mRNA Transcript Diversity Creates New Opportunities for Pharmacological Intervention (original) (raw)
Abstract
Most protein coding genes generate multiple RNA transcripts through alternative splicing, variable 3′ and 5′UTRs, and RNA editing. Although drug design typically targets the main transcript, alternative transcripts can have profound physiological effects, encoding proteins with distinct functions or regulatory properties. Formation of these alternative transcripts is tissue-selective and context-dependent, creating opportunities for more effective and targeted therapies with reduced adverse effects. Moreover, genetic variation can tilt the balance of alternative versus constitutive transcripts or generate aberrant transcripts that contribute to disease risk. In addition, environmental factors and drugs modulate RNA splicing, affording new opportunities for the treatment of splicing disorders. For example, therapies targeting specific mRNA transcripts with splice-site–directed oligonucleotides that correct aberrant splicing are already in clinical trials for genetic disorders such as Duchenne muscular dystrophy. High-throughput sequencing technologies facilitate discovery of novel RNA transcripts and protein isoforms, applications ranging from neuromuscular disorders to cancer. Consideration of a gene's transcript diversity should become an integral part of drug design, development, and therapy.
Introduction
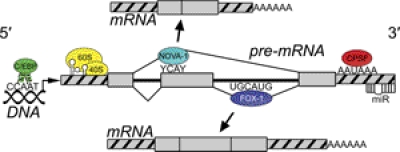
The human genome consists of 20,000 to 25,000 protein-coding genes, but the repertoire of mRNA sequences and encoded proteins is far greater as a result of multiple RNA isoforms generated from each gene. RNA transcript diversity evolves from several mechanisms, including alternative splicing, alternative transcription initiation and polyadenylation site usage, RNA editing, and _trans_-splicing over long distances from different gene loci. RNA splicing, in particular, is a major factor driving phenotypic diversity in higher eukaryotes. Alternative transcripts are created by a series of splicing events: exon skipping, intron retention, alternative 5′ and 3′ splice sites, alternative last exons, tandem 3′ untranslated regions, alternative first exons, and mutually exclusive exons. In addition, translational control elements in mRNA isoforms can affect protein levels, including internal ribosome entry sites in the 5′ UTR or degradation signals and microRNA (miR) binding sites in the 3′UTR (Fig. 1). All RNA processing events follow a tissue-specific expression pattern that varies less across individuals than among different tissues in the same individual, enabling selective drug targeting of RNA and protein isoforms to the site of action. Tissue-specific alternative splicing is greatly affected by splicing factors present in that tissue. These may be ubiquitously expressed but present at different levels in different tissues, or the factors may be preferentially expressed in certain tissues. For example, NOVA-1 is only expressed in neurons, and is present at different levels, depending on the brain region studied. Although the target sequences for many of these splicing factors are available, much is still unknown about their mechanisms. Whole-transcript screening has elucidated some tissue-specific regulatory motifs and detected patterns that cluster by tissue (Castle et al., 2008), but this area is still under investigation.
Fig. 1.
Schematics of a protein coding gene locus, showing consensus binding motifs in genomic DNA: transcription factor complexes (e.g., C/EBP); in RNA exons and introns: alternative splicing enhancers/silencers (NOVA-1 and FOX-1); and in untranslated regions (hatched): internal ribosome entry sites (60S/40S), alternative polyadenylation signals [cleavage polyadenylation specificity factor (CPSF)], and microRNA sites (miR).
RNA Splicing and Human Transcriptome Diversity: Relevance to Disease and Drug Design
Splicing events are highly prevalent, estimated to occur for 95% of all multiexon genes (Pan et al., 2008), 86% of which express a minor isoform abundance of 15% or more of the total gene expression (Wang et al., 2008). The majority of splicing events (70–88%) alter the encoded protein (Kan et al., 2001; Modrek et al., 2001), more than half causing a shift in the mRNA reading frame. Using high-throughput sequencing technologies to accurately predict transcript isoforms remains challenging because of short and less than parsimonious sequence motifs directing processing, complex effects of tertiary structure, and tissue-specific assembly of the multicomponent spliceosome (Modrek and Lee, 2002). In addition to genetically determined splicing differences, environmental factors such as diet or toxins can alter splicing patterns. Even environmental regulation of epigenetic factors typically associated with transcription can regulate splicing, with the same proteins involved in both events (Young et al., 2005). Consequently, transcript variability resulting from both genetic and environmental factors can adversely affect drug efficacy or enhance toxicity but can also be exploited for therapeutic purposes.
Common genetic variants can afford changes in alternative splicing within a “normal” physiological range. However, mutations causing aberrant splicing typically result in nonfunctional protein or nonsense-mediated RNA decay, if a codon phase shift introduces premature termination signals (Lareau et al., 2007; Wang and Cooper, 2007; Cooper et al., 2009). Up to 50% of diseases with genetic components involve splicing mutations (Faustino and Cooper, 2003; Wang and Cooper, 2007). One approach to effective therapy is to reduce formation of disease-associated aberrant RNA isoforms using oligonucleotides to suppress splicing defects (Wilton and Fletcher, 2005), further discussed below. Alternatively, targeting a specific protein or RNA isoform can increase efficacy or reduce off-target adverse effects. A drug typically binds to multiple proteins in the body involving many genes, refuting the idea of one drug/one protein target. However, the concept of multiple proteins generated from a single gene locus has not systematically affected drug design, even though protein isoforms can have distinct functions. For example, the exon 9-lacking (Δ9) cholesteryl ester transfer protein (CETP) (Inazu et al., 1992; Lira et al., 2008) and dopamine receptor D3 (Elmhurst et al., 2000; Karpa et al., 2000) act in a dominant-negative manner; the alternatively spliced transcript prevents the function of the normal transcript. CETP transfers cholesterol esters from HDL to low-density lipoprotein, thus decreasing the ratio of HDL to overall cholesterol. In an effort to protect against coronary artery disease by boosting HDL levels at the expense of low-density lipoprotein, several CETP inhibitors are currently under development. The first drug candidate acting as a CETP inhibitor (torcetrapib) was withdrawn in late phase III trials because of an unexpected rise in coronary artery disease incidence (Barter et al., 2007), suggesting a complex relationship between HDL, cardiovascular disease, and underlying genetic factors. A better understanding of the physiological regulation of CETP activity by Δ9-CETP as an “internal” inhibitor, and genetic variants potentially affecting this process, will prove critical in guiding optimal therapy with CETP inhibitors.
Although it is important to consider transcript diversity exhibited by drug targets, the enzymes that metabolize these drugs can also be affected by both normal and aberrant splicing. Variation in drug-metabolizing enzyme activity, as observed with cytochrome P450s, introduces variability in drug response, leading to increased incidence of toxicity, poor efficacy, or both (Phillips et al., 2001). A common variant allele, CYP2D6*4, introduces a mutation in the splice site of intron 3, causing the pre-mRNA of this gene to be incorrectly spliced, resulting in a nonfunctional enzyme (Kagimoto et al., 1990; Marez et al., 1997). Patients homozygous with this variant are poor metabolizers of numerous drugs; therefore, current drug design aims at avoiding CYP2D6 substrates. Another metabolizing enzyme, _N_-acetyltransferase 1 (NAT1), processes endogenous and exogenous compounds, including drugs and environmental carcinogens. NAT1 RNA transcripts arise from one coding exon (exon 9) with three possible upstream promoter/transcription start sites, several splice variants, and three polyadenylation sites, resulting in different translation yields (Boukouvala and Sim, 2005; Butcher et al., 2005; Wang et al., 2011). In contrast to the mostly hepatic expression of NAT2, NAT1 is ubiquitously expressed, but enzyme activities are differentially regulated across tissues, pharmacological and toxicological implications resulting from drug or toxin acetylation. Another example of pharmacological significance, the nuclear receptors constitutive androstane receptor and pregnane X receptor, regulators of drug-metabolizing enzyme expression with a key role in defense against xenobiotic exposure, are alternatively spliced, increasing the range of recognized ligands and downstream target genes. Phthalates, chemicals used in commercial manufacturing, and other endocrine-disrupting compounds have differential selectivity for constitutive androstane receptor isoforms (DeKeyser et al., 2011), an issue relevant to toxicology.
In Vitro and In Silico Screening for Drugs That Alter Splicing
General splicing defects occur in several complex disorders, including cancer and psychiatric disorders (Wang and Cooper, 2007; Cooper et al., 2009). For example, mutations in genes encoding auxiliary splicing factors such as methyl CpG binding protein 2 and RNA binding protein fox-1 homolog (Caenorhabditis elegans) affect multiple downstream targets in autism spectrum disorders and schizophrenia (Smith and Sadee, 2011; Voineagu et al., 2011). Discovery of drugs broadly targeting splicing has accelerated with rapid screening techniques. For example, a splicing reporter assay allows for high-throughput screening of splicing modulators (Stoilov et al., 2008). Using splicing reporter constructs, 4000 compounds were screened for their ability to inhibit spliceosome assembly (Soret et al., 2005). In silico approaches can assist in predicting the impact of sequence variation on splicing enhancer/suppressor sites (human splicing finder) (Desmet et al., 2009) or assessing secondary RNA structure with programs such as Mfold (Zuker, 2003) to reveal sequence-independent structural motifs crucial for protein-RNA binding (G-quartets, kissing complexes, etc.). These tools can guide in designing and screening compounds in drug development.
