Ubiquitin enzymes in the regulation of immune responses (original) (raw)
Abstract
Ubiquitination plays a central role in the regulation of various biological functions including immune responses. Ubiquitination is induced by a cascade of enzymatic reactions by E1 ubiquitin activating enzyme, E2 ubiquitin conjugating enzyme, and E3 ubiquitin ligase, and reversed by deubiquitinases. Depending on the enzymes, specific linkage types of ubiquitin chains are generated or hydrolyzed. Because different linkage types of ubiquitin chains control the fate of the substrate, understanding the regulatory mechanisms of ubiquitin enzymes is central. In this review, we highlight the most recent knowledge of ubiquitination in the immune signaling cascades including the T cell and B cell signaling cascades as well as the TNF signaling cascade regulated by various ubiquitin enzymes. Furthermore, we highlight the TRIM ubiquitin ligase family as one of the examples of critical E3 ubiquitin ligases in the regulation of immune responses.
Keywords: Ubiquitin, E3 ligase, deubiquitinase, T cell signaling, B cell signaling, TNF signaling, TRIM
The ubiquitin system
Ubiquitin conjugation (ubiquitination) is a type of post-translational modification mediated by an enzymatic reaction cascade (Deshaies and Joazeiro 2009, Komander and Rape 2012). Ubiquitination impacts on the protein stability, activity, and interactome, and fine-tunes the function of proteins (Ikeda et al. 2010, Swatek and Komander 2016), thereby controls many branches of cellular functions (Chen 2005, Hu and Sun 2016). The human genome encodes a remarkably high number of genes for components of the ubiquitin system; two E1 ubiquitin-activating enzymes, around 40 E2 ubiquitin-conjugating enzymes, over 700 E3 ubiquitin ligases, and approximately 100 deubiquitinases (DUBs) (Deshaies and Joazeiro 2009, Michelle et al. 2009, Reyes-Turcu et al. 2009, Schulman and Harper 2009). This makes up around 5% of the human protein-coding genes, which further exemplifies the importance of ubiquitin-based regulation of cellular processes and pathways.
Ubiquitination is a versatile signal
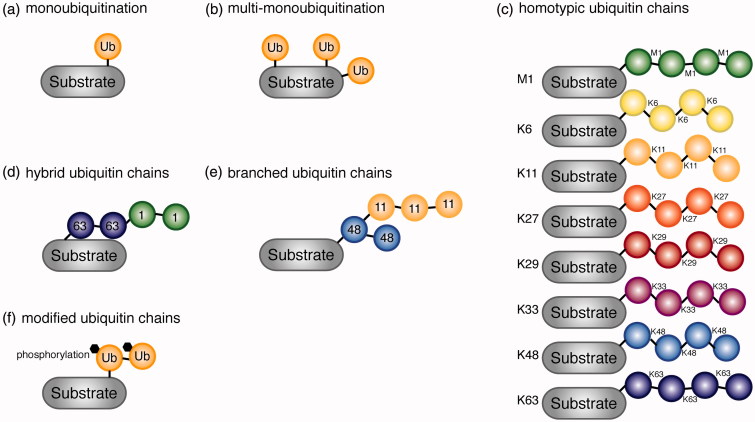
Ubiquitin is a stable, and highly conserved 76-amino acid protein used for post-translational modification of substrates (Vijay-Kumar et al. 1987). During ubiquitin conjugation of the substrate (ubiquitination), the C-terminal glycine (Gly) of ubiquitin is attached typically to a lysine (Lys) residue on the target protein. In rare cases, however, also serine (Ser), threonine (Thr) (Wang et al. 2007, Bhogaraju et al. 2016) and cysteine (Cys) (Schwartzkopff et al. 2015) were detected as target sites. Interestingly, it has been suggested that a free α-NH2 group of the N-terminal residue of the substrates is ubiquitinated in special cases such as Lys-less substrates (Ciechanover and Ben-Saadon 2004). Attachment of one ubiquitin moiety is called monoubiquitination (Figure 1(a)), whereas monoubiquitination that occurs at multiple sites in the same substrate is called multi-monoubiquitination (Figure 1(b)). Ubiquitin can also form homotypic chains by using intrinsic Lys residues (Lys 6, Lys 11, Lys 27, Lys 29, Lys 33, Lys 48, Lys 63), as well as methionine (Met) 1 (Ikeda and Dikic 2008, Iwai and Tokunaga 2009, Swatek and Komander 2016, Yau and Rape 2016) (Figure 1(c)). Among the homotypic ubiquitin chains, Lys 6-, Lys 11- and Lys 48-linked ubiquitin chains adopt a very compact three-dimensional structure (Cook et al. 1992, Varadan et al. 2002, Tenno et al. 2004, Virdee et al. 2010, Bremm et al. 2010). By contrast, Lys 63- and Met 1-linked chains have a very open, extended structure (Tenno et al. 2004, Komander et al. 2009, Weeks et al. 2009). These different three-dimensional conformations of ubiquitin chains that depend on the linkage types result in a variety of different functional outcomes (discussed in more detail in section Recognition of ubiquitin by ubiquitin binding domains (UBDs)). Furthermore, hybrid and branched ubiquitin chains have been identified (Kim et al. 2007, Boname et al. 2010, Meyer and Rape 2014, Grice et al. 2015, Yau and Rape 2016) (Figure 1(d,e)). The physiological relevance of those chain types is not yet fully established, however, an involvement of Lys 63/Met 1-hybrid chains on NF-κB essential modulator (NEMO) and receptor-interacting serine/threonine-protein kinase 1 (RIPK1) in interleukin 1 beta (IL-1β)-dependent activation of the canonical IκB kinase (IKK) complex (Emmerich et al. 2013), as well as on RIPK2 in the nucleotide-binding oligomerization domain-containing protein 2 (NOD2) signaling cascade (Hrdinka et al. 2016) have been demonstrated. Another example is the ubiquitin ligase (Ufd4p), which ubiquitinates Lys 29-linked ubiquitin chains with Lys 48-linked ubiquitin chains to form Lys 29-/Lys 48-hybrid chains, leading to proteasomal degradation of the substrate (Liu et al. 2017).
Figure 1.
Various types of ubiquitin signals are generated based on the linkage type. (a) Attachment of one ubiquitin molecule to a substrate, monoubiquitination. (b) Monoubiquitination on several lysine residues on the same substrate, multi-monoubiquitination. (c) Homotypic ubiquitin chains linked via intrinsic Met 1 and Lys residues (M1, K6, K11, K27, K29, K33, K48, and K63). (d) Hybrid ubiquitin chains consisting of multiple linkage types of chains. K63-M1 hybrid chain is shown. (e) Branched ubiquitin chain consisting of K48 and K11 linkages. (f) Modified (phosphorylated) ubiquitin moiety forming ubiquitin chains on the substrate (see color version of this figure at www.tandfonline.com/ibmg).
More recently, post-transcriptional modifications, phosphorylation, and acetylation of ubiquitin were found by mass spectrometry-based studies (Figure 1(f)) (Ohtake et al. 2015, Ordureau et al. 2015, Swatek and Komander 2016). For example, PTEN-induced putative kinase 1 (PINK1)-dependent phosphorylation at Ser 65 of ubiquitin occurs at mitochondria (Koyano et al. 2014, Ordureau et al. 2015, Yamano et al. 2016), whereas acetylation of ubiquitin at Lys 6 and Lys 48 inhibits ubiquitin chain elongation (Ohtake et al. 2015). Modification of ubiquitin further alters the site where a substrate is ubiquitinated; Ser in the substrate was shown to be a ubiquitination site, only when arginine (Arg) phosphoribosylation of ubiquitin is induced by SdeA, an effector protein of pathogenic Legionella pneumophila (Bhogaraju et al. 2016). Modifications of ubiquitin itself impact on the conventional ubiquitination and deubiquitination cascades, providing an additional layer of regulation by ubiquitination.
Ubiquitin enzymes as key players in ubiquitination
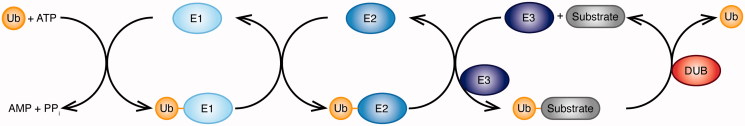
Ubiquitination is a tightly controlled three-step enzymatic process. A ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) act in a concerted manner to form a covalent bond between ubiquitin and its substrate protein, and thereby write the ubiquitin code (Figure 2). The E1 enzyme forms a thioester bond between its active site Cys and the C-terminal Gly of ubiquitin in an ATP-dependent manner. Ubiquitin is then transferred on to the Cys residue in the active site of the E2 enzyme, which cooperates with one of three types of E3 enzymes: Really Interesting New Gene (RING)-type, homologous with E6-associated protein C-terminus (HECT)-type and RING-Between-RING (RBR)-type E3 ligases. RING-type E3 ligases function as scaffolds to position the E2 enzyme and the substrate to promote direct transfer of ubiquitin on to the substrate (Deshaies and Joazeiro 2009, Metzger et al. 2014). HECT-type E3 ligases form an intermediate thioester bond with ubiquitin on their active site Cys and subsequently transfer ubiquitin to the substrate (Metzger et al. 2012, Scheffner and Kumar 2014). Some of the RBR-type E3 ligases, including the human homolog of Ariadne (HHARI), Parkin, and HOIL-1-interacting protein (HOIP) were shown to use a hybrid mechanism of RING- and HECT-type E3 ligases (Wenzel et al. 2011, Stieglitz et al. 2013, Smit and Sixma 2014, Spratt et al. 2014, Dove et al. 2016, Lechtenberg et al. 2016).
Figure 2.
Ubiquitination is a three-step enzymatic process. A ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin ligase (E3) act together to form a covalent bond between ubiquitin and its substrate protein. The E1 enzyme uses ATP to form a thioester bond between its active site cysteine and the C-terminal glycine of ubiquitin. The ubiquitin is then transferred on to the cysteine in the active site of the E2 enzyme, which cooperates with three classes of E3 enzymes to conjugate ubiquitin on the substrate. Deubiquitinases (DUBs) reverse the ubiquitination reaction and hydrolyze ubiquitin from the substrate (see color version of this figure at www.tandfonline.com/ibmg).
The specificity of the substrate is determined by E3 ligases, whereas the ubiquitin linkage type is determined by E2 enzymes as well as E3 ligases (Ye and Rape 2009, Komander and Rape 2012). The E2 enzyme complex Ubiquitin Conjugating Enzyme E2 N (UBE2N)/UBC13-Ubiquitin-conjugating enzyme E2 variant 1 A (UEV1A) generates Lys 63-linked ubiquitin chains and conjugation of the ubiquitin chains on the substrate is carried out by E3 ligases such as Tumor necrosis factor (TNF) receptor associated factor 6 (TRAF6) and the C-terminus of Hsc70-interacting protein (CHIP) (Deng et al. 2000, Zhang et al. 2005). By contrast, the HECT-RING hybrid-type E3 ligase complex called the linear ubiquitin chain assembly complex (LUBAC) specifically synthesizes Met 1-linked (linear) ubiquitin chains (Kirisako et al. 2006, Haas et al. 2009, Tokunaga et al. 2009, Ikeda et al. 2011) with various E2 enzymes including UBE2K, UBCH5A, UBCH5B, UBCH5C, and UBCH7, which typically work with HECT-type E3 ligases (Kirisako et al. 2006).
It has been recently shown that ubiquitination can occur without a classical enzymatic cascade. The SidE effector family of the pathogen Legionella pneumophila is capable of ubiquitinating multiple Rab small GTPases associated with the endoplasmic reticulum without an E1 or E2 enzyme (Qiu et al. 2016). This ATP-independent activation of ubiquitin is mediated through the formation of ADP-ribosylated ubiquitin by SdeA, which was later found to catalyze phosphoribosylation of ubiquitin on a specific Arg via an ADP-ribose-ubiquitin intermediate (Bhogaraju et al. 2016) (as discussed in section Ubiquitination is a versatile signal).
DUBs are proteases that reverse the reaction and erase the ubiquitin code by specifically cleaving ubiquitin from ubiquitin conjugates and substrates. Hydrolyzed monoubiquitins are recycled to replenish the free ubiquitin pool in cells (Wiborg et al. 1985, Wilkinson et al. 1995, Reyes-Turcu et al. 2009). DUBs also have a critical function to produce active ubiquitin monomers from the gene products of four genes (UBB, UBC, RPS27A, and UBA52) encoding ubiquitin, which are either a linear fusion protein of ubiquitin (for UBB and UBC), or a fusion protein with a ribosomal subunit (for RPS27A and UBA52) (Grou et al. 2015, Asaoka and Ikeda 2015). DUBs are specific either for their substrate or for the linkage type of ubiquitin chains (Komander and Rape 2012). Thus far, DUBs specific to each of the eight homotypic ubiquitin chains have been identified and these linkage-specific DUBs are applied to examine the linkage type of ubiquitin chains (Hospenthal et al. 2015).
Recognition of ubiquitin by ubiquitin binding domains (UBDs)
Ubiquitin tags are versatile three-dimensional signals, which are recognized by proteins harboring UBDs that bind to monoubiquitin or ubiquitin chains in a non-covalent manner. Up to now, 20 different families of UBDs have been identified with at least five different structural folds (Hicke et al. 2005, Dikic et al. 2009, Husnjak and Dikic 2012). Most UBDs interact with a monoubiquitin via a conserved hydrophobic patch surrounding isoleucine (Ile) 44. Even though most UBDs target the same surface of ubiquitin, the amino acids surrounding the hydrophobic patch differ, which highlights the requirement of various UBDs. Typically, the interaction between a single UBD and a single ubiquitin moiety does not form a stable complex, however, the binding strength can be further amplified by positioning multiple UBDs within the same protein, or by oligomerization of the UBD-containing protein (Dikic et al. 2009, Ikeda et al. 2010, Lopitz-Otsoa et al. 2010). In addition to recognizing monoubiquitin, UBDs display selectivity for different ubiquitin chains (Komander and Rape 2012). The Ubiquitin binding in ABIN and NEMO (UBAN) domain in NEMO is one example of a UBD, which has a high selectivity to linear ubiquitin chains (Rahighi et al. 2009). The varying distance between ubiquitin molecules in different chain types can further be exploited for specific recognition. Such an example of UBD includes the Ubiquitin interacting motifs (UIMs) in Rap80, which are separated by a 7-amino acid helix allowing for recognition of Lys 63-linked ubiquitin chains due to their extended structure (Sims and Cohen 2009). The challenge in the research field is to understand how the modified ubiquitin (phosphorylation or acetylation), or branched chains, which are able to provide unique protein surfaces interact with known and unknown UBDs.
The functional readouts which are regulated by the UBDs include the canonical Lys 48-linked ubiquitin chain-dependent proteasomal degradation of substrates (Chau et al. 1989). The Lys 48-linked ubiquitin tag serves as a recognition signal for ubiquitin receptors located at the regulatory particle of the proteasome. Two subunits of the regulatory particle, 26S proteasome regulatory subunit RPN10 (RPN10) and RPN13, as well as three proteasome-associated proteins, UV excision repair protein RAD23 (RAD23), Ubiquitin domain-containing protein DSK2 (DSK2) and DNA Damage Inducible 1 Homolog 1 (DDI1) are responsible for recognition of Lys 48-linked ubiquitin chains (Finley 2009). In addition to canonical Lys 48-linked ubiquitin chains, also atypical ubiquitin chains have been shown to contribute to proteasomal degradation. It was already suggested by Johnson et al. in 1995 that Lys 29-linked ubiquitin chains are involved in proteasomal degradation of the substrate (Johnson et al. 1995). Another example is the anaphase-promoting complex dependent upregulation of mitotic Lys 11-linked ubiquitin chains, which leads to proteasomal degradation of the substrate, and which was later rectified to be branched Lys 11-linked ubiquitin chains (Matsumoto et al. 2010, Meyer and Rape 2014).
The proteolytic nature of the ubiquitin signal is not just restricted to proteasomal degradation. Ubiquitinated substrate can be also specifically targeted for autophagy-dependent lysosomal degradation, mediated by UBD-containing autophagy receptors. Since autophagy receptors harbor both an LC3-interacting region (LIR) motif for the autophagosomal LC3 protein family and a UBD, a ubiquitinated substrate is targeted for auto-lysosomal degradation (van Wijk et al. 2012, Birgisdottir et al. 2013). Two of the best understood examples are p62/SQSTM1 (Bjorkoy et al. 2005, Pankiv et al. 2007) and Optineurin (Wild et al. 2011), which harbor UBA and UBAN domains, respectively.
In addition to proteolysis, UBDs are involved in various signaling cascades including the immune signaling pathways, which will be discussed in the following sections: Ubiquitination in various immune cell signaling cascades and TRIM E3 ligases in immune signaling.
Ubiquitination in various immune cell signaling cascades
Immune responses are highly regulated processes where the ubiquitin system plays an important role. In this section, we will discuss about the regulatory mechanisms of ubiquitin-dependent immune responses by focusing on different immune cell signaling cascades, namely T cell, B cell, and TNF signaling cascades. The precise regulation of T and B cell effector functions, as well as T cell-mediated autoimmunity strongly rely on ubiquitination and require a large set of ubiquitinating enzymes, which are responsible for fine-tuning the adaptive immune response. Furthermore, the TNF signaling cascades are regulated by various types of ubiquitin chains, which are generated or hydrolyzed by specific ubiquitin enzymes.
Ubiquitination in the T cell-mediated immune response
Adaptive defense against intracellular microbes is called cell-mediated immunity, which is based on the function of T lymphocytes. CD4+ helper T cells activate phagocytes to destroy ingested microbes, whereas CD8+ cytotoxic T cells are responsible to eliminate host cells harboring intracellular pathogens (Germain 2002). After maturation in the thymus, naïve CD4+ and CD8+ T cells migrate to the periphery, where they become activated upon antigen detection by the T cell receptor (TCR) (Germain 2002). Activated T cells clonally expand and differentiate into different effector T cells, mediating the adaptive immune response (Smith-Garvin et al. 2009).
Ubiquitination in TCR signal transduction
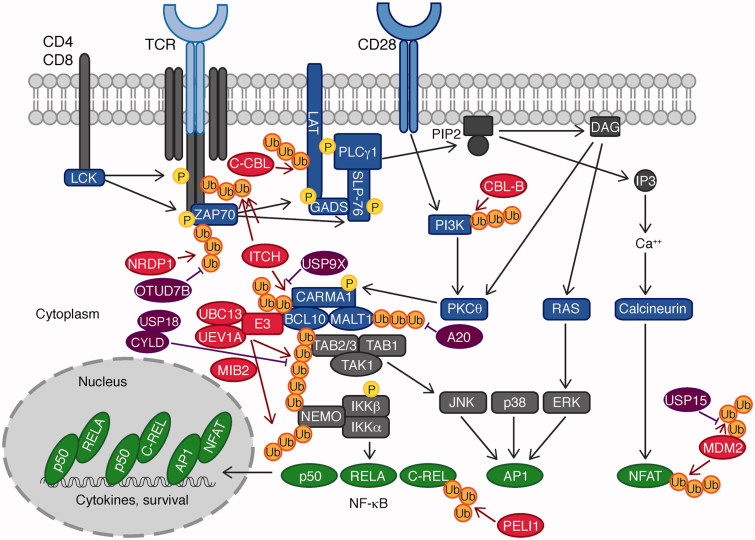
T cell activation requires binding of the TCR to an antigen presented by the major histocompatibility complex (MHC) of an antigen-presenting cell (APC). Additional co-stimulatory molecules, particularly CD28, are required for the complete activation of T cells (Figure 3). Antigen binding to the TCR leads to the recruitment and activation of the proto-oncogene tyrosine-protein kinase (LCK), which phosphorylates the TCR signaling chain CD3ζ, inducing recruitment of the protein tyrosine kinase 70 kDa ζ-chain associated protein (ZAP70) (Figure 3). The latter phosphorylates the transmembrane scaffold proteins linker for activation of T cells (LAT) and Src homology 2 domain-containing leukocyte protein of 76 kDa (SLP-76), which triggers activation of phospholipase C γ 1 (PLCγ1). PLCγ1 cleaves the membrane lipid phosphatidylinositol-4,5-biphosphate (PIP2) to inositol-1,4,5-triphosphate (IP3) and diacylglycerol (DAG). IP3 and DAG lead to the activation of the protein kinase PKCθ, RAS, and calcium pathways, which culminates in activation of the transcription factors NF-κB, AP-1, and nuclear factor of activated T cells (NFAT), for the induction of T cell proliferation and survival genes. NF-κB and AP-1 activation requires formation of the CARMA1-BCL10-MALT1 (CBM) signalosome complex, consisting of CARD-containing MAGUK protein 1 (CARMA1), B cell lymphoma/leukemia 10 (BCL10) and Mucosa-associated lymphoid tissue lymphoma translocation protein 1 homolog (MALT1), which cooperates with an E3 ligase and the Lys 63-specific E2 complex UBC13/UEV1A. Subsequent ubiquitination of BCL10 leads to the recruitment of the TGF-β-activated kinase 1 (TAK1)/TAK1 and MAP3K7-binding protein 2 (TAB2) complex via the UBD called the Npl4 zinc finger (NZF) and the hetero-trimeric inhibitor of NF-κB kinase **(**IKK) complex, which are crucial for ultimate NF-κB activation (Smith-Garvin et al. 2009, Park et al. 2014).
Figure 3.
