Regulation of the Chemokine Receptor CXCR4 by Hypoxia (original) (raw)
Abstract
Cell adaptation to hypoxia (Hyp) requires activation of transcriptional programs that coordinate expression of genes involved in oxygen delivery (via angiogenesis) and metabolic adaptation (via glycolysis). Here, we describe that oxygen availability is a determinant parameter in the setting of chemotactic responsiveness to stromal-derived factor 1 (CXCL12). Low oxygen concentration induces high expression of the CXCL12 receptor, CXC receptor 4 (CXCR4), in different cell types (monocytes, monocyte-derived macrophages, tumor-associated macrophages, endothelial cells, and cancer cells), which is paralleled by increased chemotactic responsiveness to its specific ligand. CXCR4 induction by Hyp is dependent on both activation of the Hyp-inducible factor 1 α and transcript stabilization. In a relay multistep navigation process, the Hyp–Hyp-inducible factor 1 α–CXCR4 pathway may regulate trafficking in and out of hypoxic tissue microenvironments.
Keywords: cell migration, SDF-1/CXCL12 receptor (CXCR4), low oxygen concentration, hypoxia-inducible factor 1 (HIF-1)
Introduction
Oxygen homeostasis represents an important organizing principle for human development and physiology (1). The essential requirement for oxidative phosphorylation to generate ATP is balanced by the risk of oxidative damage to cellular lipids, nucleic acids, and proteins. As a result, cellular and systemic O2 concentrations are tightly regulated via short- and long-acting response pathways that affect the activity and expression of a multitude of cellular proteins (2). Dysregulation of O2 homeostasis is found in inflammatory and cardiovascular diseases, cancer, cerebrovascular disease, and chronic obstructive pulmonary disease. Recent papers have begun to delineate the molecular basis of cellular and systemic mechanisms of O2 homeostasis and the most global regulator identified to date is the transcriptional activator hypoxia (Hyp)-inducible factor 1 (HIF-1) composed of the HIF-1α and HIF-1β subunits (1, 3).
During migration and invasion of normal and pathological tissues, cells may encounter different oxygen levels, due to poor or altered vascularization, and recent evidence has suggested that chemotaxis is a cell function which may be affected by oxygen availability (4, 5). Leukocyte trafficking, an event which plays a central role in fundamental functions of multicellular organisms, including tissue remodelling, defense, and pathology, is orchestrated by a superfamily of small proteins termed chemokines, which are essential players in immune and inflammatory reactions as well as in infections (6–8). Based on a cysteine motif, chemokines have been classified into a CXC, CC, C, and CX3C family, and >37 members are identified in humans to date. Chemokines interact with 7 transmembrane domain G-protein–coupled receptors and, so far, 10 CC (CCR1-10), 6 CXC (CXCR1-6), 1 CX3C (CXCR1), and 1 C (XCR1) receptors have been identified (9). Emerging evidence shows that the expression and function of G-protein–coupled receptors is strictly controlled by cytokines and other microenvironmental signals, such as Hyp (10–12), and that the regulation of receptor expression during cell activation and deactivation is as important as the regulation of chemokine production for tuning the chemokine system (13).
Experimental and clinical studies point to the fundamental pathophysiological role of Hyp in inflammatory (14, 15) and neoplastic diseases (16). In particular, tumor Hyp is a consequence of a structurally and functionally disturbed microcirculation and, in some cases, of a reduced O2-carrying capacity of the blood due to tumor-associated anemia. Hyp in human tumors by itself has been shown to contribute to resistance to standard anticancer therapies, and recent data suggest that O2-deprived tumor cells are predisposed to a more malignant phenotype (i.e., tumor cells are likely to be more metastatic and/or invasive; reference 16). Solid tumors consist of malignant cells and stroma, which includes new blood vessels, matrix components and cells responsible for their production, a fibrin-gel matrix, and inflammatory leukocytes (17, 18). Thus, tumor stromal cells encounter low oxygen conditions and, in particular, tumor-associated macrophages (TAMs) have been reported to localize preferentially in the hypoxic areas of tumors (4, 5, 19, 20). Based on this evidence, we investigated the effect of Hyp on the expression and function of chemokine receptors. We observed that Hyp selectively augments CXCR4 expression through HIF-1 activation in human monocytes, macrophages, endothelial cells, and cancer cells. The identification of the Hyp–HIF-1α–CXCR4 pathway provides novel insights into the mechanisms controlling cell migration in hypoxic regions, with potential relevance in the pathogenesis of human diseases. The Hyp–HIF-1α–CXCR4 pathway is likely to regulate the migration and localization of diverse cell types in tissues.
Materials and Methods
Cells and Culture Conditions.
Human monocytes were separated from peripheral blood of human healthy donors by Percoll gradient centrifugation (10). Monocytes (>98% pure as assessed by morphology) were resuspended at 107/ml in RPMI 1640 supplemented with 10% FBS, 2 mM glutamine, and antibiotics. All reagents contained <0.125 EU/ml of endotoxin as checked by limulus amebocyte lysate assay (Microbiological Associates). Monocyte-derived macrophages (MDMs) were derived from freshly isolated monocytes (3–5 × 106 cells/ml) after incubation for 5 d in RPMI 1640 medium supplemented with 10% FBS, 2 mM glutamine, antibiotics, and 40% autologous serum on hydrophobic petriperm plates (Sigma-Aldrich) as described previously (21).
TAMs were obtained from ascitic fluids collected from untreated patients with histologically confirmed epithelial ovarian carcinoma admitted to the Department of Obstetric and Gynecology, S. Gerardo Hospital, Monza, Italy. All patients had cancer classified as stage II, III, or IV. Ascitic fluid was collected and centrifuged. Cells pellet was resuspended in RPMI 1640 medium without serum and layered on top of a Ficoll-Hypaque cushion to prepare mononuclear cells. Purification of peritoneal macrophages was further conducted by two subsequent adherence steps for 45 min each in RPMI 1640 medium without serum. After adherence procedures, cells were repeatedly washed with saline to remove all nonadherent cells. The adherent cells were rested with complete medium overnight at 37°C and subsequently stimulated as indicated in the test.
Human endothelial cells were obtained from umbilical veins and cultured as described previously (22). We used routinely confluent cells at second to sixth passages. Cells at the concentration of 1.5 × 104/0.2 ml were cultured for 24 h in flat-bottomed 96-well plates (Falcon) in M199 medium with 20% FCS, supplemented with 50 μg/ml of endothelial cell growth supplement (Collaborative Research Inc.) and 100 μg/ml heparin. Cells were maintained at 37°C in a humidified incubator containing 20% O2, 5% CO2, and 75% N2. For hypoxic conditions, cells were cultured in an atmosphere-controlled culture chamber (Bellco Glass) containing a gas mixture composed of 94% N2, 5% CO2, and 1% O2.
Mouse embryonic fibroblasts (MEFs), wild type, and HIF-1α− / − were a gift from G. Semenza (The Johns Hopkins University School of Medicine, Baltimore, MD) and were routinely maintained in DMEM (Invitrogen and Life Technologies) supplemented with 10% heat-inactivated FBS (Whittaker Bioproducts), 50 IU/ml penicillin, 50 μg/ml streptomycin, and 2 mM glutamine (all purchased from Invitrogen and Life Technologies). MCF-7 (human breast cancer cells), CAOV3 (human ovarian cancer cell line), 786.0 (36), and WT2 human renal cancer cells (23) were routinely maintained in RPMI 1640 medium (Whittaker Bioproducts) supplemented with 5% heat-inactivated FBS, 50 IU/ml penicillin, 50 μg/ml streptomycin, and 2 mM glutamine. Cells were maintained at 37°C in a humidified incubator containing 20% O2, 5% CO2 in air (referred to as normoxic conditions). Hyp treatment was performed by placing cells in a modular incubator chamber (Billupus-Rothemberg Inc.) and flushing with a mixture of 1% O2, 5% CO2, and 94% nitrogen for 20 min. The chamber was placed at 37°C.
FACS® Analysis.
Cell staining was performed using mouse monoclonal anti–human CXCR4 antibody (clone 12G5; BD Biosciences) and an irrelevant isotype-specific control mouse, IgG2a, κ (UPC10; Sigma-Aldrich) followed by FITC-conjugated, isotype-matched affinity-purified, goat anti–mouse antibody (Southern Biotechnology Associates, Inc.).
Cytokines and Reagents.
Human recombinant CXCL12/stromal-derived factor 1 (SDF-1) and CCL5/regulated on activation normal T cell expressed and secreted were from PeproTech. Desferrioxamine (DFX) and actinomycin D (used at 1 mg/ml) were purchased from Sigma-Aldrich.
Migration Assay.
