Chromatin Inactivation Precedes De Novo DNA Methylation during the Progressive Epigenetic Silencing of the RASSF1A Promoter (original) (raw)
Abstract
Epigenetic inactivation of the RASSF1A tumor suppressor by CpG island methylation was frequently detected in cancer. However, the mechanisms of this aberrant DNA methylation are unknown. In the RASSF1A promoter, we characterized four Sp1 sites, which are frequently methylated in cancer. We examined the functional relationship between DNA methylation, histone modification, Sp1 binding, and RASSF1A expression in proliferating human mammary epithelial cells. With increasing passages, the transcription of RASSF1A was dramatically silenced. This inactivation was associated with deacetylation and lysine 9 trimethylation of histone H3 and an impaired binding of Sp1 at the RASSF1A promoter. In mammary epithelial cells that had overcome a stress-associated senescence barrier, a spreading of DNA methylation in the CpG island promoter was observed. When the _RASSF1A_-silenced cells were treated with inhibitors of DNA methyltransferase and histone deacetylase, binding of Sp1 and expression of RASSF1A reoccurred. In summary, we observed that histone H3 deacetylation and H3 lysine 9 trimethylation occur in the same time window as gene inactivation and precede DNA methylation. Our data suggest that in epithelial cells, histone inactivation may trigger de novo DNA methylation of the RASSF1A promoter and this system may serve as a model for CpG island inactivation of tumor suppressor genes.
Epigenetic modifications are hallmarks for changes in expression pattern. Abnormal DNA methylation of promoter is a main mechanism for the inactivation of tumor suppressor genes and therefore of fundamental importance for the understanding of the etiology of cancer (24). Gene promoters are often located in CpG-rich DNA regions, so-called CpG islands (3). Active promoters are associated with unmethylated CpG islands and open chromatin structure for transcription regulators, whereas inactive promoters are characterized by a repressed chromatin structure and hypermethylated CpGs. Acetylation of lysines at histone H3 is associated with active chromatin, and deacetylation results in a repressed chromatin structure (23). Methylation of histone lysine 9 residue of histone H3 is observed at promoters of inactive genes. Methyl-CpG binding proteins provide a link between methylated DNA and hypoacetylated histones by recruiting histone deacetylases (25, 32). DNA methyltransferases, which are responsible for the maintenance of methylated DNA and necessary for the establishment of newly methylated promoters, were shown to be associated with proteins including retinoblastoma protein (Rb), E2F1, histone deacetylases, histone methyltransferase, and a transcriptional repressor (13, 14, 35, 37). It has been proposed that the DNA methylation may be directed by alterations in the chromatin structure (2). Several studies indicate an altered DNA methylation pattern when components of the chromatin remodeling system, such as SNF2-like factors, were mutated (11, 16, 22). In Neurospora crassa and in Arabidopsis thaliana, it has been shown that a repressive chromatin modification, like histone H3 lysine 9 (H3-K9) methylation, can direct DNA methylation (21, 44). In vertebrates, it has been reported that Suv39h-mediated H3-K9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin (28). In a recent report, H3-K9 methylation and DNA methylation were excluded as the cause of silencing of a transgene (30).
Normal human mammary epithelial cells (HMEC) grown in culture exhibit two types of proliferation barriers, a stress-associated senescence barrier (stasis) and a senescence barrier resulting from telomere erosion and dysfunction (36, 42). Prestasis HMEC can grow for several population doublings prior to a proliferative arrest associated with elevated levels of p16 (4). Under certain culture conditions, a small number of cells spontaneously emerge from stasis and continue proliferation with ongoing telomere erosion (17, 42). These poststasis cells show a loss of p16 expression associated with promoter hypermethylation of the p16 CpG islands (4, 12, 19). When the telomeres become critically shortened, widespread genomic aberrations are observed (36). In the presence of functional p53, a mostly viable growth arrest (agonescence) occurs; in the absence of functional p53, the HMEC exhibit the massive cell death associated with crisis (5, 36). HMEC may provide new insights into the mechanistic basis of epigenetic silencing of tumor suppressor genes associated with neoplastic transformation.
The Ras association domain family 1 gene (RASSF1) has been located in 3p21.3, a segment which is frequently lost in a variety of human tumors (7). The RASSF1 gene consists of two major transcripts, termed RASSF1A and RASSF1C, which are expressed from two distinct CpG island promoters (9). Both transcripts are present in normal human tissues. However, the RASSF1A message is missing in a variety of human cancer cell lines and primary tumors, including lung and breast carcinomas (7, 9, 10). Silencing of RASSF1A transcript has been correlated with aberrant DNA methylation of the RASSF1A CpG island (7). Methylation profiling of breast tumors suggests that DNA methylation spreads from the first exon into the CpG island area (49). However, a functional promoter sequence has not been mapped, and the exact mechanisms of RASSF1A inactivation have not been investigated at all.
To gain insight into the mechanisms of epigenetic inactivation of RASSF1A, we analyzed the promoter sequence of RASSF1A by a luciferase reporter assay, electrophoretic mobility shift assay (EMSA), in vivo footprinting, and chromatin immunoprecipitation (ChIP). We identified four functional Sp1 sites in the promoter of RASSF1A. In proliferating HMEC and breast cancer cells, we investigated the expression of RASSF1A, DNA methylation, chromatin modification, and Sp1 binding at the RASSF1A promoter. In stasis and poststasis HMEC, drastic silencing of RASSF1A was observed and a spreading of DNA methylation occurred. Our data indicate that histone inactivation precedes DNA methylation and the repressed chromatin state is associated with occlusion of Sp1 binding at the RASSF1A promoter.
MATERIALS AND METHODS
Cells.
Four breast cancer cell lines (T47D, MDA-MB-231, MCF7, and ZR75-1), HeLa cells, and the A549 lung cancer cell line were obtained from the American Type Culture Collection and cultured in the recommended medium. Human mammary epithelial cells (HMEC-184 and HMEC-48R) were obtained from reduction mammaplasty tissue as described previously (41). Additional mammary epithelial cells (HMEC-219 and HMEC-1001) were purchased from Clonetics (BioWhittaker, Verviers, Belgium) or isolated from mammary epithelium (HMEC-141). Prestasis HMEC were grown in mammary epithelial cell growth medium and/or serum-containing medium (41). Poststasis HMEC were cultivated in serum-free mammary epithelial cell growth medium (PromoCell, Heidelberg, Germany) to no more than 80% confluence. Cells were grown at 37°C in 5% CO2, and medium was changed every 3 days. To determine the population doublings, the cells were counted at each passage.
Luciferase reporter gene analysis.
Regulatory sequences of RASSF1A and RASSF1C were cloned in the pRL-null vector (renilla luciferase; Promega, Mannheim, Germany). Promoter sequences of RASSF1 were amplified from fibroblast DNA and cloned in the pRL-null vector. All primer sequences are available in Table S1 in the supplemental material. To in vitro methylate the A-511 construct, 10 μg of plasmid DNA was treated with 30 U of SssI methylase (New England Biolabs [NEB], Frankfurt, Germany). Truncation and mutation in the promoter reporter gene were generated by site-directed mutagenesis (QuickChange XL kit; Stratagene, Amsterdam, The Netherlands) using specific primers. A-213 was cloned by creation of a novel XhoI site at position −196 of A-511 and deletion of the upstream fragment. A-Δ129 was generated through an XmaI site at −112 of A-511 and deletion of the downstream segment. All plasmids were verified by sequencing and cotransfected in HeLa S3 cells with pGL3-SV40-promoter vector control (firefly luciferase; Promega). After 24 h, the expression of luciferase reporter genes was determined and normalized by the Dual-Luciferase reporter assay (Promega).
