Regulation of homologous recombination in eukaryotes (original) (raw)
. Author manuscript; available in PMC: 2014 Jul 29.
Abstract
Homologous recombination is required for accurate chromosome segregation during the first meiotic division and constitutes a key repair and tolerance pathway for complex DNA damage including DNA double-stranded breaks, interstrand crosslinks, and DNA gaps. In addition, recombination and replication are inextricably linked, as recombination recovers stalled and broken replication forks enabling the evolution of larger genomes/replicons. Defects in recombination lead to genomic instability and elevated cancer predisposition, demonstrating a clear cellular need for active recombination. However, recombination can also lead to genome rearrangements. Unrestrained recombination causes undesired endpoints (translocation, deletion, inversion) and the accumulation of toxic recombination intermediates. Evidently, homologous recombination must be carefully regulated to match specific cellular needs. Here we review the mechanistic stages and proteins in recombination that are subject to regulation and suggest that recombination achieves flexibility and robustness by proceeding through meta-stable, reversible intermediates.
Keywords: DNA damage response, DNA repair, phosphorylation, sumoylation, ubiquitylation
INTRODUCTION
Homologous recombination (HR) is a key pathway to maintain genomic integrity between generations (meiosis) and during ontogenic development in a single organism (DNA repair). A typical diploid human cell needs to maintain about 6×109 base pairs in the correct sequence and chromosomal organization, a formidable task that is usually performed with no errors from one somatic cell generation to the next (42). On one hand, recombination is required for the repair or tolerance of DNA damage and the recovery of stalled or broken replication forks (91). On the other hand, recombination also leads to gross chromosomal rearrangements and potentially lethal intermediates (85). How does the cell achieve the balance between too little and too much recombination? There must be regulation and the answer will be complex depending on the organism, cell type, cell cycle stage, chromosomal region, as well as the type and level of genotoxic stress.
HR in meiosis is subject to specific regulation that targets recombination events to homologs, establishing crossover outcomes to assist in meiotic chromosome segregation (44, 73). Meiosis-specific aspects of recombination are covered in other contributions to this issue of Annual Reviews in Genetics (RELATED RESOURCES: Lichten & Sourijan, Hassold, Borts & deMassy). We only include examples of meiotic regulation of HR proteins that may be applicable also to somatic cells. In addition, a dedicated chapter on the RecQ helicases will provide a much more comprehensive discussion of this important class of proteins than can be achieved here (RELATED RESOURCE: Rothstein, Bernstein & Gangloff).
Here, we will review how recombinational DNA repair is regulated in somatic (vegetative) cells by focusing on the mechanistic phases of recombination (Figure 1) and the proteins that execute them (Table 1), identifying key regulatory transitions and mechanisms. We will elaborate that HR is modulated by multiple levels of positive and, primarily, negative regulation. Mechanisms that are seemingly anti-recombinogenic appear to be integral to the HR pathway. We suggest that HR gains flexibility and robustness by proceeding through reversible, meta-stable intermediates. Due to space limitations we defer the reader to recent reviews on how modulation of the DNA substrate affects HR (115, 156) including at specific nuclear territories such as telomeres (RELATED RESOURCE: Cooper & Greenwood) and the nucleolus (94). While transcriptional regulation is at the heart of the DNA damage (SOS) response in bacteria (34), there is little evidence of biologically significant transcriptional induction of HR genes by DNA damage in eukaryotes (29, 32, 82).
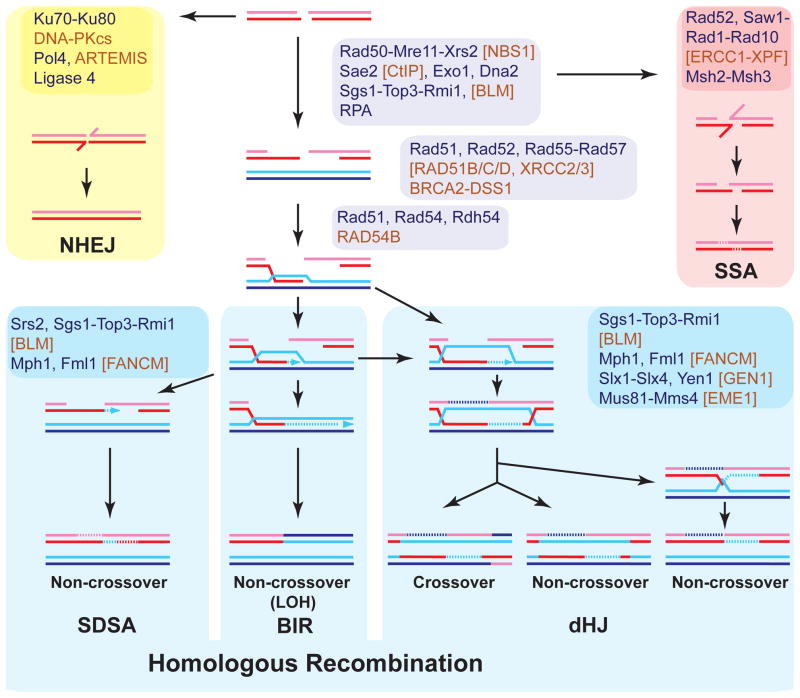
Figure 1. Pathways of DSB repair.
Protein names refer to the budding yeast S. cerevisiae (black). Where different in human, names (brown) are given in brackets. For proteins without yeast homolog brackets for human proteins are omitted. Broken lines indicate new DNA synthesis and stretches of hDNA that upon MMR can lead to gene conversion.
Table 1.
Post-translational modifications and their effects on proteins involved in homologous recombination.
| Protein | Organism | PTM | Function and PTM effect | Reference |
|---|---|---|---|---|
| BLM | H. sapiens | Multiple roles in DNA damage signaling and HR | ||
| SUMO | BLM-K317/331-SUMO required for full activity and Rad51 focus formation after HU treatment | (46, 117) | ||
| BRCA1 | H. sapiens | E3 ligase involved in HR and NHEJ | ||
| SUMO | PIAS1/4-dependent SUMO of BRCA1-K119 enhances BRCA1 UBI ligase activity | (103) | ||
| BRCA2 | H. sapiens | RAD51 filament formation | ||
| PO4 | CDK-mediated PO4 of S3291 inhibits RAD51 interaction of C-terminal RAD51 interaction site | (47) | ||
| CtIP | H. sapiens | DSB resection | ||
| PO4 | CDK-consensus site T847 required for CtIP activity (see also Sae2) | (72) | ||
| PO4 | CDK-consensus site S237 required for BRCA1 binding and HR | (169) | ||
| Exo1 | S. cerevisiae | 5′-3′ DNA exonuclease | ||
| PO4 | Rad53-mediated PO4 of S372, 567, 587, 692 inhibits Exo1 activity | (101) | ||
| hEXO1 | 5′-3′ DNA exonuclease | |||
| PO4 | ATR-dep. PO4 leads to degradation | (45) | ||
| Nej1 | S. cerevisiae | DNA ligase 4 co-factor | ||
| PO4 | Dun1-dependent PO4 of Nej1-S297/8 enhances binding to Srs2 anti-recombinase favoring NHEJ/SSA over HR | (26) | ||
| PCNA | S. cerevisiae | Processivity clamp | (119, 121) | |
| SUMO | PCNA-K164 (K127)-SUMO recruits Srs2 anti-recombinase | |||
| UBI | PCNA-K164-UBI prevents SUMO, anti-recombination effect by favoring TLS or fork regression | |||
| RAD51 | H. sapiens | Homology search and DNA strand invasion | ||
| PO4 | CHK1-dep. PO4 on T309 may be required for RAD51 focus formation | (138) | ||
| Rad52 | S. cerevisiae | Rad51 filament formation, SSA | ||
| SUMO | Sumoylation of K10,11, 2201 affects protein stability and intranuclear localization | (125) (149) | ||
| Rad54 | S. cerevisiae | Cofactor for Rad51 | ||
| PO4 | Mek1-mediated PO4 at T132 inhibits Rad51 interaction during meiosis | (112) | ||
| Rhp54 | S. pombe | Co-factor for Rad51 | ||
| UBI | APC/C mediated ubiquitylation of Rhp54-K26 leads to proteolysis in G1 cells | (152) | ||
| Rad55 | S. cerevisiae | Rad51 filament assembly/stability | ||
| PO4 | Rad55-S2,8,14 PO4 required for full Rad55 activity | (66) | ||
| Sae2 | S. cerevisiae | DSB resection | ||
| PO4 | CDK-mediated PO4 at S267 required for Sae2 activity (see also CtIP) | (71) |
MECHANISM OF HOMOLOGOUS RECOMBINATION
Significant strides have been made in identifying the proteins that catalyze HR in eukaryotes and defining their mechanisms of action (67, 87, 126, 159). HR can be conceptually divided into three stages (pre-synapsis, synapsis, post-synapsis), and we briefly discuss the principal proteins involved (Figure 1). In pre-synapsis, the DNA damage is processed to form an extended region of ssDNA, which is bound by the ssDNA binding protein RPA. For double-stranded DNA breaks (DSBs), this step involves a surprising complexity of four nuclease (Mre11-Rad50-Xrs2/NBS1 [M-R-X/N], Exo1, Dna2, Sae2/CtIP) and a helicase activity (Sgs1/BLM) (99). Binding of RPA eliminates secondary structures in ssDNA, which is needed for competent Rad51 filaments to assemble. However, RPA bound to ssDNA also forms a kinetic barrier against Rad51 filament assembly, necessitating so-called mediator proteins to allow timely Rad51 filament formation on RPA-covered ssDNA. Three different classes of mediators have been described, but their mechanisms of action and the interplay between them is poorly understood. The Rad51 paralogs constitute a first group and comprise four members (Rad55-Rad57, Shu1-Psy3) in Saccharomyces cerevisiae and five in mammals (RAD51B/C/D, XRCC2/3). These proteins share the RecA core with Rad51, but fail to form extensive filaments on DNA and are unable to perform the range of DNA pairing reactions catalyzed by Rad51. A second class is typified by the budding yeast Rad52 protein, which performs two independent roles: its mediator function, and a second, later function in strand annealing of RPA-bound ssDNA. A third class of mediator proteins is represented by BRCA2, the human breast and ovarian cancer tumor suppressor protein. The presence of ssDNA binding motifs (OB-folds), a dsDNA binding motif (tower domain), and a number of Rad51 binding sites, suggested that BRCA2 targets RAD51 filament nucleation to the dsDNA junction at the resected end (165).
During synapsis, the Rad51 filament performs homology search and DNA strand invasion, generating the D-loop where the invading strand primes DNA synthesis. Besides a function in stabilizing the Rad51 filament and enhancing D-loop formation by Rad51, the Rad54 motor protein is required for the transition from DNA strand invasion to DNA synthesis by dissociating Rad51 from heteroduplex DNA (hDNA) (68).
