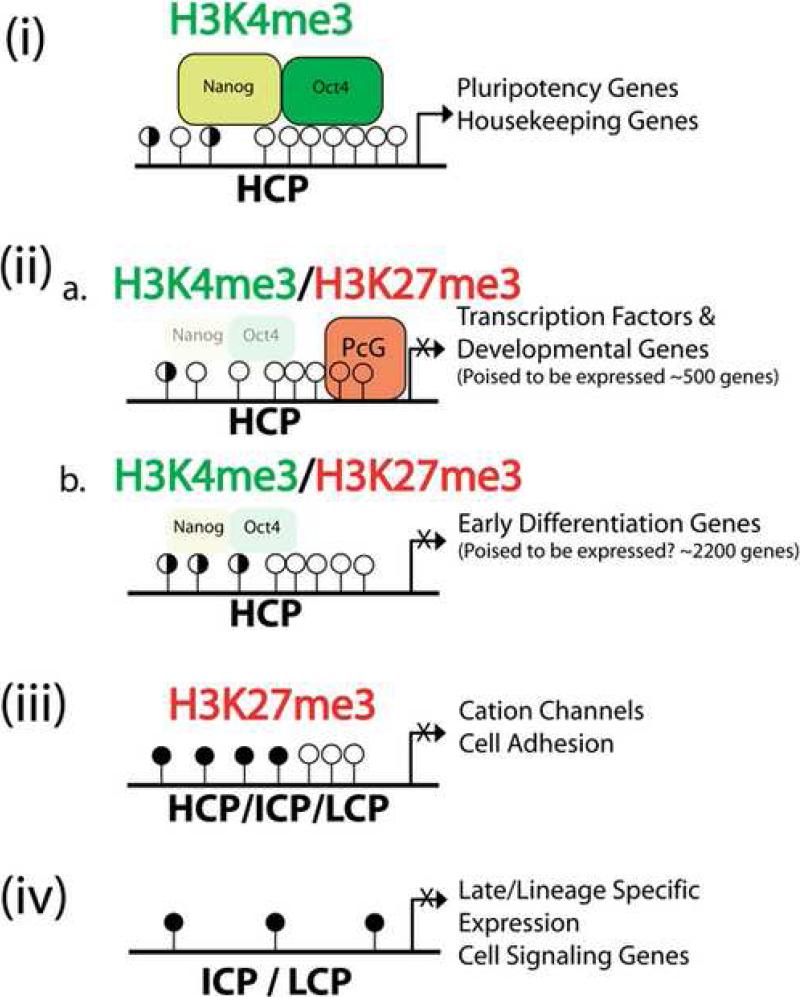
Figure 7. A Schematic Summary of Epigenetic and Transcriptional Regulation in mESCs.
The patterns of DNA methylation and histone modifications are depicted into four subclasses of gene promoters with H3K4me3 (i), bivalent genes (ii) with (a.) or without (b.) polycomb binding, H3K27me3 only (iii), and no histone marks (iv). Gene promoters are further annotated as HCP, ICP, and LCP that corresponds to different density of CpG dinucleotides and DNA methylation. Open circles designate unmethylated CpG sites, filled circles represent methylated CpG sites and half filled circles represent promoters which can be found either methylated or unmethylated. Unmethylated CpG islands are enriched in HCP promoters with H3K4me only or bivalent H3K4/K27 marks. Methylated CpGs are enriched in LCP and ICP gene promoters without H3 K4 and K27 trimethylation or with K27me3 only. The preferential interaction of Polycomb proteins and Oct4/Nanog complex with different classes of gene promoter is also illustrated. Different classes of genes that are either expressed or repressed in mESCs are listed next to each type of promoters.