Ubiquitin enzymes in the TCR signaling pathway. Antigen binding to the TCR leads to the recruitment of the tyrosine kinase LCK, which phosphorylates the TCR signaling chain CD3ζ, which recruits ZAP70. Subsequent phosphorylation of LAT and SLP-76 by ZAP70 triggers activation of PLCγ1, cleaving PIP2 to IP3, and DAG. IP3 and DAG activate NF-κB, AP-1, and NFAT via the protein kinase PKCθ, RAS, and calcium, respectively. NF-κB and AP-1 activation is mediated by the CBM complex, consisting of CARMA1, BCL10, and MALT1, which cooperates with an E3 and the Lys 63-specific E2 dimer UBC13/UEV1A. Ubiquitination of BCL10 leads to the recruitment of the TAB2/TAB1/TAK1 complex and the IKK complex for NF-κB and JNK activation. E3 ligases are indicated in red, DUBs in purple, and transcription factors in green (see color version of this figure at www.tandfonline.com/ibmg).
In fact, many of the proteins in the TCR signaling cascade are subject to regulation by the ubiquitin system, which is essential for their proper function and crucial for signal transduction to the CBM complex. The first level of regulation happens on the TCR itself, which gets ubiquitinated on its ζ chain. The HECT-type E3 ubiquitin ligase Itchy (ITCH), together with the RING-type E3 ligase Casitas B-Lineage Lymphoma Proto-Oncogene B (CBL-B) ubiquitinate TCRζ with Lys 33-linked ubiquitin chains. Lys 33-linked, non-degradative ubiquitin chains block the association of TCRζ with ZAP70 (Huang et al. 2010). This causes impaired signal transduction after antigen-mediated TCR stimulation, resulting in downregulation of TCR signaling. Thus, CBL-B and ITCH are crucial for preventing excessive TCR activation. In line with this notion, mice deficient in CBL-B and ITCH show strong spontaneous autoimmunity (Huang et al. 2010).
Following successful recruitment, phosphorylated ZAP70 is a key molecule to transduce the activation signal. ZAP70 activity is heavily regulated by the ubiquitin system. The E3 ligase CBL-B was implicated in binding and inhibiting ZAP70; however, it is not entirely understood whether this requires ubiquitin-mediated degradation or not (Lupher et al. 1996). There is evidence that ZAP70 ubiquitination leads to the recruitment of the tyrosine phosphatases suppressor of T cell receptor signaling (STS) 1 and STS2, which harbor a UBD and the SRC Homology 3 (SH3) domain in addition to their phosphatase domain. This allows STS1 and STS2 to bind to phosphorylated, ubiquitinated ZAP70 and to dephosphorylate it as a consequence of ubiquitination (Carpino et al. 2009). Dephosphorylated ZAP70 in turn is inactive in TCR signal transduction. The E3 ligase Neuregulin Receptor Degradation Protein-1 (NRDP1)/RNF41 also contributes to ZAP70 ubiquitination, by attaching Lys 33-linked ubiquitin chains, leading to STS1/2 recruitment and ZAP70 dephosphorylation, culminating in abrogation of early TCR signaling (Yang et al. 2015). Thus far, only a few DUBs have been identified to regulate the TCR signaling axis. Among them is OTU domain-containing protein 7B (OTUD7B)/CEZANNE, which is a member of the OTU family of DUBs and a DUB specific for hydrolyzing Lys 11-linked ubiquitin chains (Mevissen et al. 2016). OTUD7B has been shown to antagonize ZAP70 ubiquitination, which prevents association with STS1 and STS2 (Hu et al. 2016). Hence, OTUD7B acts as a positive regulator by facilitating TCR proximal signaling (Hu et al. 2016). Taken together, ubiquitination of ZAP70 ensures proper regulation of T cell-mediated immunity. This is exemplified by the observation that OTUD7B deficiency attenuates T cell activation, and Otud7b−/− mice are refractory to T cell-mediated autoimmune responses (Hu et al. 2016). In contrast, Nrdp1−/− mice have an increased probability to develop autoimmune disease, which involves excessive ZAP70-mediated TCR signaling (Yang et al. 2015).
The DUB probable ubiquitin carboxyl-terminal hydrolase FAF-X (USP9X) also targets ZAP70; however, ZAP70 phosphorylation is intact in USP9X-deficient T cells (Naik et al. 2014). ZAP70 substrates, LAT and SLP-76, were markedly less phosphorylated, pointing to a signal transduction defect. However, it is not known to date, whether USP9X regulates ZAP70 in a direct or indirect manner. USP9X-deficient T cells have a proliferation defect, but at the same time Usp9x−/− mice show spontaneous expansion of T cells associated with lupus-like autoimmunity, suggesting a complex role for USP9X in T cell activation (Naik et al. 2014).
Finally, the ZAP70 substrate, LAT, can also be directly targeted for ubiquitin-mediated regulation. The E3 ligase C-CBL might be responsible for targeting LAT. T cells lacking C-CBL or expressing a RING mutant C-CBL display impaired ubiquitin-mediated internalization and degradation of LAT, resulting in elevated LAT levels and aberrant TCR activation (Balagopalan et al. 2007).
Apart from TCR stimulation, the ubiquitin system also participates in regulating the co-stimulatory pathway via CD28. CD28 co-stimulation has been shown to be required for full T cell activation and serves as a backup mechanism to prevent premature activation and autoimmune reactions. The E3 ligase CBL-B has been shown to target p85, the regulatory subunit of PI3K, which is required for signal transduction from CD28 (Bachmaier et al. 2000). p85 ubiquitination prevents its recruitment to CD28, where CBL-B takes an important role in terminating CD28-mediated co-stimulation. CBL-B-deficient T cells are therefore able to be fully activated even in the absence of CD28 co-stimulation (Bachmaier et al. 2000, Chiang et al. 2000). Whole body knockout of CBL-B, as well as genetic inactivation of its E3 ligase activity, causes T cell hyper-proliferation and spontaneous autoimmune reactions in mice (Bachmaier et al. 2000, Chiang et al. 2000, Paolino et al. 2011), underpinning the importance of CBL-B in limiting aberrant T cell activation.
Regulation of the CBM complex by ubiquitination
The scaffolding protein CARMA1, the adaptor protein BCL10, and the paracaspase MALT1 form the CBM complex (Figure 3). Activating signals from the TCR are transduced and integrated via PKCθ, which phosphorylates CARMA1, resulting in assembly and activation of the CBM complex. The CBM complex mediates signal transduction of proximal TCR activation, culminating in activation of the IKK and JNK pathways (Park et al. 2014, Hu and Sun 2016).
Two components of the CBM complex itself are subject to ubiquitin-dependent regulation: BCL10 and MALT1. The adaptor protein BCL10 is targeted for Lys 48-linked ubiquitin chain-dependent degradation by the E3 ligase ITCH (Scharschmidt et al. 2004). This can be counteracted by USP9X, which interacts with BCL10 and specifically hydrolyzes Lys 48-linked ubiquitin chains. USP9X thereby facilitates CBM complex association and promotes TCR-mediated NF-κB activation (Park et al. 2013). Consequently, while ITCH causes termination of TCR signal transduction via the CBM complex, USP9X promotes CBM complex association and thereby signal transduction. In line with this, USP9X knockdown mice display reduced T cell proliferation, circumstantiating its importance in promoting TCR signal transduction (Park et al. 2013).
Both BCL10 and MALT1 have also been shown to be conjugated with Lys 63-linked ubiquitin chains, which have a positive effect on CBM signaling. By providing an interaction platform, Lys 63-linked ubiquitin chains enhance the recruitment and activation of downstream signaling molecules, like TAK1, IKK, MAPKs, JNK, and p38. The responsible ubiquitin conjugating E2 enzyme has been identified to be the E2 complex UBC13/UEV1A, which associates with the complete CBM complex (Sun et al. 2004, Zhou et al. 2004). The importance of UBC13/UEV1A in CBM regulation is further supported by the fact that a T cell-specific deletion of UBC13 leads to a strong reduction of peripheral T cells. Both JNK/p38 and NF-κB pathways are strongly impaired in UBC13-deficient T cells, which have been proposed to contribute to this reduction in T cell number (Yamamoto et al. 2006).
The E3 ligase that cooperates with UBC13 in ubiquitinating the CBM complex has not been pinpointed with certainty; however, tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) is a strong candidate. It was shown that TRAF6 physically interacts with MALT1 and ubiquitinates NEMO, which mediates TCR-stimulated IKK-dependent NF-κB activation (Sun et al. 2004). In agreement with this, TRAF6 deficiency in mice causes chronic T cell activation as well as inflammatory disorder (Chiffoleau et al. 2003). However, TCR-stimulated NF-κB activation is not significantly impaired, suggesting an additional contribution of other E3s (King et al. 2006). Using a proteomics-based approach, the E3 ligase mind bomb homolog 2 (MIB2) was found to be recruited to the CBM complex by directly binding to BCL10. Upon co-expression in HEK293T cells, MIB2 was shown to ubiquitinate NEMO, subsequently inducing activation of TAK1 as well as IKK. In agreement with these biochemical data, deletion of MIB2 in Jurkat T cells inhibits NF-κB activation (Stempin et al. 2011). However, it has not been shown yet, whether MIB2 promotes CBM-mediated NF-κB activation in vivo. Loss of the murine MIB2 homolog, MIB1, has been reported to cause defects in T cell development and marginal zone B cells. Although no direct conclusions for human MIB2 can be drawn from these mouse experiments, the fact that MIB2 is dispensable for the development of T cells suggests that there are functional differences between these two related proteins (Song et al. 2008). Thus, it is still elusive whether TRAF6 or MIB2, a cooperative action of both, or another yet unknown enzyme works together with UBC13/UEV1A.
MALT1 ubiquitination upon TCR stimulation is counteracted by A20. By targeting MALT1, A20 restricts TCR-induced NF-κB activation. RNAi-based knockdown of A20 in Jurkat T cells allows strong NF-κB activation, without the need for CD28 co-stimulation (Duwel et al. 2009). This is consistent with the findings that A20-deficient CD8+ T cells exhibit elevated cytokine production, including IL-12 and IFN-γ, through increased NF-κB activation (Giordano et al. 2014). Mechanistically, by cleaving Lys 63-linked ubiquitin chains, A20 prevents sustained interaction of MALT1 and the IKK complex, which impairs prolonged NF-κB activation (Duwel et al. 2009). Interestingly, A20 is itself cleaved and thereby negatively regulated by the paracaspase MALT1 in an incoherent feed-forward loop. Upon recruitment, A20 gets cleaved and inactivated, which exemplifies a complex functional interaction between A20 and MALT1 (Coornaert et al. 2008).
Ubiquitin-dependent regulation of the CBM downstream signaling molecule TAK1 is crucial for T cell development and homeostasis (Reiley et al. 2006, Reiley et al. 2007, Tsagaratou et al. 2010). Two DUBs, cylindromatosis (CYLD) and USP18 are responsible for hydrolyzing ubiquitin chains from TAK1, and thereby represent two crucial negative regulators of the CBM signaling axis. CYLD deletion causes accumulation of constitutively active TAK1, and its downstream kinases JNK and IKK, which results in T cells that become hyper-responsive to TCR stimulation (Reiley et al. 2007). Cyld −/− mice fail to maintain T cell homeostasis and are more susceptible to develop colitis, which likely results from uncontrolled immune responses to commensal microbiota in the gut (Zhang et al. 2006, Reiley et al. 2007). In support of this notion, adoptive transfer experiments show that Cyld − / − T cells induce colitis in Rag1 − / − mice, suggesting an important role of CYLD in terminating T cell activation (Reiley et al. 2007). Complementary to CYLD, which controls homeostatic ubiquitination of TAK1, USP18 specifically acts during differentiation of the CD4+ subset to Th17 cells (Liu et al. 2013).
In a nutshell, the ubiquitin system cooperatively works to balance TCR signaling and CBM-mediated signal transduction, enabling a healthy immune response. Removing any of the above-mentioned components causes an imbalanced immune response, often resulting in fatal pathological conditions.
Transcription factor regulation
By controlling the stability of the two downstream transcription factors C-REL and NFAT, ubiquitination is a key regulatory mechanism of the last stage of TCR signaling. Tight regulation of C-REL and NFAT levels ensures that autoimmune responses towards self-antigens is prevented and thereby constitutes a crucial layer of negative regulation.
C-REL is a subunit of the transcription factor NF-κB, which requires co-stimulation of the receptors TCR and CD28, and plays a key role in T cell activation and differentiation (Kontgen et al. 1995, Maggirwar et al. 1997, Liou et al. 1999, Zhou et al. 2002). C-REL is subject to ubiquitin-dependent degradation by the RING-type E3 ubiquitin protein ligase pellino homolog 1 (PELI1), which conjugates Lys 48-linked ubiquitin chains on C-REL (Ordureau et al. 2008, Chang et al. 2011). PELI1 is highly expressed in T cells at the steady state, and expression is even further boosted upon T cell activation, providing a feedback mechanism to terminate TCR-induced gene transcription. In line with this, loss of PELI1 causes accumulation of nuclear C-REL in T cells, causing T cells to become hyper-responsive to TCR and CD28 stimulation in vitro. As a consequence, PELI1-deficient mice have been shown to spontaneously develop autoimmune disease, characterized by multi-organ inflammation and autoantibody production, which goes in hand with an increase in the number of memory T cells, and concomitant decreased levels of naïve T cells (Chang et al. 2011).
Similarly, negative regulation of the NFAT family member NFATc2/NFAT1 is mediated by ubiquitin-dependent degradation by the E3 ligase MDM2. An additional layer of regulation is added by the fact that MDM2 undergoes auto-ubiquitination, which results in proteasomal degradation and enables nuclear accumulation of NFATc2 (Zou et al. 2014). By specifically antagonizing MDM2 auto-ubiquitination, the DUB USP15 prevents NFATc2-dependent gene activation, including IFN-γ production (Zou et al. 2014). The crucial negative regulatory role of USP15 is further exemplified by the fact that USP15 deficiency in mice promotes TCR- and CD28-stimulated production of cytokines in unstimulated, naïve CD4+ T cells, and enhances the T cell response to bacterial infection and tumor challenge in vivo (Zou et al. 2014). Concluding, ubiquitin-dependent degradation of C-REL and NFAT is pivotal in avoiding an overreacting T cell-mediated immune response to various stimuli.
Ubiquitination in T cell-mediated autoimmunity
For the defense against pathogens, T cells need to respond to foreign antigens. But at the same time, they have to tolerate self-antigens to prevent the development of autoimmune reactions. Three mechanisms are in place to prevent the formation of self-reactive T cells. First, during T cell development in the thymus, self-reactive T cells are deleted by negative selection, so-called “central tolerance”. Self-reactive T cells that escape the thymus are eliminated by a second checkpoint “peripheral tolerance” via antigen-induced cell death or inactivation (anergy) in the absence of co-stimulation. And finally, autoreactive T cells are recognized and suppressed by regulatory T cells (Tregs). All three mechanisms are heavily controlled by ubiquitination (Bhoj and Chen 2009).
Central tolerance
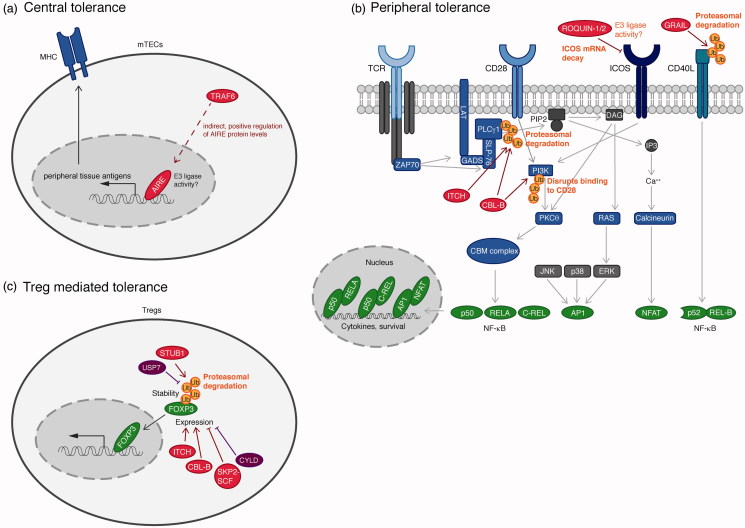
Thymic epithelial cells (TECs) play a key role in mediating central tolerance (Figure 4(a)). While cortical TECs (cTECs) are responsible for positive selection, medullary TECs (mTECs) are crucial for negative selection of self-reactive T cells (Xing and Hogquist 2012). mTECs present a broad array of self-peptides on their MHC, which is crucial for the negative selection process. T cells recognizing this array of peripheral tissue antigens (PTAs) are eliminated to avoid autoimmune reactions. Expression of PTAs is controlled by the nuclear protein autoimmune regulator (AIRE), which functions as a transcription factor in mTECs (Abramson and Goldfarb 2016). Mutations in AIRE cause the autoimmune polyendocrine syndrome type 1 (APS-1), also known as autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) (Nagamine et al. 1997, Anderson et al. 2002). AIRE contains two plant homeodomains (PHDs) that resemble RING finger domains. PHD1 has been shown to have E3 ubiquitin ligase activity, which was suggested to play a role in mediating central tolerance (Uchida et al. 2004). However, the involvement and physiological relevance of the E3 ligase activity of AIRE is still controversial, since Bottomley et al. (2005) were unable to demonstrate E3 activity. While both studies are using cell-free in vitro ubiquitination assays to prove AIRE E3 ligase activity, Uchida et al. (2004) clearly showed that AIRE has a strong preference for the E2 UBC4, while Bottomley et al. (2005) used the E2 UBCH5B, which could explain the reported controversy.
Figure 4.
Ubiquitin enzymes in the regulation of T cell-mediated autoimmunity. (a) The transcription factor AIRE, crucial for central tolerance, controls expression of peripheral tissue antigens, which are required for the negative selection process. An involvement of the E3 ligase activity of AIRE has not been clarified. The E3 ligase TRAF6 indirectly controls AIRE expression via the canonical NF-κB pathway. (b) Peripheral tolerance ensures that self-reactive T cells enter an inactive state (anergy). The E3 ligases ITCH and CBL-B ubiquitinate PLCγ1 and the regulatory subunit of PI3K, p85. Lys 48-linked ubiquitination of PLCγ1 induces proteasomal degradation, whereas ubiquitination of PI3K blocks recruitment to CD28, both of which causes termination of TCR signaling. The E3 ligase GRAIL induces ubiquitination of the co-stimulatory molecule CD40 ligand, which induces its proteasomal degradation. The E3 ligases ROQUIN-1 and ROQUIN-2 control levels of ICOS by inducing ICOS mRNA decay. (c) Treg generation is controlled by the transcription factor FOXP3. The ubiquitin E3 ligase STUB1 directly ubiquitinates FOXP3 with Lys 48-linked ubiquitin chains, leading to proteasomal degradation, which can be counteracted by the DUB USP7. CBL-B and ITCH indirectly promote Treg generation by positively regulating FOXP3 expression. Contrarily, the SKP2-SCF complex as well as the DUB CYLD induce loss of FOXP3 expression via indirect effects. E3 ligases are indicated in red, DUBs in purple, and transcription factors in green (see color version of this figure at www.tandfonline.com/ibmg).
Reduced expression of AIRE and impaired maturation of mTECs was also found in mice lacking the E3 ligase TRAF6. As a consequence, the elimination of self-reactive T cells is impaired, resulting in autoimmune reactions (Akiyama et al. 2005). Moreover, REL-B levels in TRAF6_-_deficient mTECs were reported to be strongly reduced compared to wild-type mTECs (Akiyama et al. 2005). This study indicated that TRAF6-dependent regulation of central tolerance involves activation of canonical NF-κB and thereby expression of REL-B. The functional link between TRAF6 and REL-B in mTECs is not well understood; however, Akiyama et al. found that TRAF6 is required for mTEC development, induced by receptor activator of NF-κB (RANK) and CD40 signals. A cooperative action of RANK and CD40 is essential for mTEC development by establishing the medullary microenvironment, which would explain a crucial role of TRAF6 in signal transduction for AIRE expression and ultimately, mTEC development (Aireakiyama et al. 2008).
In conclusion, central tolerance is mainly controlled by the putative transcription factor AIRE and the E3 ligase TRAF6, by indirectly affecting AIRE expression.
Peripheral tolerance
Peripheral tolerance relies on various factors that negatively regulate TCR signaling and control the threshold of T cell activation (Figure 4(b)). Normally, T cell activation requires antigen binding by the TCR as well as a co-stimulatory signal via CD28. When T cells are stimulated by self-antigens, no co-stimulatory signal is present. This causes T cells to become functionally inactive, in a state called anergy. Anergy is controlled by several ubiquitin E3 ligases. One of them is TRAF6. Cells deficient for TRAF6 are hyper-sensitive to TCR stimulation and display a diminished need for CD28-mediated co-stimulation. Moreover, anergy induction is defective and TRAF6-deficient T cells are resistant to Treg-mediated suppression (King et al. 2006, 2008).
A similar phenotype of activation in the absence of CD28 co-stimulation is observed upon deletion of the E3 ligases CBL-B, gene related to anergy in lymphocytes protein (GRAIL)/RNF128 and ITCH. Knockout of CBL-B in T cells leads to hyper-activation of PKCθ, AKT and NF-κB (Bachmaier et al. 2000). Mechanistically, two targets have been identified to be ubiquitinated by CBL-B that are relevant for anergy induction. On one hand, CBL-B ubiquitinates PLCγ1, resulting in its inactivation and degradation, which prevents T cells from becoming fully activated (Jeon et al. 2004). On the other hand, as discussed in the previous section (Ubiquitination in TCR signal transduction section), CBL-B mediates ubiquitination of p85, the regulatory subunit of PI3K. This disrupts recruitment of p85 to CD28, resulting in impaired T cell activation (Fang and Liu 2001).
