A pseudo-atomic model of the dynamin polymer identifies a hydrolysis-dependent powerstroke (original) (raw)
. Author manuscript; available in PMC: 2012 Sep 30.
Summary
The GTPase dynamin catalyzes membrane fission. Though this process requires dynamin assembly, G domain dimerization and stimulated GTP hydrolysis, the underlying structural interactions and conformational changes remain a mystery. Here we present the GMPPCP-bound structures of the truncated human dynamin 1 helical polymer at 12.2Å and a fusion protein linking human dynamin 1’s catalytic G domain to its GTPase effector domain (GG) at 2.2Å. Newly resolved density features in the polymer reconstruction and the unique conformation of GGGMPPCP allowed us to position crystallized dynamin fragments in the assembled structure and define their connectivity. The resulting model shows that G domain dimers only form between tetramers in sequential rungs of the dynamin helix. Using chemical crosslinking, we demonstrate that dynamin tetramers are dimers of domain-swapped dimers. Structural comparison of GGGMPPCP to the GG transition-state complex identifies a hydrolysis-dependent powerstroke that may play a role in membrane remodeling events necessary for fission.
INTRODUCTION
Clathrin-mediated endocytosis (CME) is a highly regulated pathway wherein nutrients, growth factors, and macromolecules are concentrated in invaginating clathrin-coated pits (CCPs) that pinch-off to form vesicles to carry these cargo into the cell (McMahon and Boucrot, 2011). The large, multidomain GTPase dynamin assembles into collars at the necks of deeply invaginated CCPs to catalyze membrane fission in the final stages of CME (Mettlen et al., 2009; Schmid and Frolov, 2011).
Purified dynamin exists as a tetramer (Muhlberg et al., 1997) that can self-assemble into helical structures reminiscent of collars observed in vivo (Hinshaw and Schmid, 1995). Dynamin encodes five domains (Figure S1A): a catalytic G domain, a middle domain involved in self-assembly and oligomerization, a membrane binding pleckstrin homology (PH) domain, a GTPase effector domain (GED), and a C-terminal proline and arginine rich domain (PRD) that binds SH3 domains of accessory proteins important for CME (Praefcke and McMahon, 2004; Mettlen et al, 2009) but is not essential for GTPase activities or oligomerization in vitro (Muhlberg et al., 1997). Aside from the PRD, structures of all of dynamin’s individual domains or their homologs have been solved by crystallography (Figure S1A). These include the human dynamin 1 PH domain (Ferguson et al., 1994; Timm et al., 1994), the G domains of rat dynamin (Reubold et al., 2005) and dictyostelium dynamin A (Niemann et al., 2001), the middle domain and GED of the related interferon-induced GTPase MxA (Gao et al., 2010), and a fusion linking the C-terminus of human dynamin 1’s GED (CGED) to its G domain (GG) (Chappie et al., 2010). Crystallographic and biochemical studies have shown that the CGED forms a three-helix bundle with the N- and C-termini of the G domain (NGTPase and CGTPase respectively) (Figure S1B) and that this module –the bundle signaling element (BSE) – transmits the conformational changes associated with dynamin assembly to the G domain (Chappie et al., 2009, 2010). However, as the BSE was structurally characterized in the context of the GG fusion, it is not known whether CGED’s interaction with the G domain occurs in cis within the same polypeptide or in trans via another polypeptide in the dynamin tetramer.
Dynamin has a low affinity for guanine nucleotides (10–100 μM) and a high basal turnover (~0.4–1 min−1) (Praefcke and McMahon, 2004). Assembly into helical oligomers stimulates dynamin’s basal GTPase activity >100 fold (Warnock et al. 1996; Stowell et al., 1999). This enhancement arises from G domain dimerization, which optimally positions dynamin’s catalytic machinery and stabilizes conformationally flexible switch regions (Chappie et al., 2010). Mutations that impair GTP binding, assembly, or stimulated GTP hydrolysis also cause defects in endocytic uptake in vivo (reviewed in Schmid and Frolov, 2011), thus establishing the importance of dynamin’s GTPase activities in CME.
Despite its essential role in CME, the mechanism of dynamin-catalyzed membrane fission remains poorly understood. Efforts to recapitulate these activities in vitro using synthetic membranes suggested dynamin functions as a mechanochemical enzyme that actively severs the membrane via hydrolysis-dependent conformational changes (Sweitzer and Hinshaw, 1998; Stowell et al., 1999; Chen et al., 2004; Mears et al., 2007; Roux et al., 2006) that generate a constricted neck and impose strain on the membrane lipids (Bashkirov et al., 2008; Roux et al., 2010). GTP hydrolysis also promotes partial dissociation of dynamin subunits from membranes (Danino et al, 2004; Ramachandran and Schmid, 2008; Pucadyil and Schmid, 2008; Bashkirov et al, 2008). Loosening of the dynamin scaffold could allow local lipid rearrangements and an energetically favorable hemifission intermediate that promotes non-leaky membrane scission (Bashkirov et al., 2008; Schmid and Frolov, 2011). The hydrolysis-dependent conformational changes that trigger these membrane-remodeling events have yet to be elucidated.
Unraveling the mechanisms governing dynamin-catalyzed membrane fission requires a detailed structural understanding of the architecture of assembled dynamin and the conformational changes induced by stimulated GTP hydrolysis. Dynamin’s propensity to form helical arrays in vitro has previously been exploited for cryo-electron microscopy (cryo-EM) structure determination. Three-dimensional reconstructions of truncated dynamin 1 (ΔPRD, Figure S1A) polymers assembled on anionic lipid-scaffolds have been obtained both in the absence of nucleotides (Chen et al., 2004) and in the presence of the non-hydrolyzable GTP analog GMPPCP (Zhang and Hinshaw, 2001). In both cases, the asymmetric unit of assembly is a dimer that adopts a T-shape when viewed in cross-section (‘T-view’). The structural differences between these maps suggest that rearrangements in the middle domain and GED mediate a nucleotide-dependent constriction of the ΔPRD assembly (Chen et al., 2004). Constriction alone, however, is not sufficient for membrane fission (Ramachandran and Schmid, 2008; Bashkirov et al., 2008), suggesting additional conformational changes are required. While it has been inferred that the middle domain and GED form a coiled-coil ‘stalk’ that connects the PH domain ‘leg’ to the G domain ‘head’ (Zhang and Hinshaw, 2001; Chen et al., 2004), neither the organization nor their connectivity in the polymer is known, owing to the low-resolution (>20Å) of the ΔPRD reconstructions and the lack of a complete, atomic-resolution dynamin structure. These limitations have also hindered our understanding of how assembly promotes G domain dimerization, leading to stimulated GTP hydrolysis and membrane fission. To address these issues, we have used cryo-EM to extend the resolution of the constricted ΔPRD polymer map and employed computational docking and biochemistry to define the underlying subunit interactions. We also present the crystal structure of GG in complex with GMPPCP, which identifies a major hydrolysis-dependent BSE conformational change. Our results provide insights into how dynamin assembly directly facilitates G domain dimerization and stimulated turnover and suggest how the energy of this dimerization and GTP hydrolysis can be converted into large structural movements that may play a role in precipitating membrane fission.
