A Mitochondrial Paradigm of Metabolic and Degenerative Diseases, Aging, and Cancer: A Dawn for Evolutionary Medicine (original) (raw)
. Author manuscript; available in PMC: 2010 Feb 12.
Abstract
Life is the interplay between structure and energy, yet the role of energy deficiency in human disease has been poorly explored by modern medicine. Since the mitochondria use oxidative phosphorylation (OXPHOS) to convert dietary calories into usable energy, generating reactive oxygen species (ROS) as a toxic by-product, I hypothesize that mitochondrial dysfunction plays a central role in a wide range of age-related disorders and various forms of cancer. Because mitochondrial DNA (mtDNA) is present in thousands of copies per cell and encodes essential genes for energy production, I propose that the delayed-onset and progressive course of the age-related diseases results from the accumulation of somatic mutations in the mtDNAs of post-mitotic tissues. The tissue-specific manifestations of these diseases may result from the varying energetic roles and needs of the different tissues. The variation in the individual and regional predisposition to degenerative diseases and cancer may result from the interaction of modern dietary caloric intake and ancient mitochondrial genetic polymorphisms. Therefore the mitochondria provide a direct link between our environment and our genes and the mtDNA variants that permitted our forbears to energetically adapt to their ancestral homes are influencing our health today.
Keywords: mitochondria; reactive oxygen species; human origins; diabetes; neurodegenerative diseases; aging; mtDNA: mitochondrial DNA; Mitochondrion (s), mitochondria (pl): cellular organelle of endosymbiotic origin that resides in the cytosol of most nucleated (eukaryotic) cells and which produces energy by oxidizing organic acids and fats with oxygen by the process oxidative phosphorylation (OXPHOS) and generates oxygen radicals (reactive oxygen species, ROS) as a toxic by-product; ROS: reactive oxygen species, oxygen radicals; OXPHOS: oxidative phosphorylation; Mitochondrial DNA (mtDNA): the portion of the mitochondrial genome that currently resides in the matrix of the mitochondrion, as a circular DNA molecule containing the mitochondrial rRNA genes, tRNA genes, and 13 subunits of the mitochondrial oxidative phosphorylation (OXPHOS) enzyme complexes; CR: mtDNA control region; TCA: mitochondrial tricarboxylic acid cycle; SDH: succinate dehydrogenase; COX: cytochrome c oxidase, complex IV; ETC: mitochondrial electron transport chain, a part of the OXPHOS system; ANT: adenine nucleotide translocator; Unc 1,2,3: uncoupling proteins 1,2,3; Reactive oxygen species (ROS): primarily superoxide anion (O2• −), hydrogen peroxide (H2O2), and hydroxyl radical (• OH), commonly referred to as oxygen radicals; generated as a toxic by-product of oxidative energy production by OXPHOS damage the mitochondrial and cellular DNA, proteins, lipids, and other molecules causing oxidative stress; Oxidative phosphorylation (OXPHOS): the process by which the mitochondrion generates energy through oxidation of organic acids and fats with oxygen to create a capacitor [electron chemical gradient (ΔP = ΔΨ + ΔpH)] across the mitochondrial inner membrane. This ΔP is used as a source of potential energy to generate adenosine triphosphate (ATP), transport substrates or ions, or produce heat. OXPHOS encompasses five multipolypepetide complexes I, II, III, IV and V. Complex I is NADH dehydrogenase or NADH:ubiquinone oxidoreductase, complex II is succinate dehydrogenase (SDH) of succinate:ubiquinone oxidoreductase, complex III is the bc1 complex or ubiquinole: cytochrome c oxidoreductase,complex IV is cytochrome c oxidase (COX) or reduced cytochrome c: oxygen oxidoreductase, and complex V is the ATP synthase or proton-translocating ATP synthase. Complexes I, III, IV, and V encompass both nDNA- and mtDNA-encoded subunits; MnSOD (Sod2): mitochondrial matrix superoxide dismutase; Cu/ZnSOD (Sod1): mitochondrial inner membrane space and cytosolic superoxide dismutase; mtPTP: mitochondrial permeability transition pore; Apoptosis: a process of programmed cell death resulting in the activation of caspase enzymes and intracellular nucleases that degrade the cellular proteins and nDNA. Apoptosis can be initiated via the mitochondrion through the activation of the mitochondrial permeability transition pore (mtPTP) in response to energy deficiency, increased oxidative stress, excessive Ca2+, and, or other factors; CPEO/KSS: chronic progressive external ophthalmoplegia, Kearn-Sayre syndrome; NARP: neurogenic muscle weaknes, ataxia, and retinitis pigmentosa; LHON: Leber’s hereditary optic neuropathy; MERRF: myoclonic epilepsy and ragged red fiber disease; MELAS: mitochondrial encephalomyopathy, lactic acidosis, and stroke-like episodes; PGC-1: PPARγ (peroxisome-proliferating-activated receptor γ) coactivator 1; FOXO: mammalian forkhead transcription factor; APP: amyloid precursor protein; Mitochondrial medicine: the new medical discipline that pertains to all clinical problems that involve the mitochondria, Evolutionary medicine: a clinical perspective that posits that many of the common clinical problems of today are rooted in adaptive genetic programs that permitted our human ancestors to survive in the environments which they confronted in the past
LEAN DIETS, LONG LIFE, AND THE MITOCHONDRIA
Age-related metabolic and degenerative diseases are increasing to epidemic proportions in all industrialized countries. Diabetes has increased sharply over the past century, with a worldwide incidence in 2000 of 151 million cases and the projected incidence for 2010 of 221 million cases (for the United States, 14.2 million in 2000 versus 17.5 million by 2010) (73, 257). Today, Alzheimer disease (AD) affects about 4.5 million Americans, projected to increase to 11 to 16 million by 2050 (6, 70). Parkinson’s disease (PD) afflicts approximately one million persons in the United States today; 60,000 new cases are diagnosed every year, and the incidence is projected to quadruple by 2040 (160, 164). The risk of cancer also increases with age. Prostate cancer is the most common malignancy among men in industrialized countries. In the United States in 2004, approximately 230,000 new cases were reported, with an annual death toll of approximately 30,000 men (31). Moreover, the incidence of prostate cancer has been increasing steadily in the United States and Canada over the past 30 years (132).
As many of these age-related diseases show some familial association, a genetic basis has been assumed. Yet, the nature and frequency of genetic variants in the human population has not changed significantly in the past 50 years, even though the incidence of these diseases has climbed continuously. Therefore, it isn’t our genes that have changed; it is our environment, and the biggest environmental change that we have experienced is in our diet.
For the first time in human history, individuals can live their entire lives free from hunger. Yet, it has been known for over 70 years that laboratory rodents, if maintained on a restricted calorie diet, will be healthier, more active, more intelligent, and will live longer and have fewer cancers (69, 126, 127, 130, 204).
So what is the link between genetics and diet for these diseases? Prodigious effort has been expended to identify chromosomal genetic loci that are responsible for these problems, but with disappointingly little success. One hint as to why nuclear DNA (nDNA) analysis has been so unfruitful is that all of these diseases have a delayed onset and a progressive course. Thus while the most important risk factor for contracting all of these diseases is age, nothing in Mendelian genetics can provide the needed aging clock. The aging clock must involve the gradual accumulation of sequential changes in thousands of copies of the same gene or genes, yet each Mendelian gene is present in only two copies.
Furthermore, certain age-related diseases are preferentially found in distinct populations, suggesting that they are influenced by regional genetic differences. Yet most ancient nDNA polymorphisms are common to all global populations. Only one human genetic system is known to be present in thousands of copies per cell, to exhibit striking regional genetic variation, and to be directly involved in calorie utilization: the mitochondrial genetic system.
The mitochondria are ancient bacterial symbionts with their own mitochondrial DNA (mtDNA), RNA, and protein synthesizing systems. Each human cell contains hundreds of mitochondria and thousands of mtD-NAs. The mtDNA is maternally inherited and shows striking regional genetic variation. This regional variation was a major factor in permitting humans to adapt to the different global environments they encountered and mastered. Moreover, the mitochondria burn the calories in our diet with the oxygen that we breathe to make chemical energy to do work and heat to maintain our body temperature. As a by-product of energy production, the mitochondria also generate most of the endogenous reactive oxygen species (ROS) of the cell, and these damage the mitochondria, mtDNAs, and cell. Hence, the mitochondria are the only human genetic system that embodies the features necessary to explain the observed characteristics of the common age-related diseases (237).
In this review, I make the case that the mitochondrial decline and mtDNA damage are central to the etiology of the age-related metabolic and degenerative diseases, aging, and cancer. This is because the rate of mitochondria and mtDNAs damage and thus decline is modulated by the extent of mitochondrial oxidative stress. Mitochondrial ROS production, in turn, is increased by the availability of excess calories, modulated by regional mtDNA genetic variation, and regulated by alterations in nDNA expression of stress response genes.
MITOCHONDRIAL BIOGENESIS AND BIOENERGETICS: AN AGING PARADIGM
The proto-mitochondrion entered the primitive eukaryotic cell between two and three billion years ago. Initially, the bacterial genome encoded all of the genes necessary for a free-living organism. However, as the symbiosis matured, many bacterial genes were transferred to extrabacterial plasmids (chromosomes), such that today the maternally inherited mtDNA retains only the genes for the 12S and 16S rRNAs and the 22 tRNAs required for mitochondrial protein synthesis plus 13 polypeptides of the mitochondrial energy generating process, oxidative phosphorylation (OXPHOS) (Figure 1). The remaining ~1500 genes of the mitochondrial genome are now scattered throughout the chromosomal DNA. These nDNA-encoded mitochondrial proteins are translated on cytosolic ribosomes and selectively imported into the mitochondrion through various mitochondrial protein import systems. For example, certain proteins destined for the mitochondrial matrix are synthesized with an amino terminal, positively charged, amphoteric targeting peptide that is cleaved off once the protein enters the mitochondrial matrix (237).
Figure 1.
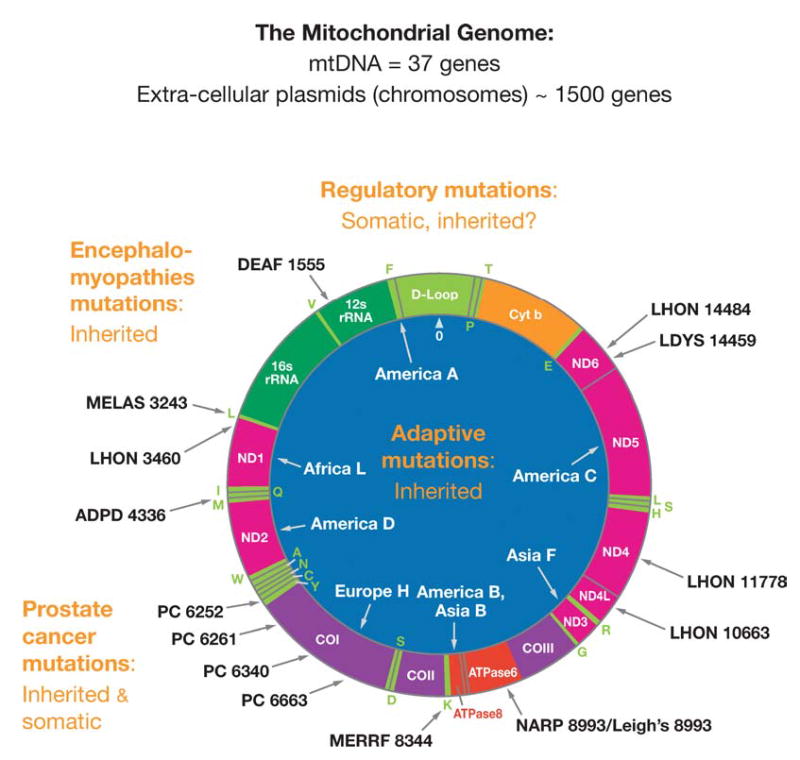
Human mitochondrial DNA map showing representative pathogenic and adaptive base substitution mutations. D-loop = control region (CR). Letters around the outside perimeter indicate cognate amino acids of the tRNA genes. Other gene symbols are defined in the text. Arrows followed by continental names and associated letters on the inside of the circle indicate the position of defining polymorphisms of selected region-specific mtDNA lineages. Arrows associated with abbreviations followed by numbers around the outside of the circle indicate representative pathogenic mutations, the number being the nucleotide position of the mutation. Abbreviations: DEAF, deafness; MELAS, mitochondrial encephalomyopathy, lactic acidosis and stroke-like episodes; LHON, Leber hereditary optic neuropathy; ADPD, Alzheimer and Parkinson disease; MERRF, myoclonic epilepsy and ragged red fiber disease; NARP, neurogenic muscle weakness, ataxia, retinitis pigmentosum; LDYS, LHON + dystonia; PC, prostate cancer.
The 13 mtDNA-encoded polypeptide genes are translated on mitochondrial ribosomes and all are structural subunits of OXPHOS enzyme complexes. These include 7 (ND1, 2, 3, 4L, 4, 5, 6) of the 46 polypeptides of complex I (NADH dehydrogenase), one (cytochrome b, cytb) of the 11 polypeptides of complex III (bc1 complex), 3 (COI, II, III) of the 13 polypeptides of complex IV (cytochrome c oxidase), and 2 (ATP 6 & 8) of the 16 proteins of complex V (ATP synthetase) (Figure 1). The nDNA codes for all other mitochondrial proteins including all four subunits of complex II (succinate dehydrogenase), the mitochondrial DNA polymerase γ (POLG) subunits, the mitochondrial RNA polymerase components, the mitochondrial transcription factor (mtTFA), the mitochondrial ribosomal proteins and elongation factors, and the mitochondrial metabolic enzymes.
The organization of the mtDNA is unique in that its structural genes have no 5′ or 3′ non-coding sequences, no introns, and no spacers between the genes. The mtDNA is symmetrically transcribed from two promoters, one for the G-rich heavy (H) stand and the other for the C-rich light (L) strand. These H- and L-strand promoters (PH and PL) are contained in the approximately 1121-np control region (CR) that encompasses four mtTFA binding sites, the H-strand origin of replication (OH), and three conserved sequence blocks (CSB I, II, and III). The L-strand origin of replication (OL) is two thirds of the way around the mtDNA from OH. Transcription is initiated at PH or PL and progresses around the mtDNA, generating a polycistronic message. The tR-NAs, which punctuate the genes, are cleaved out and the mRNAs and are then polyadenylated (237) (Figure 1).
The mtDNA gene repertoire has remained relatively constant since the formation of the fungal-animal lineage. This is because the mtDNA genetic code began to drift in the fungi such that the mtDNA genes can no longer be interpreted by the nuclear-cytosol system (233). Consequently, when a mtDNA sequence is transferred to the nDNA, it remains as a nonfunctional pseudogene (147).
The mitochondria generate energy by oxidizing hydrogen derived from our dietary carbohydrates (TCA cycle) and fats (β-oxidation) with oxygen to generate heat and ATP (Figure 2). Two electrons donated from NADH + H+ to complex I (NADH dehydrogenase) or from succinate to complex II (succinate dehydrogenase, SDH) are passed sequentially to ubiquinone (coenzyme Q or CoQ) to give ubisemiquinone (CoQH• ) and then ubiquinol (CoQH2). Ubiquinol transfers its electrons to complex III (ubiquinol: cytochrome c oxidoreductase), which transfers them to cytochrome c. From cytochrome c, the electrons flow to complex IV (cytochrome c oxidase, COX) and finally to ½O2 to give H2O. Each of these electron transport chain (ETC) complexes incorporates multiple electron carriers. Complexes I, II, and III encompass several iron-sulfur (Fe-S) centers, whereas complexes III and IV encompass the b + c1 and a + a3 cytochromes, respectively. The mitochondrial TCA cycle enzyme aconitase is also an iron-sulfur center protein (234, 235, 237).
Figure 2.
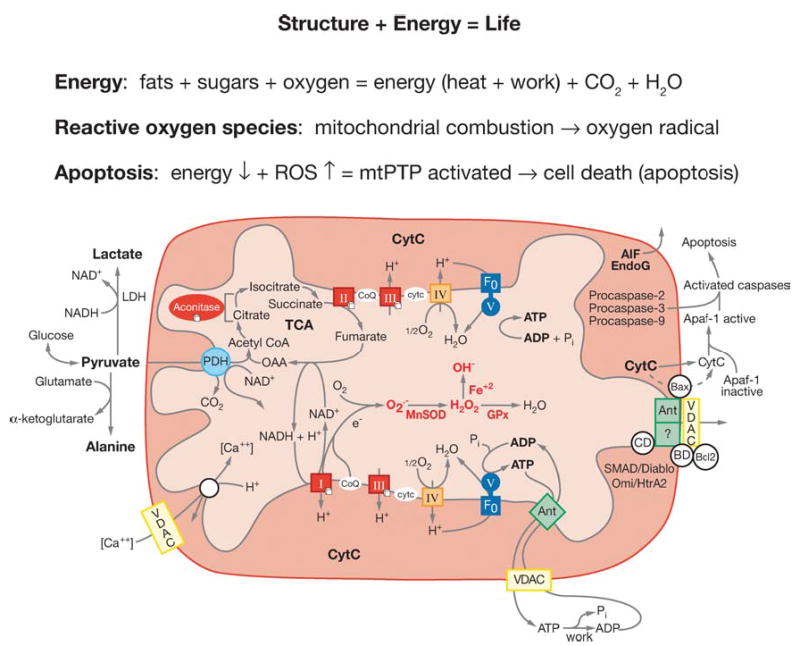
Diagram showing the relationships of mitochondrial oxidative phosphorylation (OXPHOS) to (a) energy (ATP) production, (b) reactive oxygen species (ROS) production, and (c) initiation of apoptosis through the mitochondrial permeability transition pore (mtPTP). The OXPHOS complexes, designated I to V, are complex I (NADH: ubiquinone oxidoreductase) encompassing a FMN (flavin mononucleotide) and six Fe-S centers (designated with a cube); complex II (succinate: ubiquinone oxidoreductase) involving a FAD (flavin adenine dinucleotide), three Fe-S centers, and a cytochrome b; complex III (ubiquinol: cytochrome c oxidoreductase) encompassing cytochromes b, c1 and the Rieske Fe-S center; complex IV (cytochrome c oxidase) encompassing cytochromes a + a3 and CuA and CuB; and complex V (H+-translocating ATP synthase). Pyruvate from glucose enters the mitochondria via pyruvate dehydrogenase (PDH), generating acetylCoA, which enters the TCA cycle by combining with oxaloacetate (OAA). _Cis_-aconitase converts citrate to isocitrate and contains a 4Fe-4S center. Lactate dehydrogenase (LDH) converts excess pyruvate plus NADH to lactate. Small molecules defuse through the outer membrane via the voltage-dependent anion channel (VDAC) or porin. The VDAC together with ANT, Bax, and the cyclophilin D (CD) protein are thought to come together at the mitochondrial inner and outer membrane contact points to create the mtPTP. The proapoptotic Bax of the mtPTP is thought to interact with the anti-apoptotic Bcl2 and the benzodiazepine receptor (BD). The opening of the mtPTP is associated with the release of several proapoptotic proteins. Cytochrome c (cytc) interacts with and activates cytosolic Apaf-1, which then binds to and activates procaspase-9. The activated caspase-9 then initiates the proteolytic degradation of cellular proteins. Apoptosis initiating factor (AIF) and endonuclease G (EndoG) have nuclear targeting peptides that are transported to the nucleus and degrade the chromosomal DNA. Modified from Reference (237).
The energy released by the flow of electrons through the ETC is used to pump protons out of the mitochondrial inner membrane through complexes I, III, and IV. This creates a capacitance across the mitochondrial inner membrane, the electrochemical gradient (ΔP = ΔΨ+ ΔpH). The potential energy stored in ΔP is coupled to ATP synthesis by complex V (ATP synthase). As protons flow back into the matrix through a proton channel in complex V, ADP and Pi are bound, condensed, and released as ATP. Matrix ATP is then exchanged for cytosolic ADP by the adenine nucleotide translocator (ANT) (Figure 2).
Because the ETC is coupled to ATP synthesis through ΔP, mitochondrial oxygen consumption rate is regulated by the matrix concentration of ADP. In the absence of ADP, the consumption of oxygen is regulated by proton leakage across the inner membrane and thus is slow (state IV respiration). However, when ADP is added, it binds to the ATP synthase and is rapidly converted into ATP at the expense of ΔP. As protons flow through the ATP synthase proton channel, the proton gradient is depolarized. Stored fats and carbohydrates are then mobilized to provide electrons to the ETC, which reduce oxygen to water and pump the protons back out of the mitochondrial matrix. The resulting increased oxygen consumption on addition of ADP is known as state III respiration. ΔP is also used for the import of cytosolic proteins, substrates, and Ca2+ into the mitochondrion. Drugs such as 2,4-dinitrophenol (DNP) and nDNA-encoded uncoupler proteins 1, 2, and 3 (Unc1, 2, and 3) render the mitochondrial inner membrane leaky for protons. This short-circuits the capacitor and “uncouples” electron transport from ATP synthesis. This causes the ETC to run at its maximum rate, causing maximum oxygen consumption and heat production, but diminished ATP generation (Figure 2).
The efficiency by which dietary calories are converted to ATP is determined by the coupling efficiency of OXPHOS. If the ETC is highly efficient at pumping protons out of the mitochondrial inner membrane and the ATP synthesis is highly efficient at converting the proton flow through its proton channel into ATP, then the mitochondria will generate the maximum ATP and the minimum heat per calorie consumed. These mitochondria are said to be tightly coupled. By contrast, if the efficiency of proton pumping is reduced and/or more protons are required to make each ATP by the ATP synthase, then each calorie burned will yield less ATP but more heat. Such mitochondria are said to be loosely coupled. Therefore, in an endothermic animal, the coupling efficiency determines the proportion of calories utilized by the mitochondrion to perform work versus those to maintain body temperature.