Finally in post-synapsis, the three sub-pathways of HR are distinguished (Figure 1), each with specific enzymatic requirements that have been only partially defined (67, 87, 126, 159). In the absence of a second end, the D-loop may become a full-fledged replication fork in a process termed break-induced replication (BIR). While this process restores the integrity of the chromosome, it can lead to loss-of-heterozygosity of all genetic information distal to the DSB. In the presence of a second end, the predominant pathway for DSB repair in somatic cells appears to be synthesis-dependent strand annealing (SDSA), where the extended D-loop is reversed, leading to annealing of the newly synthesized strand with the resected strand of the second end (Figure 1) (120). This pathway inherently avoids crossovers, which reduces the potential for genomic rearrangements. While generation of crossovers by double Holliday junction (dHJ) formation is the purpose of meiotic recombination, recombinational DNA repair in somatic cells is rarely associated with crossovers. Only recently have dHJs been identified as an intermediate in recombinational DNA repair in vegetative (somatic) cells (25). dHJ formation involves capture of the second end, a process that is actively blocked by Rad51 protein in vitro, suggesting an inherent mechanistic bias towards SDSA (164). The dHJ intermediate could be resolved by endonucleases in a manner described for the bacterial RuvC protein into crossover or non-crossover products (159), but the exact mechanisms and identity of proteins involved remain under debate (see Figure 1). Alternatively, dHJs can be dissolved by a complex mechanism involving a RecQ-like DNA motor protein (Sgs1/BLM), topoisomerase 3, and cofactors. The two junctions are migrated towards each other, leading to a hemi-catenane that is eliminated by Topo3. Genetically, the end point of dissolution is always a non-crossover, avoiding the potential for rearrangements associated with crossovers.
REGULATORY CONTROL POINTS AND IRREVERSIBLE COMMITMENTS
Paradigmatic studies of cell cycle control showed that regulatory molecular processes are either reversible (e.g. cyclin-dependent kinase (CDK) phosphorylation-dephosphorylation) or irreversible (e.g. cyclin proteolysis, cohesin cleavage) (106). It is attractive to apply these concepts to the regulation of HR. An increasing number of reversible post-translational modifications on HR proteins, such as phosphorylation, ubiquitylation, and sumoylation, are being identified (14, 16) (Table 1). The existence of phospho-, ubiquitin-, and SUMO-specific protein interaction motifs makes it likely that many post-translational modifications lead to novel protein interactions (131). First examples of irreversible proteolytic control of HR proteins (Rad52, Rhp54; Table 1) have been elaborated (125, 152). Several key HR intermediates, such as the Rad51-ssDNA filament, the D-loop, and the dHJ, constitute reversible intermediates and hence likely regulatory control points. Exonucleolytic degradation of DNA and endonucleolytic processing of DNA junction intermediates could be viewed as irreversible commitments (Figure 1). Below, we provide a detailed discussion of regulatory targets and processes, as well as their mechanistic consequences for HR.
DSB REPAIR: COMPETITION BETWEEN HR, SSA AND NHEJ
HR, single-strand annealing (SSA) and non-homologous end joining (NHEJ) are the principal pathways for DSB repair and the balance between them depends on the species, cell type, cell cycle stage, and type of DNA damage. NHEJ is a specialized ligation reaction with varying accuracy that depends on the end structure (Figure 1) (135). SSA is a homology-directed DNA repair pathway between repeated sequences that involves reannealing of RPA-covered ssDNA by the Rad52 protein (Figure 1) (87). SSA does not involve DNA strand invasion and is therefore independent of Rad51. This process leads to deletion of the interstitial DNA and one of the repeated homologous sequences.
How is the balance between NHEJ, SSA, and HR regulated? While SSA and NHEJ can occur intrachromosomally (Figure 1), HR is a template-dependent process and studies in S. cerevisiae have demonstrated that the sister chromatid is the preferred template over a homolog, when given the choice (80). Sister chromatid cohesion likely provides the mechanistic underpinning for this preference (108). This could suggest that HR is entirely restricted to the S- and G2 phases of the cell cycle, but HR has also been demonstrated to occur in the G1 phase of budding yeast, using the homolog as a template (48). HR in G1 can only occur in diploid cells, and most organisms, including budding yeast, are naturally diplontic. The fission yeast Schizosaccharomyces pombe, on the other hand, is a true haplont, rationalizing why in this organism HR is down-regulated in the G1 phase by targeting Rhp54 (fission yeast Rad54) for ubiquitin-mediated degradation (152).
In budding yeast, the mating-type locus provides an example of complex regulation of HR in response to ploidy (155). Haploid yeast cells are either of the a or alpha mating type, whereas diploid cells are a/alpha and contain the Mata1-Matα2 co-repressor that turns off haploid-specific genes and induces diploid-specific genes. Early radiobiological studies established that a/alpha diploid cells are more radiation-resistant than haploid cells or a/a or alpha/alpha diploid cells (105). One mechanism to explain this observation is the transcriptional repression of Nej1, a co-factor for the principal NHEJ ligase, DNA ligase 4, by the Mata1-Matα2 repressor. This shifts the balance from NHEJ to HR in diploid cells (5, 51, 89). Targeting Nej1 results in a bi-fold regulation, as Nej1 is required for NHEJ and recruits the Srs2 anti-recombinase (see below) to resected ends to favor NHEJ or SSA over HR (26).
DSB resection is a key commitment step for homology-directed repair as both SSA and HR depend on ssDNA. As discussed in detail below, DSB resection is highly regulated and low in the G1 phase, favoring NHEJ over HR and SSA (52). At least in budding and fission yeast, the NHEJ DNA end-binding factors Ku70-Ku80 inhibit DSB resection (88, 148). Interestingly, M-R-X/N complex, BRCA1, DNA PKcs, and ATM function in both NHEJ and HR (135). The M-R-X/N complex and BRCA1 are connected to resection, providing a possible regulatory target. Using elegant substrate design, it was shown that SSA and HR compete for the repair of DSBs in budding yeast and mammals (77, 140). Since SSA requires sufficient resection to expose direct repeats as single stranded DNA (Figure 1), the balance is expected to be highly locus- and assay-dependent.
How is the balance between the sub-pathways of HR (BIR, SDSA, dHJ) regulated in DSB repair?
BIR, SDSA, and the dHJ sub-pathways of HR (Figure 1) lead to repair of a DSB but are associated with different genetic consequences. BIR can lead to loss-of-heterozygosity distal to the break site, introducing the threat to expose recessive mutations with detrimental consequences. dHJ formation has the potential to create genomic rearrangements, if HR occurs in non-allelic sites. By this logic, SDSA is expected to be the favored sub-pathway, and the experimental evidence suggests that this is the case (120).
Experiments in budding yeast demonstrated that the SDSA pathway outcompetes BIR in mitotic DSB repair, because BIR is a much slower process (95). Using an ingenious experimental setup, Haber and colleagues demonstrated that BIR is suppressed at the DNA synthesis step for over 5 hrs after DSB formation (78, 142). This suppression required Sgs1, but surprisingly not its helicase activity (78). Mec1 kinase, the master regulator of the DNA damage response (DDR) in budding yeast (Figure 3) is not required to suppress BIR. Close proximity of the second end suppresses BIR (78), but it is unclear how this is communicated to the D-loop to prevent replication fork assembly. Maybe this involves the end-tethering function of the M-R-X/N complex (39)? Unlike meiotic HR, dHJs are formed only at low levels during mitotic DSB repair (25), consistent with the low association of mitotic DSB repair with a crossover outcome (75).
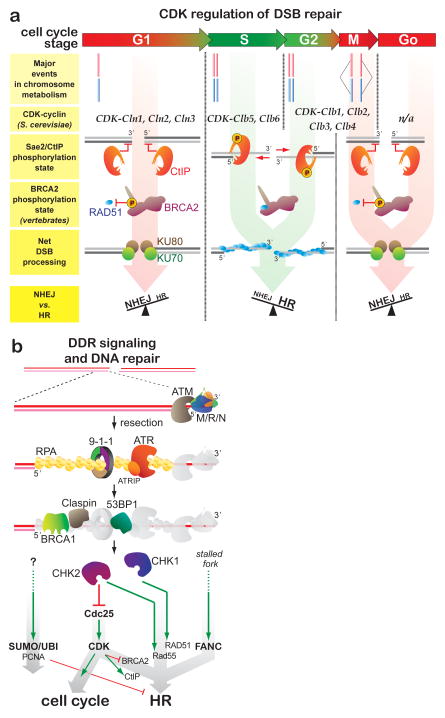
Figure 3. Homologous recombination is regulated by cell cycle control and DNA damage signaling.
(a) The cell cycle controls the competition between NHEJ and HR in DSB repair. Cell cycle stages are color-coded: Red, HR is least active and green, HR is most active. Cdc28 is the sole CDK responsible for cell cycle progression in S. cerevisiae, and partners with the indicated cyclins. In mammals, six CDKs drive cell cycle progression and their relative importance varies in different tissue types. (b) The DDR results in HR activation and inhibition of cell cycle progression. The relationship between the DDR and the FANC pathway as well as PCNA sumoylation/ubiquitylation is poorly understood.
DNA GAP REPAIR: COMPETITION BETWEEN HR, TLS, AND FORK REGRESSION
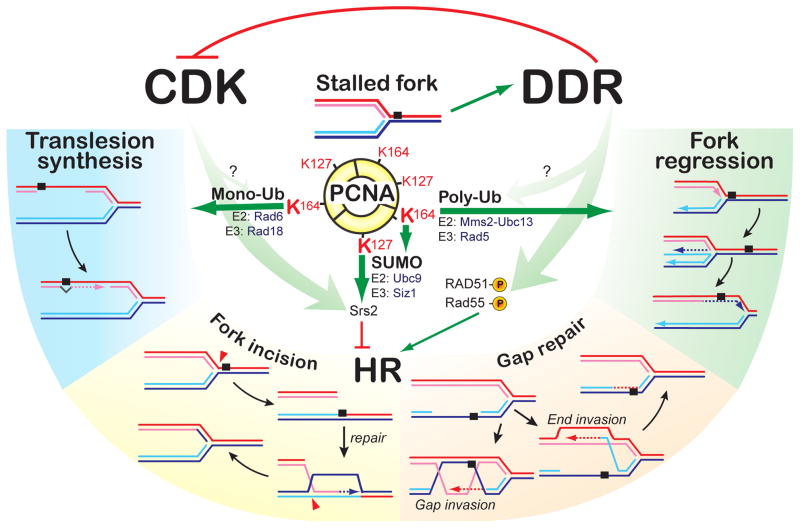
Replication fork stalling leads to gaps resulting from down stream reinitiation by DNA polymerases on the leading and lagging strands (16, 65, 93). Stalled forks and gaps can be recovered by different pathways, including translesion synthesis (TLS), template switching by fork regression, or HR (16) (Figure 2). While the accuracy of TLS is lesion- and polymerase-dependent (123), template-switching by fork regression and HR are inherently highly accurate. TLS is favored by mono-ubiquitination of PCNA on K164 by the Rad6-Rad18 E2-E3 complex (Figure 2), which enhances the intrinsic affinity of Y-family TLS polymerases (Pol eta, iota, kappa) for PCNA through their ubiquitin binding motifs (123). Subsequent poly-ubiquitylation of PCNA by Ubc13-Mms2 (E2) and Rad5 (E3) controls fork regression by a mechanism that is not understood (123). Alternatively, K164 (and K127) can be sumoylated by Ubc9, which leads to recruitment of the Srs2 anti-recombinase through its SUMO binding motif (119, 121). As discussed in more detail below, Srs2 dissociates Rad51 from ssDNA, antagonizing Rad51-ssDNA filament formation (86, 157). It is unclear whether PCNA ubiquitylation and sumoylation can co-exist in a hetero-trimeric PCNA ring, and the relationship between HR and these ubiquitylation and sumoylation pathways (Figure 2) is still poorly understood (17, 18).
