The Development and Application of Optogenetics (original) (raw)
. Author manuscript; available in PMC: 2019 Aug 19.
Abstract
Genetically encoded, single-component optogenetic tools have made a significant impact on neuroscience, enabling specific modulation of selected cells within complex neural tissues. As the optogenetic toolbox contents grow and diversify, the opportunities for neuroscience continue to grow. In this review, we outline the development of currently available single-component optogenetic tools and summarize the application of various optogenetic tools in diverse model organisms.
Keywords: channelrhodopsin, halorhodopsin, bacteriorhodopsin, electrophysiology
Introduction
In describing unrealized prerequisites for assembling a general theory of the mind, Francis Crick observed that the ability to manipulate individual components of the brain would be needed, requiring “a method by which all neurons of just one type could be inactivated, leaving the others more or less unaltered” (Crick 1979, p. 222). Extracellular electrical manipulation does not readily achieve true inactivation, and even electrical excitation, while allowing for temporal precision in stimulating within a given volume, lacks specificity for cell type. However, pharmacological and genetic manipulations can be specific to cells with certain expression profiles (in the best case) but lack temporal precision on the timescale of neural coding and signaling.
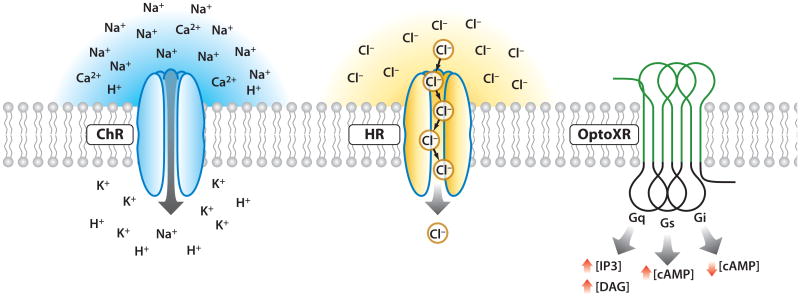
Because no prior technique has achieved both high-temporal and cellular precision within intact mammalian neural tissue, there has been strong pressure to develop a new class of technology. As a result of these efforts, neurons now may be controlled with optogenetics for fast, specific excitation or inhibition within systems as complex as freely moving mammals [for example, with microbial opsin methods, light-induced inward cation currents may be used to depolarize the neuronal membrane and positively modulate firing of action potentials, while optical pumping of chloride ions can induce outward currents and membrane hyperpolarization, thereby inhibiting spiking (Figure 1)]. These optogenetic tools of microbial origin (Figure 1) may be readily targeted to subpopulations of neurons within heterogeneous tissue and function on a temporal scale commensurate with physiological rates of spiking or critical moments in behavioral tests, with fast deactivation upon cessation of light. With these properties, microbe-derived optogenetic tools fulfill the criterion set forth by Crick in 1979 (Deisseroth 2010, 2011).
Figure 1.
Optogenetic tool families. Channelrhodopsins conduct cations and depolarize neurons upon illumination (left). Halorhodopsins conduct chloride ions into the cytoplasm upon yellow light illumination (center). OptoXRs are rhodopsin-GPCR (G protein–coupled receptor) chimeras that respond to green (500 nm) light with activation of the biological functions dictated by the intracellular loops used in the hybrid (right).
Early Efforts Toward Optical Control
The microbial opsin approach is heir to a long tradition of using light as an intervention in biology. With chromophore-assisted laser inactivation, light can be used to inhibit targeted proteins by destroying them [what a geneticist would call “loss of function” (Schmucker et al. 1994)]; conversely, lasers can be used to stimulate neurons directly in a way that could be adapted (in principle) to control fluorescently labeled, genetically targeted cells [what a geneticist would call “gain of function” (Fork 1971, Hirase et al. 2002)]. Next, various cascades of genes, and combinations of genes with chemicals, were tested as multicomponent strategies for optical control; rhodopsin and arrestin genes from Drosophila photoreceptors were combined to light-sensitize neurons (Zemelman et al. 2002); ligand-gated channels, combined with ultraviolet (UV)-light photolysis of caged agonists, were developed for Drosophila experiments (Lima & Miesenbock 2005, Zemelman et al. 2003); and UV light–isomerizable chemicals linked to genetically encoded channels were employed in cultured cells and in zebrafish (Banghart et al. 2004, Szobota et al. 2007, Volgraf et al. 2006). These efforts have been reviewed (Gorostiza & Isacoff 2008, Miesenbock & Kevrekidis 2005) and while elegant, have thus far been found to be limited to various extents in speed, targeting, tissue penetration, and/or applicability because of their multicomponent nature. Here, we review development and application efforts focused on the distinct single-component optogenetic tools, such as microbial opsins, over the past six years since they were first implemented.
Microbial Opsins
Species from multiple branches of the animal kingdom have evolved mechanisms to sense electromagnetic radiation in their environments. Likewise many microbes, in the absence of complex eye structures employed by metazoans, have developed light-activated proteins for a variety of purposes. For some, this serves as a mechanism of homeostasis to remain at a certain depth in the ocean (Beja et al. 2000, 2001); for others, this helps maintain osmotic balance in a highly saline environment (Stoeckenius 1985). These and other diverse roles are, in many cases, fulfilled by a family of seven-transmembrane, light-responsive proteins encoded by opsin genes.
Opsin genes are divided into two distinct superfamilies: microbial opsins (type I) and animal opsins (type II). Opsin proteins from both families require retinal, a vitamin A–related organic cofactor that serves as the antenna for photons; when retinal is bound, the functional opsin proteins are termed rhodopsins. Retinal covalently attaches to a conserved lysine residue of helix 7 by forming a protonated retinal Schiff base (RSBH+). The ionic environment of the RSB, defined by the residues of the binding pocket, dictates the spectral and kinetic characteristics of each individual protein. Upon absorption of a photon, retinal isomerizes and triggers a sequence of conformational changes within the opsin partner. The photoisomerized retinal is the trigger for subsequent structural rearrangements and activities performed by these proteins.
Although both opsin families encode seven-transmembrane structures, sequence homology between the two families is extremely low; homology within families, however, is high (25%–80% residue similarity) (Man et al. 2003). Whereas type I opsin genes are found in prokaryotes, algae, and fungi (Spudich 2006), type II opsin genes are present only in higher eukaryotes and are responsible mainly for vision (but also play roles in circadian rhythm and pigment regulation) (Sakmar 2002, Shichida & Yamashita 2003). Type II opsin genes encode G protein–coupled receptors (GPCRs) and, in the dark, bind retinal in the 11-cis configuration. Upon illumination, retinal isomerizes to the all-trans configuration and initiates the reactions that underlie the visual phototransduction second messenger cascade. After photoisomerization, the retinal-protein linkage is hydrolyzed; free all-trans retinal then diffuses out of the protein and is replaced by a fresh 11-cis retinal molecule for another round of signaling (Hofmann et al. 2009).
In contrast, type I opsins more typically encode proteins that utilize retinal in the all-trans configuration, which photoisomerizes upon photon absorption to the 13-cis configuration. Unlike the situation with type II rhodopsins, the activated retinal molecule in type I rhodopsins does not dissociate from its opsin protein but thermally reverts to the all-trans state while maintaining a covalent bond to its protein partner (Haupts et al. 1997). Type I opsins encode several distinct subfamilies of protein, discussed in more detail below. The central operating principle of these elegant molecular machines [established for this broad family of opsins since bacteriorhodopsin (BR) in 1971 (Oesterhelt & Stoeckenius 1971) and now including halorhodopsins and channelrhodopsins (Figure 1)] is their unitary nature. They combine the two tasks of light sensation and ion flux into a single protein (with bound small organic cofactor), encoded by a single gene. In 2005, one of these microbial opsins was brought to neuroscience as the first single-component optogenetic tool (Boyden et al. 2005), and the other microbial opsin subfamilies followed close behind. For example, channelrhodopsin-1 (ChR1) (Nagel et al. 2002) and channelrhodopsin-2 (ChR2) (Nagel et al. 2003) from Chlamydomonas reinhardtii are blue-light-activated nonspecific cation channels. In common with other type I opsins, these proteins require retinal as the photon-sensing cofactor to function. In response to light stimulation, the channel shuttles from the dark-adapted state through a stereotyped progression of functional and conformational states, eventually (in the absence of further light stimulation) reaching the dark-adapted state once again. These different states, which all have unique spectroscopic signatures, are collectively referred to as the photocycle, which (as for BR and halorhodopsin in earlier work) has been extensively investigated (Bamann et al. 2010, Berndt et al. 2010, Ernst et al. 2008, Hegemann et al. 2005, Ritter et al. 2008, Stehfest et al. 2010, Stehfest & Hegemann 2010).
The size, kinetic properties, and wavelength sensitivity of photocurrents resulting from activation of an individual protein are a direct result of its photocycle topology, ion selectivity, and activation/deactivation/inactivation time constants (Ernst et al. 2008, Hegemann et al. 2005, Ritter et al. 2008). Typically, a transient peak photocurrent, evoked at the onset of light stimulation, decays modestly to a steady-state photocurrent even in the presence of continuous light, owing in part to the desensitization of a certain population of channels (Nagel et al. 2003). The desensitized population can recover in the dark with a characteristic time constant on the order of 5 seconds, giving rise to a similar peak photocurrent if a second light pulse is applied after sufficient time has elapsed (Nagel et al. 2003). The fraction of desensitized proteins is crucial for determining the reliability of light stimulation during prolonged experiments (e.g., behavioral or long-term physiological experiments) where light is applied for extended periods. This issue is addressed in more detail below.
Optogenetic Tools for Neuronal Excitation
Many years passed after the discovery of BRs, channelrhodopsins, and halorhodopsins, prior to the development of optogenetics. As noted above, optogenetics with microbial opsins began with a channelrhodopsin, introduced into hippocampal neurons in 2005 (Boyden et al. 2005) where it was found to confer millisecond-precision control of neuronal spiking. A number of additional reports followed over the next year (Bi et al. 2006, Ishizuka et al. 2006, Li et al. 2005, Nagel et al. 2005). Moreover, this turned out to be a single-component system; through a remarkable twist of nature, investigators found that sufficient retinal is present in mammalian brains (and as later established, in all vertebrate tissues tested thus far) to enable functional expression of these optogenetic tools as single components, in the absence of any added chemical or other gene (Deisseroth et al. 2006, Zhang et al. 2006). The optogenetic toolbox has been vastly expanded since the original 2005 discovery to include dozens of single-component proteins activated by various wavelengths of light, with various ion conductance regulation properties that operate in neurons over a wide range of speeds (from milliseconds to tens of minutes), enabling broad experimental configurations and opportunities.
