Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming (original) (raw)
. Author manuscript; available in PMC: 2014 Aug 25.
Abstract
Reprogramming of somatic cells to a pluripotent embryonic stem cell-like state has been achieved by nuclear transplantation of a somatic nucleus into an enucleated egg and most recently by introducing defined transcription factors into somatic cells. Nuclear reprogramming is of great medical interest as it has the potential to generate a source of patient-specific cells. Here, we review strategies to reprogram somatic cells to a pluripotent embryonic state and discuss our understanding of the molecular mechanisms of reprogramming based on recent insights into the regulatory circuitry of the pluripotent state.
Introduction
Stem cells ---characterized by the ability to both self-renew and to generate differentiated functional cell types--- have been derived from the embryo and from various sources of the postnatal animal. It is customary to classify stem cells according to their developmental potential (Table 1). In mammals only the zygote and early blastomeres are totipotent and can generate the whole organism including extraembryonic tissues. Mouse embryonic stem (ES) cells are an example of pluripotent cells that can self-renew and generate all cell types of the body in vivo and in culture but are not able to generate the extraembryonic trophoblast lineage (see Essay by J. Rossant, page XXX of this issue). Multipotent cells such as hematopoietic stem cells can give rise to all cell types within one particular lineage (see Review by S.H. Orkin and L.I. Zon, page XXX). Spermatogonial stem cells are an example of unipotent stem cells as they can only form sperm (see Minireview by R.M. Cinalli et al,. page XXX).
Nuclear transplantation (NT), also referred to as somatic cell nuclear transfer (SCNT) denotes the introduction of a nucleus from an somatic donor cell into an enucleated oocyte to generate a cloned animals such as Dolly (Wilmut et al, 1997). The generation of live animals by nuclear transfer (NT) has demonstrated that the epigenetic state of somatic cells, including that of terminally differentiated cells, while stable, is not irreversibly fixed but can be reprogrammed to an embryonic state that is capable of directing development of a new organism. In addition to providing an exciting experimental approach for elucidating the basic epigenetic mechanisms involved in embryonic development and disease, nuclear cloning technology is of potential interest for patient-specific transplantation medicine (see Essay by S.M. Wu et al., page XXX). However, any medical application is hampered by the inefficiency of the cloning process, the lack of knowledge of the underlying mechanisms and ethical concerns. One of the key issues raised by nuclear cloning relates to the mechanism of reprogramming, i.e. how to define the “reprogramming factors” in the egg cytoplasm that convert the epigenome of a somatic cell into that of an embryonic cell.
Table 1.
Definition of some terms
| Potency | Sum of developmental options accessible to cell |
|---|---|
| Totipotent | Ability to form all lineages of organism; in mammals only thezygote and the first cleavage blastomeres are totipotent. |
| Pluripotent | Ability to form all lineages of body. Example: embryonic stemcells |
| Multipotent | Ability of adult stem cells to form multiple cell types of onelineage. Example: hematopoietic stem cells |
| Unipotent | Cells form one cell type. Example: spermatogonial stem cells(can only generate sperm) |
| Reprogramming | Increase in potency, dedifferentiation. Can be induced bynuclear transfer, cell fusion, genetic manipulation |
| Transdifferentiation plasticity | Notion that somatic stem cells have broadened potency and cangenerate cells of other lineages, a concept that is controversial inmammals. |
This article focuses on two main topics. We will discuss strategies to reprogram somatic cells to a pluripotent embryonic state and we will review recent advances in defining the molecular circuitry that maintains a pluripotent state while allowing for differentiation into more specialized states in response to particular signaling cues. We will restrict our review to mammalian systems and refer the reader to a number of recent reviews on stem cells and nuclear reprogramming in other systems such as amphibians and invertebrates (Gurdon and Byrne, 2003; Sanchez Alvarado, 2006 ; also see Review by K.D.Birnbaum et al., page XXX)
Strategies of reprogramming somatic cells
Several different strategies such as nuclear transplantation, cellular fusion and culture induced reprogramming have been employed to induce the conversion of differentiated cells into an embryonic state (Figure 1). These experimental approaches have been extensively reviewed (Hochedlinger and Jaenisch, 2006; Yamanaka, 2007) and will only be briefly summarized here. Instead, our main focus will be on the most recently established strategy that uses transduction of defined factors into somatic cells to induce reprogramming. In normal development, cells transit in a unidirectional process from the totipotent zygote to pluripotent inner cell mass (ICM) and epiblast cells and to more restricted and eventually differentiated cells. These transitions occur in the context of the embryo as a result of cell-cell interactions and are characterized by distinct epigenetic modifications (Gan et al., 2007; Surani et al., 2007). It is important to realize, however, that cells growing in tissue culture, in contrast to cells in the embryo, are exposed to different selective conditions and this will result in cell states that are unlike those seen in vivo. For example, although embryonic stem (ES) cells or embryonic germ (EG) cells are derived from the ICM or from primordial germ cells, respectively, their growth and molecular characteristics are the product of tissue culture selection for rapid in vitro proliferation and this invariably will result in cells that are epigenetically and biologically different from their corresponding cells of origin.
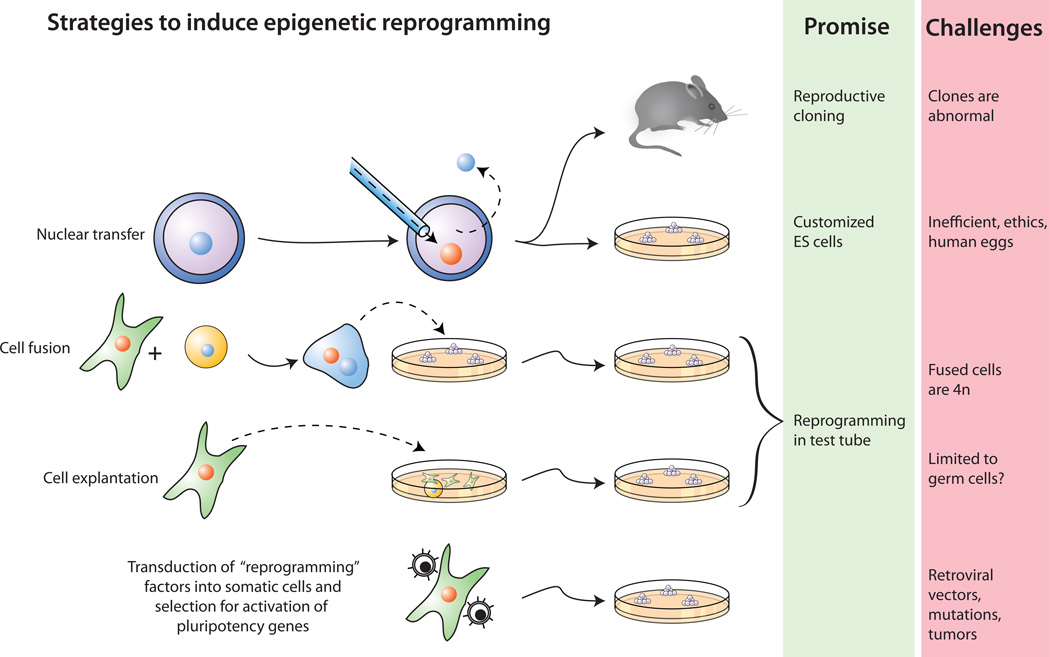
Figure 1. Four strategies to induce reprogramming of somatic cells.
(i) Nuclear transfer involves the injection of a somatic nucleus into an enucleated oocyte which, upon transfer into a surrogate mother, can give rise to a clone (“reproductive cloning”), or, upon explantation in culture, can give rise to genetically matched embryonic stem (ES) cells (“somatic cell nuclear transfer”, SCNT). (ii) Cell fusion of somatic cells with ES cells results in the generation of hybrids that show all features of pluripotent ES cells. (iii) Explantation of somatic cells in culture selects for immortal cell lines that may be pluripotent or multipotent. At present, spermatogonial stem cells are the only source of pluripotent cells that can be derived from postnatal animals. (iv) Transduction of somatic cells with defined factors can initiate reprogramming to a pluripotent state.
In this review we will focus on cells grown in vitro and it is important to emphasize that cells adapted to proliferate in tissue culture represent only a proxy for the in vivo situation and may at best approximate the properties of cells in the embryo (Gan et al., 2007; Surani et al., 2007). Consequently, concepts such as “pluripotency”, “multipotency” or “differentiation” of cultured cells rely on operational criteria and are typically assessed by different functional and molecular standards. The least stringent functional assay for the developmental potential of a cultured cell is in vitro differentiation followed, with increasing stringency, by the generation of teratomas (germ cell tumors), chimera formation and germ line contribution (Table 2). The most rigorous test for developmental potency is the injection of cells into 4n host blastocysts (Eggan et al., 2001; Nágy et al., 1990), which results in animals composed only of the injected donor cells (“all ES” embryos or animals) rather than a chimeric composite of injected and host derived cells.
Table 2.
Commonly used functional criteria to assess the developmental potential of cells
| Assay | Experimental approach | Limitations |
|---|---|---|
| _In vitro_differentiation | Differentiation induced in culturedcells and cells are assayed for theexpression of cell type specificmarkers | The expression fordifferentiation markers is notest for functionality; markerexpression can be due tocellular stress response |
| Teratomaformation | Induction of tumors demonstratingthe potential to generatedifferentiated cell types of variouslineages | Does not test for the ability ofcells to promote normaldevelopment |
| Chimeraformation | Contribution of cells to normaldevelopment following injection intohost blastocyst | Host-derived cells in chimeramay complement cell non-autonomous defects |
| Germ linecontribution | Ability of test cells to generatefunctional germ cells | Excludes genetic but notepigenetic defects that couldinterfere with promotingdevelopment |
| Tetraploidcomplementation | Injection of test cells into 4n hostblastocyst. Because 4n host cellscannot contribute to somatic lineagesembryo is exclusively composed oftest cells. | Most stringent test forpluripotency; does not test forthe ability to formtrophectoderm (placental)lineage |
Nuclear transplantation
Nuclear cloning provided proof for the notion that irreversible alterations of the genome are not required for normal development. However, because no genetic marker was available in the initial cloning experiments it remained an open question whether terminally differentiated cells could be reprogrammed to a totipotent state. The successful generation of cloned mice from genetically marked lymphoid cells (Hochedlinger and Jaenisch, 2002; Inoue et al., 2005) or from postmitotic neurons (Eggan et al., 2004; Li et al., 2004) unambiguously demonstrated that terminal differentiation does not restrict the potential of the nucleus to support development. Cloning from terminally differentiated donors cells is, however, inefficient and was in many instances successful only when a “two step” procedure, which involved the generation of cloned ES cells as an intermediate, was used. These observations suggested that the differentiation state of the donor cell affects the efficiency of producing cloned animals, with less differentiated cells being more amenable to epigenetic reprogramming. For example, the generation of cloned ES cells from neurons was less efficient than that from neural stem cells (Blelloch et al., 2006; Inoue et al., 2007) and direct cloning of mice from skin stem cells was more efficient than cloning from transiently amplifying keratinoyctes (transiently amplifying keratinocytes are non-self renewing cells derived from the skin stem cells that are on the path to generate differentiated cells; Li et al., 2007). However, because the cloning process is affected by many other parameters, such as cell cycle and the physical characteristics of the donor nucleus, it has remained unresolved whether cloning efficiency decreases with progressive cell differentiation in all cases (for discussion of this issue see Hochedlinger and Jaenisch, 2006; Oback and Wells, 2007). For example, it has been argued that nuclei from granulocytes are more efficient donors than nuclei from hematopoietic stem cells (Sung et al., 2006), but the validity of these claims has been challenged (Hochedlinger and Jaenisch, 2007).
