Structural insights into the catalysis and regulation of E3 ubiquitin ligases (original) (raw)
. Author manuscript; available in PMC: 2018 Nov 1.
Published in final edited form as: Nat Rev Mol Cell Biol. 2016 Aug 3;17(10):626–642. doi: 10.1038/nrm.2016.91
Abstract
Covalent attachment of one or more ubiquitin molecules to protein substrates governs numerous eukaryotic cellular processes including apoptosis, cell division and immune response. Ubiquitylated proteins can be targeted for degradation but ubiquitylation also mediates processes such as protein-protein interactions and cell signalling, depending on the type of ubiquitin conjugation. Ubiquitin ligases (E3s) catalyze the final step of ubiquitin conjugation by transferring ubiquitin from ubiquitin-conjugating enzymes (E2s) to substrates. In humans, over 600 E3s contribute to determining the fates of thousands of substrates; hence E3s are tightly regulated to ensure accurate substrate ubiquitylation. Recent findings illustrate how E3s are self-regulated and how they coordinate with E2s and substrates to meticulously conjugate ubiquitin.
E3s are a large family of enzymes that catalyze the covalent attachment of a small protein modifier, ubiquitin, to a plethora of substrates in eukaryotic cells. Ubiquitylation (also known as ubiquitination) plays a fundamental role in nearly all aspects of eukaryotic cellular processes. By marking substrates with ubiquitin, E3s bestow substrates with new protein-protein interaction platforms that alter substrate activity, localization and/or interactions to elicit distinct biological signals.
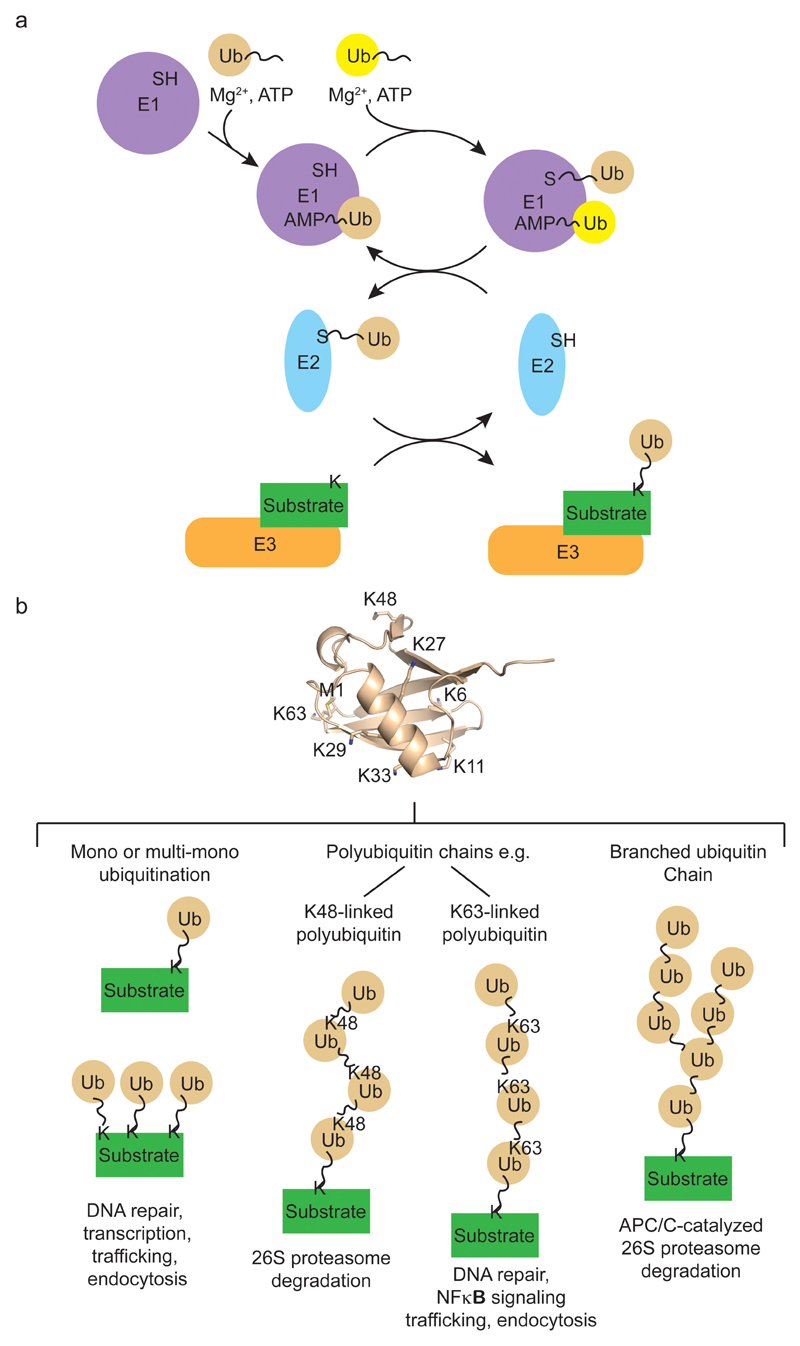
Ubiquitylation is achieved by the sequential actions of ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2) and a ubiquitin ligase (E3)1–3 (Figure 1a). E1 uses Mg2+-ATP to form a covalent thioester between its catalytic cysteine and the di-glycine motif at the C-terminus of ubiquitin. E1 then transfers ubiquitin via the di-glycine motif to E2’s catalytic cysteine to form an E2~ubiquitin thioester complex (~ indicates a thioester bond). E3s then bind E2~ubiquitin and substrate to facilitate formation of an isopeptide bond between the C-terminal carboxyl of ubiquitin and the ε-amino group of a substrate lysine sidechain or free N-terminal amino group. Successive rounds of E3-catalyzed reactions can produce substrates with polyubiquitin chains linked via one of ubiquitin’s seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48 and Lys63) or ubiquitin’s N-terminal methionine4 (Figure 1b). Recognition of different ubiquitylation patterns by effectors harboring a ubiquitin-binding domain (UBD) elicits distinct downstream signals. For example, Lys48-linked polyubiquitin targets substrate to the 26S proteasome for degradation5, 6; Lys63-linked polyubiquitin directs substrate to the endocytic pathway7 and regulates kinase activation in the NFκB pathway8, and monoubiquitin plays a role in DNA repair9, 10 and chromatin remodeling11.
Figure 1. Ubiquitin conjugation system.
a | Schematic diagram of ubiquitin conjugation cascade. E1 catalyzes two rounds of adenylation of ubiquitin (labelled Ub in figure) in the presence of Mg2+ and ATP to bind two molecules of ubiquitin: one forms a thioester with E1’s catalytic cysteine and the other is bound non-covalently via its AMP-modified C-terminus to E1’s adenylation domain. E1 then binds E2 and transfers ubiquitin from its catalytic cysteine to E2’s catalytic cysteine to form E2~ubiquitin. E2~ubiquitin dissociates from E1 and E1 re-enters its catalytic cycle. E3 recruits E2~ubiquitin and substrate to catalyze ubiquitin transfer to substrate. Upon transfer, E2 disengages from E3 and E3 binds another E2~ubiquitin to catalyze the next round of substrate ubiquitylation. b | Substrate ubiquitylation outcomes. Ubiquitylated substrates interact with binding partners that harbour ubiquitin-binding domains, thus altering protein-protein interactions and cellular functions of substrates. A single molecule of ubiquitin can be attached to substrate at one (monoubiquitylation) or multiple (multi-monoubiquitylation) lysine sites. Additionally, the seven surface lysine residues (labelled as K and number in the protein structure) and N-terminal methionine (labelled as M1) of ubiquitin can serve as acceptor sites leading to the formation of polyubiquitin chains. These chains can be homogeneous like K48-linked and K63-linked polyubiquitin, where only one site on ubiquitin is modified, or branched, where two molecules of ubiquitin are attached to different acceptor sites on a single ubiquitin molecule. Different ubiquitin linkages vary in structure and therefore recruit discrete interacting partners to produce unique downstream signals147, 154. APC/C synthesizes branched ubiquitin chains on its substrate, resulting in enhanced substrate recognition and degradation by the 26S proteasome155.
In the human genome, there are two E1s, approximately 38 E2s and over 600 E3s12–14. Within this cascade, E3s play a pivotal role in selecting substrates and E2~ubiquitin to coordinate conjugation of specific ubiquitin linkages onto substrate. In this review, we focus on the three classes of E3s identified to date: RING (Really Interesting New Gene), HECT (Homologous to E6AP C-Terminus) and RBR (RING-between-RING). We discuss recent structural insights into the mechanisms by which E3s recruit E2~ubiquitin, how they prime and position ubiquitin and the substrate lysine for transfer, and how they extend ubiquitin chains to achieve polyubiquitylation. Moreover, we discuss structural details of emerging themes in E3 regulation, including autoinhibition.
General E3 features
All E3s harbor an E2~ubiquitin binding domain and are classified on the basis of the structure of this domain and their ubiquitin transfer mechanism. RING E3s catalyze the direct transfer of ubiquitin from E2~ubiquitin to substrate. In contrast, HECT and RBR E3s contain a catalytic cysteine that first receives ubiquitin from E2~ubiquitin to form an E3~ubiquitin thioester intermediate and subsequently transfers this ubiquitin to substrate (Figure 2). How E3s target substrates varies. Some contain one or several protein-protein interaction domains to which substrate can bind directly or indirectly via an interacting partner, or both. For example, the E3 ligase CBL downregulates EGFR signaling by ubiquitylating and targeting the receptor for lysosomal degradation; CBL harbors a substrate-binding domain that recognizes phosphorylated EGFR as well as a domain that binds the EGFR-interacting protein GRB2, which facilitates indirect ubiquitylation15, 16. Some E3s contain domains that bind non-protein molecules to mediate substrate binding and ubiquitylation. In the ER-associated degradation system, the E3 Cullin-RING ligase (CRL) CRLFBS1 uses a sugar-binding domain to recruit and ubiquitylate N-linked glycoproteins17. Other E3s lack specificity in their substrate selection but instead contain a domain or binding motif that recruits them to sites within the cell where they then ubiquitylate any accessible target. For example, during the repair of DNA lesions induced by ultraviolet light, the E3 CRLDDB binds directly to pyrimidine dimer photolesions and ubiquitylates DNA-bound XPC and DDB2 to initiate the nucleotide excision repair pathway18–20. For many E3s, the substrate targeting mechanism is unknown. Here we focus on how E3s catalyze ubiquitin transfer.
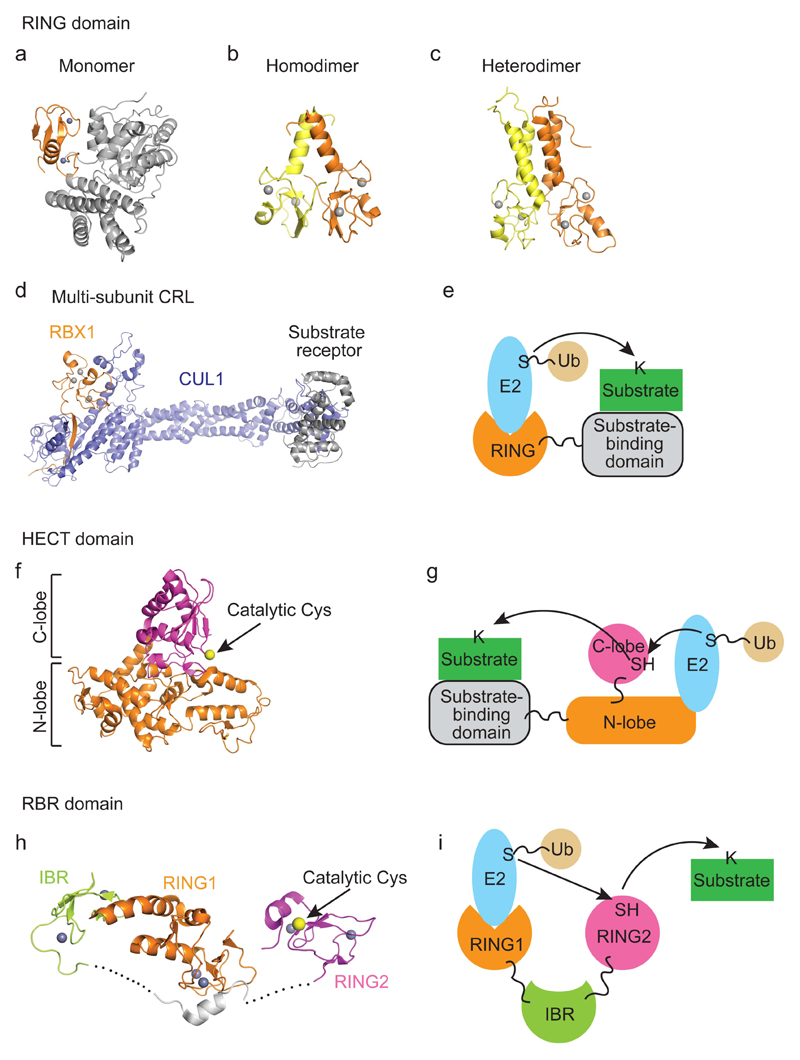
Figure 2. E3 catalytic mechanisms.
