APOBEC proteins and intrinsic resistance to HIV-1 infection (original) (raw)
Abstract
Members of the APOBEC family of cellular polynucleotide cytidine deaminases, most notably APOBEC3G and APOBEC3F, are potent inhibitors of HIV-1 infection. Wild type HIV-1 infections are largely spared from APOBEC3G/F function through the action of the essential viral protein, Vif. In the absence of Vif, APOBEC3G/F are encapsidated by budding virus particles leading to excessive cytidine (C) to uridine (U) editing of negative sense reverse transcripts in newly infected cells. This registers as guanosine (G) to adenosine (A) hypermutations in plus-stranded cDNA. In addition to this profoundly debilitating effect on genetic integrity, APOBEC3G/F also appear to inhibit viral DNA synthesis by impeding the translocation of reverse transcriptase along template RNA. Because the functions of Vif and APOBEC3G/F proteins oppose each other, it is likely that fluctuations in the Vif–APOBEC balance may influence the natural history of HIV-1 infection, as well as viral sequence diversification and evolution. Given Vif's critical role in suppressing APOBEC3G/F function, it can be argued that pharmacologic strategies aimed at restoring the activity of these intrinsic anti-viral factors in the context of infected cells in vivo have clear therapeutic merit, and therefore deserve aggressive pursuit.
Keywords: HIV-1, APOBEC3G, Vif, hypermutation, reverse transcription
1. HIV-1 regulatory/accessory proteins as critical mediators of virus–host interactions
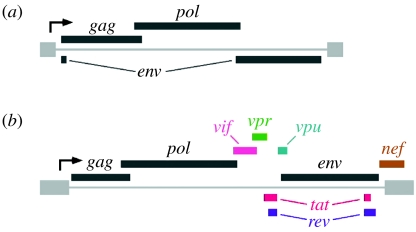
Human immunodeficiency virus type-1 (HIV-1), the retrovirus that causes AIDS, is markedly more complex than the long-studied oncogenic retroviruses that infect avian and rodent species (figure 1). In particular, and as recognized during the initial genetic characterization of HIV-1 in the mid-1980s, this virus carries six regulatory/accessory genes (tat, rev, vif, vpr, vpu and nef) that are additional to the gag, pol and env genes common to all replication competent retroviruses. Indeed, additional genetic complexity is a molecular signature shared by all members of the lentivirus genus of retroviruses—to which HIV-1 belongs and is the prototypic member. Broadly speaking, the regulatory/accessory proteins all serve as ‘adapters’ that bridge viral or cellular molecules to cellular pathways or factors in ways that benefit viral infection, replication, dissemination or persistence (Malim & Emerman 2008; Swanson & Malim 2008).
Figure 1.
The genetic complexity of HIV-1. The six regulatory/accessory genes of (b) HIV-1 are not present in oncogenic retroviruses such as (a) avian leukosis virus (ALV). The proviruses of all replication-competent retroviruses are flanked by LTRs (pale grey) and encode gag, pol and env genes (dark grey). The direction of transcription from the 5′-LTR is indicated.
The replication of all viruses is intimately linked with, and completely reliant upon, the action of multiple host cell factors. The vast majority of such virus–host interactions are beneficial to viral propagation and are therefore exploited by viruses to promote aspects of replication: good examples of host proteins that are required for replication include cell surface molecules that function as receptors for viral entry into cells, and transcription factors that mediate viral gene expression. Indeed, a recent functional RNAi-based screen identified close to 300 human genes whose action facilitates HIV-1 replication, but whose depletion was not overtly deleterious to normal human cell function (Brass et al. 2008).
Conversely, work in the last few years, particularly in the HIV-1 field, has revealed that cells may also harbour intrinsic, cell-autonomous activities that can act to suppress viral replication: these are collectively called restriction factors (Bieniasz 2004; Goff 2004). It is only by averting the inhibitory functions of these host factors that HIV-1 is able to replicate effectively. Current evidence indicates that three of the regulatory/accessory proteins of HIV-1—namely, Vif, Vpr and Vpu—are important for counteracting three very different forms of intracellular restriction (Malim & Emerman 2008). One important concept in this area is that wild-type strains of HIV-1, HIV-2 and simian immunodeficiency virus (SIV) are not affected substantially by the restriction factors present in cells of their natural host species. This is attributable to the protective effects of the accessory proteins and/or to the insensitivity of viral target molecules, and presumably reflects the acquisition of beneficial adaptive changes that evolve more rapidly in the viruses than potential counteracting genetic changes can be established in populations of susceptible hosts. By contrast, these viruses tend to be sensitive to restriction in cells from species that are not natural hosts of infection, indicating that the accessory proteins are important determinants of cross-species transmission and subsequent adaptation to new animal hosts.
2. Vif is an essential regulator of HIV-1 infection
Soon after its initial description, it was recognized that the 192 amino acid HIV-1 Vif protein is a potent regulator of viral infection—hence its name, virion infectivity factor (Fisher et al. 1987; Strebel et al. 1987; Gallo et al. 1988). Despite the efforts of a number of laboratories, the mechanistic basis for Vif function remained enigmatic for many years. Nevertheless, several key observations were made during this period that have helped enlighten future work, specifically: (i) an intact vif gene is required for pathogenic infections in the SIV/rhesus macaque system (Desrosiers et al. 1998), (ii) Vif is critical for virus replication in some cells such as primary CD4 T cells and immortalized lines such as HUT78 or CEM (non-permissive cells), yet entirely dispensable in other T-cell lines such as SupT1 or CEM-SS (permissive cells) (Gabuzda et al. 1992; von Schwedler et al. 1993), (iii) Vif acts in virus producing cells to regulate the quality, i.e. the infectivity, rather than the quantity of progeny virions (Gabuzda et al. 1992; von Schwedler et al. 1993; Simon & Malim 1996), (iv) virions produced in the absence of Vif yield reduced levels of cDNA reverse transcripts following the challenge of susceptible target cells (Sova & Volsky 1993; von Schwedler et al. 1993; Courcoul et al. 1995; Simon & Malim 1996), and (v) the stoichiometries and nature of the structural and enzymatic proteins (namely, Gag, Pol and Env) that are incorporated into nascent virus particles are not influenced by Vif (von Schwedler et al. 1993; Fouchier et al. 1996; Gaddis et al. 2003).
In light of the lack of a readily noticeable effect of Vif on virion composition, a central question was whether Vif acts upon viral or cellular substrates to regulate infectivity? Two lines of evidence pointed towards the latter possibility. First, by analysing assorted HIV and SIV strains in cells of different primate species, it was noted that Vif's ability to impact viral infectivity was species specific, implying that its immediate substrate is cellular (Simon et al. 1998_b_). Second, cell fusion experiments employing mixtures of non-permissive and permissive cells revealed that the non-permissive phenotype is dominant in that _vif_-deficient virions produced by heterokaryons are non-infectious (Madani & Kabat 1998; Simon et al. 1998_a_). Together, these results suggested that non-permissive cells express inhibitors of HIV/SIV infection, and that Vif suppresses these in a species-specific manner.
3. Identification of APOBEC3G as a target of Vif
Based on the arguments set out in §2, cDNA subtraction was used to search for human genes that are expressed in non-permissive cells but not in permissive cells. In one such effort that used the CEM and CEM-SS cell lines, a gene called APOBEC3G (A3G, initially CEM15) was identified as sufficient to confer a potent Vif-regulated anti-HIV-1 phenotype (Sheehy et al. 2002). More specifically, transient or stable ectopic expression of A3G in non-expressing cells results in decreases in infection or replication of _vif_-deficient HIV-1 of at least 99 per cent, while the effects on wild-type (Vif expressing) viruses are minimal. A3G is the initialism for apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G, and the membership of this protein within the APOBEC protein family offered some immediate clues regarding its mechanism of action (§5).
4. Vif inhibits APOBEC3G function by inducing proteasomal degradation and preventing virion incorporation
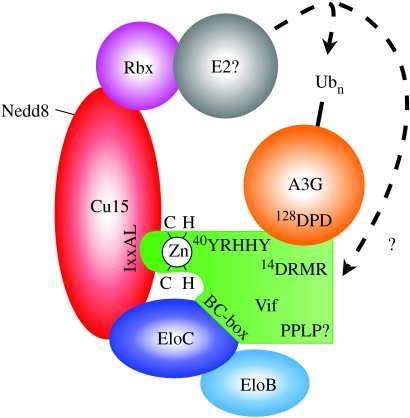
As recounted above, the anti-viral properties of A3G were first recognized through efforts to understand its virus-encoded antagonist, Vif. Importantly, and as described in §§5–7, the incorporation of A3G into virus particles is central to its anti-viral function under most circumstances. Perhaps the simplest way to prevent A3G action would, therefore, be to block virion encapsidation—and this is what Vif does. Though there have been reports of Vif directly impeding packaging (Mariani et al. 2003; Opi et al. 2007), Vif's principal function is to induce the polyubiquitylation and subsequent proteasomal degradation of A3G, thereby depleting the pool of cytosolic A3G available for incorporation into assembling virus particles (Conticello et al. 2003; Marin et al. 2003; Sheehy et al. 2003; Stopak et al. 2003; Yu et al. 2003; Mehle et al. 2004_b_; Kobayashi et al. 2005). Vif accomplishes this by simultaneously binding to both A3G and the cullin5-elongin B/C-Rbx ubiquitin ligase, thus serving as an adaptor to recruit the ligase complex to its substrate (Yu et al. 2003; figure 2). Vif itself may also be ubiquitylated and degraded, though it is not yet clear whether this is a bystander effect or important for Vif function (Mehle et al. 2004_a_).
Figure 2.
HIV-1 Vif induces the polyubiquitylation of APOBEC3G through recruitment of a cellular ubiquitin ligase. The known components of the cullin5 (Cul5)-elongin (Elo)B/C-Rbx ligase complex are indicated together with the identity of amino acid motifs that mediate important protein–protein interactions.
