Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications (original) (raw)
Abstract
Covalent histone post-translational modifications such as acetylation, methylation, phosphorylation, and ubiquitylation play pivotal roles in regulating many cellular processes, including transcription, response to DNA damage, and epigenetic control. Although positive-acting post-translational modifications have been studied in Saccharomyces cerevisiae, histone modifications that are associated with transcriptional repression have not been shown to occur in this yeast. Here, we provide evidence that histone sumoylation negatively regulates transcription in S. cerevisiae. We show that all four core histones are sumoylated and identify specific sites of sumoylation in histones H2A, H2B, and H4. We demonstrate that histone sumoylation sites are involved directly in transcriptional repression. Further, while histone sumoylation occurs at all loci tested throughout the genome, slightly higher levels occur proximal to telomeres. We observe a dynamic interplay between histone sumoylation and either acetylation or ubiquitylation, where sumoylation serves as a potential block to these activating modifications. These results indicate that sumoylation is the first negative histone modification to be identified in S. cerevisiae and further suggest that sumoylation may serve as a general dynamic mark to oppose transcription.
Keywords: Histone post-translational modifications, sumoylation, transcription regulation, telomeric silencing
Covalent post-translational modifications of lysine residues such as acetylation, methylation, and ubiquitylation play important roles in controlling protein function. In histone proteins, dynamic lysine acetylation/deacetylation was the first modification to be characterized in detail (Brownell et al. 1996; Taunton et al. 1996), thereby galvanizing the study of chromatin structure and regulation of DNA-related processes. The histones H2A, H2B, H3, and H4 comprise the core complex of the nucleosome, which is the basic repeating unit of chromatin in the eukaryotic cell. The nucleosome core serves as a paradigm for modifications and their effect on cellular processes such as transcription, repair, and epigenetic control. Various modifications of histones affect interactions within the core unit as well as with the associated DNA and other factors and complexes (Strahl and Allis 2000; Berger 2002). Further, certain protein modules, often occurring within protein complexes, recognize specific patterns of histone modifications to recruit effector proteins that change chromatin structure and dynamics (Dhalluin et al. 1999; Bannister et al. 2001; Lachner et al. 2001). Positive-acting histone modifications occur universally in eukaryotes and include acetylation and phosphorylation, as well as ubiquitylation (e.g., H2B Lys123 [K123]) and lysine methylation (e.g., H3 K4) at certain sites (Berger 2002). In contrast, negative-acting histone modifications, such as lysine methylation (including H3 K9, K27, and H4 K20) and H2A K119 ubiquitylation, occur in Schizosaccharomyces pombe and metazoans but have not been detected in Saccharomyces cerevisiae (Martin and Zhang 2005). One possibility is that the small, gene-rich S. cerevisiae genome, which is subject to rapid and wide-spread gene induction, has chromatin “poised” for transcription and hence in a constitutively open state.
Recently, it has been shown that histones are sumoylated in mammalian cells (Shiio and Eisenman 2003). SUMO (SMT3 in the yeast S. cerevisiae) is a small ubiquitin-related modifier protein of ∼100 amino acids that is conserved in all eukaryotes. It shares a similar three-dimensional structure with ubiquitin and a related, but separate pathway of activation (E1), conjugation (E2), and ligation (E3) of its processed form to the targeted lysine side chain (Johnson 2004). In yeast the heterodimer Aos1/Uba2 (E1) forms a high-energy thioester bond to activate SUMO. SUMO is then transferred to Ubc9, the only known conjugating enzyme (E2) in yeast and higher eukaryotes. While Ubc9 is capable of ligating SUMO to its target, several E3 ligases have been characterized that confer specificity to the sumoylation process. In yeast the known E3s are Siz1, Siz2 (Nfi1), and Mms21 (Johnson and Gupta 2001; Takahashi et al. 2001; Zhao and Blobel 2005).
Protein sumoylation results in a variety of effects, including changes in cellular localization or stability, modulation of protein–protein or protein–DNA interactions, or antagonizing other lysine modifications such as ubiquitylation (Verger et al. 2003; Johnson 2004). Recent genomic studies aimed at identifying cellular substrates for sumoylation indicate that many are involved in transcription regulation and include transcription factors, transcription machinery proteins, and components that modify chromatin structure (Manza et al. 2004; Wohlschlegel et al. 2004; Zhou et al. 2004; Hannich et al. 2005). In most cases, sumoylation of transcriptional activators results in transcriptional repression (Gill 2003; Hay 2005). Many sumoylation sites harbor the sequence ΨKXE (where Ψ is a hydrophobic residue, K is the attachment site lysine, and X represents any amino acid). Some consensus sites occur in negative regulatory domains of transcription factors (Holmstrom et al. 2003). Recent studies demonstrate that a few charged and conserved residues in the SUMO ubiquitin-like fold are essential for its transcriptional repressive properties (Chupreta et al. 2005).
In this study we focus on the role of sumoylation of histones. Histone H4 sumoylation in mammalian cells has been reported, although attachment sites were not determined (Shiio and Eisenman 2003). Promoter sumoylation was shown to correlate with certain events accompanying transcriptional repression, including H3 deacetylation and recruitment of the heterochromatin protein HP1 (Shiio and Eisenman 2003). However, the important question remains whether transcriptional repression mediated by the SUMO pathway is linked specifically to histone sumoylation. In this study we addressed the role of histone sumoylation in S. cerevisiae. We detect sumoylation of all four core histones and identify specific sites of sumoylation in H2A, H2B, and H4. Importantly, we observe a relationship between histone sumoylation and transcriptional repression. Our results further suggest that histone sumoylation may exist in a dynamic interplay with histone acetylation and ubiquitylation.
Results
Sumoylation of all core histones within chromatin
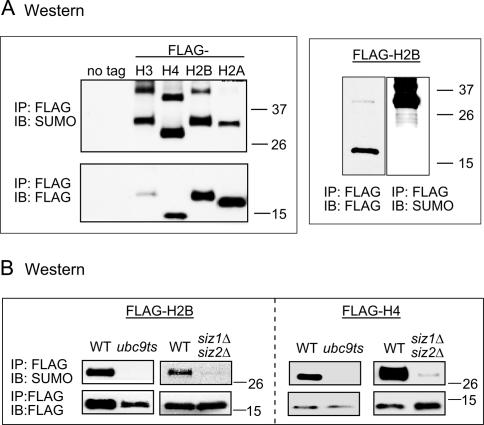
SUMO conjugates are broadly distributed on yeast chromosomes as determined by anti-SUMO ChIP, chromatin fractionation, and chromosome spreads (Biggins et al. 2001; data not shown). To determine the sumoylation status of the four core histones, we used a shuffle system in which the endogenous genes were deleted and replaced by a plasmid encoding the tagged histone. Immunoprecipitation (IP) followed by Western blot (immunoblot [IB]) analysis with anti-Flag and anti-SUMO antibodies indicated that all four histones are sumoylated in vivo (Fig. 1A, left). The anti-SUMO antibody is highly specific for sumoylated histone in this experiment, as it does not react with unmodified histone H2B, although the modified form is a small proportion of the total histone (Fig. 1A, right). At least two sumoylated bands can be seen, indicating either chain formation or multiple sumoylation sites. In general, histone sumoylation accounts for a small percentage of total histone (approximately <5%) (Figs. 1A [right], 2) and is labile under native conditions. Therefore, IPs were routinely conducted on denatured cell extracts prepared by trichloroacetic acid (TCA) precipitation (Ohashi et al. 1982).
