TALE-mediated modulation of transcriptional enhancers (original) (raw)
. Author manuscript; available in PMC: 2014 Feb 1.
Published in final edited form as: Nat Methods. 2013 Jun 30;10(8):762–767. doi: 10.1038/nmeth.2543
Abstract
We tested whether Transcription Activator-Like Effectors (TALEs) can mediate repression and activation of endogenous enhancers in the Drosophila genome. TALE-repressors (TALERs) targeting each of the five even-skipped (eve) “stripe” enhancers generated repression specifically of the focal stripes. TALE-activators (TALEAs) targeting the eve promoter or eve enhancers caused increased expression primarily in cells normally activated by the promoter or targeted enhancer, respectfully. The phenotypic effects of TALER and TALEA expression in larvae and adults are consistent with the observed modulations of eve expression. In these assays, the Hairy repression domain did not exhibit previously described long-range transcriptional repression activity. The precise effects of the TALEAs support the view that repression acts in a dominant fashion on transcriptional activators and that the activity state of an enhancer influences TALE binding or the ability of VP16 to enhance transcription. TALEs thus provide a novel tool for detection and functional modulation of transcriptional enhancers in their native genomic context.
Transcriptional enhancers encode patterns of gene expression by binding transcription factor proteins that recognize specific sequences within enhancers and enhancers often integrate the combined activity of multiple transcription factors1. Transcriptional enhancers can be located close to or up to hundreds of kilobase pairs from their respective gene promoters1. Alteration in enhancers underlie development, evolution, and disease1 and, in many eukaryotic genomes, more DNA may encode transcriptional enhancers than encodes proteins2. Despite the importance of transcriptional enhancers, we currently understand far less about the structure and function of enhancer regions than we do about protein coding regions.
Our understanding of enhancer structure and function is derived mainly from reporter gene assays, wherein putative enhancer DNA is coupled to a heterologous promoter and reporter. Reporter gene assays have provided most of our current knowledge of enhancer structure and function. These studies indicate that transcriptional regulation of some, but not all, eukaryotic genes is modulated by multiple enhancers that act independently3. Despite the insight that has been provided by reporter gene assays, these experiments suffer from several limitations. First, reporter constructs often drive incomplete and/or ectopic patterns of expression4, probably because enhancers are tested away from their native genomic context. Second, reporter constructs rarely drive expression at normal levels, which confounds quantitative studies of gene regulation. Third, some studies have failed to identify modular autonomous enhancers that recapitulate components of the complete expression pattern3,5,6. Publication bias probably has resulted in under-reporting of genes that appear to lack modular enhancers5.
To provide a method complementary to classical reporter-gene assays, we exploited Transcription Activator-Like Element (TALE) DNA-binding proteins to target transcriptional repressor and activator protein domains to specific genomic locations. TALEs can be engineered to target specific DNA sequences7,8 and TALE DNA-binding domains fused to activators and repressors and targeted specifically to promoters can modulate gene expression in plants9,10 and in human cell-culture11–16. In the present paper we demonstrate that TALEs can be targeted to enhancers to modulate specific domains of complex expression patterns.
Results
Experimental design
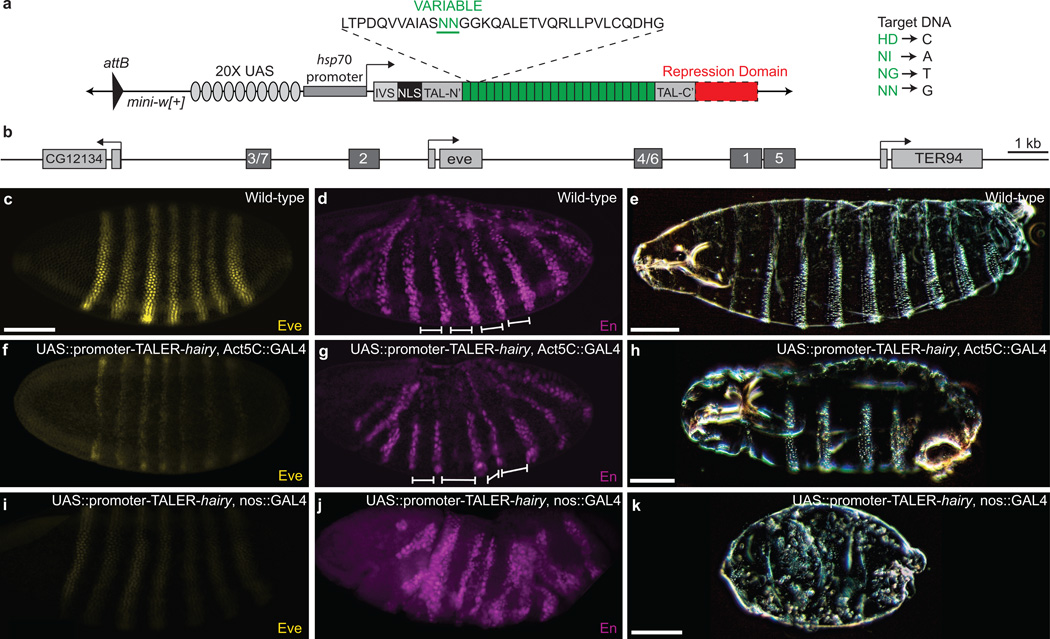
We engineered GAL4 responsive vectors for Drosophila melanogaster transgenesis that allow fusion of a TALE DNA-binding domain to regulatory domains17 (Fig. 1a, Supplementary Fig. 1, and Supplementary Text 1). In each of these fusion genes the native activator domain of the TALE C-terminus was removed. We tested the Krüppel and Hairy repression domains in TALE-mediated repressors (TALERs) and the VP64 (four tandem copies of VP16) activation domain in TALE-mediated activators (TALEAs). Estimates of repressor activity from reporter-gene assays suggest that Krüppel can repress enhancers within approximately 100 bp of a DNA binding site18, whereas Hairy can reportedly silence enhancers up to 5 kb from a DNA binding site19,20.