Monocyte migration was evaluated using a chemotaxis microchamber technique as described previously (10). 27 ml of chemoattractant solution or control medium (RPMI 1640 with 1% FCS) was added to the lower wells of a chemotaxis chamber (Neuroprobe). A polycarbonate filter (5 μm pore size; Neuroprobe) was layered onto the wells and covered with a silicon gasket and the top plate. 50 ml of cell suspension (1.5 × 106/ml fresh human monocytes) was preincubated for 16 h in the presence of 400 μM DFX and seeded in the upper chamber. The chamber was incubated at 37°C in air with 5% CO2 for 90 min. At the end of the incubation, filters were removed and stained with Diff-Quik (Baxter), and 10 high-power oil immersion fields were counted. Cancer cell migration was assayed as described previously (24) by using Falcon transwells (24-well format, 8-μM pore; BD Biosciences). 0.5 ml of media containing 5 × 105 cells was added to the upper chamber, and 0.5 ml of medium alone or media supplemented with CXCL12 was added to the lower chamber. After overnight incubation in hypoxic conditions, cells on the upper surface of the filter were removed using a cotton wool swab. Migrated cells on the lower surface were stained using DiffQuick (Dade Behring). For each transwell, the number of migrated cells in 10 medium-power fields (20×) was counted. For human umbilical vein endothelial cell (HUVEC) migration (25), polycarbonate filters (5 μm pore size, polyvinylpyrrolidine free) were soaked in 0.5 M acetic acid, washed with PBS, incubated for 24 h in 0.01% gelatin (Sigma-Aldrich), and air dried. CXCL12 and fibrinogen (Sigma-Aldrich) in M199, containing 1% FCS, were seeded in the lower compartment, and 50 μl of HUVECs (2 × 106/ml) was added to the upper compartment. After 6 h of incubation at 37°C, the upper surface of the filter was scraped with a rubber policeman. The filters were fixed and stained, and five oil immersion fields (lower surface) were counted after coding samples.
Northern Blot Analysis.
Cells were cultured in medium alone or supplemented with the indicated agents and total RNA was purified as described previously (10). 10 μg of total RNA from each sample was electrophoresed under denaturing conditions, blotted onto Nytran membranes (Schleicher & Schuell), and cross-linked by UV irradiation. Membranes were prehybridized at 42°C in hybrisol (Oncor, Inc.) and hybridized overnight with 106 cpm/ml of 32P-labeled probe. Membranes were washed three times at room temperature for 10 min in 0.2× SSC (1× SSC = 0.15 M NaCl, 0.015 M of sodium citrate, pH 7.0), 0.1% SDS, and twice at 60°C for 20 min in 0.2× SSC, 0.1% SDS before being autoradiographed using films and intensifier screens at −80°C (XAR-5; Kodak). cDNAs were labeled by random priming using a commercial kit (Boehringer) and α-[32P]dCTP (3,000 Ci/mmol; Amersham Biosciences). CCR1 and CCR5 cDNAs were obtained as described previously (11). CXCR4 cDNA was provided by T.N.C. Wells (Serono Pharmaceutical Research Institute, Geneva, Switzerland). Densitometric analysis was performed with a scanning densitometer (model GS300; Hoefer Scientific Instruments).
Transient Transfection.
DNA plasmids were prepared using a commercially available kit (Endofree Maxi-Prep; QIAGEN). Transfections were performed using effectene transfection reagents (QIAGEN) according to the manufacturer's instructions. Cells were seeded at a concentration of 5 × 104 per well in 48-well plates the day before transfection. 24 h after transfection, reagents were removed, and cells were allowed to recover for 8 h before being treated for 16–24 h. Cotransfection experiments were performed using a 1:1 ratio between the reporter plasmid and HIF-1α expression vector. Luciferase reporter assays were performed in 96-well optiplates (Packard Instrument Co.) using a luciferase assay system (Promega) according to the manufacturer's instructions. Results were normalized for the protein content using the protein assay (Bio-Rad Laboratories). Reporter gene assay pGL–Hyp responsive element (HRE) plasmid contains three copies of the canonical HRE (5′-GTGACTACGTGCTGCCTAG-3′) from the inducible nitric oxide synthase promoter (26). pCXCR4 plasmid, containing a 2.6-kb fragment from the human CXCR4 promoter upstream of the luciferase reporter gene, was obtained from A.J. Caruz (Universidad de Jaen, Madrid, Spain; reference 27). HIF-1 α (ODD−) expression vector was obtained from E. Huang (Brigham and Women's Hospital, Harvard Medical School, Boston, MA; reference 28). The pCMV(HA)–HIF-1α expression vector was obtained from D. Livingston (Dana Farber Cancer Institute, Boston, MA).
Chromatin Immunoprecipitation Assay (ChIP).
ChIP assays was performed in CAOV3 cells, transiently transfected with the p(HA)HIF-1α expression vector, and performed as described previously (29). In brief, 4 × 106 cells were fixed by adding directly to the medium formaldehyde (formaldehyde from a 37% formaldehyde/10% methanol stock; Calbiochem) to a final concentration of 1%. After 10 min, ice-cold PBS was immediately added, plates were transferred on ice and washed extensively with PBS, and cells were collected. After centrifugation, cells were lysed for 5 min in L1 buffer (50 mM Tris, pH 8.0, 2 mM EDTA, 0.1% NP-40, and 10% Glycerol) supplemented with protease inhibitors. Nuclei were pelleted at 3,000 rpm in microfuge and resuspended in L2 buffer (50 mM Tris, pH 8.0, 1% SDS, and 5 mM EDTA). Chromatin was sheared by sonication (5 × 10 s at one fifth of the maximum potency in a Sonics vibracell VC13 equipped with a 3-mm tip), centrifuged to pellet debris, and diluted ten times in dilution buffer (50 mM Tris, pH 8.0, 0.5% NP-40, 0.2 M NaCl, and 0.5 mM EDTA). Extracts were precleared for 2 h with 80 μl of a 50% suspension of salmon sperm DNA–saturated protein A. Immunoprecipitations were performed at 4°C overnight with 2 μg of polyclonal anti–human hemagglutin antibody (Santa Cruz Biotechnology, Inc.). Immune complexes were collected with salmon sperm DNA–saturated protein A, and washed three times (5 min each) with high salt buffer (washing buffer: 20 mM Tris, pH 8.0, 0.1% SDS, 1% NP-40, 2 mM EDTA, and 500 mM NaCl), two times with a 0.5 M LiCl buffer, and three times with low salt buffer (1× TE). Immune complexes were extracted in 1× TE containing 2% SDS and protein; DNA cross-links were reverted by heating at 65°C for 6 h. After proteinase K digestion (100 μg, 1–2 h at 50°C), DNA was extracted by phenol, chloroform, and ethanol precipitated. Approximately 1/20 of the immunoprecipitated DNA was used in each PCR. Sequences of promoter-specific primers included the CXCR4 promoter region −1860 to −1578 as follows: CXCR4, sense, 5′-TCGTGCCAAAGCTTGTCCCTG-3′; and CXCR4, anti-sense, 5′-GCGGTAACCAATTCGCGAATAGTGC-3′.
Real-time PCR.
Total RNA from MEFs, 786.0, and WT2 cells was obtained using an RNA mini kit (QIAGEN). RT-PCR was performed using a RT-PCR kit (PE Biosystems) as described previously (30). To measure the human vascular endothelial growth factor (VEGF), human CXCR4 and mouse CXCR4 expression real-time PCR was performed using a sequence detector (ABI-Prism, model 7700; Applied Biosystems). The following primers were used: human CXCR4, forward, 5′-GCA-TGACGGACAAGTACAGGCT-3′, reverse, 5′-AAAGTACCAGTTTGCCACGGC-3′; and mouse CXCR4, forward, 5′-TTGTCCACGCCACCAACAGTCA-3′, reverse, 5′-TGAAACACCACCATCCACAGGC-3′. Detection of 18S rRNA, used as internal control, was performed using premixed reagents from Applied Biosystems. Detection of VEGF and 18S rRNA was performed using a PCR master mix (TaqMan Universal; Applied Biosystems) and CXCR4 detection was also performed using a PCR master mix (SyBr Green; Applied Biosystems). Detection of SDF-1/CCL12 expression by the MCF7 and CAOV3 cell lines was performed by using a PCR master mix (SyBr Green; Applied Biosystem), and the following primers were used: human CXCL12, forward, 5′-ACACTCCAAACTGTGCCCTTCA-3′; and human CXCL12, reverse, 5′-CCACGTCTTTGCCCTTTCATC-3′.
Laser Confocal Microscopic Analysis of CXCR4 Expression.
Cells growing on sterile coverslips were washed with PBS after 16-h incubation in Hyp or Norm conditions as indicated in the text. Non-specific sites were blocked by 10-min incubation at room temperature with wash buffer containing 0.9% wt/vol sodium chloride, 1% vol/vol human serum, and 0.02% wt/vol sodium azide. Thereafter, the cells were stained with mouse anti–human CXCR4 antibodies (BD Biosciences) at a dilution of 1:20 for 30 min at room temperature. The cells were washed twice with 2 ml of wash buffer and incubated with goat F(ab′)2 anti–mouse Ig-FITC (human adsorbed) at a dilution of 1:20 for another 30 min at room temperature. After two changes in wash buffer, the cells were fixed in 4% wt/vol paraformaldehyde for 15 min, and the coverslips were mounted on glass slides for microscopy. Images were visualized using a system with differential interference contrast (FV500; Olympus).
Results
Hyp-increased CXCR4 Expression in Mononuclear Phagocytes.