RESULTS
12.2Å cryo-EM reconstruction of ΔPRD in the constricted state reveals new structural features of the assembled dynamin polymer
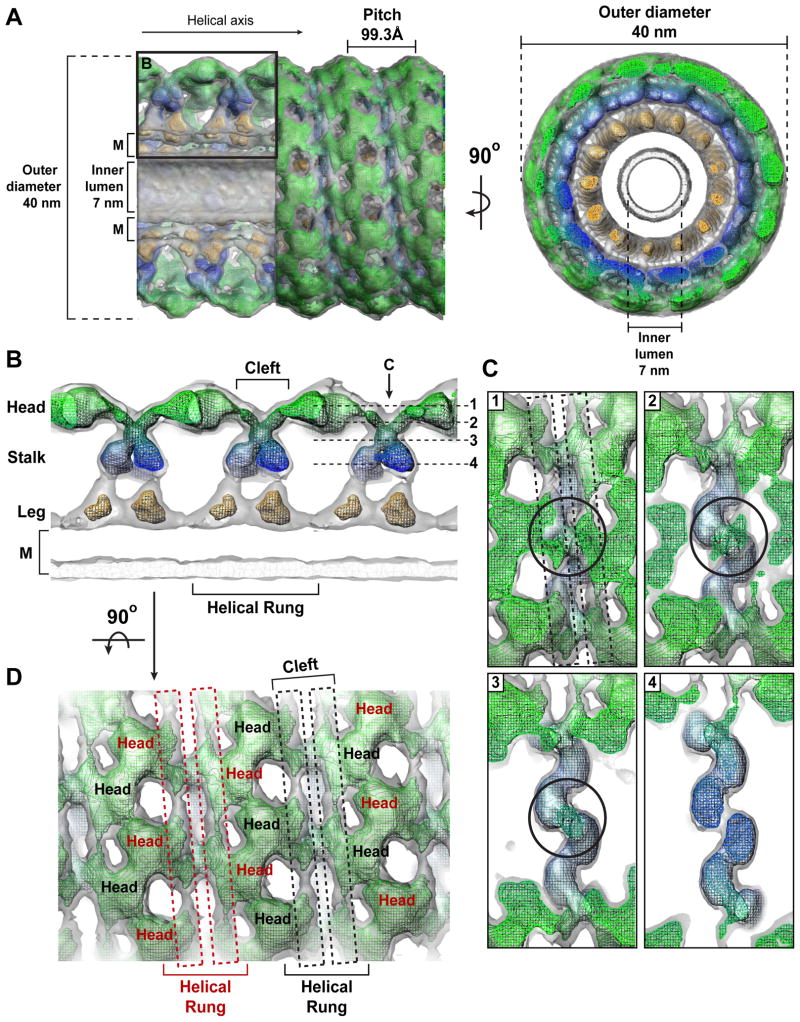
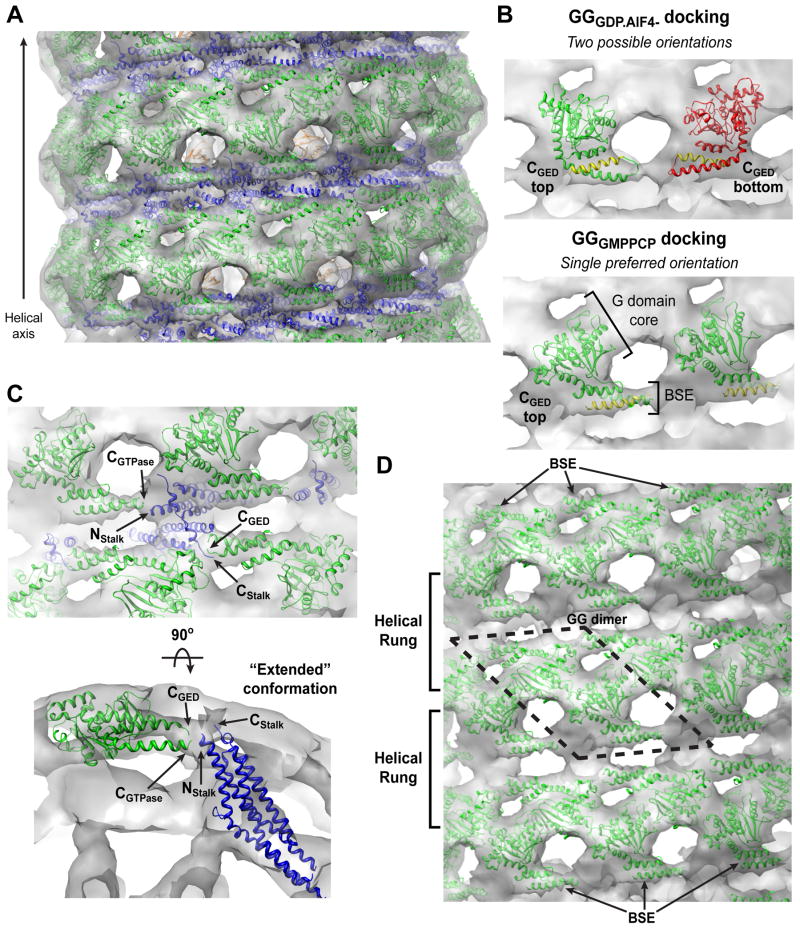
Our initial attempt to characterize GMPPCP-bound, constricted ΔPRD tubes using cryo-EM and Fourier-Bessel synthesis produced an 18Å resolution reconstruction (Wilson-Kubalek et al., 2010) that displayed only minor differences compared to previously published structures (Zhang and Hinshaw, 2001; Chen et al., 2004; Wilson-Kubalek et al., 2010). The resolution was limited by variations in the tube diameter, which produced long-range disorder and diminished the overall diffracting power. To circumvent this, we segmented the tubes into individual, overlapping particles that were then aligned, classified, sorted, and averaged with the iterative helical real-space reconstruction (IHRSR) algorithm (Egelman, 2007, Figure S2A–C). This single-particle based approach produced a 12.2Å helical map (Figure 1A, Figure S2D) that has inner luminal diameter of 7 nm, an outer diameter of 40 nm, 13.2 subunits per turn, and a pitch of 99.3Å. The improved resolution reveals new structural features of the ΔPRD polymer. First, the stalk density, which constitutes the base of the characteristic ‘T-view’ (Figure 1B, Movie S1), appears to twist in a crisscross fashion (Figure 1B and 1C), intersecting just below the cleft that separates the ‘head’ density regions along the exterior of the polymer. Second, there are two additional strips of density within the cleft that wrap around the tube (Figure 1D, highlighted with dashed boxes). Each strip forms a continuous connection with the alternating head densities of a single helical rung.
Figure 1. 12.2 Å reconstruction of ΔPRD in the constricted state reveals new structural features of the assembled dynamin polymer.
A. Structure of the ΔPRD polymer. Two density thresholds of the ΔPRD map are shown: the lower threshold is colored gray, the higher threshold is in mesh and colored radially to denote the locations of the ‘leg’ (orange), ‘stalk’ (blue), and ‘head’ (green) regions. Left panel shows a side view of the decorated helical tube oriented perpendicular to the helical axis. A section of the tube’s outer surface has been removed to show the interior of the structure in cross-section. The membrane bilayer (M), inner luminal diameter, outer tube diameter, and pitch are labeled. Black box denotes section of map highlighted in B. Right panel is an end-on view of the tube looking down the helical axis that is rotated 90° relative to the view on the left. B. Cross-section through ΔPRD polymer, the classical ‘T-view’ of dynamin subunits within individual helical rungs. The ‘leg’, ‘stalk’, ‘head’, and membrane bilayer (M) density regions are labeled and colored as in A. The cleft separating ‘head’ densities within the same helical run is labeled. Dashed lines (1–4) indicate the locations of planar slices through a single helical rung that are shown in C with orientation defined by black arrow. C. Sequential planar sections through a single helical rung shows the crisscross twisting of the dynamin stalk density. Black circles highlight intersection point of stalk density. Black dashed boxes in section 1 highlight the newly resolved strips of density visible in the cleft between G domains in the same helical rung. D. Two additional strips of density (dashed red and black boxes) are visible in the cleft along the exterior of the structure and form continuous connections with the head densities of a single helical rung. Subunits belonging to alternating helical rungs are colored as red and black for distinction. View is rotated 90° relative to B.
Docking of crystallized dynamin fragments illustrates ambiguities in structural models
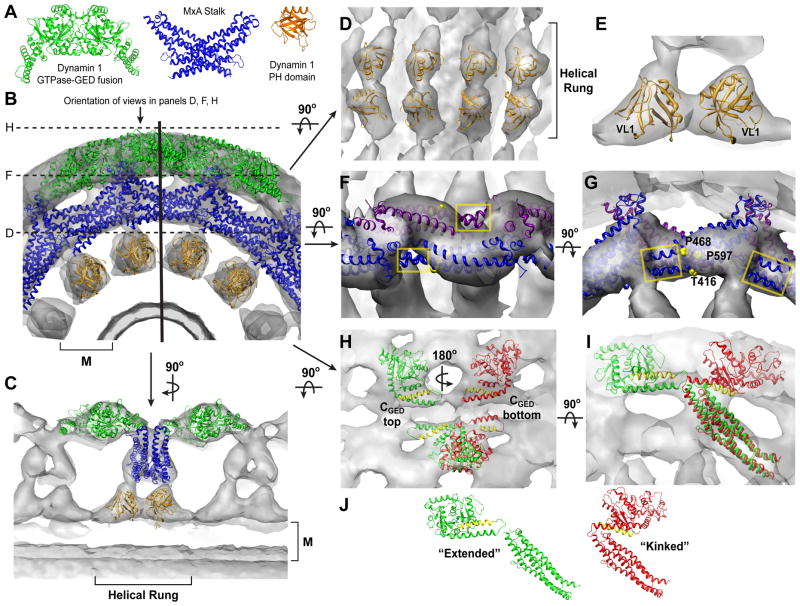
To decipher the subunit organization of the dynamin polymer, we docked the crystal structures of the GDP.AlF4−-stabilized GG dimer (GGGDP.AlF4−; PDB 2X2E), the human MxA middle/GED stalk (PDB 3LJB), and the human dynamin 1 PH domain (PDB 1DYN) into our improved ΔPRD reconstruction (Figure 2A). The MxA stalk structure shares a high degree of sequence homology (19.5% identical, 54.9% similar) with dynamin’s middle domain and GED (Figure S3) and currently represents the best structural model for these domains. Attempts to dock GGGDP.AlF4− as a dimer failed as one monomer always grossly protruded from the density, regardless of its orientation (Figure S4A). The GGGDP.AlF4- dimer from an alternate crystal form (PDB 2X2F) exhibited the same discrepancies (data not shown). We therefore selected only one monomer for docking (monomer A from PDB 2X2E), which allowed more degrees of freedom during the fitting procedures. We similarly positioned the MxA stalks individually, as the crystallized assembly could only be fit into a previously published 23Å ΔPRD map after a significant rotation between adjacent pairs of monomers (Gao et al., 2010). Fitting was carried out using YUP (Tan et al., 2006, 2008) as described in the Experimental Procedures. In total, eight GG monomers, twelve MxA monomers, and eight PH domains were positioned into the cryo-EM density. In agreement with previous biochemical data and structural modeling (Chen et al., 2004; Mears et al., 2007), the PH domain is situated in the ‘leg’ density adjacent to the plasma membrane, the middle/GED fragment inhabits the interior ‘stalk’ density, and the G domain occupies the exterior ‘head’ density of the tube (Figure 2B and 2C). It should be noted that, in our model, the density in the T-view cross-section represents the interaction of four different MxA stalk monomers (Figure S4B).
