The Epigenomics of Cancer (original) (raw)
. Author manuscript; available in PMC: 2014 Jan 17.
Abstract
Aberrant gene function and altered patterns of gene expression are key features of cancer. Growing evidence shows that acquired epigenetic abnormalities participate with genetic alterations to cause this dysregulation. Here, we review recent advances in understanding how epigenetic alterations participate in the earliest stages of neoplasia, including stem/precursor cell contributions, and discuss the growing implications of these advances for strategies to control cancer.
For decades, scientists have been engaged in dissecting the origins of human cancer, and the relative roles of genetic versus epigenetic abnormalities have been hotly debated. An explosion of data indicating the importance of epigenetic processes, especially those resulting in the silencing of key regulatory genes, has led to the realization that genetics and epigenetics cooperate at all stages of cancer development. Recent advances include the understanding that silencing is part of global epigenomic alterations in cancer, that pathways relevant to stem cell growth and differentiation become altered, and the approval of three drugs that target these defects in cancer patients.
Gene Silencing and Cancer
Epigenetics is defined as heritable changes in gene expression that are not accompanied by changes in DNA sequence. Gene silencing at the level of chromatin is necessary for the life of eukaryotic organisms and is particularly important in orchestrating key biological processes, including differentiation, imprinting, and silencing of large chromosomal domains such as the X chromosome, over the life span of female mammals. In many species, silencing can be initiated and maintained solely by processes involving the covalent modifications of histones and other chromatin components. Vertebrates, however, have taken advantage of the heritability of DNA cytosine methylation patterns to add another layer of control to these processes.
Like most biological processes, silencing can become dysregulated, resulting in the development of disease states. It can also result in the acquired inactivation of genes during normal aging. A key property of silencing is that it can spread over genomic regions in a progressive way, as perhaps best exemplified by position-effect variegation in Drosophila. It seems to involve the cooperation of multiple processes, including noncoding RNAs, covalent modifications of chromatin, physical alterations in nucleosomal positioning, and DNA methylation, among others.
It must be appreciated, as we will outline, that epigenetic abnormalities in cancer comprise a multitude of aberrations in virtually every component of chromatin involved in packaging the human genome. Since epigenetic silencing processes are mitotically heritable, they can play the same roles and undergo the same selective processes as genetic alterations in the development of a cancer. A principal tenet of Darwin’s hypotheses for the evolution of species is that most germline mutations are deleterious, or of no functional significance; mutations give rise to a specific advantage selected for in an evolving population. These same selective concepts apply for epigenetic events, which can occur at a much more increased rate compared to mutations in somatic cells. Alterations in gene expression induced by epigenetic events, which give rise to a cellular growth advantage, are therefore selected for in the host organ, resulting in the progressive uncontrolled growth of the tumor. This does not mean that all silenced genes play direct roles, since it is becoming clear, as we will discuss later, that whole groups of genes may be inactivated as part of an abnormal “program.” Epigenetic changes can collaborate with genetic changes to cause the evolution of a cancer because they are mitotically heritable. The high degree of mitotic stability of silencing coupled with the progressive nature by which it is achieved makes pathological silencing of growth controlling and other genes an essential part of the development of a human cancer.
The Importance of Chromatin Remodeling
Much is now known about the importance of promoter cytosine methylation in CpG islands and gene silencing, and it has been established beyond doubt that such methylation is intimately involved in cancer development. As discussed later, many hundreds of genes may be inactivated in a single cancer by promoter methylation. In general, methylated CpG islands are not capable of the initiation of transcription unless the methylation signal can be overridden by alterations in factors that modulate chromatin, such as removal of methylated cytosine binding proteins (Bakker et al., 2002) or the deacetylase, SIRT1 (Pruitt et al., 2006).
The driving force in DNA methylation research, particularly as it relates to cancer, has, until now, been particularly focused on CpG island promoter methylation. However, it remains true that about 40% of human genes do not contain bona fide CpG islands in their promoters (Takai and Jones, 2002). The reason for the focus on islands is because of the demonstrable ability of CpG-island promoter methylation to permanently silence genes both physiologically and pathologically in mammalian cells. The role of methylation in non-CpG island promoters has been largely overlooked because the mechanistic links have not been so well demonstrated. Recent work has shown strong correlations between tissue-specific expression and methylation of non-CpG islands, including, for example, the maspin gene (Futscher et al., 2002). Maspin has a CpG rich promoter that does not meet established criteria for a CpG island (Takai and Jones, 2002). There are other examples of genes, such as the MAGE gene family, that are commonly upregulated by epigenetic therapy in which the promoters do not satisfy recognized criteria for “island-ness.” Indeed, recent DNA methylation profiling of human chromosomes 6, 20, and 22 has shown that 17% of 873 analyzed genes are differentially methylated and that about a third of these show inverse correlations between methylation and transcription (Eckhardt et al., 2006). While it has not been rigorously established that cytosine methylation in such promoters causally blocks transcription, it is also true that this possibility has not been excluded. There is clearly room for more work on the CpG poor genes in which cytosine methylation could also play a role in normal development as well as in cancer.
The importance of DNA methylation in cancer has been established (Jones and Baylin, 2002; Jones and Laird, 1999), and the focus in the field is changing to include the mechanisms by which other chromatin modifications play a role in cancer development (Figure 1). Foremost among these are the covalent modifications of histones that can control gene activity. For example, histone deacetylation and methylation of specific lysine residues such as lysine 9 in histone H3 or lysine 27 in histone H3 clearly participate in the silencing of genes (Jenuwein, 2006; see also the Review by B. Li et al., page 707 in this issue).
Figure 1. Gene Silencing in Normal Cells.