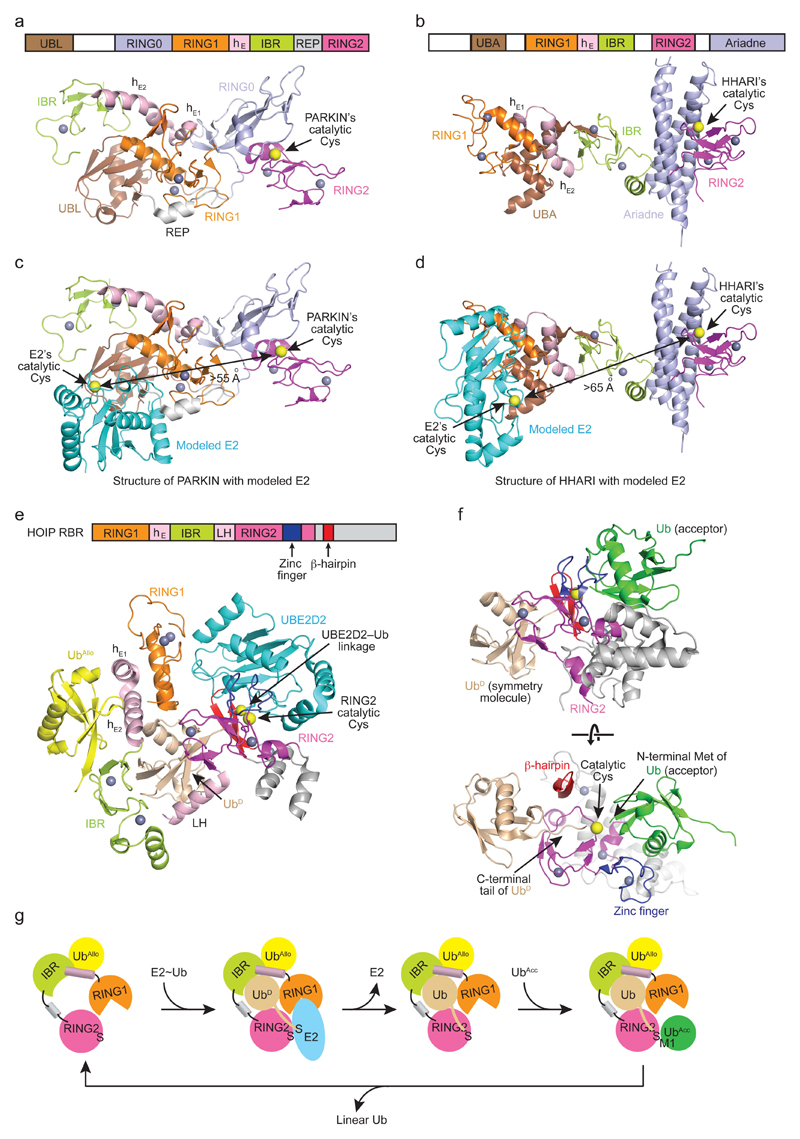
a–e | RING E3-catalyzed ubiquitin transfer. (a) Crystal structure of a monomeric RING E3 from c-CBL (PDB: 1FBV). The RING domain is in orange and the substrate-binding domain is in grey. (b) Crystal structure of a dimeric RING E3 from cIAP2 (PDB: 3EB5). One RING domain is colored orange and the other yellow. (c) NMR structure of BRCA1-BARD1 RING domain heterodimer (PDB: 1JM7). BRCA1 is colored in orange and BARD1 is in yellow. (d) Crystal structure of a multi-subunit RING E3 CRL. The RING domain is in orange, CUL1 is in blue, and the substrate receptor is in grey. (e) A schematic drawing of RING E3-mediated catalysis. The RING domain binds E2~ubiquitin and a substrate-binding domain recruits substrate. Ubiquitin is transferred directly from E2 to a substrate lysine. f,g | HECT E3-catalyzed ubiquitin transfer. (f) Crystal structure of a HECT domain from NEDD4L (PDB: 3JVZ). C- and N-lobes are indicated. C-lobe’s catalytic cysteine is shown as a yellow sphere. (g) A schematic drawing of HECT E3 catalysis. N-lobe of the HECT domain binds E2~ubiquitin and a substrate-binding domain recruits substrate. Ubiquitin is transferred from E2 to the catalytic cysteine of the C-lobe of the HECT domain and subsequently to a substrate lysine. h,i | RBR E3-catalyzed ubiquitin transfer. (h) Crystal structure of an RBR domain from autoinhibited PARKIN (PDB: 5C23). RING1, IBR and RING2 are indicated. RING2’s catalytic cysteine is shown as a yellow sphere. The grey helix is a fragment of the repressor element. Dashed lines indicate disordered regions. This autoinhibited arrangement is specific to PARKIN and may not be representative of all E3s. (i) A schematic drawing of RBR E3 catalysis. RING1 domain binds E2~ubiquitin and ubiquitin is transferred from E2 to RING2’s catalytic cysteine and then to a substrate lysine. How RBR E3s bind substrates remain elusive. PARKIN is recruited to the damaged mitochondria by PINK1 and ubiquitylates proteins bound to the outer mitochondria membrane143, 144. HOIP contains a zinc finger motif in the RING2 domain that binds ubiquitin as described later in Figure 4f. Zinc atoms, which function to stabilize domain folding, are shown as grey spheres.
RING E3s
Bioinformatic analyses predict there are around 600 RING E3s in humans, making it the largest of the three E3 classes14 (Supplementary Table 1). RING E3s are characterized by the presence of a RING domain (Figure 2a–e), which is the minimal component required to recruit E2~ubiquitin and stimulate ubiquitin transfer21. Structurally, RING domains are characterized by two zinc ions coordinated by cysteines and histidines arranged in a cross-braced configuration (Figure 2a–d). Some RING domains are active as monomers like in CBL22 (Figure 2a), CNOT423 and RNF3824, whereas others are active as oligomers. For example, cIAP225 (Figure 2b), TRAF626 and RNF427 are only active when homodimerized via their RING domains. Some RING domains, like the ones in BARD1, BMI1 and MDMX lack ligase activity but become functional upon heterodimerization with a RING domain-containing partner (BRCA1, RNF2 and MDM2, respectively28–32; Figure 2c). The TRIM family of RING E3s use their RING domains and coiled coil regions to assemble homodimers, heterodimers and oligomers33. Some RING E3s like CRLs and anaphase promoting complex/cyclosome (APC/C) are large multi-subunit complexes. CRLs comprise a RING E3 (RBX1 or RBX2), a cullin protein (CUL1, CUL2, CUL3, CUL4A/4B, CUL5 or CUL7) and a protein substrate-receptor34, 35. The cullin protein acts as scaffold that joins the RING domain and substrate-receptor (Figure 2d). There are approximately 300 different substrate-receptors in humans, making CRLs versatile at ubiquitylating a number of substrates. APC/C is a 1.2 MDa complex consisting of a 14-subunit core complex including the RING E3 APC11, the cullin-like subunit APC2 and a coactivator protein (CDC20 or CDH1)36. U-box proteins are also classified as RING E3 ligases: they use the same ubiquitin transfer mechanism and resemble the RING domain in structure but lack zinc ions37. There are 8 U-box E3s in humans14 and like the RING E3s, some function as monomers like UBE4B38 while others like PRP1939 and CHIP40 require dimerization for activity (Supplementary Table 1).
HECT E3s
There are 28 HECT E3s in humans41. In general, they comprise an N-terminal substrate-binding domain and a C-terminal HECT domain of approximately 350 amino acids containing the catalytic components for ubiquitin transfer. HECT E3s are subdivided into three groups based on their N-terminal protein-protein interaction domains: the NEDD4 family with WW domains that bind PY motifs in substrates, the HERC family with regulator of chromosome condensation 1-like domains and HECTs with other protein-protein interaction domains41 (Supplementary Table 1). HECT domains contain two lobes connected by a flexible hinge loop (Figure 2f). The N-terminal lobe binds E2~ubiquitin and the C-terminal lobe contains the catalytic cysteine42 (Figure 2g). The flexible hinge enables the lobes to rotate and support ubiquitin transfer43.
RBR E3s
In humans, there are 14 RBR E3s including PARKIN, HHARI, and HOIP, all of which possess a RING1-IBR-RING2 motif (Figure 2h and Supplementary Table 1) as well as other domains14, 44, 45. Early studies showed that the RING1-IBR-RING2 motif was essential for E2 binding and ligase activity but the mechanism remained elusive and was thought to resemble that of RING E3s46–49. However, in 2011, a conserved catalytic cysteine residue in the RING2 domain of HHARI and PARKIN was identified and found to form a thioester intermediate with ubiquitin’s C-terminus in a HECT E3-like manner50. This redefined the RBR E3 mechanism as a hybrid of RING and HECT E3 ligases, where the RING1 domain recruits E2~ubiquitin and transfers ubiquitin onto the catalytic cysteine of the RING2 domain prior to its conjugation to a substrate (Figure 2i).
Structural basis of RING E3 activity
E2s contain a highly conserved core domain of about 150 residues including a catalytic cysteine residue that forms a covalent thioester bond with the C-terminus of ubiquitin to generate E2~ubiquitin13. In the absence of E3, ubiquitin transfer from E2~ubiquitin to an amine can occur but the process is inefficient. Addition of a RING E3 massively stimulates this process24, 50–53 but until recently the mechanism underlying this stimulation was not well understood. In 2000, the first crystal structure of a RING E3-E2 complex, CBL-UBE2L3, showed that CBL’s RING domain binds the L1 and L2 loops and N-terminal α1 of UBE2L3. In this E3-E2 conformation, E2’s active site is distal from the RING domain, precluding direct participation of the RING domain in catalysis22. Although CBL is not reactive with UBE2L3, these E3-E2 interactions are conserved in subsequently determined structures of reactive RING E3-E2 pairs. Ensuing studies suggested that RING E3 binding might induce allosteric changes in E2’s active site51, 54, but details remained elusive. In 2012, the crystal structures of RNF455 and BIRC756 bound to E2–ubiquitin (en dash denotes a covalent interaction used to mimic E2~ubiquitin) and NMR analyses of E4B in complex with the E2 UBE2D3–ubiquitin57 revealed the catalytic mechanism of RING E3-mediated ubiquitin transfer (Figure 3).
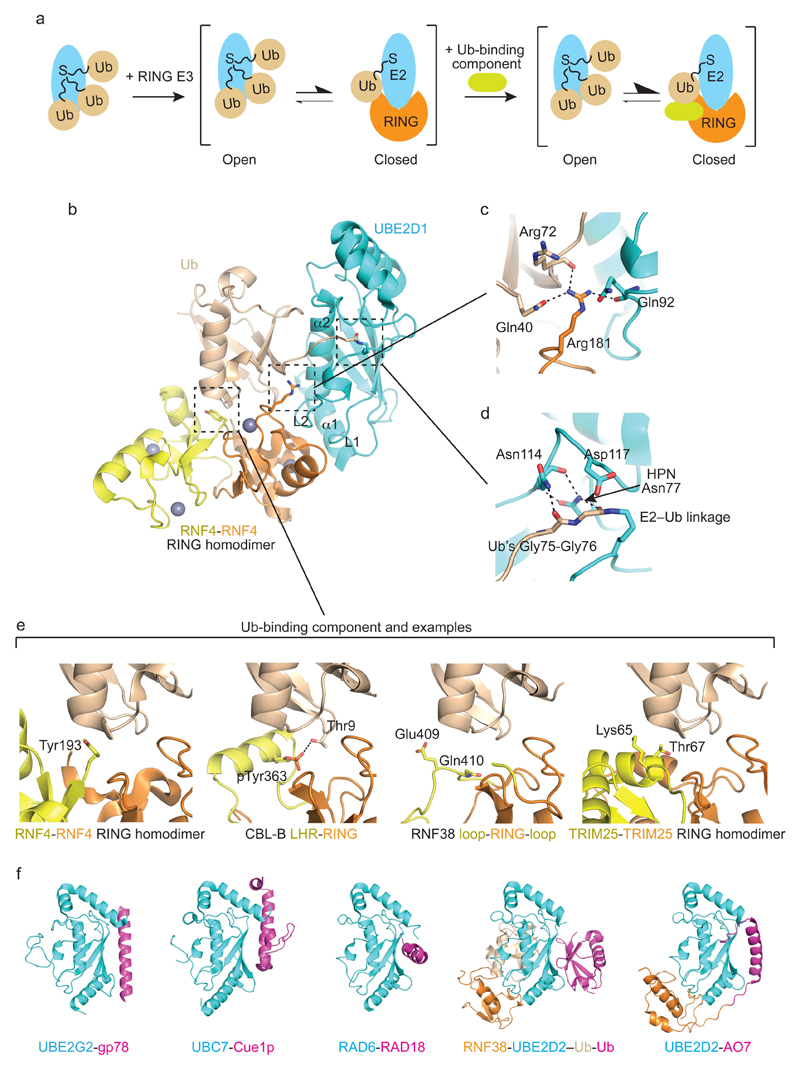
Figure 3. Mechanism of priming ubiquitin for transfer by RING E3s.