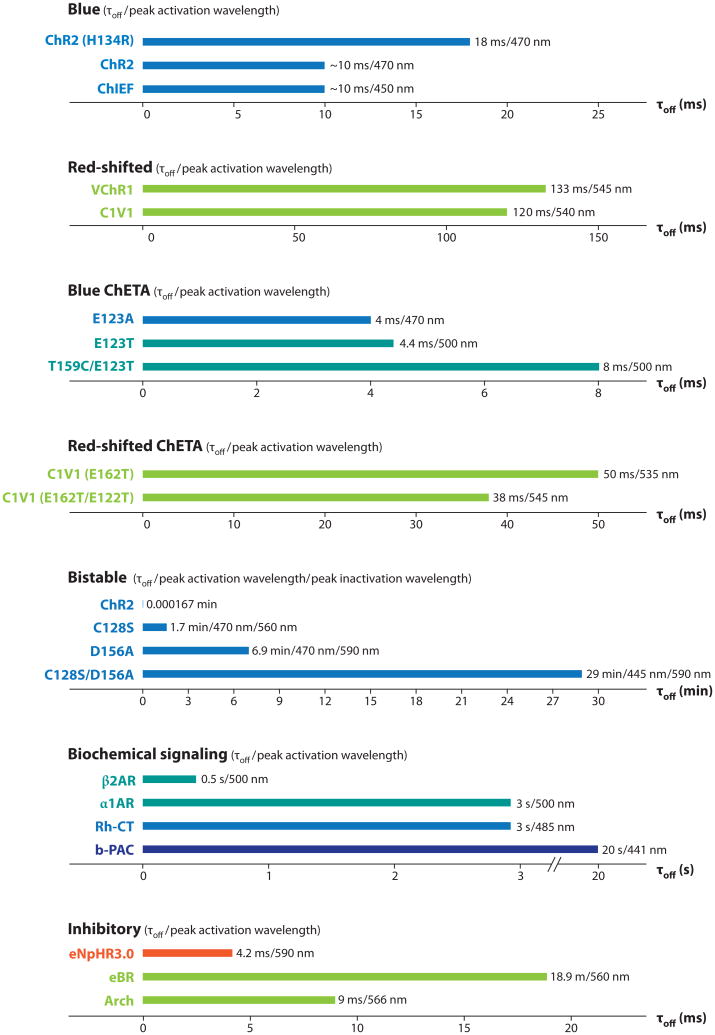
Initial improvements in ChR2 were carried out in an incremental fashion and focused on improving expression and photocurrent in mammalian systems. Human codon optimization for improved expression of ChR2 (Boyden et al. 2005, Gradinaru et al. 2007) was combined with substitution of histidine for arginine at position 134 to increase steady-state current size, although the mechanism of the latter effect (delayed channel closure) also significantly impaired temporal precision and high-speed spiking (ChR2-H134R; Nagel et al. 2005, Gradinaru et al. 2007). Better solutions were needed, and further diversification of the optogenetic toolbox via mutational engineering has proven to be challenging but highly productive (Figure 2).
Figure 2.
Spectral and kinetic diversification of optogenetic tools. Deactivation time constants (τoff) and approximate peak activation/inactivation wavelengths are shown for blue light–activated opsins, blue ChETAs (including E123T/T159C) (Berndt et al. 2011, Mattis et al. 2011), red-shifted opsins, red-shifted ChETAs, bistable (SFO) opsins, and modulatory/inhibitory tools in a compact look-up table. ChR2 is listed among the bistable group for scale purposes only. Where precise values are not available, decay kinetics were measured (courtesy of J. Mattis, personal communication) or estimated from published traces; all values were recorded at room temperature (except for optoXRs measured at 37°C), with substantial acceleration in kinetics (∼50%) expected for all at 37°C.
First, because many microbial tools do not express well in mammalian neurons, general strategies for enabling mammalian expression were required. Membrane-trafficking modifications turned out to be crucial, beginning with the observation in 2008 that endoplasmic reticulum (ER)-export motifs were helpful for achieving high, safe expression of halorhodopsins (Gradinaru et al. 2008, Zhao et al. 2008), a principle that turned out to be extendable [to other classes of membrane-trafficking motif (Gradinaru et al. 2010)] and generalizable [to most microbial opsins tested (Gradinaru et al. 2010)]. Next, chimera strategies (using hybrids of ChR1 and ChR2) were found to be helpful in giving rise to improved expression and spiking properties for channelrhodopsins (Lin et al. 2009, Wang et al. 2009).
Finally, much subsequent opsin engineering for optogenetics was carried out on the basis of hypothesized ChR structures. To date, there has been no reported crystal structure of any excitatory optogenetic tools, although structures exist for halorhodopsin and for the proton pump BR (Luecke et al. 1999a,b), which can function to inhibit neurons when heterologously expressed (Gradinaru et al. 2010); proton conductance is a property shared by ChR2, whether through channel or possible pumping mechanisms (Feldbauer et al. 2009). Capitalizing upon homology between BR and ChRs, we have introduced a number of mutations that modify various properties of the opsins (discussed below) and have described a framework by which these improvements can be applied to other novel opsins [indeed, this direction has come full circle, with insights from ChRs now used to improve the optogenetic function of BR itself (Gradinaru et al. 2010)].
These structural hypothesis-based opsin engineering efforts were spurred by inherent limitations of the channelrhodopsin system (Figure 2). Specifically, the deactivation time constant of ChR2 upon cessation of light (∼10–12 ms in neurons) imposed a limit on temporal precision, leading in some cases to artifactual multiplets of spikes after even single brief light pulses (as well as a plateau potential when pulsing blue light at frequencies above 40 hz; the next pulse of light would occur before all the ChRs had deactivated, leading eventually to failed deinactivation of native voltage-gated sodium channels and thus missed spikes and further loss of fidelity). The latter problem was compounded by the desensitization of ChR itself even in the presence of light, leading to further missed spikes late in trains. Addressing part of this challenge, the chimeric opsins noted above (e.g., ChIEF; Lin et al. 2009), demonstrated reduced desensitization in cultured neurons, allowing more robust spiking over trains in culture as well as stronger currents. In another approach addressing both desensitization and deactivation, considering the crystal structure of BR led to modification of the counterion residue E123 of ChR2 to threonine or alanine; the resulting faster opsin is referred to as ChETA (Figure 2) (Gunaydin et al. 2010). This substitution introduced two advantages over wild-type ChR2. First, it reduced desensitization during light exposure, with the result that light pulses late in high-frequency trains became as likely as early light pulses to drive spikes (a very important property referred to as temporal stationarity). Second, it destabilized the active conformation of retinal, speeding spontaneous isomerization to the inactive state after light-off and thus closing the channel much more quickly after cessation of light than wild-type or improved ChR2 variants. The resulting functional consequences of ChETA mutations are temporal stationarity, reduced extra spikes, reduced plateau potentials, and improved high-frequency spike following at 200 Hz or more over sustained trains, even within intact mammalian brain tissue (Gunaydin et al. 2010).
Whereas many experimental designs employ optogenetic tools to initiate precise spiking, alternative paradigms may instead require the researcher to simply alter the excitability of a target neuronal population. Indeed, it is often important to bias the activity of a particular neuronal population without specifically driving action potentials or synchronous activity. To facilitate experiments examining altered excitability, Berndt et al. (2009) developed the step-function opsins (SFOs). SFOs are a family of ChR mutants that display bistable behavior—orders-of-magnitude prolonged activity after termination of the light stimulus— first instantiated as mutations in position 128 in ChR2 (cysteine to serine, alanine, or threonine). Again based on homology between ChR2 and BR (Peralvarez-Marin et al. 2004), we mutated this residue to manipulate the interaction between the opsin backbone and the covalently bound retinal photon sensor. In contrast to ChETA, the SFO mutations are designed to stabilize the active retinal isomer, the functional consequence of which is prolonging the active state of the channel even after light-off. SFOs have inactivation time constants on the order of tens of seconds or more instead of milliseconds (Figure 2) and can be activated by a single 10-ms pulse of blue light (Berndt et al. 2009). The SFOs can also be deactivated by a pulse of yellow light; the yellow pulse drives isomerization of retinal back to the nonconducting state. A subsequently engineered SFO, the ChR2(D156A) opsin (Bamann et al. 2010), displays an even longer inactivation time constant, which in our hands approaches eight minutes. One potential noted use of opsins with extended time constants could be for scanning two-photon stimulation paradigms (Rickgauer & Tank 2009), during which it would be helpful to have persistent accumulating activity as a small two-photon spot scans a cell or tissue of interest.
We have now engineered a third class of SFO by combining the D156 and C128 mutations to produce a ChR2 variant that has a spontaneous deactivation time constant approaching 30 min; this stabilized SFO (SSFO) (O. Yizhar, L. Fenno, M. Prigge, K. Stehfest, J. Paz, F. Schneider, S. Tsunoda, R. Fudim, C. Ramakrishnan, J. Huguenard, P. Hegemann & K. Deisseroth, submitted) induces peak currents of _>_200 pA. An advantage of having an opsin with such a long time constant is the ability to conduct behavioral protocols in the absence of tethered external light delivery devices (e.g., optical fibers). Because a single pulse of blue light is sufficient to induce activity for a time period extending beyond that of most behavioral paradigms, the fiber may be removed before commencing the experiment. Just as with the original SFO proteins, SSFO may be inactivated by yellow light, allowing for precise control of network dynamics. A final advantage of the SFOs (which scales with their kinetic stability) is orders-of-magnitude greater light sensitivity of transduced cells, particularly for long light pulses, a direct result of the photon integration bestowed by their prolonged deactivation time constant. This property renders SFOs especially attractive as minimally invasive tools for stimulating large brain regions (for example, in primate studies) and deep within tissue.
Separate from, but synergistic with, molecular engineering is the systematic genomic identification of novel opsins. Adapting novel opsins activated by red-shifted wavelengths could enable control of two separate populations of circuit elements within the same physical volume. To this end, we launched genomic search strategies that led to identification of anopsin from Volvox carteri (VChR1) (Zhang et al. 2008), which shares homology with ChR2 and similarly functions as a cation channel. In contrast to ChR2, VChR1 is activated by red-shifted light. However, the relatively small currents due to low expression in mammalian neurons, as described in the initial report, have hampered in vivo adaptation of VChR1, even after codon optimization. To this end, we have engineered a chimeric opsin, C1V1 (O. Yizhar, L. Fenno, M. Prigge, K. Stehfest, J. Paz, F. Schneider, S. Tsunoda, R. Fudim, C. Ramakrishnan, J. Huguenard, P. Hegemann & K. Deisseroth, submitted), composed of the first two and one-half helices of ChR1 (a poorly expressing relative of ChR2 from the same organism) (Nagel et al. 2002) and the last four and one-half helices of VChR1. The resulting tool retains the red-shifted activation spectrum of VChR1, but with nanoampere-scale currents that exceed those of ChR2. The large current of C1V1 allows use in vivo and also use of off-peak (redder) light wavelengths, together enabling truly separable control of multiple populations of neurons when used in conjunction with ChR2 (O. Yizhar, L. Fenno, M. Prigge, K. Stehfest, J. Paz, F. Schneider, S. Tsunoda, R. Fudim, C. Ramakrishnan, J. Huguenard, P. Hegemann & K. Deisseroth, submitted). In addition to combinatorial experiments with ChR2, tools with red-shifted activation wavelengths such as C1V1 are also more amenable to use with simultaneous imaging of genetically encoded calcium indicators, such as GCaMP variants (Hires et al. 2008, Zhang & Oertner 2007).
