The LL-100 panel: 100 cell lines for blood cancer studies (original) (raw)
Abstract
For many years, immortalized cell lines have been used as model systems for cancer research. Cell line panels were established for basic research and drug development, but did not cover the full spectrum of leukemia and lymphoma. Therefore, we now developed a novel panel (LL-100), 100 cell lines covering 22 entities of human leukemia and lymphoma including T-cell, B-cell and myeloid malignancies. Importantly, all cell lines are unequivocally authenticated and assigned to the correct tissue. Cell line samples were proven to be free of mycoplasma and non-inherent virus contamination. Whole exome sequencing and RNA-sequencing of the 100 cell lines were conducted with a uniform methodology to complement existing data on these publicly available cell lines. We show that such comprehensive sequencing data can be used to find lymphoma-subtype-characteristic copy number aberrations, mRNA isoforms, transcription factor activities and expression patterns of NKL homeobox genes. These exemplary studies confirm that the novel LL-100 panel will be useful for understanding the function of oncogenes and tumor suppressor genes and to develop targeted therapies.
Subject terms: Cancer models, Haematological cancer
Introduction
Human cancer cell lines form a renewable resource and are vital models for studying the cellular and molecular mechanisms underlying tumorigenesis as well as for anti-cancer drug screening1,2. In particular, leukemia-lymphoma (LL) cell lines serve as convenient in vitro tool due to their world-wide accessibility, straightforward manipulability and low culture costs, providing experimental models to address a multitude of questions in the field of LL biology3. Indeed, the scientific benefits of utilizing LL cell lines have definitely boosted our knowledge on a plethora of aspects of these diseases4. Importantly, many studies contoured our appreciation of the suitability of LL cell lines as model systems, replicating faithfully most features of the primary cells5,6.
The National Cancer Institute (NCI) tumor cell line panel (known as NCI-60 as 60 cancer cell lines were assembled) was developed in the 1980s as an in vitro drug discovery tool intended to supplant animal studies in drug screening (reviewed in7). This screening tool was quickly appreciated as an invaluable source of information about the mechanisms of growth inhibition and tumor cell cytotoxicity7. Later in the 2000s, the NCI-60 panel transitioned from a drug-discovery pipeline to a more general research tool in support of the cancer research community7,8. Another panel incorporating a reduced number of cell lines of particular interest which had been derived from several solid tumor types was established in Japan9. These two cell line panels did not aim at one single cancer category but were designed to represent a variety of different tumor entities. Nevertheless, these sets have provided the framework for the use of defined panels of cell lines at the same time as keeping with the information-rich character of screens7.
The majority of studies in the arena of LL focus on a narrow number of cell lines. We realized that there is a need for a reference panel specialized on LL cell lines to facilitate hypothesis-driven research efforts10. We have assembled a panel of 100 authenticated LL cell lines that reflects the heterogeneity of the entities under the umbrella category of LL. In addition to well-known and commonly analyzed cell lines, this invaluable and publicly available platform includes additional cell lines assigned unequivocally to the various entities but with specific characteristics. It is hoped that this focused LL-100 cell lines panel may enhance the current scientific momentum, helping to fully elucidate the underlying pathology of these LL malignancies and providing an important and unique resource for the testing of novel therapeutic agents.
Based on data of the human genome project, high-throughput methods have boosted the knowledge of processes in normal and malignant cells. The microarray technology showed for the first time simultaneous activities of thousands of genes and allowed the classification of tissues and diseases11. This approach is being steadily replaced by next generation sequencing technologies which comprise the sequencing of complete transcriptomes, exomes and whole genomes. These applications are used in cancer research to identify aberrations in the genome, deregulated and mutated genes, and alternative splicing. The obtained data are helpful to classify malignancies, to improve existing therapies, and to identify new targets for novel therapeutic approaches12. Here, we present transcriptome and exome sequencing data of a panel of 100 authenticated LL cell lines (LL-100) and selected examples of their utilization.
Results and Discussion
Sequencing of exomes and transcriptomes of the LL-100 panel
We performed whole exome sequencing (WES) and mRNA-sequencing (RNA-seq) on a panel of 100 LL cell lines representing 22 subtypes (Table 1). For exomic analyses over 10 million reads (2 × 151 bases) per sample were sequenced resulting in at least 50x coverage on a 60 MB exome size. RNA-seq yielded over 29 million (2 × 151 bases) reads per sample. Sequencing data have been deposited at ENA under the accession number PRJEB30297 for WES and PRJEB30312 for RNA-seq, respectively.
Table 1.
The LL-100 panel.
| Entity | Cell line | DSMZ ACC # | Selected aberrations |
|---|---|---|---|
| Pre-B-ALL | 697 | 42 | TCF3-PBX1 |
| KOPN-8 | 552 | KMT2A-MLLT1 | |
| NALM-6 | 128 | ||
| REH | 22 | ETV6-RUNX1; RUNX1-PRDM7 | |
| SEM | 546 | KMT2A-AFF1 | |
| B-NHL: Burkitt/B-ALL | BJAB | 757 | KMT2A-CLTC* |
| DAUDI | 78 | t(8;14)(q24;q32) | |
| RAJI | 319 | IGH-MYC | |
| RAMOS | 603 | IGH-MYC | |
| VAL | 586 | t(8;14;18)(q24;q32;q21) | |
| B-NHL: CLL/PLL | HG-3 | 765 | |
| JVM-3 | 18 | ||
| JVM-13 | 19 | ||
| MEC-1 | 497 | R3HCC1L-HTRA1* | |
| PGA-1 | 766 | ||
| B-NHL: DLBCL ABC | NU-DHL-1 | 583 | t(3;8)(p25;q24); t(14;18)(q32;q21) |
| OCI-LY3 | 761 | IGH-SPIB | |
| RI-1 | 585 | t(4;8)(q22;q24); BCL2 amp | |
| U-2932 | 633 | t(8;14)(q24;q32) in clone R2 | |
| U-2946 | 795 | IGH-MYC | |
| B-NHL: DLBCL GC | DOHH-2 | 47 | t(8;14;18)(q24;q32;q21) |
| OCI-LY7 | 688 | IGH-MYC | |
| OCI-LY19 | 528 | t(14;18)(q32;q21) | |
| SU-DHL-4 | 495 | EZH2 Y646S; IGH-BCL2 | |
| SU-DHL-6 | 572 | EZH2 Y646N; IGH-BCL2 | |
| WSU-DLCL2 | 575 | EZH2 Y646F | |