Figure 1.
In vivo histone sumoylation in yeast. (A) Western blot (IB) analysis of immunoprecipitated (IP) Flag-tagged histones using anti-Flag and anti-SUMO antibodies. Flag-tagged histones migrate ∼15 kDa. Singly sumoylated histones migrate between 26 and 37 kDa. (B) Flag IP followed by Western blot (IB) analysis of histones in sumoylation pathway mutant strains ubc9ts and _siz1_Δ_siz2_Δ. Detection was carried out as in A.
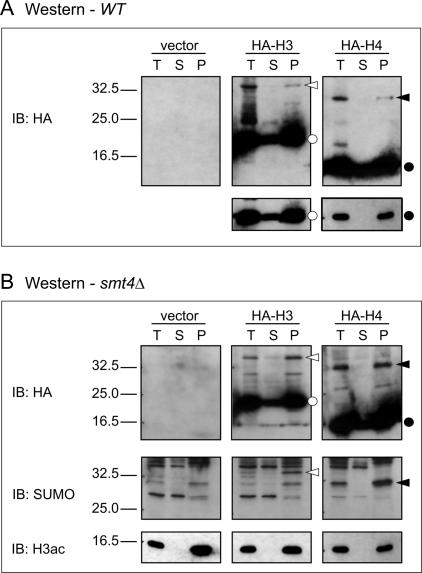
Figure 2.
Western blot analysis of histones in fractionated yeast cells. (A) Western blot analysis using anti-HA antibody of untagged (vector) fractionated wild-type cells or HA-tagged H3 or H4. Fractions are total (T), soluble (S), or pellet (P). Open circles indicate HA-H3, closed circles indicate HA-H4, open arrowheads point to sumoylated HA-H3, and closed arrowheads point to sumoylated HA-H4. A shorter exposure of the nonsumoylated histones is shown below. (B) Similar analysis as in A except extracts were obtained from a strain bearing a SMT4 (SUMO protease) deletion, which increases the cellular level of sumoylated histones. HA and SUMO immunoblots are shown, as well as H3 acetylation (H3ac) immunoblot as a control for fractionation.
Strains defective in the sumoylation process were used to check their effect on histone sumoylation by transforming the Flag-tagged histone plasmids for ectopic expression. A temperature-sensitive mutant of UBC9 (ubc9ts, which is partially defective at restrictive temperature) (Schwarz et al. 1998), as well as a double deletion of the E3 ligases SIZ1 and SIZ2 (Johnson and Gupta 2001), cause dramatic reduction of H2B and H4 sumoylation (Fig. 1B). Deletion of SIZ1 alone did not result in any changes in histone sumoylation levels (data not shown). The reduction of histone sumoylation in the mutants is unlikely to be an indirect effect on the availability of free SUMO, which is readily detected in whole-cell extracts of ubc9ts and siz mutant strains (data not shown).
We investigated the chromatin association of sumoylated H3 and H4 to determine whether sumoylation prevents chromatin incorporation. Total histones were prepared from strains bearing HA-tagged H3 or H4 and were subjected to fractionation to separate insoluble chromatin pellet from soluble supernatant (Liang and Stillman 1997). In a wild-type strain (Fig. 2A) or a strain bearing a deletion of the SUMO protease Smt4 (Ulp2) (Fig. 2B), which increases the level of sumolyated histones, sumoylated histones are detected exclusively in the pellet. The SUMO modification status of the HA-histones was confirmed using a SUMO antibody (Fig. 2B). In addition, the bands match immunoreactive species on anti-H3 and anti-H4 Western blots (data not shown). Localization of acetylated H3 to the chromatin pellet serves as a control for the fractionation procedure (Fig. 2B, bottom). Thus, sumoylated histones occur within chromatin, suggesting a role in genomic regulation.
H2B and H4 sumoylation sites are located in their N-terminal regions
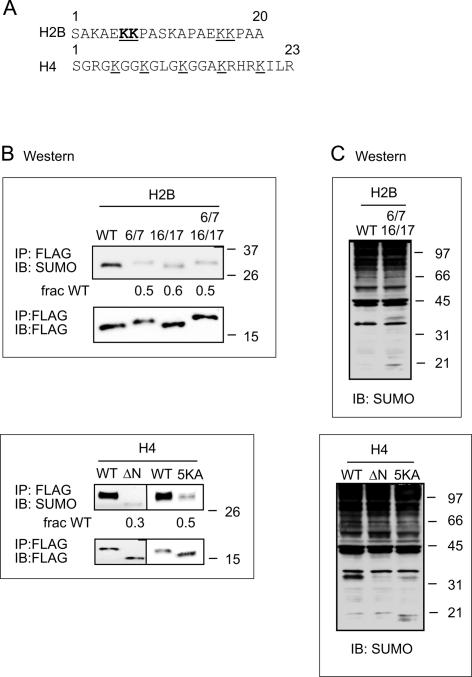
To characterize the sites of sumoylation in histone H2B, affinity chromatography of His-tagged SUMO was used to generate samples for tandem mass spectrometry (MS/MS) analysis. Initial analysis revealed the presence of H2B and H2A in the samples. Specific endopeptidases were chosen to generate maximum peptide fragments containing the branched SUMO moiety from the targeted lysine. These efforts revealed that SUMO is attached to histone H2B at Lys6 or Lys7 (K6/7) in the N-terminal tail (Fig. 3A) and also showed that one of these lysines is a novel acetylation site as well. Analysis of a limited number of peptide fragments (three) indicated that K6/7 is either singly acetylated or sumoylated, but no MS/MS spectra were observed where they were simultaneously modified. One of these peptide spectra indicated unambiguous acetylation of Lys6. The sequence surrounding K6/7 bears some similarity to the N-terminal extension of yeast SUMO, which is itself sumoylated (Bylebyl et al. 2003). In H2B this sequence with surrounding alanines (AEKKPA) is repeated, generating another putative sumoylation/acetylation site at Lys16 and Lys17 (K16/17). The proteolysis and MS/MS peptide fragmentation patterns did not allow for the identification of this second putative sumoylation site.
Figure 3.
Identification and confirmation of histone SUMO sites in vivo. (A) Regions of sumoylation in histone H2B and H4. The lysine residues for which sumoylation was confirmed by MS/MS in histone H2B are shown in bold and underlined letters. Additional putative sumoylation sites in H2B are also underlined. Lysine residues that were mutated to alanine in histone H4 are indicated by underlined letters. (B) Effect of substitution mutations in putative sumoylation sites on in vivo modification in yeast. Lys6 and Lys7 (6/7), Lys16 and Lys17 (16/17), or all four lysines (6/7 16/17) in H2B were mutated to alanine and were either shuffled in as the only source of histone H2B in the cell or transformed and kept as an ectopic copy. Histone H4 was expressed ectopically, and the entire tail was deleted (ΔN), or all five lysines in the H4 tail were mutated to alanine (5K). Sumoylation levels were detected using Flag IP followed by Western blot analysis with anti-Flag (bottom) and anti-SUMO (top) antibodies. The numbers below the SUMO blots represent quantification of the sumoylation levels compared with the signal in wild-type (WT) strain after normalization to the Flag levels in each strain. (C) Whole-cell extracts from the same strains used in B were analyzed by SDS-PAGE and immunoblotting with an anti-SUMO antibody. At higher molecular weight (above the 97-kDa marker), the lanes are smeared and distinct bands are no longer seen. The H4 wild-type (WT) lane shows a band ∼33 kDa, which corresponds to the molecular weight of sumoylated H4, and is greatly reduced in the H4 mutant lanes. The H2B genes were not on a high-copy plasmid as were the H4 genes; therefore, the sumoylated form of H2B is not visible in whole-cell extracts.
