DNA Transposons and the Evolution of Eukaryotic Genomes (original) (raw)
. Author manuscript; available in PMC: 2007 Dec 28.
Abstract
Transposable elements are mobile genetic units that exhibit broad diversity in their structure and transposition mechanisms. Transposable elements occupy a large fraction of many eukaryotic genomes and their movement and accumulation represent a major force shaping the genes and genomes of almost all organisms. This review focuses on DNA-mediated or class 2 transposons and emphasizes how this class of elements is distinguished from other types of mobile elements in terms of their structure, amplification dynamics, and genomic effect. We provide an up-to-date outlook on the diversity and taxonomic distribution of all major types of DNA transposons in eukaryotes, including Helitrons and Mavericks. We discuss some of the evolutionary forces that influence their maintenance and diversification in various genomic environments. Finally, we highlight how the distinctive biological features of DNA transposons have contributed to shape genome architecture and led to the emergence of genetic innovations in different eukaryotic lineages.
Keywords: transposable elements, transposase, molecular domestication, chromosomal rearrangements
INTRODUCTION
Dazzling advances in molecular biology, genetics, and genomics have allowed scientists to understand in great detail many aspects of transposable element (TE) biology. Significant discoveries at the interface of these fields have provided new insight into transposition mechanisms, allowed the identification of new TEs and the broadening of their taxonomic distribution, revealed relationships between TEs and viruses, and uncovered the means by which TE movement can be controlled epigenetically by their host. Coupled to these new discoveries is a greater understanding of the extent to which TEs influence the structure and dynamics of the genomes they inhabit. The focus of this review is on one specific class of TEs, the class 2 or DNA transposons. We begin by presenting key features of the structure and life cycle of these elements, with an emphasis on the factors that govern their maintenance and propagation within the genome and throughout the eukaryotic tree of life. We then shift our focus to the repercussions of DNA transposon movement and amplification on the genome, including large-scale structural changes and epigenetic modifications, and the contribution of elements of this type to the generation of allelic diversity, new genes, and biological innovations.
EVOLUTIONARY DYNAMICS OF DNA TRANSPOSONS
Classification and Distribution of DNA Transposons
Class 2 transposable elements (TEs) or DNA transposons are mobile DNA that move utilizing a single or double-stranded DNA intermediate (35). Eukaryotic DNA transposons can be divided into three major subclasses: (i) those those that excise as double-stranded DNA and reinsert elsewhere in the genome, i.e., the classic “cut-and-paste” transposons (35); (ii) those that utilize a mechanism probably related to rolling-circle replication, Helitrons (91); and (iii) Mavericks, whose mechanism of transposition is not yet well understood, but that likely replicate using a self-encoded DNA polymerase (160). Both Helitrons and Mavericks most likely rely on distinct transposition mechanisms involving the displacement and replication of a single-stranded DNA intermediate, respectively. Thus these elements probably transpose through a replicative, copy-and-paste process.
All cut-and-paste transposons are characterized by a transposase encoded by autonomous copies and, with few exceptions, by the presence of terminal inverted repeats (TIRs). Helitrons have no TIRs, but rather short conserved terminal motifs and autonomous copies encode a Rep/Helicase (91, 158). Mavericks, also known as Polintons, are very large transposons with long TIRs and coding capacity for multiple proteins, most of which are related to double-stranded DNA viruses, including a B-type DNA polymerase (52, 94, 160).
To date, ten superfamilies of cut-and-paste DNA transposons are recognized (Table 1). Elements belong to the same superfamily when they can be linked to transposases that are significantly related in sequence. Typically, transposases from the same superfamily can be confidently aligned in their core catalytic region and a monophyletic ancestry can be inferred from phylogenetic analysis (22, 164). In some cases, such as Tc1/mariner, the superfamily can be further divided into monophyletic groups that deeply diverged in eukaryotic evolution (155, 164) (Table 1). Two superfamilies (CACTA and PIF/Harbinger) are characterized by the presence of a second transposon-encoded protein required for transposition (Table 1).
Table 1.
Classification and characteristics of eukaryotic DNA transposons
| Superfamily | Related IS | TSD | Length1 (kb) | TIRs1 (bp) | Terminal motif (5′-3′) | TPase1 (aa) | Catalytic motif | DNA-binding motif | Additionnal proteins |
|---|---|---|---|---|---|---|---|---|---|
| Tc1/mariner | IS630 | TA | 1.2–5.0 | 17–1100 | variable | 300–550 | DD(30–41)D/E | HTH (cro/paired) | |
| hAT | nd | 8 bp | 2.5–5 | 10–25 | YARNG | 600–850 | D(68)D(324)E2 | ZnF (BED) | |
| P element | nd | 7/8 bp | 3–11 | 13–150 | CANRG | 800–900 | D(83)D(2)E(13)D3 | ZnF (THAP) | |
| MuDR/Foldback | IS256 | 7–10 bp | 1.3–7.4 | 0-sev. Kb | variable | 450–850 | DD(~110)E | ZnF (WRKY/GCM1) | |
| CACTA | nd | 2/3 bp | 4.5–15 | 10–54 | CMCWR | 500–1,200 | Nd | nd | TNPA (DNA-binding protein) |
| PiggyBac | IS1380 | TTAA | 2.3–6.3 | 12–19 | CCYT | 550–700 | DDE? | nd | |
| PIF/Harbinger | IS5 | TWA | 2.3–5.5 | 15–270 | GC-rich | 350–550 | DD(35–37/47–48)E | HTH | PIF2p (Myb/SANT domain) |
| Merlin | IS1016 | 8/9 bp | 1.4–3.5 | 21–462 | GGNRM | 270–330 | DD(36–38)E | nd | |
| Transib | nd | 5-bp | 3–4 | 9–60 | CACWATG | 650–700 | DD(206–214)E | nd | |
| Banshee | IS481 | 4/15 bp | 3.5 | 41–950 | TGT | 300–4004 | DD(34)E | HTH | |
| Helitron | IS91 | none | 5.5–17 | none | 5′-TC…CTAR-3′ | 1,400–3,0005 | HHYY (‘REP motif’) | ZnF-like | RPA (in Plants) |
| Maverick | none | 5/6 bp | 15–25 | 150–700 | simple repeat | 350–4504 | DD(33–35)E | ZnF (HHCC) | 4–10 DNA virus-like proteins |
The explosion of sequence data in the databases over the past decade has fueled the discovery of large numbers of elements in a wide range of organisms. These discoveries have yielded several new insights into the distribution and broad evolutionary history of eukaryotic DNA transposons. First, the taxonomic distribution of superfamilies initially believed to be restricted to a few related taxons has been significantly expanded to cover several eukaryotic kingdoms or supergroups (41, 68, 170) (e.g., P element, CACTA, PiggyBac; see Table 1 and Figure 1). Second, links have been established between superfamilies that were previously separated (e.g., union of MuDR and Foldback). Finally, novel superfamilies have been recognized (e.g., PIF/Harbinger, Merlin, Transib, Banshee) (48, 90, 93, 206) and two distinct subclasses of DNA transposons have been identified (Helitrons and Mavericks).
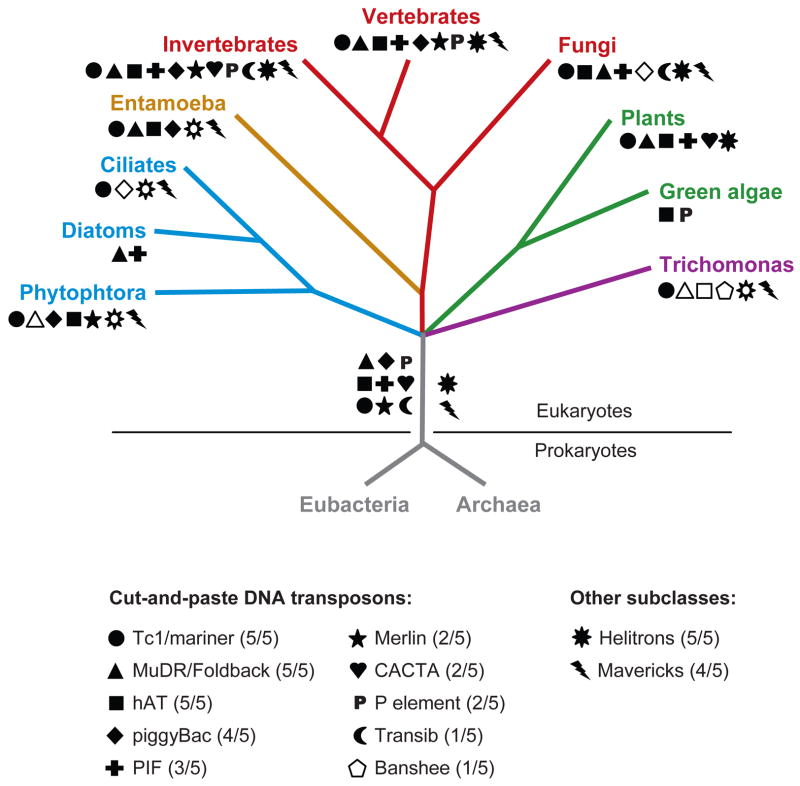
Figure 1.
Distribution of the major groups of DNA transposons across the eukaryotic tree of life. The tree depicts 4 of the 5 “supergroups” of eukaryotes (based on Keeling et al. 2005**AU: Please check: 2005 reference is listed only. 2005 is ok) where DNA transposons have been detected. The “unikonts” are represented by the opisthokonts (vertebrates, invertebrates, and fungi) and by the Ameobozoa Entamoeba, the Chromoalveolates by the oomycete Phytophtora infestans, the diatom Thalassiosira pseudonana and several ciliates, the Plantae by the unicellular green algae Chlamydomonas reinhardtii and a broad range of flowering plants, and the Excavates by the parabasalid Trichomonas vaginalis. The occurrence of each superfamily/subclass of DNA transposons is denoted by a different symbol. The data were primarily gathered from the literature (references available upon request). Open symbols denote unpublished observations gathered by the authors or from Repbase (http://www.girinst.org). The taxonomic breadth of the different groups among the 5 supergroups of eukaryotes is shown in parentheses. These data suggest that 11 of the 12 major types of DNA transposons were already diversified in the common ancestor of eukaryotes.
A superimposition of the distribution of each cut-and-paste DNA transposon superfamily on the most current representation of the eukaryotic tree of life (97) reveals that 8 of the 10 superfamilies are represented in two or more eukaryotic supergroups (Figure 1). Given that there is no convincing evidence for horizontal transfer of DNA transposons between eukaryotic supergroups, this distribution suggests that most superfamilies were already differentiated in the eukaryotic ancestor. Furthermore, alliances with prokaryotic insertion sequence families can be drawn for six of the ten eukaryotic superfamilies (Table 1), suggesting that the divergence of most superfamilies may even predate the split of eukaryotes and prokaryotes. Finally, Helitrons and Mavericks are also distributed across multiple eukaryotic supergroups (Figure 1). These data underscore the extremely ancient roots of the major types of DNA transposons and their remarkable persistence over evolutionary time.
Differential Success of DNA Transposons among Species
Eukaryotic species show enormous variation in the amount of TEs occupying their genomes (1, 111). It is now well established that these variations largely account for the wide differences in genome size observed among eukaryotes, and even between closely related species (64, 100). Retrotransposons seem to be major players in promoting rapid increase, and perhaps also decrease, in the genome size of multicellular eukaryotes (7, 10, 124, 128, 150, 171, 185). This is best exemplified by studies of maize and of the rice Oryza australiensis, showing that massive bursts of LTR retrotransposon amplification caused a concomitant doubling of the genome independently in the lineages of these two species (152, 169).
DNA transposons may also contribute substantially to genome expansion. An estimated 65% of the genome of the single-celled eukaryote Trichomonas vaginalis, which was recently sequenced, is made of repetitive DNA (23). Virtually all TEs that have been recognized in this genome are DNA transposons (94, 160, 174; E.J.P, unpublished). In fact, only a handful of retrotransposon-related proteins are recognizable in the genome (23) and it is not yet clear whether they actually belong to mobile elements (E.J.P, unpublished). Recent studies indicate that genome expansion in this species can be largely accounted for by the massive amplification of Maverick transposons (160). There are an estimated 3,000 Maverick copies per haploid genome and considering an average size of these elements in T. vaginalis of 15 to 20 kb, it can be inferred that these transposons occupy a stunning ~60 Mb of the ~160 Mb genome, that is ~37% of the genomic space.
Tremendous variation also exists among species in the relative abundance of DNA transposons and retrotransposons, regardless of their sheer numbers (Figure 2). For example, DNA transposons seem completely absent from the genomes of budding and fission yeast, although different families of LTR retrotransposons have survived in both species. Yet DNA transposons are common in filamentous fungi and occur occasionally in other yeasts, such as Candida albicans (36). Thus, two independent extinction events of DNA transposons occurred in the lineages leading to Saccharomyces cerevisiae and Schizosaccharomyces pombe.
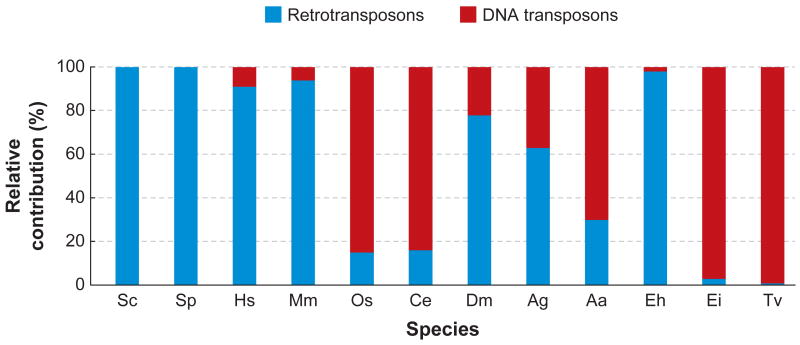
Figure 2.
The relative amount of retrotransposons and DNA transposons in diverse eukaryotic genomes. The graph shows the contribution of DNA transposons and retrotransposons in percentage relative to the total number of transposable elements in each species. The data were compiled from papers reporting draft genome sequences (references available upon request) and from the Repeatmasker output tables available at the UCSC Genome Browser (http://genome.ucsc.edu) or from the following sources: E. histolytica and E. invadens: (159); T. vaginalis: E. Pritham, unpublished data. Species abbreviations: Sc: Saccharomyces cerevisiae; Sp: Schizosaccharomyces pombe; Hs: Homo sapiens; Mm: Mus musculus; Os: Oryza sativa; Ce: Caenorhabditis elegans; Dm: Drosophila melanogaster; Ag: Anopheles gambiae, malaria mosquito; Aa: Aedes aegypti, yellow fever mosquito; Eh: Entamoeba histolytica; Ei: Entamoeba invadens; Tv: Trichomonas vaginalis.