Optogenetic Tools for Neuronal Inhibition
Complementing these tools for precise control of neural excitation, certain light-activated ion pumps may be used for inhibition, although thus far only one ion pump has shown efficacy at modulating behavior in mammals (Tye et al. 2011, Witten et al. 2010, Zhang et al. 2007a): the ER trafficking-enhanced version of a halorhodopsin called NpHR (Gradinaru et al. 2008, 2010; Zhang et al. 2007a) derived from the halobacterium Natronomonas pharaonis. In the context of optogenetic application, this yellow light–activated electrogenic chloride pump acts to hyperpolarize the targeted neuron upon activation (Figure 1) (Zhang et al. 2007a). Unlike the excitatory channelrhodopsins, NpHR is a true pump and requires constant light to move through its photocycle. Although optogenetic inhibition with NpHR was shown to operate well in freely moving worms (Zhang et al. 2007a), mammalian brain slices (Zhang et al. 2007a), and cultured neurons (Han & Boyden 2007, Zhang et al. 2007a), several years passed before final validation of this (or any) inhibitory optogenetic tool was obtained by successful application to intact mammals (Tye et al. 2011, Witten et al. 2010) because of membrane trafficking problems that required additional engineering (Gradinaru et al. 2008, 2010; Zhao et al. 2008). Indeed, a number of modifications to NpHR were required to improve its function, initially codon-optimizing the sequence followed by enhancement of its subcellular trafficking (eNpHR2.0 and eNpHR3.0) (Gradinaru et al. 2008, 2010), which resulted in improved membrane targeting and higher currents suitable even for use in human tissue (Busskamp et al. 2010), as well as activation with red-shifted wavelengths at the infrared border (680 nm). Kouyama et al. (2010) published the 2.0A crystal structure of halorhodopsin and illustrate that this protein has a high degree of homology within the retinal binding pocket with the proton pump BR. The two are proposed to distribute charge and store energy from absorbed photons in similar ways.
The diversification of the inhibitory opsin toolbox has been guided by bioinformatics approaches to screen nature for novel inhibitory ion pumps with desirable properties, just as had been successful previously for excitatory opsins (Zhang et al. 2008). In 2010, we and others explored the use of proton pumps (eBR, Mac, and Arch) as optogenetic tools (Gradinaru et al. 2010, Chow et al. 2010), finding evidence for robust efficacy but leaving open questions of long-term tolerability and functionality of proton-motive pumps in mammalian neurons. A major concern is the extent to which pumping of protons into the extracellular space (especially in juxtamembranous compartments difficult to visualize or measure) could have deleterious or noncell-type-specific effects on local tissue, which could show up as light-induced inhibition affecting all recorded units (more units than expected for a given transduction efficiency) or with a slightly slower time course than expected for the fast proton pumps.
Although the proton pumps must be treated with caution until these issues are addressed, many opportunities exist; indeed, we have improved the ability of proton pumps to hyperpolarize neurons following a methodology similar to that used for improving NpHR (Gradinaru et al. 2010), although none of the proton pumps yet described is as kinetically stable or as potent as eNpHR3.0, especially at the safe light levels (_<_20 mW/mm2 at the target cell) required for in vivo use (Gradinaru et al. 2010). Indeed, eNpHR3.0 has now delivered the loss-of-function side of the optogenetic coin for freely moving mammals (Tye et al. 2011, Witten et al. 2010), complementing the channelrhodopsins, which delivered gain of function. Witten and colleagues (2010) used eNpHR3.0 to inhibit the cholinergic neurons of the nucleus accumbens and identified a causal role for these rare cells in implementing cocaine conditioning in freely moving mice by enhancing inhibition of inhibitory striatal medium spiny neurons. Tye and colleagues (2011) used eNpHR3.0 to inhibit a specific intra-amygdala projection in freely moving mice, creating a switchable model of anxiety in mammals and implicating a highly defined neural pathway as a native circuit involved in anxiolysis.
Optogenetic Tools for Biochemical Control
The microbial (type I) opsins described up to this point serve strictly as ion flow modulators and control the excitability of a neuron by manipulating its membrane potential: either bringing the voltage nearer or above the threshold for generating an action potential or hyperpolarizing the potential and thereby inhibiting spiking. Several classes of biochemical control have now been achieved, beginning with the use of eukaryotic (type II) opsins for precise control of well-defined GPCR signaling pathways.
Rhodopsin, the light-sensing protein in the mammalian eye, is both an opsin, in that it is covalently bound to retinal and its function is modulated by the absorption of photons, and a GPCR, in that it is coupled on the intracellular side of the membrane to a G-protein, transducin. Virtually all neurons can communicate via GPCRs, which of course respond not only to neuromodulators from dopaminergic, serotonergic, and adrenergic pathways, but also to “fast” neurotransmitters such as glutamate and GABA. Building both on our finding that adequate retinal is present within mammalian brain tissue (Deisseroth et al. 2006, Zhang et al. 2006) and on a long history of elegant GPCR structure-function work from Khorana, Kobilka, Caron, Lefkowitz, and others, we have determined that GPCRs can be converted into light-activated regulators of well-defined biochemical signaling pathways that function within freely moving mammals. These proteins are referred to as optoXRs (Airan et al. 2009). OptoXRs allow for receptor-mediated intracellular signaling with temporal resolution suitable for modulation of behavior in freely moving mice (Airan et al. 2009). OptoXRs are modulated by 500-nm light and now include the alpha−1 and beta−2 adrenergic receptors (Airan et al. 2009), which are coupled to Gq and Gs signaling pathways, respectively, and the 5-HT1a receptor,which is Gi/o coupled (Oh et al.2010).
Optical control over small GTPases was next achieved in cultured cells by several different laboratories (Levskaya et al. 2009, Wu et al. 2009, Yazawa et al. 2009) using optically modulated protein-protein interactions; although these have not yet been shown to express or display single-component functionality in freely moving mammals, such capability is plausible where flavin or biliverdin chromophores are required and present. Finally, investigators have recently described microbial adenylyl cyclases with lower dark activity than earlier microbial cyclases, and because they employ a flavin chromophore, these tools appear suitable for single-component optogenetic control (Ryu et al. 2010, Stierl et al. 2010). Together, these experiments have extended optogenetic capability to essentially every cell type, even in nonexcitable tissues, in biology.
Delivering Optogenetic Tools into Neuronal Systems
Viral expression systems have the dual advantages of fast/versatile implementation and high infectivity/copy number for robust expression levels. Cellular specificity can be obtained with viruses by specific promoters (if small, specific, and strong enough), by spatial targeting of virus injection, and by restriction of opsin activation to particular cells (or projections of specific cells) via targeted light delivery (Zhang et al. 2010, Diester et al. 2011). Lenti and adeno-associated (AAV) viral vectors have been used successfully to introduce opsins into the mouse, rat, and primate brain (Zhang et al. 2010). Additionally, these have been well tolerated and highly expressed over long periods of time with no reported adverse effects. Lentivirus is easily produced using standard tissue culture techniques and an ultracentrifuge (see Zhang et al. 2010 for protocol). AAV may be produced either by individual laboratories or through core viral facilities. Neither AAV nor lentivirus were found to be highly expressed in the zebrafish, for which Sindbis and rabies are more effective (Zhu et al. 2009). Viruses have been used to target (among other cells) hypocretin neurons (Adamantidis et al. 2007), excitatory pyramidal neurons (Lee et al. 2010, Sohal et al. 2009, Zhang et al. 2007a), and astroglia (Gourine et al. 2010, Gradinaru et al. 2009). For example, one group (Gourine et al. 2010) recently described the use of AAV-delivered ChR2 to control astroglial activity in the brain stem of mice and to dissect a mechanism by which astroglia can transfer systemic information from the blood to neurons underlying homeostasis, in this case directly modulating neurons that manipulate the rate of respiration. However,amajor downside of viral expression systems is a maximum genetic payload length; only promoter fragments that are small (less than ∼4 kb), specific, and strong may be used, and these are rare. This limitation may be skirted using Cre-driver animals and Cre-dependent viruses, discussed below.
Transgenic Animals
The use of transgenic or knock-in animals obviates viral payload limitations and allows for tighter control of transgene expression using larger promoter fragments or indeed the endogenous genome in full via knock-in. The first transgenic opsin-expressing mouse line was generated using the Thy1 promoter (Arenkiel et al. 2007, Zhao et al. 2008), with widespread expression throughout neocortical layer 5 projection neurons as well as in some subcortical structures (Arenkiel et al. 2007). This mouse line has been widely used, for example, to examine the roles of inhibitory neurons on cortical information processing (Sohal et al. 2009) and the mechanism of action of deep brain stimulation for Parkinson's disease (Gradinaru et al. 2009). Several other groups have subsequently also generated transgenic mouse lines directly expressing opsin genes (Hagglund et al. 2010, Katzel et al. 2010, Thyagarajan et al. 2010).
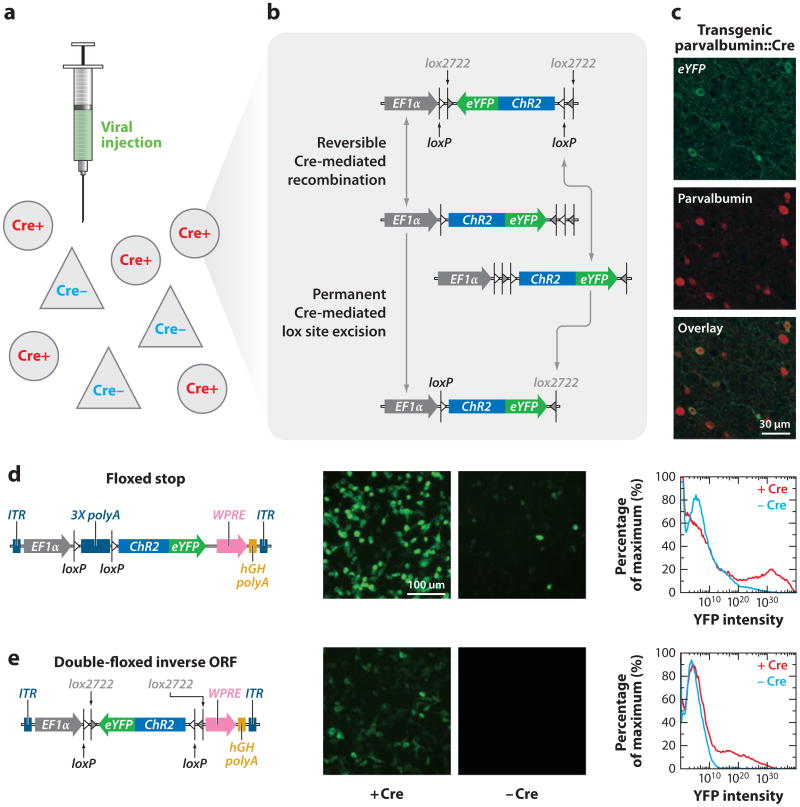
Caveats to using transgenic mouse lines to directly express optogenetic tools include the time, effort, and cost associated with their production, validation, and maintenance. To enable widespread use of the latest optogenetic tools, investigators have designed opsindelivering viruses for which opsin expression is dependent on the coexpression of Cre recombinase (Figure 3). This doublefloxed inverted open-reading-frame (DIO; reviewed in Zhang et al. 2010) strategy (Atasoy et al. 2008, Sohal et al. 2009, Tsai et al. 2009) situates the opsin gene (inhibitory or excitatory) in the inverted (meaningless) orientation, but the gene is flanked by two sets of incompatible Cre recombinase recognition sequences (Sohal et al. 2009, Tsai et al. 2009). The recombinase recognition sequences are placed such that in the presence of Cre, the ORF is inverted instead of being excised. Reversing the sequence then allows one of the Cre recognition sites to be excised (Figure 3), locking the reading frame into the correct direction and allowing for strong expression of the opsin with (for example) the elongation factor 1-alpha (EF1α) promoter. The specificity of this gene expression can then come (for example) from the targeted expression of Cre in driver rodent lines in which Cre is controlled with high specificity in the context of very large chromosomal promoter-enhancer regions; the DIO strategy thus enables versatile and widespread use of optogenetics with the many (and growing number of) experimental systems selectively expressing Cre recombinase (Geschwind 2004; Gong et al. 2003, 2007; Heintz 2004). This strategy has been used recently in many systems, for example to target dopamine-1 (D1) or dopamine-2 (D2) receptor–expressing neurons of the striatum via transgenic D1-Cre and D2-Cre mouse lines, to examine the effects of their stimulation in the classic direct/indirect-movement pathways (Kravitz et al. 2010). With these same Cre lines, Lobo and colleagues (2010) examined the roles of nucleus accumbens D1 and D2 neurons in modulating cocaine reward. The Cre-dependent optogenetic system allowed for the first time a direct examination of the relationship between neuronal activity of specific neuronal populations and animal behavior, thus paving the way for a deeper understanding of diseases such as Parkinson's disease, depression, and substance abuse.
