Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation (original) (raw)
. Author manuscript; available in PMC: 2013 Sep 16.
Published in final edited form as: Nature. 2008 Apr 30;453(7196):812–816. doi: 10.1038/nature06906
Abstract
Numerous post-translational modifications of histones have been described in organisms ranging from yeast to humans1. Growing evidence for dynamic regulation of these modifications, position- and modification-specific protein interactions, and biochemical crosstalk between modifications has strengthened the ‘histone code’ hypothesis, in which histone modifications are integral to choreographing the expression of the genome1,2. One such modification, ubiquitylation of histone H2B (uH2B) on lysine 120 (K120) in humans3, and lysine 123 in yeast4, has been correlated with enhanced methylation of lysine 79 (K79) of histone H3 (refs 5–8), by K79-specific methyltransferase Dot1 (KMT4)9–11. However, the specific function of uH2B in this crosstalk pathway is not understood. Here we demonstrate, using chemically ubiquitylated H2B, a direct stimulation of hDot1L-mediated intranucleosomal methylation of H3 K79. Two traceless orthogonal expressed protein ligation (EPL) reactions were used to ubiquitylate H2B site-specifically. This strategy, using a photolytic ligation auxiliary and a desulphurization reaction, should be generally applicable to the chemical ubiquitylation of other proteins. Reconstitution of our uH2B into chemically defined nucleosomes, followed by biochemical analysis, revealed that uH2B directly activates methylation of H3 K79 by hDot1L. This effect is mediated through the catalytic domain of hDot1L, most likely through allosteric mechanisms. Furthermore, asymmetric incorporation of uH2B into dinucleosomes showed that the enhancement of methylation was limited to nucleosomes bearing uH2B. This work demonstrates a direct biochemical crosstalk between two modifications on separate histone proteins within a nucleosome.
It has been proposed that uH2B may induce H3 K79 methylation directly, either by altering chromatin structure and therefore nucleosomal accessibility, or through the recruitment of enzymatic function12,13. However, the possibility that one or more additional factors may be required to translate the effect of uH2B into heightened Dot1 methyltransferase activity is equally likely14–16. To decipher the role of uH2B in H3 K79 methylation, it is necessary to generate or purify homogenously ubiquitylated H2B. In vivo, ubiquitin ligases catalyse the site-specific condensation of the carboxy-terminal carboxylic acid of ubiquitin and the ε-NH2 of the target lysine in H2B, forming an isopeptide bond4–6,17,18. In vitro reconstitution of H2B with these ubiquitin ligases and associated factors allows the production of uH2B in limited quantities6. Because of its natural abundance, uH2B can also be purified from endogenous sources3. However, heterogeneity due to the presence of additional modifications may complicate biochemical analyses. We decided to use EPL technology to ubiquitylate H2B regioselectively, thus bypassing the requirement for the complex cellular ubiquitylation machinery and ensuring chemical homogeneity. EPL allows the formation of an amide bond between two polypeptides of recombinant and synthetic origins, one containing a C-terminal-α-thioester, and the other, an amino-terminal cysteine (Supplementary Fig. 1) (ref. 19).
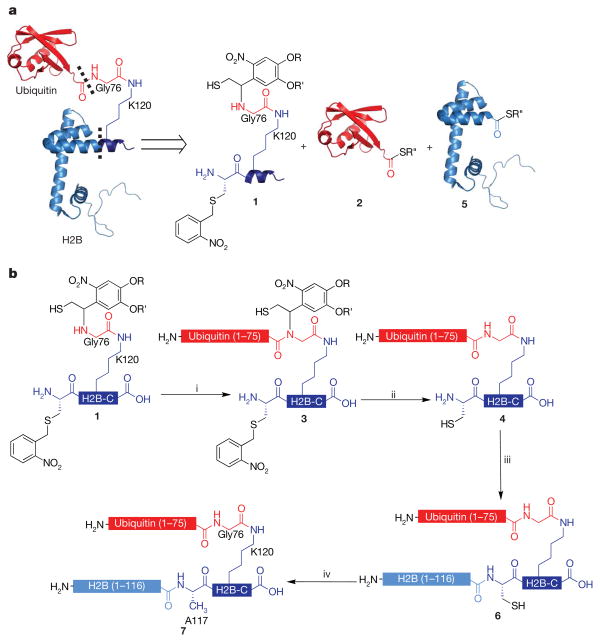
In designing a semi-synthesis of site-specifically ubiquitylated H2B, we imagined it would be necessary to link three polypeptide building blocks together covalently, one of synthetic and two of recombinant origins (Fig. 1a). Owing to the absence of native cysteines in both H2B and ubiquitin, such a scheme requires the use of two traceless ligation strategies to generate native uH2B (Fig. 1b). In the first ligation, we reasoned that a photolytically removable, thiol-bearing ligation auxiliary could be used. We have previously demonstrated that this auxiliary allows for the site-specific ubiquitylation of a lysine side chain in model peptides20. Adapting this approach to the production of a ubiquitylated protein requires the incorporation of additional functionality to facilitate a second regioselective EPL reaction. With this in mind, peptide 1 was synthesized corresponding to residues 117–125 of H2B bearing both the ligation auxiliary, attached to the ε-NH2 of K120, and an A117C mutation. Orthogonal side-chain protection of K120 allowed the ligation auxiliary to be coupled to this amino group through a glycyl linker (Supplementary Fig. 2). This linker will eventually become Gly76 of ubiquitin and, importantly, allows us to exploit the Gly-Gly sequence at the C terminus of ubiquitin as an optimal junction for auxiliary-mediated ligation (Fig. 1b). Also critical to the synthetic design was transient protection of the N-terminal cysteine in 1, which precluded unwanted double ligation of ubiquitin. After examining several possibilities, we settled on the photoremovable _S_-(_o_-nitrobenzyl) group for this purpose, as this proved to be completely stable during the course of the first ligation reaction and was easily removed by photolysis.
Figure 1. Semi-synthesis of ubiquitylated H2B.
a, Retrosynthetic analysis of uH2B synthesis. uH2B was generated by a three-piece ligation with the following polypeptides: auxiliary-linked synthetic peptide containing residues 117–125 of H2B and bearing an A117C mutation, H2B-C, 1; recombinant ubiquitin(1–75)- α-thioester, 2; and recombinant H2B(1-116)- α-thioester, 5. Dashed lines indicate junctions formed by EPL reactions. b, Synthetic scheme for the ubiquitylation of H2B. i, EPL was used to ligate peptide 1 to protein 2, forming branched protein 3. ii, Ligation product 3 was irradiated with 365-nm light, yielding protein 4. iii, Ligation of protein 4 to protein 5, forming uH2B(A117C), 6. iv, Raney nickel desulphurization of protein 6, forming uH2B, 7. R = CH2CH2CH2C(O)NH2CH3; R′ =CH3; R″ =CH2CH2SO3H.
