CELL SURFACE SIGNALING MOLECULES IN THE CONTROL OF IMMUNE RESPONSES: A TIDE MODEL (original) (raw)
. Author manuscript; available in PMC: 2012 Apr 22.
Summary
A large numbers of cell surface signaling molecules (CSSMs) have been molecularly identified and functionally characterized in recent years and, via these studies, our knowledge in the control of immune response has increased exponentially. Two major lines of evidence emerge. First, the majority of immune cells rely on one or few CSSMs to deliver a primary triggering signal to sense their environment, leading to initiation of an immune response. Second, both costimulatory CSSMs that promote the response, and coinhibitory CSSMs that inhibit the response, are required to control direction and magnitude of a given immune response. With such tight feedback, immune responses are tuned and returned to baseline. These findings extend well beyond our previous observation in the requirement for lymphocyte activation and argue a revisit of the traditional “two-signal model” for activation and tolerance of lymphocytes. Here we propose a “tide” model to accommodate and interpret current experimental findings.
INTRODUCTION
All immune cells, including those participating in the innate and adaptive immune response, have evolved to express distinct cell surface receptors or ligands to sense and respond to environmental cues. These cell surface signaling molecules (CSSMs) are vital for differentiation, recognition and cellular function. Many cell types consistently monitor the dynamic environmental stimuli through their unique receptors to recognize specific changes. For example, a specific T cell receptor (TCR) binds a major histocompatibility complex (MHC)-peptide structure on a professional antigen presenting cell (APC). This TCR transmits an antigenic signal, initiating the downstream signaling pathways of an immune response(Smith-Garvin et al., 2009). APCs also respond to changes in their surrounding environmental stimuli using toll-like receptors (TLRs) to identify potential pathogens(Palm and Medzhitov, 2009). Natural killer cells (or NK cells) utilize natural cytotoxicity receptors (NCRs) to recognize changes on target cells caused by viral infections or stress(Lanier, 2005). High affinity IgE receptors on mast cells contribute to immune detection and surveillance by identifying allergen-IgE complexes(Sayed et al., 2008). Therefore, immune cells utilize these receptors to transmit an initial signal to turn on the immune response.
For years immunologists have sought to understand the mechanisms of antigen-specific immune responses and the versatile nature of antigen receptors on the surface of lymphocytes. With the attempt to produce a cellular model incorporating the theory of self and nonself discrimination, Bretscher and Cohn were the first to propose a two-signal model to account for B cell tolerance induction (Bretscher and Cohn, 1970). Later this model was extended and applied to T lymphocytes by Lafferty, Schwartz and colleague(Lafferty and Cunningham, 1975; Mueller et al., 1989). The two signal model explains why lymphocytes are only partially activated or even unresponsive after exposure to Signal 1 from antigen-receptor. Only after exposure to a second, costimulatory cell surface signal does full activation of the lymphocyte occur (Lafferty and Cunningham, 1975; Lafferty et al., 1983). Further experiments revealed a molecular identity for the costimulatory signal (also called signal 2): the CD28–CD80 interaction(Linsley et al., 1990). Since then, the two-signal model has gained increasing experimental support, contributing to our understanding of lymphocyte activation.
The last decade has witnessed dramatic progress in the identification and characterization of CSSMs; largely due to the completion of the human genome project. More than 4000 molecules have been identified as potential CSSMs, based on similarities in their transmembrane protein structure, along with their signature intracellular and extracellular domains (Lander et al., 2001; Venter et al., 2001). A group of these CCSMs have been functionally characterized, giving new perspectives on the regulation of immune responses and translating to the clinic new therapeutic treatments of human disease. Studies characterizing the role of individual immune cells and their specific type of immune responses have revealed several large gene families which provide critical signals in immune cell activation, including tumor necrosis factor (TNF), immunoglobulin (Ig), G-protein coupled receptor (GPCR), and the lectin receptor family (i.e. Siglec, Dectin, DNGR-1, DC-SIGN, etc). In this review, we propose a comprehensive “tide” model to bridge the gap that exists between our current knowledge of specific immune cell regulation and how the complex, downstream, multi-cellular immune response is regulated and controlled by CSSMs.
RATIONALES FOR THE REVISION OF THE TWO-SIGNAL MODEL
Our understanding of T lymphocyte activation has been profoundly influenced by the “two-signal model” in which costimulation provided by “Signal 2” is necessary for optimal activation of lymphocytes. Functional analyses indicate that CSSMs are not only costimulatory but also coinhibitory(Greenwald et al., 2005). Importantly, expression of these coinhibitory molecules is often induced de novo or upregulated upon T cell activation in the presence of costimulation, indicating a negative feedback response of the immune system. Genetic ablation of these coinhibitory molecules in mouse models often leads to various autoimmune diseases associated with overactive T cell responses(Chen, 2004), demonstrating the indispensible role of coinhibitors in in vivo T cell tolerance induction. Therefore, although the TCR signal is essential and required for the selection and initial triggering of T cell responses, it is often the co-signal that dictates the outcome of a T cell response, including both activation and tolerance. Additionally, nearly all immune cells including NKT cells, NK cells, γδ T cells, macrophages, dendritic cells (DCs), neutrophils, and mast cells (besides T and B cells) also have been shown to have similar requirements for a primary triggering signal (Signal 1- like), and ample experimental data demonstrate that CSSMs are present on these cell types and are able to modulate and fine tune their responses.
THE “TIDE” MODEL
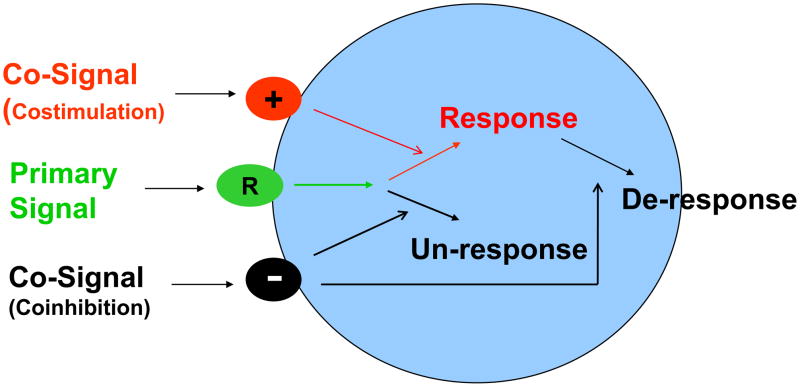
Left unchecked, uncontrolled inflammatory immune responses are dangerous to the host; therefore under this hypothesis the necessary immune components and the resulting inflammatory responses are tightly controlled to limit or avoid excess damage to the surrounding tissue. Here we propose a new tide model of immunity which incorporates our current molecular understanding of an immune response (Figure 1) and provides additional insight into both innate and adaptive immunity. In this model, we define primary signal as the initiator of specific immune cells reacting to extracellular stimuli. Meanwhile, the co-signals, either costimulatory or coinhibitory, are modulators which decide the direction and magnitude of the immune response. The characteristics of this self-feedback, tide signal model are outlined below.
Figure 1. The Tide Model for the Control of Immune Response.
We define primary signal as a triggering signal or signals which initiate specific immune cell response to extracellular stimuli. The co-signals, which could be either costimulatory or coinhibitory, are modulator of signal one, and decide the direction and magnitude of a cellular reaction, leading to activation of naïve cells (Response), deactivation of already activated cells (De-response) or unresponsiveness (Un-response).
First, the role of primary signal is extended to include a broader, more intricate function in immunity; spreading beyond its current activity through TCR and BCR to include initiating a cascade of activation or deactivation on all immune cells during both innate and adaptive immune responses.
Secondly, primary signal is the initiator, but primary signal itself does not decide the fate of the immune response. Primary signal is received by the receptors on certain immune cell types; then these cells can induce several early downstream biochemical signaling events. However, it is the co-signals that determine the overall outcome of the immune response.
Thirdly, co-signals include signals with stimulatory function (costimulator) as well as that with inhibitory function (coinhibitor). While a costimulatory signal is required to optimize an immune response, the coinhibitory signal is often induced and triggered by primary signal together with a costimulator, thus serving as a strong negative feedback signal. Therefore we use “tide” to describe the rise and fall of immune response due to interplay of these signals.
Finally, co-signaling molecules are highly diverse in abilities to monitor and control immune cell responses. The expression of co-signaling molecules is differentially and tightly regulated through every stage and location of immune cell activation. They transduce signals through a series of co-signaling pathways. In addition, the nature of individual co-signaling molecules determines its preferential participation in one or more functional aspects of immune cell activation.
SIGNAL ONE AND CO-SIGNALS FOR IMMUNE CELL SUBSETS
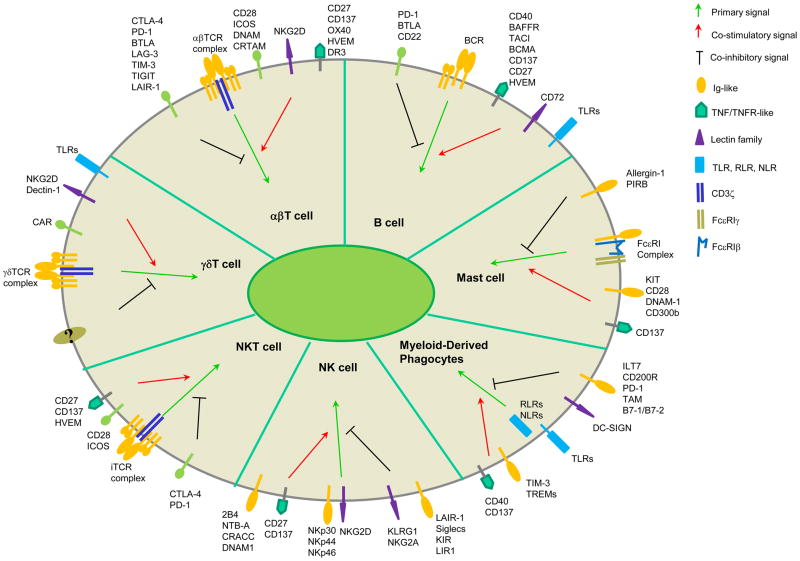
Each cell type bears different ‘recognizing’ receptors for primary signal, and signaling through these receptors is very distinct (Figure 2). As for co-signaling molecules, a lot of them are shared among several cell types while some are uniquely present on certain cell types (Table 1).
