Biogenesis, Architecture, and Function of Bacterial Type IV Secretion Systems (original) (raw)
. Author manuscript; available in PMC: 2013 Dec 26.
Abstract
Type IV secretion (T4S) systems are ancestrally related to bacterial conjugation machines. These systems assemble as a translocation channel, and often also as a surface filament or protein adhesin, at the envelopes of Gram-negative and Gram-positive bacteria. These organelles mediate the transfer of DNA and protein substrates to phylogenetically diverse prokaryotic and eukaryotic target cells. Many basic features of T4S are known, including structures of machine subunits, steps of machine assembly, substrates and substrate recognition mechanisms, and cellular consequences of substrate translocation. A recent advancement also has enabled definition of the translocation route for a DNA substrate through a T4S system of a Gram-negative bacterium. This review emphasizes the dynamics of assembly and function of model conjugation systems and the Agrobacterium tumefaciens VirB/D4 T4S system. We also summarize salient features of the increasingly studied effector translocator systems of mammalian pathogens.
Keywords: conjugation, DNA transfer, pathogenesis, protein transport, transformation
INTRODUCTION
The bacterial type IV secretion (T4S) systems mediate the translocation of macromolecules across the cell envelopes of Gram-negative and Gram-positive bacteria (23, 59). These systems are classified on the basis of an ancestral relatedness to bacterial conjugation machines (29, 88). The conjugation machines are a large subfamily of T4S systems that transmit DNA substrates among diverse species of bacteria, and some systems also deliver DNA to fungal, plant, and mammalian cells (9, 19, 130, 156, 163). Conjugation systems enable bacteria to adapt, at a population level, to changing environments. Throughout evolution, conjugation has contributed substantially to genome rearrangement and plasticity. Of more immediate interest, however, conjugation is a significant medical concern because it represents a dominant mechanism for widespread transmission of antibiotic resistance genes and virulence factors among pathogenic bacteria (109).
Although most textbooks depict conjugation as a mechanism for intercellular DNA transfer, recent studies indicate that conjugation systems actually transmit the DNA as a nucleoprotein particle composed of a protein component, the relaxase, covalently bound to the 5′ end of a molecule of ss-DNA (29). In addition, DNA transfer is directional, from 5′ to 3′, further suggesting that the relaxase serves to pilot the ssDNA through the secretion channel. Conjugation machines might therefore correspond to ancestral protein translocation systems that evolved to recognize and translocate relaxases as substrates and, only coincidentally, the “hitch-hiker” ss-DNA. With this view, it is not surprising that conjugation machines also translocate protein substrates independently of associated DNA. Moreover, a second large subfamily of T4S systems is now known to translocate only protein substrates to target cells (23, 42, 107). Studies conducted over the past 10 years have identified numerous examples in which bacteria have appropriated T4S systems for delivery of effector proteins into the cytosols of eukaryotic cells (94). Through interkingdom protein transfer, these effector translocator systems induce myriad changes in host cell physiologies to aid in the establishment of pathogenic or symbiotic relationships with the eukaryotic host (23, 42, 51).
Conjugation and most effector translocator systems transmit their substrates inter-cellularly by a cell-contact-dependent mechanism. Other type IV translocation systems deliver DNA substrates to or acquire them from the extracellular environment. In Neisseria gonorrhoeae, a T4S machine highly related to the F plasmid conjugation system exports DNA substrates to the extracellular milieu (41). Conversely, in Helicobacter pylori, a competence system mediates uptake of DNA from the milieu (68, 69). Such systems are called uptake and release systems (23, 42).
Besides a classification scheme based broadly on function, the T4S systems have been grouped as type IVA, IVB, or “other” (31). The type IVA systems are composed of subunits similar in composition and number to those of the archetypal Agrobacterium tumefaciens VirB/D4 system (29). The type IVB systems are assembled from subunits related to the archetypal Legionella pneumophila Dot/Icm system (133). The “other” systems bear little or no discernible ancestral relatedness to the types IVA or IVB systems and will be named once their subunit compositions and phylogenies are better understood. Most studies aimed at deciphering the mechanism of action of the T4S machines have focused on the A. tumefaciens VirB/D4 system and model IVA conjugation systems such as those encoded by plasmids F (IncF), RP4 (IncP), R388 (IncW), and pKM101 (IncN) (6, 29, 88). Recent studies have decidedly shifted to include type IVA effector translocator systems, several type IVB systems, and conjugation machines of Gram-positive bacteria (59, 133). At this time, however, most of the mechanistic information relevant to a review on the form and function of T4S systems derives from long-term studies of type IVA systems. Consequently, here we focus mainly on these systems and only briefly cite related findings for the IVB and “other” systems.
First, we describe available structure-function information for individual T4S subunits using the A. tumefaciens VirB/D4 T4S system as a framework for the discussion. Next, we summarize the partner subunit contacts and protein interaction networks identified to date. By incorporating this information with other biochemical findings, we describe a morphogenetic pathway for the T4S apparatus. The discussion then shifts to the secretion substrates and substrate translocation signals. Finally, we describe the translocation route for a DNA substrate through a T4S machine, made possible through the application of a novel assay system. The emphasis of this review is on the dynamic aspects of type IV machine assembly and function. Owing to space limitations, we do not provide a detailed description of T4S gene regulation or early DNA or protein substrate processing reactions (42, 90, 107). We also only briefly mention the fascinating and rapidly expanding topic of how type IV translocation contributes to pathogenic or symbiotic interactions with eukaryotic hosts (10, 23, 94, 107).
T4S ORGANELLES
The T4S systems are multisubunit cell-envelope-spanning structures (29, 88) composed of a secretion channel and often a pilus or other surface filament or protein(s). In Gram-negative bacteria, the T4S apparatus spans both membranes, the periplasm, and the cell wall. This structure likely corresponds to a transenvelope channel, through which substrates pass, sequestered from periplasmic proteases or nucleases. However, one T4S system mediates passage of its substrate exclusively across the outer membrane. In Bordetella pertussis, the Ptl T4S apparatus recognizes PT, a member of the multisubunit AB toxin family, in the periplasm and mediates its release into the milieu (20). This two-step translocation route is probably the exception rather than the rule for the T4S machines; yet it does serve to illustrate the remarkable structural and functional versatility of this translocation superfamily.
No high-resolution images currently exist for any T4S apparatus analogous to those of the needle complexes of type III secretion (T3S) systems (100), which are also utilized for interkingdom protein trafficking (65). Conjugational junctions between pairs of mating Escherichia coli cells show no discernible structures, suggesting that the transfer channel is small or forms only transiently (58, 123). Yet, T4S systems do elaborate easily visible surface structures (76, 88) (Figure 1). The conjugative pili of Gram-negative bacteria consist of flexible tube-like structures. For example, the F pilus encoded by the E. coli F plasmid has an 8- to 10-nm outer diameter and a length of 2 to 20 _μ_m, and an inner lumen 2 nm in diameter (88). F-pilus-mediated contacts linking donor and recipient cells are easily detected. By contrast, the conjugative pili of plasmids RP4 (IncP), R388 (IncW), and pKM101 (IncN), and the A. tumefaciens VirB/D4 T4S system (the T pilus), are 8 to 12 nm in diameter and less than 1 _μ_m in length (46, 76). These brittle structures are rarely detected on intact cells, but they are easily recovered from the culture supernatants of cells probably through passive breakage or an active sloughing mechanism. Such structures are thought to function as attachment devices for inducing formation of donor-recipient cell aggregates as a precondition for DNA transfer (29, 76).
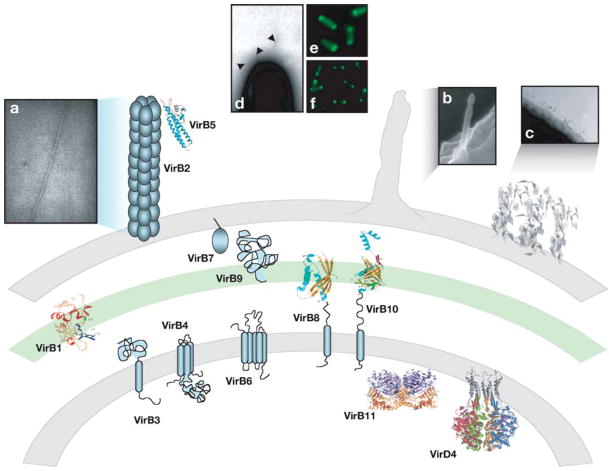
Figure 1.
Topologies, structures, and cellular localizations of T4S subunits and organelles. Locations and structures or membrane topologies of the VirB/D4-like T4S subunits at the bacterial cell envelope. Microscopy images of (a) the A. tumefaciens T pilus, (b) H. pylori sheathed structure, and (c) L. pneumophila fibrous mesh are depicted. The A. tumefaciens VirB/D4 T4S system localizes at the cell poles as represented by positioning of (d) T pilus, (e) VirD4-GFP, and (f) VirB6-GFP.
Three-dimensional structures of T4S subunits and surface organelles are reproduced with permission.
In contrast to the conjugative pili, the Cag T4S system of H. pylori elaborates a considerably larger sheathed structure (119, 141) (Figure 1). The needle portion of this structure is estimated to be 40 nm in diameter and the cross-section of the sheathed structure is estimated to be 70 nm. In L. pneumophila, the type IVB Dot/Icm system elaborates a fibrous mesh composed at least in part of the DotO and DotH subunits that covers the cell surface (154) (Figure 1). This fibrous mesh is thought to facilitate specific stages of the L. pneumophila infection cycle. The conjugation systems of Gram-positive bacteria do not elaborate pilus filaments, but instead produce surface proteins that function as nonspecific adhesins. An example is aggregation substance synthesized by the pheromone-inducible plasmid transfer systems of Enterococcus faecalis (155).
T4S GENE ARRANGEMENTS
The T4S subunits required for assembly of the channel/pilus are termed Mpf proteins, in accordance with an early nomenclature developed for components of conjugation machines (60). Genes for the Mpf subunits, numbering as many as 12 or more for a given system, are often arranged as a single operon. Though not required for assembly of the extracellular filaments, another protein designated the T4CP is required for substrate transfer by most T4S systems (95). This gene is linked either to the Mpf operon or to another operon whose products, termed the Dtr proteins, are responsible for processing DNA substrates at the oriT sequence (90). Fulfilling early predictions that the T4CP participates in DNA processing and/or transfer reactions, recent studies confirm that the T4CP functions as a DNA substrate receptor. Gene arrangements of the currently known T4S systems can be accessed through the Christie laboratory website (http://mmg.uth.tmc.edu/webpages/faculty/pchristie.html).
Many T4S systems are highly similar to the A. tumefaciens VirB/D4 T4S system. Others are assembled from a subset of VirB homologs together with subunits of a different ancestry. Such systems appear to have retained a VirB/D4 subcomplex(es) of structural or functional importance and then appropriated other proteins or subcomplexes for specialized activities. For example, the F plasmid encodes a T4S apparatus consisting of eight VirB homologs and other subunits found only among F-like plasmids (88). Several of these F-specific subunits form an interaction network that regulates F pilus extension and retraction, a dynamic property that has not been demonstrated for other conjugation systems (63, 76, 88). Other examples include the B. pertussis Ptl system and Brucella spp. VirB systems, which lack a discernible T4CP substrate receptor. As an alternative to T4CP-mediated recruitment and translocation, the B. pertussis Ptl system uses the general secretory pathway to export the monomeric constituents of PT across the inner membrane (20). At this time, it is not known how the Brucella spp. VirB systems recruit and translocate their secretion substrates across the inner membrane.
Mpf SUBUNIT STRUCTURES AND CATALYTIC ACTIVITIES
Several of the T4S subunits required for substrate transfer and pilus production have been characterized. In this section, we summarize recent findings pertaining to the structures, topologies, and catalytic functions of the VirB/D4 T4S machine components (Figure 1). Multiple alignments cited below were derived from the ProDom and Clustal W algorithms and are accessible through the Christie laboratory website (http://mmg.uth.tmc.edu/webpages/faculty/pchristie.html).
VirB1
VirB1 is a member of a large family of subunits commonly associated with macromolecular surface structures, including the T2S, T3S, and T4S systems, type IV pili and flagella, DNA-uptake systems, and bacteriophage entry systems. The signature for this protein family is a lysozyme-like structural fold (77, 104) (Figure 1). A muramidase activity for VirB1-like proteins is supported by the finding that overexpression of VirB1-like P19 of plasmid R1, but not a catalytic site mutant form of P19, results in massive lysis of E. coli cells. It was further reported that a purified MalE-P19 fusion protein degrades peptidoglycan (11). For conjugation systems null or catalytic site mutations of VirB1-like subunits diminish but do not abolish DNA transfer (11, 15). Similarly, in Brucella abortus, VirB1 is dispensable for T4S function in the mouse model of persistent infection (40). These findings indicate that the muramidase subunits facilitate, but are not required for, assembly of T4S systems across the cell wall.
VirB2
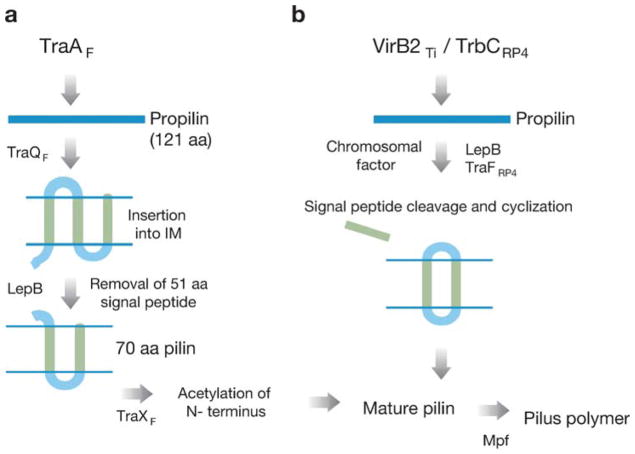
VirB2 is the major pilin subunit of the A. tumefaciens VirB/D4 T pilus and an essential component of the secretion channel (15, 71, 87). VirB2 homologs or orthologs are components of most type IVA systems (22). These pilin subunits are small, hydrophobic subunits with an unusually long signal sequence and two or more hydrophobic stretches. In general, VirB2-like proteins display low levels of sequence similarity, but a multiple alignment identifies several conserved Gly residues dispersed along the polypeptide. As yet, there is no ultrastructural information for any T4S pilin subunit, but novel processing reactions are associated with pilin maturation (Figure 2). The TraAF (e.g., TraA from the F plasmid) propilin is inserted into the inner membrane by the TraQ protein, cleaved of its signal sequence by LepB leader peptidase, and N-terminally acetylated by TraXF. Membrane-bound mature TraA is thought to form a reservoir for F pilus assembly, a process requiring the other Mpf subunits (76, 88). For the F-like plasmid R27, pilin monomers are added to the base of the nascent polymer, as shown by time-resolved phage adsorption (97). Interestingly, two other pilin proteins, TrbCRP4 and VirB2Ti, are processed to form cyclic polypeptides (45) (Figure 2). TrbCRP4 is cyclized through the action of the serine protease TraFRP4, whereas VirB2Ti is cyclized by an unknown chromosomal enzyme (46, 86). The cyclized mature pilins serve as the building blocks for pilus polymerization, again by a process requiring all cognate Mpf subunits (60, 85).