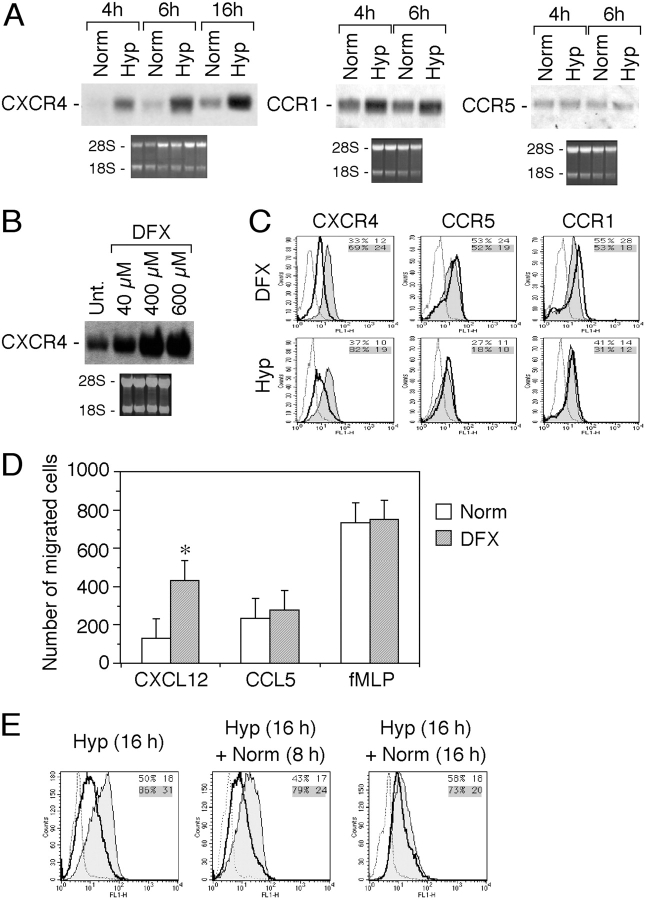
As shown in Fig. 1 A, human monocytes cultured in Hyp, for periods of 4 and 6 h, showed a strong increase in the expression of CXCR4 mRNA, as compared with normoxia (Norm)-cultured monocytes (20% oxygen). Hyp-mediated up-regulation of CXCR4 mRNA was still present at 16 h. In Hyp, a slight increase was also observed for CCR1 mRNA expression, whereas CCR5 mRNA level was unaffected. Moreover, Hyp down-regulates responsiveness of monocytes to CCR2 agonists (4, 5). Therefore, we focused our attention on the CXCR4 receptor. The iron chelator DFX is recognized as an Hyp-mimicking compound (30). Incubation of fresh human monocytes for 4 h in the presence of DFX resulted in a dose-dependent increase of the CXCR4 mRNA levels (Fig. 1 B). Together, these results show that CXCR4 mRNA expression is selectively controlled by changes in oxygen levels. Modulation of CXCR4 by Hyp was confirmed at the protein level by flow cytometry. To evaluate the functional effects of Hyp on CXCR4, monocytes were incubated in hypoxic conditions and CXCR4 surface expression was determined. As shown in Fig. 1 C, exposure of monocytes for 16 h to Hyp resulted in a strong increase of CXCR4 surface expression. Similarly, cells incubated in the presence of 400 μM DFX for the same duration showed a significant increase of CXCR4 surface expression. In contrast, neither Hyp nor DFX treatment affected CCR1 and CCR5 surface expression. The observed DFX-dependent increase of CXCR4 surface expression was paralleled by a rise in the number of monocytes migrating in response to CXCL12 (Fig. 1 D). The increased chemotactic responsiveness toward CXCL12 was specific, whereas both CCL5 (regulated on activation normal T cell expressed and secreted) and FMLP-induced migration were not significantly affected. Thus, in human monocytes, low oxygen conditions result in a specific up-regulation of the CXCR4 expression and function. As it was important to assess the reversibility of Hyp on the induction of CXCR4 expression, we determined the effect of reoxygenation on the levels of CXCR4 surface expression in fresh human monocytes. As shown in Fig. 1 E, after culture in hypoxic conditions for 16 h and reoxygenation, monocytes retained high levels of CXCR4 for 8 h. Higher levels than control were still present at 16 h, and returned to baseline by 24 h (unpublished data).
Figure 1.
Effect of hypoxia (Hyp) on CXCR4 expression by fresh human monocytes. (A) Fresh human monocytes obtained from peripheral blood of healthy donors were cultured for different times in normoxia (Norm) or hypoxic (Hyp) conditions, as indicated. Total RNA was analyzed by Northern blot for CXCR4, CCR1, and CCR5 mRNA expression. (B) Total RNA from fresh human monocytes cultured for 4 h in the presence of increasing concentration of DFX was analyzed by Northern blot. (C) Hyp- and DFX-induced CXCR4 surface expression in fresh human monocytes. Cells were cultured for 16 h in the indicated conditions and analyzed for CXCR4 surface expression. Surface expression was determined by flow cytometry using a mouse monoclonal antibody anti–human CXCR4. The results are representative of three independent experiments. (dotted line) Irrelevant antibody. (continuous line) Norm. (shaded region) Hyp or DFX. (D) Effect of DFX on the chemotactic response of monocytes to 100 ng/ml CXCL12, 100 ng/ml CCL5, and 10−8 M FMLP. Cells were cultured for 16 h in the presence of 400 μM DFX. Migration of monocytes was assayed by chemotaxis microchamber technique. Results are mean ± SD of five experiments. *, P < 0.05 versus cells cultured in normoxic conditions (paired Student's t test). (E) Effect of reoxygenation on CXCR4 surface expression. Fresh human monocytes were cultured for 16 h in Hyp and subsequently reexposed to Norm for the indicated times. CXCR4 surface expression was determined by flow cytometry.
Monocytes differentiate into macrophages during infiltration of tissues. As tissues are characterized by lower oxygen concentrations in comparison with peripheral circulation (1), it was important to determine whether CXCR4 up-regulation in response to low oxygen conditions could be applied also to macrophages. CXCR4 expression in Hyp was determined in fresh human monocytes, in vitro–differentiated MDMs, and TAMs obtained from the ascitic fluid of human ovarian carcinoma. Cells were cultured for 4 h in Norm or Hyp, and total RNA was analyzed for CXCR4 mRNA expression by Northern blot. As shown in Fig. 2 , hypoxic conditions resulted in a strong up-regulation of the CXCR4 mRNA levels in all the three cell populations (Fig. 2 A). These results were confirmed by laser confocal microscopy Fig. 2 (B and C), wherein Hyp induced a strong up-regulation of CXCR4 surface expression, in both MDMs and TAMs. Clearly, Hyp controls CXCR4 expression in mononuclear phagocytes at different stages of differentiation.
Figure 2.
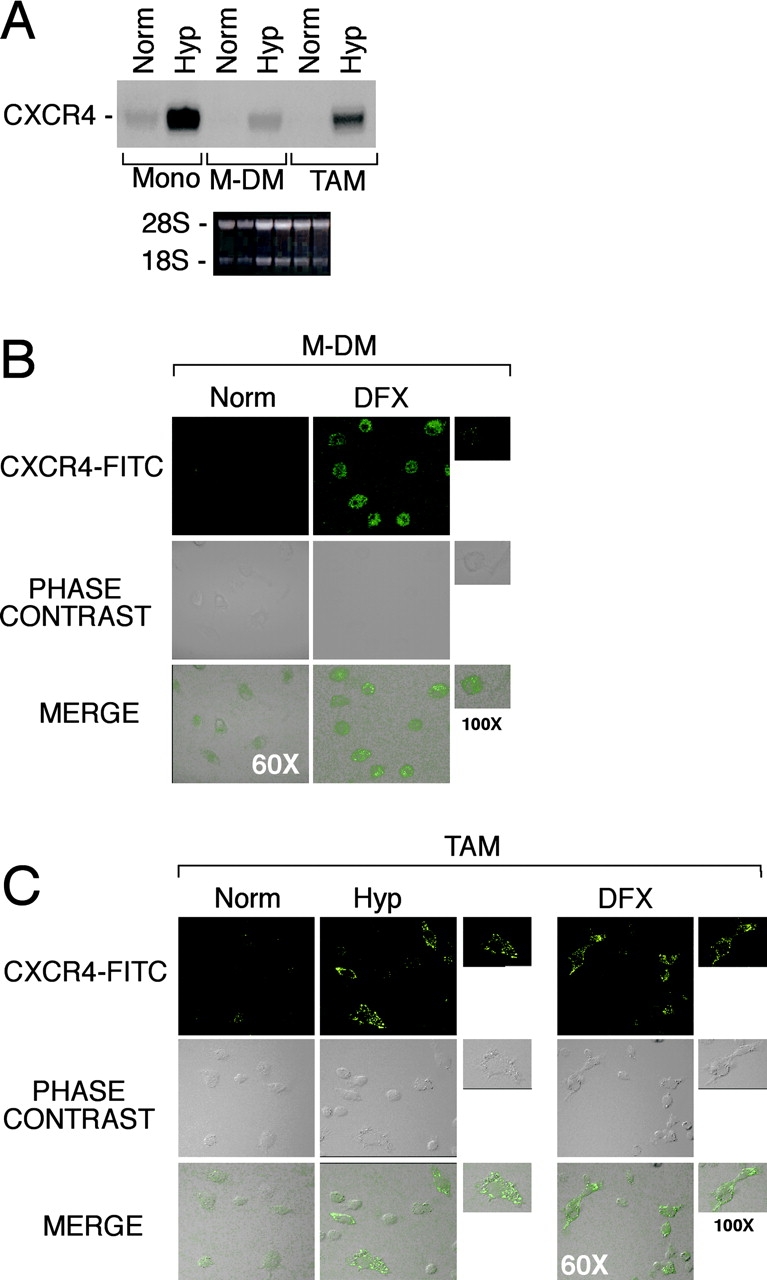
Effect of Hyp on CXCR4 expression by fresh human monocytes (mono), monocyte-derived macrophages (MDMs), and tumor-associated macrophages (TAMs). (A) Cells were cultured for 4 h in Hyp, and total RNA was analyzed by Northern blot for CXCR4 mRNA expression. MDM (B) and TAMs from ascitic fluid of human ovarian cancer (C) were cultured for 16 h in Norm, Hyp, or in the presence of 400 μM DFX, as indicated. CXCR4 surface staining was performed and detected by laser confocal microscopy. Staining with both isotype-matched control antibodies was done for all samples (not depicted).