EMSA.
Nuclear extracts were prepared as previously described with some modifications (46). Briefly, HeLa cells were washed with and incubated in lysis buffer. The isolated nuclei were resuspended in extraction buffer and dialyzed overnight. Nuclear extracts were stored at −80°C in aliquots. To label the EMSA probe, two complementary single-stranded oligonucleotides (200 pmol) were mixed in equimolar amounts, annealed in a water bath, and labeled with 20 μCi of [γ-32P]ATP and 10 U of T4 polynucleotide kinase (NEB). Methylated probes were generated with 2 U of SssI methylase. The DNA-binding assays were carried out in 20 μl of binding buffer with 5 μg of nuclear extract and 2 μl of the probe (10 pmol/μl) for 1 h on ice. If necessary, 500 pmol of cold competitor oligonucleotides or 2 μg of anti-Sp1 or anti-XPA (Santa Cruz Biotechnology, Santa Cruz, Calif.) was included. DNA-protein complexes were mixed with loading buffer and resolved on native 6% polyacrylamide gels at 100 V for 4 h in Tris-borate-EDTA.
LMPCR.
For genomic footprinting experiments, HeLa cells and genomic DNA were treated with 0.2% dimethyl sulfate (DMS). Ligation-mediated PCR (LMPCR) of the cleaved DNA was performed as previously described (8). Primer sequences are available in Table S1 in the supplemental material. The amplified fragments were separated on 8% polyacrylamide-7 M urea gels and electroblotted onto nylon membranes, and the sequences were visualized by hybridization with a single-strand gene-specific PCR probe.
ChIP.
In order to perform chromatin immunoprecipitation (ChIP) analysis, proteins were cross-linked to DNA by adding 1% formaldehyde to the cells for 10 min at 37°C. The cells were lysed in 1% sodium dodecyl sulfate, and the lysates were sonicated to shear DNA to lengths of approximately 300 bp and diluted in 2 ml of ChIP buffer with protease inhibitors. One percent of the diluted cell supernatant was kept to quantify the amount of DNA present in different samples. This probe is considered to be the input control, and DNA-protein cross-links were reversed. To reduce nonspecific background, the 2 ml of diluted cell supernatant was precleared with 75 μl of salmon sperm DNA-protein A agarose (Upstate, Charlottesville, Va.). The immunoprecipitating antibody (0.9 μg of histone H3-trimethyl K9 antibody [ab8898] from Abcam, Cambridgeshire, United Kingdom; anti-acetyl-histone H3 [Ac-K9 and Ac-K14] from Biomol, Hamburg, Germany; or 1 μg of Sp1 antibody from Santa Cruz Biotechnology) was added to the supernatant fraction and incubated overnight at 4°C. The antibody-histone complex was isolated by adding 60 μl of salmon sperm DNA-protein A agarose and pelleted. For a negative control, a no-antibody immunoprecipitation was utilized by incubating the supernatant fraction with only salmon sperm DNA-protein A agarose. The protein A agarose-antibody-protein complex was washed, and the DNA was eluted as described in the ChIP protocol of Upstate. Protein-DNA cross-links were reversed in 0.25 M NaCl at 65°C for 4 h. The DNA was purified by proteinase K digestion, phenol extraction, and ethanol precipitation. The DNA was resuspended in Tris-EDTA buffer, and the amount of histone modification and Sp1 binding were quantified by real-time PCR with the primers listed in Table S1 in the supplemental material. The input sample and no-antibody probe were used as positive (100%) and negative (0%) controls, and the bound-to-input (B/I) fraction was determined. Real-time analyses were repeated at least three times from two independent experiments.
Real-time reverse transcription-PCR.
Total cellular RNA was extracted from cells using the Trizol reagent (Invitrogen, Groningen, The Netherlands) and quantified. Total RNA of normal mammary glands was obtained from Clontech (BD Biosciences, Erembodegem, Belgium). cDNA was synthesized from 0.5 μg of RNA with the iScript cDNA synthesis kit (Bio-Rad, Munich, Germany). Real-time PCR was carried out using the Rotor Gene 2000 (Corbett Research, Sydney, Australia). Reactions were performed using the following conditions: 2 U of Taq (InViTek, Berlin, Germany), 1.5 mM MgCl2, 0.25 mM concentrations of each deoxynucleoside triphosphate, 20 pmol of each primer (see Table S1 in the supplemental material), 0.2× SYBR Green (Biozym, Germany), and 0.7 μl of cDNA. After an initial denaturing step at 95°C for 5 min, the cycling conditions were as follows: 95°C for 30 s, annealing temperature for 30 s, 72°C for 30 s, and a fluorescence measurement at the appropriate melting temperature (83°C for RASSF1A) for a total of 30 cycles. cDNA prepared from 1 μg of RNA of human fibroblast was used as an internal standard and diluted 20 times (10 relative units), 4 times (50 relative units), or 2 times (100 relative units). The relative amounts of RNA were determined with the software Rotor-Gene 4.4 in the quantification mode. Real-time analyses were repeated at least three times with independent cDNA preparations.
Methylation analysis of the RASSF1 locus.
The methylation status of the RASSF1 locus was determined by combined bisulfite restriction analysis (COBRA) (48). For COBRA, 100 ng of bisulfite-treated genomic DNA was amplified for 25 cycles with 20 pmol of each primer and conditions described in Table S1 in the supplemental material. One-fifth portions of the first PCR products were used as templates for a second PCR with internal primers for 35 cycles. Twenty to 50 ng of PCR products was digested with 2 U of restriction enzyme (NEB) as described in Table S1 in the supplemental material. DNA amplified from HeLa cells and treated with SssI methylase (NEB) was used as a control for complete digestion. The restriction products were resolved on 2% Tris-acetate-EDTA agarose gels, and data were analyzed by ImageJ 1.28v (National Institutes of Health). Amplified bisulfite PCR products were subcloned into pGEM-T vector (Promega) according to the manufacturer's instructions. After transformation, DNA of positive clones was prepared with a Qiaprep spin kit (QIAGEN, Hilden, Germany) and sequenced (SeqLab, Göttingen, Germany).
RESULTS
Characterization of regulatory sequences in the RASSF1 gene.