Heritable gene silencing involves, among other processes, the interplay between DNA methylation, histone covalent modifications, and nucleosomal remodeling. Some of the enzymes that contribute to these modifications include DNA methyltransferase (DNMTs), histone deacetylases (HDACs), histone methyltransferases (HMTs), and complex nucleosomal remodeling factors (NURFs). The interplay between these processes establishes a heritable repressive state at the start site of genes resulting in gene silencing. Physiologically, silencing is critical for development and differentiation. Pathologically, silencing leads to diseases such as cancer. Recent evidence suggests global changes in all three processes in cancer, perhaps reflecting their interrelationships.
A key link between these covalent histone modifications and DNA methylation was established by the pioneering experiments of Nan et al. (1998) and Jones et al. (1998) who showed that cytosine methylation could attract methylated DNA binding proteins and histone deacetylases to methylated CpG islands during chromatin compaction and gene silencing. More recently, however, the link between covalent histone modifications and nucleosomal remodeling is increasingly being explored. Zhang et al. (1999) showed that DNA methylation binding protein (MBD2) interacts with the nucleosomal remodeling complex (NuRD) and directs the complex to methylate DNA. Harikrishnan et al. (2005) showed that Brahma (Brm), which is a catalytic component of the SWI/SNF chromatin-remodeling complex, associates with the methylated DNA binding protein MeCP2. These experiments provide a potential link between DNA methylation and chromatin silencing. Very recent studies have shown that covalent modifications of histones couple these processes with chromatin remodeling by ATP-dependent remodeling machines (Li et al., 2006; Wysocka et al., 2006). This has led to the realization that the three processes of DNA cytosine methylation, histone modification, and nucleosomal remodeling are intimately linked and that alterations in these processes result in the permanent silencing of cancer-relevant genes (Figure 1).
The fact that nucleosomal remodeling is a key component to the epigenetic silencing in cancer has been known for some time. It has been directly shown that mutations in the SWI/SNF complex play causative roles in the development of certain kinds of human cancer. Mutations in the SNF5 gene stimulate cell-cycle progression and cooperate with p53 loss in oncogenic transformation, and they are also associated with inactivation of the p21 and p16 pathways (Chai et al., 2005; Roberts and Orkin, 2004). Once again, these changes may in fact be quite extensive in the epigenome.
Global Changes in the Cancer Epigenome
The occurrence of localized changes in chromatin structure at transcriptional start sites has been well appreciated; however, it is now emerging that the alterations are genome wide. Indeed, early studies pointed to an overall decrease in the 5-methylcytosine content of cancer genomes (Feinberg and Tycko, 2004; Riggs and Jones, 1983). The hypermethylation consistently observed in CpG islands therefore represents a change in 5-methylcytosine distribution across the genome rather than an overall increase in the total amount of methylation. Interestingly, it has recently been found that large stretches of DNA can become abnormally methylated in cancer (Frigola et al., 2006).
The changes in CpG-island methylation in single tumors can involve a group of loci and has been hypothesized to constitute a distinct phenotype, first proposed by Toyota et al. (1999) as the “CpG island methylator phenotype” or CIMP. The existence of CIMP has been challenged (Yamashita et al., 2003), but recent studies by Weisenberger et al. (2006) showing that a subset of CpG islands is coordinately methylated in tumors argues, in our opinion, for the reality of such a phenotype. As will be discussed later, it is intriguing that many of these CIMP loci are targets for polycomb group proteins (Widschwendter et al., 2007).
Surprising results have also been obtained with respect to genome-wide changes in histone modifications. Loss of acetylation at lysine 16 and trimethylation at lysine 20 of histone H4 is a common hallmark of human cancer (Fraga et al., 2005), and global histone modification patterns predict the risk of prostate cancer recurrence (Seligson et al., 2005). Other evidence for global changes being involved in carcinogenesis come from studies in the polycomb group gene family, which is highly conserved throughout evolution (Valk-Lingbeek et al., 2004). The polycomb repressor complex 2 (PRC2) is involved in the initiation of silencing and contains histone methyltransferases that can methylate histone H3 lysine 9 and 27, which are marks of silenced chromatin. The significance of these findings will be discussed later. The polycomb gene BMI1, a component of PRC1, is overexpressed in several human cancers so that it might be expected that aberrations in this system would give rise to global alterations in gene silencing in cancer (Valk-Lingbeek et al., 2004).
It is well known that certain transcription factors such as c-Myc do not bind to their recognition sequences in the methylated condition, suggesting that CpG methylation may affect the ability of Myc to bind to multiple sites within the genome. Given that Myc can influence chromatin structure (Knoepfler et al., 2006), it is certainly plausible that inappropriate methylation of its recognition sites could have profound implications on the cancer epigenome. Perhaps more attention should be paid in the future to methylation of such genomic regions.
Given the linkage between processes that regulate epigenetic silencing, it should not be surprising that such changes are observed on a genome-wide scale (Figure 1). For example, DNA methylation and histone acetylation are known to be intimately linked, so that global hypomethylation might be expected to lead to global alterations in the level of histone acetylation and vice versa. These rapidly emerging data strongly indicate that the entire epigenome is fundamentally disturbed in cancer development. While the focus on research until recently has been on silencing, more attention is now being paid to the possibility that these genome-wide changes in the structure of the epigenome can lead to the genomic instability, that is a hallmark of cancer (Cadieux et al., 2006).