| B-NHL: HCL | BONNA-12 | 150 | |
| HAIR-M | 762 | IGH-TCL1A* | |
| HC-1 | 301 | ||
| B-NHL: MCL | GRANTA-519 | 342 | t(11;14)(q13;q32) |
| JEKO-1 | 553 | t(11;14)(q13;q32) | |
| JVM-2 | 12 | t(11;14)(q13;q32) | |
| MINO | 687 | t(11;14)(q13;q32) | |
| REC-1 | 584 | t(11;14)(q13;q32) | |
| B-NHL: PEL | BC-3 | 679 | |
| BCBL-1 | 683 | MYC amp | |
| CRO-AP2 | 48 | ||
| CRO-AP5 | 215 | ||
| B-NHL: PMBL | U-2940 | 634 | biallelic SOCS1 del |
| Multiple Myeloma/PCL | KMS-12-BM | 551 | t(11;14)(q13;q32) |
| L-363 | 49 | ||
| LP-1 | 41 | IGH-WHSC1* | |
| OPM-2 | 50 | IGH-WHSC1 | |
| RPMI-8226 | 402 | ||
| U-266 | 9 | ||
| Hodgkin Lymphoma | HDLM-2 | 17 | |
| KM-H2 | 8 | CIITA-C15ORF65 | |
| L-428 | 197 | EZH2 Y646S | |
| L-1236 | 530 | SOCS1 L150V; SOCS1 L162R | |
| SUP-HD1 | 574 | ||
| T-ALL/T-LL | CCRF-CEM | 240 | NKX2.5-BCL11B |
| DND-41 | 525 | TLX3-BCL11B | |
| HPB-ALL | 483 | t(5;14)(q35;q32); CBFB-MYLPF* | |
| JURKAT | 282 | ||
| MOLT-4 | 362 | ||
| RPMI-8402 | 290 | LMO1-TRD; SIL-TAL1 | |
| Mature T-Malignancy | DERL-7 | 524 | |
| HH | 707 | FOXK2-TP63* | |
| MOTN-1 | 559 | TBL1XR1-TP63 | |
| NK Malignancy | KHYG-1 | 725 | |
| NK-92 | 488 | ||
| YT | 434 | ||
| ALCL | DEL | 338 | NPM1-ALK |
| SR-786 | 369 | NPM1-ALK | |
| SU-DHL-1 | 356 | NPM1-ALK | |
| SUP-M2 | 509 | NPM1-ALK | |
| AML myelocytic | EOL-1 | 386 | KMT2A PTD; FIP1L1-PDGFRA |
| HL-60 | 3 | ||
| KASUMI-1 | 220 | RUNX1-RUNX1T1 | |
| KG-1 | 14 | FGFR1OP2-FGFR1 | |
| NB-4 | 207 | PML-RARA | |
| OCI-AML3 | 582 | NPMcy type A, DNMT3A R882C | |
| SKNO-1 | 690 | RUNX1-RUNX1T1 | |
| AML monocytic | ME-1 | 537 | CBFB-MYH11 |
| MOLM-13 | 554 | FLT3 ITD, KMT2A-MLLT3 | |
| MONO-MAC-6 | 124 | KMT2A-MLLT3; RUNX1-ATP8A2 | |
| MUTZ-3 | 295 | ||
| THP-1 | 16 | CSNK2A1-DDX39B | |
| U-937 | 5 | MLLT10-PICALM | |
| AML erythroid | F-36P | 543 | |
| HEL | 11 | JAK2 V617F | |
| OCI-M2 | 619 | RUNX1-TSPEAR* | |
| TF-1 | 334 | CBFA2T3-ABHD12* | |
| AML megakaryocytic | CMK | 392 | JAK3 A572V |
| ELF-153 | 693 | ||
| M-07e | 104 | ANO7-DHDH* | |
| MEGAL | 719 | SET-NUP214 | |
| MKPL-1 | 697 | RBM6-CSF1R | |
| UT-7 | 137 | ||
| CML myeloid BC | EM-2 | 135 | BCR-ABL1 |
| K-562 | 10 | BCR-ABL1 | |
| KCL-22 | 519 | BCR-ABL1 | |
| KU-812 | 378 | BCR-ABL1 | |
| LAMA-84 | 168 | BCR-ABL1 | |
| MOLM-20 | 591 | KMT2A-SEPT11 | |
| CML lymphoid BC | BV-173 | 20 | BCR-ABL1 |
| CML-T1 | 7 | BCR-ABL1 | |
| NALM-1 | 131 | BCR-ABL1 | |
| TK-6 | 723 | BCR-ABL1; MAPK1-AIF1L* | |
| MPN | SET-2 | 608 | JAK2 V617F |
Based on the analysis of WES and RNA-seq data we show the usefulness of the LL-100 panel for LL research in five exemplary studies.
PEL and HL cell lines cluster separate from cell lines of other B-NHL entities
For many years, expression profiling has been applied to classify tumors including LL11. RNA-seq and microarray analyses show highly reproducible results with correlation between expression profiles13. We performed cluster analysis to test whether the two techniques yield comparable results also in the LL-100 panel. We analyzed gene expression of primary effusion lymphoma (PEL) cell lines and of cell lines from various other B-non Hodgkin lymphoma (B-NHL) entities as well as from Hodgkin lymphoma (HL).
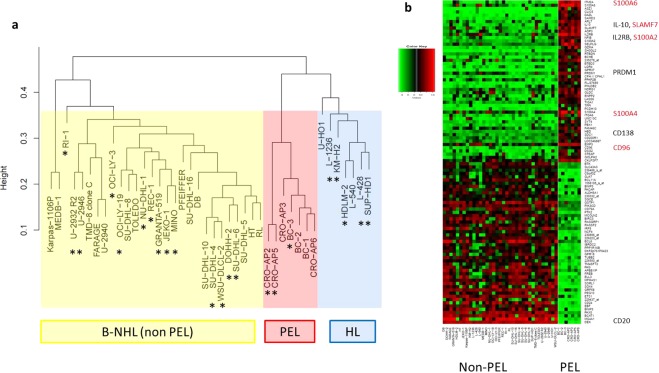
Unsupervised cluster analysis showed that all PEL cell lines grouped together, separate from cell lines derived from activated-B-cell-like (ABC) and germinal center (GC) diffuse large B-cell lymphoma (DLBCL), mantle cell lymphoma (MCL), primary mediastinal B-cell lymphoma (PMBL) and from cell lines derived from HL (Fig. 1a). Notably, PEL and HL cell lines clustered on one arm, separate from all cell lines representing the other B-NHL entities (Fig. 1a). Microarray and RNA-seq data yielded identical results, confirming the suitability of both techniques (Figs 1a, S1).
Figure 1.
Microarray analysis of HL and B-NHL cell lines including PEL. (a) PEL cell lines (in red color) cluster separately from other B-NHL (in yellow color), but on the same arm as HL cell lines (in blue color). 30% of the most variant probe sets were taken for hierarchical clustering by average linkage. (b) PEL cell lines show tumor type-specific expression of genes including SLAMF7. For highest validity, the analysis was conducted with expression array data from LL-100 cell lines and additional cell lines. LL-100 cell lines are marked with an asterisk. Differentially expressed probe sets were filtered for one probe set per gene and top 50 positive and negative fold changes. Black: previously described PEL-specific genes; red: PEL-specific genes not described hitherto.
PEL and HL cell lines are characterized by a set of common up- and downregulated genes (Fig. S2). Prominent were expression of CCND2 and the absence of B-cell markers in PEL and HL cell lines. CD19, CD20 (MS4A1), CD24, CD79A and CD79B were expressed in all tested lymphoma entities beside PEL and HL (Fig. S2). Absence, low or rare expression of these “early” B-cell markers in PEL and HL has been described for both primary lymphoma cells and cell lines14–16.
Highly expressed in PEL – but not in HL - were CD138 (SDC1), IL-10, IL2RB, and PRDM1, all described as PEL-characteristic genes (Fig. 1b)17–19. Not reported hitherto was that PEL cells expressed CD96, SLAMF7, S100A2, S100A4 and S100A6 (Fig. 1b). RT-PCR, flow cytometry and Western blot analysis confirmed the PEL-associated expression of CD138, PRDM1/BLIMP1, SLAMF7 and the three S100A family genes (Fig. S3a–c).
PRDM1/BLIMP1 is a master regulator of terminal B-cell differentiation. Originally described as repressor20, BLIMP1 can also enhance transcription of SLAMF7 in multiple myeloma21 and of IL-10 in type 1 regulatory T-cells22. Thus, coexpression of the three genes in PEL suggests a causal relationship between transcriptionally active PRDM1 and the targets SLAMF7 and IL-10 also in this B-NHL entity. Independent of its regulation, the expression of SLAMF7 in PEL is remarkable because a monoclonal antibody targeting SLAMF7 (elotuzumab) has recently been approved for treatment of patients with multiple myeloma23. RQ-PCR analysis showed that SLAMF7 is comparably expressed in PEL and multiple myeloma cell lines (Fig. S4).
PEL is a rare, aggressive form of NHL, cells typically being infected with HHV-814. With a median survival time of six months the prognosis for PEL patients is poor24. If our cell line results can be translated to primary tumor cells, PEL patients might benefit from targeted therapy with elotuzumab.