Figure 2. Cell surface signaling molecules in the control of immune responses.
Primary signal and co-signals (co-stimulatory or co-inhibitory) are defined in each immune cell type. TLR, Toll-Like Receptor; RLR, RIG-like Receptors; NLR, NOD-like Receptor.
Table 1.
Signal 1 and co-signals for immune cell subsets
| Cell type | Signal 1 | Co-signals | Receptor(s) | Ligand(s) | Intracellular signaling | Ref. |
|---|---|---|---|---|---|---|
| T cell (αβ) | TCR | Costimulator | CD28 | B7-1, B7-2, B7-H2 (h)* | PI3K, GRB2, Vav | 1–3 |
| ICOS | B7-H2 | PI3K | 1 | |||
| TLT2? | B7-H3 | - | 4,5 | |||
| - | B7-H1, B7-DC | - | 6 | |||
| CD27 | CD70 | NF-kB, Bcl2 | 7 | |||
| CD137 | CD137L | NF-kB, Bcl-2, Erk | 8 | |||
| OX40 | OX40L | NF-kB, Bcl-2, PI3K | 8 | |||
| Survivin | 8 | |||||
| HVEM | LIGHT | NF-kB | 9 | |||
| DR3 | TL1A | NF-kB | 10–13 | |||
| NKG2D | MICA, MICB | DAP-10 | 14 | |||
| ULBP-4, RAET1G | 14 | |||||
| TIM-1 | TIM-4, phosphatidylserine | - | 15–18 | |||
| TIM-2 | H-Ferritin, Sema4A? | - | 16,19,20 | |||
| DNAM-1 | PVR, PVRL2 | - | 21 | |||
| CRTAM | Necl2 | Scrib | 22 | |||
| - | BTN3 | - | 23 | |||
| Coinhibitor | CTLA-4 | B7-1, B7-2, B7-H2 (h)* | PI3K, SHP-2, PP2A | 1 | ||
| PD-1 | B7-H1, B7-DC | SHP-2 | 24 | |||
| B7-1 | B7-H1 | - | 25,26 | |||
| - | B7-H3 | - | 27 | |||
| - | B7-H4 | - | 28 | |||
| BTLA | HVEM | SHIP-1, SHIP-2 | 29 | |||
| CD160 | HVEM | - | 30 | |||
| LAG-3 | MHC II | - | 31 | |||
| TIM-3 | Galectin-9, phosphatidylserine | Ca++ | 32,33 | |||
| TIGIT | PVR, PVRL2, PVRL3 | - | 34 | |||
| LAIR-1 | Collagen | SHIP-1, SHP-2 | 35 | |||
| - | Sema3A | - | 36 | |||
| - | VSIG4 | - | 37 | |||
| - | BTNL1 | - | 38 | |||
| - | BTNL2 | - | 39 | |||
| - | BTN1A1 | - | 40 | |||
| - | BTN2A2 | - | 40 | |||
| T cell (γδ) | TCR | Costimulator | NKG2D | MICA, MICB | DAP-10 | 41 |
| CAR | JAML | PI3K | 42,43 | |||
| CD137 | CD137L | NF-kB, Bcl-2, Erk | 44 | |||
| NKT | iTCR | Costimulator | CD28 | B7-1, B7-2, B7-H2 (h) | PI3K, GRB2, Vav | 45,46 |
| ICOS | B7-H2 | PI3K | 47 | |||
| CD40 | CD40L | TRAF2, 5, 6, JNK, p38 | 48 | |||
| OX40 | OX40L | NF-kB, Bcl-2, PI3K | 49 | |||
| CD137 | CD137L | NF-kB, Bcl-2, Erk | 50 | |||
| TIM-1 | TIM-4, phosphatidylserine | - | 51 | |||
| Coinhibitor | PD-1 | B7-H1, B7-DC | SHP-2 | 52 | ||
| BTLA | HVEM | SHIP-1, SHIP-2 | 53 | |||
| GITR | GITRL | - | 54 | |||
| B cell | BCR | Costimulator | CD40 | CD40L, C4BP | TRAF2, 5, 6, JNK, p38 | 55–57 |
| HVEM | LIGHT | NF-kB? | 9 | |||
| CD137 | CD137L | NF-kB? | 58 | |||
| CD27 | CD70 | NF-kB | 7 | |||
| BAFF-R | BAFF | NF-kB2, PI3K-Akt1-mTOR | 59 | |||
| TACI | BAFF, APRIL | NF-kB1, MyD88 | 59 | |||
| BCMA | BAFF, APRIL | NF-kB | 59 | |||
| CD72 | CD100 | Grb2/CD19-PI3K | 60 | |||
| Coinhibitor | PD-1 | B7-H1, B7-DC | SHIP-2 | 24 | ||
| BTLA | HVEM | SHIP-1 | 29 | |||
| CD72 | CD100 | SHIP-1 | 60 | |||
| CD22 | Sialic acid | SHIP-1 | 61 | |||
| NK cell | NKG2D | Costimulator | DNAM-1 | PVRL2, PVR | - | 21 |
| NKp30 | CD96 | PVR | - | 62 | ||
| NKp44 | CD137 | CD137L | - | 63 | ||
| NKp46 | CD27 | CD70 | - | 64 | ||
| 2B4 | CD48 | SAP | 14,65 | |||
| NTB-A | NTB-A | SAP | 65 | |||
| CRACC | CRACC | SAP/EAT-2 | 65 | |||
| Coinhibitor | LAIR-1 | Collagen | SHIP-1, 2 | 35 | ||
| Siglec-3.7.9 | Sialic acid | SHIP-1, 2 | 66,67 | |||
| PD-1 | B7-H1, B7-DC | SHIP-2 | 68 | |||
| KLRG1 | Cadherins | SHIP-1, 2 | 69 | |||
| NKR-P1A | CLEC2D | SHIP-1, 2 | 70 | |||
| ILT2 | HLA class I | SHIP-1, 2 | 71 | |||
| KIR2DL1,2,3 | HLA-C | SHIP-1, 2 | 72 | |||
| KIR3DL1,2 | HLA-A/B | SHIP-1, 2 | 72 | |||
| CD94-NKG2A | HLA-E | SHIP-1, 2 | 72 | |||
| CEACAM1 | CEACAM1 | SHIP-1, 2 | 73 | |||
| 2B4 | CD48 | EAT-2, Csk | 14 | |||
| Myeloid cells (DCs, Macrophages, Neutrophils) | TLR, RLR | Costimulator | B7-1, B7-2 | CD28 | p38, MAPK | 74 |
| NLR | TIM-3 | galectin-9, phosphatidylserine | Erk, NF-kB | 16 | ||
| Plexin-A1 | Sema6D | TREM2-DAP12 | 75 | |||
| Plexin-A4 | Sema3A | Rac1 | 36 | |||
| CD137 | CD137L | Stat 3 | 76,77 | |||
| CD40 | CD40L | NF-kB | 78 | |||
| CD300b,e | - | DAP12, Grb2 | 79 | |||
| TREM1,2,3 | - | DAP12 | 80 | |||
| Coinhibitor | ILT7 | BST2 | FcεRIγ | 79 | ||
| ILT3,4 | - | SHIP-1,2 | 79 | |||
| TLT-1 | - | SHIP-1,2 | 80 | |||
| CD200R | CD200 | Dok1, Dok2 | 81 | |||
| PD-1 | B7-H1, B7-DC | JNK, PI3K | 82 | |||
| TAM family | GAS6/protein S | SOCS1, SOCS3 | 83 | |||
| Cd300a,f | - | SHIP-1,2 | 79 | |||
| DC-SIGN | Carbohydrate | Raf-1 | 84 | |||
| B7-1, B7-2 | CTLA-4 | Stat1, p38, MAPK | 85 | |||
| Mast Cell | FcεRI | Costimulator | Kit | SCF | SHC, Grb2, PI3K, PLCγ | 86 |
| CD137 | CD137L | Lyn | 87 | |||
| CD28 | B7-1, B7-2 | Syk | 88 | |||
| DNAM-1 | PVR, PVRL2 | Fyn, Lat, PLCγ2 | 89 | |||
| CD300b | TIM-1 | DAP12 | 90 | |||
| Coinhibitor | Allergin-1 | - | SHIP-1,2 | 91 | ||
| Pir-B | MHC class I | SHIP-1.2 | 92 | |||
| MAFA | Terminal mannose | - | 93 | |||
| Residues | 93 | |||||
| Gp49B1 | - | SHIP-1,2 | 94 | |||
| CD300A | - | SHIP-1,2 | 95 |
αβ T Lymphocytes
T lymphocytes are the essential components for adaptive immune responses. αβ T lymphocytes constitutes 98% of total T cells. T cells represent the cell type that has been most comprehensively and extensively studied with regard to the role of co-signals. In addition to CSSMs, cytokines, such as interleukin-12 (IL-12), transforming growth factor-β (TGF-β) and IL-6, are crucial for further differentiation of T cells, especially CD4+ T helper cells, though the role of cytokines as co-signals is beyond the scope of this review.
Primary signal for conventional T cells is mediated through TCR engagement. Conventional T cells carrying the αβ TCR recognize small antigenic peptides presented in the groove of the self major histocompatibility complex (MHC). As a result of this recognition, TCR complexes aggregate on T cell surfaces to form stable contacts resulting in the formation of immunological synapses on APC (Huppa and Davis, 2003). Early intracellular signaling, following TCR engagement involves the activation of Src (Lck and Fyn) protein tyrosine kinases (PTKs), leading to the phosphorylation of CD3-localized immunoreceptor tyrosine-based activation motifs (ITAMs). Subsequently, the PTK ZAP-70 is recruited, resulting in a series of phosphorylation events (Smith-Garvin et al., 2009).