Below, we discuss pharmacologically relevant examples of transcript complexity as demonstrated by the calcium channel CACNA1C and the drug-metabolizing enzyme UDP-glucuronosyltransferase (UGT) 1A. In addition, we provide examples where transcript isoforms are relevant to drug response and drug design (Table 1). Finally, we review approaches to altering specific RNA processing as a means for treating genetic disorders.
TABLE 1.
Biological functions of transcript isoforms and pharmacological implications
| Gene | Function/Role | Features | References |
|---|---|---|---|
| CETP | Lipid transport protein | Splicing affected by diet, dominant-negative | Yang et al., 1996; Dessi et al., 1997; Lira et al., 2008 |
| UGT1A | Drug metabolism | Alternative first exon usage and exon 5 splicing | Levesque et al., 2007; Gong et al., 2001 |
| CACNA1C | Target in hypertension, arrhythmia treatment | Splice variants have different expression patterns and dihydropyridines sensitivity | Welling et al., 1997,Tang et al., 2004; ; Wang et al., 2006 |
| DRD2 | Antipsychotic drug target | Splice forms have different drug sensitivity | Malmberg et al., 1993; Usiello et al., 2000; Xu et al., 2002 |
| OPRM1 | Analgesics and narcotics receptor | Alternate exon 1 affects opioid analgesic effects | Schuller et al., 1999; Pan et al., 2005a,b, 2009 |
| COX-1 | NSAID target | Three splice isoforms, relevance in humans | Chandrasekharan et al., 2002; Kis et al., 2005; Qin et al., 2005 |
| MYD88 | Inflammation | Splice form lacking exon 2 (MyD88S) decreases inflammation | Janssens et al., 2002; Burns et al., 2003; Vickers et al., 2006 |
| TNFR | Inflammation | Exon exclusion produces soluble TNF receptor | Graziewicz et al., 2008 |
| NAT1 | Drug detoxification | Isoforms with different translation efficiencies | Boukouvala and Sim, 2005; Butcher et al., 2005; Wang et al., 2011 |
| CYP2D6 | Drug metabolism | *4 allele alters splicing, no enzyme activity | Marez et al., 1997; Kagimoto et al., 1990 |
| SCN1A | Drug target in epilepsy treatment | Splice isoforms have different sensitivity to phenytoin and lamotrigine | Thompson et al., 2011 |
| CAR | Nuclear receptor, xenobiotic sensing/processing | Alternate splicing alters ligands recognized by receptor | DeKeyser et al., 2011 |
| VEGF | Growth factor | Splice isoforms can have opposite effect | Harper and Bates, 2008 |
| BCL2L | Apoptosis regulator | Splice isoforms are anti- or pro-apoptotic | Bauman et al., 2010 |
| Disease-causing genes | |||
| MSTR1 | Oncogene | SSO can increase transcript length | Ghigna et al., 2010 |
| SMN | Spinal muscular atrophy | Exon 7 skipped. Multiple approaches for inclusion | Wirth et al., 2006; Vitte et al., 2007; Singh et al., 2009; Hua et al., 2010 |
| DMD | Duchenne muscular dystrophy | Exon 51 mutation and frameshift | van Deutekom et al., 2007, Goemans et al., 2011; Cirak et al., 2011 |
| IKBKAP | Familial dysautonomia | Phosphatidylserine and kinetin correct splicing defect | Keren et al., 2010; Axelrod et al., 2011; Shetty et al., 2011 |
| NF-1 | Neurofibromatosis | Kinetin corrects splicing defect | Pros et al., 2010 |
A Voltage Dependent L-Type Calcium Channel: Example of Extreme Splicing Diversity
The L-type calcium channel α-subunit 1c (Cav1.2, CACNA1C) regulates blood pressure and cardiac function, serving as a target of Ca2+ channel blockers. An extraordinary example of transcript diversity, at least 10 of 55 exons undergo alternative splicing, with several sites yielding multiple isoforms, leading to more than 10,000 possible splice variants (Welling et al., 1997; Tang et al., 2004). In addition to displaying profound differences in channel functions, these splice variants have been associated with disease susceptibility and variable response to drugs used to treat hypertension and arrhythmias. CACNA1C is expressed in cardiac and smooth muscle, brain, and other tissues. Mutually exclusive exons E8a/8 encode part of the dihydropyridine drug-binding site. E8-containing channels expressed in smooth muscle display 10-fold greater affinity for dihydropyridine channel blockers than channels containing E8a, the cardiac form (Welling et al., 1997). We have studied interindividual variability in mRNA expression and splicing of CACNA1C in 65 heart tissue samples from recipients of heart transplants (Wang et al., 2006), demonstrating dramatic differences in the expression pattern of exon 8/8a among individuals, ranging from 9 to 87% for exon 8a usage. Two heart tissues predominantly contained the smooth muscle-specific form, raising the question as to the pharmacological response to dihydropyridines in persons with this expression pattern, likely to experience strong effects in the heart (Wang et al., 2006). This example highlights interindividual variability in alternative splicing, which drastically modulates pharmacological effects. We failed to detect any genetic factors accounting for these differences, suggesting that _trans_-effects from the splicing machinery play a key role. We propose that this type of variability should be considered in drug design, potentially targeting channel blockers to binding pockets unaffected by the 8/8a splice variation.
Transcript Diversity Affecting Multiple Drugs: UGT1A
UGTs are a family of enzymes responsible for the detoxification of endogenous and exogenous compounds through the formation of hydrophilic glucuronides excreted through urine, feces, or bile. Metabolic targets include bilirubin, steroids, and drugs such as irinotecan and clozapine. The UGT1A gene locus is under the control of tissue-specific promoters, which results in expression of unique multiple first exons paired with four constitutively expressed downstream exons via alternative splicing (Gong et al., 2001). The alternative first exons encode the N-terminal 280 residues, which are responsible for substrate specificity (Ritter et al., 1992). To date, 13 UGT1A alternative first exons have been identified that have various degrees of substrate selectivity and tissue expression. This transcript diversity has physiological and pharmacological relevance. For example, the primary function of UGT1A1, mainly expressed in the liver, is the glucuronidation of bilirubin (Ritter et al., 1992).
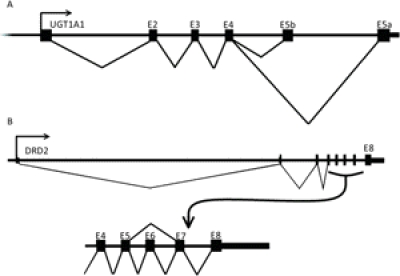
Alternative splicing also occurs at the 3′ end of the UGT1A locus. Originally identified in UGT1A1, alternative splicing of exon 5 (Lévesque et al., 2007) yields two splice variants, the active enzyme UGT1A1_i1 and a truncated isoform UGT1A1_i2, illustrated in Fig. 2A. Upon heterodimerization, UGT1A_i2 inhibits UGT1A_i1-mediated glucuronidation, another example in which a splice isoform exerts dominant-negative effects (Lévesque et al., 2007). Alternative splicing of exon 5 extends through all active UGT1A genes (UGT1A1, 1A3-10) in a tissue-specific manner, presumably creating inhibitory isoforms for each (Girard et al., 2007).
Fig. 2.
A, splice isoforms of UGT1A1. Alternative splicing of exon 5 at UGT1A gene locus creates protein isoforms capable of modulating UGT1A activity. Usage of exon 5a creates UGT1A1_ i1, responsible for enzymatic activity, whereas usage of exon 5b truncates the C terminus, creating UGT1A1_i2. Multiple alternative exon 1 transcripts with separate promoter regions are not shown. B, alternative splicing of DRD2 at exon 6 produces distinct protein isoforms D2S (short) and D2L (long) with varying pharmacological profiles.
Genetic variation contributes an additional layer of complexity to the relationship between UGT1A expression and substrate metabolism. Located in the promoter region of an alternative exon 1, a dinucleotide repeat allele, termed *28, alters UGT1A1 expression. Homozygous carriers of the minor allele have Gilbert's syndrome (hyperbilirubinemia) (Bosma et al., 1995) and reduced irinotecan inactivation, associated with irinotecan toxicity (Marsh and McLeod, 2004), making *28 a potential biomarker for guiding irinotecan therapy and reducing toxicity. Nonsynonymous mutations in UGT1A4, such as P24T or L48V, can alter metabolism of xenobiotics, steroids (Ehmer et al., 2004), and clozapine (Mori et al., 2005).
The ability of the UGT1A locus to affect endogenous and exogenous drug metabolism strongly depends on transcript variation across the many tissues expressing UGT1A. In a similar fashion, UGT1A transcript complexity via alternative first exon usage and exon 5 splicing, together with genetic factors, will have significant impact on drug efficacy in target tissues. Certainly, tissue-selective isoform expression bears upon drug design, considering the site of pharmacological action or toxicity.