Figure 3.
Low-leak Cre-dependent expression using the doublefloxed inverted open-reading-frame (DIO) strategy. The combination of a transgenic mouse expressing Cre recombinase in specific neuronal subtypes and the injection of a virally encoded DIO opsin (a) results in the physical inversion of the open reading frame (ORF) in only that population (b,c), which may be transient and revert back to the original state or undergo further recombination to be permanently anchored in the sense direction, resulting in functional expression of the opsin (c). The DIO strategy may be contrasted with the lox-stop-lox (“floxed STOP”) strategy (d,e). In the absence of Cre recombinase, lox-stop-lox (d) allows for some level of expression leak as assayed by both enhanced yellow fluorescent protein (eYFP) expression and fluorescence-activated cell sorting (FACS) analysis. Because the ORF of an opsin in DIO configuration encodes nonsense (e), there is no functional expression in the absence of Cre recombinase. Adapted with permission from Sohal et al. (2009) and F. Zhang and K. Deisseroth.
Developmental and Layer-Specific Targeting
The ability to target specific neocortical layers has been a long-sought goal of neuroscience; this can now be achieved either with layer-specific Cre driver lines or with developmental targeting strategies such as in utero electroporation (IUE). As a result, multiple laboratories have now successfully teased apart the role of layer-specific neurons in behavioral paradigms and network dynamics. Optogenetic tools have been well tolerated when electroporated in utero into mouse embryos (Adesnik & Scanziani 2010, Gradinaru et al. 2007, Lewis et al. 2009, Petreanu et al. 2007); IUE may be used to target specific layers of cortex by incorporating the DNA (with no promoter size limit) into neurons being born during a specific embryonic stage. A major advantage of using IUE or transgenic mice over viral infection is that opsins are expressed at the time of birth, allowing electrophysiological researchers to harvest acute slices at a younger stage. Counteracting this advantage is the fact that transgenic animals typically express lower levels of opsins, likely owing to the reduced gene copy number.
Circuit Targeting
Another generalizeable strategy for targeting is referred to as projection targeting, which capitalizes on the efficient membrane trafficking of engineered opsin gene products, especially down axons to axon terminals. Light can be delivered not to somata but to axons, thereby recruiting cells defined by virtue of their wiring without any genetic information about the downstream target required (Gradinaru et al. 2007, 2009; Lee et al. 2010; Petreanu et al. 2007). Neurons may also be targeted by projection using viruses that transduce axon terminals, such as herpes simplex virus (HSV) family viruses, certain serotypes of AAV, or pseudotyped lentiviruses. Trans-synaptic targeting may be achieved by exploiting the transcellular trafficking of, for example, the wheat germ agglutinin (WGA) peptide sequence (Gradinaru et al. 2010), which can deliver Cre recombinase to the site of a second Cre-dependent virus injection. Combination strategies are also possible; by crossing transgenic Drd2-GFP mice with mice expressing Cre under the control of Emx1, then injecting DIO-ChR2-mCherry into cortex, Higley & Sabatini (2010) were able to localize synapses originating from cortex (red projections) onto neurons expressing the dopamine-2 receptor (D2) in striatum (green cell bodies) and use brief pulses of blue light to elicit synaptic activity onto the D2 neurons. Combining optogenetic manipulation of the synapse with pharmacology and two-photon glutamate uncaging allowed the investigators to elaborate precisely upon the role of D2 receptors in glutamatergic synaptic transmission.
A noteworthy method pioneered by the Callaway group (Wickersham et al. 2007a,b) using a glycoprotein-deficient pseudotyped rabies virus is yet another technique for monosynaptic circuit tracing. Rabies virus is well known to travel trans-synaptically from neuron to neuron; the virus used in this technique is not able to produce viable packaged copies of itself after moving trans-synaptically and thus will be stopped after one synapse jump. Applying this method enables tracing of all neurons synaptically connected to a single neuron of interest. Two plasmids, one containing the glycoprotein-deficient rabies payload and another containing the glycoprotein, are coelectroporated into a single neuron in vivo. This neuron is then able to produce competent rabies virus; the payload, however, does not encode the coat protein. In this case, the virus is stuck after moving one synapse. By using red and green fluorophores in the two components, the targeted neuron and its synaptic partners may be identified. This system has not yet been integrated with optogenetics, however, and the extremely high levels of expression resulting from rabies virus expression may result in toxicity incompatible with typical optogenetic experiments.
Light Delivery and Readout Hardware for Optogenetics
Optogenetics fundamentally relies on light-delivery technology, the development of which has led to improved precision of modulation both in vitro and in vivo. In vitro, optogenetic tools are typically activated with filtered light from mercury arc lamps (e.g., Berndt et al. 2009, Boyden et al. 2005, Gunaydin et al. 2010), lasers (Cardin et al. 2010, Cruikshank et al. 2010, Kravitz et al. 2010, Petreanu et al. 2009), light-emitting diodes (LEDs) (e.g., Adesnik & Scanziani 2010; Grubb & Burrone 2010a,b; Wang et al. 2009), or LED arrays for multisite stimulation (Grossman et al. 2010). In vivo, stimulation of behaving animals has been conducted mostly with laser light delivered to the transduced tissue via optical fibers inserted through chronically implanted cannulas (Adamantidis et al. 2007, Aravanis et al. 2007, Zhang et al. 2010) or with fiber-coupled high-power LEDs (Wang et al. 2010). The chronic delivery of light using implanted infrared-triggered LEDs (Iwai et al. 2011) is in the early developmental stages but promises to open a new direction in optogenetic research.
To achieve rich readouts from optogenetically controlled tissue, major effort has been directed toward generating electrophysiological systems that combine high-density single-unit recordings with optogenetic stimulation in mice and other organisms. The first readouts from in vivo optogenetic modulation were obtained in anesthetized animals using a device composed of a fiberoptic cable integrated with a tungsten electrode (Gradinaru et al. 2007), called an “optrode” (Cardin et al. 2009; Gradinaru et al. 2007, 2009; Sohal et al. 2009; Zhang et al. 2009). More advanced strategies have emerged recently, employing silicone multisite electrodes (Royer et al. 2010) and movable tetrode arrays combined with optical fibers for more flexible interrogation of neural activity in vivo (Lima et al. 2009). Two-photon imaging is another avenue with which optogenetics may be integrated to stimulate and record neural activity simultaneously. Several studies have made progress toward this type of experiment (Andrasfalvy et al. 2010, Mohanty et al. 2008, Papagiakoumou et al. 2010, Rickgauer & Tank 2009, Zhu et al. 2009); it seems that the major limitation that hampers optogenetic activation with two-photon approaches is the combination of rapidly decaying opsin-mediated photocurrents in the setting of typical slow 2P raster-scanning techniques. Modifying the raster scan paradigm (Rickgauer & Tank 2009) or modulating the laser light such that fast activation is possible across wider regions (i.e., an entire cell soma; Papagiakoumou et al. 2010) can address this problem (Shoham 2010). Appropriately rich readouts, when combined with optogenetic inputs, will powerfully facilitate fundamental studies regarding the organization and function of intact, complex neural networks.
Optogenetics in Diverse Animal Models
Caenorhabditis elegans
In transgenic nematodes harboring the channelrhodopsin gene, it is possible to control muscle wall motor neuron and mechanosensory neuron activity (Nagel et al. 2005). Zhang and colleagues (2007a) controlled body wall muscle contraction bidirectionally with ChR2 and NpHR, demonstrating the power of combinatorial optogenetics. This concept has been built on with the description of three-color LCD-based multimodal light delivery (Stirman et al. 2011) and digital micromirror device (DMD)/laser light delivery (Leifer et al. 2011), each coupled with tracking software for use with the behaving specimen. The facility of quantifying body-wall contraction and elongation in C. elegans has enabled large-scale investigation of various mutant strains for synaptic protein defects (Liewald et al. 2008, Stirman et al. 2010) and nicotinic acetylcholine receptor function (Almedom et al. 2009). Finally, C. elegans was also used for combined light-based stimulation and readout of neural activity (Guo et al. 2009, Tian et al. 2009), fulfilling the promise of all-optical physiological experiments using optogenetic tools and genetically encoded activity sensors (Scanziani & Hausser 2009).
Fly
Fly lines expressing upstream activation sequence (UAS):ChR2 (Zhang et al. 2007b) have been used to investigate the neuronal basis of the nociceptive response (Hwang et al. 2007) and appetitive/aversive odorant learning at the receptor (Bellmann et al. 2010) or neurotransmitter (Schroll et al. 2006) level and to rescue photosensory mutants (Xiang et al. 2010). Additionally, Hortstein et al. (2009) demonstrated Gal4/UAS targeting of ChR2 to the larval neu-romuscular junction system. Creative uses of optogenetic tools in Drosophila include validating neurons identified in a screen to probe the proboscis extension reflex (Gordon & Scott 2009), driving monoamine release to validate fast-scanning cyclic voltammetry detection of serotonin and dopamine (Borue et al. 2009), and investigating the innate escape response (Zimmermann et al. 2009). Special considerations are required for this model organism (Pulver et al. 2009). Unlike mammals, flies and worms do not possess levels of endogenous retinal sufficient for the function of optogenetic tools, but food supplement can provide sufficient retinal to drive ChR2 function (Xiang et al. 2010).
Flies also possess innate behavioral responses to blue light that are developmentally dependent (Bellmann et al. 2010, Pulver et al. 2009, Suh et al. 2007, Xiang et al. 2010), complicating behavioral studies using opsins with blue activation spectra. This confound may be partially rectified using fly lines without vision, such as those lacking the norpA gene (Bellmann et al. 2010), although norpA-deficient lines remain sensitive to blue light (Xiang et al. 2010). These issues could, in principle, be circumvented with red-shifted optogenetic tools for excitation and inhibition. Complementing eNpHR3.0 for red-shifted inhibition, we have developed an optogenetic toolset for potent red-shifted excitation (O. Yizhar, L. Fenno, M. Prigge, K. Stehfest, J. Paz, F. Schneider, S. Tsunoda, R. Fudim, C. Ramakrishnan, J. Huguenard, P. Hegemann & K. Deisseroth, submitted). These have yet to be tested in Drosophila, but their green peak activation wavelength is outside the range of key Drosophila photosensory proteins (Xiang et al. 2010); moreover, spiking may be driven with up to 630 nm light, improving the potential for deep-penetrating excitation (Pulver et al. 2009).