Aberrant Gene Silencing during Early Neoplastic Progression
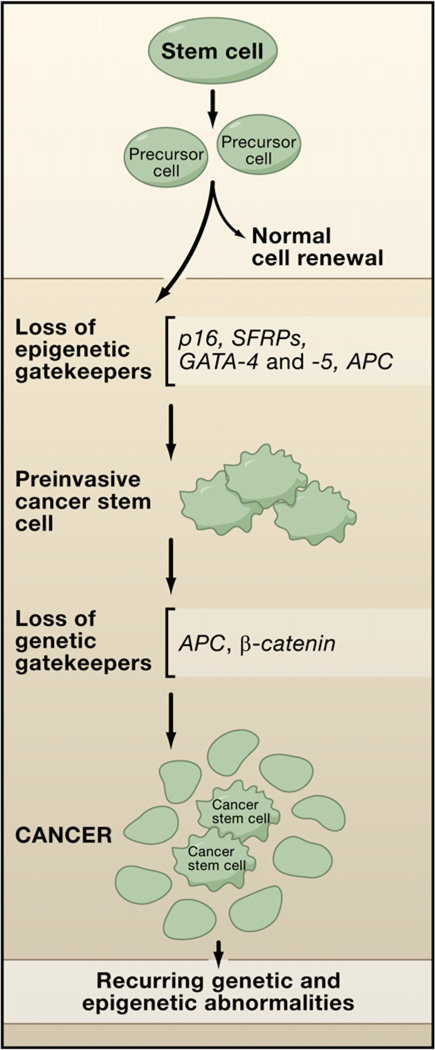
A key to understanding the contributions of aberrant epigenetic gene silencing to cancer has been to consider these in the context of their timing in cancer progression just as has been done for genetic changes. Recent reviews (Baylin and Ohm, 2006; Feinberg et al., 2006) have emphasized that epigenetic abnormalities might play a seminal role in the earliest steps in cancer initiation. Abnormal gene imprinting and/or silencing may help push the early aberrant clonal expansion of cells, providing a “substrate” for risk of subsequent genetic and epigenetic alterations that further foster tumor progression (Baylin and Ohm, 2006; Feinberg et al., 2006). This concept is shown for colon cancer in Figure 2, in which risk factors for common cancers such as aging (Sharpless and DePinho, 2005) and inflammation (Coussens and Werb, 2002; Lu et al., 2006; Nelson et al., 2004) are depicted as causing such expansions in either normal colon epithelial stem cells or precursor cells derived from them. A series of genes, all documented to exhibit DNA hypermethylation in preinvasive stages of colon and other cancers, but which are rarely mutated in such cancers, are shown and referred to as “epigenetic gatekeepers.” In other words, the normal epigenetic modulation of these genes allows them to prevent stem/precursor cells from becoming immortalized and acquiring infinite cell renewal capacity during periods of chronic stresses and renewal pressures on cell systems. It also allows these genes to be activated, as needed, when stem/precursor cells differentiate. The inappropriate silencing of these genes blocks their activation and allows for abnormal survival and clonal expansion and prevents differentiation. Depicted also are genes such as in APC or β-catenin involved as genetic gatekeepers for colon cancer, since mutations foster abnormal activation of the developmental Wnt pathway, which plays a canonical role in driving colon tumorigenesis throughout the life history of these tumors. In the paradigm shown, broaching of the epigenetic gatekeeper steps allows cell expansion and time for the genetic gatekeeper mutations to appear and even to undergo selection because the cells are now “addicted” to Wnt pathway activation (Baylin and Ohm, 2006).
Figure 2. An “Epigenetic Gatekeeper” Prevents Early Tumor Progression.
Epigenetic silencing of genes p16, _SFRP_s, GATA-4 and -5, and APC (red X) in stem/precursor cells of adult cell-renewal systems may serve to abnormally lock these cells into stem-like states that foster abnormal clonal expansion. These genes are termed “epigenetic gatekeepers” because their normal epigenetic pattern of expression should allow them to be activated during stem/precursor cell differentiation as needed to properly control adult cell renewal. The repertoire of abnormal gene silencing then allows abnormal survival of the cells in the setting of chronic stress, such as inflammation (see Figure 3). The resulting preinvasive stem cells become “addicted” to the survival pathways involved so that selection for mutations in genetic gatekeeper genes provide an even stronger stimulus for further tumor progression. The bulk of the resulting tumor is composed of a sub-population of cancer stem cells and neoplastic progeny.
How might loss of function for the epigenetic gatekeeper genes actually foster early abnormal clonal expansion? As shown in Figure 2, Wnt pathway activation may be a striking example where epigenetic events can play important roles. First, APC, the classically mutated genetic gatekeeper gene in colon cancer leading to Wnt pathway activation (Kinzler and Vogelstein, 1996), can be inactivated in sporadic tumors both by such mutations, or, occasionally, epigenetic gene silencing (Esteller et al., 2000) (Figure 2). Also, methylation on one allele can also serve as a ”second hit” for gene inactivation when paired with mutations on the opposite allele (Esteller et al., 2001). Second, four members of a family of genes, the SFRPs, which encode proteins that antagonize the action of the Wnt ligand at the cell membrane, can be hypermethylated simultaneously in the majority of preinvasive lesions for colon cancer (Suzuki et al., 2004). This silencing can foster increased Wnt pathway signaling, which may precede and addict cells toward evolving later mutations in the downstream pathway genes, APC or β-catenin, that further activate Wnt signaling to foster colon tumorigenesis (Baylin and Ohm, 2006).
The tumor suppressor gene, p16ink4A, is one of the most common, and earliest, epigenetically mediated losses of tumor suppressor function events in human cancer. This silencing begins in subsets of preinvasive stages of breast, colon, lung, and other cancers (Belinsky et al., 1998; McDermott et al., 2006; Reynolds et al., 2006). Recent studies of knockout mice have revealed that germline loss of this gene increases stem cell life span (Janzen et al., 2006; Krishnamurthy et al., 2006; Molofsky et al., 2006), consistent with a proposed role in tumorigenesis for facilitating early abnormal clonal expansion of cells at risk for cancer. Indeed, loss of this gene is permissive for allowing such expanding cells to develop genomic instability (Foster et al., 1998; Kiyono et al., 1998) and further epigenetic gene-silencing events (Reynolds et al., 2006).
The GATA-4 and -5 transcription factor genes are important for both embryonic gastrointestinal epithelial development and for maturation in adults (Gao et al., 1998; Laverriere et al., 1994; Molkentin et al., 1997) and are epigenetically silenced in virtually half of all of the preinvasive and invasive lesions for colon cancer (Akiyama et al., 2003). This can then impede differentiation and foster precursor cell expansion.