Hyp-increased CXCR4 Expression in HUVECs.
CXCL12 is a potent chemoattractant for endothelial cells of different origin and participates in angiogenesis (31, 32). We investigated the effect of Hyp on CXCR4 expression by endothelial cells. As shown in Fig. 3 , HUVECs, cultured in Hyp or in the presence of 400 μM DFX for 4–16 h, showed increased CXCR4 mRNA expression, as compared with normoxic conditions. VEGF gene expression is induced by Hyp (1) and served as an internal control. As expected, VEGF mRNA levels were up-regulated either by Hyp or DFX. Next, CXCR4 surface expression was analyzed by laser confocal microscopy. As shown in Fig. 3 B, after Hyp and DFX treatment, CXCR4 surface expression in endothelial cells was strongly increased. Endothelial cell recruitment represents an initial step of the process of angiogenesis. HUVECs cultured in Norm or Hyp were tested for their capability to migrate in response to CXCL12. As shown in Fig. 3 C, the number of migrated HUVECs in response to CXCL12 was significantly higher in hypoxic conditions, at concentrations ranging from 1 to 100 ng/ml. CXCL12 was ∼100-fold more effective at eliciting HUVEC migration under Hyp than under normoxic conditions. Migration elicited by fibrinogen, used as a control, was not affected.
Figure 3.
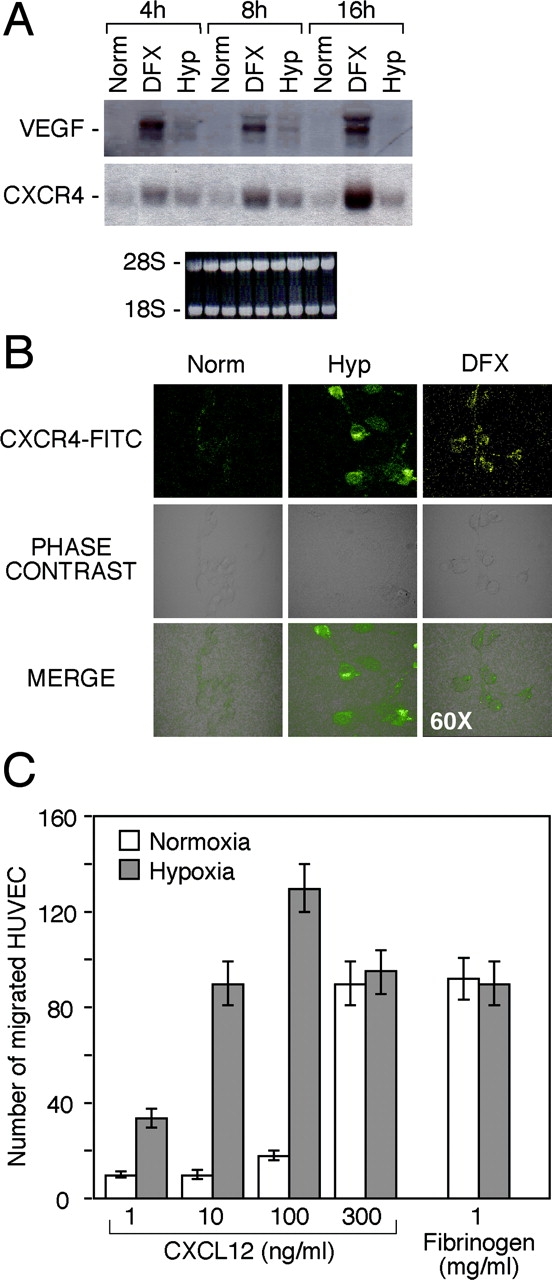
Effect of Hyp on CXCR4 expression by human endothelial venules (HUVECs). (A) Cells were cultured for 4, 8, and 16 h under Norm, Hyp, or in the presence of 400 μM DFX, respectively. Thereafter, total RNA was analyzed by Northern blot for CXCR4 and VEGF mRNAs expression. (B) HUVECs were cultured for 16 h in Norm, Hyp, and in the presence of 400 μM DFX, as indicated. CXCR4 surface staining was performed and detected by laser confocal microscopy. (C) Effect of Hyp on the chemotactic response of HUVECs to CXCL12. Cells were cultured for 16 h in hypoxic conditions. Fibrinogen was used as a reference attractant. Results are mean ± SD of three experiments.
Hyp-increased CXCR4 Expression in Cancer Cells.
Chemokine receptors may act as molecular tools exploited by cancer cells to metastasize to target organs (33). In particular, CXCR4 was shown to play a major role in the migration of breast cancer cells from the primary tumor to secondary metastatic sites, such as lung, liver, and bone. As solid tumors are often characterized by the presence of necrotic areas with low oxygen tension, we reasoned that Hyp may be a potential mechanism that up-regulates expression of chemokine receptors in cancer cells localized in poorly avascularized and oxygenated areas of tumors. To test this hypothesis, we investigated the level of CXCR4 mRNA expression in response to Hyp in the ovarian cancer cell line CAOV3 (Fig. 4) . As shown in Fig. 4 A, CAOV3 cells showed up-regulation of CXCR4 mRNA after 4 h of culture under hypoxic conditions, which was paralleled by a significant increase in the surface expression of this receptor, as demonstrated by cytofluorimetric analysis (Fig. 4 B). The Hyp-induced expression of CXCR4 in the CAOV3 cells correlated with an increased migration of these cells toward CXCL12 in the chemotaxis assay (Fig. 4 C). Similar results were obtained with the MCF-7 breast cancer cell line (unpublished data). Fig. 4 D shows the effect of Hyp on CXCL12 expression by the same cancer cell lines MCF7 and CAOV3.
Figure 4.
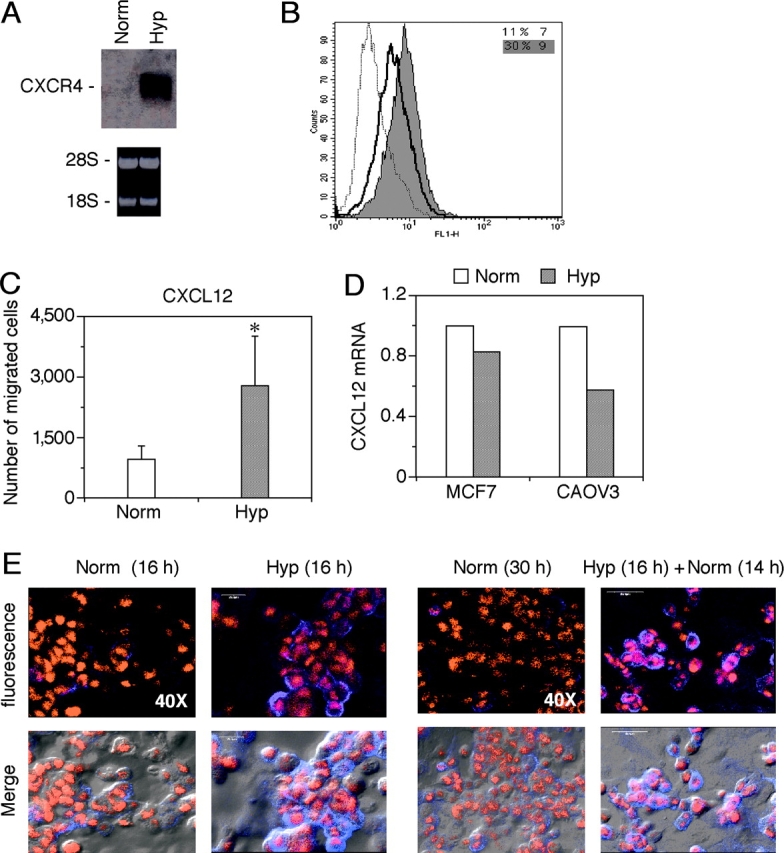
Effects of Hyp on CXCR4 expression by cancer cells. The ovarian cancer cell line CAOV3 was cultured in normoxic or hypoxic conditions and analyzed respectively for both CXCR4 mRNA expression (A) and CXCR4 surface expression (B). (A) cells were cultured for 4 h in normoxia (Norm) or hypoxic (Hyp) conditions, as indicated. Total RNA was analyzed by Northern blot for CXCR4. (B) Cells were cultured for 16 h in Norm or hypoxic (Hyp) conditions. After this period, CXCR4 surface expression was determined by flow cytometry. (dotted line) Irrelevant antibody. (continuous line) Norm. (shaded region) Hyp. (C) Effect of Hyp on the chemotactic response of CAOV3 ovarian cancer cells to 100 ng/ml CXCL12. Cells were incubated overnight in Hyp condition and migration determined by Transwells, as described in Materials and Methods. Results are mean ± SD of three experiments. *, P < 0.05 versus cells cultured in normoxic conditions (Paired Student's t test). (D) Effects of Hyp on CXCL12 mRNA expression by MCF-7 and CAOV3 cells. Cells were incubated for 4 h in Norm and Hyp as indicated, and CXCR4 gene expression was next determined by real-time PCR. Results are representative of two independent experiments. Values are expressed as fold increases relative to the reference sample (Norm). (E) Sustained Hyp-induced CXCR4 expression upon reoxygenation. CAOV3 cells were cultured under Norm or Hyp conditions for 16 h. Thereafter, the cells were exposed to Norm for a further 14 h and stained for CXCR4 expression. The figure shows a representative field using confocal microscopy. CAOV3 cells stained for CXCR4 (blue fluorescence) and nuclei (red fluorescence). The bottom panels in each group shows phase-contrast images merged with fluorescence readings.