Zebrafish
The short generational time and easy integration of foreign DNA into zebrafish are complemented by ease of optogenetic manipulation owing to transparency of the organism (McLean & Fetcho 2011, White et al. 2008). The first use of optogenetic tools in zebrafish (Douglass et al. 2008) appeared in a study examining the role of somatosensory control of escape behavior. The use of ChR2 to drive single spikes in a genetically defined population during the course of movement took advantage of a number of properties of optogenetic tools not available with traditional pharmacological or electrophysiological methods. Neurons were stimulated with a simple setup combining a dissecting scope and epifluorescence source, with light restricted by the microscope aperture. Recent advances (Arrenberg et al. 2009) reported zebrafish lines with eNpHR- enhanced yellow fluorescent protein (eYFP) and ChR2-eYFP expression controlled by the Gal4/UAS system to allow the tools to be easily targeted to specific neuronal subtypes via genetic crosses with zebrafish expressing Gal4 in various cell populations. Of note, Arrenberg et al. (2009) compared various iterations of NpHR and fluorophore and concluded that eNpHR2.0-eYFP had the most reliable and efficient expression. Photoconverting proteins Kaede and Dendra were used to approximate the upper bound of light spread with low numerical aperture, small-diameter (50 um) fibers in place of a microscope aperture, observing that the combination of small-fiber and neuronal targeting allowed for the stimulation of an approximately 30-um-diameter spot.
Other groups have reported using Gal4/ UAS systems driving optogenetic tools to examine cardiac function and development (Arrenberg et al. 2010), transduction of sensory neuron mechanoreception (Low et al. 2010), command of swim behavior (Arrenberg et al. 2009, Douglass et al. 2008), and saccade generation (Schoonheim et al. 2010). As an alternative to producing stable transgenic lines, Zhu et al. (2009) undertook a systematic examination of viral infection in zebrafish and found successful ChR2 delivery by the Sindbis and rabies viruses. Of note, they also modulated ChR2 expression using a Tet-inducible expression strategy. A specific technical consideration of implementing optogenetic tools in studies using zebrafish is the stimulation of neurons that express endogenous light-actived proteins; Arrenberg et al. (2009) found that 26% of control neurons in zebrafish had a firing rate modulated by yellow light. This percentage was reduced to 14% in congenitally blind zebrafish lines. The remaining response was postulated to be due to either expression of other optically activated proteins or a thermal response, but it was not investigated further.
Mouse
By far, the most widely published optogenetic model organism to date has been the mouse. Mice represent the majority of transgenic animals, including a vast selection of transgenic lines expressing Cre recombinase in specific subpopulations of neurons (Gong et al. 2007). Mouse embryonic stem cells have also been amenable to expression and interrogation with optogenetic technologies (Stroh et al. 2010). The first report to use channelrhodopsin in behaving mammals examined the contribution of hypothalamic hypocretin (orexin) neurons to sleep and wakefulness (Adamantidis et al. 2007).
Optogenetic modulation in mouse has also yielded control of monoaminergic systems. Recently, in a study that used ChR2, eNpHR2.0, and TH∷Cre transgenic mice to modulate the locus coeruleus neurons bidirectionally, these noradrenergic neurons strongly modulated sleep and arousal states (Carter et al. 2010). Using the same TH∷Cre transgenic mice, causal relationships were identified between activity patterns in VTA dopamine neurons and reward behavior in mice, showing that phasic dopamine release is more effective than tonic release in driving reward behavior (Tsai et al. 2009). Using optogenetic stimulation of axonal terminals in the nucleus accumbens, investigators recently discovered that dopamine neurons corelease glutamate (Stuber et al. 2010, Tecuapetla et al. 2010). DAT-Cre mice have been used in conjunction with Cre-dependent ChR2 to examine mechanisms underlying dopamine-modulated addiction (Brown et al. 2010). And ChAT-Cre transgenic mice were used in combination with Cre-dependent ChR2 and NpHR viruses to show that cholinergic interneurons of the nucleus accumbens are key regulators of medium spiny neuron activity and can modulate cocaine-based place preference (Witten et al. 2010).The connectivity of striatal medium spiny neurons themselves has been described using a tetracycline-based ChR2 transgenic system (Chuhma et al. 2011).
Direct optogenetic modulation of principal and local-circuit inhibitory neurons in mouse cortex and hippocampus has also enabled contributions to understanding the complexity of mammalian neural circuit dynamics. Reports on the functions of parvalbumin-expressing fast-spiking interneurons demonstrated directly their involvement in gamma oscillations and information processing in mouse prefrontal (Sohal et al. 2009) and somatosensory (Cardin et al. 2009, 2010) cortex. Focal stimulation of pyramidal neurons in Thy1∷ChR2 mice has enabled rapid, functional mapping of motor control across the motor cortex (Ayling et al. 2009, Hira et al. 2009), and axonal stimulation in regions contralateral to injected cortical areas has enabled the mapping of projection patterns in callosal cortical projections (Petreanu et al. 2007). Within local cortical microcircuits, ChR2 has been used to characterize the spatial receptive fields of various neuron types (Katzel et al. 2010, Petreanu et al. 2009, Wang et al. 2007) and to study the basic properties of cortical disynaptic inhibition (Hull et al. 2009). Optogenetics is also being used to discern the possible therapeutic mechanism of cortical intervention in mouse models of depression (Covington et al. 2010) and to develop novel strategies for control of peripheral nerves (Llewellyn et al. 2010).
Mice have also been used to study amygdala circuits involved in fear and anxiety. Johansen and colleagues (2010) used ChR2 to demonstrate the sufficiency of lateral amygdala pyramidal neurons in auditory cued fear conditioning. In two recent reports, functional circuits within the central amygdala were further delineated, demonstrating that distinct subpopulations of inhibitory central amygdala neurons separately gate the acquisition and expression of conditioned fear (Ciocchi et al. 2010, Haubensak et al. 2010). Finally, Tye and colleagues (2011) described the differential effects of activating lateral amygdala projections onto central amygdala neurons in regulating anxiety behaviors. These studies shed new light on fear and anxiety behaviors and demonstrate the utility of optogenetic techniques in dissecting complex local neuronal circuits.
Rat
Rats are important for neuroscience research because of their ability to perform complex behavioral tasks, the relative simplicity of their brains (compared with human and nonhuman primates), and the ability to perform highdensity recordings of neural ensembles during free behavior. Recently, virally delivered optogenetic tools were used inrats to examine blood oxygen level–dependent (BOLD) responses in functional magnetic resonance imaging (fMRI) (Lee et al. 2010). Driving ChR2 in excitatory neuronal populations was sufficient to elicit a BOLD response not only in local cortical targets (where both the virus and light delivery optical fiber were targeted) but also in downstream thalamic regions, allowing global maps of activity causally driven by defined cell populations to be obtained within intact living mammals. Optogenetic work in rats has been limited by the availability of viral promoters that are capable of driving specific expression in the absence of transgenic targeting, but the advent of transgenic rat lines expressing Cre recombinase in specific neuronal subtypes (in addition to projection targeting) will greatly expand the potential for using rat models of neural circuit function in health and disease. Outside the central nervous system, optogenetic manipulation in rodents is providing insights into diverse physiological functions. ChR2 was used to modulate rhythmic beating activity in rodent cardiomyocytes, demonstrating the potential for future applications in this field (Bruegmann et al. 2010), and several groups have used optogenetics to modulate cardiovascular function, breathing, and blood pressure (Abbott et al. 2009a,b; Alilain et al. 2008; Kanbar et al. 2010) in both anesthetized and awake rats.
Primate
Optogenetic modulation of primate neurons (Han et al. 2009, Diester et al. 2011) has been explored by ChR2 delivery to cortical neurons of macaques via lentiviral transduction, but behavioral responses have not yet been observed. eNpHR2.0 has been delivered to human neural tissue in the form of ex vivo human retinas and has shown optogenetic efficacy on physiological measures (Busskamp et al. 2010) with possible relevance to retinitis pigmentosa (RP), a disease in which light-sensing cells degenerate in the retina. By expressing eNpHR2.0 in light-insensitive cone cells, normal phototransduction was restored,as well as center/surround computational features, directional sensitivity, and light-guided behavior. Additionally, Weick et al. (2010) demonstrated the functionality of ChR2 in human embryonic stem cell–derived neurons.
Outlook
The optogenetic toolbox has broadly expanded to include proteins that are powerful and diverse in their ionic selectivity, spectral sensitivity, and temporal resolution. Combined with powerful molecular techniques for trans-genic and viral expression in rodents, zebrafish, and flies, the current generation of optogenetic tools may be adapted to an extensive landscape of questions within neuroscience. The current generation of optogenetic tools has been optimized for stronger expression, higher currents, and spectral shifts to allow combinatorial control within the same volume of space. Ongoing improvements to the toolbox will yield molecular tools targeted to subcellular compartments [such as dendrites or axons (Lewis et al. 2009)], tools for two-photon activation, and tools that further expand the optical control of biochemistry.At this moment in time, single-component optogenetics has become a staple in neuroscience laboratories, even as many opportunities remain yet untapped.