As a toxic by-product of OXPHOS, the mitochondria generate most of the endogenous ROS. ROS production is increased when the electron carriers in the initial steps of the ETC harbor excess electrons, i.e., remain reduced, which can result from either inhibition of OXPHOS or from excessive calorie consumption. Electrons residing in the electron carriers; for example, the unpaired electron of ubisemiquinone bound to the CoQ binding sites of complexes I, II, and III; can be donated directly to O2 to generate superoxide anion (O2•−). Superoxide O2•− is converted to H2O2 by mitochondrial matrix enzyme Mn superoxide dismutase (MnSOD, Sod2) or by the Cu/ZnSOD (Sod1), which is located in both the mitochondrial inner membrane space and the cytosol. Import of Cu/ZnSOD into the mitochondrial intermembrane space occurs as the apoprotein, which is metallated upon entrance into the intermembrane space by the CCS metallochaperone (166, 207). H2O2 is more stable than O2• − and can diffuse out of the mitochondrion and into the cytosol and the nucleus. H2O2 can be converted to water by mitochondrial and cytosolic glutathione peroxidase (GPx1) or by peroxisomal catalase. However, H2O2, in the presence of reduced transition metals, can be converted to the highly reactive hydroxyl radical (•OH) (Figure 2). Iron-sulfur centers in mitochondrial enzymes are particularly sensitive to ROS inactivation. Hence, the mitochondria are the prime target for cellular oxidative damage (241, 242).
For tightly coupled mitochondria, in the presence of excess calories and in the absence of exercise, the electrochemical gradient (ΔP) becomes hyperpolarization and the ETC chain stalls. This is because without ADP the ATP synthase stops turning over, thus blocking the flow of protons back across the mitochondrial inner membrane through the proton channel of the ATP synthase. However, the ETC will continue to draw on the excess calories for electrons to pump protons out of the mitochondrial inner membrane until the electrostatic potential of ΔP inhibits further proton pumping. At this point, the ETC stalls and the electron carriers become maximally occupied with electrons (maximally reduced). These electrons (reducing equivalents) can then be transferred to O2 generating increased ROS and oxidative stress.
By contrast, in individuals who actively exercise and generate ADP, the ATP synthase keeps ΔP hypopolarized, electrons flowing through the ETC to sustain ΔP, and the electron carriers retain few electrons (remain oxidized). The low electron density of the ETC electron carriers limits ROS production and reduces oxidative stress.
This same effect can be achieved if the mitochondria become partially uncoupled, either by decreasing the number of protons pumped per electron pair oxidize of by permitting protons to flow back through the inner membrane without making ATP. Uncoupling can be achieved by disconnecting electron transport from proton pumping by alterations of complexes I, III, or IV; by increasing the number of protons required by the ATP synthase to make an ATP; or by expression of an alternative proton channel such as an uncoupling protein (UCP).
The mitochondria are also the major regulators of apoptosis, accomplished via the mitochondrial permeability transition pore (mtPTP). The mtPTP is thought to be composed of the inner membrane ANT, the outer membrane voltage-dependent anion channel (VDAC) or porin, Bax, Bcl2, and cyclophilin D. The outer membrane channel is thought to be VDAC, but the identity of the inner membrane channel is unclear since elimination of the ANTs does not block the channel (98). The ANT performs a key regulatory role for the mtPTP (98). When the mtPTP opens, ΔP collapses and ions equilibrate between the matrix and cytosol, causing the mitochondria to swell. Ultimately, this results in the release of the contents of the mitochondrial intermembrane space into the cytosol. The released proteins include a number of cell death-promoting factors including cytochrome c, AIF, latent forms of caspases (possibly procaspases-2, 3, and 9), SMAD/Diablo, endonuclease G, and the Omi/HtrA2 serine protease 24. On release, cytochrome c activates the cytosolic Apaf-1, which activates the procaspase-9. Caspase 9 then initiates a proteolytic cascade that destroys the proteins of the cytoplasm. Endonuclease G and AIF are transported to the nucleus, where they degrade the chromatin. The mtPTP can be stimulated to open by the mitochondrial uptake of excessive Ca2+, by increased oxidative stress, or by deceased mitochondrial ΔP, ADP, and ATP. Thus, disease states that inhibit OXPHOS and increase ROS production increase the propensity for mtPTP activation and cell death by apoptosis (Figure 2) (241, 242).
The mtDNA is maternally inherited (63) and semi-autonomous. The genetic independence of the mtDNA has been demonstrated by enucleating cells harboring a putative mtDNA marker, such as resistance to the mitochondrial ribosome inhibitor chloramphenicol (CAPR), and fusing the cytoplasmic fragments containing the donor mitochondria and mtDNA to recipient cells that lack the genetic marker, e.g., CAPS recipient cells (26, 240). The resulting transmitochondrial cybrids have the mtDNA of the donor and thus inherit the donor’s mitochondrial phenotype (CAPR), yet they have the nDNA of the recipient, demonstrating that mtDNA inheritance can be independent of nDNA inheritance.
The mtDNAs of the donor cell can more completely repopulate the recipient cells if the recipient cells are depleted, or cured, of their resident mtDNAs. The recipient cells can be depleted of their mtDNA by treatment with the mitochondrial poison rhodamine-6-G (220) or they can be cured of their mtDNAs by previous selection in ethidium bromide plus glucose, pyruvate and uridine, resulting in cells that are permanently mtDNA-deficient (ρ0 cells) (94).
The mtDNA has a very high mutation rate, presumably due to its chronic exposure to mitochondrial ROS. When a new mtDNA mutation arises in a cell, a mixed intracellular population of mtDNAs is generated, a state known as heteroplasmy. As the heteroplasmic cell undergoes cytokinesis, the mutant and normal molecules are randomly distributed into the daughter cells. As a consequence of this replicative segregation, the proportion of mutant and normal mtDNAs can drift toward homoplasmic mutant or wild type. Furthermore, in post-mitotic tissues, mtDNAs harboring deleterious mutations have been found to be preferentially, clonally, amplified within cells. This may be a consequence of the nucleus attempting to compensate for an energy deficiency by making more mitochondria. Therefore, cells with defective mitochondria are preferentially stimulated to replicate their mitochondria and mtDNAs. As the proliferating mitochondria are turned over, presumably by autophagy (2, 116, 136), the mutant mtDNA can become enriched in some cells by genetic drift. As the percentage of mutant mtDNAs increases, the mitochondrial energetic output declines, ROS production increases, and the propensity for apoptosis increases. As cells are progressively lost through apoptosis, tissue function declines, ultimately leading to symptoms. Thus the accumulation of mutant mtDNA creates the aging clock (241, 242) (Figure 3).
Figure 3.
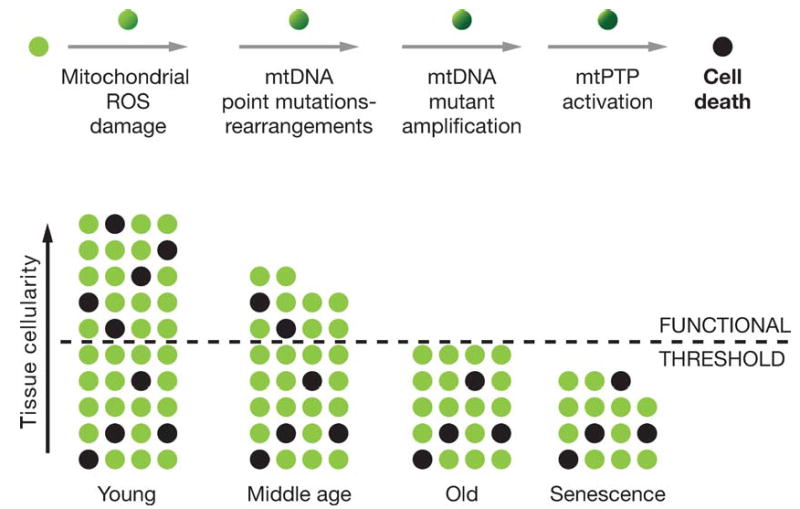
Mitochondrial and cellular model of aging. The upper line of cells diagrams the mitochondrial role in the energetic life and death of a cell. The bottom diagram represents the loss of cells in a tissue over the life of an individual through mitochondrial-mediated death, black cells. The minimum number of cells for the tissue to function normally is indicated by the dashed line.
Clinical symptoms appear when the number of cells in a tissue declines below the minimum necessary to maintain function. The time when this clinical threshold is reached is related to the rate at which mitochondrial and mtDNA damage accumulates within the cells, leading to activation of the mtPTP and cell death, and to the number of cells present in the tissue at birth in excess of the minimum required for normal tissue function. Given that the primary factor determining cell metabolism and tissue structure is reproductive success, it follows that each tissue must have sufficient extra cells at birth to make it likely that that tissue will remain functional until the end of the human reproductive period, or about 50 years. If the mitochondrial ROS production rate increases, the rate of cell loss will also increase, resulting in early tissue failure and age-related disease. However, if mitochondrial ROS production is reduced, then the tissue cells will last longer and age-related symptoms will be deferred (236, 238, 241) (Figure 2).
PATHOGENIC mtDNA MUTATIONS
The first evidence that mtDNA mutations might be a key factor in aging and age-related degenerative diseases came with the identification of systemic diseases caused by mtDNA mutations. Pathogenic mtDNA mutations fall into three categories: rearrangement mutations (74), polypeptide gene missense mutations (243), and protein synthesis (rRNA and tRNA) gene mutations (200, 244). As the number of mtDNA-associated diseases identified increased, it became clear that mitochondrial diseases commonly have a delayed onset and progressive course and that they result in the same clinical problems as observed in age-related diseases and in the elderly. Clinical manifestations that have been linked to mtDNA mutations affect the brain, heart, skeletal muscle, kidney, and endocrine system, the same tissues affected in aging. Specific symptoms include forms of blindness, deafness, movement disorders, dementias, cardiovascular disease, muscle weakness, renal dysfunction, and endocrine disorders including diabetes (51, 149, 237, 241, 242).
Pathogenic mtDNA Rearrangement Mutations
Systemically distributed mtDNA rearrangement mutations can be either inherited or spontaneous. Inherited mtDNA rearrangement mutations are primarily insertions. The first inherited insertion mutation to be identified caused maternally inherited diabetes and deafness (13, 14).
Spontaneous rearrangement mutations, primarily deletions, generally result in a related spectrum of symptoms, irrespective of the position of the deletion end points. This is because virtually all deletions remove at least one tRNA and thus inhibit protein synthesis (154). Thus the nature and severity of the symptoms from mtDNA deletion rearrangements is not a consequence of the nature of the rearrangement, but rather of the tissue distribution of the rearranged mtDNAs. Rearrangements that are widely disseminated prevent bone marrow stem cells from proliferating and lead to the frequently lethal childhood pancytopenia, known as Pearson marrow pancreas syndrome. Widely distributed mtDNA rearrangements that spare the bone marrow result in mitochondrial myopathy with ragged red fibers (RFF). Mitochon-drial myopathy is frequently associated with ophthalmoplegia and ptosis, which is referred to as chronic progressive external ophthalmoplegia (CPEO). More severe, earlier-onset cases with multisystem involvement are known as the Kearns-Sayre syndrome (KSS).
Pathogenic mtDNA missense mutations
Missense mutations in mtDNA polypeptide genes can also result in an array of clinical manifestations. A mutation in the mtDNA ATP6 gene at np 8993 (T to G) is associated with neurogenic muscle weakness, ataxia, and retinitus pigmentosum (NARP) when present at lower percentages of mutant (75) and lethal childhood Leigh syndrome when present at higher percentages of mutant (213). This mutation causes a marked inability of the ATP synthase to utilize the electrochemical gradient to make ATP (219) and an associated increase in mitochondrial ROS production (128).
Missense and nonsense mutations in the cytb gene have been increasingly linked to progressive muscle weakness (8, 52). Rare nonsense or frameshift mutants in COI have been associated with encephalomyopathies (25, 37).
Missense mutations in complex I genes have been linked to Leigh syndrome (205), generalized dystonia and deafness (88), and to Leber hereditary optic neuropathy (LHON), a form of midlife, sudden-onset blindness (22, 23, 243). Surprisingly, the phenotype of LHON can be caused by mutations in a number of ND genes that change amino acids, with very different amino acid conservation resulting in varying severities of complex I defects (22). This anomaly has been explained by the discovery that the mildest LHON mtDNA mutations, particularly those in ND6 at np 14,484 and ND4L at np 10,663, are usually found on a particular mtDNA background, the European mtDNA lineage J (21, 23). Lineage J mtDNAs have been found to harbor mtDNA missense mutations that partially uncouple OXPHOS, thus exacerbating the ATP defect of the pathogenic mutations (194).
Pathogenic mtDNA proteins synthesis mutations
Base substitutions in mtDNA protein synthesis genes can also result in multisystem disorders with a wide range of symptoms, including mitochondrial myopathy, cardiomyopathy, deafness, mood disorders, movement disorders, dementia, diabetes, intestinal dysmotility, etc. As with the ATP6 np 8993 (T > G) mutation, studies on the tRNALys np 8344 A to G mutation associated with myoclonic epilepsy and ragged red fiber (MERRF) disease have revealed that the level of mutant heteroplasmy plus the age of the patient influence the severity of the clinical symptoms (200, 244).
Perhaps the most common mtDNA protein synthesis mutation is the np 3243 (>G) in the tRNALeu(UUR) gene (64). This mutation is remarkable in the variability of its clinical manifestations. When present at relatively low levels (10%–30%) in the patient’s blood, the patient may manifest only type II diabetes with or without deafness (223). This is the most common known molecular cause of type II diabetes, purportedly accounting for between 0.5% and 1% of all type II diabetes worldwide (241). By contrast, when the A3243G mutation is present in >70% of the mtDNAs, it does not cause diabetes, but instead causes more severe symptoms including short stature, cardiomyopathy, CPEO, and mitochondrial encephalomyopathy, lactic acid and stroke-like episodes, the MELAS syndrome (64).
Mitochondrial Defects in Diabetes
The genetic linkage of mtDNA rearrangement (13, 14) and tRNA mutations (223) to type II diabetes directly implicated mitochondrial defects in the etiology of diabetes and the metabolic syndrome. Evidence that mtDNA defects are a common factor in the etiology of diabetes comes from the observation that as the age-of-onset of the proband increases, the probability that the mother was the affected parent also increases, reaching a ratio of 3:1 for patients with a mean age-of-onset of 46 years (241). These observations have been linked to the larger human metabolic syndrome through the identification of a mtDNA tRNAIle mutation at np 4291 (T > C) that causes hypertension, hyperc-holesterolemia, and hypomagnesemia (renal ductal convoluted tubule defect). This mutation was found in a maternal pedigree in association with reduced mitochondrial ATP production and the secondary clinical findings of migraine, hearing loss, hypertrophic cardiomyopathy, and mitochondrial myopathy (248).
Consistent with these genetic results, studies of patients with type II diabetes have revealed that mitochondrial function and gene expression are generally down-regulated. Insulin-resistant offspring of type II diabetic patients have been found to have impaired mitochondrial energetics, as assessed by 31P-MR spectroscopy (176). Furthermore, type II diabetes patients consistently show a down-regulation in the expression of nDNA-encoded mitochondrial genes, in association with alterations in the levels of the peroxisome-proliferation-activated receptor γ (PPARγ )-coactivator 1 (PGC-1) (153, 175), a major regulator of mitochondrial biogenesis and fat oxidation (105, 251).
Diabetes mellitus is also seen in Friedreich ataxia. Patients with Friedreich ataxia manifest cerebellar ataxia, peripheral neuropathy, hypertrophic cardiomyopathy, and diabetes as the result of inactivation of the frataxin gene on chromosome 9q13. Frataxin binds iron in the mitochondrial matrix, thus minimizing mitochondrial •OH production. The loss of the frataxin protein results in excessive ROS generation which inactivates all mitochondrial iron-sulfur-containing enzymes including complex I, II, III, and aconitase. Thus increased mitochondrial ROS production and decreased mitochondrial OXPHOS must be the cause of diabetes in Friedreich ataxia (103, 192, 249).
Type II diabetes has also been associated with a Pro121A polymorphism in the PPARγ gene (5) and with a Gly482Ser polymorphism in the PGC-1 gene in Danish (53) and Pima Indian (156) populations.
Maturity onset of diabetes in the young (MODY) is an early onset autosomal dominant form of type II diabetes that can also be linked to mitochondrial dysfunction. The molecular defects of four forms of MODY have been identified. MODY 2 is the result of mutations in glucokinase, MODY 1 is due to mutations in the hepatocyte nuclear factor (HNF)-1α, MODY 3 to mutations in HNF-4 α, and MODY 4 to mutations in insulin promoter factor (IPF)-1.
Glucokinase has a very high _K_m for glucose and is thought to be the glucose sensor. HNF-1α is a transcription factor and mutations in its gene are associated with post-pubertal diabetes, obesity, dyslipidemia, and arterial hypertension, all features of mitochondrial diseases. HNF-1α is also important in regulating nDNA-encoded mitochondrial gene expression and the expression of the GLUT 2 glucose transporter (250). HNF-4α, a member of the steroid/thyroid hormone receptor superfamily, acts as an upstream regulator of HNF-1α (231) (Figure 4).
Figure 4.
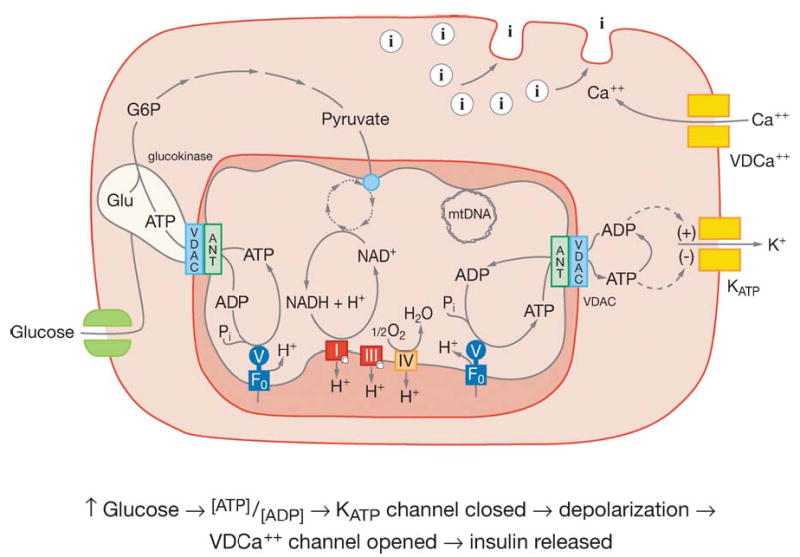
Model of pancreatic β cell showing the mitochondrial regulation of insulin secretion. The light green blob associated with VDAC represents glucokinase. The green double squares in the plasma membrane on the left side represent the glucose transporter. The two pairs of double squares on the right side plasma membrane represent: below and labeled KATP the ATP gated K+ channel and above and labeled VDCa++ the voltage-dependent Ca++ channel. The circles with the internal “I” represent insulin containing vesicles. Other abbreviations as in Figure 2. Reprinted from Reference 25.
The importance of mitochondrial defects in β cell insulin secretion deficiency has been confirmed in two mouse models. In the first, the mitochondrial transcription factor Tfam was inactivated in the pancreatic β cells. This resulted in increased blood glucose in both fasting and nonfasting states, and the progressive decline in β cell mass by apoptosis (201). In the second, the ATP-dependent K+-channel (KATP) affinity for ATP was reduced, resulting in a severe reduction in serum insulin, severe hyperglycemic with hypoinsulinemia, and elevated D-3-hydroxybutyrate levels (102). These models demonstrate that mitochondrial ATP production is critical in the signaling system of the β cell to permit insulin release (238). Thus pancreatic β cell mitochondrial defects are important in both glucose sensing through glucokinase and insulin release through the KATP channel.
Type II diabetes thus involves mutations in energy metabolism genes including the mtDNA and glucokinase; mutations in the transcriptional control elements PPARγ, PGC-1, HNF-1α, HNF-4α, and IPF-1; and alterations in insulin signaling. These seemingly disparate observations can be unified through the energetic interplay between the various organs of the body. The human and mammalian organs can be divided into four energetic categories: the energy-utilization tissues including skeletal muscle, heart, kidney and brain; the energy-storage tissues including brown adipose tissue (BAT) and white adipose tissue (WAT); the energy-homeostasis tissue, liver; and the energy-sensing tissues including the α and β cells of the pancreatic Islets of Langerhans. All of these tissues interact to coordinate the utilization and storage of energy, based on the availability of calories in the environment. For our hunter-gather ancestors, the primary variation in available dietary calories was due to the cyclic growing seasons of edible plants caused by either warm versus cold or wet versus dry seasons. During the growing season, plants convert the Sun’s energy into glucose, which the plants store as starch. When humans and animals ingest these plant tissues the concentration of glucose in their blood rises. Hence, serum glucose is the metabolic surrogate for monitoring plant calorie abundance.
When plant calories are abundant and consumed, the elevated serum glucose is detected by the energy-sensing pancreatic β cells, which respond by secreting insulin into the blood stream. The insulin signal then informs the energy-utilizing heart and muscle tissues to down-regulate mitochondrial energy utilization, since food-seeking behavior is less pressing. It also informs the energy-storing WAT and BAT tissues to store the excess calories as fat for when the season changes and plant calories again become limiting. When plant calories do become limited, insulin secretion declines and the pancreatic α cells secrete glucagon. These low blood sugar hormonal signals inform the energy-utilization tissues to up-regulate the mitochondrial OXPHOS system, thus enhancing food-seeking capacity. They also mobilize the energy-storage tissues to transfer the stored triglycerides into the blood to fuel the increased mitochondria OXPHOS. Furthermore, low blood glucose stimulates the energy-homeostasis tissue, liver, to synthesis glucose to maintain the basal level of blood sugar, which is particularly critical for brain metabolism. The molecular basis of this primeval system for adapting to energy fluctuation can now be partially understood through recent discoveries pertaining to the transcriptional regulation of mitochondrial gene expression.