CXCR4 expression by cancer cells may play an important role in the metastatic process (33). The process of metastasis formation includes the exit of cancer cells from the primary tumor site and their entrance into the circulation, where the oxygen levels will markedly increase, relative to those present in the tumor microenvironment. To assess the impact that the exposure of cancer cells to reoxygenation had on the CXCR4 surface expression, CAOV3 cells were cultured in hypoxic conditions for 16 h and subsequently exposed to Norm for up to 14 h. As shown in Fig. 4 E, Hyp-increased CXCR4 surface expression in CAOV3 cells was retained after reoxygenation for 14 h and returned to baseline by 24 h (not depicted).
Involvement of HIF-1α in Hyp-induced Expression of CXCR4.
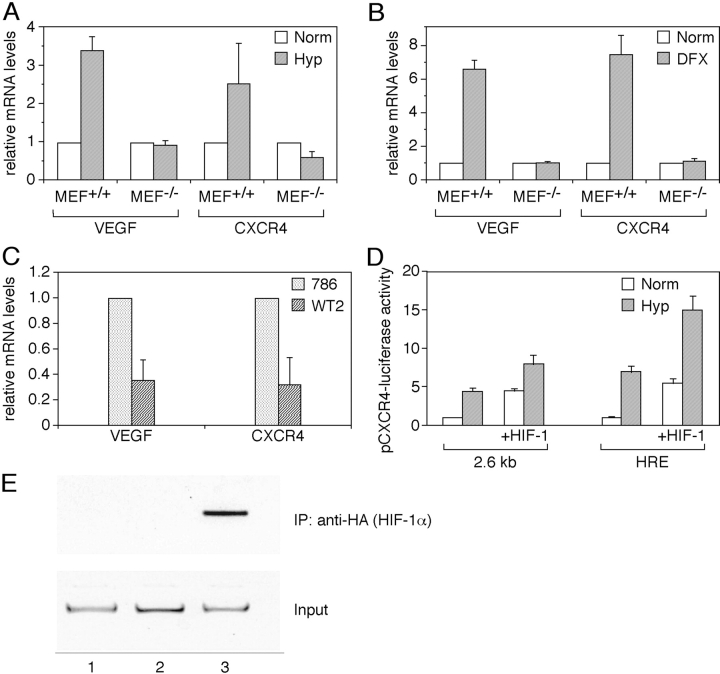
HIF-1 is a key regulator of the transcriptional response to Hyp. To investigate the role of HIF-1 in the hypoxic induction of CXCR4, MEFs from WT or deficient animals for the α subunit of HIF-1 were incubated under normoxic or hypoxic conditions for 6 h, and total RNA was tested for VEGF and CXCR4 mRNA levels by real-time PCR. As shown in Fig. 5 A, MEFs from WT animals, but not from HIF-1α KO mice, expressed 3.5-fold-higher levels of VEGF mRNA when incubated under hypoxic conditions relative to levels expressed in normoxic conditions. Likewise, MEFs from WT mice expressed 2.5-fold-higher levels of CXCR4 mRNA when cultured under hypoxic conditions relative to normoxic conditions. In contrast, in MEFs from HIF-1α−/− mice, the hypoxic induction of CXCR4 mRNA was no longer apparent. Furthermore, DFX also induced VEGF and CXCR4 mRNA in HIF-1 WT cells (6.5- and 7.2-fold, respectively) but not in HIF-1α−/− cells (Fig. 5 B). These data suggest a role for HIF-1 in the induction of CXCR4 mRNA expression by Hyp.
Figure 5.
Role of HIF-1α in the regulation of CXCR4 gene expression. (A) Expression of CXCR4 in HIF-1α KO mouse embryo fibroblast. Mouse embryo fibroblast from wild type (MEF+/+) or knockout for the α subunit of HIF-1 (MEF−/−) were incubated under normoxic or hypoxic conditions for 6 h, and total RNA was tested for VEGF and CXCR4 mRNA levels by real-time PCR. (B) DFX was used as Hyp-inducing agent. Results are the average of three independent experiments. (C) Expression of CXCR4 in VHL WT and mutated renal carcinoma cells. Expression of CXCR4 and VEGF mRNAs was tested by real-time PCR in the renal cancer cell line 786.0 (VHL mutated) and WT2 (in which a WT VHL has been reintroduced). Results are the average of three independent experiments. (D) HIF-1–dependent transcriptional activation of CXCR4 promoter. MCF-7 breast carcinoma cells were transiently transfected with a plasmid containing a 2.6 kb fragment of the CXCR4 promoter linked to the luciferase reporter gene, with or without a HIF-1α expression vector. Cells were incubated under normoxic or hypoxic conditions for 24 h and evaluated for the luciferase activity. Results are the average of three independent experiments. (E) Hyp-induced HIF-1α recruitment to the CXCR4 promoter. CAOV3 cells transfected with the p(HA)HIF-1α plasmid were cultured for 4 h in normoxic or hypoxic conditions. ChIP was performed to investigate the recruitment of HIF-1α on the CXCR4 promoter. (lane 1) Untransfected. (lane 2) Norm. (lane 3) Hyp.
The von Hippel–Lindau tumor suppressor protein (VHL) is involved in the degradation of HIF-α; mutations of VHLs are associated with high levels of HIF-1α protein and transcriptional activity (34, 35). Therefore, we tested the expression of CXCR4 mRNA in the renal cancer cell line 786.0 (VHL mutated; reference 36) and the WT2 cell line (in which a WT VHL has been reintroduced). These cells have been extensively used in several laboratories as prototype in which high levels of HIF-1 activity are associated with increased levels of expression of HIF-1–inducible genes (23). As reported previously (37), 786.0 cells expressed high constitutive levels of VEGF mRNA that were significantly lower (>60%) in WT2 cells (Fig. 5 C). Accordingly, 786.0 cells expressed higher levels of CXCR4 mRNA relative to WT2 cells, in which we observed >65% reduction in three independent experiments. These data again are consistent with the involvement of HIF-1 in the hypoxic regulation of CXCR4 mRNA expression. We investigated the role of HIF-1 in the transcriptional activation of CXCR4 promoter. MCF-7 breast carcinoma cells, in which Hyp augments expression of CXCR4 mRNA (unpublished data), were transiently transfected with a plasmids containing a 2.6-kb fragment of the CXCR4 promoter linked to the luciferase reporter gene and incubated under normoxic or hypoxic conditions for 24 h. As shown in Fig. 5 D, Hyp induced fourfold higher levels of luciferase expression relative to cells cultured under normoxic conditions. Interestingly, cotransfection of a HIF-1α expression vector significantly increased luciferase expression driven by the 2.6-kb CXCR4 promoter under normoxic conditions (approximately fourfold relative to control). Hyp further increased luciferase expression induced by HIF-1α cotransfection up to ninefold compared to untreated normoxic cells. Experiments conducted in parallel using a plasmid containing three copies of a canonical Hyp-responsive element upstream of the luciferase reporter gene showed a similar trend of luciferase expression, although levels of induction were substantially higher.
To obtain direct evidence for the interaction between HIF-1α and the CXCR4 promoter, we used the ChIP assay to measure the HIF-1α recruitment to the CXCR4 promoter. CAOV3 cells, either cultured in Norm or Hyp for different times, were fixed in formaldehyde, and subsequently analyzed by ChIP. Although no interaction between HIF-1α and the CXCR4 promoter was observed in Norm, recruitment of HIF-1α to the CXCR4 promoter was clearly detected at 4 h after Hyp, in the promoter region −1860 to −1578 (Fig. 5 E). This result is consistent with the functional data obtained in transient transfection experiments (Fig. 5 D). Moreover, the HIF-1α inhibitor Topotecan (38), was able to prevent the Hyp-induced up-regulation of CXCR4 in CAOV3 cells (unpublished data). Overall these data demonstrate the involvement of HIF-1α in the induction of CXCR4 promoter.
