Environmental epigenomics and disease susceptibility (original) (raw)
. Author manuscript; available in PMC: 2018 May 8.
Published in final edited form as: Nat Rev Genet. 2007 Apr;8(4):253–262. doi: 10.1038/nrg2045
Abstract
Epidemiological evidence increasingly suggests that environmental exposures early in development have a role in susceptibility to disease in later life. In addition, some of these environmental effects seem to be passed on through subsequent generations. Epigenetic modifications provide a plausible link between the environment and alterations in gene expression that might lead to disease phenotypes. An increasing body of evidence from animal studies supports the role of environmental epigenetics in disease susceptibility. Furthermore, recent studies have demonstrated for the first time that heritable environmentally induced epigenetic modifications underlie reversible transgenerational alterations in phenotype. Methods are now becoming available to investigate the relevance of these phenomena to human disease.
Traditional research on the combined effects of the environment and genetics on individual variation in disease risk examines the relationship between disease susceptibility, environmental exposures and germline mutations in the coding and promoter regions of genes. Such research efforts have highlighted the importance of genotype in human diseases. However, it is now becoming clear that a full understanding of environmental interactions with the genome will require that epigenetic mechanisms are also considered. Human epidemiological studies have provided evidence that prenatal and early postnatal environmental factors influence the adult risk of developing various chronic diseases, such as cancer, cardiovascular disease, diabetes, obesity (for reviews, see REFS 1–4) and even behavioural disorders such as schizophrenia5,6. One general mechanism by which prenatal and postnatal exposures could be linked to phenotypic changes later in life is the alteration of epigenetic marks, which have a central role in determining the functional output of the information that is stored in the genome.
Although the term ‘epigenetics’ literally means ‘above the genetics,’ it is now generally used to refer to changes in gene expression that take place without a change in the DNA sequence (BOX 1). Epigenetic changes encompass an array of molecular modifications to both DNA and chromatin7–10, the most extensively investigated of which are DNA methylation, which takes place at the carbon-5 position of cytosine in CpG dinucleotides, and changes to the chromatin packaging of DNA by post-translational histone modifications7–9. Other epigenetic mechanisms of gene-expression control include regulation by non-coding RNAs, such as microRNAs, and mechanisms that control the higher-level organization of chromatin within the nucleus, which have a range of effects on gene expression. Two of the most comprehensively studied epigenetically regulated phenomena in mammals are X-chromosome inactivation (for reviews, see REFS 11,12) and genomic imprinting (for reviews, see REFS 13–16). Epigenetic regulation is also involved in tissue-specific gene expression and in the silencing of repetitive (transposable) elements, inhibiting their replication and transposition, and therefore preventing insertional mutagenesis17.
Box 1. A brief history of epigenetics.
The developmental biologist Conrad Waddington107 first defined ‘epigenetics’ in the 1940s as “…the interactions of genes with their environment which bring the phenotype into being.” Holliday and Pugh108 proposed in 1975 that covalent chemical DNA modifications, including methylation of cytosine-guanine (CpG) dinucleotides, were the molecular mechanisms behind Conrad’s hypothesis. The further revelations that X inactivation in mammals and genomic imprinting are regulated by epigenetic mechanisms highlighted the heritable nature of epigenetic gene-regulation mechanisms109,110. Therefore, in the 1990s, epigenetics was described as the study of changes in gene expression that occur not by changing the DNA sequence, but by modifying DNA methylation and remodelling chromatin111.
The genomics revolution inspired the investigation of global, rather than local, gene analyses, and the term ‘epigenomics’ was coined as the study of the “…effects of chromatin structure, including the higher order of chromatin folding and attachment to the nuclear matrix, packaging of DNA around nucleosomes, covalent modifications of histone tails (acetylation, methylation, phosphorylation, ubiquitination), and DNA methylation.”112 The resistance of some gene loci to methylation reprogramming during embryogenesis revealed the possibility that epigenetic modifications are inherited not only during somatic-cell division, but also in the subsequent generation31–34.
Importantly, epigenetic changes can be inherited mitotically in somatic cells, providing a potential mechanism by which environmental effects on the epigenome can have long-term effects on gene expression. In support of the importance of such a mechanism, increasing evidence from animal studies indicates that prenatal and early postnatal environmental factors — including nutritional supplements18–22, xenobiotic chemicals23–25, behavioural cues26,27, reproductive factors28,29 and low-dose radiation30 — can result in altered epigenetic programming and subsequent changes in the risk of developing disease. In addition, epigenetic alterations might also be inherited transgenerationally, thereby potentially affecting the health of future generations — a theory for which there is also increasing evidence25,31–35. The results of these animal and human studies support the hypothesis that is referred to as the fetal basis or developmental origins of adult-onset disease. This theory posits the intriguing idea that the evolution of developmental plasticity, which enables an organism to adapt to environmental signals during early life, can also increase the risk of developing chronic diseases when there is a mismatch between the perceived environment and that which is encountered in adulthood. Developmental plasticity is evident when environmental exposure produces a broad range of adult phenotypes from a single genotype by epigenetically altering gene expression18,19,36.
Here we bring together the mounting evidence that environmental influences early in development are linked to disease phenotypes through modifications of the epigenome. We begin by discussing the epigenetic marks that are known or considered likely to be susceptible to environmental influence. In particular, we focus on metastable epialleles, for which studies in mice have provided strong evidence of an effect of prenatal environmental exposures on postnatal phenotypes. We then discuss the increasing evidence that supports the trans-generational inheritance of environmentally induced epigenetic changes. We conclude the Review by looking ahead to how these studies in animal models could be extended to determine the importance of environmental epigenetics in human disease susceptibility. Such studies might ultimately lead to new diagnostic, prognostic and therapeutic strategies.
Epigenetic targets of the environment
Environmental exposures to nutritional, chemical and physical factors have the potential to alter gene expression and modify adult disease susceptibility in various ways through changes in the epigenome. Three genomic targets that are likely to be susceptible to gene-expression changes owing to environmental perturbations of epigenetic marks are the promoter regions of some housekeeping genes, transposable elements that lie adjacent to genes with metastable epialleles, and regulatory elements of imprinted genes. These genomic targets contain regions that are rich in CpG dinucleotide sequences, which are normally unmethylated, methylated or differentially methylated, respectively, with the methylation status, and in some cases with the status of histone modifications in the same region, determining levels of gene expression.
Both DNA methylation and histone modifications are markedly altered in the promoter regions of tumour-suppressor genes and oncogenes in human cancer. Furthermore, changes in the epigenome that alter the expression of the tumour-suppressor mismatch repair genes MLH1 and MSH2 have been shown to be inherited through the germ line37,38. The importance of epigenetics in the aetiology of cancer has been discussed extensively elsewhere (for reviews, see REFS 39–41). Here we focus on imprinted genes and genes with metastable epialleles, which have the potential to link the environment, through epigenetic changes, to early developmental influences on adult susceptibility to chronic diseases and behavioural disorders.
Genes with metastable epialleles
Metastable epialleles are defined as loci that can be epigenetically modified in a variable and reversible manner, such that a distribution of phenotypes occurs from genetically identical cells. Currently, only a few genes with metastable epialleles have been identified, including the mouse Avy (viable yellow agouti)42, AxinFu (axin fused)36, and CabpIAP (CDK5 activator binding protein–intra-cisternal A particle (IAP))43 genes.
Of these examples, the Avy allele of the murine agouti gene has been the most thoroughly characterized. In this case, allelic expression is correlated with the epigenetic status of a transposon that is associated with the promoter region of the gene19,42 (FIG. 1a). Most transposable elements are silenced by CpG methylation; however, the epigenetic status of a subset of transposable elements is metastable, and varies in a stochastic manner from hypomethylated to hypermethylated44. This variable epigenetic status can affect the expression of neighbouring genes, causing epigenetic mosaicism between cells and phenotypic variability among cells in genetically identical individuals19,45 (FIGS 1,2).