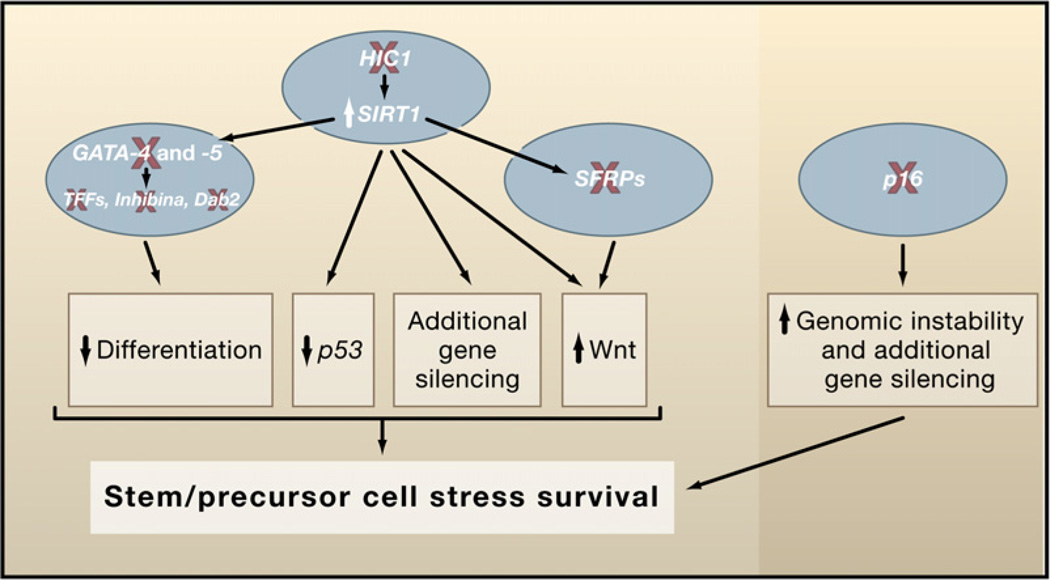
Finally, an intriguing abnormal survival circuit regulated by epigenetic gene silencing concerns upregulation of the survival protein, SIRT1, via loss of the transcription factor HIC1, which is DNA hypermethylated in early preinvasive lesions for colon tumorigenesis and many other common cancers (Baylin and Ohm, 2006). HIC1, a transcriptional repressor complexed with SIRT1, can downmodulate SIRT1 promoter activity (Chen et al., 2005). The loss of HIC1 function potentially sets off a network of survival events, which are detailed later below (Figure 3).
Figure 3. Networks of Gene-Silencing Events.
Such networks help to foster early and later steps during neoplastic progression. Examples of early gene silencing (red X) occur at multiple points in key tumor control pathways to allow abnormal cell survival after stress and early clonal expansion. These epigenetic events are shown as provoking disruptive crosstalk between the pathways facilitating this expansion. Examples of gene-silencing events that foster subsequent silencing events (green arrows linking SIRT1 to silencing of GATA-4 and -5 and _SFRP_s) are depicted.
Cancer Gene Silencing Versus Genetic Mutations
How does loss of function of cancer genes via epigenetic silencing resemble, or differ from, genetically mediated losses of gene function? Gene mutations in a single tumor are seldom multiple in a given cell pathway, since selection for one hit appears sufficient to produce full pathway disruption (Hanahan and Weinberg, 2000). Certainly, this can sometimes also be the case for gene silencing as found for classic tumor suppressor genes. Thus, in the cyclin D-Rb pathway, silencing of p16 does not appear in tumors in which the Rb gene is mutated, since both events powerfully disrupt cell-cycle control (Jones and Baylin, 2002). Similarly, mutations in the VHL gene, seminal for renal cancer pathogenesis, occur in 60% of tumors, while the gene is DNA hypermethylated in another ∼20% (Jones and Baylin, 2002). Also, mutations and epigenetic silencing of E-cadherin are mutually exclusive in the lobular and ductal forms of breast cancer, respectively (Graff et al., 1995).
Despite the above similarities between genetic and epigenetic gene loss of function in cancer, there is an important emerging theme that, unlike for mutations, multiple epigenetic events may frequently affect a single-cell pathway (Figure 3). These changes may function as networks in which multiple genes are not only affected within a pathway but can generate alterations of other key signaling pathways and even involve epigenetic events that cause other epigenetic events. Thus, these epigenetic abnormalities form a more nuanced, integrated disruption of pathways than do gene mutations to foster tumorigenesis (Figure 3). A first example is the previously mentioned silencing of the four SFRP genes and their interactions with Wnt pathway gene mutations. A second is the silencing of p16ink4A, which, in addition to disrupting the cyclin D-Rb cell-cycle control pathway, may foster recruitment of silencing complexes through other means and cause abnormal DNA methylation to the promoter of a HOX gene (Reynolds et al., 2006) (Figure 3). It may be that this loss of transcriptional activation of the downstream genes renders them susceptible to heritable silencing through adoption of the types of repressive promoter chromatin, which will be revisited below.
Perhaps one of the most complex series of events for networks of epigenetic abnormalities in cancers involves the loss of HIC1 function and the resultant potential survival events (Figure 2). As recently reviewed (Baylin and Ohm, 2006), the increases in SIRT1 may prolong cell survival through multiple mechanisms, including downmodulation of p53 function through deacetylation of this target protein. In addition, through mechanisms including deacetylation of the histone residue, H4K16, the yeast SIRT1 ortholog, Sir2, is an important participant in gene silencing (Guarente, 2000; Kimura et al., 2002; Suka et al., 2002). It appears that this is also the case in humans, including those genes that are DNA hypermethylated and silenced in cancer (Pruitt et al., 2006). Thus, increases in SIRT1 appear important for silencing of genes including the _SFRP_s, where the loss can upregulate the Wnt pathway, again facilitating cell survival in tumors such as colon (Figure 2). Finally, HIC1 itself has been shown to normally sequester, in the nucleus, transcription factors that drive the Wnt pathway. Thus, loss can free these for increased Wnt pathway function (Valenta et al., 2006).