Figure 2.
Processing of conjugative pilin monomers. Biochemical reactions leading to maturation of (a) TraAF and (b) VirB2Ti and TrbCRP4 pilin monomers and pilus formation are depicted. Pilin processing requires plasmid or chromosomal factors, and pilus polymerization requires the cognate Mpf proteins.
VirB3
VirB3 is a short polypeptide with one or two predicted TMS near the N terminus. An early study reported evidence for localization of VirB3Ti mainly at the outer membrane (74). However, other studies predict inner membrane locations for VirB3Ti, TraLF, and TrbDRP4 homologs (13, 58, 88), and the following observations support this assignment. A Clustal W alignment identifies clusters of strongly conserved residues, e.g., (GAT)L(ST)RP and P(VI)G motifs, before the predicted TMS among VirB3 family members. This degree of sequence conservation is unexpected for a signal sequence and more consistent with an integral inner membrane topology. In addition, a recent sequence analysis identified genes for related T4S systems on two large TetR plasmids, pTet and pCC31, of Campylobacter jejuni. One of the encoded subunits, CmgB3/4, is a single polypeptide composed of VirB3 and VirB4 domains fused together (8). A current BLAST search identifies eight such chimeric subunits of putative T4S machines in the database, suggestive of a distinct phylogenetic clade. The VirB4 ATPase localizes predominantly at the cytoplasmic face of the inner membrane (see below). It thus seems likely that the VirB3 subunits assemble as a functional complex with the VirB4Ti ATPase at the inner membrane.
VirB4
VirB4 subunits are large inner membrane proteins with consensus Walker A and B nucleoside triphosphate-binding domains. VirB4 homologs are extensively distributed among T4S systems of Gram-negative and Gram-positive bacteria. These subunits were reported to share four common sequence motifs (22), but the Clustal W alignment reveals more extensive sequence similarities among six domains distributed along the entire polypeptides. A VirB4Ti topology model, developed from reporter protein fusion and protease susceptibility studies, includes possible periplasmic loops, one near the N terminus and a second just N-terminal to the Walker A motif (34). These experimental findings are consistent with computer-based predictions and results of fractionation studies of the TrbERP4 homolog (114). However, evidence also exists that the TraCF and TrhCR27 homologs associate with the inner membrane through interactions with other T4S subunits (55, 126). Also of interest, a recent computer analysis identified sequence and possible structural similarities between the C-terminal residues 426 to 787 of VirB4 and the TrwBR388 T4CP (102). On the basis of an atomic structure for TrwBR388 (see below), it was postulated that the C-terminal region of VirB4 subunits might assemble as higher order homohexamers (102). Consistent with this prediction, it was recently shown that B. suis VirB4 produced in E. coli fractionates as a high-molecular-weight complex (161).
VirB5
VirB5 subunits are exported to the periplasm, and they also localize extracellularly as components of the pilus (128, 129). The X-ray crystallography structure of one family member, TraCpKM101, presents as a single-domain protein with a mostly _α_-helical, elongated structure (160) (Figure 1). Further structure-based mutagenesis studies identified regions likely involved in partner contacts. Moreover, effects of TraCpKM101 mutations on the capacity of bacteriophages to attach to pilus receptors suggested that the VirB5-like subunits might bind both at the pilus tip and base (160). In Bartonella henselae, a VirB5 ortholog is an immunodominant protein in patients diagnosed for cat-scratch disease. This protein is postulated to play a role in mediating an interaction with mammalian target cells during infection (39, 110). An early study also showed that the conjugal donor activity of E. coli cells carrying a pKM101 derivative lacking the gene for TraC was restored upon mixture with TraC-producing cells (157). The pilus-associated form of VirB5-like subunits might contribute generally to target cell attachment. However, recent findings further demonstrate that the periplasmic form of VirB5Ti is required for translocation of a DNA substrate to the cell surface (see below) (25).
VirB6
The VirB6 subunits are highly hydrophobic with multiple predicted TMS. The VirB6 family members display relatively low overall sequence similarities with the exception of a conserved region corresponding to residues ~ 170 to 205 of VirB6Ti that includes an invariant Trp residue required for protein function (22, 75, 88). A combination of reporter fusion (13, 37, 72, 75) and Cys accessibility studies of functional substitution mutants (72) support a topology model for VirB6Ti as a polytopic membrane protein with a periplasmic N terminus, five TMS, and a cytoplasmic C terminus. A particularly notable feature of the topology model is loop P2, a large central periplasmic loop whose secondary structure appears important for DNA substrate translocation (72) (see below). Intriguing similarities exist between VirB6Ti and ComEC of the Bacillus subtilis competence system. ComEC also is a polytopic subunit and possesses a large, central periplasmic loop whose secondary structure is important for DNA translocation (43).
VirB7
The VirB7 subunits are small lipoproteins found in only a subset of the T4S systems (22). However, lipoproteins common to T2S, T3S, and T4S systems might supply stabilizing functions that have been reported for VirB7Ti. VirB7Ti stabilizes several VirB sub-units, in part through formation of a disulfide bridge with VirB9Ti (3, 7, 12, 50, 140). VirB7Ti localizes predominantly at the outer membrane, although both inner-membrane-associated (49) and extracellular (120) forms also have been detected. Extracellular VirB7Ti copurifies with the T pilus but is also recovered from the supernatant of pilus-minus cells (120). H. pylori HP0532, a lipoprotein considerably larger than VirB7Ti, also localizes extracellularly, as a component of the large sheathed filament produced by the Cag T4S system (141).
VirB8
VirB8 subunits are associated with most type IVA systems. These subunits are configured as bitopic inner membrane proteins with an N-proximal TMS (37, 88). A Clustal W alignment confirms and extends an earlier identified sequence conservation between residues 100 and 143 and between residues 190 and 235 (83). A three-dimensional structure of a periplasmic fragment (residues 77 to 239) of B. suis VirB8 presents as a large extended _β_-sheet and five _α_-helices, giving rise to an overall globular fold (143) (Figure 1). Conserved residues important for protein function are buried in the hydrophobic core, where they are predicted to contribute to VirB8 structural integrity. Other conserved residues are surface exposed and might mediate contacts with VirB8 partner subunits (81, 143).
VirB9
The VirB9 subunits are generally hydrophilic and predicted to localize in the periplasm or outer membrane. Similarities exist between the C-terminal regions of VirB9-like TraKF subunit and the ring-forming outer membrane secretins of the T2S and T3S systems (88). VirB9Ti is composed of three functional domains, each approximately 80 to 100 residues (71). The N-terminal, periplasmic domain is highly conserved among VirB9 family members and is required for both channel activity and pilus biogenesis. A central domain is poorly conserved but possesses several potential _β_-sheet TMS suggestive of a possible outer membrane association. The C-terminal region also is well conserved among VirB9 family members and required for channel assembly and pilus biogenesis. Within the C-terminal region, Cys-262 forms an intermolecular disulfide bridge with the VirB7 lipoprotein (140).
VirB10
The VirB10 subunits possess several interesting features. These are bitopic inner membrane proteins situated with the bulk of the protein in the periplasm (37). After the TMS, most homologs possess a Pro-rich region, which is predicted to form an extended structure in the periplasm (24). VirB10 subunits also possess potential protein-protein interaction motifs, e.g., coiled-coils near the TMS and clusters of hydrophobic residues near their C termini (24, 53, 95). These domains probably contribute to establishment of interactions with partner proteins at both membranes. VirB10 subunits interact with inner membrane T4CPs (24, 53, 95), bitopic VirB8 (36), and VirB4 ATPase (V. Krishnamoorthy & P.J. Christie, unpublished findings), and with the outer-membrane-associated VirB7-VirB9 dimer (12, 24).
A crystal structure is available for a periplasmic fragment corresponding to residues 146 to 376 of the H. pylori ComB10 subunit (Figure 1). The structure comprises an extensively modified _β_-barrel with an _α_-helix projecting off one side and a second, flexible helix-loop-helix of 70 Å in length projecting off the top (143). Though generally compacted, it should be noted that this structure corresponds only to the central and C-terminal portions of VirB10 subunits. The overall structure for this protein family thus consists of a small cytoplasmic N-terminal domain, a TMS, a Pro-rich extended domain, and a _β_-barrel with a sizable helical projection presumably oriented toward the outer membrane. This structure is compatible with recent evidence that VirB10Ti functions in a way similar to the E. coli TonB energy coupling protein (see below) (24).
The H. pylori HP0527 subunit of the Cag T4S system is considerably larger than other VirB10 subunits and it is related in sequence only at its N terminus to other VirB10 sub-units. Unlike VirB10Ti, HP0527 is surface localized and is also antigenically variable owing to intragenic recombination among repeat sequence motifs in the middle of the protein. HP0527 also is reported to be a component of the Cag T4S sheathed structure (119).
VirB11
VirB11 is a member of a large family of AT-Pases associated with systems dedicated to secretion of macromolecules (22). Homologs are widely distributed among the Gram-negative bacteria, and they also function in secretion systems of Gram-positive bacteria as well as species of the Archaea (113). Purified homologs TrbBRP4, TrwCR388, and H. pylori HP0525Cag assemble as homohexameric rings discernible by electron microscopy, and the last also by X-ray crystallography (79, 80, 125, 159) (Figure 1). These structures present as double-stacked rings formed by the N- and C-terminal halves and a central cavity of ~50 Å in diameter. The C-terminal domain adopts a RecA fold, whereas the N-terminal domain is unique to HP0525Cag. The overall HP0525Cag structure appears highly conserved, even among distantly related ATPases associated with other transport or fimbrial biogenesis systems (16, 66, 118, 135).
The apo-, nonhydrolyzable ATP_γ_S-, and ADP-bound structures differ appreciably, such that ATP-binding correlates with a swiveling of the N-terminal domains that converts the open and asymmetric hexamer to a more compact, symmetrical hexamer (125). ATP-dependent conformational changes also were detected for the PilQR64 homohexamer, a related ATPase also required for pilus biogenesis (122). The VirB11-like ATPases fractionate as cytoplasmic or peripheral inner membrane proteins (80, 116). Mutants of VirB11Ti and L. pneumophila DotBDot/Icm bearing Walker A nucleotide-binding motif substitutions bind the inner membrane more tightly than the wild-type protein, suggestive of an ATP-regulated membrane interaction (116). Consistent with these findings, TrbBRP4 and HP0525Cag both undergo conformational changes when in contact with phospholipid membranes (80). Thus, the mechanical energy generated by nucleotide-dependent conformational changes might promote membrane and Mpf subunit contacts required for T4S biogenesis (see below) (125).
THE T4CP STRUCTURE AND BIOCHEMICAL ACTIVITY
In addition to the Mpf proteins, conjugation systems and most effector translocator systems require a T4CP for substrate transfer (93). T4CPs are dispensable for formation of conjugative pili or the H. pylori sheathed structure (85, 119). They are composed of an N-proximal periplasmic loop and a large C-terminal cytoplasmic domain (37, 89). The atomic structure for a soluble fragment of TrwBR388 presents as a molecule with two domains, a nucleotide-binding domain similar to RecA and DNA ring helicases and an all _α_-domain (56). This fragment crystallizes as a homohexameric sphere with dimensions of 110 Å in diameter and 90 Å in height and a central channel of 20 Å in diameter. The structure of the entire molecule bears a striking resemblance to the F1-ATPase _α_3_β_3 heterohexamer (Figure 1). The T4CPs carry conserved Walker motifs that are required for function and they bind ssDNA and dsDNA nonspecifically (103). Recently, the soluble fragment of TrwBR388 was shown to display a DNA-dependent ATPase activity. Moreover, this fragment of TrwBR388 oligomerized upon DNA binding but not ATP binding. The T4CPs might form higher order hexamers upon binding of a DNA substrate, and then use ATP energy to pump the bound substrate across the inner membrane (142).
BIOGENESIS OF THE T4S APPARATUS
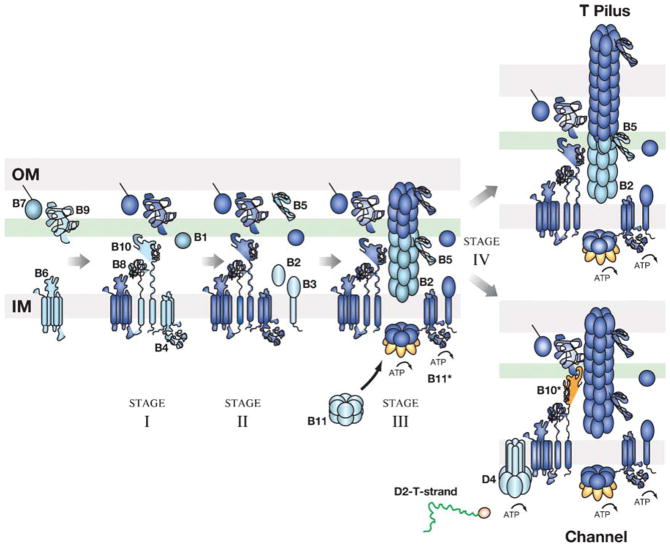
With these subunit structures and membrane topologies in mind, it is of interest to consider how T4S machines assemble as functional translocation systems. In early studies, this question was approached through definition of protein stabilization networks for the A. tumefaciens VirB/D4 T4S components (28). These and more recent biochemical studies support a general proposal that the T4S machine assembles in several discrete stages. For purposes of discussion, we depict a four-stage biogenesis pathway for the VirB/D4 T4S system (Figure 3).
Figure 3.