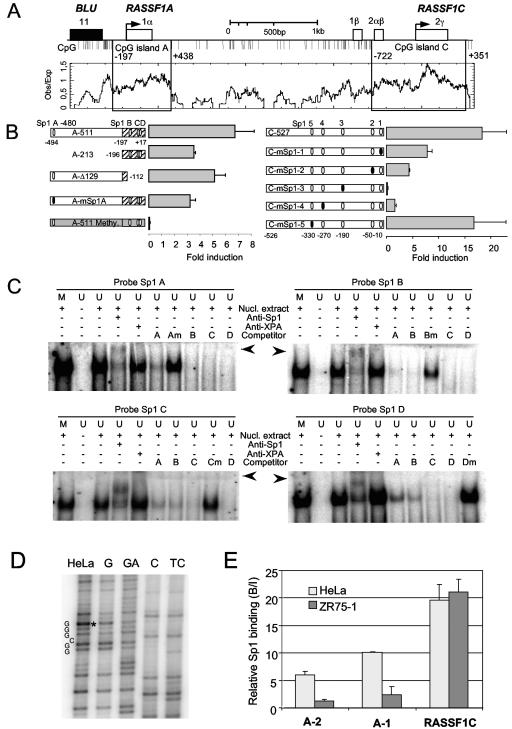
RASSF1A is frequently epigenetically inactivated in human tumors; however, transcriptional regulators have not been mapped in its promoter region. To identify functional elements of the RASSF1A promoter, we cloned 511 bp of the RASSF1A promoter region in a luciferase reporter vector and found high transcriptional activity of this construct (A-511) in HeLa cells (Fig. 1). The fragment includes four putative Sp1 binding sites (GGGCGG), the upstream CpG island of RASSF1A, and the translation start site (Fig. 1A). These Sp1 sites were the only significant transcription binding sites revealed by in silico analysis (www.cbil.upenn.edu/tess and http://genome.ucsc.edu). Deletion of an upstream Sp1 site at position −480 (Sp1 A) or mutation of this Sp1 site resulted in a 50% reduction of the promoter activity (Fig. 1B). Deletion of three Sp1 sites located in the CpG island resulted in 30% reduction. In vitro methylation of the RASSF1A promoter containing reporter plasmid reduced the expression completely. In the RASSF1C promoter, five putative Sp1 binding sites were identified (Fig. 1B). A 527-bp fragment (C-527) including these Sp1 sites was analyzed in the luciferase assay (Fig. 1B). Sequential mutation of four Sp1 sites (Sp1-1 to Sp1-4) in C-527 reduced the activity significantly. Mutational inactivation of the Sp1 site located at −330 had only minor effects. Subsequently, we analyzed the Sp1 binding sites of the RASSF1A promoter by EMSA (Fig. 1C). When the Sp1 sites containing oligonucleotides were incubated with nuclear extract, a shift was detected. A similar shift was detected with the methylated oligonucleotide (Fig. 1C). Shifts with unmethylated probes were further supershifted with Sp1 antibody but not with anti-XPA antibody. The cold Sp1 probes competed for the binding. However, competitors that harbor mutations in the Sp1 site (GTTCGG) were not able to compete significantly (Fig. 1). To verify the binding of Sp1 to the RASSF1A promoter, we performed in vivo footprinting and ChIP assays (Fig. 1D and E). Genomic footprinting of the upper Sp1 site revealed a hyperreactive G in the Sp1 consensus sequence (Fig. 1D). In ChIP assays, two fragments of the RASSF1A promoter (A-1 and A-2) and a fragment of RASSF1C were analyzed for the abundance of cross-linked Sp1 factor (Fig. 1E). The A-2 and A-1 regions contain one Sp1 binding site and three Sp1 binding sites, respectively, and the analyzed RASSF1C fragment harbored two Sp1 binding sites (at positions −270 and −190). In HeLa cells with active RASSF1A and RASSF1C genes, Sp1 binding at both RASSF1 promoters was observed (Fig. 1E). In contrast, reduced binding of Sp1 at the RASSF1A promoter was detected in the breast cancer cell line ZR75-1. In these cancer cells, the RASSF1A promoter is silenced by hypermethylation of the CpG island promoter, but RASSF1C is unmethylated (Fig. 2).
FIG. 1.
Promoter analysis of RASSF1A and RASSF1C. (A) Map of the RASSF1 locus. The locations of exons are shown. The CpG islands are indicated relative to the translational start sites. CpG islands were determined by CpGplot (http://www.ebi.ac.uk). Obs/Exp sets the minimum average observed-to-expected ratio of C plus G to CpG in a set of 10 windows that are required before a CpG island is reported. (B) Luciferase reporter assay of the RASSF1A (left panel) and RASSF1C (right panel) promoters. A 511-bp upstream fragment (A-511) including the translation start site of RASSF1A and four Sp1 binding sites was cloned in pRL-null vector and in vitro methylated (A-511 Methy.). The indicated promoter deletion (A-213 and A-Δ129) and mutation (A-mSp1) were generated. A 527-bp fragment of the RASSF1C promoter (C-527) including five Sp1 sites was cloned into pRL-null vector, and its activity was compared to that of the fragment with mutated Sp1 sites (C-mSp1). The transcriptional activities of the RASSF1A and RASSF1C fragments were determined relative to the promoterless pRL-null vector (set at 1) in three independent assays. (C) EMSA of four Sp1 sites (A, B, C, and D) located in the RASSF1A promoter. Twenty-two-bp labeled unmethylated (U) and in vitro methylated (M) oligonucleotides were incubated with nuclear extract of HeLa cells and analyzed by EMSA. Additionally, the probes were incubated with anti-Sp1 or XPA antibody (supershift is indicated by arrowheads) and competitor oligonucleotides. Mutated (m) competitors wereincluded in the assays. (D) In vivo footprint of the Sp1 A site. HeLa cells were treated with DMS in vivo, and footprints were resolved by LMPCR. Genomic DNA of HeLa was treated in vitro with DMS (lane G). A hyperreactive G in the Sp1 A site in HeLa is marked. Maxam-Gilbert control sequences are shown (GA, C, and TC). (E) HeLa and ZR75-1 cells were cross-linked with formaldehyde and analyzed by ChIP. Protein-DNA complexes were precipitated with Sp1 antibody, and the abundance was determined by real-time PCR in three independent experiments. The bound-to-input ratio (B/I) was plotted against input chromatin (100%) and no-antibody probe (0%). Two regions in the RASSF1A promoter (A-2 and A-1; see also Fig. 6) and a segment in RASSF1C are plotted.
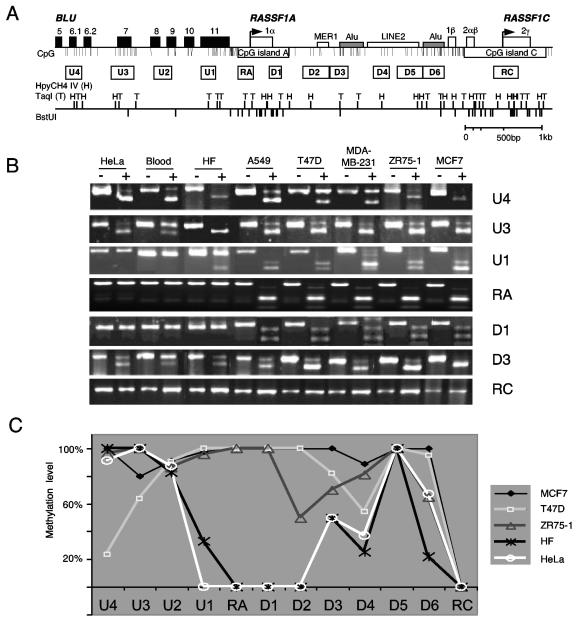
FIG. 2.