A number of studies have started to map the peptide sequences involved in the interactions between Vif, A3G and the ligase complex (figure 2). The binding of HIV-1 Vif to A3G is critically dependent on an Asp-Pro-Asp motif at positions 128–130 in A3G (Huthoff & Malim 2007). Satisfyingly, the recognition of this key element as a Vif-binding site has helped provide an explanation for the aforementioned species specificity of HIV/SIV Vif function (§2). More specifically, the African green monkey (AGM) A3G protein contains a lysine at position 128 and is unresponsive to HIV-1 Vif. Replacement of the aspartic acid found at this position in human-A3G with lysine renders the protein resistant to HIV-1 Vif but responsive to, and able to bind to, SIV-AGM Vif (Bogerd et al. 2004; Mangeat et al. 2004; Schrofelbauer et al. 2004; Xu et al. 2004). Consistent with the importance of acidic residues in human-A3G for HIV-1 Vif binding, a ‘complementary’ region harbouring positively charged residues at positions 14 and 17 in HIV-1 Vif (Asp-Arg-Met-Arg) appears to be important for the interaction (Schrofelbauer et al. 2006). This region is not, however, sufficient for the interaction, and a hydrophobic motif between residues 40 and 44 in HIV-1 Vif also plays a central role (Russell & Pathak 2007).
HIV-1 Vif has been shown to connect to the cullin5-elongin B/C-Rbx complex in at least two places, presumably stabilizing the interaction and helping confer specificity. The conserved Ser-Leu-Gln-Tyr-Leu-Ala-Leu-Ala-Ala-Leu motif between positions 144 and 153 of Vif (which is the most highly conserved sequence element among divergent Vif proteins (Oberste & Gonda 1992)) interacts with sequences in the elongin C component of the elongin B/C complex and is known as the BC-box (Mehle et al. 2004_a_; Yu et al. 2004_b_). This interaction mimics the well-described interaction between the von Hippel-Lindau tumour suppressor (pVHL), a cellular protein that inter alia targets the transcription factor hypoxia-inducible factor-α for ubiquitylation, and elongin B/C (Stebbins et al. 1999), with residues Leu-145, Gln-146, Ala-149 and Leu-150 in Vif playing central roles. In addition, and somewhat unusually for substrates of cullin-RING ubiquitin ligases, Vif also contacts the cullin5 scaffold protein via a zinc-coordinating motif (Luo et al. 2005; Mehle et al. 2006).
Though a considerable amount is known regarding the mechanism and factors involved in Vif-mediated A3G degradation, much still remains to be determined. For instance, the E2 conjugating enzyme(s) that catalyses the transfer of ubiquitin to A3G has yet to be identified; phosphorylation of Ser-144 in Vif's BC-box inhibits elongin C binding, suggesting that A3G degradation may be sensitive to regulation by as yet unidentified signalling pathways (Mehle et al. 2004_a_); and, there has been a recent report that A3G can be degraded without prior ubiquitylation, raising the possibility that polyubiquitylated Vif may be responsible for targeting A3G to the proteasome (Dang et al. 2008). Lastly, efforts employing structural biology to define various important interactions at atomic resolution will assist rational drug design efforts where the objective would be to interfere with Vif's capacity to block A3G function, thereby liberating A3G and allowing it to exert its anti-HIV-1 function.
5. APOBEC3G catalyses cytidine deamination of HIV-1 DNA
The APOBEC family of proteins comprises 11 members in humans (Harris & Liddament 2004; Conticello et al. 2005; Holmes et al. 2007_b_; Chiu & Greene 2008). The founder member, APOBEC1, was originally identified as the enzyme that is expressed in gastrointestinal tissues and post-transcriptionally edits apolipoprotein B mRNA at a unique site to create a premature stop codon (Teng et al. 1993). More specifically, APOBEC1 induces the deamination of the cytidine (C) at position 6666, resulting in its conversion to uridine (U). The two forms of apolipoprotein B have quite different functions: the longer form (apoB-100) is important for the transport of endogenously synthesized triglycerides and cholesterol, whereas the truncated version (apoB-48) is important for the absorption and transport of dietary fats (Anant & Davidson 2001). The second protein in this family to be recognized as an editing enzyme was AID (activation-induced deaminase) (Muramatsu et al. 1999), the protein that is expressed in germinal centre B cells and triggers both class switch recombination and somatic hypermutation by deaminating C residues at the immunoglobulin locus during the process of antibody diversification (Di Noia & Neuberger 2007). Thus, APOBEC proteins are defined as a collection of related polynucleotide (RNA/DNA) cytidine deaminases.
Each APOBEC protein contains one or two copies (A3G has two) of a cytidine deaminase (CDA) domain that is distinguished be the presence of a signature His/Cys-X-Glu-X23–28-Pro-Cys-X2-Cys motif: in some cases the CDA domain has deaminase activity, whereas in others it does not (discussed below) (Harris & Liddament 2004; Conticello et al. 2005; Holmes et al. 2007_b_; Chiu & Greene 2008). With respect to the likely mechanism of catalysis, the histidine and two cysteine residues are thought to coordinate a zinc ion that enables the catalytic glutamic acid to deprotonate a water molecule and generate the zinc hydroxide nucleophile that is necessary for the deamination of the pyrimidine ring of cytidine (Betts et al. 1994; Harris & Liddament 2004). Two recent structural studies, one on APOBEC2 and one on the carboxy-terminal CDA domain of A3G (the domain that is catalytically active), reveal that the core elements of the CDA domain comprise a platform of five β-strands that is flanked on either side by α-helices and connecting loops (Prochnow et al. 2007; Chen et al. 2008).
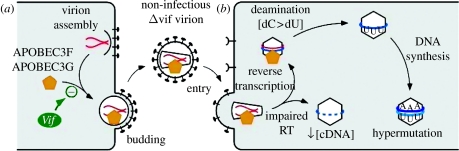
As an APOBEC protein, one obvious question that arose following the recognition of A3G as an inhibitor of HIV-1 infection was whether A3G has the capacity to edit/mutate HIV-1 sequences, particularly since DNA mutational activity had been noted using a bacteria-based assay system (Harris et al. 2002). Accordingly, it was demonstrated that the expression of A3G in virus-producing cells yields A3G-containing progeny virions (_vif_-deficient HIV-1 or murine leukaemia virus, MLV) that, following infection of fresh cells, produce reverse transcripts that are littered with guanosine (G) to adenosine (A) transition mutations on the plus (second) strand (figure 3; Harris et al. 2003_a_; Mangeat et al. 2003; Mariani et al. 2003; Zhang et al. 2003). Because the frequency of mutation can exceed 10 per cent of all G residues, this phenomenon is often called hypermutation. Since minus (first) strand DNA comprises the vast majority of the single-stranded DNA that is present during retroviral reverse transcription, and A3G selectively recognizes single-stranded substrates (Suspene et al. 2004; Yu et al. 2004_a_; Chelico et al. 2006), it is the minus strands that are subjected to cytidine deamination and then copied into plus-stranded DNA harbouring G-to-A mutations. The exceptions to this are the U3 region of the 5′-long terminal repeat (LTR) and the primer binding site (PBS), whose plus strand sequences are thought to be transiently single stranded during the later phases of reverse transcription and have been shown to be marked by C to thymine (T) mutations in the presence of A3G (Yu et al. 2004_a_). At the level of mutational burden discussed here, the disruption to viral genetic integrity is excessive and certainly sufficient to suppress infection and replication effectively: indeed, this mechanism of viral inhibition can be viewed as a form of error catastrophe.
Figure 3.
Inhibition of HIV-1 infection by APOBEC3G. (a) In the absence of Vif, A3G and A3F are packaged into progeny virions (producer) and transferred to (b) target cells. Infection is inhibited through a combination of cytidine deaminase-driven hypermutation and diminished reverse transcription. Refer to the text for further explanation.
A number of additional features of A3G-mediated mutagenesis of HIV-1 have been examined in depth. First, there are very evident local nucleotide sequence preferences for deamination, with the most favoured site (or hot spot) being 5′-CCCA (the substrate cytidine is underlined) (Harris et al. 2003_a_; Suspene et al. 2004; Bishop et al. 2004_a_; Yu et al. 2004_a_). Interestingly, this motif corresponds to 5′-TGGG in plus-stranded sequence such that mutation of a TGG tryptophan codon converts it to a TAG stop codon—a mutation that has a very high chance of disrupting the function of an encoded protein. Enzymatic studies using recombinant A3G in vitro have shown that it exhibits 3′-to-5′ processivity, perhaps explaining the preference for the 3′ C residue in the substrate consensus (Chelico et al. 2006). Second, sequence determinations across completely reverse transcribed HIV-1 sequences reveal polarity to the A3G-induced mutational frequency in that it (usually) increases in a 5′-to-3′ direction (when considering plus strand sequence) to reach peaks just upstream of the two polypurine tracts (PPTs) that prime plus strand DNA synthesis (i.e. there are two gradients per full length viral sequence) (Yu et al. 2004_a_; Suspene et al. 2006; Kijak et al. 2008). This has been attributed to the progressively longer time that the minus strand DNA exists in a single-stranded state (prior to becoming double-stranded DNA), thus offering increasingly greater opportunity for deamination to occur. This effect is probably accentuated by the aforementioned 3′-to-5′ processive character of A3G itself, which also results in preferential mutagenesis towards the 5′-terminus of single-stranded (minus sense) DNA (Chelico et al. 2006).