a | RING E3 binding induces the formation of a closed E2~ubiquitin conformation that is primed for catalysis. Left panel, in the absence of RING E3, ubiquitin samples various conformations relative to E2. Middle panel, RING E3 binding shifts the equilibrium toward the closed and primed E2~ubiquitin conformation. Right panel, the presence of an additional ubiquitin-binding component outside the RING domain further stabilizes the primed E2~ubiquitin conformation. Examples of the additional component are shown in e. E2 is colored cyan, ubiquitin wheat, the RING domain orange and the additional component yellow. b | Structure of dimeric RNF4-UBE2D1–ubiquitin complex (PDB 4AP4). Loops L1 and L2 and α1 from UBE2D1, which bind the RING domain, and α2 of UBE2D1, which binds ubiquitin, are indicated. One RNF4 subunit is colored orange, the second yellow, UBE2D1 cyan and ubiquitin wheat. Zinc atoms are shown as grey spheres. c | Close-up view of the linchpin Arg in b with colors as in b. Arg181 forms hydrogen bonds with the sidechain of Gln40 and carbonyl oxygen of Arg72 of ubiquitin and carbonyl oxygen of Gln92 of UBE2D1. This interaction stabilizes ubiquitin in the closed conformation. d | Close-up view of UBE2D1–ubiquitin active site in b. The catalytic cysteine of UBE2D1 was mutated to a lysine to generate a stable amide linkage with the C-terminus of ubiquitin to enable crystallization of the complex. e | Additional ubiquitin-binding components that stabilize E2~ubiquitin in the closed conformation. Left panel, Tyr193 from the C-terminus of the second subunit of RNF4 packs against ubiquitin. Middle left panel, phospho-Tyr363 and the linker helix region (LHR) of CBL-B contact ubiquitin. Middle right panel, loops adjacent to the RING domain in RNF38 bind ubiquitin. Right panel, residues from the C-terminal helix of the second subunit of TRIM25 pack against ubiquitin. RING domains are colored orange, non-RING components yellow and ubiquitin wheat. f | Additional E2-E3 interactions involving the backside of E2. Left to right: UBE2G2-gp78 complex (PDB: 3H8K), UBC7-Cue1p complex (PDB: 4JQU), RAD6-RAD18 complex (PDB: 2YBF), RNF38-UBE2D2–Ub-Ub complex (PDB: 4V3L) and UBE2D2-AO7 complex (PDB: 5D1K). E2 is colored in cyan, E2 backside-binding component is in pink, ubiquitin is in wheat and RING domain is in orange.
Priming ubiquitin for transfer
NMR and SAXS analyses showed that in the absence of a RING E3, UBE2D3~ubiquitin is highly dynamic and prefers to adopt open conformations where there is little or no interaction between the E2 and ubiquitin; in contrast, analyses of the E2 UBE2N~ubiquitin conjugate showed it more frequently adopts closed conformations but is still dynamic58. For the UBE2D family of E2s, addition of a RING E3 shifts the equilibrium toward closed conformations in which ubiquitin is proximal to the RING domain and more reactive toward transfer56, 57, 59, 60 (Figure 3a). When ubiquitin is optimally positioned or “primed” for transfer, the RING domain binds both the E2 and the Ile36 surface of ubiquitin, and the Ile44 surface of ubiquitin contacts α2 of the E2 (Figure 3b). The C-terminal tail of ubiquitin (residues 71-76) is braced in a conformation favorable for catalysis in which the incoming lysine can be deprotonated and the thioester oriented for attack. The acquisition of a primed ubiquitin conformation in the presence of a RING E3 appears to be conserved across different E2s and RING E3s and has been corroborated by various crystal structures24, 55, 56, 61–65 and biochemical studies53, 66. Collectively, these works show that several E2~ubiquitin conjugates are not reactive for transfer of ubiquitin onto a lysine because they adopt various inactive conformations; interactions with RING E3s favor the acquisition of a closed E2~ubiquitin conformation where ubiquitin is primed for transfer.
Key interactions in the primed conformation
The sidechain of an arginine from the RING E3 forms hydrogen bonds with both the E2 and ubiquitin, thereby functioning as a “linchpin”57 within the RING E3-UBE2D-ubiquitin interface to stabilize the primed ubiquitin conformation (Figure 3c). Direct interactions between this linchpin Arg and the carbonyl of Arg72 of ubiquitin contribute to constraining the C-terminal tail of ubiquitin in an activated conformation along E2’s active site cleft. Most E2s contain an HPN motif within their active sites that is critical for catalysis. The asparagine from this motif stabilizes the loop comprising residues 114-117 from the E2 UBE2D255, 67 (Figure 3d). This loop helps stabilize the tail of ubiquitin and, as observed in the SUMO E268, contains an aspartate that reportedly reduces the p_K_a of the incoming substrate lysine to facilitate catalysis55.
While the RING domain alone primes E2~ubiquitin57, the presence of various additional ubiquitin-binding components in some RING E3s helps stabilize E2~ubiquitin in the primed conformation and enhance activity (Figure 3e)56, 61. In CBL-B, the phosphate moiety of phospho-Tyr363 forms a hydrogen bond with ubiquitin’s Thr9 sidechain61 and in RNF38, loops adjacent to RNF38’s RING domain contact the Thr9-containing surface of ubiquitin24. In the monomeric RING E3 ARK2C, ubiquitin transfer requires binding of a second molecule of ubiquitin to the RING domain opposite the E2-binding surface; modeling and biochemical data suggest that this ubiquitin directly contacts and stabilizes the E2-conjugated ubiquitin for transfer69. For the dimeric RING E3s RNF4 and BIRC7, the RING domain of one subunit and the C-terminal tail of the second RING subunit interact with the Gly35-containing surface of ubiquitin55, 56. A conserved aromatic residue in the C-terminal tail from both of these dimers is critical for this interaction (Figure 3e). In contrast, the RING E3 TRIM25 has a different dimer interface and lacks a conserved aromatic tail. In the TRIM25-UBE2D1–ubiquitin complex structure, the N-terminal helix from one subunit and C-terminal helix from the second subunit make hydrophilic interactions with the Gly35-containing surface of ubiquitin65. For many RING E3s, these additional components are distinct features that require additional structural and biochemical studies to fully characterize their roles in RING E3-mediated ubiquitylation.
Additional E2-E3 interactions
Several RING E3s harbor an additional domain that binds to the “backside” surface of the E2 opposite the active site (Figure 3f). For example, the G2BR domain of gp78 allows such ‘backside’ interactions with the E2 UBE2G270. Similarly, U7BR of Cue1p binds UBC771, R6BD of RAD18 binds RAD672 and U5BR of AO7 binds UBE2D273. These various E3 domains that facilitate ‘backside’ binding of E2s all enhance RING E3-E2 affinity but affect activity disparately. G2BR stimulates ubiquitylation by inducing conformational changes at the RING-UBE2G2 interface that enhance UBE2G2~ubiquitin binding70. Moreover, RING domain binding reduces G2BR-UBE2G2 affinity, thereby enabling release of UBE2G2 and promoting processive ubiquitylation74. U7BR binding not only enhances RING E3-E2 affinity, but also increases the accessibility of UBC7’s active site to E1 and ubiquitin’s K48 to respectively facilitate recharging and substrate ubiquitylation71. In some E2s like RAD6 and the UBE2D family, noncovalent backside binding of ubiquitin promotes processive polyubiquitin chain formation72, 75–77 (Figure 3f). For UBE2D2, noncovalent ubiquitin binding induces allosteric changes in the E2 that increase the affinity of RING E3s for E2~ubiquitin and the catalytic efficiency of ubiquitylation leading to enhanced initial ubiquitin transfer and subsequent polyubiquitin chain formation24. U5BR enhances AO7’s affinity for UBE2D2 but blocks noncovalent ubiquitin binding, leading to decreased ubiquitylation activity73. Similarly, R6BD binding to RAD6 competes against noncovalent ubiquitin binding and limits RAD18 catalysis to monoubiquitylation72.
Transfer of ubiquitin onto substrate
To transfer ubiquitin to substrate, RING E3s juxtapose a substrate lysine and E2~ubiquitin thioester. How different RING E3s accomplish this varies, but in each case accessibility of the substrate lysine governs its capacity for ubiquitylation. This is discussed in the substrate ubiquitylation outcomes section.
Structural basis of HECT E3 activity
HECT E3s are characterized by an N-lobe that binds E2 connected by a hinge to a C-lobe containing the catalytic cysteine. The L1 and L2 loops and N-terminal α1 of E2 interact with an 80-residue subdomain in the HECT E3 N-lobe42, 78, 79. Following binding of E2~ubiquitin, ubiquitin is transferred from the E2 to the catalytic cysteine on the C-lobe of the HECT domain in a transthiolation reaction. The primed HECT~ubiquitin is then subsequently juxtaposed with and transferred to a substrate lysine.
Mechanism of transthiolation
In structures of the HECT E3s E6AP, WWP1 and SMURF2, the position of the C-lobe relative to the N-lobe varies, demonstrating that the hinge between the two lobes is flexible42, 43, 80 (Figure 4a–c). Early structures and models of HECT E3-E2 complexes revealed a large distance between the E2 and HECT E3 catalytic cysteines, suggesting conformational changes are required for the E2-E3 transthiolation reaction (Figure 4c). Subsequently, the crystal structure of NEDD4L bound to UBE2D2–ubiquitin showed that rotation about the hinge allowed the C-lobe to bind ubiquitin and position its catalytic cysteine adjacent to the UBE2D2–ubiquitin linkage to facilitate transthiolation78 (Figure 4d). In the current NEDD4L transthiolation model, the N-lobe initially recruits E2~ubiquitin, and upon rotation about the hinge, the C-lobe binds ubiquitin and juxtaposes both catalytic cysteines to promote formation of the HECT E3~ubiquitin intermediate. This mechanism is likely shared among NEDD4 family HECT E3s since residues contributing to hydrophobic interactions with Ile36 and surrounding residues of ubiquitin are conserved. These residues are not conserved in other HECT E3s; hence, further studies are required to elucidate how these other HECT E3s engage ubiquitin for transthiolation.
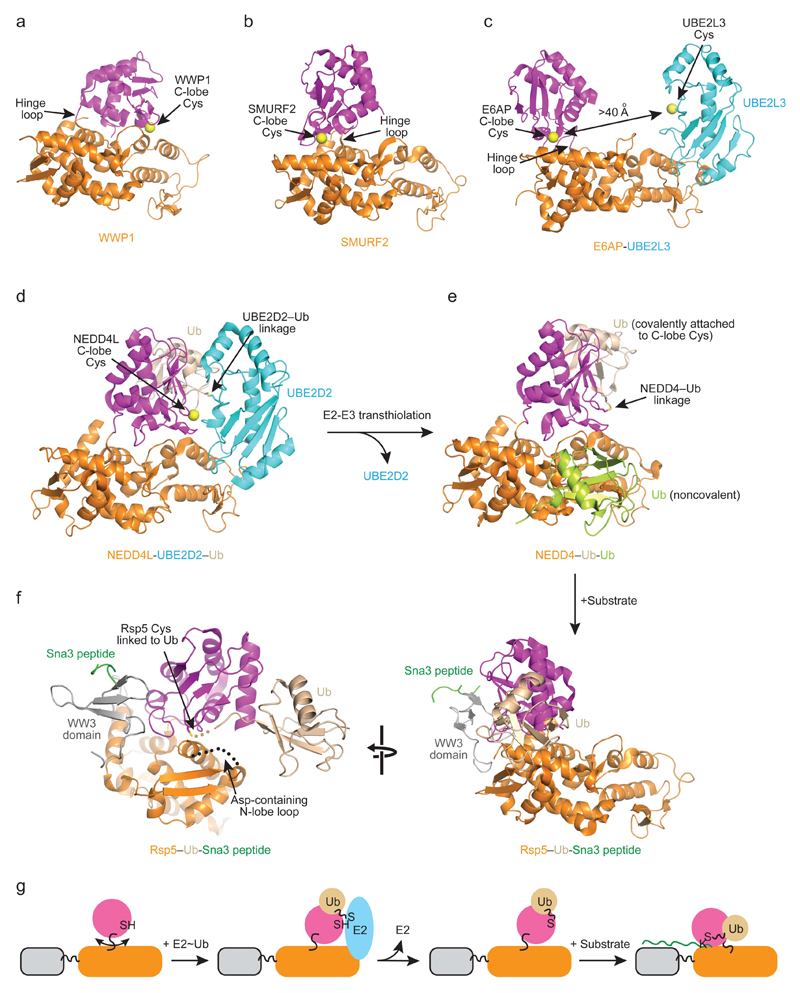
Figure 4. Mechanism of ubiquitin transfer by HECT E3s.