Splicing in the CNS Affects Drug Targets
Nowhere is mRNA transcript diversity more prevalent than in the brain (Modrek et al., 2001; Yeo et al., 2004; de la Grange et al., 2010), offering opportunities for generating selective pharmacological profiles where alternative splicing results in unique functional proteins. Less obvious effects on protein expression also arise from alternative UTR usage. For example, greater length and complexity of the 5′ UTR generally correlates with reduced protein translation efficiency (Kozak, 1989; Gray and Hentze, 1994). In heteromeric receptor complexes, receptor stoichiometry can depend on the availability of a particular subunit (Moroni et al., 2006), thus resulting in unique pharmacological profiles in which tissue-specific 5′UTRs differ in translation efficiency. A similar case can be made for polyadenylation site usage, which determines 3′ UTR length, potentially increasing the number of regulatory elements in longer UTRs that are subject to _trans_-acting factors (miRs, translational control proteins, etc.). Discussed below are select examples of transcript diversity creating distinct pharmacological profiles.
The Neuronal Nav1.1 Sodium Channel.
Voltage-gated sodium channels are heteromultimeric complexes involved in generating and propagating action potentials. Type I channels transmit signals between neurons in the brain. Defects in SCN1A cause certain types of epilepsy and migraines. Encoding Nav1.1, a large α subunit, SCN1A is alternatively spliced to include either the canonical 5A or alternative (5N) exon 5. Channels with the alternative exon are more sensitive to the effects of phenytoin and lamotrigine, common antiepileptic medications (Thompson et al., 2011). These drugs interact differently with spliced targets, thus affecting their efficacy. Epileptic patients with single-nucleotide polymorphism rs3812718, located in a splice donor site (Tate et al., 2005), have increased expression of the 5N form of the transcript, requiring lower doses of phenytoin.
The μ Opioid Receptor.
The μ opioid receptor (MOR, encoded by OPRM1) is the crucial target for analgesic compounds and narcotics, including morphine, codeine, and heroin. OPRM1 transcripts originate from two distinct first exon promoters (Pan et al., 2001; Xu et al., 2009; Xu et al., 2011), whereas the 3′ end undergoes extensive splicing to produce at least 19 distinct isoforms in humans (20 isoforms at AceView and 23 at Ensembl, 4 of which do not encode protein.) These different mRNA transcripts are regionally distributed throughout the brain (Pan et al., 2001).
Ligand-dependent MOR signaling varies depending on alternative promoter usage at the OPRM1 gene. OPRM1 transcription can initiate approximately 28 kilobases upstream of the annotated exon 1 to transcribe an alternative first exon (known as exon 11) (Xu et al., 2009). Disruption of exon 11-derived variants markedly reduces analgesic effects of heroin and morphine-6β-glucuronide, but does not affect morphine and methadone analgesia (Pan et al., 2009). Conversely, disruption of exon-1-derived spliced isoforms in mice causes ligand-dependent differences in analgesia whereby heroin and morphine-6β-glucuronide retain residual analgesic properties, whereas morphine does not (Schuller et al., 1999). The mechanisms underlying differential opioid effects on OPRM1 isoforms remain controversial.
A second critical element of MOR signaling is protein diversity generated at the carboxyl terminus through alternative splicing at the 3′ ends of OPRM1 transcripts, with significant influence on ligand-binding affinity. For example, the human MOR transcript variant hMOR-1B2 is alternatively spliced at exon 5, binding most opioids and opioid peptides with lower affinity than other C-terminal variants (Pan et al., 2005a). C-terminal splicing of OPRM1 RNA in humans (Pan et al., 2005a) and rodents (Pan et al., 2005b) also affects potency and efficacy for a range of opioid compounds. An additional layer of complexity among all G protein-coupled receptors (GPCRs), of which OPRM1 is a member, is receptor oligomerization (Bouvier, 2001; Wang et al., 2005). As it pertains to transcript splicing, ligand-affinity differences for hetero-oligomers of MORs and δ opioid receptors are dependent upon carboxyl tail length (Fan et al., 2005). Although GPCRs seem to dimerize and polymerize readily, the promiscuity of G protein-coupled receptor heteropolymerization is still under debate, as is its role in pharmacology and drug design.
The Dopamine D2 Receptor.
The D2 receptor serves as a primary target for antipsychotic compounds, with a finite window of D2 receptor occupancy linked to clinical efficacy (Nordström et al., 1993; Kapur et al., 2000) or adverse effects (Farde et al., 1992; Kapur et al., 2000). DRD2 is spliced to produce functionally distinct proteins with unique pharmacological profiles. Alternative splicing of exon 6 produces either the short (D2S) or long (D2L) protein isoforms (Giros et al., 1989) (Fig. 2B) that differ in their subcellular localization (Khan et al., 1998). D2S predominantly localizes on the presynaptic terminal, acting as an autoreceptor to inhibit synaptic dopamine release, whereas D2L acts primarily as a postsynaptic target for dopamine transmission (Khan et al., 1998; Usiello et al., 2000). We have identified two single-nucleotide polymorphisms in introns 5 and 6 that significantly shift splicing toward inclusion of exon 6, thereby reducing autoreceptor activity of D2S in the striatum and prefrontal cortex, significantly affecting cognition, neural activity (Zhang et al., 2007; Bertolino et al., 2009, 2010; Blasi et al., 2009) and risk of cocaine abuse/overdose (Moyer et al., 2011).
Differential pharmacological modulation of D2S versus D2L signaling offers an opportunity for correcting imbalanced DRD2 signaling with drugs that display differential activity for the long versus short protein (Castro and Strange, 1993; Malmberg et al., 1993; Usiello et al., 2000; Xu et al., 2002). In a neuroendocrine cell line, sarizotan acts as a full agonist at D2L but as a partial agonist at D2S (Kuzhikandathil and Bartoszyk, 2006). As another example, glutamate-mediated spontaneous excitatory postsynaptic currents are inhibited by quinpirole when the D2 ratio was altered in favor of D2S (Centonze et al., 2004). Using D2L-specific knockout mice, Xu et al. (2002) indirectly demonstrated higher D2L affinity compared with D2S for haloperidol and clozapine, whereas they observed the opposite for raclopride. Malmberg et al. (1993) also found greater D2L affinity for remoxipride, sulpiride, chlorpromazine, clozapine, and thioridazine but greater D2S affinity for raclopride (Castro and Strange, 1993; Malmberg et al., 1993). However, owing to the propensity of antipsychotics to interact with multiple receptors, most notably serotonin receptor 2A (Borroto-Escuela et al., 2010), it remains uncertain whether targeting D2S or D2L conveys higher efficacy or toxicity. However, treatment success may depend on such subtype selectivity, an area in need of further exploration.
Nonsteroidal Anti-Inflammatory Drugs.
Drug design and development benefit from knowing the molecular mechanism of action and intended target, especially when represented by multiple isoforms. Currently, neither of these issues is clear for NSAIDs, thought to act in part by inhibiting cyclooxygenases (COX). Originally, two cyclooxygenases derived from separate genes were described, COX-1 and COX-2. Varying affinities for COX-1 or COX-2 define pharmacological and toxicological NSAID properties (Chandrasekharan et al., 2002). A third isoform, COX-3, created by retention of intron 1 after alternative splicing of COX-1, was identified in canine tissues. However, the proposed selectivity of acetaminophen for COX-3 did not translate to humans (Kis et al., 2005). Alternate splicing of COX-1 actually produces at least three COX-1 isoforms in humans that differ from the canine isoforms. The most prevalent isoform, COX-1b1, encodes a truncated and nonfunctional protein. COX-1b2, the isoform most similar to COX-3, did not differ in its response to NSAIDs compared with COX-1 (Qin et al., 2005). Targeting of alternate transcripts from COX genes stimulated short-lived excitement, but failure to produce a breakthrough unfortunately quenched enthusiasm for drug design targeting splice variants.
Drug Design Directly Targeting Aberrant Splicing or Suppressing Pathogenic RNA Transcripts
Drugs that target RNA processing to treat disorders involving aberrant isoforms are an alternative approach to current therapies. On one hand, small molecules can modulate _trans_-acting splice proteins, thereby changing spliced isoform expression profiles across many genes. On the other hand, splice-switching oligonucleotides (SSOs) can be deployed to correct aberrant splicing or down-regulate unwanted RNA transcripts, as discussed below. The technique of using SSOs to correct splicing defects was originally developed by the Kole group to treat β-thalassemia (Sierakowska et al., 1996; Schmajuk et al., 1999). This disorder is caused by mutations creating aberrant splicing sites, leading to the inclusion of a stop codon and the creation of a truncated β-globin protein. In this case, the SSOs bind and cover the mutated splice site to prevent inclusion of the intron; however, SSOs can affect splicing along several mechanisms.
A component of the spliceosome required for splice donor site recognition, uridine-rich small nuclear RNAs (U1 snRNA), can be modified to correct specific splicing defects (Gorman et al., 1998, 2000). Likewise, spliceosome-adapted U7 snRNA is needed to process histone mRNA. Modified synthetic U1 and U7 RNAs were shown to be effective in cell culture at inducing exon skipping or inclusion (Pinotti et al., 2008; Geib and Hertel, 2009; Hartmann et al., 2010; Incitti et al., 2010). A means of targeting specific mRNA transcript isoforms relies on complementary antisense oligonucleotides that can restore the normal splicing process disrupted by a mutation, skip disease-associated exons (Skordis et al., 2003), degrade unwanted transcripts, or increase production of a desired transcript. RNA interference therapies employ small interfering RNA duplexes of 21 to 23 nucleotides. By incorporation into the RNA-induced silencing complex, siRNAs promote degradation of mRNA containing matched complementary sequences. siRNAs, such as miRs and their antisense counterparts, can regulate splicing, but the numerous other mechanisms by which they impart therapeutic potential goes beyond the scope of this review.