Nuclear cloning is an inherently inefficient process due to faulty reprogramming, which results in the death of most clones soon after implantation or birth of clones with serious abnormalities (Hochedlinger and Jaenisch, 2003; Yang et al., 2007). It therefore became important to determine whether faulty reprogramming would affect the therapeutic utility of patient specific or “customized” ES cells derived by NT. Subsequent experiments showed no molecular or biological differences when ES cells derived from fertilized embryos or by NT were compared (Brambrink et al., 2006; Wakayama et al., 2006), indicating that NT ES cells are as useful for therapeutic application as ES cells derived from fertilized embryos.
Based upon earlier results with mice it was postulated that cloning of mammals could be accomplished only when oocytes rather than fertilized eggs were used as nuclear recipients (McGrath and Solter, 1984). Given the difficulty of obtaining unfertilized human oocytes, this result posed a significant impediment for the potential of nuclear transplantation approaches for therapeutic application. It is of considerable interest, therefore, that cloned ES cells and mice can be generated from somatic donor nuclei transplanted into enucleated zygote recipients if drug-induced synchronization of donor cells and zygote was employed (Egli et al., 2007; Greda et al., 2006). Because fertilized human embryos are easier to obtain than unfertilized human eggs, the adaptation of this strategy to the human system would solve major practical problems that hamper the eventual application of nuclear transplantation for medicine.
Fusion of somatic cells and embryonic stem cells
Epigenetic reprogramming of somatic nuclei to an undifferentiated state has been demonstrated in murine hybrids produced by fusion of embryonic cells with somatic cells. Hybrids between various somatic cells and embryonic carcinoma cells (Solter, 2006), embryonic germ (EG) or ES cells (Zwaka and Thomson, 2005) share many features with the parental embryonic cells indicating that the pluripotent phenotype is dominant in such fusion products. As with mouse (Tada et al., 2001), human ES cells have the potential to reprogram somatic nuclei after fusion (Cowan et al., 2005; Yu et al., 2006). Activation of silent pluripotency markers such as Oct4 or reactivation of the inactive somatic X chromosome provided molecular evidence for reprogramming of the somatic genome in the hybrid cells. It has been suggested that DNA replication is essential for the activation of pluripotency markers which is first observed 2 days after fusion (Do and Scholer, 2004) and that forced over-expression of Nanog in ES cells promotes pluripotency when fused with neural stem cells (Silva et al., 2006). However, the inefficiency of the fusion process has impeded the study of molecular mechanisms involved in somatic reprogramming.
While the fusion method does not rely on nuclear transfer to generate pluripotent cells, tetraploidy of the reprogrammed cells presents a major shortcoming for using this approach for customized cell therapy. Although selective elimination of some ES cell derived chromosomes is possible (Matsumura et al., 2007) it may be difficult to generate diploid reprogrammed cells in this way due to the risk of generating large-scale genomic instability. In another approach, short-term incubation of permeabilized somatic cells with extracts of ES cells was claimed to result in genome-wide reprogramming (Taranger et al., 2005), but convincing evidence for reprogramming of the somatic cell genome is still lacking.
Culture-induced reprogramming
Pluripotent cells have been derived from embryonic sources such as blastomeres and ICM of blastocysts (ES cells), the epiblast (EpiSC cells), primordial germ cells (EG cells) and postnatal spermatogonial stem cells (“maGSCs”, “ES-like” cells) (Table 3) (see Essays by J. Rossant, page XXX; J. Silva and A. Smith, page XXX). Donor cells from the germ cell lineage such as PGCs or spermatogonial stem cells are known to be unipotent in vivo but it has been shown that pluripotent “ES-like” cells (Kanatsu-Shinohara et al., 2004) or “maGSCs” (Guan et al., 2006) can be isolated after prolonged in vitro culture. While most of these pluripotent cell types were capable of in vitro differentiation and teratoma formation, only ES, EG, EC and spermatogonial stem cell-derived maGCSs or ES-like cells were pluripotent by more stringent criteria **(**compare Table 2), as they were able to form postnatal chimeras and contribute to the germ line. Recently, multipotent adult spermatogonial stem cells (MASCs) were derived from testicular spermatogonial stem cells of adult mice and these cells had an expression profile different from that of ES cells (Seandel et al., 2007) but similar to EpiSC cells, which were derived from the epiblast of post-implantation mouse embryos (Brons et al., 2007; Tesar et al., 2007). While both MASCs and EpiSCs were able to differentiate in vitro and to generate teratomas in vivo, they were unable to form chimeras in contrast to ES, EG, EC and maGSCs cells. MASCs and EpiSCs were similar to human ES cells in many ways: they required FGF but not LIF for growth; they were able to express trophoblast markers in vitro; and they displayed expression profiles that were more typical of human than mouse ES cells (see Essay by J. Rossant, page XXX). These similarities raise the possibility that the embryonic origin of human ES cells may be the epiblast stage in contrast to that of mouse ES cells, which are derived from the inner cell mass of the blastocyst (ICM). It may be that the present isolation protocols of human ES cells using FGF and activin selects against “true” ES cells and results in cells that resemble mouse EpiSCs rather than mouse ES cells (Lovell-Badge, 2007). It is possible that the existing human ES cells, the murine EpiSCs and MASCs are multipotent cell types that are endowed with a more restricted developmental potential than pluripotent mouse ES cells.
Table 3.
Generation of pluripotent cells from various sources and the different criteria used for assessing developmental potential.
| Donor cell/tissue | Pluripotentcells | Criteria for pluripotency | ref | ||||
|---|---|---|---|---|---|---|---|
| _In vitro_differentiation | Teratoma | Postnatalchimera | Germline | 4n complementation | |||
| Murine oocyte | ParthogeneticES cells | Yes | Yes | Yes | Yes | No | 1 |
| Blastomere | ES | 2 | |||||
| ICM | Yes | 3 | |||||
| PGC | EG, EC | No | 4 | ||||
| Spermato-gonial stemcells | GMCS,maSSC,MASC | 5 | |||||
| Epiblast | EpiSC | No | No | 6 | |||
| Human oocyte | ParthogeneticES cells | 7 | |||||
| Humanblastocyst | Hu ES cells | 8 | |||||
| Bonemarrowderived cells | MAPC | No | ? | 9 | |||
| Cord bloodcells | No | 10 | |||||
| Neural cells | Neurospherederived | 11 |
It remains an open question whether somatic stem cells derived from the postnatal animal are pluripotent and whether truly pluripotent cells can be isolated from somatic tissues by expansion in culture (as can been done with unipotential PGCs or spermatogonial stem cells). At issue is whether somatic stem cells of tissues such as the hematopoietic system, the intestine or the skin that are multipotent and can generate all cell types in their respective lineages in vivo are inherently plastic and capable of “transdifferentiation” into cell types of other lineages (see table 1 for definitions of terms). Claims for cellular “plasticity” rest on two criteria: (i) in vitro differentiation to different cell types and (ii) transplantation of the cells into blastocysts or postnatal mice to assess their ability to contribute in vivo to different tissues.
As summarized in Table 3, in vitro differentiation, often used as the only criterion for transdifferentiation, is the least stringent measure for pluripotency (compare Table 2). The expression of a limited set of differentiation markers as assayed in most studies is often insufficient for concluding that a cell has been converted to a new state of differentiation and cellular function. A case in point is the activation of commonly used neural differentiation markers such as nestin, NeuroD1 and beta-III-tubulin in bone marrow or skin derived cells which can reflect a cellular stress response rather than indicating differentiation into the neural lineage (Croft and Przyborski, 2006; Neuhuber et al., 2004). Thus, the ability of aberrant responses of lineage-restricted cells to inappropriate physiological signals may be due to “cellular mimicry” (Rizzino, 2007) and may not reflect transdifferentiation to another lineage.
Claims for in vivo transdifferentiation of cells derived from bone marrow, brain or skin to cells of different lineages have remained controversial because of flaws in experimental design or interpretation (Joseph and Morrison, 2005; Wagers and Weissman, 2004). For example, the presence of genetically marked cells in non-hematopoietic tissues such as heart, muscle or brain recipients after transplantation of marked bone marrow cells may be due to the circulation and homing of the transplanted cells to the respective tissues (Balsam et al., 2004; Murry et al., 2004) rather than to transdifferentiation of the cells. Alternatively, autofluorescence may explain the detection of GFP marker expression rather than the incorporation of donor cells into tissues of transplanted mice (Jackson et al., 2004). In addition, fusion between donor and recipient cells may account for the expression of the donor marker in cells of the host (Alvarez-Dolado et al., 2003; Terada et al., 2002; Wang et al., 2003; Ying et al., 2002). For example, the detection of amniotic stem cell-derived cells in the brains of adult mice that were injected as newborns (De Coppi et al., 2007) is not a sufficiently strong criterion for transdifferentiation in the absence of stringent characterization of cell specific marker expression and functional integration of the donor-derived cells into the host tissue. While it is possible that prolonged in vitro culture induces transdifferentiation and pluripotency, this has not been clearly proven. For instance, although bone marrow derived MAPC (mulitpotential adult progenitor) cells were able to express various differentiation-specific markers upon in vitro differentiation, they were unable to generate teratomas (Jiang et al., 2002). Injection of cells into blastocysts has been used as a more stringent assay for pluripotency. Though the generation of a single high contribution postnatal chimera from MAPCs was reported (Jiang et al., 2002), this result has not been independently confirmed to date. Also, the mere detection of marked cells in midgestation embryos (Clarke et al., 2000) provides insufficient evidence to conclude that the donor cells “contributed to development”. The demonstration of functional integration of donor cells into viable late stage embryos or postnatal chimeras is needed to make this conclusion. Finally, the evaluation of plasticity and transdifferentiation is further complicated by the observation that soluble factors secreted by mesenchymal stem cells can alter the tissue microenvironment after transplantation (for recent review of this controversial field see Phinney and Prockop, 2007).
In summary, pluripotency and transdifferentiation of somatic cells remains an unproven concept. While unexpected transformation events may occur in somatic lineages, such events are exceedingly rare, are not a major force in physiological repair and may simply be due to events such as cell fusion.
