DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner (original) (raw)
Abstract
DNA methylation is a major epigenetic mechanism that has been suggested to control developmental gene regulation during embryogenesis, but its regulatory mechanisms remain unclear. In this report, we show that CpG islands associated with the X-linked homeobox gene cluster Rhox, which is highly expressed in the extraembryonic trophectoderm, are differentially methylated in a stage- and lineage-specific manner during the post-implantation development of mice. Inactivation of both Dnmt3a and Dnmt3b, DNA methyltransferases essential for the initiation of de novo DNA methylation, abolished the establishment of DNA methylation and the silencing of Rhox cluster genes in the embryo proper. The Dnmt3-dependent CpG-island methylation at the Rhox locus extended for a large genomic region (∼1 Mb) containing the Rhox cluster and surrounding genes. Complementation experiments using embryonic stem (ES) cells deficient in the DNA methyltransferases suggested that the CpG-island methylation by Dnmt3a and Dnmt3b was restricted within this large genomic region, and did not affect the neighboring genes outside it, implicating the existence of region-specific boundaries. Our results suggest that DNA methylation plays important roles in both long-range gene silencing and lineage-specific silencing in embryogenesis.
Keywords: DNA methylation, Dnmt3a, Dnmt3b, Rhox
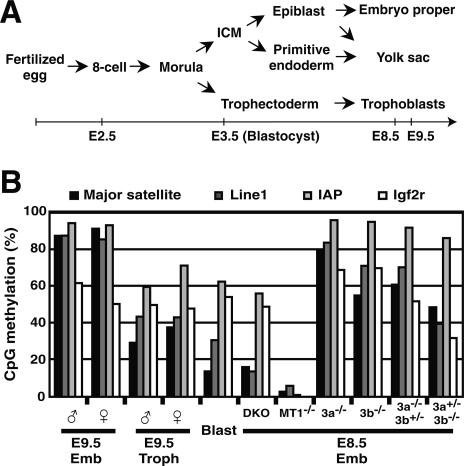
In development, epigenetic mechanisms function to maintain cellular phenotypes, in which differential gene expression is stably inherited in a clonal manner. The direct modification of DNA by methylation plays important roles in repressing gene expression and transmitting the silenced state to daughter cells (Bird 2002). DNA methylation marking can be interpreted in several ways, including the exclusion of transcriptional regulators at their DNA-binding sites, or by the attraction of transcriptional repressor complexes through a group of proteins with methyl-CpG-binding activities (Jaenisch and Bird 2003). DNA methylation profiles in mammals are reprogrammed and established during early embryogenesis through a highly orchestrated process in a cell lineage-dependent manner (Morgan et al. 2005). Following fertilization, a genome-wide and stepwise reduction of DNA methylation occurs during the cleavage cycles, causing the blastocyst to have the lowest level of genomic DNA methylation during embryogenesis. By the blastocyst stage, two rounds of cell differentiation give rise to three cell lineages: the inner cell mass (ICM)/epiblast, the trophectoderm, and the primitive endoderm (Fig. 1A). Shortly after implantation, the ICM/epiblast-lineage cells undergo a wave of de novo methylation that establishes hypermethylated genome profiles in the embryo proper, while de novo methylation occurs only weakly in the trophectoderm- and primitive endoderm-lineage cells, resulting in a relatively hypomethylated genome in the extraembryonic tissues. These dynamic changes in DNA methylation, along with a reorganization of histone modifications and chromatin regulators, are known as “epigenetic reprogramming,” which has been postulated to be required for the totipotency/pluripotency in normal embryogenesis and in somatic cell nuclear transfer cloning procedures, although its roles and mechanisms remain largely unknown (Morgan et al. 2005).
Figure 1.
(A) A summary of cell lineages in pre- and post-implantation mouse embryogenesis. (B) Dnmt3-deficient embryos retain the global DNA methylation states that existed at the blastocyst stage. DNA methylation states of the pericentromeric major satellite repeats (black), Line1 (dark gray), IAP element (light gray), and Igf2r (white) in the embryonic tissues of male and female wild-type embryos and male mutant embryos were analyzed by bisulfite sequencing. The graph indicates the percentage of total CpG sites that were methylated in the bisulfite-sequenced clones of each region. (Emb) Embryo proper; (Troph) trophoblast; (8-cell) eight-cell embryo; (Blast) blastocyst; (ICM) inner cell mass; (WT) wild type; (DKO) _Dnmt3a_−/− _Dnmt3b_−/− (double knockout); (MT1−/−) Dnmt1−/−; (3a−/−) _Dnmt3a_−/−; (3b−/−) _Dnmt3b_−/−; (3a−/−3b+/−) _Dnmt3a_−/− Dnmt3b+/−; (3a+/−3b−/−) Dnmt3a+/− _Dnmt3b_−/−. The DNA methylation patterns of bisulfite-sequenced clones for Igf2r (DMR2) are shown in Supplementary Figure 1A.
In mammals, three CpG DNA methyltransferases, Dnmt1, Dnmt3a, and Dnmt3b, coordinately regulate the DNA methylation of the genome (Chen and Li 2004). Dnmt1 associates with the replication foci and propagates DNA methylation profiles after DNA replication, probably by the preferential methylation of hemimethylated DNA (Bestor 1992; Leonhardt et al. 1992). The inactivation of mouse Dnmt1 leads to an extensive decrease in DNA methylation of the whole genome indiscriminately, in agreement with its major role in maintenance methylation (Li et al. 1992; Lei et al. 1996). A Dnmt1 deficiency causes embryonic lethality in mice, frogs, and zebrafish, indicating that the proper level of DNA methylation is essential for vertebrate development (Li et al. 1992; Lei et al. 1996; Stancheva and Meehan 2000; Rai et al. 2006). Dnmt3a and Dnmt3b are developmentally regulated enzymes that are required for the initiation of de novo methylation in mouse embryonic stem (ES) cells, for the establishment of the embryonic hypermethylated genome in post-implantation development, and for the establishment of DNA methylation imprints in the imprinted genes of male and female germ cells (Okano et al. 1999; Kaneda et al. 2004). These functions indicate that Dnmt3a and Dnmt3b are physiological determinants for the DNA methylation profiles and dynamics in mammalian development. _Dnmt3a_−/− _Dnmt3b_−/− embryos die shortly after the gastrulation stage, suggesting the importance of DNA methylation reprogramming (Okano et al. 1999), but its role in developmental gene regulation during embryogenesis remains largely unclear.
Programmed changes in higher-order chromatin structure play important roles in the coordinated gene expressions in development (Sproul et al. 2005). Long-range gene regulatory mechanisms have been shown to control the spatially and temporally coordinated expression or allele-specific expression in many gene clusters, including the globin loci, imprinted gene clusters, and Hox gene clusters (Verona et al. 2003; Sproul et al. 2005; West and Fraser 2005). Recently, a new homeobox gene cluster on the mouse X chromosome has been identified, the Rhox gene cluster, which shows temporal-collinear expression in the reproductive and reproduction-associated tissues (Maclean et al. 2005). In this study, we show that DNA methylation regulates cell lineage-specific silencing of the Rhox gene cluster in the post-implantation development of mice. The CpG islands associated with genes in a large genomic region (∼1 Mb) that contains the Rhox gene cluster and its surrounding genes were heavily methylated in the silenced embryonic tissue. Genetic analyses using knockout mice revealed that Dnmt3a and Dnmt3b were required for the establishment of the lineage-specific methylation and for long-range gene silencing in the Rhox region. Rescue experiments in which Dnmt1, Dnmt3a, or Dnmt3b was expressed in their mutant ES cells suggested that CpG-island hypermethylation was confined to the Rhox region and did not occur in the neighboring genomic regions, indicating the possibility of a specific boundary at the ends of this region.
Results
Global DNA methylation in the Dnmt3a−/− Dnmt3b−/− embryos
To elucidate the physiological regulation of DNA methylation, we examined the DNA methylation profiles in embryos deficient for Dnmt3a and Dnmt3b. In our previous study, we performed a conventional methylation analysis on these mutants by Southern hybridization, which did not, however, provide sufficient quantitative results (Okano et al. 1999). Here, for a more quantitative analysis, we performed bisulfite sequencing and examined the DNA methylation status of tandem repeats (pericentromeric major satellites), interspersed retrotransposon repeats (LINE1 and IAP), and an imprinted gene (Igf2r) in wild-type and various DNA methyltransferase-deficient embryos. In normal embryogenesis, the methylated DNA of repetitive sequences is subject to demethylation during the preimplantation period; consequently, the blastocyst genome has the lowest level of methylation (Fig. 1B, Blast; Morgan et al. 2005). Subsequently, extensive de novo methylation after implantation establishes a hypermethylation of these repetitive sequences in the embryo proper (Fig. 1B, Emb; Morgan et al. 2005). In contrast, the DNA methylation levels and allele-specific profiles of Igf2r remain unchanged during pre- and post-implantation development (Fig. 1B; Supplementary Fig. 1A; Stöger et al. 1993). We found that the DNA methylation levels of the repetitive sequences in the embryo proper of Dnmt3a_−/− Dnmt3b_−/− double-knockout (DKO) embryos were much lower than in wild-type embryos at the same stage, but were the same as those of the wild-type blastocyst (Fig. 1B, DKO). This indicates that DNA methylation of the repetitive sequences in the DKO embryo did not increase after implantation, but they maintained their methylation profiles from the blastocyst stage. The DNA methylation levels and allele-specific patterns of Igf2r were unaffected in the DKO embryo proper (Fig. 1B; Supplementary Fig. 1A). In contrast, most of the DNA methylation in the repetitive sequences and Igf2r was lost in the _Dnmt1_−/− embryos (Fig. 1B, MT1−/−; Supplementary Fig. 1A), in agreement with this enzyme’s physiological function in maintenance methylation. Consistent with our previous study (Okano et al. 1999), these results support the idea that Dnmt3a and Dnmt3b function in introducing new DNA methylation in the genome (de novo methylation), but contribute less to maintaining pre-existing DNA methylation during embryogenesis.