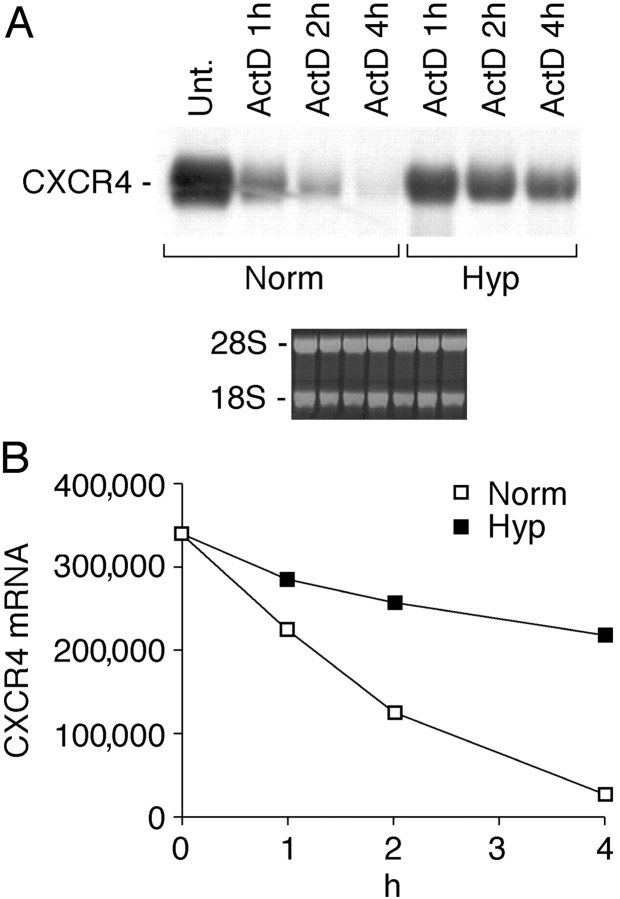
Effect of Hyp and DFX on CXCR4 mRNA Stability.
To further investigate the mechanisms of Hyp action, we estimated its effects on CXCR4 mRNA stability (Fig. 6) . Fresh human monocytes were cultured either in Norm or Hyp in the presence or absence of 1 μg/ml actinomycin D. Total RNA was extracted at different times as indicated. The mRNA decay observed in Hyp conditions was compared with the rate of mRNA degradation observed in normoxic conditions. The results indicate that Hyp increased CXCR4 mRNA stability, suggesting that Hyp-induced CXCR4 mRNA expression relies on both transcriptional and posttranscriptional mechanisms.
Figure 6.
Stabilization of CXCR4 mRNA by Hyp. (A) Fresh human monocytes were cultured for 4 h under Norm or Hyp, and in the presence or absence of 1 μg/ml actinomycin D (ActD). Thereafter, total RNA was extracted at different times as indicated and analyzed by Northern blot for CXCR4 mRNA expression. (B) Densitometric analysis: CXCR4 mRNA levels are expressed as arbitrary units. In this experiment, the basal CXCR4 mRNA level appears higher compared with other experiments, as we exposed the film for a longer period for better visualization of the blot.
Discussion
Regulation of cell migration by changes in oxygen availability is a central event during the organization of host response in inflammatory and neoplastic diseases as it may influence leukocyte recruitment and activation, angiogenesis, and metastasis formation (16). Here, we report that Hyp mediates selective up-regulation of CXCR4 in different cell types, including mononuclear phagocytes (monocytes, MDMs, and TAMs), endothelial cells, and cancer cells, and demonstrate that oxygen levels act as an important regulator of CXCR4 receptor expression. Our data also indicate that HIF-1 activation is involved in the Hyp-dependent up-regulation of CXCR4 expression and that the Hyp–HIF-1–CXCR4 circuit may participate in pathophysiological mechanisms under several conditions, ranging from inflammation to tumor angiogenesis and metastasis.
In contrast to standard cell culture conditions, characterized by 20% oxygen concentration, cells in the human body are exposed to much lower oxygen concentrations, ranging from 16% in the pulmonary alveoli to <6% in most other organs of the body. Moreover, oxygen concentration may even drop to extremely low concentrations, close to anoxia, in the presence of altered vascularization as observed at pathological sites such as tumors (1). As selective accumulation of leukocyte subpopulations is the hallmark in allergy, inflammation, and tumors (18), it was important to investigate how leukocyte recruitment is affected by changes in oxygen tension. We first observed that in response to Hyp monocytes and MDM increase, CXCR4 expression and function, as assessed by surface expression and chemotactic responsiveness to its specific ligand CXCL12. Thus, dynamic regulation of the chemotactic responsiveness of monocytes/macrophages may represent a feature of the pathophysiology of inflammatory diseases associated with Hyp. Interestingly, Hyp is present in the joint microenvironment, because articular cartilage is an avascular tissue that functions at lower oxygen tension than do most tissues. Moreover, in the setting of diseases such as rheumatoid arthritis and osteoarthritis, in which macrophages promote perpetuation of chronic inflammation (39), a further decrease in synovial fluid oxygen tension may occur (40, 41).
In solid tumors, TAMs represent a prominent component of the mononuclear leukocyte population, which displays an ambivalent relationship with tumors (the “macrophage balance hypothesis”; reference 17). Interestingly, TAMs preferentially localize at the tumor–host tissue interface, in regions often associated with low oxygen tensions. Several lines of evidence also indicate that chemokines play a pivotal role in the recruitment of monocytes in neoplastic tissues (17, 18, 42) and a variety of chemokines have been detected as products of cancer cells or tumor stromal elements. In particular, CCL2 was proposed as tumor-derived chemotactic factors, which play a major role in the recruitment of macrophages at the tumor site (18). Receptor expression is a crucial determinant of the spectrum of action of chemokines (18). It was reported that the capacity of monocytes/macrophages to migrate in response to CCL2 is decreased in low oxygen conditions (4, 5). Thus, our observation of Hyp-mediated up-regulation of CXCR4 expression in TAMs may indicate that in regions associated with oxygen decrease, a dynamic change of their receptor profile occurs, with up-regulation of functional CXCR4. In support of this hypothesis, Cramer et al. have recently shown that myeloid cell infiltration in vivo is dependent on the presence of an intact HIF-1α subunit, suggesting that oxygen gradient may be a critical factor for myeloid cells' migration in inflammatory sites (43). A relay of distinct chemokine–chemokine receptor interactions may regulate initial recruitment, tissue infiltration in hypoxic areas, and in neoplastic and non-neoplastic inflammatory sites in a multistep navigation process (44).
Angiogenesis is a prerequisite for the expansion of solid tumors and is often activated during the early, preneoplastic stages of tumor development (45, 46). Tumor angiogenesis is controlled by a number of positive and negative regulators produced by cancer cells and tumor-associated leukocytes. A number of molecules with possible impact on angiogenesis have been shown to be expressed by macrophages in low oxygen conditions, such as VEGF, TNF-α, bFGF, and CXCL8 (47). The contribution of chemokines toward angiogenesis is currently a focus of intensive investigation (48). Strikingly, it was recently reported that CXCL12 acts as a potent chemoattractant for endothelial cells of different origins bearing CXCR4 and is a participant in angiogenesis that is regulated at the receptor level by VEGF and bFGF (31, 49–51). In agreement with these observations, we observed an increased chemotactic responsiveness of HUVECs toward CXCL12, which may well be part of the Hyp-induced angiogenic program. Hyp is a well-recognized pathophysiological condition for the induction of angiogenic factors, including but not limited to VEGF (16). In agreement with these observations, our data suggest that the angiogenic program established by Hyp may rely also on the increased expression of CXCR4 by different cellular components in the tumor microenvironment, including endothelial cells, tumor cells, and TAMs.
The involvement of CXCR4 in cancer metastasis has been proposed by Muller and colleagues, who showed that this receptor and its ligand (CXCL12) together govern the pattern of breast cancer metastasis in a mouse model (33, 52). This observation is in support of the “chemoattraction” theory of metastasis, which holds that organ-specific attractant molecules stimulate the migrating tumor cells to invade the walls in blood vessels and enter the organs. However, this remarkable observation does not clarify the mechanisms of selection by which cancer cells became CXCR4 positive. Our observation that the levels of CXCR4 surface expression induced by Hyp are sustained for several hours after reoxygenation is consistent with the idea that this pathway may confer metastatic potential to cancer cells. Indeed, it was described in in vivo models of metastasis that after entering the circulation, the majority of cancer cells home to target organs in a timeframe ranging from 1 to 24 h (53). We propose that in solid tumors, in addition to genetic alterations such as mutation of VHL, PTEN, or p53 genes that are associated with increased levels of HIF-1 transcriptional activity, microenvironmental Hyp may increase CXCR4 expression and the metastatic potential of cancer cells. In line with this hypothesis, recent evidence has been provided that invasive cancer phenotype is associated with Hyp and/or HIF-1α overexpression (54). In agreement with the data and concepts described here, by using immunohistochemical analysis, we have observed that ductal carcinoma cells from breast tissue located in areas of intratumoral necrosis display nuclear HIF-1α expression and high levels of CXCR4 (unpublished data).