Mutation of either K6/7 or K16/17 to alanine resulted in reduced H2B sumoylation (Fig. 3B, upper panels). Mutating all four K6/7/16/17 to alanine did not result in further decrease in sumoylation, suggesting that sumoylation at one of the repeats may positively influence sumoylation at the adjacent repeat. Quantitative analysis of the sumoylated forms relative to total H2B indicates that the level of sumoylation decreases by ∼50% in the mutants compared with wild type (Fig. 3B). Mutating any other lysine in H2B did not result in significantly lowered SUMO–H2B levels (data not shown). The residual sumoylation observed when all four lysines in these sequences were mutated to alanine also suggests nonspecific sumoylation or a small population with an alternative sumoylation site.
Mass spectrometry studies also indicated a sumoylation site at the C-terminal K126 of histone H2A. However, substitution of this site (H2A K126R) did not cause significant changes in H2A sumoylation levels compared with wild type (data not shown).
In our efforts to identify sumoylation sites in other histones, we observed that deletion of the N-terminal tail of histone H4 resulted in a considerable reduction of sumoylation levels (Fig. 3B, lower panels). This result is in accordance with previous studies of mammalian histones, which indicated that the H4 tail is a substrate for sumoylation in vitro (Shiio and Eisenman 2003). Mutating all five lysines in the N-terminal tail yielded a similar effect (lowered to 30%–50%; Figure 3B), suggesting that these are the major sites of sumoylation in H4. However, mutating any three lysines out of the five did not reduce H4 sumoylation levels significantly (data not shown). Thus, it may be that there is a lack of specificity among these sites. Similar to the H2B multiple mutation results, residual sumoylation was observed even when the entire tail of H4 was deleted or all five lysines substituted, underscoring the apparent redundancy and/or promiscuity of histone sumoylation. These five N-terminal lysines are major sites of acetylation in H4.
To ensure that the lowered sumoylation states of the mutant histones H2B and H4 were not due to lower amounts of total SUMO synthesis/processing, we examined whole-cell extracts made from these strains and analyzed by Western with an anti-SUMO antibody. Compared with wild type, the mutant strains showed very similar intensities and patterns of sumoylated proteins (Fig. 3C). We therefore conclude that the reduced histone sumoylation levels seen in the mutants were indeed the result of eradication of the sumoylation receptor sites.
Thus, we identified a subset of histone sumoylation sites. Overall, it appears that all four core histones are sumoylated, that there are multiple potential sites within each histone, and that sumoylation sites are coincident with or near known sites of acetylation.
Sumoylation is higher at subtelomeres compared with more internal regions
We investigated genomic localization of sumoylation associated with H2B using chromatin IP (ChIP). We were unable to generate H2B–SUMO-specific antibodies using branched peptides (containing several SUMO-specific residues attached to a base of several histone-specific residues) as antigens. Instead we developed a “ChDIP” (double-affinity ChIP) method to examine sumoylation, similar to our previous analysis of ubiquitylation associated with H2B (Henry et al. 2003; Kao et al. 2004). We used a yeast strain carrying a polyhistidine tag on SUMO and a plasmid bearing Flag-tagged H2B. The ChDIP involved Flag IP followed by elution and nickel column purification, prior to de-cross-linking and quantitation by real-time PCR. A portion of the first step of the ChIP (Flag-H2B) was retained, and results from the second step (nickel column) were normalized to the Flag-H2B ChIP signal in order to evaluate the percentage of total H2B that is sumoylated.
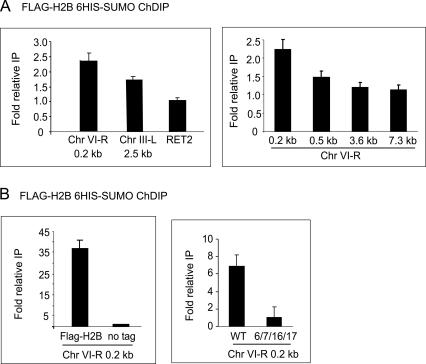
We found H2B-associated sumoylation at many genomic locations, including the RET2 (Fig. 4A) and GAL1 genes (Fig. 6B, below). We arbitrarily set the ChDIP signal for RET2 to 1.0 and found comparable levels at numerous genomic locations (data not shown). Interestingly, sequences adjacent to telomeric repeats, such as the right arm of chromosome VI (Chr VI-R) or the left arm of chromosome III (Chr III-L) are enriched in H2B-associated sumoylation by roughly twofold, compared with other regions in the genome (Fig. 4A, left). Indeed, there is a gradual decrease in this higher telomeric H2B-associated sumoylation with ChIP probes more internal to the chromosome (Fig. 4A, right). This correlation is opposite to the pattern previously observed for H2B ubiquitylation (Emre et al. 2005).
Figure 4.
Location of histone sumoylation in the yeast genome. (A) Quantitative PCR analysis of ChDIP in a His-SUMO Flag-H2B strain. Telomeric end primers are located ∼0.2 kb away from the telomeric repeats on Chr VI-R or 2.5 kb away from telomeric repeats of Chr III-L. The right panel shows ChDIP using primers to regions 0.5, 3.6, and 7.3 kb away from the telomere end on Chr VI-R. RET2 is located ∼20 kb away from the Chr VI-R telomere end. (B) Controls for quantitative PCR analysis of ChDIP samples. All strains have His-tagged SUMO. Left panel shows a comparison control stain lacking the Flag-H2B plasmid. Right panel shows a comparison control strain containing H2B substitution mutant K6/7/16/17A in the Flag-H2B plasmid.
Figure 6.
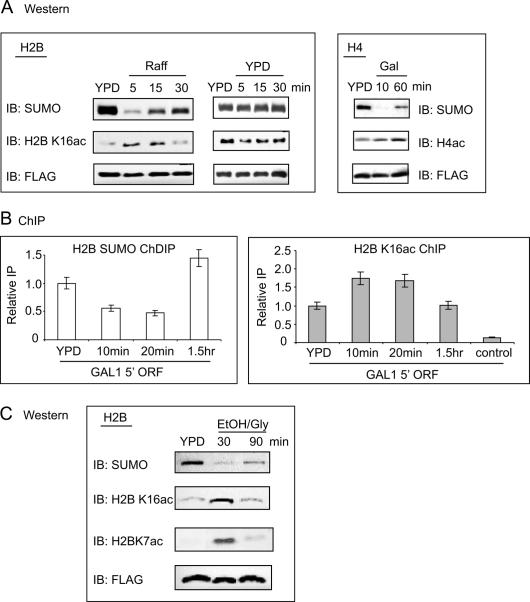
Interplay of acetylation–sumoylation in histone H2B and H4 upon changes in carbon source. (A) Flag IP followed by Western blot analysis of Flag-H2B (left and middle) or Flag-H4 (right) samples taken at the indicated time points during growth switched from glucose-to-raffinose (Raff, H2B) or glucose-to-galactose (Gal, H4). Center panels show the control Glu-to-Glu switch for Flag H2B. Antibodies are as indicated. (B) Real-time PCR analysis at the GAL1 promoter region of a ChDIP experiment (left) or H2B K16ac ChIP (right) in a His-SUMO Flag-H2B tagged strain. Samples were taken at the indicated time points after switching from glucose (YPD) to raffinose. (C) Flag IP followed by Western blot analysis of Flag-H2B samples taken at the indicated time points during glucose (YPD) to ethanol/glycerol (EtOH/Gly) carbon source change. Antibodies are as indicated.