Figure 2. Pathways and regulation at stalled replication forks.
PCNA modification regulates the choice of competing pathways for stalled replication fork recovery. A stalled fork triggers the DDR, which directly activates HR. The relationship between the DDR and cell cycle control to PCNA sumoylation/ubiquitylation has not been determined yet.
How is the balance between TLS, fork regression, and HR regulated? Genetic evidence in yeast favors the model that TLS and fork regression are primary pathways. At least initially, HR is actively repressed, but the sensitivity of HR mutants to fork stalling agents suggests that this inhibition is temporary. Mutations in RAD6 or RAD18 disable TLS and fork regression, leading to severe DNA damage sensitivity. An additional mutation in SRS2 (_s_uppressor of _r_ad _s_ix) suppresses the sensitivity to a very significant degree by relieving the inhibition of HR (2, 129). These data suggest that Rad6-Rad18 binding to RPA-covered ssDNA (37) is kinetically favored over Rad51 filament formation. Possibly, PCNA sumoylation marks a later phase, where Srs2 actively removes Rad51 filaments. What regulates PCNA ubiquitylation or sumoylation and whether DDR signaling is involved remains to be determined (Figure 2).
SIGNALING BY THE CELL CYCLE MACHINERY AND THE DNA DAMAGE RESPONSE
Two signaling systems intersect in the control of HR: the cell cycle control machinery and the DDR (Figure 3) (16). In S. cerevisiae, the Cdc28 CDK drives directional progress through the cell cycle, dependent on the expression of stage-specific cyclins that modulate CDK activity and impart substrate specificity (96, 161). As discussed below, CDK phosphorylates HR proteins to positively and negatively regulate HR. The availability of sister chromatids largely determines whether HR is a primary pathway, explaining why HR is favored in the S and G2 but not in the G0, G1 or during M phases (Figure 3a). Stalled replication forks and DNA damage trigger the DDR signaling cascade that activates DNA repair and pauses cell cycle momentum (Figure 3b) (16). The key intermediate is RPA-bound ssDNA, which serves as a platform for DDR signaling, recruitment of ubiquitylation and, likely, sumoylation factors, as well as for Rad51 filament formation. DDR signaling is required for efficient DNA damage-induced HR (13, 104). In addition, the DDR affords time for HR to be completed by leading to a transient cell cycle arrest, which in most organisms, but not S. cerevisiae, is achieved by down-regulating CDK activity (113).
DSB RESECTION AND ITS REGULATION
Resection of DNA DSB ends seems deceptively straightforward in principle, but in S. cerevisiae resection involves four nucleases (Mre11, Sae2, Exo1, Dna2), dependent on the specific chemical structure encountered at the DSB (hairpins, modified bases, covalent protein-DNA adducts) (99). A current view proposes that the Mre11 subunit of the M-R-X/N complex, recruited or supported by the endonuclease Sae2/CtIP, catalyzes initial end processing that results in the removal of about 50–100 nucleotides (99). Sae2/CtIP is thought to clip DNA ends in preparation for the more processive nucleases that catalyze the extended resection responsible for 3′-ssDNA tail generation (90). The 3′→5′ polarity of the Mre11 nuclease appears unsuited to conduct the 5′→3′ resection (153), but it could act as an endonuclease in this context. Extended resection is achieved by the 5′→3′ exonuclease activity of Exo1 or the helicase activity of Sgs1 in cooperation with the endonuclease activity of Dna2 (98, 170). How these options for extended DNA resection (Exo1 alone or Sgs1 with Dna2 or Exo1) or the extent of resection are regulated is unknown. In meiosis, the resulting 3′-tail of ssDNA is about 500 nt in length, whereas it has been argued that resection is more extensive in mitotic cells (170). Compounding the complexity associated with the collaboration of multiple nucleases to achieve end resection is the question of their regulation by post-translation modification.
Activation of DSB resection by CDK-mediated Sae2/CtIP phosphorylation
Repair of an endonuclease-mediated DSB by HR is repressed in haploid budding yeast cells in the G1 phase of the cell cycle because of limited DSB resection (7, 76). End resection is primarily regulated by CDK-dependent phosphorylation of the Sae2/CtIP nuclease (71, 72, 76) (Figure 3, Table 1), which determines whether a DSB is channeled into NHEJ or HR. The pivotal phosphorylation occurs at serine 267, located in one of three Sae2 CDK consensus sites (71). An endonuclease-mediated DSB at the MAT locus is poorly resected in an S. cerevisiae sae2-Δ mutant; a sae2 mutant in which serine 267 has been substituted with alanine (sae2-S267A) phenocopies the sae2-Δ strain for unresected DSB ends. In contrast, a Sae2 phosphomimic mutant in which serine 267 has been replaced with aspartic acid (sae2-S267E) is hypermorphic for DSB resection, sidestepping a requirement for CDK activity to sanction DSB resection.
These observations are mirrored by results from human cells, where CtIP, the human homolog of Sae2, is also required for DSB resection. Phosphorylation on threonine 847 is required for ssDNA generation and RPA phosphorylation in response to the topoisomerase I inhibitor camptothecin, laser-induced DNA damage, or ionizing radiation (IR) (72, 127). A transfected phosphomimic CtIP-T847E resects DSBs even after CDK inhibition, whereas the non-phosphorylatable CtIP-T847A mutant impairs resection (72). CDK phosphorylation of Sae2/CtIP therefore appears to be conserved in eukaryotes as a key switch in determining whether DSB ends are sanctioned for resection and HR. In addition to the conserved mechanism described for S. cerevisiae Sae2, human CtIP function also appears to be regulated by an additional CDK phosphorylation at serine 327, a modification that enhances CtIP interaction with the BRCT domain of BRCA1 and is critical for HR (167, 169). The function of BRCA1 in HR remains enigmatic. It is interesting to observe that BRCA1 is sumoylated by PIAS1/4 to enhance its ubiquitin ligase activity (Table 1, (55, 102)) and that CtIP appears to be one of its native ubiquitylation targets (168), implying a potential regulatory role of BRCA1 in resection.
Sae2-S267 is unlikely to be the exclusive target of CDK relevant to end resection. When CDK is inhibited, the sae2-S267E allele product is not quite sufficient for wild type resection levels (71). Mre11 and Xrs2 have CDK phosphorylation consensus sites, but no resection phenotype has been observed when these sites are mutated (76).
An additional level of control on Sae2 was uncovered by mutating Mec1/Tel1 consensus sites and showing that they were essential during meiotic recombination (147) and required for full Sae2 function during DNA repair in mitotic cells (12). In sum, two signaling pathways, the cell cycle control machinery and DDR signaling, converge on Sae2/CtIP to regulate end resection.
Inhibition of Exo1 activity by the DDR kinase Rad53
ssDNA is a defining intermediate of DSB processing in HR and also occurs at telomeres that are uncapped during end-replication in S-phase and in mutants (e.g. cdc13-1) that lose the protective T-loop and associated factors (RELATED RESOURCE: Cooper & Greenwood). In mutants that cause telomere uncapping, Exo1 is phosphorylated in a manner dependent on the DDR. Four serines in the Exo1 C-terminus (S372, S567, S587 and S692) are phosphorylated, presumably by Rad53, because Exo1 phosphorylation is absent in a rad53-K227A kinase-defective mutant (101). Unlike Sae2/CtIP activation by CDK, Exo1 phosphorylation by Rad53 limits extended resection of ssDNA at uncapped telomeres and consequently minimizes further activation of the DDR. Mutants in which all four serines are substituted by alanine result in hyperactivation of the DDR, as does overexpression of Exo1, suggesting that Rad53-dependent phosphorylation of Exo1 reduces its activity. The inhibition of Exo1 activity is not limited to pathological situations such as telomere uncapping, however; Exo1 is also phosphorylated in yku70-Δ mutant cells following bleomycin treatment (101). Repression of Exo1 activity by DDR signaling to avoid fork regression at stalled replication forks was also reported in budding yeast (Figure 2) (33). Another mechanism of negative regulation of EXO1 is observed in human cells challenged with the replication inhibitor hydroxyurea (HU): phosphorylation by ATR targets EXO1 for destruction (45). This may reflect a prohibition of resection at ssDNA gaps associated with stalled replication forks. These results suggest that Exo1 is not required for the generation of ssDNA to allow DDR signaling, but that fork regression and potentially HR may require more extensive stretches of ssDNA generated by Exo1.
Regulation of BLM by sumoylation
Biochemical and genetic evidence demonstrate an involvement of the RecQ helicase Sgs1 and human BLM in DSB resection (60, 98, 111, 170). BLM was found to be sumoylated on several lysine residues, and BLM lacking sumoylation on lysine K317 and K331 (Table 1) only partly complemented the genetic defects in BLM-deficient cells, suggesting that SUMO modification of these residues may exert positive regulation (46). Sgs1/BLM likely have several functions during HR (resection, joint reversal, dHJ dissolution) and in DDR signaling (RELATED RESOURCE: Rothstein, Bernstein & Gangloff). The observation that cells with SUMO-deficient BLM exhibit a defect in RAD51 focus formation after HU treatment (117) may suggest that sumoylation inhibits an early BLM function, possibly resection, congruent with the negative regulation of yeast and human Exo1 in response to replication fork stalling (45, 101).
In summary, post-translational modification of factors involved in DSB resection is paramount to the regulation of eukaryotic HR. CDK-dependent modification of Sae2/CtIP demonstrates that pathway choice for DSB repair depends to a large extent on commitment to resection.
THE RAD51 FILAMENT: A BALANCE BETWEEN FORMATION AND DISRUPTION
The Rad51-ssDNA filament performs the central function of HR, homology search and DNA strand exchange (Figure 1). Not surprisingly, this crucial role is reflected in an elaborate regulation of the balance between Rad51 filament formation and disruption.
CDK- and DDR-mediated phosphorylation of RPA
RPA functions at the nexus of all DNA metabolic processes, because it has the highest affinity to ssDNA among known proteins. Hence, RPA-covered ssDNA is the physiologically relevant target for assembly of the Rad51-ssDNA filament, which competes with other processes such as recruitment of the Rad6-Rad18 ubiquitylation complex, TLS, fork regression, or ATR signaling (Figure 3). Cell cycle-dependent and DNA damage-induced phosphorylation of RPA2, the middle subunit of RPA, were discovered about two decades ago, but the specific function of RPA phosphorylation remains to be determined. RPA phosphorylation does not appear to affect its DNA binding properties, but likely modulates protein interactions that may also affect its intranuclear localization (50).
Positive control by DDR-mediated Rad51 phosphorylation
In response to HU, human RAD51 is phosphorylated by CHK1 kinase within a consensus site at threonine 309 (Figure 2, Table 1) (138). Cells depleted for CHK1 activity by UCN-01-mediated inhibition or siRNA display a defect in RAD51 focus formation in response to HU, consistent with positive regulation of HR by CHK1. Targets other than RAD51 may be involved as well. Expression of the RAD51-T309A phosphorylation-defective mutant but not wild type protein causes dominant hypersensitivity to HU, supporting an activating role of threonine 309 phosphorylation (138).