Activities of hematopoietic transcription factors in leukemia-lymphoma cell lines
Numerous transcription factors (TF) regulate normal hematopoiesis and their activities are precisely controlled during hematopoietic stem cell self-renewal and their differentiation into the diverse blood cell lineages. Consequently, many of these TFs emerged as proto-oncogenes or tumor suppressors because deregulation of these TFs alters the cellular transcriptional program eventually impairing differentiation and thus fostering malignant transformation. Aberrant activities of TFs which can be caused by a variety of direct or indirect mutations and epigenetic alterations, are a hallmark of cancer, including hematological malignancies25,26.
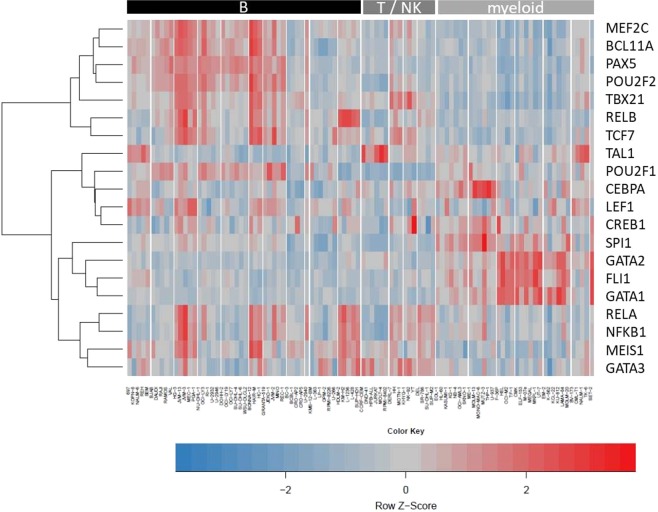
We aimed to analyze TF activities from TFs relevant for hematopoiesis across the LL-100 panel. Because the expression level of a TF itself barely gives information about its downstream activity27, activities of TFs were predicted via the expression levels of their direct target genes. TF activities were estimated via so-called consensus TF regulons (CTFRs) which have been defined recently by Garcia-Alonso et al. on the basis of diverse sources for human TF-target interactions28,29. For each cell line from the LL-100 panel relative TF activities were computed from RNA-seq data applying DoRothEA (Discriminant Regulon Expression Analysis) for CTFRs from 289 single TFs (Table S1). From these 289 TFs we selected 20 TFs based on their known role in hematopoiesis. The activities of the respective CTFRs in the LL-100 cell lines are represented in Fig. 2.
Figure 2.
Activities of hematopoietic TFs across LL-100 cell lines according to their CTFR activity scores. Hierarchical clustering heatmap (distance: Euclidian; agglomeration method: complete; centered to row) of TF activities calculated via VIPER based on DoRothEA TF-interactions for each cell line from RNA-seq data. Each column represents a single cell line. Cell lines are grouped according to entities. The color code depicts the activity for each row-wise normalized CTFR. Bars on top of the heatmap indicate cellular origin of cell lines.
Obviously, activity patterns of several TFs within the cell lines mirror their cell of origin: PAX5 and OCT-2 (encoded by POU2F2) are critical for B-cell development30. Accordingly, the CTFRs of these TFs showed strong activity in cell lines from B-cell derived malignancies but were inactive in myeloid-derived leukemias (Fig. 2). Other TF activities reflect the differentiation status of their respective normal counterparts: the strong activity of the CTFRs from GATA1 and GATA2 was highly specific for the cell lines from erythroid and megakaryocytic AML, CML and in cell line SET-2 (myeloproliferative neoplasm) (Fig. 2), which is in line with the role of GATA1 and GATA2 in the differentiation of erythroid-megakaryocytic progenitors where alterations in their dosages are related to transformation31.
Other CTFR activities indicate the mutation status of hematopoietic TFs in specific entities: C/EBPα is a TF relevant for granulopoiesis and AML patients frequently show inactivating mutations of C/EBPα impairing final differentiation of the cells32. Accordingly, the activity of the CEBPA-CTFR was diminished in erythroid and megakaryocytic AML cell lines compared to myelocytic and monocytic AML cell lines (Fig. 2).
Another subset of TF activities is characteristic for specific leukemia or lymphoma entities: TAL1 impairs T-cell differentiation and is a master oncogene in T-ALL33. Accordingly, T-ALL cell lines showed the strongest activity of the TAL1-CTFR (Fig. 2).
The lymphocyte specific TF LEF1 was primarily active in pre-B-ALL and CLL/PLL cell lines but rather inactive in T-ALL and ALCL cell lines (Fig. 2), which is in line with the current literature34–36. In addition, the LEF1-CTFR activity was moderately upregulated in MCL cell lines and some BL and GC-DLBCL cell lines (Fig. 2). This activity pattern seems to reflect the situation in patients because upregulation of LEF1 in subsets of B-NHL patients has been reported before37–39.
For 9 of the 20 hematopoietic TFs we observed a moderate positive correlation between gene expression levels of the TFs and their corresponding CTFR activities (Table 2, Fig. S5). Best correlations were detected for PU.1 (encoded by SPI1) and GATA2. However, in general hematopoietic TF activities determined via CTFRs did hardly correlate with gene expression levels of the TFs (Fig. S6). For example TAL1 expression was rather weak in cell lines from CML in blast crisis (Fig. S5), but the TAL1-CTFR activity was increased in these cell lines (Fig. 2). On the other hand TAL1 expression was detected in several AML cell lines on a comparable level to T-ALL cell lines (Fig. S5), but activity of the TAL1-CTFR was low in AML cell lines (Fig. 2). This underpins that upregulation of a TF alone is not sufficient to regulate its target genes. In some cases a defined CTFR (e.g. from TAL1) might also be regulated by further transcriptional activators or repressors.
Table 2.
Correlation analysis between CTFR activity and gene expression levels.
| TF gene name | mean CTFR activity score | mean TF expression [logCPM] | Spearman correlation coefficient |
|---|---|---|---|
| BCL11A | 2.68 | 3.34 | 0.32 |
| CEBPA | 0.16 | 1.23 | 0.28 |
| CREB1 | −0.51 | 6.19 | −0.13 |
| FLI1 | −0.56 | 6.33 | 0.31 |
| GATA1 | −1.24 | −1.31 | 0.55 |
| GATA2 | 0.11 | 0.57 | 0.74 |
| GATA3 | −1.79 | 1.09 | 0.66 |
| LEF1 | −0.22 | 3.54 | 0.05 |
| MEF2C | 3.06 | 5.49 | 0.37 |
| MEIS1 | −0.81 | 1.38 | 0.02 |
| NFKB1 | −1.37 | 6.50 | 0.55 |
| PAX5 | 2.95 | 2.82 | 0.71 |
| POU2F1 | 0.06 | 5.91 | 0.27 |
| POU2F2 | 2.77 | 4.37 | 0.64 |
| RELA | −0.49 | 6.44 | 0.06 |
| RELB | 2.12 | 4.82 | 0.70 |
| SPI1 | −0.78 | 3.31 | 0.75 |
| TAL1 | 0.98 | −0.55 | 0.28 |
| TBX21 | 2.52 | −0.64 | 0.55 |
| TCF7 | 2.11 | 1.38 | 0.43 |
In summary, activity scores of CTFRs are much more informative concerning the role of a TF than its transcript levels alone. We show that transcriptional activities in LL-100 cell lines mirror the lineage origin of hematologic malignancies for a set of specific TFs (e.g. PAX5). Other TFs (e.g. GATA1) reflect the differentiation status of the respective normal counterpart and a third group of TFs (e.g. TAL1) depicts aberrant activities highly characteristic for specific entities. In general, TF activities across all studied cell lines did rarely correlate with their gene expression levels. Thus, analyses of CTFR activities from RNA-seq data are a suitable tool to measure and evaluate the relevance of a specific TF in hematological cell lines.