It is then apparent that from the earliest stages of neoplastic development, epigenetic changes can, just as for gene mutations, perturb multiple key pathways in ways that foster cancer risk and evolution. As more and more cancer genes are discovered, which are functionally altered via epigenetic mechanisms, these pathways, and the links between them, will undoubtedly be even further appreciated.
The “Cancer Stem Cell” Hypothesis
Much of the recent work, including information derived from random screens to discover DNA hypermethylated cancer genes and the deciphering of networks of silenced genes, indicate that hundreds of epigenetically silenced genes possibly exist even in individual tumors. While it is conceivable that selection for stochastic events could account for this, it seems unlikely that all of the changes observed arise in such a random fashion and then come to dominate the tumor clone through selective advantage. Perhaps, just as multiple mutations arising in tumors secondary to central defects in genetic control programs, such as mismatch repair deficiency states, are not all directly important for tumor progression, the existence of multiple epigenetically silenced genes might reflect programs of epigenetic control abnormalities. Some might even be derived from genetic alterations that dictate abnormal chromatin regulation.
What could account for these early epigenetic silencing events and the many genes that appear to be involved, which may be key steps in the earliest phases of neoplastic evolution? As has been reviewed recently (Baylin and Ohm, 2006), one possibility concerns a role for the control of expression of groups of genes at the chromatin level, which is integral to the maintenance of cells in a stem cell state. The contribution of a stem cell state is integral to current thinking in the cancer biology field relative to an old, but still vital concept, that each patient’s tumor is a heterogeneous population of cells, some of which have more tumorigenic and metastatic potential than others (Al-Hajj et al., 2003). In recent years, this has evolved into the concept of the “cancer stem cell” that is believed to constitute the population that is ultimately responsible for perpetuating the tumor. These cells have many properties common to normal stem cells, but their exact origins remain controversial (Bjerkvig et al., 2005). Currently, most researchers seem to favor the view that a range of cells in normal cell renewing systems, from the ultimate stem cells to a subsequent series of precursor and progenitor cells, have the potential to constitute the focal transformation point for individual cancers. This could, in fact, explain the existence of many subtypes of major tumor types such as lung and breast cancers.
In the above context, if epigenetic cancer gene silencing might begin in a cancer stem cell, this would dictate that many such changes constitute early events in tumor progression and might have their molecular origins tied to stem/precursor cell population characteristics. We have discussed, in this review, a body of solid evidence for the former point and exciting concepts are emerging for the latter. We have mentioned earlier that long-term silencing of genes in embryonic stem (ES) cells is under control of the polycomb complexes of proteins (PcG), which act in concert for long-term maintenance of transcriptional repression. The PcG complex, PRC2, is involved in the initiation of silencing and contains EZH2, the histone methyltransferase that places the histone methylation modification, HeK27me (Orlando, 2003; Pirrotta and Gross, 2005; Ringrose and Paro, 2004; see also the Review by B. Schuettengruber et al., page 735 in this issue). In turn, this mark can attract the PRC1 complexes that maintain the silencing (Orlando, 2003; Pirrotta and Gross, 2005; Ringrose and Paro, 2004). The PRC1 complexes contain chromo domain proteins such as the CBX family (Bernstein et al., 2006b) that recognize the HeK27me mark, and the key stem cell protein Bmi1, which can silence the p16 gene (discussed earlier as a key gene epigenetically silenced early in cancers [Valk-Lingbeek et al., 2004; Varambally et al., 2002]). Steady-state levels of EZH2, Bmi1, and other PcG complex members are increased in cancer (Bracken et al., 2003; Varambally et al., 2002). Enrichment of EZH2 and the H3K27me mark is a property of the promoters of DNA hypermethylated and silenced genes (McGarvey et al., 2006), as is the sirtuin deactylase SIRT1 (Pruitt et al., 2006), which has been associated with PRC2 complexes found in stem and cancer cells (Kuzmichev et al., 2005).
Thus, dysregulation of the PcG system potentially links cancer formation to stem cell biology (Valk-Lingbeek et al., 2004). A large group of genes is marked by PcG control in ES cells (Boyer et al., 2006; Lee et al., 2006), as well as in other more committed stem/precursor cells (Bracken et al., 2006; see also the Review by M.A. Surani et al., page 747 in this issue). This marking appears to hold these genes at a transcription level required by the ES cell state until needed for up- or downregulation of the genes in more committed progeny (Bernstein et al., 2006a; Bracken et al., 2006). The PcG system has been incriminated in targeting DNA methylation for locking in gene silencing (Vire et al., 2006). If borne out in subsequent studies, it is appealing to consider that this targeting of silencing, in concert with other histone modifications such as H3K9 methylation (Tamaru and Selker, 2003), may be a link between stem cell biology, promoter DNA hypermethylation, and gene silencing in cancer. If, in mature cell renewal populations, survival responses to chronic inflammation and aging involve stem/precursor cells, then the set of PcG marked genes may render genes vulnerable to abnormal DNA methylation. Indeed, very recent findings have shown that stem cell PcG targets are 20-fold more likely to have cancer-specific promoter methylation than nontargets, supporting a stem cell origin for cancer (J.E. Ohm et al., submitted; Widschwendter et al., 2007; Schlesinger et al., 2007). The resultant tight heritable gene silencing for the genes we have been discussing may then abnormally hold cells in stem/precursor cell states, allowing them to participate in early steps in neoplastic progression.
The Potential of Epigenetic Therapeutics
The fact that epigenetic changes are so prevalent in cancers and play a causative role in their biologies has led to the development of an entirely new therapeutic approach in which the goal is to reverse gene silencing. It is 30 years since the first description of the remarkable effects of azanucleoside drugs on the differentiated state of cells (Constantinides et al., 1977). It is now clear that these compounds function as inhibitors of the DNA methyltransferase enzymes (Santi et al., 1983). Nevertheless, it has taken three decades for the drugs to be approved by the United States Food and Drug Administration for the treatment of myelodysplastic syndrome (MDS). Other nucleoside inhibitors of DNA methylation, including 5-fluoro-2′-deoxycytidine (Jones and Taylor, 1980) and zebularine (Cheng et al., 2004), are at an earlier stage of development.