Negative control by CDK phosphorylation of BRCA2
The tumor suppressor protein BRCA2 plays a key role in the formation of the RAD51 filament (126). CDK-cyclin A can phosphorylate BRCA2 on serine 3291 in vitro and this residue is also phosphorylated in vivo, peaking during M-phase (Figure 2, Table 1) (47). S3291 of BRCA2 is near the minimum C-terminal RAD51 interaction site (residues 3196–3226 (132)), and phosphorylation of this residue or a change from serine to alanine ablates the interaction of the BRCA2 C-terminus with RAD51 (47). These data led to the model that CDK-mediated BRCA2 phosphorylation precludes HR during M-phase, where it could interfere with chromosome segregation (47). Furthermore, BRCA2 and the RAD51 paralog, RAD51C, are also involved in nuclear transport of RAD51 after DNA damage (38, 59).
Rad52 sumoylation affects protein stability and intranuclear localization
Rad52 is the lynchpin of HR in budding yeast, essential both for HR and SSA (67, 87, 126, 159). Sumoylation of a significant fraction of Rad52 protein is induced in meiosis or after DNA damage, dependent on the M-R-X complex (125). A triple mutant at lysine residues 10, 11 and 220 ablated Rad52 sumoylation, leading to faster proteasome-dependent protein turnover (125) (Table 1). While SUMO-deficient Rad52 protein is largely proficient for HR, the mutant displayed a 2.5-fold reduction in direct repeat recombination (125). Live cell imaging revealed that sumoylation controls intra-nuclear localization of Rad52 protein (150). In wild type cells, Rad52 protein is excluded from the nucleolus, the nuclear compartment containing the rDNA repeats. The Rad52 SUMO-deficient mutant overcomes the nucleolar exclusion and forms foci within the nucleolus, resulting in slightly elevated rDNA recombination (150), opposite to the effect on nuclear repeat recombination (125).
Phosphorylation of Rad55 serines 2, 8, and 14 is required for optimal HR
The yeast Rad51 paralogs, Rad55-Rad57, facilitate the formation or stabilization of Rad51 filaments (92, 144). Rad55 is phosphorylated in response to DNA damage on multiple residues by Mec1 (serine 378), Rad53 (serine 14), and an unidentified kinase (serines 2, 8) (13, 66, 79). The N-terminal phosphorylation mutant (Rad55-S2,8,14A) displays strong defects in growth and survival in response to the alkylating agent methyl methansulfonate. These conditions lead to replication fork stalling, and Rad55 phospho-deficient mutants exhibit a defect in the recovery of stalled replication forks (66) (Table 1).
Disruption of Rad51-ssDNA filaments by anti-recombinogenic DNA helicases
Srs2 is the prototype for anti-recombination helicases capable of disrupting Rad51-ssDNA filaments (86, 157) (Figure 2), exerting biologically significant anti-recombination activity (1, 28, 129). During S-phase, Srs2 is recruited to replication forks by sumoylated PCNA (Table 1) (119, 121). Similarly, the NHEJ factor Nej1 recruits Srs2 to DSBs to repress HR and favor NHEJ or SSA, and this interaction is enhanced by DNA damage-induced phosphorylation of Nej1 by Dun1 kinase (Table 1) (26). Srs2 has no direct ortholog in mammals, but genetic studies with human FBH1 in budding yeast have led to the proposal that FBH1 is the mammalian counterpart of yeast Srs2 (31). This is consistent with the original identification of Fbh1 as a suppressor of a hypomorphic mutant in the fission yeast RAD52 (rad22) gene (116). Moreover, overexpression of human FBH1 in human cells impaired recruitment of RAD51 to ssDNA and suppressed HR, whereas FBH1 depletion caused an increase in sister chromatid exchanges (SCE) (54), consistent with an anti-recombination role of FBH1.
RECQ5, a RecQ family helicase in mammals, displaces RAD51 from ssDNA and inhibits D-loop formation in vitro (70) (RELATED RESOURCE: Rothstein, Bernstein & Gangloff). A defect in RECQ5 causes increased levels of spontaneous RAD51 foci, as well as elevated frequencies of spontaneous DSBs and HR between direct repeats. These phenotypes and the physical interaction of RECQ5 with RAD51 (70) are consistent with an anti-recombinogenic role. BLM, another RecQ family member, also interacts with RAD51 and is capable of disrupting RAD51-ssDNA filaments in vitro (19, 23). BLM only disrupts filaments in conditions containing Mg++, which has been interpreted as targeting the ADP-bound, inactive form of RAD51 and does not dissociate the ATP-bound RAD51 in the presence of Ca++ (22, 23). The biological relevance of this observation is uncertain. Unlike the helicases mentioned above that display a 3′→5′ polarity, FANCJ, a component of the Fanconi Anemia pathway (100), exhibits a 5′→3′ directionality. Similar to BLM, FANCJ dissociates only the inactive, ADP-bound form of RAD51 from ssDNA in vitro (137). No specific interaction between FANCJ and RAD51 has been reported. The biological significance of RAD51 dissociation by FANCJ remains uncertain, as a mutant in dog-1, the FANCJ homolog in C. elegans, shows no significant increase in Rad51 foci (166).
In summary, the Rad51-ssDNA filament is controlled by a balance between mediator proteins that promote assembly and anti-recombinogenic DNA helicases that promote disassembly. Cell cycle-dependent and DNA damage-inducible post-translational modifications of these factors impinge on both assembly and disassembly of the Rad51 filament.
REGULATION OF HOMOLOGY SEARCH AND DNA STRAND INVASION
Homology search and DNA strand invasion generate D-loops, a key intermediate for all sub-pathways of HR (Figure 1). These reactions are catalyzed by Rad51, which interacts with the dsDNA motor protein Rad54 (68). The meiosis-specific kinase Mek1 enforces the preference for homologs over sister chromatids in meiotic cells. Mek1-mediated Rad54-T132 phosphorylation reduces Rad54 binding to Rad51, thus decreasing Rad51-dependent recombination in vivo and in vitro (112) (Table 1). This mechanism is independent of Hed1 (112), a meiosis-specific repressor of the Rad51-Rad54 interaction that binds to Rad51 protein (24, 154). Both meiosis-specific mechanisms are thought to transiently inhibit Rad51-dependent recombination to bias meiotic HR to homologs mediated by the meiosis-specific Rad51 paralog Dmc1 (73), but both mechanisms are active in mitotic cells when Hed1 or the Rad54 phosphomimic mutant are ectopically expressed (24, 112, 154). The results show that a critical protein interaction (Rad51-Rad54) can be targeted to assert negative regulation of HR.
REVERSION OF D-LOOPS AND EXTENDED D-LOOPS: PRO- AND ANTI-RECOMBINOGENIC FUNCTIONS
D-loop reversion before extension by DNA polymerases is a potentially powerful mechanism of anti-recombination. A number of DNA helicases/translocases including FANCM/Mph1/Fml1, RTEL1, RECQ1, BLM, and Rad54 are capable of disrupting D-loops in vitro (Figure 4) (8, 11, 20, 21, 57, 143). However, reversion of an extended D-loop is also inherent to the SDSA pathway and constitutes in this context a pro-recombination activity (Figure 1). In some genetic assays such an activity can be scored to suppress crossovers, constituting another anti-crossover mechanism besides dHJ dissolution (Figure 1).
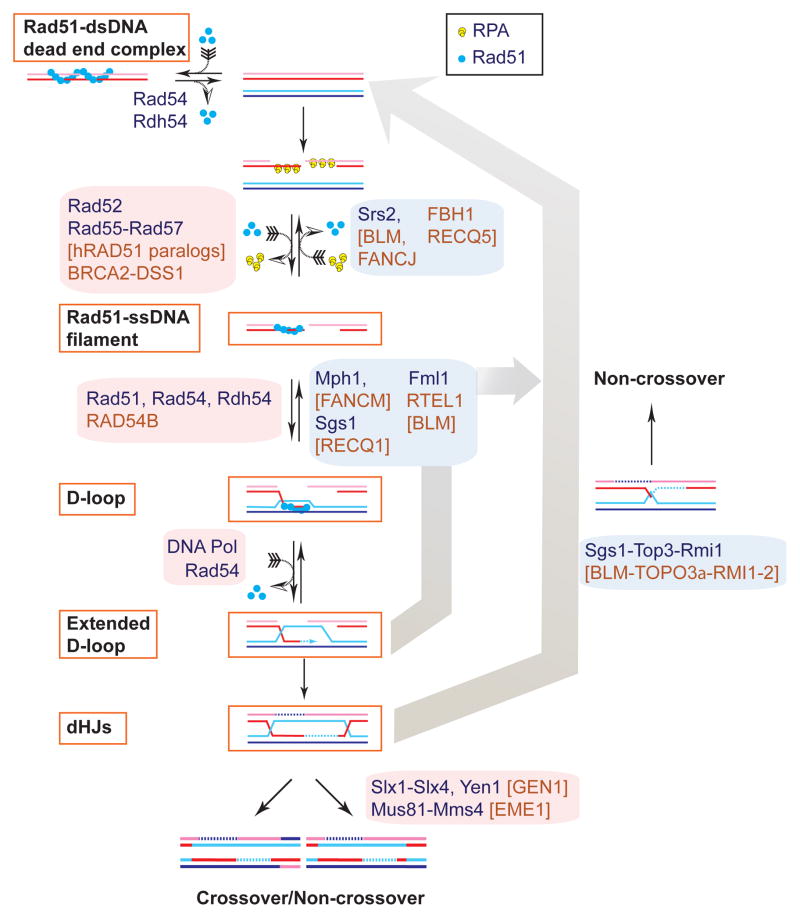
Figure 4. Model: Reversible, meta-stable intermediates in homologous recombination.
HR is proposed to involve key intermediates that are reversible and meta-stable including: (1) the Rad51–ssDNA filament, (2) the initial D-loop, (3) the extended D-loop, and (4) the double Holliday junction. The dead-end complex of Rad51/Dmc1 with dsDNA, although not an HR intermediate, can be added to this list of reversible HR protein-DNA complexes (69).
Rad54 is essential for HR in budding yeast and is required for in vitro D-loop formation by the yeast Rad51 protein (68). However, Rad54 can also dissociate D-loops in vitro (21), the very product it forms in conjunction with Rad51, making it difficult to test the biological significance of this activity.
Genetic studies on Sgs1 have provided critical insights on the cellular functions and regulation of mammalian RecQ helicases in HR (RELATED RESOURCE: Rothstein, Bernstein & Gangloff). However, the multiple functions of Sgs1 in DDR signaling, DSB resection, dHJ dissolution, and potentially other steps of HR complicate interpretation of the genetic data. Importantly, Sgs1 suppresses crossovers during mitotic and meiotic recombination (75, 114). This role of Sgs1 is most readily explained by the ability of the BLM-TOPO3alpha-RMI complex to dissolve dHJs into non-crossover products (163). dHJ dissolution by BLM also explains the elevated levels of SCE in BLM-deficient cells. BLM may also contribute to non-crossover outcome by promoting the SDSA pathway, as indicated by genetic studies in Drosophila (3). BLM interacts with RAD51 protein and can dissociate mobile D-loops (8, 19), but not D-loops during an ongoing RAD51-mediated in vitro reaction (111). This leaves open the question of how BLM may promote SDSA.