Conceptually, there are two potential fates for uracilated minus strand DNA. First, as discussed above, second strand DNA could be synthesized and the G-to-A mutations then fixed, should the viral DNA be stable maintained in a cell (i.e. as integrated proviral DNA) (figure 3). Second, it was proposed that U residues could be recognized by cellular DNA repair enzymes, ultimately leading to the degradation of edited reverse transcripts as a consequence of abortive repair of unpaired DNA strands (Harris et al. 2003_b_). Indeed, this was an attractive hypothesis to explain long-standing observations that _vif_-deficient viruses produce lower levels of cDNA following viral challenge (§2). However, a number of laboratories have demonstrated that inhibiting the cellular uracil DNA glycosidases UNG2 and/or SMUG1 neither ameliorates the inhibition of infection by A3G nor rescues the accumulation of viral cDNA in infected cells, indicating that these enzymes do not contribute to the A3G phenotype (Kaiser & Emerman 2006; Mbisa et al. 2007; Langlois & Neuberger 2008).
6. The packaging of APOBEC3G into HIV-1 virions
The packaging of A3G into newly assembled progeny virus particles is fundamental to anti-viral function. In the absence of Vif, and as illustrated in figure 3, encapsidated A3G is carried forward as HIV-1 infects target cells and must remain associated with the uncoated reverse transcription complex (RTC) to enable cytidine deamination to occur. A3G needs to interact with a number of viral components and macromolecules to accomplish this. During encapsidation, A3G must interact with the p55Gag viral polyprotein, potentially p160Gag-Pol, and with the genomic RNA of the virus. In slight contrast, following entry into target cells, A3G must interface with the mature processed viral proteins present in RTCs, but in the context of viral RNA that is being converted into cDNA by reverse transcription. Considerable effort has been devoted to understanding these aspects of A3G function, but experimentation is quite challenging, not least owing to difficulties in purifying full length A3G protein in a soluble form.
The current consensus view is that A3G packaging is determined by specific interactions between the N-terminal CDA domain of A3G and the nucleocapsid (NC) region of Gag that are also dependent upon the binding of A3G to RNA (Luo et al. 2004; Schafer et al. 2004; Svarovskaia et al. 2004; Zennou et al. 2004; Khan et al. 2005; Navarro et al. 2005; Burnett & Spearman 2007; Bogerd & Cullen 2008). Accordingly, A3G is not encapsidated by either HIV-1 virus-like particles that lack NC domains (Schafer et al. 2004; Zennou et al. 2004) or other retroviruses whose Gag proteins do not form complexes with A3G (Doehle et al. 2006). The identity of the RNA(s) that promote A3G packaging in HIV-1 infected cells remains somewhat unresolved, though HIV-1 genomic RNA and 7SL RNA (a small pol III transcript that is found in the signal recognition particle, SRP) have each been assigned stimulatory roles in the process (Luo et al. 2004; Khan et al. 2005, 2007; Wang et al. 2007). The fact that A3G is an avid RNA-binding protein (Navarro et al. 2005; Iwatani et al. 2006) raises an important question concerning packaging: specifically, since A3G is associated with an array of cellular ribonucleoprotein (RNP) complexes in an RNA-dependent manner (Chiu et al. 2005, 2006; Chelico et al. 2006; Kozak et al. 2006; Wichroski et al. 2006; Gallois-Montbrun et al. 2007), how can its RNA-binding site(s) be liberated to allow engagement with the RNA that facilitates packaging? Accordingly, elegant pulse–chase analyses revealed that it is newly synthesized A3G that is incorporated into virions, presumably averting its recruitment into cytosolic RNPs (Soros et al. 2007).
Further evidence to support the importance of RNA binding in A3G packaging is derived from the analyses of genetically engineered A3G proteins. Specifically, amino acid substitutions at Tyr-124 or Trp-127 yield proteins that are severely debilitated for packaging into HIV-1 particles, and are therefore poor inhibitors of virus infection (Huthoff & Malim 2007; Wang et al. 2007; Zhang et al. 2007). More recent results indicate that these mutant proteins are also substantially deficient for RNA binding and are unable to form A3G–A3G dimers efficiently (H. Huthoff & M. H. Malim 2008, unpublished observations). In other words, and reminiscent of its interactions with NC, the presence of RNA appears to be important for A3G's ability to interact with itself, and to be packaged into viral particles. A further inference that can be drawn from these data are that A3G dimerization involves interactions between the N-terminal CDA domains of two A3G monomers. In fact, this dimer interface can be modelled on the reported structure of the contact region between the two APOBEC2 dimers that comprise the APOBEC2 tetramer (Prochnow et al. 2007; H. Huthoff & M. H. Malim 2008, unpublished observations). Notably, the 124–127 regions lie adjacent to the Asp-Pro-Asp motif that is central to Vif binding: efforts to block this interaction pharmacologically will need to avoid perturbing function of the critical 124–127 motif as this would probably interfere with anti-viral efficacy.
Much less is currently understood concerning the nature of A3G's interaction(s) with the RTC. Presumably, it is tethered through a combination of interactions with NC, RNA/DNA and perhaps other components such as integrase (Luo et al. 2007). One feature that has been described is the activation of A3G's deaminase function by RNaseH (Soros et al. 2007). This activity may facilitate the release of RNA from A3G thereby allowing access of substrate DNA to the catalytic site; this role is distinct from RNaseH's obvious role in liberating single-stranded DNA for editing through the degradation and removal of RNA from RNA–DNA duplexes during reverse transcription.
7. Cytidine deamination independent effects of APOBEC3G on HIV-1
Though A3G is unequivocally a potent mutator of HIV-1 DNA (§5), recent work has revealed that A3G can also exert deamination-independent effects during infection. The initial impetus for this line of investigation emanated from analyses of experimentally mutated A3G proteins. Specifically, replacement of the conserved cysteine, histidine or glutamic acid residues at the heart of the C-terminal CDA domain demonstrated not only that editing is entirely mediated by this domain (Navarro et al. 2005; Newman et al. 2005), but also that these deamination-deficient mutant proteins can still inhibit virus infection to a significant extent (Newman et al. 2005). Considerable effort is being devoted towards defining the precise features of inhibition, as well as the underlying mechanism(s): this remains a controversial topic in the field, whose investigation is probably exacerbated by differences in the experimental configurations that are used in different laboratories. Nevertheless, it is a widely held view that wild-type A3G diminishes the accumulation of nascent HIV-1 reverse transcripts in newly challenged cells (§2). These effects have been described for early and late reverse transcription intermediates, and it appears that the magnitude of the defect(s) increases as later cDNA products are measured (Mangeat et al. 2003; Mariani et al. 2003; Bishop et al. 2006; Guo et al. 2006; Holmes et al. 2007_a_; Luo et al. 2007; Mbisa et al. 2007; Anderson & Hope 2008).
A confounding feature of analyses performed in living cells is that it is not possible to differentiate between effects on DNA synthesis and DNA degradation. Accordingly, in vitro systems that lack the nucleases present in cells have been employed to address such questions. Endogenous reverse transcription reactions in which purified HIV-1 particles are permeabilized with melittin, incubated with dNTPs and the levels of cDNA synthesis then measured demonstrate that A3G inhibits reverse transcription in a dose-dependent fashion (Bishop et al. 2008). As with cell-based experiments, the extent of inhibition becomes progressively greater as later reverse transcription intermediates are examined. Moreover, the addition of the first deoxynucleotide to the 3′-hydroxyl of the tRNALys primer is not discernibly inhibited, implying that tRNALys placement at the PBS of the viral RNA is unaffected by A3G. These findings, together with results obtained using reconstituted in vitro systems (Iwatani et al. 2007), indicate that A3G does not directly inhibit the biosynthetic capabilities of the reverse transcriptase enzyme but, rather, impedes its progression along template RNA (figure 3). The molecular mechanism for this effect continues to be investigated, but A3G's capacity to bind single-stranded RNA and thereby sterically hinder the translocation of reverse transcriptase would lead to the observed polarity of inhibition to cDNA accumulation.
More recent analyses of editing-deficient A3G proteins of the aforementioned genre have indicated that while they clearly possess demonstrable anti-HIV-1 phenotypes at high concentrations (e.g. as attained by transient transfection of expression plasmids), these effects titrate away at lower expression levels (Holmes et al. 2007_a_; Miyagi et al. 2007; Schumacher et al. 2008). These findings have been interpreted as indicating that A3G does not possess editing-independent anti-viral activity. However, such conclusions are not justified as endogenous reverse transcription assays show that such mutant proteins also have diminished capacity to interfere with reverse transcription (K. N. Bishop & M. H. Malim 2008, unpublished observations), implying that it is not necessarily straightforward to segregate the editing-dependent and -independent activities of A3G. Indeed, nuances as this suggest that a combination of experimental approaches and systems are needed to determine precisely how A3G inhibits HIV-1 replication.
Interestingly, the balance between the editing and non-editing effects of A3G may vary under differing circumstances. This concept is illustrated by a recent analysis of _vif_-deficient HIV-1 infection in primary blood lymphocytes (PBLs) and the human T-cell line H9, neither of which support productive replication of this virus (Knoepfel et al. 2008). Here, nascent viral cDNAs recovered from challenged cells harboured greater levels of G-to-A mutation (both in numbers of DNAs carrying mutations, and in the extent of mutational burden in the DNAs that were mutated) in the context of H9 cells compared with PBLs. This may imply that non-editing effects contribute a greater proportion of A3G's anti-HIV-1 phenotype in PBLs than in H9 cells. One can speculate that the differential regulation of A3G activity in different cell types may contribute to such phenomena (Thielen et al. 2007).