The majority of co-signaling molecules for conventional T cells stem from the B7 family and TNF superfamily of receptors (Table 1). In addition to interactions between CD28 and CTLA-4 receptors with their ligand B7-1 (CD80) and B7-2 (B70, CD86)(Greenwald et al., 2005), several recent studies add new perspectives for this classic pathway. B7-1 has been found to interact with B7-H1. In this case, B7-1 serves as a receptor to inhibit T cell responses in vitro (Butte et al., 2007) and contributes to the induction of T cell tolerance in vivo (Park et al., 2010). B7-H2, a molecule best known as the ligand for Inducible Costimulator (ICOS), is found to be a costimulatory ligand for CD28 in vitro (Yao et al., 2011). Interestingly, this interaction is only found in human, not in mouse, warranting re-evaluation of data previously obtained from mouse models. Herpesvirus entry mediator (HVEM), a member of the TNF receptor (TNFR) superfamily, is commonly recognized as a co-stimulatory receptor for LIGHT (TNFSF14) and lymphotoxin- α (Xu et al., 2007). Interestingly, HVEM has recently been found to interact with B and T lymphocyte attenuator (BTLA) and CD160, and deliver a suppressive signal to T cells (Murphy and Murphy, 2010). The complex molecular network between HVEM and its binding partners makes bidirectional signaling feasible, and reveals surprising cross-talk between TNFR superfamilies and Ig-like receptors. In addition to the B7 and TNF families, several new families of molecules have emerged with potent co-signaling function, including T-cell immunoglobulin domain and mucin domain (TIM) family, poliovirus receptor (PVR)-like proteins(Joller et al., 2011; Tahara-Hanaoka et al., 2004; Xu and Jin, 2010), semaphorins(Suzuki et al., 2008) and butyrophilin-like molecules(Nguyen et al., 2006; Smith et al., 2010; Stefferl et al., 2000).
Why do T cells need so many co-signals? One simple answer for this question is that T cell activation is an instructively programmed process and every step of a T cell response is tightly controlled by many different groups of co-signaling molecules with both costimulatory and coinhibitory functions. A typical T cell response evolves at least three steps: priming, expansion, and contraction. Co-stimulators, such as CD27, CD28 and HVEM which are constantly expressed on naive T cells, are known to be important initiators for naive T cell activation in lymphoid organs in the presence of a TCR signal (Sharpe, 2009; Watts, 2005), while the majority of coinhibitory receptors are undetectable during this period. Further expansion of such T cells requires signals through ICOS, death receptor 3 (DR3), CD137 and OX40(Sharpe, 2009; Watts, 2005). If coinhibitory ligands are available in lymphoid organs, co-inhibitory molecules like CTLA-4, PD-1, BTLA and LAG-3 are now unregulated and make activated T cells susceptible to negative control(Chen, 2004; Murphy et al., 2006; Okazaki et al., 2011). This could be considered the first level of negative control. For example, expression of B7-H1 is found to be upregulated in lymph nodes, upon interacting with PD-1, leading to tolerance induction of T cells(Tsushima et al., 2007). Upon exit from lymphoid organs and arrival into the periphery, effector T cells begin to execute their functions. OX40 and CD137 have proven to be critical for effector T cell survival and therefore memory T cell generation (Watts, 2005). During and after execution of effector functions, effector T cells are subjected to another level, and possibly the most severe negative regulation. Peripheral organs and tissues are equipped with various coinhibitory ligands including B7-H1, B7-H4, Galectin-9, PVR, Semaphorins, V-set and immunoglobulin domain containing 4 (VSIG4)(Vogt et al., 2006) and butylophines, which are ready to either tune down or terminate effector T cells. Therefore, different co-signaling molecules may form distinct groups to regulate various stages of T cell activation.
Diverse expression of co-signaling molecules also allows differential control of T cell subsets and their “personalized” characters are critical to modulating immune responses. For example, CD137 is a potent co-stimulator for CD8+ T cells, while its effects on CD4+ T cell are less profound(Watts, 2005) On the contrary, OX40 and ICOS are found to preferentially costimulate CD4+ over CD8+ T cells despite the fact that they are induced on both activated CD4+ and CD8+ T cells(Greenwald et al., 2005; Watts, 2005). These results suggest an intrinsic difference between CD4+ and CD8+ T cells in responding to co-signals. Another example is that, although both CTLA-4 and PD-1 are coinhibitory for effector T cells, blockage of PD-1 signal, but not CTLA-4, restores the function of exhausted T cells thereby reducing viral load during chronic viral infection (Barber et al., 2006). This would support the concept that PD-1 selectively controls the exhaustion phenotype. In many cases, distinct expression patterns of co-signaling molecules have selective effects on T cell subsets. Both B7-DC and B7-H1 deliver an inhibitory signal to T cells through their shared receptor PD-1 (Keir et al., 2008). The expression of B7-DC is restricted to professional APCs such as DCs (Keir et al., 2008). In contrast, B7-H1 mRNA is present in almost all peripheral tissues and cell surface protein expression is extremely sensitive to the regulation by proinflammatory cytokines (Chen, 2004; Zou and Chen, 2008). As a result, the role of B7-DC is limited to T cell priming, while the role of B7-H1 on T cells could be broad: acting on both priming and effector phases.
CD28 is constitutively present on T cells, providing instant help to potential T cell activation. Meanwhile, its counterpart- CTLA-4, is mainly found in intracellular reservoirs, which allows CTLA-4 to rapidly transport to the cell surface in response to antigenic stimuli and to exert its immunomodulatory function (Schneider et al., 2006). Interestingly, CTLA-4 is constitutively present on the surface of foxhead box protein 3 (Foxp3)+ regulatory T (Treg) cells and is required for the maintenance of Treg cell function in vivo (Wing et al., 2008). Meanwhile the co-inhibitory PD-1 protein is not expressed on naive T cells yet is transiently induced on activated T cells (Chen, 2004). However, PD-1 is highly expressed on T cells with exhausted phenotypes, induced by constant exposure to antigen stimuli during chronic viral infection or malignancy. Blockade of the PD-1 pathway restores T cell function, emphasizing the critical role of PD-1 in T cell dysfunction during chronic viral infection(Barber et al., 2006; Keir et al., 2008). Thus, the expression pattern of each co-signaling molecule contributes greatly to their differential regulatory functions.
Our understanding of how intracellular biochemical pathways transmit co-signals is still rudimentary. This is largely due to an absolute dependence of co-signal function on TCR-mediated signaling. Given the overlapping but distinctive function of co-signals, it is not difficult to understand that co-signaling molecules regulate T cell immunity utilizing both shared and unique signaling pathways. The most straightforward strategy is to consider intracellular motifs from each co-signaling receptor. One common pathway for CD28 and its family members, such as ICOS and CTLA-4, is the recruitment of class 1A forms of phosphatidylinositol 3-kinase (PI3K) to their cytoplasmic domains (Rudd and Schneider, 2003). Unlike CD28, ICOS lacks the intracellular motif to bind growth-factor receptor-bound protein 2 (GRB2), which might contribute to its ineffective induction of IL-2 production (Rudd and Schneider, 2003). Co-inhibitory receptors such as PD-1 and BTLA utilize the immunoreceptor tyrosine-based inhibitory motif (ITIM) to recruit SRC homology 2 (SH2)-domain-containing protein tyrosine phosphatase 2 (SHP-2) or SHP-1 and SHP-2, which dephosphorylates and therefore deactivates downstream signal transducers (Greenwald et al., 2005; Watanabe et al., 2003). Co-signaling receptors from the TNFR superfamily contain the intracellular regions necessary for interacting with TNFR-associated factors (TRAFs) (Croft, 2009). TRAFs can recruit inhibitors of nuclear factor-κB (NF-κB), α subunit (IκBα), IκB kinase-β (IKKβ) and NF-κB-inducing kinase (NIK), leading to the activation of both canonical and non-canonical NF-κB pathways which are essential for cell survival (Vallabhapurapu and Karin, 2009). Several TNFR co-receptors also activate signaling pathways other than NF-κB, which might not be shared among TNFR members but contribute to their regulatory functions. For instance, ligation of CD137 activates extracellular-signal-regulated kinase (ERK) and regulates the expression of cyclins (Watts, 2005). Meanwhile, signaling through the TNFR OX40, promotes the expression of survivin and aurora B kinase (Sugamura et al., 2004).
There are numerous studies detailing the coordination of co-signals with TCR signals (Sharpe, 2009; Smith-Garvin et al., 2009). Engagement of TCR regulates the expression of several co-signaling molecules on T cells, and the strength of signal one presumably affects the recruitment of co-signaling molecules to the immune synapse(Egen and Allison, 2002). TCR ligation alone can trigger many signaling pathways; however, the magnitude of the response is considerably altered in the presence of co-signaling molecules It appears that engagement of co-signaling molecules results primarily in a quantitative, rather than a qualitative change in T cell signaling parameters (Smith-Garvin et al., 2009). However, quantitative differences in signaling may lead to qualitatively distinct outcomes. Directly following TCR engagement, co-signaling molecules like CD28, CTLA-4 and PD-1 are enriched in the immune synapse (Fooksman et al., 2010). Incorporation of CD28 into T cell synapses is completely independent of this signaling, but it is perfectly positioned to enhance sustained TCR signaling (Yokosuka et al., 2008). By contrast, localization of CTLA-4 in synapses depends on ligand engagement as well as ligand dosage (Pentcheva-Hoang et al., 2004). Engagement of CTLA-4 reduces ZAP-70 recruitment to the synapse and increases the threshold for T cell activation (Schneider et al., 2005). One elegant in vivo study conducted by Helga Schneider and colleagues provides in vitro observations through two-photon laser scanning microscopy (Schneider et al., 2006); where they found that T cells normally migrate rapidly through the lymph node, but while they are continuously scanning cell surfaces for antigens they slow down. The presence of CTLA-4 increases T cell motility and reverses the TCR-induced stop signal normally required for stable interaction between the T cell and APC. A recent study also demonstrates that PD-1 signaling blocks the TCR-induced stop-signal and therefore is required for maintaining T cell tolerance (Fife et al., 2009). It remains to be seen how co-signals other than B7 family regulate the threshold for T cell activation in vivo.