Human FANCM protein is a core component in the Fanconi Anemia pathway that is critical for the repair of interstrand DNA crosslinks (100). FANCM and its homologs form an evolutionarily deeply rooted family including the archaeal Hef nuclease/helicase, budding yeast Mph1 and fission yeast Fml1. The eukaryotic enzymes either lost or degenerated their nuclease domain (160). FANCM-deficient cells display elevated levels of spontaneous SCE, consistent with the ability of FANCM protein to dissociate mobile D-loops (9, 57, 124). The FANC pathway is negatively regulated in mitosis by polo-like kinase PLK1 phosphorylation of FANCM, leading to its ubiquitin-mediated degradation (84). Similar to human FANCM, Mph1 and Fml1 dissociate D-loops in vitro (122, 143). A defect in the yeast FANCM orthologs, Mph1 or Fml1, also causes a 3–4-fold increase in crossovers. Epistasis analysis in both fission and budding yeast suggests that Mph1 and Fml1 act independently of Srs2 or Sgs1 in suppressing crossovers (10, 122, 143). Both proteins promote Rad51-dependent recombination at stalled replication forks (118, 130, 143). Using an inducible replication fork stalling system in fission yeast, Whitby and colleagues showed a requirement for Fml1 in spontaneous and fork stalling-induced HR (143). Moreover, mutants in MPH1 have the same mutator phenotype as HR mutants (rad51) and this effect is epistatic with an HR defect, suggesting that Mph1 functions in concert with HR to avoid Rev3-dependent mutagenesis (130). Mph1 appears to function late in HR, as the synthetic lethality with srs2 is suppressed by mutations in rad51, rad55, rad57, and rad52 (130). However, Mph1 has also been suggested to promote gross chromosomal rearrangements by inhibiting HR through stabilizing RPA on ssDNA (10).
RECQ1, another RecQ-like helicase, is required for genome stability in mouse and human cells, as RECQ1 deficiency leads to aneuploidy, chromosomal instability, and hypersensitivity to DNA damage (IR, camptothecin) (133, 134). RECQ1-deficient cells exhibit increased levels of spontaneous DNA damage as suggested by an increase in spontaneous gamma-H2AX foci and elevated SCE levels. In vitro, RECQ1 disrupts D-loops with significant preference for D-loops resulting from invasion of the 5′-end (20). Since DNA polymerase cannot extend such a D-loop, it constitutes a potential dead-end complex. This activity provides a potential mechanism for RecQ1 function in reversing a potentially toxic HR intermediate.
The RAD3-like helicase RTEL1 was isolated in a screen for functional analogs of Srs2 in C. elegans (11). A defect in RTEL1 causes synthetic lethality when combined with mutations in BLM, RECQ5, or MUS81, as well as a four-fold increase in meiotic crossovers and DNA damage sensitivity to interstrand crosslinks and the topoisomerase I inhibitor camptothecin. Depletion of RTEL1 in human cells causes a 4-fold increase in DSB-mediated intrachromosomal repeat recombination that is unlikely explained by a defect in crossover suppression, as well as hypersensitivity to the crosslinking agent mitomycin C but not IR. In vitro, RTEL1 dissociates D-loops, which could explain the anti-recombination and anti-crossover phenotype. However, the DNA damage sensitivity profile is more consistent with a defect in HR. Unlike Srs2, RTEL1 cannot dissociate RAD51 from ssDNA (11, 86, 157). Interestingly, Ira et al. postulated that Srs2 exerts its anti-crossover effect through a function in SDSA and suggested that Srs2 dissociates D-loops (75). While this biochemical activity has not been directly demonstrated yet (86, 157), the stimulation of Srs2 helicase activity by Rad51 bound to dsDNA suggests the possibility that the Srs2 targets two HR intermediates, Rad51-ssDNA filaments and (extended?) D-loops (43).
In summary, a number of proteins are capable of dissociating D-loops, which may function in HR to favor SDSA and suppress crossovers, or be a mechanism of anti-recombination. The mutant phenotypes suggest potentially complex roles for RTEL1, Srs2, Sgs1/BLM and the FANCM helicases in HR.
MISMATCH REPAIR EDITS RECOMBINATION FIDELITY
Mismatch repair (MMR) functions to edit replication errors, and mismatch correction in hDNA achieves gene conversion during HR (see Figure 1). More critical to the regulation of HR, however, is that MMR proteins help to discriminate homology from homeology (partial homology) (63). This MMR-mediated screening of recombination fidelity favors HR between perfectly homologous sequences and actively opposes homeologous recombination, responding to the degree of homology.
Genetic studies systematically surveyed the effects of homeology on HR using an elegant intron-based assay (35, 36). Remarkably, even a single mismatch reduced spontaneous recombination rates by four-fold relative to substrates with 100% identity. Between 99% and 74% identity, recombination rates were reduced 9- to 4600-fold. Most importantly, defects in MMR suppressed the effects on spontaneous HR rates when homeology was up to 15% sequence divergence. MMR factors therefore regulate whether HR is sanctioned over given sequences when the interacting sequences are between 85–100% similar. Three yeast complexes function in both replication-associated mismatch repair and in negative regulation of HR: MutSα (Msh2-Msh6), MutSβ (Msh2-Msh3), and MutLα (Mlh1-Pms1) (35, 36). In addition, the nucleases Rad1-Rad10 and Exo1 and the helicases Sgs1 and Srs2 function in the MMR-mediated barrier to HR between homeologous sequences (110, 158). MMR not only affects the frequency of HR but also influences the crossover/non-crossover outcome of HR (Figure 1). At merely 2% sequence divergence, crossover events were reduced 7-fold, whereas non-crossover events were reduced only 2-fold (158). This characteristic of the MMR-mediated editing of HR may be particularly useful to suppress crossovers between slightly divergent repeated DNA sequences, where crossovers would lead to genome rearrangements.
What is the mechanism of the MMR-mediated barrier to HR between divergent sequences? Suppression of homeologous recombination by MMR could function during DNA strand exchange, hDNA extension, or even later, in joint molecule resolution (Figure 1). Paradigmatic biochemical studies with bacterial RecA, MutS and MutL proteins suggest that hDNA extension may be the decisive stage (162). Similar biochemical work with eukaryotic proteins has not been reported, but genetic evidence from S. cerevisiae is consistent with this scenario. Mitotic and meiotic gene conversion tracts in msh2 msh3 mutants are ~50% longer than in wild type cells, indicating that hDNA extension may be blocked by Msh2-Msh3 binding to mismatches in vivo (30). S. cerevisiae Sgs1 and Mph1 are candidates for motor proteins active in hDNA rejection (107, 146). sgs1 mutants allowed an increased rate of homeologous HR (substrate with 91% sequence identity), synergistic with MMR mutants (139). This increase in homeologous recombination was also linked to a role for Sgs1 in suppression of gross chromosomal translocations (107). In addition, Sgs1 as well as Msh2-Msh6 suppress SSA between homeologous sequences (141).
In summary, MMR is a key regulator of HR in the distinction between allelic sites and ectopic sites. This editing function is sufficiently sensitive to discriminate allelic targets on sister chromatids from allelic targets on homologs. The importance of MMR in focusing HR to perfect sequence identity (allelic sites on sister chromatids) suggests that MMR defects in tumors not only increase the rates of point mutations, but also increase rates of inappropriate HR between homeologous sequences leading to genome rearrangements. Such association has been noted in at least some MMR-deficient colon cancers (151).
NUCLEOLYTIC PROCESSING OF STALLED REPLICATION FORKS AND DOUBLE HOLLIDAY JUNCTIONS
A number of DNA joint molecules are intermediates at which regulatory ‘decisions’ can be made, providing successive opportunities to decide whether HR is initiated, aborted, or sanctioned for a specific genetic outcome (crossover or non-crossover). The regulation of HR relevant to two specific joint molecules is elaborated here: replication forks and dHJs.
Stalled replication forks are potentially substrates for HR, but the mechanisms by which HR promotes fork restart and recovery remain unclear. Fork incision to generate a single-sided DSB end or a ssDNA gap could initiate HR (Figure 2). The relative significance of fork incision versus gap repair is uncertain, although Fabre et al. (49) suggested that breaks are rare in S-phase and that ssDNA gaps are the primary substrates for replication-associated HR. Nevertheless, Hanada et al. (61, 62) and Froget et al. (53) implicate MUS81-EME1 in fork incision. DSBs are observed after eighteen hours of chronic HU challenge, dependent on MUS81-EME1 (61). Interestingly, S. pombe Mus81 dissociates from chromatin in response to HU treatment, although Mus81 is required for resistance to HU (15, 81). It was proposed that fork incision by Mus81-Mms4/EME1 represents a last resort for fork recovery, and it may be negatively regulated under some circumstances of replication stress (81).
dHJs are intermediates during mitotic DSB repair by HR (25). Alternative mechanisms for removing dHJs determine whether the genetic products result in a crossover or non-crossover (Figure 1). A number of endonucleases have been proposed to cut Holliday junctions or their precursors in vivo, including Mus81-Mms4/EME1, Yen1/GEN1 and Slx1-Slx4 (Figure 1; (99)). In addition to endonucleolytic ‘resolution’, dHJs can be ‘dissolved’ by the concerted activities of a helicase-topoisomerase complex (Figure 1) (163). How endonucleolytic resolution of dHJs is regulated relative to dissolution is unknown. Caspari et al. (27) suggested that CDK phosphorylates Top3 in S. pombe, dependent on interaction with the DDR mediator Crb2. Loss of Top3 function results in hyper-recombination and cell death after IR, perhaps associated with an inability to optimally resolve dHJs.
In summary, nucleolytic processing of stalled replication forks and dHJs determines whether HR is initiated after fork stalling and whether the genetic outcome of HR is potentially a crossover. The processes and proteins involved, and their specific function and regulation, still needs to be determined.
MODEL: HOMOLOGOUS RECOMBINATION – A PATHWAY WITH METASTABLE, REVERSIBLE INTERMEDIATES TO ACHIEVE FLEXIBILITY AND ROBUSTNESS
DNA repair is a formidable task that requires quality control to balance accuracy of the repair event with the potential for genome rearrangements (83). It has been proposed that reversibility of HR intermediates provides robustness to the pathway (83, 145). In biology, the concept of robustness has been largely discussed in the context of mutational robustness, keeping an organism’s phenotype constant in spite of mutations (41). In the present discussion, however, the term robustness applies more in the engineering sense, where a system or algorithm does not break down easily, continues to operate despite single application failures and recovers quickly from, and holds up under, exceptional circumstances (4, 128).