Similarly, GRAIL-deficient T cells show enhanced proliferation and cytokine production as a response to TCR activation, which happens independent of CD28 co-stimulation (Nurieva et al. 2010). In line with a negative regulatory role of GRAIL in T cell activation, T cell hybridomas that ectopically express GRAIL display an upregulated anergic response, which is dependent on its E3 ligase activity. Furthermore, GRAIL levels have been found to be upregulated in anergic CD4+ T cells, as well as in CD4+CD25+ Tregs, suggesting a critical role of GRAIL in promoting anergy (Heissmeyer et al. 2004). On a molecular level, it is not well understood how GRAIL promotes anergy. A known target of GRAIL is CD40 ligand (CD40L), which gets ubiquitinated, followed by proteasomal degradation (Lineberry et al. 2008). GRAIL has also been shown to catalyze non-degradative Lys 63-linked ubiquitination of Rho guanine dissociation inhibitor (RhoGDI), which leads to inhibition of RhoA GTPase activity (Su et al. 2006). However, the functional links between CD40L and RhoGDI ubiquitination, and T cell anergy induction are not well understood.
The HECT-type E3 ligase ITCH has also been shown to be upregulated in anergic T cells (Heissmeyer et al. 2004). ITCH deficiency in humans correlates with the development of multi-system autoimmune diseases, which affects physical growth, craniofacial morphology, muscle development, and immune function (Lohr et al. 2010). The same pathology is phenocopied in ITCH-deficient mice, highlighting a crucial role for ITCH in regulating T cell anergy (Perry et al. 1998). Mechanistically, it was suggested that ITCH mediates ubiquitin-dependent degradation of several players in T cell signaling, the main target being PLCγ1. Transfection-based experiments in cell lines showed that PLCγ1 is ubiquitinated and destabilized by ITCH, whereas proteasome inhibition by MG132 inhibits PLCγ1 degradation, thereby terminating TCR signaling (Heissmeyer et al. 2004). An additional study showed that ITCH associates with and ubiquitinates Jun-B, an essential transcription factor for promoting IL-4 expression. ITCH deficiency, therefore, results in elevated IL-4 expression, causing aberrant T cell activation, which explains the observed inflammatory phenotype mentioned above (Fang et al. 2002).
Whereas ITCH regulates receptor proximal signaling in a ubiquitin-dependent manner, control can also be exerted on membrane-bound co-receptor molecules, like inducible T cell co-stimulator (ICOS). Control is not exerted at the protein level, however, but rather by controlling mRNA stability. Two RNA-binding RING-type E3 ligases, RING finger and C3H zinc finger protein (ROQUIN) 1 and 2, have been shown to regulate peripheral tolerance by controlling the expression of ICOS (Yu et al. 2007a). ROQUIN-1 and ROQUIN-2, which display functional redundancy (Pratama et al. 2013, Vogel et al. 2013), mediate decay of ICOS mRNA, but also several other mRNAs (Leppek et al. 2013). It is not known to date, whether ICOS is the sole target contributing to T cell regulation and an involvement of the E3 ligase activity has not been clarified yet. However, mice carrying a point mutation in ROQUIN-1, called sanroque mice, develop a lupus-like autoimmune disease, with excessive numbers of follicular helper T cells and germinal centers (Vinuesa et al. 2005), highlighting the importance of ROQUIN-1 in anergy induction.
In summary, several E3 ligases contribute to anergy induction and thereby peripheral tolerance, by targeting various positive TCR signaling molecules for ubiquitin-dependent degradation.
Treg-mediated tolerance
Tregs play an important role in maintaining immune homeostasis by suppressing the induction and proliferation of effector T cells (Figure 4(c)). As such, Tregs are crucial for the prevention of autoimmune diseases (Sakaguchi et al. 2008). Tregs are either generated in the thymus, called naturally occurring Tregs (nTreg), or they are induced and develop in the periphery, called induced Tregs (iTreg). Both Treg populations depend on and are characterized by a high expression of the master transcription factor forkhead box protein P3 (FOXP3). FOXP3 loss-of-function gene mutations in humans cause a severe multi-organ autoimmune and inflammatory disorder immuno-dysregulation polyendocrinopathy enteropathy X-linked syndrome (IPEX) (Bennett et al. 2001). In support of this notion, FOXP3 mutant mice scurfy display a similar fatal phenotype, which is dependent on excessive T cell activity (Blair et al. 1994, Brunkow et al. 2001, Wildin et al. 2001). Conversely, forced expression of FOXP3 in CD25−CD4+ T cells results in the acquisition of a suppressive function and Treg phenotype. Since Treg generation and maintenance mainly depend on the master regulator FOXP3, it is not surprising that its function and expression are heavily regulated by post-translational modifications, including ubiquitination. For example, it is important to diminish Treg influence during induction of e.g. pro-inflammatory responses to achieve the correct shift in dynamics away from a repressed state towards an active state. One central mechanism by which this is achieved is through ubiquitin-dependent regulation of FOXP3 stability.
In this context, the ubiquitin E3 ligase STIP1 homology and U box-containing protein 1 (STUB1) directly controls the stability of FOXP3 (Chen et al. 2013). Upon pro-inflammatory cytokine or LPS stimulation, STUB1 ubiquitinates FOXP3 with Lys 48-linked ubiquitin chains, inducing its proteasomal degradation. In agreement with this, it was observed that STUB1 overexpression causes prominent loss of FOXP3, resulting in elevated numbers of Th1 cells, ultimately culminating in autoimmunity (Chen et al. 2013). The DUB USP7 counteracts STUB1-dependent ubiquitination of FOXP3 and thereby stabilizes it (van Loosdregt et al. 2013). In fact, USP7 expression is highly upregulated in FOXP3+ Tregs, whereas USP7 downregulation or inhibition causes loss of FOXP3 and impairs Treg functions (van Loosdregt et al. 2013).
Several other (de)ubiquitinating enzymes indirectly control expression levels of FOXP3, and thereby Treg generation. CBL-B for example has been shown to be essential for transforming growth factor β (TGF-β)-mediated iTreg generation by promoting expression of FOXP3 (Harada et al. 2010). CBL-B-deficient mice are characterized to have impaired iTreg generation, both in vivo and in vitro (Wohlfert et al. 2006). Mechanistically, CBL-B negatively regulates PI3K activation and thereby ensures activation of the transcription factors FOXO3A and FOXO1, to ultimately promote expression of FOXP3.
The E3 ligase ITCH also positively controls FOXP3 expression. At the molecular level, ITCH ubiquitinates the transcription factor transforming growth factor β-inducible early growth response protein 1 (TIEG1)/KLF10 with non-degradative ubiquitin chains to activate it. Active TIEG1 in turn promotes expression of FOXP3, and, therefore, stimulates the generation and maintenance of Tregs (Venuprasad et al. 2008).
The Skp-Cullin-F-box (SCF) ubiquitin ligase complex, containing the F-box protein S-phase kinase-associated protein 2 (SKP2), negatively regulates Treg function. Overexpression of SKP2 reduces Treg function by inducing loss of FOXP3 (Wang et al. 2012). Conversely, downregulation of SKP2 induces the conversion of pathogenic T cells to FOXP3 expressing Tregs (Wang et al. 2012). In short, the SKP2 containing SCF complex controls the fate of T cells by regulating FOXP3 maintenance.
By indirectly controlling expression of FOXP3, the DUB CYLD also plays a negative regulatory part in TGFβ-mediated iTreg generation (Reissig et al. 2012). CYLD-deficient mice display a markedly increased number of Tregs in the periphery, but not in the thymus. At the molecular level, CYLD counteracts ubiquitination of mothers against decapentaplegic homolog 7 (SMAD7), which is required for the activation of TAK1 and p38 kinases. Thus, CYLD prevents excessive activation of TAK1 and p38, which results in reduced FOXP3 expression and Treg generation (Zhao et al. 2011).
Collectively, several E3 ligases as well as DUBs cooperate to control homeostatic levels of FOXP3 transcription factor and subsequently Treg generation and maintenance. STUB1 and USP7 counteract each other in directly controlling proteasome-dependent FOXP3 degradation and stability. Indirect control of FOXP3 levels is exerted by the E3 ligases CBL-B and ITCH, which positively contribute to maintain FOXP3 levels, whereas the SKP2 containing SCF complex as well as the DUB CYLD promote degradation of the FOXP3 protein and associated Treg levels.
Apart from controlling FOXP3 expression, a few ubiquitin enzymes affect Treg generation and maintenance via different mechanisms. The Lys 63-specific E2 enzyme UBC13 for example is crucial for maintaining Treg stability. UBC13-deficient Tregs lose their suppressive function, which leads to activation of effector T cells and development of autoimmune symptoms in mice (Chang et al. 2012). Lack of UBC13 further renders Tregs prone to convert to Th1 or Th17 inflammatory T cells, together with loss of FOXP3 expression. This function of UBC13 requires activation of IKK, which ensures constant expression of SOCS1, which normally prevents Tregs from acquiring inflammatory effector functions of Th1 or Th17 cells (Chang et al. 2012). Therefore, loss of UBC13 results in IKK-dependent reduced expression of SOCS1, causing pathological conversion of Tregs to inflammatory T cells.
The E3 ligase GRAIL does not only play a role in peripheral tolerance but also positively controls Treg functions, where it was found to be highly expressed (MacKenzie et al. 2007). GRAIL-deficient T cells have normal expression of FOXP3 and other markers; however, they are impaired in suppressing activation of naïve T cells. The underlying mechanism is not understood, yet GRAIL overexpression in a T cell line is sufficient to mediate a suppressor phenotype by promoting Treg activity (MacKenzie et al. 2007).
The E3 ligase Von Hippel–Lindau (VHL) further contributes to maintaining the suppressive capacity of Tregs (Lee et al. 2015). Treg-specific deletion of VHL causes strong inflammation, accompanied by excessive Treg IFN-γ production. At the molecular level, the absence of VHL leads to aberrant upregulation of hypoxia-inducible factor 1α (HIF-1α), which directly binds to the IFN-γ promoter and thereby mediates excessive production of IFN-γ in VHL-deficient Tregs (Lee et al. 2015), causing pathological inflammation.
The membrane-associated E3 ligase membrane-associated RING finger protein 1 (MARCH1) plays a crucial role in antigen presentation of dendritic cells. Since antigen presentation is critical for the development of Tregs in the thymus, MARCH1 has a direct positive influence on thymic Treg development (Oh et al. 2013). MARCH1 functions by ubiquitinating MHCII and the co-stimulatory molecule CD86, which promotes their endocytosis and lysosomal degradation (Corcoran et al. 2011). Mice lacking MARCH1 are characterized with increased surface expression of MHCII, which causes a reduction in thymus-derived T cells. This function of MARCH1 is dependent on its E3 ligase activity; however, it is not understood yet how MHCII ubiquitination regulates Treg development (Oh et al. 2013).
The E3 ligase TRAF3 has also been shown to regulate thymic T cell development, in addition to its role in B cells. Conversion of precursor T cells to mature CD25+Foxp3+ Tregs requires the action of IL-2. TRAF3 negatively regulates IL-2 signaling by promoting the association of IL-2 with T cell protein tyrosine phosphatase (TCPTP), which is a negative regulatory phosphatase (Yi et al. 2014a). Whether this requires the enzymatic activity or whether TRAF3 has an adaptor function is not clear yet. T cell-specific deletion of TRAF3 leads to an increase in the number of Tregs in the thymus as well as the periphery. Interestingly, these mice also display an increased frequency of CD4+ T cells with effector- or memory-like surface markers, suggesting a positive role for TRAF3 in regulating established Tregs (Chang et al. 2014, Yi et al. 2014b).
In summary, several enzyme members of the ubiquitin system contribute to regulate Treg-mediated tolerance, either by controlling expression of FOXP3, the Treg master transcription factor, or via other mechanisms. Only a cooperative action of these regulators ensures balanced generation and function of Tregs.
Ubiquitination in the B cell-mediated immune response
B lymphocytes mediate the humoral immune response, targeting extracellular microbes and antigens (Zinngrebe et al. 2014). Upon antigen encounter, immune-competent B cells become active in a two-step process. The first activation signal is provided by antigen binding to the B cell receptor (BCR). The antigen subsequently gets processed and complexed with MHC II molecules, which are displayed on the B cell surface. The second activation signal requires recognition of the displayed antigen by a CD4+ T helper cell. The activated T helper cell provides the second activation signal to the B cell, which initiates B cell proliferation and differentiation into memory B cells and plasma cells. Plasma cells secrete antibodies and thereby mediate the humoral immune response (Heesters et al. 2016). B cell signaling needs to be tightly controlled, to ensure a proper immune response when needed, but at the same time to avoid excessive reactions against self-antigens. Ubiquitination constitutes a major regulatory mechanism to control B cell signaling; however, its role is less well understood compared to T cell activation (Malynn and Ma 2010).
Ubiquitination in B cell signaling
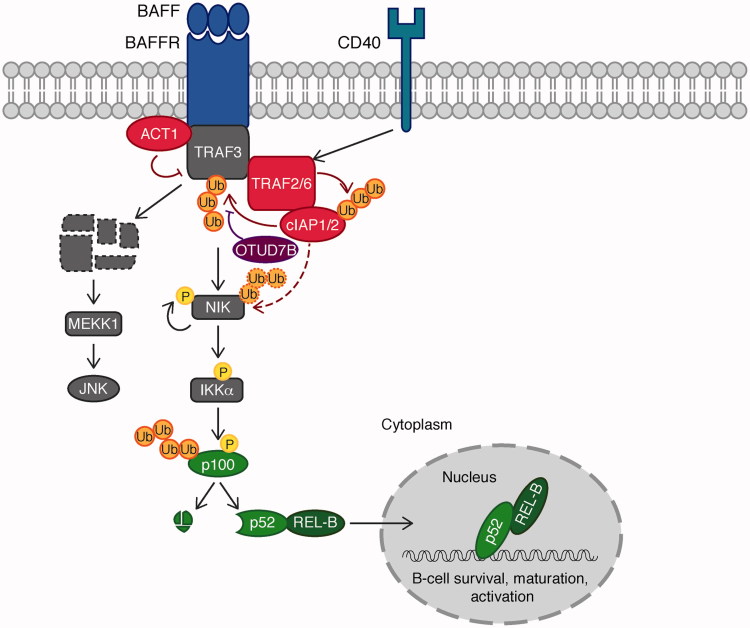
Ubiquitination-mediated regulation in B cell activation has been shown to be critical, especially for the non-canonical NF-κB pathway, mediated by CD40 and BAFF receptor (BAFFR), two proteins of the TNF receptor superfamily (Figure 5). Upon activation of CD40 or BAFFR, the TNFR-associated factor 2 (TRAF2) promotes activation of the E3 ligases cellular inhibitor of apoptosis protein-1 (cIAP1) and cIAP2, which in turn mediate Lys 48-linked ubiquitination and proteasomal degradation of TRAF3 leading to two distinct signaling events (Matsuzawa et al. 2008, Vallabhapurapu et al. 2008). First, TRAF3 is an adaptor protein, which recruits TRAF2/TRAF6-cIAP1-cIAP2 to promote ubiquitin-dependent degradation of NF-κB inducing kinase (NIK). Degradation of TRAF3 therefore results in stabilization of NIK, which is subsequently activated through auto-phosphorylation. Active NIK phosphorylates and activates IKKα, which in turn phosphorylates the NF-κB subunit p100. Phosphorylation-induced ubiquitination of p100 induces processing by the proteasome. This releases the mature NF-κB subunit p52, which associates with the other NF-κB subunit REL-B to promote expression of genes required for B cell survival, maturation, and activation (Neumann and Naumann 2007, Vallabhapurapu et al. 2008). Second, degradation of TRAF3 also results in cytosolic translocation of a signaling complex containing the MAPK kinase MEKK1. Subsequently, MEKK1 activates JNK and other MAPK cascades (Matsuzawa et al. 2008). Taken together, activation of CD40 and BAFFR leads to ubiquitin-dependent degradation of TRAF3, resulting in activation of NF-κB, JNK and MAPK signaling pathways.
Figure 5.
Ubiquitination in B cell activation. In B cells, the non-canonical NF-κB pathway mediated by two TNF receptor superfamily members, CD40 and BAFF receptor (BAFFR) is regulated by ubiquitination. Upon activation of CD40 or BAFFR, TRAF2/6 promotes activation of cIAP1-cIAP2, which in turn mediates Lys 48-linked ubiquitination and degradation of TRAF3. Degradation of TRAF3 results in stabilization of NIK. Active NIK phosphorylates and activates IKKα, which in turn phosphorylates the NF-κB subunit p100 (a precursor of p52). Phosphorylation-induced ubiquitination of p100 induces processing by the proteasome. This releases the mature p52 subunit, which associates with REL-B to promote expression of genes required for B cell survival, maturation and activation. Degradation of TRAF3 results in cytosolic translocation of a signaling complex containing the MAPK kinase kinase MEKK1. MEKK1 then activates JNK. An additional layer of regulation is provided by the E3 ligase ACT1 as well as the DUB OTUD7B. E3 ligases are indicated in red, DUBs in purple, and transcription factors in green (see color version of this figure at www.tandfonline.com/ibmg).
Thus, regulation of B cell signaling is heavily controlled by ubiquitination. Some of the signaling molecules are members of the ubiquitin system themselves, others are regulated by ubiquitination. Initial signal transduction upon CD40 ligand or BAFF binding to their respective receptors, for example, is blocked by the U-box type E3 ligase nuclear factor NF-κB activator 1 (ACT1), a negative regulator of non-canonical NF-κB activation (Qian et al. 2004). Upon ligand stimulation, ACT1 has been shown to associate with CD40, BAFFR, as well as TRAF3 and thereby blocks NF-κB activation. Specific association of ACT1 with the negative regulator TRAF3 allows us to speculate that ACT1 either competes for interaction of TRAF3 with positive regulators, or it could target TRAF3-associated positive regulators for proteasomal degradation. In support of a critical role of ACT1 in restricting B cell signaling, is the fact that ACT1-deficient B cells are characterized with elevated CD40 as well as BAFF signaling, mediating aberrant B cell survival (Qian et al. 2004). Consistently, Act1−/− mice develop systemic autoimmune responses, resulting in lymphadenopathy, splenomegaly, hyper-gamma-globulinemia and auto-antibodies (Qian et al. 2008).
Two signaling molecules, TRAF2, as well as its family member, the adaptor protein TRAF3, are two crucial negative regulators of B cell signaling. Deficiency of TRAF2 as well as TRAF3 in mice causes postnatal lethality (Xu et al. 1996, Yeh et al. 1997). The observed lethality is accompanied by a defect in development of bone marrow B cells, and an increase in the number of marginal zone B cells, exemplifying the importance of TRAF2 and TRAF3 in controlling B cell homeostasis. Interestingly, data suggest that TRAF2 and its family member TRAF6 could play redundant roles in B cell activation. B cell-specific deficiency of TRAF6 results in a reduced number of mature B cells in the bone marrow and spleen. Moreover, TRAF6-deficient mice are defective in T cell-dependent as well as T cell-independent antigen responses, suggesting a critical role for TRAF6 in B cell regulation (Kobayashi et al. 2009). However, unexpectedly, the non-canonical NF-κB pathway is intact in B cells lacking TRAF6 (Rowland et al. 2007, Kobayashi et al. 2009). This led to the hypothesis that TRAF6 and TRAF2 have partially redundant roles in CD40-mediated NF-κB activation. Indeed, only double knockout of TRAF2 and TRAF6 in the mouse B cell tumor A20.2J cells leads to a complete block of CD40-mediated NF-κB activation (Rowland et al. 2007).
As pointed out above, TRAF3 degradation is a key event in driving BCR signaling. By hydrolyzing ubiquitin chains from TRAF3, which results in its stabilization, the DUB OTUD7B is a crucial negative regulator of B cell signaling (Hu et al. 2013). Hence, deletion of OTUD7B in B cells renders them hyper-responsive to antigens, highlighting a critical role for OTUD7B in B cell homeostasis (Hu et al. 2013). Interestingly, as discussed in the previous section, OTUD7B is a positive regulator of T cell signaling, while it negatively controls B cell signaling (Hu et al. 2013). Molecular mechanisms by which OTUD7B as a specific DUB for Lys 11-linked ubiquitin chains regulates the T cell and B cell signaling cascades remain to be understood.
The ubiquitin editing enzyme A20 also plays a role in restricting B cell activation. A20-deficient B cells have a low activation threshold and are hyper-responsive to multiple stimuli, causing autoimmune reactions (Tavares et al. 2010, Chu et al. 2011). A20 functions by restricting canonical as well as non-canonical NF-κB activation. The exact molecular events are not well understood; however, B cells lacking A20 display increased phosphorylation of IκBα as well as p100 in response to anti-CD40 (Tavares et al. 2010, Chu et al. 2011).
In summary, in the case of B cell signaling, various components of the ubiquitin system cooperate to restrict B cell signaling, ensuring a tightly controlled humoral immune response. Eliminating any of the above-mentioned regulators causes severe autoimmune reactions.