Biogenesis pathway of the A. tumefaciens VirB/D4 T4S system. A four-stage assembly pathway is presented for the A. tumefaciens VirB/D4 T4S system. Stage I: Assembly of the VirB6 plus B4-B7-B8-B9-B10 core complex; stage II: recruitment of B2-B3-B5 components; stage III: recruitment of hexameric VirB11 ATPase; stage IV: pathway bifurcation to yield the pilus or secretion channel. Subunits added at each stage of the pathway are depicted in blue-green, whereas subunits previously incorporated are depicted in dark blue. Conformational changes observed for the H. pylori HP0525 (B11*) N-terminal domain and A. tumefaciens VirB10 (B10*) are depicted in orange (*). We propose that progression through the stage IV reaction to form a functional secretion channel is regulated by intrinsic (ATP, substrate engagement) and extrinsic (recipient cell contact) signals. IM, inner membrane; OM, outer membrane.
Stage I: Formation of the Core Complex
In stage I, a stable substructure, termed a core complex, assembles across the cell envelope. The core complex is composed of the VirB4, VirB7, VirB8, VirB9, and VirB10 subunits. These are among the most highly conserved subunits among T4S systems, and the virB7 through virB10 genes often overlap or are contiguous, suggestive of translational coupling (23). Although polytopic VirB6 is not as evolutionarily conserved as VirB4 and VirB7 to VirB10, VirB6Ti contributes in an unspecified way to the formation of VirB7 and VirB9 multimers (62, 73). Therefore, we also depict VirB6 positioning and a role in VirB7-VirB9 multimerization as an early morphogenetic event (Figure 3). Detected interactions between VirB8Ti and VirB1Ti transglycosylase (152), as well as the finding that B. pertussis PtlE is a chimera of VirB1- and VirB8-like domains (115), further suggest that VirB1 is recruited early on to facilitate core assembly. Reactions leading to stage I complex formation also include general secretory pathway-mediated translocation of VirB7 and VirB9, lipid modification of VirB7, formation of disulfide bridges to form VirB7 dimers and VirB7-VirB9 heterodimers, and sorting of the dimers to the outer membrane (28). At the outer membrane, VirB7-VirB9 dimers most likely assemble as a higher order multimer, which then stabilizes the complex through contacts with the bitopic inner membrane subunits VirB8 and VirB10. VirB4 also interacts with, and is stabilized through, contacts with subunits VirB8 and VirB10 (30).
The existence of this core complex is supported by demonstrated interactions between VirB6, VirB7, and VirB9, between VirB8, VirB9, and VirB10, and between VirB4, VirB8, and VirB10 (12, 35, 36, 73, 81, 152). In addition, the core subunits cofractionate as a large complex exceeding 500 kDa in gel filtration columns and blue native gels (78). There also is indirect evidence that this core substructure alone confers function. In H. pylori, homologs of VirB4 and VirB7 to VirB10 assemble as a competence system (68, 69). Similarly, during conjugation between A. tumefaciens donor and recipient cells, synthesis of core subunits in the recipient cells stimulates acquisition of DNA by several orders of magnitude (18, 92). This core complex minimally promotes or stabilizes agrobacterial mating pairs, and it might even function more directly as a channel for DNA uptake.
Stage II: Recruitment of Pilus-Associated Proteins
In the stage II reaction, the core complex recruits subunits important for pilus assembly, including VirB2, VirB3, and VirB5. The stage II structure thus corresponds to a transenvelope complex composed of VirB2 through VirB10 (Figure 3). Several lines of evidence are compatible with this stage II reaction. Most notably, biochemical fractionation studies have shown that core subunits partition aberrantly in sucrose gradients if synthesized in the absence of the latter subunits (12, 92). In addition, in bacterial matings the production of the stage II subunits in agrobacterial recipients stimulates DNA acquisition to a significantly greater extent than synthesis only of the core components (92). Studies exploring pairwise contacts among Mpf subunits of various T4S systems (1, 74, 128, 129, 136, 144) also yield evidence for a protein interaction network in which the core subunits form one interaction subgroup and VirB2, VirB3, and VirB5 form another (30). The stage II reaction proposed here would thus correspond to the productive integration of these two machine subassemblies.
Recently, a comprehensive biochemical study nicely detailed how the core complex integrates with the pilus proteins as a prerequisite to pilus assembly (161). The study identified a network of stabilizing interactions required for formation of a VirB2-VirB5 complex. VirB4 stabilized VirB3 and VirB8, and, in turn, VirB8 interacted with VirB5 in the periplasm. These stabilizing interactions plus a contribution by VirB6 enabled VirB5 complex formation with VirB2 (62, 161). The study further showed that purified forms of VirB5 interact with VirB8 and also with VirB10. Finally, and intriguingly, a Walker A mutant form of VirB4 displayed properties of the native protein with respect to stabilizing VirB3 and VirB8 and supporting pilus biogenesis (161). Thus, VirB4 appears to contribute structurally not catalytically to the integration of core and pilus components to yield the stage II substructure (Figure 3).
Stage III: Recruitment of VirB11 ATPase
In the stage III reaction, the Mpf structure recruits the VirB11 homohexameric ATPase (Figure 3). This postulated reaction is supported by findings that VirB11 is dispensable for assembly of the stage II substructure, as monitored by biochemical fractionation and the VirB-mediated DNA-uptake assay (92), but required for the secretion channel and pilus production (15, 85). As described above, cytosolic VirB11 homohexamers undergo ATP-dependent conformational changes that likely affect membrane binding and formation of Mpf subunit contacts (117, 125). Moreover, recently it was shown that VirB11Ti uses ATP energy to drive a late-stage morphogenetic reaction required for substrate transfer (see below) (24). Therefore, we propose that the VirB11 ATPases are recruited to Mpf structures, where they stimulate formation of transenvelope structures composed of an inner membrane platform, a VirB2 polymer that extends across the periplasm, and an outer membrane pore complex. The existence of a periplasmic VirB2 polymer is speculation but nevertheless compatible with experimental evidence that a cellular form of VirB2 comprises a portion of the secretion channel required for substrate passage through the periplasm (25, 71).
Stage IV: A Pathway Bifurcation
The stage IV reaction is depicted as a bifurcation in the assembly pathway to yield a pilus or a secretion channel (Figure 3). Although T4S systems are classically depicted as a single supramolecular organelle, there is accumulating support for the notion that Mpf subunits alternatively assemble as a pilus or secretion channel. First, T4CPs are completely dispensable for pilus production. However, T4CPs function in several ways as obligatory components of secretion channels. As described below in more detail, T4CPs recruit substrates to the secretion channel and they might also function as inner membrane translocases. In addition, a recent study showed that the A. tumefaciens VirD4 T4CP coordinates with the VirB11 ATPase to energize a late-stage structural transition in the Mpf structure that is required for substrate transfer across the cell envelope (see below) (24). This latter activity, in particular, strongly suggests the T4CPs configure Mpf subunits specifically for channel activity.
Genetic studies have also supplied evidence for the alternative assembly of pili or secretion channels. Mutations in Mpf subunits of at least two conjugation systems selectively arrest production of either the conjugal pilus or the secretion channel. In the RP4 transfer system, mutations in the pilin subunit TrbCRP4 block pilus production as shown by an absence of extracellular pilin protein and resistance to bacteriophage binding to RP4 pilus-specific receptors. Yet, these pilus-minus strains retain the capacity to efficiently transfer DNA substrates to recipient cells. Other pilin mutations confer the reciprocal pilus-positive transfer-minus phenotype (46). In the A. tumefaciens VirB/D4 T4S system, “uncoupling” mutations in several Mpf components, including VirB6, and VirB9, and VirB11 also arrested pilus polymerization without abolishing substrate transfer (71, 72, 121, 162). Also of interest, A. tumefaciens virB1 mutants do not produce detectable levels of pilus, but they deliver substrates intercellularly (15, 85). A transglycosylase activity thus seems essential for pilus formation but not for assembly of a functional secretion channel.
For pilus biogenesis, we propose that pilin monomers, e.g., VirB2Ti, are added to the base of a nascent polymer on a platform of inner membrane Mpf subunits (Figure 3). Such a process likely requires an energy source, especially if pilin monomers are extracted from a membrane pool for polymer formation. The recent finding that a Walker A mutant of B. suis VirB4 still supports pilus biogenesis (161) suggests VirB11 is the best candidate for supplying an ATP energy source, although it is also possible that pmf is used. For assembly of the secretion channel, the VirB4, VirB10, and VirB11 subunits and perhaps other Mpf components would recruit the T4CP (e.g., VirD4) to the stage III structure (4, 53, 95). Even at this point, however, secretion channel biogenesis is not complete. Findings summarized below indicate that at least two additional signals, ATP energy and substrate binding to the receptor, are required for channel activity.
ENERGETIC REQUIREMENTS FOR T4S MACHINE BIOGENESIS
ATP energy and the pmf are important for translocation of DNA across the bacterial cell envelope (38, 91, 98, 111, 134). In early studies of E. coli cells carrying the F or RP4 plasmids, depletion of cellular ATP levels as well as dissipation of pmf inhibited formation of stable mating aggregates as well as surface presentation of bacteriophage pilus receptors (38, 111). Moreover, electrochemical measurements showed that membranes of E. coli cells producing the RP4 Mpf proteins in the absence of the Dtr proteins (the relaxosome components) and the TraGRP4 T4CP are more permeable than membranes from cells carrying a complete RP4 transfer system (38). These findings led to the suggestion that the Dtr proteins, the DNA transfer intermediate, and/or the TraG T4CP contribute to ATP- or pmf-dependent reactions required for assembly of an intact T4S system.
Indeed, recent studies of the A. tumefaciens VirB/D4 T4S system confirm this prediction. The 48-kDa VirB10 subunit produced in wild-type cells is cleaved by a protease upon treatment of spheroplasts. However, VirB10 was insensitive to protease treatments of spheroplasts from wild-type cells depleted of cellular ATP, as well as strains deleted of virD4 or virB11, or strains producing Walker A mutant forms of VirD4 and VirB11. Moreover, VirB10 interacts with the outer-membrane-associated VirB7-VirB9 dimer, but only in strains producing native forms of VirD4 and VirB11. Together, the findings strongly indicate that VirD4 and VirB11 ATP binding or hydrolysis serves to induce a VirB10 structural transition, detected as a change in protease susceptibility, that is required for formation of an envelope-spanning bridge that links inner and outer membrane VirB/D4 machine sub-assemblies (24). Such an ATP sensing system might account for the observed effects of ATP depletion on loss of stable mating pair formation and phage receptors by E. coli cells bearing RP4 and F plasmids, as well as the increase in membrane permeability, by cells producing the corresponding Mpf subunits in the absence of T4CPs (38, 111). These new findings, including the observed structural similarities with TonB mentioned above, suggest that VirB10 subunits contribute to T4S machine biogenesis through a TonB-like energy-sensing mechanism (24).
Intriguingly, the reported VirB10 energy-sensing mechanism is dependent on ATP utilization by both VirD4 T4CP and VirB11; however, as noted above, the T4CP is dispensable for pilus production (24). It must be concluded that the detected energization reaction is required specifically for a late stage of channel morphogenesis. However, pilus biogenesis requires energetic contributions by the VirB4 and VirB11 Mpf components. VirB10 also interacts stably with VirB4 (152, V. Krishnamoorthy & P.J. Christie, unpublished findings), and thus VirB10 might be energized differently through contacts with VirD4 or with VirB4. Such an alternative energy-sensing mechanism might represent a late-stage molecular switch serving to regulate formation of the secretion channel or the extracellular pilus.
Cell-Position-Specific Assembly of T4S Machines
Various bacterial organelles, including flagella, type IV pili, and the cell division machinery, assemble at discrete locations at the cell surface (99, 137). There is also increasing evidence that at least some T4S machines also target to specific sites (see Figure 1). A. tumefaciens tends to attach polarly to plant cells (101), and more recent work has confirmed that the VirBTi-encoded pilus is polar (85). The VirD4 T4CP also localizes predominantly to the cell poles (82). Moreover, polar targeting of VirD4 has been experimentally exploited to provide evidence that the T4CPs function in recruitment of protein substrates to the T4S apparatus (5). Two groups also have detected polar localization of VirB6, though the studies reported different VirB subunit requirements for VirB6 targeting (72, 75). Even so, it can generally be concluded that VirB6 assembles at the cell poles at an early stage of T4S machine biogenesis.
Intriguingly, other T4S components, including VirB8, VirB9, and VirB10 from wild-type A. tumefaciens, form foci that are distributed throughout the cell surface (83, 94). VirB9 and VirB10 foci are detected only in cells cosynthesizing VirB8, prompting the suggestion that VirB8 nucleates assembly of the core complex (83). Given the evidence for polar localization of the VirB/D4 T4S apparatus, these surface-distributed complexes might represent pools of physiologically relevant assembly intermediates, or structures important for T4S-mediated transfer but not for channel or pilus production per se. Or they may simply correspond to off-pathway, dead-end complexes. Such distributed foci are not unique to VirB channel subunits, however. In E. coli, cells bearing plasmid R27form discernible foci composed of VirB4-like TrhC (55). These foci are detected only in cells producing 12 of 18 Tra subunits of this T4S system (54).
T4S SUBSTRATES AND CELLULAR CONSEQUENCES OF SUBSTRATE TRANSFER
With the discovery that T4S machines translocate effector macromolecules into eukaryotic cells, many laboratories have focused on identifying T4S substrates and characterizing the cell biology of substrate trafficking. The literature on this topic is too extensive to summarize here in detail. Instead, we provide an update of the known substrates of effector translocator systems and general descriptions of how these substrates alter host physiological pathways. The reader is referred to recent excellent reviews for more comprehensive information (10, 23, 39, 42, 51, 64, 107, 133).
Screens based on fusion of suspected substrates to reporter proteins have proven useful in these studies. Two powerful assays include CRAfT, which monitors intercellular translocation of Cre recombinase (149), and the adenylate cyclase assay, which monitors translocation of the Cya domain of B. pertussis Cya toxin (139). The current list of known T4S substrates is summarized in Table 1.
Table 1.