What can we learn from the analysis of regulation of HR about the mechanism of HR and how it achieves robustness? One aspect is protein interactions. The myriad of direct protein-protein interactions between HR proteins have been previously projected into a single time point and interpreted as a stable ‘recombinosome’ (64). Further analysis now suggests that these interactions can be regulated by reversible post-translational modifications, are transient, and occur sequentially (67, 87, 126, 159), which provides significantly more plasticity. A second aspect is pathway flexibility. While the HR pathway is typically portrayed as a linear sequence, Figure 1 reveals bifurcation, where identical intermediates (D-loop, dHJ) can enter different sub-pathways and fates. Finally, the abundance of motor proteins that dissociate recombination intermediates suggests that apparent anti-recombination mechanisms are integral parts of the HR pathway. Four key intermediates in HR that are reversible by the action of motor proteins include the Rad51-ssDNA filament, the initial D-loop, the extended D-loop, and the dHJ (Figure 4). These intermediates also appear to be meta-stable, because they can be visualized cytologically (Rad51 foci (92)) or identified physically (73, 74).
There is significant evidence that key HR intermediates are reversible in vivo and that this feature is important for HR. Reversal of extended D-loops is central to the SDSA model (Figure 1). There is compelling genetic evidence for multiple, sequential DNA strand invasion events during HR, implying dissociation of D-loops or extended D-loops (3, 136). Promiscuous joint formation, at least in meiotic HR, is not rare and needs active reversal by Sgs1 helicase (114) and suppression by MMR (see above). Another indication that anti-recombination mechanisms are an integral part of the HR pathway is provided by the complex phenotypes of mutations in HR motor proteins. Mutations in Sgs1 show increased spontaneous recombination that appears unrelated to crossover suppression, consistent with the anti-recombination role of Sgs1, but reduced DNA damage-induced recombination, suggesting a pro-recombination role (56). Likewise, Srs2 was shown to have anti- and pro-recombination phenotypes, as was suggested for FBH1 (6, 54, 75, 157). A defect in RTEL1 causes hyper-recombination but also a DNA damage-sensitivity profile that suggests a defect in HR (11). The FANCM related proteins (human FANCM, fission yeast Fml1, budding yeast Mph1) can reverse D-loops in vitro and depending on the assay, mutants display anti- or pro-recombination phenotypes (10, 58, 97, 118, 122, 130, 160). Furthermore, the phenomenon of recombination-dependent lethality, where the synthetic lethality of certain double mutant combinations (e.g. srs2 sgs1, mus81 sgs1) can be suppressed by an HR defect demonstrates the occurrence of potentially toxic HR intermediates that require resolution by nucleases or motor proteins (49, 56).
In summary and as depicted in Figure 4, we suggest that the HR pathway proceeds through a series of meta-stable, reversible intermediates that are under active positive and negative regulation to allow flexibility for the repair outcomes (crossover vs. non-crossover), accommodation of the unforeseen (e.g. absence of a second end and switch to BIR, Figure 1), and recovery from unwanted intermediates (e.g. independent invasions of both ends of a DSB into different targets), which are all aspects that define robustness of a well-engineered system that is essential for maintaining a stable genome. Reversibility entails the destruction of potentially normal intermediates (83) and the MMR barrier, for example, also affects HR between perfectly homologous sequences (109). While counterintuitive at first, it appears that reactions that reverse recombination intermediates are required for the optimal functioning of HR, as the stochastic nature of the process will favor accurate pathway progression.
SUMMARY POINTS.
- Recombinational DNA repair is not constitutive but highly modulated by positive and a preponderance of negative regulatory mechanisms.
- Two signaling systems intersect in the control of HR, the cell cycle control machinery and the DDR.
- In DSB repair, end resection is a major commitment point to HR, regulated by CDK-dependent phosphorylation of Sae2/CtIP.
- The Rad51 filament is a major regulatory control point of HR governed by mechanisms that favor its assembly (mediators and their post-translational modifications) or disassembly (anti-recombinogenic motor proteins).
- Several mechanisms, including MMR, extended D-loop reversion, and dHJ dissolution, enforce an anti-crossover bias during DSB repair in somatic (mitotic) cells.
- Anti-recombination mechanisms mediated by DNA motor proteins appear to be integral to the HR pathway, providing flexibility and robustness through reversible, meta-stable intermediates.
Acknowledgments
We thank Steve Kowalczykowski, Neil Hunter, and all members of the Heyer lab (Clare Fasching, Ryan Janke, Damon Meyer, Erin Schwartz, Jessica Sneeden, William Wright, Xiao-Ping Zhang) for helpful discussions and critical comments. The work was supported by NIH (GM58015, CA92776) and the DoD (BC083684). JL is supported by a TRDRP postdoctoral fellowship (17FT-0046). We apologize that not all of the outstanding work in this area could be discussed or cited because of space constraints.
Glossary
KEY TERMS
BIR
Break-induced replication; a sub-pathway of HR where a single-ended DSB invades and establishes a full-fledged replication fork
Crossover/Non-crossover
Describes outcome of HR with respect to the flanking DNA, which is either in parental (non-crossover) or non-parental (crossover) configuration
D-loop
Displacement loop; primary DNA strand invasion product of the Rad51-ssDNA filament leading to the different HR sub-pathways (BIR, SDSA, dHJ)
Homeologous recombination
Recombination between similar but not identical sequences, as found in repeated DNA
dHJ
Double Holliday junction; HR intermediate leading to crossovers. Also used here to label the HR sub-pathway that involves this intermediate
SDSA
Synthesis-dependent strand annealing; a sub-pathway of HR where the second end of the DSB anneals with the extended strand of the first end
SSA
Single-strand annealing; a mode of homology-directed DNA repair that does not involve Rad51-mediated DNA strand invasion, but DNA reannealing
TLS
Translesion synthesis performed by specialized DNA polymerases to accommodate template lesion in DNA damage tolerance
ACRONYMS
CDK
Cyclin-dependent kinase
DDR
DNA damage response
DSB
DNA double-stranded break
hDNA
heteroduplex DNA
HR
Homologous recombination
HU
Hydroxyurea
IR
Ionizing radiation
MMR
Mismatch repair
M-R-N/X
S. cerevisiae Mre11-Rad50-Xrs2 complex or human MRE11-RAD50-NBS1 complex
NHEJ
Non-homologous end joining
PCNA
Proliferating cell nuclear antigen
RPA
Replication protein A
SCE
Sister chromatid exchange
Footnotes
BRIEF ANNOTATIONS
- Ref. 52 and 53 show that sumoylated PCNA recruits the Srs2 anti-recombinase.
- Ref. 54 and 55 determine that the anti-recombinase Srs2 dissociates Rad51 from ssDNA.
- Ref. 70 and 71 demonstrate the importance of CDK activity for regulating DSB resection and commitment to HR.
- Ref. 72 and 73 identify Sae2/CtIP as the key CDK target to control DSB end resection.
- Ref. 91 identifies that CDK-mediated phosphorylation of BRCA2 inhibits its interaction with RAD51 through its C-terminal site.
- Ref. 98 demonstrates that DDR-mediated phosphorylation of Rad55 positively regulates HR.
- Ref. 27 and 95 show that Rad52 sumoylation controls protein stability and intranuclear localization.
- Ref. 138 studied the effect of mismatches on HR, showing that even a single mismatch has an effect.
DISCLOSURE STATEMENT
The authors are not aware of any affiliation, membership, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Aboussekhra A, Chanet R, Adjiri A, Fabre F. Semi-dominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA protein. Mol Cell Biol. 1992;12:3224–34. doi: 10.1128/mcb.12.7.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aboussekhra A, Chanet R, Zgaga Z, Cassier Chauvat C, Heude M, Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–19. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams MD, McVey M, Sekelsky JJ. Drosophila BLM in double-strand break repair by synthesis-dependent strand annealing. Science. 2003;299:265–67. doi: 10.1126/science.1077198. [DOI] [PubMed] [Google Scholar]
- 4.Alon U. Biological Networks: The tinkerer as an engineer. Science. 2003;301:1866–67. doi: 10.1126/science.1089072. [DOI] [PubMed] [Google Scholar]
- 5.Astrom SU, Okamura SM, Rine J. Yeast cell-type regulation of DNA repair. Nature. 1999;397:310. doi: 10.1038/16833. [DOI] [PubMed] [Google Scholar]
- 6.Aylon Y, Liefshitz B, Bitan-Banin G, Kupiec M. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2003;23:1403–17. doi: 10.1128/MCB.23.4.1403-1417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aylon Y, Liefshitz B, Kupiec M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. Embo J. 2004;23:4868–75. doi: 10.1038/sj.emboj.7600469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom’s syndrome helicase. Nucleic Acids Res. 2006;34:2269–79. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bakker ST, van de Vrugt HJ, Rooimans MA, Oostra AB, Steltenpool J, et al. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet. 2009;18:3484–95. doi: 10.1093/hmg/ddp297. [DOI] [PubMed] [Google Scholar]
- 10.Banerjee S, Smith S, Oum JH, Liaw HJ, Hwang JY, et al. Mph1p promotes gross chromosomal rearrangement through partial inhibition of homologous recombination. J Cell Biol. 2008;181:1083–93. doi: 10.1083/jcb.200711146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, et al. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008;135:261–71. doi: 10.1016/j.cell.2008.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baroni E, Viscardi V, Cartagena-Lirola H, Lucchini G, Longhese MP. The functions of budding yeast Sae2 in the DNA damage response require Mec1- and Tel1-dependent phosphorylation. Mol Biol Cell. 2004;24:4151–65. doi: 10.1128/MCB.24.10.4151-4165.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bashkirov VI, King JS, Bashkirova EV, Schmuckli-Maurer J, Heyer WD. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol Cell Biol. 2000;20:4393–404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergink S, Jentsch S. Principles of ubiquitin and SUMO modifications in DNA repair. Nature. 2009;458:461–67. doi: 10.1038/nature07963. [DOI] [PubMed] [Google Scholar]
- 15.Boddy MN, Gaillard P-HL, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–48. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 16.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nature Rev Mol Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 17.Branzei D, Sollier J, Liberi G, Zhao XL, Maeda D, et al. Ubc9-and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell. 2006;127:509–22. doi: 10.1016/j.cell.2006.08.050. [DOI] [PubMed] [Google Scholar]
- 18.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–20. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 19.Braybrooke JP, Li JL, Wu L, Caple F, Benson FE, Hickson ID. Functional interaction between the Bloom’s syndrome helicase and the RAD51 paralog, RAD51L3 (RAD51D) J Biol Chem. 2003;278:48357–66. doi: 10.1074/jbc.M308838200. [DOI] [PubMed] [Google Scholar]
- 20.Bugreev DV, Brosh RM, Mazin AV. RECQ1 possesses DNA branch migration activity. J Biol Chem. 2008;283:20231–42. doi: 10.1074/jbc.M801582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bugreev DV, Hanaoka F, Mazin AV. Rad54 dissociates homologous recombination intermediates by branch migration. Nature Struct Mol Biol. 2007;14:746–53. doi: 10.1038/nsmb1268. [DOI] [PubMed] [Google Scholar]
- 22.Bugreev DV, Mazin AV. Ca2+ activates human homologous recombination protein Rad51 by modulating its ATPase activity. Proc Natl Acad Sci USA. 2004;101:9988–93. doi: 10.1073/pnas.0402105101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 2007;21:3085–94. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev. 2008;22:786–95. doi: 10.1101/gad.1638708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010 doi: 10.1038/nature08868. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carter SD, Vigasova D, Chen J, Chovanec M, Astrom SU. Nej1 recruits the Srs2 helicase to DNA double-strand breaks and supports repair by a single-strand annealing-like mechanism. Proc Natl Acad Sci U S A. 2009;106:12037–42. doi: 10.1073/pnas.0903869106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caspari T, Murray JM, Carr AM. Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev. 2002;16:1195–208. doi: 10.1101/gad.221402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chanet R, Heude M, Adjiri A, Maloisel L, Fabre F. Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol Cell Biol. 1996;16:4782–89. doi: 10.1128/mcb.16.9.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen FQ, Nastasi A, Shen ZY, Brenneman M, Crissman H, Chen DJ. Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes Rad51 and Rad52. Mutat Res. 1997;384:205–11. doi: 10.1016/s0921-8777(97)00020-7. [DOI] [PubMed] [Google Scholar]