γδ T Lymphocytes
γδ T cells represent a small subset of T cells which possess an unique TCR and preferentially reside within epithelial-rich tissues, such as the skin, intestine and reproductive tracts. γδ T cells recognize conserved non-peptide antigens that are upregulated by stressed cells in an MHC-independent manner. The physiological roles of γδ T cells include protective immunity against pathogens, tumor surveillance, and wound healing (Bonneville et al., 2010; Carding and Egan, 2002). γδ T cells do not express CD28 or ICOS, and the role of co-signaling in γδ T cell activation has only recently been investigated. NKG2D, which is known to costimulate CD8+ αβ T cell, promotes the cytotoxicity and IL-2 secretion of γδ T cells (Whang et al., 2009). Another important co-signaling pathway for γδ T cells is the junctional adhesion molecule-like protein (JAML)-coxsackie and adenovirus receptor (CAR) pair (Verdino et al., 2010; Witherden et al., 2010). The JAML protein is selectively expressed on γδ T cell, but not on αβ T cells. Signaling through JAML costimulates γδ T cell proliferation and cytokine production, presumably through recruitment and activation of PI3K (Verdino et al., 2010). In vivo blockade of JAML-CAR interaction results in diminished γδ T cell activation, and therefore delayed wound healing (Witherden et al., 2010).
B Lymphocytes
B lymphocytes are the central mediators of humoral responses. The generation of plasma cells and long-lived memory B cells is a tightly regulated process, which involves an ordered series of molecular and cellular changes in vivo. Upon encountering antigen, B cells rapidly proliferate and undergo class switching and somatic hypermutation(McHeyzer-Williams and McHeyzer-Williams, 2005; Shapiro-Shelef and Calame, 2005). This process usually happens in the germinal centers and requires cellular coordinative interactions among follicular DCs, helper T cells and B cells(McHeyzer-Williams and McHeyzer-Williams, 2005). In addition to cytokines, CSSMs are known to be critical for B cell activation, as original proposed in the two-signal model (Bretscher and Cohn, 1970).
The primary signal for B cells is mediated through the B cell receptor (BCR), which is a multi-protein complex containing a membrane-bound Ig for antigen-binding, and two non-covalently associating elements (Ig-α and Igβ) for signal transduction (Kurosaki et al., 2010). Upon antigen binding, BCR signals are initiated by SRC family kinases, like Lyn, which phosphorylate ITAMs on the BCR complex, thereafter recruiting and phosphrylating Syk (spleen tyrosine kinase). Subsequently, a series of signaling cascades are triggered, which include the PI3K pathway, the activation of phospholipase-C γ2 (PLCγ2), and the increase of intracellular calcium; all of which change cellular metabolism, gene expression, and cytoskeletal organization. However, the complexity of BCR signaling itself permits many distinct outcomes, which include anergy, apoptosis, proliferation, and differentiation into plasma cells or memory B cells (Kurosaki et al., 2010). Similar to T cells, the ultimate outcome of the response is largely controlled by signals from CSSMs.
Currently, members of the TNFR superfamily are dominant in delivering co-stimulatory signals to B cells. Among them, many co-stimulatory pathways also provide co-signal for T cells, including CD27-CD70, CD137-CD137L and HVEM-LIGHT interactions (Duhen et al., 2004; Kobata et al., 1995; Zhang et al., 2010). The CD40-CD40L pair is the best-studied pathway and plays an indispensable role in T cell-dependent B cell responses (Banchereau et al., 1994). CD40 ligation stimulates B cell proliferation, survival, isotype switching, formation of the germinal center (GC), and memory B cell generation. Mice deficient in CD40L or CD40 are unable to generate a primary or a secondary antibody response to a T cell-dependent antigen; do not form GCs; and are deficient in generating antigen-specific memory B cells(Grewal and Flavell, 1998). Consistently, isotype switching is severely impaired in patients with CD40 or CD40L gene mutations(Lougaris et al., 2005). CD40 signaling in B cells leads to the recruitment of TRAFs, thereafter leading to the activation of NF-κB and the MAP kinases JNK and p38. BAFF (B cell activating factor belonging to the TNF family), together with its close homologue APRIL (a proliferation inducing ligand), are key regulators for B cell homeostasis (Mackay and Schneider, 2009). In addition to their crucial roles in B cell development and survival, they are also essential for peripheral B cell activation. BAFF and APRIL both bind to TACI (transmembrane activator and CAML interactor) and BCMA (B cell maturation protein A), and BAFF also interacts with BAFF receptor (BAFFR). BAFF interacts with BAFFR to control the development and survival of B2 cells and marginal zone B cells (Mackay and Schneider, 2009). APRIL binds to TACI to accelerate CD40-independent class switching (Mackay and Schneider, 2008), while also promoting plasma cell survival through the BCMA receptor (O’Connor et al., 2004). As members of the TNFR superfamily, stimulation of BAFFR, TACI and BCMA receptors triggers the recruitment of TRAF adaptor proteins, , to activate NF-κB pathways, which are critical for B cell survival. Signaling through BAFFR preferentially activates the alternative NF-κB2 pathway, whereas TACI is a potent stimulator of the classical NF-κB1 pathway. BCR signaling is required to provide substrates for sustaining the NF-κB2 pathway triggered by BAFF. In addition, BAFFR signals can activate the PI3K - AKT1-mTOR pathway to promote cellular metabolism (Mackay and Schneider, 2009). In addition, TACI triggers class switching through direct associating with the adaptor MyD88 (He et al., 2010). Similar to T lymphocytes, B cells express several co-inhibitory receptors. PD-1, now known as a key T cell checkpoint modulator, was originally thought of as a co-inhibitor for B cells (Okazaki et al., 2001). Ligation of the PD-1 intracellular domain recruits the phosphatase SHP-2, leading to the inhibition of BCR signaling by dephosphorylating several key signal transducers in vitro. Several recent studies support PD-1 as a regulator for GC B cell survival and formation of memory plasma cells because PD-1 deficiency or deficiency of its ligand B7-H1 and B7-DC results in impaired memory B cell pools, likely mediated by poor survival of follicular helper T cells (Good-Jacobson et al., 2010; Hamel et al., 2010). Similarly BTLA, another T cell co-inhibitor, attenuates BCR signaling by recruiting SHP-1 (Vendel et al., 2009).
Natural Killer (NK) Cells
Natural killer (NK) cells are cytotoxic lymphocytes that mediate innate immunity against viral infection and tumors (Cerwenka and Lanier, 2001). A major difference of NK cells from other lymphocyte lineages is that they often utilize multiple activating receptors to transmit a primary signal, leading to rapid activation of NK cells (Moretta et al., 2001). This is understandable in the context of NK cell functions as the host’s rapid reacting force to survey the tissues for abnormality. Functions of these germline-encoded receptors, however, are still tightly regulated by a great number of cell surface co-signaling molecules. The activation of NK cells can result in direct cytotoxic attack on their targets and/or secretion of array of cytokines and chemokines, which contributes to initiation of antigen-specific responses. NK cells are also armed with a large number of co-inhibitory molecules that control their activity.
NK cells use multiple surface receptors to distinguish normal healthy cells with abnormal cells undergoing various forms of stress, such as viral infection or tumor transformation. Some receptors detect viral proteins on the surface of infected cells, which are structurally similar to MHC and otherwise intend to evade immune recognition (Natarajan et al., 2002). Some receptors can identify a type of stress molecule expressed on the surface of viral-infected or malignant-transformed cells. Those stress molecules are encoded by the host’s genome, yet are rarely expressed by normal cells but upregulated by stressed or diseased cells. For example, NKG2D identifies a series of MHC-like ligands preferentially present on many tumor cells (Raulet, 2003); In addition to its ability to bind to cytomegalovirus (CMV) pp65, NKp30 has recently been shown to be a receptor for B7-H6, a protein not found on normal cells but highly expressed on varieties types of tumor cell lines (Brandt et al., 2009). These activating NK cell receptors often have short intracellular domains that lack intrinsic signaling activity. Their charged transmembrane regions, however, can associate with transmembrane adaptor molecules to transmit signals. For instance, NKp44 couples to DAP12 while NKG2D uses the adaptor protein DAP10 (Moretta et al., 2001). CD16, NKp30 and NKp46 receptors associate with FcεRIγ and CD3ξ (Vivier et al., 2004).
Since NK cells share a common progenitor with T cells and in many aspects are very closely related to T cells, it is not surprising that a lot of surface molecules are shared between NK cells and T cells. Many co-stimulators for T cells, like CD137 (Wilcox et al., 2002), CD27 (Takeda et al., 2000), CD96 (Fuchs et al., 2004), CD226 (Tahara-Hanaoka et al., 2004) and LAIR-1 (Meyaard et al., 1997), are also found to be crucial for NK cell activation, though their exact roles in two cell types might not be identical. Signaling lymphocytic activation molecule (SLAM)-related and PVR-like proteins are two main families which are important for the regulation of NK cell function. SLAM family proteins have been shown to exhibit homotypic interactions with the exception of 2B4, which recognizes CD48. Interestingly, 2B4 can act as both a co-stimulatory receptor and co-inhibitory receptor for NK cells, depending on the signaling pathways it initiates (Moretta et al., 2001). The positive role of 2B4 requires its association with SLAM-associated protein (SAP), leading to activation of numerous intracellular molecules, such as Vav-1, PLCγ and SHIP(Cannons et al., 2011). In the absence of SAP, 2B4 may deliver a negative signal to NK cells by recruitment of EAT-2 or Csk. CD96 (Tactile) and CD226 (DNAM1) are two receptors for PVR-like family ligands and promote adhesion to ligand-expressing targets and enhance the cytolytic capability of NK cells (Xu and Jin, 2010). In contrast, engagement of TIGIT on NK cells by PVR leads to an ITIM-mediated suppression (Stanietsky et al., 2009). One common feature for NK cell co-inhibitory receptors is that they contain an intracellular ITIM motif. Many co-inhibitory receptors for NK cells, such as CD94-NKG2A, KIR2DL1-3 and KIR3DL1-2, recognize MHC molecules, which allow normal cells to avoid NK cell killing (Lanier, 2005). Other important co-inhibitory pathways include LAIR1-collagen, NKR-P1A-CLEC2D (Rosen et al., 2005), KLRG1-cadherins (Ito et al., 2006) and ITIM-containing SIGLEC family members (SIGLEC3, SIGLEC7 and SIGLEC9) which recognize sialic acid-containing molecules (Avril et al., 2004; Hernandez-Caselles et al., 2006).