Footnotes
Disclosure Statement: The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Literature Cited
- Abbott SB, Stornetta RL, Fortuna MG, Depuy SD, West GH, et al. Photostimulation of retrotrape-zoid nucleus phox2b-expressing neurons in vivo produces long-lasting activation of breathing in rats. J Neurosci. 2009a;29:5806–19. doi: 10.1523/JNEUROSCI.1106-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abbott SB, Stornetta RL, Socolovsky CS, West GH, Guyenet PG. Photostimulation of channelrhodopsin-2 expressing ventrolateral medullary neurons increases sympathetic nerve activity and blood pressure in rats. J Physiol. 2009b;587:5613–31. doi: 10.1113/jphysiol.2009.177535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–24. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adesnik H, Scanziani M. Lateral competition for cortical space by layer-specific horizontal circuits. Nature. 2010;464:1155–60. doi: 10.1038/nature08935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airan RD, Thompson KR, Fenno LE, Bernstein H, Deisseroth K. Temporally precise in vivo control of intracellular signalling. Nature. 2009;458:1025–29. doi: 10.1038/nature07926. [DOI] [PubMed] [Google Scholar]
- Alilain WJ, Li X, Horn KP, Dhingra R, Dick TE, et al. Light-induced rescue of breathing after spinal cord injury. J Neurosci. 2008;28:11862–70. doi: 10.1523/JNEUROSCI.3378-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almedom RB, Liewald JF, Hernando G, Schultheis C, Rayes D, et al. An ER-resident membrane protein complex regulates nicotinic acetylcholine receptor subunit composition at the synapse. EMBO J. 2009;28:2636–49. doi: 10.1038/emboj.2009.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrasfalvy BK, Zemelman BV, Tang J, Vaziri A. Two-photon single-cell optogenetic control of neuronal activity by sculpted light. Proc Natl Acad Sci USA. 2010;107:11981–86. doi: 10.1073/pnas.1006620107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravanis AM, Wang LP, Zhang F, Meltzer LA, Mogri MZ, et al. An optical neural interface: in vivo control of rodent motor cortex with integrated fiberoptic and optogenetic technology. J Neural Eng. 2007;4:S143–56. doi: 10.1088/1741-2560/4/3/S02. [DOI] [PubMed] [Google Scholar]
- Arenkiel BR, Peca J, Davison IG, Feliciano C, Deisseroth K, et al. In vivo light-induced activation of neural circuitry in transgenic mice expressing channelrhodopsin-2. Neuron. 2007;54:205–18. doi: 10.1016/j.neuron.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrenberg AB, Del Bene F, Baier H. Optical control of zebrafish behavior with halorhodopsin. Proc Natl Acad Sci USA. 2009;106:17968–73. doi: 10.1073/pnas.0906252106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrenberg AB, Stainier DY, Baier H, Huisken J. Optogenetic control of cardiac function. Science. 2010;330:971–74. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–30. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayling OG, Harrison TC, Boyd JD, Goroshkov A, Murphy TH. Automated light-based mapping of motor cortex by photoactivation of channelrhodopsin-2 transgenic mice. Nat Methods. 2009;6:219–24. doi: 10.1038/nmeth.1303. [DOI] [PubMed] [Google Scholar]
- Bamann C, Gueta R, Kleinlogel S, Nagel G, Bamberg E. Structural guidance of the photocycle of channelrhodopsin-2 by an interhelical hydrogen bond. Biochemistry. 2010;49:267–78. doi: 10.1021/bi901634p. [DOI] [PubMed] [Google Scholar]
- Banghart M, Borges K, Isacoff E, Trauner D, Kramer RH. Light-activated ion channels for remote control of neuronal firing. Nat Neurosci. 2004;7:1381–86. doi: 10.1038/nn1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–6. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- Beja O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–89. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- Bellmann D, Richardt A, Freyberger R, Nuwal N, Schwarzel M, et al. Optogenetically induced olfactory stimulation in Drosophila larvae reveals the neuronal basis of odor-aversion behavior. Front Behav Neurosci. 2010;4:27. doi: 10.3389/fnbeh.2010.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt A, Prigge M, Gradmann D, Hegemann P. Two open states with progressive proton selectivities in the branched channelrhodopsin-2 photocycle. Biophys J. 2010;98:753–61. doi: 10.1016/j.bpj.2009.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt A, Schoenenberger P, Mattis J, Tye KM, Deisseroth K, et al. High-efficiency channelrhodopsins for fast neuronal stimulation at low light levels. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1017210108. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt A, Yizhar O, Gunaydin LA, Hegemann P, Deisseroth K. Bi-stable neural state switches. Nat Neurosci. 2009;12:229–34. doi: 10.1038/nn.2247. [DOI] [PubMed] [Google Scholar]
- Bi A, Cui J, Ma YP, Olshevskaya E, Pu M, et al. Ectopic expression of a microbial-type rhodopsin restores visual responses in mice with photoreceptor degeneration. Neuron. 2006;50:23–33. doi: 10.1016/j.neuron.2006.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borue X, Cooper S, Hirsh J, Condron B, Venton BJ. Quantitative evaluation of serotonin release and clearance in Drosophila. J Neurosci Methods. 2009;179:300–8. doi: 10.1016/j.jneumeth.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–68. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Brown MT, Bellone C, Mameli M, Labouebe G, Bocklisch C, et al. Drug-driven AMPA receptor redistribution mimicked by selective dopamine neuron stimulation. PLoS One. 2010;5:e15870. doi: 10.1371/journal.pone.0015870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruegmann T, Malan D, Hesse M, Beiert T, Fuegemann CJ, et al. Optogenetic control of heart muscle in vitro and in vivo. Nat Methods. 2010;7:897–900. doi: 10.1038/nmeth.1512. [DOI] [PubMed] [Google Scholar]
- Busskamp V, Duebel J, Balya D, Fradot M, Viney TJ, et al. Genetic reactivation of cone photoreceptors restores visual responses in retinitis pigmentosa. Science. 2010;329:413–17. doi: 10.1126/science.1190897. [DOI] [PubMed] [Google Scholar]
- Cardin JA, Carlen M, Meletis K, Knoblich U, Zhang F, et al. Driving fast-spiking cells induces gamma rhythm and controls sensory responses. Nature. 2009;459:663–67. doi: 10.1038/nature08002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardin JA, Carlén M, Meletis K, Knoblich U, Zhang F, et al. Targeted optogenetic stimulation and recording of neuronsin vivo using cell-type-specific expression of channel rhodopsin-2. Nat Protoc. 2010;5:247–54. doi: 10.1038/nprot.2009.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter ME, Yizhar O, Chikahisa S, Nguyen H, Adamantidis A, et al. Tuning arousal with optogenetic modulation of locus coeruleus neurons. Nat Neurosci. 2010;13:1526–33. doi: 10.1038/nn.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow BY, Han X, Dobry AS, Qian X, Chuong AS, et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature. 2010;463:98–102. doi: 10.1038/nature08652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuhma N, Tanaka KF, Hen R, Rayport S. Functional connectome of the striatal medium spiny neuron. J Neurosci. 2011;31:1183–92. doi: 10.1523/JNEUROSCI.3833-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciocchi S, Herry C, Grenier F, Wolff SB, Letzkus JJ, et al. Encoding of conditioned fear in central amygdala inhibitory circuits. Nature. 2010;468:277–82. doi: 10.1038/nature09559. [DOI] [PubMed] [Google Scholar]
- Covington HE, 3rd, Lobo MK, Maze I, Vialou V, Hyman JM, et al. Antidepressant effect of optogenetic stimulation of the medial prefrontal cortex. J Neurosci. 2010;30:16082–90. doi: 10.1523/JNEUROSCI.1731-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH. Thinking about the brain. Sci Am. 1979;241:219–32. doi: 10.1038/scientificamerican0979-219. [DOI] [PubMed] [Google Scholar]
- Cruikshank SJ, Urabe H, Nurmikko AV, Connors BW. Pathway-specific feedforward circuits between thalamus and neocortex revealed by selective optical stimulation of axons. Neuron. 2010;65:230–45. doi: 10.1016/j.neuron.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K. Controlling the brain with light. Sci Am. 2010;303:48–55. doi: 10.1038/scientificamerican1110-48. [DOI] [PubMed] [Google Scholar]
- Deisseroth K. Optogenetics. Nat Methods. 2011;8:26–29. doi: 10.1038/nmeth.f.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Feng G, Majewska AK, Miesenbock G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J Neurosci. 2006;26:10380–86. doi: 10.1523/JNEUROSCI.3863-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diester I, Kaufman MT, Mogri M, Pashaie R, Goo W, et al. An optogenetic toolbox designed for primates. Nat Neurosci. 2011;14:387–97. doi: 10.1038/nn.2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass AD, Kraves S, Deisseroth K, Schier AF, Engert F. Escape behavior elicited by single, channelrhodopsin-2-evoked spikes in zebrafish somatosensory neurons. Curr Biol. 2008;18:1133–37. doi: 10.1016/j.cub.2008.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst OP, Sánchez Murcia PA, Daldrop P, Tsunoda SP, Kateriya S, Hegemann P. Photoactivation of channelrhodopsin. J Biol Chem. 2008;283:1637–43. doi: 10.1074/jbc.M708039200. [DOI] [PubMed] [Google Scholar]
- Feldbauer K, Zimmermann D, Pintschovius V, Spitz J, Bamann C, Bamberg E. Channelrhodopsin-2 is a leaky proton pump. Proc Natl Acad Sci USA. 2009;106:12317–22. doi: 10.1073/pnas.0905852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fork RL. Laser stimulation of nerve cells in aplysia. Science. 1971;171:907–8. doi: 10.1126/science.171.3974.907. [DOI] [PubMed] [Google Scholar]
- Geschwind D. GENSAT: a genomic resource for neuroscience research. Lancet Neurol. 2004;3:82. doi: 10.1016/s1474-4422(03)00658-6. [DOI] [PubMed] [Google Scholar]
- Gong S, Doughty M, Harbaugh CR, Cummins A, Hatten ME, et al. Targeting Cre recombinase to specific neuron populations with bacterial artificial chromosome constructs. J Neurosci. 2007;27:9817–23. doi: 10.1523/JNEUROSCI.2707-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–25. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Gordon MD, Scott K. Motor control in a Drosophila taste circuit. Neuron. 2009;61:373–84. doi: 10.1016/j.neuron.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorostiza P, Isacoff EY. Optical switches for remote and noninvasive control of cell signaling. Science. 2008;322:395–99. doi: 10.1126/science.1166022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourine AV, Kasymov V, Marina N, Tang F, Figueiredo MF, et al. Astrocytes control breathing through pH-dependent release of ATP. Science. 2010;329:571–75. doi: 10.1126/science.1190721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Mogri M, Thompson KR, Henderson JM, Deisseroth K. Optical deconstruction of parkinsonian neural circuitry. Science. 2009;324:354–59. doi: 10.1126/science.1167093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Deisseroth K. eNpHR: a natronomonas halorhodopsin enhanced for optogenetic applications. Brain Cell Biol. 2008;36:129–39. doi: 10.1007/s11068-008-9027-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Thompson KR, Zhang F, Mogri M, Kay K, et al. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J Neurosci. 2007;27:14231–38. doi: 10.1523/JNEUROSCI.3578-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–65. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman N, Poher V, Grubb MS, Kennedy GT, Nikolic K, et al. Multi-site optical excitation using ChR2 and micro-LED array. J Neural Eng. 2010;7:16004. doi: 10.1088/1741-2560/7/1/016004. [DOI] [PubMed] [Google Scholar]