Differential DNA methylation of Rhox6 and Rhox9 occurs in a lineage-dependent manner
To elucidate the function of de novo methylation in mammalian development, we searched for target genes developmentally regulated by Dnmt3a and Dnmt3b. Suppressive subtractive PCR screening resulted in the isolation of partial cDNA fragments of Rhox6 (also known as Psx1), an X-linked homeobox gene highly expressed in the trophectodermal tissues in post-implantation development (Chun et al. 1999). The Rhox6 gene is transcriptionally repressed during post-implantation development in the ICM/epiblast lineage, which gives rise to the embryo proper.
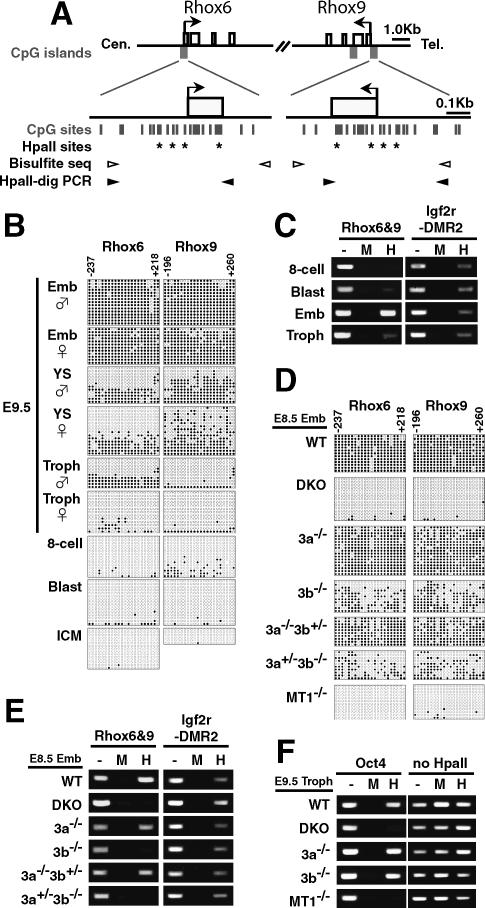
We first examined the DNA methylation states of the CpG islands of the Rhox6 gene in different embryonic tissues by bisulfite sequencing. Due to the extensive sequence similarity between Rhox6 and Rhox9 (also known as Psx2), our PCR primers for bisulfite sequencing amplified the CpG islands of both genes (Fig. 2A). This analysis revealed that the CpG islands of both Rhox6 and Rhox9 were highly methylated in the embryonic day 9.5 (E9.5) embryo proper (Fig. 2B; Supplementary Fig. 1C,D, Emb), in which Rhox6 and Rhox9 are silenced, whereas the CpG islands in the same genes were largely hypomethylated in the trophectoderm tissues (Fig. 2B; Supplementary Fig. 1D, Troph), in which these genes are highly expressed (Fig. 3A; Chun et al. 1999). The yolk sac, which consists of the extraembryonic mesoderm and extraembryonic visceral endoderm, showed a mixed DNA methylation profile of hypomethylated and hypermethylated sequences (Fig. 2B). These results suggest that Rhox6 and Rhox9 are differentially methylated in a cell lineage-dependent manner.
Figure 2.
Stage- and lineage-specific DNA methylation of Rhox6 and Rhox9 by Dnmt3a and Dnmt3b. (A) Schematic diagrams of the regions around the Rhox6 and Rhox9 transcription start sites.(Top) All the exons (open boxes) of Rhox6 and Rhox9 are represented. Regions around each transcription start site (arrows) meet the criteria of a CpG island (filled gray boxes). Expanded depictions of the regions around the promoter CpG islands are shown below, indicating the CpG sites (vertical bars) and HpaII sites (asterisks). PCR primers for bisulfite sequencing (white arrowheads) covered 22 CpG sites (from −237 to +218 for the Rhox6 CpG island, and from −196 to +260 for the Rhox9 CpG island). The PCR primers for HpaII-digestion PCR (black arrowheads) covered four HpaII sites. The numbering begins with the transcription start site as +1. (B–E) DNA methylation analysis of Rhox6 and Rhox9 in the E9.5 wild-type conceptus and blastocyst (B,C) and in the E8.5 wild-type and mutant embryo proper (D,E), by bisulfite sequencing (B,D) or by HpaII-digestion PCR (C,E). (B,D) DNA methylation patterns obtained by bisulfite sequencing are shown with filled (methylated) and open (unmethylated) circles. (YS) Yolk sac. A different set of PCR primers yielded similar results (Supplementary Fig. 1D). (C,E) DNA methylation states of Rhox6 and Rhox9 assessed by HpaII-digestion PCR. Genomic DNA digested with the CpG-methylation-sensitive restriction enzyme HpaII were amplified by PCR using primers flanking the HpaII sites. (Lane —) Undigested DNA. (Lane M) CpG-methylation-insensitive MspI-digested DNA. (Lane H) HpaII-digested DNA. _Igf2r_-DMR2 served as a control for a methylated locus (Stöger et al. 1993). (F) DNA methylation analysis of the promoter region of Oct4 in trophoblast cells by HpaII-digestion PCR (Gidekel and Bergman 2002). An arbitrarily selected genomic region lacking an HpaII site on mouse chromosome 10 (no HpaII) served as a loading control.
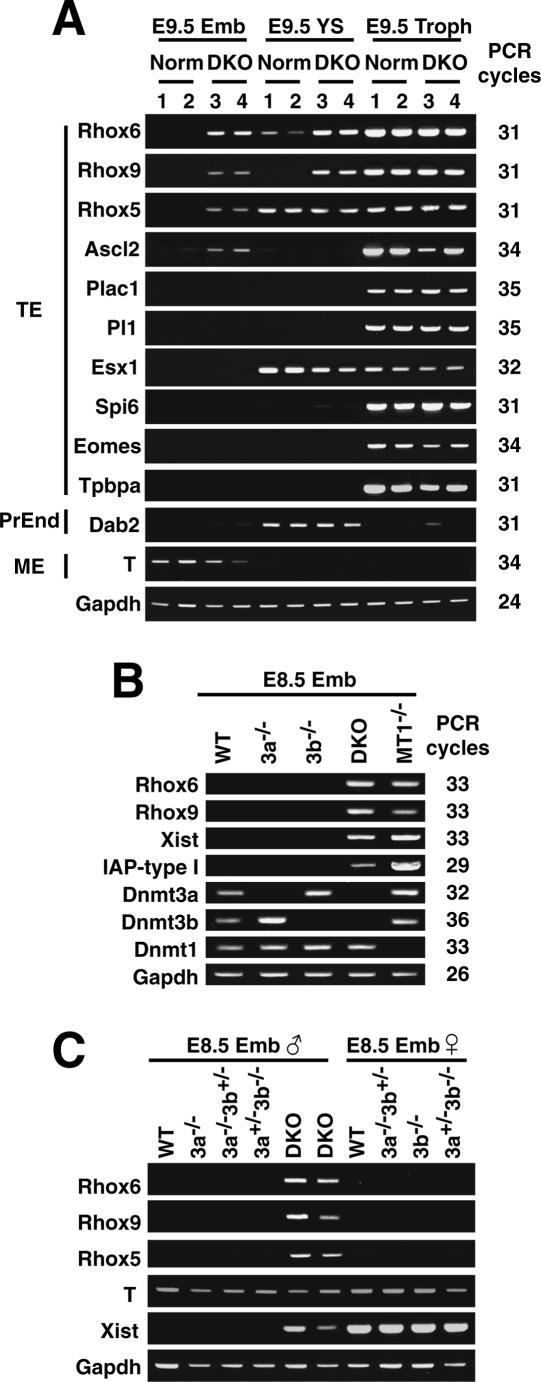
Figure 3.