The human TE landscape is clearly dominated by retrotransposons, mostly LINEs and associated SINEs (111). Nonetheless, human DNA transposons are highly diversified (120 families falling into 5 superfamilies) and they are numerically abundant (111, 151). With 300,000 copies, the human genome contains about 15 times more DNA elements than the DNA transposon-rich genome of Caenorhabditis elegans and 40 times more than Drosophila melanogaster (Table 1 and data from the UCSC Genome Browser). In addition, nearly 100,000 DNA transposon copies from 40 families and 4 different superfamilies integrated during the primate radiation (151). None of these elements, however, appears to have survived a seemingly general extinction event of DNA transposons that occurred about 40 My (million years) in an anthropoid primate ancestor. The picture emerging from the initial analyses of the mouse, rat, and dog genome sequences is strikingly similar, with no evidence for the activity of DNA transposons during the past 40–50 My (59, 118, 192; J. Pace & C.F., unpublished). At first sight, these data suggest an intriguing scenario whereby DNA transposons went extinct independently in different mammalian lineages around the same evolutionary time (Eocene, 35–55 My) and have not been maintained or reintroduced into these lineages since this epoch.
Does it mean that all mammals are now refractory to the propagation of DNA transposons? The answer, which came unexpectedly from the genome of the little brown bat, Myotis lucifigus, is no. With a haploid genome size of ~2,000 Mb, M. lucifigus is one of the smallest mammalian genomes, but it harbors a surprisingly diverse collection of DNA transposons that is also distinct from other mammalian genomes examined. In particular, the genome is packed with Helitrons (at least 3% of the genome)(158), whereas none are recognizable in any of 22 other placental species (including two other bat species) for which a substantial amount of genomic sequences is now available. In contrast to other mammals so far examined, the recent data point to a continuous colonization of the vesper bat genome(s) by various DNA transposon families (158, 161; D. Ray. J. Smith, H.J.T. Pagan, E.J.P., C.F., N.L. Craig, submitted). Several waves of amplification of different families have succeeded over the past 40 My. Moreover, the invasion seems to be ongoing because there is mounting evidence that some hAT and piggyBac families are still active in natural populations of Myotis (161; D. Ray. J. Smith, H.J.T. Pagan, E.J.P., C.F., N.L. Craig, submitted).
Hence, sharp variation in the success of DNA transposons may exist even between closely related species. This variation is also illustrated by a comparative study of TE composition in the genomes of four species of Entamoeba, a single-celled eukaryote distantly related to animals and fungi (159). The four Entamoeba species all have relatively small genomes estimated to be about 20 Mb, but their TE composition varies dramatically. The genomes of E. invadens and E. moshkovskii host many families of DNA transposons from four different superfamilies and few retrotransposons (159), whereas the genomes of E. histolytica and E. dispar contain virtually no DNA transposons but instead were colonized by several lineages of non-LTR retrotransposons (5). The genomes of Entamoeba, despite harboring completely different TE complements, are composed of the same relative proportion of TEs (5%–7%), and all four genomes contain recently active elements (5, 159). Thus these genomes seem to be similarly constrained in size, but retrotransposons and DNA transposons have experienced differential success.
Population Dynamics of DNA Transposons Within Genomes: the MITE Paradox
DNA transposons are typically grouped into families. In principle, members of the same family are all descended from a common autonomous ancestor copy, which transposed and generated copies of itself in the process. Because most DNA transposons move through a nonreplicative mechanism, these elements increase their copy numbers through indirect mechanisms that rely on the host machinery (35). The first mechanism invokes the transposition of the element during DNA replication from a newly replicated chromatid to an unreplicated site. The transposon is thereby effectively replicated twice, leading to a net gain of one transposon copy. This behavior has been documented for the maize Ac and Spm elements (106). For Ac, the timing of transposition during DNA replication is explained by the preferential binding of Ac transposase to hemimethylated binding sites (166). The second mechanism draws on the repair of the double-strand break left by excision of the element. If the element is present on the homologous chromosome, gap repair via homologous recombination results in the reintroduction of the transposon at the donor site. If transposition occurs during the S phase of the cell cycle, the sister chromatid may also be used as the template for gap repair, resulting in the restoration of the excised element. Gap repair has been demonstrated to be the mechanism by which P elements rapidly increase their copy number in D. melanogaster (46). This process operates for other transposons in various species and gives rise to various internal deletion derivatives as a result of abortion, slippage, or template switching during gap repair (46, 77, 154, 168).
Because the terminal sequences of DNA transposons are often the only requirement for transposase recognition (35), internally deleted or rearranged nonautonomous elements may still transpose by using enzymes encoded elsewhere in the genome by an autonomous copy. The frequent emergence of nonautonomous derivatives coupled to the apparent lack of _cis_-preference of eukaryotic transposases poses a major hurdle for the successful propagation of an autonomous element (70). Indeed, unless there exists a mechanism to prevent the formation of nonfunctional copies upon gap repair [see the possible case of _Tam3_ in snapdragon (198)], it can be predicted that autonomous copies will be rapidly out-numbered by nonautonomous copies (70, 112). As copy number increases, the entire family potentially faces two constraints: (i) titration of the transposase by binding to multiple nonautonomous copies and (ii) an increased chance to trigger host- or self-induced repression mechanisms, such as RNA interference (RNAi) (1, 69, 112, 173, 177). Both constraints would eventually prevent the autonomous element from replicating, leading to its elimination or inactivation from the population and to the extinction of the entire family.
Considering this disastrous sequence of events, also referred to as vertical inactivation (69), the amplification of nonautonomous copies could be viewed as a death sentence for DNA transposons. Yet we observe a paradoxical situation where the genomes that harbor the most diverse and the highest density of DNA transposons (i.e., rice, nematodes, human) are also the ones filled with the largest amount of miniature inverted-repeat transposable elements (MITEs) (56, 85, 151). Why MITEs are so prevalent and how DNA transposons can be so successfully maintained and propagated in this context are the subjects of the next section.
Mechanism and Consequences of MITE Amplification
MITEs are short transposons (100–600 bp) that are distinguished from other nonautonomous elements by high copy numbers and length homogeneity (19, 56, 205). The structural homogeneity of MITE families indicates that they arose by amplification of a single or a few progenitor copies (49, 50). Presumably, the progenitor copy arises by deletion of a larger transposon during gap repair. Yet it is sometimes difficult, if at all possible, to directly connect a given MITE family with an autonomous transposon present within the same genome (53, 207). In many cases, sequence similarity between MITEs and the closest autonomous element is restricted to the TIRs (53, 149). Two hypotheses can be put forward to explain this paradox. First, some MITEs may arise de novo from the fortuitous juxtaposition of solo TIRs or sequences resembling the TIRs of an autonomous transposon (127, 183). A second possibility is that MITE progenitors are the relics of the past invasion of transposons whose autonomous copies have been erased or have not reached fixation within the population (53).
The accumulation of MITE families over time creates a reservoir of elements ready for accidental cross-activation by newly emerged autonomous transposons, triggering new waves of MITE amplification (53). This scenario is supported by studies of rice _mariner_-like transposons and their related Stowaway MITEs, which reveal that currently active transposases can bind to the TIRs of a wide diversity of distantly related MITEs represented by thousands of copies within the same genome (51).
How then do DNA transposons replicate given such strong competition? One explanation is that MITE amplification might pass under the radar of the host defense system, either because the transposons are too small or because they fail to trigger the _trans_-silencing of the autonomous transposon providing the source of transposase (53). One mechanism of defense evasion may occur as a result of the absense of homology between MITEs and the transposase gene or its promoter region. In this model, the lack of shared sequence similarity allows the continuous expression of the transposase source, which serves to propagate the MITEs, as well as the autonomous transposon but at a lower frequency. The potential problem of titration of transposase molecules by binding to many illegitimate targets remains (38, 69), perhaps representing a major selective force favoring the emergence of transposon variants that minimize cross-interaction with MITEs present in the same genome (38, 51, 110, 126). Thus the presence of MITEs could actually benefit the long-term evolution of DNA transposons by driving their vertical diversification.
The recent isolation of active MITE-transposase systems (84, 103, 198b) has allowed most of these hypotheses to be tested in the laboratory and also in the context of natural populations. The most promising model is the mPing/Pong system of rice. mPing was identified as the first actively transposing MITE in any organism (84, 103). mPing transposition has been observed in vivo in response to various stress conditions and correlated with the coactivation of Pong, a distantly related autonomous transposon of the PIF/Harbinger superfamily (84, 116, 172). Recently, it was also shown that Pong and _Ping_-encoded proteins are necessary and sufficient to mobilize mPing in transgenic Arabidopsis plants (198c). Finally, evidence was gathered that mPing copy number has recently exploded in the field and reached approximately 1,000 copies in some cultivated rice strains (142). This situation offers an unprecedented opportunity to comprehend how MITEs attain such high copy numbers without killing the host or silencing their autonomous transposon partner.
Horizontal Transmission and Vertical Diversification
Even in the absence of MITE amplification, the vertical inactivation theory predicts that DNA transposons would ultimately go extinct unless autonomous elements can be periodically reintroduced in a genome that has not been previously exposed to the proliferation of the same element (69). The best way to achieve this is by horizontal introduction of an autonomous element to a new species (or population). Clear cases of horizontal transfer (HT) of DNA transposons have been documented, especially for Tc1/mariner and P elements among insect species (39, 164, 175). Recently, a possible HT of a MULE transposon between plants was reported (42). Thus, it is believed that all DNA transposons rely heavily on HT for their propagation and maintenance throughout evolution (69, 164).
Support for the notion that DNA transposons are well adapted to HT comes from in vitro experiments, which showed that, for all transposon systems so far examined, transposase is the only protein needed for transposition [for review, see (35)]. Consistent with the apparent lack of requirement for host-specific factors, most active transposons isolated from one species are readily functional in a wide range of heterologous species [for review, see (135, 148, 155)].
Important gaps remain in our understanding of the evolutionary dynamics of DNA transposons. Recent large-scale phylogenetic analyses of DNA transposon populations within species and in closely related species indicate that HT cannot account for the diversity and multiplicity of DNA transposons coexisting within a single genome. For example, phylogenetic analysis of 68 distinct _mariner_-like transposase sequences from 25 grass species revealed no instances of HT, but is consistent with vertical transmission and continuous diversification of multiple lineages of transposases during grass evolution (55). Likewise, distant Entamoeba species shared deeply diverged lineages of transposases, indicative of their presence in the common ancestor of the species followed by their vertical diversification (159). These data point to the existence of mechanisms allowing DNA transposons to rapidly diversify within species. Rapid diversification would limit the chances for cross-interactions between related copies and promote the speciation of new active families (1, 51, 110). One possible opportunity for diversification is during gap repair following transposon excision. The capture of filler DNA sequences at double-strand breaks owing to template switching and other aberrant repair events has been documented in various organisms (61, 117, 193, 199). These processes can readily explain the capture of new internal sequences by transposons (72, 83, 143, 168). Likewise, a frequent exchange of sequence information was recorded between actively transposing Tc1 elements dispersed in the genome of C. elegans, suggesting that gap repair processes following excision may accelerate the evolution of the elements (57). Such mechanisms could account for the great sequence variation observed in the subterminal regions of transposons that share otherwise highly conserved transposase genes (53, 207).
IMPACT OF DNA TRANSPOSONS ON GENOME EVOLUTION
Like other transposable elements, DNA transposons have the potential to influence the evolutionary trajectory of their host in three distinct ways: (i) via alterations of gene function through insertion; (ii) through the induction of chromosomal rearrangements; (iii) as a source of coding and noncoding material that allows for the emergence of genetic novelty (such as new genes and regulatory sequences). DNA transposons have properties distinct from those of retrotransposons that uniquely affect the means and propensity for participation in each of these mechanisms. Here we review how the properties of DNA transposons contribute to the generation of allelic diversity in natural populations, shape the genomic and epigenetic landscape of their hosts, and contribute to the creation of new genes.
Generation of Allelic Diversity through Insertion and Excision of DNA Transposons
Like other TEs, DNA transposons are potent insertional mutagens. The insertion of DNA transposons may affect host gene expression in myriad ways, the phenotypic consequences of which were richly illustrated by the molecular characterization of a plethora of TE-induced mutations during the first decades of TE research (40, 49, 101, 194). The most straightforward outcome of TE insertion is the disruption of the coding sequences of a gene inhibiting the production of viable gene product. However, TE insertion, for example within promoters, introns, and untranslated regions, can directly trigger the full gambit of phenotypes, ranging from subtle and epigenetic regulatory perturbations to the complete loss of gene function (101, 194).
Unlike the majority of retrotransposons, many cut-and-paste transposons exhibit a marked preference for insertion into or within the vicinity of genes, a property that has allowed their development into powerful gene-tagging tools routinely used by geneticists (9, 179, 187). P elements in Drosophila (179), Mutator elements in maize (43), and the Tc3 element in nematodes (163) have all been shown to have a bias for insertion into genic neighborhoods. Additionally, in both plant and animal genomes MITEs are typically found in low-copy-number genomic regions and gene-rich environments (19, 56, 205). A breakthrough study of a recent MITE explosion in rice demonstrated for the first time that this pattern of insertion, at least for mPing, was primarily due to targeting rather than the result of selection (142). The genic proximity of DNA transposon insertions confers on them a significant potential for generating allelic diversity in natural populations. In addition, we propose that genic proximity also facilitates the co-option of DNA transposons for gene regulation (see below).
Another important property of DNA transposon-mediated insertional mutagenesis is the ability of DNA transposons, unlike retrotransposons, to subsequently undergo spontaneous excisions (194). Therefore DNA transposons frequently generate unstable mutations with reversible phenotypes. Excisions are often imperfect, leaving behind a transposon footprint and/or alter the flanking host DNA (e.g., 103, 154, 194). The nature of these changes have been determined through the examination of the sites of DNA transposon excision and include small deletions, inversions, as well as the introduction of random filler DNA. Multiple alleles with an array of phenotypic consequences have been identified in fungi, plants, and animals (29, 60, 101, 193, 195). A striking example was recently reported involving a member of the hAT superfamily, Tol2, in the medaka fish (105). In an inbred line, a wide range of pigmentation phenotypes could be recovered, ranging from albino to wild type through partially pigmented patterns. Closer molecular examination revealed that individuals homozygous for a Tol2 insertion in the promoter region of a pigmentation gene exhibit complete albino phenotypes. Perfect Tol2 excision accounted for wild-type individuals, and imprecise excisions gave rise to new alleles with different footprints and various heritable pigmentation phenotypes. The phenotypic mutation rate induced by Tol2 excision at this locus was as high as 2% per gamete, representing a 1000-fold increase from the spontaneous mutation rates previously determined for this species (105).
The generation of new alleles and the creation of novel regulatory circuits is a major force underlying the diversification of species (14, 24, 104, 196). As DNA transposon excision can rapidly generate allelic diversity, many subtle adaptive modifications of gene and promoter sequences could conceivably have involved insertion/excision of DNA transposons, but unless the transposon is caught in the act, these would prove difficult to demonstrate (15, 60, 105). Thus, the broad range of alterations and phenotypes caused by transposon excision in the lab may just represent the tip of the iceberg of what has actually occurred in nature.