The TNF-induced NF-κB and apoptosis signaling pathways in immune cells
The TNF signaling pathway is important in various immune cells including T and B cells as well as macrophages (Aggarwal 2003). Well-known non-proteolytic ubiquitin signals including Lys 63-linked and linear (Met 1-linked) ubiquitin chains play a central role in the regulation of the TNF-induced NF-κB and apoptosis pathways (Walczak 2011, Ikeda 2015, Varfolomeev and Vucic 2016). Here, we focus on the ubiquitin enzymes including the LUBAC ligase complex, cIAP ligases, as well as DUBs such as the OTU DUB with linear linkage specificity (OTULIN) and CYLD in the regulation of the TNF-signaling pathways.
TNF-induced NF-κB activation signal
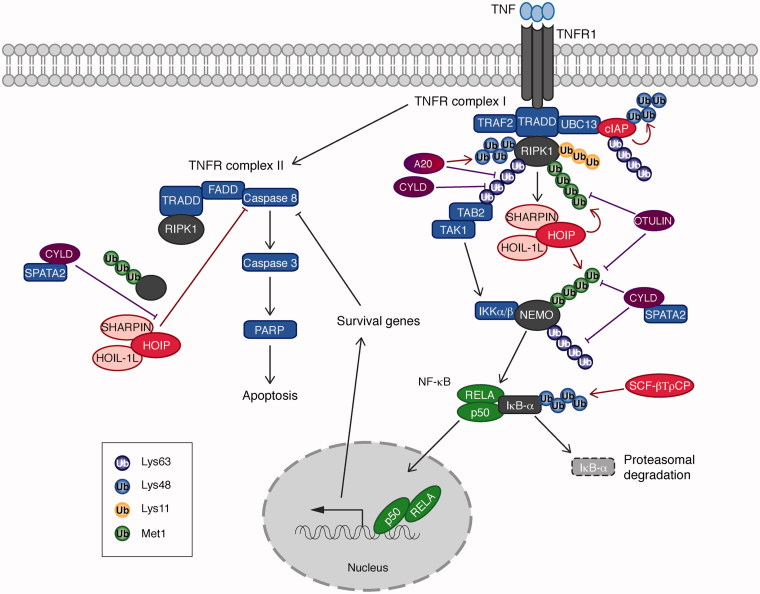
Upon TNF stimulation, TNF receptor (TNFR) complex I consisting of multiple adaptor proteins including TNFR1-associated death domain protein (TRADD) and TRAF2 as well as ubiquitin ligases such as cIAPs and LUBAC is formed (Walczak 2011, Ikeda 2015, Varfolomeev and Vucic 2016) (Figure 6). Lys 63-linked ubiquitin chains play a role upstream of this signaling pathway; cIAP ubiquitinates RIPK1 with Lys 63-linked chains that recruit the TAB2/TAK1 kinase complex and LUBAC, an E3 ligase complex composed of HOIP, HOIL-1L and SHARPIN (as discussed in section Ubiquitin enzymes as key players in ubiquitination) (Haas et al. 2009). Lys 63-linked ubiquitin chains are recognized by UBDs in various signaling molecules; for example, the TAB2-NZF domain directly interacts with Lys 63-linked ubiquitin chains (Kulathu et al. 2009, Sato et al. 2009). Recognition of ubiquitin chains by the TAB2-NZF domain is important to activate NF-κB. It was shown that Escherichia coli NleE-dependent Cys methylation in the TAB2-NZF domain abolishes binding to ubiquitin chains and NF-κB activation (Zhang et al. 2011). Another important E3 ligase is the LUBAC complex, whose recruitment to the TNFR complex I is cIAP-catalytic activity dependent (Haas et al. 2009). Each of the LUBAC component has a UBD; HOIP-NZF1, HOIL-1L-NZF, and SHARPIN-NZF (Ikeda et al. 2011), suggesting that an interaction between ubiquitin chains generated by cIAP and the UBDs in the LUBAC components may be responsible for its recruitment to the TNFR complex I. In the TNF-dependent NF-κB activation pathway, RIPK1 and NEMO are linearly ubiquitinated by LUBAC (Tokunaga et al. 2009, Gerlach et al. 2011, Ikeda et al. 2011). It has been demonstrated that an interaction between linear ubiquitin chains and the NEMO-UBAN domain, which is a linear ubiquitin chain specific interaction domain (as discussed in section Recognition of ubiquitin by ubiquitin binding domains (UBDs)), is essential to activate NF-κB (Rahighi et al. 2009). Interestingly, in the IL-1β signaling and the nucleotide-oligomerization domain-containing protein 2 (NOD2) signaling cascades, LUBAC seems to ubiquitinate pre-existing Lys 63-linked ubiquitin chains to generate hybrid ubiquitin chains (Emmerich et al. 2013, Fiil et al. 2013). Whether LUBAC contributes in the generation of Lys 63/linear hybrid ubiquitin chains in the TNF-signaling cascades remains to be clarified.
Figure 6.
Ubiquitin chains and ubiquitin enzymes in the TNF-induced NF-κB and apoptosis pathways. Different linkage types of ubiquitin chains, Met 1-, Lys 11-, Lys 48-, and Lys 63-linked ubiquitin chains play a critical role in the TNF-induced canonical NF-κB and the TNFR complex II-dependent apoptosis pathways. Ubiquitin chains are generated by the E3 ligases, cIAP, LUBAC complex, and SCF-βTrCP. These ubiquitin chains are hydrolyzed by two DUBs, OTULIN and CYLD. A20 is a hybrid of E3 ligase and DUB. Ubiquitination of the substrates, including cIAPs, RIPK1, NEMO, and IκB-α, impacts on the downstream signaling pathways. The TNFR complex II-mediated apoptosis pathway includes RIPK1, TRADD, FADD, and Caspase 8. Activation of Caspase 8 leads to Caspase 3-dependent cleavage of PARP and apoptosis. The LUBAC complex (HOIP, SHARPIN, and HOIL-1L) and the CYLD–SPATA2 complex regulate the TNFR complex II-induced apoptosis pathway. E3 ligases are indicated in red, DUBs in purple, and transcription factors in green (see color version of this figure at www.tandfonline.com/ibmg).
In this signaling cascade, DUBs play an inhibitory role; CYLD hydrolyzes Lys 63-linked and linear ubiquitin chains, and OTULIN deconjugates linear ubiquitin chains. Both CYLD and OTULIN were found to interact indirectly or directly with the HOIP-PNGase/ubiquitin-associated (PUB) domain (Elliott et al. 2014, Schaeffer et al. 2014, Takiuchi et al. 2014). A HOIP PUB mutant, which cannot interact with CYLD or OTULIN, activates NF-κB more prominently than HOIP wild type, confirming a critical role of CYLD and OTULIN as negative regulators. Furthermore, these data suggest that CYLD and OTULIN regulate the downstream signaling pathway by making a complex with HOIP, and by hydrolyzing ubiquitin chains formed on the components in close proximity, presumably those in the TNFR complex I. A20 is a hybrid enzyme of E3 ligase and DUB, thereby able to edit ubiquitin chains in a dual manner (Wertz et al. 2004, Vucic et al. 2011). A20 negatively regulates NF-κB activity by hydrolyzing Lys 63-linked chains on RIPK1, and by ubiquitinating RIPK1 with Lys 48-linked ubiquitin chains for proteasomal degradation (Wertz et al. 2004). More recently, it was found that phosphorylation of A20 regulates its own catalytic activities of DUB and E3 ligase (Wertz et al. 2015). A20 also has a linear ubiquitin chain interaction domain, zinc finger (ZF) 7, which controls NF-κB activation (Tokunaga et al. 2012, Verhelst et al. 2012).
In summary, ubiquitin ligases and DUBs play a very important role in regulating the TNF-induced NF-κB signaling pathway. Strikingly, various linkage types of ubiquitin chains (Lys 11-, Lys 48-, Lys 63-linked, and Lys 63/linear hybrid) are involved, thereby a complete understanding of the regulatory mechanisms around the ubiquitin chains requires further studies.
TNFR complex II-dependent cell death induction
Downstream of TNFR signaling cascade, the apoptosis cascade is regulated by (1) NF-κB-dependent survival gene induction and (2) TNFR complex II-dependent caspase activation (Ashkenazi and Salvesen 2014) (Figure 6). The TNFR complex II shares components with TNFR complex I, such as TRADD and RIPK1, in addition to Fas-Associated protein with Death Domain (FADD) and Caspase 8. Once Caspase 8 is activated in the TNFR complex II, an executor Caspase 3 becomes active and cleaves Poly (ADP-ribose) polymerase (PARP), leading to apoptosis induction.
In the signaling cascade mediated by the TNFR complex II, the LUBAC components, HOIP, SHARPIN, and HOIL-1L play a regulatory role. SHARPIN-deficient (Chronic proliferative dermatitis mutant (Cpdm)) mice (Gerlach et al. 2011, Ikeda et al. 2011), Hoil−/− mice (Tokunaga et al. 2009) and Hoip−/− mice (Peltzer et al. 2014) all show increased apoptosis in some tissues. For example, Cpdm mice suffer from systemic inflammation and apoptosis in various tissues (Seymour et al. 2007). Skin inflammation is especially severe in Cpdm mice and massive induction of apoptosis in keratinocytes is observed. This skin inflammatory phenotype is suppressed in Tnf−/−; Sharpincpdm/cpdm and TNFR1EKO ; Sharpincpdm/cpdm mice suggesting that the TNF pathway plays a major role (Gerlach et al. 2011, Kumari et al. 2014). By contrast, knockout of the necroptosis regulator RIPK3 or Mixed lineage kinase domain like pseudokinase (MLKL) in Cpdm has a modest effect on the skin phenotype (Rickard et al. 2014). However, additional epithelial-specific knockout of the apoptosis essential regulator FADD to RIPK3 knockout drastically rescues skin inflammation and keratinocyte apoptosis in Cpdm mice (Kumari et al. 2014). Furthermore, heterozygosity of Caspase 8 protects from the skin phenotype in Cpdm mice (Rickard et al. 2014). These data indicate that the skin inflammation in Cpdm mice largely depends on apoptosis, whereas necroptosis plays a minor role. However, the apoptotic phenotype in mice deficient for SHARPIN, HOIL-1L, and HOIP is distinct. Kazuhiro Iwai’s group demonstrated that HOIL-1L knockout mice have no major inflammatory phenotype without any challenges, but apoptosis is induced in the liver tissue of HOIL-1L knockout mice upon TNF treatment (Tokunaga et al. 2009). On the contrary, HOIP knockout mice are embryonic lethal at E10.5 and aberrant endothelial cell death is observed. Embryonic lethality of HOIP knockout mice is partially rescued by TNF ablation, and the double-knockout mice of HOIP and TNF survive until E15.5 (Peltzer et al. 2014). These observations suggest that (1) each of the LUBAC components has its own function outside of the context of LUBAC, or (2) the HOIP-HOIL-1L complex and the HOIP-SHARPIN complex have distinct substrates and functions. This is an interesting point and further studies are required to understand how TNF-dependent apoptosis is regulated by the LUBAC components at the molecular level.
At the cellular level, the E3 ligase activity of HOIP is required for the anti-apoptosis function (Peltzer et al. 2014, Rickard et al. 2014). Recently, LUBAC was found to ubiquitinate FADD, suggesting that LUBAC-dependent ubiquitination of FADD regulates TNFR complex II formation (Goto and Tokunaga 2017). Whether FADD ubiquitination is homotypic linear ubiquitination or hybrid chains is still open, and how DUBs contribute to FADD-deubiquitination would be an important point to clarify further.
TRIM E3 ligases in immune signaling
One of the main points from the previous sections in this review is that virtually all different cell signaling pathways that control immune output are extensively regulated through post-translational modification by ubiquitin. Nevertheless, the E3 ligases which control these processes are diverse and span all three-different main E3 types: HECT, RBR, and RING (as discussed in Ubiquitin enzymes as key players in ubiquitination section).
In recent years, one particular family of over between 65 and 100 RING E3 ligases has been recognized to contain many members with important roles in regulating the higher vertebrate immune system: tripartite motif proteins (TRIM) (Ozato et al. 2008, Han et al. 2011). These putative E3 ligases are found in all multi-cellular eukaryotes, except for plants. Especially for the mammalian innate immune system compelling evidence exists that individual TRIMs play key roles in most of the pathways connected to a diverse range of cellular receptors recognizing “non-self” molecules during pathogen infection (Versteeg et al. 2013, 2014, Rajsbaum et al. 2014a). The importance of these TRIMs in these pathways is underpinned by the fact that different pathogens have devised strategies to block TRIM action.
Most members of this protein family have a RING domain and are thus predicted to be E3 ligases. Indeed, E3 ligase activity has been shown in vitro for various TRIMs. Interestingly, some of them appear to also have RING-independent functions (Versteeg et al. 2014). This is of special interest considering that several TRIMs have been implicated in two or more distinct cellular processes. Detailed molecular analyses to understand how individual TRIMs can target different substrates, how TRIMs themselves are activated, and how different domains of individual TRIM proteins contribute to these processes is limited. Progress in this area has mainly been hampered by the difficulty of obtaining sufficient quantities of soluble, full-length TRIM protein for in depth biochemical, biophysical, and structural studies.
Several recent reviews itemize all TRIMs with reported functions in different cell signaling pathways, their targets, and the model systems used to address their contribution (Hatakeyama 2011, 2017, Rajsbaum et al. 2014a, Versteeg et al. 2014). In this section, we will review our current understanding of how individual domains contribute to TRIM function, with a focus on TRIMs contributing to the anti-pathogen response, and TRIMs, which are actively counteracted by pathogens. Here, we aim to exemplify for selected TRIMs how they contribute to an effective anti-pathogen response, and the diverse ways how this is achieved molecularly in ubiquitin-dependent and -independent manners.
TRIM domains determine enzymatic activity, oligomerization, and target specificity
TRIM proteins derive their name from their N-terminal domain organization consisting of a RING domain, one or two B-boxes, and a coiled-coil (Reymond et al. 2001). Together, this tripartite motif is referred to as the RING-B-box-Coiled-coil (RBCC). The most divergent components, which set the different TRIM proteins apart, are their 11 different constellations of C-terminal domains, which cluster the TRIMs in the same number of sub-groups (Ozato et al. 2008). The majority of TRIM family members indeed possess a truly tripartite RBCC. However, several family members lack one or two of these domains and are sometimes referred to as TRIM-like proteins (Rajsbaum et al. 2014a, Versteeg et al. 2014). We will not make this distinction in this review since they are part of the same evolutionary family, and proteins with complete or partial RBCCs have both been reported to be important immune regulators (Rajsbaum et al. 2014a, Versteeg et al. 2014).
To date, no full-length TRIM structures have been solved at high resolution, only individual domains, or combination of domains. However, higher order hexagonal rings have been identified by electron microscopy for the retroviral restriction factor TRIM5α. Based on these EM data, in this TRIM protein, the RBCC domain has a predominant role in higher order assembly into the hexagonal array (Javanbakht et al. 2005, Diaz-Griffero et al. 2009, Ganser-Pornillos et al. 2011, Li et al. 2016), whereas its C-terminal domain likely facilitates recognition and binding to its target: the retroviral capsid lattice (Woo et al. 2006, Li et al. 2016). Yet, this C-terminal domain is not required for hexagonal ring assembly. TRIM5α function has been studied in detail, and these recent findings have made important contributions to understanding how this TRIM exerts its antiviral activity by forming these higher-order structures. Yet, even though structural information on other TRIMs is limited, various studies indicate that the RBCC and subgroup-specific C-terminal domains may have different functions for other TRIMs. This is true even for family members, which are in the same sub-group as TRIM5α and thus share the same domain architecture. Nevertheless, various domain features have been reported to have similar function across different individual TRIMs.
RING and B-box domains can both contribute to TRIM oligomerization
From a functional perspective, it is the RING domain – a conserved docking domain for E2 ligases – which predicts most of these TRIM proteins to be E3 ligases (Deshaies and Joazeiro 2009). Almost all the TRIM RING domains are similar to their counterparts in other RING E3 ligases. The prevailing dogma is that a combination of eight Cys/histidine (His) residues coordinates a central zinc molecule, thereby folding out two protein loops which form part of a docking platform for E2 ligases.
B-box domains are also zinc-fingers similar to RING domains (Keown and Goldstone 2016). Nevertheless, their biological role for TRIM function has remained largely unclear. So far, data that B-boxes can mediate direct E3 ligase activity are minimal. Some published data suggest that TRIM16, which does not have a RING domain, has auto-ubiquitination E3 ligase activity in vitro (Bell et al. 2012). However, it will require more in depth analysis to determine the exact contribution of this to TRIM16 function in cell-free systems. Some data are more in support of a role of B-boxes in TRIM multimerization (Keown and Goldstone 2016) and suggest that it may allow for hetero-multimerization with other TRIMs. E.g. TRIM16 has been shown to associate with TRIM18/MID-1, TRIM19/PML, and TRIM24 in overexpression assays (Bell et al. 2012). Thus, RING domains have mainly been linked to E3 activity, whereas B-boxes may facilitate TRIM multimerization.
Recent biochemical analyses, combined with structural and biophysical studies of TRIM25 and TRIM32, demonstrated that their RING domains form dimers, coordinated by helices on the side of the RING core (Koliopoulos et al. 2016). This self-association enhanced E2 activity in discharge assays, consistent with what has been shown for other dimeric E2s, namely that this RING multimer stabilizes a closed-conformation of ubiquitin-loaded E2 conjugating enzymes (Dou et al. 2012, 2013, Pruneda et al. 2012). However, the B-box did not enhance TRIM multimerization or activity. This is in contrast with what has been described for the retroviral restriction factor TRIM5α, where its B-box2 domain is critical for multimerization, and thereby viral restriction (Diaz-Griffero et al. 2009). A single point mutant in an exposed Arg in B-box 2 (R121) is sufficient to prevent higher assembly and viral restriction (Li and Sodroski 2008, Diaz-Griffero et al. 2009).
Mutations in TRIM B-boxes have been associated with autoimmune diseases in humans. In this context, the etiology of Familial Mediterranean Fever (FMF) has been mapped to mutations in TRIM20 (known as PYRIN). It should be noted that TRIM20 has a unique domain architecture containing a PYRIN domain instead of a RING domain: PYRIN-B-box2-CC-SPRY. TRIM20 has been reported as an activator of the inflammasome adaptor ASC, thereby contributing to its activation and release of mature IL-1β (Yu et al. 2007b). Mechanistically, the B-box2 in TRIM20 keeps it in an auto-inhibited state by binding the adjacent PYRIN domain in TRIM20, thereby preventing association with the PYRIN domain in ASC1, and inhibiting subsequent inflammasome activation (Yu et al. 2007b). This indicates that in this particular TRIM, the B-box may mediate a repressive role in cis. Whether B-boxes are main protein-interaction sites important for the function of other TRIMs remains to be determined. However, while the PYRIN domain is present only in TRIM20 (within the TRIM family), conceptually it poises B-box domains as possible coordinators of TRIM folding in their inactive (monomeric) forms.
All in all, these studies suggest that TRIMs may in general exist in higher order structures and that these structures are required for generating their functional output. It is clear that differences exist between these higher order structures for different TRIMs, yet the lack of detailed structural information has thus far limited our understanding of how individual RING and B-box domains contribute to higher order structure, and how this itself contributes to function.
TRIM coiled-coil domains contribute to oligomer formation
Despite the limited structural information of coiled-coils in TRIM multimers, functional studies and interaction studies with domain mutants have indicated that the coiled-coil domains of most – if not all – TRIM proteins have the ability to facilitate TRIM homo-multimerization (Ciani et al. 2010, Sanchez et al. 2014), and in some cases hetero-multimerization (Bell et al. 2012). Thus, while individual TRIM domains can assemble into oligomeric structures (Koliopoulos et al. 2016), it may well be that within the context of full-length proteins, coiled-coils are indispensable for formation of biologically active oligomers. In line with this notion, for most of the functions reported for individual TRIMs, coiled-coil domains are essential for E3 ligase activity and TRIM function (Streich et al. 2013). Thus far, most evidence points towards coiled-coil domains facilitating anti-parallel higher order TRIM structures, as shown for TRIM5α, TRIM25, and TRIM32. However, whether they may have specific functions in maintaining inactive TRIM monomers, or as molecular rulers determining correct spacing between the RBCC and the C-terminal domains remains to be determined.
Subgroup-specific C-terminal domains often determine substrate specificity
The TRIM C-terminal domains are what set individual members apart. Currently, almost all studies using TRIM domain mutants to investigate which domains are required and sufficient for interaction with their substrates have indicated that this is almost always determined by the C-terminal domain. Conceptually, this suggests that most TRIMs have similar RBCCs for recruiting different ubiquitin-loaded E2 complexes, whereas it is usually the unique C-terminal domains, which determine target binding and specificity.
The most prominent C-terminal domain is found in group IV TRIMs: the PRY-SPRY (B30.2) domain. The reported functions of the PRY-SPRY domain are diverse; mostly this domain mediates protein interactions: presumably a unique substrate for every PRY-SPRY domain. This specificity notion is quite impressive considering that PRY-SPRY domains are not unique to TRIM proteins and are present in over 500 proteins encoded by most mammals (Grutter et al. 2006, Woo et al. 2006, Weinert et al. 2009).
This ability of the PRY-SPRY domain to determine precise target specificity within the TRIM context is well illustrated by the PRY-SPRY domain in TRIM5α, which is critical for its direct viral restriction activity. Few amino acid differences in this domain between TRIM5α of humans and several simian species determine whether it can bind and restrict the capsid of HIV or not, and vice versa for the simian counter-part SIV (Sawyer et al. 2005).