Figure 2. Computational docking of crystallized fragments derived from dynamin family members.
A. Crystal structures of isolated domains from different dynamin family members. From left to right: GDP.AlF4−-stabilized dimer of the GTPase-GED fusion (GG) from human dynamin 1 (PDB 2×2e, green), including the G domain and the BSE; stalk dimer from human MxA (PDB 3ljb, blue), which includes the middle domain and GED; dynamin 1 PH domain (PDB 1dyn, orange). B. Computational docking of structures in A into the ΔPRD map (gray). A section of the ΔPRD tube is shown in an end-on view looking down the helical axis to highlight the positions of the docked fragments relative to the membrane bilayer (M). Solid vertical black line indicates orientation of the cross-section plane that is rotated by 90° on panel C. Dashed black lines denote the planar sections that are rotated by 90° and shown in panels D, H, and F. C. ‘T-view’ cross-section illustrating the positioning of domains within a single helical rung. M denotes membrane bilayer. D, E. Docking of PH domain monomers. (D) shows orientation of PH domains within the same helical rung. (E) depicts asymmetry of PH domain fitting. Variable loop 1 (VL1) is labeled. Orientation is the same as in the ‘T-view’ in C. F, G. Fitting of MxA stalk monomers. (F) shows a zoomed in top view perpendicular to the membrane bilayer. Four of MxA stalk monomers (colored blue and purple) are shown. Note that portions of each MxA monomer protrude from the ΔPRD density map (yellow boxes). (G) is rotated 90° and shows a side view of the stalk monomers in the same orientation as shown in B. Residues corresponding to the putative dynamin proline hinge are labeled in the MxA structure (yellow spheres). H, I. Two possible fittings of GGGDP.AlF4− monomers (green and red) viewed either from the top (H) or the side (I). The different orientations are related by a 180° rotation about an axis parallel to the membrane surface (H) and result in the CGED helix of the BSE (yellow) facing either up (green) or down (red). Each fitting generates a different connection with the stalk (I), in subunits that are either ‘extended’ or ‘kinked’ (J).
At the membrane surface, the PH domains are arranged as dimers within the same helical rung (Figure 2C–E). The density within this region, however, is asymmetric, resulting in non-equivalent orientations for each of the neighboring monomers (Figure 2E). Our confidence in this fitting is strengthened by the fact that variable loop 1 of both PH domains – shown by fluorescence quenching experiments to penetrate the outer leaflet of PIP2-containing bilayers (Ramachandran and Schmid, 2008) – point into the lipid bilayer density as expected.
Although MxA middle/GED monomers match the overall shape of the stalk region density, a portion of these structures protrudes from the map (Figure 2F and 2G, yellow boxes). Where they diverge, the human MxA model contains two prolines (P468 and P597) and a threonine (T416) in helices α2, α4, and α1c respectively (Figure 2G, yellow spheres). Human dynamin 1 instead contains three highly conserved prolines (Figure S2) and we speculate that these residues form a flexible hinge that would allow the dynamin stalk to kink downward into the density and connect to the PH domain below.
We also observe an unfilled segment of density beneath each docked MxA stalk model that is continuous with the PH domain density below (Figure S4B). This is not unexpected as the dynamin fragment structures are missing the amino acids (58 in total) that link the middle domain to the PH domain (residues 487–517) and PH domain to the GED (residues 631–657) (Figure S1 and S2), which likely occupy this density. The absence of these connections in our model prohibits us from defining the stalk-PH domain connectivity unambiguously.
Our docking yields two equally viable fittings for the GGGDP.AlF4− monomers (Figure 2H and 2I, green versus red). Although both place the globular G domain core into the ‘head’ density and the BSE into the newly identified strips of density in the cleft (Figure 1D), their relative orientations differ by a 180° rotation around an axis parallel to the plasma membrane (Figure 2H). In one orientation the CGED helix is on top (Figure 2H, green) while in the other the CGTPase helix is on top (Figure 2H, red). Each orientation creates a different connectivity between the G domain and the stalk below (Figure 2I), producing two possible subunit arrangements (Figure 2J): long and extended (green) or short and kinked (red). Each imposes a different set of constraints on dynamin assembly and implies different structural contacts between neighboring subunits in the polymer.
Structure of GMPPCP-bound GG identifies a major BSE conformational change
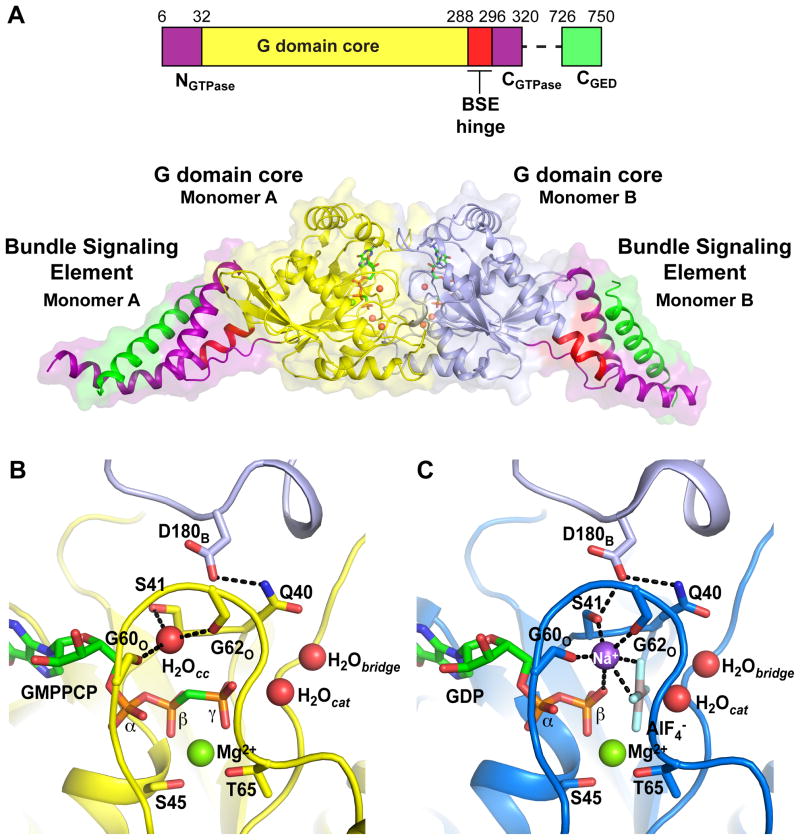
We hypothesized that the uncertainty associated with docking GGGDP.AlF4−monomers into the ΔPRD map may reflect nucleotide-dependent conformational differences between the crystallized GG dimer, stabilized by the transition-state mimic GDP.AlF4−, and ΔPRD dynamin in the assembled polymer, stabilized by the ground-state analog GMPPCP. To address this problem, we solved the crystal structure of GG in complex with GMPPCP (GGGMPPCP) at 2.2Å (Figure 3A). Although GGGMPPCP is entirely monomeric when analyzed by size exclusion chromatography (Chappie et al., 2010) and analytical ultracentrifugation (Figure S5A,B), in the crystal it forms a dimer similar to that of the transition-state complex, presumably due to the high protein concentration during crystallization.
Figure 3. Structure of GGGMPPCP.