T4S substrates and consequences of substrate translocation
| Bacterial species | T4S system | Substrate | Size (kDa) | Known or postulated role in host | Target cell manifestation | Reference (s) |
|---|---|---|---|---|---|---|
| Agrobacterium tumefaciens | VirB | VirD2 | 56 | Interacts with host proteins AtKAP_α_ CypA, PP2C, RocA, Roc4, nuclear localization and integration of T-DNA | Crown gall | 145, 146 |
| VirD5 | 92 | Host range determinant | 150 | |||
| VirE2 | 65 | Forms membrane pore, protects T-strand, interacts with host proteins VIP1 & VIP2, nuclear localization and integration of T-DNA | 32, 44, 145, 153 | |||
| VirE3 | 74 | Interacts with AtKAP_α_ and VirE2, mimics VIP1 function for T-DNA import | 146 | |||
| VirF | 23 | Host range determinant targeted proteolysis of VIP1 and VirE2 | 146, 150 | |||
| MobA | 78 | 150 | ||||
| Atu6154 | 34 | 150 | ||||
| Agrobacterium rhizogenes | VirB | GALLS | 194 | Protects T-strand, promotes nuclear localization and integration of T-DNA into plant genome | Hairy roots | 67 |
| Mesorhizobium loti | VirB | Msi059 | 197 | Deconjugates proteins stabilized by SUMOylation | Root nodulation | 70 |
| Msi061 | 32 | Target specific proteins for degradation | 70 | |||
| Legionella pneumophila | Dot/Icm | RalF | 41 | Exchange factor for ADP ribosylation factor (ARF) GTPases | Legionnaire’s pneumonia | 106 |
| LidA | 83 | Docking of vesicles to the membrane of phagosome | 33 | |||
| DotA | 125 | Membrane pore in host | 33, 107 | |||
| SidA-H, SdeC, SdcA | 46–240 | 96 | ||||
| LepA, LepB | 127, 142 | Alter exocytic pathway in protozoa | 27 | |||
| WipA, WipB | 57 | 108 | ||||
| YlfA, YlfB | Faciliate binding and fusion of vesicles to _Legionella_-containing vacuole | 21 | ||||
| VipA, D, F | 38, 68, 32 | VipA affects carboxypeptidase trafficking; VipD interferes with multivesicular body formation; VipF inhibits lysosomal protein trafficking | 138 | |||
| Helicobacter pylori | Cag | CagA | 128 | Leads to dephosphorylation of host cell proteins such as cortactin and ezrin, binds SHP2, CSK, HGF/SF receptor, ZO-1, JAM, Rho GTPases, ARP2/3, and N-WASP | Gastritis, peptic ulcer | 64 |
| Helicobacter pylori | Cag | Peptidoglycan | Induces NF-_κ_B activity in gastric epithelial cells, Nod1 recognition | Gastritis, peptic ulcer | 151 | |
| Bordetella pertussis | Ptl | AB5 toxin | 105 | Interacts with αβγ heterotrimeric Gi/o proteins | Whooping cough | 20 |
| Bartonella henselae | VirB-Trw | BepA to G | 51–111 | Cat scratch angiomatosis | 132 | |
| Burkholderia cenocepacia | VirB | Plan tissue watersoaking | 47 | |||
| Brucella sp. | VirB | Brucellosis | 10 |
To date, the A. tumefaciens VirB/D4 T4S system translocates 10 substrates. These include VirE2, VirE3, VirF, and VirD5, which are implicated as playing a role in the establishment of crown gall disease (32, 84). In addition, this T4S system translocates two substrates of a related T4S system carried by Mesorhizobium meliloti (see below) (70) and GALLS, a TraA strand transferase-like protein identified in Agrobacterium rhizogenes that substitutes for VirE2 function in A. tumefaciens (67). Other substrates include MobA relaxase either in association with or independently of its RSF1010 DNA substrate (17, 150).
M. loti strain R27 carries a VirB/D4 T4S system, highly similar to that of A. tumefaciens, that translocates the effector proteins Msi059 and Msi061. Both Msi061 and A. tumefaciens VirF possess F-box motifs that, upon delivery into the plant cell, are postulated to mimic the activities of eukaryotic F-box receptors for attracting specific proteins to the Skp1-Cdc53-F-box protein complexes for ubiquitination and proteolysis (70, 146). These related T4S systems thus have been appropriated for different purposes, induction of a pathogenic response by A. tumefaciens and establishment of a symbiotic relationship by M. loti.
The L. pneumophila Dot/Icm system secretes a repertoire of substrates to evade fusion with lysosomes and support bacterial replication in the host (27, 33, 96, 105, 106). This T4S system appears to function at different stages of the L. pneumophila infection process, as well as for different purposes in human and protozoan hosts. LidA is exported rapidly, within 5 min of L. pneumophila contact with host macrophages, indicating a possible role as a gatekeeper for the Dot/Icm system (33). RalF is a Sec7 homology domain–containing protein that functions as an exchange factor for the eukaryotic ADP ribosylation factor family of small GTPases. RalF also appears to be translocated prior to internalization of L. pneumophila by the host (106). Recently, by using the CRAfT assay, Luo & Isberg (96) identified the SidA to SidH and SdeC substrates. Many Sid paralogs also were identified and some of these were translocated, leading to the suggestion that the Dot/Icm system translocates a minimum of 24 substrates (96). Recently, additional substrates designated WipA and WipB were identified on the basis of interactions with the Dot/Icm machine subunit IcmS. By using the adenylate cyclase assay, Chen et al. (27) identified proteins LepA and LepB. These proteins are not necessary for L. pneumophila replication in mammalian cells, but they are required for survival in protozoan hosts (27). Finally, a recent genetic screen for L. pneumophila proteins that confer conditional growth and vacuole sorting defects in yeast led to the identification of new Dot/Icm system-dependent substrate proteins YlfA and HlfB (21) and VipA, VipD, and VipF (138). Functions for the latter proteins include affects on carboxypeptidase 4 trafficking, inhibition of lysosomal trafficking, and interference with multivesicular body formation at the late endosome and ER-to-Golgi body transport (138).
CagA is the only virulence protein thus far shown to be translocated by the H. pylori Cag T4S system (64). Yet, upon translocation to gastric epithelial cells, CagA is phosphorylated, and both phosphorylated and nonphosphorylated forms interact with a number of cellular factors associated with distinct signaling pathways. As a result, CagA translocation induces myriad effects on host signaling pathways that contribute to the establishment of gastric carcinomas (23, 64, 107). Recently it was reported that H. pylori activates the NF-_κ_B signaling pathway through delivery of peptidoglycan to gastric epithelial cells. Results further implicated the Cag T4S system as a peptidoglycan delivery system. If a nonspecific release mechanism can be excluded, this would be the first example of T4S-mediated trafficking of a substrate other than DNA or protein (151).
In Bartonella henselae, seven proteins, designated BepA–G, are encoded by a region located downstream of a virB locus. By use of CRAfT, the Bep proteins are translocated into human vascular endothelial cells in a VirB/D4-dependent manner. Proteins BepA–C carry a conserved N-terminal domain (Fic) of unknown function, whereas the N-terminal regions of BepD–F contain repeated EPLYA and ETIYA motifs that resemble the EPIYA tyrosine phosphorylation motif of CagA. Deletion of this bep locus results in a mutant phenotype comparable to virB4 and virD4 mutants, indicating that the Bep proteins mediate all VirB/D4-dependent host cell phenotypes (39, 110, 132).
Burkholderia cenocepacia strain K56-2 carries a plasmid-encoded Ptw T4S system and a second system encoded by chromosome II (47). Ptw is a chimeric system composed of VirB/D4 and F-specific subunits, whereas the chromosomal system closely resembles the VirB/D4 systems. Mutational studies provided evidence for the involvement of the Ptw system in eliciting the plant tissue watersoaking phenotype (47). Interestingly, this phenotype is also elicited by Erwinia amylovora and Pantoea stewartii subsp. stewartii through the export of elicitors DspE and WtsE, respectively, via T3S systems (47). Although the B. cenocepacia substrates have not yet been reported, the data suggest that these three species alternatively have coopted T3S or T4S systems for a common pathogenic outcome.
Finally, a recent study identified a Xanthomonas axonopodis pv_. citri_ T4S system. A two-hybrid analysis detected interactions between the VirD4Xa subunit and 12 previously uncharacterized proteins. By virtue of an interaction with putative T4CP substrate receptor (see below), these proteins termed XVIPs (Xanthomonas VirD4 interacting proteins) are considered candidate secretion substrates (1). Sequence motifs common to all of these proteins might comprise the basis for T4S-mediated translocation (see below).
The Bases for T4S Substrate Recognition
How do T4S systems recognize their substrates and what provides the basis for substrate specificity? A decade of genetic and biochemical studies on conjugation machines point to the T4CP as the substrate receptor (57). The T4CP recognizes DNA substrates through binding of the relaxase or another relaxosomal component bound at oriT (14, 61, 131). Similarly, T4CPs are implicated in protein substrate reception among effector translocator systems, although there is also evidence that additional T4S subunits contribute to substrate specificity (42, 108).
C-Terminal and Other Specificity Translocation Signals
Recent work has significantly advanced our knowledge of how T4S machines recognize cognate substrates. For the A. tumefaciens VirB/D4 T4S system, deletion mutation studies with the CRAfT screen have localized translocation signals to the C termini of the known secretion substrates (149, 150). All these substrates carry a consensus Arg motif and a similar hydropathy profile at their C termini, and an extensive mutational analysis has confirmed the importance of the Arg residues for VirF substrate transfer (150). These findings indicate that the VirB/D4 T4S system recognizes secretion substrates through charge-based interactions.
A study of the L. pneumophila Dot/Icm system similarly identified a translocation signal within the last 20 residues of the RalF secretion substrate. However, all 13 Dot/Icm substrates examined carried hydrophobic residues as opposed to Arg residues at the −3 or −4 positions, and a mutational analysis confirmed the importance of a Leu residue at the −3 position of RalF (105). A recently solved RalF crystal structure shows the C-terminal domain at the end of a long _α_-helix; this domain composed of hydrophobic residues is predicted to mediate interactions with Dot/Icm machine components (2).
Although the above findings suggest that clusters of positively charged or hydrophobic C-terminal residues are necessary for substrate recognition, genome-wide searches identify many proteins with such characteristics suggesting the importance of additional sequence information for substrate specificity. Supporting this view, in B. henselae, BepA–G carry positively charged C termini. Each Bep also carries at least one copy of a ~143-residue domain termed the Bep-intracellular delivery domain. Mutational analyses confirmed that both the charged C terminus and the Bep-intracellular delivery domain are necessary for T4S-mediated delivery to mammalian cells (132). In the X. axonopodis pv_. citri_ T4S system, all the potential XVIP secretion substrates carry a common ~100-residue motif near their C termini. Within these motifs, termed XVIPCDs, features of potential significance include several blocks of conserved sequences and a Glnrich C terminus. The XVIPCD sequence motifs and the Glnrich C termini might correspond to a bipartite substrate recognition signal for the XVIPs (1).
Substrate Recognition Checkpoints Along the Translocation Pathway
The above observations suggest that T4S systems recognize cognate substrates minimally through bipartite signals composed of C-terminal residues and another specificity motif. Alternatively, or in addition, there is evidence that T4S machines themselves have evolved substrate discrimination mechanisms. Recent findings suggest that a soluble IcmS-IcmW complex, the integral membrane protein DotF, and possibly other Dot/Icm subunits mediate recognition of different substrates of the L. pneumophila Dot/Icm T4S apparatus (96, 108, 148, 164). Also, the TraDF T4CP possesses a 140-residue C-terminal extension not carried by other T4CPs (124). The 38 residues at the extreme C terminus mediate an interaction with TraM, a component of the relaxosome responsible for processing at the oriT sequences of F-like plasmids. This TraD-TraM interaction thus forms a basis for recognition of F plasmid substrates (14, 48). By contrast, a strain producing a TraD C-terminal deletion mutation or one lacking TraM is still capable of efficiently translocating other DNA substrates of this T4S machine, suggestive of a distinct recognition mechanism for such substrates (14, 124).
T4S machines also translocate promiscuous substrates, the best-characterized examples are the conjugation machines, e.g., F, RP4, R388, pKM101, which translocate the nonself-transmissible IncQ plasmid RSF1010 (52, 60, 88). Machines such as the A. tumefaciens VirB/D4 T4S and L. pneumophila Dot/Icm systems also translocate the MobARSF1010 relaxase, even independently of its association with RSF1010 DNA (96, 150). MobARSF1010 carries a positively charged C terminus, which could enable recognition by the VirB/D4 T4S system (29, 150). However, such promiscuous substrates must also carry translocation signals recognizable by many different T4S systems. The characteristics of such signals are not presently defined.
The docking of substrates at the T4CP receptor clearly constitutes a major substrate specificity checkpoint. This, however, is not the only checkpoint, as shown by mutational analyses of VirB11Ti and the outer membrane protein VirB9Ti. For VirB11Ti, several mutations alternatively block the translocation of VirD2Ti-T-strand or MobARSF1010-R-strand transfer intermediates (121). More recent work with an assay described below has confirmed that these mutations do not affect T4CP contacts with either substrate. Rather, they specifically arrest formation of VirB11Ti contacts with one or the other of these substrates (E. Cascales & P.J. Christie, unpublished findings). In addition, VirB9Ti mutations were recovered that selectively block translocation of the VirD2Ti-T-strand, whereas others block transit of the MobARSF1010-R-strand. Again, these mutations do not affect upstream channel subunit contacts with either substrate. Both classes of the VirB9Ti substrate discrimination mutations map predominantly to the N-terminal region of VirB9, which is implicated in establishing contacts with the VirB10 energy sensor subunit (24, 71). Although VirD2Ti and MobARSF1010 possess C-terminal Arg motifs, the findings suggest that VirB11Ti and the N terminus of VirB9Ti could recognize substrate translocation signals unique to each relaxase (71, 121).
DNA SUBSTRATE TRANSLOCATION THROUGH THE SECRETION CHANNEL
A long-standing question of interest is how T4S systems deliver DNA substrates across the cell envelope. Progress toward answering this question has been made through the recent development of an assay termed TrIP (25). Adapted from the chromatin immunoprecipitation technique, TrIP is based on formaldehyde treatment of intact cells to crosslink channel subunits to the DNA substrate as it exits the cell. The crosslinked substrate-channel subunit complexes are then recovered from detergent-solubilized cell extracts by immunoprecipitation, and PCR amplification is used to detect the precipitated DNA substrate. With this assay, the T-DNA substrate exported to plant cells by the A. tumefaciens VirB/D4 T4S system forms close contacts with 6 of 12 machine subunits. These include the VirD4 T4CP, VirB11 ATPase, VirB6, VirB8, VirB2, pilin and VirB9. Further TrIP studies of T4S mutant strains enabled formulation of a sequentially and spatially ordered translocation pathway for this substrate. Below, we describe this postulated pathway and identify the subunit and energetic requirements for each transfer step (25) (Figure 4).