Methylation analysis of the RASSF1 locus. (A) Map of the RASSF1 locus. The arrows indicate the transcriptional start sites of the RASSF1 isoforms and the white boxes the exons of RASSF1. Black boxes represent the exons of the BLU gene. Additional DNA elements (Alu, MER1, and LINE2) were located by RepeatMasker (http://ftp.genome.washington.edu/RM/RepeatMasker.html). The indicated 12 PCR fragments of the 7-kb locus were analyzed by combined bisulfite restriction analysis (COBRA). The coding DNA strand was deaminated in silico, and the restriction cutting sites of the CpG-containing sequence are shown (HpyCH4IV, TaqI, and BstUI). Primer sequences, PCR conditions, and restriction enzymes are listed in Table S1 in the supplemental material. (B) Representative COBRA analysis of normal blood DNA, human fibroblasts (HF), and cancer cell lines. PCR products of bisulfite-treated DNA were digested (+) or mock digested (−) with the appropriate enzymes. (C) The relative methylation level of the COBRA was plotted for the indicated cancer cell lines and HF.
Epigenetic status of the RASSF1A locus.
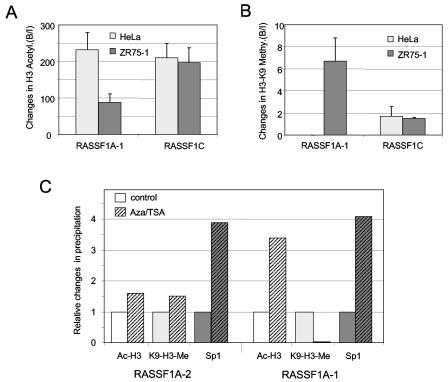
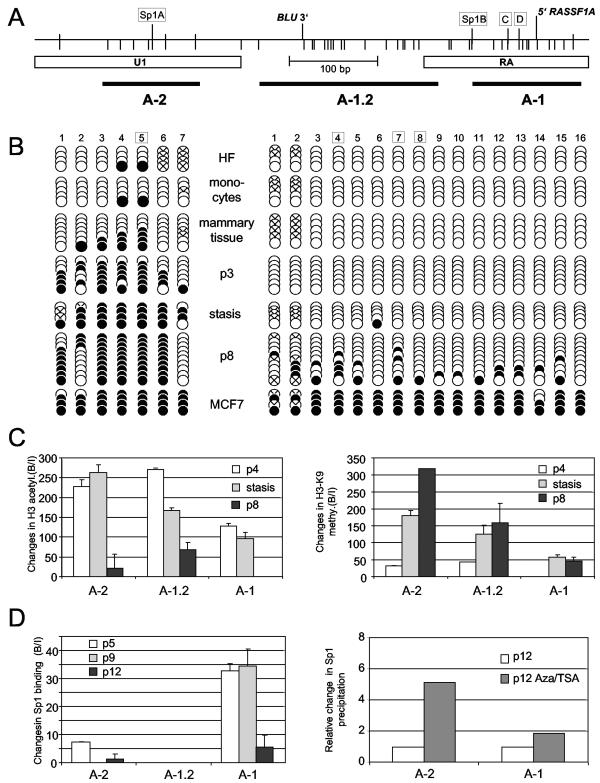
To investigate the DNA methylation pattern of the RASSF1A gene, we analyzed 12 regions of a 7-kb segment flanking the RASSF1A CpG island by combined bisulfite restriction analysis (COBRA) (Fig. 2). This analysis was performed in _RASSF1A_-expressing cells (HeLa cells, human fibroblasts, and blood) and in breast and lung cancer cell lines (T47D, MDA-MB-231, ZR75-1, MCF7, and A549), which lack RASSF1A expression. The RASSF1A (RA and D1) and RASSF1C (RC) CpG island fragments revealed differences in their methylation patterns. In human fibroblasts, blood, and cancer cell lines, the RC fragment is completely unmethylated (Fig. 2B and C). In contrast, the CpG island of RASSF1A is completely methylated in all cancer cell lines but not in fibroblasts, blood, and HeLa cells (Fig. 2B and C). Then, we analyzed the methylation pattern of the sequences flanking the RA fragment in the breast cancer cell lines. All six segments (D1 to D6) located downstream of the RASSF1A promoter were frequently methylated (Fig. 2C). In the _RASSF1A_-expressing cells, this downstream region was less methylated, but two fragments, D3 and D5, located in different Alu elements were methylated at 50 and 100%, respectively. Three upstream fragments (U2 to U4) of the RASSF1A CpG island, which are located in the BLU gene, were frequently methylated in cancer and normal cells. Analysis of the U1 fragments, which contain the upstream Sp1 binding site and the BLU termination sequence, showed low-frequency methylation in the _RASSF1A_-expressing cells (0 to 33%) compared to cancer cells (>90%). In addition to DNA methylation, chromatin modification by histone methylation and acetylation is involved in the inactive and active chromatin state. To investigate the chromatin state of the RASSF1A and RASSF1C promoter, we performed ChIP with antibodies against histone H3 lysine 9 trimethylation (H3-K9) and acetyl-histone H3 (Ac-H3) in HeLa and ZR75-1 breast cancer cells (Fig. 3). In the unmethylated RASSF1A promoter of HeLa cells, Ac-H3 is more frequently found than in the methylated RASSF1A CpG islands of ZR75-1. In contrast, H3-K9 methylation is exclusively detected in the methylated promoter of ZR75-1. At the RASSF1C promoter, no differences in histone modification between HeLa and ZR75-1 cells were observed (Fig. 3A and B). Furthermore, we treated the ZR75-1 cells with trichostatin A (TSA) and 5-aza-2′-deoxycytidine (Aza), which inhibit histone deacetylase and DNA methyltransferase, respectively. This treatment led to acetylation of H3 at the RASSF1A promoter (Fig. 3C). For the region A-1, which is located in the CpG islands, a 3.4-fold increase in Ac-H3 was observed. Additionally, a dramatic 30-fold decrease in H3-K9 methylation was revealed (Fig. 3C). For the upper A-2 region, only minor changes in histone modification were detected. Treatment with TSA and Aza enhanced the binding of Sp1 at both analyzed regions fourfold (Fig. 3C), and after Aza treatment RASSF1A expression increased threefold (Fig. 4).
FIG. 3.
Histone modification of the RASSF1A and RASSF1C CpG islands. (A) Acetylated histone H3 (Ac-H3) was analyzed in HeLa and ZR75-1 cells by ChIP. PCR products for the RASSF1A and RASSF1C promoter were analyzed by real-time PCR in three independent experiments. The bound-to-input ratio (B/I) was plotted against input chromatin (100%) and no-antibody probe (0%). (B) ChIP assay with antibody against histone H3-K9 trimethylation (H3-K9 Me) was performed in HeLa and ZR75-1 cells. (C) ZR75-1 cells were treated with 0.3 μM TSA and 10 μM Aza for 3 days. The relative changes (B/I) in histone modification (Ac-H3 and H3-K9 Me) and Sp1 binding were compared to the untreated controls (set at 1) by real-time PCR and plotted.