Transcriptional derepression of Rhox6 and Rhox9 in DKO embryos. (A) Expression profile of some lineage markers in DKO embryos by RT–PCR. Total RNA was isolated from the E9.5 embryo proper (Emb), yolk sac (YS), and trophoblast (Troph). (Lane Norm.1) A Dnmt3a+/− embryo. (Lane Norm.2) A Dnmt3b+/− embryo. (Lanes DKO 3 and 4) Two independent _Dnmt3a_−/− _Dnmt3b_−/− embryos. (TE) Trophectoderm; (PrEnd) primitive endoderm; (ME) mesoderm. Gapdh was used as a control for equal RNA loading. PCR cycles were shown on the right. (B) Expression profiles obtained by RT–PCR in different DNA methyltransferase knockout embryos. Total RNA was isolated from the embryo proper of an E8.5 male. Xist and IAP-type I are known to be derepressed in DKO embryos (male) and _Dnmt1_−/− embryos, respectively (Walsh et al. 1998; Sado et al. 2004). (C) The expression profiles in female and male embryos by RT–PCR. Total RNA was isolated from E8.5 male (left) and female (right) embryos.
X inactivation causes the methylation of CpG islands on the inactivated alleles of X-linked genes in the female (Lock et al. 1987). Using F1 hybrid embryos for Mus musculus domesticus (C57BL/6) and Mus musculus molossinus (JF1), we found that Rhox6 properly underwent imprinted X inactivation in extraembryonic tissues (Supplementary Fig. 1E; Takagi and Sasaki 1975). However, we also found that the DNA methylation patterns of Rhox6 and Rhox9 were similar between males and females in the embryonic and extraembryonic tissues (Fig. 2B), and that these genes were similarly silenced in both the male and female embryo proper (Fig. 3C). These findings indicate that the DNA methylation states of Rhox6 and Rhox9 are independent of X-chromosome inactivation.
We further found by bisulfite sequencing that the CpG islands of Rhox6 and Rhox9 were hypomethylated in the eight-cell embryo, the blastocyst, and the ICM isolated from the blastocyst (Fig. 2B; Supplementary Fig. 1D,F). We confirmed this hypomethylation of Rhox6 and Rhox9 in the eight-cell embryo and the blastocyst, as well as the differential methylation in the embryo proper and the trophectoderm, using a method based on a different principle, HpaII-digestion PCR analysis (Fig. 2C). These results indicate that establishment of the embryonic lineage-specific CpG methylation occurs after implantation of the blastocyst.
Requirement of Dnmt3a and Dnmt3b for the lineage-specific DNA methylation of Rhox6 and Rhox9
We next examined whether Dnmt3a and Dnmt3b are responsible for establishment of the lineage-specific DNA methylation of Rhox6 and Rhox9 during embryogenesis. The DNA methylation of the CpG islands of Rhox6 and Rhox9 in the embryo proper was almost completely lost in the DKO embryos, as was the case in the _Dnmt1_−/− embryos (Fig. 2D; Supplementary Fig. 1D). However, the DNA methylation profiles in the same regions were unaffected in the _Dnmt3a_−/− embryo proper (92% and 86% of the total CpG sites for Rhox6 and Rhox9, respectively), whereas those regions were slightly decreased in the _Dnmt3b_−/− embryo proper (44% and 53% of the total CpG sites). These results suggest that, although the contribution of Dnmt3b is greater, Dnmt3a and Dnmt3b are required for establishing the lineage-specific CpG-island methylation of Rhox6 and Rhox9, and their functions in this process overlap. In addition, the DNA methylation states of Rhox6 and Rhox9 in the _Dnmt3a_−/− Dnmt3b+/− and Dnmt3a+/− _Dnmt3b_−/− embryo proper were gene dosage-dependent, further supporting the close genetic interaction between these enzymes (Fig. 2D). We obtained consistent results using HpaII-digestion PCR analysis, which showed the CpG sites to be hypomethylated in the DKO embryo proper (Fig. 2E). We did not observe clear signals for the _Dnmt3b_−/− or Dnmt3a+/− _Dnmt3b_−/− embryos by HpaII-digestion PCR analysis (Fig. 2E), although we detected a significant amount of DNA methylation in these embryos by bisulfite sequencing (Fig. 2D). We speculate that the DNA methylation contents are underrepresented in our HpaII-digestion PCR analysis, which only detects fully methylated molecules at several HpaII sites (Fig. 2A), causing the apparent discrepancy.
We next asked whether Dnmt3a and Dnmt3b play a role in DNA methylation regulation in the trophectoderm lineage. The promoter region of Oct4 is methylated in the placenta (Hattori et al. 2004a). DKO trophoblast cells lost the methylation of Oct4 similar to _Dnmt1_−/− trophoblast cells, but neither _Dnmt3a_−/− nor _Dnmt3b_−/− trophoblast cells did (Fig. 2F). This suggests that Dnmt3a and Dnmt3b play some role in establishing DNA methylation in the trophoblast cells, even though the global methylation content in the trophoblast cells is much lower than in the embryo proper.
Derepression of silenced Rhox6 and Rhox9 in the DKO embryo proper
We next used RT–PCR to assess whether the loss of DNA methylation affected the tissue-specific transcription of Rhox6 and Rhox9. In wild-type embryos, the Rhox6 and Rhox9 transcripts are highly expressed in the trophectoderm tissues and are repressed in the embryo proper (Fig. 3A, Norm/Emb and Norm/Troph; Chun et al. 1999). We found that the silencing of Rhox6 and Rhox9 in the embryo proper was derepressed in the DKO (Fig. 3A, DKO/Emb), but not in the Dnmt3a_−/−, Dnmt3b_−/−, Dnmt3a_−/− Dnmt3b+/,_ or Dnmt3a+/− _Dnmt3b_−/− animals (Fig. 3B,C). We did not see any further enhancement of Rhox6 and Rhox9 expression in the DKO trophectoderm (Fig. 3A, DKO/Troph). These results suggest that DNA methylation by Dnmt3a and Dnmt3b plays a major role in the transcriptional repression of Rhox6 and Rhox9 in nonexpressing tissue. As expected, we found a similar derepression of Rhox6 and Rhox9 transcripts in _Dnmt1_−/− embryos (Fig. 3B), further supporting a causal role for DNA methylation in the transcriptional repression of Rhox6 and Rhox9.
We found that the trophectoderm markers Rhox5 (also known as Pem) and Ascl2 (also known as Mash2) were also derepressed in the DKO embryo proper, whereas other trophectoderm markers/regulators—Plac1, Pl1, Esx1, Spi6, Eomes, and _Tpbpa_—remained silenced (Fig. 3A), indicating that some, but not all, trophectoderm-specific genes are regulated by Dnmt3a and Dnmt3b. We also found comparable levels of expression of the parietal endoderm marker gene Disabled homolog 2 (Dab2) and the mesoderm marker gene Brachyury T between the DKO and wild-type embryonic tissues (Fig. 3A). In addition, we examined the transcripts of the retrotransposable element IAP, which is normally silenced but is highly activated in Dnmt1-deficient embryos (Walsh et al. 1998). We found that, although it was activated significantly, the expression of IAP transcripts in the DKO embryos was much lower than in _Dnmt1_−/− embryos (Fig. 3B), indicating that the Dnmt3 enzymes may not contribute as greatly as does Dnmt1 in the maintenance of IAP retrotransposon silencing, consistent with the DNA methylation states of IAP in their mutant embryos (Fig. 1B).
Long-range gene silencing of the Rhox gene cluster by Dnmt3a and Dnmt3b
Several homeobox genes, including Rhox5, Rhox6, and Rhox9, are densely clustered in the Reproductive homeobox (Rhox) region in mouse chromosome X A3.1 (Maclean et al. 2005; Morris et al. 2006). These Rhox cluster genes show collinear expression in the postnatal testis, implicating the existence of coordinated long-range gene regulation (Maclean et al. 2005). To test whether Dnmt3a and Dnmt3b regulate a large genomic region, we used RT–PCR to examine the expression of genes in the genomic region surrounding the Rhox cluster in embryonic tissues. We found that, whereas most genes examined in this locus were expressed normally in the trophoblast, some of them, including the Rhox cluster genes, were transcriptionally repressed in the embryo proper (Fig. 4A,B, Norm/Emb, Norm/Troph). Interestingly, we found that a cluster of genes including the Rhox cluster and neighboring genes (Gm9, Rhox1, Rhox2, Ehox/Rhox4-like, Rhox3-like, Rhox5, Rhox6, Rhox9) in a 0.7-Mb region were derepressed in the DKO embryo proper (Fig. 4A, DKO/Emb), indicating that a large genomic region at the Rhox cluster was regulated by Dnmt3a and Dnmt3b.
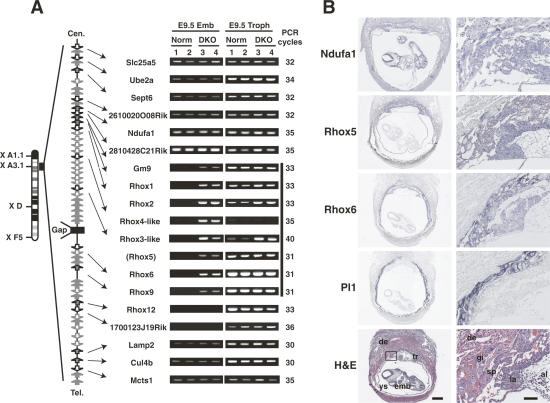
Figure 4.