Abundant delivery of oligonucleotides to the target tissue and into the cell interior presents a major challenge for developing effective therapeutics. Slow penetration of lipid bilayers and rapid degradation in vivo require chemical modifications and specific delivery strategies. Oligonucleotides can be delivered by liposomes, conjugates with cell surface recognition factors, viral vectors, or biolistic injection (plasmid DNA affixed to particles coated with heavy metal, delivered through the skin by particle bombardment); however, successful delivery remains a challenge, with only a few target tissues highly susceptible to intracellular drug delivery methods.
Small Molecules to Treat Familial Dysautonomia.
Compounds consumed in our daily diet can affect splicing, as observed with Δ9-CETP up-regulation in response to dietary components (Yang et al., 1996; Dessí et al., 1997). One can take advantage of this insight to search for drugs that modify splicing. Familial dysautonomia (FD) is caused by a splice-site mutation in the IKBKAP gene, excluding exon 20. Treatment with phosphatidylserine, an FDA-approved supplement, increases the wild-type form of IKBKAP mRNA and IKAP protein levels in a patient cell line (Keren et al., 2010). A second therapeutic option for FD treatment is the plant-derived kinetin, which has the ability to cross the blood-brain barrier. In a transgenic mouse model, kinetin corrected IKBKAP splicing defects in the brain and across tissues, resulting in increased IKAP protein levels (Shetty et al., 2011). These results were replicated in humans, with patients receiving one month of oral kinetin treatment demonstrating increased levels of wild-type IKBKAP mRNA (Axelrod et al., 2011). Kinetin's effect is not specific for the IKBAP gene; it also modulates and partially corrects aberrant splicing of neurofibromatosis type 1 (NF-1) (Pros et al., 2010). The use of amiloride in the treatment of a neuromuscular disorder and cancer to modulate splicing events is further discussed below. The number of candidate drugs in this category is still small, but they hold promise in the therapy of diseases involving aberrant splicing.
Applications in Neuromuscular Disorders.
Spinal muscular atrophy (SMA) is a neuromuscular disease caused by a mutational splicing defect in the SMN1 gene. This deficiency is only partially compensated for by the homologous SMN2 gene, because exon 7 is readily spliced out of SMN2 nascent RNA, producing an unstable protein, resulting in inadequate SMN activity (Vitte et al., 2007). Fitting to the theme of this review, mature SMN plays a role in small nuclear ribonucleoprotein assembly essential to spliceosome function (Gabanella et al., 2007) so that deficiencies lead to widespread splicing defects (Zhang et al., 2008). Modifying SMN2 splicing to include exon 7 restores the fully active protein, achievable with use of short antisense oligonucleotides (ASOs). In cells taken from a patient with SMA, an 8-mer ASO restored exon 7 inclusion and mature SMA formation, a first step toward effective therapy (Singh et al., 2009). Although encouraging, appropriate drug delivery into the CNS is particularly challenging because oligonucleotides cannot readily cross the blood-brain barrier. Getting the drug to the target tissues requires an intracranial injection with use of an osmotic pump. This approach effectively increased expression of SMN in the brain and spinal cord, in a mouse model of SMA (Williams et al., 2009; Hua et al., 2010).
Alternative therapies have also shown promise for modifying SMN splicing. The antihypertensive drug amiloride and its analog 5-(_N_-ethyl-_N_-isopropyl)amiloride increase expression of SMN2 protein in SMA cells by increasing inclusion of exon 7 (Yuo et al., 2008), as does valproic acid (Harahap et al., 2012). A number of histone deacetylase inhibitors increase the lifespan of SMA mice (Wirth et al., 2006). Modulating extracellular pH also modifies splicing of SMN2; high pH increases exon 7 inclusion and low pH increases skipping of exon 7 (Chen et al., 2008). These observations demonstrate the sensitivity of RNA splicing to environmental factors that could either aggravate disease symptoms or lead to palliative therapies.
Fukuyama-type congenital muscular dystrophy is a common recessive disorder in Japan, leading to disability and premature death. A retrotransposon insertion disrupts the fukutin gene, activating a previously inaccessible donor splice site and creating a new splice acceptor site. To correct this defect, an ASO was developed that targets the defect and blocks the detrimental splicing. It has been effective in expressing normal protein in a patient cell line and was also able to restore partial fukutin protein expression in a mouse model after intramuscular injection (Taniguchi-Ikeda et al., 2011).
Duchenne muscular dystrophy (DMD) is a lethal disorder caused by mutations in the dystrophin gene that interrupt the open reading frame. A variety of mutations can lead to DMD. Here we discuss deletion of exon 50, alone or in combination with other adjacent deletions, present in 13% of patients, with potential treatments in phase 2 clinical trials. In these patients, a new stop codon leads to a truncated, nonfunctional protein. Two oligonucleotide agents under investigation are aimed at splicing out exon 51 to restore the open reading frame, thereby producing functional dystrophin. Although the dystrophin gene is not alternatively spliced, and DMD is not caused by a splicing defect, the oligonucleotides are effective without targeting the splice site, because this therapy introduces a splicing event to overcome the genetic aberration. These oligonucleotides are complementary to a portion of the exon 51 pre-mRNA sequence and block it from being included in the processed transcript. The shift restores the reading frame, allowing a shorter yet partially functional protein to be translated. Administration of ASO PRO051 enhanced dystrophin activity in patients after either intramuscular injection (van Deutekom et al., 2007) or systemic administration (Goemans et al., 2011). Cirak et al. (2011) demonstrated the safety and efficacy of AVI-4658, a phosphorodiamidate morpholino oligomer, with increased dystrophin protein expression. These therapies all hold significant promise for treatment of neuromuscular diseases, a proof of principle indicating that correcting aberrant RNA processing can be applied to other diseases as well.
Applications in Cancer Therapy.
Both alternative and aberrant splicing play a role in oncogenesis and treatment response. Antiangiogenesis compounds that reduce the activity of vascular endothelial growth factor (VEGF) or its receptors can be effective at slowing tumor growth (Hurwitz et al., 2004). However, alternative splicing of VEGF and VEGF receptors can result in transcripts with effects opposite those traditionally attributed to VEGF signaling, inhibiting angiogenesis and slowing the growth of a wide range of cancers (Woolard et al., 2004; Varey et al., 2008). As a consequence, nonspecifically targeting VEGF signaling potentially promotes angiogenesis or cell survival; therefore, anti-VEGF drugs should target-specific spliced isoforms (for review, see Harper and Bates, 2008) or _trans_-acting splice proteins that favor expression of antiangiogenic splice variants. As an example of the latter, spliceostatin A blocks the ability of the endogenous splicing machinery to process VEGF, thereby reducing angiogenesis in vivo (Furumai et al., 2010).
Vemurafenib, a relatively new and promising treatment for metastatic melanoma, acts by inhibiting an oncogenic form of BRAF. Normally involved in cell replication and survival, BRAF is mutated in approximately half of patients with melanoma, causing increased cell proliferation via constitutive activity of BRAF(V600E). However, most patients develop resistance to the drug within a year of use, making it vital to determine the causes of drug resistance. A novel mechanism was recently discovered in vemurafenib-resistant cells containing a truncated form of BRAF, p61BRAF(V600E). Monomeric p61BRAF(V600E) is as sensitive as the full-length form, yet upon dimerization, the cells are no longer sensitive to vemurafenib. The mRNA is missing exons 4 to 8, which normally encode a region that suppresses dimerization; therefore, the deletion allows increased dimerization and bypasses the drug inhibition. BRAF variants lacking this domain were found in a subset of patient samples, confirming its clinical relevance (Poulikakos et al., 2011). Aberrant splicing of the target renders this drug ineffective, highlighting our central thesis: by understanding alternative and aberrant splicing, one can develop therapeutics tailored to the patient's genetic and biochemical characteristics.
The antihypertensive drug amiloride can regulate alternative splicing of oncogenic fusion genes, showing therapeutic promise in the treatment of chronic myelogenous leukemia by increasing the sensitivity of cancerous cells to imatinib (Chang et al., 2011). Targeting _trans_-acting splice proteins, however, lacks specificity, because these proteins direct alternative splicing of many genes and can lead to nonspecific effects. Natural compounds with antitumor activity have been discovered, and bind to components of the spliceosome, inhibiting pre-mRNA splicing (Albert et al., 2007; Kaida et al., 2007; Kotake et al., 2007; O'Brien et al., 2008). Although the development of new chemotherapeutics is promising, it is important to understand the implications of blocking splicing in general. A more direct and specific effect can be achieved with SSOs, an approach used for treatment of the neuromuscular disorders described above. Bauman et al., 2010 demonstrated successful reduction in tumor growth through lipid nanoparticle delivery of an SSO that increases splicing of the antiapoptotic B-cell lymphoma 2-like 1L transcript toward formation of the shorter proapoptotic B-cell lymphoma 2-like 1S isoform. SSOs can also be used to produce longer alternatively spliced transcripts through the inclusion of alternative exons, as demonstrated for the protooncogene Ron (MSTR1, macrophage stimulating 1 receptor) (Ghigna et al., 2010). These examples demonstrate potential utility but also reveal that the field is still in its infancy.