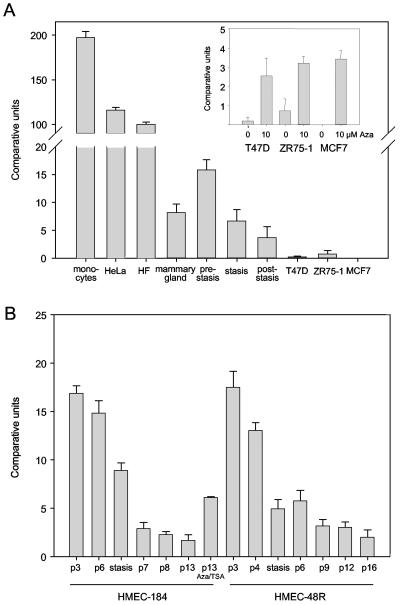
FIG. 4.
Expression analysis of RASSF1A. (A) The expression of RASSF1A was analyzed in 0.5 μg of RNA isolated from monocytes, HeLa cells, human fibroblasts (HF), normal mammary gland, HMEC (prestasis, stasis, and poststasis), and three breast cancer cell lines (T47D, MCF7, and ZR75-1) by real-time PCR. The expression data of three prestasis and stasis HMEC isolates (184, 48R, and 141) and four poststasis HMEC isolates (184, 48R, 219, and 1001) were combined. The breast cancer cells were treated for 4 days with 10 μM Aza. The expression levels were quantified in three independent experiments and plotted relative to an internal standard of 0.5 μg of fibroblast RNA (set at 100%) as described in the text. (B) RASSF1A expression in HMEC. HMEC-184 and HMEC-48R were grown for increasing passages (p), and RASSF1A expression was determined by real-time PCR. Passage 13 of HMEC-184 was treated with 10 μM Aza and 0.3 μM TSA. The expression levels were quantified in three independent experiments and plotted.
Silencing of RASSF1A in HMEC during senescence.
To analyze the epigenetic inactivation of RASSF1A, we utilized normal HMEC, which were grown for consecutive passages. It has been reported that prestasis HMEC exhibit increasing p16 expression with ongoing proliferation, while in the poststasis cells p16 is silenced (4). When we analyzed the expression of p16 in HMEC, we observed epigenetic silencing of p16 in poststasis HMEC (data not shown). Subsequently, we investigated the expression of RASSF1A in different HMEC isolates by real-time reverse transcription-PCR (Fig. 4A). As the prestasis HMEC approached the stasis barrier, RASSF1A expression was drastically reduced (Fig. 4A and B). At stasis, HMEC-48R (passage 5) showed a 70% reduction of expression, and HMEC-184 showed a 50% reduction compared to passage 3; poststasis cells showed a 90% reduction in RASSF1A expression compared to growing prestasis cells (Fig. 4B). The RASSF1C expression did not significantly change in prestasis and poststasis HMEC (data not shown). In human fibroblasts, HeLa cells, and monocytes, which are unmethylated at the RASSF1A CpG island, the highest levels of RASSF1A transcripts (Fig. 4A) were detected. In the breast cancer cell lines ZR75-1, T47D, and MCF7, RASSF1A expression was dramatically reduced, and treatment with 10 μM Aza reactivated the expression of RASSF1A significantly (Fig. 4A). In the quiescent mammary gland, the expression of RASSF1A was comparable to that in the cells at stasis. Treatment of HMEC at passage 13 with Aza and TSA increased the expression of RASSF1A 3.5 times (Fig. 4B).
Epigenetic inactivation and decrease of Sp1 binding occur during silencing of the RASSF1A promoter.
We next analyzed the DNA methylation pattern of the RASSF1A locus in different passages of HMEC by COBRA (Fig. 5). Prestasis HMEC exhibited frequent methylation of the fragments U1, D1, and D2, which flank the transcription initiation site (Fig. 5B and C), and a further increase in DNA methylation was observed in later passages of both HMEC cultures. However, the CpGs located in the RA region, which contains three Sp1 binding sites and the transcription start site, were completely unmethylated in prestasis and stasis HMEC (Fig. 5B and C). Aberrant methylation occurred only in the poststasis cells. This observation may be attributed to a spreading of de novo DNA methylation from the methylated upstream and downstream regions into the RASSF1A CpG island promoter. To verify these data at several CpG sites, single PCR fragments of bisulfite-modified DNA were subcloned and analyzed by sequencing (Fig. 6). For the RA and U1 fragments, we examined 16 and 7 CpGs, respectively, obtained from several independent clones (Fig. 6B). In the human fibroblasts and monocytes, the RA region was completely unmethylated, and in the U1 products, two methylated CpGs located in the Sp1 binding site were detected. In the breast cancer cell line MCF7, almost all analyzed CpGs were methylated (Fig. 6B). Then we investigated the methylation pattern of the U1 and RA fragments in HMEC. In concordance with the COBRA results, a spreading of DNA methylation occurred with increased passage of HMEC (Fig. 6B). The upstream fragment was heavily methylated in early passages. In contrast, the RA fragment located in the CpG island was unmethylated in prestasis cells (Fig. 6B, p3), and methylated sites were found in poststasis cells (Fig. 6B, p8). Interestingly, the methylation density was significantly lower than in the breast cancer cells.
FIG. 5.
Combined bisulfite restriction analysis (COBRA) of the RASSF1 locus in HMEC. (A) Map of the RASSF1 locus. For a description, see the legend to Fig. 4. (B) COBRA analysis of HMEC-184 grown for consecutive passages (p). PCR products of bisulfite-treated DNA were digested (+) or mock digested (−) with the appropriate enzymes (see Table S2 in the supplemental material). (C) COBRA of HMEC-48R grown for successive passages (p).
FIG. 6.
Methylation and chromatin pattern of the RASSF1A CpG island in HMEC. (A) Map of the RASSF1A promoter region and the analyzed fragments. The 5′ end and the 3′ end of the mRNA of RASSF1A and BLU are indicated. CpGs and Sp1 binding sites are marked by bars. (B) Seven and 16 CpG sites of the U1 and RA fragments, respectively, were analyzed in human fibroblasts (HF), monocytes, mammary cells, HMEC-184 (p3, stasis, and p8) cells, and MCF7 cells. Boxed CpGs indicate the Sp1 sites. Amplified PCR products were subcloned, and several independent clones were sequenced. Black and white dots represent methylated and unmethylated CpGs, respectively. Dots marked with a cross were not analyzable by sequencing. (C) For three fragments of the RASSF1A promoter (A-1, A-1.2, and A-2), the abundance of acetylated histone H3 (Ac-H3) and histone H3 K9 trimethylation (H3-K9 Me) were analyzed in consecutive passages (p) of HMEC by ChIP and real-time PCR in threeindependent experiments. (D) Binding of Sp1 to the RASSF1A promoter in mammary epithelial cells. HMEC-184 cells were grown, cross-linked with formaldehyde, and analyzed by ChIP. Protein-DNA complexes were precipitated with Sp1 antibody, and the abundance was determined by real-time PCR in three independent experiments. Three regions (A-2, A-1.2, and A-1) of the RASSF1A promoter are plotted. The bound-to-input ratio (B/I) was plotted against input chromatin (100%) and no-antibody probe (0%). HMEC-184 cells were treated with 0.3 μM TSA and 10 μM Aza for 3 days, and the relative changes (B/I) in Sp1 binding were compared to the untreated controls (set at 1) by real-time PCR in three independent experiments and plotted.