The energy-sensing tissue, pancreatic β cells, detects abundant plant calories when the blood glucose level exceeds the high _K_m of the β cell glucokinase. Since glucokinase is bound to the mitochondrial VDAC and ANT through the mitochondrial inner and outer membrane contact points, the ATP binding site of glucokinase is continuously occupied (3, 60, 125). The binding of glucose by glucok-inase immediately generates G-6-P and then pyruvate. Pyruvate is oxidized by the mitochondria to generate ATP. The elevated ATP production increases the ATP/ADP ratio, which results in the closure of the KATP channel, depolarizing the β cell plasma membrane. The depolarized plasma membrane opens the voltage-sensitive Ca2+ channel. The influx of Ca2+ leads to the formation of the exocytosis core complex and activation of protein kinases (255). The insulin-containing vesicles then fuse to the plasma membrane and release their stored insulin into the blood (Figure 4).
Thus, when blood glucose is high, the insulin concentration in serum increases. The insulin binds to the insulin receptor of target cells throughout the body, activating the insulin receptor tyrosine protein kinase to phosphorylate insulin receptor substrates. These activate the phosphotidylinositol 3-kinase (PI3K), which phosphorylates phosphotidylinositol 2-phosphate (PI2) to phophatidylinositol 3-phosphate (PI3). PI3 then activates the AKT 1/2 kinases, which phosphorylate the FOXO forkhead transcription factors FOXO1, FOXO3A, and FOXO4 in the target tissues. Phosphorylation of the FOXOs results in their transport out of the nucleus and the transcriptional inactivation of genes whose promoters contain insulin response elements (IRE) (1, 59). The FOXOs can also be removed from the nucleus by acetylation via Cbp or p300 (1) and reactivated by deacetylation via the NAD+-dependent SIRT1 (Sirtuin 1) (24).
When blood sugar is low, the pancreatic α cells secrete glucagons, which binds to the glucagon receptor in target cells. This stimulates the production of cAMP, which activates protein kinase A (PKA). PKA phosphorylates and activates the cAMP response element binding protein (CREB), which activates transcription of genes harboring a cAMP response element (CRE) (48).
The FOXOs and CREB regulate numerous genes through the IRE and CRE cis elements. The PGC-1α promoter contains three IREs that bind unphosphorylated and deacetylated FOXO1, plus one CRE that binds phosphorylated CREB (48). PGC-1α together with its companion genes PGC-1β and PRC (PERC) are transcriptional co-activators that regulate genes for mitochondrial biogenesis, thermogenesis, and fatty acid oxidation (92, 196).
PGC-1α was cloned through its interaction with PPARγ. It is strongly induced in BAT in response to cold acting through the cAMP-mediated β-adrenergic and thyroid hormone (TR) systems. The up-regulation of PGC-1α in turn induces UCP-1, which then creates a proton channel through the mitochondrial inner membrane, uncoupling OX-PHOS. This causes the rapid burning of the stored fats in BAT, generating heat to maintain body temperature (184). PGC-1α also induces mitochondrial biogenesis and UCP-2 in muscle cells (C2C12), acting through the nuclear respiratory factors 1 and 2 (NRF1 and NRF2) (251). NRF1 and NRF2, in turn, activate the transcription of a wide range of nDNA-encoded mitochondrial genes, including components of OXPHOS such as the ATP synthase β subunit (ATPsynβ) and cytochrome c (cytc) and the mtTFA, which regulates mtDNA transcription (92, 196).
The PGC-1 family of transcriptional coactivators interacts with a broad spectrum of tissue-specific transcription factors. This provides the tissue-specific response to the generalized insulin and glucagon hormonal signals. In the energy utilization tissue, muscle, PGC-1 induces mitochondrial biogenesis through the interaction with NRF1, PPARβ, PPARδ, MEF2, ERR2 (estrogen-related receptor 2), HDAC5, and Gabpa/b (GA-repeat binding protein) (47, 78, 117, 152). In heart, PGC-1α interacts with PPARα and NRF1 (78, 111). In the brain, the transcriptional partners of PGC-1α have not yet been identified (118). In the energy-storage tissues, PGC-1α interacts with PPARγ, PPARα, and the thyroid hormone receptor (TR) in BAT to induce mitochondrial biogenesis and UCP-1 during cold stress (78, 251), while in 3T3-Li preadipocytes, PGC-1α interacts with PPARα to stimulate fatty acid oxidation, including induction of the medium-chain acyl CoA dehydrogenase (MCAD) (230). Finally, in the energy-homeostasis tissue, liver, PGC- 1α interacts with FOXO1, HNF-4α, and the glucocorticoid receptor (GR) to induce gluconeogenesis enzymes, including phosphoenolpyruvate carboxylase (PEPCK) and glucose-6-phosphatase (G6Pase) to maintain basal blood glucose levels (78, 121, 187, 254).
PGC-1β is abundantly expressed in BAT, heart, kidney, and skeletal muscle but also in stomach and in white adipose tissue. In 3T3-L1 preadipocytes, PGC-1β is inducible, though PGC-1α is not, and PGC-1β then interacts with the ERRs to induce MCAD and fatty acid oxidation (89). Thus both insulin through the FOXOs and glucagon through cAMP modulate mitochondrial energy metabolism in response to the availability of carbohydrates via the regulation of members of the PGC-1 co-activator family.
Mitochondrial energy metabolism is further modulated by PGC-1α through deacety-lation by SIRT1. PC12 cells that overexpress SIRT1 experience a 25% reduction in oxygen utilization (161). Hepatocytes that over-express SIRT1 induce the gluconeogenesis genes PEPCK and G6Pase via interaction between PGC-1α and HNF-4α (187). Furthermore, upon caloric restriction, the SIRT1 in fat cells binds to the nuclear receptor corepressor (NCoR), blocking its interaction with PPARγ. This inhibits fat storage and enhances fat mobilization (178, 179).
These observations link all of the known genetic mutations associated with diabetes to defects in mitochondria bioenergetics and energy metabolism. Mutations in the mtDNA and in glucokinase would result in energetic dysfunction. Mutations in PPARγ and PGC-1α would reduce mitochondrial gene expression. Mutations in HNF-4α and its target HNF-1α would result in loss of glucose homeostasis. Thus, diabetes is an energy deficiency disease centered upon mitochondrial bioenergetics.
Given a mitochondrial etiology for type II diabetes, the various stages in the progression of type II diabetes can be understood. In individuals harboring a partial defect in OX-PHOS, the capacity of the energy-utilization cells to oxidize carbohydrates and fats to make ATP is reduced. Given a high caloric diet, individuals with partial OXPHOS defects overload their mitochondrial ETC with excessive calories (reducing equivalents), hyperpolarizing ΔP, stalling the ETC, and blocking the tissue utilization of glucose. As a result, the nonmetabolized glucose remains in the blood. The chronically high serum glucose signals the β cells to secrete insulin, creating concurrently elevated glucose and insulin: the hallmark of insulin resistance.
The excessive reduction of the mitochondrial ETC electron carriers in the energy-utilization tissues maximizes mitochondrial ROS production. The high serum insulin activates their Akt pathway, which phosphorylates the FOXOs. The departure of the FOXOs from the nucleus stops transcription of the stress response genes, including the mitochondrial antioxidant enzymes. It also suppresses PGC-1α transcription, which down-regulates mitochondrial OXPHOS, further exacerbating the mitochondrial energy deficiency. The resulting chronic mitochondrial oxidative stress erodes mitochondrial function and increases insulin resistance.
The excess of reducing equivalents also increases the NADH/NAD+ ratio. The conversion of NAD+ to NADH inhibits SIRT1, resulting in increased acetylation of the FOXOs and their removal from the nucleus. This further down-regulates PGC-1α and OXPHOS.
The sustained high serum glucose and insulin also activates the PGC-1α and PGC-1β in the energy-storage tissues BAT and WAT to store the excess calories as fats. Also, the liver accumulates lipid, and this can lead to nonalcoholic steatohepatitis (NASH), which is observed in over 30% of individuals with indications of insulin-resistance, hypertriglyc-eridemia, and hypercholesterolemia (174).
In the β cells, the excessive mitochondria ROS inhibits mitochondrial ATP production, eventually leading to a decline in insulin secretion due to inadequate ATP for glucokinase and a low ATP/ADP ratio that cannot activate the KATP channel. The resulting high glucose but reduced serum insulin is termed insulin-independent diabetes. Continued calorie overload in the pancreatic β cells and associated mitochondrial ROS production ultimately activates the β cell mtPTP, resulting in β cell death by apoptosis, producing insulin-dependent diabetes.
Chronic mitochondrial oxidative stress on the peripheral tissues subsequently damages the retina, vascular endothelial cells, peripheral neurons, and the nephrons, leading to the clinical sequelae of end-stage diabetes. Thus chronic mitochondrial dysfunction can explain all of the features of type II diabetes, and is thus the likely cause of the disease.
ANCIENT MIGRATIONS AND CLIMATIC ADAPTATION
In addition to having to adapt to changing caloric availability due to seasonal changes, ancient human hunter-gathers had to adapt to the rigors of different climatic zones. While induction of UCP-1 in BAT and UCP-2 in muscle permits acute adaptation to thermal stress in rodents, this is not an adequate mechanism for long-term cold adaptation by humans. UCP-1 induction in BAT cannot generate sufficient heat to effectively regulate the temperature of the much larger human body. Therefore, as humans migrated into the more northern latitudes they had to adapt to the chronic cold using another mitochondrial strategy. It now appears that this global climatic adaptive strategy was achieved by the acquisition of mtDNA mutations that partially uncoupled OXPHOS and thus resulted in perpetually increased mitochondrial heat production.
The first clear evidence that ancient mtDNA polymorphisms influence human physiology came from the observation that LHON patients with mild complex I mtDNA missense mutations generally encompassed the same European mtDNA lineage, J. It is now appears that lineage J mtDNAs harbor specific “uncoupling mtDNA variants” that reduce mitochondrial ATP output in favor of heat production. Thus this lineage exacerbates the partial ATP defect generated by the pathogenic mtDNA mutations.
Phylogeographic studies of the human mtDNAs have revealed a remarkable correlation between mtDNA lineages and the geographic origins of indigenous populations. These regional mtDNA lineages are groups of related individual mtDNA sequences (haplotypes) known as haplogroups. The various regional haplogroups form the branches on a single human dichotomous mtDNA phylogenetic tree, generated by the accumulation of sequential mtDNA mutations on radiating maternal lineages. The human mtDNA tree is rooted in Africa, and it has specific branches radiating into different geographic regions that we now believe to have been constrained by the climatic zones (Figure 5) (28, 86, 144, 239).
Figure 5.
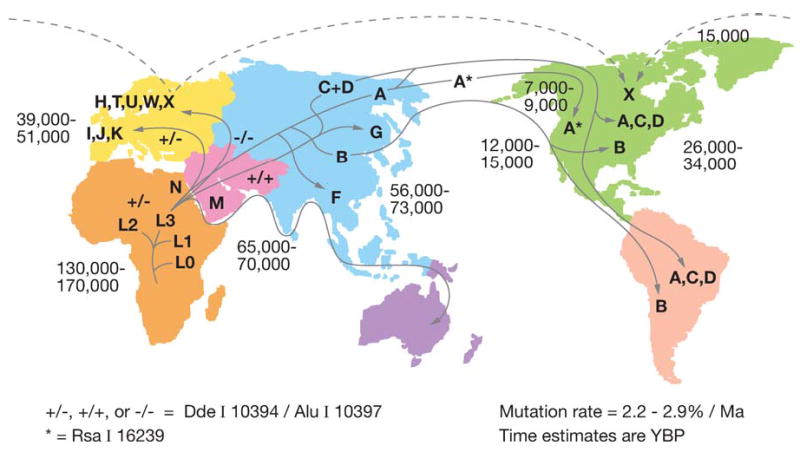
Diagram outlining the migratory history of the human mtDNA haplogroups. Homo sapiens mtDNAs arose in Africa about 150,000 to 200,000 years before present (YBP), with the first African-specific haplogroup branch being L0, followed by the appearance in Africa of lineages L1, L2, and L3. In northeastern Africa, L3 gave rise to two new lineages, M and N. Only M and N mtDNAs succeeded in leaving Africa and colonizing all of Eurasia about 65,000 YBP. In Europe, N gave rise to the H, I, J, Uk, T, U, V, W, and X haplogroups. In Asia, M and N gave rise to a diverse range of mtDNA lineages including A, B, and F from N and C, D, and G from M. A, C, and D became enriched in northeastern Siberia and crossed the Bering land bridge about 20,000 to 30,000 YBP to found the Paleo-Indians. At 15,000 YBP, haplogroup X came to central Canada either from across the frozen Atlantic or by an Asian route of which there are no clear remnants today. At 12,000 to 15,000 YBP, haplogroup B entered the Americas, bypassing Siberia and the arctic, likely by moving along the Beringian Coast. Next at 7000 to 9000 YBP, a migration bringing a modified haplogroup A moved from the northeastern Siberia into northwestern North America to found the Na-Dene (Athebaskins, Dogrib, Apaches, and Navajos). Finally, relatively recently, derivatives of A and D moved along the Arctic Circle to found the Eskimos. These observations revealed two major latitudinal discontinuities in mtDNA variation: one between the Africa L haplogroups and the Eurasia and N and M derivatives and the other between the plethora of Central Asian mtDNA lineages and the almost exclusive presence of lineages A, C, and D northeastern Siberia, the latter spawning the Native American migrations. Since these discontinuities correspond to the transitions from tropical and subtropical to temperate and from temperate to arctic, we have proposed that these discontinuities were the result of climatic selection of specific mtDNA mutations that permitted certain female lineages to prosper in the increasingly colder northern latitudes. Reprinted from http://www.mitomap.org.
African mtDNAs are the most diverse and thus most ancient, with an overall age of about 150,000 to 200,000 YBP. African mtDNAs fall into four major haplogroups: L0 (oldest), L1, L2, and L3 (youngest). L0, L1, and L2 represent about 76% of all sub-Saharan African mtDNAs and are defined by a _Hpa_I restriction site at np 3592 [macrohaplogroup L (L0, L1, and L2)] Figure 1. In northeastern Africa, two mtDNA lineages, M and N, arose from L3 about 65,000 YBP. These were the only mtDNA lineages that succeeded in leaving sub-Saharan Africa and radiating into Eurasia to give all of the Eurasian mtDNAs. In Europe, haplogroups L3 and N gave rise to haplogroups H (about 45% of European mtDNAs), H in Figure 1, T, U, V, W, and X (about 2%) as well as I, J (about 9%), and K(Uk). Europeans separated from Africans about 40,000–50,000 YBP. In Asia, lineages M and N radiated to give rise to a plethora of mtDNA lineages. These include from N, haplogroups A, B, F, and others and from M, haplogroups C, D, G, and others (239) (Figure 5).
As Asians migrated northeast into Siberia, haplogroups A, C, D (A, C, D in Figure 1) became progressively enriched, such that they became the predominant mtDNA lineages in the indigenous peoples of extreme northeastern Siberia, Chukotka. When the Bering land bridge appeared about 20,000 to 30,000 YBP, people harboring these mtDNA haplogroups were in a position to migrate into the New World, where they founded the Paleo-Indians. After the land bridge submerged, haplogroup G arose in Central Asia and moved into northeastern Siberia to populate the area around the Sea of Okhotsk. Later, a migration carrying haplogroup B (B in Figure 1) started from eastern Central Asia and moved along the coast to the New World, by-passing Siberia. Haplogroup B then mixed with A, C, and D in southern North America, Central America, and northern South America to generate the Paleo-Indians. In addition, haplogroup X was brought to the New World in a migration that took place about 15,000 YBP. These immigrants settled in the Great Lakes region, and haplogroup X mtD-NAs are found in 25% of the Ojibwa mtD-NAs today. Since haplogroup X is found primarily in northeastern Europe, it has been speculated that an ancient European migration carrying this haplogroup might also have contributed to the Paleo-Indian populations, perhaps bringing the progenitors of the Clovis lithic culture to the Americas (239).
Later migrations from northeastern Siberia, carrying a modified lineage of haplogroup A, founded the Na-Déné populations about 9500 YBP. More recently, immigrants from Siberia bearing derived lineages of haplogroups A and D moved along the Arctic Circle to found the Eskimos and Aleuts (239).
This phylogeographic distribution of mtDNA haplogroups reveals two striking discontinuities in human mtDNA diversity. The first occurs between sub-Saharan Africa and Eurasia, in which all of the sub-Saharan African mtDNA diversity remained in Africa and only derivatives of lineages M and N colonized temperate Eurasia. The second occurs between temperate Central Asia and arctic Siberia, where the plethora of Asian mtDNA types is markedly reduced to only three ancient mtDNA lineages (A, C, and D). The resulting fivefold enrichment of haplogroups A, C, D, and G in the arctic over Central Asia is unlikely to be the result of genetic bottlenecks, since there are no obvious geographic barriers that separate Asia and Siberia. It is more plausible that many Asian mtDNA lineages entered the arctic, but only a few survived the intense cold to become permanent residents. A similar logic could also apply to the African-Eurasian discontinuity.
Evidence that climatic adaptation has influenced the geographic distribution of mtDNA diversity was first obtained from analyzing the amino acid replacement (nonsynonymous, NS) (Ka) to silent (synonymous, S) (Ks) mutation ratios (Ka/Ks) from the 13 mtDNA open reading frames (83, 239). This revealed that the amino acid sequence of the ATP6 gene was highly variable in the arctic, but was strongly conserved in the tropics and temperate zone; cytb was hypervariable in temperate Europe, but conserved in the tropics and arctic; and COI was variable in the tropical Africa, but invariant in the temperate and arctic regions. Regional variation was also observed in multiple ND subunits (148). Such regional gene-specific variation would not be expected if all mtDNA mutations were random and neutral.
The geographic constraints on mtDNA protein variation were further validated by positioning all of the mtDNA variants from 1125 complete mtDNA coding sequences collected from around the world and organized into a sequentially mutational tree. This tree could be assembled because the maternally inherited mtDNA can only change by sequential mutations along radiating female lines. Therefore, mutations shared between different mtDNAs must be derived from a common female ancestor, and those that are confined to a single mtDNA haplotype must be new mutations.
Analysis of the mtDNA nucleotide variants revealed three different categories of variants: (a) neutral, including synonymous and weakly conserved nonsynonymous amino acid substitutions; (b) deleterious, altering highly conserved amino acids but located at the tips of the branches of the tree indicating they are recent; and (c) adaptive, altering highly conserved amino acids but located within the internal branches of the tree indicating that they are ancient. The mutations in class (c) must be adaptive because they alter highly conserved amino acids, yet they have persisted in the face of intense purifying selection for tens of thousands of years. Hence, as these variants cannot be neutral, they must have been adaptive.
To quantify the potential effect of selection on particular mtDNA variants, the inter-specific amino acid conservation [Conservation Index (CI)] was determined for 22 known pathogenic mtDNA replacement mutations (194). This gave an average CI for deleterious mutations of 93.3 ± 13.3%. Using two standard deviations around the mean CI of the pathogenic missense mutations to distinguish between functionally important versus neutral replacement mutations, 26% of the internal branch replacement mutations were found to be functionally significant, with a mean CI = 85.1 ± 9.2%. The remaining 74% were found to be essentially neutral, with a mean CI = 23.3 ± 14.9%.
Since these conserved internal branch missense mutations frequently initiate new limbs of the mtDNA tree, they must have permitted individuals carrying these mutations to survive and multiply in new geographic regions, generating the region-specific mtDNA haplogroups. The most obvious environmental factor that differentiates tropical and subtropical sub-Saharan Africa from temperate Eurasia and temperate Central Asia from arctic Siberia is temperature. This implies that an important functional effect of many of the conserved internal branch missense mutations was to permit adaptation to cold.
That these adaptive internal branch mutations were important in human radiation was confirmed by analyzing the ratio of NS to S missense mutations in the three major climatic zones. The internal branch NS/S ratio for the tropical and subtropical African L haplogroups was 0.31. The internal NS/S ratio for the temperate Eurasian haplogroups M and N(R) were 0.42 and 0.44, whereas the internal NS/S ratio for the primarily arctic macro-haplogroup N(nonR) was 0.62. Thus, the farther north that the mtDNAs were found, the more missense mutations they harbored.
The excess of internal branch missense mutations in the colder latitudes is particularly striking for haplogroups that reside in the arctic and subarctic. The mean internal branch NS/S ratio for northeastern Siberian-North American haplogroups A, C, D, and X was 0.61, much higher than the mean for the non-arctic haplogroups of 0.39, the mean for the African L haplogroups of 0.31, or the mean for the Native American haplogroup B, which by-passed the cold selection of Siberia, of 0.38.
The internal branch replacement mutations of the arctic and subarctic haplogroups also have a higher average CI. The internal branch CIs of haplogroups A, C, D, and X was 51%; that for the remaining global haplogroups was 39%; that of L was 36%; and that of B was 31%.
The European haplogroup internal branch replacement mutations were also higher than the African L value of 0.31. Haplogroup H was 0.48, J was 0.66, and IWX was 0.63. By contrast, haplogroup T was only 0.31. However, this is because T harbors a single highly conserved founding replacement mutation in the ND2 gene (np 4917), which contributed virtually all of the adaptive advantage to this lineage. Hence, the CI of haplogroup T was 20.3, the highest of any European haplogroup. Thus, adaptive changes fall into two categories, either the lineage accumulates multiple missense mutations, each changing a somewhat less conserved amino acid, or the lineage changes only a few amino acids, each change of which is highly evolutionarily conserved.
Examples of the adaptive mtDNA mutations that occur in the arctic mtDNAs include two replacement mutations [ND2 np 4824G (T119A) and ATP6 np 8794T (H90Y)] for haplogroup A; two replacement variants [ND4 np 11969A (A404T) and cytb np 15204C (I153T)] for haplogroup C; a ND2 np 5178A (L237M) variant for haplogroup D; and a ND5 np 13708A (A458T) variant for a sublineage of haplogroup X. This latter variant also appears in European haplogroup J.
The European sister-haplogroups J and T provide the clearest example of the two classes of adaptive mutation strategies: several less conserved mutations versus a few highly conserved mutations. J and T share a common root involving two amino acid substitutions: ND1 np 4216C (Y304H) and cytb np 15452A (L236I). J and T then diverge. Haplogroup T is founded by the single nodal adaptive mutation: ND2 np 4917G (N150D), the most conserved ND2 polymorphism found (194).