- 30.Chen WL, Jinks-Robertson S. Mismatch repair proteins regulate heteroduplex formation during mitotic recombination in yeast. Mol Cell Biol. 1998;18:6525–37. doi: 10.1128/mcb.18.11.6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiolo I, Saponaro M, Baryshnikova A, Kim JH, Seo YS, Liberi G. The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles. Mol Cell Biol. 2007;27:7439–50. doi: 10.1128/MCB.00963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cole GM, Mortimer RK. Failure to induce a DNA repair gene, RAD54, in Saccharomyces cerevisiae does not affect DNA repair or recombination phenotypes. Mol Cell Biol. 1989;9:3314–22. doi: 10.1128/mcb.9.8.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cotta-Ramusino C, Fachinetti D, Lucca C, Doksani Y, Lopes M, et al. Exo1 processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol Cell. 2005;17:153–59. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 34.Courcelle J, Khodursky A, Peter B, Brown PO, Hanawalt PC. Comparative gene expression profiles following UV exposure in wild-type and SOS-deficient Escherichia coli. Genetics. 2001;158:41–64. doi: 10.1093/genetics/158.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Datta A, Adjiri A, New L, Crouse GF, Jinks Robertson S. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:1085–93. doi: 10.1128/mcb.16.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Datta A, Hendrix M, Lipsitch M, JinksRobertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–62. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein A. Mol Cell. 2008;29:625–36. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Davies AA, Masson JY, McLlwraith MJ, Stasiak AZ, Stasiak A, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–82. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 39.de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell. 2001;8:1129–35. doi: 10.1016/s1097-2765(01)00381-1. [DOI] [PubMed] [Google Scholar]
- 40.de Mayolo AA, Lisby M, Erdeniz N, Thybo T, Mortensen UH, Rothstein R. Multiple start codons and phosphorylation result in discrete Rad52 protein species. Nucleic Acids Res. 2006;34:2587–97. doi: 10.1093/nar/gkl280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Draghi JA, Parsons TL, Wagner GP, Plotkin JB. Mutational robustness can facilitate adaptation. Nature. 2010;463:353–5. doi: 10.1038/nature08694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–86. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dupaigne P, Le Breton C, Fabre F, Giangloff S, Le Cam E, Veaute X. The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: Implications for crossover incidence during mitotic recombination. Mol Cell. 2008;29:243–54. doi: 10.1016/j.molcel.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 44.Ehmsen KT, Heyer WD. Biochemistry of meiotic recombination. In: Richard Egel DL, editor. Recombination and Meiosis. Berlin-Heidelberg: Springer-Verlag; 2008. pp. 91–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.El-Shemerly M, Hess D, Pyakurel AK, Moselhy S, Ferrari S. ATR-dependent pathways control hEXO1 stability in response to stalled forks. Nucleic Acids Res. 2008;36:511–19. doi: 10.1093/nar/gkm1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, et al. Intra-nuclear trafficking of the BLM helicase to DNA damage-induced foci is regulated by SUMO modification. Hum Mol Genet. 2005;14:1351–65. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- 47.Esashi F, Christ N, Gannon J, Liu YL, Hunt T, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 48.Fabre F. Induced intragenic recombination in yeast can occur during the G1 mitotic phase. Nature. 1978;272:795–97. doi: 10.1038/272795a0. [DOI] [PubMed] [Google Scholar]
- 49.Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci USA. 2002;99:16887–92. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–37. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frank-Vaillant M, Marcand S. NHEJ regulation by mating type is exercised through a novel protein, Lif2p, essential to the ligase IV pathway. Genes Dev. 2001;15:3005–12. doi: 10.1101/gad.206801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frank-Vaillant M, Marcand S. Transient stability of DNA ends allows nonhomologous end joining to precede homologous recombination. Mol Cell. 2002;10:1189–99. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 53.Froget B, Blaisonneau J, Lambert S, Baldacci G. Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Mol Biol Cell. 2008;19:445–56. doi: 10.1091/mbc.E07-07-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fugger K, Mistrik M, Danielsen JR, Dinant C, Falck J, et al. Human Fbh1 helicase contributes to genome maintenance via pro- and anti-recombinase activities. J Cell Biol. 2009;186:655–63. doi: 10.1083/jcb.200812138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature. 2009;462:935–U132. doi: 10.1038/nature08657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gangloff S, Soustelle C, Fabre F. Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nature Genet. 2000;25:192–94. doi: 10.1038/76055. [DOI] [PubMed] [Google Scholar]
- 57.Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci USA. 2008;105:16107–12. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008;29:141–48. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 59.Gildemeister OS, Sage JM, Knight KL. Cellular Redistribution of Rad51 in response to DNA damage. Novel role for RAD51C. J Biol Chem. 2009;284:31945–52. doi: 10.1074/jbc.M109.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–72. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14:1096–104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 62.Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. Embo J. 2006;25:4921–32. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harfe BD, Jinks-Robertson S. DNA mismatch repair and genetic instability. Annu Rev Genetics. 2000;34:359–99. doi: 10.1146/annurev.genet.34.1.359. [DOI] [PubMed] [Google Scholar]
- 64.Hays SL, Firmenich AA, Berg P. Complex formation in yeast double-strand break repair: Participation of Rad51, Rad52, Rad55, and Rad57 proteins. Proc Natl Acad Sci USA. 1995;92:6925–29. doi: 10.1073/pnas.92.15.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Heller RC, Marians KJ. Replication fork reactivation downstream of a blocked nascent leading strand. Nature. 2006;439:557–62. doi: 10.1038/nature04329. [DOI] [PubMed] [Google Scholar]
- 66.Herzberg K, Bashkirov VI, Rolfsmeier M, Haghnazari E, McDonald WH, et al. Phosphorylation of Rad55 on serines 2, 8, and 14 is required for efficient homologous recombination in the recovery of stalled replication forks. Mol Cell Biol. 2006;26:8396–409. doi: 10.1128/MCB.01317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heyer WD. Biochemistry of eukaryotic homologous recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Berlin-Heidelberg: Springer-Verlag; 2007. pp. 95–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Heyer WD, Li X, Rolfsmeier M, Zhang XP. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–25. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Holzen TM, Shah PP, Olivares HA, Bishop DK. Tid1/Rdh54 promotes dissociation of Dmc1 from nonrecombinogenic sites on meiotic chromatin. Genes Dev. 2006;20:2593–604. doi: 10.1101/gad.1447106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–84. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huertas P, Cortes-Ledesma F, Sartori AA, Aguilera A, Jackson SP. CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature. 2008;455:689–92. doi: 10.1038/nature07215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huertas P, Jackson SP. Human CtIP Mediates Cell Cycle Control of DNA End Resection and Double Strand Break Repair. J Biol Chem. 2009;284:9558–65. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hunter N. Meiotic recombination. In: Aguilera A, Rothstein R, editors. Homologous Recombination. Berlin-Heidelberg: Springer-Verlag; 2007. pp. 381–441. [Google Scholar]
- 74.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double strand break to double Holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 75.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–11. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, et al. DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1. Nature. 2004;431:1011–17. doi: 10.1038/nature02964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ivanov EL, Sugawara N, Fishmanlobell J, Haber JE. Genetic requirements for the single-strand annealing pathway of double-strand break repair in Saccharomyces cerevisiae. Genetics. 1996;142:693–704. doi: 10.1093/genetics/142.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jain S, Sugawara N, Lydeard J, Vaze M, Tanguy Le Gac N, Haber JE. A recombination execution checkpoint regulates the choice of homologous recombination pathway during DNA double-strand break repair. Genes Dev. 2009;23:291–303. doi: 10.1101/gad.1751209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Janke R, Herzberg K, Rolfsmeier M, Mar J, Bashkirov VI, et al. A truncated DNA damage signaling response is activated after DSB formation in the G1 phase of Saccharomyces cerevisiae. Nucleic Acids Res. 2010 Jan 8; doi: 10.1093/nar/gkp1222. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kadyk LC, Hartwell LH. Sister chromatids are preferred over homologs as substrates for recombinational repair in Saccharomyces cerevisiae. Genetics. 1992;132:387–402. doi: 10.1093/genetics/132.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kai M, Boddy MN, Russell P, Wang TSF. Replication checkpoint kinase Cds1 regulates Mus81 to reserve genome integrity during replication stress. Genes Dev. 2005;19:919–32. doi: 10.1101/gad.1304305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanaar R, Troelstra C, Swagemakers SMA, Essers J, Smit B, et al. Human and mouse homologs of the Saccharomyces cerevisiae RAD54 DNA repair gene: Evidence for functional conservation. Curr Biol. 1996;6:828–38. doi: 10.1016/s0960-9822(02)00606-1. [DOI] [PubMed] [Google Scholar]
- 83.Kanaar R, Wyman C, Rothstein R. Quality control of DNA break metabolism: in the ‘end’, it’s a good thing. Embo J. 2008;27:581–88. doi: 10.1038/emboj.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kee Y, Kim JM, D’Andrea A. Regulated degradation of FANCM in the Fanconi anemia pathway during mitosis. Genes Dev. 2009;23:555–60. doi: 10.1101/gad.1761309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–57. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 86.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, et al. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–09. doi: 10.1038/nature01577. [DOI] [PubMed] [Google Scholar]
- 87.Krogh BO, Symington LS. Recombination proteins in yeast. Annu Rev Genet. 2004;38:233–71. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 88.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 89.Lee SE, Paques F, Sylvan J, Haber JE. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr Biol. 1999;9:767–70. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- 90.Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 Is an Endonuclease that Processes Hairpin DNA Cooperatively with the Mre11/Rad50/Xrs2 Complex. Mol Cell. 2007;28:638–51. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li X, Heyer WD. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 2008;18:99–113. doi: 10.1038/cr.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 93.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 94.Lu SP, Lin SJ. Regulation of yeast sirtuins by NAD(+) metabolism and calorie restriction. Biochim Biophys Acta. 2009 Oct 8; doi: 10.1016/j.bbapap.2009.09.030. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Malkova A, Naylor ML, Yamauchi M, Ira G, Haber JE. RAD51-dependent break-induced replication differs in kinetics and checkpoint responses from RAD51-mediated gene conversion. Mol Biol Cell. 2005;25:933–44. doi: 10.1128/MCB.25.3.933-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–41. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 97.Mankouri HW, Ngo HP, Hickson ID. Esc2 and Sgs1 Act in Functionally Distinct Branches of the Homologous Recombination Repair Pathway in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:1683–94. doi: 10.1091/mbc.E08-08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mimitou EP, Symington LS. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770–74. doi: 10.1038/nature07312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mimitou EP, Symington LS. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Scie. 2009;34:264–72. doi: 10.1016/j.tibs.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 100.Moldovan GL, D’Andrea AD. How the Fanconi Anemia Pathway Guards the Genome. Annu Review Genet. 2009;43:223–49. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Morin I, Ngo HP, Greenall A, Zubko MK, Morrice N, Lydall D. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. Embo J. 2008;27:2400–10. doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–90. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 103.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, et al. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–90. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 104.Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage. Embo J. 2000;19:463–71. doi: 10.1093/emboj/19.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mortimer RK. Radiobiological and genetic studies on a polyploid series (haploid to hexaploid) of Saccharomyces cerevisiae. Radia Res. 1958;9:312–26. [PubMed] [Google Scholar]