A. Domain structure of the G domain-GED (GG) fusion protein from human dynamin 1 and the structure of its complex with GMPPCP. G domain cores are inyellow and light blue and the three helices of the BSE are shown as NGTPase, purple; CGTPase, purple; CGED, green. A highly conserved flexible hinge region (BSE hinge, red) connects the BSE to the G domain core. GMPPCP molecules (green) and active site waters (red spheres) are shown. B,C. Structural comparison of the GGGMPPCP (B, yellow) and GGGDP.AlF4− (C, dark blue, PDB 2X2E) active sites. The elements of the catalytic machinery are labeled in each structure along with the bound nucleotide (green), Mg2+ ion (green spheres), and catalytic and bridging waters (red spheres, H20cat and H20bridge respectively). The AlF4− (gray) and charge compensating sodium ion (purple sphere) are shown for GGGDP.AlF4−. An additional water molecule (H20cc, red sphere) occupies the ion-binding site in GGGMPPCP. The trans stabilizing loop and catalytic D180 residue from the adjacent monomer (colored light blue and labeled with subscript “B”) is shown at the top of each panel. Dashed black lines indicate hydrogen bonds.
One molecule of GMPPCP is bound to each active site along with a single Mg2+ ion that is coordinated by S45, T65, and the β- and γ-phosphates (Figure 3B). As in the GGGDP.AlF4− structure (PDB 2X2E) (Figure 3C), we resolve the catalytic water, appropriately positioned for an in-line nucleophilic attack on the γ-phosphate, and the adjacent bridging water, which contacts the conserved Q40 side chain (Figure 3B). Unlike many small G proteins, dynamin does not use an ‘arginine finger’ side chain to compensate for the developing negative charge in the transition-state (Scheffzek et al., 1998); rather, the positive charge is supplied by a monovalent cation, whose binding is stabilized by G domain dimerization (Chappie et al., 2010). Significantly, this cation is absent in GGGMPPCP as GMPPCP’s β-γ methylene connection does not provide the necessary hydrogen bonding interactions required to complete the ion coordination sphere. Instead, a water molecule (H20cc, Figure 3B) occupies the ion-binding site, but is shifted 1.7Å relative to the sodium observed in GGGDP.AlF4− (Figure 3C). H20cc is coordinated by the carbonyls of G60 and G62 and the S41 side chain, which rotates 90° to accommodate the offset from the transition- state complex. As a consequence, the hydrogen bond across the dimer interface between S41 and D180 is broken. The other facets of the nucleotide binding and catalytic machineries remain essentially unchanged.
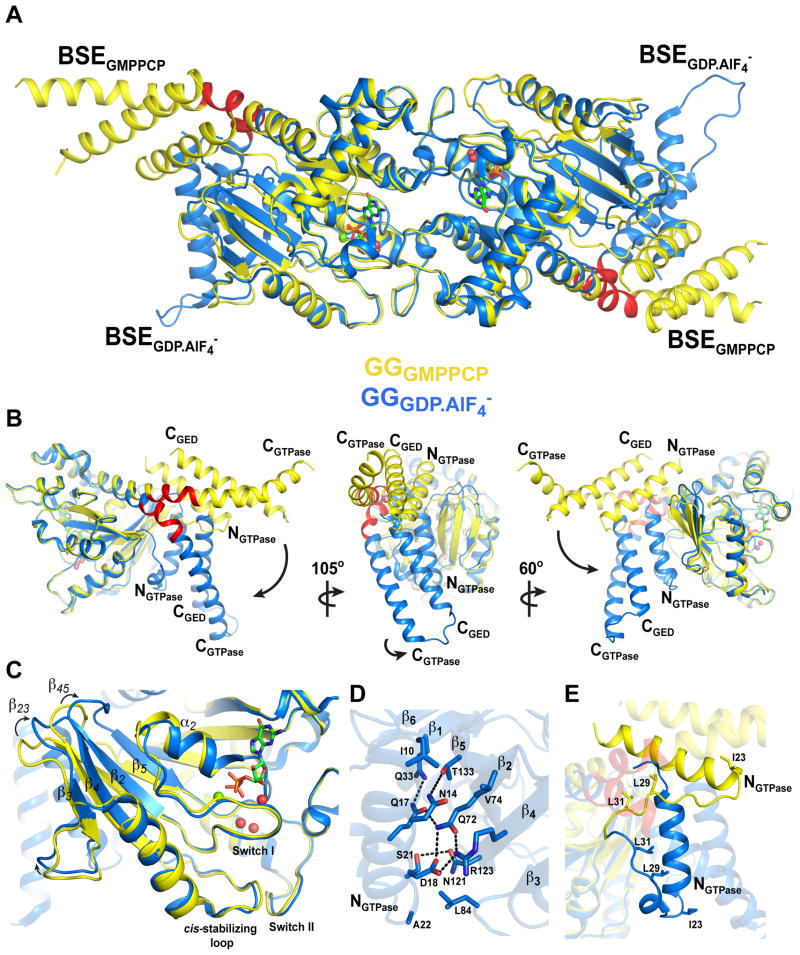
The major structural difference between the ground-state GGGMPPCP and GGGDP.AlF4− transition-state complexes (Figure 4A) is a 68.81° rigid body rotation of the BSEs downward about an axis perpendicular to the CGTPase helix coupled with a slight counter-clockwise twist (Figure 4B, Movies S2, S3). This brings each BSE close to the β-sheet of the G domain core and results in a more compact transition-state dimer, reducing its radius of gyration from 32.9Å to 30.9Å. Residues between H288 and G295 (Figure 3A and Figure 4, red) – previously identified as a flexible hinge (Chappie et al., 2010) – and residues at the start of the G domain core (P32 and Q33) serve as the pivot points for these motions.
Figure 4. Structure of GGGMPPCP identifies a hydrolysis-dependent BSE conformational change.
A. Structural superposition of GGGMPPCP (yellow) and GGGDP.AlF4− (blue, PDB: 2X2E). Note the different conformations of the BSE in each structure. The BSE hinge is colored red. B. Hydrolysis-dependent BSE conformational change. Left panel is superposition of GGGMPPCP and GGGDP.AlF4− monomers. The NGTPase, CGTPase, and CGED helices of the BSE are labeled. Black arrow depicts 68.81° downward rotation of the BSE that in the transition-state complex. Middle and right panels depict different views of the monomer superposition. Small black arrow in middle panel describes the slight counterclockwise twist that is coupled to the downward rotation of the BSE; black arrow in right panel describes combined translocation of BSE. C. BSE movement induces structural changes in the central β-sheet of the G domain core. β-strands 2–5, the α2 helix, and the β23 and β45 loops are labeled. Black arrows illustrate how these segments shift to accommodate the BSE. The GMPPCP (green), Mg2+ (green sphere), and active site waters (red spheres) from the GGGMPPCP structure are shown. D. Structural interactions between the BSE and G domain core in the GGGDP.AlF4− transition-state complex. Residues contributing to salt bridges and hydrophobic interactions are shown. Black dashed lines are hydrogen bonds. E. Structural changes of the NGTPase linker. The linker reconfigures into a short helix, allowing I23, L29, and L31 to form stabilizing interactions with the BSE hydrophobic core.
While the P loop is essentially unchanged, helix α2 tilts toward the active site (Figure 4C). The downstream end of switch 1 (residues 59–68) shifts ~1Å. The size of the changes increase toward the β sheet with a 3.5Å shift at the upstream end of switch 1 at G53 and culminating in a 4.5Å shift at the tip of the sheet affecting the connecting β23 and β45 (Figure 4C, arrows). Moving toward the transition state, the net effect of these changes is a rotation of the central β sheet (Movie S2) and tightening of the hydrophobic packing within the G domain core (Figure S5C), which brings R54, E79, and S126 into hydrogen bonding distance (Figure S5C). This may also help stabilize switch II as the _cis_-stabilizing loop (Chappie et al., 2010) shifts nearly 2Å (Figure 4C).
The repositioning of elements within the core reconfigures the outer face of the β sheet and facilitates the formation of salt bridges and hydrophobic interactions with the NGTPase helix that anchor the BSE (Figure 4D). Additional stabilization is provided by the NGTPase linker (residues 22 to 31), which partially reconfigures into a short helix and contacts the BSE’s hydrophobic core via residues I23, L29, and L31 (Figure 4E).