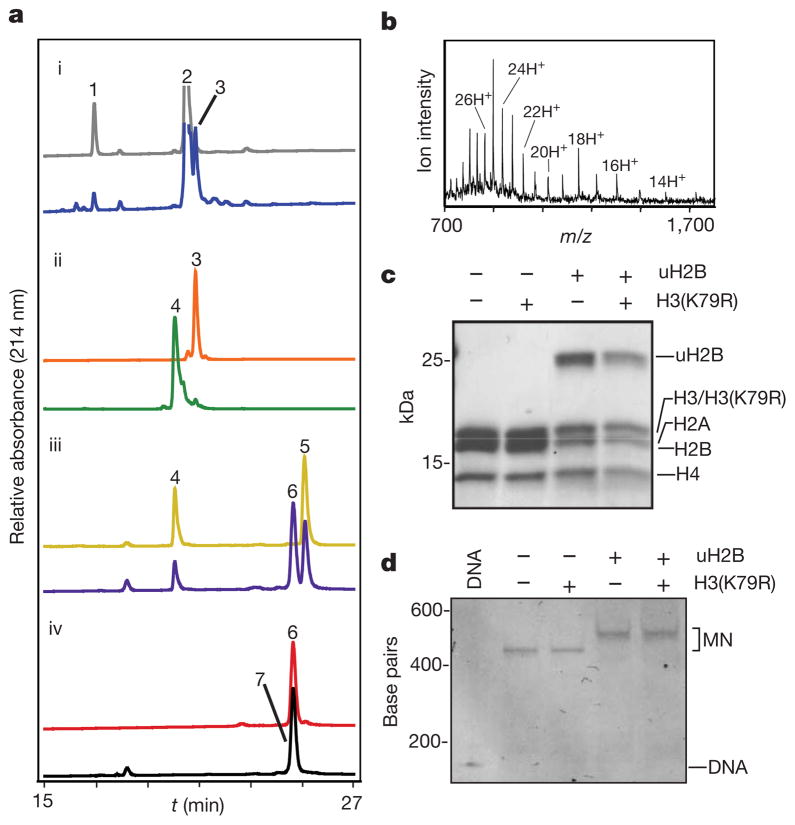
In the first step of the synthesis, peptide 1 was ligated to ubiquitin(1–75)-α-thioester, 2, produced by thiolysis of a corresponding intein fusion protein (Supplementary Fig. 3), to give ubiquitylated peptide, 3 (Fig. 2a, panel i, and Supplementary Fig. 4). Subsequent ultraviolet irradiation of 3 led to efficient removal of both the ligation auxiliary and the cysteine protecting group to give deprotected branched protein 4 (Fig. 2a, panel ii, and Supplementary Fig. 4). This intermediate product was then ligated to recombinant H2B(1–116)-α-thioester, 5 (Supplementary Fig. 3), to give uH2B(A117C), 6 (Fig. 2a, panel iii and Supplementary Fig. 4). In the final step, Raney-nickel-mediated desulphurization21 was used to convert the single cysteine residue in branched protein 6 to the native alanine residue present in uH2B (Fig. 2a, panel iv). Under optimized conditions, this reduction was found to be highly specific for cysteine desulphurization over methionine, thereby affording native ubiquitylated H2B, 7 (Fig. 2b). The overall yield of the semi-synthesis (that is, steps i–iv in Fig. 1b) was excellent (20%). After purification, semi-synthetic uH2B was successfully incorporated into core histone octamers with wild-type recombinant H2A, H3 and H4 (Fig. 2c; Supplementary Fig. 5). Additional histone octamers were formed containing a recombinant H3(K79R) mutant, either alone or in combination with uH2B. These octamers were used to reconstitute chemically defined mononucleosomes with a 147 base pair (bp) region of the 601 nucleosomal targeting sequence (Fig. 2d) (ref. 22).
Figure 2. Semi-synthesis of uH2B and incorporation into chemically defined histone octamers and nucleosomes.
a, Superimposed reverse-phase high-performance liquid chromatography (RP-HPLC) chromatograms of starting materials (top trace) and products (bottom trace) of each pair, for reactions i–iv described in Fig. 1b. b, Electrospray ionization mass spectrometry (ESI–MS) spectrum of purified uH2B, 7. Charge states are labelled. (M + H)+ observed: 22,366 ± 4 Da (s.d.); expected: 22,365 Da. c, Reconstituted octamer samples were analysed by SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and staining with Coomassie blue. d, Ethidium-bromide-stained native gel of reconstituted mononucleosomes (MN). Octamers and mononucleosomes contain recombinant, unmodified histones except where otherwise noted.
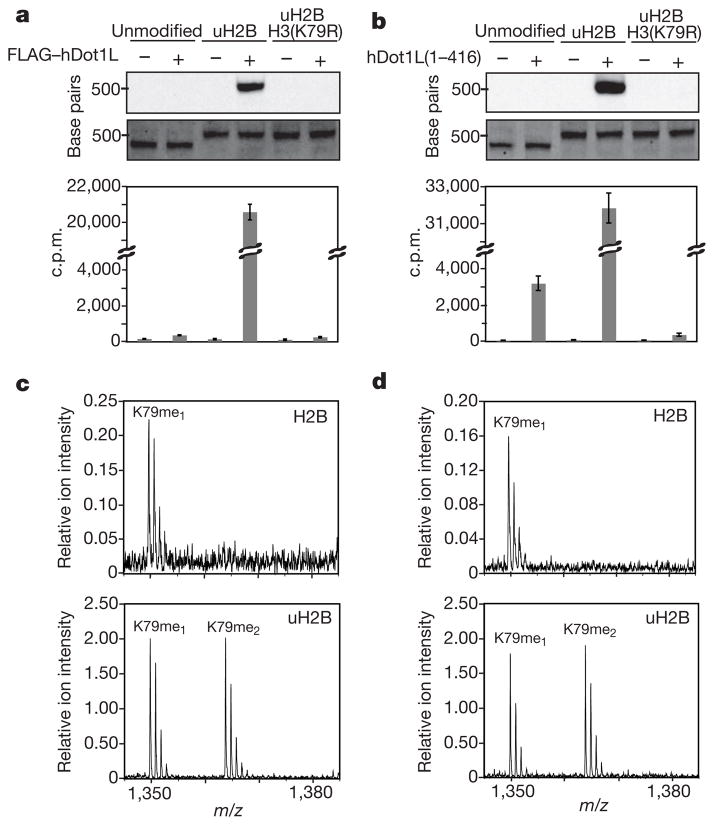
With modified nucleosomes in hand, we next turned our attention to exploring the effect of uH2B on Dot1 methyltransferase activity. A Flag-tagged version of human Dot1 (Flag–hDot1L) was isolated from an insect cell overexpression system and found to methylate unmodified histone octamers (Supplementary Figs 6 and 7). However, no activity was observed on H3 alone, (H3/H4)2 tetramers or on mononucleosomes and chromatinized plasmids assembled with recombinant unmodified histones (Fig. 3a and Supplementary Fig. 7). This suggests that the presence of nucleosomal DNA represses the activity of hDot1L in vitro. This is in stark contrast to yeast methyltransferase, Dot1, which exhibits greater activity on recombinant nucleosomes than octamers23. We predicted that ubiquitylation of H2B would stimulate hDot1L methyltransferase activity in the context of nucleosomes. To test this hypothesis, a 3H-SAM methyltransferase assay was performed using Flag–hDot1L and our chemically defined nucleosomes. Robust methyltransferase activity was observed on mononucleosomes containing uH2B, whereas no radioactivity was detected on unmodified mononucleosomes (Fig. 3a). This activity was specific for K79 of H3 as hDot1L was incapable of methylating nucleosomes containing both uH2B and H3(K79R). Importantly, the level of methylation activity observed on ubiquitylated mononucleosomes was far greater than that observed on unmodified histone octamers (Supplementary Fig. 7). hDot1L also methylated a chromatinized plasmid containing uH2B (Supplementary Fig. 8). These results establish a direct biochemical connection between ubiquitylated H2B and H3 K79 methylation by hDot1L.