Figure 1.
TALERs targeted to the promoter can repress expression of even-skipped (eve). (a) Schematic of a GAL4 responsive TALER construct. See Supplementary Fig. S1 for the complete list of Drosophila TALE constructs. (b) Schematic of the eve locus, indicating early-embryonic _cis_-regulatory stripe enhancers. (c, f, i) Stage-5 embryos stained for Eve protein. (c) Wild-type embryos. (f) Eve expression is decreased when Act5C::GAL4 drives a UAS::promoter-TALER-hairy (cf. f with c). (i) Eve expression is further diminished when the UAS::promoter-TALER-hairy is driven with a maternal nos::GAL4 driver. (d, g, j) Stage-11 embryos stained for Engrailed protein. (d) Wild-type embryo. (g) Stripes of Engrailed expression are variable in width and spacing, noted with white lines, when a Act5C::GAL4 drives the UAS::promoter-TALER-hairy (cf. g with d). (j) Stripes of Engrailed are fused and reduced in number when nos::GAL4 drives the UAS::promoter-TALER-hairy. (e, h, k) Larval cuticle preps. (e) Wild-type larva. (h) UAS::promoter-TALER-hairy expression, driven with Act5C::GAL4, causes a reduction in the number of larval segments. (k) UAS::promoter-TALER-hairy expression, driven with a nos::GAL4 driver, causes loss of most or all segmentation. Embryos in panels (c, d, f, g, i, and j) are matched in scale. Scale bars in (a, e, h, and k) equal 100 µm.
As a proof of principle, we targeted the well-studied enhancers of the gene even-skipped (eve), which encodes a transcriptional repressor required for correct segmentation and neuronal development21–23. Eve transcripts appear first in the blastoderm embryo and expression resolves rapidly into seven transverse stripes along the anterior-posterior axis (see Fig. 1). Separate enhancers drive subsets of these stripes (Fig 1b), apparently autonomously23,24.
TALER mediate repression of the eve promoter
To determine the efficiency of TALERs in the embryo, we drove ubiquitous, zygotic expression of a TALER-Hairy targeted near the eve promoter (Fig. 1b). This TALER-Hairy reduced expression of all eve stripes (Fig. 1) and resulted in abnormal expression of engrailed (en), a target of Eve22 (cf. Fig. 1). Larval cuticles of these embryos exhibited fused segments (Fig. 1). To test whether the residual eve expression in these embryos resulted from late onset of TALER expression relative to eve activation, we drove this TALER-Hairy with a maternally-expressed driver25. In these embryos, eve expression was almost undetectable (Supplementary Fig. 2), en expression was disrupted severely (Fig. 1), and outward signs of segmentation in the larval cuticle were lost (Fig. 1). These results are consistent with the effects of even-skipped hypomorphic alleles26. We also drove this TALER-Hairy using neurogenic GAL4 drivers, and, in all cases, we observed decreased Eve levels in neurons (Supplementary Fig. 3). As a control, a TALE-GFP fusion protein targeted to the same site did not alter eve expression (Supplementary Fig. 4). Promoter-targeted TALERs thus provide a complementary tool to existing conditional gene silencing technologies in Drosophila27. In addition, judicious use of GAL4 drivers may be used to allow TALERs to mimic an allelic series.