Given that nucleosides require incorporation into DNA in order to be fully effective, there have been several attempts to find other inhibitors of DNA methylation that might act without incorporation into DNA. Although procainamide (Cornacchia et al., 1988) and tea polyphenols (Fang et al., 2003) have been reported to be DNA methylation inhibitors, they are, at best, weak inhibitors in living cells (Chuang et al., 2005), and research to discover other inhibitors remains a high priority. The recent description of the drug RG101 is a promising development of a lead compound that might be effective as a DNA methylation inhibitor (Brueckner et al., 2005).
The demonstration that histone deacetylase inhibitors have antitumor potential (Marks et al., 2001; Minucci and Pelicci, 2006) has led to the development of a series of inhibitors. The first of these, SAHA, has just been approved by the FDA for the treatment of T cell cutaneous lymphoma, and several drug companies are actively pursuing new histone deacetylase inhibitors (Bolden et al., 2006).
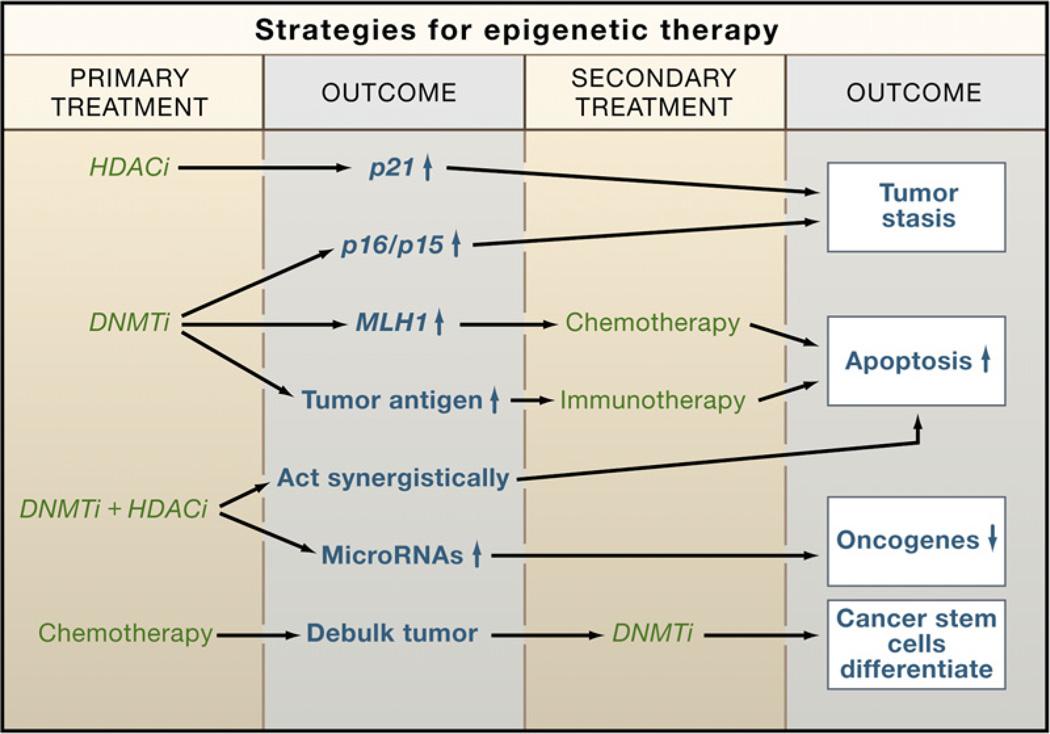
The histone methyltransferases represent another valid target for the discovery of new drugs that can reactivate silenced genes. Some lead compounds are already being investigated, and it is likely that these would activate genes either as single agents or in combination with other epigenetic drugs. In this regard, it is likely that the future of epigenetic therapy will involve the utilization of multiple drugs that individually affect epigenetic silencing but that might be expected to have synergistic effects (Figure 4). Because of the interrelationships between epigenetic processes involved in silencing (Figure 1) and the demonstrated synergistic activities of DNA methylation and HDAC inhibitors (Cameron et al., 1999; Suzuki et al., 2004; Yamashita et al., 2002), there is much interest in combination therapies (Figure 4). These are now being tested in clinical trials, as are combinations of these inhibitors with standard chemotherapeutic regimens. Knowing about the potential importance of epigenetic silencing to cancer stem cells, more innovative approaches to removing these cancer progenitors might be possible. Cancer stem cells, refractory to standard chemotherapy, might be induced to differentiate by chronic administration of epigenetic drugs, as shown in Figure 4. Also, the activation of epigenetically silenced tumor suppressor miRNAs might allow for new treatment modalities (Saito et al., 2006).
Figure 4. Strategies for Epigenetic Therapy.
Epigenetic therapy with DNA methylation inhibitors (DNMTi) and HDAC inhibitors (HDACi) is now a reality. While these agents are currently approved as single agents, combination therapies are likely to gain traction in the future because of the inherent self-reinforcing nature of silencing mechanisms (see Figure 1). Future breakthroughs could come from the use of epigenetic drugs to activate miRNAs or the use of drugs to target cancer stem cells after tumor debulking by standard chemotherapy.
A major impediment to the use of such drugs is that they are nonspecific and would be anticipated to reactivate genes nondiscriminately. However, this may not be as much of a problem as it seems, because DNA methylation inhibitors only act on dividing cells, leaving nondividing normal cells unaffected. Also, it appears that the drugs preferentially activate genes that have become abnormally silenced in cancer (Karpf et al., 1999; Liang et al., 2002). The reason for this is not clear but may be related to the fact that the chromatin structure associated with a pathologically silenced gene may be more susceptible to reactivation than the highly compacted chromatin state induced by physiological silencing. Nevertheless, the search for more specific targeted therapies remains a high priority.