In this study, we provided evidence that DNA methylation is causally linked to the silencing of a cluster of X-linked genes and a subset of developmentally regulated genes. It should be noted that the mESCs used in these experiments are male (XY) and lack X-inactivation; therefore, up-regulation of X-linked genes in TKO mESCs cannot be related to de-regulation of X-inactivation. However, it is known that genes involved in germ cell differentiation and sex development are over-represented on the X-chromosome (Wang et al., 2001), and many of these genes tend to be duplicated on the X-chromosome. Furthermore, DNA methylation is proposed to directly silence many genes involved in germ cell development (Maatouk et al., 2006). Indeed, Rhox family genes which have been shown to be duplicated extensively on the X chromosome, are repressed by DNA methylation in somatic cells and ESCs (Maclean et al., 2005; MacLean et al., 2006; Oda et al., 2006). Similarly, DNA methylation represses the Mage gene family and the Dazl gene that are related to germ cell development (Chuang et al., 2005; De Smet et al., 1999; Maatouk et al., 2006). Finally, demethylation-induced over-expression of Mage family genes is also observed in somatic cells treated with 5’azacytodine to inhibit DNMTs (Chuang et al., 2005). It is worth noting that the up-regulation of a subset of X-linked and development genes apparently does not interfere with the self-renewal of demethylated ESCs. Thus, ESCs can tolerate the over-expression of a subset of genes that serve a specialized function in germ cell or other types of somatic cells.
Although our study provides direct evidence that DNA demethylation can induce gene activation for a number of cell differentiation genes, we also found many genes are not up-regulated in the absence of DNA methylation. This result is in contrast to the result observed in demethylated primary fibroblasts, in which up to 10% of expressed genes can be up-regulated compared to wildtype primary fibroblasts (Jackson-Grusby, et al. 2001). Genome-wide mapping of histone modifications indicated that bivalent H3K4/K27me3 is more widespread than the association of Polycomb proteins (Boyer, et al., 2006; Mikkelsen, et al., 2007; Figure 7). Moreover, many methylated genes also contain H3K27me3 or bivalent H3K4/K27me3 markers in mESCs, raising the possibility that repressive histone marks can to some extent compensate for loss of DNA methylation in gene repression. It is also possible that compensatory repressive mechanisms have become activated during generation and culture of the TKO cell line. In addition, a lack of proper gene transcription activators in mESCs for those demethylated genes may also account for the continued inactive status of a majority of demethylated genes in TKO mESCs. Our results are consistent with the notion that DNA demethylation is necessary but not sufficient for gene activation. Conversely, methylation of a promoter is not always sufficient for gene repression. Using previously published gene expression data for mESCs, we found that up to 36% of genes are still expressed even if methylated in the proximal promoter. However, 80% of the expressed genes that exhibit promoter methylation are marked by the active histone H3K4me3 mark and are HCP genes. These observations are consistent with the suggestion by Weber et al. (2007) that a low density of DNA methylation in a gene promoter may not be sufficient to silence gene transcription by itself.
The mapping of promoter methylation patterns in mESCs could provide insights into why mESCs are a good cell source for somatic nuclear transfer experiments when compared to differentiated somatic cells (Yamanaka, 2007). One of the major hurdles in somatic nuclear transfer experiments is the efficiency of epigenetic reprogramming. It has been shown that a panel of genes including Oct4 and Dppa4 are only partially reactivated in somatic nuclear reconstituted blastocyst embryos, which could be attributed to the incomplete demethylation of Oct4 and Dppa family genes in somatic nuclear during reprogramming (Bortvin et al., 2003). Our methylation mapping indicated that both Oct4 and Dppa4 genes are demethylated in mESCs, supporting the notion that ESCs are more easily reprogrammed than somatic nuclei.
In summary, our comprehensive mapping of DNA methylation of gene promoters in mESCs provides a valuable resource for understanding the function of DNA methylation in the maintenance of self-renewal and pluripotency. Furthermore, the methylation patterns in gene promoters may also represent an epigenetic code that underlies the program of lineage specific differentiation.
Experimental Procedures
ES Cell Cultures
ES cells were maintained as previously described (Meissner et al., 2005). RNA was isolated using Trizol (Invitrogen) while DNA was isolated using DNA Lysis Buffer and then Phenol:Chloroform extracted.
mDIP
The methylated DNA immunoprecipitation (mDIP) method was adapted from a recent study (Zhang et al., 2007; Keshet et al., 2006). Briefly, 2ug of DNA was immunoprecipitated with 20ug of a monoclonal antibody to 5-me-cytosine. The immunoprecipitated DNA was washed, eluted and quantitated for microarray hybridization. For details, see supplemental data.