Haplogroup J has two replacement mutations at its root: ND3 np 10398G (T114A) and ND5 np 13708A (A458T), the second being the same variant found in haplogroup X. Haplogroup J then splits into to sub-haplogroups J1 and J2, each defined by a major cytb mutation. The J2 cytb variant is at np 15257A (D171N) and the J1 cytb mutation is at np 14798C (F18L). The np 14789C mutation is also found at the root of sub-haplogroup Uk. The np 15,257 and np 14,789 variants alter well-conserved amino acids with CIs of 95% and 79%, respectively. The 15,257 variant alters the outer coenzyme Q binding site (Qo) of complex III, which contacts the Rieske iron-sulfur protein, while the np 14,798 site alters the inner coenzyme Q binding site (Qi) of complex III (194). Since the Qo and Qi binding sites are essential for complex III proton pumping via the Q-cycle, the np 14,798 and 15,257 variants are both likely to have disconnected electron flow through complex III with proton pumping. This would reduce the coupling efficiency of mitochondrial OXPHOS by one third and proportionately increase heat generation.
That the internal branch mtDNA missense mutations are functionally relevant has been demonstrated by comparing the sperm mobility of males harboring the different mtDNA haplogroups. Sperm flagellar motion is driven primarily by ATP generated from the mitochondria in the mid-piece. Therefore, sperm with partially uncoupled mitochondrial should swim slower that those with coupled mitochondria. As expected, sperm from haplogroup H subjects swam significantly faster than those from T subjects (193). Thus the functional mtDNA variants that founded specific mtDNA lineages affect mitochondrial physiological functions.
ADAPTIVE mtDNA MUTATIONS IN LONGEVITY AND DEGENERATIVE DISEASES
That these adaptive mutations are clinically relevant has become apparent from studies that correlated mtDNA haplogroups with longevity and degenerative disease. In an Italian study, mtDNA haplogroup J was found to be overrepresented in centenarians (50, 189). Similarly, in an Irish centenarian study, J2 was overrepresented (191), and in a Finnish study of individuals over 90 years J2, Uk, and WIX were enriched (163). In Japanese centenarians a sublineage of haplogroup D was enriched (210, 211). Hence, specific mtDNA lineages from Europe and Asia are protective against the ravages of aging.
Some of these same mtDNA lineages have also been found to be protective against neurodegenerative diseases. Haplogroups J and Uk are underrepresented in Parkinson disease (PD) (225) and haplogroup T is underrepresented in AD patients (32, 224). The repeated association of haplogroups J1 and Uk with longevity and neuro-protection is particularly illuminating because both haplogroups encompass the same cytb mutation at np 14,798. Such convergent evolution provides strong support for the functional importance of the cytb mutations.
In other studies, haplogroup J has been found to increase the penetrance of the milder LHON mutations (20). Haplogroup T has been found to be overrepresented in bipolar affective disorder (BPAD) (133).
How can the same variants be associated with increased life span and protection against certain diseases on the one hand, yet increase the predilection of developing other degenerative diseases on the other? The answer lies with the interdependence of mitochondrial energy production, ROS production, and the mtPTP activation of apoptosis.
The cold-adapted mtDNA uncoupling mutations would generate less ATP per calorie consumed. Hence, uncoupled mitochondria would be more prone to clinical problems resulting from energy insufficiency such as LHON and bipolar mental illness. However, individuals with uncoupled mitochondria would burn calories more rapidly to generate both the required ATP plus increased heat. As a result, the ETCs of uncoupled individuals would be generally more oxidized, thus minimizing the production of mitochondrial ROS. Reduction in mitochondrial oxidative stress would reduce the activation of the mtPTP, thus preserving cells and protecting the individuals from cellular loss leading to neurological and visceral tissue degeneration and aging.
THE MITOCHONDRIAL EITIOLOGY OF AGING
Since mtDNA haplogroups that harbor adaptive mtDNA uncoupling mutations have a reduced rate of aging and neurodegenerative disease, the mitochondria must be an important factor in the aging process. Aging is the decline of structure and function over time, which is a natural consequence of entropy, the tendency for all complex systems to decay with time. To counter entropy requires energy to repair or rebuild the damaged functions.
Since uncoupling mtDNA mutations are associated with increased human life span and would have reduced mitochondrial ROS production, ROS damage to the mitochondria, mtDNA, and host cell must be one of the most important entropic factors in determining age-related cellular decline. To counter endogenous sources of oxidative damage, the nucleus must make enzymes that limit mitochondrial ROS damage, thus preserving the mitochondria and mtDNAs.
An important role of mitochondrial ROS production in aging and degenerative diseases is congruent with the life-extending capacity of caloric restriction (69, 126, 127, 130, 204). Reduction of available calories will starve the mitochondrial ETC for electrons, thus reducing ROS and protecting the mitochondria and mtDNAs. Caloric restriction can be achieved by direct dietary restriction or by blocking the retention of excess calories. This may explain why mice in which the insulin receptor gene was inactivated in adiposities had an 18% increase in life span (16).
In rodent studies, mtDNA base oxidation (8oxo-dG) and rearrangement mutation levels have been found to increase with age. Dietary restriction will inhibit the accumulation of both forms of mtDNA damage (67, 139). Furthermore, aging alters gene expression in muscle and brain, but calorie restriction prevents many of these changes (106, 107). In muscle, two classes of genes are affected in aging, those for mitochondrial energy metabolism and those for antioxidant defenses. Calorie restriction normalizes the expression of many of these genes (106).
Drosophila life span can be extended by dietary restriction, and the functional basis of the life extension appears to overlap with insulin-like growth factor defects (35). The chico (small boy) dwarf mutant of Drosophila has a defect in an insulin-like growth factor receptor substrate protein and an increased life span (36). That this relates to energy metabolism was suggested by DNA microarray analyses that revealed the down-regulation with age of genes involved in metabolism, cell growth, and reproduction (180).
Evidence for a role for mitochondrial energy metabolism in Drosophila aging also comes from the Indy (I’m not dead yet) mutant. This mutation inactivates a dicarboxylate cotransporter expressed in the midgut, fat body, and oenocytes of the fly. This enzyme probably mediates the uptake or reuptake of di- and tricarboxylic acid TCA cycle intermediates (188). Since Drosophila lives on decaying fruit, which is rich in the di- and tricarboxylic acids that are substrates for the mitochondrion, the Indy mutant could be restricting the availability of calories to the mitochondrion.
Evidence that ROS toxicity is a limiting factor for life span in Drosophila was directly demonstrated by showing that transgenic Drosophila expressing increased Cu/Zn SOD and catalase lived longer (168). Drosophila life span extension was also seen in flies in which MnSOD was overexpressed in motor neurons (173) and systemically in Drosophila adults after heat shock induction of the MnSOD trans-gene (208). These observations support the conclusion that mitochondrial ROS toxicity is an important factor in limiting Drososphila life span.
A mitochondrial role in longevity has also been observed in the nematode Caenorhabditis elegans. A number of the C. elegans mutants that extend life span specifically affect mitochondrial function. These include the clk-1 mutant, which affects a gene in the biosynthetic pathway of coenzyme Q (71, 87, 150), the isp-1 mutant, which alters the Rieske iron sulfur protein of respiratory complex III (56), a mitochondrial leucyl-tRNA synthetase mutant (110), and a mitochondrial tRNA wobble U modifying enzyme, isopentenylpyrophosphate:tRNA transferase mutant (114). Since the isp-1 mutation has been shown to both increase longevity and reduce mitochondrial ROS production, it seems likely that mitochondrial ROS must be modulating life span (56).
The importance of the mitochondria in regulating C. elegans life span was further confirmed by an RNAi scan of the genes located on chromosomes I and II. Of the genes whose inactivation extended life span, 15% were identified as having mitochondrial functions. Since only about 1.5% of the genes of the C. elegans genome have been proposed to have a mitochondrial function, mitochondrial functions must have a much greater influence on longevity than other physiological processes (109, 110).
The life span of C. elegans is also regulated by the insulin-like growth factor signal transduction pathway. The metabolic functions associated with key longevity mutations include the insulin-like receptor (IRL) (daf-2), the catalytic subunit of PI3Kinase (age-1), the PTEN-like lipid phosphatase (daf-18), the Akt-1/2 kinases, and the target of Akt 1/2 phosphorylation, the fork-head transcription factor (daf-16). Phosphorylated forkhead transcription factor is cytosolic and transcriptionally inactive. Thus mutational inactivation of the insulin-like growth factor pathway must extend life span by blocking phosphorylation of the fork-head transcription factor and rendering it constitutively transcriptionally active (65, 93, 165, 215). Two genes that are up-regulated by activation of the daf-16 forkhead transcription factor via dephosphorylation are MnSOD and catalase (122). Hence, one important function of the insulin-like signal pathway in C. elegans appears to be to the negative regulation of the stress response and antioxidant enzymes that protect the cell from the deleterious effects of mitochondrial ROS production.
Life span in C. elegans as well as in yeast can also be extended by the overexpression of the NAD+-dependent protein deacetylase Sir2. Sir2 acts, in part, through the deacetylation and activation of the forkhead transcription factors, favoring nuclear relocation and transcription activation (61, 214). In yeast, Sir2 also connects the insulin-like signaling pathway to caloric restriction. Sir2 requires NAD+ as a co-reactant. NADH will not suffice and even acts as a competitive inhibitor of yeast. Since caloric restriction would reduce the cellular NADH to NAD+ ratio, this should activate Sir2, causing the deacetylation and activation of the forkhead transcription factors and thus increasing expression of the antioxidant and stress response genes (119, 120). By contrast, defects in mitochondrial OX-PHOS should increase the NADH/NAD+ ratio, inhibit SIR2, and favor the acetylation of the forkhead transcription factors. This would turn off antioxidant and stress response gene expression, thereby reducing antioxidant defenses and life span.
This overlap between the insulin-like signaling pathway and the mitochondrion has been demonstrated in C. elegans by combining daf-2 and isp-1 mutants and showing that they do not have an additive effect on longevity. This is logical since inactivation of the daf-2 pathway would increase mitochondrial antioxidant defenses while the isp-1 mutation would reduce mitochondrial ROS production (56). Hence, the two classes of mutations would have the same effect.
That mitochondrial ROS production is an important factor affecting C. elegans life span has been confirmed by the mev-1 mutation and the effects of SOD mimetics. The mev-1 mutation affects the cytb-containing subunit of complex II. This mutation has a markedly increased rate of mitochondrial ROS production and a substantially shortened life span (84, 198). This reduced longevity can be reversed by treatment with the catalytic antiox-idant mimetic EUK134, a salen Mn complex. Furthermore, treatment of wild-type C. elegans with EUK134 can increase the mean and maximum life span of C. elegans to an extent similar to that of the long-lived C. elegans mutant age-1 (140). Therefore, the life span of C. elegans may be modulated by either decreasing mitochondrial ROS production or increasing mitochondrial antioxidant defenses.
Corroborative data that mitochondrial ROS limits the life span of mammals have come from studies on mutant and transgenic mice. Mice in which the MnSOD was genetically inactivated die at a mean age of eight days of a dilated cardiomyopathy, in association with the inactivation of iron-sulfur center containing enzymes including complex I, II, III, and aconitase (115, 137). The mitochondria of these animals have reduced state III respiration and hypersensitized mtPTPs (97), and they can be rescued from their cardiomyopathy by treatment with the SOD mimetic MnTBAP [Mn 5, 10, 15, 20-tetrakis (4-benzoic acid) porphyrin]. However, MnT-BAP does not readily cross the blood-brain barrier and whereas the treated mice avoid the cardiomyopathy, they develop movement disorders together with cortical and brainstem spongiform vacuolization and brainstem astrocytosis (142). Again, both the systemic and neurological pathology can be ameliorated by treatment with the salen Mn complex compounds EUK8, EUK134, and EUK189, which do cross the blood-brain barrier and are protective against both the visceral effects and the neurological pathologies (138). Thus mitochondrial ROS damage must be important in the age-related decline of both mammalian visceral and central nervous system cells and tissues.
Although mice heterozygous for the Mn-SOD mutation live to old age, they have a chronically reduced liver mitochondrial membrane potential, reduced state III respiration equivalent to that of an old animal, and a striking rise in mitochondrial lipid peroxidation by middle age that drops to below the wild-type levels in older mice. This striking drop in lipid peroxidation is associated with a marked increase in the sensitivity of the mtPTP to Ca2+ activation and a striking increase in number of liver apoptotic cells in older heterozygous animals. The MnSOD heterozygotes also showed a striking induction of complex IV (COX) in the older animals (97). Aging MnSOD heterozygous C57Bl/6 (B6) mice also show a significant increase in oxidative damage (8oxo-dG) to nDNA and mtDNA, and a 100% increased tumor incidence, though their life span is not diminished (226).
Proof that mitochondrial ROS limit mammalian life span has come from the targeting of the human catalase to the mouse mitochondria in transgenic mice and observing an increase in life span. To import catalase to the mitochondrial matrix, the C-terminal peroxisomal targeting peptide was inactivated, and the catalase coding sequence was fused to the N-terminal mitochondrial targeting peptide of the mitochondrial matrix enzyme ornithine transcarbamylase. This construct was then expressed in transgenic mice using an α-actin promoter and CMV enhancer. The modified catalase was shown to be imported into the mitochondrion and to strikingly increase catalase activity in heart and muscle. Moreover, heart mitochondria from the transgenic animals showed a dramatically increased resistance of mitochondrial aconitase to inactivation by H2O2, and the skeletal muscle and heart mtDNAs showed a significant reduction in the age-related accumulation of mtDNA rearrangement mutations. These protective effects to mitochondrial oxidation and mtDNA mutations were associated with a 21% increase in mean life span and a 10% increase in maximum life span in both male and female mitochondrial catalase transgenic animals (197).
The importance of mitochondrial ROS in longevity has also been supported by the selective inactivation of the p66shc splice variant of the p52shc/p46shc gene. This mutant increases mouse life span by 30%, results in increased resistance to oxidative stress caused by paraquat, and reduces mitochondrial ROS production (146). p66Shc has been shown to become incorporated within the mitochondria, where it interacts with HSP70 and affects mitochondrial function and DNA oxidative damage (169). It is also a downstream target of p53 and is required for stress-activated p53 to induce intracellular ROS production, cytochrome c release, and apoptosis (218).
That increased mitochondrial ROS production accelerates mtDNA mutation rate was shown in mice deficient in the nDNA-encoded heart-muscle (and brain) isoform of the ANT gene (Ant1). These animals exhibit severe fatigability and develop a hypertrophic cardiomyopathy. Since an ANT defect blocks the import of cytosolic ADP into the mitochondria, and the ATP synthase without ADP cannot utilize ΔP to make ATP, then ΔP becomes hyperpolarized, the ETC stalls, and the excess electrons are transferred to O2 to generate ROS. Consequently, the ANT1-deficient mice have greatly increased heart, muscle, and brain mitochondrial H2O2 production. This increased mitochondrial ROS correlates with a dramatic increase in mtDNA rearrangements in the heart by middle age (54).
That mtDNA mutations are one of the primary factors that limit mouse life span was demonstrated by the knock-in of a mutant form of the mtDNA polymerase γ (POLG) subunit A in which the proofreading function was inactivated by the mutation D257A. These mice had a shortened life span and developed a premature aging phenotype involving weight loss, reduction of subcutaneous fat, hair loss (alopecia), curvature of the spine (kyphosis), osteoporosis, anemia, reduced fertility, and heart enlargement. This was associated with an age-related decline in respiratory complexes I and IV and in mitochondrial ATP production rates in the heart. Analysis of the mtDNA revealed that the knock-in animals had a three- to fivefold increase in mtDNA base substitution mutations in brain, heart, and liver, with a higher number of mutations in the coding region cytb gene than in the CR (216). Thus, increased mtDNA mutations can directly increase the aging rate, implying that the mtDNA is one of the most important targets for mitochondrial ROS damage in aging.
The relationship between mtDNA mutations and longevity has been shown in a wide variety of animal species including humans. mtDNA rearrangement mutations have been shown to accumulate in proportion to life span in organisms as diverse as mouse, chimpanzee, and human (43, 139, 141, 143). When the level of mtDNA rearrangements was quantified in various human brain regions by the analysis of the common 5-kb mtDNA deletion, mtDNA rearrangements were found to accumulate to the highest levels in the basal ganglia, followed by the cerebral cortex, with the least mutant being found in the cerebellum (39, 206). Base substitution mutations also accumulate in human tissues with age. A mtDNA CR mutation, T414G, in the mtTFA binding site of the PL accumulates with age in diploid fibroblasts (145); a T408A promoter mutation and a A189G OH mutation accumulate with age in skeletal muscle (245); and a T150C OH mutation accumulates with age in white blood cells (256). These mtDNA CR mutations appear to be highly tissue specific. The T414G mutation can be detected at low levels in skeletal muscle by using a highly specific and sensitive protein nucleic acid (PNA)-clamping PCR method. However, this same method has been unable to detect the T414G mutation in the brains of even very elderly people (157). These mtDNA mutations can be attributed to oxidative damage, at least in the brain, since the human brain mtDNA shows a marked age-related increase in oxidative damage (135).
All of these data are consistent with the hypothesis that aging is the result of mitochondrial decline due to oxidative damage to the mitochondria and mtDNAs. This hypothesis, when combined with our current understanding of caloric regulation of mitochondrial function, can neatly explain why caloric restriction is able to extend mammalian life span. Reduction of dietary calories reduces serum glucose, thus inhibiting insulin secretion by the pancreatic β cells. This results in the dephosphorylation and activation of the FOXO forkhead transcription factors in the energy-utilizing tissues. Further, caloric restriction reduces cellular NADH/NAD+ ratios, activating the NAD+-dependent SIRT1, which further activates the FOXOs by deacetylation (24, 61, 155). The activated FOXOs then activate the transcription of the stress response genes including the antioxidant enzymes MnSOD and catalase. They also induce, along with glucagon through CREB, the transcriptional induction of PGC-1α. This increases the mitochondrial OXPHOS capacity, mobilizes the WAT triglyceride stores for mitochondrial oxidation, and activates gluconeogenesis to maintain the basal serum glucose level. Hence, caloric restriction induces the mitochondria to more completely oxidize dietary reducing equivalents and the antioxidant defense systems to more effectively eliminate ROS. Thus, dietary restriction should substantially reduce mitochondrial oxidative stress. In addition, activation of SIRT1 by reduction of the NADH/NAD+ ratio will stimulate the deacetylate p53, and deacetylation of p53 reduces its apoptotic potential. Hence, caloric restriction also protects the mtPTP from p53-mediated activation and apoptosis (82, 104, 123, 229). Therefore, caloric restriction extends life span in three ways: by decreasing mitochondrial generation of ROS, by increasing the cellular antioxidant defenses, and by inhibiting cell loss via apoptosis.
THE MITOCHONDRIAL ETIOLOGY OF DEGENERATIVE DISEASES
The level of mtDNA rearrangement mutations has also been found to be strikingly increased in a variety of age-related degenerative diseases. The 5-kb mtDNA rearrangement is increased between 7- and 2200-fold in the hearts of patients with chronic coronary artery disease (41, 42). Since coronary artery disease is associated with recurrent cardiac ischemia and reperfusion, which generates bursts on mitochondrial ROS (49), the cardiac rearrangement mutations may also be caused by mitochondrial oxidative damage.
Similarly, mtDNA rearrangements are found in patients with AD. In the brains of AD patients who died prior to age 75, the 5-kb deletion is increased on average 15-fold, whereas in the brains of patients who died after age 75, the 5-kb deletion is present at one fifth the level of that in control brains (40). The increased mtDNA rearrangement mutations in AD brains are associated with increased mtDNA oxidative damage (134). The sudden drop in the level of mtDNA deletions in older AD brains might be the result of loss of the brain cells containing the most mutated mtDNA through apoptosis. This is consistent with the increased neuronal apoptosis observed in AD brains (30).
AD, is a progressive neurodegenerative disease resulting in dementia associated with the deposition of Aβ amyloid peptide plaques and neurofibrillary tangles in the brain. Early-onset, familial AD has been associated with mutations in the Aβ precursor protein (APP) and the presenilin-containing APP peptide processing complexes (212). Although the molecular defects that cause the late-onset, sporadic AD cases have not been determined (203), the inheritance of the ApoE ɛ4 allele has been identified as a major risk factor for developing late-onset AD (190, 203).
Mitochondrial OXPHOS defects have also been detected in a variety of tissues from AD patients. The mtDNA of AD patients has been indirectly shown to harbor genetic defects by fusing blood platelets from AD patients to human ρ0 cell lines (94), with the resulting cybrids being found to have OXPHOS defects (17, 29, 45, 96, 158, 159, 217, 222, 232). The concept that mtDNA mutations might contribute to late-onset AD has been bolstered by the observation that a mtDNA tRNAGln gene mutation at np 4336 is present in about 5% of late-onset AD patients and in 7% of combined AD plus PD patients, but in 0.4% of normal controls (81, 199).
The association between mitochondrial deficiency and AD was further strengthened recently by the demonstration, using PNA-clamping PCR, that the somatic mtDNA CR T414G mutation in the PL mtTFA binding site can be detected in 65% of all AD brains but not in any control brains. Moreover, the level of somatic mtDNA CR mutations in AD brains is increased by 63% overall, with the elevation of somatic mtDNA CR mutations in AD patients over 80 years being 120%. Furthermore, in the AD brains, the somatic mtDNA mutations preferentially occur within the known mtDNA transcription and replication regulatory elements, whereas very few of the mtDNA CR mutations found in the control brains occur in functional elements. Finally, AD brains have an average 50% reduction in the ND6/ND2 mRNA ratio and a comparable reduction in the levels of the mtDNA/nDNA ratio, suggesting the inhibition of l-strand transcription and mtDNA replication (44).
These data suggest that the accumulation of somatic mtDNA CR mutations in AD brains that damage key mtDNA regulatory elements may be a central feature in the etiology of the disease. The accumulation of these mutations with age would result in the progressive decline in mitochondrial energy production and an associated increase in mitochondrial ROS production. The increased ROS would ultimately lead to the activation of the mtPTP and the loss of neuronal synapses through mitochondrial induced cytochrome c release and caspase activation.