Figure 4.
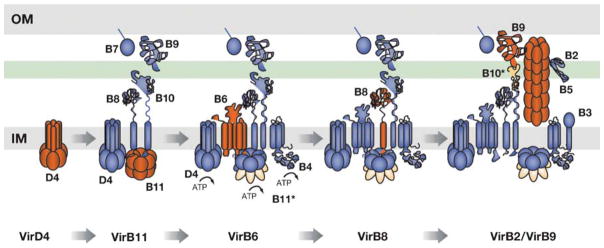
Translocation pathway for the VirD2-T-strand transfer intermediate through the A. tumefaciens VirB/D4 secretion channel. The DNA substrate forms close contacts with VirB/D4 subunits in temporal order as delineated at the bottom. Subunits interacting with substrate at each step of the pathway are depicted in red, and subunits required at each successive subunit-substrate contact are depicted in blue. Substrate passage through the channel is depicted by a change in coloration in the channel subunit from red to blue. ATP energy is required for substrate transfer to VirB6 and each subsequent transfer step. Conformational changes in VirB11 and VirB10 required for substrate translocation are depicted in orange. IM, inner membrane; OM, outer membrane.
DNA Substrate Binding to the T4CP Receptor
Using the TrIP assay, it was shown that the VirD4 T4CP forms a close contact with the T-DNA in wild-type cells as well as mutants deleted of the virB genes (25) (Figure 4). These findings confirm that the T4CP functions as a DNA substrate receptor in vivo. A VirD4 Walker A mutant also efficiently crosslinked to the DNA substrate, suggesting that ATP utilization does not contribute to substrate reception (4). Parallel investigations showed that a VirD4 Walker mutant also retains the ability to interact with the VirE2 protein substrate (5). Therefore, the VirD4 T4CP binds DNA and protein substrates independently of other Mpf components or ATP energy.
Plasmids have evolved mechanisms to inhibit the transmission of coresident plasmids. This phenomenon, termed fertility inhibition, operates at the level of tra gene transcription for some systems and posttranslationally on some aspect of T4S function for others (147, 158). In A. tumefaciens, a form of posttranslational fertility inhibition known as oncogenic suppression is mediated by the MobARSF1010-R-strand transfer intermediate and by the Osa protein of the IncW plasmid pSa (17, 26). Using the TrIP assay, it was established that both MobARSF1010-R-strand and Osa block access of the T-DNA substrate to the VirD4Ti receptor (E. Cascales, A. Krishnamohan, Z. Liu, A.N. Binns & P.J. Christie, unpublished data). In addition, both oncogenic suppressors block access of VirE2 to VirD4Ti, as shown with biochemical and cytological assays. These findings thus identify a mechanistic basis, T4S receptor interference, for a posttranslational form of fertility inhibition (E. Cascales, A. Krishnamohan, Z. Liu, A.N. Binns & P.J. Christie, unpublished data).
Transfer to the VirB11 ATPase
The VirD4 T4CP delivers the DNA substrate to VirB11, as shown by TrIP (25) (Figure 4). Further studies exploring the requirements for this transfer reaction have established the following. First, Walker A mutant forms of VirD4 and VirB11 mediate this transfer step, indicating that ATP utilization by both energetic components is dispensable for this transfer step (4). In contrast, studies of virB gene deletion mutants and strains producing subsets of VirB subunits showed that combinations of VirB7, VirB8 and VirB9, or VirB7 and VirB10 are necessary for substrate transfer from VirD4 to VirB11 (4, 25). VirD4 therefore delivers the DNA substrate to VirB11 independently of ATP energy, but only in the context of core subunits. In future studies, it will be interesting to define how the two combinations of ‘core’ subunits stimulate this transfer step.
Transfer to the Integral Inner Membrane Subunits VirB6 and VirB8
The inner membrane subunits VirB6 and VirB8 function at an intermediate point in the DNA translocation pathway, after substrate transfer to VirB11 and before transfer to VirB2 and VirB9 (25) (Figure 4). Recently, collections of VirB6 insertion and deletion mutations exerted a variety of interesting effects on substrate translocation (72). Results of these studies indicated, first, that mutations in the periplasmic loop P2 abolish substrate transfer to VirB6. Second, mutations in a multimembrane-spanning domain distal to loop P2 do not affect formation of substrate contacts with VirB6 but abolish substrate interactions with VirB8. Finally, mutations in domains at the N- and C-termini of VirB6 do not affect formation of substrate contacts with VirB6 and VirB8, but abolish substrate interactions with VirB2 and VirB9. On the basis of these findings, it is postulated that loop P2 forms part of the secretion channel structure, whereas other domains of VirB6 form critical intersubunit contacts required for assembly and function of the channel at and beyond the inner membrane (72). A combination of virB mutant analyses and in vivo reconstitution studies also established that substrate transfer to VirB6 and VirB8 requires the energetic components VirD4, VirB4, and VirB11, each with intact Walker A motifs, together with the core subunits VirB7 and either VirB9 or VirB10 (4, 25).
Transfer to the Periplasmic Outer-Membrane-Associated Subunits VirB2 and VirB9
VirB2 and VirB9 form the distal end of the proposed DNA translocation channel (25) (Figure 4). VirB2 and VirB9 appear to function together to mediate the latter step(s) of substrate transfer (25). An association of the DNA substrate with VirB2, a pilin subunit, is of special interest. Although this finding could be an indication of substrate transfer through the pilus, the isolation of uncoupling mutations described above suggests instead that the pilin monomer is a component of both the pilus and the channel that extends across the periplasm. To examine this question further, it was asked whether uncoupling mutations conferring a transfer-positive, pilus-minus phenotype also support transfer in the complete absence of VirB2 pilin synthesis. Such mutations could not compensate for a lack of pilin, strongly indicating that the cellular form of VirB2 pilin is essential for substrate transfer across the cell envelope (71).
Recently, the contribution of VirB9 to substrate transfer was assessed by examining the phenotypic consequences of small two-residue insertion mutations distributed along the length of the polypeptide. Interestingly, mutations mapping to conserved N- and C-terminal domains blocked substrate transfer to both VirB2 and VirB9, suggesting that these domains form critical contacts with the DNA substrate, VirB2, and/or other channel subunits necessary for the last stages of transfer. Also, as noted above, the mutational analyses supplied evidence that the N terminus of VirB9 functions as a substrate selection checkpoint by regulating the transfer of substrates through the distal portion of the secretion channel (71).
The TrIP studies have failed to detect substrate contacts with the VirB3, VirB5, and VirB10 subunits, VirB1 transglysolase, and VirJ, a virulence protein proposed to contribute to T4S machine function (25, 112). Interestingly, however, strains lacking each of the VirB3, VirB5, and VirB10 subunits fail to translocate the substrate from VirB6 and VirB8 to VirB2 and VirB9, indicating that VirB3, VirB5, and VirB10 are essential for substrate trafficking through the periplasm. The contributions of VirB3 and VirB5 to substrate transfer presently are unexplored. As mentioned above, TonB-like VirB10 senses ATP utilization by VirD4 and VirB11 and, in turn, forms a stable VirB9-VirB10 complex. This structural transition is strongly correlated with the capacity of substrate to pass from the portion of the channel composed of VirB6 and VirB8 to that composed of VirB2 and VirB9 (24).
It can be proposed that the VirB/D4 T4S channel forms incompletely in the absence of ATP utilization by the VirD4 and VirB11 AT-Pases. A final step(s) of assembly, subsequent to those depicted in Figure 3, would proceed only upon receipt of intrinsic (e.g., ATP binding) and, possibly, extrinsic (e.g., donor-recipient cell contact) signals. Indeed, two recent findings now point to a second intrinsic signal, the binding of a DNA substrate to the T4CP receptor, as necessary for channel morphogenesis and function. First, as mentioned above, DNA binding stimulates ATP hydrolysis by the TrwBR388 T4CP (142). Second, very recently it was determined that the observed structural transition of VirB10 as well as formation of the VirB9-VirB10 complex is dependent not only on ATP utilization by VirD4 T4CP and VirB11, but also on DNA substrate binding to the VirD4 T4CP (24; E. Cascales & P.J. Christie, unpublished findings). Thus, we propose that a combination of DNA substrate docking at the T4CP receptor with accompanying DNA-dependent stimulation of T4CP ATP hydrolysis serves to induce a structural transition in TonB-like VirB10 required for late-stage assembly (or gating) of the transenvelope secretion channel.
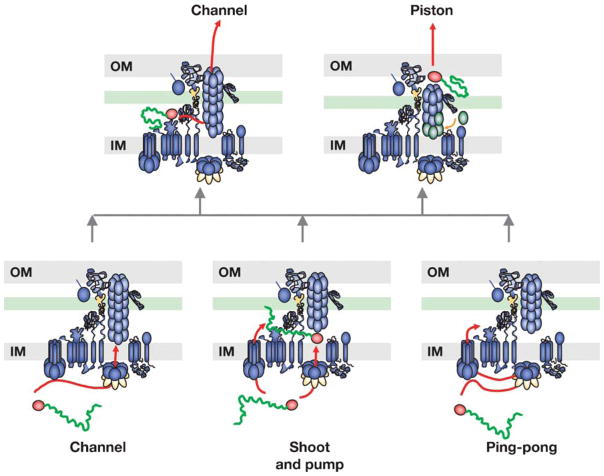
MODELS DESCRIBING THE TRANSLOCATION PATHWAY FOR A DNA SUBSTRATE THROUGH THE T4S CHANNEL
Currently, three pathways describe the possible route of DNA substrate transfer across the inner membrane (Figure 5). These are the channel, shoot-and-pump, and ping-pong models. The first, perhaps the easiest to envision, involves recruitment of secretion substrates by the T4CP and then substrate transfer through an inner membrane channel composed of the Mpf subunits. By incorporating findings from the TrIP assay, researchers postulate functions for VirD4 as a substrate receptor, VirB11 as a chaperone for unfolding the relaxase, and VirB6 and VirB8 as the inner membrane channel components (4, 72). The energetic subunits (e.g., VirB4, VirB11, VirD4) would energize conformational changes required for channel gating, and other subunits (VirB7, VirB9, VirB10) would contribute structurally to the inner membrane channel. This model accommodates the recent proposal that VirB4 hexamers engage with VirD4 and VirB11 to facilitate substrate passage through an inner membrane channel (102).
Figure 5.
Proposed models for DNA substrate translocation across the A. tumefaciens cell envelope. The A. tumefaciens VirB/D4 T4S apparatus is represented as a transenvelope organelle that mediates passage of the nucleoprotein substrate (ball-line; red arrow). The different models proposed for substrate translocation through the inner membrane (bottom level: channel, shoot and pump, ping-pong) and through the outer membrane (top level: channel, piston) are depicted. Dynamic conformational changes (VirB11 ATP utilization; VirB2 pilin polymerization; VirB10 structural transition) required for substrate passage are highlighted in orange or green.
The shoot-and-pump model is more consistent with available T4CP structure-function data (93) (Figure 5). The structures, and the recent demonstration of a DNA-dependent ATPase activity for the TrwBR388 T4CP (142), strongly suggest that the T4CPs function as DNA pumps. This model depicts the T4CP as a DNA substrate receptor. The T4CP then delivers only the relaxase component of the DNA transfer intermediate to the Mpf complex. The VirB11-like chaperone unfolds the relaxase and also coordinates with other Mpf subunits to deliver the relaxase across the membrane (shoot). Simultaneously, the T4CP uses ATP energy to independently drive the T-strand across the membrane (pump). Two dedicated DNA and protein translocases would coordinate the simultaneous delivery of a covalent protein-DNA substrate across the inner membrane (93).
The ping-pong model depicts the T4CP as the only translocase for the T4S systems (4) (Figure 5). The T4CP recruits the T-DNA to the T4S system and transfers the relaxase component to the VirB11 chaperone for relaxase unfolding (ping). At this point, VirB11 retrotransfers the unfolded substrate back to the T4CP (pong). The T4CP then uses ATP energy to translocate the DNA-protein transfer intermediate across the inner membrane. Other VirB subunits would contribute structurally to establishment of productive VirD4 and VirB11 interactions and the dynamics of substrate exchange between these ATPases. The inner membrane subunits VirB4, VirB6, and VirB8 also would participate in substrate transfer from the T4CP to the portion of the channel that extends through the periplasm (4, 72).
Although each model can explain much of the available data, in light of the T4CP structure and biochemical activities, it is hard to believe that the T4CP plays a completely passive role in mediating substrate transfer across the inner membrane, as depicted in the channel model. Moreover, unless significant changes in quaternary structures of the putative T4CP and Mpf channels are possible, it is difficult to conceptualize how a covalent protein-ssDNA complex can be delivered simultaneously through two different translocons. Therefore, at present the ping-pong model seems to best explain the available structural (57, 125), in vitro biochemical (57, 79, 80, 125), and TrIP data (4, 24, 25, 72).
Finally, alternative mechanisms have been proposed to explain how the secretion substrates pass through the periplasm and outer membrane (23) (Figure 5). In the channel model, the substrates move through the VirB2 pilus, whereas in the piston model, the substrates are delivered to the tip of the nascent pilus and extruded across the outer membrane during outgrowth. At this time, it is not possible to discriminate between these or other models. To shed light on the mechanics of substrate transfer across the outer membrane, the immediate goal must be to answer a longstanding question of whether the T4S systems assemble an outer membrane secretin complex.
PROSPECTS
Studies of the T4S systems in many laboratories are rapidly advancing our understanding of the T4S systems at many levels. For a comprehensive mechanistic picture of T4S machine assembly and function, the task for the future is to continue to generate structural information for machine subunits and substructures and to integrate this information with other biochemical data and recent findings developed through use of novel in vivo assays exploring dynamic aspects of substrate engagement and transfer. Mechanistic studies of T4S systems that are phylogenetically diverse from the well-studied model conjugation systems are needed. Equally important are the continued investigations of substrates and their roles in subverting host cell physiologies during pathogenesis. The pace of research surrounding the biology of T4S is phenomenal, as reflected by an explosion of literature on this topic over the past several years.
SUMMARY POINTS.