TALEA mediated activation of the eve promoter
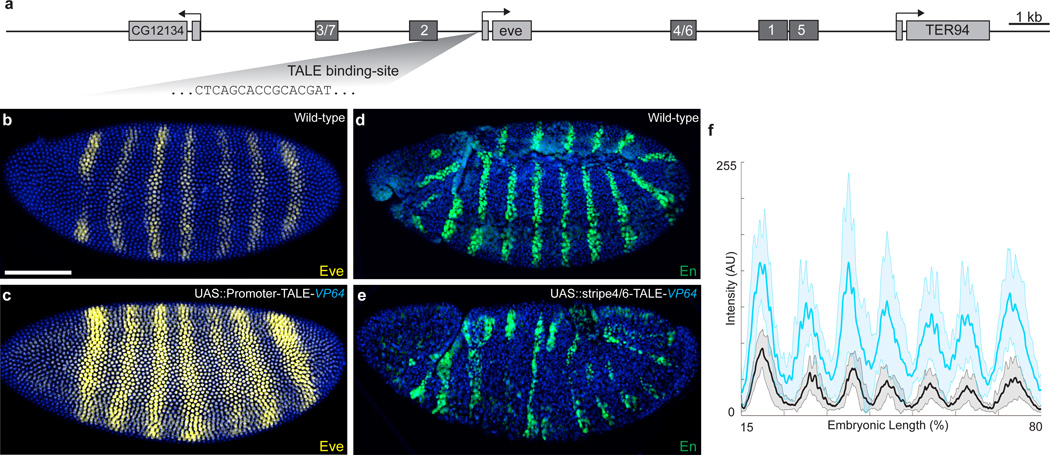
We examined next, whether TALEs could be used to selectively activate gene expression. To confirm the efficiency of the activator fusion, we drove ubiquitous, zygotic expression of a TALEA targeted near the eve promoter. Strikingly, these embryos exhibited stronger and broader patterns of expression of all seven stripes of Eve, compared to wild type (Fig. 2). While we observed low levels of Eve expression between the canonical stripes, there is still a clear 7-striped pattern of expression. Engrailed expression was disrupted in these embryos (Fig. 2), as expected28.
Figure 2.
TALE targeted activation of the eve promoter. (a) Schematic of the eve locus, indicating early-embryonic _cis_-regulatory stripe enhancers and TALE binding site. (b, c) Stage-5 wild-type embryo (b) and embryo carrying UAS::promoter-TALE-VP64, nos::GAL4 (c). (d, e) Stage-12 wild-type embryo (d) and UAS::promoter-TALE-VP64, nos::GAL4 embryo (e), stained for Engrailed (En). (f) Profiles of average expression levels of Eve in Stage-5 embryos (n=10 for each genotype). The solid gray line denotes wild-type embryos and the turquois plots denote the promoter-TALER-VP64, respectively. Lighter-shaded, bounding areas indicate ± 1 standard deviation. Signal intensity is reported in arbitrary units (AU). Embryos in panels (b–e) are matched in scale. Scale-bar in (b) equals 100 µm.
TALER mediated repression of transcriptional enhancers
Given the efficiency of TALE-mediated transcriptional repression, we tested whether TALERs could regulate specific transcriptional enhancers. We generated TALEs that targeted each of the five stripe-specific enhancers and the autoregulatory element of eve. It has been hypothesized that the regulatory autonomy of individual enhancers results from the action of short-range repressors, such as Krüppel18. It is also possible that the genomic context of eve enhancers allows enhancers to act independently.
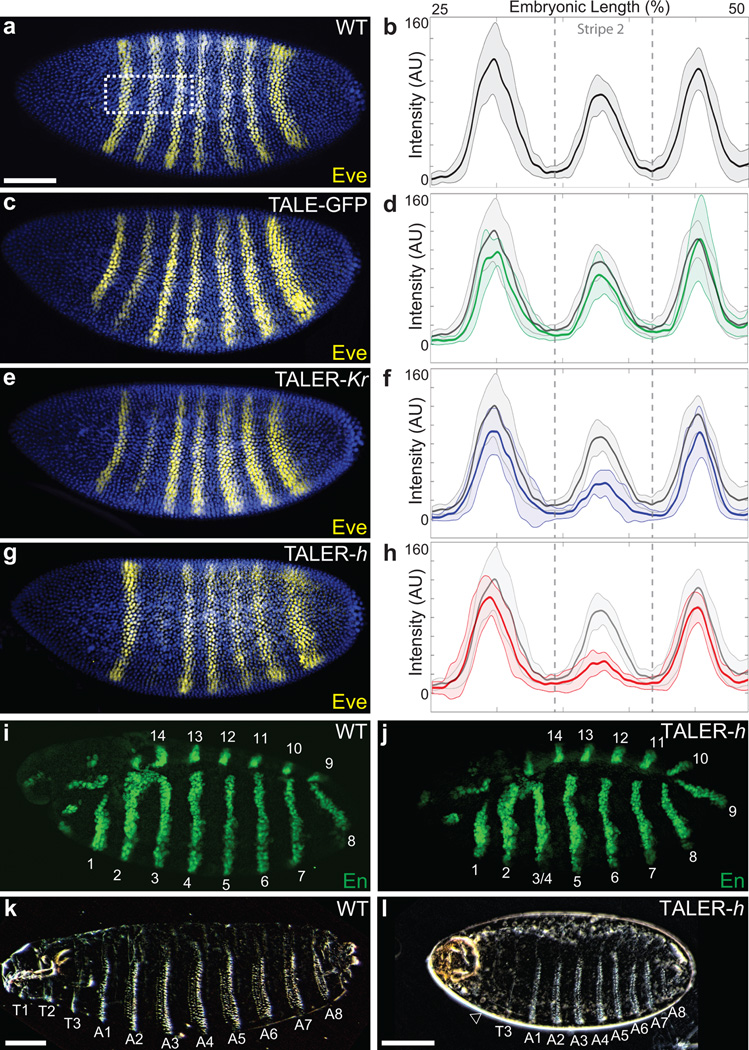
As a partial test of these alternative hypotheses—and to identify the most useful reagents—in separate experiments we drove ubiquitous expression of a TALER-Krüppel and a TALER-Hairy targeted to a 16 bp sequence within the eve stripe 2 enhancer29,30. Both TALERs repressed eve stripe 2 expression specifically, and the TALER-Hairy generated stronger repression than did the TALER-Krüppel (Fig. 3). We