By this logic, individuals harboring mtD-NAs that generate tightly coupled mitochondria would experience chronically increased ROS production on a high calorie diet. This would increase the probability of mtDNA CR mutations and thus the probability of developing AD. By contrast, individuals who harbor mtDNAs with uncoupling polymorphisms would have reduced mitochondrial ROS and be less prone to somatic mtDNA CR mutations and AD. Population studies have confirmed these predictions.
Mitochondrial ROS production might also account for the connection between the ApoE ɛ4 allele and AD, since the presence of the ApoE ɛ4 allele has been associated with increased brain oxidative damage (151, 185, 186). Aβ peptide toxicity might also be related to mitochondrial dysfunction. The Aβ peptide has recently been reported to enter the mitochondria, bind to mitochondrial alcohol dehydrogenase, and increase mitochondrial ROS production (46, 124). Moreover, incubation of rat brain mitochondria with the Aβ25–35 fragment results in a selective and dose-independent inhibition of complex IV (27). Consistent with the possibility that Aβ enters the mitochondrion, the insulin-degrading enzyme (IDE) has been discovered to have a mitochondrially directed form generated by initiating translation at an AUG codon 41 amino acids upstream from the canonical IDE peptide start codon. Since the IDE is thought to degrade both cerebral Aβ-peptide and plasma insulin, it could function in mitochondrial Aβ turnover (113).
The Aβ peptide is generated by cleavage of the APP by the presenilin-containing γ -secretase complex. Recently, the presenilin-1 (PS1) protein has been located in the inner membrane of rat brain and liver mitochondria (9). The other components of the presenilin γ -secretase including nicastrin, APH-1, and PEN-2 have also been found in a high-molecular-weight complex within the mitochondrial inner membrane. Although the import mechanism for these proteins is not fully understood, the nicastrin protein has been found to harbor both ER and mitochondrial N-terminal targeting signals (68). APP has also been found in the mitochondrion, but it is thought to be located in the outer mitochon-drial membrane, a position that would not be conducive to its processing into Aβ by the mitochondrial inner membrane γ -secretase (7). Therefore, the γ -secretase complex proteins must have other functions in the mitochondrion. Given that γ -secretase is unique in its ability to cleave peptide bonds within membranes, the PS1 complex may well be processing mitochondrial inner membrane proteins such as those of the respiratory complexes. Hence mutations in the presinilin complex associated with early onset AD might also cause mitochondrial defects.
A mitochondrial etiology of AD could also explain the near-universal association of Aβ amyloid plaques with AD. At the concentrations found in biological fluids, Aβ1–40 and Aβ1–42 act as antioxidant and anti-apoptotic polypeptides, presumably because of their tendency to chelate transition metals, particularly copper. However, when the Aβs aggregate, they become pro-oxidants, perhaps through placing the transition metal in the presence of the redox-active Met 35 (99–101). Given this perspective on the Aβ peptides, it could follow that genetic variants which chronically increase mitochondrial ROS production would overwhelm the antioxidant capacity of basal levels of Aβ. To compensate, the brain would increase Aβ production. However, this would eventually lead to excess Aβ, which would then aggregate into a pro-oxidant form (80) and thereby exacerbate the problem. Consistent with this scenario, the brains of late-onset sporadic AD patients have been observed to have increased oxidative damage, increased activated caspase activity (62, 183), and increased numbers of TUNEL-(terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling) positive cells (45). Increased mitochondrial ROS would also increase the mtDNA somatic mutation rate and exacerbate mitochondrial oxidative stress, mtPTP activation, and synaptic loss.
Early onset AD could also fit within this framework. Mutations in the Aβ processing pathway would increase the rate of Aβ production and aggregation, creating an active pro-oxidant that would increase synaptic oxidative stress, damage the mitochondria, and destroy the synapses.
A similar mitochondrial scenario could be applied to other age-related neurodegen-erative diseases. Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease that specifically affects the motor neurons. In familial cases, ALS is caused by mutations in the Cu/ZnSOD (Sod1). Transgenic mice that express a human mutant Cu/ZnSOD develop motor neuron disease associated with vacuolate mitochondria containing high concentrations of Cu/ZnSOD (85). The mutant Cu/ZnSOD mice also accumulate 8oxo-dG in cytoplasmic granules, possibly representing mtDNA nucleoids (247), and in cultured neuroblastoma cells, the toxicity of a mutant Cu/ZnSOD can be ameliorated by the increased expression of MnSOD (57). The mitochondrial uptake of the three common familial ALS Cu/ZnSOD mutants (G37R, G41D, and G93A) is blocked by the heat shock proteins Hsp70, Hsp27, and Hsp25 (167). Consequently, familial ALS appears to be due to a failure in the Cu/ZnSOD’s ability to detoxify mitochondrial superoxide anion, which is released into the mitochondrial inner membrane space from ubisemiquinone bound to the Qo site of complex III.
Idiopathic PD has also consistently been reported to be associated with mitochondrial defects, particularly in respiratory complex I. Evidence supporting a mitochondrial etiology of PD has been previously reviewed but includes the selective destruction of the dopaminergic neurons in humans and rodents by the mitochondrial complex I inhibitor 1-methyl-4-phenyl-1,2,3,6-tetrahy-dropyridine (MPTP); the detection of OX-PHOS defects and particularly complex I defects in PD patient muscle, blood, brain, and in platelet-derived cybrids; and the inheritance of PD-like symptoms to various pathogenic mtDNA mutations (241) including the common LHON np 11,778 mutation (202). Small subsets of PD patients are also found in Mendelian pedigrees. The gene for one rare autosomal recessive form of PD, which maps to chromosome 1p35-p36, has been cloned and found to code for the PINK1 (PTEN-induced kinase) protein, a kinase localized in the mitochondrion (221). A more common autosomal recessive locus, the parkin gene on chromosome 6q25.2–27, is a E3 ubiquitin ligase (228). Genetic inactivation of parkin in the mouse resulted in nigrostriatal defects associated with a decreased abundance of several mitochondrial proteins including the E1α subunit of pyruvate dehydrogenase, the 24- and 30-kDa subunits of complex I and the Vb subunit of complex IV and a general decrease in mitochondrial state III respiration. The oxidative stress proteins per-oxiredoxin 1, 2, and 6 and lactolyglutathione were also down-regulated in association with a marked increase in brain lipid peroxidation products (171). Hence, it would appear that the pathophysiology of the parkin mutation is also associated with a mitochondrial defect. Finally, the rare autosomal dominant form of PD, which maps to chromosome 4q21-q22, is due to mutations in a-synuclein (182). Expression of mutant α-synuclein in various cell systems has resulted in altered mitochondrial function (79, 108, 170). Thus, both the idiopathic and the inherited forms of PD appear to be associated with mitochondrial dysfunction. This is consistent with the protective effects of haplogroups J and Uk for PD (225).
Studies on Huntington disease have implicated a mitochondrial dysfunction in its etiology as well (172, 195, 241), and mtDNA rearrangements are elevated in the basal ganglia of patients with Huntington disease (76). Thus all of these age-related neurodegenerative diseases, as well as aging itself, seem to be directly related to the mitochondrial production of ROS and the resulting accumulation of mtDNA mutations. These observations imply that the mitochondrial paradigm for neurode-generative diseases appears to be one that can be broadly applied.
SOMATIC AND GERMLINE mtDNA MUTATIONS IN CANCER
Aging is also the most significant risk factor for most solid tumors. Moreover, caloric restriction in rodents markedly reduces cancer risk (69, 126, 127, 130, 204). Hence, the etiology of cancer might also be associated with mitochondrial dysfunction.
Gene defects in certain nDNA-encoded mitochondrial genes have been directly linked to some hereditary cancers. Mutations in the mitochondrial fumarate hydratase have been associated with uterine leiomyomas and renal cell carcinomas (112), and mutations in three of the four nDNA-encoded subunits of complex II have been linked to paragangliomas: SDHD (cytb membrane subunit) (15), SDHC (coenzyme Q-binding membrane subunit) (162), and SDHB (iron-sulfur) subunit (11, 227). In addition to the paragangliomas seen for mutations in all three subunits, mutations in the SDHB subunit are also associated with pheochromocytoma and early onset renal cell carcinoma. On the other hand, mutations in the SDHA (FAD-containing subunit) cause the mitochondrial encephalomyopathy Leigh syndrome (19) but not paraganglioma.
The striking difference in clinical effects of SDHA subunit mutations versus SDHB, C, and D mutations strongly implicates mitochondrial ROS production in the etiology of cancer. SDHA mutations would block the entry of electrons into complex II, thus depleting ATP without increasing ROS production. The result is neuromuscular disease. By contrast, mutations in subunits SDH B, C, and D would impede electron flux through complex II. This would increase ROS production, implicating ROS in cancer predisposition. This is supported by the mev-1 mutant in the C. elegans SDHC subunit, which has been shown to increase mitochondrial ROS production (84, 198).
A role for mitochondrial ROS in cancer is supported by the observations that mice heterozygous for the MnSOD knock-out locus have a 100% increase in cancer incidence (226), many types of tumors have reduced MnSOD, transformation of certain tumors with the MnSOD cDNA can reverse the malignant phenotype, and a cluster of three mutations in the MnSOD gene promoter region that alter AP-2 binding and promoter efficiency are found in a number of tumors (131, 252). Moreover, ROS production, in association with the inactivation of p16ink4a, has been hypothesized to be one of the two main mechanisms for tumorigenesis; the other is p53 deficiency (10). Hence, it would appear that mitochondrial ROS production, not ATP depletion, is the important mitochondrial factor in the etiology of cancer.
Mitochondrial DNA mutations that inhibit OXPHOS and impede electron flow down the ETC should increase ROS production and contribute to cancer. The first clear evidence that mtDNA mutations might play an important role in neoplastic transformation came with the report of a renal adenocar-cinoma that was heteroplasmic for a 294-np inframe deletion in the mtDNA ND1 gene. This deletion generated a truncated mRNA, confirming that the mutant mtDNA resulted in functional mitochondrial defect (77). Subsequently, a variety of mtDNA-coding region and CR mutations have been reported in colon cancer cells (4, 66, 181), head and neck tumors (58), astrocytic tumors (95), thyroid tumors (129, 253), breast tumors (209), and prostate tumors (33, 34, 38, 177).
Mitochondrial ROS could contribute to neoplastic transformation, both as a tumor initiator by causing nDNA mutations in proto-oncogenes and tumor-suppressor genes and as a tumor promoter through driving cellular proliferation. At low levels, ROS has been found to be an active mitogen, thought to act though interaction with various kinases (Src kinase, protein kinase C, MAPK, and receptor tyrosine kinases), as well as with different transcription factors (Fos, Jun, NF- κB) (131). Furthermore, the dual function apurinic/apyrimidinic endonuclease 1 (APE-1) functions both in the DNA base excision pathway but also in the redox regulation of the transcription factors Fos, Jun, NFκB, PAX, HIF-1a, and p53 through the redox modification of its cysteine 310 (55, 91).
Further evidence of the importance of mtDNA mutations and mitochondrial ROS production in neoplastic transformation comes from studies of complex IV (COX) mutations in prostate cancer. A proteomic survey of prostate cancer epithelium cells isolated by laser capture microscopy (LCM) revealed that the ratio of nDNA-encoded subunits (COX IV, Vb, and VIc) to mtDNA-encoded subunits (COI and II) was increased in most prostate tumors (72).
This alteration in the ratio of nDNA/mtDNA COX subunits in prostate cancer has now been shown to be due, in part, to mutations in the mtDNA COI gene (177). An analysis of the COI gene sequence from 260 prostate cancer specimens, 54 prostate cancer-negative men over age 50, and 898 European mtDNA sequences revealed that for Americans of European descent, COI mutations are found in 11% of the prostate cancer specimens, 0% of the cancer-negative prostate controls, and 5.5% of the general population, all significant differences. Furthermore, four prostate cancer COI mutations (T6253C, C6340T, G6261A, A6663G) (Figure 1) were found in multiple independent patient tumors, often on different background mtDNA haplotypes, and these changed highly conserved amino acids (CI = 69% to 97%) (177). Thus males who inherit COI missense mutations from their mother are at a greatly increased risk for developing prostate cancer.
The average amino acid CI of the COI mutants found in prostate cancers was 83% ± 25%, whereas that of the COI variants in the general population was 71% ± 35%. This indicates that somatic mutations affecting more functionally conserved amino acids also contribute to the etiology of prostate cancer. This assessment was confirmed by the observation that three of the COI mutations in prostate patients were heteroplasmic: G5949A-G16X (Stop), T6124C-M74T (CI = 95%), C6924T-A341S (CI = 100%) (177). These data demonstrate that mutant COI genes are prostate cancer oncogenes and that both germline and somatic mtDNA mutations are important factors in this etiology of prostate cancer.
A more complete assessment of the role of mtDNA mutations in prostate cancer was obtained by sequencing the entire mtDNA from the tumorigenic epithelial cells, purified using LCM, of the tumor harboring the COI chain termination (G16X). This analysis revealed three mtDNA variants: the heteroplasmic COI G16X mutation, a homoplasmic ATP6 C8932T (P136G) variant with a CI of 64%, and a homoplasmic cytb A14769G (N8S) variant with a CI of 20.5%. Of these variants, the COI and ATP6 mutations are likely to be functionally significant. The G16X mutation was found to be homoplasmic in the cancer cells and homoplasmic wild type in the adjacent normal cells, which was confirmed by immunohistochemical staining for the COI protein. Therefore, this G16X mutation must have arisen de novo at the time of neoplastic transformation and been strongly selected for in the cancerous cells (177).
The physiological significance of mtDNA mutations in cancer cells was demonstrated by introducing a known pathogenic mtDNA mutation into the prostate cancer cell line PC3 via transmitochondrial cybrid fusions (26, 240). PC3 cancer cells were cured of their resident mtDNAs by treatment with the mitochondrial poison rhodamine 6G and then fused with cytoplasts from either homoplasmic mutant (T8993G) or homoplasmic wild-type (T8993T) cell lines (177) derived from a patient who was heteroplasmic for the pathogenic ATP6 np T8993G L156R mutation (75). This mutation reduces mitochondrial ADP production (219) but also increases ROS production (128). A series of PC3 cybrids were generated that were genetically identical except for either a wild-type T or a mutant G at np 8993 in the mtDNA.
Injection of these mutant and wild-type PC3 cybrids into nude mice gave a dramatic result. The PC3 cells harboring the normal T8993T base barely grew at all. By contrast, the PC3 cybrids with the mutant T8993G base generated rapidly growing tumors that killed the mice (177). Staining the cellular nodules from the T8993T and T8993G cybrid tumors with dihydroeithidium revealed that the mutant T8993G tumors were making much more ROS that the wild-type T8993T nodules (177).
Consistent with the increased ROS production in the T8993G tumors, MnSOD has been found to be reduced in precancerous intraepithelial neoplastic lesions and in prostate cancer by immunohistochemical and in situ hybridization studies (12, 18). Prostate cancer cells also have a 42% increase in cytoplasmic staining for the redox-sensitive APE-1 (90). Thus these observations support the conclusion that increased mitochondrial ROS production is central to prostate cancer.
Therefore, mtDNA mutations appear to play an important causal role in prostate cancer, and thus may have a similar role in the etiology of other solid tumors. Moreover, the role of mitochondrial defects in the pathophysiology of cancer would appear to be the generation of increased ROS, which acts as both an nDNA mutagen and cellular mitogen. Since mitochondrial mutations that increase ROS would do so by impeding the ETC, they would also result in the accumulation within the cell of unoxidized NADH and pyruvate. The excess NADH and pyruvate would then be converted to lactate by lactate dehydrogenase. This high production of lactate by cancer cells in the presence of oxygen was first observed 70 years ago by Otto Warburg and described as aerobic-glycolysis. Warburg hypothesized that mitochondrial defects might underlie many forms of cancer (246). Modern molecular genetic analysis might yet prove him right.
Mitochondrial Medicine: An Integrated Mitochondrial Model for the Etiology of Degenerative Diseases, Aging, and Cancer
All of these observations on the pathophysiology of the common degenerative diseases, cancer, and aging reveal that these diverse clinical entities share many common underlying biochemical, gene regulation, and genetic features. Moreover, these features consistently involve energy metabolism and the mitochondrion. Therefore, we can theorize that all age-related diseases (degenerative diseases, cancer, and aging) share a common underlying mitochondrial pathophysiology which is summarized in Figure 6.
Figure 6.
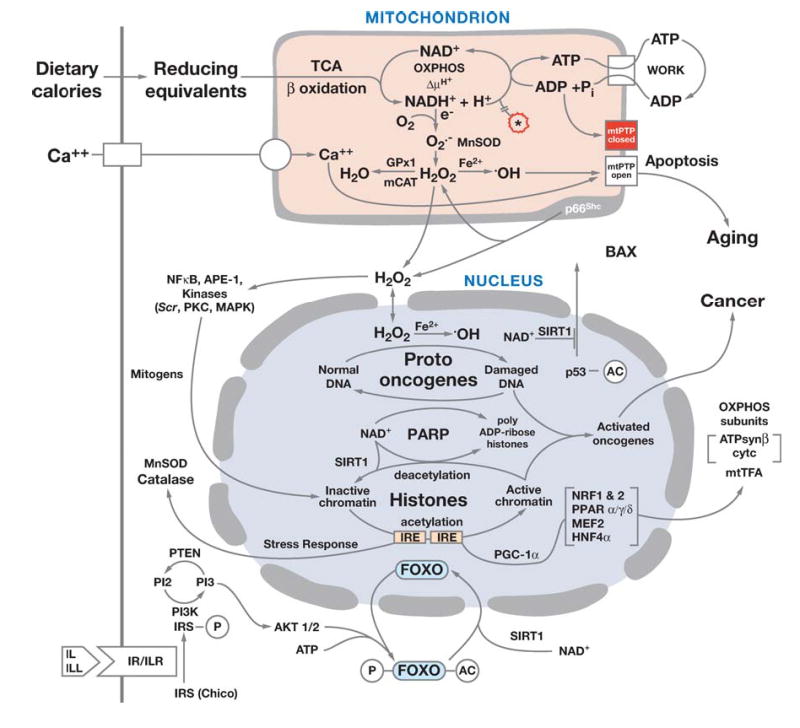
Model for the proposed role for mitochondrial dysfunction in an energy-utilization tissue cell in metabolic and degenerative diseases, aging, and cancer. The mitochondrial pathophysiology of these clinical entities is envisioned to result from the interplay between mitochondrial energy production, ROS generation, and the initiation of apoptosis through activation of the mtPTP. These components of energy metabolism are modulated by environmental constraints such as caloric availability and cold stress through the regulation of the FOXO and PGC-1α transcription factors and the SIRT1 NAD+-dependent deacetylase. The FOXO transcription factors coordinately regulate mitochondrial energy metabolism through PGC-1α as well as the antioxidant and stress response genes necessary to cope with the increased oxidative stress of oxidative metabolism. SIRT1 fine-tunes the interrelationship between energy metabolism and apoptosis through the deacetylation of PGC-1α, p53, and the histone proteins. Caloric overload or inhibition of OXPHOS perturbs the cellular mitochondrial energetic balance resulting in increased ROS. The increased ROS and decreased mitochondrial energy output sensitizes the mtPTP, ultimately driving the cell to apoptotic death. The increased ROS also diffuses into the nucleus as H2O2 where it can mutate and activate proto-oncogenes (initiation) and can interact with NFκB, APE-1 and various kinases to initiate cell division (promotion) leading to neoplastic transformation (cancer). Figure abbreviations are PARP, poly ADP-ribose polymerase; SIRT1, the mammalian homologue to Sir2; FOXO3, the most ubiquitous mammalian forkhead transcription factor; P, a phosphorylated protein; Ac, an acetylated protein; IL, insulin ligand; ILL, insulin-like ligand; IR, insulin receptor; ILR, insulin-like growth factor receptor; IRE, insulin response element; PI3K, the PI3 kinase; PI2, the membrane-bound phosphotidyl-inositol diphosphate; PI3, membrane-bound phosphatidyl-inositol triphosphate, AKT 1/2, the AKT kinases; ATPsynβ, ATP synthase β subunit; cytc, cytochrome c.
Dietary Restriction, Degenerative Diseases, Longevity, and Climatic Adaptation
In the energy-utilizing tissues (brain, heart, muscle, kidney, etc.), when calories (reducing equivalents) are plentiful and ATP is being rapidly hydrolyzed through physical activity, the mitochondrial electrochemical gradient (ΔP) is kept depolarized as the ATP synthase resynthesizes ATP. As a result, electrons flow continuously through the ETC to ½O2 to sustain ΔP, keeping the NADH/NAD+ ratio low, the ETC oxidized and mitochondrial ROS production minimized.
However, if calories are plentiful in tightly coupled mitochondria and ATP utilization is low (sedentary life style), then ΔP becomes and remains maximized. This stalls the ETC, resulting in a high NADH/NAD+ ratio, excess electrons in the ETC, and excessive ROS production. The increased ROS chronically damages the mitochondria, mtDNA, and the mtPTP, ultimately resulting in the activation of the mtPTP and cell death (Figure 6) (242).
High carbohydrate diets stimulate insulin secretion that phosphorylates the FOXOs, removing them from the nucleus. This down-regulates the cellular stress-response pathways including the mitochondrial and cytosolic antioxidant defenses and reduces the transcription of PGC-1α, thus down-regulating mitochondrial OXPHOS. Additionally, the high NADH/NAD+ ratio inhibits SIRT1 leaving the forkhead transcription factors acetylated and out of the nucleus. The suppression of SIRT1 also permits p53 to become acetylated and fully active, increasing the expression of BAX and the activation of p66Shc. BAX binds to the mitochondrial outer membrane and activates the mtPTP to initiate apoptosis. The activated mitochondrial component of p66shc increases mitochondrial ROS production and stimulates cytochrome c release and apoptosis. This destructive trend is partially countered by the ROS-mediated stimulation of the interaction of SIRT1 with the forkheads, enhancing the deacetylation of forkheads and their import into the nucleus to activate the antioxidant and stress response systems. However, continued caloric overload counters this protective effect. In the end, high calorie diets lead to the down-regulation of mitochondrial OXPHOS and antioxidant and stress response defenses, increasing mitochondrial ROPS production and the accumulation of oxidative damage to the mitochondria and mtDNA, leading to cell death.