Long-range gene silencing of the Rhox gene cluster by Dnmt3a and Dnmt3b. (A) RT–PCR analysis of genes within or surrounding the Rhox cluster in mouse chromosome X A3.1 in the E9.5 conceptus of wild-type and DKO embryos. The same total RNA samples as in Figure 3A were used for this analysis. A schematic drawing of the genomic organization of this region is shown on the left (based on NCBI Build 34). Box arrows represent genes and their transcriptional directions. There is a gap of assembled mouse genome sequences within the Rhox cluster (Gap, closed box). The location of Rhox5 was mapped between Rhox3-like and Rhox6 by referring to NCBI Build 32. The vertical bold bar on the right of the RT–PCR results represents a silenced 0.7-Mb region that is transcriptionally activated in the DKO embryo proper. The RT–PCR results of Rhox6 and Rhox9 are the same data as shown in Figure 3A. Rhox7, Rhox8, Rhox10, Rhox11, and Atp1b4 were undetectable in the examined samples. (B) Section in situ hybridization of the wild-type E9.5 conceptus. Low-magnification images (left; bar is 500 μm) and high-magnification images (right; bar is 100 μm) corresponding to a region marked with a square in the low-magnification Hematoxylin and eosin (H&E) image were shown. Rhox5 and Rhox6 expressed in three layers of trophectoderm (tr): trophoblast giant cells (gi), spongiotrophoblast (sp), and labyrinthine trophoblast (la), but not in maternal decidua (de) or embryo proper (emb). Ndufa1 expressed both in trophectoderm and embryo proper. Pl1 is a marker for trophoblast giant cells. (H&E) H&E staining; (ys) yolk sac; (al) allantoic region.
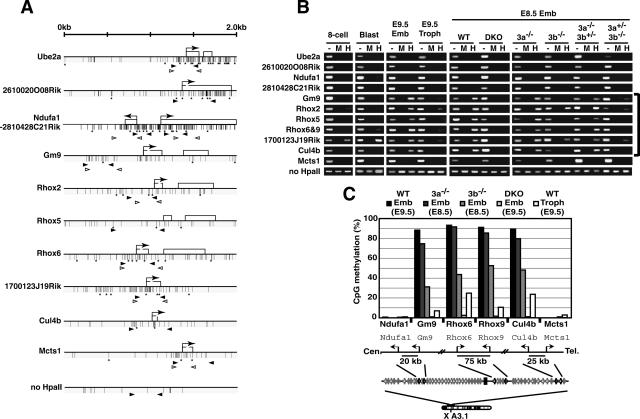
We then examined DNA methylation states in the CpG islands associated with the genes around the Rhox region by HpaII-digestion PCR (Fig. 5A,B) and bisulfite sequencing (Fig. 5C; Supplementary Fig. 2). We found that the CpG islands of the genes within the 0.7-Mb Rhox region, whose silencing was derepressed in the DKO embryos (Fig. 4A), were heavily methylated in the embryo proper, whereas they were hypomethylated in the trophectoderm (Fig. 5B,C). These CpG islands within the 0.7-Mb region were hypomethylated in the eight-cell embryo and the blastocyst (Fig. 5B, eight-cell, Blast), indicating that the establishment of lineage-specific methylation in the 0.7-Mb region occurs after implantation. These lineage-specific DNA methylations of the CpG islands were abolished in the DKO embryo proper, but not in _Dnmt3a_−/− or _Dnmt3b_−/− embryos, although the contribution of Dnmt3b was slightly greater (Fig. 5B,C; Supplementary Fig. 2). Similar tendencies to lineage-specific methylation were also found in several CpG sites other than the promoter CpG islands in the regions around Gm9 and Rhox6 (Supplementary Fig. 3). These results indicate an essential but partially redundant function of Dnmt3a and Dnmt3b in the establishment of lineage-specific DNA methylation of the genes in the 0.7-Mb region.
Figure 5.
Dnmt3-dependent CpG-island methylation of genes within a large genomic region around the Rhox cluster. (A) Schematic maps of the genes used for DNA methylation analysis on mouse chromosome X A3.1. Each map represents a 2-kb genomic region around the transcription start site (arrow) with CpG islands except for Rhox5, in which a genomic region around exon 2 (a transcription start site in the epididymis) is shown. Exons are represented as open boxes. Vertical bars and asterisks indicate CpG and HpaII sites, respectively. Black and white arrowheads represent the primers for HpaII-digestion PCR or for bisulfite sequencing, respectively. (B) DNA methylation profiles of the genomic region around the Rhox cluster obtained by HpaII-digestion PCR. Genomic DNA from the blastocyst, E8.5 or E9.5 embryo proper, or trophoblast was digested with HpaII and was subjected to PCR. (Lane —) Undigested DNA. (Lane M) MspI-digested DNA. (Lane H) HpaII-digested DNA. A region lacking an HpaII site on chromosome 10 (no HpaII) served as a loading control. (8-cell) Eight-cell embryos; (Blast) blastocyst; (Emb) embryo proper; (Troph) trophoblast; (WT) wild type; (DKO) _Dnmt3a_−/− _Dnmt3b_−/−; (3a−/−) _Dnmt3a_−/−; (3b−/−) _Dnmt3b_−/−; (3a−/−3b+/−) _Dnmt3a_−/− Dnmt3b+/−; (3a+/−3b−/−) Dnmt3a+/− _Dnmt3b_−/−. (C) DNA methylation states of Ndufa1, Gm9, Rhox6, Rhox9, Cul4b, and Mcts1 in the embryo proper of wild-type and Dnmt3-mutant embryos and wild-type trophoblast by bisulfite sequencing. The graph indicates the percentage of total CpG sites that were methylated in the bisulfite-sequenced clones of each analyzed region. The results for Rhox6 and Rhox9 are based on the data shown in Figure 2. A schematic drawing of the regions around these six genes is shown below, indicating the locations and selected distances between their transcriptional start sites. Additional and detailed results are provided in Supplementary Figure 2.
We also found that the CpG islands of two genes located proximal to the 0.7-Mb Rhox region (1700123J19Rik and Cul4b, within 300 kb from Rhox9) were heavily methylated in the embryo proper. Cul4b showed the same pattern of differential methylation in a stage-, lineage-, and Dnmt3-dependent manner as that of the genes within the 0.7-Mb region (Fig. 5B,C; Supplementary Fig. 2). Interestingly, 1700123J19Rik retained a significant amount of DNA methylation in the eight-cell embryo, the blastocyst, and trophoblast, whereas its DNA methylation level in the DKO embryo proper was significantly decreased compared with that of the wild-type embryo proper (Fig. 5B; Supplementary Fig. 2). Collectively, these results suggest that the Dnmt3-dependent regulation of CpG-island methylation extends along a 1-Mb genomic region (the 0.7-Mb Rhox region plus a 0.3-Mb region including Cul4b and 1700123J19Rik). Notably, the CpG islands associated with genes outside the 1-Mb region (Ube2a, 2610020O08Rik, Ndufa1, 2810428C21Rik, and Mcts1) were hypomethylated in the tissues and stages that we examined (Fig. 5B,C; Supplementary Fig. 2). The CpG islands of the genes closest to the 1-Mb region, Ndufa1/2810428C21Rik and Mcts1, were located 20 kb and 25 kb away from the ends of the 1-Mb region (Gm9 and Cul4b), respectively (Fig. 5C). Taken together, our results suggest that Dnmt3a and Dnmt3b regulate the expression and CpG-island methylation of genes within a large genomic region around the Rhox cluster in a lineage-specific manner during embryogenesis.
Regulation of DNA methylation in the Rhox cluster in ES cells
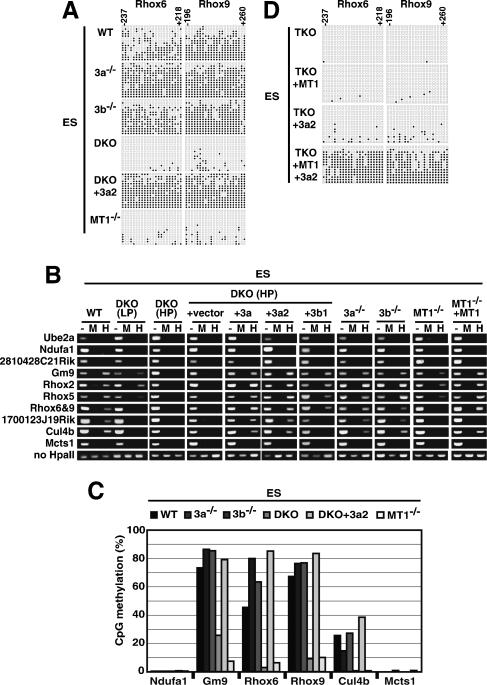
We next examined the regulation of DNA methylation in the Rhox region by Dnmt3a and Dnmt3b at the cellular level, using ES cells as a model system for the ICM/epiblast lineage. We found that the CpG islands within Rhox6 and Rhox9 were highly or moderately methylated in wild-type, _Dnmt3a_−/−, and _Dnmt3b_−/− ES cells, but largely unmethylated in DKO ES cells (under both low- and high-passage-number conditions) (Fig. 6A; Supplementary Fig. 4A). The loss of DNA methylation in the DKO ES cells was restored by the expression of exogenous Dnmt3a2 protein, indicating that DNA methylation of the CpG islands in Rhox6 and Rhox9 is regulated by the Dnmt3 enzyme in ES cells. A similar extensive loss of the DNA methylation of Rhox6 and Rhox9 was seen in _Dnmt1_−/− ES cells.