TE-Mediated Epigenetic Effects on Gene Expression
McClintock first made the observation that maize transposons could influence nearby gene expression in a heritable fashion, and therefore designated them as controlling elements (131, 132). She also realized that the regulatory influence of transposons was reversible independent of their movement, alternating phases of quiescence and reactivation. Based on these results and on a number of intricate experiments, she put forward the visionary hypothesis that the regulatory influence of transposons on nearby genes was epigenetic in nature and could be modulated by changes in the environment (133). Although this model was largely overlooked at the time, the explosion of epigenetic research over the past decade has revived these ideas and validated several aspects of the model (120, 178). There is now clear evidence that DNA transposons represent natural targets for a battery of interconnected silencing mechanisms, implicating RNAi and involving epigenetic modifications (173, 178). Of course, this intracellular defense system also operates on retrotransposons and viruses. Nonetheless, the inherent structure of DNA transposons (notably the TIRs) and the propensity for local movement apparently predispose them to elicit RNAi-based silencing mechanisms and nucleate the formation of heterochromatic islands (65, 173, 177), with latent consequences for the regulation of nearby genes.
The most direct evidence that DNA transposons play a major role in attracting the machinery responsible for formation and maintenance of heterochromatin comes from the comparative analysis of two large duplicated regions of Arabidopsis chromosome 4 using tiling microarrays (119). One region is a heterochromatic knob replete with repetitive sequences, including a high density of CACTA and MULEs, conspicuously enriched in CpG and H3K9 methylation, whereas the other region is euchromatic, almost completely free of TEs, hypomethylated, and enriched in H3K4 methylation. The heterochromatic transposons are also associated with matching siRNAs. The epigenetic marks of heterochromatin were essentially erased in plants mutant for DDM1, a chromatin-remodeling factor essential for the silencing of CACTA and MULEs in Arabidopsis (178). In the ddm1 background, the transposons become awakened as a result of the loss of transcriptional silencing. Several examples of silenced transposons inserted in the proximal promoter regions were also found to provoke the transcriptional silencing of the adjacent gene, and both transposons and the associated genes were transcriptionally reactivated in the ddm1 mutant (119, 137, 176). A tight association between the silencing status of a MULE transposon and a nearby gene was also previously reported in maize (6). These data are consistent with McClintock’s hypothesis of transposons acting as controlling elements of gene expression (131, 132). In addition, studies in Drosophila of Hoppel (71), a member of the P-element superfamily, indicate that these mechanisms are not restricted to plants, but also operate in animals and frequently implicate DNA transposons.
Together these data converge toward a model whereby DNA transposons (and other TEs) act as moving targets for local heterochromatin formation as a byproduct of their structure (TIRs) and/or simply their repetitive nature (65, 178). Together with other sequence elements such as boundary or insulator elements, which may also be derived from repeats (145, 184), and their associated _trans_-acting siRNAs, transposons actively participate in a partitioning of the genome into chromosomal domains with distinct epigenetic marks and transcriptional activity. These marks are inheritable and normally stable, but they may be subject to dynamic changes in response to environmental cues and genetic stress, such as interspecific hybridization or polyploidization (2, 31, 95, 146). These events may in turn trigger further movement and amplification of TEs, provoking a structural and epigenetic reshuffling of the genome and offering an opportunity for natural selection to establish new chromosomal domains and regulatory circuits. This scenario essentially corroborates McClintock’s genomic shock theory (133).
Large-Scale Chromosomal Rearrangements
There is a rich record implicating DNA transposons in the induction of large-scale chromosomal rearrangements in plants and animals. The transposition mechanisms of these elements, which involve multiple double-strand breaks and repair events, predispose them to actively participate in these processes. Among the initial examples reported, the Foldback (FB) elements of Drosophila stand out because of the high frequency and the amplitude of the provoked rearrangements (30). The unusually high frequency of interelement recombination between FB elements strongly suggests participation of transposase cleavage activities at the termini of one or both of the elements (63, 114). The large size (often over 10 kb) and complex inverted repeat structure of FB might also be factors contributing to the recurrent involvement of these elements in rearrangements (30).
Recently, FB elements once again took the front stage in TE-induced chromosomal rearrangements. However, this was the first time that rearrangements similar to those observed in the lab were identified in natural populations and linked to events with evolutionary consequences. In a series of elegant studies, the group of Alfredo Ruiz demonstrated that ectopic recombination between oppositely oriented _FB_-like transposons generated two independent chromosomal inversions in Drosophila buzzatii (21, 26). These inversions are geographically widespread and polymorphic in natural populations, which strongly suggests that they are selectively advantageous (8). In each case, the inversion breakpoints occur within genomic hotspots that are highly variable in sequence and structure between populations (20, 26). The inversion breakpoints are characterized by complex nesting of DNA transposons (mostly of the FB and hAT superfamilies), but strikingly, no recognizable retrotransposons. Once again, the frequency of the rearrangements and the prevalence of FB and other DNA transposons points to a transposase-triggered mechanism rather than passive ectopic recombination events.
Transposase-induced rearrangements have long been recognized as a particular class of recombination events with a strong potential for restructuring the chromosome (63, 114). These events, termed alternative transposition processes, occur typically when the termini from separate transposon copies synapse together and engage in a complete or partial cut-and-paste reaction. Depending on the orientation of the termini used for the reaction and on the chromosomal location of the elements, alternative transpositions can lead to various outcomes, including chromosomal inversions, duplications, and deletions of over 100 kb (63, 114, 157, 203). Translocations can also occur if the insertion site is on a different chromosome from that of the two elements involved in alternative transposition. The molecular ontology of each type of rearrangement has been reviewed in detail elsewhere (63). Most of the possible outcomes have been recovered experimentally in diverse model organisms, such as snapdragon, maize, Drosophila, and Fusarium oxysporum, and with members of various DNA transposon superfamilies, such as hAT, P element, Tc1/mariner, and PIF/Harbinger (47, 79, 122, 189, 203).
Local hopping is a property of many DNA transposons that may augment their propensity to create local genomic rearrangements (63). Local hopping is the preference of an element to transpose to a linked chromosomal location, a behavior exhibited by transposons from different superfamilies (66, 98, 106, 139, 182). This and other targeting activities tend to create genomic clusters of related elements (78, 86), which would further enhance the probability for alternative transposition events.
Although many of the chromosomal rearrangements observed in the laboratory would be deleterious in nature, some may even occasionally bring a selective advantage to individuals carrying them, e.g., the D. buzattii inversions (8, 21, 26). Chromosomal rearrangements and gene relocation events have been linked to speciation events (129, 144, 147). A recent study of alternative transposition pathways using reversed Ac element termini in maize showed that these events can mediate exon shuffling and create new chimeric functional genes (204). The mechanism is analogous to V(D)J recombination, a process that generates endless combinations of antibody genes in the immune system of jawed vertebrates. As discussed below in more details, this parallel makes sense in light of growing evidence that the V(D)J recombination system is actually derived from immobilized DNA transposons.
Involvement of DNA Transposons in Gene Transduction, Duplication, and Exon Shuffling
The capture of host genes as part of mobile elements was first discovered in the context of cellular oncogenes transduced by retroviruses (180). Non-LTR retrotransposons are also capable of transduction of adjacent host sequences, specifically the L1 family and related genomic parasites in human (138). Given the abundance of retrotransposons and other retroviral-like elements in some eukaryotic genomes, one might expect this process to be an evolutionarily potent mechanism for the duplication and movement of host genes. However, very few examples of host gene transduction by retrotransposition have been reported (45, 167, 197).
In contrast to these isolated examples, recent studies have shown that several types of DNA transposons have transduced hundreds to thousands of gene fragments in grass genomes. MULEs have been long suspected of capturing and carrying host gene fragments (121). The recent availability of the rice genome sequence has allowed a first quantitative appreciation of the extent of this phenomenon. Jiang et al. (83) identified over 3000 so-called PACK-MULEs containing fragments from more than 1000 cellular genes. Remarkably, about one fifth of the identified PACK-MULEs had captured exons from multiple loci, and some elements had effectively assembled chimeric genes representing novel exon combinations producing processed transcripts in planta. Although it remains to be shown whether rice PACK-MULEs have given rise to new genes with cellular function (89), the study clearly established the tremendous potential of PACK-MULEs for gene shuffling and duplication. Moreover, the tendency of MULEs to capture host sequences is not restricted to rice, but also occurred at appreciable frequency in dicot plants (75, 76). A recent example of PACK-MULE-mediated gene duplication in Arabidopsis shows that the mechanism can give rise to genes retaining functional coding capacity and likely novel function (75). Many more examples will likely soon be identified in other plant species and perhaps in other eukaryotes, given the widespread occurrence of MULEs (Table 1).
The mechanism by which PACK-MULEs capture host gene fragments is not understood. It is conceivable that it involves template switching and other aberrant events during the gap repair mechanism that followed transposon excision. Similar events of DNA capture have been reported during the repair of DSB left by excision of Drosophila P elements and maize Ac/Ds elements (61, 143, 168). Hence, not just MULEs, but other cut-and-paste DNA transposons are expected to be prone to capture. Indeed, several lines of evidence indicate that plant CACTA elements frequently transduce host sequences (96, 200). However, Helitrons, with their distinct mechanism of amplification, may raise the bar even higher in their ability to reshuffle and duplicate host sequences (140).
Ever since the discovery of Helitrons in eukaryotic genomes, their potential to act as exon shuffling machines (54) was apparent from the observation that some plant elements had seemingly captured one or multiple RPA-like proteins from the host genome to serve their own propagation (91). These proteins are involved in rolling-circle replication of other mobile elements, but normally are encoded by the host. The likelihood for Helitrons to capture genes useful for transposition implies that transduction events, regardless of the mechanism, must be extremely frequent (54). Preliminary evidence that this is indeed the case came from the isolation of the first Helitrons from maize (108, 109). These elements were very large in size (range) and were packed with fragments of seemingly unrelated genes. Most of the gene fragments were pseudogenes in various states of decomposition, and they had apparently been captured progressively from different genomic loci in the maize genome. Nonetheless, these PACK-Helitrons had clearly been recently active, as judged by their absence at orthologous position in other maize inbred lines.
Only more recently has it become clear that the first identified maize Helitrons represent only the tip of the iceberg. Elegant whole-genome analyses of gene content polymorphism between two inbred maize lines revealed ~10,000 large DNA insertions disrupting colinearity between the two lines (141). Eight of nine insertions molecularly characterized were found to be typical insertions of nonautonomous Helitrons replete with host gene fragments. It was shown that these elements and their internal gene fragments are frequently transcribed and that they transpose replicatively, peppering the genome with pieces of genes, while capturing additional gene fragments in the process (18, 67). The extrapolation of these findings to the whole maize genome revealed an unprecedented image of genome plasticity. Furthermore, if the captured fragments are indeed transcribed as was reported in the study, this could potentially create havoc considering the potential collision and interference of gene expression among the captured gene fragments and their parental copy. There must exist some mechanisms, most likely epigenetic, to keep this transcriptional burden under control.
Is the amplitude of _Helitron_-mediated transductions unique to the maize genome? Helitrons and many other TEs have clearly been unleashed recently and are probably still in an epoch of massive expansion in maize. However, it should be kept in mind that vast numbers of Helitrons have colonized the genome of a broad range of animals, including worms, mosquitoes, sea urchin, zebrafish, or bats (91, 92, 156, 158). Thus, there is no reason to assume that _Helitron_-mediated transduction events would be restricted to the maize genome. In fact, an instance of _Helitron_-mediated exon transduction and its subsequent amplification to ~1000 copies has been identified in the genome of the bat M. lucifugus (158). A more comprehensive assessment of the extent of this phenomenon in the bat genome is underway and should reveal whether this mechanism has also contributed to mammalian genome evolution.
In summary, it is becoming increasingly clear that DNA transposon-mediated transduction has been a significant mechanism contributing to the structural evolution of the genome. In fact, the maintenance of captured RPA sequences in plant Helitrons also illustrates the other side of the coin, namely that DNA transposons can take advantage of this mechanism for their own, typically modular, evolution. Likewise, it is tempting to speculate that the murdB gene that is unique to the maize MuDR element originates from a host gene fortuitously captured. Hence, there seems to be a continuous flux of sequences from the host to the DNA transposons. As we describe in the final section of this review, the flux is reversible: DNA transposons can also donate sequences to their host.
Molecular Domestication of DNA Transposons
One of the most direct contributions of TEs to host genome evolution is as a source of raw material that can be used for the assembly of new genes and functions (12, 16, 101, 102, 125, 186). TEs have numerous properties that predispose them for molecular domestication (134) or exaptation (17) by the genome for host function. For example, the palindromic structure of some MITEs may predispose them to evolve into microRNA genes (153). In this section, we focus on a particular category of exaptation events that seem to regularly implicate DNA transposons: the donation of protein-coding sequences to assemble new host genes.
Estimations of the rate at which TE-encoded proteins have been domesticated throughout evolution are necessarily conservative owing to our limited ability to recognize relationships between host genes and TEs. Indeed, many events are likely to have been erased through evolutionary time or they are so ancient that it cannot be inferred whether the TE gave rise to the host gene or vice versa [e.g., telomerase (44)]. In addition, TE genes and host genes cannot easily be distinguished in those genomes where large amounts of related and recently active TEs occur. In these genomes, very recent events of domestication will be very difficult to detect.
Different studies aimed at systematically identifying TE-derived genes have used different criteria. Some were purposely very stringent (202), whereas others were perhaps too pliant and likely yielded many false positives (11). Estimates from analyses of the human genome range widely from a few dozens to thousands (11, 13, 62, 111, 202). The reality is probably somewhere in between these estimates. Regardless of the exact count, all the studies point to a similar pattern whereby DNA transposons contribute to a proportionally large number of TE-derived genes relative to their abundance in the genome.
We have adopted a relatively conservative approach and list in Table 2 only examples of DNA transposon-derived genes in animal, fungi and plant species that have received extensive support for their transposon origin and functionality.
Table 2.