As an individual ages, the chronic oxidative stress results in the accumulation of somatic mtDNA mutations, including rearrangements and base substitutions in the regulatory elements of the CR and in the structural genes. Ultimately, the decline of the mitochondrial biosynthetic capacity renders the mitochondria unable to adequately repair the accumulating mitochondrial oxidative damage. This activates the mtPTP, p53, and p66Shc systems, increasing apoptosis rates and resulting in cell loss, tissue dysfunction, and the symptoms of age-related diseases.
These processes are ameliorated in individuals harboring partially uncoupled mitochondria. In loosely coupled mitochondria, inefficient proton pumping and(or excessive proton leakage though the ATP synthase keeps ΔP submaximal, irrespective of the caloric availability. This keeps electrons flowing through the ETC and generating heat, diminishes mitochondrial ROS production, decreases the rate of cellular apoptosis, and thus increases longevity and inhibits the progression of the age-related diseases.
The Mitochondrial Energetics and ROS Biology of Cancer
These same factors also modulate the cellular predilection to neoplastic transformation, cancer. Mutations in nDNA or mtDNA OX-PHOS genes that impede the flow of electrons through the ETC increase mitochondrial ROS production. The resulting H2O2, which is relatively stable, diffuses out of the mitochondrion, through the cytosol, and into the nucleus. In the nucleus, H2O2 can interact with transition metals and be converted to •OH, which mutagenizes the nDNA. The resulting nDNA damage activates the nDNA repair systems, including the polyADP-ribose polymerase (PARP). The activated PARP degrades the nuclear NAD+ in the process of adding poly ADP-ribose chains to histones and other nuclear proteins (Figure 6) (242).
The degradation of the nuclear NAD+, together with the high NADH/NAD+ ratio, inactivates nuclear SIRT1. SIRT1 is a histone deacetylase, and nuclear transcription is repressed by the deacetylation of histones and activated by histone acetylation. Therefore, inhibition of SIRT1 permits histone acetylation to predominate, turning on the transcription of normally inactive genes. In post-mitotic tissues, histone deacetylation permits the silencing of the genes that regulate cell replication and differentiation, the proto-oncogenes.
The diffusion of H2O2 into the nucleus and its conversion to •OH can then mutate the proto-oncogenes, converting them into functional oncogenes. Moreover, increased cytosolic and nuclear H2O2 activates a variety of cellular signal transduction factors including NFκB, APE-1, Fos, Jun, and tyrosine kinases. This drives the cell into replication. Consequently, mutations in mitochondrial genes that inhibit electron flow through the ETC result in chronically increased mitochondrial ROS production, which can act as both a tumor initiator (mutation of proto-oncogenes) and tumor promoter (activation of transcription and replication) (Figure 6) (242).
MITOCHONDRIAL ADAPTATION AND EVOLUTIONARY MEDICINE
The Interplay between Environment and Energetics
These considerations indicate that mitochondria lie at the intersection between environmental factors such as calories and cold and the human capacity to energetically cope with the environmental challenges in different regions of the globe. Our ancient ancestors had to be able to adapt to two classes of environmental changes: (a) short term changes in calories and climate associated with seasonal variation and (b) long term changes in the nature of calories and average annual temperatures defined by the latitude and geographic zone in which they lived.
The need to adapt energetically to seasonal changes is a challenge common to all mammals, including aboriginal human populations. For populations that lived in the colder temperate and arctic zones, people needed to be able to uncouple mitochondrial OXPHOS in both BAT and muscle by induction of the UCPs. However, in industrial societies, most individuals avoid cold stress through central heating. Similarly, most ancient populations had to accumulate plant carbohydrate calories and store them as fat during the plant growing season and then to efficiently use the stored fat calories to sustain their cellular energetics when the plants were dormant. This season variation in caloric availability was metabolically managed via the insulin signaling network that coordinately integrated the energy-utilization, energy-storage, and energy-homeostasis tissues, through the hormonal signals of the energy-sensing tissues, the pancreatic α and β cells. This was accomplished by cueing the energy-sensing tissues to serum glucose concentration that oscillated based on the availability of plant carbohydrate calories. When plant calories were abundant, blood sugar was high, insulin was secreted, mitochondrial OXPHOS and its attendant antioxidant defenses were down-regulated in the energy-utilization tissues, and the excess carbohydrate calories were stored as fat in energy-storage tissues. When plant calories declined at the end of the growing season, blood sugar levels declined, resulting in decreased insulin secretion and increased glucagon secretion. These hormonal changes up-regulated mitochondrial OXPHOS and the attendant antioxidant defenses in the energy-utilization tissues, mobilized the stored fats in the energy-storage tissues for use as mitochondrial fuel, and activated glucose synthesis in the energy-homeostasis liver to sustain minimal blood sugar levels.
This energetic system worked well for out ancestors who lived in an environment of periodic carbohydrate surplus and deficiency. However, in today’s developed societies, technology provides us with unlimited dietary calories including carbohydrates throughout the year. As a result, our energy signaling system remains continuously in the high carbohydrate state. Consequently, mitochondrial OXPHOS and its attendant antioxidant defenses are chronically down-regulated. At the same time, our sedentary life style means that we do not turnover ATP through sustained physical activity. Therefore, the excess of caloric reducing equivalents in our diet leads to fat accumulation and obesity and keeps the mitochondrial electron carriers of our down-regulated ETC fully reduced. This results in chronically increased mitochondrial ROS production, which damages the mitochondria and mtDNAs. The reduced mitochondrial energy output and the increased oxidative stress sensitize the mtPTP to activation, driving post-mitotic cells into premature apoptosis. The resulting loss of post-mitotic cells results in the tissue-specific symptoms associated with aging and the age-related diseases.
While the insulin and cAMP signal trans-duction systems permitted our ancestors to rapidly adapt their metabolism to sudden changes in environmental temperature and carbohydrate calorie availability, these short-term oscillations were insufficient to cope with the more general environmental differences experienced by individuals living in the tropics versus the temperate zone versus the arctic. These different climatic zones required adaptation to differences in average ambient temperature, amount of sunlight, and nature of available calories. In the tropics the predominate source of calories was plant carbohydrates, whereas in the arctic the caloric availability shifted more toward animal protein and fat.
Humans adapted to these more general regional differences through the fixation of functional mutations in their mtDNAs that changed the coupling efficiency of OXPHOS, thus shifting the energetic balance from predominantly ATP production in the tropics to increased heat production in the arctic. Functional mtDNA mutations undoubtedly permitted adaptation to many additional environmental factors as yet undiscovered. What is clear is that the random occurrence of functional mtDNA variants shifted the energetic balance in certain individuals sufficiently that they could move into and survive in more northern environments from which their warm-adapted predecessors were excluded. However, these adaptive mtDNA mutations required trade-offs. In the tropics, tightly coupled mitochondria maximized the efficiency of physical work and minimized heat production. Mutations that partially uncoupled OXPHOS decreased work efficiency, but provided essential heat for surviving chronic cold. The uncoupling mutations also partially relieved the constraints of the inner membrane electrochemical gradient (ΔP) on electron flow through the ETC, permitting the ETC to run continuously. This kept the electron carriers oxidized and minimized ROS production. Hence, in industrialized societies where calories are unlimited and exercise is minimized, individuals with uncoupled mitochondria (e.g., European hap-logroup J) are partially protected from the increased oxidative stress associated with excessive caloric intake. Therefore, these individuals are on average less prone to degenerative diseases and have increased longevity. Still, these individuals require more calories to sustain their tissue energetics. Hence, if calories become limiting again, they will be the first to starve.
CONCLUSION
Evolutionary medicine posits that the genetic variants that permitted our ancestors to adapt to diverse environments (niches) are having a profound influence on our predisposition to common diseases today. The information provided in this review now places this concept on a solid experimental foundation and shows that the major environmental challenges that confronted our hunter-gather ancestors were bioenergetics for which the mitochondria adaptations were pivotal. Thus, the adaptive mtDNA mutations of our ancestors, when combined with the environmental control and unlimited calories of our modern environment, are increasingly resulting in individual energetic imbalances. It is these imbalances between ancient mitochondrial genetics and modern caloric intake that, I believe, are driving the modern epidemics of obesity, diabetes, neurodegenerative disease, cardiovascular disease, and cancer.
Acknowledgments
The author would like to thank Ms M.T. Lott for her assistance with this manuscript. This work has been supported by NIH grants NS21328, AG13154, NS41650, AG24373, TW01366, HL64017, and an Ellison Senior Scholar Award.
References
- 1.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117:421–26. doi: 10.1016/s0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 2.Adams JU. Autophagy and longevity. How keeping house may keep one young. Scientist. 2005;19:22–24. [Google Scholar]
- 3.Adams V, Griffin L, Towbin J, Gelb B, Worley K, McCabe ER. Porin interaction with hexokinase and glycerol kinase: metabolic microcompartmentation at the outer mitochondrial membrane. Biochem Med Metab Biol. 1991;45:271–91. doi: 10.1016/0885-4505(91)90032-g. [DOI] [PubMed] [Google Scholar]
- 4.Alonso A, Martin P, Albarran C, Aquilera B, Garcia O, et al. Detection of somatic mutations in the mitochondrial DNA control region of colorectal and gastric tumors by heteroduplex and single-strand conformation analysis. Electrophoresis. 1997;18:682–85. doi: 10.1002/elps.1150180504. [DOI] [PubMed] [Google Scholar]
- 5.Altshuler D, Hirschhorn JN, Klannemark M, Lindgren CM, Vohl MC, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 6.Alzheimer’s Association. 2005. Alzheimer’s Disease Statistics http://www.alz.org/Resources/FactSheets/FSAlzheimerStats.pdf
- 7.Anandatheerthavarada HK, Biswas G, Robin MA, Avadhani NG. Mitochondrial targeting and a novel transmembrane arrest of Alzheimer’s amyloid precursor protein impairs mitochondrial function in neuronal cells. J Biol Chem. 2003;161:41–54. doi: 10.1083/jcb.200207030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreu AL, Bruno C, Dunne TC, Tanji K, Shanske S, et al. A nonsense mutation (G15059A) in the cytochrome b gene in a patient with exercise intolerance and myoglobinuria. Ann Neurol. 1999;45:127–30. doi: 10.1002/1531-8249(199901)45:1<127::aid-art20>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 9.Ankarcrona M, Hultenby K. Presenilin-1 is located in rat mitochondria. Biochem Biophys Res Commun. 2002;295:766–70. doi: 10.1016/s0006-291x(02)00735-0. [DOI] [PubMed] [Google Scholar]
- 10.Arbiser JL. Molecular regulation of angiogenesis and tumorigenesis by signal transduction pathways: evidence of predictable and reproducible patterns of synergy in diverse neoplasms. Semin Cancer Biol. 2004;14:81–91. doi: 10.1016/j.semcancer.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 11.Astuti D, Latif F, Dallol A, Dahia PL, Douglas F, et al. Gene mutations in the succinate dehydrogenase subunit SDHB cause susceptibility to familial pheochromocytoma and to familial paraganglioma. Am J Hum Genet. 2001;69:49–54. doi: 10.1086/321282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker AM, Oberley LW, Cohen MB. Expression of antioxidant enzymes in human prostatic adenocarcinoma. Prostate. 1997;32:229–33. doi: 10.1002/(sici)1097-0045(19970901)32:4<229::aid-pros1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 13.Ballinger SW, Shoffner JM, Gebhart S, Koontz DA, Wallace DC. Mitochondrial diabetes revisited. Nat Genet. 1994;7:458–59. doi: 10.1038/ng0894-458. [DOI] [PubMed] [Google Scholar]
- 14.Ballinger SW, Shoffner JM, Hedaya EV, Trounce I, Polak MA, et al. Maternally transmitted diabetes and deafness associated with a 10.4 kb mitochondrial DNA deletion. Nat Genet. 1992;1:11–15. doi: 10.1038/ng0492-11. [DOI] [PubMed] [Google Scholar]
- 15.Baysal BE, Ferrell RE, Willett-Brozick JE, Lawrence EC, Myssiorek D, et al. Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science. 2000;287:848–51. doi: 10.1126/science.287.5454.848. [DOI] [PubMed] [Google Scholar]
- 16.Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–74. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- 17.Bosetti F, Brizzi F, Barogi S, Mancuso M, Siciliano G, et al. Cytochrome c oxidase and mitochondrial F1F0-ATPase (ATP synthase) activities in platelets and brain from patients with Alzheimer’s disease. Neurobiol Aging. 2002;23:371–76. doi: 10.1016/s0197-4580(01)00314-1. [DOI] [PubMed] [Google Scholar]
- 18.Bostwick DG, Alexander EE, Singh R, Shan A, Qian J, et al. Antioxidant enzyme expression and reactive oxygen species damage in prostatic intraepithelial neoplasia and cancer. Cancer. 2000;89:123–34. [PubMed] [Google Scholar]
- 19.Bourgeron T, Rustin P, Chretien D, Birch-Machin M, Bourgeois M, et al. Mutation of a nuclear succinate dehydrogenase gene results in mitochondrial respiratory chain deficiency. Nat Genet. 1995;11:144–49. doi: 10.1038/ng1095-144. [DOI] [PubMed] [Google Scholar]
- 20.Brown MD, Starikovskaya YB, Derbeneva O, Hosseini S, Allen JC, et al. The role of mtDNA background in disease expression: a new primary LHON mutation associated with Western Eurasian haplogroup J. Hum. Genet. 2002;110:130–38. doi: 10.1007/s00439-001-0660-8. [DOI] [PubMed] [Google Scholar]
- 21.Brown MD, Sun F, Wallace DC. Clustering of Caucasian Leber hereditary optic neuropathy patients containing the 11778 or 14484 mutations on an mtDNA lineage. Am J Hum Genet. 1997;60:381–87. [PMC free article] [PubMed] [Google Scholar]
- 22.Brown MD, Trounce IA, Jun AS, Allen JC, Wallace DC. Functional analysis of lymphoblast and cybrid mitochondria containing the 3460, 11778, or 14484 Leber’s Hereditary Optic Neuropathy mtDNA mutation. J Biol Chem. 2000;275:39831–36. doi: 10.1074/jbc.M006476200. [DOI] [PubMed] [Google Scholar]
- 23.Brown MD, Zhadanov S, Allen JC, Hosseini S, Newman NJ, et al. Novel mtDNA mutations and oxidative phosphorylation dysfunction in Russion LHON families. Hum Genet. 2001;109:33–39. doi: 10.1007/s004390100538. [DOI] [PubMed] [Google Scholar]
- 24.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–15. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 25.Bruno C, Martinuzzi A, Tang Y, Andreu AL, Pallotti F, et al. A stop-codon mutation in the human mtDNA cytochrome c oxidase I gene disrupts the functional structure of complex IV. Am J Hum Genet. 1999;65:611–20. doi: 10.1086/302546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunn CL, Wallace DC, Eisenstadt JM. Cytoplasmic inheritance of chlormaphenicol resistance in mouse tissue culture cells. Proc Natl Acad Sci USA. 1974;71:1681–85. doi: 10.1073/pnas.71.5.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Canevari L, Clark JB, Bates TE. Beta-Amyloid fragment 25–35 selectively decreases complex IV activity in isolated mitochondria. FEBS Lett. 1999;457:131–34. doi: 10.1016/s0014-5793(99)01028-5. [DOI] [PubMed] [Google Scholar]
- 28.Cann RL, Stoneking M, Wilson AC. Mitochondrial DNA and human evolution. Nature. 1987;325:31–36. doi: 10.1038/325031a0. [DOI] [PubMed] [Google Scholar]
- 29.Cardoso SM, Proenca MT, Santos S, Santana I, Oliveira CR. Cytochrome c oxidase is decreased in Alzheimer’s disease platelets. Neurobiol Aging. 2004;25:105–10. doi: 10.1016/s0197-4580(03)00033-2. [DOI] [PubMed] [Google Scholar]
- 30.Castellani R, Hirai K, Aliev G, Drew KL, Nunomura A, et al. Role of mitochondrial dysfunction in Alzheimer’s disease. J Neurosci Res. 2002;70:357–60. doi: 10.1002/jnr.10389. [DOI] [PubMed] [Google Scholar]
- 31.Cent. Dis. Control Prev. 2005. Prostate Cancer: The Public Health Perspective, 2004/2005 Fact Sheet. http://www.cdc.gov/cancer/prostate/prospdf/about2004.pdf
- 32.Chagnon P, Gee M, Filion M, Robitaille Y, Belouchi M, Gauvreau D. Phylogenetic analysis of the mitochondrial genome indicates significant differences between patients with Alzheimer disease and controls in a French-Canadian founder population. Am J Med Genet. 1999;85:20–30. doi: 10.1002/(sici)1096-8628(19990702)85:1<20::aid-ajmg6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 33.Chen JZ, Gokden N, Greene GF, Mukunyadzi P, Kadlubar FF. Extensive somatic mitochondrial mutations in primary prostate cancer using laser capture microdissection. Cancer Res. 2002;62:6470–74. [PubMed] [Google Scholar]
- 34.Chinnery PF, Samuels DC, Elson J, Turnbull DM. Accumulation of mitochon-drial DNA mutations in ageing, cancer, and mitochondrial disease: Is there a common mechanism? Lancet. 2002;360:1323–25. doi: 10.1016/S0140-6736(02)11310-9. [DOI] [PubMed] [Google Scholar]
- 35.Clancy DJ, Gems D, Hafen E, Leevers SJ, Partridge L. Dietary restriction in long-lived dwarf flies. Science. 2002;296:319. doi: 10.1126/science.1069366. [DOI] [PubMed] [Google Scholar]
- 36.Clancy DJ, Gems D, Harshman LG, Oldham S, Stocker H, et al. Extension of life-span by loss of CHICO, a Drosophila insulin receptor substrate protein. Science. 2001;292:104–6. doi: 10.1126/science.1057991. [DOI] [PubMed] [Google Scholar]
- 37.Comi GP, Bordoni A, Salani S, Franceschina L, Sciacco M, et al. Cytochrome c oxidase subunit I microdeletion in a patient with motor neuron disease. Ann Neurol. 1998;43:110–16. doi: 10.1002/ana.410430119. [DOI] [PubMed] [Google Scholar]
- 38.Copeland WC, Wachsman JT, Johnson FM, Penta JS. Mitochondrial DNA alterations in cancer. Cancer Invest. 2002;20:557–69. doi: 10.1081/cnv-120002155. [DOI] [PubMed] [Google Scholar]
- 39.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992;2:324–29. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 40.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, McKee AC, et al. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics. 1994;23:471–76. doi: 10.1006/geno.1994.1525. [DOI] [PubMed] [Google Scholar]
- 41.Corral-Debrinski M, Shoffner JM, Lott MT, Wallace DC. Association of mitochondrial DNA damage with aging and coronary atherosclerotic heart disease. Mutat Res. 1992;275:169–80. doi: 10.1016/0921-8734(92)90021-g. [DOI] [PubMed] [Google Scholar]
- 42.Corral-Debrinski M, Stepien G, Shoffner JM, Lott MT, Kanter K, Wallace DC. Hypoxemia is associated with mitochondrial DNA damage and gene induction. Implications for cardiac disease. JAMA. 1991;266:1812–16. [PubMed] [Google Scholar]
- 43.Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–33. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coskun PE, Ruiz-Pesini EE, Wallace DC. Control region mtDNA variants: longevity, climatic adaptation and a forensic conundrum. Proc Natl Acad Sci USA. 2003;100:2174–76. doi: 10.1073/pnas.0630589100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cottrell DA, Borthwick GM, Johnson MA, Ince PG, Turnbull DM. The role of cytochrome c oxidase deficient hippocampal neurones in Alzheimer’s disease. Neuropathol Appl Neurobiol. 2002;28:390–96. doi: 10.1046/j.1365-2990.2002.00414.x. [DOI] [PubMed] [Google Scholar]
- 46.Crouch PJ, Blake R, Duce JA, Ciccotosto GD, Li QX, et al. Copper-dependent inhibition of human cytochrome c oxidase by a dimeric conformer of amyloidbeta1-42. J Neurosci. 2005;25:672–79. doi: 10.1523/JNEUROSCI.4276-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Czubryt MP, McAnally J, Fishman GI, Olson EN. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha) and mitochondrial function by MEF2 and HDAC5. Proc Natl Acad Sci USA. 2003;100:1711–16. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daitoku H, Yamagata K, Matsuzaki H, Hatta M, Fukamizu A. Regulation of PGC-1 promoter activity by protein kinase B and the forkhead transcription factor FKHR. Diabetes. 2003;52:642–49. doi: 10.2337/diabetes.52.3.642. [DOI] [PubMed] [Google Scholar]
- 49.Das DK, George A, Liu XK, Rao PS. Detection of hydroxyl radical in the mitochondria of ischemic-reperfused myocardium by trapping with salicylate. Biochem Biophys Res Commun. 1989;165:1004–9. doi: 10.1016/0006-291x(89)92702-2. [DOI] [PubMed] [Google Scholar]