HIF-1 activates transcription of genes that mediate adaptive responses to reduced oxygen availability and a number of HIF-1–regulated genes have been identified, whose products play key roles in angiogenesis, vascular reactivity and remodelling, and glucose and energy metabolism (1). HIF-1 is a heterodimer composed of a HIF-1β subunit that is constitutively expressed and a HIF-1α subunit that is rapidly degraded by ubiquitination via the proteasomal pathway, a process that is inhibited under hypoxic conditions. Oxygen-regulated destruction of HIF-1α requires the von Hippel–Lindau tumor suppressor protein (pVHL) (34, 35). pVHL acts as the recognition component of a ubiquitin E3 ligase complex, which binds HIF-1α, and loss of pVHL function results in constitutive activation of the hypoxic response (35, 55). A role for HIF-1 in the regulation of CXCR4 mRNA expression is suggested by the following findings: (a) mouse embryonal fibroblast lacking the α subunit of HIF-1 had impaired hypoxic induction of CXCR4 mRNA; (b) CXCR4 mRNA was differentially expressed in the renal cancer cell lines 786.0 and WT2 bearing a VHL-mutated and WT phenotype, respectively; (c) Hyp or cotransfection of a HIF-1α expression vector–induced transcriptional activation of a 2.6-kb CXCR4 promoter luciferase reporter construct; (d) ChIP analysis demonstrated that after Hyp, HIF-1α is specifically recruited to the CXCR4 promoter, in the nucleotide region −1860 to −1578; and (e) the HIF-1 inhibitor Topotecan inhibited the Hyp-induced expression of CXCR4 in CAOV3 cells. A sequence homology search of the region −1860 to −1578 of the CXCR4 promoter revealed the presence of a putative HIF-1 binding site at position −1725 (5′-GCGTG-3′). Ongoing studies in the laboratory will provide a full biochemical and functional characterization of the HRE. In addition to transcriptional activation, we found that Hyp further contributes to increased CXCR4 gene expression by stabilization of CXCR4 transcripts, suggesting that Hyp-regulated RNA binding factors may interact with and stabilize the CXCR4 mRNA at the posttranscriptional level.
Our data identify the Hyp–HIF-1–CXCR4 pathway as a relevant molecular circuit in the functional tuning of the chemokine system. The validity of this observation in different cell types (mononuclear phagocytes, HUVECs, fibroblasts, and cancer cells), consistent with the virtually universal expression of the Hyp–HIF-1 pathway in mammalian cells, argues in favor of its potential involvement in the pathophysiology of diverse conditions. In a multistep navigation process, the Hyp–HIF-1α–CXCR4 pathway may regulate trafficking and localization in hypoxic tissues and represents a target for novel therapeutic strategies.
Acknowledgments
We thank Dr. P. Allavena for critical revision of the manuscript. We thank M. Sironi, N. Polentarutti, and G. Balconi for technical support.
This work was supported by European Community and the Ministero Istruzione Università e Ricerca, Fondi per gli Investimenti della Ricerca di Base project. The contribution of the Italian Association for Cancer Research is acknowledged. This work was supported by the scholarship in memory of Maria Teresa Pignatelli, by a fellowship from the Alfredo Leonardi Fund, and G.L. Pfieffer Foundation. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organization imply endorsement by the U.S. government.
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; DFX, desferrioxamine; HIF-1, Hyp-inducible factor 1; HRE, Hyp responsive element; HUVEC, human umbilical vein endothelial cell; Hyp, hypoxia; MDM, monocyte-derived macrophage; MEF, mouse embryonic fibroblast; Norm, normoxia; pVHL, von Hippel–Lindau tumor suppressor protein; SDF-1, stromal-derived factor 1; TAM, tumor-associated macrophage; VEGF, vascular endothelial growth factor; VHL, von Hippel–Lindau tumor suppressor protein.
References
- 1.Semenza, G.L. 2001. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 7:345–350. [DOI] [PubMed] [Google Scholar]
- 2.Semenza, G.L. 1999. Perspectives on oxygen sensing. Cell. 98:281–284. [DOI] [PubMed] [Google Scholar]
- 3.Wang, G.L., B.H. Jiang, E.A. Rue, and G.L. Semenza. 1995. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA. 92:5510–5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimshaw, M.J., and F.R. Balkwill. 2001. Inhibition of monocyte and macrophage chemotaxis by hypoxia and inflammation–a potential mechanism. Eur. J. Immunol. 31:480–489. [DOI] [PubMed] [Google Scholar]
- 5.Turner, L., C. Scotton, R. Negus, and F. Balkwill. 1999. Hypoxia inhibits macrophage migration. Eur. J. Immunol. 29:2280–2287. [DOI] [PubMed] [Google Scholar]
- 6.Baggiolini, M. 1998. Chemokines and leukocyte traffic. Nature. 392:565–568. [DOI] [PubMed] [Google Scholar]
- 7.Hedrick, J.A., and A. Zlotnik. 1996. Chemokines and lymphocyte biology. Curr. Opin. Immunol. 8:343–347. [DOI] [PubMed] [Google Scholar]
- 8.Rollins, B.J. 1997. Chemokines. Blood. 90:909–928. [PubMed] [Google Scholar]
- 9.Murphy, P.M. 2002. International Union of Pharmacology. XXX. Update on chemokine receptor nomenclature. Pharmacol. Rev. 54:227–229. [DOI] [PubMed] [Google Scholar]
- 10.Sica, A., A. Saccani, A. Borsatti, C.A. Power, T.N. Wells, W. Luini, N. Polentarutti, S. Sozzani, and A. Mantovani. 1997. Bacterial lipopolysaccharide rapidly inhibits expression of C-C chemokine receptors in human monocytes. J. Exp. Med. 185:969–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sica, A., A. Saccani, B. Bottazzi, S. Bernasconi, P. Allavena, B. Gaetano, F. Fei, G. LaRosa, C. Scotton, F. Balkwill, and A. Mantovani. 2000. Defective expression of the monocyte chemotactic protein-1 receptor CCR2 in macrophages associated with human ovarian carcinoma. J. Immunol. 164:733–738. [DOI] [PubMed] [Google Scholar]
- 12.Sica, A., A. Saccani, and A. Mantovani. 2002. Tumor-associated macrophages: a molecular perspective. Int. Immunopharmacol. 2:1045–1054. [DOI] [PubMed] [Google Scholar]
- 13.Mantovani, A. 1999. The chemokine system: redundancy for robust outputs. Immunol. Today. 20:254–257. [DOI] [PubMed] [Google Scholar]
- 14.Madjdpour, C., U.R. Jewell, S. Kneller, U. Ziegler, R. Schwendener, C. Booy, L. Klausli, T. Pasch, R.C. Schimmer, and B. Beck-Schimmer. 2003. Decreased alveolar oxygen induces lung inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 284:360–367. [DOI] [PubMed] [Google Scholar]
- 15.Mapp, P.I., M.C. Grootveld, and D.R. Blake. 1995. Hypoxia, oxidative stress and rheumatoid arthritis. Br. Med. Bull. 51:419–436. [DOI] [PubMed] [Google Scholar]
- 16.Semenza, G.L. 2002. HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol. Med. 8:S62–S67. [DOI] [PubMed] [Google Scholar]
- 17.Mantovani, A., B. Bottazzi, F. Colotta, S. Sozzani, and L. Ruco. 1992. The origin and function of tumor-associated macrophages. Immunol. Today. 13:265–270. [DOI] [PubMed] [Google Scholar]
- 18.Mantovani, A., S. Sozzani, M. Locati, P. Allavena, and A. Sica. 2002. Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23:549–555. [DOI] [PubMed] [Google Scholar]
- 19.Leek, R.D., C.E. Lewis, R. Whitehouse, M. Greenall, J. Clarke, and A.L. Harris. 1996. Association of macrophage infiltration with angiogenesis and prognosis in invasive breast carcinoma. Cancer Res. 56:4625–4629. [PubMed] [Google Scholar]
- 20.Leek, R.D., R.J. Landers, A.L. Harris, and C.E. Lewis. 1999. Necrosis correlates with high vascular density and focal macrophage infiltration with angiogenesis and prognosis in invasive carcinoma of the breast. Br. J. Cancer. 79:991–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantovani, A., V. Caprioli, P. Gritti, and F. Spreafico. 1977. Human mature macrophages mediate antybody-dependent cellular cytotoxicity on tumor cells. Transplantation. 24:291–293. [DOI] [PubMed] [Google Scholar]