Applications in Immune Disorders
Novel approaches modulating RNA processes have also emerged in therapy of immune disorders. Facilitated drug delivery to immune cells ranks as a key advantage for this approach over targeting CNS disorders.
MyD88 is an adapter molecule involved in signal transduction by toll-like receptors (Akira et al., 2001) and interleukin-1 receptor (IL-1R) family members (Mitcham et al., 1996). By activating the NF-κB signaling cascade, MyD88 leads to activation of inflammatory cytokines, such as tumor necrosis factor (TNF). A naturally occurring spliced isoform lacking exon 2 (MyD88S) is unable to activate the NF-κB pathway (Janssens et al., 2002) and acts as a dominant-negative regulator of toll-like receptor and IL-1R activity (Burns et al., 2003). This alternate isoform is expressed primarily in spleen and weakly in brain, whereas MyD88L is broadly expressed in most tissues (Janssens et al., 2002). To ameliorate inflammatory diseases associated with overactive IL-1R signaling, directing MyD88 splicing from MyD88L toward MyD88S represents a promising therapeutic approach. Screening different ASOs revealed that the RNA target sequence has a significant effect on their activity: those targeted to the exon 2 donor splice site were more effective than ASOs targeted to the acceptor site (Vickers et al., 2006). A 20-mer antisense oligonucleotide was shown to induce switching to MyD88S in both murine and human liver, intestine, and adipose cells, but clinical implementation is still distant.
TNF is released by activated immune cells in response to infection. This cytokine can be pathogenic when overactive in autoimmune conditions, where high concentrations can be found in the joints of patients with rheumatoid arthritis (RA) (Palladino et al., 2003) or the cerebrospinal fluid of patients with multiple sclerosis (Sharief and Hentges, 1991). Drugs that block TNF or signaling at TNFR2 receptors, such as etanercept, infliximab, and adalimumab, are currently used to treat these conditions. TNFR2 is membrane-bound, and when activated by TNF, leads to downstream gene expression and apoptosis. The gene encoding TNFR2 (TNFRSF1B) is alternatively spliced to exclude exons 7 and 8 (Δ7/8TNFR2), resulting in a protein lacking the transmembrane domain (Lainez et al., 2004). An artificially generated isoform, Δ7TNFR2 (lacking only exon 7), can act as a soluble decoy receptor by binding and inactivating TNFα, thereby antagonizing TNFα signaling (Graziewicz et al., 2008). SSO 3274 acts by blocking the 5′ splice site of exon 7 to encourage formation of the Δ7TNFR2 isoform in an attempt to prevent TNFα damage. In a mouse model of collagen-induced RA, SSO 3274 treatment delayed disease progression and produced detectable Δ7TNFR2 protein levels in serum. The SSO effects were sequence-specific, dose-dependent, and persistent, lasting up to 35 days (Graziewicz et al., 2008). Treatment with SSO 3274, also prevented TNFα-induced liver damage in mice, proving more effective than etanercept (Graziewicz et al., 2008).
Interindividual differences in these splicing events can affect anti-TNF therapy. In a clinical trial, patients with RA who have high levels of soluble protein derived from an alternatively spliced TNFR2 transcript demonstrated prolonged responsiveness and sensitivity to anti-TNF therapy (Cañete et al., 2011). Anti-inflammatory SSO treatments may allow for less frequent dosing and avoid immune resistance observed with other currently prescribed anti-TNF medications.
Summary
Functionally relevant splicing events affect disease risk or treatment outcomes conditional on internal and external factors. The CACNA1C and UGT1A gene loci exemplify how transcriptional regulation and alternative splicing/RNA processing can generate diversity in protein structure and function with pharmacological consequences. Differential tissue distribution and functional variations are hallmarks of this transcript diversity, with relevance to drug design and clinical application. Examples in the brain include DRD2 and OPRM1 spliced isoforms with unique pharmacological properties that should be considered in drug design. Targeting of RNA processing is another therapeutic approach. Small molecules that produce global splicing effects or oligonucleotides that directly block or redirect splicing from one form to another are promising therapies. Preliminary studies have shown efficacy of these molecules in therapy of inflammatory diseases. Targeting specific RNA and protein isoforms has the potential to improve drug efficacy and reduce side effects. Moreover, environmental and genetic contributions affecting the spectrum of transcripts expressed must be considered for achieving optimal therapeutic benefits. Applications in personalized medicine may include measuring a patient's transcriptome in target tissues, thereby optimizing individual therapies against cancer, immune disorders, and more. With genomics concepts and tools moving into mainstream biomedical sciences, drug design and therapy must reflect not only multiple protein targets, but also multiple protein isoforms at each gene locus.
This work was supported in part by the National Institutes of Health National Institute of General Medical Sciences [Grant U01-GM092655].
ABBREVIATIONS:
Δ9
exon 9-lacking
ASO
antisense oligonucleotide
CACNA1C
voltage dependent L-type calcium channel α-subunit 1c
CETP
cholesterylester transfer protein
CNS
central nervous system
COX
cyclooxygenase
D2L
dopamine D2 receptor long form
D2S
dopamine D2 receptor short form
DMD
Duchenne muscular dystrophy
DRD2
dopamine D2 receptor
FD
familial dysautonomia
HDL
high-density lipoprotein
IL
interleukin
IL-1R
interleukin-1 receptor
miR
microRNA
MOR
μ opioid receptor
MyD88
myeloid differentiation primary response
NAT1
_N_-acetyltransferase 1
NF-κB
nuclear factor κ-light-chain enhancer of activated B cells
NSAID
nonsteroidal anti-inflammatory drug
OPRM1
μ opioid receptor 1
RA
rheumatoid arthritis
SCN1A
neuronal Nav1.1 sodium channel
SMA
spinal muscular atrophy
SMN
survival of motor neuron
snRNA
small nuclear RNA
SSO
splice-switching oligonucleotide
TNF
tumor necrosis factor
UGT
UDP-glucuronosyltransferase
UTR
untranslated region
VEGF
vascular endothelial growth factor.
Authorship Contributions
Wrote or contributed to the writing of the manuscript: Barrie, Smith, Sanford and Sadee.
References
- Akira S, Takeda K, Kaisho T. (2001) Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2:675–680 [DOI] [PubMed] [Google Scholar]
- Albert BJ, Sivaramakrishnan A, Naka T, Czaicki NL, Koide K. (2007) Total syntheses, fragmentation studies, and antitumor/antiproliferative activities of FR901464 and its low picomolar analogue. J Am Chem Soc 129:2648–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod FB, Liebes L, Gold-Von Simson G, Mendoza S, Mull J, Leyne M, Norcliffe-Kaufmann L, Kaufmann H, Slaugenhaupt SA. (2011) Kinetin improves IKBKAP mRNA splicing in patients with familial dysautonomia. Pediatr Res 70:480–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barter PJ, Caulfield M, Eriksson M, Grundy SM, Kastelein JJ, Komajda M, Lopez-Sendon J, Mosca L, Tardif JC, Waters DD, et al. (2007) Effects of torcetrapib in patients at high risk for coronary events. N Engl J Med 357:2109–2122 [DOI] [PubMed] [Google Scholar]
- Bauman JA, Li SD, Yang A, Huang L, Kole R. (2010) Anti-tumor activity of splice-switching oligonucleotides. Nucleic Acids Res 38:8348–8356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino A, Fazio L, Caforio G, Blasi G, Rampino A, Romano R, Di Giorgio A, Taurisano P, Papp A, Pinsonneault J, et al. (2009) Functional variants of the dopamine receptor D2 gene modulate prefronto-striatal phenotypes in schizophrenia. Brain 132:417–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino A, Taurisano P, Pisciotta NM, Blasi G, Fazio L, Romano R, Gelao B, Lo Bianco L, Lozupone M, Di Giorgio A, et al. (2010) Genetically determined measures of striatal D2 signaling predict prefrontal activity during working memory performance. PLoS One 5:e9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasi G, Lo Bianco L, Taurisano P, Gelao B, Romano R, Fazio L, Papazacharias A, Di Giorgio A, Caforio G, Rampino A, et al. (2009) Functional variation of the dopamine D2 receptor gene is associated with emotional control as well as brain activity and connectivity during emotion processing in humans. J Neurosci 29:14812–14819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroto-Escuela DO, Romero-Fernandez W, Tarakanov AO, Marcellino D, Ciruela F, Agnati LF, Fuxe K. (2010) Dopamine D2 and 5-hydroxytryptamine 5-HT(2A) receptors assemble into functionally interacting heteromers. Biochem Biophys Res Commun 401:605–610 [DOI] [PubMed] [Google Scholar]
- Bosma PJ, Chowdhury JR, Bakker C, Gantla S, de Boer A, Oostra BA, Lindhout D, Tytgat GN, Jansen PL, Oude Elferink RP. (1995) The genetic basis of the reduced expression of bilirubin UDP-glucuronosyltransferase 1 in Gilbert's syndrome. N Engl J Med 333:1171–1175 [DOI] [PubMed] [Google Scholar]