Reprogramming by defined transcription factors
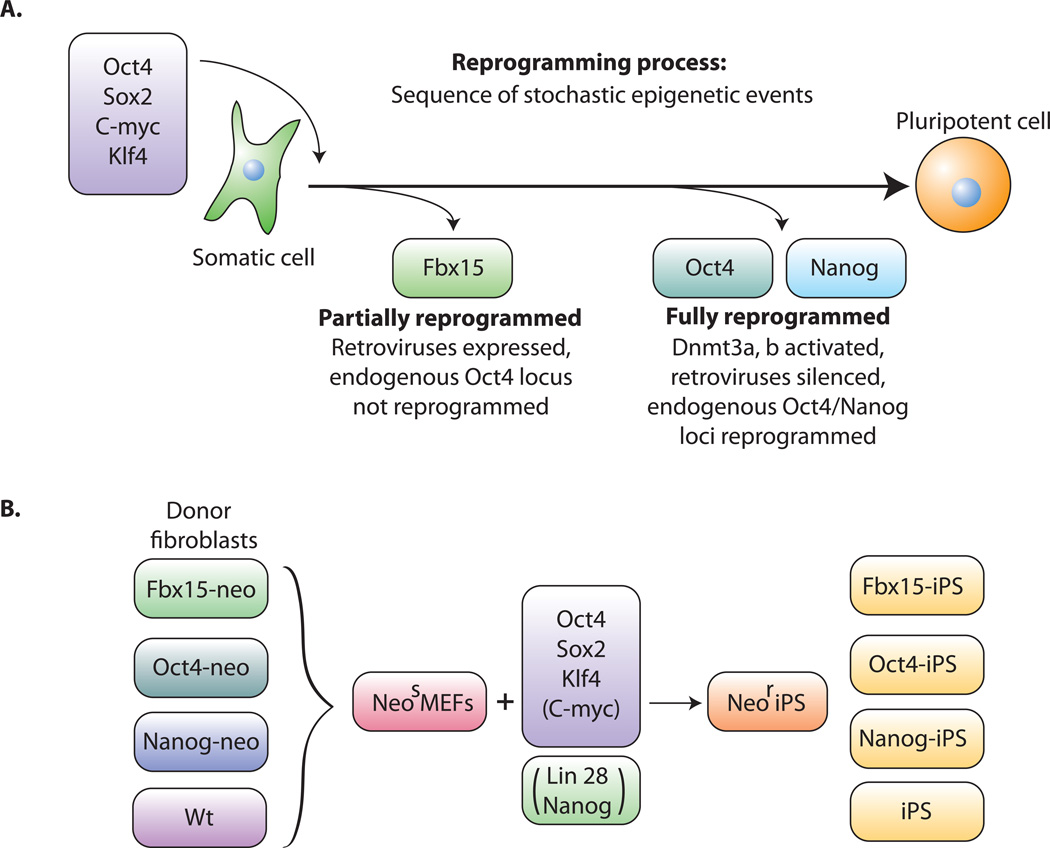
Takahashi and Yamanaka recently achieved a significant breakthrough in reprogramming somatic cells back to an ES-like state (Takahashi and Yamanaka, 2006). They successfully reprogrammed mouse embryonic fibroblasts (MEFs) and adult fibroblasts to pluripotent ES-like cells after viral-mediated transduction of the four transcription factors Oct4, Sox2, c-myc and Klf4 followed by selection for activation of the Oct4 target gene Fbx15 (Figure 2a) Cells that had activated Fbx15 were coined iPS (induced pluripotent stem) cells and were shown to be pluripotent by their ability to form teratomas although they were unable to generate live chimeras. This pluripotent state was dependent on the continuous viral expression of the transduced Oct4 and Sox2 genes, whereas the endogenous Oct4 and Nanog genes were either not expressed or were expressed at a lower level than in ES cells and their respective promoters were found to be largely methylated. This is consistent with the conclusion that the Fbx15-iPS cells did not correspond to ES cells but may have represented an incomplete state of reprogramming. While genetic experiments had established that Oct4 and Sox2 are essential for pluripotency (Chambers and Smith, 2004; Ivanova et al., 2006; Masui et al., 2007), the role of the two oncogenes c-myc and Klf4 in reprogramming is less clear. Some of these oncogenes may, in fact, be dispensable for reprogramming as both mouse and human iPS cells have been obtained in the absence of c-myc transduction, although with low efficiency (Nakagawa et al., 2008; Wernig et al., 2008; Yu et al., 2007). ).
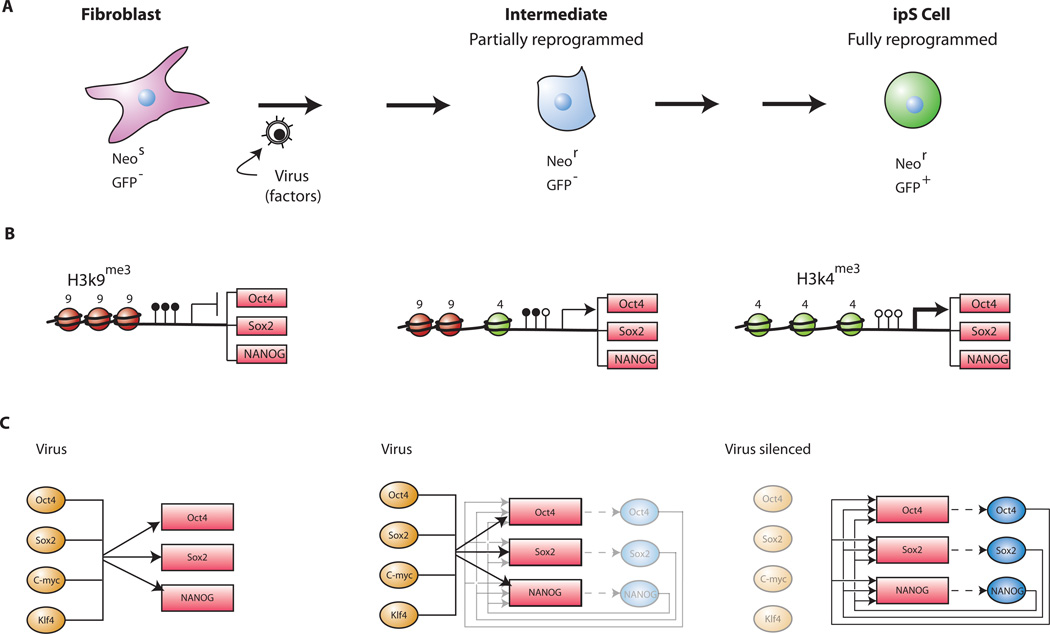
Figure 2. Reprogramming of somatic cells to a pluripotent state.
A. Transduction of the four transcription factors Oct4, Sox2, c-myc and Klf4 into fibroblasts initiates the conversion to partially reprogrammed cells that express Fbx15 or to fully reprogrammed iPS cells that express Oct4 or Nanog. The process involves a sequence of stochastic epigenetic events. B. Selection schemes. Cells carrying a drug resistance marker in the Fbx15, the Oct4 or the Nanog gene are transduced with the four factors and selected for drug resistance.
When activation of the endogenous Oct4 or Nanog genes was used as a more stringent selection criterion for pluripotency (Figure 2b), the resulting Oct4-iPS or Nanog-iPS cells, in contrast to Fbx15-iPS cells, were fully reprogrammed to a pluripotent, ES cell state by molecular and biological criteria (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007). (i) Global gene expression and the chromatin configuration of Oct4 or Nanog-selected iPS cells were indistinguishable from those of ES cells. (ii) In contrast to Fbx15-iPS cells, the pluripotent state in Oct4 or Nanog-iPS cells depended on the activity of the fully reprogrammed and hypomethylated endogenous Oct4 and Nanog promoters and not on the virally transduced factors. This is because the Moloney virus vectors, while highly expressed in the infected fibroblasts, were inactive in Oct4- and Nanog-iPS cells, consistent with the well-established evidence that Moloney viruses are strong targets for silencing in embryonic cells (Jähner et al., 1982; Okano et al., 1999) due to activation of the de novo methyltransferases Dnmt3a and Dnmt3b during the reprogramming process. (iii) Similar to somatic cell-ES cell fusion hybrid cell-mediated reprogramming, the inactive X chromosome of the somatic donor cells was reactivated in iPS cells (Maherali et al., 2007). (iv) Importantly, Oct4 and Nanog iPS cells generated postnatal chimeras, contributed to the germ line (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007) and generated late gestation embryos through tetraploid complementation (Wernig et al., 2007), the most stringent test for developmental potency (Table 2). Thus, all molecular and biological evidence indicated that Oct4 and Nanog iPS cells were indistinguishable from, if not identical to, ES cells. (v) Pluripotency markers such as alkaline phosphatase (AP), SSEA1 and Oct4 or Nanog appear sequentially during the reprogramming process (Fig 3a); (Brambrink et al., 2008; Wernig et al., 2007). (vi) Finally, evidence obtained by using inducible lentivirus-based vectors indicated that the four factors must be expressed in the infected MEFs for more than 12 days in order to generate iPS cells (Brambrink et al., 2008) (Figure 3a).
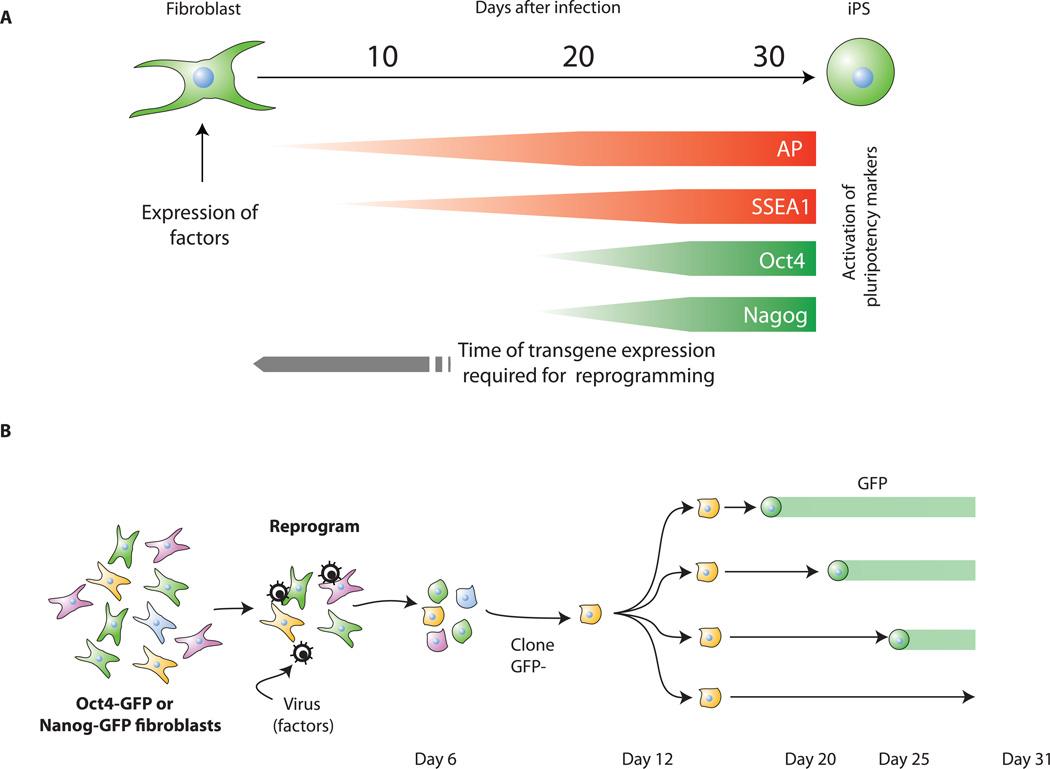
Figure 3. Reprogramming involves sequential activation of pluripotency markers and stochastic epigenetic events.
A. Kinetics of pluripotency-marker appearance. Alkaline phosphatase (AP) and SSEA1 positive cells are already detected 3 and 9 days, respectively, after factor transduction whereas GFP expressed from the endogenous Oct4 or Nanog loci first appear only after 2 weeks. The virally transduced factors need to be expressed for about 2 weeks to initiate the reprogramming process (Brambrink et al., 2008).
B. Oct4-GFP or Nanog-GFP fibroblasts were transduced with the four factors. Colonies displaying a transformed phenotype, were GFP negative and were cloned a few days after infection. Further cloning yielded subclones that activated GFP at different times (Meissner et al., 2007). Because the subclones were derived from the same infected cell, stochastic epigenetic events must be important for reprogramming.
Expression of the reprogramming factors in fibroblasts appears to initiate a sequence of stochastic events that eventually leads to a small fraction of iPS cells. This is supported by clonal analyses demonstrating that the activation of pluripotency markers can occur at different times after infection in individual mitotic daughter cells of the same infected fibroblast (Meissner et al., 2007). Thus, ectopic expression of Oct4, Sox2, c-myc and Klf4 may trigger a sequence of epigenetic events such as chromatin modifications or changes in DNA methylation that eventually result in the pluripotent state of some infected cells but not others even though they carry the identical combination of proviruses (Figure 3b). These experiments also suggested that the frequency of reprogramming increased with time, resulting in up to 0.5% of the input MEFs giving rise to iPS cells at 3 to 4 weeks after infection (Meissner et al., 2007). An unresolved issue is whether the partially reprogrammed iPS cells selected for the activation of Fbx15 (Takahashi and Yamanaka, 2006) correspond to stable intermediates in the reprogramming process to fully reprogrammed iPS cells (Figure 2a) or whether they represent a genetically homogeneous population of cells that are at different stages of stochastic reprogramming (Figure 3b).