Figure 3. Effects of uH2B on intranucleosomal methylation of H3 K79 by hDot1L.
a, b, hDot1L methyltransferase assay using chemically defined nucleosomes. Assays were performed on mononucleosomes with 3H-SAM and either full-length Flag–hDot1L, a, or a glutathione _S_-transferase (GST) fusion of the catalytic domain, hDot1L(1–416), b. Assay samples were separated on a native gel and stained with ethidium bromide (middle panel) before probing for 3H-methyl incorporation by fluorography (top panel). Quantification of methylation was performed by filter binding followed by liquid scintillation counting (bottom panel). Error bars, s.e.m (n =4–7). c, d, Characterization of degree of methylation by mass spectrometry. In-gel trypsin digestion of H3 from methyltransferase assays using Flag–hDot1L, c, and hDot1L(1–416), d, followed by MALDI-mass spectrometry. Ion intensities of methylated peptides (residues 73–83) are scaled relative to an internal standard. Assays were performed using unmodified mononucleosomes (top panel) and nucleosomes containing uH2B (bottom panel).
Although H3 K79 and H2B K120 reside in separate polypeptides, their side chains are located in close proximity to one another on the face of the nucleosome24, posing a structural basis for a crosstalk pathway. Indeed, docking of the structure of the catalytic domain of hDot1L onto the mononucleosome structure, positions the catalytic domain adjacent to the site of H2B ubiquitylation25. Therefore, the catalytic domain of hDot1L, containing residues 1–416, was purified from an Escherichia coli expression system to interrogate its role in uH2B-dependent H3 methylation (Supplementary Fig. 6). Like our observations with full-length hDot1L, a significant enhancement in activity of the catalytic domain of hDot1L was measured on mononucleosomes containing uH2B, compared with unmodified mononucleosomes (Fig. 3b). However, unlike the full-length enzyme, hDot1L(1–416) also exhibited some, albeit minimal, methyltransferase activity on unmodified mononucleosomes.
H2B ubiquitylation has been correlated with increased levels of di- and trimethylation of H3 K79 in humans5,6 and yeast26, respectively. Therefore, we examined the degree of methylation occurring in our assays. In-gel trypsin digestion of H3, followed by matrix-assisted laser desorption/ionization (MALDI)-mass spectrometry showed some monomethylation and no di- and trimethylation of unmodified mononucleosomes, in assays performed with either full-length hDot1L or the catalytic domain alone (Fig. 3c, d, top panels). However, robust mono- and dimethylation of nucleosomes containing uH2B was observed in both cases (Fig. 3c, d, bottom panels). No evidence of trimethylation was observed is our assays, which is consistent with mass spectrometry analysis of H3 K79 methylation in human cell lines27.
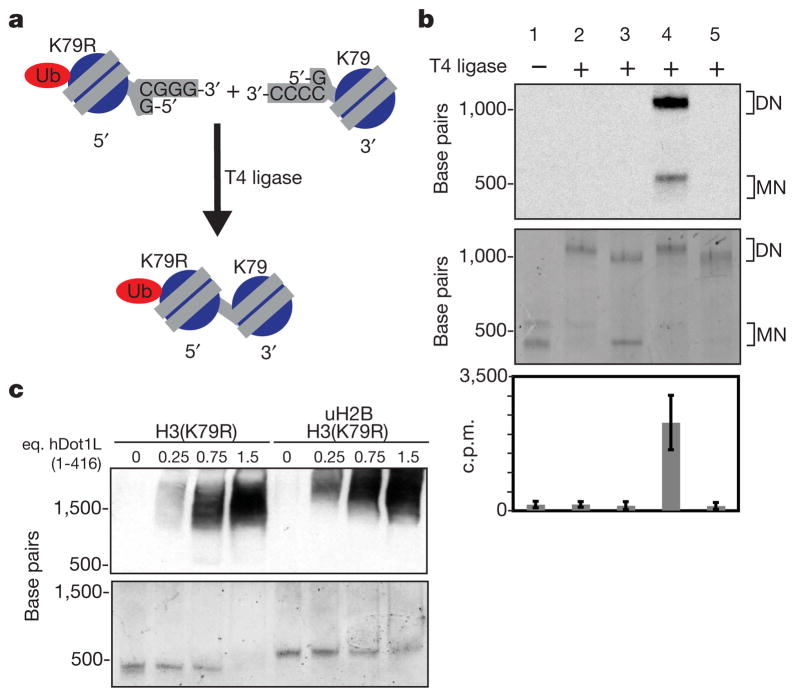
We have demonstrated that uH2B is required for effective methylation of mononucleosomes by hDot1L. However, in vivo levels of H3 K79 methylation vastly exceed those of H2B ubiquitylation3,27. This raises the question: can uH2B enhance hDot1L-mediated methylation in an internucleosomal fashion? To address this possibility, a strategy for the creation of asymmetric dinucleosomes28 was adopted. Dinucleosomes with asymmetric incorporation of uH2B and H3(K79R) were produced by ligating uniquely assembled 5′ and 3′ mononucleosomes with complementary DNA overhangs (Fig. 4a). The use of non-palindromic complementary overhangs resulted in only the desired heterodinucleosome products being formed (Supplementary Fig. 9). Unmodified, wild-type nucleosomes ligated to nucleosomes containing uH2B and H3(K79R) showed no increased hDot1L-mediated methylation compared with unmodified, wild-type nucleosomes ligated to nucleosomes bearing H3(K79R) alone (Fig. 4b, lanes 2 and 3). However, hDot1L was capable of methylating dinucleosomes, as evidenced by methylation of nucleosomes containing uH2B ligated to nucleosomes containing uH2B and H3(K79R) (Fig. 4b, lane 4). It could be demonstrated that no significant histone shuffling was occurring during these assays because an unligated mixture of nucleosomes containing both uH2B and H3(K79R) and unmodified, wild-type nucleosomes showed no methylation above that observed with dinucleosomes containing only H3(K79R) (Fig. 4b, lanes 1 and 5). These results strongly suggest that efficient methylation of H3 K79 requires the presence of uH2B in the same nucleosome. Therefore, we propose that most H3 K79 methylated nucleosomes in vivo are likely to have at one time carried uH2B, which has since been removed by deubiquitylating enzymes or histone replacement.