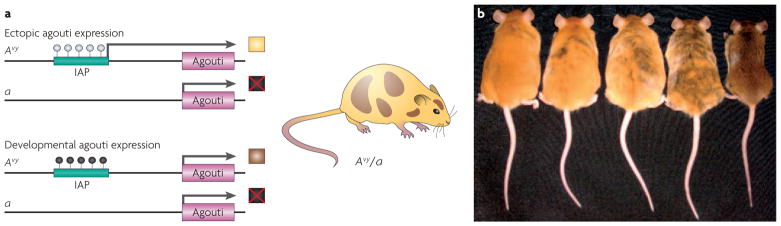
Figure 1. Epigenetic regulation of metastable epialleles.
a | Epigenetic regulation of the agouti gene in Avy/a mice. White-filled circles indicate unmethylated CpG sites and black-filled circles indicate methylated CpG sites. Phaeomelanin (the product of the agouti gene) is not produced from the a allele because the agouti gene is mutated (shown as a box marked with a red cross). Two potential epigenetic states of the Avy allele can occur within cells of Avy/a mice. The IAP (intracisternal A particle) that lies upstream of the agouti gene can remain unmethylated, allowing ectopic expression of the gene from the IAP and resulting in a yellow coat colour (top). Alternatively, the IAP can be methylated, so that the gene is expressed under its normal developmental controls, leading to a brown coat colour (bottom). If the IAP methylation event occurs later in development and does not affect all embryonic cells, the offspring will have a mottled appearance (illustrated on the right). b | Genetically identical week 15 Avy/a mouse littermates are shown, representing five coat-colour phenotypes. Mice that are predominately yellow are also clearly more obese than the brown mice. Part b reproduced with permission from REF. 20 © (2006) National Institute of Environmental Health Sciences.
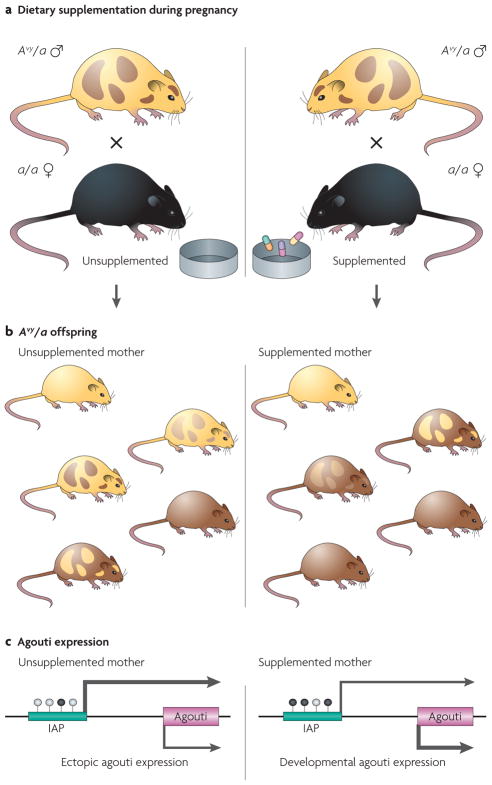
Figure 2. Effect of maternal dietary supplementation on the phenotype and epigenotype of Avy/a offspring.
a | Dietary supplementation of female mice during pregnancy. The diets of female a/a mice are supplemented with methyl-donating substances (that is, folic acid, choline, vitamin B12 and betaine)19 or the phytoestrogen genistein20 2 weeks before mating with male Avy/a agouti mice, and throughout pregnancy and lactation. b | Maternal dietary supplementation and coat-colour distribution in Avy/a offspring. The coat colour is primarily yellow in the offspring that are born to unsupplemented mothers, whereas it is mainly brown in the offspring from mothers that were supplemented with methyl-donating compounds or genistein. Approximately 50% of the offspring from these matings are black (a/a) but, as they do not contain an Avy allele, they are not shown here. c | DNA methylation and agouti gene expression. Maternal hypermethylating dietary supplementation shifts the average coat-colour distribution of the offspring to brown by causing an IAP (intracisternal A particle, shown as a green bar) upstream of the agouti gene to be more methylated on average than in offspring that are born to mothers fed an unsupplemented diet. Arrow size is directly proportional to the amount of ectopic and developmental agouti gene expression. White-filled circles indicate unmethylated CpG sites and black-filled circles indicate methylated CpG sites.
Agouti encodes a paracrine signalling molecule that promotes follicular melanocytes to produce yellow phaeomelanin rather than black eumelanin pigment. Transcription is normally initiated from a developmentally regulated hair-cycle-specific promoter in exon 2 of the agouti (A) allele. Transcription of the A allele normally occurs only in the skin, where transient expression in hair follicles during a specific stage of hair growth results in a subapical yellow band on each black hair shaft, causing the brown (agouti) coat colour of wild-type mice42. The Avy allele is the result of the insertion of a murine IAP transposable element about 100 kb upstream of the transcriptional start site of the agouti gene19,42 (FIG. 1a). A cryptic promoter in the proximal end of the Avy IAP promotes constitutive ectopic agouti transcription, leading to yellow fur, diabetes, obesity and tumorigenesis31,46. The degree of IAP methylation varies dramatically among individual isogenic Avy/a mice, causing a wide distribution in coat colour, ranging from yellow (unmethylated) to brown (methylated) (FIG. 1b).
Maternal dietary methyl-donor supplementation of mice with folic acid, vitamin B12, choline and betaine shifts the coat colour distribution of the offspring towards the brown pseudoagouti phenotype18,19,47 (FIGS 1,2). This methyl-donor-induced shift in coat-colour distribution was shown to result from an increase in DNA methylation at CpG sites in the upstream IAP transposable element19. Thus, for the first time, the effect of a mother’s diet during pregnancy on the adult phenotype of her offspring was directly linked to DNA-methylation changes in the epigenome. This observation marked the advent of studying the role of environmental epigenomic changes during early development in the aetiology of adult diseases.
Methylation profiles at these IAP CpG sites are highly correlated in tissues that are derived from the ectodermal (brain and tail), endodermal (liver) and mesodermal (kidney) lineages, providing evidence that methylation profiles in response to dietary supplementation are established before embryonic stem cell differentiation19,20. These epigenetic changes are also stable, as the amount of methylation in tissues from d21 animals is similar to that in tissues from d100 animals. The effects of maternal methyl-donor supplementation on coat-colour distribution in Avy offspring can also be inherited in the F2 generation by germline epigenetic modifications48.
Maternal dietary exposure to the phytoestrogen genistein during gestation also shifts the coat-colour distribution of viable yellow Avy/a offspring towards brown, owing to hypermethylation of the IAP in the _A_vy allele20. Furthermore, the genistein-induced hypermethylation protects the Avy/a offspring from obesity in adulthood. These findings are particularly interesting because genistein, when given at a level that is comparable to that consumed by humans with high soy diets, increases DNA methylation even though it is not a methyl-donating compound; the mechanism of its action at the _A_vy allele is unknown. These results suggest the interesting possibility that hypermethylating dietary supplements could reduce the effect of environmental toxicants that cause DNA hypomethylation, thereby protecting the epigenome from their deleterious effects.
Imprinted genes
The vast majority of autosomal genes are expressed from both parental alleles; however, approximately 1% of autosomal genes are imprinted, with expression from only one parental allele (for reviews, see REFS 13–16)(FIG. 3). Genomic imprinting is a non-Mendelian, germline-inherited, epigenetic form of gene regulation that involves heritable DNA methylation and histone modifications13,14. Expression of the single functional allele of an imprinted gene in both male and female offspring is parent-of-origin dependent, with the imprinted epigenetic marks being established in the parental gametes (FIG. 4). So, allelic expression of an imprinted gene in the present generation depends on the parental environment in which it resided in the previous generation. On the basis of the knowledge that imprinting defects cause several disease phenotypes, and the likely susceptibility of the methylation marks that underlie imprinting to alterations at certain stages of development, we argue here that imprinted loci are likely to be targets of disease-causing environmentally induced epigenetic abnormalities.
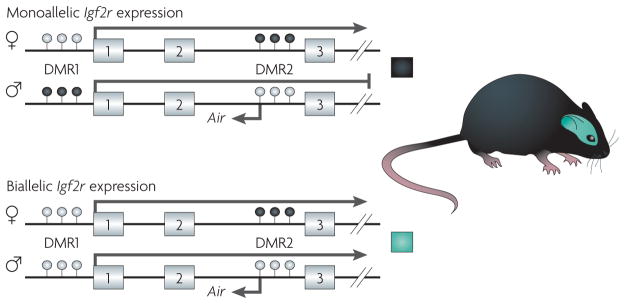
Figure 3. Epigenetic regulation of imprinted alleles.