Aberrant NKL homeobox gene activities in lymphoid malignancies
Homeobox genes encode TFs which show basic impacts in developmental processes including embryonal development and cell differentiation in the adult. Therefore, deregulation of homeobox genes generates developmental disturbances or cancer40. These genes are classified according to differences in their conserved homeobox and ordered into classes and subclasses41. The NKL subclass comprises 48 members which are involved in fundamental differentiation processes like NKX2-1 in that of the lung and the thyroid, and NKX2-5 in the heart42,43. Normal expression patterns of nine NKL homeobox genes in early hematopoiesis and subsequent lymphocyte development have been identified and termed hematopoietic NKL-code44,45. According to this code, T-cells silence all NKL homeobox genes during their thymic development while mature NK-cells maintain expression of MSX1 and mature B-cells HHEX or NKX6-344–46. Alterations of the NKL-code may underlie the generation of particular hematopoietic malignancies. According to this notion, 24 NKL homeobox genes are reported to date for aberrant activity in T-cell acute lymphoblastic leukemia (T-ALL), mediating differentiation arrest and transformation44,47,48.
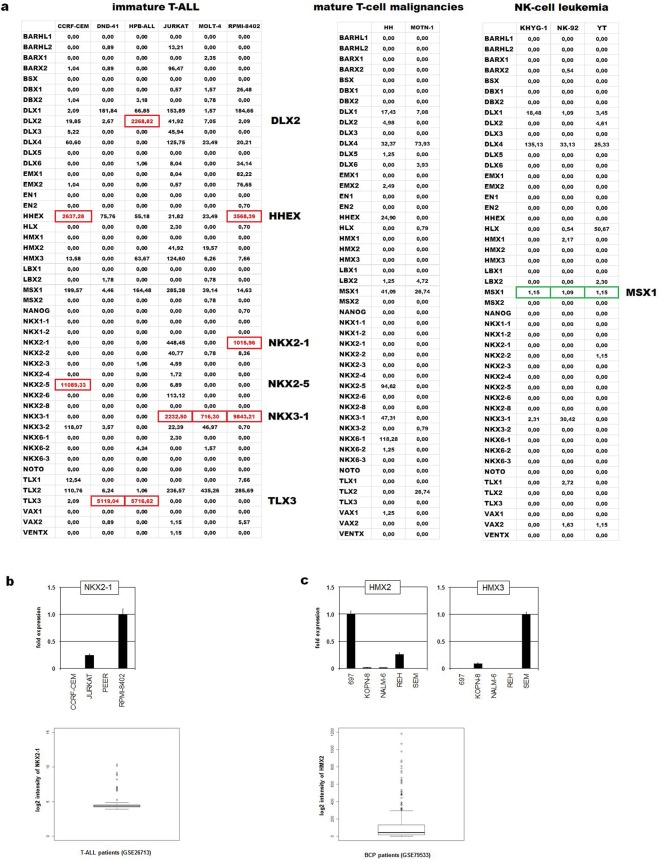
Using the LL-100 transcriptome dataset we here screened NKL homeobox gene activities in cell lines and show some results for selected lymphoid entities. To discriminate active and inactive genes we have set a cutoff at 500 normalized counts. Accordingly, aberrant activation of particular subclass members was detected in immature T-ALL but not in mature T-cell lines (Fig. 3a). This finding supported the observation that NKL homeo-oncogenes provoke an arrest in differentiation which plays a role in immature thymocytes but obviously not in mature T-cells. Furthermore, MSX1 is an oncogene in T-ALL and a tumor suppressor in NK-cells46,49, showing accordingly reduced activity in NK-cell leukemia cell lines (Fig. 3a). Thus, these data confirm published deregulated NKL homeobox genes including MSX1, NKX2-5, NKX3-1 and TLX3 in T-cell and NK-cell leukemia. Moreover, our RNA-seq data indicated elevated NKX2-1 expression in T-ALL cell line RPMI-8402 (Fig. 3a). Aberrant activation of NKX2-1 has been identified in T-ALL patients by chromosomal translocation, representing thus an additional clinically relevant oncogene50. Subsequent RQ-PCR analysis confirmed NKX2-1 activity in this cell line (Fig. 3b). Of note, chromosomal and genomic analyses indicated absence of a translocation or an amplification targeting NKX2-1 in RPMI-8402 cells (data not shown). Therefore, this cell line may represent a model to examine alternative upstream and novel downstream factors of NKX2-1 in T-ALL.
Figure 3.
Expression data of 48 NKL homeobox genes in selected cell lines. (a) The indicated tables show expression levels of 48 NKL homeobox genes in six immature T-ALL cell lines (left), in two cell lines derived from mature T-cell malignancies (middle), and three NK-cell leukemia cell lines (right). The listed values are normalized counts (reads) obtained by DESeq2 calculation. Overexpressed or downregulated gene activities are highlighted by red or green boxes, respectively. (b) RQ-PCR analysis of selected T-ALL cell lines shows elevated NKX2-1 expression levels in RPMI-8402 cells (above). The indicated box plot (below) shows NKX2-1 expression levels in 117 T-ALL patient samples obtained from GSE26713. Outliers indicate samples with NKX2-1 overexpression. (c) RQ-PCR analysis in selected BCP-ALL cell lines shows elevated expression levels of HMX2 in 697 cells and of HMX3 in SEM cells (above). The indicated box plot (below) shows HMX2 expression levels in 229 BCP-ALL patient samples obtained from GSE79533. Outliers indicate samples with HMX2 overexpression.
In normal B-cell development NKL homeobox genes HHEX and NKX6-3 are the only subclass members active in developing naïve and germinal center B-cells, and in mature memory B-cells and plasma cells while B-cell progenitor (BCP) cells additionally express HLX and MSX144,45. Our data show that malignant BCP-ALL cell lines lack activity of HLX, MSX1 and NKX6-3 (except SEM) (Fig. S7), showing fundamental changes in the normal expression pattern of NKL homeobox genes. Furthermore, three of five cell lines aberrantly expressed HMX2 or HMX3. The activity of these genes was confirmed by RQ-PCR in the indicated cell lines (Fig. 3c). Moreover, HMX2 overexpression was detected in 13% of 229 BCP-ALL patients by analysis of public dataset GSE79533 (Fig. 3c), supporting the clinical relevance of this finding. Thus, HMX2 (and HMX3) may represent NKL homeobox genes primarily deregulated in this type of B-cell malignancy, serving as diagnostic marker and/or therapeutic target.
In DLBCL cell lines we detected silencing of HHEX (in OCI-LY7 and RI-1) and NKX6-3 (except DOHH-2) and aberrant activation of HLX (NU-DHL-1) and NKX3-1 (OCI-LY3) (Fig. S7). Of note, these data did not show significant differences between ABC- and GC-DLBCL cell lines, suggesting that NKL homeobox genes do not play a role in the discrimination of these disease subtypes. Surprisingly, PEL and MM cell lines (except RPMI-8226 expressing BARX2) demonstrated complete absence of NKL homeobox gene activity (Fig. S7). Therefore, NKL subclass members may operate as basic tumor suppressors in these particular B-cell lymphoma types. The malignant cells of both PEL and MM are derived from mature B-cells suggesting that in final stages of development NKL homeobox genes lose their oncogenic potential. Finally, the lack of B-cell specific NKL homeobox gene activity in PEL is in accordance with reported downregulation of general B-cell factors as indicated above (Fig. S2)15. Together, deregulation of NKL homeobox genes in B-cell malignancies is more important than hitherto expected. The identified cell lines may serve as models to investigate the role of these genes in the indicated tumor entities. Thus, the LL-100 datasets allow the identification of cell line models for the examination of deregulated NKL homeobox genes in particular disease entities. The expression patterns of this fundamental gene subclass in cell lines reflect the situation observed in normal lymphocytes and in primary tumor cells, highlighting the significance of these cell line data for cancer research.
Copy number alterations and their effect on gene expression in DLBCL
DLBCL shows a high degree of genetic diversity with unique molecular patterns including varying occurrences of copy number alterations (CNAs), resembling the different states of B-cell maturation they derive from and which is also reflected by the diverse clinical outcome51–54.