observed no notable changes in the levels of expression of other eve stripes (Fig. 3), even though the enhancer for stripes three and seven is located only 1.6 kb upstream from the targeted binding site (Fig. 1). These embryos lost a single stripe of engrailed expression (Fig. 3), which is consistent with the engrailed phenotype produced by a deletion of eve stripe 230. Furthermore, these embryos failed to hatch and larval cuticles exhibited an altered gnathal segment (Fig. 3), as expected26. As a control, ubiquitous expression of a TALEGFP fusion protein targeted to the same 16 bp sequence in eve stripe 2 did not alter eve expression (Fig. 3). All together, these results suggest that both Krüppel and Hairy can generate local repression of an enhancer in its native genomic location, although Hairy appears to drive stronger repression than does Krüppel. We therefore used TALER-Hairy fusion proteins for all other repression experiments.
Figure 3.
TALER targeted repression of the eve stripe 2 enhancer. (a, c, e, g), Stage-5 wild-type embryo (a), and embryos carrying UAS::enhancer-TALER-GFP, nos::GAL4 (c), UAS::enhancer-TALER-Krüppel, nos::GAL4 (e), or UAS::enhancer-TALER-hairy, nos::GAL4 (g) stained for Eve protein. (b, d, f, h) Profiles of average expression levels in region highlighted in white-dashed box in panel (a), of Eve in Stage-5 wild-type embryos (b), and embryos carrying UAS::enhancer-TALER-GFP, nos::GAL4 (d), UAS::enhancer-TALER-Krüppel, nos::GAL4 (f), or UAS::enhancer-TALER-hairy, nos::GAL4 (h) (n=10 for each genotype). In all plots, the solid gray line denotes wildtype embryos, with the green (d), blue (f), and red (h) plots denoting enhancer-TALER_GFP_, enhancer-TALER-Krüppel, and enhancer-TALER-hairy, respectively. Lighter-shaded, bounding areas indicate one standard deviation. AU indicates Arbitrary Units of fluorescence intensity. i, j, En protein staining in Stage-12 wild-type embryo (i) and Stage-12 UAS::enhancer-TALER-hairy, nos::Gal4 embryo (j). Parasegments are labeled, with parasegments 3 and 4 fused in the enhancer-TALER-hairy embryos. (k, l) Cuticle preps of wild-type first instar larva (k) and UAS::enhancer-TALER-hairy, nos::Gal4 larvae (l), with denticle belts labeled. The UAS::enhancer-TALER-hairy, nos::Gal4 larva failed to hatch and possessed fused thoracic segments (empty arrowhead). Embryos in panels (a, c, e, g, i, and j) are matched in scale. Scale bars in (a, k, and l) equal 100 µm.