Taken together, data from predominantly cell-based studies have provided a wealth of insight into possible functions of individual TRIM proteins, what some of their targets are, which TRIM domains are required for substrate-binding and function, and lastly whether this function is dependent on ubiquitin E3 ligase activity of these TRIM proteins (Rajsbaum et al. 2014a, Versteeg et al. 2014). However, details on how TRIMs themselves are activated, which higher order structures are adopted in inactive and active states, and the stoichiometry of TRIM substrates under these conditions remains limited. It may be that only in certain states or combinations with substrates TRIM proteins will lend themselves for purification in quantities, which will facilitate the biochemical and structure approaches required to address these issues.
TRIM proteins regulate the immune response
Three observations have been recognized for some time now, which together have indicated that a substantial number of TRIM proteins may have immune-related functions. First, based on publicly available mRNA expression data from various tissues and cell types, we estimate that about a third of the TRIMs are above average or predominantly expressed in immune cell types, suggesting a possible biological role in these cells. Moreover, about 20–30% of the TRIMs are transcriptionally induced by immune cytokines (Rajsbaum et al. 2008, Carthagena et al. 2009). In fact, to date, mostly the effect of antiviral type I and type II interferons on TRIM expression has been systematically investigated, but it is conceivable that some of the many other immune modulatory cytokines or activation of specific immune receptors control TRIM expression (Martinez et al. 2006). In line with this notion, activation of Fc receptors on macrophages by immunoglobulin complexes, specifically induce mRNA expression of Trim9 and Trim54 (Carthagena et al. 2009), two TRIMs which are not induced by interferons.
Second, the number of Trim genes dramatically expanded recently in evolution in the same time frame during which the adaptive immune system arose, and the innate immune system increased in complexity (Versteeg et al. 2014). Lower invertebrates, such as flies and sea squirts, have between seven and ten Trim genes. This number slightly increased in non-jawed vertebrates such as lampreys, yet substantially increased to 35–40 Trim genes in puffer fish and birds, and ≥60 Trim genes in mammals. Together, this observation suggests that TRIM proteins may have evolved and expanded to regulate other systems which heavily evolved in that evolutionary time frame, such as the immune system and the vertebrate brain.
Lastly, recent computational analysis of Trim gene evolution has indicated that a substantial number of Trim genes – 16 out of 67 – have been under positive selection pressure in primates (Han et al. 2011, Malfavon-Borja et al. 2013). Such selection pressure is thought to predominantly occur in immune-related genes, enforced by pathogens targeting these proteins, resulting in adaptations on both the host and the pathogen side, known as the Red Queen hypothesis (Van Valen 1973).
Although circumstantial, together these three points, have contributed to the notion that at least a subset of TRIM proteins may have evolved to facilitate immune-related functions. In recent years, systematic TRIM expression and ablation screens have indeed further substantiated this hypothesis. Two expression screens in HEK-293 T cells with cDNAs for all human TRIMs and two TRIMs unique to rodents identified that 40–50% of these TRIM protein enhanced induction of NF-κB- and type I IFN-responsive reporters (Uchil et al. 2013, Versteeg et al. 2013). A substantial part of the hits reported in these screens have since then been validated by knockdown approaches (Rajsbaum et al. 2014a, Versteeg et al. 2014).
Although these exogenous expression approaches are more prone to artifacts compared with ablation studies, they overcome the issue that most cell types only express substantial mRNA levels for about half of the TRIMs (Versteeg et al. 2014). Thus, exogenous expression may have a higher discovery rate at the price of potential artifacts, whereas systematic analysis of TRIM function by ablation may have lower discovery rates by missing functions of TRIMs not expressed in the investigated cell type. Nevertheless, recent knockdown studies from Vojo Deretic’s group in HeLa cells and the monocyte cell line THP-1 indicate that a substantial proportion of TRIM proteins in these cells may play important roles in mediating autophagy in these cell types (Mandell et al. 2014a, Kimura et al. 2016). A link between this TRIM-dependent autophagy and immune-dependent functions has been described in a recent paper from the same group, which suggests that some of the well-studied immune-regulatory TRIMs may dampen immune-related cell signaling by targeting their substrates for lysosomal degradation (Kimura et al. 2015).
Together, these findings indeed implicate a considerable number of TRIMs in immune-related functions. Yet, how some of these TRIM proteins achieve this mechanistically, how stimulatory and repressive cell signaling are negotiated, and how these TRIMs themselves are activated remains largely unknown.
Pathogens actively antagonize TRIM proteins
Of the points raised above, particularly the fact that pathogens may target some of the TRIMs is a strong indicator that these TRIMs are important for fighting these pathogens by either (1) restricting them directly through anti-pathogen effector functions, or (2) indirectly through boosting immune-signaling. This stems from the idea that pathogens dedicating genome space to encoding such antagonists, and actively competing with the host resulting in positive selection in both the pathogen (antagonist) and host (Trim) genes, are strong indicators that these host factors are biologically important for the anti-pathogen response. Below we will highlight two well-studied TRIMs – TRIM5α and TRIM25 – which both have been under positive selection pressure and exemplify TRIMs with direct antiviral effector potential and immune-regulatory functions, respectively.
TRIM5 mediates direct viral restriction, inflammatory signaling, and autophagy
TRIM5α is one of the best-studied TRIMs and recognized for a long time as a factor with intrinsic retroviral restriction activity (Figure 7(a)). It was first identified as the major factor which renders rhesus macaque (Rh) cells resistant to infection by HIV, using a screen for Rh cDNA clones which converted infection-permissive human HeLa cells to being resistant to HIV infection (Stremlau et al. 2004). Subsequently, human TRIM5α was identified to render human cell lines resistant to infection with SIV from Rh and various other animal species (Hatziioannou et al. 2004, Keckesova et al. 2004, Perron et al. 2004, Stremlau et al. 2004). TRIM5 knockdown in these systems rescued virus restriction, indicating that TRIM5 is the major restriction factor responsible.
Figure 7.
TRIMs in the regulation of the immune response. (a) TRIM5α restricts retroviral infection. The TRIM5α SPRY domain interacts with the retroviral capsid lattice, which has two distinct consequences. First, TRIM5α recruits the autophagy machinery for degradation of the capsid. Second, TRIM5α in conjunction with UEV1A/UBC13 generates unanchored Lys 63-linked ubiquitin chains, which mediates TAK1 transactivation and subsequent NF-κB activation. (b) TRIM25 positively controls IFNβ production. By generating Lys 63-linked ubiquitin chains, TRIM25 releases RIG-I from its repressed conformation, which results in RIG-I tetramerization and subsequent IFNβ production. (c) TRIM6 controls type I interferon production and signaling, by generating Lys 48-linked, unanchored ubiquitin chains, which activate IKKɛ. Nipah virus antagonizes interferon signaling by targeting TRIM6 for degradation. (d) TRIM65 activates the cytoplasmic dsRNA sensor MDA5 by conjugation of Lys 63-linked ubiquitin chains on MDA5 Lys 743. TRIM65 is critical for MDA5 oligomerization and activation. Similar to TRIM56, Salmonella SopA can also interact with TRIM65 and mediate its degradation. However, unlike TRIM56, SopA does not interfere with TRIM65 E3 activity. (e) TRIM56 controls the STING-dependent cytosolic dsDNA response pathway by ubiquitinating STING with Lys 63-linked ubiquitin chains on Lys 150. Ubiquitination allows for STING dimerization, which is crucial for its activation. Salmonella SopA has been shown to bind and ubiquitinate TRIM56, thereby inhibiting it through preventing E3 ligase activity and degradation, respectively (see color version of this figure at www.tandfonline.com/ibmg).
Interestingly, four major TRIM5 isoforms have been identified, but from overexpression studies it has become clear that only the longest isoform – TRIM5α – is able to block retroviral infection. This is the only isoform containing a C-terminal SPRY domain, underscoring the importance of this domain for restriction (Stremlau et al. 2004). Subsequent detailed evolutionary analysis of the differences in human and simian TRIM5α SPRY domains, combined with mutagenesis approaches, identified that a 13 aa stretch in the SPRY domain determines the ability to interact with the viral capsid lattice (Sawyer et al. 2005). This region has been under strong positive selection in different primate species, and thus explains why humans can restrict SIV from Rh, and vice versa, but are antagonized by mutations in this SPRY region in viruses from the matching species. Importantly, TRIM5α and the viral capsid do not associate through standard one-to-one protein interactions. As described in the section above, several studies indicate that TRIM5α forms a hexameric ringed “net”, and that this is required for viral restriction (Ganser-Pornillos et al. 2011, Li et al. 2016) (Figure 7(a)). Interestingly, the SPRY domain – which facilitates capsid binding – is not required for these higher order structures to form, although the presence of a viral capsid enhances TRIM5α oligomer formation (Ganser-Pornillos et al. 2011, Li et al. 2016). Combined, these observations suggest that both, binding of the capsid lattice by the SPRY domain, in combination with formation of a higher order TRIM5α “net”, are essential for restriction factor functionality.
The technical challenge imposed by the fact that TRIM5α and the viral capsid only interact in higher order structures has hampered detailed analysis on the mechanism by which TRIM5α achieves viral restriction. However, infection experiments indicate that the restriction occurs in a step post-entry, yet before reverse transcription, indicating that TRIM5α targets the incoming capsid (Keckesova et al. 2004, Stremlau et al. 2004). It is known that individual retrovirus species have different uncoating dependencies and timings, some releasing their capsid early after infection, whereas others retaining it up to reverse transcription. Also, studies with capsid mutants that form normal virus particles, but fail to support reverse transcription (Craven et al. 1995, Alin and Goff 1996, Cairns and Craven 2001) suggest that structural rearrangements in the viral core may be a commonly required step to proceed to DNA synthesis. One popular model by which TRIM5α is thought to restrict the incoming viral particle is to alter the speed of uncoating and capsid disassembly, thereby altering the kinetics of structural rearrangements in the capsid required to proceed to reverse transcription. It has been reported that both increased and decreased capsid disassembly kinetics can be achieved, which are both detrimental for viral infectivity (Li et al. 2009).
Moreover, the recent studies which identified TRIM proteins as important autophagy facilitators have put forward enticing evidence that TRIM5α recruits the autophagy machinery to the viral capsid and targets it for degradation, thereby imposing viral restriction (Mandell et al. 2014a,b). Further studies will be required to determine what the contribution is of these described mechanisms in HIV restriction. Yet, what they all have in common is that their function is independent of TRIM5α RING activity, which has raised the question what the function of TRIM5α E3 activity is.
The TRIM5 RING domain has been shown to confer E3 ligase activity for auto-ubiquitination (Xu et al. 2003), which targets TRIM5 itself to the proteasome for degradation (Diaz-Griffero et al. 2006, Rold and Aiken 2008). Mutation of E2 binding residues in the RING domain prevented this (Lienlaf et al. 2011). In contrast, to date ubiquitination of viral capsids has not been detected, although one should bear in mind that this may stem from technical difficulties imposed by the rapid turnover of TRIM5 and capsids, and the low number of capsids per cell. Although various studies have addressed whether this auto-ubiquitination, and/or putative ubiquitination of the incoming viral capsid is important for viral restriction, conflicting results have been reported. This has hampered formulating a unifying model of the role of RING activity for TRIM5α function.
Mutants in the critical zinc-coordinating cysteine residues in the TRIM5 RING domain have been reported to have substantially reduced antiviral activity compared with wild-type, yet upon overexpression retains substantial ability to restrict viral infection (Xu et al. 2003, Stremlau et al. 2004, Javanbakht et al. 2005). One possible explanation for these findings may be that TRIM5α facilitates viral restriction at multiple steps of the viral life cycle. In agreement with this notion, RING mutants have been reported to disrupt restriction by premature uncoating and blocking reverse transcription, but do not affect restriction in later stages of infection that culminate in integration in the host genome (Wu et al. 2006, Roa et al. 2012). In contrast, RING domain mutants of TRIM5α have been reported which did not have diminished restrictive capabilities (Stremlau et al. 2006). Although the exact reason for this remains to be seen, it is feasible that binding with certain capsids is so strong that this fully restricts them independently of E3 ligase activity.
In addition to a role in viral restriction, E3 ligase activity of TRIM5α has been implicated in establishing NF-κB-dependent pro-inflammatory cell signaling upon viral capsid recognition and formation of the above-described TRIM5α multi-hexameric net on the viral capsid lattice (Pertel et al. 2011). Interestingly, for this, TRIM5α has been reported to work in conjunction with the UBC13/UEV1A E2 ligase complex to form Lys 63-linked ubiquitin chains. These chains are currently thought to recruit the kinase TAK1, ultimately resulting in TAK1 transactivation (Pertel et al. 2011). Interestingly, cell-free ubiquitination assays suggest that the Lys 63-linked ubiquitin chains may be unanchored, and thus trans-activate TAK1 by generating a local high concentration of these chains to facilitate TAK1 complex formation and subsequent activation.
No good animal models for HIV infection in vivo exist, thus making it difficult to determine what the impact of this NF-κB-dependent response is for viral infection. One should bear in mind that the HIV LTR contains two NF-κB response sites important for transcription; inhibition of the NF-κB response by a dominant negative form of its inhibitor IκBα has been reported to inhibit virus infection in T cells (Kwon et al. 1998, Quinto et al. 1999). Thus, it could be that signaling initiated from TRIM5α could be beneficial for HIV transcription. However, it should be noted that knockdown of TAK1 in a monocyte cell line abrogated TRIM5α-dependent viral restriction, suggesting that TAK1 and NF-κB signaling in this system are required for antiviral function.
All in all, these findings firmly establish TRIM5 as a key viral restriction factor. Although TRIM5 has E3 ligase activity, the contribution of this to direct viral restriction has remained under debate. In addition, this E3 activity has been shown to facilitate inflammatory cell signaling, which could in part contribute to antiviral activity. Lastly, TRIM5 has been suggested to target viral capsids for autophagy-dependent degradation, thereby conferring restriction. Since TAK1 has an important role in autophagy induction (Dai et al. 2012), it is tempting to speculate that TRIM5 activation, and subsequent TAK1 activation in an E3-dependent manner, contributes to autophagy induction, and capsid destruction (Figure 7(a)). While it remains unknown for most TRIMs how they themselves are activated during innate immune triggering, this example of TRIM5 indicates that some of them may in fact have pattern-recognition receptor-like mechanisms, which directly couple PAMP binding to activation.
TRIM25 is critical for activation of the 5′-ppp-RNA sensor RIG-I
RIG-I is a cytoplasmic sensor for viral 5′-ppp-RNA, which upon activation induces cell signaling resulting in antiviral type I interferon and NF-κB-dependent pro-inflammatory cytokine production (Yoneyama et al. 2004, Hornung et al. 2006, Pichlmair et al. 2006). The critical nature of this receptor has been demonstrated in knockout mice, which are hyper-susceptible to infection with various RNA viruses (Kato et al. 2006). It was recognized early on that constitutively active forms of RIG-I are ubiquitinated by Lys 63-linked ubiquitin chains (Gack et al. 2007). Proteomics approaches by Michaela Gack and Jae Jung identified TRIM25 as the E3 ligase essential for the synthesis of these chains (Gack et al. 2007).
Experiments in MEFs from Trim25−/− mice demonstrated that TRIM25 is critical for RIG-I ubiquitination, and that this is indispensable for generating an antiviral state in cell culture infections (Gack et al. 2007) (Figure 7(b)). Mutation of Lys 172 in the RIG-I N-terminus abrogated its ubiquitination and activation. This led to the conclusion that covalently attached Lys 63-linked ubiquitin chains – the synthesis of which is dependent on TRIM25 – are responsible for RIG-I activation. Yet, whether TRIM25 directly synthesized these chains remained to be determined.
Follow-up work from the group of James Chen reported for the first time full reconstitution in vitro of RIG-I activation and all downstream signaling up to activation of the transcription factor IRF3, which allow for detailed study of the molecular mechanism of RIG-I activation (Zeng et al. 2010). This work demonstrated that in a cell-free system, TRIM25 directly synthesizes Lys 63-linked ubiquitin chains, which activate RIG-I. However, unexpectedly in this cell-free system, these chains were determined not to be covalently attached to RIG-I, which led the authors to conclude that unanchored Lys 63-linked ubiquitin chains facilitate RIG-I activation (Zeng et al. 2010). It has remained debated whether these chains exist in cells and fulfill the same function, or whether these chains are only observed and functional in an in vitro setting. Additional biochemical studies demonstrated that unanchored Lys 63-linked ubiquitin chains synthesized by TRIM25 could confer RIG-I tetramerization, which was determined to be the active form able to mediate downstream cell signaling (Jiang et al. 2012).
Recently, the crystal structure of the tetrameric RIG-I N-terminal domains bound by three unanchored Lys 63-linked di-ubiquitins was solved (Peisley et al. 2014). This structure and additional biochemical analyses indicated that RIG-I represented the activated state, adopting a “lock-washer” arrangement, where the four tetramer subunits form a helical structure in a way that the first and last subunit are off-set along the axis perpendicular to their turn, and three Lys 63-linked ubiquitin chains decorate the outer rim of the tetrameric helix (Peisley et al. 2014) (Figure 7(b)). Further mutational analysis indicated that RIG-I signaling activity can be equally achieved by unanchored and covalently attached Lys 63-linked ubiquitin chains, yet supported the notion that covalent Lys 63-linked ubiquitin chains near its binding sites in the tetramer increases the stability of the RIG-I oligomer (Peisley et al. 2014).
As for most other TRIMs, mutagenesis studies have identified the TRIM25 coiled-coil to be essential for multimerization and activity (Gack et al. 2007). This is exemplified by the observation that a TRIM25 mutant lacking the coiled-coil cannot confer an antiviral state and fails to protect from viral infection (Gack et al. 2007). In further support of a critical role of TRIM25 in RIG-I activation and establishing an antiviral state, influenza A virus has been shown to antagonize TRIM25 directly by interfering with coiled-coil-dependent oligomerization (Gack et al. 2009).
It is thought that all currently circulating viruses have antagonists targeting the type I interferon system (Versteeg and Garcia-Sastre 2010). In fact, many viruses encode multiple antagonists, and/or target different steps in the IFN induction and/or signaling cascade (Versteeg and Garcia-Sastre 2010). Influenza A viruses encode a dedicated IFN-antagonist: non-structural protein 1 (NS1). From studies with recombinant viruses lacking NS1 it has been long known that this viral protein is essential for infection in IFN-competent cells, yet dispensable in IFN-deficient cells (Garcia-Sastre et al. 1998). The mechanism of action has been previously attributed to sequestration of dsRNA (Lu et al. 1995), and in most influenza A virus strains by interfering with host pre-mRNA processing through blocking function of Cleavage and Polyadenylation Specificity Factor 30 (CPSF30) (Nemeroff et al. 1998). These two distinct antagonist functions critically rely on binding of NS1 to its two targets, involving different non-overlapping residues in the NS1 N- and C-terminus (Hale et al. 2008).
Influenza A virus mutants without NS1 are strong type I interferon inducers, which is dependent on recognition of viral RNA by RIG-I (Kato et al. 2006). Thus, it is not surprising that NS1 was found to antagonize RIG-I activation. The molecular mechanism is independent from the previously described modes of blocking the innate immune response, and involves NS1 binding to the TRIM25 coiled-coil, thereby preventing its multimerization and ability to activate RIG-I (Gack et al. 2009). Binding to the TRIM25 coiled-coil is critically dependent on glutamate residues 96 and 97 in NS1, which are different from the key residues for dsRNA and CPSF30 binding (Hale et al. 2008). The importance of the ability to antagonize TRIM25 activity is exemplified by the fact that recombinant influenza A virus harboring E96/97A substitutions in its NS1 protein is non-pathogenic (Gack et al. 2009). Mice infected with this mutant virus did not lose weight or develop disease, and were indistinguishable from non-infected animals (Gack et al. 2009). In contrast, mice infected with the wild-type virus counterpart rapidly lost weight after day 2, and succumbed by day 5 post-infection. Together, these studies demonstrate that TRIM25 is an E3 ligase, which is a key for innate immune activation. This is strengthened by the discovery that influenza A viruses specifically target TRIM25 as a means to antagonize the type I interferon response.
TRIM6 controls the type I interferon signaling response
The short arm of human chromosome 11 harbors a hotspot of Trim genes, many of which have been implicated in immune-related functions. This locus includes e.g. the Trim5 gene, but also Trim6, which until recently had remained uncharacterized. The location of the Trim6 gene in this _Trim_-cluster had suggested that it may have immune-related functions. In agreement with this, we identified TRIM6 in an exogenous expression screen as a family member, which potently induced interferon-responsive reporters (Versteeg et al. 2013).
Follow-up studies in mice, cell culture, and in cell-free systems demonstrated that Trim6 ablation attenuates signaling downstream of the type I interferon receptor, abrogates proper antiviral responses, and increases susceptibility to viral infection (Rajsbaum et al. 2014b). Mechanistically, TRIM6 was demonstrated to synthesize Lys 48-linked ubiquitin chains, and that these were essential for function. However, unexpectedly these chains did not result in proteasomal degradation, but were in fact sufficient to activate the IKKɛ kinase in vitro, which is important for the antiviral response downstream of the type I interferon receptor (Tenoever et al. 2007, Rajsbaum et al. 2014b; Figure 7(c)). Unexpectedly, the Lys 48-linked ubiquitin chains were unanchored, and not covalently attached to the IKKɛ kinase (Rajsbaum et al. 2014b). To the best of our knowledge, this is the first evidence that breaks with the long-standing dogma that Lys 48-linked ubiquitin chains are exclusively involved in proteasomal degradation. Interestingly, recent evidence indicates that TRIM6 is targeted by certain members of the Paramyxoviridae family, adding to the notion that TRIM6 is important for the antiviral response (Bharaj et al. 2016).