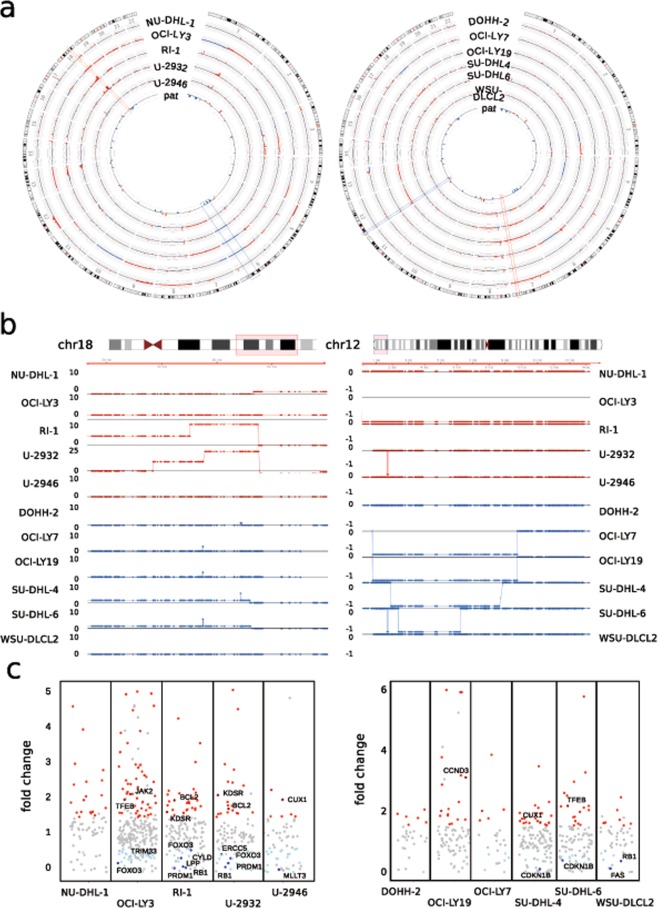
To evaluate such alterations and to test if differences in the molecular subtypes are maintained in culture, we used WES data generated in our study to call CNAs in DLBCL derived cell lines. In ABC and GC DLBCL, we identified on average 157 (+/−29) and 129 (+/−12) CNAs, respectively, with a size >10 kb (Fig. 4a and Table 3). While amplification of both arms of chr7 occurs frequently in both subtypes, certain events like the 6q-deletion seem to occur more often in the ABC-subtype. Intending to compare the identified events to primary tumors, we took a recently published set of significantly recurrent CNAs in DLBCL which described a total of 45 recurring focal events (14 amplifications and 31 deletions) in 304 patients55. Of those CNAs, we find 38 (84%) in at least one cell line, including all focal amplifications (Fig. 4a and Table S2).
Figure 4.
Copy Number Alterations (CNAs) and their impact on gene expression in DLBCL cell lines. (a) Circos plots depict the landscape of CNAs in cell lines of DLBCL subtypes ABC-DLBCL (left) and GC-DLBCL (right). Chromosome 1–22 are shown with the copy number gains in red and deletions in blue, the cell lines analyzed are indicated. The inner ring displays significant CNAs identifed in patients from Chapuy et al.55. Red and blue boxes highlight individual regions with gains or deletions, respectively that are common to patients and cell lines. (b) Examples of DLBCL-subtype specific alterations. Region of 18q22-q23 preferentially amplified in ABC-DLBCL (left) and 6q deletion in the GC-subtype (right) are shown for the indicated cell lines. DLBCL cell lines of the ABC-subtype are shown in red, those of GC-origin in blue, the respective copy number is shown on the left. Each dot resembles one exon from WES, the black line denotes 2n – normal copy number state. (c) Expression of COSMIC cancer genes affected by CNAs in ABC- and GC-DLBCL cell lines, left and right, respectively. All genes with expression change >1.5 are shown either in red or light blue, labeled are those that have been described as deregulated in a patient cohort by Chapuy et al.55.
Table 3.
Chromosomal gains and losses in ABC and GC DLBCL.
| ABC subtype | Gains | Deletions | Total events |
|---|---|---|---|
| NU-DHL-1 | 109 | 20 | 129 |
| OCI-LY3 | 139 | 44 | 183 |
| RI-1 | 133 | 27 | 160 |
| U-2932 | 150 | 37 | 187 |
| U-2946 | 94 | 32 | 126 |
| GC subtype | Gains | Deletions | Total events |
| DOHH-2 | 94 | 14 | 108 |
| OCI-LY7 | 104 | 22 | 126 |
| OCI-LY19 | 100 | 28 | 128 |
| SU-DHL-4 | 123 | 21 | 144 |
| SU-DHL-6 | 110 | 29 | 139 |
| WSU-DLCL2 | 95 | 32 | 127 |
Similarly, of the 20 arm-level alterations described, 10 out of 18 amplifications and both recurrent deletions are present in one or more cell lines (Table S2). Of particular interest are subtype specific alterations, possibly reflecting different mutational processes during tumor development. Therefore, we assessed such specific events by integrating another set of ABC- or GC-related CNAs56 and found 83% of patient-derived events in our cell lines (15 out of 18; Table S2). In addition to the previously observed 6q-deletion, we could confirm a preferentially occurring gain of 18q22-q23 in ABC-DLBCL cell lines (3/5 ABC-DLBCL cell lines and 0/6 cell lines of the GC-subtype). Also, we find the deletion of the far end of 1p36 exclusively in 4/6 GC-DLBCL cell lines (Fig. 4b).
While CNAs can serve as diagnostic markers, their main impact results from associated changes in gene expression. We therefore compared the expression of all affected genes in a DLBCL-subtype specific manner, identifying around 20,000 genes to be affected by CNAs across all cell lines. Although we observe little overall changes for the majority of genes, several outliers are present in each cell line (Fig. S8). To identify CNA-deregulated genes important for disease progression, we filtered for those genes included in the COSMIC Cancer Gene Census database57. Depending on the cell line, we find 25 (OCI-LY7) to 252 (OCI-LY3) of the COSMIC genes located in regions of CNA (Table S3), with 10 (DOHH-2) to 121 (OCI-LY3) showing changes in expression >1.5, concordant with the respective CN change (Fig. 4c). Several of these genes have been confirmed deregulated in patient derived RNA-seq data55, which we also find associated with the corresponding CNA, e.g. BCL2 and KDSR on chr18. Interestingly, we also find PRDM1 and FOXO3 to show reduced expression in ABC-DLBCL cell lines harboring a deletion of 6q21. This deletion has been described to be ABC-subtype related in an earlier study53. Nevertheless, these authors did not identify potential tumor-relevant genes in this deleted region53.
In summary, we exemplarily identified CNAs in cell lines derived from both DLBCL-subtypes and characterized the associated expressional changes. We find a high degree of similarity towards data from primary tumors and highlighted which cancer-relevant genes become deregulated in the individual cell lines.
This analysis (i) characterizes CNAs in cell lines of the two major DLBCL-subtypes and shows how they recapitulate recurring events from patients, (ii) allows the identification of those genes that are deregulated by CNAs and likely have a disease-relevant function and (iii) by doing so enables the selection of appropriate models for further molecular research related to DLBCL. Furthermore, we believe that this kind of analysis is applicable to other entities and that thereby valuable models for those entities can be obtained.
Tissue-specific RNA isoforms
Allowing different combinations of exons, alternative splicing leads to the production of multiple mRNA isoforms of the same gene, often resulting in proteins of different functionality58. More than 90% of human genes are affected by alternative splicing59,60. Tissue-specific splice factors together with ubiquitious RNA binding factors cooperate to generate tissue-specific RNA isoforms59. The existence of different promoters can also lead to different N-terminal RNA variants.