a–c | Conformational flexibility of HECT domains. Structural alignment based on the N-lobe of HECT domains from WWP1 (a, PDB: 1ND7), SMURF2 (b, PDB: 1ZVD) and E6AP bound to the E2 UBE2L3 (c, PDB: 1C4Z) reveals different C-lobe conformations. N-lobes are colored orange, C-lobes magenta and UBE2L3 cyan. Arrows indicate the flexible hinge loop. Catalytic cysteines are shown as yellow spheres. The distance between E2 and E3 catalytic cysteines observed in the structure is indicated in c. d | Structure of NEDD4L HECT-UBE2D2–ubiquitin complex (PDB: 3JVZ). UBE2D2–ubiquitin linkage (indicated) and C-lobe’s catalytic cysteine are juxtaposed allowing transthiolation of ubiquitin from the active site of the E2 to the catalytic cysteine of E3. Colors are as in a–c with ubiquitin colored wheat. e | Structure of NEDD4 HECT domain following transthiolation (PDB: 4BBN). In this case NEDD4 is bound to two ubiquitin molecules, one which is a covalently attached to the catalytic cysteine in the C-lobe following transthiolation (shown in wheat) and a second one which is non-covalently bound to the N-lobe that enhances the processivity of substrate ubiquitylation (shown in lime). Colors are as in a–c. f | Structure of Rsp5(E3)–ubiquitin-Sna3 peptide(substrate) complex (PDB: 4LCD). The catalytic Cys777 of Rsp5, Gly75 of ubiquitin and Lys125 of the Sna3 peptide are covalently linked via a three-way chemical cross-linker to mimic substrate ubiquitylation. Colors are as in a–d with Sna3 peptide colored green and the WW3 domain grey. The disordered N-lobe loop harboring Asp495 is depicted as a dashed line. In d, e and f right panel, alignment of the three structures using the N-lobe reveals different C-lobe conformations in E2-E3 transthiolation and substrate ubiquitylation reactions. g | Schematic diagrams showing the HECT E3 catalytic cycle. In the absence of any binding partner, the C-lobe can rotate relative to the N-lobe about the hinge loop. Upon encountering E2~ubiquitin, the N-lobe binds E2 and the C-lobe rotates to bind ubiquitin, thereby juxtaposing the catalytic cysteines from E2 and E3. Upon ubiquitin transfer, E2 is released. E3 binds substrate and the C-lobe undergoes rotation to juxtapose E3’s catalytic cysteine and substrate lysine for ligation. Colors are as in a–f.
Priming ubiquitin for transfer
The step immediately following the transthiolation reaction was captured in a crystal structure of a NEDD4–ubiquitin complex81 (Figure 4e). The interactions between ubiquitin and the C-lobe are identical to those observed in the structure of NEDD4L-UBE2D2–ubiquitin complex prior to transfer. Notably, ubiquitin’s C-terminal tail is positioned in an extended conformation by a hydrogen bond network involving backbone carbonyl oxygens and amides from the C-lobe. Constraining the position of ubiquitin’s C-terminus appears to be a common thioester-activating mechanism for RING and HECT E3s in which E3-E2~ubiquitin structural data is available.
Transfer of ubiquitin to substrate
In the subsequent step, HECT E3~ubiquitin juxtaposes the E3~ubiquitin thioester and substrate lysine. This process was recently captured in the crystal structure of Rsp5–ubiquitin-Sna3 complex where a three-way chemical cross-linker was used to link Rsp5’s catalytic cysteine, ubiquitin’s C-terminus and a lysine sidechain of Sna3 substrate peptide82 (Figure 4f). The C-lobe undergoes a 130° rotation about the flexible linker relative to its conformation in the NEDD4L-UBE2D2–ubiquitin and NEDD4–ubiquitin complexes (Figure 4e–g). Almost half of the N-lobe contacts the C-lobe to stabilize this conformation. Crucially, Phe806 from the C-lobe of Rsp5, also known as the “-4 phenylalanine”, sits in an N-lobe pocket containing Phe505 and Leu506. This -4 phenylalanine is conserved in all types of HECT E3s and is required for substrate ubiquitylation and polyubiquitin chain formation but not HECT~ubiquitin formation83, 84. Interestingly, mutations of this Phe to Leu (Rsp5 F806L) can be rescued by a complementary substitution of Leu506 to Trp (L506W) or to Phe (L506F), suggesting that Phe806 anchors the two HECT domain lobes in an orientation suitable for substrate ubiquitylation82. It is noteworthy that the amino acid composition of this N-lobe pocket is conserved in the NEDD4 E3s, but diverges in other HECT E3s. Furthermore the extreme C-terminus of HECT E3s has been shown to be important for ligation81, 83 but this region is disordered in all HECT E3 structures. Future studies are required to investigate these differences in the HECT E3 subfamilies and the role of the C-terminus in ligation.
The Rsp5–ubiquitin-Sna3 structure does not capture a substrate lysine poised for ligation but modeling and mutational analyses suggest that residues surrounding Rsp5’s catalytic cysteine contribute to Sna3 ubiquitylation82. The Rsp5~ubiquitin thioester sits adjacent to an N-lobe loop (residues 490-495) harboring Asp495 (Figure 4f), an essential residue for ligation82. Unfortunately this loop is disordered in the Rsp5–ubiquitin-Sna3 structure. Modeling based on other HECT E3 structures, in which the loop is ordered and contains a structurally conserved aspartate, suggests that this loop may function in positioning the C-terminus of ubiquitin. Additionally, in the model Asp495 juxtaposes with the E3~ubiquitin thioester, where it can potentially guide a substrate lysine into the active site or contribute to substrate lysine deprotonation82. Future studies are required to determine how the aspartate-containing N-lobe loop and active site residues participate in substrate ubiquitylation and chain elongation in HECT E3s.
The NEDD4-family HECT E3s harbor a noncovalent ubiquitin-binding site on the N-lobe80, 81, 85–87 (Figure 4e). Disruption of this interaction impairs polyubiquitin chain elongation but not E2-E3 transthiolation or the conjugation of the first ubiquitin to substrate86, 87. This noncovalent ubiquitin is distal from the E3~ubiquitin thioester in all available structures, suggesting it does not serve as the acceptor ubiquitin. Biochemical data suggest this site might bind ubiquitin-modified substrate to promote processive polyubiquitin chain formation87. The exact mechanism remains to be determined.
Structural basis of RBR E3 activity
Structures of PARKIN, HHARI and HOIP show that RING1, IBR and RING2 domains from different RBR E3s share a common structural fold88–97. RING1 adopts a cross-braced Zn2+-coordination configuration found in typical RING domains whereas IBR and RING2 each have two zinc ions coordinated in a linear fashion. RING1 recruits E2~ubiquitin and transfers ubiquitin to the catalytic cysteine on RING2 to form a RING2~ubiquitin intermediate; ubiquitin is subsequently transferred to substrate. The structures of PARKIN, HHARI and HOIP show distinct RING1-IBR-RING2 arrangements (Figure 5). In the structure of the RBR region from HOIP bound to E2–ubiquitin, the RBR adopts an active conformation92. In the full-length structures of PARKIN and HHARI, both adopt autoinhibited conformations where the arrangement of the RBR motif is determined by other domains unique to each RBR E393–97. In PARKIN, a RING0 domain is sandwiched between RING1 and RING2 with IBR placed adjacent to RING1; in HHARI, a C-terminal Ariadne domain is stacked between IBR and RING2 and RING1 is secluded by an ubiquitin associated (UBA) domain at the opposite end of RING2 (Figure 5a,b).
Figure 5. Mechanism of ubiquitin transfer by RBR E3s.
a,b | Structures of autoinhibited PARKIN (PDB: 5C23) and HHARI (PBD: 4KBL), respectively. Schematic diagrams above show domain architecture with structural domains colored according to the diagrams. Zinc atoms and RING2’s catalytic cysteines are shown as grey and yellow spheres, respectively. c,d | Structures of PARKIN and HHARI in a and b, respectively, with a modeled E2 in cyan generated by overlaying their RING1 domain with the structure of RNF38 RING domain bound to E2 (PDB: 4V3L). Colors are as in a. Distances between E2 and E3’s catalytic cysteines are indicated. e | Structure of HOIP RBR domain bound to UBE2D2–ubiquitin (PDB: 5EDV). HOIP was crystallized as a domain swapped dimer, but the biological unit of HOIP-UBE2D2-ubiquitin complex is shown here. A schematic diagram shows crystallized HOIP construct’s domain architecture. Structural domains are colored according to this diagram. UBE2D2 is colored cyan, ubiquitin wheat and ubiquitinAllo yellow. E2–ubiquitin linkage and RING2’s catalytic cysteines are shown as yellow spheres. In a, b and e, the helical extension (hE) comprises two segments, hE1 and hE2 (indicated in the structures), which are involved in binding ubiquitin. f | Structure of HOIP together with acceptor ubiquitin (green, serving as substrate) and a second donor ubiquitin molecule (wheat) generated via crystallographic symmetry (PDB: 4LJO). Top panel is in the same orientation as in e. g | HOIP catalytic cycle. Binding of a ubiquitin molecule to the allosteric site (UbAllo) primes HOIP’s RBR conformation for E2~ubiquitin recruitment. The E2~ubiquitin thioester and RING2’s catalytic cysteine are juxtaposed for RING2~ubiquitin formation. Upon formation of the RING2~ubiquitin, E2 is released. Subsequently, RING2 binds an acceptor ubiquitin (UbAcc) and catalyzes linear ubiquitin chain formation. To catalyze additional rounds of ubiquitin transfer, linear ubiquitin must be released to enable E2~ubiquitin recruitment.
Formation of RING2~ubiquitin
Given the structural similarity between the RING1 and canonical RING domains, modeling of the RING1 domain from PARKIN and HHARI onto RING E3-E2~ubiquitin complexes reveals that all RING1 domains harbor similar hydrophobic cores that interact with hydrophobic residues in the L1 and L2 loops of E2 (Figure 5c,d). These hydrophobic interactions were observed in the structure of HOIP’s RBR bound to UBE2D2–ubiquitin92. However, RING1 domains lack the linchpin arginine that promotes the closed, primed E2~ubiquitin conformation required for ubiquitin transfer to a lysine. In contrast to RING E3s, RBR E3s function with UBE2L3, an intrinsically cysteine-reactive E250. Without RING2 or with catalytically dead RING2, HHARI cannot promote transfer of UBE2L3~ubiquitin50, 93. Taken together, these works suggest that the RING1 domain does not activate E2~ubiquitin like a canonical RING E3; instead, data indicate the RBR E3s resemble HECT E3s in function. Depending on the class of E3, either the N-lobe or RING1 recruit E2~ubiquitin and juxtaposition with the C-lobe or RING2 promotes formation of E3~ubiquitin.
The recent HOIP-UBE2D2-ubiquitin complex structure reveals how these catalytic cysteines are juxtaposed92. Multiple interactions with RING1, RING2, IBR and regions between these components stabilize UBE2D2–ubiquitin in an open conformation92 (Figure 5e). The Ile44 surface of ubiquitin contacts a helix between IBR and RING2, thereby enabling RING2 to bind UBE2D2 and stabilize ubiquitin’s C-terminal tail in an extended conformation such that the UBE2D2 and RING2 catalytic cysteines are juxtaposed. The RING1-IBR module adopts an elongated conformation where RING1 and IBR are connected by two extension helices, hE1 and hE2. RING1 binds UBE2D2 and hE2-IBR binds the surface of ubiquitin containing Lys11 and Glu34. There is an additional ubiquitin-binding site on the RING1-IBR region opposite the UBE2D2–ubiquitin-binding site. This ubiquitin (hereby referred to as ubiquitinAllo) functions as an allosteric activator by enhancing UBE2D2~ubiquitin binding and RING2~ubiquitin formation. Like HOIP, PARKIN and HHARI both have similar RING1-IBR-RING2 module and two helices between RING1 and the IBR region (Figure 5a,b,e), suggesting they share a similar mechanism of E2~ubiquitin binding. How PARKIN and HHARI undergo conformational changes to juxtapose E2 and RING2’s cysteines and how other domains influence this transition remain to be determined.
Transfer of ubiquitin to substrate
Our current understanding of how RBR E3s recognize their substrates is limited to HOIP-mediated linear ubiquitin chain formation. This process is facilitated by the presence of a linear ubiquitin chain determining domain (LDD) in HOIP that binds and orients the acceptor ubiquitin98, 99. In the crystal structure of HOIP RING2 and LDD bound to acceptor ubiquitin, the C-terminus of a second ubiquitin molecule is oriented with its carboxylate positioned next to the active site cysteine of RING2, thereby mimicking a HOIP~ubiquitin intermediate (Figure 5f). Thus the structure captures both donor and acceptor ubiquitin in conformations poised for linear ubiquitin chain synthesis88. The C-terminal tail of the donor ubiquitin lies along a hydrophobic groove surrounding the catalytic cysteine in the core of the RING2 domain; interactions with this groove and a β-hairpin from the RING2 domain stabilize the tail in an extended conformation (Figure 5f). The RING2 domains from PARKIN and HHARI also contain hydrophobic grooves proximal to the catalytic cysteine, suggesting they may use an E3~ubiquitin-binding mode similar to HOIP when mediating ubiquitin transfer to substrate. However, they lack the β-hairpin present in HOIP and have different domains outside of the RBR motif implicated in substrate binding; hence, further studies are required to elucidate how PARKIN and HHARI juxtapose RING2~ubiquitin and substrate.