- Grubb MS, Burrone J. Activity-dependent relocation of the axon initial segment fine-tunes neuronal excitability. Nature. 2010a;465:1070–74. doi: 10.1038/nature09160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb MS, Burrone J. Channelrhodopsin-2 localised to the axon initial segment. PloS One. 2010b;5:e13761. doi: 10.1371/journal.pone.0013761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Yizhar O, Berndt A, Sohal VS, Deisseroth K, Hegemann P. Ultrafast optogenetic control. Nat Neurosci. 2010;13:387–92. doi: 10.1038/nn.2495. [DOI] [PubMed] [Google Scholar]
- Guo ZV, Hart AC, Ramanathan S. Optical interrogation of neural circuits in Caenorhabditis elegans. Nat Methods. 2009;6:891–96. doi: 10.1038/nmeth.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagglund M, Borgius L, Dougherty KJ, Kiehn O. Activation of groups of excitatory neurons in the mammalian spinal cord or hindbrain evokes locomotion. Nat Neurosci. 2010;13:246–52. doi: 10.1038/nn.2482. [DOI] [PubMed] [Google Scholar]
- Han X, Boyden ES. Multiple-color optical activation, silencing, and desynchronization of neural activity, with single-spike temporal resolution. PloS One. 2007;2:e299. doi: 10.1371/journal.pone.0000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Qian X, Bernstein JG, Zhou HH, Franzesi GT, et al. Millisecond-timescale optical control of neural dynamics in the nonhuman primate brain. Neuron. 2009;62:191–98. doi: 10.1016/j.neuron.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubensak W, Kunwar PS, Cai H, Ciocchi S, Wall NR, et al. Genetic dissection of an amygdala microcircuit that gates conditioned fear. Nature. 2010;468:270–76. doi: 10.1038/nature09553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupts U, Tittor J, Bamberg E, Oesterhelt D. General concept for ion translocation by halobacterial retinal proteins: the isomerization/switch/transfer (IST) model. Biochemistry. 1997;36:2–7. doi: 10.1021/bi962014g. [DOI] [PubMed] [Google Scholar]
- Hegemann P, Ehlenbeck S, Gradmann D. Multiple photocycles of channelrhodopsin. Biophys J. 2005;89:3911–18. doi: 10.1529/biophysj.105.069716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz N. Gene expression nervous system atlas (GENSAT) Nat Neurosci. 2004;7:483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- Higley MJ, Sabatini BL. Competitive regulation of synaptic Ca2+ influx by D2 dopamine and A2A adenosine receptors. Nat Neurosci. 2010;13:958–66. doi: 10.1038/nn.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira R, Honkura N, Noguchi J, Maruyama Y, Augustine GJ, et al. Transcranial optogenetic stimulation for functional mapping of the motor cortex. J Neurosci Methods. 2009;179:258–63. doi: 10.1016/j.jneumeth.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Hirase H, Nikolenko V, Goldberg JH, Yuste R. Multiphoton stimulation of neurons. J Neurobiol. 2002;51:237–47. doi: 10.1002/neu.10056. [DOI] [PubMed] [Google Scholar]
- Hires SA, Tian L, Looger LL. Reporting neural activity with genetically encoded calcium indicators. Brain Cell Biol. 2008;36:69–86. doi: 10.1007/s11068-008-9029-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann KP, Scheerer P, Hildebrand PW, Choe HW, Park JH, et al. A G protein-coupled receptor at work: the rhodopsin model. Trends Biochem Sci. 2009;34:540–52. doi: 10.1016/j.tibs.2009.07.005. [DOI] [PubMed] [Google Scholar]
- Hornstein NJ, Pulver SR, Griffith LC. Channelrhodopsin2 mediated stimulation of synaptic potentials at Drosophila neuromuscular junctions. J Vis Exp. 2009;25 doi: 10.3791/1133. http://www.jove.com/details.stp?id=1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull C, Adesnik H, Scanziani M. Neocortical disynaptic inhibition requires somatodendritic integration in interneurons. J Neurosci. 2009;29:8991–95. doi: 10.1523/JNEUROSCI.5717-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, et al. Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Curr Biol. 2007;17:2105–16. doi: 10.1016/j.cub.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T, Kakuda M, Araki R, Yawo H. Kinetic evaluation of photosensitivity in genetically engineered neurons expressing green algae light-gated channels. Neurosci Res. 2006;54:85–94. doi: 10.1016/j.neures.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Honda S, Ozeki H, Hashimoto M, Hirase H. A simple head-mountable LED device for chronic stimulation of optogenetic molecules in freely moving mice. Neurosci Res. 2011 doi: 10.1016/j.neures.2011.01.007. In press. [DOI] [PubMed] [Google Scholar]
- Johansen JP, Hamanaka H, Monfils MH, Behnia R, Deisseroth K, et al. Optical activation of lateral amygdala pyramidal cells instructs associative fear learning. Proc Natl Acad Sci USA. 2010;107:12692–97. doi: 10.1073/pnas.1002418107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanbar R, Stornetta RL, Cash DR, Lewis SJ, Guyenet PG. Photostimulation of Phox2b medullary neurons activates cardiorespiratory function in conscious rats. Am J Respir Crit Care Med. 2010;182:1184–94. doi: 10.1164/rccm.201001-0047OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzel D, Zemelman BV, Buetfering C, Wolfel M, Miesenbock G. The columnar and laminar organization of inhibitory connections to neocortical excitatory cells. Nat Neurosci. 2010;14:100–7. doi: 10.1038/nn.2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouyama T, Kanada S, Takeguchi Y, Narusawa A, Murakami M, Ihara K. Crystal structure of the light-driven chloride pump halorhodopsin from Natronomonas pharaonis. J Mol Biol. 2010;396:564–79. doi: 10.1016/j.jmb.2009.11.061. [DOI] [PubMed] [Google Scholar]
- Kravitz AV, Freeze BS, Parker PR, Kay K, Thwin MT, et al. Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature. 2010;466:622–26. doi: 10.1038/nature09159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Durand R, Gradinaru V, Zhang F, Goshen I, et al. Global and local fMRI signals driven by neurons defined optogenetically by type and wiring. Nature. 2010;465:788–92. doi: 10.1038/nature09108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer AM, Fang-Yen C, Gershow M, Alkema MJ, Samuel AD. Optogenetic manipulation of neural activity in freely moving Caenorhabditis elegans. Nat Methods. 2011;8:147–52. doi: 10.1038/nmeth.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis TL, Jr, Mao T, Svoboda K, Arnold DB. Myosin-dependent targeting of transmembrane proteins to neuronal dendrites. Nat Neurosci. 2009;12:568–76. doi: 10.1038/nn.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gutierrez DV, Hanson MG, Han J, Mark MD, et al. Fast noninvasive activation and inhibition of neural and network activity by vertebrate rhodopsin and green algae channelrhodopsin. Proc Natl Acad Sci USA. 2005;102:17816–21. doi: 10.1073/pnas.0509030102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liewald JF, Brauner M, Stephens GJ, Bouhours M, Schultheis C, et al. Optogenetic analysis of synaptic function. Nat Methods. 2008;5:895–902. doi: 10.1038/nmeth.1252. [DOI] [PubMed] [Google Scholar]
- Lima SQ, Hromadka T, Znamenskiy P, Zador AM. PINP: a new method of tagging neuronal populations for identification during in vivo electrophysiological recording. PloS One. 2009;4:e6099. doi: 10.1371/journal.pone.0006099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SQ, Miesenbock G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–52. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- Lin JY, Lin MZ, Steinbach P, Tsien RY. Characterization of engineered channelrhodopsin variants with improved properties and kinetics. Biophys J. 2009;96:1803–14. doi: 10.1016/j.bpj.2008.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn ME, Thompson KR, Deisseroth K, Delp SL. Orderly recruitment of motor units under optical control in vivo. Nat Med. 2010;16:1161–65. doi: 10.1038/nm.2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Covington HE, 3rd, Chaudhury D, Friedman AK, Sun H, et al. Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science. 2010;330:385–90. doi: 10.1126/science.1188472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low SE, Ryan J, Sprague SM, Hirata H, Cui WW, et al. touche is required for touch-evoked generator potentials within vertebrate sensory neurons. J Neurosci. 2010;30:9359–67. doi: 10.1523/JNEUROSCI.1639-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structural changes in bacteriorhodopsin during ion transport at 2 angstrom resolution. Science. 1999a;286:255–61. doi: 10.1126/science.286.5438.255. [DOI] [PubMed] [Google Scholar]
- Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 A resolution. J Mol Biol. 1999b;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- Man D, Wang W, Sabehi G, Aravind L, Post AF, et al. Diversification and spectral tuning in marine proteorhodopsins. EMBO J. 2003;22:1725–31. doi: 10.1093/emboj/cdg183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattis J, Tye KM, Ramakrishnan C, O'Shea D, Gunaydin LA, et al. Clarifying the optogenetics toolbox: a comprehensive analysis of new and existing opsins for scientific application. Nat Meth. 2011 Submitted. [Google Scholar]
- McLean DL, Fetcho JR. Movement, technology and discovery in the zebrafish. Curr Opin Neurobiol. 2011;21:110–15. doi: 10.1016/j.conb.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miesenbock G, Kevrekidis IG. Optical imaging and control of genetically designated neurons in functioning circuits. Annu Rev Neurosci. 2005;28:533–63. doi: 10.1146/annurev.neuro.28.051804.101610. [DOI] [PubMed] [Google Scholar]
- Mohanty SK, Reinscheid RK, Liu X, Okamura N, Krasieva TB, Berns MW. In-depth activation of channelrhodopsin 2-sensitized excitable cells with high spatial resolution using two-photon excitation with a near-infrared laser microbeam. Biophys J. 2008;95:3916–26. doi: 10.1529/biophysj.108.130187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagel G, Brauner M, Liewald JF, Adeishvili N, Bamberg E, Gottschalk A. Light activation of channelrhodopsin-2 in excitable cells of Caenorhabditis elegans triggers rapid behavioral responses. Curr Biol. 2005;15:2279–84. doi: 10.1016/j.cub.2005.11.032. [DOI] [PubMed] [Google Scholar]
- Nagel G, Ollig D, Fuhrmann M, Kateriya S, Musti AM, et al. Channelrhodopsin-1: a light-gated proton channel in green algae. Science. 2002;296:2395–98. doi: 10.1126/science.1072068. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc Natl Acad Sci USA. 2003;100:13940–45. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol. 1971;233:149–52. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- Oh E, Maejima T, Liu C, Deneris E, Herlitze S. Substitution of 5-HT1A receptor signaling by a light-activated G protein-coupled receptor. J Biol Chem. 2010;285:30825–36. doi: 10.1074/jbc.M110.147298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiakoumou E, Anselmi F, Begue A, de Sars V, Gluckstad J, et al. Scanless two-photon excitation of channelrhodopsin-2. Nat Methods. 2010;7:848–54. doi: 10.1038/nmeth.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perálvarez-Marín A, Márquez M, Bourdelande JL, Querol E, Padrós E. Thr-90 plays a vital role in the structure and function of bacteriorhodopsin. J Biol Chem. 2004;279:16403–9. doi: 10.1074/jbc.M313988200. [DOI] [PubMed] [Google Scholar]
- Petreanu L, Huber D, Sobczyk A, Svoboda K. Channelrhodopsin-2-assisted circuit mapping of long-range callosal projections. Nat Neurosci. 2007;10:663–68. doi: 10.1038/nn1891. [DOI] [PubMed] [Google Scholar]