Most – if not all – members of the Paramyxoviridae encode within their P gene antagonists, which interfere with signal transduction downstream of the type I interferon receptor. Recently, the Rajsbaum lab discovered that a member of this virus family – the zoonotic, highly fatal Nipah virus – antagonizes interferon signaling by targeting TRIM6 for degradation (Bharaj et al. 2016). However, this antagonism was not mediated by one of the P gene-encoded proteins, but unexpectedly by the matrix protein. This antagonism is exemplified by the observation that infection with wild-type Nipah virus resulted in TRIM6 degradation and perturbed interferon signaling, whereas this was not the case with a virus harboring a mutant matrix-encoding gene. Interestingly, cell culture experiments showed that related henipaviruses can also bind and antagonize TRIM6, suggesting that this antagonism strategy may well be shared within this virus genus.
Together, these studies underpin that TRIM proteins may regulate immune signaling in thus far unique mechanisms, as illustrated by the non-degradative unanchored Lys 48-linked ubiquitin chains synthesized by TRIM6. The fact that viruses encode specific antagonists targeting this TRIM member, highlights its biological importance in the innate immune response.
TRIM65 activates the cytoplasmic dsRNA sensor MDA5
Even though it has been known for a while that RIG-I requires TRIM25-dependent poly-ubiquitination for activation, the dependence on ubiquitination for activation had remained unclear for the related, other major cytoplasmic RNA sensor MDA5. Studies with RIG-I and MDA5 knockouts previously established that while RIG-I is critical for interferon induction by some viruses (e.g. most Ortho- and Paramyxoviridae), other viruses (e.g. Picornavididae such as encephalo-myocarditis virus (EMCV)) are exclusively recognized by MDA5 (Kato et al. 2006).
Recent studies identified TRIM65 to interact with MDA5 and facilitate Lys 63-linked ubiquitin chain formation on Lys 743 in the MDA5 helicase domain, which is essential for MDA5 oligomerization and activation (Lang et al. 2017; Figure 7(d)). EMCV infections in Trim65−/− mice and bone marrow macrophages derived from it, convincingly demonstrated loss of proper type I interferon induction, underpinning the importance of this TRIM for innate immune induction. This is further solidified by the observation that Trim65−/− mice did not mount a substantial innate immune response and succumbed significantly faster to EMCV infection (Lang et al. 2017). The specificity of the effect of TRIM65 on MDA5 is exemplified by the fact that RIG-I-specific induction was not affected in the knock outs, and infections with a virus recognized by RIG-I induced similar type I interferon responses in wild-type and Trim65−/− mice.
To date, no viral antagonist targeting MDA5 has been reported. However, two groups independently identified the SopA effector from Salmonella typhimurium to specifically interact with the RING domain of TRIM65, but not other TRIMs such as TRIM5, TRIM25, or TRIM62 (Kamanova et al. 2016, Fiskin et al. 2017) (Figure 7(d)). This Salmonella effector molecule is a HECT-like E3 ubiquitin ligase, which is injected into host cells, and there acts as a virulence factor and contributes to pathogenicity. Both studies consistently reported ubiquitination of TRIM65 by SopA, yet the biological relevance of this SopA-interaction and ubiquitination will require further investigation, as both reports suggest seemingly opposite effects.
Interferon reporter assays in HEK293T cells in one study indicated that wild-type SopA enhanced MDA5- and TRIM65-dependent reporter activation by two-fold (Kamanova et al. 2016). In addition, transfection in HEK293T cells of neither wild-type nor a catalytically inactive SopA mutant affected TRIM65 expression, from which the authors concluded that SopA does not mediate TRIM65 degradation.
In contrast, an independent study by Ivan Dikic’s team found that doxycycline inducible expression in HeLa cells of SopA, but not of a catalytic mutant, degraded endogenous TRIM65 (Fiskin et al. 2017). The fact that TRIM65 protein levels were stabilized by MG132 treatment suggested that this degradation is likely proteasome-dependent. In agreement with this, infection of HCT116 cells with wild-type SopA-expressing Salmonella degraded endogenous TRIM65, whereas a bacterial SopA HECT mutant did not. All in all, these studies indicate that TRIM65 is critical for MDA5 ubiquitination, activation, and antiviral control. Moreover, Salmonella may target this TRIM through SopA injection, although additional work is required to determine whether SopA can indeed inhibit MDA5 activation, and what the implications of this would be for bacterial infection. In the same studies, very similar results were reported for SopA and TRIM56. This TRIM has been identified as an activator of the cellular response to cytosolic dsDNA (Tsuchida et al. 2010).
TRIM56 the STING-dependent cytosolic dsDNA response pathway
On one hand, multiple cytosolic receptors detecting intra-cellular dsDNA have been described (Wu and Chen 2014). Inflammasome activation and IL1β production is one of the major outcomes of recognition by certain receptors (Wu and Chen 2014). On the other hand, cyclic GMP-AMP (cGAMP) synthase (cGAS) is a cytoplasmic sensor for viral and bacterial DNA which triggers activation of the kinase TBK1, and subsequent type I interferon production, and NF-κB-dependent pro-inflammatory cytokine production (Sun et al. 2013). While TBK1 activation and cytokine output are identical to the RIG-I pathway, the signaling between cGAS and TBK1 is unique and critically relies on the ER-resident transmembrane protein stimulator of interferon genes (STING) (Ishikawa and Barber 2008).
STING can be directly activated by binding bacterial cyclic di-nucleotides (Burdette et al. 2011). Moreover, upon DNA recognition cGAS synthesizes the second messenger cGAMP, which is subsequently recognized by STING. These di-nucleotides are recognized by STING only in its dimeric form (Shu et al. 2012). Hence, STING dimer formation is critical for activation.
In agreement with a critical role for this pathway in pathogen defense, STING-deficient mice have increased susceptibility to DNA viruses such as herpes simplex virus (Ishikawa et al. 2009), and cells from these mice fail to induce an interferon response upon infection with this virus or the intra-cellular bacterium Listeria monocytogenes (Lm).
STING has been implicated in autoimmune disease development (Ahn et al. 2014), which underpins that under physiological conditions it is important to tightly regulate STING activation. Such control is often exerted at various levels by post-translational modifications. In this context, TRIM56 has been shown to ubiquitinate STING with Lys 63-linked ubiquitin chains on Lys 150, which is in close proximity to the start of the cyclic-di-nucleotide binding domain (Tsuchida et al. 2010; Figure 7(e)). Substitution of this residue by arginine abolished STING ubiquitination, and its ability to induce type I interferon.
Mechanistically, STING ubiquitination by TRIM56 was reported to be required for STING dimerization, since differentially tagged STING K150R mutants were no longer able to interact upon overexpression in HEK293T cells (Tsuchida et al. 2010). As expected, these mutants were no longer able to recruit TBK1, indicating that TRIM56-dependent ubiquitination is required for kinase activation.
The crystal structure of the dimeric C-terminal half of STING (aa 152–343) has been solved by two independent groups (Ouyang et al. 2012, Shu et al. 2012). Although these structures did not include the ubiquitinated Lys 150 residue, mutagenesis studies revealed that in contrast to what was reported previously in the context of TRIM56 (Tsuchida et al. 2010), mutations in Lys 150 did not affect STING dimerization or activity in interferon reporters, whereas a mutation preventing dimerization, as determined by size exclusion chromatography (G158L), did negate STING activity (Ouyang et al. 2012). Further detailed studies will be required to elucidate exactly how TRIM56-dependent ubiquitination on Lys 150 facilitates STING activation. These recent biochemical analyses may indicate that ubiquitination is not essential for dimerization per se, but could indicate that TRIM56-dependent control of STING is more complex than just Lys 150 ubiquitination.
Interestingly, Salmonella SopA has been shown to bind and ubiquitinate TRIM56 (Kamanova et al. 2016, Fiskin et al. 2017; Figure 7(e)). As is the case for TRIM65, the reported effects of this on TRIM56 stability and function remains debated. Of note, the co-crystal structure of the TRIM56 RING domain in complex with SopA indicated that SopA occupies in part residues important for E2 interaction with the TRIM56 RING domain. In agreement with the hypothesis that SopA could be a competitive inhibitor of TRIM56 E3 activity, molar excess of SopA inhibited TRIM56-dependent ubiquitin chain formation in vitro.
In conclusion, current data indicate that TRIM56 is required for STING activation. Since STING signaling is critical for mounting a cytokine response upon infection of certain bacterial species (e.g. Listeria monocytogenes and Mycobacterium tuberculosis), it is interesting that the SopA effector of Salmonella can interact, and possibly inhibit TRIM56 action. Yet, thus far neither cGAS, nor STING have been reported as required for the cellular response to Salmonella (Owen et al. 2016), and it thus remains to be determined what the role of SopA is in relation to TRIM56 for Salmonella infection.
TRIM21 is a cytoplasmic antibody receptor
TRIM21/Ro52 has been long implicated in immune regulation since auto-antibodies against TRIM21 have been detected in patients with autoimmune diseases including Systemic lupus erythematosus and Sjögren’s syndrome (Wahren et al. 1998). Subsequently, biochemical and cell-based studies determined that TRIM21 has IgG Fc-binding properties in its PRY-SPRY domain, which allows it to act as a cytoplasmic Fc antibody receptor (James et al. 2007, Rhodes and Trowsdale 2007, Keeble et al. 2008, Mallery et al. 2010, Fletcher et al. 2015, Rakebrandt et al. 2014, Watkinson et al. 2015).
Experiments in Trim21−/− mice and cells indeed showed that TRIM21 is required for efficient response to antibody-opsonized bacteria and viruses (Mallery et al. 2010). Mechanistically, TRIM21 was shown in these studies to ubiquitinate viral proteins and target them for proteasomal degradation. This restriction appears to occur on incoming particles before translation of viral genes, suggesting that humoral immunity can provide protection through targeting viruses for degradation in the cytoplasm.
Additionally, TRIM21-dependent recognition of antibody-opsonized pathogens has been suggested to trigger cytokine expression by TRIM21-dependent synthesis of Lys 63-linked ubiquitin chains which facilitate TAK1-depdent NF-κB activation (McEwan et al. 2013). However, this reported function has been debated since two independently generated Trim21−/− mouse lines showed highly dissimilar effects on cytokine production (Espinosa et al. 2009, Yoshimi et al. 2009, Vaysburd et al. 2013). Thus far, it has remained unclear where these differences stem from, although differences in knockout alleles (Ozato et al. 2009), stimuli, and cell culture versus responses in whole animals have been proposed (Espinosa et al. 2009, Yoshimi et al. 2009).
For example, one study reported that Trim21−/− MEFs produced increased levels of pro-inflammatory cytokines, whereas bone marrow macrophages and dendritic cells from the same Trim21−/− mice did not (Yoshimi et al. 2009). In contrast, a systemic autoimmune phenotype consistent with an exacerbated Th17 profile was reported in independently generated Trim21−/− mice (Espinosa et al. 2009).
Together, these studies position TRIM21 as a critical cytoplasmic immune-globulin receptor. Knockout mice consistently indicate TRIM21 as a critical factor for restricting antibody-opsonized pathogens which retain these antibodies upon entry into the cytoplasm. However, whether TRIM21 also has a secondary effect on cytokine output remains debated, and the data currently suggest that if this role exists, it may be specific to certain cell types, or for certain stimuli and associated pathways.
It should be noted that TRIM21 shows interesting parallels with TRIM5α as described above: (1) it operates directly as a pattern recognition receptor and a pathogen restriction factor, and (2) it may additionally control pro-inflammatory output by synthesis of Lys 63-linked ubiquitin chains as a means of TAK1-dependent NF-κB activation. All in all, this indicates that additional studies to determine the role of TRIM21 in different cell types and signaling pathways in vivo are essential, but equally so in vitro studies to address how TRIM21 activation could be mechanistically coupled to driving an inflammatory response.
TRIM62 regulates the anti-fungal response downstream of C-type lectin receptors
Until recently, available data had predominantly implicated different TRIM proteins as regulators of pathways classically recognized as important for antiviral control, and direct viral restriction activity (Rajsbaum et al. 2014a, Versteeg et al. 2014). This had raised the question whether Trim genes have predominantly expanded to counter virus infection. However, a recent study identified TRIM62 as a critical regulator of anti-fungal activity through C-type lectin receptors (Cao et al. 2015), thereby demonstrating that at least some TRIMs regulate the response to classes of pathogens other than viruses.
Genome-wide association studies have indicated that alleles of CARD9, a critical adaptor for CLR signaling, exist that can be either protective, or pre-disposing to inflammatory bowel disease (Rivas et al. 2011, Beaudoin et al. 2013). This has suggested that different functional states of CARD9 could drive differential expression of pro-inflammatory cytokines. By proteomics approaches, TRIM62 was discovered to ubiquitinate Lys 125 in CARD9 with Lys 27-linked chains. Consistent with a critical role for this residue, reconstitution of Card9−/− bone marrow dendritic cells with CARD9 bearing a K125R mutation did not rescue CLR-induced cytokine production (Cao et al. 2015). Likewise, Trim62−/− mice had increased susceptibility to Candida albicans fungal infection. In this context, Trim62 ablation increased fungal titers in various organs such as kidney, spleen and liver in the face of decreased CARD9-dependent systemic IL-6 levels.
Together, these findings position TRIM62 as a critical regulator of CARD9-dependent anti-fungal responses. It remains to be investigated whether more TRIMs regulate the CLR pathway, yet this finding indicates that TRIM-dependent regulation extends beyond control of just antiviral pathways. Moreover, although this study demonstrated the importance of Lys 27-linked ubiquitin chains on CARD9, how these non-canonical chains facilitate CARD9 activation at the molecular level remains to be established. Interestingly, other TRIMs predominantly rely on synthesis of Lys 48- or Lys 63-linked ubiquitin chains for function, which may indicate that Lys 27-linked ubiquitin chains synthesized by TRIM62 underlie a unique mechanism of target activation. However, it is not the only TRIM that produces this chain type: also TRIM23 has been reported to control NEMO in a Lys 27-linked ubiquitin chain-dependent manner (Arimoto et al. 2010).
Outlook: TRIMs can be dual-edged swords with an impact on immune and cancer biology
In this section of the review, we focused on TRIM-dependent immune regulation, and means by which these proteins are targeted by certain pathogens. However, many TRIMs have additionally reported functions in immune-unrelated processes, which suggests that some of these family members may be multi-functional (Ozato et al. 2008, Rajsbaum et al. 2014a, Versteeg et al. 2014). For example, various members of the TRIM family have also been implicated in cancer biology (Hatakeyama 2011, 2017). There are some TRIMs, which have exclusively been implicated in either immune regulation or cancer, yet several of them have been implicated in both (Hatakeyama 2011, 2017). There are several possibilities, which may underlie this seeming dichotomy, which exemplify that our current knowledge on how TRIMs determine target specificity, and affect signaling in different cell types is still limited.
First, TRIMs can control multiple distinct molecular targets in RING-dependent and independent manners, and by these means affect distinct cellular pathways. In this context, RING-less TRIM29 has been reported to dampen NF-κB activation in the cytoplasm by degrading the essential adaptor NEMO (Xing et al. 2016), whereas it can also fulfill a role in the nucleus by marking DNA damage sites for repair (Masuda et al. 2015).
Second, several TRIMs control cell signaling molecules which are critical for immune signaling, but which can also contribute to cancer biology if deregulated. In this context, NF-κB and STAT3 are both regulated by TRIMs, which depending on the cell type can mediate the pro-inflammatory response (Hatakeyama 2011, 2017).
Third, TRIM isoforms can have differential functions. This is well illustrated by TRIM19/PML, which has seven different isoforms. Mainly the nuclear isoforms contribute to leukemia by regulating transcriptional output (Nisole et al. 2013). Moreover, viral restriction by nuclear PML bodies has been attributed to these isoforms (Scherer and Stamminger 2016). In contrast, a shorter cytoplasmic isoform regulates TGFβ signaling (de Figueiredo-Pontes et al. 2011).
Lastly, autophagy has been implicated in both cancer biology and immune regulation (Hatakeyama 2017). Since recent evidence suggests that various TRIMs may be context-specific autophagy adapters, it may well be that some of the processes regulated by autophagy in this context could contribute to deregulated cell control with impact on cell division (Mandell et al. 2016), or cytokine output (Kimura et al. 2015, 2016).
Taken together, the combined body of work on TRIM proteins indicates that many of them have E3 ligase activity and by that feature regulate various cellular pathways. Many of them have reported functions in activating or dampening immune signaling. Some of these functions could be dependent on their ability to regulate autophagy, or to be regulated in autophagy-dependent manners. Exactly how these TRIMs ultimately determine cellular output at the molecular level remains in many cases unknown and will require more in depth biochemical, structural, and bio-physical analysis with recombinant TRIM proteins and their substrates. Importantly, some TRIMs may also contribute to cell signaling in E3 ligase-independent capacities, which may suggest that these RING-containing proteins may have conceptual parallels with many kinases, which can exert kinase domain-dependent and -independent roles in various cell types (Rauch et al. 2011).
Concluding remarks
Ubiquitination plays a critical role in the regulation of immune responses. We understand by now that different types of ubiquitination impact on the substrate’s fate, leading to control various downstream signaling pathways. Since ubiquitin enzymes are key players in the ubiquitination process, it is important to further elucidate the underlying regulatory mechanisms. Especially how each type of ubiquitin chain is generated, hydrolyzed, and recognized are the key points. An additional layer of regulation is put forward by post-translational modifications of ubiquitin itself by phosphorylation and acetylation; more studies are required to clarify if these types of ubiquitin signals are involved in the immune response, which is not understood to date.
Acknowledgements
The authors thank all the members in Ikeda Lab at IMBA (Vienna, Austria) for scientific discussions on the topics. The authors apologize to the authors whose papers we could not cite due to space limitations.
Funding Statement
F. I. is supported by the ERC consolidator grant (LUbi, 614711), the FWF standalone grant (P 25508), COST (European Cooperation in Science and Technology, PROTEOSTASIS BM1307), and Austrian Academy of Sciences. G.A.V. is supported by the FWF Stan-Alone grant (P 30231), and the FWF Doctoral Programme (W1261).