HOIP RING2-ubiquitin and HOIP-UBE2D2–ubiquitin complexes have identical donor ubiquitin-binding modes, indicating that the ubiquitin-binding mode is maintained throughout E2-E3 ubiquitin transfer and substrate ubiquitylation92. The acceptor ubiquitin-RING2 interaction overlaps with the UBE2D2-RING2 interface in the HOIP-UBE2D2–ubiquitin complex, suggesting acceptor ubiquitin and E2~ubiquitin binding are mutually exclusive. Thus, for successive rounds of substrate ubiquitylation, acceptor ubiquitin must be released from RING2 after transfer to allow additional rounds of E2~ubiquitin recruitment and RING2~ubiquitin formation (Figure 5g). How such arrangements promote ubiquitin chain elongation is unclear.
Substrate ubiquitylation outcomes
Once ubiquitin is primed, an E3 positions a lysine sidechain from a substrate next to the E2~ubiquitin or E3~ubiquitin thioester for ligation. Subsequently, the E3 may release the substrate, resulting in monoubiquitylation. Alternatively, the E3 may catalyze consecutive ubiquitin transfers in which multiple lysine sites on a substrate are modified with a single ubiquitin resulting in multi-monoubiquitylation or polyubiquitin chains are formed with a selective linkage at a single or multiple lysine sites. Polyubiquitylation involves two events: chain initiation where ubiquitin is transferred to a lysine on the substrate and chain elongation where ubiquitin is transferred to a lysine on a ubiquitin moiety from ubiquitylated substrate. For some RING E3s, a single E2 is reportedly responsible for both events75 while in others one E2 monoubiquitylates substrate and a second E2 performs chain elongation100–102.
To mediate this array of ubiquitylation outcomes, diverse mechanisms are required. For monoubiquitinylation, E3s require a method to prioritize or isolate a single lysine on a substrate. On the other hand, for multi-monoubiquitylation and chain elongation, E3s must be flexible to bridge the varying distances between the E3~ubiquitin or E2~ubiquitin thioester and the target lysine. Flexibility is also required in E3s that interact with multiple substrates via discrete domains. Currently, no consensus ubiquitylation motif has been determined. For RING E3s, it is widely accepted that the E2 specifies the ubiquitin chain type or lysine site that is modified; however, it is the RING E3 that regulates the accessibility of substrate lysines to the E2~ubiquitin thioester. The emerging theme from recent structural studies on E2/E2~ubiquitin-RING E3-substrate and HECT E3~ubiquitin-substrate complexes illustrates that spatial arrangements and surface complementarity prioritize lysine site(s) for ubiquitylation64, 82, 103–105. Because E2~ubiquitin-E3-substrate and E3~ubiquitin-substrate complexes use different mechanisms to bind substrate, how they specify substrate lysine site(s) for ubiquitylation will vary. Here we discuss some examples.
Lys125 prioritization on Sna3 by the HECT E3 Rsp5
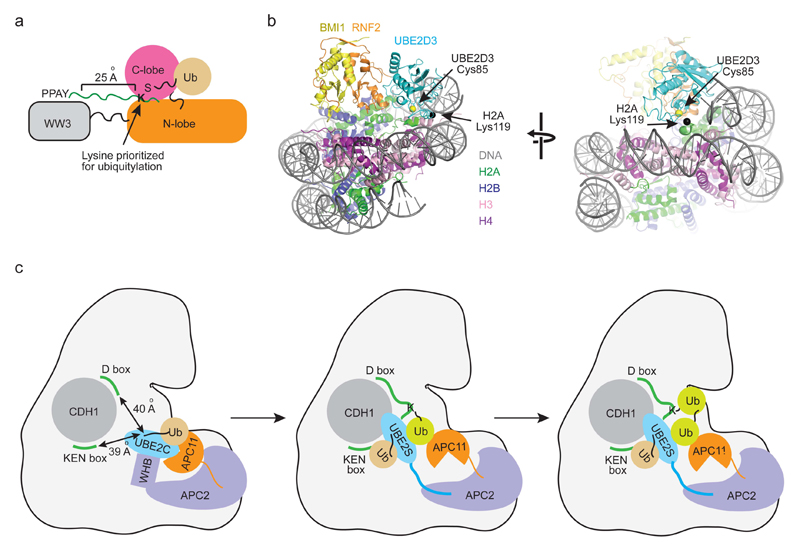
The HECT E3 Rsp5 mediates ubiquitylation and sorting of most multivesicular body cargo proteins in yeast, including Sna3106–109. Rsp5 interacts directly with Sna3 to ubiquitinylate Lys125: the WW3 domain of Rsp5 binds to the PY motif in Sna3, which is separated from Lys125 by a 15-residue linker82. This linker is sufficient to bridge the gap between Rsp5’s catalytic cysteine and Sna3’s PY motif in the Rsp5-ubiquitin-Sna3 complex (Figure 6a). Deletion of the linker to less than 10 residues strongly reduces ubiquitylation of Sna3, suggesting that Rsp5 domains do not rotate freely during ligation. The distance between Rsp5’s active site and Sna3’s PY motif seemingly selects the lysine ubiquitylation site82.
Figure 6. Mechanisms of substrate lysine selection.
a | A schematic drawing of Rsp5-catalyzed substrate ubiquitylation. Rsp5~ubiquitin presents its N- and C-lobes and WW3 domain in a conformation where the distance is 25 Å between the Rsp5~ubiquitin thioester and the substrate binding site in the WW3 domain. In this way, lysines at least 10 residues away from the WW3-binding motif PY in the substrate are prioritized for ubiquitylation. The N-lobe is colored orange, the C-lobe magenta, the WW3 domain grey, ubiquitin wheat and substrate in green. b | Structure of RNF2-BMI1-UBE2D3 complex bound to nucleosome core particle (NCP) (PDB: 4R8P). RNF2 is colored orange, BMI1 yellow, UBE2D3 cyan, H2A green, H2B blue, H3 pink, H4 purple and DNA grey. H2A’s Lys119, the target for monoubiquitylation in this reaction, is shown as a black sphere and UBE2D3’s catalytic cysteine is shown as a yellow sphere. The interactions between the RNF2-BMI1-UBE2D3 complex and NCP restrain UBE2D3 and H2A movement such that UBE2D3’s catalytic cysteine can only be juxtaposed with H2A’s Lys119 for site-specific ubiquitylation. c | Schematic drawings of APC/C-catalyzed substrate ubiquitylation. APC11 binds UBE2C~ubiquitin via the canonical RING E3-E2~ubiquitin interaction shown in Figure 3 and APC2’s WHB domain contacts UBE2C’s backside. These bipartite interactions fix the UBE2C~ubiquitin orientation such that the thioester bond is ~40 Å away from the D and KEN box binding sites on the substrate-receptor CDH1, thereby prioritizing substrate lysines located 10 residues away from these motifs for ubiquitylation. Once the substrate is modified with a single ubiquitin, APC/C tracks the tip of the growing ubiquitin chain. APC11 binds the acceptor ubiquitin (colored in lime) using a surface distinct from its E2~ubiquitin-binding site and presents the acceptor ubiquitin to a UBE2S~ubiquitin complex for ubiquitin chain elongation. APC/C is shown as an outline. APC11 is colored orange, UBE2C and UBE2S cyan, ubiquitin wheat, APC2 and WHB light blue and CDH1 grey. Green ribbons depict binding sites on CDH1 for substrates containing a D- or KEN-box.
Site-specific monoubiquitylation of H2A by RNF2-BMI1
Monoubiquitinylation of Lys119 in H2A by the heterodimeric RING E3 RNF2-BMI1 complex is important for Polycomb-group proteins to mediate gene-silencing110. In the crystal structure of RNF2-BMI1-UBE2D3 complex bound to the nucleosome core particle (NCP)103 (Figure 6b), the RING domain is anchored onto the NCP through interactions with all four histones and UBE2D3 forms a complementary surface with the nucleosomal DNA and H2A’s C-terminus, thereby restricting RING mobility and the orientation of the E2. Usually, substrate lysine modification by RING E3s with the UBE2D family of E2s is promiscuous, but in this configuration, the accessibility of the catalytic cysteine from the E2 is limited to a specific substrate lysine. For RING E3s to build ubiquitin chains, either the RING-E2 module or the ubiquitylated segment of substrate must rotate. In the RNF2-BMI1-UBE2D3-NCP complex, UBE2D3, nucleosomal DNA and histones H3 and H4 surround H2A’s C-terminus, thereby restricting movement of the substrate. Because both the RING-E2 and substrate elements are constrained in the RNF2-BMI1-UBE2D3-NCP complex, only monoubiquitylation occurs.
Lysine selection with a mobile RING domain
In E3s such as CBL61, 111, 112, CRL64, 113–115 and APC/C104, 105, 116 (Figure 6c), a flexible linker between the RING and other domains allows the E2~ubiquitin-bound RING domain to sample multiple conformations. APC/C facilitates all known degradation events controlling mitotic exit. More than 50 substrates have been identified in genome-wide studies117, yet this E3 tightly regulates lysine selectivity and chain elongation. How does APC/C select preferred lysine sites when flexible? For APC/C, CDH1 binding induces a conformational change in the APC2 and APC11 subunits to stimulate activity116. This enables bipartite interactions with UBE2C where the RING domain APC11 binds UBE2C via canonical E2-RING interactions and the winged-helix B domain of APC2 interacts with UBE2C’s backside105 (Figure 6c). This bipartite UBE2C binding restricts the orientation of the APC11-UBE2C module such that UBE2C’s active site prioritizes lysines residing ten residues beyond CDH1’s D-box and KEN-box substrate-binding motifs104, 105.
Positioning of ubiquitin for chain elongation
To build a specific ubiquitin linkage, a particular lysine from a ubiquitin molecule ligated to substrate must be poised for ligation with the donor ubiquitin. Ubiquitin chain linkage specificity depends on the E2 pairing for RING E3s and the E3 itself for HECT and RBR E3s4, 13, 88,118. Current structural understanding of acceptor ubiquitin positioning for generating specific ubiquitin–ubiquitin linkages is limited to UBE2N-UBE2V2-catalyzed Lys63-linked ubiquitin chains, in which UBE2V2 determines chain specificity,62, 119 and HOIP-catalyzed linear ubiquitin chains, in which the E3 determines chain specificity. Both UBE2V2 and HOIP harbor a ubiquitin-binding domain that binds acceptor ubiquitin and orients Lys63 (in the case of UBE2V2) or Met1 (in the case of HOIP) toward the catalytic cysteine for ligation. However, it remains unclear how E3s position ubiquitylated substrate for chain elongation.
Recent studies of APC/C reveal a novel mechanism whereby acceptor ubiquitin at the tip of a growing substrate ubiquitin chain is tracked and E3 and E2 function together to determine chain specificity120, 121. APC/C uses a coactivator, CDC20 or CDH1, to recognize and bind substrates containing D- or KEN-box motifs and initially recruits UBE2C~ubiquitin via the canonical APC11 RING-E2 binding surface to transfer ubiquitin to substrate122 (Figure 6c). Subsequently it recruits UBE2S~ubiquitin via a distinct APC2 surface that binds the C-terminal LRRL motif of UBE2S116, 123, 124 and guides UBE2S-mediated Lys11-linked ubiquitin chain elongation on substrate120, 121 (Figure 6c). APC/C stabilizes UBE2S’s core domain and binds the acceptor ubiquitin via a distinct hydrophobic patch on the APC11 RING domain opposite the UBE2C-binding surface. These abilities massively enhance the catalytic efficiency of the APC/C-UBE2S complex, presumably by enabling APC/C to juxtapose UBE2S’s active site and acceptor ubiquitin lysine on substrate to promote Lys11-linked polyubiquitylation.
These examples illustrate a common mechanism whereby an E2-E3 or E3 catalytic module is secured in a conformation that juxtaposes the ubiquitin thioester with select substrate lysine site(s) for ligation and chain elongation. For each example, structural features of the E3, E2 and substrate determine lysine selectivity and subsequent ubiquitin chain formation. Future studies on other E3-substrate systems will reveal how different structural features regulate ubiquitylation.