Epigenetic regulation of imprinting in the murine Igf2r (insulin-like growth factor 2 receptor). Exons 1–3 of the 48-exon Igf2r gene are shown. Igf2r is imprinted in murine peripheral tissues (shown in the upper panel), and expressed from the maternal allele but not from the paternal allele52. By contrast, Igf2r is biallelically expressed in neuronal cells in the brain (shown in the lower panel)113. The mechanism of imprinting at this locus is complex, but involves two differentially methylated regions, DMR1 and DMR2, and the expression status of the Air antisense transcript. DMR1 is differentially methylated during early development in peripheral tissues, but not in neuronal cells113. In the case of DMR2, the methylation status is inherited through the germ line. Monoallelic expression of Igf2r is not only tissue-dependent113, but also species-dependent53,57. See REF. 114 for more information on the role of DNA methylation and histone modification in regulating Igf2r expression in mice and humans. White-filled circles indicate unmethylated CpG sites and black-filled circles indicate methylated CpG sites.
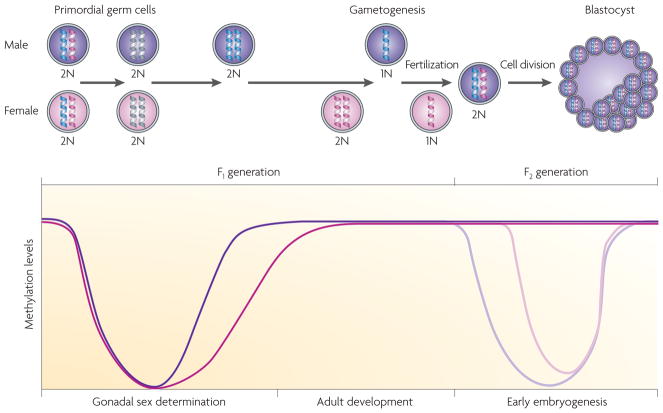
Figure 4. Alterations in methylation status during development.
During embryonic development and gonadal sex determination, primordial germ cells undergo genome-wide demethylation, which erases previous parental-specific methylation marks that regulate imprinted gene expression. In the male (coloured purple) germ line, paternal methylation marks in imprinted genes are laid down in developing gonocytes that will develop into spermatogonia. The female (coloured pink) germ line establishes maternal methylation marks in imprinted genes at a later stage. After fertilization, the paternal genome is actively demethylated (indicated by the lighter purple line in the graph), whereas the maternal genome undergoes passive demethylation (indicated by the lighter pink line in the graph)115. Genome-wide remethylation occurs on both parental genomes before implantation. However, imprinted genes maintain their methylation marks throughout this reprogramming, allowing for the inheritance of parental-specific monoallelic expression in somatic tissues throughout adulthood.
The most widely debated theory of why imprinting evolved, the ‘parental conflict hypothesis’, views imprinting not as a beneficial adaptation of the species, but rather as a deleterious consequence of a genetic battle between the sexes to control the amount of resources that are extracted from the mother by her offspring49,50. This ancestral genetic conflict is purported to have occurred because of a reproductive scenario involving polygamy, viviparity and substantial maternal investments in the offspring, in the absence of a similar level of investment by the father. This theory predicts that imprinted genes that increase the extraction of resources from the mother by the offspring will be paternally expressed (that is, maternally silenced). By contrast, imprinted genes that decrease the offspring’s extraction of resources from the mother are predicted to be maternally expressed (that is, paternally silenced). For example, in mice, the growth factor gene Igf2 (insulin-like growth factor 2), is paternally expressed51, whereas the growth inhibitor, Igf2r (insulin-like growth factor 2 receptor), is maternally expressed52.
Although genomic imprinting seems to have first evolved in mammals with the development of the placenta and the advent of viviparity53, the process of imprinted-gene formation is dynamic, allowing for both its establishment and loss during evolution. For example, NNAT (neuronatin), MEG3 (maternally expressed gene 3), DLK1 (delta-like 1 homologue) and CDKN1C (cyclin-dependent kinase inhibitor 1C) are imprinted in eutherians but not in marsupials54–56. By contrast, IGF2R imprinting, which was initiated 180 million years ago, was subsequently lost approximately 75 million years ago with the evolution of the near primates that ultimately gave rise to humans53,57. Consequently, IGF2R is imprinted and expressed from only the maternal allele in mice, but is biallelically expressed in humans. Because the IGF2R functions as a tumour suppressor58, the incidence of cancers that result from its inactivation is predicted to be higher in mice than humans. This has important human-health ramifications, because the mouse is often used as a surrogate for humans in toxicological risk assessments.
Several developmental disorders are associated with specific imprinted regions and genes — for example, Angelman syndrome (AS), Prader–Willi syndrome (PWS), and Beckwith–Wiedemann syndrome (BWS) (for reviews, see REFS 59,60). Germline or somatic-cell deregulation of imprinting control during early development has an important causative role in these disorders. Badcock and Crespi61 recently put forth the intriguing idea that human behavioural disorders such as autism also result from imbalanced genomic imprinting during brain development. In addition, numerous linkage studies of other complex disorders such as bipolar disorder, Silver–Russell syndrome, Tourette syndrome and schizophrenia show a parent-of-origin inheritance preference62, suggesting that as yet unidentified imprinted genes that are involved in human diseases reside outside known imprinted gene clusters.
Imprinted-gene deregulation or mutation can also occur in somatic cells in adulthood, thereby increasing cancer risk (for reviews, see REFS 63,64). Furthermore, only a single mutational or epigenetic event is required to completely inactivate an imprinted tumour-suppressor gene because one allele is already functionally inactivated by imprinting. Consequently, the silenced allele of an imprinted gene has been equated to the ‘first hit’, as proposed by Knudson65 in his two-step model for carcinogenesis. Imprinted oncogenes can also be inappropriately overexpressed in somatic cells through loss of imprinting (LOI; for reviews, see REFS 63,64). Importantly, Feinberg and colleagues66 demonstrated that lymphocytes in approximately 10% of the human population have LOI at the IGF2 locus. Moreover, biallelic expression in peripheral lymphocytes is strongly correlated with IGF2 LOI in normal colonic mucosa and a personal history of colorectal cancer. The mechanism for this apparent systemic epigenetic alteration is currently unknown; however, IGF2 LOI must be either inherited or acquired early in life in a subset of individuals67,68.
The functional haploid state of imprinted genes eliminates the protection that diploidy normally affords against the deleterious effects of recessive mutations. Furthermore, the DNA-methylation marks that are required for parental-specific expression of imprinted genes are particularly vulnerable to epigenetic deregulation by environmental agents during the early stage of gametogenesis, and the genome-wide demethylation and remethylation that occurs in the zygote soon after fertilization (FIG. 4). Thus, imprinted genes are candidate susceptibility loci for environmentally induced diseases with parental-inheritance bias.
Epigenetics and transgenerational effects
Germline transmission
Transgenerational inheritance involves the transmission of a biological trait to subsequent generations through the germ line. An epigenetic transgenerational effect requires that epigenetic modifications in the germ line cause the inheritance of a phenotype. Because environmental factors can alter the epigenome, their ability to influence disease risk might involve epigenetic transgenerational inheritance.
For the transgenerational inheritance of environmental effects on the epigenome to be considered as a plausible mechanism for the cause of phenotypic changes, these changes must be maintained in at least the F3 generation when an embryonic exposure is involved25,34,69 (FIG. 5). When an F0 gestating female is exposed, both the F1 embryo and the F2 generation germ line are also directly exposed. Therefore, disease phenotypes in the F1 and F2 generations might still be due to the toxicology of direct exposure to the environmental factor. Similarly, after postnatal or adult exposure to environmental toxicants70–72, because the F1 generation germ line is directly exposed, the observed phenotypes in these generations cannot unequivocally demonstrate a transgenerational phenomenon, and the F2 generation must be assessed.