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Abramson J. and Goldfarb Y., 2016. AIRE: from promiscuous molecular partnerships to promiscuous gene expression. European journal of immunology, 46, 22–33. [DOI] [PubMed] [Google Scholar]
- Aggarwal B.B., 2003. Signalling pathways of the TNF superfamily: a double-edged sword. Nature reviews immunology, 3, 745–756. [DOI] [PubMed] [Google Scholar]
- Ahn J., Ruiz P., and Barber G.N., 2014. Intrinsic self-DNA triggers inflammatory disease dependent on STING. Journal of immunology, 193, 4634–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aireakiyama T., et al, 2008. The tumor necrosis factor family receptors RANK and CD40 cooperatively establish the thymic medullary microenvironment and self-tolerance. Immunity, 29, 423–437. [DOI] [PubMed] [Google Scholar]
- Akiyama T., et al, 2005. Dependence of self-tolerance on TRAF6-directed development of thymic stroma. Science, 308, 248–251. [DOI] [PubMed] [Google Scholar]
- Alin K. and Goff S.P., 1996. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology, 222, 339–351. [DOI] [PubMed] [Google Scholar]
- Anderson M.S., et al, 2002. Projection of an immunological self shadow within the thymus by the aire protein. Science, 298, 1395–1401. [DOI] [PubMed] [Google Scholar]
- Arimoto K., et al, 2010. Polyubiquitin conjugation to NEMO by triparite motif protein 23 (TRIM23) is critical in antiviral defense. Proceedings of the national academy of sciences of the United States of America, 107, 15856–15861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka T. and Ikeda F., 2015. New insights into the role of ubiquitin networks in the regulation of antiapoptosis pathways. International review of cell and molecular biology, 318, 121–158. [DOI] [PubMed] [Google Scholar]
- Ashkenazi A. and Salvesen G., 2014. Regulated cell death: signaling and mechanisms. Annual review of cell and developmental biology, 30, 337–356. [DOI] [PubMed] [Google Scholar]
- Bachmaier K., et al, 2000. Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature, 403, 211–216. [DOI] [PubMed] [Google Scholar]
- Balagopalan L., et al, 2007. c-Cbl-mediated regulation of LAT-nucleated signaling complexes. Molecular cell biology, 27, 8622–8636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin M., et al, 2013. Deep resequencing of GWAS loci identifies rare variants in CARD9, IL23R and RNF186 that are associated with ulcerative colitis. PLoS genetics, 9, e1003723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell J.L., et al, 2012. TRIM16 acts as an E3 ubiquitin ligase and can heterodimerize with other TRIM family members. PLoS One, 7 , e37470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C.L., et al, 2001. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nature genetics, 27, 20–21. [DOI] [PubMed] [Google Scholar]
- Bharaj P., et al, 2016. The matrix protein of Nipah virus targets the E3-ubiquitin ligase TRIM6 to inhibit the IKKepsilon kinase-mediated Type-I IFN antiviral response. PLoS pathogens, 12, e1005880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogaraju S., et al, 2016. Phosphoribosylation of ubiquitin promotes serine ubiquitination and impairs conventional ubiquitination. Cell, 167, 1636–1649.e13. [DOI] [PubMed] [Google Scholar]
- Bhoj V.G. and Chen Z.J., 2009. Ubiquitylation in innate and adaptive immunity. Nature, 458, 430–437. [DOI] [PubMed] [Google Scholar]
- Birgisdottir A.B., Lamark T., and Johansen T., 2013. The LIR motif – crucial for selective autophagy. Journal of cell science, 126, 3237–3247. [DOI] [PubMed] [Google Scholar]
- Bjorkoy G., et al, 2005. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. Journal of cellular biology , 171, 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair P.J., et al, 1994. CD4 + CD8− T cells are the effector cells in disease pathogenesis in the scurfy (sf) mouse. Journal of immunology, 153, 3764–3774. [PubMed] [Google Scholar]
- Boname J.M., et al, 2010. Efficient internalization of MHC I requires lysine-11 and lysine-63 mixed linkage polyubiquitin chains. Traffic, 11, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley M.J., et al, 2005. NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease. Journal of biological chemistry, 280, 11505–11512. [DOI] [PubMed] [Google Scholar]
- Bremm A., Freund S.M., and Komander D., 2010. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nature structural & molecular biology, 17, 939–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunkow M.E., et al, 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nature genetics, 27, 68–73. [DOI] [PubMed] [Google Scholar]
- Burdette D.L., et al, 2011. STING is a direct innate immune sensor of cyclic di-GMP. Nature, 478, 515–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns T.M. and Craven R.C., 2001. Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants. Journal of virology, 75, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z., et al, 2015. Ubiquitin ligase TRIM62 regulates CARD9-mediated anti-fungal immunity and intestinal inflammation. Immunity, 43, 715–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpino N., et al, 2009. The Sts proteins target tyrosine phosphorylated, ubiquitinated proteins within TCR signaling pathways. Molecular immunology, 46, 3224–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthagena L., et al, 2009. Human TRIM gene expression in response to interferons. PLoS One, 4 , e4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.H., et al, 2012. Ubc13 maintains the suppressive function of regulatory T cells and prevents their conversion into effector-like T cells. Nature immunology, 13, 481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J.H., et al, 2014. TRAF3 regulates the effector function of regulatory T cells and humoral immune responses. Journal of experimental medicine, 211, 137–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M., et al, 2011. The ubiquitin ligase Peli1 negatively regulates T cell activation and prevents autoimmunity. Nature immunology, 12, 1002–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V., et al, 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science, 243, 1576–1583. [DOI] [PubMed] [Google Scholar]
- Chen Z., et al, 2013. The ubiquitin ligase Stub1 negatively modulates regulatory T cell suppressive activity by promoting degradation of the transcription factor Foxp3. Immunity, 39, 272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.J., 2005. Ubiquitin signalling in the NF-kappaB pathway. Nature cell biology, 7, 758–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang Y.J., et al, 2000. Cbl-b regulates the CD28 dependence of T-cell activation. Nature, 403, 216–220. [DOI] [PubMed] [Google Scholar]
- Chiffoleau E., et al, 2003. TNF receptor-associated factor 6 deficiency during hemopoiesis induces Th2-polarized inflammatory disease. Journal of immunology, 171, 5751–579. [DOI] [PubMed] [Google Scholar]
- Chu Y., et al, 2011. B cells lacking the tumor suppressor TNFAIP3/A20 display impaired differentiation and hyperactivation and cause inflammation and autoimmunity in aged mice. Blood, 117, 2227–2236. [DOI] [PubMed] [Google Scholar]
- Ciani B., et al, 2010. Molecular basis of coiled-coil oligomerization-state specificity. Proceedings of the national academy of sciences of the United States of America, 107, 19850–19855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A. and Ben-Saadon R., 2004. N-terminal ubiquitination: more protein substrates join in. Trends in cell biology, 14, 103–106. [DOI] [PubMed] [Google Scholar]
- Cook W.J., et al, 1992. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2). Journal of biological chemistry, 267, 16467–16471. [DOI] [PubMed] [Google Scholar]
- Coornaert B., et al, 2008. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Natutre immunology, 9, 263–271. [DOI] [PubMed] [Google Scholar]
- Corcoran K., et al, 2011. Ubiquitin-mediated regulation of CD86 protein expression by the ubiquitin ligase membrane-associated RING-CH-1 (MARCH1). Journal of biological chemistry, 286, 37168–37180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven R.C., et al, 1995. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. Journal of virology, 69, 4213–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., et al, 2012. TAK1, more than just innate immunity. IUBMB life, 64, 825–834. [DOI] [PubMed] [Google Scholar]
- de Figueiredo-Pontes L.L., et al, 2011. Halofuginone has anti-proliferative effects in acute promyelocytic leukemia by modulating the transforming growth factor beta signaling pathway. PLoS One, 6, e26713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., et al, 2000. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell, 103, 351–361. [DOI] [PubMed] [Google Scholar]
- Deshaies R.J. and Joazeiro C.A., 2009. RING domain E3 ubiquitin ligases. Annual review of biochemistry, 78, 399–434. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero F., et al, 2006. Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology, 349, 300–315. [DOI] [PubMed] [Google Scholar]
- Diaz-Griffero F., et al, 2009. A B-box 2 surface patch important for TRIM5alpha self-association, capsid binding avidity, and retrovirus restriction. Journal of virology, 83, 10737–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikic I., Wakatsuki S., and Walters K.J., 2009. Ubiquitin-binding domains – from structures to functions. Nature reviews molecular cell biology, 10, 659–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H., et al, 2012. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nature structural & molecular biology, 19, 876–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou H., et al, 2013. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nature structural & molecular biology, 20, 982–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove K.K., et al, 2016. Molecular insights into RBR E3 ligase ubiquitin transfer mechanisms. EMBO reports, 17, 1221–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duwel M., et al, 2009. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. The journal of immunology, 182, 7718–7728. [DOI] [PubMed] [Google Scholar]
- Elliott P.R., et al, 2014. Molecular basis and regulation of OTULIN-LUBAC interaction. Molecular cell, 54, 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerich C.H., et al, 2013. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. Proceedings of the national academy of sciences of the United States of America, 110, 15247–15252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa A., et al, 2009. Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. Journal of experimental medicine, 206, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang D. and Liu Y.C., 2001. Proteolysis-independent regulation of PI3K by Cbl-b-mediated ubiquitination in T cells. Nature immunology, 2, 870–875. [DOI] [PubMed] [Google Scholar]
- Fang D., et al, 2002. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nature immunology, 3, 281–287. [DOI] [PubMed] [Google Scholar]
- Fiil B.K., et al, 2013. OTULIN restricts met1-linked ubiquitination to control innate immune signaling. Molecular cell, 50, 818–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annual review of biochemistry, 78, 477–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiskin E., et al, 2017. Structural basis for the recognition and degradation of host TRIM proteins by Salmonella effector SopA. Nature communications, 8 , 14004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher A.J., et al, 2015. Sequential ubiquitination and deubiquitination enzymes synchronize the dual sensor and effector functions of TRIM21. Proceedings of the national academy of sciences of the United States of America, 112, 10014–10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack M.U., et al, 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature, 446, 916–920. [DOI] [PubMed] [Google Scholar]
- Gack M.U., et al, 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell host & microbe, 5, 439–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos B.K., et al, 2011. Hexagonal assembly of a restricting TRIM5alpha protein. Proceedings of the national academy of sciences of the United States of America, 108, 534–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A., et al, 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology, 252, 324–330. [DOI] [PubMed] [Google Scholar]
- Gerlach B., et al, 2011. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature, 471, 591–596. [DOI] [PubMed] [Google Scholar]
- Germain R.N., 2002. T-cell development and the CD4-CD8 lineage decision. Nature reviews immunology, 2, 309–322. [DOI] [PubMed] [Google Scholar]
- Giordano M., et al, 2014. The tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) imposes a brake on antitumor activity of CD8 T cells. Proceedings of the national academy of sciences of the United States of America, 111, 11115–11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto E. and Tokunaga F., 2017. Decreased linear ubiquitination of NEMO and FADD on apoptosis with caspase-mediated cleavage of HOIP. Biochemical and biophysical research communications, 485, 152–159. [DOI] [PubMed] [Google Scholar]
- Grice G.L., et al, 2015. The proteasome distinguishes between heterotypic and homotypic lysine-11-linked polyubiquitin chains. Cell reports, 12, 545–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grou C.P., et al, 2015. The de novo synthesis of ubiquitin: identification of deubiquitinases acting on ubiquitin precursors. Scientific reports, 5 , 12836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grutter C., et al, 2006. Structure of the PRYSPRY-domain: implications for autoinflammatory diseases. FEBS letters, 580, 99–106. [DOI] [PubMed] [Google Scholar]
- Haas T.L., et al, 2009. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Molecular cell, 36, 831–844. [DOI] [PubMed] [Google Scholar]
- Hale B.G., et al, 2008. The multifunctional NS1 protein of influenza A viruses. Journal of general virology, 89, 2359–2376. [DOI] [PubMed] [Google Scholar]
- Han K., LOU D.I., and SAWYER S.L., 2011. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS genetics, 7 , e1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada Y., et al, 2010. Transcription factors Foxo3a and Foxo1 couple the E3 ligase Cbl-b to the induction of Foxp3 expression in induced regulatory T cells. Journal of experimental medicine, 207, 1381–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatakeyama S., 2011. TRIM proteins and cancer. Nature reviews cancer, 11, 792–804. [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., 2017. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends in biochemical sciences, 42, 297–311. [DOI] [PubMed] [Google Scholar]
- Hatziioannou T., et al, 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proceedings of the national academy of sciences of the United States of America, 101, 10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heesters B.A., et al, 2016. Antigen presentation to B cells. Trends in immunology, 37, 844–854. [DOI] [PubMed] [Google Scholar]
- Heissmeyer V., et al, 2004. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nature immunology, 5, 255–265. [DOI] [PubMed] [Google Scholar]
- Hicke L., Schubert H.L., and Hill C.P., 2005. Ubiquitin-binding domains. Nature reviews molecular cell biology, 6, 610–621. [DOI] [PubMed] [Google Scholar]
- Hornung V., et al, 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science, 314, 994–997. [DOI] [PubMed] [Google Scholar]
- Hospenthal M.K., Mevissen T.E., and Komander D., 2015. Deubiquitinase-based analysis of ubiquitin chain architecture using Ubiquitin Chain Restriction (UbiCRest). Nature protocols, 10, 349–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdinka M., et al, 2016. CYLD limits Lys63- and Met1-linked ubiquitin at receptor complexes to regulate innate immune signaling. Cell reports, 14, 2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., et al, 2013. OTUD7B controls non-canonical NF-kappaB activation through deubiquitination of TRAF3. Nature, 494, 371–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H. and Sun S.C., 2016. Ubiquitin signaling in immune responses. Cell research, 26, 457–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H., et al, 2016. Otud7b facilitates T cell activation and inflammatory responses by regulating Zap70 ubiquitination. Journal of experimental medicine, 213, 399–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., et al, 2010. K33-linked polyubiquitination of T cell receptor-zeta regulates proteolysis-independent T cell signaling. Immunity, 33, 60–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husnjak K. and Dikic I., 2012. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annual review of biochemistry, 81, 291–322. [DOI] [PubMed] [Google Scholar]
- Ikeda F., 2015. Linear ubiquitination signals in adaptive immune responses. Immunological reviews, 266, 222–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F., Crosetto N., and Dikic I., 2010. What determines the specificity and outcomes of ubiquitin signaling? Cell, 143, 677–681. [DOI] [PubMed] [Google Scholar]
- Ikeda F. and Dikic I., 2008. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO reports, 9, 536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda F., et al, 2011. SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature, 471, 637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H. and Barber G.N., 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature, 455, 674–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa H., Ma Z., and Barber G.N., 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature, 461, 788–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai K. and Tokunaga F., 2009. Linear polyubiquitination: a new regulator of NF-kappaB activation. EMBO reports, 10, 706–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James L.C., et al, 2007. Structural basis for PRYSPRY-mediated tripartite motif (TRIM) protein function. Proceedings of the national academy of sciences of the United States of America, 104, 6200–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javanbakht H., et al, 2005. The contribution of RING and B-box 2 domains to retroviral restriction mediated by monkey TRIM5alpha. Journal of biological chemistry, 280, 26933–26940. [DOI] [PubMed] [Google Scholar]
- Jeon M.S., et al, 2004. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity, 21, 167–177. [DOI] [PubMed] [Google Scholar]
- Jiang X., et al, 2012. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity, 36, 959–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., et al, 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. Journal of biological chemistry, 270, 17442–17456. [DOI] [PubMed] [Google Scholar]
- Kamanova J., et al, 2016. The Salmonella effector protein SopA modulates innate immune responses by targeting TRIM E3 ligase family members. PLoS pathogens, 12, e1005552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., et al, 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature, 441, 101–105. [DOI] [PubMed] [Google Scholar]
- Keckesova Z., Ylinen L.M., and Towers G.J., 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proceedings of the national academy of sciences of the United States of America, 101, 10780–10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeble A.H., et al, 2008. TRIM21 is an IgG receptor that is structurally, thermodynamically, and kinetically conserved. Proceedings of the national academy of sciences of the United States of America, 105, 6045–6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keown J.R. and Goldstone D.C., 2016. Crystal structure of the Trim5alpha Bbox2 domain from rhesus macaques describes a plastic oligomerisation interface. Journal of structural biology, 195, 282–285. [DOI] [PubMed] [Google Scholar]
- Kim H.T., et al, 2007. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. Journal of biological chemistry, 282, 17375–17386. [DOI] [PubMed] [Google Scholar]
- Kimura T., et al, 2015. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunity. Journal of cell biology, 210, 973–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., et al, 2016. TRIM-directed selective autophagy regulates immune activation. Autophagy [online]. Available from: http://dx.doi.org/10.1080/15548627.2016.1154254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King C.G., et al, 2006. TRAF6 is a T cell-intrinsic negative regulator required for the maintenance of immune homeostasis. Nature medicine, 12, 1088–1092. [DOI] [PubMed] [Google Scholar]
- King C.G., et al, 2008. Cutting edge: requirement for TRAF6 in the induction of T cell anergy. Journal of immunology, 180, 34–38. [DOI] [PubMed] [Google Scholar]
- Kirisako T., et al, 2006. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO journal, 25, 4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T., et al, 2009. TRAF6 is required for generation of the B-1a B cell compartment as well as T cell-dependent and -independent humoral immune responses. PLoS One, 4, e4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliopoulos M.G., et al, 2016. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO journal, 35, 1204–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D. and Rape M., 2012. The ubiquitin code. Annual review of biochemistry, 81, 203–229. [DOI] [PubMed] [Google Scholar]
- Komander D., et al, 2009. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO reports, 10, 466–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontgen F., et al, 1995. Mice lacking the c-rel proto-oncogene exhibit defects in lymphocyte proliferation, humoral immunity, and interleukin-2 expression. Genes development, 9, 1965–1977. [DOI] [PubMed] [Google Scholar]
- Koyano F., et al, 2014. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature, 510, 162–166. [DOI] [PubMed] [Google Scholar]
- Kulathu Y., et al, 2009. Two-sided ubiquitin binding explains specificity of the TAB2 NZF domain. Nature structural & molecular biology, 16, 1328–1330. [DOI] [PubMed] [Google Scholar]
- Kumari S., et al, 2014. Sharpin prevents skin inflammation by inhibiting TNFR1-induced keratinocyte apoptosis. Elife, 3, e03422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon H., et al, 1998. Inducible expression of IkappaBalpha repressor mutants interferes with NF-kappaB activity and HIV-1 replication in Jurkat T cells. Journal of biological chemistry, 273, 7431–7440. [DOI] [PubMed] [Google Scholar]
- Lang X., et al, 2017. TRIM65-catalized ubiquitination is essential for MDA5-mediated antiviral innate immunity. Journal of experimental medicine, 214, 459–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtenberg B.C., et al, 2016. Structure of a HOIP/E2∼ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature, 529, 546–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., et al., 2015. E3 ubiquitin ligase VHL regulates hypoxia-inducible factor-1alpha to maintain regulatory T cell stability and suppressive capacity. Immunity, 42, 1062–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leppek K., et al, 2013. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell, 153, 869–881. [DOI] [PubMed] [Google Scholar]
- Li X. and Sodroski J., 2008. The TRIM5alpha B-box 2 domain promotes cooperative binding to the retroviral capsid by mediating higher-order self-association. Journal of virology, 82, 11495–11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kar A.K., and Sodroski J., 2009. Target cell type-dependent modulation of human immunodeficiency virus type 1 capsid disassembly by cyclophilin A. Journal of virology, 83, 10951–10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.L., et al, 2016. Primate TRIM5 proteins form hexagonal nets on HIV-1 capsids. *Elife, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienlaf M., et al, 2011. Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5alpha(rh): structure of the RING domain of TRIM5alpha. Journal of virology, 85, 8725–8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lineberry N.B., et al, 2008. Cutting edge: the transmembrane E3 ligase GRAIL ubiquitinates the costimulatory molecule CD40 ligand during the induction of T cell anergy. Journal of immunology, 181, 1622–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou H.C., et al, 1999. c-Rel is crucial for lymphocyte proliferation but dispensable for T cell effector function. International immunology, 11, 361–371. [DOI] [PubMed] [Google Scholar]
- Liu C., et al, 2017. Ufd2p synthesizes branched ubiquitin chains to promote the degradation of substrates modified with atypical chains. Nature communications, 8 , 14274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., et al, 2013. USP18 inhibits NF-κB and NFAT activation during Th17 differentiation by deubiquitinating the TAK1-TAB1 complex. Journal of experimental medicine, 210, 1575–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr N.J., et al, 2010. Human ITCH E3 ubiquitin ligase deficiency causes syndromic multisystem autoimmune disease. American journal of human genetics, 86, 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopitz-Otsoa F., Rodriguez M.S., and Aillet F., 2010. Properties of natural and artificial proteins displaying multiple ubiquitin-binding domains. Biochemical society transactions, 38, 40–45. [DOI] [PubMed] [Google Scholar]
- Lu Y., et al., 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology, 214, 222–228. [DOI] [PubMed] [Google Scholar]
- Lupher M.L., Jr, et al, 1996. A novel phosphotyrosine-binding domain in the N-terminal transforming region of Cbl interacts directly and selectively with ZAP-70 in T cells. Journal of biological chemistry, 271, 24063–24068. [DOI] [PubMed] [Google Scholar]
- Mackenzie D.A., et al, 2007. GRAIL is up-regulated in CD4+ CD25+ T regulatory cells and is sufficient for conversion of T cells to a regulatory phenotype. Journal of biological chemistry, 282, 9696–96702. [DOI] [PubMed] [Google Scholar]
- Maggirwar S.B., Harhaj E.W., and Sun S.C., 1997. Regulation of the interleukin-2 CD28-responsive element by NF-ATp and various NF-kappaB/Rel transcription factors. Molecular and cellular biology, 17, 2605–2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfavon-Borja R., et al, 2013. An evolutionary screen highlights canonical and noncanonical candidate antiviral genes within the primate TRIM gene family. Genome biology and evolution ,5, 2141–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallery D.L., et al, 2010. Antibodies mediate intracellular immunity through tripartite motif-containing 21 (TRIM21). Proceedings of the national academy of sciences of the United States of America, 107, 19985–19990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malynn B.A. and Ma A., 2010. Ubiquitin makes its mark on immune regulation. Immunity, 33, 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell M.A., et al, 2014a. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Developmental cell, 30, 394–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell M.A., et al, 2014b. TRIM proteins regulate autophagy: TRIM5 is a selective autophagy receptor mediating HIV-1 restriction. Autophagy, 10, 2387–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandell M.A., et al, 2016. TRIM17 contributes to autophagy of midbodies while actively sparing other targets from degradation. Journal of cell science, 129, 3562–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez F.O., et al, 2006. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. Journal of immunology, 177, 7303–7311. [DOI] [PubMed] [Google Scholar]