Regulation of E3 activity
Over the last 15 years, numerous mechanisms including post-translational modifications of E2, E3 and substrate as well as binding of protein partners and small molecules, exemplified how E3s are regulated throughout their catalytic cycle21. Recent studies demonstrated that all three classes of E3s are also regulated by autoinhibitory mechanisms involving catalytically incompetent E3 conformations in which E2~ubiquitin cannot be recruited or E3 flexibility is limited. In every study, domains other than those that define each class of E3 are involved; hence, how autoinhibition and activation are mediated depends on the E3. Here we describe an example from each class of E3s.
cIAP1 autoinhibition and activation
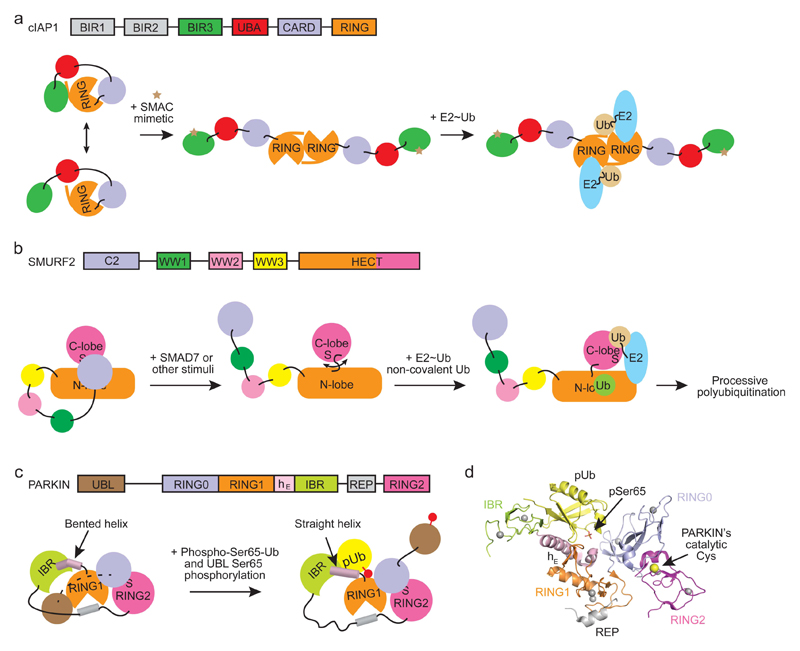
Cellular inhibitor of apoptosis 1 (cIAP1 or BIRC2) is a dimeric RING E3 that plays a central role in the NF-κB signaling cascade and programmed cell death125. cIAP1 contains three N-terminal baculoviral IAP repeat (BIR1-3) domains followed by a UBA domain, a caspase activation and recruitment domain (CARD) and a RING domain. In the crystal structure of cIAP1 lacking the BIR1 and 2 domains, cIAP1 is in an inactive conformation where the RING domain is sequestered and the protein is a monomer (Figure 7a). RING sequestration prevents dimerization, thereby impairing E2~ubiquitin recruitment126, 127. SAXS analyses showed that cIAP1 is dynamic and undergoes global conformational changes whereby the BIR3 peptide-binding groove is exposed128. Binding of second mitochondrial activator of caspases (SMAC) peptide or a SMAC peptide mimetic to the BIR3 domain disrupts the autoinhibited conformation, thereby activating ligase activity by enabling RING domain dimerization and subsequent E2~ubiquitin recruitment126, 129 (Figure 7a).
Figure 7. Mechanisms of autoinhibition and activation of E3s.
a | Schematic showing domain composition of cIAP1 (top) and the mechanism of its autoinhibition and activation (bottom). In solution, cIAP1 adopts a closed conformation where the E2~ubiquitin binding and dimerization surfaces of the RING domain are occluded. SMAC mimetic compound binds the BIR3 domain and shifts cIAP1 to an extended conformation. This enables RING domain dimerization which is critical for binding and priming E2~ubiquitin for catalysis. b | Schematic showing domain composition of SMURF2 (top) and the mechanism of its autoinhibition and activation (bottom). The C2 domain of SMURF2 binds both the N- and C-lobes of the HECT domain, thereby preventing rotation of the two lobes and blocking non-covalent ubiquitin binding. Addition of SMAD7 or other stimuli causes the release of the C2 domain, thereby facilitating E2-E3 transthiolation and non-covalent ubiquitin binding important for processive substrate ubiquitylation. c | Schematic drawings showing domain composition of PARKIN (top) and the mechanism of its autoinhibition and activation. PARKIN adopts an autoinhibited conformation where the E2~ubiquitin binding site on RING1 and catalytic cysteine of RING2 are blocked. Moreover, in this conformation the catalytic cysteine of RING2 is distal from the modeled catalytic cysteine of E2 as shown in Figure 5c. Addition of phospho-Ser65-ubiquitin (shown as pUb in the drawing) and phosphorylation of PARKIN’s Ser65 in the UBL domain (shown as a red ball and stick) synergistically activate PARKIN. The phosphorylated UBL domain is released from the RING1 domain, partially freeing the E2~ubiquitin binding surface. Phospho-Ser65-ubiquitin then binds a pocket created by RING0, RING1, hE and IBR and induces straightening of hE similar to that observed in the HOIP-UBE2D2–ubiquitin structure (Figure 5e,g). Notably, PARKIN’s RING1-hE-IBR-phospho-Ser65-ubiquitin complex adopts a conformation that resembles RING1-hE-IBR-ubiquitinAllo in the HOIP-UBE2D2–ubiquitin structure, suggesting a similar mechanism in E2~ubiquitin recruitment of both enzymes, though this remains to be investigated. For all panels, domain architectures are shown and colored according to the schematic. d | Crystal structure of PARKIN bound to phospho-Ser65-ubiquitin (PDB: 5CAW). Structural domains are colored according to the schematic drawing in c. Phospho-Ser65-ubiquitin is in yellow with pSer65 indicated. RING2’s catalytic cysteine is shown as a yellow sphere.
SMURF2 autoinhibition and activation
The NEDD4 family of HECT E3s including SMURF1, SMURF2, ITCH, NEDD4_1 and NEDD4_2 share a similar domain architecture containing an N-terminal C2 domain followed by two to four WW domains and a C-terminal HECT domain and are regulated by autoinhibition130–135. SMURF2 is better characterized and is autoinhibited by interactions between the C2 and HECT domains135. NMR analyses reveal that interactions between the C2 domain and the N-lobe of the HECT domain restrict the C-lobe to an inactive conformation in which transthiolation is unsupported135, 136 (Figure 7b). Moreover, this interaction partially buries the N-lobe noncovalent ubiquitin-binding surface essential for E3 processivity136. SMURF2 is activated upon binding of the adaptor protein SMAD7. Interactions between the PY motif of SMAD7 and SMURF2’s WW domain and SMAD7’s N-terminal domain and SMURF2’s HECT domain release the C2 domain135, 137 (Figure 7b). Moreover, SMAD7 assists E2 binding by SMURF280.
Autoinhibition and activation of PARKIN
In RBR E3s with characterized autoinhibitory mechanisms, domains outside of the RBR motif mediate autoinhibition93–99, 138. Of the three RBR E3s that have structural data available, PARKIN is a better-characterized example. PARKIN plays an important role in mitochondria quality control. When mitochondria are damaged, PINK1 accumulates on the outer mitochondrial membrane and recruits PARKIN139. PINK1 phosphorylates Ser65 in PARKIN’s UBL domain and ubiquitin’s Ser65 on mitochondrial polyubiquitin chains. Phosphorylation of PARKIN’s Ser65 enhances its ligation activity, which is further stimulated by phospho-Ser65 ubiquitin140. Binding of phospho-Ser65 ubiquitin increases the activity of unphosphorylated PARKIN and enhances PINK1-dependent phosphorylation of PARKIN’s Ser65118, 140–142. This synergistic effect establishes a feed-forward mechanism118 to ensure that PARKIN is activated and able to efficiently ubiquitylate mitochondrial outer membrane proteins143, 144 and promote mitophagy. In the autoinhibited state, there is a large gap between the RING1 domain and the RING2 catalytic cysteine in which E3~ubiquitin formation is unsupported94–97, 138, 145 (Figure 5c). Moreover, the RING0 domain binds the RING2 domain and masks the catalytic cysteine (Figures 5a and 7c) and an additional repressor element (REP) within the RBR motif and the N-terminal UBL domain occlude the E2~ubiquitin-binding site on the RING1 domain94–97, 145. Phosphorylation of PARKIN’s Ser65 releases the UBL domain from RING1 to allow E2~ubiquitin binding and alters the RING0/RING1 interface to improve phospho-Ser65 ubiquitin binding97, 118, 140, 145, 146. Phospho-Ser65 ubiquitin binding to the RING1-IBR surface opposite the E2~ubiquitin binding site induces straightening of a “bent” RING1-IBR helix that promotes conformational changes in the IBR-REP region, thereby leading to the release of the UBL domain from PARKIN146 (Figures 7c,d). Activated PARKIN and HOIP have similar RING1-IBR conformations, suggesting that straightening of the RING1-IBR linker helix may be a prerequisite for E2~ubiquitin binding to RBR E3s92.
Conclusions and future perspectives
Our understanding of the pivotal role E3s play in affecting cellular processes has grown exponentially. The capacity of E3s to mediate ubiquitin chain formation depends on the ability to recruit E2~ubiquitin and promote ubiquitin transfer. Spatial arrangements of E3-E2-substrate complexes dictate substrate ubiquitylation patterns. Technologies have only advanced enough in the last decade to support the preparation of sufficient quantities of E2~ubiquitin and E3~ubiquitin suitable for structural and biochemical analyses. Consequently, we have gained several insights into the mechanisms underlying E3-mediated ubiquitylation and regulation. However, given that current structural studies encompass only a fraction of all E3s, future studies are required to uncover the functions and mechanisms of other uncharacterized E3s. Chemical cross-linkers such as those used in the studies of Rsp582 and APC/C105 will facilitate structural studies of intermediates in E3-catalyzed reactions, allowing us to investigate mechanisms of E3-catalyzed substrate ubiquitylation and polyubiquitylation. Furthermore, recent methods for introducing site-specific ubiquitin modifications to substrate147–153 will be useful tools for further dissecting E3-catalyzed polyubiquitylation. Lastly, there is a growing interest in targeting E3s with cancer therapeutics. A better understanding of the mechanisms of E3-catalyzed reactions and regulation will greatly aid the development of such E3 inhibitors.
Supplementary Material
Supplementary Table 1
Key points.
- E3s recruit E2~ubiquitin (~ denotes a thioester linkage between E2’s catalytic cysteine and the C-terminus of ubiquitin) and substrate to promote ubiquitin transfer. Mechanisms of E3 catalysis and regulation are summarized here.
- Crystal structures and NMR studies reveal that RING E3s prime E2~ubiquitin for transfer by promoting a closed E2~ubiquitin conformation where the thioester is activated toward nucleophilic attack.
- Crystal structures of HECT E3s in different stages of catalysis reveal that conformational changes juxtapose reactants to prime ubiquitin transfer.
- Crystal structures reveal how RBR E3s are autoinhibited. Upon activation, crystal structures of RBR E3s bound to E2~ubiquitin show how ubiquitin is transferred from E2 to E3 and subsequently to substrate.
- Substrate lysine selection and the outcome of ubiquitylation depends on spatial arrangements within E2-E3-substrate complexes. Substrate lysines that are in proximity to the active sites of E2 or E3 are prioritized for ligation.
- E3s are regulated by various autoinhibitory mechanisms that hinder their catalytic cycle. Activation frequently requires post-translational modification or binding of protein partners or substrates.
Glossary.
ε-amino group
Refers to the NH3+ group in a lysine sidechain. It has a high pKa and often the active site environment(s) of E2, E3 or substrate can assist in lowering its pKa to increase its reactivity.
26S proteasome
A large protein complex that degrades proteins modified with ubiquitin chains. It contains a 20S core particle that accommodates the protease catalytic subunits and two 19S regulatory subunits that recognize ubiquitylated substrates and facilitate substrate unfolding prior to entry into the 20S core particle.
Anaphase promoting complex/cyclosome (APC/C)
A multi-subunit RING E3 complex that plays a central role in cell cycle progression by ubiquitylating cell cycle proteins and targeting them for degradation.
PY motif
A short proline-rich motif containing a proline-proline-x-tyrosine (x is any amino acid) sequence that is recognized by a WW domain.
NMR
(Nuclear magnetic resonance). A technique that provides detailed information about protein structure and dynamics in solution.
Allosteric change
Refers to changes induced in one part of a molecule, usually an enzyme’s active site, by a distal binding event.
SAXS
(Small angle X-ray scattering). A technique where the elastic scattering of X-rays by a protein in solution is used to provide information about the protein’s shape and size.
C-terminal tail of ubiquitin
Residues 71-76 of ubiquitin
SUMO
A ubiquitin-like protein modifier that has its own dedicated E1-E2-E3 enzymatic cascade for modification of protein substrates.
α-amino group
Refers to the NHM3+ group connected to the α-carbon in an amino acid. In proteins, it is only present at the N-terminus because it is otherwise involved in forming an amide link with an adjacent amino acid. Under cellular condition, it exists in the protonated NH3+ form and requires deprotonation to increase its reactivity.
Nucleosome core particle (NCP)
A basic repeating unit of DNA packaging found in eukaryotes. A single NCP contains 147 base pairs of DNA wrapped around a histone octamer consisting two histone H2A-H2B dimers and a histone H3-H4 tetramer.
NF-κB signalling
A signalling pathway that activates NF-κB in response to stimuli including proinflammatory cytokines, bacterial and viral infections, and antigen receptor binding. Activated NF-κB leads to expression of genes that are regulators of apoptosis, cell survival, proliferation and immunity.