Transposase-derived genes and their functions
| Related TE Superfamily/subgroup | Gene ID | Full name | Original Species/Distribution | Functions and activities | Protein domains derived from Tpase | Other domains fused | Reference |
|---|---|---|---|---|---|---|---|
| Tc1/mariner/pogo | CENP-B | Centromere protein B | H. sapiens/Mammals | centromeric chromatin assembly, binds CENP-B box in alphoid satellite | DBD (CENPB) + core | 148b | |
| Tc1/mariner/pogo | JRK | Jerky | H. sapiens/Mammals | probable translational regulator in neurons (mutant mouse epileptic), DNA- and RNA-binding activity | DBD (CENPB) + core | 124b | |
| Tc1/mariner/pogo | JRKL | Jerky-like | H. sapiens/Mammals | unknown | DBD (CENPB) + core | a | |
| Tc1/mariner/pogo | TIGD1 | Tigger transposable element derived 1 | H. sapiens/Primates? | unknown | DBD (CENPB) + core | a | |
| Tc1/mariner/pogo | TIGD2, 3, 5–7 | Tigger transposable element derived 2, 3, 5–7 | H. sapiens/Mammals | unknown | DBD (CENPB) + core | a | |
| Tc1/mariner/pogo | TIGD4 | Tigger transposable element derived 4 | H. sapiens/Amniotes | unknown | DBD (CENPB) + core | a | |
| Tc1/mariner/Tc2 | POGK | pogo transposable element with KRAB domain | H. sapiens/Mammals | KRAB domain typically functions in transcriptional repression | DBD (CENPB) + core | KRAB | a |
| Tc1/mariner/Tc2 | POGZ | pogo transposable element with ZNF domain | H. sapiens/Vertebrates | unknown | DBD (CENPB) + core | ZnF | a |
| Tc1/mariner/mariner | SETMAR | SET domain and mariner transposase fusion gene | H. sapiens/Anthropoid Primates | binds DNA specifically, methylates histone H3 at K36 and faciltates DSB repair | DBD (HTH) + core | SET | 113, 33 |
| Tc1/mariner/pogo | rib | ribbon | D. melanogaster | promotes epithelial cell migration and morphogenesis | DBD (HTH_psq) | BTB | 172b |
| Tc1/mariner/pogo | pfk | piefke | D. melanogaster | binds polytene chromosome, nuclear protein present in larval salivary glands and ovaries | DBD (HTH_psq x3) | BTB | 172c |
| Tc1/mariner/pogo | psq | pipsqueak | D. melanogaster | developmental regulator with pleiotropic functions during oogenesis, embryonic pattern formation, and adult development, binds GAGAG consensus motif, involved in formation of repressive chromatin | DBD (HTH_psq x4) | BTB | 172c |
| Tc1/mariner/pogo | bab1 | bric-a-brac1 | D. melanogaster | homeotic and morphogenetic regulator in development of ovaries, appendages and abdomen, binds to A/T-rich regions with TA or TAA repeats | DBD (HTH_psq) | BTB | 125b |
| Tc1/mariner/pogo | bab2 | bric-a-brac2 | D. melanogaster | synergistic, distinct and redundant functions with Bab1 during imaginal development | DBD (HTH_psq) | BTB | 125b |
| Tc1/mariner/pogo | BtbVII | BTB-protein-VII | D. melanogaster | unknown | DBD (HTH_psq) | BTB | 172c |
| Tc1/mariner/pogo | Eip93F | Drosophila cell death protein E93 | D. melanogaster | directs steroid-triggered programmed cell death | DBD (HTH_psq) | 172c | |
| Tc1/mariner/pogo | Abp1 | (ars)-binding protein 1 | S. pombe/Schizosaccharomycetale | chromosome segregation and centromeric heterochromatin assembly, binds outer repeat of centromere, also required for efficient DNA replication through interaction with Cdc23 | DBD (CENPB) + core | 142b, 124c | |
| Tc1/mariner/pogo | Cbh1 | CENP-B homolog 1 | S. pombe/Schizosaccharomycetale | chromosome segregation and centromeric heterochromatin assembly, binds outer repeat of centromere | DBD (CENPB) + core | 142b | |
| Tc1/mariner/pogo | Cbh2 | CENP-B homolog 2 | S. pombe/Schizosaccharomycetale | chromosome segregation, binds inner core region of centromere | DBD (CENPB) + core | 142b | |
| Tc1/mariner/pogo | Pdc2 | Pyruvate decarboxylase 2 | S. cerevisiae/Saccharomycetales | transcription activator of pyruvate decarboxylase and thiamin metabolism | DBD (CENPB) + core | 137b | |
| piggyBac | PGBD1–5 | PiggyBac-derived 1–5 | H. sapiens/Mammals-Primates | unknown | DBD? + core | 170 | |
| Transib | RAG1 | recombination-activating gene 1 | H. sapiens/Jawed vertebrates | interacts with RAG2 to catalyze V(D)J recombination in immune B and T cells | DBD? + core | RING, NBR x2 | 93 |
| Mutator/Foldback/MULE | FAR1, FHY3 | far-red impaired response protein 1, far-red elongated hypocotyl 3 | A. thaliana/Eudicots | FAR1 and FHY3 are transcriptional activators binding upstream of FHY1 gene and triggering signaling cascade for far-red light sensing | DBD (WRKY) + core + SWIM | 80, 115 | |
| Mutator/Foldback/MULE | FRS1–11 | FAR1-related sequences 1–11 | A. thaliana/Angiosperms | unknown | DBD (WRKY) + core + SWIM | 115 | |
| Mutator/Foldback/MULE | MUG1 | Mustang1 | A. thaliana/Angiosperms | unknown | DBD (WRKY) + core +SWIM | PB1 | 34 |
| Mutator/Foldback/MULE | Aft1, Rcs1, Rbf1 | activator of ferrous transport 1, 2 | S. cerevisiae/Saccharomycetales | transcription factor involved in iron utilization and homeostasis; binds the consensus site PyPuCACCCPu and activates the expression of target genes in response to changes in iron availability | DBD (WRKY) | 4 | |
| hAT | DREF | DNA replication-related element-binding factor | D. melanogaster/Drosophilidae | transcription factor, positive regulator of DNA replication, cell proliferation, growth and differentiation, binds DRE motif | DBD (BED) + core + hATC | 74b | |
| hAT | ZBED1 (hDREF, Tramp) | human homolog of DREF, Zinc finger BED domain containing protein 1 | H. sapiens/Vertebrates | transcription factor, positive regulator of cell proliferation and ribosomal proteins, binds hDRE motif, homodimerizes via hATC domain | DBD (BED) + core + hATC | 197b | |
| hAT | ZBED4 (KIAA0637) | Zinc finger BED domain containing protein 4 | H. sapiens/Vertebrates | homodimerizes via hATc domain | DBD (BED) + core + hATC | 197b | |
| hAT | BEAF-32 | boundary element-associated factor of 32 kDa | D. melanogaster/Drosophilidae | binds scs and other chromatin boundary elements and hundreds of sites on polyene chromosome, homodimerizes through C-term domain | DBD (BED) + hATC? | 207b | |
| hAT/Charlie1 | Buster1 (ZBED5) | Zinc finger BED domain containing protein 5 | H. sapiens/Mammals | translational repressor modulating interferon--induced apoptosis, fused with part of eIF4G2 protein | DBD (BED) + core + hATC | 178b | |
| hAT | Daysleeper | Daysleeper | A. thaliana/Unknown | essential for plant development, binds to motif upstream of Ku70 repair gene, likely transcription factor | DBD (BED) + core + hATC | 18b | |
| hAT | Gary | Gary | Grasses (Poaceae) | unknown | DBD? + core | 141b | |
| hAT/Charlie8 | GTF2IRD2 | GTF2I repeat domain containing 2 | H. sapiens/Mammals | fused to GTF2I domain of TFII-I transcription factor family with essential function in vertebrate development | DBD (BED) + core | LZ, TFII-I x2 | 181b |
| hAT (+ P element?) | LIN-15B | abnormal cell LINeage family member 15B | C. elegans/Unknwon | key developmental regulator, interacts genetically with C. elegans homolog of mammalian retinoblastoma, inhibits G1-S cell-cycle transition | DBD (BED) + core +THAP | 27b | |
| hAT (+ P element?) | GON-14 | gonadogenesis deficient, lin15b familiy member | C. elegans/Unknwon | required for gonadogenesis and probably a pleiotropic transcriptional regulator of development (mutant shows gonad, vulval, growth, and cell division defects) | DBD (THAP)+hAT core+hATC | 27b | |
| P element (+ hAT?) | THAP0 (p52riPK) | interferon-induced protein kinase-interacting protein, death-associated protein DAP4 | H. sapiens/Vertebrates | upstream regulator of interferon-induced translational repressor PKR by interaction and inhibition of p58IPK | DBD (THAP) + hATC | 178b | |
| P element | THAP1 | nuclear proapoptotic factor THAP1 | H. sapiens/Vertebrates | nuclear proapoptotic factor, binds DNA specifically and regulates endothelial cell proliferation through modulation of pRB/E2F cell-cycle target genes | DBD (THAP) | 28b | |
| P element | THAP7 | Thanatos-Associated Protein 7 | H. sapiens/Vertebrates | transcriptional repressor, binds histone H4 tail and recruits HDAC3 and NcoR to specific DNA sites, associates with template activating factor-Ibeta and inhibits H3 acetylation | DBD (THAP) | 126c | |
| P element | THAP2–5, 8, 10, 11 | THAP-containing proteins | H. sapiens/Mammals-Vertebrates | unknown | DBD (THAP) | 28b | |
| P element | HIM-17 | High Incidence of Males 7 | C. elegans | chromatin-associated protein required for initiation of meiotic recombination and chromosome segregation | DBD (THAP x6) | coiled-coil | 161c |
| P element | LIN-36 | abnormal cell LINeage family member 36 | C. elegans | functions in vulval development as inhibitor of the G1-to-S-phase cell-cycle transition, regulates cell proliferation | DBD (THAP) | C2H2 | 10b |
| P element | CDC-14B | cell-cycle regulator tyrosine phosphatase, isoform B | C. elegans | cell cycle G1/S inhibitor, required for genome stability | DBD (THAPx2) | cdc14 | 28b |
| P element | CTB-1 | homolog of CtBP transcriptional corepressor | C. elegans | vertebrate homolog CtBP is a global transcription corepressor critical for development and oncogenesis | DBD (THAP) | NAD_b | 28b |
| P element | Phsa (THAP9) | P element-homologous gene | H. sapiens/Mammals-Amniotes | unknown | DBD (THAP) + core | 68 | |
| P element | THAP–E2F6 | fusion THAP and cell-cycle transcription factor E2F6 | D. rerio/Fish+Amphibians | mammalian E2F6 interacts with Polycomb (PcG) group proteins and functions as a repressor of E2F-dependent transcription during S phase | DBD (THAP) | E2F_TDP | 28b |
| P element | P-tsa | tsacasi stationary truncated P-neogene | D. tsacasi | unknown, but binds DNA | DBD (THAP) + partial core | 160b | |
| P element | P-neo G and A type | obscura amplified P-neogene | D. subobscura/subobscura subgroup | unknown | DBD (THAP) + partial core | 134 | |
| P element | P-neo montium | montium P-neogene | D. montium/montium subgroup | unknown | DBD (THAP) + partial core | 162b | |
| PIF/Harbinger | HARBI1 (FLJ32675) | Harbinger derived-gene 1 | H. sapiens/Vertebrates | unknown, but interacts with NAIF | DBD (HTH) + core | 92b,b | |
| PIF/Harbinger | NAIF1 (c9orf90) | nuclear apoptosis-inducing factor 1 | H. sapiens/Vertebrates | unknown, but inhibits cell growth and induces apoptosis when overexpressed, interacts with and mediates nuclear translocation of HARBI1 | DBD (Myb/SANT) | 126b | |
| PIF/Harbinger | DPLG1–7 | Drosophila PIF-like gene 1–7 | Drosophila | unknown | DBD (HTH) + core | 26b | |
| PIF/Harbinger | DPM7 | Drosophila PIF MADF-like gene | Drosophila | unknown, but probable interactor of DPLG7 protein | DBD (Myb/MADF) | 26b | |
| Maverick? | KRBA, ZNF452 | c-integrases | H. sapiens/Mammals | unknown | RVE core | KRAB/ZnF | 57b,c |
Specifically, these genes fulfill at least three of the following criteria:
- Absence of flanking transposon hallmarks (such as TIRs) and no evidence for recent mobility;
- Phylogenetic placement of the encoded protein within a cluster of transposon-encoded proteins;
- Intact coding capacity and evolution under functional constraints (as opposed to TE coding regions, which typically evolve under neutral evolution);
- Detection of intact orthologs in syntenic genomic regions of distantly related species (TE genes are not expected to be maintained intact for extended period of time at orthologous positions between two distantly related species, such as human and mouse)
- Evidence of transcription (in contrast, TE genes are often transcriptionally silenced);
- Genetic evidence for a critical biological function in vivo.
Most of the genes listed in Table 2 encode transposase-related proteins, since this is the only protein encoded by most DNA transposons. Exceptions include the c-integrases of mammals, which appears to derive from a Maverick transposon (52, 57b), and a MADF domain-containing protein in Drosophila that was domesticated from the accessory protein of a _PIF_-like transposon (26b). In addition, several proteins listed in Table 2 are chimeric proteins that result from the fusion of transposase-derived domains to domains of other origins. This process is consistent with the modular evolution of proteins in general and the concept of evolutionary tinkering introduced by François Jacob (82).
Only a small fraction of the genes listed in Table 2 have been studied functionally. Thus, in most cases, the functional contribution of the transposase domain(s) to the corresponding protein remains a matter of speculation. However, one can draw several predictions based on the functional analyses of related transposases. All eukaryotic transposases that have been biochemically characterized possess two functionally separable domains: a N-terminal region that binds to the ends of the cognate transposons (generally the TIRs) and a central or C-terminal core region that catalyzes the cleavage and transfer steps of the transposition reaction (35). Any of these activities can be potentially co-opted to serve cellular function(s) and, as we outline below, there is now evidence that these activities have been differentially retained in different transposase-derived proteins. Nonetheless, a recurrent theme is the recycling of transposase DNA-binding domain (DBD) to build transcription factors (Table 1).
As long-term genomic residents coevolving with their host, transposases are expected to have developed a number of interactions with host proteins, even though these interactions may not be strictly required for transposition (107). For example, the Sleeping Beauty transposase interacts directly with the Ku70 repair protein, the DNA-bending high-mobility group protein HMGB1 and the transcription factor Miz-1 (81, 188, 201). Each of these proteins has a large number of interacting partners, and interaction with the Sleeping Beauty transposase may influence and modulate their cellular function. Similarly, the pogo transposase of D. melanogaster interacts with PCNA, the proliferating cell nuclear antigen, a key protein for DNA replication and repair (191). A functional PCNA-binding motif is also present in Tigger1, the human relative of pogo transposase (191), and a similar motif is present at a comparable position in the _Arabidopsis pogo_-like transposase Lemi1 (50), suggesting that PCNA interaction with _pogo_-like transposases is evolutionary conserved. The association of transposons with DNA repair and replication factors appears as a recurrent theme. It is easy to conceive how this association could benefit both transposon and host. In turn, these interactions may predispose the transposase to domestication and be preserved in transposase-derived proteins. This might explain why several transposase-derived proteins appear to be involved in cell cycle control, recombination, and other aspects of chromosome dynamics (Table 2).
Below we recount three tales of transposase domestication and discuss the evolutionary consequences of these innovations.