Conclusions
As the role of epigenetics in cancer becomes clearer and the interrelationships between chromatin components are increasingly understood, we are at a good point to reevaluate our approaches to cancer prevention, detection, and therapy. It is clear that cancer cells have global changes in chromatin constitution involving the whole epigenome and that entire pathways relevant to cell renewal are subject to epigenetic dysregulation. The exciting links between epigenetics and stem cell behavior are just becoming manifest and are involved at the very earliest stages of tumor progression. This gives an important window on therapeutic intervention through prevention strategies. The approval of three, albeit nonspecific, drugs for therapy of established tumors gives new promise not only in this arena but also for new prevention strategies as well. Perhaps, as we continue to explore the molecular regulation of chromatin in both normal and neoplastic settings, we will become smarter in the use of agents to target cancer stem cells or miRNAs, for example, to make further inroads in resetting epigenetic abnormalities and achieving control of cancer.
ACKNOWLEDGMENTS
Supported by grants R01 CA083867/R01 CA082422 (P.A.J.) and R01 CA04318/R01 CA116160 (S.B.B.) from the National Cancer Institute and R01 ES011858 from the National Institute of Environmental Health Sciences (S.B.B.). The authors thank G. Liang for help with the figures.
REFERENCES
- Akiyama Y, Watkins N, Suzuki H, Jair KW, van Engeland M, Esteller M, Sakai H, Ren CY, Yuasa Y, Herman JG, Baylin SB. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol. Cell. Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker J, Lin X, Nelson WG. Methyl-CpG binding domain protein 2 represses transcription from hypermethylated pi-class glutathione S-transferase gene promoters in hepatocellular carcinoma cells. J. Biol. Chem. 2002;277:22573–22580. doi: 10.1074/jbc.M203009200. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Sacco-manno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc. Natl. Acad. Sci. USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006a;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol. Cell. Biol. 2006b;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerkvig R, Tysnes BB, Aboody KS, Najbauer J, Terzis AJ. Opinion: the origin of the cancer stem cell: current controversies and new insights. Nat. Rev. Cancer. 2005;5:899–904. doi: 10.1038/nrc1740. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Poly-comb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brueckner B, Boy RG, Siedlecki P, Musch T, Kliem HC, Zielen-kiewicz P, Suhai S, Wiessler M, Lyko F. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65:6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- Cadieux B, Ching TT, Vandenberg SR, Costello JF. Genome-wide hypomethylation in human glioblastomas associated with specific copy number alteration, methylenetetrahydrofolate reductase allele status, and increased proliferation. Cancer Res. 2006;66:8469–8476. doi: 10.1158/0008-5472.CAN-06-1547. [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21:103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- Chai J, Charboneau AL, Betz BL, Weissman BE. Loss of the hSNF5 gene concomitantly inactivates p21CIP/WAF1 and p16INK4a activity associated with replicative senescence in A204 rhabdoid tumor cells. Cancer Res. 2005;65:10192–10198. doi: 10.1158/0008-5472.CAN-05-1896. [DOI] [PubMed] [Google Scholar]
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Cheng JC, Yoo CB, Weisenberger DJ, Chuang J, Wozniak C, Liang G, Marquez VE, Greer S, Orntoft TF, Thykjaer T, Jones PA. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6:151–158. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- Chuang JC, Yoo CB, Kwan JM, Li TW, Liang G, Yang AS, Jones PA. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol. Cancer Ther. 2005;4:1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- Constantinides PG, Jones PA, Gevers W. Functional striated muscle cells from non-myoblast precursors following 5-azacytidine treatment. Nature. 1977;267:364–366. doi: 10.1038/267364a0. [DOI] [PubMed] [Google Scholar]
- Cornacchia E, Golbus J, Maybaum J, Strahler J, Hanash S, Richardson B. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J. Immunol. 1988;140:2197–2200. [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M, Sparks A, Toyota M, Sanchez-Cespedes M, Capella G, Peinado MA, Gonzalez S, Tarafa G, Sidransky D, Meltzer SJ, et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000;60:4366–4371. [PubMed] [Google Scholar]
- Esteller M, Fraga MF, Guo M, Garcia-Foncillas J, Hedenfalk I, Godwin AK, Trojan J, Vaurs-Barriere C, Bignon YJ, Ramus S, et al. DNA methylation patterns in hereditary human cancers mimic sporadic tumorigenesis. Hum. Mol. Genet. 2001;10:3001–3007. doi: 10.1093/hmg/10.26.3001. [DOI] [PubMed] [Google Scholar]
- Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–7570. [PubMed] [Google Scholar]
- Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- Foster SA, Wong DJ, Barrett MT, Galloway DA. Inactivation of p16 in human mammary epithelial cells by CpG island methylation. Mol. Cell. Biol. 1998;18:1793–1801. doi: 10.1128/mcb.18.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat. Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- Futscher BW, Oshiro MM, Wozniak RJ, Holtan N, Hanigan CL, Duan H, Domann FE. Role for DNA methylation in the control of cell type specific maspin expression. Nat. Genet. 2002;31:175–179. doi: 10.1038/ng886. [DOI] [PubMed] [Google Scholar]
- Gao X, Sedgwick T, Shi YB, Evans T. Distinct functions are implicated for the GATA-4, −5, and −6 transcription factors in the regulation of intestine epithelial cell differentiation. Mol. Cell. Biol. 1998;18:2901–2911. doi: 10.1128/mcb.18.5.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- Guarente L. Sir2 links chromatin silencing, metabolism, and aging. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Harikrishnan KN, Chow MZ, Baker EK, Pal S, Bassal S, Bra-sacchio D, Wang L, Craig JM, Jones PL, Sif S, El-Osta A. Brahma links the SWI/SNF chromatin-remodeling complex with MeCP2-dependent transcriptional silencing. Nat. Genet. 2005;37:254–264. doi: 10.1038/ng1516. [DOI] [PubMed] [Google Scholar]