Acknowledgements
This work was supported by Cancer Research UK and European Research Council (grant number 647849).
Biographies
Author biographies
Lori Buetow received her Ph.D. at University of California, San Diego, USA, working with Partho Ghosh. She is currently working as an associate scientist in Danny Huang’s lab at the Beatson Institute for Cancer Research, Glasgow, UK. Her work focuses on elucidating E3 mechanisms.
Danny T. Huang is a Senior Group Leader at the Beatson Institute for Cancer Research, Glasgow, UK and a Professor of cancer biology at the University of Glasgow, UK. His research focuses on using structural and biochemical studies to understand how E3s function.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 2.Dye BT, Schulman BA. Structural mechanisms underlying posttranslational modification by ubiquitin-like proteins. Annu Rev Biophys Biomol Struct. 2007;36:131–50. doi: 10.1146/annurev.biophys.36.040306.132820. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;1695:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Komander D, Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–29. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 5.Chau V, et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science. 1989;243:1576–83. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 6.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haglund K, Dikic I. The role of ubiquitylation in receptor endocytosis and endosomal sorting. J Cell Sci. 2012;125:265–75. doi: 10.1242/jcs.091280. [DOI] [PubMed] [Google Scholar]
- 8.Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–86. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Ulrich HD, Walden H. Ubiquitin signalling in DNA replication and repair. Nat Rev Mol Cell Biol. 2010;11:479–89. doi: 10.1038/nrm2921. [DOI] [PubMed] [Google Scholar]
- 10.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–41. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 11.Robzyk K, Recht J, Osley MA. Rad6-dependent ubiquitination of histone H2B in yeast. Science. 2000;287:501–4. doi: 10.1126/science.287.5452.501. [DOI] [PubMed] [Google Scholar]
- 12.Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–8. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- 13.Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nat Rev Mol Cell Biol. 2009;10:755–64. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle's dynamics and signaling. PLoS One. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Levkowitz G, et al. Ubiquitin ligase activity and tyrosine phosphorylation underlie suppression of growth factor signaling by c-Cbl/Sli-1. Mol Cell. 1999;4:1029–40. doi: 10.1016/s1097-2765(00)80231-2. [DOI] [PubMed] [Google Scholar]
- 16.Waterman H, et al. A mutant EGF-receptor defective in ubiquitylation and endocytosis unveils a role for Grb2 in negative signaling. EMBO J. 2002;21:303–13. doi: 10.1093/emboj/21.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima T, et al. Structural basis for the selection of glycosylated substrates by SCF(Fbs1) ubiquitin ligase. Proc Natl Acad Sci U S A. 2007;104:5777–81. doi: 10.1073/pnas.0610312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scrima A, et al. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–23. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El-Mahdy MA, et al. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J Biol Chem. 2006;281:13404–11. doi: 10.1074/jbc.M511834200. [DOI] [PubMed] [Google Scholar]
- 20.Sugasawa K, et al. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell. 2005;121:387–400. doi: 10.1016/j.cell.2005.02.035. [DOI] [PubMed] [Google Scholar]
- 21.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 22.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–9. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 23.Dominguez C, et al. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–44. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 24.Buetow L, et al. Activation of a primed RING E3-E2-ubiquitin complex by non-covalent ubiquitin. Mol Cell. 2015;58:297–310. doi: 10.1016/j.molcel.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Mace PD, et al. Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment. J Biol Chem. 2008;283:31633–40. doi: 10.1074/jbc.M804753200. [DOI] [PubMed] [Google Scholar]
- 26.Yin Q, et al. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009;16:658–66. doi: 10.1038/nsmb.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plechanovova A, et al. Mechanism of ubiquitylation by dimeric RING ligase RNF4. Nat Struct Mol Biol. 2011;18:1052–9. doi: 10.1038/nsmb.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1-BARD1 heterodimeric RING-RING complex. Nat Struct Biol. 2001;8:833–7. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 29.Buchwald G, et al. Structure and E3-ligase activity of the Ring-Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–74. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Badciong JC, Haas AL. MdmX is a RING finger ubiquitin ligase capable of synergistically enhancing Mdm2 ubiquitination. J Biol Chem. 2002;277:49668–75. doi: 10.1074/jbc.M208593200. [DOI] [PubMed] [Google Scholar]
- 31.Linares LK, Hengstermann A, Ciechanover A, Muller S, Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci U S A. 2003;100:12009–14. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z, et al. Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase complex. J Biol Chem. 2006;281:20643–9. doi: 10.1074/jbc.M602461200. [DOI] [PubMed] [Google Scholar]
- 33.Micale L, Chaignat E, Fusco C, Reymond A, Merla G. The tripartite motif: structure and function. Adv Exp Med Biol. 2012;770:11–25. [PubMed] [Google Scholar]
- 34.Lydeard JR, Schulman BA, Harper JW. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14:1050–61. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 36.Chang L, Barford D. Insights into the anaphase-promoting complex: a molecular machine that regulates mitosis. Curr Opin Struct Biol. 2014;29:1–9. doi: 10.1016/j.sbi.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Ohi MD, Vander Kooi CW, Rosenberg JA, Chazin WJ, Gould KL. Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat Struct Biol. 2003;10:250–5. doi: 10.1038/nsb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tu D, Li W, Ye Y, Brunger AT. Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc Natl Acad Sci U S A. 2007;104:15599–606. doi: 10.1073/pnas.0701369104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vander Kooi CW, et al. The Prp19 U-box crystal structure suggests a common dimeric architecture for a class of oligomeric E3 ubiquitin ligases. Biochemistry. 2006;45:121–30. doi: 10.1021/bi051787e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M, et al. Chaperoned ubiquitylation--crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell. 2005;20:525–38. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 41.Rotin D, Kumar S. Physiological functions of the HECT family of ubiquitin ligases. Nat Rev Mol Cell Biol. 2009;10:398–409. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 42.Huang L, et al. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade. Science. 1999;286:1321–6. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 43.Verdecia MA, et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol Cell. 2003;11:249–59. doi: 10.1016/s1097-2765(02)00774-8. [DOI] [PubMed] [Google Scholar]
- 44.Smit JJ, Sixma TK. RBR E3-ligases at work. EMBO Rep. 2014;15:142–54. doi: 10.1002/embr.201338166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spratt DE, Walden H, Shaw GS. RBR E3 ubiquitin ligases: new structures, new insights, new questions. Biochem J. 2014;458:421–37. doi: 10.1042/BJ20140006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J Biol Chem. 2000;275:35661–4. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, et al. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A. 2000;97:13354–9. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moynihan TP, et al. The ubiquitin-conjugating enzymes UbcH7 and UbcH8 interact with RING finger/IBR motif-containing domains of HHARI and H7-AP1. J Biol Chem. 1999;274:30963–8. doi: 10.1074/jbc.274.43.30963. [DOI] [PubMed] [Google Scholar]
- 49.Shimura H, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–5. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 50.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature. 2011;474:105–8. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ozkan E, Yu H, Deisenhofer J. Mechanistic insight into the allosteric activation of a ubiquitin-conjugating enzyme by RING-type ubiquitin ligases. Proc Natl Acad Sci U S A. 2005;102:18890–5. doi: 10.1073/pnas.0509418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petroski MD, Deshaies RJ. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell. 2005;123:1107–20. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 53.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell. 2011;42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Benirschke RC, et al. Molecular basis for the association of human E4B U box ubiquitin ligase with E2-conjugating enzymes UbcH5c and Ubc4. Structure. 2010;18:955–65. doi: 10.1016/j.str.2010.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plechanovova A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature. 2012;489:115–20. doi: 10.1038/nature11376. [First crystal structure of a dimeric RING E3 bound to E2–ubiquitin in a primed conformation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol. 2012;19:876–83. doi: 10.1038/nsmb.2379. [First crystal structure of a dimeric RING E3 bound to E2–ubiquitin in a primed conformation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pruneda JN, et al. Structure of an E3:E2~Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell. 2012;47:933–42. doi: 10.1016/j.molcel.2012.07.001. [Shows that RING/U-box E3s drive E2~ubiquitin into a closed conformation essential for catalysis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pruneda JN, Stoll KE, Bolton LJ, Brzovic PS, Klevit RE. Ubiquitin in motion: structural studies of the ubiquitin-conjugating enzyme approximately ubiquitin conjugate. Biochemistry. 2011;50:1624–33. doi: 10.1021/bi101913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soss SE, Klevit RE, Chazin WJ. Activation of UbcH5c~Ub is the result of a shift in interdomain motions of the conjugate bound to U-box E3 ligase E4B. Biochemistry. 2013;52:2991–9. doi: 10.1021/bi3015949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Page RC, Pruneda JN, Amick J, Klevit RE, Misra S. Structural insights into the conformation and oligomerization of E2~ubiquitin conjugates. Biochemistry. 2012;51:4175–87. doi: 10.1021/bi300058m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. Essentiality of a non-RING element in priming donor ubiquitin for catalysis by a monomeric E3. Nat Struct Mol Biol. 2013;20:982–6. doi: 10.1038/nsmb.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Branigan E, Plechanovova A, Jaffray EG, Naismith JH, Hay RT. Structural basis for the RING-catalyzed synthesis of K63-linked ubiquitin chains. Nat Struct Mol Biol. 2015;22:597–602. doi: 10.1038/nsmb.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435:687–92. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Scott DC, et al. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell. 2014;157:1671–84. doi: 10.1016/j.cell.2014.04.037. [Crystal structure of a RING E3-E2~ubiquitin like protein-substrate complex revealing the mechanism of substrate ligation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koliopoulos MG, Esposito D, Christodoulou E, Taylor IA, Rittinger K. Functional role of TRIM E3 ligase oligomerization and regulation of catalytic activity. EMBO J. 2016 doi: 10.15252/embj.201593741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell. 2011;144:769–81. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Berndsen CE, Wiener R, Yu IW, Ringel AE, Wolberger C. A conserved asparagine has a structural role in ubiquitin-conjugating enzymes. Nat Chem Biol. 2013;9:154–6. doi: 10.1038/nchembio.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yunus AA, Lima CD. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol. 2006;13:491–9. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]
- 69.Wright JD, Mace PD, Day CL. Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity. Nat Struct Mol Biol. 2016;23:45–52. doi: 10.1038/nsmb.3142. [DOI] [PubMed] [Google Scholar]
- 70.Das R, et al. Allosteric activation of E2-RING finger-mediated ubiquitylation by a structurally defined specific E2-binding region of gp78. Mol Cell. 2009;34:674–85. doi: 10.1016/j.molcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Metzger MB, et al. A structurally unique E2-binding domain activates ubiquitination by the ERAD E2, Ubc7p, through multiple mechanisms. Mol Cell. 2013;50:516–27. doi: 10.1016/j.molcel.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hibbert RG, Huang A, Boelens R, Sixma TK. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc Natl Acad Sci U S A. 2011;108:5590–5. doi: 10.1073/pnas.1017516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li S, et al. Insights into Ubiquitination from the Unique Clamp-Like Binding of the RING E3 AO7 to the E2 UbcH5B. J Biol Chem. 2015 doi: 10.1074/jbc.M115.685867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Das R, et al. Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine. EMBO J. 2013;32:2504–16. doi: 10.1038/emboj.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE. A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell. 2006;21:873–80. doi: 10.1016/j.molcel.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 76.Sakata E, et al. Crystal structure of UbcH5b~ubiquitin intermediate: insight into the formation of the self-assembled E2~Ub conjugates. Structure. 2010;18:138–47. doi: 10.1016/j.str.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 77.Ranaweera RS, Yang X. Auto-ubiquitination of Mdm2 enhances its substrate ubiquitin ligase activity. J Biol Chem. 2013;288:18939–46. doi: 10.1074/jbc.M113.454470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kamadurai HB, et al. Insights into ubiquitin transfer cascades from a structure of a UbcH5B approximately ubiquitin-HECT(NEDD4L) complex. Mol Cell. 2009;36:1095–102. doi: 10.1016/j.molcel.2009.11.010. [First crystal structure of a HECT E3 bound to E2~ubiquitin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nuber U, Scheffner M. Identification of determinants in E2 ubiquitin-conjugating enzymes required for hect E3 ubiquitin-protein ligase interaction. J Biol Chem. 1999;274:7576–82. doi: 10.1074/jbc.274.11.7576. [DOI] [PubMed] [Google Scholar]