Figure 6.
Dnmt3-dependent DNA methylation of the Rhox cluster genes in ES cells. Genomic DNA from wild-type (WT), _Dnmt3a_−/− (3a−/−), _Dnmt3b_−/− (3b−/−), _Dnmt3a_−/− _Dnmt3b_−/− (DKO), _Dnmt1_−/− (MT1−/−), and _Dnmt1_−/− _Dnmt3a_−/− _Dnmt3b_−/− (TKO) ES cells, and from DKO and TKO ES cells expressing the Dnmt1 (MT1), Dnmt3a (3a), Dnmt3a2 (3a2), or Dnmt3b1 (3b1) proteins were used for DNA methylation analysis. (A) DNA methylation patterns in Rhox6 and Rhox9 obtained by bisulfite sequencing are shown with filled (methylated) and open (unmethylated) circles. (B) DNA methylation profiles of the Rhox cluster genes in the ES cells obtained by HpaII-digestion PCR. (Lane —) Undigested DNA. (Lane M) MspI-digested DNA. (Lane H) HpaII-digested DNA. (C) DNA methylation states of Ndufa1, Gm9, Rhox6, Rhox9, Cul4b, and Mcts1 in ES cells. The graph indicates the percentage of total CpG sites that were methylated in bisulfite-sequenced clones of each analyzed region. Additional and detailed results are provided in Supplementary Figure 4. (D) DNA methylation patterns of Rhox6 and Rhox9 obtained by bisulfite sequencing in TKO ES cells expressing Dnmt1, Dnmt3a2, or both proteins.
To test whether the Dnmt3 enzymes regulate the large genomic region at the Rhox cluster in ES cells, we examined the DNA methylation states of ES cells with different DNA methyltransferase knockout backgrounds by HpaII-digestion PCR and bisulfite sequencing analyses (Fig. 6B,C). In wild-type ES cells, the CpG islands of the genes in the 1-Mb region (Gm9, Rhox2, Rhox5, Rhox6, Rhox9, 1700123J19Rik, and Cul4b) were highly methylated, whereas those of the genes located outside this region (Ube2a, Ndufa1, 2810428C21Rik, and Mcts1) were hypomethylated (Fig. 6B,C), in agreement with the DNA methylation profiles in the wild-type embryo proper (Fig. 5B,C). Although the DNA methylation states of Gm9 and Rhox2 were not changed in the low-passage DKO ES cells [Fig. 6B, DKO(LP)], the DNA methylation of Gm9, Rhox2, Rhox5, Rhox6, Rhox9, 1700123J19Rik, and Cul4b was decreased in the high-passage DKO ES cells [Fig. 6B, DKO(HP)]. The loss of the DNA methylation in the CpG islands of these genes was restored by the expression of either one of the Dnmt3 enzymes in the DKO ES cells and the Dnmt1 enzyme in _Dnmt1_−/− ES cells, although the expression of Dnmt3b1 and Dnmt1 did not completely restore the methylation in the experimental conditions used in this study. Notably, the expression of Dnmt3a2 protein in the high-passage DKO ES cells did not increase the CpG-island methylation in genes located outside the 1-Mb region (Ube2a, Ndufa1, 2810428C21Rik, and Mcts1). This implies the existence of boundaries that restrict the regulation by the Dnmt3 enzyme at the edge of the 1-Mb region.
To elucidate the role of DNA methyltransferases in regulating the DNA methylation of Rhox6 and Rhox9 in ES cells, we examined whether Dnmt1 or Dnmt3a alone could establish and maintain the DNA methylation of these genes by using ES cells lacking all three functional DNA methyltransferases: Dnmt1, Dnmt3a, and Dnmt3b (TKO ES cells) (Tsumura et al. 2006). We found that neither Dnmt1 nor Dnmt3a alone could efficiently restore the methylation of Rhox6 and Rhox9 in the TKO ES cells (Fig. 6D). However, the coexpression of both Dnmt1 and Dnmt3a restored the methylation of the Rhox6 and Rhox9 genes in the TKO ES cells (Fig. 6D) This indicates that the coordinated regulation of both Dnmt1 and Dnmt3, possibly by a one-step or two-step mechanism, is required to complete the establishment and maintenance of the DNA methylation of Rhox6 and Rhox9.
Discussion
Tissue-specific gene expression is coordinately regulated by both positive transcriptional activation mechanisms in expressing tissues and negative repressive mechanisms in nonexpressing tissues (Chen et al. 1998). In this study, we show that the establishment of DNA methylation by Dnmt3a and Dnmt3b plays an essential role in the lineage-specific silencing of the Rhox cluster genes in the embryo proper during the post-implantation development of mice. This and previous studies (Stancheva and Meehan 2000; Takizawa et al. 2001; Futscher et al. 2002; Lunyak et al. 2002; Fan et al. 2005) support the idea that DNA methylation plays regulatory roles in developmental gene expression in vertebrate embryogenesis.
The inactivation of Dnmt1 in vivo causes the early onset of cell type-specific gene expression in the respective expressing cell types in mice and frogs (Stancheva and Meehan 2000; Fan et al. 2005) and organ-specific defects in zebrafish (Rai et al. 2006). This indicates a causal relationship of DNA methylation in developmental gene regulation. However, Dnmt1 inactivation also causes extensive loss of the bulk DNA methylation to a nonphysiological level; this loss results in global alterations in the whole transcription profile and the abnormal activation of a silenced retrotransposon and p53-dependent apoptosis in somatic cells (Walsh et al. 1998; Jackson-Grusby et al. 2001; Stancheva et al. 2001). These global abnormalities complicate the interpretation of the phenotype of Dnmt1-deficient animals. In contrast, Dnmt3a and Dnmt3b have been shown to have target preferences in the genome (Okano et al. 1999; Hattori et al. 2004b; Sado et al. 2004). In this study, we found that DNA methylation of the Rhox cluster genes was developmentally regulated in a lineage- and stage-specific manner, and that the inactivation of both Dnmt3a and Dnmt3b resulted in a specific loss of methylation in the Rhox cluster (Fig. 5B,C), whereas it caused only a mild decrease in the bulk DNA methylation, which remained at a physiological level (Fig. 1). Thus, our results support the idea that the Rhox cluster is physiologically regulated by DNA methylation. Previous studies showed that when ES cells in which both Dnmt3a and Dnmt3b were inactivated were subjected to long-term culture with high passage numbers, a global reduction of DNA methylation was seen (Chen et al. 2003), indicating roles for Dnmt3a and Dnmt3b in the stable inheritance of bulk DNA methylation. In the present study, however, this function of Dnmt3a and Dnmt3b probably contributed little to the regulation of DNA methylation in the embryos, as it was restricted to a very short period of embryogenesis.
It has been shown that the Dnmt1, Dnmt3a, and Dnmt3b proteins themselves have transcriptional repression activities that are independent of their enzymatic activities (Fuks et al. 2000, 2001; Robertson et al. 2000; Rountree et al. 2000; Bachman et al. 2001). Thus, it is possible that the nonenzymatic transcriptional repression activities of the Dnmt molecules were responsible for the silencing of the Rhox cluster genes. However, two different knockout mice (Dnmt1-KO mice and Dnmt3-DKO mice) showed the same results—that is, the loss of methylation and transcriptional derepression in the Rhox cluster genes in the embryo proper (Figs. 2D, Figs. 3B)—indicating that the DNA methylation markings, rather than the Dnmt proteins themselves, play the major role in the transcriptional repression of Rhox cluster genes. It is less likely, however, that DNA methylation is the only factor controlling Rhox gene expression. Unlike in embryos, loss of DNA methylation of Rhox6 and Rhox9 in DKO and _Dnmt1_−/− ES cells (Fig. 6A) did not cause derepression of Rhox6 and Rhox9 in undifferentiated cells, but it did after their differentiation (data not shown), indicating either that transcriptional activators of these genes may not function, or that other mechanisms may silence these genes in this cell type. These results indicate that multiple mechanisms, in addition to DNA methylation, may regulate the Rhox cluster, and that contribution of these mechanisms may vary in certain cell types or at different developmental stages.