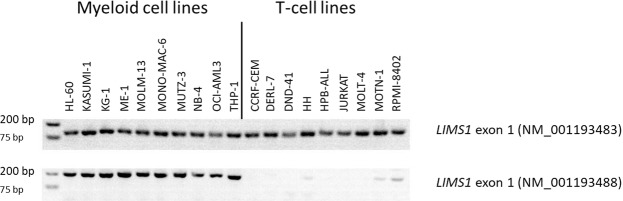
The RNA-seq data of the LL-100 panel allowed us to find RNA isoforms that specify different hematopoetic lineages. Bioinformatic analysis identified genes with tissue-specific exons, e.g. LIMS1 in myeloid vs T-cell lines (Fig. S9). Two N-terminal variants of LIMS1 were expressed in myeloid cell lines, only one of them in T-cell lines (Fig. S9). These results were confirmed by RT-PCR and validated with a second cell lines cohort (Figs 5, S10). Altogether, data from 18 AML and from 17 T-ALL cell lines revealed that the two groups could be distinguished on the basis of LIMS1 exon 1 expression (NM_001193488) with a sensitivity of 1 and a specificity of 1.
Figure 5.
Expression of N-terminal LIMS1 exons in myeloid and T-cell lines. RT-PCR analysis reveals expression of LIMS1 (NM_001193488) exon 1 in myeloid cell lines only, expression of LIMS1 (NM_001193483) exon 1 in myeloid cell lines and in T-cell lines. Data were confirmed in a second validation cohort (Fig. S10).
In sum, our data show that RNA-seq analysis allows detection of cell lines from different lineages on the basis of alternatively expressed exons.
Conclusion
One goal of personalized medicine in cancer medicine is the development of targeted therapies aiming to reverse detrimental effects of mutated or deregulated genes. The costs of sequencing technologies will presumably soon be low enough to allow routine diagnostics detecting genetic alterations for classification of the patient’s tumor and determining treatment strategies.
Immortalized tumor cell lines have been used for a long time to understand the molecular and cellular function of mutated genes and to develop new drugs. However, the cell line panels established hitherto did not represent most forms of leukemia and lymphoma7,9. Thus, the NCI-60 human cell line panel developed for use in drug development comprises sixty human cancer cell lines derived from nine different tissues7. Only six cell lines (CCRF-CEM, HL-60, K-562, MOLT-4, RPMI-8226, SR-786) represent tumors of the blood.
Covering 22 leukemia and lymphoma entities we present the novel LL-100 panel, 100 cell lines for use in basic research and drug development. The selected cell lines of this panel are authenticated and free of contamination by mycoplasma or non-inherent viruses. Furthermore, the methods of RNA- and DNA isolation and sequencing are identical in all cell lines. Therefore, this dataset allows comparative studies without methodical impact. We performed WES and RNA-seq analysis for all 100 cell lines. In exemplary studies, we show that lymphoma entities can be identified by gene expression analysis and splice variant analysis. WES analysis documented that copy number aberrations in DLBCL cell lines reflects the situation in primary tumor cells and may lead to the identification of potential oncogenes. RNA-seq analysis identified tumor entity-specific activities of CTFRs, demonstrating the usefulness of cell lines as model systems for transcription factor research. Finally, RNA-seq analysis specified aberrant activities of NKL homeobox genes.
All data and cell lines are publicly available. As demonstrated exemplarily in this study, the sequencing data can be used for various approaches. We hope that the novel LL-100 panel described here will stimulate many studies in the field of leukemia and lymphoma research.
Methods
Cell lines
Cell lines were taken from the stock of the cell lines bank (Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures). Cell lines were authenticated by DNA profiling and cytogenetics. Detailed references and cultivation protocols have been described previously3.
RNA-sequencing analysis
Total RNA was extracted via miRNeasy Mini Kit (Qiagen, Hilden, Germany) including DNase digestion. Library preparation and sequencing steps were commissioned to GATC Biotech (Cologne, Germany). The GATC pipeline included the production of strand-specific (fr-first strand) mRNA libraries, quality control via Applied Biosystems Fragment Analyzer and Nanodrop, concentration measurement via Qubit fluorometer. The libraries were sequenced on Illumina HiSeq2500 (2 × 151 cycles, paired end run, 8 bp dual indices) with >29 million reads per sample and deposited at ENA (PRJEB30312). Reads were trimmed via fastq-mcf (ea-utils 1.04.807). Reads were quality controlled via FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc). Reads were aligned by STAR (2.5.3a)61 to the Gencode Homo sapiens genome (v26) and converted/sorted via samtools (0.1.19)62. Counting the reads to each gene was done via HTSeq-count python script (0.8.0)63. Data was processed and analyzed in the R/Bioconductor environment (3.3.2/3.3, www.bioconductor.org). Normalization, estimation of dispersions, and testing for differentially expressed genes based on a test assuming negative binomial data distribution was computed via DESeq264.
Differential isoforms between given cell line groups were detected by JunctionSeq (JunctionSeq_1.4.0)65.
Whole exome sequencing analysis
DNA was isolated with the High Pure PCR Template Preparation Kit (Roche Diagnostics, Mannheim, Germany). Library preparation (Agilent SureSelect Human All Exon V6, 60 MB) and sequencing steps (2 × 151 bp + 8 bp barcoding, HiSeqX) were commissioned to Genewiz (Leipzig, Germany) and deposited at ENA (PRJEB30297). Insert lengths were aimed to be higher than 250 bp in order to increase coverage and uniformity in coding regions66.
Reads were aligned by STAR (2.5.3a)61 to the human gencode genome (v26). Subsequently, alignment files were processed (samtools 0.1.19), duplicates removed (picard 2.9.2, www.broadinstitute.github.io/picard/), and variants called via GATK tools (3.7, Haplotypecaller)67 and overlapping VarScan (v2.4.3)68 results. Mutation effects were annotated via Ensembl VEP (release-84, GRCh38)69. Data were processed and analyzed in the R/Bioconductor environment (3.3.2/3.3). Overlapping single nucleotide variations via Haplotypecaller and VarScan were filtered for >=20 quality, >=10 depth, >=0.2 allele frequency, <0.01 MAF, missense/frameshift/stop gained mutations.
For identifying copy number alterations (CNA) in DLBCL cell lines control-FREEC70 (v11.0) was applied on the duplicate-clean alignment files with NC-NC as normal cell control. Neighboring regions with identical CN in the disperse whole exome data were fused to one region and CN regions below 10 kb were omitted and visualized via circos71 (0.67–7). Individual regions and genes of interest were plotted with the R/bioconductor packages ggplot2 (3.1.0) and Gviz (1.22.3).
Expression array analysis
Profiling of gene expression was commissioned to the Genome Analytics Research Group, Helmholtz Centre for Infection Research, Braunschweig, Germany. 500 ng total RNA were used for biotin labelling according to the 3′ IVT Express Kit (Affymetrix, Santa Clara, CA, USA). 7.5 µg of biotinylated cDNA were fragmented and placed in a hybridization cocktail containing four biotinylated hybridization controls (BioB, BioC, BioD, and Cre). Samples were hybridized to an identical lot of Affymetrix GeneChip HG-U133 Plus 2.0 for 16 h at 45 °C. Steps for washing and SA-PE staining were processed on the fluidics station 450 using the recommended FS450 protocol (Affymetrix). Image analysis was performed on GCS3000 Scanner and GCOS1.2 Software Suite (Affymetrix). For data analysis spot intensities were RMA-background corrected and quantile normalized. Data processing was done via R/Bioconductor (3.2.2) using limma and affy packages72,73. Differentially expressed genes between PEL and Non-PEL cell lines were identified via moderated t-statistics (R, limma) setting p < 0.05 and log fold changes >1. For hierarchical cluster analysis (average linkage) 30% of the most varying probe sets served as basis for the distance matrix (1-correlation) and visualised via the R package hdust.
Public expression profiling datasets used in this study were generated by U133 Plus 2.0 gene chips from Affymetrix and obtained from Gene Expression Omnibus (GEO, www.ncbi.nlm.nih.gov). This type of profiling gene chip contains 37 probes of the 48 known human NKL homeobox genes, excluding HMX3. We analyzed datasets GSE26713 and GSE79533 for T-ALL and BCP-ALL patients, respectively. Expression values were given as boxplots using R-packages. Outliers were interpreted as samples showing aberrant overexpression.