- Janzen V, Forkert R, Fleming HE, Saito Y, Waring MT, Domb-kowski DM, Cheng T, DePinho RA, Sharpless NE, Scad-den DT. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443:421–426. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. The epigenetic magic of histone lysine methylation. FEBS J. 2006;273:3121–3135. doi: 10.1111/j.1742-4658.2006.05343.x. [DOI] [PubMed] [Google Scholar]
- Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20:85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- Jones PA, Laird PW. Cancer epigenetics comes of age. Nat. Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- Karpf AR, Peterson PW, Rawlins JT, Dalley BK, Yang Q, Albertsen H, Jones DA. Inhibition of DNA methyltransferase stimulates the expression of signal transducer and activator of transcription 1, 2, and 3 genes in colon tumor cells. Proc. Natl. Acad. Sci. USA. 1999;96:14007–14012. doi: 10.1073/pnas.96.24.14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A, Umehara T, Horikoshi M. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 2002;32:370–377. doi: 10.1038/ng993. [DOI] [PubMed] [Google Scholar]
- Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- Kiyono T, Foster SA, Koop JI, McDougall JK, Galloway DA, Klingelhutz AJ. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, Bonner-Weir S, Sharpless NE. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc. Natl. Acad. Sci. USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverriere AC, MacNeill C, Mueller C, Poelmann RE, Burch JB, Evans T. GATA-4/5/6, a subfamily of three transcription factors transcribed in developing heart and gut. J. Biol. Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang G, Gonzales FA, Jones PA, Orntoft TF, Thykjaer T. Analysis of gene induction in human fibroblasts and bladder cancer cells exposed to the methylation inhibitor 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:961–966. [PubMed] [Google Scholar]
- Lu H, Ouyang W, Huang C. Inflammation, a key event in cancer development. Mol. Cancer Res. 2006;4:221–233. doi: 10.1158/1541-7786.MCR-05-0261. [DOI] [PubMed] [Google Scholar]
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nat. Rev. Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- McDermott KM, Zhang J, Holst CR, Kozakiewicz BK, Singla V, Tlsty TD. p16(INK4a) prevents centrosome dysfunction and genomic instability in primary cells. PLoS Biol. 2006;4:e51. doi: 10.1371/journal.pbio.0040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGarvey KM, Fahrner JA, Greene E, Martens J, Jenuwein T, Baylin SB. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006;66:3541–3549. doi: 10.1158/0008-5472.CAN-05-2481. [DOI] [PubMed] [Google Scholar]
- Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11:1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- Molofsky AV, Slutsky SG, Joseph NM, He S, Pardal R, Krishnamurthy J, Sharpless NE, Morrison SJ. Increasing p16INK4a expression decreases forebrain progenitors and neurogenesis during ageing. Nature. 2006;443:448–452. doi: 10.1038/nature05091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- Nelson WG, De Marzo AM, DeWeese TL, Isaacs WB. The role of inflammation in the pathogenesis of prostate cancer. J. Urol. 2004;172:S6–S12. doi: 10.1097/01.ju.0000142058.99614.ff. [DOI] [PubMed] [Google Scholar]
- Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. doi: 10.1016/s0092-8674(03)00157-0. [DOI] [PubMed] [Google Scholar]
- Pirrotta V, Gross DS. Epigenetic silencing mechanisms in budding yeast and fruit fly: different paths, same destinations. Mol. Cell. 2005;18:395–398. doi: 10.1016/j.molcel.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Pruitt K, Zinn RL, Ohm JE, McGarvey KM, Kang SH, Watkins DN, Herman JG, Baylin SB. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. doi: 10.1371/journal.pgen.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds PA, Sigaroudinia M, Zardo G, Wilson MB, Benton GM, Miller CJ, Hong C, Fridlyand J, Costello JF, Tlsty TD. Tumor suppressor p16INK4A regulates polycomb-mediated DNA hypermethylation in human mammary epithelial cells. J. Biol. Chem. 2006;281:24790–24802. doi: 10.1074/jbc.M604175200. [DOI] [PubMed] [Google Scholar]
- Riggs AD, Jones PA. 5-methylcytosine, gene regulation, and cancer. Adv. Cancer Res. 1983;40:1–30. doi: 10.1016/s0065-230x(08)60678-8. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Roberts CW, Orkin SH. The SWI/SNF complex-chromatin and cancer. Nat. Rev. Cancer. 2004;4:133–142. doi: 10.1038/nrc1273. [DOI] [PubMed] [Google Scholar]
- Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Santi DV, Garrett CE, Barr PJ. On the mechanism of inhibition of DNA-cytosine methyltransferases by cytosine analogs. Cell. 1983;33:9–10. doi: 10.1016/0092-8674(83)90327-6. [DOI] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Sharpless NE, DePinho RA. Cancer: crime and punishment. Nature. 2005;436:636–637. doi: 10.1038/436636a. [DOI] [PubMed] [Google Scholar]
- Suka N, Luo K, Grunstein M. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 2002;32:378–383. doi: 10.1038/ng1017. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat. Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Selker EU. Synthesis of signals for de novo DNA methylation in Neurospora crassa. Mol. Cell. Biol. 2003;23:2379–2394. doi: 10.1128/MCB.23.7.2379-2394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc. Natl. Acad. Sci. USA. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenta T, Lukas J, Doubravska L, Fafilek B, Korinek V. HIC1 attenuates Wnt signaling by recruitment of TCF-4 and beta-catenin to the nuclear bodies. EMBO J. 2006;25:2326–2337. doi: 10.1038/sj.emboj.7601147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat. Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat. Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Upadhyay S, Osada M, Hoque MO, Xiao Y, Mori M, Sato F, Meltzer SJ, Sidransky D. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2:485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Dai T, Dai Y, Yamamoto F, Perucho M. Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell. 2003;4:121–131. doi: 10.1016/s1535-6108(03)00190-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]