Subsequently, we investigated the acetylated histone H3 and trimethylated K9 histone H3 at three regions of the RASSF1A promoter in different passages of HMEC (Fig. 6C). At all three fragments, we detected high levels of acetylated histone H3 in passage 4. We observed that the histone H3 acetylation decreased at these fragments in the poststasis passage 8 and was not detected at the downstream A-1 fragment. In contrast, trimethylated K9-H3 was frequently found at the A-1 fragment in stasis and poststasis HMEC but not observed in passage 4 (Fig. 6C). The levels of H3-K9 trimethylation increased in stasis and poststasis at all three analyzed regions compared to the preceding passage 4. This indicates that the silencing of RASSF1A occurs together with deacetylation and methylation of histone H3. Interestingly, these histone modifications were already found at the unmethylated CpGs of the promoter, suggesting that altered chromatin structure at the RASSF1A promoter may precede de novo DNA methylation. Interestingly, we observed that levels of H3-K9 methylation were more than 10 times higher during the progressive inactivation of RASSF1A in HMEC (stasis and poststasis) than levels found in the breast cancer cell line ZR75-1. Thus, H3-K9 methylation may play a crucial role in the establishment of inactive chromatin at the RASSF1A promoter.
To investigate whether these epigenetic modifications are responsible for the occlusion of Sp1 binding at the RASSF1A promoter, we analyzed the abundance of Sp1 at the RASSF1A promoter in consecutive passages of HMEC by ChIP (Fig. 6D). At the A-1 fragment, which contains three Sp1 binding sites, a decrease of Sp1 binding was detected in passage 12 compared to passages 5 and 9. The occlusion of Sp1 binding at the upstream-located site A-2 occurred earlier in passage 9. Treatment of poststasis HMEC with Aza and TSA increased the binding of Sp1 at both analyzed regions (A-2 and A-1), and the expression of RASSF1A increased 3.5-fold (Fig. 4 and 6D). Interestingly, we identified similar binding of Sp1 in the A-2 fragments in HeLa cells and prestasis HMEC (Fig. 1 and 6D). However, the A-2 sequence (U1) is frequently methylated in HMEC but not in HeLa. These data indicate that DNA methylation per se is not responsible for the occlusion of Sp1 and suggest that a repressed chromatin state at the RASSF1A promoter is primarily responsible for the decrease in Sp1 binding.
DISCUSSION
In our previous work, we found frequent epigenetic inactivation of the RASSF1A CpG island in human cancers, and this silencing was associated with a hypermethylation of the CpG islands (7). However, the epigenetic mechanisms, which are responsible for the silencing of the RASSF1A promoter, were unknown. Our data indicate that histone H3 deacetylation and histone H3-K9 trimethylation precede de novo DNA methylation during the progressive inactivation of RASSF1A in HMEC. This observation is consistent with results from studies of Neurospora crassa and Arabidopsis and with results obtained in the pericentric heterochromatin of mice, which indicated that H3-K9 methylation can direct DNA methylation (21, 28, 45). Interestingly, we observed that levels of H3-K9 methylation were higher during the progressive inactivation of RASSF1A in HMEC than in the breast cancer cell line ZR75-1. Thus, H3-K9 methylation may play a crucial role in the establishment of inactive chromatin at the RASSF1A promoter. A recent study has suggested that during the silencing of a transgene, H3-K9 methylation and DNA methylation occur late and follow histone deacetylation and loss of H3-K4 methylation (30). Here, we analyzed a bona fide CpG island promoter of a tumor suppressor gene, and therefore our results may represent the mechanism of epigenetic silencing which occurs in carcinogenesis. In order to identify crucial transcriptional activators of RASSF1A, we determined four Sp1 binding sites in the RASSF1A promoter. Three of these Sp1 sites were located in the unmethylated RASSF1A CpG island, and an additional Sp1 site was detected upstream in a region with low CpG content. This upper Sp1 binding sequence can be methylated in normal mammary cells and fibroblasts, and this methylation does not affect the binding of Sp1 directly. This observation confirms that the binding of Sp1 is DNA methylation insensitive (18, 34). We proposed that the altered binding of Sp1 at the RASSF1A promoter is mediated by the repressed chromatin state and not by DNA methylation per se.
It has been shown that methylated CpG sites attract methyl-CpG binding domain proteins (MBDs) that interact with the corepressor complex Sin3, including histone deacetylases (25, 32, 33). Other studies indicate that DNA methyltransferase and MBDs interact with histone methyltransferase (14, 15). The methylated CpGs located upstream and downstream of the RASSF1A CpG island may attract histone methyltransferase and deacetylase and lead to the repressed chromatin. For the glutathione _S_-transferase (GSTP1) promoter, neither removal of the Sp1 binding sites nor seeds of DNA methylation alone are sufficient to achieve promoter hypermethylation (39). Recently, it has been proposed that gene silencing of GSTP1 promotes DNA hypermethylation through a sequential change in chromatin modification (43). It has been suggested that a dramatic stop in transcription is the critical precursor in cancer, which is followed by de novo DNA methylation and ends with a complete cessation of gene expression (6, 47). Our results for RASSF1A are consistent with this hypothesis. In HMEC, a drastic decrease in RASSF1A expression was detected, and this was associated with a repressed chromatin state. In these cells, the methylation pattern of the RASSF1A CpG island is completely different from that in breast cancer cell lines. We propose that the epigenetic inactivation of RASSF1A occurs in distinct steps (Fig. 7). In senescent cells, silencing of RASSF1A transcription may occur through a DNA methylation-independent mechanism involving mainly histone deacetylation and methylation (Fig. 7). In cancer cells, inactivation of RASSF1A is manifested by de novo DNA methylation of its promoter (Fig. 7). This may lead to silencing of RASSF1A, whichis irreversible, since the presence of an active DNA demethylase in mammalian cells is not evident. Thus, in the proliferating poststasis HMEC, a new transcriptional pattern is established by a repressed chromatin state, and then in tumor cells, this aberrant expression profile is locked by DNA methylation and the heterochromatic state is maintained by DNA methyltransferase, methyl-CpG binding proteins, and heterochromatin protein 1 (Hp1) (Fig. 7). It has been shown that the Hp1 isoforms bind to methylated H3-K9 residues (1, 26, 31). Recently, a link between the Suv39h-Hp1 histone methylation system and the DNA methyltransferase 3b in mammals was demonstrated (28). Hp1 and MBD may be responsible for locking and maintaining the repressed chromatin together with the DNA methyltransferases in silenced cells (Fig. 7). It will be interesting to analyze other histone modification and chromatin components, such as methylation of histone H3 lysine 4, MBD, and Hp1, during the inactivation of RASSF1A.
FIG. 7.