In addition to the editing-independent activity of virion encapsidated A3G, a non-editing anti-HIV-1 effect can also be exhibited by A3G that is present in cells that are targets for infection and is, therefore, neither reliant on virion packaging nor susceptible to regulation by Vif. More specifically, the experimental alleviation of A3G expression in resting PBLs (which are ordinarily refractive to productive HIV-1 infection (Stevenson et al. 1990; Zack et al. 1990)) via RNA interference has been shown to render such cells susceptible to infection (Chiu et al. 2005), indicating that A3G expression is essential for the barrier to infection in these cells. Interestingly, only approximately10 per cent of the incomplete reverse transcripts, whose accumulation is a hallmark of resting T-cell infection, are modified by G-to-A hypermutation, implying that A3G's contribution to inefficient reverse transcription in these cells is not linked to cytidine deamination (Chiu et al. 2005). The degree to which the mechanisms of editing-independent anti-viral effects that are conferred by A3G that is virion associated, or is residing in target cells, are related to each other remains to be established, though certain parallels are clearly evident.
8. Anti-HIV-1 phenotypes of diverse human APOBEC proteins
As noted in §5, A3G is but one out of the 11 members of the human APOBEC protein family. Accordingly, multiple groups have examined the anti-HIV-1 properties of all the APOBEC proteins, and the results have been catalogued in recent review papers (Holmes et al. 2007_b_; Chiu & Greene 2008). The most significant other human protein with respect to HIV-1 is APOBEC3F (A3F): it is a strong inhibitor of _vif_-deficient HIV-1, though not as potent as A3G (Bishop et al. 2004_a_; Liddament et al. 2004; Wiegand et al. 2004; Zheng et al. 2004; Zennou & Bieniasz 2006), and is the family member that is most closely related to A3G in that they share approximately 50 per cent sequence identity. A3F is also efficiently inhibited by HIV-1 Vif via the recruitment of the cullin5-elongin B/C-Rbx ubiquitin ligase and ensuing proteasomal degradation (Liu et al. 2005). These shared features of A3G and A3F seemingly accord with the view that these are the most significant APOBEC enzymes in terms of natural HIV-1 infection (namely in humans). Interestingly, the sequence elements within HIV-1 Vif required for inhibition of A3G versus A3F differ from each other (Simon et al. 2005; Tian et al. 2006; Russell & Pathak 2007), implying that evolutionary pressures have selected for the ability to suppress both proteins. Moreover, A3G and A3F are both expressed in human CD4 T cells (the principal target for HIV-1 infection in vivo), further supporting the notion that both enzymes naturally encounter HIV-1, and that the virus must have evolved to evade the anti-viral activities of each of them.
As with A3G, A3F also induces G-to-A hypermutation, though the degree of mutagenesis has been noted to be lower than for A3G in some cultured cell experiments (Bishop et al. 2004_a_; Zheng et al. 2004; Zennou & Bieniasz 2006; Holmes et al. 2007_a_). The consensus dinucleotide target for deamination is also different with 5′-TC (substrate cytidine underlined) serving as the preferred site for A3F (Bishop et al. 2004_a_; Liddament et al. 2004; Wiegand et al. 2004), rather than the 5′-CC defined for A3G (§5). Numerous analyses of HIV-1 sequences recovered from HIV-1 infected persons reveal the frequent presence of G-to-A hypermutated viral DNA (Fitzgibbon et al. 1993; Vartanian et al. 1994; Janini et al. 2001; Kieffer et al. 2005; Pace et al. 2006; Kijak et al. 2008; Land et al. 2008). Further inspection of the local sequence context of such mutations reveals 5′-GG (plus sense sequence) as the predominant target and 5′-GA as the second most likely substrate. These observations suggest strongly that A3G and A3F both contribute to hypermutation in vivo, but that A3G is responsible for the greater proportion. Accordingly, A3G appears to be the more significant anti-HIV-1 factor, a view that is consistent with its stronger phenotype in cultured cells, and its higher level of expression in human CD4 T cells (F. A. Koning, E. N. Newman, K. J. Kunstman, S. M. Wolinsky & M. H. Malim 2008, unpublished observations).
There has also been some limited structure–function dissection of A3F. As with A3G, the C-terminal CDA domain mediates deamination whereas the N-terminal domain is non-enzymatic but important for efficient packaging into virions (Jonsson et al. 2006; Holmes et al. 2007_a_). Careful analyses of editing-deficient mutant proteins shows that the magnitudes of their anti-HIV-1 phenotypes mirror more closely that of wild-type A3F than is the case for A3G (Holmes et al. 2007_a_). Moreover, these A3F mutants also appear to inhibit reverse transcription as effectively as the wild-type protein, both during infection and in endogenous assays (Holmes et al. 2007_a_; K. N. Bishop & M. H. Malim 2008, unpublished observations). Together with the lower mutagenic activity of A3F, these findings may indicate that the non-editing activities of A3F play a substantial role in the anti-viral phenotype of this protein.
Among the remaining human APOBEC proteins, some intriguing observations have been made regarding their effects on HIV-1, though the significance for in vivo infection continues to be investigated. APOBEC3B (A3B) inhibits HIV-1 in cultured cell assays, is resistant to inhibition by Vif (A3B can therefore inhibit wild-type HIV-1), has two CDA domains that are each capable of catalysing deamination, but is barely expressed in human CD4 T cells so is unlikely to impact HIV-1 substantially during natural infection (Bishop et al. 2004_a_; Doehle et al. 2005; Bogerd et al. 2007). APOBEC3A (A3A) contains a single CDA domain, is expressed in immature monocytes, and has been reported to restrict HIV-1 infection (Peng et al. 2007). APOBEC3C (A3C) also contains a single CDA domain, is expressed in CD4 T cells, and can act in target cells to introduce infrequent G-to-A mutations (i.e. at sublethal levels) during infection by some HIV-1 strains (discussed further in §9) (Bourara et al. 2007). Lastly, a third single domain protein, APOBEC3D/E (A3D/E), can also modestly inhibit HIV-1 infection in a Vif-sensitive manner when overexpressed in virus producing cells (Dang et al. 2006).
In addition to the examination of HIV-1's susceptibility to human APOBEC proteins (discussed herein), there has been extensive interrogation of the effects of these enzymes (from multiple vertebrate species) on other retroviruses, endogenous retroviruses, transposable elements (long terminal repeat (LTR)-containing as well as non-LTR-containing autonomous and non-autonomous types) and some DNA viruses that do not include reverse transcription in their replication strategy such as adeno-associated virus (AAV) and human papillomavirus (HPV). There are many instances of inhibitory effects on infection and transposition, as well as the induction of DNA mutations, and much of this work is summarized elsewhere (Chen et al. 2006; Holmes et al. 2007_b_; Chiu & Greene 2008; Vartanian et al. 2008). Nevertheless, there is still a great deal to be understood regarding these APOBEC–pathogen/parasite interactions including: the determinants of substrate specificity for particular APOBEC proteins, mechanisms of inhibition (editing and editing-independent effects), mechanisms and routes of evasion, and assessments of the physiological significance of these interactions in the context of animal hosts (rather than in cultured cell experiments). To illustrate the last point, the examination of endogenous retrovirus sequences in the mice and human genomes indicates that some ‘ancient’ retroviral infections were inactivated by APOBEC protein-mediated mutations, thereby attesting to the biological relevance of interactions with APOBEC proteins (Esnault et al. 2005; Jern et al. 2007; Lee et al. 2008).
Somewhat provocatively, A3F and rat-APOBEC1, but not A3G, also appear to be capable of inducing low levels of C-to-T mutations in plus-stranded HIV-1 cDNA (Bishop et al. 2004_a_,b). Since double-stranded DNA is not a substrate for these enzymes (§5), this indicates that APOBEC proteins other than APOBEC1 can edit RNA substrates and that RNA viruses that replicate exclusively via RNA intermediates have the potential to be subjected to cytidine deamination: whether such events can contribute to the inhibition or sequence diversification of certain viruses remains to be determined.
9. APOBEC proteins and natural HIV-1 infection
Several clear lines of evidence combine to make a convincing case for HIV-1 encountering the APOBEC proteins during natural infection. First, the virus encodes the Vif protein to counteract A3G and A3F: without this protein, the virus cannot replicate in human CD4 T cells (§2). Second, investigation of numerous HIV and SIV vif alleles from human and non-human primate hosts strongly suggests that Vif is a critical determinant of zoonotic transmission (Simon et al. 1998_b_; Mariani et al. 2003; Gaddis et al. 2004; Hatziioannou et al. 2006). More specifically, Vif proteins derived from SIVs whose ancestors have not established themselves in humans are inactive (or weakly active) against human-A3G/F whereas Vifs encoded by SIVs from chimpanzees or sooty mangabeys, whose ancestors infected humans and evolved to become HIV-1 and HIV type-2, respectively, are functional in human T cells. Third, G-to-A hypermutated HIV-1 sequences are readily detected in infected persons (§8).
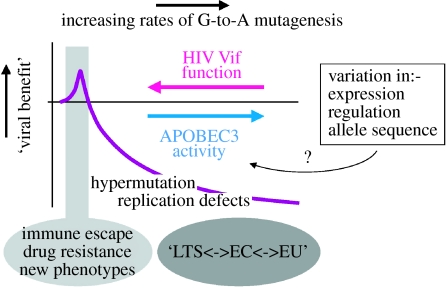
On the basis of these arguments, it is predicted that there is a balance between Vif function and the APOBEC proteins, that this is variable in its extent, and that the Vif-mediated inhibition of A3G/F activity is incomplete at times. Indeed, laboratory-based experiments where overexpression of A3G can modestly suppress wild-type HIV-1 infection and induce the accumulation of G-to-A mutations illustrate this point (Sheehy et al. 2002; Mangeat et al. 2003; Zhang et al. 2003). Conceivably, APOBEC proteins that escape suppression by Vif have the potential to be either beneficial or detrimental to the virus in the context of natural infection (cartooned in figure 4). Towards one end of the spectrum, low levels of A3G/F (or other family members such as A3B, A3C or A3D/E) may induce sublethal levels of editing that help drive sequence diversification by augmenting the copying errors (made by reverse transcriptase and RNA polymerase II) that characterize retroviral replication. In instances where such mutations are advantageous, they could facilitate either immune escape, phenotypic changes in the virus, or, in this era of anti-retroviral therapy, the acquisition of drug resistance: as an example, an extensive analysis of recently transmitted HIV-1 strains indicates that A3G/F contributes to single nucleotide sequence variation (Keele et al. 2008). One can speculate, perhaps, that the potential to acquire additional mutations may explain why the HIVs and SIVs counteract the A3G/F enzymes by the comparatively elaborate strategy of encoding a specific inhibitory protein, Vif. More straightforward alternatives might be to avert APOBEC proteins by not encapsidating them into virions (e.g. human T-cell leukaemia virus type-I, HTLV-I, and Mason–Pfizer monkey virus, M-PMV) (Doehle et al. 2006; Derse et al. 2007) or, perhaps, by simply replicating in non-expressing cells (Delebecque et al. 2006).