Myeloid-Derived Phagocytes
Monocytes and macrophages and dendritic cells represent two subgroups of the mononuclear phagocyte system originally described as a population of bone marrow–derived myeloid cells (van Furth and Cohn, 1968). Monocytes are those circulating in the blood while macrophages reside in tissues in the steady state as well as during inflammation. Monocytes and macrophages are critical effectors and regulators of inflammation (Dale et al., 2008). Dendritic cells specialize in initiating and regulating pathogen-specific adaptive immunity and are central to the development of adaptive immune response(Mellman and Steinman, 2001). A series of pattern recognition receptors (PRRs) are utilized to receive primary signal to execute their distinctive functions, including inflammation, opsonization, activation of complement and coagulation cascades and phagocytosis (Janeway and Medzhitov, 2002). These PRRs can be expressed on the cell surface, in intracellular compartments, or secreted into the bloodstream. Several classes of PRRs involved in different aspects of immune functions for those myeloid cells have been illustrated recently (Palm and Medzhitov, 2009). Here we will focus on the inflammatory response as an example to discuss the decision-making process for these myeloid cells, that is, how inflammation is regulated by co-signals.
Pathogen infection is the most common way of triggering an inflammatory response, as phagocytes express PRRs to recognize molecular motifs conserved within a class of microbes, which are often named as pathogen-associated molecular patterns (PAMPs) (Janeway, 1989). These receptors can be considered to transduce a primary signal. Recent studies indicate that PRRs are also responsible for recognizing endogenous molecules released from damaged cells, termed damage-associated molecular patterns (DAMPs) (Seong and Matzinger, 2004). Currently, at least three types of PRR families have been identified(Takeuchi and Akira, 2010). These families include the Toll-like receptors (TLRs), the Retinoic acid-inducible gene (RIG)-I-like receptors (RLRs) and NOD-like receptors (NLRs). TLRs, including ten functional members in human, are the first and best characterized PRR family which sense invading pathogens outside of the cell as well as in intracellular endosomes and lysosomes. Both RLRs and NLRs families are cytoplasmic proteins, with RLRs recognizing short double-stranded RNA derived from viruses while NLRs sensing DAMPs caused by tissue injury (Bowie and Unterholzner, 2008; Philpott and Girardin, 2010).
The activation of these PRRs involves distinctive signaling cascades, leading to the secretion of different patterns of pro-inflammatory cytokines. TLR activation leads to the direct interactions of the TLR Toll–IL-1 receptor (TIR) domain with a cytoplasmic TIR-containing adaptive molecule, such as MyD88 or TRIF (Takeda and Akira, 2004). Activation of the MyD88-dependent signaling pathway results in the activation of the classic NF-κB pathway, which leads to the expression of numerous proinflammatory cytokines, such as IL-6, IL-12 and TNF-α. TRIF is required for the MyD88-independent pathway in TLR3- and TLR4-mediated responses. The recruitment of TRIF leads to the activation of transcription factor IRF-3, thereby inducing type I interferon (IFN) secretion. The RLR family has at least three members: RIG-I, MDA5 and LGP2 (Nakhaei et al., 2009). They are composed of two N-terminal caspase recruitment domains (CARDs), a central DEAD box helicase domain(Linder, 2006), and a C-terminal regulatory domain. RLRs recognize dsRNA from RNA viruses in cytoplasm. Activation of RIG-I and MDA5 leads to its homophilic interaction with IPS-1through CARD domains, turning on signaling cascades resulting into the expression of type I IFN genes. The NLR family contains more than twenty members in human and their domain architecture consists of a variable N-terminal effector domain, a central nucleotide-binding domain (NBD) and C-terminal leucine-rich repeats (LRRs) (Schroder and Tschopp, 2010). NOD2 sense bacterial infection and interact with the receptor-interacting serine-threonine protein kinase 2 (RIPK2) to activate NF-κB and MAPK, therefore promoting the expression of pro-inflammatory molecules (Strober et al., 2006).The NLR family members NLRP1, NLRP3 and NLRC4 assemble large protein complexes known as inflammasomes, which respond to DAMPs and are responsible for the activation of caspase-1 and hence the production of IL-1β and IL-18 (Martinon et al., 2009).
Unlike T cells, the innate immune response mediated by DCs, monocytes and macrophages is a rapid process and does not require any antigen processing. Yet unrestrained signaling by PRRs in DCs and macrophages would generate a chronic inflammatory milieu or cytokine storm that can lead to sepsis. As for any dynamic system, the innate immune response must be carefully regulated so that turning it on must be followed with a mechanism that can turn it off. In fact, many co-receptors on these myeloid cells function as pivotal regulators that can either positively or negatively control inflammation, which here we call as co-signals. Similar to T cell co-signals, co-signaling molecules for DCs, macrophages and monocytes belong to many molecular families, and many of them have preferential roles on these cell types. For example, CD200R, a member of the Ig superfamily, is a co-inhibitory receptor mainly on tissue macrophages (Hoek et al., 2000). In contrast, DC-SIGN is a C-type lectin molecule preferentially found on DCs (Geijtenbeek et al., 2000).
Each co-signal utilizes different intracellular machinery to modulate TLR signaling. Many members of the triggering receptor expressed on myeloid cells (TREM) family modulate myeloid cell function through their association with DAP12 (Ford and McVicar, 2009). Another co-stimulatory pathway for myeloid cells is the plexin-A4–sema3A pair (Wen et al., 2010). Plexin-A4 genetically targeted mice are highly resistant to septic shock induced by TLR agonists, and its ligand, Sema3A, promotes LPS-induced cytokine production through plexin-A4. Signaling studies indicate that Plexin-A4 is required for TLR-induced activation of Ras-related C3 botulinum toxin substrate 1(Rac1), c-Jun N-terminal kinase (JNK) and NF-κB. In contrast, Sema6D-plexin-A1, another pair belonging to the plexin-semaphorin family, induces DCs maturation through a different pathway (Takegahara et al., 2006). Sema6D promotes the association between plexin-A1 and Trem-2, therefore recruiting adaptor DAP12 to activate downstream signaling.
One common pathway for co-inhibitory molecules to dampen TLR-mediated inflammatory pathways in myeloid cells is through the ITIM motif within the cytoplasmic domain that is used to recruit and interact with the phosphatases SHP-1 and SHP-2. Those proteins are mainly Ig superfamily members, including inhibitory members of Ig-like transcripts (ILTs), CD300 family (Clark et al., 2009) and TREM-like transcript-1 (TLT-1) (Ford and McVicar, 2009). CD200R does not have an ITIM but instead contains an NPxY motif in its cytoplasmic domain to recruit inhibitory adaptor proteins Dok1 and Dok2 (Minas and Liversidge, 2006). Another good example of co-inhibitory molecules for DCs is DC-SIGN, which recognizes the carbohydrate motifs on its ligands to tailor TLR signaling on DCs. DC-SIGN engagement modifies TLR signaling by activating the serine-threonine kinase Raf-1, leading to acetylation of the p65 subunit of NF-κB (Gringhuis and Geijtenbeek, 2010).
Mast Cells
The mast cell is a major cell type playing a key role in allergic inflammatory responses. Mast cells bind to aggregated IgE induced by allergen, and rapidly release numerous pro-inflammatory mediators, a process referred as degranulation. In the past decade mast cells have been recognized as immune cells that not only act as key effector cells in allergic responses, but also execute regulatory functions in innate as well as adaptive immune responses(Sayed et al., 2008).
The primary signal to initiate mast cell activation is triggered by allergen-induced aggregation of high-affinity receptors for IgE (FcεRIs) (Gilfillan and Tkaczyk, 2006). The FcεRI receptor is a tetrameric complex which comprises an α-chain, which is responsible for IgE binding, a β-chain and a disulphide-linked γ-chain homodimer, which are responsible for signaling. Following FcεRI aggregation, the protein tyrosine kinase FYN and Syk become activated, which results in tyrosine phosphorylation of the adaptor molecule GAB2 and subsequently the activation of phosphatidylinositol 3-kinase (PI3K) and PLCγ (Rivera et al., 2008). The transmembrane adaptor molecules LAT and NTAL are crucial for coordination of the downstream signaling pathways that are required for the release of the various pro-inflammatory mediators.
CD28, CD226 (DNAM-1) and CD137 are T cell co-stimulatory receptors known to regulate mast cell function as well(Bachelet et al., 2006; Nishimoto et al., 2005; Tashiro et al., 1997). SCF-KIT is the most well-studied costimulatory pathway for mast cells (Gilfillan and Tkaczyk, 2006). SCF alone does not induce mast cell degranulation while simultaneous addition of SCF and antigen markedly increases the secretion of multiple cytokines in both human and mouse mast cells. SCF signaling results into the activation numerous signaling pathways, including the activation of PI3K, PLCγ, calcium mobilization and MAPK-cascade activation, which are also triggered by FcεRI stimulation. However, SCF stimulation fails to induce tyrosine phosphorylation or to activate PKC, which might explain its inability to stimulate mast cell degranulation by itself(Gilfillan and Tkaczyk, 2006). The co-inhibitory receptors for mast cell include PIR-B(Uehara et al., 2001), CD300a (Bachelet et al., 2005), MAFA (Jurgens et al., 1996), gp49B1 (Katz et al., 1996), and allergin-1 (Hitomi et al., 2010), which all contain ITIMs within their cytoplasmic domains. During the initiation of mast cell activation, the phosphorylation of ITIMs recruits the tyrosine phosphatases SHP-1 and SHP-2 to block early signals mediated by FcεRI cross-linking. However, ligands for many of these receptors are yet to be identified.