We created two different strains to serve as controls for each ChIP step. For the first ChIP step using Flag antibody, a strain lacking the tag on H2B was used as a control, resulting in a nearly undetectable ChDIP signal for all regions tested (Fig. 4B, left; data not shown). As a control for the second SUMO purification step, we prepared an alanine substitution mutant of the four sumoylation sites in H2B, yielding K6/7/16/17A. This strain gave a dramatic decrease in signal intensity compared with the wild-type strain for all regions tested (Fig. 4B, right; data not shown), which indicates that the ChDIP signal largely represents SUMO attached to the tagged H2B, rather than to other chromatin proteins present at the same locus. The difference between these controls (i.e., there is a greater decrease in signal in the no-tag control than in the mutant H2B control) may represent other histones that are sumoylated and cross-linked to H2B. Overall, the ChDIP results indicate histone sumolyation exists at similar levels for many tested regions of the genome, with slightly higher subtelomeric signals.
Histone sumoylation represses transcription
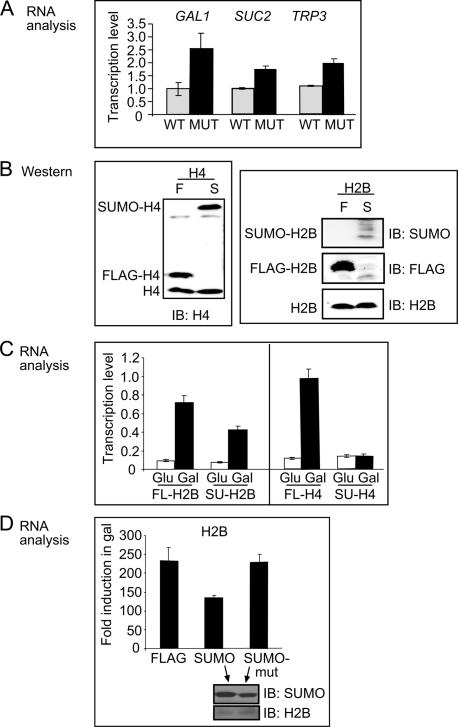
We examined a possible role of histone sumoylation in transcriptional repression by comparing gene expression in the H2B K6/7/16/17A mutant strain (in which H2B sumoylation is attenuated) to that in wild-type strain. Under noninducing conditions, basal expression levels of TRP3, SUC2, and GAL1 are increased between two- and threefold in the K6/7/16/17A substitution mutant compared with wild type (Fig. 5A). This suggests that the relatively low basal level of transcription of these three genes is maintained in part by histone H2B sumoylation.
Figure 5.
Sumoylation of histones regulates transcription. (A) Quantitative PCR analysis of TRP3, SUC2, and GAL1 RNA levels in noninducing (YPD) conditions for wild-type (WT) and H2B K6/7/16/17A substitution mutant (MUT). (B) Western blot analysis of whole-cell extracts probed with anti-histone H4 (left) or anti-histone H2B, anti-Flag, and anti-SUMO antibodies (panels on right). Indicated are the positions of endogenous (bottom), Flag-tagged histones (middle), and SUMO-fused histones (top). (C) Quantitative PCR analysis showing transcript levels of GAL1 for SUMO–H2B (SU-H2B) or Flag-H2B (FL-H2B) fusions when switching from glucose (Glu) to galactose (Gal) media and the parallels in histone H4. (D) Quantitative PCR analysis as in C comparing Flag-H2B and SUMO–H2B with a strain with a SUMO–H2B fusion containing substitution mutations in SUMO at Lys37 and Arg46 to glutamic acid (SUMO-mut). Lower insert shows Western analysis of whole-cell extracts from SUMO–H2B and SUMO-mut–H2B fusion strains using anti-SUMO and anti-H2B antibodies.
This effect on transcriptional repression by substitution mutations in the H2B sumoylation sites is modest. This might reflect redundancy due to the large number of sumoylation sites on the histones, or the inability of the substituted sites to be acetylated to achieve maximum derepression. We hypothesized that if sumoylation is indeed repressive to transcription, then direct fusion of SUMO to the histone should cause more severe and dominant repression. This approach has been frequently used to test a role of sumoylation or ubiquitylation of other factors; e.g., the transcription factor Sp3 (Ross et al. 2002) or GAL4-VP16 (Salghetti et al. 2001). We prepared SUMO–H4 and SUMO–H2B genetic fusions (or Flag-H4 and Flag-H2B as controls) in which the C terminus of SUMO (or Flag) is fused to the N terminus of histone H4 or histone H2B. This fusion places SUMO close to the N-terminal residues where it is naturally attached (see above). The fusion proteins were designed to lack the natural glycine–glycine motif that forms the isopeptide bond with the target lysine side chain, and thus cannot be cleaved by SUMO peptidases. Having a SUMO–histone conjugate as the only source of histone was toxic to the cell (data not shown), hence the fusions were expressed ectopically with a high-copy (H4) or low-copy (H2B) plasmid. Flag- or SUMO–H4 was detected by H4 antibody and was present in similar amounts to each other and to untagged H4 (Fig. 5B, left). Flag-H2B or SUMO–H2B could not be detected by the H2B antibody, and therefore, whole-cell extract was separately probed with anti-Flag and anti-SUMO antibodies (Fig. 5B, right). Thus, stable histone fusions were generated with SUMO close to the endogenous sumoylation site, potentially mimicking the sumoylated states but not susceptible to desumoylation like the native conjugates.
We tested GAL1 induction upon switching to galactose media in cells carrying the Flag versus the SUMO–histone fusion. GAL1 transcription is induced sevenfold to 10-fold by galactose; the induction is reduced twofold in the presence of SUMO–H2B and fivefold in the presence of SUMO–H4 compared with the Flag-histone fusion (Fig. 5C). As a second control, we created a SUMO–H2B fusion containing substitution mutations in SUMO that decrease the repressive effects of SUMO (Chupreta et al. 2005). The fusion containing these mutations completely reversed the repression seen with SUMO–H2B (Fig. 5D), and Western analysis indicates that the wild-type and mutant SUMO–H2B fusions are present at comparable levels (Fig. 5D, lower insert). Taken together with the increased basal transcription exhibited by the substitution mutations in the natural sumoylation sites in the histones, these results provide further support for a role of histone sumoylation in transcriptional repression.
Histone sumoylation opposes activating histone acetylation or ubiquitylation and is low in histone variant H2AZ compared with H2A
The preceding experiments indicated that the GAL1 gene is under negative control by histone sumoylation. We hypothesized that histone sumoylation might decline during gene activation when positive histone modifications are induced. To investigate this, we first tested whether the overall level of histone sumoylation changes under conditions that affect GAL1 expression. We examined H2B and H4 sumoylation by IP-Western at regular intervals following a change in carbon source from repressing glucose to nonrepressing raffinose or to inducing galactose. Overall histone sumoylation levels drop dramatically during the first 5–10 min after a shift from glucose to raffinose or from glucose to galactose and then appear to gradually increase to a new “steady-state” level (Fig. 6A, left and right top panels). The transient decline in histone sumoylation is not detected in mock-treated samples “shifted” from glucose to glucose (Fig. 6A, center top panel) or in samples shifted from raffinose to glucose (data not shown), supporting the view that it may be related to global gene activation and not to an artifact of cell manipulation or histone IP procedure.