Figure 4.
Hypothetical representation of the balance between HIV-1 Vif and the APOBEC3 proteins. Viral benefit is represented by the purple line and the rate of mutation increases from left to right. It is possible that variations in Vif–A3G/F interactions (as well as in other APOBEC proteins) may contribute to viral sequence evolution, be beneficial for viral control in infected persons (for instances of robust viral control this could manifest as long-term survivor (LTS) or elite controller (EC) phenotypes), or alter rates of viral transmission (exposed uninfected person, EU). Effects on virus replication may, or may not, require editing. Potential causes of variation in cellular APOBEC3 protein activity are indicated in the box.
How, then, might the balance between the APOBEC proteins and HIV-1 Vif vary during natural infection of humans? A number of population-based studies in HIV-1 infected/exposed cohorts provide some important clues. First, one obvious possibility is that there are variations in the functionality of different vif alleles, and observations supporting this have been described (Simon et al. 2005). A second possibility is that A3G/F expression may vary, and, indeed, A3G/F mRNA levels have been associated with HIV-1 infection status, as well as with differences in rates of disease progression, plasma virus loads, and transmission (Jin et al. 2005; Cho et al. 2006; Biasin et al. 2007). A third possibility is genetic variation among individuals, and polymorphisms in A3G or cullin5 have been correlated with differences in the rates of disease progression (An et al. 2004, 2007). Finally, though not addressing cause versus effect, increased CD4 T-cell counts and decreased virus loads have each been correlated with the burden of hypermutated (inactivated) HIV-1 sequences (Pace et al. 2006; Land et al. 2008). It is worth noting, however, that not only does a good deal of controversy remain concerning the conclusions of some of these studies (Do et al. 2005; Gandhi et al. 2008) but also unambiguously establishing causality for these in vivo phenotypes can be expected to be challenging. Accordingly, it will require substantial future investigation of longitudinally collected specimens to determine accurately the influence of APOBEC variation (in terms of expression, activity, regulation and allelic differences) on the susceptibility and resistance to HIV-1 infection, the rate of progression to AIDS following infection, as well as the diversification and evolution of viral sequences.
Acknowledgements
I wish to thank Michael Neuberger, Pat Gearhart and Tomas Lindahl for organizing the 2008 Discussion Meeting on DNA Deamination in Immunity, Virology and Cancer. I am also indebted to the members of my laboratory and our collaborators who have contributed to our work on Vif and APOBEC proteins.
Footnotes
One contribution of 17 to a Discussion Meeting Issue ‘DNA deamination in immunity, virology and cancer’.
References
- An P., et al. APOBEC3G genetic variants and their influence on the progression to AIDS. J. Virol. 2004;78:11 070–11 076. doi: 10.1128/JVI.78.20.11070-11076.2004. doi:10.1128/JVI.78.20.11070-11076.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An P., et al. Polymorphisms of CUL5 are associated with CD4+T cell loss in HIV-1 infected individuals. PLoS Genet. 2007;3:e19. doi: 10.1371/journal.pgen.0030019. doi:10.1371/journal.pgen.0030019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anant S., Davidson N.O. Molecular mechanisms of apolipoprotein B mRNA editing. Curr. Opin. Lipidol. 2001;12:159–165. doi: 10.1097/00041433-200104000-00009. doi:10.1097/00041433-200104000-00009 [DOI] [PubMed] [Google Scholar]
- Anderson J.L., Hope T.J. APOBEC3G restricts early HIV-1 replication in the cytoplasm of target cells. Virology. 2008;375:1–12. doi: 10.1016/j.virol.2008.01.042. doi:10.1016/j.virol.2008.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts L., Xiang S., Short S.A., Wolfenden R., Carter C.W., Jr Cytidine deaminase. The 2.3 A crystal structure of an enzyme: transition-state analog complex. J. Mol. Biol. 1994;235:635–656. doi: 10.1006/jmbi.1994.1018. doi:10.1006/jmbi.1994.1018 [DOI] [PubMed] [Google Scholar]
- Biasin M., et al. Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3G: a possible role in the resistance to HIV of HIV-exposed seronegative individuals. J. Infect. Dis. 2007;195:960–964. doi: 10.1086/511988. doi:10.1086/511988 [DOI] [PubMed] [Google Scholar]
- Bieniasz P.D. Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 2004;5:1109–1115. doi: 10.1038/ni1125. doi:10.1038/ni1125 [DOI] [PubMed] [Google Scholar]
- Bishop K.N., Holmes R.K., Sheehy A.M., Davidson N.O., Cho S.J., Malim M.H. Cytidine deamination of retroviral DNA by diverse APOBEC proteins. Curr. Biol. 2004;14:1392–1396. doi: 10.1016/j.cub.2004.06.057. doi:10.1016/j.cub.2004.06.057 [DOI] [PubMed] [Google Scholar]
- Bishop K.N., Holmes R.K., Sheehy A.M., Malim M.H. APOBEC-mediated editing of viral RNA. Science. 2004;305:645. doi: 10.1126/science.1100658. doi:10.1126/science.1100658 [DOI] [PubMed] [Google Scholar]
- Bishop K.N., Holmes R.K., Malim M.H. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. doi:10.1128/JVI.00839-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, K. N., Verma, M., Kim, E.-Y., Wolinsky, S. M. & Malim, M. H. 2008 APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog 4 (doi:10.1371/journal.ppat.1000231) [DOI] [PMC free article] [PubMed]
- Bogerd H.P., Cullen B.R. Single-stranded RNA facilitates nucelocapsid: APOBEC3G complex formation. RNA. 2008;14:1228–1236. doi: 10.1261/rna.964708. doi:10.1261/rna.964708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P., Doehle B.P., Wiegand H.L., Cullen B.R. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl Acad. Sci. USA. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. doi:10.1073/pnas.0307713101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P., Wiegand H.L., Doehle B.P., Cullen B.R. The intrinsic antiretroviral factor APOBEC3B contains two enzymatically active cytidine deaminase domains. Virology. 2007;364:486–493. doi: 10.1016/j.virol.2007.03.019. doi:10.1016/j.virol.2007.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourara K., Liegler T.J., Grant R.M. Target cell APOBEC3C can induce limited G-to-A mutation in HIV-1. PLoS Pathog. 2007;3:1477–1485. doi: 10.1371/journal.ppat.0030153. doi:10.1371/journal.ppat.0030153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A.L., Dykxhoorn D.M., Benita Y., Yan N., Engelman A., Xavier R.J., Lieberman J., Elledge S.J. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. doi:10.1126/science.1152725 [DOI] [PubMed] [Google Scholar]
- Burnett A., Spearman P. APOBEC3G multimers are recruited to the plasma membrane for packaging into human immunodeficiency virus type 1 virus-like particles in an RNA-dependent process requiring the NC basic linker. J. Virol. 2007;81:5000–5013. doi: 10.1128/JVI.02237-06. doi:10.1128/JVI.02237-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelico L., Pham P., Calabrese P., Goodman M.F. APOBEC3G DNA deaminase acts processively 3′-5′ on single-stranded DNA. Nat. Struct. Mol. Biol. 2006;13:392–399. doi: 10.1038/nsmb1086. doi:10.1038/nsmb1086 [DOI] [PubMed] [Google Scholar]
- Chen H., Lilley C.E., Yu Q., Lee D.V., Chou J., Narvaiza I., Landau N.R., Weitzman M.D. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. doi:10.1016/j.cub.2006.01.031 [DOI] [PubMed] [Google Scholar]
- Chen K.M., Harjes E., Gross P.J., Fahmy A., Lu Y., Shindo K., Harris R.S., Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. doi:10.1038/nature06638 [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Greene W.C. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 2008;26:317–353. doi: 10.1146/annurev.immunol.26.021607.090350. doi:10.1146/annurev.immunol.26.021607.090350 [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Soros V.B., Kreisberg J.F., Stopak K., Yonemoto W., Greene W.C. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. doi:10.1038/nature03493 [DOI] [PubMed] [Google Scholar]
- Chiu Y.L., Witkowska H.E., Hall S.C., Santiago M., Soros V.B., Esnault C., Heidmann T., Greene W.C. High-molecular-mass APOBEC3G complexes restrict Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103:15 588–15 593. doi: 10.1073/pnas.0604524103. doi:10.1073/pnas.0604524103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.J., Drechsler H., Burke R.C., Arens M.Q., Powderly W., Davidson N.O. APOBEC3F and APOBEC3G mRNA levels do not correlate with human immunodeficiency virus type 1 plasma viremia or CD4+T-cell count. J. Virol. 2006;80:2069–2072. doi: 10.1128/JVI.80.4.2069-2072.2006. doi:10.1128/JVI.80.4.2069-2072.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conticello S.G., Harris R.S., Neuberger M.S. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. doi:10.1016/j.cub.2003.10.034 [DOI] [PubMed] [Google Scholar]
- Conticello S.G., Thomas C.J., Petersen-Mahrt S.K., Neuberger M.S. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol. Biol. Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. doi:10.1093/molbev/msi026 [DOI] [PubMed] [Google Scholar]