The origin of the adaptive immune system of jawed vertebrates
V(D)J recombination is the process by which a virtually infinite population of distinct antibodies can be generated in B and T lymphocytes. The acquisition of V(D)J recombination is often regarded as a key step in the evolution of the adaptive immune system of jawed vertebrates (32, 88). The two essential components of V(D)J recombination are (i) the RAG1 and RAG2 proteins, which interact to form the recombinase responsible for the joining and transfer activities; and (ii) the recombination signal sequences (RSS) flanking the V (variable), D (diversity), and J (joining) segments, which define the specific sequences bound, cleaved, and joined by the RAG1/2 protein complex (58). The analogy of the process of V(D)J recombination to a transposition reaction is striking. RAG1/2 can catalyze transposition of a DNA segment flanked by RSS in vitro (3, 74) and in vivo in yeast (28) and mammalian cells (27, 162). Additionally, it has been observed that several eukaryotic transposases utilize a cleavage chemistry similar to that seen in V(D)J recombination (37, 208). However, until recently no transposase directly related to RAG1/2 had been identified.
Evidence of this relationship came from the discovery that RAG1, which provides the catalytic core for the reaction, is closely related in sequence to transposases encoded by Transib elements, a group of DNA transposons recently identified in the genomes of diverse invertebrates (93). Additional support for the relationship came from comparisons of the structure of the RSS to the TIRs of Transib transposons and the conservation of spatial and sequence characteristics [the so-called 12/23 rule; (58)]. Finally, Transib elements provoke a 5-bp TSD upon transposition, as do most cut-and-paste reactions mediated by RAG1 in vitro (3, 74). Together the data leave little doubt that V(D)J recombination is the product of a fortuitous event of DNA transposon domestication, an event that may be viewed as a crucial step in vertebrate evolution.
Light-sensing in plants and the FAR1/FHY3 family of transcription factors
As sessile organisms, higher plants have evolved a network of photoreceptors to sense light changes in the environment (130). Among the photoreceptors, the phytochrome A (phyA) pathway has been extensively characterized. Photoactivation leads to the conversion of phyA into an active form allowing its import to the nucleus from the cytoplasm (190). Once in the nucleus, phyA is thought to directly activate a set of transcription factors, which in turn induce a molecular cascade resulting in light-mediated photomorphogenic responses (87, 181, 190). PhyA accumulation in the nucleus is dependent on the presence of two homologous proteins, FHY1 and FHL (73). A recent series of genetic and biochemical studies established that transcription of FHY1 and FHL is directly modulated by two transcription factors FHY3 and FAR1 that bind to the proximal promoter regions of FHY1 and FHL (R. Lin, C. Casola, D. Ripoll, F. Nagy, C.F., H. Wang, submitted). Unexpectedly, it turns out that FHY3 and FAR1 are members of an ancient gene family that is related to MULE transposases (80, 115). Evolutionary analyses indicate that the entire FHY3/FAR1 family is most likely derived from a single domestication event of a MULE transposase at the dawn of angiosperm evolution (R. Lin, C. Casola, D. Ripoll, F. Nagy, C.F., H. Wang, submitted). This domestication event would coincide with the origin of FHY1 and FHL and with the early evolution of the phyA pathway (130).
FAR1 and FHY3 have a specific DBD located in the N-terminal region of the protein. This region is conserved in the transposases encoded by the most closely related MULEs found in modern plant genomes (4). It is tempting to speculate that the binding sites of FHY3/FAR1 are themselves derived from the TIRs of ancient MULE transposons that integrated upstream of the target genes regulated by FHY3 and FAR1, including FHY1 and possibly other targets (80). In this model, not only the transposase but also its unlinked binding sites, dispersed in the genome as the result of the past propagation of MULEs, could have been codomesticated to establish a regulatory network (see below). Finally, note that FHY3 possesses intrinsic transcriptional activation ability that is separable from its DNA-binding activity (R. Lin, C. Casola, D. Ripoll, F. Nagy, C.F., H. Wang, submitted). This activity requires residues located within the predicted catalytic domain of the ancestral transposase that are also conserved in distant MULE transposases. This observation indicates that many MULE transposases might have intrinsic transcription factor activity, and it would explain why several MULE transposases seem to have been domesticated repeatedly during eukaryotic evolution (4, 34, 75).
The primate SETMAR fusion gene
The two examples described above show that transposase domestication events have been instrumental in the emergence of key innovations both in vertebrates and flowering plants, respectively. In both cases, it seems that not only the transposase but also sequences present at unlinked transposon copies were codomesticated. We hypothesize that the fundamental property of transposase molecules to recognize and act in trans on multiple DNA elements dispersed throughout the genome is a major factor contributing to their recurrent domestication in eukaryotes. Indeed, transposase domestication can be instantly accompanied by the selective recruitment of a ready-to-use network of binding sites in the genome (Figure 3).
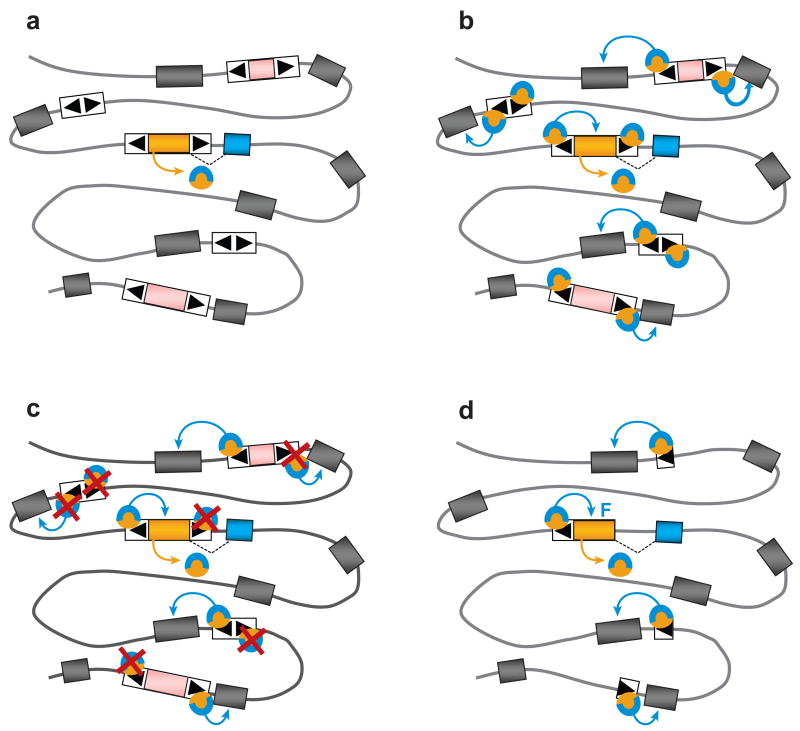
Figure 3. Model for the assembly of a regulatory network by domestication of a transposase and its binding sites.
A: Initial transposase domestication event. A family of DNA transposon is shown with autonomous and nonautonomous copies dispersed in the genome. Each TIR (black arrowhead) contains a binding site for a transposase encoded by autonomous copies (pink/yellow boxes). Flanking host genes are shown as grey boxes. One of the transposase genes (yellow box) is recruited. In this example, recruitment is promoted by transcriptional fusion of the transposase to a flanking host gene (blue box) encoding a regulatory domain, leading to the expression of a fusion protein with transpoase (yellow) and regulatory domains (blue). This is similar to the emergence of SETMAR, which arose by fusion of a mariner transposase with an adjacent gene encoding a SET domain. Note however that transposase domestication does not need to involve fusion with another domain, particularly if the transposase itself possesses regulatory activity, as demonstrated for FHY3, a transcription factor in Arabidopsis entirely derived from a Mutator transposase. B: Immediate consequences of transposase domestication. The translational fusion immediately allows the regulatory domain to be tethered to all the sites in the genome recognized by the transposase, i.e. the TIRs of all the transposon copies previously dispersed in the genome. Depending on the genomic environment of the transposons, binding of the fusion protein might have various effects on the expression of the surrounding genes: activation, repression or no effect. These effects are symbolized by the blue arrow acting on adjacent gene. C: Binding sites selection. Natural selection will retain interactions that provide an immediate benefit to the host and will eliminate deleterious interactions. Site elimination (red cross) may occur through substitutions or deletion driven by positive selection. Sites that are selectively neutral (with no positive or negative impact on adjacent genes) are expected to evolve neutrally and most will eventually disappear. Mobility of the transposons at this stage (if it persists) might accelerate the shaping of the network through transposon excision events and/or fixation of new advantageous insertions. D: A regulatory network is born. The end result is the assembly of a regulatory network, where the domesticated transposase and a subset of its ancestral binding sites conferring beneficial interactions are evolving under purifying selection, while the rest of the transposons are eroded by mutations. Note that the system also provides an intuitive opportunity for the establishment of a feedback loop “F” (positive or negative) through domestication of binding sites that were originally linked to the domesticated transposase.
To test this model and better understand the early steps of transposase domestication, it is necessary to study relatively recent exaptation events, where the transposase and its associated binding sites would be still readily recognizable as being derived from the same transposon family. We believe that the example of SETMAR described in the next section provides the ideal system to study the early steps of this model.
SETMAR is a human gene first identified by Hugh Robertson as a particular copy of the Hsmar1 family, one of two _mariner_-like families found in the human genome (165). SETMAR originates from the transcriptional fusion of a SET domain to a mariner transposase. The function of the SETMAR protein is currently unknown, but in vitro experiments have shown that the SET portion of SETMAR has specific histone methyltransferase activity for lysine 36 of histone H3 (113). The function of this epigenetic mark is not understood in mammals, but studies in yeast indicate that it may act as a repressive chromatin mark to prevent spurious intragenic transcription (25, 99). In addition, overexpression of SETMAR in human cells facilitate DNA repair via the non-homologous end-joining pathway (113). However, the contribution of the transposase domain to this activity and to the function of SETMAR remains unclear.
In order to gain further insights, comparative genome sequencing was used to trace the origin of SETMAR and delineate the steps leading to the fusion of SET and MAR domains (33). The results show that SETMAR has emerged between 58 and 40 Mya in an anthropoid primate ancestor, through an intricate process involving transposition of a mariner transposon downstream of a pre-existing gene encoding a stand-alone SET domain, followed by genomic deletion of intervening DNA and creation of a new intron. The transposase region of SETMAR has been subject to strong evolutionary constraint in all extant major lineages of anthropoid primates, suggesting that the addition of a transposase domain to the pre-existing SET domain led to the advent of a beneficial new function in primates. The signature of purifying selection has been particularly intense on the N-terminal region of the transposase containing the predicted DBD, whereas the catalytic domain appears to evolve essentially neutrally (33).
Consistent with these predictions, biochemical studies indicate that SETMAR is deficient for cleavage at the 3′ ends of the element (123, 136), but has retained the ability to bind specifically DNA through its N-terminal DBD, assemble a paired-ends complex, and inflict single-strand nicks on adjacent DNA (33, 123, 136). Furthermore, SETMAR has retained strong specificity for binding to a 19-bp site located within the TIR of the related Hsmar1 or MADE1 transposons (33). The binding site is dispersed in over 1500 conserved copies throughout the human genome and nearly all of these sites map within the TIRs of the related transposons. Together these data support a model whereby the specific DNA-binding activity of the transposase region now provides a means to target the SET domain to specific sites within the human genome (33, Figure 3). For this model to be validated, it will be necessary to pinpoint the DNA targets of SETMAR and determine the effect of tethering the protein to specific chromosomal sites.
Summary Points
- The great diversity of DNA transposons can be organized into 3 major subclasses: the cut-and-paste transposons, with ten major superfamilies; rolling-circle transposons (Helitrons); and self-replicating transposons (Mavericks).
- Almost all subclasses and superfamilies are represented in a wide range of eukaryotes, including various protozoans. Thus, DNA transposons diversified very early in evolution and have been maintained in all major branches of the eukaryotic tree of life.
- Vast variations occur among species in the level of amplification of their DNA transposon populations. Different amplification of DNA transposons among species may or may not translate into substantial differences in genome size, but probably reflect a complex combination of intrinsic (host- or self-mediated) and extrinsic (environmental, ecological) factors modulating the activity and retention of transposon activity over evolutionary time.
- The evolutionary success and astonishing diversity of eukaryotic DNA transposons offers an intriguing paradox because their amplification dynamics seems to represent an evolutionary dead end favoring the proliferation of non-autonomous derivatives (MITEs) to the detriment of the autonomous copies. We propose a more subtle vision whereby the accidental amplification of MITEs passes under the radar of the host defense system and drives the diversification of autonomous copies.
- Like other TEs, DNA transposons play a significant role in shaping eukaryotic genomes, but they possess specific features that enhance or accentuate some of their influence on the host. These features include their capacity to excise imprecisely, jump locally, inflict multiple double-strand breaks, and undergo alternative transposition.
- DNA transposons have been a recurrent source of coding sequences for the emergence of new genes. We propose that this is a pervasive pathway to create a genetic network as the recruitment of transposase DNA-binding domains opens the door for selection to instantly retain a set of unlinked binding sites previously dispersed in the genome and/or co-opt their interactions with host proteins.