The original isolation of iPS cells was based upon retrovirus-mediated transduction of oncogenes and on drug dependent selection for Fbx15, Oct4 or Nanog activation. These two experimental requirements seriously hinder the eventual application of the in vitro reprogramming approach for therapeutic use in humans because mice derived from iPS cells frequently developed cancer (Okita et al., 2007) and because the isolation of human iPS cells cannot be based on genetically modified donor cells. Some of these limitations have been overcome in recent experiments. First, in an effort to reduce the risk of tumors in iPS cell-derived chimeras, more recent experiments showed that c-myc is dispensable for reprogramming (Nakagawa et al., 2008; Wernig et al., 2008; Yu et al., 2007) though the reprogramming process was significantly delayed and less efficient in the absence of this oncogene. While mice derived from these iPS cells will not develop c-myc-induced tumors (Nakagawa et al., 2008; Wernig et al., 2008), it is not clear whether the transduction of other retrovirus-transduced transcription factors such as Oct4 (Hochedlinger et al., 2005) will cause tumors at later stages. Secondly, fully reprogrammed, genetically unmodified mouse fibroblasts were isolated based only on morphological criteria, as reprogramming occurred frequently enough to be detectable in culture (Blelloch et al., 2007; Meissner et al., 2007). Subsequent to these studies, human iPS cells were isolated from genetically unmodified fibroblasts (Takahashi et al., 2007; Yu et al., 2007; Park et al, 2008) indicating that combinations of factors similar to those used for reprogramming of mouse cells was also effective for human cells.
Which of the original factors are essential for the reprogramming process? It appears that c-myc significantly enhances and accelerates the process but is dispensable. Also, human iPS cells have been obtained by exposing fibroblasts only to Oct4, Sox2 and lin28 (Yu et al., 2007), an RNA binding protein, suggesting that alternative combinations of factors may initiate reprogramming. It is possible that Oct4, which in normal development is already expressed in the oocyte and may be the most upstream gene in the molecular circuitry of pluripotency (see later) is the only obligatory factor to initiate reprogramming and that other factors serve to accelerate the process and to increase efficiency.
One of the promises of patient-specific ES cells is the potential for customized therapy of diseases (see Essay by S.M. Wu et al., page XXX and Review by C.E. Murry and G. Keller, page XXX). Previous studies have shown that disease-specific ES cells produced by nuclear cloning in combination with gene correction can be used to correct an immunological disorder in a proof of principle experiment in mice (Rideout et al., 2002). In a similar approach we recently demonstrated that iPS cells derived from skin cells of a mouse with sickle cell anemia were able to fully restore normal blood function when transplanted into diseased mice (Hanna et al., 2007).
Molecular circuitry of pluripotency
The gene expression program of pluripotent ES cells is a product of regulation by specific transcription factors, chromatin modifying enzymes, regulatory RNA molecules and signal transduction pathways (Figure 4). Recent studies have provided new insights into how the key ES cell regulators work together to produce the pluripotent state.
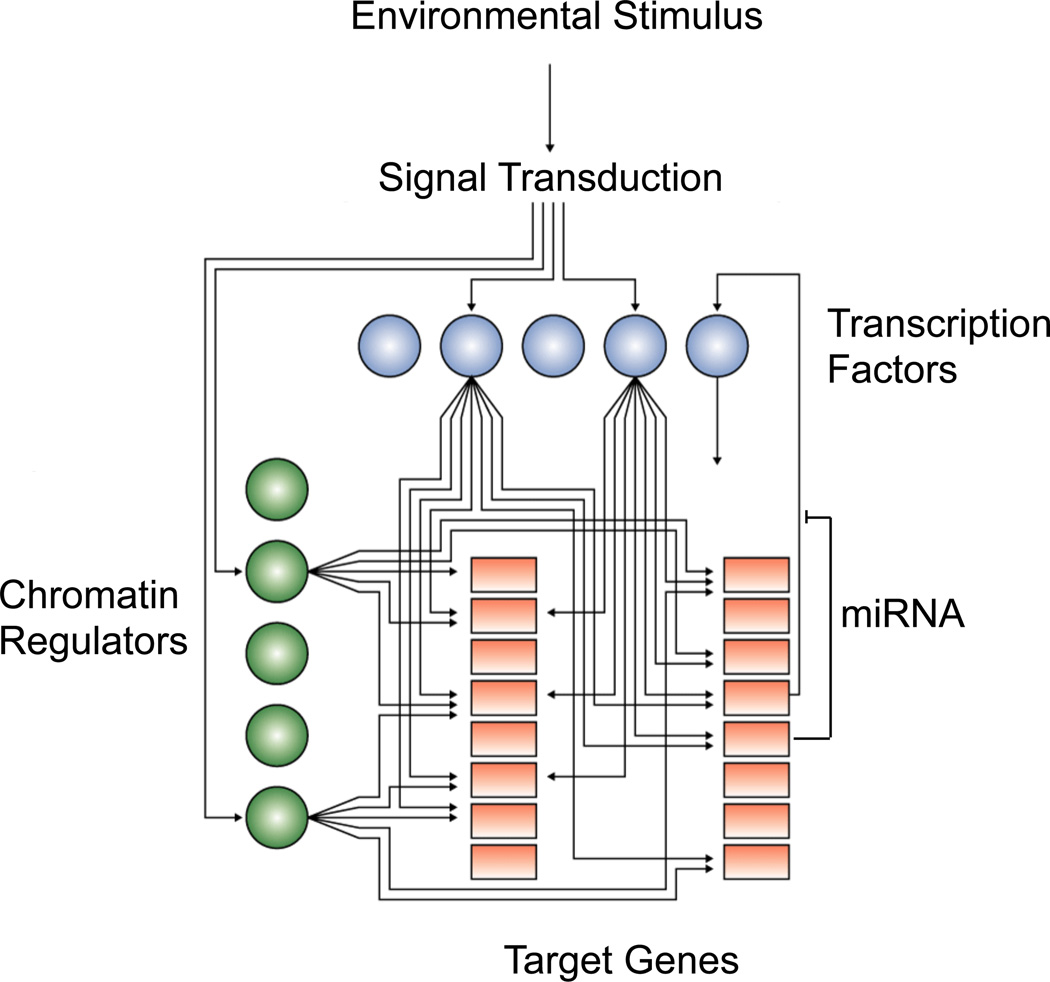
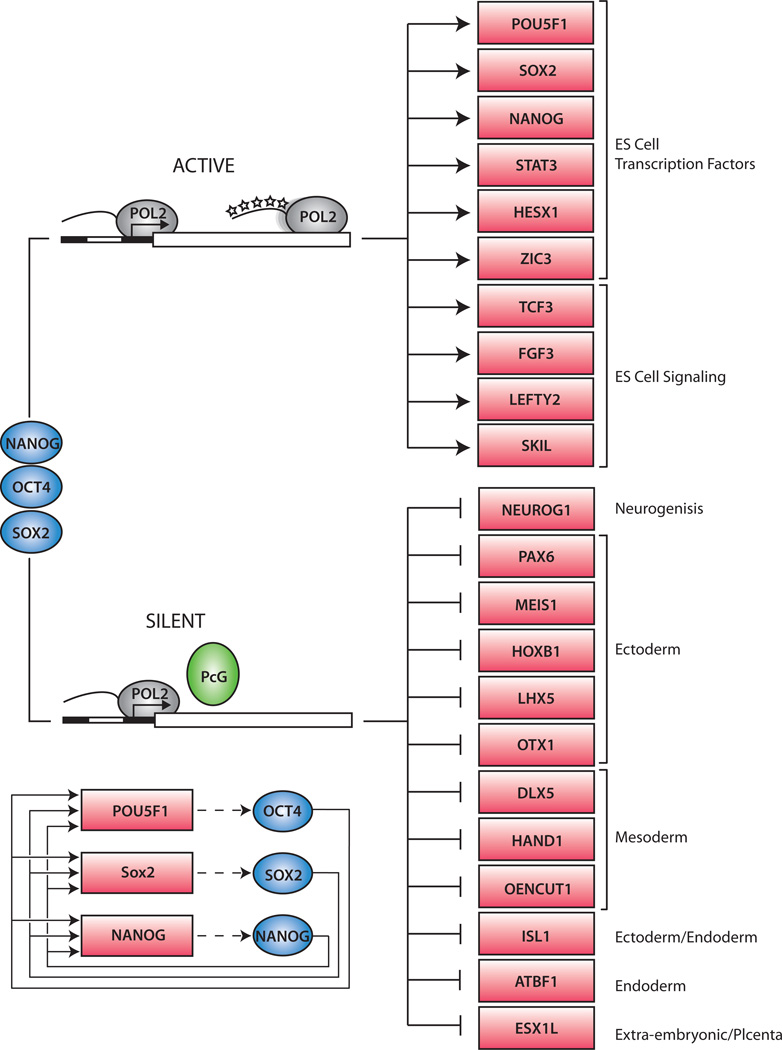
Figure 4. Pluripotency and the transcriptional regulatory circuitry.
Cartoon showing hypothetic connections between signal transduction pathways, transcription factors (blue balls), chromatin regulators (green balls) and their target genes (orange squares) to form an image of transcriptional regulatory circuitry. Some target genes produce miRNAs, which function at post-transcriptional levels.
ES cell transcription factors
Genetic studies first showed that the homeodomain transcription factors Oct4 and Nanog are essential regulators of early development and ES cell identity (Chambers et al., 2003; Chambers and Smith, 2004; Mitsui et al., 2003; Nichols et al., 1998). These transcription factors are expressed both in pluripotent ES cells and in the inner cell mass (ICM) of the blastocyst from which ES cells are derived. Disruption of Oct4 and Nanog causes loss of pluripotency and inappropriate differentiation of ICM and ES cells to trophectoderm and extra-embryonic endoderm, respectively (Chambers et al., 2003; Nichols et al., 1998; Ying et al., 2002). However, recent evidence suggests that Nanog may function to stabilize the pluripotent state rather than being essential for maintaining pluripotency of ES cells (Chambers et al., 2007) (see Essay by J. Silva and A. Smith, page XXX). Oct4 can heterodimerize with the HMG-box transcription factor Sox2 in ES cells and Sox2 contributes to pluripotency, at least in part, by regulating Oct4 levels (Masui et al., 2007). Oct4 is rapidly and apparently completely silenced during early cellular differentiation. The key roles played by Oct4, Sox2, and Nanog during early development and their unique expression pattern (Chambers et al., 2003; Hart et al., 2004; Nichols et al., 1998) make it likely that these regulators are central to the transcriptional regulatory hierarchy that specifies embryonic stem cell identity.
Identification of the genes occupied by Oct4, Sox2 and Nanog through genome-wide location analysis has provided insights into the molecular mechanisms by which these transcription factors contribute to pluripotency in human and murine ES cells (Boyer et al., 2005; Loh et al., 2006). These experiments yielded three key findings (Figure 5): 1) Oct4, Sox2 and Nanog bind together at their own promoters to form an interconnected autoregulatory loop, 2) the three factors often co-occupy their target genes, and 3) Oct4, Sox2 and Nanog collectively target two sets of genes, one that is actively expressed and another that is silent in ES cells, but remains poised for subsequent expression during cellular differentiation (Boyer et al., 2005). These observations, described in more detail below, revealed key features of the genetic logic of pluripotency and provided clues as to how multiple transcription regulators can coordinately control cell identity.
Figure 5. Model of core ES cell regulatory circuitry.