Microarray Hybridization
Gene expression microarrays were done with Agilent Whole Genome microarrays (G4122A) using the suggested protocol. These arrays were performed in triplicate Methylation microarray hybridizations were done with Agilent custom mouse promoter microarrays that covered approximately -800 to +200bp of 15,561 genes. We labeled 250ng of J1 and TKO mESC DNA for each array. We performed 7 replicates of the methylation arrays. For details, see supplemental data
Bisulfite Conversion and Sequencing
Bisulfite conversion was performed as described (Shen et al., 2006) Briefly, we digested genomic DNA with BglII overnight. Digested DNAs were then incubated with a sodium bisulfite solution for 16 hours. Bisulfite treated DNA was then desalted and precipitated. We used 1/10 of precipitated DNA for each PCR. For PCR, we used nested primers to generate our products. PCR products were gel purified and used for either Topo Cloning (Invitrogen) or direct PCR sequencing.
Quantitative Reverse Transcription PCR
RNA was DNase I treated (Invitrogen) and then quantified again. cDNA conversion was done using the iScript kit (BioRad). Quantitative PCR was done on a MyIQ Thermocycler (BioRad) using the Sybr Green Supermix (BioRad).
Statistical Methods
Detailed descriptions are found in the Supplemental Data
UCSC Genome Tracks
Methylation data can be viewed at http://epigenomics.mcdb.ucla.edu/mESC/
Supplementary Material
01
02
03
04
05
06
07
08
09
Acknowledgements
We thank Steve Smale and Leah Hutnick for critical reading of the manuscript. We also acknowledge the initial technical help from Xiaoyu Zhang, Steve Jacobsen, and Howard Cedar in the mDIP protocol, and from Stan Nelson at UCLA for help with the microarray hybridization. We also express our gratitude to Tim Baxter, David Hirschberg & Garrick Peters at Agilent for their initial support. Funding for this work is from NIH RO1 grants (NS044405 and NS051411) and California Institute for Regenerative Medicine (CIRM RC1-0111-01) to GF. SF was supported by pre-doctoral NRSA 1F31MH070204. YS was supported by a CIRM predoctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Allegrucci C, Wu YZ, Thurston A, Denning CN, Priddle H, Mummery CL, Ward-van Oostwaard D, Andrews PW, Stojkovic M, Smith N, et al. Restriction Landmark Genome Scanning identifies culture-induced DNA methylation instability in the human embryonic stem cell epigenome. Hum Mol Genet. 2007;16:1253–1268. doi: 10.1093/hmg/ddm074. [DOI] [PubMed] [Google Scholar]
- Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, Mastrangelo MA, Jun Z, Walter J, Jaenisch R. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol. 2002;22:2124–2135. doi: 10.1128/MCB.22.7.2124-2135.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, Yanagimachi R, Page DC, Jaenisch R. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130:1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr Top Dev Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang JC, Yoo CB, Kwan JM, Li TW, Liang G, Yang AS, Jones PA. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2'-deoxycytidine. Mol Cancer Ther. 2005;4:1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- Cole SW, Galic Z, Zack JA. Controlling false-negative errors in microarray differential expression analysis: a PRIM approach. Bioinformatics. 2003;19:1808–1816. doi: 10.1093/bioinformatics/btg242. [DOI] [PubMed] [Google Scholar]
- Cross SH, Charlton JA, Nan X, Bird AP. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol. 1999;19:7327–7335. doi: 10.1128/mcb.19.11.7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Fouse S, Fan G. Epigenetic regulation of neural gene expression and neuronal function. Pediatr Res. 2007;61:58R–63R. doi: 10.1203/pdr.0b013e3180457635. [DOI] [PubMed] [Google Scholar]