The finding that sumoylation sites are at, or adjacent to, acetylation sites in H2B and H4 suggests that histone sumoylation and acetylation may be in competition. We examined possible interplay between sumoylation and acetylation during the change in carbon source. Western blot analysis (Fig. 6A), as well as ChIP of the GAL1 promoter (Fig. 6B), shows that the histone acetylation pattern partially opposes sumoylation during the time course in altered carbon source. A reciprocal pattern for sumoylation/acetylation is also apparent in the switch from glucose to ethanol/glycerol media, including both H2B K7 and K16 acetylation (Fig. 6C).
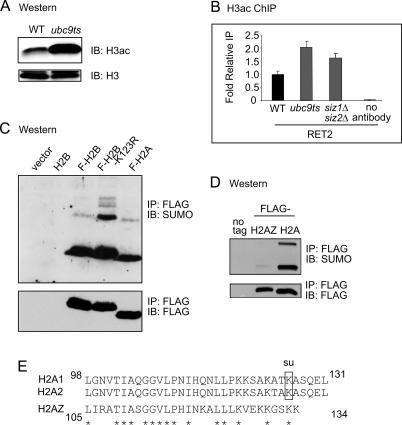
We examined whether reduced sumoylation in ubc9ts and _siz1_Δ_siz2_Δ double-deletion strains, which exhibit dramatically lower histone sumoylation compared with wild type (Fig. 1B), leads to increased histone acetylation. Western analysis shows higher levels of H3 acetylation in the ubc9ts strain (Fig. 7A), and ChIP analysis shows higher gene-associated H3 acetylation in either strain (Fig. 7B). In addition, cellular levels of H2B sumoylation are increased in a strain bearing a substitution mutation at K123 of H2B (Fig. 7C), which is a ubiquitylation site involved in gene activation (Henry et al. 2003; Kao et al. 2004). Thus, histone sumoylation and activation-linked histone modifications, including acetylation and ubiquitylation, oppose one another.
Figure 7.
Relationship between sumoylation and postive histone mechanisms. (A) Western blot of histones prepared from wild-type (WT) or ubc9ts strain probed with H3ac (K9ac, K14ac) antibody (top) or unmodified histone H3 antibody (bottom). (B) Quantitative PCR analysis of H3ac ChIP of the RET2 gene in the wild-type (WT), ubc9ts, and _siz1_Δ_siz2_Δ strains. (C) Western blot of histones prepared from wild-type (WT) strain or strains bearing Flag-H2B, Flag-H2B-K123R substitution mutation, or Flag-H2A. (D) Flag IP followed by Western blot analysis of Flag-tagged histone H2A and histone variant H2AZ. (E) Alignment of the C-terminal tails of yeast canonical H2A and variant histone H2AZ. The sumoylation site is indicated for H2A. Asterisks indicate identical residues.
The histone variant H2AZ has been localized to heterochromatic boundaries that protect euchromatin from spreading of silencing proteins (Meneghini et al. 2003) or to promoter regions that poise genes for transcriptional activation (Santisteban et al. 2000; Guillemette et al. 2005; Raisner et al. 2005). We therefore hypothesized that a repressive mark, such as sumoylation, may not be prominent on this histone variant. We noted that H2AZ exhibits conserved amino acid sequence with canonical H2A, except near to the end of the C-terminal tail (Fig. 7E), where we had mapped one sumoylation site on H2A (K126) as described above. Western analysis showed that the overall level of sumoylation on H2AZ is low compared with sumoylation of H2A (Fig. 7D).
Discussion
Prior to the present study, the yeast S. cerevisiae appeared to be devoid of typical histone modifications known to negatively regulate higher eukaryotic genomes. While eukaryotes ranging from S. pombe to humans contain negative-acting lysine methylation (Lachner and Jenuwein 2002; Grewal and Moazed 2003; Martin and Zhang 2005), this class of modification is not detected in S. cerevisiae. It thus appeared that the S. cerevisiae genome is “poised” for transcription and may not be subject to negative regulation through chromatin modifications (e.g., see White et al. 2001). In apparent contrast to this point of view, our results suggest that histone sumoylation has a negative regulatory role in S. cerevisiae. There appear to be a large number of sumoylation sites, with multiple sites on each of the core histones; we have apparently identified most sites on H2B and H4, a subset of sites on H2A, while sites on H3 have not yet been identified.
One repressive role we detect for histone sumoylation is in gene-specific negative regulation, where sumoylation helps to maintain a low basal level of transcription. Although there are numerous histone sumoylation sites, substitution mutation of only four sumoylation acceptor sites in H2B results in an increase in the uninduced transcriptional level of three genes that are subject to distinct pathways of inducible regulation (Fig. 5A). It may be that combinatorial substitutions of more sumoylation sites on the same histone/different histones will result in stronger increases in basal transcription. A similar or even stronger inhibitory effect, on induced GAL1 transcription, correlates with direct fusion of SUMO to histone H2B or H4 (Fig. 5C). Genes representing a wide range of regulatory pathways were tested in our study (GAL1 is induced by galactose, SUC2 is induced by sucrose, and TRP3 is induced by low levels of certain amino acids) and revealed to be under negative control by histone sumoylation. These results suggest that the negative role of histone sumoylation may be quite general.
A second negative regulatory role suggested by our data is that histone sumoylation may reinforce telomeric silencing. Telomeric silencing in S. cerevisiae has long been associated with reduced levels of positive-acting histone modifications. This includes low acetylation, maintained through the action of the histone deacetylase Sir2 (Grunstein 1997; Rusche et al. 2003), and low ubiquitylation and methylation (Bryk et al. 2002; Ng et al. 2003), maintained via the histone deubiquitylase Ubp10, which also serves to keep methylation levels low (Emre et al. 2005). We detect slightly higher H2B-associated sumoylation at telomeres than at more internal chromosomal sites or at genes (Fig. 4A). Strikingly, this is the reverse correlation as previously observed for both H2B-associated ubiquitylation and H3 methylation (Ng et al. 2003; Emre et al. 2005). Further, we found that telomeric silencing of URA3 cassette expression (Aparicio et al. 1991) is dramatically lowered in the ubc9ts strain (data not shown), which may indicate a role of histone sumoylation in promoting telomeric silencing.
Third, histone sumoylation appears to correlate negatively with positive-acting histone acetylation and ubiquitylation. Reduction of histone sumoylation results in increased histone acetylation (Fig. 7A,B). Further, changes in carbon source correlate with loss of histone sumoylation in parallel with increased histone acetylation (Fig. 6). Acetylation occurs broadly in the genome and can be globally altered by manipulating the levels of histone acetyltransferases or deacetylases (Vogelauer et al. 2000; Boudreault et al. 2003). Thus, genome-wide acetylation and sumoylation, as shown in our study, may broadly regulate. In addition to alternative sumoylation/acetylation, substitution mutation of the ubiquitylation site in histone H2B leads to higher H2B sumoylation (Fig. 7C) indicating an interplay between these modifications as well.