Analysis of TF activities
Relative TF activities were computed from RNA-seq data with DoRothEA v2 via VIPER74 for CTFRs with a TF-target score of high confidence (A), likely confidence (B), and medium confidence (C)29. For this, gene expression data were prepared similarly as described28: (i) lowly expressed genes were discarded, defined as those with an average CPM < 1 (counts per million) and prior count of 3; (ii) data were TMM (trimmed mean of M-values) normalized using calcNormFactors function described in limma; (iii) fitted log2 CPM were extracted and finally tested for transcriptional activity via VIPER in R/Bioconductor (3.4.4/3.6). Transcriptional activity and gene expression heatmaps were vizualised via gplots (R-package).
Quantitative real-time PCR analysis and detection of splice variants
RNA was prepared using the RNeasy Mini kit (Qiagen). For mRNA quantification, reverse transcription was performed using the SuperScript II reverse transcriptase kit (Invitrogen, Karlsruhe, Germany). PCR was performed on a 7500 Applied Biosystems (Darmstadt, Germany). TaqMan probes (Applied Biosystems) were used to quantify human CCND2 (Hs00153380_m1) CD20/MS4A1 (Hs00544819_m1), CD96 (Hs00175524_m1), CD138 (Hs00896424_g1), HMX2 (Hs01394375_m1), HMX3 (Hs01392772_m1), IL-10 (Hs00961622_m1), NKX2-1 (Hs00163037_m1), PRDM1 (Hs00153357_m1), S100A2 (Hs00195582_m1), S100A4 (Hs00243202_m1), S100A6 (Hs00170953_m1) and SLAMF7 (Hs00900280_m1) using TBP as endogenous control. Relative expression levels were calculated using the ΔΔCt method.
Primers and RT-PCR conditions for detection of splice variants of LIMS1, RABGAP1L and TFEC are shown in Table S4.
Cell surface marker analysis
For immunophenotyping, cells were washed and incubated for 30 min at 4 °C with antibodies (Ab) against CD319/SLAMF7 (#331802, BioLegend, Koblenz, Germany), CD138 Ab (#551902, Becton Dickinson, Heidelberg, Germany) and appropriate isotype controls (BD Biosciences). Subsequently, cells were treated with FITC conjugated anti-mouse secondary Ab (Biozol, Eching, Germany) and propidium iodide (PI) (Sigma-Aldrich). Labeled cells were analyzed on a FACSCalibur (BD Biosciences) using CellQuest Pro software.
Western blot analysis
Samples were prepared as described previously75. Anti BLIMP1 (#9115), CCND2 (#3741S), S100A4 (#13018S), S100A6 (#13162S) Abs were purchased from Cell Signaling (New England Biolabs, Frankfurt, Germany). Anti GAPDH (ab8245) and anti S100A2 (ab 109494) Abs were from Abcam (Cambridge, UK). Specific bands on nitrocellulose membranes were visualized with the biotin/streptavidin-horseradish peroxidase system (GE Healthcare, Little Chalfont, UK) in combination with the “Renaissance Western Blot Chemoluminescence Reagent” protocol (Perkin Elmer, Waltham, MA, USA).
Supplementary information
Author Contributions
H.G.D. developed the LL-100 panel; H.Q., S.E., M.K. and S.N. designed, performed and analyzed the experiments; C.P. performed bioinformatic analyses; K.S., W.G.D. and C.C.U. performed quality and identity control for the sequenced cell lines. R.A.F.M. performed cytogenetic analyses. All authors read and approved this manuscript.
Data Availability
Sequencing data have been deposited at ENA under the accession number PRJEB30297 for WES and PRJEB30312 for RNA-seq, respectively. The ArrayExpress accession IDs are E-MTAB-7722 (WES) and E-MTAB-7721 (RNA-seq).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-44491-x.
References
- 1.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haverty PM, et al. Reproducible pharmacogenomic profiling of cancer cell line panels. Nature. 2016;533:333–337. doi: 10.1038/nature17987. [DOI] [PubMed] [Google Scholar]
- 3.Drexler, H. G. Guide to Leukemia-Lymphoma Cell Lines. 2nd Edition Braunschweig, eBook on CD (2010).
- 4.MacLeod RA, et al. Human leukemia and lymphoma cell lines as models and resources. Curr. Med. Chem. 2008;15:339–359. doi: 10.2174/092986708783497319. [DOI] [PubMed] [Google Scholar]
- 5.Ross DT, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat. Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 6.Sandberg R, Ernberg I. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI) Proc. Natl. Acad. Sci. USA. 2005;102:2052–2057. doi: 10.1073/pnas.0408105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat. Rev. Cancer. 2006;6:813–823. doi: 10.1038/nrc1951. [DOI] [PubMed] [Google Scholar]
- 8.Abaan OD, et al. The exomes of the NCI-60 panel: a genomic resource for cancer biology and systems pharmacology. Cancer Res. 2013;73:4372–4382. doi: 10.1158/0008-5472.CAN-12-3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakatsu N, et al. Chemosensitivity profile of cancer cell lines and identification of genes determining chemosensitivity by an integrated bioinformational approach using cDNA arrays. Mol. Cancer Ther. 2005;4:399–412. doi: 10.4161/cbt.4.4.1616. [DOI] [PubMed] [Google Scholar]
- 10.Drexler HG, MacLeod RA. Leukemia-lymphoma cell lines as model systems for hematopoietic research. Ann. Med. 2003;35:404–412. doi: 10.1080/07853890310012094. [DOI] [PubMed] [Google Scholar]
- 11.Golub TR, Slonim DK, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 12.Cieślik M, Chinnaiyan AM. Cancer transcriptome profiling at the junction of clinical translation. Nat. Rev. Genet. 2018;19:93–109. doi: 10.1038/nrg.2017.96. [DOI] [PubMed] [Google Scholar]
- 13.Zhao S, et al. Comparison of RNA-seq and microarray in transcriptome profiling of activated T cells. Plos One. 2014;9:e78644. doi: 10.1371/journal.pone.0078644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cesarman E, Knowles DM. The role of Kaposi´s sarcoma-associated herpesvirus (KSHV HHV-8) in lymphoproliferative diseases. Semin. Cancer Biol. 1999;9:165–174. doi: 10.1006/scbi.1998.0118. [DOI] [PubMed] [Google Scholar]
- 15.Carbone A, et al. Understanding pathogenetic aspects and clinical presentation of primary effusion lymphoma through its derived cell lines. AIDS. 2010;24:479–490. doi: 10.1097/QAD.0b013e3283365395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzankov A, et al. Expression of B-cell markers in classical Hodgkin lymphoma: a tissue microarray analysis of 330 cases. Mod. Pathol. 2003;16:1141–1147. doi: 10.1097/01.MP.0000093627.51090.3F. [DOI] [PubMed] [Google Scholar]
- 17.Jones KD, et al. Involvement of interleukin-10 (IL-10) and viral IL-6 in the spontaneous growth of Kaposi´s sarcoma herpesvirus-associated infected primary effusion lymphoma cells. Blood. 1999;94:2871–2879. [PubMed] [Google Scholar]
- 18.Jenner RG, et al. Kaposi’s sarcoma-associated herpesvirus-infected primary effusion lymphoma has a plasma cell gene expression profile. Proc. Natl. Acad. Sci. USA. 2003;100:10399–10404. doi: 10.1073/pnas.1630810100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chadburn A, et al. Immunophenotypic analysis of the Kaposi sarcoma herpesvirus (KSHV; HHV-8)-infected B cells in HIV+ multicentric Castleman disease (MCD) Histopathology. 2008;53:513–524. doi: 10.1111/j.1365-2559.2008.03144.x. [DOI] [PubMed] [Google Scholar]
- 20.Yu J, et al. Transcriptional repression by Blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 2000;20:2592–2603. doi: 10.1128/MCB.20.7.2592-2603.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim JR, Mathew SO, Mathew PA. Blimp-1/PRDM1 regulates the transcription of human CS1 (SLAMF7) gene in NK and B cells. Immunobiology. 2016;221:31–39. doi: 10.1016/j.imbio.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Montes de Oca M, et al. Blimp-1-dependent IL-10 production by Tr1 cells regulates TNF-mediated tissue pathology. PLOS Pathogens. 2016;12:e1005398. doi: 10.1371/journal.ppat.1005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Touzeau C, Moreau P, Dumontet C. Monoclonal antibody therapy in multiple myeloma. Leukemia. 2017;31:1039–1047. doi: 10.1038/leu.2017.60. [DOI] [PubMed] [Google Scholar]
- 24.Chen YB, Rahemtullah A, Hochberg E. Primary effusion lymphoma. The Oncologist. 2007;12:569–576. doi: 10.1634/theoncologist.12-5-569. [DOI] [PubMed] [Google Scholar]
- 25.Bradner JE, Hnisz D, Young RA. Transcriptional addiction in cancer. Cell. 2017;168:629–643. doi: 10.1016/j.cell.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crans HN, Sakamoto KM. Transcription factors and translocations in lymphoid and myeloid leukemia. Leukemia. 2001;15:313–331. doi: 10.1038/sj.leu.2402033. [DOI] [PubMed] [Google Scholar]
- 27.Falco MM, et al. The pan-cancer pathological landscape. Sci. Rep. 2016;6:e39709. doi: 10.1038/srep39709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia-Alonso L, et al. Transcription factor activities enhance markers of drug sensitivity in cancer. Cancer Res. 2018;78:769–780. doi: 10.1158/0008-5472.CAN-17-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Alonso, L. et al. Benchmark and integration of resources for the estimation of human transcription factor activities. bioRxiv, 10.1101/337915 (preprint) (2018).