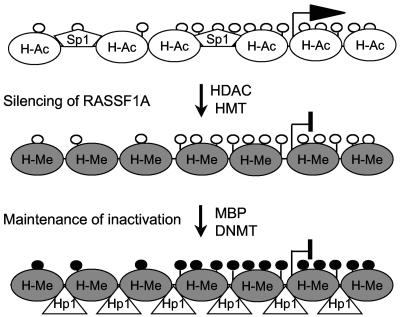
Model of the progressive epigenetic silencing of RASSF1A. In normal epithelial cells, the RASSF1A promoter is transcriptionally active, histones are acetylated (H-Ac), and the CpG island is unmethylated (white dots). The open chromatin structure allows binding of the transcription factor Sp1. In senescent cells, silencing of RASSF1A is associated with histone deacetylation and H3-K9 methylation (H-Me), which is accomplished by histone deacteylase (HDAC) and histone methyltransferase (HMT), respectively. The repressed chromatin structure triggers the de novo methylation of CpGs (black dots) by DNA methyltransferase (DNMT). In cancer cells, the inactive state of RASSF1A is locked and maintained by methyl-CpG binding domain proteins (MBP) and heterochromatin protein 1 (Hp1).
Epigenetic inactivation of tumor suppressor gene promoters plays a fundamental role in the etiology of cancer (24). We and others have analyzed RASSF1A inactivation in primary breast carcinoma and found 49 to 65% RASSF1A CpG island methylation (9, 10). Recently, DNA hypermethylation of RASSF1A was associated with poor prognosis for breast cancer patients (29). RASSF1A inactivation has also been demonstrated in epithelial hyperplasia and intraductal papillomas but has not been detected in lymphocytes, stromal tissue, normal breast epithelium, lactating breast tissue, or apocrine metaplasia (27). It has been reported that RASSF1A blocks cell cycle progression by engaging the Rb cell cycle checkpoint and by inhibiting the anaphase-promoting complex (38, 40). Therefore, silencing of RASSF1A may play a role in the ability of epithelial cells to escape the Rb-mediated stasis barrier. This is the first study which analyzes the inactivation of RASSF1A in the proliferation of HMEC before and after overcoming stasis. Here, we detected intense DNA methylation of the RASSF1A CpG island in breast cancer cell lines, but the methylation pattern was less pronounced in poststasis HMEC. Similar to this observation, it has been postulated that an age-related CpG island methylation leads to a hyperproliferative epithelium, but further hypermethylation needs to occur for tumor formation (20). Thus, proliferating HMEC may serve as an epigenetic model system for the steps leading to aging or transformation of epithelial cells.
In summary, our data show that the aberrant transcriptional silencing of RASSF1A preceded its CpG island promoter hypermethylation, and this may be triggered by inactivating chromatin modification, including histone deacetylation and H3-K9 methylation. Since RASSF1A blocks cell cycle progression, the silencing of RASSF1A may be a critical step in tumorigenesis. It will be interesting to analyze the influence of inhibitors of DNA methylation and histone deacetylase on the reactivation of the RASSF1A gene in silenced cells.
Supplementary Material
[Supplemental material]
Acknowledgments
We thank Gerd P. Pfeifer for helpful suggestions and Christel Trümpler for technical support.
This work was supported by grants DE-AC03-76SF00098 and DAMD17-02-1-0443 to M.R.S. and by grants from BMBF (FKZ01ZZ0104), Land Sachsen Anhalt, and DFG (DA-552/1) to R.D.
Footnotes
REFERENCES
- 1.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410**:**120-124. [DOI] [PubMed] [Google Scholar]
- 2.Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16**:**6-21. [DOI] [PubMed] [Google Scholar]
- 3.Bird, A. P. 1986. CpG-rich islands and the function of DNA methylation. Nature 321**:**209-213. [DOI] [PubMed] [Google Scholar]
- 4.Brenner, A. J., M. R. Stampfer, and C. M. Aldaz. 1998. Increased p16 expression with first senescence arrest in human mammary epithelial cells and extended growth capacity with p16 inactivation. Oncogene 17**:**199-205. [DOI] [PubMed] [Google Scholar]
- 5.Chin, K., C. O. de Solorzano, D. Knowles, A. Jones, W. Chou, E. G. Rodriguez, W. L. Kuo, B. M. Ljung, K. Chew, K. Myambo, M. Miranda, S. Krig, J. Garbe, M. Stampfer, P. Yaswen, J. W. Gray, and S. J. Lockett. 2004. In situ analyses of genome instability in breast cancer. Nat. Genet. 36**:**984-988. [DOI] [PubMed] [Google Scholar]
- 6.Clark, S. J., and J. Melki. 2002. DNA methylation and gene silencing in cancer: which is the guilty party? Oncogene 21**:**5380-5387. [DOI] [PubMed] [Google Scholar]
- 7.Dammann, R., C. Li, J. H. Yoon, P. L. Chin, S. Bates, and G. P. Pfeifer. 2000. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat. Genet. 25**:**315-319. [DOI] [PubMed] [Google Scholar]
- 8.Dammann, R., and G. P. Pfeifer. 1997. Lack of gene- and strand-specific DNA repair in RNA polymerase III-transcribed human tRNA genes. Mol. Cell. Biol. 17**:**219-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dammann, R., U. Schagdarsurengin, M. Strunnikova, M. Rastetter, C. Seidel, L. Liu, S. Tommasi, and G. P. Pfeifer. 2003. Epigenetic inactivation of the Ras-association domain family 1 (RASSF1A) gene and its function in human carcinogenesis. Histol. Histopathol. 18**:**665-677. [DOI] [PubMed] [Google Scholar]
- 10.Dammann, R., G. Yang, and G. P. Pfeifer. 2001. Hypermethylation of the CpG island of Ras association domain family 1A (RASSF1A), a putative tumor suppressor gene from the 3p21.3 locus, occurs in a large percentage of human breast cancers. Cancer Res. 61**:**3105-3109. [PubMed] [Google Scholar]
- 11.Dennis, K., T. Fan, T. Geiman, Q. Yan, and K. Muegge. 2001. Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15**:**2940-2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foster, S. A., D. J. Wong, M. T. Barrett, and D. A. Galloway. 1998. Inactivation of p16 in human mammary epithelial cells by CpG island methylation. Mol. Cell. Biol. 18**:**1793-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuks, F., W. A. Burgers, N. Godin, M. Kasai, and T. Kouzarides. 2001. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 20**:**2536-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuks, F., P. J. Hurd, R. Deplus, and T. Kouzarides. 2003. The DNA methyltransferases associate with HP1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 31**:**2305-2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuks, F., P. J. Hurd, D. Wolf, X. Nan, A. P. Bird, and T. Kouzarides. 2003. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 278**:**4035-4040. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons, R. J., T. L. McDowell, S. Raman, D. M. O'Rourke, D. Garrick, H. Ayyub, and D. R. Higgs. 2000. Mutations in ATRX, encoding a SWI/SNF-like protein, cause diverse changes in the pattern of DNA methylation. Nat. Genet. 24**:**368-371. [DOI] [PubMed] [Google Scholar]