- 80.Ogunjimi AA, et al. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol Cell. 2005;19:297–308. doi: 10.1016/j.molcel.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 81.Maspero E, et al. Structure of a ubiquitin-loaded HECT ligase reveals the molecular basis for catalytic priming. Nat Struct Mol Biol. 2013;20:696–701. doi: 10.1038/nsmb.2566. [First crystal structure of a primed HECT E3~ubiquitin complex.] [DOI] [PubMed] [Google Scholar]
- 82.Kamadurai HB, et al. Mechanism of ubiquitin ligation and lysine prioritization by a HECT E3. Elife. 2013;2:e00828. doi: 10.7554/eLife.00828. [First crystal structure of a HECT E3~ubiquitin-substrate peptide complex.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Salvat C, Wang G, Dastur A, Lyon N, Huibregtse JM. The -4 phenylalanine is required for substrate ubiquitination catalyzed by HECT ubiquitin ligases. J Biol Chem. 2004;279:18935–43. doi: 10.1074/jbc.M312201200. [DOI] [PubMed] [Google Scholar]
- 84.Ronchi VP, Klein JM, Haas AL. E6AP/UBE3A ubiquitin ligase harbors two E2~ubiquitin binding sites. J Biol Chem. 2013;288:10349–60. doi: 10.1074/jbc.M113.458059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.French ME, Kretzmann BR, Hicke L. Regulation of the RSP5 ubiquitin ligase by an intrinsic ubiquitin-binding site. J Biol Chem. 2009;284:12071–9. doi: 10.1074/jbc.M901106200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maspero E, et al. Structure of the HECT:ubiquitin complex and its role in ubiquitin chain elongation. EMBO Rep. 2011;12:342–9. doi: 10.1038/embor.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim HC, Steffen AM, Oldham ML, Chen J, Huibregtse JM. Structure and function of a HECT domain ubiquitin-binding site. EMBO Rep. 2011;12:334–41. doi: 10.1038/embor.2011.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stieglitz B, et al. Structural basis for ligase-specific conjugation of linear ubiquitin chains by HOIP. Nature. 2013;503:422–6. doi: 10.1038/nature12638. [First crystal structure of a RBR RING2 bound to donor and acceptor ubiquitin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spratt DE, et al. A molecular explanation for the recessive nature of parkinlinked Parkinson's disease. Nat Commun. 2013;4 doi: 10.1038/ncomms2983. 1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beasley SA, Hristova VA, Shaw GS. Structure of the Parkin inbetween-ring domain provides insights for E3-ligase dysfunction in autosomal recessive Parkinson's disease. Proc Natl Acad Sci U S A. 2007;104:3095–100. doi: 10.1073/pnas.0610548104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Spratt DE, Mercier P, Shaw GS. Structure of the HHARI catalytic domain shows glimpses of a HECT E3 ligase. PLoS One. 2013;8:e74047. doi: 10.1371/journal.pone.0074047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lechtenberg BC, et al. Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation. Nature. 2016 doi: 10.1038/nature16511. [First crystal structure of RBR domain bound to E2~ubiquitin.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duda DM, et al. Structure of HHARI, a RING-IBR-RING ubiquitin ligase: autoinhibition of an Ariadne-family E3 and insights into ligation mechanism. Structure. 2013;21:1030–41. doi: 10.1016/j.str.2013.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Riley BE, et al. Structure and function of Parkin E3 ubiquitin ligase reveals aspects of RING and HECT ligases. Nat Commun. 2013;4 doi: 10.1038/ncomms2982. 1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wauer T, Komander D. Structure of the human Parkin ligase domain in an autoinhibited state. EMBO J. 2013;32:2099–112. doi: 10.1038/emboj.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Trempe JF, et al. Structure of parkin reveals mechanisms for ubiquitin ligase activation. Science. 2013;340:1451–5. doi: 10.1126/science.1237908. [First crystal structure of full-length PARKIN in an autoinhibited conformation.] [DOI] [PubMed] [Google Scholar]
- 97.Kumar A, et al. Disruption of the autoinhibited state primes the E3 ligase parkin for activation and catalysis. EMBO J. 2015;34:2506–21. doi: 10.15252/embj.201592337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K. LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep. 2012;13:840–6. doi: 10.1038/embor.2012.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Smit JJ, et al. The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J. 2012;31:3833–44. doi: 10.1038/emboj.2012.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Christensen DE, Brzovic PS, Klevit RE. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–8. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 101.Rodrigo-Brenni MC, Morgan DO. Sequential E2s drive polyubiquitin chain assembly on APC targets. Cell. 2007;130:127–39. doi: 10.1016/j.cell.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 102.Jin L, Williamson A, Banerjee S, Philipp I, Rape M. Mechanism of ubiquitin-chain formation by the human anaphase-promoting complex. Cell. 2008;133:653–65. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.McGinty RK, Henrici RC, Tan S. Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature. 2014;514:591–6. doi: 10.1038/nature13890. [Crystal structure showing how RNF2-BMI1 monoubiquitinates histone.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Atomic structure of the APC/C and its mechanism of protein ubiquitination. Nature. 2015;522:450–4. doi: 10.1038/nature14471. [First atomic structure of APC/C revealing the mechanism of ubiquitination.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brown NG, et al. RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proc Natl Acad Sci U S A. 2015;112:5272–9. doi: 10.1073/pnas.1504161112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.McNatt MW, McKittrick I, West M, Odorizzi G. Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway. Mol Biol Cell. 2007;18:697–706. doi: 10.1091/mbc.E06-08-0663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oestreich AJ, et al. Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5. Mol Biol Cell. 2007;18:707–20. doi: 10.1091/mbc.E06-08-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stawiecka-Mirota M, et al. Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic. 2007;8:1280–96. doi: 10.1111/j.1600-0854.2007.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Watson H, Bonifacino JS. Direct binding to Rsp5p regulates ubiquitination-independent vacuolar transport of Sna3p. Mol Biol Cell. 2007;18:1781–9. doi: 10.1091/mbc.E06-10-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang H, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 111.Dou H, et al. Structural basis for autoinhibition and phosphorylation-dependent activation of c-Cbl. Nat Struct Mol Biol. 2012;19:184–92. doi: 10.1038/nsmb.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kobashigawa Y, et al. Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1110712108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Duda DM, et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Calabrese MF, et al. A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nat Struct Mol Biol. 2011;18:947–9. doi: 10.1038/nsmb.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zheng N, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 116.Chang L, Zhang Z, Yang J, McLaughlin SH, Barford D. Molecular architecture and mechanism of the anaphase-promoting complex. Nature. 2014;513:388–93. doi: 10.1038/nature13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Merbl Y, Kirschner MW. Large-scale detection of ubiquitination substrates using cell extracts and protein microarrays. Proc Natl Acad Sci U S A. 2009;106:2543–8. doi: 10.1073/pnas.0812892106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ordureau A, et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol Cell. 2014;56:360–75. doi: 10.1016/j.molcel.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol. 2006;13:915–20. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 120.Brown NG, et al. Mechanism of polyubiquitination by human anaphase-promoting complex: RING repurposing for ubiquitin chain assembly. Mol Cell. 2014;56:246–60. doi: 10.1016/j.molcel.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kelly A, Wickliffe KE, Song L, Fedrigo I, Rape M. Ubiquitin chain elongation requires E3-dependent tracking of the emerging conjugate. Mol Cell. 2014;56:232–45. doi: 10.1016/j.molcel.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 122.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 123.Frye JJ, et al. Electron microscopy structure of human APC/C(CDH1)-EMI1 reveals multimodal mechanism of E3 ligase shutdown. Nat Struct Mol Biol. 2013;20:827–35. doi: 10.1038/nsmb.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wang W, Kirschner MW. Emi1 preferentially inhibits ubiquitin chain elongation by the anaphase-promoting complex. Nat Cell Biol. 2013;15:797–806. doi: 10.1038/ncb2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fulda S, Vucic D. Targeting IAP proteins for therapeutic intervention in cancer. Nat Rev Drug Discov. 2012;11:109–24. doi: 10.1038/nrd3627. [DOI] [PubMed] [Google Scholar]
- 126.Dueber EC, et al. Antagonists induce a conformational change in cIAP1 that promotes autoubiquitination. Science. 2011;334:376–80. doi: 10.1126/science.1207862. [DOI] [PubMed] [Google Scholar]
- 127.Lopez J, et al. CARD-mediated autoinhibition of cIAP1's E3 ligase activity suppresses cell proliferation and migration. Mol Cell. 2011;42:569–83. doi: 10.1016/j.molcel.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 128.Phillips AH, et al. Internal motions prime cIAP1 for rapid activation. Nat Struct Mol Biol. 2014;21:1068–74. doi: 10.1038/nsmb.2916. [DOI] [PubMed] [Google Scholar]
- 129.Feltham R, et al. Smac mimetics activate the E3 ligase activity of cIAP1 protein by promoting RING domain dimerization. J Biol Chem. 2011;286:17015–28. doi: 10.1074/jbc.M111.222919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Mund T, Pelham HR. Control of the activity of WW-HECT domain E3 ubiquitin ligases by NDFIP proteins. EMBO Rep. 2009;10:501–7. doi: 10.1038/embor.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Riling C, et al. Itch WW Domains Inhibit Its E3 Ubiquitin Ligase Activity by Blocking E2-E3 Ligase Trans-thiolation. J Biol Chem. 2015;290:23875–87. doi: 10.1074/jbc.M115.649269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gallagher E, Gao M, Liu YC, Karin M. Activation of the E3 ubiquitin ligase Itch through a phosphorylation-induced conformational change. Proc Natl Acad Sci U S A. 2006;103:1717–22. doi: 10.1073/pnas.0510664103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wan L, et al. Cdh1 regulates osteoblast function through an APC/Cindependent modulation of Smurf1. Mol Cell. 2011;44:721–33. doi: 10.1016/j.molcel.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang J, et al. Calcium activates Nedd4 E3 ubiquitin ligases by releasing the C2 domain-mediated auto-inhibition. J Biol Chem. 2010;285:12279–88. doi: 10.1074/jbc.M109.086405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wiesner S, et al. Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain. Cell. 2007;130:651–62. doi: 10.1016/j.cell.2007.06.050. [DOI] [PubMed] [Google Scholar]
- 136.Mari S, et al. Structural and functional framework for the autoinhibition of Nedd4-family ubiquitin ligases. Structure. 2014;22:1639–49. doi: 10.1016/j.str.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 137.Aragon E, et al. Structural basis for the versatile interactions of Smad7 with regulator WW domains in TGF-beta Pathways. Structure. 2012;20:1726–36. doi: 10.1016/j.str.2012.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chaugule VK, et al. Autoregulation of Parkin activity through its ubiquitin-like domain. EMBO J. 2011;30:2853–67. doi: 10.1038/emboj.2011.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ordureau A, et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc Natl Acad Sci U S A. 2015;112:6637–42. doi: 10.1073/pnas.1506593112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kane LA, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–53. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kazlauskaite A, et al. Binding to serine 65-phosphorylated ubiquitin primes Parkin for optimal PINK1-dependent phosphorylation and activation. EMBO Rep. 2015;16:939–54. doi: 10.15252/embr.201540352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sarraf SA, et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature. 2013;496:372–6. doi: 10.1038/nature12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Bingol B, et al. The mitochondrial deubiquitinase USP30 opposes parkinmediated mitophagy. Nature. 2014;510:370–5. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 145.Sauve V, et al. A Ubl/ubiquitin switch in the activation of Parkin. EMBO J. 2015;34:2492–505. doi: 10.15252/embj.201592237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wauer T, Simicek M, Schubert A, Komander D. Mechanism of phospho-ubiquitin-induced PARKIN activation. Nature. 2015;524:370–4. doi: 10.1038/nature14879. [Crystal structure showing how phospho-ubiquitin activates PARKIN.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kulathu Y, Komander D. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat Rev Mol Cell Biol. 2012;13:508–23. doi: 10.1038/nrm3394. [DOI] [PubMed] [Google Scholar]
- 148.Rosner D, Schneider T, Schneider D, Scheffner M, Marx A. Click chemistry for targeted protein ubiquitylation and ubiquitin chain formation. Nat Protoc. 2015;10:1594–611. doi: 10.1038/nprot.2015.106. [DOI] [PubMed] [Google Scholar]
- 149.Eger S, et al. Generation of a mono-ubiquitinated PCNA mimic by click chemistry. Chembiochem. 2011;12:2807–12. doi: 10.1002/cbic.201100444. [DOI] [PubMed] [Google Scholar]
- 150.McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–6. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Chen J, Ai Y, Wang J, Haracska L, Zhuang Z. Chemically ubiquitylated PCNA as a probe for eukaryotic translesion DNA synthesis. Nat Chem Biol. 2010;6:270–2. doi: 10.1038/nchembio.316. [DOI] [PubMed] [Google Scholar]
- 152.Sommer S, Weikart ND, Brockmeyer A, Janning P, Mootz HD. Expanded click conjugation of recombinant proteins with ubiquitin-like modifiers reveals altered substrate preference of SUMO2-modified Ubc9. Angew Chem Int Ed Engl. 2011;50:9888–92. doi: 10.1002/anie.201102531. [DOI] [PubMed] [Google Scholar]
- 153.Oualid FE, Hameed DS, Atmioui DE, Hilkmann H, Ovaa H. Synthesis of atypical diubiquitin chains. Methods Mol Biol. 2012;832:597–609. doi: 10.1007/978-1-61779-474-2_42. [DOI] [PubMed] [Google Scholar]
- 154.Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi: 10.1146/annurev-biochem-051810-094654. [DOI] [PubMed] [Google Scholar]
- 155.Meyer HJ, Rape M. Enhanced protein degradation by branched ubiquitin chains. Cell. 2014;157:910–21. doi: 10.1016/j.cell.2014.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1