- 30.Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat. Rev. Immunol. 2005;5:497–508. doi: 10.1038/nri1633. [DOI] [PubMed] [Google Scholar]
- 31.Shimizu R, Yamamoto M. GATA-related hematologic disorders. Exp. Hematol. 2016;44:696–705. doi: 10.1016/j.exphem.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Koschmieder S, et al. Role of transcription factors C/EBPα and PU.1 in normal hematopoiesis and leukemia. Int. J. Hematol. 2005;81:368–377. doi: 10.1532/IJH97.05051. [DOI] [PubMed] [Google Scholar]
- 33.Sanda T, Leong WZ. TAL1 as a master oncogenic transcription factor in T-cell acute lymphoblastic leukemia. Exp. Hematol. 2017;53:7–15. doi: 10.1016/j.exphem.2017.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Hassler MR, et al. Insights into the pathogenesis of anaplastic large-cell lymphoma through genome-wide DNA methylation profiling. Cell Reports. 2016;17:596–608. doi: 10.1016/j.celrep.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kühnl A, et al. Overexpression of LEF1 predicts unfavorable outcome in adult patients with B-precursor acute lymphoblastic leukemia. Blood. 2011;118:6362–6367. doi: 10.1182/blood-2011-04-350850. [DOI] [PubMed] [Google Scholar]
- 36.Gutierrez A, et al. Inactivation of LEF1 in T-cell acute lymphoblastic leukemia. Blood. 2010;115:2845–2851. doi: 10.1182/blood-2009-07-234377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walther N, et al. Aberrant lymphocyte enhancer-binding factor 1 expression is characteristic for sporadic Burkitt’s lymphoma. Am. J. Pathol. 2013;182:1092–1098. doi: 10.1016/j.ajpath.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 38.O’Malley DP, Lee JP, Bellizzi AM. Expression of LEF1 in mantle cell lymphoma. Ann. Diagn. Pathol. 2017;26:57–59. doi: 10.1016/j.anndiagpath.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 39.Cubedo E, et al. Identification of LMO2 transcriptome and interactome in diffuse large B-cell lymphoma. Blood. 2012;119:5478–5491. doi: 10.1182/blood-2012-01-403154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat. Rev. Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- 41.Holland PW, Booth HA, Bruford EA. Classification and nomenclature of all human homeobox genes. BMC Biol. 2007;5:47. doi: 10.1186/1741-7007-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lazzaro D, et al. The transcription factor TTF-1 is expressed at the onset of thyroid and lung morphogenesis and in restricted regions of the foetal brain. Development. 1991;113:1093–1104. doi: 10.1242/dev.113.4.1093. [DOI] [PubMed] [Google Scholar]
- 43.Komuro I, Izumo S. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc. Natl. Acad. Sci. USA. 1993;90:8145–8149. doi: 10.1073/pnas.90.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagel S, et al. NKL homeobox gene activities in hematopoietic stem cells, T-cell development and T-cell leukemia. PLOS One. 2017;12:e0171164. doi: 10.1371/journal.pone.0171164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagel S, et al. NKL heomeobox gene activities in B-cell development and lymphomas. PLOS One. 2018;13:e0205537. doi: 10.1371/journal.pone.0205537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagel S, et al. NKL homeobox gene MSX1 acts like a tumor suppressor in NK-cell leukemia. Oncotarget. 2017;8:66815–66832. doi: 10.18632/oncotarget.18609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Villarese P, et al. TCRα rearrangements identify a subgroup of NKL-deregulated adult T-ALLs associated with favorable outcome. Leukemia. 2018;32:61–71. doi: 10.1038/leu.2017.176. [DOI] [PubMed] [Google Scholar]
- 48.Ferrando AA, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1:75–87. doi: 10.1016/S1535-6108(02)00018-1. [DOI] [PubMed] [Google Scholar]
- 49.Nagel S, et al. Repressed BMP signaling reactivates NKL homeobox gene MSX1 in a T-ALL subset. Leuk. Lymphoma. 2015;56:480–491. doi: 10.3109/10428194.2014.924119. [DOI] [PubMed] [Google Scholar]
- 50.Homminga I, et al. Integrated transcript and genome analyses reveal NKX2-1 and MEF2C as potential oncogenes in T cell acute lymphoblastic leukemia. Cancer Cell. 2011;19:484–497. doi: 10.1016/j.ccr.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 51.Basso K, Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 2015;15:172–184. doi: 10.1038/nri3814. [DOI] [PubMed] [Google Scholar]
- 52.Staudt LM, et al. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv. Immunol. 2005;87:163–208. doi: 10.1016/S0065-2776(05)87005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lenz G, et al. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. USA. 2008;105:13520–13525. doi: 10.1073/pnas.0804295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reddy A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171:481–494. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chapuy B, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018;24:679–690. doi: 10.1038/s41591-018-0016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karube K, et al. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia. 2018;32:675–684. doi: 10.1038/leu.2017.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Futreal PA, et al. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Badr E, El Hefnawi M, Heath LS. Computational identification of tissue-specific splicing regulatory elements in human genes from RNA-seq data. PLOS One. 2016;11:e0166978. doi: 10.1371/journal.pone.0166978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell. Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hartley SW, Mullikin JC. Detection and visualization of differential splicing in RNA-seq data with JunctionSeq. Nucleic Acids Res. 2016;44:e127. doi: 10.1093/nar/gkw501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pommerenke C, et al. Enhanced whole exome sequencing by higher DNA insert lengths. BMC Genomics. 2016;17:399. doi: 10.1186/s12864-016-2698-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McKenna A, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP effect predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boeva V, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ritchie ME, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gautier L, et al. Affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 74.Alvarez JJ, et al. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 2016;48:838–847. doi: 10.1038/ng.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Quentmeier H, et al. SET-NUP214 fusion in acute myeloid leukemia- and T-cell acute lymphoblastic leukemia-derived cell lines. J. Hematol. Oncol. 2009;2:3. doi: 10.1186/1756-8722-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data have been deposited at ENA under the accession number PRJEB30297 for WES and PRJEB30312 for RNA-seq, respectively. The ArrayExpress accession IDs are E-MTAB-7722 (WES) and E-MTAB-7721 (RNA-seq).