- 17.Hammond, S. L., R. G. Ham, and M. R. Stampfer. 1984. Serum-free growth of human mammary epithelial cells: rapid clonal growth in defined medium and extended serial passage with pituitary extract. Proc. Natl. Acad. Sci. USA 81**:**5435-5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holler, M., G. Westin, J. Jiricny, and W. Schaffner. 1988. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 2**:**1127-1135. [DOI] [PubMed] [Google Scholar]
- 19.Huschtscha, L. I., J. R. Noble, A. A. Neumann, E. L. Moy, P. Barry, J. R. Melki, S. J. Clark, and R. R. Reddel. 1998. Loss of p16INK4 expression by methylation is associated with lifespan extension of human mammary epithelial cells. Cancer Res. 58**:**3508-3512. [PubMed] [Google Scholar]
- 20.Issa, J. P. 1999. Aging, DNA methylation and cancer. Crit. Rev. Oncol. Hematol. 32**:**31-43. [DOI] [PubMed] [Google Scholar]
- 21.Jackson, J. P., A. M. Lindroth, X. Cao, and S. E. Jacobsen. 2002. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416**:**556-560. [DOI] [PubMed] [Google Scholar]
- 22.Jeddeloh, J. A., T. L. Stokes, and E. J. Richards. 1999. Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat. Genet. 22**:**94-97. [DOI] [PubMed] [Google Scholar]
- 23.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293**:**1074-1080. [DOI] [PubMed] [Google Scholar]
- 24.Jones, P. A., and S. B. Baylin. 2002. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3**:**415-428. [DOI] [PubMed] [Google Scholar]
- 25.Jones, P. L., G. J. Veenstra, P. A. Wade, D. Vermaak, S. U. Kass, N. Landsberger, J. Strouboulis, and A. P. Wolffe. 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 19**:**187-191. [DOI] [PubMed] [Google Scholar]
- 26.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410**:**116-120. [DOI] [PubMed] [Google Scholar]
- 27.Lehmann, U., F. Langer, H. Feist, S. Glockner, B. Hasemeier, and H. Kreipe. 2002. Quantitative assessment of promoter hypermethylation during breast cancer development. Am. J. Pathol. 160**:**605-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehnertz, B., Y. Ueda, A. A. Derijck, U. Braunschweig, L. Perez-Burgos, S. Kubicek, T. Chen, E. Li, T. Jenuwein, and A. H. Peters. 2003. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr. Biol. 13**:**1192-1200. [DOI] [PubMed] [Google Scholar]
- 29.Muller, H. M., A. Widschwendter, H. Fiegl, L. Ivarsson, G. Goebel, E. Perkmann, C. Marth, and M. Widschwendter. 2003. DNA methylation in serum of breast cancer patients: an independent prognostic marker. Cancer Res. 63**:**7641-7645. [PubMed] [Google Scholar]
- 30.Mutskov, V., and G. Felsenfeld. 2004. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 23**:**138-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292**:**110-113. [DOI] [PubMed] [Google Scholar]
- 32.Nan, X., H. H. Ng, C. A. Johnson, C. D. Laherty, B. M. Turner, R. N. Eisenman, and A. Bird. 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393**:**386-389. [DOI] [PubMed] [Google Scholar]
- 33.Ng, H. H., Y. Zhang, B. Hendrich, C. A. Johnson, B. M. Turner, H. Erdjument-Bromage, P. Tempst, D. Reinberg, and A. Bird. 1999. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 23**:**58-61. [DOI] [PubMed] [Google Scholar]
- 34.Pieper, R. O., S. Patel, S. A. Ting, B. W. Futscher, and J. F. Costello. 1996. Methylation of CpG island transcription factor binding sites is unnecessary for aberrant silencing of the human MGMT gene. J. Biol. Chem. 271**:**13916-13924. [DOI] [PubMed] [Google Scholar]
- 35.Robertson, K. D., S. Ait-Si-Ali, T. Yokochi, P. A. Wade, P. L. Jones, and A. P. Wolffe. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25**:**338-342. [DOI] [PubMed] [Google Scholar]
- 36.Romanov, S. R., B. K. Kozakiewicz, C. R. Holst, M. R. Stampfer, L. M. Haupt, and T. D. Tlsty. 2001. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature 409**:**633-637. [DOI] [PubMed] [Google Scholar]
- 37.Rountree, M. R., K. E. Bachman, and S. B. Baylin. 2000. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 25**:**269-277. [DOI] [PubMed] [Google Scholar]
- 38.Shivakumar, L., J. Minna, T. Sakamaki, R. Pestell, and M. A. White. 2002. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol. Cell. Biol. 22**:**4309-4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song, J. Z., C. Stirzaker, J. Harrison, J. R. Melki, and S. J. Clark. 2002. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene 21**:**1048-1061. [DOI] [PubMed] [Google Scholar]
- 40.Song, M. S., S. J. Song, N. G. Ayad, J. S. Chang, J. H. Lee, H. K. Hong, H. Lee, N. Choi, J. Kim, H. Kim, J. W. Kim, E. J. Choi, M. W. Kirschner, and D. S. Lim. 2004. The tumour suppressor RASSF1A regulates mitosis by inhibiting the APC-Cdc20 complex. Nat. Cell Biol. 6**:**129-137. [DOI] [PubMed] [Google Scholar]
- 41.Stampfer, M. R. 1985. Isolation and growth of human mammary epithelial cells. J. Tissue Cult. Methods 9**:**107-116. [Google Scholar]
- 42.Stampfer, M. R., and P. Yaswen. 2003. Human epithelial cell immortalization as a step in carcinogenesis. Cancer Lett. 194**:**199-208. [DOI] [PubMed] [Google Scholar]
- 43.Stirzaker, C., J. Z. Song, B. Davidson, and S. J. Clark. 2004. Transcriptional gene silencing promotes DNA hypermethylation through a sequential change in chromatin modifications in cancer cells. Cancer Res. 64**:**3871-3877. [DOI] [PubMed] [Google Scholar]
- 44.Tamaru, H., and E. U. Selker. 2001. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414**:**277-283. [DOI] [PubMed] [Google Scholar]
- 45.Tamaru, H., X. Zhang, D. McMillen, P. B. Singh, J. Nakayama, S. I. Grewal, C. D. Allis, X. Cheng, and E. U. Selker. 2003. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat. Genet. 34**:**75-79. [DOI] [PubMed] [Google Scholar]
- 46.Tommasi, S., and G. P. Pfeifer. 1995. In vivo structure of the human cdc2 promoter: release of a p130-E2F-4 complex from sequences immediately upstream of the transcription initiation site coincides with induction of cdc2 expression. Mol. Cell. Biol. 15**:**6901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turker, M. S. 2002. Gene silencing in mammalian cells and the spread of DNA methylation. Oncogene 21**:**5388-5393. [DOI] [PubMed] [Google Scholar]
- 48.Xiong, Z., and P. W. Laird. 1997. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 25**:**2532-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan, P. S., H. Shi, F. Rahmatpanah, T. H. Hsiau, A. H. Hsiau, Y. W. Leu, J. C. Liu, and T. H. Huang. 2003. Differential distribution of DNA methylation within the RASSF1A CpG island in breast cancer. Cancer Res. 63**:**6178-6186. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
[Supplemental material]