- Masuda Y., et al, 2015. TRIM29 regulates the assembly of DNA repair proteins into damaged chromatin. Nature communications, 6 , 7299. [DOI] [PubMed] [Google Scholar]
- Matsumoto M.L., et al, 2010. K11-linked polyubiquitination in cell cycle control revealed by a K11 linkage-specific antibody. Molecular cell, 39, 477–484. [DOI] [PubMed] [Google Scholar]
- Matsuzawa A., et al, 2008. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science, 321, 663–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcewan W.A., et al, 2013. Intracellular antibody-bound pathogens stimulate immune signaling via the Fc receptor TRIM21. Nature immunology, 14, 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger M.B., Hristova V.A., and Weissman A.M., 2012. HECT and RING finger families of E3 ubiquitin ligases at a glance. Journal of cellular science, 125, 531–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger M.B., et al, 2014. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochimica et biophysica acta, 1843, 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevissen T.E., et al, 2016. Molecular basis of Lys11-polyubiquitin specificity in the deubiquitinase Cezanne. Nature, 538, 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H.J. and Rape M., 2014. Enhanced protein degradation by branched ubiquitin chains. Cell, 157, 910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelle C., et al, 2009. What was the set of ubiquitin and ubiquitin-like conjugating enzymes in the eukaryote common ancestor? Journal of molecular evolution, 68, 616–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagamine K., et al, 1997. Positional cloning of the APECED gene. Nature genetics, 17, 393–398. [DOI] [PubMed] [Google Scholar]
- Naik E., et al, 2014. Regulation of proximal T cell receptor signaling and tolerance induction by deubiquitinase Usp9X. Journal of experimental medicine, 211, 1947–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff M.E., et al, 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Molecular cell, 1, 991–1000. [DOI] [PubMed] [Google Scholar]
- Neumann M. and Naumann M., 2007. Beyond IkappaBs: alternative regulation of NF-kappaB activity. FASEB journal, 21, 2642–2654. [DOI] [PubMed] [Google Scholar]
- Nisole S., et al, 2013. Differential Roles of PML Isoforms. Frontiers in oncology, 3 , 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., et al, 2010. The E3 ubiquitin ligase GRAIL regulates T cell tolerance and regulatory T cell function by mediating T cell receptor-CD3 degradation. Immunity, 32, 670–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., et al, 2013. MARCH1-mediated MHCII ubiquitination promotes dendritic cell selection of natural regulatory T cells. Journal of experimental medicine, 210, 1069–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtake F., et al, 2015. Ubiquitin acetylation inhibits polyubiquitin chain elongation. EMBO reports, 16, 192–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A., Munch C., and Harper J.W., 2015. Quantifying ubiquitin signaling. Molecular cell, 58, 660–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A., et al, 2008. The IRAK-catalysed activation of the E3 ligase function of Pellino isoforms induces the Lys63-linked polyubiquitination of IRAK1. Biochemical journal, 409, 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S., et al, 2012. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity, 36, 1073–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen K.A., Anderson C.J., and Casanova J.E., 2016. Salmonella suppresses the TRIF-dependent type I interferon response in macrophages. MBio, 7, e02051–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozato K., et al., 2008. TRIM family proteins and their emerging roles in innate immunity. Nature reviews immunology, 8, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozato K., et al, 2009. Comment on "Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappa B-dependent cytokine expression in fibroblasts". Journal of immunology, 183, 7619. 3RD author reply 720-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv S., et al, 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. Journal of biological chemistry, 282, 24131–24145. [DOI] [PubMed] [Google Scholar]
- Paolino M., et al, 2011. Essential role of E3 ubiquitin ligase activity in Cbl-b-regulated T cell functions. Journal of immunology, 186, 2138–2147. [DOI] [PubMed] [Google Scholar]
- Park Y., et al, 2014. The ubiquitin system in immune regulation. Advances in immunology, 124, 17–66. [DOI] [PubMed] [Google Scholar]
- Park Y., Jin H.S., and Liu Y.C., 2013. Regulation of T cell function by the ubiquitin-specific protease USP9X via modulating the Carma1-Bcl10-Malt1 complex. Proceedings of the national academy of sciences of the United States of America, 110, 9433–9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A., et al, 2014. Structural basis for ubiquitin-mediated antiviral signal activation by RIG-I. Nature, 509, 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltzer N., et al, 2014. HOIP deficiency causes embryonic lethality by aberrant TNFR1-mediated endothelial cell death. Cell reports, 9, 153–165. [DOI] [PubMed] [Google Scholar]
- Perron M.J., et al, 2004. TRIM5alpha mediates the postentry block to N-tropic murine leukemia viruses in human cells. Proceedings of the national academy of sciences of the United States of America, 101, 11827–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry W.L., et al, 1998. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nature genetics, 18, 143–146. [DOI] [PubMed] [Google Scholar]
- Pertel T., et al, 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature, 472, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A., et al, 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science, 314, 997–1001. [DOI] [PubMed] [Google Scholar]
- Pratama A., et al, 2013. Roquin-2 shares functions with its paralog Roquin-1 in the repression of mRNAs controlling T follicular helper cells and systemic inflammation. Immunity, 38, 669–680. [DOI] [PubMed] [Google Scholar]
- Pruneda J.N., et al, 2012. Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Molecular cell, 47, 933–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., et al, 2008. Deficiency of Act1, a critical modulator of B cell function, leads to development of Sjögren's syndrome. European journal of immunology, 38, 2219–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y., et al, 2004. Act1, a negative regulator in CD40- and BAFF-mediated B cell survival. Immunity, 21, 575–587. [DOI] [PubMed] [Google Scholar]
- Qiu J., et al, 2016. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature, 533, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinto I., et al, 1999. Potent and stable attenuation of live-HIV-1 by gain of a proteolysis-resistant inhibitor of NF-kappaB (IkappaB-alphaS32/36A) and the implications for vaccine development. Journal of biological chemistry, 274, 17567–17572. [DOI] [PubMed] [Google Scholar]
- Rahighi S., et al, 2009. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell, 136, 1098–1109. [DOI] [PubMed] [Google Scholar]
- Rajsbaum R., Garcia-Sastre A., and Versteeg G.A., 2014a. TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. Journal of molecular biology, 426, 1265–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R., Stoye J.P., and O'Garra A., 2008. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. European journal of immunology, 38, 619–630. [DOI] [PubMed] [Google Scholar]
- Rajsbaum R., et al, 2014b. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKepsilon kinase-mediated antiviral response. Immunity, 40, 880–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakebrandt N., et al, 2014. Antibody- and TRIM21-dependent intracellular restriction of Salmonella enterica. Pathogens and disease, 72, 131–137. [DOI] [PubMed] [Google Scholar]
- Rauch J., et al, 2011. The secret life of kinases: functions beyond catalysis. Cell communication and signaling, 9 , 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley W.W., et al, 2007. Deubiquitinating enzyme CYLD negatively regulates the ubiquitin-dependent kinase Tak1 and prevents abnormal T cell responses. Journal of experimental medicine, 204, 1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiley W.W., et al, 2006. Regulation of T cell development by the deubiquitinating enzyme CYLD. Nature immunology, 7, 411–417. [DOI] [PubMed] [Google Scholar]
- Reissig S., et al, 2012. The tumor suppressor CYLD controls the function of murine regulatory T cells. Journal of immunology, 189, 4770–4776. [DOI] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Ventii K.H., and Wilkinson K.D., 2009. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annual review of biochemistry , 78, 363–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond A., et al, 2001. The tripartite motif family identifies cell compartments. EMBO journal, 20, 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D.A. and Trowsdale J., 2007. TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain. Molecular immunology, 44, 2406–2414. [DOI] [PubMed] [Google Scholar]
- Rickard J.A., et al, 2014. TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. Elife, 3, e03464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas M.A., et al, 2011. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nature genetics, 43, 1066–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roa A., et al, 2012. RING domain mutations uncouple TRIM5alpha restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating. Journal of virology, 86, 1717–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rold C.J. and Aiken C., 2008. Proteasomal degradation of TRIM5alpha during retrovirus restriction. PLoS pathogens, 4 , e1000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland S.L., et al, 2007. A novel mechanism for TNFR-associated factor 6-dependent CD40 signaling. Journal of immunology, 179, 4645–4653. [DOI] [PubMed] [Google Scholar]
- Sakaguchi S., et al, 2008. Regulatory T cells and immune tolerance. Cell, 133, 775–787. [DOI] [PubMed] [Google Scholar]
- Sanchez J.G., et al, 2014. The tripartite motif coiled-coil is an elongated antiparallel hairpin dimer. Proceedings of the national academy of sciences of the United States of America, 111, 2494–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., et al, 2009. Structural basis for specific recognition of Lys 63-linked polyubiquitin chains by NZF domains of TAB2 and TAB3. EMBO journal, 28, 3903–3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer S.L., et al, 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proceedings of the national academy of sciences of the United States of America, 102, 2832–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer V., et al, 2014. Binding of OTULIN to the PUB domain of HOIP controls NF-κB signaling. Molecular cell, 54, 349–361. [DOI] [PubMed] [Google Scholar]
- Scharschmidt E., et al, 2004. Degradation of Bcl10 induced by T-cell activation negatively regulates NF-kappa B signaling. Molecular and cellular biology, 24, 3860–3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffner M. and Kumar S., 2014. Mammalian HECT ubiquitin–protein ligases: biological and pathophysiological aspects. Biochimica et biophysica acta, 1843, 61–74. [DOI] [PubMed] [Google Scholar]
- Scherer M. and Stamminger T., 2016. Emerging role of PML nuclear bodies in innate immune signaling. Journal of virology, 90, 5850–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman B.A. and Harper J.W., 2009. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nature reviews molecular cell biology, 10, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzkopff B., Platta H.W., and Girzalsky W., 2015. Cystein-specific ubiquitination protects the peroxisomal import receptor Pex5p against proteasomal degradation. Bioscience reports, 35, e00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour R.E., et al, 2007. Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes immunology, 8, 416–421. [DOI] [PubMed] [Google Scholar]
- Shu C, et al, 2012. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nature structural & molecular biology, 19, 722–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims J.J. and Cohen R.E., 2009. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Molecular cell, 33, 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit J.J. and Sixma T.K., 2014. RBR E3-ligases at work. EMBO reports, 15, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Garvin J.E., Koretzky G.A., and Jordan M.S., 2009. T cell activation. Annual review immunology, 27, 591–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song R., et al, 2008. Mind bomb 1 in the lymphopoietic niches is essential for T and marginal zone B cell development. Journal of experimental medicine, 205, 2525–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt D.E., Walden H., and Shaw G.S., 2014. RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochemical journal, 458, 421–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stempin C.C., et al, 2011. The E3 ubiquitin ligase mind bomb-2 (MIB2) protein controls B-cell CLL/lymphoma 10 (BCL10)-dependent NF-κB activation. Journal of biological chemistry, 286, 37147–37157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stieglitz B., et al, 2013. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature, 503, 422–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streich FC, Jr., et al, 2013. Tripartite motif ligases catalyze polyubiquitin chain formation through a cooperative allosteric mechanism. Journal of biological chemistry, 288, 8209–8221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M., et al, 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature, 427, 848–853. [DOI] [PubMed] [Google Scholar]
- Stremlau M., et al, 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proceedings of the national academy of sciences of the United States of America, 103, 5514–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L., et al, 2006. A novel E3 ubiquitin ligase substrate screen identifies Rho guanine dissociation inhibitor as a substrate of gene related to anergy in lymphocytes. Journal of immunology, 177, 7559–7566. [DOI] [PubMed] [Google Scholar]
- Sun L., et al, 2004. The TRAF6 ubiquitin ligase and TAK1 kinase mediate IKK activation by BCL10 and MALT1 in T lymphocytes. Molecular cell, 14, 289–301. [DOI] [PubMed] [Google Scholar]
- Sun L., et al, 2013. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science, 339, 786–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek K.N. and Komander D., 2016. Ubiquitin modifications. Cell research, 26, 399–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiuchi T., et al, 2014. Suppression of LUBAC-mediated linear ubiquitination by a specific interaction between LUBAC and the deubiquitinases CYLD and OTULIN. Genes cells, 19, 254–272. [DOI] [PubMed] [Google Scholar]
- Tavares R.M., et al, 2010. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity, 33, 181–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenno T., et al, 2004. Structural basis for distinct roles of Lys63-and Lys48-linked polyubiquitin chains. Genes cells, 9, 865–875. [DOI] [PubMed] [Google Scholar]
- Tenoever B.R., et al, 2007. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science, 315, 1274–1278. [DOI] [PubMed] [Google Scholar]
- Tokunaga F., et al, 2009. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nature cell biology, 11, 123–132. [DOI] [PubMed] [Google Scholar]
- Tokunaga F., et al, 2012. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO journal, 31, 3856–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagaratou A., et al, 2010. Thymocyte-specific truncation of the deubiquitinating domain of CYLD impairs positive selection in a NF-kappaB essential modulator-dependent manner. Journal of immunology, 185, 2032–2043. [DOI] [PubMed] [Google Scholar]
- Tsuchida T., et al, 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity, 33, 765–776. [DOI] [PubMed] [Google Scholar]
- Uchida D., et al, 2004. AIRE functions as an E3 ubiquitin ligase. Journal of experimental medicine, 199, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchil P.D., et al, 2013. TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. Journal of virology, 87, 257–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallabhapurapu S., et al, 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nature immunology, 9, 1364–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdregt J., et al, 2013. Stabilization of the transcription factor Foxp3 by the deubiquitinase USP7 increases Treg-cell-suppressive capacity. Immunity, 39, 259–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Valen L., 1973. A new evolutionary law. Evolutionary theory, 1, 1–30. [Google Scholar]
- van Wijk S.J., et al, 2012. Fluorescence-based sensors to monitor localization and functions of linear and K63-linked ubiquitin chains in cells. Molecular cell, 47, 797–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadan R., et al, 2002. Structural properties of polyubiquitin chains in solution. Journal of molecular biology, 324, 637–647. [DOI] [PubMed] [Google Scholar]
- Varfolomeev E. and Vucic D., 2016. Intracellular regulation of TNF activity in health and disease. Cytokine [online]. Available from: http://dx.doi.org/10.1016/j.cyto.2016.08.035 [DOI] [PubMed] [Google Scholar]
- Vaysburd M., et al, 2013. Intracellular antibody receptor TRIM21 prevents fatal viral infection. Proceedings of the national academy of sciences of the United States of America, 110, 12397–12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venuprasad K., et al, 2008. The E3 ubiquitin ligase Itch regulates expression of transcription factor Foxp3 and airway inflammation by enhancing the function of transcription factor TIEG1. Nature immunology, 9, 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhelst K., et al, 2012. A20 inhibits LUBAC-mediated NF-kappaB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO journal, 31, 3845–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A. and Garcia-Sastre A., 2010. Viral tricks to grid-lock the type I interferon system. Current opinion microbiology, 13, 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., et al, 2013. The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity, 38, 384–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Versteeg G.A., et al, 2014. InTRIMsic immunity: positive and negative regulation of immune signaling by tripartite motif proteins. *Cytokine Growth Factor Review, 25, 563–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg C.E., and Cook W.J., 1987. Structure of ubiquitin refined at 1.8 A resolution. Journal of molecular biology, 194, 531–544. [DOI] [PubMed] [Google Scholar]
- Vinuesa C.G., et al, 2005. Follicular B helper T cells in antibody responses and autoimmunity. Nature reviews immunology, 5, 853–865. [DOI] [PubMed] [Google Scholar]
- Virdee S., et al, 2010. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nature chemical biology, 6, 750–757. [DOI] [PubMed] [Google Scholar]
- Vogel K.U., et al, 2013. Roquin paralogs 1 and 2 redundantly repress the Icos and Ox40 costimulator mRNAs and control follicular helper T cell differentiation. Immunity, 38, 655–668. [DOI] [PubMed] [Google Scholar]
- Vucic D., Dixit V.M., and Wertz I.E., 2011. Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nature reviews molecular cell biology, 12, 439–452. [DOI] [PubMed] [Google Scholar]
- Wahren M., et al, 1998. Ro/SS-A and La/SS-B antibody level variation in patients with Sjögren's syndrome and systemic lupus erythematosus. Journal of autoimmunology, 11, 29–38. [DOI] [PubMed] [Google Scholar]
- Walczak H., 2011. TNF and ubiquitin at the crossroads of gene activation, cell death, inflammation, and cancer. Immunological reviews, 244, 9–28. [DOI] [PubMed] [Google Scholar]
- Wang D., et al, 2012. Inhibition of S-phase kinase-associated protein 2 (Skp2) reprograms and converts diabetogenic T cells to Foxp3+ regulatory T cells. Proceedings of the national academy of sciences of the United States of America, 109, 9493–9498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al, 2007. Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3. Journal of cell biology, 177, 613–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkinson R.E., et al, 2015. TRIM21 promotes cGAS and RIG-I sensing of viral genomes during infection by antibody-opsonized virus. PLoS pathogens, 11, e1005253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks S.D., et al, 2009. Crystal structures of Lys-63-linked tri- and di-ubiquitin reveal a highly extended chain architecture. Proteins, 77, 753–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert C., et al, 2009. The crystal structure of human pyrin b30.2 domain: implications for mutations associated with familial Mediterranean fever. Journal of molecular biology, 394, 226–236. [DOI] [PubMed] [Google Scholar]
- Wenzel D.M., et al, 2011. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature, 474, 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I.E., et al, 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature, 430, 694–699. [DOI] [PubMed] [Google Scholar]
- Wertz I.E., et al, 2015. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature, 528, 370–375. [DOI] [PubMed] [Google Scholar]
- Wiborg O., et al, 1985. The human ubiquitin multigene family: some genes contain multiple directly repeated ubiquitin coding sequences. EMBO journal, 4, 755–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild P., et al, 2011. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science, 333, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildin R.S., et al, 2001. X-linked neonatal diabetes mellitus, enteropathy and endocrinopathy syndrome is the human equivalent of mouse scurfy. Nature genetics, 27, 18–20. [DOI] [PubMed] [Google Scholar]
- Wilkinson K.D., et al, 1995. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry, 34, 14535–14546. [DOI] [PubMed] [Google Scholar]
- Wohlfert E.A., et al, 2006. Cutting edge: deficiency in the E3 ubiquitin ligase Cbl-b results in a multifunctional defect in T cell TGF-beta sensitivity in vitro and in vivo. Journal of immunology, 176, 1316–1320. [DOI] [PubMed] [Google Scholar]
- Woo J.S., et al, 2006. Structural and functional insights into the B30.2/SPRY domain. EMBO journal, 25, 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. and Chen Z.J., 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annual reviews immunology, 32, 461–488. [DOI] [PubMed] [Google Scholar]
- Wu X., et al, 2006. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proceedings of the national academy of sciences of the United States of America, 103, 7465–7470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J., et al, 2016. Identification of a role for TRIM29 in the control of innate immunity in the respiratory tract. Nature immunology, 17, 1373–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y. and Hogquist K.A., 2012. T-cell tolerance: central and peripheral. Cold spring harbor perspectives in biology, 4, a006957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., et al, 2003. BTBD1 and BTBD2 colocalize to cytoplasmic bodies with the RBCC/tripartite motif protein, TRIM5delta. Experimental cell research, 288, 84–93. [DOI] [PubMed] [Google Scholar]
- Xu Y., Cheng G., and Baltimore D., 1996. Targeted disruption of TRAF3 leads to postnatal lethality and defective T-dependent immune responses. Immunity, 5, 407–415. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., et al, 2006. Cutting edge: pivotal function of Ubc13 in thymocyte TCR signaling. Journal of immunology, 177, 7520–7524. [DOI] [PubMed] [Google Scholar]
- Yamano K., Matsuda N., and Tanaka K., 2016. The ubiquitin signal and autophagy: an orchestrated dance leading to mitochondrial degradation. EMBO reports, 17, 300–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., et al, 2015. K33-linked polyubiquitination of Zap70 by Nrdp1 controls CD8(+) T cell activation. Nature immunology, 16, 1253–1262. [DOI] [PubMed] [Google Scholar]
- Yau R. and Rape M., 2016. The increasing complexity of the ubiquitin code. Nature cell biology, 18, 579–586. [DOI] [PubMed] [Google Scholar]
- Ye Y. and Rape M., 2009. Building ubiquitin chains: E2 enzymes at work. Nature reviews molecular cell biology, 10, 755–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh W.C., et al, 1997. Early lethality, functional NF-kappaB activation, and increased sensitivity to TNF-induced cell death in TRAF2-deficient mice. Immunity, 7, 715–725. [DOI] [PubMed] [Google Scholar]
- Yi Z., et al, 2014a. The adaptor TRAF3 restrains the lineage determination of thymic regulatory T cells by modulating signaling via the receptor for IL-2. Nature immunology, 15, 866–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Z., et al, 2014b. TRAF3 regulates homeostasis of CD8+ central memory T cells. PLoS One, 9 , e102120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M., et al, 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nature immunology, 5, 730–737. [DOI] [PubMed] [Google Scholar]
- Yoshimi R., et al, 2009. Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-kappaB-dependent cytokine expression in fibroblasts. Journal of immunology, 182, 7527–7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., et al, 2007a. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature, 450, 299–303. [DOI] [PubMed] [Google Scholar]
- Yu J.W., et al, 2007b. Pyrin activates the ASC pyroptosome in response to engagement by autoinflammatory PSTPIP1 mutants. Molecular cell, 28, 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W., et al, 2010. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell, 141, 315–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al, 2006. Impaired regulation of NF-kappaB and increased susceptibility to colitis-associated tumorigenesis in CYLD-deficient mice. Journal of clinical investigation, 116, 3042–3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., et al, 2011. Cysteine methylation disrupts ubiquitin-chain sensing in NF-kappaB activation. Nature, 481, 204–208. [DOI] [PubMed] [Google Scholar]
- Zhang M., et al, 2005. Chaperoned ubiquitylation–crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Molecular cell, 20, 525–538. [DOI] [PubMed] [Google Scholar]
- Zhao Y., et al, 2011. The deubiquitinase CYLD targets Smad7 protein to regulate transforming growth factor β (TGF-β) signaling and the development of regulatory T cells. Journal of biological chemistry, 286, 40520–40530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H., et al, 2004. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature, 427, 167–171. [DOI] [PubMed] [Google Scholar]
- Zhou X.Y., et al, 2002. Molecular mechanisms underlying differential contribution of CD28 versus non-CD28 costimulatory molecules to IL-2 promoter activation. Journal of immunology, 168, 3847–3854. [DOI] [PubMed] [Google Scholar]
- Zinngrebe J., et al, 2014. Ubiquitin in the immune system. EMBO reports, 15, 28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Q., et al, 2014. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nature immunology, 15, 562–570. [DOI] [PMC free article] [PubMed] [Google Scholar]