- Updated information is presented for structures, membrane topologies, and catalytic activities of the T4S subunits.
- A biogenesis pathway for an archetypal T4S system is depicted on the basis of subunit-subunit interaction data and other biochemical findings.
- Energetic requirements are specified for T4S machine assembly and specific stages of substrate transfer through the secretion channel.
- Studies exploring the positioning of T4S machine subunits and pilus structures at the cell surface are reviewed.
- A table summarizes the available information on T4S protein substrates and the cellular consequences of transfer to eukaryotic cells.
- Mechanisms for recognition of substrates and regulating substrate passage through the secretion channel are discussed.
- The route by which a DNA substrate translocates through a T4S channel is delineated and the genetic requirements for steps of the pathway are presented.
- Models depicting the dynamics of substrate translocation are compared.
Acknowledgments
We gratefully acknowledge contributions of published materials by M. Coll, F. de la Cruz, R. Haas, R. Isberg, G. Koraimann, and G. Waksman. We also thank C. Baron, A. Bernadac, E. Cabezon, M. Coll, F. de la Cruz, C. Dehio, L. Terradot, G. Waksman, and P. Zambryski for sharing unpublished data included in this review. We are grateful to X. Zhang and L. Coutte for helpful discussions. Space limitations have prevented citation of many primary manuscripts and we apologize to the authors whose reports were not referenced. Research in the Christie laboratory is supported by NIH grant GM48746.
Glossary
Conjugation
a mechanism for transfer of DNA substrates from a bacterial donor cell to a recipient cell through establishment of mating junctions
Relaxase
a transesterase that binds the oriT sequence of a mobile DNA element and generates a site- and strand-specific nick to initiate conjugal processing of substrate DNA
Competence
a physiological condition permitting the uptake of DNA from the environment
Pilus
an extracellular filament composed of pilin monomers
PT
pertussis toxin
Mpf
mating pair formation subunits comprise the cell-envelope-spanning channel and extracellular pilus of the T4S systems
T4CP
type IV coupling protein; links the DNA processing machinery assembled at the oriT sequence of a mobile element to the Mpf structure
Dtr
DNA transfer and replication; function in initiating conjugal processing of DNA substrates through binding to the oriT sequence of a mobile element
oriT
origin of transfer
TMS
transmembrane segment; a contiguous stretch of ~20 hydrophobic residues that spans the inner membrane of Gram-negative bacteria.
Walker A and B motifs
canonical sequences carried by a superfamily of proteins that bind or hydrolyze nucleoside triphosphates; the Walker A motif is also designated as the phosphate binding or P-loop
pmf
proton-motive force
CRAfT
Cre reporter assay for translocation
Bep
_Bartonella_-translocated effector protein protein
XVIPCDs
XVIP conserved domains
TrIP
transfer DNA immunoprecipitation; a chemical crosslinking assay for recovery of complexes composed of T4S machine subunits and DNA substrates from intact cells
LITERATURE CITED
- 1.Alegria MC, Docena C, Khater L, Ramos CHI, da Silva ACR, Farah CS. Identification of new protein-protein interactions involving products of the chromosome- and plasmid-encoded type IV secretion loci of the phytopathogen Xanthomonas axonopodis pv.citri. J Bacteriol. 2005;187:2315–25. doi: 10.1128/JB.187.7.2315-2325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amor JC, Swails J, Zhu X, Roy CR, Nagai H, et al. The structure of RalF, an ADP-ribosylation factor guanine nucleotide exchange factor from Legionella pneumophila, reveals the presence of a cap over the active site. J Biol Chem. 2005;280:1392–400. doi: 10.1074/jbc.M410820200. [DOI] [PubMed] [Google Scholar]
- 3.Anderson LB, Hertzel AV, Das A. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc Natl Acad Sci USA. 1996;93:8889–94. doi: 10.1073/pnas.93.17.8889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atmakuri K, Cascales E, Christie PJ. Energetic components VirD4, VirB11 and VirB4 mediate early DNA transfer reactions required for bacterial type IV secretion. Mol Microbiol. 2004;54:1199–211. doi: 10.1111/j.1365-2958.2004.04345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Atmakuri K, Ding Z, Christie PJ. VirE2, a type IV secretion substrate, interacts with the VirD4 transfer protein at cell poles of Agrobacterium tumefaciens. Mol Microbiol. 2003;49:1699–713. doi: 10.1046/j.1365-2958.2003.03669.x. This study nicely integrates novel cytological and classical approaches to demonstrate that T4CP functions as a protein receptor for T4S machines. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baron C, O’Callaghan D, Lanka E. Bacterial secrets of secretion: Euroconference on the biology of type IV secretion processes. Mol Microbiol. 2002;43:1359–65. doi: 10.1046/j.1365-2958.2002.02816.x. [DOI] [PubMed] [Google Scholar]
- 7.Baron C, Thorstenson YR, Zambryski PC. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J Bacteriol. 1997;179:1211–18. doi: 10.1128/jb.179.4.1211-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Batchelor RA, Pearson BM, Friis LM, Guerry P, Wells JM. Nucleotide sequences and comparisons of two large conjugative plasmids from different Campylobacter species. Microbiology. 2004;153(Pt. 10):3507–17. doi: 10.1099/mic.0.27112-0. [DOI] [PubMed] [Google Scholar]
- 9.Bates S, Cashmore AM, Wilkins BM. IncP plasmids are unusually effective in mediating conjugation of Escherichia coli and Saccharomyces cerevisiae. J Bacteriol. 1998;180:6538–43. doi: 10.1128/jb.180.24.6538-6543.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batut J, Andersson SG, O’Callaghan D. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat Rev Microbiol. 2004;2:933–45. doi: 10.1038/nrmicro1044. [DOI] [PubMed] [Google Scholar]
- 11.Bayer M, Iberer R, Bischof K, Rassi E, Stabentheiner E, et al. Functional and mutational analysis of p19, a DNA transfer protein with muramidase activity. J Bacteriol. 2001;183:3176–83. doi: 10.1128/JB.183.10.3176-3183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaupre CE, Bohne J, Dale EM, Binns AN. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beijersbergen A, Smith SJ, Hooykaas PJ. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid. 1994;32:212–18. doi: 10.1006/plas.1994.1057. [DOI] [PubMed] [Google Scholar]
- 14.Beranek A, Zettl M, Lorenzoni K, Schauer A, Manhart M, Koraimann G. Thirty-eight C-terminal amino acids of the coupling protein TraD of the F-like conjugative resistance plasmid R1 are required and sufficient to confer binding to the substrate selector protein TraM. J Bacteriol. 2004;186:6999–7006. doi: 10.1128/JB.186.20.6999-7006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J Bacteriol. 1994;176:3646–60. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhattacharjee MK, Kachlany SC, Fine DH, Figurski DH. Nonspecific adherence and fibril biogenesis by Actinobacillus actinomycetemcomitans: TadA protein is an ATPase. J Bacteriol. 2001;183:5927–36. doi: 10.1128/JB.183.20.5927-5936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Binns A, Beaupre C, Dale E. Inhibition of VirB-mediated transfer of diverse substrates from Agrobacterium tumefaciens by the IncQ plasmid RSF1010. J Bacteriol. 1995;177:4890–99. doi: 10.1128/jb.177.17.4890-4899.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bohne J, Yim A, Binns AN. The Ti plasmid increases the efficiency of Agrobacterium tumefaciens as a recipient in virB-mediated conjugal transfer of an IncQ plasmid. Proc Natl Acad Sci USA. 1998;95:8057–62. doi: 10.1073/pnas.95.12.7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bundock P, den DRA, Beijersbergen A, Hooykaas PJ. Trans-kingdom T-DNA transfer from Agrobacterium tumefaciens. EMBO J. 1995;14:3206–14. doi: 10.1002/j.1460-2075.1995.tb07323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burns DL. Type IV transporters of pathogenic bacteria. Curr Opin Microbiol. 2003;6:1–6. doi: 10.1016/s1369-5274(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 21.Campodonico EM, Chesnel L, Roy CR. A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol Microbiol. 2005;56:918–33. doi: 10.1111/j.1365-2958.2005.04595.x. [DOI] [PubMed] [Google Scholar]
- 22.Cao TB, Saier MH., Jr Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology. 2001;147(Pt. 12):3201–14. doi: 10.1099/00221287-147-12-3201. [DOI] [PubMed] [Google Scholar]
- 23.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–49. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cascales E, Christie PJ. Agrobacterium VirB10, an ATP energy sensor required for type IV secretion. Proc Natl Acad Sci USA. 2004;101:17228–33. doi: 10.1073/pnas.0405843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–73. doi: 10.1126/science.1095211. This study uses TrIP to define for the first time the translocation route for a DNA substrate through the T4S channel. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CY, Kado CI. Inhibition of Agrobacterium tumefaciens oncogenicity by the osa gene of pSa. J Bacteriol. 1994;176:5697–703. doi: 10.1128/jb.176.18.5697-5703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, de Felipe KS, Clarke M, Lu H, Anderson OR, et al. Legionella effectors that promote nonlytic release from protozoa. Science. 2004;303:1358–61. doi: 10.1126/science.1094226. [DOI] [PubMed] [Google Scholar]
- 28.Christie PJ. The Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–94. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christie PJ. Bacterial type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochem Biophys Acta. 2004;1694:219–34. doi: 10.1016/j.bbamcr.2004.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christie PJ, Cascales E. Structural and dynamic properties of bacterial type IV secretion [Review] Mol Membr Biol. 2005;22:51–61. doi: 10.1080/09687860500063316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christie PJ, Vogel JP. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–60. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Citovsky V, Kapelnikov A, Oliel S, Zakai N, Rojas MR, et al. Protein interactions involved in nuclear import of the Agrobacterium VirE2 protein in vivo and in vitro. J Biol Chem. 2004;279:29528–33. doi: 10.1074/jbc.M403159200. [DOI] [PubMed] [Google Scholar]
- 33.Conover GM, Derre I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: a translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–21. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- 34.Dang TA, Christie PJ. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J Bacteriol. 1997;179:453–62. doi: 10.1128/jb.179.2.453-462.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Das A, Anderson LB, Xie YH. Delineation of the interaction domains of Agrobacterium tumefaciens VirB7 and VirB9 by use of the yeast two-hybrid assay. J Bacteriol. 1997;179:3404–9. doi: 10.1128/jb.179.11.3404-3409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das A, Xie Y-H. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J Bacteriol. 2000;182:758–63. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das A, Xie YH. Construction of transposon Tn3phoA: its application in defining the membrane topology of the Agrobacterium tumefaciens DNA transfer proteins. Mol Microbiol. 1998;27:405–14. doi: 10.1046/j.1365-2958.1998.00688.x. [DOI] [PubMed] [Google Scholar]
- 38.Daugelavicius R, Bamford JKH, Grahn AM, Lanka E, Bamford D. The IncP plasmid-encoded cell envelope-associated DNA transfer complex increases cell permeability. J Bacteriol. 1997;179:5195–202. doi: 10.1128/jb.179.16.5195-5202.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dehio C. Molecular and cellular basis of Bartonella pathogenesis. Annu Rev Microbiol. 2004;58:365–90. doi: 10.1146/annurev.micro.58.030603.123700. [DOI] [PubMed] [Google Scholar]
- 40.den Hartigh AB, Sun YH, Sondervan D, Heuvelmans N, Reinders MO, et al. Differential requirements for VirB1 and VirB2 during Brucella abortus infection. Infect Immun. 2004;72:5143–49. doi: 10.1128/IAI.72.9.5143-5149.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41:263–77. doi: 10.1046/j.1365-2958.2001.02520.x. This study and those cited in References 68 and 69 report the discovery of novel T4S systems that release DNA substrates into or acquire them from the milieu. [DOI] [PubMed] [Google Scholar]
- 42.Ding Z, Atmakuri K, Christie PJ. The outs and ins of bacterial type IV secretion substrates. Trends Microbiol. 2003;11:527–35. doi: 10.1016/j.tim.2003.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Draskovic I, Dubnau D. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol Microbiol. 2005;55:881–96. doi: 10.1111/j.1365-2958.2004.04430.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duckely M, Hohn B. The VirE2 protein of Agrobacterium tumefaciens: the yin and yang of T-DNA transfer. FEMS Microbiol Lett. 2003;223:1–6. doi: 10.1016/S0378-1097(03)00246-5. [DOI] [PubMed] [Google Scholar]
- 45.Eisenbrandt R, Kalkum M, Lai EM, Lurz R, Kado CI, Lanka E. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J Biol Chem. 1999;274:22548–55. doi: 10.1074/jbc.274.32.22548. This study describes a novel processing reaction that culminates in head-to-tail cyclization of pilin monomers. [DOI] [PubMed] [Google Scholar]
- 46.Eisenbrandt R, Kalkum M, Lurz R, Lanka E. Maturation of IncP pilin precursors resembles the catalytic dyad-like mechanism of leader peptidases. J Bacteriol. 2000;182:6751–61. doi: 10.1128/jb.182.23.6751-6761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Engledow AS, Medrano EG, Mahenthiralingam E, LiPuma JJ, Gonzalez CF. Involvement of a plasmid-encoded type IV secretion system in the plant tissue watersoaking phenotype of Burkholderia cenocepacia. J Bacteriol. 2004;186:6015–24. doi: 10.1128/JB.186.18.6015-6024.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fekete RA, Frost LS. Mobilization of chimeric oriT plasmids by F and R100–1: role of relaxosome formation in defining plasmid specificity. J Bacteriol. 2000;182:4022–27. doi: 10.1128/jb.182.14.4022-4027.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fernandez D, Dang TA, Spudich GM, Zhou XR, Berger BR, Christie PJ. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J Bacteriol. 1996;178:3156–67. doi: 10.1128/jb.178.11.3156-3167.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fernandez D, Spudich GM, Zhou XR, Christie PJ. The Agrobacterium tumefaciens VirB7 lipoprotein is required for stabilization of VirB proteins during assembly of the T-complex transport apparatus. J Bacteriol. 1996;178:3168–76. doi: 10.1128/jb.178.11.3168-3176.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fischer W, Haas R, Odenbreit S. Type IV secretion systems in pathogenic bacteria. Int J Med Microbiol. 2002;292:159–68. doi: 10.1078/1438-4221-00199. [DOI] [PubMed] [Google Scholar]