It remains unclear how Dnmt3a and Dnmt3b control the differential methylation of the Rhox cluster in the ICM/epiblast and trophectoderm lineages. Epigenetic asymmetry between the ICM and trophectoderm lineages has been shown in the regulation of genomic imprinting and X inactivation (Sado et al. 2000; Lewis et al. 2004; Umlauf et al. 2004). De novo methylation of the bulk genome predominantly occurs in the ICM/epiblast lineage, resulting in a hypermethylated genome in the embryo proper, whereas the level of DNA methylation in the trophectoderm remains low (Morgan et al. 2005). An immunofluorescence study showed that the Dnmt3b protein is consistently highly expressed in the epiblast at E4.5, but is not detectable in the trophectoderm or primitive endoderm (Watanabe et al. 2002). These results raise the possibility that the Dnmt3 enzymes may not function in the trophectoderm lineage and the lineage-specific methylation in the ICM lineage may be the default state. However, we found that CpG methylation of the Oct4 promoter in trophectoderm tissues requires Dnmt3a and Dnmt3b (Fig. 2F), indicating that the targeting mechanism of Dnmt3a and Dnmt3b is dependent on both the genomic locus and cell type. We speculate that molecules that associate with Dnmt3a and Dnmt3b or specific chromatin structures such as histone modifications associated with the Rhox cluster may recruit Dnmt3a and Dnmt3b in the ICM/epiblast lineage or restrict their accessibility to the CpG islands of the Rhox cluster genes in the trophectoderm lineage.
In this study, we show that Dnmt3a and Dnmt3b control the methylation and silencing of a large genomic region (∼1 Mb) that includes the Rhox cluster and neighboring genes (Figs. 4, 5, 6). DNA methylation has been shown to be involved in the long-range gene regulation associated with genomic imprinting and X inactivation (Sado et al. 2000; Verona et al. 2003). It is possible that similar mechanisms are involved in regulation of the Rhox cluster. Indeed, Dnmt3a is required for the establishment of the DNA methylation markings of imprinted genes in both male and female germ cells (Kaneda et al. 2004), whereas Xist, a noncoding RNA molecule essential for X inactivation, is a physiological target of both Dnmt3a and Dnmt3b (Sado et al. 2004). On the other hand, the DNA methylation of Rhox cluster genes, which was lost by the inactivation of Dnmts, could be restored by the re-expression of DNA methyltransferases in ES cells (Fig. 6), whereas that of imprinted genes, once they are lost, cannot be restored, even in the presence of both Dnmt1 and Dnmt3 in ES cells (Tucker et al. 1996; Chen et al. 2003). Unlike X inactivation, which occurs only in the female, Rhox5, Rhox6, and Rhox9 are heavily methylated and silenced in the embryo proper at E9.5 regardless of the animal’s sex (Figs. 2B, Figs. 3C). Interestingly, a recent study showed that Rhox5 is predominantly expressed in female blastocysts and mainly from its paternal allele specifically at the preimplantation stage, but not in the post-implantation stage (Kobayashi et al. 2006). This gene may be differentially regulated in a stage-specific manner. These findings suggest that a novel mechanism may control the establishment of DNA methylation and silencing in the large genomic region of the Rhox cluster.
Several mechanisms of long-range gene regulation have been shown. A number of _cis_-regulatory elements have been identified that control the transcription of large genomic regions in various ways, such as by chromatin-loop formation or noncoding RNA-dependent mechanisms (Verona et al. 2003; West and Fraser 2005). Homology-dependent silencing mechanisms, often associated with tandem repeats, have been shown to cause transcriptional repression in various organisms (Dorer and Henikoff 1994; Garrick et al. 1998; Lippman and Martienssen 2004). The Rhox domain contains tandem-repetitive arrays, probably caused by genome duplication, at Rhox2/3/4 (Morris et al. 2006). This repetitive genomic structure or an unknown _cis_-regulatory element may induce DNA hypermethylation or heterochromatin formation and may spread CpG-island methylation in cis in the large Rhox region. We also located boundaries of the CpG-island methylation at the ends of the 1-Mb Rhox region in both the embryo proper and ES cells; a clear boundary at the centromeric end between Ndufa1 and Gm9, and a less clear boundary at the telomeric end between Cul4b and Mcts1 (Figs. 5B,C, Figs. 6B,C). These boundaries of CpG-island methylation were restored after exogenous DNA methyltransferases were introduced into their knockout ES cells (Fig. 6B,C). These results indicate that there may be a mechanism that determines the range of CpG-island methylation, such as barrier elements (West and Fraser 2005).
Although our results suggest that Rhox cluster genes overall were differentially methylated in a lineage-specific manner and were transcriptionally repressed in the embryo proper, there may be variations in the regulation of each gene in certain cell types or at different developmental stages. 1700123J19Rik, unlike the other genes examined in the Rhox region, retained a significant amount of DNA methylation in the eight-cell embryo, the blastocyst, and the trophoblast; thus, its methylation was less lineage-specific than that of the other genes (Fig. 5B; Supplementary Fig. 2). This methylation profile suggests that 1700123J19Rik, like IAP (Fig. 1; Lane et al. 2003), is resistant to the genome-wide reduction in DNA methylation that occurs during the cleavage period and is less affected by inactivation of the Dnmt3 enzymes. However, Dnmt3a and Dnmt3b were still required for the hypermethylation of 1700123J19Rik in the embryo proper (Fig. 5B; Supplementary Fig. 2). Our RT–PCR analysis indicated that Ehox/Rhox4 was transcriptionally repressed in a Dnmt3a- and Dnmt3b-dependent manner in the embryo proper (Fig. 3A). However, a more sensitive in situ hybridization study has shown that Ehox/Rhox4 is expressed in a part of the pharyngeal region at E9.5 (Morris et al. 2006). This indicates that DNA methylation may not contribute to transcriptional repression in these specific cell types, or alternatively that cell type-specific demethylation may be involved (Takizawa et al. 2001).
The aberrant hypermethylation of promoter CpG islands is a hallmark of human cancers, where it associates with the epigenetic silencing of tumor suppressor genes (Jones and Baylin 2002). Most of the promoter CpG islands in the mammalian genome remain unmethylated in normal somatic tissues regardless of their transcriptional status (Bird 2002). Inhibitory mechanisms preventing the methylation of CpG islands have been shown, but their molecular natures remain poorly understood (Brandeis et al. 1994; Macleod et al. 1994). A recent study showed that promoter CpG islands clustered in 4-Mb genomic regions are aberrantly hypermethylated in colorectal cancer, leading to the proposal that a long-range epigenetic silencing mechanism occurs in cancer (Frigola et al. 2006). In this study, promoter CpG islands clustered in the large Rhox region were hypermethylated in a cell type- and region-specific manner. It would be interesting to determine whether similar mechanisms are involved in these physiological and pathological processes.
Our results using ES cells deficient for three DNA methyltransferases suggest that the coordinated regulation of both the Dnmt1 and Dnmt3 enzymes is required for completion of the CpG-island hypermethylation of Rhox6 and Rhox9 (Fig. 6), possibly either through a direct interaction between two enzymes or through two subsequent but independent processes by Dnmt1 and Dnmt3 (Fatemi et al. 2002; Kim et al. 2002). We found that neither Dnmt1 nor Dnmt3a2 alone could complete this process in the cellular environment (Fig. 6), although we do not rule out the possibility that the transgene expression may not be fully functional as a result of heterologous promoter usage or clonal effects of the cell lines. Our results are consistent with a previous study in which the forced expression of Dnmt1 failed to restore the methylation of hypomethylated repetitive sequences in the absence of both Dnmt3a and Dnmt3b in mouse ES cells (Chen et al. 2003). However, it was recently reported that the overexpression of Dnmt1 induces the methylation of integrated mammalian CpG islands in insect cells (Jair et al. 2006). These results suggest that the process by which methylation is established may vary in different sequences and/or cell types.
The inactivation of both Dnmt3a and Dnmt3b results in embryonic lethality shortly after gastrulation, but the cause of lethality remains unknown. In this study, we showed that Rhox cluster genes, which encode paired-like homeobox transcription factors, were transcriptionally derepressed in the embryo proper of Dnmt3-DKO embryos (Fig. 3). The ectopic expression of Rhox cluster gene products may partially contribute to the developmental defects of the Dnmt3-DKO embryos (Fan et al. 1999). The developmental roles of the Rhox cluster genes and their functional overlap remain unknown (Maclean et al. 2005). It is also possible that the dysregulation of other genes that are important for mammalian development cause developmental abnormalities in the DKO embryos. Indeed, Ascl2, which encodes a trophectoderm-specific basic helix–loop–helix (bHLH) transcription factor, was derepressed in the embryo proper of Dnmt3-DKO animals (Fig. 3). The loss of DNA methylation in the IAP retrotransposon causes extensive activation of its transcription in Dnmt1-KO embryos, indicating that the lethality of Dnmt1-KO may be caused by the abnormal activation of silenced parasitic elements and secondary transcriptional dysregulation in its genome (Walsh et al. 1998). In this study, we showed that the DNA methylation of IAP in the DKO embryos was equivalent to the physiological level observed in the normal blastocyst (Fig. 1), and that the IAP transcription in DKO embryos was slightly increased but much lower than in the Dnmt1-KO embryos (Fig. 3B). These results suggest that IAP dysregulation may not impact development in the DKO embryos as greatly as in the Dnmt1-KO embryos. Further embryological studies, as well as genome-wide analyses of transcription and DNA methylation profiles, will be necessary to elucidate the roles of developmental DNA methylation control by Dnmt3a and Dnmt3b in embryogenesis.