Perspectives
Cell-cell communication is a crucial mode for multi-cellular organisms to accomplish complex biological functions and various signaling molecules have evolved to meet ever complicated demands to communicate extracellular stimuli with intracellular components. Our tide model incorporates the majority, if not all, of immune cells in the context of an initiator and modulator concept to describe the rise of immune response to environmental stimuli due to transmission of primary and costimulatory signals while this response subsequently falls owing to the presence of coinhibitory signals. This process is a reminiscence of the rise and fall of sea levels due to gravity forces by the Sun, the Moon and the Earth rotation. In addition to adaptive immunity, our model might better describe how innate cells trigger an inflammatory response and how these responses are regulated. Co-signaling molecules, whose expression is responsive to local environments; balance the communication between host innate cells and microorganisms. The intimate interaction between microorganisms and the host immune system covers a wide range of contacts, which are far beyond those between DAMPs and PRRs. Compared with infectious pathogens, commensal bacteria preferentially trigger inhibitory co-signals or fail to induce stimulatory co-signals directly or indirectly to host immune cells. It remains to be seen how each co-signal is induced or dampened to cooperate with PRRs, and in so doing combat infectious pathogens, or induce tolerance to commensal bacteria.
The same co-signaling molecule could be found on various immune cell types and execute the same or similar functions, dependent on receptor(s) or ligand(s) with which it could interact. This would allow maximal efficiency of immune responses to be initiated and expanded, leading to a highly coordinated response of multiple cell types. In this context, tight control from coinhibition becomes critical to tune down such responses in a certain level to prevent tissue or organ damage spanning from an acute inflammation to chronic autoimmunity.
It appears also important that one cell type possesses multiple co-signaling molecules. Co-signaling molecules for certain cell types usually come from one or two protein families, and execute overlapping but not redundant roles. This co-signal mode could ensure that dysfunction of one co-signal could be offset by other co-signals, so that one co-signal defect would not lead to extreme immune dysfunction. At the same time, each co-signal with similar function could specialize in a certain step or type of immune response, thereby exploiting its unique expression profile, in terms of expression location, timing and sensitivity to induction. On the other hand, the types of co-receptors are far broader than we originally thought, and many pathways from unrelated families could have similar regulatory function but use distinct signaling machineries. In that way, these co-signals could operate in parallel without any signaling disruption or conflict. Finally, the multiplicity of co-signals ensures that one pathogen cannot elude immune response simply by targeting one pathway.
Acknowledgments
We thank Beth Cadugan for editing the manuscript. The study has been partially supported by National Institutes of Health grant CA98731, CA106861, CA142779, AI72592, CA97085, CA85721 and Melanoma Research Alliance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Avril T, Floyd H, Lopez F, Vivier E, Crocker PR. The membrane-proximal immunoreceptor tyrosine-based inhibitory motif is critical for the inhibitory signaling mediated by Siglecs-7 and -9, CD33-related Siglecs expressed on human monocytes and NK cells. J Immunol. 2004;173:6841–6849. doi: 10.4049/jimmunol.173.11.6841. [DOI] [PubMed] [Google Scholar]
- Bachelet I, Munitz A, Mankutad D, Levi-Schaffer F. Mast cell costimulation by CD226/CD112 (DNAM-1/Nectin-2): a novel interface in the allergic process. J Biol Chem. 2006;281:27190–27196. doi: 10.1074/jbc.M602359200. [DOI] [PubMed] [Google Scholar]
- Bachelet I, Munitz A, Moretta A, Moretta L, Levi-Schaffer F. The inhibitory receptor IRp60 (CD300a) is expressed and functional on human mast cells. J Immunol. 2005;175:7989–7995. doi: 10.4049/jimmunol.175.12.7989. [DOI] [PubMed] [Google Scholar]
- Banchereau J, Bazan F, Blanchard D, Briere F, Galizzi JP, van Kooten C, Liu YJ, Rousset F, Saeland S. The CD40 antigen and its ligand. Annu Rev Immunol. 1994;12:881–922. doi: 10.1146/annurev.iy.12.040194.004313. [DOI] [PubMed] [Google Scholar]
- Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- Bonneville M, O’Brien RL, Born WK. Gammadelta T cell effector functions: a blend of innate programming and acquired plasticity. Nat Rev Immunol. 2010;10:467–478. doi: 10.1038/nri2781. [DOI] [PubMed] [Google Scholar]
- Bowie AG, Unterholzner L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat Rev Immunol. 2008;8:911–922. doi: 10.1038/nri2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt CS, Baratin M, Yi EC, Kennedy J, Gao Z, Fox B, Haldeman B, Ostrander CD, Kaifu T, Chabannon C, et al. The B7 family member B7-H6 is a tumor cell ligand for the activating natural killer cell receptor NKp30 in humans. J Exp Med. 2009;206:1495–1503. doi: 10.1084/jem.20090681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher P, Cohn M. A theory of self-nonself discrimination. Science. 1970;169:1042–1049. doi: 10.1126/science.169.3950.1042. [DOI] [PubMed] [Google Scholar]
- Butte MJ, Keir ME, Phamduy TB, Sharpe AH, Freeman GJ. Programmed death-1 ligand 1 interacts specifically with the B7-1 costimulatory molecule to inhibit T cell responses. Immunity. 2007;27:111–122. doi: 10.1016/j.immuni.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannons JL, Tangye SG, Schwartzberg PL. SLAM Family Receptors and SAP Adaptors in Immunity. Annu Rev Immunol. 2011;29:665–705. doi: 10.1146/annurev-immunol-030409-101302. [DOI] [PubMed] [Google Scholar]
- Carding SR, Egan PJ. Gammadelta T cells: functional plasticity and heterogeneity. Nat Rev Immunol. 2002;2:336–345. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol. 2001;1:41–49. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- Chen L. Co-inhibitory molecules of the B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol. 2004;4:336–347. doi: 10.1038/nri1349. [DOI] [PubMed] [Google Scholar]
- Clark GJ, Ju X, Azlan M, Tate C, Ding Y, Hart DN. The CD300 molecules regulate monocyte and dendritic cell functions. Immunobiology. 2009;214:730–736. doi: 10.1016/j.imbio.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Croft M. The role of TNF superfamily members in T-cell function and diseases. Nat Rev Immunol. 2009;9:271–285. doi: 10.1038/nri2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale DC, Boxer L, Liles WC. The phagocytes: neutrophils and monocytes. Blood. 2008;112:935–945. doi: 10.1182/blood-2007-12-077917. [DOI] [PubMed] [Google Scholar]
- Duhen T, Pasero C, Mallet F, Barbarat B, Olive D, Costello RT. LIGHT costimulates CD40 triggering and induces immunoglobulin secretion; a novel key partner in T cell-dependent B cell terminal differentiation. Eur J Immunol. 2004;34:3534–3541. doi: 10.1002/eji.200425598. [DOI] [PubMed] [Google Scholar]
- Egen JG, Allison JP. Cytotoxic T lymphocyte antigen-4 accumulation in the immunological synapse is regulated by TCR signal strength. Immunity. 2002;16:23–35. doi: 10.1016/s1074-7613(01)00259-x. [DOI] [PubMed] [Google Scholar]
- Fife BT, Pauken KE, Eagar TN, Obu T, Wu J, Tang Q, Azuma M, Krummel MF, Bluestone JA. Interactions between PD-1 and PD-L1 promote tolerance by blocking the TCR-induced stop signal. Nat Immunol. 2009;10:1185–1192. doi: 10.1038/ni.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fooksman DR, Vardhana S, Vasiliver-Shamis G, Liese J, Blair DA, Waite J, Sacristan C, Victora GD, Zanin-Zhorov A, Dustin ML. Functional anatomy of T cell activation and synapse formation. Annu Rev Immunol. 2010;28:79–105. doi: 10.1146/annurev-immunol-030409-101308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JW, McVicar DW. TREM and TREM-like receptors in inflammation and disease. Curr Opin Immunol. 2009;21:38–46. doi: 10.1016/j.coi.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Cella M, Giurisato E, Shaw AS, Colonna M. Cutting edge: CD96 (tactile) promotes NK cell-target cell adhesion by interacting with the poliovirus receptor (CD155) J Immunol. 2004;172:3994–3998. doi: 10.4049/jimmunol.172.7.3994. [DOI] [PubMed] [Google Scholar]
- Geijtenbeek TB, Kwon DS, Torensma R, van Vliet SJ, van Duijnhoven GC, Middel J, Cornelissen IL, Nottet HS, KewalRamani VN, Littman DR, et al. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- Gilfillan AM, Tkaczyk C. Integrated signalling pathways for mast-cell activation. Nat Rev Immunol. 2006;6:218–230. doi: 10.1038/nri1782. [DOI] [PubMed] [Google Scholar]
- Good-Jacobson KL, Szumilas CG, Chen L, Sharpe AH, Tomayko MM, Shlomchik MJ. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nat Immunol. 2010;11:535–542. doi: 10.1038/ni.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald RJ, Freeman GJ, Sharpe AH. The B7 family revisited. Annu Rev Immunol. 2005;23:515–548. doi: 10.1146/annurev.immunol.23.021704.115611. [DOI] [PubMed] [Google Scholar]
- Grewal IS, Flavell RA. CD40 and CD154 in cell-mediated immunity. Annu Rev Immunol. 1998;16:111–135. doi: 10.1146/annurev.immunol.16.1.111. [DOI] [PubMed] [Google Scholar]