DEFINITION LIST
Epigenetic
related to modification of the chromatin or the DNA that affects the biology of the organism and is stable over rounds of cell division but does not involve changes in the underlying DNA sequence of the organism
Horizontal transfer
the transmission of genetic material between the genomes of two individuals (that may belong to different species) by nonvertical inheritance
Insertion sequences
prokaryotic mobile elements that resemble eukaryotic DNA transposons in their structure and transposition mechanism
RNA interference (RNAi)
a posttranscriptional mechanism of gene silencing triggered by the formation of double-stranded RNA that is processed into small interfering RNAs mediating the degradation of matching mRNAs
Vertical diversification
the emergence (speciation) of a slightly different variant of an autonomous transposon from another element, either within the same species or during the radiation of a species, given rise to a new transposon family
Transposon footprint
a short stretch of the transposon terminal sequences left behind after excision of the transposon
Boundary or insulator elements
DNA sequences that block the spread of heterochromatin and partition the genome into distinct functional chromosomal domains
Exaptation
utilization of a sequence or structural feature for a function other than that for which it was originally developed through the process of natural selection
LITERATURE CITED
- 1.Abrusan G, Krambeck HJ. Competition may determine the diversity of transposable elements. Theor Popul Biol. 2006;70:364–75. doi: 10.1016/j.tpb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Adams KL, Wendel JF. Novel patterns of gene expression in polyploid plants. Trends Genet. 2005;21:539–43. doi: 10.1016/j.tig.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 3.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–51. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 4.Babu MM, Iyer LM, Balaji S, Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 2006;34:6505–20. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakre AA, Rawal K, Ramaswamy R, Bhattacharya A, Bhattacharya S. The LINEs and SINEs of Entamoeba histolytica: comparative analysis and genomic distribution. Exp Parasitol. 2005;110:207–13. doi: 10.1016/j.exppara.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 6.Barkan A, Martienssen RA. Inactivation of maize transposon Mu suppresses a mutant phenotype by activating an outward-reading promoter near the end of Mu1. Proc Natl Acad Sci USA. 1991;88:3502–6. doi: 10.1073/pnas.88.8.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bennetzen JL, Ma J, Devos KM. Mechanisms of recent genome size variation in flowering plants. Ann Bot. 2005;95:127–32. doi: 10.1093/aob/mci008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betran E, Santos M, Ruiz A. Antagonistic pleiotropic effect of second-chromosome inversions on body size and early life-history traits in Drosophila buzzatii. Evolution. 1998;52:144–54. doi: 10.1111/j.1558-5646.1998.tb05147.x. [DOI] [PubMed] [Google Scholar]
- 9.Boeke JD. Putting mobile DNA to work: the toolbox. Washington, DC: Am. Soc. Microbiol. Press; 2002. See Ref. 35, pp. 24–37. [Google Scholar]
- 10.Boulesteix M, Weiss M, Biemont C. Differences in genome size between closely related species: the Drosophila melanogaster species subgroup. Mol Biol Evol. 2006;23:162–67. doi: 10.1093/molbev/msj012. [DOI] [PubMed] [Google Scholar]
- 10b.Boxem M, van den Heuvel S. C. elegans class B synthetic multivulva genes act in G(1) regulation. Curr Biol. 2002;12:906–911. doi: 10.1016/s0960-9822(02)00844-8. [DOI] [PubMed] [Google Scholar]
- 11.Britten R. Transposable elements have contributed to thousands of human proteins. Proc Natl Acad Sci USA. 2006;103:1798–803. doi: 10.1073/pnas.0510007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Britten RJ. DNA sequence insertion and evolutionary variation in gene regulation. Proc Natl Acad Sci USA. 1996;93:9374–77. doi: 10.1073/pnas.93.18.9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Britten RJ. Coding sequences of functioning human genes derived entirely from mobile element sequences. Proc Natl Acad Sci USA. 2004;101:16825–30. doi: 10.1073/pnas.0406985101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–57. doi: 10.1126/science.165.3891.349. [DOI] [PubMed] [Google Scholar]
- 15.Brookfield JF. Evolutionary genetics: mobile DNAs as sources of adaptive change? Curr Biol. 2004;14:R344–45. doi: 10.1016/j.cub.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 16.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–34. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 17.Brosius J, Gould SJ. On “genomenclature”: a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA. Proc Natl Acad Sci USA. 1992;89:10706–10. doi: 10.1073/pnas.89.22.10706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brunner S, Pea G, Rafalski A. Origins, genetic organization and transcription of a family of nonautonomous helitron elements in maize. Plant J. 2005;43:799–810. doi: 10.1111/j.1365-313X.2005.02497.x. [DOI] [PubMed] [Google Scholar]
- 18b.Bundock P, Hooykaas P. An Arabidopsis hAT-like transposase is essential for plant development. Nature. 2005;436:282–284. doi: 10.1038/nature03667. [DOI] [PubMed] [Google Scholar]
- 19.Bureau TE, Wessler SR. Tourist: a large family of inverted-repeat element frquently associated with maize genes. Plant Cell. 1992;4:1283–94. doi: 10.1105/tpc.4.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caceres M, Puig M, Ruiz A. Molecular characterization of two natural hotspots in the Drosophila buzzatii genome induced by transposon insertions. Genome Res. 2001;11:1353–64. doi: 10.1101/gr.174001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caceres M, Ranz JM, Barbadilla A, Long M, Ruiz A. Generation of a widespread Drosophila inversion by a transposable element. Science. 1999;285:415–18. doi: 10.1126/science.285.5426.415. [DOI] [PubMed] [Google Scholar]
- 22.Capy P, Bazin C, Higuet D, Langin T. Dynamics and Evolution of Transposable Elements. Austin, TX: Springer-Verlag; 1998. [Google Scholar]
- 23.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, et al. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–12. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carroll SB. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–92. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 26.Casals F, Caceres M, Ruiz A. The foldback-like transposon Galileo is involved in the generation of two different natural chromosomal inversions of Drosophila buzzatii. Mol Biol Evol. 2003;20:674–85. doi: 10.1093/molbev/msg070. [DOI] [PubMed] [Google Scholar]
- 26b.Casola C, Lawing AM, Betran E, Feschotte C. PIF-like Transposons Are Common in Drosophila and Have Been Repeatedly Domesticated to Generate New Host Genes. Mol Biol Evol. 2007 doi: 10.1093/molbev/msm116. in press. [DOI] [PubMed] [Google Scholar]
- 27.Chatterji M, Tsai CL, Schatz DG. Mobilization of RAG-generated signal ends by transposition and insertion in vivo. Mol Cell Biol. 2006;26:1558–68. doi: 10.1128/MCB.26.4.1558-1568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27b.Chesney MA, Kidd AR, 3rd, Kimble J. gon-14 functions with class B and class C synthetic multivulva genes to control larval growth in Caenorhabditis elegans. Genetics. 2006;172:915–928. doi: 10.1534/genetics.105.048751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clatworthy AE, Valencia MA, Haber JE, Oettinger MA. V(D)J recombination and RAG-mediated transposition in yeast. Mol Cell. 2003;12:489–99. doi: 10.1016/s1097-2765(03)00305-8. [DOI] [PubMed] [Google Scholar]
- 28b.Clouaire T, Roussigne M, Ecochard V, Mathe C, Amalric F, Girard JP. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc Natl Acad Sci U S A. 2005;102:6907–6012. doi: 10.1073/pnas.0406882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coen ES, Carpenter R, Martin C. Transposable elements generate novel spatial patterns of gene expression in Antirrhinum majus. Cell. 1986;47:285–96. doi: 10.1016/0092-8674(86)90451-4. [DOI] [PubMed] [Google Scholar]
- 30.Collins M, Rubin GM. Structure of chromosomal rearrangements induced by the FB transposable element in Drosophila. Nature. 1984;308:323–27. doi: 10.1038/308323a0. [DOI] [PubMed] [Google Scholar]
- 31.Comai L, Madlung A, Josefsson C, Tyagi A. Do the different parental ‘heteromes’ cause genomic shock in newly formed allopolyploids? Philos Trans R Soc London Ser B. 2003;358:1149–55. doi: 10.1098/rstb.2003.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cooper MD, Alder MN. The evolution of adaptive immune systems. Cell. 2006;124:815–22. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci USA. 2006;103:7941–42. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cowan RK, Hoen DR, Schoen DJ, Bureau TE. MUSTANG is a novel family of domesticated transposase genes found in diverse angiosperms. Mol Biol Evol. 2005;22:2084–89. doi: 10.1093/molbev/msi202. [DOI] [PubMed] [Google Scholar]
- 35.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, DC: Am. Soc. Microbiol. Press; 2002. [Google Scholar]
- 36.Daboussi MJ, Capy P. Transposable elements in filamentous fungi. Annu Rev Microbiol. 2003;57:275–99. doi: 10.1146/annurev.micro.57.030502.091029. [DOI] [PubMed] [Google Scholar]
- 37.Dawson A, Finnegan DJ. Excision of the Drosophila mariner transposon Mos1. Comparison with bacterial transposition and V(D)J recombination. Mol Cell. 2003;11:225–35. doi: 10.1016/s1097-2765(02)00798-0. [DOI] [PubMed] [Google Scholar]
- 38.De Aguiar D, Hartl DL Usda-Ars CfMA, Veterinary Entomology GFLUSA. Regulatory potential of nonautonomous mariner elements and subfamily crosstalk. Genetica. 1999;107:79–85. [PubMed] [Google Scholar]
- 39.de Almeida LM, Carareto CM. Multiple events of horizontal transfer of the Minos transposable element between Drosophila species. Mol Phylogenet Evol. 2005;35:583–94. doi: 10.1016/j.ympev.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 40.Deininger PL, Batzer MA. Alu repeats and human disease. Mol Genet Metab. 1999;67:183–93. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- 41.DeMarco R, Venancio TM, Verjovski-Almeida S. SmTRC1, a novel Schistosoma mansoni DNA transposon, discloses new families of animal and fungi transposons belonging to the CACTA superfamily. BMC Evol Biol. 2006;6:89. doi: 10.1186/1471-2148-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Diao X, Freeling M, Lisch D. Horizontal transfer of a plant transposon. PLoS Biol. 2006;4:e5. doi: 10.1371/journal.pbio.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dietrich CR, Cui F, Packila ML, Li J, Ashlock DA, et al. Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics. 2002;160:697–716. doi: 10.1093/genetics/160.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eickbush TH. Telomerase and retrotransposons: Which came first? Science. 1997;277:911–12. doi: 10.1126/science.277.5328.911. [DOI] [PubMed] [Google Scholar]
- 45.Ejima Y, Yang L. Trans mobilization of genomic DNA as a mechanism for retrotransposon-mediated exon shuffling. Hum Mol Genet. 2003;12:1321–28. doi: 10.1093/hmg/ddg138. [DOI] [PubMed] [Google Scholar]
- 46.Engels WR, Johnson-Schlitz DM, Eggleston WB, Sved J. High-frequency P element loss in Drosophila is homolog dependent. Cell. 1990;62:515–25. doi: 10.1016/0092-8674(90)90016-8. [DOI] [PubMed] [Google Scholar]
- 47.Engels WR, Preston CR. Formation of chromosome rearrangements by P factors in Drosophila. Genetics. 1984;107:657–78. doi: 10.1093/genetics/107.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feschotte C. Merlin, a new superfamily of DNA transposons identified in diverse animal genomes and related to bacterial IS1016 insertion sequences. Mol Biol Evol. 2004;21:1769–80. doi: 10.1093/molbev/msh188. [DOI] [PubMed] [Google Scholar]
- 49.Feschotte C, Jiang N, Wessler SR. Plant transposable elements: Where genetics meets genomics. Nat Rev Genet. 2002;3:329–41. doi: 10.1038/nrg793. [DOI] [PubMed] [Google Scholar]
- 50.Feschotte C, Mouchès C. Evidence that a family of miniature inverted-repeat transposable elements (MITEs) from the Arabidopsis thaliana genome has arisen from a pogo-like DNA transposon. Mol Biol Evol. 2000;17:730–37. doi: 10.1093/oxfordjournals.molbev.a026351. [DOI] [PubMed] [Google Scholar]
- 51.Feschotte C, Osterlund MT, Peeler R, Wessler SR. DNA-binding specificity of rice mariner-like transposases and interactions with Stowaway MITEs. Nucleic Acids Res. 2005;33:2153–65. doi: 10.1093/nar/gki509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Feschotte C, Pritham EJ. Non-mammalian c-integrases are encoded by giant transposable elements. Trends Genet. 2005;21:551–52. doi: 10.1016/j.tig.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Feschotte C, Swamy L, Wessler SR. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with Stowaway MITEs. Genetics. 2003;163:747–58. doi: 10.1093/genetics/163.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Feschotte C, Wessler SR. Treasures in the attic: rolling circle transposons discovered in eukaryotic genomes. Proc Natl Acad Sci USA. 2001;98:8923–24. doi: 10.1073/pnas.171326198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Feschotte C, Wessler SR. Mariner-like transposases are widespread and diverse in flowering plants. Proc Natl Acad Sci USA. 2002;99:280–85. doi: 10.1073/pnas.022626699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Feschotte C, Zhang X, Wessler S. Miniature inverted-repeat transposable elements (MITEs) and their relationship with established DNA transposons. 2002. See Ref. 35, pp. 1147–58. [Google Scholar]
- 57.Fischer SE, Wienholds E, Plasterk RH. Continuous exchange of sequence information between dispersed Tc1 transposons in the Caenorhabditis elegans genome. Genetics. 2003;164:127–34. doi: 10.1093/genetics/164.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57b.Gao X, Voytas DF. A eukaryotic gene family related to retroelement integrases. Trends Genet. 2005;21:133–7. doi: 10.1016/j.tig.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 58.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–32. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 59.Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 60.Girard L, Freeling M. Regulatory changes as a consequence of transposon insertion. Dev Genet. 1999;25:291–96. doi: 10.1002/(SICI)1520-6408(1999)25:4<291::AID-DVG2>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 61.Gorbunova V, Levy AA. Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucleic Acids Res. 1997;25:4650–57. doi: 10.1093/nar/25.22.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gotea V, Makalowski W. Do transposable elements really contribute to proteomes? Trends Genet. 2006;22:260–67. doi: 10.1016/j.tig.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 63.Gray YH. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–68. doi: 10.1016/s0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- 64.Gregory TR. Synergy between sequence and size in large-scale genomics. Nat Rev Genet. 2005;6:699–708. doi: 10.1038/nrg1674. [DOI] [PubMed] [Google Scholar]
- 65.Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- 66.Guimond N, Bideshi DK, Pinkerton AC, Atkinson PW, O’Brochta DA. Patterns of Hermes transposition in Drosophila melanogaster. Mol Genet Genomics. 2003;268:779–90. doi: 10.1007/s00438-002-0800-4. [DOI] [PubMed] [Google Scholar]
- 67.Gupta S, Gallavotti A, Stryker GA, Schmidt RJ, Lal SK. A novel class of Helitron-related transposable elements in maize contain portions of multiple pseudogenes. Plant Mol Biol. 2005;57:115–27. doi: 10.1007/s11103-004-6636-z. [DOI] [PubMed] [Google Scholar]
- 68.Hammer SE, Strehl S, Hagemann S. Homologs of Drosophila P transposons were mobile in zebrafish but have been domesticated in a common ancestor of chicken and human. Mol Biol Evol. 2005;22:833–44. doi: 10.1093/molbev/msi068. [DOI] [PubMed] [Google Scholar]