- 106.Murray AW, Hunt T. The Cell Cycle. New York, Oxford: Oxford University Press; 1993. [Google Scholar]
- 107.Myung K, Datta A, Chen C, Kolodner RD. SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet. 2001;27:113–6. doi: 10.1038/83673. [DOI] [PubMed] [Google Scholar]
- 108.Nasmyth K, Haering CH. Cohesin: Its Roles and Mechanisms. Annu Review Genet. 2009;43:525–58. doi: 10.1146/annurev-genet-102108-134233. [DOI] [PubMed] [Google Scholar]
- 109.Negritto MT, Wu XL, Kuo T, Chu S, Bailis AM. Influence of DNA sequence identity on efficiency of targeted gene replacement. Mol Cell Biol. 1997;17:278–86. doi: 10.1128/mcb.17.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nicholson A, Hendrix M, Jinks-Robertson S, Crouse GF. Regulation of mitotic homeologous recombination in yeast: Functions of mismatch repair and nucleotide excision repair genes. Genetics. 2000;154:133–46. doi: 10.1093/genetics/154.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci USA. 2008;105:16906–11. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Niu H, Wan L, Busygina V, Kwon Y, Allen JA, et al. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell. 2009;36:393–404. doi: 10.1016/j.molcel.2009.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Review Genet. 2002;36:617–56. doi: 10.1146/annurev.genet.36.060402.113540. [DOI] [PubMed] [Google Scholar]
- 114.Oh SD, Lao JP, Hwang PYH, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–72. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Osley MA, Tsukuda T, Nickoloff JA. ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res. 2007;618:65–80. doi: 10.1016/j.mrfmmm.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Osman F, Dixon J, Barr AR, Whitby MC. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol. 2005;25:8084–96. doi: 10.1128/MCB.25.18.8084-8096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ouyang KJ, Woo LL, Zhu JM, Huo DZ, Matunis MJ, Ellis NA. SUMO Modification Regulates BLM and RAD51 Interaction at Damaged Replication Forks. Plos Biology. 2009;7 doi: 10.1371/journal.pbio.1000252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Panico ER, Ede C, Schildmann M, Schurer KA, Kramer W. Genetic evidence for a role of Saccharomyces cerevisiae Mph1 in recombinational DNA repair under replicative stress. Yeast. 2010;27:11–27. doi: 10.1002/yea.1727. [DOI] [PubMed] [Google Scholar]
- 119.Papouli E, Chen SH, Davies AA, Huttner D, Krejci L, et al. Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol Cell. 2005;19:123–33. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 120.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature. 2005;436:428–33. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 122.Prakash R, Satory D, Dray E, Papusha A, Scheller J, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: Specificity of structure and function. Annu Rev Biochem. 2005;74:317–53. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 124.Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ. The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res. 2009;37:4360–70. doi: 10.1093/nar/gkp365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sacher M, Pfander B, Hoege C, Jentsch S. Control of Rad52 recombination activity by double-strand break-induced SUMO modification. Nature Cell Biol. 2006;8:1284–U59. doi: 10.1038/ncb1488. [DOI] [PubMed] [Google Scholar]
- 126.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–57. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 127.Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–14. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Savageau M. Parameter sensitivity as a criterion for evaluating and comparing the performance of biochemical systems. Nature. 1971;229:542–44. doi: 10.1038/229542a0. [DOI] [PubMed] [Google Scholar]
- 129.Schiestl RH, Prakash S, Prakash L. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics. 1990;124:817–31. doi: 10.1093/genetics/124.4.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schurer KA, Rudolph C, Ulrich HD, Kramer W. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from homologous recombination, but not from postreplicative repair. Genetics. 2004;166:1673–86. doi: 10.1534/genetics.166.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Seet BT, Dikic I, Zhou MM, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–83. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 132.Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–10. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 133.Sharma S, Brosh RM., Jr Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS One. 2007;2:e1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, et al. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol Cell Biol. 2007;27:1784–94. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shrivastav M, De Haro LP, Nickoloff JA. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008;18:134–47. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 136.Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–05. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- 137.Sommers JA, Rawtani N, Gupta R, Bugreev DV, Mazin AV, et al. FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J Biol Chem. 2009;284:7505–17. doi: 10.1074/jbc.M809019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, et al. The cell-cycle checkpoint kinase Chk1 is required for mammalian homolgous recombination repair. Nat Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 139.Spell RM, Jinks-Robertson S. Examination of the roles of Sgs1 and Srs2 helicases in the enforcement of recombination fidelity in Saccharomyces cerevisiae. Genetics. 2004;168:1855–65. doi: 10.1534/genetics.104.032771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Stark JM, Pierce AJ, Oh J, Pastink A, Jasin M. Genetic steps of mammalian homologous repair with distinct mutagenic consequences. Mol Cell Biol. 2004;24:9305–16. doi: 10.1128/MCB.24.21.9305-9316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sugawara N, Goldfarb T, Studamire B, Alani E, Haber JE. Heteroduplex rejection during single-strand annealing requires Sgs1 helicase and mismatch repair proteins Msh2 and Msh6 but not Pms1. Proc Natl Acad Sci USA. 2004;101:9315–20. doi: 10.1073/pnas.0305749101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Sugawara N, Wang X, Haber JE. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell. 2003;12:209–19. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 143.Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, et al. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell. 2008;32:118–28. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–21. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 145.Symington LS, Heyer WD. Some disassembly required: role of DNA translocases in the disruption of recombination intermediates and dead-end complexes. Genes Dev. 2006;20:2479–86. doi: 10.1101/gad.1477106. [DOI] [PubMed] [Google Scholar]
- 146.Tay YD, Sidebotham JM, Wu L. Mph1 requires mismatch repair-independent and -dependent functions of MutS{alpha} to regulate crossover formation during homologous recombination repair. Nucleic Acids Res. 2010 Jan 4; doi: 10.1093/nar/gkp1199. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Terasawa M, Ogawa T, Tsukamoto Y, Ogawa H. Sae2p phosphorylation is crucial for cooperation with Mre11p for resection of DNA double-strand break ends during meiotic recombination in Saccharomyces cerevisiae. Genes Genetic Syst. 2008;83:209–17. doi: 10.1266/ggs.83.209. [DOI] [PubMed] [Google Scholar]
- 148.Tomita K, Matsuura A, Caspari T, Carr AM, Akamatsu Y, et al. Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double-strand breaks but not telomeres. Mol Biol Cell. 2003;23:5186–97. doi: 10.1128/MCB.23.15.5186-5197.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nature Cell Biol. 2007;9:923–U72. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 150.Torres-Rosell J, Sunjevaric I, De Piccoli G, Sacher M, Eckert-Boulet N, et al. The Smc5-Smc6 complex and SUMO modification of Rad52 regulates recombinational repair at the ribosomal gene locus. Nat Cell Biol. 2007;9:923–31. doi: 10.1038/ncb1619. [DOI] [PubMed] [Google Scholar]
- 151.Trautmann K, Terdiman JP, French AJ, Roydasgupta R, Sein N, et al. Chromosomal instability in microsatellite-unstable and stable colon cancer. Clin Cancer Res. 2006;12:6379–85. doi: 10.1158/1078-0432.CCR-06-1248. [DOI] [PubMed] [Google Scholar]
- 152.Trickey M, Grimaldi M, Yamano H. The anaphase-promoting complex/cyclosome controls repair and recombination by ubiquitylating Rhp54 in fission yeast. Mol Cell Biol. 2008;28:3905–16. doi: 10.1128/MCB.02116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Trujillo KM, Yuan SSF, Lee E, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–50. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- 154.Tsubouchi H, Roeder GS. Budding yeast Hed1 down-regulates the mitotic recombination machinery when meiotic recombination is impaired. Genes Dev. 2006;20:1766–75. doi: 10.1101/gad.1422506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Valencia-Burton M, Oki M, Johnson J, Seier TA, Kamakaka R, Haber JE. Different mating-type-regulated genes affect the DNA repair defects of Saccharomyces RAD51, RAD52 and RAD55 mutants. Genetics. 2006;174:41–55. doi: 10.1534/genetics.106.058685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol. 2009;19:207–17. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 157.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–12. doi: 10.1038/nature01585. [DOI] [PubMed] [Google Scholar]
- 158.Welz-Voegele C, Jinks-Robertson S. Sequence divergence impedes crossover more than noncrossover events during mitotic gap repair in yeast. Genetics. 2008;179:1251–62. doi: 10.1534/genetics.108.090233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.West SC. Molecular views of recombination proteins and their control. Nature Rev Mol Cell Biol. 2003;4:435–45. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 160.Whitby MC. The FANCM family of DNA helicases/translocases. DNA Repair. 2010;9:224–36. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 161.Wohlbold L, Fisher RP. Behind the wheel and under the hood: Functions of cyclin-dependent kinases in response to DNA damage. DNA Repair. 2009;8:1018–24. doi: 10.1016/j.dnarep.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Worth L, Clark S, Radman M, Modrich P. Mismatch Repair Proteins MutS and MutL Inhibit RecA-Catalyzed Strand Transfer Between Diverged DNAs. Proc Natl Acad Sci USA. 1994;91:3238–41. doi: 10.1073/pnas.91.8.3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wu LJ, Hickson ID. The Bloom’s syndrome helicase suppresses crossing-over during homologous recombination. Nature. 2003;426:870–74. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 164.Wu Y, Kantake N, Sugiyama T, Kowalczykowski SC. Rad51 protein controls Rad52-mediated DNA annealing. J Biol Chem. 2008;283:14883–92. doi: 10.1074/jbc.M801097200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Yang HJ, Li QB, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–57. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 166.Youds JL, Barber LJ, Ward JD, Collis SJ, O’Neil NJ, et al. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol Cell Biol. 2008;28:1470–79. doi: 10.1128/MCB.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–86. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yu X, Fu S, Lai M, Baer R, Chen J. BRCA1 ubiquitinates its phosphorylation-dependent binding partner CtIP. Genes Dev. 2006;20:1721–6. doi: 10.1101/gad.1431006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–63. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhu Z, Chung WH, Shim EY, Lee SE, Ira G. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell. 2008;134:981–94. doi: 10.1016/j.cell.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]