Finally, there are lower levels of SUMO on the histone variant H2AZ compared with canonical H2A (Fig. 7D). As described above, H2AZ localizes both to genes promoters, potentiating them for activation, and to genes near silenced regions, protecting them from heterochromatic spreading. This observation is reminiscent of recent findings in Drosophila that higher levels of positive-acting modifications and lower levels of negative-acting modifications decorate the transcription-linked histone variant H3.3, compared with the replication-linked canonical H3.1 (McKittrick et al. 2004). In accordance to H2AZ correlation with active chromatin, our results of low sumoylation levels on H2AZ lend further support to a repressive role for sumoylation.
Thus, we speculate that histone sumoylation may interfere with or counteract activating histone mechanisms. There are several mechanisms that may account for alternative acetylation and sumoylation. There may be a direct competition between the modifications since many of these lysines bear both modifications. Sumoylation and other lysine modifications occur on the same residues in other proteins. Proteins that can be either sumoylated or ubiquitylated include IκBα in the NF-κB transcription pathway (Desterro et al. 1998), pathogenic Huntington’s disease protein Huntingtin (Steffan et al. 2004), and the DNA polymerase auxiliary factor, PCNA (Hoege et al. 2002). Further, sumoylation–deacetylation interplay has been suggested for the transcription factor Sp3 (Braun et al. 2001; Ross et al. 2002).
A second possible mechanism is that histone sumoylation may recruit HDACs (histone deacetylases) to deacetylate at neighboring sites. This has been demonstrated for the transcription factor Elk-1, where the sumoylated form recruits HDAC-2 to the promoter, resulting in histone deacetylation and transcriptional repression (Yang and Sharrocks 2004). The sumoylation machinery interacts with HDACs, specifically with the HDAC II protein subfamily (Colombo et al. 2002; David et al. 2002; Kirsh et al. 2002; Verdin et al. 2003; Gregoire and Yang 2005) and may be part of a regulatory mechanism that controls their repression activity. Another study showed that a SUMO–H4 fusion associates with endogenous HDAC1 as well as HP1, suggesting that histone sumoylation is involved in recruitment of deacetylases (Shiio and Eisenman 2003).
A third possible mechanism would involve activation of HDACs by sumoylation, which was previously observed (David et al. 2002; Cheng et al. 2004); thus, lowered HDAC sumoylation (particularly in the SUMO pathway mutant strains) may indirectly lead to higher histone acetylation. However, while HDAC sumoylation regulates a number of gene regulatory pathways in mammalian cells, in S. cerevisiae, proteomic analyses of sumoylated proteins indicate that HDACs are not a robust target (Wohlschlegel et al. 2004; Zhou et al. 2004; Hannich et al. 2005), although Hda1 was detected in one SUMO proteomic analysis (Panse et al. 2004). Our findings that substitution mutations in histone sumoylation sites increase transcription, and that SUMO fused to histones is repressive to transcription, strengthen the first two models of direct effect of histone sumoylation through either competition with acetylation or recruitment of HDACs (i.e., the first and second proposed mechanisms), rather than an indirect effect through altering HDAC activity (i.e., the third mechanism).
Our study confirms and extends in S. cerevisiae the initial observation of histone sumoylation made in mammalian cells. We demonstrate a role for histone sumoylation in transcriptional repression in S. cerevisiae. There is abundant evidence linking sumoylation of factors with deacetylation, which provides a potential mechanism to reinforce the antagonism that we detect between sumoylation and acetylation. The redundancy or multiplicity of sumoylation sites in histones suggest that SUMO is attached to a large number of histone lysines and exerts its regulatory effect (either steric blocking or recruitment of other factors, such as silencing proteins or HDACs) independent of a specific residue. Thus, sumoylation appears to be the first evolutionarily conserved histone modification exhibiting a negative genomic regulatory role in chromatin.
Materials and methods
Yeast strains and plasmids
Yeast strains used in this study are listed in Table 1. Histone Flag tagging and amino acid substitution and deletion (QuikChange kit, Stratagene) of H3 and H4 were performed with pRM204 (Mann and Grunstein 1992) or a high-copy URA3 plasmid containing Flag H4 (provided by M.A. Osley, Molecular Genetics and Microbiology, University of New Mexico HSC, Albuquerque, NM). For manipulations of H2A and H2B, plasmids FB1251 (Hirschhorn et al. 1995) or pRS314 and pRS316 HTB1-HTA1 (Sikorski and Hieter 1989) were used. The SUMO–H4 fusion plasmid (pRS424) was constructed by first creating a pRS424-based Flag-H4 plasmid (pTKS791), then replacing the Flag coding sequence with a PCR-amplified fragment encoding mature Smt3 (amino acids 1–98; pPM505). To prevent cleavage of the fusion protein by SUMO proteases, the C-terminal Gly–Gly motif of Smt3 was mutated to Asp–Val. SUMO–H2B was cloned by standard methods using SUMO–H4 and Flag-H2B (RS314) into pRS314. EJ362 expressed a His6-tagged version of Smt3 with a His5 sequence (VKPETHHHHHHIN) inserted after amino acid 22. The His6- and Flag-containing N-terminally truncated version of SMT3 in EJ363 has the N-terminal sequence MTSHHHHHHMHDYKDDDDKMGST22HINLK. These alleles were constructed by assembly PCR and transformation into yeast as described (Johnson and Blobel 1999).
Table 1.
Yeast strains used in this study
Sumoylated histone H2B purification
EJ363 was grown in 6 L of YPD to an A600 of ∼1.0, and cells were spheroplasted and lysed as described (Li and Hochstrasser 2000) except that cells were treated with 0.5 mg/mL Zymolyase 20T (ICN) instead of 100T and disrupted using an Emulsiflex C-5 (Avestin) instead of by sonication. Clarified lysate was bound in batch overnight at 4°C to ∼50 μL of anti-Flag M2 agarose (Sigma). Beads were washed several times with 50 mM Tris (pH 8.0), 1 M NaCl, 0.2% Triton X-100, and 2 mM N-ethylmaleimide. Proteins were eluted at room temperature using 6 M guanidine HCl, 100 mM Na·PO4, and 10 mM Tris (pH 8.0), and the eluate was bound in batch to ∼20 μL of Ni-NTA agarose (Qiagen) for 1 h at room temperature. The Ni-NTA beads were washed several times with 8 M urea, 50 mM Na·PO4 (pH 8.0), and 0.2% Triton X-100 and were eluted with a small volume of the same buffer containing 250 mM imidazole. Proteins were TCA precipitated, separated by SDS-PAGE, and stained with Coomassie Blue, and bands were excised.
SUMO identification by MS/MS
Based on the predicted C-terminal proteolytic products of SUMO by endoproteinase GluC (QIGG) and trypsin (EQIGG), excised protein bands were split in half and subjected to in gel reduction, carboxyamidomethylation, and separate digestion with GluC and trypsin. The two digests were pooled immediately before injection. Peptide sequences were determined using a 75-μm reverse-phase microcolumn terminating in a custom nanoelectrospray source (New Objective) directly coupled to an LCQ DECA XP Plus linear quadrupole ion trap mass spectrometer (Thermo Electron). The instrument was operated in data-dependent mode fragmenting (relative collision energy, 30%; isolation width, 2.5 Da; dynamic exclusion) on the four most abundant ions in each survey scan. Preliminary sequencing of peptides was facilitated by NCBI nr database correlation with the algorithm SEQUEST (Eng et al. 1994) and an in-house spectrum review workbench, FuzzyIons, and GraphMod, a program for analyzing the position(s) of a modification within a peptide sequence for a given MS/MS spectrum. All spectra were manually inspected for completeness of ion assignments and intensity-based signatures (e.g., neutral loss[es], proline ions, etc.).