- Gringhuis SI, Geijtenbeek TB. Carbohydrate signaling by C-type lectin DC-SIGN affects NF-kappaB activity. Methods Enzymol. 2010;480:151–164. doi: 10.1016/S0076-6879(10)80008-4. [DOI] [PubMed] [Google Scholar]
- Hamel KM, Cao Y, Wang Y, Rodeghero R, Kobezda T, Chen L, Finnegan A. B7-H1 expression on non-B and non-T cells promotes distinct effects on T- and B-cell responses in autoimmune arthritis. Eur J Immunol. 2010 doi: 10.1002/eji.201040690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Santamaria R, Xu W, Cols M, Chen K, Puga I, Shan M, Xiong H, Bussel JB, Chiu A, et al. The transmembrane activator TACI triggers immunoglobulin class switching by activating B cells through the adaptor MyD88. Nat Immunol. 2010;11:836–845. doi: 10.1038/ni.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Caselles T, Martinez-Esparza M, Perez-Oliva AB, Quintanilla-Cecconi AM, Garcia-Alonso A, Alvarez-Lopez DM, Garcia-Penarrubia P. A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: two isoforms of CD33 are generated by alternative splicing. J Leukoc Biol. 2006;79:46–58. doi: 10.1189/jlb.0205096. [DOI] [PubMed] [Google Scholar]
- Hitomi K, Tahara-Hanaoka S, Someya S, Fujiki A, Tada H, Sugiyama T, Shibayama S, Shibuya K, Shibuya A. An immunoglobulin-like receptor, Allergin-1, inhibits immunoglobulin E-mediated immediate hypersensitivity reactions. Nat Immunol. 2010;11:601–607. doi: 10.1038/ni.1886. [DOI] [PubMed] [Google Scholar]
- Hoek RM, Ruuls SR, Murphy CA, Wright GJ, Goddard R, Zurawski SM, Blom B, Homola ME, Streit WJ, Brown MH, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200) Science. 2000;290:1768–1771. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- Huppa JB, Davis MM. T-cell-antigen recognition and the immunological synapse. Nat Rev Immunol. 2003;3:973–983. doi: 10.1038/nri1245. [DOI] [PubMed] [Google Scholar]
- Ito M, Maruyama T, Saito N, Koganei S, Yamamoto K, Matsumoto N. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. J Exp Med. 2006;203:289–295. doi: 10.1084/jem.20051986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Janeway CA, Jr, Medzhitov R. Innate immune recognition. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- Joller N, Hafler JP, Brynedal B, Kassam N, Spoerl S, Levin SD, Sharpe AH, Kuchroo VK. Cutting edge: TIGIT has T cell-intrinsic inhibitory functions. J Immunol. 2011;186:1338–1342. doi: 10.4049/jimmunol.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurgens L, Arndt-Jovin D, Pecht I, Jovin TM. Proximity relationships between the type I receptor for Fc epsilon (Fc epsilon RI) and the mast cell function-associated antigen (MAFA) studied by donor photobleaching fluorescence resonance energy transfer microscopy. Eur J Immunol. 1996;26:84–91. doi: 10.1002/eji.1830260113. [DOI] [PubMed] [Google Scholar]
- Katz HR, Vivier E, Castells MC, McCormick MJ, Chambers JM, Austen KF. Mouse mast cell gp49B1 contains two immunoreceptor tyrosine-based inhibition motifs and suppresses mast cell activation when coligated with the high-affinity Fc receptor for IgE. Proc Natl Acad Sci U S A. 1996;93:10809–10814. doi: 10.1073/pnas.93.20.10809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobata T, Jacquot S, Kozlowski S, Agematsu K, Schlossman SF, Morimoto C. CD27-CD70 interactions regulate B-cell activation by T cells. Proc Natl Acad Sci U S A. 1995;92:11249–11253. doi: 10.1073/pnas.92.24.11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki T, Shinohara H, Baba Y. B cell signaling and fate decision. Annu Rev Immunol. 2010;28:21–55. doi: 10.1146/annurev.immunol.021908.132541. [DOI] [PubMed] [Google Scholar]
- Lafferty KJ, Cunningham AJ. A new analysis of allogeneic interactions. Aust J Exp Biol Med Sci. 1975;53:27–42. doi: 10.1038/icb.1975.3. [DOI] [PubMed] [Google Scholar]
- Lafferty KJ, Prowse SJ, Simeonovic CJ, Warren HS. Immunobiology of tissue transplantation: a return to the passenger leukocyte concept. Annu Rev Immunol. 1983;1:143–173. doi: 10.1146/annurev.iy.01.040183.001043. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- Linder P. Dead-box proteins: a family affair--active and passive players in RNP-remodeling. Nucleic Acids Res. 2006;34:4168–4180. doi: 10.1093/nar/gkl468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linsley PS, Clark EA, Ledbetter JA. T-cell antigen CD28 mediates adhesion with B cells by interacting with activation antigen B7/BB-1. Proc Natl Acad Sci U S A. 1990;87:5031–5035. doi: 10.1073/pnas.87.13.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lougaris V, Badolato R, Ferrari S, Plebani A. Hyper immunoglobulin M syndrome due to CD40 deficiency: clinical, molecular, and immunological features. Immunol Rev. 2005;203:48–66. doi: 10.1111/j.0105-2896.2005.00229.x. [DOI] [PubMed] [Google Scholar]
- Mackay F, Schneider P. TACI, an enigmatic BAFF/APRIL receptor, with new unappreciated biochemical and biological properties. Cytokine Growth Factor Rev. 2008;19:263–276. doi: 10.1016/j.cytogfr.2008.04.006. [DOI] [PubMed] [Google Scholar]
- Mackay F, Schneider P. Cracking the BAFF code. Nat Rev Immunol. 2009;9:491–502. doi: 10.1038/nri2572. [DOI] [PubMed] [Google Scholar]
- Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- Meyaard L, Adema GJ, Chang C, Woollatt E, Sutherland GR, Lanier LL, Phillips JH. LAIR-1, a novel inhibitory receptor expressed on human mononuclear leukocytes. Immunity. 1997;7:283–290. doi: 10.1016/s1074-7613(00)80530-0. [DOI] [PubMed] [Google Scholar]
- Minas K, Liversidge J. Is the CD200/CD200 receptor interaction more than just a myeloid cell inhibitory signal? Crit Rev Immunol. 2006;26:213–230. doi: 10.1615/critrevimmunol.v26.i3.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretta A, Bottino C, Vitale M, Pende D, Cantoni C, Mingari MC, Biassoni R, Moretta L. Activating receptors and coreceptors involved in human natural killer cell-mediated cytolysis. Annu Rev Immunol. 2001;19:197–223. doi: 10.1146/annurev.immunol.19.1.197. [DOI] [PubMed] [Google Scholar]
- Mueller DL, Jenkins MK, Schwartz RH. Clonal expansion versus functional clonal inactivation: a costimulatory signalling pathway determines the outcome of T cell antigen receptor occupancy. Annu Rev Immunol. 1989;7:445–480. doi: 10.1146/annurev.iy.07.040189.002305. [DOI] [PubMed] [Google Scholar]
- Murphy KM, Nelson CA, Sedy JR. Balancing co-stimulation and inhibition with BTLA and HVEM. Nat Rev Immunol. 2006;6:671–681. doi: 10.1038/nri1917. [DOI] [PubMed] [Google Scholar]
- Murphy TL, Murphy KM. Slow down and survive: Enigmatic immunoregulation by BTLA and HVEM. Annu Rev Immunol. 2010;28:389–411. doi: 10.1146/annurev-immunol-030409-101202. [DOI] [PubMed] [Google Scholar]
- Nakhaei P, Genin P, Civas A, Hiscott J. RIG-I-like receptors: sensing and responding to RNA virus infection. Semin Immunol. 2009;21:215–222. doi: 10.1016/j.smim.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Natarajan K, Dimasi N, Wang J, Mariuzza RA, Margulies DH. Structure and function of natural killer cell receptors: multiple molecular solutions to self, nonself discrimination. Annu Rev Immunol. 2002;20:853–885. doi: 10.1146/annurev.immunol.20.100301.064812. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Liu XK, Zhang Y, Dong C. BTNL2, a butyrophilin-like molecule that functions to inhibit T cell activation. J Immunol. 2006;176:7354–7360. doi: 10.4049/jimmunol.176.12.7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto H, Lee SW, Hong H, Potter KG, Maeda-Yamamoto M, Kinoshita T, Kawakami Y, Mittler RS, Kwon BS, Ware CF, et al. Costimulation of mast cells by 4-1BB, a member of the tumor necrosis factor receptor superfamily, with the high-affinity IgE receptor. Blood. 2005;106:4241–4248. doi: 10.1182/blood-2005-04-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor BP, Raman VS, Erickson LD, Cook WJ, Weaver LK, Ahonen C, Lin LL, Mantchev GT, Bram RJ, Noelle RJ. BCMA is essential for the survival of long-lived bone marrow plasma cells. J Exp Med. 2004;199:91–98. doi: 10.1084/jem.20031330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T, Maeda A, Nishimura H, Kurosaki T, Honjo T. PD-1 immunoreceptor inhibits B cell receptor-mediated signaling by recruiting src homology 2-domain-containing tyrosine phosphatase 2 to phosphotyrosine. Proc Natl Acad Sci U S A. 2001;98:13866–13871. doi: 10.1073/pnas.231486598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki T, Okazaki IM, Wang J, Sugiura D, Nakaki F, Yoshida T, Kato Y, Fagarasan S, Muramatsu M, Eto T, et al. PD-1 and LAG-3 inhibitory co-receptors act synergistically to prevent autoimmunity in mice. J Exp Med. 2011;208:395–407. doi: 10.1084/jem.20100466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227:221–233. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- Park JJ, Omiya R, Matsumura Y, Sakoda Y, Kuramasu A, Augustine MM, Yao S, Tsushima F, Narazaki H, Anand S, et al. B7-H1/CD80 interaction is required for the induction and maintenance of peripheral T-cell tolerance. Blood. 2010;116:1291–1298. doi: 10.1182/blood-2010-01-265975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentcheva-Hoang T, Egen JG, Wojnoonski K, Allison JP. B7–1 and B7-2 selectively recruit CTLA-4 and CD28 to the immunological synapse. Immunity. 2004;21:401–413. doi: 10.1016/j.immuni.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Philpott DJ, Girardin SE. Nod-like receptors: sentinels at host membranes. Curr Opin Immunol. 2010;22:428–434. doi: 10.1016/j.coi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- Rivera J, Fierro NA, Olivera A, Suzuki R. New insights on mast cell activation via the high affinity receptor for IgE. Adv Immunol. 2008;98:85–120. doi: 10.1016/S0065-2776(08)00403-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DB, Bettadapura J, Alsharifi M, Mathew PA, Warren HS, Lanier LL. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol. 2005;175:7796–7799. doi: 10.4049/jimmunol.175.12.7796. [DOI] [PubMed] [Google Scholar]