- 69.Hartl DL, Lohe AR, Lozovskaya ER. Modern thoughts on an ancyent marinere: function, evolution, regulation. Annu Rev Genet. 1997;31:337–58. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- 70.Hartl DL, Lozovskaya ER, Lawrence JG. Nonautonomous transposable elements in prokaryotes and eukaryotes. Genetica. 1992;86:47–53. doi: 10.1007/BF00133710. [DOI] [PubMed] [Google Scholar]
- 71.Haynes KA, Caudy AA, Collins L, Elgin SC. Element 1360 and RNAi components contribute to HP1-dependent silencing of a pericentric reporter. Curr Biol. 2006;16:2222–27. doi: 10.1016/j.cub.2006.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Heslip TR, Williams JA, Bell JB, Hodgetts RB. A P element chimera containing captured genomic sequences was recovered at the vestigial locus in Drosophila following targeted transposition. Genetics. 1992;131:917–27. doi: 10.1093/genetics/131.4.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hiltbrunner A, Viczian A, Bury E, Tscheuschler A, Kircher S, et al. Nuclear accumulation of the phytochrome A photoreceptor requires FHY1. Curr Biol. 2005;15:2125–30. doi: 10.1016/j.cub.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 74.Hiom K, Melek M, Gellert M. DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell. 1998;94:463–70. doi: 10.1016/s0092-8674(00)81587-1. [DOI] [PubMed] [Google Scholar]
- 74b.Hirose F, Yamaguchi M, Kuroda K, Omori A, Hachiya T, et al. Isolation and characterization of cDNA for DREF, a promoter-activating factor for Drosophila DNA replication-related genes. J Biol Chem. 1996;271:3930–3937. doi: 10.1074/jbc.271.7.3930. [DOI] [PubMed] [Google Scholar]
- 75.Hoen DR, Park KC, Elrouby N, Yu Z, Mohabir N, et al. Transposon-mediated expansion and diversification of a family of ULP-like genes. Mol Biol Evol. 2006;23:1254–68. doi: 10.1093/molbev/msk015. [DOI] [PubMed] [Google Scholar]
- 76.Holligan D, Zhang X, Jiang N, Pritham EJ, Wessler SR. The transposable element landscape of the model legume Lotus japonicus. Genetics. 2006;174:2215–28. doi: 10.1534/genetics.106.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hsia AP, Schnable PS. DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. Genetics. 1996;142:603–18. doi: 10.1093/genetics/142.2.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hua-Van A, Daviere JM, Kaper F, Langin T, Daboussi MJ. Genome organization in Fusarium oxysporum: clusters of class II transposons. Curr Genet. 2000;37:339–47. doi: 10.1007/s002940050537. [DOI] [PubMed] [Google Scholar]
- 79.Hua-Van A, Langin T, Daboussi MJ. Aberrant transposition of a Tc1-mariner element, impala, in the fungus Fusarium oxysporum. Mol Genet Genomics. 2002;267:79–87. doi: 10.1007/s00438-002-0638-9. [DOI] [PubMed] [Google Scholar]
- 80.Hudson ME, Lisch DR, Quail PH. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 2003;34:453–71. doi: 10.1046/j.1365-313x.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- 81.Izsvak Z, Stuwe EE, Fiedler D, Katzer A, Jeggo PA, Ivics Z. Healing the wounds inflicted by Sleeping Beauty transposition by double-strand break repair in mammalian somatic cells. Mol Cell. 2004;13:279–90. doi: 10.1016/s1097-2765(03)00524-0. [DOI] [PubMed] [Google Scholar]
- 82.Jacob F. Evolution and tinkering. Science. 1977;196:1161–66. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 83.Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–73. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 84.Jiang N, Bao Z, Zhang X, McCouch SR, Eddy SR, Wessler SR. An active DNA transposon in rice. Nature. 2003;421:163–67. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- 85.Jiang N, Feschotte C, Zhang XY, Wessler SR. Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs) Curr Opin Plant Biol. 2004;7:115–19. doi: 10.1016/j.pbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 86.Jiang N, Wessler SR. Insertion preference of maize and rice miniature inverted repeat transposable elements as revealed by the analysis of nested elements. Plant Cell. 2001;13:2553–64. doi: 10.1105/tpc.010235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Jiao Y, Lau OS, Deng XW. Light-regulated transcriptional networks in higher plants. Nat Rev Genet. 2007;8:217–30. doi: 10.1038/nrg2049. [DOI] [PubMed] [Google Scholar]
- 88.Jones JM, Gellert M. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev. 2004;200:233–48. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 89.Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15:1292–97. doi: 10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Jurka J, Kapitonov VV. PIFs meet Tourists and Harbingers: a superfamily reunion. Proc Natl Acad Sci USA. 2001;98:12315–16. doi: 10.1073/pnas.231490598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kapitonov VV, Jurka J. Rolling-circle transposons in eukaryotes. Proc Natl Acad Sci USA. 2001;98:8714–19. doi: 10.1073/pnas.151269298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci USA. 2003;100:6569–74. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92b.Kapitonov VV, Jurka J. Harbinger transposons and an ancient HARBI1 gene derived from a transposase. DNA Cell Biol. 2004;23:311–24. doi: 10.1089/104454904323090949. [DOI] [PubMed] [Google Scholar]
- 93.Kapitonov VV, Jurka J. RAG1 Core and V(D)J recombination signal sequences were derived from transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kapitonov VV, Jurka J. Self-synthesizing DNA transposons in eukaryotes. Proc Natl Acad Sci USA. 2006;103:4540–45. doi: 10.1073/pnas.0600833103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kashkush K, Feldman M, Levy AA. Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet. 2003;33:102–6. doi: 10.1038/ng1063. [DOI] [PubMed] [Google Scholar]
- 96.Kawasaki S, Nitasaka E. Characterization of Tpn1 family in the Japanese morning glory: En/Spm-related transposable elements capturing host genes. Plant Cell Physiol. 2004;45:933–44. doi: 10.1093/pcp/pch109. [DOI] [PubMed] [Google Scholar]
- 97.Keeling PJ, Burger G, Durnford DG, Lang BF, Lee RW, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20:670–76. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 98.Keng VW, Yae K, Hayakawa T, Mizuno S, Uno Y, et al. Region-specific saturation germline mutagenesis in mice using the Sleeping Beauty transposon system. Nat Methods. 2005;2:763–6-9. doi: 10.1038/nmeth795. [DOI] [PubMed] [Google Scholar]
- 99.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, et al. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 100.Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- 101.Kidwell MG, Lisch D. Transposable elements as sources of variation in animals and plants. Proc Natl Acad Sci USA. 1997;94:7704–11. doi: 10.1073/pnas.94.15.7704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA, and genome evolution. Evol Int J Org Evol. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 103.Kikuchi K, Terauchi K, Wada M, Hirano HY. The plant MITE mPing is mobilized in anther culture. Nature. 2003;421:167–70. doi: 10.1038/nature01218. [DOI] [PubMed] [Google Scholar]
- 104.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–16. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 105.Koga A, Iida A, Hori H, Shimada A, Shima A. Vertebrate DNA transposon as a natural mutator: the medaka fish Tol2 element contributes to genetic variation without recognizable traces. Mol Biol Evol. 2006;23:1414–19. doi: 10.1093/molbev/msl003. [DOI] [PubMed] [Google Scholar]
- 106.Kunze R, Weil CF. The hAT and CACTA superfamilies of plant transposons. 2002 See Ref. 35, pp. 565–610. [Google Scholar]
- 107.Labrador M, Corces V. Interactions between transposable elements and the host genome. 2002. See Ref. 35, pp. 1008–23. [Google Scholar]
- 108.Lai J, Li Y, Messing J, Dooner HK. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc Natl Acad Sci USA. 2005;102:9068–73. doi: 10.1073/pnas.0502923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lal SK, Giroux MJ, Brendel V, Vallejos CE, Hannah LC. The maize genome contains a Helitron insertion. Plant Cell. 2003;15:381–91. doi: 10.1105/tpc.008375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lampe DJ, Walden KK, Robertson HM. Loss of transposase-DNA interaction may underlie the divergence of mariner family transposable elements and the ability of more than one mariner to occupy the same genome. Mol Biol Evol. 2001;18:954–61. doi: 10.1093/oxfordjournals.molbev.a003896. [DOI] [PubMed] [Google Scholar]
- 111.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 112.Le Rouzic A, Capy P. Population genetics models of competition between transposable element subfamilies. Genetics. 2006;174:785–93. doi: 10.1534/genetics.105.052241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee SH, Oshige M, Durant ST, Rasila KK, Williamson EA, et al. The SET domain protein Metnase mediates foreign DNA integration and links integration to nonhomologous end-joining repair. Proc Natl Acad Sci USA. 2005;102:18075–80. doi: 10.1073/pnas.0503676102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lim JK, Simmons MJ. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. BioEssays. 1994;16:269–75. doi: 10.1002/bies.950160410. [DOI] [PubMed] [Google Scholar]
- 115.Lin R, Wang H. Arabidopsis FHY3/FAR1 gene family and distinct roles of its members in light control of Arabidopsis development. Plant Physiol. 2004;136:4010–22. doi: 10.1104/pp.104.052191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lin X, Long L, Shan X, Zhang S, Shen S, Liu B. In planta mobilization of mPing and its putative autonomous element Pong in rice by hydrostatic pressurization. J Exp Bot. 2006;57:2313–23. doi: 10.1093/jxb/erj203. [DOI] [PubMed] [Google Scholar]
- 117.Lin Y, Waldman AS. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–74. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–19. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 119.Lippman Z, Gendrel AV, Black M, Vaughn MW, Dedhia N, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–76. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 120.Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–70. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- 121.Lisch D. Mutator transposons. Trends Plant Sci. 2002;7:498–504. doi: 10.1016/s1360-1385(02)02347-6. [DOI] [PubMed] [Google Scholar]
- 122.Lister C, Jackson D, Martin C. Transposon-induced inversion in Antirrhinum modifies nivea gene expression to give a novel flower color pattern under the control of Cycloidea (Radialis) Plant Cell. 1993;5:1541–53. doi: 10.1105/tpc.5.11.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu D, Bischerour J, Siddique A, Buisine N, Bigot Y, Chalmers R. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol Cell Biol. 2007;27:1125–32. doi: 10.1128/MCB.01899-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu G, Zhao S, Bailey JA, Sahinalp SC, Alkan C, et al. Analysis of primate genomic variation reveals a repeat-driven expansion of the human genome. Genome Res. 2003;13:358–68. doi: 10.1101/gr.923303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124b.Liu W, Seto J, Sibille E, Toth M. The RNA binding domain of Jerky consists of tandemly arranged helix-turn-helix/homeodomain-like motifs and binds specific sets of mRNAs. Mol Cell Biol. 2003;23:4083–4093. doi: 10.1128/MCB.23.12.4083-4093.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124c.Locovei AM, Spiga MG, Tanaka K, Murakami Y, D’Urso G. The CENP-B homolog, Abp1, interacts with the initiation protein Cdc23 (MCM10) and is required for efficient DNA replication in fission yeast. Cell Div. 2006;1:27. doi: 10.1186/1747-1028-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Long M, Betran E, Thornton K, Wang W. The origin of new genes: glimpses from the young and old. Nat Rev Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 125b.Lours C, Bardot O, Godt D, Laski FA, Couderc JL. The Drosophila melanogaster BTB proteins bric a brac bind DNA through a composite DNA binding domain containing a pipsqueak and an AT-Hook motif. Nucleic Acids Res. 2003;31:5389–5198. doi: 10.1093/nar/gkg724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Loot C, Santiago N, Sanz A, Casacuberta JM. The proteins encoded by the pogo-like Lemi1 element bind the TIRs and subterminal repeated motifs of the Arabidopsis Emigrant MITE: consequences for the transposition mechanism of MITEs. Nucleic Acids Res. 2006;34:5238–5246. doi: 10.1093/nar/gkl688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126b.Lv B, Shi T, Wang X, Song Q, Zhang Y, et al. Overexpression of the novel human gene, nuclear apoptosis-inducing factor 1, induces apoptosis. Int J Biochem Cell Biol. 2006;38:671–683. doi: 10.1016/j.biocel.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 126c.Macfarlan T, Kutney S, Altman B, Montross R, Yu J, Chakravarti D. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. J Biol Chem. 2005;280:7346–7358. doi: 10.1074/jbc.M411675200. [DOI] [PubMed] [Google Scholar]
- 127.MacRae AF, Clegg MT. Evolution of Ac and Ds1 elements in select grasses (Poaceae) Genetica. 1992;86:55–66. doi: 10.1007/BF00133711. [DOI] [PubMed] [Google Scholar]
- 128.Marracci S, Batistoni R, Pesole G, Citti L, Nardi I. Gypsy/Ty3-like elements in the genome of the terrestrial Salamander hydromantes (Amphibia, Urodela) J Mol Evol. 1996;43:584–593. doi: 10.1007/BF02202106. [DOI] [PubMed] [Google Scholar]
- 129.Masly JP, Jones CD, Noor MA, Locke J, Orr HA. Gene transposition as a cause of hybrid sterility in Drosophila. Science. 2006;313:1448–1450. doi: 10.1126/science.1128721. [DOI] [PubMed] [Google Scholar]
- 130.Mathews S. Phytochrome-mediated development in land plants: red light sensing evolves to meet the challenges of changing light environments. Mol Ecol. 2006;15:3483–3503. doi: 10.1111/j.1365-294X.2006.03051.x. [DOI] [PubMed] [Google Scholar]
- 131.McClintock B. Chromosome organization and genic expression. Cold Spring Harbor Symp Quant Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- 132.McClintock B. Controlling elements and the gene. Cold Spring Harbor Symp Quant Biol. 1956;21:197–216. doi: 10.1101/sqb.1956.021.01.017. [DOI] [PubMed] [Google Scholar]
- 133.McClintock B. The significances of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 134.Miller WJ, Hagemann S, Reiter E, Pinsker W. P-element homologous sequences are tandemly repeated in the genome of Drosophila guanche. Proc Natl Acad Sci USA. 1992;89:4018–4022. doi: 10.1073/pnas.89.9.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Miskey C, Izsvak Z, Kawakami K, Ivics Z. DNA transposons in vertebrate functional genomics. Cell Mol Life Sci. 2005;62:629–641. doi: 10.1007/s00018-004-4232-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miskey C, Papp B, Mates L, Sinzelle L, Keller H, et al. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol Cell Biol. 2007;27:4589–4600. doi: 10.1128/MCB.02027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Miura A, Yonebayashi S, Watanabe K, Toyama T, Shimada H, Kakutani T. Mobilization of transposons by a mutation abolishing full DNA methylation in Arabidopsis. Nature. 2001;411:212–214. doi: 10.1038/35075612. [DOI] [PubMed] [Google Scholar]
- 137b.Mojzita D, Hohmann S. Pdc2 coordinates expression of the THI regulon in the yeast Saccharomyces cerevisiae. Mol Genet Genomics. 2006;276:147–161. doi: 10.1007/s00438-006-0130-z. [DOI] [PubMed] [Google Scholar]
- 138.Moran JV, DeBerardinis RJ, Kazazian HH., Jr Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 139.Moreno MA, Chen J, Greenblatt I, Dellaporta SL. Reconstitutional mutagenesis of the maize P gene by short-range Ac transpositions. Genetics. 1992;131:939–956. doi: 10.1093/genetics/131.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Morgante M. Plant genome organisation and diversity: the year of the junk! Curr Opin Biotechnol. 2006;17:168–173. doi: 10.1016/j.copbio.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 141.Morgante M, Brunner S, Pea G, Fengler K, Zuccolo A, Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- 141b.Muehlbauer GJ, Bhau BS, Syed NH, Heinen S, Cho S, et al. A hAT superfamily transposase recruited by the cereal grass genome. Mol Genet Genomics. 2006;275:553–563. doi: 10.1007/s00438-006-0098-8. [DOI] [PubMed] [Google Scholar]