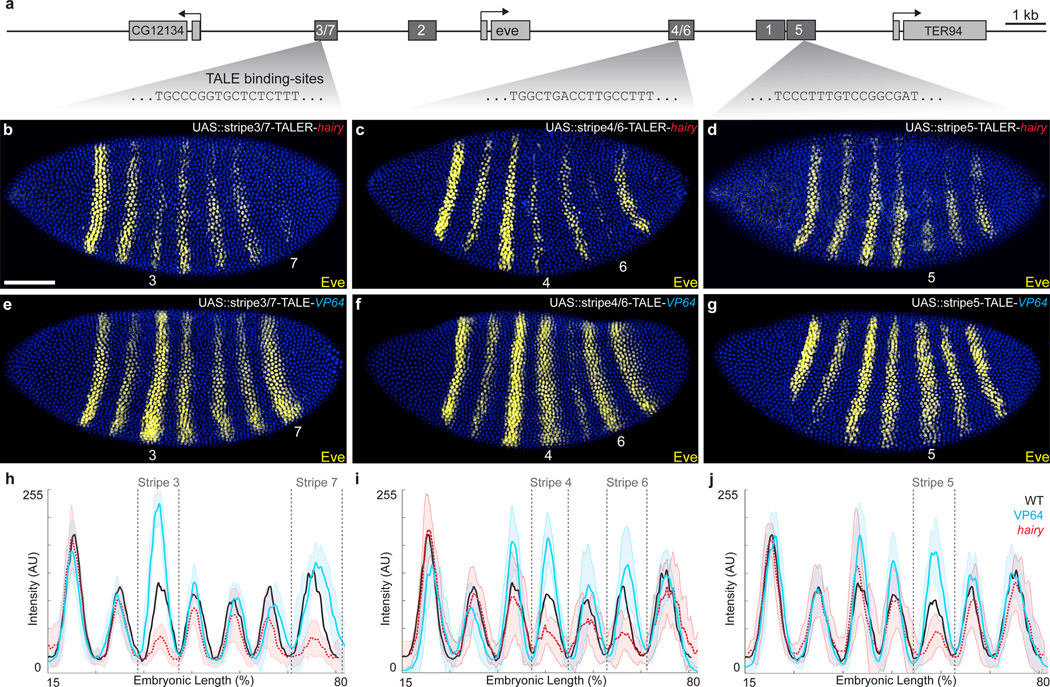
Ubiquitous expression of TALER-Hairy fusion proteins targeting each of the remaining eve stripe enhancers (Fig. 4a) caused reduced expression primarily of those stripes corresponding to the previously reported expression domain of each enhancer (Fig. 4b–d and Supplementary Fig. 5). In multiple cases, TALER-Hairy repressed stripes of eve are expressed in fewer cell rows, consistent with previous observations that eve enhancers are sensitive to repressor concentrations31. A TALER-Hairy targeted to the minimal autoregulatory sequence (MAS), located approximately 5 kb upstream of the eve promoter, caused strong reduction in expression of all eve stripes after embryonic stage 5, as expected32 (Supplementary Fig. 6). We found that a TALER-Hairy construct targeting the stripe 4/6 enhancer caused a slight reduction also in eve stripe 5 expression (Fig. 4c). However, TALERs targeting two different binding sites within the 4/6 enhancer produced similar patterns of repression of stripes four and six (Supplementary Fig. 5), while only one of these TALERs reduced expression of stripe 5. While this is an interesting observation, we cannot rule out the possibility that repression of stripe 5 by one TALER represents an experimental artifact. Each TALER-Hairy construct generated precise and predicted patterns of disruption of en and phenotypic effects in larval cuticles (Supplementary Fig. 7). We observed no evidence for ‘long-range’ repression by the TALER-Hairy constructs, suggesting that, in a native genomic context, Hairy may function at a more limited range, or with greater specificity, than suggested previously24.
Figure 4.
TALE targeted repression and activation of eve stripe enhancers. (a) Schematic of the eve locus, indicating early-embryonic _cis_-regulatory stripe enhancers and TALE binding sites. (b–g) Stage-5 embryos stained for Eve protein and carrying nos::GAL4 and either UAS::stripe 3/7-TALER-hairy (b), UAS:: stripe 4/6 TALER-hairy (c), UAS::stripe 3/7-TALER-hairy (d), UAS::stripe 3/7-TALE-VP64 (e), UAS:: stripe 4/6 TALE-VP64 (f), or UAS::stripe 3/7-TALE-VP64 (g). (h–j) Profiles of average expression levels of Eve in stage-5 embryos carrying nos::GAL4 and either UAS::stripe 3/7-TALE, nos::GAL4 (h), UAS::stripe 4/6-TALE, nos::GAL4 (i), UAS::stripe 5-TALE, nos::GAL4 (h) (n=10 for each genotype). In all plots, the solid gray line denotes wild-type embryos, with the turquois and red-dashed plots denoting enhancer-TALER-VP64 and enhancer-TALER-hairy, respectively. Lighter-shaded, bounding areas indicate one standard deviation. Embryos in panels (b–g) are matched in scale. Scale bar in (b) equals 100 µm.
TALEA mediated activation of transcriptional enhancers
The precise spatial and temporal domains of enhancer activity are believed to result, primarily, from the activity of repressors that limit the activity of more broadly expressed activators31. While the quantitative level of activators is clearly important for determining the final level of gene expression33, it is thought that most activators are unable to overcome the limiting effects of repressors31. If this is true, then targeting an additional activator to an enhancer should influence gene expression only, or mainly, in an expression domain that is already active. We tested this idea by targeting TALEAs to multiple eve enhancers.