The Oct4, Sox2 and Nanog transcription factors (blue) occupy actively transcribed genes, including transcription factors and signaling components necessary to maintain the ES cell state. The three regulators also occupy silent genes encoding transcription factors that, if expressed, would promote other more differentiated cell states. At this latter set of genes, RNA polymerase II initiates transcription but does not produce complete transcripts due to the repressive action of PcG proteins. The PcG proteins prevent RNA polymerase from transitioning into a fully modified transcription elongation apparatus (represented by phosphorylated “stars” on the tail of the POL2 enzyme at the top of the figure). The interconnected autoregulatory loop, where Oct4, Nanog and Sox2 bind together at each of their own promoters, is shown in the bottom left of the figure.
Autoregulatory circuitry
Oct4, Sox2 and Nanog all bind to their own promoters, as well the promoters of the genes encoding the two other factors (Figure 5) (Boyer et al., 2005). This autoregulatory circuitry suggests that the three factors function collaboratively to maintain their own expression. Autoregulation is thought to enhance the stability of gene expression (Alon, 2007), which may facilitate maintenance of the pluripotent state. Autoregulatory loops appear to be a general feature of master regulators of cell state (Odom et al., 2006). Functional studies have confirmed that Oct4 and Sox2 co-occupy and activate the Oct4 and Nanog genes (Kuroda et al., 2005; Okumura-Nakanishi et al., 2005) and experiments with an inducible Sox2-null murine ES-cell line have provided compelling evidence for the existence of this interconnected autoregulatory loop and its role in the maintenance of pluripotency (Masui et al., 2007).
The interconnected autoregulatory loop formed by Oct4, Sox2 and Nanog also suggests how the core regulatory circuitry of iPS cells might be jump-started when Oct4, Sox2 and other transcription factors are over-expressed in fibroblasts (Maherali et al., 2007; Okita et al., 2007; Takahashi and Yamanaka, 2006; Wernig et al., 2007). When these factors are exogenously over-expressed, they may contribute directly to the activation of endogenous Oct4, Sox2 and Nanog, the products of which in turn contribute to the maintenance of their own gene expression.
Co-occupancy and coordinate activity of pluripotency factors
Oct4, Sox2 and Nanog co-occupy several hundred genes, often at apparently overlapping genomic sites (Boyer et al., 2005; Loh et al., 2006). This suggests that these pluripotency factors generally do not control their target genes independently, but rather act coordinately to maintain the transcriptional program required for pluripotency. A large multi-protein complex containing Oct4 and Nanog can be obtained by iterative immunoprecipitation in ES cells, providing further evidence that multiple interacting proteins likely coordinately control pluripotency (Wang et al., 2006). The possibility that multiple pluripotency factors function in a complex to coordinately control their target genes may help explain why efficient iPS cell generation appears to require the combinatorial over-expression of multiple transcription factors. Not all components of this putative complex are required to initiate the process of reprogramming, however, because exogenous Nanog is not necessary for iPS generation. It seems likely that exogenous Oct4 and other factors induce expression of endogenous Nanog to levels sufficient to accomplish full reprogramming.
Regulation of developmental regulators
The master regulators of pluripotency occupy the promoters of active genes encoding transcription factors, signal transduction components and chromatin modifying enzymes that promote ES cell self-renewal (Figure 5) (Boyer et al., 2005; Loh et al., 2006). However, these transcriptionally active genes account for only about half of the targets of Oct4, Sox2 and Nanog in ES cells. These master regulators also co-occupy the promoters of a large set of developmental transcription factors that are silent in ES cells, but whose expression is associated with lineage commitment and cellular differentiation (Boyer et al., 2005; Loh et al., 2006). Silencing of these developmental regulators is almost certainly a key feature of pluripotency, because expression of these developmental factors is associated with commitment to particular lineages. MyoD, for example, is a transcription factor capable of inducing a muscle gene expression program in a variety of cells (Davis et al., 1987). Therefore Oct4, Sox2 and Nanog likely help maintain the undifferentiated state of ES cells by contributing to repression of lineage specification factors.
Most of the transcriptionally silent developmental regulators targeted by Oct4, Sox2 and Nanog are also occupied by the Polycomb group (PcG) proteins (Bernstein et al., 2006; Boyer et al., 2006; Lee et al., 2006), which are epigenetic regulators that facilitate maintenance of cell state through gene silencing. The PcG proteins form multiple Polycomb Repressive Complexes (PRCs), the components of which are conserved from Drosophila to humans (Schuettengruber et al., 2007). PRC2 catalyzes histone H3 lysine-27 (H3K27) methylation, an enzymatic activity required for PRC2-mediated epigenetic gene silencing. H3K27 methylation is thought to provide a binding surface for PRC1, which facilitates oligomerization, condensation of chromatin structure, and inhibition of chromatin remodeling activity in order to maintain silencing. PRC1 also contains a histone ubiquitin ligase, Ring1b, whose activity appears likely to contribute to silencing in ES cells (Stock et al., 2007). How the PcG proteins are recruited to genes encoding developmental regulators in ES cells is not yet understood. Some of the most conserved vertebrate sequences are associated with genes encoding developmental regulators and some of these may be sites for DNA-binding proteins that recruit PcG proteins.
Recent studies revealed that the silent developmental genes that are occupied by Oct4, Sox2 and Nanog and PcG proteins experience an unusual form of transcriptional regulation (Guenther et al., 2007). These genes undergo transcription initiation but not productive transcript elongation in ES cells (Figure 5). The transcription initiation apparatus is recruited to the promoters of genes encoding developmental regulators, where histone modifications associated with transcription initiation and the initial step of elongation (such as H3K4 methylation) are found, but RNA polymerase is incapable of fully transcribing these genes, presumably because of repression mediated by the PcG proteins. These observations explain why the silent genes encoding developmental regulators are generally organized in “bivalent” domains that are occupied by nucleosomes with histone H3K4me3, which is associated with gene activity, and by nucleosomes with histone H3K27me3, which is associated with repression (Azuara et al., 2006; Bernstein et al., 2006; Guenther et al., 2007).
The presence of RNA polymerase at the promoters of genes encoding developmental regulators (Guenther et al., 2007) may explain why these genes are especially poised for transcription activation during differentiation (Boyer et al., 2006; Lee et al., 2006). Polycomb complexes and associated proteins may serve to pause RNA polymerase machinery at key regulators of development in pluripotent cells and in lineages where they are not expressed. At genes that are activated in a given cell type, PcG proteins and nucleosomes with H3K27 methylation are lost (Bernstein et al., 2006; Boyer et al., 2006; Lee et al., 2006; Mikkelsen et al., 2007), allowing the transcription apparatus to full transcribe these genes. The mechanisms that lead to selective activation of genes encoding specific developmental regulators are not yet understood, but they almost certainly involve signals brought to the genome by signal transduction pathways and likely involve H3K27 demethylation by enzymes such as the JmjC-domain-containing UTX and JMJD3 proteins (Lan et al., 2007).
Regulatory RNAs in pluripotency and early development
Targeted deletions in mice suggest that micro-RNAs (miRNAs) are likely to play key roles in ES cell gene regulation (Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007), but little is known about how miRNAs function to control the developmental potential of ES cells. Several lines of evidence indicate that miRNAs contribute to the control of pluripotency and early development. A subset of miRNAs is preferentially expressed in ES cells or mammalian embryonic tissue (Houbaviy et al., 2003; Mineno et al., 2006; Suh et al., 2004) Dicer-deficient mice fail to develop (Bernstein et al., 2003) and ES cells deficient in miRNA processing enzymes show defects in differentiation, self-renewal and perhaps viability (Kanellopoulou et al., 2005; Murchison et al., 2005; Wang et al., 2007). Specific miRNAs have been shown to participate in mammalian cellular differentiation as well as developmental patterning and morphogenesis (Chen et al., 2004; Chen et al., 2006; Harfe et al., 2005; Hornstein et al., 2005; Krichevsky et al., 2006; Mansfield et al., 2004; Yekta et al., 2004). However, how transcription factors and miRNAs function together in the regulatory circuitry that controls pluripotency and early development is not yet understood.
Signaling and epigenetic modifications during differentiation
Pluripotent embryonic stem cells can be maintained in an undifferentiated state in culture, but are poised to rapidly differentiate. Extra-cellular signals have been identified that contribute to the maintenance of ES cell pluripotency or that stimulate differentiation down defined lineages. One such signaling molecule is LIF, which can help maintain murine ES cells in an undifferentiated state in vitro, although it is not necessary for pluripotency in vivo (Smith et al., 1988). Other soluble factors, including Wnt, activin/nodal, and bFGF, have also been shown to contribute to maintenance of pluripotency, at least under certain culture conditions (Ogawa et al., 2006). Furthermore, human ES cells, and the human fibroblasts on which they were plated, have been reported to send reciprocal paracrine signals of FGF and IGF, respectively, sufficient to maintain the pluripotency of the ES cells (Bendall et al., 2007). These findings suggest that various signals help to establish a local microenvironment, in vitro and presumably in vivo, that helps to maintain pluripotency (see Essays by J. Rossant, page XXX, and J. Silva and A. Smith, page XXX; see Review by C.E. Murry and G. Keller, page XXX).
Signaling pathways also play key roles in promoting directed cellular differentiation. For example, activation of the Notch and BMP4 pathways can promote differentiation of ES cells (Chambers and Smith, 2004; Lowell et al., 2006). The Notch pathway has been shown to promote neural differentiation in both human and mouse embryonic stem cells. BMP4, on the other hand, can under certain conditions prevent neural cell differentiation while inducing differentiation into other cell types (Chambers and Smith, 2004).
When cell lineage commitment occurs, Oct4 is rapidly silenced and the appropriate regulators of development lose Polycomb-mediated repression and are activated. Oct4 and other regulators of pluripotency are highly restricted in their expression pattern to ES cells, cells of the inner cell mass and to cells of the germ line (Lengner et al., 2007). Ectopic expression of Oct4 has been shown to lead to rapid and massive expansion of poorly differentiated cells, especially in the intestine, and rapid fatality, highlighting the strong evolutionary pressure to ensure complete silencing of pluripotency regulators in somatic cells (Hochedlinger et al., 2005). Indeed, deletion of the gene in stem cells of adult mice has no functional consequences (Lengner et al., 2007) consistent with the notion that Oct4 has no role in the self renewal of somatic stem cells or in tissue homeostasis as has been suggested in many reports. Retinoic acid, a particularly well-characterized inducer of differentiation, has been shown to directly contribute to silencing of the Oct4 locus (Okamoto et al., 1990; Pikarsky et al., 1994). In addition, a set of nuclear repressors has been identified that are induced in differentiating cells and are required for proper silencing of Oct4, including ARP-1, COUP-TF1 and GCNF (also referred to as Nr6a1) (Ben-Shushan et al., 1995; Fuhrmann et al., 2001; Gu et al., 2006; Gu et al., 2005). Histone modifications associated with gene activity, including H3K4me3 and H3K7 and H3K9 acetylation, are lost at Oct4. Histone modifications associated with heterochromatin, H3K9me2 and me3, are gained in a G9a histone methyltransferase-dependent manner (Feldman et al., 2006). Finally, in a process dependent on de novo DNA methyltransferases DNMT3a/3b, which are recruited directly or indirectly by G9a, the Oct4 promoter undergoes CpG DNA methylation. Thus Oct4, and other ES cell-specific genes including Rex1, but not Nanog or Sox2, undergo a multi-step, tightly regulated form of silencing, during which they adopt an epigenetic state characteristic of heterochromatin (Feldman et al., 2006). These epigenetic changes appear to enforce a more stable form of silencing compared to the more labile epigenetic silencing associated with H3K27 methylation at genes that must be dynamically regulated during development. As discussed below, these multilayered marks of epigenetic silencing, including H3K9 methylation and DNA methylation, must be progressively removed in the process of generating iPS cells from somatic cells.