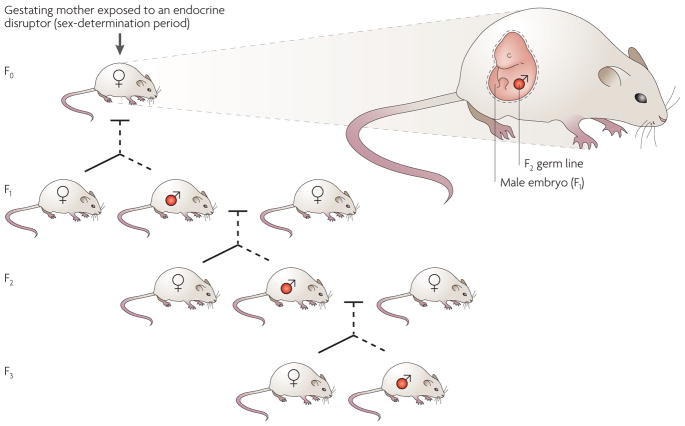
Figure 5. Germline transmission of epigenetically regulated transgenerational phenotypes.
In a gestating mother, there is multiple-generation exposure of the F0 female, the F1 embryo and the F2 generation germ line to environmental factors. The transgenerational transmission of disease phenotypes through the male germ line (labelled red) is indicated. Both male and female offspring develop disease, but the transgenerational phenotype is transmitted only paternally after exposure to vinclozolin96.
Exposure to several environmental factors during embryonic development has been shown to influence disease susceptibility in the F1 generation; these factors include heavy metals causing cancer73, abnormal nutrition (for example, caloric restriction) causing diabetic and uterine defects35,74,75, chemical exposure (for example, benzo(a)pyrene) causing brain and endocrine defects76,77, and endocrine disruptors causing reproductive and endocrine defects78–82. These environmental effects on the F1 germ line have been observed in species ranging from insects to mammals83–88. Both human35 and animal76,86,89–93 studies demonstrate that an embryonic exposure (F0 generation mother) can produce an F2 generation phenotype (FIG. 5). For example, nutritional deficiency during gestation affects the incidence of diabetes and growth defects in the F2 generation rodents93,94. Several environmental chemical exposures also affect the F2 generation (including benzo(a)pyrene90,95, orthoaminoasotoluol92 and dioxin70), and the endocrine disruptor diethylstilbestrol (DES) has a negative impact on organ (for example, mammary gland) and reproductive tract development and function in the F2 generation71. However, in all these cases, further analysis involving the F3 generation is needed to eliminate the potential problem of direct F2 generation germline exposure.
Several studies have now shown marked effects of environmental toxicants on the F3 generation through germline alterations in the epigenome34,95–98. The endocrine disruptor vinclozolin, which is an anti-androgenic compound, induces transgenerational pathogenesis in rats, resulting in spermatogenic defects, male infertility, breast cancer, kidney disease, prostate disease and immune abnormalities at frequencies ranging from 20% to 90% (REFS 34,96). These transgenerational disease phenotypes are transmitted to the majority of progeny for four generations and, although the transgenerational effect is transmitted through only the male germ line, both males and females are affected96.
The frequency and reproducibility of the vinclozolin-induced transgenerational pathologies, and the finding that most of them occur in adulthood, indicate that genetic DNA-sequence mutations are not the most likely cause34,96. The frequency of a DNA-sequence mutation, even with ionizing radiation, is normally less than 0.01%, and ranges from only 1% to 5% for hot-spot mutations72,99. The most parsimonious explanation for these findings is therefore that the vinclozolin-induced transgenerational phenotypes result from the epigenetic reprogramming of the male germ line at the stage of gonadal sex determination34,97 (FIGS 4,6).
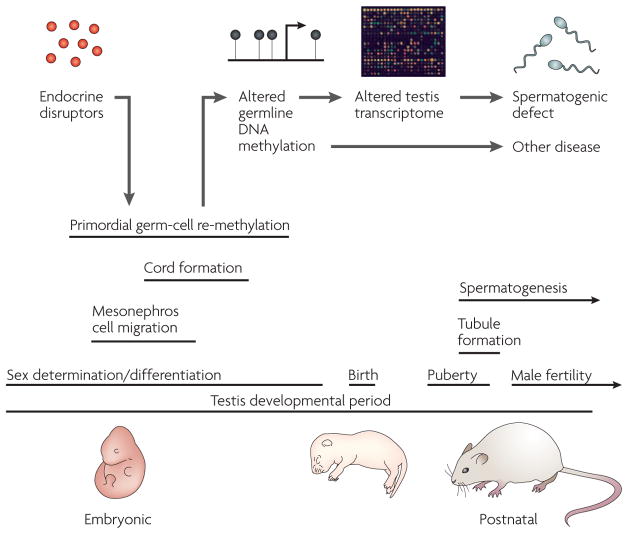
Figure 6. A model for endocrine-disruptor-induced epigenetic transgenerational disease.
Endocrine-disruptor action reprogrammes the epigenome of the developing germ cell during embryonic sex determination, leading to genes and other DNA sequences with altered DNA methylation97. These changes are proposed to alter the transcriptomes of the testis and other organs, thereby promoting adult pathologies, some of which are inherited transgenerationally. Epigenetic mechanisms might therefore have a role in the induction of adult-onset disease through environmental exposures early in development.
The ability of a postnatal environmental exposure to promote a transgenerationally inherited phenotype requires that the process of gametogenesis (that is, germ-cell differentiation in the adult) is altered in such a way as to reprogramme the germ line (FIG. 6). Epigenetic reprogramming of the germ line is known to take place during gametogenesis98,100–102, but the mechanisms of inheritance of epigenetically directed transgenerational phenotypes have yet to be defined. To experimentally address this fundamentally important issue, methylation-sensitive restriction-enzyme analysis followed by PCR amplification have been used to identify a set of genes and other DNA sequences with altered DNA methylation that are potentially epigenetically reprogrammed in the male germ line as a result of the exposure to vinclozolin, and that are associated with the transgenerational inheritance of pathological phenotypes34,96. Whether the genes that were identified are causal factors or simply markers of the transgenerational epigenetic phenotype remains to be determined.
Behavioural transmission
Not all epigenetic multi-generational effects are transmitted through the germ line. Elegant studies in mice have demonstrated that the generation-to-generation acquisition of the nurturing behaviours of pup licking and grooming and arch-back nursing are not germline inherited but, rather, are passed on to the offspring directly from the mother during the first week of postnatal life26. The adult female offspring of dams that show an increase in these maternal nurturing behaviours exhibit reduced fearfulness and more modest hypothalamic–pituitary–adrenal (HPA)-axis responses to stress.
Recently, these maternal programming effects have been shown to involve DNA methylation and histone modifications of the NGFIA (nerve growth factor inducible protein A) transcription-factor-binding motif that is present in the exon 17 promoter of the brain-specific GR (glucocorticoid receptor)26. These epigenetic changes are correlated with alterations in GR expression, and HPA and behavioural responses to stress. Moreover, infusion of the histone deacetylase inhibitor trichostatin A (TSA) or the essential amino acid L-methionine into the brain ventricles in adulthood reverses the epigenetic modifications in the hippocampus, and the GR expression and HPA response in the female offspring of dams with low and high maternal nurturing ability, respectively26,27,103. Together, these findings indicate that early postnatal life experiences can modify behaviour by altering the epigenome, and that the inherent plasticity of the epigenome potentially allows for reversal in adulthood — an important finding in terms of possible therapeutic strategies.
Identification of epigenetically labile genes
Environmental epigenomics is a newly emerging field of investigation, and many fundamental questions remain to be answered (BOX 2). A question of central importance concerns which human genes are likely to be involved in enhanced disease susceptibility when they are epigenetically deregulated by environmental factors. To address this question, novel techniques are being used to identify subsets of epigenetically labile genes in both animals and humans.
Box 2. Key questions for environmental epigenetics.
- Which human genes result in enhanced disease susceptibility when they are epigenetically deregulated by environmental factors?
- What environmental factors deleteriously alter the epigenome, and at what doses?
- What role does the epigenome have in reproduction, development and disease aetiology?