Docking of GGGMPPCP reveals putative G domain-stalk connectivity
We next asked whether docking GGGMPPCP into our ΔPRD cryo-EM map could distinguish between the two possibilities for the G domain-stalk connection (Figure 2J). The fitting approach described above was expanded to include 48 GGGMPPCP monomers, 24 MxA stalk monomers, and 24 PH domains – nearly two complete turns of the ΔPRD helix (Figure 5A). The ambiguity we previously encountered when fitting the GGGDP.AlF4− monomers (Figure 2J) is now absent in the resulting model, as GGGMPPCP adopts a single preferred orientation in the ΔPRD map (Figure 5B). This is due to the different BSE conformations relative to the G domain core in the two GG structures. The BSEs are oriented with the CGED helices on top (Figure 5B) and occupy the cleft density strips (Figure 5B–D) that encircle the exterior of the map within each rung of the dynamin helix (Figure 1D). This positions the ends of the CGTPase and CGED helices close to N- and C-termini of the stalk (Figure 5C, Nstalk and Cstalk), allowing these segments to connect via two short stretches of amino acids that are missing from the docked crystal structures – residues 311–320 and residues 722–725. The physical constraints of these connections and the docking indicate that the underlying dynamin subunits must adopt an extended conformation within the assembled polymer (Figure 5C and 2J). In this configuration, the G domains in adjacent helical rungs are poised to form the productive dimers that were identified by crystallography and are needed for dynamin’s stimulated GTPase activity (Chappie et al., 2010) (Figure 5D). Unlike the crystallized GG dimers, these docked GGGMPPCP monomers are slightly separated consistent with our findings that G domain dimerization only occurs in the presence of transition-state mimics and not with ground-state analogs such as GMPPCP (Figure S5A,B; Chappie et al., 2010). A similar docking procedure using a homology model for the dynamin 1 middle/GED stalk rather than the MxA structure yielded the same overall fitting and extended subunit arrangement (data not shown).
Figure 5. Docking of GGGMPPCP reveals putative G domain-stalk connectivity.
A. Docked model of assembled ΔPRD polymer in the constricted state. GGGMPPCP monomers are colored green, middle/GED stalk monomers are colored blue, PH domains are colored orange, and the GMPPCP-bound ΔPRD reconstruction is rendered in gray. A side view is shown perpendicular to the helical axis. B. Comparison of GGGDP.AlF4− (top) and GGGMPPCP (bottom) monomer dockings. GGGMPPCP yields a single, preferred orientation where the CGED helix of the BSE (yellow) is on top. Monomers are shown in the same orientation as A. C. Zoomed side (upper panel, perpendicular to helical axis) and end (lower panel, looking down helical axis) views highlighting the putative G domain-stalk connectivity. Labels: CGTPase, C-terminus of G domain; Nstalk, N-terminus of the middle domain; Cstalk, C-terminal portion of GED present in MxA crystal structure; CGED, beginning of C-terminal GED helix present in GGGMPPCP structure. The proximity of CGTPase to Nstalk and Cstalk to CGED suggests that dynamin subunits adopt an ‘extended’ conformation. D. GGGMPPCP monomers in adjacent helical rungs are poised for dimerization (dashed black lines). The BSEs occupy the cleft densities above the stalk on the exterior of the map (black arrows).
CGED is domain swapped in full-length dynamin
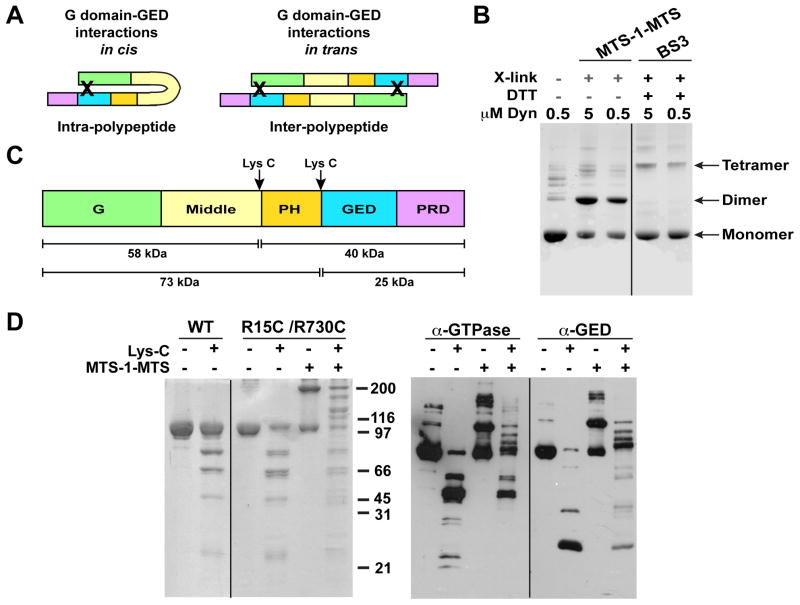
Although GG’s CGED helix mimics dynamin’s G domain-GED interactions, its minimal nature does not distinguish whether GED’s association with the G domain in the dynamin tetramer occurs in cis within the same polypeptide or is contributed by another polypeptide in trans (Figure 6A). We therefore used chemical crosslinking to resolve this ambiguity. Two cysteine mutations (R15C in NGTPase/R730C in CGED) – previously shown to enable efficient crosslinking of GG’s N- and C-termini by a short, (3.6 Å) cysteine-reactive bifunctional crosslinker (MTS-1-MTS) (Chappie et al., 2009) – were introduced into DynRCL to examine G domain-GED interactions in the tetramer. The resulting protein (DynRCL R15C/R730C) shows normal GTPase activity (Figure S6A,B) and migrates similar to WT-Dyn when analyzed by non-reducing SDS-PAGE (Figure 6D). Like WT-Dyn (Muhlberg et al., 1997), DynRCL R15C/R730C predominantly generates a tetrameric species when incubated with the general amine-reactive bifunctional crosslinker BS3 (Figure 6B). In contrast, specific G domain-GED crosslinking of DynRCL R15C/R730C by cysteine-reactive MTS-1-MTS predominantly generates a dimer (Figure 6B). Importantly, we did not detect any faster migrating species indicative of intra-polypeptide or in cis crosslinking. For both reagents, the crosslinking efficiency of the predominant species was unaffected by protein concentration (Figure 6B), consistent with intra-tetramer or in trans crosslinking. This was confirmed by size exclusion chromatography of the crosslinked species, which eluted as a tetramer (Figure S6C–E). Finally, DynRCL R15C/R730C was subjected to limited proteolysis with LysC, which cleaves sites bordering the PH domain (Figure 6C and 6D) (Muhlberg et al., 1997). Western blotting with G domain- or GED-specific antibodies confirmed that each of the higher molecular weight crosslinked species, but none of the lower molecular weight bands, contains both the G domain and GED (Figure 6D, α-G domain and α-GED respectively). Together these data establish that the GED from one polypeptide docks on the G domain of an adjacent polypeptide to form a domain-swapped full-length dynamin dimer, two of which associate through middle/GED stalk interactions to form the dynamin tetramer.
Figure 6. Dynamin tetramer is a dimer of domain-swapped dimers.
A. Cartoons illustrating possible G domain-GED interactions in full-length dynamin. Domains are colored as in C. Black ‘X’s denote expected crosslinks for each scenario. B. Chemical crosslinking of DynRCL R15C/R730C. Targeted crosslinking of the engineered cysteine residues in the NGTPase and CGED by MTS-1-MTS produces a prominent dimeric species while non-specific crosslinking of surface-reactive amines by BS3 primarily yields a tetramer. Crosslinking efficiency of the predominant species was unaffected by protein concentration. C. Cleavage products of Lys-C limited proteolysis (Muhlberg et al., 1997). D. Lys-C limited proteolysis and MTS-1-MTS crosslinking of WT and DynRCL R15C/R730C. Left panel shows Coomassie-stained gel of proteolyzed and/or crosslinked products; right panel shows western blotting of the same species. α-GTPase and α-GED are primary antibodies recognizing dynamin’s N-terminus (residues 2–17) and GED respectively.