- 142.Naito K, Cho E, Yang G, Campbell MA, Yano K, et al. Dramatic amplification of a rice transposable element during recent domestication. Proc Natl Acad Sci USA. 2006;103:17620–17625. doi: 10.1073/pnas.0605421103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142b.Nakagawa H, Lee JK, Hurwitz J, Allshire RC, Nakayama J, et al. Fission yeast CENP-B homologs nucleate centromeric heterochromatin by promoting heterochromatin-specific histone tail modifications. Genes Dev. 2002;16:1766–1778. doi: 10.1101/gad.997702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Navarro A, Barton NH. Chromosomal speciation and molecular divergence---accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- 145.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–872. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 146.Noor MA, Chang AS. Evolutionary genetics: jumping into a new species. Curr Biol. 2006;16:R890–892. doi: 10.1016/j.cub.2006.09.022. [DOI] [PubMed] [Google Scholar]
- 147.Noor MA, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci USA. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.O’Brochta DA, Atkinson PW. Transformation systems in insects. Methods Mol Biol. 2004;260:227–254. doi: 10.1385/1-59259-755-6:227. [DOI] [PubMed] [Google Scholar]
- 148b.Ohzeki J, Nakano M, Okada T, Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Oosumi T, Garlick B, Belknap WR. Identification of putative nonautonomous transposable elements associated with several transposon families in Caenorhabditis elegans. J Mol Evol. 1996;43:11–18. doi: 10.1007/BF02352294. [DOI] [PubMed] [Google Scholar]
- 150.Organ CL, Shedlock AM, Meade A, Pagel M, Edwards SV. Origin of avian genome size and structure in nonavian dinosaurs. Nature. 2007;446:180–184. doi: 10.1038/nature05621. [DOI] [PubMed] [Google Scholar]
- 151.Pace JK, 2nd, Feschotte C. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 2007;17:422–432. doi: 10.1101/gr.5826307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Piegu B, Guyot R, Picault N, Roulin A, Saniyal A, et al. Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16:1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Piriyapongsa J, Jordan IK. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS ONE. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Plasterk RH. The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J. 1991;10:1919–1925. doi: 10.1002/j.1460-2075.1991.tb07718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Plasterk RHA, Izsvák Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- 156.Poulter RT, Goodwin TJ, Butler MI. Vertebrate helentrons and other novel Helitrons. Gene. 2003;313:201–312. doi: 10.1016/s0378-1119(03)00679-6. [DOI] [PubMed] [Google Scholar]
- 157.Preston CR, Sved JA, Engels WR. Flanking duplications and deletions associated with P-induced male recombination in Drosophila. Genetics. 1996;144:1623–1638. doi: 10.1093/genetics/144.4.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pritham EJ, Feschotte C. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci USA. 2007;104:1895–1900. doi: 10.1073/pnas.0609601104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pritham EJ, Feschotte C, Wessler SR. Unexpected diversity and differential success of DNA transposons in four species of entamoeba protozoans. Mol Biol Evol. 2005;22:1751–1763. doi: 10.1093/molbev/msi169. [DOI] [PubMed] [Google Scholar]
- 160.Pritham EJ, Putliwala T, Feschotte C. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene. 2007;390:3–17. doi: 10.1016/j.gene.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 160b.Quesneville H, Nouaud D, Anxolabehere D. Recurrent recruitment of the THAP DNA-binding domain and molecular domestication of the P-transposable element. Mol Biol Evol. 2005;22:741–746. doi: 10.1093/molbev/msi064. [DOI] [PubMed] [Google Scholar]
- 161.Ray DA, Pagan HJ, Thompson ML, Stevens RD. Bats with hATs: evidence for recent DNA transposon activity in genus Myotis. Mol Biol Evol. 2007;24:632–639. doi: 10.1093/molbev/msl192. [DOI] [PubMed] [Google Scholar]
- 161b.Reddy KC, Villeneuve AM. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell. 2004;118:439–452. doi: 10.1016/j.cell.2004.07.026. [DOI] [PubMed] [Google Scholar]
- 162.Reddy YV, Perkins EJ, Ramsden DA. Genomic instability due to V(D)J recombination-associated transposition. Genes Dev. 2006;20:1575–1582. doi: 10.1101/gad.1432706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162b.Reiss D, Nouaud D, Ronsseray S, Anxolabehere D. Domesticated P elements in the Drosophila montium species subgroup have a new function related to a DNA binding property. J Mol Evol. 2005;61:470–480. doi: 10.1007/s00239-004-0324-0. [DOI] [PubMed] [Google Scholar]
- 163.Rizzon C, Martin E, Marais G, Duret L, Segalat L, Biemont C. Patterns of selection against transposons inferred from the distribution of Tc1, Tc3 and Tc5 insertions in the mut-7 line of the nematode Caenorhabditis elegans. Genetics. 2003;165:1127–1135. doi: 10.1093/genetics/165.3.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Robertson HM. Evolution of DNA transposons. 2002. See Ref. 35, pp. 1093–110. [Google Scholar]
- 165.Robertson HM, Zumpano KL. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene. 1997;205:203–217. doi: 10.1016/s0378-1119(97)00472-1. [DOI] [PubMed] [Google Scholar]
- 166.Ros F, Kunze R. Regulation of activator/dissociation transposition by replication and DNA methylation. Genetics. 2001;157:1723–1733. doi: 10.1093/genetics/157.4.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Rozmahel R, Heng HH, Duncan AM, Shi XM, Rommens JM, Tsui LC. Amplification of CFTR exon 9 sequences to multiple locations in the human genome. Genomics. 1997;45:554–561. doi: 10.1006/geno.1997.4968. [DOI] [PubMed] [Google Scholar]
- 168.Rubin E, Levy AA. Abortive gap repair: underlying mechanism for Ds element formation. Mol Cell Biol. 1997;17:6294–6302. doi: 10.1128/mcb.17.11.6294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.SanMiguel P, Gaut BS, Tikhonov A, Nakajima Y, Bennetzen JL. The paleontology of intergene retrotransposons of maize. Nat Genet. 1998;20:43–45. doi: 10.1038/1695. [DOI] [PubMed] [Google Scholar]
- 170.Sarkar A, Sim C, Hong YS, Hogan JR, Fraser MJ, et al. Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol Genet Genomics. 2003;270:173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- 171.Sen SK, Han K, Wang J, Lee J, Wang H, et al. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Shan X, Liu Z, Dong Z, Wang Y, Chen Y, et al. Mobilization of the active MITE transposons mPing and Pong in rice by introgression from wild rice (Zizania latifolia Griseb) Mol Biol Evol. 2005;22:976–990. doi: 10.1093/molbev/msi082. [DOI] [PubMed] [Google Scholar]
- 172b.Shim K, Blake KJ, Jack J, Krasnow MA. The Drosophila ribbon gene encodes a nuclear BTB domain protein that promotes epithelial migration and morphogenesis. Development. 2001;128:4923–4933. doi: 10.1242/dev.128.23.4923. [DOI] [PubMed] [Google Scholar]
- 172c.Siegmund T, Lehmann M. The Drosophila Pipsqueak protein defines a new family of helix-turn-helix DNA-binding proteins. Dev Genes Evol. 2002;212:152–157. doi: 10.1007/s00427-002-0219-2. [DOI] [PubMed] [Google Scholar]
- 173.Sijen T, Plasterk RH. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature. 2003;426:310–314. doi: 10.1038/nature02107. [DOI] [PubMed] [Google Scholar]
- 174.Silva JC, Bastida F, Bidwell SL, Johnson PJ, Carlton JM. A potentially functional Mariner transposable element in the protist Trichomonas vaginalis. Mol Biol Evol. 2005;22:126–34. doi: 10.1093/molbev/msh260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Silva JC, Kidwell MG. Horizontal transfer and selection in the evolution of P elements. Mol Biol Evol. 2000;17:1542–57. doi: 10.1093/oxfordjournals.molbev.a026253. [DOI] [PubMed] [Google Scholar]
- 176.Singer T, Yordan C, Martienssen RA. Robertson’s Mutator transposons in A. thaliana are regulated by the chromatin-remodeling gene Decrease in DNA Methylation (DDM1) Genes Dev. 2001;15:591–602. doi: 10.1101/gad.193701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Slotkin RK, Freeling M, Lisch D. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat Genet. 2005;37:641–644. doi: 10.1038/ng1576. [DOI] [PubMed] [Google Scholar]
- 178.Slotkin RK, Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- 178b.Smit AF. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr Opin Genet Devel. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- 179.Spradling AC, Stern DM, Kiss I, Roote J, Laverty T, Rubin GM. Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc Natl Acad Sci USA. 1995;92:10824–10830. doi: 10.1073/pnas.92.24.10824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260:170–173. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 181.Tepperman JM, Zhu T, Chang HS, Wang X, Quail PH. Multiple transcription-factor genes are early targets of phytochrome A signaling. Proc Natl Acad Sci USA. 2001;98:9437–9442. doi: 10.1073/pnas.161300998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181b.Tipney HJ, Hinsley TA, Brass A, Metcalfe K, Donnai D, Tassabehji M. Isolation and characterisation of GTF2IRD2, a novel fusion gene and member of the TFII-I family of transcription factors, deleted in Williams-Beuren syndrome. Eur J Hum Genet. 2004;12:551–560. doi: 10.1038/sj.ejhg.5201174. [DOI] [PubMed] [Google Scholar]
- 182.Tower J, Karpen GH, Craig N, Spradling AC. Preferential transposition of Drosophila P elements to nearby chromosomal sites. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tsubota SI, Huong DV. Capture of flanking DNA by a P element in Drosophila melanogaster: creation of a transposable element. Proc Natl Acad Sci USA. 1991;88:693–697. doi: 10.1073/pnas.88.3.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Valenzuela L, Kamakaka RT. Chromatin insulators. Annu Rev Genet. 2006;40:107–138. doi: 10.1146/annurev.genet.39.073003.113546. [DOI] [PubMed] [Google Scholar]
- 185.Vitte C, Bennetzen JL. Analysis of retrotransposon structural diversity uncovers properties and propensities in angiosperm genome evolution. Proc Natl Acad Sci USA. 2006;103:17638–17643. doi: 10.1073/pnas.0605618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Volff JN. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. BioEssays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- 187.Walbot V. Saturation mutagenesis using maize transposons. Curr Opin Plant Biol. 2000;3:103–107. doi: 10.1016/s1369-5266(99)00051-5. [DOI] [PubMed] [Google Scholar]
- 188.Walisko O, Izsvak Z, Szabo K, Kaufman CD, Herold S, Ivics Z. Sleeping Beauty transposase modulates cell-cycle progression through interaction with Miz-1. Proc Natl Acad Sci USA. 2006;103:4062–4067. doi: 10.1073/pnas.0507683103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Walker EL, Robbins TP, Bureau TE, Kermicle J, Dellaporta SL. Transposon-mediated chromosomal rearrangements and gene duplications in the formation of the maize R-r complex. EMBO J. 1995;14:2350–2363. doi: 10.1002/j.1460-2075.1995.tb07230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang H, Deng XW. Dissecting the phytochrome A-dependent signaling network in higher plants. Trends Plant Sci. 2003;8:172–178. doi: 10.1016/S1360-1385(03)00049-9. [DOI] [PubMed] [Google Scholar]
- 191.Warbrick E, Heatherington W, Lane DP, Glover DM. PCNA binding proteins in Drosophila melanogster: the analysis of a conserved PCNA binding domain. Nucleic Acids Res. 1998;26:3925–3932. doi: 10.1093/nar/26.17.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 193.Wessler S, Tarpley A, Purugganan M, Spell M, Okagaki R. Filler DNA is associated with spontaneous deletions in maize. Proc Natl Acad Sci USA. 1990;87:8731–8735. doi: 10.1073/pnas.87.22.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wessler SR. Phenotypic diversity mediated by the maize transposable elements Ac and Spm. Science. 1988;242:399–405. doi: 10.1126/science.2845581. [DOI] [PubMed] [Google Scholar]
- 195.Wessler SR, Baran G, Varagona M, Dellaporta SL. Excision of Ds produces waxy proteins with a range of enzymatic activities. EMBO J. 1986;5:2427–2432. doi: 10.1002/j.1460-2075.1986.tb04517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 197.Xing J, Wang H, Belancio VP, Cordaux R, Deininger PL, Batzer MA. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc Natl Acad Sci USA. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197b.Yamashita D, Sano Y, Adachi Y, Okamoto Y, Osada H, et al. hDREF regulates cell proliferation and expression of ribosomal protein genes. Mol Cell Biol. 2007;27:2003–2013. doi: 10.1128/MCB.01462-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Yamashita S, Takano-Shimizu T, Kitamura K, Mikami T, Kishima Y. Resistance to gap repair of the transposon Tam3 in Antirrhinum majus: a role of the end regions. Genetics. 1999;153:1899–908. doi: 10.1093/genetics/153.4.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198b.Yang G, Weil CF, Wessler SR. A rice Tc1/mariner-like element transposes in yeast. Plant Cell. 2006;18:2469–2478. doi: 10.1105/tpc.106.045906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198c.Yang G, Zhang F, Hancock N, Wessler SR. Transposition of the rice miniature inverted repeat transposable element mPing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007 doi: 10.1073/pnas.0702080104. (in press, published online 19 June 2007, 10.1073/pnas.0702080104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 200.Zabala G, Vodkin LO. The wp mutation of Glycine max carries a gene-fragment-rich transposon of the CACTA superfamily. Plant Cell. 2005;17:2619–2632. doi: 10.1105/tpc.105.033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zayed H, Izsvak Z, Khare D, Heinemann U, Ivics Z. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 2003;31:2313–2322. doi: 10.1093/nar/gkg341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Zdobnov EM, Campillos M, Harrington ED, Torrents D, Bork P. Protein coding potential of retroviruses and other transposable elements in vertebrate genomes. Nucleic Acids Res. 2005;33:946–954. doi: 10.1093/nar/gki236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Zhang J, Peterson T. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics. 2004;167:1929–1937. doi: 10.1534/genetics.103.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Zhang J, Zhang F, Peterson T. Transposition of reversed Ac element ends generates novel chimeric genes in maize. PLoS Genet. 2006;2:e164. doi: 10.1371/journal.pgen.0020164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zhang Q, Arbuckle J, Wessler SR. Recent, extensive and preferential insertion of members of the miniature inverted-repeat transposable element family Heartbreaker (Hbr) into genic regions of maize. Proc Natl Acad Sci USA. 2000;97:1160–1165. doi: 10.1073/pnas.97.3.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Zhang X, Feschotte C, Zhang Q, Jiang N, Eggleston WB, Wessler SR. P Instability Factor: an active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases. Proc Natl Acad Sci USA. 2001;98:12572–12577. doi: 10.1073/pnas.211442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Zhang X, Jiang N, Feschotte C, Wessler SR. Distribution and evolution of PIF- and Pong-like transposons and their relationships with Tourist-like MITEs. Genetics. 2004;166:971–986. doi: 10.1534/genetics.166.2.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207b.Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 208.Zhou L, Mitra R, Atkinson PW, Hickman AB, Dyda F, Craig NL. Transposition of hAT elements links transposable elements and V(D)J recombination. Nature. 2004;432:995–1001. doi: 10.1038/nature03157. [DOI] [PubMed] [Google Scholar]