- Courcoul M., Patience C., Rey F., Blanc D., Harmache A., Sire J., Vigne R., Spire B. Peripheral blood mononuclear cells produce normal amounts of defective Vif- human immunodeficiency virus type 1 particles which are restricted for the preretrotranscription steps. J. Virol. 1995;69:2068–2074. doi: 10.1128/jvi.69.4.2068-2074.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Wang X., Esselman W.J., Zheng Y.H. Identification of APOBEC3DE as another antiretroviral factor from the human APOBEC family. J. Virol. 2006;80:10 522–10 533. doi: 10.1128/JVI.01123-06. doi:10.1128/JVI.01123-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Siew L.M., Zheng Y.H. APOBEC3G is degraded by the proteasomal pathway in a Vif-dependent manner without being polyubiquitylated. J. Biol. Chem. 2008;283:13 124–13 131. doi: 10.1074/jbc.M708728200. doi:10.1074/jbc.M708728200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delebecque F., et al. Restriction of foamy viruses by APOBEC cytidine deaminases. J. Virol. 2006;80:605–614. doi: 10.1128/JVI.80.2.605-614.2006. doi:10.1128/JVI.80.2.605-614.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derse D., Hill S.A., Princler G., Lloyd P., Heidecker G. Resistance of human T cell leukemia virus type 1 to APOBEC3G restriction is mediated by elements in nucleocapsid. Proc. Natl Acad. Sci. USA. 2007;104:2915–2920. doi: 10.1073/pnas.0609444104. doi:10.1073/pnas.0609444104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrosiers R.C., Lifson J.D., Gibbs J.S., Czajak S.C., Howe A.Y., Arthur L.O., Johnson R.P. Identification of highly attenuated mutants of simian immunodeficiency virus. J. Virol. 1998;72:1431–1437. doi: 10.1128/jvi.72.2.1431-1437.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. doi:10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Do H., et al. Exhaustive genotyping of the CEM15 (APOBEC3G) gene and absence of association with AIDS progression in a French cohort. J. Infect. Dis. 2005;191:159–163. doi: 10.1086/426826. doi:10.1086/426826 [DOI] [PubMed] [Google Scholar]
- Doehle B.P., Schafer A., Cullen B.R. Human APOBEC3B is a potent inhibitor of HIV-1 infectivity and is resistant to HIV-1 Vif. Virology. 2005;339:281–288. doi: 10.1016/j.virol.2005.06.005. doi:10.1016/j.virol.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Doehle B.P., Bogerd H.P., Wiegand H.L., Jouvenet N., Bieniasz P.D., Hunter E., Cullen B.R. The betaretrovirus Mason-Pfizer monkey virus selectively excludes simian APOBEC3G from virion particles. J. Virol. 2006;80:12 102–12 108. doi: 10.1128/JVI.01600-06. doi:10.1128/JVI.01600-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C., Heidmann O., Delebecque F., Dewannieux M., Ribet D., Hance A.J., Heidmann T., Schwartz O. APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature. 2005;433:430–433. doi: 10.1038/nature03238. doi:10.1038/nature03238 [DOI] [PubMed] [Google Scholar]
- Fisher A.G., Ensoli B., Ivanoff L., Chamberlain M., Petteway S., Ratner L., Gallo R.C., Wong-Staal F. The sor gene of HIV-1 is required for efficient virus transmission in vitro. Science. 1987;237:888–893. doi: 10.1126/science.3497453. doi:10.1126/science.3497453 [DOI] [PubMed] [Google Scholar]
- Fitzgibbon J.E., Mazar S., Dubin D.T. A new type of G→ A hypermutation affecting human immunodeficiency virus. AIDS Res. Hum. Retroviruses. 1993;9:833–838. doi: 10.1089/aid.1993.9.833. [DOI] [PubMed] [Google Scholar]
- Fouchier R.A., Simon J.H., Jaffe A.B., Malim M.H. Human immunodeficiency virus type 1 Vif does not influence expression or virion incorporation of gag-, pol-, and env-encoded proteins. J. Virol. 1996;70:8263–8269. doi: 10.1128/jvi.70.12.8263-8269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabuzda D.H., Lawrence K., Langhoff E., Terwilliger E., Dorfman T., Haseltine W.A., Sodroski J. Role of vif in replication of human immunodeficiency virus type 1 in CD4+T lymphocytes. J. Virol. 1992;66:6489–6495. doi: 10.1128/jvi.66.11.6489-6495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddis N.C., Chertova E., Sheehy A.M., Henderson L.E., Malim M.H. Comprehensive investigation of the molecular defect in vif-deficient human immunodeficiency virus type 1 virions. J. Virol. 2003;77:5810–5820. doi: 10.1128/JVI.77.10.5810-5820.2003. doi:10.1128/JVI.77.10.5810-5820.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddis N.C., et al. Further investigation of simian immunodeficiency virus Vif function in human cells. J. Virol. 2004;78:12 041–12 046. doi: 10.1128/JVI.78.21.12041-12046.2004. doi:10.1128/JVI.78.21.12041-12046.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo R., Wong-Staal F., Montagnier L., Haseltine W.A., Yoshida M. HIV/HTLV gene nomenclature. Nature. 1988;333:504. doi: 10.1038/333504a0. doi:10.1038/333504a0 [DOI] [PubMed] [Google Scholar]
- Gallois-Montbrun S., Kramer B., Swanson C.M., Byers H., Lynham S., Ward M., Malim M.H. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. doi:10.1128/JVI.02287-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi S.K., Siliciano J.D., Bailey J.R., Siliciano R.F., Blankson J.N. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J. Virol. 2008;82:3125–3130. doi: 10.1128/JVI.01533-07. doi:10.1128/JVI.01533-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.P. Genetic control of retrovirus susceptibility in mammalian cells. Annu. Rev. Genet. 2004;38:61–85. doi: 10.1146/annurev.genet.38.072902.094136. doi:10.1146/annurev.genet.38.072902.094136 [DOI] [PubMed] [Google Scholar]
- Guo F., Cen S., Niu M., Saadatmand J., Kleiman L. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 2006;80:11 710–11 722. doi: 10.1128/JVI.01038-06. doi:10.1128/JVI.01038-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S., Liddament M.T. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004;4:868–877. doi: 10.1038/nri1489. doi:10.1038/nri1489 [DOI] [PubMed] [Google Scholar]
- Harris R.S., Petersen-Mahrt S.K., Neuberger M.S. RNA editing enzyme APOBEC1 and some of its homologs can act as DNA mutators. Mol. Cell. 2002;10:1247–1253. doi: 10.1016/s1097-2765(02)00742-6. doi:10.1016/S1097-2765(02)00742-6 [DOI] [PubMed] [Google Scholar]
- Harris R.S., Bishop K.N., Sheehy A.M., Craig H.M., Petersen-Mahrt S.K., Watt I.N., Neuberger M.S., Malim M.H. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003a;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. doi:10.1016/S0092-8674(03)00423-9 [DOI] [PubMed] [Google Scholar]
- Harris R.S., Sheehy A.M., Craig H.M., Malim M.H., Neuberger M.S. DNA deamination: not just a trigger for antibody diversification but also a mechanism for defense against retroviruses. Nat. Immunol. 2003b;4:641–643. doi: 10.1038/ni0703-641. doi:10.1038/ni0703-641 [DOI] [PubMed] [Google Scholar]
- Hatziioannou T., Princiotta M., Piatak M., Jr, Yuan F., Zhang F., Lifson J.D., Bieniasz P.D. Generation of simian-tropic HIV-1 by restriction factor evasion. Science. 2006;314:95. doi: 10.1126/science.1130994. doi:10.1126/science.1130994 [DOI] [PubMed] [Google Scholar]
- Holmes R.K., Koning F.A., Bishop K.N., Malim M.H. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation. Comparisons with APOBEC3G. J. Biol. Chem. 2007a;282:2587–2595. doi: 10.1074/jbc.M607298200. doi:10.1074/jbc.M607298200 [DOI] [PubMed] [Google Scholar]
- Holmes R.K., Malim M.H., Bishop K.N. APOBEC-mediated viral restriction: not simply editing? Trends Biochem. Sci. 2007b;32:118–128. doi: 10.1016/j.tibs.2007.01.004. doi:10.1016/j.tibs.2007.01.004 [DOI] [PubMed] [Google Scholar]
- Huthoff H., Malim M.H. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J. Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. doi:10.1128/JVI.02795-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y., Takeuchi H., Strebel K., Levin J.G. Biochemical activities of highly purified, catalytically active human APOBEC3G: correlation with antiviral effect. J. Virol. 2006;80:5992–6002. doi: 10.1128/JVI.02680-05. doi:10.1128/JVI.02680-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwatani Y., et al. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 2007;35:7096–7108. doi: 10.1093/nar/gkm750. doi:10.1093/nar/gkm750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janini M., Rogers M., Birx D.R., McCutchan F.E. Human immunodeficiency virus type 1 DNA sequences genetically damaged by hypermutation are often abundant in patient peripheral blood mononuclear cells and may be generated during near-simultaneous infection and activation of CD4(+) T cells. J. Virol. 2001;75:7973–7986. doi: 10.1128/JVI.75.17.7973-7986.2001. doi:10.1128/JVI.75.17.7973-7986.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P., Stoye J.P., Coffin J.M. Role of APOBEC3 in genetic diversity among endogenous murine leukemia viruses. PLoS Genet. 2007;3:2014–2022. doi: 10.1371/journal.pgen.0030183. doi:10.1371/journal.pgen.0030183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Brooks A., Chen H., Bennett R., Reichman R., Smith H. APOBEC3G/CEM15 (hA3G) mRNA levels associate inversely with human immunodeficiency virus viremia. J. Virol. 2005;79:11 513–11 516. doi: 10.1128/JVI.79.17.11513-11516.2005. doi:10.1128/JVI.79.17.11513-11516.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson S.R., Hache G., Stenglein M.D., Fahrenkrug S.C., Andresdottir V., Harris R.S. Evolutionarily conserved and non-conserved retrovirus restriction activities of artiodactyl APOBEC3F proteins. Nucleic Acids Res. 2006;34:5683–5694. doi: 10.1093/nar/gkl721. doi:10.1093/nar/gkl721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser S.M., Emerman M. Uracil DNA glycosylase is dispensable for human immunodeficiency virus type 1 replication and does not contribute to the antiviral effects of the cytidine deaminase Apobec3G. J. Virol. 