- Boukouvala S, Sim E. (2005) Structural analysis of the genes for human arylamine N-acetyltransferases and characterisation of alternative transcripts. Basic Clin Pharmacol Toxicol 96:343–351 [DOI] [PubMed] [Google Scholar]
- Bouvier M. (2001) Oligomerization of G-protein-coupled transmitter receptors. Nat Rev Neurosci 2:274–286 [DOI] [PubMed] [Google Scholar]
- Burns K, Janssens S, Brissoni B, Olivos N, Beyaert R, Tschopp J. (2003) Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J Exp Med 197:263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher NJ, Arulpragasam A, Goh HL, Davey T, Minchin RF. (2005) Genomic organization of human arylamine N-acetyltransferase type I reveals alternative promoters that generate different 5′-UTR splice variants with altered translational activities. Biochem J 387:119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañete JD, Albaladejo C, Hernández MV, Laínez B, Pinto JA, Ramírez J, López-Armada MJ, Rodríguez-Cros JR, Engel P, Blanco FJ, et al. (2011) Clinical significance of high levels of soluble tumour necrosis factor-alpha receptor-2 produced by alternative splicing in rheumatoid arthritis: a longitudinal prospective cohort study. Rheumatology (Oxford) 50:721–728 [DOI] [PubMed] [Google Scholar]
- Castle JC, Zhang C, Shah JK, Kulkarni AV, Kalsotra A, Cooper TA, Johnson JM. (2008) Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 40:1416–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro SW, Strange PG. (1993) Differences in the ligand binding properties of the short and long versions of the D2 dopamine receptor. J Neurochem 60:372–375 [DOI] [PubMed] [Google Scholar]
- Centonze D, Gubellini P, Usiello A, Rossi S, Tscherter A, Bracci E, Erbs E, Tognazzi N, Bernardi G, Pisani A, et al. (2004) Differential contribution of dopamine D2S and D2L receptors in the modulation of glutamate and GABA transmission in the striatum. Neuroscience 129:157–166 [DOI] [PubMed] [Google Scholar]
- Chandrasekharan NV, Dai H, Roos KL, Evanson NK, Tomsik J, Elton TS, Simmons DL. (2002) COX-3, a cyclooxygenase-1 variant inhibited by acetaminophen and other analgesic/antipyretic drugs: cloning, structure, and expression. Proc Natl Acad Sci USA 99:13926–13931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WH, Liu TC, Yang WK, Lee CC, Lin YH, Chen TY, Chang JG. (2011) Amiloride modulates alternative splicing in leukemic cells and resensitizes Bcr-AblT315I mutant cells to imatinib. Cancer Res 71:383–392 [DOI] [PubMed] [Google Scholar]
- Chen YC, Yuo CY, Yang WK, Jong YJ, Lin HH, Chang YS, Chang JG. (2008) Extracellular pH change modulates the exon 7 splicing in SMN2 mRNA. Mol Cell Neurosci 39:268–272 [DOI] [PubMed] [Google Scholar]
- Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, Abbs S, Garralda ME, Bourke J, Wells DJ, et al. (2011) Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet 378:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TA, Wan L, Dreyfuss G. (2009) RNA and disease. Cell 136:777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. (2010) Splicing factor and exon profiling across human tissues. Nucleic Acids Res 38:2825–2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeKeyser JG, Laurenzana EM, Peterson EC, Chen T, Omiecinski CJ. (2011) Selective phthalate activation of naturally occurring human constitutive androstane receptor splice variants and the pregnane X receptor. Toxicol Sci 120:381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C. (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37:e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessí M, Motti C, Cortese C, Leonardis E, Giovannini C, Federici G, Piemonte F. (1997) Alternative splicing of human plasma cholesteryl ester transfer protein mRNA in Caco-2 cells and its modulation by oleic acid. Mol Cell Biochem 177:107–112 [DOI] [PubMed] [Google Scholar]
- Ehmer U, Vogel A, Schütte JK, Krone B, Manns MP, Strassburg CP. (2004) Variation of hepatic glucuronidation: Novel functional polymorphisms of the UDP-glucuronosyltransferase UGT1A4. Hepatology 39:970–977 [DOI] [PubMed] [Google Scholar]
- Elmhurst JL, Xie Z, O'Dowd BF, George SR. (2000) The splice variant D3nf reduces ligand binding to the D3 dopamine receptor: evidence for heterooligomerization. Brain Res Mol Brain Res 80:63–74 [DOI] [PubMed] [Google Scholar]
- Fan T, Varghese G, Nguyen T, Tse R, O'Dowd BF, George SR. (2005) A role for the distal carboxyl tails in generating the novel pharmacology and G protein activation profile of mu and delta opioid receptor hetero-oligomers. J Biol Chem 280:38478–38488 [DOI] [PubMed] [Google Scholar]
- Farde L, Nordström AL, Wiesel FA, Pauli S, Halldin C, Sedvall G. (1992) Positron emission tomographic analysis of central D1 and D2 dopamine receptor occupancy in patients treated with classical neuroleptics and clozapine. Relation to extrapyramidal side effects. Arch Gen Psychiatry 49:538–544 [DOI] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. (2003) Pre-mRNA splicing and human disease. Genes Dev 17:419–437 [DOI] [PubMed] [Google Scholar]
- Furumai R, Uchida K, Komi Y, Yoneyama M, Ishigami K, Watanabe H, Kojima S, Yoshida M. (2010) Spliceostatin A blocks angiogenesis by inhibiting global gene expression including VEGF. Cancer Sci 101:2483–2489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabanella F, Butchbach ME, Saieva L, Carissimi C, Burghes AH, Pellizzoni L. (2007) Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One 2:e921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geib T, Hertel KJ. (2009) Restoration of full-length SMN promoted by adenoviral vectors expressing RNA antisense oligonucleotides embedded in U7 snRNAs. PLoS One 4:e8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigna C, De Toledo M, Bonomi S, Valacca C, Gallo S, Apicella M, Eperon I, Tazi J, Biamonti G. (2010) Pro-metastatic splicing of Ron proto-oncogene mRNA can be reversed: therapeutic potential of bifunctional oligonucleotides and indole derivatives. RNA Biol 7:495–503 [DOI] [PubMed] [Google Scholar]
- Girard H, Lévesque E, Bellemare J, Journault K, Caillier B, Guillemette C. (2007) Genetic diversity at the UGT1 locus is amplified by a novel 3′ alternative splicing mechanism leading to nine additional UGT1A proteins that act as regulators of glucuronidation activity. Pharmacogenet Genomics 17:1077–1089 [DOI] [PubMed] [Google Scholar]
- Giros B, Sokoloff P, Martres MP, Riou JF, Emorine LJ, Schwartz JC. (1989) Alternative splicing directs the expression of two D2 dopamine receptor isoforms. Nature 342:923–926 [DOI] [PubMed] [Google Scholar]
- Goemans NM, Tulinius M, van den Akker JT, Burm BE, Ekhart PF, Heuvelmans N, Holling T, Janson AA, Platenburg GJ, Sipkens JA, et al. (2011) Systemic administration of PRO051 in Duchenne's muscular dystrophy. N Engl J Med 364:1513–1522 [DOI] [PubMed] [Google Scholar]
- Gong QH, Cho JW, Huang T, Potter C, Gholami N, Basu NK, Kubota S, Carvalho S, Pennington MW, Owens IS, et al. (2001) Thirteen UDPglucuronosyltransferase genes are encoded at the human UGT1 gene complex locus. Pharmacogenetics 11:357–368 [DOI] [PubMed] [Google Scholar]
- Gorman L, Mercatante DR, Kole R. (2000) Restoration of correct splicing of thalassemic beta-globin pre-mRNA by modified U1 snRNAs. J Biol Chem 275:35914–35919 [DOI] [PubMed] [Google Scholar]
- Gorman L, Suter D, Emerick V, Schümperli D, Kole R. (1998) Stable alteration of pre-mRNA splicing patterns by modified U7 small nuclear RNAs. Proc Natl Acad Sci USA 95:4929–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NK, Hentze MW. (1994) Regulation of protein synthesis by mRNA structure. Mol Biol Rep 19:195–200 [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Tarrant TK, Buckley B, Roberts J, Fulton L, Hansen H, Ørum H, Kole R, Sazani P. (2008) An endogenous TNF-alpha antagonist induced by splice-switching oligonucleotides reduces inflammation in hepatitis and arthritis mouse models. Mol Ther 16:1316–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harahap IS, Saito T, San LP, Sasaki N, Gunadi, Nurputra DK, Yusoff S, Yamamoto T, Morikawa S, Nishimura N, et al. (2012) Valproic acid increases SMN2 expression and modulates SF2/ASF and hnRNPA1 expression in SMA fibroblast cell lines. Brain Dev 34:213–222 [DOI] [PubMed] [Google Scholar]
- Harper SJ, Bates DO. (2008) VEGF-A splicing: the key to anti-angiogenic therapeutics? Nat Rev Cancer 8:880–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann L, Neveling K, Borkens S, Schneider H, Freund M, Grassman E, Theiss S, Wawer A, Burdach S, Auerbach AD, et al. (2010) Correct mRNA processing at a mutant TT splice donor in FANCC ameliorates the clinical phenotype in patients and is enhanced by delivery of suppressor U1 snRNAs. Am J Hum Genet 87:480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Sahashi K, Hung G, Rigo F, Passini MA, Bennett CF, Krainer AR. (2010) Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev 24:1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, et al. (2004) Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med 350:2335–2342 [DOI] [PubMed] [Google Scholar]