Molecular mechanisms of reprogramming
Our initial understanding of the molecular circuitry of the pluripotent state provides insight into the mechanisms involved in reprogramming. The loss of most cloned embryos after implantation has been correlated with the faulty activation of the silent endogenous pluripotency genes such as Oct4, Nanog and Sox2 (Boiani et al., 2002; Bortvin et al., 2003). Similarly, the maintenance of the pluripotent state in iPS cells following transcription factor transduction has been shown to be dependent on the activation of the silent endogenous pluripotency genes. Any molecular understanding of the reprogramming process needs to explain why the process, initiated by ectopic expression of Oct4, Sox2 and other factors, is gradual and proceeds over several weeks and why only a small fraction of the infected fibroblasts eventually become pluripotent iPS cells. As outlined in Figure 3b, sequential stochastic events seem to be important in the process and this could contribute to the low overall efficiency of generating iPS cells and for the prolonged duration of the reprogramming process. In addition, genetic differences between individual infected cells may add significant variability to the frequency of iPS cells. For example, because iPS cells carry multiple proviral copies (Wernig et al., 2007), it is possible that only those rare cells that carry a high copy number of proviral inserts and thus express the transcripts at high levels or at appropriate relative levels are selected for epigenetic reprogramming. Alternatively, reprogramming may require the additional activation of as yet unidentified cellular genes by insertional mutagenesis similar to oncogene activation in leukemia or mammary carcinogenesis. Finally, it is possible that only specific cells such as the rare somatic stem cells present in the donor cell population rather than any cell are susceptible to reprogramming.
Furthermore, any mechanistic explanation of reprogramming has to take into account the following observations. (i) The process of reprogramming involves intermediate cell states. For example, drug-resistant clones can be obtained in cultures of cells carrying a neomycin gene in the Oct4 or Nanog locus by 3 to 6 days after infection (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007) whereas the GFP marker carried in the same genes becomes detectable only weeks after infection (Brambrink et al., 2008; Meissner et al., 2007) (Figure 6A). This discrepancy in timing between the appearance of antibiotic resistance and protein detection could be due to a low level of initial expression that is sufficient to render the partially reprogrammed cells drug resistant, but below the level required to visualize GFP and maintain pluripotency (Figure 6B). (ii) The pluripotent state is dominant in ES – somatic cell hybrids. (iii) Multiple DNA replication cycles and cell divisions are required for in vitro reprogramming. (iv) DNA de novo methylation and hypomethylation are likely to be important for the process of reprogramming and maintaining the pluripotent state. (v) The oncogenes c-myc and Klf4, while not essential for reprogramming, seem to enhance the efficiency and speed of the process. (vi) PcG proteins and histone modifications play key roles in the silencing of developmental regulators in ES cells (Fig 5) and need to be re-expressed in reprogrammed cells.
Figure 6. Model of sequential steps in the reprogramming of somatic cells.
A. Sequential changes of phenotype and activation of Oct4, Nanog and Sox2. Fibroblasts do not express Oct4 or Nanog. After transduction with the four transcription factors Oct4, Sox2, c-myc and Klf4 the infected fibroblasts assume a transformed phenotype. The endogenous Oct4 or Nanog genes become transcribed at a low level that is sufficient for drug resistance in cells carrying the neo gene in either locus (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007) but not sufficient to produce GFP expression in Oct4-GFP or Nanog-GFP cells. In a stochastic sequence of epigenetic events the endogenous Oct4 and Nanog genes become fully activated only after 2 to 3 weeks as indicated by the appearance of GFP+ iPS cells in Oct4-GFP or Nanog-GFP fibroblasts (Brambrink et al., 2008; Meissner et al., 2007) (compare Figure 3b).
B. Sequential changes of histone and DNA modification. In fibroblasts the promoters of Oct4, Nanog and Sox2 are methylated (filled lollipops) and histone H3 is methylated at K9. During the reprogramming process the repressive H3K9me3 histone marks may be gradually replaced by the transcriptionally active H3K4me3 histone marks and the CpG sites become gradually demethylated (open lollipops). This transient epigenetic state may permit an expression level that is sufficient to give drug resistance (driving the inserted neo gene) but not sufficient to give GFP expression. Additional epigenetic changes alter the histone modification and the DNA methylation conformation to one that is fully derepressed allowing normal factor expression and resulting in GFP+ iPS cells.
C. Molecular circuitry during reprogramming. The transduced factors may interact with the endogenous pluripotency genes encoding Oct4, Nanog and Sox2 and gradually activate the autoregulatory loop that sustains normal factor expression. During the reprogramming process the de novo methyltransferases Dnmt3a and Dnmt3b become activated and in turn de novo methylate and silence the virally transduced factors. The pluripotent state is now maintained by the autoregulatory expression of the three transcription factors Oct4, Nanog and Sox2.
The observation that the pluripotent state is dominant in ES – somatic cell hybrids is consistent with the idea that the ES cell transcription factors, when present, will generate a pluripotent state by silencing genes that produce various differentiated states. Oct4, Nanog and Sox2 generate the ES cell gene expression program, at least in part, by binding and silencing genes encoding developmental regulators, thereby repressing various differentiated states. These pluripotency regulators likely participate, directly or indirectly, in the recruitment of PcG proteins, which are spread over large portions of these developmental genes in ES cells, thus preventing their transcription. Indeed, the PcG-catalyzed H3K27me3 chromatin modifications that are lacking in MEFs are reestablished in iPS cells (Maherali et al., 2007; Wernig et al., 2007).
Chromatin changes associated with reprogramming appear to happen progressively over an extended period of time and do not occur homogenously in all cells transduced with reprogramming factors. Multiple DNA replication cycles and cell divisions seem to be required for in vitro reprogramming, possibly because the heterochromatin-like silencing that occurs at the Oct4 promoter must be reversed and the interconnected autoregulatory loop generated by Oct4, Sox2 and Nanog must be re-established (Figure 6C). In somatic cells, the Oct4 gene is hypermethylated and occupied by nucleosomes containing methylated histone H3K9 (Feldman et al., 2006). Bisulfite sequencing has revealed that DNA methylation is lost at the promoters of pluripotency regulators in iPS cells (Maherali et al., 2007; Okita et al., 2007; Wernig et al., 2007). The methylation status of histone H3K9 has not yet been examined at these promoters in iPS cells, but it seems likely that methylation of H3K9 will be lost at these promoters (Loh et al., 2007) (Figure 6B). Ectopic expression of Oct4 and Sox2 may lead to activation of the endogenous loci when, during DNA replication, one or more alleles of the Oct4 promoter fails to be CpG methylated and is assembled into nucleosomes lacking repressive modifications. Similar events may lead to activation of genes encoding the other endogenous pluripotency factors, and once levels of endogenous Oct4, Nanog and Sox2 proteins are appropriate for generating an auto-regulatory loop, the endogenous factors can then maintain the activated state of their own genes.
While knowledge of the roles of Oct4 and Sox2 in control of ES cell transcriptional circuitry suggests how these pluripotency regulators contribute to reprogramming, we have less understanding of the functions of Klf4 and c-Myc in reprogramming. One possibility is that these two factors together allow for a cancer-like transformation of somatic cells, conferring on MEFs the immortal growth potential and rapid proliferative phenotype associated with ES cells (Yamanaka, 2007). A second model posits that c-Myc modifies the chromatin state of MEFs to allow the reprogramming factors to more efficiently access genes necessary for reprogramming (Yamanaka, 2007). When expressed at high levels, the Myc protein can occupy a large population of genes and may stimulate chromatin modifications that may provide increased access for transcription factors (Fernandez et al., 2003; Li et al., 2003). This idea is consistent with the observation that reprogramming can be accomplished in the absence of c-Myc or Klf4, albeit at significantly decreased efficiency (Nakagawa et al., 2008; Wernig et al., 2008; Yu et al., 2007). A third model emerges from recent evidence that c-Myc associates with pre-replication complexes and promotes DNA synthesis independent of transcriptional regulation (Dominguez-Sola et al., 2007). DNA replication, promoted by Myc overexpression, could provide an opportunity for the somatic genome to reset its epigenetic state in the presence of exogenous reprogramming factors.
Klf4’s function in reprogramming may involve regulation of specific ES cell genes and may thus be independent of its oncogene potential. Oct4 and Sox2 bind to overlapping sites in the promoter of Lefty1, an ES-cell specific gene, but are not sufficient to transcriptionally activate the gene in somatic cells (Nakatake et al., 2006). Addition of Klf4 stimulates transcription of Lefty1 in somatic cells, suggesting that Klf4 may assist Oct4 and Sox2 to initiate expression of key ES cell genes in somatic cells.
As discussed above, reprogramming of somatic cells can be achieved without c-myc and Klf4 albeit with significantly lower efficiency (Nakagawa et al., 2008; Wernig et al., 2008; Yu et al., 2007). This is consistent with the notion that these oncogenes are nonessential for the reprogramming process and may merely promote the epigenetic remodeling and activation of essential endogenous genes such as Oct4 and Sox2, which then establish the autoregulatory loop that maintains the pluripotent state (Figure 6c). Because over-expression of Oct4 can override the need for Sox2 (Masui et al., 2007) it is possible that Oct4 is the only transcription factor that is indispensable for reprogramming and that other factors or signaling molecules can serve to facilitate activation of the “pluripotency circuitry”.
Nuclear cloning versus in vitro reprogramming
Because reprogramming by either nuclear transplantation or in vitro by over-expression of defined transcription factors result in pluripotent ES-like cells that are indistinguishable, the question can be raised of whether similar molecular mechanisms may be responsible to produce the same epigenetic state. This is unlikely for the following considerations. (i) It has been well established that a quiescent state of the donor cells is crucial for SCNT to be successful (Wilmut et al., 1997) while in vitro reprogramming seems to depend on active proliferation of the somatic cells. (ii) Reprogramming by SCNT likely occurs in a shorter time span than in vitro reprogramming. For example, Oct4 is activated at the 2 to 4 cell stage in cloned embryos (Boiani et al., 2002) and major chromatin modifications are detected early after nuclear transfer (Santos et al., 2003) suggesting that the epigenetic state of the somatic donor genome is reset within a few cell divisions. In contrast, in vitro reprogramming has been shown to be a protracted process that proceeds over many weeks (Figure 3a). (iii) Though the molecular mechanisms that initiate SCNT–mediated reprogramming have not been clarified, it is unlikely that high expression of oncogenes such as c-myc and Klf4 play a crucial role as required for efficient in vitro reprogramming.
As discussed above, in vitro culture selects for the fastest dividing cells but not for pluripotency. Thus, the generation of ES, EG or MASC or of iPS cells derived from explanted blastocysts, from primordial or from spermatogonial stem cells or from in vitro reprogrammed somatic cells, respectively, may be driven by selection for the fastest dividing cells that outgrow the corresponding slowly proliferating parental cells. It may be that the pluripotent ES-like cells, initiated by as diverse mechanisms as SCNT or in vitro reprogramming, represent a “default” state of cells growing in tissue culture. Their seemingly identical pluripotent state may represent the default epigenetic state assumed by cells that have been selected for proliferation after exposure to treatments as different as NT, transduction with reprogramming factors or explantation into culture.
Outlook
In vitro reprogramming of fibroblasts raises a number of interesting questions. For example, can cells of other lineages such as endoderm-derived epithelial cells or ectoderm-derived keratinocytes be dedifferentiated to a pluripotent state by the same combination of factors as MEFs? Is the efficiency of reprogramming somatic stem cells higher than that of terminally differentiated cells as has been seen in nuclear transplantation experiments? An important aspect of in vitro reprogramming is the induction of cell proliferation, a fact that will complicate the reprogramming of post-mitotic cells such as neurons. An interesting question is whether terminally differentiated lymphoid cells that can be induced to divide can be reprogrammed.