Membrane-bound structure of the dynamin tetramer
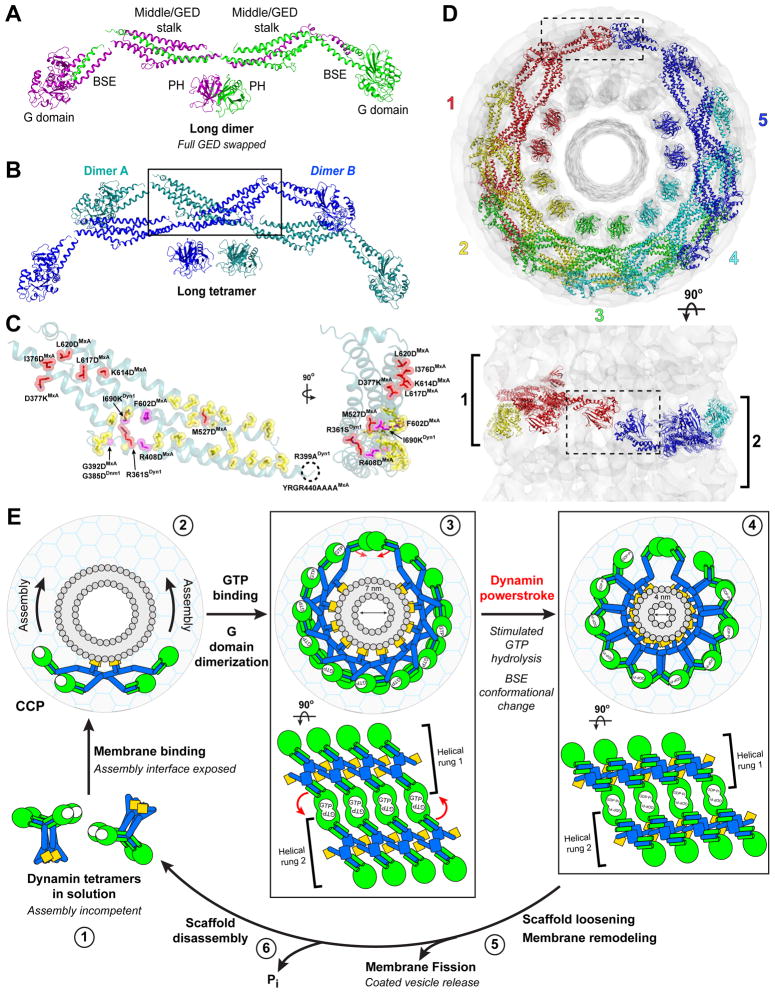
Our docking suggests two possible architectures for this full-length domain-swapped dynamin dimer (Figure 7A, Figure S7A). Swapping the entire GED would produce a long, m-shaped dimer (Figure 7A). Alternatively, exchanging only the CGED helix would result in a short, x-shaped dimer (Figure S7A). The two dimers differ in the relative placement of the PH domains and the inter-monomer interfaces. In the long dimer, the PH domains are close enough to allow complete GED exchange while the stalks are separated from their partner in the other monomer (Figure 7A). In the short dimer, this situation is reversed: the structure is stabilized by a back-to-back stalk interaction that forces the PH domains to be splayed apart (Figure S7A). Despite these differences, both dimers use the same stalk interface to form a tetramer (Figure 7B, Figure S7B,C). Mutations in this ‘assembly interface’ (Figure 7B, Table S1) – including R399 and I690 in dynamin 1 (Sever et al., 2006; Ramachandran et al., 2007), R408, G392, and Y440-R444 in human MxA (Gao et al., 2010), and G385 in S. cerevisiae Dnm1 (Ingerman et al., 2005) – shift the tetrameric state of these dynamin family members to stable dimers. This interface also provides stabilizing interactions between tetramers in our polymer structure, which may explain the cooperativity observed for membrane-mediated dynamin assembly (Stowell et al., 1999) and the assembly defects exhibited by dynamin mutant dimers (Song et al., 2004; Ramachandran et al., 2007) (Table S1).
Figure 7. Structural constraints of G domain dimerization and the dynamin powerstroke.
A. Model for the domain-swapped dynamin dimer. In this configuration, a long dimer is formed by a full GED domain swap. Monomers are colored purple and green. An alternative model also consistent with our crosslinking data is shown in Figure S6. B. Putative arrangement of membrane-bound long dynamin tetramer derived from the docked model of assembled ΔPRD polymer (Figure 5). The tetramer is comprised of two of the domain-swapped dimers shown in A (colored blue and teal, labeled A and B). Black box indicates assembly interface between these dimers (see also Figure S7B). C. Structural mapping of mutations that impair dynamin oligomerization. A stalk monomer (teal) is shown in two orientations. Mutations within the putative assembly interface (yellow) are labeled and colored magenta; mutations that also produce assembly defects but are localized outside this interface are also shown and colored red. Dashed black circle defines assumed location of R399ADyn1 and YRGR440AAAAMxA, which are disordered in the crystal structure. Phenotypes are in Table S1. D. Assembly of long dynamin tetramer models within the GMPPCP-stabilized constricted ΔPRD map (gray). The numbering and rainbow coloring (red to purple) denotes the sequential addition of tetramers and terminates when the first G domain dimer is formed. Upper panel depicts end view of the long assembly looking down the helical axis, lower panel is a side view perpendicular to this axis. The sequential rungs of the dynamin helix are marked in the lower panel with black brackets and numbered as ‘1’ and ‘2’. Dashed black box highlights the partnering helical rungs facilitating G domain dimerization. E. Proposed pathway of dynamin-catalyzed membrane fission. Dynamin tetramers exist in an assembly incompetent conformation in solution (1). Membrane binding causes a conformational change in the tetramer that exposes the assembly interface, inducing the rapid, cooperative assembly of a helical dynamin collar at the neck of an invaginated clathrin-coated pit (CCP) (2). Initial constriction of the neck, triggered by GTP binding and structural changes in middle/GED stalk (Chen et al., 2004), promotes G domain dimerization between tetramers in adjacent helical rungs to optimally position dynamin’s catalytic machinery (3). Assembly-stimulated GTP hydrolysis drives a major rotation of the BSE in the transition-state that constitutes the dynamin powerstroke (4). Propagation of this change through multiple turns of the helical dynamin collar causes further constriction of the neck (4). The resulting structural rearrangements might play a role in loosening the dynamin scaffold from the membrane surface, facilitating the membrane remodeling events that contribute to membrane fission (5). The detached dynamin scaffold disassembles upon release of the hydrolyzed γ-phosphate (6), which stabilized the dimer interface. Coloring: G domains, green; middle/GED stalk, blue; PH domains, orange; membrane bilayer, gray; lipid head groups, gray circles. The large gray circle with cyan meshwork is the CCP. The assemblies are shown in the same orientations as in D, (upper panels 3, 4) and in side view on the lower panels. Red arrows indicate movements associated with the dynamin powerstroke. The 7 nm measurement (3) corresponds to the inner luminal diameter of our GMPPCP-stabilized ΔPRD reconstruction, which is poised for G domain dimerization and represents an intermediate along the fission pathway; the 4 nm measurement (4) indicates the theoretical inner luminal diameter of the neck that would allow the spontaneous formation of a hemifission intermediate following partial detachment of the dynamin scaffold.
While both of these configurations are consistent with our crosslinking data and with mutagenesis studies defining assembly interfaces, we favor the long dimers for two reasons. First, its shape closely resembles the low-resolution structure of the R399A/I690K mutant dimer revealed by small angle x-ray scattering (Kenniston and Lemmon, 2010). Second, recent crystallographic studies of the intact ΔPRD molecule show no indication of an inter-polypeptide exchange of the CGED helix at the top of the molecule (M. Ford & J. Nunnari, personal communication), arguing against the short dimer configuration.
Structural constraints of G domain dimerization
Dynamin’s stimulated GTPase activity arises from the transition-state-dependent dimerization of its G domains (Chappie et al., 2010). This association has been proposed to occur between two dynamin tetramers and be driven by dynamin assembly on the plasma membrane (Chappie et al., 2010; Gao et al., 2010). Our docking model supports this hypothesis. The connectivity we derive from computational fitting (Figure 5A) precludes G domain interactions within a single tetramer (Figure S7C) and between tetramers in the same helical rung; instead, G domain dimers can only form between tetramers in adjacent rungs, regardless of the underlying subunit architecture (Figure 7C, Figure S7D). Assembly of the helical collar beyond a single rung thus primes the dynamin subunits for stimulated turnover. Surprisingly, only five long tetramers (10 subunits) (Figure 7C) or six short tetramers (12 subunits) (Figure S7D) are needed to partner the G domains across helical rungs in the constricted ΔPRD polymer, indicating that a complete turn of the helix (13 subunits) is not required to facilitate G domain dimerization and stimulated GTPase activity. This observation may explain the inability to detect dynamin collars in vivo unless GTP hydrolysis has been inhibited (Marks et al., 2001; Takei et al., 1995).
DISCUSSION
The building blocks of dynamin assembly
Here we have combined cryo-EM, X-ray crystallography, computational docking, and biochemistry to provide new insights into the structure of assembled dynamin. Our 12.2Å reconstruction of ΔPRD dynamin in the GMPPCP-bound constricted state revealed novel density features not observed in previous lower resolution maps (Zhang and Hinshaw, 2001; Chen et al., 2004), which served as an improved structural framework for computational docking. Guided by this molecular envelope, we successfully localized the G domain, the BSE, the middle/GED stalk, and PH domain within the polymer assembly. The resulting pseudo-atomic model, which incorporates our new 2.2Å GGGMPPCP crystal structure, reveals the putative G domain-stalk connectivity and suggests that the individual dynamin subunits are extended rather than kinked when assembled on a lipid membrane. We cannot yet define the linkages between the middle/GED stalk and the PH domain, as the intervening sequences are absent from currently available crystallographic models.
Our chemical crosslinking demonstrates that the CGED helix from one dynamin polypeptide interacts in trans with the G domain of a second polypeptide, resulting in a domain-swapped dimer. Two of these domain-swapped dimers would then associate via their stalks to form a tetramer. Such an arrangement is consistent with mutations in the middle domain (R361S, R399A) and GED (I690K) that destabilize the dynamin tetramer but generate soluble dimers (Sever et al., 2006; Ramachandran et al., 2007). An underlying domain-swapped dimer also explains how assembling tetramer subunits could generate a helical structure in which the asymmetric unit is a dimer (Zhang and Hinshaw, 2001). We therefore propose that a domain-swapped dimer is the minimal unit of dynamin assembly, serving as the basic building block for the tetramer in solution and, by extension, the helical assembly on the membrane.