- 52.Fullner KJ. Role of Agrobacterium virB genes in transfer of T complexes and RSF1010. J Bacteriol. 1998;180:430–34. doi: 10.1128/jb.180.2.430-434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gilmour MW, Gunton JE, Lawley TD, Taylor DE. Interaction between the IncHI1 plasmid R27 coupling protein and type IV secretion system: TraG associates with the coiled-coil mating pair formation protein TrhB. Mol Microbiol. 2003;49:105–16. doi: 10.1046/j.1365-2958.2003.03551.x. [DOI] [PubMed] [Google Scholar]
- 54.Gilmour MW, Taylor DE. A subassembly of R27-encoded transfer proteins is dependent on TrhC nucleoside triphosphate-binding motifs for function but not formation. J Bacteriol. 2004;186:1606–13. doi: 10.1128/JB.186.6.1606-1613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gilmour MW, Lawley TD, Rooker MM, Newnham PJ, Taylor DE. Cellular location and temperature-dependent assembly of IncHI1 plasmid R27-encoded TrhC-associated conjugative transfer protein complexes. Mol Microbiol. 2001;42:705–15. doi: 10.1046/j.1365-2958.2001.02682.x. [DOI] [PubMed] [Google Scholar]
- 56.Gomis-Ruth FX, Moncalian G, Perez-Luque R, Gonzalez A, Cabezon E, et al. The bacterial conjugation protein TrwB resembles ring helicases and F1-ATPase. Nature. 2001;409:637–41. doi: 10.1038/35054586. This study reports on a structure of the type IV coupling protein. [DOI] [PubMed] [Google Scholar]
- 57.Gomis-Ruth FX, Sola M, de la Cruz F, Coll M. Coupling factors in macromolecular type-IV secretion machineries. Curr Pharm Des. 2004;10:1551–65. doi: 10.2174/1381612043384817. [DOI] [PubMed] [Google Scholar]
- 58.Grahn AM, Haase J, Bamford DH, Lanka E. Components of the RP4 conjugative transfer apparatus form an envelope structure bridging inner and outer membranes of donor cells: implications for related macromolecule transport systems. J Bacteriol. 2000;182:1564–74. doi: 10.1128/jb.182.6.1564-1574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in Gram-positive bacteria. Microb Mol Biol Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haase J, Lurz R, Grahn AM, Bamford DH, Lanka E. Bacterial conjugation mediated by plasmid RP4: RSF1010 mobilization, donor-specific phage propagation, and pilus production require the same Tra2 core components of a proposed DNA transport complex. J Bacteriol. 1995;177:4779–91. doi: 10.1128/jb.177.16.4779-4791.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hamilton CM, Lee H, Li PL, Cook DM, Piper KR, et al. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J Bacteriol. 2000;182:1541–48. doi: 10.1128/jb.182.6.1541-1548.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hapfelmeier S, Domke N, Zambryski PC, Baron C. VirB6 is required for stabilization of VirB5 and VirB3 and formation of VirB7 homodimers in Agrobacterium tumefaciens. J Bacteriol. 2000;182:4505–11. doi: 10.1128/jb.182.16.4505-4511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harris RL, Silverman PM. Tra proteins characteristic of F-like type IV secretion systems constitute an interaction group by yeast two-hybrid analysis. J Bacteriol. 2004;186:5480–85. doi: 10.1128/JB.186.16.5480-5485.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hatakeyama M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat Rev Cancer. 2004;4:688–94. doi: 10.1038/nrc1433. [DOI] [PubMed] [Google Scholar]
- 65.He SY, Nomura K, Whittam TS. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim Biophys Acta. 2004;1694:181–206. doi: 10.1016/j.bbamcr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 66.Herdendorf TJ, McCaslin DR, Forest KT. Aquifex aeolicus PilT, homologue of a surface motility protein, is a thermostable oligomeric NTPase. J Bacteriol. 2002;184:6465–71. doi: 10.1128/JB.184.23.6465-6471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hodges LD, Cuperus J, Ream W. Agrobacterium rhizogenes GALLS protein substitutes for Agrobacterium tumefaciens single-stranded DNA-binding protein VirE2. J Bacteriol. 2004;186:3065–77. doi: 10.1128/JB.186.10.3065-3077.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hofreuter D, Karnholz A, Haas R. Topology and membrane interaction of Helicobacter pylori ComB proteins involved in natural transformation competence. Int J Med Microbiol. 2003;293:153–65. doi: 10.1078/1438-4221-00258. [DOI] [PubMed] [Google Scholar]
- 69.Hofreuter D, Odenbreit S, Haas R. Natural transformation competence in Helicobacter pylori is mediated by the basic components of a type IV secretion system. Mol Microbiol. 2001;41:379–91. doi: 10.1046/j.1365-2958.2001.02502.x. [DOI] [PubMed] [Google Scholar]
- 70.Hubber A, Vergunst AC, Sullivan JT, Hooykaas PJ, Ronson CW. Symbiotic phenotypes and translocated effector proteins of the Mesorhizobium loti strain R7A VirB/D4 type IV secretion system. Mol Microbiol. 2004;54:561–74. doi: 10.1111/j.1365-2958.2004.04292.x. [DOI] [PubMed] [Google Scholar]
- 71.Jakubowski S, Cascales E, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J Bacteriol. 2005;187:3486–95. doi: 10.1128/JB.187.10.3486-3495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a type IV secretion system. J Mol Biol. 2004;341:961–77. doi: 10.1016/j.jmb.2004.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jakubowski SJ, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB6 protein participates in formation of VirB7 and VirB9 complexes required for type IV secretion. J Bacteriol. 2003;185:2867–78. doi: 10.1128/JB.185.9.2867-2878.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jones AL, Shirasu K, Kado CI. The product of the virB4 gene of Agrobacterium tumefaciens promotes accumulation of VirB3 protein. J Bacteriol. 1994;176:5255–61. doi: 10.1128/jb.176.17.5255-5261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Judd PK, Kumar RB, Das A. The type IV secretion apparatus protein VirB6 of Agrobacterium tumefaciens localizes to a cell pole. Mol Microbiol. 2005;55:115–24. doi: 10.1111/j.1365-2958.2004.04378.x. [DOI] [PubMed] [Google Scholar]
- 76.Kalkum M, Eisenbrandt R, Lanka E. Protein circlets as sex pilus subunits. Curr Protein Pept Sci. 2004;5:417–24. doi: 10.2174/1389203043379639. [DOI] [PubMed] [Google Scholar]
- 77.Koraimann G. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci. 2003;60:2371–88. doi: 10.1007/s00018-003-3056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krall L, Wiedemann U, Unsin G, Weiss S, Domke N, Baron C. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc Natl Acad Sci USA. 2002;99:11405–10. doi: 10.1073/pnas.172390699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Krause S, Barcena M, Pansegrau W, Lurz R, Carazo JM, Lanka E. Sequence-related protein export NTPases encoded by the conjugative transfer region of RP4 and by the cag pathogenicity island of Helicobacter pylori share similar hexameric ring structures. Proc Natl Acad Sci USA. 2000;97:3067–72. doi: 10.1073/pnas.050578697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Krause S, Pansegrau W, Lurz R, de la Cruz F, Lanka E. Enzymology of type IV macromolecule secretion systems: The conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J Bacteriol. 2000;182:2761–70. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kumar RB, Das A. Functional analysis of the Agrobacterium tumefaciens T-DNA transport pore protein VirB8. J Bacteriol. 2001;183:3636–41. doi: 10.1128/JB.183.12.3636-3641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumar RB, Das A. Polar location and functional domains of the Agrobacterium tumefaciens DNA transfer protein VirD4. Mol Microbiol. 2002;43:1523–32. doi: 10.1046/j.1365-2958.2002.02829.x. This is the first evidence for the polar localization of a T4S secretion channel. [DOI] [PubMed] [Google Scholar]
- 83.Kumar RB, Xie YH, Das A. Subcellular localization of the Agrobacterium tumefaciens T-DNA transport pore proteins: VirB8 is essential for the assembly of the transport pore. Mol Microbiol. 2000;36:608–17. doi: 10.1046/j.1365-2958.2000.01876.x. [DOI] [PubMed] [Google Scholar]
- 84.Lacroix B, Vaidya M, Tzfira T, Citovsky V. The VirE3 protein of Agrobacterium mimics a host cell function required for plant genetic transformation. EMBO J. 2005;24:428–37. doi: 10.1038/sj.emboj.7600524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lai EM, Chesnokova O, Banta LM, Kado CI. Genetic and environmental factors affecting T-pilin export and T-pilus biogenesis in relation to flagellation of Agrobacterium tumefaciens. J Bacteriol. 2000;182:3705–16. doi: 10.1128/jb.182.13.3705-3716.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lai EM, Eisenbrandt R, Kalkum M, Lanka E, Kado CI. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J Bacteriol. 2002;184:327–30. doi: 10.1128/JB.184.1.327-330.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lai EM, Kado CI. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J Bacteriol. 1998;180:2711–17. doi: 10.1128/jb.180.10.2711-2717.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lawley TD, Klimke WA, Gubbins MJ, Frost LS. F factor conjugation is a true type IV secretion system. FEMS Microbiol Lett. 2003;224:1–15. doi: 10.1016/S0378-1097(03)00430-0. [DOI] [PubMed] [Google Scholar]
- 89.Lee MH, Kosuk N, Bailey J, Traxler B, Manoil C. Analysis of F factor TraD membrane topology by use of gene fusions and trypsin-sensitive insertions. J Bacteriol. 1999;181:6108–13. doi: 10.1128/jb.181.19.6108-6113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;77:321–24. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 91.Letellier L, Boulanger P, de Frutos M, Jacquot P. Channeling phage DNA through membranes: from in vivo to in vitro. Res Microbiol. 2003;154:283–87. doi: 10.1016/S0923-2508(03)00072-X. [DOI] [PubMed] [Google Scholar]
- 92.Liu Z, Binns AN. Functional subsets of the VirB type IV transport complex proteins involved in the capacity of Agrobacterium tumefaciens to serve as a recipient in virB-mediated conjugal transfer of plasmid RSF1010. J Bacteriol. 2003;185:3259–69. doi: 10.1128/JB.185.11.3259-3269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Llosa M, Gomis-Ruth FX, Coll M, de la Cruz F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45:1–8. doi: 10.1046/j.1365-2958.2002.03014.x. [DOI] [PubMed] [Google Scholar]
- 94.Llosa M, O’Callaghan D. Euroconference on the biology of type IV secretion processes: bacterial gates into the outer world. Mol Microbiol. 2004;53:1–8. doi: 10.1111/j.1365-2958.2004.04168.x. [DOI] [PubMed] [Google Scholar]
- 95.Llosa M, Zunzunegui S, de la Cruz F. Conjugative coupling proteins interact with cognate and heterologous VirB10-like proteins while exhibiting specificity for cognate relaxosomes. Proc Natl Acad Sci USA. 2003;100:10465–70. doi: 10.1073/pnas.1830264100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841–46. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Maher D, Sherburne R, Taylor DE. H-pilus assembly kinetics determined by electron microscopy. J Bacteriol. 1993;175:2175–83. doi: 10.1128/jb.175.8.2175-2183.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maier B, Chen I, Dubnau D, Sheetz MP. DNA transport into Bacillus subtilis requires proton motive force to generate large molecular forces. Nat Struct Mol Biol. 2004;11:643–49. doi: 10.1038/nsmb783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Margolin W. Bacterial division: the fellowship of the ring. Curr Biol. 2003;13:R16–18. doi: 10.1016/s0960-9822(02)01381-7. [DOI] [PubMed] [Google Scholar]
- 100.Marlovits TC, Kubori T, Sukhan A, Thomas DR, Galan JE, Unger VM. Structural insights into the assembly of the type III secretion needle complex. Science. 2004;306:1040–42. doi: 10.1126/science.1102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matthysse AG. Characterization of nonattaching mutants of Agrobacterium. J Bacteriol. 1987;169:313–23. doi: 10.1128/jb.169.1.313-323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Middleton R, Sjolander K, Krishamurthy N, Foley J, Zambryski P. Predicted hexameric structure of the Agrobacterium VirB4 C terminus suggests VirB4 acts as a docking site during type IV secretion. Proc Natl Acad Sci USA. 2005;102:1685–90. doi: 10.1073/pnas.0409399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Moncalian G, Cabezon E, Alkorta I, Valle M, Moro F, et al. Characterization of ATP and DNA binding activities of TrwB, the coupling protein essential in plasmid R388 conjugation. J Biol Chem. 1999;274:36117–24. doi: 10.1074/jbc.274.51.36117. [DOI] [PubMed] [Google Scholar]
- 104.Mushegian AR, Fullner KJ, Koonin EV, Nester EW. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc Natl Acad Sci USA. 1996;93:7321–26. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nagai H, Cambronne ED, Kagan JC, Amor JC, Kahn RA, Roy CR. A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci USA. 2005;102:826–31. doi: 10.1073/pnas.0406239101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nagai H, Kagan JC, Zhu X, Kahn RA, Roy CR. A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science. 2002;295:679–82. doi: 10.1126/science.1067025. [DOI] [PubMed] [Google Scholar]
- 107.Nagai H, Roy CR. Show me the substrates: modulation of host cell function by type IV secretion systems. Cell Microbiol. 2003;5:373–83. doi: 10.1046/j.1462-5822.2003.00285.x. [DOI] [PubMed] [Google Scholar]
- 108.Ninio S, Zuckman-Cholon DM, Cambronne ED, Roy CR. The Legionella IcmS-IcmW protein complex is important for Dot/Icm-mediated protein translocation. Mol Microbiol. 2005;55:912–26. doi: 10.1111/j.1365-2958.2004.04435.x. [DOI] [PubMed] [Google Scholar]
- 109.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 110.Padmalayam I, Karem K, Baumstark B, Massung R. The gene encoding the 17-kDa antigen of Bartonella henselae is located within a cluster of genes homologous to the virB virulence operon. DNA Cell Biol. 2000;19:377–82. doi: 10.1089/10445490050043344. [DOI] [PubMed] [Google Scholar]