2006;80:875–882. doi: 10.1128/JVI.80.2.875-882.2006. doi:10.1128/JVI.80.2.875-882.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele B.F., et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl Acad. Sci. USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. doi:10.1073/pnas.0802203105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A., et al. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. doi:10.1128/JVI.79.9.5870-5874.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A., Goila-Gaur R., Opi S., Miyagi E., Takeuchi H., Kao S., Strebel K. Analysis of the contribution of cellular and viral RNA to the packaging of APOBEC3G into HIV-1 virions. Retrovirology. 2007;4:48. doi: 10.1186/1742-4690-4-48. doi:10.1186/1742-4690-4-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer T.L., Kwon P., Nettles R.E., Han Y., Ray S.C., Siliciano R.F. G→A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J. Virol. 2005;79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. doi:10.1128/JVI.79.3.1975-1980.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijak G.H., Janini L.M., Tovanabutra S., Sanders-Buell E., Arroyo M.A., Robb M.L., Michael N.L., Birx D.L., McCutchan F.E. Variable contexts and levels of hypermutation in HIV-1 proviral genomes recovered from primary peripheral blood mononuclear cells. Virology. 2008;376:101–111. doi: 10.1016/j.virol.2008.03.017. doi:10.1016/j.virol.2008.03.017 [DOI] [PubMed] [Google Scholar]
- Knoepfel S.A., Salisch N.C., Huelsmann P.M., Rauch P., Walter H., Metzner K.J. Comparison of G-to-A mutation frequencies induced by APOBEC3 proteins in H9 cells and peripheral blood mononuclear cells in the context of impaired processivities of drug-resistant human immunodeficiency virus type 1 reverse transcriptase variants. J. Virol. 2008;82:6536–6545. doi: 10.1128/JVI.00554-08. doi:10.1128/JVI.00554-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Takaori-Kondo A., Miyauchi Y., Iwai K., Uchiyama T. Ubiquitination of APOBEC3G by an HIV-1 Vif-Cullin5-Elongin B-Elongin C complex is essential for Vif function. J. Biol. Chem. 2005;280:18 573–18 578. doi: 10.1074/jbc.C500082200. doi:10.1074/jbc.C500082200 [DOI] [PubMed] [Google Scholar]
- Kozak S.L., Marin M., Rose K.M., Bystrom C., Kabat D. The anti-HIV-1 editing enzyme APOBEC3G binds HIV-1 RNA and messenger RNAs that shuttle between polysomes and stress granules. J. Biol Chem. 2006;281:29 105–29 119. doi: 10.1074/jbc.M601901200. doi:10.1074/jbc.M601901200 [DOI] [PubMed] [Google Scholar]
- Land A.M., Ball T.B., Luo M., Pilon R., Sandstrom P., Embree J.E., Wachihi C., Kimani J., Plummer F.A. HIV-1 proviral hypermutation correlates with CD4 count in HIV infected women from Kenya. J. Virol. 2008;82:8172–8182. doi: 10.1128/JVI.01115-08. doi:10.1128/JVI.01115-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlois M.A., Neuberger M.S. Human APOBEC3G can restrict retroviral infection in avian cells and acts independently of both UNG and SMUG1. J. Virol. 2008;82:4660–4664. doi: 10.1128/JVI.02469-07. doi:10.1128/JVI.02469-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.N., Malim M.H., Bieniasz P.D. Hypermutation of an ancient human retrovirus by APOBEC3G. J. Virol. 2008;82:8762–8770. doi: 10.1128/JVI.00751-08. doi:10.1128/JVI.00751-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddament M.T., Brown W.L., Schumacher A.J., Harris R.S. APOBEC3F properties and hypermutation preferences indicate activity against HIV-1 in vivo. Curr. Biol. 2004;14:1385–1391. doi: 10.1016/j.cub.2004.06.050. doi:10.1016/j.cub.2004.06.050 [DOI] [PubMed] [Google Scholar]
- Liu B., Sarkis P.T., Luo K., Yu Y., Yu X.F. Regulation of Apobec3F and human immunodeficiency virus type 1 Vif by Vif-Cul5-ElonB/C E3 ubiquitin ligase. J. Virol. 2005;79:9579–9587. doi: 10.1128/JVI.79.15.9579-9587.2005. doi:10.1128/JVI.79.15.9579-9587.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Liu B., Xiao Z., Yu Y., Yu X., Gorelick R., Yu X.F. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 2004;78:11 841–11 852. doi: 10.1128/JVI.78.21.11841-11852.2004. doi:10.1128/JVI.78.21.11841-11852.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Xiao Z., Ehrlich E., Yu Y., Liu B., Zheng S., Yu X.F. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl Acad. Sci. USA. 2005;102:11 444–11 449. doi: 10.1073/pnas.0502440102. doi:10.1073/pnas.0502440102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Wang T., Liu B., Tian C., Xiao Z., Kappes J., Yu X.F. Cytidine deaminases APOBEC3G and APOBEC3F interact with human immunodeficiency virus type 1 integrase and inhibit proviral DNA formation. J. Virol. 2007;81:7238–7248. doi: 10.1128/JVI.02584-06. doi:10.1128/JVI.02584-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madani N., Kabat D. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 1998;72:10 251–10 255. doi: 10.1128/jvi.72.12.10251-10255.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M.H., Emerman M. HIV-1 accessory proteins: ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. doi: 10.1016/j.chom.2008.04.008. doi:10.1016/j.chom.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Mangeat B., Turelli P., Caron G., Friedli M., Perrin L., Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. doi:10.1038/nature01709 [DOI] [PubMed] [Google Scholar]
- Mangeat B., Turelli P., Liao S., Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 2004;279:14 481–14 483. doi: 10.1074/jbc.C400060200. doi:10.1074/jbc.C400060200 [DOI] [PubMed] [Google Scholar]
- Mariani R., Chen D., Schrofelbauer B., Navarro F., Konig R., Bollman B., Munk C., Nymark-McMahon H., Landau N.R. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. doi:10.1016/S0092-8674(03)00515-4 [DOI] [PubMed] [Google Scholar]
- Marin M., Rose K.M., Kozak S.L., Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003;9:1398–1403. doi: 10.1038/nm946. doi:10.1038/nm946 [DOI] [PubMed] [Google Scholar]
- Mbisa J.L., et al. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 2007;81:7099–7110. doi: 10.1128/JVI.00272-07. doi:10.1128/JVI.00272-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A., Goncalves J., Santa-Marta M., McPike M., Gabuzda D. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev. 2004a;18:2861–2866. doi: 10.1101/gad.1249904. doi:10.1101/gad.1249904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A., Strack B., Ancuta P., Zhang C., McPike M., Gabuzda D. Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J. Biol. Chem. 2004b;279:7792–7798. doi: 10.1074/jbc.M313093200. doi:10.1074/jbc.M313093200 [DOI] [PubMed] [Google Scholar]
- Mehle A., Thomas E.R., Rajendran K.S., Gabuzda D. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J. Biol. Chem. 2006;281:17 259–17 265. doi: 10.1074/jbc.M602413200. doi:10.1074/jbc.M602413200 [DOI] [PubMed] [Google Scholar]
- Miyagi E., Opi S., Takeuchi H., Khan M., Goila-Gaur R., Kao S., Strebel K. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 2007;81:13 346–13 353. doi: 10.1128/JVI.01361-07. doi:10.1128/JVI.01361-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Sankaranand V.S., Anant S., Sugai M., Kinoshita K., Davidson N.O., Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J. Biol Chem. 1999;274:18 470–18 476. doi: 10.1074/jbc.274.26.18470. doi:10.1074/jbc.274.26.18470 [DOI] [PubMed] [Google Scholar]
- Navarro F., Bollman B., Chen H., Konig R., Yu Q., Chiles K., Landau N.R. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. doi:10.1016/j.virol.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Newman E.N., Holmes R.K., Craig H.M., Klein K.C., Lingappa J.R., Malim M.H., Sheehy A.M. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. doi:10.1016/j.cub.2004.12.068 [DOI] [PubMed] [Google Scholar]
- Oberste M.S., Gonda M.A. Conservation of amino-acid sequence motifs in lentivirus Vif proteins. Virus Genes. 1992;6:95–102. doi: 10.1007/BF01703760. doi:10.1007/BF01703760 [DOI] [PubMed] [Google Scholar]
- Opi S., Kao S., Goila-Gaur R., Khan M.A., Miyagi E., Takeuchi H., Strebel K. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 2007;81:8236–8246. doi: 10.1128/JVI.02694-06. doi:10.1128/JVI.02694-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace C., Keller J., Nolan D., James I., Gaudieri S., Moore C., Mallal S. Population level analysis of human immunodeficiency virus type 1 hypermutation and its relationship with APOBEC3G and vif genetic variation. J. Virol. 2006;80:9259–9269. doi: 10.1128/JVI.00888-06. doi:10.1128/JVI.00888-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G., Greenwell-Wild T., Nares S., Jin W., Lei K.J., Rangel Z.G., Munson P.J., Wahl S.M. Myeloid differentiation and susceptibility to HIV-1 are linked to APOBEC3 expression. Blood. 2007;110:393–400. doi: 10.1182/blood-2006-10-051763. doi:10.1182/blood-2006-10-051763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochnow C., Bransteitter R., Klein M.G., Goodman M.F., Chen X.S. The APOBEC-2 crystal structure and functional implications for the deaminase AID. Nature. 2007;445:447–451. doi: 10.1038/nature05492. doi:10.1038/nature05492 [DOI] [PubMed] [Google Scholar]