We identified two possible configurations for the domain-swapped dimers and their resulting tetramer counterparts that are consistent with all available data. A caveat of these models is that they represent a membrane-bound, assembly-competent conformation that may be distinct from the conformation of the free tetramer in solution. It is possible that dynamin undergoes a major conformational change upon membrane binding that exposes the ‘assembly interface’, allowing the rapid and cooperative association of multiple tetramers. Structural studies suggest that the bacterial dynamin-like protein (BDLP) undergoes a self-propagating transition, where GTP- and membrane-induced expansion of compact diamond-shaped BDLP dimers promotes polymerization (Low and Löwe, 2006; Low et al., 2009). Interestingly, a subset of PH domain mutations linked to centronuclear myopathy – S619L, S619W, and V625del – have been shown to promote higher order assembly in the absence of a lipid scaffold (Kenniston and Lemmon, 2010). These changes also result in stimulated GTP hydrolysis (Kenniston and Lemmon, 2010), suggesting that they alleviate the inherent auto-inhibition associated with the assembly-incompetent conformation of the tetramer in solution. Conversion between assembly-incompetent and assembly-competent conformations may thus represent a conserved regulatory mechanism common to dynamin family members.
Implications for dynamin-catalyzed membrane fission
Dynamin assembly and constriction generates high curvature and localized stress (Bashkirov et al., 2008; Ramachandran et al., 2009; Roux et al., 2010) that impose a greater strain on the inner monolayer lipids of a tightly squeezed neck than those of the outer monolayer (Bashkirov et al., 2008; Schmid and Frolov, 2011). PH domain interactions with the phospholipid head groups and the membrane insertion of variable loop 1 maintain this energetically unfavorable configuration (Ramachandran and Schmid, 2008; Ramachandran et al., 2009), which can be relaxed by partial detachment and/or disassembly of dynamin subunits following stimulated GTP hydrolysis (Ramachandran and Schmid, 2008; Pucadyil and Schmid, 2008; Bashkirov et al., 2008). Theoretical modeling indicates that a hemifission intermediate will form at this stage if the luminal diameter of the neck is equivalent to the bilayer thickness (~4 nm) (Bashkirov et al., 2008). Our GMPPCP stabilized ΔPRD polymer reconstruction has an inner luminal diameter of 7 nm, indicating that it is an intermediate along the fission pathway and that additional constriction is necessary to constrain the membrane neck in a manner that allows fission to occur spontaneously once it is released from the dynamin scaffold. Further compression of the polymer also favors G domain dimerization, as the longitudinal proximity between adjacent helical rungs would be increased as the inner luminal diameter decreases.
Our structural data raises the tantalizing possibility of a BSE-mediated dynamin powerstroke (Figure 7D) that converts the energy of G domain dimerization and GTP hydrolysis into rearrangements affecting the entire dynamin collar. These changes could provide the mechanochemical force needed for constriction down to ~4nm and subsequent loosening of the dynamin scaffold from the membrane, thus precipitating the membrane remodeling events required for fission (Figure 7D). Recently, large GTP hydrolysis-dependent conformational changes were also observed for the yeast mitochondrial dynamin-like protein Dnm1 (Mears et al., 2011) that did not occur upon the addition of GMPPCP, suggesting that the formation of a G domain transition-state complex may also play an important role in mitochondrial fission. It remains to be seen whether this system exhibits a similar BSE conformational change.
It has been proposed that the assembly-dependent positioning of dynamin’s PH domains helps catalyze the lipid rearrangements needed for fission (Schmid and Frolov, 2011). The PH domains are asymmetrically distributed in the long tetramer assembly with part of the membrane surface unoccupied (Figure 7B) and arranged uniformly around the neck in the short assembly (Figure S7D). As the number of turns required to catalyze fission has yet to be established, the significance of this differential distribution remains to be determined.
Intramolecular conformational coupling
Fluorescence studies have shown that PH domain binding to/dissociation from the plasma membrane is coupled to structural changes in the G domain’s nucleotide-binding pocket (Solomaha and Palfrey, 2005; Ramachandran and Schmid, 2008). The large distance between these two domains (Figure 2) suggests that a mechanism exists for long-range communication within the dynamin molecule. We previously showed that the BSE senses and transmits assembly-dependent conformational changes to the G domain in a back-to-front manner, i.e., from the membrane to the G domain (Chappie et al., 2009). The hydrolysis-dependent BSE conformational change described here (Figure 4) illustrates that this module can also function front-to-back (i.e., from the G domain to the membrane), amplifying nucleotide-dependent changes in the active site and relaying them through the stalk. These properties make the BSE an ideal regulator of intramolecular crosstalk. Recent evidence suggests that the C-terminal α-helix of the PH domain (CPH) also plays a role in conformational coupling, as mutations in this region can indirectly modulate dynamin’s GTPase activity (Kenniston and Lemmon, 2010). Being situated at opposing ends of the GED, the CPH and BSE could communicate back and forth via structural fluctuations in the stalk to coordinate membrane binding, dynamin assembly, stimulated GTP hydrolysis, and the subsequent disassembly of the polymer.
Experimental Procedures
Protein purification and biochemical assays
See Supplemental Experimental Procedures for detailed protocols describing the purification of dynamin and GG constructs, the preparation of ΔPRD tubes, GTPase assays, chemical crosslinking, and sedimentation velocity experiments.
Cryo-electron microscopy and image processing
Samples were visualized using a Phillips Technai F20 electron microscope operating at 120 kV and images were collected using Leginon (Potter et al., 1999; Suloway et al., 2005) in manual mode at 1.0–2.0 μm underfocus with a 4K × 4K Gatan CCD camera at a nominal magnification of 50,000×, corresponding to a resolution of 2.26Å per pixel. Images were individually CTF corrected using ACE2 (Mallick et al., 2005). Ordered, straight ΔPRD tubes were manually selected for processing by the iterative helical real space reconstruction (IHRSR) methodology (Egelman, 2007; See Supplementary Experimental Procedures for details). The resolution of the final map was determined to be 12.2Å by Fourier shell correlation (FSC=0.5) (Figure S2D).
X-ray data collection, structure solution, and refinement
Native data on a GGGMPPCP crystal were collected at 95 K on a rotating anode source equipped with multilayer focusing optics using Cu Kα radiation and a Saturn A200 CCD detector. All data were integrated and scaled using XDS (Kabsch, 2010). The GGGMPPCP structure was solved by molecular replacement using PHASER (McCoy et al., 2007) and refined with CNS v1.3 (Brünger et al., 1998). See Supplemental Experimental Procedures for details. X-ray data collection and refinement statistics can be found in Table S2.
Computational docking
All-atom structures were refined using the YUP.SCX method (Tan et al., 2008) of the YUP software package (Tan et al., 2006). See Supplemental Experimental Procedures for details. Initial fitting was performed using GGGDPAlF4− monomers (PDB 2X2E), MxA middle/GED stalk monomers (PDB 3LJB), and PH domain monomers (PDB 1DYN), representing ~93% of the ΔPRD sequence. A similar procedure was used to fit GGGMPPCP. Orientations of the middle/GED and PH monomers were largely unchanged, and the GGGMPPCP placement refined to a single orientation that best matched the ΔPRD cryo-EM structure.
Supplementary Material
01
02
03
04
05
Highlights.
- 12.2Å structure of ΔPRD dynamin polymer and 2.2 Å structure of GMPPCP-bound GG fusion
- Productive G domain dimers form between dynamin tetramers in adjacent helical rungs
- Dynamin tetramer is a dimer of domain-swapped dimers
- Movement of the bundle signaling element mediates a hydrolysis-dependent powerstroke
Acknowledgments
We thank Vasyl Lukiyanchuk and Sharmistha Acharya for assistance in cloning and purification, Rodolfo Ghirlando for assistance with sedimentation velocity experiments, Juha-Pekka Mattila for communication of unpublished data, Joshua Zimmerberg for insightful discussions, Alison B. Hickman for technical advice and critical reading of the manuscript, and Yihong Ye for critical reading of the manuscript. We especially thank Marijn Ford and Jodi Nunnari for the ongoing open dialogue regarding the intricacies of dynamin structure and assembly. This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and NIH grants GM52468 and GM75820 (to R.A.M) and GM42455 (to S.L.S.). J.S.C. was supported by a Nancy Nossal Fellowship award from NIDDK. A portion of the work presented here was conducted at the National Resource for Automated Molecular Microscopy, which is supported by the National Institutes of Health through the National Center for Research Resources’ P41 program (RR017573).