Figure 4. Methyltransferase assays on dinucleosomes using full-length hDot1L.
a, 5′ and 3′ mononucleosomes (MN) were reconstituted separately using DNA containing complementary overhangs. Assembly of 5′ and 3′ nucleosomes with different histones, followed by ligation, resulted in asymmetric dinucleosome (DN) formation. b, 3H-SAM methyltransferase assays performed with Flag–hDot1L and dinucleosomes were separated on a native gel and stained with ethidium bromide (middle panel) before 3H-methyl detection by fluorography (top panel). Filter-binding and liquid scintillation counting was used to quantify 3H-methyl incorporation (bottom panel). Lanes (nucleosomes listed as [5′ ] − [3′] and [5′] + [3′], for ligated and unligated nucleosomes, respectively): 1 = [uH2B/H3(K79R)] + [unmodified]; 2 = [uH2B/H3(K79R)] − [unmodified]; 3 = [H3(K79R)] − [unmodified]; 4 = [uH2B/H3(K79R)] − [uH2B]; 5 = [uH2B/H3(K79R)] − [H3(K79R)]. Error bars, s.e.m. (n = 8). c, Electrophoretic-mobility shift assay performed with hDot1L(1–416) and indicated nucleosomes. Nucleosomes were incubated with increasing amounts of hDot1L(1–416), separated by native gel electrophoresis and stained with ethidium bromide (bottom panel). A western blot was performed against hDot1L using an anti-GST antibody (top panel; eq., molar equivalent). H3(K79R) was used in these assays to eliminate potential affinity differences introduced by disproportionate methylation.
The simplest explanation that accounts for the intranucleosomal stimulatory effect of uH2B on hDot1L-mediated methylation is that uH2B recruits hDot1L to nucleosomes. However, the presence of a large excess of free ubiquitin had only a modest effect on the extent of hDot1L-mediated H3 K79 methylation in uH2B containing nucleosomes (Supplementary Fig. 10). The catalytic domain of hDot1L has previously been demonstrated to bind to unmodified nucleosomes in electrophoretic mobility shift assays25. We observed no significant difference in the recruitment of hDot1L(1–416) to mononucleosomes as a function of ubiquitylation, based on gel-shift analysis, and only a modest difference in gel-shift competition assays, over a broad range of concentrations (Fig. 4c and Supplementary Fig. 11). This establishes that hDot1L(1–416) is able to bind to both ubiquitylated and unmodified nucleosomes under the conditions of our assays, even though efficient methyltransferase activity is only observed on ubiquitylated nucleosomes. Thus it is likely that uH2B, possibly through an allosteric mechanism, allows hDot1L to bind nucleosomes in a catalytically competent manner. Additional investigations, perhaps involving structural analysis, will be necessary to reveal more about this phenomenon.
In summary, we have reported the chemical synthesis of a site-specifically ubiquitylated H2B. This strategy will allow the attachment of ubiquitin and Gly-Gly containing ubiquitin-like modifiers to other histones and non-histone proteins, at native and non-native sites. Chemical control around the isopeptide bond presents the option of incorporating chemical probes to study context-specific ubiquitin attachment and removal. Incorporation of our uH2B into chemically defined mono- and dinucleosomes revealed a direct stimulation of intranucleosomal methylation of H3 K79 by hDot1L. We anticipate that this combination of semi-synthetic protein chemistry to create engineered chromatin substrates, and their subsequent biochemical analysis, will play an important role in the hypothesis-driven dissection of the mechanisms underlying the ‘histone code’2.
METHODS
General methods
Amino-acid derivatives, pre-loaded Wang resin and coupling reagents were purchased from Novabiochem. E. coli BL21(DE3) and pLysS cells were purchased from Novagen. 3H-_S_-adenosyl methionine, goat anti-GST antibody, Amplify solution, pGex2T vector, Sephacryl S-200 resin, GSTPrep FF 16/10 column and HiTrap SP HP 1 ml column were obtained from GE Healthcare. HRP-conjugated rabbit anti-goat antibody was obtained from Dako. Restriction enzymes, polynucleotide kinase, T4 ligase, pTXB1 vector and chitin resin were obtained from New England Biolabs. Criterion 15% and 18% Tris HCl and Criterion 5% TBE gels were purchased from Biorad. Centricons were purchased from Sartorius. PCR purification and gel extraction kits were purchased from Qiagen. All other chemical reagents were purchased from Sigma-Alrich or Fisher Scientific. Analytical and semi-preparative scale RP-HPLC were performed on a Hewlett-Packard 1100 series instrument using Vydac C18 columns (4 mm × 150 mm; 10 mm × 250 mm) at 1 and 4 ml min−1, respectively. Unless otherwise noted, all analytical gradients were 0–73% B over 30 min (A: 0.1% trifluoroacetic acid (TFA) in water; B: 90% acetronitrile, 0.1% TFA in water). Preparative and process scale RP-HPLC were performed on a Waters DeltaPrep 4000 system connected to a Waters 486 tunable detector using Vydac C18 columns (22 × 250 mm; 50 × 250 mm) at 15 and 30 ml min−1, respectively. Size-exclusion and ion-exchange chromatography were performed on an AKTA FPLC system from GE Healthcare equipped with a P-920 pump and a UPC-900 monitor. ESI-MS was performed on a Sciex API-100 single quadrupole mass spectrometer. Primer synthesis and DNA sequencing were performed by Integrated DNA Technologies and Genewiz, respectively.
Peptide synthesis
The sequence corresponding to residues 117–125 of Xenopus H2B with an A117C replacement was synthesized on pre-loaded Wang resin using manual solid-phase peptide synthesis with an Fmoc Nα protection strategy and using 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) for amino-acid activation. Standard tbutyl side-chain protection was used throughout with the following exceptions: the ε-amino group of K120 was protected with the Mtt group, and the thiol group of C117 was protected with an _o_-nitrobenzyl group (Supplementary Fig. 2). _N_α-(tbutoxycarbonyl)-_S_-(_o_-nitrobenzyl)-L-cysteine (Boc-Cys(ONB)) used in the peptide synthesis was prepared as previously described29,30. The glycyl linker and ligation auxiliary were installed on the solid phase as follows (Supplementary Fig. 2): (i) the Mtt group on K120 was deprotected by successive incubations of the peptidyl-resin with 1% TFA in DCM containing 1% triisopropylsilane (TIS) for 10 min intervals, until no yellow colour evolved; (ii) bromoacetic acid (222 mg, 1.6 mmol) was triple coupled to the ε-NH2 of K120 with DIC (1.6 mmol) for 3 × 1 h to yield peptidyl-resin 9; (iii) the ligation auxiliary (4-[4-(1-amino-2-_t_-butyldisulphanyl-ethyl)-2-methoxy-5-nitro-phenoxy]-N-methylbutyramide)20 (32.6 mg, 76 μmol) and DIEA (1.4 mmol) were added to resin 9 (120 mg, 76 μmol) suspended in 600 μl DMF. The alkylation was allowed to proceed for 72 h at room temperature, after which the resin was dried. After cleavage from the resin with TFA:TIS:H2O (95:2.5:2.5) for 3 h, peptide 10 was purified by RP-HPLC on a preparative scale using a 28–38% B gradient over 45 min, yielding 10.8 mg peptide. Peptide 10 was characterized by ESI-MS ((M + H)+ observed: 1,606.8 Da; expected: 1,607.9 Da) (Supplementary Fig. 2).