Three peptide spectra within histone H2B defined acetylation and sumoylation at K6 or K7 or both. The N-terminal tryptic peptide Ac-SSAAEKKPASK unambiguously defined the presence of an acetylation at Lys6. The GluC-derived peptide KKPASKAPAE was observed with two independent MS/MS spectra. In one, the data supported either acetylated K6 or K7. A second spectrum for this peptide revealed sumoylation at either of these lysines. Thus, the population was a mix of both acetylation and sumoylation, but the modifications were never observed in a single peptide MS/MS spectrum at the same time.
IP and Western blot analysis
Cells (50 mL/IP) were grown to mid-log phase, and pellets were washed once with 1 mL of 20% TCA and frozen. For IP, cells were resuspended in 1 mL of 20% TCA and lysed in a mini bead beater by three repeats of 1.5 min with 2-min intervals on ice. The lysate was separated from beads and spun down for 10 min (13,000 rpm, 4°C). TCA was aspirated, and the pellet was resuspended in Laemmli buffer (50 mM Tris at pH 6.8, 2% SDS, 10% glycerol, 2% β-mercaptoethanol) and boiled for 3 min. Cells were spun for 10 min at room temperature (13,000 rpm), and the supernatant was used for IP. Typically, 100 μL were added to 400 μL of IP buffer (50 mM Tris at pH 7.4, 150 mM NaCl, 0.5% NP-40), and 60 μL of anti-Flag M2-agarose beads from mouse (Sigma) were added to the mixture. IP was allowed to continue for 6 h to overnight at 4°C. Beads were washed three times with IP buffer and eluted 6 h in 40 μL IP buffer containing 2.5 μg of 3xFlag peptide (Sigma). Normally, 8 μL of the eluate was loaded for WB analysis. The SUMO antibody has been described (Johnson and Blobel 1999).
In vivo expression analysis
RNA was isolated and DNase treated using RNeasy kit (Qiagen) according to the manufacturer’s instructions. cDNA was prepared (TaqMan Reverse Transcriptase, Roche) and RT–PCR was carried out in ABI PRISM 700 sequence detection system (Applied Biosystems) using either ACT1 or 18S rDNA for normalization. Typically, 3 mL of cells (in mid-log phase) were used per data point. Results (average of duplicate or triplicate data points on plate) are representative of two or three independent experiments.
ChIP experiments and chromatin fractionation
ChIP was performed as described previously (Henry et al. 2003) except that cells (A600 of ∼0.6–0.8) were cross-linked with 1% (v/v) formaldehyde for 5 min. Typically, 10 mg total cellular protein was used for the ChDIP, and 1 mg for normal ChIP analysis. For normal ChIP, following lysis and sonication as described, cells were precleared with protein A sepharose (Amersham) for 1 h prior to overnight precipitation with 1 μL/mg anti acetyl H3 antibody (Upstate Biotechnology) or anti acetyl 16 H2B (Upstate Biotechnology). For ChDIP after overnight precipitation (in IP buffer lacking EDTA), Flag elution was performed for 4–6 h, and 90% of the eluate was incubated with 1 column volume (cv) of Ni-NTA Agarose (Qiagen) for 1 h at 4°C. The slurry was loaded on a small yellow Bio-Rad column, washed with 4 cv of buffer A (100 mM sodium phosphate at pH 7.5, 150 mM NaCl, 30 mM imidazole) and eluted with 2 cv of buffer B (100 mM sodium phosphate at pH 7.5, 150 mM NaCl, 250 mM imidazole). All samples were eluted in 100 μL. Samples were de-cross-linked followed by DNA purification with a PCR purification kit (Qiagen). All samples were diluted 1:5 prior to real time PCR analysis. Input was further diluted 1:6 and 10% Flag input was diluted 1:3. Results were normalized to 10% Flag input in strains carrying the Flag-histone plasmid. In the control ChDIP experiments comparing the wild-type Flag-H2B strain with strains containing either no Flag tag or the Flag-H2B substitution mutant at K6/7/16/17, values were normalized to total input.
Chromatin fractionation was as described (Liang and Stillman 1997) except that NEM was added to buffers at a final concentration of 15 or 20 mM.
Acknowledgments
We thank J.A. Iniguez-Lluhi and G. Moore for valuable discussions. We acknowledge J. Dorsey, T. Sasaki, and R. Heller for technical assistance. Research was supported by grants from NIH (GM55360 and RR020839 to S.L.B., GM62268 to E.S.J., and GM60464 to P.B.M.).
Footnotes
References
- Aparicio O.M., Billington B.L., Gottschling D.E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Berger S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Biggins S., Bhalla N., Chang A., Smith D.L., Murray A.W. Genes involved in sister chromatid separation and segregation in the budding yeast Saccharomyces cerevisiae. Genetics. 2001;159:453–470. doi: 10.1093/genetics/159.2.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreault A.A., Cronier D., Selleck W., Lacoste N., Utley R.T., Allard S., Savard J., Lane W.S., Tan S., Cote J. Yeast enhancer of polycomb defines global Esa1-dependent acetylation of chromatin. Genes & Dev. 2003;17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun H., Koop R., Ertmer A., Nacht S., Suske G. Transcription factor Sp3 is regulated by acetylation. Nucleic Acids Res. 2001;29:4994–5000. doi: 10.1093/nar/29.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J.E., Zhou J., Ranalli T., Kobayashi R., Edmondson D.G., Roth S.Y., Allis C.D. Tetrahymena histone acetyltransferase A: A transcriptional co-activator linking gene expression to histone acetylation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Bryk M., Briggs S.D., Strahl B.D., Curcio M.J., Allis C.D., Winston F. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr. Biol. 2002;12:165–170. doi: 10.1016/s0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- Bylebyl G.R., Belichenko I., Johnson E.S. The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J. Biol. Chem. 2003;278:44113–44120. doi: 10.1074/jbc.M308357200. [DOI] [PubMed] [Google Scholar]
- Cheng J., Wang D., Wang Z., Yeh E.T. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol. Cell. Biol. 2004;24:6021–6028. doi: 10.1128/MCB.24.13.6021-6028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chupreta S., Holmstrom S., Subramanian L., Iniguez-Lluhi J.A. A small conserved surface in SUMO is the critical structural determinant of its transcriptional inhibitory properties. Mol. Cell. Biol. 2005;25:4272–4282. doi: 10.1128/MCB.25.10.4272-4282.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo R., Boggio R., Seiser C., Draetta G.F., Chiocca S. The adenovirus protein Gam1 interferes with sumoylation of histone deacetylase 1. EMBO Rep. 2002;3:1062–1068. doi: 10.1093/embo-reports/kvf213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David G., Neptune M.A., DePinho R.A. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 2002;277:23658–23663. doi: 10.1074/jbc.M203690200. [DOI] [PubMed] [Google Scholar]
- Desterro J.M., Rodriguez M.S., Hay R.T. SUMO-1 modification of IκBα inhibits NF-κB activation. Mol. Cell. 1998;2:233–239. doi: 10.1016/s1097-2765(00)80133-1. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson J.E., Zeng L., He C., Aggarwal A.K., Zhou M.M. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dohmen R.J., Stappen R., McGrath J.P., Forrova H., Kolarov J., Goffeau A., Varshavsky A. An essential yeast gene encoding a homolog of ubiquitin-activating enzyme. J. Biol. Chem. 1995;270:18099–18109. doi: 10.1074/jbc.270.30.18099. [DOI] [PubMed] [Google Scholar]