Footnotes
Accession numbers
Atomic coordinates for the GGGMPPCP structure have been deposited in the Protein Data Bank under the accession number XXXX. The reconstructed density of GMPPCP-stabilized ΔPRD lipid tubes has been deposited in the EM Data Bank with accession code EMD-XXXX. Coordinates for the complete docked model consisting of GGGMPPCP, the human MxA stalk, and human dynamin 1 PH domain have been deposited in the Protein Data Bank with accession code XXXX.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bashkirov PV, Akimov SA, Evseev AI, Schmid SL, Zimmerberg J, Frolov VA. GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135:1276–1286. doi: 10.1016/j.cell.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT, et al. Crystallography & NMR System: a new software suite for macromolecular structure determination. Acta Cryst. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Liu YW, Leonard M, Pucadyil TJ, Schmid SL. An intramolecular signaling element that modulates dynamin function in vitro and in vivo. Mol Biol Cell. 2009;20:3561–3571. doi: 10.1091/mbc.E09-04-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin’s assembly-stimulated GTPase activity. Nature. 2010;465:435–40. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YJ, Zhang P, Egelman EH, Hinshaw JE. The stalk region of dynamin drives the constriction of dynamin tubes. Nat Struct Mol Biol. 2004;11:574–575. doi: 10.1038/nsmb762. [DOI] [PubMed] [Google Scholar]
- Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72:175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- Danino D, Moon KH, Hinshaw JE. Rapid constriction of lipid bilayers by the mechanochemical enzyme dynamin. J Struct Biol. 2004;147:259–267. doi: 10.1016/j.jsb.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Egelman EH. The iterative helical real space reconstruction method: surmounting problems posed byreal polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Crystal structure at 2.2Å resolution of the pleckstrin homology domain from human dynamin. Cell. 1994;79:199–209. doi: 10.1016/0092-8674(94)90190-2. [DOI] [PubMed] [Google Scholar]
- Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O. Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature. 2010;465:502–506. doi: 10.1038/nature08972. [DOI] [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL. Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature. 1995;374:190–192. doi: 10.1038/374190a0. [DOI] [PubMed] [Google Scholar]
- Ingerman E, Perkings EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS Acta Cryst. 2010;D66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenniston JA, Lemmon MA. Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients. EMBO J. 2010;29:3054–3067. doi: 10.1038/emboj.2010.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlovsky Y, Kozlov MM. Membrane fission: model for intermediate structures. Biophys J. 2003;85:85–96. doi: 10.1016/S0006-3495(03)74457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Löwe J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- Low HH, Sachse C, Amos LA, Lowe J. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 2009;139:1342–1352. doi: 10.1016/j.cell.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: Automated CTF estimation. Ultramicroscopy. 2005;104:8–29. doi: 10.1016/j.ultramic.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Marks B, Stowell MHB, Vallis Y, Mills IG, Gibson A, Hopkins CR, McMahon HT. GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature. 2001;410:231–235. doi: 10.1038/35065645. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- Mears JA, Ray P, Hinshaw JE. A corkscrew model for dynamin constriction. Structure. 2007;15:1190–1202. doi: 10.1016/j.str.2007.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears JA, Lackner LL, Fang S, Ingerman E, Nunnari J, Hinshaw JE. Conformational changes in Dnm1 support a contractile mechanism for mitochondrial fission. Nat Struct Mol Biol. 2011;18:20–6. doi: 10.1038/nsmb.1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettlen M, Pucadyil TJ, Ramachandran R, Schmid SL. Dissecting dynamin’s role in clathrin-mediated endocytosis. Bioc Soc Trans. 2009;37:1022–1026. doi: 10.1042/BST0371022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlberg AB, Warnock DE, Schmid SL. Domain structure and intramolecular regulation of dynamin GTPase. EMBO J. 1997;16:6676–6683. doi: 10.1093/emboj/16.22.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann HH, Knetsch MLW, Scherer A, Manstein DJ, Kull FJ. Crystal structure of a dynamin GTPase domain in both nucleotide-free and GDP-bound forms. EMBO J. 2001;20:5813–5821. doi: 10.1093/emboj/20.21.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, McMahon HT. The dynamin superfamily: universal membrane tubulation and ssion molecules? Nat Rev Mol Cell Biol. 2004;5:133–147. doi: 10.1038/nrm1313. [DOI] [PubMed] [Google Scholar]
- Potter CS, Chu H, Frey B, Green C, Kisseberth N, Madden TJ, Miller KL, Nahrstedt K, Pulokas J, Reilein A, et al. 0. Leginon: a system for fully automated acquisition of 1000 electron micrographs a day. Ultramicroscopy. 1999;77:153–161. doi: 10.1016/s0304-3991(99)00043-1. [DOI] [PubMed] [Google Scholar]
- Pucadyil TJ, Schmid SL. Real-time visualization of dynamin-catalyzed membrane fission and vesicle release. Cell. 2008;135:1263–1275. doi: 10.1016/j.cell.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Surka M, Chappie JS, Fowler DS, Foss TR, Song BD, Schmid SL. The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J. 2007;26:559–566. doi: 10.1038/sj.emboj.7601491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Schmid SL. Real-time detection reveals that effectors couple dynamin’s GTP-dependent conformational changes to the membrane. EMBO J. 2008;27:27–37. doi: 10.1038/sj.emboj.7601961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Pucadyil TJ, Liu YW, Acharya S, Leonard M, Lukiyanchuk V, Schmid SL. Membrane Insertion of the Pleckstrin Homology Domain Variable Loop 1 Is Critical for Dynamin-catalyzed Vesicle Scission. Mol Biol Cell. 2009;20:4630–4639. doi: 10.1091/mbc.E09-08-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Eschenburg S, Becker A, Leonard M, Schmid SL, Vallee RB, Kull FJ, Manstein DJ. Crystal structure of the GTPase domain of rat dynamin 1. Proc Natl Acad Sci USA. 2005;102:13093–13098. doi: 10.1073/pnas.0506491102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux A, Uyhazi K, Frost A, De Camilli P. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- Roux A, Koster G, Lenz M, Sorre B, Manneville JB, Nassoy P, Bassereau P. Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci USA. 2010;107:4141–4146. doi: 10.1073/pnas.0913734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL, Frolov VA. Dynamin: Functional design of a membrane fission catalyst. Ann Rev Cell Dev Biol. 2011 doi: 10.1146/annurev-cellbio-100109-104016. in press. [DOI] [PubMed] [Google Scholar]
- Sever S, Skoch J, Newmyer S, Ramachandran R, Ko D, McKee M, Bouley R, Ausiello D, Hyman BT, Bacskai J. Physical and functional connection between auxilin and dynamin during endocytosis. EMBO J. 2006;25:4163–4174. doi: 10.1038/sj.emboj.7601298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomaha E, Palfrey HC. Conformational changes in dynamin on GTP binding and oligomerization reported by intrinsic and extrinsic fluorescence. Biochem J. 2005;391:601–611. doi: 10.1042/BJ20050707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BD, Yarar D, Schmid SL. An assembly-incompetent mutant establishes a requirement for dynamin self-assembly in clathrin-mediated endocytosis in vivo. Mol Biol Cell. 2004;15:2243–2252. doi: 10.1091/mbc.E04-01-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell MH, Marks B, Wigge P, McMahon HT. Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat Cell Biol. 1999;1:27–32. doi: 10.1038/8997. [DOI] [PubMed] [Google Scholar]
- Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Sweitzer SM, Hinshaw JE. Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell. 1998;93:1021–1029. doi: 10.1016/s0092-8674(00)81207-6. [DOI] [PubMed] [Google Scholar]
- Tan RKZ, Petrov AS, Harvey SC. YUP: A molecular simulation program for coarse-grained and multi-scale models. J Chem Theory Comput. 2006;2:529–540. doi: 10.1021/ct050323r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan RK, Devkota B, Harvey SC. YUP. SCX: coaxing atomic models into medium resolution electron density maps. J Struct Biol. 2008;163:163–174. doi: 10.1016/j.jsb.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature. 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Timm D, Salim K, Gout I, Guruprasad L, Waterfield M, Blundell T. Crystal structure of the pleckstrin homology domain from dynamin. Nat Struct Mol Biol. 1994;1:782–788. doi: 10.1038/nsb1194-782. [DOI] [PubMed] [Google Scholar]
- Warnock DE, Hinshaw JE, Schmid SL. Dynamin self-assembly stimulates its GTPase activity. J Biol Chem. 1996;271:22310–22314. doi: 10.1074/jbc.271.37.22310. [DOI] [PubMed] [Google Scholar]
- Wilson-Kubalek L, Chappie JS, Arthur CP. Helical crystallization of soluble and membrane binding proteins. Meth Enzymol. 2010;481:45–62. doi: 10.1016/S0076-6879(10)81002-X. [DOI] [PubMed] [Google Scholar]
- Zhang P, Hinshaw JE. Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol. 2001;3:922–926. doi: 10.1038/ncb1001-922. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
01
02
03
04
05