- 50.De Benedictis G, Rose G, Carrieri G, De Luca M, Falcone E, et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–36. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- 51.DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–68. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 52.Dumoulin R, Sagnol I, Ferlin T, Bozon D, Stepien G, Mousson B. A novel gly290asp mitochondrial cytochrome b mutation linked to a complex III deficiency in progressive exercise intolerance. Mol Cell Probes. 1996;10:389–91. doi: 10.1006/mcpr.1996.0053. [DOI] [PubMed] [Google Scholar]
- 53.Ek J, Andersen G, Urhammer SA, Gaede PH, Drivsholm T, et al. Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to Type II diabetes mellitus. Diabetologia. 2001;44:2220–26. doi: 10.1007/s001250100032. [DOI] [PubMed] [Google Scholar]
- 54.Esposito LA, Melov S, Panov A, Cottrell BA, Wallace DC. Mitochondrial disease in mouse results in increased oxidative stress. Proc Natl Acad Sci USA. 1999;96:4820–25. doi: 10.1073/pnas.96.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 56.Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev Cell. 2001;1:633–44. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 57.Flanagan SW, Anderson RD, Ross MA, Oberley LW. Overexpression of manganese superoxide dismutase attenuates neuronal death in human cells expressing mutant (G37R) Cu/Zn-superoxide dismutase. J Neurochem. 2002;81:170–77. doi: 10.1046/j.1471-4159.2002.00812.x. [DOI] [PubMed] [Google Scholar]
- 58.Fliss MS, Usadel H, Caballero OL, Wu L, Buta MR, et al. Facile detection of mitochondrial DNA mutations in tumors and bodily fluids. Science. 2000;287:2017–19. doi: 10.1126/science.287.5460.2017. [DOI] [PubMed] [Google Scholar]
- 59.Furuyama T, Kitayama K, Yamashita H, Mori N. Forkhead transcription factor FOXO1 (FKHR)-dependent induction of PDK4 gene expression in skeletal muscle during energy deprivation. Biochem J. 2003;375:365–71. doi: 10.1042/BJ20030022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gelb BD, Adams V, Jones SN, Griffin LD, MacGregor GR, McCabe ER. Targeting of hexokinase 1 to liver and hepatoma mitochondria. Proc Natl Acad Sci USA. 1992;89:202–6. doi: 10.1073/pnas.89.1.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Giannakou ME, Partridge L. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 2004;14:408–12. doi: 10.1016/j.tcb.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 62.Gibson GE, Huang HM. Oxidative processes in the brain and non-neuronal tissues as biomarkers of Alzheimer’s disease. Front Biosci. 2002;7:d1007–d15. doi: 10.2741/A827. [DOI] [PubMed] [Google Scholar]
- 63.Giles RE, Blanc H, Cann HM, Wallace DC. Maternal inheritance of human mitochondrial DNA. Proc Natl Acad Sci USA. 1980;77:6715–19. doi: 10.1073/pnas.77.11.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goto Y, Nonaka I, Horai S. A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature. 1990;348:651–53. doi: 10.1038/348651a0. [DOI] [PubMed] [Google Scholar]
- 65.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–62. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 66.Habano W, Sugai T, Yoshida T, Nakamura S. Mitochondrial gene mutation, but not large-scale deletion, is a feature of colorectal carcinomas with mitochondrial microsatellite instability. Int J Cancer. 1999;83:625–29. doi: 10.1002/(sici)1097-0215(19991126)83:5<625::aid-ijc10>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 67.Hamilton ML, Van Remmen H, Drake JA, Yang H, Guo ZM, et al. Does oxidative damage to DNA increase with age? Proc Natl Acad Sci USA. 2001;98:10469–74. doi: 10.1073/pnas.171202698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hansson CA, Frykman S, Farmery MR, Tjernberg LO, Nilsberth C, et al. Nicastrin, presenilin, APH-1 and PEN-2 form active gamma-secretase complexes in mitochondria. J Biol Chem. 2004;279:51654–60. doi: 10.1074/jbc.M404500200. [DOI] [PubMed] [Google Scholar]
- 69.Harrison DE, Archer JR. Genetic differences in effects of food restriction on aging in mice. J Nutr. 1987;117:376–82. doi: 10.1093/jn/117.2.376. [DOI] [PubMed] [Google Scholar]
- 70.Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population: prevalence estimates using the 2000 census. Arch Neurol. 2003;60:1119–22. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- 71.Hekimi S, Burgess J, Bussiere F, Meng Y, Benard C. Genetics of lifespan in C. elegans: molecular diversity, physiological complexity, mechanistic simplicity. Trends Genet. 2001;17:712–18. doi: 10.1016/s0168-9525(01)02523-9. [DOI] [PubMed] [Google Scholar]
- 72.Herrmann PC, Gillespie JW, Charboneau L, Bichsel VE, Paweletz CP, et al. Mitochondrial proteome: altered cytochrome c oxidase subunit levels in prostate cancer. Proteomics. 2003;3:1801–10. doi: 10.1002/pmic.200300461. [DOI] [PubMed] [Google Scholar]
- 73.Hogan P, Dall T, Nikolov P. Economic costs of diabetes in the US in 2002. Diabetes Care. 2003;26:917–32. doi: 10.2337/diacare.26.3.917. [DOI] [PubMed] [Google Scholar]
- 74.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature. 1988;331:717–19. doi: 10.1038/331717a0. [DOI] [PubMed] [Google Scholar]
- 75.Holt IJ, Harding AE, Petty RK, Morgan-Hughes JA. A new mitochondrial disease associated with mitochondrial DNA heteroplasmy. Am J Hum Genet. 1990;46:428–33. [PMC free article] [PubMed] [Google Scholar]
- 76.Horton TM, Graham BH, Corral-Debrinski M, Shoffner JM, Kaufman AE, et al. Marked increase in mitochondrial DNA deletion levels in the cerebral cortex of Huntington’s disease patients. Neurology. 1995;45:1879–83. doi: 10.1212/wnl.45.10.1879. [DOI] [PubMed] [Google Scholar]
- 77.Horton TM, Petros JA, Heddi A, Shoffner J, Kaufman AE, et al. Novel mitochondrial DNA deletion found in a renal cell carcinoma. Genes Chromosomes Cancer. 1996;15:95–101. doi: 10.1002/(SICI)1098-2264(199602)15:2<95::AID-GCC3>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 78.Houten SM, Auwerx J. PGC-1alpha: turbocharging mitochondria. Cell. 2004;119:5–7. doi: 10.1016/j.cell.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 79.Hsu LJ, Sagara Y, Arroyo A, Rockenstein E, Sisk A, et al. Alpha-synuclein promotes mitochondrial deficit and oxidative stress. Am J Pathol. 2000;157:401–10. doi: 10.1016/s0002-9440(10)64553-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Huang X, Atwood CS, Hartshorn MA, Multhaup G, Goldstein LE, et al. The A beta peptide of Alzheimer’s disease directly produces hydrogen peroxide through metal ion reduction. Biochemistry. 1999;38:7609–16. doi: 10.1021/bi990438f. [DOI] [PubMed] [Google Scholar]
- 81.Hutchin T, Cortopassi G. A mitochondrial DNA clone is associated with increased risk for Alzheimer disease. Proc Natl Acad Sci USA. 1995;92:6892–95. doi: 10.1073/pnas.92.15.6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 83.Ingman M, Kaessmann H, Paabo S, Gyllensten U. Mitochondrial genome variation and the origin of modern humans. Nature. 2000;408:708–13. doi: 10.1038/35047064. [DOI] [PubMed] [Google Scholar]
- 84.Ishii N, Fujii M, Hartman PS, Tsuda M, Yasuda K, et al. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature. 1998;394:694–97. doi: 10.1038/29331. [DOI] [PubMed] [Google Scholar]
- 85.Jaarsma D, Rognoni F, van Duijn W, Verspaget HW, Haasdijk ED, Holstege JC. CuZn superoxide dismutase (SOD1) accumulates in vacuolated mitochondria in transgenic mice expressing amyotrophic lateral sclerosis-linked SOD1 mutations. Acta Neuropathol. 2001;102:293–305. doi: 10.1007/s004010100399. [DOI] [PubMed] [Google Scholar]
- 86.Johnson MJ, Wallace DC, Ferris SD, Rattazzi MC, Cavalli-Sforza LL. Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. J Mol Evol. 1983;19:255–71. doi: 10.1007/BF02099973. [DOI] [PubMed] [Google Scholar]
- 87.Jonassen T, Larsen PL, Clarke CF. A dietary source of coenzyme Q is essential for growth of long-lived Caenorhabditis elegans clk-1 mutants. Proc Natl Acad Sci USA. 2001;98:421–26. doi: 10.1073/pnas.021337498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jun AS, Brown MD, Wallace DC. A mitochondrial DNA mutation at np 14459 of the ND6 gene associated with maternally inherited Leber’s hereditary optic neuropathy and dystonia. Proc Natl Acad Sci USA. 1994;91:6206–10. doi: 10.1073/pnas.91.13.6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kamei Y, Ohizumi H, Fujitani Y, Nemoto T, Tanaka T, et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc Natl Acad Sci USA. 2003;100:12378–83. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kelley MR, Cheng L, Foster R, Tritt R, Jiang J, et al. Elevated and altered expression of the multifunctional DNA base excision repair and redox enzyme Ape1/ref-1 in prostate cancer. Clin Cancer Res. 2001;7:824–30. [PubMed] [Google Scholar]
- 91.Kelley MR, Parsons SH. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxid Redox Signal. 2001;3:671–83. doi: 10.1089/15230860152543014. [DOI] [PubMed] [Google Scholar]
- 92.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–68. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 93.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. Daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–46. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 94.King MP, Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–3. doi: 10.1126/science.2814477. [DOI] [PubMed] [Google Scholar]
- 95.Kirches E, Michael M, Woy C, Schneider T, Warich-Kirches M, et al. Loss of heteroplasmy in the displacement loop of brain mitochondrial DNA in astrocytic tumors. Genes Chromosomes Cancer. 1999;26:80–83. doi: 10.1002/(sici)1098-2264(199909)26:1<80::aid-gcc11>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 96.Kish SJ, Mastrogiacomo F, Guttman M, Furukawa Y, Taanman JW, et al. Decreased brain protein levels of cytochrome oxidase subunits in Alzheimer’s disease and in hereditary spinocerebellar ataxia disorders: a nonspecific change? J Neurochem. 1999;72:700–7. doi: 10.1046/j.1471-4159.1999.0720700.x. [DOI] [PubMed] [Google Scholar]
- 97.Kokoszka JE, Coskun P, Esposito L, Wallace DC. Increased mitochondrial oxidative stress in the Sod2 (+/−) mouse results in the age-related decline of mitochondrial function culminating in increased apoptosis. Proc Natl Acad Sci USA. 2001;98:2278–83. doi: 10.1073/pnas.051627098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kokoszka JE, Waymire KG, Levy SE, Sligh JE, Cai J, et al. The ADP/ATP translocator is not essential for the mitochondrial permeability transition pore. Nature. 2004;427:461–65. doi: 10.1038/nature02229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kontush A. Amyloid-beta: an antioxidant that becomes a pro-oxidant and critically contributes to Alzheimer’s disease. Free Radic Biol Med. 2001;31:1120–31. doi: 10.1016/s0891-5849(01)00688-8. [DOI] [PubMed] [Google Scholar]
- 100.Kontush A, Atwood CS. Amyloid-beta: phylogenesis of a chameleon. Brain Res Brain Res Rev. 2004;46:118–20. doi: 10.1016/j.brainresrev.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 101.Kontush A, Berndt C, Weber W, Akopyan V, Arlt S, et al. Amyloid-beta is an antioxidant for lipoproteins in cerebrospinal fluid and plasma. Free Radic Biol Med. 2001;30:119–28. doi: 10.1016/s0891-5849(00)00458-5. [DOI] [PubMed] [Google Scholar]
- 102.Koster JC, Marshall BA, Ensor N, Corbett JA, Nichols CG. Targeted overactivity of beta cell K(ATP) channels induces profound neonatal diabetes. Cell. 2000;100:645–54. doi: 10.1016/s0092-8674(00)80701-1. [DOI] [PubMed] [Google Scholar]
- 103.Koutnikova H, Campuzano V, Foury F, Dolle P, Cazzalini O, Koenig M. Studies of human, mouse and yeast homologues indicate a mitochondrial function for frataxin. Nat Genet. 1997;16:345–51. doi: 10.1038/ng0897-345. [DOI] [PubMed] [Google Scholar]
- 104.Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–96. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee CH, Olson P, Evans RM. Minireview: lipid metabolism, metabolic diseases, and peroxisome proliferator-activated receptors. Endocrinology. 2003;144:2201–7. doi: 10.1210/en.2003-0288. [DOI] [PubMed] [Google Scholar]
- 106.Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–93. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 107.Lee CK, Weindruch R, Prolla TA. Gene-expression profile of the ageing brain in mice. Nat Genet. 2000;25:294–97. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 108.Lee HJ, Shin SY, Choi C, Lee YH, Lee SJ. Formation and removal of alpha-synuclein aggregates in cells exposed to mitochondrial inhibitors. J Biol Chem. 2002;277:5411–17. doi: 10.1074/jbc.M105326200. [DOI] [PubMed] [Google Scholar]
- 109.Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–4. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- 110.Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat Genet. 2003;33:40–48. doi: 10.1038/ng1056. [DOI] [PubMed] [Google Scholar]
- 111.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–56. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lehtonen R, Kiuru M, Vanharanta S, Sjoberg J, Aaltonen LM, et al. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am J Pathol. 2004;164:17–22. doi: 10.1016/S0002-9440(10)63091-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Leissring MA, Farris W, Wu X, Christodoulou DC, Haigis MC, et al. Alternative translation initiation generates a novel isoform of insulin-degrading enzyme targeted to mitochondria. Biochem J. 2004;383:439–46. doi: 10.1042/BJ20041081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lemieux J, Lakowski B, Webb A, Meng Y, Ubach A, et al. Regulation of physiological rates in Caenorhabditis elegans by a tRNA-modifying enzyme in the mitochondria. Genetics. 2001;159:147–57. doi: 10.1093/genetics/159.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li Y, Huang TT, Carlson EJ, Melov S, Ursell PC, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11:376–81. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 116.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–76. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 117.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 118.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, et al. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–35. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 119.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lin SJ, Guarente L. Nicotinamide adenine dinucleotide, a metabolic regulator of transcription, longevity and disease. Curr Opin Cell Biol. 2003;15:241–46. doi: 10.1016/s0955-0674(03)00006-1. [DOI] [PubMed] [Google Scholar]
- 121.Louet JF, Hayhurst G, Gonzalez FJ, Girard J, Decaux JF. The coactivator PGC-1 is involved in the regulation of the liver carnitine palmitoyltransferase I gene expression by cAMP in combination with HNF4 alpha and cAMP-response element-binding protein (CREB) J Biol Chem. 2002;277:37991–8000. doi: 10.1074/jbc.M205087200. [DOI] [PubMed] [Google Scholar]
- 122.Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12:1566–73. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 123.Luo J, Nikolaev AY, Imai S, Chen D, Su F, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–48. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 124.Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science. 2004;304:448–52. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 125.Malaisse-Lagae F, Malaisse WJ. Hexose metabolism in pancreatic islets: regulation of mitochondrial hexokinase binding. Biochem Med Metab Biol. 1988;39:80–89. doi: 10.1016/0885-4505(88)90061-8. [DOI] [PubMed] [Google Scholar]
- 126.Masoro EJ. Dietary restriction and aging. J Am Geriatr Soc. 1993;41:994–99. doi: 10.1111/j.1532-5415.1993.tb06767.x. [DOI] [PubMed] [Google Scholar]
- 127.Masoro EJ, McCarter RJ, Katz MS, McMahan CA. Dietary restriction alters characteristics of glucose fuel use. J Gerontol. 1992;47:B202–8. doi: 10.1093/geronj/47.6.b202. Erratum. 1993. J Gerontol. 48(2):B73. [DOI] [PubMed] [Google Scholar]
- 128.Mattiazzi M, Vijayvergiya C, Gajewski CD, DeVivo DC, Lenaz G, et al. The mtDNA T8993G (NARP) mutation results in an impairment of oxidative phosphorylation that can be improved by antioxidants. Hum Mol Genet. 2004;13:869–79. doi: 10.1093/hmg/ddh103. [DOI] [PubMed] [Google Scholar]
- 129.Maximo V, Soares P, Lima J, Cameselle-Teijeiro J, Sobrinho-Simoes M. Mitochondrial DNA somatic mutations (point mutations and large deletions) and mitochondrial DNA variants in human thyroid pathology: a study with emphasis on Hurthle cell tumors. Am J Pathol. 2002;160:1857–65. doi: 10.1016/S0002-9440(10)61132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.McCarter RJ, Palmer J. Energy metabolism and aging: a lifelong study of Fischer 344 rats. Am J Physiol. 1992;263:E448–52. doi: 10.1152/ajpendo.1992.263.3.E448. [DOI] [PubMed] [Google Scholar]
- 131.McCord JM. The evolution of free radicals and oxidative stress. Am J Med Genet. 2000;108:652–59. doi: 10.1016/s0002-9343(00)00412-5. [DOI] [PubMed] [Google Scholar]
- 132.McDavid K, Lee J, Fulton J, Tonita J, Thompson T. Prostate cancer incidence and mortality rates and trends in the United States and Canada. Public Health Rep. 2004;119:174–86. doi: 10.1177/003335490411900211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.McMahon FJ, Chen YS, Patel S, Kokoszka J, Brown MD, et al. Mitochondrial DNA sequence diversity in bipolar affective disorder. Am J Psychiatr. 2000;157:1058–64. doi: 10.1176/appi.ajp.157.7.1058. [DOI] [PubMed] [Google Scholar]
- 134.Mecocci P, MacGarvey U, Beal MF. Oxidative damage to mitochondrial DNA is increased in Alzheimer’s disease. Ann Neurol. 1994;36:747–51. doi: 10.1002/ana.410360510. [DOI] [PubMed] [Google Scholar]
- 135.Mecocci P, MacGarvey U, Kaufman AE, Koontz D, Shoffner JM, et al. Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann Neurol. 1993;34:609–16. doi: 10.1002/ana.410340416. [DOI] [PubMed] [Google Scholar]
- 136.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 137.Melov S, Coskun P, Patel M, Tunistra R, Cottrell B, et al. Mitochondrial disease in superoxide dismutase 2 mutant mice. Proc Natl Acad Sci USA. 1999;96:846–51. doi: 10.1073/pnas.96.3.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Melov S, Doctrow SR, Schneider JA, Haberson J, Patel M, et al. Lifespan extension and rescue of spongiform encephalopathy in superoxide dismutase 2 nullizygous mice treated with superoxide dismutase-catalase mimetics. J Neurosci. 2001;21:8348–53. doi: 10.1523/JNEUROSCI.21-21-08348.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Melov S, Hinerfeld D, Esposito L, Wallace DC. Multiorgan characterization of mitochondrial genomic rearrangements in ad libitum and caloric restricted mice show striking somatic mitochondrial DNA rearrangements with age. Nucleic Acids Res. 1997;25:974–82. doi: 10.1093/nar/25.5.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Melov S, Ravenscroft J, Malik S, Gill MS, Walker DW, et al. Extension of life-span with superoxide dismutase/catalase mimetics. Science. 2000;289:1567–69. doi: 10.1126/science.289.5484.1567. [DOI] [PubMed] [Google Scholar]
- 141.Melov S, Schneider JA, Coskun PE, Bennett DA, Wallace DC. Mitochondrial DNA rearrangements in aging human brain and in situ PCR of mtDNA. Neurobiol Aging. 1999;20:565–71. doi: 10.1016/s0197-4580(99)00092-5. [DOI] [PubMed] [Google Scholar]
- 142.Melov S, Schneider JA, Day BJ, Hinerfeld D, Coskun P, et al. A novel neurological phenotype in mice lacking mitochondrial manganese superoxide dismutase. Nat Genet. 1998;18:159–63. doi: 10.1038/ng0298-159. [DOI] [PubMed] [Google Scholar]
- 143.Melov S, Shoffner JM, Kaufman A, Wallace DC. Marked increase in the number and variety of mitochondrial DNA rearrangements in aging human skeletal muscle. Nucleic Acids Res. 1995;23:4122–26. doi: 10.1093/nar/23.20.4122. Erratum. 1995. Nucleic Acids Res 23(23):4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Merriwether DA, Clark AG, Ballinger SW, Schurr TG, Soodyall H, et al. The structure of human mitochondrial DNA variation. J Mol Evol. 1991;33:543–55. doi: 10.1007/BF02102807. [DOI] [PubMed] [Google Scholar]
- 145.Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G. Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science. 1999;286:774–79. doi: 10.1126/science.286.5440.774. [DOI] [PubMed] [Google Scholar]
- 146.Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, et al. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature. 1999;402:309–13. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 147.Mishmar D, Ruiz-Pesini E, Brandon M, Wallace DC. Mitochondrial DNA-like sequences in the nucleus (NUMTs): insights into our African origins and the mechanism of foreign DNA integration. Hum Mutat. 2004;23:125–33. doi: 10.1002/humu.10304. [DOI] [PubMed] [Google Scholar]
- 148.Mishmar D, Ruiz-Pesini EE, Golik P, Macaulay V, Clark AG, et al. Natural selection shaped regional mtDNA variation in humans. Proc Natl Acad Sci USA. 2003;100:171–76. doi: 10.1073/pnas.0136972100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.MITOMAP. 2005. A Human Mitochondrial Genome Database http://www.mitomap.org
- 150.Miyadera H, Amino H, Hiraishi A, Taka H, Murayama K, et al. Altered quinone biosynthesis in the long-lived clk-1 mutants of Caenorhabditis elegans. J Biol Chem. 2001;276:7713–16. doi: 10.1074/jbc.C000889200. [DOI] [PubMed] [Google Scholar]
- 151.Miyata M, Smith JD. Apolipoprotein E allele-specific antioxidant activity and effects on cytotoxicity by oxidative insults and beta-amyloid peptides. Nat Genet. 1996;14:55–61. doi: 10.1038/ng0996-55. [DOI] [PubMed] [Google Scholar]