- Petreanu L, Mao T, Sternson SM, Svoboda K. The subcellular organization of neocortical excitatory connections. Nature. 2009;457:1142–45. doi: 10.1038/nature07709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulver SR, Pashkovski SL, Hornstein NJ, Garrity PA, Griffith LC. Temporal dynamics of neuronal activation by channelrhodopsin-2 and TRPA1 determine behavioral output in Drosophila larvae. J Neurophysiol. 2009;101:3075–88. doi: 10.1152/jn.00071.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickgauer JP, Tank DW. Two-photon excitation of channelrhodopsin-2 at saturation. Proc Natl Acad Sci USA. 2009;106:15025–30. doi: 10.1073/pnas.0907084106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter E, Stehfest K, Berndt A, Hegemann P, Bartl FJ. Monitoring light-induced structural changes of channelrhodopsin-2 by UV-visible and Fourier transform infrared spectroscopy. J Biol Chem. 2008;283:35033–41. doi: 10.1074/jbc.M806353200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer S, Zemelman BV, Barbic M, Losonczy A, Buzsaki G, Magee JC. Multi-array silicon probes with integrated optical fibers: light-assisted perturbation and recording of local neural circuits in the behaving animal. Eur J Neurosci. 2010;31:2279–91. doi: 10.1111/j.1460-9568.2010.07250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu MH, Moskvin OV, Siltberg-Liberles J, Gomelsky M. Natural and engineered photoactivated nucleotidyl cyclases for optogenetic applications. J Biol Chem. 2010;285:41501–8. doi: 10.1074/jbc.M110.177600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakmar TP. Structure of rhodopsin and the superfamily of seven-helical receptors: the same and not the same. Curr Opin Cell Biol. 2002;14:189–95. doi: 10.1016/s0955-0674(02)00306-x. [DOI] [PubMed] [Google Scholar]
- Scanziani M, Hausser M. Electrophysiology in the age of light. Nature. 2009;461:930–39. doi: 10.1038/nature08540. [DOI] [PubMed] [Google Scholar]
- Schmucker D, Su AL, Beermann A, Jackle H, Jay DG. Chromophore-assisted laser inactivation of patched protein switches cell fate in the larval visual system of Drosophila. Proc Natl Acad Sci USA. 1994;91:2664–68. doi: 10.1073/pnas.91.7.2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonheim PJ, Arrenberg AB, Del Bene F, Baier H. Optogenetic localization and genetic perturbation of saccade-generating neurons in zebrafish. J Neurosci. 2010;30:7111–20. doi: 10.1523/JNEUROSCI.5193-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroll C, Riemensperger T, Bucher D, Ehmer J, Voller T, et al. Light-induced activation of distinct modulatory neurons triggers appetitive or aversive learning in Drosophila larvae. Curr Biol. 2006;16:1741–47. doi: 10.1016/j.cub.2006.07.023. [DOI] [PubMed] [Google Scholar]
- Shichida Y, Yamashita T. Diversity of visual pigments from the viewpoint of G protein activation—comparison with other G protein-coupled receptors. Photochem Photobiol Sci. 2003;2:1237–46. doi: 10.1039/b300434a. [DOI] [PubMed] [Google Scholar]
- Shoham S. Optogenetics meets optical wavefront shaping. Nat Methods. 2010;7:798–99. doi: 10.1038/nmeth1010-798. [DOI] [PubMed] [Google Scholar]
- Sohal VS, Zhang F, Yizhar O, Deisseroth K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature. 2009;459:698–702. doi: 10.1038/nature07991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JL. The multitalented microbial sensory rhodopsins. Trends Microbiol. 2006;14:480–87. doi: 10.1016/j.tim.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Stehfest K, Hegemann P. Evolution of the channelrhodopsin photocycle model. ChemPhysChem. 2010;11:1120–26. doi: 10.1002/cphc.200900980. [DOI] [PubMed] [Google Scholar]
- Stehfest K, Ritter E, Berndt A, Bartl F, Hegemann P. The branched photocycle of the slow-cycling channelrhodopsin-2 mutant C128T. J Mol Biol. 2010;398:690–702. doi: 10.1016/j.jmb.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Stierl M, Stumpf P, Udwari D, Gueta R, Hagedorn R, et al. Light modulation of cellular cAMP by a small bacterial photoactivated adenylyl cyclase, bPAC, of the soil bacterium Beggiatoa. J Biol Chem. 2010;286:1181–88. doi: 10.1074/jbc.M110.185496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirman JN, Brauner M, Gottschalk A, Lu H. High-throughput study of synaptic transmission at the neuromuscular junction enabled by optogenetics and microfluidics. J Neurosci Methods. 2010;191:90–93. doi: 10.1016/j.jneumeth.2010.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirman JN, Crane MM, Husson SJ, Wabnig S, Schultheis C, et al. Real-time multimodal optical control of neurons and muscles in freely behaving Caenorhabditis elegans. Nat Methods. 2011;8:152–58. doi: 10.1038/nmeth.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckenius W. The rhodopsin-like pigments of halobacteria: light-energy and signal transducers in an archaebacterium. Trends Biochem Sci. 1985;10:483–86. doi: 10.1016/0968-0004(85)90210-5. [DOI] [PubMed] [Google Scholar]
- Stroh A, Tsai HC, Ping Wang L, Zhang F, Kressel J, et al. Tracking stem cell differentiation in the setting of automated optogenetic stimulation. Stem Cells. 2010;29(1):78–88. doi: 10.1002/stem.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuber GD, Hnasko TS, Britt JP, Edwards RH, Bonci A. Dopaminergic terminals in the nucleus accumbens but not the dorsal striatum corelease glutamate. J Neurosci. 2010;30:8229–33. doi: 10.1523/JNEUROSCI.1754-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh GS, Ben-Tabou de Leon S, Tanimoto H, Fiala A, Benzer S, Anderson DJ. Light activation of an innate olfactory avoidance response in Drosophila. Curr Biol. 2007;17:905–8. doi: 10.1016/j.cub.2007.04.046. [DOI] [PubMed] [Google Scholar]
- Szobota S, Gorostiza P, Del Bene F, Wyart C, Fortin DL, et al. Remote control of neuronal activity with a light-gated glutamate receptor. Neuron. 2007;54:535–45. doi: 10.1016/j.neuron.2007.05.010. [DOI] [PubMed] [Google Scholar]
- Tecuapetla F, Patel JC, Xenias H, English D, Tadros I, et al. Glutamatergic signaling by mesolimbic dopamine neurons in the nucleus accumbens. J Neurosci. 2010;30:7105–10. doi: 10.1523/JNEUROSCI.0265-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan S, van Wyk M, Lehmann K, Lowel S, Feng G, Wassle H. Visual function in mice with photoreceptor degeneration and transgenic expression of channelrhodopsin 2 in ganglion cells. J Neurosci. 2010;30:8745–58. doi: 10.1523/JNEUROSCI.4417-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Hires SA, Mao T, Huber D, Chiappe ME, et al. Imaging neural activity in worms, flies and mice with improved GCaMP calcium indicators. Nat Methods. 2009;6:875–81. doi: 10.1038/nmeth.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai HC, Zhang F, Adamantidis A, Stuber GD, Bonci A, et al. Phasic firing in dopaminergic neurons is sufficient for behavioral conditioning. Science. 2009;324:1080–84. doi: 10.1126/science.1168878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tye K, Prakash R, Kim SY, Fenno LE, Grosenick L, et al. Amygdala circuitry mediating reversible and bidirectional control of anxiety. Nature. 2011 doi: 10.1038/nature09820. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volgraf M, Gorostiza P, Numano R, Kramer RH, Isacoff EY, Trauner D. Allosteric control of an ionotropic glutamate receptor with an optical switch. Nat Chem Biol. 2006;2:47–52. doi: 10.1038/nchembio756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Peca J, Matsuzaki M, Matsuzaki K, Noguchi J, et al. High-speed mapping of synaptic connectivity using photostimulation in channelrhodopsin-2 transgenic mice. Proc Natl Acad Sci USA. 2007;104:8143–48. doi: 10.1073/pnas.0700384104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sugiyama Y, Hikima T, Sugano E, Tomita H, et al. Molecular determinants differentiating photocurrent properties of two channelrhodopsins from chlamydomonas. J Biol Chem. 2009;284:5685–96. doi: 10.1074/jbc.M807632200. [DOI] [PubMed] [Google Scholar]
- Wang J, Borton DA, Zhang J, Burwell RD, Nurmikko AV. A neurophotonic device for stimulation and recording of neural microcircuits. Conf Proc IEEE Eng Med Biol Soc. 2010;1:2935–38. doi: 10.1109/IEMBS.2010.5626296. [DOI] [PubMed] [Google Scholar]
- Weick JP, Johnson MA, Skroch SP, Williams JC, Deisseroth K, Zhang SC. Functional control of transplantable human ESC-derived neurons via optogenetic targeting. Stem Cells. 2010;28:2008–16. doi: 10.1002/stem.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Sessa A, Burke C, Bowman T, LeBlanc J, et al. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell. 2008;2:183–89. doi: 10.1016/j.stem.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletionmutant rabies virus. Nat Methods. 2007a;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJ, Mori T, Finke S, et al. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron. 2007b;53:639–47. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witten IB, Lin SC, Brodsky M, Prakash R, Diester I, et al. Cholinergic interneurons control local circuit activity and cocaine conditioning. Science. 2010;330:1677–81. doi: 10.1126/science.1193771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–8. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, Jan YN. Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature. 2010;468:921–26. doi: 10.1038/nature09576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27:941–45. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- Zemelman BV, Lee GA, Ng M, Miesenbock G. Selective photostimulation of genetically chARGed neurons. Neuron. 2002;33:15–22. doi: 10.1016/s0896-6273(01)00574-8. [DOI] [PubMed] [Google Scholar]
- Zemelman BV, Nesnas N, Lee GA, Miesenbock G. Photochemical gating of heterologous ion channels: remote control over genetically designated populations of neurons. Proc Natl Acad Sci USA. 2003;100:1352–57. doi: 10.1073/pnas.242738899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, et al. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc. 2010;5:439–56. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Prigge M, Beyriere F, Tsunoda SP, Mattis J, et al. Red-shifted optogenetic excitation: a tool for fast neural control derived from Volvox carteri. Nat Neurosci. 2008;11:631–33. doi: 10.1038/nn.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Boyden ES, Deisseroth K. Channelrhodopsin-2 and optical control of excitable cells. Nat Methods. 2006;3:785–92. doi: 10.1038/nmeth936. [DOI] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, et al. Multimodal fast optical interrogation of neural circuitry. Nature. 2007a;446:633–39. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zhang J, Laiwalla F, Kim JA, Urabe H, Van Wagenen R, et al. Integrated device for optical stimulation and spatiotemporal electrical recording of neural activity in light-sensitized brain tissue. J Neural Eng. 2009;6:055007. doi: 10.1088/1741-2560/6/5/055007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ge W, Wang Z. A toolbox for light control of Drosophila behaviors through channel-rhodopsin 2-mediated photoactivation of targeted neurons. Eur J Neurosci. 2007b;26:2405–16. doi: 10.1111/j.1460-9568.2007.05862.x. [DOI] [PubMed] [Google Scholar]
- Zhang YP, Oertner TG. Optical induction of synaptic plasticity using a light-sensitive channel. Nat Methods. 2007;4:139–41. doi: 10.1038/nmeth988. [DOI] [PubMed] [Google Scholar]
- Zhao S, Cunha C, Zhang F, Liu Q, Gloss B, et al. Improved expression of halorhodopsin for light-induced silencing of neuronal activity. Brain Cell Biol. 2008;36:141–54. doi: 10.1007/s11068-008-9034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Narita Y, Bundschuh ST, Fajardo O, Schärer YP, et al. Optogenetic dissection of neuronal circuits in zebrafish using viral gene transfer and the Tet system. Front Neural Circuits. 2009;3:21. doi: 10.3389/neuro.04.021.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann G, Wang LP, Vaughan AG, Manoli DS, Zhang F, et al. Manipulation of an innate escape response in Drosophila: photoexcitation of acj6 neurons induces the escape response. PloS One. 2009;4:e5100. doi: 10.1371/journal.pone.0005100. [DOI] [PMC free article] [PubMed] [Google Scholar]