- Are there nutritional supplements that can reduce the harmful effects of chemical and physical factors on the epigenome?
- Can epigenetic biomarkers be identified that will allow for the detection of early-stage diseases?
- Can detection technologies be developed that will allow for a quick and accurate genome-wide assessment of the epigenome?
- Can epigenetics be integrated into systems biology as an important regulatory mechanism?
Genes with metastable epialleles
The repertoire of genes with metastable epialleles is anticipated to be species dependent, because such alleles depend on the presence of nearby transposable elements and the patterns of insertion of these elements vary from species to species. Although the human genome is riddled with transposable elements104,105, it is currently unknown whether any of them regulate gene expression in a manner that is comparable to that seen at the agouti locus in Avy mice19,20 and, if so, whether they affect human disease susceptibility. One way to address this question is to look for potential metastable epialleles in humans with a methylation and expression profile that is similar to that of the agouti gene, which we refer to as the ‘agouti expression fingerprint’.
The agouti expression fingerprint is defined by a large variability in gene expression between individuals, combined with a low variability in gene expression between tissues from the three germ layers in an individual19,20. The ratio of these two gene-expression variances is expected to be large for a gene with an expression pattern that is epigenetically established in a manner that is comparable to that of the agouti gene in Avy mice. Genome-wide expression chips should be useful in detecting such genes, not only in inbred mice, but also in monozygotic human twins, in which genetic variation is controlled. However, only genes with expression patterns that are epigenetically established before embryonic stem cell differentiation, as for the Avy locus in mice, will be detectable with this experimental approach.
Genes with altered DNA methylation
Similar studies to those described above for vinclozolin need be carried out in humans that have inadvertently been exposed to environmental contaminants in order to identify genes and other DNA sequences that show alterations in DNA-methylation and allelic expression patterns. The genome-wide identification of such sequences will require the use of tiling arrays, methylation-sensitive restriction-enzyme analysis, bisulphite conversion of unmethylated CpG sites and high-throughput DNA sequencing. Although these procedures are available and rapidly improving, the high levels of both genetic and epigenetic variation in the human population will be a potential limitation in the application of these procedures to humans until genome-wide individual variation in the epigenome is ultimately assessed.
Imprinted genes
To date, most efforts to identify imprinted genes have been experimental, focusing on small regions of a chromosome. A robust method for genome-wide identification of imprinted genes could involve the use of machine learning algorithms, which are trained to identify genomic motifs that are predictive of imprinted genes. Luedi et al.106 recently developed such a bioinformatic approach for interrogating the entire mouse genome for genes that are highly likely to be imprinted. This imprinted-gene prediction algorithm identified 600, or 2.5%, of the annotated autosomal mouse genes as being potentially imprinted, 64% of which are predicted to exhibit maternal expression. Future studies are now needed to directly confirm that these candidate genes are imprinted.
However, the real power of this imprinted-gene prediction algorithm lies in its ability to readily interrogate the genomes of any eutherian species for which complete genomic sequence is available. With the use of a similar bioinformatic approach, the human genome was predicted to contain fewer imprinted genes than the mouse, and the repertoire of imprinted genes was also found to be strongly species dependent (R.L.J., unpublished observations). Thus, despite the immense popularity of using the mouse as a model for human disease, it might not be a suitable choice for studying diseases and behavioural disorders that result principally from the epigenetic deregulation of imprinted genes, or for assessing human risk from environmental factors that alter the epigenome.
Conclusions
The word ‘environment’ means vastly different things to different people. For sociologists and psychologists, it conjures up visions of social group interactions, family dynamics and maternal nurturing. Nutritionists might envision food pyramids and dietary supplements, whereas toxicologists think of water, soil and air pollutants. This Review has highlighted the evidence that these vastly different environments are all able to alter gene expression and change phenotype, in part by impinging on and modifying the epigenome. Moreover, if these environmentally induced epigenetic adaptations occur at crucial stages of life, they can potentially change behaviour, disease susceptibility and survival.
Epigenetic modifications do not alter gene sequence but, rather, gene expression. Therefore, characterizing the expression profiles of epigenetically labile genes that are susceptible to environmental dysregulation will ultimately identify epigenetic biomarkers for disease and environmental exposure. These epigenetic biomarkers will hopefully allow for the early diagnosis of individuals with a propensity for adult-onset disease. They could also be used in novel preventative and therapeutic approaches before disease symptoms develop. Such an approach to human disease management could revolutionize medical care, which now mainly treats diseases only after they develop. Understanding how the environment influences human health and disease will ultimately require a comprehensive knowledge of the human epigenome, because the epigenome is not only tissue and stage-of-life dependent, but also varies markedly between species. As simply stated by the English poet Alexander Pope in the early eighteenth century, “The proper study of Mankind is Man.”
Acknowledgments
The authors wish to thank D. Dolinoy for critically reading the manuscript and for her helpful suggestions. We thank J. Griffin for assistance in the preparation of this manuscript. This work was supported by grants from the US Department of Energy and the US National Institutes of Health.
Glossary
Epigenetic
Refers to mitotically or meiotically heritable changes in gene expression that do not involve a change in DNA sequence.
DNA methylation
DNA methylation occurs predominantly in repetitive genomic regions that contain CpG residues. DNA methylation represses transcription directly by inhibiting the binding of specific transcription factors, and indirectly by recruiting methyl-CpG-binding proteins and their associated repressive chromatin-remodelling activities.
Histone modifications
Histones undergo post-translational modifications that alter their interaction with DNA and nuclear proteins. In particular, the tails of histones H3 and H4 can be covalently modified at several residues. Modifications of the tail include methylation, acetylation, phosphorylation and ubiquitination, and influence several biological processes, including gene expression, DNA repair and chromosome condensation.
MicroRNAs
Endogenous small RNAs of ~22 nucleotides in length that act as a cellular rheostat for fine-tuning gene expression during development and differentiation. They target the 3′ UTRs of mRNAs with which they share partial sequence complementarity, leading to post-transcriptional gene silencing through translational repression. When a microRNA has complete sequence complementarity with a target mRNA, it instead directs cleavage of the transcript.
X-chromosome inactivation
The process that occurs in female mammals by which gene expression from one of the pair of X chromosomes is downregulated to match the levels of gene expression from the single X chromosome that is present in males. The inactivation process involves a range of epigenetic mechanisms on the inactivated chromosome, including changes in DNA methylation and histone modifications.
Genomic imprinting
The epigenetic marking of a locus on the basis of parental origin, which results in monoallelic gene expression.
Epigenome
The global epigenetic patterns that distinguish or are variable between cell types. These patterns include DNA methylation, histone modifications and chromatin-associated proteins.
Melanocyte
A specialized cell type, lying at the boundary between the dermis and epidermis, in which the pigment melanin is synthesized.
Embryonic stem cell
A type of pluripotent stem cell that is derived from the inner cell mass of the early embryo. Pluripotent cells are capable of generating virtually all cell types of the organism.
Angelman syndrome
A defect that is caused by the loss of expression of a maternally expressed gene, UBE3A, which is imprinted only in the brain and encodes an E3 ubiquitin ligase. Angelman syndrome occurs in ~1 in 15,000 births and its main characteristics include mental retardation, speech impairment and behavioural abnormalities.
Prader–Willi syndrome
The molecular defect that causes this syndrome is complex and involves defects that affect an ~2 Mb imprinted domain that contains both paternally and maternally expressed genes. Prader–Willi syndrome occurs in ~1 in 20,000 births and is characterized by a failure to thrive during infancy, hyperphagia and obesity during early childhood, mental retardation and behavioural problems.
Beckwith–Weidemann syndrome
A predominantly maternally transmitted disorder, involving fetal and postnatal overgrowth and a predisposition to embryonic tumours. The Beckwith–Weidemann syndrome locus includes several imprinted genes, including IGF2, H19 and KCNQ1, and loss of imprinting at IGF2 is seen in ~20% of cases.
Bisulphite conversion
A technique that is used to identify methylcytosines. The approach depends on the relative resistance of the conversion of methylcytosine to uracil compared with cytosine. Conversion can be followed by PCR amplification and sequencing of the DNA. The persistence of a cytosine, instead of a thymine, being detected, reflects the methylation of the cytosine in the starting DNA sample.