- 22.Romano, M., M. Sironi, C. Toniatti, N. Polentarutti, P. Fruscella, P. Ghezzi, R. Faggioni, W. Luini, V. van Hinsbergh, S. Sozzani, et al. 1997. Role of IL-6 and its soluble receptor in induction of chemokines and leukocyte recruitment. Immunity. 6:315–325. [DOI] [PubMed] [Google Scholar]
- 23.Ivanov, S.V., I. Kuzmin, M.H. Wei, S. Pack, L. Geil, B.E. Johnson, E.J. Stanbridge, and M.I. Lerman. 1998. Down-regulation of transmembrane carbonic anhydrases in renal cell carcinoma cell lines by wild-type von Hippel-Lindau transgenes. Proc. Natl. Acad. Sci. USA. 95:12596–12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scotton, C.J., J.L. Wilson, D. Milliken, G. Stamp, and F.R. Balkwill. 2001. Epithelial cancer cell migration: a role for chemokine receptors? Cancer Res. 61:4961–4965. [PubMed] [Google Scholar]
- 25.Colotta, F., F. Bussolino, N. Polentarutti, A. Guglielmetti, M. Sironi, E. Bocchietto, M. De Rossi, and A. Mantovani. 1993. Differential expression of the common b and specific a chains of the receptors for GM-CSF, IL-3, and IL-5 in endothelial cells. Exp. Cell Res. 206:311–317. [DOI] [PubMed] [Google Scholar]
- 26.Melillo, G., T. Musso, A. Sica, L.S. Taylor, G.W. Cox, and L. Varesio. 1995. A hypoxia-responsive element mediates a novel pathway of activation of the inducible nitric oxide synthase promoter. J. Exp. Med. 182:1683–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caruz, A., M. Samsom, J.M. Alonso, J. Alcami, F. Baleux, J.L. Virelizier, M. Parmentier, and F. Arenzana-Seisdedos. 1998. Genomic organization and promoter characterization of human CXCR4 gene. FEBS Lett. 426:271–278. [DOI] [PubMed] [Google Scholar]
- 28.Huang, E., J.M. Gu, M. Schau, and H.F. Bunn. 1998. Regulation of hypoxia-inducible factor 1 is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc. Natl. Acad. Sci. USA. 95:7987–7992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saccani, S., S. Pantano, and G. Natoli. 2002. p38-Dependent marking of inflammatory genes for increased NF-kappa B recruitment. Nat. Immunol. 3:69–75. [DOI] [PubMed] [Google Scholar]
- 30.Wang, G.L., and G.L. Semenza. 1993. Desferrioxamine induces erythropoietin gene expression and hypoxia-inducible factor 1 DNA-binding activity: implications for models of hypoxia signal transduction. Blood. 82:3610–3615. [PubMed] [Google Scholar]
- 31.Salcedo, R., K. Wasserman, H.A. Young, M.C. Grimm, O.M. Howard, M.R. Anver, H.K. Kleinman, W.J. Murphy, and J.J. Oppenheim. 1999. Vascular endothelial growth factor and basic fibroblast growth factor induce expression of CXCR4 on human endothelial cells: in vivo neovascularization induced by stromal-derived factor-1alpha. Am. J. Pathol. 154:1125–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagasawa, T., S. Hirota, K. Tachibana, N. Takakura, S. Nishikawa, Y. Kitamura, N. Yoshida, H. Kikutani, and T. Kishimoto. 1996. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 382:635–638. [DOI] [PubMed] [Google Scholar]
- 33.Muller, A., B. Homey, H. Soto, N. Ge, D. Catron, M.E. Buchanan, T. McClanahan, E. Murphy, W. Yuan, S.N. Wagner, et al. 2001. Involvement of chemokine receptors in breast cancer metastasis. Nature. 410:50–56. [DOI] [PubMed] [Google Scholar]
- 34.Hon, W.C., M.I. Wilson, K. Harlos, T.D. Claridge, C.J. Schofield, C.W. Pugh, P.H. Maxwell, P.J. Ratcliffe, D.I. Stuart, and E.Y. Jones. 2002. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 417:975–978. [DOI] [PubMed] [Google Scholar]
- 35.Min, J.H., H. Yang, M. Ivan, F. Gertler, W.G. Jr. Kaelin, and N.P. Pavletich. 2002. Structure of an HIF-1alpha-pVHL complex: hydroxyproline recognition in signaling. Science. 296:1886–1889. [DOI] [PubMed] [Google Scholar]
- 36.Lieubeau-Teillet, B., J. Rak, S. Jothy, O. Iliopoulos, W. Kaelin, and R.S. Kerbel. 1998. von Hippel-Lindau gene-mediated growth suppression and induction of differentiation in renal cell carcinoma cells grown as multicellular tumor spheroids. Cancer Res. 58:4957–4962. [PubMed] [Google Scholar]
- 37.Stratman, R., M. Krieg, R. Haas, and K.H. Plate. 1997. Putative control of angiogenesis in hemangioblastomas by the von Hippel–Lindau tumor supressor gene. J. Neuropathol. Exp. Neurol. 56:1142–1152. [DOI] [PubMed] [Google Scholar]
- 38.Rapisarda, A., B. Uranchimeg, D.A. Scudiero, M. Selby, E.A. Sausville, R.H. Shoemaker, and G. Melillo. 2002. Identification of small molecule inhibitors of hypoxia-inducible factor 1 transcriptional activation pathway. Cancer Res. 62:4316–4324. [PubMed] [Google Scholar]
- 39.Liew, F.Y., and I.B. McInnes. 2002. The role of innate mediators in inflammatory response. Mol. Immunol. 38:887–890. [DOI] [PubMed] [Google Scholar]
- 40.Blake, D.R., P.G. Winyard, and R. Marok. 1994. The contribution of hypoxia-reperfusion injury to inflammatory synovitis: the influence of reactive oxygen intermediates on the transcriptional control of inflammation. Ann. NY Acad. Sci. 723:308–317. [PubMed] [Google Scholar]
- 41.Cernanec, J., F. Guilak, J.B. Weinberg, D.S. Pisetsky, and B. Fermor. 2002. Influence of hypoxia and reoxygenation on cytokine-induced production of proinflammatory mediators in articular cartilage. Arthritis Rheum. 46:968–975. [DOI] [PubMed] [Google Scholar]
- 42.Gerard, C., and B.J. Rollins. 2001. Chemokines and disease. Nat. Immunol. 2:108–115. [DOI] [PubMed] [Google Scholar]
- 43.Cramer, T., Y. Yamanishi, B.E. Clausen, I. Forster, R. Pawlinski, N. Mackman, V.H. Haase, R. Jaenisch, M. Corr, V. Nizet, et al. 2003. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 112:645–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Foxman, E.F., J.J. Campbell, and E.C. Butcher. 1997. Multistep navigation and the combinatorial control of leukocyte chemotaxis. J. Cell Biol. 139:1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Folkman, J. 1995. Angiogenesis in cancer, vascular, rheumatoid and other disease. Nat. Med. 1:27–31. [DOI] [PubMed] [Google Scholar]
- 46.Hanahan, D., and J. Folkman. 1996. Patterns of emerging mechanisms of the angiogenic switch during tumorigenesis. Cell. 86:353–364. [DOI] [PubMed] [Google Scholar]
- 47.Crowther, M., N.J. Brown, E.T. Bishop, and C.E. Lewis. 2001. Microenvironmental influence on macrophage regulation of angiogenesis in wounds and malignant tumors. J. Leukoc. Biol. 70:478–490. [PubMed] [Google Scholar]
- 48.Hirani, N., F. Antonicelli, R.M. Strieter, M.S. Wiesener, P.J. Ratcliffe, C. Haslett, and S.C. Donnelly. 2001. The regulation of interleukin-8 by hypoxia in human macrophage–a potential role in the pathogenesis of the Acute Respiratory Distress Syndrome (ARDS). Mol. Med. 10:685–697. [PMC free article] [PubMed] [Google Scholar]
- 49.Payne, A.S., and L.A. Cornelius. 2002. The role of chemokines in melanoma tumor growth and metastasis. J. Invest. Dermatol. 118:915–922. [DOI] [PubMed] [Google Scholar]
- 50.Nagasawa, T. 2001. Role of chemokine SDF-1/PBSF and its receptor CXCR4 in blood vessel development. Ann. NY Acad. Sci. 947:112–115. [DOI] [PubMed] [Google Scholar]
- 51.Strieter, R.M., P.J. Polverini, D.A. Arenberg, and S.L. Kunkel. 1995. The role of CXC chemokines as regulators of angiogenesis. Shock. 4:155–160. [DOI] [PubMed] [Google Scholar]
- 52.Liotta L.A. 2001. An attractive force in metastasis. Nature. 410:24–25. [DOI] [PubMed] [Google Scholar]
- 53.Aoudjit, F., E.F. Potworowski, and Y. St-Pierre. 1998. The metastatic characteristics of murine lymphoma cell lines in vivo are manifested after target organ invasion. Blood. 91:623–629. [PubMed] [Google Scholar]
- 54.Krishnamachary, B., S. Berg-Dixon, B. Kelly, F. Agani, D. Feldser, G. Ferreira, N. Iyer, J. LaRusch, B. Pak, P. Taghavi, and G.L. Semenza. 2003. Regulation of colon carcinoma cell invasion by hypoxia-inducible factor 1. Cancer Res. 63:1138–1143. [PubMed] [Google Scholar]
- 55.Pugh, C.W., and P.J. Ratcliffe. 2003. The von Hippel-Lindau tumor suppressor, hypoxia-inducible factor-1 (HIF-1) degradation, and cancer pathogenesis. Semin. Cancer Biol. 13:83–89. [DOI] [PubMed] [Google Scholar]