Ubiquitously expressed TALEAs targeted to the stripe 3/7, stripe 4/6, and stripe 5 eve enhancers each caused an increase in the level of expression specifically in the stripe driven by the native enhancer (Fig. 4e–g). In several cases, the targeted eve stripe was expressed in more cell rows than in wild-type embryos. In two cases, TALEAs influenced primarily one stripe of an enhancer that was previously reported to regulate two stripes; the TALEA targeting the stripe 3/7 enhancer mainly increased stripe 3 expression and the TALEA targeting the 4/6 enhancer mainly increased stripe 4 expression (Fig. 4h and 4j). There are several possible explanations for these observations. First, while these composite enhancers cannot be divided cleanly by reporter assays into fragments that drive separate stripes, the regulatory information encoded in these enhancers may be sufficiently spatially segregated that a TALEA can influence mainly one stripe. Alternatively, the VP64 activator may be less efficient at activating some enhancers, depending on interactions with other repressive and activating factors occupying a given enhancer.
Each of the TALEAs we tested resulted in the fusion of en stripes that flanked the altered eve stripes (Fig. 5a–d). Remarkably, adult flies developed from embryos treated with all three TALEAs: TALEA stripe 4/6 adults displayed reduced abdominal segments one and six (compare Fig. 5e and Fig. 5f); TALEA stripe 3/7-adults displayed fusion of the T2 and T3 segments, including loss of a pair of legs, and reduced abdominal segment seven (Fig. 5g); and TALEA stripe 5 adults exhibited a reduced abdominal segment five (Fig. 5h). These results also reinforce that although we observed weak activation of eve stripes 7 and 6 (see above), these manipulations were sufficient to disrupt normal development of these body regions.
Figure 5.

Phenotypes resulting from enhancer-TALE-VP64 activation of eve enhancers. (a–d), Stage-15 wild-type embryo (a), and embryos carrying UAS::stripe4/6-TALE-VP64, nos::Gal4 (b), UAS::stripe3/7-TALE-VP64, nos::Gal4 (c), or UAS::stripe5-TALE_VP64, nos_::Gal4 (d) stained for En protein. Altered and fused parasegments are labeled with white brackets. Embryos in panels (a–d) are matched in scale. Scale bar in (a) equals 100 µm. (e–h), Adult wild type fly (e), and adults carrying UAS::stripe4/6-TALE-VP64, nos::Gal4 (f), UAS::stripe3/7-TALE-VP64, nos::Gal4 (g), or UAS::stripe5-TALE-VP64, nos::Gal4 (h). Adult abdominal segments are labeled with fused and altered segments labeled in red. Scale bars in (e–h) equal 400 µm.
TALER specificity for a minimal transcriptional enhancer
All together, these observations indicate that ubiquitously expressed TALEs fused to a repressor or an activator and targeted to single regulatory elements can generate specific effects. As a further test of the specificity of the TALEs, we compared the effect of the TALER-Hairy targeted to eve stripe 2 on a synthetic D. melanogaster eve stripe 2 construct and the homologous D. pseudoobscura eve stripe 2 construct, which differs by 3 bp from the D. melanogaster construct at the target sequence (Supplementary Fig. 8). When the TALER-Hairy was expressed ubiquitously, we observed lower expression of the D. melanogaster reporter gene, but no change in expression of the D. pseudoobscura reporter gene34 (Supplementary Fig. 8), suggesting that this TALE, at least, displays high specificity for its target site.
Discussion
These results indicate that individual regulatory elements in the genome can be targeted in situ with single transcriptional repressors or activators using TALEs. We were surprised that a single TALE could provide robust repression and we hypothesize that the protein-DNA interaction for TALEs is more specific than binding observed for metazoan transcription factors, which seem to have evolved relatively low specificity protein-DNA interactions to enable cooperative and synergistic binding35. The relatively local effects of the enhancer-TALER-hairy constructs that we observed are inconsistent with previous reports of long-range repression by hairy36. We suggest two hypotheses to explain this discrepancy. First, enhancers may bind proteins—either directly by DNA-protein interactions or indirectly through protein-protein interactions—that prevent interactions between neighboring enhancers. If DNA regions responsible for this hypothetical “antisocial” behavior of enhancers do not promote transcription on their own, then these DNA regions may have been trimmed from “minimal” enhancer fragments that have been used widely in classical reporter gene assays. Second, the DNA between transcriptional enhancers may encode boundary elements that limit the spread of repressor activity. This second hypothesis is consistent with the observation that deleting DNA outside of the “minimal” _eve_S2 leads to decreased transcriptional robustness30.
Perhaps the most interesting finding is that none of the ubiquitously expressed TALEAs disrupt all seven stripes of eve expression or drive expression in other ectopic locations. Even the TALEA targeted to the promoter drives increased expression mainly in the seven-stripe region. There are several distinct possibilities for these results. First, TALEAs may bind to their respective targets in all embryonic cells, but their activating signals may be overridden by repressive cues. Alternatively, the TALEA binding sites may be inaccessible to TALEA binding in cells where the enhancers are not normally active. This second hypothesis is consistent with the view that chromatin accessibility is responsible for directing the widespread patterns of Drosophila transcription factor binding37,38.