- Inazu A, Quinet EM, Wang S, Brown ML, Stevenson S, Barr ML, Moulin P, Tall AR. (1992) Alternative splicing of the mRNA encoding the human cholesteryl ester transfer protein. Biochemistry 31:2352–2358 [DOI] [PubMed] [Google Scholar]
- Incitti T, De Angelis FG, Cazzella V, Sthandier O, Pinnarò C, Legnini I, Bozzoni I. (2010) Exon skipping and duchenne muscular dystrophy therapy: selection of the most active U1 snRNA antisense able to induce dystrophin exon 51 skipping. Mol Ther 18:1675–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens S, Burns K, Tschopp J, Beyaert R. (2002) Regulation of interleukin-1- and lipopolysaccharide-induced NF-kappaB activation by alternative splicing of MyD88. Curr Biol 12:467–471 [DOI] [PubMed] [Google Scholar]
- Kagimoto M, Heim M, Kagimoto K, Zeugin T, Meyer UA. (1990) Multiple mutations of the human cytochrome P450IID6 gene (CYP2D6) in poor metabolizers of debrisoquine. Study of the functional significance of individual mutations by expression of chimeric genes. J Biol Chem 265:17209–17214 [PubMed] [Google Scholar]
- Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, Watanabe H, Kitahara T, Yoshida T, Nakajima H, et al. (2007) Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 3:576–583 [DOI] [PubMed] [Google Scholar]
- Kan Z, Rouchka EC, Gish WR, States DJ. (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res 11:889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur S, Zipursky R, Jones C, Remington G, Houle S. (2000) Relationship between dopamine D(2) occupancy, clinical response, and side effects: a double-blind PET study of first-episode schizophrenia. Am J Psychiatry 157:514–520 [DOI] [PubMed] [Google Scholar]
- Karpa KD, Lin R, Kabbani N, Levenson R. (2000) The dopamine D3 receptor interacts with itself and the truncated D3 splice variant d3nf: D3–D3nf interaction causes mislocalization of D3 receptors. Mol Pharmacol 58:677–683 [DOI] [PubMed] [Google Scholar]
- Keren H, Donyo M, Zeevi D, Maayan C, Pupko T, Ast G. (2010) Phosphatidylserine increases IKBKAP levels in familial dysautonomia cells. PLoS One 5:e15884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan ZU, Mrzljak L, Gutierrez A, de la Calle A, Goldman-Rakic PS. (1998) Prominence of the dopamine D2 short isoform in dopaminergic pathways. Proc Natl Acad Sci USA 95:7731–7736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis B, Snipes JA, Busija DW. (2005) Acetaminophen and the cyclooxygenase-3 puzzle: sorting out facts, fictions, and uncertainties. J Pharmacol Exp Ther 315:1–7 [DOI] [PubMed] [Google Scholar]
- Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y, Shimizu H, Uesugi M, Ishihama Y, Iwata M, Mizui Y. (2007) Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol 3:570–575 [DOI] [PubMed] [Google Scholar]
- Kozak M. (1989) Circumstances and mechanisms of inhibition of translation by secondary structure in eucaryotic mRNAs. Mol Cell Biol 9:5134–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzhikandathil EV, Bartoszyk GD. (2006) The novel antidyskinetic drug sarizotan elicits different functional responses at human D2-like dopamine receptors. Neuropharmacology 51:873–884 [DOI] [PubMed] [Google Scholar]
- Lainez B, Fernandez-Real JM, Romero X, Esplugues E, Cañete JD, Ricart W, Engel P. (2004) Identification and characterization of a novel spliced variant that encodes human soluble tumor necrosis factor receptor 2. Int Immunol 16:169–177 [DOI] [PubMed] [Google Scholar]
- Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE. (2007) The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol 623:190–211 [DOI] [PubMed] [Google Scholar]
- Lévesque E, Girard H, Journault K, Lépine J, Guillemette C. (2007) Regulation of the UGT1A1 bilirubin-conjugating pathway: role of a new splicing event at the UGT1A locus. Hepatology 45:128–138 [DOI] [PubMed] [Google Scholar]
- Lira ME, Loomis AK, Paciga SA, Lloyd DB, Thompson JF. (2008) Expression of CETP and of splice variants induces the same level of ER stress despite secretion efficiency differences. J Lipid Res 49:1955–1962 [DOI] [PubMed] [Google Scholar]
- Malmberg A, Jackson DM, Eriksson A, Mohell N. (1993) Unique binding characteristics of antipsychotic agents interacting with human dopamine D2A, D2B, and D3 receptors. Mol Pharmacol 43:749–754 [PubMed] [Google Scholar]
- Marez D, Legrand M, Sabbagh N, Lo Guidice JM, Spire C, Lafitte JJ, Meyer UA, Broly F. (1997) Polymorphism of the cytochrome P450 CYP2D6 gene in a European population: characterization of 48 mutations and 53 alleles, their frequencies and evolution. Pharmacogenetics 7:193–202 [DOI] [PubMed] [Google Scholar]
- Marsh S, McLeod HL. (2004) Pharmacogenetics of irinotecan toxicity. Pharmacogenomics 5:835–843 [DOI] [PubMed] [Google Scholar]
- Mitcham JL, Parnet P, Bonnert TP, Garka KE, Gerhart MJ, Slack JL, Gayle MA, Dower SK, Sims JE. (1996) T1/ST2 signaling establishes it as a member of an expanding interleukin-1 receptor family. J Biol Chem 271:5777–5783 [DOI] [PubMed] [Google Scholar]
- Modrek B, Lee C. (2002) A genomic view of alternative splicing. Nat Genet 30:13–19 [DOI] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C. (2001) Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res 29:2850–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori A, Maruo Y, Iwai M, Sato H, Takeuchi Y. (2005) UDP-glucuronosyltransferase 1A4 polymorphisms in a Japanese population and kinetics of clozapine glucuronidation. Drug Metab Dispos 33:672–675 [DOI] [PubMed] [Google Scholar]
- Moroni M, Zwart R, Sher E, Cassels BK, Bermudez I. (2006) alpha4beta2 nicotinic receptors with high and low acetylcholine sensitivity: pharmacology, stoichiometry, and sensitivity to long-term exposure to nicotine. Mol Pharmacol 70:755–768 [DOI] [PubMed] [Google Scholar]
- Moyer RA, Wang D, Papp AC, Smith RM, Duque L, Mash DC, Sadee W. (2011) Intronic polymorphisms affecting alternative splicing of human dopamine D2 receptor are associated with cocaine abuse. Neuropsychopharmacology 36:753–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström AL, Farde L, Wiesel FA, Forslund K, Pauli S, Halldin C, Uppfeldt G. (1993) Central D2-dopamine receptor occupancy in relation to antipsychotic drug effects: a double-blind PET study of schizophrenic patients. Biol Psychiatry 33:227–235 [DOI] [PubMed] [Google Scholar]
- O'Brien K, Matlin AJ, Lowell AM, Moore MJ. (2008) The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J Biol Chem 283:33147–33154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palladino MA, Bahjat FR, Theodorakis EA, Moldawer LL. (2003) Anti-TNF-alpha therapies: the next generation. Nat Rev Drug Discov 2:736–746 [DOI] [PubMed] [Google Scholar]
- Pan L, Xu J, Yu R, Xu MM, Pan YX, Pasternak GW. (2005a) Identification and characterization of six new alternatively spliced variants of the human mu opioid receptor gene, Oprm. Neuroscience 133:209–220 [DOI] [PubMed] [Google Scholar]
- Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. (2008) Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 40:1413–1415 [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Bolan E, Moskowitz HS, Xu M, Pasternak GW. (2005b) Identification of four novel exon 5 splice variants of the mouse mu-opioid receptor gene: functional consequences of C-terminal splicing. Mol Pharmacol 68:866–875 [DOI] [PubMed] [Google Scholar]
- Pan YX, Xu J, Mahurter L, Bolan E, Xu M, Pasternak GW. (2001) Generation of the mu opioid receptor (MOR-1) protein by three new splice variants of the Oprm gene. Proc Natl Acad Sci USA 98:14084–14089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YX, Xu J, Xu M, Rossi GC, Matulonis JE, Pasternak GW. (2009) Involvement of exon 11-associated variants of the mu opioid receptor MOR-1 in heroin, but not morphine, actions. Proc Natl Acad Sci USA 106:4917–4922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips KA, Veenstra DL, Oren E, Lee JK, Sadee W. (2001) Potential role of pharmacogenomics in reducing adverse drug reactions: a systematic review. Jama 286:2270–2279 [DOI] [PubMed] [Google Scholar]
- Pinotti M, Rizzotto L, Balestra D, Lewandowska MA, Cavallari N, Marchetti G, Bernardi F, Pagani F. (2008) U1-snRNA-mediated rescue of mRNA processing in severe factor VII deficiency. Blood 111:2681–2684 [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. (2011) RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 480:387–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pros E, Fernández-Rodríguez J, Benito L, Ravella A, Capellá G, Blanco I, Serra E, Lázaro C. (2010) Modulation of aberrant NF1 pre-mRNA splicing by kinetin treatment. Eur J Hum Genet 18:614–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin N, Zhang SP, Reitz TL, Mei JM, Flores CM. (2005) Cloning, expression, and functional characterization of human cyclooxygenase-1 splicing variants: evidence for intron 1 retention. J Pharmacol Exp Ther 315:1298–1305 [DOI] [PubMed] [Google Scholar]