Preparation of ubiquitin(1–75) and H2B(1–116)-α-thioesters
Ubiquitin (1–75)-MES (2) was prepared as previously described20. A truncated Xenopus H2B gene, containing residues 1–116, was amplified by PCR using primers H2B-FP (5′-GGAATTCCATATGCCTGAGCCAGCCAAGTCCGCTCCAGCC-CCG-3′) and H2B116-RP (5′-GGTGGTTGCTCTTCCGCACTTGGTGCCCT-CGGACAC-3′) and a Xenopus H2B expression plasmid31 as a template. After digestion by _Nde_I and _Sap_I, the fragment was ligated into a similarly digested pTXB1 vector, and the resulting plasmid, pRMH2B-N, which encodes H2B (1–116) fused at its C terminus to the GyrA intein and a chitin-binding domain, was verified by DNA sequencing.
E. coli BL21(DE3) cells transformed with pRMH2B-N were grown in Luria-Bertani medium at 37 °C until mid-log phase, and protein expression was induced by the addition of 0.5 mM IPTG and allowed to continue at 25 °C for 16 h. After harvesting the cells by centrifugation at 6.8 kg for 15 min, the cell pellet was resuspended in lysis buffer (50 mM Tris, 200 mM NaCl, 1 mM EDTA, pH 7.5) and frozen at −80 °C. Thawed cells were lysed by passage through a French press and the insoluble material was removed by centrifugation at 26,000_g_ for 30 min. The supernatant was filtered and incubated overnight at 4 °C with chitin resin (35 ml) pre-equilibrated in lysis buffer. The resin was washed with 200 ml of wash buffer 1 (50 mM Tris, 200 mM NaCl, 1 mM EDTA, pH 7.2) and 700 ml wash buffer 2 (50 mM Tris, 200 mM NaCl, 1 mM EDTA, pH 7.4). The resin was then incubated with cleavage buffer (50 mM Tris, 200 mM NaCl, 1 mM EDTA, 100 mM MESNa, pH 7.4) for 70 h, resulting in thiolysis of the intein fusion, forming H2B(1–116)-MES, 5. The column was eluted and the resin was washed with 2 × 25 ml cleavage buffer. The thiolysis reaction was repeated and the combined elution fractions further purified by preparative RP-HPLC using a 42–52% B gradient over 45 min, yielding 4–5 mg of lyophilized protein per litre of culture. The identity of the purified protein was verified as 5 by ESI-MS ((M + H)+ observed: 12,991 ± 3 Da; expected: 12,991 Da) (Supplementary Fig. 3). Mass spectrometry indicated that the non-native N-terminal methionine used for expression of 5 was processed during recombinant expression, leaving the native N-terminal sequence.
Expressed protein ligation reaction 1
The ligation reaction between peptide 1 and ubiquitin(1–75)-MES, 2, was performed using conditions similar to those previously optimized for model reactions20. In a typical reaction, purified peptide 10 (1.1 mg, 0.69 μmol) was dissolved in 180 μl of buffer containing 3 M guanidinium chloride, 300 mM sodium phosphate pH 7.8, 50 mM tris(2-carboxyethyl)phosphine (TCEP), and incubated at room temperature for 30 min to remove the S-tbutyl protection group on the ligation auxiliary. The resulting reduced peptide 1 was added to ubiquitin(1–75)-MES, 2 (17.1 mg, 1.98 μmol) and dissolved in ligation buffer (3 M guanidinium chloride, 300 mM sodium phosphate, 100 mM MES, pH 7.8). The reaction volume was increased to 950 μl with ligation buffer and the pH was adjusted to 7.8 using NaOH. After 120 h at 4 °C, the reaction was quenched with 1 ml of 50% HPLC buffer B containing 100 mM TCEP. Ligation product 3 was purified using preparative HPLC with a 32–42% B gradient over 45 min, yielding 4.0 mg of lyophilized protein. ESI-MS was used to verify the identity of the ligation product ((M + H)+ observed: 10,008 ± 1 Da; expected: 10,009 Da) (Supplementary Fig. 4).
Photolysis
In a typical reaction, ligation product 3 (3.5 mg, 0.35 mmol) was dissolved in 1.5 ml of photolysis buffer (25% HPLC buffer B containing 10 mM semicarbazide, 10 mM DTT, 10 mM ascorbic acid and 2 mM cysteine)30. The resulting solution was irradiated at 365 nm for 4 h using a collimated light source from Oriel equipped with a 200 W Hg lamp. Irradiance (4 mW cm−2) was measured by using a model 840-c monochromic photometer. Selective irradiation at 365 nm was achieved by using an analytical line filter (9.4 nm bandwidth) obtained from Oriel. Irradiation effected removal of the ligation auxiliary and the _o_-nitrobenzyl protecting group, forming branched protein 4. Semi-preparative RP-HPLC purification of protein 4 was accomplished using a 0–73% B gradient over 45 min, yielding 2.5 mg of lyophilized protein 4. Removal of the two photolytic groups was verified by ESI-MS ((M + H)+ observed: 9,547 ± 2 Da; expected: 9,548 Da) (Supplementary Fig. 4).
Expressed protein ligation reaction 2
In a typical reaction, photolysis product, 4 (2.5 mg, 0.25 μmol), and H2B(1–116)-MES, 5 (5.0 mg, 0.38 μmol), were dissolved in ligation buffer to a final volume of 225 μl. The pH of the resulting solution was increased to 7.8 with NaOH and the reaction was allowed to proceed for 78 h before quenching with 225 μl of 50% HPLC buffer B containing 100 mM TCEP. The ligation product, 6, was purified using semi-preparative RP-HPLC with a 42–52% B gradient over 45 min, yielding 3.0 mg of uH2B(A117C), 6. The identity of the ligation product was verified by ESI-MS ((M + H)+ observed: 22,395 ± 3 Da; expected: 22,397 Da) (Supplementary Fig. 4).