Machine learning
The ability of a program to learn from experience — that is, to modify its execution on the basis of newly acquired information. In bioinformatics, neural networks and Monte Carlo Markov chains are well-known examples.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
References
- 1.Yajnik CS. Early life origins of insulin resistance and type 2 diabetes in India and other Asian countries. J Nutr. 2004;134:205–210. doi: 10.1093/jn/134.1.205. [DOI] [PubMed] [Google Scholar]
- 2.Barker DJ. The developmental origins of chronic adult disease. Acta Paediatr Suppl. 2004;93:26–33. doi: 10.1111/j.1651-2227.2004.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 3.Painter RC, Roseboom TJ, Bleker OP. Prenatal exposure to the Dutch famine and disease in later life: an overview. Reprod Toxicol. 2005;20:345–352. doi: 10.1016/j.reprotox.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Gluckman PD, Hanson MA, Beedle AS. Early life events and their consequences for later disease: a life history and evolutionary perspective. Am J Hum Biol. 2007;19:1–19. doi: 10.1002/ajhb.20590. References 1–4 discuss evidence for the early origins of the adult disease susceptibility hypothesis. [DOI] [PubMed] [Google Scholar]
- 5.St Clair D, et al. Rates of adult schizophrenia following prenatal exposure to the Chinese famine of 1959–1961. JAMA. 2005;294:557–562. doi: 10.1001/jama.294.5.557. [DOI] [PubMed] [Google Scholar]
- 6.van Os J, Selten JP. Prenatal exposure to maternal stress and subsequent schizophrenia. The May 1940 invasion of The Netherlands. Br J Psychiatry. 1998;172:324–326. doi: 10.1192/bjp.172.4.324. References 5 and 6 discuss epidemiological evidence that the adult incidence of schizophrenia is significantly increased in humans who were exposed prenatally to famine conditions. [DOI] [PubMed] [Google Scholar]
- 7.Li E. Chromatin modification and epigenetic reprogramming in mammalian development. Nature Rev Genet. 2002;3:662–673. doi: 10.1038/nrg887. [DOI] [PubMed] [Google Scholar]
- 8.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nature Rev Genet. 2006;7:793–803. doi: 10.1038/nrg1920. [DOI] [PubMed] [Google Scholar]
- 10.Richards EJ. Inherited epigenetic variation — revisiting soft inheritance. Nature Rev Genet. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 11.Thorvaldsen JL, Verona RI, Bartolomei MS. X-tra! X-tra! News from the mouse X chromosome. Dev Biol. 2006;298:344–353. doi: 10.1016/j.ydbio.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Huynh KD, Lee JT. X-chromosome inactivation: a hypothesis linking ontogeny and phylogeny. Nature Rev Genet. 2005;6:410–418. doi: 10.1038/nrg1604. [DOI] [PubMed] [Google Scholar]
- 13.Lewis A, Reik W. How imprinting centres work. Cytogenet Genome Res. 2006;113:81–89. doi: 10.1159/000090818. [DOI] [PubMed] [Google Scholar]
- 14.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nature Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 15.Falls JG, Pulford DJ, Wylie AA, Jirtle RL. Genomic imprinting: implications for human disease. Am J Pathol. 1999;154:635–647. doi: 10.1016/S0002-9440(10)65309-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy SK, Jirtle RL. Imprinting evolution and the price of silence. BioEssays. 2003;25:577–588. doi: 10.1002/bies.10277. [DOI] [PubMed] [Google Scholar]
- 17.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nature Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 18.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 19.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. This study demonstrates that maternal methyl donor supplementation during gestation can alter offspring phenotype by methylating the epigenome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waterland RA, et al. Maternal methyl supplements increase offspring DNA methylation at Axin fused. Genesis. 2006;44:401–406. doi: 10.1002/dvg.20230. [DOI] [PubMed] [Google Scholar]
- 22.Waterland RA, Lin JR, Smith CA, Jirtle RL. Post-weaning diet affects genomic imprinting at the insulin-like growth factor 2 (IGF2) locus. Hum Mol Genet. 2006;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]
- 23.Li S, et al. Neonatal diethylstilbestrol exposure induces persistent elevation of c-fos expression and hypomethylation in its exon-4 in mouse uterus. Mol Carcinog. 2003;38:78–84. doi: 10.1002/mc.10147. [DOI] [PubMed] [Google Scholar]
- 24.Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology. 2006;147:S43–S49. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- 26.Weaver ICG, et al. Epigenetic programming by maternal behavior. Nature Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 27.Weaver IC, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niemitz EL, Feinberg AP. Epigenetics and assisted reproductive technology: a call for investigation. Am J Hum Genet. 2004;74:599–609. doi: 10.1086/382897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rossignol S, et al. The epigenetic imprinting defect of patients with Beckwith–Wiedemann syndrome born after assisted reproductive technology is not restricted to the 11p15 region. J Med Genet. 2006;43:902–907. doi: 10.1136/jmg.2006.042135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koturbash I, et al. Epigenetic dysregulation underlies radiation-induced transgenerational genome instability in vivo. Int J Radiat Oncol Biol Phys. 2006;66:327–330. doi: 10.1016/j.ijrobp.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Morgan HD, Sutherland HGE, Martin DIK, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nature Genet. 1999;23:314–318. doi: 10.1038/15490. This study demonstrates the maternal inheritance of an epigenetic modification at the agouti locus in mice. [DOI] [PubMed] [Google Scholar]
- 32.Lane N, et al. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 33.Rakyan VK, et al. Transgenerational inheritance of epigenetic states at the murine Axin(Fu) allele occurs after maternal and paternal transmission. Proc Natl Acad Sci USA. 2003;100:2538–2543. doi: 10.1073/pnas.0436776100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. This study demonstrates the ability of environmental factors to induce an epigenetic transgenerational disease phenotype for four generations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pembrey ME, et al. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. This study demonstrates an inherited disease phenotype in humans that is potentially induced by an epigenetic phenomena. [DOI] [PubMed] [Google Scholar]
- 36.Vasicek TJ, et al. Two dominant mutations in the mouse fused gene are the result of transposon insertions. Genetics. 1997;147:777–786. doi: 10.1093/genetics/147.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suter CM, Martin DI, Ward RL. Germline epimutation of MLH1 in individuals with multiple cancers. Nature Genet. 2004;36:497–501. doi: 10.1038/ng1342. [DOI] [PubMed] [Google Scholar]
- 38.Chan TL, et al. Heritable germline epimutation of MSH2 in a family with hereditary nonpolyposis colorectal cancer. Nature Genet. 2006;38:1178–1183. doi: 10.1038/ng1866. [DOI] [PubMed] [Google Scholar]
- 39.Esteller M. Epigenetics provides a new generation of oncogenes and tumour-suppressor genes. Br J Cancer. 2006;94:179–183. doi: 10.1038/sj.bjc.6602918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nature Rev Drug Discov. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 41.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer — a mechanism for early oncogenic pathway addiction? Nature Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 42.Duhl DM, Vrieling H, Miller KA, Wolff GL, Barsh GS. Neomorphic agouti mutations in obese yellow mice. Nature Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. These authors show that the Avy allele results from the insertion of an intracisternal A particle upstream of the agouti gene. [DOI] [PubMed] [Google Scholar]