- Ritter JK, Chen F, Sheen YY, Tran HM, Kimura S, Yeatman MT, Owens IS. (1992) A novel complex locus UGT1 encodes human bilirubin, phenol, and other UDP-glucuronosyltransferase isozymes with identical carboxyl termini. J Biol Chem 267:3257–3261 [PubMed] [Google Scholar]
- Schmajuk G, Sierakowska H, Kole R. (1999) Antisense oligonucleotides with different backbones. Modification of splicing pathways and efficacy of uptake. J Biol Chem 274:21783–21789 [DOI] [PubMed] [Google Scholar]
- Schuller AG, King MA, Zhang J, Bolan E, Pan YX, Morgan DJ, Chang A, Czick ME, Unterwald EM, Pasternak GW, et al. (1999) Retention of heroin and morphine-6 beta-glucuronide analgesia in a new line of mice lacking exon 1 of MOR-1. Nat Neurosci 2:151–156 [DOI] [PubMed] [Google Scholar]
- Sharief MK, Hentges R. (1991) Association between tumor necrosis factor-alpha and disease progression in patients with multiple sclerosis. N Engl J Med 325:467–472 [DOI] [PubMed] [Google Scholar]
- Shetty RS, Gallagher CS, Chen YT, Hims MM, Mull J, Leyne M, Pickel J, Kwok D, Slaugenhaupt SA. (2011) Specific correction of a splice defect in brain by nutritional supplementation. Hum Mol Genet 20:4093–4101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierakowska H, Sambade MJ, Agrawal S, Kole R. (1996) Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc Natl Acad Sci USA 93:12840–12844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN. (2009) A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol 6:341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordis LA, Dunckley MG, Yue B, Eperon IC, Muntoni F. (2003) Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci USA 100:4114–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RM, Sadee W. (2011) Synaptic signaling and aberrant RNA splicing in autism spectrum disorders. Front Synaptic Neurosci 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soret J, Bakkour N, Maire S, Durand S, Zekri L, Gabut M, Fic W, Divita G, Rivalle C, Dauzonne D, et al. (2005) Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc Natl Acad Sci USA 102:8764–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. (2008) A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc Natl Acad Sci USA 105:11218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang ZZ, Liang MC, Lu S, Yu D, Yu CY, Yue DT, Soong TW. (2004) Transcript scanning reveals novel and extensive splice variations in human l-type voltage-gated calcium channel, Cav1.2 alpha1 subunit. J Biol Chem 279:44335–44343 [DOI] [PubMed] [Google Scholar]
- Taniguchi-Ikeda M, Kobayashi K, Kanagawa M, Yu CC, Mori K, Oda T, Kuga A, Kurahashi H, Akman HO, DiMauro S, et al. (2011) Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature 478:127–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate SK, Depondt C, Sisodiya SM, Cavalleri GL, Schorge S, Soranzo N, Thom M, Sen A, Shorvon SD, Sander JW, et al. (2005) Genetic predictors of the maximum doses patients receive during clinical use of the anti-epileptic drugs carbamazepine and phenytoin. Proc Natl Acad Sci USA 102:5507–5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CH, Kahlig KM, George AL., Jr (2011) SCN1A splice variants exhibit divergent sensitivity to commonly used antiepileptic drugs. Epilepsia 52:1000–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usiello A, Baik JH, Rougé-Pont F, Picetti R, Dierich A, LeMeur M, Piazza PV, Borrelli E. (2000) Distinct functions of the two isoforms of dopamine D2 receptors. Nature 408:199–203 [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, Goemans NM, et al. (2007) Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med 357:2677–2686 [DOI] [PubMed] [Google Scholar]
- Varey AH, Rennel ES, Qiu Y, Bevan HS, Perrin RM, Raffy S, Dixon AR, Paraskeva C, Zaccheo O, Hassan AB, et al. (2008) VEGF 165 b, an antiangiogenic VEGF-A isoform, binds and inhibits bevacizumab treatment in experimental colorectal carcinoma: balance of pro- and antiangiogenic VEGF-A isoforms has implications for therapy. Br J Cancer 98:1366–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers TA, Zhang H, Graham MJ, Lemonidis KM, Zhao C, Dean NM. (2006) Modification of MyD88 mRNA splicing and inhibition of IL-1beta signaling in cell culture and in mice with a 2′-O-methoxyethyl-modified oligonucleotide. J Immunol 176:3652–3661 [DOI] [PubMed] [Google Scholar]
- Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S, Roblot N, Brahe C, Saugier-Veber P, Bonnefont JP, Melki J. (2007) Refined characterization of the expression and stability of the SMN gene products. Am J Pathol 171:1269–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. (2011) Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature 474:380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Papp AC, Binkley PF, Johnson JA, Sadée W. (2006) Highly variable mRNA expression and splicing of L-type voltage-dependent calcium channel alpha subunit 1C in human heart tissues. Pharmacogenet Genomics 16:735–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Para MF, Koletar SL, Sadee W. (2011) Human N-acetyltransferase 1 *10 and *11 alleles increase protein expression through distinct mechanisms and associate with sulfamethoxazole-induced hypersensitivity. Pharmacogenet Genomics 21:652–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Sun X, Bohn LM, Sadée W. (2005) Opioid receptor homo- and heterodimerization in living cells by quantitative bioluminescence resonance energy transfer. Mol Pharmacol 67:2173–2184 [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. (2008) Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GS, Cooper TA. (2007) Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8:749–761 [DOI] [PubMed] [Google Scholar]
- Welling A, Ludwig A, Zimmer S, Klugbauer N, Flockerzi V, Hofmann F. (1997) Alternatively spliced IS6 segments of the alpha 1C gene determine the tissue-specific dihydropyridine sensitivity of cardiac and vascular smooth muscle L-type Ca2+ channels. Circ Res 81:526–532 [DOI] [PubMed] [Google Scholar]
- Williams JH, Schray RC, Patterson CA, Ayitey SO, Tallent MK, Lutz GJ. (2009) Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci 29:7633–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilton SD, Fletcher S. (2005) RNA splicing manipulation: strategies to modify gene expression for a variety of therapeutic outcomes. Curr Gene Ther 5:467–483 [DOI] [PubMed] [Google Scholar]
- Wirth B, Brichta L, Hahnen E. (2006) Spinal muscular atrophy: from gene to therapy. Semin Pediatr Neurol 13:121–131 [DOI] [PubMed] [Google Scholar]
- Woolard J, Wang WY, Bevan HS, Qiu Y, Morbidelli L, Pritchard-Jones RO, Cui TG, Sugiono M, Waine E, Perrin R, et al. (2004) VEGF165b, an inhibitory vascular endothelial growth factor splice variant: mechanism of action, in vivo effect on angiogenesis and endogenous protein expression. Cancer Res 64:7822–7835 [DOI] [PubMed] [Google Scholar]
- Xu J, Xu M, Hurd YL, Pasternak GW, Pan YX. (2009) Isolation and characterization of new exon 11-associated N-terminal splice variants of the human mu opioid receptor gene. J Neurochem 108:962–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Xu M, Rossi GC, Pasternak GW, Pan YX. (2011) Identification and characterization of seven new exon 11-associated splice variants of the rat mu opioid receptor gene, OPRM1. Mol Pain 7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Hranilovic D, Fetsko LA, Bucan M, Wang Y. (2002) Dopamine D2S and D2L receptors may differentially contribute to the actions of antipsychotic and psychotic agents in mice. Mol Psychiatry 7:1075–1082 [DOI] [PubMed] [Google Scholar]
- Yang TP, Agellon LB, Walsh A, Breslow JL, Tall AR. (1996) Alternative splicing of the human cholesteryl ester transfer protein gene in transgenic mice. Exon exclusion modulates gene expression in response to dietary or developmental change. J Biol Chem 271:12603–12609 [DOI] [PubMed] [Google Scholar]
- Yeo G, Holste D, Kreiman G, Burge CB. (2004) Variation in alternative splicing across human tissues. Genome Biol 5:R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, Kang D, Richman R, Johnson JM, Berget S, et al. (2005) Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci USA 102:17551–17558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuo CY, Lin HH, Chang YS, Yang WK, Chang JG. (2008) 5-(N-ethyl-N-isopropyl)-amiloride enhances SMN2 exon 7 inclusion and protein expression in spinal muscular atrophy cells. Ann Neurol 63:26–34 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Bertolino A, Fazio L, Blasi G, Rampino A, Romano R, Lee ML, Xiao T, Papp A, Wang D, et al. (2007) Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci USA 104:20552–20557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. (2008) SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133:585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]