Desulphurization
Raney nickel reduction was used to convert C117 of protein 6 to the native alanine21,32. In a typical reaction, uH2B(A117C), 6, (1.3 mg, 58 nmol) was dissolved in 3 ml desulphurization buffer (6 M guanidinium chloride, 200 mM sodium phosphate, 35 mM TCEP). Raney nickel was prepared by adding 200 mg NaBH4 to a stirred solution of 1.2 g of nickel acetate in 6 ml of water. After 5 min, the Raney nickel was filtered, washed with 200 ml water and added to the solution of uH2B(A117C), 6. The reaction progress was followed by HPLC and ESI-MS. An identical amount of fresh Raney nickel was added after 6 h and the reaction was found to be complete at 8.5 h. (Note: much longer reaction times (over 24 h) led to a second desulphurization reaction on methionine.) The Raney nickel was pelleted by centrifugation and washed with 4 × 0.5 ml desulphurization buffer. The reaction supernatant and washes were combined, added to an equivalent volume of 50% HPLC buffer B, and purified using semi-preparative RP-HPLC with a 42–52% B gradient over 45 min, yielding 1.1 mg of uH2B, 7. uH2B was characterized by ESI-MS ((M + H)+ observed: 22,366 ± 4 Da; expected: 22,365 Da).
Recombinant histone preparation
Recombinantly expressed Xenopus histones H2A, H2B, H3 and H4 were prepared similarly to previously described33. The three N-terminal residues of H2B (PEP) were added to the Xenopus H2B expression plasmid31. The DNA encoding residues 1–125 of H2B was amplified by PCR using primers H2B-FP described above and H2B-RP (5′-CGGGATCCTTACTTGGCGCTGGTGTACTTG-3′) and the H2B plasmid described above as a template. After digestion with _Nde_I and _Bam_HI, the H2B gene was ligated into a similarly digested pET vector from the Xenopus H2A expression plasmid31. An H3(K79R) point mutation was generated with a QuikChange II XL kit (Stratagene) using primers H3K79R-FP (5′-GCTCAGGACTTCAGGACCGACCTGCGC-3′) and H3K79R-RP (5′-GCGCAGGTCGGTCCTGAAGTCCTGAGC-3′) and the Xenopus H3 expression plasmid31 as a template. The resulting histone expression plasmids were verified by DNA sequencing.
For protein expression, E. coli BL21(DE3)pLysS cells, transformed with the appropriate histone expression plasmid, were grown in 6 l 2 × TY media at 37 °C until mid-log phase, and protein expression was induced by the addition of 0.5 mM IPTG at 37 °C for 2–3 h. Cells were harvested and lysed as described above for proteins 2 and 5. The insoluble fractions of the bacterial lysates were washed twice with 20 ml wash buffer (20 mM Tris, 200 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, pH 7.5) and once with 20 ml of Triton wash buffer (20 mM Tris, 200 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 1% triton, pH 7.5). DMSO (1 mL) was then added to the pellets and after 15 min, 15–50 ml of extraction buffer (7 M guanidinium chloride, 20 mM Tris, 200 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, pH 7.5) was added and the suspension stirred for a further 15–30 min before clearing by centrifugation at 26,000_g_ for 30 min. The supernatants were purified using a Sephacryl S-200 column (approximately 1 l bed volume), eluting with extraction buffer. Purified fractions were combined, dialysed into water containing 2 mM DTT and lyophilized in aliquots suitable for octamer formation. H3(K79R) was further purified using process RP-HPLC with a 45–65% B gradient over 60 min. The identity of all purified histones was verified by ESI-MS (Supplementary Fig. 5).
Preparation of DNA for nucleosome and dinucleosome formation
A plasmid containing 12 copies of 177 bp of the 601 sequence34 was purified using a Qiagen Plasmid Giga kit. The 177-bp repeat was digested from the vector using Eco_RV sites flanking the segment. The desired segment was selectively precipitated by incrementally increasing the concentration of PEG-6000 from 4% to 8.5%, followed by centrifugation at 26,000_g to separate precipitated DNA. _Sca_I digestion and gel purification using a QIAquick Gel Extraction Kit afforded a 177-bp fragment of the 601 sequence, 1_177_601. PCR amplification of the central 147-bp region of 1_177_601 was accomplished using primers 147-FP (5′-CTGGAGAATCCCGGTGCCGAGG-3′) and 147-RP (5′-ACAGGATGTATATATCTGACACG-3′) and gel-purified 1_177_601 as a template. Purification of the PCR product using a QIAquick PCR purification kit yielded 1_147_601.
5′ and 3′ DNA fragments for dinucleosome formation were generated using PCR amplification as described above. The 5′ and 3′ fragments were designed to contain 3′ and 5′ _Dra_III restriction sites, respectively, allowing for the formation of complementary overhangs. The DNA segment for 5′ mononucleosome formation, 601-5′, was generated using the primers 147-FP and 5′-RP (5′-ATTGAGCACCCCGTGGGATCTTACATGCACAGGATG-3′) and 1_177_601 DNA as the template. The DNA segment for 3′ mononucleosome formation, 601-3′, was generated using 3′-FP (5′-ATTGAGCACGGGGTGCGGCCGCCCTGGAG-3′) and 147-RP primers. The PCR-amplified fragments (110 μg) were digested with _Dra_III (700 U) using NEB Buffer 3 (1 ml) for 6 h at 37 °C, followed by purification using a QIAquick PCR purification kit. When ligated, the resulting 601-5′ and 601-3′ sequences connect two identical 147-bp regions of the 601 sequence, separated by a 30-bp linker.
Nucleosome formation
Histone octamers were formed as previously described33. Briefly, individual histones were dissolved in unfolding buffer (7 M guanidinium chloride, 20 mM Tris, 10 mM DTT, pH 7.5) at approximately 4 mg ml−1. Histones were combined in equimolar amounts (combined protein ranged from 0.75 to 12 mg), and the solution was diluted to 1 mg ml−1 with unfolding buffer. The resulting mixture was dialysed into refolding buffer (2 M NaCl, 10 mM Tris, 1 mM EDTA, 1 mM DTT, pH 7.5) (three changes of 2 l each). Crude octamer assemblies were concentrated in Vivaspin 2 and 20 centricons (3–10 kDa molecular-weight cutoff (MWCO)) and purified using a Superdex 200 10/300 column, eluted with refolding buffer. Octamer quality was verified by 18% SDS–PAGE, followed by staining with Coomassie blue. Octamer samples were stored at −20 °C in 50% glycerol.