- 43.Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Complex patterns of transcription at the insertion site of a retrotransposon in the mouse. Nucl Acids Res. 2004;32:5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Metastable epialleles in mammals. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 45.Druker R, Whitelaw E. Retrotransposon-derived elements in the mammalian genome: a potential source of disease. Inherit Metab Dis. 2004;27:319–330. doi: 10.1023/B:BOLI.0000031096.81518.66. [DOI] [PubMed] [Google Scholar]
- 46.Miltenberger RJ, Mynatt RL, Wilkinson JE, Woychik RP. The role of the agouti gene in the Yellow Obese Syndrome. J Nutr. 1997;127:1902S–1907S. doi: 10.1093/jn/127.9.1902S. [DOI] [PubMed] [Google Scholar]
- 47.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 48.Cropley JE, Suter CM, Beckman KB, Martin DI. Germ-line epigenetic modification of the murine Avy allele by nutritional supplementation. Proc Natl Acad Sci USA. 2006;103:17308–17312. doi: 10.1073/pnas.0607090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haig D, Graham C. Genomic imprinting and the strange case of the insulin-like growth factor II receptor. Cell. 1991;64:1045–1046. doi: 10.1016/0092-8674(91)90256-x. These authors propose that genomic imprinting evolved because of a parental genetic battle to control the amount of nutrients that is extracted from the mother by the offspring. [DOI] [PubMed] [Google Scholar]
- 50.Wilkins JF, Haig D. What good is genomic imprinting: the function of parent-specific gene expression. Nature Rev Genet. 2003;4:359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- 51.DeChiara TM, Robertson E, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. doi: 10.1016/0092-8674(91)90513-x. [DOI] [PubMed] [Google Scholar]
- 52.Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. doi: 10.1038/349084a0. References 51 and 52 report the first-identified imprinted genes. [DOI] [PubMed] [Google Scholar]
- 53.Killian JK, et al. M6p/IGF2R imprinting evolution in mammals. Mol Cell. 2000;5:707–716. doi: 10.1016/s1097-2765(00)80249-x. This paper demonstrates that genomic imprinting evolved approximately 180 million years ago with the advent of live birth in therian mammals. [DOI] [PubMed] [Google Scholar]
- 54.Evans HK, Weidman JR, Cowley DO, Jirtle RL. Comparative phylogenetic analysis of Blcap/Nnat reveals eutherian-specific imprinted gene. Mol Biol Evol. 2005;22:1740–1748. doi: 10.1093/molbev/msi165. [DOI] [PubMed] [Google Scholar]
- 55.Weidman JR, Maloney KA, Jirtle RL. Comparative phylogenetic analysis reveals multiple non-imprinted isoforms of opossum DLK1. Mamm Genome. 2006;17:157–167. doi: 10.1007/s00335-005-0116-x. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki S, et al. Genomic imprinting of IGF2, p57(KIP2) and PEG1/MEST in a marsupial, the tammar wallaby. Mech Dev. 2005;122:213–222. doi: 10.1016/j.mod.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 57.Killian JK, et al. Divergent evolution in M6P/IGF2R imprinting from the Jurassic to the Quaternary. Hum Mol Genet. 2001;10:1721–1728. doi: 10.1093/hmg/10.17.1721. [DOI] [PubMed] [Google Scholar]
- 58.De Souza AT, Hankins GR, Washington MK, Orton TC, Jirtle RL. M6P/IGF2R gene is mutated in human hepatocellular carcinomas with loss of heterozygosity. Nature Genet. 1995;11:447–449. doi: 10.1038/ng1295-447. [DOI] [PubMed] [Google Scholar]
- 59.Weksberg R, Shuman C, Smith AC. Beckwith–Wiedemann syndrome. Am J Med Genet C Semin Med Genet. 2005;137:12–23. doi: 10.1002/ajmg.c.30058. [DOI] [PubMed] [Google Scholar]
- 60.Kantor B, Shemer R, Razin A. The Prader-Willi–Angelman imprinted domain and its control center. Cytogenet Genome Res. 2006;113:300–305. doi: 10.1159/000090845. [DOI] [PubMed] [Google Scholar]
- 61.Badcock C, Crespi B. Imbalanced genomic imprinting in brain development: an evolutionary basis for the aetiology of autism. J Evol Biol. 2006;19:1007–1032. doi: 10.1111/j.1420-9101.2006.01091.x. These authors propose that human neurological disorders, such as autism, result from an imbalanced expression of imprinted genes during development. [DOI] [PubMed] [Google Scholar]
- 62.Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends Genet. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 63.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nature Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 64.Feinberg AP. A genetic approach to cancer epigenetics. Cold Spring Harb Symp Quant Biol. 2005;70:335–341. doi: 10.1101/sqb.2005.70.027. [DOI] [PubMed] [Google Scholar]
- 65.Knudson AG. Two genetic hits (more or less) to cancer. Nature Rev Cancer. 2001;1:157–162. doi: 10.1038/35101031. [DOI] [PubMed] [Google Scholar]
- 66.Cui H, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. This paper reports that some humans have IGF2 LOI in peripheral lymphocytes, which is correlated with biallelic expression in normal colonic mucosa and a personal history of colorectal cancer. [DOI] [PubMed] [Google Scholar]
- 67.Cruz-Correa M, et al. Loss of imprinting of insulin growth factor II gene: a potential heritable biomarker for colon neoplasia predisposition. Gastroenterology. 2004;126:964–970. doi: 10.1053/j.gastro.2003.12.051. [DOI] [PubMed] [Google Scholar]
- 68.Jirtle RL. IGF2 loss of imprinting: a potential heritable risk factor for colorectal cancer. Gastroenterology. 2004;126:1190–1193. doi: 10.1053/j.gastro.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 69.Oates NA, et al. Increased DNA methylation at the AXIN1 gene in a monozygotic twin from a pair discordant for a caudal duplication anomaly. Am J Hum Genet. 2006;79:155–162. doi: 10.1086/505031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ikeda M, Tamura M, Yamashita J, Suzuki C, Tomita T. Repeated in utero and lactational 2,3,7,8-tetrachlorodibenzo-p-dioxin exposure affects male gonads in offspring, leading to sex ratio changes in F2 progeny. Toxicol Appl Pharmacol. 2005;206:351–355. doi: 10.1016/j.taap.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 71.Blatt J, Van Le L, Weiner T, Sailer S. Ovarian carcinoma in an adolescent with transgenerational exposure to diethylstilbestrol. J Pediatr Hematol Oncol. 2003;25:635–636. doi: 10.1097/00043426-200308000-00009. [DOI] [PubMed] [Google Scholar]
- 72.Dubrova YE. Radiation-induced transgenerational instability. Oncogene. 2003;22:7087–7093. doi: 10.1038/sj.onc.1206993. [DOI] [PubMed] [Google Scholar]
- 73.Cheng RY, Hockman T, Crawford E, Anderson LM, Shiao YH. Epigenetic and gene expression changes related to transgenerational carcinogenesis. Mol Carcinog. 2004;40:1–11. doi: 10.1002/mc.20022. [DOI] [PubMed] [Google Scholar]
- 74.Hemmings DG, Veerareddy S, Baker PN, Davidge ST. Increased myogenic responses in uterine but not mesenteric arteries from pregnant offspring of diet-restricted rat dams. Biol Reprod. 2005;72:997–1003. doi: 10.1095/biolreprod.104.035675. [DOI] [PubMed] [Google Scholar]
- 75.Ferguson LR, Karunasinghe N, Philpott M. Epigenetic events and protection from colon cancer in New Zealand. Environ Mol Mutagen. 2004;44:36–43. doi: 10.1002/em.20029. [DOI] [PubMed] [Google Scholar]
- 76.Csaba G, Karabelyos C. Transgenerational effect of a single neonatal benzpyrene treatment (imprinting) on the sexual behavior of adult female rats. Hum Exp Toxicol. 1997;16:553–556. doi: 10.1177/096032719701601001. [DOI] [PubMed] [Google Scholar]
- 77.Fujii T. Transgenerational effects of maternal exposure to chemicals on the functional development of the brain in the offspring. Cancer Causes Control. 1997;8:524–528. doi: 10.1023/a:1018477809755. [DOI] [PubMed] [Google Scholar]
- 78.Brucker-Davis F. Effects of environmental synthetic chemicals on thyroid function. Thyroid. 1998;8:827–856. doi: 10.1089/thy.1998.8.827. [DOI] [PubMed] [Google Scholar]