In conclusion, our findings point to Dnmt3a and Dnmt3b as key regulators of developmental genes that are targets of dynamic changes in DNA methylation in early embryogenesis. Elucidation of the de novo methylation mechanism may provide insight not only into epigenetic reprogramming in embryogenesis but also into the abnormal methylation of the CpG islands in tumor suppressor genes in cancer.
Materials and methods
Mice
Dnmt1 (_c_-allele)-, Dnmt3a-, and Dnmt3b-mutant mice (Lei et al. 1996; Okano et al. 1999) were maintained in a 129SvJae-C57BL/6 hybrid background. The mutant embryos and their normal littermates at E8.5 and E9.5 were obtained from intercrosses of double heterozygotes via in vitro fertilization. The DKO embryos obtained by this procedure showed the same morphological phenotype as those produced by natural mating. The trophectoderm, yolk sac, and embryo proper were dissected under a microscope. Our RT–PCR analysis failed to detect several extraembryonic markers in the embryo proper (Fig. 3A), thus it was unlikely that extraembryonic tissues contaminated the embryo proper during dissection. The genotype and sex of post-implantation embryos were determined by PCR of genomic DNA from the yolk sac, using specific primers. If not specifically described, male embryos were used for the analyses of post-implantation embryos. Blastocysts were obtained from superovulated C57BL/6 females at E3.5 or by 24-h culture of eight-cell embryos (E2.5) in KSOM medium. We obtained the same DNA methylation results using blastocysts obtained from these two procedures. Eight-cell embryos for DNA methylation analysis were obtained by 2-d culture of two-cell embryos in KSOM medium. ICM was isolated by immunosurgery of blastocysts using anti-mouse red blood cell antibody (Inter-Cell Technologies) and guinea pig complement (Calbiochem) (Supplementary Fig. 1F). All animal experiments and maintenance were carried out according to the institutional guidelines for animal experimentation with approval from the Committee of the RIKEN Center for Developmental Biology.
ES cell culture
Wild-type J1 and Dnmt-mutant ES cells were maintained as described previously (Lei et al. 1996; Okano et al. 1999; Tsumura et al. 2006). The cells were grown on gelatin-coated culture dishes without feeder cells. Plasmid vectors expressing the Dnmt3a, Dnmt3a2, Dnmt3b1, or Dnmt1 proteins were generated by subcloning the corresponding cDNAs into pCAG-IRESzeo or pCAG-IRESblas, expressing vectors that contained CAG promoters (a synthetic promoter that includes the chicken-β-actin promoter and the human cytomegalovirus immediate early enhancer). These constructs were individually electroporated into _Dnmt3a_−/− _Dnmt3b_−/− ES cells (line 7aabb), _Dnmt1_−/− ES cells (line 36), or _Dnmt1_−/− Dnmt3a−/− _Dnmt3b_−/− ES cells (line 19), which were subsequently selected in zeocin- or blasticidin-containing medium for 9 d.
Total RNA isolation, RT–PCR analyses, and in situ hybridization
The total RNA of individual embryos was isolated using an RNA/DNA minikit (Qiagen), and that of ES cells was isolated by Trizol Reagent (Invitrogen), according to the manufacturer’s protocol. For RT–PCR, cDNA was synthesized from 1–5 μg of total RNA with random hexamers and SuperScript II reverse transcriptase (Invitrogen), and was subjected to PCR with primers for each gene. Primer sequences and PCR conditions are available from the authors on request. Section in situ hybridization was performed for 5-μm paraffin sections of E9.5 deciduas using FANTOM3 cDNA clones (Carninci et al. 2005) as probes.
Genomic DNA preparation and DNA methylation assay by bisulfite sequencing
The genomic DNA of embryos and ES cells was isolated using an RNA/DNA minikit (Qiagen) or by a standard protocol with Proteinase K treatment. Two-hundred or more blastocysts, 2600 eight-cell embryos, or 120 ICMs were pooled for the single genomic DNA preparations. A small number of morulas was included in the pools of eight-cell embryos. Sodium bisulfite treatment of genomic DNA digested with EcoRV or HindIII was performed as described previously (Clark et al. 1994) or using an EZ DNA methylation kit (Zymoresearch). PCR amplification of each region from the bisulfite-treated genomic DNA was carried out using specific primers. The primer sequences and PCR conditions are available from the authors on request.
DNA methylation analysis by HpaII-digestion PCR
Genomic DNA (50–200 ng) digested with a CpG methylation-sensitive restriction enzyme, HpaII, was subjected to PCR amplification using appropriate primers (primer sequences and PCR conditions are available on request). If the HpaII sites in the analyzed region were fully methylated, the expected fragments were amplified by PCR. As a control for complete digestion, genomic DNA was digested with a CpG-methylation-insensitive isoschizomer, MspI. Representative data from two independent sample preparations are shown for blastocysts and DKO embryos. PCR was performed more than three times for each sample with similar results.
Acknowledgments
We thank S. Takebayashi for cell lines and the genomic DNA of _Dnmt1_-knockout embryos, H. Niwa for plasmids, T. Chen for the Dnmt3a2 cDNA, C. Matsuoka for ICM and blastocyst preparation, Y. Hayashizaki for FANTOM3 cDNA clones, and M. Royle and D. Sipp for critical reading. We are grateful to the Laboratory for Animal Resources and Genetic Engineering for the housing and reproduction support of mice, and to the Genome Resources and Analysis Subunit for DNA sequencing, RIKEN CDB. This work was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to M. Okano.
Footnotes
References
- Bachman, K.E., Rountree, M.R., Baylin, S.B. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 2001;276:32282–32287. doi: 10.1074/jbc.M104661200. [DOI] [PubMed] [Google Scholar]
- Bestor, T.H. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J. 1992;11:2611–2617. doi: 10.1002/j.1460-2075.1992.tb05326.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Brandeis, M., Frank, D., Keshet, I., Siegfried, Z., Mendelsohn, M., Nemes, A., Temper, V., Razin, A., Cedar, H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M.C., Maeda, N., Oyama, R., Ravasi, T., Lenhard, B., Wells, C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Chen, T., Li, E. Structure and function of eukaryotic DNA methyltransferases. Curr. Top. Dev. Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- Chen, Z.F., Paquette, A.J., Anderson, D.J. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Chen, T., Ueda, Y., Dodge, J.E., Wang, Z., Li, E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 2003;23:5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun, J.Y., Han, Y.J., Ahn, K.Y. Psx homeobox gene is X-linked and specifically expressed in trophoblast cells of mouse placenta. Dev. Dyn. 1999;216:257–266. doi: 10.1002/(SICI)1097-0177(199911)216:3<257::AID-DVDY4>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Clark, S.J., Harrison, J., Paul, C.L., Frommer, M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorer, D.R., Henikoff, S. Expansions of transgene repeats cause heterochromatin formation and gene silencing in Drosophila. Cell. 1994;77:993–1002. doi: 10.1016/0092-8674(94)90439-1. [DOI] [PubMed] [Google Scholar]
- Fan, Y., Melhem, M.F., Chaillet, J.R. Forced expression of the homeobox-containing gene Pem blocks differentiation of embryonic stem cells. Dev. Biol. 1999;210:481–496. doi: 10.1006/dbio.1999.9279. [DOI] [PubMed] [Google Scholar]
- Fan, G., Martinowich, K., Chin, M.H., He, F., Fouse, S.D., Hutnick, L., Hattori, D., Ge, W., Shen, Y., Wu, H., et al. DNA methylation controls the timing of astrogliogenesis through regulation of JAK–STAT signaling. Development. 2005;132:3345–3356. doi: 10.1242/dev.01912. [DOI] [PubMed] [Google Scholar]
- Fatemi, M., Hermann, A., Gowher, H., Jeltsch, A. Dnmt3a and Dnmt1 functionally cooperate during de novo methylation of DNA. Eur. J. Biochem. 2002;269:4981–4984. doi: 10.1046/j.1432-1033.2002.03198.x. [DOI] [PubMed] [Google Scholar]
- Frigola, J., Song, J., Stirzaker, C., Hinshelwood, R.A., Peinado, M.A., Clark, S.J. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat. Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- Fuks, F., Burgers, W.A., Brehm, A., Hughes-Davies, L., Kouzarides, T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat. Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Fuks, F., Burgers, W.A., Godin, N., Kasai, M., Kouzarides, T. Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription. EMBO J. 2001;20:2536–2544. doi: 10.1093/emboj/20.10.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futscher, B.W., Oshiro, M.M., Wozniak, R.J., Holtan, N., Hanigan, C.L., Duan, H., Domann, F.E. Role for DNA methylation in the control of cell type specific maspin expression. Nat. Genet. 2002;31:175–179. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- Garrick, D., Fiering, S., Martin, D.I., Whitelaw, E. Repeat-induced gene silencing in mammals. Nat. Genet. 1998;18:56–59. doi: 10.1038/ng0198-56. [DOI] [PubMed] [Google Scholar]