Our results strongly support a model for combinatorial activation of independent, modular Drosophila eve enhancers4,23,24. The precise effects of the TALEAs supports the view that repression acts in a dominant fashion on transcriptional activators31,39. Because TALERs and TALEAs provide experimental access specifically to active enhancers, they may allow functional dissection of non-modular enhancer architectures that have confounded reporter gene assays.
Methods
Construction of TALE plasmids
TALE constructs were based on the JFRC-7 vector26 and modified for use with the Golden-Gate method11 by mutating all Esp3I sites. TALE-C terminus fusion proteins were synthesized by GeneScript and subcloned into JFRC-7 at the XhoI/XbaI sites, removing the mCD8 and GFP domains. The following domains were added in separate constructs: GFP40; Kr repression domain, amino acids 402–502; hairy repression domain 255–337, VP64 activation domain40. Plasmids will be deposited at AddGene (www.addgene.org).
Construction of TALEs
TALE target sites were identified using the TAL Effector-Nucleotide Targeter, TALE-NT17. TALEs were subsequently assembled using the Golden-Gate method17.
Fly Strains and Crosses
Drosophila melanogaster strains were maintained under standard laboratory conditions. Transgenic TALE constructs were created by Rainbow Transgenic Flies Inc., and were integrated at the attP2-landing site. The following GAL4 drivers were used: Actin5C-GAL4; NGT4025 (Bloomington stock 4442); and rhomboid-GAL4, (Bloomington stock 26871).
Embryo Manipulations
For each respective GAL4 line, virgins were collected and crossed with male, TALE-bearing lines. Embryos were raised at 28°C and collected, and fixed according to standard protocols. Antibody staining was carried out according to standard procedures. Briefly, primary antibodies obtained from the Developmental Studies Hybridoma Bank were used to detect Eve (3C10 used 1:20) and En (4D9 used 1:20) proteins, followed by detection of primary antibodies using secondary antibodies labeled with Alexa Fluor dyes (used 1:500, Invitrogen). Cuticle preps were performed using standard protocols.
Microscopy
Confocal images were obtained on a Leica DM5500 Q Microscope, using an ACS APO 20×/0.60 IMM CORR lens, with Leica Microsystems LAS AP software. Sum projections of confocal stacks were assembled, embryos were scaled to match sizes, background was subtracted using 50 pixel rolling ball radius, and plot profiles of fluorescence intensity were analyzed using ImageJ software (rsb.info.nih.gov/ij/). Data from the plot profiles were further analyzed in Matlab.
Supplementary Material
1
2
Acknowledgements
We thank T. Martin and A. DePace, Department of Systems Biology, Harvard Medical School, for the _eve_S2-lacZ enhancer flies, E. Preger-Ben Noon and C. Standley for comments on the manuscript, B. Pfeiffer, M. Schroeder, R. Mann and the entire Stern lab for discussion. We thank the anonymous reviews for comments that significantly improved this manuscript. Finally, we thank the Janelia Farm Research Campus community for facilitating this work and for providing an inspiring scientific environment.
Footnotes
Author Contributions
JC conceived of, designed, and executed the experiments and analyzed the data, with mentorship of DLS. JC and DLS wrote the manuscript.
The authors declare they have no competing financial interests.
References
- 1.Spitz F, Furlong EEM. Transcription factors: from enhancer binding to developmental control. Nature reviews. Genetics. 2012;13:613–26. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frankel N. Multiple layers of complexity in cis-regulatory regions of developmental genes. Developmental dynamics : an official publication of the American Association of Anatomists. 2012 doi: 10.1002/dvdy.23871. [DOI] [PubMed] [Google Scholar]
- 4.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. The EMBO journal. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davis GK, Srinivasan DG, Wittkopp PJ, Stern DL. The function and regulation of Ultrabithorax in the legs of Drosophila melanogaster. Developmental biology. 2007;308:621–631. doi: 10.1016/j.ydbio.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klingler M, Soong J, Butler B, Gergen JP. Disperse versus compact elements for the regulation of runt stripes in Drosophila. Developmental biology. 1996;177:73–84. doi: 10.1006/dbio.1996.0146. [DOI] [PubMed] [Google Scholar]
- 7.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science (New York, N.Y.) 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 8.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science (New York, N.Y.) 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 9.Mahfouz MM, et al. Targeted transcriptional repression using a chimeric TALE-SRDX repressor protein. Plant molecular biology. 2012;78:311–321. doi: 10.1007/s11103-011-9866-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morbitzer R, Römer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeder ML, et al. Robust, synergistic regulation of human gene expression using TALE activators. Nature methods. 2013 doi: 10.1038/nmeth.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perez-Pinera P, et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nature methods. 2013 doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cong L, Zhou R, Kuo Y-C, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nature communications. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic acids research. 2012;40:7584–7595. doi: 10.1093/nar/gks404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Moore R, Guinn M, Bleris L. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Scientific reports. 2012;2:897. doi: 10.1038/srep00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geissler R, et al. Transcriptional activators of human genes with programmable DNA-specificity. PloS one. 2011;6:e19509. doi: 10.1371/journal.pone.0019509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cermak T, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic acids research. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray S, Levine M. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes & development. 1996;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- 19.Li LM, Arnosti DN. Long- and short-range transcriptional repressors induce distinct chromatin states on repressed genes. Current biology : CB. 2011;21:406–412. doi: 10.1016/j.cub.2011.01.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai HN, Arnosti DN, Levine M. Long-range repression in the Drosophila embryo. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9309–9314. doi: 10.1073/pnas.93.18.9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harding K, Rushlow C, Doyle HJ, Hoey T, Levine M. Cross-regulatory interactions among pair-rule genes in Drosophila. Science (New York, N.Y.) 1986;233:953–959. doi: 10.1126/science.3755551. [DOI] [PubMed] [Google Scholar]
- 22.Frasch M, Warrior R, Tugwood J, Levine M. Molecular analysis of even-skipped mutants in Drosophila development. Genes & development. 1988;2:1824–1838. doi: 10.1101/gad.2.12b.1824. [DOI] [PubMed] [Google Scholar]
- 23.Fujioka M, Emi-Sarker Y, Yusibova GL, Goto T, Jaynes JB. Analysis of an even-skipped rescue transgene reveals both composite and discrete neuronal and early blastoderm enhancers, and multi-stripe positioning by gap gene repressor gradients. Development (Cambridge, England) 1999;126:2527–2538. doi: 10.1242/dev.126.11.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Small S, Arnosti DN, Levine M. Spacing ensures autonomous expression of different stripe enhancers in the even-skipped promoter. Development (Cambridge, England) 1993;119:762–772. [PubMed] [Google Scholar]
- 25.Tracey WD, Ning X, Klingler M, Kramer SG, Gergen JP. Quantitative analysis of gene function in the Drosophila embryo. Genetics. 2000;154:273–284. doi: 10.1093/genetics/154.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nüsslein-Volhard C, Wieschaus E. Mutations affecting segment number and polarity in Drosophila. Nature. 1980;287:795–801. doi: 10.1038/287795a0. [DOI] [PubMed] [Google Scholar]
- 27.Staller MV, et al. Depleting gene activities in early Drosophila embryos with the “maternal-Gal4-shRNA” system. Genetics. 2013;193:51–61. doi: 10.1534/genetics.112.144915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manoukian AS, Krause HM. Concentration-dependent activities of the even-skipped protein in Drosophila embryos. Genes & development. 1992;6:1740–1751. doi: 10.1101/gad.6.9.1740. [DOI] [PubMed] [Google Scholar]
- 29.Small S, Blair A, Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. The EMBO journal. 1992;11:4047–4057. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig MZ, Manu, Kittler R, White KP, Kreitman M. Consequences of eukaryotic enhancer architecture for gene expression dynamics, development, and fitness. PLoS genetics. 2011;7:e1002364. doi: 10.1371/journal.pgen.1002364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Clyde DE, et al. A self-organizing system of repressor gradients establishes segmental complexity in Drosophila. Nature. 2003;426:849–853. doi: 10.1038/nature02189. [DOI] [PubMed] [Google Scholar]
- 32.Jiang J, Hoey T, Levine M. Autoregulation of a segmentation gene in Drosophila: combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes & development. 1991;5:265–277. doi: 10.1101/gad.5.2.265. [DOI] [PubMed] [Google Scholar]
- 33.Wunderlich Z, et al. Dissecting sources of quantitative gene expression pattern divergence between Drosophila species. Molecular systems biology. 2012;8:604. doi: 10.1038/msb.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ludwig MZ, et al. Functional evolution of a cis-regulatory module. PLoS biology. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 1. Models and theory. Biochemistry. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 36.Barolo S, Levine M. hairy mediates dominant repression in the Drosophila embryo. The EMBO journal. 1997;16:2883–2891. doi: 10.1093/emboj/16.10.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kaplan T, et al. Quantitative models of the mechanisms that control genome-wide patterns of transcription factor binding during early Drosophila development. PLoS genetics. 2011;7:e1001290. doi: 10.1371/journal.pgen.1001290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li X-Y, et al. The role of chromatin accessibility in directing the widespread, overlapping patterns of Drosophila transcription factor binding. Genome biology. 2011;12:R34. doi: 10.1186/gb-2011-12-4-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davidson EH. The Regulatory Genome: Gene Regulatory Networks In Development And Evolution. Academic Press; 2006. p. 304. [Google Scholar]
- 40.Pfeiffer BD, et al. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010;186:735–755. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1
2