The translation of in vitro reprogramming to patient-specific transplantation therapy (Takahashi et al., 2007; Yu et al., 2007) faces several challenges. Though it appears likely that the requirement for oncogenes such as c-myc and Klf4 can be eliminated (Nakagawa et al., 2007; Wernig et al., 2007; Yu et al., 2007; Park et al, 2007), transduction of Oct4 raises concerns as it was shown to be a powerful oncogene when expressed in somatic cells (Hochedlinger et al., 2005). Also, the use of retroviral vectors harbors the risk of insertional mutations that could activate genes such as oncogenes that accelerate proliferation. Thus, to overcome these technical barriers for the eventual application of this approach in the clinic we need to understand the molecular pathways of in vitro reprogramming. This may allow the development of alternative methods of in vitro reprogramming that do not rely on the use of potentially harmful agents such as transcription factors and retroviruses.
Acknowledgements
We thank Caroline Beard, Tobi Brambrink, Menno Creyghton, Jacob Hanna, Dirk Hockemeyer, Konrad Hochedlinger, Chris Lengner, Alex Marson, Frank Soldner and Grant Welstaed for their contributions and their comments on the manuscript. We thank Tom DeCesare, Stuart Levine and Julia Zeitlinger for contributions to figures.
References
- Alon U. Network motifs: theory and experimental approaches. Nat Rev Genet. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Alvarez-Dolado M, Pardal R, Garcia-Verdugo JM, Fike JR, Lee HO, Pfeffer K, Lois C, Morrison SJ, Alvarez-Buylla A. Fusion of bone-marrow-derived cells with Purkinje neurons, cardiomyocytes and hepatocytes. Nature. 2003;425:968–973. doi: 10.1038/nature02069. [DOI] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jorgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- Balsam LB, Wagers AJ, Christensen JL, Kofidis T, Weissman IL, Robbins RC. Haematopoietic stem cells adopt mature haematopoietic fates in ischaemic myocardium. Nature. 2004;428:668–673. doi: 10.1038/nature02460. [DOI] [PubMed] [Google Scholar]
- Ben-Shushan E, Sharir H, Pikarsky E, Bergman Y. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct-3/4 expression in embryonal carcinoma cells. Mol Cell Biol. 1995;15:1034–1048. doi: 10.1128/mcb.15.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall SC, Stewart MH, Menendez P, George D, Vijayaragavan K, Werbowetski-Ogilvie T, Ramos-Mejia V, Rouleau A, Yang J, Bosse M, et al. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature. 2007;448:1015–1021. doi: 10.1038/nature06027. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Blelloch R, Venere M, Yen M, M R-S. Generation of Induced Pluripotent Stem Cells in the Absence of Drug Selection. Cell Stem Cell. 2007;1:245–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blelloch R, Wang Z, Meissner A, Pollard S, Smith A, Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem cells (Dayton, Ohio) 2006;24:2007–2013. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiani M, Eckardt S, Scholer HR, McLaughlin KJ. Oct4 distribution and level in mouse clones: consequences for pluripotency. Genes Dev. 2002;16:1209–1219. doi: 10.1101/gad.966002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortvin A, Eggan K, Skaletsky H, Akutsu H, Berry DL, Yanagimachi R, Page DC, Jaenisch R. Incomplete reactivation of Oct4-related genes in mouse embryos cloned from somatic nuclei. Development. 2003;130:1673–1680. doi: 10.1242/dev.00366. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brambrink T, Foreman R, Welstead G, Lengner C, Wernig M, Suh H, Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of somatic cells. Cell Stem Cell Submitted. 2008 doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambrink T, Hochedlinger K, Bell G, Jaenisch R. ES cells derived from cloned and fertilized blastocysts are transcriptionally and functionally indistinguishable. Proc Natl Acad Sci U S A. 2006;103:933–938. doi: 10.1073/pnas.0510485103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brons IG, Smithers LE, Trotter MW, Rugg-Gunn P, Sun B, Chuva de Sousa Lopes SM, Howlett SK, Clarkson A, Ahrlund-Richter L, Pedersen RA, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature. 2007;448:191–195. doi: 10.1038/nature05950. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chambers I, Smith A. Self-renewal of teratocarcinoma and embryonic stem cells. Oncogene. 2004;23:7150–7160. doi: 10.1038/sj.onc.1207930. [DOI] [PubMed] [Google Scholar]
- Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibelli JB, Grant KA, Chapman KB, Cunniff K, Worst T, Green HL, Walker SJ, Gutin PH, Vilner L, Tabar V, et al. Parthenogenetic stem cells in nonhuman primates [In Process Citation] Science. 2002;295:819. doi: 10.1126/science.1065637. [DOI] [PubMed] [Google Scholar]
- Clarke DL, Johansson CB, Wilbertz J, Veress B, Nilsson E, Karlstrom H, Lendahl U, Frisen J. Generalized potential of adult neural stem cells. Science. 2000;288:1660–1663. doi: 10.1126/science.288.5471.1660. [DOI] [PubMed] [Google Scholar]
- Cowan CA, Atienza J, Melton DA, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. doi: 10.1126/science.1116447. [DOI] [PubMed] [Google Scholar]
- Croft AP, Przyborski SA. Formation of neurons by non-neural adult stem cells: potential mechanism implicates an artifact of growth in culture. Stem cells (Dayton, Ohio) 2006;24:1841–1851. doi: 10.1634/stemcells.2005-0609. [DOI] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- De Coppi P, Bartsch G, Jr, Siddiqui MM, Xu T, Santos CC, Perin L, Mostoslavsky G, Serre AC, Snyder EY, Yoo JJ, et al. Isolation of amniotic stem cell lines with potential for therapy. Nat Biotechnol. 2007;25:100–106. doi: 10.1038/nbt1274. [DOI] [PubMed] [Google Scholar]
- Do JT, Scholer HR. Nuclei of embryonic stem cells reprogram somatic cells. Stem cells (Dayton, Ohio) 2004;22:941–949. doi: 10.1634/stemcells.22-6-941. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, Galloway DA, Gu W, Gautier J, Dalla-Favera R. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- Eggan K, Akutsu H, Loring J, Jackson-Grusby L, Klemm M, Rideout WM, 3rd, Yanagimachi R, Jaenisch R. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc Natl Acad Sci U S A. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggan K, Baldwin K, Tackett M, Osborne J, Gogos J, Chess A, Axel R, Jaenisch R. Mice cloned from olfactory sensory neurons. Nature. 2004;428:44–49. doi: 10.1038/nature02375. [DOI] [PubMed] [Google Scholar]
- Egli D, Rosains J, Birkhoff G, Eggan K. Developmental reprogramming after chromosome transfer into mitotic mouse zygotes. Nature. 2007;447:679–685. doi: 10.1038/nature05879. [DOI] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a–mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann G, Chung AC, Jackson KJ, Hummelke G, Baniahmad A, Sutter J, Sylvester I, Scholer HR, Cooney AJ. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev Cell. 2001;1:377–387. doi: 10.1016/s1534-5807(01)00038-7. [DOI] [PubMed] [Google Scholar]
- Gan Q, Yoshida T, McDonald OG, Owens GK. Concise review: epigenetic mechanisms contribute to pluripotency and cell lineage determination of embryonic stem cells. Stem cells (Dayton, Ohio) 2007;25:2–9. doi: 10.1634/stemcells.2006-0383. [DOI] [PubMed] [Google Scholar]
- Greda P, Karasiewicz J, Modlinski JA. Mouse zygotes as recipients in embryo cloning. Reproduction. 2006;132:741–748. doi: 10.1530/rep.1.01204. [DOI] [PubMed] [Google Scholar]
- Gu P, Le Menuet D, Chung AC, Cooney AJ. Differential recruitment of methylated CpG binding domains by the orphan receptor GCNF initiates the repression and silencing of Oct4 expression. Mol Cell Biol. 2006;26:9471–9483. doi: 10.1128/MCB.00898-06. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gu P, LeMenuet D, Chung AC, Mancini M, Wheeler DA, Cooney AJ. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol Cell Biol. 2005;25:8507–8519. doi: 10.1128/MCB.25.19.8507-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Guan K, Nayernia K, Maier LS, Wagner S, Dressel R, Lee JH, Nolte J, Wolf F, Li M, Engel W, et al. Pluripotency of spermatogonial stem cells from adult mouse testis. Nature. 2006;440:1199–1203. doi: 10.1038/nature04697. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Byrne JA. The first half-century of nuclear transplantation. Proc Natl Acad Sci U S A. 2003;100:8048–8052. doi: 10.1073/pnas.1337135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Wernig M, Markoulaki S, Sun C, Meissner A, Cassady J, Beard C, Wu L, Brambrink T, Townes T, et al. Therapy of Sickle Cell Anemia by Autologous Skin Derived Induced Pluripotent Stem Cells. Science. 2007 In press. [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AH, Hartley L, Ibrahim M, Robb L. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev Dyn. 2004;230:187–198. doi: 10.1002/dvdy.20034. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature. 2002;415:1035–1038. doi: 10.1038/nature718. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Nuclear transplantation, embryonic stem cells, and the potential for cell therapy. N Engl J Med. 2003;349:275–286. doi: 10.1056/NEJMra035397. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–1067. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. On the cloning of animals from terminally differentiated cells. Nat Genet. 2007;39:136–137. doi: 10.1038/ng0207-136. [DOI] [PubMed] [Google Scholar]
- Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- Hornstein E, Mansfield JH, Yekta S, Hu JK, Harfe BD, McManus MT, Baskerville S, Bartel DP, Tabin CJ. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- Inoue K, Noda S, Ogonuki N, Miki H, Inoue S, Katayama K, Mekada K, Miyoshi H, Ogura A. Differential developmental ability of embryos cloned from tissue-specific stem cells. Stem cells (Dayton, Ohio) 2007;25:1279–1285. doi: 10.1634/stemcells.2006-0747. [DOI] [PubMed] [Google Scholar]
- Inoue K, Wakao H, Ogonuki N, Miki H, Seino K, Nambu-Wakao R, Noda S, Miyoshi H, Koseki H, Taniguchi M, et al. Generation of cloned mice by direct nuclear transfer from natural killer T cells. Curr Biol. 2005;15:1114–1118. doi: 10.1016/j.cub.2005.05.021. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jackson KA, Snyder DS, Goodell MA. Skeletal muscle fiber-specific green autofluorescence: potential for stem cell engraftment artifacts. Stem cells (Dayton, Ohio) 2004;22:180–187. doi: 10.1634/stemcells.22-2-180. [DOI] [PubMed] [Google Scholar]
- Jähner D, Stuhlmann H, Stewart CL, Harbers K, Löhler J, Simon I, Jaenisch R. De Novomethylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Jahagirdar BN, Reinhardt RL, Schwartz RE, Keene CD, Ortiz-Gonzalez XR, Reyes M, Lenvik T, Lund T, Blackstad M, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- Joseph NM, Morrison SJ. Toward an understanding of the physiological function of Mammalian stem cells. Dev Cell. 2005;9:173–183. doi: 10.1016/j.devcel.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, et al. Generation of pluripotent stem cells from neonatal mouse testis. Cell. 2004;119:1001–1012. doi: 10.1016/j.cell.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem cells (Dayton, Ohio) 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda T, Tada M, Kubota H, Kimura H, Hatano SY, Suemori H, Nakatsuji N, Tada T. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengner C, Camargo F, Hochedlinger K, Zaidi S, Welstead G, Gokhale S, Scholer H, Tomilin A, Jaenisch R. Oct4 has no role in somatic stem cell self-renewal Cell Stem Cell. 2007;1:403–415. doi: 10.1016/j.stem.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Greco V, Guasch G, Fuchs E, Mombaerts P. Mice cloned from skin cells. Proc Natl Acad Sci U S A. 2007;104:2738–2743. doi: 10.1073/pnas.0611358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ishii T, Feinstein P, Mombaerts P. Odorant receptor gene choice is reset by nuclear transfer from mouse olfactory sensory neurons. Nature. 2004;428:393–399. doi: 10.1038/nature02433. [DOI] [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci U S A. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545–2557. doi: 10.1101/gad.1588207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell-Badge R. Many ways to pluripotency. Nat Biotechnol. 2007;25:1114–1116. doi: 10.1038/nbt1007-1114. [DOI] [PubMed] [Google Scholar]