- Gidekel, S., Bergman, Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J. Biol. Chem. 2002;277:34521–34530. doi: 10.1074/jbc.M203338200. [DOI] [PubMed] [Google Scholar]
- Hattori, N., Nishino, K., Ko, Y.G., Ohgane, J., Tanaka, S., Shiota, K. Epigenetic control of mouse Oct-4 gene expression in embryonic stem cells and trophoblast stem cells. J. Biol. Chem. 2004a;279:17063–17069. doi: 10.1074/jbc.M309002200. [DOI] [PubMed] [Google Scholar]
- Hattori, N., Abe, T., Suzuki, M., Matsuyama, T., Yoshida, S., Li, E., Shiota, K. Preference of DNA methyltransferases for CpG islands in mouse embryonic stem cells. Genome Res. 2004b;14:1733–1740. doi: 10.1101/gr.2431504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson-Grusby, L., Beard, C., Possemato, R., Tudor, M., Fambrough, D., Csankovszki, G., Dausman, J., Lee, P., Wilson, C., Lander, E., et al. Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat. Genet. 2001;27:31–39. doi: 10.1038/83730. [DOI] [PubMed] [Google Scholar]
- Jaenisch, R., Bird, A. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat. Genet. 2003;33 (Suppl.):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jair, K.W., Bachman, K.E., Suzuki, H., Ting, A.H., Rhee, I., Yen, R.W., Baylin, S.B., Schuebel, K.E. De novo CpG island methylation in human cancer cells. Cancer Res. 2006;66:682–692. doi: 10.1158/0008-5472.CAN-05-1980. [DOI] [PubMed] [Google Scholar]
- Jones, P.A., Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Kaneda, M., Okano, M., Hata, K., Sado, T., Tsujimoto, N., Li, E., Sasaki, H. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature. 2004;429:900–903. doi: 10.1038/nature02633. [DOI] [PubMed] [Google Scholar]
- Kim, G.D., Ni, J., Kelesoglu, N., Roberts, R.J., Pradhan, S. Co-operation and communication between the human maintenance and de novo DNA (cytosine-5) methyltransferases. EMBO J. 2002;21:4183–4195. doi: 10.1093/emboj/cdf401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, S., Isotani, A., Mise, N., Yamamoto, M., Fujihara, Y., Kaseda, K., Nakanishi, T., Ikawa, M., Hamada, H., Abe, K., et al. Comparison of gene expression in male and female mouse blastocysts revealed imprinting of the X-linked gene, Rhox5/Pem, at preimplantation stages. Curr. Biol. 2006;16:166–172. doi: 10.1016/j.cub.2005.11.071. [DOI] [PubMed] [Google Scholar]
- Lane, N., Dean, W., Erhardt, S., Hajkova, P., Surani, A., Walter, J., Reik, W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35:88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- Lei, H., Oh, S.P., Okano, M., Juttermann, R., Goss, K.A., Jaenisch, R., Li, E. De novo DNA cytosine methyltransferase activities in mouse embryonic stem cells. Development. 1996;122:3195–3205. doi: 10.1242/dev.122.10.3195. [DOI] [PubMed] [Google Scholar]
- Leonhardt, H., Page, A.W., Weier, H.U., Bestor, T.H. A targeting sequence directs DNA methyltransferase to sites of DNA replication in mammalian nuclei. Cell. 1992;71:865–873. doi: 10.1016/0092-8674(92)90561-p. [DOI] [PubMed] [Google Scholar]
- Lewis, A., Mitsuya, K., Umlauf, D., Smith, P., Dean, W., Walter, J., Higgins, M., Feil, R., Reik, W. Imprinting on distal chromosome 7 in the placenta involves repressive histone methylation independent of DNA methylation. Nat. Genet. 2004;36:1291–1295. doi: 10.1038/ng1468. [DOI] [PubMed] [Google Scholar]
- Li, E., Bestor, T.H., Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Lippman, Z., Martienssen, R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- Lock, L.F., Takagi, N., Martin, G.R. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Lunyak, V.V., Burgess, R., Prefontaine, G.G., Nelson, C., Sze, S.H., Chenoweth, J., Schwartz, P., Pevzner, P.A., Glass, C., Mandel, G., et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Maclean, J.A., II, Chen, M.A., Wayne, C.M., Bruce, S.R., Rao, M., Meistrich, M.L., Macleod, C., Wilkinson, M.F. Rhox: A new homeobox gene cluster. Cell. 2005;120:369–382. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Macleod, D., Charlton, J., Mullins, J., Bird, A.P. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes & Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- Morgan, H.D., Santos, F., Green, K., Dean, W., Reik, W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;14 (Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Morris, L., Gordon, J., Blackburn, C.C. Identification of a tandem duplicated array in the Rhox α locus on mouse chromosome X. Mamm. Genome. 2006;17:178–187. doi: 10.1007/s00335-005-0138-4. [DOI] [PubMed] [Google Scholar]
- Okano, M., Bell, D.W., Haber, D.A., Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Rai, K., Nadauld, L.D., Chidester, S., Manos, E.J., James, S.R., Karpf, A.R., Cairns, B.R., Jones, D.A. Zebra fish Dnmt1 and Suv39h1 regulate organ-specific terminal differentiation during development. Mol. Cell. Biol. 2006;26:7077–7085. doi: 10.1128/MCB.00312-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, K.D., Ait-Si-Ali, S., Yokochi, T., Wade, P.A., Jones, P.L., Wolffe, A.P. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000;25:338–342. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- Rountree, M.R., Bachman, K.E., Baylin, S.B. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat. Genet. 2000;25:269–277. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Sado, T., Fenner, M.H., Tan, S.S., Tam, P., Shioda, T., Li, E. X inactivation in the mouse embryo deficient for Dnmt1: Distinct effect of hypomethylation on imprinted and random X inactivation. Dev. Biol. 2000;225:294–303. doi: 10.1006/dbio.2000.9823. [DOI] [PubMed] [Google Scholar]
- Sado, T., Okano, M., Li, E., Sasaki, H. De novo DNA methylation is dispensable for the initiation and propagation of X chromosome inactivation. Development. 2004;131:975–982. doi: 10.1242/dev.00995. [DOI] [PubMed] [Google Scholar]
- Sproul, D., Gilbert, N., Bickmore, W.A. The role of chromatin structure in regulating the expression of clustered genes. Nat. Rev. Genet. 2005;6:775–781. doi: 10.1038/nrg1688. [DOI] [PubMed] [Google Scholar]
- Stancheva, I., Meehan, R.R. Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes & Dev. 2000;14:313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stancheva, I., Hensey, C., Meehan, R.R. Loss of the maintenance methyltransferase, xDnmt1, induces apoptosis in Xenopus embryos. EMBO J. 2001;20:1963–1973. doi: 10.1093/emboj/20.8.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stöger, R., Kubicka, P., Liu, C.G., Kafri, T., Razin, A., Cedar, H., Barlow, D.P. Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell. 1993;73:61–71. doi: 10.1016/0092-8674(93)90160-r. [DOI] [PubMed] [Google Scholar]
- Takagi, N., Sasaki, M. Preferential inactivation of the paternally derived X chromosome in the extraembryonic membranes of the mouse. Nature. 1975;256:640–642. doi: 10.1038/256640a0. [DOI] [PubMed] [Google Scholar]
- Takizawa, T., Nakashima, K., Namihira, M., Ochiai, W., Uemura, A., Yanagisawa, M., Fujita, N., Nakao, M., Taga, T. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell. 2001;1:749–758. doi: 10.1016/s1534-5807(01)00101-0. [DOI] [PubMed] [Google Scholar]
- Tsumura, A., Hayakawa, T., Kumaki, Y., Takebayashi, S., Sakaue, M., Matsuoka, C., Shimotohno, K., Ishikawa, F., Li, E., Ueda, H.R., et al. Maintenance of self-renewal ability of mouse embryonic stem cells in the absence of DNA methyltransferases Dnmt1, Dnmt3a and Dnmt3b. Genes Cells. 2006;11:805–814. doi: 10.1111/j.1365-2443.2006.00984.x. [DOI] [PubMed] [Google Scholar]
- Tucker, K.L., Beard, C., Dausmann, J., Jackson-Grusby, L., Laird, P.W., Lei, H., Li, E., Jaenisch, R. Germ-line passage is required for establishment of methylation and expression patterns of imprinted but not of nonimprinted genes. Genes & Dev. 1996;10:1008–1020. doi: 10.1101/gad.10.8.1008. [DOI] [PubMed] [Google Scholar]
- Umlauf, D., Goto, Y., Cao, R., Cerqueira, F., Wagschal, A., Zhang, Y., Feil, R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Verona, R.I., Mann, M.R., Bartolomei, M.S. Genomic imprinting: Intricacies of epigenetic regulation in clusters. Annu. Rev. Cell Dev. Biol. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- Walsh, C.P., Chaillet, J.R., Bestor, T.H. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet. 1998;20:116–117. doi: 10.1038/2413. [DOI] [PubMed] [Google Scholar]
- Watanabe, D., Suetake, I., Tada, T., Tajima, S. Stage- and cell-specific expression of Dnmt3a and Dnmt3b during embryogenesis. Mech. Dev. 2002;118:187–190. doi: 10.1016/s0925-4773(02)00242-3. [DOI] [PubMed] [Google Scholar]
- West, A.G., Fraser, P. Remote control of gene transcription. Hum. Mol. Genet. 2005;14 (Spec No 1):R101–R111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]