- Russell R.A., Pathak V.K. Identification of two distinct human immunodeficiency virus type 1 Vif determinants critical for interactions with human APOBEC3G and APOBEC3F. J. Virol. 2007;81:8201–8210. doi: 10.1128/JVI.00395-07. doi:10.1128/JVI.00395-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer A., Bogerd H.P., Cullen B.R. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. doi:10.1016/j.virol.2004.08.006 [DOI] [PubMed] [Google Scholar]
- Schrofelbauer B., Chen D., Landau N.R. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc. Natl Acad. Sci. USA. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. doi:10.1073/pnas.0307132101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B., Senger T., Manning G., Landau N.R. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 2006;80:5984–5991. doi: 10.1128/JVI.00388-06. doi:10.1128/JVI.00388-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A.J., Hache G., Macduff D.A., Brown W.L., Harris R.S. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. doi:10.1128/JVI.02391-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis N.C., Choi J.D., Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. doi:10.1038/nature00939 [DOI] [PubMed] [Google Scholar]
- Sheehy A.M., Gaddis N.C., Malim M.H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9:1404–1407. doi: 10.1038/nm945. doi:10.1038/nm945 [DOI] [PubMed] [Google Scholar]
- Simon J.H., Malim M.H. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.H., Gaddis N.C., Fouchier R.A., Malim M.H. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 1998;4:1397–1400. doi: 10.1038/3987. doi:10.1038/3987 [DOI] [PubMed] [Google Scholar]
- Simon J.H., Miller D.L., Fouchier R.A., Soares M.A., Peden K.W., Malim M.H. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 1998;17:1259–1267. doi: 10.1093/emboj/17.5.1259. doi:10.1093/emboj/17.5.1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon V., Zennou V., Murray D., Huang Y., Ho D.D., Bieniasz P.D. Natural variation in Vif: differential impact on APOBEC3G/3F and a potential role in HIV-1 diversification. PLoS Pathog. 2005;1:e6. doi: 10.1371/journal.ppat.0010006. doi:10.1371/journal.ppat.0010006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soros V.B., Yonemoto W., Greene W.C. Newly synthesized APOBEC3G is incorporated into HIV virions, inhibited by HIV RNA, and subsequently activated by RNase H. PLoS Pathog. 2007;3:e15. doi: 10.1371/journal.ppat.0030015. doi:10.1371/journal.ppat.0030015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sova P., Volsky D.J. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J. Virol. 1993;67:6322–6326. doi: 10.1128/jvi.67.10.6322-6326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins C.E., Kaelin W.G., Jr., Pavletich N.P. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. doi:10.1126/science.284.5413.455 [DOI] [PubMed] [Google Scholar]
- Stevenson M., Stanwick T.L., Dempsey M.P., Lamonica C.A. HIV-1 replication is controlled at the level of T cell activation and proviral integration. EMBO J. 1990;9:1551–1560. doi: 10.1002/j.1460-2075.1990.tb08274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopak K., de Noronha C., Yonemoto W., Greene W.C. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. doi:10.1016/S1097-2765(03)00353-8 [DOI] [PubMed] [Google Scholar]
- Strebel K., Daugherty D., Clouse K., Cohen D., Folks T., Martin M.A. The HIV ‘A’ (sor) gene product is essential for virus infectivity. Nature. 1987;328:728–730. doi: 10.1038/328728a0. doi:10.1038/328728a0 [DOI] [PubMed] [Google Scholar]
- Suspene R., et al. APOBEC3G is a single-stranded DNA cytidine deaminase and functions independently of HIV reverse transcriptase. Nucleic Acids Res. 2004;32:2421–2429. doi: 10.1093/nar/gkh554. doi:10.1093/nar/gkh554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suspene R., Rusniok C., Vartanian J.P., Wain-Hobson S. Twin gradients in APOBEC3 edited HIV-1 DNA reflect the dynamics of lentiviral replication. Nucleic Acids Res. 2006;34:4677–4684. doi: 10.1093/nar/gkl555. doi:10.1093/nar/gkl555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svarovskaia E.S., Xu H., Mbisa J.L., Barr R., Gorelick R.J., Ono A., Freed E.O., Hu W.S., Pathak V.K. Human apolipoprotein B mRNA-editing enzyme-catalytic polypeptide-like 3G (APOBEC3G) is incorporated into HIV-1 virions through interactions with viral and nonviral RNAs. J. Biol. Chem. 2004;279:35 822–35 828. doi: 10.1074/jbc.M405761200. doi:10.1074/jbc.M405761200 [DOI] [PubMed] [Google Scholar]
- Swanson C.M., Malim M.H. SnapShot: HIV-1 proteins. Cell. 2008;133:742. doi: 10.1016/j.cell.2008.05.005. doi:10.1016/j.cell.2008.05.005 [DOI] [PubMed] [Google Scholar]
- Teng B., Burant C.F., Davidson N.O. Molecular cloning of an apolipoprotein B messenger RNA editing protein. Science. 1993;260:1816–1819. doi: 10.1126/science.8511591. doi:10.1126/science.8511591 [DOI] [PubMed] [Google Scholar]
- Thielen B.K., Klein K.C., Walker L.W., Rieck M., Buckner J.H., Tomblingson G.W., Lingappa J.R. T cells contain an RNase-insensitive inhibitor of APOBEC3G deaminase activity. PLoS Pathog. 2007;3:1320–1334. doi: 10.1371/journal.ppat.0030135. doi:10.1371/journal.ppat.0030135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C., Yu X., Zhang W., Wang T., Xu R., Yu X.F. Differential requirement for conserved tryptophans in human immunodeficiency virus type 1 Vif for the selective suppression of APOBEC3G and APOBEC3F. J. Virol. 2006;80:3112–3115. doi: 10.1128/JVI.80.6.3112-3115.2006. doi:10.1128/JVI.80.6.3112-3115.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian J.P., Meyerhans A., Sala M. G→A hypermutation of the human immunodeficiency virus type 1 genome: evidence for dCTP pool imbalance during reverse transcription. Proc. Natl Acad. Sci. USA. 1994;91:3092–3096. doi: 10.1073/pnas.91.8.3092. doi:10.1073/pnas.91.8.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian J.P., Guetard D., Henry M., Wain-Hobson S. Evidence for editing of human papillomavirus DNA by APOBEC3 in benign and precancerous lesions. Science. 2008;320:230–233. doi: 10.1126/science.1153201. doi:10.1126/science.1153201 [DOI] [PubMed] [Google Scholar]
- von Schwedler U., Song J., Aiken C., Trono D. Vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 1993;67:4945–4955. doi: 10.1128/jvi.67.8.4945-4955.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Tian C., Zhang W., Luo K., Sarkis P.T., Yu L., Liu B., Yu Y., Yu X.F. 7SL RNA mediates virion packaging of the antiviral cytidine deaminase APOBEC3G. J. Virol. 2007;81:13 112–13 124. doi: 10.1128/JVI.00892-07. doi:10.1128/JVI.00892-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichroski M.J., Robb G.B., Rana T.M. Human retroviral host restriction factors APOBEC3G and APOBEC3F localize to mRNA processing bodies. PLoS Pathog. 2006;2:e41. doi: 10.1371/journal.ppat.0020041. doi:10.1371/journal.ppat.0020041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand H.L., Doehle B.P., Bogerd H.P., Cullen B.R. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. doi:10.1038/sj.emboj.7600246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Svarovskaia E.S., Barr R., Zhang Y., Khan M.A., Strebel K., Pathak V.K. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl Acad. Sci. USA. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. doi:10.1073/pnas.0400830101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Yu Y., Liu B., Luo K., Kong W., Mao P., Yu X.F. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. doi:10.1126/science.1089591 [DOI] [PubMed] [Google Scholar]
- Yu Q., Konig R., Pillai S., Chiles K., Kearney M., Palmer S., Richman D., Coffin J.M., Landau N.R. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat. Struct. Mol. Biol. 2004a;11:435–442. doi: 10.1038/nsmb758. doi:10.1038/nsmb758 [DOI] [PubMed] [Google Scholar]
- Yu Y., Xiao Z., Ehrlich E.S., Yu X., Yu X.F. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004b;18:2867–2872. doi: 10.1101/gad.1250204. doi:10.1101/gad.1250204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack J.A., Arrigo S.J., Weitsman S.R., Go A.S., Haislip A., Chen I.S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. doi:10.1016/0092-8674(90)90802-L [DOI] [PubMed] [Google Scholar]
- Zennou V., Bieniasz P.D. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006;349:31–40. doi: 10.1016/j.virol.2005.12.035. doi:10.1016/j.virol.2005.12.035 [DOI] [PubMed] [Google Scholar]
- Zennou V., Perez-Caballero D., Gottlinger H., Bieniasz P.D. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J. Virol. 2004;78:12 058–12 061. doi: 10.1128/JVI.78.21.12058-12061.2004. doi:10.1128/JVI.78.21.12058-12061.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Yang B., Pomerantz R.J., Zhang C., Arunachalam S.C., Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. doi:10.1038/nature01707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K.L., Mangeat B., Ortiz M., Zoete V., Trono D., Telenti A., Michielin O. Model structure of human APOBEC3G. PLoS ONE. 2007;2:e378. doi: 10.1371/journal.pone.0000378. doi:10.1371/journal.pone.0000378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y.H., Irwin D., Kurosu T., Tokunaga K., Sata T., Peterlin B.M. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J. Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. doi:10.1128/JVI.78.11.6073-6076.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]