Mononucleosomes were formed using a previously described stepwise dilution procedure35. Briefly, octamers and DNA (1_147_601, 601-5′ or 601-3′) were combined in 10 μl high salt refolding buffer to a final concentration of 3 μM. After incubation at 37 °C for 15 min, 3.3 μl of dilution buffer 1 (10 mM HEPES, 1 mM EDTA, 0.5 mM PMSF, pH 7.9) was added and the temperature was dropped to 30 °C. Further dilutions of 6.7, 5, 3.6, 4.7, 6.7, 10, 30 and 20 μl, respectively, were then performed every 15 min. A final dilution with 100 μl of dilution buffer 2 (10 mM Tris, 1 mM EDTA, 0.1% NP-40, 5 mM DTT, 0.5 mM PMSF, 20% glycerol, pH 7.5) was performed. After an additional 15 min, the nucleosomes were concentrated using Vivaspin 500 centricons (3–10 kDa MWCO) at 4 °C. Nucleosome formation was verified by separation on a Criterion 5% TBE gel run in 0.5× TBE, followed by staining with ethidium bromide. The chromatinized plasmid was assembled using a previously described procedure36 and plasmid37. Micrococcal nuclease assays were used to verify equivalent chromatin reconstitution with uH2B and H2B (Supplementary Fig. 8).
Dinucleosome formation
Dinucleosome ligations were performed as previously described28. In a typical ligation reaction, 5′ and 3′ mononucleosomes (6.25 pmol) were combined in 200 μl of 1× ligation buffer (New England Biolabs) and 3,600 U of T4 ligase were added. After 1 h at room temperature, the ligated nucleosomes were concentrated using Vivaspin 500 centricons (3–10 kDa MWCO) and dinucleosome formation was verified using a 5% TBE gel as described above.
Preparation of Flag–hDot1L and hDot1L(1–416)
The cDNA for hDot1L was a gift from Y. Zhang. After modification to include an N-terminal Flag-epitope, the gene was subcloned into the pFASTBAC1 vector (Invitrogen) and the baculovirus was generated according to the manufacturer’s instructions. Sf9 cells were infected with the baculovirus and the resulting cell extracts were subjected to standard purification procedures as described below. Infected sf9 cells were collected, resuspended in lysis buffer (20 mM Tris, 500 mM NaCl, 4 mM MgCl2, 0.4 mM EDTA, 20% glycerol, 1 mM PMSF, 2 mM DTT, pH 7.9), and disrupted with a dounce homogenizer. After removal of cell debris by centrifugation, the supernatant was adjusted to 0.1% NP-40 and 300 mM NaCl by dilution with 20 mM Tris containing 10% glycerol. The resulting solution was incubated with M2-agarose beads (Sigma). After washing extensively (20 mM Tris-HCl, 150 mM NaCl, 2 mM MgCl2, 0.2 mM EDTA, 15% glycerol, 0.1% NP-40, 1 mM PMSF, 1 mM DTT, pH 7.9), the bound Flag–hDot1L was eluted with Flag peptide (Supplementary Fig. 6).
The catalytic domain of hDot1L encoding residues 1–416 was amplified from MSCN-hDot1L (a gift from Y. Zhang) using primers 416-FP (5′-CGGGATCCCATCACCATCATCATCACATGGGGGAGAAGCTGGAGCTG-3′) and 416-RP (5′-GGAATTCCTACTTCTTGGGGCGCCCGCGC-3′). The resulting sequence was digested with _Bam_HI and _Eco_RI and ligated into a similarly digested pGexTEV vector (pGex2T with the thrombin recognition sequence replaced with a TEV sequence). The resulting vector, pGexTEV-hDot1L416, was verified by DNA sequencing. E. coli BL21(DE3) cells transformed pGexTEV-hDot1L416, were grown in 6 l LB media at 37 °C until mid-log phase, and protein expression of the hDot1L(1–416) GST fusion protein was induced by the addition of 0.5 mM IPTG at 18 °C for 18 h (ref. 25). Cells were harvested and lysed as described above. Cleared cell lysates were continuously loaded onto a GST-Prep FF 16/10 column at 1 ml min−1 for 3 h. The column was washed with 200 ml of 20 mM Tris, 150 mM NaCl, 1 mM EDTA, 2 mM DTT, pH 7.5 and eluted with 200 ml of the same buffer containing 5 mM glutathione. Fractions deemed pure by SDS–PAGE were stored in 20% glycerol at −80 °C. Impure fractions were diluted with an equal volume of start buffer (20 mM Tris, 1 mM EDTA, 2 mM DTT, pH 7.5) and loaded onto a HiTrap SP HP 1-ml column. After washing with 10 ml start buffer, hDot1L(1–416) was eluted with a 10-ml linear gradient from start buffer to end buffer (20 mM Tris, 1 mM EDTA, 2 mM DTT, 1 M NaCl, pH 7.5). Pure fractions were stored at −80 °C in 20% glycerol (Supplementary Fig. 6).
Methyltransferase assays
Mono- and dinucleosomes (4.25 pmol) were combined with Flag–hDot1L (0.12 pmol) or hDot1L(1–416) (0.18 pmol) in 25 μl of assay buffer (20 mM Tris, 4 mM EDTA, 0.5 mM DTT, 1 mM PMSF, pH 7.9). To initiate the reaction, 3H-_S_-adenosyl methionine (SAM) (1 μCi, 14.25 pmol) was added and the reaction was allowed to proceed at 30 °C for 1 h. After 1 h, 20 μl of each sample were separated on a 5% TBE gel as described above and stained with ethidium bromide. After visualization, the gel was fixed with 25% isopropanol, 10% acetic acid in water, followed by a 30 min incubation with Amplify solution. Dried gels were visualized by fluorography. The remaining 5 μl of reaction mixture were spotted on Whatman p81 filter paper, washed 3 × 10 min with sodium carbonate, pH 9, and dried. Filter papers were added to 5 ml Ready Safe Liquid Scintillation Cocktail (Beckman Coulter). The samples were vortexed for 10 s and counted using a LKB Wallac 1209 RackBeta Primo Liquid scintillation counter.
Mass spectrometric analysis of methyltransferase assays
Methyltransferase assays were performed similar to that described above but on twice the scale using cold SAM (1 mM). Assays were incubated at 30 °C for 3 h before concentration in a Vivaspin 500 centricon (10 kDa MWCO), separated on a Criterion 15% Tris HCl gel, and stained with Coomassie blue. The Coomassie-stained H3 bands were excised from the gel and destained with 200 mM ammonium bicarbonate. Tryptic digestion was initiated with the addition of 25 ng μl−1 Sequence Grade Modified Trypsin (Promega) in ammonium bicarbonate buffer. The protein was digested for at least 16 h at 37 °C. The digestion products were mixed with 0.5 μl of 10 mg ml−1 α-cyano-4-hydroxysuccinnamic acid in 50% acetonitrile containing 0.1% TFA, and applied to a MALDI plate. MALDI mass spectra were recorded with a PerSeptive Voyager-DE STR MALDI time-of-flight mass spectrometer operated in the reflectron mode. Ion intensity of the tryptic fragment of H3 containing residues 40–49 was used to scale the intensities of the methylated peptides for comparison of separate samples.