- 111.Palmen R, Driessen AJ, Hellingwerf KJ. Bioenergetic aspects of the translocation of macromolecules across bacterial membranes. Biochim Biophys Acta. 1994;1203:417–51. doi: 10.1016/0005-2728(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 112.Pantoja M, Chen L, Chen Y, Nester EW. Agrobacterium type IV secretion is a two-step process in which export substrates associate with the virulence protein VirJ in the periplasm. Mol Microbiol. 2002;45:1325–35. doi: 10.1046/j.1365-2958.2002.03098.x. [DOI] [PubMed] [Google Scholar]
- 113.Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci USA. 2001;98:2503–8. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rabel C, Grahn AM, Lurz R, Lanka E. The VirB4 family of proposed traffic nucleoside triphosphatases: Common motifs in plasmid RP4 TrbE are essential for conjugation and phage adsorption. J Bacteriol. 2003;185:1045–58. doi: 10.1128/JB.185.3.1045-1058.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rambow-Larsen AA, Weiss AA. The PtlE protein of Bordetella pertussis has peptidoglycanase activity required for Ptl-mediated pertussis toxin secretion. J Bacteriol. 2002;184:2863–69. doi: 10.1128/JB.184.11.2863-2869.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rashkova S, Spudich GM, Christie PJ. Mutational analysis of the Agrobacterium tumefaciens VirB11 ATPase: identification of functional domains and evidence for multimerization. J Bacteriol. 1997;179:583–89. doi: 10.1128/jb.179.3.583-591.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rashkova S, Zhou X, Christie P. Self-assembly of the Agrobacterium tumefaciens VirB11 traffic ATPase. J Bacteriol. 2000;182:413–45. doi: 10.1128/jb.182.15.4137-4145.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Robien MA, Krumm BE, Sandkvist M, Hol WG. Crystal structure of the extracellular protein secretion NTPase EpsE of Vibrio cholerae. J Mol Biol. 2003;333:657–74. doi: 10.1016/j.jmb.2003.07.015. [DOI] [PubMed] [Google Scholar]
- 119.Rohde M, Puls J, Buhrdorf R, Fischer W, Haas R. A novel sheathed surface organelle of the Helicobacter pylori cag type IV secretion system. Mol Microbiol. 2003;49:219–34. doi: 10.1046/j.1365-2958.2003.03549.x. [DOI] [PubMed] [Google Scholar]
- 120.Sagulenko V, Sagulenko E, Jakubowski S, Spudich E, Christie PJ. VirB7 lipoprotein is exocellular and associates with the Agrobacterium tumefaciens T pilus. J Bacteriol. 2001;183:3642–51. doi: 10.1128/JB.183.12.3642-3651.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sagulenko Y, Sagulenko V, Chen J, Christie PJ. Role of Agrobacterium VirB11 ATPase in T-pilus assembly and substrate selection. J Bacteriol. 2001;183:5813–25. doi: 10.1128/JB.183.20.5813-5825.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sakai D, Horiuchi T, Komano T. ATPase activity and multimer formation of PilQ protein are required for thin pilus biogenesis in plasmid R64. J Biol Chem. 2001;276:17968–75. doi: 10.1074/jbc.M010652200. [DOI] [PubMed] [Google Scholar]
- 123.Samuels AL, Lanka E, Davies JE. Conjugative junctions in RP4-mediated mating of Escherichia coli. J Bacteriol. 2000;182:2709–15. doi: 10.1128/jb.182.10.2709-2715.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Sastre JI, Cabezon E, de la Cruz F. The carboxyl terminus of protein TraD adds specificity and efficiency to F-plasmid conjugative transfer. J Bacteriol. 1998;180:6039–42. doi: 10.1128/jb.180.22.6039-6042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Savvides SN, Yeo HJ, Beck MR, Blaesing F, Lurz R, et al. VirB11 ATPases are dynamic hexameric assemblies: new insights into bacterial type IV secretion. EMBO J. 2003;22:1969–80. doi: 10.1093/emboj/cdg223. This study describes novel structural changes imparted through nucleotide binding by the hexameric VirB11 ATPases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schandel KA, Muller MM, Webster RE. Localization of TraC, a protein involved in assembly of the F conjugative pilus. J Bacteriol. 1992;174:3800–6. doi: 10.1128/jb.174.11.3800-3806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schmid MC, Schulein R, Dehio M, Denecker G, Carena I, Dehio C. The VirB type IV secretion system of Bartonella henselae mediates invasion, proinflammatory activation and antiapoptotic protection of endothelial cells. Mol Microbiol. 2004;52:81–92. doi: 10.1111/j.1365-2958.2003.03964.x. [DOI] [PubMed] [Google Scholar]
- 128.Schmidt-Eisenlohr H, Domke N, Angerer C, Wanner G, Zambryski PC, Baron C. Vir proteins stabilize VirB5 and mediate its association with the T pilus of Agrobacterium tumefaciens. J Bacteriol. 1999;181:7485–92. doi: 10.1128/jb.181.24.7485-7492.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schmidt-Eisenlohr H, Domke N, Baron C. TraC of IncN plasmid pKM101 associates with membranes and extracellular high-molecular-weight structures in Escherichia coli. J Bacteriol. 1999;181:5563–71. doi: 10.1128/jb.181.18.5563-5571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schrammeijer B, den Dulk-Ras A, Vergunst AC, Jurado Jacome E, Hooykaas PJ. Analysis of Vir protein translocation from Agrobacterium tumefaciens using Saccharomyces cerevisiae as a model: evidence for transport of a novel effector protein VirE3. Nucleic Acids Res. 2003;31:860–68. doi: 10.1093/nar/gkg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schroder G, Krause S, Zechner EL, Traxler B, Yeo HJ, et al. TraG-like proteins of DNA transfer systems and of the Helicobacter pylori type IV secretion system: inner membrane gate for exported substrates? J Bacteriol. 2002;184:2767–79. doi: 10.1128/JB.184.10.2767-2779.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schulein R, Guye P, Rhomberg TA, Schmid MC, Schroder G, et al. A bipartite signal mediates the transfer of type IV secretion substrates of Bartonella henselae into human cells. Proc Natl Acad Sci USA. 2005;102:856–61. doi: 10.1073/pnas.0406796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Segal G, Feldman M, Zusman T. The Icm/Dot type-IV secretion systems of Legionella pneumophila and Coxiella burnetii. FEMS Microbiol Rev. 2005;29:65–81. doi: 10.1016/j.femsre.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 134.Serwer P. Models of bacteriophage DNA packaging motors. J Struct Biol. 2003;141:179–88. doi: 10.1016/s1047-8477(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 135.Sexton JA, Pinkner JS, Roth R, Heuser JE, Hultgren SJ, Vogel JP. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J Bacteriol. 2004;186:1658–66. doi: 10.1128/JB.186.6.1658-1666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shamaei-Tousi A, Cahill R, Frankel G. Interaction between protein subunits of the type IV secretion system of Bartonella henselae. J Bacteriol. 2004;186:4796–801. doi: 10.1128/JB.186.14.4796-4801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Shapiro L, McAdams HH, Losick R. Generating and exploiting polarity in bacteria. Science. 2002;298:1942–46. doi: 10.1126/science.1072163. [DOI] [PubMed] [Google Scholar]
- 138.Shohdy N, Efe J, Emr SD, Shuman RA. Pathogen effector protein screening in yeast identifies Legionella factors that interfere with membrane trafficking. Proc Natl Acad Sci USA. 2005;102:4866–71. doi: 10.1073/pnas.0501315102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sory MP, Cornelis GR. Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol. 1994;14:583–94. doi: 10.1111/j.1365-2958.1994.tb02191.x. [DOI] [PubMed] [Google Scholar]
- 140.Spudich GM, Fernandez D, Zhou XR, Christie PJ. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc Natl Acad Sci USA. 1996;93:7512–17. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tanaka J, Suzuki T, Mimuro H, Sasakawa C. Structural definition on the surface of Helicobacter pylori type IV secretion apparatus. Cell Microbiol. 2003;5:395–404. doi: 10.1046/j.1462-5822.2003.00286.x. [DOI] [PubMed] [Google Scholar]
- 142.Tato I, Zunzunegui S, de la Cruz F, Cabezon E. TrwB, the coupling protein involved in DNA transport during bacterial conjugation, is a DNA dependent ATPase. Proc Natl Acad Sci USA. 2005 doi: 10.1073/pnas.0503402102. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Terradot L, Bayliss R, Oomen C, Leonard G, Baron C, Waksman G. Structures of two core subunits of the bacterial type IV secretion system, VirB8 from Brucella suis and ComB10 from Helicobacter pylori. Proc Natl Acad Sci USA. 2005;102:4596–601. doi: 10.1073/pnas.0408927102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Terradot L, Durnell N, Li M, Ory J, Labigne A, et al. Biochemical characterization of protein complexes from the Helicobacter pylori protein interaction map: strategies for complex formation and evidence for novel interactions within type IV secretion systems. Mol Cell Proteomics. 2004;3:809–19. doi: 10.1074/mcp.M400048-MCP200. [DOI] [PubMed] [Google Scholar]
- 145.Tzfira T, Citovsky V. Partners-in-infection: host proteins involved in the transformation of plant cells by Agrobacterium. Trends Cell Biol. 2002;12:121–29. doi: 10.1016/s0962-8924(01)02229-2. [DOI] [PubMed] [Google Scholar]
- 146.Tzfira T, Vaidya M, Citovsky V. Involvement of targeted proteolysis in plant genetic transformation by Agrobacterium. Nature. 2004;431:87–92. doi: 10.1038/nature02857. [DOI] [PubMed] [Google Scholar]
- 147.van Biesen T, Frost LS. The FinO protein of IncF plasmids binds FinP antisense RNA and its target, traJ mRNA, and promotes duplex formation. Mol Microbiol. 1994;14:427–36. doi: 10.1111/j.1365-2958.1994.tb02177.x. [DOI] [PubMed] [Google Scholar]
- 148.VanRheenen SM, Dumenil G, Isberg RR. IcmF and DotU are required for optimal effector translocation and trafficking of the Legionella pneumophila vacuole. Infect Immun. 2004;72:5972–82. doi: 10.1128/IAI.72.10.5972-5982.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Vergunst AC, Schrammeijer B, den Dulk-Ras A, de Vlaam CM, Regensburg-Tuink TJ, Hooykaas PJ. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science. 2000;290:979–82. doi: 10.1126/science.290.5493.979. This report and studies cited in References 70, 96, 105, 132, and 150 illustrate the power of Cre recombinase and other reporter protein fusion assays for identifying T4S substrates and defining their translocation signals. [DOI] [PubMed] [Google Scholar]
- 150.Vergunst AC, van Lier MC, den Dulk-Ras A, Grosse Stuve TA, Ouwehand A, Hooykaas PJ. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc Natl Acad Sci USA. 2005;102:832–37. doi: 10.1073/pnas.0406241102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Viala J, Chaput C, Boneca IG, Cardona A, Girardin SE, et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat Immunol. 2004;5:116–74. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- 152.Ward DV, Draper O, Zupan JR, Zambryski PC. Peptide linkage mapping of the Agrobacterium tumefaciens vir-encoded type IV secretion system reveals protein subassemblies. Proc Natl Acad Sci USA. 2002;99:11493–500. doi: 10.1073/pnas.172390299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ward DV, Zupan JR, Zambryski PC. Agrobacterium VirE2 gets the VIP1 treatment in plant nuclear import. Trends Plant Sci. 2002;7:1–3. doi: 10.1016/s1360-1385(01)02175-6. [DOI] [PubMed] [Google Scholar]
- 154.Watarai M, Andrews HL, Isberg R. Formation of a fibrous structure on the surface of Legionella pneumophila associated with exposure of DotH and DotO proteins after intracellular growth. Mol Microbiol. 2000;39:313–29. doi: 10.1046/j.1365-2958.2001.02193.x. [DOI] [PubMed] [Google Scholar]
- 155.Waters CM, Hirt H, McCormick JK, Schlievert PM, Wells CL, Dunny GM. An amino-terminal domain of Enterococcus faecalis aggregation substance is required for aggregation, bacterial internalization by epithelial cells and binding to lipoteichoic acid. Mol Microbiol. 2004;52:1159–71. doi: 10.1111/j.1365-2958.2004.04045.x. [DOI] [PubMed] [Google Scholar]
- 156.Waters VL. Conjugation between bacterial and mammalian cells. Nat Genet. 2001;29:375–76. doi: 10.1038/ng779. [DOI] [PubMed] [Google Scholar]
- 157.Winans SC, Walker GC. Conjugal transfer system of the IncN plasmid pKM101. J Bacteriol. 1985;161:402–10. doi: 10.1128/jb.161.1.402-410.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Winans SC, Walker GC. Fertility inhibition of RP1 by IncN plasmid pKM101. J Bacteriol. 1985;161:425–27. doi: 10.1128/jb.161.1.425-427.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yeo H-J, Savvides SN, Herr AB, Lanka E, Waksman G. Crystal structure of the hexameric traffic ATPase of the Helicobacter pylori type IV system. Mol Cell. 2000;6:1461–72. doi: 10.1016/s1097-2765(00)00142-8. The authors characterize first structure for a member of the VirB11-like ATPase superfamily. [DOI] [PubMed] [Google Scholar]
- 160.Yeo H-J, Yuan Q, Beck MR, Baron C, Waksman G. Structural and functional characterization of the VirB5 protein from the type IV secretion system encoded by the conjugative plasmid pKM101. Proc Natl Acad Sci USA. 2003;100:15947–52. doi: 10.1073/pnas.2535211100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yuan Q, Carle A, Gao C, Sivanesan D, Aly KA, et al. Identification of the VirB4-VirB8-VirB5-VirB2 pilus assembly sequence of type IV secretion systems. J Biol Chem. 2005 doi: 10.1074/jbc.M502347200. In press. An excellent biochemical study that defines subunit interactions and energetic requirements for pilus biogenesis by a T4S system. [DOI] [PubMed] [Google Scholar]
- 162.Zhou X-R, Christie PJ. Suppression of mutant phenotypes of the Agrobacterium tumefaciens VirB11 ATPase by overproduction of VirB proteins. J Bacteriol. 1997;179:5835–42. doi: 10.1128/jb.179.18.5835-5842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhu J, Oger PM, Schrammeijer B, Hooykaas PJ, Farrand SK, Winans SC. The bases of crown gall tumorigenesis. J Bacteriol. 2000;182:3885–95. doi: 10.1128/jb.182.14.3885-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zusman T, Feldman M, Halperin E, Segal G. Characterization of the icmH and icmF genes required for Legionella pneumophila intracellular growth, genes that are present in many bacteria associated with eukaryotic cells. Infect Immun. 2004;72:3398–409. doi: 10.1128/IAI.72.6.3398-3409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]