- Emre N.C., Ingvarsdottir K., Wyce A., Wood A., Krogan N.J., Henry K.W., Li K., Marmorstein R., Greenblatt J.F., Shilatifard A., et al. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol. Cell. 2005;17:585–594. doi: 10.1016/j.molcel.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Eng J., McCormack A.L., Yates III J.R. An approach to correlate tandem mass specral data of peptides with amino acid sequences in a protein database. J. Am. Mass Spectrum. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- Gill G. Post-translational modification by the small ubiquitin-related modifier SUMO has big effects on transcription factor activity. Curr. Opin. Genet. Dev. 2003;13:108–113. doi: 10.1016/s0959-437x(03)00021-2. [DOI] [PubMed] [Google Scholar]
- Gregoire S., Yang X.J. Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell. Biol. 2005;25:2273–2287. doi: 10.1128/MCB.25.6.2273-2287.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal S.I., Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- Guillemette B., Bataille A.R., Gevry N., Adam M., Blanchette M., Robert F., Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannich J.T., Lewis A., Kroetz M.B., Li S.J., Heide H., Emili A., Hochstrasser M. Defining the SUMO-modified proteome by multiple approaches in Saccharomyces cerevisiae. J. Biol. Chem. 2005;280:4102–4110. doi: 10.1074/jbc.M413209200. [DOI] [PubMed] [Google Scholar]
- Hay R.T. SUMO: A history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Henry K.W., Wyce A., Lo W.S., Duggan L.J., Emre N.C., Kao C.F., Pillus L., Shilatifard A., Osley M.A., Berger S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn J.N., Bortvin A.L., Ricupero-Hovasse S.L., Winston F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol. Cell. Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- Holmstrom S., Van Antwerp M.E., Iniguez-Lluhi J.A. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc. Natl. Acad. Sci. 2003;100:15758–15763. doi: 10.1073/pnas.2136933100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- Johnson E.S., Blobel G. Cell cycle-regulated attachment of the ubiquitin-related protein SUMO to the yeast septins. J. Cell Biol. 1999;147:981–994. doi: 10.1083/jcb.147.5.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., Gupta A.A. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- Kao C.F., Hillyer C., Tsukuda T., Henry K., Berger S., Osley M.A. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes & Dev. 2004;18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsh O., Seeler J.S., Pichler A., Gast A., Muller S., Miska E., Mathieu M., Harel-Bellan A., Kouzarides T., Melchior F., et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]
- Lachner M., O’Carroll D., Rea S., Mechtler K., Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Li S.J., Hochstrasser M. The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol. Cell. Biol. 2000;20:2367–2377. doi: 10.1128/mcb.20.7.2367-2377.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C., Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes & Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann R., Grunstein M. Histone H3 N-terminal mutations allow hyperactivation of the yeast GAL1 gene in vivo. EMBO J. 1992;11:3297–3306. doi: 10.1002/j.1460-2075.1992.tb05408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manza L.L., Codreanu S.G., Stamer S.L., Smith D.L., Wells K.S., Roberts R.L., Liebler D.C. Global shifts in protein sumoylation in response to electrophile and oxidative stress. Chem. Res. Toxicol. 2004;17:1706–1715. doi: 10.1021/tx049767l. [DOI] [PubMed] [Google Scholar]
- Martin C., Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- McKittrick E., Gafken P.R., Ahmad K., Henikoff S. Histone H3.3 is enriched in covalent modifications associated with active chromatin. Proc. Natl. Acad. Sci. 2004;101:1525–1530. doi: 10.1073/pnas.0308092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini M.D., Wu M., Madhani H.D. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Ng H.H., Ciccone D.N., Morshead K.B., Oettinger M.A., Struhl K. Lysine-79 of histone H3 is hypomethylated at silenced loci in yeast and mammalian cells: A potential mechanism for position-effect variegation. Proc. Natl. Acad. Sci. 2003;100:1820–1825. doi: 10.1073/pnas.0437846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi A., Gibson J., Gregor I., Schatz G. Import of proteins into mitochondria: The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J. Biol. Chem. 1982;257:13042–13047. [PubMed] [Google Scholar]
- Panse V.G., Hardeland U., Werner T., Kuster B., Hurt E. A proteome-wide approach identifies sumoylated substrate proteins in yeast. J. Biol. Chem. 2004;279:41346–41351. doi: 10.1074/jbc.M407950200. [DOI] [PubMed] [Google Scholar]
- Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robzyk K., Recht J., Osley M.A. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–504. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- Ross S., Best J.L., Zon L.I., Gill G. SUMO-1 modification represses Sp3 transcriptional activation and modulates its subnuclear localization. Mol. Cell. 2002;10:831–842. doi: 10.1016/s1097-2765(02)00682-2. [DOI] [PubMed] [Google Scholar]
- Rusche L.N., Kirchmaier A.L., Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Salghetti S.E., Caudy A.A., Chenoweth J.G., Tansey W.P. Regulation of transcriptional activation domain function by ubiquitin. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- Santisteban M.S., Kalashnikova T., Smith M.M. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- Schwarz S.E., Matuschewski K., Liakopoulos D., Scheffner M., Jentsch S. The ubiquitin-like proteins SMT3 and SUMO-1 are conjugated by the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. 1998;95:560–564. doi: 10.1073/pnas.95.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seufert W., Futcher B., Jentsch S. Role of a ubiquitin-conjugating enzyme in degradation of S- and M-phase cyclins. Nature. 1995;373:78–81. doi: 10.1038/373078a0. [DOI] [PubMed] [Google Scholar]
- Shiio Y., Eisenman R.N. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. 2003;100:13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., Illes K., Lukacsovich T., Zhu Y.Z., Cattaneo E., et al. SUMO modification of Huntingtin and Huntington’s disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Allis C.D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Toh-e A., Kikuchi Y. A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene. 2001;275:223–231. doi: 10.1016/s0378-1119(01)00662-x. [DOI] [PubMed] [Google Scholar]
- Taunton J., Hassig C.A., Schreiber S.L. A mammalian histone deacetylase related to the yeast transcription regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Verdin E., Dequiedt F., Kasler H.G. Class II histone deacetylases: Versatile regulators. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- Verger A., Perdomo J., Crossley M. Modification with SUMO: A role in transcriptional regulation. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M., Wu J., Suka N., Grunstein M. Global histone acetylation and deacetylation in yeast. Nature. 2000;408:495–498. doi: 10.1038/35044127. [DOI] [PubMed] [Google Scholar]
- White C.L., Suto R.K., Luger K. Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Johnson E.S., Reed S.I., Yates J.R., III Global analysis of protein sumoylation in Saccharomyces cerevisiae. J. Biol. Chem. 2004;279:45662–45668. doi: 10.1074/jbc.M409203200. [DOI] [PubMed] [Google Scholar]
- Yang S.H., Sharrocks A.D. SUMO promotes HDAC-mediated transcriptional repression. Mol. Cell. 2004;13:611–617. doi: 10.1016/s1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- Zhao X., Blobel G. A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Ryan J.J., Zhou H. Global analyses of sumoylated proteins in Saccharomyces cerevisiae: Induction of protein sumoylation by cellular stresses. J. Biol. Chem. 2004;279:32262–32268. doi: 10.1074/jbc.M404173200. [DOI] [PMC free article] [PubMed] [Google Scholar]