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. doi: 10.1146/annurev.biochem.74.010904.153721. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori N, Abe T, Hattori N, Suzuki M, Matsuyama T, Yoshida S, Li E, Shiota K. Preference of DNA methyltransferases for CpG islands in mouse embryonic stem cells. Genome Res. 2004;14:1733–1740. doi: 10.1101/gr.2431504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura M, Miura K, Iwabuchi K, Ichisaka T, Nakagawa M, Lee J, Kanatsu-Shinohara M, Shinohara T, Yamanaka S. Transcriptional repression and DNA hypermethylation of a small set of ES cell marker genes in male germline stem cells. BMC Dev Biol. 2006;6:34. doi: 10.1186/1471-213X-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, Jaenisch R. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev. 2005;19:1129–1155. doi: 10.1101/gad.1303605. [DOI] [PubMed] [Google Scholar]
- Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, Pikarski E, Young RA, Niveleau A, Cedar H, Simon I. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- Kremenskoy M, Kremenska Y, Ohgane J, Hattori N, Tanaka S, Hashizume K, Shiota K. Genome-wide analysis of DNA methylation status of CpG islands in embryoid bodies, teratomas, and fetuses. Biochem Biophys Res Commun. 2003;311:884–890. doi: 10.1016/j.bbrc.2003.10.078. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei H, Oh SP, Okano M, Juttermann R, Goss KA, Jaenisch R, Li E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Maatouk DM, Kellam LD, Mann MR, Lei H, Li E, Bartolomei MS, Resnick JL. DNA methylation is a primary mechanism for silencing postmigratory primordial germ cell genes in both germ cell and somatic cell lineages. Development. 2006;133:3411–3418. doi: 10.1242/dev.02500. [DOI] [PubMed] [Google Scholar]
- Maclean JA, 2nd, Chen MA, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C, Wilkinson MF. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–382. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- MacLean JA, 2nd, Lorenzetti D, Hu Z, Salerno WJ, Miller J, Wilkinson MF. Rhox homeobox gene cluster: recent duplication of three family members. Genesis. 2006;44:122–129. doi: 10.1002/gene.20193. [DOI] [PubMed] [Google Scholar]
- Meissner A, Gnirke A, Bell GW, Ramsahoye B, Lander ES, Jaenisch R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 2005;33:5868–5877. doi: 10.1093/nar/gki901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Oda M, Yamagiwa A, Yamamoto S, Nakayama T, Tsumura A, Sasaki H, Nakao K, Li E, Okano M. DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev. 2006;20:3382–3394. doi: 10.1101/gad.1470906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Pan G, Tian B, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-Genome Analysis of Histone H3 Lysine 4 and Lysine 27 Methylation in Human Embryonic Stem Cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- Panning B, Jaenisch R. DNA hypomethylation can activate Xist expression and silence X-linked genes. Genes Dev. 1996;10:1991–2002. doi: 10.1101/gad.10.16.1991. [DOI] [PubMed] [Google Scholar]
- Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- Shen Y, Chow J, Wang Z, Fan G. Abnormal CpG island methylation occurs during in vitro differentiation of human embryonic stem cells. Hum Mol Genet. 2006;15:2623–2635. doi: 10.1093/hmg/ddl188. [DOI] [PubMed] [Google Scholar]
- Tsumura A, Hayakawa T, Kumaki Y, Takebayashi S, Sakaue M, Matsuoka C, Shimotohno K, Ishikawa F, Li E, Ueda HR, et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Walker E, Ohishi M, Davey RE, Zhang W, Cassar PA, Tanaka TS, Der SD, Morris Q, Hughes TR, Zandstra PW, Stanford WL. Prediction and Testing of Novel Transcription Networks Regulation Embryonic Stem Cell Self-Renewal and Commitment. Cell Stem Cell. 2007;1:71–86. doi: 10.1016/j.stem.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and New Developments in the Generation of Patient-Specific Pluripotent Stem Cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, Ecker JR. Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell. 2006;126:1189–1201. doi: 10.1016/j.cell.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng HH, Wei CL. Whole-Genome Mapping of Histone H3 Lys4 and 27 Trimethylations Reveals Distinct Genomic Compartments in Human Embryonic Stem Cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet. 2007;39:61–69. doi: 10.1038/ng1929. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05
06
07
08
09