Electrophoretic mobility-shift assay
Mononucleosomes (3.0 pmol), hDot1L (1–416) (0–4.5 pmol) and cold SAM (10 nmol) were combined in 30 μl of assay buffer. After 1 h at 30 °C, samples were separated on a 5% TBE gel as described above and visualized with ethidium bromide staining. Proteins were transferred to a PVDF membrane and a western blot was performed against GST (goat anti-GST used at 1:1,000).
For radiolabelled gel-shift competition experiments, 1_147_601 was endlabelled with 32P using 32P-ATP (Perkin Elmer) and polynucleotide kinase. Radiolabelled nucleosomes were prepared as described above at 2 μM starting concentrations. Radiolabelled nucleosomes (0.64 pmol), hDot1L(1–416) (3.2 pmol) and cold nucleosomes (0–3.2 pmol) were incubated and separated as described above. Dried gels were imaged by phosphorimaging on a Typhoon 8400 (GE Healthcare).
Supplementary Material
Supplemental_material
Acknowledgments
We acknowledge H. Deng and J. Fernandez at The Rockefeller University Proteomics Resource Center for mass spectrometric analysis of methylated peptides. We thank T. J. Richmond for donating the 12_177_601 plasmid. We thank C. D. Allis for contributing the Xenopus histone plasmids for recombinant histone expression. We thank Y. Zhang for donating a plasmid containing hDot1L. We thank B. R. Rosenberg for assistance with phosphorimaging. We thank C. D. Allis, J. Tanny, and K. P. Chiang for discussions. This work was funded by the US National Institutes of Health. R.K.M. was supported by National Institutes of Health MSTP grant GM07739. J.K. was supported by the LLS SCOR grant.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Author Contributions R.K.M., J.K. and C.C. did the experimental work; all authors performed project planning, data analysis and manuscript preparation.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 3.West MHP, Bonner WM. Histone 2B can be modified by the attachment of ubiquitin. Nucleic Acids Res. 1980;8:4671–4680. doi: 10.1093/nar/8.20.4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 5.Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Zhu B, et al. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 7.Ng HH, Xu RM, Zhang Y, Struhl K. Ubiquitination of histone H2B by Rad6 is required for efficient Dot1-mediated methylation of histone H3 lysine 79. J Biol Chem. 2002;277:34655–34657. doi: 10.1074/jbc.C200433200. [DOI] [PubMed] [Google Scholar]
- 8.Briggs SD, et al. Trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- 9.Feng Q, et al. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 10.Ng HH, et al. Lysine methylation within the globular domain of histone H3 by Dot1 is important for telomeric silencing and Sir protein association. Genes Dev. 2002;16:1518–1527. doi: 10.1101/gad.1001502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Leeuwen F, Gafken PR, Gottschling DE. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell. 2002;109:745–756. doi: 10.1016/s0092-8674(02)00759-6. [DOI] [PubMed] [Google Scholar]
- 12.Henry KW, Berger SL. Trans-tail histone modifications: wedge or bridge? Nature Struct Biol. 2002;9:565–566. doi: 10.1038/nsb0802-565. [DOI] [PubMed] [Google Scholar]
- 13.Sun ZW, Allis CD. Ubiquitylation of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 14.Krogan NJ, et al. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- 15.Ezhkova E, Tansey WP. Proteasomal ATPases link ubiquitylation of histone H2B to methylation of H3. Mol Cell. 2004;13:435–442. doi: 10.1016/s1097-2765(04)00026-7. [DOI] [PubMed] [Google Scholar]
- 16.Lee JS, et al. Histone crosstalk between H2B monoubiquitylation and H3 methylation mediated by COMPASS. Cell. 2007;131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 17.Hwang WW, et al. A conserved RING finger protein required for histone H2B monoubiquitination and cell size control. Mol Cell. 2003;11:251–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 18.Wood A, et al. Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol Cell. 2003;11:267–274. doi: 10.1016/s1097-2765(02)00802-x. [DOI] [PubMed] [Google Scholar]
- 19.Muralidharan V, Muir TW. Protein ligation: an enabling technology for the biophysical analysis of proteins. Nature Methods. 2006;3:429–438. doi: 10.1038/nmeth886. [DOI] [PubMed] [Google Scholar]
- 20.Chatterjee C, McGinty RK, Pellois JP, Muir TW. Auxiliary-mediated site-specific peptide ubiquitylation. Angew Chem Int Edn Engl. 2007;46:2814–2818. doi: 10.1002/anie.200605155. [DOI] [PubMed] [Google Scholar]
- 21.Yan LZ, Dawson PE. Synthesis of peptides and proteins without cysteine residues by native chemical ligation combined with desulfurization. J Am Chem Soc. 2001;123:526–533. doi: 10.1021/ja003265m. [DOI] [PubMed] [Google Scholar]
- 22.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 23.Sawada K, et al. Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase. J Biol Chem. 2004;279:43296–43306. doi: 10.1074/jbc.M405902200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 25.Min J, Feng Q, Li Z, Zhang Y, Xu RM. Structure of the catalytic domain of human Dot1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 26.Shahbazian MD, Zhang K, Grunstein M. Histone H2B ubiquitylation controls processive methylation but not monomethylation by Dot1 and Set1. Mol Cell. 2005;19:271–277. doi: 10.1016/j.molcel.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Garcia BA, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- 28.Zheng C, Hayes JJ. Intra- and inter-nucleosomalprotein–DNA interactionsofthe core histone tail domains in a model system. J Biol Chem. 2003;26:24217–24224. doi: 10.1074/jbc.M302817200. [DOI] [PubMed] [Google Scholar]
- 29.Wang SS, et al. Facile synthesis of amino acid and peptide esters under mild conditions via cesium salts. J Org Chem. 1977;42:1286–1290. doi: 10.1021/jo00428a004. [DOI] [PubMed] [Google Scholar]
- 30.Smith AB, Savinov SN, Manjappara UV, Chaiken IM. Peptide-small molecule hybrids via orthogonal deprotection-chemoselective conjugation to cysteine-anchored scaffolds. Org Lett. 2002;4:4041–4044. doi: 10.1021/ol026736d. [DOI] [PubMed] [Google Scholar]
- 31.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 32.Pentelute BL, Kent SB. Selective desulfurization of cysteine in the presence of Cys(Acm) in polypeptides obtained by native chemical ligation. Org Lett. 2007;9:687–690. doi: 10.1021/ol0630144. [DOI] [PubMed] [Google Scholar]
- 33.Luger K, Rechsteiner TJ, Richmond TJ. Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol. 1999;119:1–16. doi: 10.1385/1-59259-681-9:1. [DOI] [PubMed] [Google Scholar]
- 34.Dorigo B, Schalch T, Bystricky K, Richmond TJ. Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 35.Owen-Hughes T, et al. Analysis of nucleosome disruption by ATP-driven chromatin remodelling complexes. Methods Mol Biol. 1999;119:319–331. doi: 10.1385/1-59259-681-9:319. [DOI] [PubMed] [Google Scholar]
- 36.Ito T, et al. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental_material