- 79.Giusti RM, Iwamoto K, Hatch EE. Diethylstilbestrol revisited: a review of the long-term health effects. Ann Intern Med. 1995;122:778–788. doi: 10.7326/0003-4819-122-10-199505150-00008. [DOI] [PubMed] [Google Scholar]
- 80.Klip H, et al. Hypospadias in sons of women exposed to diethylstilbestrol in utero: a cohort study. Lancet. 2002;359:1102–1107. doi: 10.1016/S0140-6736(02)08152-7. [DOI] [PubMed] [Google Scholar]
- 81.Parks LG, et al. The plasticizer diethylhexyl phthalate induces malformations by decreasing fetal testosterone synthesis during sexual differentiation in the male rat. Toxicol Sci. 2000;58:339–349. doi: 10.1093/toxsci/58.2.339. [DOI] [PubMed] [Google Scholar]
- 82.Steinhardt GF. Endocrine disruption and hypospadias. Adv Exp Med Biol. 2004;545:203–215. doi: 10.1007/978-1-4419-8995-6_13. [DOI] [PubMed] [Google Scholar]
- 83.Ruden DM, Xiao L, Garfinkel MD, Lu X. Hsp90 and environmental impacts on epigenetic states: a model for the trans-generational effects of diethylstibesterol on uterine development and cancer. Hum Mol Genet. 2005;14:R149–R155. doi: 10.1093/hmg/ddi103. [DOI] [PubMed] [Google Scholar]
- 84.Matta MB, Linse J, Cairncross C, Francendese L, Kocan RM. Reproductive and transgenerational effects of methylmercury or Aroclor 1268 on Fundulus heteroclitus. Environ Toxicol Chem. 2001;20:327–335. [PubMed] [Google Scholar]
- 85.Omholt SW, Amdam GV. Epigenetic regulation of aging in honeybee workers. Sci Aging Knowledge Environ. 2004;2004:pe28. doi: 10.1126/sageke.2004.26.pe28. [DOI] [PubMed] [Google Scholar]
- 86.Ottinger MA, et al. Assessing the consequences of the pesticide methoxychlor: neuroendocrine and behavioral measures as indicators of biological impact of an estrogenic environmental chemical. Brain Res Bull. 2005;65:199–209. doi: 10.1016/j.brainresbull.2004.11.019. [DOI] [PubMed] [Google Scholar]
- 87.Seidl MD, Paul RJ, Pirow R. Effects of hypoxia acclimation on morpho-physiological traits over three generations of Daphnia magna. J Exp Biol. 2005;208:2165–2175. doi: 10.1242/jeb.01614. [DOI] [PubMed] [Google Scholar]
- 88.Foran CM, Peterson BN, Benson WH. Transgenerational and developmental exposure of Japanese medaka (Oryzias latipes) to ethinylestradiol results in endocrine and reproductive differences in the response to ethinylestradiol as adults. Toxicol Sci. 2002;68:389–402. doi: 10.1093/toxsci/68.2.389. [DOI] [PubMed] [Google Scholar]
- 89.Anderson CM, Lopez F, Zimmer A, Benoit JN. Placental insufficiency leads to developmental hypertension and mesenteric artery dysfunction in two generations of Sprague–Dawley rat offspring. Biol Reprod. 2006;74:538–544. doi: 10.1095/biolreprod.105.045807. [DOI] [PubMed] [Google Scholar]
- 90.Csaba G, Inczefi-Gonda A. Transgenerational effect of a single neonatal benzpyrene treatment on the glucocorticoid receptor of the rat thymus. Hum Exp Toxicol. 1998;17:88–92. doi: 10.1177/096032719801700203. [DOI] [PubMed] [Google Scholar]
- 91.Newbold RR, Padilla-Banks E, Jefferson WN. Adverse effects of the model environmental estrogen diethylstilbestrol are transmitted to subsequent generations. Endocrinology. 2006;147:S11–S17. doi: 10.1210/en.2005-1164. [DOI] [PubMed] [Google Scholar]
- 92.Popova NV. Transgenerational effect of orthoaminoasotoluol in mice. Cancer Lett. 1989;46:203–206. doi: 10.1016/0304-3835(89)90131-6. [DOI] [PubMed] [Google Scholar]
- 93.Zambrano E, et al. Sex differences in transgenerational alterations of growth and metabolism in progeny (F2) of female offspring (F1) of rats fed a low protein diet during pregnancy and lactation. J Physiol. 2005;566:225–236. doi: 10.1113/jphysiol.2005.086462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cesani MF, et al. Effect of undernutrition on the cranial growth of the rat. An intergenerational study. Cells Tissues Organs. 2003;174:129–135. doi: 10.1159/000071153. [DOI] [PubMed] [Google Scholar]
- 95.Turusov VS, Nikonova TV, Parfenov Y. Increased multiplicity of lung adenomas in five generations of mice treated with benz(a)pyrene when pregnant. Cancer Lett. 1990;55:227–231. doi: 10.1016/0304-3835(90)90123-f. [DOI] [PubMed] [Google Scholar]
- 96.Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chang HS, Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic imprinting of the male germline by endocrine disruptor exposure during gonadal sex determination. Endocrinology. 2006;147:5524–5541. doi: 10.1210/en.2006-0987. This report demonstrates the ability of vinclozolin to induce the reprogramming of the germ line, and the formation of genes and DNA sequences that contain paternal-allele alterations in DNA methylation associated with transgenerational disease. [DOI] [PubMed] [Google Scholar]
- 98.Durcova-Hills G, et al. Influence of sex chromosome constitution on the genomic imprinting of germ cells. Proc Natl Acad Sci USA. 2006;103:11184–11188. doi: 10.1073/pnas.0602621103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Forum TC. News and Information. J Radiol Prot. 2005;25:499–502. [Google Scholar]
- 100.Allegrucci C, Thurston A, Lucas E, Young L. Epigenetics and the germline. Reproduction. 2005;129:137–149. doi: 10.1530/rep.1.00360. [DOI] [PubMed] [Google Scholar]
- 101.McCarrey JR, Geyer CB, Yoshioka H. Epigenetic regulation of testis-specific gene expression. Ann NY Acad Sci. 2005;1061:226–242. doi: 10.1196/annals.1336.025. [DOI] [PubMed] [Google Scholar]
- 102.Trasler JM. Origin and roles of genomic methylation patterns in male germ cells. Semin Cell Dev Biol. 1998;9:467–474. doi: 10.1006/scdb.1998.0225. [DOI] [PubMed] [Google Scholar]
- 103.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci USA. 2006;103:3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hurst GD, Werren JH. The role of selfish genetic elements in eukaryotic evolution. Nature Rev Genet. 2001;2:597–606. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- 105.Bestor TH. Cytosine methylation mediates sexual conflict. Trends Genet. 2003;19:185–190. doi: 10.1016/S0168-9525(03)00049-0. [DOI] [PubMed] [Google Scholar]
- 106.Luedi PP, Hartemink AJ, Jirtle RL. Genome-wide prediction of imprinted murine genes. Genome Res. 2005;15:875–884. doi: 10.1101/gr.3303505. These authors demonstrate that imprinted genes and their parental expression bias can be predicted genome-wide with the use of machine learning algorithms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Waddington CH. Organisers and Genes. Cambridge Univ. Press; Cambridge: 1940. [Google Scholar]
- 108.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 109.Willard HF, Brown CJ, Carrel L, Hendrich B, Miller AP. Epigenetic and chromosomal control of gene expression: molecular and genetic analysis of X chromosome inactivation. Cold Spring Harb Symp Quant Biol. 1993;58:315–322. doi: 10.1101/sqb.1993.058.01.037. [DOI] [PubMed] [Google Scholar]
- 110.Monk M. Genomic imprinting. Genes Dev. 1988;2:921–925. doi: 10.1101/gad.2.8.921. [DOI] [PubMed] [Google Scholar]
- 111.Wolffe AP, Matzke MA. Epigenetics: regulation through repression. Science. 1999;286:481–486. doi: 10.1126/science.286.5439.481. [DOI] [PubMed] [Google Scholar]
- 112.Murrell A, Rakyan VK, Beck S. From genome to epigenome. Hum Mol Genet. 2005;14:R3–R10. doi: 10.1093/hmg/ddi110. [DOI] [PubMed] [Google Scholar]
- 113.Kishino T. Imprinting in neurons. Cytogenet Genome Res. 2006;113:209–214. doi: 10.1159/000090834. [DOI] [PubMed] [Google Scholar]
- 114.Vu TH, Jirtle RL, Hoffman AR. Cross-species clues of an epigenetic imprinting regulatory code for the IGF2R gene. Cytogenet Genome Res. 2006;113:202–208. doi: 10.1159/000090833. [DOI] [PubMed] [Google Scholar]
- 115.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–552. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]