- 152.Mootha VK, Handschin C, Arlow D, Xie X, St Pierre J, et al. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc Natl Acad Sci USA. 2004;101:6570–75. doi: 10.1073/pnas.0401401101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately down-regulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 154.Moraes CT, DiMauro S, Zeviani M, Lombes A, Shanske S, et al. Mitochondrial DNA deletions in progressive external ophthalmoplegia and Kearns-Sayre syndrome. N Engl J Med. 1989;320:1293–99. doi: 10.1056/NEJM198905183202001. [DOI] [PubMed] [Google Scholar]
- 155.Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–63. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 156.Muller YL, Bogardus C, Pedersen O, Baier L. A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes. 2003;52:895–98. doi: 10.2337/diabetes.52.3.895. [DOI] [PubMed] [Google Scholar]
- 157.Murdock DG, Christacos NC, Wallace DC. The age-related accumulation of a mitochondrial DNA control region mutation in muscle, but not brain, detected by a sensitive PNA-directed PCR clamping based method. Nucleic Acids Res. 2000;28:4350–55. doi: 10.1093/nar/28.21.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Mutisya EM, Bowling AC, Beal MF. Cortical cytochrome oxidase activity is reduced in Alzheimer’s disease. J Neurochem. 1994;63:2179–84. doi: 10.1046/j.1471-4159.1994.63062179.x. [DOI] [PubMed] [Google Scholar]
- 159.Nagy Z, Esiri MM, LeGris M, Matthews PM. Mitochondrial enzyme expression in the hippocampus in relation to Alzheimer-type pathology. Acta Neuropathol. 1999;97:346–54. doi: 10.1007/s004010050997. [DOI] [PubMed] [Google Scholar]
- 160.Natl. Parkinson Found. 2004. About Parkinson Disease http://www.parkinson.org/site/pp.asp?c=9dJFJLPwB&b=71125
- 161.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J Biol Chem. 2005;280:16456–60. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 162.Niemann S, Muller U. Mutations in SDHC cause autosomal dominant paragan-glioma, type 3. Nat Genet. 2000;26:268–70. doi: 10.1038/81551. [DOI] [PubMed] [Google Scholar]
- 163.Niemi AK, Hervonen A, Hurme M, Karhunen PJ, Jylha M, Majamaa K. Mitochondrial DNA polymorphisms associated with longevity in a Finnish population. Hum Genet. 2003;112:29–33. doi: 10.1007/s00439-002-0843-y. [DOI] [PubMed] [Google Scholar]
- 164.Obeso JA, Olanow CW, Nutt JG. Levodopa motor complications in Parkinson’s disease. Trends Neurosci. 2000;23:S2–7. doi: 10.1016/s1471-1931(00)00031-8. [DOI] [PubMed] [Google Scholar]
- 165.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–99. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 166.Okado-Matsumoto A, Fridovich I. Subcellular distribution of superoxide dismutases (SOD) in rat liver: Cu,Zn-SOD in mitochondria. J Biol Chem. 2001;276:38388–93. doi: 10.1074/jbc.M105395200. [DOI] [PubMed] [Google Scholar]
- 167.Okado-Matsumoto A, Fridovich I. Amyotrophic lateral sclerosis: a proposed mechanism. Proc Natl Acad Sci USA. 2002;99:9010–14. doi: 10.1073/pnas.132260399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Orr WC, Sohal RS. Extension of life-span by overexpression of superoxide dismutase and catalase in Drosophila melanogaster. Science. 1994;263:1128–30. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- 169.Orsini F, Migliaccio E, Moroni M, Contursi C, Raker VA, et al. The life span determinant p66Shc localizes to mitochondria where it associates with mitochondrial heat shock protein 70 and regulates trans-membrane potential. J Biol Chem. 2004;279:25689–95. doi: 10.1074/jbc.M401844200. [DOI] [PubMed] [Google Scholar]
- 170.Orth M, Tabrizi SJ, Schapira AH, Cooper JM. Alpha-synuclein expression in HEK293 cells enhances the mitochondrial sensitivity to rotenone. Neurosci Lett. 2003;351:29–32. doi: 10.1016/s0304-3940(03)00941-8. [DOI] [PubMed] [Google Scholar]
- 171.Palacino JJ, Sagi D, Goldberg MS, Krauss S, Motz C, et al. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J Biol Chem. 2004;279:18614–22. doi: 10.1074/jbc.M401135200. [DOI] [PubMed] [Google Scholar]
- 172.Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, et al. Early mitochondrial calcium defects in Huntington’s disease are a direct effect of polyglutamines. Nat Neurosci. 2002;5:731–36. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]
- 173.Parkes TL, Elia AJ, Dickinson D, Hilliker AJ, Phillips JP, Boulianne GL. Extension of Drosophila lifespan by overexpression of human SOD1 in motorneurons. Nat Genet. 1998;19:171–74. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 174.Patrick L. Nonalcoholic fatty liver disease: relationship to insulin sensitivity and oxidative stress. Treatment approaches using vitamin E, magnesium, and betaine. Alt Med Rev. 2002;7:276–91. [PubMed] [Google Scholar]
- 175.Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc Natl Acad Sci USA. 2003;100:8466–71. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004;350:664–71. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci USA. 2005;102:719–24. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Picard F, Guarente L. Molecular links between aging and adipose tissue. Int J Obes Relat Metab Disord. 2005;29(Suppl 1):S36–39. doi: 10.1038/sj.ijo.0802912. [DOI] [PubMed] [Google Scholar]
- 179.Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–76. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, et al. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 2002;12:712–23. doi: 10.1016/s0960-9822(02)00808-4. [DOI] [PubMed] [Google Scholar]
- 181.Polyak K, Li Y, Zhu H, Lengauer C, Willson JK, et al. Somatic mutations of the mitochondrial genome in human colorectal tumours. Nat Genet. 1998;20:291–93. doi: 10.1038/3108. [DOI] [PubMed] [Google Scholar]
- 182.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276:2045–47. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 183.Pratico D. Alzheimer’s disease and oxygen radicals: new insights. Biochem Pharmacol. 2002;63:563–67. doi: 10.1016/s0006-2952(01)00919-4. [DOI] [PubMed] [Google Scholar]
- 184.Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 185.Ramassamy C, Averill D, Beffert U, Theroux L, Lussier-Cacan S, et al. Oxidative insults are associated with apolipoprotein E genotype in Alzheimer’s disease brain. Neurobiol Dis. 2000;7:23–37. doi: 10.1006/nbdi.1999.0273. [DOI] [PubMed] [Google Scholar]
- 186.Ramassamy C, Krzywkowski P, Averill D, Lussier-Cacan S, Theroux L, et al. Impact of apoE deficiency on oxidative insults and antioxidant levels in the brain. Brain Res Mol Brain Res. 2001;86:76–83. doi: 10.1016/s0169-328x(00)00268-0. [DOI] [PubMed] [Google Scholar]
- 187.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–18. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 188.Rogina B, Reenan RA, Nilsen SP, Helfand SL. Extended life-span conferred by cotransporter gene mutations in Drosophila. Science. 2000;290:2137–40. doi: 10.1126/science.290.5499.2137. [DOI] [PubMed] [Google Scholar]
- 189.Rose G, Passarino G, Carrieri G, Altomare K, Greco V, et al. Paradoxes in longevity: sequence analysis of mtDNA haplogroup J in centenarians. Eur J Hum Genet. 2001;9:701–7. doi: 10.1038/sj.ejhg.5200703. [DOI] [PubMed] [Google Scholar]
- 190.Roses AD, Einstein G, Gilbert J, Goedert M, Han SH, et al. Morphological, biochemical, and genetic support for an apolipoprotein E effect on microtubular metabolism. Ann NY Acad Sci. 1996;777:146–57. doi: 10.1111/j.1749-6632.1996.tb34413.x. [DOI] [PubMed] [Google Scholar]
- 191.Ross OA, McCormack R, Curran MD, Duguid RA, Barnett YA, et al. Mitochondrial DNA polymorphism: its role in longevity of the Irish population. Exp Gerontol. 2001;36:1161–78. doi: 10.1016/s0531-5565(01)00094-8. [DOI] [PubMed] [Google Scholar]
- 192.Rotig A, de Lonlay P, Chretien D, Foury F, Koenig M, et al. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat Genet. 1997;17:215–17. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 193.Ruiz-Pesini E, Diez C, Lapena AC, Perez-Martos A, Montoya J, et al. Correlation of sperm motility with mitochondrial enzymatic activities. Clin Chem. 1998;44:1616–20. [PubMed] [Google Scholar]
- 194.Ruiz-Pesini E, Mishmar D, Brandon M, Procaccio V, Wallace DC. Effects of purifying and adaptive selection on regional variation in human mtDNA. Science. 2004;303:223–26. doi: 10.1126/science.1088434. [DOI] [PubMed] [Google Scholar]
- 195.Sawa A, Wiegand GW, Cooper J, Margolis RL, Sharp AH, et al. Increased apoptosis of Huntington disease lymphoblasts associated with repeat length-dependent mitochondrial depolarization. Nat Med. 1999;5:1194–98. doi: 10.1038/13518. [DOI] [PubMed] [Google Scholar]
- 196.Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 197.Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, et al. 2005. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science: publ. online 5 May 2005 (10.1126/science.1106653) [DOI] [PubMed]
- 198.Senoo-Matsuda N, Yasuda K, Tsuda M, Ohkubo T, Yoshimura S, et al. A defect in the cytochrome b large subunit in complex II causes both superoxide anion overproduction and abnormal energy metabolism in Caenorhabditis elegans. J Biol Chem. 2001;276:41553–58. doi: 10.1074/jbc.M104718200. [DOI] [PubMed] [Google Scholar]
- 199.Shoffner JM, Brown MD, Torroni A, Lott MT, Cabell MR, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–84. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 200.Shoffner JM, Lott MT, Lezza AM, Seibel P, Ballinger SW, Wallace DC. Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNALys mutation. Cell. 1990;61:931–37. doi: 10.1016/0092-8674(90)90059-n. [DOI] [PubMed] [Google Scholar]
- 201.Silva JP, Kohler M, Graff C, Oldfors A, Magnuson MA, et al. Impaired insulin secretion and beta-cell loss in tissue-specific knockout mice with mitochondrial diabetes. Nat Genet. 2000;26:336–40. doi: 10.1038/81649. [DOI] [PubMed] [Google Scholar]
- 202.Simon DK, Pulst SM, Sutton JP, Browne SE, Beal MF, Johns DR. Familial multisystem degeneration with parkinsonism associated with the 11778 mitochondrial DNA mutation. Neurology. 1999;53:1787–93. doi: 10.1212/wnl.53.8.1787. [DOI] [PubMed] [Google Scholar]
- 203.Smith MA, Casadesus G, Joseph JA, Perry G. Amyloid-beta and tau serve antioxidant functions in the aging and Alzheimer brain. Free Radic Biol Med. 2002;33:1194–99. doi: 10.1016/s0891-5849(02)01021-3. [DOI] [PubMed] [Google Scholar]
- 204.Sohal RS, Ku HH, Agarwal S, Forster MJ, Lal H. Oxidative damage, mitochondrial oxidant generation and antioxidant defenses during aging and in response to food restriction in the mouse. Mech Ageing Dev. 1994;74:121–33. doi: 10.1016/0047-6374(94)90104-x. [DOI] [PubMed] [Google Scholar]
- 205.Solano A, Roig M, Vives-Bauza C, Hernandez-Pena J, Garcia-Arumi E, et al. Bilateral striatal necrosis associated with a novel mutation in the mitochondrial ND6 gene. Ann Neurol. 2003;54:527–30. doi: 10.1002/ana.10682. [DOI] [PubMed] [Google Scholar]
- 206.Soong NW, Hinton DR, Cortopassi G, Arnheim N. Mosaicism for a specific somatic mitochondrial DNA mutation in adult human brain. Nat Genet. 1992;2:318–23. doi: 10.1038/ng1292-318. [DOI] [PubMed] [Google Scholar]
- 207.Sturtz LA, Diekert K, Jensen LT, Lill R, Culotta VC. A fraction of yeast Cu,Zn-superoxide dismutase and its metallochaperone, CCS, localize to the intermembrane space of mitochondria. A physiological role for SOD1 in guarding against mitochondrial oxidative damage. J Biol Chem. 2001;276:38084–89. doi: 10.1074/jbc.M105296200. [DOI] [PubMed] [Google Scholar]
- 208.Sun J, Folk D, Bradley TJ, Tower J. Induced overexpression of mitochondrial Mn-superoxide dismutase extends the life span of adult Drosophila melanogaster. Genetics. 2002;161:661–72. doi: 10.1093/genetics/161.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Tan DJ, Bai RK, Wong LJ. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res. 2002;62:972–76. [PubMed] [Google Scholar]
- 210.Tanaka M, Gong J, Zhang J, Yamada Y, Borgeld HJ, Yagi K. Mitochondrial genotype associated with longevity and its inhibitory effect on mutagenesis. Mech Ageing Dev. 2000;116:65–76. doi: 10.1016/s0047-6374(00)00149-4. [DOI] [PubMed] [Google Scholar]
- 211.Tanaka M, Gong JS, Zhang J, Yoneda M, Yagi K. Mitochondrial genotype associated with longevity. Lancet. 1998;351:185–86. doi: 10.1016/S0140-6736(05)78211-8. [DOI] [PubMed] [Google Scholar]
- 212.Tandon A, Rogaeva E, Mullan M, St George-Hyslop PH. Molecular genetics of Alzheimer’s disease: the role of beta-amyloid and the presenilins. Curr Opin Neurol. 2000;13:377–84. doi: 10.1097/00019052-200008000-00003. [DOI] [PubMed] [Google Scholar]
- 213.Tatuch Y, Christodoulou J, Feigenbaum A, Clarke JTR, Wherret J, et al. Heteroplasmic mtDNA mutation (T-G) at 8993 can cause Leigh disease when the percentage of abnormal mtDNA is high. Am J Hum Genet. 1992;50:852–58. [PMC free article] [PubMed] [Google Scholar]
- 214.Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–30. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 215.Tissenbaum HA, Guarente L. Model organisms as a guide to mammalian aging. Dev Cell. 2002;2:9–19. doi: 10.1016/s1534-5807(01)00098-3. [DOI] [PubMed] [Google Scholar]
- 216.Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–23. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- 217.Trimmer PA, Keeney PM, Borland MK, Simon FA, Almeida J, et al. Mitochondrial abnormalities in cybrid cell models of sporadic Alzheimer’s disease worsen with passage in culture. Neurobiol Dis. 2004;15:29–39. doi: 10.1016/j.nbd.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 218.Trinei M, Giorgio M, Cicalese A, Barozzi S, Ventura A, et al. A p53-p66Shc signalling pathway controls intracellular redox status, levels of oxidation-damaged DNA and oxidative stress-induced apoptosis. Oncogene. 2002;21:3872–78. doi: 10.1038/sj.onc.1205513. [DOI] [PubMed] [Google Scholar]
- 219.Trounce I, Neill S, Wallace DC. Cytoplasmic transfer of the mtDNA nt 8993 TG (ATP6) point mutation associated with Leigh syndrome into mtDNA-less cells demonstrates cosegregation with a decrease in state III respiration and ADP/O ratio. Proc Natl Acad Sci USA. 1994;91:8334–38. doi: 10.1073/pnas.91.18.8334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Trounce IA, Kim YL, Jun AS, Wallace DC. Assessment of mitochondrial oxidative phosphorylation in patient muscle biopsies, lymphoblasts, and transmitochondrial cell lines. Methods Enzymol. 1996;264:484–509. doi: 10.1016/s0076-6879(96)64044-0. [DOI] [PubMed] [Google Scholar]
- 221.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 222.Valla J, Berndt JD, Gonzalez-Lima F. Energy hypometabolism in posterior cin-gulate cortex of Alzheimer’s patients: superficial laminar cytochrome oxidase associated with disease duration. J Neurosci. 2001;21:4923–30. doi: 10.1523/JNEUROSCI.21-13-04923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.van den Ouweland JM, Lemkes HH, Trembath RC, Ross R, Velho G, et al. Maternally inherited diabetes and deafness is a distinct subtype of diabetes and associates with a single point mutation in the mitochondrial tRNALeu(UUR) gene. Diabetes. 1994;43:746–51. doi: 10.2337/diab.43.6.746. [DOI] [PubMed] [Google Scholar]
- 224.van der Walt JM, Dementieva YA, Martin ER, Scott WK, Nicodemus KK, et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- 225.van der Walt JM, Nicodemus KK, Martin ER, Scott WK, Nance MA, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003;72:804–11. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, et al. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- 227.Vanharanta S, Buchta M, McWhinney SR, Virta SK, Peczkowska M, et al. Early-onset renal cell carcinoma as a novel extraparaganglial component of SDHB-associated heritable paraganglioma. Am J Hum Genet. 2004;74:153–59. doi: 10.1086/381054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Vaughan JR, Davis MB, Wood NW. Genetics of Parkinsonism: a review. Ann Hum Genet. 2001;65:111–26. doi: 10.1017/S0003480001008557. [DOI] [PubMed] [Google Scholar]
- 229.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 230.Vega RB, Huss JM, Kelly DP. The coactivator PGC-1 cooperates with peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000;20:1868–76. doi: 10.1128/mcb.20.5.1868-1876.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Velho G, Froguel P. Genetic, metabolic and clinical characteristics of maturity onset diabetes of the young. Eur J Endocrinol. 1998;138:233–39. doi: 10.1530/eje.0.1380233. [DOI] [PubMed] [Google Scholar]
- 232.Verwer RW, Jansen KA, Sluiter AA, Pool CW, Kamphorst W, Swaab DF. Decreased hippocampal metabolic activity in Alzheimer patients is not reflected in the immunoreactivity of cytochrome oxidase subunits. Exp Neurol. 2000;163:440–51. doi: 10.1006/exnr.2000.7385. [DOI] [PubMed] [Google Scholar]
- 233.Wallace DC. Structure and evolution of organelle genomes. Microbiol Rev. 1982;46:208–40. doi: 10.1128/mr.46.2.208-240.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 235.Wallace DC. Mitochondrial genetics: a paradigm for aging and degenerative diseases? Science. 1992;256:628–32. doi: 10.1126/science.1533953. [DOI] [PubMed] [Google Scholar]
- 236.Wallace DC. 1997. Mitochondrial DNA mutations and bioenergetic defects in aging and degenerative diseases. In The Molecular and Genetic Basis of Neurological Disease, ed. RN Rosenberg, SB Prusiner, S DiMauro, RL Barchi, pp. 237–69. Boston: Butterworth-Heinemann
- 237.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–88. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 238.Wallace DC. Mouse models for mitochondrial disease. Am J Med Genet. 2001;106:71–93. doi: 10.1002/ajmg.1393. [DOI] [PubMed] [Google Scholar]
- 239.Wallace DC, Brown MD, Lott MT. Mitochondrial DNA variation in human evolution and disease. Gene. 1999;238:211–30. doi: 10.1016/s0378-1119(99)00295-4. [DOI] [PubMed] [Google Scholar]
- 240.Wallace DC, Bunn CL, Eisenstadt JM. Cytoplasmic transfer of chloramphenicol resistance in human tissue culture cells. J Cell Biol. 1975;67:174–88. doi: 10.1083/jcb.67.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Wallace DC, Lott MT. 2002. Mitochondrial genes in degenerative diseases, cancer and aging. In Emery and Rimoin’s Principles and Practice of Medical Genetics, ed. DL Rimoin, JM Connor, RE Pyeritz, BR Korf, pp. 299–409. London: Churchill Livingstone
- 242.Wallace DC, Lott MT, Brown MD, Kerstann K. 2001. Mitochondria and neuro-ophthalmological diseases. In The Metabolic and Molecular Basis of Inherited Disease, ed. CR Scriver, AL Beaudet, WS Sly, D Valle, pp. 2425–512. New York: McGraw-Hill
- 243.Wallace DC, Singh G, Lott MT, Hodge JA, Schurr TG, et al. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–30. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- 244.Wallace DC, Zheng X, Lott MT, Shoffner JM, Hodge JA, et al. Familial mitochondrial encephalomyopathy (MERRF): genetic, pathophysiological, and biochemical characterization of a mitochondrial DNA disease. Cell. 1988;55:601–10. doi: 10.1016/0092-8674(88)90218-8. [DOI] [PubMed] [Google Scholar]
- 245.Wang Y, Michikawa Y, Mallidis C, Bai Y, Woodhouse L, et al. Muscle-specific mutations accumulate with aging in critical human mtDNA control sites for replication. Proc Natl Acad Sci USA. 2001;98:4022–27. doi: 10.1073/pnas.061013598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Warburg O. 1931. The Metabolism of Tumors New York: R.R. Smith
- 247.Warita H, Hayashi T, Murakami T, Manabe Y, Abe K. Oxidative damage to mitochondrial DNA in spinal motoneurons of transgenic ALS mice. Brain Res Mol Brain Res. 2001;89:147–52. doi: 10.1016/s0169-328x(01)00029-8. [DOI] [PubMed] [Google Scholar]
- 248.Wilson FH, Hariri A, Farhi A, Zhao H, Petersen KF, et al. A cluster of metabolic defects caused by mutation in a mitochondrial tRNA. Science. 2004;306:1190–94. doi: 10.1126/science.1102521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Wilson RB, Roof DM. Respiratory deficiency due to loss of mitochondrial DNA in yeast lacking the frataxin homologue. Nat Genet. 1997;16:352–57. doi: 10.1038/ng0897-352. [DOI] [PubMed] [Google Scholar]
- 250.Wollheim CB. Beta-cell mitochondria in the regulation of insulin secretion: a new culprit in type II diabetes. Diabetologia. 2000;43:265–77. doi: 10.1007/s001250050044. [DOI] [PubMed] [Google Scholar]
- 251.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–24. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 252.Xu Y, Krishnan A, Wan XS, Majima H, Yeh CC, et al. Mutations in the promoter reveal a cause for the reduced expression of the human manganese superoxide dismutase gene in cancer cells. Oncogene. 1999;18:93–102. doi: 10.1038/sj.onc.1202265. [DOI] [PubMed] [Google Scholar]
- 253.Yeh JJ, Lunetta KL, van Orsouw NJ, Moore FD, Jr, Mutter GL, et al. Somatic mitochondrial DNA (mtDNA) mutations in papillary thyroid carcinomas and differential mtDNA sequence variants in cases with thyroid tumours. Oncogene. 2000;19:2060–66. doi: 10.1038/sj.onc.1203537. [DOI] [PubMed] [Google Scholar]
- 254.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–38. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 255.Yoon JC, Xu G, Deeney JT, Yang SN, Rhee J, et al. Suppression of beta cell energy metabolism and insulin release by PGC-1alpha. Dev Cell. 2003;5:73–83. doi: 10.1016/s1534-5807(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 256.Zhang J, Asin-Cayuela J, Fish J, Michikawa Y, Bonafe M, et al. Strikingly higher frequency in centenarians and twins of mtDNA mutation causing remodeling of replication origin in leukocytes. Proc Natl Acad Sci USA. 2003;100:1116–21. doi: 10.1073/pnas.242719399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Zimmet P, Alberti KG, Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–87. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]