- Lowell S, Benchoua A, Heavey B, Smith AG. Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 2006;4:e121. doi: 10.1371/journal.pbio.0040121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko Y, Tchieu J, Jaenisch R, et al. Global epigenetic remodeling in directly reprogrammed fibroblasts. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, et al. MicroRNA-responsive 'sensor' transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui S, Nakatake Y, Toyooka Y, Shimosato D, Yagi R, Takahashi K, Okochi H, Okuda A, Matoba R, Sharov AA, et al. Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol. 2007;9:625–635. doi: 10.1038/ncb1589. [DOI] [PubMed] [Google Scholar]
- Matsui Y, Zsebo K, Hogan BLM. Derivation of pluripotential embryonic stem cells from murine primordial germ cells in culture. Cell. 1992;70:841–847. doi: 10.1016/0092-8674(92)90317-6. [DOI] [PubMed] [Google Scholar]
- Matsumura H, Tada M, Otsuji T, Yasuchika K, Nakatsuji N, Surani A, Tada T. Targeted chromosome elimination from ES-somatic hybrid cells. Nat Methods. 2007;4:23–25. doi: 10.1038/nmeth973. [DOI] [PubMed] [Google Scholar]
- McGrath J, Solter D. Inability of mouse blastomere nuclei transferred to enucleated zygotes to support development in vitro. Science. 1984;226:1317–1319. doi: 10.1126/science.6542249. [DOI] [PubMed] [Google Scholar]
- Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M, et al. The expression profile of microRNAs in mouse embryos. Nucleic Acids Res. 2006;34:1765–1771. doi: 10.1093/nar/gkl096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, Soonpaa MH, Reinecke H, Nakajima H, Nakajima HO, Rubart M, Pasumarthi KB, Ismail Virag J, Bartelmez SH, Poppa V, et al. Haematopoietic stem cells do not transdifferentiate into cardiac myocytes in myocardial infarcts. Nature. 2004 doi: 10.1038/nature02446. [DOI] [PubMed] [Google Scholar]
- Nágy A, Gócza E, Diaz EM, Prideaux VR, Iványi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Devel. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- Nakagawa M, Koyanagi M, Tanabe K, Takahashi K, Ichisaka T, Aoi T, Okita K, Mochiduki Y, Takizawa N, Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008 doi: 10.1038/nbt1374. in press. [DOI] [PubMed] [Google Scholar]
- Nakatake Y, Fukui N, Iwamatsu Y, Masui S, Takahashi K, Yagi R, Yagi K, Miyazaki J, Matoba R, Ko MS, et al. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol Cell Biol. 2006;26:7772–7782. doi: 10.1128/MCB.00468-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimha M, Barton SC, Surani MA. The role of the paternal genome in the development of the mouse germ line. Curr Biol. 1997;7:881–884. doi: 10.1016/s0960-9822(06)00377-0. [DOI] [PubMed] [Google Scholar]
- Neuhuber B, Gallo G, Howard L, Kostura L, Mackay A, Fischer I. Reevaluation of in vitro differentiation protocols for bone marrow stromal cells: disruption of actin cytoskeleton induces rapid morphological changes and mimics neuronal phenotype. J Neurosci Res. 2004;77:192–204. doi: 10.1002/jnr.20147. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Oback B, Wells DN. Donor cell differentiation, reprogramming, and cloning efficiency: elusive or illusive correlation? Mol Reprod Dev . 2007;74:646–654. doi: 10.1002/mrd.20654. [DOI] [PubMed] [Google Scholar]
- Odom DT, Dowell RD, Jacobsen ES, Nekludova L, Rolfe PA, Danford TW, Gifford DK, Fraenkel E, Bell GI, Young RA. Core transcriptional regulatory circuitry in human hepatocytes. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100059. 2006 0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa K, Nishinakamura R, Iwamatsu Y, Shimosato D, Niwa H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem Biophys Res Commun. 2006;343:159–166. doi: 10.1016/j.bbrc.2006.02.127. [DOI] [PubMed] [Google Scholar]
- Okamoto K, Okazawa H, Okuda A, Sakai M, Muramatsu M, Hamada H. A novel octamer binding transcription factor is differentially expressed in mouse embryonic cells. Cell. 1990;60:461–472. doi: 10.1016/0092-8674(90)90597-8. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007 Jun 6; doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- Okumura-Nakanishi S, Saito M, Niwa H, Ishikawa F. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J Biol Chem. 2005;280:5307–5317. doi: 10.1074/jbc.M410015200. [DOI] [PubMed] [Google Scholar]
- Park I-H, Zhao R, West J, Yabuuchi A, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008 doi: 10.1038/nature06534. published EOP December 27. [DOI] [PubMed] [Google Scholar]
- Phinney DG, Prockop DJ. Concise Review: Mesenchymal Stem/Multi-Potent Stromal Cells (MSCs): The State of Transdifferentiation and Modes of Tissue Repair - Current Views. Stem cells (Dayton, Ohio) 2007 doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- Pikarsky E, Sharir H, Ben-Shushan E, Bergman Y. Retinoic acid represses Oct-3/4 gene expression through several retinoic acid-responsive elements located in the promoter-enhancer region. Mol Cell Biol. 1994;14:1026–1038. doi: 10.1128/mcb.14.2.1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick JL, Bixler LS, Cheng L, Donovan PJ. Long-term proliferation of mouse primordial germ cells in culture. Nature. 1992;359:550–551. doi: 10.1038/359550a0. [DOI] [PubMed] [Google Scholar]
- Revazova ES, Turovets NA, Kochetkova OD, Kindarova LB, Kuzmichev LN, Janus JD, Pryzhkova MV. Patient-specific stem cell lines derived from human parthenogenetic blastocysts. Cloning Stem Cells. 2007;9:432–449. doi: 10.1089/clo.2007.0033. [DOI] [PubMed] [Google Scholar]
- Rideout WM, 3rd, Hochedlinger K, Kyba M, Daley GQ, Jaenisch R. Correction of a genetic defect by nuclear transplantation and combined cell and gene therapy. Cell. 2002;109:17–27. doi: 10.1016/s0092-8674(02)00681-5. [DOI] [PubMed] [Google Scholar]
- Rizzino A. A challenge for regenerative medicine: Proper genetic programming, not cellular mimicry. Dev Dyn. 2007 doi: 10.1002/dvdy.21285. [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado A. Planarian regeneration: its end is its beginning. Cell. 2006;124:241–245. doi: 10.1016/j.cell.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Santos F, Zakhartchenko V, Stojkovic M, Peters A, Jenuwein T, Wolf E, Reik W, Dean W. Epigenetic marking correlates with developmental potential in cloned bovine preimplantation embryos. Curr Biol. 2003;13:1116–1121. doi: 10.1016/s0960-9822(03)00419-6. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Seandel M, James D, Shmelkov SV, Falciatori I, Kim J, Chavala S, Scherr DS, Zhang F, Torres R, Gale NW, et al. Generation of functional multipotent adult stem cells from GPR125+ germline progenitors. Nature. 2007;449:346–350. doi: 10.1038/nature06129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. doi: 10.1038/nature04914. [DOI] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Solter D. From teratocarcinomas to embryonic stem cells and beyond: a history of embryonic stem cell research. Nat Rev Genet. 2006;7:319–327. doi: 10.1038/nrg1827. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, Cha KY, Chung HM, Yoon HS, Moon SY, et al. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- Sung LY, Gao S, Shen H, Yu H, Song Y, Smith SL, Chang CC, Inoue K, Kuo L, Lian J, et al. Differentiated cells are more efficient than adult stem cells for cloning by somatic cell nuclear transfer. Nat Genet. 2006;38:1323–1328. doi: 10.1038/ng1895. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Taranger CK, Noer A, Sorensen AL, Hakelien AM, Boquest AC, Collas P. Induction of Dedifferentiation, Genome-wide Transcriptional Programming, and Epigenetic Reprogramming by Extracts of Carcinoma and Embryonic Stem Cells. Mol Biol Cell. 2005 doi: 10.1091/mbc.E05-06-0572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada N, Hamazaki T, Oka M, Hoki M, Mastalerz DM, Nakano Y, Meyer EM, Morel L, Petersen BE, Scott EW. Bone marrow cells adopt the phenotype of other cells by spontaneous cell fusion. Nature. 2002;416:542–545. doi: 10.1038/nature730. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- van de Ven C, Collins D, Bradley MB, Morris E, Cairo MS. The potential of umbilical cord blood multipotent stem cells for nonhematopoietic tissue and cell regeneration. Exp Hematol. 2007 doi: 10.1016/j.exphem.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Wagers AJ, Weissman IL. Plasticity of adult stem cells. Cell. 2004;116:639–648. doi: 10.1016/s0092-8674(04)00208-9. [DOI] [PubMed] [Google Scholar]
- Wakayama S, Hikichi T, Suetsugu R, Sakaide Y, Bui HT, Mizutani E, Wakayama T. Efficient establishment of mouse embryonic stem cell lines from single blastomeres and polar bodies. Stem cells (Dayton, Ohio) 2007;25:986–993. doi: 10.1634/stemcells.2006-0615. [DOI] [PubMed] [Google Scholar]
- Wakayama S, Jakt ML, Suzuki M, Araki R, Hikichi T, Kishigami S, Ohta H, Van Thuan N, Mizutani E, Sakaide Y, et al. Equivalency of nuclear transfer-derived embryonic stem cells to those derived from fertilized mouse blastocysts. Stem cells (Dayton, Ohio) 2006;24:2023–2033. doi: 10.1634/stemcells.2005-0537. [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Wang X, Willenbring H, Akkari Y, Torimaru Y, Foster M, Al-Dhalimy M, Lagasse E, Finegold M, Olson S, Grompe M. Cell fusion is the principal source of bone-marrow-derived hepatocytes. Nature. 2003;422:897–901. doi: 10.1038/nature01531. [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Cassady J, Jaenisch R. C-Myc Is Dispensable for Direct Reprogramming of Mouse Fibroblasts. Cell Stem Cell. 2008 doi: 10.1016/j.stem.2007.12.001. in press. [DOI] [PubMed] [Google Scholar]
- Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- Wilmut I, Schnieke AE, McWhir J, Kind AJ, Campbell KH. Viable offspring derived from fetal and adult mammalian cells. Nature. 1997;385:810–813. doi: 10.1038/385810a0. [DOI] [PubMed] [Google Scholar]
- Yamanaka S. Strategies and New Developments in the Generation of Patient-Specific Pluripotent Stem Cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Yang X, Smith SL, Tian XC, Lewin HA, Renard JP, Wakayama T. Nuclear reprogramming of cloned embryos and its implications for therapeutic cloning. Nat Genet. 2007;39:295–302. doi: 10.1038/ng1973. [DOI] [PubMed] [Google Scholar]
- Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Evans EP, Smith AG. Changing potency by spontaneous fusion. Nature. 2002;416:545–548. doi: 10.1038/nature729. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, He P, Slukvin II, Thomson JA. Human embryonic stem cells reprogram myeloid precursors following cell-cell fusion. Stem cells (Dayton, Ohio) 2006;24:168–176. doi: 10.1634/stemcells.2005-0292. [DOI] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007 doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- Zwaka TP, Thomson JA. A germ cell origin of embryonic stem cells? Development. 2005;132:227–233. doi: 10.1242/dev.01586. [DOI] [PubMed] [Google Scholar]