- Rudd CE, Schneider H. Unifying concepts in CD28, ICOS and CTLA4 co-receptor signalling. Nat Rev Immunol. 2003;3:544–556. doi: 10.1038/nri1131. [DOI] [PubMed] [Google Scholar]
- Sayed BA, Christy A, Quirion MR, Brown MA. The master switch: the role of mast cells in autoimmunity and tolerance. Annu Rev Immunol. 2008;26:705–739. doi: 10.1146/annurev.immunol.26.021607.090320. [DOI] [PubMed] [Google Scholar]
- Schneider H, Downey J, Smith A, Zinselmeyer BH, Rush C, Brewer JM, Wei B, Hogg N, Garside P, Rudd CE. Reversal of the TCR stop signal by CTLA-4. Science. 2006;313:1972–1975. doi: 10.1126/science.1131078. [DOI] [PubMed] [Google Scholar]
- Schneider H, Valk E, da Rocha Dias S, Wei B, Rudd CE. CTLA-4 up-regulation of lymphocyte function-associated antigen 1 adhesion and clustering as an alternate basis for coreceptor function. Proc Natl Acad Sci U S A. 2005;102:12861–12866. doi: 10.1073/pnas.0505802102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- Seong SY, Matzinger P. Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nat Rev Immunol. 2004;4:469–478. doi: 10.1038/nri1372. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Calame K. Regulation of plasma-cell development. Nat Rev Immunol. 2005;5:230–242. doi: 10.1038/nri1572. [DOI] [PubMed] [Google Scholar]
- Sharpe AH. Mechanisms of costimulation. Immunol Rev. 2009;229:5–11. doi: 10.1111/j.1600-065X.2009.00784.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith IA, Knezevic BR, Ammann JU, Rhodes DA, Aw D, Palmer DB, Mather IH, Trowsdale J. BTN1A1, the mammary gland butyrophilin, and BTN2A2 are both inhibitors of T cell activation. J Immunol. 2010;184:3514–3525. doi: 10.4049/jimmunol.0900416. [DOI] [PubMed] [Google Scholar]
- Stanietsky N, Simic H, Arapovic J, Toporik A, Levy O, Novik A, Levine Z, Beiman M, Dassa L, Achdout H, et al. The interaction of TIGIT with PVR and PVRL2 inhibits human NK cell cytotoxicity. Proc Natl Acad Sci U S A. 2009;106:17858–17863. doi: 10.1073/pnas.0903474106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefferl A, Schubart A, Storch M, Amini A, Mather I, Lassmann H, Linington C. Butyrophilin, a milk protein, modulates the encephalitogenic T cell response to myelin oligodendrocyte glycoprotein in experimental autoimmune encephalomyelitis. J Immunol. 2000;165:2859–2865. doi: 10.4049/jimmunol.165.5.2859. [DOI] [PubMed] [Google Scholar]
- Strober W, Murray PJ, Kitani A, Watanabe T. Signalling pathways and molecular interactions of NOD1 and NOD2. Nat Rev Immunol. 2006;6:9–20. doi: 10.1038/nri1747. [DOI] [PubMed] [Google Scholar]
- Sugamura K, Ishii N, Weinberg AD. Therapeutic targeting of the effector T-cell co-stimulatory molecule OX40. Nat Rev Immunol. 2004;4:420–431. doi: 10.1038/nri1371. [DOI] [PubMed] [Google Scholar]
- Suzuki K, Kumanogoh A, Kikutani H. Semaphorins and their receptors in immune cell interactions. Nat Immunol. 2008;9:17–23. doi: 10.1038/ni1553. [DOI] [PubMed] [Google Scholar]
- Tahara-Hanaoka S, Shibuya K, Onoda Y, Zhang H, Yamazaki S, Miyamoto A, Honda S, Lanier LL, Shibuya A. Functional characterization of DNAM-1 (CD226) interaction with its ligands PVR (CD155) and nectin-2 (PRR-2/CD112) Int Immunol. 2004;16:533–538. doi: 10.1093/intimm/dxh059. [DOI] [PubMed] [Google Scholar]
- Takeda K, Akira S. TLR signaling pathways. Semin Immunol. 2004;16:3–9. doi: 10.1016/j.smim.2003.10.003. [DOI] [PubMed] [Google Scholar]
- Takeda K, Oshima H, Hayakawa Y, Akiba H, Atsuta M, Kobata T, Kobayashi K, Ito M, Yagita H, Okumura K. CD27-mediated activation of murine NK cells. J Immunol. 2000;164:1741–1745. doi: 10.4049/jimmunol.164.4.1741. [DOI] [PubMed] [Google Scholar]
- Takegahara N, Takamatsu H, Toyofuku T, Tsujimura T, Okuno T, Yukawa K, Mizui M, Yamamoto M, Prasad DV, Suzuki K, et al. Plexin-A1 and its interaction with DAP12 in immune responses and bone homeostasis. Nat Cell Biol. 2006;8:615–622. doi: 10.1038/ncb1416. [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- Tashiro M, Kawakami Y, Abe R, Han W, Hata D, Sugie K, Yao L, Kawakami T. Increased secretion of TNF-alpha by costimulation of mast cells via CD28 and Fc epsilon RI. J Immunol. 1997;158:2382–2389. [PubMed] [Google Scholar]
- Tsushima F, Yao S, Shin T, Flies A, Flies S, Xu H, Tamada K, Pardoll DM, Chen L. Interaction between B7-H1 and PD-1 determines initiation and reversal of T-cell anergy. Blood. 2007;110:180–185. doi: 10.1182/blood-2006-11-060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Blery M, Kang DW, Chen CC, Ho LH, Gartland GL, Liu FT, Vivier E, Cooper MD, Kubagawa H. Inhibition of IgE-mediated mast cell activation by the paired Ig-like receptor PIR-B. J Clin Invest. 2001;108:1041–1050. doi: 10.1172/JCI12195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- van Furth R, Cohn ZA. The origin and kinetics of mononuclear phagocytes. J Exp Med. 1968;128:415–435. doi: 10.1084/jem.128.3.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendel AC, Calemine-Fenaux J, Izrael-Tomasevic A, Chauhan V, Arnott D, Eaton DL. B and T lymphocyte attenuator regulates B cell receptor signaling by targeting Syk and BLNK. J Immunol. 2009;182:1509–1517. doi: 10.4049/jimmunol.182.3.1509. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Verdino P, Witherden DA, Havran WL, Wilson IA. The molecular interaction of CAR and JAML recruits the central cell signal transducer PI3K. Science. 2010;329:1210–1214. doi: 10.1126/science.1187996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Nunes JA, Vely F. Natural killer cell signaling pathways. Science. 2004;306:1517–1519. doi: 10.1126/science.1103478. [DOI] [PubMed] [Google Scholar]
- Vogt L, Schmitz N, Kurrer MO, Bauer M, Hinton HI, Behnke S, Gatto D, Sebbel P, Beerli RR, Sonderegger I, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116:2817–2826. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe N, Gavrieli M, Sedy JR, Yang J, Fallarino F, Loftin SK, Hurchla MA, Zimmerman N, Sim J, Zang X, et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat Immunol. 2003;4:670–679. doi: 10.1038/ni944. [DOI] [PubMed] [Google Scholar]
- Watts TH. TNF/TNFR family members in costimulation of T cell responses. Annu Rev Immunol. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- Wen H, Lei Y, Eun SY, Ting JP. Plexin-A4-semaphorin 3A signaling is required for Toll-like receptor- and sepsis-induced cytokine storm. J Exp Med. 2010;207:2943–2957. doi: 10.1084/jem.20101138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whang MI, Guerra N, Raulet DH. Costimulation of dendritic epidermal gammadelta T cells by a new NKG2D ligand expressed specifically in the skin. J Immunol. 2009;182:4557–4564. doi: 10.4049/jimmunol.0802439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox RA, Tamada K, Strome SE, Chen L. Signaling through NK cell-associated CD137 promotes both helper function for CD8+ cytolytic T cells and responsiveness to IL-2 but not cytolytic activity. J Immunol. 2002;169:4230–4236. doi: 10.4049/jimmunol.169.8.4230. [DOI] [PubMed] [Google Scholar]
- Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, Nomura T, Sakaguchi S. CTLA-4 control over Foxp3+ regulatory T cell function. Science. 2008;322:271–275. doi: 10.1126/science.1160062. [DOI] [PubMed] [Google Scholar]
- Witherden DA, Verdino P, Rieder SE, Garijo O, Mills RE, Teyton L, Fischer WH, Wilson IA, Havran WL. The junctional adhesion molecule JAML is a costimulatory receptor for epithelial gammadelta T cell activation. Science. 2010;329:1205–1210. doi: 10.1126/science.1192698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Flies AS, Flies DB, Zhu G, Anand S, Flies SJ, Xu H, Anders RA, Hancock WW, Chen L, Tamada K. Selective targeting of the LIGHT-HVEM costimulatory system for the treatment of graft-versus-host disease. Blood. 2007;109:4097–4104. doi: 10.1182/blood-2006-09-047332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Jin B. A novel interface consisting of homologous immunoglobulin superfamily members with multiple functions. Cell Mol Immunol. 2010;7:11–19. doi: 10.1038/cmi.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S, Zhu Y, Zhu G, Augustine M, Zheng L, Goode DJ, Broadwater M, Ruff W, Flies S, Xu H, et al. B7-H2 is a costimulatory ligand for CD28 in human. Immunity. 2011 doi: 10.1016/j.immuni.2011.03.014. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokosuka T, Kobayashi W, Sakata-Sogawa K, Takamatsu M, Hashimoto-Tane A, Dustin ML, Tokunaga M, Saito T. Spatiotemporal regulation of T cell costimulation by TCR-CD28 microclusters and protein kinase C theta translocation. Immunity. 2008;29:589–601. doi: 10.1016/j.immuni.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Voskens CJ, Sallin M, Maniar A, Montes CL, Zhang Y, Lin W, Li G, Burch E, Tan M, et al. CD137 promotes proliferation and survival of human B cells. J Immunol. 2010;184:787–795. doi: 10.4049/jimmunol.0901619. [DOI] [PubMed] [Google Scholar]
- Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8:467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]